Abstract

Cancer is characterized by altered cellular metabolism, and metabolic enzymes are considered as a promising target for anticancer therapy. Pyrimidine metabolism dysregulation is associated with various types of cancer, particularly lung cancer, which is one of the leading causes of cancer-related mortality worldwide. Recent studies have shown that small-cell lung cancer cells are particularly reliant on the pyrimidine biosynthesis pathway and are sensitive to its disruption. DHODH, the rate-limiting enzyme of the de novo pyrimidine production pathway, is essential in the production of RNA and DNA and is overexpressed in malignancies such as AML, skin cancer, breast cancer, and lung cancer, thereby highlighting DHODH as a viable target for developing drugs to combat lung cancer. Herein, rational drug design and computational techniques were used to discover novel DHODH inhibitors. A small combinatorial library was generated, and the top hits were synthesized and tested for anticancer activity against three lung cancer cell lines. Among the tested compounds, compound 5c possessed a stronger cytotoxicity (TC50 of 11 μM) compared to the standard FDA-approved drug (Regorafenib, TC50 of 13 μM) on the A549 cell line. Furthermore, compound 5c demonstrated potent inhibitory activity against hDHODH at a nanomolar level of 421 nM. DFT, molecular docking, molecular dynamic simulations, and free energy calculations were also carried out to understand the inhibitory mechanisms of the synthesized scaffolds. These in silico studies identified key mechanisms and structural features that will be crucial for future studies.
1. Introduction
Cellular metabolism is the base of all biological functions as it encompasses every aspect of biology.1 Disruption or dysregulation of the normal functions of cellular metabolism leads to serious conditions such as cancer. These dysregulations are characterized by persistent cell migration, proliferation, and differentiation of tumors, which are considered as hallmarks of cancer.2 Since metabolic enzymes play a crucial role in this process, they are increasingly deemed a promising target for the development of new anticancer therapies.3,4 Among the various metabolic pathways involved in the genesis of cancer, dysregulation of pyrimidine metabolism has been strongly associated with many types of cancer.5
Pyrimidine is produced via two pathways: the salvage synthesis pathway and the de novo synthesis pathway.6 The salvage synthesis route is prevalent during resting periods or in fully differentiated cells, accounting for the majority of Pyrimidines.7,8 Yet, in highly proliferating cells, such as tumor cells, the de novo synthesis pathway (Figure 1) becomes extremely active to meet the increased demand for nucleic acid precursors and other biological components.7,9 Compared to normal proliferous cells, cancer cells have a considerable imbalance in pyrimidine metabolism, linking pyrimidine imbalance to tumor transformation and progression.5 Dihydroorotate dehydrogenase (DHODH) is the rate-limiting enzyme of the de novo pyrimidine production pathway which is essential in the production of RNA and DNA.10 DHODH, which is housed in the inner membrane of the mitochondria, catalyzes a redox reaction that converts dihydroorotate into orotate.11
Figure 1.
Pyrimidine de novo synthesis pathway.
Among the various types of cancers, lung cancer is one of the cancers that is characterized by the overexpression of DHODH. A recent study employed CRISPR screening to find that small-cell lung cancer cells are particularly reliant on the pyrimidine biosynthesis pathway and thus sensitive to disruption, highlighting its potential as a therapeutic target,12 while another study identified that DHODH inhibition suppressed SCLC tumor growth and boosted mice survival in vivo.13 Accordingly, DHODH is regarded as a promising target for the development of novel therapeutics for the treatment of lung cancer.12,14 These findings coupled with the fact that to date Leflunomide remains to be the only FDA-approved DHODH inhibitor (for the treatment of rheumatoid arthritis in 1998) highlights the unmet critical need for developing novel DHODH inhibitors with anticancer activity.15 However, developing new drugs is a lengthy and costly process. Herein, rational drug design was utilized to expedite the identification process by generating a novel scaffold, which was further enumerated to create a small library. This library was virtually screened, and the top compounds were synthesized and subjected to in vitro and in silico studies identifying their cytotoxic and DHODH inhibitory activity as well as characterizing their key structural features for future studies.
2. Results and Discussion
2.1. Design of the Small Virtual Combinatorial Library
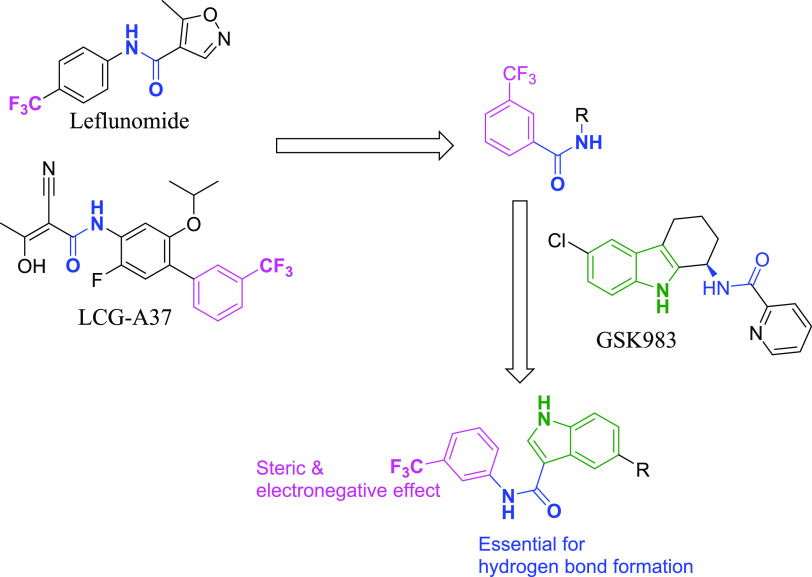
Employing rational drug design approach, a small virtual combinatorial library was generated in silico to speed up the identification process. The virtual library was established on a hybridization-based strategy of incorporating several structural features of known DHODH potent inhibitors to produce novel structures with readily available chemical structures, thereby reducing the synthetic cost as demonstrated in Figure 2. The hybridization strategy was centered on the amide coupling of the indole moiety of GSK983 with trifluoromethyl-substituted aniline to produce the hybrid scaffold illustrated in Figure 2.16 The trifluoromethyl moiety was chosen as it was present in several reported potent DHODH inhibitors such as Leflunomide and LCG-A37 and is reported to be of significance in providing both steric interaction and electron-withdrawing effect,16,17 while the amide linker was reported to be key for hydrogen bond formation.18
Figure 2.
Schematic representation of the rational design strategy employed in the design of the 5-substituted-N-(3-(trifluoromethyl)phenyl)-1H-indole-3-carboxamide scaffold, which was utilized for the virtual library enumeration.
In an effort to reduce the cost of synthesis, only the compounds that were already available in the laboratory were used to generate the virtual library. By only utilizing the available compounds, a small combinatorial library was designed with a diverse set of analogues. While this may limit the diversity of the library, it allowed the generation of a focused set of compounds that could be synthesized at a low cost. This led to the generation of a virtual library comprising 20 compounds that were easily synthesizable by reacting the proposed scaffold using Suzuki coupling and Buchwald reactions with different aromatic substitutions. To prioritize the most promising compounds, the virtual library was then virtually screened, and the structures were ordered by their docking scores.
2.2. Screening of the Virtual Library
To accelerate the identification process and reduce costs, a virtual combinatorial library was generated in silico.19 This library was based on a hybridization-based strategy that incorporated various substituted indole moieties with trifluoromethyl-substituted aniline to generate a diverse range of derivatives. To construct the library, all available aromatic amines and cyclic boronic acid derivatives from our laboratory chemicals were utilized, resulting in a virtual library of 20 hit compounds. To identify the most promising compounds for synthesis, a molecular docking simulation was performed using the extra precision module of the Maestro Schrodinger software, as a structure-based approach.20 The virtual library, along with Leflunomide (the only FDA-approved DHODH inhibitor), was docked into the active site of the DHODH crystal structure (PDB: 6LP7). The compounds were ranked based on their affinity to the target protein using the GlideScore, where the more negative the score, the higher the affinity. The seven compounds that exhibited a higher docking score than Leflunomide (GlideScore = −9.35 kcal/mol) were chosen for synthesis and further testing for their anticancer and DHODH inhibitory activity. This approach allowed for a fast and cost-effective selection of the most promising compounds for further study.
