Summary
In this review article, we discuss the model for end-stage liver disease (MELD) score and its dual purpose in general and transplant hepatology. As the landscape of liver disease and transplantation has evolved considerably since the advent of the MELD score, we summarise emerging concepts, methodologies, and technologies that may improve mortality prognostication in the future. Finally, we explore how these novel concepts and technologies may be incorporated into clinical practice.
Keywords: MELD, Prognostication, Allocation, Frailty, Sarcopenia, EHR, OMOP, Clinical, Decision Support
Introduction
The deficit of available donor organs in relation to the number of patients in need of liver transplantation necessitates systems to allocate organs in an efficient yet equitable manner. The current principles of liver allocation in the United States,1 the Eurotransplant region,2,3 and elsewhere include determination of priority through objective and measurable medical criteria, ordered from most to least medically urgent.1,4 Urgency has been represented primarily by the model for end-stage liver disease (MELD) score, rather than the Child-Pugh score, to avoid subjective variables such as ascites and encephalopathy and to expand the scale (to reduce the number of candidates with identical scores).5,6
The MELD score, which is comprised of serum bilirubin, creatinine, and the international normalised ratio, has since served a dual purpose in general and transplant hepatology. It effectively predicts short-term (e.g., over 90 days) mortality among patients with chronic liver disease, thereby providing clinicians with a critical tool to prognosticate liver-related and waitlist mortality. It has been used to determine medical urgency (and hence priority) for liver transplant candidates since 2002 in the United States and 2006 in the Eurotransplant region, making it an essential tool for transparent and equitable organ allocation.7,8
The landscape of chronic liver disease and liver transplantation has evolved considerably in the last two decades. Both waitlist mortality prediction and transplant organ allocation require ongoing re-evaluation to ensure accurate prognostication and appropriate distribution of donor organs. In 2016, the MELD score was updated to include serum sodium, an objective biomarker that is often a surrogate indicator for ascites.9 A new update to and recalibration of the MELD score, MELD 3.0, was recently published with the inclusion of sex and serum albumin.10 At the same time, a substantial proportion of liver transplants are allocated by MELD “exception”, representing indications where the mortality risk and need for transplant are not well-represented by the MELD score.11
In addition, emerging technologies, new methodologies, and evolving conceptual frameworks for liver disease may improve clinicians’ ability to prognosticate and manage patients with end-stage liver disease. In this article, we present emerging tools and techniques “beyond MELD” for improvement in liver allocation, prognostication, and outcomes in patients with end-stage liver disease.
Beyond MELD – for liver allocation
Improving the MELD score
Over the past two decades of MELD score-based liver allocation, the demographics of chronic liver disease and indications for liver transplantation have changed dramatically worldwide. The widespread availability of effective direct-acting antiviral therapy for hepatitis C and the increasing prevalence of alcohol-associated liver disease and non-alcoholic steatohepatitis has fundamentally changed the population of patients awaiting liver transplantation.11,12 Throughout these changes, however, the MELD score has continued to provide robust predictions of short-term waitlist mortality that outperform most other clinical scoring systems, with c-statistics that exceed 0.80 in various cohorts.9,10,13 Still, it has been perceived that the predictive power of the MELD score may have diminished in recent years.14,15 The MELD score may not represent mortality risk as accurately for patients with some of the most severe clinical complications of cirrhosis, such as acute-on-chronic liver failure (ACLF), refractory ascites/hepatic hydrothorax, recurrent variceal bleeding, and hepatocellular carcinoma.14,16
In addition, the MELD score has historically underpredicted mortality risks for women.17,18 This sex disparity is multifactorial but in part stems from the reliance of the MELD score on the measurement of serum creatinine, which can vary by sex for the same degree of renal dysfunction.17,19,20 Women on average have lower muscle mass compared to men, leading to systematic underestimation of renal function by serum creatinine.21 Alternatives to the creatinine component of the MELD score have been proposed, including MDRD (modification of diet in renal disease),18,22 GRAIL (glomerular filtration rate assessment in liver disease),23,24 and cystatin C,25,26 but are still less-than-ideal owing to the lack of improvement in model performance, inclusion of age and/or race-based equations, or clinical availability (cystatin C) (Table 1). The most recent iteration of the MELD score, MELD 3.0, incorporates sex as an independent variable to correct for the sex disparity due to creatinine, while also updating coefficients, adding serum albumin and adjusting the creatinine to a lower cap of 3.0 mg/dl.10 Other factors contributing to the sex disparity, including anthropometric differences and thus fewer opportunities for size-appropriate organs or the allocation of exception points, may require other types of adjustments to fully address the differences in outcomes and access to transplant between sexes.17,19,27,28
Table 1.
Limitations of existing and proposed waitlist mortality risk scores to be used in liver allocation.
| Score | Components | Strengths | Limitations |
|---|---|---|---|
|
| |||
| Child-Pugh score139 | Bilirubin, INR, albumin, ascites, encephalopathy | • Established minimal listing criteria for liver transplant candidates | • Inclusion of potentially subjective variables i.e. ascites and encephalopathy |
| MELD7 | Bilirubin, INR, creatinine | • Adequate discriminative
ability • Use of objective and widely available tests • Improved waitlist mortality, equity in liver allocation |
• Underestimation of renal dysfunction
in women compared to men • Does not accurately represent transplant urgency for certain disease etiologies such as hepatocellular carcinoma |
| MELD-Na9 | Bilirubin, INR, creatinine, sodium | • Addition of sodium as a surrogate for ascites | • May not accurately represent mortality risk for complications such as hepatic encephalopathy or acute-on-chronic liver failure |
| MELD-Plus140 | Bilirubin, INR, creatinine, sodium, albumin, total cholesterol, WBC, age, length of stay | • Improved mortality prediction compared to MELD-Na after hospital admission | • Only calculated after a cirrhosis-related hospital admission |
| MELD-lactate141 | Bilirubin, INR, creatinine, sodium, lactate | • Improved in-hospital mortality prediction compared to MELD or MELDNa in patients hospitalised for infection or MELD ≤15 | • Only calculated during a hospital admission |
| MELD-Na-MDRD18, 22 | Bilirubin, INR, creatinine, age, sex, race | • More accurate estimation of renal function accounting for potential differences in muscle mass | • Did not improve mortality prediction |
| MELD-GRAIL-Na23, 24 | Bilirubin, INR, creatinine, blood urea nitrogen, age, sex, race, albumin, sodium | • Estimation of renal function
developed for liver disease with better accuracy and precision compared
to standard eGFR calculations • Improved mortality prediction in MELD >32 |
• Inclusion of age and race could lead to bias in allocation |
| MELD-Cystatin C25, 26 | Bilirubin, INR, creatinine, cystatin C | • Biomarker of renal function less susceptible to differences in muscle mass | • Lack of clinical
availability • Mitigated sex differences but no improvement in predictive power |
| MELD-Na-Shift28 | Bilirubin, INR, creatinine, sodium | • Adds 0–1 MELD points for
women • Modelled to eliminate sex disparity in transplant rates |
• Addition of points for women at arbitrary levels |
| MELD 3.010 | Bilirubin, INR, creatinine, sodium, sex, albumin | • Addition of 1.33 points for
women • Updated coefficients and interactions; adjusted upper bound for serum creatinine • Improved mortality prediction compared to MELD-Na |
• Calculation somewhat more complex |
eGFR, estimated glomerular filtration rate; GRAIL, glomerular filtration rate assessment in liver disease; INR, international normalised ratio; MDRD, modification of diet in renal disease; MELD, model for end-stage liver disease; WBC, white blood cell.
While the MELD score remains a reliable indicator of mortality risk in liver disease, it can certainly benefit from further refinement. In so doing, the selection of variables should be carefully considered. Older age, medical co-morbidities, or certain aetiologies of liver disease may be associated with increased mortality risk, yet there is no consensus that these variables should influence waitlist priority or access to liver transplantation. Race may also be predictive, but this variable in clinical prediction scores can be problematic, as racial differences among populations in large datasets are often not genetic or biological, but rather reflect socioeconomics and healthcare policy.29 Race adjustment in these situations, while well-intentioned, can exacerbate inequity. Lastly, variables should be objective, verifiable, and readily available. Although addition of such variables may generate better prediction of waitlist mortality, they are not necessarily appropriate for use in organ allocation. Systems for organ distribution also need to be interpretable and transparent with regards to how changes of a specific variable would impact allocation.
Emerging concepts to improve allocation
The rationale behind organ allocation systems is to maximise the use of available organs and reduce deaths on the waiting list. Organ allocation may be driven by 3 important principles:
- Urgency – Allocation to the patient estimated to have the shortest survival without a transplant.
- Utility – Allocation to the patient estimated to have the longest post-transplant survival.
- Transplant benefit – Allocation based on the difference between the mean survival estimates with and without a transplant.
In the past two decades, liver allocation in the United States and parts of Europe has been based almost entirely on the principle of urgency – in other words, by risk of death as determined by the MELD score.7,8 Although the Final Rule instituted by the Department of Health and Human Services in the United States also provides for consideration of utility and survival benefit – to make the best use of donated organs, to avoid wasting organs, and to avoid futile transplants.1 However, acceptable standards and thresholds for post-transplant longevity and futility have been challenging to define,30 and current models for post-transplant survival do not perform well enough alone to be used in allocation.31–33 Moreover, the net benefit of liver transplant, defined by the difference between survival with and without transplant, is largely driven by waitlist mortality, where the candidates with the highest MELD score gained the most life-years from transplant.34,35
In many MELD-based liver allocation systems, exception points grant waitlist priority and thus access to transplant for patients whose mortality risk and need for transplant is not well-represented by the MELD score, the most common exception being for hepatocellular carcinoma.36 Calibration of these exception points to approximate the mortality risk and urgency for transplant and to equitably allocate organs has turned out to be a moving target as patient characteristics and management of various conditions have shifted over time. Ensuring equitable allocation for this population may require additional solutions, including integration of transplant benefit and flexibility for donor-recipient matching in certain cases.37 For example, the United States allocation system does consider utility in the specific contexts of hepatocellular carcinoma or cholangiocarcinoma, by which patients exceeding certain criteria do not receive standard priority for liver transplant, owing to the excess risk of post-transplant recurrence and thus lower transplant benefit.37 Such rules may set a precedent for utility to be considered in future liver allocation policies.
