Abstract
The developmental biology underlying the morphogenesis of mushrooms remains poorly understood despite the essential role of fungi in the terrestrial environment and global carbon cycle. The mushroom Coprinopsis cinerea is a leading model system for the molecular and cellular basis of fungal morphogenesis. The dikaryotic vegetative hyphae of this fungus grow by tip growth with clamp cell formation, conjugate nuclear division, septation, subapical peg formation, and fusion of the clamp cell to the peg. Studying these processes provides many opportunities to gain insights into fungal cell morphogenesis. Here, we report the dynamics of five septins, as well as the regulators CcCla4, CcSpa2, and F-actin, visualized by tagging with fluorescent proteins, EGFP, PA-GFP or mCherry, in the growing dikaryotic vegetative hyphae. We also observed the nuclei using tagged Sumo proteins and histone H1. The five septins colocalized at the hyphal tip in the shape of a dome with a hole (DwH). CcSpa2-EGFP signals were observed in the hole, while CcCla4 signals were observed as the fluctuating dome at the hyphal tip. Before septation, CcCla4-EGFP was also occasionally recruited transiently around the future septum site. Fluorescent protein-tagged septins and F-actin together formed a contractile ring at the septum site. These distinct specialized growth machineries at different sites of dikaryotic vegetative hyphae provide a foundation to explore the differentiation program of various types of cells required for fruiting body formation.
Subject terms: Fungal biology, Cellular imaging
Introduction
Coprinopsis cinerea is a model mushroom and grows vegetatively as typical dikaryotic hyphae, which usually produce fruiting bodies consisting of an obvious cap and stipe1–3. Fruiting body formation begins with the aggregation of aerial vegetative hyphae, producing hyphal knots of approximately 0.2 mm or less in diameter. In the hyphal knots, cells divide rapidly and differentiate into a compact core composed of highly branched short cells and a layer of veil cells covering the core4. Following differentiation of the primordial shaft, the rudimentary pileus (cap) differentiates at the upper region of the primordial shaft, forming a tiny fruiting body primordium1. The fruiting body is composed of cells that exhibit different growth modes: hyphae growing at the hyphal tip, cells that rapidly divide without directional polarized growth, and cells displaying diffuse extension growth, which is characterized by expansion of the overall cell surface. Stipe and veil cells exhibit typical diffuse extension growth and provide an opportunity to study fungal cell elongation5,6.
While there is limited understanding of the molecular basis of mushroom body assembly in any system, the septin family of proteins have been linked to the process. A defect in the promoter region of a septin gene of C. cinerea, Cccdc3, resulted in a failure of stipe cell elongation at the maturation stage of fruiting7. Septins were originally identified as cdc (cell division cycle) mutants in Saccharomyces cerevisiae8. They are GTP-binding proteins conserved from fungi to mammals and some algae species9, and form heteromeric complexes that assemble into filaments10 and various higher-order structures in cells11,12. Since septins have a polybasic domain involved in membrane binding13, various higher-order structures of septins are frequently associated with the cell cortex. Subcellular localization of septins has been observed and investigated in yeasts, suggesting that septins play roles in recruitment of cytokinetic proteins, morphogenesis checkpoint, and membrane curvature sensing11,14,15. The subcellular localizations of septins have also been observed in a variety of filamentous fungi16–20.
Consistent with CcCdc3 being essential for stipe cell elongation, EGFP-tagged CcCdc3 septin assembles into abundant thin filaments at the cortex of stipe cells7. Notably, the EGFP-CcCdc3 signals were observed in the stipe cells and the vegetative hyphae, suggesting that CcCdc3 also functions in vegetative hyphal growth. The C. cinerea genome contains at least 5 septin genes encoding CcCdc3, CcCdc10, CcCdc11a, CcCdc11b and CcCdc1221. CcCdc11a and CcCdc11b are included in a clade of Shs1/Sep7 septin found in S. cerevisiae7. Expression of the septin genes except for Cccdc11a are upregulated during stipe elongation7,22.
A variety of proteins have been implicated in septin organization of S. cerevisiae23,24. The Cla4 kinase has been shown to have synthetic lethal interactions with various genes, which encode a septin and proteins involved in cell polarity and cell wall synthesis25,26, and has been demonstrated to interact directly with and phosphorylate certain septins24. The Cla4 kinase activity has been reported to be regulated by the Rho family GTPase Cdc4227, which has also been implicated in the recruitment of the yeast septins28 and regulation of the polarisome including Spa223,29. In various filamentous fungi, Cla4 functions have also been implicated in cell morphogenesis30–34. Septins in S. cerevisiae have been reported to be modified not only by phosphorylation but also by sumoylation35–37.
Growth of the dikaryotic vegetative hyphae in model mushrooms includes various cellular events and provides many opportunities to analyze fungal cell morphogenesis38–40. Tip growth is observed in the hyphal tip and the tip of the clamp cell, which is formed as a protrusion from the preexisting hyphal cell wall under which the two conjugate nuclei overlap and determine to divide38. In C. cinerea, the leading nucleus of the conjugate nuclei enters the clamp cell and divides. The clamp cell fuses to the subapical cell to release the nucleus trapped in the clamp cell into the main hypha. Before the clamp cell fuses to the subapical cell, the subapical cell responds to form a subapical peg41.
Here, we report the dynamics of fluorescent protein-tagged septins, CcCla4, CcSpa2, two CcSumo proteins and Lifeact peptide, as well as histone H1 in the dikaryotic vegetative hyphae of C. cinerea. Live cell imaging allows us to observe the transient assembly of tagged proteins to specific sites at a specific time in cell cycle progression. Two color tagging of proteins in interest provides insights into relationships between septins and the related proteins in hyphal tip growth, clamp cell formation, septum formation, and subapical peg formation. These studies delineate an atlas of key molecular assemblies for a major model mushroom, providing a foundation for molecular understanding of the developmental biology of mushrooms.
Results
Dynamics of septins tagged with fluorescent proteins at the hyphal tip
The C. cinerea septin CcCdc3 tagged with EGFP localized at the cortex of stipe cells and in dikaryotic vegetative hyphae7. The C. cinerea has at least five septins, CcCdc3, CcCdc10, CcCdc11a, CcCdc11b and CcCdc12. Septins often form a complex with each other and assemble into non-polar filaments10. However, there are also examples of septins acting independently of a complex such as with AspE or SEPT942,43. To examine the degree to which septins colocalize at certain time points or places in C. cinerea, two different septins were tagged with different color fluorescent proteins, EGFP or mCherry, and observed in the growing dikaryotic vegetative hyphae with a fluorescence microscope to analyze through time.
The five septins tagged with fluorescent proteins accumulated as two spots near the hyphal tip. They did not occupy the very tip of the growing hypha, suggesting that septins form a ring at the hyphal tip region or are excluded in some way from the very apex (Fig. 1; Supplementary Movies 1–4). The fluorescent signals from the tags fused to CcCdc3, CcCdc10, CcCdc11a and CcCdc12 were observed to expand from the septin ring towards the subapical region on the plasma membrane, resulting in a septin dome with a hole (DwH) at the growing hyphal tip region.
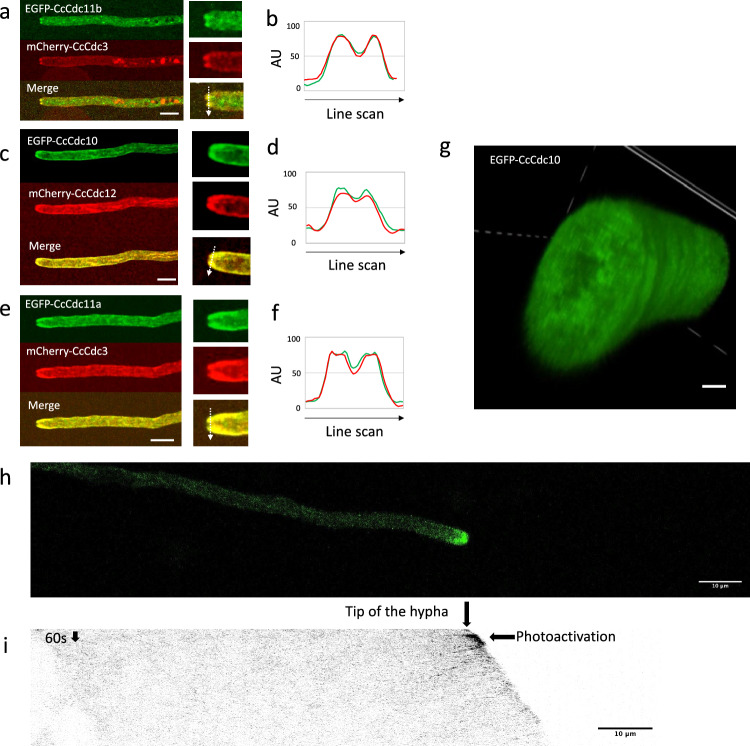
Figure 1.
