Significance
The search for signs of life beyond Earth is a motivator in modern-day planetary exploration. While our “sister” planet Venus has a surface too hot for life, scientists have speculated that the much cooler atmosphere at 48 to 60 km above the surface might host life in Venus’ perpetual cloud cover, as Earth’s clouds do. The Venus clouds, however, are composed of concentrated sulfuric acid—an aggressive chemical that destroys most of Earth life’s biochemicals and are thought to be sterile to life of any kind. Here, we show that key molecules needed for life (nucleic acid bases) are stable in concentrated sulfuric acid, advancing the notion that the Venus atmosphere environment may be able to support complex chemicals needed for life.
Keywords: Venus, habitability, NMR, nucleic acid bases, sulfuric acid
Abstract
What constitutes a habitable planet is a frontier to be explored and requires pushing the boundaries of our terracentric viewpoint for what we deem to be a habitable environment. Despite Venus’ 700 K surface temperature being too hot for any plausible solvent and most organic covalent chemistry, Venus’ cloud-filled atmosphere layers at 48 to 60 km above the surface hold the main requirements for life: suitable temperatures for covalent bonds; an energy source (sunlight); and a liquid solvent. Yet, the Venus clouds are widely thought to be incapable of supporting life because the droplets are composed of concentrated liquid sulfuric acid—an aggressive solvent that is assumed to rapidly destroy most biochemicals of life on Earth. Recent work, however, demonstrates that a rich organic chemistry can evolve from simple precursor molecules seeded into concentrated sulfuric acid, a result that is corroborated by domain knowledge in industry that such chemistry leads to complex molecules, including aromatics. We aim to expand the set of molecules known to be stable in concentrated sulfuric acid. Here, we show that nucleic acid bases adenine, cytosine, guanine, thymine, and uracil, as well as 2,6-diaminopurine and the “core” nucleic acid bases purine and pyrimidine, are stable in sulfuric acid in the Venus cloud temperature and sulfuric acid concentration range, using UV spectroscopy and combinations of 1D and 2D 1H 13C 15N NMR spectroscopy. The stability of nucleic acid bases in concentrated sulfuric acid advances the idea that chemistry to support life may exist in the Venus cloud particle environment.
The search for signs of life is a key motivator in modern-day planetary exploration. Our “sister” planet Venus has long been relegated to the uninhabitable category because its surface at 735 K is too hot for any plausible solvent and for most organic covalent chemistry. Yet for over five decades, a small but growing group of scientists have continued to support the speculative idea that microbial-type life might permanently occupy the temperate cloud layers of Venus (1–14). The cloud layers are permanent, continuous across the planet, and vertically extensive (48- to 60-km altitude above the surface). The cloud layers have the main requirements for life (15): suitable temperatures for covalent bonds; a liquid environment; and an energy source. So the Venus cloud layers could, in principle, host systems with the key characteristics of life, including their ability to undergo Darwinian evolution, even if the specifics of their chemistry are very different from those of terrestrial life.
The concept of life in the clouds of Venus is both motivated and supported by the fact that Earth has an aerial biosphere. A diverse set of microbes are transported up from the Earth’s surface (16–19) and remain aloft for on average 3 to 7 d (20) before being rained out. Earth’s clouds are both transient and fragmented—a challenging ecological niche for permanent habitation. Venus, in contrast, has a permanent and continuous cover from vertically extensive clouds. Venus microbial type life residing inside cloud particles could remain aloft indefinitely via a life cycle of droplet growth, sedimentation, sporulation to a dormant hibernation in a haze layer beneath the clouds, followed by upward mixing via gravity waves followed by cloud nucleation (6).
The Venus cloud particles are made of concentrated sulfuric acid as inferred by Pioneer Venus in situ measurements of the backscattered polarized radiation (21). Technically, the measurements yield the particle refractive index and particle size assuming spherical particles. These measurements confirmed earlier polarization observations from Earth-based telescopes (22). The concentration of sulfuric acid in the Venus cloud particles has not been directly measured, but inferred from Pioneer Venus measurements of gases by the mass spectrometer. The gases evolved from cloud particles that clogged the inlet and are consistent with cloud droplets composed of 85% w/w H2SO4 and 15% w/w H2O (23). Based on models, it is likely that the sulfuric acid concentration of the cloud particles varies with altitude, in the cloud tops reaching 79% w/w while in the lower clouds the sulfuric acid concentration could reach 98% w/w (24).
Two potential show-stoppers exist for the survival of life in the clouds of Venus. First, there is nearly no water available, as the atmosphere is extremely dry and the cloud sulfuric acid droplets have very low water activity. Any water in the sulfuric acid droplets is locked away in strong hydrogen bonds to sulfuric acid. Second, the concentrated sulfuric acid is an aggressive solvent and is thought to destroy most biochemicals (25). Indeed, concentrated sulfuric acid is orders of magnitude more acidic than the most acidic environments on Earth that host acid-adapted microorganisms (6). The general accepted viewpoint is that concentrated sulfuric acid is sterile for any interesting chemistry and for any life (26).
1. Motivation to Study Venus Sulfuric Acid Organic Chemistry
We promote the concept that the concentrated sulfuric acid droplets may support a rich organic chemistry of a kind that might be able to support life different from Earth life. Spacek and Benner (27–29) have shown that a rich organic chemistry in sulfuric acid evolves from a simple organic seed molecule such as formaldehyde [and propose that such organic chemistry may be the origin of the perplexing and unidentified material in Venus’ atmosphere that absorbs 50% of all incident ultraviolet (UV) sunlight (30), the so-called “unknown UV absorber” (31)]. Even gas phase CO and CO2 can act as seed molecules, themselves originating from atmospheric photochemical processes. The experiments by Spacek and Benner use a precursor molecule containing only C, H, and O elements and yet even such limited elemental composition can yield complex organic molecules. The Venus clouds may have a much more diverse organic chemistry than these laboratory models, because the clouds also contain molecules that have N, S, or even P atoms (32).
The idea of a rich organic chemistry in concentrated sulfuric acid has long been known outside of planetary science. The oil refinement industry, for example, uses concentrated sulfuric acid to process crude oil. As a byproduct, “red oil” waste product is generated which includes a diversity of organic compounds including aromatic ringed molecules dissolved in concentrated sulfuric acid (33–35).
Complex organic chemistry is of course not life, but there is no life without it. We can consider the potential stability of complex organic chemistry in an environment as a prerequisite to habitability. Thus, we are motivated to study the stability of complex molecules that may make up a biochemistry of non-Earth-like life in concentrated sulfuric acid to explore whether sulfuric acid is inherently uninhabitable to all possible life, rather than just to terrestrial life. While the Venus cloud droplets likely contain dissolved substances that might influence reactivity, such as atmospheric gases and metal ions, pure concentrated sulfuric acid is a foundational starting point for investigating stability of chemicals needed for life to function.
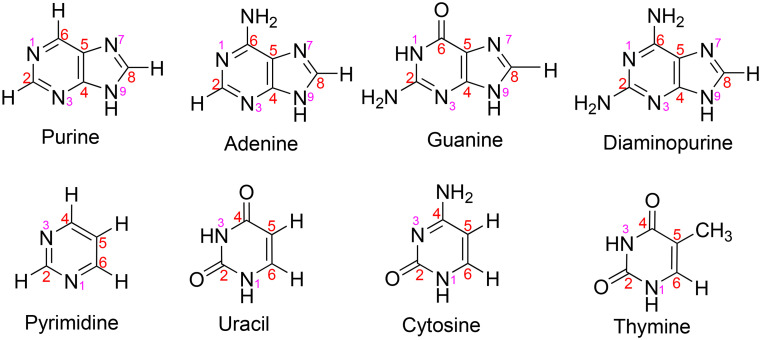
Specifically, we are motivated to study the stability of the components that could make up an informational polymer, one that would be very different from DNA or RNA on Earth due to the overall instability of DNA or RNA in concentrated sulfuric acid. We focus on nucleic acid bases and related molecules (Fig. 1) as a starting point and consider sulfuric acid concentrations and temperatures found in the Venus cloud layers. Studies from decades ago explored not only stability and chemistry of aromatics in concentrated sulfuric acid (e.g., refs. 36–38) but also, curiously, some isolated experiments on protonation of a few of the nucleic acid bases in acidic conditions (39–42).
Fig. 1.
The nucleic acid bases and related molecules studied in concentrated sulfuric acid in this work. Red numbers indicate carbon atoms and pink numbers indicate nitrogen atoms in the ring.
We describe our experimental results followed by contextual discussion, then present the methods.
