Abstract
Oncotype Dx Recurrence Score (RS) has been validated in patients with ER + /HER2 − invasive breast carcinoma to estimate patient risk of recurrence and guide the use of adjuvant chemotherapy. We investigated the role of MRI-based radiomics features extracted from the tumor and the peritumoral tissues to predict the risk of tumor recurrence. A total of 62 patients with biopsy-proved ER + /HER2 − breast cancer who underwent pre-treatment MRI and Oncotype Dx were included. An RS > 25 was considered discriminant between low-intermediate and high risk of tumor recurrence. Two readers segmented each tumor. Radiomics features were extracted from the tumor and the peritumoral tissues. Partial least square (PLS) regression was used as the multivariate machine learning algorithm. PLS β-weights of radiomics features included the 5% features with the largest β-weights in magnitude (top 5%). Leave-one-out nested cross-validation (nCV) was used to achieve hyperparameter optimization and evaluate the generalizable performance of the procedure. The diagnostic performance of the radiomics model was assessed through receiver operating characteristic (ROC) analysis. A null hypothesis probability threshold of 5% was chosen (p < 0.05). The exploratory analysis for the complete dataset revealed an average absolute correlation among features of 0.51. The nCV framework delivered an AUC of 0.76 (p = 1.1∙10−3). When combining “early” and “peak” DCE images of only T or TST, a tendency toward statistical significance was obtained for TST with an AUC of 0.61 (p = 0.05). The 47 features included in the top 5% were balanced between T and TST (23 and 24, respectively). Moreover, 33/47 (70%) were texture-related, and 25/47 (53%) were derived from high-resolution images (1 mm). A radiomics-based machine learning approach shows the potential to accurately predict the recurrence risk in early ER + /HER2 − breast cancer patients.
Keywords: Artificial intelligence, Breast cancer, Oncotype DX, Machine learning, Magnetic resonance imaging
Introduction
Breast cancer is a leading cause of death and the most common cancer in women [1]. It consists of four main subtypes, classified by tumor genotype and molecular characterization in luminal A, luminal B, HER2-enriched, and basal-like cancer [1–3]. Luminal tumors represent the most invasive breast cancer (70%) in western countries. They are usually estrogen-receptor (ER) and/or progesterone-receptor (PR) positive and HER2-receptor (HER2) negative.
Hormone therapy represents a mainstay for patient management [4]. About 15% of luminal B cancers will develop a recurrence within 10 years from the diagnosis if treated with hormonal therapy alone. Although the risk of recurrence could be lowered by adjuvant chemotherapy, selecting patients who might benefit from adjuvant chemotherapy is debated [5–7].
Oncotype Dx Recurrence Score (RS) 21-gene expression assay (Genomic Health Inc., Redwood City, CA) produces a score based on the quantitative expression level of 21 genes in ribonucleic acid extracted from formalin-fixed, paraffin-embedded breast tumor tissues. This assay has been validated in patients with ER + /HER2 − invasive breast carcinoma to estimate patient risk of distant breast cancer recurrence and guide the use of adjuvant chemotherapy [8–11]. Recent studies demonstrated that RS correlates with recurrence rates and adjuvant treatment response [5, 9, 12–14]. The National Comprehensive Cancer Network (NCCN) and the American Society of Clinical Oncology (ASCO) both recommended the use of Oncotype Dx RS testing in patients with ER + /HER2 − breast cancers [15, 16]. Moreover, the Guidelines Development Group of the European Commission Initiative on Breast Cancer recently prioritized a clinical question on the use of multigene test to guide the use of adjuvant chemotherapy in ER + , HER2 − , and lymph node–negative or up to 3 lymph node–positive invasive breast cancer [17]. For these reasons, this assay is now incorporated into clinical practice guidelines for treatment decisions [12]. However, the technique is costly and is performed on surgical breast tumor specimen. These limitations prompted researchers to investigate new imaging-based biomarkers [18–24]. Multiparametric magnetic resonance imaging (MRI) is the most sensitive modality to diagnose and assess treatment response in breast cancer patients [25–28]. In this regard, recently developed imaging-based methods, such as radiomics, allow analyzing imaging data and extracting many quantitative features, thereby adding a whole tumor volume of extra information to the conventional qualitative visual assessment [18–24, 29–35]. MRI-based predictors of tumor recurrence allow the non-invasive selection of patients at high risk of recurrence, with significant improvements on patient healthcare and overall costs. Few studies investigated the use of MRI-based radiomics for the prediction of breast cancer recurrence having the Oncotype Dx RS as reference standard [36–44]. Compared to previous studies on Oncotype Dx, ours not only was focused on the tumor but also investigated the peritumoral tissues. In fact, as in other recent studies, not only on breast cancer, radiomics features extracted from the tumor site and the peritumoral environment showed a potential role in terms of prediction of treatment response [45–48]. Moreover, the American Society of Clinical Oncology (ASCO) recently revised the breast cancer recurrence risk by addressing the use of Oncotype Dx in guiding decisions on the use of adjuvant systemic therapy. In detail, they divided high risk and low-intermedium risk based on a RS cut-off of 25 [9]. In this regard, only one study was recently published on the potential correlation between radiomics and RS adopting this cut-off, but it did not assess the role of peritumoral tissues [49].
This study investigated the ability of MRI-based radiomics features extracted from the tumor and the peritumoral tissues to predict the risk of tumor recurrence in ER + /HER2 − breast cancer patients. Thus, by demonstrating the presence of imaging-based biomarkers, we could non-invasively identify patients who are more likely to benefit from adjuvant therapy.
