Abstract
Phages and their bacterial hosts together constitute a vast and diverse ecosystem. Facing the infection of phages, prokaryotes have evolved a wide range of antiviral mechanisms, and phages in turn have adopted multiple tactics to circumvent or subvert these mechanisms to survive. An in-depth investigation into the interaction between phages and bacteria not only provides new insight into the ancient coevolutionary conflict between them but also produces precision biotechnological tools based on anti-phage systems. Moreover, a more complete understanding of their interaction is also critical for the phage-based antibacterial measures. Compared to the bacterial antiviral mechanisms, studies into counter-defense strategies adopted by phages have been a little slow, but have also achieved important advances in recent years. In this review, we highlight the numerous intracellular immune systems of bacteria as well as the countermeasures employed by phages, with an emphasis on the bacteriophage strategies in response to host antiviral immunity.
Keywords: bacteriophage, prokaryotic immune system, abortive infection, phage-host interactions, anti-immunity measures
1. Introduction
The typical life cycle of bacteriophages (or simply phages) is a complex process that involves various stages, including adsorption, DNA entry, DNA replication, gene transcription and translation, phage assembly, host lysis, and phage burst (Olszak et al., 2017). During the adsorption stage, phages attach themselves to the specific receptor sites present on the surface of bacterial cells. Once bound, DNA entry occurs as the virus injects its genetic material into the host cell. This is followed by DNA replication, transcription and translation of viral genes responsible for enabling the assembly of new phage particles. In the final stages, host lysis and phage burst take place, where the infected host cell is destroyed, resulting in the release of new phages that can go on to infect neighboring cell.
The host deploys a series of immune measures both on the cell membrane and inside the cell to prevent phage infection. The inhibition of adsorption and DNA entry depends on the components of the cell membrane, and the subsequent phage process is mainly countered by the intracellular immune system. Adsorption of phages to the bacterial cell surface is the initial step of infection, which depends on the recognition of specific phage receptors on the cell surface (Labrie et al., 2010). Bacteria have evolved at least three types of blocking mechanisms to prevent phage adsorption, including blocking of phage receptors, production of extracellular matrix, and encoding competitive inhibitors.
After the phage is successfully adsorbed, the injected nucleic acid needs to pass through the outer membrane, peptidoglycan layer, and inner membrane to enter the cell. Notably, some phages do not proliferate immediately after infecting the host bacteria, but integrate their nucleic acids into the host chromosome, replicate during the replication of host nucleic acids, and passage with host cell division. Such phages are called temperate or lysogenic phages, and the phage genomes incorporated into the host chromosome are referred to as prophages (Olszak et al., 2017). Interestingly, these prophages integrated into the host genome may lead to superinfection exclusion, which means a preexisting viral infection prevents a secondary infection with the same or a closely related virus (Folimonova, 2012). Specifically, some prophages can encode membrane-anchored or membrane-component-associated proteins, which prevent other phage DNA from entering host cells, conferring host immunity to the specific phages (Lu and Henning, 1994; Domingo-Calap et al., 2020).
When the phage nucleic acids manage to enter the cell, it is the turn of the intracellular immune system to become the main weapons against the phage. During the past years, restriction-modification systems and CRISPR-Cas systems have been extensively studied as innate and adaptive immune systems respectively, and both have been successfully developed as widely used biotechnology tools (Barrangou and Horvath, 2017; Felice et al., 2019). The great potential for developing biotechnology tools has attracted lots of researchers’ attention to bacterial immune systems. Identifying new prokaryotic defense systems and characterizing their biochemical activities and working mechanisms have become increasingly important research efforts. Interestingly, many antiviral defense genes in bacterial and archaeal genomes show a unique tendency to cluster together to form the so-called “defense islands” (Makarova et al., 2011; Doron et al., 2018). This phenomenon inspired that unknown functional genes around known defense genes in the genome may also participate in the anti-phage defense. Based on this notion, the exploration of genes next to known defense genes led to the discovery of 59 new systems that protect bacteria from phages (Doron et al., 2018; Gao et al., 2020; Millman et al., 2022). In addition, the identification of antiviral systems outside the defense island has also made some progress recently. Results from two groups unveil 31 new defense systems, none of which has been previously detected as enriched in defense islands (Rousset et al., 2022; Vassallo et al., 2022). And some other researches do not specifically emphasize the comprehensive identification of multiple defense systems on a large scale but rather characterize an interesting novel immune system individually (Ofir et al., 2018; Cohen et al., 2019; Xiong et al., 2020; Bernheim et al., 2021; Jaskólska et al., 2022; Johnson et al., 2022; LeRoux et al., 2022; Tal et al., 2022; Zaremba et al., 2022; Zhang et al., 2022). In fact, over 100 novel immune systems have been reported in the past 5 years, highlighting the growing interest in elucidating their functional mechanisms and developing their applications as biological tools.
In turn, phages have also adopted various strategies to antagonize host antiviral immunity. In recent years, significant progress has been made both in the identification of novel prokaryotic immune systems and countermeasures against these defense systems by phages. Here, we review the numerous intracellular immune systems of bacteria as well as the countermeasures employed by phages, with an emphasis on the bacteriophage strategies used to evade these intracellular immune systems. The main strategies will be discussed in two parts, respectively: (1) inactivation of the immune systems by encoding protein inhibitors; (2) bacteriophage gene modifications or mutations.
2. Inactivation of the immune system by encoding protein inhibitors
In the case of pathogenic bacteria, effectors refer to proteins that enable the pathogen to modify the host normal cellular processes, thereby promoting the infection process. This pattern is similarly observed in phage-prokaryote interactions, where phage-encoded proteins can inactivate the host immune systems through various mechanisms such as directly binding immune proteins, post-translational modifications of immune proteins, and targeting second messengers (Table 1).
Table 1.
Summary of the counter-defense strategies using encoding protein inhibitors.
| Strategy | Anti-type | Targeting | Mechanism | Reference |
|---|---|---|---|---|
| Binding immune proteins | Anti-CRISPR proteins | Cascade Cas protein Cas-crRNA |
Inhibit the binding or cleavage of nucleic acids, or inhibit the assembly of Cas-crRNA complex | Jia and Patel (2021) and Yin et al. (2023) |
| Anti-restriction proteins | REase MTase |
Inhibit the cleavage of nucleic acids, stimulate the DNA methylation | Krüger et al. (1983) and King and Murray (1995) | |
| Anti-RecBCD proteins | RecBCD | Inhibit the cleavage of nucleic acids | Murphy (2000) and Court et al. (2007) | |
| Anti-TA proteins | Toxin Proteases of antitoxins |
Inhibit the toxin, or inhibit the degradation of antitoxin by proteases | Otsuka and Yonesaki (2012) | |
| Anti-SIR2-containing-system protein | DSR2 | Inhibits DSR2 activation | Garb et al. (2022) | |
| Anti-Gabija protein | GajAB complex | Inhibits the binding and cleavage of nucleic acids | Antine et al. (2023) | |
| PTM of immune proteins | Anti-CRISPR proteins | Csy complex Cas12a |
Inhibit the recognition of nucleic acids | Dong et al. (2019) and Niu et al. (2020) |
| Anti-TA protein | MazF | Inhibits the toxin | Alawneh et al. (2016) | |
| Targeting second messengers | Anti-CBASS proteins | cGAMP | Cleave or sequester signal molecules | Hobbs et al. (2022), Huiting et al. (2023), and Jenson et al. (2023) |
| Anti-Pycsar protein | Cyclic pyrimidine mononucleotide | Cleaves signal molecules | Hobbs et al. (2022) | |
| Anti-CRISPR protein | Cyclic oligo-adenylate | Cleaves signal molecules | Athukoralage et al. (2020) | |
| Anti-Thoeris proteins | gcADPR | Sequester signal molecules | Leavitt et al. (2022) and Yirmiya et al. (2023) | |
| Other | Anti-CRISPR protein | DNA | Modifies DNA topology | Forsberg et al. (2021) |
| Anti-restriction proteins | DNA SAM |
Occlude restriction sites, hydrolysis of SAM | Studier and Movva (1976), Iida et al. (1987), and Krüger et al. (1989) | |
| Anti-RecBCD proteins | DNA | Protects the injected DNA | Appasani et al. (1999) |
2.1. Directly binding immune proteins
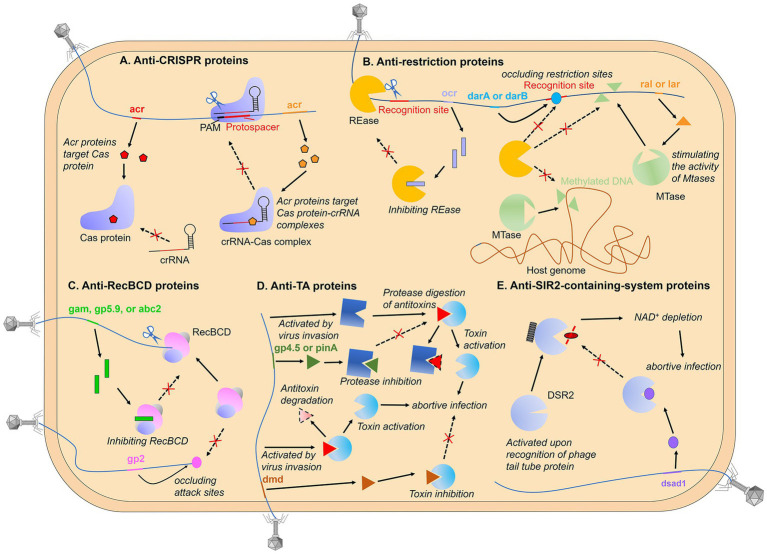
Anti-CRISPR proteins. Direct binding to host immune protein is the most common inhibition mode among phage protein inhibitors identified at present (Figure 1), which has been widely characterized in the investigation of the anti-CRISPR/Cas mechanisms (Borges et al., 2017). CRISPR-Cas system is an adaptive immune system of prokaryotes, which can obtain foreign nucleic acids as “immune memory” and destroy previously encountered non-self nucleic acid sequences. For more mechanistic information on CRISPR-Cas systems, please see references (Marraffini, 2015; Barrangou and Horvath, 2017). The identification of anti-CRISPR proteins in phages can be traced back to the year 2013 (Bondy-Denomy et al., 2013). Bondy-Denomy et al. found five genes that can inhibit the type I-F CRISPR-Cas system in the prophage sequence of Pseudomonas aeruginosa for the first time, called anti-CRISPR genes (Acr genes). Later, the research group found that there was always an Aca (anti-CRISPR-associated) gene downstream of the anti-CRISPR gene, which inspired the identification of numerous Acr genes (Pawluk et al., 2016; Borges et al., 2017). Subsequent structural and biological characterization found that most Acr proteins directly target Cas protein-crRNA complexes, such as Cascade (Yang et al., 2021; Gao Z. et al., 2022; Wang et al., 2022; Yang et al., 2022), Cas9-sgRNA (Dong et al., 2017; Rauch et al., 2017), Cas12-crRNA (Marino et al., 2018; Zhang et al., 2019), Cas13-crRNA (Lin et al., 2020; Meeske et al., 2020), and play a role by shielding the domain of recognition or binding nucleic acid substrate. A small amount of Acr proteins target apo Cas proteins, for instance, AcrIIC2 targets Cas9 and impedes the loading of sgRNA onto Cas9 proteins and the subsequent assembly of complexes (Zhu et al., 2019). Meanwhile, AcrIF3 and AcrIF23 directly inhibit Cas2/3 nucleases, with AcrIF3 mimicking Cas8f-HB (the structural domain responsible for recruiting Cas2/3 nucleases in the Cascade) and binding directly to Cas2/3 to prevent its recruitment (Wang et al., 2016; Sun et al., 2019), while AcrIF23 only suppresses the enzymatic activity of Cas2/3 on DNA and does not prevent its recruitment to the Cascade-dsDNA complex (Ren et al., 2022). And it is noteworthy that the AcrIF5 protein specifically targets Cascade-dsDNA and competes for the binding site of Cas8f-HB, thereby inhibiting Cas2/3 recruitment (Xie et al., 2022). AcrIF5 exhibits a noteworthy complementary relationship with the acrIF3 gene, with all the AcrIF5-encoding genes coexisting upstream of the acrIF3 gene in the same operon. This complementary functional relationship enables the targeting of Cascade-DNA and Cas2/3, respectively, thereby achieving a thoroughly negative effect on the process of Cas2/3 recruitment by Cascade-DNA.
Figure 1.
Phage-encoded proteins that direct bind immune proteins. (A) the host CRISPR-Cas system targets and cleaves the exogenously invading nucleic acid under the guidance of crRNA upon recognizing the PAM of foreign nucleic acid. Phage-encoded Acr proteins directly target Cas proteins or Cas protein-crRNA complexes, inhibiting recognition or cleavage of phage nucleic acids; (B) the restriction-modification (R-M) system is a mechanism that modifies specific sequence motifs in the host genome and degrades unmodified foreign DNA. Phage proteins can circumvent the R-M system through various strategies, such as inhibiting restriction endonucleases (REases), shielding unmodified restriction sites, and stimulating host protein modifications to the phage genome; (C) RecBCD degrades DNA lacking Chi and containing free DNA ends, and phage-encoded proteins can directly bind to RecBCD, preventing cleavage of the phage genome, or they can bind to the ends of linear phage genomes to protect the injected DNA from RecBCD attack; (D) the antitoxin functions to neutralize the toxin during normal bacterial growth and activate toxins during phage propagation. Phage-encoded proteins can mimic antitoxins or prevent the degradation of antitoxins, thereby avoiding the release of toxins; (E) bacterial defense systems with SIR2 domains deplete NAD+ upon phage infection, thus triggering abortive infection. Phage-encoded proteins can bind to SIR2-domain proteins and inactivate their enzymatic activity.
