Abstract
Aesthetics for the visualization of biomolecular structures have evolved over the years according to technological advances, user needs, and modes of dissemination. In this article, we explore the goals, challenges, and solutions that have shaped the current landscape of biomolecular imagery from the overlapping perspectives of computer science, structural biology, and biomedical illustration. We discuss changing approaches to rendering, color, human-computer interface, and narrative in the development and presentation of biomolecular graphics. With this historical perspective on the evolving styles and trends in each of these areas, we identify opportunities and challenges for future aesthetics in biomolecular graphics that encourage continued collaboration from multiple intersecting fields.
The structural biology community were early adopters of computer graphics, driven by the need to visualize and explore the complex three-dimensional shapes of biological molecules such as proteins and DNA. This is no less true today, and structural biologists rely on an advanced suite of molecular graphics tools as a central part of their research pipeline, as well as for dissemination and outreach. Over the 50 or so years from the first biomolecular visualization to the rich graphics environment today, the aesthetics of biomolecular graphics have changed and matured, driven by multiple orthogonal demands. In the 1960s, the hardware was often difficult to access and software was limited, so the field of biomolecular visualization was limited to a small community of experts who developed software and provided access to methods for the production of visual materials. Imagery was strongly influenced by the hardware: lines and points in the interactive Evans and Sutherland MultiPicture System, monochrome raster screens, and pen plotters. As consumer hardware continued to improve and the structural biology community became increasingly convinced of the utility of biomolecular graphics, an explosion of method development ensued, and the best techniques were adopted and made more user friendly. Throughout this period of development, the applications and modes of dissemination strongly shaped the aesthetics of the methods and the visualizations that were produced (Figure 1). In this article, we explore several aspects of visualization aesthetics and how the changing computer graphics environment has shaped the imagery that we see today.
FIGURE 1. Biomolecular Graphics Development.
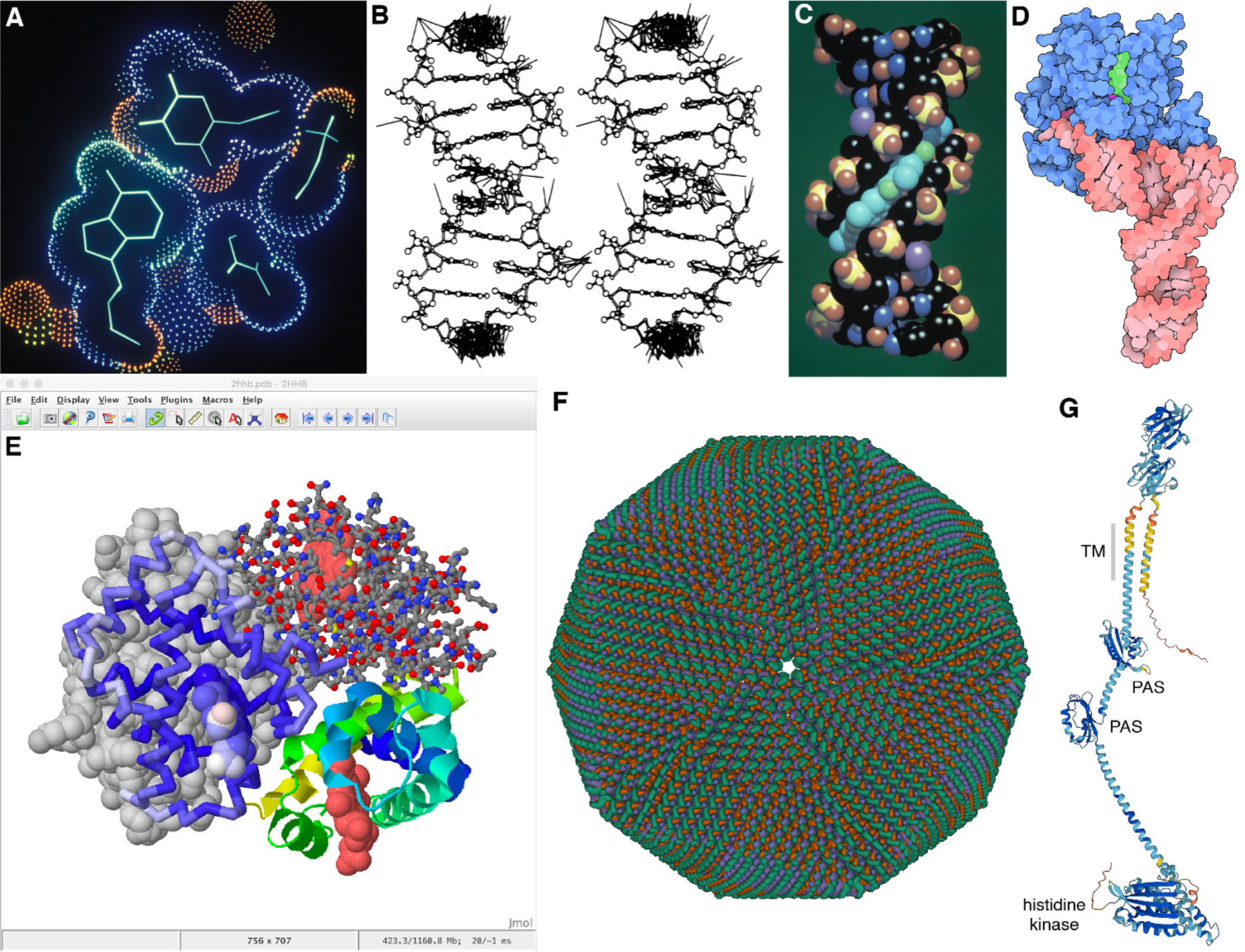
Images created over a span of three decades. (A) Dot surface and wireframe for a complex of DNA with an inhibitor, displayed interactively on an Evans and Sutherland MultiPicture System with custom software (PDB ID 6bna). (B) Wireframe with extra lines to depict lattice contacts, created with a modified version of ORTEP. Printed using a pen plotter and presented in publication as a stereo pair for 3D viewing (PDB ID 126d). (C) Raster space-filling image of DNA (atomic colors) and inhibitor (cyan) with Phong shading, created with several hours of computation with custom software (PDB ID 6bna). (D) Non-photorealistic rendering of tRNA and elongation factor created with custom software (PDB ID 1ttt). (E) Natural language scripting of Jmol allows nimble, interactive access to multiple rendering options, as seen in this fanciful image of hemoglobin in four styles created with ten minutes of effort (PDB ID 2hhb). (F) Mol* is leading a new generation of web-based biomolecular viewers, here providing interactive exploration of faustovirus (PDB ID 5j7v), currently the largest structure in the Protein Data Bank. (G) Predicted structure of PleC, a protein involved in formation of a bacterial microdomain, with most confident regions in dark blue and least confident regions in orange (AlphaFold2 ID AF-P37894-F1).
