Abstract
Artificial neural networks (ANN) are artificial intelligence (AI) techniques used in the automated recognition and classification of pathological changes from clinical images in areas such as ophthalmology, dermatology, and oral medicine. The combination of enterprise imaging and AI is gaining notoriety for its potential benefits in healthcare areas such as cardiology, dermatology, ophthalmology, pathology, physiatry, radiation oncology, radiology, and endoscopic. The present study aimed to analyze, through a systematic literature review, the application of performance of ANN and deep learning in the recognition and automated classification of lesions from clinical images, when comparing to the human performance. The PRISMA 2020 approach (Preferred Reporting Items for Systematic Reviews and Meta-analyses) was used by searching four databases of studies that reference the use of IA to define the diagnosis of lesions in ophthalmology, dermatology, and oral medicine areas. A quantitative and qualitative analyses of the articles that met the inclusion criteria were performed. The search yielded the inclusion of 60 studies. It was found that the interest in the topic has increased, especially in the last 3 years. We observed that the performance of IA models is promising, with high accuracy, sensitivity, and specificity, most of them had outcomes equivalent to human comparators. The reproducibility of the performance of models in real-life practice has been reported as a critical point. Study designs and results have been progressively improved. IA resources have the potential to contribute to several areas of health. In the coming years, it is likely to be incorporated into everyday life, contributing to the precision and reducing the time required by the diagnostic process.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10278-023-00775-3.
Keywords: Diagnosis, Computer-assisted, Artificial intelligence, Convolutional neural network, Photography, Automated classification
Introduction
In recent years, the potential of technological resources involving artificial intelligence (AI) to assist in the processes of health diagnoses has been discussed. Its incorporation is based on pattern recognition from the analysis of clinical images [1]. Static images, video clips, and sound multimedia are routinely captured in cardiology, dermatology, ophthalmology, pathology, physiatry, radiological oncology, radiology, endoscopies, and other medical specialties [2].
The increasing interest on the insertion of AI technologies in the health area is based on the fact that many people have limited access to specialized health services, as well as a shortage of specialists working in rural areas or far from large centers [3–15]. It may be assumed that the incorporation of these resources could offer clinical support to health professionals to speed up decision-making regarding diagnosis and management of different diseases [1, 11, 16–18]. The combination of interactive documentation with metadata, image annotations with text, tables, graphics, and hyperlinks optimize communication between medical professionals [2, 19]. The incorporation of these resources with artificial intelligence has been gaining prominence in the health area [2].
Studies have produced promising evidence from the application of AI and deep learning (DL) technologies. The results were equivalent or even superior to those of human experts in the identification of visual patterns and automated classification of clinic images [20, 21], including the classification of ophthalmic diseases [16, 17, 22–25], skin lesions [4, 5, 21, 26–29], and change genetic/phenotypic lesions [30], among others, with high sensitivity and specificity.
The incorporation of AI capabilities in ophthalmology has shown promising impacts in mass screening programs [16, 25]. Diabetic retinopathy (DR) is a common complication of diabetes mellitus and the leading cause of blindness [16], while glaucoma can cause irreversible blindness, with worldwide projections of 111.8 million by 2040 [22]. Early diagnosis is essential and can be done by analyzing fundus photographs [16, 31, 32]. Automated image classification can become an enabler to support screening services in resource-limited environments [16, 25, 31, 32].
The literature demonstrates relevance in the use of artificial intelligence for the early diagnosis of skin diseases that affect 1.9 billion people worldwide [5]. There are an estimated 6480 melanoma diagnoses in the USA in 2019, resulting in 7230 deaths [33]. On the other hand, basal cell carcinoma (BCC) is the most common skin cancer, and although not usually fatal, it imposes a great burden on health services [26, 33]. Due to the shortage of dermatologists, general practitioners see many cases, with less diagnostic accuracy [5]. Deep learning systems demonstrate equivalent performance to specialists and superior to general practitioners, highlighting the great potential to aid in the diagnosis of other skin pathologies [4], such as psoriasis [28, 34], onychomycosis [27], acne [35], and melanoma [36].
Other studies have been conducted with the use of AI and machine learning (ML) in the field of oral medicine, for different purposes, like predict the survival of patients diagnosed with oral cancer to support early diagnosis and definition of the treatment plan. Many studies describe the association of AI with resources, such as computed tomography [37, 38], autofluorescence [39, 40], hyperspectral images [41, 42], intraoral probes [43], and photodynamic therapy [44]. Even if incipient, the great potential of technology [4, 5, 16, 17, 22–28, 45–47] with performance comparable to that of specialists is demonstrated [1, 3, 48].
Therefore, the objective of this review was to perform a systematic review about the performance of artificial neural networks and deep learning in automated recognition and classification of lesions from clinic images in different health fields, comparing them with human performance.
Materials and Methods
The review was conducted using a predetermined protocol which followed the recommendations of the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) statement and checklist [49]. The protocol was registered in the International Prospective Register of Systematic Review (PROSPERO) database, CRD42021243521.
Review Question and Search Strategy
The research question of the study was: “How effective is the use of deep artificial neural networks in the automated recognition and classification of lesions and pathological changes, from clinical images in the areas of ophthalmology, dermatology and oral medicine when compared to human evaluation?”.
Electronic searches without publication date or language restriction were made in four databases in January 2021 (and updated on January 2022), as the following: PubMed (National Library of Medicine), Web of Science (Thomson Reuters), Scopus (Elsevier), and Embase (Elsevier). Keywords were defined according to Medline (MESH), Embase (Emtree), and Capes (DECs), with combinations of the following terms: “Dermatology,” “skin diseases,” “Ophthalmology,” “eyes diseases,” “oral medicine,” “stomatology,” “artificial intelligence,” “machine learning,” “neural network,” “computational intelligence,” “machine intelligence,” “computer reasoning,” “AI (Artificial Intelligence),” “computer vision systems,” “computer vision system,” “convolutional neural network,” “photography,” “photographies.” In addition to the electronic search, a manual search was performed using references cited in the identified articles and also a search in Gray Literature (Google Scholar and CAPES Bank of Theses).
Eligibility Criteria
Studies evaluating the effectiveness of using deep artificial neural networks to automated classification of pathological changes pertaining to ophthalmology, dermatology, and oral medicine based on their clinical images (photos) were included. There was no restriction by language or year of publication. Reviews, ratings, letters, comments, personal or expert opinions, follow-up studies, and event summaries were excluded.
Study Selection and Data Extraction
Reference management was performed using the EndNote X7.4 software (Clarivate Analytics, Toronto, Canada). Duplicates were removed upon identification. After duplicate removal, two authors (RFTG and EFH) reviewed titles and abstracts of all studies. If the title and abstract met the eligibility criteria, the study was included. A third research group member solved possible disagreements between the two authors (RMF).
The following items were extracted from the articles: name of the author(s) and year of publication; country, health, and pathology area to which it refers; details of the artificial neural network technique used; standardization in image capture; use of public databases for training and validation; pre-processing of images; metrics used to measure performance; main results; and reported limitations.
Assessments of the Risk of Bias and Quality
Risk of bias and study quality analyses were performed independently by two authors (RFTG and EFH). This evaluation was based on the Quality Assessment of Studies of Diagnostic Accuracy (QUADAS) questionnaire adapted [50], where only the items that applied to the study design were considered, whose criteria are depicted in Table 1. The table demonstrating the quality assessment of the studies is included as Supplementary Material 1.
Table 1.
