Lei et al. uncover that a subpopulation of breast cancer cells in the lymph node is marked by MHC-II expression in the absence of costimulatory molecules, which causes the expansion of immunosuppressive regulatory T cells.
Abstract
Tumor-draining lymph nodes (TDLNs) are important for tumor antigen–specific T cell generation and effective anticancer immune responses. However, TDLNs are often the primary site of metastasis, causing immune suppression and worse outcomes. Through cross-species single-cell RNA-Seq analysis, we identified features defining cancer cell heterogeneity, plasticity, and immune evasion during breast cancer progression and lymph node metastasis (LNM). A subset of cancer cells in the lymph nodes exhibited elevated MHC class II (MHC-II) gene expression in both mice and humans. MHC-II+ cancer cells lacked costimulatory molecule expression, leading to regulatory T cell (Treg) expansion and fewer CD4+ effector T cells in TDLNs. Genetic knockout of MHC-II reduced LNM and Treg expansion, while overexpression of the MHC-II transactivator, Ciita, worsened LNM and caused excessive Treg expansion. These findings demonstrate that cancer cell MHC-II expression promotes metastasis and immune evasion in TDLNs.
Graphical Abstract
Introduction
In multiple solid tumors—including breast carcinomas, head and neck carcinomas, colon cancer, and melanoma—the first sites of metastasis are often the tumor-draining lymph nodes (TDLNs; Farnsworth et al., 2018; Jones et al., 2018; du Bois et al., 2021; Zhou et al., 2021). The presence of lymph node metastasis (LNM) is strongly correlated with poor prognosis and guides treatment strategies (Giuliano et al., 2011; Hernandez-Aya et al., 2011; Miranda et al., 2011; Donker et al., 2015; David Nathanson et al., 2020). Recently, we and others independently showed that cancer cells in some LNM could escape the lymph nodes and disseminate to distant organs (Brown et al., 2018; Pereira et al., 2018), which suggests a critical need to target LNM to prevent local and systemic metastasis in some patients.
As metastatic cancer cells arrive in the lymph node, they experience and respond to a new microenvironment. Studies have shown that the lymph node microenvironment promotes the metabolic remodeling of cancer cells by upregulation of genes related to lipid and fatty acid metabolism, which could lead to the selection of more aggressive cancer cells in metastatic lymph nodes (metLNs) compared with the primary tumor (Pascual et al., 2017; Lee et al., 2019). Several studies have also shown that cancer cells disseminating from lymph nodes can acquire properties that allow them to better cope with oxidative stresses in the blood during subsequent metastasis (Ubellacker et al., 2020) and make themselves less susceptible to natural killer (NK) cell killing (Reticker-Flynn et al., 2022). Despite the cancer cell heterogeneity that is generated by the process of metastasis, currently, most metastatic tumors are diagnosed and treated based on pathological characteristics of the primary tumor growth in its native microenvironment. This can lead to differential treatment responses of primary tumors and metastases that grow in lymph nodes or distant sites. Gaining a comprehensive understanding of the biological differences between primary tumors and LNM that occur during breast cancer progression is crucial for developing optimal strategies for treating metastatic disease.
The TDLNs are also the first opportunity for the immune system to experience tumor antigens and generate anticancer immune responses. Emerging studies have shown that TDLNs contain TCF1+ stem-like CD8 T cells and tumor-specific resident memory CD8 T cells that maintain active antitumor immune responses (Connolly et al., 2021; Molodtsov et al., 2021). Further, tumor-specific PD-1+ T cells are enriched in TDLNs, PD-1/PD-L1 interactions occur frequently in TDLNs, and blocking PD-L1 in TDLNs elicited effective antitumor immunity (Dammeijer et al., 2020). Paradoxically, metastatic cancer cells can survive and grow in TDLNs, which should generate an immune response trained to attack these cancer cells. The mechanisms underlying the growth and survival of metastatic cancer cells in lymph nodes are still not well understood.
Recent studies have shown that TDLNs are immunosuppressed by cancer (Alonso et al., 2018; Binnewies et al., 2018; Sahovaler et al., 2019; Reticker-Flynn et al., 2020). We have shown that there is a lack of lymphocyte infiltration into metastatic lesions in lymph nodes, which may limit immune activation (Jones et al., 2021). In addition, the upregulation of MHC class I (MHC-I) on some cancer cells and the presence of regulatory T cells (Tregs) provide additional evidence for an immunosuppressed environment in metLNs (Reticker-Flynn et al., 2022). However, the mechanisms for the increase in Tregs in metLNs have not been clearly characterized. Here, we will characterize one mechanism of Treg expansion in metLNs.
Results
The single-cell atlas of murine breast cancer progression to LNM
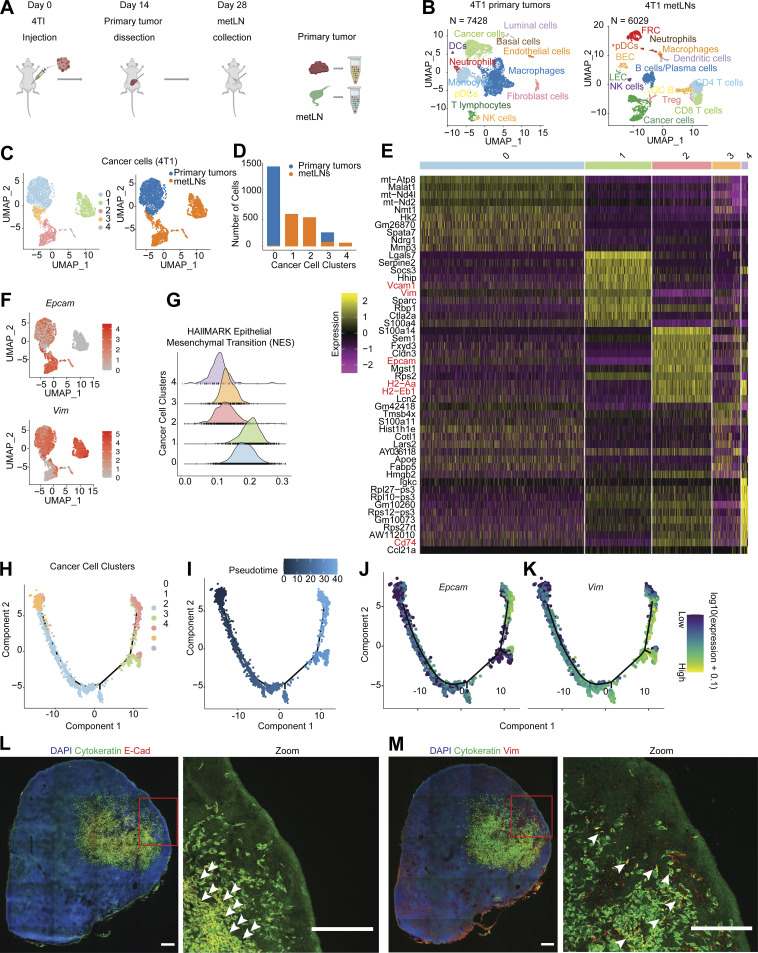
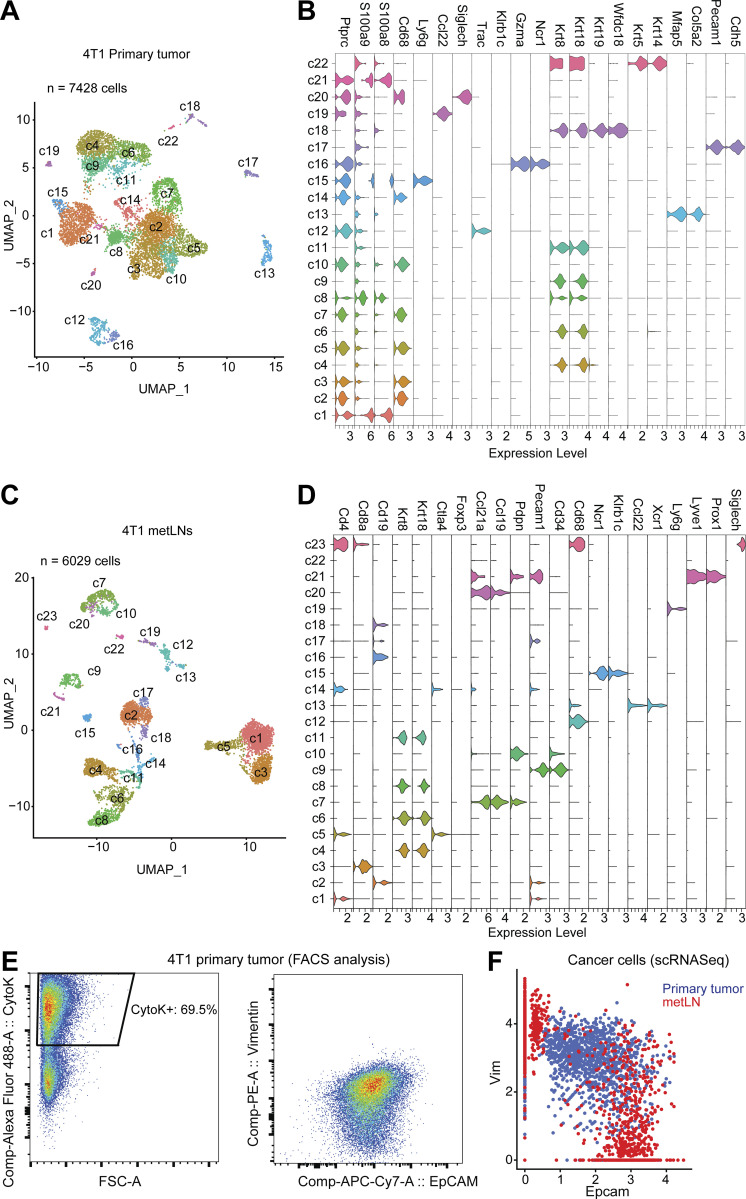
To understand the mechanism of how breast cancer cells invade and survive in the lymph nodes, we characterized cancer cells in both primary tumors and metLNs using 4T1 basal-like triple-negative murine breast cancer that develops spontaneous LNM from orthotopically implanted tumors in immunocompetent, syngeneic hosts (Pulaski and Ostrand-Rosenberg, 2001). We collected the primary tumor and metLN from the same mouse and performed single-cell RNA sequencing (RNA-Seq; n = 3; Fig. 1 A). In the primary tumor samples, we identified 7,428 single cells that grouped into 22 unique cell clusters (Fig. S1 A). Next, we identified the most upregulated genes in each cluster to annotate the individual cell types. Around 70% of the cells in the primary tumor microenvironment (TME) are immune cells (Fig. 1 B and Fig. S1 B), as shown by the expression of Ptprc—the gene encoding CD45. The myeloid-derived monocytes and macrophages are the largest population of immune cells in the TME. In the metLNs, we identified 6,029 single cells that grouped into 23 cell clusters. According to these marker genes, we found a rich diversity of cells in the metLNs, including cancer cells, conventional CD4 T cells, CD8 T cells, Treg cells, B cells, NK cells, macrophages, dendritic cells, plasmacytoid dendritic cells (pDCs), and lymph node stromal cells (Fig. 1 B and Fig. S1, C and D).
Figure 1.
Single-cell RNA-Seq identified EMT-plasticity of cancer cells in 4T1 primary tumors and metLNs. (A) Pipeline for single-cell RNA-Seq library preparation. 4T1 cells (1 × 105) were injected into the fourth mammary fat pad of female Balb/c mice (6–10 wk old). Primary tumors were dissected and dissociated into single cells for 10X Genomics Chromium Single Cell 3′ library preparation when they reached 500 mm3 or on day 14. 14 d after primary tumor resection, tumor metLNs were collected, digested into single cells, and processed by the 10X Genomics platform. The Illumina NextSeq platform was used for sequencing. This experiment was repeated with triplicates, and samples from four mice were combined in each cohort. (B) UMAP plot of distinct cell types in the microenvironment of 4T1 primary tumors (left) and metLNs (right). (C) UMAP plot of aggregated cancer cells (n = 3,007) from primary tumors and metLNs after cell cycle regression. The plot on the left is colored by original cluster names, and the plot on the right is colored by samples. The top 20 PCs were chosen for UMAP analysis, and the minimum distance is 0.5. (D) Bar plot showing the number of cancer cells in each cluster from C. (E) Heatmap of single-cell gene expression of the top 10 differentially expressed genes in cancer cells. The color bar above the heatmap is the same color code as in C. In the heatmap, purple represents low expression, and yellow represents high expression. (F) UMAP plot of gene expression of epithelial cell marker gene (EpCAM) and mesenchymal cell marker gene (Vim) in cancer cells. Gene expression values are log-normalized, gray represents low expression, and red represents high expression. (G) Histogram of the single-cell enrichment score of MSigDB Hallmark EMT gene set. (H) The single-cell trajectory of cancer cells colored by original cluster identities. (I) The pseudotime trajectory of cancer cells, with the early stage represented by dark color and the late stage represented by bright color. (J and K) Expression of EpCAM and Vim projected onto the single-cell trajectories. Gene expression values are scaled and log-normalized. (L and M) Representative confocal microscopy images of immunofluorescence staining of cytokeratin, E-cadherin, and vimentin in 4T1 metLNs. Two adjacent tissue sections from the same lymph node were used for staining. The tissue section thickness is 10 μm. Scale bars represent 200 μm in all the images, and the red rectangle indicates the enlarged view. The white arrowheads indicate the cytokeratin and E-Cadherin–positive cells in L, and cytokeratin and vimentin–positive cells in M.
Figure S1.
The single-cell landscape of the 4T1 breast tumor and metLNs microenvironment. (A) The UMAP displays the clustering of 7,428 cells from 4T1 primary tumor samples, colored by the clusters. We used the top 50 PCs for the UMAP analysis with a minimum distance of 0.5 for the display. The resolution for the UMAP clustering is 1.0. (B) The violin plots show the log-normalized gene expression levels of selected marker genes in each cluster of cells in primary tumors. (C) The UMAP displays the clustering of 6,029 cells from metLNs after correction for batch bias, colored by the clusters. metLN samples from three independent sequencing datasets were aggregated and normalized to correct for batch bias. We used the top 30 significant PCs for UMAP analysis with a minimum distance of 0.5, and resolution for the UMAP is 1.0. (D) The violin plots show the log-normalized gene expression levels of selected marker genes in each cluster of cells in metLNs. (E) Flow cytometry analysis of EMT markers in cancer cells from 4T1 primary tumors. (F) The single-cell gene co-expression of EMT-marker in cancer cells from primary tumors and metLNs.
Lineage analysis of epithelial-to-mesenchymal transition (EMT) remodeling during breast cancer progression to LNM
To examine the heterogeneity of intratumoral gene expression during LNM, we integrated cancer cells from primary tumors and metLNs and analyzed them using Uniform Manifold Approximation and Projection (UMAP) algorithm. Our analysis revealed that the cancer cells could be classified into five distinct clusters, with the cells in primary tumors clearly distinguishable from those found in metLNs (Fig. 1 C). Further analysis showed that the cancer cells from metLNs were divided into two major populations, cluster 1 and cluster 2 (Fig. 1 D).
To further investigate the cancer cell heterogeneity, we compared the differentially expressed genes of the five cancer cell clusters. Our analysis showed that the mesenchymal cell marker genes Vcam1 and Vim were highly expressed in cluster 1 cancer cells, while the epithelial cell marker gene Epcam was highly expressed in cluster 2 and cluster 4 cancer cells (Fig. 1 E). We also projected the single-cell gene expression of Epcam and Vim into the cancer cell UMAP and found that the cancer cells in primary tumors expressed both Epcam and Vim (cluster 0 and part of cluster 3), suggesting the presence of EMT-hybrid cells (Fig. 1 F and Fig. S1 E), while cancer cells in metLNs expressed either Epcam or Vim, with a small population of Epcam and Vim double-positive cells (Fig. 1 F and Fig. S1 F).
In addition, we examined the single-cell gene set enrichment score of the cancer cells based on the Molecular Signatures Database (MSigDB) Hallmark genesets. We found that the cancer cells in cluster 0 and cluster 1 had a higher EMT score, with cluster 1 exhibiting the highest enrichment score of the EMT signature. The clusters 2, 3, and 4 cancer cells showed lower levels of EMT signature, indicating a more epithelial-like phenotype (Fig. 1 G). Interestingly, we also found that Cd74, H2-Eb1, and H2-Aa, key components of the MHC-II complex needed for antigen presentation, were highly expressed in the epithelial-like cancer cells (cluster 2 and cluster 4; Fig. 1 E).
Increasing evidence suggests that EMT status is a “spectrum” rather than a binary status (Pastushenko et al., 2018). Furthermore, this transition is reversible as mesenchymal-like cells can also transition to an epithelial phenotype (MET; Pei et al., 2019). To investigate EMT/MET during LNM, we examined the lineage of cancer cells during LNM by single-cell trajectory analysis (Trapnell et al., 2014; Qiu et al., 2017). We found that most of the cancer cells in cluster 2 were in the late stage of the pseudotime trajectory (Fig. 1, H and I). Next, we projected the gene expression of Epcam and Vim into the pseudotime trajectory. Of note, we found that epithelial cancer cells in the metLNs were derived from mesenchymal cancer cells (Fig. 1, J and K), suggesting MET of cancer cells in the metLNs.
We further investigated the EMT phenotype of cancer cells and the metastatic burden in individual lymph nodes by flow cytometry (Fig. S2 A). We found that in the metLNs in which cancer cells account for >5% of the cell population, nearly 80% of the cancer cells were epithelial cell adhesion molecule (EpCAM) positive. In contrast, in the metLNs in which cancer cells account for <1% of the cell population, on an average around 14% of the cancer cells were mesenchymal phenotype (EpCAM−vimentin+) and 19% of the cancer cells were epithelial phenotype (EpCAM+vimentin−; Fig. S2, B and C). Using immunostaining of EMT hallmarks in metLNs, we found both E-cadherin–positive (epithelial) cancer cells and vimentin-positive (mesenchymal) cancer cells in LNM, with most of the E-cadherin–positive cancer cells in the center of the metastatic lesion (Fig. 1, L and M). These data demonstrate that the metastatic cancer cells in metLNs displayed heterogeneous and spatially organized epithelial and mesenchymal phenotypes in metastatic lesions.
