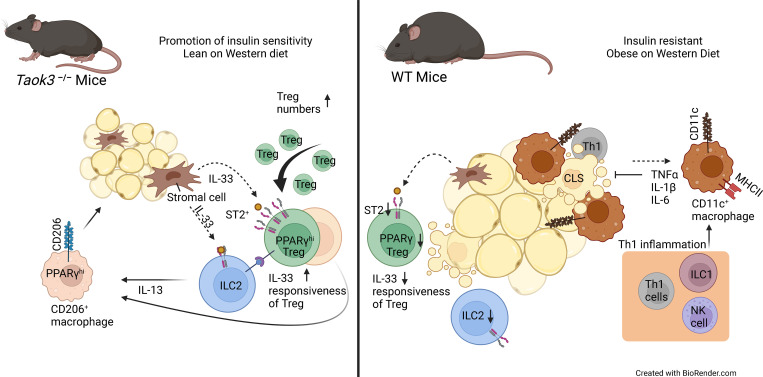
Maes et al. reveal an unexpected role of TAOK3 in regulating ST2+ regulatory T cells in mouse adipose tissue. Absence of TAOK3 sustains Tregs in obesity and improves metabolic dysfunction.
Abstract
Healthy adipose tissue (AT) contains ST2+ Tregs, ILC2s, and alternatively activated macrophages that are lost in mice or humans on high caloric diet. Understanding how this form of type 2 immunity is regulated could improve treatment of obesity. The STE20 kinase Thousand And One amino acid Kinase-3 (TAOK3) has been linked to obesity in mice and humans, but its precise function is unknown. We found that ST2+ Tregs are upregulated in visceral epididymal white AT (eWAT) of Taok3−/− mice, dependent on IL-33 and the kinase activity of TAOK3. Upon high fat diet feeding, metabolic dysfunction was attenuated in Taok3−/− mice. ST2+ Tregs disappeared from eWAT in obese wild-type mice, but this was not the case in Taok3−/− mice. Mechanistically, AT Taok3−/− Tregs were intrinsically more responsive to IL-33, through higher expression of ST2, and expressed more PPARγ and type 2 cytokines. Thus, TAOK3 inhibits adipose tissue Tregs and regulates immunometabolism under excessive caloric intake.
Graphical Abstract
Introduction
Obesity is a complex metabolic disorder determined by environment, behavior, age, sex, and (epi)genetics (Cani, 2019; Burgio et al., 2015; Upadhyay et al., 2018). Almost 2 billion adults have overweight or obesity, with prevalence increasing at an alarming rate (World Health Organization, 2016). The obesity epidemic is paralleled by an increase in type 2 diabetes mellitus (T2DM), hyperlipidemia, hypertension, and certain cancers. Obesity profoundly impacts public health care due to high morbidity, mortality, and socioeconomic costs (Biener et al., 2020; Tobias and Hu, 2018; Scully et al., 2021; Must et al., 1999). Despite impressive scientific and clinical progress, there is an unmet need for innovative prevention and treatment.
White adipose tissue (WAT) stores excess nutrients as triglycerides, releases free fatty acids during fasting, and acts as an endocrine source of various adipokines that regulate fat homeostasis, whole body energy expenditure, and metabolic health (Rosen and Spiegelman, 2014). WAT depots are often separated conceptually in subcutaneous and visceral pools. Visceral WAT in humans is mainly localized to the large abdominal fat depot of the omentum, whereas in mice it is mainly localized around the gonads and is therefore called gWAT or epididymal (e)WAT in male mice (Rosen and Spiegelman, 2014). Even in homeostasis, WAT is densely populated by immune cells. These are recovered in the stromal vascular fraction (SVF) that also contains endothelial and stromal cells (Kanneganti and Dixit, 2012). Increasing evidence implicates the immune system as a key node in maintaining AT homeostasis by controlling the immunometabolic set point of inflammation (Goldberg et al., 2021). Lean, healthy WAT is populated by type 2 “alternatively activated” CD206+ macrophages, eosinophils, type 2 innate lymphoid cells (ILC2s), and T regulatory cells (Tregs; Goldberg et al., 2021). These innate and adaptive immune cells interact with stromal cells to maintain a type 2 cytokine milieu, rich in IL-4 and IL-13, that promotes insulin sensitivity and “beiging” of WAT (Brestoff and Artis, 2015).
Foxp3hiCD25hi Tregs play a special role in fat tissue homeostasis, specifically in male mice. They are abundant in eWAT and express a unique set of genes including the IL-33 receptor ST2 (encoded by Il1rl1), the activation marker Klrg1, and the cytokine Il10 (Li et al., 2020). Expression of these genes is regulated by the transcription factor peroxisome proliferator-activated receptor γ (PPARγ), expressed in up to 80% of WAT Tregs, and typically associated with T helper (Th)2 effector cytokine production in conventional T cells and AT ILC2s (Li et al., 2018; Chen et al., 2017; Fali et al., 2021; Cipolletta et al., 2012). Development and activation of eWAT Tregs is highly dependent on the pro-type 2 alarmin IL-33, released from AT mesenchymal stromal cells, like PDGFRα+ fibroblasts and mesothelial cells (Goldberg et al., 2021; Mahlakõiv et al., 2019; Spallanzani et al., 2019). Genetic variants in IL-33 have been linked to risk for obesity in humans, and circulating IL-33 levels are negatively correlated with body mass index, suggesting that IL-33 is a key regulator of metabolic health (Hasan et al., 2014; Melén et al., 2010).
In obesity, excess lipid storage in WAT leads to adipocyte hypertrophy and hyperplasia, hypoxia, ER- and biomechanical stress, adipocyte cell death, and release of free fatty acids (Reilly and Saltiel, 2017). These processes lead to low-grade inflammation in eWAT and release of chemokines and cytokines that promote systemic insulin resistance and T2DM (Lee et al., 2018; Hotamisligil, 2017). There is a shift of a type 2 to a type 1–dominated immune response, characterized by influx of inflammatory macrophages, B cells, CD8+ T cells, Th1 T cells, and ILC1s which propagate inflammation by producing cytokines (Brestoff and Artis, 2015; Man et al., 2022). Strikingly, obese WAT is devoid of eosinophils, ILC2s, and ST2+ Tregs, further aggravating type 1 inflammation and promoting metabolic dysfunction (Brestoff and Artis, 2015; Man et al., 2022). Whereas high fat diet (HFD)–induced obesity leads to reduced eWAT Tregs, their restoration to normal numbers improves inflammation and glucose tolerance (Vasanthakumar et al., 2015; Feuerer et al., 2009; Eller et al., 2011). This led to the view that Tregs in healthy WAT adopt a Th2-like phenotype to control homeostasis, whereas their loss in obesity contributes to metabolic dysfunction (Vasanthakumar et al., 2015; Spallanzani et al., 2019). Understanding the key factors that determine the presence and function of AT Tregs in homeostasis and their demise at times of caloric excess could have a major impact on treatment and prevention of obesity.
STE20 kinases are serine/threonine kinases that regulate the cell cycle, apoptosis, and cell stress responses through MAPK cascade signaling following nutrient sensing (Delpire, 2009; Strange et al., 2006). The Tao kinase subfamily has three members in mammals: TAOK1 (also known as MAP3K16, PSK2, or MARKK), TAOK2 (MAP3K17 or PSK1) and TAOK3 (MAP3K18, JIK, or DPK; Yustein et al., 2003) whose function is increasingly implicated in immune homeostasis (Fang et al., 2020; Vanderkerken et al., 2020; Hammad et al., 2017; Ormonde et al., 2018). Interestingly, DNA methylation profiles on whole blood obtained from children has revealed that methylation of the TAOK3 gene locus (and thus inhibition of transcription) was associated with reduced risk of obesity (Huang et al., 2015). As a MAP3 kinase, TAOK3 was shown to regulate p38 kinase and c-Jun N terminal kinase, pathways that have previously been linked to metabolic inflammation and T2DM (Yustein et al., 2003; Yoneda et al., 2001; Nikolic et al., 2020). Here, we show that TAOK3 suppresses ST2+ Tregs in WAT and contributes to metabolic dysfunction at times of nutrient excess.
Results and discussion
