Abstract
Amelogenin, the dominant organic component (>90%) in the early stages of amelogenesis, orchestrates the mineralization of apatite crystals into enamel. The self-association properties of amelogenin as a function of pH and protein concentration have been suggested to play a central role in this process; however, the large molecular weight of the self-assembled quaternary structures has largely prevented structural studies of the protein in solution under physiological conditions using conventional approaches. Here, using perdeuterated murine amelogenin (0.25 mM, 5 mg/mL) and TROSY-based NMR experiments to improve spectral resolution, we assigned the 1H-15N spectra of murine amelogenin over a pH range (5.5 to 8.0) where amelogenin is reported to exist as oligomers (pH ≤~6.8) and nanospheres (pH ≥~7.2). The disappearance or intensity reduction of amide resonances in the 1H-15N HSQC spectra was interpreted to reflect changes in dynamics (intermediate millisecond-to-microsecond motion) and/or heterogenous interfaces of amide nuclei at protein–protein interfaces. The intermolecular interfaces were concentrated toward the N-terminus of amelogenin (L3-G8, V19-G38, L46-Q49, and Q57-L70) at pH 6.6 (oligomers) and at pH 7.2 (nanospheres) including the entire N-terminus up to Q76 and regions distributed through the central hydrophobic region (Q82-Q101, S125-Q139, and F151-Q154). At all pH levels, the C-terminus appeared disordered, highly mobile, and not involved in self-assembly, suggesting nanosphere structures with solvent-exposed C-termini. These findings present unique, residue-specific insights into the intermolecular protein–protein interfaces driving amelogenin quaternary structure formation and suggest that nanospheres in solution predominantly contain disordered, solvent-exposed C-termini.
Graphical Abstract
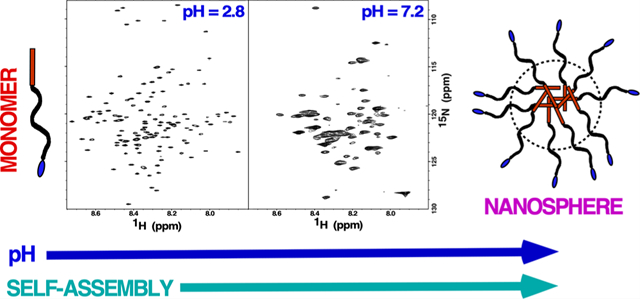
INTRODUCTION
The hardest and most mineralized tissue in vertebrates is enamel, the 1−2 mm of material deposited over the surface of dentin in the latter stages of tooth development.1 Unable to self-repair, enamel evolved to be extraordinarily resilient because it needs to survive a lifetime of repeated masticatory and parafunctional activity within an oral cavity harboring a dynamic microflora that includes over 700 species of bacteria.2 The exceptional strength and robust mechanical properties of enamel arise from the weaving of long and narrow hydroxyapatite (HAP) crystals into a unique lattice architecture composed of closely packed parallel arrays (enamel rods).3–6 The predominant matrix protein (>90%) present during enamel formation is amelogenin,7–9 secreted in the first of three stages (secretory, transition, and maturation) in the biomineralization process (amelogenesis).8,10 By the end of the third stage, the enamel is largely devoid of amelogenin or any other type of organic matter, having been progressively reabsorbed, assisted by digestion with the proteases enamelysin (matrix metalloproteinase-20, MMP-20)11,12 and kallikrein (KLK-4).13
While amelogenin’s central role in enamel formation is unquestionable, how this low molecular weight protein (~20 kDa) controls HAP crystal growth at a molecular level is still poorly understood.5,6,14 The primary amino acid sequence of murine amelogenin is compared to the porcine and human sequences in Figure 1A. An analysis of these and other vertebrate tetrapod sequences illustrates a conserved distribution of hydrophilic and hydrophobic regions that can be divided into three parts (Figure 1B): the N-terminal, hydrophilic tyrosine-rich region (TRAP, red); the hydrophobic central region heavily populated with histidine, glutamine, and proline residues (HR, black); and the mineral binding C-terminal hydrophilic region (CTHR, blue).15 Variations in the length of amelogenin among species is largely due to differences in the length of the central HR region.15 On the other hand, the length of both termini is more uniform across species and this feature, along with the conserved charged amino acid residues, indirectly suggests that both ends of the protein play significant functional roles in mediating biomineralization.16
Figure 1.
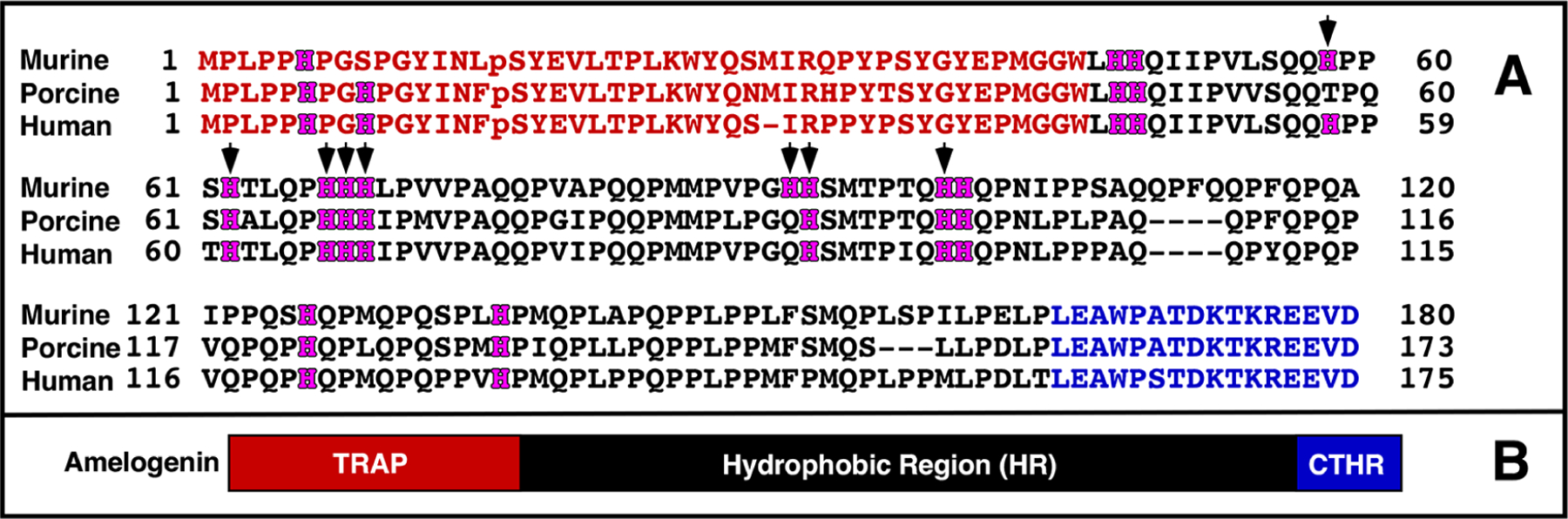
(A) Primary amino acid sequences of murine, porcine, and human amelogenin. The amelogenin primary amino acid sequence is highly conserved across species with the variations usually observed in the length of the hydrophobic region. The pS at position 16 is a phosphoserine observed in native protein. While S16 is phosphorylated in amelogenins isolated from natural sources, it is not phosphorylated in proteins generated recombinantly in Escherichia coli. The histidine residues are highlighted in magenta with arrows indicating the site of the eight histidine residues deleted in the M179Δ8H construct. (B) Cartoon representation of the three regions present in amelogenin: N-terminal tyrosine-rich region (TRAP; red), hydrophobic region (HR; black), and C-terminal hydrophilic region (CTHR; blue).
Changes in solution pH trigger the self-assembly of amelogenin into different quaternary structures that are likely essential to function.16–19 Starting at low pH (<~3.5), amelogenin is primarily monomeric.20,21 As physiological pH levels are approached (pH ~6.8), it forms oligomers of increasing size, maxing out at an average size of about eight monomers.22,23 At pH values greater than 7, nanospheres composed of 20 to 100 monomers form23–25 and, under the right conditions, nanosphere chains19 and nanoribbons.26,27 Nanospheres have been observed both in vivo11 and in vitro,28 and the consensus is that supramolecular structures are responsible for the templating of enamel.11,29,30
Factors other than pH influence the equilibrium between the various amelogenin quaternary states, including protein concentration, amelogenin degradation products, temperature, and properties of the solution such as ionic strength and solutes.6,16,31 These conditions change over the course of amelogenesis as HAP formation releases protons into solution and amelogenin is progressively degraded into smaller and smaller fragments by proteases. Mutations to amelogenin have also been observed to perturb its quaternary states.32,33 Amelogenesis imperfecta describes a group of hereditary conditions that affect the quantity and quality of enamel.34,35 The majority of these genetic disorders are associated with the amelogenin gene AMELX and include two separate missense mutations that result in a single amino acid substitution, T21I or P40T.36 In vitro circular dichroism32 and NMR33 studies identified errant and altered self-assembly properties, respectively, for full-length amelogenins containing these single amino acid substitutions, suggesting that perturbations to the self-assembly properties of amelogenin may be the biophysical basis for the phenotypes associated with these two AMELX mutations.
Given the importance of amelogenin self-assembly during amelogenesis, we previously mapped the intermolecular interface of amelogenin oligomerization by first assigning its 1H-15N HSQC spectrum under conditions where it was predominately monomeric (low concentration, no salt, pH 2.8)20,37 and then followed perturbations to the 1H-15N HSQC spectrum under conditions that promoted self-assembly. With amelogenin, such perturbations primarily took the form of intensity reductions, often beyond detection limits, to the backbone amide resonances, making it possible to follow self-association at the residue level. This is because the disappearance of amide resonances in 1H-15N HSQC spectra were interpreted to reflect changes in dynamics (intermediate millisecond-to-microsecond motion) and/or multiple chemical environments (heterogenous interfaces) of amide nuclei at protein–protein interfaces.20,33,38 Starting under conditions with no salt at low pH (2.8), increasing concentrations of NaCl were observed to promote dimer formation starting at a region near the N-terminus of murine amelogenin (T21–R31) that extended to a larger region (Y12–I51) near the N-terminus along with a region near the C-terminus (L141–T171) at higher NaCl concentrations.20 However, pH 2.8 is far from physiological, and therefore here, using perdeuterated39,40 amelogenin and TROSY-based NMR experiments41 to improve spectral resolution, we assigned the 1H-15N spectra of murine amelogenin over a pH range (5.5 to 8.0) where amelogenin is reported to exist as oligomers (pH ≤6.8) and nanospheres (pH ≥7.2).24 This allowed us to map the amelogenin–amelogenin interresidue surfaces in these different large molecular weight quaternary structures. In addition, perturbations to 1H-15N spectra were used to follow the self-assembly of amelogenin into octamers as a function of increasing amelogenin concentration at a single pH value (5.5). Finally, because histidine residues, with a pKa of 6.6 and distributed throughout the HR region (magenta, Figure 1B), have been postulated to temper pH-driven oligomerization transitions (at pH >6.6, the histidine side chain is neutral and more hydrophobic),24 a murine amelogenin construct was prepared with eight out of the 13 histidine residues in the HR region removed and 1H-15N HSQC TROSY spectra collected as a function of pH to monitor self-assembly.
