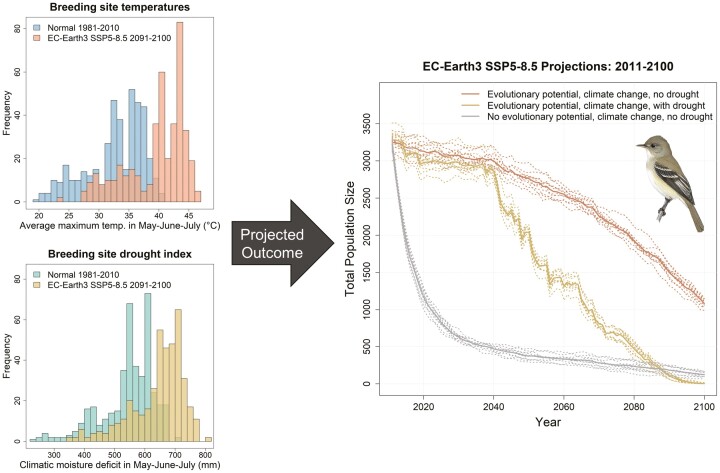
Abstract
The complexity of global anthropogenic change makes forecasting species responses and planning effective conservation actions challenging. Additionally, important components of a species’ adaptive capacity, such as evolutionary potential, are often not included in quantitative risk assessments due to lack of data. While genomic proxies for evolutionary potential in at-risk species are increasingly available, they have not yet been included in extinction risk assessments at a species-wide scale. In this study, we used an individual-based, spatially explicit, dynamic eco-evolutionary simulation model to evaluate the extinction risk of an endangered desert songbird, the southwestern willow flycatcher (Empidonax traillii extimus), in response to climate change. Using data from long-term demographic and habitat studies in conjunction with genome-wide ecological genomics research, we parameterized simulations that include 418 sites across the breeding range, genomic data from 225 individuals, and climate change forecasts spanning 3 generalized circulation models and 3 emissions scenarios. We evaluated how evolutionary potential, and the lack of it, impacted population trajectories in response to climate change. We then investigated the compounding impact of drought and warming temperatures on extinction risk through the mechanism of increased nest failure. Finally, we evaluated how rapid action to reverse greenhouse gas emissions would influence population responses and species extinction risk. Our results illustrate the value of incorporating evolutionary, demographic, and dispersal processes in a spatially explicit framework to more comprehensively evaluate the extinction risk of threatened and endangered species and conservation actions to promote their recovery.
Keywords: CDMetaPOP, computer simulations, conservation genomics, dynamic eco-evolutionary simulation model, Empidonax trailii extimus, local adaptation
Graphical Abstract
Graphical Abstract.
Introduction
In a period of unprecedented transformation of the Earth’s climate and ecosystems by human actions, forecasting environmental change is challenging due to the novelty and interaction of the drivers, such as habitat degradation and loss, climate change, and invasive species (Sage 2020). For conservation biologists, this challenge complicates our ability to understand and predict species responses to change and implement conservation actions to mitigate extinction risk. Species responses are multifaceted and can include 1) dispersal to track changing conditions, 2) plastic responses, such as changes in behavior or physiology, and 3) rapid evolutionary adaptation mediated by evolutionary potential (Dawson et al. 2011). Here, we define evolutionary potential as the capacity to evolve genetically based changes in traits that increase population-level fitness in response to novel or changing environmental conditions (Forester et al. 2022). Together, these 3 responses constitute a species’ adaptive capacity—its ability to accommodate, cope with, or respond to dynamic environmental conditions (Foden et al. 2019).
The prospect for evolutionary potential to mitigate declines in at-risk species has been difficult to evaluate due to a lack of informative proxies for evolutionary potential in these species (Forester et al. 2022). However, the increasing availability of genomic-scale data in species of conservation concern is improving our ability to incorporate evolutionary potential into quantitative models of vulnerability. Quantifying extinction risk is itself challenging, however, and there have been few evaluations of the impact of evolutionary potential on population-level extirpation risk (e.g. Bay et al. 2017b) or species-wide extinction. Evaluating extinction risk in a context that includes evolutionary potential requires the integration of both evolutionary and demographic dynamics in response to environmental change, i.e. eco-evolutionary modeling. While genomic proxies for evolutionary potential account for only 1 potential mechanism of a species’ complex response to multivariate environmental change, their incorporation into eco-evolutionary simulation models can help us better understand how adaptive evolutionary change interacts with demographic and environmental parameters across space and time (Chevin et al. 2010; Pierson et al. 2015; Bay et al. 2017a; Xuereb et al. 2021). Specifically, spatially explicit, individual-based models can be used to evaluate how heterogeneity in individual genotypes, behaviors, and fine-scale interactions between individuals and the landscape scale up to produce emergent properties of the larger population (DeAngelis and Mooij 2005; Bach et al. 2006). This process produces models that are more mechanistic and therefore transferable to novel scenarios or environments (Radchuk et al. 2019). These models, in turn, can be useful in conservation planning, providing a more comprehensive assessment of how species will respond to complex, interacting environmental change drivers (Forester et al. 2022; Funk et al. 2019).
In this study, we investigate the interaction of evolutionary potential, dispersal, demographic processes, climate change, and environmental stochasticity in driving population dynamics of an endangered songbird. The southwestern willow flycatcher (Empidonax trailii extimus) is a neotropical migrant that was listed as endangered under the United States Endangered Species Act (ESA) in 1995 (USFWS 1995). This small songbird winters in Costa Rica and Nicaragua (Ruegg et al. 2021) and breeds in the desert southwest of the United States and extreme northern part of Mexico. Southwestern willow flycatchers nest adjacent to water in dense riparian habitats with spatially complex vegetation structure. They will nest in native and non-native (e.g. saltcedar and Russian olive) vegetation, in both contiguous vegetation and mosaics of vegetation and open areas, and along rivers, reservoirs, marshy seeps, or areas with saturated soil (Sogge et al. 2010). Patch disturbance history (i.e. from flooding) and vegetation successional status influences patch quality, occupancy, and demographic patterns for breeding flycatchers (Theimer et al. 2018). The patchy distribution of riparian habitats across the breeding range in combination with variable patch quality dynamics leads to metapopulation structure on the breeding grounds, with birds often moving among patches within drainages (Paxton et al. 2007).
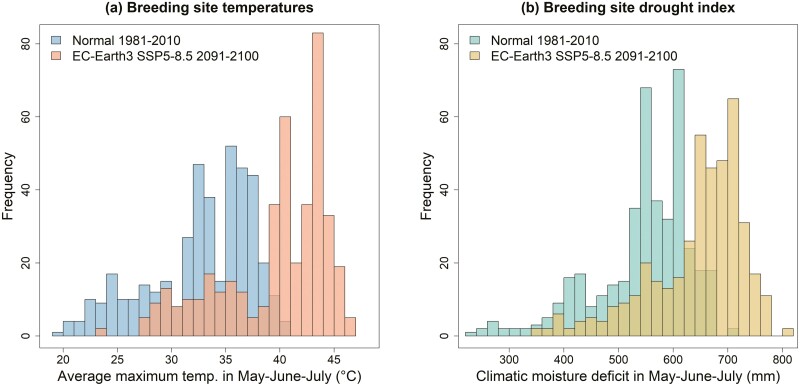
Factors triggering the listing of southwestern willow flycatchers under the ESA included negative effects of invasive species and brood parasites, demographic and genetic effects of small population sizes, and stressors on the migration and wintering grounds (USFWS 2002). However, the most important historical factor driving population declines has been the widespread loss, modification, and fragmentation of riparian breeding habitats, including extensive loss of cottonwood-willow riparian habitats (Unitt 1987; Sogge et al. 2010). Ongoing climate change is compounding the impacts of this historical and current habitat loss. First, increasing summer temperatures in the desert southwest are already impacting bird communities through direct physiological mechanisms such as dehydration and increased cooling costs (Albright et al. 2017; Smith et al. 2017; Riddell et al. 2019). Secondly, the quality of riparian vegetation and habitats continues to be negatively impacted by increasing temperatures and more frequent and intense droughts, along with associated changes in hydrologic regimes (Archer and Predick 2008; Perry et al. 2012; Zhang et al. 2021). In particular, the increasing magnitude, frequency, and duration of drought is directly affecting productivity of desert birds (Saracco et al. 2018), and is linked to nest failure in southwestern willow flycatchers (Paxton et al. 2007). Finally, ecological genomics research has identified the southwestern willow flycatcher as highly vulnerable to climate change due to a substantial mismatch between current adaptive genotypes and those predicted to be needed to adapt to future climate conditions (Ruegg et al. 2018). Future projections of breeding season maximum temperatures and drought conditions at flycatcher breeding sites indicate that these ongoing stressors will only increase in magnitude and severity over the next century (Fig. 1).
