Abstract
The COVID-19 pandemics has made sparkly evident the importance of acute inflammation and its timely resolution to protect humans from pathogenic viruses while sparing them from collateral damages due to an uncontrolled immune response.
It is clear now that resolution of inflammation is an active process regulated by endogenous specialized proresolving lipid mediators (SPM) biosynthesized from essential polyunsaturated fatty acids. Accruing evidence indicates that SPM are produced during viral infections and play key roles in controlling the magnitude and duration of the inflammatory response and in regulating adaptive immunity.
Here, we reviewed biosynthesis and bioactions of SPM in virus-mediated human diseases. Harnessing SPM and their proresolutive actions can help in providing new therapeutic approaches to current and future human viral diseases by controlling infection, stimulating host immunity, and protecting from organ damage.
Keywords: inflammation, lipid mediators, immunity, GPCR, Neutrophils, Macrophages, leukocytes, COVID-19, respiratory viruses, viral infections, Resolution
1. Inflammation and viral infections
Viral infectious diseases represent a major threat for human health, especially those sustained by emerging viruses, such as avian influenza, Ebola, and coronaviruses. In viral infections, the acute inflammatory response is meant to be a primordial, necessary protective mechanism to restrain microorganisms, adequately initiate the adaptive cellular and humoral immune response, and to allow tissue repair [1], [2]. The importance of acute inflammation during infections is evident in neutropenic individuals, who typically succumb of disseminated infections [3].
Acute inflammation can be divided into 2 general phases: initiation and resolution. The classical cardinal signs of the initiation phase identified by Celsius, i.e., rubor (redness), tumor (swelling), calor (heat), and dolor (pain) are gross manifestations of molecular and cellular responses. Following infections, increased blood flow and microvascular permeability result into tissue edema, mediated by lipid mediators (eg., cysteinyl leukotrienes and prostaglandins) and other vasoactive mediators. Subsequently, polymorphonuclear neutrophils (PMN) are among the first white blood cell that accumulate – sensing leukotriene (LT) B4 and other chemo attractants - in inflamed tissues. Monocytes enter as a second wave and differentiate into macrophages (MФs). Infiltrated PMN and monocyte-MΦs are crucial for killing microbes, infected cells, and contain the spread of infection. Once the inciting cause is removed, leukocytes play also key roles in clearing the site from dead cells through non-phlogistic phagocytosis and repair the damaged tissue [2], [4]. Countless times every day, viral infections remain unnoticed because acute inflammation protects us and these challenges are timely eliminated and inflammation resolves.
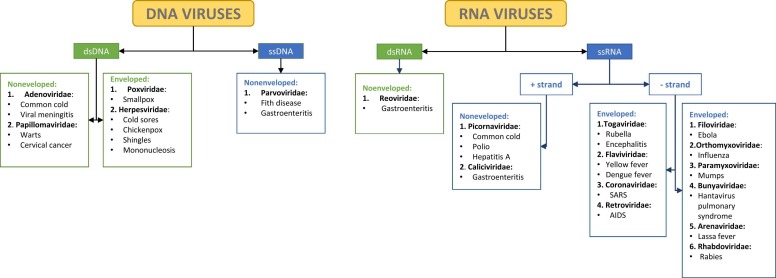
Viruses are obligate intracellular parasites that infect and replicate exclusively within cells of many living organisms, including bacteria, fungi, protozoa, plants, and animal. Their identification dates back to late XIX century, when Dmitrii Iwanowski (1864-1920) proposed that the disease Adolph Mayer (1843-1942) named tobacco mosaic disease was caused by an infectious agent several times smaller than bacteria [5], [6]. Almost contemporaneously, Martinus Beijerinck (1851-1931) replicated Iwanoski’s findings and called the pathogenic agent of the tobacco mosaic disease “contagium vivum fluidum” (contagious living fluid) [7]. It was not until the Nobel laureate Wendell M Stanley (1904-1971) obtained the first crystal of the tobacco mosaic virus that viruses were proven to be particulate microorganisms [8]. Their discoveries marked the beginning of virology and made possible to understand the etiology and pathophysiology of diseases that were described much earlier [9]. Viruses are divided according to the Baltimore classification based on the structure of their genome, strandedness, sense, and method of replication into 7 classes encompassing > 30,000 isolates ( Fig. 1), most of which do not cause serious illness to the human population. However, many viruses can cause common, severe, or even life-threatening diseases involving brain, hearth, blood, liver, pancreas, gut, lungs, skin and mucous membranes.
Fig. 1.
Baltimore classification of viruses The figure represents the Baltimore classification of DNA and RNA viruses. This classification was originally proposed by the Nobel laureate David Baltimore as a scheme for organizing known viruses based on the nature of their genome and replication strategy [92].
As an arm of innate immunity, acute inflammation represents a formidable barrier mechanism to suppress viral replication and spread. It is also important for activating adaptive immunity and, hence, coordinating the overall host immune system. Acute inflammation is activated upon recognition of viral pathogen associated molecular patterns (PAMPs) by the host pattern recognition receptors (PRRs), which encompass toll-like receptors (TLRs), Nod-like receptors (NLRs), and RIG-I-like receptors (RLRs). These PRRs sense specific viral molecules and signal downstream pathways that culminate with recruitment and activation of leukocytes, enhancement of cytokines and chemokines, and induction of antiviral genes like type I and III interferons [10]. Innate immune responses mediated by acute inflammation normally can clear virally infected cells and resolve virosis. On the contrary, inability to mount a timely and effective pro-resolution and antiviral responses can lead to virus persistence, pathogenic excessive inflammation, and fatal outcomes. Influenza viruses and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) offer clear examples of this [11], [12], emphasizing the crucial role and intriguing therapeutic functions of pro-resolving mechanisms during viral infections.
2. Resolution of inflammation and SPM
One of the major strides in our understanding of inflammation was the discovery that resolution is an active process introduced by the biochemical synthesis of specific proresolving molecules [13], among which are lipid mediators derived from ω-6 and ω-6 fatty acids that were dubbed “Specialized Pro-resolving lipid Mediators (SPM) by its discoverer CN Serhan.
Using a system lipidomics-informatics approach to self-resolving inflammation, pioneering research from the laboratory of Dr. Serhan led to the discovery of SPM in inflammatory exudates during resolution [14], [24]. SPM act through specific receptors to halt excessive PMN infiltration and activation, counter pro-inflammatory signals, enhance the active clearance of pathogens and death cells by MΦ, protect organ from loss of function, and stimulate tissue regeneration.
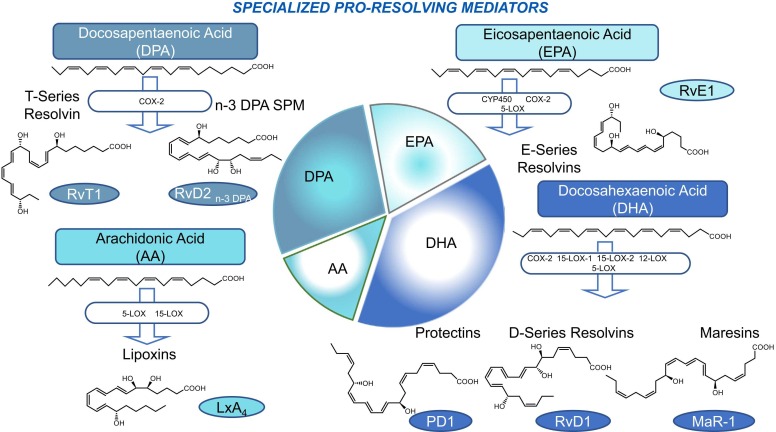
Notably, SPM biosynthesis is impinged by pro-inflammatory mediators, for instance prostaglandin E2, generated during the onset of the inflammatory response [15], indicating that “the beginning of inflammation programs its end”. The main biosynthetic pathways and members of SPM families are shown in Fig. 2.
Fig. 2.
Illustration of SPM biosynthesis Precursors AA, EPA, DHA and n-3 DPA polyunsaturated fatty acids (PUFA) are converted via biosynthetic enzymes to SPM. The pie chart visualizes the members of SPM accordingly to they precursor. The size of each slice is proportionate to the number of SPM produced by the specific precursor. See text for details on each SPM structure and biosynthetic mechanisms.
The eicosanoid lipoxins (LX) A4 and B4 are the SPM derived from arachidonic acid (AA) metabolism [16], [17], [18]. The regio- and stereoselective oxidation catalized by 5- and 15-lipoxygenase (LO) constitutes the first pathway for the formation of LX [16], [17], [18]. This pathway occurs in 15-LO expressing epithelial cells or MФ and leukocyte 5-LO. A second pathway relies on the LX synthase activity of 12-LO in platelets during cell-cell interactions with PMN [19], [20]. A the third pathway produces LX epimers, i.e., 15-epi-LXA4 and 15-epi-LXB4, which are formed in the presence of acetylated cyclooxygenase (COX)-2. Since this pathways was originally described with aspirin-treated endothelial cells expressing COX-2, 15-epi-LX are also called “aspirin-triggered lipoxins” (ATL) [21], [22].
RvE1 was the first SPM isolated from eicosapentaenoic acid (EPA)[23]. The current members of the E-series resolvins include RvE1, RvE2, and RvE3, with the recent addition and elucidation of RvE4. They are produced through transcellular biosynthesis with human neutrophils by acetylated cyclooxygenase-2 (COX-2) or microbial cytochrome P450 [24].
Docosahexaenoic acid (DHA)-SPM include D-series resolvins, protectins, and maresins.
The D-series resolvins (RvD1-6) are biosynthesized from the sequential oxygenation of precursor ω-3 fatty acid docosahexaenoic acid (DHA) [25], either via aspirin-triggered cyclooxygenase catalysis (17(R) AT-RvDs) or via the lipoxygenase pathway (15-LOX-1 and 15-LOX-2) forming the epimeric 17(S)-RvD1-6 resolvins [26].
Protectin D1 (PD1) is biosynthesized by DHA via 15-LOX and is produced enzymatically by human leucocytes from 16,17-epoxide-intermediates, PMN, macrophages, and eosinophils [27].
The third group produced by DHA biosynthesis is the Maresins (MaR-1 and MaR-2). Maresin biosynthesis occurs from carbon-14 via human 12-LOX, producing a 13(14)epoxide-intermediate (eMaR) that stimulates the conversion of M1 to M2 macrophages and blocks LTA4 hydrolase [28], [29].
Another precursor substrate for SPM formation is n-3 docosapentaenoic acid (n-3 DPA). n-3 DPA is converted into new SPM, including RvDn-3 DPA, MaRn-3 DPA, and PDn-3 DPA, as well as into series 13 resolvins (RvTs). SPM from n-3 DPA are characterized by the presence of an -OH group at position C13 in the PUFA chain [30], [31].
