Abstract
This review discusses the possible involvement of infections-associated cancers in humans, with virus infections contributing 15% to 20% of total cancer cases in humans. DNA virus encoded proteins interact with host cellular signaling pathways and control proliferation, cell death and genomic integrity viral oncoproteins are known to bind cellular Deubiquitinates (DUBs) such as cyclindromatosis tumor suppressor, ubiquitin-specific proteases 7, 11, 15 and 20, and A-20 to improve their intracellular stability and cellular signaling pathways and finally transformation. Human papillomaviruses (cervical carcinoma, oral cancer and laryngeal cancer); human polyomaviruses (mesotheliomas, brain tumors); Epstein-Barr virus (B-cell lymphoproliferative diseases and nasopharyngeal carcinoma); Kaposi’s Sarcoma Herpesvirus (Kaposi’s Sarcoma and primary effusion lymphomas); hepatitis B (hepatocellular carcinoma (HCC)) cause up to 20% of malignancies around the world.
Keywords: DNA viruses, cancer, oncogenic viruses
Introduction
Cancer is a devastating disease which affects people and their families as well as health care systems by 10.9 million new cases and 6.7 million deaths annually. 1 The cell-free transmission of oral dog warts was described by M’Fadyan and Hobday in 1907, and similar findings were published in 1898 by Ciuffo for human warts.2,3 The Epstein-Barr virus (EBV) discovery from Burkitt’s lymphoma (BL) in 1964 and the hepatitis B discovery in human sera in 1970, resulted new studies in the roles of viruses in human cancer. 4 Human papillomavirus (HPV) 16 and 18 DNAs were isolated from human cervical cancer specimens by nucleic acid hybridization. 5 DNA tumor viruses in permissive cells, carried on replication leading to cell lysis and cell death and in nonpermissive cells mostly integrated into the cell chromosomes.6,7 DNA viruses and other cofactors such as immune suppression and the environment (smoking, drinking, and genetic susceptibility) have effects on the development of viral-associated malignancies.6,8-10
Human Oncogenic DNA Viruses
Carcinogenic process contributed by oncogenic viruses with the association of 15% to 20% viral infection. 11 Oncogenic viruses have used different strategies for evading the host immune response to establish long-term persistent infections.12-14 Human tumor viruses belong to a number of DNA virus families such as Hepadnaviridae (HBV), Herpesviridae, Papillomaviridae (HPV), human polyomaviruses (HPyV), human herpes virus 8 (HHV8), Epstein-Barr Virus (EBV) are involved in various cancers.15-17 DNA oncogenes of viruses are necessary for replication of the virus and also controlling mammalian cell growth. The retinoblastoma gene product (pRB) is the first cellular tumor suppressor, which interact with The E2F genes, which play roles in cell cycle progression, which targeted by DNA virus oncoproteins. The second suppressor tumor is p53. These modulations, which are further interpreted in later chapters, eventually cause S-phase induction. Tumor suppressor genes participate in cell growth and mutation in one allele causes great risk for malignancy development (Figure 1).18-20
Figure 1.
Schematic depiction of the effect of some viral oncoproteins on apoptosis, cell arrest, and cell proliferation.
Oncogenesis
Oncogenic viruses comprise of both DNA and RNA viruses. Virus oncogenes are responsible for encoding the viral proteins that are necessary for the virus to replicate. Oncogenic viruses lead cell transformation and development of malignant tumors. 21 Protooncogenes (c-onc genes) are the cellular genes such as protein kinases, growth factors, growth factor receptors, DNA binding proteins, and their activation with mutation leads to uncontrolled cell growth. Point mutations, deletions, or chromosomal translocations transform C-onc genes into V-onc 22 (Table 1).
Table 1.
DNA virus oncoproteins and cellular protein interactions.
| Virus | Viral oncoprotein | Cellular targets |
|---|---|---|
| SV40 | Large T-antigen | P53, pRb |
| Small antigen | PP2A | |
| HPV | E6 | P53 via E6AP, DLG, MAGI-1, MUPP1 |
| E7 | pRb | |
| Adeno | E1A | pRb |
| E1B-55K | P53 | |
| Adeno9 | E4ORF1 | DLG, MAGI-1, MUPP1 |
| BPV | E5 | PDGFβ receptor |
| HBV | HBx | P53, DDB1 |
| Polyma | Large T-antigen | pRb |
| Middle T-antigen | c-Src, PI3-k, PLC-γ, Shc | |
| Small T-antigen | PP2A |
Abbreviations: Adeno, adenovirus; BPV, bovine papillomavirus; PDGF, platelet-derived growth factor; PI3-K, phosphatidyl inositol-3; PLC-γ, phospholipase C-γ; Polyoma, polyomavirus; PP2A, protein phosphatase 2A.
We will describe each of the DNA viruses that are involved in cancer development and progression as follows:
Human papillomavirus (HPV)
The Papillomaviridae family of viruses includes the small, non-enveloped, double-stranded DNA viruses known as Papillomaviruses (PVs). HPV virus is divided into 2 groups: mucosal and cutaneous, with a range of epithelial hyperplastic lesions. It can also be classified into group A and group B, based on the incidence of malignant lesions. In 1842, the Rigoni Stern and colleagues mentioned that cervical cancer and genital warts in women were associated with sexual contacts but the contagious nature of genital warts were not recognized until 1907 when Ciuffo observed the transmission of warts using cell-free extracts.23-25 Based on the consistency of their association with cervical and anal cancer, the high-risk strains HPV-16, HPV-18, HPV-31, and HPV-45 are also found in these cancers. 26 HPV virus does not have a polymerase gene so the replication of the viral genome depends on the stimulation of cellular DNA synthesis. As low-risk HPV, strains 5 and 8 may be associated genetically skin disease and epidermodysplasia verruciformis (EV). More recent studies have revealed that HPV5 and 8 in immunocompromised patients might contribute to the development of non-melanoma skin cancers (NMSC). 27 The HPV 6 and 11 as low-risk mucosal HPVs, make genital warts. 28 The incidence of cervical carcinoma is about 400 000 patients per year in worldwide. About 85% of invasive squamous cell cancers are caused by HPV 31, 33, 35, and 51, as well as HPV 16 and 18 less commonly. HPV 16 and 18 and less commonly, HPV 31, 33, 35, and 51 are found in about 85% of invasive squamous cell cancers.29,30
Some papilloma proteins have oncoprotein proteins such as the carboxyl terminal of E2 proteins binds to consensus sequences within the regulatory region. A short E2R protein has homology to the c-mos proto-oncogene. 31 Like c-mos, the E2R represses the transcription when it binds to regulatory sequences. Full-length E2 proteins, trans activates by binding to the sequences in the NCR. E4 early is the major extractable viral protein in warts. It is thought to be involved in viral maturation and is therefore probably a late protein. In BPV-1, the E5 protein as a transforming protein has not been identified in human infections. The repetitive cys-X-X-cys motifs in the E6 and E7 proteins resemble zinc fingers that important for secondary structure formation and necessary for splicing and DNA. Full-length E6 and a shorten E6 transcripts have been shown in HPV 16 and 18, but not in HPV 6 or 11. The shorten E6 is produced by splice/donor acceptor sites within the E6 gene. Shorten E6 is the most abundant transcript in vitro but has not been detected in vivo. The E7 protein maintains the number of DNA copies in the establishment phase of plasmid replication.31,32 It has been shown that HPV-16 and 18 -genome and the BPV-1 E5 and E6 proteins can be capable of transforming immortalized cells and immortalizing primary cells and Focus formation is not seen with HPV 6 or 11, which are associated with benign proliferations. 33 Crook et al indicated that The proteins were expressed and the cells doubled in number if the E7 gene was under the control of a hormone-inducible promoter, but when the hormone was absent the E7 protein wasn’t expressed and the cells grew. The HPV E7 protein is structurally and functionally similar to the adeno virus El A protein. It can transactivation the adenovirus E2 promoter and RAS oncogene to induce transformation of primary cells. 34
Persistent expression of E6 and E7 is important for maintenance of the transformed phenotype of cervical carcinoma cells. E6 and E7 can immortalize cells and induce skin tumors. Among these, E6 binds p53 and causes its degradation, whereas E7 binds retinoblastoma family members. Both actions lead to inhibition of apoptosis. E6 can bind to an ubiquitin ligase, AP proteins. Moreover, E6 can stimulate telomerase activity and immortalize cells and it interacts with PDZ domains which are located in tight junctions of epithelial cells. The E6 proteins of cutaneous HPV can degrade the proapoptotic Bcl-2 family member and Bak. Bak plays an important role in signaling apoptosis in response the UV irradiation.35,36 E7 inactivates INK-4A, which is ordinarily upregulated when E2F is released and it can induce cyclins A and E and inactivates the cyclin-dependent kinase inhibitors. E7 binds to the hypo-phosphorylated form of Rb and blocks it from binding to the transcription factor E2F. In addition, E7 inactivates the cyclin dependent kinase inhibitors p21/WAF1 and p27/KIP1 by inducing cyclin-A and cyclin dependent kinase activity. E7 can also cause abnormal centriole synthesis and aneuploidy early in the oncogenic process.37-39 Wang et al showed that E7 elevates levels of c-Myc in murine C127 cells infected with bovine papillomavirus type 1 and E7 increases c-Myc binding to the hTERT promoter, E6 and E7 can increase transcription from TERT promoters in a synergistic manner.40,41 Early protein 5 (E5) can affect receptor tyrosine kinases by associating with the cell membrane. HPV DNA has been reported to be found in human breast milk, indicating that the virus can certainly infect and accumulate in breast tissue,42,43 while some studies44,45 indicated that HPV DNA was not found in breast milk. Understanding how breast milk is infected with HPV and clarifying the effect of this infection on HPV transmission and its possible involvement in HPV-induced carcinogenesis are still unknown, but there are potent suggestions to explain how breast milk is infected. The oral HPV positivity of the father was significantly associated with the breast milk HPV positivity already at the baseline and remained significant. Importantly, this association of breast milk-to-spouse oral HPV showed a feasible temporal relationship supporting the view that breast milk is the vehicle for vertical HPV transmission. 42
Cervical cancer is the second most common cancer among women worldwide but it has been become less in the United States because the Pap test has been widely available for many years (Figure 2). 2
Figure 2.
