Abstract
Background.
VTP-50469 is a potent inhibitor of the menin-MLL1 interaction and is implicated in signaling downstream of EWSR1-FLI1.
Procedure.
VTP-50469 was evaluated against 7 Ewing sarcoma (EwS) xenograft models and in vitro against EwS cell lines.
Results.
VTP-50469 showed limited antitumor activity, statistically significantly slowing tumor progression in 4 tumor models but with no evidence of tumor regression. In vitro, the IC50 concentration was 10 nM for the MLL-rearranged leukemia cell line MV4;11, but > 3μM for EwS cell lines.
Conclusions.
In contrast to its high level of activity against MLL1-rearranged leukemia xenografts, VTP-50469 shows little activity against EwS models.
Keywords: Ewing sarcoma, Pediatric Oncology, Xenograft
Introduction
MLL1, the mammalian homolog of drosophila Trithorax (trx), positively regulates HOX gene expression during development.1 The activity of trx group proteins is balanced by the repressive activity of Polycomb group (PcG) genes.2 MLL1 (KMT2A), a lysine methyltransferase, binds promoters of HOX genes resulting in H3 Lys 4 methylation and H3 and H4 acetylation.3 Dysregulation of HOX genes occurs in several cancers, suggesting a critical role for developmental programs in transformation.4–6 While the role of MLL1 translocations is well established for several leukemias1, less is known regarding the role of MLL1 in solid tumors. In leukemias, the oncogenic activity of MLL1 fusion proteins is dependent on association with menin, a scaffolding protein that binds MLL1 and MLL4 (KMT2B) in the context of TrxG COMPASS complexes.7 Consequently, small molecules that inhibit the Menin-MLL1 interaction have potential therapeutic value for treatment of MLL1 rearranged leukemia.8 Posterior HOXD genes are overexpressed in Ewing sarcoma (EwS).9 Promoter regions for these genes are characterized by MLL-mediated H3K4me3 marks and are devoid of recessive H3K27me3 marks.9 Recently, evidence has been presented that the tumorigenicity of EwS cells is dependent on the Menin-MLL1 interaction.10 Here we have evaluated VTP-50469, a potent inhibitor of Menin-MLL1 interactions that shows profound activity against Pediatric Preclinical Testing Consortium (PPTC) MLL-rearranged leukemia models.11
Materials and Methods
In vivo testing:
C.B.17SC scid−/− (C.B-Igh-1b/IcrTac-Prkdcscid) female mice were maintained under barrier conditions and experiments were conducted using protocols and conditions approved by the institutional animal care and use committee at UTHSCSA as previously described12 Details of the statistical analytic methods are provided in Appendix 1.
Drugs and Formulation:
VTP-50469 was provided to the Pediatric Preclinical Testing Consortium (PPTC) by Syndax Pharmaceuticals Inc., through the Cancer Therapy Evaluation Program (NCI). VTP-50469 was suspended in the required amount of vehicle (0.5% Natrosol + 1% Polysorbate-80). The resulting formulation was sonicated in a water bath set to 37°C until completely dissolved or visually uniform suspensions were achieved. Formulated drug was stored at 4°C for up to one month. VTP-50469 was administered by oral gavage (PO) at 120 mg/kg, twice daily (BID) for a planned 28 consecutive days.
In vitro testing:
The potency of VTP-50469 was evaluated against four EwS cell lines (ES-1, ES-4, ES-6, EW-8). Each line has the EWSR1-FLI1 type 1 translocation. VTP-50469 was also tested against the MV4;11 cell line that has the MLL-AF4 (KMT2A-AFF1) fusion. Cells were exposed to VTP-50469 for 96h and viability assessed by Alamar Blue staining, as described previously.13
Results
For CHLA-258 and EW-8 cell lines the VTP-50469 dose was reduced to 100 mg/kg BID because of excessive toxicity observed in models treated at 120 mg/kg BID. For the five Ewing sarcoma models treated at 120 mg/kg BID, excessive mortality (18 of 50) was observed in the absence of antecedent weight loss. For the remaining two models (CHLA-258 and EW-8) treated at 100 mg/kg BID, 1 of 20 mice in the treatment group died (5%). VTP-50469 caused a statistically significant growth delay in 4 EwS models, Table 1. Among models with significant slowing of tumor growth, the ratio of median time to event for the treated versus control groups (EFS T/C) ranged from 1.24 to 1.74. There were no tumor regressions, and the mean minimum relative tumor volumes (RTV) for treated groups ranged from 1.2 to 3.5 (Table 1). The overall objective response classification for all models was Progressive Disease 1 (PD1). Kaplan-Meier EFS for each tumor line is shown in Figure 1A. The expression (mRNA) and mutation status of KMT2A and Menin (MEN1) in the PPTC leukemia and solid tumor models is shown in Supplemental Figures 1 and 2.