Figure S2.
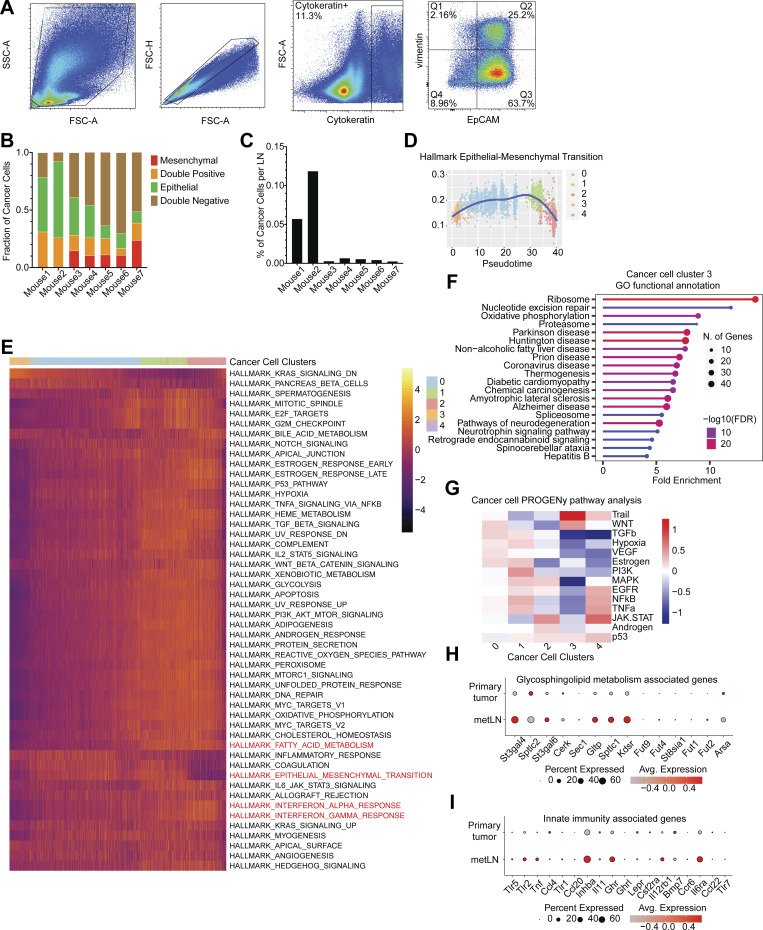
The plasticity of cancer cells during LNM. (A) The representative flow cytometry gating strategy of EMT profiles of 4T1 cancer cells in metLNs. Cytokeratin (Alexa-488), EpCAM (APC-Cy7), vimentin (PE). (B) Percentage of EpCAM+vimentin− (epithelial), EpCAM−vimentin+ (mesenchymal), EpCAM−vimentin− (double-negative), and EpCAM+vimentin+ (double-positive) cancer cells compared with all cancer cells (cytokeratin+) in metLNs. (C) Percentage of cancer cells in metLNs measured by FACS. (D) The single-cell enrichment score of MSigDB Hallmark EMT gene set. Cancer cells were ranked by pseudotime, and the blue line represents the loess regression of the enrichment score. (E) The single-cell gene set enrichment analysis of cancer cells in 4T1 primary tumors and metLNs. The gene sets are from GSEA MSigDB hallmark gene sets. Each column represents a single cell, and each row represents a gene set. Cancer cells were ranked by pseudotime. Yellow represents high, and purple represents low. (F) GO functional annotation of upregulated genes in cluster 3 4T1 cancer cells. (G) PROGENy pathway analysis of differentially expressed genes in cancer cells from our murine datasets. (H and I) Single-cell gene expression of glycosphingolipid metabolism (H) and innate immunity associated genes (I) was examined using data from a previous publication on human TNBCs (Karaayvaz et al., 2018). Gene expression levels were log-normalized, and gray indicates low expression while red indicates high expression. The size of the circle represents the percentage of cells in each cluster that expressed the labeled genes.
Taken together, we hypothesize that the process of EMT of cancer cells in the primary tumors generates mesenchymal-like cells that subsequently metastasize to the lymph nodes. However, in the metLN, a portion of the mesenchymal cancer cells underwent MET, particularly those in the center of the lesion where they are less exposed to the native lymph node microenvironment.
Dynamic transcriptomic alterations during breast cancer LNM
EMT/MET is a frequently observed phenotype during cancer progression and represents an appealing target for clinical interventions. Nonetheless, the direct targeting of EMT molecules is still a challenging task. Understanding how cancer cells adjust to the lymph node microenvironment during EMT/MET could potentially identify novel signaling pathways involved in EMT and discover innovative approaches to target this process indirectly. As EMT/MET phenotype seemed to change with disease progression, we ranked cancer cells based on the pseudotime trajectory and compared the differentially expressed gene signatures among them. Besides the elevation of EMT and MET gene signatures (Fig. S2 D), we also found that type I and II IFN signaling, fatty acid metabolism, and IL6/JAK/STAT3 signaling were elevated in cancer cells in LNM (Fig. S2 E). It is worth noting that a subcluster of cancer cells from the LNM (cluster 3) was ranked at an early stage in the pseudotime trajectory. Gene ontology (GO) functional annotation analysis revealed that this population of cells exhibited elevated levels of genes associated with ribosomes, nucleotide excision repair, proteasome, and spliceosome (Fig. S2 F). The PROGENy pathway analysis further showed that genes associated with the Trail signaling pathway, Wnt signaling pathway, and p53 signaling pathway were upregulated in cluster 3 (Fig. S2 G). These pathways suggest that these cells, along with a similar population of cells in the primary tumor, maybe more genetically unstable or attempting DNA repair. Therefore, they cluster together in the pseudotime analysis.
Triple-negative breast cancer (TNBC) reveals extensive intratumoral heterogeneity. A study of six human TNBC primary tumors using single-cell RNA-Seq uncovered that a small population of cancer cells present across all patients had upregulated glycosphingolipid metabolism and an associated innate immunity pathway (Karaayvaz et al., 2018). The elevation of glycosphingolipid metabolism and innate immunity pathway-associated gene signatures revealed worsened prognosis. In our single-cell dataset, we also observed the upregulation of glycosphingolipid metabolism and innate immunity-associated genes in the LNMs (Fig. S2, H and I). Of note, the previous study (Karaayvaz et al., 2018) focused on the phenotypes of primary tumors. Here, our data suggest that the upregulation of the glycosphingolipid metabolism and innate immunity-associated genes in cancer cells may play a role in promoting LNM.
The elevation of MHC-II on cancer cells during LNM
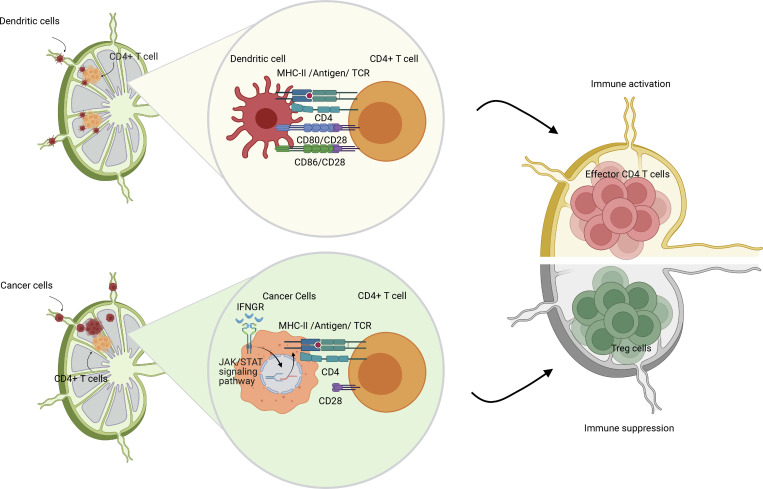
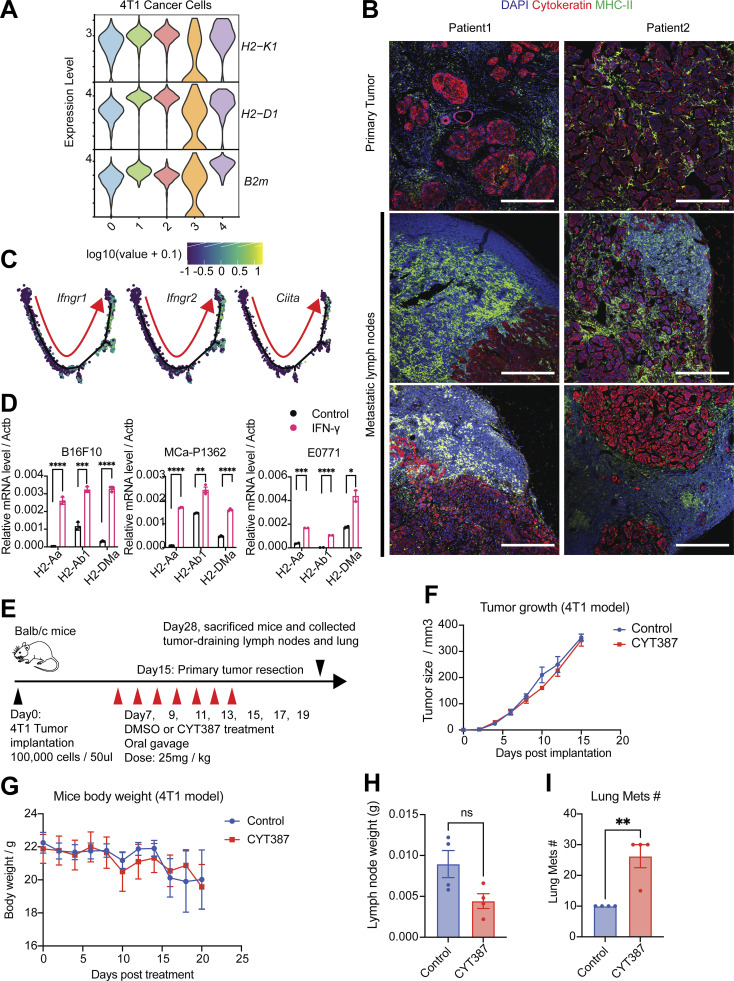
The elevation of IL6/JAK/STAT3 signaling pathway in cancer cells induces a more invasive and advanced phenotype in breast cancer, ovarian cancer, and prostate cancer (Izar et al., 2020; Siersbæk et al., 2020; Chan et al., 2022). IFN-γ and JAK/STAT-induced gene activation also account for the induction of MHC-I and MHC-II on antigen-presenting cells (van den Elsen, 2011). Published data suggest that EMT processes protect mesenchymal cancer cells from immune surveillance in the primary tumor by downregulation of MHC-I molecules (Dongre et al., 2017). However, in our data, we found that MHC-I molecules were generally highly expressed in all cancer cells (Fig. S3 A). In contrast, MHC-II molecules were expressed in a fraction of primary cancer cells, particularly in epithelial-like metastatic cancer cells (cluster 2 and cluster 4; Fig. 2 A). Single-cell pseudotime trajectory results suggest the elevation in gene expression of MHC-II molecules (H2-Aa and H2-Ab1) on late-stage cancer cells that have undergone MET (Fig. 2, B and C). Although MHC-II is predominantly displayed on professional antigen-presenting cells, we and others have recently demonstrated expression and functional significance of MHC-II in both malignant (Wosen et al., 2018; Axelrod et al., 2019; Izar et al., 2020) and normal epithelial (Biton et al., 2018; Beyaz et al., 2021) as well as lymphatic endothelial cells (Santambrogio et al., 2019). CD4+ T cell recognition of cognate antigen bound to MHC-II in the absence of costimulation leads to T cell tolerance through anergy (Wosen et al., 2018) or induction of Treg differentiation (Nadafi et al., 2020). Accordingly, we found that the LNM cancer cells lacked expression of costimulatory molecules Cd80, Cd86, and Icosl (Fig. 2 D). The elevation of the type II IFN signaling pathway in cancer cells has been shown to mediate the expression of the coinhibitory molecule PD-L1 to enhance immune evasion (Garcia-Diaz et al., 2017). However, the expression of Cd274—the gene that encodes PD-L1—was almost undetectable in LNM cancer cells (Fig. 2 D), suggesting cancer cells might rely on PD-L1–independent strategies to escape immune surveillance in the lymph node. Consistent with this, our published work has shown that metastatic lesions in lymph nodes did not respond to anti–PD-1 therapy (Jones et al., 2021).
Figure S3.
The elevation of the IFN-γ signaling pathway drives MHC-II expression in cancer cells. (A) The violin plot shows the gene expression of MHC-I molecules in 4T1 cancer cells from all clusters. (B) Immunofluorescence staining of nuclei (DAPI, blue), pan-cytokeratin (red), and MHC-II (green) in human breast tumors and metLNs in formalin-fixed paraffin-embedded specimens. The thickness of the sections is 5 μm. The scale bar represents 500 μm in all images. The deidentified breast tumor samples were provided by the Massachusetts General Hospital pathology department. (C) The gene expression of Ifngr1, Ifngr2, and Ciita were projected to the single-cell pseudotime trajectories. Gene expression values are scaled and log-normalized. The red arrow indicates the pseudotime trajectory of cancer cells progression. (D) IFN-γ induces the expression of MHC-II molecules H2-Aa, H2-Ab1, and H2-DMa in vitro. B16F10 (melanoma), MCa-P1362 (breast cancer), and E0771 (breast cancer) cells were treated with or without IFN-γ (10 ng/ml) for 24 h. Student’s t test was used for statistical analysis; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (E) The experimental design of the JAK/STAT inhibitor CYT387 in vivo assay. (F) 4T1 tumor growth in control and CYT387 treatment groups (n = 4). (G) Mice body weight change in control and CYT387 treatment groups (n = 4). (H) The weight of the TDLNs at day 28. (I) The number of lung metastases at day 28. Student’s t test was used for the statistical analysis.
Figure 2.
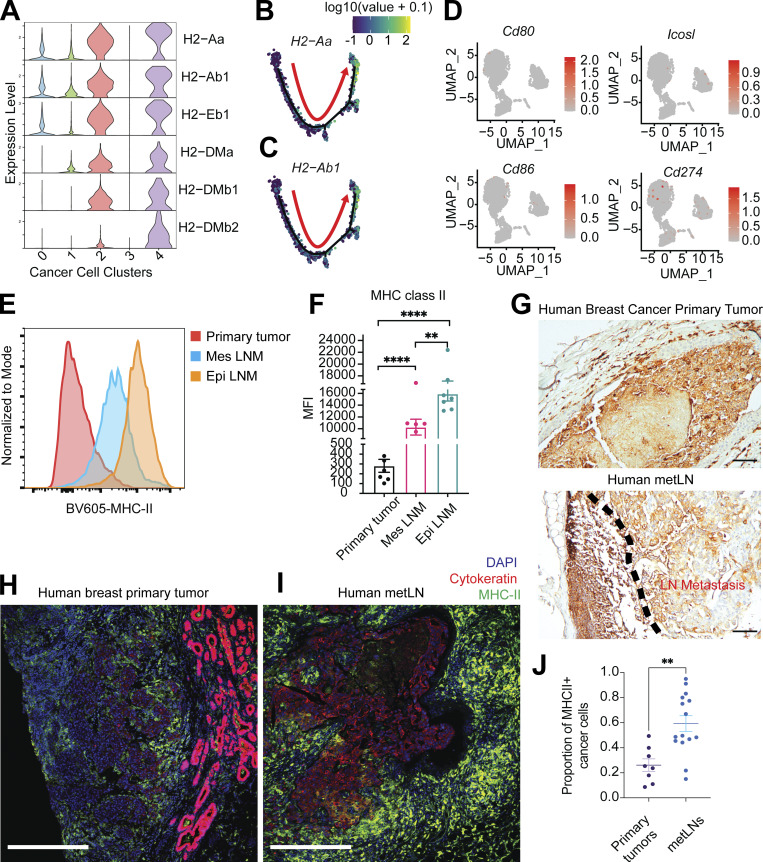
The presence of MHC-II on cancer cells during LNM. (A) Violin plot represents the gene expression of MHC-II molecules in 4T1 cancer cells. (B and C) Expression of MHC-II molecules H2-Aa and H2-Ab1 were projected to the single-cell trajectories. Gene expression values are scaled and log-normalized. The red arrow indicates the pseudotime trajectory of cancer cells’ progression. (D) UMAP of single-cell gene expression of costimulatory and coinhibitory molecules in cancer cells. (E) Surface MHC-II presence on cancer cells measured by flow cytometry. (F) The mean fluorescent intensity (MFI) of MHC-II on cancer cells from 4T1 primary tumors and metLNs. Mes LNM, EpCAM−vimentin+; Epi LNM, EpCAM+vimentin−. One-way ANOVA was used for statistical analysis; **, P < 0.01; ****, P < 0.0001. Tukey’s multiple comparisons test was used for the post-hoc test. (G) Immunohistochemistry staining of MHC-II (HLA-D) in human breast cancer primary tumor and metLN. The scale bar represents 100 μm in all images. (H and I) Immunofluorescence staining of nuclei (DAPI, blue), pan-cytokeratin (red), and MHC-II (green) in human breast cancer primary tumor (H) and matched metLN (I). The thickness of the tissue section is 5 μm. The scale bar represents 500 μm in all images. (J) The proportion of MHC-II–positive cancer cells in human breast tumors (n = 8) and LNM (n = 15). Cell annotation was performed using QuPath software. **, P < 0.01. Student’s t test was used for the statistical analysis.