Schematic depiction of the major biological activities that contribute to the transforming activities of high-risk mucosal HPVs.
High-risk HPV infection is related with activated expression of the Id-1 transcription factor (a family of helix–loop–helix transcription factors) in aggressive breast cancer tissues, and suggests that the virus can induce cell invasion and metastasis via Id-1. 46 Frega et al did not observe expression of E6 and E7 in HPV-positive breast cancer tissues but Dimri et al were able to immortalize human mammary epithelial cells in culture using E6 and E7 oncogenes.47,48 There is an association between c-Myc and HPV 16 and 18 infections in which, over expression of c-MYC gene is a signature of breast cancers. It was reported that HPV 16 and 18 integrates its genome near c-myc in cervical carcinoma and activates c-Myc. 49
It is also possible that sex hormones can activate latent viruses. Aceto et al showed that some regions in the HPV genome control region (LCR) are targets of steroid hormones that increase the expression of E6 and E7 of high-risk HPVs. Poor T cell responses may lose to clear HPV infected cells. AIDS patients, renal transplant patients receiving immunosuppressive therapy, and individuals with T cell deficiencies have increased rates of HPV persistence, anogenital lesions and cervical cancers. 41 There is no treatment for HPV other than destroying infected cells. But in most people, the body’s immune system can control the HPV infection. The development of a promising papillomavirus vaccine against cervical cancer based on HPV-16 L1 capsid protein has been a prominent development in the papillomavirus field. In recent years, vaccines have been developed against high-risk HPV, such as Gardasil, which is produced by Merck Sharp & Dohme and Cervarix, which is produced by GlaxoSmithKline. Vaccines that prevent high risk HPVs consist of recombinant L1 proteins that form virus-like particles (VLPs).40,41 Epidermodysplasia verruciformis is a hereditary disease infecting the skin linked to chronic HPV infection and also genetic predisposition play a part in cervical cancer. Anal cancer by HPV is more frequent in HIV-infected persons with immunodeficiency which increases the risk of high-grade cervical dysplasia and cancer. 50 Smoking is associated with tobacco carcinogen accumulation in cervical mucus. 51 Sunlight and genetic are cofactors for skin carcinomas caused by HPV5 and 8 in patients. Chronic immunosuppression increases the risk cervical dysplasia and cancer. 52
Hepatitis B virus (HBV)
Hepatitis B virus (HBV) is in the family of viruses called Hepadnaviridae, and belongs to the genus Orthohepadnavirus. Over 400 million people worldwide are chronic carriers of HBV and the most of infections are asymptomatic and the non-cytopathic.53,54
HBV will develop chronic active hepatitis (CAH) which can lead to cirrhosis, liver failure and HCC69,72 CAH is introduced by liver cell necrosis, inflammation, fibrosis, and it leads to HCC which is a regeneration of hepatocytes. The necrosis may lead to the accumulation of mutations.55,56 HBV DNA integration into the host chromosome is not necessary for viral replication, but it allow for persistence of the viral genome. Integrated HBV genomes can be found in the host chromosome of CAH and HCC patients. Transpositions of viral sequences, chromosomal deletions, and proto-oncogene activation may result from this integration.57-59 HBV DNA integration takes place at multiple sites on various chromosomes at early stages in acute infection and the integration sites have been mapped in the HBV genome within the “cohesive ends” region. Fifteen of 22 cellular genes targeted by HBV were important regulators of cell proliferation and viability including integration into the hTERT gene encoding the catalytic subunit of telomerase. The spliced HBV RNA can be reverse-transcribed and encapsidated in defective HBV particles or expressed as HBSP. The HBSP promotes apoptosis without blocking cell cycle, and a significant proportion of chronic hepatitis patients have HBSP antibodies in their serum. 60
The HBVX protein (HBx) and envelope PreS2/S viral proteins are expressed in the majority of HCC tumor cells. HBx is a 154 amino acid regulatory protein plays an important role in the viral life cycle. The expression of HBx is prolonged in all stages of carcinogenesis, including in cells with integrated HBV genomes. HBx stimulates signal transduction MAPK/ERK pathways and induces the expression of genes TATA binding protein, NF-κB, c-Jun, c-Myc, Ap-2, AP-1, RPB5 subunit of RNA polymerase II. HBx has some effects on NF-κB and AP-1 by activating protein kinase C. The HBx protein inactivates p53 and interacts with the DNA repairing protein DDB1, which may affect repair functions and allow genetic changes.61,62 HBx protein can affect cytosolic proteasome function and MHC expression and leads proteasome degradation in MHC function. HBx influences intracellular calcium mobilization By activating calcium-dependent kinases, which can affect multiple cell regulatory pathways such as NF-κB activation. This protein interacts and interferes with several transcription factors including CREB, ATF, EGR1, OCT1 RXR, and the p53 tumor suppressor, all of which may participate in HCC progression.63-65 Second group of regulatory proteins (PreS2) is derived from the HBV surface gene ORF. PreS2 enhances COX-2 and cyclin-A and induces cell cycle progression. It has been shown that transgenic mice expressing MHBs in the liver have increased hepatocyte proliferation rate and an increased occurrence of liver tumors. The preS2 protein activates c-Raf-1/Erk2 signaling and increases proliferation rate of hepatocytes (Figure 3).66,67
Figure 3.
Schematic depiction of the major biological activities that contribute to the transforming activities of HBV.
In the late 1970s, an HBV vaccine was developed to prevent the spread of HBV infection and the progression of HBV-associated HCC. A number of HBV vaccines are available including Engerix-B R (GlaxoSmithKline), Shanvac-B R (Shantha Biotech), and Genevac BTM (Serum Institute). HBV vaccine as a recombinant subunit vaccine has had a remarkable public health impact. 68
Epstein–Barr virus (EBV)
EBV is a double-stranded DNA virus classified in γ-herpesviruses subfamily and Lymphocryptovirus (LCV) genus. EBV, also known as human herpes virus 4 (HHV-4), and more than 95% of the population in Worldwide is infected with EBV. 69 Denis Burkitt revealed a childhood B-cell malignancy in Uganda in 1958, now known as Burkitt’s lymphoma. 70 In 1965, Tony Epstein and Yvonne Barr identified herpesvirus particle and was named the Epstein–Barr virus (EBV). 71 The EBV infection occurs without symptoms during childhood and post adolescent infection results in mononucleosis, a self-limiting lymphoproliferative disease. EBV can cause Burkitt’s lymphoma, nasopharyngeal carcinomas, Hodgkin’s lymphoma, post-transplantation lymphoproliferative disease, B-cell lymphoma in immunocompromised patients, gastric carcinoma, pharyngeal epithelium infection, and transform lymphocytes to an immortal phenotype. EBV also is found in tongue lesion known as hairy cell leukoplaki that is common in individuals that are infected with the human immunodeficiency virus. EBV also associated lymphoproliferative diseases may develop in organ-transplant recipients, patients with AIDS, X-linked lymphoproliferative syndrome, and Wiscott-Aldrich syndrome.72,73
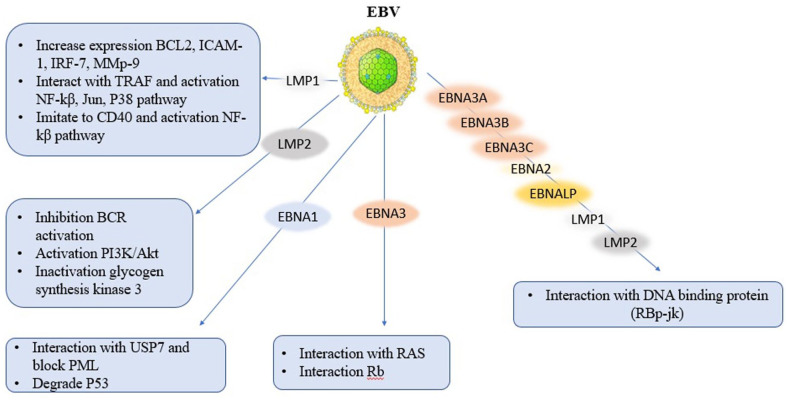