Fig. 1.
Frequency of (a) average maximum temperatures and (b) climatic moisture deficit during the southwestern willow flycatcher breeding season (May, June, and July) at 418 breeding sites for 1981 to 2010 (30-year climate normal) and 2091 to 2100, projected using the EC-Earth3 (middle impact) GCM under climate scenario SSP5-8.5 (high emissions scenario).
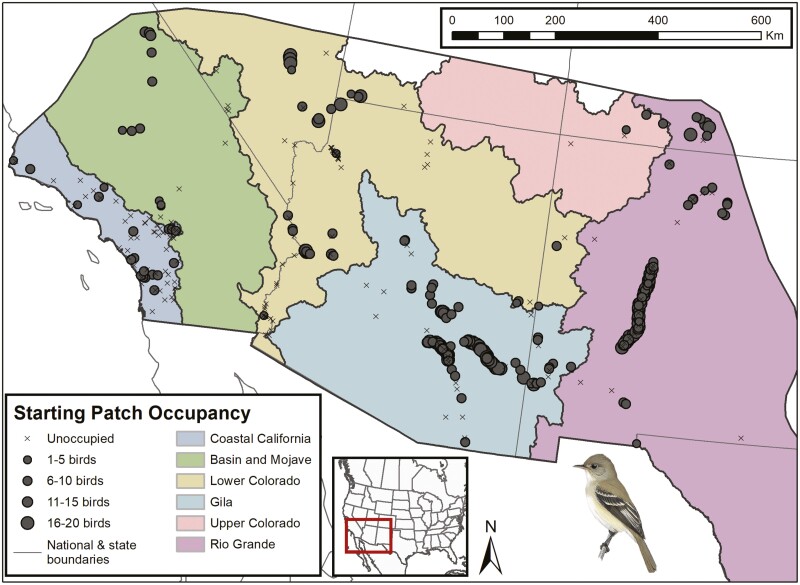
The interaction of these climate-driven impacts with overall small population sizes and metapopulation dynamics of southwestern willow flycatchers, in addition to their expansive and spatially disjunct distribution on the breeding grounds (Fig. 2) makes assessment of extinction risk challenging for this endangered species. The potential for flycatcher populations to adapt to warming and drying conditions from standing genetic variation across this complex landscape adds another challenge to projecting population trajectories (Razgour et al. 2019).
Fig. 2.
Southwestern willow flycatcher breeding grounds in the southwestern United States showing initial patch occupancy of 418 breeding sites used for the simulation study and USFWS Recovery Units (colored polygons representing major watersheds).
In this study, we tackle this complexity using an individual-based, spatially explicit, dynamic eco-evolutionary simulation model parameterized with data from long-term demographic research and a genome-wide ecological genomics study. We first evaluate how existing adaptive genetic variation associated with temperature across the breeding range may facilitate adaptive responses (via selection and dispersal) to increasing summer temperatures due to climate change. We investigate how population trajectories would be impacted if existing evolutionary potential were fixed, with no capacity to evolve in response to rising temperatures. We then assess the compounding impact of drought and warming temperatures on population trajectories through the mechanism of increased nest failure. Finally, we evaluate how rapid action to reverse greenhouse gas emissions would influence population responses and species extinction risk. Our results illustrate the importance, when data are available, of incorporating evolutionary processes in extinction risk assessments for threatened and endangered species. Even with incomplete estimates of evolutionary potential, we can use available information in conjunction with demographic and spatial data to better understand how evolutionary processes may play a role in facilitating population persistence under changing environmental conditions.
Methods
Simulation model
To simulate southwestern willow flycatcher population dynamics, dispersal, and evolutionary potential, we used CDMetaPOP, a dynamic eco-evolutionary simulation program that incorporates spatially explicit modeling of demographic, genetic, and environmental variability (Landguth et al. 2017). CDMetaPOP uses forward-in-time modeling of individual dispersal, demography, and genetic variability as a function of temporally dynamic selection surfaces, habitat variability, and cost distances. Briefly, individuals interact across patches, which are areas sharing common environmental conditions. Within patches, parameters related to class structure (e.g. age, mortality, fecundity) are used to simulate demographic processes, while individual movement between patches is determined by species-relevant dispersal surfaces (e.g. Euclidean distance or an effective distance generated from a resistance surface). Genetic processes are simulated at the individual level, where neutral and/or adaptive population structure emerges as a function of demographic properties, dispersal, and (for adaptive genotypes) natural selection as a function of environmental conditions in patches. In the willow flycatcher case, we focus on the impacts to overall species persistence and occupancy on the breeding grounds as a function of adaptive evolution in response to temporally dynamic environmental conditions and demographic change.
Study system
The simulation extent of our study includes the entire occupied breeding range for southwestern willow flycatchers in the United States (Fig. 2). This includes 6 recovery units representing major watersheds that are used to assess recovery status under the United States Fish and Wildlife Service’s (USFWS) recovery plan for the species (USFWS 2002). Management units (referenced below) are smaller watershed units nested within recovery units. Occupancy of nesting sites across the breeding range is highly variable and can shift over time in response to riparian vegetation dynamics and environmental conditions (Paxton et al. 2007; Theimer et al. 2018). In any given year, as many as half of potential breeding sites will not be occupied by any nesting willow flycatchers (Durst 2017). Based on survey data collected between 1993 and 2012, the largest and most consistent aggregations of willow flycatcher breeding sites include those along the Gila River (Gila Recovery Unit in blue, Fig. 2) and Rio Grande River (Rio Grande Recovery Unit in purple, Fig. 2). By contrast, other breeding sites are more spatially disjunct and have temporally variable occupancy.
Demographic parameterization
We used a modified Leslie matrix (Miller and Ankley 2004) to produce an age-structured logistic model of population growth represented by 6 age classes, hatch year through sixth year. We used age-specific survivorship and seasonal fecundity data derived from a long-term study at 2 locations in central Arizona (Paxton et al. 2007) to parameterize the Leslie matrix (Supplementary Table S1). Survivorship estimates were based on return rates to the breeding grounds, integrating mortality across the annual migratory cycle (Paxton et al. 2007, 2017). Birds matured and were able to reproduce in their second year on the breeding grounds. We calculated the proportion of birds in each age class in our simulations and compared the distributions to empirical data from the Arizona study as a check on the demographic parameters.
Patch attributes
We parameterized 418 patches on the breeding grounds based on compiled breeding site data from Durst (2017) and genetic sampling locations from Ruegg et al. (2018). Due to inconsistency in survey reporting between 1993 and 2012, Durst (2017) defined sites as generalized locations where birds establish a territory, where a territory is an area defended by birds within sites. Sites therefore ranged from small discrete areas of riparian habitat holding a few territories up to long riparian corridors with several hundred territories. For each patch, we set a carrying capacity (K) and initialized the starting number of birds (N0) to reflect compiled data from Durst (2017) on the number of sites per management unit and the number of territories per site, where each territory is assumed to include 2 birds (Fig. 2). In order to more realistically represent habitat availability and dispersal behavior along riparian corridors, sites identified in Durst (2017) with >10 territories were split into multiple patches distributed within 3 km of each other (a distance with a high probability of connectivity, see Dispersal parameterization). Patches were distributed within suitable riparian habitat, identified using an updated (2017) satellite-derived model of southwestern willow flycatcher breeding habitat (Hatten and Paradzick 2003; Hatten 2016). All simulations were initialized with an overall breeding ground carrying capacity of 5,860 individuals and a starting number of birds of 3,258 (roughly equivalent to the total estimated number of individuals species wide as of 2012, Durst 2017). We simulated birds migrating to their wintering grounds in Central America each year by moving all birds to a single patch with sufficient carrying capacity for all individuals (e.g. K = 1,000,000).