Moreover, there is a more specific class of Acr proteins, such as AcrIIA1, AcrIIC1, and AcrIIC3, that target Cas9 in all states (free Cas9, Cas9-gRNA, or Cas9-gRNA-DNA). Nonetheless, their distinct mechanisms of action are noteworthy: AcrIIA1 induces Cas9 degradation in a specific intracellular environment (Osuna et al., 2020); AcrIIC1 interacts with the HNH structural domain in Cas9, resulting in the inhibition of substrate cleavage by Cas9, while preserving its recognition and binding to the substrate (Harrington et al., 2017); AcrIIC3 facilitates Cas9 dimerization, which significantly impairs its ability to bind to target DNA and completely eliminates its nuclease cleavage activity (Sun et al., 2019).
Anti-restriction proteins. Restriction-modification (R-M) systems exist widely in bacteria and constitute a so-called restriction barrier that prevents interspecies horizontal gene transfer. In these systems, restriction endonucleases (REases) are used to cut invading foreign DNA at specific recognition sites, while the host DNA would not be digested as it is modified at the recognition site by a partner modification enzyme, usually a methyltransferase (MTase; Bickle and Krüger, 1993; Murray, 2002). Protein inhibitors (anti-restriction proteins) are also widespread as anti-R-M strategies. Encoding DNA mimics interacting with the restriction complex can effectively inhibit the activity of R-M systems. A prototypical protein of this type is the coliphage T7 protein Ocr (overcome classical restriction) produced from the gene 0.3. It forms a dimer with each monomer structurally mimicking one helical turn of B-form DNA while the negatively charged surface imitates the distribution of negatively charged phosphate groups along the DNA double helix (Krüger et al., 1983; Walkinshaw et al., 2002). Intriguingly, T7 Ocr is also active against the type I BREX (Bacteriophage Exclusion) system encoded by Escherichia coli, a novel defense system that possesses a six-gene cassette consisting of an ATPase-domain protein BrxC, a SAM-dependent N6-adenine methyltransferase BrxX, an alkaline phosphatase domain protein BrxZ, a putative Lon-like protease BrxL, a putative RNA-binding protein BrxB and a small protein BrxA with unknown function (Isaev et al., 2020). Like the R-M system, the BREX system distinguishes self from non-self through methylation of specific motifs in the bacterial genome by BrxX. However, it prevents phage amplification by an unknown mechanism without cleavage of phage DNA. Pull-down assay shows that Ocr binds the BrxX methyltransferase, thus neutralizing the ability of BREX to both methylate and exclude foreign phage DNA (Goldfarb et al., 2015; Gordeeva et al., 2019; Isaev et al., 2020). Similar to Ocr, the coliphage Т4 protein Arn (anti-restriction endonuclease) uses the same principle as an anti-restriction protein (Kim et al., 1997; Ho et al., 2014; Isaev et al., 2020). Not only bacteriophages, but some transmissible plasmids also adopt the same strategy to inhibit R-M systems, which is called Ard (alleviation of restriction of DNA) protein (Zavil'gel'skiĭ et al., 1984; Zavil'gel'skiĭ and Rastorguev, 2009).
Another anti-restriction inhibitor is Stp, an alleviator of DNA restriction encoded by phage T4 that can bind the type I restriction complex EcoprrI, inhibiting the restriction but not the modification activity (Amitsur et al., 1989; Penner et al., 1995). Unexpectedly, a bacterial tRNALys-specific anticodon nuclease PrrC is associated with EcoprrI and activated by the binding of Stp to EcoprrI, causing the cleavage of tRNAlys and abolishment of protein synthesis (Amitsur et al., 1992). In this way, even if the R-M system is collapsed by phage inhibitor as the first defense line, the PrrC can still provide a secondary line of defense.
Some phages avoid being targeted by the restriction complex through genome modification (see more details below), but bacteria have also acquired the ability to recognize and cleave modified phage DNA in the co-evolutionary arms race. These MDSs (modification-dependent systems) that are contrasted to classical R-M systems, are classified as the type IV R-M system, such as DpnI, Mcr (modified cytosine restriction), Mrr (methylation requiring restriction) and GmrSD (glucose-modified restriction), etc., (Raleigh and Wilson, 1986; de la Campa et al., 1988; Tesfazgi Mebrhatu et al., 2011; Loenen and Raleigh, 2014). Here, the GmrS–GmrD (or GmrSD) system is taken as an example to introduce the battle between phages and bacteria. Two proteins encoded by the GmrSD system form a complex to specifically target and attack glucosylated hydroxymethylcytosine (glc-HMC) modified DNA, in which GmrS protein is responsible for restriction cleavage and GmrD binds to DNA (Bair and Black, 2007). The GmrSD system is inhibited by the T4 phage internal protein I* (IPI*). Immunoprecipitation assays show that IPI* interacts with both GmrD and GmrS, inactivating restriction activity to phage DNA (Bair et al., 2007; Bair and Black, 2007; Rifat et al., 2008). In yet another twist, a single polypeptide produced from a gmrSD gene fusion can resist IPI* binding (Rifat et al., 2008).
Finally, it is worth mentioning that not all anti-restriction proteins directly interact with the immune complex, and some phages use indirect, passive strategies to bypass bacterial R-M systems, here we only give a brief introduction. The coliphage P1 proteins DarA (defense against restriction, Dar) and DarB can bind to the phage genome and occlude restriction sites, thus avoiding restriction by the R-M systems (Iida et al., 1987). Modulating cellular methylation processes by phage-encoded effector proteins causes a positive impact on the invasion of viruses. Coliphage λ protein Ral and Lar can alleviate restriction by stimulating the activity of MTases, which allows the phage genome to be rapidly methylated to escape from REases (Zabeau et al., 1980; Loenen and Murray, 1986; King and Murray, 1995). Coliphage T3 encodes an Ado-Met hydrolase that is contributed to overcoming host restriction. As an essential R–M cofactor, S-adenosyl-L-methionine (AdoMet, or SAM) plays a role in both methylation and DNA cleavage reaction (Sistla and Rao, 2004). The hydrolase degrades the methyl donor, preventing DNA methylation and inhibiting SAM-dependent restriction enzymes (Studier and Movva, 1976; Krüger et al., 1989). Recently, a preprint article also demonstrated that this SAMase can also help the host overcome the BREX system through SAM cleavage and inhibition of SAM synthesis (Andriianov et al., 2023).
Anti-RecBCD proteins. The linear fragments caused by restriction enzymes acting on foreign DNA will further become substrates for more extensive degradation by the RecBCD, an enzyme cascade complex that contains both helicase and nuclease activities. RecBCD unwinds and digests the blunt-ended DNA until a specific octamer motif that modulates the nuclease activity of RecBCD and results in the subsequent repair of double-strand breaks by homologous recombination, Chi, is encountered. That is, RecBCD degrades DNA lacking Chi and containing free DNA ends, including linear viral DNA present during phage replication or resulting from the action of restriction endonucleases, thus acting as a defense mechanism against phages (Dillingham and Kowalczykowski, 2008; Bobay et al., 2013; Cheng et al., 2020). As a survival guarantee of those phages exposing free DNA ends during the life cycle, anti-RecBCD inhibitors came into being. Phage lambda possesses the gene gam encoding a competitive inhibitor, which mimics the structure of a duplex DNA end to prevent cleavage of the phage genome (Murphy, 1991; Court et al., 2007). Additionally, both phage T7 protein gp5.9 and enterobacteria phage P22 protein Abc2 can directly bind RecBCD to prevent and inactivate it (Poteete et al., 1988; Murphy, 2000; Millman et al., 2020a). Notably, similar to the mechanism of PrrC that is activated only when the first defensive lines have collapsed, RecBCD inhibition by phage protein activates the Retron, an anti-phage defense system comprised of reverse transcriptase (RT) and a non-coding RNA (ncRNA), leading to abortive infection and cell death (Millman et al., 2020a; Bobonis et al., 2022). Besides, in addition to inhibiting RecBCD directly, T4 phage protein gp2 binds the ends of the linear phage genomes and protects the injected DNA from attack by the RecBCD complex (Silverstein and Goldberg, 1976; Appasani et al., 1999).
Anti-Abi proteins. The systems described above successfully defend by attacking the nucleic acid of the virus directly, however, bacteria have also developed a defense mechanism called Abi (abortive infection) systems, where infected cells commit suicide or dormancy thus allowing the uninfected bacterial population to survive from phage infection (Lopatina et al., 2020). These Abi systems are usually inactivated to ensure normal bacterial growth and are triggered only when specific substances during phage infection are recognized. Some TA (toxin–antitoxin) systems are well-characterized Abi models, typically, the antitoxin neutralizes the toxin during normal bacterial growth and activates toxins during phage propagation. Toxins are mostly proteins while the corresponding antitoxins are proteins or non-coding RNAs. In addition to directly binding toxins, antitoxins can also inhibit toxin toxicity in an indirect way, such as inhibiting the translation of toxin mRNA or competing for binding to the cellular target, and some antitoxins can regulate transcription of the TA operon with an additional DNA-binding domain. TA systems have been categorized into six types according to the mechanism by which the antitoxin neutralizes the toxin (Schuster and Bertram, 2013; Unterholzner et al., 2013; Chan et al., 2016).
The strategy of phage against TA systems is mainly to make the systems lose their recognized protein targets through gene mutations (to be discussed in detail below). Some phages encode antitoxin mimics that combine with the toxin to re-neutralize the toxin. P. atrosepticum Phage ϕTE produces a pseudo-RNA that mimics the antitoxin ToxI of the type III TA system, a repeats-containing RNA that binds and neutralizes the toxic Rnase ToxN (Blower et al., 2012). Phage T4 expresses a protein mimic named Dmd that can replace two antitoxins and bind directly to the RnlA or LsoA toxins (Otsuka and Yonesaki, 2012). Notably, at the position relatively close to dmd homologs in the genomes of T-even phages, a gene gp61.2 encoding anti-DarT protein was identified there. The gp61.2 protein and its homologs help phage evade the DarTG toxin-antitoxin system that ADP-ribosylates phage DNA to disrupt its replication (LeRoux et al., 2022). The phenomenon of anti-defense genes cluster has also been described in the distribution of anti-CRISPR and anti-RM genes, indicating a potential means to identify anti-defense genes (Pinilla-Redondo et al., 2020). Besides, two genes of phage T4 called rIIA and rIIB can help escape a type II TA system, the RexAB system, through unclear mechanisms (Shinedling et al., 1987). Finally, lots of TA systems are protease-dependent, that is, toxin release needs the degradation of antitoxins through proteases (Lehnherr and Yarmolinsky, 1995; Christensen et al., 2001, 2004; Wang et al., 2011), like Lon protease and ClpP protease, suggesting that inactivating these proteases may be an effective anti-TA strategy. This mode of action was demonstrated for phage T7 protein gp4.5 and phage T4 PinA protein (Hilliard et al., 1998; Sberro et al., 2013), which both directly target Lon protease, and for the phage lambda RexB protein, which blocks ClpP protease (Engelberg-Kulka et al., 1998).
Anti-SIR2-containing-system proteins. As mentioned above, with the research into “defense islands,” more and more novel prokaryotic immune systems have been identified. Next, we focus on defense systems containing a Sirtuin (SIR2)-domain protein. SIR2 proteins have been characterized as the family of NAD+-dependent histone deacetylases in eukaryotes, participating in the regulation of multiple important cellular processes (Imai et al., 2000; North and Verdin, 2004; Schwer and Verdin, 2008). In bacteria, SIR2 proteins have been previously detected as PIWI-associated proteins among the prokaryotic argonautes (pAgos) neighbors (Makarova et al., 2009). As pAgos are involved in nucleic acid-guided cleavage of the phage genome, PIWI-associated SIR2 is presumed to participate in prokaryotic antiviral and confirmed by a recent report (Zaremba et al., 2022). Unsurprisingly, besides pAgo, SIR2-domain proteins were recently shown to be associated with multiple bacterial anti-phage defense systems including Thoeris, AVAST (antiviral ATPases/NTPases of the STAND), DSR (defense-associated sirtuin), and additional unnamed SIR2-containing systems, in which SIR2-domain plays a role of the “effector” in biological conflict, and domains playing similar roles include nucleases, peptidases, phospholipase, etc., (Doron et al., 2018; Burroughs and Aravind, 2020; Gao et al., 2020). Avs (antiviral STAND) proteins have a similar domain architecture to eukaryotic NLR (nucleotide-binding oligomerization domain-like receptors)-related proteins (Platnich and Muruve, 2019), which belongs to STAND (signal transduction ATPases with numerous associated domains) NTPases superfamily (Leipe et al., 2004). Bacterial NLRs have also been described elsewhere (Kibby et al., 2022; Rousset et al., 2022) and these proteins are highly conserved with a central NTPase domain that is flanked by an N-terminal effector and a C-terminal sensor domain. DSR proteins possess an N-terminal SIR2 domain and an uncharacterized C-terminal domain, but lack a central NTPase module, indicating its differences from that of the Avs proteins (Gao et al., 2020). The mechanisms of DSR and AVAST were both described as abortive infection strategies that trigger cell death upon the pattern recognition of phage proteins, such as terminase, portal, or tail tube protein (Garb et al., 2022; Gao L. A. et al., 2022). And it has been confirmed that currently reported various bacterial defense systems with SIR2 domains all deplete NAD+ upon infection (Garb et al., 2022). But there is no doubt that phages have also developed resistance strategies against this kind of prokaryotic innate immunity. B. subtilis phages phi3T and Spbeta encode genes that inhibit DSR2, called DSAD1 (DSR anti-defense 1), which directly binds to DSR2 competitively with tail tube protein, the activator of DSR2 (Garb et al., 2022). At least three anti-defense proteins can help phages fight against AVAST in vivo by an unclear mechanism at present (Gao L. A. et al., 2022). Finally, another SIR2-containing system, Thoeris, and its resistance mechanisms will be discussed below because of its particularity.