Materials and Methods
Subjects
This study received formal approval from the Ethical Committee of the University G. d’Annunzio of Chieti-Pescara, Italy; informed consent was waived by the same ethics committee that approved the study (Comitato Etico per la Ricerca Biomedica delle Province di Chieti e Pescara e dell’Università degli Studi “G. d’Annunzio” di Chieti e Pescara). The study was conducted according to ethical principles laid down by the latest version of the Declaration of Helsinki. A total of 62 patients who underwent clinically indicated breast MRI between January 2016 and May 2020 at our institution were retrospectively included. Inclusion criteria were as follows: [1] ER + /HER2 − early breast cancer confirmed via biopsy, [2] MRI performed on a 1.5-T scanner, and [3] availability of Oncotype DX RS.
MRI Protocol
All patients in this cohort underwent a clinically indicated breast MRI consisting of a standard T1-weighted (T1w), T2-weighted (T2w), diffusion-weighted imaging (DWI), and dynamic contrast enhancement (DCE) acquisition performed using a 1.5-T MR scanner (Achieva, Philips Medical System, Best, the Netherlands) equipped with a dedicated phased-array breast coil. Detailed information regarding the DCE acquisition is described in Table 1.
Table 1.
MRI protocol parameters
| T1-weighted post-contrast 3D-FFE |
|
|---|---|
| Repetition time (msec) | 3000–5000 |
| Echo time (msec) | 80 |
| Section thickness (mm) | 2 |
| Section gap (mm) | 0 |
| Acquisition matrix size | 340 × 340 |
| No. of signals acquired | 2 |
| Field of view (mm) | 340 × 340 |
| Sensitivity encoding (SENSE) | Yes |
| Acquisition time (sec) | 54.3, 90** |
| No. of sections | 167 |
FFE, Fast Field Echo
**First (“early”) and second (“peak”) DCE acquisition after the endovenous administration of contrast agent (gadolinium chelate)
Imaging Analysis
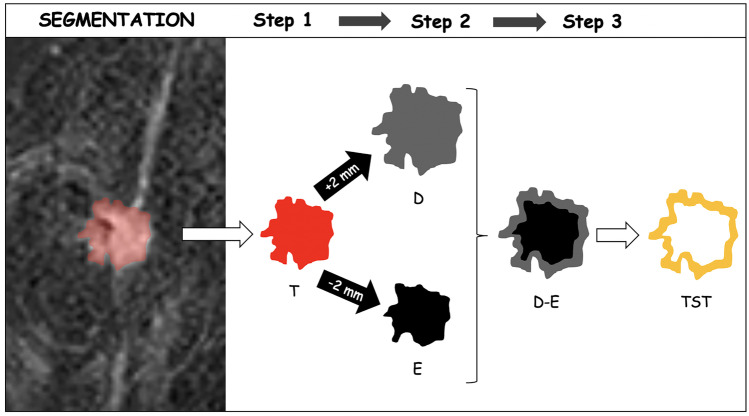
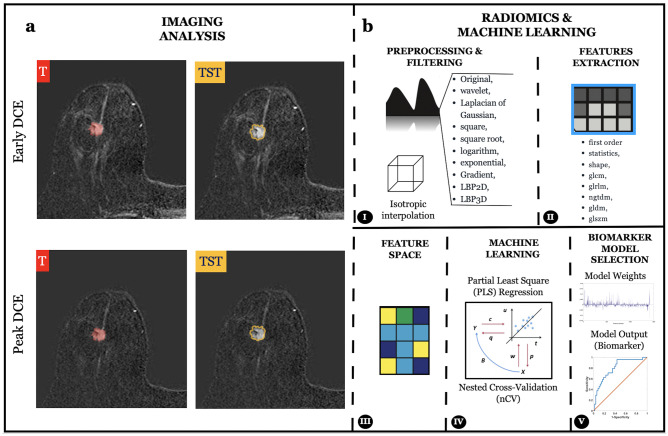
Whole volume tumor manual segmentation of the tumor (T) was performed on the first (“early”) and second (“peak”) contrast-enhanced dynamic T1w images for each patient by two independent senior radiology residents. The software used for the segmentation was an open source medical image computing platform, 3D Slicer Version 4.8 (www.3dslicer.org). To create the “tissue surrounding tumor” segmentations (TST), a “3dmask_tool” (AFNI) was used [50]. First, a 2-mm dilatation (“dilate”) and a 2-mm erosion (“erode”) were obtained from the CT of each patient. Then, the two masks were subtracted (“dilate” − “erode”) to obtain the TST which was 4 mm thick (Fig. 2) [45]. All the TST segmentations were then checked by the two readers and manually adjusted if necessary to include only the outer border of the tumor and the adjacent perivisceral tissue. T and TST are shown in Fig. 1a.
Fig. 2.