We further examined cell surface MHC-II in primary tumor and LNM cancer cells via flow cytometry (Fig. 2 E). We found that EpCAM+/vimentin− (epithelial) cancer cells in metLNs exhibited the highest MHC-II on their cell surface (Fig. 2 F). Consistent with these findings, we also found the presence of MHC-II+ cancer cells in metLNs from patients with invasive ductal breast carcinoma (Fig. 2 G and Fig. S3 B). The proportion of MHC-II+ cancer cells in metLNs was significantly (P < 0.01) higher than in the primary tumors in these patient samples (Fig. 2, H–J). Altogether, these results suggest that cancer cells upregulate MHC-II during the progression of breast cancer to metLNs in both mice and humans. We hypothesized that cancer cell MHC-II expression likely contributes to immune evasion by eliciting CD4 T cell tolerance.
IFN-γ signaling pathway induces MHC-II expression in cancer cells
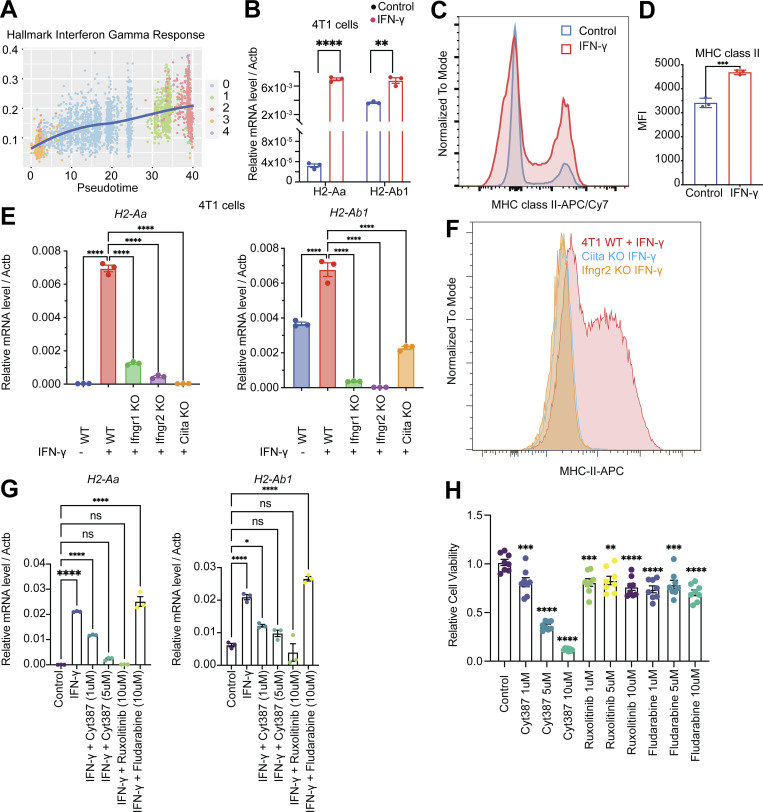
IFN-γ has been shown to regulate MHC-I/II expression in epithelial cells via the JAK/STAT signaling pathway (van den Elsen, 2011; Axelrod et al., 2019; Beyaz et al., 2021). The elevation of the IFN-γ response gene signature (Fig. 3 A) and the increase in expression of IFN-γ receptors Ifngr1/2 and Ciita, the transactivator that controls the activation of MHC-II, during breast cancer progression to LNM (Fig. S3 C) led us to hypothesize that this pathway is involved in the elevation of MHC-II on LNM cancer cells. In vitro administration of IFN-γ to 4T1 cells led to a profound increase in the mRNA expression of the key MHC-II molecules H2-Aa and H2-Ab1 (Fig. 3 B). Of note, we also observed a significant (P < 0.001) increase in MHC-II+ cells and cell surface MHC-II molecules after IFN-γ stimulation compared with the control group (Fig. 3, C and D). Further, we found that IFN-γ was able to induce mRNA expression of MHC-II genes (H2-Aa, H2-Ab1, and H2-DMa) in additional cancer models, B16F10 (melanoma), E0771 (breast), and MCa-P1362 (breast; Jones et al., 2021; Fig. S3 D). CIITA is a general regulator of both inducible and constitutive MHC-II expression (van den Elsen, 2011). To understand whether the IFN-γ–induced expression of MHC-II on cancer cells is CIITA dependent, we generated the Ifngr1/2 and Ciita knockout 4T1 cells using CRISPR/Cas9 systems. In Ifngr1/2 and Ciita knockout cells, the administration of IFN-γ was not able to induce mRNA expression of H2-Aa and H2-Ab1 (Fig. 3 E) and cell surface MHC-II (Fig. 3 F), confirming CIITA is essential for the IFN-γ–induced MHC-II expression on 4T1 cancer cells.
Figure 3.
The induction of IFN-γ signaling pathway drives MHC-II expression on cancer cells. (A) The single-cell enrichment score of MSigDB Hallmark IFN-γ Response gene set. Cancer cells were ranked by pseudotime, and the blue line represents the loess regression of the enrichment score. (B) IFN-γ–induced expression of MHC-II molecules H2-Aa and H2-Ab1 in vitro. 4T1 cells were treated with or without IFN-γ (10 ng/ml) for 24 h. Student’s t test for statistical analysis; **, P < 0.01; ****, P < 0.0001. (C) IFN-γ–induced cell surface MHC-II on 4T1 cells. 4T1 cells were treated with or without IFN-γ (10 ng/ml) for 24 h and cell surface MHC-II was measured by flow cytometry. (D) The mean fluorescent intensity (MFI) of MHC-II on MHC-II–positive 4T1 cancer cells. (E) Expression of MHC-II molecules H2-Aa and H2-Ab1 in 4T1 WT, Ifngr1, Ifngr2, and Ciita knockout cells with or without IFN-γ (10 ng/ml) treatment for 24 h. One-way ANOVA was used for statistical analysis; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Tukey’s multiple comparisons test was used for the post-hoc test. (F) The cell surface MHC-II on 4T1 WT, Ifngr2, and Ciita knockout cells after IFN-γ (10 ng/ml) treatment for 24 h. (G) Expression of MHC-II molecules H2-Aa and H2-Ab1 mRNA in 4T1 cells with IFN-γ or IFN-γ combined with JAK/STAT inhibitor treatment. One-way ANOVA was used for statistical analysis; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Tukey’s multiple comparisons test was used for the post-hoc test. (H) Cell viability under the treatment with different JAK/STAT inhibitors. 4T1 cells were treated with DMSO, Cyt387, ruxolitinib, and fludarabine for 72 h. Cell viability was measured by MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) tetrazolium reduction assay.
We previously showed that downstream of IFN-γ signaling, the JAK/STAT pathway mediates the induction of epithelial MHC-II expression in the small intestine (Beyaz et al., 2021). As the IL6/JAK/STAT signaling pathway was upregulated in LNM cancer cells (Fig. S2 E), we tested whether targeting the JAK/STAT pathway is capable of suppressing MHC-II expression. Administration of JAK/STAT inhibitors Cyt387 (momelotinib), ruxolitinib, and fludarabine in combination with IFN-γ to 4T1 cancer cells for 48 h showed that JAK1/2 inhibitors Cyt387 and ruxolitinib suppressed IFN-γ–induced MHC-II mRNA expression on 4T1 cancer cells (Fig. 3 G). Together, these results suggest that 4T1 cancer cells can upregulate MHC-II in response to IFN-γ.
We next tested whether pharmacologic inhibition of JAK/STAT signaling influences tumor progression in the 4T1 orthotopic breast cancer model. Treatment of 4T1 cancer cells with a JAK1/2 inhibitor Cyt387 showed direct anticancer cell activity at 5 and 10 μM concentration in vitro, whereas ruxolitinib and fludarabine did not (Fig. 3 H). Next, we tested the anticancer response with in vivo administration of JAK1/2 inhibitor. Briefly, we injected 100,000 4T1 cancer cells into the mammary fat pad of Balb/c mice, and on day 7, we started the treatment with JAK/STAT inhibitor Cyt387 every 2 d for seven doses (Fig. S3 E). The growth of tumors (Fig. S3 F) and weight of mice (Fig. S3 G) did not show a significant difference between the control and Cyt387 treatment group. The weight of the TDLNs also did not show a significant difference (Fig. S3 H), and the Cyt387 treatment group had more metastatic nodules in the lung (Fig. S3 I). These results could be due to the fact that Cyt387 also will inhibit JAK/STAT signaling in immune cells, which are necessary for antitumor immune responses.
Invasion of cancer cells in metLNs enhances Treg activation and elicits an immunosuppressive microenvironment
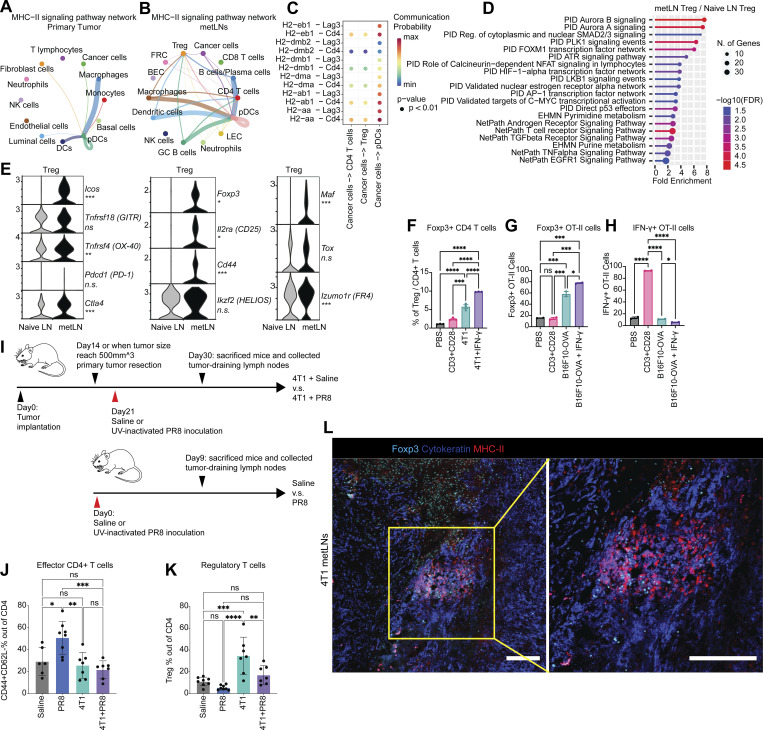
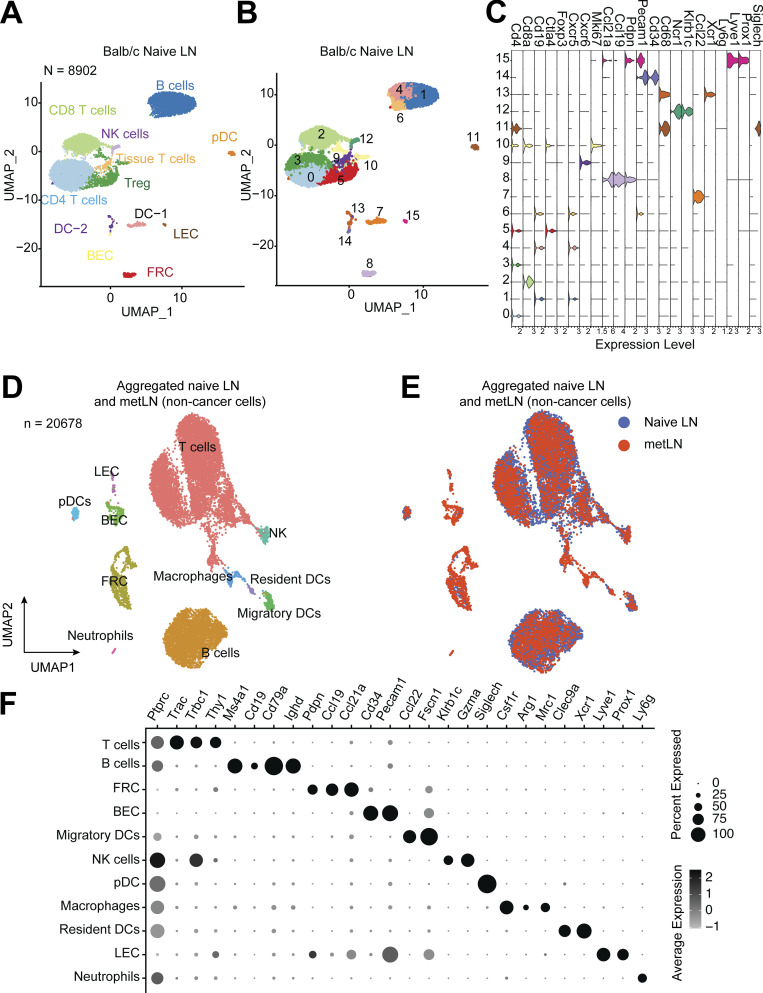
Induction of T cell anergy is a key mechanism of self-tolerance. Anergic T cells can also further expand the CD4+Foxp3+ Treg repertoire (Kuczma et al., 2021). To investigate whether MHC-II on epithelial-like cancer cells leads to CD4+ T cell tolerance and enhances the expansion of Tregs in metLNs, we investigated the MHC-II signaling pathway cell–cell interaction network in primary tumors and metLNs (Jin et al., 2021). Cancer cells in the primary tumor interacted with pDCs more frequently (Fig. 4 A), while cancer cells in the metLNs interacted with CD4+ T cells and Treg cells as well as pDCs (Fig. 4 B). The ligand–receptor pair analysis predicted that MHC-II molecules on cancer cells interact with CD4 on CD4+ T cells, Treg, and pDCs in metLNs as well as LAG3 on pDCs (Fig. 4 C). To understand the impact of cancer cell invasion on the lymph node microenvironment, we performed single-cell RNA-Seq in the inguinal lymph nodes from naive Balb/c mice (Fig. S4, A–C). We aggregated single-cell mRNA expression data from naive lymph nodes with non-cancer cells from metLNs by UMAP analysis to be able to compare expression profiles of specific T cell populations from naive lymph nodes and metLNs (Fig. S4, D–F). GO functional annotation of the differentially expressed genes in Treg cells between metLN and naive LN demonstrated that Aurora A/B signaling, PLK1 signaling, and proliferation-associated transcription factor FOXM1 network were upregulated in Tregs in metLNs, suggesting they were highly proliferative (Fig. 4 D). Additionally, androgen receptor signaling and TGF-β receptor signaling pathways were also upregulated in Tregs in metLNs (Fig. 4 D). The T cell–intrinsic androgen receptor activity limits antitumor immunity and T cell reinvigoration (Guan et al., 2022; Vellano et al., 2022). TGF-β induces Foxp3+ Treg cells from CD4+CD25− precursors (Fu et al., 2004) and helps maintain Treg suppressor function (Marie et al., 2005). Furthermore, we found that Tregs in both naive lymph nodes and metLNs expressed the inhibitory checkpoint molecule Ctla4, but not Pdcd1 (PD-1; Fig. 4 E). The costimulatory molecules Tnfrsf18 (GITR) and Tnfrsf4 (OX-40) were prevalently expressed in Tregs; however, only the Tregs in metLNs expressed Icos (Fig. 4 E). A recent study demonstrated that Icos+ Treg cells exhibited a superior suppressive function (Mair et al., 2022), suggesting that these Icos+ Treg cells in the metLNs may be more suppressive. Moreover, Treg activation–associated genes as well as T cell anergy and exhaustion-associated genes (Akimova et al., 2011; Trefzer et al., 2021), such as Foxp3, Il2ra (CD25), Cd44, Izumo1r, and Maf, were profoundly elevated in Tregs in metLNs (Fig. 4 E).
Figure 4.
MHC-II–positive cancer cells mediated the enrichment of tolerogenic Treg cells in lymph nodes through energy. (A and B) Single-cell MHC-II signaling pathway network in (A) 4T1 primary tumor and (B) metLNs. The cell–cell interaction analysis was performed by CellChat. (C) MHC-II signaling pathway ligand–receptor interactions between cancer cells and CD4+ T cells, Treg, and pDCs. (D) Functional annotation of upregulated genes in Tregs in metLNs by ShinyGO. (E) The single-cell gene expression of Treg cell–associated genes. (F) The percentage Tregs when cocultured with or without 4T1 cancer cells. 4T1 cells were pretreated with or without IFN-γ (10 ng/ml) for 48 h before the coculture assay. (G) The percentage of Foxp3+ OT-II T cells in the in vitro co-culture assay. (H) The percentage of IFN-γ+ OT-II T cells in the in vitro co-culture assay. B16F10-OVA cells were pretreated with or without IFN-γ (10 ng/ml) for 48 h before the coculture assay. (I) The experiment design of UV-inactivated influenza virus PR8 inoculation assay. (J and K) The percentage of (J) effector CD4+ T cells and (K) Tregs in the draining lymph nodes from UV-inactivated PR8 influenza virus inoculation assay. (L) Immunofluorescence staining of Foxp3 (cyan), pan-cytokeratin (blue), and MHC-II (red) in 4T1 metLNs. The scale bar represents 200 μm in all images. The yellow rectangle indicates the region of interest at a higher magnification.
Figure S4.
The single-cell atlas of inguinal lymph nodes in naive Balb/c mice. (A and B) The UMAP of 8,902 cells from inguinal lymph nodes in tumor-free Balb/c mice (naive mice), (A) colored by the cell types, (B) colored by the clusters. We selected the top 30 PCs for the UMAP analysis with a minimum distance of 0.5 for the display. The resolution for the UMAP clustering is 0.8. (C) Violin plots show the gene expression levels of selected marker genes in each cluster of cells from naive lymph node. Gene expression values are log-normalized. (D) The UMAP of aggregated cells in naive lymph nodes and 4T1 metLNs (n = 20,678). Top 30 PCs were chosen for the UMAP analysis, with a minimum distance of 0.5 and clustering resolution of 0.8, and cells were colored by main cell types in the lymph nodes. (E) The UMAP of aggregated cells in naive lymph nodes and 4T1 metLNs grouped by the samples. (F) The gene expression pattern of selected marker genes in the main cell types in the lymph nodes. Black represents high expression, and gray represents low expression. The size of the circle represents the proportion of cells expressing the indicated genes in each cluster.