2.3. Chemistry
Scheme 1 illustrates the synthetic route utilized in synthesizing the target compounds 4a–b, 5a–d. Synthesis of the intermediate compound 2 involved the reaction of 5-bromoindole (1) with trifluoroacetic anhydride in the presence of N,N-dimethylformamide (DMF), which was followed with 20% NaOH. Compound 2 was converted to its corresponding acyl chloride and then immediately coupled with 3-(trifluoromethyl)aniline to afford compound 3 in moderate yield. Suzuki cross-coupling of compound 3 and the appropriate boronic acid derivative was performed using [1′1′-Bis(diphenylphosphino)ferrocene]-dichloropalladium (II) as a catalyst to obtain compounds 4a–b in moderate to good yields. Meanwhile, reacting compound 3 with the appropriate anilines via Buchwald–Hartwig amination in the presence of a palladium catalyst afforded compounds 5a–d. The structures of the target compounds were verified by checking the spectral NMR, HRMS, and HPLC data.
Scheme 1. Reagents and Conditions.

(a) (i) trifluoroacetic anhydride, DMF, 0 °C, 3.5 h; (ii) 20% NaOH, reflux, overnight, 100% yield; (b). (i) oxalyl chloride, 50 °C, 1 h; (ii) DCM, 3-(trifluoromethyl) aniline, rt, overnight; 49% yield; (c) Th appropriate boronic acid derivatives, [1′1′-Bis(diphenylphosphino)ferrocene]-dichloropalladium (II), K2CO3, dioxane, water, 110 °C, 4 h, seal tube, 21–77% yield; (d) appropriate aniline derivatives, t-butyl Xphos, Pd2dba3, K2CO3, toluene, 100 °C, 12 h, seal tube, 17–31% yield
Compound 2 synthesis was confirmed by the use of nuclear magnetic resonance (NMR) spectroscopy, specifically the presence of an indole singlet NH peak at 12.07 ppm and an acid peak at 12.14 ppm in the 1H NMR spectra. The successful synthesis of compound 3 was confirmed by the disappearance of the acid peak and the appearance of a new singlet peak corresponding to the amide NH at 10.10 ppm in the 1H NMR spectra. The success of the Suzuki cross-coupling reaction in yielding compound 4a was validated by the presence of three aliphatic peaks at 3.61–3.57, 3.56, and 2.40 ppm in the 1H NMR spectra, which are attributable to the 10 hydrogens of the morpholinomethyl moiety. Compound 4b was characterized by the appearance of a new singlet peak at 3.81 ppm, which is attributable to the methoxyphenyl group in the 1H NMR spectra.
Compounds 5a-d were all characterized by the presence of an additional singlet NH peak in the range of 7.69–8.26 ppm in the 1H NMR spectra. Additionally, compounds 5a-d showed an indole singlet NH peak between 11.56 and 11.89 ppm and an amide singlet NH peak between 9.90 and 10.05 ppm, which were inherited from the intermediate compound 3. Compound 5a was characterized by the appearance of a new singlet peak at 8.85 ppm, which is attributable to the hydroxy group of the phenol moiety, while compound 5b was confirmed by the appearance of two singlet peaks at 3.71 and 3.69 ppm, which are attributable to the dimethoxyphenyl moiety in the 1H NMR spectra.
2.4. Biological Evaluation
2.4.1. WST-1 Cell Viability Assay
In this study, cell assay studies were carried out first to select the compounds with good anticancer activity for the DHODH assay. The rationale behind this approach was that the primary objective of the study was to identify potential inhibitors of DHODH for the treatment of lung cancer. Therefore, it was deemed more cost-effective and practical to first focus on compounds that exhibited good anticancer activity, and then further evaluate their inhibitory activity against DHODH. This approach allowed the prioritization of the compounds with a higher likelihood of exhibiting both anticancer and DHODH inhibitory activities, thus optimizing the resources and efforts toward the primary objective of the study.
Accordingly, the synthesized compounds were subjected to preliminary screening for their effects on cell viability using the WST-1 assay. The assay was performed by treating the lung cancer cell lines, A549, H1299, and H1975 with 100 μM of each compound for 24 h. The results showed that all of the compounds were able to decrease cell viability by more than 50% (Figure S1). The positive control, Regorafenib, was similarly able to decrease cell viability by more than 50%. Regorafenib was chosen as the positive control as it is an FDA-approved anticancer drug that has been reported in several studies to be effective for the treatment of lung cancers such as NSCLC and lung squamous cell carcinoma.21−23
Next, the concentration dependency of cytotoxicity was examined, and the half-maximal toxic concentration (TC50) was calculated with these seven chemicals (Table 1). All values are expressed as mean ± standard error (n = 6–10). The study tested the seven synthesized compounds for their cytotoxic effect on three different cancer cell lines: A549, H1299, and H1975. Among the compounds tested, compounds 4a and 5c were found to have the best overall cytotoxic effect (with TC50 of 13 and 11 μM, respectively) compared to the standard drug (Regorafenib, with a TC50 of 13 μM) on the A549 cell line. Similarly, both compounds 4a and 5c displayed similar levels of activity (with TC50 values of 13 and 11 μM, respectively) to the standard (with a TC50 of 10 μM) on the H1299 cell line. However, only compound 5c (with a TC50 of 15 μM) exhibited better activity than the standard (with a TC50 of 16 μM) on the H1975 cells. Additionally, the only FDA-approved DHODH inhibitor (Leflunomide) was tested against the three lung cancer cell lines where it showed extremely weak cytotoxicity activity indicating its inability to combat lung cancer. When the top cytotoxic compounds (compounds 4a and 5c), their intermediate (compound 3), and the positive standard (Regorafenib) were tested against normal lung cell lines (MRC-5), their cytotoxicity profile was found to be comparable (Table 1 and Figure S2). Overall, most of the compounds tested showed strong cytotoxicity on the A549 and H1299 cell lines but relatively lower cytotoxicity on the H1975 cells.
Table 1. TC50 of Tested Compounds on A549, H1299, H1975, and MRC-5 Cellsa.
| TC50 (μM) |
||||
|---|---|---|---|---|
| compound | A549 | H1299 | H1975 | MRC-5 |
| 3 | 22 ± 1.0 | 27 ± 0.8 | 28 ± 2.0 | 45 ± 8.0 |
| 4a | 13 ± 0.3 | 13 ± 0.1 | 23 ± 2.0 | 17 ± 3.0 |
| 4b | 17 ± 0.5 | 17 ± 0.3 | 35 ± 4.0 | |
| 5a | 32 ± 0.9 | 33 ± 0.7 | 33 ± 2.0 | |
| 5b | 19 ± 0.4 | 16 ± 0.6 | 25 ± 5.0 | |
| 5c | 11 ± 0.5 | 11 ± 0.3 | 15 ± 1.0 | 17 ± 0.3 |
| 5d | 16 ± 0.4 | 18 ± 0.5 | 29 ± 4.0 | |
| regorafenib | 13 ± 0.8 | 10 ± 0.4 | 16 ± 0.5 | 18 ± 2.0 |
| Leflunomide | 100 < TC50 <300 | 300 < | 100 < | 300 < |
All values are expressed as mean ± standard error (n = 6).