Key point.
While the MELD score remains a reliable indicator of mortality risk in liver disease, further refinements, exception points, and continuous distribution are required as we move toward truly fair and equitable organ allocation.
Disparities in waitlist outcomes also arise from unequal access to transplant. Patients with the same medical urgency should have an equal opportunity of receiving a liver transplant, yet this is currently not the case. Upcoming changes in allocation in the United States include not only optimisation of the MELD score but also continuous distribution, a composite point scoring system that will enable the consideration of additional variables, including height, body surface area, blood type, geography, paediatric status, and travel efficiency, and indication for transplant (i.e. exceptions), to move closer to fair and equitable organ allocation. Under the proposed framework defined by the Organ Procurement and Transplantation Network (OPTN) in the United States, continuous distribution will attempt to balance 5 goals: medical urgency, post-transplant survival, candidate biology, patient access, and placement efficiency, although the specific attributes ultimately included and their respective weighting will depend on feedback from the transplant community and modelling and analysis. The system is envisioned to provide a more dynamic reflection of patient-related factors and thereby improve access.38–40 Consensus processes, such as that described by the Italian liver transplant community, may help to develop allocation policy that fairly balances the various priorities of liver transplantation, including urgency, utility, and transplant benefit.37
Key point.
Factors not traditionally reflected by the MELD score, such as malnutrition, frailty, and sarcopenia, have improved prognostication in patients with cirrhosis.
Beyond MELD – For prognostication
Muscle dysfunction as a clinical marker for assessing disease severity in patients with cirrhosis
Emerging factors that have not classically been reflected by the MELD score, such as malnutrition, frailty, and sarcopenia, have improved our ability to dynamically characterise the morbidity and mortality associated with cirrhosis.41 Malnutrition represents a spectrum of nutritional deficiencies that cause adverse effects on physiologic function or clinical outcomes.42 It contributes to and is interdependent with measurable clinical manifestations of muscle dysfunction: frailty and sarcopenia.41
Frailty is classically defined as the clinical state of decreased physiologic reserve and increased vulnerability to health stressors.43 In patients with cirrhosis, this manifests as the phenotypic representation of impaired muscle contractile function.44 Frailty is estimated to be present in 17% to 43% of patients with cirrhosis based on different measurement standards;45–48 it worsens in patients with cirrhosis over time and has been strongly associated with waitlist and post-transplant mortality. For instance, frailty was associated with a nearly 2-fold higher adjusted risk of death in 1,044 ambulatory patients with cirrhosis awaiting liver transplantation in a multicentre study in the United States.45 Moreover, frailty is linked with increased healthcare utilisation both in the ambulatory and hospitalised settings. Given frailty’s strong association with post-transplant outcomes, the concept of “prehabilitation” or intervening to modify physical reserve prior to surgery has gained traction in both transplant and non-transplant surgical fields.49,50 Arrest or reversal of the progression of frailty is thought to be a clinically relevant achievement that should incentivise liver transplantation.49 As such, the American Association for the Study of Liver Diseases now recommends all patients with cirrhosis should be assessed for frailty with a standardised tool at baseline and longitudinally;41 and the American Society of Transplantation recommends the same for patients awaiting liver transplantation.49
arcopenia is defined as the progressive and generalised loss of skeletal muscles associated with increased likelihood of adverse outcomes.51 Sarcopenia is also common in adults with cirrhosis, affecting 30% to 70% of patients with strong sex-based differences in prevalence.52,53 The gold standard for sarcopenia assessment is computed tomography imaging; since abdominal imaging is commonly performed for clinical reasons, muscle mass measurements are often obtainable.54,55 Sarcopenia has a robust association with waitlist mortality before and after transplant, as well as with hepatic decompensation.52,56,57 Sarcopenia is progressive in patients with cirrhosis, and serial/longitudinal measures of muscle loss have been associated with clinical outcomes including waitlist mortality.58
Electronic health data and multicentre electronic consortiums
Recent advances in computing power in conjunction with the availability of large databases and analytical methodologies have dramatically increased the tools available for clinical research in hepatology. Historically, the predominant forms of large clinical research databases in the United States and Europe have been based on either patient registries, such as the Scientific Registry of Transplant Recipients or Eurotransplant databases,11,59,60 multicentre curated cohorts, or administrative claims databases.61–63 Beyond these large databases, there has been a growing movement towards aggregation of longitudinal electronic health records (EHRs) across multiple institutions and health systems.64–66
In the United States and the European Union, EHRs now have greater than 96% penetration in acute care hospital and physicians’ offices.67,68 EHR data, gathered as the transactional record of health care delivery and operations, are now viewed as a key resource to generate unique insights.69 Novel applications of data science and clinical informatics on EHR data have the potential to accelerate clinical research and improve patient care. One of the key advantages of EHR data is its dynamic longitudinal nature with data acquisition occurring at every interaction that the patient has with the healthcare system. Correctly harnessed, integration of longitudinal data could produce more comprehensive reflections of patients’ clinical trajectory.
For instance, incorporation of time-variant variables, such as laboratory values and vital signs, captured in EHRs have enabled continuous prediction of the development of acute kidney injury during inpatient admissions.70,71 Moreover, the use of longitudinal and sequential data elements gathered from EHR flowsheets, medication administrations, physician notes, and radiology reports have enabled the construction of deep-learning models to more accurately predict in-hospital mortality, 30-day readmissions, and prolonged length of stay.72 In clinical hepatology, the integration of longitudinal EHR elements, such as structured flowsheet entries, medication administration, procedure orders, vital signs, and laboratory values, has enabled dynamic calculations of the North American Consortium for the Study of End-Stage Liver Disease-ACLF and Chronic Liver Failure Consortium-ACLF prognostication scores in hospitalised patients with ACLF.73
Despite the potential for longitudinal EHR data to improve outcome prediction, the lack of standards, lack of semantic interoperability, and disparate EHR systems/implementations have historically limited large multi-institution collaborations.74 Early regional-based EHR consortiums, such as HealthLNK based in the Chicago area, have demonstrated the value of multicentre EHR data in predicting factors associated with mortality in patients with cirrhosis.75
Key point.
Longitudinal electronic health records hold great promise for dynamic outcome prediction, particularly with the application of common data models and the centralisation of data.
The development and wider availability of common data models, such as the observational medical outcomes partnership (OMOP) model and the fast healthcare interoperability resources (FHIR) model, may now facilitate larger EHR-based collaboratives.64,76 Examples of such large EHR-based research collaboratives include the Observational Health Data Sciences and Informatics group based in the United States and the European Health Data and Evidence Network based in the European Union.64,77 While the trend towards common data models and centralised EHR data for observational research had already been underway, the COVID-19 pandemic drastically accelerated this movement with the creation of the National COVID Cohort Collaborative (N3C).65,78
N3C is a novel, centralised, and harmonised repository of EHR data from more than 64 sites from across the United States built on the OMOP platform, formed in response to the need for rapid accrual and analyses of clinical data during the COVID-19 pandemic.65,78 Its effective use has allowed for the rapid generation of insights into the mortality risk of SARS-CoV-2 infection among patients with cirrhosis.79 The work highlights the prospect of transplant hepatology-specific multicentre EHR collaboratives with deep clinical content expertise, which may accelerate the development of comprehensive models for mortality prediction in patients with end-stage liver disease.
Novel modelling methodologies for mortality risk prediction
While high-dimension multicentre EHR data has tremendous potential, their “big data” nature may require the use of novel analytical techniques.80,81 “Big data” is an amorphous term that is classically defined in terms of the 5 “Vs” (volume, velocity, variety, veracity, and value) to describe large datasets that may be more effectively analysed using 82,83 artificial intelligence-based methods, such as machine learning (ML), which permit data-driven rather than hypothesis-driven discovery.84,85 The most prevalent ML algorithms are divided into supervised (classification) and unsupervised (sorting) methods (Table 2).84,86,87
Table 2.
