Abstract
Tumor phenotypes can be characterized by radiomics features extracted from images. However, the prediction accuracy is challenged by difficulties such as small sample size and data imbalance. The purpose of the study was to evaluate the performance of machine learning strategies for the prediction of cancer prognosis. A total of 422 patients diagnosed with non-small cell lung carcinoma (NSCLC) were selected from The Cancer Imaging Archive (TCIA). The gross tumor volume (GTV) of each case was delineated from the respective CT images for radiomic features extraction. The samples were divided into 4 groups with survival endpoints of 1 year, 3 years, 5 years, and 7 years. The radiomic image features were analyzed with 6 different machine learning methods: decision tree (DT), boosted tree (BT), random forests (RF), support vector machine (SVM), generalized linear model (GLM), and deep learning artificial neural networks (DL-ANNs) with 70:30 cross-validation. The overall average prediction performance of the BT, RF, DT, SVM, GLM and DL-ANNs was AUC with 0.912, 0.938, 0.793, 0.746, 0.789 and 0.705 respectively. The RF and BT gave the best and second performance in the prediction. The DL-ANN did not show obvious advantage in predicting prognostic outcomes. Deep learning artificial neural networks did not show a significant improvement than traditional machine learning methods such as random forest and boosted trees. On the whole, the accurate outcome prediction using radiomics serves as a supportive reference for formulating treatment strategy for cancer patients.
Keywords: Radiomics, Prognostic prediction, Machine learning
Introduction
Cancer is the abnormal uncontrolled growth of cells. Unlike biopsy, medical imaging approaches are non-invasive for the assessment and grading of cancer and are able to assess the heterogeneity and functional information of the tumor using radiomics other than geometrical and morphological information [1]. At the same time, suitable treatment strategies like surgery, radiation therapy (RT), or chemotherapy are important for proper management of a patient with cancer. For the RT treatment in non-small cell lung carcinoma (NSCLC), there are numerous fractionation schemes like normal fractionation or hyperfractionated scheme, which depend on the cancer staging [2].
Radiomics
Radiomics is the approach that extracts the radiographic image features for the tumor phenotype information. To analyze the prognosis using radiomics, medical images are obtained from one or more imaging modalities, such as Computed Tomography (CT), Magnetic Resonance Imaging (MRI), and Positron Emission Tomography (PET). The regions of interest (ROIs) are delineated manually or automatically to define the macroscopic tumor. Manual delineation is usually performed by oncologists or experienced radiologists [1], while computer-aided delineation is implemented by automatic or semi-automatic segmentation methods [3]. The radiomic features are then extracted as quantitative image features (such as intensity, texture, shape, and size of the tumor) and it can help to correlate prognostic imaging phenotypic information of the tumor, including tumor heterogeneity and the protein expression [4]. The clinical outcomes such as disease recurrence or patient survival can be used as “gold standards” for machine learning to establish the predictive models [5].
Machine Learning
Machine learning is a computational method that learns experience from the finite sample data in order to predict the outcome of the unseen data. The use of radiomics in cancer prognosis prediction has received a lot of attention recently. Machine learning with radiomics has been used to predict the outcome of rectal cancer, head and neck cancer, and NSCLC [6, 7]. Feature classification is a step of deducing the hypothesis from the training data to predict the outcome of the unseen testing data [8]. The feature classifiers include support vector machine (SVM), decision trees, linear model, etc. [9, 10]. There were studies comparing different feature classifiers on radiomics which indicated that random forest (RF) produces the highest prediction performance on the overall survival [11]. Several studies on esophageal cancer and glioma concluded that deep learning improved the prediction performance on radiomics, when compared with the statistic-based methods such as SVM, RF, gradient boosting, etc. [12].
There are diverse machine learning methods for radiomics while the deep learning method, a more special type of machine learning that comprises complicated layers of algorithms, appears to be more promising for the treatment outcome prediction [13] and image feature extraction [14–16]. There are concerns that when the number of radiomic features is comparable with sample size (e.g., about 100–200 features against 200–300 sample size), the performance of machine learning methods will be adversely affected [9].