2.4.2. hDHODH Evaluation
Compounds 4a and 5c, due to their potent cytotoxic activity, were chosen to be subjected to an in vitro screening over hDHODH enzyme in a fluorescence-based assay. Moreover, to test the designed scaffold potential, the intermediate compound 3 was also tested against hDHODH. The results of the inhibitory hDHODH assay are displayed in Table 2, while the IC50 curve is illustrated in Figure 3.
Table 2. Inhibitory IC50 Values of Compounds 3, 4a, and 5c over hDHODH.
| compound | IC50 (μM) |
|---|---|
| 3 | 0.195 |
| 4a | 1.210 |
| 5c | 0.421 |
Figure 3.
IC50 curve of compounds 3, 4a, and 5c over hDHODH.
Compounds 3 and 5c were found to possess a potent DHODH inhibitory activity in the fluorescence-based assay, with IC50 values of 195 and 421 nM respectively. This activity is 3- and 1.5-fold the reported DHODH inhibitory activity of Leflunomide (600 nM),16 while compound 4a displayed a moderate DHODH inhibitory activity of 1.2 μM. This is coupled with the fact that compound 5c possessed the most potent cytotoxic activity over all three tested lung cancer cell lines making it an attractive candidate for lung cancer treatment and worthy of further investigations.
2.5. DFT Calculations
Density functional theory (DFT) is a quantum mechanical approach that accurately characterizes the structural and electronic features of small molecules.24 Herein, DFT was utilized to compute the orbital energies, providing information about the electronic distribution of two potential inhibitors selected through virtual screening. This information can help to enhance the comprehension of the interaction between the protein and ligand and the inhibitory potential of the compounds. DFT calculations can reinforce the outcomes obtained from molecular docking and in vitro enzyme inhibition experiments.25 It is crucial to localize the HOMO and LUMO orbitals since electrons from the HOMO orbitals take part in reactions, and both orbitals are involved in charge transfer during chemical reactions.24 The smaller the energy gap between HOMO and LUMO, the more reactive the molecule, due to the delicate nature of bound electrons, which can lead to rapid electron transfer and exchange when the energy gap is small.24,26
The HOMO and LUMO energy levels were computed to determine the regions of high and low electron density in the inhibitors, as shown in Table 3. The seven synthesized compounds had similar energy gaps between HOMO and LUMO, which justifies their comparable reactivity against cancer cell lines. Compound 5c had a lower energy gap (ΔE = −0.1395) between HOMO and LUMO compared to Leflunomide (ΔE = −0.1705). Meanwhile, both compounds 3 and 4a displayed a comparable energy gap to Leflunomide, indicating similar reactivity profiles.
Table 3. Frontier Orbital Energies of the Seven Synthesized Compounds and Leflunomide.
| compound | HOMO (ev) | LUMO (ev) | energy gap (ΔE) |
|---|---|---|---|
| 3 | –0.223650 | –0.046990 | –0.17666 |
| 4a | –0.209278 | –0.043641 | –0.16564 |
| 4b | –0.198182 | –0.039350 | –0.15883 |
| 5a | –0.177838 | –0.035467 | –0.14237 |
| 5b | –0.184413 | –0.032434 | –0.15198 |
| 5c | –0.210149 | –0.070595 | –0.13955 |
| 5d | –0.205535 | –0.081115 | –0.12442 |
| Leflunomide | –0.348842 | –0.178321 | –0.17052 |
2.6. Molecular Docking
Molecular docking is an important component in computer-aided drug design, with numerous applications. For instance, it can predict the binding modes of a ligand and its target, rank a series of compounds based on their docking scores, and draw a correlation between the scores and potential activity.27 The interactions visualized from the docking study can also aid in enhancing the affinity features of the studied ligands.28 Subsequently, a comprehensive molecular docking analysis was performed in conjunction with the results obtained from density functional theory calculations, on the compounds investigated for their potential binding affinity toward DHODH (namely, compounds 3, 4a, and 5c) and Leflunomide.
Overall, the synthesized compounds exhibited increased binding cavity occupation in comparison to Leflunomide, due to their larger molecular size. This was especially the case for compounds 4a and 5c, which were able to occupy the majority of the available binding cavity due to their size and orientation. Compounds 3, 4a, and 5c established several π–π stacking and hydrophobic interactions with the amino acid residues of the binding cavity. Moreover, both 4a and 5c established a π–sulfur bond with the Met111 amino acid residue indicating a shared binding mechanism.
Furthermore, the presence of two hydrogen bonds between compound 5c and Thr357 and Asn145 amino acid residues of the DHODH binding cavity helps to explain its stronger DHODH inhibitory activity relative to compound 4a. Similarly, both compounds 3 and Leflunomide were able to form hydrogen bonds with the DHODH binding cavity. Compound 3 established one hydrogen bond with Arg136, while Leflunomide formed two hydrogen bonds with Tyr38 and Leu67 amino acid residues of the DHODH binding cavity, which explains their potent inhibitory activity.
Consequently, the orientation within the binding cavity, degree of binding site occupation, and the number and strength of the bonds established with the binding cavity’s amino acid residues are all crucial factors in determining the DHODH inhibitory activity. The 3D binding orientation of compounds 3, 4a, 5c, and Leflunomide within the binding cavity of DHODH and their associated 2D interactions are represented in Figure 4.
Figure 4.
Depiction of the 3D binding orientation of compounds 3, 4a, 5c, and Leflunomide within the binding cavity of dihydroorotate dehydrogenase (DHODH) and the corresponding 2D interactions established with the binding cavity’s amino acid residues. Favorable interactions represented as dashed lines: green—hydrogen bonds, yellow—π–sulfur, dark pink—π–π stacking interactions, light pink—hydrophobic interactions, turquoise—halogen interaction.
2.7. MD Simulations
The synthesized compounds showed high activity and favorable interactions based on molecular docking studies. However, it is important to note that this method only considers flexible ligand conformations while keeping the protein in a rigid state.29 To assess binding pose stability and protein conformation dynamics, molecular dynamics (MD) simulations were performed on protein–ligand complexes of compounds 3, 4a, 5c, and Leflunomide for 100 ns. These simulations were compared to the DHODH unbound protein state. In total, five 100 ns MD simulations were conducted.
The stability of the ligand–target complex and the validity of the simulation protocol were assessed by calculating the root-mean-square deviation (RMSD). The RMSD values provide a measure of the structural similarity between the initial and final structures of the protein–ligand complexes, allowing for an assessment of the stability of the complexes over the course of the simulation.27,28 Additionally, by comparing the simulation results to the unbound protein state and a known complex, the validity of the simulation protocol can be evaluated. This approach provides a more accurate picture of the binding interactions and stability of the complexes, taking into account the dynamic nature of the protein and the ligand in solution.30
The unbound protein and bound complexes exhibited fluctuations during the initial 40 ns of the MD simulation, after which the protein stabilized. The unbound protein exhibited an average RMSD of 1.86 Å. The RMSD values of the protein–ligand complexes of compounds 3, 4a, 5c, and Leflunomide were calculated to be 1.58, 1.41, 1.52, and 1.29 Å, respectively. The RMSD values of the complexes are lower than that of the unbound protein, showing that all investigated complexes were more stable than the unbound protein. This is consistent with both the hypothesis that the compounds bind to the protein and stabilize it, as well as the exhibited DHODH inhibitory data. Furthermore, the lower RMSD values of the complexes indicate that the complexes are more similar to the initial structures of the complexes than the unbound protein, which suggests that the complexes are less affected by fluctuations and are more stable over the course of the simulation. Accordingly, the MD simulation results validate the chosen docking approach as well as indicate that the compounds 3, 4a, 5c, and Leflunomide are able to effectively inhibit the DHODH protein and form stable complexes with the protein. The RMSD of the unbound protein and investigated ligand–protein complexes are depicted in Figure 5.