2. Results
We find that nucleic acid bases adenine, cytosine, guanine, thymine, and uracil are stable in sulfuric acid ranging from 81 to 98% concentration by weight (the rest being water) at room temperature (18 to 21 °C). We also find the stability under the same conditions of the core structure of the nucleic acid bases purine and pyrimidine. We additionally tested the adenine-like compound 2,6-diaminopurine, used as a genetic base substitute for adenine by specialized viruses (43–46), and found it stable as well.
By stable, we mean no detectable reactivity and degradation of the tested compound after incubation in concentrated sulfuric acid up to 2 wk. We first analyzed the compounds after 18 to 24 h in concentrated sulfuric acid and next repeated one of the analyses after a 2-wk incubation period for an additional stability test. By stable we also mean long-term stability of the heavy atom bonding topology in the bases, i.e. the bonding of nonhydrogen atoms to each other, and not rapid, reversible exchange of protons between the bases and the solvent.
We begin with evidence that the aromatic compound ring structure is not broken, using data from both UV spectroscopy (Section 2.1, Fig. 2) and 13C NMR (Section 2.2, Figs. 3 and 4). In order to demonstrate stability of a given molecule in concentrated sulfuric acid, we elucidate the molecular structure using a combination of 1H NMR, 13C NMR, 15N NMR, and/or 2D heteronuclear multiple-quantum correlation spectroscopy (HMQC) and heteronuclear multiple bond correlation spectroscopy (HMBC) (1H-13C and 1H-15N) NMR (Section 2.3 and Figs. 5 and 6).
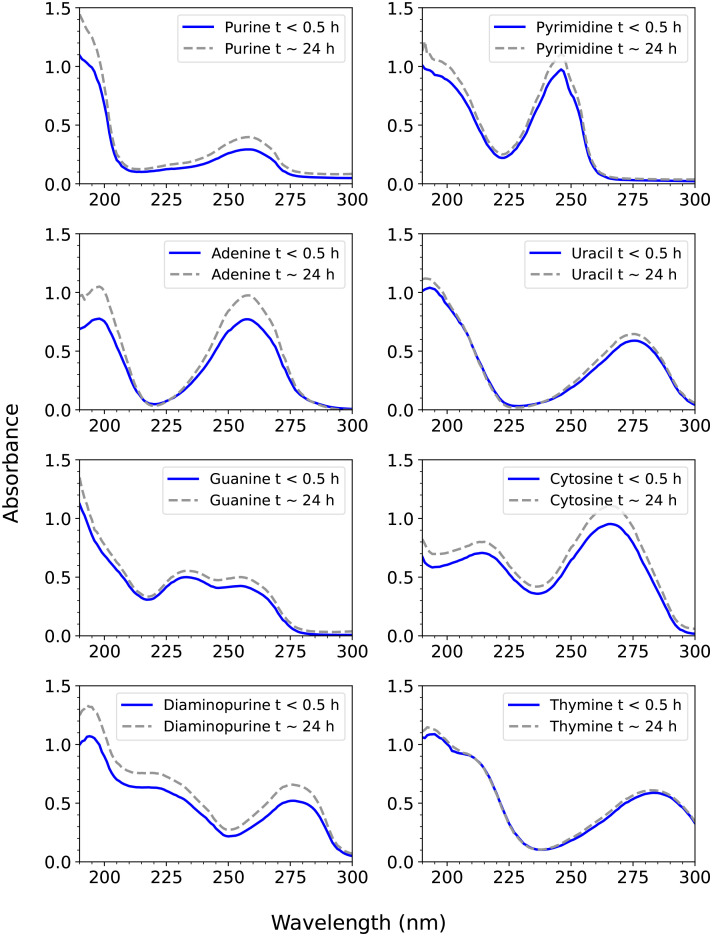
Fig. 2.
UV spectroscopy of the eight compounds studied in 98% w/w sulfuric acid. The absorbance, defined as A = εLc, where A is absorbance (dimensionless), ε is the molar absorption coefficient (in units of M−1 cm1), L is pathlength (in cm), and c is concentration (in units of M), as a function of wavelength. Each compound shows two characteristic UV peaks, due to π-π conjugated bonds. The blue line shows the UV spectrum measured within about 15 to 20 min after mixing of the compound in 98% w/w H2SO4 in H2O and the gray dashed line is the same compound measured after about 24 h. While some compounds have a higher absorbance due to more dissolution over the 24 h, the same peak wavelength maximum and peak shape demonstrates stability of each compound in 98% w/w sulfuric acid.
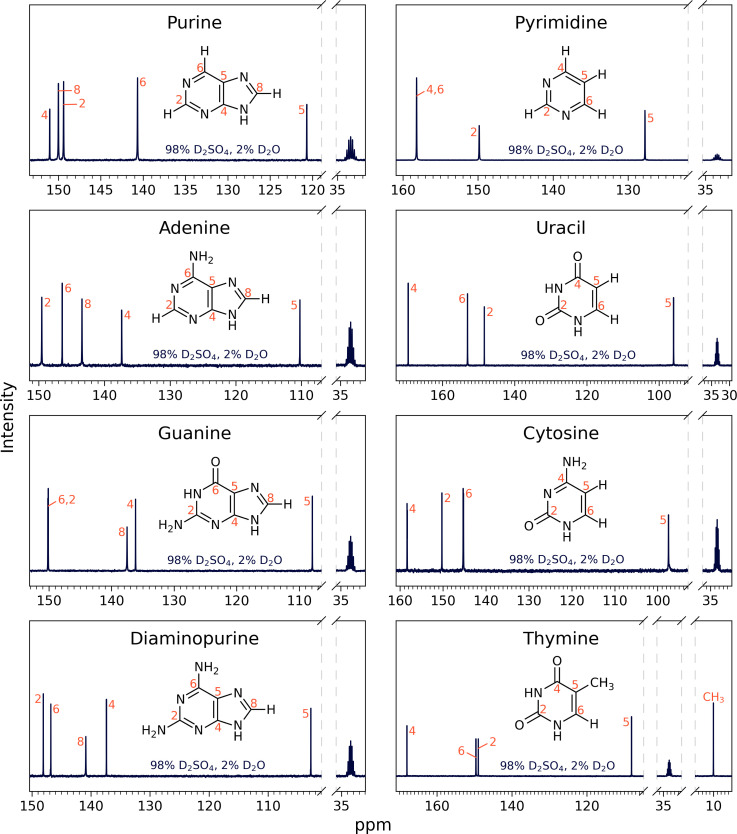
Fig. 3.
13C NMR spectra for eight nucleic acid bases: purine, adenine, guanine, diaminopurine, pyrimidine, uracil, cytosine, and thymine in 98% D2SO4/2% D2O (by weight) with DMSO-d6 as a reference, at room temperature. The labeled NMR carbon peaks match the number of carbon atoms in the known molecular structure for a given compound. All peaks are consistent with the molecules being stable and the structure not being affected by the concentrated sulfuric acid solvent. For a description of peak assignments, see Section 2.3., Figs. 5 and 6, and SI Appendix, Figs. S1–S10.
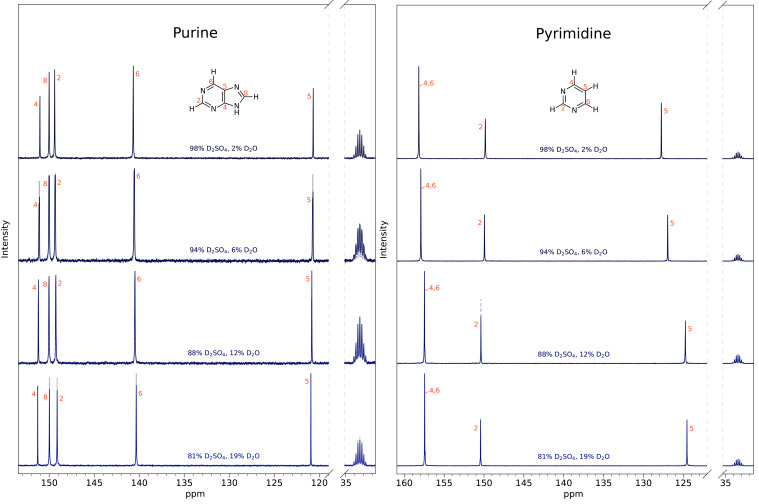
Fig. 4.