EBV infects B lymphocytes and oral epithelium in the oral pharynx become latently infected and circularization form via Terminal Repeats (TR). Those infected B lymphocytes resemble antigen activated B cells and the gene expression in these cells is restricted to a B cell growth program, termed Latency III, that includes LMP-1, LMP-2a/b, EBNAs 1, 2, 3a, 3b, 3c, LP, miRNAs, BARTs, and EBERs.74,75 These cells are killed by immune response to EBNA3 proteins, resulting in Latency I, a reservoir of latently infected resting memory B cells expressing only EBNA-1 and LMP-2. 76 some viral oncoprotein such as LMP-1 can transform cells like rodent fibroblasts and is necessary for B cells immortalization that can increase the expression of numerous anti-apoptotic (bcl-2), adhesion genes (ICAM-1). In cells expressing LMP1, the intermembrane domains and carboxyl terminal interact with several tumor necrosis factor receptors associated factors (TRAFs) to activate NF-κB, Jun, and p38. The LMP-1 receptor has the ability to mimic CD40 to activate NF-κB, the LMP-2 receptor mimics the B antigen receptor, and EBNA-2, LP, 3A, 3B, and 3C receptors are components of activated Notch.77-79 all these pathways are active and cause proliferation in normal cell cycle. Another viral oncoprotein is LMP-2. It phosphorylated by the cellular tyrosine kinases such as fyn and lyn. LMP-2 separates Fyn and Lyn in the Src family and prevents its translocation into lipid rafts with BCR and inhibits its activation. also, LMP-2 interacts with Lyn and Syk to mimic B cell receptor (BCR) signaling, including activation of the PI3K/AKT survival pathway. LMP-2 inhibits differentiation in epithelial cells and induces cell proliferation through activation of PI-3 kinase and Akt activation and inactivates glycogen synthesis kinase-3 leading to increased cytoplasmic and nuclear catenin signaling which is an important pathway in carcinoma. 80 Like LMP-1, EBNA-2 is essential for B-cell transformation and can immortalize B lymphocytes. EBNA-2 with EBNA-LP is transcriptional activator of both cellular and viral genes and are essential for B cell transformation. EBNA-3A, 3B, and 3C are hydrophilic nuclear transcriptional regulators. EBNA-3A and EBNA-3C are necessary for B cell transformation in vitro, whereas EBNA-3B is inessential. All 3 EBNA-3 proteins can interact with EBNA-2 activation by blocking its interaction with the DNA-binding protein RBP-Jk, thereby suppressing EBNA-2 mediated trans-activation. EBNA-3C can cooperate with the proto-oncogene Ras to immortalize and transform rodent fibroblasts. It can interact with the Rb tumor suppressor protein and promotes tumor progression. 81 Binding of the EBNA-1oncoprotein to the family of repeats (FR) on Ori-P leads the replication of episomal viral DNA in the latently infected cells. EBNA-1 blocks PML aggregation in nucleus by interaction with USP-7 and separation of USP-7 to Ori-P in the cytoplasm. At Ori-P, USP-7 deubiquitinates the monoubiquitinated H2B leading to the transcriptional activation of FR-controlled LMP-1and Cp promoter necessary for the virus life cycle. EBNA-1 by binding to the N-terminal of TRAF-like domain of USP-7, also threatens the stability of p53 leading to proliferation of EBV infected cells (Figure 4).82,83 environmental and genetic factors are important for EBV infections. For example, Tumor-promoting chemicals in salted fish and other food products in Southern China can be helpful for neoplastic process. In the case of BL, the incidence of malarial infection in certain regions of Africa results in an expansion of the germinal centers. 84 The major surface glycoprotein, gp-350/220, binds to the CD-21 receptor on B cells. Most of the vaccine researches have been done on gp350/220 subunit vaccines, since it is one of the most abundant proteins on the virus coat and also the protein which the human neutralizing antibody response against it. However, there is no vaccine to prevent EBV infection.96-98
Figure 4.
Schematic depiction of the major biological activities that contribute to the transforming activities of EBV.
Polyomaviruses
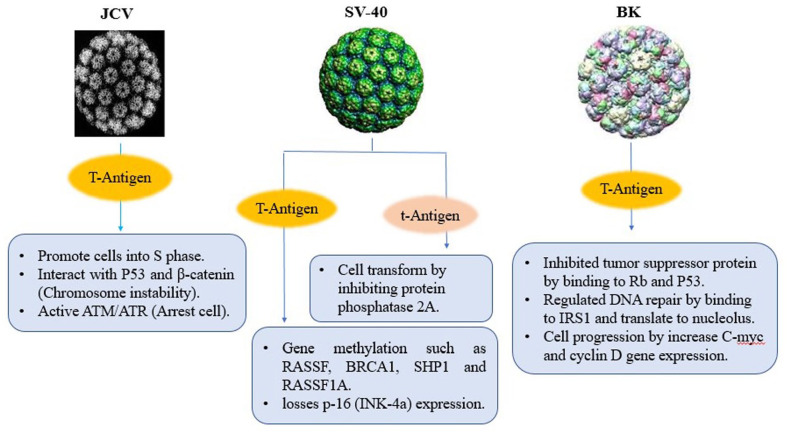
Polyoma virus is a non-enveloped, small double-stranded DNA and classified in Polyomaviridae family which is a group of viruses that have been discovered in humans, monkeys, rodents and birds. in 1971, two human polyomaviruses (BKV and JCV) are isolated from patients with progressive multifocal leukoencephalopathy (PML) and renal transplant patient.85-87 B-cell neoplasms and colon cancers have also been linked to JCV, as well as human fetal glial cells and primary hamster brain cells. From 85 tumors of glial origin, 57% to 83% of glial tumors were positive for JCV DNA and expression of viral T-Ag depending on tumor type. Because polyomavirus genomes do not encode replication proteins, it must promote cells into S-phase whenever host replication proteins are made. JCV large T-antigen affects wild type p53, stabilizes b-catenin and causes chromosomal instability and also it activates ATM- and ATR-mediated G2 checkpoint pathways and causes G2 cell cycle arrest.88,89 In rhesus monkey, SV40 establishes a low-level infection and persists in the kidneys without any noticeable affects. SV40 replicates in rhesus monkeys without producing lesions and also can replicate in human diploid fibroblasts and transform them. It has been shown that SV40 is closely associated with mesothelioma, an aggressive cancer of mesothelial cells as well as brain, lung, colon, breast and prostate tumors, bone cancers, and lymphomas. A recent study links SV40 to non-Hodgkin’s lymphomas, some brain tumors, and osteosarcomas. No human neoplasms have been related to SV40 infection, but the ability to induce tumors in animal models. 90 SV40 large T antigen, harboring potent oncogenic activity. Transgenic Mice and rat expressing SV40 T/t-antigen in mammary epithelium progress invasive and metastatic cancers. SV40 large T-antigen has an Rb-binding domain which alters gene expression and losses p-16 (INK-4a) expression. Moreover, RASSF-1A, SHP-1, BRCA-1, and TIMP-3 methylation have been reported higher in SV40-positive cases, with higher levels of P53 protein. Small T-Ag of SV40 can transform cells by binding and inhibiting protein phosphatase 2A. It has been proposed that Polyomavirus Agno protein also binds directly to p53 and represses it.91-93
BKV persistently infects epithelial cells in the urinary tract and have linked to prostate cancer. Large T-antigen and small t-antigen push the host cell into the cell cycle. Rb and p53-binding domains are present in large T-antigens, which inhibit the function of these tumor suppressor proteins. As a result, E2F transcription factor releases from Rb suppression and activates cyclin promoters. Moreover, the interaction of large T-antigen with p53 blocks its apoptotic function and the activation of inhibitors of the cellular cyclins prevented. Besides pRb and p53 regulation, large T-Ag can bind to insulin receptor substrate 1 (IRS-1) and make it translocated to the nucleus where it regulates Rad-51 and homologous recombination-directed DNA repair. Also, large T-Ag can bind to catenin to translocate it to the nucleus where it increases the expression of genes such as c-myc and cyclin D1.94,95 Small T-antigen cooperates with large T-antigen in the transformation of differentiated cells and also can develop demyelinating disease, adrenal neuroblastomas and neural tumors (Figure 5).96,97
Figure 5.
Schematic depiction of the major biological activities that contribute to the transforming activities of Polyomaviruses.