Quality assessment of studies of diagnostic accuracy [50]
| Item | Description | Aspects considered in the evaluation | Score quality* |
|---|---|---|---|
| 1 | Patient representation | This criterion was evaluated considering the importance of a large volume of data for the development and validation of AI models (n = clinical image) | 57.6% |
| 2 | Clear selection criteria | Evaluation of the division of the database into training (70%) and validation (30%) groups | 91.5% |
| 3 | Sample receives verification using a reference standard | Considering that the studies were based on the interpretation of a clinical image, and not on the gold standard, to evaluate this aspect, considering the randomization processes of the training and validation bank | 45.7% |
| 4 | Independent reference standard | In this criterion, it was considered that the index test using AI was not part of the reference standard | 100.0% |
| 5 | Index test described in detail | We consider the description developments of AI models, architectures, equations, training and adjustment parameters, data processing and pre-processing | 69.5% |
| 6 | Reference standard described in detail | Evaluation of the sample selection method, data labeling, description of classification categories, and agreement between researchers | 66.1% |
| 7 | Results interpreted without knowledge of index test | Professional performance in the data classification process was independent of the automated classification | 100.0% |
| 8 | Reference standard results interpreted without knowledge of index test | Automated classification performance of the data was independent of human classification | 100.0% |
| 9 | Uninterpretable/intermediate results reported | Description of data that was misclassified in the output of AI | 64.4% |
| 10 | Withdrawals explained | Description of data inclusion and exclusion criteria | 69.5% |
*Quality score: Percentage of articles that satisfactorily meet the evaluated item
Results
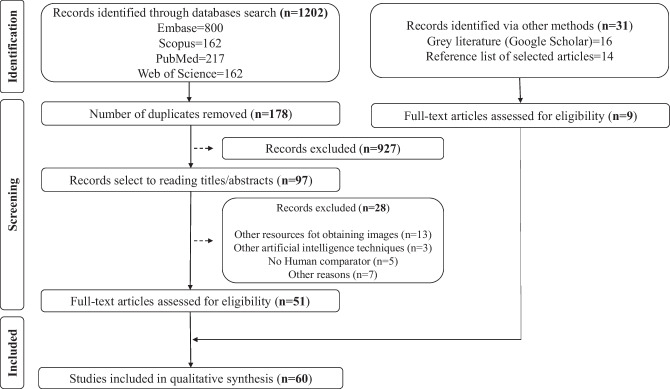
A total of 1024 articles were found, excluding articles that were repeated in all researched locations. After reading the titles and abstracts, 97 articles were selected for reading in full. After reviewing the 97 studies, 60 articles met the inclusion criteria and were included in the review (Fig. 1).
Fig. 1.
Flowchart summarizing the search strategy
Characteristics of the Selected Studies
Fifty percent of the studies were carried out in the Asian continent (50.8%), with China being the country that had the most productions with this theme (25.4%), followed by Singapore (8.4%) and Korea (6.7%). The other studies are distributed among the American (13.5%) and European (13.5%) continents, and some studies were intercontinental multicenter (13.5%).
The publication dates of the 60 selected articles varied between the years 2003 and 2021. At the end of 2018, we identified ten studies (16.9%), in 2003 [51], in 2016 [25], in 2017 [24, 52, 53], and in 2018 [23, 26, 54–56]. The predominance of publications occurred from the year 2019, where we identified 14 studies (23.7%) [6–8, 18, 22, 28, 34, 35, 57–62]; in 2020 there were 17 publications (27.11%) [3–5, 9, 10, 16, 17, 26, 31, 32, 36, 48, 63–67]; and in 2021, there were 18 studies (32.2%) [1, 10–15, 33, 68–78].
Resources involving AI can be applied in several areas of health. In this study, specialties with potential similarities were considered, which use clinical images as a complementary registration document in the routine of the services. The articles were in the field of ophthalmology (66.6%) (Table Supplementary 2) [6–18, 22–25, 31–33, 51, 53, 54, 56, 58–61, 63, 64, 68–78], dermatology (15%) (Table Supplementary 3) [4, 5, 26–28, 34–36, 71], and oral medicine (18.3%) (Table Supplementary 4) [1, 3, 48, 52, 55, 57, 62, 65–67, 79]. The use of instruments that make it possible to obtain images was admitted, such as the ophthalmoscope in ophthalmology and the dermatoscope in dermatology.
To enable the analysis of advances obtained with the use of AI in the automated classification of lesions using clinical images in ophthalmology, dermatology, and oral medicine, it was opted for leaving aside the established criteria. In the specific case of oral medicine, studies were included when the automated classification of clinical images was tested, although without the comparison between the performance of the machine and the human comparator, which resulted in the inclusion of 8 additional studies (15%) [48, 52, 55, 57, 62, 65, 67, 79].
Approximately 40 types of architectures, used alone or in combination, were found in the studies. Most frequent were (a) InceptionV3 [6, 25, 34, 54, 56, 58, 61, 68, 69, 73], U-Net20 [11, 17, 23, 59, 79], and ResNet-50 [22, 78] in ophthalmology; (b) DenseNet alone [17, 63, 64, 72] or in combination with InceptionV3 [34] in dermatology; and (c) U-Net4 ResNet-50 [57, 79] in oral medicine. Some technologies, which undergo updates to meet new demands, were Inception V1 [11, 23, 59], ResNetV2 initiation [34], InceptionV3 [6, 34, 54, 56, 58, 61, 68, 69, 73], and Inception-v44 [5, 62, 68].
A great diversity can be seen in the composition of the research teams that carried out the collection and labeling of the data and the architecture training. The composition of the research teams ranged from one [24, 51], two [17, 31, 32, 61, 71], three [7, 11, 16, 35, 53, 60, 64, 68, 77], four [14, 15], six [22, 63, 75], and eight researchers [10, 23]. Collections and labels were performed by nurses, technicians [8], or students [74] but supervised by ophthalmologists. The performance of AI was compared with specialists [8, 74], students [12, 69, 73], trained graders [13], and ophthalmologists [12, 69, 73]. The experienced ophthalmologists did the discrepancy assessment [12, 13, 71]. Large teams ranging from 20 to 67 professionals with varying degrees have been described [3–6, 10, 25, 26, 34, 36, 54, 56]. Some studies did not describe the research team [59, 70, 76].
A frequent technique in the studies was the pre-processing of the database images before development and testing the AI architectures. The description of pre-processing in 60% of ophthalmology studies, 55% of dermatology studies, and 90.9% of oral medicine studies was observed (Table 2).
Table 2.
Artificial intelligence model performance
| Specialty | Ophthalmology (n = 40) | Dermatology (n = 9) | Oral medicine (n = 11) | |
|---|---|---|---|---|
| Performance | Superior to experts | 1 | 1 | 1 |
| Equivalent | 30 | 5 | 1 | |
| Equivalent and superior to non-expert | 2 | 2 | 1 | |
| Equivalent and inferior to expert | 2 | 1 | 0 | |
| Inferior to experts | 5 | 0 | 0 | |
| Do not compare with humans | 0 | 0 | 8 | |
| Pre-processing | Yes | 24 | 5 | 10 |
| No report | 16 | 4 | 1 | |
| Image criteria | Yes | 36 | 6 | 10 |
| No report | 4 | 3 | 1 | |
| Public Bank | Yes | 15 | 4 | 0 |
| No | 25 | 5 | 11 |
Most studies (86.6%) describe the existence of protocols for collecting clinical images, and regarding the use of images obtained from public databases, 37.5% of the studies of ophthalmology and 44.4% of the dermatology studies used these sources to build their research database. Among the oral medicine studies, 100% used their own databases for the development of the studies (Table 2).