Purine (Left) and pyrimidine (Right) are stable for 2 wk in a range of sulfuric acid concentrations found in the Venus clouds. We have incubated 30 to 40 mg of each base in 81 to 98% w/w D2SO4 for 2 wk. After the 2-wk incubation, we measured 1D 13C NMR spectra (solid line spectra), at each of the tested acid concentrations, and compared them to the original 1D 13C NMR spectra collected after ~30 to 48 h (dashed line spectra and SI Appendix, Fig. S11). The 2-wk spectra and the ~30 to 48-h spectra look virtually identical for all tested concentrations, confirming long-term stability of the compounds in concentrated sulfuric acid solvent. From top to bottom are different concentrations (by weight) of sulfuric acid in water: 98% D2SO4/2% D2O; 94% D2SO4/6% D2O; 88% D2SO4/12% D2O; 81% D2SO4/19% D2O with DMSO-d6 as a reference and at room temperature. All peaks are consistent with the molecules being stable and the structure not being affected by the concentrated sulfuric acid solvent. For 2-wk stability of other compounds, see SI Appendix, Figs. S12–S14.
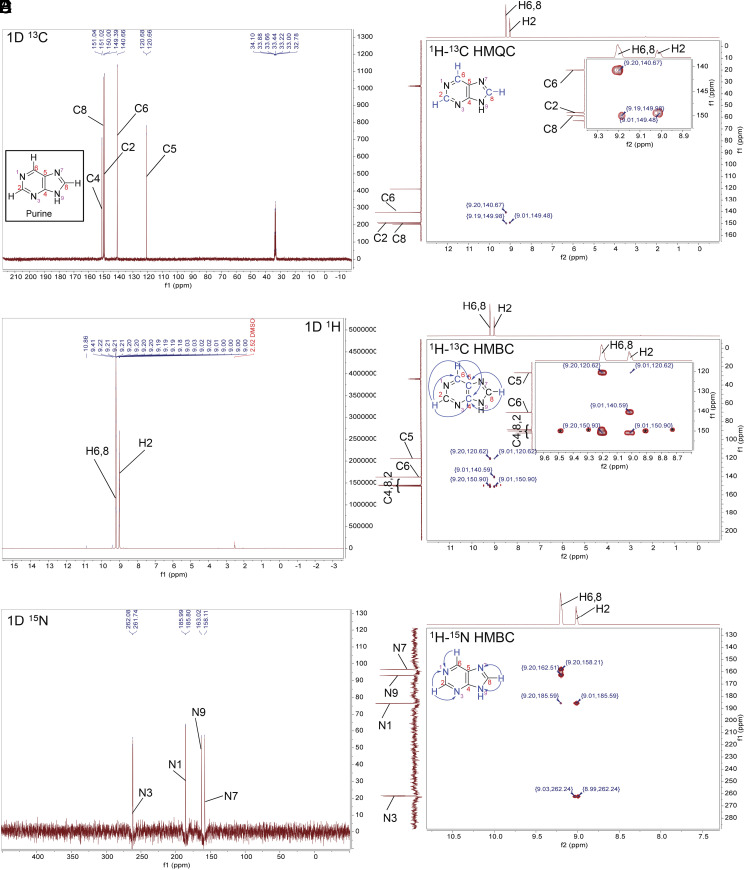
Fig. 5.
NMR spectra for purine in concentrated sulfuric acid (98% D2SO4 and 2% D2O, by weight, with reference DMSO-d6) at room temperature. The NMR experiments confirm the stability of purine in concentrated sulfuric acid. (A) 1D 13C NMR. (B) 1D 1H NMR. The solvent peak is suppressed for clarity. (C) 1D 15N NMR. (D) The 2D 1H-13C HMQC NMR shows direct bonding between H and C atoms in the purine ring structure. (E) The 2D 1H-13C HMBC NMR shows signals that correspond to hydrogen and carbon atoms separated from each other by the distance of 2 to 4 chemical bonds in the purine ring structure (blue arrows). The HMBC “one-bond artifacts” are marked with an asterisk (*). (F) The 2D 1H-15N HMBC NMR shows 2 bond distances between hydrogen and nitrogen atoms (blue arrows).
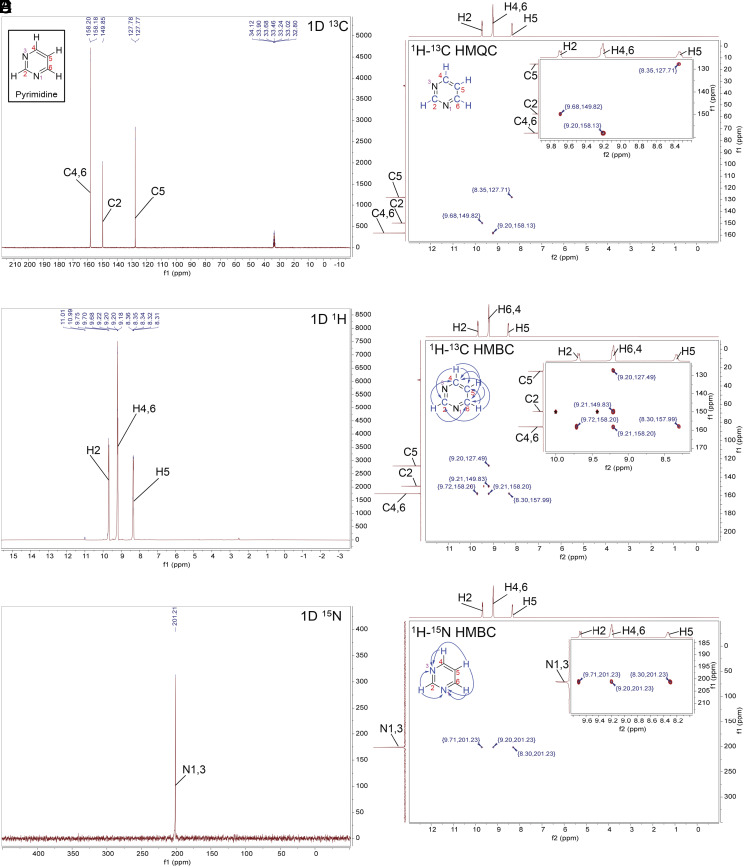
Fig. 6.
NMR spectra for pyrimidine in concentrated sulfuric acid (98% D2SO4 and 2% D2O, by weight, with reference DMSO-d6) at room temperature. The NMR experiments confirm the stability of pyrimidine in concentrated sulfuric acid. (A) 1D 13C NMR. (B) 1D 1H NMR. The solvent peak is suppressed for clarity. (C) 1D 15N NMR. (D) The 2D 1H-13C HMQC NMR shows direct bonding between H and C atoms in the pyrimidine ring structure. (E) The 2D 1H-13C HMBC NMR shows signals that correspond to hydrogen and carbon atoms separated from each other by the distance of 2 or 3 chemical bonds in the pyrimidine ring structure (blue arrows). The HMBC one-bond artifacts are marked with an asterisk (*). (F) The 2D 1H-15N HMBC NMR shows 2 or 3 bond distances between hydrogens attached to carbon and nitrogen atoms (blue arrows).
2.1. UV Spectroscopy.
For an initial investigation of the compounds’ stability in concentrated sulfuric acid, we used UV spectroscopy (190 to 300 nm; Fig. 2). The two absorption peaks at ~180-nm (in the range 160 to 210 nm) and at ~260-nm wavelengths or longer are characteristic of nucleic acid bases and are due to the presence of conjugated π bonds in the aromatic rings (e.g., refs. 47). In other words, absorption is caused when π or nonbonding (nb) electrons jump from π→π* or nb →π* molecular orbitals, and so these absorptions are dependent on the presence of conjugated π systems. The wavelength of the two peaks for each nucleic acid base may slightly differ in concentrated sulfuric acid as compared to other solvents (e.g., refs. 42 and 48 and SI Appendix, Table S9) due to the change in pH, which would cause a change to the conjugation on the nucleic acid bases (including likely protonation), thus leading to the wavelength shifts.
For each compound, the maximum wavelength and shape of each of the two major peaks did not change after about 24 h in concentrated sulfuric acid (Fig. 2), indicating stability of each compound, i.e., that the aromatic ring remained stable and intact. We do note a slight increase in absorbance for the sample after 24 h due to increased dissolution of the tested compound over time. We emphasize that the shape of the curve stays the same over time, meaning that the rings themselves remain stable as the compound dissolves. We note that we do not detect any features beyond 300 nm (apart from a very low-amplitude signal from contamination around 320 nm), because the electron excitation is confined to shorter wavelengths.
Because we do not see any dramatically different absorption characteristics, we can further rule out a change in the substituent groups (apart from likely protonation). Such a substitution would preserve the ring structures but would alter the shape of the spectrum. We can also rule out any reaction to link two rings which would produce a multiring conjugated molecule, which would still have conjugated rings but be expected to have different absorption characteristics.
2.2. 13C NMR.
