Abstract
Previous studies have shown the essential roles of O-GlcNAc transferase (Ogt) and O-GlcNAcylation in neuronal development, function and neurologic diseases. However, the function of Ogt and O-GlcNAcylation in the adult cerebellum has not been well elucidated. Here, we have found that cerebellum has the highest level of O-GlcNAcylation relative to cortex and hippocampus of adult male mice. Specific deletion of Ogt in granule neuron precursors (GNPs) induces abnormal morphology and decreased size of the cerebellum in adult male Ogt deficient [conditional knock-out (cKO)] mice. Adult male cKO mice show the reduced density and aberrant distribution of cerebellar granule cells (CGCs), the disrupted arrangement of Bergman glia (BG) and Purkinje cells. In addition, adult male cKO mice exhibit aberrant synaptic connection, impaired motor coordination, and learning and memory abilities. Mechanistically, we have identified G-protein subunit α12 (Gα12) is modified by Ogt-mediated O-GlcNAcylation. O-GlcNAcylation of Gα12 facilitates its binding to Rho guanine nucleotide exchange factor 12 (Arhgef12) and consequently activates RhoA/ROCK signaling. RhoA/ROCK pathway activator LPA can rescue the developmental deficits of Ogt deficient CGCs. Therefore, our study has revealed the critical function and related mechanisms of Ogt and O-GlcNAcylation in the cerebellum of adult male mice.
SIGNIFICANCE STATEMENT Cerebellar function are regulated by diverse mechanisms. To unveil novel mechanisms is critical for understanding the cerebellar function and the clinical therapy of cerebellum-related diseases. In the present study, we have shown that O-GlcNAc transferase gene (Ogt) deletion induces abnormal cerebellar morphology, synaptic connection, and behavioral deficits of adult male mice. Mechanistically, Ogt catalyzes O-GlcNAcylation of Gα12, which promotes the binding to Arhgef12, and regulates RhoA/ROCK signaling pathway. Our study has uncovered the important roles of Ogt and O-GlcNAcylation in regulating cerebellar function and cerebellum-related behavior. Our results suggest that Ogt and O-GlcNAcylation could be potential targets for some cerebellum-related diseases.
Keywords: Ogt, O-GlcNAcylation, cerebellum, cerebellar granule cells, Gα12, Arhgef12
Introduction
Cerebellar development is a highly orchestrated process including the proliferation and differentiation of progenitor cells, and migration and maturation of newborn cells, which are intensively regulated by diverse mechanisms such as genetics and epigenetics during the embryonic and early postnatal stages (L. Chen et al., 2022a; X. Chen et al., 2022b; Hatten and Roussel, 2011; Penas et al., 2019; Takahashi et al., 2022; ten Donkelaar et al., 2003; Zanin and Friedman, 2022). Cerebellum plays important function in motor control, decision-making, rewarding, social interaction and cognition (Beuriat et al., 2020; Carta et al., 2019; X. Chen et al., 2022b,c; Haldipur et al., 2022; Hibi and Shimizu, 2012; Takahashi et al., 2022). The dysregulation of cerebellar development leads to abnormal morphogenesis and involves in neurodevelopmental disorders including schizophrenia, autism spectrum disorder (ASD), attention deficit hyperactivity disorder (ADHD) and intellectual disability (ID; X. Chen et al., 2022b; Haldipur et al., 2022; Mackie et al., 2007; Pinchefsky et al., 2019; ten Donkelaar et al., 2003; Tobe et al., 2010; Veleanu et al., 2022).
During cerebellar development, granule neuron progenitors (GNPs) generate cerebellar granule cells (CGCs), the most abundant cells in the cerebellum. Multiple signaling pathways have been showed to regulate cerebellar neurogenesis (Anne et al., 2013; H. Yang et al., 2019; L. Chen et al., 2022a; Takahashi et al., 2022). Casein kinase 1δ (CK1δ) is regulated by proteolysis mediated by anaphase-promoting complex/cyclosome (APC/CCdh1) ubiquitin ligase, and CK1δ deficiency inhibited the proliferation of GNPs (Penas et al., 2015). Autism spectrum disorders-related gene CHD8 is a member of the chromodomain helicase (CHD) family, and the loss of CHD8 in GNPs induces the cerebellar hypoplasia, decreased proliferation and excess production of Purkinje neurons (X. Chen et al., 2022b). However, it remains unknown regarding the effects of Ogt deficiency in GNP for the adult cerebellum, which is important to understand the mechanisms regulating cerebellar function.
Ogt and O-GlcNAcase (Oga) catalyze the addition and removal of single O-GlcNAc moieties to serine and/or threonine residues of intracellular proteins, respectively (Torres and Hart, 1984; G.W. Hart et al., 2007). Dynamic O-GlcNAcylation regulates gene expression, energy metabolism and signal transduction in response to various cellular stresses (G.W. Hart et al., 2007; Hanover et al., 2012; Levine and Walker, 2016; X. Yang and Qian, 2017). Ogt and O-GlcNAcylation are abundant and dynamic during the development and aging of the nervous system. Our previous studies found that Ogt could regulate embryonic and adult neurogenesis by regulating Wnt and Notch signaling, respectively (J. Chen et al., 2021; Shen et al., 2021b). Ogt deficiency in neurons led to reduced dendritic spine density, increased the proportion of immature dendritic spines, altered feeding behavior, impaired synaptic development and neurodegeneration (Lagerlöf et al., 2016, 2017). A recent study has shown that Ogt deficiency resulted in abnormal proliferation and differentiation of GNPs, and medulloblastoma growth via disrupting Shh signaling during the cerebellar development (L. Chen et al., 2022a). However, the function and related mechanisms of Ogt and O-GlcNAcylation in regulating cerebellar function of adult mice are still under extensive study.
In the present study, we showed that Ogt deficiency [conditional knock-out (cKO)] in GNPs significantly reduced the level of O-GlcNAcylation in the CGCs and adult cerebellum, respectively. Adult cKO mice showed cerebellar atrophy and abnormal structure including the aberrant density and distribution of CGCs, the disorganized Bergman glia (BG) and Purkinje cells, and the impaired synaptogenesis. Ogt deficiency also impaired motor coordination, learning and memory abilities of adult mice. Mechanistically, we uncovered that Ogt directly interacted with and catalyzed O-GlcNAcylation of G-protein subunit α 12 (Gα12). Decreased O-GlcNAcylation of Gα12 inhibited its interaction with Rho guanine nucleotide exchange factor 12 (Arhgef12), and subsequently inhibited RhoA/ROCK signaling pathway. The treatment with Oleoyl-L-α-lysophosphatidic acid (LPA), an activator of RhoA/ROCK pathway, consistently rescued the deficits of cKO CGCs. Collectively, our findings suggest the essential roles of Ogt and O-GlcNAcylation in regulating the cerebellum function of adult mice.
Materials and Methods
Animals
Ogtloxp/Y mice line was obtained from the Jackson Laboratory (#004860). Ogt conditional knock-out mice (cKO) were generated by crossing Ogtloxp/Y mice with Math1-Cre mice. All animals were in the C57BL/6 genetic background and maintained at the Experimental Animal Center of Zhejiang University under a 12/12 h light/dark cycle with free access to food and water. All experiments were performed following the protocol approved by the Animal Experimental Ethics Committee of Zhejiang University. The genotypes were confirmed using primers showed in Table 1. Only adult male mice (8–10 weeks old) were adopted in this study.
Table 1.
The list of used primers
| Gene | Application | Organism | Length (bp) | Reference Sequence | Sequence (5′ >3′) |
|---|---|---|---|---|---|
| Gα12-HA | Plasmid cloning | Mus musculus | 1167 | NM_010302.2 | Fw: AACGGGCCCTCTAGACTCGAGTCAAGCGTAGTCTGGGACGTCGTATGGGTACTGCAGCATGATGTCTTT |
| Rv: GCACAGTGGCGGCCGCTCGAGGCCACCATGTCCGGGGTGGTGCGGACCC | |||||
| RGSL-Flag | Plasmid cloning | Mus musculus | 669 | NM_027144.2 | Fw: AACGGGCCCTCTAGACTCGAGTTACTTATCGTCGTCATCCTTGTAATCCACTTTTACTCCCAAATACT |
| Rv: GCACAGTGGCGGCCGCTCGAGGCCACCATGGCCCCTCACATCATTGGA | |||||
| Ogt | qRT-PCR | Mus musculus | 88 | NM_139144.4 | Fw: CTGGGGAACCTGCTCAAAGC |
| Rv: CTGCAAAGTTTGGTTGCGTCT | |||||
| Gapdh | qRT-PCR | Mus musculus | 83 | NM_001289726.1 | Fw: ACCCTTAAGAGGGATGCTGC |
| Rv: CGGGACGAGGAAACACTCTC | |||||
| Arhgef12 | qRT-PCR | Mus musculus | 116 | NM_027144.2 | Fw: TGGGGGTGGGGTGTG |
| Rv: TGAGGGGAAACCTGTCCG | |||||
| ROCK2 | qRT-PCR | Mus musculus | 104 | NM_009072.2 | Fw: TGCCCGATCATCCCCTAGAA |
| Rv: TAGAAGGCAGTTAGCTTGGC | |||||
| GNA12 | qRT-PCR | Mus musculus | 113 | NM_010302.2 | Fw: GACTCGGGGATCAGGGAAGC |
| Rv: CTGGGGAAGTAGTTCAGCTGGC | |||||
| Map2 | qRT-PCR | Mus musculus | 111 | NM_001310634.1 | Fw: CGGAAAACCACAGCAGCAAG |
| Rv: GGAAGAACGTTTCTCTGGGC | |||||
| PlxnA2 | qRT-PCR | Mus musculus | 109 | NM_008882.2 | Fw: CTGCTTCTCCGCAGGACTGA |
| Rv: GCATGAAGAGTGGTTCCCCA | |||||
| Grin2a | qRT-PCR | Mus musculus | 153 | NM_008170.4 | Fw: AGACCTTAGCAGGCCCTCTC |
| Rv: CTCTTGCTGTCCTCCAGACC | |||||
| Cacnb4 | qRT-PCR | Mus musculus | 167 | NM_001037099.3 | Fw: GCAAGAGTAGGAACCGCTTG |
| Rv: GCAGCCTCAAAGCCTATGTC | |||||
| Scn1b | qRT-PCR | Mus musculus | 129 | NM_001402334.1 | Fw: ACCAGCGTCGTCAAGAAGAT |
| Rv: CATCTCTGCCACAAGCCATA | |||||
| Ogtfloxp/Y | Genotyping | Mus musculus | Ogtfloxp = 338; Mutant = 487 | Fw: CATCTCTCCAGCCCCACAAACTG | |
| Rv: GACGAAGCAGGAGGGGAGAGCAC | |||||
| Math1-Cre | Genotyping | Mus musculus | Mutant = 400 | Fw: TCGATGCAACGAGTGATGAG | |
| Rv: TCCATGAGTGAACGAACCTG | |||||
This table contains the primers for plasmids cloning, qRT-PCR and genotyping PCR.
Isolation and culture of cerebellar granule cells
The isolation and culture of cerebellar granule cells (CGCs) was adapted from a previous study (Lee et al., 2009). Briefly, mouse pups [postnatal day (P)4–P7] were deeply anesthetized with ice before decapitation, and cerebellar tissues were dissected. Meninges of cerebella were carefully removed in precold 1× HBSS (14185052, Invitrogen) supplemented with 3 mg/ml bovine serum albumin (BSA). Cerebellar tissues were dissected out and digested with 10 ml 1× Earle's balanced salt solution (EBSS; E7510, Sigma-Aldrich) containing 100 U/ml papain (9001-73-4, Sangon Biotech) and 2.5 U/ml DNase 1 (9003-98-9, Sangon Biotech) at 37°C for 15 min, then triturated with fetal bovine serum (FBS)-rinsed pipette tips and centrifuged at 1000 rpm for 5 min. After discarding the supernatant, cell pellets were resuspended with Neurobasal medium (21103-049, Invitrogen) supplement with 10% FBS, 1% 2 m potassium chloride, 1% GlutaMAX (35050061, Invitrogen), 0.5% L-Glutamine (5030-149, Invitrogen) and 0.2% antibiotic-antimycotic (15070063, Invitrogen), dissociated into single-cell suspension with FBS-rinsed glass Pasteur pipettes and preplated on a dish coated with poly-D-lysine (P6407, Sigma-Aldrich) for 25 min to remove glial cells. Unattached cells were collected and filtered through 70 μm nylon meshes and centrifuged at 1500 rpm for 2 min. The pellet was resuspended with Neurobasal medium supplement with 2% B27 (17504-044, Invitrogen), 1% 2 m potassium chloride, 1% GlutaMAX (35050061, Invitrogen), 0.5% L-glutamine, and 0.2% antibiotic-antimycotic. CGCs were seeded onto poly-D-lysine-coated cell culture plates and coverslips at a certain density, respectively, and cultured in a humidified incubator supplemented with 5% CO2 at 37°C. The medium was completely replaced after 24 h, and later on, the medium was half replaced every 2–3 d.
Plasmid construction
Scramble shRNA (5′-ttctccgaacgtgtcacgt-3′) and shRNAs targeting mouse Ogt (5′-atagcatgctgtaccctcttt-3′) were cloned into Lenti-U6 vector, respectively. For the overexpression of Gα12, Ogt and RGS-like domain, the mouse cDNA was amplified and cloned to pcDNA3.1(+) vector, respectively. The used primers can be found in Table 1.
Neuro2a cell culture and transfection
Neuro2a (N2a) cells were cultured in DMEM medium (D6429, Sigma-Aldrich) containing 10% FBS and 1% antibiotic-antimycotic, and incubated in a humidified incubator supplemented with 5% CO2 at 37°C. Plasmids transfection was conducted with Lipofectamine 2000 following the manufacturer's instructions (11668500, Thermo Fisher Scientific) and cells were collected 48 h after transfection for assays.
Immunofluorescence staining and quantitative analysis
Mice were deeply anesthetized by intraperitoneal injection of chloral hydrate and transcardially perfused with cold PBS followed by 4% paraformaldehyde (PFA). Brains were dissected, postfixed with 4% PFA overnight and completely dehydrated with 30% sucrose at 4°C. Brain samples were embedded with O.C.T (4583, SAKURA) at −20°C, and 20-µm sagittal sections were prepared with a cryostat (Leica CM1950) and stored in cryoprotectant solution (glycerol, ethylene glycol, and 0.2 m phosphate buffer, pH 7.4, 1:1:2 in volume) at −20°C. For CGCs immunostaining, coverslips in 24-well plates were fixed with 4% PFA for 30 min.
Brain sections and cell samples were blocked with PBS containing 3% goat serum and 0.3% Triton X-100 at room temperature for 1 h and incubated with proper primary antibodies at 4°C overnight. Samples were then washed with PBS and incubated with fluorescence-conjugated secondary Alexa antibodies and DAPI for nuclei staining at room temperature for 1 h. After washed with PBS for three times, sections and slides were mounted onto glass slides with mounting medium. Images were captured with a confocal microscope (Olympus FV3000) and analyzed with ImageJ software. All images were taken with identical settings.
RNA extraction and real-time PCR assay
Total RNA was extracted by TRIzol reagent following the manufacturer's protocol (15596018, Invitrogen) and quantified with a Nanodrop spectrophotometer 2000 (Thermo Fisher Scientific). 500 ng of total RNA was used for reverse transcription and the relative gene expression level was measured by Applied Biosystems Viiia 7 with SYBR Green qPCR master mix (Q711, Vazyme) in triplicate. The primers for qRT-PCR were shown in Table 1. The relative expression levels were calculated by the 2-ΔΔCT method.
Western blot assay
Total proteins were extracted with lysis buffer containing RIPA lysis buffer (ab156034, Abcam) and 1× protease inhibitor cocktail (P8340, Sigma). Cerebella were dissected in ice-cold PBS and quickly frozen with liquid nitrogen. For protein extraction, cerebellar tissues and cell pellets were treated with lysis buffer on ice, respectively, and centrifuged for 30 min at 12,000 rpm at 4°C. Supernatants were collected and quantified by Bradford assay. Ten- to 30-μg proteins were subjected to SDS-PAGE, and then transferred onto nitrocellulose membranes. Membranes were blocked with 5% nonfat milk in TBST at room temperature for 1 h and incubated with primary antibodies at 4°C overnight. After washed with TBST for three times, membranes were incubated with HRP-labeled secondary antibodies at room temperature for 1 h. Membranes were washed for three times with TBST and incubated with UltraSignal ECL (4AW011, 4A Biotech). Protein bands were visualized by Tanon Detection System (Tanon 5200) and the signal intensity was quantified with ImageJ software.
