Significance
mRNA translation is required for memory consolidation, as its inhibition causes impairment of long-term memory formation. How mRNA translation impacts cognition is not fully understood. In the current study, we show that eIF4G1, a eukaryotic translation initiation factor, promotes the synthesis of mitochondrial proteins involved in energy metabolism. eIF4G1 deficiency results in a reduction in mitochondrial energy production, leading to abnormal morphology of in vitro cultured primary hippocampal neurons. eIF4G1 haploinsufficient mice display impaired hippocampus-dependent learning and memory. Our findings demonstrate a critical role of eIF4G1-mediated translational control in memory-associated early neurodevelopment via the control of mitochondrial energy metabolism.
Keywords: translational control, mitochondria, oxidative phosphorylation, neuronal morphogenesis, learning and memory
Abstract
mRNA translation initiation plays a critical role in learning and memory. The eIF4F complex, composed of the cap-binding protein eIF4E, ATP-dependent RNA helicase eIF4A, and scaffolding protein eIF4G, is a pivotal factor in the mRNA translation initiation process. eIF4G1, the major paralogue of the three eIF4G family members, is indispensable for development, but its function in learning and memory is unknown. To study the role of eIF4G1 in cognition, we used an eIF4G1 haploinsufficient (eIF4G1-1D) mouse model. The axonal arborization of eIF4G1-1D primary hippocampal neurons was significantly disrupted, and the mice displayed impairment in hippocampus-dependent learning and memory. Translatome analysis showed that the translation of mRNAs encoding proteins of the mitochondrial oxidative phosphorylation (OXPHOS) system was decreased in the eIF4G1-1D brain, and OXPHOS was decreased in eIF4G1-silenced cells. Thus, eIF4G1-mediated mRNA translation is crucial for optimal cognitive function, which is dependent on OXPHOS and neuronal morphogenesis.
Control of eukaryotic translation initiation, a rate-limiting step in protein synthesis, has been extensively studied in many biological processes, including brain function, such as synaptic plasticity and memory (1). Translation initiation is primarily regulated by two initiation factor complexes: eIF2-GTP-tRNAiMet ternary complex and the eIF4F complex (2). eIF4F facilitates eukaryotic translation by recruiting the 43S preinitiation complex to the messenger RNA (mRNA) 5′ cap structure (m7GpppN, where N is any nucleotide). eIF4F consists of three subunits: the cap-binding protein eIF4E, ATP-dependent DEAD-box RNA helicase eIF4A, and scaffold protein eIF4G. The formation of eIF4F is suppressed by the eIF4E-binding proteins (4E-BPs); when unphosphorylated, the latter binds eIF4E and prevents its interaction with eIF4G to form the eIF4F complex, resulting in the inhibition of cap-dependent mRNA translation (3). Translational control by eIF4E and 4E-BPs is critical for brain function (4–7). However, how translational regulation mediated by other eIF4F components, such as eIF4G, is associated with cognition is not understood (see also SI Appendix, Supporting Text).
eIF4G1, the most abundant form of three eIF4G family proteins, is indispensable for development since the homozygous depletion causes lethality in yeast and during embryogenesis in mice (8, 9). Reduction in eIF4G1 amount activates catabolic pathways and inhibits anabolic pathways, which phenocopies nutrient depletion or inhibition of a key nutrient sensor, mechanistic target of rapamycin complex 1 (mTORC1) (10). Moreover, as one of the targets of E3 ubiquitin ligase culin3 (CUL3), eIF4G1 protein levels are increased in the brain of CUL3-deficient mice showing social deficits and anxiety-like behaviors (11). eIF4G1 contains a microexon that recruits the translation repressor FMRP-containing neuronal granule to suppress the translation of synaptic mRNAs (12). When the splicing out of the eIF4G1 microexon is disrupted, it engenders excessive synaptic translation and deficits in social behavior and memory associated with autism (12). These observations suggest that a subset of mRNAs is translationally regulated in an eIF4G1-sensitive manner, but the direct targets of eIF4G1-mediated translational control and their role in brain function are not known.