The performance of the model in the automated classification of images was evaluated, compared to human classification. Categories were created to report the findings, as can be seen in Table 2. It was observed that in 5% of studies [34, 61, 66], the AI model surpassed the experts, 60% of studies showed AI performance equivalent to expert performance, and in 8.3% of cases [3–5, 58, 78], AI overcame only non-specialized professionals. In 13.3% of cases, AI did not surpass the human experts. In another 13.3% of the studies, there was no comparison with humans; all studies were of oral medicine [48, 52, 55, 57, 62, 65, 67, 79].
Discussion
The present study carried out a systematic review of the literature on the effectiveness of AI techniques based on DL and artificial neural networks for automated classification of clinical images, when compared to human performance. Despite the evident potential to help health professionals to identify different diseases, the preliminary results indicate that more studies are needed to resolve some critical points and make present sufficient security for the clinical application of these technologies in clinical practice.
The results of the review show that only 5% of the studies demonstrate an AI model performance that outperforms human comparators and in 60% of cases the performance of the model that equals human. Among the studies with performance equivalent to the human comparator, 24 of the 36 studies (66.6%) reported protocols for image collection, as well as all those that surpassed the human comparator. Despite this encouraging result, it is necessary to observe the research scenario that must reproduce the real-life scenario to be generalized.
Image quality is highlighted as a critical point for automatic classification technologies to get high precision [23, 27, 31, 36, 60, 63, 72, 76, 78]. Evidence point the need to maintain the image quality control mechanisms in the development of technologies, for the applicability of the model in clinical practice [23]. Deshmukh et al. [70] demonstrated a semiautomatic model that allows manual corrections when the technology performs imperfect segmentations recognizing the importance of human participation in checking the automated process.
Some AI models are designed to work online with the proposal of a preliminary classification in real time. If the image obtained does not meet the minimum criteria established for the good performance of the technology, a new image is requested [12, 15, 71]. However, the performance of AI models may require greater computational power and server for hosting deployment and operation data; this was a reported limitation in a country with few technological resources [8]. This is an important finding, since many studies idealize the benefit of this resource for remote regions with limited structures and lack of professionals [3–7, 10, 12, 33, 66, 69, 71, 74, 77]. Performance validation of real-time AI software still requires studies and testing [56].
Despite good results in image classification, generalizability to clinical practice may be compromised if the training database does not include the heterogeneity of lesions [3, 28]. The ophthalmology has a standardized protocol for obtaining clinical images. Although this presents a restricted anatomical field, there are many structural variations and visually similar pathologies that can manifest in the same region of interest [16, 32, 59]. Despite this, the performance of the models used in the field of ophthalmology was predominantly equivalent to human comparators, only 7 of the 40 (17.5%) studies had inferior performance but still with high sensitivity and specificities [13, 54, 60, 64].
A superficial analysis could consider that clinical dermatology images configure easy access and present visual patterns easily recognized by AI architectures, as they are often well defined and homogeneous. However, in addition to the various skin pathologies [4, 5], it was noticed that the technology was greatly influenced by aspects such as background variations, lighting, distance, and image capture angle [7, 27, 35]. They have also been reported due to the presence of skin appendages, skin tones in different ethnicities, and even the anatomical region involved [28, 34, 71].
When considering images of visually similar lesions, Zhen et al. [34] observed confounding factors, demonstrating that AI has a limit for classifying similar lesions. To overcome this barrier, the model by Liu et al. [5] established three outputs as hypotheses for diagnosing the lesion analyzed, greatly improving the accuracy of the AI. According to the authors, this method can alert professionals to differential diagnoses that they may not have considered, which can be significant in clinical decision-making [5]. Stereophotogrammetry is a tool that has been proposed for the accurate assessment of skin lesions that project volume, such as those induced by HPV. Future studies should explore the examination of 3D photography to assess lesion size and response to treatment [80].
Multimedia reports and integration with health records remain evolving, such as Digital Imaging and Communications in Medicine (DICOM). Standards can be adopted to facilitate work steps such as creating, viewing, and exchanging reports, with markings and annotations that can contribute to communication between professionals and to monitoring the evolution of identified lesions [19].
A process widely accepted to homogenize data is pre-processing. This step makes it possible for standardization of input data in the AI model, the manipulation, or removal of artifacts [33]; edge clipping [9, 16, 25, 57, 63, 69, 72, 74, 75], color manipulation, lighting, contrast, and sharpness [3, 16, 26, 31, 33, 69, 70, 73, 74, 78]; data augmentation through techniques such as rotational scaling and spatial variations [3, 5, 16, 31, 33, 57, 59, 61, 63, 64, 70, 72, 74, 75, 78]; transfer of learning [48, 57, 79]; standardize the resolution of images [3, 6, 9, 26, 33, 57, 59, 69, 70, 73]; resizing [3, 5, 6, 9, 16, 31, 33, 54, 69, 72, 74, 75]; and also metadata encoding [5].
Regarding the oral medicine, 90% of the studies describe standardization methods of data collection and pre-processing techniques. To standardize the collection of images, the use of a camera with a specific configuration, protocols defining the degrees of inclination in capturing images [3, 66], and drying of the mucosa to reduce the reflex [65] were identified. Among the pre-processing techniques cited are cropping the area of interest [57, 62, 65–67, 79] to avoid confounding factors such as teeth and dental instruments [79], resizing [55, 65, 66, 79], contrast adjustment, reflection removal [62, 65], and resolution standardization [55]. Considering the application of these technologies in real-life practice, it seems feasible to standardize the pre-processing step. On the other hand, optimal image capture setup is a critical issue and is generally more difficult to implement in real-life applications.
In oral medicine, there is a variety of designs. The studies usually have a relatively high accuracy rates in automated classification, but some aspects should be highlighted. The study by Fu et al. was the only one that had a larger development sample, with 5775 images of benign and malignant lesions in different stages and in various anatomical sites of the oral cavity as well as controls without lesion, obtained from 11 Chinese hospitals. Classification into cancer, non-cancer, and healthy mucosa reached an AUC that ranged from 0.93 to 0.99 [3]. Tanriver et al. used a mixed database, in which part of the data had histopathological confirmation and the rest was obtained from the web. Its sample consisted of 652 images of benign, potentially malignant, and malignant lesions with variations in image quality and anatomical points. Based on the selection of an area of interest, it obtained an average accuracy of 0.87 [79].
In other previous study, the database contained 2155 images with and without injury, part received from specialists and part obtained from the Web, with no restriction regarding the standard and quality of the images, demonstrating the difficulty of obtaining a broad database. In a binary analysis, they achieved a precision of 84.77 for identifying the existence of a lesion, 67.15 for the need for referral to a specialist, and 52.13 for classifying it as benign, malignant, or potentially malignant. There was a significant reduction in the multiclass analysis, precision of 46.61 for identifying the existence of a lesion, 32.97 for the need for referral to a specialist, and 17.71 for classifying it as benign, malignant, or potentially malignant, demonstrating that a critical point is the heterogeneity of the lesions that appear in the oral cavity [48].
In general, studies demonstrated high precision, but with specific pathological conditions, not very comprehensive and reduced databases, with part of the data obtained on the internet. Shamim et al. used a sample of 200 images. Classifying conditions that manifest themselves in the tongue, it obtained an accuracy of 0.93 [57]; Jurczyszyn et al. used a sample of 63 patients, divided into leukoplakia, lichen planus, and healthy. Clinical images were obtained using an image capture and histological confirmation protocol. It obtained the best results in the comparison between lesion and normal mucosa, however, with little expressive result in the differentiation between leukoplakia and lichen planus (sensitivity of 57%, 38%, and 94% and specificity of 74%, 81%, and 88% for leukoplakia, lichen planus, and healthy mucosa, respectively) [62].