METHODS
Full-length recombinant murine amelogenin (M179; P63278 UniProt; P2-D180; the N-terminal methionine is removed in E. coli by methionine aminopeptidase42) without an affinity tag was expressed with either single- (15N-) or triple- (2H-, 13C-, 15N-) labels using minimal media and purified from E. coli following a previously described protocol using 2% acetic acid at 70 °C to lyse the cells.43,44 Similarly, a murine amelogenin construct with eight of the 14 histidine residues deleted (M179Δ8H; H58, H62, H67-H69, H91, H92, H99 deleted) was prepared using a synthetic oligonucleotide that was inserted in the pJexpress414 expression vector by ATUM (Newark, California). Nitrogen-15 labeled M179 was expressed using autoinduction protocols45 while 2H-, 13C-, and 15N-labeled M179 was expressed using isopropyl β-d-1- thiogalactopyranoside (IPTG) induction (described below). The purity of the proteins used for all the experiments was greater than 95% as determined by SDS-PAGE (Figure S1). Intact mass spectral analysis of unlabeled M179Δ8H prepared using unlabeled media further confirmed the eight deletions in the construct with the major deconvoluted monoisotopic peak having an m/z equal to 19051.65 (expected 19051.77) (Figure S2). Most of the common chemicals were purchased from Research Products International Corporation (Mount Prospect, Illinois) except glacial acetic acid and HPLC-grade acetonitrile (Fisher Scientific, Hampton, New Hampshire).
Expression of Perdeuterated M179 and M179Δ8H.
A 1 mL, ~15% glycerol stock of E. coli containing an expression vector for M179 or M179Δ8H, previously prepared from a single colony grown in LB media (100% H2O) and stored at −80 °C, was thawed and added to 20 mL of minimal media (Miller) prepared in 99.8% D2O containing the following: 15NH4Cl (1 mg/mL, 99% enriched), d-[13C6]glucose (2.0 mg/mL; 99% enriched), NaCl (50 μg/mL), MgSO4 (120 μg/mL), CaCl2 (11 μg/mL), FeCl3 (10 ng/mL), and ampicillin (100 μg/mL). Following ~8 h of growth (OD600 ~ 0.8), the cells were spun down by mild centrifugation and resuspended into 100 mL of minimal media of the same composition as the starter culture. After ~6 h of growth (OD600 ~ 0.6), these cells were directly added to 750 mL of similarly prepared minimal media. Protein expression was induced by the addition of IPTG (0.026 μg/mL) roughly 10 h later (OD600 ~ 0.8). Approximately 12 h later, the cells were harvested by mild centrifugation and frozen (–80 °C). All incubations were at 37 °C in a shaker operating at 200 rpm.
pH Titrations.
To prepare NMR samples at different pH values, a solution of 2H-, 13C-, and 15N-labeled M179 (~20 mg/mL) was prepared by gently shaking in purified water for 2 days. The solution was filtered through a 0.2 μm microspin filter unit (Lida, Kenosha, Wisconsin) and the concentration measured by UV absorption spectroscopy using a 280 nm extinction coefficient of 25,400 M–1 cm–1 (calculated). Through dilution with water, a final stock solution (10 mg/mL) was made that was used to prepare the following five samples at approximately 0.25 mM M179: (1) pH 2.8 in 2% deuterated acetic acid, (2) pH 5.4 in 25 mM sodium acetate, (3) pH 6.6 in 25 mM Tris–HCl, (4) pH 7.2 in 25 mM Tris–HCl, and (5) 8.0 in 25 mM Tris–HCl. For each sample above pH 3, 125 μL of the stock M179 solution and 125 μL of buffer 1–2 pH units higher than the target pH and twice the salt concentration were used. The pH was then adjusted to the final value with the addition of microliter amounts of 0.1 M NaOH or 0.1 M HCl to the solution (vigorously vortexing the solutions after each adjustment) and the solutions placed into 5 mm Shigemi deuterium-matched NMR tubes that were parafilm sealed.
Two-dimensional 1H-15N TROSY-HSQC spectra41 were recorded for each pH sample at 20 °C on four-channel Varian-600, −750, or −800 NMR spectrometers equipped with triple-resonance probes and pulse field gradients. Three-dimensional TROSY-enhanced, HNCA, HNCO, HNCACO, HNCACB, and HNCOCACB data (Varian Biopack pulse programs) were collected to assign the 1H-15N TROSY-HSQC spectra at each pH point. From this data, it was possible to extract side-chain 13Cα and 13Cβ chemical shifts at each pH. A tauCC value of 7 ms was used in the HNCACB experiments to optimize for the detection of β-carbons. Using a mixing time of 90 ms, 3-D 15N-edited NOESY spectra were collected at 20 °C on 15N-labeled M179 samples (0.25 mM) prepared at a pH of 5.5 and 7.8 as described above. The proton and nitrogen chemical shifts in all the TROSY 1H-15N HSQC spectra were referenced against their non-TROSY 1H-15N HSQC spectrum.
Following the same protocol described for triple-labeled M179, five 2H-, 13C-, and 15N-labeled M179Δ8H samples (~0.25 mM) were prepared at similar pH values (2.8, 5.5, 6.5, 7.2, 8.0) and an 1H-15N TROSY-HSQC spectrum recorded.
Concentration Titration.
A 0.5 mM 15N-labeled M179 sample was prepared in 25 mM sodium acetate buffer at pH 5.5. An 1H-15N HSQC spectrum was collected (20 °C) at this concentration and after a series of dilutions in the same buffer at M179 concentrations of 0.25, 0.125, and 0.05 mM. There was no need for pH adjustment after preparing the 0.5 mM sample using the 25 mM sodium acetate, pH 5.5 buffer.
RESULTS AND DISCUSSION
Defining the Species Represented by the 1H-15N HSQC Spectra.
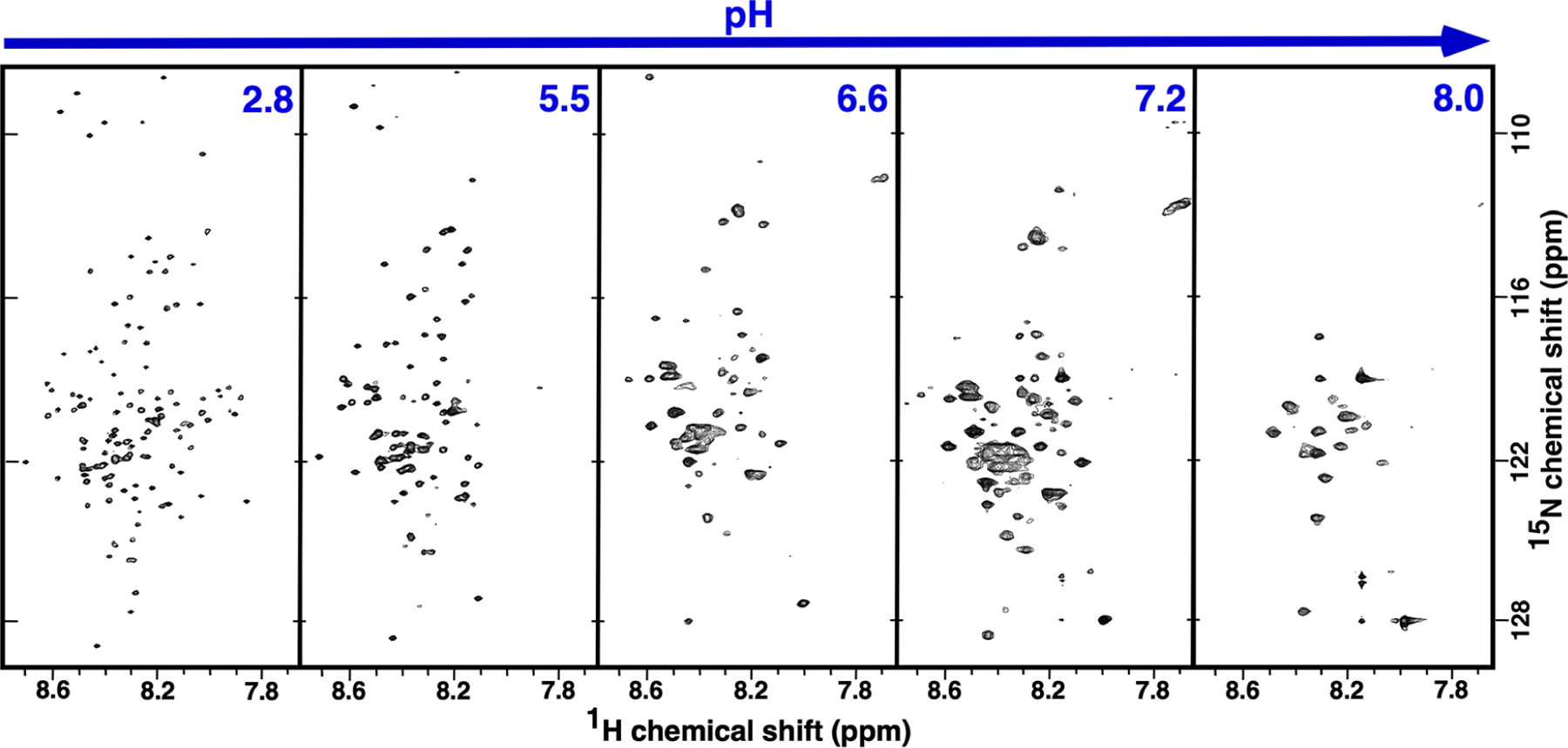
Figure 2 compares the 1H-15N TROSY HSQC spectra for 2H-, 13C-, and 15N-labeled M179 at approximately the same protein concentration, ~0.25 mM, over a range of pH values. Across this pH range, amelogenin monomers (pH ~ 3) self-assemble into oligomers (pH 5.5–6.8) and then nanospheres (pH >7.2) with precipitation observed in between at approximately pH 6.8. Prominent features of the 1H-15N HSQC spectra support this progression in size as a function of increasing pH. First, and most noticeable, is the decrease in the number of amide cross peaks in the spectra as the pH of the solution is increased. Relative to the 1H-15N HSQC spectrum in 2% acetic acid where 0.25 mM amelogenin is primarily monomeric and cross peaks are observed for each non-proline amide residue (Figure 3), there are 7, 42, 80, and 97 fewer amide cross peaks at pH 5.4, 6.6, 7.2, and 8.0, respectively. Previous NMR studies employed to follow the self-assembly of amelogenin at the residue level equated such disappearances to intermediate dynamics (ms–μs) and/or heterogenicity at intermolecular interfaces.20,33 Such an interpretation here would be consistent with the formation of larger self-assembled units with increasing pH. Second, amide resonances whose intensities do not disappear as the pH is increased generally become broader. An increase in resonance linewidths is typically observed with an increase in molecular weight due to slower isotropic tumbling.46 Third, there is no increase in the chemical shift dispersion of the visible amide proton and nitrogen resonances over this pH range, indicating that there is no significant change in secondary structure as the pH is increased; these regions of amelogenin that remain observable are just as disordered as those regions observed at pH 2.8.
Figure 2.