In this study, we attempted to evaluate the prediction performance of different machine learning methods including deep learning ANNs for prediction of prognosis for patients with NSCLC, with the survival endpoints of 1 year, 3 years, 5 years, and 7 years after RT treatment.
Methodology
Data Acquisition and Case Selection
Data Acquisition
Since cases with validated clinical data are limited, we tried to retrieve the eligible cases as many as possible. The criteria are (1) cases with confirmed diagnosis, (2) cases with more than one primary tumor were excluded, and (3) image dataset should include radiotherapy structure sets, DICOM Segmentation with Gross Tumor Delineation. In this retrospective study, 422 patients with stage I, II, and III non-small cell lung carcinomas of various histological types were acquired from The Cancer Imaging Archive (TCIA). TCIA is an online public database with cancer medical images for cancer research hosted by the Department of Biomedical Informatics at the University of Arkansas for Medical Sciences and contracted with National Cancer Institute. Data were reviewed and approved by the TCIA Advisory Group before allowing for public access. Our dataset consisted of pretreatment CT scans with 3-dimensional Gross Tumor Volume (GTV) delineation, and clinical outcome data. Data that we obtained for this study included CT image sets, the radiotherapy structure sets (RTSTUCT), DICOM Segmentation (SEG) with the GTV delineation, organs at risk delineation, and clinical data of patients including histology and survival status. The DICOM structure set ensured the same region was used in the radiomics analysis. The GTV and anatomical delineation were performed manually by radiation oncologists who contributed to TCIA [17].
Case Selection
In the 422 cases collected, 28 cases with more than one primary tumor inside the lung (multiple GTVs in the lung) were excluded. However, for the cases with multiple GTVs due to lymph nodes involvement or distal metastasis (one GTV in the lung, others locate at other body sites such as bronchus, mediastinal lymph nodes, etc.) were included, because the lymph node GTV neighboring the lung were considered as the same tumor originated from the primary tumor due to the cancer spread. The primary tumor lesion was able to be distinguished with the radiomics features extraction. Finally, 394 cases were used for this study.
Radiomics Feature Extraction
A total of 107 radiomic features of each sample were utilized for machine learning in this study (Table 1). These include the following: 6 groups of features, namely the shape, gray level dependence matrix (gldm), gray level co-occurrence matrix (glcm), first-order feature, gray level run length matrix (glrlm), gray level size zone (glszm), and neighborhood gray tone difference matrix (ngtdm). The 3D slicer (4.10.2 version) with the Pyradiomics extension (Computational Imaging and Bioinformatics Lab, Harvard medical School) [18] was utilized to extract the radiomic features.
Table 1.
Radiomic features summary
| Feature group | Number of features | Feature description |
|---|---|---|
| Shape | 14 | Morphological features |
| Gray level dependence matrix | 14 | Textual features |
| Gray level co-occurrence matrix | 24 | Textual features |
| First-order feature | 18 | Region of Interest (ROI) statistics features |
| Gray level run length matrix | 32 | Textual features |
| Neighborhood gray tone difference matrix | 5 | Textual features |
| total | 107 |
Machine Learning Analysis and Predictive Performance
Radiomics model used quantitative imaging features to generate predictions with classifications of “survival” or “death” as outcome. In our study, 6 machine learning classification algorithms were used: decision tree (DT), boosted tree (BT), random forest (RF), support vector machine (SVM), generalized linear model (GLM), and deep learning neural networks (DL-ANNs) (see Table 7 in Appendix for the characteristics of common machine learning algorithms). The predictive performance was evaluated by Area Under Curve (AUC) from Receiver Operating Characteristic (ROC) analysis. ROC curves are used to evaluate the accuracy of a diagnostic test. The technique is used when there is a criterion variable which will be used to make a binary outcome (yes or no decision) based on the value of this variable. The area under the ROC curve (AUC) is a summary index of an ROC curve and is the probability that an observer correctly determines which of the disease positive or negative subjects are more likely to have the disease [19]. In this study, survival status was dichotomized for prediction: patients survived beyond the time endpoint were denoted as “1”, whereas the patients died were denoted by “ 0 “(survived=1, death=0). There were 4 endpoints for survival: 1 years, 3 years 5 years and 7 years.
Table 7.