Common machine learning algorithms used in clinical research.
| Algorithm | Summary | Application example in hepatology |
|---|---|---|
|
| ||
| Supervised learning | ||
| Linear regression | Relationship modelling between a response variable and one or more explanatory variables | Prediction of liver fat fraction from the presence of metabolic syndrome, type 2 diabetes, and laboratory markers142 |
| Logistic regression | Prediction of the probability of a target variable in binary classification | Prediction of 30-day readmissions for acute-on-chronic liver failure patients100 |
| Decision tree | Classification or regression of data based on simple rules splitting values of input variables | Prediction of acute kidney injury after liver transplantation utilising scoring systems143 |
| Random forest | Ensemble of multiple decision trees operating as a committee | Personalised surveillance model for development of hepatocellular carcinoma in patients with hepatitis C cirrhosis144 |
| Gradient boosted trees | Ensemble method of building weaker prediction models sequentially where each model predicts leftover error | Risk stratification of mortality for patients with cirrhosis in the United States Veteran Health Administration88 |
| Support vector machine | Linear classification by finding the hyperplane that maximises the margins between 2 classes | Prediction of 30-day readmissions for acute-on-chronic liver failure patients100 |
| K-Nearest neighbor | Classification of new data or cases based on similarity or distance between input features | Identification of molecular signature associated with development of hepatocellular carcinoma145 |
| Naïve Bayes | Use of Bayes theorem to predict membership probability assuming independence among predictors | Prediction of hepatitis B cirrhosis utilising serum biomarkers146 |
|
| ||
| Unsupervised learning | ||
| K-Means | Partition observations into k clusters in each observation belongs to the cluster with nearest center | Classification of cirrhosis based on un-labelled MRI data147 |
| Principal component analysis | Reduce dimensionality by converting correlated variables into a set of uncorrelated variables | Identification of splanchnic and clinical characteristics associated with hyperdynamic circulation in patients with cirrhosis148 |
| Gaussian mixture | Probabilistic model that assumes all data are generated from a finite set of Gaussian distributions | Detection of hepatocellular carcinoma from computed tomography images149 |
| Hidden Markov | System is assumed to be a Markov model with unobservable states | Progression from cirrhosis to hepatocellular carcinoma based on clinical covariates and diagnostic codes150 |
|
| ||
| Neural network algorithms | ||
| Artificial neural networks | Group of interconnected nodes/computing units that form a network | Quantification of skeletal muscle mass from computed tomography scans54 |
| Convolutional neural networks | Neural network with nodes designed to resemble visual cortices | Prediction of hepatocellular carcinoma development among patients with hepatitis C cirrhosis95 |
| Recurrent neural networks | Neural network where connections between nodes are based on temporal sequences | Prediction of 1-year mortality in patients with cirrhosis utilising EHR data92 |
| Deep neural networks | Multiple layers between the input and output layers | Longitudinal analyses of EHR data elements to predict hospitalisation outcomes72 |
EHR, electronic health record.
There is often some overlap between traditional statistical and ML approaches: Logistic regression is such an example of a methodology common to both. In general, classification trees and neural network-based methods have generally been the predominant ML algorithms applied to contemporary hepatology research. The cirrhosis mortality model, developed from the United States Veterans Affairs Corporate Data Warehouse (VHACDW) using a combination of gradient boosting and logistic regression methods, offered significantly improved discrimination compared to the MELD score.88 Of particular interest are artificial neural networks (ANNs), which are learning algorithms that can be employed for both supervised and unsupervised tasks. Neural networks are inspired by neuroanatomy – each neuron is a computing unit, and all neurons are connected to build a network. Signals travel from input layer to the output layer going through multiple hidden layers – which represent higher complexity.89–91 Deep neural networks, characterised by multiple layers between the input and output layers,91 have been utilised for longitudinal analyses of EHR data to predict outcomes of cirrhosis.92
In liver transplant, ML methodologies have been used to explore waitlist mortality and organ allocation.87,88,92–96 One of the first ML models in transplant hepatology developed in 2003 was an ANN model to predict 1-year mortality in a cohort of 92 patients. While limited in scale, this ANN model outperformed logistic regression and the Child-Pugh score.93 Similarly, an ANN-based mortality model derived from patients awaiting liver transplantation in Italy and validated in the United Kingdom showed better predictive ability than the original MELD score.94 The optimised prediction of mortality model – developed in 2019 and trained on OPTN data using ML optimal classification trees – demonstrated superior mortality prediction vs. the MELD score, and led to decreased mortality and increased survival benefit across all candidate demographics, diagnoses, and geographic regions in liver simulated allocation model simulations.97
Despite these encouraging results, ML models for waitlist mortality have several limitations, including interoperability and complexity. In addition, many early applications of ML methodologies have only considered binary outcomes rather than a time-dependent survival function which is key in the accurate determination of transplant urgency and waitlist priority. Due to these limitations and challenges in practical implementation, waitlist mortality models based on ML have yet to gain much traction in organ allocation.98,99 ML models have the potential to better predict post-transplant outcomes through the real-time considerations of longitudinal candidate variables, donor variables, and the interaction of donor-candidate matching, which may play a role in continuous distribution.38,39
Potential pitfalls of algorithms for clinical prediction
While there is substantial potential for ML to influence clinical practice in transplant hepatology and potentially improve patient outcomes, limitations of these technologies must be recognised.85 First, additional complexity may not improve predictive performance if underlying data and variables are the same. When comparing the ability of ML models (support vector classification and random forest) vs. logistic regression to predict readmission and death in 2,179 North American patients with ACLF, ML model accuracies were equivalent to models generated using only the MELD score. The performance of future ML modelling may improve if higher density data incorporating novel variables, such as sarcopenia and frailty, are available.100
Second, despite harmonisation and rationalisation of different ontologies and semantics, data quality, shift, and reproducibility are still ongoing issues in the modelling of EHR data.80,101 Dataset shift describes the changes in model performance due to temporal or spatial shifts between the population used for training and the population upon which the algorithm is deployed.102,103 One prominent recent example is the University of Michigan’s deactivation of a proprietary sepsis-alert model due to shifts in patient populations during the COVID-19 pandemic.104 Dataset shift is not exclusive to ML algorithms but also to other clinical prediction scoring systems. Periodic audits and updating of scoring systems, such as the update of MELD to MELD 3.0,10 are necessary to adapt our clinical tools to changing conditions.
Third, underlying bias can be amplified by clinical prediction and ML-based algorithms.105,106 The most prominent example in transplantation is the incorporation of race in estimated glomerular filtration rate (eGFR) calculations, which have disadvantaged racial minorities in listing practices and allocation for kidney transplant.29,107 In transplant hepatology, eGFR has been avoided in clinical prognostication modelling due to its potential for exacerbating race- and sex-based disparities. Human intelligence, in addition to artificial intelligence, remains critically important for the thoughtful and deliberate selection of data features, variables and analytic methodologies.
Fourth, structured data, which forms the basis for most classical models and ML algorithms at this time, are limited by coding. For example, efforts to diagnose Fontan-associated liver disease were limited by the lack of specific structured diagnostic codes across multiple clinical databases.108 The volume of unstructured data far exceeds structured data, with an estimated 90% of digital data in healthcare being unstructured. Incorporating or converting unstructured data elements in the EHR, such as imaging reports, pathology reports, and clinical documentation, into structured or tagged features remains challenging. Transformation of such data into structured data requires substantial cleaning, splitting, merging, validating, and sorting, but does improve clinical representation in predictive analytics.109
Finally, algorithms are not anticipated to completely replace the “subjective” judgment of clinicians involved in the care of the peritransplant patient.110 For instance, significant technical expertise is required to conduct split liver transplantation,111 to use donor organs with technical variants or higher risk features,112 or to successfully transplant patients with complex surgical histories.113 These institution- and clinicianspecific knowledge and skills are often illcaptured and ill-evaluated by algorithms.
Key point.
There is an increasing push to develop data-driven machine learning-based algorithms to further improve outcome prediction in patients with liver disease.
For these reasons, the application of ML-based artificial intelligence has received a mixed reception from both clinicians and the general population.114–116 Among clinicians, there are latent fears that algorithms may ultimately replace their skills or functions.116,117 In addition, many clinicians are uncomfortable with “black box” ML tools, even though examples of similar opacity abound in other diagnostic and therapeutic areas of clinical medicine.118 Among providers and patients, there is a concern about the loss of patient-provider relationships, privacy in data use, and accountability – namely who is responsible for adverse outcomes due to clinical decisions influenced or augmented by artificial intelligence.114,115,119 There is an increasing recognition that transparency, interpretability, and explainability are necessary for long-term acceptance of artificial intelligence tools. Ante hoc systems, which are interpretable by design, and post hoc systems, which provide local and reproducible explanations for algorithm outputs, are now commonly utilised to enable greater trust in ML algorithms.116,120 Similarly, active incorporation of human knowledge, or expert-augmentation, in the algorithm construction process is another strategy to improve “explainability.”121 To begin to address these concerns, the development of standardised tools and evaluations on transplant reporting and assessments of bias in applied ML techniques is currently underway.102,122
Key point.
Clinical decision support and prospective risk modelling are emerging areas of research that are hoped to lead to improvements in the management of patients with cirrhosis and those on the liver transplant waiting list.
Beyond MELD – for improvement in patient outcomes
Emerging technologies to actively manage waitlist mortality risk
One technology to overcome issues with unstructured data is natural language processing (NLP), which is a suite of related techniques to convert unstructured or narrative text into tagged or structured elements for analysis.123,124 There has been particular interest in utilising NLP for the diagnosis of non-alcoholic fatty liver disease as this condition is poorly documented in structured EHR data.125,126 NLP has been used on abdominal ultrasound, computerised tomography, and magnetic resonance imaging reports from the VHACDW to rapidly screen patients with radiographic evidence of fatty liver disease.126 In an analysis of clinical notes available for 38,575 patients enrolled in the Mount Sinai BioMe cohort, NLP methods outperformed ICD codes and text search.125
Real-time clinical decision support (CDS) and prospective risk modelling are also emerging areas of research/implementation in the management of patients with cirrhosis. Simple decision support tools have been implemented to support targeted quality improvement efforts, such as the proper use of ceruloplasmin in liver disease evaluation,127 improving hepatitis C screening,128 and albumin utilisation.129 The substitutable medical applications and reusable technologies on FHIR (SMART-on-FHIR) application programming interface allows for the development of more complex and prospective CDS systems by securely and automatically pulling in relevant patient data from disparate locations in the EHR.130,131 Previous SMART-on-FHIR CDS applications created to support the American Academic of Pediatrics guideline on management of neonatal hyperbilirubinemia were shown to have excellent usability and improved order rates for clinically appropriate phototherapy.132 SMART-on-FHIR CDS applications have yet to be widely pilot tested or implemented in the care of patients with cirrhosis.
