Abstract
Three prevalent SARS-CoV-2 variants of concern (VOCs) emerged and caused epidemic waves. It is essential to uncover advantageous mutations that cause the high transmissibility of VOCs. However, viral mutations are tightly linked, so traditional population genetic methods, including machine learning–based methods, cannot reliably detect mutations conferring a fitness advantage. In this study, we developed an approach based on the sequential occurrence order of mutations and the accelerated furcation rate in the pandemic-scale phylogenomic tree. We analyzed 3,777,753 high-quality SARS-CoV-2 genomic sequences and the epidemiology metadata using the Coronavirus GenBrowser. We found that two noncoding mutations at the same position (g.a28271−/u) may be crucial to the high transmissibility of Alpha, Delta, and Omicron VOCs although the noncoding mutations alone cannot increase viral transmissibility. Both mutations cause an A-to-U change at the core position −3 of the Kozak sequence of the N gene and significantly reduce the protein expression ratio of ORF9b to N. Using a convergent evolutionary analysis, we found that g.a28271−/u, S:p.P681H/R, and N:p.R203K/M occur independently on three VOC lineages, suggesting that coordinated changes of S, N, and ORF9b proteins are crucial to high viral transmissibility. Our results provide new insights into high viral transmissibility co-modulated by advantageous noncoding and nonsynonymous changes.
Keywords: SARS-CoV-2, VOCs, noncoding mutation, Kozak sequence, transmissibility, a28271−/u
Introduction
Three prevalent SARS-CoV-2 variants of concern (VOCs) emerged in 2 years and caused the epidemic waves that have drawn the most attentions of people (fig. 1A). These VOCs were named as the Alpha, Delta, and Omicron VOCs. The Alpha VOC, also known as SARS-CoV-2 lineage B.1.1.7, is a variant first detected in the United Kingdom in September 2020 (Rambaut et al. 2020) and has higher transmissibility than preexisting variants (Volz et al. 2021). Its high transmissibility remains similar across different ages, genders, and socioeconomic strata (Davies et al. 2021). The Delta VOC was first identified in India in October 2020 (WHO 2021) and has higher transmissibility than the Alpha VOC (Campbell et al. 2021; Dhar et al. 2021; Meng et al. 2021). The Omicron VOC was first detected in South Africa in late November 2021 (Viana et al. 2022). Then, it spread rapidly into 76 countries in <1 month (Hui et al. 2022; Suzuki et al. 2022). Therefore, it is essential to uncover advantageous mutations that cause the high transmissibility of VOCs (Kang et al. 2021; Obermeyer et al. 2022).
Fig. 1.
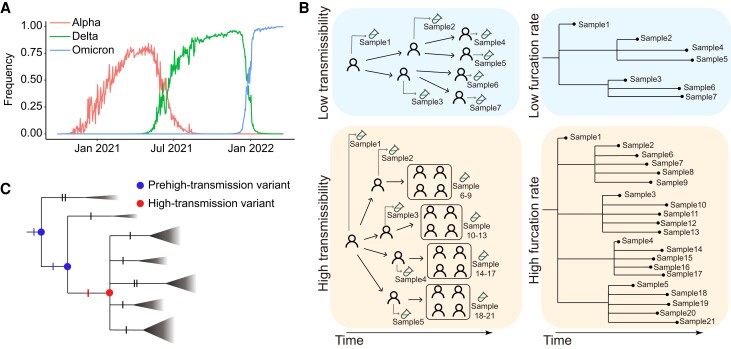
COVID-19 waves and a model for the sudden appearance of a VOC. (A) Three COVID-19 waves caused by the Alpha, Delta, and Omicron VOCs. (B) Schematic diagram of the relationship between furcation rate and transmissibility. (C) Schematic diagram of a model for the sudden appearance of a high transmissibility variant. The triangles represent collapsed clades, and the size of the triangle represents the size of the clade. Each notch of branches represents a mutation.
All three VOCs carry a large number of mutations. Compared with the reference genomic sequence of SARS-CoV-2 (GenBank accession number: NC_045512.2) (Wu et al. 2020), the Alpha, Delta, Omicron BA.1, and Omicron BA.2 variants have at least 21, 27, 52, and 51 amino acid change mutations, respectively (WHO 2021), together with a number of noncoding mutations (Yu, Yang, et al. 2022). These viral mutations are tightly linked to each other because the virus lacks recombination that commonly occurs in sexual species during the stage of meiosis. Recombination in coronaviruses occurs by a copy-choice mechanism that requires coinfection of the same cell within a host by two distinct viral strains (Simon-Loriere and Holmes 2011). Only a limited number of recombination events have been revealed after analyzing millions of SARS-CoV-2 genomes (Jackson et al. 2021; VanInsberghe et al. 2021; Turakhia et al. 2022). Without sufficient recombination events to break the linkage between neutral and selected loci, traditional population genetic methods, such as selective sweeps and machine learning–based methods (Kaplan et al. 1989; Kim and Stephan 2002; Li and Stephan 2005, 2006; Lin et al. 2011; Schrider and Kern 2018; Stephan 2019), are not suitable for SARS-CoV-2 and may not reliably detect advantageous mutations causing the high transmissibility of Alpha, Delta, and Omicron VOCs. Therefore, a novel approach is urgently needed.
In this study, we proposed a pandemic-scale phylogenomic approach based on the sequential occurrence order of mutations and the accelerated furcation rate to detect mutations crucial to viral high transmissibility (fig. 1B and C). Let us assume that the high transmissibility of a VOC is due to the emergence of an advantageous haplotype with multiple crucial mutations. The last (i.e., the most recent) crucial mutation significantly increases viral transmissibility and accelerates the furcation rate in the phylogenetic tree. To identify the crucial coding and noncoding mutations for three VOCs, the Coronavirus GenBrowser (CGB) was used to analyze a tip-dated tree with 3,777,753 high-quality SARS-CoV-2 genomic sequences and the associated epidemiology metadata. The CGB is a free platform offering a panoramic vision of the transmission and evolution of SARS-CoV-2 (Yu, Yang, et al. 2022; Yu, Zhu, et al. 2022). The analysis revealed that two noncoding mutations at the position of 28,271 (g.a28271−/u) might be crucial to the high transmissibility of three VOCs. Both mutations caused an A-to-U change in the Kozak translation initiation context of the N gene (Kozak 1987) and reduced protein expression of the ORF9b gene, which shares the same transcript as N gene. This caused a reduced ratio of ORF9b to N proteins. Further analyses revealed that the noncoding mutations alone could not cause high viral transmissibility. The noncoding mutations may comodulate the viral transmissibility together with other coding mutations, such as S:p.P681H/R and N:p.R203K/M.
Results
Accelerated Furcation Rate due to High Transmissibility
Different evolutionary tree shapes reflect different transmission patterns (Colijn and Gardy 2014). Furcation, as the process of lineage splitting (Oakley et al. 2007), may reflect transmissibility. An infected patient represents at least one transmission event, and the strain collected from the patient forms a new furcation in an evolutionary tree (De Maio et al. 2018). If the tree has n leaves, it must have (n − 1) furcation events. Since the CGB evolutionary tree is tip dated and the date of each internal node has been estimated, the furcation rate along each lineage was measured by the number of furcation events per day (see the Materials and methods). A variant with high transmissibility could cause more infections and is detectable by examining its accelerated rate of furcation during a long period of time (fig. 1B). Superspreader transmission can also result in a multifurcation node although it only affects the furcation of the node, indicating that the high furcation rate due to superspreader transmission only lasts for a very short period of time. Thus, superspreader transmission can be easily distinguished from cases with high transmissibility.