Summary of Machine Learning Methods
| Methods | Mechanism | Characteristics |
|---|---|---|
| Decision Trees [30] | Inverted-tree-like graph that with input root nodes, internal nodes with the classification based on the feature value, and output as leaf nodes. | Simplifies the complex relationship and allows easier interpretation However, it has limited robustness and the strong correlation of the input variables may have the probability of inaccurate results |
| Boosted Trees [31] | Ensemble of multiple decision trees, which the trees are built dependent with the results of the previous tree. | It can adjust the errors made by the previous trees. The combining of numerous trees increases the robustness. |
| Random Forests [27] | Ensemble of numerous independent decision trees which composed of randomly selected samples, the results of each tree are averaged out to become the final result. | It has lower susceptibility of the biased sample and a higher robustness |
| Support Vector Machine [9] | A hyperplane which can separate the data into two categories with the margin on the either side of the hyperplane. | The maximized margins can reduce the generalization error, yet the misclassified data that not lied within the margin may lower the accuracy. |
| Generalized Linear Model [32] | The linear relationship of the observed features and the real output is drawn with similar number of samples lying on both sides of the regression line. Then the output can be predicted by the linear regression line with the known feature | The results may be affected when the numbers of feature are much more than the numbers of output |
| Artificial Neural Networks [33] | Feed-Forward. Neural Network consists of input hidden and output layer. | Ability to learn for nonlinear and complex relationship; yet it is difficult to interpret It works well with the tabular data. |
| Convolution Neural Networks [33] | The convolution filter is applied on the image forming kernels to form the features map. Then extracting the relevant features from the input data automatically | It captures the spatial feature of the image well |
| Recurrent Neural Networks [33] | It consists of the ANN-like structure but with a recurrent loop in the hidden layer. | The recurrent loop can well capture the sequential feature & it works well with the text data. |
Since in radiomics studies, the number of radiomics features as the input nodes or input parameters is comparable with the sample size, in order to evaluate the effects of distribution of positive and negative outcomes, the data is further divided into two groups. In group A (all data group), the entire sample (n=394) was used as input data for training, validation, and testing. In group B (balanced data group), equal numbers of survived and death cases were randomly selected from the 394 cases respectively for machine learning, validation and testing. In each group A or B, there were 4 subgroups: 1-year survival with 286 samples (143 survived and 143 deaths), 3-year survival with 236 samples (118 survived and 118 deaths), 5-year survival with 136 samples (68 survived and 68 deaths), and 7-year survival with 86 samples (43 survived, 43 deaths).
The machine learning analyses (DT, BT, RF, SVM, GLM) were performed using R software (R Core Team, Vienna, Austria) version 4.0.4 implemented through Rattle (the graphic user interface using R) version 5.4.0 [20]. The AUC values were used to evaluate the performance of each machine learning algorithm with the 70:30 cross-validation, which used 70 of the samples for training and the rest 30 for validation and testing (70/15/15). This is the hold-out validation technique by stratified sampling in which the sample is randomly partitioned into subsamples as the training, validation and test dataset. Part of data is set aside for training the model. Another set is held out for testing and evaluating the model. The deep learning neural networks used in this study was implemented by Python programming language using a convolutional model with a two 1D convolutional layers (the numbers of units are 64 and 128) followed by two fully connected layer with output size 2. The model is regularized by dropout with probability of 0.5 between each layer. Rectified linear unit (ReLU) function was deployed after each layer except the last fully connected layer. We adopt the stochastic gradient descent (SGD) optimizer with a learning rate of 0.01 and a weight decay of 1e-6 and 10 epochs of training.
Results
Patient Demographics
A total of 394 patients with NSCLC were included in this study (Table 2). Among the patients, 70 were male and 30 were female, mostly aged above 50. For the overall staging of the patients, a high proportion of patients were diagnosed with stage III NSCLC: 27 patients with stage IIIa and 42 patients with stage IIIb. For the histology of the NSCLC, 37 of patients diagnosed with squamous cell carcinoma, 26 patients with large cell carcinoma, and 12 patients had adenocarcinoma.
Table 2.