Localization of septins in the tip region of the dikaryotic vegetative hypha. (a) EGFP-CcCdc11b and mCherry-CcCdc3 localizations at the growing hyphal tip. The right panels indicate the enlarged tip. See Supplementary Movie 1. (b) Line scan analysis along an arrow in the enlarged merged image in panel (a). (c) EGFP-CcCdc10 and mCherry-CcCdc12 localizations at the growing hyphal tip. The right panels indicate the enlarged tip. See Supplementary Movie 2. (d) Line scan analysis along an arrow in the enlarged merged image in panel (c). (e) EGFP-CcCdc11a and mCherry-CcCdc3 localizations at the growing hyphal tip. The right panels indicate the enlarged tip. See Supplementary Movie 3. (f) Line scan analysis along an arrow in the merged image in panel (e). (g) 3D image of the growing apical cell expressing EGFP-CcCdc10, showing a septin dome with a hole (DwH) at the hyphal tip. Scale bar: 1 µm. See Supplementary Movie 4 (Z stack slice step: 0.2 µm). (h) PA-GFP-CcCdc3 showing a backward movement of CcCdc3 from the hyphal tip after photoactivation. Scale bar: 10 µm. See Supplementary Movie 5. (i) Kymograph of panel h along a line placed at the center of the hypha. Scale bar: 10 µm.
In contrast, EGFP-CcCdc11b signals appeared to display somewhat different behaviors than other tagged septins. EGFP-CcCdc11b colocalized with other tagged septins as the ring at the hyphal tip, at the basal part of the protruding clamp cell and branch and at the septum. However, the cortical localization of EGFP-CcCdc11b was reduced compared with other cortical septins (Fig. 1a,c,e). In Supplementary Movies 1–3, the nuclei were observed as relatively large dark areas and EGFP-CcCdc11b signals were observed as puncta in the front of the nuclei. The structures with EGFP-CcCdc11b signals dynamically changed in shape to tubular particles. These tubular particles were observed more in the posterior region of the two nuclei (Supplementary Movie 1).
To confirm the dynamic movement of cortical septins, we replaced EGFP with PA-GFP and observed it after photo-activation (Fig. 1h; Supplementary Movie 5). PA-GFP-CcCdc3 signals were observed on the membrane at the hyphal tip region after the photo-activation of the hyphal tip, and the intensity of the signals diminished over time. While the diminished signals would be due, at least in part, to photobleaching or exchange, the PA-GFP signal was observed as a particle moving backward after 2–3 min from the photo-activation (Supplementary Movie 5). Kymograph shows that the hypha can grow continuously before and after the photo-activation and the PA-GFP-CcCdc3 signal moves backward after 2–3 min from the photo-activation from the tip after the photo-activation (Fig. 1i).
Dynamics of the septin-related cell polarity proteins at the hyphal tip
Septin behaviors in S. cerevisiae have been found to be regulated by a variety of proteins23. The Cla4 kinase has been implicated in regulating septin organization in S. cerevisiae26,44,45. To examine the relationship between CcCla4 and Cc-septins (C. cinerea septins), CcCla4 was tagged with EGFP. CcCla4-EGFP signals were observed as a fluctuating dome at the hyphal tip. The posterior edge of the CcCla4 dome appeared to overlap with the septin DwH at the subapical region (Fig. 2a,b; Supplementary Movies 6 and 7). The overlapping of their localizations indicates that CcCla4 may function with and/or regulate Cc-septins at the growing hyphal tip.
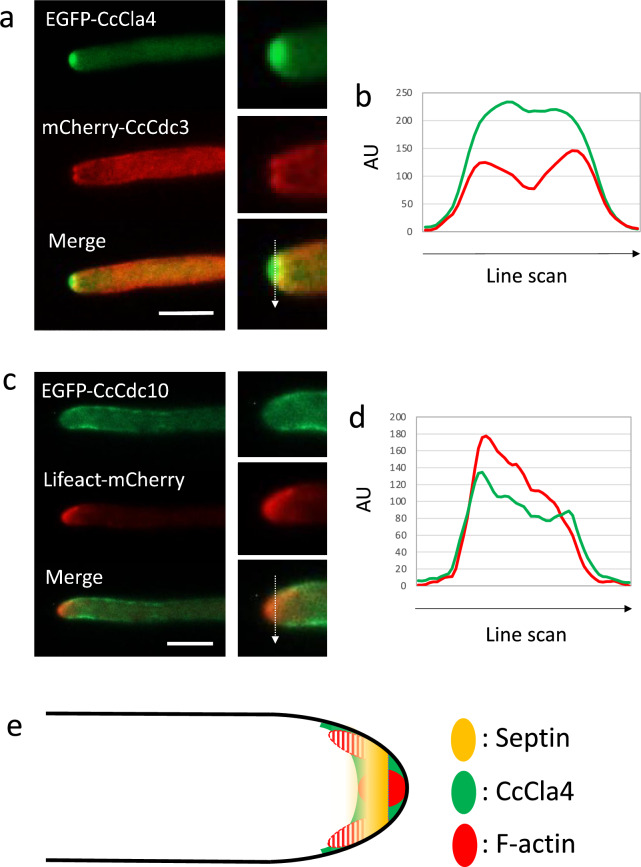
Figure 2.
Localization of septin-related cell polarity proteins in the tip region of the dikaryotic vegetative hypha. (a) CcCla4-EGFP and mCherry-CcCdc3 localizations at the growing hyphal tip. The right panels indicate the enlarged tip. See Supplementary Movie 6. Localization of CcCla4-EGFP at the growing hyphal tip was also observed in Supplementary Movie 7. (b) Line scan analysis along an arrow in the enlarged merged image in panel (a). (c) EGFP-CcCdc10 and Lifeact-mCherry localizations at the growing hyphal tip. See Supplementary Movie 8. (d) Line scan analysis along an arrow in the enlarged merged image in panel (c). (e) A schematic diagram showing the septin DwH and localizations of CcCla4-EGFP and Lifeact-mCherry at the hyphal tip region. The fluctuated actin patches in the subapical region are shown by red-striped areas surrounded by a dashed line. AU arbitrary units. Scale bar: 10 µm.
To observe dynamics of the F-actin in a growing dikaryotic vegetative hypha of C. cinerea, the mCherry-tagged Lifeact peptide was expressed by the actin promoter. At the growing hypha in Aspergillus nidulans, a subapical endocytic ring has been observed, at which actin/actin binding protein A (ABPA) was localized as endocytic patches46. The actin distributions have been observed and investigated in many fungal species, including C. cinerea using antibodies40, S. commune, N. crassa and A. nidulans using Lifeact peptide38,47–49. At the hyphal tip in C. cinerea, Lifeact-mCherry signals were observed as patches, which appeared to be classified into two types: one that occupied the apex of the growing hypha and the other that localized and fluctuated at the subapical region (Fig. 2c; Supplementary Movie 8). The fluorescent protein-tagged septins were observed as a ring at the subapical region: within 1 μm behind the apex of the hypha (Fig. 1). At the subapical region, Lifeact-mCherry signals were observed at the inner side of EGFP-CcCdc10 signals on the plasma membrane (Fig. 2d). The colocalizations of Lifeact-mCherry and EGFP-CcCdc10 signals suggested that F-actin and septins are coordinately assembled in the subapical region and could function there in maintenance of the apical compartment (summarized in Fig. 2e).
The polarisome has been observed at the apex of the growing hypha in many filamentous fungi and has been considered to nucleate F-actin at the hyphal tip: Ashbya gossypii50, Candida albicans51, Aspergillus nidulans52, Ustilago maydis53, and Neurospora crassa54. To observe the polarisome in C. cinerea, CcSpa2 was tagged with EGFP. CcSpa2-EGFP signals were observed constitutively as a spherical body at the growing hyphal tip. The rim of the polarisome appeared to overlap with the septin DwH (Fig. 3a; Supplementary Movie 9).
Figure 3.