2.2. Post-translational modification of immune proteins
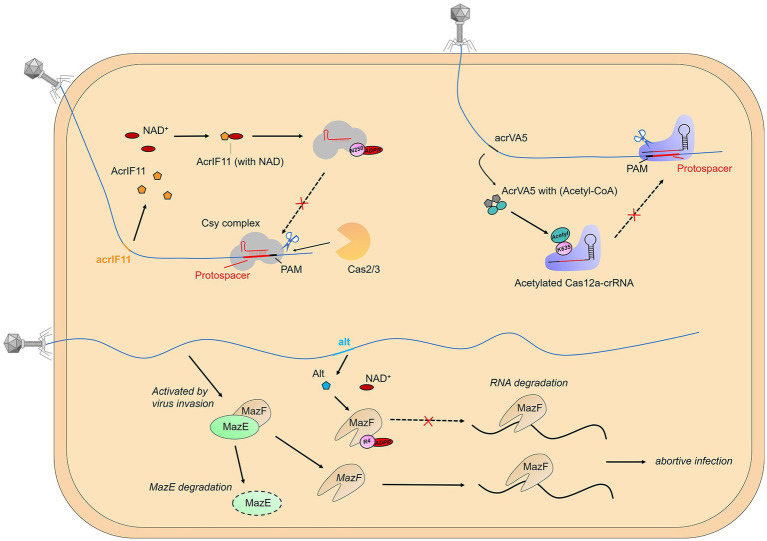
Post-translational modifications (PTMs) increase the diversity and complexity of proteomes and have vital roles in various cellular processes. In eukaryotic immunity, PTMs of innate sensors and downstream signaling molecules regulate the innate inflammatory response and maintain cellular coordination and balance by inducing their covalent linkage to new functional groups, such as phosphorylation, ubiquitination, SUMOylation, etc. (Liu et al., 2016; Zhou et al., 2017). Protein PTMs are also widespread in bacteria and implicated in all significant physiological processes known to be regulated by PTMs in eukaryotes (Macek et al., 2019). Recently, bacteria have also been reported to use PTMs to regulate the immune response. Millman et al. reported a phage-resistant system encoded by bacteria, which comprises a homolog of ISG15 (Interferon-stimulated gene 15). ISG15 is a ubiquitin-like protein that can be conjugated onto many viruses and host proteins during infection by particular ubiquitin-conjugating enzymes, called ISGylation, like ubiquitination. Notably, a point mutation at the conserved site for ISGylation on the bacterial ISG15-like gene abolished defense, indicating that the ISGylation may also be involved in bacterial immunity (Millman et al., 2022). And about 39% of the CBASS (an Abi immune system present in prokaryotes, see more details below) reported encode Cap2 and Cap3 (CBASS-associated protein 2 and 3) that are homologous to ubiquitin-conjugating (E1/E2) and deconjugating (DUB) enzymes, respectively (Cohen et al., 2019; Millman et al., 2020b). Two groups have reported that E1 catalytic cysteine of the E1-E2 fusion protein Cap2 thioester-linked to the C-terminal glycine of bacterial cGAS that finally leads to the conjugation of the cGAS C-terminus to target proteins in a way similar to ubiquitin transferase-like mechanism. These processes increase the production of cGAMP, which is conducive to resistance to phages. Similarly, the mutation of C-terminal glycine of cGAS eliminated the antiviral activity, highlighting the role of PTMs in CBASS immunity (Jenson et al., 2023; Ledvina et al., 2023).
Remarkably, phages have also been found to encode proteins that aid in their invasion of bacterial hosts by modifying bacterial immune proteins through PTMs (Figure 2). This phenomenon has been mainly observed in the mechanisms of anti-toxin-antitoxin (TA) and anti-CRISPR. For example, the phage T4 protein Alt has been described as an ADP-ribosyltransferase that can modify and inactivate the toxin MazF, an RNAase that blocks protein translation and is inhibited by antitoxin MazE (Alawneh et al., 2016). Moreover, biochemical and structural data have revealed that the AcrIF11 protein, encoded by the P. aeruginosa prophage, functions as a novel mono-ADP-ribosyltransferase (mART). This mART modifies N250 of the Cas8f subunit located within the Cascade of the type I-F CRISPR-Cas system, a residue that is essential for the recognition of the protospacer-adjacent motif (PAM) (Niu et al., 2020). And a comparable mechanism has been observed in the type V-A CRISPR-Cas system, in which the Moraxella bovoculi prophage-encoded AcrVA5 protein acetylates K635 of the Cas12a protein of the V-K CRISPR-Cas system. This residue is also essential for the recognition of the protospacer-adjacent motif, and acetylation results in the complete loss of double-stranded DNA (dsDNA) cleavage activity of Cas12a (Dong et al., 2019).
Figure 2.
Post-translational modification of immune proteins. Phages have also been found to encode proteins that aid in their invasion of bacterial hosts by modifying bacterial immune proteins through PTMs, mainly observed in the mechanisms of anti-toxin-antitoxin (TA) and anti-CRISPR.
2.3. Targeting secondary messengers
Secondary messengers (or second messengers) are nonprotein molecules or ions that bind to specific target proteins, and disseminate information received by cellular receptors (Newton et al., 2016). In addition to regulating the enzymatic activity of the intracellular metabolic system and controlling the life activities of cells, second messengers are also involved in the antiviral immune process by eukaryotic cells, which use second messengers to transmit viral invasion signals and activate immunity (Wu et al., 2013). For example, the cGAS-STING pathway has a central antiviral effect in the cellular innate immune system. The cGAS (cyclic GMP–AMP synthase) can recognize foreign nucleic acids and utilize GTP and ATP to synthesize cGAMP (cyclic GMP-AMP). As a second messenger, cGAMP binds and activates the endoplasmic reticulum protein STING (stimulator of interferon genes). Through the STING pathway, the signal is transferred to the nucleus, regulates gene transcription, and starts the immune response (Ablasser et al., 2013; Wu et al., 2013; Ablasser and Chen, 2019). Studies into the prokaryotic immune systems show that prokaryotic genes that protect bacteria from bacteriophages have the evolutionary roots of the core components of the eukaryotic innate immune system, including the cGAS-STING pathway and Toll/IL-1 receptor (TIR) domain-containing pathogen receptors, which use second messengers to transmit signals of virus invasion (Whiteley et al., 2019; Ofir et al., 2021; Wein and Sorek, 2022).
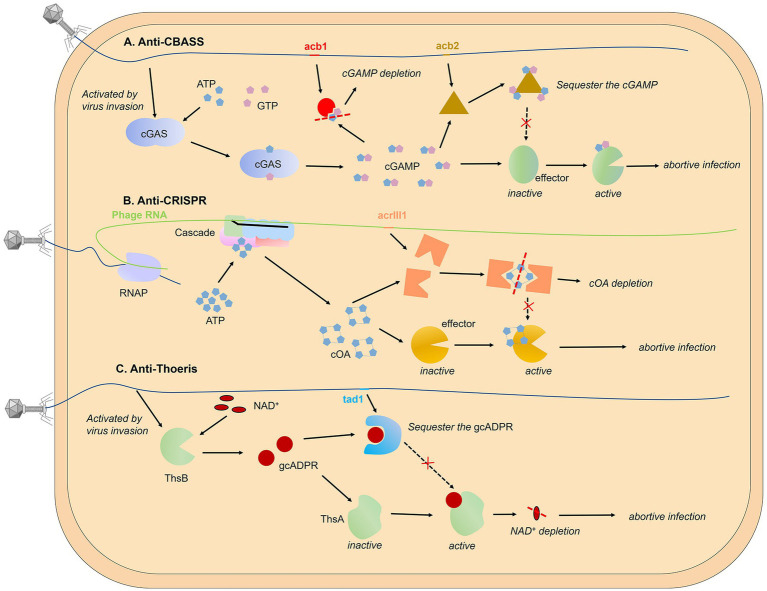
Bacterial CD-NTases (the cGAS/DncV-like nucleotidyltransferases) are a large family of phage-defensive oligonucleotide cyclases that share the structural architecture of cGAS but have different product specificities, such as cyclic GMP–AMP, cyclic UMP–AMP, cyclic UMP–UMP, cyclic AMP–AMP–GMP and many more (Cohen et al., 2019; Whiteley et al., 2019; Lau et al., 2020; Ye et al., 2020). These cyclic-oligonucleotide-based anti-phage signaling systems (CBASS) rely on synthesizing cyclic oligonucleotides to activate the abortive infection (Abi) response (Cohen et al., 2019; Lopatina et al., 2020). Effector protein activated by the cyclic oligonucleotides triggers premature cell death through various mechanisms, such as membrane impairment (Cohen et al., 2019), DNA cleavage (Lowey et al., 2020; Fatma et al., 2021), NAD+ depletion (Ko et al., 2022; Morehouse et al., 2022), etc. Sacrificing the infected cell deprives the phage of resources for multiplication, thereby protecting against phage infection. Such systems are found in about 13% of prokaryotic genomes and are present in all major bacterial phyla as well as in archaea (Whiteley et al., 2019; Millman et al., 2020b). Diverse operon organization, signaling molecules, and effector function of CBASS classify them into four major types (type I–type IV) (Millman et al., 2020b). Recent studies have shown that in addition to cyclic-oligonucleotide, cyclic pyrimidines are also used as a second messenger to transmit signals in prokaryotic antiviral immunity, which has not been reported in eukaryotic cells. Pyrimidine cyclase systems for anti-phage resistance, called Pycsar, are widespread in prokaryotes and work in similar ways as the CBASS (Tal et al., 2021).
The Toll/Interleukin-1 receptor (TIR) domain is a canonical component of animal and plant immune systems (Fitzgerald and Kagan, 2020). Bacteria have also been found to encode TIR domain-containing proteins, and subsequent studies have shown that the generation of intracellular signaling molecules is a conservative function of the TIR domain in both plant and bacterial immunity (Wan et al., 2019; Ofir et al., 2021; Yu et al., 2022). A representative example is the bacterial Thoeris system, an anti-phage defense system composed of ThsA and ThsB proteins. Phage infection triggers TIR domain-containing protein ThsB to produce cyclic ADP ribose isomer, a second messenger binding to the SIR2 NADase domain-containing protein ThsA specifically and resulting in the activation of NADase activity, which then depletes the cells of the essential molecule nicotinamide adenine dinucleotide (NAD+), leading to abortion infection and cell death (Ofir et al., 2021).
It seems that phages can always find counter-strategies against bacterial immunity, with no exception for CBASS, Pycsar, and Thoeris (Figure 3).
Figure 3.
Targeting secondary messenger. The use of second messengers to signal viral invasion has been reported in various prokaryotic systems, including CBASS (A), CRISPR-Cas system (B), and Thoeris system (C). Phage-encoded proteins can degrade second messengers in an enzymatically active form or bind and sequester them in a non-enzymatically active form without cleaving small molecules. Pyrimidine cyclase systems, referred to as Pycsar, exhibit analogous functionality to the CBASS mechanism. The anti-Pycsar protein Apyc1 and anti-CBASS protein Acb1 act as inhibitors of anti-phage defense through the cleavage of second messengers, demonstrating a similar mechanism. For reference, please refer to Figure 3A for the mechanism models of both Pycsar and Apyc1.
Cleave signal molecules. Hobbs et al. have reported the first anti-CBASS protein Acb1 and anti-Pycsar protein Apyc1 that degrade the cyclic nucleotide messengers to inhibit anti-phage defense (Hobbs et al., 2022). Structural analyses of Acb1 in complex with 3′,3′-cGAMP reveal the mechanism of metal-independent hydrolysis 3′ of adenosine bases that allows broad recognition and degradation of cyclic dinucleotide and trinucleotide signals. Crystal structures of Apyc1 define a metal-dependent cNMP phosphodiesterase, which targets and hydrolyzes a wide range of cyclic pyrimidine mononucleotide signals with relaxed specificity. In fact, the arms race between bacteria and phage around the second messenger has long been found in the type III CRISPR-Cas system. The coupling with the abortive infection mechanism defines the specificity of the type III CRISPR/Cas system. After recognizing the target RNA strand, this system not only shows the ability to cut the nucleic acid substrate but also activates the synthesis of cyclic oligo-adenylate (cAn) second messengers, an activator of downstream effectors that results in cell death and prevents viral propagation (Molina et al., 2020; Kolesnik et al., 2021). Athukoralage et al. identified a type III anti-CRISPR protein (AcrIII1) that rapidly degrades cyclic tetra-adenylate (cA4). Crystal structure of AcrIII1 in complex with cA4 show that a molecule of cA4 bound at the dimer interface of two AcrIII1 and is cleaved by a conserved active site (Athukoralage et al., 2020).
Sequester the second messenger. Besides cleaving signal molecules, there is a class of phage proteins that bind and sequester the second messenger without enzymatic activity. This kind of protein was first found in the gene that helps phages escape from the Thoeris system, called Tad1 (Thoeris anti-defence 1). Tad1 protein shows broad-spectrum inhibition as it can sequester molecules derived from a TIR-domain protein of both plants and bacteria (Leavitt et al., 2022). A similar mechanism has been found in the anti-CBASS mechanism, Huiting et al. revealed that the Acb2 protein encoded by P. aeruginosa phage displays a high binding affinity towards 3′, 3’-cGAMP molecules, but lacks enzymatic activity (Huiting et al., 2023). Through structural analyses of the Acb2 protein and its complexes with 3′, 3’-cGAMP, and c-di-AMP, it was determined that the Acb2 protein exists as a compact hexameric structure, with each 3′, 3’-cGAMP molecule residing within a “binding pocket” formed by the N-terminal ends of two Acb2 monomers, thereby facilitating stable interactions. By selectively adsorbing and isolating 3′, 3’-cGAMP molecules, Acb2 effectively disrupts the immune action of the CBASS encoded by P. aeruginosa. Furthermore, Acb2 has also been discovered and characterized in coliphage T4 (called Vs.4 in T4 phage), exhibiting a similar binding mechanism (Jenson et al., 2023).
