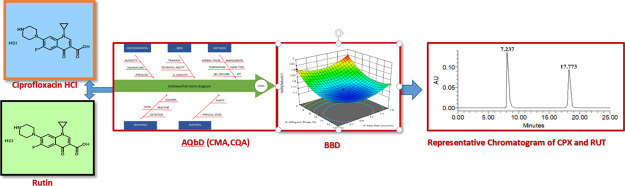
Abstract
In the given study, a new reverse-phase high-performance liquid chromatography (RP-HPLC) method has been reported for the simultaneous estimation of ciprofloxacin hydrochloride (CPX) and rutin (RUT) using quality by design (QbD) approach. The analysis was carried out by applying the Box–Behnken design having fewer design points and less experimental runs. It relates between factors and responses and gives statistically significant values, along with enhancing the quality of the analysis. CPX and RUT were separated on the Kromasil C18 column (4.6 × 150 mm, 5 μm) using an isocratic mobile phase combination of phosphoric acid buffer (pH 3.0) and acetonitrile with the ratio of 87:13% v/v at a flow rate of 1.0 mL/min. CPX and RUT were detected at their respective wavelengths of 278 and 368 nm using a photodiode array detector. The developed method was validated according to guideline ICH Q2 R (1). The validation parameters taken were linearity, system suitability, accuracy, precision, robustness, sensitivity, and solution stability which were in the acceptable range. The findings suggest that the developed RP-HPLC method can be successfully applied to analyze novel CPX-RUT-loaded bilosomal nanoformulation prepared by thin-film hydration technique.
1. Introduction
A biofilm is a cluster of microbes encapsulated in the self-produced extracellular polymeric substance (EPS) adhered on living or nonliving surfaces,1 making infections challenging to treat and causing the emergence of drug-resistant infections.2 Reportedly, 90% of nosocomial infections are caused by bacterial biofilms, posing significant challenges to the healthcare sector.3 Several studies have reported that bacteria in biofilms can be 10–1000 fold more resistant to antibiotics than free-floating single-celled (planktonic) bacteria.3,4 Quorum sensing is an inter and intrabacterial communication channel which plays a crucial role in the development of bacterial biofilms. It controls various metabolic activities of the planktonic compartment and shows vulnerable adaptive antibiotic resistance in biofilms which leads to hindrance in biofilm-associated treatment and chronic infections globally. Many research studies reported that phytoconstituents act as quorum-sensing inhibitors which result in the remarkable degradation of biofilms when combined with antibiotics5,6 The main factor which makes biofilms resistant to antibiotics is the composition of EPS, which inhibits penetration and reduces drug diffusion.7
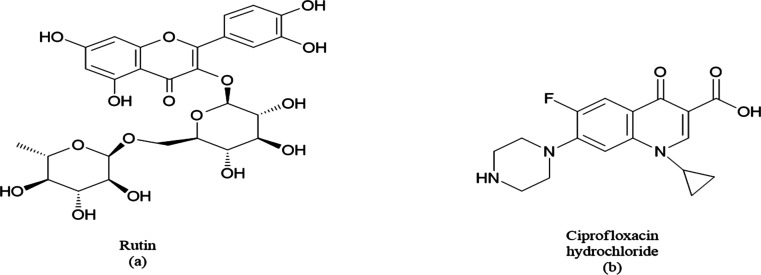
Ciprofloxacin hydrochloride (CPX) is a broad-spectrum fluoroquinolone second-generation antibiotic (Figure 1b) that targets the DNA gyrase of bacteria. While most medicines typically have trouble piercing biofilms, CPX has been found to diffuse properly, especially against Escherichia coli and Klebsiella pneumoniae biofilms.8 Irrational and improper usage of these antimicrobials leads to resistance, resulting in higher doses of antimicrobial therapy.9
Figure 1.
Chemical structure of (a) RUT and (b) CPX.
Several studies have reported that plant extracts or their active phytoconstituents could also inhibit bacterial biofilm development, check viable bacteria number in biofilms, and reduce biomass. Rutin (RUT) is a citrus flavonoid glycoside and a low-molecular-weight polyphenolic compound (Figure 1a), which shows different pharmacological properties like antiprotozoal,10 antitumor,11 anti-inflammatory12 antiallergic,13 antiviral,13 and antibacterial.14 Recent studies demonstrated RUT as a potential antibiofilm agent against Klebsella Pneumonia and Staphylococcus xylosus strains isolated from hospitalized patients.15 By using antibacterial agents CPX and RUT, a complete eradication of bacterial infection can be expected. Furthermore, by using nanoformulations, the efficacy can further be enhanced. It results in increased bioavailability, enhanced uptake, biofilm disruption, and improved pharmacokinetic profile of the drug.
Effectiveness, safety, and quality are crucial for a pharmaceutical product. Scientific strategies such as, quality by design (QbD) and process analytical technology are used to improve product quality.16−20 ICH defines QbD as a systematic approach to the development that begins with predefined objectives and emphasizes product and process understanding and control based on sound science and quality risk management. Analytical QbD is used in the analytical process. The analytical method is developed using AQbD, which is equivalent to QbD that helps in the development of a robust and cost-effective analytical method which leads to establishment of method-operable design region by considering the method factor. There are some factorial experimental designs such as Box–Behnken design (BBD) and central composite design (CCD), which are frequently used in pharmaceutical analysis.21 In the given manuscript, BBD experimental runs were used having fewer design points and less experimental runs. It relates between factors and responses and gives statistically significant values. In the experimental runs, organic phase ratio, flow rate, and pH of mobile phase were selected as independent variables, whereas retention time (RT), theoretical plate, and tailing factor were considered as dependent factors. Validation of the developed method was carried out as per ICH guideline ICH Q2 R (1). A degradation test was also performed to evaluate the proposed HPLC method stability.22
There are several literature reports of HPLC analytical method of CPX, for example, in bulk drugs HPLC-UV23,24 and in human plasma HPLC-flourescence.25 Similarly, for RUT quantitative analysis, different HPLC methods have been reported.26 Quantitative analyses of RUT in different species of solanum plant extracts HPLC-UV,27 in solid dosage forms,28 in human plasma,29 and in cigarette tobacco30 have been reported. The above-reported methods are for the analysis of a single drug; no analytical method has been reported for the simultaneous analysis of CPX and RUT in any application. Therefore, this method has been developed to simultaneously analyze CPX and RUT by HPLC using a PDA detector. The developed method was successfully applied to analyze the novel CPX-RUT-loaded bilosomal nanoformulation.