Dispersal parameterization
In the following section, we discuss 3 classes of dispersal: patch fidelity, where birds return to the patch they occupied the previous breeding season, local dispersal, where birds move within management units (smaller scale watersheds), and long-distance dispersal, where birds move between recovery units (large scale watersheds, Fig. 2). We used data from the long-term study in central Arizona to parameterize dispersal on the breeding grounds (Paxton et al. 2007). Juvenile birds show low natal site fidelity (i.e. only ~2% return to their natal patch), with most juvenile birds (93%) dispersing locally (mean distance = 9 km, range = 0.3 to 40 km). About 6% of juveniles show long-distance dispersal (mean distance = 214 km, range = 52 to 444 km). By comparison, adult birds show higher patch fidelity when they return to the breeding grounds each season (~33% of adults). About 64% of adult birds exhibit local dispersal (mean distance = 11.3 km, range = 0.3 to 44 km), while long-distance dispersal was less common in adults (3% of birds, average distance = 108 km, range = 59 to 179 km).
In each simulation year, birds travel from the wintering to the breeding grounds, returning to the patch they left in the previous breeding season. When the birds arrive, we apply a dispersal parameterization as follows. Reflecting the empirical data discussed above, only 2% of the returning juvenile birds will show patch fidelity (no dispersal), while 33% of adult birds will show patch fidelity. For birds who do not show patch fidelity (i.e. dispersing individuals), the dispersal location is selected using a random draw from a cost matrix that defines the relative probability of moving a given Euclidean distance between 2 patches. Generally speaking, and reflecting the empirical data presented above, birds that do not exhibit patch fidelity are more likely to disperse locally (i.e. move within management units) than exhibit long-distance dispersal (i.e. move between recovery units). To parameterize the cost matrix representing these probabilities of dispersal, we first set Euclidean distances less than 100 m (representing only 0.2% of dispersal distances in the matrix) to a probability of zero (meaning no dispersal), since these distances are within the average southwestern willow flycatcher territory size of 1 ha. For distances between 100 m and 3 km, we assigned the maximum dispersal probability of 0.95, reflecting a very high probability of local dispersal within 3 km. We then used a truncated normal distribution to set dispersal probabilities for distances between 3 and 44 km (reflecting average local, within-drainage dispersal distances in the empirical data). The truncated normal distribution was calculated in R (v. 4.0.3; R Core Team 2020) and scaled from a dispersal probability of 0.95 (3 km) to 0.001 (44 km), with a mean of 2.6 km (the median of local dispersal distances across juveniles and adults) and SD of 11.4 km (average of SDs of local dispersal distances across juveniles and adults).
At distances greater than 44 km (long-distance dispersal), dispersal probabilities in the cost matrix were much lower as a function of Euclidean distance: probability of 0.001 between 44 and 100 km; probability of 0.0009 between 100 and 200 km; probability of 0.0008 between 200 and 300 km; and probability of 0.0005 for very long distances between 300 and 444 km (with zero probability at distances greater than the maximum recorded movement of 444 km). We calculated the distribution of dispersal distances across juvenile and adult age classes and compared them to empirical data from the Arizona study to evaluate this dispersal parameterization. Finally, we used a simplified approach for the mating cost distance matrix, which is used to identify all possible male mates for a given female in a patch. For distances less than 3 km the probability of selecting an available male as a mate was set to 0.95, while distances between 3 and 50 km used the same truncated normal distribution as for dispersal above; all distances greater than 50 km were set to zero.
Climate data
All climate data were derived from ClimateNA v7.20 (Wang et al. 2016). Three downscaled CMIP6 general circulation models (GCMs) were selected using the CMIP6-NA model ensemble selection tool (Mahony et al. 2022) for western North America: EC-Earth3, GFDL-EDM4, and MRI-ESM2-0. These GCMs meet stringent quality control criteria while spanning the range of climate change projections within Shared Socioeconomic Pathway (SSP) scenarios for western North America (Mahony et al. 2022). EC-Earth3 generally represents a middle impact GCM (Fig. 1), bracketed by GFDL-EDM4 (lower impact, Supplementary Fig. S1), and MRI-ESM2-0 (higher impact, Supplementary Fig. S2). For all applications of climate data, we used the following 2 parameters: average breeding season maximum temperature (the average of maximum temperatures in May, June, and July, Tmax_MJJ) and the sum of Hargreave’s climatic moisture deficit (CMD, a measure of drought) during the breeding season (the sum of CMD for May, June, and July, CMD_MJJ). We used the 1981 and 2010 climate normal as a reference, and annual data from 2011 to 2100 for Tmax_MJJ and CMD_MJJ for the following SSP scenarios: SSP5-8.5, SSP2-4.5, and SSP1-2.6 (IPCC 2021). SSP5-8.5 is a high greenhouse gas emissions scenario with no additional climate policy implemented; CO2 emissions roughly double from current levels by 2050, leading to average global warming of ~4.4 °C (range 3.3 to 5.7 °C) by 2100. SSP2-4.5 is an intermediate greenhouse gas emissions scenario with CO2 emissions remaining at current levels up to 2050 and socioeconomic factors following historical trends; average global warming is ~2.7 °C (range 2.1 to 3.5 °C) by 2100. SSP1-2.6 is a scenario with strong greenhouse gas emissions mitigation; warming stays at ~1.8 °C globally (range 1.3 to 2.4 °C) by 2100 with implied net zero CO2 emissions after 2050, followed by net negative CO2 emissions.
Parameterization of evolutionary potential
In each simulation year, birds return to the breeding grounds and exhibit either site fidelity or dispersal (see Dispersal parameterization). Under climate change scenarios (see Simulation scenarios), each bird will then experience maximum breeding season temperatures (Tmax_MJJ) in their breeding season patch for that year. Depending on the genotype of individual birds at 2 adaptive loci (discussed below), each bird will experience a fitness penalty, in the form of an increased chance of mortality, as a function of Tmax_MJJ in their patch. Thus, evolutionary potential across the species as a whole is mediated by the dispersal of birds on the breeding grounds and the resulting movement of and selection on adaptive alleles at the 2 loci under selection.
To parameterize evolutionary potential, we compiled allele frequency data for 20 candidate adaptive single nucleotide polymorphisms (SNPs) for 15 southwestern willow flycatcher sites from Ruegg et al. (2018). The total sample size was 239 individuals, with sites averaging 15.7 individuals (median = 11, range 4 to 31). Because 7 of the 8 top environmental predictors used to identify candidate loci across the willow flycatcher species complex were related to temperature (Ruegg et al. 2018), we used southwestern willow flycatcher breeding season maximum temperature (Tmax_MJJ) for the 1981 to 2010 climate normal to identify the candidate SNPs with the strongest relationships to biologically relevant temperature across the southwestern willow flycatcher range. We calculated linear models of allele frequencies and Tmax_MJJ at each site for the 20 candidate adaptive loci, retaining 2 SNPs with P values <0.1. We then used these models to predict allele frequencies for the 418 breeding sites at the retained SNPs as a function of Tmax_MJJ for each site (again using the 1981 to 2010 climate normal) to establish starting adaptive allele frequencies for individuals at each location.
We fit a relationship between the count of warm-associated alleles at the 2 selected loci and fitness, based on the observed distribution of alleles across sites. Histograms of allele counts across site temperatures (Supplementary Fig. S3) indicate that high counts of warm-associated alleles are found at all sites but are at higher frequency in the hottest locations. By contrast, low counts of warm-associated alleles are at low frequency across sites. Based on these relationships, we used a Gompertz function (analogous to a logistic curve with an asymmetrical inflection point) to fit a relationship describing a mortality penalty for each genotype (i.e. count of 0, 1, 2, 3, or 4 warm-associated alleles) across current and future maximum breeding season temperatures, using the R package aomisc (Onofri 2020). For each count, we set the lower and upper asymptotes of the curve (lowest and highest fitness penalties) to 0 and 25, respectively. The slope of the curve at the inflection point was set to 0.6, and the temperature value of the inflection point varied by allele count (Supplementary Fig. S4). All genotypes experienced increased mortality (up to the maximum of 25%) at 37 °C, which approximates an upper critical limit for temperature in similarly sized desert songbirds (Owen et al. 2005; Smith et al. 2017). For simulations evaluating no capacity for evolutionary response, we fixed all individuals for either 2 or 3 warm-adapted alleles for the simulation period of 2011 to 2100.