Rendering approaches leverage the strengths of graphics hardware
Rendering approaches for biomolecular data have evolved with advancements and expansions in capabilities of graphics hardware and software. ORTEP (Oak Ridge Thermal-Ellipsoid Plot Program1), developed in 1965 out of Oak Ridge National Laboratory, dominated protein crystallography for many years due to widespread use of pen plotters to create publication-quality images. These images have a beautiful economy of line (Figure 1B).
With higher-powered graphics hardware, more photorealistic rendering styles were increasingly used for biomolecular graphics. Photorealistic rendering techniques aim to mimic the look of real-life objects as closely as possible. Interestingly, biomolecular structures measure below the wavelength of visible light, so in some sense the term “photorealism” is, strictly speaking, a misnomer in this context. However, more advanced simulation of illumination serves to make biomolecular structures possibly more relatable by embedding them in a larger space with light and shadow.
During the 1990s, amidst a large body of work in graphics on exploring non-photorealistic rendering techniques which mimic the aesthetics of hand-drawn artwork, these approaches also became popular for depicting biomolecular structures. Toon shading, also known as cel shading, for instance, refers to a class of non-photorealistic rendering approaches inspired by cartoons and comics and their use of shades and tints. This is a relatively inexpensive rendering style from a computational perspective, and is visually simplistic while conveying the information necessary to understand the structural arrangement of the molecules in question. Goodsell’s illustrative style is an attempt to capture some of the visual effectiveness of ORTEP images (Figure 1D). Today, toon shading is readily accessible in interactive graphics, and provided by, for example, Mol* and Protein Explorer.
Other rendering approaches have also been developed to mirror the appearance of structures under a given acquisition method, i.e., scanning electron microscopy (SEM) (Figure 2, Left). This method applies a fresnel effect to a basic Phong shader, where structures appear to have a white outline that becomes thicker as the view angle becomes more oblique to the eye. This “SEM” shading technique has largely fallen out of fashion. However, when in use, the fresnel effect is usually more subtle, e.g., the CDC’s visualization of the COVID-19 virion, crafted by Alissa Eckert and Dan Higgins2. Ambient occlusion (AO) in rendering output provides helpful depth cues to biomolecular structures and has become more prevalent with better and more efficient hardware and software for computer graphics (Figure 2, Middle). With ambient occlusion, a separate rendering pass calculates the exposure of each point on an object to ambient light. For example, the cavity of a molecule appears darker because it is more occluded from light. With improved hardware and software solutions enabling physically-based rendering in most 3D applications, computationally “expensive” features like global illumination, subsurface scattering, and clearcoat are more accessible and frequently used (Figure 2, Right). Molecules with this treatment can have a “gummy bear” appearance that is more editorial than strictly educational, and has been especially popular in editorial graphics, for example, in stylized illustrations for journal covers or in marketing materials for pharmaceutical products.
FIGURE 2.