Mitochondria is the cellular powerhouse generating adenosine 5′-triphosphate (ATP) as the energy source for most cellular processes. ATP is synthesized through mitochondrial oxidative phosphorylation (OXPHOS) or glycolysis. Notably, energy metabolism in the brain is highly specialized in a cell-type–dependent manner–neurons are predominantly oxidative, while astrocytes are largely glycolytic (13, 14). Neurons consume a large part of the brain’s energy to generate action potentials and mediate synaptic transmission and neuronal morphogenesis during early brain development (15). Morphogenesis of axons and dendrites is critical for neurons to form a fundamental neural network structure for synaptic plasticity and cognition (16, 17).
We studied a previously reported animal model, eIF4G1 haploinsufficient mouse, heterozygously harboring a premature termination codon in the Eif4g1 gene caused by a CRISPR/Cas9-generated frameshift (1-nucleotide deletion, hereafter referred to as 1D) (9). Translatome analysis showed that translation of a subset of mRNAs involved in mitochondrial OXPHOS was decreased in the eIF4G1-1D brain. Accordingly, mitochondrial respiration was reduced in eIF4G1-silenced cells. eIF4G1-1D primary hippocampal neurons exhibited defective axonal arborization that caused a disruption in neuronal connectivity. eIF4G1-1D mice were impaired in hippocampus-dependent learning and memory. The study elucidates the mechanism by which eIF4G1 impacts cognitive function by coupling energy metabolism and neurodevelopment.
Results
eIF4G1 is Expressed in the Mouse Brain.
eIF4G1 protein in the mouse brain was examined by immunoblotting and immunofluorescence (SI Appendix, Fig. S1 A and B). Importantly, eIF4G1 protein levels in the brain are high from the embryonic (E13.5) to early postnatal (P14) period and then rapidly decrease (SI Appendix, Fig. S1C), implying a possible role in early brain development. eIF4G1 is mainly localized to the cytosol and synaptosomes (SI Appendix, Fig. S1D). Immunofluorescence displayed strong coexpression of eIF4G1 with the neuronal marker NeuN, including excitatory (CaMKIIα+) and inhibitory (GAD67+) neurons in the hippocampus (SI Appendix, Fig. S1E).
eIF4G1 protein levels in the hippocampus and cortex in eIF4G1 haploinsufficient (eIF4G1-1D) mice are approximately 50% decreased at two different ages – P14 and 10 wk (SI Appendix, Fig. S2 A and B). The levels of eIF4G2 (DAP5), eIF4G3, and other eIF4F components are not significantly altered in the eIF4G1-1D brain (SI Appendix, Fig. S2C), indicating that the reduction of eIF4G1 is compensated by neither the eIF4G paralogs nor eIF4F components.
Translation of mRNAs Encoding Mitochondrial OXPHOS Genes Is Decreased in the eIF4G1-1D Brain.
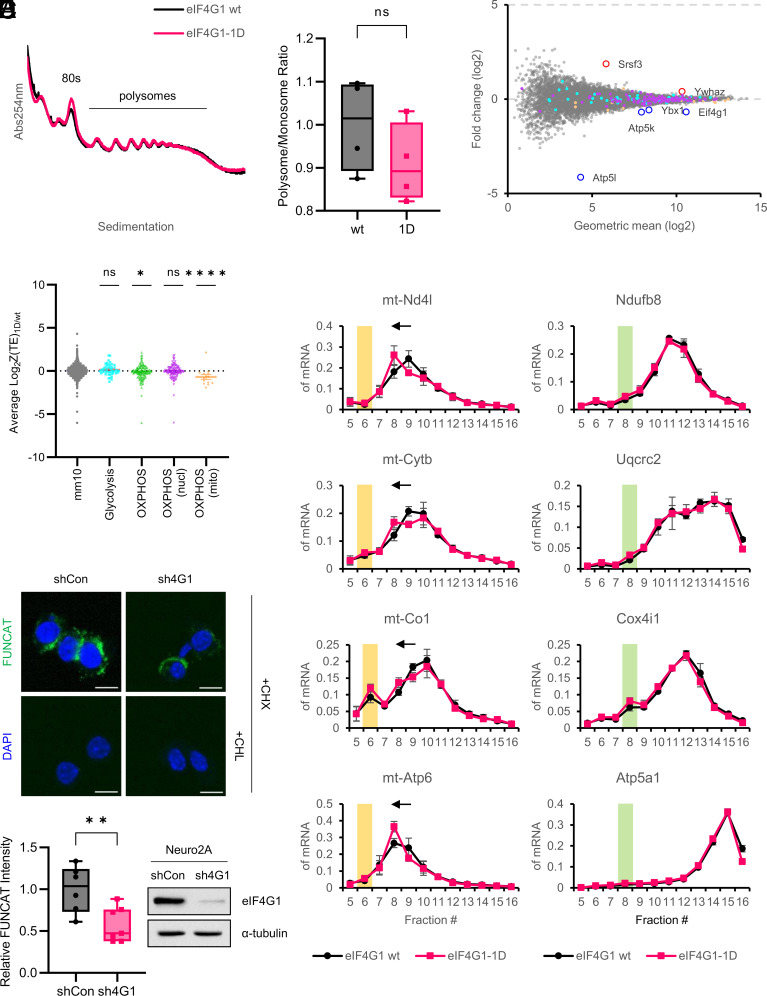
There is no difference in the polysome profiles of P14 forebrains between the genotypes (Fig. 1A), and the ratio of polysome to monosome, an indicator of the rate of global translation, is not significantly changed in the eIF4G1-1D brain (Fig. 1B). This result is consistent with a previous study, which showed that global translation in MCF10A, a human mammary epithelial cell line, is not dramatically affected upon 90% reduction of eIF4G1 levels (10).