Localization of the polarisome and conjugate nuclear division. (a) The growing hypha expressing CcSpa2-EGFP and mCherry-CcSumo1. mCherry-CcSumo1 was accumulated in the nucleus. At time point 00:00, CcSpa2-EGFP signals were observed at the growing hyphal tip. At time point 44:40, the posterior nucleus approached to the leading nucleus. From time points 47:00 to 57:40, CcSpa2-EGFP signals were observed at the tip of the clamp cell growing backward. At time point 57:40, the leading nucleus began to enter the clamp cell. At time point 59:40, CcSpa2-EGFP signals disappeared from the tip of the clamp cell. At time point 1:02:00, mCherry-CcSumo1 signals disappeared from the nuclei during mitosis. See Supplementary Movie 9. (b,c) The nuclear dynamics in mitosis was observed by tagging histone H1 with EGFP. The anterior and posterior nuclei before mitosis are indicated by blue and red arrows, respectively. In panel (b), a kymograph for 40 min between the top and bottom photos is shown. A white arrowhead in the kymograph indicates the point of the release of the nucleus from the clamp cell to the subapical cell. In panel c, the nuclear behavior is shown every 40 s. The anterior nucleus entered the clamp cell, in which the nucleus divided. The posterior nucleus divided into two daughter nuclei in the main hyphal cell. The anterior daughter nucleus occurring in the main hypha passed through the nucleus returned from the clamp cell to the hyphal cell and ran toward the hyphal tip. Scale bar: 10 µm. See Supplementary Movies 10, 11 and schematic diagrams of the nuclear behavior in Fig. 6.
Clamp formation and conjugate nuclear division
Dikaryotic hyphae of some species in basidiomycetes have clamp connections between cells. A clamp cell is formed to give a space where one nucleus divides during conjugate nuclear division. In Supplementary Movies 1–3, in which the septins were tagged with EGFP or mCherry, the shape of the nucleus was observed as dark area, suggesting that septins did not enter the nucleus. The dark area disappeared during mitosis, suggesting that the nuclear envelope opened.
In S. cerevisiae, sumoylation of some septins have been reported36. To examine sumoylation of Cc-septins, two sumo proteins, CcSumo1 and CcSumo2, were tagged with mCherry. The mCherry signals from tagged Sumo proteins were not observed in the region where Cc-septins were accumulated, suggesting that most Cc-septins were not sumoylated or that tag interfered with their linkage to the septins. The mCherry signals from tagged CcSumo1 and CcSumo2 were observed only in the interphase nucleus (Fig. 3a; Supplementary Movies 9 and 10) and disappeared during mitosis, consistent with the reports of localizations of sumoylated proteins in A. nidulans55. When the clamp protruded from the preexisting cell wall, CcSpa2-EGFP signals emerged at the tip of the protrusion (Fig. 3a, time point: 47:00). CcSpa2-EGFP signals stayed at the tip of the growing clamp cell. The leading nucleus visualized by mCherry-CcSumo1 started to enter the growing clamp cell whose tip had CcSpa2-EGFP signals (Fig. 3a, time point: 57:40). During the nuclear entry into the clamp cell, mCherry-CcSumo1 signals disappeared (Fig. 3a, time point: 1:02:00).
To observe the nuclear behavior during mitosis, histone H1 was tagged with EGFP. The leading nucleus entered the clamp cell, in which the nucleus divided (Fig. 3c; Supplementary Movies 10 and 11). After mitosis in the clamp cell, the new anterior nucleus returned to the apical cell and the new posterior nucleus remained trapped in the clamp cell. The nucleus returning from the clamp cell to the apical cell was overtaken by the new anterior sister nucleus in the main hypha. This nuclear behavior is consistent with a previous report in C. cinerea56. About thirty minutes after trapping the nucleus in the clamp cell, the clamp cell fused to the subapical cell to release the trapped nucleus into the subapical cell, leading to the two nuclei in the subapical cell (Fig. 3b, a white arrowhead).
The clamp protrusion started between the two conjugate nuclei or at the site near the anterior edge of the posterior nucleus. The fluorescent signals of the tagged septins were observed around the protrusion of the clamp cell (Fig. 4a–c,h–j; Supplementary Movies 2, 3 and 12). As the clamp cell protruded, the fluorescent signals of the tagged septins were accumulated at the basal part of the protrusion and the tip of the clamp cell (Fig. 4d). During clamp cell elongation, the fluorescent signals of the tagged septins stayed on the cell cortex (Fig. 4d). However, as the septin-actin ring was formed for septum formation (see the next section), these cortical septins disappeared (Fig. 4e–g). CcSpa2-EGFP signals emerged at the tip of the clamp cell (Fig. 3a, time point 47:00–57:40). At the early stage of the clamp protrusion, CcCla4-EGFP signals emerged at the tip of the protrusion (Fig. 5a). The CcCla4-EGFP signals stayed at the tip of the clamp cell growing backward (Fig. 5b,c). The clamp cell protrusion can be considered as the emergence of a new hyphal tip linked to the conjugate nuclear division cycle.
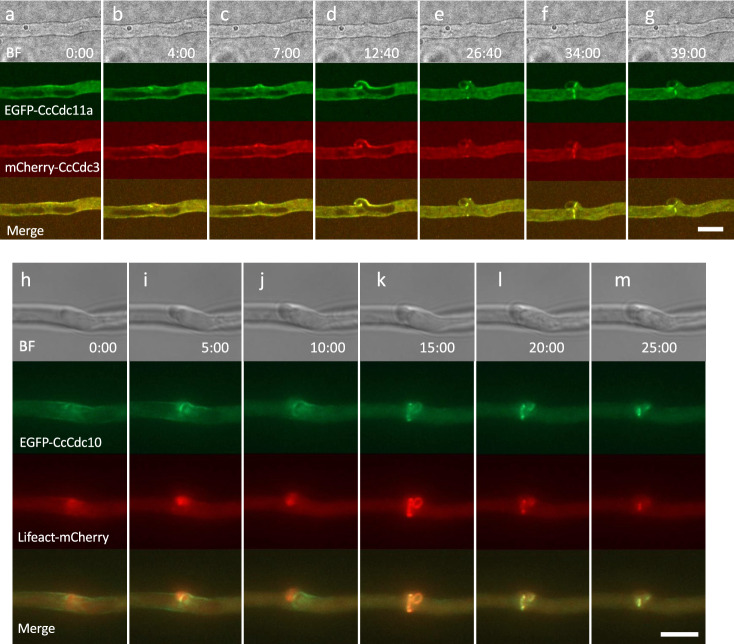
Figure 4.
EGFP-CcCdc11a, mCherry-CcCdc3, EGFP-CcCdc10 and Lifeact-mCherry dynamics during clamp formation and septation. (a–g) Dynamics of EGFP-CcCdc11a and mCherry-CcCdc3 during clamp formation and septation. (a) EGFP-CcCdc11a and mCherry-CcCdc3 signals were observed at the cell cortex around the approached nuclei. (b) Both septins began to assemble at the one side of the hyphal tube between the two conjugate nuclei. (b,c) The cell wall began to protrude to form the clamp cell. Both septins are assembled at the basal part of the protrusion. (d) Both septins are assembled at the tip of the clamp cell. (e) mCherry-CcCdc3 disappeared from the basal part and the surface of the clamp cell. And both septins emerged as spots, which indicate a cross section of a ring, both at the main hypha and the clamp cell to separate the nucleus after mitosis. (e–g) The diameters of septin rings became narrower. (g) The EGFP-CcCdc11a and mCherry-CcCdc3 signals were observed at the septal pore and the subapical peg. See Supplementary Movie 3. (h–m) The clamp region of the hypha expressing EGFP-CcCdc10 and Lifeact-mCherry. (h–j) EGFP-CcCdc10 signals became observed around the protrusion of the clamp cell and Lifeact-mCherry signals became localized at the tip of the protrusion. (k) After clamp cell formation, EGFP-CcCdc10 and Lifeact-mCherry signals were observed as two rings: one formed in the main hypha and the other at the clamp cell’s anterior side. (k–m) The diameters of septin rings became narrower. See Supplementary Movie 12 and schematic diagrams of clamp cell formation in Fig. 6. Scale bar: 10 μm.