2. Materials and Methods
2.1. Chemicals and Reagents
RUT (purity ≥98.0%) was procured from Sisco Research Laboratories Pvt. Ltd. (Mumbai, India), and CPX (percentage purity; 99.0%) was obtained ex-gratia from Sun pharmaceutical industries Ltd. Gurugram, India. Acetonitrile (99.8%) and methanol (99.8%) HPLC grade were purchased from CDH fine chemical (New Delhi, India). Orthophosphoric acid (85%) was purchased from CDH fine chemical (New Delhi, India). Milli-Q water (Gradient A10R Millipore, Moscheim Cedex, France) was used for sample preparation.
2.1.1. HPLC Instrumental Attributes and Analytical Setup
The chromatographic analysis of the APIs was performed on the Waters Alliance HPLC system (model no e2695 Waters Co, MA, U.S.A.) having a PDA detector. The instrument consisted of a quaternary solvent manager, an auto-sampler and a column oven. Empower software ver.2.0 was used. The analysis was carried out using a reversed-phase C18 column (100-5-C18 4.6 × 150 mm, 5 μm) from Kromasil Electron Corporation, Nouryon, Sweden. Different wavelengths were used for detection (278 nm for CPX and 368 nm for RUT).31 The mobile phase combination of phosphoric acid buffer (pH 3.0) and acetonitrile with a ratio of 87:13% v/v was eluted at a flow rate of 1.0 mL/min. The run time for the analysis was 20 min at 40 °C column temperature. Injection volume was taken as 10 μL. A 0.45 μm membrane filter (Millipore) was used for the mobile phase, and a 0.22 μm syringe filter was used (Avantor performance materials India limited) for chromatographic samples.
2.1.2. Preparation of Stock Solution, Working Solution, and Quality Control Solution
The stock solutions of CPX and RUT were separately prepared in a volumetric flask (10 mL), taking the accurate weight of their solid standard to obtain 1 mg/mL concentration. The standard stock solution of CPX was prepared in a buffer solution of the mobile phase, while RUT was dissolved in methanol and volume adjusted by the mobile phase, followed by sonication (6 min). The working solution was prepared by taking 1 mL from the stock solution and diluting with the mobile phase to obtain 100 μg/mL of concentration. The working solution of CPX and RUT was further serially diluted to prepare the calibration standard, with concentration 1–15 μg/mL28 for analytes. A similar procedure was followed to prepare quality control samples of 4, 8, and 12 μg/mL, which were considered as lower quality control (LQC), middle quality control (MQC), and higher quality control (HQC) solutions, respectively. All solutions were filtered and freshly prepared and stored in the refrigerator.
2.1.3. Preliminary Method Development Studies
Based on the literature on the physicochemical properties of the drug, the solubility of both drugs was screened to identify the maximum solubility profiles in organic solvents (methanol acetonitrile). However, the method development was carried out using an equal volume of organic phase and aqueous phase in combination, where aqueous phase with different pH (3–5) values was adjusted by orthophosphoric acid, formic acid, and glacial acetic acid. All possible mobile phase ratios at different pH values were studied to evaluate the solubility profile and established better chromatographic estimation.
2.1.4. Assessment of the Analytical Method Using the QbD Approach
The AQbD approach is categorized into different steps. The initial step is an analytical target product profile, which is an essential element of an AQbd system that ensures how and what to be measured for the desired quality process. The second one is the critical analytical attribute (CAA), which refers to the analytical method and parameter attributes like product identification and separation of peak, robustness, method accuracy, and their precision. Critical method attributes (CMAs) that determine analytical technique performance include mobile phase pH, injection volume, column temperature, and so forth. CMAs influence method execution factors like sample concentration and reagent and their grades. The DoE method is employed to identify a central process and produce design space based on statistical significance. Method validation provides credible verification of quality outcomes. The control approach ensures that the technique performs as expected.32
2.1.5. Chemometrics-Assisted RP-HPLC Method Development
The factorial design BBD is a type of DoE software (version 11.1) that has been used for the optimization of the HPLC method consisting of independent and dependent variables. As compared to CCD, this design gives high responses of independent variables with fewer probable runs. In this study, three independent variables were taken that is, percentage organic phase (A), flow rate (B), and mobile phase pH (C). The dependent variables for CPX were RT (Y1), theoretical plates (Y2), and tailing factor (Y3), and the dependent variables for RUT were RT (Y4), theoretical plates (Y5), and tailing factor (Y6). Polynomial equations were used to analyze the results of 17 test runs as shown in Table 2. The details of the method validation study, polynomial equation, and coefficient have been mentioned in the Section 3.
Table 2. Experimental Runs, Variables, and Measured Responses Using Factorial Design (Box–Behnken) for the Optimization Studya.
| run | factor A | factor B | factor C | Y1 RT | Y2 theoretical plate | Y3 tailing factor | Y4 RT | Y5 theoretical plate | Y6 tailing factor |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 13 | 0.5 | 3.45 | 7.78 | 6126.9 | 1.47 | 17.78 | 7136.6 | 1.61 |
| 2 | 10 | 1 | 3.45 | 7.79 | 4462.7 | 1.36 | 17.79 | 5442.6 | 1.56 |
| 3 | 10 | 0.5 | 4 | 8.93 | 5390.8 | 1.59 | 18.93 | 6360.4 | 1.61 |
| 4 | 10 | 1 | 3.45 | 7.87 | 4387.5 | 1.33 | 17.87 | 5337.7 | 1.53 |
| 5 | 10 | 1 | 3.45 | 7.9 | 4566.2 | 1.32 | 17.9 | 5586.3 | 1.52 |
| 6 | 10 | 0.5 | 2.9 | 8.71 | 5171.6 | 1.43 | 18.71 | 6181.4 | 1.61 |
| 7 | 7 | 0.5 | 3.45 | 8.98 | 3926.7 | 1.58 | 18.98 | 4966.7 | 1.72 |
| 8 | 7 | 1.5 | 3.45 | 8.17 | 4790.8 | 1.76 | 18.17 | 5750.8 | 1.74 |
| 9 | 13 | 1.5 | 3.45 | 6.79 | 6987.9 | 1.42 | 16.79 | 7947.9 | 1.62 |
| 10 | 10 | 1.5 | 4 | 7.88 | 5958.1 | 1.58 | 17.88 | 6978.3 | 1.68 |
| 11 | 10 | 1 | 3.45 | 7.98 | 4790.8 | 1.36 | 17.98 | 5780 4 | 1.56 |
| 12 | 7 | 1 | 4 | 7.99 | 4280.8 | 1.69 | 17.99 | 5240.8 | 1.75 |
| 13 | 13 | 1 | 2.9 | 6.09 | 6858.1 | 1.39 | 16.09 | 7878.1 | 1.59 |
| 14 | 7 | 1 | 2.9 | 7.88 | 4136.4 | 1.59 | 17.88 | 5156.4 | 1.67 |
| 15 | 13 | 1 | 4 | 6.59 | 7161.7 | 1.48 | 16.59 | 8141.6 | 1.58 |
| 16 | 10 | 1.5 | 2.9 | 7.88 | 5890.8 | 1.56 | 17.88 | 6880.8 | 1.62 |
| 17 | 10 | 1 | 3.45 | 7.99 | 4493.9 | 1.34 | 17.99 | 5473.6 | 1.54 |
Abbreviations: A, organic phase %; B, flow rate; C, mobile phase.