Table 1.
Summary of Antitumor Activity of VTP-50469 against EwS Xenograft Models
| Tumor Line | Treatment | KM Median (days) | EFS T-C (days) | EFS T/C | p-value Gehan-Wilcoxan | Median Response | Minimum Relative Tumor Volume |
|---|---|---|---|---|---|---|---|
| CHLA258 | Control | 16.2 | 1.971±0.771 | ||||
| VTP-50469 | 20.1 | 3.9 | 1.24 | 0.036 | PD1 | 1.553±0.430 | |
| ES-1 | Control | 10.4 | 3.047±0.816 | ||||
| VTP-50469 | 14.0 | 3.6 | 1.35 | 0.046 | PD1 | 2.612±0.991 | |
| ES-4 | Control | 10.7 | 2.494±0.616 | ||||
| VTP-50469 | 12.2 | 1.5 | 1.14 | 0.186 | PD1 | 2.092±0.460 | |
| ES-6 | Control | 12.4 | 2.066±0.280 | ||||
| VTP-50469 | 11.5 | −0.9 | 0.93 | 0.084 | PD1 | 2.333±0.953 | |
| EW-5 | Control | 13.1 | 1.709±0.370 | ||||
| VTP-50469 | 20.8 | 7.8 | 1.6 | <0.001 | PD1 | 1.241±0.394 | |
| EW-8 | Control | 6.1 | 5.014±0.931 | ||||
| VTP-50469 | 10.6 | 4.5 | 1.74 | 0.015 | PD1 | 3.497±1.177 | |
| NCH-EWS-1 | Control | 10.3 | 2.741±1.078 | ||||
| VTP-50469 | 10.4 | 0.1 | 1.01 | 0.665 | PD1 | 3.112±0.823 |
Figure 1.
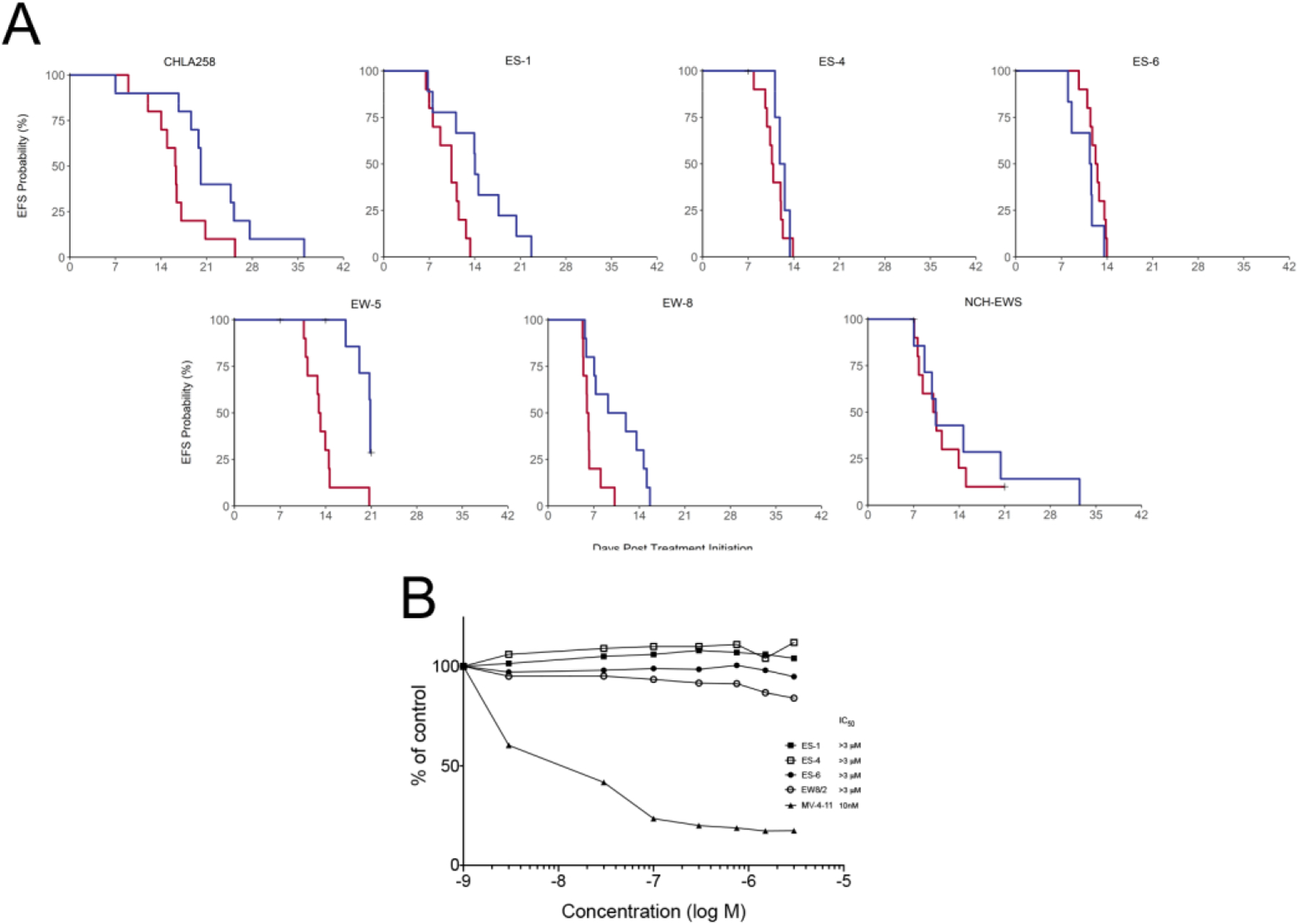
A, Kaplan-Meier plots showing probability of being Event-Free with time. Red line: Controls; Blue line VTP-50469 treatment; B, Sensitivity of EwS cell lines compared to MV4;11 a cell line that has the t(4;11)(q21;q23) chromosomal translocation leading to fusion of the mixed lineage leukemia (MLL) gene located on 11q23 and AF4 located on 4q21 (MLL/AF4; or KMT2A/AFF1). Cells were exposed to VTP-50469 for 96h and viability assessed by Alamar Blue staining.
Because VTP-50469 demonstrated limited activity against EwS xenograft models at a dose and schedule that was highly active against MLL-rearranged leukemias, we determined the sensitivity of EwS cell lines in comparison with the MLL-rearranged leukemia cell line MV4;11. Cells were exposed to VTP-50469 at concentrations from 3 nM to 3 μM for 96h. The MV4;11 cell line was very sensitive to VTP-50469 (IC50 =10 nM), whereas for all EwS cell lines the IC50 was >3 μM, (Figure1B).
Discussion
MLL translocations occur in 5–10% of B-cell ALL with a similar frequency observed for AML. Rearrangements are more frequent in infant leukemias (~70% of infant ALL). Menin inhibitors are highly active in preclinical models of MLL-rearranged ALL,8,11,14 and clinical trials for this class of agents have been initiated (SNDX-5613 in NCT04065399 and KO-539 in NCT04067336).