To further confirm the stability for each compound and validate the compound’s structure, we used 13C NMR spectroscopy (Fig. 3). We mixed 10 to 40 mg of each compound in 500 to 600 µL of a combination of D2SO4 and D2O, ranging from 81 to 98% D2SO4 by weight, with 10% of DMSO-d6 (deuterated dimethyl sulfoxide) added as a chemical shift reference and took a 13C NMR spectrum 30 to 48 h after sample preparation. We used D2O instead of H2O for all NMR experiments, so that we could use the same sample and minimize the intensity of the hydrogen peaks in any 1H NMR experiment. To confirm the stability of the bases over time, the 13C NMR measurements for all compounds in 98% w/w D2SO4 were additionally measured after 2 wk later and the peak positions remain unchanged (Fig. 4 and SI Appendix, Figs. S12–S14).
For each compound, the number of carbon peaks and their chemical shift position in the 13C NMR spectrum show that the aromatic ring structure and number of carbon atoms of the original compound is preserved over extended time periods in concentrated sulfuric acid (Figs. 3 and 4). The carbon peak assignments shown in Figs. 3 and 4 are described in Section 2.3 and in the SI Appendix. We made the assignment for a given compound in 98% w/w sulfuric acid and since the peaks in lower concentration of sulfuric acid are the same, we conclude that the bases in lower acid concentrations (i.e., 81 to 94% w/w) are also stable.
2.3. Molecular Structure Determination by NMR in 98% Concentrated Sulfuric Acid.
To confirm that the molecular structure of nucleic acid bases does not change in concentrated sulfuric acid, we use results from a series of 1D and 2D NMR experiments of nucleic acid bases dissolved in 98% D2SO4 with 2% D2O (by weight). In this section, we choose two exemplar nucleic acid bases to describe fully, purine and pyrimidine. Details of the assignments of the remaining six compounds that we investigated are presented in SI Appendix.
Our aim in this section is to confirm the known chemical structure. A key point is that while the approximate chemical shifts of 1H and 13C are predictable from the proposed structure, their exact positions will vary with the solvent, and hence will not be known in a rarely studied solvent such as concentrated sulfuric acid. To identify NMR spectral peaks, we rely on general rules for chemical shifts for 1H, 13C, and 15N as well as known chemical shifts in solvents other than concentrated sulfuric acid (SI Appendix, Tables S7 and S8), and combined with 2D NMR.
2.3.1. Purines in concentrated sulfuric acid.
For purine, in the 1D 13C NMR spectrum (Figs. 3 and 5A), we find five peaks for carbon that correspond to the five carbons in the purine ring. Each of the five peaks are found in the region of the NMR spectra associated with aromatic compounds.
We assign the carbon peaks by comparison with literature data (SI Appendix, Table S7) and by our 2D NMR experiments (Fig. 5 D and E). We can assign C5 because it is the most magnetically shielded atom in the ring structure with a chemical shift distinctly upfield from the other four carbon peaks (SI Appendix, Table S7). To assign C2, C6, and C8 we use our 2D NMR spectra where we correlate the positions of H and C within the ring. Purine has three protonated carbons, at C2, C6, and C8. Our 2D 1H-13C HMQC shows three signals that correspond to C2 (at 149.39 ppm) attached to H2 (at 9.01 ppm), C6 (at 140.66 ppm) attached to H6 (at 9.20 ppm), and the C8 carbon peak at 150.00 ppm attached to H8 at 9.20 ppm. The two peaks corresponding to H6 and H8 hydrogens (at 9.20 ppm) on the 1D 1H NMR spectrum are overlapping (Fig. 5B).
We assign C4 on the basis of 1H-13C HMBC data (Fig. 5E). As expected, the 1H-13C HMBC correlates the chemical shift of carbon C4 at 151.04 ppm to the nearby H2, at 9.01 ppm, and H6, H8 at 9.20 ppm (all separated from C4 by three chemical bonds) (Fig. 5E).
The 1H-13C HMBC spectra also further confirms the assignments of C2, C5, C6, and C8. The C6 (at 140.66 ppm) correlates with the chemical shift of H2, at 9.02 ppm. C6 and H2 are at the right separation of three chemical bonds, supporting the assignment of 140.66 ppm and 9.02 ppm chemical shifts to C6 and H2, respectively. Further supporting these assignments and the integrity of the imidazole ring, the 1H-13C HMBC correlates the distinct 13C chemical shift of C5 to the nearby H8 which in turn is correlated with C4. Note that there is also a weak signal on the 1H-13C HMBC spectra (coordinates: 9.01 ppm, 120.62 ppm) (Fig. 5E). The signal corresponds to the carbon atom C5 at the separation of four bonds from the H2 hydrogen.
Finally, we use 1D 15N NMR to confirm the presence of the N atoms in the purine ring (Fig. 5C). We see four peaks corresponding to four nitrogen atoms of the purine ring and make assignments of N1 and N3 to 185.99 ppm and 262.08 ppm, respectively based on the 1H-15N HMBC experiment (Fig. 5F). We assign N7 and N9 to 158.11 ppm and 163.02 ppm peaks, respectively, based on the known N7 and N9 chemical shifts of purine in concentrated sulfuric acid (40), where N7 is more magnetically shielded (shifted toward lower ppm) in acidic conditions than N9 (Fig. 7 and SI Appendix, Table S7). Taken together the NMR data confirm that the purine ring structure remains intact in 98% w/w D2SO4 in D2O.
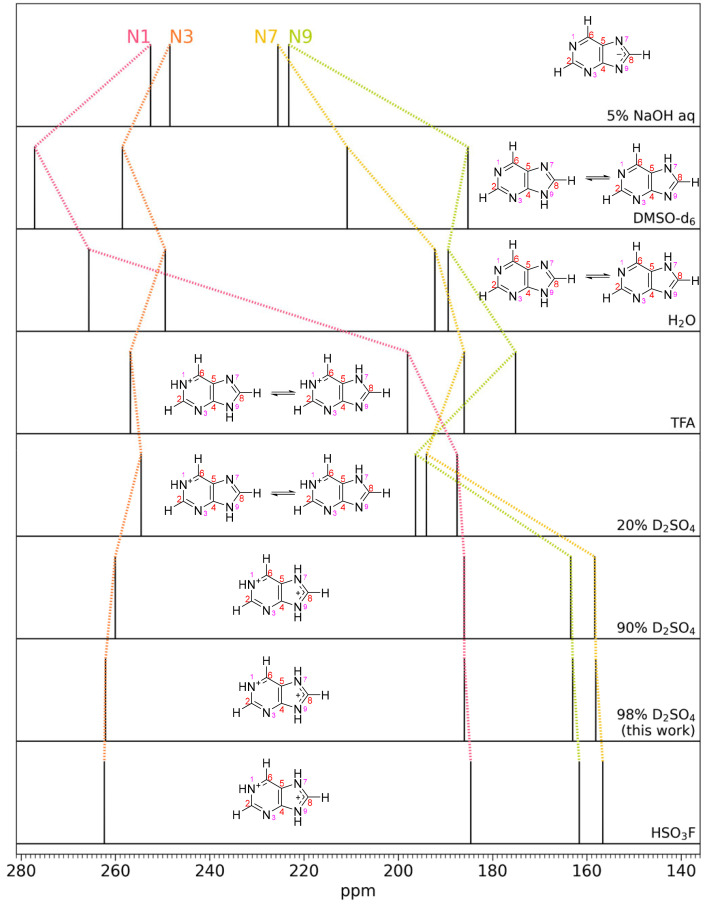
Fig. 7.
Purine 15N NMR spectral peaks for different solvent acidities. The ordering from top to bottom is in increasing acidity of the solvent. The change in chemical shifts of different N atoms indicate different protonation states of the N atoms; each of the four peaks correspond to a different N atom as indicated. The second and third row shows purine in DMSO-d6 and H2O, which can be considered well-known standards. Top: in the basic aqueous solution of 5% NaOH, all of the N atoms are deprotonated (i.e., magnetically deshielded) and the N atom spectral peaks are shifted downfield as compared to the H2O and DMSO solutions. With increasing solvent acidity, N atoms sequentially get protonated, causing dramatic upfield spectral peak migration as added protons provide more magnetic shielding. In some cases (e.g., DMSO-d6 and H2O) two tautomeric structures exist in equilibrium due to fast proton exchange; the relative abundance controls the spectral peak positions. See text for more details. Data and figure adapted from ref. 40. Our data at 98% D2SO4 match the high-acidity solvents 90% H2SO4 and FSO3H from ref. 40, demonstrating protonation of the N1, N7, and N9 nitrogen atoms in the purine molecule. TFA is trifluoroacetic acid.