Three novel series of SPM conjugated with peptide-lipids have been recently introduced.
They include maresin conjugates for tissue regeneration (MCTR), protectin conjugates for tissue regeneration (PCTR) and resolvin conjugates for tissue regeneration (RCTR), collectively referred to as cysteinyl-SPM (cys-SPM)[30], [31]. Recent studies confirm their pro-resolution action and organ protection in many organs, including lungs [32], [33], [34], [35] (and reviewed in [23]).
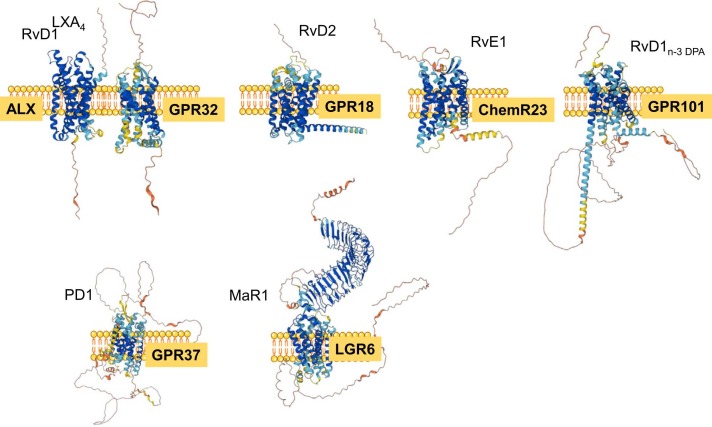
Several SPM G-protein coupled receptors (GPCRs) have been identified to date, using robust pharmacological approaches including library screening, specific binding with labeled ligands, engineered GPCR-β-arrestin cell for monitoring receptor engagement, and gain and loss of function strategies(recent reviewed in [36], [37]) ( Fig. 3). These GPCRs convey SPM actions transmitting signals to activate intracellular pathways and cell responses
Fig. 3.
SPM receptors The figure shows molecular graphics and protein structure of identified SPM GPCR. Analyses were carried out with UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from National Institutes of Health R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases.
Readers interested in cell- and tissue/organ-specific SPM bioactions are directed to excellent recent papers [36], [38].
Substantial evidence has accumulated that pro-resolving endogenous mediators also encompass proteins and peptides. One of the first polypeptide identified playing crucial biological roles in regulating acute inflammation is the glucocorticoid-regulated protein annexin (Anx) A1. AnxA1 is a 37 kDa protein initially recognized as an inhibitor of phospholipase A2 and, therefore, as a cellular mediator of glucocorticoid pharmacological anti-inflammatory actions, including the inhibition of prostaglandin and leukotriene biosynthesis [39], [40], [41], [42]. However, it is now clear that the biology of AnxA1 and N-teminal peptides with glucocorticoids is much more complex than initially thought. Readers interested can refer to [43] for excellent coverage of this topic. The development of recombinant human AnxA1 (hr-AnxA1) has helped to understand its biological activities, including the control of leukocyte migration, the promotion of neutrophil apoptosis, and induction efferocytosis, which underlie the therapeutic potential of the AnxA1-centered proresolving pathway that has been demonstrated in various experimental models [43]. The discovery of the lipoxin A4 receptor (ALX/FPR2) as a receptor for AnxA1 actions was also crucial in decoding mechanisms underlying resolution of inflammation, since ALX was the first GPCR identified that binds proresolving ligands of lipid and peptide structure[44]. The AnxA1- ALX/FPR2 axis is associated with key events in the resolution of inflammation, such as decreased neutrophil recruitment, induction of noninflammatory monocyte recruitment, promotion of neutrophil apoptosis and efferocytosis, contribution to tissue repair and resolution program amplification [43].
3. SPM and viral infection
In addition to their well-characterized roles in tissue homeostasis described above, several studies highlighted beneficial functions of SPM in the modulation of host responses to various infectious diseases triggered by viruses ( Table 1). Indeed, a large body of evidence shows that SPM decrease the inflammatory response by 1) promoting resolution and clearance of infection through modulation of host cell activities and 2) directly affecting the life cycle of viruses. These combined actions reduce viral replication in target cells thus providing greater ability of the host to deal with harmful infection. Along these lines, defective SPM production is suppressed during viral challenges and inversely correlated with virus pathogenicity [45], [46]. Therefore, SPM may represent a viable strategy for controlling the viral load and the excessive inflammation during viral infections.
Table 1.
Main bioactions of SPM in virus-mediated diseases.
| Virus | SPM | Function | References |
|---|---|---|---|
| Influenza | AnxA1 PD-1 and PDX AT-RvD1 MCTR |
|
[45], [48] [50] [53] [56] |
| Respiratory syncytial virus | LXA4, RvE1 RvD1 PCTR1, PD1 |
|
[59] [60] [61] |
| SARS-CoV-2 | RvD1, RvD2 |
|
[67], [75] [75] [67], [70] |
| Herpes viruses | RvE1 PD1 AT-RvD1 |
|
[82] [83] [84] |
| Kaposi’s Sarcoma-Associated Herpesvirus | LXA4 |
|
[86] [87] |
3.1. Influenza A virus
Respiratory viruses are among the most frequent causative agents of disease in humans, causing illness in nose, throat and breathing passages including lungs, with significant impact on morbidity and mortality. Respiratory viruses include rhinoviruses and enteroviruses (Picornaviridae), influenza viruses (Orthomyxoviridae), parainfluenza, metapneumoviruses and respiratory syncytial viruses (Paramyxoviridae), coronaviruses (Coronaviridae), and several adenoviruses.
A number of studies evaluated the relevance of SPM in the context of viral infections of the lung, especially influenza A (IAV), a negative-sense RNA viruses that causes seasonal epidemics of disease in people, particularly harmful in fragile individuals [47].
Using lung tissues lipidomics in mice subjected to intratracheal inoculation of the H1N1 PR8 strain, the lipid protectin D1 (PD1) isomer PDX was identified as one of the most reduced lipid mediators in the lungs of PR8-infected mice. Mechanistically, this reduction could be ascribed to a viral-induced defect of the 12/15-LOX enzyme, a key component of PD biosynthesis. Importantly, treatment of mice with exogenous PD increased survival and improved pulmonary injury trough reduction of viral titers in lungs of PR8 challenged mice. In vitro and in vivo experiments demonstrated that, rather than altering the host inflammatory response, PDX dampened IAV life cycle via attenuation of viral RNA nuclear export, a key step for virus replication [45], [48]. Similarly, the highly pathogenic H5N1 IAV altered the gene expression levels of the lipoxin pathway machinery in lungs to disseminate in multiple organs after infection in mice lungs [46]. Among the most affected genes, these included the suppressor of cytokine signaling (SOCS) 2 gene, an intracellular lipoxin mediator regulating cytokine and immune cells dynamics [49], thus indicating that this reduction could crucially impair pro-resolutive actions against viral infection.
Therefore, IAV hijack key pathways of SPM biosynthesis to reduce the production of crucial anti-viral and pro-resolutive SPM that would impair viral proliferation and dissemination.
Opposite to these beneficial effects of SPM supplementation, the pro-resolving AnxA1 enhanced IAV infectivity. Indeed, AnxA1 deficient (-/-) mice are protected against IAV infection due to an enhanced leukocyte infiltration, thus suggesting a sustained inflammatory response against viral infection. In addition, AnxA1 silencing and overexpression experiments in vitro suggested that, in addition to regulate host immunity, the presence of AnxA1 promoted viral replication, binding at the host cell membrane, viral uptake by host cells, viral transport to the nucleus and viral-induced apoptosis of target A549 lung cells, all key steps leading to greater virus production. Mechanistically, AnxA1 was incorporated within IAV and co-localized with the IAV protein NS1 in endosomes, indicating that AnxA1 facilitated endosomal trafficking and IAV infection life cycle [50]. These effects could be also due, at least in part, to ALX. As discussed, ALX is a plastic receptor able to sense and to activate a variety of pro-inflammatory and pro-resolving stimulus, such as AnxA1, LXA4 and RvD1 among the latter. IAV infection up-regulated ALX in murine lungs and lungs human cell lines [51], thus suggesting the needed of the virus to exploit the receptor to support the viral cycle. Indeed, activation of ALX with the agonists WKYMVm-NH2 and IAV harboring AnxA1 increased viral replication in vitro and in vivo and altered cytokine release in lungs of infected mice [51]. These effects of AnxA1 on IAV are in sharp contrast with those demonstrated in viral dengue fever, a potentially lethal hemorrhagic disease caused by one of the 4 serotypes of dengue virus (DENV1-4) transmitted through mosquitos that can result in fatal exacerbation of innate and adaptive immune responses. Indeed, Costa and Sugimoto recently demonstrated that therapeutic administration of an AnxA1 derived peptide to DENV-infected mice improves clinical signs of the disease (e.g., reduction in blood platelets and hematocrit), liver damage, and inflammatory markers. Strickingly, the absence of AnxA1 or its receptor ALX in knockout mice resulted in more severe illness of DENV-infected animals, signifying the important protective roles of AnxA1 and ALX in dengue fever [52].
Therapeutic treatment with AT-RvD1, another agonist of ALX, during an acute co-infection pneumonia in mice co-infected with Streptococcus pneumoniae and IAV, markedly reduced PMN infiltration and pneumonia severity promoting pro-resolution pathways [53]. Therefore, even in in a co-infection model, these results signified that diverse stimuli (peptide vs lipid) may differentially fuel ALX to translate pro-inflammatory and pro-viral or pro-resolutive signals that could be explored for therapeutic purposes.
SPM also hold the potential to activate adaptive immunity as well. In particular, the DHA-derived SPM 17-hydroxydocosahexaenoic acid (17-HDHA) enhanced plasma cell differentiation and production of specific antibodies (Abs) directed against the recombinant H1N1 hemagglutinin (HA) used to immunize mice. Importantly, 17-HDHA–mediated HA-specific Abs protected mice live influenza infection, indicating that 17-HDHA increase a defensive humoral response sustaining a specific B-cell differentiation and Ab-secreting phenotype [54]. Similarly, studies showing that LXB4 enhances the production of IgG in B lymphocytes derived from donors vaccinated against influenza [55] and others demonstrating that MCTR protect from bacterial pneumonia post-IAV acting on MΦ [56] confirm that SPM could be used to stimulate host immunity against IAV and collateral bacterial infections.
3.2. Respiratory syncytial virus
Respiratory syncytial virus (RSV) is a common respiratory virus infects the lungs and respiratory tract causing mild, cold-like symptoms except in infants, older adults, and fragile people where severe infections lead to pneumonia and bronchiolitis [57]. After epithelia infection, RSV elicits a potent inflammatory response mainly sustained by pro-inflammatory (M1) lung MΦ that, as expected, is dampened after MΦ skew to M2 polarization [58]. Along these lines, in vitro treatment with LXA4 or RvE1 induced gene expression of arginase-1 and mannose receptor in mouse MΦ from 5LO-/- transgenic mice, suggestive of M2 alternative activation that stimulate RSV resolution [59].