TAOK3 regulates the homeostasis of immune cells in AT
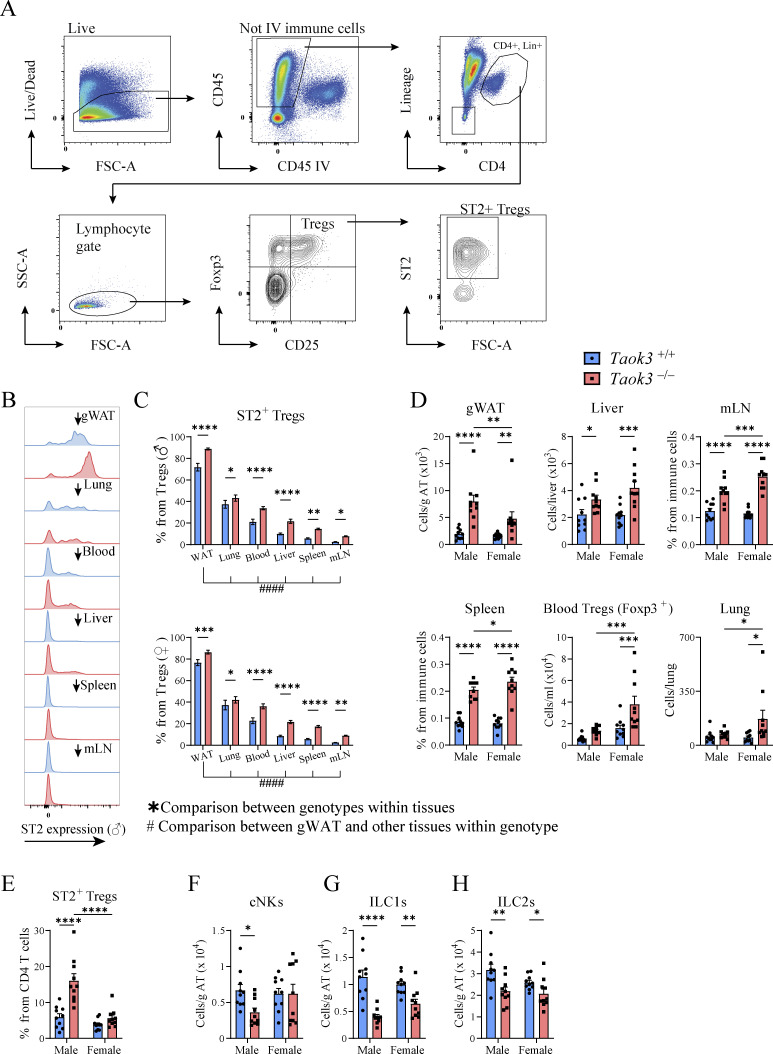
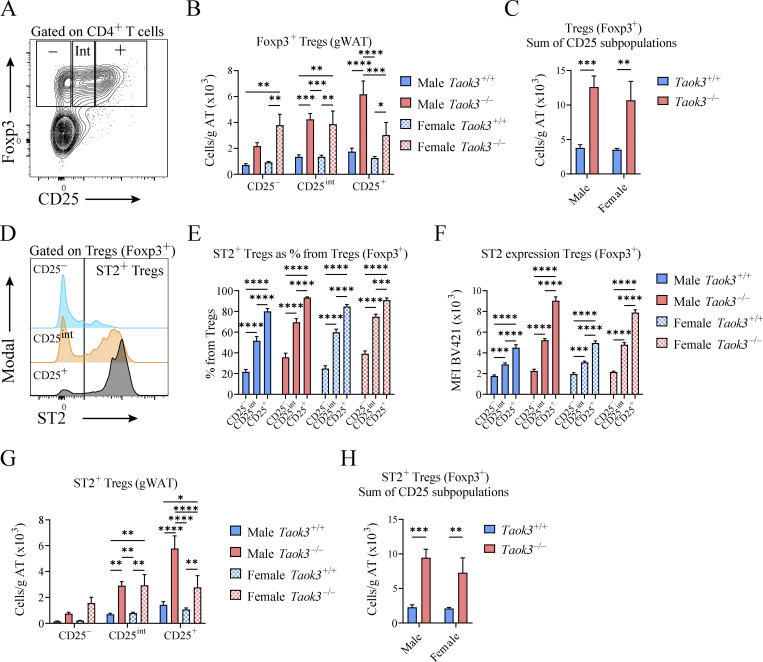
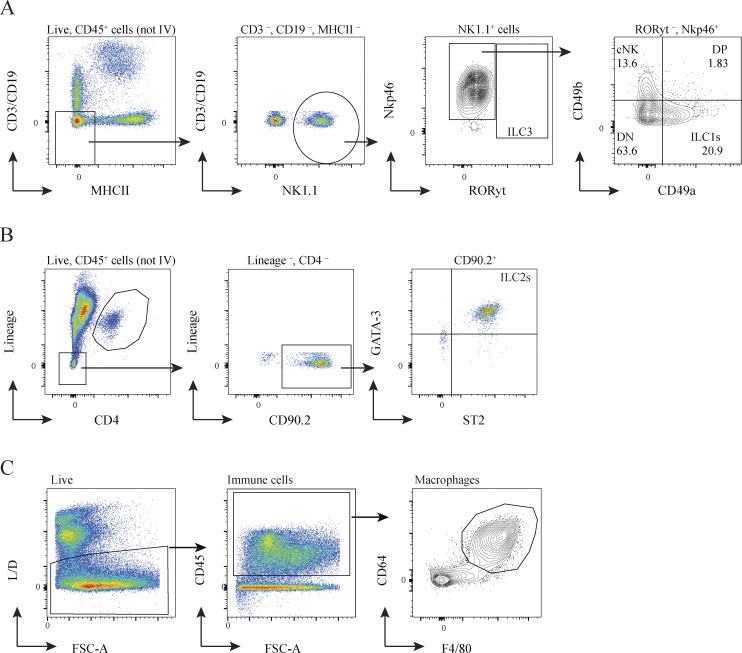
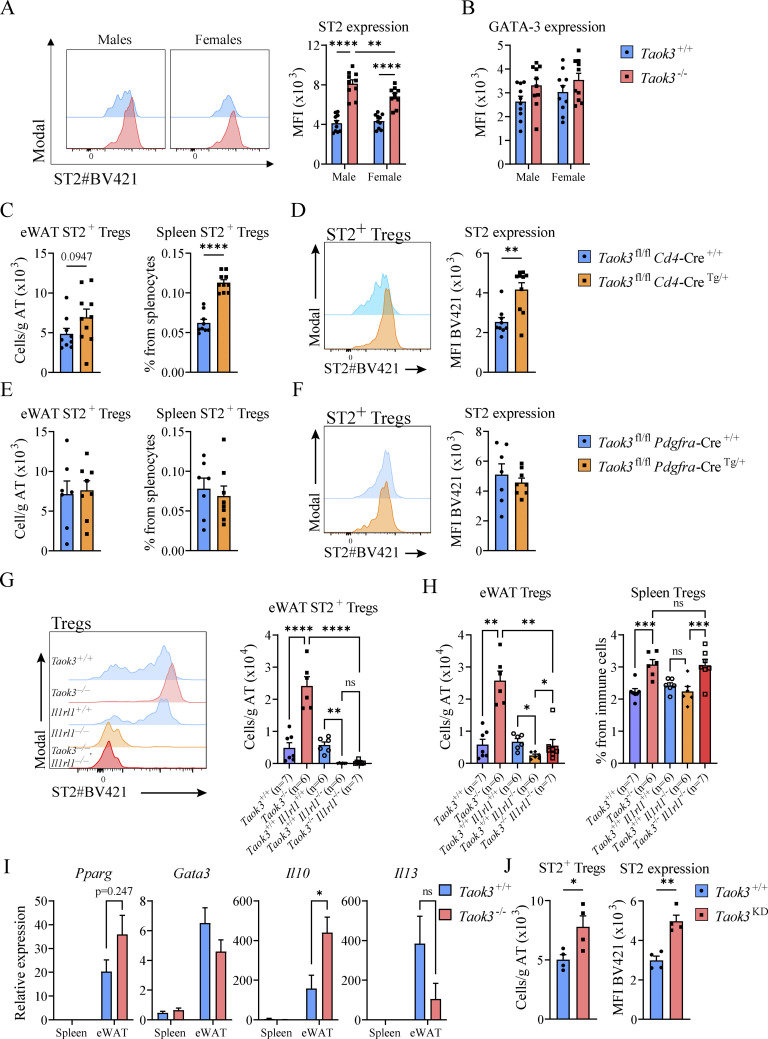
In visceral WAT, IL-33 responsive ST2+ Tregs are emerging as powerful controllers of metabolic health. These Tregs originate in the thymus and acquire a specific visceral WAT transcriptional signature in central lymphoid organs to then accumulate as PPARγ+ ST2+ Tregs in fat tissues (Li et al., 2020; Sivasami and Li, 2020). Since silencing of TAOK3 has been linked to obesity in humans, and TAOK3 is increasingly implicated in adaptive immunity (Huang et al., 2015; Ormonde et al., 2018; Vanderkerken et al., 2020; Hammad et al., 2017), we studied how TAOK3 influenced Treg biology in AT. In male and female mice aged 8–12 wk, detailed immunophenotyping was performed on gWAT, liver, mediastinal lymph node (mLN), spleen, blood, and lung (Fig. 1, A–D). Intravascular labeling ensured focus on tissue-resident cells, and Tregs were defined as CD25+Foxp3+ cells within the CD4+ lymphocyte gate (Fig. 1 A). In WT Taok3+/+ mice, ST2+ Tregs made up almost 70% of the resident Treg pool in gWAT, whereas in other peripheral tissues and peripheral lymphoid tissues, this percentage varied between 5 and 35% (Fig. 1, B and C). Strikingly, in the absence of Taok3, ST2+ Tregs were overrepresented in most of the tissues analyzed. In fat tissue, close to 90% of Tregs expressed ST2 (Fig. 1, C and D). This ST2+ Treg bias was apparent in both sexes, and reflected by a relative and absolute increase of these cells (Fig. 1, C and D), despite earlier reports that WT male mice preferentially accumulate Tregs in gWAT, although mice in these reports were generally analyzed at 20–25 wk of age (Li et al., 2018; Vasanthakumar et al., 2020). When expressed as percentage of ST2+CD25+Foxp3+ Tregs among the CD4 lymphocyte pool, ST2+ Tregs made up on average 15% of the CD4+ T cell population in eWAT from male Taok3−/− mice, compared with only 5% in WT eWAT (Fig. 1 E). Concordantly, total Treg numbers (Foxp3+CD25+) were also higher in gWAT taken from male and female Taok3−/− mice (Fig. S1, A and B). Similarly, when only taking into account Foxp3 as a definitive marker for Tregs, we found that total Treg numbers were upregulated in gWAT of male and female Taok3−/− mice compared to WT littermates (Fig. S1, A–C). Interestingly, both ST2 expression and percentage of ST2+ Tregs increased with a higher level of CD25 expression in Foxp3+ Tregs independent from sex or genotype in gWAT (Fig. S1, D–F), and, consequently, we found higher numbers of ST2+ Tregs within Foxp3+ Tregs with intermediate or high CD25 expression in Taok3−/− mice compared to Taok3+/+ mice (Fig. S1 G). Total ST2+ Tregs numbers were upregulated in gWAT taken from Taok3−/− mice when all Foxp3+ Tregs were taken into account, including CD25–Foxp3+ cells (Fig. S1 H). We next studied whether other innate immune cells related to immunometabolism (Molofsky et al., 2013) were also regulated by TAOK3 (gating strategy Fig. S2, A and B). In contrast to the increase in adaptive Treg numbers per gram AT (Fig. 1 D), male Taok3−/− mice had significantly less conventional natural kills (cNK) cells, ILC1s, and ILC2s in gWAT compared with WT Taok3+/+ mice (Fig. 1, F–H). In female mice, only ILC1s and ILC2s were reduced, and effects of Taok3 deficiency were generally less pronounced. There were very few RORγt+ ILC3s in WAT and these were not further analyzed between genotypes. These data indicate that TAOK3 relays a suppressive signal on ST2+ Tregs in AT homeostasis.
Figure 1.
TAOK3 regulates the homeostasis of immune cells in AT. (A) Representative gating strategy for ST2+CD25+Foxp3+ Tregs in male eWAT. This gating strategy is similar for the other tissues, except for blood, where only intravascular Foxp3+ Tregs were taken into account. Intravascular (IV) immune cells are excluded from CD45+ immune cells, except for liver, spleen, and blood. (B) Plot depicting mean fluorescent intensities (MFI) as a measure for ST2 expression on CD25+Foxp3+ Tregs (male mice, but representative for female mice). (C) ST2+ Tregs as a percentage from the total Treg population in the tissues depicted, upper figure shows percentages for male mice, figure below shows percentages for female mice. (D) ST2+ Treg numbers in gWAT (per gram AT), liver, mLN, spleen, blood, and lung taken from male and female Taok3+/+ and Taok3−/− mice. (E) Proportion of ST2+ Tregs from the total CD4+ T cell population present in gWAT. (F) cNK cell numbers in eWAT. (G) ILC1 numbers in gWAT. (H) ILC2 numbers in gWAT. Data are pooled from two independent experiments (n = 10 mice/group, 8–12-wk-old mice). Statistical analysis: Panel C: Comparisons between genotypes within the different tissues was analyzed by two-way ANOVA with correction for multiple comparisons (*). For both Taok3+/+ and Taok3−/− mice, ST2+ Treg percentages in eWAT were compared to cell percentages in other tissues by one-way ANOVA with correction for multiple comparisons (#). All other data were analyzed by two-way ANOVA with correction for multiple comparisons. Statistical significant differences between genotypes within mice from the same sex (F–H) and differences between sexes within mice from the same genotype were depicted (D–E). Data are shown as means ± SEMs. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Figure S1.
CD25 expression on Foxp3+ Tregs is associated with ST2 surface levels. (A) Representative plot for CD25–, CD25 intermediate (Int), or CD25+Foxp3+ Tregs. (B) Foxp3+ Treg numbers in gWAT (per gram AT) stratified by CD25 expression (CD25–, CD25int, or CD25+). (C) Total Foxp3+ Treg numbers (calculated as the sum of CD25–, CD25int, and CD25+ Tregs) in gWAT (per gram AT). (D) Representative plot depicting MFI as a measure for ST2 expression on CD25–, CD25int, or CD25+Foxp3+ Tregs. (E) Percentage of Foxp3+ Tregs that express ST2 stratified by CD25 expression (CD25–, CD25int, or CD25+) in gWAT taken from male and female Taok3+/+ and Taok3−/− mice. (F) ST2 expression (MFI) in Foxp3+ Tregs stratified by CD25 expression (CD25–, CD25int, or CD25+) in gWAT taken from male and female Taok3+/+ and Taok3−/− mice. (G) Total ST2+ Foxp3+ Treg numbers stratified by CD25 expression (CD25–, CD25int, or CD25+) in gWAT (per gram AT) taken from male and female Taok3+/+ and Taok3−/− mice. (H) Total ST2+Foxp3+ Treg numbers (calculated as the sum of CD25–, CD25int, and CD25+ST2+ Tregs) in gWAT (per gram AT). Data are pooled from two independent experiments (n = 10 mice/group, 8–12-wk-old mice). Statistical analysis: all data were analyzed by two-way ANOVA with correction for multiple comparisons. For B and G, statistical significant differences between groups within CD25 strata were analyzed; for C and H, statistical significant differences between genotypes within mice of the same sex were analyzed; for E and F, statistical significant differences between CD25 strata within mice of the same sex and genotype were analyzed. Data are shown as means ± SEMs. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Figure S2.