Next, we tested whether the induction of MHC-II on 4T1 cancer cells in the absence of costimulatory signals can lead to CD4+ T cell anergy and expansion of Tregs. We treated 4T1 cancer cells with or without IFN-γ for 48 h, then isolated CD4+ T cells from 12-wk-old female Balb/c mice and cocultured them in vitro for 72 h. PBS and anti-CD3/anti-CD28 coated wells served as T cell activation negative and positive controls, respectively. When CD4+ T cells were cultured alone in the presence of both anti-CD3 (signal 1) and anti-CD28 (signal 2), we observed around 2% of Treg cells. In contrast, we observed a twofold increase in the percentage of Treg cells when cocultured with 4T1 cells and a fourfold increase in the percentage of Treg cells in the presence of IFN-γ–pretreated 4T1 cells (MHC-II+) compared with the anti-CD3+CD28 group monoculture group (Fig. 4 F). We further examined the interaction between cancer cell MHC-II and TCRs on CD4+ T cells with an antigen-specific B16F10-OVA/OT-II coculture assay. Very similarly, in the presence of IFN-γ–pretreated B16F10-OVA cancer cells, there was a threefold to fourfold increase of Foxp3+ OT-II cells compared with the anti-CD3+CD28 group (Fig. 4 G). In addition, we observed the lowest population of IFN-γ + OT-II cells in the presence of IFN-γ–pretreated B16F10-OVA cancer cells (Fig. 4 H).
To further evaluate the immune consequences of tumor metastasis in the draining lymph nodes in vivo, we tested the immune responses of metastatic LNs against UV-inactivated PR8 influenza viruses, a well-recognized model to study the immune activation in the lymph nodes (Jayasekera et al., 2007; Fernandez Gonzalez et al., 2008). We compared the T cell phenotype in normal lymph nodes, UV-inactivated PR8 influenza virus inoculated lymph nodes, and 4T1 metLNs as well as 4T1 metLNs inoculated with UV-inactivated PR8 influenza virus (Fig. 4 I). We found that the population of effector CD4+ T cells remained similar between unstimulated normal lymph nodes and metLNs. However, the immune-activated naive lymph nodes had higher effector CD4+ T cells (Fig. 4 J). In contrast, metLNs inoculated with UV-inactivated PR8 influenza virus did not have an increase in effector CD4+ T cells. Further, there was a substantial increase in Tregs in 4T1 metLNs (Fig. 4 K). We also confirmed the colocalization of MHC-II+ cancer cells and Foxp3+ T cells in the metLNs using immunofluorescent staining (Fig. 4 L). These findings collectively suggest that the invasion of MHC-II+ cancer cells into the lymph node triggers the generation and activation of Tregs, ultimately leading to the establishment of an immunosuppressive microenvironment in the metLNs that inhibits the generation of immune responses to even potent antigen stimulation.
Preventing the expression of MHC-II in cancer cells decreases LNM and improves overall survival
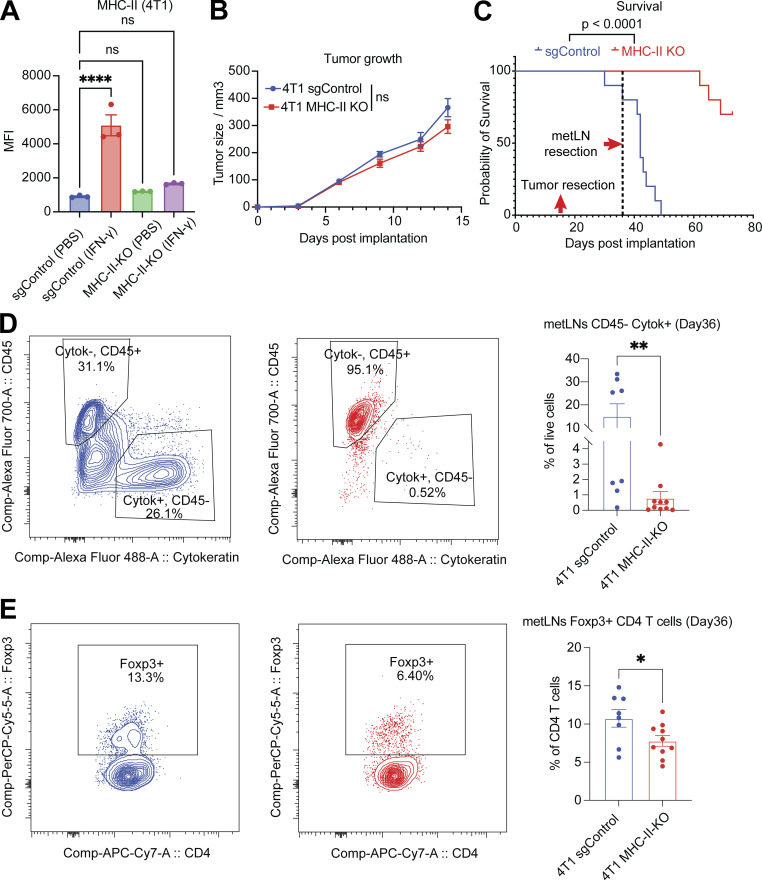
To determine the role of MHC-II expression in cancer cells in tumor progression and LNM, we generated MHC-II knockout 4T1 cancer cells using guide RNAs that target H2-Aa, H2-Ab1, and H2-Eb1. The efficacy of MHC-II deletion was confirmed using flow cytometry (Fig. 5 A). The MHC-II knockout cancer cells were able to form tumors in Balb/c mice. The tumor growth of MHC-II knockout cancer cells did not exhibit a significant difference compared with control cancer cells (Fig. 5 B). After the resection of primary tumors, mice were monitored for LNM and survival. The MHC-II knockout group had significantly (P < 0.0001) better survival (Fig. 5 C) and fewer cancer cells in the tumor-draining inguinal lymph nodes (Fig. 5 D), suggesting that MHC-II expression in cancer cells promotes metastatic growth. Additionally, there were fewer Treg cells in the lymph nodes of the MHC-II knockout group (Fig. 5 E), suggesting that MHC-II expression in cancer cells leads to Treg cell expansion and immune suppression. These results support our hypothesis that cancer cell MHC-II expression in lymph nodes drives tumor progression and immune evasion.
Figure 5.
The loss of MHC-II in cancer cells diminishes LNM. (A) The mean fluorescence intensity of cell surface MHC-II molecules in 4T1 sgControl and MHC-II knockout cells, with or without IFN-γ (10 ng/ml) treatment for 24 h, measured by flow cytometry. One-way ANOVA was used for statistical analysis, and Tukey’s multiple comparisons test was used for the post-hoc test. (B) The tumor growth of 4T1 sgControl (n = 10) and MHC-II knockout cells (n =10). Student’s t test was used for the statistical analysis. (C) Knockout of MHC-II in cancer cells increases the overall survival of mice. The primary tumors were removed on day 15, and the draining lymph nodes were removed on day 36. (D) Knockout of MHC-II in cancer cells led to a significant decrease in the proportion of cancer cells in the TDLNs. (E) Knockout of MHC-II in cancer cells resulted in a significant reduction in the proportion of Treg cells in the TDLNs. Student’s t test was used for statistical analysis; *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Overexpression of Ciita in cancer cells accelerates LNM
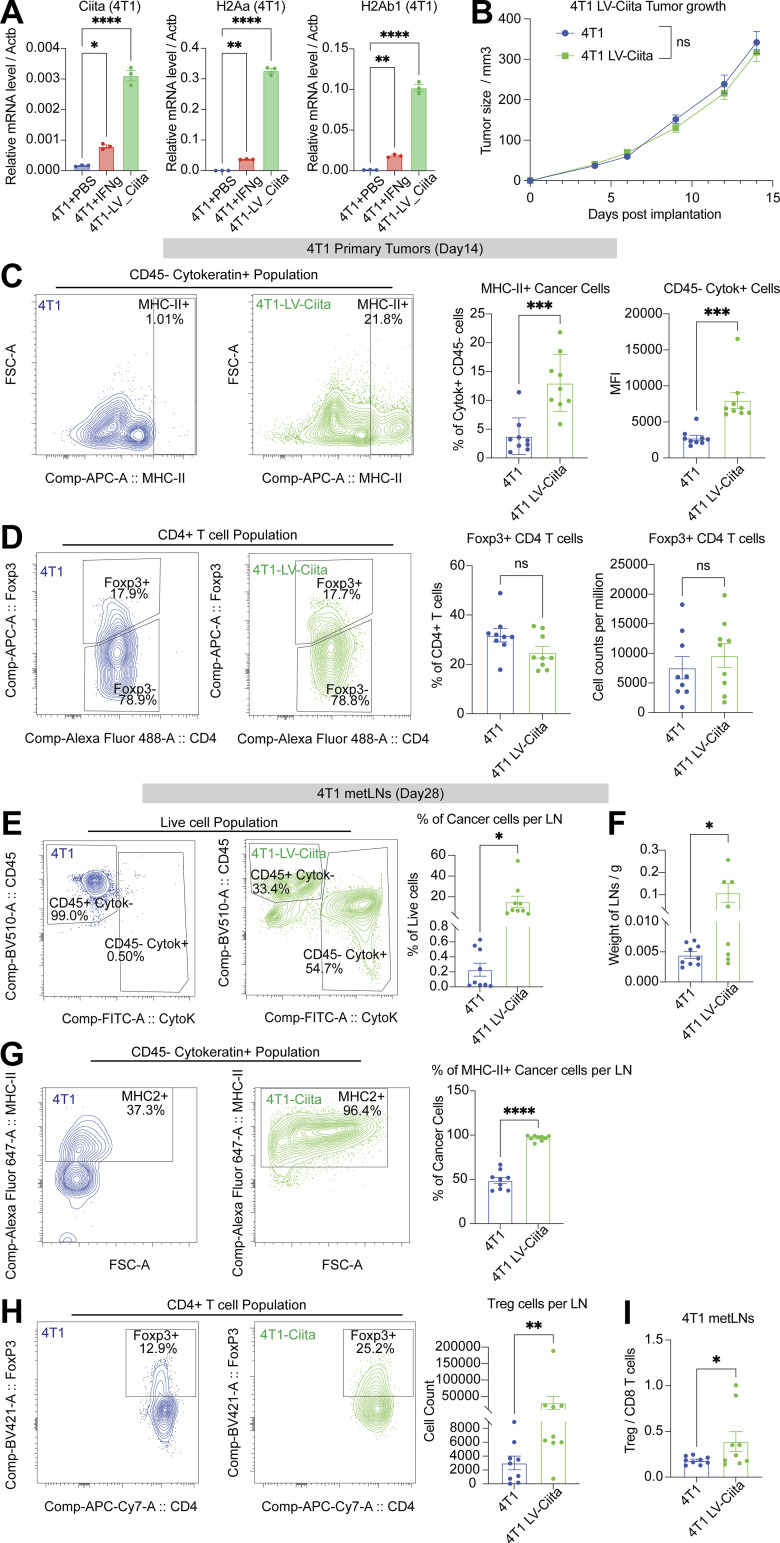
To determine if overexpression of MHC-II in cancer cells accelerates LNM, we generated 4T1 cancer cells that overexpressed Ciita. Ciita mRNA expression was measured using real-time PCR assay (Fig. 6 A), and the Ciita overexpression cancer cells had higher levels of MHC-II gene expression even in the absence of IFN-γ stimulation (Fig. 6 A). We observed no significant difference in primary tumor growth between the 4T1 WT cancer cells and the Ciita overexpression cancer cells (Fig. 6 B). On day 14, we collected primary tumors to investigate the MHC-II cancer cell phenotypes and the tumor microenvironment. We found that cancer cells from Ciita overexpressing primary tumors showed significantly higher levels of MHC-II expression (Fig. 6 C). Notably, the proportion of Treg cells remained unchanged between the 4T1 WT tumors and the Ciita overexpressing primary tumors (Fig. 6 D).
Figure 6.
Overexpression of Ciita in cancer cells accelerates LNM. (A) Ciita overexpression significantly enhances MHC-II gene expression in cancer cells compared with control cells. One-way ANOVA was used for statistical analysis, and Tukey’s multiple comparisons test was used for the post-hoc test. (B) Ciita overexpression in 4T1 cancer cells does not significantly alter primary tumor growth in Balb/c mice compared to WT 4T1 cells. (n = 9 per group). Student’s t test was used for the statistical analysis. (C) Flow cytometry analysis demonstrates increased cell surface expression of MHC-II in cancer cells of primary tumors with Ciita overexpression compared to control tumors. (D) Flow cytometry analysis shows no significant difference in the proportion of Treg cells in primary tumors with Ciita overexpression compared with control tumors. Student’s t test was used for the statistical analysis. (E) Flow cytometry analysis shows an increase in the proportion of cancer cells in metLNs with Ciita overexpression compared to control tumors. (F) metLNs from mice injected with Ciita-overexpressing cancer cells in the mammary fat pad have significantly more weight than metLNs from control tumor-bearing mice. (G) Flow cytometry analysis demonstrates an increase in the proportion of MHC-II+ cancer cells in metLNs with Ciita overexpression compared to control tumors. (H) Ciita overexpression leads to an increase in the number of Treg cells in metLNs compared with control tumors, as demonstrated by flow cytometry analysis. (I) Flow cytometry analysis shows a significant increase in the ratio of Treg cells to CD8 T cells in metLNs with Ciita overexpression compared to control tumors. Student’s t test was used for statistical analysis; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
On day 28, we collected the TDLNs to examine the metastatic burden and immune microenvironment. In the 4T1 WT metLNs, the average proportion of cancer cells was 0.23%, while the average proportion of cancer cells in the Ciita-overexpressing metLNs was 15.09%, showing 65 times more cancer cells in the lymph nodes from Ciita overexpressing tumors (Fig. 6 E). Moreover, the average weight of lymph nodes in the 4T1 WT group was 0.0045 g, whereas, in the Ciita-overexpressing group, it was 0.1086 g, a 24-fold increase (Fig. 6 F). Almost all cancer cells in the Ciita overexpressing metLNs exhibited high levels of MHC-II expression (Fig. 6 G), whereas ∼50% of cancer cells in the 4T1 WT group displayed lower levels of MHC-II expression. Although only 5–20% of cancer cells in Ciita overexpressing primary tumors were MHC-II positive (Fig. 6 C), our findings indicate that >95% of cancer cells in metLNs from Ciita overexpressing tumors express MHC-II molecules (Fig. 6 G). This suggests that cancer cells that express MHC-II have a higher tendency to establish metastasis in the lymph nodes. In addition, we observed a large population of Treg cells and a higher ratio of Treg/CD8 in the Ciita-overexpressing metLNs (Fig. 6, H and I). Taken together, our data indicate that the expression of Ciita in 4T1 cancer cells promotes tumor progression and LNM.
Invasion of cancer cells remodels human breast cancer sentinel lymph node microenvironment
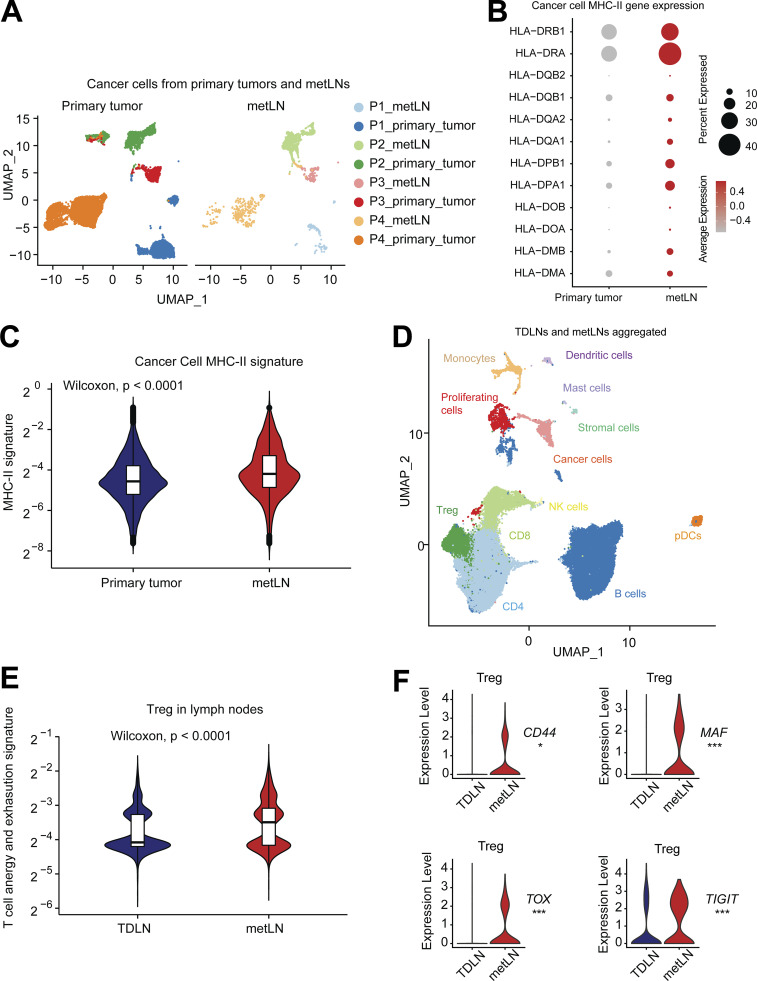
To understand the clinical significance of our findings, we utilized a human breast cancer single-cell transcriptome dataset with paired primary tumors and axillary lymph nodes from the same side (Xu et al., 2021). This dataset consists of specimens from five patients, including one Luminal B breast cancer patient, two Her2+ breast cancer patients, and two TNBC patients. For each of these patients, there were two lymph nodes collected for single-cell sequencing, including one metastasis-positive lymph node and one cancer-negative TDLN.