Patient Demographic
| Number of patients | Number of patients | ||
|---|---|---|---|
| Gender | Histology | ||
| Female | 118 (30%) | Adenocarcinoma | 47 (12%) |
| Male | 276 (70%) | Large Cell | 104 (26%) |
| Total | 394 | ||
| Overall Staging | Squamous Cell Carcinoma | 145 (37%) | |
| Stage I | 86 (22%) | NA | 39 (10%) |
| Stage II | 38 (10%) | Not Otherwise Specified | 59 (15%) |
| Stage IIIa | 105 (27%) | Age | |
| Stage IIIb | 164 (42%) | <50 | 17 (4%) |
| NA | 1 (0%) | >50 | 355 (90%) |
| NA | 22 (6%) |
Prognostic Performance of Machine Learning Methods
Receiver Operating Characteristic (ROC) was used to evaluate the prognostic performance of different machine learning algorithms. For boosted trees (BT), random forests (RF), decision trees (DT), support vector machine (SVM), and generalized linear model (GLM) the area under the curve (AUC) of ROC was generated by Rattle. For Deep Learning Artificial Neural Networks (DL-ANNs), the AUC is generated by scikit-learn in Python. All machine learning methods were performed with 70:30 cross-validation.
The overall average prediction performance of the BT, RF, DT, SVM, GLM and DL-ANNs were AUC of 0.912, 0.938, 0.793, 0.746, 0.789 and 0.705 respectively (Table 3). The highest performance in prediction were from RF (1st) and BT (2nd). The DL-ANN did not show obvious advantage in prediction of prognostic outcomes.
Table 3.
Summary of predictive performance of ML methods. (AUC)
| Survival Year(s) | Sample Types | Sample Size | DT | BT | RF | SVM | GLM | CNN |
|---|---|---|---|---|---|---|---|---|
| 1 | Balanced | 286 | 0.817 | 0.947 | 0.947 | 0.781 | 0.737 | 0.717 |
| (“0”=143, | [0.768, | [0.921, | [0.921, | [0.728, | [0.679, | [0.658, | ||
| “1”=143) | 0.866] | 0.974] | 0.974] | 0.834] | 0.795] | 0.776] | ||
| All | 394 | 0.814 | 0.951 | 0.961 | 0.815 | 0.829 | 0.709 | |
| (“0”=143, | [0.767, | [0.938, | [0.938, | [0.768, | [0.784, | [0.654, | ||
| “1”=251) | 0.861] | 0.976] | 0.983] | 0.862] | 0.874] | 0.764] | ||
| 3 | Balanced | 236 | 0.781 | 0.947 | 0.974 | 0.821 | 0.832 | 0.777 |
| (“0”=118, | [0.722, | [0.917, | [0.977, | [0.767, | [0.780, | [0.718, | ||
| “1”=118) | 0.840] | 0.977] | 0.995] | 0.875] | 0.884] | 0.836] | ||
| All | 394 | 0.814 | 0.951 | 0.961 | 0.815 | 0.829 | 0.709 | |
| (“0”=276, | [0.785, | [0.905, | [0.917, | [0.792, | [0.789, | [0.652, | ||
| “1”=118) | 0.865] | 0.953] | 0.961] | 0.870] | 0.869] | 0.757] | ||
| 5 | Balanced | 136 | 0.846 | 0.904 | 0.921 | 0.771 | 0.697 | 0.795 |
| (“0”=68, | [0.780, | [0.851, | [0.873, | [0.692, | [0.609, | [0.728, | ||
| “1”=68) | 0.912] | 0.957] | 0.969] | 0.850] | 0.785] | 0.862] | ||
| All | 394 | 0.685 | 0.782 | 0.884 | 0.200 | 0.740 | 0.626 | |
| (“0”=326, | [0.622, | [0.731, | [0.850, | [0.134, | [0.683, | [0.558, | ||
| “1”=68) | 0.748] | 0.833] | 0.918] | 0.266] | 0.797] | 0.694] | ||
| 7 | Balanced | 86 | 0.825 | 0.970 | 0.973 | 0.923 | 0.799 | 0.619 |
| (“0”=43, | [0.736, | [0.933, | [0.938, | [0.863, | [0.705, | [0.525, | ||