Drought effects on egg mortality
To investigate the interaction of climate-mediated selection and environmental stochasticity on population trajectories, we evaluated the impact of severe drought events on southwestern willow flycatcher nest success. A severe drought in 2002 caused almost complete reproductive failure at Roosevelt Lake sites in central Arizona, with an average female nest failure rate of 94% (95% CI 86% to 100%) (Paxton et al. 2007). Using breeding season CMD (CMD_MJJ) as our indicator of drought (where higher CMD_MJJ indicates more severe drought), we set a threshold for drought effects in our simulations by subtracting the 2002 CMD_MJJ data for the 12 Roosevelt Lake sites from 1981 to 2010 average CMD_MJJ values to produce an index of severe drought. We selected the highest value (59, range: 46 to 59) as our severe drought indicator, which sets a higher (more conservative) threshold for initiating drought effects on nesting success. For each year, if the difference between CMD_MJJ and the 1981 to 2010 average for that site was greater than or equal to 59, we imposed an 86% egg mortality penalty, equivalent to the lower CI of nest failure identified at Roosevelt Lake sites in 2002.
Simulation scenarios
For every simulation scenario, we ran 10 replicates (Monte Carlo, MC), each for 120 years, with 15 years of demographic burn-in (no selection on adaptive genotypes), 15 years of selection burn-in (temperatures in each patch held at the 1981 to 2010 normal), followed by 90 years of climate change (2011 to 2100). We ran the following scenarios:
Demography only: selection module turned off; this scenario was used to evaluate demographic and dispersal parameterization.
Constant temperature (no climate change): selection module turned on, with maximum breeding season temperature held at its 1981 to 2010 value in every patch from 2011 to 2100; this scenario was used as a baseline to compare against climate change scenarios.
Climate change with evolutionary potential: evolutionary change in response to climate change. This scenario was run across 3 GCM projections of climate change, each of which used 2 climate change scenarios (SSP2-4.5 and SSP5-8.5).
Climate change with no evolutionary potential: all populations (species-wide) are fixed for the same number of warm-associated alleles and have no capacity to adapt. We consider both 2 and 3 warm-associated allele fixation scenarios. Uses the same GCMs and SSPs as #3.
Climate change with evolutionary potential interacting with drought effects on nest failure: evolutionary change in response to climate change (as in #3), with additional drought impacts on egg mortality as a function of CMD.
Rapid action to reduce climate change: changes in flycatcher population trajectories under a climate change scenario reflecting substantial and rapid mitigation efforts to keep warming below 1.8 °C (SSP1-2.6). Uses the same 3 GCMs and includes both evolutionary potential alone and evolutionary potential interacting with drought effects on nest failure.
Results
Simulation model evaluation
Using the demography only simulation (selection module turned off), we evaluated demographic and dispersal parameters across MC replicates. Age class structure was similar between the simulations and empirical data collected in central Arizona (Paxton et al. 2007), with a slight bias toward older age classes in the simulations compared with empirical data (Supplementary Table S2). Natal and adult dispersal in the simulations were similar to empirical data from central Arizona, with the largest difference being a 3% underestimate of long-distance dispersal in juvenile birds (Supplementary Table S3). For parameterization of evolutionary potential, we retained 2 climate-associated SNPs (climate_01 and climate_14 from Ruegg et al. 2018) with the strongest relationships (P < 0.10) with average breeding season maximum temperature (Tmax_MJJ) across southwestern willow flycatcher genomic sampling sites (climate_01 R2 = 0.689, P value = 0.0001; climate_14 R2 = 0.241, P value = 0.063).
Climate change with evolutionary potential
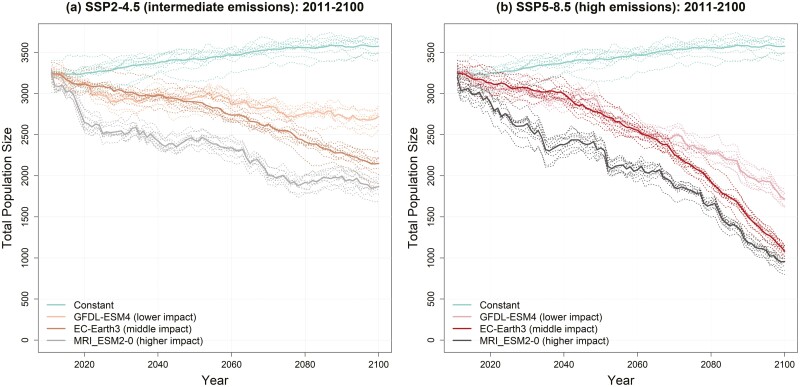
A baseline scenario where maximum breeding season temperatures were held at their 1981 to 2010 levels showed increasing population size trajectories (Fig. 3, constant line), likely due to predictable environmental conditions that facilitated local adaptation and associated increasing fitness and population sizes (Senner et al. 2018). By contrast, both intermediate (Fig. 3a) and high (Fig. 3b) greenhouse gas emission scenarios showed population declines due to fitness impacts of increasing maximum breeding season temperatures over time. As expected based on GCM selection (bracketing the range of GCM projections for the region), there was substantial variability across GCMs within emissions scenarios, including some overlap across scenarios (e.g. population size at 2100 for SSP2-4.5 scenario for MRI_ESM2-0 and SSP5-8.5 scenario for GFDL-ESM4, Fig. 3, Supplementary Table S4). Consistent with no additional climate policies, SSP5-8.5 GCMs show increasing climate change impacts on flycatcher population trends relative to SSP2-4.5 starting around 2050.
Fig. 3.
Population trajectories for southwestern willow flycatchers under constant selection (1981 to 2010 average maximum breeding season temperature) and variable selection due to climate change under (a) an intermediate emissions scenario (SSP2-4.5), and (b) a high emissions scenario (SSP5-8.5). Each scenario shows responses to 3 generalized circulation models (GCMs) that bracket GCM projections across western North America. Solid lines are the mean of 10 MC replicates (dotted lines).
Climate change with no evolutionary potential
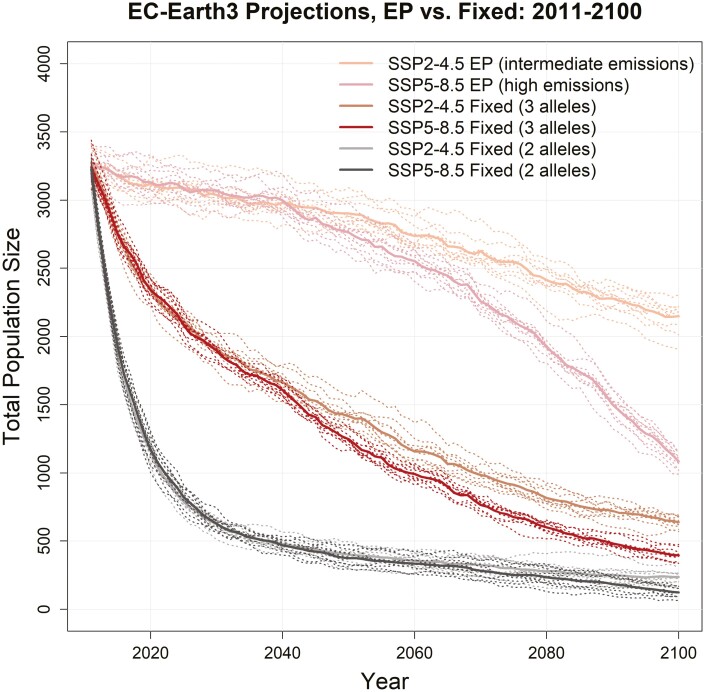
To evaluate how evolutionary potential may mitigate population declines under climate change, we ran 2 sets of simulations where flycatchers were unable to evolve in response to warming conditions. Flycatchers were provided at least some resilience to warming temperatures by fixing populations at either 3 (out of 4) warm-adapted alleles (better case) or 2 warm-adapted alleles (worse case). In both cases, population declines relative to evolutionary potential runs were rapid, with total population sizes falling below 700 birds for EC-Earth3 (middle impact GCM) projections (Fig. 4), and total population declines ranging from 70% to 96% across all fixed (no evolutionary potential) runs (Supplementary Table S4). Population declines in fixed runs for the GFDL-ESM4 (lower impact) GCM were similar to EC-Earth3 (Supplementary Fig. S5a) and were much steeper for the MRI_ESM2-0 (higher impact) GCM (Supplementary Fig. S5b).