Lastly, of the various immunosuppressive agents discussed above, anti-CRISPR proteins exhibit the greatest diversity in terms of number, variety, and mechanism of action. This can be attributed to the stringent regulation by the aca proteins, which significantly facilitates the identification and characterization of anti-CRISPR proteins. However, not all anti-defense genes have such trans-acting factors downstream. In numerous instances, anti-defense genes are discovered by comparing genomic variations between host-sensitive and non-sensitive phages. Typically, host-insensitive phages are repelled by the host immune system, whereas host-sensitive phages are more likely to have evaded immunity by encoding antagonistic proteins, enabling successful infection of the host. In this way, in a recent preprint article, Yirmiya et al. identified suppressor proteins of three novel bacterial immune systems, namely Gabija, Thoeris and Hachiman (Antine et al., 2023; Yirmiya et al., 2023). Specifically, Gad1 (Gabija anti-defense 1) binds the Gabija complex as an octamer and impedes its ability to bind and cleave DNA. Tad2 (Thoeris anti-defense 2) exhibits a mechanism similar to Tad1, inhibiting Thoeris defense by binding and sequestering bacterial immune signaling molecules, thereby preventing the activation of the Thoeris immune effector. While Had1 (Hachiman anti-defense 1) was identified, its functional mechanism remains unknown.
3. Genes modifications or mutations that help phages evade the immune system
3.1. Mutations or modifications of non-coding sequences
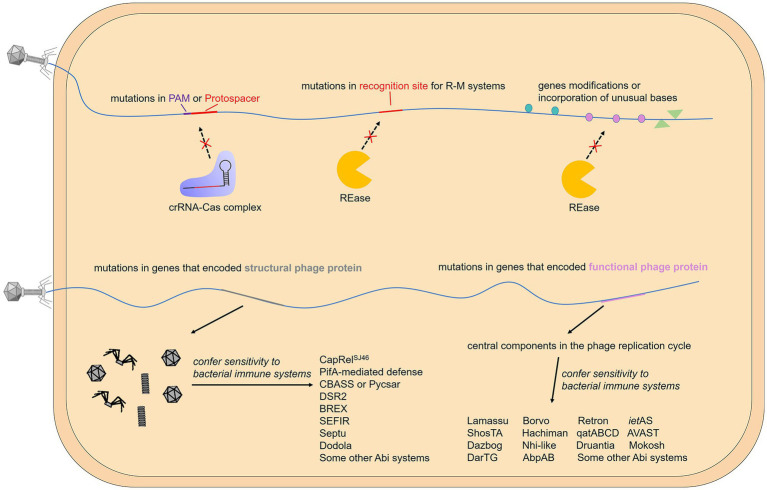
Differences in genome modifications between a host and its phage allow self from non-self-discrimination, which means that the same DNA modification as the host may help phages overcome the restriction–modification systems and related defense, such as the BREX system (Gordeeva et al., 2019). In fact, phage DNA has been found to become methylated by methyltransferases from the host or phages themselves. For example, certain phage proteins mentioned earlier can stimulate the activity of host MTases, thereby facilitating prompt methylation of the phage genome. However, upon infecting methyltransferase-deficient bacteria, newly-formed virions will be unmethylated and once again become phenotypically sensitive to REases. In order to enhance their chances of survival, some phages have acquired a competitive edge by integrating a cognate orphan MTase, which allows them to encode their own MTases. This phenomenon is observed in various phages such as B. subtilis phage SPR and L. lactis phage φ50 (Günthert and Reiners, 1987; Hill et al., 1991; Murphy et al., 2013). Notably, another RM-like system recently identified, called DISARM (defense island associated with restriction–modification), still protected against the phages propagated from drmMII-expressing strain, in which genome has been hypermethylation by DrmMII, a C-5 cytosine-specific DNA methyltransferase of DISARM system (Ofir et al., 2018). These results indicate an unusual mechanism to identify invading phage DNA that differs from classic R-M systems. In addition to the same methylation modification as the host DNA, other modifications and incorporation of unusual bases can also protect the phage genome from cleavage by the host REases (Figure 4). One example is the gene mom of coliphage Mu, which encodes an enzyme converting adenine to N6-(1-acetamido)-adenine in the phage genome (Hattman, 1980; Drozdz et al., 2012). Moreover, the genome of several B. subtilis phages possess the unusual base hydroxymethyl uracil that replaces thymine, and coliphage T4 contains hydroxymethyl cytosine (HMC) or glucosylated HMC (glc-HMC) instead of cytosines to make these sites unrecognizable and thus circumvent the R-M, and even some CRISPR-Cas mechanisms (Warren, 1980; Weigele and Raleigh, 2016; Vlot et al., 2018; Table 2).
Figure 4.
Gene modifications or mutations that help phages evade the immune system. The activation of some bacterial immune systems relies on the recognition of specific phage motifs, while others rely on the recognition of specific phage proteins. When specific motifs are targeted by the host, mutations, modifications, and base substitutions can effectively help the phage evade immunity. On the other hand, when the expressed protein is targeted, the phage usually mutates the gene encoding it.
Table 2.
Summary of the counter-defense strategies by genes modifications or mutations.
| Strategy | Position/type | Bypassed system | Reference |
|---|---|---|---|
| Genes mutations | PAM or protospacer | CRISPR-Cas systems | Deveau et al. (2008), Sapranauskas et al. (2011), and Semenova et al. (2011) |
| Restriction recognition sites | R-M systems | Krüger et al. (1995) and Pleška and Guet (2017) | |
| Structural phage proteins (capsids, tail tube, tail fiber, head fiber, etc.) | CapRelSJ46 PifA-mediated defense CBASS or Pycsar DSR2 BREX SEFIR Septu Dodola Some other Abi systems |
Champness and Snyder (1982), Molineux et al. (1989), Schmitt et al. (1991), Bingham et al. (2000), Labrie et al. (2012), Tal et al. (2021), Garb et al. (2022), Zhang et al. (2022), Huiting et al. (2023), and Stokar-Avihail et al. (2023) | |
| Functional phage proteins (DNA polymerase, ssDNA binding protein, helicase, terminase, etc.) | DarTG system dNTP-depletion-mediated defense AbpAB Lamassu Borvo Retron ietAS ShosTA Hachiman qatABCD AVAST Dazbog Nhi-like Druantia Mokosh Some other Abi systems |
Bouchard and Moineau (2004), Bidnenko et al. (2009), Haaber et al. (2009), Haaber et al. (2010), Samson et al. (2013a), Yasui et al. (2014), Chen et al. (2017), Montgomery et al. (2019), LeRoux et al. (2022), Tal et al. (2022), and Stokar-Avihail et al. (2023) | |
| Genes modifications | Methylation | R-M systems BREX system |
Loenen and Murray (1986), Labrie et al. (2010), Murphy et al. (2013), and Gordeeva et al. (2019) |
| Glucosylation | CRISPR-Cas systems BREX system |
Vlot et al. (2018) and Gordeeva et al. (2019) | |
| Other modified bases | R-M systems | Warren (1980) and Weigele and Raleigh (2016) |
Different R-M systems have unique preferences for the recognition motifs of viral DNA, and the antiviral efficiency is influenced by the number, and the orientation of the specific recognition motif and even the distance between them (Meisel et al., 1992; Krüger et al., 1995; Pleška and Guet, 2017). These factors result in a selective advantage for those phages whose restriction recognition sites in their genomes are congenital insufficiency or underrepresented via the accumulation of point mutations. This strategy is also used by phages against CRISPR-Cas systems. The guide crRNA comes from the transcription and processing of the CRISPR array, in which the spacers separated by the repeats come from the adaptive acquisition of foreign nucleic acid, while the protospacer on the genome of bacteriophages or other MGEs (mobile genetic elements) does not have a bacterial conserved repeat region next to it. Therefore, CRISPR-Cas system distinguishes self from non-self through the recognition of PAM (Marraffini and Sontheimer, 2010). For phages, in addition to encoding Acr proteins, mutations in protospacer or PAM can also help them escape the recognition and targeting of the bacterial CRISPR-Cas system (Deveau et al., 2008). Notably, RNA-guided Cas nuclease can cut DNA sequences that do not match the spacer incompletely, which causes off-target effects (Fu et al., 2013). And a requirement for a complete pairing between the spacer on crRNA with the sequence close to PAM on the protospacer called “seed regions” is essential for target strand binding, which means substitutions in the protospacer region that are close to PAM can more effectively avoid targeting of CRISPR-Cas system (Sapranauskas et al., 2011; Semenova et al., 2011). Interestingly, AcrIIA22 and its homologs, discovered in clostridial bacteria and their prophage, are reported to function as a DNA nickase that can relieve the torsional stress of DNA and thus convert it into a Cas9-resistant conformation (Forsberg et al., 2021), as the torsional stress in DNA modulates the formation of R-loop and further efficiency of Cas9 cleavage (Szczelkun et al., 2014; Farasat and Salis, 2016; Ivanov et al., 2020). This indicates that modifying DNA topology, like DNA modification or mutation, is also an effective way for MGEs to escape bacterial CRISPR-Cas system.
Recently, a novel defense system involving SMC proteins called Lamassu is reported to protect against phage via abortive infection (Doron et al., 2018; Millman et al., 2022). While Lamassu systems were originally speculated to be activated by recognizing phage DNA replication intermediates (Jaskólska et al., 2022; Millman et al., 2022), the actual activator is more like to be protein–DNA complexes that are produced uniquely by either palindromic sequences or certain types of DNA damage (Robins et al., 2022). As the palindromic DNA sequences can trigger Lamassu-dependent cell death, introducing mutations artificially in palindromic sequence can avoid Lamassu sensing, suggesting that mobile genetic elements such as phages may also be able to bypass the Lamassu system by mutations.
3.2. Mutations of coding sequences
The strategies mentioned above are based on the fact that foreign nucleic acids activate the immune system. In other cases, similar to pathogen-associated molecular patterns (PAMPs) recognized by eukaryotic innate immune systems (Medzhitov, 2001), some specific phage proteins are utilized by bacteria to sense viral invasion. Therefore, mutations or deletions of genes encoding these proteins can effectively help bacteriophages escape from bacterial immunity (Figure 4). A typical example is phage capsid protein, and multiple prokaryotic immune systems are triggered by this unique type of structural phage protein. Viral capsids are the protein shell in a shape of a polygon-like sphere or a helix that encases the nucleic acid. Targeted by a variety of defense systems, the huge evolutionary pressure results in inevitable mutations of capsid-coding genes. To our knowledge, T4 phages carrying mutations in gp23 (encode peptide Gol that derived from T4 capsid) escape the Lit Abi system (Champness and Snyder, 1982; Bingham et al., 2000); Phage SECΦ27 capsid protein gp57 triggers a TA system named CapRelSJ46 and mutations in the corresponding gene help escape (Zhang et al., 2022); Simultaneous mutations in gp1.2 and gp10 (T7 capsid protein) allow phage T7 to escape the PifA-mediated defense (Molineux et al., 1989; Schmitt et al., 1991); Phages PaMx41and T5 can escape CBASS and Pycsar, respectively, through mutations in the major capsid gene but there is no direct evidence that capsid activates CBASS or Pycsar (Tal et al., 2021; Huiting et al., 2023); Comparative genomic analyses of AbiT-insensitive phages showed that mutations in the major capsid protein or other two early-expressed phage genes help L. lactis virulent phages evade AbiT, an Abi system which molecular mechanism remains unknown (Labrie et al., 2012).
In addition to structural phage proteins, some functional phage proteins expressed in the cell during infection are also sensed by defense systems, and these proteins are related to intracellular processes including DNA replication, recombination, repair, and host transcription shut-off, etc. For example, in the DarTG TA system mentioned above, mutations on gene mga42 encoding a DNA polymerase and an unknown functional gene mga32 allow SECϕ18 phages to escape DarTG-mediated defense (LeRoux et al., 2022). Genome analysis of phage mutants that evade certain defense systems shows that the gene involved in sensitivity to the AbiK system (sak gene) is related to single-strand annealing proteins involved in homologous recombination or DNA repair (Bouchard and Moineau, 2004); genes involved in sensitivity to AbiQ system (saq gene) also have been proven to participate in DNA replication, repair or recombination (Samson et al., 2013a); mutations in gene 41 of phage T4 that avoid AbpAB system, encodes a replicative DNA helicase that plays an essential role for DNA replication (Yasui et al., 2014). In addition, genes involved in sensitivity to AbiV and AbiD, and some other Abi systems, have been located on the phage genome, but their function is unknown (Bidnenko et al., 2009; Haaber et al., 2009, 2010; Chen et al., 2017; Montgomery et al., 2019).
Interestingly, phage proteins with counter-defense functions can also be sensed by the host immune system. For example, the Ocr protein, a phage-encoded protein inhibitor that can inactivate the BREX and R-M systems, has recently been shown to as a trigger for multiple defense systems, including PARIS, Gabija, and Zorya type II (Rousset et al., 2022; Wu et al., 2022). This clever strategy, similar to PrrC and Retrons, can activate additional anti-phage mechanisms when the first line of defense fails. Mutations in the ocr gene could help phage evade these systems (Wu et al., 2022), but also re-expose phage to immune threats that could otherwise be overcome by the Ocr protein.
Modulating host RNA polymerase is required to make it serve viral needs, and covalent modifications or RNA polymerase-binding proteins result in host transcription shut-off, which would otherwise hamper phage multiplication (Nechaev and Severinov, 2003). Some defense systems can sense phage-mediated inhibition of host transcription. For example, host gene expression is immediately shut off caused by the invasion of phage T4, leading to an insufficient supply of antitoxin RnlB which then triggers the consequent activation of toxin RnlA (Svenson and Karlström, 1976; Koga et al., 2011; Otsuka and Yonesaki, 2012). Recently, a novel defense system has been identified, which can block phage replication by depleting deoxynucleotides, and is also considered to be activated by sensing transcriptional inhibition. The reason is that mutation of a gene gp5.7 in the phage T7 that is responsible for shutting down RpoS-dependent RNAP transcription, despite slight growth defects, is not interfered with by the dNTP-depletion-mediated defense (Tabib-Salazar et al., 2018; Tal et al., 2022). Very recently, Rotem Sorek’s group studied the activation factors of more than 50 defense systems by analyzing phage escape mutants, almost including all the newly identified prokaryotic immune systems. This study proposed that central components in the phage replication cycle commonly confer sensitivity to bacterial defense systems, which greatly promoted the decoding of the activation mode of the bacterial defense system (Stokar-Avihail et al., 2023).