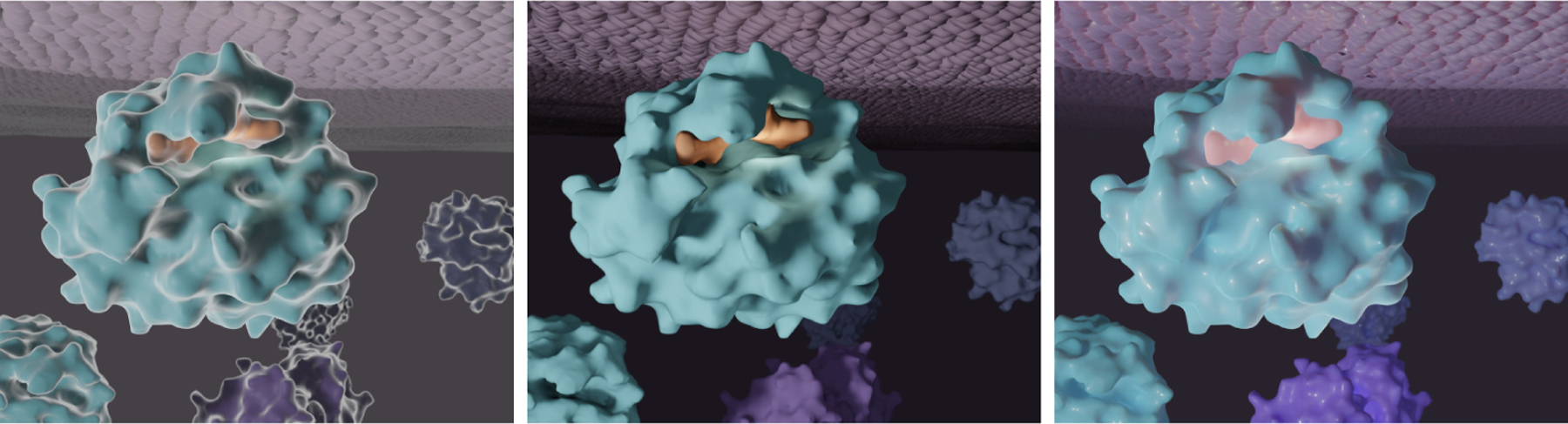
Demonstration of three common historical and contemporary biomolecular graphics rendering styles on a basic biomolecular scene with a ligand bound to a receptor on the interior of a membrane: (Left) “Scanning electron microscope (SEM)-look” material, (Middle) Ambient occlusion (AO) layered with a matte material, and (Right) Subsurface-scatter and clearcoat layers applied to a physics-based rendering material.
Today, advancements in science and technology pose an entirely new set of rendering challenges, providing new capabilities and requiring new aesthetic decisions. Virtual reality and augmented reality are finally becoming a useful tool in research and education, driven by the increased affordability of consumer-level hardware. Currently, the need for seamless response during navigation imposes limitations on the complexity of VR scenes, requiring artists and developers to pare down the rendering options for objects being depicted. 3D printing has also benefited from affordable options, now available to hobbyists and classrooms. Most commercial machines are limited to rigid, monochrome builds, but clever designs with snap-together parts and magnets are used to expand the types of stories that can be captured in the models. 3D models also impose strict limitations on the types of representations that may be used, to ensure that the model is buildable and strong enough to handle.
Researchers are utilizing an increasingly nuanced approach to color
Choice of color is arguably one of the most impactful decisions made during the creation of a biomolecular visualization. Coloring strategies can draw viewer attention to key features or molecules of interest, or encode physical or functional features of the molecule to aid in exploration and analysis, such as a ligand in a pathway or binding site of a receptor molecule. These perceptual tricks may include coloring key molecules in light or highly saturated colors that contrast with the surrounding molecules and environment. Here, the use of color is more often about drawing attention to the intended structures rather than encoding particular structural or functional properties of the molecules, for example, hydrophobicity.
Early approaches to color were largely driven by technology and nascent traditions. Color printing was expensive and predominant technologies were pen plotters and monochrome or 8-bit raster screens. Much of early computer-generated imagery was produced and published in black-and-white. As color became increasingly feasible, published imagery was often colored using default settings, leading to a predominance of saturated colors. The CPK coloring scheme popularized by Linus Pauling and his physical molecular models (carbon black, oxygen red, nitrogen blue, hydrogen white, Figure 1C), was the de facto standard for most research graphics. Interestingly, developers immediately encountered a problem as color interactive hardware became available: how to deal with black carbons on a black screen. The most common approach at the time, which was still quite wedded to saturated color, was to use green.
As color screens became more common in laboratories, an explosion of experimentation followed, leading to multiple color palettes for specific needs. Some of these are now only rarely-used, such as amino-acid-specific colors based on the “Shapely” physical models, whereas others showed widespread utility and are provided as hard-wired options in most current packages. These include coloration of entire biomolecular subunits (Figure 1D), coloring of properties such as electrostatics and hydrophobicity, and coloration that highlights local structural features such as protein secondary structure or position in a polymer chain (Figure 1E).