The segmentation process included three steps. Firstly, the whole breast tumor (T) was manually segmented on contrast-enhanced dynamic T1w images. In the second step, the edge of T was dilated (D) and eroded (E) by 2 mm, respectively. In the third step, we overlapped the dilated and eroded masks and subsequently subtracted them to include the most peripheral portion of the tumor and the surrounding tissues (TST)
Fig. 1.
a Tumor (T) and tissue surrounding tumor (TST) segmentation on the first (“early”) and second (“peak”) contrast-enhanced dynamic (DCE) T1w images. b Schematic representation of the radiomics features extraction and the machine learning framework implemented
Radiomic Features Extraction
The extraction of radiomics features from the masked (T and TST) T1w images was performed using PyRadiomics [51]. Reproducibility assessments of the features extracted by the two readers from the segmentations of all patients were performed (Fig. 2). To avoid data heterogeneity bias and minimize acquisition-related radiomics variability, MR images and masks were resampled using 3 isotropic voxel dimensions (1 × 1 × 1 mm, 2 × 2 × 2 mm, and 3 × 3 × 3 mm). For each segmentation and for each image resolution (1 mm, 2 mm, and 3 mm), ten built-in filters (Original, wavelet, Laplacian of Gaussian (LoG), square, square root, logarithm, exponential, Gradient, LBP2D, and LBP3D) were applied, and seven feature classes (first-order statistics, shape descriptors, glcm, glrlm, ngtdm, gldm, and glszm) were calculated, which resulted in a total of 1409 radiomics features for each image (Fig. 1b) [52–54]. Prior to the machine learning analysis, all features were converted into z-scores relying on their subject distribution.
Machine Learning Analysis
A machine learning approach was used to exploit the radiomics features’ multidimensionality and infer the risk of recurrence (high vs. low-intermedium). Two main strategies were implemented to address the large number of features extracted [55, 56]. The first approach reduced the number of used features by selecting only highly repeatable features between the masks delineated by the two radiologists (r > 0.95). The second approach leveraged the high collinearity among radiomics features which was evaluated through an initial exploratory analysis. It then used a linear regression analysis to infer the risk of recurrence, thus employing a space dimension reduction procedure, namely, the partial least square (PLS) regression [55–58]. PLS has one hyperparameter, namely, the number of uncorrelated components to be used in the regression. Leave-one-out nested cross-validation (nCV) was used to achieve hyperparameter optimization and evaluate the generalizable performance of the procedure [58–60]. In nCV, data are divided into folds, and the model is trained on all data except one-fold in an iterative, nested manner. Whereas the outer loop estimates the model’s performances among iterations (test), the inner loop evaluates the optimal hyperparameter (validation). If the number of folds equals the number of samples (one-fold per sample), the procedure is defined as leave-one-out nCV, an approach highly suited for medical applications where samples represent subjects [61–63]. The whole leave-one-out nCV PLS analysis was repeated multiple times for the following group of masks: (a) DCE images (“early” and “peak”) in both T and TST, (b) “peak” DCE in both T and TST, (c) “early” DCE in both T and TST, (d) “peak” DCE in T, (e) “peak” DCE in TST, (f) “early” DCE in T, and (g) “early” DCE in TST.
Reference Standard
A recurrence score > 25 was considered to discriminate between low-intermediate (≤ 25) and high risk ( >) of tumor recurrence [9, 11, 64, 65].
Calculation
The classification performances were assessed through receiver operating characteristic (ROC) analyses considering the inferred (out-of-training-sample) recurrence risk in the outer loop fold of the machine learning framework. Patients with low-intermediate recurrence risk were attributed to the “negative” group, whereas patients with high recurrence risk were attributed to the “positive” group. The ROC analyses were also performed on random shuffled outcomes to simulate the null hypothesis and evaluate its confidence interval (repeated 106 times). The ROC analysis delivered an area under the curve (AUC), which, using the random shuffled outcomes, could be transformed into a z-score for assessing its statistical significance. The statistical analysis was performed in MATLAB.
Results
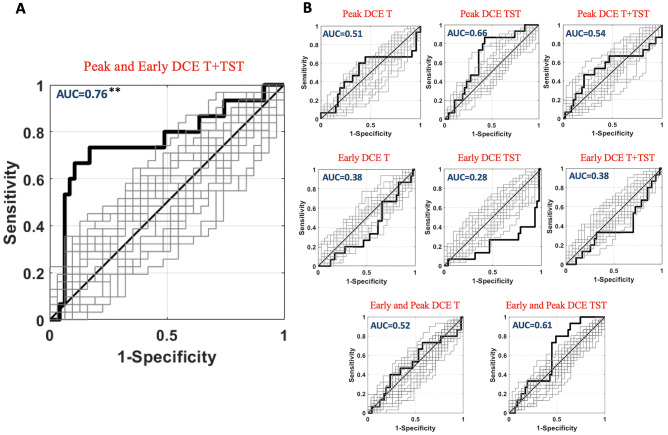
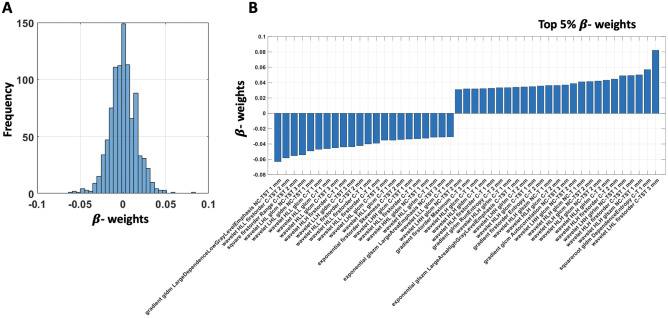
Out of the 62 women included in the study, the mean age was 49 (interquartile range: 44.25–53) years. Forty-seven (75.8%) patients showed a RS ≤ 25 (low-intermediate recurrence risk) and 15 (24.2%) showed a RS > 25 (high recurrence risk) (Table 2). In total, 1409 radiomics features were extracted for each image. Each MRI included “early” and “peak” DCE images. We extracted two masks (T and TST) from each set of images (“early” DCE T, “early” DCE TST, “peak” DCE T, and “peak” DCE TST). All MRI images were resampled at 3 resolutions, for a total of 33,816 features per patients. In detail, the number of features selected (based on inter-read repeatability of r > 0.95) for each repetition of the analysis was as follows: n = 940 from “early” and “peak” DCE T + TST images, n = 644 from “peak” DCE T + TST images, n = 296 from “early” DCE T + TST images, n = 315 from “peak” DCE T, n = 329 from “peak” DCE TST, n = 230 from “early” DCE T, and n = 66 from “early” DCE TST. The exploratory analysis for the complete dataset revealed an average absolute correlation among features of 0.51. The high average absolute correlation among