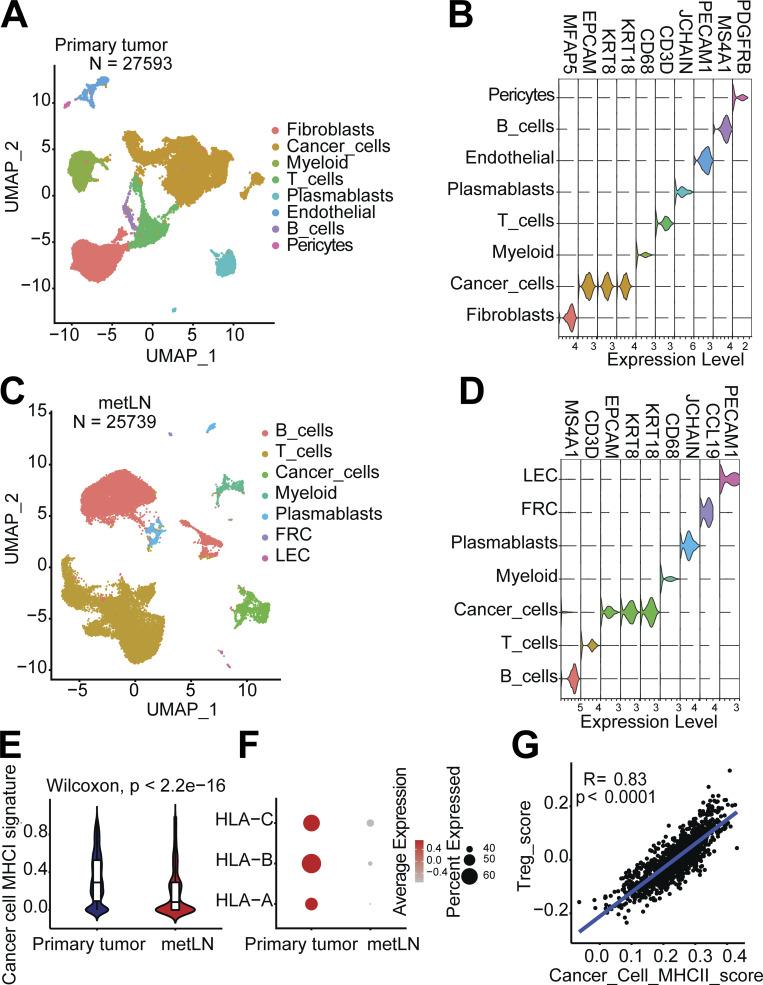
In the breast cancer primary tumors, we characterized 27,593 single cells that group into fibroblasts, cancer cells, myeloid cells, T cells, plasmablasts, endothelial cells, B cells, and pericytes (Fig. S5, A and B). In the metLNs, we characterized 25,739 single cells and identified B cells, T cells, cancer cells, myeloid cells, plasmablasts, fibroblastic reticular cells, and lymphatic endothelial cells based on their marker genes (Fig. S5, C and D). We integrated only the cancer cells from primary tumors and metLNs from patients with LNM. Interestingly, we found that cancer cells from the same patient cluster closer to each other (Fig. 7 A), showing the expected interpatient cancer cell heterogeneity. Next, we examined the expression of MHC-II molecules and gene signatures in all cancer cells. We found that both Her2+ and TNBC breast cancer metastatic LNs showed expression of MHC-II molecules and their MHC-II signatures were higher compared with primary tumors (Fig. 7, B and C). In contrast, the MHC-I gene signature and expression of MHC-I molecules were reduced in metLNs compared to the primary tumor in these human data (Fig. S5, E and F). These results suggest that the upregulation of MHC-II is a more universal phenotype in breast cancer that can lead to cancer cell immune evasion in lymph nodes.
Figure S5.
The single-cell atlas of human breast cancer and metLNs, and TCGA BRCA dataset analysis. (A) The UMAP of 27,593 cells from human breast tumors from NCBI GEO database GSE180286. We selected the top 30 PCs for the UMAP analysis with a minimum distance of 0.5 for the display. The resolution for the clustering is 0.5. (B) Violin plots show the gene expression levels of cell type-specific marker genes in breast tumors. (C) The UMAP of 25,739 cells from breast cancer metLNs from NCBI GEO database GSE180286. We selected the top 30 PCs for the UMAP analysis with a minimum distance of 0.5 for the display. The resolution for the clustering is 0.5. (D) Violin plots show the gene expression levels of cell type–specific marker genes in metLNs. (E) Violin plots show the MHC-I gene signature in cancer cells. (F) The expression profiling of MHC-I genes in cancer cells. (G) The correlation between Treg signature score and cancer cell MHC-II score in the TCGA breast cancer dataset. We retrieved the TCGA BRCA gene expression dataset using the TCGAbiolinks R package (Colaprico et al., 2016). The Treg cell signature score was defined based on the expression of CD4, FOXP3, and CTLA4, while the cancer cell MHC-II signature score was defined based on the expression of cytokeratin (KRT8 and KRT18) and MHC-II molecules. To evaluate the gene signature scores, we used the Singscore R package (Foroutan et al., 2018; Bhuva et al., 2020). Wilcoxon rank-sum test was used for the statistical analysis.
Figure 7.
Plasticity of T cell states in human breast cancer progression and metastasize to lymph nodes. (A) The UMAP of 14,646 aggregated cancer cells from human primary breast tumors and metLNs. The aggregated cancer cell UMAP is separated into primary tumors (left) and metLNs (right) for visualization. We selected the top 30 PCs for the UMAP analysis with a minimum distance of 0.5 for the display. (B) The expression profiling of MHC-II genes in cancer cells from primary tumors and metLNs. Red represents high expression, and gray represents low expression. The size of the circle represents the proportion of cells expressing the indicated genes in each cluster. (C) The MHC-II gene signature in cancer cells from primary tumors and metLNs. Single-cell MHC-II signature score was measured by UCell. (D) The UMAP of cells from TDLNs and metLNs, colored by cell types. (E) Violin plot shows the anergy and exhaustion gene signature in Tregs in TDLNs and metLNs from all the patients. Gene expression signature score was measured by UCell. The CD4+ T cell anergy and exhaustion genes were generated by Trefzer et al. (2021). (F) Violin plots show the gene expression of CD44, MAF, TOX, and TIGIT in Treg cells in the lymph nodes. Wilcoxon rank-sum test was used for statistical analysis; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To understand the impact of cancer cell invasion on T cells in patient lymph nodes, we integrated the lymph node single-cell datasets from patients with LNM to be able to compare the T cell phenotypes between TDLNs and metLNs (Fig. 7 D). Using a regulatable antigen presentation system, a recent study characterized a comprehensive CD4+ T cell anergy and exhaustion gene signature after exposure to persistent antigen stimulation (Trefzer et al., 2021). Using this CD4+ T cell anergy and exhaustion gene signature as a reference, we found that Treg cells in TDLN and metLN from the same patient exhibited very different phenotypes. In metLNs, the Tregs cells showed increasing in T cell anergy and exhaustion gene signatures (Fig. 7 E; Akimova et al., 2011; Neumann et al., 2019; Amoozgar et al., 2021). Of note, in one of the patients—who had the largest numbers of cancer cells in their metLN (Fig. 7 A)—we found the elevation of CD44, TOX, MAF, and TIGIT in Treg cells in metLNs (Fig. 7 F), which is consistent with metLNs in the mouse models (Fig. 4 E). Of note, we also calculated the Treg cell signature score and cancer cell MHC-II signature score in individual patients in The Cancer Genome Atlas (TCGA) BRCA dataset. The result indicates a strong positive correlation (R = 0.83, P < 0.0001) between cancer cell MHC-II score and Treg cell signature in primary tumors in breast cancer patients (Fig. S5 G).
Altogether, our study shows that in the progression of breast cancer cells to the lymph node, metastatic breast cancer cells displayed significant plasticity. Further, a subset of LNM cancer cells upregulates MHC-II in metLNs to induce immune tolerance and evade antitumor immunity.
Discussion
During metastasis, many epithelial-origin cancer cells undergo EMT (Puram et al., 2017; Tsoukalas et al., 2017; Karaayvaz et al., 2018; Loret et al., 2019; Dominguez et al., 2020), which involves reprogramming the expression of tight junction and cell membrane proteins. This results in a decrease in E-cadherin and EpCAM expression, and an increase in N-cadherin and vimentin expression (Yang et al., 2004; Mani et al., 2008; Shaul et al., 2014). These changes facilitate cancer cell detachment, which is a key step in dissemination. EMT of cancer cells in the primary tumor has been shown to result in increased LNM (Karlsson et al., 2017). A recent study showed mixed populations of malignant cells and various states of cancer cell EMT in human triple-negative breast tumors by using single-cell RNA-Seq (Karaayvaz et al., 2018). Several studies have shown that the transition of fully epithelial cells into fully mesenchymal cells occurs gradually (Pastushenko et al., 2018), increasing the probability of identifying cells along a continuum of the EMT process when looking at a single snapshot in time. Cancer cells in an EMT hybrid state exhibited increased invasion and tumor propagation (Ruscetti et al., 2015). In our study, most of the cancer cells in 4T1 primary tumors were triple-positive for Krt8, Krt18, and EpCAM. The mesenchymal cell marker genes, Twist1 and Vim, were also expressed in the EpCAM+ cancer cells, revealing these cells were in a hybrid state along the EMT continuum (Pastushenko and Blanpain, 2019).
In metLNs, we observed the presence of both mesenchymal-like and epithelial-like cancer cells (Fig. 1, F, L, and M). The epithelial-like cancer cells appeared round and tightly clustered in the center of the LNM, while some were observed at the tumor margins (Fig. 1 L). In contrast, the mesenchymal-like cancer cells were spindle-shaped and localized toward the margins of the metastases, near the subcapsular sinus of the lymph nodes (Fig. 1 M). Previous studies have demonstrated that metastatic cancer cells enter the lymph nodes through the subcapsular sinus and then invade the cortex and medullary region (Das et al., 2013; Jeong et al., 2015). Additionally, it has been shown that metastatic cancer cells can exit the lymph node through local blood vessels and spread to distant sites (Brown et al., 2018; Pereira et al., 2018). Based on our data, we propose that the LNM mesenchymal cancer cells are “seeds” from the primary tumor, some of which undergo MET and differentiate back to an epithelial phenotype in the center of the lesion where they are shielded from the native lymph node microenvironment and can better recreate a microenvironment that resembles the primary tumor. Supporting this concept, single-cell trajectory analysis revealed that LNM epithelial cancer cells were derived from LNM mesenchymal cancer cells in lymph nodes (Fig. 1, H–K).
A recent study using single-cell RNA-Seq to analyze six primary TNBC tumors revealed that a small population of cancer cells present in all patients had upregulated glycosphingolipid metabolism and an associated innate immunity pathway (Karaayvaz et al., 2018). The upregulation of these pathways was associated with a worse prognosis. In our own single-cell dataset, we observed similar upregulation of glycosphingolipid metabolism and innate immunity-associated genes in LNM (Fig. S2, H and I). This suggests that the upregulation of these pathways in cancer cells may contribute to the development or survival of LNM. It is worth noting that the previous study only looked at the phenotypes of primary tumors, whereas our data indicate that these pathways may play a role in the context of LNM.
In addition to cancer cell EMT plasticity, we uncovered an increase in the IFN-γ signaling pathway during metastatic breast cancer progression to lymph nodes (Fig. 3 A), which was associated with the upregulation of MHC-II molecules in cancer cells (Fig. 2 A). Specifically, we observed elevated expression of genes encoding MHC-II molecules in epithelial cancer cells from LNM (Fig. 2 E), suggesting a potential role for these cells in antigen presentation to CD4+ T cells. While MHC-II molecules are mainly found on professional antigen-presenting cells, epithelial cells can also express them and act as non-professional antigen-presenting cells, especially at mucosal sites, to preserve immune tolerance or surveillance (Rescigno et al., 2008; Biton et al., 2018; Wosen et al., 2018; Beyaz et al., 2021). However, we observed that MHC-II+ LNM cancer cells lacked costimulatory molecules Cd80 and Cd86 (Fig. 2 D), which are essential for CD4+ T cell activation. This suggests that MHC-II presence may promote tolerance through T cell anergy or induction of Treg differentiation (Nadafi et al., 2020). Consistent with this, we found high expression of Foxp3, Il2ra (CD25), Icos, Maf, and Tox in Tregs from the metLNs (Fig. 4 E). In vitro coculture of T cells with 4T1 cancer cells also showed a decrease of IFN-γ+ CD4+ T cells and an increase of Tregs (Fig. 4, F–H). Moreover, we measured impaired CD4+ T cell activation and higher numbers of Tregs in metLNs in response to strong antigens (Fig. 4, J and K) and confirmed the colocalization of MHC-II+ cancer cells and Foxp3+ T cells in the metLNs (Fig. 4 L). These findings highlight a previously unrecognized mechanism by which cancer cells interact with the CD4 T cells to induce immune tolerance during LNM.
Of note, we also found the presence of MHC-II+ cancer cells in human breast cancer metastatic LNs (Fig. 2, G and I; and Fig. 7 B). Cancer cells upregulated MHC-II across their progression trajectory from the primary tumor to LNM (Fig. 2, B and C). Notably, ablation of cancer cell MHC-II expression inhibited LNM and significantly extended mouse survival (Fig. 5, C and D). However, overexpressing Ciita in 4T1 cancer cells significantly accelerated LNM and promoted the expansion of Treg cells in the lymph nodes (Fig. 6, E–H). Emerging evidence suggests that cancer cells utilize MHC-II–induced tolerance mechanisms to evade antitumor immune responses, but the underlying mechanisms are not yet clear. For example, a recent single-cell analysis in pancreatic cancer identified a subset of cancer-associated fibroblasts that express high levels of MHC-II without costimulatory molecules, which may contribute to the immune evasion of cancer cells (Elyada et al., 2019). In contrast to these observations, we recently demonstrated that epithelial stem cell MHC-II expression plays a critical role in promoting antitumor immune surveillance during intestinal tumor initiation (Beyaz et al., 2021). Unlike breast cancer cells, intestinal stem cells exhibit the expression of costimulatory-like molecules (Biton et al., 2018; Beyaz et al., 2021). Furthermore, the differences in the microenvironment of lymph nodes and intestine, including the microbiome, may account for the contrasting functional outcomes on antitumor immunity elicited by epithelial MHC-II expression. Nonetheless, the functional significance of cancer MHC-II expression is context-dependent. The presence of MHC-II in melanoma cancer cells showed resistance to anti-CTLA4 treatment but not anti–PD-1 treatment (Rodig et al., 2018). In both melanoma and breast tumors, the presence of MHC-II+ cancer cells in the primary tumor is associated with LAG3+ and FCRL6+ tumor-infiltrating lymphocytes. MHC-II on cancer cells could interact with LAG3 and FCRL6 on lymphocytes, thus leading to immune evasion (Johnson et al., 2018). Furthermore, the elevation of the IFN-γ signaling pathway and the presence of MHC-II in cancer cells is associated with advanced high-grade serous ovarian cancer (Izar et al., 2020). Future studies are needed to delineate the precise kinetics of cancer-intrinsic MHC-II expression, cancer–immune cell interactions, and induction of tolerance mechanisms to develop actionable strategies that prevent immune evasion during the progression of metastatic breast cancer to lymph nodes.
IFN-γ is predominantly produced by NK cells, CD4+ T helper cells, and CD8+ cytotoxic T cells in the tumor microenvironment (Gocher et al., 2022). IFN-γ has pleiotropic effects in the tumor microenvironment and the consequences of IFN-γ exposure depend on the duration and magnitude of IFN-γ–induced signaling. The prolonged growth of cancer cells in lymph nodes accompanied by the ineffective T cell response could lead to persistent exposure to IFN-γ. The sustained IFN-γ signaling induces PD-L1–independent immune evasion and resistance (Benci et al., 2016). In a recent study using a syngeneic melanoma mouse model, cancer cells were serially selected for their preferential dissemination to the lymph node, revealing that the lymph node microenvironment can reprogram cancer cells to exhibit IFN-induced gene signatures (Reticker-Flynn et al., 2022). The study demonstrated that the induction of MHC-I and PD-L1 in cancer cells promoted LNM. In addition, this study showed an increase of Foxp3+CD25+ Treg cells in the metLNs. Using ovalbumin-expressing B16 melanoma cells, the study proved that IFN-licensed cancer cells induce profound antigen-specific Treg cells in the lymph nodes. Our study also demonstrated that in vitro administration of IFN-γ enhanced the expression of MHC-II on B16F10 cancer cells (Fig. S3 D), suggesting that these cancer cells may also have higher MHC-II gene expression enabling them to present antigens to conventional CD4 T cells and induce the formation of Tregs, providing a mechanism for the observations in Reticker-Flynn et al. (2022). In addition to cancer cell MHC-II induction of Tregs, it is important to acknowledge that other mechanisms, such as TGF-β–induced Treg expansion, may contribute to Treg cell accumulation in the TDLNs.
Previous research has demonstrated that pDCs are a major source of IFN-γ. Single-cell ligand–receptor analysis revealed frequent pDCs interactions with both cancer cells and Tregs in the lymph nodes (Fig. 4 A). It is plausible that pDCs are concurrently interacting with cancer cells and Treg cells in vivo, resulting in Treg cell expansion. Further investigations are required to prove this hypothesis.