Merkel cell polyomavirus (MCV or MCPyV)
Merkel cell polyoma virus (MCV or MCPyV) is one of human oncoviruses that has been classified in polyomaviridae family and orthopolyomavirus genus and belongs to the murine polyomavirus group. In 1972, Merkel cell carcinoma was described by Cyril Toker as a highly aggressive type of skin cancer in older individuals, called neuroectodermal tumor. MCV is an oncogenic virus and causes Merkel cell carcinoma (MCC), a highly aggressive cancer and approximately 80% of MCC patients have the integrated genome of MCV. The MCV encodes large T-antigen, small T-antigen, VP1, and VP2/3 genes. From them, the T-antigen of MCV has been known as oncoproteins and expressed in human tumor. Both large and small T-antigen oncoproteins are needed to transform cells into cancer cells by targeting tumor suppressor proteins, such as retinoblastoma protein. MCV small T-antigen transforms cells in vitro by cap-dependent translation.98,99
Adenoviruses
Adenoviruses are viruses with a double-stranded DNA genome and a icosahedral nucleocapsid and without enveloped. In 1953 initial isolation of adenovirus from human adenoids was discovered. More than 50 different adenovirus serotypes be divided into 6 species, designated A to F. 100 Adenovirus is a member of Adenoviridae family and cause lytic and persistent infection in a range of mammalian and avian hosts. Adenoviruses can be routinely isolated from the mouth and some groups of adenoviruses can transform cells and can induce tumors in newborn rodents. An adenovirus has 2 stages of its life cycle: the early stage, the late stage and the viral DNA replication separates the early and late phases. The early genes express non-structural, regulatory proteins to change the expression of host proteins to activate virus genes and to avoid viral inactivation by the host-immune defenses such as blockage of apoptosis, interferon activity and MHC class I translocation and expression. Although adenovirus is not thought to cause cancer in humans, its early gene products are particularly effective at transforming mammalian cells in vitro. 101 Some adenoviruses can transform cells using their early gene products. The adenoviral takes cells into S-phase by E1A gene which is responsible for inactivation of several proteins, including retinoblastoma. The adenovirus prevents apoptosis by E1B-55kDa cooperating with E4ORF-6, to inactivate p53.102,103
Kaposi’s sarcoma-associated herpesvirus (KSHV)
Human herpes virus 8 (HHV8), also known as KSHV, is a DNA double-stranded classified in rhadinovirus genus and γ-herpesvirus subfamily. The viral genome codes for more than 80 open reading frames (ORFs). In 1872, Moritz Kaposi described Kaposi’s sarcoma as an Angio proliferative tumor developing in connective tissues typically under the skin. KSHV transforms endothelial cells and related with KS, primary effusion lymphomas (PELs), and multicentric Castleman’s disease (MCD).104,105
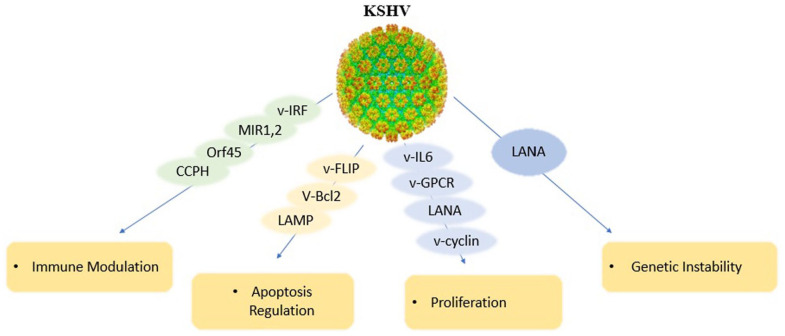
KS is a slow growing cancer that appears as reddish-purple or blue-brown tumors just underneath the skin. KS exists in central Africa and the Middle East.106,107 KS lesions can be fatal if they develop in internal organs such as the lungs, liver or gastrointestinal tract. 108 The KSHV malignancy mediated by latently expressed viral proteins together with a mechanism that is exerted by the lytically expressed v-cytokines and viral G-protein coupled receptor (vGPCR). As an Angio proliferative disease, KS uses angiogenic factors, endothelial cell (EC) growth factors, and proinflammatory cytokines including viral factors such as vIL-6, vCCL-1, 2, 3, and viral G-protein-coupled receptor (vGPCR). VEGF is stimulated by vIL-6 and promotes angiogenesis. 109 Latency expressed KSHV proteins such as the latency associated antigen (LANA, ORF-73), viral cyclin (v-cyclin, ORF-72), viral FLICE inhibitory protein (vFLIP, ORF-71), viral interferon regulatory factor 1 (vIRF-1), and the Kaposin/K-12 gene can promote cell proliferation and cellular transformation. The main function of LANA is induction of cellular proliferation and v-cyclin activates cell cycle progression vFLIP and vIRF-3 mediate pro-survival signaling and kaposins enhances cytokine expression and cell growth.110-113
There are also 2 signaling membrane proteins involved in KSHV malignancy, the variable ITAM-containing protein (VIP) and the latency associated membrane protein (LAMP). When VIP encoded by the K1 ORF injected in to nude mice induces multiple tumors and can transform rodent fibroblasts, which K1 can induce B-cell signaling and proliferation through its immune receptor tyrosine-based activation motif (ITAM) and by blocking Fas induced apoptosis of these cells. Furthermore, K1 can activate the NF-κB and PI3K pathways. VIP can induce angiogenic factors and inflammatory cytokines. The KSHV K3 and K5 proteins, MIR1 and MIR2, blocks MHC I expression, so reduces viral antigen presentation. LAMP encoded by the K15 ORF, shows mitogenic and survival signaling by Src family kinases and NF-κB activation and leads to cell survival via interaction with the Bcl-2 related anti-apoptotic protein HAX-1.114,115 Viral (vFlip) is a homolog of the human FLICE inhibitory protein, acts as an inhibitor for procaspase 8 activation during FasL activated apoptosis. vFlip can also induce the IKK complex leading to NF-kB activation, a process that has a key role in the survival of PEL cells. vFlip is expressed in latent KS infection in spindle cells and it is an ideal candidate for the growth-promoting role of KSHV. LANA blocks some cellular pathways involving the tumor suppressors like pRb and p53. LANA can help with H-Ras to transform fibroblasts and also interacts with GSK3b, leading to cytoplasmatic and nuclear b-catenin accumulation and abnormal c-Myc stabilization. Kaposin B, encoded by KSHV latent gene K12, has the potential to cooperate with the major latent gene to induce sarcomagenesis, as its gene product binds and activates MK2 and thus could stabilize mRNAs containing ARE sequences in their 3′ UTRs, many of which are suspected to have a role in KS, such as IL-8 or TNF-α.112,116 KSHV miRNAs regulate gene expression by mediating post transcriptional target specific RNA interference. It has been identified that 17 distinct miRNAs encoded by 12 miRNA genes are present in the latency associated transcript, but they are not required for viral replication. Viral Bcl-2 blocks apoptosis in diverse cellular systems as effectively as do Bcl-2 and Bcl-xL. 117
KSHV encodes a homolog of human cytokines and chemokines. Viral IL-6 is induced by IFN-α in PEL cells and blocks cell cycle arrest and apoptosis induced by IFN-α. 118 The genes vCCL-1, vCCL-2, and vCCL-3 (vMIPs) are expressed during viral replication and appear to be involved in Th-2 lymphocyte chemoattraction. vCCL-2 is a non-signaling ligand with affinity for CC and CXC receptors expressed on Th-1 cells. Viral G protein coupled receptor (GPCR) can promote the tumor growth leading to cellular transforming and proangiogenesis. The KSHV GPCR is encoded by the ORF-74 and its closest human homologs are CXCR-1 and CXCR-2. The transforming effect of vGPCR involves the activation of multiple mitogens activated protein kinases and small GTPases of the Rho family. Their activities assemble in the nucleus to control transcription factors such as hypoxia-inducible factor 1a, AP-1, and NF-κB, thereby promoting the expression and secretion of proinflammatory cytokines such as VEGF, IL-6, IL-8/CXCL8, and MIP-1/CCL3.119-121
KSHV has immune evasion genes such as MIR1, MIR2, vIRFs, Orf-45, and complement control protein homolog (CCPH). CCPH inhibits complement-mediated lysis of infected cells, while the vIRFs of KSHV prevent interferon response. Anti-apoptotic genes encoded by KSHV such as viral Bcl-2 (vBcl-2), viral FADD-like interleukin-1 converting enzyme inhibitory protein (vFLIP/Orf-71), and viral inhibitor of apoptosis (vIAP/K-7) survive the virus by apoptosis inhibition in infected cells. All these immune evasion proteins cause life-long viral persistence in the host and contribute to KSHV pathogenesis. In latency, the limited viral genes are expressed and the viral DNA is retained as a circular episome in the nucleus, thus often virus evades from immune detection.122,123 USP-7 has a physical interaction with the viral interferon regulatory factor 4 (vIRF-4) of KSHV to inhibit its function. The inhibitory functions of vIRF-4 are dependent to 2 peptides regions, Vif-1 and Vif-2 of vIRF4 which can bind to USP-37 and inhibit its function as a p53 deubiquitinase. So, vIRF-4 can interfere with p53-HAUSP Mdm2 to break the cell cycle pause and apoptosis activation by p53 and all together promote uncontrolled cell proliferation and transformation in host cells. 124 The tegument protein ORF-64 of KSHV is a viral cysteine protease with DUB activity that interacts with IFN signaling. KSHV replication is sensitive to interferon alpha (IFN-α) and ORF-64 interacts with retinoic acid inducible gene-I (RIG-I) to weaken IFN signaling. ORF-64 deubiqiutinates the K63 ubiquitin present on the CARD domain of RIG-I and blocks RIG-I interaction with MAVS-CARD signaling (Figure 6).125,126
Figure 6.
Schematic depiction of the major biological activities that contribute to the transforming activities of HHV-8/KSHV.