Figure 5.
RMSD plots of the unbound protein (black), compound 3–protein complex (blue), compound 4a–protein complex (magenta), compound 5c–protein complex (orange), and Leflunomide–protein complex (red).
RMSF is another useful tool that can be used in interpreting the MD simulation results. RMSF is calculated by measuring the deviation of each atom’s position from its average position throughout the MD simulation.31 The more an atom deviates from its average position, the more flexible or mobile it is. RMSF can be used to identify the regions of the protein that are flexible and mobile, which may be important for binding and activity.30 Furthermore, RMSF can be used to identify regions of the protein that are important for specificity. Since RMSF shows the flexibility of atoms in the protein complex, the regions that are rigid and specific to the ligand can be identified.32 Overall, RMSF can help in identifying regions of the protein that are important for binding, activity, stability, and specificity.
The unbound protein backbone exhibited the highest peak limit of ∼5 Å, while the compound–protein complexes exhibited lower maximum fluctuations. The RMSF calculations (Figure 6) showed that there are increased fluctuations in the residue regions of 180–198 and 252–270, indicating that these regions are highly flexible. Generally, the complexes of the investigated compounds exhibited significantly lower RMSF fluctuation compared to the unbound protein, which explains their higher biological activity.
Figure 6.
RMSF fluctuations of the unbound protein, compounds 3, 4a, 5c, and Leflunomide–protein complexes.
2.8. Molecular Mechanics-Generalized Born Surface Area (MM-GBSA) Calculations
The MM-GBSA method is a rigorous and widely used method for predicting binding free energy (ΔGBind) following simulation that takes protein flexibility, entropy, and polarizability into account. It identifies ligands that bind to receptors efficiently and is essential in biomolecular research for understanding molecular activities. MM-GBSA binding free energy calculations were utilized to confirm the validity of compounds identified by docking and molecular dynamics simulations.33,34 From frame 0–1001, the post-simulation MM-GBSA was calculated at every 10th frame, yielding a total of 100 conformations of each simulated complex.33,34 The MM-GBSA binding energy statistics in Table 4 show the average cumulative contributions of coulombic, hydrogen bonding, lipophilic, and van der Waals interactions that had a significant impact on ΔGBind.
Table 4. MM-GBSA Calculations for the Bound Complexes of Compounds 3, 4a, 5c, and Leflunomide.
| MM-GBSA (kcal/mol) |
||||||
|---|---|---|---|---|---|---|
| complex name | average ΔGBind | average ΔGCoulomb | average ΔGH_bond | average ΔGLipo | average ΔGSolv_GB | average ΔGvdW |
| compound 3-DHODH | –37.25 | –5.34 | –0.19 | –10.35 | 13.45 | –34.23 |
| compound 4a-DHODH | –16.61 | –5.62 | –0.19 | –4.72 | 9.68 | –15.02 |
| compound 5c-DHODH | –29.99 | –6.44 | –0.50 | –8.65 | 8.09 | –23.60 |
| Leflunomide-DHODH | –23.72 | –4.52 | –0.09 | –7.38 | 6.36 | –17.60 |
In MM-GBSA calculations, a more negative average free energy (ΔGBind) indicates stronger binding. The MM-GBSA calculation results showed a direct correlation between the average free energy and the DHODH inhibitory activity. This was highlighted by compound 3, which possessed the strongest DHODH inhibitory activity, displaying the highest free energy of −37.25 kcal/mol. This high free energy is attributable to its high average lipophilic and van der Waals interaction energies of −10.35 and −34.23, respectively, due to the presence of the highly lipophilic and bulky bromine moiety. Similarly, the second-best DHODH inhibitor (compound 5c) exhibited the second-best average free energy of −29.99 kcal/mol, while the compound with the lowest DHODH inhibitory activity, compound 4a, exhibited the lowest average free energy of −16.61 kcal/mol. In summary, both compounds 3 and 5c displayed stronger binding than Leflunomide (−23.72 kcal/mol), which corresponds with their DHODH inhibitory activity. This suggests that MD-based free energy calculations can accurately predict DHODH inhibitory activity, making it a useful tool for future DHODH drug optimization.
3. Conclusions
Recent studies have identified dihydroorotate dehydrogenase (DHODH) as a viable target for drug development in lung cancer. In this study, a virtual library was created using a structure-based design and screened using molecular docking to efficiently identify new DHODH inhibitors. The compounds with higher docking scores than Leflunomide were chosen for synthesis and further testing for anticancer and DHODH inhibitory activity. Among the compounds tested, compounds 4a and 5c were found to have a higher cytotoxic effect compared to the standard drug, Regorafenib on the A549 cell line. Additionally, both compounds 4a and 5c displayed similar levels of cytotoxic activity to the standard on the H1299 cell line. The intermediate compound 3 along with compounds 4a and 5c were tested for their DHODH inhibitory activity, and both compounds 3 and 5c exhibited potent inhibitory activity against DHODH. Moreover, given that normal tissue cells normally employ the alternate salvage pathway for pyrimidine synthesis, our results indicate that compound 5c may be a viable treatment option for the treatment of lung cancer therapy without causing side effects.
Overall, this study demonstrates that even with a small library of compounds, significant results can be achieved by the proper integration of computational methods with rational design. Our laboratory is currently conducting safety and pharmacokinetic tests on the identified hit compounds and further optimizing them based on the findings mentioned. The results of these studies will be reported in future work.
4. Methodology
4.1. Chemistry
All solvents and reagents were used without further purification. A Varian 400 MHz spectrometer (Varian Medical Systems, Inc., Palo Alto, CA) was used to calculate the 1H NMR spectra with chemical shifts and coupling constants measured in ppm (parts per million) and Hz, respectively. HR-ESIMS data were analyzed with a JMS-700 mass spectrometer or G2 QTOF mass spectrometer. Reaction observation was performed using TLC on 0.25 mm silica plates (E. Merck; silica gel 60 F254). Reversed-phase high-performance liquid chromatography (RP-HPLC) with a UV detector set at 254 nm was used to test the purity of the products. Mobile phases included H2O with 0.05% TFA and CH3CN, and a gradient of 75% B or 100% B was used for 45 min. Melting points were measured with a Fisher brand digital melting point instrument to determine the purity of the final product.
4.1.1. 5-Bromo-1H-indole-3-carboxylic acid (2)
Gray powder, yield: 100%, 1H NMR (400 MHz, DMSO-d6) δ 12.14 (s, 1H), 12.07 (s, 1H), 8.13 (d, J = 1.8 Hz, 1H), 8.05 (d, J = 3.0 Hz, 1H), 7.45 (d, J = 8.6 Hz, 1H), 7.32 (dd, J = 8.6, 1.9 Hz, 1H); 13C NMR (101 MHz, DMSO-d6) δ 149.82, 130.35, 130.11, 130.05, 126.34, 123.64, 117.65, 111.84, 109.83.
4.1.2. 5-Bromo-N-(3-(trifluoromethyl)phenyl)-1H-indole-3-carboxamide (3)
Yellow powder, yield: 48.68%, mp: 205.9 °C, HPLC purity: 20.173 min, 100%, 1H NMR (400 MHz, DMSO-d6) δ 12.01 (s, 1H), 10.10 (s, 1H), 8.39 (d, J = 2.9 Hz, 1H), 8.36 (d, J = 1.7 Hz, 1H), 8.26 (s, 1H), 8.02 (d, J = 8.5 Hz, 1H), 7.58 (t, J = 8.0 Hz, 1H), 7.48 (d, J = 8.6 Hz, 1H), 7.40 (d, J = 7.6 Hz, 1H), 7.34 (dd, J = 8.6, 1.9 Hz, 1H). 13C NMR (101 MHz, DMSO-d6) δ 163.55, 140.87, 135.41, 130.85, 130.25, 129.94, 129.62, 128.61, 125.35, 123.64, 123.50, 119.46, 116.10, 114.63, 114.17, 110.04. HRMS (ESI) m/z calcd for C16H11BrF3N2O [M + H]+: 383.0007, found, 383.0013.