Today, we enjoy an environment that is filled with options for coloring, and a great freedom to customize coloring based on our personal ideas and preferences. Current molecular graphics software typically provide a menu of traditional options with more-or-less standardized color choices, paired with flexible methods for selecting atom sets and assigning custom colors to them. Given this ability, color is frequently used, especially by biomolecular animators and designers, to evoke a variety of moods or feelings when viewing a molecule or scene, and the choice of palette may be tuned to appeal to the intended audience and use case. Current research in coloring aesthetics is layering advanced capabilities on these basic coloring choices. For example, the Viola laboratory is exploring methods to transition smoothly from molecule-based coloring to atomic coloration in multiscale systems as viewers transition from whole-cell views to individual molecules (Figure 3).
FIGURE 3. Adaptive multiscale representation and coloring.
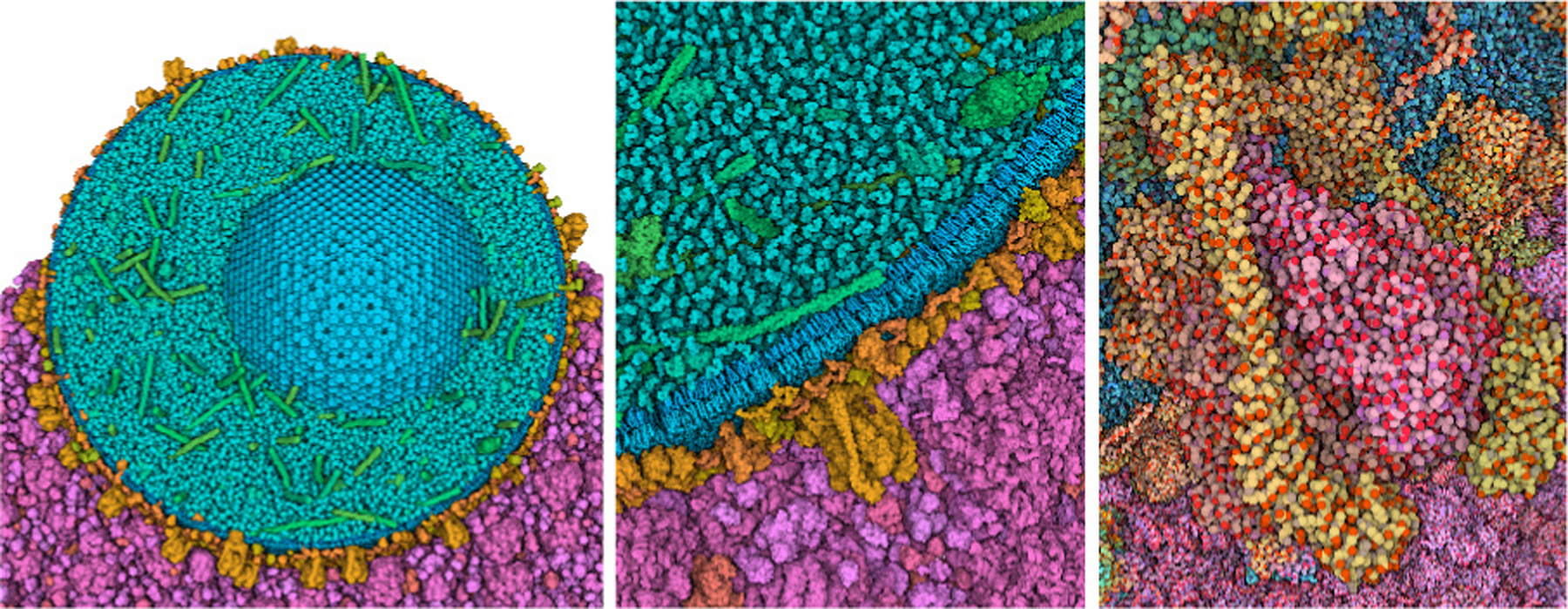
Model of an insulin secretory granule (blue, green and orange) and cytoplasm (magenta) is displayed with subunit colors and coarse surfaces at left. As the user zooms in, the view progressively changes to a full atomic representation with atomic color (right). Images by Ludovic Autin.
We have observed a growing interest in the biomolecular visualization community to standardize coloring schemes. This has great advantages. For example, it would greatly aid with issues of accessibility, by promoting the availability and use of colorblind-friendly palettes. Standards also unify a field. For example, the CPK coloring scheme, given its excellent provenance and widespread use, is instantly recognizable by most viewers, allowing facile comparison when viewing figures from multiple labs. The confidence coloring scheme currently used in AlphaFold2 structures (Figure 1G) is rapidly becoming a similar de facto standard. Standards, however, can only codify current knowledge and thus may potentially inhibit creative exploration as the field of structural biology continues to grow. Current molecular graphics tools typically express both of these views, providing turnkey methods to apply the (currently) most useful standards while also streamlining the ability to develop customized palettes.