Let us consider a SARS-CoV-2 VOC as a multimutation haplotype conferring a fitness advantage. To understand the sudden appearance of its high transmissibility, the fitness advantage of VOC is assumed to be gained as soon as the last crucial mutation occurs (fig. 1C). In this model, a variant with a subset of the crucial mutations was named as the prehigh-transmission variant, the furcation rate of which remains low. As soon as the last crucial mutation occurs, the advantageous haplotype emerges, and the viral transmissibility increases suddenly, causing an instantaneously accelerated furcation rate in the phylogenetic tree.
The furcation rate is difficult to compare among different time points since it is affected by viral genome sequencing capacities that frequently change over time. Because of similar reasons, the furcation rate should not be used to compare among different countries/regions. To avoid these confounding factors in the following analyses, the furcation rate was taken to compare between prehigh-transmission and high-transmission variants collected from the same geographic region during the same time period.
Crucial Noncoding Deletion in Alpha
The evolution of the Alpha lineage was examined using the CGB (Yu, Yang, et al. 2022). The sequential occurrence of Alpha characteristic mutations is shown (fig. 2A). To confirm the evolutionary path of the Alpha VOC, the VENAS (Ling et al. 2022) was applied to obtain an evolution network of SARS-CoV-2 major haplotypes (supplementary fig. S1 and table S1, Supplementary Material online). The VENAS results are consistent with the CGB evolutionary tree.
Fig. 2.
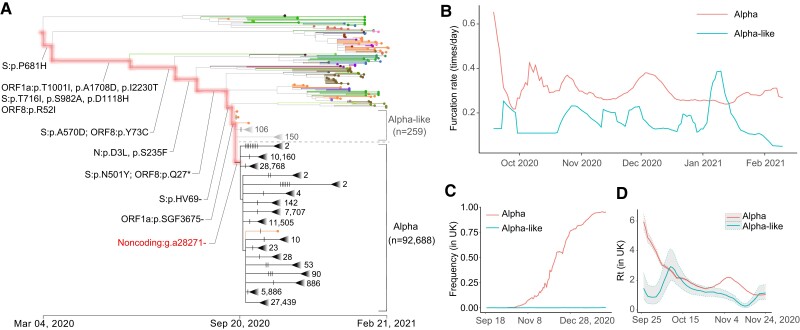
High transmissibility of Alpha compared with Alpha like. (A) CGB evolutionary tree of SARS-CoV-2 lineage Alpha. The analysis was performed on 400,051 high-quality SARS-CoV-2 genomic sequences (data version “data.2021-03-06”) using the CGB (Yu, Yang, et al. 2022). The searchable CGB ID of the internal node with g.a28271− is CGB84017.91425, assigned by the CGB binary nomenclature system. The triangles represent collapsed clades. The size of collapsed clades was labeled after the collapse triangle. The mutations on the highlighted branches were labeled, and the mutations on the Alpha and Alpha-like lineages were marked by notches. The number of Alpha-like strains is 259, and the number of Alpha strains is 92,688. (B) The furcation rate of Alpha and Alpha like. To remove the factor of sequencing capacity, the furcation rates should be compared during the same period of time. (C) The frequency trajectory of Alpha and Alpha like in the United Kingdom. (D) The Rt of Alpha and Alpha like in the United Kingdom. The shadowed area represents the 95% confidence interval.
Most of the nodes were bifurcating () on the Alpha evolutionary path where Alpha characteristic mutations occurred (fig. 2A). However, multifurcating nodes were frequently observed after a noncoding deletion (g.a28271−) occurred. To examine whether the noncoding deletion is crucial to the high transmissibility of the Alpha VOC, sister lineages without or with the mutation were compared (fig. 2A). Two sister lineages were defined as Alpha-like and Alpha according to whether strains were derived from an ancestral node without or with the noncoding deletion. Viral strains in the Alpha-like lineage generally lack the noncoding deletion but carry all Alpha characteristic amino acid mutations found in previous studies (Chand et al. 2020), including all Alpha spike mutations (Gobeil et al. 2021). Viral strains in the Alpha lineage generally carry the noncoding deletion and all Alpha characteristic amino acid mutations. Therefore, the noncoding deletion was studied by comparing two sister lineages since the two lineages have the same genetic background and circulate in the same geographic area during the same time period.
The furcation rate was calculated for two sister lineages (fig. 2B). After considering the factor of sequencing capacity, the furcation rate of the Alpha lineage is generally higher than that of the Alpha-like lineage. Such higher furcation rate lasted for 5 months, indicating that the accelerated furcation rate of the Alpha lineage is not due to superspreader transmission. The distribution of mutation rate was also examined in the two lineages. The two distributions were largely overlapped, indicating that the accelerated furcation rate of the Alpha lineage should not be due to the difference in mutation rate between the two lineages (supplementary fig. S2A, Supplementary Material online).
The frequencies of Alpha and Alpha-like strains were calculated (fig. 2C). It was found that, within 6 months, the global frequency of Alpha strains increased rapidly to over 80% while that of Alpha-like strains remained no >1%. Moreover, the reproduction numbers (Rt) of Alpha and Alpha-like strains in the United Kingdom were estimated using the EpiEstim (Cori et al. 2013). The Rt of Alpha strains is significantly larger than that of Alpha-like strains (4.79 vs. 1.00; P value < 2.2 ×10−16) during the first examined week (September 26 to October 2, 2020) (fig. 2D). The results remain similar during the next two months (2.20 vs. 1.25; P value < 2.2 ×10−16). These results indicated that the Alpha lineage has a transmission advantage over the Alpha like. Therefore, the noncoding deletion g.a28271− may be crucial for viral transmissibility.
The higher transmissibility of the Alpha lineage causes the number of descendants in the Alpha lineage to be highly significantly larger than that in the Alpha-like lineage (n = 92,688 vs. 259, P value < 4.9 ×10−324). Pooling data of viral sequences from different countries are likely to be biased due to complex differences in sampling, such as viral genome sequencing capacities or the anticontagion policies of the targeted countries during the pandemic (Hsiang et al. 2020). To address this problem, the numbers of descendants in the Alpha and Alpha-like lineages were compared for different countries and continents, that is, England (76,871 vs. 27), Spain (712 vs. 30), Switzerland (1,332 vs. 8), Germany (570 vs. 2), the United States (1,028 vs. 8), Australia (58 vs. 1), South America (22 vs. 1), Africa (86 vs. 1), and Asia (642 vs. 3). The transmissibility of strains with or without the noncoding deletion is significantly unequal (supplementary table S2, Supplementary Material online, P value ≤ 2.74 ×10−6). The highly significant differences were also observed in 10 more countries, such as India and Italy. Moreover, the same conclusion holds when considering different gender and age groups (supplementary tables S3 and S4, Supplementary Material online). Therefore, the significant difference in transmissibility between the Alpha and Alpha-like lineages has been observed, which is likely due to the noncoding deletion g.a28271−.