Similarly to IAV, SPM modulate the adaptive harm of immunity during RSV infection. In particular, exposure of RSV infected mice with RvD1 increased the frequency of specific memory precursors CD8 T cells against virus in the lung, and modulate memory CD8 T cells gene expression by increasing transcript of anti-inflammatory genes II-4, II-10, and Ifng [60].
Recent work shows that intranasal administration of PCTR1 and PD1 in RVS-infected mice decrease viral load and leukocyte infiltration while raising IFN-responses [61].
Collectively, these results highlight the critical role of SPM in the immune and inflammatory host response to RSV.
3.3. SARS-CoV-2
The coronavirus disease (COVID-19) caused by the severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) has been an unprecedented global threat for human health. SARS-CoV-2 is a RNA virus that infects the lungs, that encompass a wide range of symptoms and variable clinical outcomes, with many people developing severe, often lethal, pneumonia, sepsis, and respiratory failure, and others showing only a mild illness that resolves in few days [62]. SARS-CoV-2 can also determine blood disturbancies, including clotting and formation of NETs (neutrophil extracellular traps) [12]. Evidence indicates that viral load is not correlated with the worsening of the symptoms, while cytokine storm, increase in inflammatory mediators, and an imbalance in immunity are associated with poor prognosis [12], [63], [64], [65].
Indeed, it is now clear that failure in resolution of inflammation is a key determinant of SARS-CoV-2 infection. Serum and bronchoalveolar lavage fluids of symptomatic SARS-CoV-2 infected patients ad significantly higher concentrations of both omega-6–derived proinflammatory lipids and omega-6– and omega-3–derived SPM age- and sex-matched SARS-CoV-2–negative group, suggestive of an unrelenting inflammation that failed to resolve [66], [67], [68].
Importantly, failure in resolution also characterized worsen outcomes in COVID-19 disease. Patients who recovered from disease showed upregulation of the pro-resolution peptide AnxA1 in peripheral blood monocytes, indicating increased resolution in people that mount an active response against SARS-CoV-2 [69]. Consistent with this, lipid mediator profiling demonstrated that plasma SPM concentrations were downregulated in people with severe COVID-19 disease [67], [70], [71], [72]. More in details, an overall downregulation in PCTR3, MCTR3 [67] and RvD3 [70] were observed in patients with severe disease, with an upregulation of arachidonic acid-derived LM, including LTB4 and LTE4, LTC4 and LTD4. Importantly, in addition to reduced SPM levels, patients with severe disease demonstrated lowered expression of SPM biosynthetic enzymes (ALOX15B in neutrophils, COX-2 and ALOX5 in classical monocytes, and ALOX5 in nonclassical monocytes) [67], [70] and receptors (GPR18 on neutrophils, classical monocytes, and nonclassical monocytes, ChemR23, on classical and intermediate monocytes, GPR32 on intermediate and nonclassical monocytes, GPR101 on classical monocytes) on circulating leukocytes [67]. Collectively, these results pointed to defects in SPM biosynthesis and production as critical determinant for disease severity and suggested that restoration of adequate pro-resolution programs may be beneficial. Indeed, patients treated with dexamethasone, a corticosteroid proved to upregulate SPM formation [73], [74], reduced plasma pro-inflammatory eicosanoids while increasing SPM concentration, along with upregulation of ALOX15, ALOX15B, ALOX12, GPR18 and GPR37 in circulating leukocyte subsets [67]. Exposure of PMN, monocytes and monocyte-derived MΦ to MCTR3, PCTR3, 17R-RvD3, RvD1 and RvD2 restored phagocytic ability of these cells and reprogrammed MΦ toward a pro-resolutive phenotype characterized by lowered production of pro-inflammatory cytokines [67], [75]. Along these lines, we recently reported that RvD1 and RvD2 treatment abated the inflammatory responses induced by SARS-CoV-2 virion spike 1 glycoprotein (S1) by dampening the release of IL-8 and TNF-α and modulating the expression of the inflammatory microRNAs (miRNA) miR-16, miR-29a, miR-223 and miR-125a [75]. These effects are of paramount importance, since the imbalanced pro-inflammatory MΦ-derived cytokine storm may cause severe pulmonary edema, acute respiratory distress, and multi-organ failure [76]. Thus, by broadly inhibiting proinflammatory cytokine production by MΦ and other cells, SPM proved valuable as potential therapeutics to limit SARS-CoV-2-induced inflammation [77]. Finally, since SPM reduce NETosis (e.g., RvD4, RvD1, RvT1, RvT2, RvT3, RvT4) [78], [79], [80], they can also have roles in reducing the severity of COVID-19.
3.4. Herpes viruses
Severe infections with ocular Herpes simplex virus (HSV) can lead to scarring of the cornea or blindness mainly due to a chronic inflammatory reaction within cornea [81]. Thus, stimulation of pro-resolution pathways could be an attractive strategy to reduce the incidence of eye defects. Topical treatment with RvE1 of HSV-induced ocular disease reduced PMN and pathological CD4 T cells infiltration, levels of pro-inflammatory cytokines such as IFN-g, IL-6, KC while increasing anti-inflammatory IL-10 in corneas of infected mice [82]. Similar findings were also reported in eyes of HSV-infected mice treated neuro PD1 [83] and AT-RvD1 [84]. These results highlight that SPM could be harnessed as novel approach to control virally-induced immunopathological disease in the eye.
3.5. Kaposi’s Sarcoma-Associated Herpesvirus
The Kaposi sarcoma herpesvirus (KSHV) is the causative agents of Kaposi sarcoma, a form of multicentric Castleman disease, and primary effusion lymphoma [85]. In vitro experiments showed that exposure to LXA4 of KSHV positive cell lines or de novo KSHV infected cells not only reduced levels of key pro-inflammatory mediators (PGE2, LTB4, IL-6, IL-8) [86] but also critically impact on reactivation from latency of dormant KSHV. Indeed, LXA4 physically interact with chromatin-remodeling proteins finally leading to viral gene lytic replication and viral progression. These events, together with the decreased expression of the immunomodulatory PD-L1 protein triggered by LXA4 in infected cells, should unleash cellular immunity against active KSHV [87].
4. Summary and Future Directions
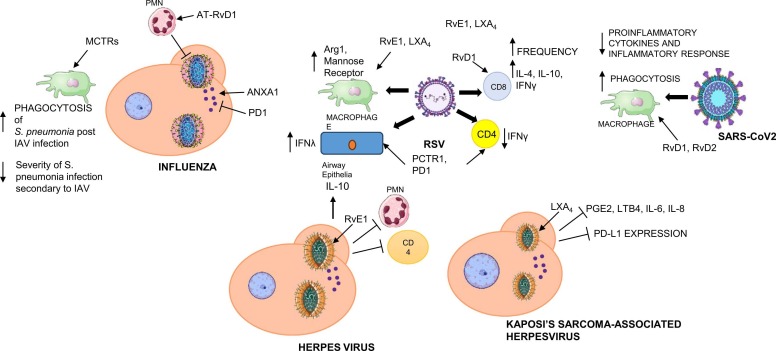
Considerable research effort has been made to decipher the underlying mechanisms of active resolution of inflammation. It is becoming clear that failure in specific resolution pathways can contribute to a worse clinical outcome of viral infectious diseases. Therefore, harnessing endogenous proresolution mechanisms is gaining traction as a new therapeutic approach to treating viral diseases given their proresolving actions ( Fig. 4). Conventional anti-inflammatory strategies stop the inception phase of inflammation by inhibiting prostaglandin and/or leukotrienes biosynthesis. However, in viral diseases, this approach may undermine the beneficial effects of inflammation to restrain viral diffusion, lead to immune suppression, or delay resolution. SPM proved to enhance host defenses and lower threshold for antibiotic therapies in bacterial infections [88], [89]. Their roles in virus-mediated infections are of timely paramount importance in view of possible future outbreaks caused by highly pathogenic viruses (e.g., new SARS variants, Ebola and Crimean-Congo hemorragic fever viruses, and zoonotic Nipah viruses) that under surveillance by the WHO [90]. The latest COVID-19 pandemics has shown our unpreparedness to face viruses that had no vaccines or therapeutics available to regulate host immunity. As a result COVID-19 has claimed ~ 7,000,000 human lives worldwide [91]. Further studies on how viruses hijack SPM production, as well as on SPM functions will contribute towards understanding the pathogenesis of viral diseases and finding new ways to encompass resolution of inflammation to protect human health.
Fig. 4.
General bioactions of SPM in virus-driven infectious diseases. Shown here are main effects of SPM demonstrated in vitro and in vivo. See text for further description and references.
Acknowledgements
The support of the Cystic Fibrosis Foundation (Grant RECCHIG1) to A.R is gratefully acknowledged.