Recent work has suggested that Menin-MLL1 interactions may be important for tumorigenesis induced by the EWSR1-FLI1 fusion oncogene in EwS and that this effect may be in part mediated through effects on serine biosynthesis.10,15 MI-503, a small molecule that disrupts the Menin-MLL1 interaction,8 reduced proliferation and suppressed anchorage independent growth of EwS cell lines.10 In vitro, MI-503 induced loss of both MLL1 and menin, and it also reduced tumorigenicity of EwS cells that were pre-treated with MI-503 prior to inoculation into athymic nude mice.10 mRNA expression in EwS models for KMT2A and Menin (MEN1) show that expression of KMT2A is lower than in MLL models that have fusions. The only EwS model to have a mutation (missense; E2419K) is SK-NEP-1, a model not used in the current study. Expression of MEN1 was similar in leukemias and EwS models, and no mutations in the EwS models were detected (https://pedcbioportal.org/study?id=pptc#summary).16
VTP-50469, like MI-503, potently disrupts the Menin-MLL1 interaction and is highly active over a broad range of doses in infant leukemias that have MLL1 translocations. Against EwS xenografts, VTP-50469 showed little antitumor activity, statistically significantly slowing tumor growth in several models. However, extension of EFS in these models was modest, and tumor regression was not observed. Consistent with the in vivo results, EwS cell lines were >300-fold less sensitive to VTP-50469 than the MLL leukemia cell line MV4;11. Pharmacodynamic studies were not undertaken to determine target inhibition, as VTP-50469 administered at these and lower dose levels on the same schedule has robust activity in several infant leukemia models with MLL1 translocations. Our results are consistent with those of a recent report that found that while MI-503 had in vitro activity against a range of leukemia and solid tumor cell lines, the more selective menin inhibitor BAY-155 was primarily active in AML and ALL models.17
Our results and an examination of existing literature for menin inhibitors suggest that EwS cells are less dependent on the Menin-MLL1 interaction for survival in comparison to MLL-rearranged leukemias. First, the IC50 values for EwS cell lines are approximately one log greater than those for MLL-rearranged leukemia cell lines for the menin inhibitor MI-503.8,10 Similarly, our in vitro results show much greater sensitivity for a MLL-rearranged leukemia cell line to VTP-50469 in comparison to that observed for EwS cell lines. Second, the effect of menin inhibitors on the expression of the MLL1 gene fusion target genes (e.g., Hox9 and Meis1) is much greater than the effect of menin inhibition on expression of HoxD genes in EwS cell lines.8,10 Finally, VTP-50469 shows remarkably high in vivo activity against MLL-rearranged leukemia xenograft lines, but shows minimal levels of in vivo activity against EwS models.
