ABSTRACT
The biological carbon pump (BCP) in the Southern Ocean is driven by phytoplankton productivity and is a significant organic matter sink. However, the role of particle-attached (PA) and free-living (FL) prokaryotes (bacteria and archaea) and their diversity in influencing the efficiency of the BCP is still unclear. To investigate this, we analyzed the metagenomes linked to suspended and sinking marine particles from the Sub-Antarctic Southern Ocean Time Series (SOTS) by deploying a Marine Snow Catcher (MSC), obtaining suspended and sinking particulate material, determining organic carbon and nitrogen flux, and constructing metagenome-assembled genomes (MAGs). The suspended and sinking particle-pools were dominated by bacteria with the potential to degrade organic carbon. Bacterial communities associated with the sinking fraction had more genes related to the degradation of complex organic carbon than those in the suspended fraction. Archaea had the potential to drive nitrogen metabolism via nitrite and ammonia oxidation, altering organic nitrogen concentration. The data revealed several pathways for chemoautotrophy and the secretion of recalcitrant dissolved organic carbon (RDOC) from CO2, with bacteria and archaea potentially sequestering particulate organic matter (POM) via the production of RDOC. These findings provide insights into the diversity and function of prokaryotes in suspended and sinking particles and their role in organic carbon/nitrogen export in the Southern Ocean.
IMPORTANCE The biological carbon pump is crucial for the export of particulate organic matter in the ocean. Recent studies on marine microbes have shown the profound influence of bacteria and archaea as regulators of particulate organic matter export. Yet, despite the importance of the Southern Ocean as a carbon sink, we lack comparable insights regarding microbial contributions. This study provides the first insights regarding prokaryotic contributions to particulate organic matter export in the Southern Ocean. We reveal evidence that prokaryotic communities in suspended and sinking particle fractions harbor widespread genomic potential for mediating particulate organic matter export. The results substantially enhance our understanding of the role played by microorganisms in regulating particulate organic matter export in suspended and sinking marine fractions in the Southern Ocean.
KEYWORDS: Southern Ocean, carbon export, functional capacity, marine fractions, Marine Snow Catcher, metagenomics, particulate organic matter, prokaryotes
INTRODUCTION
The Southern Ocean plays a significant role in carbon cycling, buffering the impacts of climate change by accounting for 50% of the total oceanic uptake of CO2 (1–3). Phytoplankton primary production and carbon export to the deep ocean (i.e., the biological carbon pump [BCP]) (4, 5), are considered a major contributor to the sink of natural CO2, removing approximately 33% of the global organic carbon flux as particulate organic carbon (POC) (6, 7). However, only a small fraction of the organic carbon fixed by phytoplankton in surface waters ultimately reaches the ocean interior (8, 9). The factors that control the fraction of production, which is exported, or how effectively this material is transferred to depth remain unclear. Factors that regulate phytoplankton growth, particle formation, rates of sinking, and remineralization all modify the extent to which fixed POC is effectively exported and transformed into dissolved organic carbon (DOC) and CO2. Altogether, these factors determine the efficiency of the BCP (10, 11).
Particulate organic matter (POM), derived from phytoplankton, in addition to detritus and zooplankton fecal pellets, typically aggregates at the surface and may be separated into suspended and sinking particles based on their sinking velocity (12–14). The particle composition of the suspended and sinking fractions is highly variable in time and space, with different efficiencies in POM export and attenuation (4, 15). Earlier studies focused on the role of sinking particles and their importance in enhancing POM export (16). Recently, the suspended particles have also been observed in mesopelagic sediment traps (17), suggesting that they play a large role in exporting carbon to depth by physical processes such as the particle injection pumps (e.g., ocean mixing and migrant pump) (18, 19). In addition, the suspended particles are produced at depth from degradation of large sinking particles (20). Therefore, both particle types (suspended and sinking) are important for determining the role of the BCP in exporting POM to depth (14, 17, 21, 22).
Marine prokaryotes (bacteria and archaea), in particular those associated with suspended and sinking particles, have been shown to mediate key processes linked to the BCP (23–26). Prokaryotic diversity influences the composition of DOC, which includes a diverse range of molecules, which may be biologically labile (e.g., amino acids and glucose). These molecules are rapidly remineralized by microbes in the surface ocean to produce dissolved inorganic carbon (DIC), thus reducing export efficiencies (the microbial loop) (24, 27). Alternatively, prokaryotic-produced recalcitrant dissolved organic carbon (RDOC) (e.g., lignin and lipids) may be exported to the deep ocean, facilitating longer-term storage (28–30). A small percentage of prokaryotic DOC production is refractory (rDOC) (e.g., ~5 to 7% derived from glucose), which resists rapid remineralization and further degradation (31–34). The rDOM produced by prokaryotic degradation of complex organic carbon accumulates in the ocean interior, accounting for >95% of the large DOC pool (35, 36). This long-lived reservoir plays an important role in shaping global climate by sequestering CO2 from the atmosphere (37). Prokaryotic production of RDOC involves complex compounds that are difficult to manage, while rDOC represents labile compounds that are resistant to further degradation; both of these components of DOC form part of the microbial carbon pump (MCP) (34). The MCP sequestrates organic carbon by aiding the transfer of DOC to the deep ocean (38–40). Indeed, in instances where the microbial loop dominates (i.e., a system with small-celled, nonsinking particles and low POC flux) the MCP can be considered the prevailing mechanism for carbon sequestration (37). Despite the intricate role of prokaryote diversity and activity in regulating both the BCP and MCP, we lack genomic information regarding the phylogeny and function of prokaryotes linked with suspended and sinking particle-pools in the ocean.
The composition of the suspended and sinking particle pool (either labile, semi-labile, semi-recalcitrant, or recalcitrant) (41) may also determine the change in phylogenetic and genomic potential of the prokaryotic community (42, 43). Prokaryotes associated with suspended and sinking particles utilize and/or change in response to labile and semi-labile POM (44), in addition to having the genomic potential to slowly degrade complex high-molecular-weight organic compounds such as carbohydrates or lipids from phytoplankton cells (45). As such, genomic potential differentiation is expected in prokaryotic distribution based on their ability to degrade particulate organic matter (POM) (46) in either the suspended or the sinking particle pool. In addition, the prokaryotic community can both attach and detach from particles, thus having the capacity to enrich the surrounding water with free-living (FL) prokaryotes with the genomic potential to degrade complex material (42).
Here, we present the first assessment of prokaryotic genomic potential in suspended and sinking marine particle fractions collected with a Marine Snow Catcher (MSC) at five stations from the Southern Ocean Time Series (SOTS) site in the sub-Antarctic zone (SAZ) during austral autumn (Fig. 1). In addition to determining organic carbon and nitrogen flux, we specifically elucidate carbon and nitrogen cycling metabolic pathways linked to prokaryotes collected from the suspended and sinking particle fractions. By using metagenome-assembled genomes (MAGs) to link bacterial and archaeal genomes to POM sequestration, we provide insights into the role of prokaryotic community in organic matter export in the Southern Ocean.
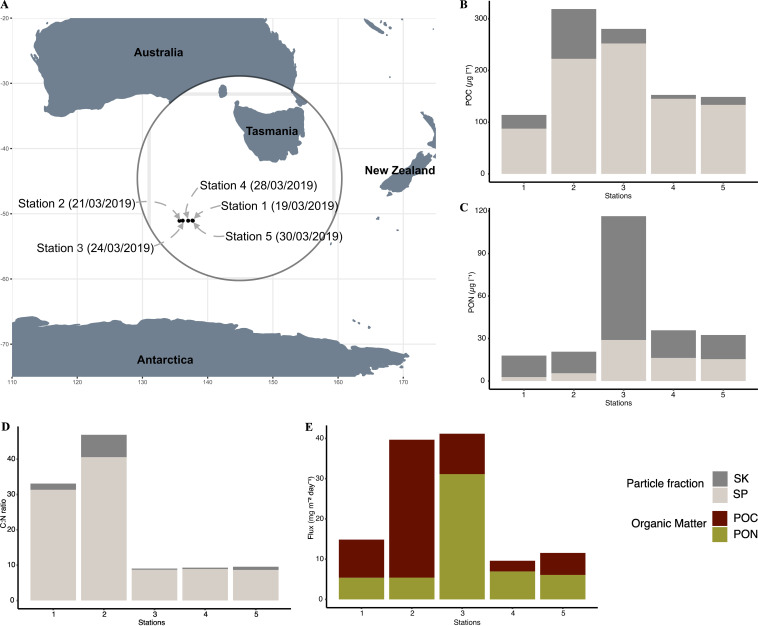
FIG 1.
Marine Snow Catcher (MSC) deployment at Southern Ocean Time Series sites during the IN2019_V02 using RV Investigator collecting POC and PON from the suspended (SP) and sinking (SK) particle pool. (A) The sampling locations at the SOTS site stations 1 and 5 are slightly different in latitude and longitude (see Data Set S1, Tab 1). (B) POC concentrations for SP (light gray bars) and SK (dark gray bars); (C) PON concentration for SP and SK; (D) POC/PON ratio for SP and SK; (E) POC and PON export flux at five stations.
(Tab 1) Detailed sampling of the Marine Snow Catcher from five Southern Ocean Time Series (SOTS) stations, including the date, time, and depth where the sample was collected. Sampling stations are numbered based on sampling date and time. (Tab 2) Predicted functional genes from unbinned bacterial contigs with gene description, module/pathways, and header. Each unbinned contig has a number that represent gene counts or copies. Gene counts greater than 1 are colored pink, while the gene counts equal to 0 are colored white. (Tab 3) Predicted functional genes from unbinned archaea contigs with gene description, module/pathways, and header. Each unbinned contig has a number that represents gene counts or copies. Gene counts greater than 1 are colored pink, while gene counts equal to 0 are colored white. (Tab 4) Classification of reconstructed MAGs from GTDB-Tk with completeness, contamination percentages, GC content, genome size, and number of rRNA genes. (Tab 5) Average nucleotide score of the reconstructed MAGs against RefSeq obtained from NCBI. A score above 95% represents similar taxa. (Tab 6) Predicted functional genes from reconstructed bacterial MAGs with gene description, module/pathways, and header. Each MAG has a number that represent gene counts or copies. Gene counts greater than 1 are colored pink, while gene counts equal to 0 are colored white. (Tab 7) Predicted functional genes from reconstructed archaeal MAGs with gene description, module/pathways, and header. Each MAG has a number that represent gene counts or copies. Gene counts greater than 1 are colored pink, while gene counts equal to 0 are colored white. Download Data Set S1, XLSX file, 0.1 MB (127.9KB, xlsx) .
Copyright © 2023 Dithugoe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RESULTS
Taxonomic profiling of raw reads.