Gating strategy. (A) Representative gating strategy for cNK cells (defined as lineage negative, NK1.1+, Nkp46+, CD49b+) and ILC1s (defined as lineage negative, NK1.1+, Nkp46+, CD49a+) in eWAT. Lineage includes CD3, CD19. (B) Representative gating strategy for ILC2s in eWAT. ILC2s are defined as CD45+, lineage–, CD4–, CD90.2+, GATA-3+, ST2+. Intravascular (IV) immune cells are excluded from CD45+ immune cells. Lineage includes CD8, CD11c, B220, TCRβ, TCRγδ, Ly-6C/G, Ter119, NK1.1, CD19, CD3e, CD11b. (C) Representative gating strategy for macrophages in eWAT. ILC1/2, type 1/2 innate lymphoid cell; DN, double negative; DP, double positive; L/D, live/dead.
TAOK3 controls eWAT Treg homeostasis through cell-intrinsic and cell-extrinsic IL-33 signaling
IL-33 is an alarmin stored in the nucleus of epithelial, endothelial, and stromal cells that acts as the natural ligand for ST2 (a.k.a. Interleukin 1 Receptor Like 1, encoded by Il1rl1). In eWAT, IL-33 is produced by subsets of AT stromal cells and interacts with the ST2 receptor on Tregs to induce and sustain a unique transcriptional profile (Spallanzani et al., 2019; Goldberg et al., 2021). In support for an increased IL-33 axis driving eWAT Tregs, we observed that the degree of expression of the ST2 receptor for IL-33 was consistently higher on ST2+Foxp3+CD25+ Tregs in Taok3−/− mice. This effect was most pronounced in young male mice (Fig. 2 A). In many cell types, ST2 expression is under control of GATA-3 (Nawijn et al., 2001; Hayakawa et al., 2005), yet intracellular GATA-3 protein expression was similar in ST2+ Tregs in eWAT (Fig. 2 B). Next, we investigated whether T cell intrinsic Taok3 expression is required to suppress ST2+ Tregs. We crossed Taok3fl/fl mice (Vanderkerken et al., 2020), to mice expressing the Cre recombinase under control of the Cd4-promotor (Lee et al., 2001), deleting TAOK3 from T cells starting from the CD4/CD8 double positive stage onwards. Interestingly, we found that in normal chow-fed Cd4-Cre transgenic mice, aged 8–12 wk, splenic ST2+ Treg numbers were increased. There was only a trend for an increase in eWAT Treg numbers in these young mice (Fig. 2 C), but we did observe consistently higher ST2 expression on eWAT-resident ST2+ Tregs in Cd4-Cre transgenic mice (Fig. 2 D). Although this suggests T cell–intrinsic effects of loss of Taok3 on Tregs, clearly T cell–extrinsic effects were also at play.
Figure 2.
TAOK3 controls eWAT Treg homeostasis through cell-intrinsic and cell-extrinsic IL-33 signaling. (A) Representative plot for MFI as a measure for ST2 expression on ST2+CD25+Foxp3+ Tregs in eWAT from lean male and female Taok3+/+ and Taok3−/− mice (left). Right, ST2 expression (MFI) on ST2+ AT Tregs. (B) GATA-3 expression (MFI) in ST2+ Tregs in eWAT. (C) ST2+ Tregs numbers in eWAT and ST2+ Tregs as percentage of splenocytes in Taok3fl/fl Cd4-Cre WT and transgenic mice. (D) Representative plot for MFI as a measure for ST2 expression on ST2+CD25+Foxp3+ Tregs in eWAT from Taok3fl/fl Cd4-Cre WT and transgenic mice (left). Right, ST2 expression (MFI) on ST2+ Tregs in eWAT. (E) ST2+ Tregs numbers in eWAT and ST2+ Tregs as percentage of splenocytes in Taok3fl/fl Pdgfra-Cre WT and transgenic mice. (F) Representative plot for MFI as a measure for ST2 expression on ST2+CD25+Foxp3+ Tregs in eWAT from Taok3fl/fl Pdgfra-Cre WT and transgenic mice (left). Right, ST2 expression (MFI) on ST2+ AT Tregs. (G) Representative plot for MFI as a measure for ST2 expression on CD25+Foxp3+ Tregs in eWAT from Taok3+/+, Taok3−/−, Il1rl1+/+, Il1rl1−/−, and Taok3−/− Il1rl1−/− mice (left). Right, ST2 expression (MFI) on ST2+ Tregs in eWAT. (H) Total Treg numbers in eWAT (left) and total Tregs as percentage from immune cells in spleen (right). (I) Gene expression in CD3+CD4+CD25+ T cells sorted from spleen and eWAT from Taok3+/+ and Taok3−/− mice. (J) ST2+ Tregs numbers in eWAT (left). Right, ST2 expression (MFI) on ST2+ Tregs in eWAT in Taok3+/+ or Taok3KD mice. A–H, data pooled from two independent experiments (n = 6–10 mice/group). I, data from one large cohort of mice (n = 9–10 mice/group). For spleen, sufficient numbers of CD4+CD25+ T cells were sorted/mouse to perform qPCR. For eWAT, two samples from the same genotype were pooled to reach a sufficient number of CD4+CD25+ T cells for qPCR. J, data from one experiment (n = 4 mice/group). Statistical analysis: two-way ANOVA with correction for multiple comparisons (A and B). For comparisons between two groups, data were analyzed with Shapiro-Wilk normality test to assess whether data were normally distributed. Parametric data were analyzed with an unpaired two-tailed t test and nonparametric data with a two-tailed Mann-Whitney test (C–F and J). For comparisons between multiple groups and one continuous variable, data were analyzed with Shapiro-Wilk normality test to assess whether data were normally distributed. Parametric data were analyzed with an ordinary one-way ANOVA and nonparametric data with a Kruskal–Wallis test (G and H). Data representing mRNA transcript expression ratios were log transformed before statistical analysis and an unpaired t test was subsequently performed to compare genotypes within spleen/eWAT (I). Data are shown as means ± SEMs. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
We next searched for T cell–extrinsic effects on ST2+ Tregs in WAT, and since IL-33 is the cytokine driving these cells, we directed our interest at the cells producing IL-33. In lean eWAT three distinct mesenchymal stromal cell subsets regulate Treg numbers through IL-33 secretion. In young mice, PDGFRα+Podoplanin (PDPN)+ stromal cells are the main source of functional IL-33, whereas with increasing age, mesothelial cells become a dominant source (Spallanzani et al., 2019; Mahlakõiv et al., 2019; Goldberg et al., 2021). We therefore crossed Pdgfra-Cre mice (Roesch et al., 2008) to Taok3fl/fl mice, thus deleting TAOK3 from IL-33 producing PDGFRα+ stromal cells in WAT. However, in Pdgfra-Cre Taok3fl/fl transgenic mice numbers of ST2+ Tregs and ST2 expression were similar to WT mice, ruling out a dominant role for TAOK3 in these stromal cells to promote ST2+ Treg numbers in WAT (Fig. 2, E and F). Potentially, other stromal cells expressing mesothelin might be more important in controlling Treg numbers, but we did not evaluate this possibility further.
Another possibility is that expansion of ST2+ Tregs in Taok3−/− mice is less dependent on IL-33 derived from stromal cells, despite higher expression of the IL-33 receptor on these cells in Taok3−/− mice. To investigate whether the accumulation of eWAT-dwelling Tregs was controlled by TAOK3 in an IL-33–dependent manner, we crossed whole body Taok3−/− mice to mice lacking Il1rl1 (Taok3−/−Il1rl1−/−; Fig. 2 G). In alignment with previous reports (Vasanthakumar et al., 2015; Kolodin et al., 2015), we found that Taok3+/+Il1rl1−/− mice lacking ST2 had reduced numbers of eWAT Tregs (Fig. 2 H). As before, total and ST2+Foxp3+CD25+ Tregs were upregulated in eWAT from Taok3−/−Il1rl1+/+ mice (Fig. 2, G and H). However, Taok3−/−Il1rl1−/− double KO mice had severely reduced numbers of Foxp3+CD25+ Tregs, showing that the Tregs of Taok3−/− mice were still dependent on IL-33 (Vasanthakumar et al., 2015; Kolodin et al., 2015). Interestingly, for splenic Tregs, there were no differences between Il1rl1+/+ and Il1rl1−/− mice on a Taok3−/− background, and Tregs were relatively increased in Taok3−/− mice compared to Taok3+/+ mice regardless of Il1rl1 genotype, again suggesting a tissue-specific role for TAOK3 in controlling Treg accumulation in eWAT (Fig. 2 H).
ST2 signaling controls the Treg transcriptional profile in visceral AT, specifically in the regulation of genes associated with a type 2 immune signature (Cipolletta et al., 2012, 2015). A key transcriptional regulator downstream of ST2 is the master lipid metabolism transcription factor and nuclear receptor PPARγ (Cipolletta et al., 2012) which acts in concert with Foxp3 to drive type 2 immune genes in eWAT Tregs (Li et al., 2018). Cell-intrinsic loss of PPARγ expression in Tregs in Ppargfl/fl x Foxp3-Cre mice or Ppargfl/fl x Lck-Cre mice leads to loss of eWAT Tregs and metabolic derangements, while the selective PPARγ agonist pioglitazone expands eWAT Tregs (Cipolletta et al., 2012; Bapat et al., 2015). Indeed, CD4+CD25+ sorted eWAT Tregs had abundant expression of I10, Il13, Pparg, and Gata3 compared to splenic Tregs (Fig. 2 I). Additionally, we found statistically significant increased Il10 expression in Taok3−/− Tregs in eWAT. However, Il13, Pparg, and Gata3 expression were similar between genotypes, suggesting that Taok3−/− Tregs have a globally preserved type 2 signature in eWAT in homeostatic conditions and normal chow feeding. Finally, since TAOK3 has previously been shown to facilitate the JNK signaling cascade as a scaffold protein rather than by its kinase activity (Yoneda et al., 2001), we sought to address whether TAOK3 kinase activity was required to suppress eWAT Tregs and ST2 expression. To do so, TAOK3 kinase dead mice (Taok3KD mice) were generated, where a lysine is substituted with an alanine at amino acid position 53, inhibiting ATP binding to the kinase domain (Maes et al., 2022). We found that Taok3KD mice phenocopied Taok3−/− mice with increased eWAT ST2+ Tregs numbers and increased ST2 expression compared to WT littermates (Fig. 2 J). Together, these data support a role for T cell–intrinsic kinase activity of TAOK3 in controlling eWAT-resident Tregs through ST2 signaling. More research is needed to pinpoint the stromal cells or hematopoietic cells that further contribute to some of the T cell–extrinsic effects of Taok3 deficiency on Treg numbers, phenotype, and function.