Due to the heterogeneity of oral lesions and the clinical similarity between some pathological entities [3, 48, 62], the diagnostic process in oral medicine goes far beyond the analysis of the characteristics of an image. This process requires the search for strategies that allow the combination of information that can be recognized through imaging and inclusion of additional information obtained through clinical examination. No study about oral diseases describes the use of public databases, which indicates the inexistence of this type of database and justifies the decision to Welikala et al. [48] in creating a database with several oral pathological entities, together with the respective clinical information [48]. Due to lack of public databases, or even lack of collection, some oral medicine studies used web mined images for the development of AI models, obtaining a small and unrepresentative volume of data [48, 52, 55, 57, 79].
The understanding of the automated classification process is an important step, especially in the health area. Although the connections occur in the intermediate layers of the DL models and are unknown [81], the error analysis can give clues to the data that generate confusion. This analysis can improve the understanding of the parameters that the models use to make their decisions [81–83]. Of the studies analyzed, 36.6% identified errors [3, 6, 8, 9, 13, 15, 17, 18, 22, 23, 54, 56, 57, 63, 66, 69, 70, 73, 74, 76–79], but only 23.3% perform some analysis of these errors [6, 9, 13, 17, 18, 22, 56, 63, 69, 70, 73, 74, 78, 79].
Although studies consider the use of AI as an important and promising tool to aid decision-making in clinical practice [48, 57, 79], it does not seem realistic to consider the replacement of specialist professionals. So far, studies show that in the future, this technology can help healthcare networks [57, 65, 79], enabling the screening process in areas of difficult access and optimizing time and the work process of different professionals.
Our review has limitations that should be recognized. Although most of the studies were of good quality when evaluated individually in this study (60%), the heterogeneity of the methodologies used does not allow their use in a meta-analysis. Owing to the heterogeneity of the methodologies used in the studies included in this review, in a rapidly changing field, it is also difficult to generalize the results. Taking the lack of robust evidence into account, further studies are necessary, particularly in different countries, to strengthen the evidence about the application of these features in real life.
Conclusion
Recent studies have generated remarkable advances. The knowledge produced so far has determined the factors that need to be explored to improve the performance of the architectures. In the coming years, the great challenges will be to achieve a satisfactory performance of AI in the identification of lesions at an early stage and the interpretability of the automated classification process for the understanding of the parameters that the models use to make their decisions. Above all, it is essential to be aware that, at least until this moment, one should not create the expectation of creating a tool that will replace human evaluation but may be an adjunct for the clinical setting, particularly for health professionals with little training for recognition of a specific disease. The pre-processing seems to be a predictable technical step, which allows the reduction of the variability of the data presented to the artificial intelligence, enhancing the performance of automated classification in the real-life multicentric scenario.
Supplementary Information
Below is the link to the electronic supplementary material.
Author Contribution
RFTG, RMF, LFS, and VCC conceived, designed, guided, and coordinated the study and the writing. RFTG and EFH identified publication records from MEDLINE and screened the titles, abstracts, and full text of the articles. Rita identified additional articles that were not retrieved in MEDLINE. RFTG and VCC prepared the figures and tables. All the authors contributed to the writing of the article. The Abstract, Introduction, Materials and Methods, Discussion, and Conclusion sections were written jointly by RFTG and VCC. RFTG and VCC performed thorough editing of the article. All the authors revised and approved the final article.
Funding
No funding was obtained for this study.
Data Availability
Not applicable.
Declarations
Ethics Approval
Ethical approval for this type of study is not required by our institute.
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Warin K, Limprasert W, Suebnukarn S, Jinaporntham S, Jantana P. Automatic classification and detection of oral cancer in photographic images using deep learning algorithms. J Oral Pathol Med. 2021;50(9):911–918. doi: 10.1111/jop.13227. [DOI] [PubMed] [Google Scholar]
- 2.Roth CJ, Clunie DA, Vining DJ, Berkowitz SJ, Berlin A, Bissonnette JP, Clark SD, Cornish TC, Eid M, Gaskin CM, Goel AK, Jacobs GC, Kwan D, Luviano DM, McBee MP, Miller K, Hafiz AM, Obcemea C, Parwani AV, Rotemberg V, Silver EL, Storm ES, Tcheng JE, Thullner KS, Folio LR. Multispecialty Enterprise Imaging Workgroup Consensus on Interactive Multimedia Reporting Current State and Road to the Future: HIMSS-SIIM Collaborative White Paper. J Digit Imaging. 2021;34(3):495–522. doi: 10.1007/s10278-021-00450-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu, Q., Chen, Y., Li, Z., Jing, Q., Hu, C., Liu, H., Bao, J., Hong, Y., Shi, T., Li, K., Zou, H., Song, Y.Y., Wang, H., Wang, X., Wang, Y., Liu, J., Liu, H., Chen, S., Chen, R., Zhang, M., Zhao, J., Xiang, J., Liu, B., Jia, J., Wu, H., Zhao, Y., Wan, L., & Xiong, X. (2020). A deep learning algorithm for detection of oral cavity squamous cell carcinoma from photographic images: A retrospective study. EClinicalMedicine, 27. [DOI] [PMC free article] [PubMed]
- 4.Han SS, Moon IJ, Lim W, Suh IS, Lee SY, Na JI, Kim SH, Chang SE. Keratinocytic Skin Cancer Detection on the Face Using Region-Based Convolutional Neural Network. JAMA Dermatol. 2020;156(1):29–37. doi: 10.1001/jamadermatol.2019.3807.PMID:31799995;PMCID:PMC6902187.(a). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Jain A, Eng C, Way DH, Lee K, Bui P, Kanada K, de Oliveira Marinho G, Gallegos J, Gabriele S, Gupta V, Singh N, Natarajan V, Hofmann-Wellenhof R, Corrado GS, Peng LH, Webster DR, Ai D, Huang SJ, Liu Y, Dunn RC, Coz D. A deep learning system for differential diagnosis of skin diseases. Nat Med. 2020;26(6):900–908. doi: 10.1038/s41591-020-0842-3. [DOI] [PubMed] [Google Scholar]
- 6.Keel S, Li Z, Scheetz J, Robman L, Phung J, Makeyeva G, Aung K, Liu C, Yan X, Meng W, Guymer R, Chang R, He M. Development and validation of a deep-learning algorithm for the detection of neovascular age-related macular degeneration from colour fundus photographs. Clin Exp Ophthalmol. 2019;47(8):1009–1018. doi: 10.1111/ceo.13575. [DOI] [PubMed] [Google Scholar]