4.1.3. 5-(3-(Morpholinomethyl)phenyl)-N-(3-(trifluoromethyl)phenyl)-1H-indole-3-carboxamide (4a)
Yellow crystal, yield: 21.02%, mp: 94.2 °C, HPLC purity: 7.969 min, 98.92%, 1H NMR (400 MHz, DMSO-d6) δ 11.97–11.88 (m, 1H), 10.10 (s, 1H), 8.46 (s, 1H), 8.39 (s, 1H), 8.29 (s, 1H), 8.03 (d, J = 8.5 Hz, 1H), 7.60–7.54 (m, 4H), 7.51 (d, J = 8.4 Hz, 1H), 7.41 (dd, J = 15.5, 7.9 Hz, 2H), 7.27 (d, J = 7.7 Hz, 1H), 3.61–3.57 (m, 4H), 3.56 (s, 2H), 2.40 (s, 4H). 13C NMR (101 MHz, CD3OD-d4) δ 165.22, 142.31, 140.01, 137.11, 136.28, 134.38, 131.58, 129.15, 128.87, 128.61, 128.49, 128.30, 128.23, 127.49, 126.88, 125.98, 123.19, 122.02, 119.15, 116.51, 111.73, 110.59, 66.27, 63.10, 53.24. HRMS (ESI) m/z calcd for C27H25F3N3O2 [M + H]+: 480.1899, found, 480.1893.
4.1.4. 5-(4-Methoxyphenyl)-N-(3-(trifluoromethyl)phenyl)-1H-indole-3-carboxamide (4b)
White crystal, yield: 77.07%, mp: 175.2 °C, HPLC purity: 20.578 min, 99.14%, 1H NMR (400 MHz, DMSO-d6) δ 11.86 (s, 1H), 10.07 (s, 1H), 8.41 (d, J = 1.1 Hz, 1H), 8.36 (s, 1H), 8.30 (s, 1H), 8.03 (d, J = 8.3 Hz, 1H), 7.61 (d, J = 8.8 Hz, 2H), 7.59–7.52 (m, 2H), 7.47 (dd, J = 8.5, 1.7 Hz, 1H), 7.39 (d, J = 7.7 Hz, 1H), 7.04 (d, J = 8.8 Hz, 2H), 3.81 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 163.98, 158.72, 141.07, 135.88, 134.44, 133.66, 130.19, 129.90, 129.59, 128.26, 127.40, 126.07, 123.43, 123.36, 121.91, 119.22, 118.92, 116.00, 115.96, 114.74, 112.86, 110.62, 55.56. HRMS (ESI) m/z calcd for C23H18F3N2O2 [M + H]+: 411.1320, found, 411.1313.
4.1.5. 5-((4-Hydroxyphenyl)amino)-N-(3-(trifluoromethyl)phenyl)-1H-indole-3-carboxamide (5a)
Gray powder, yield: 17.23%, mp: 194.9 °C, HPLC purity: 14.114 min, 94.84%, 1H NMR (400 MHz, DMSO-d6) δ 11.56 (s, 1H), 9.90 (s, 1H), 8.85 (s, 1H), 8.25–8.17 (m, 2H), 8.02 (d, J = 8.2 Hz, 1H), 7.80 (s, 1H), 7.55 (t, J = 8.0 Hz, 1H), 7.45 (s, 1H), 7.35 (d, J = 7.4 Hz, 1H), 7.30 (d, J = 8.6 Hz, 1H), 6.92 (d, J = 8.7 Hz, 2H), 6.86 (d, J = 8.6 Hz, 1H), 6.67 (d, J = 8.7 Hz, 2H). 13C NMR (101 MHz, CD3OD-d4) δ 165.53, 151.03, 140.29, 140.04, 137.58, 131.85, 130.75, 130.43, 129.10, 128.30, 127.19, 123.15, 119.77, 119.21, 119.17, 116.42, 116.38, 115.31, 115.21, 111.79, 109.53, 107.45. HRMS (ESI) m/z calcd for C22H17F3N3O2 [M + H]+: 412.1273, found, 412.1267.
4.1.6. 5-((3,4-Dimethoxyphenyl)amino)-N-(3-(trifluoromethyl)phenyl)-1H-indole-3-carboxamide (5b)
Yellow crystal, yield: 21.56%, mp: 99.7 °C, HPLC purity:17.027 min, 97.03%, 1H NMR (400 MHz, DMSO-d6) δ 11.61 (s, 1H), 9.93 (s, 1H), 8.23 (d, J = 3.1 Hz, 2H), 8.02 (d, J = 8.5 Hz, 1H), 7.94 (d, J = 1.7 Hz, 1H), 7.69 (s, 1H), 7.55 (t, J = 8.0 Hz, 1H), 7.38–7.32 (m, 2H), 6.93 (dd, J = 8.7, 2.1 Hz, 1H), 6.83 (d, J = 8.7 Hz, 1H), 6.71 (d, J = 2.4 Hz, 1H), 6.59 (dd, J = 8.5, 2.4 Hz, 1H), 3.71 (s, 3H), 3.69 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 164.09, 149.95, 142.59, 141.20, 139.70, 138.95, 131.70, 130.15, 129.85, 129.45, 129.44, 127.71, 123.29, 123.27, 115.86, 115.82, 114.07, 112.89, 109.72, 108.24, 107.92, 102.94, 56.66, 55.69. HRMS (ESI) m/z calcd for C24H21F3N3O3 [M + H]+: 456.1535, found, 456.1533.
4.1.7. 5-((4-Nitrophenyl)amino)-N-(3-(trifluoromethyl)phenyl)-1H-indole-3-carboxamide (5c)
Red crystal, yield: 31.25%, mp: 109.4 °C, HPLC purity:19.368 min, 97.48%, 1H NMR (400 MHz, DMSO-d6) δ 11.89 (s, 1H), 10.05 (s, 1H), 9.29 (s, 1H), 8.37 (s, 1H), 8.24 (s, 1H), 8.12–8.01 (m, 4H), 7.56 (dd, J = 16.9, 8.3 Hz, 2H), 7.38 (d, J = 7.5 Hz, 1H), 7.12 (d, J = 8.5 Hz, 1H), 6.96 (d, J = 9.3 Hz, 2H). 13C NMR (101 MHz, DMSO-d6) δ 163.55, 155.29, 152.15, 140.87, 135.40, 135.25, 130.85, 130.26, 130.24, 128.60, 125.35, 123.86, 123.63, 123.50, 123.49, 121.33, 119.46, 117.44, 116.05, 114.64, 114.17, 110.03. HRMS (ESI) m/z calcd for C22H16F3N4O3 [M + H]+: 441.1174, found, 441.1172.
4.1.8. 5-((3-Nitrophenyl)amino)-N-(3-(trifluoromethyl)phenyl)-1H-indole-3-carboxamide (5d)
Red crystal. yield: 30.49%, mp: 73.5 °C, HPLC purity: 20.123 min, 99.26%, 1H NMR (400 MHz, DMSO-d6) δ 11.82 (s, 1H), 10.02 (s, 1H), 8.59 (s, 1H), 8.34 (s, 1H), 8.24 (s, J = 9.1 Hz, 1H), 8.06–8.01 (m, 2H), 7.69 (d, J = 2.4 Hz, 1H), 7.56 (t, J = 8.0 Hz, 1H), 7.50 (d, J = 8.4 Hz, 2H), 7.43 (t, J = 8.1 Hz, 1H), 7.36 (dd, J = 13.4, 8.3 Hz, 2H), 7.11–7.06 (m, 1H). 13C NMR (101 MHz, DMSO-d6) δ 165.92, 163.94, 149.26, 148.07, 141.08, 138.05, 135.39, 133.43, 130.79, 130.21, 127.72, 123.39, 123.38, 120.43, 120.39, 118.29, 118.16, 115.93, 113.38, 112.21, 110.12, 107.56. HRMS (ESI) m/z calcd for C22H16F3N4O3 [M + H]+: 441.1174, found, 441.1175.