Potential applications of encounter-level CDS include improving adherence to guideline-recommended care in cirrhosis, promoting timely intervention before anticipated/forecasted clinical decompensation,133,134 or aiding immunosuppression surveillance in the post-transplant setting.135 On a patient or precision-level, CDS could allow for the calculation of “personalised” risk models for progression of fibrosis to cirrhosis, development of hepatocellular carcinoma, and risk of waitlist dropout.136 The use of these models and CDS systems may help inform decisions surrounding organ allocation and acceptance in the future. Prospective implementation of such CDS systems could allow for real-world “electronic” experiments or clinical trials (Fig. 1).137,138 These concepts remain unexplored in chronic liver disease and liver transplantation, but may generate significant real-world evidence that could be used to optimise organ allocation and reduce waitlist mortality.
Fig. 1. Rapid-cycle testing in ‘electronic’ randomised controlled trials.
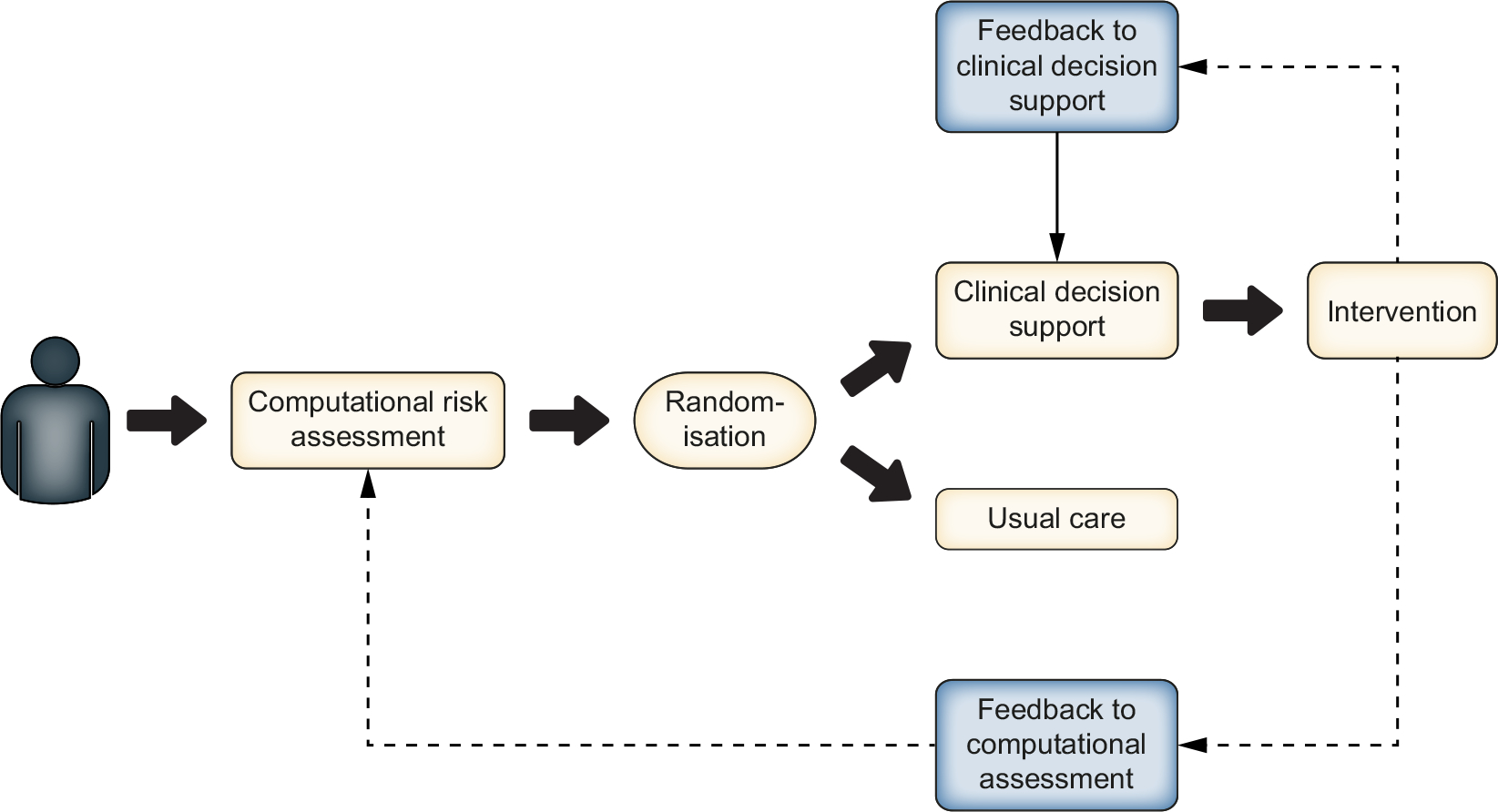
Schematic of rapid-cycle ‘electronic’ randomised controlled trials could be implemented using CDS systems: computational risk assessment allows a patient to be randomised for an intervention associated with a CDS, the results of which could then be used to iteratively modify the risk stratification algorithm or the CDS system. CDS, clinical decision support.
Conclusions
While the demographics and epidemiology of chronic liver diseases have changed dramatically in the past two decades, the MELD score and its successors have continued to provide robust predictions of short-term waitlist mortality. Continued refinements of the MELD score, such as MELD 3.0, improve its predictive ability and actively address deficiencies such as sex-based differences in waitlist mortality. Continuous distribution has emerged as a conceptual framework to optimise organ allocation by weighing factors beyond waitlist mortality. The selection of variables for changes to the liver allocation system, however, remains fraught with challenges, requiring careful consideration of objectivity, verifiability, and availability.
In the management of patients with cirrhosis and hepatic decompensation, more accurate, comprehensive, and real-time prediction of mortality, based on availability of the large amounts of information in EHRs, has the potential to dramatically change how we approach the clinical care of patients with cirrhosis and its complications. In addition, novel concepts and emerging technologies may play a major role in refining mortality prediction in an individual patient. For example, the prognosis of a patient with cirrhosis may be accurately assessed by deep neural network-based algorithms incorporating past clinical data in the EHR, current MELD 3.0, frailty measurements, and muscle mass volume derived from a computed tomography scan on an integrated SMART-on-FHIR application in the EHR system. We hope that, sometime in the near future, these novel tools will provide clinically actionable information to alter a patient’s outcome, well beyond determining a patient’s priority ranking for liver allocation.
Financial support
The authors of this study were supported by the AASLD Anna S. Lok Advanced/Transplant Hepatology Award (AASLD Foundation, Ge), P30DK026743 (UCSF Liver Center Grant, Ge and Lai), R01DK127224 (National Institute of Diabetes and Digestive and Kidney Diseases, Kim), R01AG059183 (National Institute on Aging, Lai), the Clinical, Translational, and Outcomes Research Award (AASLD Foundation, Kwong), and K23AA029197 (National Institute on Alcohol Abuse and Alcoholism, Kwong). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or any other funding agencies. The funding agencies played no role in the analysis of the data or the preparation of this manuscript.
Abbreviations
- ACLF
acute-on-chronic liver failure
- ANNs
artificial neural networks
- CDS
clinical decision support
- eGFR
estimated glomerular filtration rate
- EHR
electronic health record
- FHIR
fast healthcare interoperability resources
- GRAIL
glomerular filtration rate assessment in liver disease
- MELD
model for end-stage liver disease
- MDRD
modification of diet in renal disease
- ML
machine learning
- N3C
National COVID Cohort Collaborative
- NLP
natural language processing
- OMOP
observational medical outcomes partnership
- OPTN
Organ Procurement and Transplantation Network
- SMART-on-FHIR
substitutable medical applications and reusable technologies on fast health interoperability resources
- VHACDW
Veterans Affairs Corporate Data Warehouse
Footnotes
Conflict of interest
The authors of this manuscript have no conflicts of interest to disclose as described by the Journal of Hepatology.
Please refer to the accompanying ICMJE disclosure forms for further details.
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhep.2022.03.003.