features justified the use of PLS. Using the nCV machine learning PLS framework, a significant inference on the risk of recurrence was obtained when including all features in the analysis (“early” and “peak” DCE T + TST, optimal number of PLS components, n = 19), with an AUC = 0.76, z = 3.01, p = 1.1∙10−3 (Fig. 3A). Standalone combinations of “early” and “peak” DCE images of T and TST did not deliver a significant multivariate inference of the risk (p > 0.05; Fig. 3B). When combining “early” and “peak” DCE images of only T or TST, a tendency toward statistical significance was obtained for TST with an AUC of 0.61 (p = 0.05). Figure 4A reports the nCV β-weight distribution depicting the strength and sign of the effect of the original radiomics features in the inference of the outcome. Since the larger labeling value of “1” was associated with an increased risk of tumor recurrence, the positive β-weight suggested a higher risk at increasing feature value and vice-versa for negative weights. Figure 4B reports the top 5% (n = 47) β-weights associated with the most relevant features involved in the prediction (those with the largest β-weight magnitudes). These features were balanced between T and TST (23 and 24, respectively). In detail, 25 of the top 5% features were associated with images at 1 mm resolution, 15 at 2 mm resolution, and 7 at 3 mm resolution. Most (33/47) of those features were related to the texture analysis. Thirty-three (70%) top 5% weights were associated with the second-order analysis of the images (e.g., features computed using the gray-level co-occurrence matrix (GLCM), or the gray-level dependence matrix (GLDM)), whereas only 14 features were related to first-order analysis. In addition, a larger number of “peak” (n = 33) versus “early” DCE (n = 14) features were present.
Table 2.
Demographics and baseline features of the included patients
| Value | |
|---|---|
| Gender | |
| Female | 62 (100%) |
| Mean age (IQR) | 49.4 (44.25–53) |
| MRI exam (n) | 62 |
| Mean Oncotype Recurrence Score | 20.4 |
| Primary pT stage | |
| T1 (T1a; T1b; T1c) | 41 |
| T2 | 21 |
| T3 and T4 | 0 |
| Primary pN stage | |
| N0 | 31 |
| N1 (N1mi; N1a; N1b) | 31 |
| N2 and N3 | 0 |
| Recurrence score (RS) | |
| ≤ 25 | 47 (75.8%) |
| > 25 | 15 (24.2%) |
Fig. 3.
ROC analysis of the machine learning (PLS) classification performance. Patients with RS ≤ 25 were attributed to the “negative” group, whereas patients with RS > 25 were attributed to the “positive” group
Fig. 4.
A Partial least square analysis showing β-weights associated with reliable (r > 0.95) radiomics features. B β-weights are associated to the top 5% of features with the largest β-weights in magnitude
Discussion
Our results showed that MRI-based radiomics can predict the risk of recurrence in ER + /HER2 − early breast cancer patients. These findings confirmed the promising preliminary results showing a significant association between radiomics signatures and risk of breast cancer recurrence [38, 40]. For example, Li et al. reported that radiomics features including tumor size and tumor heterogeneity predicted multigene assay recurrence scores [38]. A recent study generated a radiomics signature based on dynamic contrast-enhanced MRI to distinguish between low (recurrence score < 18) and non-low (recurrence score > 18) Oncotype DX risk groups in estrogen receptor (ER)–positive invasive breast cancer [40]. The authors obtained a Rad score based on 10 radiomics features reaching an AUC of 0.759 [40].
Of note, we distinguished low-intermediate risk (recurrence score < 25) and high-risk (recurrence score > 25) patients according to the last American Society of Clinical Oncology (ASCO) clinical practice guideline update. In this regard, the panel of experts referred to the publication of the Trial Assigning Individualized Options for Treatment (TAILORx) evaluating noninferiority of endocrine therapy alone versus chemoendocrine therapy for invasive disease-free survival in women with Oncotype DX scores. Based on informal consensus, the panel recommended that oncologists offer chemoendocrine therapy to patients with recurrence scores of 26 to 30 (9). Only one study adopting this cut-off was recently published in literature for assessing ER + /HER2 − breast cancer patients’ 21-gene RS using a multiparametric MRI-based radiomics model [49]. They obtained an AUC of 0.82 from DCE of the tumor that improved to 0.92 when adding DWI and T2-weighted images. Compared to this study, ours analyzed not only the tumor but also the tissues surrounding the tumor. In detail, the machine learning framework delivered a significant inference on the risk of recurrence when including radiomics features from the tumor and the peritumoral tissues. On the other hand, the standalone combinations of radiomics features did not deliver a significant multivariate inference of the risk. These results are in line with other recent studies, not only focused on breast cancer, showing a potential predictive role of radiomics features extracted from the peritumoral environment [45–48]. For example, Braman et al. investigated the role of MRI-based radiomics signatures to characterize HER2-positive tumor biological factors and estimate tumor response to HER2-targeted neoadjuvant therapy [46]. The authors indicated a classifier performance with an AUC of 0.89 when combining peritumoral and intratumoral features [47]. Other authors investigated the predictive role of DCE-based quantitative features to distinguish molecular subtypes (luminal A/B or basal). They showed that DCE-based features of background parenchymal enhancement were statistically significant in separating luminal A versus nonluminal A cancers and distinguishing basal subtypes [48].