4.2. Biological Evaluation
4.2.1. WST-1 Cell Viability Assay
The American Type Culture Collection supplied the human lung adenocarcinoma cell line A549 and the non-small-cell lung cancer cell lines NCI-H1299 and NCI-H1975 (Manassas, VA). H1299 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 g/mL streptomycin. A549 and H1975 cells were grown in Roswell Park Memorial Institute (RPMI) 1640 media. After all cells reached 80–90% confluence, they were subcultured at 37 °C and 5% CO2 in a humidified incubator. The cells were exposed to the indicated concentrations of the compounds and the positive control regorafenib. Following 24 hours, each well received an addition of 10% WST-1 solution (Takara Bio, Shiga, Japan) and was then incubated for 1 additional hour.
4.2.2. hDHODH Evaluation
Compounds 3, 4a, and 5c were screened using the fluorescence-based assay to detect the fluorescent signal from the resorufin method by Reaction Biology Co. (http://www.reactionbiology.com, accessed on 27-12-2022). hDHODH catalyzes the oxidation of L-DHO to orotate, which is followed by resazurin catalysis to resorufin.35 Stop and detection step was employed for orotate addition to stop the reaction and detect the fluorescent signal from resorufin. The assay involved two steps: (i) Enzymatic step for hDHODH catalyzes the oxidation of L-DHO to orotate and followed by resazurin catalyzed to resorufin. The compounds were tested in 10-dose IC50 singlet with a 3-fold serial dilution starting at 10 μM. The base reaction buffer included: 100 mM Hepes pH 7.0, 150 mM NaCl, 0.3% CHAPS, 0.5 mg/mL BSA, 0.1 μM FMN, and 1% DMSO. The reaction was incubated for 60 minutes at room temperature, and a stop mixture composed of 100 mM Hepes pH 7.0 and 10 mM Orotate (5 mM as the final concentration) was used. IC50 Curve fits were performed when the activity at the highest concentration of compound was less than 65%.
4.3. DFT Calculations
The seven synthesized structures were exported from ChemDraw and optimized in Schrödinger 2021.2 using B3LYP theory and a 6-31G** basis set. The HOMO and LUMO energies were calculated by keeping an eye on the surfaces and atomic electrostatic potential charges.36,37 As a complex is formed, a molecule’s ability to donate electrons is indicated by its HOMO energy, whereas its ability to take electrons from its partner protein is indicated by its LUMO energy. The electronic excitation energy required to evaluate the stability and reactivity of the compounds is determined by the HOMO-LUMO gap energy, which is the difference between HOMO and LUMO energy.38
4.4. Molecular Docking
The crystal structure of human DHODH in complex with inhibitor 0944 was downloaded from the Protein Data Bank (PDB ID: 6LP7, resolution: 1.80 Å).39 The crystal structure was then prepared using Maestro Schrodinger’s Ligprep module by removing the water molecules and the 0944 co-crystal and adding any missing residues or hydrogen atoms.27 A molecular docking study was carried out for the synthesized compounds to predict their possible binding modes and understand their biological activity.19 Maestro Schrodinger Glide extra precision module was employed to dock each ligand in the binding site resulting in 32 poses for each ligand. The pose with the most negative energy score was then chosen and displayed using the using BIOVIA Discovery Studio Visualizer 2022 package.28
4.5. MD Calculations
DESMOND MD simulations were performed with the following parameters: The simulation system was built using an auto-calculated orthorhombic box solvated with explicit Single Point Charge (SPC) water molecules. The temperature was set to 300 K, and the pressure was set to 1 bar for the MD run.40 The simulation length was set at 100 ns, with a relaxation time of 1 ps for the selected positions. OPLS 2005 force parameters were used to minimize the energy of the solvated system.41 Particle mesh Ewald (PME) was used to handle electrostatic interactions, whereas periodic boundary conditions (PBC) and a 9.0 Å (Angstrom) cutoff were used for nonbond interactions.30
Each MD simulation was prepared employing a six-step relaxation protocol, which included 2000 steps of LBFGS minimization where the solute was restricted and a 50 kcal mol–1 Å–1 loose convergence criteria was applied. This was followed by two quick 12 ns simulations of temperature and pressure at T = 10 K (thermostat relaxation constant = 0.1 ps) and P = 1 atm (barostat relaxation constant = 50 ps) where nonhydrogen solute atoms were restrained. Finally, a third 24-ps NPT ensemble simulation where T = 300 K (thermostat relaxation constant = 0.1 ps; P = 1 atm) concluded the preparation processes.28 After relaxation, the systems underwent a 5 ns molecular dynamics (MD) simulation in the NPT ensemble, using a Nose–Hoover thermostat and Martyna–Tobias–Klein barostat (T = 300 K, thermostat relaxation time = 1.0 ps; P = 1 atm; barostat relaxation time = 2.0 ps). The MD simulation results were analyzed using QtGrace and Microsoft Excel.30
4.6. MM-GBSA Calculations
The binding free energy of the ligand–protein complexes was determined using the MM-GBSA method, which combines molecular mechanics and the Generalized Born surface area method. Using the 0–100 ns MD simulation trajectory, the VSGB solvation model, the OPLS3e force field, and a sampling rate of 10 steps per ns, the calculations were carried out using the thermal mmgbsa.py script.42 Applying the law of additivity, which took into account a variety of energy components including hydrogen bonding, van der Waals interactions, coloumbic interactions, lipophilic interactions, covalent interactions, solvation, stacking, and self-contact of the ligand and protein, allowed for the determination of the binding free energy.33,43
Acknowledgments
This study was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) [No. 2018R1A5A2023127] & [No. 2023R1A2C3004599]. This work was also supported by the BK21 FOUR program, which was funded by the Ministry of Education of Korea through NRF.
Data Availability Statement
All data used in this study has been publicly available. The raw data for the small combinatorial library, in vitro biological studies, molecular docking, molecular dynamic simulations (MD), density functional theory (DFT), and MM-GBSA calculations can be found in the data in a brief file shared in the OSF data repository: https://osf.io/wfrzh/?view_only=519abe6aeb464e9ba04e77f8b16c5827.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c01323.