Biomolecular imagery has grown to encompass larger narratives and personal styles
Biological stories are growing larger to span new experimental results from atoms to cells, and visualization options provide myriad opportunities for building new and effective visual explorations. When designing and executing these stories, we are always faced with three orthogonal challenges: (1) technical capabilities of turning data into images, (2) the needs of the intended audience, and (3) our own personal artistry. Much of the early history of biomolecular visualization was centered around the creation of figures for research publications. These were effectively “molecular portraits” that presented the structure and, hopefully, some aspects of their function. These portraits were created with a handful of programs, and most often did not stray far from the default coloring and rendering options.
Today, the audiences for biological stories have expanded. Education and more general outreach, e.g., public exhibitions, have pushed content authors to incorporate narrative devices to make molecules and their environments more engaging and comprehensible to broader audiences. These tend to require artistic expertise and knowledge of advanced 3D software, such as Blender, Autodesk Maya, or Maxon Cinema 4D. Pharmaceutical company growth and marketing initiatives for new drug developments have helped drive cinematic storytelling approaches to biomolecular reactions and pathways. New software solutions, like Molecumentary [1], allow content authors to semi-automatically create tailored narratives to disseminate scientific content. Tools like this enable users without deep expertise in 3D animation and design to create narratives for education and outreach.
The narratives themselves have also grown larger, with an explosion of new big data and methods for accessing and visualizing these data. Continued advances in structure determination methods, such as the current resolution revolution in cryoelectron microscopy, and recent advances in protein structure prediction, such as AlphaFold2 and RoseTTAFold, are radically increasing the number and complexity of atomic structures that are available for detailed depiction of biomolecular structures. Visualization methods are faced with depiction of larger and larger data sets, interactively, on the web. We no longer can limit ourselves to presenting only a basic portrait of a molecule. We now need to explore uncertainty in predicted structure models (Figure 1G) and the dynamic aspects of single molecules, assemblies, and ensembles. Biomolecular dynamics simulations capture a vast amount of information where only a few time slices may be of interest to the viewer. Choosing how to display this information in a digestible way requires multiscale thinking in both time and space. Connections to sequence and functional annotations need to be at our fingertips to explore bioinformatics data related to our subjects.
The current software and hardware environment for biomolecular visualization is robust and provides nimble opportunities for building a personal aesthetic within this data-rich environment. Figure 4 includes four pioneering artists who have helped shape current trends in biomolecular graphics design and animation. Drew Berry pioneered a cinematic style combining dynamic scene design, a unique approach to biomolecular motion, and immersive sound in his animations for general audiences. Gaël McGill has perfected an editorial style with design decisions that produce arresting images for textbook and commercial applications. For example, the image in Figure 4 has circulated for a decade on social media as “the most detailed depiction of a cell.” Janet Iwasa creates data-heavy imagery in collaboration with researchers for use in publication and presentations, and has developed a direct style that is true to the science. Veronica Falconieri Hays creates dynamic portraits of molecules using contemporary methods for scientific illustration, often framed within larger stories that show their cellular context.