Immunoprecipitation (IP) and co-immunoprecipitation (Co-IP) assay
For immunoprecipitation, cell pellets and cerebellar tissues were lysed with RIPA buffer containing 1× protease inhibitor cocktail on ice for 30 min. The supernatants were diluted using Co-IP lysis buffer, and incubated with proper primary antibodies overnight at 4°C. For co-immunoprecipitation, cells and cerebellum were lysed with Co-IP lysis buffer containing 1× protease inhibitor cocktail on ice for 1 h. The supernatants were directly incubated with primary antibodies at 4°C overnight. Samples were then mixed with Protein A Magnetic Beads (10002D, Thermo Fisher Scientific), and incubated at 4°C for 4 h. After washed with washing buffer for 3 times (10 min each time at 4°C), samples were added with 1× loading buffer and were denatured at 100°C for 5 min followed by immunoblotting assays. For Flag-tag and HA-tag, anti-FLAG M2 Magnetic Beads (M8823, Sigma) and anti-HA Magnetic Beads (88 836, Thermo Fisher Scientific) were used, respectively.
Antibodies
The following primary antibodies were used in this study: Mouse anti-O-GlcNAcylation (ab2739, Abcam), Rabbit anti-Ogt (61 355, Active motif), Mouse anti-NeuN (MAB377, Millipore), Rabbit anti-Calbindin (ET1702-54, Huabio), Mouse anti-Calbindin (C9848, Sigma), Mouse anti-VGluT1 (135011, Synaptic systems), Mouse anti-VGluT2 (MA5-31378, Invitrogen), Rabbit anti-Gfap (Z0334, Dako), Mouse anti-Map2 (MA5-12826, Invitrogen), Rabbit anti-SMI32 (SMI-32R-100, Covance), Rabbit anti-Tbr2 (ab23345, Abcam), Rabbit anti-Tubulin (ab15246, Abcam), Mouse anti-β-actin (AC004, Abclonal), Mouse anti-Gα12 (sc-515445, Santa Cruz), Rabbit anti-ROCK2 (9029S, Cell Signaling Technology), Rabbit anti-RhoA (10749-1-AP, Proteintech), Rabbit anti-Arhgef12 (22441-1-AP, Proteintech), Rabbit anti-GFP (50430-2-AP, Proteintech), Mouse anti-HA Tag (26183, Thermo Fisher Scientific), Mouse anti-Flag Tag (MA1-91878, Thermo Fisher Scientific) and Mouse anti-Gapdh (MA5-15738, Thermo Fisher Scientific). The used secondary antibodies were included: AlexaFluor488 goat anti-mouse (A11001, Thermo Fisher Scientific), AlexaFluor488 goat anti-rabbit (A11008, Thermo Fisher Scientific), AlexaFluor568 goat anti-mouse (A11004, Thermo Fisher Scientific), AlexaFluor568 goat anti-rabbit (A11036, Thermo Fisher Scientific), HRP-labeled goat anti-mouse (GAM007, Multi Science), and HRP-labeled goat anti-mouse (GAR007, Multi Science).
RhoA activation assay
RhoA activation was detected by RhoA Pulldown Activation Assay kit as manufacturer's protocol (NewEast Bioscience). In brief, N2a cells and cerebellar tissues were lysed with ice-cold 1× assay/lysis buffer for 20 min, respectively, and supernatants were collected. 1 mg of total protein was diluted to 1 ml with 1× assay/lysis buffer, then 1 μl anti-RhoA-GTP antibody (26904) and 20 μl of resuspended Protein A/G Agarose beads (30 301) were added to protein sample. Samples were incubated at 4°C for 1 h with gentle agitation. Discard the supernatant through centrifugation and wash the beads with 0.5 ml 1× assay/lysis buffer for three times. After three times washes, beads were resuspended with 1× loading buffer and denatured at 100°C for 5 min, followed by immunoblotting assays.
Morphologic analysis of cerebellar granule cells
Cerebellar granule cells (CGCs) were cultured fixed with 4% PFA at days in vitro (DIV)9 and morphologic analysis was performed. The Fiji plugin NeuronJ (Meijering, 2010; Meijering et al., 2004) was used to manually trace dendritic arbors and analyze the length and branch number of individual CGCs. The soma center and neurite termination points were marked using the straight tool.
Click-It O-GlcNAcylation analysis
CGCs at DIV9 were collected for Click-It reaction assay followed by mass spectrometry analysis. Chemoenzymatic labeling and biotinylating were conducted as described previously (J. Chen et al., 2021). Proteins with O-GlcNAcylation were labeled with GalNAz according to the Click-iT O-GlcNAc Enzymatic Labeling System protocol (Invitrogen), and conjugated with an alkyne-biotin compound according to the Click-iT Protein Analysis Detection kit protocol (Invitrogen). Control samples were treated without the enzyme GalT or UDP-GalNAz in parallel. Biotinylated proteins were precipitated as described in the Click-iT Protein Analysis Detection Kit protocol, resolubilized in 1% SDS, neutralized with an equal volume of neutralization buffer (6% NP-40, 100 mm Na2HPO4, 150 mm NaCl) and incubated with streptavidin beads (Thermo Fisher Scientific) at 4°C overnight. After washed five times with low-salt buffer (100 mm Na2HPO4, 150 mm NaCl, 0.1% SDS, 1% Triton X-100, 0.5% sodium deoxycholate) and five times with high-salt buffer (100 mm Na2HPO4, 500 mm NaCl, 0.2% Triton X-100), the bound proteins were eluted in the boiling elution buffer containing 50 mm Tris-HCl pH 6.8, 2.5% SDS, 100 mm DTT, 10% glycerol, and 20 mm biotin for 10 min.
Mass spectrometry assay was conducted on an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) operated in data-dependent scan mode. Survey scan (m/z 375–1300) was performed at a resolution of 60,000 followed by MS2 scans to fragment the 50 most abundant precursors with collision-induced dissociation. The activation time was set at 30 ms, the isolation width was 1.5 amu, the normalized activation energy was 35%, and the activation q was 0.25. Mass spectrometry raw data were searched by Proteome Discovery version 1.3 using MASCOT search engine with a percolator against mouse ref-sequence protein database. The mass tolerance was set to be 20 ppm for precursor and 0.5 Da for production. The identification data were filtered to a 1% false discovery rate (FDR) on both peptide and protein levels using a target-decoy strategy. The intensity of identified peptides was represented by the peak area of the extracted ion chromatogram of their monoisotopic peaks. Protein abundances were estimated by iBAQ algorithm.
Behavioral tests
Behavioral tests were performed with adult male mice. Apparatus was cleaned with diluted sodium hypochlorite solution before the test of each animal. All data were recorded by an experimenter who was blinded to the mice genotypes.
Wire-hang test
Mice were placed individually on a wire-mesh grid (10 × 10 cm) and then were inverted to induce animals to grip the wire. The latency of falling was recorded within 60 s.
Elevated plus-maze test
An elevated plus-maze apparatus was used to assess the anxiety of mice. The apparatus consisted of two open arms and two enclosed arms of the same sizes (25 × 5 × 15 cm). The arms and central square were made of white plastic plates and were elevated to a height of 55 cm above the floor. Arms of the same type were arranged on opposite sides. Each animal was placed in the central square of the maze (5 × 5 cm) facing one of the closed arms, and was recorded over 10 min. The total distance, the time and distance spent in the open arms and the number of entries into the open arms were measured with the ViewPoint software.
Open-field test
Each animal was placed at the center of an open-field apparatus (48 × 48 × 40 cm). Total traveled distance, distance traveled and time spent in the central area (24 × 24 cm) were recorded over 10 min, respectively. Data acquisition for mice locomotion was performed and analysis was performed automatically with the ViewPoint software.
Rotarod test
On the first day, each animal was placed on a constantly rotating drum (20 rpm) for up to 5 min. From the next day, mice were placed on an accelerating rotating (from 4 to 40 rpm in 5 min) drum for three times per day during four constant days. The average latency to fall in three trials at each day was recorded.
Balance-beam test
Each mouse was placed at one end of a straight beam (100 × 1.5 cm), which is 50 cm above the floor. On the first day, mice were trained to cross the beam for three times. On the second day, the time to pass 90 cm of the beam and the percentage of steps with hind paw slips on the beam were recorded manually (Rexach et al., 2012).
Y-maze test
Y-maze test was performed as described previously (Lainiola et al., 2014). A Y-maze consists of three arms (each arm: 50 × 9 × 25 cm) connected with a triangular central platform. Each arm contained an opaque removable door at the connection and a certain pattern symbol at the end. Y-maze test consisted of a training phase and a testing phase. Each animal was placed at the distal end of the start arm and allowed to explore freely for 5 min with the novel arm closed. After 2 h of the training phase, each mouse was placed for another 5 min at the distal end of the start arm with all three arms opened. All data were videotaped and analyzed with the infrared tracking software (ViewPoint, Life Sciences).
Morris water maze test
Morris water maze (MWM) test was conducted as described previously (J. Chen et al., 2021). MWM test was performed in a round, opaque water-filled (36 cm in depth) pool (120 cm in diameter) with a detachable platform (10 cm in diameter, submerged 2 cm below the water surface). Maze was made opaque by covering its surface with food-grade titanium dioxide and divided into four virtual quadrants (NE, NW, SE, SW). MWM test consists of a training phase and a testing phase. Each mouse was trained four times per day for four constant days, with an interval of 15 min between each training. The order of entry into each quadrant was pseudo-randomized. In each trial, mice were allowed to find the hidden platform within 60 s or gently guided to the platform if failed, and allowed to rest on the platform for 10 s. The latency to find the platform was recorded. During the probe trial phase (day 5), the platform was removed from the pool to measure any spatial bias toward the original platform location. Each mouse was placed in the water at the quadrant opposite the platform and allowed to swim freely for 60 s. All trials were monitored and analyzed with MazeScan software (Acimetrics).
Transmission electron microscopy (TEM) assay
Brain samples were dissected out after perfusion, and sagittal slices (1 mm) were prepared from the cerebellar vermis and rectangular molecular layer of lobules V and VI were separated. Cerebellar sections were stored in fixative solution (0.1 m phosphate buffer with 2.5% glutaraldehyde) at 4°C before further processing. After three-time washes with 0.1 m PBS (10 min for each time), samples were postfixed with 1% OsO4 for 1.5 h. Samples were washed for three times with purified water and stained with 2% uranyl acetate for 30 min. Samples were then subjected to gradient dehydration: 50%, 70%, 90% ethanol for 10 min, 100% ethanol for 15 min, 100% acetone for 15 min, respectively. Cerebellar tissues were embedded in epoxy resin with analar acetone (volume ratio = 3:1) at room temperature overnight, then transferred to pure epoxy resin and embedded at 37°C. Ultrathin sections (100 nm) were cut using an ultra-microtome (Leica UC7), stained with Lead Citrate solution, and mounted onto grids. Images were captured at 18,500× and 30,000× magnification using a TEM (FEI), respectively. Parallel fiber-Purkinje cell synapses were identified by asymmetrical and short contacts at the outside of molecular layers (Ichikawa et al., 2016a, b; X.T. Wang et al., 2020). ImageJ was used to quantify the numbers of synapse and presynaptic vesicles.
RNA-seq analysis
Total RNA of CGCs from P7 Ctrl and cKO mice pups were extracted for RNA-seq analysis, respectively. All samples used for the cDNA library were assessed with NanoDrop 2000 (Thermo Fisher Scientific), and the RNA integrity value (RIN) was determined with the RNA Nano 6000 Assay kit of the Bioanalyzer 2100 system (Agilent Technologies Inc.). A total amount of 2 µg of mRNA per sample was used as input for each RNA sample preparation. Sequencing libraries were generated following the protocol of NEBNext UltraTM RNA Library Prep kit for Illumina (NEB). The library fragments were purified with AMPure XP system (Beckman Coulter) to preferentially select cDNA fragments of 250–300 bp. The library was sequenced with an Illumina Hiseq platform (Illumina NovaSeq6000).
Raw reads of fastq format were processed, and clean reads were obtained by removing low-quality reads and adapter. The retained clean reads were aligned to the Mus musculus reference genome mm10. To quantify the gene expression level, feature counts v1.5.0-p3 was used to count the read numbers mapped to each gene. Expected number of fragments per kilobase of transcript sequence per millions base pairs sequenced (FPKM) of each transcript was calculated based on the length of the gene and the count of reads mapped to this gene. Differential expression analysis was performed using the DESeq2 R package (1.20.0). The resulting p-values were adjusted using the Benjamini and Hochberg's approach for controlling the false discovery rate. Genes with an adjusted p-value <0.05 found by DESeq2 and absolute value of fold change >1.5 were assigned as differentially expressed genes (DEGs).
Gene ontology and KEGG analysis
Gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) was performed were performed with the cluster Profiler R package. Each enriched GO functional term and KEGG pathway are represented by a node, and the node size is proportional to the number of genes in its corresponding functional term in the enrichment maps.
Quantification and statistical analysis
All data are expressed as mean ± SEM GraphPad Prism 7.0 (GraphPad Software Inc.) was used for statistical analysis. Unpaired Student's t test was used to assess the differences between two groups; one-way and two-way ANOVA analysis followed by Tukey's post hoc analysis were used to determine differences between multiple groups. p < 0.05 was considered statistic significance. Replicates information is indicated in the figure legends.
Data and code availability
The accession number for the RNA-seq data reported in this paper is SRA: PRJNA884808.
Results
Cerebellum has abundant O-GlcNAcylation and adult Math1-Cre::Ogtloxp/Y mice display no change of ogt and O-GlcNAcylation in cortex and hippocampus
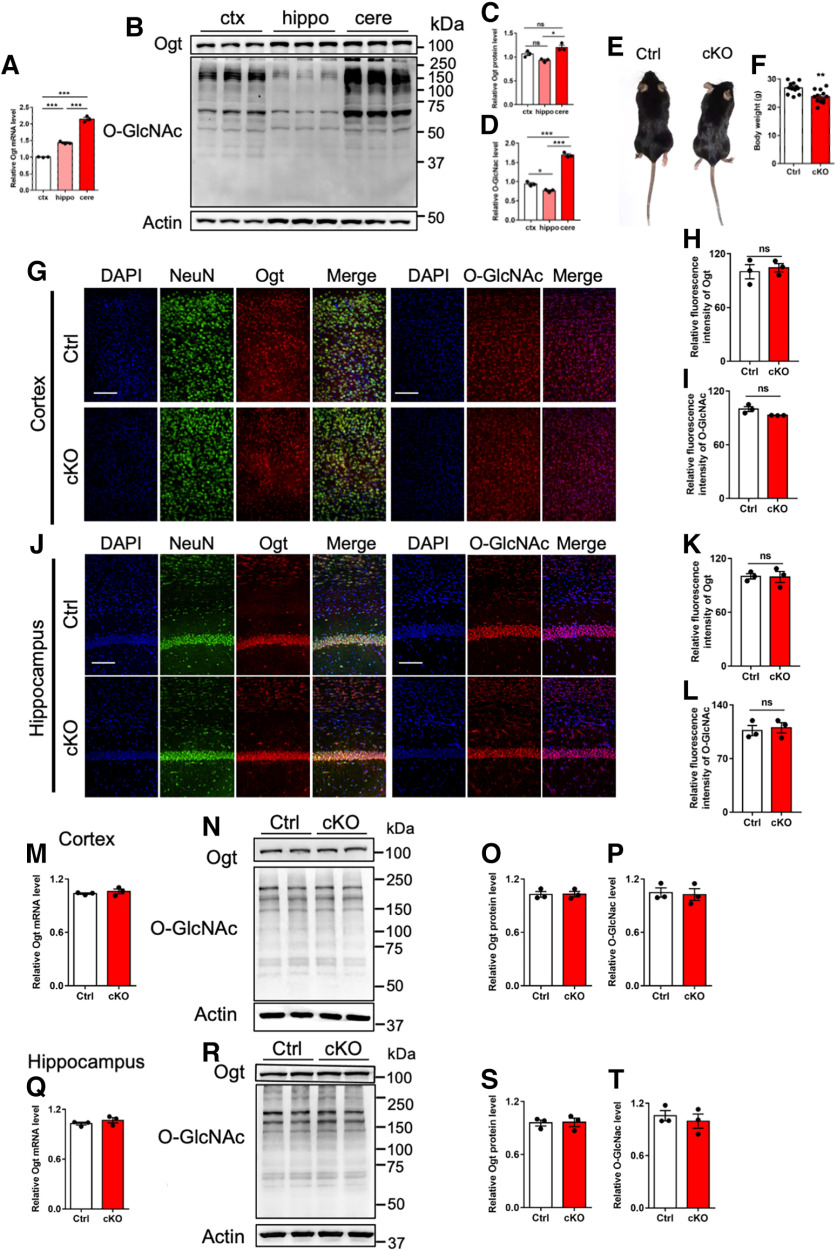
To examine the function of Ogt in cerebellum, we first compared the levels of Ogt and O-GlcNAcylation among cortex, hippocampus, and cerebellum of adult (postnatal eight-week) male mice. qRT-PCR assay results showed that the level of Ogt in cerebellum is higher than that of hippocampus and cortex (Fig. 1A). In addition, Western blot (WB) assay and quantification results showed that cerebellum has the highest levels of Ogt and O-GlcNAcylation relative to hippocampus and cortex (Fig. 1B–D).