References
- 1.Medzhitov R. Inflammation 2010: new adventures of an old flame. Cell. 2010;140:771–776. doi: 10.1016/j.cell.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Majno G., Joris I. Oxford University Press; 2004. Cells, tissues, and disease: principles of general pathology. [Google Scholar]
- 3.Gudiol C., Bodro M., Simonetti A., Tubau F., González-Barca E., Cisnal M., Domingo-Domenech E., Jiménez L., Carratalà J. Changing aetiology, clinical features, antimicrobial resistance, and outcomes of bloodstream infection in neutropenic cancer patients. Clin Microbiol Infect. 2013;19:474–479. doi: 10.1111/j.1469-0691.2012.03879.x. [DOI] [PubMed] [Google Scholar]
- 4.Serhan C.N., Savill J. Resolution of inflammation: the beginning programs the end. Nat Immunol. 2005;6:1191–1197. doi: 10.1038/ni1276. [DOI] [PubMed] [Google Scholar]
- 5.Mayer A. Concerning the mosaic disease of tobbacco. Über die Mosaikkrankheit des Tabaks. Die Landwirtschaftliche Versuchsstationen. 1886;32:451–467. [Google Scholar]
- 6.Iwanowski D.I. Über die Mosaikkrankheit der Tabakspflanze. Bulletin de l’Académie Impériale Des Sciences de St.-Pétersbourg. 1892;35:67–70. [Google Scholar]
- 7.Beijerinck M. Concerning a contagium viwm fluidum as cause of the spot disease of tobacco leaves. Phytopathol Class. 1898;7:33–52. [Google Scholar]
- 8.Stanley W.M. Crystalline Tobacco-Mosaic Virus Protein. American Journal of Botany. 1937;24:59–68. doi: 10.2307/2436720. [DOI] [Google Scholar]
- 9.Lustig A., Levine A. One hundred years of virology. J Virol. 1992;66:4629–4631. doi: 10.1128/jvi.66.8.4629-4631.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farag N.S., Breitinger U., Breitinger H.G., Azizi M.A.El. Viroporins and inflammasomes: A key to understand virus-induced inflammation. The International Journal of Biochemistry & Cell Biology. 2020;122 doi: 10.1016/j.biocel.2020.105738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwasaki A., Pillai P.S. Innate immunity to influenza virus infection. Nat Rev Immunol. 2014;14:315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merad M., Blish C.A., Sallusto F., Iwasaki A. The immunology and immunopathology of COVID-19. Science. 2022;375:1122–1127. doi: 10.1126/science.abm8108. [DOI] [PubMed] [Google Scholar]
- 13.Ortega-Gómez A., Perretti M., Soehnlein O. Resolution of inflammation: an integrated view. EMBO Molecular Medicine. 2013;5:661–674. doi: 10.1002/emmm.201202382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bannenberg G.L., Chiang N., Ariel A., Arita M., Tjonahen E., Gotlinger K.H., Hong S., Serhan C.N. Molecular circuits of resolution: formation and actions of resolvins and protectins. J Immunol. 2005;174:4345–4355. doi: 10.4049/jimmunol.174.7.4345. [DOI] [PubMed] [Google Scholar]
- 15.Levy B.D., Clish C.B., Schmidt B., Gronert K., Serhan C.N. Lipid mediator class switching during acute inflammation: signals in resolution. Nature Immunology. 2001;2:612–619. doi: 10.1038/89759. [DOI] [PubMed] [Google Scholar]
- 16.Serhan C.N., Hamberg M., Samuelsson B. Trihydroxytetraenes: a novel series of compounds formed from arachidonic acid in human leukocytes. Biochem Biophys Res Commun. 1984;118:943–949. doi: 10.1016/0006-291x(84)91486-4. [DOI] [PubMed] [Google Scholar]
- 17.Serhan C.N., Hamberg M., Samuelsson B. Lipoxins: novel series of biologically active compounds formed from arachidonic acid in human leukocytes. Proc Natl Acad Sci U S A. 1984;81:5335–5339. doi: 10.1073/pnas.81.17.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serhan C.N., Hamberg M., Samuelsson B., Morris J., Wishka D.G. On the stereochemistry and biosynthesis of lipoxin B. Proc Natl Acad Sci U S A. 1986;83:1983–1987. doi: 10.1073/pnas.83.7.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romano M., Serhan C.N. Lipoxin generation by permeabilized human platelets. Biochemistry. 1992;31:8269–8277. doi: 10.1021/bi00150a021. [DOI] [PubMed] [Google Scholar]
- 20.Romano M., Chen X.S., Takahashi Y., Yamamoto S., Funk C.D., Serhan C.N. Lipoxin synthase activity of human platelet 12-lipoxygenase. Biochem J. 1993;296(Pt 1):127–133. doi: 10.1042/bj2960127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aspirin triggers previously undescribed bioactive eicosanoids by human endothelial cell-leukocyte interactions., (n.d.). https://doi.org/10.1073/pnas.92.21.9475.
- 22.Clària J., Lee M.H., Serhan C.N. Aspirin-triggered lipoxins (15-epi-LX) are generated by the human lung adenocarcinoma cell line (A549)-neutrophil interactions and are potent inhibitors of cell proliferation. Mol Med. 1996;2:583–596. [PMC free article] [PubMed] [Google Scholar]
- 23.Serhan C.N., Chiang N. Resolvins and cysteinyl-containing pro-resolving mediators activate resolution of infectious inflammation and tissue regeneration. Prostaglandins & Other Lipid Mediators. 2023;166 doi: 10.1016/j.prostaglandins.2023.106718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serhan C.N., Clish C.B., Brannon J., Colgan S.P., Chiang N., Gronert K. Novel Functional Sets of Lipid-Derived Mediators with Antiinflammatory Actions Generated from Omega-3 Fatty Acids via Cyclooxygenase 2–Nonsteroidal Antiinflammatory Drugs and Transcellular Processing. Journal of Experimental Medicine. 2000;192:1197–1204. doi: 10.1084/jem.192.8.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serhan C.N. Treating inflammation and infection in the 21st century: new hints from decoding resolution mediators and mechanisms. FASEB j. 2017;31:1273–1288. doi: 10.1096/fj.201601222R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serhan C.N., Hong S., Gronert K., Colgan S.P., Devchand P.R., Mirick G., Moussignac R.-L. Resolvins. Journal of Experimental Medicine. 2002;196:1025–1037. doi: 10.1084/jem.20020760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong S., Gronert K., Devchand P.R., Moussignac R.-L., Serhan C.N. Novel Docosatrienes and 17S-Resolvins Generated from Docosahexaenoic Acid in Murine Brain, Human Blood, and Glial Cells. Journal of Biological Chemistry. 2003;278:14677–14687. doi: 10.1074/jbc.M300218200. [DOI] [PubMed] [Google Scholar]
- 28.Dalli J., Zhu M., Vlasenko N.A., Deng B., Haeggström J.Z., Petasis N.A., Serhan C.N. The novel 13 S,14 S ‐epoxy‐maresin is converted by human macrophages to maresin 1 (MaR1), inhibits leukotriene A 4 hydrolase (LTA 4 H), and shifts macrophage phenotype. FASEB j. 2013;27:2573–2583. doi: 10.1096/fj.13-227728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serhan C.N., Yang R., Martinod K., Kasuga K., Pillai P.S., Porter T.F., Oh S.F., Spite M. Maresins: novel macrophage mediators with potent antiinflammatory and proresolving actions. Journal of Experimental Medicine. 2009;206:15–23. doi: 10.1084/jem.20081880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dalli J., Chiang N., Serhan C.N. Identification of 14-series sulfido-conjugated mediators that promote resolution of infection and organ protection. Proc. Natl. Acad. Sci. U.S.A. 2014;111 doi: 10.1073/pnas.1415006111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalli J., Ramon S., Norris P.C., Colas R.A., Serhan C.N. Novel proresolving and tissue-regenerative resolvin and protectin sulfido-conjugated pathways. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology. 2015;29:2120–2136. doi: 10.1096/fj.14-268441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levy B.D., Abdulnour R.-E.E., Tavares A., Brüggemann T.R., Norris P.C., Bai Y., Ai X., Serhan C.N. Cysteinyl maresins regulate the prophlogistic lung actions of cysteinyl leukotrienes. J Allergy Clin Immunol. 2020;145:335–344. doi: 10.1016/j.jaci.2019.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Säfholm J., Abma W., Bankova L.G., Boyce J.A., Al-Ameri M., Orre A.-C., Wheelock C.E., Dahlén S.-E., Adner M. Cysteinyl-maresin 3 inhibits IL-13 induced airway hyperresponsiveness through alternative activation of the CysLT1 receptor. Eur J Pharmacol. 2022;934 doi: 10.1016/j.ejphar.2022.175257. [DOI] [PubMed] [Google Scholar]
- 34.Pistorius K., Ly L., Souza P.R., Gomez E.A., Koenis D.S., Rodriguez A.R., Foster J., Sosabowski J., Hopkinson M., Rajeeve V., Spur B.W., Pitsillides A., Pitzalis C., Dalli J. MCTR3 reprograms arthritic monocytes to upregulate Arginase-1 and exert pro-resolving and tissue-protective functions in experimental arthritis. EBioMedicine. 2022;79 doi: 10.1016/j.ebiom.2022.103974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiang N., de la Rosa X., Libreros S., Pan H., Dreyfuss J.M., Serhan C.N. Cysteinyl-specialized proresolving mediators link resolution of infectious inflammation and tissue regeneration via TRAF3 activation. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2013374118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiang N., Serhan C.N. Specialized pro-resolving mediator network: an update on production and actions. Essays in Biochemistry. 2020;64:443–462. doi: 10.1042/EBC20200018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romano M., Patruno S., Pomilio A., Recchiuti A. Proresolving Lipid Mediators and Receptors in Stem Cell Biology: Concise Review. Stem Cells Transl Med. 2019;8:992–998. doi: 10.1002/sctm.19-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dyall S.C., Balas L., Bazan N.G., Brenna J.T., Chiang N., da Costa Souza F., Dalli J., Durand T., Galano J.-M., Lein P.J., Serhan C.N., Taha A.Y. Polyunsaturated fatty acids and fatty acid-derived lipid mediators: Recent advances in the understanding of their biosynthesis, structures, and functions. Prog Lipid Res. 2022;86 doi: 10.1016/j.plipres.2022.101165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goulding N.J., Godolphin J.L., Sharland P.R., Peers S.H., Sampson M., Maddison P.J., Flower R.J. Anti-inflammatory lipocortin 1 production by peripheral blood leucocytes in response to hydrocortisone. Lancet. 1990;335:1416–1418. doi: 10.1016/0140-6736(90)91445-g. [DOI] [PubMed] [Google Scholar]