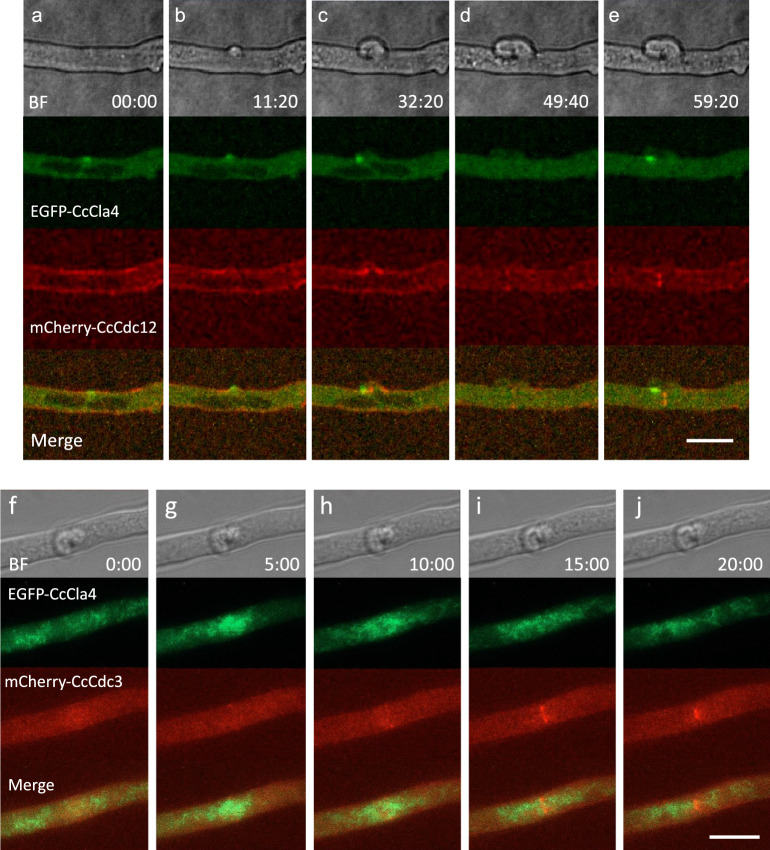
Figure 5.
Dynamics of CcCla4-EGFP kinase, mCherry-tagged CcCdc12 and CcCdc3 septins from clamp formation to septation. (a–e) Dynamics of CcCla4-EGFP and mCherry-CcCdc12 from clamp formation to septation. (a) CcCla4-EGFP signals emerged as a spot at the plasma membrane between two conjugate nuclei. (b) The site where CcCla4-EGFP assembled began to protrude. (c) CcCla4-EGFP signals continued to exist at the tip of the growing clamp cell. mCherry-CcCdc12 signals were observed at the basal part of the clamp cell protrusion. (d) CcCla4-EGFP signals disappeared and mCherry-CcCdc12 signals were observed as a ring from which septum formation occurs. (e) After the tip of the clamp cell attached to the subapical cell, CcCla4-EGFP signals emerged at the subapical peg site, where mCherry-CcCdc12 signals also assembled. The diameter of the ring visualized by mCherry-CcCdc12 became narrower. See Supplementary Movie 13. (f–j) Dynamics of CcCla4-EGFP and mCherry-CcCdc3 from clamp formation to septation. After clamp cell formation, CcCla4-EGFP assembled over the broad region around the clamp cell. The assembly of CcCla4-EGFP before septum formation was also observed in Supplementary Movie 14. (i–j) As CcCla4-EGFP signals dispersed, mCherry-CcCdc3 signals became assembled as a ring to form the septum. (j) The diameter of the septin ring visualized by mCherry-CcCdc3 became narrower. Scale bar: 10 μm. See schematic diagrams of clamp cell formation in Fig. 6.
Septum formation
When the clamp cell protrusion and the conjugate nuclear division were completed, CcCla4-EGFP signals were observed occasionally around the future septum site (Fig. 5g; Supplementary Movie 14). Immediately after the transient assembly of CcCla4-EGFP, fluorescent protein-tagged septins and Lifeact-mCherry began to assemble as a ring between the sister nuclei, suggesting the septin-actin ring formation (Fig. 4h–k). Then, the diameter of the rings became narrower, indicating contraction of the rings (Figs. 4e–g,k–m, 5d,e,i,j). The contraction rates were 0.104 μm/min on average; max: 0.125 μm/min at 30 °C (Supplementary Movie 12). Thus, septum formation was accomplished by the contractile ring containing the septin and actin.
Subapical peg formation
When the tip of the clamp cell is attached to the subapical cell, CcCla4-EGFP signals emerge at the site where the clamp cell fuses to the subapical hypha, called “subapical peg”38,41. The attachment of the clamp tip to the subapical cell appeared to induce the assembly of CcCla4 at the side of the subapical hypha. The CcCla4 assembly in the subapical hypha occurred after the beginning of the contraction of the septin-actin ring at the septum site (Fig. 5d,e; Supplementary Movie 13). When the CcCla4-EGFP signals emerged at the subapical peg, mCherry-CcCdc12 was also assembled at the peg region (Fig. 5e; Supplementary Movie 13). EGFP-CcCdc11a and mCherry-CcCdc3 signals also assembled at the subapical peg region (Fig. 4g). These assemblies suggested that polarized growth occurred to form the subapical peg in the subapical cell.
Branching
After clamp formation, branching occurred behind the clamp connection (Supplementary Movies 1–3, 7, 9, and 13). At the curved basal part of the branch hypha, fluorescent protein-tagged septins accumulated (Supplementary Movies 1–3), consistent with septin assembly on micron-scale curved membranes. CcCla4-EGFP was assembled at the tip of the branch hypha. Thus, the protrusion of the branch hypha appeared to resemble that of the clamp cell, although the direction of the protrusion was opposite, suggesting a similar molecular mechanism for such protrusion.
Discussion
Tagging proteins of interest with dual color fluorescent proteins and live cell imaging enabled the examination of the dynamics and relationships between septins and other cell polarity proteins in the growing dikaryotic vegetative hyphae of C. cinerea. Different events in the growing dikaryotic vegetative hypha of this model mushroom allow us to dissect and discuss molecular mechanisms by which the fungal cell shape changes in their life cycle. The localizations of Cc-septins, CcCla4, CcSpa2, and F-actin are summarized in Fig. 6.
Figure 6.
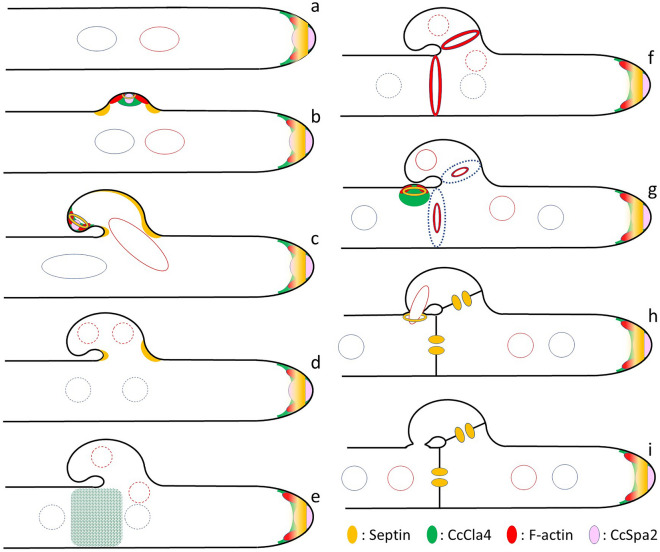
Schematic diagrams showing subcellular localization of septins, CcCla4, F-actin, and CcSpa2 in the growing dikaryotic vegetative hypha of C. cinerea. (a) The hyphal tip of C. cinerea has a tip growth apparatus, which contains a CcCla4 crescent dome, a septin DwH, a polarisome, and fluctuated actin patches. In this diagram, the hyphal tip is depicted not to elongate, but actually elongates. The polarisome contains F-actin, which is not depicted. The two compatible nuclei are depicted by red and blue circles and shown by dashed circles when the nuclear envelope appears to open. (b) The protrusion occurs to form the clamp cell. At the tip of the protrusion, another tip growth apparatus emerges. (c) The clamp cell grows backward, and the leading nucleus enters the growing clamp cell. (d) The conjugate nuclear division occurs. (e) After nuclear division and before septation, CcCla4 occasionally assembles around the future septum site transiently. (f) The actin and septin rings emerge at the future septum sites. (g) The subapical peg is formed in the subapical cell. (h) The clamp cell fuses to the peg formed in the subapical cell to release the nucleus trapped in the clamp cell. (i) In the new apical cell, the order of the two nuclei derived from (a) is exchanged.
Assemblies of fluorescent protein-tagged septins at the growing hyphal tip revealed that the septins form a dome with a hole (DwH) at the hyphal tip (Supplementary Movies 1, 2, and 3). EGFP-CcSpa2 visualized the polarisome at the growing hyphal tip (Fig. 3a, Supplementary Movie 9). CcCla4-EGFP signals were observed as a fluctuating dome at the hyphal tip and appeared to be overlapped with both the polarisome and the septin DwH. F-actin visualized by Lifeact-mCherry was also overlapped with the polarisome and the septin DwH. It has been reported that in S. cerevisiae, Cla4 kinase phosphorylates certain septins to form septin collar24 and a cla4 mutant is synthetically lethal with a variety of genes, including the genes encoding the polarisome components: Spa2, Pea2, Bni1 and Bud625. The spa2 mutant is also synthetically lethal with a septin mutant, cdc10-1057. These findings in S. cerevisiae suggest that CcCla4 also exerts influences on both septins and the polarisome at the hyphal tip of the dikaryotic vegetative hypha. In Bipolaris maydis, deletion of cla4 showed hyper-blanching and swelling hyphae31. In A. gossypii, AgCla4-EGFP localized at the hyphal tip30. These findings also suggest a role of Cla4 at the hyphal tip. In S. cerevisiae, Hof1 links septins to actin58. In future study, it will be interesting to examine whether Hof1 homologs (CC1G_06830, CC1G_04571) localize at the hyphal tip region to link septins to actin.