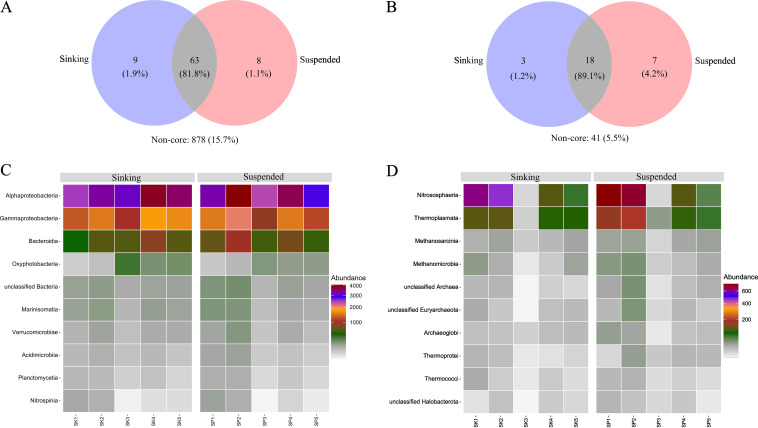
To elucidate bacterial and archaeal communities in suspended and sinking particle fractions, we assigned taxonomy to raw metagenomic reads. The operational taxonomic units (OTUs) within the top frequency cutoff of 80% were regarded as core OTUs, while those below 80% were excluded from further analysis as non-core OTUs. Additionally, core OTUs, with abundances below 0.1%, were excluded from downstream analysis. Bacterial communities shared 81.8% OTUs, while approximately 1.1% (suspended) and 1.9% (sinking) were unique, and 15.7% were non-core OTUs (Fig. 2A). Similarly, the archaea shared 89.1% OTUs, while ~4.2% (suspended) and about 1.2% (sinking) were unique, and 5.5% were non-core OTUs (Fig. 2B). Classification at the class level revealed that bacterial communities were dominated by Alphaproteobacteria, Gammaproteobacteria, and Bacteroidia in all stations for both the sinking and suspended fractions (Fig. 2C). Additionally, Pelagibacteraceae (Alphaproteobacteria) and Flavobacteriaceae (Bacteroidia), at the family level, were the most dominant taxa at all station in both suspended and sinking particle-pools (see Fig. S2A in the supplemental material). Very little difference was observed in bacterial community distribution when comparing the suspended and sinking particle-pools across all stations at the class level. However, at the family level, bacterial communities were diverse, with Nitrospinaceae (Nitrospina) only represented at station 1 and Cyanobiaceae (Oxyphotobacteria) only found at stations 3, 4, and 5. Interstation differences, at both class and family levels, were more apparent in the distribution of archaeal lineages (Fig. 2D; Fig. S2B). In general, archaeal communities were less dominant than bacteria at all stations. We observed Nitrososphaeria (Nitrosopumilaceae), Thermoplasmata (Thermoplasmata and Thermoplasmataceae), and Methanosarcinia (unclassified Methanosarcinia) in the suspended and sinking fractions (Fig. 2D; Fig. S2B). Stations 1 and 2 were dominated by Nitrosophaeria, Thermoplasmata, and Methanomicrobia. The station 3 suspended fraction was dominated by Thermoplasmata, Thermoprotei, and Methanomicrobia, while the sinking fraction was dominated by Methanosarcinia, Archaeoglobi, and Thermoplasmata. Stations 4 and 5 were also dominated by members of Nitrosophaeria, Thermoplasmata, and Methanosarcinia. Similar to bacterial communities, very few differences were observed when comparing archaea in the suspended and sinking particle-pools, at both class and family levels, with the exception of samples from station 3. The archaeal community at station 3 showed the most diverse composition and displayed the largest difference between suspended and sinking fractions.
FIG 2.
Taxonomic composition and distribution of the Southern Ocean Time Series prokaryotic communities, determined using single-copy marker genes (ribosomal protein genes) with the SingleM pipeline. (A) Venn diagram showing the core OTUs shared by suspended and sinking bacterial taxonomic composition; (B) Venn diagram showing the core OTU percentage of read abundance shared by suspended and sinking archaeal taxonomic composition; (C) heat map showing the percentages of abundance of the bacterial class composition in the suspended and sinking fractions at each station. Taxa with low abundance are colored blue, and those in higher abundance are white, while the highest are red. (D) Heat map showing the percentages of abundance of the archaeal class in the suspended and sinking communities at each station.
Ancillary data from the Southern Ocean Time Series sites obtained from CTD data. (A) Temperature (degree Celsius) and salinity plot showing the five sampling stations. The colored circles represent the depths at which the samples were collected. (B) Chlorophyll (milligrams per cubic meter) depth profile for five stations determined by CTD fluorescence sensor and (C) the MLD integrated chlorophyll (milligrams per square meter). Download FIG S1, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2023 Dithugoe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Overview of the prokaryotic community composition for five stations at the class and family level for the SP and SK particle-pool. The color scale represents read abundance. (A) Bacterial community composition; (B) archaeal community composition at both the class and family levels. Download FIG S2, TIF file, 1.9 MB (1.9MB, tif) .
Copyright © 2023 Dithugoe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Functional annotation of unbinned metagenomic contigs.
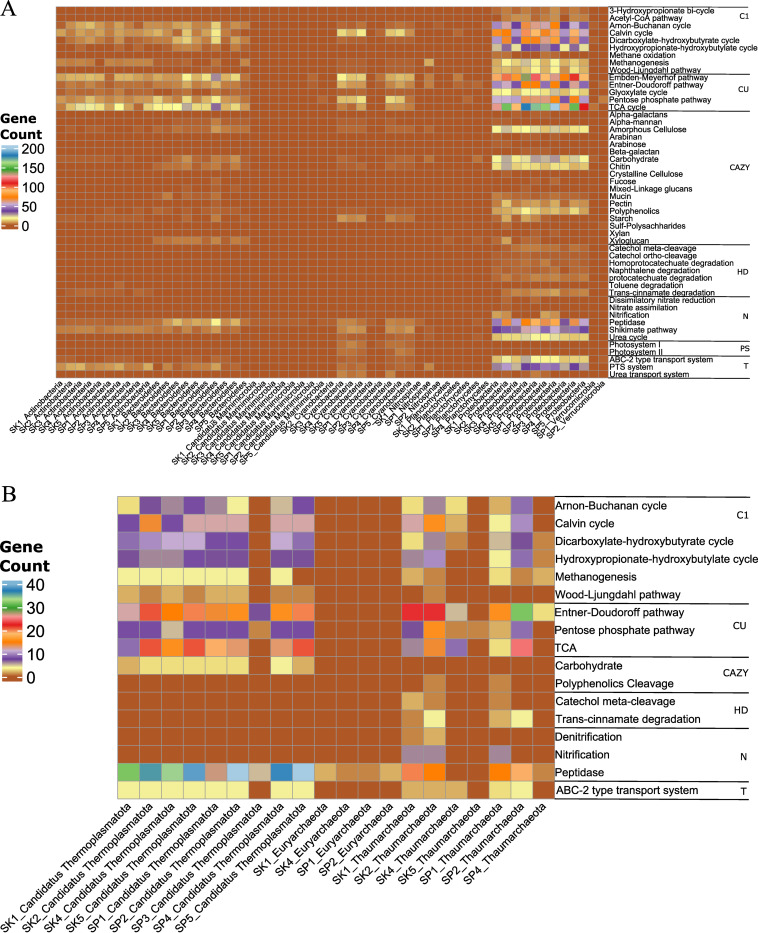
Proteobacteria, the most dominant bacterial phyla (see Text S1 in the supplemental material), were predicted to possess more genes with genomic potential for degrading high-molecular weight organic compounds (CAZymes and hydrocarbon degradation) (Fig. 3A). Several bacteria, recovered from all stations, including members of the Actinobacteria, had more genes involved in the degradation of carbohydrates (acetyl xylan esterase), amorphous cellulose (β-glucosidase), and chitin (lysozyme type G), with more gene copies in the sinking fraction—mostly at stations 1 and 2 (see Data Set S1, Tab 2, in the supplemental material). Interestingly, only archaea from the class Thaumarchaeota and “Candidatus Thermoplasmatota” had more gene copies involved in the degradation of high-molecular-weight compounds such as carbohydrates (esterases), polyphenolics (laccase/p-diphenol:oxygen oxidoreductase), catechol (4-oxalocrotonate tautomerase), and trans-cinnamate (3-phenylpropionate/cinnamic acid dioxygenase). These archaea were found mostly in samples retrieved from suspended and sinking fractions of stations 1 and 2, respectively (Fig. 3B; Data Set S1, Tab 3).
FIG 3.
Predicted genes for both complex, central, and chemoautotrophic prokaryotic community from unbinned contigs based on the DRAM tool in both the suspended (SP) and sinking (SK) particle pool. The predicted genes are involved in chemoautotrophs (C1), carbon utilization (CU), carbohydrate active enzymes (CAZymes [CAZY]), hydrocarbon degradation (HD), nitrogen metabolism (N), photosynthesis (PS), and transporter systems (T). The color scale represents predicted gene counts, including gene copies. Detailed gene names and copies are given in Data Set S1: Tab 2 for bacteria and Tab 3 for archaea. (A) Bacterial community functional annotation; (B) archaeal community functional annotation.
Detailed methodology and results section. The section includes additional data collected from a conductivity, temperature, and depth (CTD) instrument and the Marine Snow Catcher (MSC). The methods include further information regarding sample site descriptions, MSC deployment, and the sequencing protocols. The supplemental results section outlines the analyzed data from ancillary data, including water mass, phytoplankton biomass, and the particulate organic carbon (PON) and particulate organic nitrogen (PON) concentrations. This section also includes details regarding the taxonomic profile and genomic potential of unbinned metagenomic contigs, as well as the reconstructed metagenome-assembled genomes (MAGs). Download Text S1, DOCX file, 0.03 MB (33.1KB, docx) .
Copyright © 2023 Dithugoe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Archaea had a higher diversity of genes implicated in organic nitrogen degradation (e.g., protein degradation to amino acids) than bacteria. These genes include amino-, carboxy-, endo-, exo-, and isopeptidases, and most were ascribed to bacteria in suspended fractions (Data Set S1, Tab 3). Bacteria had more aminopeptidase and carboxypeptidase genes, mostly in the sinking fraction, and endopeptidase genes were found mainly in the suspended fraction (Data Set S1, Tab 2). On the other hand, archaea in stations 1 (both fractions) and 2 (sinking fraction) had genes involved in nitrification (ammonia monooxygenase). In addition to these stations, samples from the sinking fraction had several genes involved in denitrification (nitrite reductase). We identified genes implicated in the storage of nitrogen from bacterial taxa. These genes include those involved in the Shikimate pathway (3-deoxy-7-phosphoheptulonate synthase, chorismite synthase, and shikimate kinase) and urea cycle (argininosuccinate lyase and synthase). In general, we found more metabolic genes in the sinking fraction (Fig. 3A).
Moreover, bacterial and archaeal contigs from all stations were predicted to have genes involved in CO2 fixation and to produce RDOC via chemoautotrophic pathways (Text S1). These chemoautotrophic pathways, and related gene copies, were higher at stations 1 and 2 (suspended), station 3 (both suspended and sinking), and stations 4 and 5 (sinking). Our analyses suggest that bacteria harbored more genes or chemoautotroph-related pathways than archaea. In addition, these bacteria had several transporter systems, including ABC-2 type (ABC-2.A/P), phosphoenolpyruvate(PEP):carbohydrate phosphotransferase (PTS; PTS-Gut-EIIA) and urea (urtA-E) transport systems in both size fractions. In contrast, archaea had genes involved in only the ABC-2 type (ABC-2.A/P) transport system, mostly in the sinking fraction.
Functional profiling of bacterial and archaeal genomes from the suspended and sinking particle-pools.