We now turn to evidence of protonation of purine in concentrated sulfuric acid. We summarize pioneering work by Schumacher and Günther (40) and show that our NMR 15N chemical shifts agree in acidic solvents, thus demonstrating protonation of N atoms in the purine ring (SI Appendix, Fig. S15). Schumacher and Günther show that the purine ring is protonated in 90% w/w concentrated sulfuric acid by analyzing the chemical shift change of 15N NMR peaks of purine in different solvents ranging from strongly basic to strongly acidic (Fig. 7). Purine 15N NMR spectra in DMSO-d6 and H2O can be considered well-known standards. In the basic aqueous solution of 5% NaOH, the N7 and N9 atom spectral peaks are shifted downfield as compared to their peaks in H2O and DMSO-d6 solutions indicating deprotonation of N atoms. With increasing solvent acidity, N atoms sequentially get protonated, causing dramatic upfield spectral peak migration. In some cases (e.g., H2O and DMSO-d6) two tautomeric structures exist in equilibrium due to fast proton exchange; the relative abundance controls the spectral peak positions. For example, in H2O, the N7 and N9 spectral peaks are relatively close to each other, indicating a similar chemical environment, including a similar time-averaged protonation state, i.e., a relatively equal abundance of an N7 and N9 protonated tautomeric structures (40). In comparison, in DMSO vs. H2O, the purine tautomeric structure with protonated N9 is more abundant than that with protonated N7 (49). Note that the N3 atom is not protonated even in 90% H2SO4 (40), as shown by the insignificant migration of its spectral peak. However, N3 does get protonated in superacids (41). Our data at 98% D2SO4 match the high acidity solvents 90% H2SO4 and FSO3H from ref. 40, demonstrating protonation of the N1, N7, and N9 nitrogen atoms in the purine molecule (Fig. 7).
For other purines, adenine, guanine, and diaminopurine, we demonstrate stability in concentrated sulfuric acid as follows. In the 1D 13C NMR spectra (Fig. 3), we find five peaks for carbon that correspond to the five carbons in the adenine, guanine, and diaminopurine ring. Each of the five peaks are found in the region of the NMR spectra associated with aromatic compounds. To further confirm the integrity and stability of the adenine, guanine, and diaminopurine nucleic acid bases in concentrated sulfuric acid we employ 1H and 15N NMR, combined with HMQC and HMBC 2D NMR (SI Appendix, Figs. S1, S3, and S5). The results of the 15N NMR show the correct number of nitrogen atoms in the adenine, guanine, and diaminopurine rings. The 2D NMR spectra further confirm the 13C, 15N, and 1H peak assignments. Taken together, the results show that adenine, guanine, and diaminopurine are stable in concentrated sulfuric acid for a prolonged time. For detailed justification and description of the results, please see SI Appendix.
We now turn to assessment of the protonation state of adenine, guanine, and diaminopurine. Apart from purine itself, the protonation of other purines in concentrated sulfuric acid has not been previously studied. Comparison of the 13C NMR chemical shift changes in different solvents suggests that guanine, adenine, and diaminopurine are protonated in concentrated sulfuric acid (SI Appendix, Tables S1, S3, and S5). Protonation of nitrogen atoms results in greater shielding of the nearby carbon atoms and the upfield shift (toward lower ppm) of peaks corresponding to carbon atoms directly adjacent to the nitrogen atoms that are protonated in the acidic solvent. This effect is uniform among all tested purines, where the upfield shift of 13C NMR carbon peaks in concentrated sulfuric acid is particularly pronounced for C2, C4, and C6 peaks (SI Appendix, Tables S1, S3, and S5). All nitrogen atoms of adenine, guanine, and diaminopurine appear to be protonated in concentrated sulfuric acid, while the protonation of the oxygen atom in the carbonyl group of guanine is a possibility.
2.3.2. Pyrimidines in concentrated sulfuric acid.
We now turn to our second example, pyrimidine, again as in the purine case aiming to confirm the known structure to demonstrate stability in concentrated sulfuric acid. Also as for purine, while the chemical shifts are known for pyrimidine, they are different in concentrated sulfuric acid as compared to other solvents (SI Appendix, Table S8).
For pyrimidine, the 1D 13C NMR spectrum (Figs. 3 and 6A) shows three peaks for carbon that correspond to the four carbons in the pyrimidine ring. Since carbons C4 and C6 are distributed symmetrically within the pyrimidine ring, they present as a single NMR peak. All peaks are found in the region of the NMR spectra associated with aromatic compounds.
We assign the carbon peaks by comparison with literature data (SI Appendix, Table S8) and by our 2D NMR experiments (Fig. 6 D and E). We can assign C5 because it is the most magnetically shielded atom in the ring structure with a chemical shift distinctly upfield from the other two carbon peaks (SI Appendix, Table S8). To assign C2, C4, and C6 we use our 2D NMR spectra where we correlate the positions of H and C within the ring (Fig. 6 D and E). Pyrimidine has four carbons with directly attached hydrogen atoms. Our 2D 1H-13C HMQC shows three distinct signals that we use to assign C2, C4, C6, and C5 (Fig. 6D). The first signal corresponds to the C2 carbon peak at 149.85 ppm and H2 (bonded to C2) at 9.68 ppm. We assign the peak at 158.18 ppm to symmetric carbon atoms C4 and C6. The 2D 1H-13C HMQC also confirms the assignment of C5 at 127.77 ppm.
We further confirm our carbon atoms peak assignments on the basis of 1H-13C HMBC data (Fig. 6E).
Finally, we use 1D 15N NMR to confirm the presence of the symmetric N atoms in the pyrimidine ring (Fig. 6C). We detect a single peak at 201.21 ppm corresponding to two symmetrical nitrogen atoms, N1 and N3, of the pyrimidine ring. The 1H-15N HMBC experiment (Fig. 6F) further confirms this assignment.
Taken together the NMR data confirm that the pyrimidine ring structure remains intact in 98% w/w D2SO4 in D2O.
We now describe evidence of protonation of pyrimidine in concentrated sulfuric acid. Work by Wagner and von Philipsborn (39) shows that pyrimidine is protonated in strong acid solvents, as compared to less acidic solvents. While Wagner and von Philipsborn do not use 15N NMR, they instead analyze 1H NMR data to demonstrate protonation in fluorosulfuric acid (FSO3H). The effects of the two acids, concentrated sulfuric acid and fluorosulfuric acid, are shown to be the same as demonstrated by purine showing the same chemical shift changes and protonation state in fluorosulfuric acid as in sulfuric acid (40) (Fig. 7).
In our 13C NMR spectra, we see a significant spectral peak upfield shift of C2 as compared to C2 spectral peaks in lower acidity solvents such as DMSO-d6 and D2O (SI Appendix, Table S8). Protonation of the adjacent ring N atom results in shielding of the nearby C2 nucleus, causing the upfield shift. For pyrimidine, we see shifts in 13C peaks as the concentration of acid increases from 81 to 98% (Fig. 4). For the same reasons, carbon C5 becomes significantly deshielded by the protonation of N atoms in the pyrimidine ring.
For other pyrimidines, cytosine, uracil, and thymine, we demonstrate stability in concentrated sulfuric acid as follows. In the 1D 13C NMR spectra (Figs. 3 and 6A), we find four peaks for carbon that correspond to the four carbons in the pyrimidine ring. Each of the four peaks are found in the region of the NMR spectra associated with aromatic compounds. To further confirm the integrity and stability of the cytosine, uracil, and thymine nucleic acid bases in concentrated sulfuric acid, we employ 1H and 15N NMR, combined with 2D HMQC and HMBC NMR. The results of the 15N NMR show the correct number of nitrogen atoms in the cytosine, uracil, and thymine rings (SI Appendix, Figs. S2, S4, and S6). The 2D NMR further confirms the 13C, 15N, and 1H peak assignments. Taken together the results show that cytosine, uracil, and thymine are stable in concentrated sulfuric acid for a prolonged time. For detailed justification and the description of the results, please see SI Appendix.
We now turn to assessment of the protonation state of other pyrimidines cytosine, uracil, and thymine. For cytosine, uracil, and thymine, the N atoms and the carbonyl group O atoms are likely protonated in concentrated sulfuric acid (SI Appendix, Fig. S15). The 13C NMR carbon chemical shifts of cytosine, uracil, and thymine in concentrated sulfuric acid reported by Benoit and Frechette agree with ours (SI Appendix, Table S2, S4, and S6) and are consistent with protonation of nitrogen atoms and oxygens of the carbonyl groups (42). The protonation of cytosine, uracil, and thymine in strong acid is further supported by early studies on protonation of pyrimidines in FSO3H (39).