Response surface methodology examines the correlation between the variables, and variance analysis was used to confirm the model relevance (ANOVA). Here, the range of dependent parameters was chosen between 7 and 13% for the organic phase for pH of the mobile phase 2.9 to 4 and for flow rate 0.5 to 1.5 mL/min as given in Table 1.
Table 1. Selected Full Factorial Design 33 22 Variables and Their Constraints.
| independent variable | levels |
||
|---|---|---|---|
| low (−1) | medium (0) | high (+1) | |
| A = organic phase %(v/v) | 7.0 | 10 | 13 |
| B = flow rate (mL/min) | 0.5 | 1.0 | 1.5 |
| C = mobile phase pH | 2.9 | 3.45 | 4 |
| independent variable | constraint | importance | |
|---|---|---|---|
| A = organic phase %(v/v) | in range | +++ | |
| B = flow rate (mL/min) | in range | +++ | |
| C = mobile phase pH | in range | +++ |
| dependent variable | |||
|---|---|---|---|
| Y1 = RT of CPX (min) | minimum | +++ | |
| Y2 = number of theoretical plates of CPX | maximum | +++ | |
| Y3 = tailing factor of CPX | minimum | +++ | |
| Y4 = RT of RUT (min) | minimum | +++ | |
| Y5 = number of theoretical plates of RUT | maximum | +++ | |
| Y6 = tailing factor of RUT | minimum | +++ | |
| independent variable | constraint | importance | |
|---|---|---|---|
| A = organic phase %(v/v) | in range | +++ | |
| B = flow rate (mL/min) | in range | +++ | |
| C = mobile phase pH | in range | +++ |
| dependent variable | |||
|---|---|---|---|
| Y1 = RT of CPX (min) | minimum | +++ | |
| Y2 = number of theoretical plates of CPX | maximum | +++ | |
| Y3 = tailing factor of CPX | minimum | +++ | |
| Y4 = RT of RUT (min) | minimum | +++ | |
| Y5 = number of theoretical plates of RUT | maximum | +++ | |
| Y6 = tailing factor of RUT | minimum | +++ | |
2.2. Method Validation
As per ICH Q2(R1) guidelines, the proposed method was validated with different parameters, system suitability, specificity linearity, accuracy, precision, limit of detection (LOD), limit of quantification (LOQ), robustness, as well as stability studies at respective storage conditions.33−36
2.2.1. System Suitability Test
System suitability is an integral part of chromatography technique to check method reproducibility and to ensure that the procedure is adequate for the intended use. The test was conducted at MQC concentration (8 μg/mL) to inject six replicas, and the outcome was evaluated by the RT, the peak area, the theoretical column plate. and tailing factors. To ensure the system’s suitability, the considerable approved criterion is the relative standard deviation (% RSD) of RT. The range of peak area should be ≤2%. The range of tailing factor should not surpass 2. The range of theoretical plates of the column should not be more than 2000 (N > 2000) (Table 2).
2.2.2. Specificity
Specificity is an important parameter of HPLC analytical method which refers to the ability of the analytical method to identify the analytes from the heterogeneous mixture. Specificity has been evaluated by comparing the chromatograms of individual drug solutions and their complex mixture with their blank solution (without adding the drugs CPX and RUT) at their MQC concentration.
2.2.3. Linearity Range
Calibration plots of RUT and CPX mixture solution were plotted at concentrations ranging from 1 to 15 μg/mL. These concentrations cover 40, 60, 80, 100, 120, 140, and 160% of the target concentration. The regression equation of the respective drug was calculated by plotting the individual calibration curve using the concentration and the peak area, respectively. The response factor was also calculated.
2.2.4. Robustness
The robustness was assessed by the impact on RT and peak area by a slight variation in chromatographic properties. Samples were analyzed by slight changes in the mobile phase composition ratio (85:15, 87:13, and 89:11), pH (2.9, 3.0, and 3.1), and column oven temperature (35, 40, and 45 °C). The effect caused by the method change was investigated, and the result was analyzed. The analysis was performed at MQC concentration (8 μg/mL). The experiment was performed in triplicate for each sample and each variation. The % RSD evaluates the robustness of RUT and CPX, ≤2% range for acceptance of the method.37
2.2.5. Accuracy
Accuracy means the closeness between the expected and actual values. It was evaluated by measuring the % recovery of both the analytes (CPX and RUT) at three QC concentrations (4, 8, and 12 μg/mL). Both the analytes were injected at all three QC concentration levels in triplicate, and % recovery and their %RSD were measured. The percentage variation in recovery should be limited to 90–110% with %RSD ≤ 2% for the acceptance of the method.
| 1 |
2.2.6. Precision
The capacity to which a method is repeatedly utilized to examine several replicates in different circumstances is referred to as precision. The samples of both the analytes were subjected to interday and intraday QC analyses for the determination of precision. Lay out of the study for intraday was three replicas of the same concentration in a day for both analytes at all three QC concentrations (LQC, MQC, and HQC), while for interday, same study was on different days (3 days). For the calculation of peak area of both the analytes, their % recovery were noted. The limit of % RSD calculation is ≤2%.
2.2.7. Sensitivity (LOD and LOQ)
Assay sensitivity of analytes was evaluated by determining the LOD and LOQ using the developed analytical technique. The LOD is considered the lowest concentration of the calibration curve, of which the signal-to-noise ratio (S/N) is ≥3. In contrast, LOQ is the concentration that delivers approximately S/N ≥ 10 with a % RSD (n = 3) of >10. LOD was determined by analyzing the lowest possible concentration likely to be reliably distinguished from the limit of blank and the concentration at which detection is feasible. In contrast, LOQ is the concentration at which quantification is possible with acceptable accuracy and precision. The LOD and LOQ of both analytes were calculated using the formula38A = kr/S, where A denotes the detection and quantification value, r represents response’s SD, and S is the slope of the calibration curve. k is a constant the value of which LOQ = 10 and LOD = 3.3.