In summary, our study provides a comprehensive analysis of the progression of breast cancer to LNM at a single-cell resolution. These findings help to characterize the disease progression from the primary tumor to the lymph node and shed light on how cancer cells evade immune surveillance and survive in the lymph node, a critical organ for priming an anticancer immune response (Alonso et al., 2018; Fransen et al., 2018). However, whether reprogramming of the lymph node microenvironment by cancer cells affects the anticancer immune function, response to immunotherapy, and clinical outcomes of patients with LNM remains to be determined. Further studies are needed to address these questions.
Materials and methods
Mice and in vivo study
6- to 12-wk-old female Balb/c mice were housed in animal facilities at Massachusetts General Hospital. 1 × 105 4T1 cells were injected into the fourth mammary fat pad (draining to the inguinal lymph node) in a volume of 50 μl per mouse. CYT387 (SelleckChem) was reconstituted in DMSO at 10 mg/ml and diluted in injection buffer (5% PEG400, 5% Tween80, and 90% H2O) for oral gavage every 2 d at 25 mg/kg for seven doses. All animal experimental protocols were reviewed and approved by the Institutional Animal Care and Use Committee of the Massachusetts General Hospital, Boston, MA.
UV-inactivated influenza virus PR8 inoculation
Influenza A virus (H1N1) A/Puerto Rico/8/34 (PR8) VR-95 and VR-95PQ were purchased from ATCC. Viruses were inactivated by UV light for 15 min at room temperature prior to injection. Mice were inoculated with 1 × 105 PFU viruses + 2 μg anti-CD40 (BioXCell, Cat: BE0016-2) + 2 μg poly I:C (InvivoGen, Cat: VAC-PIC) subcutaneously. On days 9–10, mice were euthanized and draining lymph nodes were collected.
Cell lines
4T1, 4T1 sgRNA control cells, 4T1 Ifngr1−/−, Ifngr2−/−, Ciita−/−, 4T1 Ciita overexpression (4T1-LV-Ciita), B16F10, E0771, and MCa-P1362 cell lines were used in this study and tested to be mycoplasma free (MycoAlert-Lonza). Cells were cultured in vitro in Dulbecco’s modified Eagle’s medium (GIBCO, Invitrogen Life Technologies) supplemented with 10% (vol/vol) fetal bovine serum (Atlanta Biologicals). Cells were maintained in a 5% CO2-humidified incubator at 37°C.
Immunohistochemical (IHC) staining
Mouse inguinal lymph nodes were collected and embedded in optimal cutting temperature compound (Tissue-Tek) and frozen immediately on dry ice. For immunofluorescence staining, the lymph nodes were cut into 10-μm serial sections using a cryostat. Several tissue sections representing different depths of the lymph node were selected for staining. A hydrophobic pen was used to draw a circle around the tissues and then air-dried. Tissues were fixed with 4% formaldehyde at room temperature for 5 min. Slides were washed with 1× PBS twice and then blocked with 5% normal donkey serum + 0.5% TritonX-100 (Millipore Sigma) for 30 min at room temperature. Cytokeratin-FITC (Cat: #F3418, 1:200 dilution; Sigma-Aldrich) was used for the staining of all the cancer cells in metLNs. E-Cadherin (Cat: #3195, rabbit anti-mouse, 1:100 dilution; CST) and vimentin (Cat: #3932, rabbit anti-mouse, 1:100 dilution; CST) were used for staining of epithelial and mesenchymal cancer cells in the lymph node, respectively. These primary antibodies were diluted in IHC blocking buffer (1× PBS, 5% normal donkey serum, 0.5% TritonX-100) and incubated at 4°C overnight. After primary antibody staining, slides were washed with IHC washing buffer (1× PBS + 0.5% TritonX-100) three to five times for 5 min each. Anti-rabbit-647 (1:200 dilution) was used for secondary antibody staining and incubated at room temperature for 1 h. In all the slides, DAPI (1:1,000) was used for nuclear staining at room temperature for 10–20 min. For cancer cells MHC-II staining, Cytokeratin-FITC (Cat: #F3418, 1:200 dilution; Sigma-Aldrich) was used for the staining of cancer cells in metLNs. Anti-mouse I-A/I-E-APC (Cat: #107614, clone M5/114.15.2, 1:100 dilution; BioLegend) was used for the MHC-II staining.
Formalin-fixed, paraffine-embedded deidentified human invasive ductal carcinoma specimens were obtained from the Massachusetts General Hospital Pathology Department. The human sample–related study was reviewed and approved by the Mass General Brigham Institutional Review Board (protocol #2021P001804). For immunofluorescence and immunohistochemistry staining, the tissues were cut into 5-μm thickness sections and mounted on glass slides, followed by deparaffinization and heat-induced epitope retrieval in citrate-based antigen retrieval solution. MHC-II (LGII-612.14, Cat: #68258S; CST) and Anti-human pan-Cytokeratin-Alexa647 (Cat: #628604; BioLegend) were used for the staining. For MHC-II IHC staining, we used 1:800 dilution. For Cytokeratin staining, we used 1:100 dilution and incubated the primary antibody at 4°C overnight.
Real-time PCR
Cells were administered IFN-γ (10 ng/ml, Cat: #575302; BioLegend) in vitro for 12 and 48 h. mRNA was extracted by RNeasy plus kit (Qiagen) according to the manufacturer’s instructions. RNA reverse transcription was performed by iScript cDNA synthesis kits (Bio-Rad). Real-time PCR mix with SYBR was purchased from Bio-Rad Laboratories. The primers for real-time PCR are listed in Table 1.
Table 1.
Primers for real-time PCR
| Primer targets | Primers |
|---|---|
| H2-Aa-F | 5′-TGGGAGTCTTGACTAAGAGGTC-3′ |
| H2-Aa-R | 5′-CTGACTTGCTATTTCTGAGCCAT-3′ |
| H2-Ab1-F | 5′-TGAACAGCCCAATGTCGTCAT-3′ |
| H2-Ab1-R | 5′-CAGCGCACTTTGATCTTGGC-3′ |
| Ciita-F | 5′-CAGGCTCCCACGGTAGAGA-3′ |
| Ciita-R | 5′-GGTAGAGATGTAGGGGGTCGG-3′ |
| Actb-F | 5′-GGCTGTATTCCCCTCCATCG-3′ |
| Actb-R | 5′-CCAGTTGGTAACAATGCCATGT-3′ |
CRISPR/Cas9-mediated gene disruption
sgRNA oligonucleotides for mouse Ifngr1, Ifngr2, Ciita, H2-Aa, H2-Ab1, and H2-Eb1 genes were cloned into lentiCRISPRv2 according to the published protocol (Ran et al., 2013). For each gene disrupted, 1 mg of the plasmid was transfected to 293T cells (#CRL-11268; ATCC) using FuGENE 6 Transfection Reagent (Promega) to produce the retroviruses. The viral supernatants were harvested at 48 and 72 h after transfection. To infect 1–2 × 105 cells in a 6-well plate, we added 1 ml of viral supernatants with 2 mg/ml polybrene and added 1 ml fresh DMEM medium. Knockout clones were identified either by Western blot or by flow cytometry analysis (Table 2).
Table 2.
Guide-RNAs for CRISPR/Cas9 gene knockout assays
| sgRNA | Original sequence 5′–3′ | BsmBI site added |
|---|---|---|
| Ifngr1 sgRNA 1 | 5′-CCAGGAAAGGTCGGGCCCCC-3′ | 5′-CACCGCCAGGAAAGGTCGGGCCCCC-3′ |
| Complementary | 5′-GGGGGCCCGACCTTTCCTGG-3′ | 5′-AAACGGGGGCCCGACCTTTCCTGGC-3′ |
| Ifngr1 sgRNA 2 | 5′-TGGTATTCCCAGCATACGAC-3′ | 5′-CACCGTGGTATTCCCAGCATACGAC-3′ |
| Complementary | 5′-GTCGTATGCTGGGAATACCA-3′ | 5′-AAACGTCGTATGCTGGGAATACCAC-3′ |
| Ifngr1 sgRNA 3 | 5′-GATCCTACATACGAAACATA-3′ | 5′-CACCGGATCCTACATACGAAACATA-3′ |
| Complementary | 5′-TATGTTTCGTATGTAGGATC-3′ | 5′-AAACTATGTTTCGTATGTAGGATCC-3′ |
| Ifngr2 sgRNA 1 | 5′-TCTGTGATGTCCGTACAGTT-3′ | 5′-CACCGTCTGTGATGTCCGTACAGTT-3′ |
| Complementary | 5′-AACTGTACGGACATCACAGA-3′ | 5′-AAACAACTGTACGGACATCACAGAC-3′ |
| Ifngr2 sgRNA 2 | 5′-TTCAGTATCTTGTCCACTAC-3′ | 5′-CACCGTTCAGTATCTTGTCCACTAC-3′ |
| Complementary | 5′-GTAGTGGACAAGATACTGAA-3′ | 5′-AAACGTAGTGGACAAGATACTGAAC-3′ |
| Ifngr2 sgRNA 3 | 5′-CACTGGTCTTGGGTCATTGC-3′ | 5′-CACCGCACTGGTCTTGGGTCATTGC-3′ |
| Complementary | 5′-GCAATGACCCAAGACCAGTG-3′ | 5′-AAACGCAATGACCCAAGACCAGTGC-3′ |
| Ciita sgRNA 1 | 5′-ATAGAGATCCCTGTAGAAGC-3′ | 5′-CACCGATAGAGATCCCTGTAGAAGC-3′ |
| Complementary | 5′-GCTTCTACAGGGATCTCTAT-3′ | 5′-AAACGCTTCTACAGGGATCTCTATC-3′ |
| Ciita sgRNA 2 | 5′-CCAGTTCAGCAAGCTGTTGC-3′ | 5′-CACCGCCAGTTCAGCAAGCTGTTGC-3′ |
| Complementary | 5′-GCAACAGCTTGCTGAACTGG-3′ | 5′-AAACGCAACAGCTTGCTGAACTGGC-3′ |
| Ciita sgRNA 3 | 5′-TACTGAAGAGGTCCTTGCTC-3′ | 5′-CACCGTACTGAAGAGGTCCTTGCTC-3′ |
| Complementary | 5′-GAGCAAGGACCTCTTCAGTA-3′ | 5′-AAACGAGCAAGGACCTCTTCAGTAC-3′ |
| H2-Aa sgRNA1 | 5′-ATAGCAAGTCAGTCGCAGA-3′ | 5′-CACCGATAGCAAGTCAGTCGCAGA-3′ |
| Complementary | 5′-TCTGCGACTGACTTGCTAT-3′ | 5′-AAACTCTGCGACTGACTTGCTATC-3′ |
| H2-Aa sgRNA2 | 5′-TATACCATAGGTGCCTACG-3′ | 5′-CACCGTATACCATAGGTGCCTACG-3′ |
| Complementary | 5′-CGTAGGCACCTATGGTATA-3′ | 5′-AAACCGTAGGCACCTATGGTATAC-3′ |
| H2-Ab1 sgRNA1 | 5′-CGTATGCGCTGCGTCCCGT-3′ | 5′-CACCGCGTATGCGCTGCGTCCCGT-3′ |
| Complementary | 5′-ACGGGACGCAGCGCATACG-3′ | 5′-AAACACGGGACGCAGCGCATACGC-3′ |
| H2-Ab1 sgRNA2 | 5′-GCGGCCAGACGCCGAGTAC-3′ | 5′-CACCGGCGGCCAGACGCCGAGTAC-3′ |
| Complementary | 5′-GTACTCGGCGTCTGGCCGC-3′ | 5′-AAACGTACTCGGCGTCTGGCCGCC-3′ |
| H2-Eb1 sgRNA1 | 5′-CAACGGGACGCAGCGCGTG-3′ | 5′-CACCGCAACGGGACGCAGCGCGTG-3′ |
| Complementary | 5′-CACGCGCTGCGTCCCGTTG-3′ | 5′-AAACCACGCGCTGCGTCCCGTTGC-3′ |
| H2-Eb1 sgRNA2 | 5′-GACGTGGGCGAGTTCCGCG-3′ | 5′-CACCGGACGTGGGCGAGTTCCGCG-3′ |
| Complementary | 5′-CGCGGAACTCGCCCACGTC-3′ | 5′-AAACCGCGGAACTCGCCCACGTCC-3′ |
Flow cytometry (cultured cells)
Cells were administrated IFN-γ (10 ng/ml, Cat: #575302; BioLegend) in vitro for 24–48 h. Cells were dissociated into single cells by 0.05% Trypsin-EDTA (Gibco) at 37°C for 10 min. After dissociation, cells were filtered by a 35-μm strainer cap twice. Fc block was added to the pellet (1:500) and kept on ice for 15 min. After blocking, cells were stained with MHC-I (H-2Kd/Dd-Alexa-647, clone 34-1-25, 1:200 dilution; BioLegend) or MHC-II (I-A/I-E-APC-Cy7, clone M5/114.15.2, 1:200; BioLegend) in FACS buffer (PBS with 1% BSA) on ice for 20 min. After staining, cells were fixed and spun down, and the cell pellet was resuspended in 150 μl FACS buffer and analyzed by a BD LSR-II flow cytometer.
Flow cytometry (mouse tissue)
We collected 4T1 metastatic inguinal lymph nodes from Balb/c mice. The lymph nodes were dissociated into single cells in digestion buffer (collagenase P 0.2 mg/ml, dispase II 0.8 mg/ml, DNase 0.1 mg/ml, RPMI with 1% FBS) at 37°C. Every 5–8 min, tubes were agitated and then the contents were allowed to resettle. Next, the supernatant was transferred to the collecting buffer (RPMI with 1% FBS and 2 mM EDTA). After 45–60 min, the lymph nodes were completely dissociated and centrifuged at 4°C, at 1,200 rpm for 5 min. After discarding the supernatant, Fc block was added to the pellet (1:500) and kept on ice for 15 min. After blocking, we stained with EpCAM-APC-eFluor780 (1:200 dilution) with FACS buffer (PBS with 1% BSA), leaving samples on ice in the dark for 20 min. After EpCAM staining, samples were washed with FACS buffer twice and then 1 ml of fixation/permeabilization working solution was added to each sample at room temperature for 60 min. After fixation, 2 ml of permeabilization buffer was added and the solution was centrifuged at 1,400 rpm for 5 min at room temperature. Antibodies for intracellular epitopes (cytokeratin-FITC, 1:100; vimentin-PE, 1:100) were then added and cells were incubated at room temperature in the dark for 60 min. After intracellular staining, cells were washed two to three times by adding 2 ml permeabilization buffer followed by centrifugation at 1,400 rpm for 5 min at room temperature. The cell pellet was resuspended in 150 µl FACS buffer and analyzed by a BD LSR-II flow cytometer.
A separate cohort of mice was used to measure the presence of MHC-II molecules on cancer cells in 4T1 primary tumors and metLNs. Briefly, 14 d after tumor implantation, we dissected the primary tumor and dissociated it into single cells as above. The mice were kept to further grow LNM. On day 28, we sacrificed the mice and collected inguinal lymph nodes for flow cytometry. After dissociation, the red blood cells were lysed by ACK buffer (Thermo Fisher Scientific) if needed. Then, cells were filtered by a 35-μm strainer cap twice. Fc block was added to the pellet (1:500) and kept on ice for 15 min. After blocking, cells were stained with MHC-II (I-A/I-E-BV605, 1:200; BioLegend) and EpCAM-FITC (1:200) in FACS buffer (PBS with 1% BSA) on ice for 20 min. After cell surface marker staining, the cells were fixed and permeabilized for intracellular staining of vimentin-PE (1:100). After intracellular staining, cells were washed two to three times by adding 2 ml permeabilization buffer followed by centrifugation at 1,400 rpm for 5 min at room temperature. The cell pellet was resuspended in 150 µl FACS buffer and analyzed by a BD flow cytometer.
Immune activation of lymph node
To measure T cell phenotype in metLNs, we set up four arms: saline, PR8, 4T1, and 4T1+PR8. In the saline group, we subcutaneously injected 10 ml saline into the mice thigh of tumor-naive mice. In the PR8 group, we inoculated 1 × 105 PFU UV-inactivated PR8 influenza virus + 2 μg anti-CD40 (Cat: BE0016-2; BioXCell) + 2 μg poly I:C (Cat: VAC-PIC; InvivoGen) via subcutaneous injection into the thigh of tumor-naive mice. 9–10 d later, we collected the draining lymph nodes in both groups for flow cytometry. In the 4T1 and 4T1+PR8 groups, we implanted 4T1 cancer cells into the fourth mammary fat pad. When the tumor reached 250 mm3, we dissected the primary tumor and randomized the mice into two groups. 5 d later, we inoculated 1 × 105 PFU UV-inactivated PR8 influenza virus + 2 μg anti-CD40 (Cat: BE0016-2; BioXCell) + 2 μg poly I:C (Cat: VAC-PIC; InvivoGen) in half of the mice (4T1+PR8 group) and saline in the other half of the mice (4T1 group) via thigh injection. Another 9 d later, draining lymph nodes were collected for flow cytometry.