4. Conclusion
This review summarizes prokaryotic anti-phage measures and corresponding counter-defense strategies around a process described as an ‘arms race’ between bacteria and their bacteriophages. Research in this field used to focus on R-M, CRISPR-Cas, and some Abi mechanisms. Many excellent reviews have covered this topic in depth (Labrie et al., 2010; Samson et al., 2013b; Hampton et al., 2020). However, since 2018, researches into defense islands and revenge of the phages have been keeping rapid growth (Doron et al., 2018; Gao et al., 2020; Millman et al., 2022; Tal and Sorek, 2022), which inspired our expanded discussion on new discoveries over past few years. These studies have brought at least two important potential values. First, fundamental research into interactions between phages and bacterial hosts expanded biotechnology tools and helped evolve and screen phages or strains with designated resistance, which have underpinned the development of many fields, including but not limited to gene editing (Adli, 2018; Manghwar et al., 2019), clinical therapy (Abedon, 2019; Dedrick et al., 2019), food industry, and agriculture (Nakai and Park, 2002; O'Sullivan et al., 2019). Secondly, another exciting discovery is that a variety of defense proteins of the human innate immune system have direct homologs in bacteria (Wein and Sorek, 2022). Evolutionary conservatism has brought some functional similarities, enabling researchers to decipher the eukaryotic immune mechanism in a relatively simple bacteria-phage experimental model and reveal unprecedented potential therapeutic targets.
One area that requires substantial attention is how bacterial defense systems sense phage infection. Characterization of the activation mode of the immune system lags far behind the identification of new immune systems. Understanding how the immune systems sense invasive MGEs is of great significance for phage-based antibacterial treatments, which will help to genetically modify phages and help them bypass the immune recognition of pathogens. Moreover, most of the studies on defense systems have rarely or limited consideration of other co-existing anti-phage mechanisms. A variety of defense systems are clustered in the defense islands and provide abundant resistance against phages. The priority of their activation must be strictly regulated and the conditions for bacteria to prefer different resistance mechanisms are not fully studied. For example, the systems that respond to the invasion through the Abi mechanism make biological sense only when the phage reaches a stage as late as possible of its infection cycle or when other mechanisms are insufficient to deal with the threat, just like PrrC and Retrons (Amitsur et al., 1992; Bobonis et al., 2022). How do the first lines of defense, non-Abi systems work synergistically in the same bacterial cell (Dupuis et al., 2013; Silas et al., 2017)? The effects of conjoint resistance mechanisms on phage population and evolution have rarely been assessed. Furthermore, beyond the individual range, from the perspective of bacteria and bacteriophage communities, the cooperation between individuals should be further studied (Borges et al., 2018; Bernheim and Sorek, 2020), which will further promote the more accurate use of microbial resources.
Finally, more researches should be conducted on ‘anti-defense islands’ (Pinilla-Redondo et al., 2020; LeRoux et al., 2022). Genes without functional annotations but clustered next to genes with clear anti-defense functions may also encode anti-defense proteins. These unknown genes within these regions may provide a wide range of regulatory tools to help us better tame the bacterial defense systems.
Author contributions
ZG: writing – original draft. YF: writing – review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Key Research and Development Program of China (2022YFC3401500 and 2022YFC2104800) to YF. Beijing Nova Program (20220484160), the Fundamental Research Funds for the Central Universities (QNTD2023-01).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We apologize to colleagues whose work could not be cited due to space limitations.
References
- Abedon S. T. (2019). Use of phage therapy to treat long-standing, persistent, or chronic bacterial infections. Adv. Drug Deliv. Rev. 145, 18–39. doi: 10.1016/j.addr.2018.06.018, PMID: [DOI] [PubMed] [Google Scholar]
- Ablasser A., Chen Z. J. (2019). cGAS in action: expanding roles in immunity and inflammation. Science 363:eaat8657. doi: 10.1126/science.aat8657, PMID: [DOI] [PubMed] [Google Scholar]
- Ablasser A., Goldeck M., Cavlar T., Deimling T., Witte G., Röhl I., et al. (2013). cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384. doi: 10.1038/nature12306, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adli M. (2018). The CRISPR tool kit for genome editing and beyond. Nat. Commun. 9:1911. doi: 10.1038/s41467-018-04252-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alawneh A. M., Qi D., Yonesaki T., Otsuka Y. (2016). An ADP-ribosyltransferase alt of bacteriophage T4 negatively regulates the Escherichia coli MazF toxin of a toxin-antitoxin module. Mol. Microbiol. 99, 188–198. doi: 10.1111/mmi.13225, PMID: [DOI] [PubMed] [Google Scholar]
- Amitsur M., Morad I., Chapman-Shimshoni D., Kaufmann G. (1992). HSD restriction-modification proteins partake in latent anticodon nuclease. EMBO J. 11, 3129–3134. doi: 10.1002/j.1460-2075.1992.tb05385.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitsur M., Morad I., Kaufmann G. (1989). In vitro reconstitution of anticodon nuclease from components encoded by phage T4 and Escherichia coli CTr5X. EMBO J. 8, 2411–2415. doi: 10.1002/j.1460-2075.1989.tb08371.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriianov A., Triguis S., Drobiazko A., Sierro N., Ivanov N. V., Selmer M., et al. (2023). Phage T3 overcomes the BREX defence through SAM cleavage and inhibition of SAM synthesis. bioRxiv:2023.2002.2027.530186. doi: 10.1101/2023.02.27.530186 [DOI] [PubMed] [Google Scholar]
- Antine S. P., Johnson A. G., Mooney S. E., Leavitt A., Mayer M. L., Yirmiya E., et al. (2023). Structural basis of Gabija anti-phage defense and viral immune evasion. bioRxiv. doi: 10.1101/2023.05.01.538945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appasani K., Thaler D. S., Goldberg E. B. (1999). Bacteriophage T4 gp2 interferes with cell viability and with bacteriophage lambda red recombination. J. Bacteriol. 181, 1352–1355. doi: 10.1128/JB.181.4.1352-1355.1999, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athukoralage J. S., Mcmahon S. A., Zhang C., Grüschow S., Graham S., Krupovic M., et al. (2020). An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity. Nature 577, 572–575. doi: 10.1038/s41586-019-1909-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bair C. L., Black L. W. (2007). A type IV modification dependent restriction nuclease that targets glucosylated hydroxymethyl cytosine modified DNAs. J. Mol. Biol. 366, 768–778. doi: 10.1016/j.jmb.2006.11.051, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bair C. L., Rifat D., Black L. W. (2007). Exclusion of glucosyl-hydroxymethylcytosine DNA containing bacteriophages is overcome by the injected protein inhibitor IPI*. J. Mol. Biol. 366, 779–789. doi: 10.1016/j.jmb.2006.11.049, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R., Horvath P. (2017). A decade of discovery: CRISPR functions and applications. Nat. Microbiol. 2:17092. doi: 10.1038/nmicrobiol.2017.92 [DOI] [PubMed] [Google Scholar]
- Bernheim A., Millman A., Ofir G., Meitav G., Avraham C., Shomar H., et al. (2021). Prokaryotic viperins produce diverse antiviral molecules. Nature 589, 120–124. doi: 10.1038/s41586-020-2762-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernheim A., Sorek R. (2020). The pan-immune system of bacteria: antiviral defence as a community resource. Nat. Rev. Microbiol. 18, 113–119. doi: 10.1038/s41579-019-0278-2, PMID: [DOI] [PubMed] [Google Scholar]
- Bickle T. A., Krüger D. H. (1993). Biology of DNA restriction. Microbiol. Rev. 57, 434–450. doi: 10.1128/mr.57.2.434-450.1993, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidnenko E., Chopin A., Ehrlich S. D., Chopin M. C. (2009). Activation of mRNA translation by phage protein and low temperature: the case of Lactococcus lactis abortive infection system AbiD1. BMC Mol. Biol. 10:4. doi: 10.1186/1471-2199-10-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham R., Ekunwe S. I., Falk S., Snyder L., Kleanthous C. (2000). The major head protein of bacteriophage T4 binds specifically to elongation factor Tu. J. Biol. Chem. 275, 23219–23226. doi: 10.1074/jbc.M002546200, PMID: [DOI] [PubMed] [Google Scholar]
- Blower T. R., Evans T. J., Przybilski R., Fineran P. C., Salmond G. P. (2012). Viral evasion of a bacterial suicide system by RNA-based molecular mimicry enables infectious altruism. PLoS Genet. 8:e1003023. doi: 10.1371/journal.pgen.1003023, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobay L. M., Touchon M., Rocha E. P. (2013). Manipulating or superseding host recombination functions: a dilemma that shapes phage evolvability. PLoS Genet. 9:e1003825. doi: 10.1371/journal.pgen.1003825, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobonis J., Mitosch K., Mateus A., Karcher N., Kritikos G., Selkrig J., et al. (2022). Bacterial retrons encode phage-defending tripartite toxin-antitoxin systems. Nature 609, 144–150. doi: 10.1038/s41586-022-05091-4, PMID: [DOI] [PubMed] [Google Scholar]
- Bondy-Denomy J., Pawluk A., Maxwell K. L., Davidson A. R. (2013). Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432. doi: 10.1038/nature11723, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges A. L., Davidson A. R., Bondy-Denomy J. (2017). The discovery, mechanisms, and evolutionary impact of anti-CRISPRs. Ann Rev Virol 4, 37–59. doi: 10.1146/annurev-virology-101416-041616, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges A. L., Zhang J. Y., Rollins M. F., Osuna B. A., Wiedenheft B., Bondy-Denomy J. (2018). Bacteriophage cooperation suppresses CRISPR-Cas3 and Cas9 immunity. Cells 174, 917–925.e910. doi: 10.1016/j.cell.2018.06.013, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard J. D., Moineau S. (2004). Lactococcal phage genes involved in sensitivity to AbiK and their relation to single-strand annealing proteins. J. Bacteriol. 186, 3649–3652. doi: 10.1128/JB.186.11.3649-3652.2004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs A. M., Aravind L. (2020). Identification of uncharacterized components of prokaryotic immune systems and their diverse eukaryotic reformulations. J. Bacteriol. 202:e00365-20. doi: 10.1128/JB.00365-20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champness W. C., Snyder L. (1982). The gol site: a cis-acting bacteriophage T4 regulatory region that can affect expression of all the T4 late genes. J. Mol. Biol. 155, 395–407. doi: 10.1016/0022-2836(82)90478-8, PMID: [DOI] [PubMed] [Google Scholar]
- Chan W. T., Espinosa M., Yeo C. C. (2016). Keeping the wolves at bay: antitoxins of prokaryotic type II toxin-antitoxin systems. Front. Mol. Biosci. 3:9. doi: 10.3389/fmolb.2016.00009, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Akusobi C., Fang X., Salmond G. P. C. (2017). Environmental T4-family bacteriophages evolve to escape abortive infection via multiple routes in a bacterial host employing "altruistic suicide" through type III toxin-antitoxin systems. Front. Microbiol. 8:1006. doi: 10.3389/fmicb.2017.01006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K., Wilkinson M., Chaban Y., Wigley D. B. (2020). A conformational switch in response to chi converts RecBCD from phage destruction to DNA repair. Nat. Struct. Mol. Biol. 27, 71–77. doi: 10.1038/s41594-019-0355-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S. K., Maenhaut-Michel G., Mine N., Gottesman S., Gerdes K., Van Melderen L. (2004). Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin-antitoxin system. Mol. Microbiol. 51, 1705–1717. doi: 10.1046/j.1365-2958.2003.03941.x, PMID: [DOI] [PubMed] [Google Scholar]
- Christensen S. K., Mikkelsen M., Pedersen K., Gerdes K. (2001). RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. U. S. A. 98, 14328–14333. doi: 10.1073/pnas.251327898, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D., Melamed S., Millman A., Shulman G., Oppenheimer-Shaanan Y., Kacen A., et al. (2019). Cyclic GMP-AMP signalling protects bacteria against viral infection. Nature 574, 691–695. doi: 10.1038/s41586-019-1605-5, PMID: [DOI] [PubMed] [Google Scholar]
- Court R., Cook N., Saikrishnan K., Wigley D. (2007). The crystal structure of lambda-gam protein suggests a model for RecBCD inhibition. J. Mol. Biol. 371, 25–33. doi: 10.1016/j.jmb.2007.05.037, PMID: [DOI] [PubMed] [Google Scholar]
- de La Campa A. G., Springhorn S. S., Kale P., Lacks S. A. (1988). Proteins encoded by the DpnI restriction gene cassette. Hyperproduction and characterization of the DpnI endonuclease. J. Biol. Chem. 263, 14696–14702. doi: 10.1016/S0021-9258(18)68093-7, PMID: [DOI] [PubMed] [Google Scholar]