Interestingly, most of the top 5% features derived from 1-mm slice thickness images. This result suggests that high-resolution imaging was a relevant parameter for the prediction performance, and it is in line with Chen et al. that used a cubic spline interpolation algorithm resizing DCE images at 0.9 × 0.9 × 2.2 mm. Most of those features were texture-related, reflecting the degree of heterogeneity in breast tissue.
The role of contrast-enhanced imaging was also relevant in our study. This type of imaging assesses the permeability of blood vessels by using an intravenous contrast agent (gadolinium chelate) that shortens the local T1 time leading to a higher signal on T1-weighted images. The neoplastic neoangiogenesis produces leaky vessels allowing for faster extravasation of contrast agents. This leads to a rapid local enhancement which makes the tumor detectable on post-contrast images [25, 26]. In this regard, 70% of the top 5% features were extracted by contrast-enhanced images obtained at the “peak” of contrast enhancement. These results align with the current state-of-the-art breast MRI that recommends the acquisition of images approximately 60–90 s after the administration of contrast [26]. Although breast MRI without intravenous contrast administration has been proposed as a screening procedure, current techniques, such as DWI, are not sensitive enough to replace DCE-MRI [66, 67].
Our study has some limitations. First of all, it included a relatively low number of patients and lacked a validation cohort. This is due in part to the extraordinary cost of genetic testing that limited the study population size. However, our analysis is set to be a proof-of-concept study, and the nCV implemented in our study minimized the effect driven by the reduced number of samples and overfitting (59). Second, ours is a retrospective single-center study. Further studies, possibly with a prospective design and multicentric, are warranted to confirm our findings and better define the role of radiomics as a predictive biomarker in breast cancer. Third, we only analyzed dynamic contrast-enhanced MRI images, thereby excluding T2-weighted or diffusion-weighted images. Further studies are needed to clarify the potential role of these sequences in tumor recurrence prediction.
Conclusions
In conclusion, a radiomics-based machine learning approach showed the potential to accurately predict the recurrence risk in early ER + /HER2 − breast cancer patients. Most of the discriminant radiomics features were extracted from high-resolution images obtained at the “peak” of the contrast enhancement. They were mainly related to texture analysis from the tumor and peritumoral environment.
Acknowledgements
The authors thank Daniele Petrucci and Darien Calvo Garcia for their insightful contribution on the MRI protocol settings and the acquisition of data.
Author Contribution
A.D.P. conceived and developed the research idea with the assistance of D.M., M.M., A.D.C., and M.C. The study design was performed by A.D.P., D.M., P.C.H., S.S., and M.C. R.L. and M.S. performed the segmentation task. C.H.M., D.A., C.A.M. A.R., C.T., C.D.E., and G.C. performed data collection. P.C.R., P.C.H., and A.M.C. performed the computational experiments with guidance from A.D.P. The paper was written by A.D.P. with technical content from A.M.C. and P.CH and extensive editorial input from all authors. All authors have read and approved the final version submitted.
Funding
Open access funding provided by Università degli Studi G. D'Annunzio Chieti Pescara within the CRUI-CARE Agreement.
Data Availability
The datasets generated and/or analyzed during the current study are not publicly available due to the clinical and confidential nature of the material but can be made available from the corresponding author on reasonable request.
Declarations
Ethics Approval
This study received formal approval (protocol number 2190, 23/09/2020) from the Ethical Committee of the University G. d’Annunzio of Chieti-Pescara, Italy; informed consent was waived by the same ethics committee that approved the study (Comitato Etico per la Ricerca Biomedica delle Province di Chieti e Pescara e dell’Università degli Studi “G. d’Annunzio” di Chieti e Pescara). The study was conducted according to ethical principles laid down by the latest version of the Declaration of Helsinki.
Consent to Participate
Informed consent was obtained from all individual participants included in the study.
Consent for Publication
No individual person’s data were published in the manuscript.
Competing Interests
Domenico Mastrodicasa: activities related to the present article: none; activities not related to the present article: shareholder of Segmed, Inc. and consultant for Segmed, Inc. Andrea Delli Pizzi: member of the Scientific Editorial Board for the journal Insights Into Imaging. The other authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Piero Chiacchiaretta, Email: p.chiacchiaretta@unich.it.
Antonio Corvino, Email: an.cor@hotmail.it.