- 7.Kim MC, Okada K, Ryner AM, Amza A, Tadesse Z, Cotter SY, Gaynor BD, Keenan JD, Lietman TM, Porco TC. Sensitivity and specificity of computer vision classification of eyelid photographs for programmatic trachoma assessment. PLoS One. 2019 Feb 11;14(2):e0210463. 10.1371/journal.pone.0210463. PMID: 30742639; PMCID: PMC6370195. [DOI] [PMC free article] [PubMed]
- 8.Bellemo V, Lim ZW, Lim G, Nguyen QD, Xie Y, Yip MYT, Hamzah H, Ho J, Lee XQ, Hsu W, Lee ML, Musonda L, Chandran M, Chipalo-Mutati G, Muma M, Tan GSW, Sivaprasad S, Menon G, Wong TY, Ting DSW. Artificial intelligence using deep learning to screen for referable and vision-threatening diabetic retinopathy in Africa: a clinical validation study. Lancet Digit Health. 2019;1(1):e35–e44. doi: 10.1016/S2589-7500(19)30004-4. [DOI] [PubMed] [Google Scholar]
- 9.Lu L, Ren P, Lu Q, Zhou E, Yu W, Huang J, He X, Han W. Analyzing fundus images to detect diabetic retinopathy (DR) using deep learning system in the Yangtze River delta region of China. Ann Transl Med. 2021;9(3):226. doi: 10.21037/atm-20-3275.PMID:33708853;PMCID:PMC7940941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Son J, Shin JY, Kim HD, Jung KH, Park KH, Park SJ. Development and Validation of Deep Learning Models for Screening Multiple Abnormal Findings in Retinal Fundus Images. Ophthalmology. 2020;127(1):85–94. doi: 10.1016/j.ophtha.2019.05.029. [DOI] [PubMed] [Google Scholar]
- 11.Redd TK, Campbell JP, Brown JM, Kim SJ, Ostmo S, Chan RVP, Dy J, Erdogmus D, Ioannidis S, Kalpathy-Cramer J, Chiang MF; Imaging and Informatics in Retinopathy of Prematurity (i-ROP) Research Consortium. Evaluation of a deep learning image assessment system for detecting severe retinopathy of prematurity. Br J Ophthalmol. 2018 Nov 23: bjophthalmol-2018–313156. 10.1136/bjophthalmol-2018-313156. Epub ahead of print. PMID: 30470715; PMCID: PMC7880608. [DOI] [PMC free article] [PubMed]
- 12.Jain A, Krishnan R, Rogye A, Natarajan S. Use of offline artificial intelligence in a smartphone-based fundus camera for community screening of diabetic retinopathy. Indian J Ophthalmol. 2021;69(11):3150–3154. doi: 10.4103/ijo.IJO_3808_20.PMID:34708760;PMCID:PMC8725118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wintergerst MWM, Bejan V, Hartmann V, Schnorrenberg M, Bleckwenn M, Weckbecker K, Finger RP. Telemedical Diabetic Retinopathy Screening in a Primary Care Setting: Quality of Retinal Photographs and Accuracy of Automated Image Analysis. Ophthalmic Epidemiol. 2021 Jun 20:1–10. 10.1080/09286586.2021.1939886. Epub ahead of print. PMID: 34151725. [DOI] [PubMed]
- 14.Pawar B, Lobo SN, Joseph M, Jegannathan S, Jayraj H. Validation of Artificial Intelligence Algorithm in the Detection and Staging of Diabetic Retinopathy through Fundus Photography: An Automated Tool for Detection and Grading of Diabetic Retinopathy. Middle East Afr J Ophthalmol. 2021;28(2):81–86. doi: 10.4103/meajo.meajo_406_20.PMID:34759664;PMCID:PMC8547660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baget-Bernaldiz M, Pedro R-A, Santos-Blanco E, Navarro-Gil R, Valls A, Moreno A, Rashwan HA, Puig D. Testing a Deep Learning Algorithm for Detection of Diabetic Retinopathy in a Spanish Diabetic Population and with MESSIDOR Database. Diagnostics. 2021;11(8):1385. doi: 10.3390/diagnostics11081385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turk LA, Wang S, Krause P, Wawrzynski J, Saleh GM, Alsawadi H, Alshamrani AZ, Peto T, Bastawrous A, Li J, Tang HL. Evidence Based Prediction and Progression Monitoring on Retinal Images from Three Nations. Transl Vis Sci Technol. 2020;9(2):44. doi: 10.1167/tvst.9.2.44.PMID:32879754;PMCID:PMC7443119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biousse V, Newman NJ, Najjar RP, Vasseneix C, Xu X, Ting DS, Milea LB, Hwang JM, Kim DH, Yang HK, Hamann S, Chen JJ, Liu Y, Wong TY, Milea D; BONSAI (Brain and Optic Nerve Study with Artificial Intelligence) Study Group. Optic Disc Classification by Deep Learning versus Expert Neuro-Ophthalmologists. Ann Neurol. 2020 Oct;88(4):785–795. 10.1002/ana.25839. Epub 2020 Aug 7. PMID: 32621348. [DOI] [PubMed]
- 18.Lin H, Li R, Liu Z, Chen J, Yang Y, Chen H, Lin Z, Lai W, Long E, Wu X, Lin D, Zhu Y, Chen C, Wu D, Yu T, Cao Q, Li X, Li J, Li W, Wang J, Yang M, Hu H, Zhang L,Yu Y, Chen X, Hu J, Zhu K, Jiang S, Huang Y, Tan G, Huang J, Lin X, Zhang X, Luo L, Liu Y, Liu X, Cheng B, Zheng D, Wu M, Chen W, Liu Y. Diagnostic Efficacy and Therapeutic Decision-making Capacity of an Artificial Intelligence Platform for Childhood Cataracts in Eye Clinics: A Multicentre Randomized Controlled Trial. Volume 9, P52–59, March 01, 2019. 10.1016/j.eclinm.2019.03.001 [DOI] [PMC free article] [PubMed]
- 19.Berkowitz SJ, Kwan D, Cornish TC, Silver EL, Thullner KS, Aisen A, Bui MM, Clark SD, Clunie DA, Eid M, Hartman DJ, Ho K, Leontiev A, Luviano DM, O'Toole PE, Parwani AV, Pereira NS, Rotemberg V, Vining DJ, Gaskin CM, Roth CJ, Folio LR. Interactive Multimedia Reporting Technical Considerations: HIMSS-SIIM Collaborative White Paper. J Digit Imaging. 2022;35(4):817–833. doi: 10.1007/s10278-022-00658-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Waal I, de Bree R, Brakenhoff R, Coebergh JW. Early diagnosis in primary oral cancer: is it possible? Med Oral Patol Oral Cir Bucal. 2011;16(3):e300–e305. doi: 10.4317/medoral.16.e300. [DOI] [PubMed] [Google Scholar]
- 21.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 22.Al-Aswad LA, Kapoor R, Chu CK, Walters S, Gong D, Garg A, Gopal K, Patel V, Sameer T, Rogers TW, Nicolas J, De Moraes GC, Moazami G. Evaluation of a Deep Learning System For Identifying Glaucomatous Optic Neuropathy Based on Color Fundus Photographs. J Glaucoma. 2019;28(12):1029–1034. doi: 10.1097/IJG.0000000000001319. [DOI] [PubMed] [Google Scholar]
- 23.Brown JM, Campbell JP, Beers A, Chang K, Ostmo S, Chan RVP, Dy J, Erdogmus D, Ioannidis S, Kalpathy-Cramer J, Chiang MF; Imaging and Informatics in Retinopathy of Prematurity (i-ROP) Research Consortium. Automated Diagnosis of Plus Disease in Retinopathy of Prematurity Using Deep Convolutional Neural Networks. JAMA Ophthalmol. 2018 Jul 1;136(7):803–810. 10.1001/jamaophthalmol.2018.1934. PMID: 29801159; PMCID: PMC6136045. [DOI] [PMC free article] [PubMed]
- 24.Burlina PM, Joshi N, Pekala M, Pacheco KD, Freund DE, Bressler NM. Automated Grading of Age-Related Macular Degeneration From Color Fundus Images Using Deep Convolutional Neural Networks. JAMA Ophthalmol. 2017;135(11):1170–1176. doi: 10.1001/jamaophthalmol.2017.3782.PMID:28973096;PMCID:PMC5710387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, Kim R, Raman R, Nelson PC, Mega JL, Webster DR. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA. 2016;316(22):2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 26.Han SS, Kim MS, Lim W, Park GH, Park I, Chang SE. Classification of the Clinical Images for Benign and Malignant Cutaneous Tumors Using a Deep Learning Algorithm. J Invest Dermatol. 2018;138(7):1529–1538. doi: 10.1016/j.jid.2018.01.028. [DOI] [PubMed] [Google Scholar]
- 27.Kim, Y. J., Han, S. S., Yang, H. J., & Chang, S. E. (2020). Prospective, comparative evaluation of a deep neural network and dermoscopy in the diagnosis of onychomycosis. PloS one, 15(6), e0234334. 10.1371/journal.pone.0234334 [DOI] [PMC free article] [PubMed]