TAOK3 contributes to diet-induced weight gain and loss of Tregs in obesity
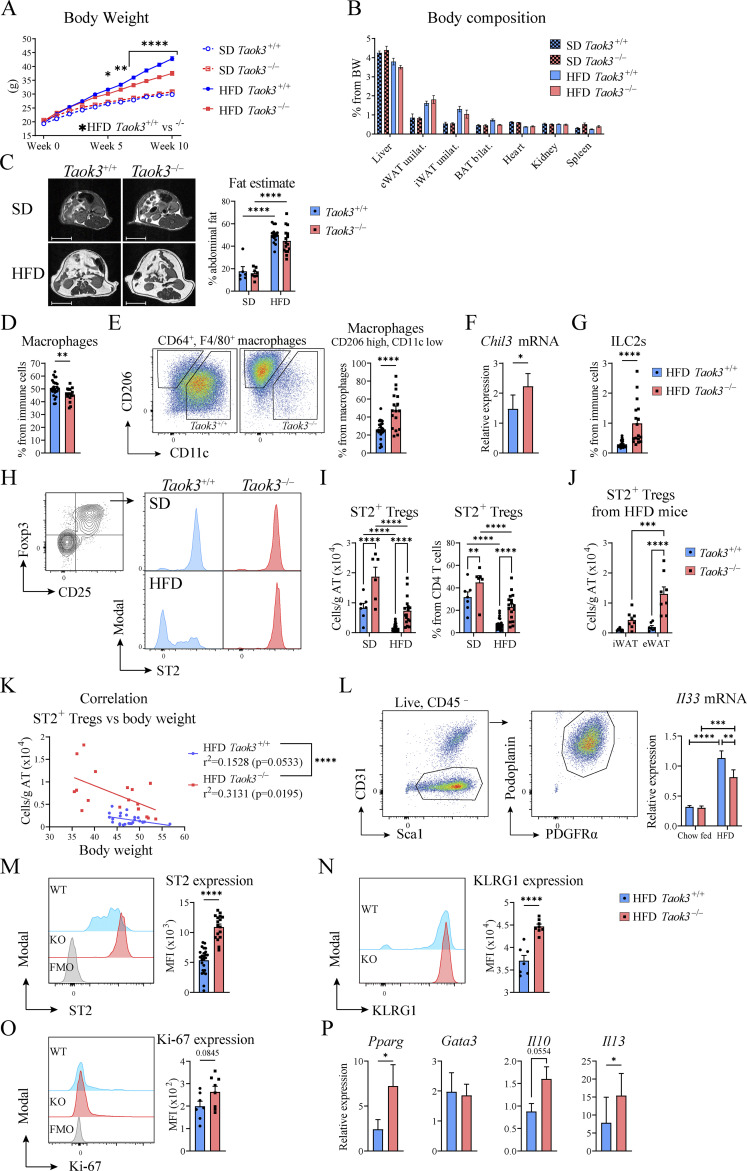
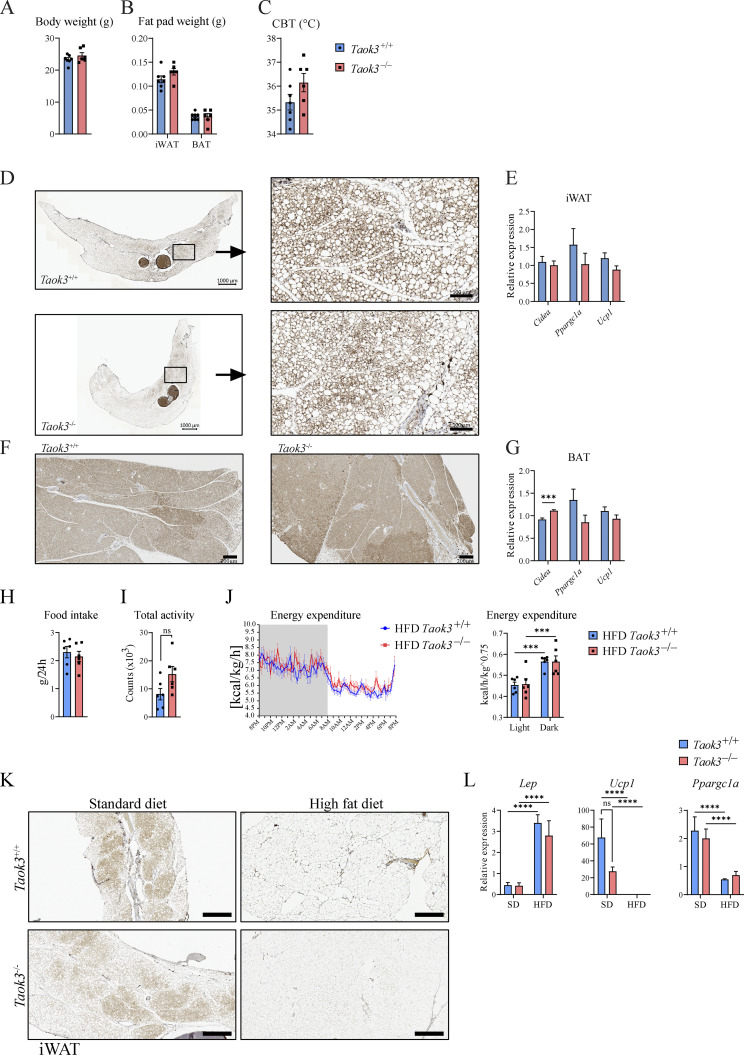
Previous studies have shown that Treg numbers in male eWAT are severely reduced at times of HFD feeding, and that depletion of Tregs from fat tissues leads to metabolic dysfunction (Feuerer et al., 2009). To address the effects of diet-induced obesity (DIO), Taok3−/− mice and Taok3+/+ littermates were fed a standard diet (SD) or HFD for at least 10 wk. Considering that males had the highest percentage of ST2+ Tregs in eWAT in homeostasis, we restricted experiments to male mice, and confirmed that mice on HFD gained more weight compared to SD mice. We observed that HFD-induced weight gain was less pronounced in Taok3−/− mice compared with Taok3+/+ littermates (Fig. 3 A). Despite attempts to localize this difference in weight gain using weight measurements of individual organs and AT depots and use of MRI to localize fat, we could not reliably pinpoint differences in body composition, or abdominal fat content between genotypes on either SD or HFD (Fig. 3, B and C). We found no differences in AT beiging in lean or HFD-fed Taok3−/− or Taok3+/+ mice (Fig. S3, A–G, K and L), nor did we observe differences in food intake or whole body energy expenditure between genotypes on HFD (Fig. S3, H–J).
Figure 3.
TAOK3 is required for HFD to induce gain weight and to deplete ST2+ Tregs in eWAT. (A) Body weight curves of Taok3−/− and WT control mice placed on a SD or HFD. (B) Organ weight as a percentage of total body weight (BW) as a measure for body composition. (C) Representative MRI images of the abdominal region of Taok3+/+ and Taok3−/− mice after 12 wk of SD/HFD (left; scale bar = 1 cm). Right, abdominal AT content estimates as calculated by MRI in SD/HFD-fed Taok3+/+ and Taok3−/− mice. (D) Macrophages as a percentage from immune cells in eWAT from HFD-fed Taok3+/+ or Taok3−/− mice. (E) Representative plot for CD206 and CD11c expression in AT macrophages (left). Right, percentages of CD206high, CD11clow macrophages from total AT macrophages. (F) Chil3 expression within the SVF isolated from HFD-fed mice. (G) ILC2s, as a percentage of immune cells in eWAT. ILC2s are defined as lineage negative, CD45+ST2+Ly-6A/E+CD127+KLRG1+. Lineage includes CD4, CD8, CD11c, B220, TCRβ, TCRγδ, Ly-6C/G, Ter119, NK1.1, CD19, CD3e, CD11b. (H) Representative plots for MFI as a measure for ST2 expression in eWAT Foxp3+CD25+ Tregs taken from SD and HFD-fed Taok3+/+ and Taok3−/− mice. (I) ST2+ Treg numbers (per gram AT; left) and as percentage of CD4 T cells in eWAT (right). (J) ST2+ Treg numbers (per gram AT) in iWAT or eWAT taken from HFD-fed Taok3+/+ and Taok3−/− mice. (K) Plot depicting body weight versus ST2+ Tregs per gram AT. R squared (r2) values are calculated for correlation between body weight and cell numbers for both Taok3+/+ and Taok3−/− mice. Next, simple linear regression was used to calculate the slopes and to determine if elevations (Y intercepts) were statistically significantly different. Data pooled from three independent experiments (n = 17–25). (L) Stromal cells were sorted from normal chow–fed or HFD-fed Taok3+/+ and Taok3−/− and Il33 mRNA levels were determined by qPCR. (M) Representative plot for MFI as a measure for ST2 expression on ST2+ Tregs in eWAT of HFD-fed mice (left). Right, ST2 expression (MFI) in ST+ Tregs in eWAT of HFD-fed mice. (N) Representative plot showing KLRG1 expression on ST2+ Tregs in eWAT of HFD-fed mice (left). Right, KLRG1 expression (MFI) in ST+ Tregs in eWAT of HFD-fed mice. (O) Representative plot showing Ki-67 expression in ST2+ Tregs in eWAT of HFD-fed mice (left). Right, Ki-67 expression (MFI) in ST+ Tregs in eWAT of HFD-fed mice. (P) mRNA expression for Pparg, Gata3, Il10, and Il13 in sorted CD3+CD4+CD25+ lymphocytes from eWAT from HFD-fed mice. Data from A are pooled from four independent experiments (n = 27–32 mice/group). B and C, data pooled from one (SD, n = 6–8 mice/group) or two (HFD, n = 16 mice/group) independent experiment(s). For flow experiments, a different cohort of mice was used. D, E, G, and M, data pooled from three independent experiments (n = 17–25 mice/group). H and I, data pooled from one experiment (SD, n = 6–7 mice/group) or three (HFD, n = 17–25 mice/group) independent experiments. J, data from one experiment (n = 7 mice/group). L, N, and O, data representative for two independent experiments (n = 6–7 mice/group). P, data pooled from two independent experiments (n = 9 samples/group). Statistical analysis: two-way ANOVA with correction for multiple comparisons was used (A–C, I, J, and L). Statistical significant differences between genotypes within mice on the same diet, and differences between SD vs. HFD within mice from the same genotype were depicted. For comparisons between two groups, data were analyzed with Shapiro-Wilk normality test to assess whether data were normally distributed. Parametric data were analyzed with an unpaired two-tailed t test and nonparametric data with a two-tailed Mann-Whitney test (D, E, G, M–O). Data representing mRNA transcript expression ratios were log transformed before statistical analysis (F, L–P). Data are shown as means ± SEMs. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Figure S3.
TAOK3 does not control AT beiging, food intake, or whole body energy expenditure. (A) Body weight from 15-wkp-old Taok3+/+ and Taok3−/− mice fed a chow diet (g). (B) iWAT and brown adipose tissue (BAT) fat pad weight (g). (C) Core body temperature (°C). (D) Representative images from Ucp1 staining of iWAT taken from Taok3+/+ and Taok3−/− mice; scale bar, 1,000 µm (left) and 100 µm (right). Note that we focus on subcutaneous iWAT, since beiging is mainly restricted to this compartment and not eWAT. (E) mRNA transcript expression of browning genes (Cidea, Ppargc1a, and Ucp1) in iWAT. (F) Representative images from Ucp1 staining of BAT taken from Taok3+/+ and Taok3−/− mice; scale bar, 200 µm. (G) mRNA transcript expression of browning genes in BAT. (H) Food intake from mice fed HFD for 5 wk. (I) Total activity (day and night combined) measured in mice fed HFD for 5 wk. (J) Plot depicting energy expenditure (left) and energy expenditure quantification (right). (K) Representative microscopy images from Ucp1 staining of iWAT taken from Taok3+/+ and Taok3−/− mice, fed an SD or HFD. Scale bar, 500 µm. (L) mRNA transcript expression of Lep, Ucp1, Ppargc1a in iWAT. A–G, data from one experiment (n = 7 and six mice/group). H, data representative for two independent experiments (n = 6 mice/group). I and J, data from one experiment (n = 6 mice/group). K and L, data representative for two independent experiments (SD, n = 6, 8 mice/group; HFD, n = 8 mice/group). Data are shown as means ± SEMs. ***P < 0.001, ****P < 0.0001.