Tissue collection and single-cell sequencing
4T1 cells were implanted into the left-side (supine position) fourth mammary fat pad of adult (6–10 wk) female Balb/c mice. All analyzed lymph nodes were dissociated into single cells with digestion buffer (200 μg/ml collagenase I, 800 μg/ml dispase, and 100 μg/ml DNase I). All samples used ACK buffer (Thermo Fisher Scientific) to lyse red blood cells and had the cell concentration and viability tested using a Nadia instrument (Dolomite Bio). The loading quantity of viable cells for 10X Genomics platform was estimated to capture around 5,000 cells. In metLN1 and metLN2 dataset, we harvested the inguinal lymph nodes for single-cell sequencing from tumor-bearing mice that had the primary tumor resected ∼2 wk after implantation or when the tumors reached ∼500 mm3. We harvested the inguinal lymph nodes from naive mice as a control. Naive LN1 and metLN1 were prepared and sequenced at the same time. Naive LN2 and metLN2 were prepared and sequenced at the same time. In the metLN2 dataset, we further enriched the stromal cells by FACS cell sorting for CD45− population. Naive LN1, naive LN2, metLN1, and metLN2 samples were sequenced with Illumina NextSeq High Output SE75 kit, with a format of 26 × 8 × 56. In the metLN3 dataset, we collected paired primary tumors and draining lymph nodes from the same mice (n = 4). The primary tumors were collected on day 14 or when tumor size reached ∼250 mm3. After resection, we sutured the wound and allowed these mice to grow spontaneous LNM. The primary tumors were cut into 1–2 mm pieces and dissociated into single cells. To decrease bias from individual mice, we collected primary tumors from 4 mice and pooled them together for single-cell library preparation. To avoid sequencing reagent and sample loading bias, the primary tumor single-cell RNA-Seq library was stored at −80°C until the metLN3 sample was available so that all sequencing occurred in the same flow-cell. On day 28, we sacrificed the mice and collected the inguinal lymph nodes (n = 4) to create the sample for the metLN3 dataset. In the metLN3 dataset, we enriched for tumor and stromal cells by MACS magnetic beads separation into CD45+ cells and CD45− cells with a CD45 Biotin antibody (10 μl CD45-biotin antibody in 500 μl MACS buffer [1× PBS, 1% FBS, 2 mM EDTA]). We estimated the proportion of cells from the lymph node and spiked-in 15,000 CD45+ cells into 1,000,000 CD45− cells. For 10X Genomics library preparation, we aimed to capture 5,000 cells from the combined metLN3 sample. The metLN1 and metLN2 samples were sequenced independently. The paired primary tumor and metLN3 libraries were combined and loaded in the same flow cell for sequencing on the Illumina HiSeq platform with High-Output mode PE26/98. Collectively, metLN1, metLN2, and metLN3 datasets after confirming the presence of cancer cells formed the data for the metLNs analyzed in this manuscript.
Bioinformatic data analysis
UMAP and clustering
Cellranger (version 3.0.2; https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome) was used to pre-process the fastq-format raw data. Cellranger aggr was used to combine data from different runs. After that, Seurat (Butler et al., 2018; Stuart et al., 2019; version 3.1.3, https://satijalab.org/seurat/) was used to perform the graph-based clustering and analysis of differentially expressed genes. For quality control, we depleted cells with <200 genes detected as well as genes expressed in less than three cells. We also removed cells that had >10,000 featured RNAs and >20% mitochondrial genes. Cleaned data were normalized by NormalizeData function with the method LogNormalize. The most variable genes were detected by FindVariableFeatures function with the selection method “vst.” After normalization, data were scaled by ScaleData, and RunPCA function was used to find the most significant principal components (PCs). For the primary tumor sample, we selected the top 50 PCs for UMAP analysis and clustering. Using different clustering resolutions for the UMAP analysis, we found that cluster 4 and cluster 9 were deemed as one group at a resolution of 0.8 and 0.9, while cluster 12 and cluster 16 were grouped together at a resolution of 0.8. When the resolution increased from 1.0 to 1.3, we did not see additional cancer cell clusters. In contrast, additional myeloid populations were found when increasing resolution. We decided to use resolution 1.0 for the downstream analysis as the cancer cell populations were well resolved. To normalize the batch bias of metLN samples, we employed Seurat integrated strategy (Stuart et al., 2019). We used the “FindIntegrationAnchors” function in the Seurat R package to obtain the anchors and subsequently applied the “IntegrateData” function to integrate the datasets and correct for technical differences between them. After integration, we selected the top 30 PCs for UMAP analysis. The min.dist for UMAP analysis is 0.5 for both primary tumors and metLNs. For the cancer cell–only analysis, we selected all cancer cell clusters in the primary tumor and metLNs, aggregated, and integrated them. The top 20 PCs were chosen for UMAP analysis and clustering.
Single-cell trajectory
Monocle (Trapnell et al., 2014; Qiu et al., 2017; version 2.10.1) was used to perform the trajectory analysis of cancer cells. Cell cycle regression of cancer cells was performed according to the Seurat cell cycle scoring and regression method based on the canonical marker genes from Nestorowa et al. (2016). The 2,000 most variable genes were selected for the trajectory analysis.
Gene set variation analysis (GSVA)
For the GSVA analysis, msigdbr (Liberzon et al., 2015; version 7.7.7) R package was used to retrieve the C2 curated gene sets from MSigDB (https://www.gsea-msigdb.org/gsea/msigdb). GSVA (Hänzelmann et al., 2013; version 1.30.0) R package was used for the single-cell gene set enrichment analysis. The limma (Ritchie et al., 2015; version 3.38.3) package was used to identify the differentially enriched gene sets across single-cell clusters.
Human breast cancer single-cell sequencing analysis
The human breast cancer single-cell dataset GSE180286 was downloaded from National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). Seurat was used for the single-cell UMAP analysis. For quality control, we depleted cells with <200 genes detected as well as genes expressed in less than three cells. We also removed cells that had >10,000 featured RNAs and >20% mitochondrial genes. Sctransform (https://github.com/ChristophH/sctransform/) was used for data normalization and cell cycle regression. The cancer cell in each of these samples was identified by scGate (https://github.com/carmonalab/scGate) with gene signature (KRT8+KRT8+). We used UCell R package (https://github.com/carmonalab/UCell) to measure the single-cell gene signatures (Andreatta and Carmona, 2021).
GO annotation and cell-–cell interaction analysis
ShinyGO version 0.61 (http://bioinformatics.sdstate.edu/go/; Ge et al., 2020) was used for the GO functional annotation, with P value cutoff of 0.05 and the top 30 most significant GO terms returned. CellChat (Jin et al., 2021; version 1.4.0) R package was used for ligand–receptor analysis with the manually curated database of literature-supported ligand–receptor interactions in CellChatDB mouse.
Imaging processing
Stained lymph node sections were imaged by confocal microscopy (Olympus 1X81) using 10× air and 20× air objectives (Olympus). ImageJ (https://imagej.net/ImageJ) was used to process the multi-tiff format raw image data. Human breast cancer immunofluorescence images were analyzed in QuPath (Bankhead et al., 2017). For human breast cancer image data analysis, cells were identified based on a positive DAPI signal, and each of the cell populations was classified as positive or negative based on a single intensity threshold on mean expression within the cell. MHC-II+ cells located in both the tumor (Cytokeratin+) and stromal (Cytokeratin−) compartments were included in the quantitative analysis. The mean proportion of MHC-II+ Cytokeratin+ cells was subsequently calculated and reported.
Statistical analysis
The unpaired t test and one-way ANOVA was conducted across each group. An alpha value of 0.05 was considered statistically significant. All analyses were performed using Prism version 9 Software (GraphPad).
Online supplemental material
Fig. S1 shows the clusters of cells and the expression of selected marker genes for each cell type in the 4T1 primary tumors and metLN microenvironment. Fig. S2 displays the EMT plasticity of cancer cells in the metLNs and the alteration of signaling pathways during breast cancer progression to the lymph nodes. Fig. S3 demonstrates that IFN-γ can activate the expression of MHC-II+ in various types of cancer cells, and the in vivo experiment of Cyt387, a JAK1/2 inhibitor. Fig. S4 reveals the single-cell microenvironment of naive lymph nodes in Balb/c mice. Fig. S5 presents the single-cell microenvironment of human breast cancer and metLNs, the expression of MHC-I in cancer cells, and includes an analysis of TCGA breast cancer data. Table S1 provides a summary of the antibodies and reagents used in this study.
Supplementary Material
lists antibodies and reagents.
Acknowledgments
The authors would like to acknowledge Dr. Rakesh K. Jain, Dr. Yves Boucher, and Dr. Michael Carroll for critical discussions. The authors would also like to thank Cold Spring Harbor Laboratory Cancer Center Shared Resources (Single Cell Biology and Sequencing Technologies), supported in part by the National Cancer Institute Cancer Center Support Grant 5P30CA045508, as well as Ulandt Kim from the NextGen core at Massachusetts General Hospital for the help with single-cell sequencing. We would also like to acknowledge Dr. Naxerova Kamila for material support.
This work was financially supported by grants from the National Institutes of Health (R21AI097745, R01CA214913, R01HL128168, R01CA284372, and R01CA284603 to T.P. Padera; P30CA045508-33 to S. Beyaz; F32CA275298 to M.J. O'Melia; K00CA234940 to H. Zhou; K22CA230315 to D. Jones; and T32HL007627 and T32CA251062 to J.M. Posada), the Rullo Family Massachusetts General Hospital Research Scholar Award (T.P. Padera), the Oliver S. and Jennie R. Donaldson Charitable Trust (S. Beyaz), the G. Harold and Leila Y. Mathers Charitable Foundation (S. Beyaz), the Mark Foundation for Cancer Research (20-028-EDV to S. Beyaz), Chan Zuckerberg Initiative/Silicon Valley Community Foundation (2021-239862 to S. Beyaz), STARR Cancer Consortium (I13-0052 to S. Beyaz), the Cold Spring Harbor Laboratory and Northwell Health Affiliation (S. Beyaz), New York Genome Center Polyethnic-1000 Initiative (S. Beyaz), Walter Benjamin Programme, Deutsche Forschunsgemeinschaft number ME5486/1-2 (L. Menzel), and an Agency for Science, Technology and Research (A*STAR) graduate scholarship (A.S. Kumar and W.W. Ho). D. Jones was supported by a METAvivor Early Career Investigator Grant, American Association for Cancer Research/Breast Cancer Research Foundation Career Development Award, and the Karin Grunebaum Cancer Research Foundation.
Author contribution: P.J. Lei, E.R. Pereira, T.P. Padera, and S. Beyaz conceived and designed the study and analyzed data. P.J. Lei, S. Beyaz, and T.P. Padera wrote the manuscript, which all coauthors commented on. E.R. Pereira, D. Jones, P.J. Lei, P. Andersson, J. Wijnbergen, S. Chatterjee, Z. Amoozgar, W.W. Ho, J.M. Posada, M.J. O’Melia, H. Zhou, L. Menzel, and S. Morita designed and performed experiments and analyzed data. P.J. Lei, M.J. O’Melia, L. Menzel, S. Chatterjee, Z. Amoozgar, and W.W. Ho performed and analyzed flow cytometry experiments. C. Chung, I. Ergin, and S. Beyaz helped with the single-cell sequencing experiment. P. Andersson and J. Wijnbergen helped with the animal work. D. Jones advised and helped with the manuscript. J.M. Posada helped with the breast cancer pathology data. A.S. Kumar helped with human breast cancer pathology data quantification. P. Huang helped with the animal experiment. All authors read and accepted the manuscript.
Data availability
Single-cell mRNA sequencing data generated to support this study have been deposited in NCBI GEO under accession number GSE168181. The authors declare that all other data supporting the findings of this study are available within the paper and its supplementary information files.
References
- Akimova, T., Beier U.H., Wang L., Levine M.H., and Hancock W.W.. 2011. Helios expression is a marker of T cell activation and proliferation. PLoS One. 6:e24226. 10.1371/journal.pone.0024226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, R., Flament H., Lemoine S., Sedlik C., Bottasso E., Péguillet I., Prémel V., Denizeau J., Salou M., Darbois A., et al. 2018. Induction of anergic or regulatory tumor-specific CD4+ T cells in the tumor-draining lymph node. Nat. Commun. 9:2113. 10.1038/s41467-018-04524-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoozgar, Z., Kloepper J., Ren J., Tay R.E., Kazer S.W., Kiner E., Krishnan S., Posada J.M., Ghosh M., Mamessier E., et al. 2021. Targeting Treg cells with GITR activation alleviates resistance to immunotherapy in murine glioblastomas. Nat. Commun. 12:2582. 10.1038/s41467-021-22885-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreatta, M., and Carmona S.J.. 2021. UCell: Robust and scalable single-cell gene signature scoring. Comput. Struct. Biotechnol. J. 19:3796–3798. 10.1016/j.csbj.2021.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod, M.L., Cook R.S., Johnson D.B., and Balko J.M.. 2019. Biological consequences of MHC-II expression by tumor cells in cancer. Clin. Cancer Res. 25:2392–2402. 10.1158/1078-0432.CCR-18-3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankhead, P., Loughrey M.B., Fernández J.A., Dombrowski Y., McArt D.G., Dunne P.D., McQuaid S., Gray R.T., Murray L.J., Coleman H.G., et al. 2017. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 7:16878. 10.1038/s41598-017-17204-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benci, J.L., Xu B., Qiu Y., Wu T.J., Dada H., Twyman-Saint Victor C., Cucolo L., Lee D.S.M., Pauken K.E., Huang A.C., et al. 2016. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell. 167:1540–1554.e12. 10.1016/j.cell.2016.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyaz, S., Chung C., Mou H., Bauer-Rowe K.E., Xifaras M.E., Ergin I., Dohnalova L., Biton M., Shekhar K., Eskiocak O., et al. 2021. Dietary suppression of MHC class II expression in intestinal epithelial cells enhances intestinal tumorigenesis. Cell Stem Cell. 28:1922–1935.e5. 10.1016/j.stem.2021.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuva, D.D., Cursons J., and Davis M.J.. 2020. Stable gene expression for normalisation and single-sample scoring. Nucleic Acids Res. 48:e113–e113. 10.1093/nar/gkaa802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnewies, M., Roberts E.W., Kersten K., Chan V., Fearon D.F., Merad M., Coussens L.M., Gabrilovich D.I., Ostrand-Rosenberg S., Hedrick C.C., et al. 2018. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 24:541–550. 10.1038/s41591-018-0014-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biton, M., Haber A.L., Rogel N., Burgin G., Beyaz S., Schnell A., Ashenberg O., Su C.W., Smillie C., Shekhar K., et al. 2018. T helper cell cytokines modulate intestinal stem cell renewal and differentiation. Cell. 175:1307–1320.e22. 10.1016/j.cell.2018.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, M., Assen F.P., Leithner A., Abe J., Schachner H., Asfour G., Bago-Horvath Z., Stein J.V., Uhrin P., Sixt M., and Kerjaschki D.. 2018. Lymph node blood vessels provide exit routes for metastatic tumor cell dissemination in mice. Science. 359:1408–1411. 10.1126/science.aal3662 [DOI] [PubMed] [Google Scholar]
- Butler, A., Hoffman P., Smibert P., Papalexi E., and Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J.M., Zaidi S., Love J.R., Zhao J.L., Setty M., Wadosky K.M., Gopalan A., Choo Z.N., Persad S., Choi J., et al. 2022. Lineage plasticity in prostate cancer depends on JAK/STAT inflammatory signaling. Science. 377:1180–1191. 10.1126/science.abn0478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaprico, A., Silva T.C., Olsen C., Garofano L., Cava C., Garolini D., Sabedot T.S., Malta T.M., Pagnotta S.M., Castiglioni I., et al. 2016. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Research. 44:e71–e71. 10.1093/nar/gkv1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly, K.A., Kuchroo M., Venkat A., Khatun A., Wang J., William I., Hornick N.I., Fitzgerald B.L., Damo M., Kasmani M.Y., et al. 2021. A reservoir of stem-like CD8+ T cells in the tumor-draining lymph node preserves the ongoing antitumor immune response. Sci. Immunol. 6:eabg7836. 10.1126/sciimmunol.abg7836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammeijer, F., van Gulijk M., Mulder E.E., Lukkes M., Klaase L., van den Bosch T., van Nimwegen M., Lau S.P., Latupeirissa K., Schetters S., et al. 2020. The PD-1/PD-L1-checkpoint restrains T cell immunity in tumor-draining lymph nodes. Cancer Cell. 38:685–700.e8. 10.1016/j.ccell.2020.09.001 [DOI] [PubMed] [Google Scholar]
- Das, S., Sarrou E., Podgrabinska S., Cassella M., Mungamuri S.K., Feirt N., Gordon R., Nagi C.S., Wang Y., Entenberg D., et al. 2013. Tumor cell entry into the lymph node is controlled by CCL1 chemokine expressed by lymph node lymphatic sinuses. J. Exp. Med. 210:1509–1528. 10.1084/jem.20111627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David Nathanson, S., Leonard-Murali S., Burmeister C., Susick L., and Baker P.. 2020. Clinicopathological evaluation of the potential anatomic pathways of systemic metastasis from primary breast cancer suggests an orderly spread through the regional lymph nodes. Ann. Surg. Oncol. 27:4810–4818. 10.1245/s10434-020-08904-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez, C.X., Müller S., Keerthivasan S., Koeppen H., Hung J., Gierke S., Breart B., Foreman O., Bainbridge T.W., Castiglioni A., et al. 2020. Single-cell RNA sequencing reveals stromal evolution into LRRC15+ myofibroblasts as a determinant of patient response to cancer immunotherapy. Cancer Discov. 10:232–253. 10.1158/2159-8290.CD-19-0644 [DOI] [PubMed] [Google Scholar]