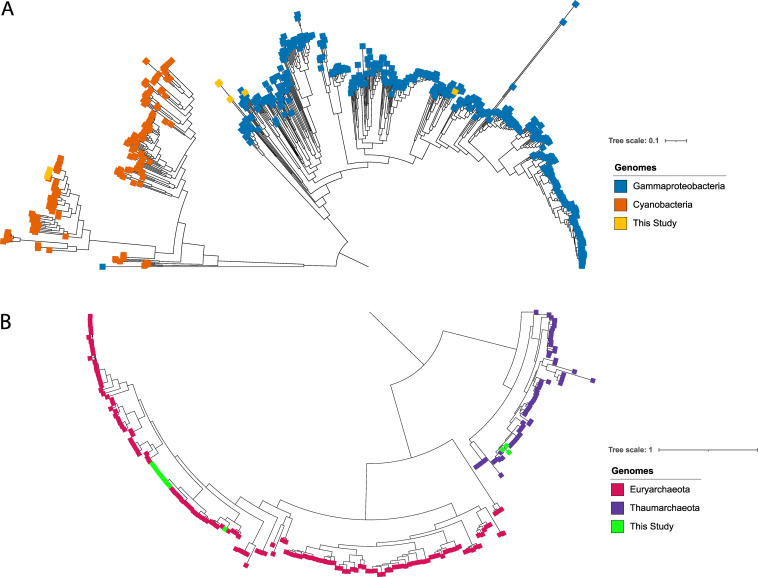
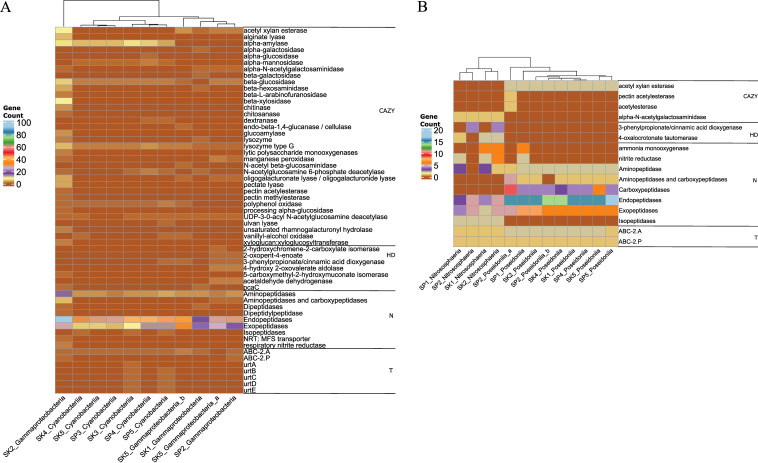
The average nucleotide identity (ANI) scores of all 11 bacterial and 13 archaeal MAGs were below 90% against 1,254 bacterial (Cyanobacteriia and Gammaproteobacteria) (Fig. 4A) and 4,957 archaeal (Thermoplasmatota and Thaumarchaeota) (Fig. 4B) RefSeq complete genomes, respectively. Furthermore, the genomic potential suggests that bacterial genomes reconstructed from the sinking fraction had more CAZymes involved in the degradation of carbohydrates (acetyl xylan esterase), starch (α-amylase), amorphous cellulose (β-glucosidase), and chitin (lysozyme type G). The analyses revealed that gammaproteobacterial MAGs, from station 2, had more CAZymes (Fig. 5A; Data Set S1, Tab 6). We also found that cyanobacterial MAGs appear to have unique CAZymes, including genes involved in the degradation of α-mannan (α-mannosidase), arabinan (dextranase), and sulf-polysaccharides (ulvan lyase). These CAZymes were mostly retrieved in samples from the suspended fraction. Bacterial genomes from station 2 (mostly suspended fraction) and station 5 (mostly sinking fraction) had genes involved in the degradation of hydrocarbon compounds, including catechol (2-oxopent-4-enoate), naphthalene (2-hydroxychromene-2-carboxylate isomerase), and trans-cinnamate (4-hydroxy 2-oxovalerate aldolase). On the other hand, Poseidoniia harbored CAZymes involved in the degradation of carbohydrates (acetyl xylan esterase). Only one genome (SP2_Poseidoniia_a) had CAZyme genes, including those coding for pectin acetylesterase and acetylesterase (Fig. 5B; Data Set S1, Tab 7), whereas, Nitrososphaeria MAGs had CAZyme genes and hydrocarbon degradation genes involved in the degradation of mucin (α-N-acetylgalactosaminidase), catechol (4-oxalocrotonate tautomerase), and trans-cinnamate (3-phenylpropionate/cinnamic acid dioxygenase). These were mostly recovered from the station 1 and 2 sinking fractions, respectively.
FIG 4.
Phylogenomic inference of our 24 MAGs. The phylogenomic tree was based on alignment of 40% of the marker gene present in our MAGs. (A) Bacterial MAGs (yellow) against Gammaproteobacteria (blue) and Cyanobacteria (orange); (B) archaeal MAGs (green) against Euryarchaeota (red) and Thaumarchaeota (purple).
FIG 5.
Predicted and identified genes from the 24 MAGs involved in carbohydrate-active enzymes (CAZymes [CAZY]), hydrocarbon degradation (HD), nitrogen metabolism (N), and transporter systems (T) based on the DRAM pipeline. The color scale represents the gene counts/copies. (A) Bacterial functional annotation; (B) archaeal functional annotation.
A Gammaproteobacteria genome affiliated with Alteromonadaceae, from the sinking fraction of station 2, had a respiratory nitrite reductase gene. This gene plays a key role in dissimilatory nitrate reduction. However, several bacterial MAGs had genes involved in the degradation of proteins to amino acids using aminopeptidases and endo- and exopeptidases. All archaeal MAGs had genes involved in the degradation of peptides, including amino-, carboxy-, endo-, and exopeptidases. Samples from stations 1 and 2 (both fractions) had nitrite reductase genes involved in the denitrification process and also had ammonia monooxygenase genes implicated in nitrification—mostly in the sinking fraction. Evidence suggests that these bacterial and archaeal genomes may be chemoautotrophs (Text S1). All bacterial and the archaeal genome (Poseidoniia) had genes implicated in photorespiration via the glycine cleavage system, whereas only Cyanobacteria MAGs were photoautotrophs with genes related to photosynthesis (psa A-F and psb A-F genes) (Fig. S4; Data Set S1, Tabs 6 and 7). On the other hand, all bacterial and archaeal genomes had the ABC-2.A transporter system (Fig. 5; Data Set S1, Tabs 6 and 7). Additionally, gammaproteobacterial genomes from station 2 (suspended and sinking fractions) and all archaeal MAGs had the ABC-2.P transporter system (Fig. 5; Data Set S1, Tabs 6 and 7). Only Cyanobacterial MAGs, from the station 3 sinking fraction and station 5 suspended fraction, had genes implicated in the urea transport system (urtA-E) (Fig. 5A; Data Set S1, Tab 6).
Taxonomic classification of the prokaryotic community from unbinned contigs at the phylum level. The color scale represents read abundance. (A) Bacterial community; (B) archaeal community presented as relative abundance. Download FIG S3, TIF file, 0.9 MB (914KB, tif) .
Copyright © 2023 Dithugoe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Predicted genes involved in carbon utilization and chemoautotrophic genomic potential. The color scale represents gene counts/copies. (A) Bacterial central metabolism and electron transport chain (ETC) complexes; (B) Archaeal central metabolism and ETC complexes. Download FIG S4, TIF file, 1.7 MB (1.7MB, tif) .
Copyright © 2023 Dithugoe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Organic carbon and nitrogen are the main resource supporting diverse microbial communities in both suspended and sinking particle-pools (42). The microbial genomic potential subsequently alters the nature of both the particulate and dissolved organic pools. Microbial communities, therefore, contribute to the quantity and quality of organic carbon/nitrogen that is effectively exported below the seasonal mixed layer. Knowledge of prokaryotic community composition and their genomic potential on both sinking and suspended particles is thus required to facilitate a mechanistic understanding of prokaryotic contributions to POC/particulate organic nitrogen (PON) degradation/synthesis, which impacts the efficiency of the BCP. While limited to 24 MAGs, this study addresses this knowledge gap by providing the first metagenome-assembled genomes from suspended and sinking particle-pools in the Southern Ocean. The limited number of stations does, however, constrain our ability to interpret the complete functional potential within the context of POC/PON flux variability at the SOTS.
Differences in prokaryotes may explain divergence in POC and PON contents of sinking and suspended particle-pools.
There is a complex interplay among several factors (e.g., particle composition, sinking rates and prokaryotic activity, etc.) that influences the flux of POM from the surface layer (47, 48). For example, recent studies have demonstrated a positive relationship between phytoplankton biomass and the magnitude of POM export (49, 50), with our data corroborating these findings. In addition, POM content (labile, semi-labile, recalcitrant, or refractory) rather than concentration is considered the main driver of prokaryotic community structure (51, 52), with sinking POM subject to degradation by several FL and particle-attached (PA) prokaryotes (44) that alter its chemical and biological properties (53). Prokaryotic communities (i.e., PA and FL) may in turn contribute as secondary drivers of change, by altering their community structure and/or associated function in response to the altered POM (38, 54, 55). For example, prokaryotes can degrade polysaccharides from POM into labile or semi-labile DOM, while also producing RDOM or rDOM (30, 34). The RDOM compounds act as “sticky polysaccharides,” which then aggregate into POM (28, 30, 56).
Since sinking POC flux is insufficient to meet the carbon demands of prokaryotes, suspended particles are considered a major sustaining source of organic carbon for microbes in the mesopelagic (44). While the concentration of POC in sinking particles decreases exponentially with depth, the concomitant POC concentration in suspended particles remains largely constant and is typically ~1 to 2 orders of magnitude higher than that of sinking particles (44). Our data support these findings, with substantially higher POC concentrations in the suspended than the sinking fraction. This was, however, not true for PON, which typically had more PON (or similar concentrations) in the sinking material than in the suspended material.
Several factors may account for variability in POC and PON concentration, which alters POC/PON ratios below or above the Redfield ratio (6.6) (57). Factors that increase POC/PON ratios include (i) preferential degradation of nitrogen-rich POM, (ii) synthesis of refractory POC resistant to further degradation, and (iii) chemoautotrophic microbial activity on POM, including exopolymer substances (EPSs). Whereas factors that decrease POC/PON include (i) the presence of PA diazotrophs (N2-fixing prokaryotes) (58, 59), (ii) oxidation of sinking POC that drives preferential reduction in POC relative to PON, and (iii) nutrient limitation (60). Despite large differences in the distribution of POC/PON ratios between suspended and sinking samples and between stations, the bacterial community compositions were very similar. This similarity at the class level suggests that variability in POC/PON ratios may be influenced by prokaryotic functional potential. On the other hand, differences were observed in archaeal communities at the class level, such that PON contents might have been different between stations. Alternatively, the compositions of source material may have been similar, while degradation by bacteria and archaea may have driven secondary changes in their community structure at the family level (25, 38, 61). There is evidence for this argument in the abundance of bacteria from the Proteobacteria and Bacteroidota phyla, which suggests variability in bacterial potential between suspended and sinking pools, and between stations, despite similarities in community composition at the class level. As such, differences in genomic potential rather than diversity, particularly in the case of bacteria, might impact the signature of POC and PON (62–64) in both sinking and suspended material at the class level. Nevertheless, the impact of archaeal diversity on POC/PON variability is evident when observing Nitrososphaeria (Nitrosopumilaceae), which were highest at stations with the highest POC/PON ratios and POC flux. This is likely due to Nitrososphaeria having more genes coding for peptidases that degrade polypeptides (reducing PON concentration) releasing ammonia, which was further oxidized to nitrous oxide nitrification (nitrite reductase) and denitrification (ammonia monooxygenase) processes. Conversely, station 3 had the lowest abundance of Nitrososphaeria, coincident with particularly high PON flux relative to other stations.
Prokaryotic genomic potential based on POC and PON content.