- 28.Meienberger N, Anzengruber F, Amruthalingam L, Christen R, Koller T, Maul JT, Pouly M, Djamei V, Navarini AA. Observer-independent assessment of psoriasis-affected area using machine learning. JEADV 2020, 34, 1362–1368 European Academy of Dermatology and Venereology. 10.1111/jdv.16002 [DOI] [PubMed]
- 29.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017 Feb 2;542(7639):115–118. 10.1038/nature21056. Epub 2017 Jan 25. Erratum in: Nature. 2017 Jun 28;546(7660):686. PMID: 28117445; PMCID: PMC8382232. [DOI] [PMC free article] [PubMed]
- 30.Gurovich Y, Hanani Y, Bar O, et al. Identifying facial phenotypes of genetic disorders using deep learning. Nat Med. 2019;25:60–64. doi: 10.1038/s41591-018-0279-0. [DOI] [PubMed] [Google Scholar]
- 31.He J, Cao T, Xu F, Wang S, Tao H, Wu T, Sun L, Chen J. Artificial intelligence-based screening for diabetic retinopathy at community hospital. Eye (Lond). 2020 Mar;34(3):572–576. 10.1038/s41433-019-0562-4. Epub 2019 Aug 27. PMID: 31455902; PMCID: PMC7042314. [DOI] [PMC free article] [PubMed]
- 32.Hsieh, Yi-Ting, Lee‐Ming Chuang, Yi-Der Jiang, Tien-Jyun Chang, Chung-May Yang, Chang-Hao Yang, Li-Wei Chan, Tzu-Yun Kao, Ta-Ching Chen, Hsuan-Chieh Lin, Chin-Han Tsai and Mingke Chen. “Application of deep learning image assessment software VeriSee™ for diabetic retinopathy screening.” Journal of the Formosan Medical Association = Taiwan yi zhi (2020): n. pag. 10.1016/j.jfma.2020.03.024. [DOI] [PubMed]
- 33.Soenksen LR, Kassis T, Conover ST, Marti-Fuster B, Birkenfeld JS, Tucker-Schwartz J, Naseem A, Stavert RR, Kim CC, Senna MM, Avilés-Izquierdo J, Collins JJ, Barzilay R, Gray ML. Using deep learning for dermatologist-level detection of suspicious pigmented skin lesions from wide-field images. Sci Transl Med. 2021 Feb 17;13(581):eabb3652. 10.1126/scitranslmed.abb3652. PMID: 33597262. [DOI] [PubMed]
- 34.Zhao S, Xie B, Li Y, Zhao X, Kuang Y, Su J, He X, Wu X, Fan W, Huang K, Su J, Peng Y, Navarini AA, Huang W, Chen X. Smart identification of psoriasis by images using convolutional neural networks: a case study in China. J Eur Acad Dermatol Venereol. 2020;34(3):518–524. doi: 10.1111/jdv.15965. [DOI] [PubMed] [Google Scholar]
- 35.Seité S, Khammari A, Benzaquen M, Moyal M, Dréno B. Development and accuracy of an artificial intelligence algorithm for acne grading from smartphone photographs. Experimental Dermatology. 2019;28:1252–1257. doi: 10.1111/exd.14022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han SS, Moon IJ, Kim SH, Na JI, Kim MS, Park GH, Park I, Kim K, Lim W, Lee JH, Chang SE. Assessment of deep neural networks for the diagnosis of benign and malignant skin neoplasms in comparison with dermatologists: A retrospective validation study. PLoS Med. 2020 Nov 25;17(11):e1003381. 10.1371/journal.pmed.1003381. PMID: 33237903; PMCID: PMC7688128. (b) [DOI] [PMC free article] [PubMed]
- 37.Pan X, Zhang T, Yang Q, Yang D, Rwigema JC, Qi XS. Survival prediction for oral tongue cancer patients via probabilistic genetic algorithm optimized neural network models. The British journal of radiology. 2020;93(1112):20190825. doi: 10.1259/bjr.20190825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stadler TM, Hüllner MW, Broglie MA, Morand GB. Predictive value of suvmax changes between two sequential post-therapeutic FDG-pet in head and neck squamous cell carcinomas. Scientific reports. 2020;10(1):16689. doi: 10.1038/s41598-020-73914-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song B, Sunny S, Uthoff RD, Patrick S, Suresh A, Kolur T, Keerthi G, Anbarani A, Wilder-Smith P, Kuriakose MA, Birur P, Rodriguez JJ, Liang R. Automatic classification of dual-modalilty, smartphone-based oral dysplasia and malignancy images using deep learning. Biomedical optics express. 2018;9(11):5318–5329. doi: 10.1364/BOE.9.005318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uthoff, R. D., Song, B., Sunny, S., Patrick, S., Suresh, A., Kolur, T., Keerthi, G., Spires, O., Anbarani, A., Wilder-Smith, P., Kuriakose, M. A., Birur, P., & Liang, R. (2018). Point-of-care, smartphone-based, dual-modality, dual-view, oral cancer screening device with neural network classification for low-resource communities. PloS one, 13(12), e0207493. 10.1371/journal.pone.0207493 [DOI] [PMC free article] [PubMed]
- 41.Uthoff RD, Song B, Birur P, Kuriakose MA, Sunny S, Suresh A, Patrick S, Anbarani A, Spires O, Wilder-Smith P, ; Liang R. Development of a dual-modality, dual-view smartphone-based imaging system for oral cancer detection. In R. Raghavachari, & R. Liang (Eds.), Design and Quality for Biomedical Technologies XI [104860V] Biomedical Optics and Imaging - Proceedings of SPIE 2018;10486). DOI:10.1117/12.2296435.
- 42.Jeyaraj PR, Samuel Nadar ER. Computer-assisted medical image classification for early diagnosis of oral cancer employing deep learning algorithm. Journal of cancer research and clinical oncology. 2019;145(4):829–837. doi: 10.1007/s00432-018-02834-7. [DOI] [PubMed] [Google Scholar]
- 43.Uthoff RD, Song B, Sunny S, Patrick S, Suresh A, Kolur T, Gurushanth K, Wooten K, Gupta V, Platek ME, Singh AK, Wilder-Smith P, Kuriakose MA, Birur P, Liang R. Small form factor, flexible, dual-modality handheld probe for smartphone-based, point-of-care oral and oropharyngeal cancer screening. Journal of biomedical optics. 2019;24(10):1–8. doi: 10.1117/1.JBO.24.10.106003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jurczyszyn, K., Kazubowska, K., Kubasiewicz-Ross, P., Ziółkowski, P., & Dominiak, M. (2018). Application of fractal dimension analysis and photodynamic diagnosis in the case of differentiation between lichen planus and leukoplakia: A preliminary study. Advances in clinical and experimental medicine : official organ Wroclaw Medical University, 27(12), 1729–1736. 10.17219/acem/80831 [DOI] [PubMed]
- 45.Alhazmi A, Alhazmi Y, Makrami A, Masmali A, Salawi N, Masmali K, Patil S. Application of artificial intelligence and machine learning for prediction of oral cancer risk. J Oral Pathol Med. 2021;50(5):444–450. doi: 10.1111/jop.13157. [DOI] [PubMed] [Google Scholar]
- 46.Hung M, Park J, Hon ES, Bounsanga J, Moazzami S, Ruiz-Negrón B, Wang D. Artificial intelligence in dentistry: Harnessing big data to predict oral cancer survival. World J Clin Oncol. 2020;11(11):918–934. doi: 10.5306/wjco.v11.i11.918.PMID:33312886;PMCID:PMC7701911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alabi RO, Elmusrati M, Sawazaki-Calone I, Kowalski LP, Haglund C, Coletta RD, Mäkitie AA, Salo T, Leivo I, Almangush A. Machine learning application for prediction of locoregional recurrences in early oral tongue cancer: a Web-based prognostic tool. Virchows Arch. 2019 Oct;475(4):489–497. 10.1007/s00428-019-02642-5. Epub 2019 Aug 17. PMID: 31422502; PMCID: PMC6828835. [DOI] [PMC free article] [PubMed]
- 48.Welikala RA, Remagnino P, Lim JH, Chan CS, Rajendran S, Kallarakkal TG, Zain RB, Jayasinghe RD, Rimal J, Kerr AR, Amtha R, Patil K, Tilakaratne WM, Gibson J, Cheong SC, Barman AS. Automated Detection and Classification of Oral Lesions Using Deep Learning for Early Detection of Oral Cancer, in IEEE Access, vol.