References
Author names in bold designate shared co-first authorship
- [1].Organ Procurement and Transplantation Network (OPTN). Final Rule as revised by amendments. 1999. [Google Scholar]
- [2].Jochmans I, van Rosmalen M, Pirenne J, Samuel U. Adult liver allocation in eurotransplant. Transplantation 2017;101:1542–1550. 10.1097/TP.0000000000001631. [DOI] [PubMed] [Google Scholar]
- [3].Goudsmit BFJ, Putter H, Tushuizen ME, Vogelaar S, Pirenne J, Alwayn IPJ, et al. Refitting the model for end-stage liver disease for the eurotransplant region. Hepatology 2021;74:351–363. 10.1002/hep.31677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Trotter JF. Liver transplantation around the world. Curr Opin Organ Transpl 2017;22:123–127. 10.1097/MOT.0000000000000392. [DOI] [PubMed] [Google Scholar]
- [5].Wiesner R, Edwards E, Freeman R, Harper A, Kim R, Kamath P, et al. Model for end-stage liver disease (MELD) and allocation of donor livers. Gastroenterology 2003;124:91–96. 10.1053/gast.2003.50016. [DOI] [PubMed] [Google Scholar]
- [6].Freeman RB, Wiesner RH, Harper A, McDiarmid SV, Lake J, Edwards E, et al. The new liver allocation system: moving toward evidence-based transplantation policy. Liver Transpl 2002;8:851–858. 10.1053/jlts.2002.35927. [DOI] [PubMed] [Google Scholar]
- [7].Kamath PS, Wiesner RH, Malinchoc M, Kremers W, Therneau TM, Kosberg CL, et al. A model to predict survival in patients with end-stage liver disease. Hepatology 2001;33:464–470. 10.1053/jhep.2001.22172. [DOI] [PubMed] [Google Scholar]
- [8].Quante M, Benckert C, Thelen A, Jonas S. Experience since MELD implementation: how does the new system deliver? Int J Hepatol 2012;2012:264015. 10.1155/2012/264015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kim WR, Biggins SW, Kremers WK, Wiesner RH, Kamath PS, Benson JT, et al. Hyponatremia and mortality among patients on the liver-transplant waiting list. N Engl J Med 2008;359:1018–1026. 10.1056/NEJMoa0801209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kim WR, Mannalithara A, Heimbach JK, Kamath PS, Asrani SK, Biggins SW, et al. MELD 3.0: the model for end-stage liver disease updated for the modern era. Gastroenterology 2021. 10.1053/j.gastro.2021.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kwong AJ, Kim WR, Lake JR, Smith JM, Schladt DP, Skeans MA, et al. OPTN/SRTR 2019 annual data report: liver. Am J Transpl 2021;21(Suppl 2):208–315. 10.1111/ajt.16494. [DOI] [PubMed] [Google Scholar]
- [12].Younossi ZM, Stepanova M, Younossi Y, Golabi P, Mishra A, Rafiq N, et al. Epidemiology of chronic liver diseases in the USA in the past three decades. Gut 2020;69:564–568. 10.1136/gutjnl-2019-318813. [DOI] [PubMed] [Google Scholar]
- [13].Leise MD, Kim WR, Kremers WK, Larson JJ, Benson JT, Therneau TM. A revised model for end-stage liver disease optimizes prediction of mortality among patients awaiting liver transplantation. Gastroenterology 2011;140:1952–1960. 10.1053/j.gastro.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Godfrey EL, Malik TH, Lai JC, Mindikoglu AL, Galván NTN, Cotton RT, et al. The decreasing predictive power of MELD in an era of changing etiology of liver disease. Am J Transpl 2019;19:3299–3307. 10.1111/ajt.15559. [DOI] [PubMed] [Google Scholar]
- [15].Kwong A, Mannalithara A, Kim WR. Reply to: “The decreasing predictive power of MELD in an era of changing etiology of liver disease”. Am J Transpl 2020;20:901–902. 10.1111/ajt.15733. [DOI] [PubMed] [Google Scholar]
- [16].Hernaez R, Liu Y, Kramer JR, Rana A, El-Serag HB, Kanwal F. Model for end-stage liver disease-sodium underestimates 90-day mortality risk in patients with acute-on-chronic liver failure. J Hepatol 2020;73:1425–1433. 10.1016/j.jhep.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Allen AM, Heimbach JK, Larson JJ, Mara KC, Kim WR, Kamath PS, et al. Reduced access to liver transplantation in women: role of height, MELD exception scores, and renal function underestimation. Transplantation 2018;102:1710–1716. 10.1097/TP.0000000000002196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Myers RP, Shaheen AAM, Aspinall AI, Quinn RR, Burak KW. Gender, renal function, and outcomes on the liver transplant waiting list: assessment of revised MELD including estimated glomerular filtration rate. J Hepatol 2011;54:462–470. 10.1016/j.jhep.2010.07.015. [DOI] [PubMed] [Google Scholar]
- [19].Mathur AK, Schaubel DE, Gong Q, Guidinger MK, Merion RM. Sexbased disparities in liver transplant rates in the United States. Am J Transpl 2011;11:1435–1443. 10.1111/j.1600-6143.2011.03498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Verna EC, Lai JC. Time for action to address the persistent sex-based disparity in liver transplant access. JAMA Surg 2020;155:545–547. 10.1001/jamasurg.2020.1126. [DOI] [PubMed] [Google Scholar]
- [21].Cholongitas E, Marelli L, Kerry A, Goodier DW, Nair D, Thomas M, et al. Female liver transplant recipients with the same GFR as male recipients have lower MELD scores–a systematic bias. Am J Transpl 2007;7:685–692. 10.1111/j.1600-6143.2007.01666.x. [DOI] [PubMed] [Google Scholar]
- [22].Leithead JA, MacKenzie SM, Ferguson JW, Hayes PC. Is estimated glomerular filtration rate superior to serum creatinine in predicting mortality on the waiting list for liver transplantation? Transpl Int 2011;24:482–488. 10.1111/j.1432-2277.2011.01231.x. [DOI] [PubMed] [Google Scholar]
- [23].Asrani SK, Jennings LW, Kim WR, Kamath PS, Levitsky J, Nadim MK, et al. MELD-GRAIL-Na: glomerular filtration rate and mortality on liver-transplant waiting list. Hepatology 2020;71:1766–1774. 10.1002/hep.30932. [DOI] [PubMed] [Google Scholar]
- [24].Asrani SK, Jennings LW, Trotter JF, Levitsky J, Nadim MK, Kim WR, et al. A model for glomerular filtration rate assessment in liver disease (GRAIL) in the presence of renal dysfunction. Hepatology 2019;69:1219–1230. 10.1002/hep.30321. [DOI] [PubMed] [Google Scholar]
- [25].Finkenstedt A, Dorn L, Edlinger M, Prokop W, Risch L, Griesmacher A, et al. Cystatin C is a strong predictor of survival in patients with cirrhosis: is a cystatin C-based MELD better? Liver Int 2012;32:1211–1216. 10.1111/j.1478-3231.2012.02766.x. [DOI] [PubMed] [Google Scholar]
- [26].De Souza V, Hadj-Aissa A, Dolomanova O, Rabilloud M, Rognant N, Lemoine S, et al. Creatinine- versus cystatine C-based equations in assessing the renal function of candidates for liver transplantation with cirrhosis. Hepatology 2014;59:1522–1531. 10.1002/hep.26886. [DOI] [PubMed] [Google Scholar]
- [27].Nephew LD, Goldberg DS, Lewis JD, Abt P, Bryan M, Forde KA. Exception points and body size contribute to gender disparity in liver transplantation. Clin Gastroenterol Hepatol 2017;15:1286–1293.e2. 10.1016/j.cgh.2017.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wood NL, VanDerwerken D, Segev DL, Gentry SE. Correcting the sex disparity in MELD-Na. Am J Transpl 2021;21:3296–3304. 10.1111/ajt.16731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Vyas DA, Eisenstein LG, Jones DS. Hidden in plain sight - reconsidering the use of race correction in clinical algorithms. N Engl J Med 2020;383:874–882. 10.1056/NEJMms2004740. [DOI] [PubMed] [Google Scholar]
- [30].Linecker M, Krones T, Berg T, Niemann CU, Steadman RH, Dutkowski P, et al. Potentially inappropriate liver transplantation in the era of the “sickest first” policy - a search for the upper limits. J Hepatol 2018;68:798–813. 10.1016/j.jhep.2017.11.008. [DOI] [PubMed] [Google Scholar]
- [31].Rana A, Hardy MA, Halazun KJ, Woodland DC, Ratner LE, Samstein B, et al. Survival outcomes following liver transplantation (SOFT) score: a novel method to predict patient survival following liver transplantation. Am J Transpl 2008;8:2537–2546. 10.1111/j.1600-6143.2008.02400.x. [DOI] [PubMed] [Google Scholar]
- [32].Dutkowski P, Oberkofler CE, Slankamenac K, Puhan MA, Schadde E, Müllhaupt B, et al. Are there better guidelines for allocation in liver transplantation? A novel score targeting justice and utility in the model for end-stage liver disease era. Ann Surg 2011;254:745–753. 10.1097/SLA.0b013e3182365081. discussion 753. [DOI] [PubMed] [Google Scholar]
- [33].Goldberg D, Mantero A, Newcomb C, Delgado C, Forde K, Kaplan D, et al. Development and validation of a model to predict long-term survival after liver transplantation. Liver Transpl 2021;27:797–807. 10.1002/lt.26002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Luo X, Leanza J, Massie AB, Garonzik-Wang JM, Haugen CE, Gentry SE, et al. MELD as a metric for survival benefit of liver transplantation. Am J Transpl 2018;18:1231–1237. 10.1111/ajt.14660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kwong AJ, Kim WR. Predicting survival after liver transplantation: a noble pursuit or a fool’s errand? Liver Transpl 2021;27:789–790. 10.1002/lt.26057. [DOI] [PubMed] [Google Scholar]
- [36].Freeman RB, Gish RG, Harper A, Davis GL, Vierling J, Lieblein L, et al. Model for end-stage liver disease (MELD) exception guidelines: results and recommendations from the MELD Exception Study Group and Conference (MESSAGE) for the approval of patients who need liver transplantation with diseases not considered by the standard MELD formula. Liver Transpl 2006;12:S128–S136. 10.1002/lt.20979. [DOI] [PubMed] [Google Scholar]
- [37].Cillo U, Burra P, Mazzaferro V, Belli L, Pinna AD, Spada M, et al. A multistep, consensus-based approach to organ allocation in liver transplantation: toward a “blended principle model”. Am J Transpl 2015;15:2552–2561. 10.1111/ajt.13408. [DOI] [PubMed] [Google Scholar]
- [38].Kasiske BL, Pyke J, Snyder JJ. Continuous distribution as an organ allocation framework. Curr Opin Organ Transpl 2020;25:115–121. 10.1097/MOT.0000000000000733. [DOI] [PubMed] [Google Scholar]
- [39].Continuous Distribution - OPTN n.d. https://optn.transplant.hrsa.gov/governance/key-initiatives/continuous-distribution/ (accessed October 3, 2021).