- Dedrick R. M., Guerrero-Bustamante C. A., Garlena R. A., Russell D. A., Ford K., Harris K., et al. (2019). Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 25, 730–733. doi: 10.1038/s41591-019-0437-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H., Barrangou R., Garneau J. E., Labonté J., Fremaux C., Boyaval P., et al. (2008). Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190, 1390–1400. doi: 10.1128/JB.01412-07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham M. S., Kowalczykowski S. C. (2008). RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72, 642–671, Table of Contents. doi: 10.1128/mmbr.00020-08, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo-Calap P., Mora-Quilis L., Sanjuán R. (2020). Social bacteriophages. Microorganisms 8:533. doi: 10.3390/microorganisms8040533, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Guan X., Li N., Zhang F., Zhu Y., Ren K., et al. (2019). An anti-CRISPR protein disables type V Cas12a by acetylation. Nat. Struct. Mol. Biol. 26, 308–314. doi: 10.1038/s41594-019-0206-1, PMID: [DOI] [PubMed] [Google Scholar]
- Dong D., Guo M., Wang S., Zhu Y., Wang S., Xiong Z., et al. (2017). Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein. Nature 546, 436–439. doi: 10.1038/nature22377, PMID: [DOI] [PubMed] [Google Scholar]
- Doron S., Melamed S., Ofir G., Leavitt A., Lopatina A., Keren M., et al. (2018). Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359:eaar4120. doi: 10.1126/science.aar4120, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdz M., Piekarowicz A., Bujnicki J. M., Radlinska M. (2012). Novel non-specific DNA adenine methyltransferases. Nucleic Acids Res. 40, 2119–2130. doi: 10.1093/nar/gkr1039, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis M., Villion M., Magadán A. H., Moineau S. (2013). CRISPR-Cas and restriction-modification systems are compatible and increase phage resistance. Nat. Commun. 4:2087. doi: 10.1038/ncomms3087 [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka H., Reches M., Narasimhan S., Schoulaker-Schwarz R., Klemes Y., Aizenman E., et al. (1998). rexB of bacteriophage lambda is an anti-cell death gene. Proc. Natl. Acad. Sci. U. S. A. 95, 15481–15486. doi: 10.1073/pnas.95.26.15481, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farasat I., Salis H. M. (2016). A biophysical model of CRISPR/Cas9 activity for rational design of genome editing and gene regulation. PLoS Comput. Biol. 12:e1004724. doi: 10.1371/journal.pcbi.1004724, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatma S., Chakravarti A., Zeng X., Huang R. H. (2021). Molecular mechanisms of the CdnG-Cap5 antiphage defense system employing 3′,2'-cGAMP as the second messenger. Nat. Commun. 12:6381. doi: 10.1038/s41467-021-26738-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felice F., Micheli G., Camilloni G. (2019). Restriction enzymes and their use in molecular biology: an overview. J. Biosci. 44:38. doi: 10.1007/s12038-019-9856-8 [DOI] [PubMed] [Google Scholar]
- Fitzgerald K. A., Kagan J. C. (2020). Toll-like receptors and the control of immunity. Cells 180, 1044–1066. doi: 10.1016/j.cell.2020.02.041, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folimonova S. Y. (2012). Superinfection exclusion is an active virus-controlled function that requires a specific viral protein. J. Virol. 86, 5554–5561. doi: 10.1128/JVI.00310-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg K. J., Schmidtke D. T., Werther R., Uribe R. V., Hausman D., Sommer M. O. A., et al. (2021). The novel anti-CRISPR AcrIIA22 relieves DNA torsion in target plasmids and impairs SpyCas9 activity. PLoS Biol. 19:e3001428. doi: 10.1371/journal.pbio.3001428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Foden J. A., Khayter C., Maeder M. L., Reyon D., Joung J. K., et al. (2013). High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826. doi: 10.1038/nbt.2623, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Altae-Tran H., Böhning F., Makarova K. S., Segel M., Schmid-Burgk J. L., et al. (2020). Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science 369, 1077–1084. doi: 10.1126/science.aba0372, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L. A., Wilkinson M. E., Strecker J., Makarova K. S., Macrae R. K., Koonin E. V., et al. (2022). Prokaryotic innate immunity through pattern recognition of conserved viral proteins. Science 377:eabm4096. doi: 10.1126/science.abm4096, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Zhang L., Ge Z., Wang H., Yue Y., Jiang Z., et al. (2022). Anti-CRISPR protein AcrIF4 inhibits the type I-F CRISPR-Cas surveillance complex by blocking nuclease recruitment and DNA cleavage. J. Biol. Chem. 298:102575. doi: 10.1016/j.jbc.2022.102575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garb J., Lopatina A., Bernheim A., Zaremba M., Siksnys V., Melamed S., et al. (2022). Multiple phage resistance systems inhibit infection via SIR2-dependent NAD+ depletion. Nat. Microbiol. 7, 1849–1856. doi: 10.1038/s41564-022-01207-8, PMID: [DOI] [PubMed] [Google Scholar]
- Goldfarb T., Sberro H., Weinstock E., Cohen O., Doron S., Charpak-Amikam Y., et al. (2015). BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 34, 169–183. doi: 10.15252/embj.201489455, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordeeva J., Morozova N., Sierro N., Isaev A., Sinkunas T., Tsvetkova K., et al. (2019). BREX system of Escherichia coli distinguishes self from non-self by methylation of a specific DNA site. Nucleic Acids Res. 47, 253–265. doi: 10.1093/nar/gky1125, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günthert U., Reiners L. (1987). Bacillus subtilis phage SPR codes for a DNA methyltransferase with triple sequence specificity. Nucleic Acids Res. 15, 3689–3702. doi: 10.1093/nar/15.9.3689, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaber J., Rousseau G. M., Hammer K., Moineau S. (2009). Identification and characterization of the phage gene sav, involved in sensitivity to the lactococcal abortive infection mechanism AbiV. Appl. Environ. Microbiol. 75, 2484–2494. doi: 10.1128/AEM.02093-08, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaber J., Samson J. E., Labrie S. J., Campanacci V., Cambillau C., Moineau S., et al. (2010). Lactococcal abortive infection protein AbiV interacts directly with the phage protein SaV and prevents translation of phage proteins. Appl. Environ. Microbiol. 76, 7085–7092. doi: 10.1128/AEM.00093-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton H. G., Watson B. N. J., Fineran P. C. (2020). The arms race between bacteria and their phage foes. Nature 577, 327–336. doi: 10.1038/s41586-019-1894-8, PMID: [DOI] [PubMed] [Google Scholar]
- Harrington L. B., Doxzen K. W., Ma E., Liu J. J., Knott G. J., Edraki A., et al. (2017). A broad-spectrum inhibitor of CRISPR-Cas9. Cells 170, 1224–1233.e1215. doi: 10.1016/j.cell.2017.07.037, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S. (1980). Specificity of the bacteriophage mu mom+ −controlled DNA modification. J. Virol. 34, 277–279. doi: 10.1128/jvi.34.1.277-279.1980, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Miller L. A., Klaenhammer T. R. (1991). In vivo genetic exchange of a functional domain from a type II a methylase between lactococcal plasmid pTR2030 and a virulent bacteriophage. J. Bacteriol. 173, 4363–4370. doi: 10.1128/jb.173.14.4363-4370.1991, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard J. J., Maurizi M. R., Simon L. D. (1998). Isolation and characterization of the phage T4 PinA protein, an inhibitor of the ATP-dependent lon protease of Escherichia coli. J. Biol. Chem. 273, 518–523. doi: 10.1074/jbc.273.1.518, PMID: [DOI] [PubMed] [Google Scholar]
- Ho C. H., Wang H. C., Ko T. P., Chang Y. C., Wang A. H. (2014). The T4 phage DNA mimic protein Arn inhibits the DNA binding activity of the bacterial histone-like protein H-NS. J. Biol. Chem. 289, 27046–27054. doi: 10.1074/jbc.M114.590851, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs S. J., Wein T., Lu A., Morehouse B. R., Schnabel J., Leavitt A., et al. (2022). Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity. Nature 605, 522–526. doi: 10.1038/s41586-022-04716-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huiting E., Cao X., Ren J., Athukoralage J. S., Luo Z., Silas S., et al. (2023). Bacteriophages inhibit and evade cGAS-like immune function in bacteria. Cells 186, 864–876.e821. doi: 10.1016/j.cell.2022.12.041, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida S., Streiff M. B., Bickle T. A., Arber W. (1987). Two DNA antirestriction systems of bacteriophage P1, darA, and darB: characterization of darA- phages. Virology 157, 156–166. doi: 10.1016/0042-6822(87)90324-2, PMID: [DOI] [PubMed] [Google Scholar]
- Imai S., Johnson F. B., Marciniak R. A., Mcvey M., Park P. U., Guarente L. (2000). Sir2: an NAD-dependent histone deacetylase that connects chromatin silencing, metabolism, and aging. Cold Spring Harb. Symp. Quant. Biol. 65, 297–302. doi: 10.1101/sqb.2000.65.297, PMID: [DOI] [PubMed] [Google Scholar]
- Isaev A., Drobiazko A., Sierro N., Gordeeva J., Yosef I., Qimron U., et al. (2020). Phage T7 DNA mimic protein Ocr is a potent inhibitor of BREX defence. Nucleic Acids Res. 48, 7601–7602. doi: 10.1093/nar/gkaa510, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I. E., Wright A. V., Cofsky J. C., Aris K. D. P., Doudna J. A., Bryant Z. (2020). Cas9 interrogates DNA in discrete steps modulated by mismatches and supercoiling. Proc. Natl. Acad. Sci. U. S. A. 117, 5853–5860. doi: 10.1073/pnas.1913445117, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskólska M., Adams D. W., Blokesch M. (2022). Two defence systems eliminate plasmids from seventh pandemic Vibrio cholerae. Nature 604, 323–329. doi: 10.1038/s41586-022-04546-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenson J. M., Li T., Du F., Ea C.-K., Chen Z. J. (2023). Ubiquitin-like conjugation by bacterial cGAS enhances anti-phage defence. Nature 616, 326–331. doi: 10.1038/s41586-023-05862-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N., Patel D. J. (2021). Structure-based functional mechanisms and biotechnology applications of anti-CRISPR proteins. Nat. Rev. Mol. Cell Biol. 22, 563–579. doi: 10.1038/s41580-021-00371-9, PMID: [DOI] [PubMed] [Google Scholar]
- Johnson A. G., Wein T., Mayer M. L., Duncan-Lowey B., Yirmiya E., Oppenheimer-Shaanan Y., et al. (2022). Bacterial gasdermins reveal an ancient mechanism of cell death. Science 375, 221–225. doi: 10.1126/science.abj8432, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibby E. M., Conte A. N., Burroughs A. M., Nagy T. A., Vargas J. A., Aravind L., et al. (2022). Bacterial NLR-related proteins protect against phage. Cell 186, 2410–2424.e2418. doi: 10.1016/j.cell.2023.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. C., Kim K., Park E. H., Lim C. J. (1997). Nucleotide sequence and revised map location of the arn gene from bacteriophage T4. Mol. Cells 7, 694–696. PMID: [PubMed] [Google Scholar]
- King G., Murray N. E. (1995). Restriction alleviation and modification enhancement by the Rac prophage of Escherichia coli K-12. Mol. Microbiol. 16, 769–777. doi: 10.1111/j.1365-2958.1995.tb02438.x, PMID: [DOI] [PubMed] [Google Scholar]
- Ko T. P., Wang Y. C., Yang C. S., Hou M. H., Chen C. J., Chiu Y. F., et al. (2022). Crystal structure and functional implication of bacterial STING. Nat. Commun. 13:26. doi: 10.1038/s41467-021-26583-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga M., Otsuka Y., Lemire S., Yonesaki T. (2011). Escherichia coli rnlA and rnlB compose a novel toxin-antitoxin system. Genetics 187, 123–130. doi: 10.1534/genetics.110.121798, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnik M. V., Fedorova I., Karneyeva K. A., Artamonova D. N., Severinov K. V. (2021). Type III CRISPR-Cas systems: deciphering the Most complex prokaryotic immune system. Biochemistry (Mosc) 86, 1301–1314. doi: 10.1134/S0006297921100114, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger D. H., Hansen S., Reuter M. (1983). The ocr+ gene function of bacteriophages T3 and T7 counteracts the Salmonella typhimurium DNA restriction systems SA and SB. J. Virol. 45, 1147–1149. doi: 10.1128/jvi.45.3.1147-1149.1983, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger D. H., Kupper D., Meisel A., Reuter M., Schroeder C. (1995). The significance of distance and orientation of restriction endonuclease recognition sites in viral DNA genomes. FEMS Microbiol. Rev. 17, 177–184. doi: 10.1016/0168-6445(94)00066-2, PMID: [DOI] [PubMed] [Google Scholar]
- Krüger D. H., Schroeder C., Santibanez-Koref M., Reuter M. (1989). Avoidance of DNA methylation. A virus-encoded methylase inhibitor and evidence for counterselection of methylase recognition sites in viral genomes. Cell Biophys. 15, 87–95. doi: 10.1007/BF02991582, PMID: [DOI] [PubMed] [Google Scholar]
- Labrie S. J., Samson J. E., Moineau S. (2010). Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327. doi: 10.1038/nrmicro2315, PMID: [DOI] [PubMed] [Google Scholar]
- Labrie S. J., Tremblay D. M., Moisan M., Villion M., Magadán A. H., Campanacci V., et al. (2012). Involvement of the major capsid protein and two early-expressed phage genes in the activity of the lactococcal abortive infection mechanism AbiT. Appl. Environ. Microbiol. 78, 6890–6899. doi: 10.1128/aem.01755-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau R. K., Ye Q., Birkholz E. A., Berg K. R., Patel L., Mathews I. T., et al. (2020). Structure and mechanism of a cyclic trinucleotide-activated bacterial endonuclease mediating bacteriophage immunity. Mol. Cell 77, 723–733.e726. doi: 10.1016/j.molcel.2019.12.010, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt A., Yirmiya E., Amitai G., Lu A., Garb J., Herbst E., et al. (2022). Viruses inhibit TIR gcADPR signalling to overcome bacterial defence. Nature 611, 326–331. doi: 10.1038/s41586-022-05375-9, PMID: [DOI] [PubMed] [Google Scholar]