Herpes simplex virus
Herpes simplex virus 1 and 2 or human herpesvirus 1 and 2 (HHV-1 and HHV-2), are classified in the herpesvirus genus and herpesviridae family. Herpes simplex virus makes watery blisters in the skin or mucous membranes of the mouth, lips, or genitals. HSV-1 produces most cold sores and it is not considered an oncogenic virus itself but may increase risk of malignant progression. Cancer cells are vulnerable to superimposed viral infections, including HSV-1, which likely led to the findings in this case. 127 HSV-2 causes most genital herpes and has been epidemiologically associated with cervical cancer. It has been found in prostate cancer cells. In a hybridization experiment with DNA from cervical cancer cells, DNA from type 2 herpes simplex virus was found, but 60% of the viral DNA molecule was missing. 128 HSV induces mutations by spontaneous mutations in several ways such as (1) HSV-1 stimulates more than one mutation in several plasmids, including substitutions, insertions, and deletions, (2) Number of HSV-1 related mutations happen in a small region called hot-spot sequence in the target site, (3) Shorter length and spontaneous deletions mutants following by HSV-1 infection have been revealed, and (4) Several point mutations were found in some HSV-1 related infection. Chromosomal rearrangements are typical of cancers that are associated with DNA tumor viruses have been reported in HSV-1 infections.129,130
The region of the genome of HSV-1 that encodes MUT has not been analyzed but MUT it has been shown that MUT increases the frequency of histidine reversion mutation.131,132 The transforming region of HSV-1 also encodes the gB, ICP-18.5, ICP-8, and DNA pol genes. However, there is no evidence to link any of them to transformation, and so the MUT protein is a candidate for transformation as any other.133,134 HSV Infection in immunosuppressed patients is responsible for development of oral or genital tumors. HSV induces the cytokine immune response by Toll-like receptors (TLRs) which entail NF-kB and MAPK signaling. The immediate early viral protein (ICP-0), engages a deubiquitinating enzyme, USP-7 known as herpesvirus-associated ubiquitin-specific protease (HAUSP). In cytoplasm, USP-7 binds with TRAF-6 and Iκκ-γ and cuts their K-63 ubiquitin residues. The deubiquitination restrains the release of NF-κB from the Iκ-Bα inhibitory complex and blocks the cytokine secretion. 135 The HSV-1 tegument protein UL-36 (belongs to a unique class of viral DUB) formed during the late stage of viral replication, act as a ubiquitin-specific cysteine protease. UL-36 has a K-48 influenced DUB activity which is restrained to the N-terminal 500 amino acids. 136
In HSV latent infection, Latency Associated Transcript (LAT) is expressed to regulate the host cell genome and interrupts natural cell death mechanisms and also herpes virus ICP-4 gene is an important transactivator for lytic infection in HSV-1. HSV blocks the immune system interference with MHC class I presentation of antigen with TAP by secretion of ICP-47. ICP-47 prevents initiation of a CTL-response against HSV, allowing the virus to live for a long period in the host.137,138
Human cytomegalovirus (HCMV)
CMV, Human herpesvirus 5 (HHV-5) belongs to the Betaherpesvirinae subfamily, Herpesviridae family and herpesviruses genus. Humans and other mammals often contract CMV infections through their salivary glands. in human, it can be life-threatening for the immunocompromised, such as HIV-infected persons, organ transplant recipients or new born infants. HCMV has been detected in 90% to 100% of breast, colon, prostate, and ovarian cancer, in sarcomas and in neural derived cancers such as glioblastoma, neuroblastoma and medulloblastoma, malignant glioma, prostate, skin, and colorectal cancers. 139 The viral infection has been revealed that there is an association between serum CMV IgG levels and breast cancer. 140
As a result of HCMV proteins altering cell cycle regulation, inhibiting apoptosis, activating angiogenesis and causing metastatic phenotypes, the amount of mutations increases, which leads to cancer cells being established in the body. Some studies have been indicated that multiple viral proteins can interfere with MHC class I presentation of viral antigens. Moreover, HCMV shows immunosuppressive character by helping tumor cells to escape from immune surveillance mechanisms. HCMV can express a viral analog of human IL-10 leading to breast cancer promotion by the virus. It has been shown that IL-10 was expressed differentially in breast tumor cells and infiltrating lymphocytes.141,142 The HCMV viral UL-48 as a DUB is a homolog of UL-36 of HSV-1. UL48 has a ubiquitin C-terminal hydrolase/isopeptidase dual activity and can cut both K-48 and K-63 linked ubiquitin although the K-63 linked ubiquitin is preferred. 143
Torque teno virus (TTV)
Transfusion Transmitted Virus or Torque teno virus (TTV) is a single-stranded circular DNA virus of the Circoviridae family. recently, it has been classified in the member of new family Anelloviridae. In 1997, TTV virus reported by T. Nishizawa in a Japanese patient with non-A-E hepatitis. Approximately 10% of blood donors in the UK and the US and patients with liver disease show TTV virus. TTV virus infection happens in early childhood and remains prevalent in adults. Cancers of the gastrointestinal tract, lung cancers, breast cancers, and myelomas have all been associated with TTV-related DNA sequences. TTV viral loads have been associated with severe idiopathic inflammatory myopathies, aplastic anemia, cancer, and lupus.144,145
In summary, carcinogenesis can be influenced by several factors such as viruses. Best established human DNA oncogenic viruses are HPV (cervical cancer), EBV (B-cell lymphoproliferative diseases), KSHV (Kaposi’s sarcoma and primary effusion lymphoma PEL), and HBV (hepatocellular carcinoma). The ability to detect and analyze viral gene products can implicate more viruses in cancers of human beings. Accordingly, in comparison to traditional therapies such as chemotherapy and radiation, antiviral therapies have greater advantages for some of viruses. The treatment of HIV infection has decreased the risk of KS and also vaccination for HBV is effective to prevent liver cancer and HPV vaccine promises to be able to prevent cervical cancer.
Footnotes
Correction (October 2023): Article updated to include Mehdi Fazlalipour in the author byline along with his affiliations 1 and 2. The “Author Contributions” section is amended.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: Study concept: Fazlalipour M and Arefinia N; Study design: Molaei HR; Manuscript drafting: Fazlalipour M; Critical revision of the manuscript: Arefinia N and Ghoreshi Z. All authors read and approved the final manuscript.
References
- 1. Balakumar P, Maung-U K, Jagadeesh G. Prevalence and prevention of cardiovascular disease and diabetes mellitus. Pharmacol Res. 2016;113:600-609. [DOI] [PubMed] [Google Scholar]
- 2. zur Hausen H. Papillomaviruses in anogenital cancer as a model to understand the role of viruses in human cancers. Cancer Res. 1989;49:4677-4681. [PubMed] [Google Scholar]
- 3. zur Hausen H. Infections Causing Human Cancer. John Wiley & Sons; 2007. [Google Scholar]
- 4. Lok AS. Hepatitis B: 50 years after the discovery of Australia antigen. J Viral Hepat. 2016;23:5-14. [DOI] [PubMed] [Google Scholar]
- 5. Fuchs PG, Girardi F, Pfister H. Human papillomavirus DNA in normal, metaplastic, preneoplastic and neoplastic epithelia of the cervix uteri. Int J Cancer. 1988;41:41-45. [DOI] [PubMed] [Google Scholar]
- 6. Collin V, Flamand L. HHV-6A/B integration and the pathogenesis associated with the reactivation of chromosomally integrated HHV-6A/B. Viruses. 2017;9:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sanyal A. Cellular Factor ND10 Complex is Involved in Repression of Lytic Replication of Human Herpesvirus 6A; 2019. [Google Scholar]
- 8. Vyshenska D, Lam KC, Shulzhenko N, Morgun A. Interplay between viruses and bacterial microbiota in cancer development. Semin Immunol. 2017;32:14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Setiawan VW, Monroe K, Lugea A, Yadav D, Pandol S. Uniting epidemiology and experimental disease models for alcohol-related pancreatic disease. Alcohol Res Curr Rev. 2017;38:173. [PMC free article] [PubMed] [Google Scholar]
- 10. Hyndman IJ. Review: the contribution of both nature and nurture to carcinogenesis and progression in solid tumours. Cancer Microenviron. 2016;9:63-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McLaughlin-Drubin ME, Munger K. Viruses associated with human cancer. Biochim Biophys Acta (BBA)-Molecular Basis Dis. 2008;1782:127-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cai Q, Chen K, Young KH. Epstein-Barr virus-positive T/NK-cell lymphoproliferative disorders. Exp Mol Med. 2015;47:e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jha HC, Banerjee S, Robertson ES. The role of gammaherpesviruses in cancer pathogenesis. Pathogens. 2016;5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chiantore MV, Mangino G, Zangrillo MS, et al. Role of the microenvironment in tumourigenesis: focus on virus-induced tumors. Curr Med Chem. 2015;22:958-974. [DOI] [PubMed] [Google Scholar]