Fig. 4.
Population trajectories for southwestern willow flycatchers under variable selection due to climate change under the intermediate (SSP2-4.5) and high (SSP5-8.5) emissions scenarios for the EC-Earth3 (middle impact) GCM. Each scenario compares responses where populations can adapt to warming conditions (“EP” or evolutionary potential runs), and 2 cases where populations cannot adapt and are fixed for either 3 or 2 warm-associated alleles. Solid lines are the mean of 10 MC replicates (dotted lines).
Climate change with evolutionary potential interacting with drought effects
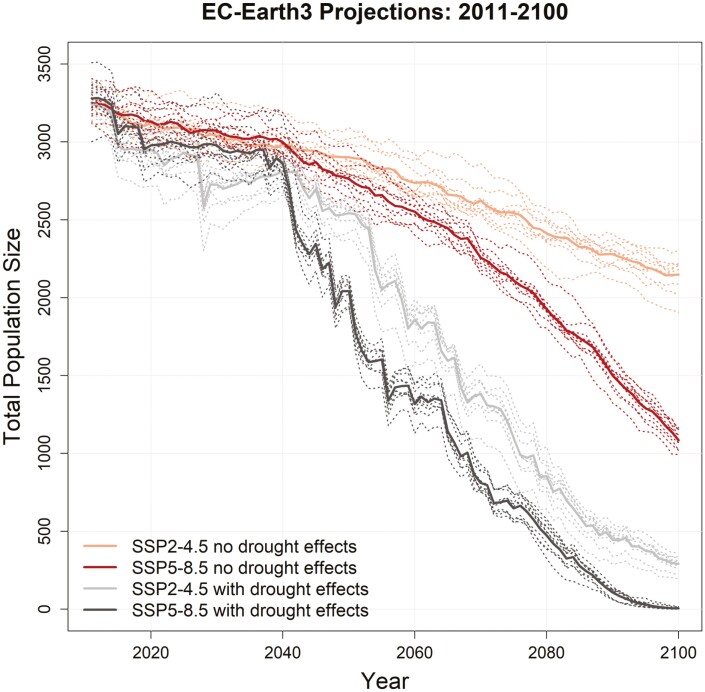
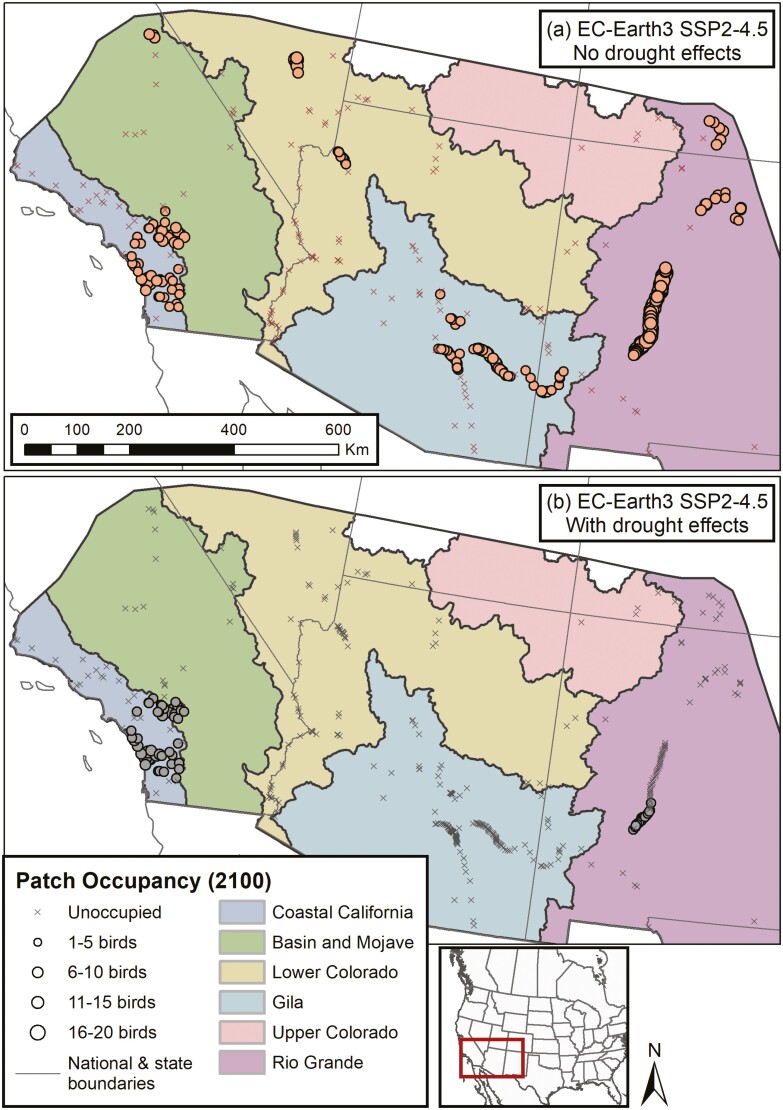
We simulated the interactive effects of rising temperatures and increasing drought from climate change on flycatcher population trajectories. Temperature effects were modeled through selection as a function of adaptive genotype (as above), while drought was modeled through egg mortality (i.e. nest failure) when moisture deficits reached a critical threshold. The impact of drought-induced nest failure on population trajectories (gray lines in Fig. 5 and Supplementary Fig. S6) was pronounced relative to climate change only simulations (red lines) across all climate change scenarios and GCMs. The maximum total population size at 2100 across all drought scenarios was only 491 individuals, with an average across GCMs for SSP2-4.5 (intermediate emissions) of 214 birds, and only 40 birds for SSP5-8.5 (high emissions, Supplementary Table S4). Spatial assessment of population extirpation across simulations with and without drought effects (Fig. 6) illustrates how drought impacts are spatially heterogeneous across the breeding range, with greatest impacts in recovery units that show the largest increases in breeding season temperatures and drought (Supplementary Figs. S7 and S8).
Fig. 5.
Population trajectories for southwestern willow flycatchers under variable selection due to climate change with and without drought effects on egg mortality under intermediate (SSP2-4.5) and high (SSP5-8.5) emissions scenarios for the EC-Earth3 (middle impact) GCM. Solid lines are the mean of 10 MC replicates (dotted lines).
Fig. 6.
Patch occupancy at simulation year 2100 across Recovery Units contrasting climate change scenarios with (a) no drought effect on nesting success and (b) a drought effect on nesting success for the EC-Earth3 (middle impact) GCM under the intermediate emissions scenario (SSP2-4.5).