In our NMR analysis we have not fully addressed protonation of the nucleic acid bases, likely in the high acid solvent concentrated sulfuric acid. In this paper, we are concerned with the long-term stability of the heavy atom bonding topology in the bases, i.e., the bonding of nonhydrogen atoms to each other, and not with rapid, reversible exchange of protons between the nucleic acid bases and the solvent. Thus, while protonation of the ring N atoms is formally a chemical change, it will at least not destabilize the structure of the bases as conventionally drawn (i.e. without explicit hydrogens), and may well stabilize it to certain reactivity (e.g., once protonated, the bases will react only as electrophiles).
In this assessment of stability, we follow convention, as illustrated by the common understanding that carboxylic acids and amines are stable to pH difference; even though reducing the pH causes the molecules to lose or gain a proton respectively, this change does not alter how the nonhydrogen atoms are bonded to each other, and losing or gaining a proton is readily reversed by reversing the pH change. The stability of DNA bases shown in this paper is in contrast to the instability of other species in concentrated sulfuric acid, such as some aliphatic carbonyl compounds, where protonation of the oxygen leads to rapid and irreversible rearrangement of the carbon skeleton (50).
3. Discussion
For life to exist, complex organic chemistry has to be capable of forming an informational biopolymer. On Earth those polymers are DNA and RNA. The complementarity of the nucleic acid bases is the key to the DNA and RNA structure and to its informational function. The stability of nucleic acid bases in concentrated sulfuric acid therefore goes far beyond the finding of a new category of molecules stable in this aggressive solvent—and toward the design of an informational biopolymer that employs the canonical or other, modified nucleic acid bases (e.g., if protonation of the nitrogen atoms in the ring prevents their pairing). While it is already well known that DNA’s phosphate ester backbone and ribose linker are unstable in concentrated sulfuric acid, we can aim to find a stable substitute, with similar structural characteristics, size, flexibility, shape, etc. By designing a stable informational biopolymer in concentrated sulfuric acid, we can support the idea that concentrated sulfuric acid can support some kind of life different from Earth’s, because such a polymer can in principle carry information and support genetics and Darwinian evolution.
One key gap in understanding a path from sulfuric acid-stable bases to a sulfuric acid-stable informational polymer is on the likely protonation of the nucleic acid bases. Because concentrated sulfuric acid is such a strong acid, it may be that every atom that could serve as a hydrogen bond acceptor is protonated. Protonation will change the nucleic acid base’s physical shape and hydrogen bonding potential, and hence is relevant for formation of macromolecular structures such as a double helix. Specifically, protonation of the N atoms in the ring would interfere with the canonical Watson–Crick base pairing between the bases (SI Appendix, Fig. S16). Understanding the protonation of N atoms in the canonical DNA/RNA nucleic acid bases at different concentrations of sulfuric acid is essential for progress. As relevant to a sulfuric acid-stable informational polymer, we emphasize that alternative nucleic acid bases (e.g., ref. 51), such as 4-methylbenzimidazole and 2,4-difluorotoluene, are known to form functional nucleic acid base pairs (52) that will not be subjected to N protonation in concentrated sulfuric acid and hence the N protonation will not affect their potential base pair complementarity. Whether such nucleic acid bases can form pairs in solution in concentrated sulfuric acid is a subject for future work.
We do not know if the origin of life in concentrated sulfuric acid is possible, but such a possibility cannot be excluded a priori. Life could use concentrated sulfuric acid as a solvent instead of water and could have originated in the cloud droplets in liquid concentrated sulfuric acid. This scenario does not require Venus to be canonically habitable in the past, i.e., with liquid surface water, and is relevant because some researchers think Venus was always too warm for surface liquid water oceans to condense and may have looked like it does today for most of its geological history (53). In this scenario, the Venus atmosphere could still support the strictly aerial concentrated sulfuric acid-based life.
Another hypothetical scenario for how life might have come to be in the Venusian clouds involves life changing its solvent in the course of evolution (i.e., undergo solvent replacement). In this scenario Venus had early water oceans, that later evaporated (54, 55), and life originated in water on the early Venus billions of years ago. As water became very scarce, life would have adapted its biochemistry to the concentrated sulfuric acid environment, eventually substituting water for concentrated sulfuric acid. The solvent replacement might appear as an impossible adaptation as most of Earth’s life biochemicals that are stable in water are not stable in concentrated sulfuric acid (25). We do not however know for sure if such gradual adaptation to another solvent is possible or impossible, if the change would have been gradual enough (over millions, if not billions of years). It may have been, considering the evolutionary pressure would have been constant and strong. However, we definitely have no example of such a mechanism and adaptation on Earth.
For completeness, we also mention a third possibility for how life could be in the Venusian clouds, in which the Venus cloud-based life originated in water on the early Venus billions of years ago and as water became very scarce, life would have adapted its biochemistry to the concentrated sulfuric acid environment but never to the point of using it as a solvent. Such a scenario involves “neutralization” of sulfuric acid droplets with biologically produced ammonia (4, 5) and other specialized adaptations to the hostile sulfuric acid environment (56). Many unexplained atmospheric gases detected in the clouds of Venus and other cloud properties are consistent with this hypothesis (32). We also emphasize that if life exists in the clouds of Venus, it has to be very different than life on Earth, with biochemical and evolutionary adaptations that have no precedent here on Earth.
All life needs a liquid solvent and concentrated sulfuric acid is one of only three liquids known to exist on rocky solar system planets and moons. Liquid water is present on the surface of the Earth and in the subsurface of several icy moons (e.g., Europa and Enceladus). Liquid hydrocarbons have been identified on Saturn’s moon Titan. Concentrated sulfuric acid as a planetary solvent is not only relevant to Venus, but also for exoplanets; while we know rocky exoplanets are common, we do not know if Venus-like planets are more prevalent than Earth-like exoplanets. Ballesteros et al. (57) postulate that concentrated sulfuric acid is one of the most common liquids in the Galaxy, although the stability of concentrated sulfuric acid as a surface liquid is unknown. Further studies of concentrated sulfuric acid organic chemistry can therefore move the search for habitable worlds forward.
Venus is right “next door” and the cloud particles can be directly probed by space missions. NASA’s Pioneer Venus in situ missions in 1978, and several Soviet descent craft, established the particle size distribution in the clouds, but did not unambiguously determine the composition of all types of cloud particles or search for organic chemicals. Today NASA and ESA have plans to send missions to Venus at the end of this decade, though none of the three planned missions will probe the cloud particles directly (58–60).
Our findings show that complex organic chemistry, including DNA nucleic acid bases, can be stable in concentrated sulfuric acid and motivates us to design missions that directly probe the cloud particles for the presence of organic material. As a first mission in our “Morning Star Missions to Venus” program, our Rocket Lab Mission to Venus is planned for launch in January 2025 (61). The Rocket Lab mission will deliver a probe containing one instrument, the autofluorescence nephelometer, to search for autofluorescence (and backscattered polarized radiation) in the Venus cloud droplets, indicative of organic molecules, especially aromatic ring systems (62). More sophisticated future missions (63–65), can then identify organics if they are present (66). Ultimately a sample return from the Venus atmosphere may be needed to robustly identify life, if present (67).
4. Materials and Methods
4.1. Materials.
We purchased chemical compounds from Millipore-Sigma, with pyrimidine ordered from Thermo Scientific. The compounds were used without further purification. The catalog numbers and purities are as follows: adenine A8626-5G 99%; cytosine C3506-1G 99%; 2,6-diaminopurine 247847-1G 98%; guanine G11950-10G 98%; purine P55805-1G 98%; pyrimidine 289-95-2 99%; thymine T0376-10G 99%; and uracil U0750-25G 99%. We used H2SO4 from Sigma Aldrich (used for UV spectroscopy) and D2SO4 from ACROS Organics (sulfuric acid-d2 for NMR, 98 wt.% in D2O, 99.5+ atom % D) and D2O (deuteration degree min 99.9%) from MagniSolv.
4.2. UV and Visible Wavelength Spectroscopy.
We used UV and visible (UV-Vis) wavelength spectrometry from 190–600 nm for our initial investigation of compound stability in 98% w/w H2SO4 in H2O. We prepared our samples by mixing each compound into a small stock solution and diluted the solution until the measurement reached a desired absorbance value near A = 1, where, from the Beer–Lambert law A = εLc, where A is the absorbance (dimensionless), ε is the molar absorption coefficient in units of M−1 cm−1 [on order of 6,000 to 13,000 in water (68)], L is pathlength in cm (the cuvette width was 1 cm), and c is concentration in molarity. We stored the solution in a sealed glass vial and after about 24 h put the solution back into a quartz cuvette for remeasurement. Our UV-Vis spectrometer is a HELIOS OMEGA, 244001, v8.01 with a scan rate of 1,200 nm/min and data interval of 1 nm. No data analysis was required other than the baseline correction from an initially measured sample of 98% w/w H2SO4 with 2% H2O.