Crucial Noncoding Deletion in Delta
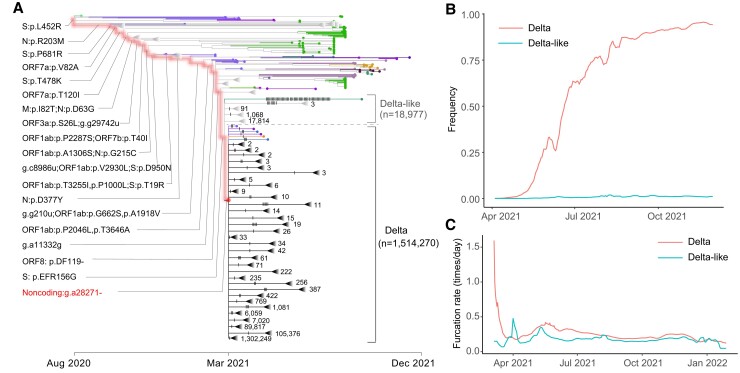
The noncoding deletion (g.a28271−) was found to occur independently in the Delta VOC (fig. 3A). A recent CGB datum (data.2022-04-14) was used to analyze this VOC since it became prevalent globally in the middle of 2021. It was observed that the number of furcation events increased as soon as the noncoding deletion (g.a28271−) occurred on the Delta lineage (fig. 3A), similar to the case observed in the Alpha lineage. Similarly, viral strains generally lack the noncoding deletion but carry all Delta characteristic amino acid mutations were defined as Delta like (fig. 3A). The furcation rate of the Delta lineage is also generally higher than that of the Delta-like lineage (fig. 3C). The distribution of mutation rate was also largely overlapped between the Delta and Delta-like lineages (supplementary fig. S2B, Supplementary Material online). Therefore, the accelerated furcation rate of the Delta lineage is not due to the change in mutation rate.
Fig. 3.
High transmissibility of Delta compared with Delta like. (A) CGB evolutionary tree of SARS-CoV-2 lineage Delta. The analysis was performed on 3,777,753 high-quality SARS-CoV-2 genomic sequences (data version “data.2022-04-14”) using the CGB (Yu, Yang, et al. 2022). The searchable CGB ID of the internal node with g.a28271− is CGB531065.736525, assigned by the CGB binary nomenclature system. The triangles represent collapsed clades. The size of collapsed clades was labeled after the collapse triangle. The mutations on the highlighted branches were labeled, and the mutations on the Delta and Delta-like lineages were marked by notches. The number of Delta-like strains is 18,977, and the number of Delta strains is 1,514,270. (B) The frequency trajectory of Delta and Delta like. (C) The furcation rate of Delta and Delta like. To remove the factor of sequencing capacity, the furcation rates should be compared during the same period of time.
The global frequencies of Delta and Delta-like strains were calculated (fig. 3B). It was found that the global frequency of Delta strains increased rapidly to over 90% within six months while that of Delta-like strains remained ∼1%. The higher transmissibility of the Delta lineage also caused the number of its descendants highly significantly larger than that of the Delta-like lineage (n = 1,514,270 vs. 18,977, P value ≤ 4.9 ×10−324). All these results indicate the modulation of noncoding deletion g.a28271− on viral transmissibility.
Crucial Noncoding Mutation in Omicron
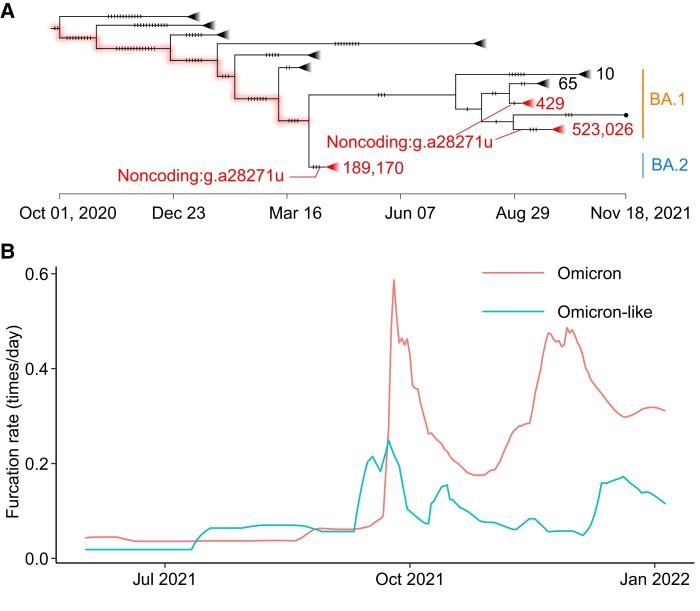
The Omicron VOC was first reported in South Africa in November 2021 (Viana et al. 2022), and then multiple sublineages evolved, such as BA.1 and BA.2. It was found that most of Omicron strains carry another noncoding mutation at position 28271 (g.a28271u) (fig. 4A). The mutation occurred independently on two sublineages (BA.1 and BA.2). Therefore, to examine the effects of the mutation on the viral transmissibility, the furcation rate was calculated for the lineages with and without the mutation. To keep the consistency, the term Omicron stands for strains with the mutation g.a28271u, whereas Omicron like indicates strains without the mutation on the considered lineage. It was found that the Omicron lineage has a higher furcation rate than the Omicron-like lineage, indicating that the mutation g.a28271u plays a key role in viral transmissibility. Unlike the cases observed in the Alpha and Delta VOCs, a delayed increase of furcation rate was observed in Omicron (fig. 4B), indicating that g.a28271u may coact with other mutations to form a haplotype conferring a fitness advantage. It was found again that the distribution of mutation rate overlapped between the Omicron and Omicron-like lineages (supplementary fig. S2C, Supplementary Material online).
Fig. 4.
High transmissibility of Omicron compared with Omicron like. (A) CGB evolutionary tree of SARS-CoV-2 lineage Omicron. The analysis was performed on 3,777,753 high-quality SARS-CoV-2 genomic sequences (data version “data.2022-04-14”) using the CGB (Yu, Yang, et al. 2022). The CGB IDs of the internal nodes with g.a28271u are CGB3145480.3251694 and CGB2437061.2958631 for BA.1 and CGB3053091.3265861 for BA.2. The triangles represent collapsed clades. The size of collapsed clades was labeled after the collapse triangle. The mutations were marked by notches. (B) The furcation rate of Omicron and Omicron like. To remove the factor of sequencing capacity, the furcation rates should be compared during the same period.
Noncoding Mutations at the Core Kozak Site of N Gene
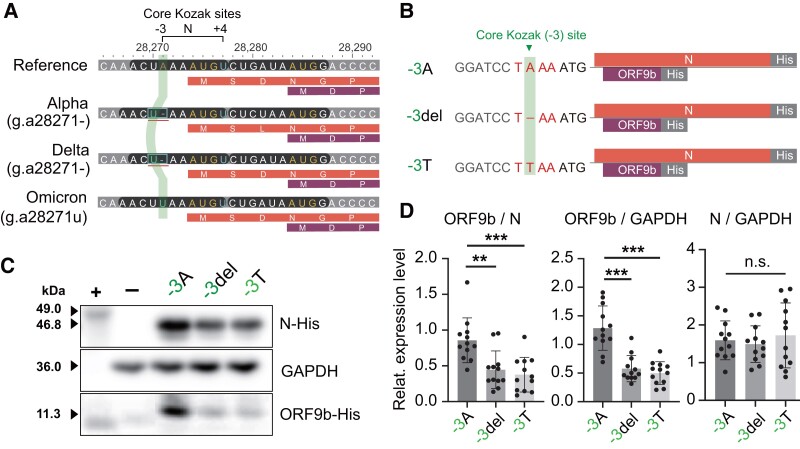
The base 28271 is located at the third base upstream of the start codon of the N gene. It is the core Kozak site of the gene (fig. 5A). The Kozak sequence is a short sequence around the start codon and functions as the protein translation initiation site in higher eukaryotes (Kozak 1987; Xu et al. 2010). Nucleotides in each site of the Kozak sequence influence the translational efficiency, especially the positions −3 and +4. The position 28271 is the core position −3 of the Kozak sequence of the N gene. The g.a28271− deletion makes u28,270 slip one base and changes the Kozak context of the N gene from a suboptimal Kozak context (A at −3 and U at +4) to an undesirable one (U at −3 and U at +4). The mutation g.a28271u produces a similar Kozak-related change by mutating the −3A to −3U. Therefore, both noncoding mutations may have a similar effect on the translation initiation of N protein.