- [40].Snyder JJ, Salkowski N, Wey A, Pyke J, Israni AK, Kasiske BL. Organ distribution without geographic boundaries: a possible framework for organ allocation. Am J Transpl 2018;18:2635–2640. 10.1111/ajt.15115. [DOI] [PubMed] [Google Scholar]
- [41].Lai JC, Tandon P, Bernal W, Tapper EB, Ekong U, Dasarathy S, et al. Malnutrition, frailty, and sarcopenia in patients with cirrhosis: 2021 practice guidance by the american association for the study of liver diseases. Hepatology 2021;74:1611–1644. 10.1002/hep.32049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lochs H, Allison SP, Meier R, Pirlich M, Kondrup J, Schneider S, et al. Introductory to the ESPEN guidelines on enteral nutrition: terminology, definitions and general topics. Clin Nutr 2006;25:180–186. 10.1016/j.clnu.2006.02.007. [DOI] [PubMed] [Google Scholar]
- [43].Morley JE, Vellas B, van Kan GA, Anker SD, Bauer JM, Bernabei R, et al. Frailty consensus: a call to action. J Am Med Dir Assoc 2013;14:392–397. 10.1016/j.jamda.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lai JC, Covinsky KE, Dodge JL, Boscardin WJ, Segev DL, Roberts JP, et al. Development of a novel frailty index to predict mortality in patients with end-stage liver disease. Hepatology 2017;66:564–574. 10.1002/hep.29219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lai JC, Rahimi RS, Verna EC, Kappus MR, Dunn MA, McAdams-DeMarco M, et al. Frailty associated with waitlist mortality independent of ascites and hepatic encephalopathy in a multicenter study. Gastroenterology 2019;156:1675–1682. 10.1053/j.gastro.2019.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lai JC, Feng S, Terrault NA, Lizaola B, Hayssen H, Covinsky K. Frailty predicts waitlist mortality in liver transplant candidates. Am J Transpl 2014;14:1870–1879. 10.1111/ajt.12762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Tandon P, Tangri N, Thomas L, Zenith L, Shaikh T, Carbonneau M, et al. A rapid bedside screen to predict unplanned hospitalization and death in outpatients with cirrhosis: a prospective evaluation of the clinical frailty scale. Am J Gastroenterol 2016;111:1759–1767. 10.1038/ajg.2016.303. [DOI] [PubMed] [Google Scholar]
- [48].Tapper EB, Konerman M, Murphy S, Sonnenday CJ. Hepatic encephalopathy impacts the predictive value of the Fried Frailty Index. Am J Transpl 2018;18:2566–2570. 10.1111/ajt.15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lai JC, Sonnenday CJ, Tapper EB, Duarte-Rojo A, Dunn MA, Bernal W, et al. Frailty in liver transplantation: an expert opinion statement from the American Society of Transplantation Liver and Intestinal Community of Practice. Am J Transpl 2019;19:1896–1906. 10.1111/ajt.15392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lai JC, Segev DL, McCulloch CE, Covinsky KE, Dodge JL, Feng S. Physical frailty after liver transplantation. Am J Transpl 2018;18:1986–1994. 10.1111/ajt.14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cruz-Jentoft AJ, Bahat G, Bauer J, Boirie Y, Bruyère O, Cederholm T, et al. Sarcopenia: revised European consensus on definition and diagnosis. Age Ageing 2019;48:16–31. 10.1093/ageing/afy169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Carey EJ, Lai JC, Wang CW, Dasarathy S, Lobach I, Montano-Loza AJ, et al. A multicenter study to define sarcopenia in patients with end-stage liver disease. Liver Transpl 2017;23:625–633. 10.1002/lt.24750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Mazurak VC, Tandon P, Montano-Loza AJ. Nutrition and the transplant candidate. Liver Transpl 2017;23:1451–1464. 10.1002/lt.24848. [DOI] [PubMed] [Google Scholar]
- [54].Paris MT, Tandon P, Heyland DK, Furberg H, Premji T, Low G, et al. Automated body composition analysis of clinically acquired computed tomography scans using neural networks. Clin Nutr 2020;39:3049–3055. 10.1016/j.clnu.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Carey EJ, Lai JC, Sonnenday C, Tapper EB, Tandon P, Duarte-Rojo A, et al. A north american expert opinion statement on sarcopenia in liver transplantation. Hepatology 2019;70:1816–1829. 10.1002/hep.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Englesbe MJ, Patel SP, He K, Lynch RJ, Schaubel DE, Harbaugh C, et al. Sarcopenia and mortality after liver transplantation. J Am Coll Surg 2010;211:271–278. 10.1016/j.jamcollsurg.2010.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kaido T, Ogawa K, Fujimoto Y, Ogura Y, Hata K, Ito T, et al. Impact of sarcopenia on survival in patients undergoing living donor liver transplantation. Am J Transpl 2013;13:1549–1556. 10.1111/ajt.12221. [DOI] [PubMed] [Google Scholar]
- [58].Welch N, Dasarathy J, Runkana A, Penumatsa R, Bellar A, Reen J, et al. Continued muscle loss increases mortality in cirrhosis: impact of aetiology of liver disease. Liver Int 2020;40:1178–1188. 10.1111/liv.14358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Leppke S, Leighton T, Zaun D, Chen S-C, Skeans M, Israni AK, et al. Scientific Registry of Transplant Recipients: collecting, analyzing, and reporting data on transplantation in the United States. Transpl Rev 2013;27:50–56. 10.1016/j.trre.2013.01.002. [DOI] [PubMed] [Google Scholar]
- [60].Langer RM, Cohen B, Rahmel A. History of eurotransplant. Transpl Proc 2012;44:2130–2131. 10.1016/j.transproceed.2012.07.125. [DOI] [PubMed] [Google Scholar]
- [61].Mahmud N, Goldberg DS, Bittermann T. Best practices in large database clinical epidemiology research in hepatology: barriers and opportunities. Liver Transpl 2021. 10.1002/lt.26231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Okafor PN, Chiejina M, de Pretis N, Talwalkar JA. Secondary analysis of large databases for hepatology research. J Hepatol 2016;64:946–956. 10.1016/j.jhep.2015.12.019. [DOI] [PubMed] [Google Scholar]
- [63].Hirode G, Saab S, Wong RJ. Trends in the burden of chronic liver disease among hospitalized US adults. JAMA Netw Open 2020;3:e201997. 10.1001/jamanetworkopen.2020.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].OHDSI – Observational Health Data Sciences and Informatics n.d. https://ohdsi.org/ (accessed February 21, 2021). [Google Scholar]
- [65].Haendel MA, Chute CG, Bennett TD, Eichmann DA, Guinney J, Kibbe WA, et al. The National COVID Cohort Collaborative (N3C): rationale, design, infrastructure, and deployment. J Am Med Inform Assoc 2021;28:427–443. 10.1093/jamia/ocaa196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Peterson TA, Fontil V, Koliwad SK, Patel A, Butte AJ. Quantifying variation in treatment utilization for type 2 diabetes across five major university of California health systems. Diabetes Care 2021. 10.2337/dc20-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Adoption of Electronic Health Record Systems among U.S. Non-Federal Acute Care Hospitals: 2008–2015 | HealthIT.gov n.d. https://www.healthit.gov/data/data-briefs/adoption-electronic-health-record-systems-among-us-non-federal-acute-care-1 (accessed October 3, 2021). [Google Scholar]
- [68].Villanueva FL, Folkvord F, Fauli C. Benchmarking deployment of eHealth among general practitioners. RAND org; 2018.
- [69].Obermeyer Z, Emanuel EJ. Predicting the future - big data, machine learning, and clinical medicine. N Engl J Med 2016;375:1216–1219. 10.1056/NEJMp1606181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Chua H-R, Zheng K, Vathsala A, Ngiam K-Y, Yap H-K, Lu L, et al. Health care analytics with time-invariant and time-variant feature importance to predict hospital-acquired acute kidney injury: observational longitudinal study. J Med Internet Res 2021;23:e30805. 10.2196/30805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Weisenthal SJ, Quill C, Farooq S, Kautz H, Zand MS. Predicting acute kidney injury at hospital re-entry using high-dimensional electronic health record data. PLoS One 2018;13:e0204920. 10.1371/journal.pone.0204920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Rajkomar A, Oren E, Chen K, Dai AM, Hajaj N, Hardt M, et al. Scalable and accurate deep learning with electronic health records. Npj Digital Med 2018;1:18. 10.1038/s41746-018-0029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Ge J, Najafi N, Zhao W, Somsouk M, Fang M, Lai JC. A methodology to generate longitudinally updated acute-on-chronic liver failure prognostication scores from electronic health record data. Hepatol Commun 2021;5:1069–1080. 10.1002/hep4.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Haendel MA, Chute CG, Robinson PN. Classification, ontology, and precision medicine. N Engl J Med 2018;379:1452–1462. 10.1056/NEJMra1615014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Atiemo K, Skaro A, Maddur H, Zhao L, Montag S, VanWagner L, et al. Mortality risk factors among patients with cirrhosis and a low model for End-Stage Liver Disease Sodium score (<−15): an analysis of liver transplant allocation policy using aggregated electronic health record data. Am J Transpl 2017;17:2410–2419. 10.1111/ajt.14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Health Level Seven International - Homepage | HL7 International n.d. https://www.hl7.org/ (accessed October 3, 2021).
- [77].European Health Data Evidence Network – ehden.eu n.d. https://www.ehden.eu/ (accessed November 20, 2021).