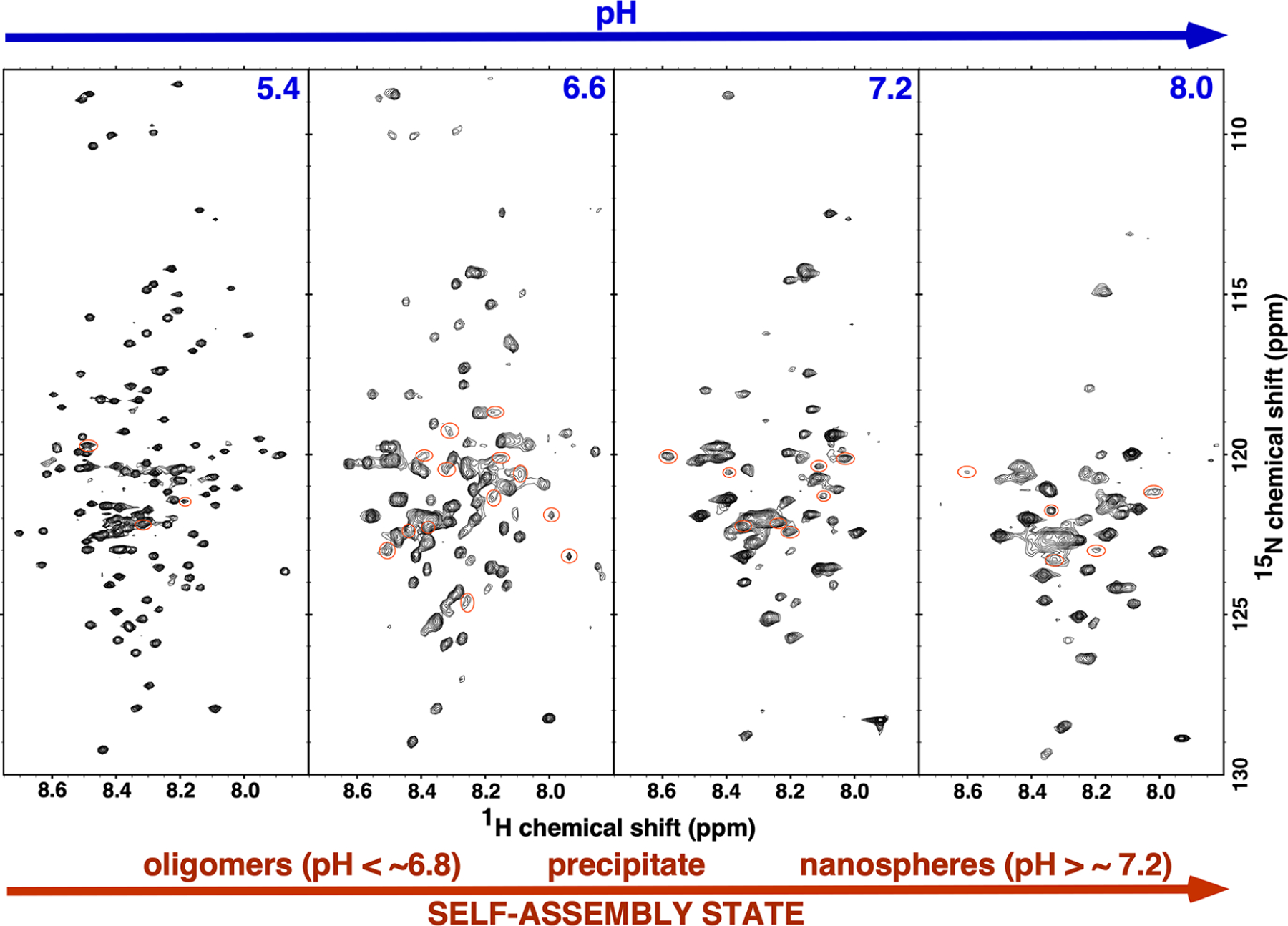
The 1H-15N TROSY-HSQC spectrum for triple labeled (2H-, 13C-, and 15N-) M179 at four different pH values. Spectra were collected on ~0.25 mM samples, at 20 °C, with a 1H resonance frequency of 750 or 800 MHz. Aside from small differences in the gradients due to the different magnetic field strengths, the basic spectral parameters for all the spectra were identical (sw(1H) = 6600 Hz, sw1(15N) = 1500 Hz, nt = 16, ni = 128, np = 2048). All the samples were in 25 mM Tris except the pH 5.4 sample, which was in 25 mM sodium acetate. Amide resonances circled red could not be unambiguously assigned. Note that the width of the 1H dimension in all the spectra is less than 1 ppm and any chemical shift changes as a function of pH were small.
Figure 3.
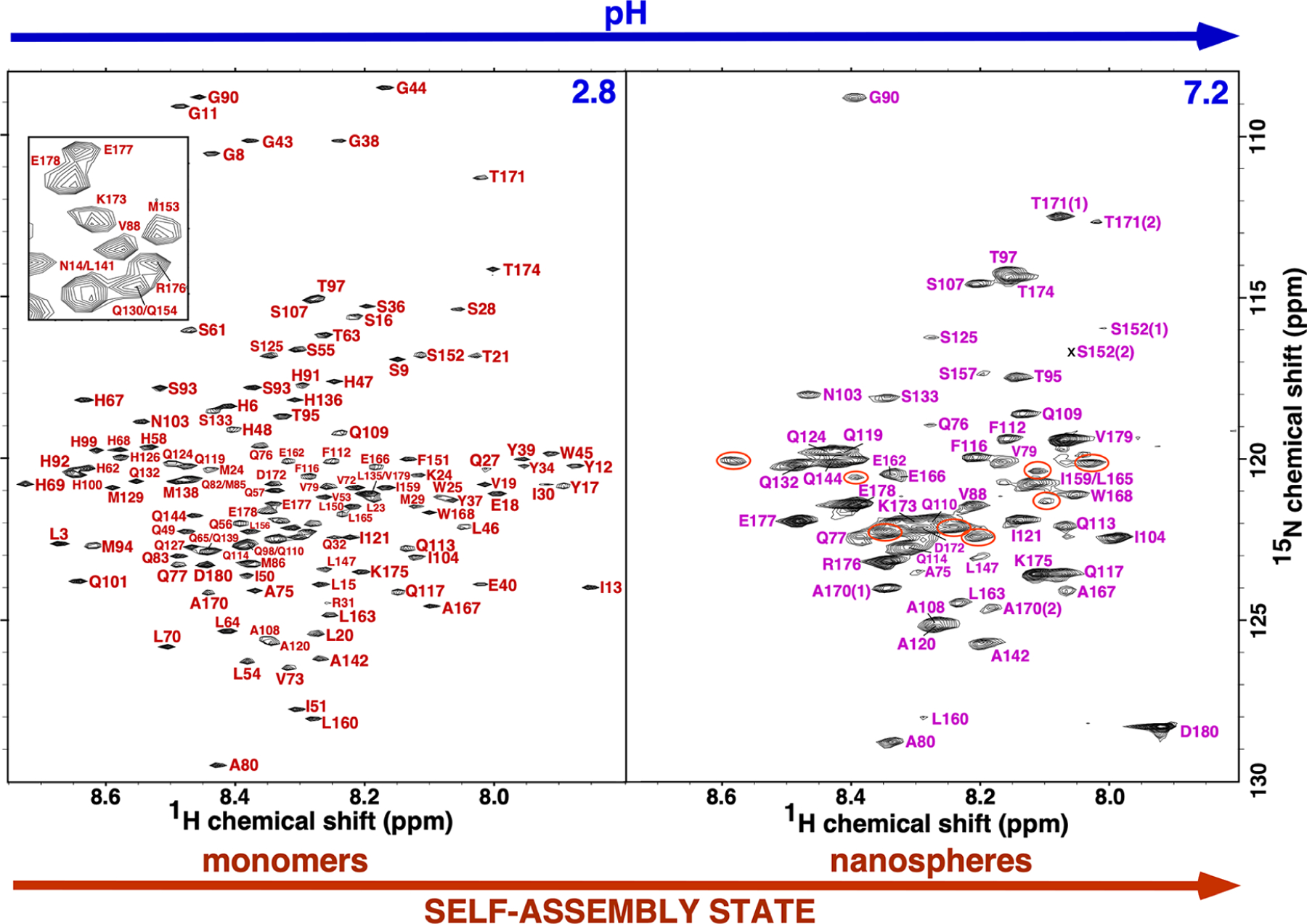
The assigned 1H-15N TROSY-HSQC spectrum of M179 (~0.25 mM) at pH 2.8 and 7.2 collected at 20 °C with a 1H resonance frequency of 600 or 800 MHz, respectively. Not shown are the downfield tryptophan ring amides (three at pH 2.8 and one at pH 7.2) and the Y26 amide at pH 2.8. The inset in the pH 2.8 spectrum is an expansion of a congested region in the middle of the spectrum. In the spectrum at pH 7.2, significant cross peaks that could not be unambiguously assigned are circled orange. Two sets of cross peaks, labeled (1) and (2), were observed for S152, A170, and T171 in the pH 7.2 spectrum.
Before discussing the NMR data from pH and concentration titrations in detail, it is necessary to first explain what species the 1H-15N HSQC spectra are sampling because the 1H-15N HSQC spectrum depends on the dynamic equilibrium between all the species in solution and the molecular weight of these species.
If the self-association properties of murine amelogenin are similar to porcine amelogenin, at the 0.25 mM (5 mg/mL) concentration used for our pH titration study, previous dynamic light scattering-based studies suggest that the average M179 oligomer should contain between 3 and 15 monomers at pH 5.4 with the average containing eight.24 In addition, the 5 mg/mL used here is above the reported 4 mg/mL plateau above which no changes in the dynamic radius or the polydispersity of the samples was observed.24 At pH 7.2 and above, nanospheres containing 20–100 monomers (depending on the conditions) predominate.5,11,22,23 The M179 octamers have a molecular weight of over 160 kDa while nanosphere molecular weights range from 400 to 2000 kDa, species that should all result in broad spectral linewidths. Such broad linewidths in the 1H-15N HSQC spectra of M179 are observed at pH ≥7.2 and at pH 6.6, but not at pH 5.4 (Figure 2). At pH 5.4, the linewidths are only marginally broader than at pH 2.8, with the 1H-15N HSQC spectrum at pH 5.4 looking more like a 20 kDa protein than a 160 kDa protein. Relative to the 1H-15N HSQC spectrum at pH 2.8, only seven amide cross peaks were missing in the spectrum at pH 5.4 (Figure 4). Also note that it was not necessary to increase the NMR data collection time to acquire 1H-15N HSQC with the same signal to noise as the pH was increased, all the spectra in Figure 2 were collected using similar parameters, suggesting that the 1H-15N HSQC spectra are capturing the major species in solution.
Figure 4.

Summary of the amide resonances whose intensities change marginally (dark circles), change significantly (gray-filled circles), or completely disappear (open circles) in the 1H-15N HSQC spectra of triple-labeled M179 (~0.25 mM) as a function of pH. Amide cross peaks that could not be unambiguously assigned are indicated by blue circles. The primary amino acid sequence of full-length murine amelogenin is shown with the proline and histidine residues highlighted in magenta and red, respectively.
There is evidence, over a wide pH range and above a critical concentration, that monomeric amelogenin species are in dynamic equilibrium between oligomers and nano-spheres.6,24,28,47 For example, a size exclusion study with murine amelogenin at a concentration of 0.2 mM (4 mg/mL) at pH 8 showed two major bands of roughly equal intensity corresponding to an excluded peak (nanospheres) and included peak (monomers).28 Assuming this monomer–oligomer/nanosphere equilibrium, there are three possible scenarios for the NMR observations depending on the timescale of the exchange between state A (monomers) and state B (oligomers or nanospheres): slow exchange–unique chemical shifts observed for state A and B nuclei; intermediate exchange—no chemical shifts observed for states A and B nuclei because they all broaden out beyond detection; fast exchange—the average chemical shift observed for state A and B nuclei.48 While the situation for amelogenin is more complicated than a two-state system because state B contains a range of monomers (3 to 15 reported at pH 5.5), it is possible to identify the timescale of the exchange observed in the amelogenin 1H-15N HSQC spectra. The slow exchange scenario is eliminated because (1) a large number of new (unassignable) cross peaks were not observed in the 1H-15N HSQC spectra as the pH was increased (the total number of cross peaks decreased as the pH was increased) and (2) it was not necessary to increase the NMR data acquisition time to obtain spectra with similar signal to noise as the pH was increased (the cross peaks for the larger molecular weight complexes would be much broader than the monomer and perhaps not even visible above the noise). The intermediate exchange scenario is eliminated because signals were observed in the spectra. This leaves the fast exchange scenario (KD of 100 mM and larger) to account for the spectra in Figure 2, the average chemical shift for the monomeric and self-associated species in solution.