Rapid action to reduce climate change impacts
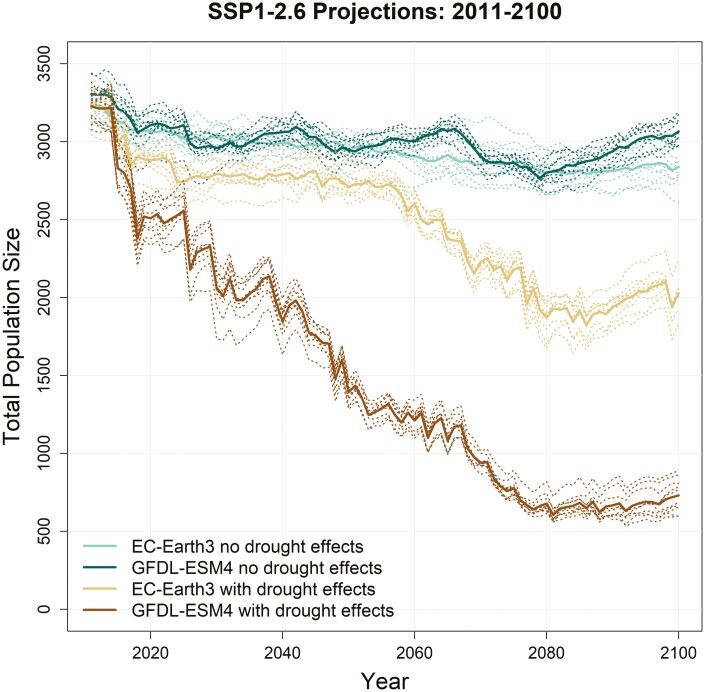
Under very strong greenhouse gas emissions reduction measures (SSP1-2.6), climate-driven threats to southwestern willow flycatchers are substantially reduced, though not completely eliminated. When considering only climate change, populations remained relatively stable (Fig. 7, MRI-ESM2-0 [high emissions scenario] plot provided in Supplementary Fig. S9). Relative to a starting population size of 3,258, the average population decline at the year 2100 across all SSP1-2.6 GCMs was 17%. However, drought impacts were highly variable across GCMs, with overall declines ranging from 38% (EC-Earth3, middle impact) to 97% (MRI_ESM2-0, higher impact, more analogous to results from the intermediate emissions scenario, SSP2-4.5, using the EC-Earth3 [middle impact] GCM).
Fig. 7.
Population trajectories for southwestern willow flycatchers under variable selection due to climate change with and without drought effects on egg mortality under a strong emissions mitigation scenario (SSP1-2.6) for the EC-Earth3 (middle impact) and GFDL-ESM4 (lower impact) GCMs. Solid lines are the mean of 10 MC replicates (dotted lines).
Discussion
Evolutionary potential is an important component of adaptive capacity in response to changing environmental conditions, providing a reservoir of genetic variation that can underlie rapid adaptive responses that increase population-level fitness. Genomic assessments of adaptive variation in at-risk species can provide a proxy for evolutionary potential, however incorporation of these data into extinction risk assessments has been limited to date. This is largely due to 2 major challenges: the difficulty of identifying genotype–fitness relationships in wild populations of at-risk species, and the complexity of integrating ecological and evolutionary drivers of species persistence across spatially and temporally shifting environmental conditions. In the first case, genomic proxies for evolutionary potential are still relatively uncommon in threatened and endangered species, and studies linking candidate adaptive markers to fitness-relevant traits are even more scarce. However, even when we have direct estimates of genotype–fitness relationships, the complexity of selective and genomic landscapes in wild systems makes prediction extremely difficult (e.g. Fournier-Level et al. 2016). In this context, viewing genomic proxies of evolutionary potential as hypotheses to be assessed in a larger context of demographic change, environmental variability, and species vulnerability will be more useful in a management context than a focus on targeted management of specific loci (e.g. Kardos and Shafer 2018; Kardos et al. 2021). This is the approach taken in this study, where empirical genotype–environment relationships and their potential fitness impacts are evaluated across a range of alternative scenarios of climate change, assessing the interaction of selection, gene flow, demographic factors, and spatially heterogeneous environmental change in overall extinction risk trajectories.
In the second case, eco-evolutionary simulation models can provide a comprehensive framework for incorporating genomic proxies of evolutionary potential with other information, such as demographic, dispersal, habitat, climatic, and behavioral data, to evaluate extinction risk under scenarios of environmental change (Forester et al. 2022). For example, a novel study of a single population of brush coral (Acropora hyacinthus) in the South Pacific evaluated extirpation risk under climate change by integrating demographic parameters and genomic data related to thermal tolerance as a proxy for evolutionary potential in response to warming ocean temperatures (Bay et al. 2017b). This study did not include a spatial component however, which is essential to determining how dispersal and resulting gene flow across populations might facilitate evolutionary responses to warming conditions. Here, we build off of this work by evaluating species-level extinction risk across temporally and spatially dynamic environments in the endangered southwestern willow flycatcher using an individual-based, spatially explicit, eco-evolutionary simulation model. Efforts such as these that link evolutionary potential and extinction risk with a focus on conservation applications are still uncommon, making this an important area for additional development in at-risk species (reviewed in Forester et al. 2022).
In our simulations, evolutionary potential across the species as a whole was mediated by the dispersal of birds on the breeding grounds and the resulting movement of and selection on adaptive alleles across the landscape. We found that this capacity to adapt in response to climate change mitigated extinction risk relative to scenarios where populations could not evolve. Even when all individuals were fixed for 3 of 4 adaptive alleles (providing some resilience to warming conditions), population declines were rapid and precipitous (e.g. Fig. 4). By contrast, the capacity to evolve in response to temperature change mitigated population declines even under high greenhouse gas emissions scenarios (e.g. Fig. 3). Despite rapid adaptation, however, declines were significant across intermediate and high emissions scenarios, averaging 31% for SSP2-4.5 and 62% for SSP5-8.5. Breeding season maximum temperatures under these emissions scenarios (e.g. Fig. 1) frequently surpassed the ~37 °C upper critical limit for small desert passerines like southwestern willow flycatchers (Owen et al. 2005; Smith et al. 2017), potentially contributing to increased mortality through both direct physiological mechanisms (Albright et al. 2017; Riddell et al. 2019) and indirect trade-offs associated with behavioral thermoregulation (Ricklefs and Hainsworth 1968; Austin 1976).
When considering the compounding effects of drought on nest failure, population declines were even more significant, averaging 93% for the intermediate (SSP2-4.5) and 99% for the high (SSP5-8.5) emissions scenarios (Supplementary Table S4). These overall population declines provide an incomplete assessment of flycatcher extinction risk, however, as climate-driven impacts play out across a demographic and spatial template of small population sizes and metapopulation dynamics. For example, under climate change without drought effects, many isolated breeding sites remained occupied due to maintenance of larger core populations (e.g. in the Rio Grande and Gila Recovery Units) that act as sources for the colonization of more isolated patches (Fig. 6a, USFWS 2002). However, stochastic nest failure due to drought reduces population sizes at these larger core sites, resulting in disrupted metapopulation dynamics and extirpation of isolated breeding sites (e.g. Fig. 6b). Extirpation of large populations in the central portion of the breeding grounds will also effectively eliminate gene flow that otherwise facilitates the movement of adaptive alleles across the breeding range. These results illustrate how interacting demographic, spatial, and genetic impacts of climate change can substantially alter population vulnerability.
Previous ecological genomics research identified the southwestern willow flycatcher as highly vulnerable to climate change due to a mismatch between current genotypes and future climatic conditions (Ruegg et al. 2018). Our results largely agree with the spatial distribution of vulnerability identified in that study. For example, we found that the southern part of the Coastal California Recovery Unit, identified as a low-mismatch region, persisted in most simulation scenarios, including climate change coupled with drought effects (e.g. Fig. 6b). By contrast, areas of the Rio Grande Recovery Unit, largely identified as high-mismatch regions, showed reduced occupancy, though not complete extirpation under most climate change simulation scenarios (e.g. Fig. 6a). Population persistence in the face of high mismatch between current genotypes and future climate is likely due to the movement of adaptive alleles into this recovery unit facilitated by dispersal that can be explicitly modeled in our simulations. This ability to incorporate evolutionary change due to gene flow illustrates an advantage over static mismatch assessments (Capblancq et al. 2020; Rellstab et al. 2021) when using a spatially and temporally dynamic framework for assessing climate change vulnerability.
Our results also provide evidence that flycatcher declines can be substantially reduced by immediate and deep reductions in greenhouse gas emissions (i.e. emissions scenario SSP1-2.6, Fig. 7). While it is currently still feasible to implement the mitigation efforts required to meet emissions reductions targets as laid out in SSP1-2.6, it will require major transitions in the energy, industry, and land use sectors, as well as accelerated investment to meet sustainable development goals (IPCC 2022). Given high uncertainty in the likelihood of meeting these targets, additional mitigation and conservation efforts will be needed to ensure the recovery of the southwestern willow flycatcher. Though not included in this study, vegetation restoration efforts coupled with water conservation policies could buffer populations under climate change by increasing the quality of available breeding habitat and providing thermal refugia to reduce exposure during extreme heat events (Wolf et al. 1996; Albright et al. 2017). Our results indicate that targeting restoration and habitat rehabilitation efforts in recovery units that will maintain lower temperatures and drought indices under climate change will be most effective. However, these efforts must be implemented under a broader consideration of maintaining sufficient connectivity across the breeding range to facilitate metapopulation dynamics and gene flow for evolutionary adaptation and the maintenance of genome-wide genetic diversity (USFWS 2002). Future simulations will focus on testing the relative effectiveness of these and other conservation actions beyond climate change mitigation.