FIGURE 4. Large-scale narratives and personal aesthetics.
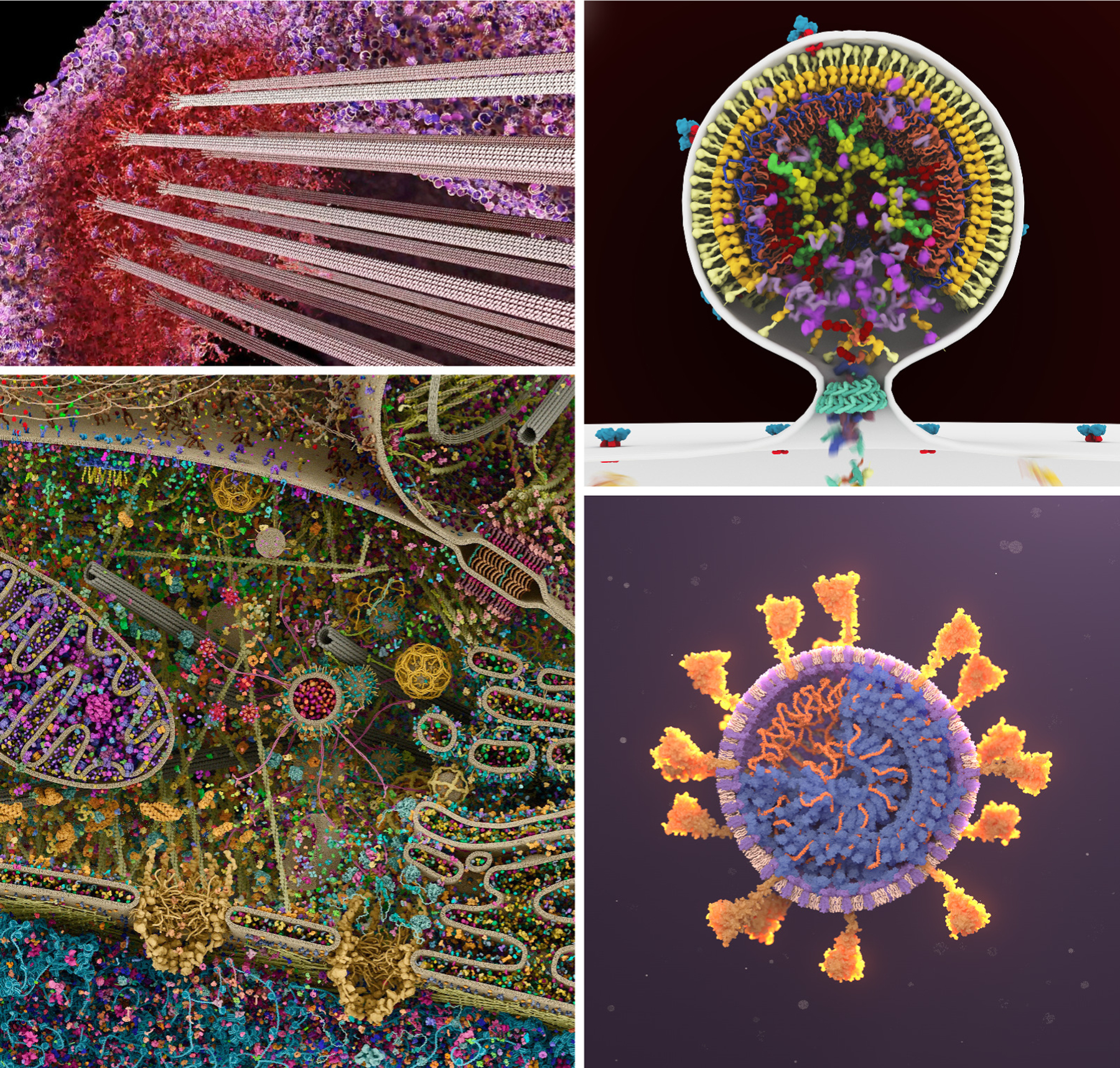
Work from four contemporary artists shows state-of-the-art design decisions in complex biomolecular systems. Notice in each case how the approach to color and lighting is tuned for the audience, and representations are chosen to depict complexity at a level appropriate for the scene. (Upper left) Drew Berry creates groundbreaking video animations for general audiences with station WEHI.TV, such as this still image from a video animation of the kinetochore. (Lower left) Gaël McGill of Digizyme and Harvard Medical School creates dynamic editorial images and animations for textbooks and commercial clients, such as this complex image of the interior of a living cell created with Digizyme team member Evan Ingersoll. (Upper right) Janet Iwasa is a pioneer in the creation of data-rich animations for dissemination of information in research settings, such as this animation of budding of HIV-1 from an infected cell. (Lower right) Veronica Falconieri Hays is a Certified Medical Illustrator who creates captivating editorial and educational imagery and animations with beautiful interplay of color and light, as in this SARS-CoV-2 illustration.
Outlook and Challenges
Molecular graphics is mature, but still offers ample opportunities for research and software development in graphics and visualization, to address current limitations and challenges. Amazingly, most widely-used tools with a significant visualization component still focus on fairly basic rendering/aesthetic approaches, and the vast majority of users will employ the default rendering and coloring when using these tools. As the field of biomolecular graphics becomes more mature with tools for creating content more widely available, our community could consider careful attention to defaults and presets provided by common methods. Additional guidance could support users with limited artistic skills to navigate the vast design space when creating biomolecular graphics. This is particularly important, as methods and tools are increasingly used by diverse user communities outside of structural biology, which also necessitates more extensive conventions that facilitate our understanding of molecules and their environments. To address these challenges, programs like Chimera and Mol* currently provide one-click preset views for different applications. The VIS and graphics community have begun building dedicated applications (such as MegaMol, CellPAINT, and Marion) that are more oriented to the needs of artists and are more specialized than generic graphics applications like Blender and Maya.
Accessibility of images is also becoming a stronger design specification when creating imagery, particularly in settings for non-technical audiences. Colorblind-friendly color palettes have emerged as a low-hanging fruit, and visualization tools are increasingly adopting default options that are distinguishable to those with limited vision. While this is a good first step, we need to do more to make biomolecular graphics accessible to segments of the population. Sound engineering can help engage and immerse viewers in an environment, but this can go further to encode real meaning to molecules for those with low or limited vision. Thinking about how to integrate audio and other sensory modalities to enhance the accessibility of biomolecular graphics–sound, haptics, physical models–can be critical to advance public understanding of molecules and their relevance in society.