After establishing that the 1H-15N HSQC spectra in Figure 2 likely represent the average amide chemical shifts for the monomeric and self-assembled species, it is possible to identify which species dominates the equilibrium by the linewidths of amide resonances. As mentioned, relative to the M179 1H-15N HSQC spectrum at pH 2.8, the linewidths are only marginally broader and only seven resonances appear to be missing in the 1H-15N HSQC spectrum at pH 5.4, suggesting that the monomeric species dominates the equilibrium at this pH. At pH values of 6.6 and greater, the linewidths are considerably broader and fewer cross peaks are progressively observed as the pH is increased, suggesting that the observed 1H-15N HSQC spectra are more representative of the larger species. Note that the scattering intensity in DLS measurements depends on the sixth power of the diameter, and at concentrations of 5 mg/mL, detection of the smaller species is likely beyond the dynamic range limits of the instruments even if the smaller species is the dominant species in solution.49
M179 pH Titration.
Despite the poor chemical shift dispersion of the amelogenin amide resonances in both the 1H- and 15N-dimensions in the 1H-15N HSQC spectrum at pH 2.8, 143 out of the 146 expected amide cross peaks (the recombinant protein contained a 12-residue metal-affinity tag) were previously assigned with the assistance of residue-specific 15N-labeled amino acid samples.37 The 21.6 kDa protein (including the N-terminal tag) was predominately monomeric (~1 mM, 20 mg/mL) in solution at this pH, a feature that assisted chemical shift assignments.21,37 In general, chemical shift assignments become progressively more difficult as the molecular weight of a protein increases due to unfavorable relaxation mechanisms that broaden spectral resonances and restrict magnetization in three-dimensional NMR experiments.50 These issues can be countered to some extent through the use of TROSY-based NMR experiments41 and deuteration of the non-exchangeable protons.39,50 Using these two techniques and an amelogenin construct containing no N-terminal “scar” residues,43 it was possible to assign most of the observable M179 amide resonances at all five pH levels. Such assignments are illustrated for M179 at pH 2.8 and 7.2 in Figure 3. At pH 2.8, the amide cross-peak assignments were complete and included the unambiguous assignment of L3 and L15 based on NMR data for a M179 construct containing a proline to cysteine substitution at P2 (data not shown). At pH 7.2, the assignments were incomplete with seven amide cross peaks, circled orange, unassigned. Moreover, two cross peaks were assigned to S152, A170, and T171 suggesting that part of the C-terminal region may exist in two different conformational states.
Figure 4 summarizes the amide resonances whose intensity change marginally (dark circles), change significantly (gray-filled circles), or completely disappear (open circles) in the 1H-15N HSQC spectra shown in Figure 2. Amide cross peaks were not identified for 7, 43, 60, and 73 residues at pH 5.4, 6.6, 7.2, and 8.0, respectively, and are among the 3, 13, 8, and 5 unassigned cross peaks (orange circles) at these respective pH values (although a few may also be duplicate peaks for assigned residues as illustrated for three residues at pH 7.2 in Figure 3). At pH 5.4, amide intensity perturbations (significant reduction or complete disappearance) were largely confined to three short regions, L23-S28, H67-H69, and H92-S93; however, this spectrum is likely mostly representative of the monomeric state. At pH 6.6, these regions expanded in length, V19-G38, Q57-L70, and H91-Q101, and included two additional regions, L3-G8 and L46-Q49. After the transition into nanospheres at pH >7.2, amide cross peaks disappeared for the entire N-terminus up to V73, along with most residues between Q82-Q101, S125-Q139, and F151-Q154. Increasing the pH to 8.0 resulted in a further expansion of the regions with missing amide resonances to include most of L3-N103, Q124-Q139, and L150-L156. Note that the increase in missing amide resonances from pH 7.2 to 8.0 may not reflect structural changes but instead result from NMR unfavorable exchange rates with solvent water at the higher pH.
The 13Cα and 13Cβ chemical shifts are sensitive to their secondary structure environment.51,52 For M179 at pH 2.8, these chemical shifts are near random coil values with perhaps a slight propensity in the direction characteristic of β-strand structure.20,21 For M179 at pH 6.6 and 7.2, there was little change in the 13Cα and 13Cβ values for the 92 and 55 tractable amide resonances, respectively, relative to pH 2.8 indicating that ~70% (92/135, oligomers) and ~40% (55/135; nano-spheres) of the protein still remained largely unfolded/unstructured during these transitions. Note that while the analysis of the 13Cα and 13Cβ chemical shifts for the “NMR visible” part of the M179 1H-15N HSQC spectra indicates these regions of the protein remain largely unstructured, we cannot make any conclusions regarding the structure for the “NMR invisible” regions. Structural insights into these “NMR invisible” regions may be made using optical spectroscopy, such as circular dichroism (CD) or Fourier transform infrared (FTIR) spectroscopy. Previous CD spectroscopy studies showed no significant concomitant increase in porcine amelogenin folding as measured by the [Θ]200:[Θ]220 ratios; however, these studies were conducted at lower (~0.01 mM; 0.25 mg/mL) protein concentrations than the studies reported here.24 Circular dichroism wavelength spectra at pH 5.8 (oligomers) from 0.01 to 0.08 mM with porcine amelogenin did suggest an increase in β-strand structure as the concentration of the protein was increased.18 Analysis of FTIR data collected on porcine amelogenin at 0.5 mM (10 mg/mL) as a function of pH showed the protein in an unfolded disordered state at pH 3.0 with significant fractions of random coil, β-turn, and polyproline II (PPII) secondary structure elements. As the pH was increased to pH 7.2, the amount of random coil structure decreased and intramolecular β-sheet and PPII secondary structure elements increased.53 Our analysis of the 13Cα and 13Cβ chemical shifts for the “NMR visible” data show that these regions remained largely random coil at all pH levels. Moreover, the “NMR visible” data decreased for M179 as the pH was increased (fewer amide cross peaks were present in the 1H-15N HSQC spectra) in agreement with the FTIR data that also suggested less random coil elements as the pH was increased. Hence, the intra-molecular β-sheet and PPII secondary structure identified by optical spectroscopy must be in regions of amelogenin for which amide chemical shifts disappear as a function of increasing pH—regions attributed to intermolecular interfaces. Such an increase in ordered structure upon interacting with their target, in this case itself, is also one of the characteristic features of intrinsically disordered proteins.54
M179 Concentration Titration.
Figure 5 summarizes the perturbations to the 1H-15N HSQC spectrum of M179 as a function of protein concentration (0.05 to 0.5 mM) at pH 5.5. For porcine amelogenin at this pH, a previous DLS study showed that this concentration change (1 to 10 mg/mL) resulted in a transition from trimers (1 mg/mL) to octamers (10 mg/mL).24 Given the high sequence similarity between murine and porcine amelogenin (Figure 1A), it is expected that murine amelogenin should behave similarly. Note that our NMR experiments were performed with non-deuterated, 15N-labeled M179 so that the results could be compared to earlier data studying the effect of NaCl on non-deuterated M179.20
Figure 5.
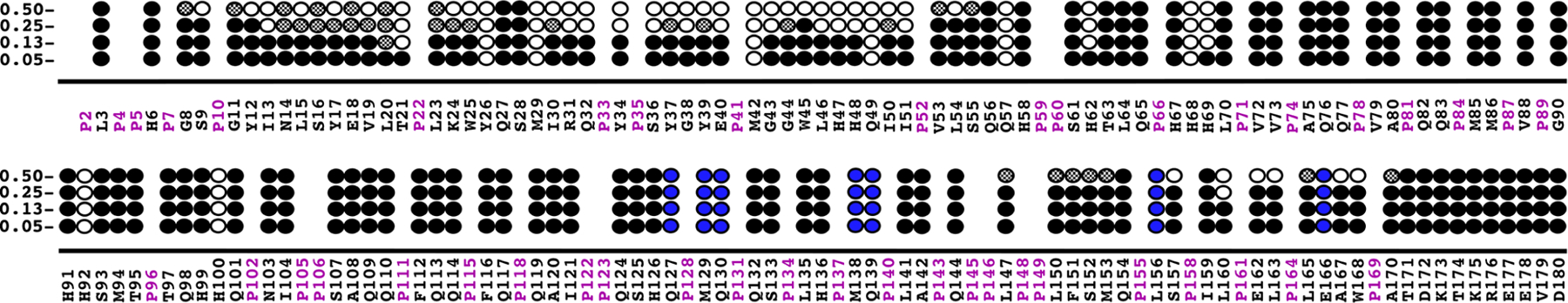
Summary of the amide resonances whose intensities change marginally (dark circles), change significantly (gray-filled circles), or completely disappear (open circles) in the 1H-15N HSQC spectra of 15N-labeled M179 as a function of protein concentration at pH 5.5. Amide cross peaks that could not be unambiguously assigned are indicated by blue circles. The primary amino acid sequence of full-length murine amelogenin is shown with the proline residues highlighted in magenta.
The first thing to note is that for deuterated M179, amide resonances could not be assigned for only seven resonances at a 0.25 mM concentration at pH 5.4. On the other hand, for non-deuterated M179 at essentially the same pH (5.5), amide resonances could not be observed for 24 residues, clearly illustrating the advantage of using deuterated amelogenin to better follow perturbations to the 1H-15N HSQC spectrum of M179 due to self-association. As the concentration of amelogenin was increased from 0.05 to 0.13 mM (1 to 2.5 mg/mL), all the amide cross peaks could still be followed despite a reported transition from trimers to tetramers over this concentration range for porcine amelogenin.24 At a 0.25 mM (5 mg/mL) concentration, porcine amelogenin was reported to form octamers and retain octamers at 0.5 mM (10 mg/mL). Our concentration study shows amide cross peaks disappearing (16 new) and decreasing in intensity (15) at the 0.25 mM (5 mg/mL) concentration. This trend continued at the 0.50 mM (10 mg/mL) concentration with further increases in the number of disappearing amide cross peaks (32 new total) and reduced intensity cross peaks (17). Furthermore, there was also a broadening of most of the amide cross peaks as the M179 concentration was increased from 0.13 to 0.50 mM. Together, all these observations are consistent with an increase in molecular weight of the complex due to self-association as the concentration was increased. While previous DLS data report that the average size of the complex reaches a plateau at a size of eight monomers at 0.25 mM (5 mg/mL), the NMR data presented here suggests that the population of large molecular weight species continues to increase with increasing concentrations above 0.25 mM because the amide cross peaks become broader and additional cross peaks disappear in the 1H-15N HSQC spectrum at 0.50 mM (10 mg/mL).
As the concentration of amelogenin increases, amide cross peaks disappear or decrease in intensity starting with a region toward the N-terminal of the protein and later a region toward the C-terminal. At a 0.5 mM concentration of M179, this corresponds to approximately G8-S55 and L156-A170. A concentration effect was previously observed for M179 in 2% acetic acid (pH 2.8) over the concentration range of 0.1 to 1.8 mM; however, chemical shift perturbations were only observed in the 1H-15N HSQC spectra and these were largest at a region near the N-terminus (I13-E40).33 Figure 6 summarizes the chemical shift perturbation studies conducted here as a function of pH and M179 concentration (Conc) along with an earlier study with increasing concentrations of salt (NaCl). The patterns for the concentration and NaCl titrations are similar and may be due to a “salting-out” effect of NaCl on amelogenin.55 On the other hand, the patterns for the pH titration are different from the concentration and NaCl titrations, suggesting that different factors, at least at pH values under 6.6, are driving the self-association. Amide cross peaks disappear or decrease in intensity starting with regions more toward the N-terminal as the pH is increased to 6.6 and expands to include the entire N-terminal region and larger swaths of the HR region as the pH is increased to 8.0. It has been proposed that the 14 histidine residues in M179 (13 in the HR region, Figure 1) are physically responsible for the pH-driven self-assembly properties of amelogenin.24,56 At low pH, the histidine residues are protonated and charged. As the pH approaches physiological values of around 7, the histidine side chain loses the proton,56 becoming neutral and hydrophobic, allowing hydrophobic interactions to drive nanosphere formation. The pH titration supports this hypothesis as the amide cross peaks for 12 out of the 13 histidine residues in the HR region disappear at pH 6.6 and all disappear at pH 7.2 and 8.0, suggesting these residues are at protein–protein interfaces. To further explore the influence of the histidine residues on the pH-dependent self-assembly properties of amelogenin, we prepared a construct with eight out of the 13 histidines in the HR region removed (Figure 1A) and conducted a pH titration with a perdeuterated sample.