- 40.Perretti M., Flower R.J. Modulation of IL-1-induced neutrophil migration by dexamethasone and lipocortin 1. J Immunol. 1993;150:992–999. [PubMed] [Google Scholar]
- 41.Flower R.J., Blackwell G.J. Anti-inflammatory steroids induce biosynthesis of a phospholipase A2 inhibitor which prevents prostaglandin generation. Nature. 1979;278:456–459. doi: 10.1038/278456a0. [DOI] [PubMed] [Google Scholar]
- 42.Blackwell G.J., Carnuccio R., Di Rosa M., Flower R.J., Parente L., Persico P. Macrocortin: a polypeptide causing the anti-phospholipase effect of glucocorticoids. Nature. 1980;287:147–149. doi: 10.1038/287147a0. [DOI] [PubMed] [Google Scholar]
- 43.Perretti M., Dalli J. Resolution Pharmacology: Focus on Pro-Resolving Annexin A1 and Lipid Mediators for Therapeutic Innovation in Inflammation. Annu. Rev. Pharmacol. Toxicol. 2023;63:449–469. doi: 10.1146/annurev-pharmtox-051821-042743. [DOI] [PubMed] [Google Scholar]
- 44.Perretti M., Chiang N., La M., Fierro I.M., Marullo S., Getting S.J., Solito E., Serhan C.N. Endogenous lipid- and peptide-derived anti-inflammatory pathways generated with glucocorticoid and aspirin treatment activate the lipoxin A4 receptor. Nat Med. 2002;8:1296–1302. doi: 10.1038/nm786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morita M., Kuba K., Ichikawa A., Nakayama M., Katahira J., Iwamoto R., Watanebe T., Sakabe S., Daidoji T., Nakamura S., Kadowaki A., Ohto T., Nakanishi H., Taguchi R., Nakaya T., Murakami M., Yoneda Y., Arai H., Kawaoka Y., Penninger J.M., Arita M., Imai Y. The Lipid Mediator Protectin D1 Inhibits Influenza Virus Replication and Improves Severe Influenza. Cell. 2013;153:112–125. doi: 10.1016/j.cell.2013.02.027. [DOI] [PubMed] [Google Scholar]
- 46.Cilloniz C., Pantin-Jackwood M.J., Ni C., Goodman A.G., Peng X., Proll S.C., Carter V.S., Rosenzweig E.R., Szretter K.J., Katz J.M., Korth M.J., Swayne D.E., Tumpey T.M., Katze M.G. Lethal Dissemination of H5N1 Influenza Virus Is Associated with Dysregulation of Inflammation and Lipoxin Signaling in a Mouse Model of Infection. J Virol. 2010;84:7613–7624. doi: 10.1128/JVI.00553-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar D., Michaels M.G., Morris M.I., Green M., Avery R.K., Liu C., Danziger-Isakov L., Stosor V., Estabrook M., Gantt S., Marr K.A., Martin S., Silveira F.P., Razonable R.R., Allen U.D., Levi M.E., Lyon G.M., Bell L.E., Huprikar S., Patel G., Gregg K.S., Pursell K., Helmersen D., Julian K.G., Shiley K., Bono B., Dharnidharka V.R., Alavi G., Kalpoe J.S., Shoham S., Reid G.E., Humar A. Outcomes from pandemic influenza A H1N1 infection in recipients of solid-organ transplants: a multicentre cohort study. The Lancet Infectious Diseases. 2010;10:521–526. doi: 10.1016/S1473-3099(10)70133-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Imai Y. Role of omega-3 PUFA-derived mediators, the protectins, in influenza virus infection. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2015;1851:496–502. doi: 10.1016/j.bbalip.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 49.Machado F.S., Johndrow J.E., Esper L., Dias A., Bafica A., Serhan C.N., Aliberti J. Anti-inflammatory actions of lipoxin A4 and aspirin-triggered lipoxin are SOCS-2 dependent. Nat Med. 2006;12:330–334. doi: 10.1038/nm1355. [DOI] [PubMed] [Google Scholar]
- 50.Arora S., Lim W., Bist P., Perumalsamy R., Lukman H.M., Li F., Welker L.B., Yan B., Sethi G., Tambyah P.A., Fairhurst A.-M., Alonso S., Lim L.H.K. Influenza A virus enhances its propagation through the modulation of Annexin-A1 dependent endosomal trafficking and apoptosis. Cell Death Differ. 2016;23:1243–1256. doi: 10.1038/cdd.2016.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ampomah P.B., Moraes L.A., Lukman H.M., Lim L.H.K. Formyl peptide receptor 2 is regulated by RNA mimics and viruses through an IFN‐β‐STAT3–dependent pathway. FASEB j. 2018;32:1468–1478. doi: 10.1096/fj.201700584RR. [DOI] [PubMed] [Google Scholar]
- 52.Costa V.V., Sugimoto M.A., Hubner J., Bonilha C.S., Queiroz-Junior C.M., Gonçalves-Pereira M.H., Chen J., Gobbetti T., Libanio Rodrigues G.O., Bambirra J.L., Passos I.B., Machado Lopes C.E., Moreira T.P., Bonjour K., Melo R.C., Oliveira M.A., Andrade M.V.M., Sousa L.P., Souza D.G., da H., Santiago C., Perretti M., Teixeira M.M. Targeting the Annexin A1-FPR2/ALX pathway for host-directed therapy in dengue disease. ELife. 2022;11 doi: 10.7554/eLife.73853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H., Anthony D., Yatmaz S., Wijburg O., Satzke C., Levy B., Vlahos R., Bozinovski S. Aspirin-triggered resolvin D1 reduces pneumococcal lung infection and inflammation in a viral and bacterial coinfection pneumonia model. Clinical Science. 2017;131:2347–2362. doi: 10.1042/CS20171006. [DOI] [PubMed] [Google Scholar]
- 54.Ramon S., Baker S.F., Sahler J.M., Kim N., Feldsott E.A., Serhan C.N., Martínez-Sobrido L., Topham D.J., Phipps R.P. The Specialized Proresolving Mediator 17-HDHA Enhances the Antibody-Mediated Immune Response against Influenza Virus: A New Class of Adjuvant? The Journal of Immunology. 2014;193:6031–6040. doi: 10.4049/jimmunol.1302795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim N., Lannan K.L., Thatcher T.H., Pollock S.J., Woeller C.F., Phipps R.P. Lipoxin B4 Enhances Human Memory B Cell Antibody Production via Upregulating Cyclooxygenase-2 Expression. The Journal of Immunology. 2018;201:3343–3351. doi: 10.4049/jimmunol.1700503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tavares L.P., Brüggemann T.R., Rezende R.M., Machado M.G., Cagnina R.E., Shay A.E., Garcia C.C., Nijmeh J., Teixeira M.M., Levy B.D. Cysteinyl Maresins Reprogram Macrophages to Protect Mice from Streptococcus pneumoniae after Influenza A Virus Infection. MBio. 2022;13 doi: 10.1128/mbio.01267-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Welliver R.C. Review of epidemiology and clinical risk factors for severe respiratory syncytial virus (RSV) infection. The Journal of Pediatrics. 2003;143:112–117. doi: 10.1067/S0022-3476(03)00508-0. [DOI] [PubMed] [Google Scholar]
- 58.Shirey K.A., Pletneva L.M., Puche A.C., Keegan A.D., Prince G.A., Blanco J.C.G., Vogel S.N. Control of RSV-induced lung injury by alternatively activated macrophages is IL-4Rα-, TLR4-, and IFN-β-dependent. Mucosal Immunology. 2010;3:291–300. doi: 10.1038/mi.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shirey K.A., Lai W., Pletneva L.M., Karp C.L., Divanovic S., Blanco J.C.G., Vogel S.N. Role of the lipoxygenase pathway in RSV-induced alternatively activated macrophages leading to resolution of lung pathology. Mucosal Immunology. 2014;7:549–557. doi: 10.1038/mi.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.do D., de Freitas N., Marinho Franceschina C., Muller D., Hilario G.T., Gassen R.B., Fazolo T., de Lima Kaminski V., Bogo Chies J.A., Maito F., Antunes K.H., Zanin R.F., Rodrigues L.C., Jr, Duarte de Souza A.P. RvD1 treatment during primary infection modulates memory response increasing viral load during respiratory viral reinfection. Immunobiology. 2021;226 doi: 10.1016/j.imbio.2021.152151. [DOI] [PubMed] [Google Scholar]