Figure 1.
Abundant O-GlcNAcylation in cerebellum and Math1-Cre::Ogtloxp/Y not affecting the levels of Ogt and O-GlcNAcylation in the cortex and hippocampus of adult mice. A, qRT-PCR assays of Ogt mRNA expression in cortex, hippocampus and cerebellum of eight-week-old (adult) mice, respectively. The primers used can be found in Table 1. B–D, Representative images of Western blot (WB) assays, and quantification of Ogt (C) and O-GlcNAcylation (D) levels in cortex, hippocampus and cerebellum of adult mice. n = 3 independent experiments for each group. Data are presented as mean ± SEM. One-way ANOVA analysis followed by Tukey's post hoc analysis, *p < 0.05, **p < 0.001, ***p < 0.0001. E, Gross appearance of adult Ctrl (Ogtloxp/Y) and cKO (Math1-Cre::Ogtloxp/Y) mice. F, Body weight of adult Ctrl and cKO mice. n = 12 mice per genotype. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. G–I, Immunostaining (G) and quantification of the relative fluorescence intensity of Ogt (H) and O-GlcNAcylation (I) in the cortex of adult Ctrl and cKO mice. n = 3 mice for each group, and 3 sections from each animal. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001, ns, not significant. Scale bar, 100 μm. J–L, Immunostaining (J) and quantification of the relative fluorescence intensity of Ogt (K) and O-GlcNAcylation (L) in the hippocampus of adult Ctrl and cKO mice. n = 3 mice for each group, and 3 sections from each animal. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001, ns, not significant. Scale bar, 100 μm. M, qRT-PCR assay of Ogt mRNA expression in the cortex of adult Ctrl and cKO mice. n = 3 independent experiments for each group. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. N–P, WB assay (N) and quantification results of Ogt (O) and O-GlcNAcylation (P) levels in the cortex of adult Ctrl and cKO mice. n = 3 independent experiments for each group. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. Q, qRT-PCR assay of Ogt mRNA expression in the cortex of adult Ctrl and cKO mice. n = 3 independent experiments for each group. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. R–T, WB assay (R) and quantification results of Ogt (S) and O-GlcNAcylation (T) levels in the cortex of adult Ctrl and cKO mice. n = 3 independent experiments for each group. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001.
Given that cerebellar granule cells (CGCs) are the major type of cells in the cerebellum, we speculated that Ogt and O-GlcNAcylation could play important roles in CGCs. We then generated Math1-Cre::Ogtloxp/Y mouse (cKO) to specifically delete Ogt in Math1+ granule neuron precursors (GNPs). Compared with control (Ctrl) littermates, adult Math1-Cre::Ogtloxp/Y mice displayed decreased body weight (Fig. 1E,F). Immunofluorescence staining showed no difference of the signal intensities of Ogt and O-GlcNAcylation in cortex (Fig. 1G–I) and hippocampus (Fig. 1J–L) between adult Ctrl and cKO mice. In addition, qRT-PCR and WB assays showed no change of Ogt and O-GlcNAcylation levels in cortex (Fig. 1M–P) and hippocampus (Fig. 1Q–T) between adult Ctrl and cKO mice. These results suggested that cerebellum has the highest level of O-GlcNAcylation compared with cortex and hippocampus, and Ogt deficiency in Math1+ GNPs did not affect the levels of Ogt and O-GlcNAcylation in the cortex and hippocampus of adult mice.
Ogt specific deficiency in granule neuron precursors leads to abnormal cerebellar cytoarchitecture
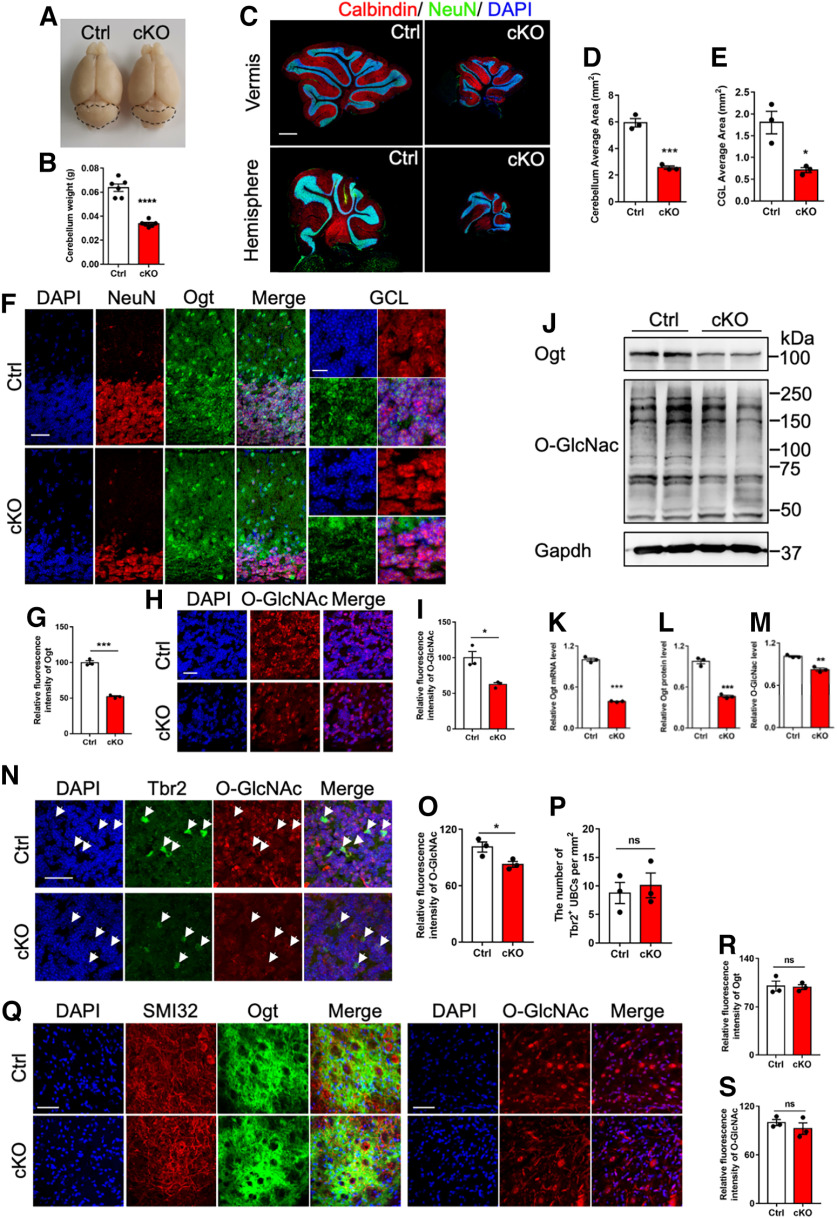
Next, we examined the effects of Ogt deficiency in Math1+ GNPs on the cerebellum. At adult stage, we observed that the overall size and weight of cerebellum were both significantly decreased in cKO mice (Fig. 2A,B). Immunofluorescence staining showed the significantly decreased area of whole cerebellum and granule cell layer (GCL) in cKO mice compared with Ctrl mice (Fig. 2C–E). The immunostaining signal intensities of Ogt (Fig. 2F,G) and O-GlcNAcylation (Fig. 2H,I) also significantly reduced in GCL area of cerebellum of cKO mice (Fig. 2E,F). qRT-PCR and WB assays showed that the levels of Ogt and O-GlcNAcylation were significantly decreased in the cerebellum of cKO mice compared with Ctrl mice (Fig. 2J–M). Of note, immunofluorescence signal intensities of O-GlcNAcylation also decreased in the unipolar brush cells (UBCs) of adult cKO mice (Fig. 2N,O), whereas we did not observe a significant alteration of the density of Tbr2+ UBCs (Fig. 2P). The signal intensities of Ogt and O-GlcNAcylation in deep cerebellar nuclei (DCNs) neurons showed no difference between adult cKO and Ctrl mice (Fig. 2Q–S). These results suggested that cerebellar Ogt deficiency in Math1+ GNPs reduces the level of O-GlcNAcylation and induces abnormal morphology of cerebellum.
Figure 2.
cKO mice display cerebellar atrophy and the reduced Ogt and O-GlcNAcylation. A, Representative brain images of adult Ctrl and cKO mice. The dashed lines outline the area of cerebellum. B, Weight of the cerebellum of adult Ctrl and cKO mice. n = 6 mice per genotype. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. C, Representative images of NeuN and Calbindin immunostaining in the cerebellum of adult Ctrl and cKO mice. Scale bar, 500 μm. D, E, Quantification of the cerebellum area (D) and cerebellar granule cell layer (CGL) area (E) of adult Ctrl and cKO mice. n = 3 mice for each group, and 6 sections from each animal. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. F, Representative images of NeuN and Ogt immunostaining in the cerebellum of adult Ctrl and cKO mice. Scale bars, 50 μm (left, lower magnification) and 10 μm (right, higher magnification). G, Quantification of the fluorescence intensity of Ogt in (F). n = 3 mice for each group, and 3 sections from each animal. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. H, Representative images of O-GlcNAcylation in the granular cell layer (GCL) of cerebellum of adult Ctrl and cKO mice. Scale bar, 20 μm. I, Quantification of the relative fluorescence intensity of O-GlcNAcylation in H. n = 3 mice for each group, and 3 sections from each animal. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. J–L, WB assays (J) and quantification results of Ogt (K) and O-GlcNAcylation (L) levels in the cerebellum of adult Ctrl and cKO mice, respectively. n = 3 independent experiments for each group. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. M, qRT-PCR assay of Ogt mRNA expression in the cerebellum of adult Ctrl and cKO mice. n = 3 independent experiments for each group. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. N–P, Representative images of Tbr2 and Ogt immunostaining (N) and quantification of the fluorescence intensity of Ogt in Tbr2+ cells (O) and the density of Tbr2+ UBCs (P) in the lobule VI of cerebellum of adult Ctrl and cKO mice. n = 3 mice for each group, 3 sections from each animal and 6 visual fields per section. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001, ns, not significant. Scale bar, 20 μm. Tbr2, a marker for unipolar brush cells. Q–S, Representative images of Ogt and SMI32 (left), and O-GlcNAcylation (right; Q) and quantification of the fluorescence intensity of Ogt (R) and O-GlcNAcylation (S) in the deep cerebellum nuclei (DCN) of adult Ctrl and cKO mice. n = 3 mice for each group, and 3 sections from each animal. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001, ns, not significant. SMI32, a marker for deep cerebellum nuclei. Scale bar, 50 μm.
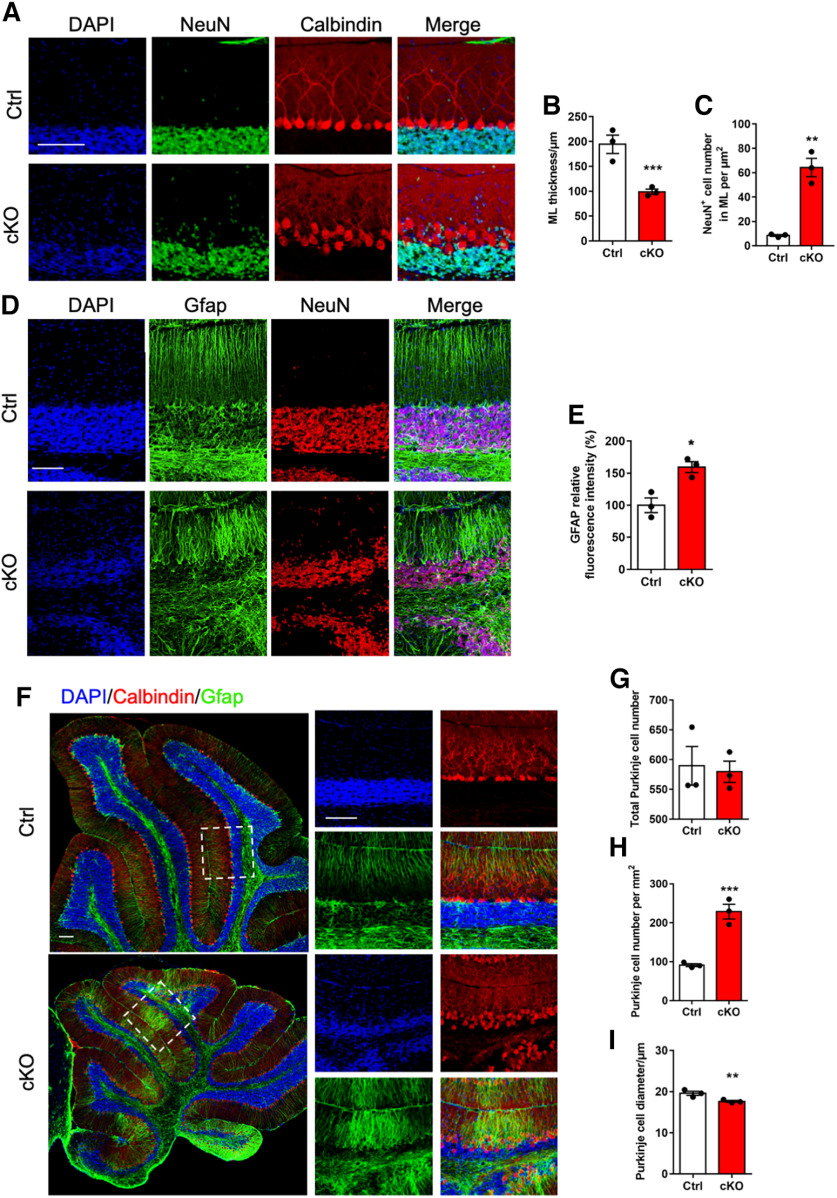
Next, we analyzed the effects of Ogt deficiency on the cytoarchitecture of cerebellum. We observed that adult cKO mice showed the decreased thickness of molecular layer (ML) and the diminished CGCs (Fig. 3A,B). Notably, the number of CGCs aberrantly localized in ML significantly increased in adult cKO mice (Fig. 3A,C). Immunofluorescence staining showed that adult cKO mice displayed the disorganized Bergman glia (BG) scaffold (Fig. 3D,E). In addition, the arrangement of Purkinje cells was also disrupted in the cerebellum of adult cKO mice (Fig. 3F). Although the total number of Calbindin+ Purkinje cells showed no significant difference between adult Ctrl and cKO mice (Fig. 3G), the density of Purkinje cells was remarkably increased (Fig. 3H), and the diameter of Purkinje cells was decreased in adult cKO mice (Fig. 3I). Collectively, these results suggested that Ogt deficiency leads to abnormal cerebellar cytoarchitecture of adult mice.
Figure 3.
Adult Ogt cKO mice exhibit the disorganized glia and Purkinje cells in the cerebellum. A, Representative images of NeuN and Calbindin immunostaining in the lobule VI of cerebellum of adult Ctrl and cKO mice. Scale bar, 100 μm. B, C, Quantification of the thickness of molecular layer (ML; B) and the number of NeuN+ cell in ML (C) of cerebellum in adult Ctrl and cKO mice, respectively. n = 3 mice for each group, and 3 sections from each animal. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. D, E, Representative images of NeuN and Gfap immunostaining (D) quantification of the relative fluorescence intensity of Gfap (E) in the lobule VI of cerebellum of adult Ctrl and cKO mice. n = 3 mice for each group, and 3 sections from each animal. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. Scale bar, 100 μm. F, Representative images of Gfap and Calbindin immunostaining in the lobule VI of cerebellum of adult Ctrl and cKO mice. Scale bar, 100 μm. G–I, Quantification of total number (G), density (H) and diameter (I) of Calbindin+ Purkinje cells (PCs) in the cerebellum of adult Ctrl and cKO mice. n = 3 mice for each group, and 6 sections from each animal. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001.