The crucial role for IL-33 in adipocyte homeostasis is reflected by improvement in glucose homeostasis and body weight in mice treated with IL-33, or conversely, development of insulin resistance and obesity in mice lacking IL-33 or its receptor (Mahlakõiv et al., 2019). These effects of IL-33 can be mediated by Tregs, but partially also by IL-13 that has the potential to promote accumulation of metabolically favorable M2 macrophages, eosinophils, and ILC2s in AT (Vasanthakumar et al., 2015; Brestoff et al., 2015; Han et al., 2015; Miller et al., 2010; Duffen et al., 2018). We therefore looked for evidence of type 2 immunity and IL-33 bioactivity in WT and Taok3−/− mice fed an HFD. We found that there were slightly less macrophages in eWAT from HFD-fed Taok3−/− mice, compared with Taok3+/+ mice (Fig. 3 D), yet macrophages within eWAT from HFD-fed Taok3−/− mice were strongly skewed toward a CD206high, CD11clow phenotype, that is typically associated with lean AT (Fig. 3 E; gating strategy depicted in Fig. S2 C). These macrophages also express more mRNA encoding the chitinase like protein Chil3 (Ym1), a marker of type 2 skewed macrophages in the SVF (Fig. 3 F). Additionally, we noticed that ILC2s, another IL-33 responsive population, were upregulated in Taok3−/− eWAT taken from HFD-fed mice, again indicative for a type 2 skewed immune environment in Taok3−/− mice (Fig. 3 G).
We next assessed whether IL-33 responsive ST2+ Tregs were also spared during DIO in eWAT taken from Taok3−/− mice. The number and percentage of ST2+ Tregs in eWAT harvested from WT mice was strongly reduced upon DIO, but this was not the case in Taok3−/− mice (Fig. 3, H and I). Since these mice were already at least 18 wk old, absolute number and the percentage of ST2+ Tregs among CD4 T cells (Fig. 3 I) was higher than in younger mice (Fig. 1, D and E) fed a regular chow diet, yet further enhanced by lack of Taok3 in mice on an SD. The number of ST2+ Tregs per gram eWAT in HFD-fed Taok3−/− mice (around 10 × 103 cells per gram) approached the numbers seen in young Taok3−/− mice fed a normal chow diet (Fig. 1 D), suggesting lack of Taok3 maintains the fat tissue in a healthy state. The effects of Taok3 deficiency on ST2+ Tregs in the HFD state were less pronounced in the inguinal (i)WAT depot (Fig. 3 J).
Since body weight confounds immune cell distribution in eWAT from obese mice, and Taok3−/− mice were leaner compared with WT littermates upon DIO (Fig. 3 A), we also studied the relationship between body weight and Treg abundance in eWAT. Upon HFD feeding, body weight was inversely correlated with the number of ST2+ Tregs in eWAT in Taok3+/+ and Taok3−/− mice alike (P values of P = 0.0533 and P = 0.0195, respectively; Fig. 3 K). However, the elevation (Y-interception) of the Taok3−/− slope was significantly higher compared with the slope of Taok3+/+ mice. Consequently, mice with the same body weight consistently had higher numbers of ST2+ Tregs in Taok3−/− mice compared to WTs, demonstrating that differences in body weight were not solely responsible for the sparing of ST2+ Tregs in Taok3−/− mice. Together this indicates that lack of Taok3 attenuated DIO and maintained a type 2 polarized immune environment rich in ST2+ Tregs.
TAOK3 controls sensitivity to IL-33, suppressing PPARγ-expressing Tregs in obesogenic conditions
To study if IL-33 production in eWAT was altered by TAOK3 in lean or obesogenic conditions, we FACS sorted PDGFRα+PDPN+ mesenchymal stromal cells from eWAT taken from normal chow-fed or HFD-fed Taok3 WT and KO mice and analyzed Il33 mRNA levels (Fig. 3 L). In lean mice, there were no differences in Il33 mRNA between genotypes. Consistent with previous reports, we found that obesity was associated with increased Il33 expression in AT stromal cells. Interestingly, rather than observing increased Il33 expression in Taok3−/− stromal cells, Il33 expression was reduced in Taok3−/− stromal cells in obesogenic conditions. This reduction might reflect the negative feedback loop controlled by eWAT ST2+ Tregs on IL-33–expressing stromal cells that was previously proposed by Mathis and colleagues (Spallanzani et al., 2019). Obesity is a potent suppressor of the typical beneficial AT phenotype of visceral adipose tissue Tregs, characterized by high expression of ST2, KLRG1, and PPARγ. However, in DIO, we also observed higher ST2 expression (Fig. 3 M) and higher expression of the activation marker KLRG1 (Fig. 3 N) on eWAT Tregs of Taok3−/− mice, suggesting that these cells were activated, potentially via IL-33. IL-33 is also a well-known proliferation signal for ST2+ Tregs (Vasanthakumar et al., 2015; Kolodin et al., 2015), yet Ki-67 expression was similar between ST2+ Tregs from HFD-fed Taok3−/− mice and their WT controls, suggesting equal proliferation (Fig. 3 O). We measured the mRNA transcripts of key AT Treg transcription factors in CD4+CD25+ Tregs sorted from HFD-fed Taok3+/+ and Taok3−/− mice, and found that Taok3−/− Tregs had double the amount of Pparg mRNA transcripts, and equal amounts of Gata3 transcripts (Fig. 3 P), a clear difference compared with younger Taok3−/− mice on standard chow (Fig. 2 I). In conventional T cells and in AT ILC2s, PPARγ was shown to control expression of type 2 cytokines, including IL-13 (Chen et al., 2017; Fali et al., 2021). Accordingly, in eWAT Tregs of Taok3−/− mice, there were also more Il13 transcripts compared with Taok3+/+ mice (Fig. 3 P), and more Il10 transcripts although this failed to reach statistical significance (Fig. 3 P). In conclusion, in DIO the Taok3-deficient mice show signs of enhanced sensitivity to IL-33 in visceral AT, leading to accumulation of activated ST2+ Tregs, that express higher amounts of PPARγ and type 2 cytokines. Enhanced IL-13 in eWAT Tregs of Taok3−/− mice could be the explanation for the maintenance of CD206+ alternatively activated macrophages in these mice under HFD, since these cells accumulate in an IL4Rα-dependent manner (Wu et al., 2011).
TAOK3 promotes metabolic dysfunction in HFD-induced obesity
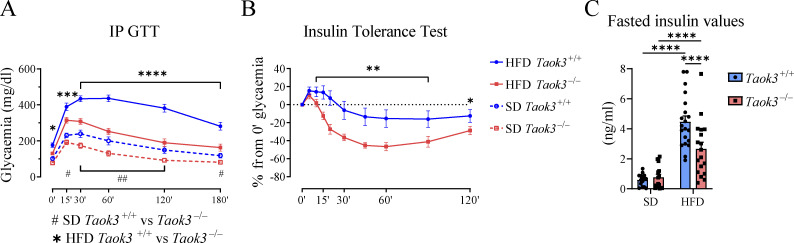
Tissue-resident ST2+ Tregs in eWAT contribute to a type 2 cytokine environment in lean AT that keeps inflammation in check and promotes insulin sensitivity, particularly in young mice (Vasanthakumar et al., 2015; Cipolletta et al., 2012). Tregs have also been shown to attenuate WAT inflammation and improve metabolic homeostasis in obesity (Feuerer et al., 2009; Winer et al., 2009; Eller et al., 2011). Given the accumulation of eWAT Tregs in lean and obese conditions, we studied the metabolic consequences of Taok3 deficiency in mice on an SD and HFD, using the intraperitoneal glucose tolerance test (IP GTT). In WT mice, HFD led to abnormal glucose tolerance, as reflected by higher and more sustained blood glucose concentration after IP injection, compared with SD-fed mice (Fig. 4 A). Glucose tolerance was, however, markedly better in HFD-fed Taok3−/− mice, compared with littermate Taok3+/+ mice, suggesting TAOK3 is a mediator of the metabolic derangement caused by excessive caloric intake (Fig. 4 A). Improved glucose tolerance was also observed Taok3−/− mice on an SD diet. This could be due to their accumulation of ST2+ Tregs in lean eWAT promoting insulin sensitivity. Depleting Tregs from lean mice has indeed been shown to induce insulin resistance (Feuerer et al., 2009). To probe for this, we measured insulin sensitivity. After an injection of a bolus of insulin, HFD-fed Taok3−/− mice displayed vastly improved insulin sensitivity, as reflected by a more marked and sustained drop in the blood glucose concentration (Fig. 4 B). We also studied insulin resistance more directly, by studying fasting serum insulin concentration (Fig. 4 C). In Taok3+/+ mice, an HFD feeding led to higher fasted serum insulin concentrations compared with SD-fed mice, reflective of HFD-induced insulin resistance in this model. Fasted serum insulin concentration was, however, significantly lower in HFD-fed Taok3−/− mice, indicating reduced insulin resistance compared to WTs (Fig. 4 C). The improved insulin sensitivity did not merely reflect mild differences in body weight, as weight-matched Taok3−/− animals on HFD were also protected from metabolic dysfunction (data not shown). Thus, our data show that TAOK3 controls the levels of type 2 immunity and ST2+ Tregs in AT and is a key mediator of HFD-induced metabolic dysfunction.
Figure 4.
TAOK3 promotes metabolic dysfunction in HFD-induced obesity. (A) IP GTT in Taok3+/+ and Taok3−/− mice fed an SD or HFD for 15 wk. (B) Intraperitoneal insulin tolerance test in mice fed an HFD for 15 wk. (C) Serum insulin values (fasted) at 15 wk of special diet. Data is pooled from at least four independent experiments (SD, n = 15–17, HFD, n = 25–27 mice/group). Data were analyzed by two-way ANOVA with correction for multiple comparisons. Statistical significant differences between genotypes within mice on the same diet (A and B), and differences between diets within mice from the same genotype were depicted (C). Data are shown as means ± SEMs. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
In this paper, we show that Taok3−/− mice are protected from developing HFD-induced metabolic dysfunction and gain less weight, accompanied by an increase in eWAT ST2+ Tregs that expressed higher levels of type 2 cytokines, and by increases in alternatively activated CD206high macrophages and ILC2s, reflecting a general type 2 bias. Disentangling the relative contributions of AT Tregs versus ILC2s and the precise effector functions of the CD206+ alternatively activated macrophages (cell types that can all be stimulated by IL-33 and/or IL-13) has proven to be challenging, as these cell types often depend on reciprocal cooperation (Brestoff and Artis, 2015; Molofsky et al., 2015; Man et al., 2022). The underlying mechanisms driving anti-inflammatory and insulin-sensitizing effects of eWAT Tregs are generally poorly understood. Interestingly, the PPARγ agonist pioglitazone partially exerts its insulin sensitizing effects through PPARγ-expressing visceral AT Tregs upon DIO (Cipolletta et al., 2012). Importantly, Taok3−/− Tregs in obese eWAT had more mRNA transcripts of Pparg and might as such improve insulin sensitivity. Tregs could also regulate fat homeostasis via release of IL-10. IL-10 has been shown to dampen TNFα-induced pro-inflammatory cytokine production in adipocytes and regulate Glut4 expression in vitro (Feuerer et al., 2009). Although we saw higher levels of IL-10 in Taok3−/− Tregs, we did not investigate the contribution of IL-10 in immunometabolism further.
We found Taok3 controls ST2 expression and Treg numbers in a T cell–intrinsic and tissue-specific manner, dependent on IL-33. Given that T cell–specific deletion of Taok3 expression was effective in boosting splenic ST2+ Tregs, but not eWAT ST2+ Tregs, we speculated that additional tissue-specific cues in eWAT, like IL-33 production by stromal cells, are needed to promote their accumulation in Taok3−/− mice. However, IL-33 expression was not increased in PDGFRα+ stromal cells isolated from Taok3−/− mice, and selectively targeting Taok3 in stromal cells did not fully phenocopy the Taok3−/− phenotype. The importance of IL-33 derived from eWAT stromal cells in driving Tregs in eWAT clearly deserves more study. Even in the first description of this pathway, deletion of IL-33 under control of the Pdgfra-promotor only led to a percentual reduction of eWAT Tregs, whereas total cell number was unaffected. This implied from the start that other sources of IL-33 or other AT cues also contribute to eWAT Treg maintenance (Spallanzani et al., 2019). Adding to the complexity, it was recently reported that AT Treg accumulation by exogenous IL-33 administration did not require Treg-intrinsic ST2 expression, indicating that IL-33 acts on an unidentified third-party cell type that controls eWAT-dwelling Tregs (Hemmers et al., 2021).