Despite extensive studies that focus on prokaryotes from sinking particles from sediment traps (14, 65–67), few focus on prokaryotes from suspended and sinking particles collected from an MSC (42, 44). Based on amplicon sequencing data, M. T. Duret et al. (44) reported niche differentiation between sinking and suspended pools below the epipelagic zone (44). However, prokaryotes may detach from sinking particles, thereby enriching the suspended particle pool with microbes that are similar to those found on sinking particles (42). Our data suggest clear functional differences in PA and FL prokaryotes associated with suspended and sinking POM. For instance, Proteobacteria (Gammaproteobacteria) MAGs and Bacteroidota unbinned contigs had the genomic potential to degrade polysaccharides produced by phytoplankton with specific CAZymes, which is in line with previous findings (68), while archaeal functional predictions suggest capacity to degrade proteins, secondary metabolites, and carbohydrates from POM, also in agreement with previous reports (69). Therefore, despite the phylogenetic similarities between our MAGs, there is evidence that prokaryotes exhibit diverse functional traits. Most specifically, we found higher potential for degrading complex organic carbon in the sinking fraction, consistent with previous reports (67). In addition, taxonomic classification from unbinned contigs and MAGs suggests that the bacterial community (e.g., Proteobacteria) may be more prevalent in the sinking particle pool, with more genes involved in the degradation of complex POM. This particular finding is in contrast to previous studies, which have instead shown these degradation genes to be more abundant in the suspended fraction (44). A. O. Leu et al. (67) reported that PA microbes had more diverse CAZymes, extracellular peptidases, and substrate-specific transporters than their FL counterparts. That said, it is likely that any genomic potential to use labile DOM, resulting in the formation of polymers, may consequently initiate aggregation (38), which may subsequently enhance POM export flux, thus accounting for the presence of PA and FL prokaryotes in both sinking and suspended material. Additionally, our results suggests that labile DOM (e.g., glucose) uptake, via the PTS-EIIA transport system (70), might be used in the synthesis of building blocks for key biopolymers (71) that are then exported out of the cell using the ABC2 transport system (72). In such instances, bacterial communities associated with sinking POM may lead to the formation and release of complex organic compounds (i.e., RDOM) from labile DOM (14, 62). Bacteroidota contigs and gammaproteobacterial MAGs from this study appear to be more prevalent in the sinking particle pool, which is in agreement with previous reports (14, 73). On the other hand, the sinking POM may be colonized by FL prokaryotes, which use oligosaccharides and other organic compounds released from particle degradation (74, 75).
Prokaryotic genomes and unbinned contigs in both suspended and sinking fractions revealed genomic capacity to fix CO2, including phosphoenolpyruvate carboxylase (Arnon-Buchanan cycle), ribulose-bisphosphate carboxylase (Calvin cycle) and acetyl coenzyme A (acetyl-CoA) synthetase (methanogenesis). Prokaryotes that exhibit this genomic potential are typically chemoautotrophs, which synthesize polysaccharide polymers (e.g., RDOM) from CO2 (76) to form smaller particles secreted through the ABC2 transport system (72, 77). These small particles are produced at depth via RDOM aggregation or the disaggregation of larger particles (20) and may form part of the particle injection pump through physical processes (18, 19). In addition to fixing CO2, the members of the PA chemoautotrophic bacterial community resemble FL bacteria; however, this is not surprising since bacteria constantly attach or detach from particles enriching either the particle or suspended pool with chemoautotrophic taxa that degrade high-molecular-weight compounds (42, 78).
Indeed, there has been some debate regarding the similarity of FL and PA prokaryotic communities, with some studies suggesting that these taxa may be similar (79, 80), while others suggest stark dissimilarities (81, 82). These characteristics may be affected by many factors, including particle sources and substrate availability (83), particle residence times, geographic location (84), and nutrient gradients (85). Here, we collected metagenomic samples in both suspended and sinking fractions to elucidate both FL and PA prokaryotic contributions to BCP efficiency. Communities from suspended and sinking particles showed some evidence of chemoautotrophic potential, which is consistent with previous findings on FL prokaryotes (86). These chemoautotrophic communities also have the genetic capacity to produce labile DOC, thereby sustaining the microbial food web when organic carbon content in the water column is mostly rDOM (87, 88). The chemoautotrophic responses to carbon sources (either inorganic or organic carbon), due to environmental and biological stimuli, nonetheless remain unclear, with ongoing studies needed to investigate the fate of the POM produced by chemoautotrophs (87).
There was also a discernible difference in genomic potentials between prokaryotic communities from MAGs associated with suspended material and those associated with sinking material (notably at station 2). For instance, bacteria from the Bacteroidetes and Proteobacteria phyla (Gammaproteobacteria MAGs) were present in both sinking and suspended samples at station 2, and both possessed various CAZyme genes. However, there were more genes involved in the degradation of labile and complex POM found in the sinking pool. In addition to the gene copies being higher, we found several genes implicated in diatom-derived POM (89, 90), grass POM, and virus-induced POC from picocyanobacterial and polysaccharides (91) in the sinking pool. It is expected that these mechanisms may result in reduced POC flux via particle degradation while sinking into the mesopelagic. All bacterial communities from both MAGs and unbinned contigs were associated with the degradation of chitin, regardless of their association with suspended or sinking material. Chitin is rich in both carbon and nitrogen that can be reintegrated into biomass-forming polysaccharide polymers or remineralized to enrich the water column (92), thus reducing the export flux of both POC and PON.
In addition, the taxonomic classification of prokaryotes and genomic potential (on either the suspended or sinking fraction) was insufficient to infer preference, due to functional evolution involving gene gain and loss (20, 93). Gene gain and loss may occur through the deletion or insertion of genes, including genomic islands, via nonhomologous recombination mechanisms and mobile genetic elements (94). For instance, the rate of bacterial growth and organic matter consumption of ~0.1 to 0.7 day−1 (95, 96) occurs within the settling time of 0.833 day−1 for the separation of particles. This may explain the difference observed in genomic potential between closely related taxa (Cyanobiaceae), complicating the changes observed in bacterial genomic potential in suspended and sinking fractions (14, 26).
Our chemoautotrophic archaeal MAGs from Marine Group I (e.g., Nitrososphaeria), specifically those associated with Nitrosopumilaceae, were prevalent at stations 1 and 2 while virtually absent in both suspended and sinking fractions at stations 4 and 5. Chemoautotrophic archaeal MAGs are predicted to have genes involved in the degradation of hydrocarbons (4-oxalocrotonate tautomerase), notably in the sinking particle pool (97), which was indeed the case for our samples at stations 1 and 2. This was in-line with predicted archaeal MAG distribution known to be prevalent in the sinking particle-pool scavenging complex POM (26, 67, 98). Previous studies have also shown that archaeal communities may colonize and degrade complex POM (99, 100), with their genomic potential thus more commonly appearing in the sinking particle-pool. Marine Group II (e.g., Poseidoniia), associated with Thalassoarchaeaceae, are also predicted to play a major role in PON transformations based on the protein degradation pathways recovered (100, 101). These heterotrophs typically use low-molecular-weight PON (102) and, in this study, were more dominant in the suspended particle pool. Our results suggest that Poseidoniia use peptidases to degrade protein to amino acids, while Nitrososphaeria utilize ammonia released from the degradation of amino acids, which are subsequently converted to nitrous oxide and removed from the system, thereby reducing PON flux at stations 1 and 2. There is, however, no direct evidence (e.g., mechanistic experiments or metatranscriptomic data) of the role of archaea in utilizing PON (e.g., protein) and interactions with RDOC in the ocean.
Ammonia-oxidizing archaeal (AOA) MAGs with the ammonia monooxygenase gene were present in the sinking fraction, at stations 1 and 2 (Nitrososphaeria), and the suspended particles, at station 1 (Poseidoniia). The AOA have the capacity to use labile and complex organic nitrogen as their main source of ammonia and nitrite and may therefore influence the POC/PON ratio (103). These AOA harbor peptidases that enable the degradation of nitrogen-rich POM, resulting in low concentrations of PON relative to POC in the suspended fraction. Several studies have shown that members of Marine Group I typically dominate the particle-associated AOA community (104) and are involved in the uptake and assimilation of ammonia (98) and the release of DON via the degradation of particles (105, 106). However, Proteobacteria contigs and MAGs from the sinking samples at station 2 had the genomic potential for dissimilatory nitrate reduction via respiratory nitrite reductase. Interestingly, this pathway was also found at the sinking particle fraction at station 5, from unbinned Proteobacteria contigs. Proteobacteria from unbinned contigs and gammaproteobacterial MAGs indicate potential genetic capacity for decreasing nitrogen-rich POM, thus potentially driving an elevated POC/PON ratio. Nitrite reduction (nitrite reductase) was also more prevalent in archaeal than bacterial MAGs. The presence of AOA (Nitrososphaeria and Poseidoniia), nitrite-oxidizing bacteria (NOB) (Gammaproteobacteria), and nitrite-oxidizing archaea (NOA) (Nitrososphaeria) MAGs at station 1 (suspended and sinking) and station 2 (suspended) had the highest POC/PON ratio, the highest POC flux, and the lowest PON flux. These NOB/NOA and AOA are obligatory partners where the AOA catalyze the oxidized ammonia released from PON to nitrite and NOB/NOA, which further oxidize the nitrate to nitrate (107), thus possibly explaining the decrease in PON flux at these stations. NOB/NOA and AOA are also key players in the removal of nitrogen from PON, increasing the POC/PON ratio at stations 1 and 2 and subsequently increasing POC export flux relative to PON. In addition to preferential degradation of PON, archaeal MAGs may also be involved in the synthesis and secretion of RDOC via ABC2.A and P transport systems, enriching the water column with organic carbon particles, thus increasing POC relative to PON flux (103).
Although Cyanobacteria (Cyanobiaceae) are well-known photosynthetic microbes involved in nitrogen fixation (45, 108), our unbinned contigs and MAGs showed no evidence for nitrogen fixation, with no diazotrophic Cyanobacteria. This is not unexpected as many Cyanobiaceae (Synechococcus) do not have genes for nitrogen fixation (109–111). Nonetheless, noncyanobacterial diazotrophs (NCDs), such as dinitrogen (N2)-fixing bacteria and archaea, might be present (112). Indeed, Gammaproteobacteria (on the sinking sample at station 2) were the only bacterial MAG containing nitrogen metabolism, while Nitrososphaeria were also present at stations 1 and 2. The Poseidoniia MAG at station 1 (suspended) also had the genomic potential for nitrogen metabolism. Coincidentally, these were the two stations with the highest POC/PON ratio (and highest carbon flux), indicative of preferential nitrogen uptake by the prokaryotic community. Since phytoplankton biomass accounts for only ~6% of the nitrogen flux, the high PON flux observed at stations 3 to 5 may be due to prokaryotic activity on PON by assimilating the available inorganic nitrogen into its biomass (113). The dissimilation of inorganic nitrogen, from PON (114), favors PON export and is thus more likely to be a result of prokaryotic activity.
Conclusion.
Particulate organic matter (POM) signatures in suspended and sinking particles are influenced by both bacterial and archaeal communities as PA and FL, respectively. Data from genomic potential suggest that the variability in archaeal communities can impact PON flux, whereas the bacterial communities may impact POC flux. Archaeal MAGs were consistent with the predicted dominance of taxa known to scavenge PON in the suspended particle-pool. The relationship between phytoplankton and prokaryotes in POM export is complex. Mechanistic studies may shed light regarding the precise mechanisms governing the relationship between phytoplankton and prokaryotes. These studies may provide further insights into trophic interactions between phytoplankton and prokaryotes and their respective contribution to POM export.
MATERIALS AND METHODS
Site description and Marine Snow Catcher sample collection.
The Southern Ocean Time Series (SOTS) site is located at 47°S and 142°E, ~530 km southwest of Tasmania, in the Indian/Australian sector of the SAZ (115). Samples were collected from five Marine Snow Catcher (MSC) stations (Fig. 1A) over the course of 2 weeks between March and April 2019 aboard the RV Investigator. The Marine Snow Catcher (MSC) was deployed at 10 m below the mixed-layer depth (MLD) (see Data Set S1, Tab 1, in the supplemental material) following the method described by J. S. Riley et al. (12) (see Text S1 in the supplemental material). The POC/PON samples collected from the MSC were analyzed in the Department of Archaeology at the University of Cape Town. The POC/PON concentration determination and flux calculations were adjusted based on J. S. Riley et al. (12) (Text S1).