| “1”=43) | 0.914] | 1.010] | 1.010] | 0.983] | 0.893] | 0.713] | ||
| All | 394 | 0.751 | 0.869 | 0.902 | 0.823 | 0.842 | 0.692 | |
| (“0”=351, | [0.687, | [0.828, | [0.868, | [0.772, | [0.795, | [0.619, | ||
| “1”=43) | 0.815] | 0.910] | 0.936] | 0.874] | 0.889] | 0.765] | ||
| Average | 0.793 | 0.912 | 0.938 | 0.746 | 0.789 | 0.705 |
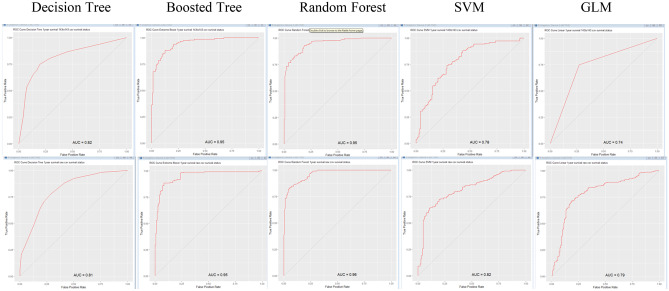
The best AUC was 0.974 for the balanced sample sets of RF with 3 year survival endpoint, while the moderate AUC was 0.829 for sample set of GLM with 3 year survival endpoint and lowest AUC was 0.685 for DT with all sample set with 5 year survival endpoint (Fig. 1). The AUC with 0.200 for all sample sets of SVM with 5 year endpoint was considered as outlier. The paired t-test did not demonstrate any significant difference between the “Balanced” and “All” groups endpoints with different numbers of survival years as endpoint (p>0.05, paired t-test, Table 4). In summary, the best predictive values of the algorithms in C-index or AUC (for survival after 1, 3, 5, 7 years) are noted in Table 5. All ROC curves (Figs. 2, 3, 4, 5, 6) are shown in Appendix.
Fig. 1.
ROC curves showing best, immediate and lowest results. a Random Forest (3-year survival, balanced sample data), b Generalized Linear Model (3-year survival, all sample data), c Decision Tree (5-year survival, all sample data)
Table 4.
Test for Balanced and All sample datasets
| Survival Year(s) | Sample Types | paired t-test value | Significance |
|---|---|---|---|
| 1 | Balanced | 0.078 | p>0.05 |
| All | |||
| 3 | Balanced | 0.501 | p>0.05 |
| All | |||
| 5 | Balanced | 0.148 | p>0.05 |
| All | |||
| 7 | Balanced | 0.050 | p>0.05 |
| All |
Table 5.
Summary of best predictive value. (*The samples are tested with 70:30 cross-validation.)
| Survival Years endpoint | Algorithm* | C-index (AUC) |
|---|---|---|
| 1 | RF (all sample) | 0.961, [0.938, 0.983] |
| 3 | RF (balanced sample) | 0.974, [0.977, 0.995] |
| 5 | RF (balanced sample) | 0.921, [0.873, 0.959] |
| 7 | BT (balanced sample) | 0.970, [0.933, 1.010] |
Fig. 2.
ROC of DT, BT, RF, SVM, and GLM for 1-year balanced data and all data
Fig. 3.
ROC of DT, BT, RF, SVM, and GLM for 3-year balanced data and all data
Fig. 4.
ROC of DT, BT, RF, SVM, and GLM for 5-year balanced data and all data
Fig. 5.
ROC of DT, BT, RF, SVM, and GLM for 7-year balanced data and all data
Fig. 6.
ROC of CNN for 1-year, 3-year, 5-year and 7-year balanced data and all data
Discussion
Radiomics is becoming a clinically effective tool for personalized medicine and prognostic prediction of cancer treatment. However, the large number of radiomics features and limited number for each category of observations with unbalanced datasets posed a challenge for radiomics-based machine learning model. In this study, we attempted to document the effect of small datasets for the performance of machine learning methods in radiomics prediction.
