Figure 1.
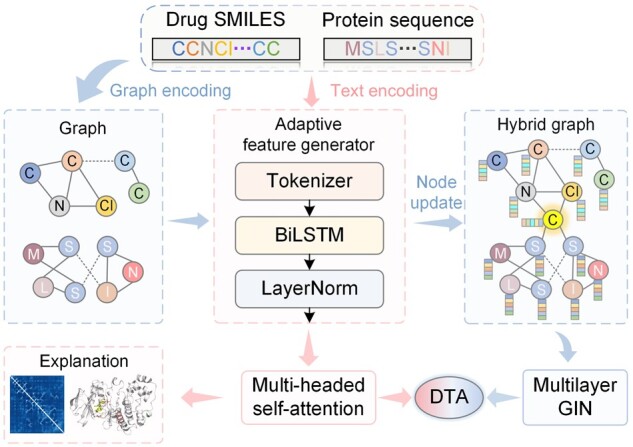
The flowchart of NHGNN-DTA for drug design. First, the graphs of drugs and proteins and the features of drug atoms and protein amino acids are obtained through graph and text encoding. The text embeddings are fed into the adaptive feature generator, processed through the tokenizer, BiLSTM and LayerNorm layers, and fed into the multi-head self-attention layer, which outputs sequence-based DTA predictions and interpretability. During this process, the feature generator adaptively obtains normalized node feature updates. We combine the graphs of drugs and proteins into a hybrid graph with a unique central node, and blend in adaptive node features, which are then fed into a multilayer GIN to obtain graph-based DTA predictions. Finally, the sequence-based and graph-based prediction results are integrated to obtain the final DTA prediction.