Spectra data (1H NMR, 13C NMR, mass, and HPLC) for the synthesized compounds (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- DeBerardinis Ralph J.; Thompson Craig B. Cellular Metabolism and Disease: What Do Metabolic Outliers Teach Us?. Cell 2012, 148, 1132–1144. 10.1016/j.cell.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia I.; Haigis M. C. Metabolites and the tumour microenvironment: from cellular mechanisms to systemic metabolism. Nature Metabolism 2021, 3, 21–32. 10.1038/s42255-020-00317-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L.; Kepp O.; Heiden M. G. V.; Kroemer G. Metabolic targets for cancer therapy. Nat. Rev. Drug Discovery 2013, 12, 829–846. 10.1038/nrd4145. [DOI] [PubMed] [Google Scholar]
- Zhang W.; Zhang S.-L.; Hu X.; Tam K. Y. Targeting Tumor Metabolism for Cancer Treatment: Is Pyruvate Dehydrogenase Kinases (PDKs) a Viable Anticancer Target?. International Journal of Biological Sciences 2015, 11, 1390–1400. 10.7150/ijbs.13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deberardinis R. J. Is cancer a disease of abnormal cellular metabolism? New angles on an old idea. Genetics in Medicine 2008, 10, 767–777. 10.1097/GIM.0b013e31818b0d9b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shambaugh G. E. III Pyrimidine biosynthesis. The American Journal of Clinical Nutrition 1979, 32, 1290–1297. 10.1093/ajcn/32.6.1290. [DOI] [PubMed] [Google Scholar]
- Dimitrova P.; Skapenko A.; Herrmann M. L.; Schleyerbach R.; Kalden J. R.; Schulze-Koops H. Restriction of De Novo Pyrimidine Biosynthesis Inhibits Th1 Cell Activation and Promotes Th2 Cell Differentiation1. J. Immunol. 2002, 169, 3392–3399. 10.4049/jimmunol.169.6.3392. [DOI] [PubMed] [Google Scholar]
- Echizenya S.; Ishii Y.; Kitazawa S.; Tanaka T.; Matsuda S.; Watanabe E.; Umekawa M.; Terasaka S.; Houkin K.; Hatta T.; Natsume T.; Maeda Y.; Watanabe S.-I.; Hagiwara S.; Kondo T. Discovery of a new pyrimidine synthesis inhibitor eradicating glioblastoma-initiating cells. Neuro-Oncology 2019, 22, 229–239. 10.1093/neuonc/noz170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D. R.; Guy H. I. Mammalian Pyrimidine Biosynthesis: Fresh Insights into an Ancient Pathway *. J. Biol. Chem. 2004, 279, 33035–33038. 10.1074/jbc.R400007200. [DOI] [PubMed] [Google Scholar]
- Pal S.; Kaplan J. P.; Nguyen H.; Stopka S. A.; Savani M. R.; Regan M. S.; Nguyen Q.-D.; Jones K. L.; Moreau L. A.; Peng J.; Dipiazza M. G.; Perciaccante A. J.; Zhu X.; Hunsel B. R.; Liu K. X.; Alexandrescu S.; Drissi R.; Filbin M. G.; McBrayer S. K.; Agar N. Y. R.; Chowdhury D.; Haas-Kogan D. A. A druggable addiction to de novo pyrimidine biosynthesis in diffuse midline glioma. Cancer Cell 2022, 40, 957–972.e10. 10.1016/j.ccell.2022.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee R.; Purhonen J.; Kallijärvi J. The mitochondrial coenzyme Q junction and complex III: biochemistry and pathophysiology. FEBS J. 2022, 289, 6936–6958. 10.1111/febs.16164. [DOI] [PubMed] [Google Scholar]
- Li L.; Ng S. R.; Colón C. I.; Drapkin B. J.; Hsu P. P.; Li Z.; Nabel C. S.; Lewis C. A.; Romero R.; Mercer K. L.; Bhutkar A.; Phat S.; Myers D. T.; Muzumdar M. D.; Westcott P. M. K.; Beytagh M. C.; Farago A. F.; Vander Heiden M. G.; Dyson N. J.; Jacks T. Identification of DHODH as a therapeutic target in small cell lung cancer. Sci. Transl. Med. 2019, 11, eaaw7852 10.1126/scitranslmed.aaw7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He T.; Haapa-Paananen S.; Kaminskyy V. O.; Kohonen P.; Fey V.; Zhivotovsky B.; Kallioniemi O.; Perälä M. Inhibition of the mitochondrial pyrimidine biosynthesis enzyme dihydroorotate dehydrogenase by doxorubicin and brequinar sensitizes cancer cells to TRAIL-induced apoptosis. Oncogene 2014, 33, 3538–3549. 10.1038/onc.2013.313. [DOI] [PubMed] [Google Scholar]
- Molina J. R.; Yang P.; Cassivi S. D.; Schild S. E.; Adjei A. A. Non-Small Cell Lung Cancer: Epidemiology, Risk Factors, Treatment, and Survivorship. Mayo Clin. Proc. 2008, 83, 584–594. 10.1016/S0025-6196(11)60735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann M. L.; Schleyerbach R.; Kirschbaum B. J. Leflunomide: an immunomodulatory drug for the treatment of rheumatoid arthritis and other autoimmune diseases. Immunopharmacology 2000, 47, 273–289. 10.1016/S0162-3109(00)00191-0. [DOI] [PubMed] [Google Scholar]
- Zhou Y.; Tao L.; Zhou X.; Zuo Z.; Gong J.; Liu X.; Zhou Y.; Liu C.; Sang N.; Liu H.; Zou J.; Gou K.; Yang X.; Zhao Y. DHODH and cancer: promising prospects to be explored. Cancer Metab. 2021, 9, 22 10.1186/s40170-021-00250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.; Zhou Y.; Xu J.; Zhou X.; Liu S.; Huang Z.; Qiu Z.; Zeng T.; Gou K.; Tao L.; Zhong X.; Yang X.; Zhou Y.; Su N.; Chen Q.; Zhao Y.; Luo Y. Discovery of potent human dihydroorotate dehydrogenase inhibitors based on a benzophenone scaffold. Eur. J. Med. Chem. 2022, 243, 114737 10.1016/j.ejmech.2022.114737. [DOI] [PubMed] [Google Scholar]
- Vyas V. K.; Variya B.; Ghate M. D. Design, synthesis and pharmacological evaluation of novel substituted quinoline-2-carboxamide derivatives as human dihydroorotate dehydrogenase (hDHODH) inhibitors and anticancer agents. Eur. J. Med. Chem. 2014, 82, 385–393. 10.1016/j.ejmech.2014.05.064. [DOI] [PubMed] [Google Scholar]
- Lee K.; Nada H.; Byun H. J.; Lee C. H.; Elkamhawy A. Hit Identification of a Novel Quinazoline Sulfonamide as a Promising EphB3 Inhibitor: Design, Virtual Combinatorial Library, Synthesis, Biological Evaluation, and Docking Simulation Studies. Pharmaceuticals 2021, 14, 1247. 10.3390/ph14121247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner R. A.; Murphy R. B.; Repasky M. P.; Frye L. L.; Greenwood J. R.; Halgren T. A.; Sanschagrin P. C.; Mainz D. T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein–Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Hu X.; Wu L.-w.; Zhang Z.-y.; Chen M.-l.; Li Y.-l.; Zhang C. The anti-tumor effect of regorafenib in lung squamous cell carcinoma in vitro. Biochem. Biophys. Res. Commun. 2018, 503, 1123–1129. 10.1016/j.bbrc.2018.06.129. [DOI] [PubMed] [Google Scholar]
- Kies M. S.; Blumenschein G. R. Jr.; Christensen O.; Lin T.; Tolcher A. W.. Phase I study of regorafenib (BAY 73-4506), an inhibitor of oncogenic and angiogenic kinases, administered continuously in patients (pts) with advanced refractory non-small cell lung cancer (NSCLC). 2010; Vol. 28 (15_suppl), , pp 7585.