Ogt deficiency induces abnormal synaptic connection and formation in the cerebellum of adult mice
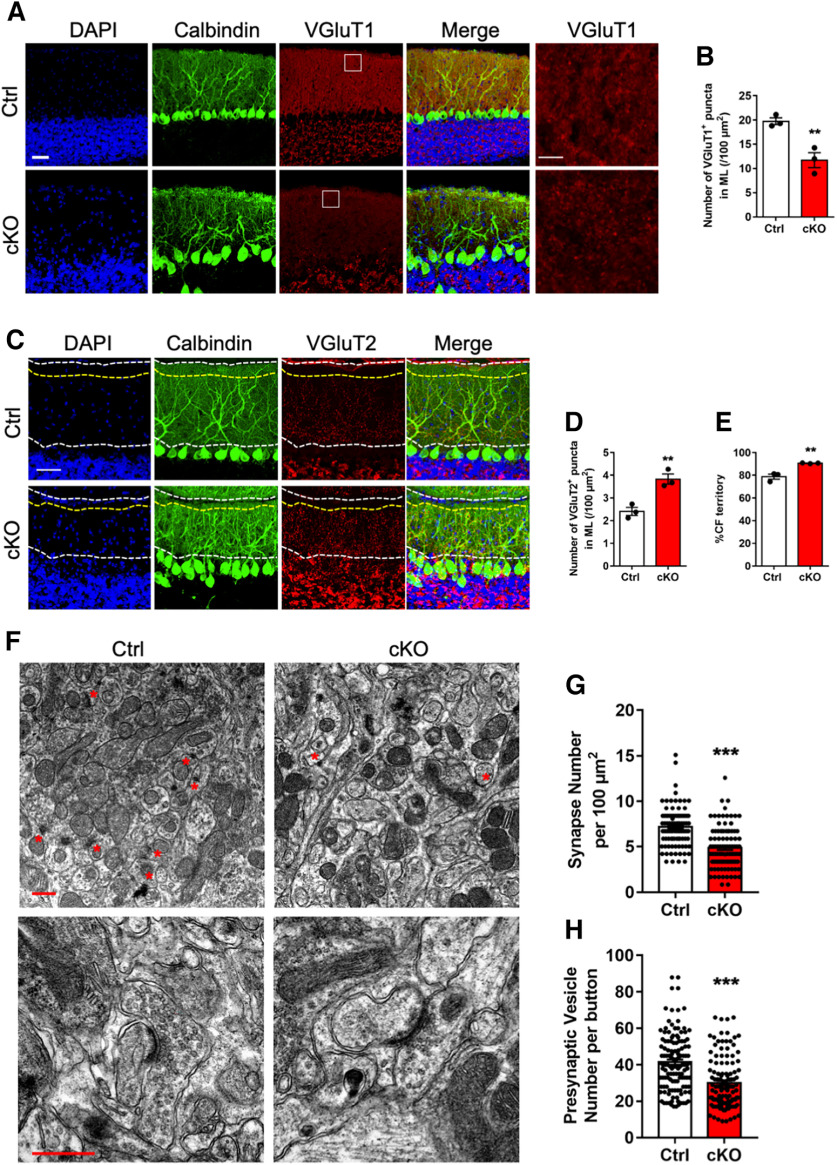
In cerebellum, Purkinje cells receive excitatory inputs primarily from parallel fibers from CGCs and climbing fibers from the inferior olive. Parallel fibers and climbing fibers were marked by the expression of VGluT1 and VGluT2, respectively. In adult cKO mice, we observed that the puncta density of VGluT1 at molecular layer was significantly decreased (Fig. 4A,B). Meanwhile, cKO mice displayed the increased VGluT2+ puncta density and territory of Purkinje cells (Fig. 4C–E). These results suggested that Ogt deficiency induces abnormal synaptic connection in the cerebellum of adult cKO mice.
Figure 4.
Adult Ogt cKO mice show abnormal synaptogenesis in cerebellum. A, B, Representative images of Calbindin and VGluT1 immunostaining (A) and quantification of the number of VGluT1-positive puncta (B) in the lobule VI of cerebellum of adult Ctrl and cKO mice. Left, Lower magnification. Right, Higher magnification. n = 3 mice for each group, 3 sections from each animal and 6 visual fields per section. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. Scale bar, 50 μm. C–E, Representative images of Calbindin and VGluT2 immunostaining (C) and quantification of the number of VGluT2-positive puncta (D) and the proportion of CF territory in the molecular layer (E) in the lobule VI of cerebellum of adult Ctrl and cKO mice. n = 3 mice for each group, and 3 sections from each animal. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. Scale bar, 50 μm. CF, climbing fibers. F, Representative transmission electron microscope images showing PF-PC synapses (top, lower magnification) and PF-PC synapses (bottom, higher magnification) in the molecular layer of lobules V and VI of adult Ctrl and cKO cerebellum. Red asterisks marked PF-PC synapses. Scale bar, 500 nm. PF, prallel fibers. PC, Purkinje cells. G, Quantification of the average number of synapses in F per 100 μm2 in the molecular layer of lobules V and VI of cerebellum. n = 97 and n = 98 visual fields from 4 Ctrl/cKO mice, respectively. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. H, Quantification of the average number of presynaptic vesicles per synapse in F. n = 129 and n = 121 synapses were analyzed in the molecular layer of lobules V and VI of the cerebellum from 4 Ctrl/cKO mice, respectively. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001.
Next, using a transmission electron microscope (TEM), we analyzed the parallel fiber-Purkinje cell synapses in the cerebellum of adult Ctrl and cKO mice (Fig. 4F). The density of parallel fiber-Purkinje cell synapses was significantly decreased in cKO mice (4.871 ± 0.2324, n = 98) compared with Ctrl mice (7.175 ± 0.217, n = 97; Fig. 4G). The total number of presynaptic vesicles was also significantly decreased in cKO mice (29.94 ± 1.827, n = 121) compared with Ctrl mice (41.44 ± 1.868, n = 129; Fig. 4H). These results suggested that Ogt deficiency induces abnormal synaptic formation in the cerebellum of adult cKO mice.
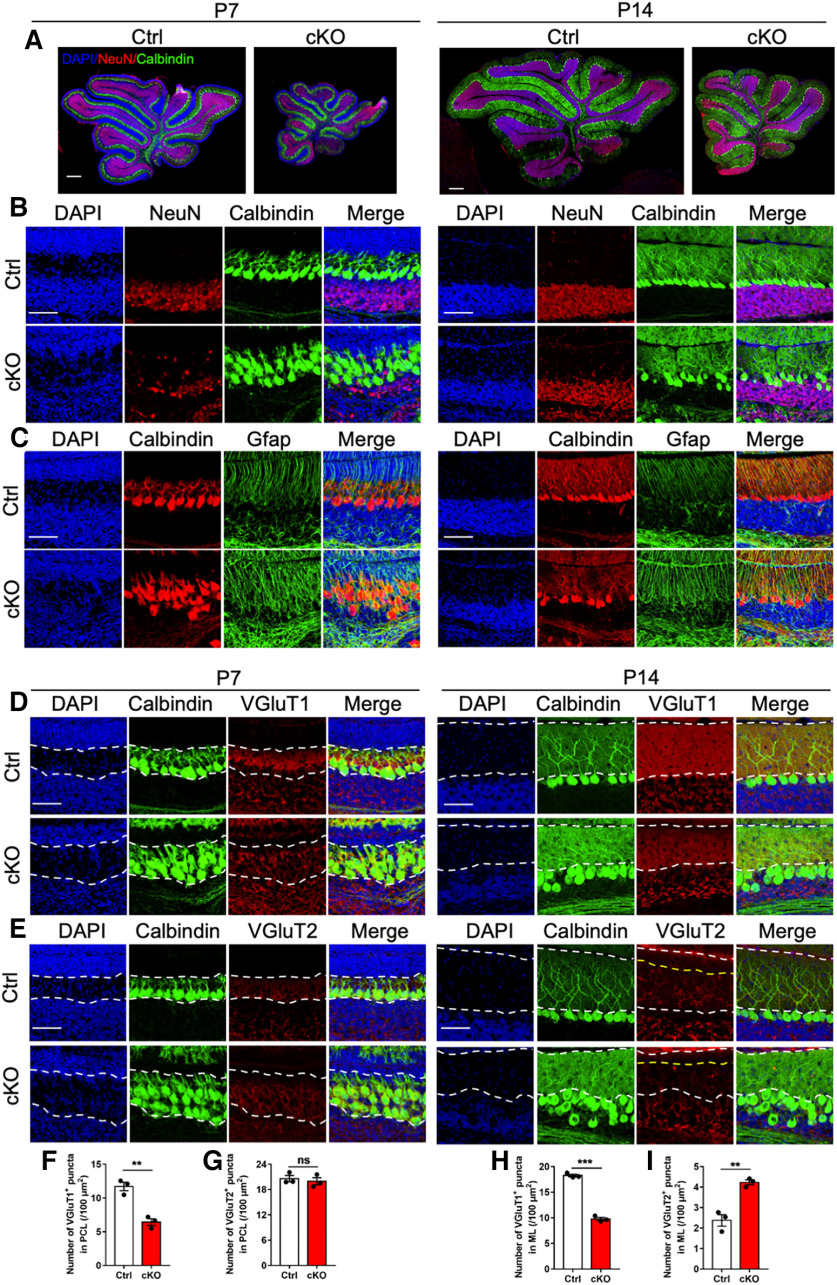
Given adult cKO mice displayed abnormal architecture and synaptic connection, we aim to examine when these abnormities appear. We collected cerebella from postnatal day 7 (P7) and day 17 (P14) Ctrl and cKO mice, respectively. Immunofluorescence staining showed that cKO mice already exhibited the disorganized Bergman glia and Purkinje cells as early as P7 (Fig. 5A–C). At P7, cKO mice showed the reduced number of VGluT1 puncta, whereas the number of VGluT2 puncta showed no difference (Fig. 5D–G). At P14 time point, cKO mice showed the decreased numbers of VGluT1, but the increased VGluT2 puncta (Fig. 5D,E,H,I). These results suggested that cKO mice have the aberrant architecture and synaptic connection during developmental stage.
Figure 5.
cKO mice show abnormal cytoarchitecture and synaptogenesis at early postnatal stage. A, Representative images of NeuN and Calbindin immunostaining in the whole cerebellum of Ctrl and cKO mice at postnatal day 7 (P7) and P14, respectively. Scale bar, 200 μm. B, Representative images of NeuN and Calbindin immunostaining in the lobule VI of cerebellum of Ctrl and cKO mice at P7 (left) and P14 (right), respectively. Scale bar, 50 μm. C, Representative images of Calbindin and Gfap immunostaining in the lobule VI of cerebellum of Ctrl and cKO mice at P7 (left) and P14 (right), respectively. Scale bar, 50 μm. D, Representative images of Calbindin and VGluT1 immunostaining in the lobule VI of cerebellum of Ctrl and cKO mice at P7 (left) and P14 (right), respectively. Scale bar, 50 μm. E, Representative images of Calbindin and VGluT2 immunostaining in the lobule VI of cerebellum of Ctrl and cKO mice at P7 (left) and P14 (right), respectively. Scale bar, 50 μm. F, G, Quantification of the numbers of VGluT1-positive (F) and VGluT2-positive (G) puncta in the Purkinje cell layer (PCL) of P7 mice. n = 3 mice per genotype, and 3 sections from each animal. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001, ns, not significant. H, I, Quantification of the number of VGluT1-positive puncta (H) and VGluT2-positive puncta (I) in the molecular layer (ML) of P14 mice. n = 3 mice for each group, and 3 sections from each animal. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001.
Adult cKO mice display multiple behavioral deficits
Given adult cKO mice displayed abnormal architecture and synaptic connection, we next examined whether Ogt deficiency impaired the behavioral performance of adult mice. Wire-hang test (Fig. 6A), elevated plus maze test (Fig. 6B–F) and open-field test (Fig. 6G–L) showed no difference between adult Ctrl and cKO mice. Rotarod test showed that cKO mice had the shorter latency to fall compared with Ctrl mice (Fig. 6M,N). Balance-beam test showed that cKO mice had longer time to pass the balance beam and a higher number of hind-paw slips (Fig. 6O,P). These data indicated the impairment in motor coordination and balance in cKO mice.
Figure 6.
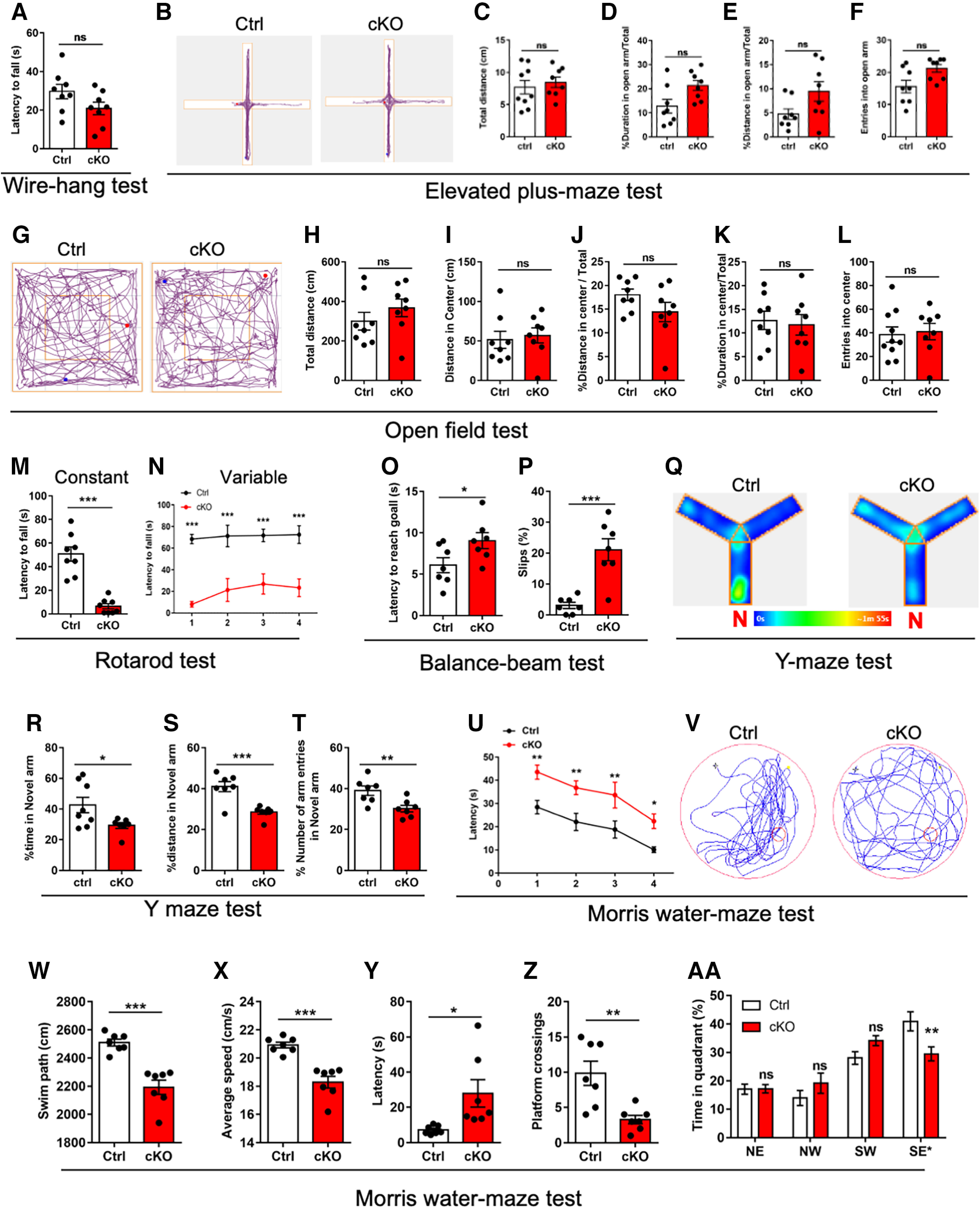
Adult cKO mice exhibit abnormal motor coordination, and learning and memory abilities. A, Latency to falling in the wire-hang test for adult Ctrl and cKO mice. n = 8 mice per genotype. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. B–F, Representative images of traveled path (B), total traveled distance (C), percentage of duration (D) and distance in open arm (E), and entries into open arm (F) in the elevated plus-maze test for adult Ctrl and cKO mice. n = 8 mice per genotype. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. G–L, Representative images of traveled path (G), total traveled distance (H), traveled distance in center (I), percent of traveled distance in center relative to total traveled distance (J), and percent duration in center (K), and entries into central area (L) in the open-field test for adult Ctrl and cKO mice. n = 8 mice per genotype. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. M, N, Latency to falling in the constant (M) and variable (N) rotarod test for adult Ctrl and cKO mice. n = 8 mice per genotype. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. O, P, The latency to reach the goal (O) and the percentages of hind-paw slips during passing the beam (P) in the balance-beam test. n = 7 mice per genotype. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. Q, Representative images of traveled path for the Y-maze test of adult Ctrl and cKO mice. N, the novel arm. R–T, Time spent (R), percentage of traveled distance in total traveled distance (S), number of entries (T) into the novel arm of Y-maze test for adult Ctrl and cKO mice. n = 7 mice per genotype. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. U, Escape latency during the training phase for Morris water maze (MWM) test of adult Ctrl and cKO mice. n = 7 mice per genotype. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. V, Representative images of the swimming path of adult Ctrl and cKO mice during the test phase in MWM test. W–Z, Swimming path (W), average speed (X), latency to platform (Y) and platform-crossing times (Z) of adult Ctrl and cKO mice during the test phase of MMW test. n = 7 mice per genotype. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. AA, Percentage of time spending in four quadrants of adult Ctrl and cKO mice during the test phase of MMW test. n = 7 mice per genotype. Data are presented as mean ± SEM. Two-way ANOVA analysis followed by Tukey's post hoc analysis, *p < 0.05, **p < 0.001, ***p < 0.0001, ns, not significant.