Molecular analysis and sequencing.
To explore the composition and function of the entire prokaryotic community associated with the suspended and sinking particles, including the FL community, 2 L of water from both fractions was filtered using 0.2-μm-pore-size polycarbonate membrane filters (47-mm diameter); Millipore (Burlington, MA, USA) (115). The filters were stored at −80°C until further processing. DNA was extracted using the PowerSoil kit (Qiagen, Hilden, Germany) as described by M. Hirai et al. (116). The resultant DNA was sequenced by Admera Health Biopharma Services (South Plainfield, NJ, USA) (Text S1).
Taxonomic classification and MAG reconstruction.
The quality of raw metagenomic data was assessed using FastQC (https://github.com/s-andrews/FastQC). The reads were processed to remove sequencing adapters and low-quality reads using Trimmomatic v.0.36 (117). These reads were used for taxonomic classification, using the default parameters in SingleM v.0.13.2, concentrating on 14 single-copy marker genes, ribosomal protein genes for differentiation of species (https://github.com/wwood/singlem). The ATLAS workflow (118) was used to assemble raw reads and for generating MAGs using the default parameter settings. Unbinned metagenomic contigs with lengths of ≥2.5 kb were assigned taxonomy using Contig Annotation Tool (CAT) with the default settings (119). The top phyla from CAT output were extracted and concatenated for DRAM annotation. CheckM v.1.1.3 was used to assess the quality of MAGs as detailed previously (120). Following genome reporting standards, MAGs with genome completeness scores of >50% and <10% contamination were selected for downstream analysis (121). About 24 of our MAGs were medium-quality drafts with ≥50% completeness and <10% contamination. The Genome Taxonomy Database toolkit (GTDB-Tk) v.1.5.0 was used to assign taxonomy to all MAGs. Phylogenetic diversity of the reconstructed MAGs was inferred against complete archaeal and bacterial genomes acquired from the NCBI RefSeq database using the FastANI v.1.32 tool (122). To estimate the abundance of each taxon, MAGs from the suspended and sinking particle-pools were mapped using default parameters in CoverM v.0.6.1 (https://github.com/wwood/CoverM). Gene calling of the unbinned metagenomic contigs retrieved from CAT and the MAGs was obtained by using DRAM v.1.2.0 with 6 databases, including UniRef90, Pfam, dbCAN, RefSeq viral, VOGDB, and MEROPS, with E value of <10−15 (123).
Data availability.
All ten raw metagenomic sequences collected from the Southern Ocean Time Series stations are available at the NCBI (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA749920).
ACKNOWLEDGMENTS
We thank Australia’s Integrated Marine Observing System (IMOS), which is enabled by the National Collaborative Research Infrastructure Strategy (NCRIS). We also acknowledge Philip Boyd (Institute for Marine and Antarctic Studies, University of Tasmania), Thomas Trull (CSIRO, Tasmania, Australia), and Mathew Bressac (Institute for Marine and Antarctic Studies, University of Tasmania) for organizing the sampling of the MSC. In addition, we acknowledge Laique Djeutchouang, Natasha Van Horsten, and Thomas Ryan-Keogh for assistance with Python scripts. We acknowledge the Technology Innovation Agency for supporting the sequencing costs. We thank the Centre for High Performance Computing (Cape Town, South Africa) and the Centre for Bioinformatics and Computational Biology, University of Pretoria, for providing computational resources. We thank the Technology Innovation Agency (TIA) for supporting this project.
The research voyage was operated by a consortium of institutions as an unincorporated joint venture, with the University of Tasmania as Lead Agent. The MSC sampling at SOTS was made possible by support to Philip Boyd (Institute for Marine and Antarctic Studies, University of Tasmania), Thomas Trull (CSIRO, Tasmania, Australia), and Mathew Bressac (Institute for Marine and Antarctic Studies, University of Tasmania) using the RV Investigator IN2019_V02 (GIpr08) research ship. This work was supported through the CSIR’s Southern Ocean and Carbon Climate Observatory (SOCCO) Program (http://socco.org.za/) funded by the Department of Science and Innovation (DST/CON 0182/2017) and the CSIR’s Parliamentary Grant, together with National Research Foundation (NRF) UID ID 110729, 110717, 129225, 136491, and 148867. These funding bodies were responsible for funding the design of the study, data collection, analysis, and interpretation and supported the authors through the manuscript preparation.
S.J.T. and T.P.M. conceived the study, supervised the research, and funded the analysis. C.D.D. performed the experiments, analyzed the data, contributed to experimental design, and wrote the manuscript. O.K.I.B. assisted with the bioinformatic analysis. E.L.C., W.P.F., and S.J.T. contributed to the analysis of oceanographic data. All authors edited, read, and approved the final version of the manuscript.
We declare no conflict of interest.
Contributor Information
Sandy J. Thomalla, Email: sandy.thomalla@gmail.com.
Thulani P. Makhalanyane, Email: thulani.makhalanyane@up.ac.za.
Barbara J. Campbell, Clemson University Department of Biological Sciences
REFERENCES
- 1.DeVries T, Holzer M, Primeau F. 2017. Recent increase in oceanic carbon uptake driven by weaker upper-ocean overturning. Nature 542:215–218. doi: 10.1038/nature21068. [DOI] [PubMed] [Google Scholar]
- 2.Friedlingstein P, Jones MW, O'Sullivan M, Andrew RM, Hauck J, Peters GP, Peters W, Pongratz J, Sitch S, Le Quéré C, Bakker DCE, Canadell JG, Ciais P, Jackson RB, Anthoni P, Barbero L, Bastos A, Bastrikov V, Becker M, Bopp L, Buitenhuis E, Chandra N, Chevallier F, Chini LP, Currie KI, Feely RA, Gehlen M, Gilfillan D, Gkritzalis T, Goll DS, Gruber N, Gutekunst S, Harris I, Haverd V, Houghton RA, Hurtt G, Ilyina T, Jain AK, Joetzjer E, Kaplan JO, Kato E, Klein Goldewijk K, Korsbakken JI, Landschützer P, Lauvset SK, Lefèvre N, Lenton A, Lienert S, Lombardozzi D, Marland G, et al. 2019. Global carbon budget 2019. Earth Syst Sci Data 11:1783–1838. doi: 10.5194/essd-11-1783-2019. [DOI] [Google Scholar]
- 3.Castillo DJ, Dithugoe CD, Bezuidt OK, Makhalanyane TP. 2022. Microbial ecology of the Southern Ocean. FEMS Microbiol Ecol 98:fiac123. doi: 10.1093/femsec/fiac123. [DOI] [PubMed] [Google Scholar]
- 4.Boyd PW, Claustre H, Levy M, Siegel DA, Weber T. 2019. Multi-faceted particle pumps drive carbon sequestration in the ocean. Nature 568:327–335. doi: 10.1038/s41586-019-1098-2. [DOI] [PubMed] [Google Scholar]
- 5.Henson S, Le Moigne F, Giering S. 2019. Drivers of carbon export efficiency in the global ocean. Global Biogeochem Cycles 33:891–903. doi: 10.1029/2018GB006158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlitzer R. 2002. Carbon export fluxes in the Southern Ocean: results from inverse modeling and comparison with satellite-based estimates. Deep Sea Res Part 2 Top Stud Oceanogr 49:1623–1644. doi: 10.1016/S0967-0645(02)00004-8. [DOI] [Google Scholar]
- 7.Siegenthaler U, Sarmiento JL. 1993. Atmospheric carbon dioxide and the ocean. Nature 365:119–125. doi: 10.1038/365119a0. [DOI] [Google Scholar]
- 8.Martin JH, Knauer GA, Karl DM, Broenkow WW. 1987. Vertex: carbon cycling in the Northeast Pacific. Deep Sea Res Part A Oceanogr Res Pap 34:267–285. doi: 10.1016/0198-0149(87)90086-0. [DOI] [Google Scholar]
- 9.Giering SL, Sanders R, Lampitt RS, Anderson TR, Tamburini C, Boutrif M, Zubkov MV, Marsay CM, Henson SA, Saw K, Cook K, Mayor DJ. 2014. Reconciliation of the carbon budget in the ocean's twilight zone. Nature 507:480–483. doi: 10.1038/nature13123. [DOI] [PubMed] [Google Scholar]
- 10.De La Rocha CL, Passow U. 2007. Factors influencing the sinking of POC and the efficiency of the biological carbon pump. Deep Sea Res Part 2 Top Stud Oceanogr 54:639–658. doi: 10.1016/j.dsr2.2007.01.004. [DOI] [Google Scholar]
- 11.Talmy D, Martiny AC, Hill C, Hickman AE, Follows MJ. 2016. Microzooplankton regulation of surface ocean POC:PON ratios. Global Biogeochem Cycles 30:311–332. doi: 10.1002/2015GB005273. [DOI] [Google Scholar]
- 12.Riley JS, Sanders R, Marsay C, Le Moigne FAC, Achterberg EP, Poulton AJ. 2012. The relative contribution of fast and slow sinking particles to ocean carbon export. Global Biogeochem Cycles doi: 10.1029/2011GB004085. [DOI] [Google Scholar]
- 13.Arístegui J, Gasol JM, Duarte CM, Herndld GJ. 2009. Microbial oceanography of the dark ocean's pelagic realm. Limnol Oceanogr 54:1501–1529. doi: 10.4319/lo.2009.54.5.1501. [DOI] [Google Scholar]
- 14.Boeuf D, Edwards BR, Eppley JM, Hu SK, Poff KE, Romano AE, Caron DA, Karl DM, DeLong EF. 2019. Biological composition and microbial dynamics of sinking particulate organic matter at abyssal depths in the oligotrophic open ocean. Proc Natl Acad Sci USA 116:11824–11832. doi: 10.1073/pnas.1903080116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner JT. 2015. Zooplankton fecal pellets, marine snow, phytodetritus and the ocean’s biological pump. Prog Oceanogr 130:205–248. doi: 10.1016/j.pocean.2014.08.005. [DOI] [Google Scholar]
- 16.Fowler SW, Knauer GA. 1986. Role of large particles in the transport of elements and organic compounds through the oceanic water column. Prog Oceanogr 16:147–194. doi: 10.1016/0079-6611(86)90032-7. [DOI] [Google Scholar]
- 17.Durkin CA, Estapa ML, Buesseler KO. 2015. Observations of carbon export by small sinking particles in the upper mesopelagic. Mar Chem 175:72–81. doi: 10.1016/j.marchem.2015.02.011. [DOI] [Google Scholar]
- 18.Omand MM, D'Asaro EA, Lee CM, Perry MJ, Briggs N, Cetinic I, Mahadevan A. 2015. Eddy-driven subduction exports particulate organic carbon from the spring bloom. Science 348:222–225. doi: 10.1126/science.1260062. [DOI] [PubMed] [Google Scholar]
- 19.Stukel MR, Aluwihare LI, Barbeau KA, Chekalyuk AM, Goericke R, Miller AJ, Ohman MD, Ruacho A, Song H, Stephens BM, Landry MR. 2017. Mesoscale ocean fronts enhance carbon export due to gravitational sinking and subduction. Proc Natl Acad Sci USA 114:1252–1257. doi: 10.1073/pnas.1609435114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Briggs N, Dall'Olmo G, Claustre H. 2020. Major role of particle fragmentation in regulating biological sequestration of CO2 by the oceans. Science 367:791–793. doi: 10.1126/science.aay1790. [DOI] [PubMed] [Google Scholar]
- 21.Durkin CA, Buesseler KO, Cetinic I, Estapa ML, Kelly RP, Omand M. 2021. A visual tour of carbon export by sinking particles. Global Biogeochem Cycles 35:e2021GB006985. doi: 10.1029/2021GB006985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Estapa M, Buesseler K, Durkin CA, Omand M, Benitez-Nelson CR, Roca-Martí M, Breves E, Kelly RP, Pike S. 2021. Biogenic sinking particle fluxes and sediment trap collection efficiency at Ocean Station Papa. Elementa Sci Anthropocene 9:00122. doi: 10.1525/elementa.2020.00122. [DOI] [Google Scholar]