- 15. Nordfors C. Studies on Human Papillomavirus and Molecular Markers in Head-Neck Cancer. Inst för onkologi-patologi/Dept of Oncology-Pathology; 2015. [Google Scholar]
- 16. Mui UN, Haley CT, Tyring SK. Viral oncology: molecular biology and pathogenesis. J Clin Med. 2017;6:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burrell CJ, Howard CR, Murphy FA. Viral syndromes. In: Burrell CJ, Howard CR, Murphy FA. (eds) Fenner and White’s Medical Virology. 5th ed. Academic Press; 2017:537. [Google Scholar]
- 18. Wallace NA, Galloway DA. Chapter 8 - Viral oncogenesis: infections that can lead to cancer. In: Katze MG, Korth MJ, Law GL, Nathanson N. (eds) Viral Pathogenesis. 3rd ed. Academic Press; 2016:95-105. doi: 10.1016/B978-0-12-800964-2.00008-2. [DOI] [Google Scholar]
- 19. Lamsisi M, Ennaji MM. Chapter 6 - Involvement and Roles of Long Noncoding RNAs in the Molecular Mechanisms of Emerging and Reemerging Viral Infections. In: Ennaji MM. ed. Emerging and Reemerging Viral Pathogens. Academic Press; 2020:71-92. doi: 10.1016/B978-0-12-814966-9.00006-8 [DOI] [Google Scholar]
- 20. Eisenreich W, Rudel T, Heesemann J, Goebel W. How viral and intracellular bacterial pathogens reprogram the metabolism of host cells to allow their intracellular replication. Front Cell Infect Microbiol. 2019;9:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krump NA, You J. Molecular mechanisms of viral oncogenesis in humans. Nat Rev Microbiol. 2018;16:684-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Varmus H. How tumor virology evolved into cancer biology and transformed oncology. Annu Rev Cancer Biol. 2017;1:1-18. [Google Scholar]
- 23. Irabor G. Prevalence of high risk human papillomavirus in cervical cancer specimens at the University of Calabar Teaching Hospital, Calabar: histopathological and molecular analysis (2009–2014). Fac Pathol. Published online April 16, 2019. [Google Scholar]
- 24. Vashisht S, Mishra H, Mishra PK, Ekielski A, Talegaonkar S. Structure, genome, infection cycle and clinical manifestations associated with human papillomavirus. Curr Pharm Biotechnol. 2019;20:1260-1280. [DOI] [PubMed] [Google Scholar]
- 25. Bogdanovska L, Velickova N. The importance of pap smear as cytological screening methods. Cytopathology. 2018;29:S1-30. [Google Scholar]
- 26. Sousa H, Tavares A, Campos C, et al. High-risk human papillomavirus genotype distribution in the Northern region of Portugal: data from regional cervical cancer screening program. Papillomavirus Res. 2019;8:100179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Biliris KA, Koumantakis E, Dokianakis DN, Sourvinos G, Spandidos DA. Human papillomavirus infection of non-melanoma skin cancers in immunocompetent hosts. Cancer Lett. 2000;161:83-88. [DOI] [PubMed] [Google Scholar]
- 28. Lacey CJN, Guimera N, Garland SM. Chapter 10 - Low-risk Human Papillomavirus: Genital Warts, Cancer and Respiratory Papillomatosis. In: Jenkins D, Bosch FX. (eds) Human Papillomavirus. Academic Press; 2020:165-178. doi: 10.1016/B978-0-12-814457-2.00010-6 [DOI] [Google Scholar]
- 29. Rakislova N, Saco A, Sierra A, Del Pino M, Ordi J. Role of human papillomavirus in vulvar cancer. Adv Anat Pathol. 2017;24:201-214. [DOI] [PubMed] [Google Scholar]
- 30. von Knebel Doeberitz M, Cubie H, Broker TR, Jenkins D. Chapter 2 - Linking Human Papillomavirus to Human Cancer and Understanding Its Carcinogenic Mechanisms. In: Jenkins D, Bosch FX. (eds) Human Papillomavirus. Academic Press; 2020:17-39. doi: 10.1016/B978-0-12-814457-2.00002-7 [DOI] [Google Scholar]
- 31. Steele C, Shillitoe EJ. Viruses and oral cancer. Crit Rev Oral Biol Med. 1991;2:153-175. [DOI] [PubMed] [Google Scholar]
- 32. Van Doorslaer K, Li Z, Xirasagar S, et al. The Papillomavirus Episteme: a major update to the papillomavirus sequence database. Nucleic Acids Res. 2017;45:D499-D506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Van Doorslaer K, Burk RD. Evolution of human papillomavirus carcinogenicity. Adv Virus Res. 2010;77:41-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barbaresi S, Cortese MS, Quinn J, Ashrafi GH, Graham SV, Campo MS. Effects of human papillomavirus type 16 E5 deletion mutants on epithelial morphology: functional characterization of each transmembrane domain. J Gen Virol. 2010;91:521-530. [DOI] [PubMed] [Google Scholar]
- 35. Cortés Gutiérrez EI, García-Vielma C, Aguilar-Lemarroy A, et al. Expression of the HPV18/E6 oncoprotein induces DNA damage. Eur J Histochem. 2017;61:2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marsh EK, Delury CP, Davies NJ, et al. Mitotic control of human papillomavirus genome-containing cells is regulated by the function of the PDZ-binding motif of the E6 oncoprotein. Oncotarget. 2017;8:19491-19506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vaquero J, Nguyen Ho-Bouldoires TH, Clapéron A, Fouassier L. Role of the PDZ-scaffold protein NHERF1/EBP50 in cancer biology: from signaling regulation to clinical relevance. Oncogene. 2017;36:3067-3079. [DOI] [PubMed] [Google Scholar]
- 38. Parfenov M, Pedamallu CS, Gehlenborg N, et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc Natl Acad Sci U S A. 2014;111:15544-15549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hoppe-Seyler K, Bossler F, Braun JA, Herrmann AL, Hoppe-Seyler F. The HPV E6/E7 oncogenes: key factors for viral carcinogenesis and therapeutic targets. Trends Microbiol. 2018;26:158-168. [DOI] [PubMed] [Google Scholar]
- 40. Arshad B, Khan S, Adhikari VP, et al. Role of various types of viruses in development of early aged breast cancer: a brief review. Int J Endors Health Sci Res. 2016;4:1-11. [Google Scholar]
- 41. Isok-Paas H. Viral-Host Interactions in the Life Cycle of Human Papillomaviruses; 2017. [Google Scholar]
- 42. Louvanto K, Sarkola M, Rintala M, Syrjänen K, Grenman S, Syrjänen S. Breast Milk is a potential vehicle for human papillomavirus transmission to oral mucosa of the spouse. Pediatr Infect Dis J. 2017;36:627-630. [DOI] [PubMed] [Google Scholar]
- 43. Diaz S, Boulle N, Molès JP, et al. Human papillomavirus (HPV) shedding in breast milk from African women living with HIV. J Clin Virol. 2018;106:41-43. [DOI] [PubMed] [Google Scholar]
- 44. Hedau S, Kumar U, Hussain S, et al. Breast cancer and human papillomavirus infection: no evidence of HPV etiology of breast cancer in Indian women. BMC Cancer. 2011;11:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vernet-Tomas M, Mena M, Alemany L, et al. Human papillomavirus and breast cancer: no evidence of association in a Spanish set of cases. Anticancer Res. 2015;35:851-856. [PubMed] [Google Scholar]
- 46. Wu Z, Li J, Zhang Y, Hu L, Peng X. Synchronous co-expression of id-1 and nuclear NF-κB p65 promotes cervical cancer progression and malignancy, and is associated with a poor prognosis and chemosensitivity. Oncol Rep. 2019;42:2075-2086. [DOI] [PubMed] [Google Scholar]
- 47. Salih DAMAM. Molecular Detection of Human Papilloma Virus Type 16 and 18 DNA in Breast Cancer Biopsies from Sudanese Women Attending Omdurman Military Hospital; 2017. [Google Scholar]
- 48. Liu Z, Wang L, Yang J, et al. Estrogen receptor alpha inhibits senescence-like phenotype and facilitates transformation induced by oncogenic RAS in human mammary epithelial cells. Oncotarget. 2016;7:39097-39107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tuna M, Amos CI. Next generation sequencing and its applications in HPVassociated cancers. Oncotarget. 2017;8:8877-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tulay P, Serakinci N. The role of human papillomaviruses in cancer progression. J Cancer Metastasis Treat. 2016;2:201. [Google Scholar]
- 51. Nersesyan A, Muradyan R, Kundi M, Fenech M, Bolognesi C, Knasmueller S. Smoking causes induction of micronuclei and other nuclear anomalies in cervical cells. Int J Hyg Environ Health. 2020;226:113492. [DOI] [PubMed] [Google Scholar]
- 52. Wallace NA. Cutaneous Human Papillomaviruses as Cofactors in Nonmelanoma Skin Cancer. Kansas State University Manhattan United States; 2018. [Google Scholar]
- 53. Suslov A, Boldanova T, Wang X, Wieland S, Heim MH. Hepatitis B virus does not interfere with innate immune responses in the human liver. Gastroenterology. 2018;154:1778-1790. [DOI] [PubMed] [Google Scholar]
- 54. Rana D, Menachery J, Chawla Y, Duseja A, Dhiman R, Arora S. HBV specific T-cell responses in hepatitis B. Trop Gastroenterol. 2011;32:273-278. [PubMed] [Google Scholar]
- 55. Kanwal F, Hoang T, Kramer JR, et al. Increasing prevalence of HCC and cirrhosis in patients with chronic hepatitis C virus infection. Gastroenterology. 2011;140:1182-1188.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Marrero CR, Marrero JA. Viral hepatitis and hepatocellular carcinoma. Arch Med Res. 2007;38:612-620. [DOI] [PubMed] [Google Scholar]