- [78].Bennett TD, Moffitt RA, Hajagos JG, Amor B, Anand A, Bissell MM, et al. The national COVID cohort collaborative: clinical characterization and early severity prediction. medRxiv 2021. 10.1101/2021.01.12.21249511. [DOI] [Google Scholar]
- [79].Ge J, Pletcher MJ, Lai JC, N3C Consortium. Outcomes of SARS-CoV-2 infection in patients with chronic liver disease and cirrhosis: a national COVID cohort collaborative study. Gastroenterology 2021. 10.1053/j.gastro.2021.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Rumsfeld JS, Joynt KE, Maddox TM. Big data analytics to improve cardiovascular care: promise and challenges. Nat Rev Cardiol 2016;13:350–359. 10.1038/nrcardio.2016.42. [DOI] [PubMed] [Google Scholar]
- [81].Genta RM, Sonnenberg A. Big data in gastroenterology research. Nat Rev Gastroenterol Hepatol 2014;11:386–390. 10.1038/nrgastro.2014.18. [DOI] [PubMed] [Google Scholar]
- [82].Favaretto M, De Clercq E, Schneble CO, Elger BS. What is your definition of Big Data? Researchers’ understanding of the phenomenon of the decade. PLoS One 2020;15:e0228987. 10.1371/journal.pone.0228987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Asri H, Mousannif H, Al Moatassime H, Noel T. Big data in healthcare: Challenges and opportunities. In: 2015 International Conference on Cloud Technologies and Applications (CloudTech). IEEE; 2015. p. 1–7. 10.1109/CloudTech.2015.7337020. [DOI] [Google Scholar]
- [84].Wiens J, Shenoy ES. Machine learning for healthcare: on the verge of a major shift in healthcare epidemiology. Clin Infect Dis 2018;66:149–153. 10.1093/cid/cix731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Obermeyer Z, Weinstein JN. Adoption of artificial intelligence and machine learning is increasing, but irrational exuberance remains. NEJM Catal 2020;1. 10.1056/CAT.19.1090. [DOI] [Google Scholar]
- [86].Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med 2019;380:1347–1358. 10.1056/NEJMra1814259. [DOI] [PubMed] [Google Scholar]
- [87].Spann A, Yasodhara A, Kang J, Watt K, Wang B, Goldenberg A, et al. Applying machine learning in liver disease and transplantation: a comprehensive review. Hepatology 2020;71:1093–1105. 10.1002/hep.31103. [DOI] [PubMed] [Google Scholar]
- [88].Kanwal F, Taylor TJ, Kramer JR, Cao Y, Smith D, Gifford AL, et al. Development, validation, and evaluation of a simple machine learning model to predict cirrhosis mortality. JAMA Netw Open 2020;3:e2023780. 10.1001/jamanetworkopen.2020.23780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Jain AK, Mao Jianchang, Mohiuddin KM. Artificial neural networks: a tutorial. Computer (Long Beach Calif) 1996;29:31–44. 10.1109/2.485891. [DOI] [Google Scholar]
- [90].Schuster M, Paliwal KK. Bidirectional recurrent neural networks. IEEE Trans Signal Process 1997;45:2673–2681. 10.1109/78.650093. [DOI] [Google Scholar]
- [91].LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436–444. 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- [92].Guo A, Mazumder NR, Ladner DP, Foraker RE. Predicting mortality among patients with liver cirrhosis in electronic health records with machine learning. PLoS One 2021;16:e0256428. 10.1371/journal.pone.0256428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Banerjee R, Das A, Ghoshal UC, Sinha M. Predicting mortality in patients with cirrhosis of liver with application of neural network technology. J Gastroenterol Hepatol 2003;18:1054–1060. 10.1046/j.1440-1746.2003.03123.x. [DOI] [PubMed] [Google Scholar]
- [94].Cucchetti A, Vivarelli M, Heaton ND, Phillips S, Piscaglia F, Bolondi L, et al. Artificial neural network is superior to MELD in predicting mortality of patients with end-stage liver disease. Gut 2007;56:253–258. 10.1136/gut.2005.084434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Ioannou GN, Tang W, Beste LA, Tincopa MA, Su GL, Van T, et al. Assessment of a deep learning model to predict hepatocellular carcinoma in patients with hepatitis C cirrhosis. JAMA Netw Open 2020;3:e2015626. 10.1001/jamanetworkopen.2020.15626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Ferrarese A, Sartori G, Orrù G, Frigo AC, Pelizzaro F, Burra P, et al. Machine learning in liver transplantation: a tool for some unsolved questions? Transpl Int 2021;34:398–411. 10.1111/tri.13818. [DOI] [PubMed] [Google Scholar]
- [97].Bertsimas D, Kung J, Trichakis N, Wang Y, Hirose R, Vagefi PA. Development and validation of an optimized prediction of mortality for candidates awaiting liver transplantation. Am J Transpl 2019;19:1109–1118. 10.1111/ajt.15172. [DOI] [PubMed] [Google Scholar]
- [98].Kwong AJ, Asrani SK. Artificial neural networks and liver transplantation: are we ready for self-driving cars? Liver Transpl 2018;24:161–163. 10.1002/lt.24993. [DOI] [PubMed] [Google Scholar]
- [99].Miller PE, Pawar S, Vaccaro B, McCullough M, Rao P, Ghosh R, et al. Predictive abilities of machine learning techniques may be limited by dataset characteristics: insights from the UNOS database. J Card Fail 2019;25:479–483. 10.1016/j.cardfail.2019.01.018. [DOI] [PubMed] [Google Scholar]
- [100].Hu C,Anjur V, Saboo K, Reddy KR, OʼLeary J, Tandon P, et al. Low predictability of readmissions and death using machine learning in cirrhosis. Am J Gastroenterol 2020. 10.14309/ajg.0000000000000971. [DOI] [PubMed] [Google Scholar]
- [101].Raghupathi W, Raghupathi V. Big data analytics in healthcare: promise and potential. Health Inf Sci Syst 2014;2:3. 10.1186/2047-s2501-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Bates DW, Auerbach A, Schulam P, Wright A, Saria S. Reporting and implementing interventions involving machine learning and artificial intelligence. Ann Intern Med 2020;172:S137–S144. 10.7326/M19-0872. [DOI] [PubMed] [Google Scholar]
- [103].Finlayson SG, Subbaswamy A, Singh K, Bowers J, Kupke A, Zittrain J, et al. The clinician and dataset shift in artificial intelligence. N Engl J Med 2021;385:283–286. 10.1056/NEJMc2104626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Wong A, Otles E, Donnelly JP, Krumm A, McCullough J, DeTroyer-Cooley O, et al. External validation of a widely implemented proprietary sepsis prediction model in hospitalized patients. JAMA Intern Med 2021;181:1065–1070. 10.1001/jamainternmed.2021.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].What Do We Do About the Biases in AI? n.d. https://hbr.org/2019/10/what-do-we-do-about-the-biases-in-ai (accessed October 3, 2021).
- [106].Gianfrancesco MA, Tamang S, Yazdany J, Schmajuk G. Potential biases in machine learning algorithms using electronic health record data. JAMA Intern Med 2018;178:1544–1547. 10.1001/jamainternmed.2018.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Kuppachi S, Norman SP, Lentine KL, Axelrod DA. Using race to estimate glomerular filtration and its impact in kidney transplantation. Clin Transpl 2021;35:e14136. 10.1111/ctr.14136. [DOI] [PubMed] [Google Scholar]
- [108].Kim MH, Nguyen A, Lo M, Kumar SR, Bucuvalas J, Glynn EF, et al. Big data in transplantation practice-the devil is in the detail-Fontan-associated liver disease. Transplantation 2021;105:18–22. 10.1097/TP.0000000000003308. [DOI] [PubMed] [Google Scholar]
- [109].Adnan K, Akbar R, Khor SW, Ali ABA. Role and challenges of unstructured big data in healthcare. In: Sharma N, Chakrabarti A, Balas VE, editors. Data management, analytics and innovation: proceedings of ICDMAI 2019, volume 1, vol. 1042. Singapore: Springer Singapore; 2020. p. 301–323. 10.1007/978-981-32-9949-8_22. [DOI] [Google Scholar]
- [110].Kuo RYL, Harrison CJ, Jones BE, Geoghegan L, Furniss D. Perspectives: a surgeon’s guide to machine learning. Int J Surg 2021;94:106133. 10.1016/j.ijsu.2021.106133. [DOI] [PubMed] [Google Scholar]
- [111].Cauley RP, Vakili K, Fullington N, Potanos K, Graham DA, Finkelstein JA, et al. Deceased-donor split-liver transplantation in adult recipients: is the learning curve over? J Am Coll Surg 2013;217:672–684.e1. 10.1016/j.jamcollsurg.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Feng S, Goodrich NP, Bragg-Gresham JL, Dykstra DM, Punch JD, DebRoy MA, et al. Characteristics associated with liver graft failure: the concept of a donor risk index. Am J Transpl 2006;6:783–790. 10.1111/j.1600-6143.2006.01242.x. [DOI] [PubMed] [Google Scholar]
- [113].Trapero-Marugán M, Little EC, Berenguer M. Stretching the boundaries for liver transplant in the 21st century. Lancet Gastroenterol Hepatol 2018;3:803–811. 10.1016/S2468-1253(18)30213-9. [DOI] [PubMed] [Google Scholar]
- [114].Bartoletti I AI in healthcare: ethical and privacy challenges. In: Riaño D, Wilk S, ten Teije A, editors. Artificial intelligence in medicine, vol. 11526. Cham: Springer International Publishing; 2019. p. 7–10. 10.1007/978-3-030-21642-9_2. [DOI] [Google Scholar]
- [115].DeCamp M, Lindvall C. Latent bias and the implementation of artificial intelligence in medicine. J Am Med Inform Assoc 2020;27:2020–2023. 10.1093/jamia/ocaa094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Wang F, Kaushal R, Khullar D. Should health care demand interpretable artificial intelligence or accept “black box” medicine? Ann Intern Med 2020;172:59–60. 10.7326/M19-2548. [DOI] [PubMed] [Google Scholar]