There are still abundant areas that would benefit from creative development. For example, on the data-in side, cryo-EM is currently providing structural views of biomolecular assemblies of unprecedented complexity. Often these structures have a hierarchical organization, with multiple biologically-relevant entities in one assembly. Currently, this hierarchical structure is difficult to define within existing graphics methods, requiring laborious manual selection and coloring of individual chains (Figure 5, Top). A more facile connection to functional annotations will be needed to address this problem. Another challenge lies in visual depiction of the underlying uncertainty of the model(s) responsible for the biomolecular graphic, and displaying the provenance of the data. While color-coding areas of confidence is a common approach to the former (Figure 1G), aesthetic and easily identifiable display of such information is by no means a solved problem. Data provenance for dynamic data is another area of great need and current creative effort. Successful current approaches often use a dashboard-style visualization, where a surface model of the molecule is paired with other views that show key aspects of the structure and underlying data at each time step of the simulation (Figure 5, Bottom). On the data-out side, the fields of virtual reality and 3D printing are still very much the Wild West, and creative approaches to aesthetics and design can lead quickly to effective results.
FIGURE 5. (Top) Hierarchical nature of large biomolecular assemblies.
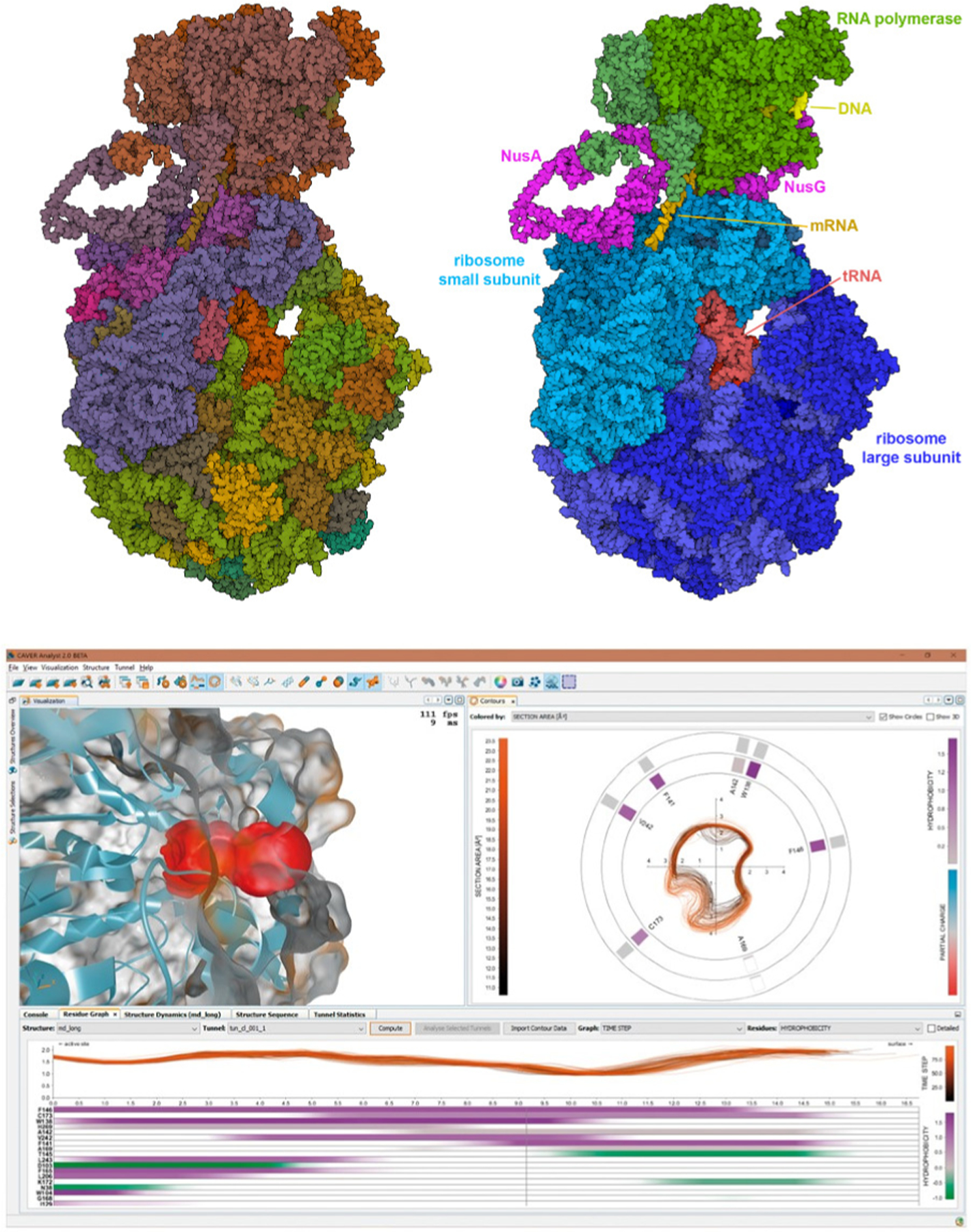
The expressome (PDB ID 6×9q) includes RNA polymerase and a ribosome tethered together by two processivity factors, NusA and NusG. This cryo-EM structure also includes a small fragment of DNA, a messenger RNA, and two tRNA molecules. Both figures are rendered interactively with Mol* at the RCSB PDB website, with user-tunable options for ambient occlusion, outlines, and flat shading. The left figure is colored using a default option based on chain instances in the coordinate file. The right figure is colored manually by selecting individual chains and choosing colors to highlight the structural hierarchy of biologically-relevant sub-assemblies. (Bottom) CAVER Analyst 2.0 software application [2]. This system enables real-time visualization of tunnels and channels in biomolecular structures. The upper left panel depicts a molecular structure with a highlighted region of interest, in this case, a tunnel, with the accompanying right panel showing a cross-cut contour of the highlighted tunnel. Each contour indicates a time step from the underlying long MD trajectory. The bottom panel depicts the tunnel radius in profile along its length, with variation over time (each line is one time step) and the amino acids that form the boundary of the tunnel, ranked according to their influence on the tunnel’s boundary. A consistent color design is used throughout to help researchers understand connections between data in the separate panels.