We recently reported that Taok3 deficiency leads to a defect in lymphotoxin-β−dependent ESAM+ type 2 conventional dendritic cells (cDC2s) in the spleen, and discrete abnormalities in gut cDC2s (Vanderkerken et al., 2020). Interestingly, a closely related population of lymphotoxin-β−dependent cDC2s controls lipid uptake in the gut, by driving expression of lipid transporters in the jejunum (Guendel et al., 2020). Future experiments will address if this population of gut cDC2s and their influence on dietary lipid uptake or more broad effects on gut physiology are affected in Taok3−/− mice (Vanderkerken et al., 2020). We have indeed noticed that the occurrence of a protective metabolic phenotype in Taok3−/− males is dependent on the microbial status of the animal facility. Although this suggests an important impact of gut or other microbiome, high abundance of ST2+ Tregs in Taok3−/− mice was observed across all experiments performed in different animal facilities and different background strains (data not shown). Another aspect of the immunophenotype of Taok3−/− mice is that they lack Notch2-dependent immune cells like marginal zone B cells and ESAM+ cDC2, effects that are reversed by cell-specific overexpression of constitutively active Notch intracellular domain (Vanderkerken et al., 2020; Hammad et al., 2017). Given that Notch signaling also profoundly affects metabolism (Pajvani et al., 2011, 2013; Bi et al., 2014), we also studied if Notch target genes were altered in eWAT of Taok3−/− mice, but did not find evidence of such regulation (data not shown).
The kinase domain of TAOK3 is highly conserved and structurally related to the kinase domain of TAOK1 and TAOK2, possibly entailing redundancy (Fang et al., 2020). Nevertheless, selective and cell-intrinsic loss of Taok3 causes distinct cellular phenotypes in vivo, suggesting cell context-dependent unique roles for the kinase activity (Hammad et al., 2017; Vanderkerken et al., 2020). TAOK3 has also been proposed to function as a scaffold protein on which other signaling intermediates convene to mediate activation or regulation, particularly during the unfolded protein response (Yoneda et al., 2001). Importantly, we have previously shown that disruption of the catalytic kinase domain of the TAOK3 protein was sufficient to protect these mice from developing eosinophilic asthma. Likewise, in the current report, we show that TAOK3 kinase activity is required to control ST expression and ST2+ Tregs in eWAT. This finding opens the possibility for targeting the TAOK3 pathway using highly specific small-molecule kinase inhibitors for immunomodulation in obesity and insulin resistance. Despite important differences between mice and men, our findings could also explain how “loss of function” methylation of the TAOK3 locus in humans is associated with reduced obesity risk (Huang et al., 2015). Since TAOK3 is a druggable STE20-kinase, selectively inhibiting its kinase activity could be pursued as an innovative angle to tackle obesity-associated metabolic derangement.
Materials and methods
Mice
Taok3−/− mice were described previously (Hammad et al., 2017). In brief, a loxP-flanked splicing acceptor was inserted as a gene trap in the intronic region between exon 1 and exon 2 of the Taok3 gene, leading to premature transcriptional termination. Alternatively, an additional whole body knock-out Taok3−/− was generated by the Transgenic Core Facility (TCF) of the Inflammation Research Center of VIB-UGent by injecting C57BL/6J zygotes with RNP complexes with gRNAs targeting a region in intron 5 (5′-GGGTAACTGTGGTGACTTTG-3′) and a region in intron 6 (5′-GGAGGCTGAGGCGGAACCAA-3′), resulting in the deletion of exon 6 (Vanderkerken et al., 2020). Conditional Taok3fl/fl mice were generated in house by the TCF with the Easi-CRISPR method (Quadros et al., 2017) and were described previously (Vanderkerken et al., 2020). Briefly, a long single-stranded DNA repair template containing exon 6, with upstream and downstream intronic region loxP sites inserted, was coinjected with the RNP complexes in zygotes of C57BL/6J mice. Using similar technology, Taok3KD were generated in house by the TCF by injecting along single-stranded DNA repair template containing exon 4 with an AAG-to-GCC mutation, causing a lysine-to-alanine mutation in position 53 of the Taok3 protein, predicted to be the catalytic site of Taok3 kinase domain (Maes et al., 2022). Mice lacking the IL-33-receptor ST2 [Il1rl1−/− mice (B6J-IL1rl1em1IRC)] were generated in house by the TCF by electroporating Cas9 RNP complex with guide sequence 5′-GTCGAGTACATCAAATAATC-3′ (gRNA1) and guide sequence 5′-CAGGTGCTGTCCAATTATAC-3′ (gRNA2), which resulted in a 1 bp insertion near the gRNA1 target site (chr1+: 40481539) and a 2 bp deletion near the gRNA2 target site (chr1+: 40481608-40481609) in exon4 ENSMUSE00000309818 of the Il1rl1 gene (ENSMUSG00000026069). This is predicted to cause amino acid sequence changes and a premature stop codon resulting in nonsense-mediated decay. Cd4-Cre [B6.Cg-Tg(Cd4-cre)1Cwi/BfluJ] mice (Lee et al., 2001) and Pdgfra-Cre [C57BL/6-Tg(Pdgfra-cre)1Clc/J] mice (Roesch et al., 2008) were acquired from the Jackson Laboratory. All experimental mice were on a C57BL/6J background and were housed in the VIB-UGent animal facility under specific pathogen–free conditions. Housing conditions entailed individually ventilated cages in a controlled day–night cycle, and food and water intake was ad libitum. For baseline immunophenotyping experiments (Fig. 1 and Fig. 2, A and B), both male and female mice, between 8–12 wk of age, were used. For all other experiments exclusively male mice were used. Mice were age- and sex-matched and randomly assigned to experimental groups. Taok3+/+ and Taok3−/− mice breedings are housed in separate cages, for economic and ethical reasons (i.e., avoiding a surplus of heterozygous mice that are not used in experiments). Every 2 yr WT and KO mice are put into breeding to generate heterozygous litter that is used to produce a new generation of WT and KO littermates that are used to set up fresh breedings. Doing so, genetic drift is avoided. In experiments using Taok3fl/fl mice crossed to Cre-lines, controls are Cre-negative littermates. No commercial source of C57BL/6 mice was used as controls. All animal experiments and procedures were approved by the animal ethics committee of the VIB Center for Inflammation Research and were in accordance with the Belgian animal protection law.
Metabolic phenotyping
Male mice were randomly assigned to rodent diet with 10% kcal% fat (#D12450Bi; SD, Research Diets Inc.) or rodent diet with 60% kcal% fat (#D12492i; HFD, Research Diets Inc.) at 5–6 wk of age. Glucose tolerance and insulin sensitivity were assessed at 10–15 wk of special diet. For IP GTT, mice were starved for food overnight and were then injected with D+ glucose (2 g/kg body weight, G7528-250G; Sigma-Aldrich). For insulin tolerance testing, mice were starved for food for 4 h and were then injected with human insulin Actrapid Penfill (0.75 mU/g body weight; Novo Nordisk). Glucose concentrations were measured from a drop of blood from the tail vein using FreeStyle Lite measuring strips (Abbott) and Freedom Freestyle Lite Sensor glucometer (Abbott). To measure serum insulin, mice were starved for food overnight and blood was taken from the tail vein. Serum was further analyzed with Ultra-Sensitive Mouse Insulin ELISA kit (Crystal Chem) according to the manufacturer’s instructions. Abdominal AT percentage estimation was assessed by 7 Tesla small animal MRI (Bruker BioSpin PharmaScan 70/16) at 10–13 wk of special diet feeding.
Isolation of tissue leukocytes
Cell suspensions from mLN and spleen were obtained through mechanical disruption and filtering over a 100-μm cell strainer. Lungs and WAT were cut with scissors. Lungs were then digested for 30 min in RPMI-1640 (Gibco, Thermo Fisher Scientific) containing 20 μg/ml liberase TM (Roche), 10 U/ml DNase I (Roche), and 10% of FCS (Bodinco) at 37°C. WAT was digested in DMEM (Gibco) containing 10 µg/ml liberase TM (Roche), 10 μ/ml DNase I (Roche), and 2% bovine serum albumin (A2153; Sigma-Aldrich) for 40–50 min. Next, lungs and WAT were filtered through a 100 µm cell strainer. Livers were smashed over a 100 µg cell strainer. Next, cells were washed and then the lymphocyte fraction was purified by using a 37.5% Percoll (GE Healthcare) gradient. For bone marrow, isolated tibia and femur bones were first cut from one end. Then bones were placed with the cut edge down in a 0.5 ml tube containing an opening in the bottom. Next, the 0.5 ml tube was placed inside a 1.5 ml tube and bone marrow was obtained by centrifugation. Blood was obtained from the inguinal artery in Microvette (Sarstedt) tubes containing EDTA. All samples were washed, followed by an incubation in ammonium chloride buffer (10 mM KHCO3, 155 mM NH4Cl, 0.1 mM EDTA in milliQ water) for erythrocyte lysis. Total cell counts were obtained by adding counting beads (123count eBeads, Thermo Fisher Scientific).
Treg/mesenchymal stromal cell sorting
Mice were euthanized with an overdose pentobarbital, eWAT depots and/or spleen were isolated, and tissue single-cell suspensions were obtained as described above. Cell suspensions were stained and Tregs (CD45+CD3+CD4+CD25+) or stromal cells (CD45–CD31–Sca1+PDPN+PDGFRα+) were fluorescence-activated cell sorted directly in into RNeasy Plus lysis buffer, and RNA was extracted with the RNeasy Plus Micro kit (Qiagen) according to the manufacturer’s protocol. RNA was amplified using WT-OVATION RNA amplification kit (#2210-24; NuGEN Technologies).
RNA isolation, cDNA synthesis, and quantitative PCR (qPCR)
For sorted cells RNA was isolated as described above. For determination of Chil3 expression on SVFs, mRNA was isolated by using Aurum total RNA mini kit (Bio-Rad). In Fig. S2, AT pads were disrupted using Qiagen TissueLyser beads and bead mill. RNA was isolated using TRIzol LS Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. For additional RNA purity, AT samples were additionally processed by using Aurum total RNA mini kit (Bio-Rad), according to the manufacturer’s instructions. The cDNA was synthesized with the Sensifast cDNA synthesis kit (Bioline, Meridian Bioscience). qRT-PCR reactions were conducted in a LightCycler 480 (Roche) using the SensiFAST sybr no-ROX mix (Bioline). Primer pairs are listed in Table S1. Data were analyzed using qBASE software (BioGazelle).
Indirect calorimetry
Mice, fed HFD for 5 wk, were individually housed in automated cages for indirect calorimetry (TSE Phenomaster Calocages) in a room with 22°C ambient temperature and a 12 h dark/light cycle, as described (Dubois et al., 2016). All mice had ad libitum access to water and food. Food intake, oxygen consumption, carbon dioxide production, and ambulatory activity were recorded over a 48-h period, but only the last 24 h were used for calculations, to exclude new cage environment bias. Respiratory exchange ratio and heat production were calculated as described (Ferrannini, 1988).
Microscopy and immunohistochemistry
After overnight fixation in 4% PFA solution, tissues were embedded in paraffin, cut into 5-µm slices, and processed for immunochemistry. Cut paraffin slides from ATs were first dehydrated and dewaxed; next, antigen was retrieved using Dako Real Target retrieval Solution (# S169984-2; Dako) and pressure cooker. Slides were washed in PBS and endogenous peroxidase was blocked by incubating the slides with 3% hydrogen peroxidase in PBS. After washing, tissue slides were blocked using 5% goat serum in 1% bovine serum albumin in PBS. Then slides were stained against Ucp1 (# ab23841; Abcam) at 4°C overnight. Immunoreactivity was detected by means of sequential incubations of AT sections with a secondary HRP antibody (P0448; Dako) and 3,3-diaminobenzidine chromogen (# K3468; Dako). Sections were mounted with Entellan mounting medium (Merck). Whole slides were acquired with ZEISS AxioScanner (ZEISS).