- Ledvina H. E., Ye Q., Gu Y., Sullivan A. E., Quan Y., Lau R. K., et al. (2023). An E1-E2 fusion protein primes antiviral immune signalling in bacteria. Nature 616, 319–325. doi: 10.1038/s41586-022-05647-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnherr H., Yarmolinsky M. B. (1995). Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 92, 3274–3277. doi: 10.1073/pnas.92.8.3274, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe D. D., Koonin E. V., Aravind L. (2004). STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J. Mol. Biol. 343, 1–28. doi: 10.1016/j.jmb.2004.08.023, PMID: [DOI] [PubMed] [Google Scholar]
- Leroux M., Srikant S., Teodoro G. I. C., Zhang T., Littlehale M. L., Doron S., et al. (2022). The DarTG toxin-antitoxin system provides phage defence by ADP-ribosylating viral DNA. Nat. Microbiol. 7, 1028–1040. doi: 10.1038/s41564-022-01153-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P., Qin S., Pu Q., Wang Z., Wu Q., Gao P., et al. (2020). CRISPR-Cas13 inhibitors block RNA editing in Bacteria and mammalian cells. Mol. Cell 78, 850–861.e855. doi: 10.1016/j.molcel.2020.03.033, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Qian C., Cao X. (2016). Post-translational modification control of innate immunity. Immunity 45, 15–30. doi: 10.1016/j.immuni.2016.06.020, PMID: [DOI] [PubMed] [Google Scholar]
- Loenen W. A., Murray N. E. (1986). Modification enhancement by the restriction alleviation protein (Ral) of bacteriophage lambda. J. Mol. Biol. 190, 11–22. doi: 10.1016/0022-2836(86)90071-9, PMID: [DOI] [PubMed] [Google Scholar]
- Loenen W. A., Raleigh E. A. (2014). The other face of restriction: modification-dependent enzymes. Nucleic Acids Res. 42, 56–69. doi: 10.1093/nar/gkt747, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopatina A., Tal N., Sorek R. (2020). Abortive infection: bacterial suicide as an antiviral immune strategy. Annu Rev Virol 7, 371–384. doi: 10.1146/annurev-virology-011620-040628, PMID: [DOI] [PubMed] [Google Scholar]
- Lowey B., Whiteley A. T., Keszei A. F. A., Morehouse B. R., Mathews I. T., Antine S. P., et al. (2020). CBASS immunity uses CARF-related effectors to sense 3′-5′- and 2′-5′-linked cyclic oligonucleotide signals and protect bacteria from phage infection. Cells 182, 38–49.e17. doi: 10.1016/j.cell.2020.05.019, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M. J., Henning U. (1994). Superinfection exclusion by T-even-type coliphages. Trends Microbiol. 2, 137–139. doi: 10.1016/0966-842X(94)90601-7, PMID: [DOI] [PubMed] [Google Scholar]
- Macek B., Forchhammer K., Hardouin J., Weber-Ban E., Grangeasse C., Mijakovic I. (2019). Protein post-translational modifications in bacteria. Nat. Rev. Microbiol. 17, 651–664. doi: 10.1038/s41579-019-0243-0 [DOI] [PubMed] [Google Scholar]
- Makarova K. S., Wolf Y. I., Snir S., Koonin E. V. (2011). Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J. Bacteriol. 193, 6039–6056. doi: 10.1128/JB.05535-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K. S., Wolf Y. I., Van Der Oost J., Koonin E. V. (2009). Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol. Direct 4:29. doi: 10.1186/1745-6150-4-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manghwar H., Lindsey K., Zhang X., Jin S. (2019). CRISPR/Cas system: recent advances and future prospects for genome editing. Trends Plant Sci. 24, 1102–1125. doi: 10.1016/j.tplants.2019.09.006, PMID: [DOI] [PubMed] [Google Scholar]
- Marino N. D., Zhang J. Y., Borges A. L., Sousa A. A., Leon L. M., Rauch B. J., et al. (2018). Discovery of widespread type I and type V CRISPR-Cas inhibitors. Science 362, 240–242. doi: 10.1126/science.aau5174, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini L. A. (2015). CRISPR-Cas immunity in prokaryotes. Nature 526, 55–61. doi: 10.1038/nature15386 [DOI] [PubMed] [Google Scholar]
- Marraffini L. A., Sontheimer E. J. (2010). Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463, 568–571. doi: 10.1038/nature08703, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. (2001). Toll-like receptors and innate immunity. Nat. Rev. Immunol. 1, 135–145. doi: 10.1038/35100529, PMID: [DOI] [PubMed] [Google Scholar]
- Meeske A. J., Jia N., Cassel A. K., Kozlova A., Liao J., Wiedmann M., et al. (2020). A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity. Science 369, 54–59. doi: 10.1126/science.abb6151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel A., Bickle T. A., Krüger D. H., Schroeder C. (1992). Type III restriction enzymes need two inversely oriented recognition sites for DNA cleavage. Nature 355, 467–469. doi: 10.1038/355467a0, PMID: [DOI] [PubMed] [Google Scholar]
- Millman A., Bernheim A., Stokar-Avihail A., Fedorenko T., Voichek M., Leavitt A., et al. (2020a). Bacterial Retrons function in anti-phage defense. Cells 183, 1551–1561.e1512. doi: 10.1016/j.cell.2020.09.065 [DOI] [PubMed] [Google Scholar]
- Millman A., Melamed S., Amitai G., Sorek R. (2020b). Diversity and classification of cyclic-oligonucleotide-based anti-phage signalling systems. Nat. Microbiol. 5, 1608–1615. doi: 10.1038/s41564-020-0777-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millman A., Melamed S., Leavitt A., Doron S., Bernheim A., Hör J., et al. (2022). An expanded arsenal of immune systems that protect bacteria from phages. Cell Host Microbe 30, 1556–1569.e1555. doi: 10.1016/j.chom.2022.09.017, PMID: [DOI] [PubMed] [Google Scholar]
- Molina R., Sofos N., Montoya G. (2020). Structural basis of CRISPR-Cas type III prokaryotic defence systems. Curr. Opin. Struct. Biol. 65, 119–129. doi: 10.1016/j.sbi.2020.06.010, PMID: [DOI] [PubMed] [Google Scholar]
- Molineux I. J., Schmitt C. K., Condreay J. P. (1989). Mutants of bacteriophage T7 that escape F restriction. J. Mol. Biol. 207, 563–574. doi: 10.1016/0022-2836(89)90465-8, PMID: [DOI] [PubMed] [Google Scholar]
- Montgomery M. T., Guerrero Bustamante C. A., Dedrick R. M., Jacobs-Sera D., Hatfull G. F. (2019). Yet more evidence of collusion: a new viral defense system encoded by gordonia phage CarolAnn. mBio:10. doi: 10.1128/mBio.02417-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morehouse B. R., Yip M. C. J., Keszei A. F. A., Mcnamara-Bordewick N. K., Shao S., Kranzusch P. J. (2022). Cryo-EM structure of an active bacterial TIR-STING filament complex. Nature 608, 803–807. doi: 10.1038/s41586-022-04999-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K. C. (1991). Lambda gam protein inhibits the helicase and chi-stimulated recombination activities of Escherichia coli RecBCD enzyme. J. Bacteriol. 173, 5808–5821. doi: 10.1128/jb.173.18.5808-5821.1991, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K. C. (2000). Bacteriophage P22 Abc2 protein binds to RecC increases the 5′ strand nicking activity of RecBCD and together with lambda bet, promotes chi-independent recombination. J. Mol. Biol. 296, 385–401. doi: 10.1006/jmbi.1999.3486, PMID: [DOI] [PubMed] [Google Scholar]
- Murphy J., Mahony J., Ainsworth S., Nauta A., Van Sinderen D. (2013). Bacteriophage orphan DNA methyltransferases: insights from their bacterial origin, function, and occurrence. Appl. Environ. Microbiol. 79, 7547–7555. doi: 10.1128/AEM.02229-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N. E. (2002). Immigration control of DNA in bacteria: self versus non-self. Microbiology 148, 3–20. doi: 10.1099/00221287-148-1-3, PMID: [DOI] [PubMed] [Google Scholar]
- Nakai T., Park S. C. (2002). Bacteriophage therapy of infectious diseases in aquaculture. Res. Microbiol. 153, 13–18. doi: 10.1016/S0923-2508(01)01280-3, PMID: [DOI] [PubMed] [Google Scholar]
- Nechaev S., Severinov K. (2003). Bacteriophage-induced modifications of host RNA polymerase. Annu. Rev. Microbiol. 57, 301–322. doi: 10.1146/annurev.micro.57.030502.090942, PMID: [DOI] [PubMed] [Google Scholar]
- Newton A. C., Bootman M. D., Scott J. D. (2016). Second messengers. Cold Spring Harb. Perspect. Biol. 8:a005926. doi: 10.1101/cshperspect.a005926, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y., Yang L., Gao T., Dong C., Zhang B., Yin P., et al. (2020). A type I-F anti-CRISPR protein inhibits the CRISPR-Cas surveillance complex by ADP-Ribosylation. Mol. Cell 80, 512–524.e515. doi: 10.1016/j.molcel.2020.09.015, PMID: [DOI] [PubMed] [Google Scholar]
- North B. J., Verdin E. (2004). Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 5:224. doi: 10.1186/gb-2004-5-5-224, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofir G., Herbst E., Baroz M., Cohen D., Millman A., Doron S., et al. (2021). Antiviral activity of bacterial TIR domains via immune signalling molecules. Nature 600, 116–120. doi: 10.1038/s41586-021-04098-7, PMID: [DOI] [PubMed] [Google Scholar]
- Ofir G., Melamed S., Sberro H., Mukamel Z., Silverman S., Yaakov G., et al. (2018). DISARM is a widespread bacterial defence system with broad anti-phage activities. Nat. Microbiol. 3, 90–98. doi: 10.1038/s41564-017-0051-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszak T., Latka A., Roszniowski B., Valvano M. A., Drulis-Kawa Z. (2017). Phage life cycles behind bacterial biodiversity. Curr. Med. Chem. 24, 3987–4001. doi: 10.2174/0929867324666170413100136, PMID: [DOI] [PubMed] [Google Scholar]
- O'sullivan L., Bolton D., Mcauliffe O., Coffey A. (2019). Bacteriophages in food applications: from foe to friend. Annu. Rev. Food Sci. Technol. 10, 151–172. doi: 10.1146/annurev-food-032818-121747, PMID: [DOI] [PubMed] [Google Scholar]
- Osuna B. A., Karambelkar S., Mahendra C., Christie K. A., Garcia B., Davidson A. R., et al. (2020). Listeria phages induce Cas9 degradation to protect lysogenic genomes. Cell Host Microbe 28, 31–40.e39. doi: 10.1016/j.chom.2020.04.001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka Y., Yonesaki T. (2012). Dmd of bacteriophage T4 functions as an antitoxin against Escherichia coli LsoA and RnlA toxins. Mol. Microbiol. 83, 669–681. doi: 10.1111/j.1365-2958.2012.07975.x, PMID: [DOI] [PubMed] [Google Scholar]
- Pawluk A., Staals R. H., Taylor C., Watson B. N., Saha S., Fineran P. C., et al. (2016). Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nat. Microbiol. 1:16085. doi: 10.1038/nmicrobiol.2016.85 [DOI] [PubMed] [Google Scholar]
- Penner M., Morad I., Snyder L., Kaufmann G. (1995). Phage T4-coded Stp: double-edged effector of coupled DNA and tRNA-restriction systems. J. Mol. Biol. 249, 857–868. doi: 10.1006/jmbi.1995.0343, PMID: [DOI] [PubMed] [Google Scholar]
- Pinilla-Redondo R., Shehreen S., Marino N. D., Fagerlund R. D., Brown C. M., Sørensen S. J., et al. (2020). Discovery of multiple anti-CRISPRs highlights anti-defense gene clustering in mobile genetic elements. Nat. Commun. 11:5652. doi: 10.1038/s41467-020-19415-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platnich J. M., Muruve D. A. (2019). NOD-like receptors and inflammasomes: a review of their canonical and non-canonical signaling pathways. Arch. Biochem. Biophys. 670, 4–14. doi: 10.1016/j.abb.2019.02.008, PMID: [DOI] [PubMed] [Google Scholar]
- Pleška M., Guet C. C. (2017). Effects of mutations in phage restriction sites during escape from restriction-modification. Biol. Lett. 13:20170646. doi: 10.1098/rsbl.2017.0646, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteete A. R., Fenton A. C., Murphy K. C. (1988). Modulation of Escherichia coli RecBCD activity by the bacteriophage lambda gam and P22 Abc functions. J. Bacteriol. 170, 2012–2021. doi: 10.1128/jb.170.5.2012-2021.1988, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh E. A., Wilson G. (1986). Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc. Natl. Acad. Sci. U. S. A. 83, 9070–9074. doi: 10.1073/pnas.83.23.9070, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch B. J., Silvis M. R., Hultquist J. F., Waters C. S., Mcgregor M. J., Krogan N. J., et al. (2017). Inhibition of CRISPR-Cas9 with bacteriophage proteins. Cells 168, 150–158.e110. doi: 10.1016/j.cell.2016.12.009, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Wang H., Yang L., Li F., Wu Y., Luo Z., et al. (2022). Structural and mechanistic insights into the inhibition of type I-F CRISPR-Cas system by anti-CRISPR protein AcrIF23. J. Biol. Chem. 298:102124. doi: 10.1016/j.jbc.2022.102124, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifat D., Wright N. T., Varney K. M., Weber D. J., Black L. W. (2008). Restriction endonuclease inhibitor IPI* of bacteriophage T4: a novel structure for a dedicated target. J. Mol. Biol. 375, 720–734. doi: 10.1016/j.jmb.2007.10.064, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins W. P., Meader B. T., Toska J., Mekalanos J. J. (2022). Cell density-dependent death triggered by viral palindromic DNA sequences. bioRxiv. doi: 10.1101/2022.11.18.517080 [DOI] [PubMed] [Google Scholar]