- 49.Page MJ, McKenzie JE, Bossuyt PM , Boutron I , Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM , Akl EA, Brennan SE, Chou R, Glanville J, Grimshaw JM, Hróbjartsson A, Lalu MM, Li T , Loder EW, Mayo-Wilson E, McDonald E, McGuinness LA, Stewart LA, Thomas J, Tricco AC, Welch VA, Whiting P, Moher D. A declaração PRISMA 2020: uma diretriz atualizada para relatar revisões sistemáticas BMJ 2021 ; 372 10.1136/bmj.n71. Cite isso como: BMJ 2021;372:n7.
- 50.Moher, D., Liberati, A., Tetzlaff, J., Altman, D. G., & PRISMA Group (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS medicine, 6(7), e1000097. 10.1371/journal.pmed.1000097 [DOI] [PMC free article] [PubMed]
- 51.Usher D, Dumskyj M, Himaga M, Williamson TH, Nussey S, Boyce J. Automated detection of diabetic retinopathy in digital retinal images: a tool for diabetic retinopathy screening. Diabet Med. 2004;21(1):84–90. doi: 10.1046/j.1464-5491.2003.01085.x. [DOI] [PubMed] [Google Scholar]
- 52.Anantharaman R, Anantharaman V, Lee Y. Oro Vision: Deep Learning for Classifying Orofacial Diseases. IEEE International Conference on Healthcare Informatics (ICHI) 2017;2017:39–45. doi: 10.1109/ICHI.2017.69. [DOI] [Google Scholar]
- 53.Ting DSW, Cheung CY, Lim G, Tan GSW, Quang ND, Gan A, Hamzah H, Garcia-Franco R, San Yeo IY, Lee SY, Wong EYM, Sabanayagam C, Baskaran M, Ibrahim F, Tan NC, Finkelstein EA, Lamoureux EL, Wong IY, Bressler NM, Sivaprasad S, Varma R, Jonas JB, He MG, Cheng CY, Cheung GCM, Aung T, Hsu W, Lee ML, Wong TY. Development and Validation of a Deep Learning System for Diabetic Retinopathy and Related Eye Diseases Using Retinal Images From Multiethnic Populations With Diabetes. JAMA. 2017;318(22):2211–2223. doi: 10.1001/jama.2017.18152.PMID:29234807;PMCID:PMC5820739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Z, He Y, Keel S, Meng W, Chang RT, He M. Efficacy of a Deep Learning System for Detecting Glaucomatous Optic Neuropathy Based on Color Fundus Photographs. Ophthalmology. 2018;125(8):1199–1206. doi: 10.1016/j.ophtha.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 55.Anantharaman R, Velazquez M, Lee Y. Utilizing Mask R-CNN for Detection and Segmentation of Oral Diseases. IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 2018;2018:2197–2204. doi: 10.1109/BIBM.2018.8621112. [DOI] [Google Scholar]
- 56.Li Z, Keel S, Liu C, He Y, Meng W, Scheetz J, Lee PY, Shaw J, Ting D, Wong TY, Taylor H, Chang R, He M. An Automated Grading System for Detection of Vision-Threatening Referable Diabetic Retinopathy on the Basis of Color Fundus Photographs. Diabetes Care. 2018;41(12):2509–2516. doi: 10.2337/dc18-0147. [DOI] [PubMed] [Google Scholar]
- 57.Shamim MZ, Syed S, Shiblee M, Usman M, Ali SJ, Hussein HS, Farrag M. Automated Detection of Oral Pre-Cancerous Tongue Lesions Using Deep Learning for Early Diagnosis of Oral Cavity Cancer. Comput. J. 2022;65:91–104. doi: 10.1093/comjnl/bxaa136. [DOI] [Google Scholar]
- 58.Phene S, Dunn RC, Hammel N, Liu Y, Krause J, Kitade N, Schaekermann M, Sayres R, Wu DJ, Bora A, Semturs C, Misra A, Huang AE, Spitze A, Medeiros FA, Maa AY, Gandhi M, Corrado GS, Peng L, Webster DR. Deep Learning and Glaucoma Specialists: The Relative Importance of Optic Disc Features to Predict Glaucoma Referral in Fundus Photographs. Ophthalmology. 2019;126(12):1627–1639. doi: 10.1016/j.ophtha.2019.07.024. [DOI] [PubMed] [Google Scholar]
- 59.Ting DSW, Cheung CY, Nguyen Q, Sabanayagam C, Lim G, Lim ZW, Tan GSW, Soh YQ, Schmetterer L, Wang YX, Jonas JB, Varma R, Lee ML, Hsu W, Lamoureux E, Cheng CY, Wong TY. Deep learning in estimating prevalence and systemic risk factors for diabetic retinopathy: a multi-ethnic study. NPJ Digit Med. 2019;10(2):24. doi: 10.1038/s41746-019-0097-x.PMID:31304371;PMCID:PMC6550209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang WH, Zheng B, Wu MN, Zhu SJ, Fei FQ, Weng M, Zhang X, Lu PR. An Evaluation System of Fundus Photograph-Based Intelligent Diagnostic Technology for Diabetic Retinopathy and Applicability for Research. Diabetes Ther. 2019 Oct;10(5):1811–1822. 10.1007/s13300-019-0652-0. Epub 2019 Jul 9. PMID: 31290125; PMCID: PMC6778552. [DOI] [PMC free article] [PubMed]
- 61.Zhen Y, Chen H, Zhang X, Meng X, Zhang J, Pu J. ASSESSMENT OF CENTRAL SEROUS CHORIORETINOPATHY DEPICTED ON COLOR FUNDUS PHOTOGRAPHS USING DEEP LEARNING. Retina. 2020;40(8):1558–1564. doi: 10.1097/IAE.0000000000002621. [DOI] [PubMed] [Google Scholar]
- 62.Jurczyszyn K, Kozakiewicz M. Differential diagnosis of leukoplakia versus lichen planus of the oral mucosa based on digital texture analysis in intraoral photography. Adv Clin Exp Med. 2019;28(11):1469–1476. doi: 10.17219/acem/104524. [DOI] [PubMed] [Google Scholar]
- 63.Kuo, MT., Hsu, B.WY., Yin, YK. et al. A deep learning approach in diagnosing fungal keratitis based on corneal photographs. Sci Rep 10, 14424 (2020). 10.1038/s41598-020-71425-9 [DOI] [PMC free article] [PubMed]
- 64.Ludwig CA, Perera C, Myung D, Greven MA, Smith SJ, Chang RT, Leng T. Automatic Identification of Referral-Warranted Diabetic Retinopathy Using Deep Learning on Mobile Phone Images. Transl Vis Sci Technol. 2020;9(2):60. doi: 10.1167/tvst.9.2.60.PMID:33294301;PMCID:PMC7718806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jurczyszyn K, Gedrange T, Kozakiewicz M. Theoretical Background to Automated Diagnosing of Oral Leukoplakia: A Preliminary Report. J Healthc Eng. 2020;13(2020):8831161. doi: 10.1155/2020/8831161.PMID:33005316;PMCID:PMC7509569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.You W, Hao A, Li S, Wang Y, Xia B. Deep learning-based dental plaque detection on primary teeth: a comparison with clinical assessments. BMC Oral Health. 2020;20(1):141. doi: 10.1186/s12903-020-01114-6.PMID:32404094;PMCID:PMC7222297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thomas B, Kumar V, Saini S. Texture analysis based segmentation and classification of oral cancer lesions in color images using ANN, 2013 IEEE International Conference on Signal Processing, Computing and Control (ISPCC), 2013, pp. 1–5, 10.1109/ISPCC.2013.6663401.