At the hole of the septin dome at the hyphal tip, there must be the Spitzenkörper, originally described in two Coprinus species59 and is thought to function as a vesicle supply center to promote hyphal tip growth60–65. The Spitzenring of GS-1, which is involved in the synthesis of β-1,3-glucan, has also been reported at the hyphal tip in N. crassa65. It has been reported in S. cerevisiae that when the bud emerges, interplay of Cdc42, septins, and exocysts occurs to confine the Cdc42 activity and exocytosis at the growing bud site66. The situation that the septin ring encloses the exocytic region resembles the septin localization observed at the hyphal tip of C. cinerea. In Ustilago maydis, septin function has been implicated in exocytosis using septin-deletion mutants19. The Spitzenkörper also colocalizes with the polarisome and has been discussed in relation to polarisome function67. The polarisome has been recognized in S. cerevisiae and observed in a variety of filamentous fungi50,53,67,68. The localization of C. cinerea septins suggested that the septins function at the periphery of the Spitzenkörper and polarisome.
CcCla4-EGFP signals were also observed at the hyphal tip. The region where CcCla4-EGFP signals emerged appeared to be broader than the diameter of the hole of the septin DwH and the polarisome. In addition, the dome of the CcCla4-EGFP signals fluctuated much more than the septin DwH and the polarisome. Similar fluctuation was also observed in actin patches localized at the subapical region. At the subapical region of the hypha, the subapical endocytic ring has been reported in A. nidulans46, and endocytosis at the hyphal tip has been considered to be an essential process for tip growth69. Fluctuated actin patches at the subapical region might be relevant to the subapical endocytic ring. The relationships between endosomes and septin have been investigated in U. maydis70. In the hyphal tip of C. cinerea, septins appeared to exist at the place where both endocytosis and exocytosis should be balanced. Thus, in the subapical region behind the hyphal tip, the relationships between relatively stable Cc-septin DwH and the polarisome, fluctuating CcCla4 and actin patches, and endosomes remain to be elucidated in terms of endocytosis and exocytosis.
As the first indication of clamp cell formation, CcCla4 appeared to assemble at the plasma membrane between two compatible nuclei (Figs. 5a, 6b). This CcCla4 assembly might result from the change in lipid composition of the plasma membrane, which received a signal from the nucleus that decided to start mitosis because it has been reported that the PH domain of Cla4 in S. cerevisiae can interact with specific lipids27. Then, cortical septins might be accumulated around the CcCla4 assembly site at the clamp cell protrusion. CcSpa2-EGFP and F-actin assembled at the tip of the clamp cell protrusion, resulting in the outgrowth of the clamp cell from the preexisting cell wall. The mechanism by which the clamp cell grows backward to the subapical cell would depend on the position of the anterior nucleus, from which microtubules might elongate and enter the protruding clamp cell39. Since CcSpa2-EGFP signals emerged at the tip of the growing clamp cell, the Spitzenkörper and polarisome might form at the tip of the growing clamp cell. While the polarisome stayed at the tip of the clamp cell, the tip might be able to grow continuously (Fig. 6c).
During clamp cell formation, localizations of the Cc-septins dramatically change. The emergence of the protrusion of the clamp cell brings about the plasma membranes with both positive and negative curvatures. The septin DwH in the tip growth apparatus shows the positive curvature of the plasma membrane. The negative curvature emerges at the base of the protrusion, and the septins accumulate at the base of the growing clamp cell. Immediately after nuclear division, the Cc-septins staying on the membrane with positive and negative curvatures are sequestered from the plasma membrane and reassembled as the rings for septation. Septin filaments can sense micrometer-scale membrane curvature71,72, and consistent with this, septins are enriched at the curved bases of clamp cells. The cortical septin filaments on the plasma membrane of growing cell regions might enable septin filaments to monitor the membrane curvature all over the cell surface continuously. Further study will be needed to understand dynamic changes in localization of the Cc-septins during clamp cell formation.
When mitosis started, both sumo proteins, CcSumo1 and CcSumo2, disappeared from the nucleus (Fig. 3a; Supplementary Movies 9 and 10), as described in A. nidulans55. Although some of septins in S. cerevisiae have been reported to be sumoylated36,37, the C. cinerea two sumo proteins, CcSumo1 and CcSumo2, did not show obvious colocalization with Cc-septins. In Candida albicans, septins have been reported not to be sumoylated73. The proteins that are abundant in cytoplasm and do not enter the nucleus, such as septins, allow us to recognize the nuclear shape as a dark area in cytoplasm (Supplementary Movies 1–3). The dark area disappeared during mitosis, suggesting that the nuclear envelope opened. It is possible that signals to prepare the following events are released from the nucleus by opening the nuclear envelope. Various types of nuclear envelope behaviors in mitosis have been observed in fungal cells: from open mitosis to closed mitosis38,74,75. Further study will be required to determine whether “open mitosis” actually occurs in C. cinerea.
Following the nuclear division, the actin and septin rings emerged at the septum site. The diameters of these rings together became concurrently narrow (Supplementary Movie 12), indicating contraction of these rings. In Ashbya gossypii, when the septum forms, Sep7, Hof1, actin, Myo1, Bud3, and Cyk1 colocalized at the sites of septation76. In S. cerevisiae, Hof1 interacts with Chs4, which is an activator for the catalytic subunit of chitin synthase-III77,78. In experiments using S. cerevisiae actin and septins in vitro, Hof1 can physically link actin and septin filaments58. The colocalization of septins and actin at the inner edge of the growing septum in our observations, combined with the results in other fungi, suggest collaborative processes of septins and actin in cell wall synthesis during septation. As animal septins are also assembled into the contractile actomyosin ring during cytokinesis79, septin functions in cytokinesis may, at least in part, be conserved among fungal and animal cells.
CcCla4 did not assemble at the septum, despite the septin ring forming and contracting at the septum site (Fig. 5; Supplementary Movies 13 and 14), consistent with the observations in Ashbya gossypii30. However, CcCla4-EGFP signals were occasionally observed around the future septum region after clamp cell formation and before septum formation (Fig. 5g; Supplementary Movie 14). This CcCla4 assembly might be interpreted as preparation for septum formation, because cla4Δ A. gossypii mutants lose the ability for septum assembly30. This interpretation appears to be consistent with one possible explanation in A. gossypii that AgCla4 acts early in establishing the future site of septation30. Alternatively, it might be relevant to mitosis, because Cla4 has been identified as a protein encoded by the gene responsible for synthetic lethality with cyclin deletion in S. cerevisiae80,81. CcCla4 contains a PH domain, which has been reported to interact with specific lipids27. The broad assembly of CcCla4 around the future septum site might be related to the emergence of such lipids in the plasma membrane.
In the subapical cell, the subapical peg protrudes from the site near the septum41. After clamp cell formation, CcCla4 and Cc-septins assembled at the subapical peg region (Figs. 5e, 6g; Supplementary Movie 13). These assemblies would mean polarized growth to form the subapical peg, as described in S. commune38. Fusion of the clamp cell to the subapical cell in the mating reaction is regulated by B-mating factors, which encode pheromones and pheromone receptors82,83. Therefore, the response to form the subapical peg in the subapical cell likely occurs downstream of the pathway controlled by B-mating factors41,84,85.
C. cinerea has two Cdc11 orthologs, CcCdc11a and 11b. Expression of Cccdc11b is up-regulated in fruiting, compared to that of Cccdc11a7. Live cell imaging of CcCdc11b showed that this septin can colocalize with other septins at the hyphal tip, the septum and the basal parts of the protrusion of clamp cells and branching, suggesting that CcCdc11b can form septin complexes with other septins. However, in cytoplasm, CcCdc11b showed a different localization from other septins. The localization suggests that CcCdc11b can associate with tubular structures, such as mitochondria or the endoplasmic reticulum, in the posterior region of the apical cell. In some organisms, septins have been observed at mitochondria86,87, which notably present positive membrane curvature. It is possible that CcCdc11b can associate with mitochondria in certain situations. Recent work in A. gossypii shows that different Cdc11 variants lead to filaments with different average lengths88, potentially consistent with the different dome sizes of CcCdc11a and CcCdc11b. Since Cdc11 occupies the terminal subunit of the septin heterooligomer, it also may have the capacity to control filament flexibility in a manner that may impact the septin assembly on intracellular organelle membranes. Further studies will be required to determine the relationship between organelles and tubular structures associated with septins in the cytoplasm.