Molecular graphics, now and throughout its history, has been a multidisciplinary effort bringing together the talents of molecular biologists, computer scientists, and artists to build methods and imagery. This collaboration is continuing to expand to encompass growing fields of knowledge, for example, leveraging expert annotations from bioinformatics, best practices from perceptual science and science historians, and direct user feedback from educational evaluation experts.
Acknowledgments
This work was supported in part by grant GM120604 from the National Institutes of Health and the RCSB Protein Data Bank (National Science Foundation DBI-1832184, National Institutes of Health GM133198, and US Department of Energy DE-SC0019749).
Footnotes
Contributor Information
Laura A. Garrison, Mohn Medical Imaging & Visualization Centre, Haukeland University Hospital, 5021 Bergen, Norway; Bouvet ASA, Solheimsgaten 15, 5058 Bergen, Norway; Department of Informatics, University of Bergen, 5020 Bergen, Norway
David S. Goodsell, Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA; Research Collaboratory for Structural Bioinformatics Protein Data Bank, Institute for Quantitative Biomedicine, Rutgers Cancer Institute of New Jersey, Rutgers, The State University of New Jersey, New Brunswick, NJ 08903, USA
Stefan Bruckner, Institute for Visual and Analytic Computing, University of Rostock, Albert-Einstein-Straße 22, 18059 Rostock, Germany.
References
- [1].Kouril D, Strnad O, Mindek P, et al. , “Molecumentary: Adaptable narrated documentaries using molecular visualization,” IEEE Transactions on Visualization and Computer Graphics, 2021. [DOI] [PubMed] [Google Scholar]
- [2].Jurčík A, Bednar D, Byška J, et al. , “Caver analyst 2.0: Analysis and visualization of channels and tunnels in protein structures and molecular dynamics trajectories,” Bioinformatics, vol. 34, no. 20, pp. 3586–3588, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Olson AJ, “Perspectives on structural molecular biology visualization: From past to present,” Journal of Molecular Biology, vol. 430, no. 21, pp. 3997–4012, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Martinez X, Krone M, Alharbi N, et al. , “Molecular graphics: Bridging structural biologists and computer scientists,” Structure, vol. 27, no. 11, pp. 1617–1623, 2019. [DOI] [PubMed] [Google Scholar]
- [5].Kozlíková B, Krone M, Falk M, et al. , “Visualization of biomolecular structures: State of the art revisited,” in Computer Graphics Forum, Wiley Online Library, vol. 36, 2017, pp. 178–204. [Google Scholar]
- [6].Johnson GT and Hertig S, “A guide to the visual analysis and communication of biomolecular structural data,” Nature Reviews Molecular Cell Biology, vol. 15, no. 10, pp. 690–698, 2014. [DOI] [PubMed] [Google Scholar]
- [7].O’Donoghue SI, Goodsell DS, Frangakis AS, et al. , “Visualization of macromolecular structures,” Nature Methods, vol. 7, no. Suppl 3, S42–S55, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Goddard TD and Ferrin TE, “Visualization software for molecular assemblies,” Current Opinion in Structural Biology, vol. 17, no. 5, pp. 587–595, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Goodsell DS, “Visual methods from atoms to cells,” Structure, vol. 13, no. 3, pp. 347–354, 2005. [DOI] [PubMed] [Google Scholar]
- [10].Iwasa JH, “Bringing macromolecular machinery to life using 3d animation,” Current Opinion in Structural Biology, vol. 31, pp. 84–88, 2015. [DOI] [PubMed] [Google Scholar]