Fig. 5.
Noncoding A-to-U Kozak changes modulate the translation of N and ORF9b proteins. (A) Mutations at the genomic position 28271 change the core Kozak (−3) site of the N gene. The two Kozak positions −3 and +4 have the dominant influence on the protein translation. The translucent vertical strip indicates the Kozak −3 site of the N gene on different strains. For each variant, the first bar is the nucleotide sequence, the second and the third bars are the protein sequences of dual-coding genes (N and ORF9b). (B) Schematic diagram of three cDNA constructs. The construct −3A represents the wild-type (i.e., the reference), the construct −3del represents the g.a28271− mutation, and the construct −3T represents the g.a28271u mutation. (C) Western blotting of N and ORF9b proteins in cells transfected with different expression constructs (−3A, −3del, and −3T). Purified His-tagged ATP4B antigen (49.0 kDa) was used as the positive control (+) and cells transfected with pcDNA3.1-EGFP vector as the negative control (−). (D) Relative abundance of N and ORF9b proteins from N gene transcripts.
The transcript of the N gene is dual coding (Xu et al. 2010). There is an alternative open reading frame (ORF) within the N gene. The alternative ORF encodes the ORF9b protein, and the protein was found to be translated via a leaky ribosomal scanning mechanism in SARS-CoV (Xu et al. 2009). To investigate whether the mutations (g.a28271−/u) have a similar effect on the expression of N and ORF9b proteins, HEK-293FT cells were transfected with different recombinant plasmids carrying the ancestral allele (−3A) or Kozak mutations (−3del and −3T) along with the downstream ORFs tagged with six-histidine codon (fig. 5B). After 48-h transfection, the expression levels of C-terminal His-tagged N and ORF9b proteins were analyzed using western blotting (fig. 5C).
When the core site −3A of the Kozak sequence of the N gene was changed to −3U, the abundance of N protein remained unchanged (fig. 5D). However, the expression level of ORF9b protein was significantly reduced (P value < 0.0001). The expression ratio of ORF9b to N proteins was also reduced (P value = 0.0024 and 0.0004 for g.a28271− and g.a28271u). Overall, these results showed that both g.a28271− and g.a28271u have similar effects on the expression of ORF9b and N proteins.
Noncoding Mutations Alone Do Not Increase Viral Transmissibility
The noncoding mutations g.a28271− and g.a28271u were found to be crucial for the high transmissibility of SARS-CoV-2. It was investigated whether these two mutations function alone or co-act with other crucial mutations. The noncoding mutations occurred independently many times due to recurrent mutations or recombination in the evolution of SARS-CoV-2 (supplementary fig. S3, Supplementary Material online). The frequency trajectories of the two mutations were checked when Alpha, Delta, and Omicron strains were excluded. The first sample with high sequencing quality that carries one of the mutations was collected in Germany on June 16, 2020 (EPI_ISL_732537 with g.a28271−). However, the frequency of the two mutations remained <1% in the next 6 months (supplementary fig. S4, Supplementary Material online), indicating that the two mutations alone are not advantageous. Therefore, the mutated core Kozak site of the N gene requires interaction with other mutated loci to increase the viral transmissibility, consistent with the cases observed in the Omicron VOC.
Convergent Evolution of Alpha, Delta, and Omicron
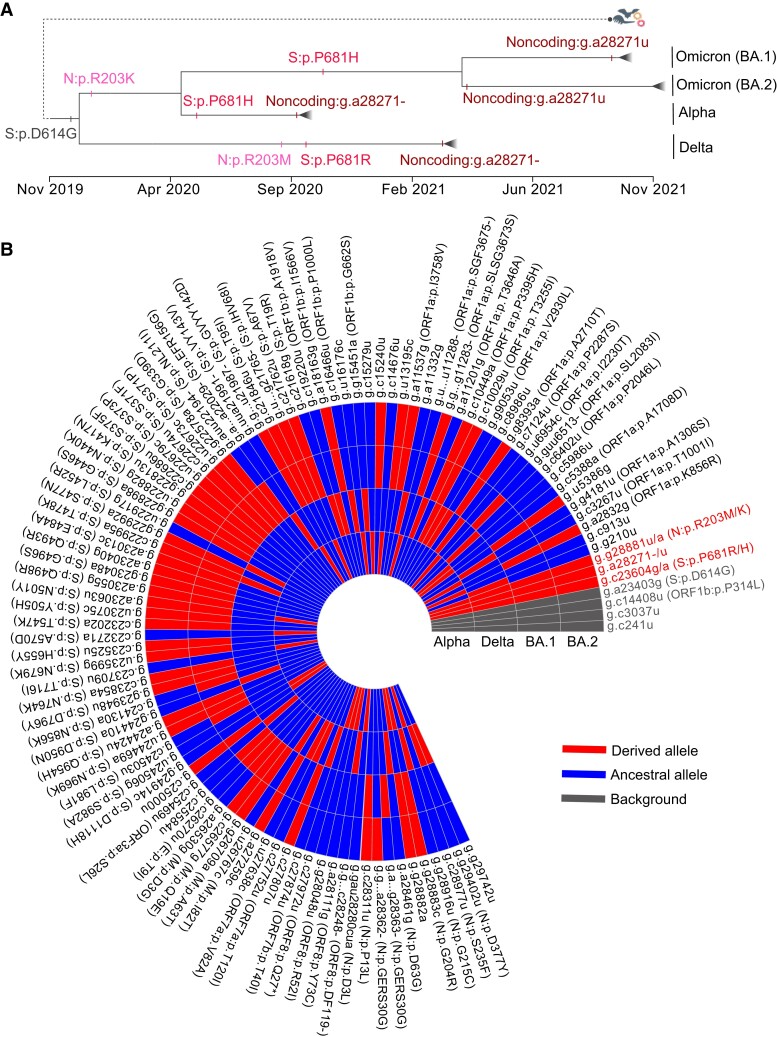
To identify mutations that interact with g.a28271−/u, the convergent evolution of the Alpha, Delta, and Omicron VOCs may provide important clues. The three VOCs diverged in January 2020 and evolved independently in the background of the spike D614G substitution (fig. 6A). Thus, the mutations of the three VOCs were screened in the CGB phylogenetic tree. The two noncoding mutations (g.a28271− and g.a28271u) evolved convergently in the three VOCs. Moreover, another two nonsynonymous mutations (S:p.P681H/R and N:p.R203K/M) were found to occur independently on the lineages (fig. 6B).
Fig. 6.
The convergent evolution of the Alpha, Delta, and Omicron VOCs. (A) Schematic diagram of convergent evolution of Alpha, Delta, and Omicron VOCs. Mutations under convergent evolution are labeled, and the occurring time of mutations was inferred using the CGB (Yu, Yang, et al. 2022). (B) Heatmap showing the presence of mutations in Alpha, Delta, and Omicron (BA.1 and BA.2) VOCs. The heatmap was prepared using the eGPS software (Yu et al. 2019).
Position 681 in the spike protein is immediately upstream of the furin cleavage site (Huang, Yang, et al. 2020; Zuckerman et al. 2021). The mutation S:p.P681H occurred independently in the Alpha and Omicron VOCs (fig. 6A). It has been proposed that the mutated spike (681H) increases furin cleavage (Lista et al. 2022; Lubinski, Fernandes, et al. 2022). The mutation S:p.P681R, which occurred in the Delta VOC, also has the same effect (Liu et al. 2022; Lubinski, Frazier, et al. 2022; Saito et al. 2022). It has also been found that P681H is essential for resistance to IFN-β(Lista et al. 2022), and P681R enhances viral replication and confers the pseudovirus relatively resistant to neutralizing antibodies (Liu et al. 2022; Saito et al. 2022).