- 61.Walker K.H., Krishnamoorthy N., Brüggemann T.R., Shay A.E., Serhan C.N., Levy B.D. Protectins PCTR1 and PD1 Reduce Viral Load and Lung Inflammation During Respiratory Syncytial Virus Infection in Mice. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.704427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., Rosain J., Bilguvar K., Ye J., Bolze A., Bigio B., Yang R., Arias A.A., Zhou Q., Zhang Y., Onodi F., Korniotis S., Karpf L., Philippot Q., Chbihi M., Bonnet-Madin L., Dorgham K., Smith N., Schneider W.M., Razooky B.S., Hoffmann H.-H., Michailidis E., Moens L., Han J.E., Lorenzo L., Bizien L., Meade P., Neehus A.-L., Ugurbil A.C., Corneau A., Kerner G., Zhang P., Rapaport F., Seeleuthner Y., Manry J., Masson C., Schmitt Y., Schlüter A., Le Voyer T., Khan T., Li J., Fellay J., Roussel L., Shahrooei M., Alosaimi M.F., Mansouri D., Al-Saud H., Al-Mulla F., Almourfi F., Al-Muhsen S.Z., Alsohime F., Al Turki S., Hasanato R., van de Beek D., Biondi A., Bettini L.R., D’Angio’ M., Bonfanti P., Imberti L., Sottini A., Paghera S., Quiros-Roldan E., Rossi C., Oler A.J., Tompkins M.F., Alba C., Vandernoot I., Goffard J.-C., Smits G., Migeotte I., Haerynck F., Soler-Palacin P., Martin-Nalda A., Colobran R., Morange P.-E., Keles S., Çölkesen F., Ozcelik T., Yasar K.K., Senoglu S., Karabela Ş.N., Rodríguez-Gallego C., Novelli G., Hraiech S., Tandjaoui-Lambiotte Y., Duval X., Laouénan C., COVID-STORM Clinicians, COVID Clinicians, Imagine COVID Group. French COVID Cohort Study Group. CoV-Contact Cohort, Amsterdam UMC Covid-19 Biobank, COVID Human Genetic Effort, NIAID-USUHS/TAGC COVID Immunity Group. Snow A.L., Dalgard C.L., Milner J.D., Vinh D.C., Mogensen T.H., Marr N., Spaan A.N., Boisson B., Boisson-Dupuis S., Bustamante J., Puel A., Ciancanelli M.J., Meyts I., Maniatis T., Soumelis V., Amara A., Nussenzweig M., García-Sastre A., Krammer F., Pujol A., Duffy D., Lifton R.P., Zhang S.-Y., Gorochov G., Béziat V., Jouanguy E., Sancho-Shimizu V., Rice C.M., Abel L., Notarangelo L.D., Cobat A., Su H.C., Casanova J.-L., Foti G., Bellani G., Citerio G., Contro E., Pesci A., Valsecchi M.G., Cazzaniga M., Abad J., Aguilera-Albesa S., Akcan O.M., Darazam I.A., Aldave J.C., Ramos M.A., Nadji S.A., Alkan G., Allardet-Servent J., Allende L.M., Alsina L., Alyanakian M.-A., Amador-Borrero B., Amoura Z., Antolí A., Arslan S., Assant S., Auguet T., Azot A., Bajolle F., Baldolli A., Ballester M., Feldman H.B., Barrou B., Beurton A., Bilbao A., Blanchard-Rohner G., Blanco I., Blandinières A., Blazquez-Gamero D., Bloomfield M., Bolivar-Prados M., Borie R., Bosteels C., Bousfiha A.A., Bouvattier C., Boyarchuk O., Bueno M.R.P., Bustamante J., Cáceres Agra J.J., Calimli S., Capra R., Carrabba M., Casasnovas C., Caseris M., Castelle M., Castelli F., de Vera M.C., Castro M.V., Catherinot E., Chalumeau M., Charbit B., Cheng M.P., Clavé P., Clotet B., Codina A., Colkesen F., Çölkesen F., Colobran R., Comarmond C., Dalmau D., Darley D.R., Dauby N., Dauger S., de Pontual L., Dehban A., Delplancq G., Demoule A., Diehl J.-L., Dobbelaere S., Durand S., Eldars W., Elgamal M., Elnagdy M.H., Emiroglu M., Erdeniz E.H., Aytekin S.E., Euvrard R., Evcen R., Fabio G., Faivre L., Falck A., Fartoukh M., Faure M., Arquero M.F., Flores C., Francois B., Fumadó V., Fusco F., Solis B.G., Gaussem P., Gil-Herrera J., Gilardin L., Alarcon M.G., Girona-Alarcón M., Goffard J.-C., Gok F., González-Montelongo R., Guerder A., Gul Y., Guner S.N., Gut M., Hadjadj J., Haerynck F., Halwani R., Hammarström L., Hatipoglu N., Hernandez-Brito E., Heijmans C., Holanda-Peña M.S., Horcajada J.P., Hoste L., Hoste E., Hraiech S., Humbert L., Iglesias A.D., Íñigo-Campos A., Jamme M., Arranz M.J., Jordan I., Jorens P., Kanat F., Kapakli H., Kara I., Karbuz A., Yasar K.K., Keles S., Demirkol Y.K., Klocperk A., Król Z.J., Kuentz P., Kwan Y.W.M., Lagier J.-C., Lambrecht B.N., Lau Y.-L., Le Bourgeois F., Leo Y.-S., Lopez R.L., Leung D., Levin M., Levy M., Lévy R., Li Z., Linglart A., Loeys B., Lorenzo-Salazar J.M., Louapre C., Lubetzki C., Luyt C.-E., Lye D.C., Mansouri D., Marjani M., Pereira J.M., Martin A., Pueyo D.M., Martinez-Picado J., Marzana I., Mathian A., Matos L.R.B., Matthews G.V., Mayaux J., Mège J.-L., Melki I., Meritet J.-F., Metin O., Meyts I., Mezidi M., Migeotte I., Millereux M., Mirault T., Mircher C., Mirsaeidi M., Melián A.M., Martinez A.M., Morange P., Mordacq C., Morelle G., Mouly S., Muñoz-Barrera A., Naesens L., Nafati C., Neves J.F., Ng L.F.P., Medina Y.N., Cuadros E.N., Ocejo-Vinyals J.G., Orbak Z., Oualha M., Özçelik T., Pan-Hammarström Q., Parizot C., Pascreau T., Paz-Artal E., Pellegrini S., de Diego R.P., Philippe A., Philippot Q., Planas-Serra L., Ploin D., Poissy J., Poncelet G., Pouletty M., Quentric P., Raoult D., Rebillat A.-S., Reisli I., Ricart P., Richard J.-C., Rivet N., Rivière J.G., Blanch G.R., Rodrigo C., Rodriguez-Gallego C., Rodríguez-Palmero A., Romero C.S., Rothenbuhler A., Rozenberg F., Ruiz del Prado M.Y., Riera J.S., Sanchez O., Sánchez-Ramón S., Schluter A., Schmidt M., Schweitzer C.E., Scolari F., Sediva A., Seijo L.M., Sene D., Senoglu S., Seppänen M.R.J., Ilovich A.S., Shahrooei M., Slabbynck H., Smadja D.M., Sobh A., Moreno X.S., Solé-Violán J., Soler C., Soler-Palacín P., Stepanovskiy Y., Stoclin A., Taccone F., Tandjaoui-Lambiotte Y., Taupin J.-L., Tavernier S.J., Terrier B., Thumerelle C., Tomasoni G., Toubiana J., Alvarez J.T., Trouillet-Assant S., Troya J., Tucci A., Ursini M.V., Uzunhan Y., Vabres P., Valencia-Ramos J., Van Braeckel E., Van de Velde S., Van Den Rym A.M., Van Praet J., Vandernoot I., Vatansev H., Vélez-Santamaria V., Viel S., Vilain C., Vilaire M.E., Vincent A., Voiriot G., Vuotto F., Yosunkaya A., Young B.E., Yucel F., Zannad F., Zatz M., Belot A., Bole-Feysot C., Lyonnet S., Masson C., Nitschke P., Pouliet A., Schmitt Y., Tores F., Zarhrate M., Abel L., Andrejak C., Angoulvant F., Bachelet D., Basmaci R., Behillil S., Beluze M., Benkerrou D., Bhavsar K., Bompart F., Bouadma L., Bouscambert M., Caralp M., Cervantes-Gonzalez M., Chair A., Coelho A., Couffignal C., Couffin-Cadiergues S., D’Ortenzio E., Da Silveira C., Debray M.-P., Deplanque D., Descamps D., Desvallées M., Diallo A., Diouf A., Dorival C., Dubos F., Duval X., Eloy P., Enouf V.V., Esperou H., Esposito-Farese M., Etienne M., Ettalhaoui N., Gault N., Gaymard A., Ghosn J., Gigante T., Gorenne I., Guedj J., Hoctin A., Hoffmann I., Jaafoura S., Kafif O., Kaguelidou F., Kali S., Khalil A., Khan C., Laouénan C., Laribi S., Le M., Le Hingrat Q., Le Mestre S., Le Nagard H., Lescure F.-X., Lévy Y., Levy-Marchal C., Lina B., Lingas G., Lucet J.C., Malvy D., Mambert M., Mentré F., Mercier N., Meziane A., Mouquet H., Mullaert J., Neant N., Noret M., Pages J., Papadopoulos A., Paul C., Peiffer-Smadja N., Petrov-Sanchez V., Peytavin G., Picone O., Puéchal O., Rosa-Calatrava M., Rossignol B., Rossignol P., Roy C., Schneider M., Semaille C., Mohammed N.S., Tagherset L., Tardivon C., Tellier M.-C., Téoulé F., Terrier O., Timsit J.-F., Trioux T., Tual C., Tubiana S., van der Werf S., Vanel N., Veislinger A., Visseaux B., Wiedemann A., Yazdanpanah Y., Alavoine L., Amat K.K.A., Behillil S., Bielicki J., Bruijning P., Burdet C., Caumes E., Charpentier C., Coignard B., Costa Y., Couffin-Cadiergues S., Damond F., Dechanet A., Delmas C., Descamps D., Duval X., Ecobichon J.-L., Enouf V., Espérou H., Frezouls W., Houhou N., Ilic-Habensus E., Kafif O., Kikoine J., Le Hingrat Q., Lebeaux D., Leclercq A., Lehacaut J., Letrou S., Lina B., Lucet J.-C., Malvy D., Manchon P., Mandic M., Meghadecha M., Motiejunaite J., Nouroudine M., Piquard V., Postolache A., Quintin C., Rexach J., Roufai L., Terzian Z., Thy M., Tubiana S., van der Werf S., Vignali V., Visseaux B., Yazdanpanah Y., van Agtmael M., Algera A.G., van Baarle F., Bax D., Beudel M., Bogaard H.J., Bomers M., Bos L., Botta M., de Brabander J., de Bree G., Brouwer M.C., de Bruin S., Bugiani M., Bulle E., Chouchane O., Cloherty A., Elbers P., Fleuren L., Geerlings S., Geerts B., Geijtenbeek T., Girbes A., Goorhuis B., Grobusch M.P., Hafkamp F., Hagens L., Hamann J., Harris V., Hemke R., Hermans S.M., Heunks L., Hollmann M.W., Horn J., Hovius J.W., de Jong M.D., Koning R., van Mourik N., Nellen J., Paulus F., Peters E., van der Poll T., Preckel B., Prins J.M., Raasveld J., Reijnders T., Schinkel M., Schultz M.J., Schuurman A., Sigaloff K., Smit M., Stijnis C.S., Stilma W., Teunissen C., Thoral P., Tsonas A., van der Valk M., Veelo D., Vlaar A.P.J., de Vries H., van Vugt M., Wiersinga W.J., Wouters D., (Koos) Zwinderman A.H., van de Beek D., Abel L., Aiuti A., Al Muhsen S., Al-Mulla F., Anderson M.S., Arias A.A., Feldman H.B., Bogunovic D., Bolze A., Bondarenko A., Bousfiha A.A., Brodin P., Bryceson Y., Bustamante C.D., Butte M., Casari G., Chakravorty S., Christodoulou J., Cirulli E., Condino-Neto A., Cooper M.A., Dalgard C.L., David A., DeRisi J.L., Desai M., Drolet B.A., Espinosa S., Fellay J., Flores C., Franco J.L., Gregersen P.K., Haerynck F., Hagin D., Halwani R., Heath J., Henrickson S.E., Hsieh E., Imai K., Itan Y., Karamitros T., Kisand K., Ku C.-L., Lau Y.-L., Ling Y., Lucas C.L., Maniatis T., Mansouri D., Marodi L., Meyts I., Milner J., Mironska K., Mogensen T., Morio T., Ng L.F.P., Notarangelo L.D., Novelli A., Novelli G., O’Farrelly C., Okada S., Ozcelik T., de Diego R.P., Planas A.M., Prando C., Pujol A., Quintana-Murci L., Renia L., Renieri A., Rodríguez-Gallego C., Sancho-Shimizu V., Sankaran V., Barrett K.S., Shahrooei M., Snow A., Soler-Palacín P., Spaan A.N., Tangye S., Turvey S., Uddin F., Uddin M.J., van de Beek D., Vazquez S.E., Vinh D.C., von Bernuth H., Washington N., Zawadzki P., Su H.C., Casanova J.-L., Jing H., Tung W., Luthers C.R., Bauman B.M., Shafer S., Zheng L., Zhang Z., Kubo S., Chauvin S.D., Meguro K., Shaw E., Lenardo M., Lack J., Karlins E., Hupalo D.M., Rosenberger J., Sukumar G., Wilkerson M.D., Zhang X. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.