References
- 1.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5(1):5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howlader N, Altekruse SF, Li CI, Chen VW, Clarke CA, Ries LAG, et al. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. JNCI: Journal of the National Cancer Institute. 2014;106(dju055). [DOI] [PMC free article] [PubMed]
- 4.Hancock MJ. Appraisal of clinical practice guideline: early and locally advanced breast cancer: diagnosis and management. NICE guideline [NG101]. J Physiother. 2019;65(1):57. [DOI] [PubMed]
- 5.Giorgi Rossi P, Lebeau A, Canelo-Aybar C, Saz-Parkinson Z, Quinn C, Langendam M, et al. Recommendations from the european commission initiative on breast cancer for multigene testing to guide the use of adjuvant chemotherapy in patients with early breast cancer, hormone receptor positive, HER-2 negative. British Journal of Cancer. 2021:1–10. [DOI] [PMC free article] [PubMed]
- 6.Harris LN, Ismaila N, McShane LM, Andre F, Collyar DE, Gonzalez-Angulo AM, et al. Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: American Society of Clinical Oncology Clinical Practice Guideline. JCO. 2016;34(10):1134–1150. doi: 10.1200/JCO.2015.65.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Group EBCTC. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. The Lancet. 2011;378(9793):771–84. [DOI] [PMC free article] [PubMed]
- 8.Albain KS, Barlow WE, Shak S, Hortobagyi GN, Livingston RB, Yeh IT, et al. Prognostic and predictive value of the 21-gene Recurrence Score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 2010;11(1):55–65. doi: 10.1016/S1470-2045(09)70314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andre F, Ismaila N, Henry NL, Somerfield MR, Bast RC, Barlow W, et al. Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: ASCO clinical practice guideline update-integration of results from TAILORx. J Clin Oncol. 2019;37(22):1956–1964. doi: 10.1200/JCO.19.00945. [DOI] [PubMed] [Google Scholar]
- 10.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 11.Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N Engl J Med. 2018;379(2):111–121. doi: 10.1056/NEJMoa1804710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardoso F, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, Rubio IT, et al. Early breast cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2019;30(10):1674. doi: 10.1093/annonc/mdz189. [DOI] [PubMed] [Google Scholar]
- 13.Duffy MJ, Harbeck N, Nap M, Molina R, Nicolini A, Senkus E, et al. Clinical use of biomarkers in breast cancer: updated guidelines from the European Group on Tumor Markers (EGTM) Eur J Cancer. 2017;75:284–298. doi: 10.1016/j.ejca.2017.01.017. [DOI] [PubMed] [Google Scholar]
- 14.McVeigh TP, Kerin MJ. Clinical use of the Oncotype DX genomic test to guide treatment decisions for patients with invasive breast cancer. Breast Cancer (Dove Med Press). 2017;9:393–400. doi: 10.2147/BCTT.S109847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iles K, Roberson ML, Spanheimer P, Gallagher K, Ollila DW, Strassle PD, et al. The impact of age and nodal status on variations in Oncotype DX testing and adjuvant treatment. NPJ Breast Cancer. 2022;8(1). [DOI] [PMC free article] [PubMed]
- 16.Gradishar WJ, Moran MS, Abraham J, Aft R, Agnese D, Allison KH, et al. NCCN Guidelines® Insights: Breast Cancer, Version 4.2021. Journal of the National Comprehensive Cancer Network. 2021;19(5):484–93. [DOI] [PubMed]
- 17.Giorgi Rossi P, Lebeau A, Canelo-Aybar C, Saz-Parkinson Z, Quinn C, Langendam M, et al. Recommendations from the European Commission Initiative on Breast Cancer for multigene testing to guide the use of adjuvant chemotherapy in patients with early breast cancer, hormone receptor positive, HER-2 negative. British Journal of Cancer. 2021;124(9):1503–1512. doi: 10.1038/s41416-020-01247-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avanzo M, Stancanello J, El Naqa I. Beyond imaging: the promise of radiomics. Phys Med. 2017;38:122–139. doi: 10.1016/j.ejmp.2017.05.071. [DOI] [PubMed] [Google Scholar]
- 19.Liu Z, Wang S, Dong D, Wei J, Fang C, Zhou X, et al. The applications of radiomics in precision diagnosis and treatment of oncology: opportunities and challenges. Theranostics. 2019;9(5):1303–1322. doi: 10.7150/thno.30309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogers W, Thulasi Seetha S, Refaee TAG, Lieverse RIY, Granzier RWY, Ibrahim A, et al. Radiomics: from qualitative to quantitative imaging. The British Journal of Radiology. 2020;93(1108):20190948. doi: 10.1259/bjr.20190948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tagliafico AS, Piana M, Schenone D, Lai R, Massone AM, Houssami N. Overview of radiomics in breast cancer diagnosis and prognostication. Breast. 2020;49:74–80. doi: 10.1016/j.breast.2019.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valdora F, Houssami N, Rossi F, Calabrese M, Tagliafico AS. Rapid review: radiomics and breast cancer. Breast Cancer Res Treat. 2018;169(2):217–229. doi: 10.1007/s10549-018-4675-4. [DOI] [PubMed] [Google Scholar]
- 23.Yip SSF, Aerts HJWL. Applications and limitations of radiomics. Phys Med Biol. 2016;61(13):R150–R166. doi: 10.1088/0031-9155/61/13/R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 25.Mann RM, Balleyguier C, Baltzer PA, Bick U, Colin C, Cornford E, et al. Breast MRI: EUSOBI recommendations for women’s information. European Radiology. 2015;25(12):3669–3678. doi: 10.1007/s00330-015-3807-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mann RM, Cho N, Moy L. Breast MRI: state of the art. Radiology. 2019;292(3):520–536. doi: 10.1148/radiol.2019182947. [DOI] [PubMed] [Google Scholar]
- 27.Marino MA, Helbich T, Baltzer P, Pinker-Domenig K. Multiparametric MRI of the breast: a review. J Magn Reson Imaging. 2018;47(2):301–315. doi: 10.1002/jmri.25790. [DOI] [PubMed] [Google Scholar]
- 28.Menezes GL, Knuttel FM, Stehouwer BL, Pijnappel RM, van den Bosch MA. Magnetic resonance imaging in breast cancer: a literature review and future perspectives. World J Clin Oncol. 2014;5(2):61–70. doi: 10.5306/wjco.v5.i2.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crivelli P, Ledda RE, Parascandolo N, Fara A, Soro D, Conti M. A new challenge for radiologists: radiomics in breast cancer. BioMed Research International. 2018;2018:e6120703. doi: 10.1155/2018/6120703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaheed K, Mao A, Qureshi I, Kumar M, Hussain S, Ullah I, et al. DS-CNN: a pre-trained Xception model based on depth-wise separable convolutional neural network for finger vein recognition. Expert Systems with Applications. 2022;191.