The N protein is required for replication and packaging. Position 203 is in the linker region of the N protein. The mutation N:p.R203K occurred on the common ancestor of Alpha and Omicron VOCs in February 2020 (fig. 6A). Its adjacent mutation N:G204R also occurs on the two VOCs (fig. 6B). It has been found that the double-mutated strain (203K/204R) has a replication advantage over the wildtype (R203/G204) (Wu et al. 2021) and increase the expression of subgenomic RNA (Leary et al. 2021). Similarly, the mutation N:p.R203M occurred in September 2020, before the Delta VOC emerged (fig. 6A). The mutation N:p.R203M can enhance replication in lung epithelial cells (Syed Abdullah et al. 2021). Therefore, both the mutations N:p.R203K/M may be crucial for replication.
Discussion
It is highly challenging to map mutations conferring a fitness advantage along the SARS-CoV-2 genome using traditional population genetic methods, such as selective sweep (Kaplan et al. 1989; Kim and Stephan 2002; Li and Stephan 2005, 2006; Stephan 2019). These methods assume that a new mutation with selective advantage is surrounded by neutral loci. All loci are partially linked to the locus with an advantageous mutation. Due to the hitchhiking effect, a locally reduced genetic diversity can be observed (i.e., selective sweep). However, recombination events in SARS-CoV-2 are generally insufficient (Simon-Loriere and Holmes 2011; Jackson et al. 2021; VanInsberghe et al. 2021; Turakhia et al. 2022) to break the linkage between neutral and selected loci. Therefore, selective sweep-based methods may reveal false candidates of positive selection and should not be applied to analyze SARS-CoV-2 genomes. Machine learning–based approaches (Lin et al. 2011; Schrider and Kern 2018) also face the same challenge because of the same reason.
In this study, we analyzed the sequential occurrence order of mutations and the change of furcation rate in the pandemic-scale phylogenetic tree of SARS-CoV-2 to map advantageous mutations. We found that the noncoding g.a28271− mutation may play a crucial role in the high transmissibility of the Alpha and Delta VOCs (figs. 2 and 3) and g.a28271u for the Omicron VOC (fig. 4). The two mutations caused the same A-to-U change at the core Kozak (−3) site of the N gene, which may affect the translation efficiency of the downstream N and ORF9b genes. It was also found that the amount of ORF9b protein and the expression ratio of ORF9b to N proteins were reduced in both constructs (−3del for g.a28271− and −3T for g.a28271u) while the amount of N protein remained nearly unchanged (fig. 5D). Moreover, we normalized peptide intensity of the N gene to the level of its sgRNA using proteomic data (Thorne et al. 2022) and found no significant difference in the expression efficiency of N protein between the Alpha and an early strain VIC at 10-h postinfection (Alpha/VIC = 1.18; P value = 0.052). Thorne et al. (2022) also speculated that the change in Kozak sequence may affect the N and ORF9b translation, consistent with our findings. Overall, these pieces of evidence suggest that the relative abundance between N and ORF9b proteins may be crucial to high viral transmissibility.
Our analyses also revealed that the noncoding mutations alone do not increase viral transmissibility. Convergent evolution of high transmissibility in the three VOCs may be due to the recurrent advantageous noncoding mutations, S:p.P681H/R, and N:p.R203K/M (fig. 6). It has been found that S:p.P681H/R mutation facilitates cleavage of the spike protein and may enhance viral entry (Mohammad et al. 2021; Saito et al. 2022) and that N:p.R203K/M enhances viral replication (Syed Abdullah et al. 2021; Wu et al. 2021). As ORF9b plays multiple roles in modulating the host immune response to aid virus replication and spread (Zandi et al. 2022), it was proposed that g.a28271−/u and N:p.R203K/M may comodulate the abundance of ORF9b during viral infection and facilitate viral transmissibility. The coevolved mutations related to host entry, virus replication, and host immune response in three VOCs indicated that the emergence of a variant conferring the fitness advantage requires coordinated changes in stages of virus life cycle, consistent with the established hypothesis (Markov et al. 2023).
Several new questions also rise, such as whether N:p.R203K/M affects the expression of N and ORF9b proteins, how g.a28271−/u and N:p.R203K/M regulate protein expression in vivo, and how mutations involved in different virus life stages co-modulate viral transmissibility. All these questions need to be further explored. Overall, our findings provide new insights into the evolution of SARS-CoV-2 and the modulation of the S, N, and ORF9b genes in viral transmissibility.
Materials and Methods
Data Sources
The annotated evolutionary tree and evolutionary network data were obtained from the CGB (Yu, Yang, et al. 2022) and VENAS (Ling et al. 2022). All sequence data of SARS-CoV-2 were obtained from the 2019nCoVR database (Gong et al. 2020; Zhao et al. 2020), which is an integrated resource based on Global Initiative on Sharing All Influenza Data (GISAID) (Elbe and Buckland-Merrett 2017; Shu and McCauley 2017), National Center for Biotechnology Information (NCBI) GenBank (Sayers et al. 2021), China National GeneBank DataBase (CNGBdb) (Chen et al. 2020), the Genome Warehouse (GWH) (Zhang et al. 2020), and the National Microbiology Data Center (NMDC, https://nmdc.cn/). Two data versions of CGB (“data.2021-03-06” and “data.2022-04-14”) were used in this study, which contains 400,051 and 3,777,753 genomic sequences, respectively.
Furcation Rate to Evaluate Virus Transmissibility
To evaluate virus transmissibility, it was proposed to calculate the furcation rate (FR) of nodes in a considered phylogenetic lineage (fig. 1C). Furcation rate of the i-th node () can be calculated using the following equation:
where is the number of child nodes and is the branch length of the ith node (with time in units of days). Since represents the furcation rate during the spanning time period of the branch of the i-th node, the furcation rate at time t () was calculated by averaging the of branches that overlap with the point of time t in the considered phylogenetic lineage. The estimated furcation rate over time was smoothed by a sliding window of size equal to 7 days.
The Fitness Effect of Mutations in Improving Viral Transmissibility
To examine the fitness effect of a mutation, it was tested whether two sister lineages with or without the mutation have similar viral transmissibility (fig. 1C). When two sister lineages circulate in the same geographic area during the same period of time, it is expected that the one with higher transmissibility infects more persons and thus relatively more samples of this lineage are collected. Therefore, the null hypothesis is that the two sister lineages have the same viral transmissibility. The number of genetic differences between the two sister lineages is two mutations in the example shown in figure 1C. The alternative hypothesis is that the lineage with the mutation (red) has higher viral transmissibility than the sister lineage without the mutation. The binomial probability was applied to test the null hypothesis and the test was one tailed. Because the three VOCs occurred at different time points, the analysis for Alpha was based on the data version “data.2021-03-06” (), and the analysis of Delta and Omicron was based on the data version “data.2022-04-14” (), where n is the number of viral strains. The two data sets were obtained from the CGB (Yu, Yang, et al. 2022).
Calculation of Rt
Because sequenced strains were randomly sampled from infected patients, the frequency of each variant at time t was estimated from the CGB data sets. The number of new infections of a variant at time t () was approximated using the following equation:
where is the frequency of the variant at time t and is the total number of new infections at time t. in the United Kingdom was downloaded from the UK Coronavirus Dashboard (https://coronavirus.data.gov.uk/).
Moreover, because of the low frequency of the Alpha-like variant, it was smoothed to reduce the effects of random errors before calculating Rt. The smoothing window size is 35 days. Rt was then calculated using the EpiEstim (Cori et al. 2013) with a mean (±SD) serial interval of 7.5 ± 3.4 days (Li et al. 2020) and time steps of 7 days. A permutation test with 1,000 permutations was used to test whether the Rt of the two lineages is significantly different.