- 57. Tarocchi M, Polvani S, Marroncini G, Galli A. Molecular mechanism of hepatitis B virus-induced hepatocarcinogenesis. World J Gastroenterol. 2014;20:11630-11640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hai H, Tamori A, Kawada N. Role of hepatitis B virus DNA integration in human hepatocarcinogenesis. World J Gastroenterol. 2014;20:6236-6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hsieh YH, Hsu JL, Su IJ, Huang W. Genomic instability caused by hepatitis B virus: into the hepatoma inferno. Front Biosci. 2011;16:2586-2597. [DOI] [PubMed] [Google Scholar]
- 60. Wu SX, Chen WN, Jing ZT, Liu W, Lin XJ, Lin X. Hepatitis B spliced protein (HBSP) suppresses Fas-mediated hepatocyte apoptosis via activation of PI3K/Akt signaling. J Virol. 2018;92:e01273-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bréchot C. Pathogenesis of hepatitis B virus-related hepatocellular carcinoma: old and new paradigms. Gastroenterology. 2004;127:S56-S61. [DOI] [PubMed] [Google Scholar]
- 62. Brechot C, Kremsdorf D, Soussan P, et al. Hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC): molecular mechanisms and novel paradigms. Pathol Biol. 2010;58:278-287. [DOI] [PubMed] [Google Scholar]
- 63. Mirzaei H, Khodadad N, Karami C, Pirmoradi R, Khanizadeh S. The AP-1 pathway; A key regulator of cellular transformation modulated by oncogenic viruses. Rev Med Virol. 2020;30:e2088. [DOI] [PubMed] [Google Scholar]
- 64. Ausserer WA, Bourrat-Floeck B, Green CJ, Laderoute KR, Sutherland RM. Regulation of c-jun expression during hypoxic and low-glucose stress. Mol Cell Biol. 1994;14:5032-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Staib F, Hussain SP, Hofseth LJ, Wang XW, Harris CC. TP53 and liver carcinogenesis. Hum Mutat. 2003;21:201-216. [DOI] [PubMed] [Google Scholar]
- 66. Rojas J, Teran-Angel G, Barbosa L, Peterson DL, Berrueta L, Salmen S. Activation-dependent mitochondrial translocation of foxp3 in human hepatocytes. Exp Cell Res. 2016;343:159-167. [DOI] [PubMed] [Google Scholar]
- 67. Zheng Y, Qian YY, Fan H. Pre-S2 and HBV associated hepatocellular carcinoma. Hepatoma Res. 2018;4:17. [Google Scholar]
- 68. Sultanik P, Pol S. Infectious diseases & therapy hepatitis delta virus: epidemiology, natural course and treatment. J Infect Dis Ther. 2016;4:2-7. [Google Scholar]
- 69. Wegner F. Genomic Studies on the Impact of Host/Virus Interaction in EBV Infection Using Massively Parallel High Throughput Sequencing; 2017. [Google Scholar]
- 70. Schmitz R, Ceribelli M, Pittaluga S, Wright G, Staudt LM. Oncogenic mechanisms in Burkitt lymphoma. Cold Spring Harb Perspect Med. 2014;4:a014282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kliszczewska E, Jarzyński A, Boguszewska A, Pasternak J, Polz-Dacewicz M. Epstein-barr virus – pathogenesis, latency and cancers. J Pre Clin Clin Res. 2017;11:142-146. [Google Scholar]
- 72. Fukayama M, Abe H, Kunita A, et al. Thirty years of Epstein-Barr virus-associated gastric carcinoma. Virchows Arch. 2020;476:353-365. [DOI] [PubMed] [Google Scholar]
- 73. Liu XW, Xie CM, Mo YX, et al. Magnetic resonance imaging features of nasopharyngeal carcinoma and nasopharyngeal non-Hodgkin’s lymphoma: are there differences? Eur J Radiol. 2012;81:1146-1154. [DOI] [PubMed] [Google Scholar]
- 74. Odongo J, Janani MK. PCR based DNA sequencing targeting LMP, EBNA2, and EBNA3C genes of Epstein Barr virus for genotyping. Int J Res Innov Appl Sci. 2019;4:126-129. [Google Scholar]
- 75. Montes-Moreno S, Odqvist L, Diaz-Perez JA, et al. EBV-positive diffuse large B-cell lymphoma of the elderly is an aggressive post-germinal center B-cell neoplasm characterized by prominent nuclear factor-kB activation. Mod Pathol. 2012;25:968-982. [DOI] [PubMed] [Google Scholar]
- 76. Styles CT, Paschos K, White RE, Farrell PJ. The cooperative functions of the EBNA3 proteins are central to EBV persistence and latency. Pathogens. 2018;7:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Migliavacca M, Assanelli A, Ponzoni M, et al. First occurrence of plasmablastic lymphoma in adenosine deaminase-deficient severe combined immunodeficiency disease patient and review of the literature. Front Immunol. 2018;9:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Elfeky RA, Furtado-Silva JM, Chiesa R, et al. One hundred percent survival after transplantation of 34 patients with Wiskott-Aldrich syndrome over 20 years. J Allergy Clin Immunol. 2018;142:1654-1656.e7. [DOI] [PubMed] [Google Scholar]
- 79. Peng H, Chen Y, Gong P, et al. Higher methylation intensity induced by EBV LMP1 via NF-κB/DNMT3b signaling contributes to silencing of PTEN gene. Oncotarget. 2016;7:40025-40037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Si Y, Deng Z, Lan G, et al. The safety and immunological effects of rAd5-EBV-LMP2 vaccine in nasopharyngeal carcinoma patients: a phase I clinical trial and two-year follow-up. Chem Pharm Bull. 2016;64:1118-1123. [DOI] [PubMed] [Google Scholar]
- 81. Lao TD, Nguyen DH, Nguyen TM, Le TAH. Molecular screening for Epstein-Barr virus (EBV): detection of genomic EBNA-1, EBNA-2, LMP-1, LMP-2 among Vietnamese patients with nasopharyngeal brush samples. Asian Pacific J cancer Prev. 2017;18:1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Al-Eidan A, Wang Y, Skipp P, Ewing RM. The USP7 protein interaction network and its roles in tumorigenesis. Genes Dis. 2022;9:41-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wang Z, Kang W, You Y, et al. USP7: novel drug target in cancer therapy. Front Pharmacol. 2019;10:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Richardo T, Prattapong P, Ngernsombat C, et al. Epstein-Barr virus mediated signaling in nasopharyngeal carcinoma carcinogenesis. Cancers. 2020;12:2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Marcel V, Nguyen Van Long F, Diaz JJ. 40 years of research put p53 in translation. Cancers. 2018;10:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Cason C, Monasta L, Zanotta N, et al. Antibody response to polyomavirus primary infection: high seroprevalence of Merkel cell polyomavirus and lymphoid tissue involvement. J Neurovirol. 2018;24:314-322. [DOI] [PubMed] [Google Scholar]
- 87. Barth H, Solis M, Lepiller Q, et al. 45 years after the discovery of human polyomaviruses BK and JC: time to speed up the understanding of associated diseases and treatment approaches. Crit Rev Microbiol. 2017;43:178-195. [DOI] [PubMed] [Google Scholar]
- 88. Frisque RJ, Hofstetter C, Tyagarajan SK. Transforming activities of JC virus early proteins. Adv Exp Med Biol. 2006;577:288-309. [DOI] [PubMed] [Google Scholar]
- 89. Henriksen S, Tylden GD, Dumoulin A, Sharma BN, Hirsch HH, Rinaldo CH. The human fetal glial cell line SVG p12 contains infectious BK polyomavirus. J Virol. 2014;88:7556-7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Rotondo JC, Mazzoni E, Bononi I, Tognon M, Martini F. Association between simian virus 40 and human tumors. Front Oncol. 2019;9:670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kellogg C, Kouznetsova VL, Tsigelny IF. Interactions of large T-Antigen (LT) protein of polyomaviruses with p53 unfold their cancerogenic potential. J Biomol Struct Dyn. 2022;40:5243-5252. [DOI] [PubMed] [Google Scholar]
- 92. Starrett GJ, Buck CB. The case for BK polyomavirus as a cause of bladder cancer. Curr Opin Virol. 2019;39:8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Carbone M, Gazdar A, Butel JS. SV40 and human mesothelioma. Transl Lung Cancer Res. 2020;9:S47-S59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. de Kort H, Heutinck KM, Ruben JM, et al. Primary human renal-derived tubular epithelial cells fail to recognize and suppress BK virus infection. Transplantation. 2017;101:1820-1829. [DOI] [PubMed] [Google Scholar]
- 95. An P, Sáenz Robles MT, Duray AM, Cantalupo PG, Pipas JM. Human polyomavirus BKV infection of endothelial cells results in interferon pathway induction and persistence. PLoS Pathog. 2019;15:e1007505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ajuh ET, Wu Z, Kraus E, et al. Novel human polyomavirus noncoding control regions differ in bidirectional gene expression according to host cell, large T-antigen expression, and clinically occurring rearrangements. J Virol. 2018;92:e02231-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Nwogu N, Ortiz LE, Kwun HJ. Surface charge of Merkel cell polyomavirus small T antigen determines cell transformation through allosteric FBW7 WD40 domain targeting. Oncogenesis. 2020;9:53-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Pietropaolo V, Prezioso C, Moens U. Merkel cell polyomavirus and Merkel cell carcinoma. Cancers. 2020;12:1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Verhaegen ME, Mangelberger D, Harms PW, et al. Merkel cell polyomavirus small T antigen initiates Merkel cell carcinoma-like tumor development in mice. Cancer Res. 2017;77:3151-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Pettersson U. Encounters with adenovirus. Ups J Med Sci. 2019;124:83-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Chakraborty AA, Tansey WP. Adenoviral E1A function through myc. Cancer Res. 2009;69:6-9. [DOI] [PubMed] [Google Scholar]