Figure 6.
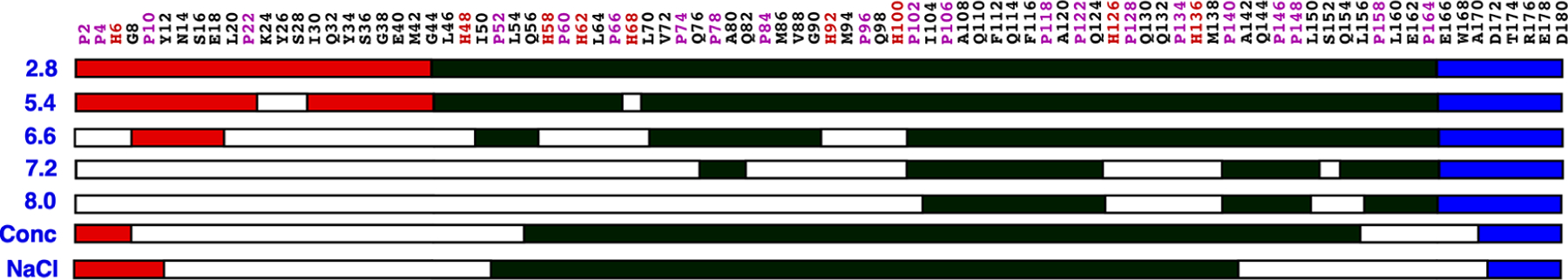
A summary of the general regions of amide intensity perturbations observed in the 1H-15N HSQC spectra of M179 as a function of pH (2.5 to 8.0) and increasing concentrations of M179 (Conc) and NaCl (NaCl).20 The three regions in the primary amino acid sequence of amelogenin is shown in a linear cartoon representation: N-terminal tyrosine-rich region (TRAP; red), hydrophobic region (HR; black), and C-terminal hydrophilic region (CTHR; blue). Regions of more than two consecutive amide residues where the intensity of the signal disappears or changes significantly in intensity in the 1H-15N HSQC spectra are colored white.
M179Δ8H pH Titration.
The M179Δ8H 1H-15N HSQC spectrum is consistent with the deletion of the targeted eight histidine residues in the primary amino acid sequence of M179, and the similarities to the M179 1H-15N HSQC spectrum suggest these deletions do not dramatically alter the structural properties of the construct (Figure S3) (e.g., M179Δ8H is still an intrinsically disordered protein under acidic conditions). Figure 7 illustrates the 1H-15N HSQC TROSY spectra collected for M179Δ8H (0.25 mM) using perdeuterated samples at similar pH values as collected for M179. There are 30, 69, 65, and 96 fewer amide cross peaks in the M179Δ8H 1H-15N HSQC spectrum at pH 5.5, 6.6, 7.2, and 8.0, respectively, in comparison to the 7, 42, 80, and 97 that disappeared over this pH range for M179 (Figure 2). The spectra for M179Δ8H changes more significantly between pH 2.8 and 6.6 than it did for M179, with more amide cross peaks disappearing over this pH range. This observation suggests that M179Δ8H has a greater tendency to self-assemble at the lower pH levels than M179, which may be predicted due to the removal of eight histidine residues that would be positively charged in M179 under these conditions. Between pH 6.6 and 7.2, there is little change in the number of cross peaks that disappear for M179Δ8H (a few return), which contrasts with M179 where almost double the number of cross peaks disappeared. This observation suggests small changes in the quaternary structure of M179Δ8H over this pH transition range, which also may be predicted due to the eight fewer histidine residues in M179Δ8H that lose their proton and become more hydrophobic (neutral) in M179. In the transition to pH 8.0, the spectra for M179Δ8H and M179 are most similar with both spectra losing over 90 cross peaks. As suggested earlier for M179, the increase in missing amide resonances from pH 7.2 to 8.0 may not reflect structural changes but instead result from NMR unfavorable exchange rates with solvent water at the higher pH.
Figure 7.
The 1H-15N TROSY-HSQC spectrum for 15N-labeled M179Δ8H at five different pH values. Spectra were collected on ~0.25 mM samples, at 20 °C, with an 1H resonance frequency of 600 MHz. The basic spectral parameters for all the spectra were identical (sw(1H) = 6600 Hz, sw1(15N) = 1500 Hz, nt = 16, ni = 128, np = 2048). All the samples were in 25 mM Tris solution except the pH 5.5 (25 mM sodium acetate) and pH 2.8 (2% acetic acid) samples.
In summary, the pattern of disappearing amide resonances for M179Δ8H as a function of pH can be interpreted in terms of the protonation state of histidine residues over this pH range. Although the spectral linewidths for M179Δ8H are generally broader at pH 7.2 than at 6.6, suggesting a larger molecular weight species at the higher pH, the spectra at the two pH values are otherwise similar with little change in the number of cross peaks that disappear. Given that the isoelectric point of M179Δ8H (6.0) is lower than M179 (6.5), it may be that M179Δ8H has already largely transitioned into a nanosphere quaternary state at pH 6.6. Indeed, the M179 1H-15N HSQC spectrum at pH 7.2 (Figure 2) looks most similar to the M179Δ8H 1H-15N HSQC spectrum at pH 6.6 (Figure 7). Note that previous DLS studies for M179Δ8H and M179 at pH 8, a condition where amelogenin forms nanospheres, showed that M179Δ8H formed nanospheres with diameters that were over twice the size of M179, 47 versus 22 nm.57 If the M179Δ8H nanospheres observed at pH 7.2, and possibly at 6.6, are also of the same size as observed at pH 8 (there is little change in the size of M179 nanospheres between pH 7.2 and 8),16 then it appears that the dynamic/exposed regions of M179Δ8H is similar to those observed in M179 (the 1H-15N HSQC spectra are generally similar for both proteins at the higher pH values) despite a nanosphere diameter that is twice as large.
Amelogenin’s N-Terminal—The Main Attraction.
The acquisition of 1H-15N HSQC spectra over a range of protein concentrations is a set of NMR experiments routinely performed to assess concentration-dependent self-association or transient aggregation. Many proteins self-associate at high protein concentrations, and the results of the concentration-dependent experiments reported here for M179 at pH 5.5 further corroborate amelogenin’s self-assembly properties. Moreover, by tracking the concentration-dependent disappearance and intensity reduction of individual amide resonances due to intermediate dynamics (ms–μs) and/or heterogenicity at protein–protein interfaces,20,33,38 it is possible to map the self-association to specific regions on the protein. For M179, self-association as a function of protein concentration first starts at a region near the N-terminus and then a small region near the C-terminus: G8-S55 and L156-A170. This pattern is similar to previous observations with M179 at pH 2.8 as a function of increasing NaCl concentration (Figure 6),20 suggesting that the NaCl “salts-out” M179, influencing it to self-associate in a pattern similar to that observed at high protein concentrations.
The N-terminal region of amelogenin has been identified as playing an essential role in amelogenesis,58 and our pH titration study (summarized in Figures 4 and 6) provides further biophysical evidence for this role. At pH 5.4, the 1H-15N HSQC spectra for M179 are likely more representative of the monomeric species in solution than the oligomeric species, and therefore, little can be learned about the protein–protein interface. However, at pH 6.6 and above, the increased amide line widths and disappearance or intensity reduction of cross peaks suggests that the 1H-15N HSQC spectra are more representative of the oligomeric (pH 6.6) or nanosphere (pH ≥7.2) state, and hence, the disappearance and intensity reduction of amide resonances provide a potential map of the protein–protein interfaces in these quaternary structures. At pH 6.6, the disappearing and intensity-reduced resonances suggest that self-association is confined to intermittent locations in the TRAP region of the protein (L3-G8 and V19-G38) and the N-terminal end of the HR region (L46-Q49, Q57-L70, and H91-Q101). Upon transition into nanospheres at pH ≥7.2, amide cross peaks disappeared for the entire TRAP region and extended to V73 in the HR region together with most residues between Q82-Q101, S125-Q139, and F151-Q154. Increasing the pH to 8.0 resulted in a further expansion of the regions with missing amide resonances to include the entire N-terminal of the protein up to N103 and two patches, Q124-Q139 and L150-L156, in the HR region. Hence, the pH titration study suggests that N-terminal protein–protein interactions dominate in both the oligomeric (pH 6.6) and nanospheric (pH ≥7.2) states, as observed in both the concentration and NaCl titration studies, although it is likely the mechanism responsible is different (change in the number of water molecules available to interact with the protein55 versus change in the protein’s protonation state).
Amelogenin’s C-Terminal—Nanospheres with Solvent Exposed and Disordered C-Termini.
While the pH titration results at pH ≥7.2 suggest that amelogenin’s N-terminal plays a role in nanosphere assembly, the entire CTHR region and the C-terminus of the HR region do not appear to be at a protein–protein interface. Indeed, if the major species represented by the resonances in the 1H-15N HSQC spectra of M179 at these pH values is the nanosphere, the presence of resonances at all is likely because they represent a region with local dynamics much different from the core of the 400 kDa plus nanosphere structure. This C-terminal region is disordered, as corroborated by 13Cα and 13Cβ chemical shifts characteristic of a random coil for the “NMR visible” amides. Nanospheres (and oligomers) with surface-exposed C-termini are supported by porcine amelogenin single tryptophan fluorescence studies (constructs with only a single tryptophan, W25, W45, or W161) showing the N-terminal tryptophans (W25 and W45) were buried while the C-terminal tryptophan (W161) was solvent exposed.24 These NMR observations are at odds with a cryoelectron microscopy (cryo-EM) study of murine amelogenin self-assembly where hollow dodecamer ring structures were observed that then aggregate in a heterogenous manner to form nanospheres, as illustrated in Figure 8.22 The dodecamers are believed to form by the prior assembly of dimers stabilized by anti-parallel interactions between C-termini. If the 1H-15N HSQC spectra at pH ≥7.2 represent nanospheres, it should be possible to verify such a structure by the observation of inter-residue 1H-1H NOEs using NMR spectroscopy. Such NOEs were not detected in three-dimensional 15N-edited NOESY data collected on a protonated sample at pH 7.8 or 5.5 (data not shown).
Figure 8.