2.2.8. Stability Studies of Analytes
The stability of both the analytes (CPX and RUT) in an aqueous solution was studied to ensure the accuracy of the concentration at different storage levels. Both LQC and HQC concentration were measured in triplicate and analyzed in % recovery±SD by storing the samples at 25 ± 2 °C (ambient temperature) for 24, 48, and 72 h and 4 °C (refrigerator) for 15 days. The concentrations of each QC and the variation in the chromatographic pattern were measured against prepared calibration curves. The obtained data must rely in the range.
2.3. Application of the Simultaneous Analytical Method in CPX-RUT-Loaded Bilosomal Nanoformulation
The developed analytical method was utilized to quantify CPX and RUT in various novel bilosomal nanoformulations. The drug-release profile in dissolution studies and the amount of drug entrapped in the bilosomes were analyzed by using the given assay. Briefly, the thin-film hydration technique39 was used to prepare CPX-RUT-loaded bilosomes with some significant modifications. A 25 mL round bottom flask (RBF) was taken in which an accurately weighed amount of surfactant (Span 60), hydrophobic drug, and cholesterol was dissolved in an organic solvent (chloroform) followed by sonication up to 15 min (Ultrasonic bath sonicator, model SH 150-41; U.S.A.). The sample solution was evaporated using a Rota evaporator at a temperature of 65 ± 2 °C under reduced pressure (Rotavapor, Heidolph VV 2000; Germany) till all organic solvents evaporated and a thin dry film formed in RBF. Weighed amounts of bile salt and hydrophilic drug were dissolved in Milli-Q water and poured in a thin film-containing RBF for their hydration. The hydration process lasted for 45 min. A suspension of bilosome was obtained. The formed opalescent formulation was stored at 4 °C until further use.40
3. Results and Discussion
3.1. Preliminary Method Development Studies
This study was performed to analyze the initial easement and suitability for different chromatographic conditions. The mobile phase contains acetonitrile as an organic phase, and the aqueous phase (pH 3) contains orthophosphoric acid as the pH modifier with a ratio of 13:87% v/v. Initial trials performed using the said solvent system at 1.0 mL/min flow rate, the column oven temperature of 25 °C, and the sample injection volume of 10 μL give the characteristic peak of both drugs. Initially, the peaks were not highly symmetric, and some tailing was also observed. To overcome these conditions, detailed screening of the different key method variables was performed which critically impacts the method performance.
3.2. Assessment of the Analytical Method Using the QbD Approach
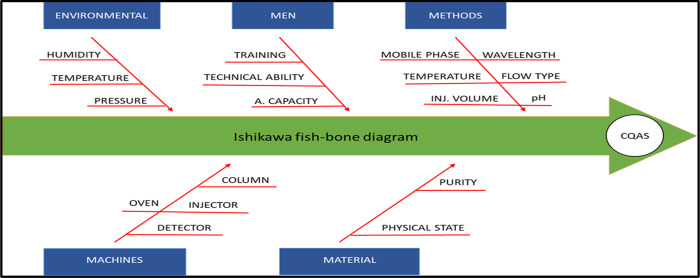
In the AQbD approach, the analytical target profile (ATP) plays a similar role as the quality target product profile (QTPP) in specifying the objectives for method development (Table 3). ATP serves as the intended characteristic of the CAA of the method’s intended purpose and regulatory constraints. The ATP specifies a collection of attributes and target analytes that must be measured, such as the technique and concentration range that will be employed as per the required characteristics of the method. The analytical method monitoring is associated with critical method parameters (CMPs). CMP and critical quality attributes are connected in a cause-and-effect manner, and it has the ability to influence the selected CAAs. The three most important technical factors affecting the HPLC procedure are the column temperature, the organic phase concentration, and the mobile phase pH. Column aging (CMP), for example, can have an effect on the tailing factor and plate counts (CAA). In this experimental design, tailing factor, theoretical plates, peak area, and RT have been considered as CAAs as shown in Table 4. The fish-bone diagram, commonly called as an Ishikawa diagram, has been designed for the current study to investigate the risk assessment to identify the CMPs as shown in Figure 2.
Table 3. Analytical Target Profile Parameters of the Method Development.
| parameters | target | justification |
|---|---|---|
| sample | API in developed bilosome | developing an analytical method for the simultaneous quantification of CPX and RUT in developed Bilosomes. |
| instrument | HPLC | both CPX and RUT are nonvolatile and show absorbance in the UV range. So, the HPLC method with a UV detector was selected. |
| type of method | reverse phase-HPLC | nonpolar stationary phase tends to offer improved retention of molecules. |
| nature of sample | aqueous | analyte must be in the aqueous phase, ensuring complete miscibility |
| standard and sample preparation | methanol | based on the pKa and solubility of CPX and RUT, buffer and ACN were selected as the diluent. |
| application of method | assessment of CPX and RUT | the method is applicable to assess CPX and RUT in the bilosome and in the biological samples. |
Table 4. CMP and CAA and Their Relationship in the Method Development.
| S.N. | critical method parameters (CMP) | critical analytical attributes (CAA) |
|---|---|---|
| 1 | HPLC column (dimensions, stationary phase, make) | peak area |
| 2 | column flow rate | RT |
| 3 | column oven temperature | tailing factor |
| 4 | buffer for mobile phase | plate counts |
| 5 | buffer concentration pH of mobile phase buffer | |
| 6 | mobile phase gradient | |
| 7 | column flow rate |
Figure 2.
Ishikawa (fish-bone diagram) explains the risk assessment parameter responsible for the quality attributes of the method.