In this study, dynamics of C. cinerea homologs of players identified in cellular morphogenesis of S. cerevisiae were observed in the dikaryotic vegetative hyphae. This fungus can produce fruiting bodies consisting of the obvious cap and stipe, like edible mushrooms, such as Lentinus edodes and Tricholoma matsutake. Some kinds of cells in fruiting bodies, such as veil cells and stipe cells, show diffuse extension growth, characterized by expansion all over the cell surface6,89,90, in contrast to hyphal tip growth in the vegetative hyphae. However, some components of the molecular machinery underlying vegetative hyphal growth should be utilized in fruiting body morphogenesis, as demonstrated in stipe cell elongation7. How modification of the vegetative hyphal growth machinery occurs to produce fruiting bodies is still mysterious, and further studies would be needed to unravel the relationship between hyphal tip growth and diffuse extension growth in fruiting body morphogenesis of this mushroom. Since edible mushrooms should share in part the molecular mechanisms for fruiting in C. cinerea, identification and regulation of key steps in the transition from vegetative hyphal growth to diffuse extension growth would improve the edible mushroom cultivation conditions.
Methods
Strains, culture conditions and cloning
The strains used are listed in Supplementary Table S1. The mycelia were grown on MYG (1% malt extract, 0.4% yeast extract, 0.4% glucose, 1.5% agar) medium for usual culture, or in TM (2% glucose, 10 mM (NH4)2HPO4, 0.12 µg/ml thiamine, 0.05% MgSO4·7H2O, 10 mM potassium phosphate, pH7.0, in autoclaved tap water) liquid medium for observations of the growing vegetative hyphae.
Twelve proteins were tagged with EGFP, PA-GFP and/or mCherry, as described in Supplementary Table S2. Primers, plasmids, and cloning methods used to construct expression vectors are listed in Supplementary Table S3. For CcCdc3 tagging see our previous report7. Gene sawing PCR and FastCloning91 were mainly used for cloning. pLifeact-mCherry-7 was purchased from Addgene and driven by actin promoter and terminator.
Transformation
The protoplasts of strain #292 (A3B1 trp1-1, 1–6) or #8 (A2B2 trp1-1, 1–6) were transformed with a mixture of pCc1003 (trp1+) and the constructed expression vectors, as described previously92. The trp+ transformants were recovered on minimal medium to purify the transformed cells. The purified trp+ transformants were observed by a fluorescence microscope to check expression of tagged proteins and selected for mating to construct dikaryons. Transformation in C. cinerea results in the random insertion of the expression vectors into the genome, causing the situation where both native and tagged proteins are expressed in a cell.
Microscopy and image processing
A small agar (1 mm cube) with hyphae was inoculated in 100–200 µl of a liquid TM medium on a coverslip and incubated at 28–30 °C for 16–24 h. After removing the liquid medium, the inverted coverslip with the hyphae was placed on 4% agarose pad, in which a hole was made and filled with the removed medium from the coverslip to set the inoculum. After 1 h or more, the vegetative hyphae were observed by fluorescence microscopes. The fluorescence microscopes used and image capturing conditions are listed in Supplementary Table S4. Arduino was used for exchanging filters and opening shutters when the microscope used did not equip such functions, as described93. Images were processed with ImageJ and/or iMovie and exported as MOV files by PowerPoint. The MOV files were compressed by VideoSmaller (https://www.videosmaller.com/) to less than 10 Mb.
Supplementary Information
Acknowledgements
The authors would like to acknowledge the support of Akita Prefectural University Biotechnology Center for sequencing. We also thank Toshiaki Nose (Faculty of Systems Science and Technology, Akita Prefectural University) for his advice on microscopy using Arduino.
Author contributions
T.K., H.M., K.S.C., B.L.W., A.G. and N.T. conceived and designed the study. T.K., H.M., Y.K., M.M, M.S., C.H., K.T., S.S. T.S., H.N. and N.T. performed the experiments. T.K., H.M and N.T. analyzed and edited the data. T.K. and H.M. wrote the manuscript. N.T., K.S.C., B.L.W., A.G. and H.A. reviewed the manuscript.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author upon reasonable request. All data generated or analysed during this study are included in this published article [and its supplementary information files].
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-37115-y.
References
- 1.Muraguchi H, Kamada T. The ich1 gene of the mushroom Coprinus cinereus is essential for pileus formation in fruiting. Development. 1998;125:3133–3141. doi: 10.1242/dev.125.16.3133. [DOI] [PubMed] [Google Scholar]
- 2.Kües U. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol. Mol. Biol. Rev. 2000;64:316–353. doi: 10.1128/MMBR.64.2.316-353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pukkila PJ. Coprinopsis cinerea. Curr. Biol. 2011;21:R616–R617. doi: 10.1016/j.cub.2011.05.042. [DOI] [PubMed] [Google Scholar]
- 4.van der Valk P, Marchant R. Hyphal ultrastructure in fruit-body primordia of the basidiomycetes Schizophyllum commune and Coprinus cinereus. Protoplasma. 1978;95:57–72. doi: 10.1007/BF01279695. [DOI] [Google Scholar]
- 5.Gooday GW. In: Developmental Biology of Higher Fungi. Moore D, Casselton LA, Wood DA, Frankland JC, editors. Cambridge University Press; 1985. pp. 311–331. [Google Scholar]
- 6.Kamada T. In: The Mycota I. Esser K, Lemke PA, editors. Springer; 1994. pp. 367–379. [Google Scholar]
- 7.Shioya T, et al. The Coprinopsis cinerea septin Cc.Cdc3 is involved in stipe cell elongation. Fungal Genet. Biol. 2013;58–59:80–90. doi: 10.1016/j.fgb.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Hartwell LH. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp. Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- 9.Yamazaki T, et al. Localization and evolution of septins in algae. Plant J. 2013;74:605–614. doi: 10.1111/tpj.12147. [DOI] [PubMed] [Google Scholar]
- 10.Bertin A, et al. Saccharomyces cerevisiae septins: Supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc. Natl. Acad. Sci. U.S.A. 2008;105:8274–8279. doi: 10.1073/pnas.0803330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woods BL, Gladfelter AS. The state of the septin cytoskeleton from assembly to function. Curr. Opin. Cell Biol. 2021;68:105–112. doi: 10.1016/j.ceb.2020.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeMay BS, Meseroll RA, Occhipinti P, Gladfelter AS. Regulation of distinct septin rings in a single cell by Elm1p and Gin4p kinases. Mol. Biol. Cell. 2009;20:2311–2326. doi: 10.1091/mbc.e08-12-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casamayor A, Snyder M. Molecular dissection of a yeast septin: Distinct domains are required for septin interaction, localization, and function. Mol. Cell Biol. 2003;23:2762–2777. doi: 10.1128/MCB.23.8.2762-2777.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gladfelter AS, Kozubowski L, Zyla TR, Jew DJ. Interplay between septin organization, cell cycle and cell shape in yeast. J. Cell Sci. 2005;118:1617–1628. doi: 10.1242/jcs.02286. [DOI] [PubMed] [Google Scholar]
- 15.Woods BL, Cannon KS, Vogt EJD, Crutchley JM, Gladfelter AS. Interplay of septin amphipathic helices in sensing membrane-curvature and filament bundling. Mol. Biol. Cell. 2021 doi: 10.1091/mbc.E20-05-0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernández-Rodríguez Y, Hastings S, Momany M. The septin AspB in Aspergillus nidulans forms bars and filaments and plays roles in growth emergence and conidiation. Eukaryot. Cell. 2012;11:311–323. doi: 10.1128/EC.05164-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berepiki A, Read ND. Septins are important for cell polarity, septation and asexual spore formation in Neurospora crassa and show different patterns of localisation at germ tube tips. PLoS One. 2013;8:e63843. doi: 10.1371/journal.pone.0063843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warenda AJ, Konopka JB. Septin function in Candida albicans morphogenesis. Mol. Biol. Cell. 2002;13:2732–2746. doi: 10.1091/mbc.e02-01-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarez-Tabarés I, Pérez-Martín J. Septins from the phytopathogenic fungus Ustilago maydis are required for proper morphogenesis but dispensable for virulence. PLoS One. 2010;5:e12933. doi: 10.1371/journal.pone.0012933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozubowski L, Heitman J. Septins enforce morphogenetic events during sexual reproduction and contribute to virulence of Cryptococcus neoformans. Mol. Microbiol. 