- 31.Chhabra P, Garg NK, Kumar M. Content-based image retrieval system using ORB and SIFT features. Neural Computing and Applications. 2018;32(7):2725–2733. doi: 10.1007/s00521-018-3677-9. [DOI] [Google Scholar]
- 32.Gupta S, Kumar M, Garg A. Improved object recognition results using SIFT and ORB feature detector. Multimedia Tools and Applications. 2019;78(23):34157–34171. doi: 10.1007/s11042-019-08232-6. [DOI] [Google Scholar]
- 33.Bansal M, Kumar M, Kumar M, Kumar K. An efficient technique for object recognition using Shi-Tomasi corner detection algorithm. Soft Computing. 2020;25(6):4423–4432. doi: 10.1007/s00500-020-05453-y. [DOI] [Google Scholar]
- 34.Kumar M, Gupta S, Gao X-Z, Singh A. Plant Species Recognition Using Morphological Features and Adaptive Boosting Methodology. IEEE Access. 2019;7:163912–163918. doi: 10.1109/ACCESS.2019.2952176. [DOI] [Google Scholar]
- 35.Bansal M, Kumar M, Kumar M. 2D object recognition: a comparative analysis of SIFT, SURF and ORB feature descriptors. Multimedia Tools and Applications. 2021;80(12):18839–18857. doi: 10.1007/s11042-021-10646-0. [DOI] [Google Scholar]
- 36.Cui Y, Fan M, Peng W, Liu L, Bai Q, Li L, editors. Prediction of Oncotype DX Recurrence Score in breast cancer by integration of DCE-MRI radiomics and clinicopathologic data. Medical Imaging 2021: Imaging Informatics for Healthcare, Research, and Applications; 2021 2021/02/15/: International Society for Optics and Photonics.
- 37.Ha R, Chang P, Mutasa S, Karcich J, Goodman S, Blum E, et al. Convolutional neural network using a breast MRI tumor dataset can predict Oncotype Dx Recurrence Score. J Magn Reson Imaging. 2019;49(2):518–524. doi: 10.1002/jmri.26244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Zhu Y, Burnside ES, Drukker K, Hoadley KA, Fan C, et al. MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of MammaPrint, Oncotype DX, and PAM50 Gene Assays. Radiology. 2016;281(2):382–391. doi: 10.1148/radiol.2016152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazurowski MA, Zhang J, Grimm LJ, Yoon SC, Silber JI. Radiogenomic analysis of breast cancer: luminal B molecular subtype is associated with enhancement dynamics at MR imaging. Radiology. 2014;273(2):365–372. doi: 10.1148/radiol.14132641. [DOI] [PubMed] [Google Scholar]
- 40.Nam KJ, Park H, Ko ES, Lim Y, Cho H-H, Lee JE. Radiomics signature on 3T dynamic contrast-enhanced magnetic resonance imaging for estrogen receptor-positive invasive breast cancers: preliminary results for correlation with Oncotype DX Recurrence Scores. Medicine (Baltimore). 2019;98(23):e15871. doi: 10.1097/MD.0000000000015871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saha A, Harowicz MR, Wang W, Mazurowski MA. A study of association of Oncotype DX Recurrence Score with DCE-MRI characteristics using multivariate machine learning models. J Cancer Res Clin Oncol. 2018;144(5):799–807. doi: 10.1007/s00432-018-2595-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sutton EJ, Oh JH, Dashevsky BZ, Veeraraghavan H, Apte AP, Thakur SB, et al. Breast cancer subtype intertumor heterogeneity: MRI-based features predict results of a genomic assay. J Magn Reson Imaging. 2015;42(5):1398–1406. doi: 10.1002/jmri.24890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wan T, Bloch BN, Plecha D, Thompson CL, Gilmore H, Jaffe C, et al. A radio-genomics approach for identifying high risk estrogen receptor-positive breast cancers on DCE-MRI: preliminary results in predicting Oncotype DX Risk Scores. Scientific Reports. 2016;6(1):21394. doi: 10.1038/srep21394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woodard GA, Ray KM, Joe BN, Price ER. Qualitative radiogenomics: association between Oncotype DX Test Recurrence Score and BI-RADS mammographic and breast MR imaging features. Radiology. 2018;286(1):60–70. doi: 10.1148/radiol.2017162333. [DOI] [PubMed] [Google Scholar]
- 45.Delli Pizzi A, Chiarelli AM, Chiacchiaretta P, d’Annibale M, Croce P, Rosa C, et al. MRI-based clinical-radiomics model predicts tumor response before treatment in locally advanced rectal cancer. Scientific Reports. 2021;11(1). [DOI] [PMC free article] [PubMed]
- 46.Braman N, Prasanna P, Whitney J, Singh S, Beig N, Etesami M, et al. Association of peritumoral radiomics with tumor biology and pathologic response to preoperative targeted therapy for HER2 (ERBB2)-positive breast cancer. JAMA Netw Open. 2019;2(4):e192561. doi: 10.1001/jamanetworkopen.2019.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Braman NM, Etesami M, Prasanna P, Dubchuk C, Gilmore H, Tiwari P, et al. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res. 2017;19(1):57. doi: 10.1186/s13058-017-0846-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu J, Sun X, Wang J, Cui Y, Kato F, Shirato H, et al. Identifying relations between imaging phenotypes and molecular subtypes of breast cancer: model discovery and external validation. J Magn Reson Imaging. 2017;46(4):1017–1027. doi: 10.1002/jmri.25661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y, Tang W, Liu W, Li R, Wang Q, Shen X, et al. Multiparametric MR imaging radiomics signatures for assessing the recurrence risk of ER + /HER2 − breast cancer quantified with 21‐Gene Recurrence Score. Journal of Magnetic Resonance Imaging. 2022. [DOI] [PubMed]