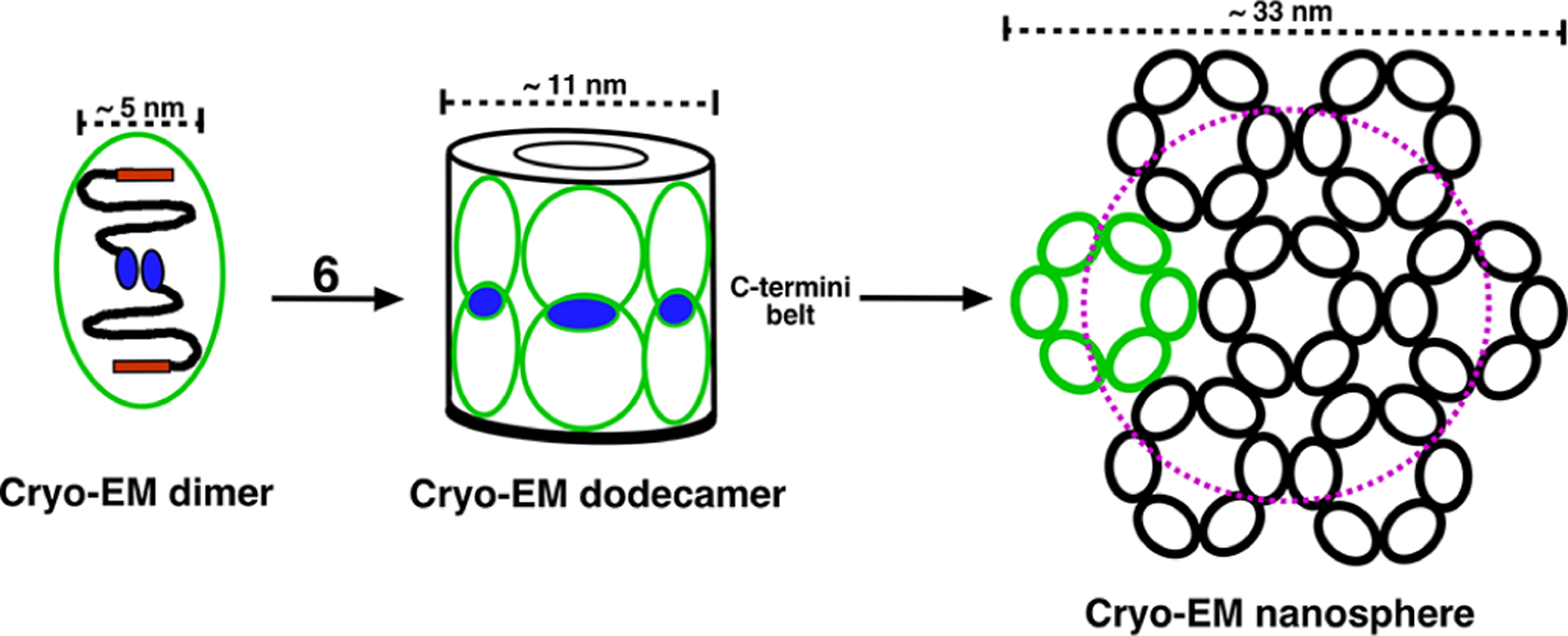
Cryo-EM-based model for the self-assembly of M179 into higher-order quaternary structures in PBS solution at pH 8.22 Upon stimulation by an increase in pH, amelogenin transitions from a disordered into a more ordered state, which triggers the self-assembly of dimer subunits. These dimers (green ovals), held together by anti-parallel interactions postulated to be electrostatic in nature between a foot-like extension at opposing C-termini (CTHR region, blue ovals), are the building blocks for higher-order quaternary structures. Six of these dimers assemble to form a dodecamer, a double-ring barrel-like structure that is hollow in the middle with all 12 C-termini aligned to form a “belt” in the equatorial plane. The resolution of the cryo-EM structures does not allow identification of the location of the N-termini (TRAP region, red rectangle); however, experiments with solution nanospheres suggest the N-termini are buried. These dodecamers are postulated to assemble into nanospheres resembling a hockey puck if assemblage is confined to one plane. The magenta dashed circle represents the DLS measured diameter (22 nm) reported for nanospheres in 25 mM Tris buffer.24
There are many possible explanations for why the cryo-EM nanospheres were not observed in the NMR data. Perhaps foremost, the self-association properties of amelogenin are sensitive to the protein concentration and the cryo-EM data was collected on ~0.005 mM samples (0.1 mg/mL) while the NMR data was collected at a 50-fold higher concentration (0.25 mM, 5 mg/L). Higher concentrations of amelogenin may favor NMR nanosphere formation. Furthermore, regardless of the concentration of the cryo-EM nanospheres, stabilization of the dimer units by C-terminal interactions would also likely make this region of the protein “NMR invisible” due to intermediate dynamics (ms–μs) and/or heterogenicity at this protein–protein interface. Consequently, cryo-EM nanospheres are likely not NMR visible. Note that in this nanosphere co-existence scenario, the population of NMR nanospheres is much greater than the cryo-EM nanospheres because there was no detectable loss in the signal intensity at the higher pH values (e.g., it was not necessary to collect more transients to achieve the same signal-to-noise ratio observed at lower pH values). Alternatively, after the dodecamers associate with form nanospheres, perhaps the C-termini dis-assemble to become dynamic and disordered? Another possible explanation is that the cryo-EM nanospheres were obtained by placing the solutions on TEM meshed grids. Perhaps a solid surface is necessary to initiate dimer assembly that leads to dodecamer formation? Indirect support for a solid surface requirement is a set of solid-state NMR experiments conducted in our laboratory. Using M179 with specific 13C- and 15N-labeled residues (arginine and threonine) mineralized on a HAP surface, we were able to detect intermolecular interactions using solid-state NMR spectroscopy that could be understood in terms of an anti-parallel C-terminal structure defined in the cryo-EM structure.59 Moreover, in vitro atomic force microscopy (AFM) studies show that the quaternary structure of amelogenin changes upon interacting with a surface, transitioning from nanospheres in solution to oligomers adsorbed onto HAP surfaces.60–63 Perhaps the oligomers observed in the AFM studies may be the cryo-EM dodecamers; however, these oligomers were reported to have a larger average size (~25 monomers). Another possibility is the use of “4 mM PBS” to prepare the cryo-EM samples22 while our buffers, along with those of Bromley et al.,24 used 25 mM Tris at pH values >7. The self-assembly properties of amelogenin are known to be sensitive to solution conditions, and indeed, nanospheres are not observed if calcium and phosphate ions (e.g., mineralization) are present in solution from the start, only oligomers of a size similar to the cryo-EM dodecamers.22,64,65
Our previous solid-state NMR experiments with mineralized (dodecamer/oligomer) and non-mineralized (nanosphere) M179 containing 13C- and 15N-labeled lysine residues (K24, K173, and K175) at pH >7.2 suggested differences in dynamics and structure between the two samples at these three sites with the former spectra consistent with dodecamers (Figure 8) and the latter spectra more consistent with NMR instead of cryo-EM nanospheres.66 It was possible to collect interpretable NMR data for the mineralized sample at 37 °C, while on the other hand, it was necessary to cool the non-mineralized sample to −35 °C to see interpretable data, and even at this temperature, significant dynamics and limited structure were detected. These observations not only suggest that the nanospheres are not bundles of dodecamers because the solid-state NMR spectra for the mineralized and non-mineralized samples should be similar but also that the nanospheres are more heterogenous/dynamic than the species present in the mineralized sample.
Some nanosphere models place amelogenin’s hydrophilic CTHR region on the outside of these complexes, solvent-exposed, to guide calcium phosphate crystallization into HAP. 23,24 Solvent exposure of the C-terminus would also facilitate proteolytic degradation of amelogenin by metal-loproteinase-20 (MMP20),12 a protein shown to be essential for proper enamel formation in MMP20 knockout mice67,68 and associated with amelogenesis imperfecta.69,70 Various studies show that MMP20, under conditions favorable for amelogenin nanosphere formation, more rapidly attack sites near the C-terminus than the N-terminus.44,71 While the C-termini are proposed to sit on the outside of the dodecamer double barrels in the cryo-EM nanosphere model, these termini are predicted to form an intermolecular interface and, as such, may be resistant to proteolysis.
Refined Models for Amelogenin Self-Assembly.
At pH 6.6, previous DLS and single tryptophan fluorescence studies led to a proposed model for pH 6.6 oligomers that contained an average of eight monomers where the N-termini were all buried and the C-termini were all surface exposed.24 Our NMR data shows that the N-terminal TRAP region is only partially buried in these oligomers, with the region between G8 and E18 dynamic and disordered. Hence, as modeled in Figure 9, we suggest instead that amelogenin oligomers at pH <~6.8 are composed of a dense, heterogenous core with a diameter of approximately 8 nm with long, dynamic, and disordered C-termini extending from this core from N103-D180. Because DLS measurements extract diameter information by measuring the diffusion of particles,49 particles with dynamic surfaces will diffuse more rapidly than particles with rigid surfaces and effectively appear to have smaller diameters. At pH >~7.2, the core size enlarges to accommodate more monomers and now contains the complete N-terminal region out to Q76 and additional short regions in the HR domain (Figure 6). In these NMR nanospheres, the continuous C-terminal dynamic and disordered region is largely the same as observed at pH 6.6 aside for two clusters (S125-Q139 and F151-Q154) between N103 and D180 (note that eight resonances were unassignable in the 1H-15N HSQC spectrum at pH 7.2 and these could be residues within these two clusters). Little is known about how the monomers pack in the core region in both the oligomers and nanospheres except that optical studies show an increase in the β-sheet and PPII secondary structure, suggesting that individual monomers may be extended.
Figure 9.
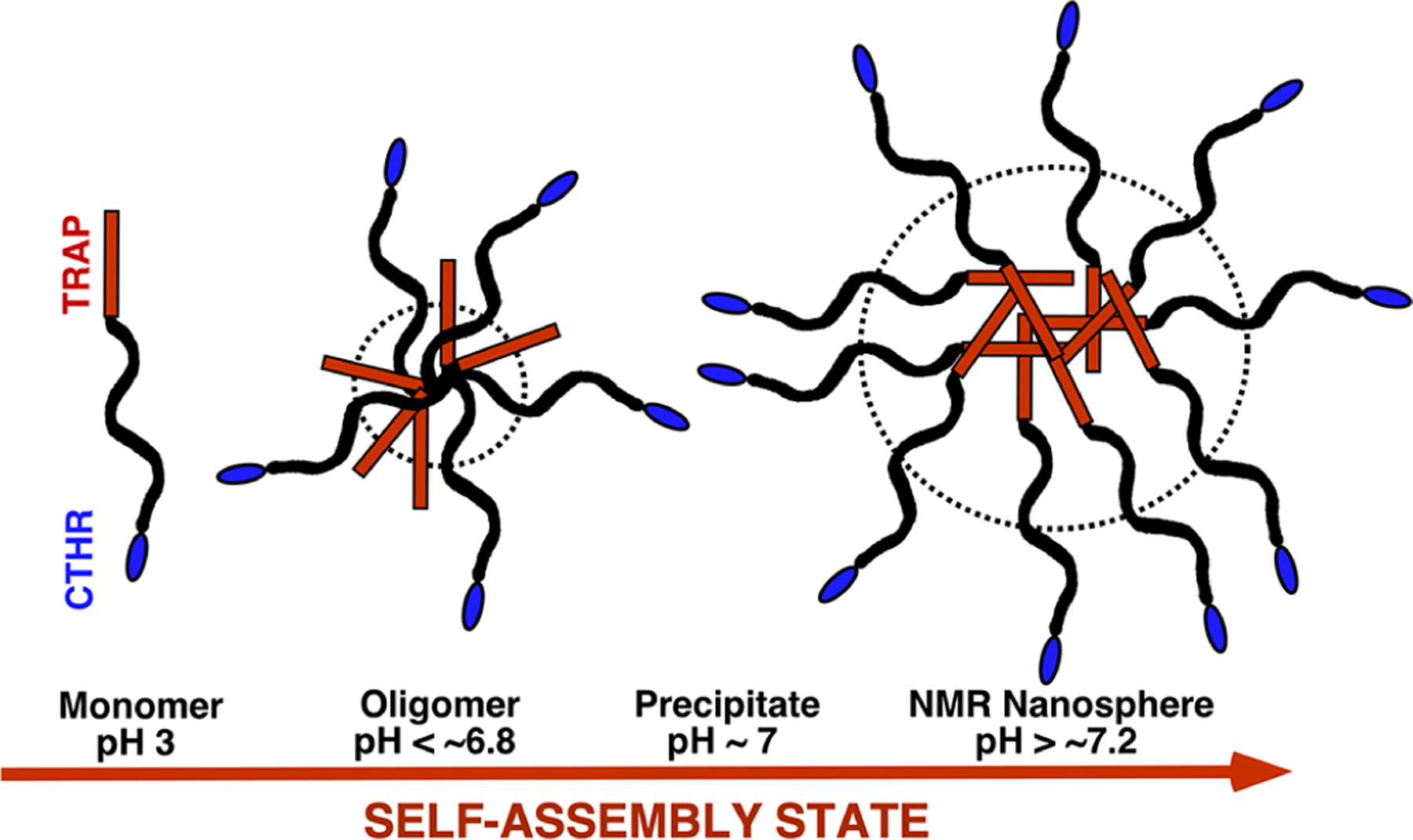
Crude, solution NMR-based, modified model for the self-assembly of M179 into higher order quaternary structures in Tris buffer as a function of increasing pH at a concentration of ~0.25 mM. Upon stimulation by an increase in pH, amelogenin self-associates into larger complexes in a process likely driven by an increase in hydrophobic interactions. At low pH, especially in dilute solution, M179 is an extended (~20 nm), intrinsically disordered protein.21 At pH 6.6 interactions between regions toward the N-terminal, half of the protein forms a core with a diameter of ~8 nm (dashed circle) with most of the C-terminus from N103 to D180 dynamic and disordered around this core. At pH >~7.2, the size of the core increases to a diameter of ~22 nm (dashed circle) and includes the entire N-terminus up to Q76 and the majority of the adjacent region up to Q101. Much of the region from N103 to D180 also remains dynamic and disordered at pH 7.2 aside from a couple of clusters (S125-Q139 and F151-Q154). The oligomer and nanosphere models are drawn approximating the C-terminus from N103 outward outside the core (dashed circle), but note that clusters within this region, especially in the nanospheres, may either fold back toward the core or self-associate outside the core. While optical methods suggest that the core region in both oligomers and nanospheres contains more elements of β-sheet and PPII secondary structure, there is no evidence that the inter- and intramolecular structures in the core are homogenous. The TRAP and CTHR regions of amelogenin are colored red and blue, respectively.