We further performed Y-maze and Morris water maze tests to detect the short-term and long-term memories in cKO mice. Y-maze test showed that the spent time, the traveled distance and the number of entries in the novel arm were significantly decreased in cKO mice compared with Ctrl mice (Fig. 6Q–T). Morris water maze test showed that cKO mice required longer time to reach the platform during the training phase (Fig. 6U). During the probe trial phase, cKO mice displayed the reduced swimming path and speed compared with Ctrl mice (Fig. 6V–X). In addition, cKO mice exhibited longer latency, decreased numbers of crossing the platform and decreased time in the target quadrant (Fig. 6Y–AA). These results collectively suggested that cerebellar Ogt deficiency results in the behavioral deficits of adult mice.
Ogt deficiency alters neuronal gene expression of cerebellum
To reveal the mechanism how Ogt regulates the cerebellar function and mice behavior, we performed RNA-seq with the cultured CGCs from Ctrl and cKO mice. qRT-PCR results showed the significant decrease of Ogt in cKO CGCs (Fig. 7A). RNA-seq data analysis identified 1925 differentially expressed genes (DEGs; Extended Data Table 7-1): 634 upregulated genes and 1291 downregulated genes (fold change > 1.5, p-values < 0.05; Fig. 7B). Among the genes specifically expressed in CGCs (Rosenberg et al., 2018), 50 genes were downregulated revealed by RNA-seq analysis (Fig. 7C). Gene ontology (GO) analysis revealed that upregulated genes were enriched for terms including mitochondrion organization and ribose phosphate metabolic process, and downregulated genes were enriched for terms including synapse organization and axon development (Fig. 7D,E). Enrichment analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways revealed that upregulated DEGs were enriched in the pathways related to neurodegeneration diseases including Alzheimer's disease, Parkinson's disease, and downregulated DEGs were enriched in the pathways of cGMP-PKG signaling pathway (Fig. 7F,G). In addition, Gene Set Enrichment Analysis (GSEA) analysis revealed that the expression of genes related to axonogenesis, synapse organization and Rho protein signal transduction were significantly decreased in cKO CGCs (Fig. 7H–J). The expression of some genes revealed by GSEA was determined by qRT-PCR assay (Fig. 7K–M).
Figure 7.
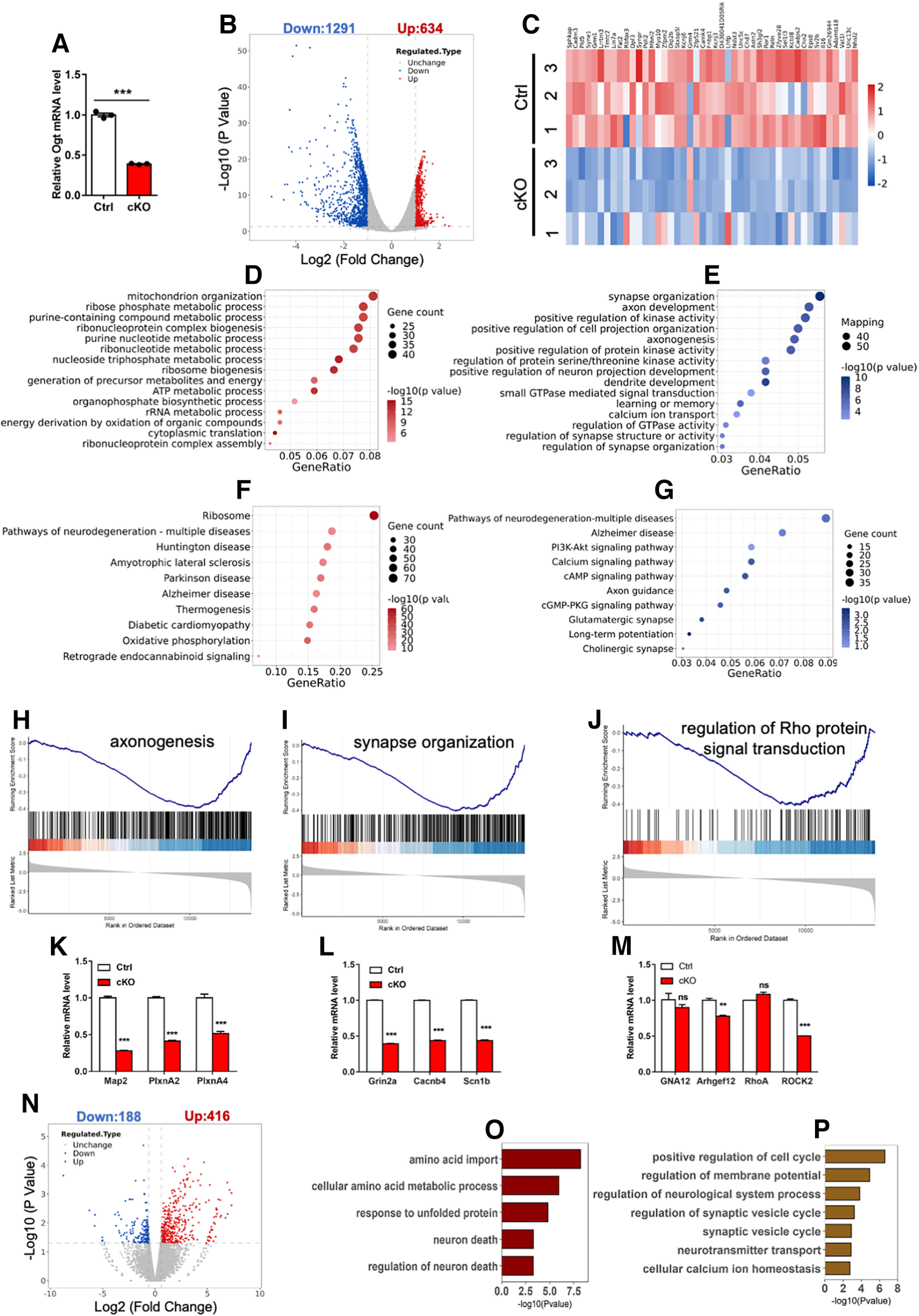
Ogt deficiency alters gene expression in CGCs. A, qRT-PCR analysis of Ogt mRNA expression in Ctrl and cKO CGCs. n = 3 independent experiments for each group. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. B, Volcano plot for the differentially expressed genes in cKO CGCs compared with Ctrl. The dashed line indicates the Log2(fold change) = 1.5 and p = 0.05. Three biological repeating samples of Ctrl and cKO CGCs were adopted for sequencing. The list of DEGs can be found in Extended Data Table 7-1. C, Heatmap for the Log2 fold changes of 50 CGC-specific genes in Ctrl and cKO CGCs. n = 3 biological repeats for each group. D, E, GO analysis of the upregulated (D) and downregulated (E) for biological process in cKO CGCs. F, G, KEGG analysis of the upregulated (F) and downregulated (G) genes in cKO CGCs. H–J, GSEA plots of differentially expressed genes (DEGs) among genes related to axonogenesis (H), synapse organization (I) and Rho protein signal transduction (J) in cKO CGCs. K–M, qRT-PCR analysis of the mRNA expression of axonogenesis (K), synapse organization (L) and Rho protein signal transduction (M)-related genes in Ctrl and cKO CGCs. n = 3 independent experiments for each group. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001, ns, not significant. N, Volcano plot for the differentially expressed genes in Ogt acute knock-down CGCs compared with Ctrl CGCs. The dashed line indicates the Log2(fold change) = 1.5 and p = 0.05. Two biological repeating samples of Ctrl and cKO CGCs were adopted for sequencing. O, P, GO analysis of the upregulated (O) and downregulated (P) genes in Ogt knock-down CGCs.
The list of differentially expressed genes in cKO CGGs. RNA-seq data analysis identified 1925 differentially expressed genes in cKO CGGs. Download Table 7-1, XLSX file (339.6KB, xlsx) .
Next, we performed RNA-seq with the cultured Ctrl CGCs infected with lentivirus expressing scramble and shOgt, respectively. RNA-seq data analysis identified 604 differentially expressed genes (DEGs): 416 upregulated and 188 downregulated in Ogt knock-down (KD) CGCs (Fig. 7N). The upregulated DEGs were enriched for GO terms including neuronal death, and the downregulated DEGs were enriched for GO terms including axon guidance and synaptic vesicle cycle (Fig. 7O,P). Collectively, these results suggest that Ogt deficiency altered the expression of neuronal genes in CGCs.
The reduced O-GlcNAcylation of Gα12 inhibits RhoA/ROCK signaling pathway
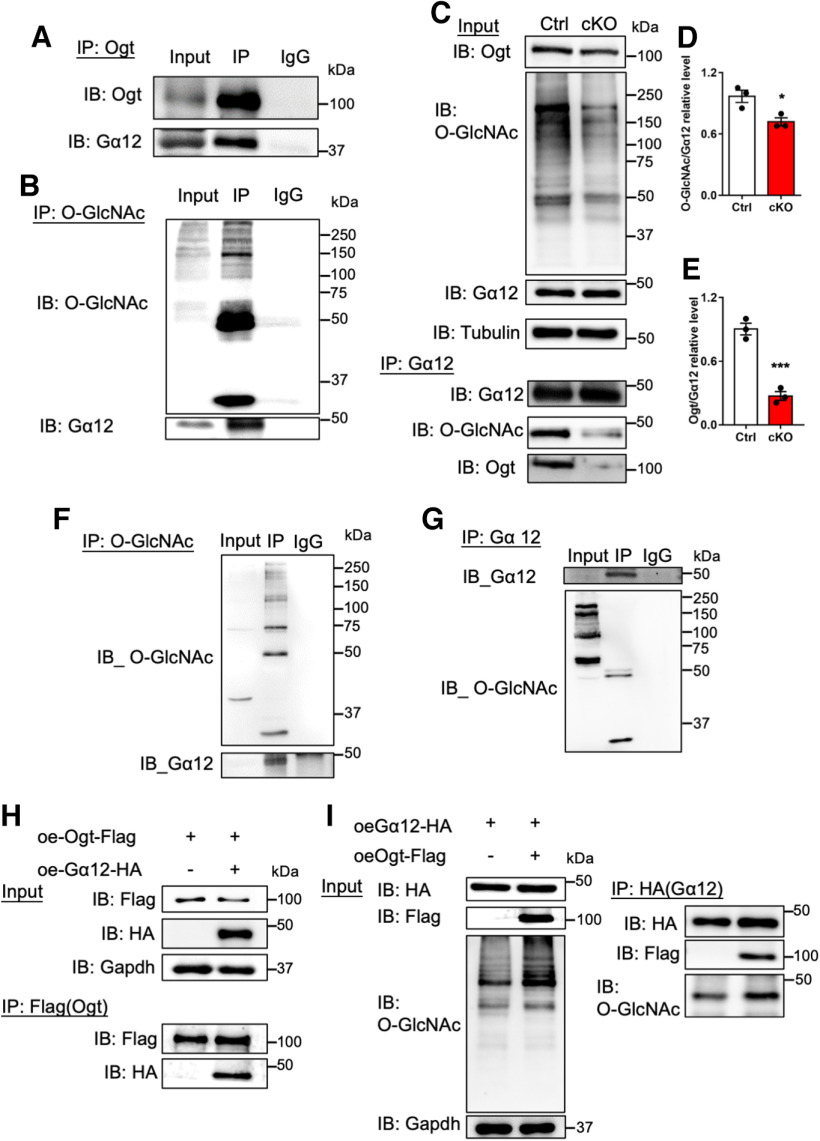
To reveal the proteins modified with O-GlcNAcylation in CGCs, we performed Click-IT assay with wild-type CGCs and identified 57 proteins with O-GlcNAcylation including G-protein subunit α 12 (Gα12), an up-stream regulator of RhoA/ROCK signaling pathway (Extended Data Table 8-1). RhoA/ROCK is a critical component of cGMP-PKG signaling pathway, which is important for synaptic development and cerebellar function (Gulisano et al., 2019; Niewiadomska-Cimicka et al., 2021). Immunoprecipitation (IP) followed by WB assays with antibodies against Ogt and O-GlcNAcylation could precipitate Gα12 in the cerebellum of adult mice, respectively, suggesting that Ogt directly interacted with Gα12 and catalyzed O-GlcNAcylation of Gα12 (Fig. 8A,B). Consistently, Gα12 IP followed by WB assays showed that the level of O-GlcNAcylation on Gα12 significantly decreased in the cerebellum of adult cKO mice compared with Ctrl (Fig. 8C–E). IP-WB assays validated O-GlcNAcylation of Gα12 in N2a cells (Fig. 8F,G). Ectopic expression of Ogt (Flag) and Gα12 (HA) followed by Co-IP further confirmed the physical interaction of Ogt and Gα12, and O-GlcNAcylation of Gα12 in N2a cells (Fig. 8H,I). These results together suggest that Ogt physically interacts with Gα12 and catalyzes O-GlcNAcylation of Gα12.
Figure 8.
Ogt interacts with Gα12 and catalyzes the O-GlcNAc modification on Gα12 in cerebellum. A, B, Immunoprecipitation (IP) followed by WB (IP-WB) assays with Ogt (A) and O-GlcNAcylation (B) antibodies, respectively, showed that Ogt precipitated Gα12 and Gα12 had O-GlcNAcylation in the cerebellum of adult Ctrl mice. The list of O-GlcNAcylation modified proteins can be found in Extended Data Table 8-1. C–E, IP-WB assays showed the significantly reduced level of Ogt bound to Gα12 (C, D) and the remarkable decrease of O-GlcNAcylation level on Gα12 (C, E). n = 3 independent experiments for each group. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. F, G, IP-WB with O-GlcNAcylation antibody precipitated Gα12 (F), and vice versa (G) in N2a cells. H, I, IP-WB assays showed that exogenous Ogt (Flag) precipitated Gα12 (H) and vice versa (I) in N2a cells. Ectopic Ogt (Flag) remarkably increased the signal intensities of total O-GlcNAcylation and O-GlcNAcylation on Gα12.
The list of proteins modified with O-GlcNAcylation in CGCs. Click-IT assay followed by Mass spectrometry assay identified 57 proteins with O-GlcNAcylation in wild-type CGCs. Download Table 8-1, XLSX file (14KB, xlsx) .
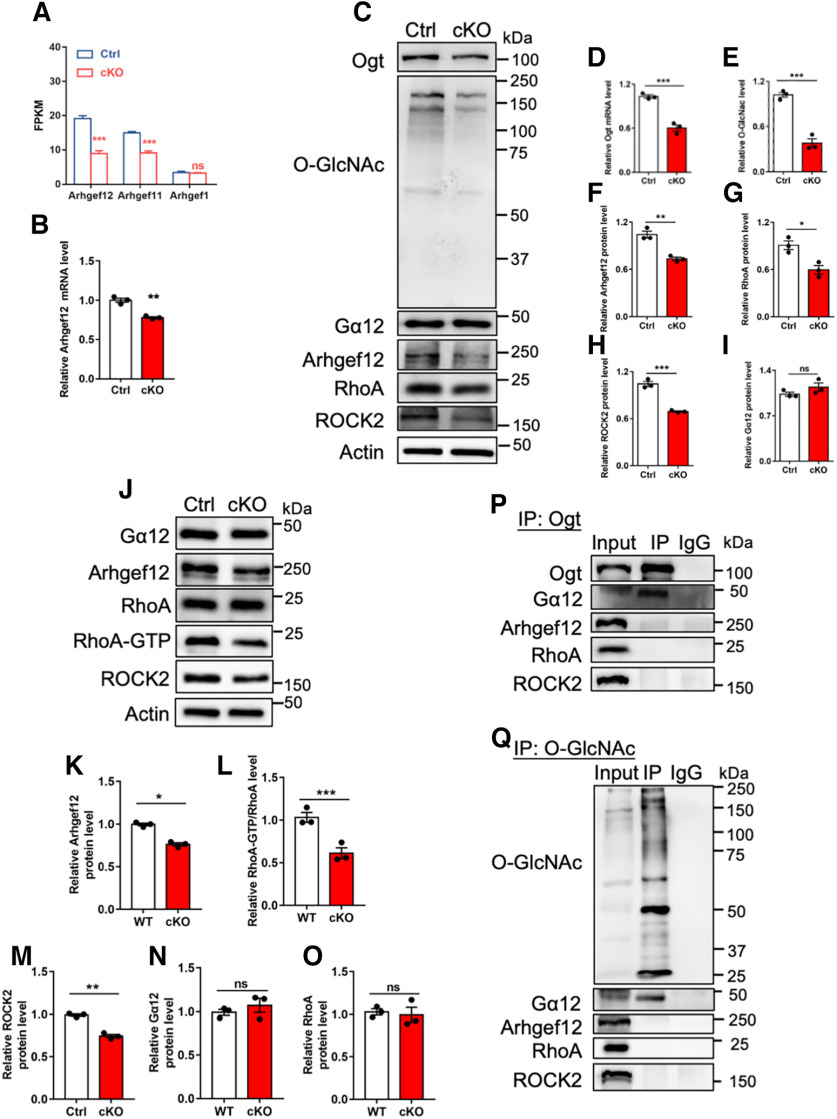
Previous studies have shown that Gα12 can interact with Rho-specific guanine nucleotide exchange factor (GEF) and activate RhoA/ROCK pathway (M.J. Hart et al., 1998; Ross and Wilkie, 2000; Chikumi et al., 2002; Suzuki et al., 2003). Next, we aim to examine whether the decreased O-GlcNAcylation of Gα12 affected RhoA/ROCK pathway. RNA-seq data analysis and qRT-PCR results showed that the expression of Arhgef12, a Rho-specific GEF, remarkably decreased in the cerebellum of cKO mice (Fig. 9A,B). cKO CGCs showed the reduced levels of Ogt, O-GlcNAcylation, Arhgef12, RhoA and ROCK2 compared with Ctrl cells, whereas the level of Gα12 was not affected (Fig. 9C–I). WB assays and quantification results with cerebellar tissues showed that the levels of Arhgef12, ROCK2 and the activation of RhoA (RhoA-GTP/RhoA) significantly decreased in cKO mice compared with Ctrl mice, but the levels of Gα12 and RhoA were not affected (Fig. 9J–O). These results suggested that RhoA/ROCK signaling was repressed under Ogt deficient condition in vitro and in vivo.