- 23.Cavan EL, Le Moigne FAC, Poulton AJ, Tarling GA, Ward P, Daniels CJ, Fragoso GM, Sanders RJ. 2015. Attenuation of particulate organic carbon flux in the Scotia Sea, Southern Ocean, is controlled by zooplankton fecal pellets. Geophys Res Lett 42:821–830. doi: 10.1002/2014GL062744. [DOI] [Google Scholar]
- 24.Azam F, Smith DC, Hollibaugh JT. 2016. The role of the microbial loop in Antarctic pelagic ecosystems. Polar Res 10:239–244. doi: 10.3402/polar.v10i1.6742. [DOI] [Google Scholar]
- 25.Manganelli M, Malfatti F, Samo TJ, Mitchell BG, Wang H, Azam F. 2009. Major role of microbes in carbon fluxes during Austral winter in the Southern Drake Passage. PLoS One 4:e6941. doi: 10.1371/journal.pone.0006941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poff KE, Leu AO, Eppley JM, Karl DM, DeLong EF. 2021. Microbial dynamics of elevated carbon flux in the open ocean’s abyss. Proc Natl Acad Sci USA 118:e2018269118. doi: 10.1073/pnas.2018269118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fenchel T. 2008. The microbial loop—25 years later. J Exp Mar Biol Ecol 366:99–103. doi: 10.1016/j.jembe.2008.07.013. [DOI] [Google Scholar]
- 28.Romera-Castillo C, Álvarez M, Pelegrí JL, Hansell DA, Álvarez-Salgado XA. 2019. Net additions of recalcitrant dissolved organic carbon in the deep Atlantic Ocean. Global Biogeochem Cycles 33:1162–1173. doi: 10.1029/2018GB006162. [DOI] [Google Scholar]
- 29.Ki B, Park S, Choi JH. 2014. Production of recalcitrant organic matter under the influence of elevated carbon dioxide and temperature. Water Environ Res 86:779–787. doi: 10.2175/106143014x13975035526347. [DOI] [PubMed] [Google Scholar]
- 30.Goto S, Tada Y, Suzuki K, Yamashita Y. 2020. Evaluation of the production of dissolved organic matter by three marine bacterial strains. Front Microbiol 11:584419. doi: 10.3389/fmicb.2020.584419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogawa H, Amagai Y, Koike I, Kaiser K, Benner R. 2001. Production of refractory dissolved organic matter by bacteria. Science 292:917–920. doi: 10.1126/science.1057627. [DOI] [PubMed] [Google Scholar]
- 32.Koch BP, Kattner G, Witt M, Passow U. 2014. Molecular insights into the microbial formation of marine dissolved organic matter: recalcitrant or labile? Biogeosciences 11:4173–4190. doi: 10.5194/bg-11-4173-2014. [DOI] [Google Scholar]
- 33.Gruber DF, Simjouw JP, Seitzinger SP, Taghon GL. 2006. Dynamics and characterization of refractory dissolved organic matter produced by a pure bacterial culture in an experimental predator-prey system. Appl Environ Microbiol 72:4184–4191. doi: 10.1128/AEM.02882-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landry Z, Swan BK, Herndl GJ, Stepanauskas R, Giovannoni SJ. 2017. SAR202 genomes from the dark ocean predict pathways for the oxidation of recalcitrant dissolved organic matter. mBio 8:e00413-17. doi: 10.1128/mBio.00413-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen CT. 2011. Microbial carbon pump: additional considerations. Nat Rev Microbiol 9:555. doi: 10.1038/nrmicro2386-c4. [DOI] [PubMed] [Google Scholar]
- 36.Jiao N, Herndl GJ, Hansell DA, Benner R, Kattner G, Wilhelm SW, Kirchman DL, Weinbauer MG, Luo T, Chen F, Azam F. 2010. Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat Rev Microbiol 8:593–599. doi: 10.1038/nrmicro2386. [DOI] [PubMed] [Google Scholar]
- 37.Zhang C, Dang H, Azam F, Benner R, Legendre L, Passow U, Polimene L, Robinson C, Suttle CA, Jiao N. 2018. Evolving paradigms in biological carbon cycling in the ocean. Natl Sci Rev 5:481–499. doi: 10.1093/nsr/nwy074. [DOI] [Google Scholar]
- 38.Jiao N, Robinson C, Azam F, Thomas H, Baltar F, Dang H, Hardman-Mountford NJ, Johnson M, Kirchman DL, Koch BP, Legendre L, Li C, Liu J, Luo T, Luo YW, Mitra A, Romanou A, Tang K, Wang X, Zhang C, Zhang R. 2014. Mechanisms of microbial carbon sequestration in the ocean—future research directions. Biogeosciences 11:5285–5306. doi: 10.5194/bg-11-5285-2014. [DOI] [Google Scholar]
- 39.Jiao N, Herndl GJ, Hansell DA, Benner R, Kattner G, Wilhelm SW, Kirchman DL, Weinbauer MG, Luo T, Chen F, Azam F. 2011. The microbial carbon pump and the oceanic recalcitrant dissolved organic matter pool. Nat Rev Microbiol 9:555. doi: 10.1038/nrmicro2386-c5. [DOI] [PubMed] [Google Scholar]
- 40.Legendre L, Rivkin RB, Weinbauer MG, Guidi L, Uitz J. 2015. The microbial carbon pump concept: potential biogeochemical significance in the globally changing ocean. Prog Oceanogr 134:432–450. doi: 10.1016/j.pocean.2015.01.008. [DOI] [Google Scholar]
- 41.Kharbush JJ, Close HG, Van Mooy BAS, Arnosti C, Smittenberg RH, Le Moigne FAC, Mollenhauer G, Scholz-Böttcher B, Obreht I, Koch BP, Becker KW, Iversen MH, Mohr W. 2020. Particulate organic carbon deconstructed: molecular and chemical composition of particulate organic carbon in the ocean. Front Mar Sci 7:518. doi: 10.3389/fmars.2020.00518. [DOI] [Google Scholar]
- 42.Baumas CMJ, Le Moigne FAC, Garel M, Bhairy N, Guasco S, Riou V, Armougom F, Grossart HP, Tamburini C. 2021. Mesopelagic microbial carbon production correlates with diversity across different marine particle fractions. ISME J 15:1695–1708. doi: 10.1038/s41396-020-00880-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu S, Deng Y, Jiang Z, Wu Y, Huang X, Macreadie PI. 2020. Nutrient loading diminishes the dissolved organic carbon drawdown capacity of seagrass ecosystems. Sci Total Environ 740:140185. doi: 10.1016/j.scitotenv.2020.140185. [DOI] [PubMed] [Google Scholar]
- 44.Duret MT, Lampitt RS, Lam P. 2019. Prokaryotic niche partitioning between suspended and sinking marine particles. Environ Microbiol Rep 11:386–400. doi: 10.1111/1758-2229.12692. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Zhang Y, Li Y, Luo Y-L, Pan Y-R, Liu J, Butler D. 2021. Alkaline environments benefit microbial K-strategists to efficiently utilize protein substrate and promote valorization of protein waste into short-chain fatty acids. Chem Eng J 404:127147. doi: 10.1016/j.cej.2020.127147. [DOI] [Google Scholar]
- 46.Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, Kassabgy M, Huang S, Mann AJ, Waldmann J, Weber M, Klindworth A, Otto A, Lange J, Bernhardt J, Reinsch C, Hecker M, Peplies J, Bockelmann FD, Callies U, Gerdts G, Wichels A, Wiltshire KH, Glockner FO, Schweder T, Amann R. 2012. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336:608–611. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- 47.Garcia CA, Baer SE, Garcia NS, Rauschenberg S, Twining BS, Lomas MW, Martiny AC. 2018. Nutrient supply controls particulate elemental concentrations and ratios in the low latitude eastern Indian Ocean. Nat Commun 9:4868. doi: 10.1038/s41467-018-06892-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moutin T, Raimbault P. 2002. Primary production, carbon export and nutrients availability in western and eastern Mediterranean Sea in early summer 1996 (MINOS cruise). J Mar Syst 33-34:273–288. doi: 10.1016/S0924-7963(02)00062-3. [DOI] [Google Scholar]
- 49.Dybwad C, Assmy P, Olsen LM, Peeken I, Nikolopoulos A, Krumpen T, Randelhoff A, Tatarek A, Wiktor JM, Reigstad M. 2021. Carbon export in the seasonal sea ice zone north of Svalbard from winter to late summer. Front Mar Sci doi: 10.3389/fmars.2020.525800. [DOI] [Google Scholar]
- 50.Hung J-J, Tung C-H, Lin Z-Y, Chen Y-l, Peng S-H, Lin Y-H, Tsai L-S. 2021. Active and passive fluxes of carbon, nitrogen, and phosphorus in the northern South China Sea. Biogeosciences 18:5141–5162. doi: 10.5194/bg-18-5141-2021. [DOI] [Google Scholar]
- 51.Hernandez-Magana AE, Liu Y, Debeljak P, Crispi O, Marie B, Koedooder C, Obernosterer I. 2021. Prokaryotic diversity and activity in contrasting productivity regimes in late summer in the Kerguelen region (Southern Ocean). J Mar Syst 221:103561. doi: 10.1016/j.jmarsys.2021.103561. [DOI] [Google Scholar]
- 52.Liu S, Parsons R, Opalk K, Baetge N, Giovannoni S, Bolaños LM, Kujawinski EB, Longnecker K, Lu Y, Halewood E, Carlson CA. 2020. Different carboxyl-rich alicyclic molecules proxy compounds select distinct bacterioplankton for oxidation of dissolved organic matter in the mesopelagic Sargasso Sea. Limnol Oceanogr 65:1532–1553. doi: 10.1002/lno.11405. [DOI] [Google Scholar]
- 53.Alldredge AL, Silver MW. 1988. Characteristics, dynamics and significance of marine snow. Prog Oceanogr 20:41–82. doi: 10.1016/0079-6611(88)90053-5. [DOI] [Google Scholar]
- 54.Kamalanathan M, Doyle SM, Xu C, Achberger AM, Wade TL, Schwehr K, Santschi PH, Sylvan JB, Quigg A. 2020. Exoenzymes as a signature of microbial response to marine environmental conditions. mSystems 5:e00290-20. doi: 10.1128/mSystems.00290-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manna V, Malfatti F, Banchi E, Cerino F, De Pascale F, Franzo A, Schiavon R, Vezzi A, Del Negro P, Celussi M. 2020. Prokaryotic response to phytodetritus-derived organic material in epi- and mesopelagic Antarctic waters. Front Microbiol 11:1242. doi: 10.3389/fmicb.2020.01242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hwang J, Druffel ERM, Eglinton TI. 2010. Widespread influence of resuspended sediments on oceanic particulate organic carbon: insights from radiocarbon and aluminum contents in sinking particles. Global Biogeochem Cycles doi: 10.1029/2010GB003802. [DOI] [Google Scholar]
- 57.Redfield AC. 2014. Eighty years of Redfield. Nat Geosci 7:849. [Google Scholar]
- 58.Farnelid H, Turk-Kubo K, Ploug H, Ossolinski JE, Collins JR, Van Mooy BAS, Zehr JP. 2019. Diverse diazotrophs are present on sinking particles in the North Pacific Subtropical Gyre. ISME J 13:170–182. doi: 10.1038/s41396-018-0259-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chakraborty S, Andersen KH, Visser AW, Inomura K, Follows MJ, Riemann L. 2021. Quantifying nitrogen fixation by heterotrophic bacteria in sinking marine particles. Nat Commun 12:4085. doi: 10.1038/s41467-021-23875-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taucher J, Boxhammer T, Bach LT, Paul AJ, Schartau M, Stange P, Riebesell U. 2020. Changing carbon-to-nitrogen ratios of organic-matter export under ocean acidification. Nat Clim Chang 11:52–57. doi: 10.1038/s41558-020-00915-5. [DOI] [Google Scholar]