3.3. Chemometrics-Assisted RP-HPLC Method Development
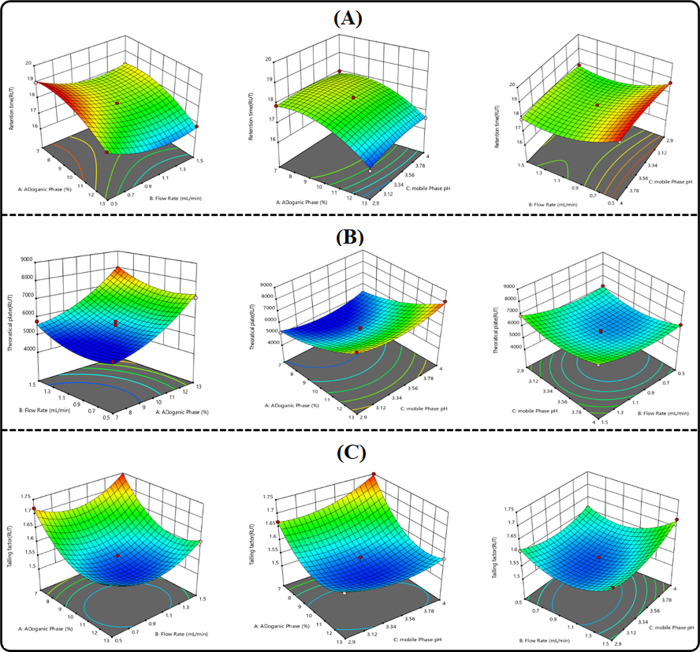
The experimental settings were optimized using BBD. 3D graphs illustrating the impact of variables were obtained. Notable disparities in values were evident in every response. Table 2 shows all dependent variables, expected and actual r2 values. The corrected r2 value and the projected r2 value agreed reasonably well. A strong correlation associated with the experimental data and the fitted model is predicted by the increased adjusted r2 value. The polynomial equation has been used to forecast how the variables and responses relate to one another. The effect that promotes optimization is indicated by a positive value, whereas an opposition interaction between the elements is indicated by a negative value. The two interacting components from equations (AB and BC) were discovered to be negative, whereas the effect of the AC interaction on answer Y1 was positive. For response Y4, it was determined from eq 4 that the two interactions, BC and AC, were negative, while one AB was positive. Responses Y2 and Y5 from the polynomial eqs 2 and 5 revealed that the three interaction terms, AB, BC, and AC, were all antagonistic to one another. The two interaction terms AB and AC were shown to be favorable for responses Y3 and Y6, while the correlation of BC showed the negative result, as shown in eqs 3 and 7
| 2 |
| 3 |
| 4 |
| 5 |
| 6 |
| 7 |
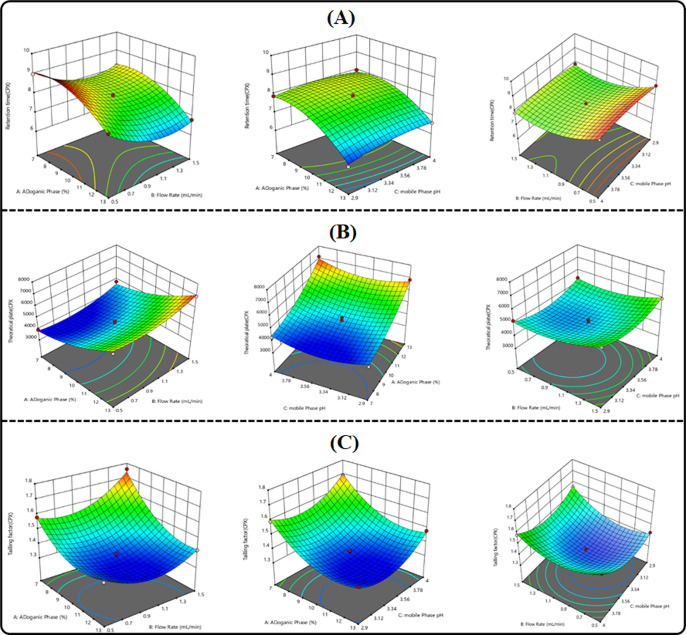
In the 3D plots, three factors were taken; out of three factors, the effect of two factors was considered and the third one was kept constant. The effects of independent factors on the RT of both analytes are depicted in Figure 3A and Figure 4A Y1 & Y4. The flow rate and the organic-to-aqueous phase ratio had a noticeable impact on the RT of both analytes, whereas the pH of the aqueous mobile phase had not shown any effect on the RT of both analytes. The effects of independent variables on the theoretical plates are depicted in Figure 3B and Figure 4B Y2 & Y5. This finding demonstrated that the organic phase ratio and the flow rate unfavorably impact the theoretical plates.
Figure 3.
Factorial design response surface plots describing the effect of independent variables on the dependent variables. (A) RT, (B) theoretical plates, and (C) tailing factor of CPX.
Figure 4.
Factorial design response surface plots describe the effect of independent variables on the dependent variables. (A) RT, (B) theoretical plates, and (C) tailing factor of RUT.
Theoretical plates were somewhat raised by a rise in the % organic ratio, where as it was found to be increased by a decrease in the flow rate. With the increase in the pH of the mobile phase, the theoretical plates increased in proportion and vice versa, which demonstrates the significant impact of pH of CPX and RUT on the theoretical plates. The impact of independent variables on the tailing factor is depicted in Figure 3C and Figure 4C Y3, Y6 in factorial graphs. With the increase in the percentage of the organic phase concentration and flow rate, the decreases in the tailing effect of the analytes have been observed.
The tailing factor is slightly increased by the pH of the aqueous phase. The Design-Expert Stat-Ease, Inc. (version 11.1) software gives the optimal value for independent parameters such as the organic phase (13%), the mobile phase pH (2.9), and the flow rate (1 mL/min) at these concentrations, and it was discovered that every response was good. A negligible prediction error (± 5%) was found between the predicted and observed values, indicating the strong predicted capability of the model for their response variables. Considering the optimized conditions, the method shows excellent chromatographic separation of the drug, as shown in the figure.
3.4. Method Validation
3.4.1. System Suitability Test
The % RSD of the peak area and RT of both analytes was calculated and was found to be within the range, that is, ±2%, as depicted in supplementary Tables S1 and S2. The mean ± %RSD of the number of theoretical plates in the column and of the tailing factor in the six replicate injections were found to be 6829.16 ± 1.4 and 1.38 ± 0.74% for CPX and 7795.87 ± 1.21 and 1.57 ± 0.66% for RUT, respectively. In the system suitability, the number of theoretical plates exceeded 2000 considered for the test. The tailing factor % RSD of both analytes was found to be in the limit as mentioned in ICH guidelines. These outcomes suggest that the HPLC method was adequate, and the data are in an acceptable range.