- 102. Radke JR, Cook JL. Human adenovirus infections: update and consideration of mechanisms of viral persistence. Curr Opin Infect Dis. 2018;31:251-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Greber UF. Adenoviruses – infection, pathogenesis and therapy. FEBS Lett. 2020;594:1818-1827. [DOI] [PubMed] [Google Scholar]
- 104. Hendrikse LD, Kambli A, Kayko C, et al. Identification of a novel gammaherpesvirus in Canada lynx (Lynx canadensis). Viruses. 2019;11:363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Vangipuram R, Tyring SK. Epidemiology of Kaposi sarcoma: review and description of the nonepidemic variant. Int J Dermatol. 2019;58:538-542. [DOI] [PubMed] [Google Scholar]
- 106. Bedaiwi M, Al-Homood IA, El-Garf A, et al. Disease burden and treatment challenges of psoriatic arthritis in Africa and the Middle East. Rheumatol Int. 2019;39:1321-1329. [DOI] [PubMed] [Google Scholar]
- 107. Geme B, Duarte-Chavez R, Stone L, et al. Kaposi’s sarcoma of the colon presenting with diarrhea and weight loss. Int J Acad Med. 2018;4:295. [Google Scholar]
- 108. Galanina N, Goodman AM, Cohen PR, Frampton GM, Kurzrock R. Successful treatment of HIV-associated Kaposi sarcoma with immune checkpoint blockade. Cancer Immunol Res. 2018;6:1129-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Medina MV, Agostino AD, Ma Q, et al. KSHV G-protein coupled receptor vGPCR oncogenic signaling upregulation of cyclooxygenase-2 expression mediates angiogenesis and tumorigenesis in Kaposi’s sarcoma. PLoS Pathog. 2020;16:e1009006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Vo MT, Smith BJ, Nicholas J, Choi YB. Activation of NIX-mediated mitophagy by an interferon regulatory factor homologue of human herpesvirus. Nat Commun. 2019;10:3203-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Nakayama R, Ueno Y, Ueda K, Honda T. Latent infection with Kaposi’s sarcoma-associated herpesvirus enhances retrotransposition of long interspersed element-1. Oncogene. 2019;38:4340-4351. [DOI] [PubMed] [Google Scholar]
- 112. Ueda K. KSHV genome replication and maintenance in latency. Adv Exp Med Biol. 2018;1045:299-320. [DOI] [PubMed] [Google Scholar]
- 113. Mariggiò G, Koch S, Zhang G, et al. Kaposi sarcoma herpesvirus (KSHV) latency-associated nuclear antigen (LANA) recruits components of the MRN (Mre11-Rad50-NBS1) repair complex to modulate an innate immune signaling pathway and viral latency. PLoS Pathog. 2017;13:e1006335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kopparthy V, Erickson D. Ultra-portable, spectrometer based nucleic acid quantification system for point-of-care applications (Conference presentation). In: Optics and Biophotonics in Low-Resource Settings IV. International Society for Optics and Photonics; 2018:104850E. [Google Scholar]
- 115. Ballard Z, Ozcan A. Nucleic acid quantification in the field. Nat Biomed Eng. 2018;2:629-630. [DOI] [PubMed] [Google Scholar]
- 116. Ye X, Zhao Y, Karijolich J. The landscape of transcription initiation across latent and lytic KSHV genomes. PLoS Pathog. 2019;15:e1007852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Hussein HAM, Alfhili MA, Pakala P, et al. miRNAs and their roles in KSHV pathogenesis. Virus Res. 2019;266:15-24. [DOI] [PubMed] [Google Scholar]
- 118. Qin J, Li W, Gao SJ, Lu C. KSHV microRNAs: tricks of the devil. Trends Microbiol. 2017;25:648-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Watanabe T, Fujimuro M. [Replication machinery of Kaposi’s sarcoma-associated herpesvirus and drug discovery research]. Yakugaku Zasshi. 2019;139:69-73. [DOI] [PubMed] [Google Scholar]
- 120. Gay LA, Sethuraman S, Thomas M, Turner PC, Renne R. Modified Cross-Linking, ligation, and sequencing of hybrids (qCLASH) identifies Kaposi’s sarcoma-associated herpesvirus MicroRNA targets in endothelial cells. J Virol. 2018;92:e02138-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Gottwein E. Kaposi’s sarcoma-associated herpesvirus microRNAs. Front Microbiol. 2012;3:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Bukur T. RNA-Seq Based Decomposition of Human Cell Lines and Primary Tumors for the Identification and Quantification of Viral Expression; 2017. [Google Scholar]
- 123. Mishra R, Kumar A, Ingle H, Kumar H. The interplay between viral-derived miRNAs and host immunity during infection. Front Immunol. 2019;10:3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Nininahazwe L, Liu B, He C, Zhang H, Chen ZS. The emerging nature of Ubiquitin-specific protease 7 (USP7): a new target in cancer therapy. Drug Discov Today. 2021;26:490-502. [DOI] [PubMed] [Google Scholar]
- 125. Brychtová V, Hrabal V, Vojtěšek B. Oncogenic viral protein interactions with p53 family proteins. Klin Onkol. 2019;32:72-77. [DOI] [PubMed] [Google Scholar]
- 126. Hui KF, Tam KP, Chiang AKS. Therapeutic strategies against Epstein-barr virus-associated cancers using proteasome inhibitors. Viruses. 2017;9:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Brown SH, States VAR, Afghan AK, Satyanarayana G. Herpes simplex virus-infected squamous cell carcinoma: a case report. BMC Infect Dis. 2022;22:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Rafferty Ka., Jr. Herpes viruses and cancer. Sci Am. 1973;229:26-33. [DOI] [PubMed] [Google Scholar]
- 129. Mostafa HH, Thompson TW, Konen AJ, et al. Herpes simplex virus 1 mutant with point mutations in UL39 is impaired for acute viral replication in mice, establishment of latency, and explant-induced reactivation. J Virol. 2018;92:e01654-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Liu L, Cheng J, Mou T, et al. The mutation of the genes related to neurovirulence in HSV-2 produces an attenuated phenotype in mice. Viruses. 2020;12:770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Kato A, Adachi S, Kawano S, et al. Identification of a herpes simplex virus 1 gene encoding neurovirulence factor by chemical proteomics. Nat Commun. 2020;11:4894-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Paintsil E, Cheng YC. Antiviral agents. Encycl Microbiol. 2019;2019:176-225. [Google Scholar]
- 133. Garg N, Boyle D, Randall A, et al. Rapid immunodiagnostics of multiple viral infections in an acoustic microstreaming device with serum and saliva samples. Lab Chip. 2019;19:1524-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Goodwin CM, Schafer X, Munger J. U(L)26 attenuates IKKβ-mediated induction of interferon-stimulated gene (ISG) expression and enhanced protein ISGylation during human cytomegalovirus infection. J Virol. 2019;93:e01052-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Pozhidaeva A, Bezsonova I. USP7: structure, substrate specificity, and inhibition. DNA Repair. 2019;76:30-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Yuan H, You J, You H, Zheng C. Herpes simplex virus 1 UL36USP antagonizes type I interferon-mediated antiviral innate immunity. J Virol. 2018;92:e01161-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Velusamy T. Role of Herpes Simplex Virus 1 Protein ICP47 in Antigen Presentation and Pathogenesis; 2020. [Google Scholar]
- 138. Petti S, Lodi G. The controversial natural history of oral herpes simplex virus type 1 infection. Oral Dis. 2019;25:1850-1865. [DOI] [PubMed] [Google Scholar]
- 139. Söderberg-Nauclér C. New mechanistic insights of the pathogenicity of high-risk cytomegalovirus (CMV) strains derived from breast cancer: hope for new cancer therapy options. EBioMedicine. 2022;81:104103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Raval AD, Kistler KD, Tang Y, Murata Y, Snydman DR. Epidemiology, risk factors, and outcomes associated with cytomegalovirus in adult kidney transplant recipients: a systematic literature review of real-world evidence. Transpl Infect Dis. 2021;23:e13483. [DOI] [PubMed] [Google Scholar]
- 141. Quan H, Kim J, Na YR, et al. Human cytomegalovirus-induced interleukin-10 production promotes the proliferation of Mycobacterium massiliense in macrophages. Front Immunol. 2020;11:518605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Joseph GP, McDermott R, Baryshnikova MA, Cobbs CS, Ulasov IV. Cytomegalovirus as an oncomodulatory agent in the progression of glioma. Cancer Lett. 2017;384:79-85. [DOI] [PubMed] [Google Scholar]
- 143. Kwon KM, Oh SE, Kim YE, Han TH, Ahn JH. Cooperative inhibition of RIP1-mediated NF-κB signaling by cytomegalovirus-encoded deubiquitinase and inactive homolog of cellular ribonucleotide reductase large subunit. PLoS Pathog. 2017;13:e1006423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Reshetnyak VI, Maev IV, Burmistrov AI, Chekmazov IA, Karlovich TI. Torque teno virus in liver diseases: on the way towards unity of view. World J Gastroenterol. 2020;26:1691-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Doberer K, Schiemann M, Strassl R, et al. Torque teno virus for risk stratification of graft rejection and infection in kidney transplant recipients-a prospective observational trial. Am J Transplant. 2020;20:2081-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]