The importance of genetic factors in both the extinction risk and recovery of threatened and endangered species has been debated for decades (e.g. Lande 1988; Schemske et al. 1994; Bürger and Lynch 1995; Soulé and Mills 1998; Frankham 2005). However, novel insights into neutral and adaptive components of genetic diversity in at-risk species, driven by the increasing ease of genomic data production and decreased sequencing costs, have provided growing evidence for the importance of genetic factors, such as inbreeding depression and loss of evolutionary potential, in the decline of at-risk species (Kardos et al. 2016, 2021; Allendorf 2017). These data have also provided new opportunities for the integration of genetic factors, including evolutionary potential, into extinction risk assessments, creating a frontier in eco-evolutionary research and applied conservation science (Forester et al. 2022). Uncertainty about the rate and magnitude of anthropogenic change, the complexity of multivariate environmental change, novel selection pressures, and incomplete knowledge of species responses means that eco-evolutionary simulation models will be most valuable as problem-solving tools, rather than predictors of a highly uncertain future (Starfield 1997). Mechanistic models that focus on fine-scale interactions are especially robust to simulating alternative scenarios of global change or conservation actions, and are therefore valuable for understanding relative scenario outcomes and quantifying outcome uncertainty (Johnston et al. 2019). As we continue to gain insight into the genomic landscape of evolutionary potential in at-risk species, the integration of these data into models of extinction risk will provide more comprehensive insights into the adaptive capacity of threatened and endangered species, and its importance in conservation planning to mitigate declines and facilitate population recovery.
Supplementary Material
Acknowledgments
We are grateful for helpful discussions with Eben Paxton, Clark Rushing, Scott Durst, Mary Whitfield, Barbara Kus, and Greg Beatty.
Contributor Information
Brenna R Forester, Department of Biology, Colorado State University, Fort Collins, CO, United States.
Casey C Day, Computational Ecology Lab, School of Public and Community Health Sciences, University of Montana, Missoula, MT, United States.
Kristen Ruegg, Department of Biology, Colorado State University, Fort Collins, CO, United States.
Erin L Landguth, Computational Ecology Lab, School of Public and Community Health Sciences, University of Montana, Missoula, MT, United States; Center for Population Health Research, School of Public and Community Health Sciences, University of Montana, Missoula, MT, United States.
Funding
This work was supported by a David H. Smith Conservation Research Fellowship to BRF and a National Science Foundation CAREER award to KR (#1942313). Eco-evolutionary modeling work was supported in part by Seattle City Light (ELL, CCD) and National Institute of General Medical Sciences of the National Institutes of Health grant P20GM130418 (ELL).
Data availability
Genomic data used in this study are archived in the NCBI Sequence Read Archive under accession number PRJNA453612 (https://www.ncbi.nlm.nih.gov/bioproject/453612). Code for data processing used in Ruegg et al. 2018 is available at https://github.com/eriqande/ruegg-et-al-wifl-genoscape.
References
- Albright TP, Mutiibwa D, Gerson AR, Smith EK, Talbot WA, O’Neill JJ, McKechnie AE, Wolf BO.. Mapping evaporative water loss in desert passerines reveals an expanding threat of lethal dehydration. Proc Natl Acad Sci USA. 2017;114:2283–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allendorf FW. Genetics and the conservation of natural populations: allozymes to genomes. Mol Ecol. 2017;26:420–430. [DOI] [PubMed] [Google Scholar]
- Archer SR, Predick KI.. Climate change and ecosystems of the southwestern United States. Rangelands. 2008;30:23–28. [Google Scholar]
- Austin GT. Behavioral adaptations of the Verdin to the desert. Auk. 1976;93:245–262. [Google Scholar]
- Bach LA, Thomsen R, Pertoldi C, Loeschcke V.. Kin competition and the evolution of dispersal in an individual-based model. Ecol Model. 2006;192:658–666. [Google Scholar]
- Bay RA, Rose N, Barrett R, Bernatchez L, Ghalambor CK, Lasky JR, Brem RB, Palumbi SR, Ralph P.. Predicting responses to contemporary environmental change using evolutionary response architectures. Am Nat. 2017a:189:463–473. [DOI] [PubMed] [Google Scholar]
- Bay RA, Rose NH, Logan CA, Palumbi SR.. Genomic models predict successful coral adaptation if future ocean warming rates are reduced. Sci Adv. 2017b:3:e1701413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger R, Lynch M.. Evolution and extinction in a changing environment: a quantitative-genetic analysis. Evolution. 1995;49:151–163. [DOI] [PubMed] [Google Scholar]
- Capblancq T, Fitzpatrick MC, Bay RA, Exposito-Alonso M, Keller SR.. Genomic prediction of (mal)adaptation across current and future climatic landscapes. Annu Rev Ecol Evol Syst. 2020;51:245–269. [Google Scholar]
- Chevin L-M, Lande R, Mace GM.. Adaptation, plasticity, and extinction in a changing environment: towards a predictive theory. PLoS Biol. 2010;8:e1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson TP, Jackson ST, House JI, Prentice IC, Mace GM.. Beyond predictions: biodiversity conservation in a changing climate. Science. 2011;332:53–58. [DOI] [PubMed] [Google Scholar]
- DeAngelis DL, Mooij WM.. Individual-based modeling of ecological and evolutionary processes. Annu Rev Ecol Evol Syst. 2005;36:147–168. [Google Scholar]
- Durst SL. Southwestern willow flycatcher breeding site and territory summary—2012. Albuquerque (NM): U.S. Fish and Wildlife Service; 2017. p. 42. [Google Scholar]
- Foden WB, Young BE, Akçakaya HR, Garcia RA, Hoffmann AA, Stein BA, Thomas CD, Wheatley CJ, Bickford D, Carr JA, et al. Climate change vulnerability assessment of species. WIREs Clim Change. 2019;10:e551. [Google Scholar]
- Forester BR, Beever EA, Darst C, Szymanski J, Funk WC.. Linking evolutionary potential to extinction risk: applications and future directions. Front Ecol Environ. 2022; 20:507–515. doi: 10.1002/fee.2552 [DOI] [Google Scholar]
- Fournier-Level A, Perry EO, Wang JA, Braun PT, Migneault A, Cooper MD, Metcalf CJ, Schmitt J.. Predicting the evolutionary dynamics of seasonal adaptation to novel climates in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2016;113:E2812–E2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R. Genetics and extinction. Biol Conserv. 2005;126:131–140. [Google Scholar]
- Funk WC, Forester BR, Converse SJ, Darst C, Morey S.. Improving conservation policy with genomics: a guide to integrating adaptive potential into U.S. Endangered Species Act decisions for conservation practitioners and geneticists. Conserv Genet. 2019;20:115–134. [Google Scholar]
- Hatten JR. A satellite model of Southwestern Willow Flycatcher (Empidonax traillii extimus) breeding habitat and a simulation of potential effects of tamarisk leaf beetles (Diorhabda spp.), southwestern United States. Reston (VA): U.S. Geological Survey; 2016. p. 98. [Google Scholar]
- Hatten JR, Paradzick CE.. A multiscaled model of southwestern willow flycatcher breeding habitat. J Wildl Manag. 2003;67:774–788. [Google Scholar]
- IPCC. Climate Change 2021: The physical science basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge (UK): Cambridge University Press; 2021. [Google Scholar]
- IPCC. Climate Change 2022: Mitigation of climate change. Contribution of Working Group III to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge (UK): Cambridge University Press; 2022. [Google Scholar]