Recurrent g.a28271−/u mutations
The noncoding mutations (g.a28271− and g.a28271u) were searched to examine these recurrent mutations in the CGB evolutionary tree (Yu, Yang, et al. 2022). The string “A28271−” was used to search g.a28271− and “A28271T” to search g.a28271u. To present the reappearance patterns of mutations, the data version “data.2021-04-14” () of the CGB (Yu, Yang, et al. 2022) was used. These data were also used to examine the frequency trajectory of g.a28271−/u when the Alpha, Delta, and Omicron lineages were excluded.
Design, Construction, and the Expression of Recombinant Plasmid
The cDNA of the N and ORF9b gene was synthesized according to the modified reference genomic sequence of SARS-CoV-2 (GenBank accession number: NC_045512) (fig. 5B). To investigate the function of mutants, the synthesized DNA included the Kozak context (extending to the −4 position) of the N gene. Three DNA fragments were obtained: the wild-type (−3A), g.a28271− mutant (−3del), and g.a28271u mutant (−3 T). To detect the expression of the two proteins, a hexa-histidine (6xHis) was tagged at their C-termini. The synthesized DNA was then cloned into the mammalian expression vector pcDNA3.1, using cloning sites BamHI/XbaI. The three constructs are shown in figure 5B. The constructs were prepared commercially by Sangon Biotech (Shanghai, China). A pcDNA3.1-EGFP vector (HG-VPH0002) was purchased from HonorGene (Changsha, China).
Cell Culture and Transfections
HEK-293FT cells were donated by Dr Yu Sun. Cells were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM; Sangon Biotech) with 10% fetal bovine serum (FBS; Gibco) and 0.1 mM MEM Non-Essential Amino Acids (NEAA; Gibco) at 37 °C in a humidified atmosphere containing 10% CO2. Cells were subcultured every 2 or 3 days using 0.06% Trypsin-EDTA solution. For transfection, HEK-293FT cells in exponential growth phase were seeded into 6-well plates at a density of 6 × 105 cells/well 24 h before transfection. ViaFect Transfection Reagent (Promega) was applied for transient transfection according to the manufacturer's instructions. Three hundred microliters per well transfected cell culture was prepared by addition of 18 µl ViaFect transfection reagent to Opti-MEM Reduced Serum Medium (Gibco) containing 3 µg of plasmid DNA. Transfection complexes were incubated at room temperature for 12 min and added dropwise to each well. The cells were further incubated for 48 h. Empty vector cells, cells transfected with pcDNA3.1-EGFP vector, and empty vector cells treated with transfection reagent were all used as negative controls.
Immunoblotting Assay
After 48 h of transfection, the amount of N and ORF9b proteins was investigated in different transfected cell lines (fig. 5B). Total protein samples from each transfected cell line (n = 3, two wells for each n) were separated by SDS–PAGE and transferred onto a 0.2-μm polyvinylidene difluoride (PVDF) membrane (Millipore, USA) using an electroblotting apparatus. The PVDF membranes were then blocked in TBST blocking buffer and probed with anti-His mAb (cat. no. D191001, Sangon Biotech, Shanghai, China) or anti-GAPDH (Proteintech, USA) antibodies (supplementary table S5, Supplementary Material online). Following thorough washing, the blots were incubated with appropriate secondary antibodies (supplementary table S5, Supplementary Material online) and visualized using a chemiluminescent horseradish peroxidase (HRP) kit (Millipore, USA). Purified His-tagged ATP4B antigen (49.0 kDa, cat. no. D620298, Sangon Biotech, Shanghai, China) was used as the positive control. Images of the blots were captured using ImageQuant LAS-4000 (Amersham Biosciences, USA), and the density of all protein bands detected was quantified using ImageQuant TL software (version 7.0, Amersham Biosciences, USA). The relative quantity of a protein was determined by dividing the density value of the protein band on the western blot by the density value of the GAPDH protein band in the same lane (Huang, Liao, et al. 2020; Li et al. 2022). Four independent runs of Western blotting were performed for each cell line sample (supplementary fig. S5, Supplementary Material online). Statistical significance between different transfected cell lines was determined using one-way analysis of variance (ANOVA) with post hoc Holm–Šídák test. P value < 0.01 was considered statistically significant.
Protein Structure Prediction
Since the N and ORF9b proteins are translated from the dual-coding transcript of N gene, the ORF9b 6xHis-tagged changes 101 M of the N protein to 101TSSPSPL (fig. 5B). To study whether the inserted short peptide affects the stability of the N protein, the N-terminal domain of the N protein was first examined (PDB code: 7CDZ) (Peng et al. 2020). The N-terminal domain structure was depicted using Chimera 1.16 (Pettersen et al. 2004). It was then found that the 101 M of the N protein locates in the middle of a loop (supplementary fig. S6, Supplementary Material online). Moreover, the structure of the altered N-terminal domain (i.e., with 101TSSPSPL) was predicted using AlphaFold2 (Jumper et al. 2021). The predicted structure remains very similar to that of the unchanged N protein (supplementary fig. S6, Supplementary Material online), indicating that the ORF9b 6xHis-tagged should not affect the stability of the N protein.
Supplementary Material
Acknowledgments
We thank Wolfgang Stephan for discussing on the issue of selective sweep, Yu Sun for his donation of HEK-293FT cells, and the researchers who generated and deposited sequence data of SARS-CoV-2 in GISAID, GenBank, CNGBdb, GWH, and NMDC. This work was supported by grants from the National Key Research and Development Project (Grant Nos. 2022YFF1203202, 2021YFC0863300, and 2020YFC0845900), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDPB17), the National Natural Science Foundation of China (Grant Nos. 31100273, 32270674, and 91531306), and the Shandong Academician Workstation Program #170401 (to G.P.Z.).
Contributor Information
Jianing Yang, Bio-Med Big Data Center, CAS Key Laboratory of Computational Biology, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai, China.
Yingmin Cui, Key Laboratory of Brain Functional Genomics of Ministry of Education, School of Life Science, East China Normal University, Shanghai, China.
Dalang Yu, Bio-Med Big Data Center, CAS Key Laboratory of Computational Biology, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai, China.
Guoqing Zhang, Bio-Med Big Data Center, CAS Key Laboratory of Computational Biology, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai, China.
Ruifang Cao, Bio-Med Big Data Center, CAS Key Laboratory of Computational Biology, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai, China.
Zhili Gu, Bio-Med Big Data Center, CAS Key Laboratory of Computational Biology, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai, China.
Guangyi Dai, Bio-Med Big Data Center, CAS Key Laboratory of Computational Biology, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai, China.
Xiaoxian Wu, Key Laboratory of Synthetic Biology, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, China.
Yunchao Ling, Bio-Med Big Data Center, CAS Key Laboratory of Computational Biology, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai, China.
Chunyan Yi, Laboratory of Cell Biology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, Shanghai, China.
Xiaoyu Sun, Laboratory of Cell Biology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, Shanghai, China.
Bing Sun, Laboratory of Cell Biology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, Shanghai, China.
Xin Lin, Bio-Med Big Data Center, CAS Key Laboratory of Computational Biology, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai, China.
Yu Zhang, Key Laboratory of Synthetic Biology, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, China.
Guo-Ping Zhao, Bio-Med Big Data Center, CAS Key Laboratory of Computational Biology, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai, China; Key Laboratory of Synthetic Biology, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, China; School of Life and Health Sciences, Hangzhou Institute for Advanced Study, University of Chinese Academy of Sciences, Hangzhou, China.
Yixue Li, Bio-Med Big Data Center, CAS Key Laboratory of Computational Biology, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai, China; Guangzhou Laboratory, Guangzhou, China.