- 50.Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29(3):162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 51.Griethuysen JJMv, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Research. 2017;77(21):e104-e7. [DOI] [PMC free article] [PubMed]
- 52.Delli Pizzi A, Chiarelli AM, Chiacchiaretta P, d’Annibale M, Croce P, Rosa C, et al. MRI-based clinical-radiomics model predicts tumor response before treatment in locally advanced rectal cancer. Sci Rep. 2021;11(1):5379. doi: 10.1038/s41598-021-84816-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Delli Pizzi A, Chiarelli AM, Chiacchiaretta P, Valdesi C, Croce P, Mastrodicasa D, et al. Radiomics-based machine learning differentiates “ground-glass” opacities due to COVID-19 from acute non-COVID-19 lung disease. Scientific Reports. 2021;11(1). [DOI] [PMC free article] [PubMed]
- 54.Trebeschi S, Drago SG, Birkbak NJ, Kurilova I, Calin AM, Delli Pizzi A, et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann Oncol. 2019;30(6):998–1004. doi: 10.1093/annonc/mdz108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Magidson J, editor Correlated component regression: re-thinking regression in the presence of near collinearity2013 2013. New York, NY: Springer.
- 56.Traverso A, Wee L, Dekker A, Gillies R. Repeatability and reproducibility of radiomic features: a systematic review. International Journal of Radiation Oncology, Biology, Physics. 2018;102(4):1143–1158. doi: 10.1016/j.ijrobp.2018.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abdi H, Williams LJ. Partial least squares methods: partial least squares correlation and partial least square regression. Computational Toxicology. Methods in Molecular Biology2013. p. 549–79. [DOI] [PubMed]
- 58.Wold S, Ruhe A, Wold H, Dunn IWJ. The collinearity problem in linear regression. the partial least squares (PLS) approach to generalized inverses. SIAM Journal on Scientific and Statistical Computing. 1984;5(3):735–43.
- 59.Filzmoser P, Liebmann B, Varmuza K. Repeated double cross validation. Journal of Chemometrics. 2009;23(4):160–171. doi: 10.1002/cem.1225. [DOI] [Google Scholar]
- 60.Liu R, Gillies DF. Overfitting in linear feature extraction for classification of high-dimensional image data. Pattern Recognition. 2016;53:73–86. doi: 10.1016/j.patcog.2015.11.015. [DOI] [Google Scholar]
- 61.Chiarelli AM, Croce P, Assenza G, Merla A, Granata G, Giannantoni NM, et al. Electroencephalography-derived prognosis of functional recovery in acute stroke through machine learning approaches. International Journal of Neural Systems. 2020;30(12):2050067. doi: 10.1142/S0129065720500677. [DOI] [PubMed] [Google Scholar]
- 62.Kearns M, Ron D. Algorithmic stability and sanity-check bounds for leave-one-out cross-validation. Neural Computation. 1999;11(6):1427–1453. doi: 10.1162/089976699300016304. [DOI] [PubMed] [Google Scholar]
- 63.Krstajic D, Buturovic LJ, Leahy DE, Thomas S. Cross-validation pitfalls when selecting and assessing regression and classification models. Journal of Cheminformatics. 2014;6(1):10. doi: 10.1186/1758-2946-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Geyer CE, Tang G, Mamounas EP, Rastogi P, Paik S, Shak S, et al. 21-gene assay as predictor of chemotherapy benefit in HER2-negative breast cancer. NPJ Breast Cancer. 2018;4:37. doi: 10.1038/s41523-018-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mamounas EP, Russell CA, Lau A, Turner MP, Albain KS. Clinical relevance of the 21-gene Recurrence Score® assay in treatment decisions for patients with node-positive breast cancer in the genomic era. NPJ Breast Cancer. 2018;4:27. doi: 10.1038/s41523-018-0082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baltzer PA, Benndorf M, Dietzel M, Gajda M, Camara O, Kaiser WA. Sensitivity and specificity of unenhanced MR mammography (DWI combined with T2-weighted TSE imaging, ueMRM) for the differentiation of mass lesions. Eur Radiol. 2010;20(5):1101–1110. doi: 10.1007/s00330-009-1654-5. [DOI] [PubMed] [Google Scholar]
- 67.Pinker K, Moy L, Sutton EJ, Mann RM, Weber M, Thakur SB, et al. Diffusion-weighted imaging with apparent diffusion coefficient mapping for breast cancer detection as a stand-alone parameter: comparison with dynamic contrast-enhanced and multiparametric magnetic resonance imaging. Invest Radiol. 2018;53(10):587–595. doi: 10.1097/RLI.0000000000000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are not publicly available due to the clinical and confidential nature of the material but can be made available from the corresponding author on reasonable request.