- 61.Bacosa HP, Kamalanathan M, Chiu MH, Tsai SM, Sun L, Labonte JM, Schwehr KA, Hala D, Santschi PH, Chin WC, Quigg A. 2018. Extracellular polymeric substances (EPS) producing and oil degrading bacteria isolated from the northern Gulf of Mexico. PLoS One 13:e0208406. doi: 10.1371/journal.pone.0208406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilson B, Müller O, Nordmann E-L, Seuthe L, Bratbak G, Øvreås L. 2017. Changes in marine prokaryote composition with season and depth over an Arctic polar year. Front Mar Sci doi: 10.3389/fmars.2017.00095. [DOI] [Google Scholar]
- 63.Yamada N, Fukuda H, Ogawa H, Saito H, Suzumura M. 2012. Heterotrophic bacterial production and extracellular enzymatic activity in sinking particulate matter in the western North Pacific Ocean. Front Microbiol 3:379. doi: 10.3389/fmicb.2012.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caruso G, Madonia A, Bonamano S, Miserocchi S, Giglio F, Maimone G, Azzaro F, Decembrini F, La Ferla R, Piermattei V, Piazzolla D, Marcelli M, Azzaro M. 2020. Microbial abundance and enzyme activity patterns: response to changing environmental characteristics along a transect in Kongsfjorden (Svalbard Islands). J Mar Sci Eng 8:824. doi: 10.3390/jmse8100824. [DOI] [Google Scholar]
- 65.Belcher A, Iversen M, Giering S, Riou V, Henson SA, Berline L, Guilloux L, Sanders R. 2016. Depth-resolved particle-associated microbial respiration in the northeast Atlantic. Biogeosciences 13:4927–4943. doi: 10.5194/bg-13-4927-2016. [DOI] [Google Scholar]
- 66.Belcher A, Iversen M, Manno C, Henson SA, Tarling GA, Sanders R. 2016. The role of particle associated microbes in remineralization of fecal pellets in the upper mesopelagic of the Scotia Sea, Antarctica. Limnol Oceanogr 61:1049–1064. doi: 10.1002/lno.10269. [DOI] [Google Scholar]
- 67.Leu AO, Eppley JM, Burger A, DeLong EF. 2022. Diverse genomic traits differentiate sinking-particle-associated versus free-living microbes throughout the oligotrophic open ocean water column. mBio 13:e01569-22. doi: 10.1128/mbio.01569-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Unfried F, Becker S, Robb CS, Hehemann JH, Markert S, Heiden SE, Hinzke T, Becher D, Reintjes G, Kruger K, Avci B, Kappelmann L, Hahnke RL, Fischer T, Harder J, Teeling H, Fuchs B, Barbeyron T, Amann RI, Schweder T. 2018. Adaptive mechanisms that provide competitive advantages to marine bacteroidetes during microalgal blooms. ISME J 12:2894–2906. doi: 10.1038/s41396-018-0243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jain A, Krishnan KP. 2021. Marine Group-II archaea dominate particle-attached as well as free-living archaeal assemblages in the surface waters of Kongsfjorden, Svalbard, Arctic Ocean. Antonie Van Leeuwenhoek 114:633–647. doi: 10.1007/s10482-021-01547-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deutscher J, Ake FM, Derkaoui M, Zebre AC, Cao TN, Bouraoui H, Kentache T, Mokhtari A, Milohanic E, Joyet P. 2014. The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol Mol Biol Rev 78:231–256. doi: 10.1128/MMBR.00001-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pontiller B, Martinez-Garcia S, Lundin D, Pinhassi J. 2020. Labile dissolved organic matter compound characteristics select for divergence in marine bacterial activity and transcription. Front Microbiol 11:588778. doi: 10.3389/fmicb.2020.588778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cuthbertson L, Kos V, Whitfield C. 2010. ABC transporters involved in export of cell surface glycoconjugates. Microbiol Mol Biol Rev 74:341–362. doi: 10.1128/MMBR.00009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martinez-Garcia M, Swan BK, Poulton NJ, Gomez ML, Masland D, Sieracki ME, Stepanauskas R. 2012. High-throughput single-cell sequencing identifies photoheterotrophs and chemoautotrophs in freshwater bacterioplankton. ISME J 6:113–123. doi: 10.1038/ismej.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gralka M, Szabo R, Stocker R, Cordero OX. 2020. Trophic interactions and the drivers of microbial community assembly. Curr Biol 30:R1176–R1188. doi: 10.1016/j.cub.2020.08.007. [DOI] [PubMed] [Google Scholar]
- 75.Ebrahimi A, Schwartzman J, Cordero OX. 2019. Cooperation and spatial self-organization determine rate and efficiency of particulate organic matter degradation in marine bacteria. Proc Natl Acad Sci USA 116:23309–23316. doi: 10.1073/pnas.1908512116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kusch S, Wakeham SG, Dildar N, Zhu C, Sepulveda J. 2021. Bacterial and archaeal lipids trace chemo(auto)trophy along the redoxcline in Vancouver Island fjords. Geobiology 19:521–541. doi: 10.1111/gbi.12446. [DOI] [PubMed] [Google Scholar]
- 77.Silver RP, Prior K, Nsahlai C, Wright LF. 2001. ABC transporters and the export of capsular polysaccharides from Gram-negative bacteria. Res Microbiol 152:357–364. doi: 10.1016/s0923-2508(01)01207-4. [DOI] [PubMed] [Google Scholar]
- 78.Bachmann J, Heimbach T, Hassenruck C, Kopprio GA, Iversen MH, Grossart HP, Gardes A. 2018. Environmental drivers of free-living vs. particle-attached bacterial community composition in the Mauritania upwelling system. Front Microbiol 9:2836. doi: 10.3389/fmicb.2018.02836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ortega-Retuerta E, Joux F, Jeffrey WH, Ghiglione JF. 2013. Spatial variability of particle-attached and free-living bacterial diversity in surface waters from the Mackenzie River to the Beaufort Sea (Canadian Arctic). Biogeosciences 10:2747–2759. doi: 10.5194/bg-10-2747-2013. [DOI] [Google Scholar]
- 80.Hollibaugh JT, Wong PS, Murrell MC. 2000. Similarity of particle-associated and free-living bacterial communities in northern San Francisco Bay, California. Aquat Microb Ecol 21:103–114. doi: 10.3354/ame021103. [DOI] [Google Scholar]
- 81.Zhang Y, Xiao W, Jiao N. 2016. Linking biochemical properties of particles to particle-attached and free-living bacterial community structure along the particle density gradient from freshwater to open ocean. J Geophys Res Biogeosci 121:2261–2274. doi: 10.1002/2016JG003390. [DOI] [Google Scholar]
- 82.Rieck A, Herlemann DP, Jurgens K, Grossart HP. 2015. Particle-associated differ from free-living bacteria in surface waters of the Baltic Sea. Front Microbiol 6:1297. doi: 10.3389/fmicb.2015.01297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grossart H-P. 2010. Ecological consequences of bacterioplankton lifestyles: changes in concepts are needed. Environ Microbiol Rep 2:706–714. doi: 10.1111/j.1758-2229.2010.00179.x. [DOI] [PubMed] [Google Scholar]
- 84.Fuhrman JA, Steele JA, Hewson I, Schwalbach MS, Brown MV, Green JL, Brown JH. 2008. A latitudinal diversity gradient in planktonic marine bacteria. Proc Natl Acad Sci USA 105:7774–7778. doi: 10.1073/pnas.0803070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carlson CA, Morris R, Parsons R, Treusch AH, Giovannoni SJ, Vergin K. 2009. Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J 3:283–295. doi: 10.1038/ismej.2008.117. [DOI] [PubMed] [Google Scholar]
- 86.Robinson JJ, Cavanaugh CM. 1995. Expression of form I and form II Rubisco in chemoautotrophic symbioses: implications for the interpretation of stable carbon isotope values. Limnol Oceanogr 40:1496–1502. doi: 10.4319/lo.1995.40.8.1496. [DOI] [Google Scholar]
- 87.Middelburg JJ. 2011. Chemoautotrophy in the ocean. Geophys Res Lett doi: 10.1029/2011GL049725. [DOI] [Google Scholar]
- 88.Taylor GT, Iabichella M, Ho T-Y, Scranton MI, Thunell RC, Muller-Karger F, Varela R. 2001. Chemoautotrophy in the redox transition zone of the Cariaco Basin: a significant midwater source of organic carbon production. Limnol Oceanogr 46:148–163. doi: 10.4319/lo.2001.46.1.0148. [DOI] [Google Scholar]
- 89.Landa M, Cottrell MT, Kirchman DL, Kaiser K, Medeiros PM, Tremblay L, Batailler N, Caparros J, Catala P, Escoubeyrou K, Oriol L, Blain S, Obernosterer I. 2014. Phylogenetic and structural response of heterotrophic bacteria to dissolved organic matter of different chemical composition in a continuous culture study. Environ Microbiol 16:1668–1681. doi: 10.1111/1462-2920.12242. [DOI] [PubMed] [Google Scholar]
- 90.Baltar F, Herndl GJ. 2019. Ideas and perspectives: is dark carbon fixation relevant for oceanic primary production estimates? Biogeosciences 16:3793–3799. doi: 10.5194/bg-16-3793-2019. [DOI] [Google Scholar]
- 91.Zhao Z, Gonsior M, Schmitt-Kopplin P, Zhan Y, Zhang R, Jiao N, Chen F. 2019. Microbial transformation of virus-induced dissolved organic matter from picocyanobacteria: coupling of bacterial diversity and DOM chemodiversity. ISME J 13:2551–2565. doi: 10.1038/s41396-019-0449-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Beier S, Bertilsson S. 2013. Bacterial chitin degradation-mechanisms and ecophysiological strategies. Front Microbiol 4:149. doi: 10.3389/fmicb.2013.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Iranzo J, Wolf YI, Koonin EV, Sela I. 2019. Gene gain and loss push prokaryotes beyond the homologous recombination barrier and accelerate genome sequence divergence. Nat Commun 10:5376. doi: 10.1038/s41467-019-13429-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Koonin EV, Wolf YI. 2008. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res 36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Moran XAG, Garcia FC, Rostad A, Silva L, Al-Otaibi N, Irigoien X, Calleja ML. 2022. Diel dynamics of dissolved organic matter and heterotrophic prokaryotes reveal enhanced growth at the ocean's mesopelagic fish layer during daytime. Sci Total Environ 804:150098. doi: 10.1016/j.scitotenv.2021.150098. [DOI] [PubMed] [Google Scholar]
- 96.Kirchman DL. 2016. Growth rates of microbes in the oceans. Annu Rev Mar Sci 8:285–309. doi: 10.1146/annurev-marine-122414-033938. [DOI] [PubMed] [Google Scholar]
- 97.Liu J, Zheng Y, Lin H, Wang X, Li M, Liu Y, Yu M, Zhao M, Pedentchouk N, Lea-Smith DJ, Todd JD, Magill CR, Zhang WJ, Zhou S, Song D, Zhong H, Xin Y, Yu M, Tian J, Zhang XH. 2019. Proliferation of hydrocarbon-degrading microbes at the bottom of the Mariana Trench. Microbiome 7:47. doi: 10.1186/s40168-019-0652-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li J, Gu L, Bai S, Wang J, Su L, Wei B, Zhang L, Fang J. 2021. Characterization of particle-associated and free-living bacterial and archaeal communities along the water columns of the South China Sea. Biogeosciences 18:113–133. doi: 10.5194/bg-18-113-2021. [DOI] [Google Scholar]
- 99.Tully BJ. 2019. Metabolic diversity within the globally abundant Marine Group II Euryarchaea offers insight into ecological patterns. Nat Commun 10:271. doi: 10.1038/s41467-018-07840-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Orsi WD, Smith JM, Wilcox HM, Swalwell JE, Carini P, Worden AZ, Santoro AE. 2015. Ecophysiology of uncultivated marine euryarchaea is linked to particulate organic matter. ISME J 9:1747–1763. doi: 10.1038/ismej.2014.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Iverson V, Morris RM, Frazar CD, Berthiaume CT, Morales RL, Armbrust EV. 2012. Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science 335:587–590. doi: 10.1126/science.1212665. [DOI] [PubMed] [Google Scholar]