2010 doi: 10.1111/j.1365-2958.2009.06983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stajich JE, et al. Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus) Proc. Natl. Acad. Sci. U.S.A. 2010;107:11889–11894. doi: 10.1073/pnas.1003391107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muraguchi H, et al. Strand-specific RNA-Seq analyses of fruiting body development in Coprinopsis cinerea. PLoS One. 2015;10:e0141586. doi: 10.1371/journal.pone.0141586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park HO, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Versele M, Thorner J. Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4. J. Cell Biol. 2004;164:701–715. doi: 10.1083/jcb.200312070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goehring AS, et al. Synthetic lethal analysis implicates Ste20p, a p21-activated protein kinase, in polarisome activation. Mol. Biol. Cell. 2003;14:1501–1516. doi: 10.1091/mbc.e02-06-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt M, Varma A, Drgon T, Bowers B, Cabib E. Septins, under Cla4p regulation, and the chitin ring are required for neck integrity in budding yeast. Mol. Biol. Cell. 2003;14:2128–2141. doi: 10.1091/mbc.e02-08-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wild AC, Yu JW, Lemmon MA, Blumer KJ. The p21-activated protein kinase-related kinase Cla4 is a coincidence detector of signaling by Cdc42 and phosphatidylinositol 4-phosphate. J. Biol. Chem. 2004;279:17101–17110. doi: 10.1074/jbc.M314035200. [DOI] [PubMed] [Google Scholar]
- 28.Iwase M, et al. Role of a Cdc42p effector pathway in recruitment of the yeast septins to the presumptive bud site. Mol. Biol. Cell. 2006;17:1110–1125. doi: 10.1091/mbc.e05-08-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bidlingmaier S, Snyder M. Regulation of polarized growth initiation and termination cycles by the polarisome and Cdc42 regulators. J. Cell Biol. 2004;164:207–218. doi: 10.1083/jcb.200307065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ayad-Durieux Y, Knechtle P, Goff S, Dietrich F, Philippsen P. A PAK-like protein kinase is required for maturation of young hyphae and septation in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 2000;113(Pt 24):4563–4575. doi: 10.1242/jcs.113.24.4563. [DOI] [PubMed] [Google Scholar]
- 31.Kitade Y, Sumita T, Izumitsu K, Tanaka C. Cla4 PAK-like kinase is required for pathogenesis, asexual/sexual development and polarized growth in Bipolaris maydis. Curr. Genet. 2019;65:1229–1242. doi: 10.1007/s00294-019-00977-9. [DOI] [PubMed] [Google Scholar]
- 32.Rolke Y, Tudzynski P. The small GTPase Rac and the p21-activated kinase Cla4 in Claviceps purpurea: Interaction and impact on polarity, development and pathogenicity. Mol. Microbiol. 2008;68:405–423. doi: 10.1111/j.1365-2958.2008.06159.x. [DOI] [PubMed] [Google Scholar]
- 33.Frieser SH, Hlubek A, Sandrock B, Bölker M. Cla4 kinase triggers destruction of the Rac1-GEF Cdc24 during polarized growth in Ustilago maydis. Mol. Biol. Cell. 2011;22:3253–3262. doi: 10.1091/mbc.E11-04-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leveleki L, Mahlert M, Sandrock B, Bölker M. The PAK family kinase Cla4 is required for budding and morphogenesis in Ustilago maydis. Mol. Microbiol. 2004;54:396–406. doi: 10.1111/j.1365-2958.2004.04296.x. [DOI] [PubMed] [Google Scholar]
- 35.Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol. 1999;147:981–993. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi Y, et al. Smt3, a SUMO-1 homolog, is conjugated to Cdc3, a component of septin rings at the mother-bud neck in budding yeast. Biochem. Biophys. Res. Commun. 1999;259:582–587. doi: 10.1006/bbrc.1999.0821. [DOI] [PubMed] [Google Scholar]
- 37.Hernández-Rodríguez Y, Momany M. Posttranslational modifications and assembly of septin heteropolymers and higher-order structures. Curr. Opin. Microbiol. 2012;15:660–668. doi: 10.1016/j.mib.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 38.Jung EM, Kothe E, Raudaskoski M. The making of a mushroom: Mitosis, nuclear migration and the actin network. Fungal Genet. Biol. 2018;111:85–91. doi: 10.1016/j.fgb.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 39.Tanabe S, Kamada T. The role of astral microtubules in conjugate division in the dikaryon of Coprinus cinereus. Exp. Mycol. 1994;18:338–348. doi: 10.1016/S0147-5975(06)80007-1. [DOI] [Google Scholar]
- 40.Tanabe S, Kamada T. Dynamics of the actin cytoskeleton, hyphal tip growth and the movement of the two nuclei in the dikaryon of Coprinus cinereus. Mycoscience. 1996;37:339–344. doi: 10.1007/BF02461306. [DOI] [Google Scholar]
- 41.Badalyan SM, Polak E, Hermann R, Aebi M, Kües U. Role of peg formation in clamp cell fusion of homobasidiomycete fungi. J. Basic Microbiol. 2004;44:167–177. doi: 10.1002/jobm.200310361. [DOI] [PubMed] [Google Scholar]
- 42.Golan M, Mabjeesh NJ. SEPT9_i1 is required for the association between HIF-1α and importin-α to promote efficient nuclear translocation. Cell Cycle. 2013;12:2297–2308. doi: 10.4161/cc.25379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hernández-Rodríguez Y, et al. Distinct septin heteropolymers co-exist during multicellular development in the filamentous fungus Aspergillus nidulans. PLoS One. 2014;9:e92819. doi: 10.1371/journal.pone.0092819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blanco N, Reidy M, Arroyo J, Cabib E. Crosslinks in the cell wall of budding yeast control morphogenesis at the mother-bud neck. J. Cell Sci. 2012;125:5781–5789. doi: 10.1242/jcs.110460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kadota J, Yamamoto T, Yoshiuchi S, Bi E, Tanaka K. Septin ring assembly requires concerted action of polarisome components, a PAK kinase Cla4p, and the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:5329–5345. doi: 10.1091/mbc.e04-03-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teheri-Talesh N, et al. The tip growth apparatus of Aspergillus nidulans. Mol. Biol. Cell. 2008;19:1439–1449. doi: 10.1091/mbc.e07-05-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Delgado-Alvarez DL, et al. Visualization of F-actin localization and dynamics with live cell markers in Neurospora crassa. Fungal Genet. Biol. 2010;47:573–586. doi: 10.1016/j.fgb.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 48.Berepiki A, Lichius A, Shoji JY, Tilsner J, Read ND. F-actin dynamics in Neurospora crassa. Eukaryot. Cell. 2010;9:547–557. doi: 10.1128/EC.00253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bergs A, Ishitsuka Y, Evangelinos M, Nienhaus GU, Takeshita N. Dynamics of actin cables in polarized growth of the filamentous fungus Aspergillus nidulans. Front. Microbiol. 2016;7:682. doi: 10.3389/fmicb.2016.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knechtle P, Dietrich F, Philippsen P. Maximal polar growth potential depends on the polarisome component AgSpa2 in the filamentous fungus Ashbya gossypii. Mol. Biol. Cell. 2003;14:4140–4154. doi: 10.1091/mbc.e03-03-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng XD, Wang YM, Wang Y. CaSPA2 is important for polarity establishment and maintenance in Candida albicans. Mol. Microbiol. 2003;49:1391–1405. doi: 10.1046/j.1365-2958.2003.03646.x. [DOI] [PubMed] [Google Scholar]
- 52.Virag A, Harris SD. Functional characterization of Aspergillus nidulans homologues of Saccharomyces cerevisiae Spa2 and Bud6. Eukaryot. Cell. 2006;5:881–895. doi: 10.1128/EC.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carbó N, Pérez-Martín J. Spa2 is required for morphogenesis but it is dispensable for pathogenicity in the phytopathogenic fungus Ustilago maydis. Fungal Genet. Biol. 2008;45:1315–1327. doi: 10.1016/j.fgb.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 54.Araujo-Palomares CL, Riquelme M, Castro-Longoria E. The polarisome component SPA-2 localizes at the apex of Neurospora crassa and partially colocalizes with the Spitzenkörper. Fungal Genet. Biol. 2009;46:551–563. doi: 10.1016/j.fgb.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 55.Wong KH, et al. Sumoylation in Aspergillus nidulans: sumO inactivation, overexpression and live-cell imaging. Fungal Genet. Biol. 2008;45:728–737. doi: 10.1016/j.fgb.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iwasa M, Tanabe S, Kamada T. The two nuclei in the dikaryon of the Homobasidiomycete Coprinus cinereus change position after each conjugate division. Fungal Genet. Biol. 1998;23:110–116. doi: 10.1006/fgbi.1997.1019. [DOI] [PubMed] [Google Scholar]