- Rousset F., Depardieu F., Miele S., Dowding J., Laval A. L., Lieberman E., et al. (2022). Phages and their satellites encode hotspots of antiviral systems. Cell Host Microbe 30, 740–753.e745. doi: 10.1016/j.chom.2022.02.018, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson J. E., Bélanger M., Moineau S. (2013a). Effect of the abortive infection mechanism and type III toxin/antitoxin system AbiQ on the lytic cycle of Lactococcus lactis phages. J. Bacteriol. 195, 3947–3956. doi: 10.1128/jb.00296-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson J. E., Magadán A. H., Sabri M., Moineau S. (2013b). Revenge of the phages: defeating bacterial defences. Nat. Rev. Microbiol. 11, 675–687. doi: 10.1038/nrmicro3096 [DOI] [PubMed] [Google Scholar]
- Sapranauskas R., Gasiunas G., Fremaux C., Barrangou R., Horvath P., Siksnys V. (2011). The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 39, 9275–9282. doi: 10.1093/nar/gkr606, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sberro H., Leavitt A., Kiro R., Koh E., Peleg Y., Qimron U., et al. (2013). Discovery of functional toxin/antitoxin systems in bacteria by shotgun cloning. Mol. Cell 50, 136–148. doi: 10.1016/j.molcel.2013.02.002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt C. K., Kemp P., Molineux I. J. (1991). Genes 1.2 and 10 of bacteriophages T3 and T7 determine the permeability lesions observed in infected cells of Escherichia coli expressing the F plasmid gene pifA. J. Bacteriol. 173, 6507–6514. doi: 10.1128/jb.173.20.6507-6514.1991, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster C. F., Bertram R. (2013). Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol. Lett. 340, 73–85. doi: 10.1111/1574-6968.12074, PMID: [DOI] [PubMed] [Google Scholar]
- Schwer B., Verdin E. (2008). Conserved metabolic regulatory functions of sirtuins. Cell Metab. 7, 104–112. doi: 10.1016/j.cmet.2007.11.006, PMID: [DOI] [PubMed] [Google Scholar]
- Semenova E., Jore M. M., Datsenko K. A., Semenova A., Westra E. R., Wanner B., et al. (2011). Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U. S. A. 108, 10098–10103. doi: 10.1073/pnas.1104144108, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinedling S., Parma D., Gold L. (1987). Wild-type bacteriophage T4 is restricted by the lambda rex genes. J. Virol. 61, 3790–3794. doi: 10.1128/jvi.61.12.3790-3794.1987, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silas S., Lucas-Elio P., Jackson S. A., Aroca-Crevillén A., Hansen L. L., Fineran P. C., et al. (2017). Type III CRISPR-Cas systems can provide redundancy to counteract viral escape from type I systems. elife 6:e27601. doi: 10.7554/eLife.27601, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein J. L., Goldberg E. B. (1976). T4 DNA injection. II. Protection of entering DNA from host exonuclease V. Virology 72, 212–223. doi: 10.1016/0042-6822(76)90324-X, PMID: [DOI] [PubMed] [Google Scholar]
- Sistla S., Rao D. N. (2004). S-Adenosyl-L-methionine-dependent restriction enzymes. Crit. Rev. Biochem. Mol. Biol. 39, 1–19. doi: 10.1080/10409230490440532 [DOI] [PubMed] [Google Scholar]
- Stokar-Avihail A., Fedorenko T., Hör J., Garb J., Leavitt A., Millman A., et al. (2023). Discovery of phage determinants that confer sensitivity to bacterial immune systems. Cells 186, 1863–1876.e1816. doi: 10.1016/j.cell.2023.02.029, PMID: [DOI] [PubMed] [Google Scholar]
- Studier F. W., Movva N. R. (1976). SAMase gene of bacteriophage T3 is responsible for overcoming host restriction. J. Virol. 19, 136–145. doi: 10.1128/jvi.19.1.136-145.1976, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Yang J., Cheng Z., Amrani N., Liu C., Wang K., et al. (2019). Structures of Neisseria meningitidis Cas9 complexes in catalytically poised and anti-CRISPR-inhibited states. Mol. Cell 76, 938–952.e935. doi: 10.1016/j.molcel.2019.09.025, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenson S. B., Karlström O. H. (1976). Bacteriophage T4-induced shut-off of host-specific translation. J. Virol. 17, 326–334. doi: 10.1128/jvi.17.2.326-334.1976, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczelkun M. D., Tikhomirova M. S., Sinkunas T., Gasiunas G., Karvelis T., Pschera P., et al. (2014). Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc. Natl. Acad. Sci. U. S. A. 111, 9798–9803. doi: 10.1073/pnas.1402597111, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabib-Salazar A., Liu B., Barker D., Burchell L., Qimron U., Matthews S. J., et al. (2018). T7 phage factor required for managing RpoS in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 115, E5353–e5362. doi: 10.1073/pnas.1800429115, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal N., Millman A., Stokar-Avihail A., Fedorenko T., Leavitt A., Melamed S., et al. (2022). Bacteria deplete deoxynucleotides to defend against bacteriophage infection. Nat. Microbiol. 7, 1200–1209. doi: 10.1038/s41564-022-01158-0, PMID: [DOI] [PubMed] [Google Scholar]
- Tal N., Morehouse B. R., Millman A., Stokar-Avihail A., Avraham C., Fedorenko T., et al. (2021). Cyclic CMP and cyclic UMP mediate bacterial immunity against phages. Cells 184, 5728–5739.e5716. doi: 10.1016/j.cell.2021.09.031, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal N., Sorek R. (2022). SnapShot: bacterial immunity. Cells 185, 578–578.e571. doi: 10.1016/j.cell.2021.12.029, PMID: [DOI] [PubMed] [Google Scholar]
- Tesfazgi Mebrhatu M., Wywial E., Ghosh A., Michiels C. W., Lindner A. B., Taddei F., et al. (2011). Evidence for an evolutionary antagonism between Mrr and type III modification systems. Nucleic Acids Res. 39, 5991–6001. doi: 10.1093/nar/gkr219, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner S. J., Poppenberger B., Rozhon W. (2013). Toxin-antitoxin systems: biology, identification, and application. Mob Genet Elements 3:e26219. doi: 10.4161/mge.26219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassallo C. N., Doering C. R., Littlehale M. L., Teodoro G. I. C., Laub M. T. (2022). A functional selection reveals previously undetected anti-phage defence systems in the E. coli pangenome. Nat. Microbiol. 7, 1568–1579. doi: 10.1038/s41564-022-01219-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot M., Houkes J., Lochs S. J. A., Swarts D. C., Zheng P., Kunne T., et al. (2018). Bacteriophage DNA glucosylation impairs target DNA binding by type I and II but not by type V CRISPR-Cas effector complexes. Nucleic Acids Res. 46, 873–885. doi: 10.1093/nar/gkx1264, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkinshaw M. D., Taylor P., Sturrock S. S., Atanasiu C., Berge T., Henderson R. M., et al. (2002). Structure of Ocr from bacteriophage T7, a protein that mimics B-form DNA. Mol. Cell 9, 187–194. doi: 10.1016/S1097-2765(02)00435-5, PMID: [DOI] [PubMed] [Google Scholar]
- Wan L., Essuman K., Anderson R. G., Sasaki Y., Monteiro F., Chung E. H., et al. (2019). TIR domains of plant immune receptors are NAD+-cleaving enzymes that promote cell death. Science 365, 799–803. doi: 10.1126/science.aax1771, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Gao T., Zhou Y., Ren J., Guo J., Zeng J., et al. (2022). Mechanistic insights into the inhibition of the CRISPR-Cas surveillance complex by anti-CRISPR protein AcrIF13. J. Biol. Chem. 298:101636. doi: 10.1016/j.jbc.2022.101636, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Kim Y., Hong S. H., Ma Q., Brown B. L., Pu M., et al. (2011). Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 7, 359–366. doi: 10.1038/nchembio.560, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Yao D., Xu J.-G., Li A.-R., Xu J., Fu P., et al. (2016). Structural basis of Cas3 inhibition by the bacteriophage protein AcrF3. Nat. Struct. Mol. Biol. 23, 868–870. doi: 10.1038/nsmb.3269, PMID: [DOI] [PubMed] [Google Scholar]
- Warren R. A. (1980). Modified bases in bacteriophage DNAs. Annu. Rev. Microbiol. 34, 137–158. doi: 10.1146/annurev.mi.34.100180.001033, PMID: [DOI] [PubMed] [Google Scholar]
- Weigele P., Raleigh E. A. (2016). Biosynthesis and function of modified bases in Bacteria and their viruses. Chem. Rev. 116, 12655–12687. doi: 10.1021/acs.chemrev.6b00114, PMID: [DOI] [PubMed] [Google Scholar]
- Wein T., Sorek R. (2022). Bacterial origins of human cell-autonomous innate immune mechanisms. Nat. Rev. Immunol. 22, 629–638. doi: 10.1038/s41577-022-00705-4, PMID: [DOI] [PubMed] [Google Scholar]
- Whiteley A. T., Eaglesham J. B., De Oliveira Mann C. C., Morehouse B. R., Lowey B., Nieminen E. A., et al. (2019). Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 567, 194–199. doi: 10.1038/s41586-019-0953-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Sun L., Chen X., Du F., Shi H., Chen C., et al. (2013). Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830. doi: 10.1126/science.1229963, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Van Den Hurk A., Aparicio-Maldonado C., Kushwaha S. K., King C. M., Ou Y., et al. (2022). Defence systems provide synergistic anti-phage activity in E. coli. bioRxiv:2022.2008.2021.504612. doi: 10.1101/2022.08.21.504612 [DOI] [Google Scholar]
- Xie Y., Zhang L., Gao Z., Yin P., Wang H., Li H., et al. (2022). AcrIF5 specifically targets DNA-bound CRISPR-Cas surveillance complex for inhibition. Nat. Chem. Biol. 18, 670–677. doi: 10.1038/s41589-022-00995-8, PMID: [DOI] [PubMed] [Google Scholar]
- Xiong X., Wu G., Wei Y., Liu L., Zhang Y., Su R., et al. (2020). SspABCD-SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities. Nat. Microbiol. 5, 917–928. doi: 10.1038/s41564-020-0700-6, PMID: [DOI] [PubMed] [Google Scholar]
- Yang L., Zhang L., Yin P., Ding H., Xiao Y., Zeng J., et al. (2022). Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24. Nat. Commun. 13:1931. doi: 10.1038/s41467-022-29581-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Zhang Y., Yin P., Feng Y. (2021). Structural insights into the inactivation of the type I-F CRISPR-Cas system by anti-CRISPR proteins. RNA Biol. 18, 562–573. doi: 10.1080/15476286.2021.1985347, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui R., Washizaki A., Furihata Y., Yonesaki T., Otsuka Y. (2014). AbpA and AbpB provide anti-phage activity in Escherichia coli. Genes Genet. Syst. 89, 51–60. doi: 10.1266/ggs.89.51, PMID: [DOI] [PubMed] [Google Scholar]
- Ye Q., Lau R. K., Mathews I. T., Birkholz E. A., Watrous J. D., Azimi C. S., et al. (2020). HORMA domain proteins and a Trip13-like ATPase regulate bacterial cGAS-like enzymes to mediate bacteriophage immunity. Mol. Cell 77, 709–722.e707. doi: 10.1016/j.molcel.2019.12.009, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P., Zhang Y., Yang L., Feng Y. (2023). Non-canonical inhibition strategies and structural basis of anti-CRISPR proteins targeting type I CRISPR-Cas systems. J. Mol. Biol. 435:167996. doi: 10.1016/j.jmb.2023.167996, PMID: [DOI] [PubMed] [Google Scholar]
- Yirmiya E., Leavitt A., Lu A., Avraham C., Osterman I., Garb J., et al. (2023). Phages overcome bacterial immunity via diverse anti-defense proteins. bioRxiv. doi: 10.1101/2023.05.01.538930 [DOI] [PubMed] [Google Scholar]
- Yu D., Song W., Tan E. Y. J., Liu L., Cao Y., Jirschitzka J., et al. (2022). TIR domains of plant immune receptors are 2′,3'-cAMP/cGMP synthetases mediating cell death. Cells 185, 2370–2386.e2318. doi: 10.1016/j.cell.2022.04.032, PMID: [DOI] [PubMed] [Google Scholar]
- Zabeau M., Friedman S., Van Montagu M., Schell J. (1980). The ral gene of phage lambda. I. Identification of a non-essential gene that modulates restriction and modification in E. coli. Mol. Gen. Genet. 179, 63–73. doi: 10.1007/BF00268447, PMID: [DOI] [PubMed] [Google Scholar]
- Zaremba M., Dakineviciene D., Golovinas E., Zagorskaitė E., Stankunas E., Lopatina A., et al. (2022). Short prokaryotic Argonautes provide defence against incoming mobile genetic elements through NAD+ depletion. Nat. Microbiol. 7, 1857–1869. doi: 10.1038/s41564-022-01239-0, PMID: [DOI] [PubMed] [Google Scholar]
- Zavil'gel'skiĭ G. B., Mershavka V., Iusifov T. N., Belogurov A. A. (1984). Weakening of bacteriophage lambda EcoK DNA restriction in the presence of plasmid pKM101 ard+. I. General characteristics and genetic localization. Mol. Biol. (Mosk) 18, 1590–1596. [PubMed] [Google Scholar]
- Zavil'gel'skiĭ G. B., Rastorguev S. M. (2009). Antirestriction proteins ardA and Ocr as effective inhibitors of the type I restriction-modification enzymes. Mol. Biol. (Mosk) 43, 264–273. doi: 10.1134/S0026893309020071 PMID: [DOI] [PubMed] [Google Scholar]
- Zhang H., Li Z., Daczkowski C. M., Gabel C., Mesecar A. D., Chang L. (2019). Structural basis for the inhibition of CRISPR-Cas12a by anti-CRISPR proteins. Cell Host Microbe 25, 815–826.e814. doi: 10.1016/j.chom.2019.05.004, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang T., Tamman H., Coppieters ‘t Wallant K., Kurata T., LeRoux M., Srikant S., et al. (2022). Direct activation of a bacterial innate immune system by a viral capsid protein. Nature 612, 132–140. doi: 10.1038/s41586-022-05444-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., He C., Wang L., Ge B. (2017). Post-translational regulation of antiviral innate signaling. Eur. J. Immunol. 47, 1414–1426. doi: 10.1002/eji.201746959, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Gao A., Zhan Q., Wang Y., Feng H., Liu S., et al. (2019). Diverse mechanisms of CRISPR-Cas9 inhibition by type II-C anti-CRISPR proteins. Mol. Cell 74, 296–309.e297. doi: 10.1016/j.molcel.2019.01.038, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]