Flow cytometry
Single-cell suspensions were incubated with a fixable viability dye (eFluor506 or eFluor780 from eBioscience, Thermo Fisher Scientific) to identify dead cells and with an FcγRII/III antibody (2.4G2; Bioceros) for 30 min to counter aspecific binding. After washing, the cells were incubated for 30 min at 4°C with a mixture of fluorochrome-labeled antibodies (listed in Table S2). For some panels a separate staining step with biotinylated ST2-antibody was performed, followed by staining step with streptavidin.
For intracellular staining of Ki-67, GATA-3, Rorγt, and Foxp3 cells were fixed using the Foxp3 fixation/permeabilization kit (eBioscience, Thermo Fisher Scientific) according to the manufacturer’s protocol. Samples were acquired on an LSR Fortessa or BD Symphony cytometer with FACSDiva software (BD Biosciences) and analyzed using FlowJo software (TreeStar). Cell sorting was performed using a FACS Aria II (BD Biosciences).
Statistical analysis
When two groups were compared, the normality of each group was first checked by using the Shapiro-Wilk statistical test. Parametric data were analyzed with an unpaired two-tailed t test and nonparametric data with a two-tailed Mann–Whitney test. When more than two groups were analyzed parametric data were analyzed with an ordinary one-way ANOVA. Nonparametric data were analyzed with Kruskal–Wallis test. When two independent variables (diet or sex and genotype) were involved a two-way ANOVA was used. Correction for multiple testing was applied (two-stage step-up method of Benjamini, Krieger, Yekutieli). To analyze expression ratios (mRNA transcripts), ratios were first log transformed, and then the same statistical approach was used as described above. Statistical significance was accepted at a P value of <0.05. P values are represented as *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. These statistical tests were performed in GraphPad Prism software (GraphPad Software).
Online supplemental material
Fig. S1 describes an alternative gating strategy for Tregs and absolute and relative numbers of CD25+Foxp3+ Tregs in ATs of WT and Taok3−/− mice. Fig. S2 contains the gating strategy of AT immune cells of WT and Taok3−/− mice. Fig. S3 contains data on food intake, whole body metabolism, energy expenditure, and fat tissue adaptations of WT and Taok3−/− mice.
Supplementary Material
shows qPCR primers.
shows flow cytometry antibodies.
Acknowledgments
We thank the technicians of the Sophie Janssens lab, Hammad and Lambrecht lab. We thank the VIB Flow Core for training, support, and access to the instrumental park. We also like to thank the collaborators of the Small Animal Imaging facility (INFINITY) of Ghent University. We also thank the TCF of the Center for Inflammation Research (VIB-Ghent University) for the generation of several of the mouse lines.
B. Maes is supported by a Research Foundation–Flanders (FWO) grant (3F015615). S. Janssens is supported by a European Research Council Consolidator Grant (DCRIDDLE-819314), FWO Program Grants (G086015, G063218 and G050622N) and FWO Excellence of Science research grant (G0G7318N). B.N. Lambrecht is supported by a European Research Council advanced grant (789384 ASTHMA CRYSTAL CLEAR), a concerted research initiative grant from Ghent University (GOA, 01 G010C9), an FWO Methusalem grant (01M01521), and an FWO Excellence of Science research grant (3 G0H1222). L. Catrysse was supported as a PhD fellow by the Instituut voor Innovatie door Wetenschap en Technologie and by a research grant from Kom op tegen Kanker.
Author contributions: B. Maes and F. Fayazpour performed the bulk of the experiments. B. Maes analyzed the data and wrote the manuscript. L. Catrysse provided ideas and helped with several experiments. G. Lornet, E. Van De Velde, C. De Wolf, S. De Prijck, J. Van Moorleghem, M. Vanheerswynghels, and K. Deswarte provided help during several experiments. B. Descamps and C. Vanhove performed MRI imaging. R. Vangoitsenhoven and B. Van der Schueren performed metabolic cage experiments and provided tools and ideas. H. Hammad provided tools and ideas. S. Janssens and B.N. Lambrecht provided ideas, analyzed the data, coordinated the project, wrote the manuscript, and supervised the final draft of the manuscript.
Data availability
The data are available from the corresponding author upon reasonable request (bart.lambrecht@ugent.be).
References
- Bapat, S.P., Myoung Suh J., Fang S., Liu S., Zhang Y., Cheng A., Zhou C., Liang Y., LeBlanc M., Liddle C., et al. 2015. Depletion of fat-resident Treg cells prevents age-associated insulin resistance. Nature. 528:137–141. 10.1038/nature16151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biener, A.I., Cawley J., and Meyerhoefer C.. 2020. The medical care costs of obesity and severe obesity in youth: An instrumental variables approach. Health Econ. 29:624–639. 10.1002/hec.4007 [DOI] [PubMed] [Google Scholar]
- Bi, P., Shan T., Liu W., Yue F., Yang X., Liang X.R., Wang J., Li J., Carlesso N., Liu X., and Kuang S.. 2014. Inhibition of Notch signaling promotes browning of white adipose tissue and ameliorates obesity. Nat. Med. 20:911–918. 10.1038/nm.3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brestoff, J.R., and Artis D.. 2015. Immune regulation of metabolic homeostasis in health and disease. Cell. 161:146–160. 10.1016/j.cell.2015.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brestoff, J.R., Kim B.S., Saenz S.A., Stine R.R., Monticelli L.A., Sonnenberg G.F., Thome J.J., Farber D.L., Lutfy K., Seale P., and Artis D.. 2015. Group 2 innate lymphoid cells promote beiging of white adipose tissue and limit obesity. Nature. 519:242–246. 10.1038/nature14115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgio, E., Lopomo A., and Migliore L.. 2015. Obesity and diabetes: From genetics to epigenetics. Mol. Biol. Rep. 42:799–818. 10.1007/s11033-014-3751-z [DOI] [PubMed] [Google Scholar]
- Cani, P.D.. 2019. Microbiota and metabolites in metabolic diseases. Nat. Rev. Endocrinol. 15:69–70. 10.1038/s41574-018-0143-9 [DOI] [PubMed] [Google Scholar]
- Chen, T., Tibbitt C.A., Feng X., Stark J.M., Rohrbeck L., Rausch L., Sedimbi S.K., Karlsson M.C.I., Lambrecht B.N., Karlsson Hedestam G.B., et al. 2017. PPAR-γ promotes type 2 immune responses in allergy and nematode infection. Sci. Immunol. 2:eaal5196. 10.1126/sciimmunol.aal5196 [DOI] [PubMed] [Google Scholar]
- Cipolletta, D., Cohen P., Spiegelman B.M., Benoist C., and Mathis D.. 2015. Appearance and disappearance of the mRNA signature characteristic of Treg cells in visceral adipose tissue: Age, diet, and PPARγ effects. Proc. Natl. Acad. Sci. USA. 112:482–487. 10.1073/pnas.1423486112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolletta, D., Feuerer M., Li A., Kamei N., Lee J., Shoelson S.E., Benoist C., and Mathis D.. 2012. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 486:549–553. 10.1038/nature11132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpire, E. 2009. The mammalian family of sterile 20p-like protein kinases. Pflugers Arch. 458:953–967. 10.1007/s00424-009-0674-y [DOI] [PubMed] [Google Scholar]
- Dubois, V., Laurent M.R., Jardi F., Antonio L., Lemaire K., Goyvaerts L., Deldicque L., Carmeliet G., Decallonne B., Vanderschueren D., et al. 2016. Androgen Deficiency Exacerbates High-Fat Diet-Induced Metabolic Alterations in Male Mice. Endocrinology. 157:648–665. 10.1210/en.2015-1713 [DOI] [PubMed] [Google Scholar]
- Duffen, J., Zhang M., Masek-Hammerman K., Nunez A., Brennan A., Jones J.E.C., Morin J., Nocka K., and Kasaian M.. 2018. Modulation of the IL-33/IL-13 Axis in obesity by IL-13Rα2. J. Immunol. 200:1347–1359. 10.4049/jimmunol.1701256 [DOI] [PubMed] [Google Scholar]
- Eller, K., Kirsch A., Wolf A.M., Sopper S., Tagwerker A., Stanzl U., Wolf D., Patsch W., Rosenkranz A.R., and Eller P.. 2011. Potential role of regulatory T cells in reversing obesity-linked insulin resistance and diabetic nephropathy. Diabetes. 60:2954–2962. 10.2337/db11-0358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fali, T., Aychek T., Ferhat M., Jouzeau J.Y., Busslinger M., Moulin D., and Eberl G.. 2021. Metabolic regulation by PPARγ is required for IL-33-mediated activation of ILC2s in lung and adipose tissue. Mucosal Immunol. 14:585–593. 10.1038/s41385-020-00351-w [DOI] [PubMed] [Google Scholar]
- Fang, C.Y., Lai T.C., Hsiao M., and Chang Y.C.. 2020. The diverse roles of tao kinases in health and diseases. Int. J. Mol. Sci. 21:1–21. 10.3390/ijms21207463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrannini, E. 1988. The theoretical bases of indirect calorimetry: a review. Metabolism. 37:287–301. 10.1016/0026-0495(88)90110-2 [DOI] [PubMed] [Google Scholar]
- Feuerer, M., Herrero L., Cipolletta D., Naaz A., Wong J., Nayer A., Lee J., Goldfine A.B., Benoist C., Shoelson S., and Mathis D.. 2009. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat. Med. 15:930–939. 10.1038/nm.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, E.L., Shchukina I., Youm Y.H., Ryu S., Tsusaka T., Young K.C., Camell C.D., Dlugos T., Artyomov M.N., and Dixit V.D.. 2021. IL-33 causes thermogenic failure in aging by expanding dysfunctional adipose ILC2. Cell Metab. 33:2277–2287.e5. 10.1016/j.cmet.2021.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guendel, F., Kofoed-Branzk M., Gronke K., Tizian C., Witkowski M., Cheng H.W., Heinz G.A., Heinrich F., Durek P., Norris P.S., et al. 2020. Group 3 innate lymphoid cells Program a distinct subset of IL-22BP-producing dendritic cells demarcating solitary intestinal lymphoid tissues. Immunity. 53:1015–1032.e8. 10.1016/j.immuni.2020.10.012 [DOI] [PubMed] [Google Scholar]
- Hammad, H., Vanderkerken M., Pouliot P., Deswarte K., Toussaint W., Vergote K., Vandersarren L., Janssens S., Ramou I., Savvides S.N., et al. 2017. Transitional B cells commit to marginal zone B cell fate by Taok3-mediated surface expression of ADAM10. Nat. Immunol. 18:313–320. 10.1038/ni.3657 [DOI] [PubMed] [Google Scholar]
- Han, J.M., Wu D., Denroche H.C., Yao Y., Verchere C.B., and Levings M.K.. 2015. IL-33 reverses an obesity-induced deficit in visceral adipose tissue ST2+ T regulatory cells and ameliorates adipose tissue inflammation and insulin resistance. J. Immunol. 194:4777–4783. 10.4049/jimmunol.1500020 [DOI] [PubMed] [Google Scholar]
- Hasan, A., Al-Ghimlas F., Warsame S., Al-Hubail A., Ahmad R., Bennakhi A., Al-Arouj M., Behbehani K., Dehbi M., and Dermime S.. 2014. IL-33 is negatively associated with the BMI and confers a protective lipid/metabolic profile in non-diabetic but not diabetic subjects. BMC Immunol. 15:19. 10.1186/1471-2172-15-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa, M., Yanagisawa K., Aoki S., Hayakawa H., Takezako N., and Tominaga S.. 2005. T-helper type 2 cell-specific expression of the ST2 gene is regulated by transcription factor GATA-3. Biochim. Biophys. Acta. 1728:53–64. 10.1016/j.bbaexp.2005.01.012 [DOI] [PubMed] [Google Scholar]