The concentration of each compound in 98% w/w H2SO4 in H2O is given in SI Appendix, Table S9, and comes from the mass (or volume in the case of pyrimidine) in 3.2 mL H2SO4. The concentrations are low, less than one mM, because the compounds are strong UV absorbers. The original data for all UV-Vis experiments are available for download as SI Appendix, Dataset S1, available from Zenodo (69).
4.3. 1D and 2D NMR Spectroscopy.
We prepared our NMR samples by dissolving 10 to 80 mg of the aromatic compounds into 500 to 600 µL of solvent D2SO4 in D2O in glass vials. We added DMSO-d6, used as a chemical shift reference compound, to a final concentration of 10% by volume. We used 10 to 40 mg of compounds for the 1D 1H and 13C NMR. We used 80 mg for the 15N NMR due to low sensitivity (except for adenine for which we used 50 mg) and also used 80 mg for the 2D NMR (though using the lower 30 to 40 mg concentrations did not change the results). For 2,6-diaminopurine we used only 10 mg for the 6 to 19% D2O solutions due to limited solubility. We heated sealed glass vials in a hot water bath (~80 °C) when this was needed to dissolve the compounds. Out of all the compounds analyzed only 2,6-diaminopurine partially precipitated back out on cooling to room temperature. We stored the sealed glass vials for 12 to 24 h before transferring the solution to 5-mm NMR tubes. After NMR measurements, we stored the solutions in the NMR tubes, where the storage room temperature varied from about 18 to 24 °C.We used the same tubes to acquire 13C NMR spectra after two weeks.
To acquire NMR data, we used a Bruker Avance III-HD 400 MHz spectrometer equipped with a Prodigy liquid nitrogen cryoprobe (BBO) at 25 °C. We acquired 1D 1H, 13C, 15N, 2D 1H-13C HMBC and HMQC, and 1H-15N HMBC NMR spectra to confirm the structures and hence stability of the compounds in 98% w/w D2SO4 in D2O. For our solutions of 94%, 88%, and 81% D2SO4 in D2O (by weight) we acquired 1D 13C NMR spectra. In all cases, we locked on DMSO-d6 for consistency, where we found the DMSO-d6 peak to be at 33.44 ppm ± 0.02 ppm in our 98% w/w D2SO4 solutions. For solutions of different acidity, the DMSO-d6 varied by about ± 0.1 ppm. We note that for uracil, cytosine, and pyrimidine in 81% w/w sulfuric acid the DMSO-d6 was significantly shifted to about 26.70 ppm. For Figs. 3 and 4, we set the DMSO-d6 reference to 33.44 ppm in all cases.
We used MNova software (Mestrelab Research) to process and analyze the NMR data (70). The original data for all NMR experiments are available for download as SI Appendix Dataset S2, available from Zenodo (69).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank the MIT Department of Chemistry Instrumentation Facility NMR Consultant Bruce Adams and Director Walter Massefski. We thank Adam Jost for experimental assistance with the mass microbalance. We thank Jingcheng Huang for help with the compilation of the spectroscopic data from the literature. We thank Steven Benner and Jan Spacek for useful discussion. This work was partially funded by the Massachusetts Institute of Technology and Nanoplanet Consulting.
Author contributions
S.S., J.J.P., M.D.S., J.H.G., H.R.V.-S., M.K.A.E.-R., D.S.W., and B.L.L. designed research; S.S., J.J.P., M.D.S., J.H.G., Z.Z., M.K.A.E.-R., B.L.L., and C.D. performed research; S.S., J.J.P., M.D.S., J.H.G., Z.Z., D.S.W., B.L.L., V.G., and L.H. analyzed data; M.D.S., J.H.G., Z.Z., H.R.V.-S., D.S.W., B.L.L., W.B., and L.H. edited the paper; and S.S. and J.J.P. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Preprint Servers: On acceptance a version of the manuscript will be submitted to arXiv (https://arxiv.org/archive/astro-ph).
Reviewers: S.S.L., University of Wisconsin-Madison; and R.M., California State Polytechnic University.
Data, Materials, and Software Availability
Ascii files and zip files data have been deposited in Zenodo (69).
Supporting Information
References
- 1.Morowitz H., Sagan C., Life in the clouds of venus? Nature 215, 1259–1260 (1967). [Google Scholar]
- 2.Grinspoon D. H., Venus Revealed: A New Look Below the Clouds of Our Mysterious Twin Planet (Basic Books, New York City, NY, 1997). [Google Scholar]
- 3.Patel M. R., Mason J. P., Nordheim T. A., Dartnell L. R., Constraints on a potential aerial biosphere on Venus: II. Ultraviolet radiation. Icarus 373, 114796 (2021). [Google Scholar]
- 4.Mogul R., Limaye S. S., Lee Y. J., Pasillas M., Potential for phototrophy in venus’ clouds. Astrobiology 21, 1237–1249 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Bains W., Petkowski J. J., Rimmer P. B., Seager S., Production of ammonia makes venusian clouds habitable and explains observed cloud-level chemical anomalies. Proc. Natl. Acad. Sci. U.S.A. 118, e2110889118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seager S., et al. , The venusian lower atmosphere haze as a depot for desiccated microbial life: A proposed life cycle for persistence of the venusian aerial biosphere. Astrobiology 21, 1206–1223 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Cockell C. S., Life on venus. Planet. Space Sci. 47, 1487–1501 (1999). [Google Scholar]
- 8.Schulze-Makuch D., Irwin L. N., Reassessing the possibility of life on venus: Proposal for an astrobiology mission. Astrobiology 2, 197–202 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Schulze-Makuch D., Irwin L. N., The prospect of alien life in exotic forms on other worlds. Naturwissenschaften 93, 155–172 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Schulze-Makuch D., Grinspoon D. H., Abbas O., Irwin L. N., Bullock M. A., A sulfur-based survival strategy for putative phototrophic life in the Venusian atmosphere. Astrobiology 4, 11–18 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Grinspoon D. H., Bullock M. A., Astrobiology and venus exploration. Geophys. Monogr. Geophys. Union 176, 191 (2007). [Google Scholar]
- 12.Limaye S. S., et al. , Venus’ spectral signatures and the potential for life in the clouds. Astrobiology 18, 1181–1198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotsyurbenko O. R., et al. , Exobiology of the venusian clouds: New insights into habitability through terrestrial models and methods of detection. Astrobiology 21, 1186–1205 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dartnell L. R., et al. , Constraints on a potential aerial biosphere on Venus: I. Cosmic rays. Icarus 257, 396–405 (2015). [Google Scholar]
- 15.Baross J., et al. , The Limits of Organic Life in Planetary Systems (National Academies Press, 2007). [Google Scholar]
- 16.Vaïtilingom M., et al. , Long-term features of cloud microbiology at the puy de Dôme (France). Atmos. Environ. 56, 88–100 (2012). [Google Scholar]
- 17.Amato P., et al. , Active microorganisms thrive among extremely diverse communities in cloud water. PLoS One 12, e0182869. (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amato P., et al. , Metatranscriptomic exploration of microbial functioning in clouds. Sci. Rep. 9, 4383 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bryan N. C., Christner B. C., Guzik T. G., Granger D. J., Stewart M. F., Abundance and survival of microbial aerosols in the troposphere and stratosphere. ISME J. 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burrows S. M., et al. , Bacteria in the global atmosphere–Part 2: Modeling of emissions and transport between different ecosystems. Atmos. Chem. Phys. 9, 9281–9297 (2009). [Google Scholar]
- 21.Knollenberg R., et al. , The clouds of Venus: A synthesis report. J. Geophys. Res. Sp. Phys. 85, 8059–8081 (1980). [Google Scholar]
- 22.Hansen J. E., Hovenier J. W., Interpretation of the polarization of Venus. J. Atmos. Sci. 31, 1137–1160 (1974). [Google Scholar]
- 23.Hoffman J. H., Oyama V. I., Von Zahn U., Measurements of the Venus lower atmosphere composition: A comparison of results. J. Geophys. Res. Sp. Phys. 85, 7871–7881 (1980). [Google Scholar]
- 24.Krasnopolsky V. A., Vertical profiles of H2O, H2SO4, and sulfuric acid concentration at 45–75 km on Venus. Icarus 252, 327–333 (2015). [Google Scholar]