- 68.Rêgo S, Dutra-Medeiros M, Soares F, Monteiro-Soares M. Screening for Diabetic Retinopathy Using an Automated Diagnostic System Based on Deep Learning: Diagnostic Accuracy Assessment. Ophthalmologica. 2021;244(3):250–257. doi: 10.1159/000512638. [DOI] [PubMed] [Google Scholar]
- 69.Wang Y, Shi D, Tan Z, Niu Y, Jiang Y, Xiong R, Peng G, He M. Screening Referable Diabetic Retinopathy Using a Semi-automated Deep Learning Algorithm Assisted Approach. Front Med (Lausanne). 2021 Nov 25;8:740987. 10.3389/fmed.2021.740987. PMID: 34901058; PMCID: PMC8656222. [DOI] [PMC free article] [PubMed]
- 70.Deshmukh M, Liu YC, Rim TH, Venkatraman A, Davidson M, Yu M, Kim HS, Lee G, Jun I, Mehta JS, Kim EK. Automatic segmentation of corneal deposits from corneal stromal dystrophy images via deep learning. Comput Biol Med. 2021 Oct;137:104675. 10.1016/j.compbiomed.2021.104675. Epub 2021 Jul 27. PMID: 34425417. [DOI] [PubMed]
- 71.Ming S, Xie K, Lei X, Yang Y, Zhao Z, Li S, Jin X, Lei B. Evaluation of a novel artificial intelligence-based screening system for diabetic retinopathy in community of China: a real-world study. Int Ophthalmol. 2021;41(4):1291–1299. doi: 10.1007/s10792-020-01685-x. [DOI] [PubMed] [Google Scholar]
- 72.Hung JY, Perera C, Chen KW, Myung D, Chiu HK, Fuh CS, Hsu CR, Liao SL, Kossler AL. A deep learning approach to identify blepharoptosis by convolutional neural networks. Int J Med Inform. 2021 Apr;148:104402. 10.1016/j.ijmedinf.2021.104402. Epub 2021 Jan 28. PMID: 33609928; PMCID: PMC8191181. [DOI] [PMC free article] [PubMed]
- 73.Zheng C, Xie X, Wang Z, Li W, Chen J, Qiao T, Qian Z, Liu H, Liang J, Chen X. Development and validation of deep learning algorithms for automated eye laterality detection with anterior segment photography. Sci Rep. 2021;11(1):586. doi: 10.1038/s41598-020-79809-7.PMID:33436781;PMCID:PMC7803760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Malerbi FK, Andrade RE, Morales PH, Stuchi JA, Lencione D, de Paulo JV, Carvalho MP, Nunes FS, Rocha RM, Ferraz DA, Belfort R Jr. Diabetic Retinopathy Screening Using Artificial Intelligence and Handheld Smartphone-Based Retinal Camera. J Diabetes Sci Technol. 2021 Jan 12:1932296820985567. 10.1177/1932296820985567. Epub ahead of print. PMID: 33435711. [DOI] [PMC free article] [PubMed]
- 75.Li F, Wang Y, Xu T, et al. Deep learning-based automated detection for diabetic retinopathy and diabetic macular edema in retinal fundus photographs. Eye. 2021 doi: 10.1038/s41433-021-01552-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Katz O, Presil D, Cohen L, Nachmani R, Kirshner N, Hoch Y, Lev T, Hadad A, Hewitt RJ, Owens DR. Evaluation of a New Neural Network Classifier for Diabetic Retinopathy. J Diabetes Sci Technol. 2021 Sep 22:19322968211042665. 10.1177/19322968211042665. Epub ahead of print. PMID: 34549633. [DOI] [PMC free article] [PubMed]
- 77.Shah A, Clarida W, Amelon R, Hernaez-Ortega MC, Navea A, Morales-Olivas J, Dolz-Marco R, Verbraak F, Jorda PP, van der Heijden AA, Peris Martinez C. Validation of Automated Screening for Referable Diabetic Retinopathy With an Autonomous Diagnostic Artificial Intelligence System in a Spanish Population. J Diabetes Sci Technol. 2021 May;15(3):655–663. 10.1177/1932296820906212. Epub 2020 Mar 16. PMID: 32174153; PMCID: PMC8120039. [DOI] [PMC free article] [PubMed]
- 78.Chiang M, Guth D, Pardeshi AA, Randhawa J, Shen A, Shan M, Dredge J, Nguyen A, Gokoffski K, Wong BJ, Song B, Lin S, Varma R, Xu BY. Glaucoma Expert-Level Detection of Angle Closure in Goniophotographs With Convolutional Neural Networks: The Chinese American Eye Study. Am J Ophthalmol. 2021 Jun;226:100–107. 10.1016/j.ajo.2021.02.004. Epub 2021 Feb 9. PMID: 33577791; PMCID: PMC8286291. [DOI] [PMC free article] [PubMed]
- 79.Tanriver G, Soluk Tekkesin M, Ergen O. Automated Detection and Classification of Oral Lesions Using Deep Learning to Detect Oral Potentially Malignant Disorders. Cancers (Basel). 2021;13(11):2766. doi: 10.3390/cancers13112766.PMID:34199471;PMCID:PMC8199603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rijsbergen M, Pagan L, Niemeyer-van der Kolk T, Rijneveld R, Hogendoorn G, Lemoine C, Meija Miranda Y, Feiss G, Bouwes Bavink JN, Burggraaf J, van Poelgeest MIE, Rissmann R. Stereophotogrammetric three-dimensional photography is an accurate and precise planimetric method for the clinical visualization and quantification of human papilloma virus-induced skin lesions. J Eur Acad Dermatol Venereol. 2019 Aug;33(8):1506–1512. 10.1111/jdv.15474. Epub 2019 Mar 12. PMID: 30720900; PMCID: PMC6767777. [DOI] [PMC free article] [PubMed]
- 81.Alzubaidi L, Zhang J, Humaidi AJ, Al-Dujaili A, Duan Y, Al-Shamma O, Santamaría J, Fadhel MA, Al-Amidie M, Farhan L. Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. Journal of big data. 2021;8(1):53. doi: 10.1186/s40537-021-00444-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ker J, Wang L, Rao J, Lim T. Deep Learning Applications in Medical Image Analysis. IEEE Access. 2018;6:9375–9389. doi: 10.1109/ACCESS.2017.2788044. [DOI] [Google Scholar]
- 83.Li Y, Huang C, Ding L, Li Z, Pan Y, Gao X. Deep learning in bioinformatics: introduction, application, and perspective in big data era. Methods. 2019 doi: 10.1016/j.ymeth.2019.04.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.