Supplementary Material
Acknowledgements.
This work was supported by NCI grants: UO1 CA199297, U01 CA199222, and P30 CA054174.
Abbreviation
- EwS
Ewing sarcoma
- PPTC
Pediatric Preclinical Testing Consortium
Footnotes
Conflict of Interest: Dr. McGeehan is an employee of Syndax Pharmaceuticals. The other authors have no conflicts.
References
- 1.Hess JL. Mechanisms of transformation by MLL. Crit Rev Eukaryot Gene Expr. 2004;14(4):235–254. [DOI] [PubMed] [Google Scholar]
- 2.Kassis JA, Kennison JA, Tamkun JW. Polycomb and Trithorax Group Genes in Drosophila. Genetics. 2017;206(4):1699–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakamura T, Mori T, Tada S, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10(5):1119–1128. [DOI] [PubMed] [Google Scholar]
- 4.Gallo M, Ho J, Coutinho FJ, et al. A tumorigenic MLL-homeobox network in human glioblastoma stem cells. Cancer Res. 2013;73(1):417–427. [DOI] [PubMed] [Google Scholar]
- 5.Muntean AG, Hess JL. The pathogenesis of mixed-lineage leukemia. Annu Rev Pathol. 2012;7:283–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu B, Li SH, Zheng R, et al. Menin promotes hepatocellular carcinogenesis and epigenetically up-regulates Yap1 transcription. Proc Natl Acad Sci U S A. 2013;110(43):17480–17485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123(2):207–218. [DOI] [PubMed] [Google Scholar]
- 8.Borkin D, He S, Miao H, et al. Pharmacologic Inhibition of the Menin-MLL Interaction Blocks Progression of MLL Leukemia In Vivo. Cancer cell. 2015;27(4):589–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Svoboda LK, Harris A, Bailey NJ, et al. Overexpression of HOX genes is prevalent in Ewing sarcoma and is associated with altered epigenetic regulation of developmental transcription programs. Epigenetics. 2014;9(12):1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Svoboda LK, Bailey N, Van Noord RA, et al. Tumorigenicity of Ewing sarcoma is critically dependent on the trithorax proteins MLL1 and menin. Oncotarget. 2017;8(1):458–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krivtsov AV, Evans K, Gadrey JY, et al. A Menin-MLL Inhibitor Induces Specific Chromatin Changes and Eradicates Disease in Models of MLL-Rearranged Leukemia. Cancer cell. 2019;36(6):660–673 e611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houghton PJ, Morton CL, Tucker C, et al. The pediatric preclinical testing program: Description of models and early testing results. Pediatr Blood Cancer. 2006. [DOI] [PubMed] [Google Scholar]
- 13.Bid HK, Phelps DA, Xaio L, et al. The Bromodomain BET Inhibitor JQ1 Suppresses Tumor Angiogenesis in Models of Childhood Sarcoma. Molecular cancer therapeutics. 2016;15(5):1018–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lock RB, Evans K, Prichard T, et al. Pediatric Preclinical Testing Consortium evaluation of the menin inhibitor, VTP-50469, against xenograft models of MLL-rearanged infant acute lymphoblastic leukemia. Proc Am Assoc Cancer Research. 2018;Abstract 3187. [Google Scholar]
- 15.Svoboda LK, Teh SSK, Sud S, et al. Menin regulates the serine biosynthetic pathway in Ewing sarcoma. The Journal of pathology. 2018;245(3):324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rokita JL, Rathi KS, Cardenas MF, et al. Genomic Profiling of Childhood Tumor Patient-Derived Xenograft Models to Enable Rational Clinical Trial Design. Cell reports. 2019;29(6):1675–1689 e1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brzezinka K, Nevedomskaya E, Lesche R, et al. Characterization of the Menin-MLL Interaction as Therapeutic Cancer Target. Cancers. 2020;12(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