Figure 9.
Ogt depletion inhibits RhoA/ROCK signaling activation through regulating Gα12. A, FPKM values of Arhgef12, Arhgef11 and Arhgef1 in Ctrl and cKO CGCs. B, qRT-PCR assay results of Arhgef12 in the cerebellum of adult Ctrl and cKO mice. n = 3 independent experiments for each group. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001, ns, not significant. C–I, Representative images of WB assays (C) and quantification results showed the reduced levels of Ogt (D), O-GlcNAcylation (E), Arhgef12 (F), RhoA (G), and ROCK2(H) while no difference of Gα12 (I) in Ctrl and cKO CGCs. n = 3 independent experiments for each group. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001, ns, not significant. J–O, Representative images of WB assays (J) and quantification results showed the significant decrease of Arhgef12 (K), RhoA-GTP (L) and ROCK2 (M) while no difference of Gα12 (N) and RhoA (O) in the cerebellum of adult cKO mice compared with Ctrl. n = 3 independent experiments for each group. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001, ns, not significant. P, IP-WB assays showed that endogenous Ogt directly interacted with endogenous Gα12, but not Arhgef12, RhoA and ROCK2 in the cerebellum of adult mice. Q, IP-WB assays showed that Gα12, but not Arhgef12, RhoA and ROCK2, was modified with O-GlcNAcylation in the cerebellum of adult mice.
To exclude the possibility that Ogt directly interacted with Arhgef12, RhoA and ROCK2, and catalyzed the O-GlcNAcylation of those proteins. We performed IP with cerebellar tissues followed by WB assay. We observed that IP with Ogt antibody could precipitate Gα12, but not RhoA, Arhgef12 and ROCK2, respectively (Fig. 9P). In addition, IP with O-GlcNAcylation could precipitate Gα12, but not RhoA, Arhgef12 and ROCK2, respectively (Fig. 9Q). These results suggested that Ogt could directly interact with Gα12 and catalyze its O-GlcNAcylation, but not RhoA, Arhgef12, and ROCK2.
The supplement of O-GlcNAcylation substrate GlcNAc enhances the activity of RhoA/ROCK signaling pathway under ogt deficient condition
Rho-specific GEFs have been shown to interact with Gα12 through its RGS-like (RGSL) domain and thus active RhoA (M.J. Hart et al., 1998; Chikumi et al., 2002; Suzuki et al., 2003). We hypothesized that the O-GlcNAcylation of Gα12 promoted RhoA/ROCK signaling by maintaining the binding of Gα12 to RGSL domain. To test this hypothesis, we co-transfected exogenous RGSL (Flag) and Gα12 (HA) under scramble (Ctrl) and shOgt conditions in N2a cells, respectively. IP followed by WB assays showed that Ogt depletion significantly reduced the interaction between Gα12 (HA) and Arhgef12 (Flag; Fig. 10A–C).
Figure 10.
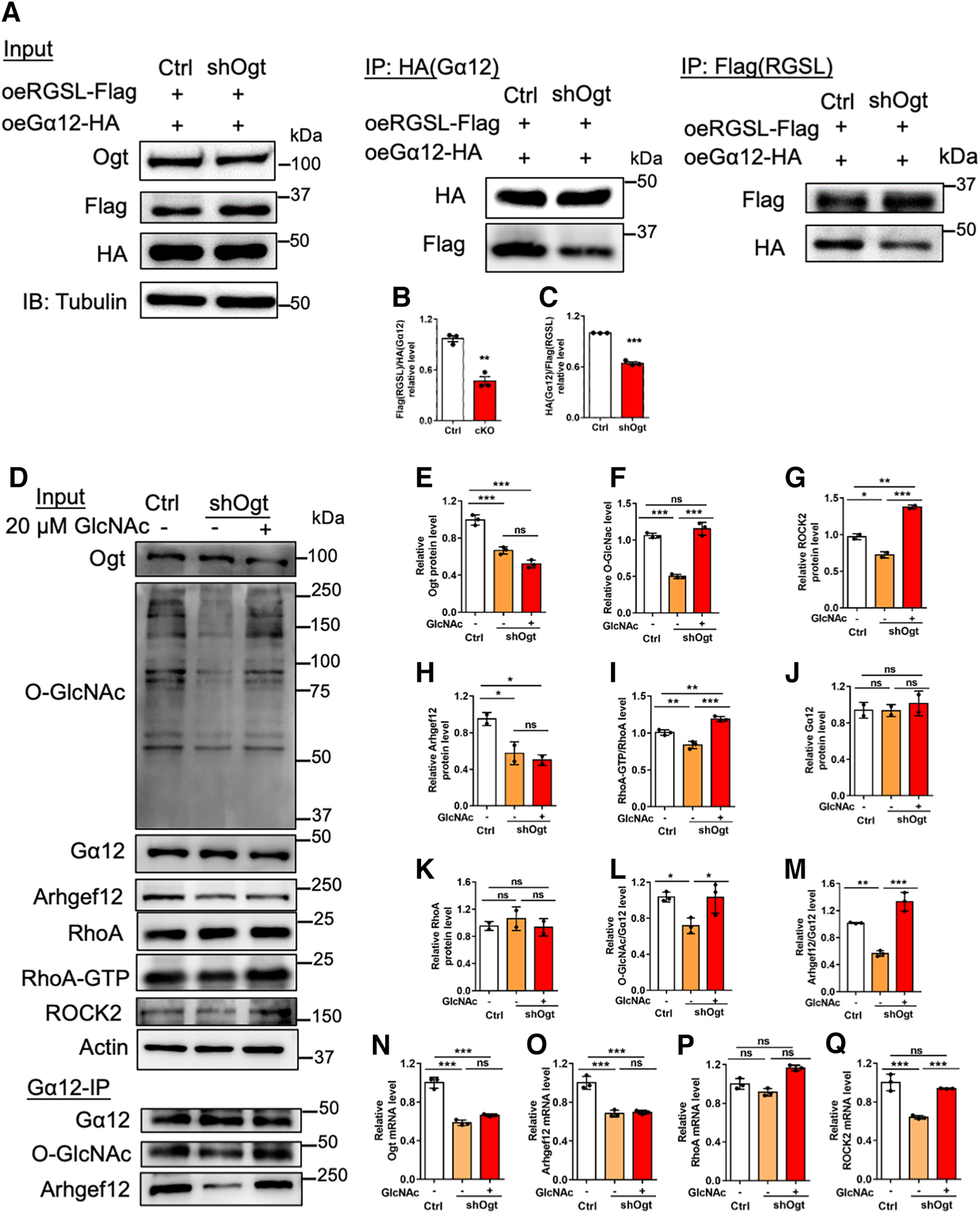
O-GlcNAcylation on Gα12 regulates the interaction between Gα12 and Arhgef12, and RhoA/ROCK signaling activation. A, IP-WB assays of exogenous Arhgef12 (RGSL-Flag) and Gα12 (HA) transfected with scramble and shOgt in N2a cells, respectively. RGSL, RGS-like domain. B, C, Quantification of Arhgef12 (RGSL-Flag) IP (A) and Gα12 (HA) IP (A) showed that the interaction between Arhgef12 (RGSL-Flag) and Gα12 (HA) significantly decreased. n = 3 independent experiments for each group. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.001, ***p < 0.0001. D, Representative images of WB assays of Ogt, O-GlcNAcylation, Gα12, Arhgef12, RhoA, RhoA-GTP and ROCK2 (input) and Gα12 IP followed by WB assays with Gα12, O-GlcNAcylation and Arhgef12 antibody in N2a cells, respectively. Cells were transfected with scramble and shOgt, respectively, and medium was replaced 12 h later. Cells were collected for assay 48 h later. Ogt substrate GlcNAc was dissolved with PBS and was supplemented for 48 h (20 μm) under Ogt knock-down condition. E–K, Quantification of the levels of Ogt (E), O-GlcNAcylation (F), Gα12 (G), Arhgef12 (H), total RhoA (I), RhoA-GTP (J), and ROCK2 (K) level of the Input of D. n = 3 independent experiments for each group. Data are presented as mean ± SEM. One-way ANOVA analysis followed by Tukey's post hoc analysis, *p < 0.05, **p < 0.001, ***p < 0.0001, ns, not significant. L, M, Quantification of O-GlcNAcylation level on Gα12 (O-GlcNAcylation/Gα12; L) and interaction between Gα12-Arhgef12 (Arhgef12/Gα12; M) of the Gα12-IP of D. n = 3 independent experiments for each group. Data are presented as mean ± SEM. One-way ANOVA analysis followed by Tukey's post hoc analysis, *p < 0.05, **p < 0.001, ***p < 0.0001. N–Q, qRT-PCR assays of Ogt (N), Arhgef12 (O), RhoA (P), and ROCK2 (Q) mRNA expression in N2a cells. n = 3 independent experiments for each group. Data are presented as mean ± SEM. One-way ANOVA analysis followed by Tukey's post hoc analysis, *p < 0.05, **p < 0.001, ***p < 0.0001, ns, not significant.
Next, we aim to examine whether the rescue of O-GlcNAcylation could promote RhoA/ROCK signaling pathway. Scramble (Ctrl) and shOgt plasmids were transfected into N2a cells, respectively, and O-GlcNAcylation substrate GlcNAc was supplemented under shOgt condition. WB assay and quantification results showed that Ogt depletion significantly reduced the levels of Ogt, O-GlcNAcylation, ROCK2, Arhgef12, and the activation of RhoA (RhoA-GTP/RhoA; Fig. 10D–I), but not affected the levels of Gα12 and RhoA (Fig. 10D,J,K). Gα12-IP followed by WB assays showed that Ogt knock-down reduced the O-GlcNAcylation of Gα12 and the interaction between Gα12-Arhgef12 (Fig. 10D,L,M). The supplement of GlcNAc did not affect the levels of Ogt, Gα12, RhoA and Arhgef12, but significantly enhanced the levels of total O-GlcNAcylation, ROCK2 and the activation of RhoA (RhoA-GTP/RhoA; Fig. 10D–K). The supplement of GlcNAc also increased the O-GlcNAcylation level of Gα12 and the interaction between Gα12-Arhgef12 (Fig. 10D,L,M). In addition, GlcNAc supplement increased the mRNA level of ROCK2, but not affected the levels of Ogt, RhoA, and Arhgef12 (Fig. 10N–Q). Taken together, these results suggested that O-GlcNAcylation of Gα12 is essential for the interaction between Gα12-Arhgef12 and the activation of RhoA/ROCK signaling pathway.
Modulation of RhoA signaling regulates the development of CGCs
Finally, we aimed to examine whether the modulation of RhoA/ROCK signaling pathway affected the development of CGCs. CGCs were isolated from the cerebella of P7 Ctrl and cKO mice, respectively, and the morphology of CGCs was analyzed based on the immunofluorescence staining signal of neuronal marker Map2. We observed that cKO CGCs had the reduced length of neurite and number of intersections (Fig. 11A–C). The treatment of RhoA signaling activator Oleoyl-L-α-lysophosphatidic acid (LPA; 10 μm, 24 h) could significantly rescue neuronal deficits of cKO CGCs, while LPA treatment did not show observable effects on morphologic development of Ctrl CGCs (Fig. 11A–C). In addition, WB assay and quantification results showed that LPA treatment significantly increased the levels of RhoA and ROCK2 in both Ctrl and cKO CGCs (Fig. 11D–G).
Figure 11.
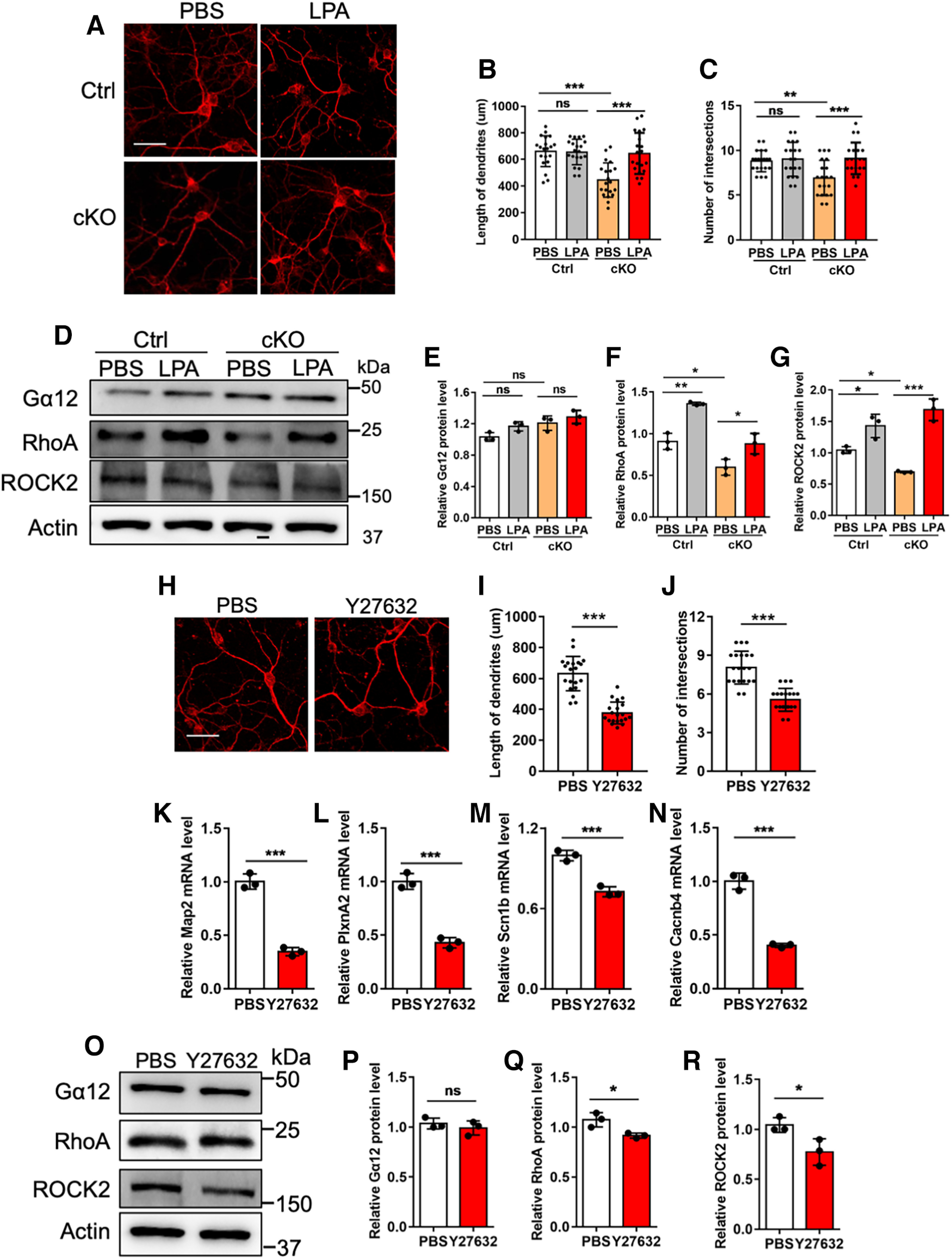
Modulation of RhoA signaling pathway regulates the development of CGCs. A, Representative images of Map2 immunostaining with Ctrl and cKO CGCs treated with PBS and 10 μm LPA, respectively. CGCs were isolated and treated with PBS and 10 μm LPA at DIV8, respectively. Twenty-four hours later, cells were harvested for assay. Scale bar, 50 μm. B, C, Quantification of the length of dendrites (B) and intersection numbers (C) in A. n = 20 cells for each group from 2 independent experiments. One-way ANOVA analysis followed by Tukey's post hoc analysis, *p < 0.05, **p < 0.001, ***p < 0.0001, ns, not significant. All values shown as mean ± SEM. D, Representative images of WB assays of Gα12, RhoA, ROCK2 level in Ctrl and cKO CGCs. CGCs were isolated and treated with PBS and 10 μm LPA at DIV8, respectively. Twenty-four hours later, cells were harvested for assay. E–G, Quantification results of Gα12 (E), RhoA (F), ROCK2 (G) levels of Ctrl and cKO CGCs treated with PBS and 10 μm LPA, respectively. n = 3 independent experiments for each group. Data are presented as mean ± SEM. One-way ANOVA analysis followed by Tukey's post hoc analysis, *p < 0.05, **p < 0.001, ***p < 0.0001, ns, not significant. H, Representative images of Map2 immunostaining with Ctrl CGCs. CGCs were treated with PBS and Y27632 (10 μm) at DIV8, respectively, and harvested for assay 24 h later. Scale bar, 50 μm. I, J, Quantification of the dendritic length (I) and intersection numbers (J) in H. n = 20 cells for each group from 2 independent experiments. Data are presented as mean ± SEM. One-way ANOVA analysis followed by Tukey's post hoc analysis, *p < 0.05, **p < 0.001, ***p < 0.0001. K–N, qRT-PCR assays of the levels of neuronal genes Map2, PlxnA2, Scn1b and Cacnb4 in Ctrl CGCs treated with PBS and Y27632, respectively. n = 3 independent experiments for each group. Data are presented as mean ± SEM. Unpaired Student's t test, *p < 0.05, **p < 0.01, ***p < 0.001. O, Representative images of WB assays of Gα12, RhoA, ROCK2. CGCs were treated with PBS and Y27632 (10 μm) at DIV8, respectively, and harvested for assay 24 h later. P–R, Quantification of Gα12 (P), RhoA (Q), ROCK2 (R) levels in (O). n = 3 independent experiments for each group. Data are presented as mean ± SEM. Unpaired Student's t test; ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001.