3.4.2. Specificity
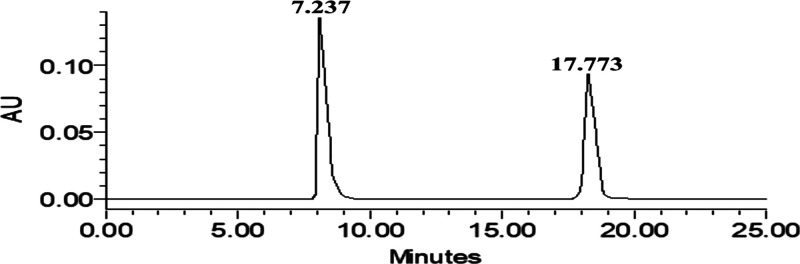
Specificity can be measured by comparing the chromatograms of CPX and RUT individually and simultaneously at a predetermined concentration (Figure S1). When a different set of sample (containing CPX and RUT) equivalent to 20 μL was injected into the HPLC, its RT was found to be 7.237 ± 0.073 min and 17.773 ± 0.034 for CPX and RUT. The results obtained show that the RT of CPX and RUT remains the same when samples have the individual drug as well as when the samples containing a mixture of drugs are injected. Furthermore, no other interfering peaks were obtained other than that of drugs. All this indicates that the proposed method is highly specific for the estimation of CPX and RUT.
3.4.3. Linearity
The linear relation was obtained in the form of a calibration curve by plotting a graph between the mean peak areas of the analytes (RUT, CPX) and their respective concentrations (1–15 μg/mL). The linear equation and r2 of both the analytes (CPX and RUT) were calculated (y = 91,541x + 37,280; 0.999 and y = 80,945x + 16,633; 0.998, respectively). These results show a linear relationship for both the analytes.
3.4.4. Robustness
Analytical process robustness was assessed by slightly changing the HPLC parameters, that is, changes in the pH of both the analytes, the column oven temperature, and the mobile phase ratio. The %RSD of the peak area and RT of both the analytes analyzed at MQC concentration were within the acceptable limits of ≤2%. Similarly, the tailing factor and the number of theoretical plates also did not show not any notable effect. Slight change in the method indicated that the proposed method is optimized and robust for estimation of both the analytes.
3.4.5. Accuracy
The accuracy measured by % recovery is regarded as closeness toward the actual value. At all QC concentrations, the % recovery range is 98.45–101.83 and 99.58–103.17% for CPX and RUT, as shown in Table S3. These results are in the limits and show that the method is accurate.
3.4.6. Precision
At all QC levels, the % RSD of % recovery for intraday and interday precisions (intermittent) was calculated, and the value was in the range of 0.93–1.04 and 1.12–1.69% for CPX and 0.77–1.97 and 1.54–1.66% for RUT, respectively. The data show that the method gives good precision and reproducibility with % RSD, found to be less than 2%, as given in Table S4. This suggests that the methods are repeatable and reliable.
3.4.7. Sensitivity (LOD and LOQ)
The signal-to-noise ratios for LOD and LOQ were obtained for both the analytes at 3:1 and 1:10. LOD and LOQ were calculated as 0.5 and 0.9 μg/mL for RUT and 0.44 and 1.1 μg/mL for CPX, respectively. The result shows that the proposed method is sufficiently sensitive than the previously reported method.23,27,28
3.4.8. Stability Studies of Analytes
The stability of both the analytes (CPX and RUT) in aqueous solution was assessed in all QC levels at different storage conditions. Short-term stability study also called as the accelerated stability study was performed at room temperature, whereas long-term stability study was performed at 4 °C for a period of 2 weeks in the refrigerator. The results were in the range of 90.4–101.4% for RUT and 87.4–100.2% for CPX, as shown in Table S5. These findings suggest that neither drug degraded under various storage circumstances. This means that the aqueous QC samples were stable up to 72 h at ambient temperature. Additionally, its standard solutions remained stable for at least 15 days at 4 °C (refrigerator).
3.5. Developed Optimized Method and Their Pharmaceutical Application
The optimized method was successfully used to determine CPX and RUT simultaneously on prepared bilosomal nanoformulations for drug release and entrapment studies. The calibration range (1–15 μg/mL) was sufficient to cover the quantification of different drug samples obtained in in vitro and ex vivo studies. The formulation characterization outcome did not show any significant difference in the RT of both the analytes. No extra peaks were observed at the RT of CPX and RUT, which further confirmed the assay’s specificity (Figure 5).
Figure 5.
Representative chromatogram of CPX and RUT obtained during estimation of bilosome formulation.
4. Conclusions
The present research proposes a novel RP-HPLC-PDA method, which was successfully developed and validated to simultaneously estimate CPX and RUT in bilosomal nanoformulation and shows therapeutic efficacy against antibiotic resistance by disrupting bacterial biofilms. The use of AQbD and the Box–Banken experimental design facilitates the significant improvement of performance and the robustness of the method for the successful separation and estimation of two analytes. The developed method was found to be efficient and effective for routine analysis and offered a quality result. The use of Design-Expert Stat-Ease, Inc. (version 11.1) approach reduces the number of experimental runs, and validation was carried out using ICH Q2 R (1) guidelines. All parameters were considered for method validation, which were found to be in the acceptable range and confirm that the proposed method is affordable, precise, robust, and sensitive. These outcomes reveal that the proposed analytical HPLC method can be used to simultaneously estimate CPX and RUT in combined dosage forms for future investigations.
Acknowledgments
No support of this research was provided by governmental, private, or non-profit funding sources.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c00956.
System suitability parameters for CPX; system suitability parameters for RUT; accuracy of CPX and RUT; repeatability and intermediate precision of CPX and RUT; stability study of both analytes CPX and RUT at all QC levels under different storage conditions; chromatograms of RUT, CPX, and simultaneous RUT and CPX at MQC concentration (PDF)
Author Contributions
Conceptualization, methodology, investigation, data curation, and writing, A.S.; project administration, A.A. and Z.I.; resources and supervision, A.M. and M.A.; software and validation, M.I.; writing—review and editing and formal analysis, A.A.; supervision, resources, and visualization, M.J.A. All authors have read and agreed to the published version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Erratum to: Dynamic Remodeling of Microbial Biofilms by Functionally Distinct Exopolysaccharides: MBio: Volume 5, No. 4, 10.1128/MBio.01536-14, 2014. mBio. 2015. 10.1128/mBio.00688-15.