We stress that the amelogenin “oligomers” observed at pH <~6.8 characterized by DLS,24 single tryptophan fluorescence,24 and our NMR studies are different than the “oligomers” observed at pH >~7.2 characterized by cryo-EM studies22,72 (summarized in Figure 8). The oligomers at pH <~6.8 contain an average of eight monomers while the oligomers at pH >~7.2 contain an average of 12 monomers and exist as bundles of dimers. Moreover, the pH >~7.2 oligomers are short-lived, quickly assembling into larger nanosphere structures (unless calcium and phosphate ions are present) while the pH <~6.8 oligomers are stable and do not form nanospheres. Nanospheres only exist above pH ~ 7.2 and, once formed, become unstable in the presence of calcium and phosphate ions.65 Our NMR data does not support a dodecamer-based nanosphere model as a dominant species in solution (Figure 8).
Precipitates are observed in transitioning between approximately pH 6.8 to 7.2 around the isoelectric point of amelogenin (Figure 2).56 It is not known if the quaternary structures observed on either side of this “pH fence” have any influence on each other, but all in vitro experiments involve taking recombinant amelogenin from low pH to high pH across the pH fence. In vivo, the pH change, at least initially, is in the opposite direction. Could this play a role in the quaternary structure adopted by amelogenin? Regardless, pH appears to be important as it is tightly regulated during the secretory stage in ameloblasts, starting on the high pH side of this fence at approximately pH 7.2. As HAP crystals form, protons are released, and this results in local oscillations of the pH between near neutral and acidic values during the maturation stage.73 Some studies record a crossing of the pH fence during this stage, with pH values as low as 5.8–6.0 reported.74
CONCLUSIONS
We have shown here that it is possible to gather information on the nature of amelogenin quaternary structures in solution using solution-state NMR spectroscopy. This is noteworthy given that the size of the average amelogenin oligomer at pH ≤6.8 (octamers) is ~160 kDa and the smallest nanospheres at pH ≥7.2 is ~400 kDa. Based on our best interpretations, we are led to conclude that both pH- and concentration-dependent self-assemblies are initiated by protein–protein interactions toward the N-terminus of the protein, though the details of those interaction interfaces appear to vary as a function of self-assembly stimulus. At all pH values, cross-peak resonances for the CTHR region can be assigned in the 1H-15N HSQC spectra, suggesting that the C-terminal CTHR region is solvent-exposed, is dynamic, and does not occupy a protein–protein interface in the major soluble complexes. This information enabled us to refine previously proposed quaternary structure models for amelogenin self-assembly in the absence of calcium and phosphate ions as summarized in Figure 9. While these solution-state NMR experiments provided additional details regarding the nature of the amelogenin quaternary structures in solution, it only probed the dynamic and disordered NMR-visible parts of these structures. The next challenging step is to determine the molecular details for the structure of the NMR-invisible parts of these quaternary structures as these regions must contain the secondary structure elements suggested by various types of optical spectroscopy. Such solid-state experiments will not only assist in the generation of better models for amelogenin oligomer and nanosphere self-assembly but also advance our understanding of the mechanism used by amelogenin to control the growth of enamel at the molecular level.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by NIH-NIDCH Grant number DE-015347 and performed at the Pacific Northwest National Laboratory (PNNL), a facility operated by Battelle for the U.S. Department of Energy, including access to the W.R. Wiley Environmental Molecular Sciences Laboratory (EMSL), a national scientific user facility sponsored by the U.S. DOE Biological and Environmental Research program. Battelle operates PNNL for the U.S. Department of Energy under contract DE-AC05-76RL01830. We thank Dr. Mowei Zhou (PNNL) for collecting and analyzing mass spectral data for M179Δ8H.
Footnotes
The authors declare no competing financial interest.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.biochem.2c00522
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.2c00522.
(Figure S1) The SDS-PAGE gel for M179 and M179Δ8H; (Figure S2) deconvoluted monoisotopic intact mass spectrum for unlabeled M179Δ8H; and (Figure S3) comparison of the 1H-15N HSQC spectra of M179 and M179Δ8H (PDF)
Contributor Information
Garry W. Buchko, Earth and Biological Sciences Directorate, Pacific Northwest National Laboratory, Richland, Washington 99352, United States; School of Molecular Biosciences, Washington State University, Pullman, Washington 99164, United States;
Sebastian T. Mergelsberg, Physical and Computational Sciences Directorate, Pacific Northwest National Laboratory, Richland, Washington 99352, United States;
Barbara J. Tarasevich, Physical and Computational Sciences Directorate, Pacific Northwest National Laboratory, Richland, Washington 99352, United States
Wendy J. Shaw, Physical and Computational Sciences Directorate, Pacific Northwest National Laboratory, Richland, Washington 99352, United States;
REFERENCES
- (1).Ten Cate AR Oral histology: Developement, structure, and function; 4th ed., Mosby: St. Louis, 1994. [Google Scholar]
- (2).Aas JA; Paster BJ; Stokes LN; Olsen I; Dewhirst FE Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol 2005, 43, 5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Daculsi G; Menanteau J; Kerebel LM; Mitre D Length and shape of enamel crystals. Calcif. Tissue Int 1984, 36, 550–555. [DOI] [PubMed] [Google Scholar]
- (4).Hunter GK Interfacial aspects of biomineralization. Curr. Opin. Solid State Mater. Sci 1996, 1, 430–435. [Google Scholar]
- (5).Margolis HC; Beniash E; Fowler CE Role of macromolecular assembly of enamel matrix proteins in enamel formation. J. Dent. Res 2006, 85, 775–793. [DOI] [PubMed] [Google Scholar]
- (6).Shaw WJ; Tarasevich BJ; Buchko GW; Arachchige RMJ; Burton SD Controls of nature: Secondary, tertiary, and quaternary structure of the enamel protein amelogenin in solution and on hydroxyapatite. J. Struct. Biol 2020, 212, 107630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Termine JD; Belcourt AB; Christner PJ; Conn KM; Nylen MU Properties of dissociatively extracted fetal tooth matrix proteins. I. Principal molecular species in developing bovine enamel. J. Biol. Chem 1980, 255, 9760–9768. [PubMed] [Google Scholar]
- (8).Simmer JP; Fincham AG Molecular mechanims of dental enamel formation. Crit. Rev. Oral Biol. Med 1995, 6, 84–108. [DOI] [PubMed] [Google Scholar]
- (9).Gibson CW; Yaun ZA; Hall B; Longenecker G; Chen E; Thyagarajan T; Sreenath T; Wright JT; Decker S; Piddington R; Harrison G; Kulkarni AB Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J. Biol. Chem 2001, 276, 31871. [DOI] [PubMed] [Google Scholar]
- (10).Reith EJ The stages of amelogenesis as observed in molar teeth of young rats. J. Ultrastruct. Res 1970, 30, 111–151. [DOI] [PubMed] [Google Scholar]
- (11).Fincham AG; Moradian-Oldak J; Diekwisch TGH; Lyaruu DM; Wright JT; Bringas P Jr.; Slavkin HC Evidence for amelogenin ″nanospheres″ as functional components of secretory-stage enamel matrix. J. Struct. Biol 1995, 115, 50–59. [DOI] [PubMed] [Google Scholar]
- (12).Bartlett JD; Simmer JP Proteinases in developing dental enamel. Crit. Rev. Oral Biol. Med 1999, 10, 425–441. [DOI] [PubMed] [Google Scholar]
- (13).Simmer JP; Hu JC-C Expression, structure, and function of enamel proteinases. Connect. Tissue Res 2002, 43, 441–449. [DOI] [PubMed] [Google Scholar]
- (14).Brookes SJ; Robinson C; Kirkham J; Bonass WA Biochemistry and molecular biology of amelogenin proteins of developing dental enamel. Arch. Oral Biol 1995, 40, 1–14. [DOI] [PubMed] [Google Scholar]
- (15).Toyosawa S; O’hUigin C; Figueroa F; Tichy H; Klein J Identification and characterization of amelogenin genes in monotremes, reptiles, and amphibians. Proc. Natl. Acad. Sci. U. S. A 1998, 95, 13056–13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Moradian-Oldak J; Leung W; Fincham AG Temperature and pH-dependence of amelogin self-assembly: A particle size distribution study. J. Struct. Biol 1998, 122, 320–327. [DOI] [PubMed] [Google Scholar]
- (17).Moradian-Oldak J; Simmer JP; Lau EC; Sarte PE; Slavkin HC; Fincham AG Detection of monodisperse aggregates of a recombinant amelogenin by dynamic light scattering. Biopolymers 1994, 34, 1339–1347. [DOI] [PubMed] [Google Scholar]
- (18).Lakshminarayanan R; Fan D; Du C; Moradian-Oldak J The role of secondary structure in the entropically driven amelogenin self-assembly. Biophys. J 2007, 93, 3664–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wiedemann-Bidlack FB; Beniash E; Yamakoshi Y; Simmer JP; Margolis HC pH triggered self-assembly of native and recombinant amelogenins under physiological pH and termperature in vitro. J. Struct. Biol 2007, 160, 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Buchko GW; Tarasevich BJ; Bekhazi J; Snead ML; Shaw WJ A solution NMR investigation into the early events of amelogenin nanosphere self-assembly initiated with sodium chloride or calcium chloride. Biochemistry 2008, 47, 6571–6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Delak K; Harcup C; Lakshminarayanan R; Sun Z; Fan Y; Moradian-Oldak J; Evans JS The tooth enamel protein, porcine amelogenin, is an intrinsically disordered protein with an extended molecular configuration in the monomeric form. Biochemistry 2009, 48, 2272–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Fang PA; Conway JF; Margolis HC; Simmer JP; Beniash E Hierarchical self-assembly of amelogenin and the regulation of biomineralization at the nanoscale. Proc. Natl. Acad. Sci. U. S. A 2011, 108, 14097–14102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Du C; Falini G; Fermani S; Abbott C; Moradian-Oldak J Supramolecular assembly of amelogenin nanospheres into birefringent microribbons. Science 2005, 307, 1450–1454. [DOI] [PubMed] [Google Scholar]
- (24).Bromley KM; Kiss AS; Lokappa SB; Lakshminarayanan R; Fan D; Ndao M; Evans JS; Moradian-Oldak J Dissecting amelogenin protein nanospheres: characterization of metastable oligomers. J. Biol. Chem 2011, 286, 34643–34653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Zhang X; Ramirez BE; Liao X; Diekwisch TGH Amelogenin supramolecular assembly in nanospheres defined by a complex helix-coil-PPII helix 3D-structure. PLoS One 2011, 6, No. e24952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).He X; Wu S; Martinez-Avila O; Cheng Y; Habelitz S Self-aligning amelogenin nanoribbons in oil-water system. J. Struct. Biol 2011, 174, 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Carneiro KMM; Zhai H; Zhu L; Horst JA; Sitlin M; Nguyen M; Wagner M; Simpliciano C; Milder M; Chen C-L; Ashby P; Bonde J; Li W; Habelitz S Amyloid-like ribbons of amelogenins in enamel mineralization. Sci. Rep 2016, 6, 23105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Fincham AG; Moradian-Oldak J; Simmer JP; Sarte P; Lau EC; Diekwisch T; Slavkin HC Self-assembly of a recombinant amelogenin protein generates supramolecular structures. J. Struct. Biol 1994, 112, 103–109. [DOI] [PubMed] [Google Scholar]