Next, we examined whether the inhibition of RhoA/ROCK signaling pathway could suppress the development of CGCs. Ctrl CGCs were treated with RhoA inhibitor Y27632 and morphology of CGCs was analyzed. We observed that Y27632 treatment significantly reduced the length of neurites and number of intersections (Fig. 11H–J). mRNA levels of some neuronal genes including Map2, PlxnA2, Scn1b, and Cacnb4 were also significantly decreased (Fig. 11K–N). In addition, WB assay and quantification results showed that Y27632 treatment significantly reduced the levels of RhoA and ROCK2 in Ctrl CGCs (Fig. 11O–R). Taken together, these results suggested that the modulation of RhoA/ROCK pathway can regulate the development of CGCs.
Discussion
Ogt and O-GlcNAcylation are enriched in the brain, and play essential roles in neuronal functions and neurologic disorders (Okuyama and Marshall, 2003; Levine and Walker, 2016; A.C. Wang et al., 2016; Ma et al., 2017; Su and Schwarz, 2017; Wani et al., 2017; X. Yang and Qian, 2017). Here, we have found that Ogt deficiency in GNPs significantly reduced the level of O-GlcNAcylation in the cerebellum and CGCs of adult mice. Ogt deficient mice display cerebellar atrophy, aberrant cytoarchitecture and synaptic connection, the impaired motor coordination and cognitive ability. Our mechanistic study revealed that Ogt catalyzed O-GlcNAcylation of Gα12, which is essential for its binding to Arhgef12. Under Ogt deficient condition, the reduced O-GlcNAcylation of Gα12 inhibits its binding to Arhgef12 and consequently repressed RhoA/ROCK signaling. The activation of RhoA/ROCK signaling can rescue neuronal deficits of CGCs induced by Ogt deficiency. Collectively, our study has revealed the critical roles and related mechanisms of Ogt and O-GlcNAcylation in regulating cerebellar function.
Ogt and O-GlcNAcylation regulate the normal establishment of cytoarchitecture of cerebellum
Previous studies have shown the important function of Ogt and O-GlcNAcylation in brain. Ogt deficiency in neural stem/progenitor cells impaired embryonic and adult neurogenesis through modulating Wnt and Notch pathways (J. Chen et al., 2021; Shen et al., 2021a). Specific deletion of Ogt in neurons reduced the survival of neurons, synaptic plasticity and maturation, and impaired learning and memory of mice (Taylor et al., 2014; Lagerlöf et al., 2017; Su and Schwarz, 2017). In addition, Ogt regulates myelination development, Glucose and lipid metabolism (Ruan et al., 2014; Kim et al., 2016; Lagerlöf et al., 2016).
A very recent study showed that at the early postnatal cerebellum of mice, the deficiency of Ogt in granule precursor cells (GNPs) disrupted cerebellar neurogenesis via modulating Shh signaling (L. Chen et al., 2022a). Our present study examined the function of Ogt in the cerebellum of adult mice. Our results showed that Ogt deficiency induced multiple cerebellar deficits including abnormal cytoarchitecture, atrophy, abnormal distribution of CGCs, the disrupted arrangement of Bergman glia and Purkinje cells of adult mice. These data indicated the extensive effects of Ogt-mediated O-GlcNAcylation in cerebellum of adult mice. A previous study showed that Ogt deficiency in sensory neurons of adult mice could also reduce nerve fiber endings and cell bodies (Uriarte et al., 2017). Meanwhile, we observed that some cerebellar deficits already emerged as early as postnatal day 7, suggesting that abnormal development of cerebellum induces, at least partially, the aberrant morphology and function of adult cerebellum. Collectively, these studies suggest that Ogt and O-GlcNAcylation are essential for both developmental and adult cerebellum of mice.
Ogt and O-GlcNAcylation in cerebellar function and neurologic disorders
Ogt and O-GlcNAcylation regulate neuronal activity, feeding behavior, obesity and neurodegeneration (Ruan et al., 2014; Lagerlöf et al, 2016, 2017; A.C. Wang et al., 2016; Su and Schwarz, 2017). Some human patients with intellectual disability have mutations and catalytic deficiency of OGT (Vaidyanathan et al., 2017; Willems et al., 2017; Selvan et al., 2018; Pravata et al., 2019). The global level of O-GlcNAcylation decreased along with brain aging and contributes to neurodegenerative diseases including Alzheimer's disease and Parkinson's disease (Rex-Mathes et al., 2001; Marotta et al., 2015; Wani et al., 2017; Wheatley et al., 2019).
Our results showed that Ogt ablation in GNPs caused the deficiency of Ogt in CGCs. In cerebellum, CGCs regulated the formation of Bergmann fiber through secreting growth factors, and Bergmann glia provided structural support for the migration and dendritic extension of PCs (Lin et al., 2009; Araujo et al., 2019). In addition, PCs receive information from parallel fibers (the mossy fiber→granule cell→parallel fiber→Purkinje cell pathway) and climbing fibers (the climbing fiber→Purkinje cell pathway), and form circuits with Bergmann glia (Galliano et al., 2013; Leto et al., 2016; De Zeeuw et al., 2021; Joyner and Bayin, 2022). These studies suggested that the interplay between types of cells and multiple components in the cerebellar microenvironment could regulate the acquisition and maintenance of Bergmann glia phenotype and the organization of PCs.
Although Galliano et al., found that significant reduction of parallel fiber (PF)-Purkinje cell (PC) transmission did not lead to observable behavioral phenotypes of mice (Galliano et al., 2013), our present study showed that adult cKO mice displayed severely impaired motor coordination, learning and memory capabilities, suggesting that these behavioral deficits could be induced not only by the reduced number and abnormal distribution of CGCs, also by the aberrant synaptic transmission. In addition, the level of O-GlcNAcylation was also decreased in unipolar brush cells (UBCs) of cKO mice; we speculated that aberrant UBCs could also contribute to the abnormal cerebellar function.
In addition, functional interplay between cerebellum and other brain regions including hippocampus has been identified (Watson et al., 2019; Rondi-Reig et al., 2022; Torres-Herraez et al., 2022). Although our results show the specific alteration of Ogt and O-GlcNAcylation in cerebellum, but not in cortex and hippocampus, we could not exclude the possibility that the abnormal connection between cerebellum and other brain regions such as hippocampus and cortex contributes to the observed behavioral deficits especially the impaired learning and memory. Collectively, our study provided novel insights regarding the critical roles of Ogt and O-GlcNAcylation in regulating cerebellar function of adult mice.
Ogt-mediated O-GlcNAcylation regulates RhoA/ROCK signaling in cerebellum
As an abundant posttranslational modification (PTM), O-GlcNAcylation interacts with other PTMs and epigenetic modifications, and regulates a number of biological processes at multiple levels (Hanover et al., 2012; Levine and Walker, 2016; X. Yang and Qian, 2017). Ogt and O-GlcNAcylation regulate gene expression including Notch, Wnt and Shh signaling pathways and involves in development, aging and diseases (Hanover et al., 2012; Taylor et al., 2014; Levine and Walker, 2016; Su and Schwarz, 2017; X. Yang and Qian, 2017; Wheatley et al., 2019; J. Chen et al., 2021; Shen et al., 2021b; L. Chen et al., 2022a).
In the present study, we identified 57 O-GlcNAcylation-modified proteins including G-protein family member Gα12. Previous studies have showed the important function of Gα12, and RhoA/ROCK signaling in neuronal development and function (Sayas et al., 2002; Hiley et al., 2006; Moers et al., 2008; Akiyama et al., 2016; Acquarone et al., 2019; Wong et al., 2019). Gα12 can interact with RhoGEFs and subsequently activates RhoA/ROCK signaling (Kranenburg et al., 1999; Hiley et al., 2006; Meyer et al., 2008). Deletion of Gα12 in neuronal system results in neuronal ectopia in cerebellar cortices (Moers et al., 2008). RhoA regulates active transport of synaptic vesicles and coordinates neurotransmission (H.G. Wang et al., 2005; Chenouard et al., 2020). The decreased O-GlcNAcylation of Gα12 reduced the interaction between Gα12 and Arhgef12, which inhibited RhoA/ROCK signaling. Our results showed that the activation of RhoA/ROCK signaling partially rescued neuronal deficits of Ogt deficient CGCs. Of note, we observed that the supplement of O-GlcNAc did not affect mRNA and protein levels of Arhgef12, but significantly enhanced the interaction between Gα12 and Arhgef12. Meanwhile, Arhgef12 was not modified with O-GlcNAcylation, suggesting that Ogt could regulate the expression of Arhgef12 at transcriptional level. Therefore, our results on the one hand suggested that the O-GlcNAcylation of Gα12 is critical for RhoA/ROCK signaling, which is essential for normal cerebellar function. On the other hand, our results suggested that Ogt regulates cerebellar function at transcriptional and post-translational levels.
In conclusion, our present study has revealed the important function and related mechanisms of Ogt-mediated O-GlcNAcylation in regulating cerebellar function and behavior of adult mice. Our results suggested that Ogt and O-GlcNAcylation could be a potential target for some cerebella-related diseases such as the OGT congenital disorder of glycosylation (OGT-CDG) and cerebellar dysplasia.
Footnotes
This work was supported in part by the National Natural Science Foundation of China Grants 92049108 and 31571518 (to X.L.) and the National Key Research and Development Program of China Grant 2017YFE0196600 (to X.L.). S.X. is supported by Shandong Provincial Natural Science Foundation China #ZR2015HM024 and #2019GSF108066.
The authors declare no competing financial interests.
References
- Acquarone E, Argyrousi EK, van den Berg M, Gulisano W, Fa M, Staniszewski A, Calcagno E, Zuccarello E, D'Adamio L, Deng SX, Deng SX, Puzzo D, Arancio O, Fiorito J (2019) Synaptic and memory dysfunction induced by tau oligomers is rescued by up-regulation of the nitric oxide cascade. Mol Neurodegener 14:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama H, Fukuda T, Tojima T, Nikolaev VO, Kamiguchi H (2016) Cyclic nucleotide control of microtubule dynamics for axon guidance. J Neurosci 36:5636–5649. 10.1523/JNEUROSCI.3596-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anne SL, Govek EE, Ayrault O, Kim JH, Zhu X, Murphy DA, Van Aelst L, Roussel MF, Hatten ME (2013) WNT3 inhibits cerebellar granule neuron progenitor proliferation and medulloblastoma formation via MAPK activation. PLoS One 8:e81769. 10.1371/journal.pone.0081769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo APB, Carpi-Santos R, Gomes FCA (2019) The role of astrocytes in the development of the cerebellum. Cerebellum 18:1017–1035. 10.1007/s12311-019-01046-0 [DOI] [PubMed] [Google Scholar]
- Beuriat PA, Cohen-Zimerman S, Smith GNL, Krueger F, Gordon B, Grafman J (2020) A new insight on the role of the cerebellum for executive functions and emotion processing in adults. Front Neurol 11:593490. 10.3389/fneur.2020.593490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carta I, Chen CH, Schott AL, Dorizan S, Khodakhah K (2019) Cerebellar modulation of the reward circuitry and social behavior. Science 363:eaav0581. 10.1126/science.aav0581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Dong X, Cheng X, Zhu Q, Zhang J, Li Q, Huang X, Wang M, Li L, Guo W, Sun B, Shu Q, Yi W, Li X (2021) Ogt controls neural stem/progenitor cell pool and adult neurogenesis through modulating Notch signaling. Cell Rep 34:108905. 10.1016/j.celrep.2021.108905 [DOI] [PubMed] [Google Scholar]
- Chen L, Li Y, Song Z, Xue S, Liu F, Chang X, Wu Y, Duan X, Wu H (2022a) O-GlcNAcylation promotes cerebellum development and medulloblastoma oncogenesis via SHH signaling. Proc Natl Acad Sci U S A 119:e2202821119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Chen T, Dong C, Chen H, Dong X, Yang L, Hu L, Wang H, Wu B, Yao Y, Xiong Y, Xiong M, Lin Y, Zhou W (2022b) Deletion of CHD8 in cerebellar granule neuron progenitors leads to severe cerebellar hypoplasia, ataxia, and psychiatric behavior in mice. J Genet Genomics 49:859–869. 10.1016/j.jgg.2022.02.011 [DOI] [PubMed] [Google Scholar]
- Chen X, Du Y, Broussard GJ, Kislin M, Yuede CM, Zhang S, Dietmann S, Gabel H, Zhao G, Wang SS, Zhang X, Bonni A (2022c) Transcriptomic mapping uncovers Purkinje neuron plasticity driving learning. Nature 605:722–727. 10.1038/s41586-022-04711-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenouard N, Xuan F, Tsien RW (2020) Synaptic vesicle traffic is supported by transient actin filaments and regulated by PKA and NO. Nat Commun 11:5318. 10.1038/s41467-020-19120-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikumi H, Fukuhara S, Gutkind JS (2002) Regulation of G protein-linked guanine nucleotide exchange factors for Rho, PDZ-RhoGEF, and LARG by tyrosine phosphorylation: evidence of a role for focal adhesion kinase. J Biol Chem 277:12463–12473. 10.1074/jbc.M108504200 [DOI] [PubMed] [Google Scholar]
- De Zeeuw CI, Lisberger SG, Raymond JL (2021) Diversity and dynamism in the cerebellum. Nat Neurosci 24:160–167. 10.1038/s41593-020-00754-9 [DOI] [PubMed] [Google Scholar]
- Galliano E, Gao Z, Schonewille M, Todorov B, Simons E, Pop AS, D'Angelo E, van den Maagdenberg AM, Hoebeek FE, De Zeeuw CI (2013) Silencing the majority of cerebellar granule cells uncovers their essential role in motor learning and consolidation. Cell Rep 3:1239–1251. 10.1016/j.celrep.2013.03.023 [DOI] [PubMed] [Google Scholar]
- Gulisano W, Melone M, Ripoli C, Tropea MR, Li Puma DD, Giunta S, Cocco S, Marcotulli D, Origlia N, Palmeri A, Arancio O, Conti F, Grassi C, Puzzo D (2019) Neuromodulatory action of picomolar extracellular Aβ42 oligomers on presynaptic and postsynaptic mechanisms underlying synaptic function and memory. J Neurosci 39:5986–6000. 10.1523/JNEUROSCI.0163-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldipur P, Millen KJ, Aldinger KA (2022) Human cerebellar development and transcriptomics: implications for neurodevelopmental disorders. Annu Rev Neurosci 45:515–531. 10.1146/annurev-neuro-111020-091953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover JA, Krause MW, Love DC (2012) Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat Rev Mol Cell Biol 13:312–321. 10.1038/nrm3334 [DOI] [PubMed] [Google Scholar]
- Hart GW, Housley MP, Slawson C (2007) Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446:1017–1022. 10.1038/nature05815 [DOI] [PubMed] [Google Scholar]
- Hart MJ, Jiang X, Kozasa T, Roscoe W, Singer WD, Gilman AG, Sternweis PC, Bollag G (1998) Direct stimulation of the guanine nucleotide exchange activity of p115 RhoGEF by Galpha13. Science 280:2112–2114. 10.1126/science.280.5372.2112 [DOI] [PubMed] [Google Scholar]
- Hatten ME, Roussel MF (2011) Development and cancer of the cerebellum. Trends Neurosci 34:134–142. 10.1016/j.tins.2011.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibi M, Shimizu T (2012) Development of the cerebellum and cerebellar neural circuits. Dev Neurobiol 72:282–301. 10.1002/dneu.20875 [DOI] [PubMed] [Google Scholar]