- Dongre, A., Rashidian M., Reinhardt F., Bagnato A., Keckesova Z., Ploegh H.L., and Weinberg R.A.. 2017. Epithelial-to-mesenchymal transition contributes to immunosuppression in breast carcinomas. Cancer Res. 77:3982–3989. 10.1158/0008-5472.CAN-16-3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donker, M., van Tienhoven G., Straver M.E., Meijnen P., van de Velde C.J., Mansel R.E., Cataliotti L., Westenberg A.H., Klinkenbijl J.H., Orzalesi L., et al. 2015. Radiotherapy or surgery of the axilla after a positive sentinel node in breast cancer (EORTC 10981-22023 AMAROS): A randomised, multicentre, open-label, phase 3 non-inferiority trial. Lancet Oncol. 15:1303–1310. 10.1016/S1470-2045(14)70460-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Bois, H., Heim T.A., and Lund A.W.. 2021. Tumor-draining lymph nodes: At the crossroads of metastasis and immunity. Sci. Immunol. 6:eabg3551. 10.1126/sciimmunol.abg3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elyada, E., Bolisetty M., Laise P., Flynn W.F., Courtois E.T., Burkhart R.A., Teinor J.A., Belleau P., Biffi G., Lucito M.S., et al. 2019. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 9:1102–1123. 10.1158/2159-8290.CD-19-0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnsworth, R.H., Achen M.G., and Stacker S.A.. 2018. The evolving role of lymphatics in cancer metastasis. Curr. Opin. Immunol. 53:64–73. 10.1016/j.coi.2018.04.008 [DOI] [PubMed] [Google Scholar]
- Fernandez Gonzalez, S., Jayasekera J.P., and Carroll M.C.. 2008. Complement and natural antibody are required in the long-term memory response to influenza virus. Vaccine. 26:I86–I93. 10.1016/j.vaccine.2008.11.057 [DOI] [PubMed] [Google Scholar]
- Foroutan, M., Bhuva D.D., Lyu R., Horan K., Cursons J., and Davis M.J.. 2018. Single sample scoring of molecular phenotypes. BMC Bioinformatics. 19(1):404–404. 10.1186/s12859-018-2435-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen, M.F., Schoonderwoerd M., Knopf P., Camps M.G., Hawinkels L.J., Kneilling M., van Hall T., and Ossendorp F.. 2018. Tumor-draining lymph nodes are pivotal in PD-1/PD-L1 checkpoint therapy. JCI Insight. 3:1–7. 10.1172/jci.insight.124507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, S., Zhang N., Yopp A.C., Chen D., Mao M., Chen D., Zhang H., Ding Y., and Bromberg J.S.. 2004. TGF-beta induces Foxp3 + T-regulatory cells from CD4 + CD25 - precursors. Am. J. Transplant. 4:1614–1627. 10.1111/j.1600-6143.2004.00566.x [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz, A., Shin D.S., Moreno B.H., Saco J., Escuin-Ordinas H., Rodriguez G.A., Zaretsky J.M., Sun L., Hugo W., Wang X., et al. 2017. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 19:1189–1201. 10.1016/j.celrep.2017.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, S.X., Jung D., and Yao R.. 2020. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics. 36:2628–2629. 10.1093/bioinformatics/btz931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano, A.E., Hunt K.K., Ballman K.V., Beitsch P.D., Whitworth P.W., Blumencranz P.W., Leitch A.M., Saha S., McCall L.M., and Morrow M.. 2011. Axillary dissection vs no axillary dissection in women with invasive breast cancer and sentinel node metastasis: A randomized clinical trial. JAMA. 305:569–575. 10.1001/jama.2011.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocher, A.M., Workman C.J., and Vignali D.A.A.. 2022. Interferon-γ: Teammate or opponent in the tumour microenvironment? Nat. Rev. Immunol. 22:158–172. 10.1038/s41577-021-00566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, X., Polesso F., Wang C., Sehrawat A., Hawkins R.M., Murray S.E., Thomas G.V., Caruso B., Thompson R.F., Wood M.A., et al. 2022. Androgen receptor activity in T cells limits checkpoint blockade efficacy. Nature. 606:791–796. 10.1038/s41586-022-04522-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänzelmann, S., Castelo R., and Guinney J.. 2013. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 14:7. 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Aya, L.F., Chavez-Macgregor M., Lei X., Meric-Bernstam F., Buchholz T.A., Hsu L., Sahin A.A., Do K.A., Valero V., Hortobagyi G.N., and Gonzalez-Angulo A.M.. 2011. Nodal status and clinical outcomes in a large cohort of patients with triple-negative breast cancer. J. Clin. Oncol. 29:2628–2634. 10.1200/JCO.2010.32.1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izar, B., Tirosh I., Stover E.H., Wakiro I., Cuoco M.S., Alter I., Rodman C., Leeson R., Su M.J., Shah P., et al. 2020. A single-cell landscape of high-grade serous ovarian cancer. Nat. Med. 26:1271–1279. 10.1038/s41591-020-0926-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasekera, J.P., Moseman E.A., and Carroll M.C.. 2007. Natural antibody and complement mediate neutralization of influenza virus in the absence of prior immunity. J. Virol. 81:3487–3494. 10.1128/JVI.02128-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, H.S., Jones D., Liao S., Wattson D.A., Cui C.H., Duda D.G., Willett C.G., Jain R.K., and Padera T.P.. 2015. Investigation of the lack of angiogenesis in the formation of lymph node metastases. J. Natl. Cancer Inst. 107:1–11. 10.1093/jnci/djv155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.H., Myung P., Plikus M.V., and Nie Q.. 2021. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12:1088. 10.1038/s41467-021-21246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, D.B., Nixon M.J., Wang Y., Wang D.Y., Castellanos E., Estrada M.V., Ericsson-Gonzalez P.I., Cote C.H., Salgado R., Sanchez V., et al. 2018. Tumor-specific MHC-II expression drives a unique pattern of resistance to immunotherapy via LAG-3/FCRL6 engagement. JCI Insight. 3:e120360. 10.1172/jci.insight.120360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D., Pereira E.R., and Padera T.P.. 2018. Growth and immune evasion of lymph node metastasis. Front. Oncol. 8:36. 10.3389/fonc.2018.00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D., Wang Z., Chen I.X., Zhang S., Banerji R., Lei P.J., Zhou H., Xiao V., Kwong C., van Wijnbergen J.W.M., et al. 2021. Solid stress impairs lymphocyte infiltration into lymph-node metastases. Nat. Biomed. Eng. 5:1426–1436. 10.1038/s41551-021-00766-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaayvaz, M., Cristea S., Gillespie S.M., Patel A.P., Mylvaganam R., Luo C.C., Specht M.C., Bernstein B.E., Michor F., and Ellisen L.W.. 2018. Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nat. Commun. 9:3588. 10.1038/s41467-018-06052-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson, M.C., Gonzalez S.F., Welin J., and Fuxe J.. 2017. Epithelial-mesenchymal transition in cancer metastasis through the lymphatic system. Mol. Oncol. 11:781–791. 10.1002/1878-0261.12092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczma, M.P., Szurek E.A., Cebula A., Ngo V.L., Pietrzak M., Kraj P., Denning T.L., and Ignatowicz L.. 2021. Self and microbiota-derived epitopes induce CD4+ T cell anergy and conversion into CD4+Foxp3+ regulatory cells. Mucosal Immunol. 14:443–454. 10.1038/s41385-020-00349-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.K., Jeong S.H., Jang C., Bae H., Kim Y.H., Park I., Kim S.K., and Koh G.Y.. 2019. Tumor metastasis to lymph nodes requires YAP-dependent metabolic adaptation. Science. 363:644–649. 10.1126/science.aav0173 [DOI] [PubMed] [Google Scholar]
- Liberzon, A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., and Tamayo P.. 2015. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1:417–425. 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loret, N., Denys H., Tummers P., and Berx G.. 2019. The role of epithelial-to-mesenchymal plasticity in ovarian cancer progression and therapy resistance. Cancers. 11:838. 10.3390/cancers11060838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair, F., Erickson J.R., Frutoso M., Konecny A.J., Greene E., Voillet V., Maurice N.J., Rongvaux A., Dixon D., Barber B., et al. 2022. Extricating human tumour immune alterations from tissue inflammation. Nature. 605:728–735. 10.1038/s41586-022-04718-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani, S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., et al. 2008. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 133:704–715. 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie, J.C., Letterio J.J., Gavin M., and Rudensky A.Y.. 2005. TGF-β1 maintains suppressor function and Foxp3 expression in CD4+CD25+ regulatory T cells. J. Exp. Med. 201:1061–1067. 10.1084/jem.20042276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda, K., Pace D.A., Cintron R., Rodrigues J.C., Fang J., Smith A., Rohloff P., Coelho E., de Haas F., de Souza W., et al. 2011. Characterization of a novel organelle in Toxoplasma gondii with similar composition and function to the plant vacuole. Mol. Microbiol. 76:1358–1375. 10.1111/j.1365-2958.2010.07165.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molodtsov, A.K., Khatwani N., Vella J.L., Lewis K.A., Zhao Y., Han J., Sullivan D.E., Searles T.G., Preiss N.K., Shabaneh T.B., et al. 2021. Resident memory CD8+ T cells in regional lymph nodes mediate immunity to metastatic melanoma. Immunity. 54:2117–2132.e7. 10.1016/j.immuni.2021.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadafi, R., Gago de Graça C., Keuning E.D., Koning J.J., de Kivit S., Konijn T., Henri S., Borst J., Reijmers R.M., van Baarsen L.G.M., and Mebius R.E.. 2020. Lymph node stromal cells generate antigen-specific regulatory T cells and control autoreactive T and B cell responses. Cell Rep. 30:4110–4123.e4. 10.1016/j.celrep.2020.03.007 [DOI] [PubMed] [Google Scholar]
- Nestorowa, S., Hamey F.K., Pijuan Sala B., Diamanti E., Shepherd M., Laurenti E., Wilson N.K., Kent D.G., and Göttgens B.. 2016. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood. 128:e20–e31. 10.1182/blood-2016-05-716480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann, C., Blume J., Roy U., Teh P.P., Vasanthakumar A., Beller A., Liao Y., Heinrich F., Arenzana T.L., Hackney J.A., et al. 2019. c-Maf-dependent Treg cell control of intestinal TH17 cells and IgA establishes host-microbiota homeostasis. Nat. Immunol. 20:471–481. 10.1038/s41590-019-0316-2 [DOI] [PubMed] [Google Scholar]
- Pascual, G., Avgustinova A., Mejetta S., Martín M., Castellanos A., Attolini C.S., Berenguer A., Prats N., Toll A., Hueto J.A., et al. 2017. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature. 541:41–45. 10.1038/nature20791 [DOI] [PubMed] [Google Scholar]
- Pastushenko, I., and Blanpain C.. 2019. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 29:212–226. 10.1016/j.tcb.2018.12.001 [DOI] [PubMed] [Google Scholar]
- Pastushenko, I., Brisebarre A., Sifrim A., Fioramonti M., Revenco T., Boumahdi S., Van Keymeulen A., Brown D., Moers V., Lemaire S., et al. 2018. Identification of the tumour transition states occurring during EMT. Nature. 556:463–468. 10.1038/s41586-018-0040-3 [DOI] [PubMed] [Google Scholar]
- Pei, D., Shu X., Gassama-Diagne A., and Thiery J.P.. 2019. Mesenchymal-epithelial transition in development and reprogramming. Nat. Cell Biol. 21:44–53. 10.1038/s41556-018-0195-z [DOI] [PubMed] [Google Scholar]
- Pereira, E.R., Kedrin D., Seano G., Gautier O., Meijer E.F.J., Jones D., Chin S.M., Kitahara S., Bouta E.M., Chang J., et al. 2018. Lymph node metastases can invade local blood vessels, exit the node, and colonize distant organs in mice. Science. 359:1403–1407. 10.1126/science.aal3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulaski, B.A., and Ostrand-Rosenberg S.. 2001. Mouse 4T1 breast tumor model. Curr. Protoc. Immunol. 20:Unit 20.2. 10.1002/0471142735.im2002s39 [DOI] [PubMed] [Google Scholar]
- Puram, S.V., Tirosh I., Parikh A.S., Patel A.P., Yizhak K., Gillespie S., Rodman C., Luo C.L., Mroz E.A., Emerick K.S., et al. 2017. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 171:1611–1624.e24. 10.1016/j.cell.2017.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, X., Hill A., Packer J., Lin D., Ma Y.A., and Trapnell C.. 2017. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods. 14:309–315. 10.1038/nmeth.4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rescigno, M., Lopatin U., and Chieppa M.. 2008. Interactions among dendritic cells, macrophages, and epithelial cells in the gut: Implications for immune tolerance. Curr. Opin. Immunol. 20:669–675. 10.1016/j.coi.2008.09.007 [DOI] [PubMed] [Google Scholar]
- Reticker-Flynn, N.E., Basto P.A., Zhang W., Bejnood A., Kenkel J.A., Martins M.M., Chang S., Gentles A.J., Sunwoo J.B., Plevritis S.K., et al. 2020. Abstract PR04: Lymph node colonization promotes distant tumor metastasis through the induction of tumor-specific immunosuppression. Cancer Res. 80:PR04-PR04. 10.1158/1538-7445.CAMODELS2020-PR04 [DOI] [Google Scholar]
- Reticker-Flynn, N.E., Zhang W., Belk J.A., Basto P.A., Escalante N.K., Pilarowski G.O.W., Bejnood A., Martins M.M., Kenkel J.A., Linde I.L., et al. 2022. Lymph node colonization induces tumor-immune tolerance to promote distant metastasis. Cell. 185:1924–1942.e23. 10.1016/j.cell.2022.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., and Smyth G.K.. 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodig, S.J., Gusenleitner D., Jackson D.G., Gjini E., Giobbie-Hurder A., Jin C., Chang H., Lovitch S.B., Horak C., Weber J.S., et al. 2018. MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. Sci. Transl. Med. 10:1–14. 10.1126/scitranslmed.aar3342 [DOI] [PubMed] [Google Scholar]
- Ruscetti, M., Quach B., Dadashian E.L., Mulholland D.J., and Wu H.. 2015. Tracking and functional characterization of epithelial–mesenchymal transition and mesenchymal tumor cells during prostate cancer metastasis. Cancer Res. 75:2749–2759. 10.1158/0008-5472.CAN-14-3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahovaler, A., Krishnan R.J., Yeh D.H., Zhou Q., Palma D., Fung K., Yoo J., Nichols A., and MacNeil S.D.. 2019. Outcomes of cutaneous squamous cell carcinoma in the head and neck region with regional lymph node metastasis: A systematic review and meta-analysis. JAMA Otolaryngol. Head Neck Surg. 145:352–360. 10.1001/jamaoto.2018.4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santambrogio, L., Berendam S.J., and Engelhard V.H.. 2019. The antigen processing and presentation machinery in lymphatic endothelial cells. Front. Immunol. 10:1033. 10.3389/fimmu.2019.01033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul, Y.D., Freinkman E., Comb W.C., Cantor J.R., Tam W.L., Thiru P., Kim D., Kanarek N., Pacold M.E., Chen W.W., et al. 2014. Dihydropyrimidine accumulation is required for the epithelial-mesenchymal transition. Cell. 158:1094–1109. 10.1016/j.cell.2014.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbæk, R., Scabia V., Nagarajan S., Chernukhin I., Papachristou E.K., Broome R., Johnston S.J., Joosten S.E.P., Green A.R., Kumar S., et al. 2020. IL6/STAT3 signaling hijacks estrogen receptor α enhancers to drive breast cancer metastasis. Cancer Cell. 38:412–423.e9. 10.1016/j.ccell.2020.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. III, Hao Y., Stoeckius M., Smibert P., and Satija R.. 2019. Comprehensive integration of single-cell data. Cell. 177:1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., and Rinn J.L.. 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32:381–386. 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trefzer, A., Kadam P., Wang S.H., Pennavaria S., Lober B., Akçabozan B., Kranich J., Brocker T., Nakano N., Irmler M., et al. 2021. Dynamic adoption of anergy by antigen-exhausted CD4+ T cells. Cell Rep. 34:108748. 10.1016/j.celrep.2021.108748 [DOI] [PubMed] [Google Scholar]
- Tsoukalas, N., Aravantinou-Fatorou E., Tolia M., Giaginis C., Galanopoulos M., Kiakou M., Kostakis I.D., Dana E., Vamvakaris I., Korogiannos A., et al. 2017. Epithelial-mesenchymal transition in non small-cell lung cancer. Anticancer Res. 37:1773–1778. 10.21873/anticanres.11510 [DOI] [PubMed] [Google Scholar]
- Ubellacker, J.M., Tasdogan A., Ramesh V., Shen B., Mitchell E.C., Martin-Sandoval M.S., Gu Z., McCormick M.L., Durham A.B., Spitz D.R., et al. 2020. Lymph protects metastasizing melanoma cells from ferroptosis. Nature. 585:113–118. 10.1038/s41586-020-2623-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Elsen, P.J. 2011. Expression regulation of major histocompatibility complex class I and class II encoding genes. Front. Immunol. 2:48. 10.3389/fimmu.2011.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellano, C.P., White M.G., Andrews M.C., Chelvanambi M., Witt R.G., Daniele J.R., Titus M., McQuade J.L., Conforti F., Burton E.M., et al. 2022. Androgen receptor blockade promotes response to BRAF/MEK-targeted therapy. Nature. 606:797–803. 10.1038/s41586-022-04833-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosen, J.E., Mukhopadhyay D., Macaubas C., and Mellins E.D.. 2018. Epithelial MHC class II expression and its role in antigen presentation in the gastrointestinal and respiratory tracts. Front. Immunol. 9:2144. 10.3389/fimmu.2018.02144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, K., Wang R., Xie H., Hu L., Wang C., Xu J., Zhu C., Liu Y., Gao F., Li X., et al. 2021. Single-cell RNA sequencing reveals cell heterogeneity and transcriptome profile of breast cancer lymph node metastasis. Oncogenesis. 10:66. 10.1038/s41389-021-00355-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., Mani S.A., Donaher J.L., Ramaswamy S., Itzykson R.A., Come C., Savagner P., Gitelman I., Richardson A., and Weinberg R.A.. 2004. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 117:927–939. 10.1016/j.cell.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Zhou, H., Lei P.J., and Padera T.P.. 2021. Progression of metastasis through lymphatic system. Cells. 10:627. 10.3390/cells10030627 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists antibodies and reagents.
Data Availability Statement
Single-cell mRNA sequencing data generated to support this study have been deposited in NCBI GEO under accession number GSE168181. The authors declare that all other data supporting the findings of this study are available within the paper and its supplementary information files.