- 57.Flescher EG, Madden K, Snyder M. Components required for cytokinesis are important for bud site selection in yeast. J. Cell Biol. 1993;122:373–386. doi: 10.1083/jcb.122.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garabedian MV, et al. A septin-Hof1 scaffold at the yeast bud neck binds and organizes actin cables. Mol. Biol. Cell. 2020;31:1988–2001. doi: 10.1091/mbc.E19-12-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brunswick H. Untersuchungen über die Geschlechts- und Kernverhältnisse bei der Hymenomyzetengattung Coprinus. Botanische Abhandlung; 1924. [Google Scholar]
- 60.Steinberg G. Hyphal growth: A tale of motors, lipids, and the Spitzenkörper. Eukaryot. Cell. 2007;6:351–360. doi: 10.1128/EC.00381-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bartnicki-Garcia S, Bracker CE, Gierz G, López-Franco R, Lu H. Mapping the growth of fungal hyphae: Orthogonal cell wall expansion during tip growth and the role of turgor. Biophys. J. 2000;79:2382–2390. doi: 10.1016/S0006-3495(00)76483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.López-Franco R, Bartnicki-Garcia S, Bracker CE. Pulsed growth of fungal hyphal tips. Proc. Natl. Acad. Sci. U.S.A. 1994;91:12228–12232. doi: 10.1073/pnas.91.25.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Riquelme M, et al. Fungal morphogenesis, from the polarized growth of hyphae to complex reproduction and infection structures. Microbiol. Mol. Biol. Rev. 2018 doi: 10.1128/MMBR.00068-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verdín J, et al. Off the wall: The rhyme and reason of. Cell Surf. 2019;5:100020. doi: 10.1016/j.tcsw.2019.100020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Verdín J, Bartnicki-Garcia S, Riquelme M. Functional stratification of the Spitzenkörper of Neurospora crassa. Mol. Microbiol. 2009;74:1044–1053. doi: 10.1111/j.1365-2958.2009.06917.x. [DOI] [PubMed] [Google Scholar]
- 66.Okada S, et al. Daughter cell identity emerges from the interplay of Cdc42, septins, and exocytosis. Dev. Cell. 2013;26:148–161. doi: 10.1016/j.devcel.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harris SD, et al. Polarisome meets spitzenkorper: Microscopy, genetics, and genomics converge. Eukaryot. Cell. 2005;4:225–229. doi: 10.1128/EC.4.2.225-229.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Köhli M, Galati V, Boudier K, Roberson RW, Philippsen P. Growth-speed-correlated localization of exocyst and polarisome components in growth zones of Ashbya gossypii hyphal tips. J. Cell Sci. 2008;121:3878–3889. doi: 10.1242/jcs.033852. [DOI] [PubMed] [Google Scholar]
- 69.Steinberg G. Endocytosis and early endosome motility in filamentous fungi. Curr. Opin. Microbiol. 2014;20:10–18. doi: 10.1016/j.mib.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zander S, Baumann S, Weidtkamp-Peters S, Feldbrügge M. Endosomal assembly and transport of heteromeric septin complexes promote septin cytoskeleton formation. J. Cell Sci. 2016;129:2778–2792. doi: 10.1242/jcs.182824. [DOI] [PubMed] [Google Scholar]
- 71.Cannon KS, Woods BL, Crutchley JM, Gladfelter AS. An amphipathic helix enables septins to sense micrometer-scale membrane curvature. J. Cell Biol. 2019;218:1128–1137. doi: 10.1083/jcb.201807211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bridges AA, Jentzsch MS, Oakes PW, Occhipinti P, Gladfelter AS. Micron-scale plasma membrane curvature is recognized by the septin cytoskeleton. J. Cell Biol. 2016;213:23–32. doi: 10.1083/jcb.201512029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martin SW, Konopka JB. SUMO modification of septin-interacting proteins in Candida albicans. J. Biol. Chem. 2004;279:40861–40867. doi: 10.1074/jbc.M406422200. [DOI] [PubMed] [Google Scholar]
- 74.De Souza CP, Osmani AH, Hashmi SB, Osmani SA. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr. Biol. 2004;14:1973–1984. doi: 10.1016/j.cub.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 75.Theisen U, Straube A, Steinberg G. Dynamic rearrangement of nucleoporins during fungal "open" mitosis. Mol. Biol. Cell. 2008;19:1230–1240. doi: 10.1091/mbc.e07-02-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaufmann A, Philippsen P. Of bars and rings: Hof1-dependent cytokinesis in multiseptated hyphae of Ashbya gossypii. Mol. Cell. Biol. 2009;29:771–783. doi: 10.1128/MCB.01150-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oh Y, et al. Hof1 and Chs4 interact via F-BAR domain and Sel1-like repeats to control extracellular matrix deposition during cytokinesis. Curr. Biol. 2017;27:2878–2886.e2875. doi: 10.1016/j.cub.2017.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meitinger F, Palani S, Hub B, Pereira G. Dual function of the NDR-kinase Dbf2 in the regulation of the F-BAR protein Hof1 during cytokinesis. Mol. Biol. Cell. 2013;24:1290–1304. doi: 10.1091/mbc.E12-08-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Founounou N, Loyer N, Le Borgne R. Septins regulate the contractility of the actomyosin ring to enable adherens junction remodeling during cytokinesis of epithelial cells. Dev. Cell. 2013;24:242–255. doi: 10.1016/j.devcel.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 80.Cvrcková F, Nasmyth K. Yeast G1 cyclins CLN1 and CLN2 and a GAP-like protein have a role in bud formation. EMBO J. 1993;12:5277–5286. doi: 10.1002/j.1460-2075.1993.tb06223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cvrcková F, De Virgilio C, Manser E, Pringle JR, Nasmyth K. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 1995;9:1817–1830. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- 82.Wendland J, et al. The mating-type locus B alpha 1 of Schizophyllum commune contains a pheromone receptor gene and putative pheromone genes. EMBO J. 1995;14:5271–5278. doi: 10.1002/j.1460-2075.1995.tb00211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O'Shea SF, et al. A large pheromone and receptor gene complex determines multiple B mating type specificities in Coprinus cinereus. Genetics. 1998;148:1081–1090. doi: 10.1093/genetics/148.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tymon AM, Kües U, Richardson WV, Casselton LA. A fungal mating type protein that regulates sexual and asexual development contains a POU-related domain. EMBO J. 1992;11:1805–1813. doi: 10.1002/j.1460-2075.1992.tb05232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kües U, et al. The combination of dissimilar alleles of the A alpha and A beta gene complexes, whose proteins contain homeo domain motifs, determines sexual development in the mushroom Coprinus cinereus. Genes Dev. 1992;6:568–577. doi: 10.1101/gad.6.4.568. [DOI] [PubMed] [Google Scholar]
- 86.Pagliuso A, et al. A role for septin 2 in Drp1-mediated mitochondrial fission. EMBO Rep. 2016;17:858–873. doi: 10.15252/embr.201541612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wloga D, Strzyzewska-Jówko I, Gaertig J, Jerka-Dziadosz M. Septins stabilize mitochondria in Tetrahymena thermophila. Eukaryot. Cell. 2008;7:1373–1386. doi: 10.1128/EC.00085-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cannon KS, et al. A gene duplication of a septin reveals a developmentally regulated filament length control mechanism. J. Cell Biol. 2023 doi: 10.1083/jcb.202204063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mol PC, Wessels JGH. Differences in wall structure between substrate hyphae and hyphae of fruit-body stipes in Agaricus bisporus. Mycol. Res. 1990;94:472–479. doi: 10.1016/S0953-7562(10)80006-1. [DOI] [Google Scholar]
- 90.Mol PC, Vermeulen CA, Wessels JGH. Diffuse extension of hyphae in stipes of Agaricus bisporus may be based on a unique wall structure. Mycol. Res. 1990;94:480–488. doi: 10.1016/S0953-7562(10)80007-3. [DOI] [Google Scholar]
- 91.Li C, et al. FastCloning: A highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol. 2011;11:92. doi: 10.1186/1472-6750-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Binninger DM, Skrzynia C, Pukkila PJ, Casselton LA. DNA-mediated transformation of the basidiomycete Coprinus cinereus. EMBO J. 1987;6:835–840. doi: 10.1002/j.1460-2075.1987.tb04828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Muraguchi H, Matsubuchi M, Sugo M, Nose T. Septin dynamics of Coprinopsis cinerea. Akita Pref. Univ. Web J. B. 2017;1:1–5. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author upon reasonable request. All data generated or analysed during this study are included in this published article [and its supplementary information files].