- 102.Alderkamp AC, Sintes E, Herndl GJ. 2006. Abundance and activity of major groups of prokaryotic plankton in the coastal North Sea during spring and summer. Aquat Microb Ecol 45:237–246. doi: 10.3354/ame045237. [DOI] [Google Scholar]
- 103.Kim JG, Gazi KS, Awala SI, Jung MY, Rhee SK. 2021. Ammonia-oxidizing archaea in biological interactions. J Microbiol 59:298–310. doi: 10.1007/s12275-021-1005-z. [DOI] [PubMed] [Google Scholar]
- 104.Cai X, Yao L, Hu Y, Jiang H, Shen M, Hu Q, Wang Z, Dahlgren RA. 2019. Particle-attached microorganism oxidation of ammonia in a hypereutrophic urban river. J Basic Microbiol 59:511–524. doi: 10.1002/jobm.201800599. [DOI] [PubMed] [Google Scholar]
- 105.Qin W, Amin SA, Martens-Habbena W, Walker CB, Urakawa H, Devol AH, Ingalls AE, Moffett JW, Armbrust EV, Stahl DA. 2014. Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc Natl Acad Sci USA 111:12504–12509. doi: 10.1073/pnas.1324115111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Alonso-Saez L, Waller AS, Mende DR, Bakker K, Farnelid H, Yager PL, Lovejoy C, Tremblay JE, Potvin M, Heinrich F, Estrada M, Riemann L, Bork P, Pedros-Alio C, Bertilsson S. 2012. Role for urea in nitrification by polar marine Archaea. Proc Natl Acad Sci USA 109:17989–17994. doi: 10.1073/pnas.1201914109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lam P, Kuypers MM. 2011. Microbial nitrogen cycling processes in oxygen minimum zones. Annu Rev Mar Sci 3:317–345. doi: 10.1146/annurev-marine-120709-142814. [DOI] [PubMed] [Google Scholar]
- 108.Momper LM, Reese BK, Carvalho G, Lee P, Webb EA. 2015. A novel cohabitation between two diazotrophic cyanobacteria in the oligotrophic ocean. ISME J 9:882–893. doi: 10.1038/ismej.2014.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Blank CE, Sanchez-Baracaldo P. 2010. Timing of morphological and ecological innovations in the cyanobacteria—a key to understanding the rise in atmospheric oxygen. Geobiology 8:1–23. doi: 10.1111/j.1472-4669.2009.00220.x. [DOI] [PubMed] [Google Scholar]
- 110.Chandra N, Mallick N. 2022. Co-production of bioethanol and commercially important exopolysaccharides from the marine cyanobacterium Synechococcus elongatus BDU10144 in a novel low-cost seawater-fertilizer-based medium. Intl J Energy Res 46:13487–13510. doi: 10.1002/er.8069. [DOI] [Google Scholar]
- 111.Dominguez-Martin MA, Lopez-Lozano A, Melero-Rubio Y, Gomez-Baena G, Jimenez-Estrada JA, Kukil K, Diez J, Garcia-Fernandez JM. 2022. Marine Synechococcus sp. strain WH7803 shows specific adaptative responses to assimilate nanomolar concentrations of nitrate. Microbiol Spectr 10:e00187-22. doi: 10.1128/spectrum.00187-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Moisander PH, Benavides M, Bonnet S, Berman-Frank I, White AE, Riemann L. 2017. Chasing after non-cyanobacterial nitrogen fixation in marine pelagic environments. Front Microbiol 8:1736. doi: 10.3389/fmicb.2017.01736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Polerecky L, Masuda T, Eichner M, Rabouille S, Vancová M, Kienhuis MVM, Bernát G, Bonomi-Barufi J, Campbell DA, Claquin P, Červený J, Giordano M, Kotabová E, Kromkamp J, Lombardi AT, Lukeš M, Prášil O, Stephan S, Suggett D, Zavřel T, Halsey KH. 2021. Temporal patterns and intra- and inter-cellular variability in carbon and nitrogen assimilation by the unicellular cyanobacterium Cyanothece sp. ATCC 51142. Front Microbiol 12:620915. doi: 10.3389/fmicb.2021.620915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mettam C, Zerkle AL, Claire MW, Prave AR, Poulton SW, Junium CK. 2019. Anaerobic nitrogen cycling on a Neoarchaean ocean margin. Earth Planet Sci Lett 527:115800. doi: 10.1016/j.epsl.2019.115800. [DOI] [Google Scholar]
- 115.Silva GG, Green KT, Dutilh BE, Edwards RA. 2016. SUPER-FOCUS: a tool for agile functional analysis of shotgun metagenomic data. Bioinformatics 32:354–361. doi: 10.1093/bioinformatics/btv584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hirai M, Nishi S, Tsuda M, Sunamura M, Takaki Y, Nunoura T. 2017. Library construction from subnanogram DNA for pelagic sea water and deep-sea sediments. Microbes Environ 32:336–343. doi: 10.1264/jsme2.ME17132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kieser S, Brown J, Zdobnov EM, Trajkovski M, McCue LA. 2020. ATLAS: a Snakemake workflow for assembly, annotation, and genomic binning of metagenome sequence data. BMC Bioinformatics 21:257. doi: 10.1186/s12859-020-03585-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.von Meijenfeldt FAB, Arkhipova K, Cambuy DD, Coutinho FH, Dutilh BE. 2019. Robust taxonomic classification of uncharted microbial sequences and bins with CAT and BAT. Genome Biol 20:217. doi: 10.1186/s13059-019-1817-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bowers RM, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Doud D, Reddy TBK, Schulz F, Jarett J, Rivers AR, Eloe-Fadrosh EA, Tringe SG, Ivanova NN, Copeland A, Clum A, Becraft ED, Malmstrom RR, Birren B, Podar M, Bork P, Weinstock GM, Garrity GM, Dodsworth JA, Yooseph S, Sutton G, Glockner FO, Gilbert JA, Nelson WC, Hallam SJ, Jungbluth SP, Ettema TJG, Tighe S, Konstantinidis KT, Liu WT, Baker BJ, Rattei T, Eisen JA, Hedlund B, McMahon KD, Fierer N, Knight R, Finn R, Cochrane G, Karsch-Mizrachi I, Tyson GW, Rinke C, Genome Standards Consortium, Lapidus A, Meyer F, Yilmaz P, Parks DH, et al. 2017. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol 35:725–731. doi: 10.1038/nbt.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jain C, Rodriguez RL, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shaffer M, Borton MA, McGivern BB, Zayed AA, La Rosa SL, Solden LM, Liu P, Narrowe AB, Rodriguez-Ramos J, Bolduc B, Gazitua MC, Daly RA, Smith GJ, Vik DR, Pope PB, Sullivan MB, Roux S, Wrighton KC. 2020. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res 48:8883–8900. doi: 10.1093/nar/gkaa621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(Tab 1) Detailed sampling of the Marine Snow Catcher from five Southern Ocean Time Series (SOTS) stations, including the date, time, and depth where the sample was collected. Sampling stations are numbered based on sampling date and time. (Tab 2) Predicted functional genes from unbinned bacterial contigs with gene description, module/pathways, and header. Each unbinned contig has a number that represent gene counts or copies. Gene counts greater than 1 are colored pink, while the gene counts equal to 0 are colored white. (Tab 3) Predicted functional genes from unbinned archaea contigs with gene description, module/pathways, and header. Each unbinned contig has a number that represents gene counts or copies. Gene counts greater than 1 are colored pink, while gene counts equal to 0 are colored white. (Tab 4) Classification of reconstructed MAGs from GTDB-Tk with completeness, contamination percentages, GC content, genome size, and number of rRNA genes. (Tab 5) Average nucleotide score of the reconstructed MAGs against RefSeq obtained from NCBI. A score above 95% represents similar taxa. (Tab 6) Predicted functional genes from reconstructed bacterial MAGs with gene description, module/pathways, and header. Each MAG has a number that represent gene counts or copies. Gene counts greater than 1 are colored pink, while gene counts equal to 0 are colored white. (Tab 7) Predicted functional genes from reconstructed archaeal MAGs with gene description, module/pathways, and header. Each MAG has a number that represent gene counts or copies. Gene counts greater than 1 are colored pink, while gene counts equal to 0 are colored white. Download Data Set S1, XLSX file, 0.1 MB (127.9KB, xlsx) .
Copyright © 2023 Dithugoe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Ancillary data from the Southern Ocean Time Series sites obtained from CTD data. (A) Temperature (degree Celsius) and salinity plot showing the five sampling stations. The colored circles represent the depths at which the samples were collected. (B) Chlorophyll (milligrams per cubic meter) depth profile for five stations determined by CTD fluorescence sensor and (C) the MLD integrated chlorophyll (milligrams per square meter). Download FIG S1, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2023 Dithugoe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Overview of the prokaryotic community composition for five stations at the class and family level for the SP and SK particle-pool. The color scale represents read abundance. (A) Bacterial community composition; (B) archaeal community composition at both the class and family levels. Download FIG S2, TIF file, 1.9 MB (1.9MB, tif) .
Copyright © 2023 Dithugoe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detailed methodology and results section. The section includes additional data collected from a conductivity, temperature, and depth (CTD) instrument and the Marine Snow Catcher (MSC). The methods include further information regarding sample site descriptions, MSC deployment, and the sequencing protocols. The supplemental results section outlines the analyzed data from ancillary data, including water mass, phytoplankton biomass, and the particulate organic carbon (PON) and particulate organic nitrogen (PON) concentrations. This section also includes details regarding the taxonomic profile and genomic potential of unbinned metagenomic contigs, as well as the reconstructed metagenome-assembled genomes (MAGs). Download Text S1, DOCX file, 0.03 MB (33.1KB, docx) .
Copyright © 2023 Dithugoe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Taxonomic classification of the prokaryotic community from unbinned contigs at the phylum level. The color scale represents read abundance. (A) Bacterial community; (B) archaeal community presented as relative abundance. Download FIG S3, TIF file, 0.9 MB (914KB, tif) .
Copyright © 2023 Dithugoe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Predicted genes involved in carbon utilization and chemoautotrophic genomic potential. The color scale represents gene counts/copies. (A) Bacterial central metabolism and electron transport chain (ETC) complexes; (B) Archaeal central metabolism and ETC complexes. Download FIG S4, TIF file, 1.7 MB (1.7MB, tif) .
Copyright © 2023 Dithugoe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All ten raw metagenomic sequences collected from the Southern Ocean Time Series stations are available at the NCBI (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA749920).