- Johnston AS, Boyd RJ, Watson JW, Paul A, Evans LC, Gardner EL, Boult VL.. Predicting population responses to environmental change from individual-level mechanisms: towards a standardized mechanistic approach. Proc R Soc B. 2019;286:20191916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardos M, Armstrong EE, Fitzpatrick SW, Hauser S, Hedrick PW, Miller JM, Tallmon DA, Funk WC.. The crucial role of genome-wide genetic variation in conservation. PNAS. 2021;118:e2104642118. doi: 10.1073/pnas.2104642118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardos M, Shafer ABA.. The peril of gene-targeted conservation. Trends Ecol Evol. 2018;33:827–839. [DOI] [PubMed] [Google Scholar]
- Kardos M, Taylor HR, Ellegren H, Luikart G, Allendorf FW.. Genomics advances the study of inbreeding depression in the wild. Evol Appl. 2016;9:1205–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. Genetics and demography in biological conservation. Science. 1988;241:1455–1460. [DOI] [PubMed] [Google Scholar]
- Landguth EL, Bearlin A, Day CC, Dunham J.. CDMetaPOP: an individual-based, eco-evolutionary model for spatially explicit simulation of landscape demogenetics. Methods Ecol Evol. 2017;8:4–11. [Google Scholar]
- Mahony, C. R., Wang, T., Hamann, A., & Cannon, A. J.. A global climate model ensemble for downscaled monthly climate normals over North America. Int J Climatol. 2022;42:5871–5891. doi: 10.1002/joc.7566 [DOI] [Google Scholar]
- Miller DH, Ankley GT.. Modeling impacts on populations: fathead minnow (Pimephales promelas) exposure to the endocrine disruptor 17β-trenbolone as a case study. Ecotoxicol Environ Saf. 2004;59:1–9. [DOI] [PubMed] [Google Scholar]
- Onofri A. The broken bridge between biologists and statisticians: a blog and R package. 2020. Accessed April 6, 2022. https://www.statforbiology.com
- Owen JC, Sogge MK, Kern MD.. Habitat and sex differences in physiological condition of breeding southwestern willow flycatchers (Empidonax traillii extimus). Auk. 2005;122:1261–1270. [Google Scholar]
- Paxton EH, Durst SL, Sogge MK, Koronkiewicz TJ, Paxton KL.. Survivorship across the annual cycle of a migratory passerine, the willow flycatcher. J Avian Biol. 2017;48:1126–1131. [Google Scholar]
- Paxton EH, Sogge MK, Durst SL, Theimer TC, Hatten JR.. The ecology of the Southwestern Willow Flycatcher in central Arizona—a 10-year synthesis report. Open-File Report 2007-1381. Reston (VA):U.S. Geological Survey; 2007. p. 149. [Google Scholar]
- Perry LG, Andersen DC, Reynolds LV, Nelson SM, Shafroth PB.. Vulnerability of riparian ecosystems to elevated CO2 and climate change in arid and semiarid western North America. Glob Change Biol. 2012;18:821–842. [Google Scholar]
- Pierson JC, Beissinger SR, Bragg JG, Coates DJ, Oostermeijer JGB, Sunnucks P, Schumaker NH, Trotter MV, Young AG.. Incorporating evolutionary processes into population viability models. Conserv Biol. 2015;29:755–764. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2020. Accessed April 15, 2022. https://www.R-project.org/ [Google Scholar]
- Radchuk V, Kramer-Schadt S, Grimm V.. Transferability of mechanistic ecological models is about emergence. Trends Ecol Evol. 2019;34:487–488. [DOI] [PubMed] [Google Scholar]
- Razgour O, Forester BR, Taggart JB, Bekaert M, Juste J, Ibáñez C, Puechmaille SJ, Novella-Fernandez R, Alberdi A, Manel S.. Considering adaptive genetic variation in climate change vulnerability assessment reduces species range loss projections. PNAS. 2019;20:605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rellstab C, Dauphin B, Exposito-Alonso M.. Prospects and limitations of genomic offset in conservation management. Evol Appl. 2021;14:1202–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklefs RE, Hainsworth FR.. Temperature dependent behavior of the cactus wren. Ecology. 1968;49:227–233. [Google Scholar]
- Riddell EA, Iknayan KJ, Wolf BO, Sinervo B, Beissinger SR.. Cooling requirements fueled the collapse of a desert bird community from climate change. PNAS. 2019;116:21609–21615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegg K, Anderson EC, Somveille M, Bay RA, Whitfield M, Paxton EH, Smith TB.. Linking climate niches across seasons to assess population vulnerability in a migratory bird. Glob Change Biol. 2021;27:3519–3531. [DOI] [PubMed] [Google Scholar]
- Ruegg K, Bay RA, Anderson EC, Saracco JF, Harrigan RJ, Whitfield M, Paxton EH, Smith TB.. Ecological genomics predicts climate vulnerability in an endangered southwestern songbird. Ecol Lett. 2018;21:1085–1096. [DOI] [PubMed] [Google Scholar]
- Sage RF. Global change biology: a primer. Glob Change Biol. 2020;26:3–30. [DOI] [PubMed] [Google Scholar]
- Saracco JF, Fettig SM, San Miguel GL, Mehlman DW, Thompson BE, Albert SK.. Avian demographic responses to drought and fire: a community-level perspective. Ecol Appl. 2018;28:1773–1781. [DOI] [PubMed] [Google Scholar]
- Schemske DW, Husband BC, Ruckelshaus MH, Goodwillie C, Parker IM, Bishop JG.. Evaluating approaches to the conservation of rare and endangered plants. Ecology. 1994;75:585–606. [Google Scholar]
- Senner NR, Stager M, Cheviron ZA.. Spatial and temporal heterogeneity in climate change limits species’ dispersal capabilities and adaptive potential. Ecography. 2018;41:1428–1440. [Google Scholar]
- Smith EK, O’Neill JJ, Gerson AR, McKechnie AE, Wolf BO.. Avian thermoregulation in the heat: resting metabolism, evaporative cooling and heat tolerance in Sonoran Desert songbirds. J Exp Biol. 2017;220:3290–3300. [DOI] [PubMed] [Google Scholar]
- Sogge MK, Ahlers D, and Sferra SJ.. A natural history summary and survey protocol for the southwestern willow flycatcher. U.S. Geological Survey Techniques and Methods 2A-10. Reston (VA):U.S. Geological Survey; 2010. p. 38. [Google Scholar]
- Soulé ME, Mills LS.. No need to isolate genetics. Science. 1998;282:1658–1659. [Google Scholar]
- Starfield AM. A pragmatic approach to modeling for wildlife management. J Wildl Manag. 1997;61:261–270. [Google Scholar]
- Theimer TC, Sogge MK, Paxton EH.. Patch age since disturbance drives patch dynamics for flycatchers breeding in both reservoir and riverine habitat. Ecosphere. 2018;9:e02425. [Google Scholar]
- Unitt P. Empidonax traillii extimus: an endangered subspecies. West Birds. 1987;18:137–162. [Google Scholar]
- USFWS. Final rule determining endangered status for the Southwestern Willow Flycatcher. Fed Reg. 1995;60:10694–10715. [Google Scholar]
- USFWS. Southwestern Willow Flycatcher recovery plan. Albuquerque (NM):Region 2 USFWS; 2002. p. 229. [Google Scholar]
- Wang T, Hamann A, Spittlehouse D, Carroll C.. Locally downscaled and spatially customizable climate data for historical and future periods for North America. PLoS One. 2016;11:e0156720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf BO, Wooden KM, Walsberg GE.. The use of thermal refugia by two small desert birds. Condor. 1996;98:424–428. [Google Scholar]
- Xuereb A, Rougemont Q, Tiffin P, Xue H, Phifer-Rixey M.. Individual-based eco-evolutionary models for understanding adaptation in changing seas. Proc R Soc B Biol Sci. 2021;288:20212006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Biederman JA, Dannenberg MP, Yan D, Reed SC, Smith WK.. Five decades of observed daily precipitation reveal longer and more variable drought events across much of the western United States. Geophys Res Lett. 2021;48:e2020GL092293. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic data used in this study are archived in the NCBI Sequence Read Archive under accession number PRJNA453612 (https://www.ncbi.nlm.nih.gov/bioproject/453612). Code for data processing used in Ruegg et al. 2018 is available at https://github.com/eriqande/ruegg-et-al-wifl-genoscape.