- Hall-Stoodley L.; Costerton J. W.; Stoodley P.; Hall-Stoodley L.; Costerton J. W.; Stoodley P. Bacterial Biofilms: From the Natural Environment to Infectious Diseases. Nat. Rev. Microbiol. 2004, 2004, 95–108. 10.1038/Nrmicro821. [DOI] [PubMed] [Google Scholar]
- Oliveira A. S.; Rolo J.; Gaspar C.; Cavaleiro C.; Salgueiro L.; Palmeira-de-Oliveira R.; Ferraz C.; Coelho S.; Pastorinho M. R.; Sousa A. C.; Teixeira J. P.; Martinez-de-Oliveira J.; Palmeira-de-Oliveira A. Chemical Characterization and Bioactive Potential of Thymus × Citriodorus (Pers.) Schreb. Preparations for Anti-Acne Applications: Antimicrobial, Anti-Biofilm, Anti-Inflammatory and Safety Profiles. J. Ethnopharmacol. 2022, 287, 114935 10.1016/j.jep.2021.114935. [DOI] [PubMed] [Google Scholar]
- Penesyan A.; Gillings M.; Paulsen I. T. Antibiotic Discovery: Combatting Bacterial Resistance in Cells and in Biofilm Communities. Molecules 2015, 20, 5286. 10.3390/molecules20045286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju D. V.; Nagarajan A.; Pandit S.; Nag M.; Lahiri D.; Upadhye V. Effect of Bacterial Quorum Sensing and Mechanism of Antimicrobial Resistance. Biocatal. Agric. Biotechnol. 2022, 43, 102409 10.1016/j.bcab.2022.102409. [DOI] [Google Scholar]
- Shamim A.; Ali A.; Iqbal Z.; Mirza M. A.; Aqil M.; Kawish S. M.; Siddiqui A.; Kumar V.; Naseef P. P.; Alshadidi A. A. F.; Saheer Kuruniyan M. Natural Medicine a Promising Candidate in Combating Microbial Biofilm. Antibiotics 2023, 12, 299. 10.3390/antibiotics12020299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega N. M.; Gore J. Collective Antibiotic Resistance: Mechanisms and Implications. Curr. Opin. Microbiol. 2014, 21, 28. 10.1016/j.mib.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebreyohannes G.; Nyerere A.; Bii C.; Sbhatu D. B. Challenges of Intervention, Treatment, and Antibiotic Resistance of Biofilm-Forming Microorganisms. Heliyon 2019, 5, e02192 10.1016/j.heliyon.2019.e02192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verderosa A. D.; Totsika M.; Fairfull-Smith K. E. Bacterial Biofilm Eradication Agents: A Current Review. Front. Chem. 2019, 7, 824. 10.3389/fchem.2019.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman F.; Tabrez S.; Ali R.; Alqahtani A. S.; Ahmed M. Z.; Rub A. Molecular Docking Analysis of Rutin Reveals Possible Inhibition of SARS-CoV-2 Vital Proteins. J. Tradit. Complement. Med. 2021, 11, 173. 10.1016/j.jtcme.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Castro A. J.; Domínguez F.; García-Carrancá A. Rutin Exerts Antitumor Effects on Nude Mice Bearing SW480 Tumor. Arch. Med. Res. 2013, 44, 346. 10.1016/j.arcmed.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Guardia T.; Rotelli A. E.; Juarez A. O.; Pelzer L. E. Anti-Inflammatory Properties of Plant Flavonoids. Effects of Rutin, Quercetin and Hesperidin on Adjuvant Arthritis in Rat. Farmaco 2001, 56, 683. 10.1016/S0014-827X(01)01111-9. [DOI] [PubMed] [Google Scholar]
- Yong D. O. C.; Saker S. R.; Chellappan D. K.; Madheswaran T.; Panneerselvam J.; Choudhury H.; Pandey M.; Chan Y. L.; Collet T.; Gupta G.; Oliver B. G.; Wark P.; Hansbro N.; Hsu A.; Hansbro P. M.; Dua K.; Zeeshan F. Molecular and Immunological Mechanisms Underlying the Various Pharmacological Properties of the Potent Bioflavonoid, Rutin. Endocrine, Metab. Immune Disord. Drug Targets 2020, 20, 1590. 10.2174/1871530320666200503053846. [DOI] [PubMed] [Google Scholar]
- Motallebi M.; Khorsandi K.; Sepahy A. A.; Chamani E.; Hosseinzadeh R. Effect of Rutin as Flavonoid Compound on Photodynamic Inactivation against P. Aeruginosa and S. Aureus. Photodiagn. Photodyn. Ther. 2020, 32, 102074 10.1016/j.pdpdt.2020.102074. [DOI] [PubMed] [Google Scholar]
- Qu Q.; Cui W.; Xing X.; Zou R.; Huang X.; Wang X.; Wu T.; Bello-Onaghise G.; Yuan S.; Li Y. Rutin, A Natural Inhibitor of IGPD Protein, Partially Inhibits Biofilm Formation in Staphylococcus Xylosus ATCC700404 in Vitro and in Vivo. Front. Pharmacol. 2021, 12, 728354 10.3389/fphar.2021.728354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K. Y.; Dedania Z. R.; Dedania R. R.; Patel U. QbD Approach to HPLC Method Development and Validation of Ceftriaxone Sodium. Futur. J. Pharm. Sci. 2021, 7, 141. 10.1186/s43094-021-00286-4. [DOI] [Google Scholar]
- Dedania Z. R.; Dedania R. R.; Sheth N. R.; Patel J. B.; Patel B. Stability Indicating HPLC Determination of Risperidone in Bulk Drug and Pharmaceutical Formulations. Int. J. Anal. Chem. 2011, 2011, 124917 10.1155/2011/124917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talele S. G.; Derle D. V. Stability-Indicating High-Performance Liquid Chromatography (HPLC) Method Development and Validation for the Determination of Quetiapine Fumarate in Bulk and Dosage Form by HPLC. Int. J. Green Pharm. 2018, 12, S188. [Google Scholar]
- Nuchtavorn N.; Leanpolchareanchai J.; Chanton D.; Supapsophon P.; Chongruchiroj S.; Chatmapanrangsee J.; Suksiriworapong J. A Rapid Stability Indicating HPLC Method for Determination of Quetiapine Fumarate in Tablets and Extemporaneous Formulations. Pharm. Chem. J. 2021, 13, 845. 10.1007/s11094-021-02505-x. [DOI] [Google Scholar]
- Vijaya Kumar M.; Muley P. R.. Stability Indicating Hplc Method for Determination of Quetiapine Fumarate in Bulk Drug and Solid Dosage Forms. Indian Drugs 2004. [Google Scholar]