- Vanwynsberghe H.; Verbeke X.; Coolen J.; Van Cutsem E. Predictive Value of Early Tumor Shrinkage and Density Reduction of Lung Metastases in Patients With Metastatic Colorectal Cancer Treated With Regorafenib. Clin. Colorectal Cancer 2017, 16, 377–380. 10.1016/j.clcc.2017.03.017. [DOI] [PubMed] [Google Scholar]
- LaPointe S. M.; Weaver D. F. A Review of Density Functional Theory Quantum Mechanics as Applied to Pharmaceutically Relevant Systems. Current Computer - Aided Drug Design 2007, 3, 290–296. 10.2174/157340907782799390. [DOI] [Google Scholar]
- Banavath H. N.; Sharma O. P.; Kumar M. S.; Baskaran R. Identification of novel tyrosine kinase inhibitors for drug resistant T315I mutant BCR-ABL: a virtual screening and molecular dynamics simulations study. Sci. Rep. 2014, 4, 6948 10.1038/srep06948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Porta F. A.; Ramalho T. C.; Santiago R. T.; Rocha M. V. J.; da Cunha E. F. F. Orbital Signatures as a Descriptor of Regioselectivity and Chemical Reactivity: The Role of the Frontier Orbitals on 1,3-Dipolar Cycloadditions. J. Phys. Chem. A 2011, 115, 824–833. 10.1021/jp108790w. [DOI] [PubMed] [Google Scholar]
- Nada H.; Lee K.; Gotina L.; Pae A. N.; Elkamhawy A. Identification of novel discoidin domain receptor 1 (DDR1) inhibitors using E-pharmacophore modeling, structure-based virtual screening, molecular dynamics simulation and MM-GBSA approaches. Comput. Biol. Med. 2022, 142, 105217 10.1016/j.compbiomed.2022.105217. [DOI] [PubMed] [Google Scholar]
- Nada H.; Elkamhawy A.; Lee K. Identification of 1H-purine-2,6-dione derivative as a potential SARS-CoV-2 main protease inhibitor: molecular docking, dynamic simulations, and energy calculations. PeerJ 2022, 10, e14120 10.7717/peerj.14120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa S. F.; Fernandes P. A.; Ramos M. J. Protein–ligand docking: Current status and future challenges. Proteins: Struct., Funct., Bioinf. 2006, 65, 15–26. 10.1002/prot.21082. [DOI] [PubMed] [Google Scholar]
- Nada H.; Kim S.; Godesi S.; Lee J.; Lee K. Discovery and optimization of natural-based nanomolar c-Kit inhibitors via in silico and in vitro studies. J. Biomol. Struct. Dyn. 2023, 1–12. 10.1080/07391102.2022.2164061. [DOI] [PubMed] [Google Scholar]
- Bornot A.; Etchebest C.; de Brevern A. G.. Predicting protein flexibility through the prediction of local structures Proteins: Struct., Funct., Bioinf. 2011, 79 (3), 839–852. 10.1002/prot.22922. [DOI] [PMC free article] [PubMed]
- Kuzmanic A.; Zagrovic B. Determination of Ensemble-Average Pairwise Root Mean-Square Deviation from Experimental B-Factors. Biophys. J. 2010, 98, 861–871. 10.1016/j.bpj.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmaaty A. A.; Alnajjar R.; Hamed M. I. A.; Khattab M.; Khalifa M. M.; Al-Karmalawy A. A. Revisiting activity of some glucocorticoids as a potential inhibitor of SARS-CoV-2 main protease: theoretical study. RSC Adv. 2021, 11, 10027–10042. 10.1039/D0RA10674G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsagaby S. A.; Iqbal D.; Ahmad I.; Patel H.; Mir S. A.; Madkhali Y. A.; Oyouni A. A. A.; Hawsawi Y. M.; Alhumaydhi F. A.; Alshehri B.; Alturaiki W.; Alanazi B.; Mir M. A.; Al Abdulmonem W. In silico investigations identified Butyl Xanalterate to competently target CK2α (CSNK2A1) for therapy of chronic lymphocytic leukemia. Sci. Rep. 2022, 12, 17648 10.1038/s41598-022-21546-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero I.; Lafuente M. J.; Gamo F.-J.; Cid C. A high-throughput fluorescence-based assay for Plasmodium dihydroorotate dehydrogenase inhibitor screening. Anal. Biochem. 2016, 506, 13–21. 10.1016/j.ab.2016.04.013. [DOI] [PubMed] [Google Scholar]
- Bochevarov A. D.; Harder E.; Hughes T. F.; Greenwood J. R.; Braden D. A.; Philipp D. M.; Rinaldo D.; Halls M. D.; Zhang J.; Friesner R. A. Jaguar: A high-performance quantum chemistry software program with strengths in life and materials sciences. Int. J. Quantum Chem. 2013, 113, 2110–2142. 10.1002/qua.24481. [DOI] [Google Scholar]
- Parcha P.; Sarvagalla S.; Madhuri B.; Pajaniradje S.; Baskaran V.; Coumar M. S.; Rajasekaran B. Identification of natural inhibitors of Bcr-Abl for the treatment of chronic myeloid leukemia. Chem. Biol. Drug Des. 2017, 90, 596–608. [DOI] [PubMed] [Google Scholar]
- Ke Y.-Y.; Singh V. K.; Coumar M. S.; Hsu Y. C.; Wang W.-C.; Song J.-S.; Chen C.-H.; Lin W.-H.; Wu S.-H.; Hsu J. T. A.; Shih C.; Hsieh H.-P. Homology modeling of DFG-in FMS-like tyrosine kinase 3 (FLT3) and structure-based virtual screening for inhibitor identification. Sci. Rep. 2015, 5, 11702 10.1038/srep11702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z.; Liu X.; Qian X.; Zeng T.; Sang N.; Liu H.; Zhou Y.; Tao L.; Zhou X.; Su N.; Yu Y.; Chen Q.; Luo Y.; Zhao Y. Bifunctional Naphtho[2,3-d][1,2,3]triazole-4,9-dione Compounds Exhibit Antitumor Effects In Vitro and In Vivo by Inhibiting TDihydroorotate Dehydrogenase and Inducing Reactive Oxygen Species Production. J. Med. Chem. 2020, 63, 7633–7652. 10.1021/acs.jmedchem.0c00512. [DOI] [PubMed] [Google Scholar]
- Christensen N. J.; Kepp K. P. Stability Mechanisms of a Thermophilic Laccase Probed by Molecular Dynamics. PLoS One 2013, 8, e61985 10.1371/journal.pone.0061985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar D.; Williams J.; Wu Y.; Damm W.; Shelley J.; Sherman W. Prediction of Absolute Solvation Free Energies using Molecular Dynamics Free Energy Perturbation and the OPLS Force Field. J. Chem. Theory Comput. 2010, 6, 1509–1519. 10.1021/ct900587b. [DOI] [PubMed] [Google Scholar]
- Wang L.; Wu Y.; Deng Y.; Kim B.; Pierce L.; Krilov G.; Lupyan D.; Robinson S.; Dahlgren M. K.; Greenwood J.; Romero D. L.; Masse C.; Knight J. L.; Steinbrecher T.; Beuming T.; Damm W.; Harder E.; Sherman W.; Brewer M.; Wester R.; Murcko M.; Frye L.; Farid R.; Lin T.; Mobley D. L.; Jorgensen W. L.; Berne B. J.; Friesner R. A.; Abel R. Accurate and Reliable Prediction of Relative Ligand Binding Potency in Prospective Drug Discovery by Way of a Modern Free-Energy Calculation Protocol and Force Field. J. Am. Chem. Soc. 2015, 137, 2695–2703. 10.1021/ja512751q. [DOI] [PubMed] [Google Scholar]
- Zrieq R.; Ahmad I.; Snoussi M.; Noumi E.; Iriti M.; Algahtani F. D.; Patel H.; Saeed M.; Tasleem M.; Sulaiman S.; Aouadi K.; Kadri A.. Tomatidine and Patchouli Alcohol as Inhibitors of SARS-CoV-2 Enzymes (3CLpro, PLpro and NSP15) by Molecular Docking and Molecular Dynamics Simulations Int. J. Mol. Sci. 2021, 22 (19), 10693. 10.3390/ijms221910693. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this study has been publicly available. The raw data for the small combinatorial library, in vitro biological studies, molecular docking, molecular dynamic simulations (MD), density functional theory (DFT), and MM-GBSA calculations can be found in the data in a brief file shared in the OSF data repository: https://osf.io/wfrzh/?view_only=519abe6aeb464e9ba04e77f8b16c5827.