- 25.Bains W., Petkowski J. J., Zhan Z., Seager S., Evaluating alternatives to water as solvents for life: The example of sulfuric acid. Life 11, 400 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hallsworth J. E., et al. , Water activity in Venus’s uninhabitable clouds and other planetary atmospheres. Nat. Astron. 1–11 (2021). [Google Scholar]
- 27.Spacek J., Organic carbon cycle in the atmosphere of venus. arXiv [Preprint] (2021). 10.48550/arXiv.2108.02286 (Accessed 20 February 2023). [DOI]
- 28.Benner S. A., Spacek J., The limits to organic life in the solar system: From cold titan to hot venus. LPI Contrib. 2629, 4003 (2021). [Google Scholar]
- 29.Spacek J., Benner S. A., The organic carbon cycle in the atmosphere of venus and evolving red oil. LPI Contrib. 2629, 4052 (2021). [Google Scholar]
- 30.Spacek J., et al. , Organics produced in the clouds of Venus resemble the spectrum of the unknown absorber. LPI Contrib. 2807, 8060 (2023). [Google Scholar]
- 31.Titov D. V., Ignatiev N. I., McGouldrick K., Wilquet V., Wilson C. F., Clouds and hazes of Venus. Space Sci. Rev. 214, 1–61 (2018). [Google Scholar]
- 32.Petkowski J. J., et al. , Astrobiological potential of venus atmosphere chemical anomalies and other unexplained cloud properties. Astrobiology in press, (2023). [DOI] [PubMed] [Google Scholar]
- 33.Miron S., Lee R. J., Molecular structure of conjunct polymers. J. Chem. Eng. Data 8, 150–160 (1963). [Google Scholar]
- 34.Albright L. F., Houle L., Sumutka A. M., Eckert R. E., Alkylation of isobutane with butenes: Effect of sulfuric acid compositions. Ind. Eng. Chem. Process Des. Dev. 11, 446–450 (1972). [Google Scholar]
- 35.Huang Q., Zhao G., Zhang S., Yang F., Improved catalytic lifetime of H2SO4 for isobutane alkylation with trace amount of ionic liquids buffer. Ind. Eng. Chem. Res. 54, 1464–1469 (2015). [Google Scholar]
- 36.Cerfontain H., Telder A., The solubility of toluene and benzene in concentrated aqueous sulfuric acid; implications to the kinetics of aromatic sulfonation. Recl. des Trav. Chim. des Pays-Bas 84, 545–550 (1965). [Google Scholar]
- 37.Cerfontain H., Sixma F. L. J., Vollbracht L., Aromatic sulphonation IX heterogeneous sulphonation of toluene with aqueous sulphuric acid. Recl. des Trav. Chim. des Pays-Bas 83, 226–232 (1964). [Google Scholar]
- 38.Cerfontain H., Solubility of aromatic hydrocarbons in aqueous sulfuric acid. Recl. des Trav. Chim. des Pays-Bas 84, 491–502 (1965). [Google Scholar]
- 39.Wagner R., von Philipsborn W., Protonierung von Amino-und Hydroxypyrimidinen NMR-Spektren und Strukturen der Mono-und Dikationen. Helv. Chim. Acta 53, 299–320 (1970). [DOI] [PubMed] [Google Scholar]
- 40.Schumacher M., Günther H., Beiträge zur 15N-NMR-Spektroskopie Protonierung und Tautomerie in Purinen: Purin und 7-und 9-Methylpurin. Chem. Ber. 116, 2001–2014 (1983). [Google Scholar]
- 41.Wagner R., von Philipsborn W., Protonierung von Purin, Adenin und Guanin NMR.-Spektren und Strukturen der Mono-, Di-und Tri-Kationen. Helv. Chim. Acta 54, 1543–1558 (1971). [DOI] [PubMed] [Google Scholar]
- 42.Benoit R. L., Frechette M., 1H and 13C nuclear magnetic resonance and ultraviolet studies of the protonation of cytosine, uracil, thymine, and related compounds. Can. J. Chem. 64, 2348–2352 (1986). [Google Scholar]
- 43.Zhou Y., et al. , A widespread pathway for substitution of adenine by diaminopurine in phage genomes. Science (80-.) 372, 512–516 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Kirnos M. D., Khudyakov I. Y., Alexandrushkina N. I., Vanyushin B. F., 2-Aminoadenine is an adenine substituting for a base in S-2L cyanophage DNA. Nature 270, 369–370 (1977). [DOI] [PubMed] [Google Scholar]
- 45.Pezo V., et al. , Noncanonical DNA polymerization by aminoadenine-based siphoviruses. Science (80-.) 372, 520–524 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Sleiman D., et al. , A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes. Science (80-.) 372, 516–520 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Akash M. S. H., Rehman K., “Ultraviolet-visible (UV-VIS) spectroscopy” in Essentials of Pharmaceutical Analysis (Springer, 2020), pp. 29–56. [Google Scholar]
- 48.Cavalieri L. F., Bendich A., Tinker J. F., Brown G. B., Ultraviolet absorption spectra of purines, pyrimidines and triazolopyrimidines. J. Am. Chem. Soc. 70, 3875–3880 (1948). [DOI] [PubMed] [Google Scholar]
- 49.Schumacher M., Guenther H., Carbon-13-proton spin-spin coupling. 9. Purine. J. Am. Chem. Soc. 104, 4167–4173 (1982). [Google Scholar]
- 50.Bains W., Petkowski J. J., Seager S., A Data Resource for Sulfuric Acid Reactivity of Organic Chemicals. Data 6, 24 (2021). [Google Scholar]
- 51.Dhami K., et al. , Systematic exploration of a class of hydrophobic unnatural base pairs yields multiple new candidates for the expansion of the genetic alphabet. Nucleic Acids Res. 42, 10235–10244 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morales J. C., Kool E. T., Efficient replication between non-hydrogen-bonded nucleoside shape analogs. Nat. Struct. Biol. 5, 950–954 (1998). [DOI] [PubMed] [Google Scholar]
- 53.Turbet M., et al. , Day–night cloud asymmetry prevents early oceans on Venus but not on Earth. Nature 598, 276–280 (2021). [DOI] [PubMed] [Google Scholar]
- 54.Way M. J., et al. , Was Venus the first habitable world of our solar system?. Geophys. Res. Lett. 43, 8376–8383 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Way M. J., Del Genio A. D., Venusian habitable climate scenarios: Modeling venus through time and applications to slowly rotating venus-like exoplanets. J. Geophys. Res. Planets 125, e2019JE006276 (2020). [Google Scholar]
- 56.Bains W., Petkowski J. J., Seager S., Venus’ atmospheric chemistry and cloud characteristics are compatible with Venusian life. Astrobiology (2023), in press. [DOI] [PubMed] [Google Scholar]
- 57.Ballesteros F. J., Fernandez-Soto A., Martínez V. J., Diving into exoplanets: Are water seas the most common? Astrobiology 19, 642–654 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Garvin J. B., et al. , Revealing the Mysteries of Venus: The DAVINCI Mission. Planet. Sci. J. 3, 117 (2022). [Google Scholar]
- 59.Freeman A., et al. , Veritas: A discovery-class Venus surface geology and geophysics mission (2016).
- 60.de Oliveira M. R. R., Gil P. J. S., Ghail R., A novel orbiter mission concept for venus with the EnVision proposal. Acta Astronaut. 148, 260–267 (2018). [Google Scholar]
- 61.French R., et al. , Rocket lab mission to venus. Aerospace 9, 445 (2022). [Google Scholar]
- 62.Baumgardner D., et al. , Deducing the composition of venus cloud particles with the autofluorescence nephelometer (AFN). Aerospace 9, 492 (2022). [Google Scholar]
- 63.Seager S., et al. , Venus life finder habitability mission: Motivation, science objectives, and instrumentation. Aerospace 9, 733 (2022). [Google Scholar]
- 64.Agrawal R., et al. , Mission architecture to characterize habitability of venus cloud layers via an aerial platform. Aerospace 9, 359 (2022). [Google Scholar]
- 65.Buchanan W. P., et al. , Aerial platform design options for a life-finding mission at venus. Aerospace 9, 363 (2022). [Google Scholar]
- 66.Ligterink N. F. W., et al. , The ORIGIN space instrument for detecting biosignatures and habitability indicators on a venus life finder mission. Aerospace 9, 312 (2022). [Google Scholar]
- 67.Seager S., et al. , Venus life finder missions motivation and summary. Aerospace 9, 385 (2022). [Google Scholar]
- 68.Taniguchi M., Lindsey J. S., Database of absorption and fluorescence spectra of> 300 common compounds for use in photochem CAD. Photochem. Photobiol. 94, 290–327 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Seager S., Stability of nucleic acid bases in concentrated sulfuric acid: implications for the habitability of Venus’ clouds. Zenodo. https://zenodo.org/record/7808571. Deposited 18 May 2023. [DOI] [PMC free article] [PubMed]
- 70.Willcott M. R., MestRe Nova. J. Am. Chem. Soc. 131, 13180 (2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Ascii files and zip files data have been deposited in Zenodo (69).