- Ostovan A.; Ghaedi M.; Arabi M.; Yang Q.; Li J.; Chen L. Hydrophilic Multitemplate Molecularly Imprinted Biopolymers Based on a Green Synthesis Strategy for Determination of B-Family Vitamins. ACS Appl. Mater. Interfaces 2018, 10, 4140. 10.1021/acsami.7b17500. [DOI] [PubMed] [Google Scholar]
- Krishna M. V.; Dash R. N.; Jalachandra Reddy B.; Venugopal P.; Sandeep P.; Madhavi G. Quality by Design (QbD) Approach to Develop HPLC Method for Eberconazole Nitrate: Application Oxidative and Photolytic Degradation Kinetics. J. Saudi Chem. Soc. 2016, 20, S313. 10.1016/j.jscs.2012.12.001. [DOI] [Google Scholar]
- Pascual-Reguera M. I.; Parras G. P.; Díaz A. M. Solid-Phase UV Spectrophotometric Method for Determination of Ciprofloxacin. Microchem. J. 2004, 77, 79. 10.1016/j.microc.2004.01.003. [DOI] [Google Scholar]
- Emami J.; Rezazadeh M. A Simple and Sensitive High-Performance Liquid Chromatography Method for Determination of Ciprofloxacin in Bioavailability Studies of Conventional and Gastroretentive Prolonged-Release Formulations. Adv. Biomed. Res. 2016, 5, 163. 10.4103/2277-9175.190995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imre S.; Dogaru M. T.; Vari C. E.; Muntean T.; Kelemen L. Validation of an HPLC Method for the Determination of Ciprofloxacin in Human Plasma. J. Pharm. Biomed. Anal. 2003, 33, 125. 10.1016/S0731-7085(03)00151-1. [DOI] [PubMed] [Google Scholar]
- Alajmi M. F.; Alam P.; Rehman M. T.; Husain F. M.; Khan A. A.; Siddiqui N. A.; Hussain A.; Kalam M. A.; Parvez M. K. Interspecies Anticancer and Antimicrobial Activities of Genus Solanum and Estimation of Rutin by Validated UPLC-PDA Method. Evidence-based Complement. Altern. Med. 2018, 2018, 6040815 10.1155/2018/6040815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attia T. Z. Simultaneous Determination of Rutin and Ascorbic Acid Mixture in Their Pure Forms and Combined Dosage Form. Spectrochim. Acta, Part A 2016, 169, 82. 10.1016/j.saa.2016.06.030. [DOI] [PubMed] [Google Scholar]
- Kuntić V.; Pejić N.; Ivković B.; Vujić Z.; Ilić K.; Mićić S.; Vukojević V. Isocratic RP-HPLC Method for Rutin Determination in Solid Oral Dosage Forms. J. Pharm. Biomed. Anal. 2007, 43, 718. 10.1016/j.jpba.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Ishii K.; Furuta T.; Kasuya Y. Determination of Rutin in Human Plasma by High-Performance Liquid Chromatography Utilizing Solid-Phase Extraction and Ultraviolet Detection. J. Chromatogr. B: Biomed. Sci. Appl. 2001, 759, 161. 10.1016/S0378-4347(01)00224-9. [DOI] [PubMed] [Google Scholar]
- Sun Y.; Li W.; Wang J.; Bi J.; Su S. Determination of Rutin in Cigarette Tobacco, Filters, Mainstream Smoke and Burned Ash of Different Branded Cigarettes by High Performance Liquid Chromatography. Molecules 2012, 17, 3751. 10.3390/molecules17043751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kicel A.; Owczarek A.; Michel P.; Skalicka-Woźniak K.; Kiss A. K.; Olszewska M. A. Application of HPCCC, UHPLC-PDA-ESI-MS3 and HPLC-PDA Methods for Rapid, One-Step Preparative Separation and Quantification of Rutin in Forsythia Flowers. Ind. Crops Prod. 2015, 76, 86. 10.1016/j.indcrop.2015.06.019. [DOI] [Google Scholar]
- Tome T.; Žigart N.; Časar Z.; Obreza A. Development and Optimization of Liquid Chromatography Analytical Methods by Using AQbD Principles: Overview and Recent Advances. Org. Process Res. Dev. 2019, 23, 1784. 10.1021/acs.oprd.9b00238. [DOI] [Google Scholar]
- Thi Thuy Linh D.; Thanh Duong H.; Tuan Hiep N.; Thanh Huyen P.; Minh Khoi N.; Doan Long D. Simultaneous Quantification of Hederacoside C and α-Hederin in Hedera Nepalensis K.Koch Using HPLC-UV. VNU J. Sci. Med. Pharm. Sci. 2020, 36, 4227. 10.25073/2588-1132/vnumps.4227. [DOI] [Google Scholar]
- Serfaty D.25 Years ESC. Eur. J. Contracept. Reprod. Health Care 2014. [Google Scholar]
- Tsuang M. T. 1983 United States Pharmacopia Dispensing Information. J. Urol. 1983, 129, 683. 10.1016/s0022-5347(17)52316-2. [DOI] [Google Scholar]
- Beyea L.Big Savings & Fast Paybacks in Existing Laboratory Buildings. In 33rd West Coast Energy Management Congress, EMC 2015, 2015.
- Ganorkar S. B.; Dhumal D. M.; Shirkhedkar A. A. Development and Validation of Simple RP-HPLC-PDA Analytical Protocol for Zileuton Assisted with Design of Experiments for Robustness Determination. Arab. J. Chem. 2017, 10, 273. 10.1016/j.arabjc.2014.03.009. [DOI] [Google Scholar]
- Beg S.; Panda S. S.; Katare O. P.; Singh B. Applications of Monte-Carlo Simulation and Chemometric Techniques for Development of Bioanalytical Liquid Chromatography Method for Estimation of Rosuvastatin Calcium. J. Liq. Chromatogr. Relat. Technol. 2017, 40, 907. 10.1080/10826076.2017.1382377. [DOI] [Google Scholar]
- Dai Y.; Zhou R.; Liu L.; Lu Y.; Qi J.; Wu W. Liposomes Containing Bile Salts as Novel Ocular Delivery Systems for Tacrolimus (FK506): In Vitro Characterization and Improved Corneal Permeation. Int. J. Nanomed. 2013, 8, 1921. 10.2147/IJN.S44487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Mahallawi A. M.; Abdelbary A. A.; Aburahma M. H. Investigating the Potential of Employing Bilosomes as a Novel Vesicular Carrier for Transdermal Delivery of Tenoxicam. Int. J. Pharm. 2015, 485, 329. 10.1016/j.ijpharm.2015.03.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.