-H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., Manry J., Shaw E., Haljasmägi L., Peterson P., Lorenzo L., Bizien L., Trouillet-Assant S., Dobbs K., de Jesus A.A., Belot A., Kallaste A., Catherinot E., Tandjaoui-Lambiotte Y., Le Pen J., Kerner G., Bigio B., Seeleuthner Y., Yang R., Bolze A., Spaan A.N., Delmonte O.M., Abers M.S., Aiuti A., Casari G., Lampasona V., Piemonti L., Ciceri F., Bilguvar K., Lifton R.P., Vasse M., Smadja D.M., Migaud M., Hadjadj J., Terrier B., Duffy D., Quintana-Murci L., van de Beek D., Roussel L., Vinh D.C., Tangye S.G., Haerynck F., Dalmau D., Martinez-Picado J., Brodin P., Nussenzweig M.C., Boisson-Dupuis S., Rodríguez-Gallego C., Vogt G., Mogensen T.H., Oler A.J., Gu J., Burbelo P.D., Cohen J.I., Biondi A., Bettini L.R., D’Angio M., Bonfanti P., Rossignol P., Mayaux J., Rieux-Laucat F., Husebye E.S., Fusco F., Ursini M.V., Imberti L., Sottini A., Paghera S., Quiros-Roldan E., Rossi C., Castagnoli R., Montagna D., Licari A., Marseglia G.L., Duval X., Ghosn J., HGID Lab, NIAID-USUHS Immune Response to COVID Group. COVID Clinicians, COVID-STORM Clinicians, Imagine COVID Group. French COVID Cohort Study Group, The Milieu Intérieur Consortium, CoV-Contact Cohort. Amsterdam UMC Covid-19 Biobank. COVID Human Genetic Effort. Tsang J.S., Goldbach-Mansky R., Kisand K., Lionakis M.S., Puel A., Zhang S.-Y., Holland S.M., Gorochov G., Jouanguy E., Rice C.M., Cobat A., Notarangelo L.D., Abel L., Su H.C., Casanova J.-L., Arias A.A., Boisson B., Boucherit S., Bustamante J., Chbihi M., Chen J., Chrabieh M., Kochetkov T., Le Voyer T., Liu D., Nemirovskaya Y., Ogishi M., Papandrea D., Patissier C., Rapaport F., Roynard M., Vladikine N., Woollett M., Zhang P., Kashyap A., Ding L., Bosticardo M., Wang Q., Ochoa S., Liu H., Chauvin S.D., Stack M., Koroleva G., Bansal N., Dalgard C.L., Snow A.L., Abad J., Aguilera-Albesa S., Akcan O.M., Darazam I.A., Aldave J.C., Ramos M.A., Nadji S.A., Alkan G., Allardet-Servent J., Allende L.M., Alsina L., Alyanakian M.-A., Amador-Borrero B., Amoura Z., Antolí A., Arslan S., Assant S., Auguet T., Azot A., Bajolle F., Baldolli A., Ballester M., Feldman H.B., Barrou B., Beurton A., Bilbao A., Blanchard-Rohner G., Blanco I., Blandinières A., Blazquez-Gamero D., Bloomfield M., Bolivar-Prados M., Borie R., Bousfiha A.A., Bouvattier C., Boyarchuk O., Bueno M.R.P., Bustamante J., Cáceres Agra J.J., Calimli S., Capra R., Carrabba M., Casasnovas C., Caseris M., Castelle M., Castelli F., de Vera M.C., Castro M.V., Catherinot E., Chalumeau M., Charbit B., Cheng M.P., Clavé P., Clotet B., Codina A., Colkesen F., Colkesen F., Colobran R., Comarmond C., Corsico A.G., Dalmau D., Darley D.R., Dauby N., Dauger S., de Pontual L., Dehban A., Delplancq G., Demoule A., Di Sabatino A., Diehl J.-L., Dobbelaere S., Durand S., Eldars W., Elgamal M., Elnagdy M.H., Emiroglu M., Erdeniz E.H., Aytekin S.E., Euvrard R., Evcen R., Fabio G., Faivre L., Falck A., Fartoukh M., Faure M., Arquero M.F., Flores C., Francois B., Fumadó V., Fusco F., Solis B.G., Gaussem P., Gil-Herrera J., Gilardin L., Alarcon M.G., Girona-Alarcón M., Goffard J.-C., Gok F., González-Montelongo R., Guerder A., Gul Y., Guner S.N., Gut M., Hadjadj J., Haerynck F., Halwani R., Hammarström L., Hatipoglu N., Hernandez-Brito E., Holanda-Peña M.S., Horcajada J.P., Hraiech S., Humbert L., Iglesias A.D., Íñigo-Campos A., Jamme M., Arranz M.J., Jordan I., Kanat F., Kapakli H., Kara I., Karbuz A., Yasar K.K., Keles S., Demirkol Y.K., Klocperk A., Król Z.J., Kuentz P., Kwan Y.W.M., Lagier J.-C., Lau Y.-L., Le Bourgeois F., Leo Y.-S., Lopez R.L., Leung D., Levin M., Levy M., Lévy R., Li Z., Linglart A., Lorenzo-Salazar J.M., Louapre C., Lubetzki C., Luyt C.-E., Lye D.C., Mansouri D., Marjani M., Pereira J.M., Martin A., Pueyo D.M., Martinez-Picado J., Marzana I., Mathian A., Matos L.R.B., Matthews G.V., Mayaux J., Mège J.-L., Melki I., Meritet J.-F., Metin O., Meyts I., Mezidi M., Migeotte I., Millereux M., Mirault T., Mircher C., Mirsaeidi M., Melián A.M., Martinez A.M., Morange P., Mordacq C., Morelle G., Mouly S., Muñoz-Barrera A., Nafati C., Neves J.F., Ng L.F.P., Medina Y.N., Cuadros E.N., Ocejo-Vinyals J.G., Orbak Z., Oualha M., Özçelik T., Hammarström Q.P., Parizot C., Pascreau T., Paz-Artal E., de Diego R.P., Philippe A., Philippot Q., Planas-Serra L., Ploin D., Poissy J., Poncelet G., Pouletty M., Quentric P., Raoult D., Rebillat A.-S., Reisli I., Ricart P., Richard J.-C., Rivet N., Rivière J.G., Blanch G.R., Rodrigo C., Rodriguez-Gallego C., Rodríguez-Palmero A., Romero C.S., Rothenbuhler A., Rozenberg F., Ruiz del Prado M.Y., Riera J.S., Sanchez O., Sánchez-Ramón S., Schluter A., Schmidt M., Schweitzer C.E., Scolari F., Sediva A., Seijo L.M., Sene D., Senoglu S., Seppänen M.R.J., Ilovich A.S., Shahrooei M., Smadja D., Sobh A., Moreno X.S., Solé-Violán J., Soler C., Soler-Palacín P., Stepanovskiy Y., Stoclin A., Taccone F., Tandjaoui-Lambiotte Y., Taupin J.-L., Tavernier S.J., Terrier B., Thumerelle C., Tomasoni G., Toubiana J., Alvarez J.T., Trouillet-Assant S., Troya J., Tucci A., Ursini M.V., Uzunhan Y., Vabres P., Valencia-Ramos J., Van Den Rym A.M., Vandernoot I., Vatansev H., Vélez-Santamaria V., Viel S., Vilain C., Vilaire M.E., Vincent A., Voiriot G., Vuotto F., Yosunkaya A., Young B.E., Yucel F., Zannad F., Zatz M., Belot A., Foti G., Bellani G., Citerio G., Contro E., Pesci A., Valsecchi M.G., Cazzaniga M., Bole-Feysot C., Lyonnet S., Masson C., Nitschke P., Pouliet A., Schmitt Y., Tores F., Zarhrate M., Abel L., Andrejak C., Angoulvant F., Bachelet D., Basmaci R., Behillil S., Beluze M., Benkerrou D., Bhavsar K., Bompart F., Bouadma L., Bouscambert M., Caralp M., Cervantes-Gonzalez M., Chair A., Coelho A., Couffignal C., Couffin-Cadiergues S., D’ortenzio E., Da Silveira C., Debray M.-P., Deplanque D., Descamps D., Desvallées M., Diallo A., Diouf A., Dorival C., Dubos F., Duval X., Eloy P., Enouf V.V.E., Esperou H., Esposito-Farese M., Etienne M., Ettalhaoui N., Gault N., Gaymard A., Ghosn J., Gigante T., Gorenne I., Guedj J., Hoctin A., Hoffmann I., Jaafoura S., Kafif O., Kaguelidou F., Kali S., Khalil A., Khan C., Laouénan C., Laribi S., Le M., Le Hingrat Q., Le Mestre S., Le Nagard H., Lescure F.-X., Lévy Y., Levy-Marchal C., Lina B., Lingas G., Lucet J.C., Malvy D., Mambert M., Mentré F., Mercier N., Meziane A., Mouquet H., Mullaert J., Neant N., Noret M., Pages J., Papadopoulos A., Paul C., Peiffer-Smadja N., Petrov-Sanchez V., Peytavin G., Picone O., Puéchal O., Rosa-Calatrava M., Rossignol B., Rossignol P., Roy C., Schneider M., Semaille C., Mohammed N.S., Tagherset L., Tardivon C., Tellier M.-C., Téoulé F., Terrier O., Timsit J.-F., Treoux T., Tual C., Tubiana S., van der Werf S., Vanel N., Veislinger A., Visseaux B., Wiedemann A., Yazdanpanah Y., Abel L., Alcover A., Aschard H., Astrom K., Bousso P., Bruhns P., Cumano A., Demangel C., Deriano L., Di Santo J., Dromer F., Eberl G., Enninga J., Fellay J., Gomperts-Boneca I., Hasan M., Hercberg S., Lantz O., Mouquet H., Patin E., Pellegrini S., Pol S., Rausell A., Rogge L., Sakuntabhai A., Schwartz O., Schwikowski B., Shorte S., Tangy F., Toubert A., Touvier M., Ungeheuer M.-N., Albert M.L., Duffy D., Quintana-Murci L., Alavoine L., Amat K.K.A., Behillil S., Bielicki J., Bruijning P., Burdet C., Caumes E., Charpentier C., Coignard B., Costa Y., Couffin-Cadiergues S., Damond F., Dechanet A., Delmas C., Descamps D., Duval X., Ecobichon J.-L., Enouf V., Espérou H., Frezouls W., Houhou N., Ilic-Habensus E., Kafif O., Kikoine J., Le Hingrat Q., Lebeaux D., Leclercq A., Lehacaut J., Letrou S., Lina B., Lucet J.-C., Malvy D., Manchon P., Mandic M., Meghadecha M., Motiejunaite J., Nouroudine M., Piquard V., Postolache A., Quintin C., Rexach J., Roufai L., Terzian Z., Thy M., Tubiana S., van der Werf S., Vignali V., Visseaux B., Yazdanpanah Y., van Agtmael M., Algera A.G., van Baarle F., Bax D., Beudel M., Bogaard H.J., Bomers M., Bos L., Botta M., de Brabander J., Bree G., Brouwer M.C., de Bruin S., Bugiani M., Bulle E., Chouchane O., Cloherty A., Elbers P., Fleuren L., Geerlings S., Geerts B., Geijtenbeek T., Girbes A., Goorhuis B., Grobusch M.P., Hafkamp F., Hagens L., Hamann J., Harris V., Hemke R., Hermans S.M., Heunks L., Hollmann M.W., Horn J., Hovius J.W., de Jong M.D., Koning R., van Mourik N., Nellen J., Paulus F., Peters E., van der Poll T., Preckel B., Prins J.M., Raasveld J., Reijnders T., Schinkel M., Schultz M.J., Schuurman A., Sigaloff K., Smit M., Stijnis C.S., Stilma W., Teunissen C., Thoral P., Tsonas A., van der Valk M., Veelo D., Vlaar A.P.J., de Vries H., van Vugt M., Wiersinga W.J., Wouters D., (Koos) Zwinderman A.H., van de Beek D., Abel L., Aiuti A., Al Muhsen S., Al-Mulla F., Anderson M.S., Arias A.A., Feldman H.B., Bogunovic D., Bolze A., Bondarenko A., Bousfiha A.A., Brodin P., Bryceson Y., Bustamante C.D., Butte M., Casari G., Chakravorty S., Christodoulou J., Cirulli E., Condino-Neto A., Cooper M.A., Dalgard C.L., DeRisi J.L., Desai M., Drolet B.A., Espinosa S., Fellay J., Flores C., Franco J.L., Gregersen P.K., Haerynck F., Hagin D., Halwani R., Heath J., Henrickson S.E., Hsieh E., Imai K., Itan Y., Karamitros T., Kisand K., Ku C.-L., Lau Y.-L., Ling Y., Lucas C.L., Maniatis T., Mansouri D., Marodi L., Meyts I., Milner J.D., Mironska K., Mogensen T., Morio T., Ng L.F.P., Notarangelo L.D., Novelli G., Novelli A., O’Farrelly C., Okada S., Ozcelik T., de Diego R.P., Planas A.M., Prando C., Pujol A., Quintana-Murci L., Renia L., Renieri A., Rodríguez-Gallego C., Sancho-Shimizu V., Sankaran V., Barrett K.S., Shahrooei M., Snow A., Soler-Palacín P., Spaan A.N., Tangye S., Turvey S., Uddin F., Uddin M.J., van de Beek D., Vazquez S.E., Vinh D.C., von Bernuth H., Washington N., Zawadzki P., Su H.C., Casanova J.-L. Vol. 370. 2020. Autoantibodies against type I IFNs in patients with life-threatening COVID-19; p. eabd4585. (Science). [DOI] [Google Scholar]