Yi-Hsuan Pan, Key Laboratory of Brain Functional Genomics of Ministry of Education, School of Life Science, East China Normal University, Shanghai, China.
Haipeng Li, Bio-Med Big Data Center, CAS Key Laboratory of Computational Biology, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai, China.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
Conceptualization: J.Y., G.Z., D.Y., Y.H.P., B.S., Y.Z., G.P.Z., Y.L., and H.L.; data analysis: J.Y., Y.C., Y.H.P., G.Z., D.Y., R.C., X.W., Y.L., C.Y., and X.S.; experiment: Y.C., Z.G., G.D., X.L., and Y.H.P.; writing: J.Y., Y.C., G.Z., D.Y., R.C., Y.L., Y.H.P., C.Y., X.S., Y.Z., G.P.Z., and H.L.; and supervision & funding acquisition: Y.H.P., G.Z., Y.Z., G.P.Z., Y.L., and H.L.
Data Availability
All the SARS-CoV-2 data can be obtained from the Coronavirus GenBrowser (https://ngdc.cncb.ac.cn/ncov/apis/) and VENAS. Raw data were also deposited on Mendeley (https://data.mendeley.com/datasets/tbbxjy3gyr/1). Other data are available from the corresponding authors upon request.
References
- Campbell F, Archer B, Laurenson-Schafer H, Jinnai Y, Konings F, Batra N, Pavlin B, Vandemaele K, Van Kerkhove MD, Jombart T, et al. 2021. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill. 26:2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chand M, Hopkins S, Dabrera G, Achison C, Barclay W, Ferguson N, Volz E, Loman N, Rambaut A, Barrett J. 2020. Investigation of novel SARS-CoV-2 variant of concern 202012/01.https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/959438/Technical_Briefing_VOC_SH_NJL2_SH2.pdf, last accessed June 2, 2023.
- Chen F, You L, Yang F, Wang L, Guo X, Gao F, Hua C, Tan C, Fang L, Shan R, et al. 2020. CNGBdb: China National GeneBank DataBase. Hereditas (Beijing). 42:799–809. [DOI] [PubMed] [Google Scholar]
- Colijn C, Gardy J. 2014. Phylogenetic tree shapes resolve disease transmission patterns. Evol Med Public Health. 2014:96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cori A, Ferguson NM, Fraser C, Cauchemez S. 2013. A new framework and software to estimate time-varying reproduction numbers during epidemics. Am J Epidemiol. 178:1505–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, Pearson CAB, Russell TW, Tully DC, Washburne AD, et al. 2021. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 372:eabg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maio N, Worby CJ, Wilson DJ, Stoesser N. 2018. Bayesian Reconstruction of transmission within outbreaks using genomic variants. PLoS Comput Biol. 14:e1006117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar MS, Marwal R, Radhakrishnan VS, Ponnusamy K, Jolly B, Bhoyar RC, Sardana V, Naushin S, Rophina M, Mellan TA, et al. 2021. Genomic characterization and epidemiology of an emerging SARS-CoV-2 variant in Delhi, India. Science 374:995–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S, Buckland-Merrett G. 2017. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Chall. 1:33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobeil SM-C, Janowska K, McDowell S, Mansouri K, Parks R, Stalls V, Kopp MF, Manne K, Li D, Wiehe K, et al. 2021. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Science 373:eabi6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Zhu J-W, Li C-P, Jiang S, Ma L-N, Tang B-X, Zou D, Chen M-L, Sun Y-B, Song S-H, et al. 2020. An online coronavirus analysis platform from the National Genomics Data Center. Zool Res. 41:705–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang S, Allen D, Annan-Phan S, Bell K, Bolliger I, Chong T, Druckenmiller H, Huang LNY, Hultgren A, Krasovich E, et al. 2020. The effect of large-scale anti-contagion policies on the COVID-19 pandemic. Nature 584:262–267. [DOI] [PubMed] [Google Scholar]
- Huang W, Liao C-C, Han Y, Lv J, Lei M, Li Y, Lv Q, Dong D, Zhang S, Pan Y-H, et al. 2020. Co-activation of Akt, Nrf2, and NF-κB signals under UPRER in torpid Myotis ricketti bats for survival. Commun Biol. 3:658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Yang C, Xu XF, Xu W, Liu SW. 2020. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 41:1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui KPY, Ho JCW, Cheung MC, Ng KC, Ching RHH, Lai KL, Kam TT, Gu HG, Sit KY, Hsin MKY, et al. 2022. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature 603:715–720. [DOI] [PubMed] [Google Scholar]
- Jackson B, Boni MF, Bull MJ, Colleran A, Colquhoun RM, Darby AC, Haldenby S, Hill V, Lucaci A, McCrone JT, et al. 2021. Generation and transmission of interlineage recombinants in the SARS-CoV-2 pandemic. Cell 184:5179–5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L, He GJ, Sharp AK, Wang XF, Brown AM, Michalak P, Weger-Lucarelli J. 2021. A selective sweep in the spike gene has driven SARS-CoV-2 human adaptation. Cell 184:4392–4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan NL, Hudson RR, Langley CH. 1989. The “hitchhiking effect” revisited. Genetics 123:887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Stephan W. 2002. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics 160:765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1987. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol. 196:947–950. [DOI] [PubMed] [Google Scholar]
- Leary S, Gaudieri S, Parker M, Chopra A, James I, Pakala S, Alves E, John M, Lindsey B, Keeley A, et al. 2021. Generation of a novel SARS-CoV-2 sub-genomic RNA due to the R203K/G204R variant in nucleocapsid: homologous recombination has potential to change SARS-CoV-2 at both protein and RNA level. Pathog Immun. 6:27–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, et al. 2020. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med. 382:1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Lv QY, Zheng GT, Liu D, Ma J, He G-M, Zhang L-B, Zheng S, Li H, Pan YH. 2022. Unexpected expression of heat-activated transient receptor potential (TRP) channels in winter torpid bats and cold-activated TRP channels in summer active bats. Zool Res. 43:52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Stephan W. 2005. Maximum likelihood methods for detecting recent positive selection and localizing the selected site in the genome. Genetics 171:377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Stephan W. 2006. Inferring the demographic history and rate of adaptive substitution in Drosophila. PLoS Genet. 2:e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Li H, Schlotterer C, Futschik A. 2011. Distinguishing positive selection from neutral evolution: boosting the performance of summary statistics. Genetics 187:229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling YC, Cao RF, Qian JQ, Li JF, Zhou HK, Yuan LY, Wang Z, Ma LX, Zheng GY, Zhao GP, et al. 2022. An interactive viral genome evolution network analysis system enabling rapid large-scale molecular tracing of SARS-CoV-2. Sci Bull. 67:665–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lista MJ, Winstone H, Wilson HD, Dyer A, Pickering S, Galao RP, De Lorenzo G, Cowton VM, Furnon W, Suarez N, et al. 2022. The P681H mutation in the spike glycoprotein of the Alpha variant of SARS-CoV-2 escapes IFITM restriction and is necessary for type I interferon resistance. J Virol. 96:e0125022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Liu J, Johnson BA, Xia H, Ku Z, Schindewolf C, Widen SG, An Z, Weaver SC, Menachery VD, et al. 2022. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. Cell Rep. 39:110829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubinski B, Fernandes MHV, Frazier L, Tang T, Daniel S, Diel DG, Jaimes JA, Whittaker GR. 2022. Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant B.1.1.7 (Alpha) spike. iScience. 25:103589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubinski B, Frazier L, Phan M, Bugumbe D, Cunningham JL, Tang T, Daniel S, Cotten M, Jaimes JA, Whittaker G. 2022. Spike protein cleavage-activation in the context of the SARS-CoV-2 P681R mutation: an analysis from its first appearance in lineage A.23.1 identified in Uganda. Microbiol Spectr. 10:e0151422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov PV, Ghafari M, Beer M, Lythgoe K, Simmonds P, Stilianakis NI, Katzourakis A. 2023. The evolution of SARS-CoV-2. Nat Rev Microbiol. 21:361–379. [DOI] [PubMed] [Google Scholar]