Feature selection was an issue for radiomics. Li et al. used 671 radiomics features for prediction of low-grade glioma [12]. Kotsiantis et al. [9] mentioned about 14 features in their selection methods. To address the problem of a lack of standardization in feature selections, Pyradiomic, an open-source python package developed mainly by researchers from the Department of Radiation, Harvard Medical School [18], was used for the extraction of radiomics features from medical images as a reference standard for radiomics analysis. The features needed to be preprocessed with a set of filters (including wavelet, Laplacian of gaussian, square, square root, logarithm and exponential [18]) to reduce noises before calculation for radiomics features used in this study. For different machine learning algorithms, random forests had the highest accuracy (average AUC= 0.94) throughout different survival years and sample sizes. Boosted tree (with average AUC= 0.91) also showed a good prediction performance. This performance was consistent with those in other studies [11, 25]. It appears that random forest can have a lower susceptibility to the biased samples. Also, the out-of-bag method helps to validate the trees and achieved a higher accuracy with more robust results [26]. Random forest was formed by many decision trees. The results of all trees were averaged out to generate the result, since the samples were randomly selected to form a decision tree by many training subsets [27]. However, it was noted that random trees and boosted trees were not good for regression prediction as they did not predict precise continuous data beyond the range in the training data, and they suffered from overfitting in noisy datasets. As random forest used a black box approach, there was little control on the model from the users’ perspective.
Our results indicated that the balanced datasets performed slightly better than the whole sample data (average AUC= 0.80 against average AUC= 0.76). However, the paired t-test did not indicate any significant difference for our datasets, although the balanced data could foster a higher accuracy [28].
Xu et al. [29] used transfer learning convolution neural networks (CNN) and recurrent neural networks (RNN) for patients with NSCLC and obtained AUC=0.74 (n=268), while Joost [18] used CNN with transfer learning obtained AUC=0.7 (n=771) for NSCLC patients with CT images. In our study, we obtained a comparable result of AUC=0.795 for balanced data with 5-year survival endpoint (n=136) for 1D CNN DL-ANN. It appears that small sample size with large input parameters may affect the predictive performance of neural networks for radiomics data. It is noted that the relative performance of the algorithms with other studies in radiomics was similar to our study using deep learning algorithm. However, random forest in our study performed better than other studies (Table 6).
Table 6.
Performance of machine learning
| Authors | Study task | Method | Result |
|---|---|---|---|
| Park et al. [21] | To investigate whether CT slice thickness influences the performance of radiomics prognostic models | Patients with NSCLC with development set (n=185) and validation set (n=126), using radiomic prediction with CT slices thickness 1, 3, 5 mm | AUC with development set=0.68 to 0.7, AUC with validation set=0.73 to 0.76 |
| Lao et al. [22] | To investigate if deep features extracted via transfer learning can generate radiomics signatures for prediction of overall survival in patients with Glioblastoma Multiforme | LASSO Cox regression model was applied for 75 patient set for training and 37 patient set for validation | AUC =0.71 to 0.739 |
| Liu et al. [23] | Prediction of distant metastasis through deep learning radiomics | 235 patients with neoadjuvant chemotherapy using deep learning radiomics signature | AUC =0.747 to 0.775 |
| Vils et al. [24] | Evaluation of radiomics in recurrent glioblastoma | Multivariable models using radiomic feature selection for patients (n=69 training, n=49 validation) | AUC=0.67 to 0..673 |
| Our study | Different machine learning algorithms to evaluate radiomics prediction of NSCLC | 6 machine algorithms used for 422 patients with 70:30 cross-validation | Average AUC=0.67 to 0.91 |
Conclusions
Our study attempted to evaluate the prediction performance of different machine learning methods for radiomics prognosis prediction. DL-ANNs with 70:30 cross-validation did not show a significant improvement compared with other traditional machine learning methods such as random forest and boosted trees. On the whole, the accurate outcome prediction using machine learning serves as a supportive reference for formulating treatment strategy for cancer patients. This helps to facilitate personalized treatment for cancer patients in the clinical settings.
Appendix
Summary of machine learning methods and ROC curves for all the methods.
Author Contributions
Conceptualization, F.-H.T.; methodology, C.X., C.Y.W., T.H.C., C.K.L; data acquisition, C.K.L.; writing—original draft preparation, C.Y.W. and T.H.C; writing—review and editing, F.-H.T., C.X. and M.Y.L; All authors have read and agreed to the published version of the manuscript.