- 65.Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., tenOever B.R. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Turnbull J., Jha R.R., Ortori C.A., Lunt E., Tighe P.J., Irving W.L., Gohir S.A., Kim D.-H., Valdes A.M., Tarr A.W., Barrett D.A., Chapman V. Serum Levels of Proinflammatory Lipid Mediators and Specialized Proresolving Molecules Are Increased in Patients With Severe Acute Respiratory Syndrome Coronavirus 2 and Correlate With Markers of the Adaptive Immune Response. The Journal of Infectious Diseases. 2022;225:2142–2154. doi: 10.1093/infdis/jiab632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koenis D.S., Beegun I., Jouvene C.C., Aguirre G.A., Souza P.R., Gonzalez-Nunez M., Ly L., Pistorius K., Kocher H.M., Ricketts W., Thomas G., Perretti M., Alusi G., Pfeffer P., Dalli J. Disrupted Resolution Mechanisms Favor Altered Phagocyte Responses in COVID-19. Circ Res. 2021;129 doi: 10.1161/CIRCRESAHA.121.319142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Archambault A., Zaid Y., Rakotoarivelo V., Turcotte C., Doré É., Dubuc I., Martin C., Flamand O., Amar Y., Cheikh A., Fares H., El Hassani A., Tijani Y., Côté A., Laviolette M., Boilard É., Flamand L., Flamand N. High levels of eicosanoids and docosanoids in the lungs of intubated COVID‐19 patients. The FASEB Journal. 2021;35 doi: 10.1096/fj.202100540R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wen W., Su W., Tang H., Le W., Zhang X., Zheng Y., Liu X., Xie L., Li J., Ye J., Dong L., Cui X., Miao Y., Wang D., Dong J., Xiao C., Chen W., Wang H. Immune cell profiling of COVID-19 patients in the recovery stageby single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwarz B., Sharma L., Roberts L., Peng X., Bermejo S., Leighton I., Casanovas-Massana A., Minasyan M., Farhadian S., Ko A.I., Yale IMPACT Team. Dela Cruz C.S., Bosio C.M. Cutting Edge: Severe SARS-CoV-2 Infection in Humans Is Defined by a Shift in the Serum Lipidome, Resulting in Dysregulation of Eicosanoid Immune Mediators. The Journal of Immunology. 2021;206:329–334. doi: 10.4049/jimmunol.2001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Palmas F., Clarke J., Colas R.A., Gomez E.A., Keogh A., Boylan M., McEvoy N., McElvaney O.J., McElvaney O., Alalqam R., McElvaney N.G., Curley G.F., Dalli J. Dysregulated plasma lipid mediator profiles in critically ill COVID-19 patients. PLoS ONE. 2021;16 doi: 10.1371/journal.pone.0256226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Biagini D., Franzini M., Oliveri P., Lomonaco T., Ghimenti S., Bonini A., Vivaldi F., Macera L., Balas L., Durand T., Oger C., Galano J.-M., Maggi F., Celi A., Paolicchi A., Di Francesco F. MS-based targeted profiling of oxylipins in COVID-19: A new insight into inflammation regulation. Free Radical Biology and Medicine. 2022;180:236–243. doi: 10.1016/j.freeradbiomed.2022.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pyrillou K., Chairakaki A.-D., Tamvakopoulos C., Andreakos E. Dexamethasone induces ω3-derived immunoresolvents driving resolution of allergic airway inflammation. Journal of Allergy and Clinical Immunology. 2018;142:691–695. doi: 10.1016/j.jaci.2018.04.004. e4. [DOI] [PubMed] [Google Scholar]
- 74.Andreakos E., Papadaki M., Serhan C.N. Dexamethasone, pro‐resolving lipid mediators and resolution of inflammation in COVID‐19. Allergy. 2021;76:626–628. doi: 10.1111/all.14595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Recchiuti A., Patruno S., Mattoscio D., Isopi E., Pomilio A., Lamolinara A., Iezzi M., Pecce R., Romano M. Resolvin D1 and D2 reduce SARS-CoV-2-induced inflammatory responses in cystic fibrosis macrophages. FASEB J. 2021;35 doi: 10.1096/fj.202001952R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dockrell D.H., Russell C.D., McHugh B., Fraser R. Does autonomous macrophage-driven inflammation promote alveolar damage in COVID-19. Eur Respir J. 2022;60 doi: 10.1183/13993003.01521-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Panigrahy D., Gilligan M.M., Huang S., Gartung A., Cortés-Puch I., Sime P.J., Phipps R.P., Serhan C.N., Hammock B.D. Inflammation resolution: a dual-pronged approach to averting cytokine storms in COVID-19? Cancer Metastasis Rev. 2020;39:337–340. doi: 10.1007/s10555-020-09889-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cherpokova D., Jouvene C.C., Libreros S., DeRoo E.P., Chu L., de la Rosa X., Norris P.C., Wagner D.D., Serhan C.N. Resolvin D4 attenuates the severity of pathological thrombosis in mice. Blood. 2019;134:1458–1468. doi: 10.1182/blood.2018886317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chiang N., Sakuma M., Rodriguez A.R., Spur B.W., Irimia D., Serhan C.N. Resolvin T-series reduce neutrophil extracellular traps. Blood. 2022;139:1222–1233. doi: 10.1182/blood.2021013422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Recchiuti A., Federti E., Matte A., Mazzi F., Ceolan J., Porreca A., Di Nicola M., Menotti S., Alivernini S., De Franceschi L. Impaired pro-resolving mechanisms promote abnormal NETosis, fueling autoimmunity in sickle cell disease. Am J Hematol. 2023;98:E45–E48. doi: 10.1002/ajh.26797. [DOI] [PubMed] [Google Scholar]
- 81.Liesegang T.J. Herpes Simplex Virus Epidemiology and Ocular Importance. Cornea. 2001;20:1–13. doi: 10.1097/00003226-200101000-00001. [DOI] [PubMed] [Google Scholar]
- 82.Rajasagi N.K., Reddy P.B.J., Suryawanshi A., Mulik S., Gjorstrup P., Rouse B.T. Controlling Herpes Simplex Virus-Induced Ocular Inflammatory Lesions with the Lipid-Derived Mediator Resolvin E1. The Journal of Immunology. 2011;186:1735–1746. doi: 10.4049/jimmunol.1003456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rajasagi N.K., Reddy P.B.J., Mulik S., Gjorstrup P., Rouse B.T. Neuroprotectin D1 Reduces the Severity of Herpes Simplex Virus–Induced Corneal Immunopathology. Invest. Ophthalmol. Vis. Sci. 2013;54:6269. doi: 10.1167/iovs.13-12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rajasagi N.K., Bhela S., Varanasi S.K., Rouse B.T. Frontline Science: Aspirin-triggered resolvin D1 controls herpes simplex virus-induced corneal immunopathology. Journal of Leukocyte Biology. 2017;102:1159–1171. doi: 10.1189/jlb.3HI1216-511RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goncalves P.H., Ziegelbauer J., Uldrick T.S., Yarchoan R. Kaposi sarcoma herpesvirus-associated cancers and related diseases. Current Opinion in HIV and AIDS. 2017;12:47–56. doi: 10.1097/COH.0000000000000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marginean A., Sharma-Walia N. Lipoxins exert antiangiogenic and anti-inflammatory effects on Kaposi’s sarcoma cells. Translational Research. 2015;166:111–133. doi: 10.1016/j.trsl.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 87.Asha K., Balfe N., Sharma-Walia N. Concurrent Control of the Kaposi’s Sarcoma-Associated Herpesvirus Life Cycle through Chromatin Modulation and Host Hedgehog Signaling: a New Prospect for the Therapeutic Potential of Lipoxin A4. J Virol. 2020;94 doi: 10.1128/JVI.02177-19. e02177-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chiang N., la Rosa X., Libreros S., Serhan C.N. Novel Resolvin D2 Receptor Axis in Infectious Inflammation. Journal of Immunology. 2017;198:842–851. doi: 10.4049/jimmunol.1601650. (Baltimore, Md.: 1950) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Codagnone M., Cianci E., Lamolinara A., Mari V.C., Nespoli A., Isopi E., Mattoscio D., Arita M., Bragonzi A., Iezzi M., Romano M., Recchiuti A. Resolvin D1 enhances the resolution of lung inflammation caused by long-term Pseudomonas aeruginosa infection. Mucosal Immunol. 2018;11:35–49. doi: 10.1038/mi.2017.36. [DOI] [PubMed] [Google Scholar]
- 90.WHO to identify pathogens that could cause future outbreaks and pandemics, (n.d.). 〈https://www.who.int/news/item/21-11-2022-who-to-identify-pathogens-that-could-cause-future-outbreaks-and-pandemics〉 (accessed March 15, 2023).
- 91.WHO, WHO Coronavirus Disease (COVID-19) Dashboard, (n.d.). 〈https://covid19.who.int/〉.
- 92.Baltimore D. Expression of animal virus genomes. Bacteriol Rev. 1971;35:235–241. doi: 10.1128/br.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]