- Meng Z, Jianpeng X, Aiping D, Yingtao Z, Yali Z, Ting H, Jiansen L, Hongwei T, Bosheng L, Yan Z, et al. 2021. Transmission dynamics of an outbreak of the COVID-19 Delta variant B.1.617.2—Guangdong Province, China, May–June 2021. China CDC Weekly. 3:584–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad A, Abubaker J, Al-Mulla F. 2021. Structural modelling of SARS-CoV-2 alpha variant (B.1.1.7) suggests enhanced furin binding and infectivity. Virus Res. 303:198522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley TH, Plachetzki DC, Rivera AS. 2007. Furcation, field-splitting, and the evolutionary origins of novelty in arthropod photoreceptors. Arthropod Struct Dev. 36:386–400. [DOI] [PubMed] [Google Scholar]
- Obermeyer F, Jankowiak M, Barkas N, Schaffner SF, Pyle JD, Yurkovetskiy L, Bosso M, Park DJ, Babadi M, MacInnis BL, et al. 2022. Analysis of 6.4 million SARS-CoV-2 genomes identifies mutations associated with fitness. Science 376:1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Du N, Lei YQ, Dorje S, Qi JX, Luo TR, Gao GF, Song H. 2020. Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design. Embo J. 39:e105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Loman N, Pybus O, Barclay W, Barrett J, Carabelli A, Connor T, Peacock T, Robertson DL, Volz E, et al. 2020. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations.https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563, last accessed June 2, 2023
- Saito A, Irie T, Suzuki R, Maemura T, Nasser H, Uriu K, Kosugi Y, Shirakawa K, Sadamasu K, Kimura I, et al. 2022. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 602:300–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Beck J, Bolton EE, Bourexis D, Brister JR, Canese K, Comeau DC, Funk K, Kim S, Klimke W, et al. 2021. Database resources of the national center for biotechnology information. Nucleic Acids Res. 49:D10–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Kern AD. 2018. Supervised machine learning for population genetics: a new paradigm. Trends Genet. 34:301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu YL, McCauley J. 2017. GISAID: global initiative on sharing all influenza data—from vision to reality. Euro Surveill. 22:30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Loriere E, Holmes EC. 2011. Why do RNA viruses recombine? Nat Rev Microbiol. 9:617–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan W. 2019. Selective sweeps. Genetics 211:5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Yamasoba D, Kimura I, Wang L, Kishimoto M, Ito J, Morioka Y, Nao N, Nasser H, Uriu K, et al. 2022. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature 603:700–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed Abdullah M, Taha Taha Y, Tabata T, Chen Irene P, Ciling A, Khalid Mir M, Sreekumar B, Chen P-Y, Hayashi Jennifer M, Soczek Katarzyna M, et al. 2021. Rapid assessment of SARS-CoV-2–evolved variants using virus-like particles. Science 374:1626–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne LG, Bouhaddou M, Reuschl A-K, Zuliani-Alvarez L, Polacco B, Pelin A, Batra J, Whelan MVX, Hosmillo M, Fossati A, et al. 2022. Evolution of enhanced innate immune evasion by SARS-CoV-2. Nature 602:487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turakhia Y, Thornlow B, Hinrichs A, McBroome J, Ayala N, Ye C, Smith K, De Maio N, Haussler D, Lanfear R, et al. 2022. Pandemic-scale phylogenomics reveals the SARS-CoV-2 recombination landscape. Nature 609:994–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanInsberghe D, Neish AS, Lowen AC, Koelle K. 2021. Recombinant SARS-CoV-2 genomes circulated at low levels over the first year of the pandemic. Virus Evol. 7:veab059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana R, Moyo S, Amoako DG, Tegally H, Scheepers C, Althaus CL, Anyaneji UJ, Bester PA, Boni MF, Chand M, et al. 2022. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in Southern Africa. Nature 603:679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, Hinsley WR, Laydon DJ, Dabrera G, O'Toole A, et al. 2021. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 593:266–269. [DOI] [PubMed] [Google Scholar]
- WHO . 2021. Tracking SARS-CoV-2 variants.https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/, last accessed June 2, 2023
- Wu H, Xing N, Meng K, Fu B, Xue W, Dong P, Tang W, Xiao Y, Liu G, Luo H, et al. 2021. Nucleocapsid mutations R203K/G204R increase the infectivity, fitness, and virulence of SARS-CoV-2. Cell Host Microbe. 29:1788–1801.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, et al. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Wang P, Fu Y, Zheng Y, Tang Q, Si L, You J, Zhang Z, Zhu Y, Zhou L, et al. 2010. Length of the ORF, position of the first AUG and the Kozak motif are important factors in potential dual-coding transcripts. Cell Res. 20:445–457. [DOI] [PubMed] [Google Scholar]
- Xu K, Zheng BJ, Zeng R, Lu W, Lin YP, Xue L, Li L, Yang LL, Xu C, Dai J, et al. 2009. Severe acute respiratory syndrome coronavirus accessory protein 9b is a virion-associated protein. Virology 388:279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Dong L, Yan F, Mu H, Tang B, Yang X, Zeng T, Zhou Q, Gao F, Wang Z, et al. 2019. eGPS 1.0: comprehensive software for multi-omic and evolutionary analyses. Natl Sci Rev. 6:867–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Yang X, Tang B, Pan Y-H, Yang J, Duan G, Zhu J, Hao Z-Q, Mu H, Dai L, et al. 2022. Coronavirus GenBrowser for monitoring the transmission and evolution of SARS-CoV-2. Brief Bioinform. 23:bbab583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Zhu J, Yang J, Pan YH, Mu HL, Cao RF, Tang BX, Duan GY, Hao ZQ, Dai L, et al. 2022. Global cold-chain related SARS-CoV-2 transmission identified by pandemic-scale phylogenomics. Zool Res. 43:871–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandi M, Shafaati M, Kalantar-Neyestanaki D, Pourghadamyari H, Fani M, Soltani S, Kaleji H, Abbasi S. 2022. The role of SARS-CoV-2 accessory proteins in immune evasion. Biomed Pharmacother. 156:113889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhao W, Xiao J, Bao Y, He S, Zhang G, Li Y, Zhao G, Chen R, Gao Y, et al. 2020. Database resources of the National Genomics Data Center in 2020. Nucleic Acids Res. 48:D24–D33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W-M, Song S-H, Chen M-L, Zou D, Ma L-N, Ma Y-K, Li R-J, Hao L-L, Li C-P, Tian D-M, et al. 2020. The 2019 novel coronavirus resource. Hereditas (Beijing) 42:212–221. [DOI] [PubMed] [Google Scholar]
- Zuckerman NS, Fleishon S, Bucris E, Bar-Ilan D, Linial M, Bar-Or I, Indenbaum V, Weil M, Lustig Y, Mendelson E, et al. 2021. A unique SARS-CoV-2 spike protein P681H variant detected in Israel. Vaccines (Basel) 9:616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the SARS-CoV-2 data can be obtained from the Coronavirus GenBrowser (https://ngdc.cncb.ac.cn/ncov/apis/) and VENAS. Raw data were also deposited on Mendeley (https://data.mendeley.com/datasets/tbbxjy3gyr/1). Other data are available from the corresponding authors upon request.