Funding
We acknowledged the University Grants Council Faculty Development Scheme grant UGC/FDS17/M10/19 for support of model development of this project and Tung Wah College for the support of article publication charge. On behalf of all authors, the corresponding author states that there is no conflict of interest.
Data Availability
The data is available upon request to: Professor F.H. Tang by email: fhtang@twc.edu.hk
Declarations
Ethics Approval
This study used public dataset; no ethics approval is required.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Fuk-hay Tang, Email: fhtang@twc.edu.hk.
Cheng Xue, Email: xchengjlu@gmail.com.
Maria YY Law, Email: marialaw@twc.edu.hk.
Chui-ying Wong, Email: tiffany041299@gmail.com.
Tze-hei Cho, Email: thcho1998@yahoo.com.hk.
Chun-kit Lai, Email: keithsofk@gmail.com.
References
- 1.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, Van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. European journal of cancer. 2012;48(4):441–6. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zierhut D, Bettscheider C, Schubert K, Van Kampen M, Wannenmacher M. Radiation therapy of stage I and II non-small cell lung cancer (NSCLC) Lung Cancer. 2001;34:39–43. doi: 10.1016/S0169-5002(01)00381-6. [DOI] [PubMed] [Google Scholar]
- 3.Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, et al. Radiomics: the process and the challenges. Magnetic resonance imaging. 2012;30(9):1234–48. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.GGillies R, Kinahan P, Hricak H. Radiomics: Images Are More than Pictures. They Are Data Radiology. 2016;278(2):563-77. [DOI] [PMC free article] [PubMed]
- 5.FH T, CYW C, EYW C. Radiomics AI prediction for head and neck squamous cell carcinoma (HNSCC) prognosis and recurrence with target volume approach. BJR| Open. 2021;3:20200073. [DOI] [PMC free article] [PubMed]
- 6.Liu Z, Zhang XY, Shi YJ, Wang L, Zhu HT, Tang Z, et al. Radiomics analysis for evaluation of pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Clinical Cancer Research. 2017;23(23):7253–62. doi: 10.1158/1078-0432.CCR-17-1038. [DOI] [PubMed] [Google Scholar]
- 7.Bogowicz M, Riesterer O, Ikenberg K, Stieb S, Moch H, Studer G, et al. Computed tomography radiomics predicts HPV status and local tumor control after definitive radiochemotherapy in head and neck squamous cell carcinoma. International Journal of Radiation Oncology* Biology* Physics. 2017;99(4):921-8. [DOI] [PubMed]
- 8.Singh A. Foundations of Machine Learning. International Journal of Radiation Oncology* Biology* Physics. 2019.
- 9.Kotsiantis SB, Zaharakis ID, Pintelas PE. Machine learning: a review of classification and combining techniques. Artificial Intelligence Review. 2006;26(3):159–90. doi: 10.1007/s10462-007-9052-3. [DOI] [Google Scholar]
- 10.Osisanwo F, Akinsola J, Awodele O, Hinmikaiye J, Olakanmi O, Akinjobi J. Supervised machine learning algorithms: classification and comparison. International Journal of Computer Trends and Technology (IJCTT). 2017;48(3):128–38. doi: 10.14445/22312803/IJCTT-V48P126. [DOI] [Google Scholar]
- 11.Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJ. Machine learning methods for quantitative radiomic biomarkers. Scientific reports. 2015;5(1):1–11. doi: 10.1038/srep13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Wang Y, Yu J, Guo Y, Cao W. Deep learning based radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Scientific reports. 2017;7(1):1–11. doi: 10.1038/s41598-017-05848-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avanzo M, Wei L, Stancanello J, Vallieres M, Rao A, Morin O, et al. Machine and deep learning methods for radiomics. Medical physics. 2020;47(5):e185–202. doi: 10.1002/mp.13678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue C, Zhu L, Fu H, Hu X, Li X, Zhang H, et al. Global guidance network for breast lesion segmentation in ultrasound images. Medical image analysis. 2021;70:101989. doi: 10.1016/j.media.2021.101989. [DOI] [PubMed] [Google Scholar]
- 15.Xue C, Dou Q, Shi X, Chen H, Heng PA. Robust learning at noisy labeled medical images: Applied to skin lesion classification. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019). IEEE; 2019. p. 1280-3.