- Hiley E, McMullan R, Nurrish SJ (2006) The Galpha12-RGS RhoGEF-RhoA signalling pathway regulates neurotransmitter release in C. elegans. EMBO J 25:5884–5895. 10.1038/sj.emboj.7601458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa R, Hashimoto K, Miyazaki T, Uchigashima M, Yamasaki M, Aiba A, Kano M, Watanabe M (2016a) Territories of heterologous inputs onto Purkinje cell dendrites are segregated by mGluR1-dependent parallel fiber synapse elimination. Proc Natl Acad Sci U S A 113:2282–2287. 10.1073/pnas.1511513113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa R, Sakimura K, Watanabe M (2016b) GluD2 endows parallel fiber-Purkinje cell synapses with a high regenerative capacity. J Neurosci 36:4846–4858. 10.1523/JNEUROSCI.0161-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner AL, Bayin NS (2022) Cerebellum lineage allocation, morphogenesis and repair: impact of interplay amongst cells. Development 149:dev185587. 10.1242/dev.185587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Maynard JC, Sasaki Y, Strickland A, Sherman DL, Brophy PJ, Burlingame AL, Milbrandt J (2016) Schwann cell O-GlcNAc glycosylation is required for myelin maintenance and axon integrity. J Neurosci 36:9633–9646. 10.1523/JNEUROSCI.1235-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranenburg O, Poland M, van Horck FP, Drechsel D, Hall A, Moolenaar WH (1999) Activation of RhoA by lysophosphatidic acid and Galpha12/13 subunits in neuronal cells: induction of neurite retraction. Mol Biol Cell 10:1851–1857. 10.1091/mbc.10.6.1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagerlöf O, Slocomb JE, Hong I, Aponte Y, Blackshaw S, Hart GW, Huganir RL (2016) The nutrient sensor OGT in PVN neurons regulates feeding. Science 351:1293–1296. 10.1126/science.aad5494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagerlöf O, Hart GW, Huganir RL (2017) O-GlcNAc transferase regulates excitatory synapse maturity. Proc Natl Acad Sci U S A 114:1684–1689. 10.1073/pnas.1621367114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lainiola M, Procaccini C, Linden AM (2014) mGluR3 knockout mice show a working memory defect and an enhanced response to MK-801 in the T- and Y-maze cognitive tests. Behav Brain Res 266:94–103. 10.1016/j.bbr.2014.03.008 [DOI] [PubMed] [Google Scholar]
- Lee HY, Greene LA, Mason CA, Manzini MC (2009) Isolation and culture of post-natal mouse cerebellar granule neuron progenitor cells and neurons. J Vis Exp (23):990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leto K, et al. (2016) Consensus paper: cerebellar development. Cerebellum 15:789–828. 10.1007/s12311-015-0724-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ZG, Walker S (2016) The biochemistry of O-GlcNAc transferase: which functions make it essential in mammalian cells? Annu Rev Biochem 85:631–657. 10.1146/annurev-biochem-060713-035344 [DOI] [PubMed] [Google Scholar]
- Lin Y, Chen L, Lin C, Luo Y, Tsai RY, Wang F (2009) Neuron-derived FGF9 is essential for scaffold formation of Bergmann radial fibers and migration of granule neurons in the cerebellum. Dev Biol 329:44–54. 10.1016/j.ydbio.2009.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Li H, He Y, Hao J (2017) The emerging link between O-GlcNAcylation and neurological disorders. Cell Mol Life Sci 74:3667–3686. 10.1007/s00018-017-2542-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie S, Shaw P, Lenroot R, Pierson R, Greenstein DK, Nugent TF 3rd, Sharp WS, Giedd JN, Rapoport JL (2007) Cerebellar development and clinical outcome in attention deficit hyperactivity disorder. Am J Psychiatry 164:647–655. 10.1176/ajp.2007.164.4.647 [DOI] [PubMed] [Google Scholar]
- Marotta NP, Lin YH, Lewis YE, Ambroso MR, Zaro BW, Roth MT, Arnold DB, Langen R, Pratt MR (2015) O-GlcNAc modification blocks the aggregation and toxicity of the protein α-synuclein associated with Parkinson's disease. Nat Chem 7:913–920. 10.1038/nchem.2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E (2010) Neuron tracing in perspective. Cytometry A 77:693–704. 10.1002/cyto.a.20895 [DOI] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M (2004) Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A 58:167–176. 10.1002/cyto.a.20022 [DOI] [PubMed] [Google Scholar]
- Meyer BH, Freuler F, Guerini D, Siehler S (2008) Reversible translocation of p115-RhoGEF by G(12/13)-coupled receptors. J Cell Biochem 104:1660–1670. 10.1002/jcb.21732 [DOI] [PubMed] [Google Scholar]
- Moers A, Nürnberg A, Goebbels S, Wettschureck N, Offermanns S (2008) Galpha12/Galpha13 deficiency causes localized overmigration of neurons in the developing cerebral and cerebellar cortices. Mol Cell Biol 28:1480–1488. 10.1128/MCB.00651-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomska-Cimicka A, et al. (2021) SCA7 mouse cerebellar pathology reveals preferential downregulation of key Purkinje cell-identity genes and shared disease signature with SCA1 and SCA2. J Neurosci 41:4910–4936. 10.1523/JNEUROSCI.1882-20.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama R, Marshall S (2003) UDP-N-acetylglucosaminyl transferase (OGT) in brain tissue: temperature sensitivity and subcellular distribution of cytosolic and nuclear enzyme. J Neurochem 86:1271–1280. 10.1046/j.1471-4159.2003.01939.x [DOI] [PubMed] [Google Scholar]
- Penas C, Govek EE, Fang Y, Ramachandran V, Daniel M, Wang W, Maloof ME, Rahaim RJ, Bibian M, Kawauchi D, Finkelstein D, Han JL, Long J, Li B, Robbins DJ, Malumbres M, Roussel MF, Roush WR, Hatten ME, Ayad NG (2015) Casein kinase 1δ is an APC/C(Cdh1) substrate that regulates cerebellar granule cell neurogenesis. Cell Rep 11:249–260. 10.1016/j.celrep.2015.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penas C, Maloof ME, Stathias V, Long J, Tan SK, Mier J, Fang Y, Valdes C, Rodriguez-Blanco J, Chiang CM, Robbins DJ, Liebl DJ, Lee JK, Hatten ME, Clarke J, Ayad NG (2019) Time series modeling of cell cycle exit identifies Brd4 dependent regulation of cerebellar neurogenesis. Nat Commun 10:3028. 10.1038/s41467-019-10799-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinchefsky EF, Accogli A, Shevell MI, Saint-Martin C, Srour M (2019) Developmental outcomes in children with congenital cerebellar malformations. Dev Med Child Neurol 61:350–358. 10.1111/dmcn.14059 [DOI] [PubMed] [Google Scholar]
- Pravata VM, Muha V, Gundogdu M, Ferenbach AT, Kakade PS, Vandadi V, Wilmes AC, Borodkin VS, Joss S, Stavridis MP, van Aalten DMF (2019) Catalytic deficiency of O-GlcNAc transferase leads to X-linked intellectual disability. Proc Natl Acad Sci U S A 116:14961–14970. 10.1073/pnas.1900065116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach JE, Clark PM, Mason DE, Neve RL, Peters EC, Hsieh-Wilson LC (2012) Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation. Nat Chem Biol 8:253–261. 10.1038/nchembio.770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex-Mathes M, Werner S, Strutas D, Griffith LS, Viebahn C, Thelen K, Schmitz B (2001) O-GlcNAc expression in developing and ageing mouse brain. Biochimie 83:583–590. 10.1016/s0300-9084(01)01305-0 [DOI] [PubMed] [Google Scholar]
- Rondi-Reig L, Paradis AL, Fallahnezhad M (2022) A liaison brought to light: cerebellum-hippocampus, partners for spatial cognition. Cerebellum 21:826–837. 10.1007/s12311-022-01422-3 [DOI] [PubMed] [Google Scholar]
- Rosenberg AB, Roco CM, Muscat RA, Kuchina A, Sample P, Yao Z, Graybuck LT, Peeler DJ, Mukherjee S, Chen W, Pun SH, Sellers DL, Tasic B, Seelig G (2018) Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360:176–182. 10.1126/science.aam8999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross EM, Wilkie TM (2000) GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem 69:795–827. 10.1146/annurev.biochem.69.1.795 [DOI] [PubMed] [Google Scholar]
- Ruan HB, Dietrich MO, Liu ZW, Zimmer MR, Li MD, Singh JP, Zhang K, Yin R, Wu J, Horvath TL, Yang X (2014) O-GlcNAc transferase enables AgRP neurons to suppress browning of white fat. Cell 159:306–317. 10.1016/j.cell.2014.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayas CL, Avila J, Wandosell F (2002) Glycogen synthase kinase-3 is activated in neuronal cells by Galpha12 and Galpha13 by Rho-independent and Rho-dependent mechanisms. J Neurosci 22:6863–6875. 10.1523/JNEUROSCI.22-16-06863.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvan N, George S, Serajee FJ, Shaw M, Hobson L, Kalscheuer V, Prasad N, Levy SE, Taylor J, Aftimos S, Schwartz CE, Huq AM, Gecz J, Wells L (2018) O-GlcNAc transferase missense mutations linked to X-linked intellectual disability deregulate genes involved in cell fate determination and signaling. J Biol Chem 293:10810–10824. 10.1074/jbc.RA118.002583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Zhao X, Chen J, Qu W, Huang X, Wang M, Shao Z, Shu Q, Li X (2021a) O-GlcNAc transferase Ogt regulates embryonic neuronal development through modulating Wnt/β-catenin signaling. Hum Mol Genet 31:57–68. 10.1093/hmg/ddab223 [DOI] [PubMed] [Google Scholar]
- Shen H, Zhao X, Chen J, Qu W, Huang X, Wang M, Shao Z, Shu Q, Li X (2021b) O-GlcNAc transferase Ogt regulates embryonic neuronal development through modulating Wnt/beta-catenin signaling. Hum Mol Genet 31:57–68. [DOI] [PubMed] [Google Scholar]
- Su C, Schwarz TL (2017) O-GlcNAc transferase is essential for sensory neuron survival and maintenance. J Neurosci 37:2125–2136. 10.1523/JNEUROSCI.3384-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Nakamura S, Mano H, Kozasa T (2003) Galpha 12 activates Rho GTPase through tyrosine-phosphorylated leukemia-associated RhoGEF. Proc Natl Acad Sci U S A 100:733–738. 10.1073/pnas.0234057100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Stoiljkovic M, Song E, Gao XB, Yasumoto Y, Kudo E, Carvalho F, Kong Y, Park A, Shanabrough M, Szigeti-Buck K, Liu ZW, Kristant A, Zhang Y, Sulkowski P, Glazer PM, Kaczmarek LK, Horvath TL, Iwasaki A (2022) LINE-1 activation in the cerebellum drives ataxia. Neuron 110:3278–3287.e8. 10.1016/j.neuron.2022.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor EW, Wang K, Nelson AR, Bredemann TM, Fraser KB, Clinton SM, Puckett R, Marchase RB, Chatham JC, McMahon LL (2014) O-GlcNAcylation of AMPA receptor GluA2 is associated with a novel form of long-term depression at hippocampal synapses. J Neurosci 34:10–21. 10.1523/JNEUROSCI.4761-12.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Donkelaar HJ, Lammens M, Wesseling P, Thijssen HO, Renier WO (2003) Development and developmental disorders of the human cerebellum. J Neurol 250:1025–1036. 10.1007/s00415-003-0199-9 [DOI] [PubMed] [Google Scholar]
- Tobe RH, Bansal R, Xu D, Hao X, Liu J, Sanchez J, Peterson BS (2010) Cerebellar morphology in Tourette syndrome and obsessive-compulsive disorder. Ann Neurol 67:479–487. 10.1002/ana.21918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres CR, Hart GW (1984) Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem 259:3308–3317. [PubMed] [Google Scholar]
- Torres-Herraez A, Watson TC, Rondi-Reig L (2022) Delta oscillations coordinate intracerebellar and cerebello-hippocampal network dynamics during sleep. J Neurosci 42:2268–2281. 10.1523/JNEUROSCI.1479-21.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uriarte M, Ogundele OM, Pardo J (2017) Long-lasting training in the Barnes maze prompts hippocampal spinogenesis and habituation in rats. Neuroreport 28:307–312. 10.1097/WNR.0000000000000755 [DOI] [PubMed] [Google Scholar]
- Vaidyanathan K, Niranjan T, Selvan N, Teo CF, May M, Patel S, Weatherly B, Skinner C, Opitz J, Carey J, Viskochil D, Gecz J, Shaw M, Peng Y, Alexov E, Wang T, Schwartz C, Wells L (2017) Identification and characterization of a missense mutation in the O-linked β-N-acetylglucosamine (O-GlcNAc) transferase gene that segregates with X-linked intellectual disability. J Biol Chem 292:8948–8963. 10.1074/jbc.M116.771030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veleanu M, Urrieta-Chávez B, Sigoillot SM, Paul MA, Usardi A, Iyer K, Delagrange M, Doyle JP, Heintz N, Bécamel C, Selimi F (2022) Modified climbing fiber/Purkinje cell synaptic connectivity in the cerebellum of the neonatal phencyclidine model of schizophrenia. Proc Natl Acad Sci U S A 119:e2122544119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AC, Jensen EH, Rexach JE, Vinters HV, Hsieh-Wilson LC (2016) Loss of O-GlcNAc glycosylation in forebrain excitatory neurons induces neurodegeneration. Proc Natl Acad Sci U S A 113:15120–15125. 10.1073/pnas.1606899113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HG, Lu FM, Jin I, Udo H, Kandel ER, de Vente J, Walter U, Lohmann SM, Hawkins RD, Antonova I (2005) Presynaptic and postsynaptic roles of NO, cGK, and RhoA in long-lasting potentiation and aggregation of synaptic proteins. Neuron 45:389–403. 10.1016/j.neuron.2005.01.011 [DOI] [PubMed] [Google Scholar]
- Wang XT, Zhou L, Cai XY, Xu FX, Xu ZH, Li XY, Shen Y (2020) Deletion of Mea6 in cerebellar granule cells impairs synaptic development and motor performance. Front Cell Dev Biol 8:627146. 10.3389/fcell.2020.627146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani WY, Chatham JC, Darley-Usmar V, McMahon LL, Zhang J (2017) O-GlcNAcylation and neurodegeneration. Brain Res Bull 133:80–87. 10.1016/j.brainresbull.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson TC, Obiang P, Torres-Herraez A, Watilliaux A, Coulon P, Rochefort C, Rondi-Reig L (2019) Anatomical and physiological foundations of cerebello-hippocampal interaction. Elife 8:e41896. 10.7554/eLife.41896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley EG, Albarran E, White CW 3rd, Bieri G, Sanchez-Diaz C, Pratt K, Snethlage CE, Ding JB, Villeda SA (2019) Neuronal O-GlcNAcylation improves cognitive function in the aged mouse brain. Curr Biol 29:3359–3369.e4. 10.1016/j.cub.2019.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems AP, Gundogdu M, Kempers MJE, Giltay JC, Pfundt R, Elferink M, Loza BF, Fuijkschot J, Ferenbach AT, van Gassen KLI, van Aalten DMF, Lefeber DJ (2017) Mutations in N-acetylglucosamine (O-GlcNAc) transferase in patients with X-linked intellectual disability. J Biol Chem 292:12621–12631. 10.1074/jbc.M117.790097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LW, Tann JY, Ibanez CF, Sajikumar S (2019) The p75 neurotrophin receptor is an essential mediator of impairments in hippocampal-dependent associative plasticity and memory induced by sleep deprivation. J Neurosci 39:5452–5465. 10.1523/JNEUROSCI.2876-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Zhu Q, Cheng J, Wu Y, Fan M, Zhang J, Wu H (2019) Opposite regulation of Wnt/β-catenin and Shh signaling pathways by Rack1 controls mammalian cerebellar development. Proc Natl Acad Sci U S A 116:4661–4670. 10.1073/pnas.1813244116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Qian K (2017) Protein O-GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol 18:452–465. 10.1038/nrm.2017.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanin JP, Friedman WJ (2022) p75NTR prevents the onset of cerebellar granule cell migration via RhoA activation. Elife 11:e79934. 10.7554/eLife.79934 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The list of differentially expressed genes in cKO CGGs. RNA-seq data analysis identified 1925 differentially expressed genes in cKO CGGs. Download Table 7-1, XLSX file (339.6KB, xlsx) .
The list of proteins modified with O-GlcNAcylation in CGCs. Click-IT assay followed by Mass spectrometry assay identified 57 proteins with O-GlcNAcylation in wild-type CGCs. Download Table 8-1, XLSX file (14KB, xlsx) .
Data Availability Statement
The accession number for the RNA-seq data reported in this paper is SRA: PRJNA884808.