- (29).Fincham AG; Moradian-Oldak J; Simmer JP The structural biology of the developing dental enamel matrix. J. Struct. Biol 1999, 126, 270–299. [DOI] [PubMed] [Google Scholar]
- (30).Lacruz RS; Habelitz S; Wright JT; Paine ML Dental enamel formation and implications for oral health and disease. Physiol. Rev 2017, 97, 939–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Engelberth SA; Bacino MS; Sandhu S; Li W; Bonde J; Habelitz S Progression of self-assembly of amelogenin protein supramolecular structures in simulated enamel fluid. Biomacromolecules 2018, 19, 3917–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Lakshminarayanan R; Bromley KM; Lei YP; Snead ML; Moradian-Oldak J Perturbed amelogenin secondary structure leads to uncontrolled aggregation in amelogenesis imperfecta mutant proteins. J. Biol. Chem 2010, 285, 40593–40603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Buchko GW; Lin G; Tarasevich BJ; Shaw WJ A solution NMR investigation into the impaired self-assembly properties of two murine amelogenins containing the point mutations T21→I or P41→T. Arch. Biochem. Biophys 2013, 537, 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Witkop CJ Jr.; Kuhlmann W; Sauk J Autosomal recessive pigmented hypomaturation amelogenesis imperfecta: Report of a kindred. Oral Surg. Oral Med. Oral Pathol 1973, 36, 367–382. [DOI] [PubMed] [Google Scholar]
- (35).Ravassipour DB; Hart PS; Hart TC; Ritter AV; Yamauchi M; Gibson C; Wright JT Unique enamel phenotype associated with amelogenin gene (AMELX) codon 41 point mutation. J. Dent. Res 2000, 79, 1476–1481. [DOI] [PubMed] [Google Scholar]
- (36).Collier PM; Sauk JJ; Rosenbloom J; Yaun ZA; Gibson CW An amelogenin gene defect associated with human X-linked amelogenesis imperfecta. Arch. Oral Biol 1997, 42, 235–242. [DOI] [PubMed] [Google Scholar]
- (37).Buchko GW; Bekhazi J; Cort JR; Valentine NB; Snead ML; Shaw WJ 1H 13C and 15 N resonance assignments of murine amelogenin, an enamel biomineralization protein. Biomol. NMR Assignments 2008, 2, 89–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Buchko GW; Zhou M; Craig JK; Van Voorhis WC; Myler PJ Backbone chemical shift assignments for the SARS-CoV-2 non-structural protein nsp9: Intermediate (ms – μs) dynamics in the C-terminal helix at the dimer interface. Biomol. NMR Assignments 2021, 15, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Lemaster DM; Richards FM NMR sequential assignment of Escherichia-coli thioredoxin utilizing random fractional deuteriation. Biochemistry 1988, 27, 142–150. [DOI] [PubMed] [Google Scholar]
- (40).Grzesiek S; Anglister J; Ren H; Bax A Carbon-13 line narrowing by deuterium decoupling in deuterium/carbon-13/nitrogen-15 enriched proteins. Application to triple resonance 4D J connectivity of sequential amides. J. Am. Chem. Soc 1993, 115, 4369–4370. [Google Scholar]
- (41).Pervushin K; Riek R; Wider G; Wüthrich K Attenuated t2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. U. S. A 1997, 94, 12366–12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Bonde JS; Bulow L One-step purification of recombinant human amelogenin and use of amelogenin as a fusion protein. PLoS One 2012, 7, No. e33269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Buchko GW; Shaw WJ Improved protocol to purify untagged amelogenin - application to murine amelogenin containing the equivalent P70 → T point mutation observed in human amelogenesis imperfecta. Protein Expression Purif. 2015, 105, 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Buchko GW; Jayasinha Arachchige R; Tao J; Tarasevich BJ; Shaw WJ Identification of major matrix metalloproteinase-20 proteolytic processing products of murine amelogenin and tyrosine-rich amelogenin peptide using a nuclear magnetic resonance spectroscopy based method. Arch. Oral Biol 2018, 93, 187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Studier FW Protein Production by auto-induction in high-density shaking cultures. Protein Expression Purif. 2005, 41, 207–234. [DOI] [PubMed] [Google Scholar]
- (46).Frueh DP; Goodrich AC; Mishra SH; Nichols SR NMR methods for structural studies of large monomeric and multimeric proteins. Curr. Opin. Struct. Biol 2013, 23, 734–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Limeback H; Simic A Biochemical characterization of stable high molecular-weight aggregates of amelogenins formed during porcine enamel development. Arch. Oral Biol 1990, 35, 459–468. [DOI] [PubMed] [Google Scholar]
- (48).Kovermann M; Rogne P; Wolf-Watz M Protein dynamics and function from solution state NMR spectroscopy. Q. Rev. Biophys 2016, 49, No. e6. [DOI] [PubMed] [Google Scholar]
- (49).Stetefeld J; McKenna SA; Patel TR Dynamic light scattering: A practical guide and applications in biomedical sciences. Biophys. Rev 2016, 8, 409–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Gardner KH; Kay LE The use of 2H, 13C, 15N multidimensional NMR to study the structure and dynamics of proteins. Annu. Rev. Biophys. Biomol. Struct 1998, 27, 357–406. [DOI] [PubMed] [Google Scholar]
- (51).Wishart DS; Sykes BD The 13C chemical-shift index: A simple method for the identification of protein secondary structure using 13C chemical shift data. J. Biomol. NMR 1994, 4, 171–180. [DOI] [PubMed] [Google Scholar]
- (52).Hafsa NE; Arndt D; Wishart DS CSI 3.0: A web server for identifying secondary and super-secondary structure in proteins using NMR chemical shifts. Nucleic Acids Res. 2015, 43, W370–W377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Beniash E; Simmer JP; Margolis HC Structural changes in amelogenin upon self-assembly and mineral interactions. J. Dent. Res 2012, 91, 967–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Dyson HJ; Wright PE Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol 2002, 12, 54–60. [DOI] [PubMed] [Google Scholar]
- (55).Arakawa T; Timasheff SN Mechanism of protein salting in and salting out by divalent-cation salts - balance between hydration and salt binding. Biochemistry 1984, 23, 5912–5923. [DOI] [PubMed] [Google Scholar]
- (56).Uskoković V; Castiglione Z; Cubas P; Zhu L; Li W; Habelitz S Zeta-potential and particle size analysis of human amelogenins. J. Dent. Res 2010, 89, 149–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Tao J; Hanson E; Dohnalkova A; Buchko GW; Jin B; Shaw WJ; Tarasevich BJ Changes in the C-terminal, N-terminal, and histidine regions of amelogenin reveal the role of oligomer quaternary structure on adsorption and hydroxyapatite mineralization. Front. Physiol 2022, 13, 1034662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Paine ML; Snead ML Protein interactions during assembly of the enamel organic extracellular matrix. J. Bone Miner. Res 1997, 12, 221–227. [DOI] [PubMed] [Google Scholar]
- (59).Arachchige RJ; Burton SD; Lu JX; Ginovska B; Harding LK; Taylor ME; Tao J; Dohnalkova A; Tarasevich BJ; Buchko GW; Shaw WJ Solid-state NMR identification of intermolecular interactions in amelogenin bound to hydroxyapatite. Biophys. J 2018, 115, 1666–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Tarasevich BJ; Lea S; Bernt W; Engelhard MH; Shaw WJ Changes in the quaternary structure of amelogenin when adsorbed onto surfaces. Biopolymers 2009, 91, 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Chen CL; Bromley KM; Moradian-Oldak J; DeYoreo JJ In situ AFM study of amelogenin assembly and disassembly dynamics on charged surfaces provides insights on matrix protein self-assembly. J. Am. Chem. Soc 2011, 133, 17406–17413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Tao J; Buchko GW; Shaw WJ; De Yoreo JJ; Tarasevich BJ Sequence-defined energetic shifts control the disassembly kinetics and microstructure of amelogenin adsorbed onto hydroxyapatite (100). Langmuir 2015, 31, 10451–10460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Tao J; Shin Y; Jayasinha R; Buchko GW; Burton SD; Dohnalkova AC; Wang Z; Shaw WJ; Tarasevich BJ The energetic basis for hydroxyapatite mineralization by amelogenin variants provides insights into the origin of amelogenesis imperfecta. Proc. Natl. Acad. Sci. U. S. A 2019, 116, 13867–13872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Beniash E; Simmer JP; Margolis HC The effect of recombinant mouse amelogenins on the formation and organization of hydroxyapatite crystals in vitro. J. Struct. Biol 2005, 149, 182–190. [DOI] [PubMed] [Google Scholar]
- (65).Martinez-Avila O; Wu S; Kim SJ; Cheng Y; Khan F; Samudrala R; Sali A; Horst JA; Habelitz S Self-assembly of filamentous amelogenin requires calcium and phosphate: From dimers via nanoribbons to fibrils. Biomacromolecules 2012, 13, 3494–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Lu J-X; Xu YS; Buchko GW; Shaw WJ Mineral association changes the secondary structure and dynamics of murine amelogenin. J. Dent. Res 2013, 92, 1000–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Caterina JJ; Skobe Z; Shi J; Ding Y; Simmer JP; Birkedal-Hansen H; Bartlett JD Enamelysin (matrix metal-loproteinase 20)-deficient mice display an amelogenesis imperfecta phenotype. J. Biol. Chem 2002, 277, 49598–49604. [DOI] [PubMed] [Google Scholar]
- (68).Yamakoshi Y; Richardson AS; Nunez SM; Yamakoshi F; Milkovich RN; Hu JC-C; Bartlett JD; Simmer JP Enamel proteins and proteases in MMP20 and KLK4 null and double-null mice. Eur. J. Oral Sci 2011, 119, 206–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Wright JT; Torain M; Long K; Seow K; Crawford P; Aldred MJ; Hart PS; Hart TC Amelogenesis imperfecta: Genotype-phenotype studies in 71 families. Cells Tissues Organs 2011, 194, 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Bartlett JD Dental enamel development: Proteinases and their enamel matrix substrates. ISRN Dent. 2013, 2013, 684607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Ryu OH; Fincham AG; Hu CC; Zhang C; Qian Q; Bartlett JD; Simmer JP Characterization of recombinant pig enamelysin activity and cleavage of recombinant pig and mouse amelogenins. J. Dent. Res 1999, 78, 743–750. [DOI] [PubMed] [Google Scholar]
- (72).Fang PA; Margolis HC; Conway JF; Simmer JP; Beniash E CryoTEM study of effects of phosphorylation on the hierarchical assembly of porcine amelogenin and its regulation of mineralization in vitro. J. Struct. Biol 2013, 183, 250–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Lacruz RS; Nanci A; Kurtz I; Wright JT; Paine ML Regulation of pH during amelogenesis. Calcif. Tissue Int 2010, 86, 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Sasaki S; Takagi T; Suzuki M Cyclical changes in pH in bovine developing enamel as sequential bands. Arch. Oral Biol 1991, 36, 227–231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