- [117].Blease C, Bernstein MH, Gaab J, Kaptchuk TJ, Kossowsky J, Mandl KD, et al. Computerization and the future of primary care: a survey of general practitioners in the UK. PLoS One 2018;13:e0207418. 10.1371/journal.pone.0207418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Pazzani MJ, Mani S, Shankle WR. Acceptance of rules generated by machine learning among medical experts. Methods Inf Med 2001;40:380–385. [PubMed] [Google Scholar]
- [119].Yakar D, Ongena YP, Kwee TC, Haan M. Do people favor artificial intelligence over physicians? A survey among the general population and their view on artificial intelligence in medicine. Value Health 2021. 10.1016/j.jval.2021.09.004. [DOI] [PubMed] [Google Scholar]
- [120].Holzinger A, Langs G, Denk H, Zatloukal K, Müller H. Causability and explainability of artificial intelligence in medicine. Wiley Interdiscip Rev Data Min Knowl Discov 2019;9:e1312. 10.1002/widm.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Gennatas ED, Friedman JH, Ungar LH, Pirracchio R, Eaton E, Reichmann LG, et al. Expert-augmented machine learning. Proc Natl Acad Sci USA 2020;117:4571–4577. 10.1073/pnas.1906831117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Collins GS, Dhiman P, Andaur Navarro CL, Ma J, Hooft L, Reitsma JB, et al. Protocol for development of a reporting guideline (TRIPOD-AI) and risk of bias tool (PROBAST-AI) for diagnostic and prognostic prediction model studies based on artificial intelligence. BMJ Open 2021;11:e048008. 10.1136/bmjopen-2020-048008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. J Am Med Inform Assoc 2011;18:544–551. 10.1136/amiajnl-2011-000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Chowdhury GG. Natural language processing. Ann Rev Info Sci Tech 2005;37:51–89. 10.1002/aris.1440370103. [DOI] [Google Scholar]
- [125].Van Vleck TT, Chan L, Coca SG, Craven CK, Do R, Ellis SB, et al. Augmented intelligence with natural language processing applied to electronic health records for identifying patients with non-alcoholic fatty liver disease at risk for disease progression. Int J Med Inform 2019;129:334–341. 10.1016/j.ijmedinf.2019.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Redman JS, Natarajan Y, Hou JK, Wang J, Hanif M, Feng H, et al. Accurate identification of fatty liver disease in data warehouse utilizing natural language processing. Dig Dis Sci 2017;62:2713–2718. 10.1007/s10620-017-4721-9. [DOI] [PubMed] [Google Scholar]
- [127].Tapper EB, Sengupta N, Lai M, Horowitz G. Understanding and reducing ceruloplasmin overuse with a decision support intervention for liver disease evaluation. Am J Med 2016;129. 10.1016/j.amjmed.2015.07.019. 115.e17–22. [DOI] [PubMed] [Google Scholar]
- [128].Sidlow R, Msaouel P. Improving hepatitis C virus screening rates in primary care: a targeted intervention using the electronic health record. J Healthc Qual 2015;37:319–323. 10.1097/JHQ.0000000000000010. [DOI] [PubMed] [Google Scholar]
- [129].Mudireddy PR, Mull NK, Williams K, Bushen JL, Kasbekar N, Krok K, et al. Impact of a clinical decision support intervention on albumin utilization and appropriateness of use in an academic healthcare system. medRxiv 2021. 10.1101/2021.04.05.21254943. [DOI] [Google Scholar]
- [130].Mandel JC, Kreda DA, Mandl KD, Kohane IS, Ramoni RB. SMART on FHIR: a standards-based, interoperable apps platform for electronic health records. J Am Med Inform Assoc 2016;23:899–908. 10.1093/jamia/ocv189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Bloomfield RA, Polo-Wood F, Mandel JC, Mandl KD. Opening the Duke electronic health record to apps: implementing SMART on FHIR. Int J Med Inform 2017;99:1–10. 10.1016/j.ijmedinf.2016.12.005. [DOI] [PubMed] [Google Scholar]
- [132].Kawamoto K, Kukhareva P, Shakib JH, Kramer H, Rodriguez S, Warner PB, et al. Association of an electronic health record add-on app for neonatal bilirubin management with physician efficiency and care quality. JAMA Netw Open 2019;2:e1915343. 10.1001/jamanetworkopen.2019.15343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Debnath S, Barnaby DP, Coppa K, Makhnevich A, Kim EJ, Chatterjee S, et al. Machine learning to assist clinical decision-making during the COVID-19 pandemic. Bioelectron Med 2020;6:14. 10.1186/s42234-020-00050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Kipnis P, Turk BJ, Wulf DA, LaGuardia JC, Liu V, Churpek MM, et al. Development and validation of an electronic medical record-based alert score for detection of inpatient deterioration outside the ICU. J Biomed Inform 2016;64:10–19. 10.1016/j.jbi.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Jacobs J, Weir C, Evans RS, Staes C. Assessment of readiness for clinical decision support to aid laboratory monitoring of immunosuppressive care at U.S. liver transplant centers. Appl Clin Inform 2014;5:988–1004. 10.4338/ACI-2014-08-RA-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Kao D, Larson C, Fletcher D, Stegner K. Clinical decision support may link multiple domains to improve patient care: viewpoint. JMIR Med Inform 2020;8:e20265. 10.2196/20265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Pletcher MJ, Flaherman V, Najafi N, Patel S, Rushakoff RJ, Hoffman A, et al. Randomized controlled trials of electronic health record interventions: design, conduct, and reporting considerations. Ann Intern Med 2020;172:S85–S91. 10.7326/M19-0877. [DOI] [PubMed] [Google Scholar]
- [138].Horwitz LI, Kuznetsova M, Jones SA. Creating a learning health system through rapid-cycle, randomized testing. N Engl J Med 2019;381:1175–1179. 10.1056/NEJMsb1900856. [DOI] [PubMed] [Google Scholar]
- [139].Pugh RN, Murray-Lyon IM, Dawson JL, Pietroni MC, Williams R. Transection of the oesophagus for bleeding oesophageal varices. Br J Surg 1973;60:646–649. 10.1002/bjs.1800600817. [DOI] [PubMed] [Google Scholar]
- [140].Kartoun U, Corey KE, Simon TG, Zheng H, Aggarwal R, Ng K, et al. The MELD-Plus: a generalizable prediction risk score in cirrhosis. PLoS One 2017;12:e0186301. 10.1371/journal.pone.0186301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Mahmud N, Asrani SK, Kaplan DE, Ogola GO, Taddei TH, Kamath PS, et al. The predictive role of model for end-stage liver disease-lactate and lactate clearance for in-hospital mortality among a national cirrhosis cohort. Liver Transpl 2021;27:177–189. 10.1002/lt.25913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Kotronen A, Peltonen M, Hakkarainen A, Sevastianova K, Bergholm R, Johansson LM, et al. Prediction of non-alcoholic fatty liver disease and liver fat using metabolic and genetic factors. Gastroenterology 2009;137:865–872. 10.1053/j.gastro.2009.06.005. [DOI] [PubMed] [Google Scholar]
- [143].Xin W, Yi W, Liu H, Haixia L, Dongdong L, Ma Y, et al. Early prediction of acute kidney injury after liver transplantation by scoring system and decision tree. Ren Fail 2021;43:1137–1145. 10.1080/0886022X.2021.1945462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Audureau E, Carrat F, Layese R, Cagnot C, Asselah T, Guyader D, et al. Personalized surveillance for hepatocellular carcinoma in cirrhosis - using machine learning adapted to HCV status. J Hepatol 2020;73:1434–1445. 10.1016/j.jhep.2020.05.052. [DOI] [PubMed] [Google Scholar]
- [145].Kim JW, Ye Q, Forgues M, Chen Y, Budhu A, Sime J, et al. Cancer-associated molecular signature in the tissue samples of patients with cirrhosis. Hepatology 2004;39:518–527. 10.1002/hep.20053. [DOI] [PubMed] [Google Scholar]
- [146].Cao Y, He K, Cheng M, Si H-Y, Zhang H-L, Song W, et al. Two classifiers based on serum peptide pattern for prediction of HBV-induced liver cirrhosis using MALDI-TOF MS. Biomed Res Int 2013;2013:814876. 10.1155/2013/814876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Lee GN, Fujita H. K-means clustering for classifying unlabelled MRI data. In: 9th Biennial Conference of the Australian Pattern Recognition Society on Digital Image Computing Techniques and Applications (DICTA 2007). IEEE; 2007. p. 92–98. 10.1109/DICTA.2007.4426781. [DOI] [Google Scholar]
- [148].Møller S, Hobolth L, Winkler C, Bendtsen F, Christensen E. Determinants of the hyperdynamic circulation and central hypovolaemia in cirrhosis. Gut 2011;60:1254–1259. 10.1136/gut.2010.235473. [DOI] [PubMed] [Google Scholar]
- [149].Das A, Rajendra Acharya U, Panda SS, Sabut S. Deep learning based liver cancer detection using watershed transform and Gaussian mixture model techniques. Cogn Syst Res 2018;54:165–175. 10.1016/j.cogsys.2018.12.009. [DOI] [Google Scholar]
- [150].Bartolomeo N, Trerotoli P, Serio G. Progression of liver cirrhosis to HCC: an application of hidden Markov model. BMC Med Res Methodol 2011;11:38. 10.1186/1471-2288-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]