- Hemmers, S., Schizas M., and Rudensky A.Y.. 2021. T reg cell-intrinsic requirements for ST2 signaling in health and neuroinflammation. J. Exp. Med. 218:e20201234. 10.1084/jem.20201234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil, G.S.. 2017. Foundations of immunometabolism and implications for metabolic health and disease. Immunity. 47:406–420. 10.1016/j.immuni.2017.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, R.C., Garratt E.S., Pan H., Wu Y., Davis E.A., Barton S.J., Burdge G.C., Godfrey K.M., Holbrook J.D., and Lillycrop K.A.. 2015. Genome-wide methylation analysis identifies differentially methylated CpG loci associated with severe obesity in childhood. Epigenetics. 10:995–1005. 10.1080/15592294.2015.1080411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti, T.D., and Dixit V.D.. 2012. Immunological complications of obesity. Nat. Immunol. 13:707–712. 10.1038/ni.2343 [DOI] [PubMed] [Google Scholar]
- Kolodin, D., van Panhuys N., Li C., Magnuson A.M., Cipolletta D., Miller C.M., Wagers A., Germain R.N., Benoist C., and Mathis D.. 2015. Antigen- and cytokine-driven accumulation of regulatory T cells in visceral adipose tissue of lean mice. Cell Metabol. 21:543–557. 10.1016/j.cmet.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, P.P., Fitzpatrick D.R., Beard C., Jessup H.K., Lehar S., Makar K.W., Pérez-Melgosa M., Sweetser M.T., Schlissel M.S., Nguyen S., et al. 2001. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 15:763–774. 10.1016/S1074-7613(01)00227-8 [DOI] [PubMed] [Google Scholar]
- Lee, Y.S., Wollam J., and Olefsky J.M.. 2018. An integrated view of immunometabolism. Cell. 172:22–40. 10.1016/j.cell.2017.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., DiSpirito J.R., Zemmour D., Spallanzani R.G., Kuswanto W., Benoist C., and Mathis D.. 2018. TCR transgenic mice reveal stepwise, multi-site acquisition of the distinctive fat-treg phenotype. Cell. 174:285–299.e12. 10.1016/j.cell.2018.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., Spallanzani R.G., and Mathis D.. 2020. Visceral adipose tissue Tregs and the cells that nurture them. Immunol. Rev. 295:114–125. 10.1111/imr.12850 [DOI] [PubMed] [Google Scholar]
- Maes, B., Smole U., Vanderkerken M., Deswarte K., Van Moorleghem J., Vergote K., Vanheerswynghels M., De Wolf C., De Prijck S., Debeuf N., et al. 2022. The STE20 kinase TAOK3 controls the development of house dust mite-induced asthma in mice. J. Allergy Clin. Immunol. 149:1413–1427.e2. 10.1016/j.jaci.2021.08.020 [DOI] [PubMed] [Google Scholar]
- Mahlakõiv, T., Flamar A.L., Johnston L.K., Moriyama S., Putzel G.G., Bryce P.J., and Artis D.. 2019. Stromal cells maintain immune cell homeostasis in adipose tissue via production of interleukin-33. Sci. Immunol. 4:eaax0416. 10.1126/sciimmunol.aax0416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man, K., Kallies A., and Vasanthakumar A.. 2022. Resident and migratory adipose immune cells control systemic metabolism and thermogenesis. Cell. Mol. Immunol. 19:421–431. 10.1038/s41423-021-00804-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melén, E., Himes B.E., Brehm J.M., Boutaoui N., Klanderman B.J., Sylvia J.S., and Lasky-Su J.. 2010. Analyses of shared genetic factors between asthma and obesity in children. J. Allergy Clin Immunol. 126:631–7.e1-8. 10.1016/j.jaci.2010.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, A.M., Asquith D.L., Hueber A.J., Anderson L.A., Holmes W.M., McKenzie A.N., Xu D., Sattar N., McInnes I.B., and Liew F.Y.. 2010. Interleukin-33 induces protective effects in adipose tissue inflammation during obesity in mice. Circ. Res. 107:650–658. 10.1161/CIRCRESAHA.110.218867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky, A.B., Van Gool F., Liang H.E., Van Dyken S.J., Nussbaum J.C., Lee J., Bluestone J.A., and Locksley R.M.. 2015. Interleukin-33 and interferon-γ counter-regulate group 2 innate lymphoid cell activation during immune perturbation. Immunity. 43:161–174. 10.1016/j.immuni.2015.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky, A.B., Nussbaum J.C., Liang H.E., Van Dyken S.J., Cheng L.E., Mohapatra A., Chawla A., and Locksley R.M.. 2013. Innate lymphoid type 2 cells sustain visceral adipose tissue eosinophils and alternatively activated macrophages. J. Exp. Med. 210:535–549. 10.1084/jem.20121964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Must, A., Spadano J., Coakley E.H., Field A.E., Colditz G., and Dietz W.H.. 1999. The disease burden associated with overweight and obesity. JAMA. 282:1523–1529. 10.1001/jama.282.16.1523 [DOI] [PubMed] [Google Scholar]
- Nawijn, M.C., Dingjan G.M., Ferreira R., Lambrecht B.N., Karis A., Grosveld F., Savelkoul H., and Hendriks R.W.. 2001. Enforced expression of GATA-3 in transgenic mice inhibits Th1 differentiation and induces the formation of a T1/ST2-expressing Th2-committed T cell compartment in vivo. J. Immunol. 167:724–732. 10.4049/jimmunol.167.2.724 [DOI] [PubMed] [Google Scholar]
- Nikolic, I., Leiva M., and Sabio G.. 2020. The role of stress kinases in metabolic disease. Nat. Rev. Endocrinol. 16:697–716. 10.1038/s41574-020-00418-5 [DOI] [PubMed] [Google Scholar]
- Ormonde, J.V.S., Li Z., Stegen C., and Madrenas J.. 2018. TAOK3 regulates canonical TCR signaling by preventing early SHP-1-Mediated inactivation of LCK. J. Immunol. 201:3431–3442. 10.4049/jimmunol.1800284 [DOI] [PubMed] [Google Scholar]
- Pajvani, U.B., Qiang L., Kangsamaksin T., Kitajewski J., Ginsberg H.N., and Accili D.. 2013. Inhibition of Notch uncouples Akt activation from hepatic lipid accumulation by decreasing mTorc1 stability. Nat. Med. 19:1054–1060. 10.1038/nm.3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajvani, U.B., Shawber C.J., Samuel V.T., Birkenfeld A.L., Shulman G.I., Kitajewski J., and Accili D.. 2011. Inhibition of Notch signaling ameliorates insulin resistance in a FoxO1-dependent manner. Nat. Med. 17:961–967. 10.1038/nm.2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadros, R.M., Miura H., Harms D.W., Akatsuka H., Sato T., Aida T., Redder R., Richardson G.P., Inagaki Y., Sakai D., et al. 2017. Easi-CRISPR: A robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol. 18:92. 10.1186/s13059-017-1220-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly, S.M., and Saltiel A.R.. 2017. Adapting to obesity with adipose tissue inflammation. Nat. Rev. Endocrinol. 13:633–643. 10.1038/nrendo.2017.90 [DOI] [PubMed] [Google Scholar]
- Roesch, K., Jadhav A.P., Trimarchi J.M., Stadler M.B., Roska B., Sun B.B., and Cepko C.L.. 2008. The transcriptome of retinal Müller glial cells. J. Comp. Neurol. 509:225–238. 10.1002/cne.21730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen, E.D., and Spiegelman B.M.. 2014. What we talk about when we talk about fat. Cell. 156:20–44. 10.1016/j.cell.2013.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully, T., Ettela A., LeRoith D., and Gallagher E.J.. 2021. Obesity, type 2 diabetes, and cancer risk. Front. Oncol. 10:615375. 10.3389/fonc.2020.615375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasami, P., and Li C.. 2020. Derivation and differentiation of adipose-tissue regulatory T cells: A stepwise, multi-site process. Front. Immunol. 11:599277. 10.3389/fimmu.2020.599277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spallanzani, R.G., Zemmour D., Xiao T., Jayewickreme T., Li C., Bryce P.J., Benoist C., and Mathis D.. 2019. Distinct immunocyte-promoting and adipocyte-generating stromal components coordinate adipose tissue immune and metabolic tenors. Sci. Immunol. 4:eaaw3658. 10.1126/sciimmunol.aaw3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange, K., Denton J., and Nehrke K.. 2006. Ste20-type kinases: Evolutionarily conserved regulators of ion transport and cell volume. Physiology. 21:61–68. 10.1152/physiol.00139.2005 [DOI] [PubMed] [Google Scholar]
- Tobias, D.K., and Hu F.B.. 2018. The association between BMI and mortality: Implications for obesity prevention. Lancet Diabetes Endocrinol. 6:916–917. 10.1016/S2213-8587(18)30309-7 [DOI] [PubMed] [Google Scholar]
- Upadhyay, J., Farr O., Perakakis N., Ghaly W., and Mantzoros C.. 2018. Obesity as a disease. Med. Clin. North Am. 102:13–33. 10.1016/j.mcna.2017.08.004 [DOI] [PubMed] [Google Scholar]
- Vanderkerken, M., Maes B., Vandersarren L., Toussaint W., Deswarte K., Vanheerswynghels M., Pouliot P., Martens L., Van Gassen S., Arthur C.M., et al. 2020. TAO-kinase 3 governs the terminal differentiation of NOTCH2-dependent splenic conventional dendritic cells. Proc. Natl. Acad. Sci. USA. 117:31331–31342. 10.1073/pnas.2009847117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasanthakumar, A., Chisanga D., Blume J., Gloury R., Britt K., Henstridge D.C., Zhan Y., Torres S.V., Liene S., Collins N., et al. 2020. Sex-specific adipose tissue imprinting of regulatory T cells. Nature. 579:581–585. 10.1038/s41586-020-2040-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasanthakumar, A., Moro K., Xin A., Liao Y., Gloury R., Kawamoto S., Fagarasan S., Mielke L.A., Afshar-Sterle S., Masters S.L., et al. 2015. The transcriptional regulators IRF4, BATF and IL-33 orchestrate development and maintenance of adipose tissue-resident regulatory T cells. Nat. Immunol. 16:276–285. 10.1038/ni.3085 [DOI] [PubMed] [Google Scholar]
- Winer, S., Chan Y., Paltser G., Truong D., Tsui H., Bahrami J., Dorfman R., Wang Y., Zielenski J., Mastronardi F., et al. 2009. Normalization of obesity-associated insulin resistance through immunotherapy. Nat. Med. 15:921–929. 10.1038/nm.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2016. WHO | Obesity and overweight fact sheet. Word Health Organization [Google Scholar]
- Wu, D., Molofsky A.B., Liang H.E., Ricardo-Gonzalez R.R., Jouihan H.A., Bando J.K., Chawla A., and Locksley R.M.. 2011. Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science. 332:243–247. 10.1126/science.1201475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda, T., Imaizumi K., Oono K., Yui D., Gomi F., Katayama T., and Tohyama M.. 2001. Activation of caspase-12, an endoplastic reticulum (ER) resident caspase, through tumor necrosis factor receptor-associated factor 2-dependent mechanism in response to the ER stress. J. Biol. Chem. 276:13935–13940. 10.1074/jbc.M010677200 [DOI] [PubMed] [Google Scholar]
- Yustein, J.T., Xia L., Kahlenburg J.M., Robinson D., Templeton D., and Kung H.J.. 2003. Comparative studies of a new subfamily of human ste20-like kinases: Homodimerization, subcellular localization, and selective activation of MKK3 and p38. Oncogene. 22:6129–6141. 10.1038/sj.onc.1206605 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows qPCR primers.
shows flow cytometry antibodies.
Data Availability Statement
The data are available from the corresponding author upon reasonable request (bart.lambrecht@ugent.be).