- 16.Xue C, Deng Q, Li X, Dou Q, Heng PA. Cascaded robust learning at imperfect labels for chest x-ray segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2020. p. 579-88.
- 17.Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. Journal of digital imaging. 2013;26(6):1045–57. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Griethuysen JJ, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. Computational radiomics system to decode the radiographic phenotype. Cancer research. 2017;77(21):e104–7. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cortes C, Mohri M. Confidence intervals for the area under the ROC curve. Advances in neural information processing systems. 2004;17.
- 20.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria; 2021. Available from: https://www.R-project.org/.
- 21.Park S, Lee SM, Kim S, Choi S, Kim W, Do KH, et al. Performance of radiomics models for survival prediction in non-small-cell lung cancer: influence of CT slice thickness. European Radiology. 2021;31(5):2856–65. doi: 10.1007/s00330-020-07423-2. [DOI] [PubMed] [Google Scholar]
- 22.Lao J, Chen Y, Li ZC, Li Q, Zhang J, Liu J, et al. A deep learning-based radiomics model for prediction of survival in glioblastoma multiforme. Scientific reports. 2017;7(1):1–8. doi: 10.1038/s41598-017-10649-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Zhang D, Liu Z, Li Z, Xie P, Sun K, et al. Deep learning radiomics-based prediction of distant metastasis in patients with locally advanced rectal cancer after neoadjuvant chemoradiotherapy: A multicentre study. EBioMedicine. 2021;69:103442. doi: 10.1016/j.ebiom.2021.103442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vils A, Bogowicz M, Tanadini-Lang S, Vuong D, Saltybaeva N, Kraft J, et al. Radiomic Analysis to Predict Outcome in Recurrent Glioblastoma Based on Multi-Center MR Imaging From the Prospective DIRECTOR Trial. Frontiers in oncology. 2021;11:1097. doi: 10.3389/fonc.2021.636672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Oikonomou A, Wong A, Haider MA, Khalvati F. Radiomics-based prognosis analysis for non-small cell lung cancer. Scientific reports. 2017;7(1):1–8. doi: 10.1038/srep46349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oshiro TM, Perez PS, Baranauskas JA. How many trees in a random forest? In: International workshop on machine learning and data mining in pattern recognition. Springer; 2012. p. 154-68.
- 27.Boulesteix AL, Janitza S, Kruppa J, König IR. Overview of random forest methodology and practical guidance with emphasis on computational biology and bioinformatics. Wiley Interdisciplinary Reviews: Data Mining and Knowledge Discovery. 2012;2(6):493–507. [Google Scholar]
- 28.Wei Q, Dunbrack RL., Jr The role of balanced training and testing data sets for binary classifiers in bioinformatics. PloS one. 2013;8(7):e67863. doi: 10.1371/journal.pone.0067863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Y, Hosny A, Zeleznik R, Parmar C, Coroller T, Franco I, et al. Deep learning predicts lung cancer treatment response from serial medical imaging. Clinical Cancer Research. 2019;25(11):3266–75. doi: 10.1158/1078-0432.CCR-18-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song YY, Ying L. Decision tree methods: applications for classification and prediction. Shanghai archives of psychiatry. 2015;27(2):130. doi: 10.11919/j.issn.1002-0829.215044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen T, Guestrin C. Xgboost: A scalable tree boosting system. In: Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining; 2016. p. 785-94.
- 32.Bandos TV, Bruzzone L, Camps-Valls G. Classification of hyperspectral images with regularized linear discriminant analysis. IEEE Transactions on Geoscience and Remote Sensing. 2009;47(3):862–73. doi: 10.1109/TGRS.2008.2005729. [DOI] [Google Scholar]
- 33.ANN vs CNN vs RNN: Types of Neural Networks;. Accessed: 2021-05-15. https://www.analyticsvidhya.com/blog/2020/02/cnn-vs-rnn-vs-mlp-analyzing-3-types-of-neural-networks-in-deep-learning/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data is available upon request to: Professor F.H. Tang by email: fhtang@twc.edu.hk