Fig. 1.
Mitochondrial OXPHOS translation is decreased by reduction of eIF4G1. (A) Representative polysome profiles of eIF4G1 wt (black) and eIF4G1-1D (red) P14 forebrain. 80S monosome and polysomes are indicated. (B) Relative ratio of polysome to monosome. The graph is shown as the mean ± SEM of four biological replicates for each genotype. wt (1.000 ± 0.054) vs. 1D (0.909 ± 0.046); P = 0.249; ns, not significant (two-tailed Student’s t-test). (C) Differential gene expression analysis. The scatter plot shows a log2 fold change of translation efficiency (TE) of eIF4G1-1D over eIF4G1 wt vs. geometric mean (log2). Differentially translated genes (|Z-score| > 2.576) are indicated in red (up-regulated) or blue (down-regulated) outlined circles with their name. Genes involved in glycolysis are indicated in cyan, and nuclear- and mitochondria-encoded OXPHOS genes are indicated in purple and orange, respectively. (D) Z-score analysis. The scatter plot shows a log2 fold change of TE of eIF4G1-1D over eIF4G1 wt of groups of genes involved in the indicated biological function; ns, not significant; *P < 0.05; ****P < 0.0001 (one-way ANOVA followed by Dunnett’s post hoc test). (E) Polysome-qPCR analysis of mitochondria-encoded (E, Left) and nuclear-encoded (E, Right) OXPHOS genes for OXPHOS complex I, III, IV, and V (from top to bottom). The graph is shown as the mean ± SEM of five biological replicates for each genotype. The fractions corresponding to mitochondrial monosome (55S) or cytosolic monosome (80S) are highlighted in yellow or green boxes, respectively. (F) Mitochondria-specific fluorescent noncanonical amino acid tagging (mito-FUNCAT) in eIF4G1 stable knockdown cells. (F, Upper) Neuro2A cells stably expressing control shRNA (shCon) or eIF4G1 shRNA (sh4G1) were treated with cycloheximide (CHX) and with or without chloramphenicol (CHL). The mitochondrial FUNCAT signal is shown in green. Nuclei were stained by DAPI (blue). (Scale bar, 10 µm.) (F, Lower, Left) Relative FUNCAT intensity. Each point indicates the intensity of FUNCAT signal from a single image shCon (n = 6; 1.000 ± 0.114) vs. sh4G1 (n = 7; 0.577 ± 0.078); **P < 0.01 (two-tailed Student’s t-test). (F, Lower, Right) Immunoblot validation of knockdown of eIF4G1 in sh4G1 cells. α-tubulin was used as a loading control.
To investigate the eIF4G1-sensitive brain translatome, we performed ribosome profiling using P14 forebrains (SI Appendix, Fig. S3A). Ribosome-protected footprints (RPFs) for Ribo-Seq were generated by collecting ribosome-protected mRNA fragments from brain lysates treated with micrococcal nuclease (MNase), which provides a better yield of monosomes than RNase I (18, 19). RNA fragments for RNA-Seq were generated by alkaline fragmentation of total RNA. Multiplex sequencing generated approximately 2.5 to 9 million reads uniquely assigned to the mouse genome (UCSC.mm10) after rRNA and tRNA removal and PCR deduplication (SI Appendix, Fig. S3B). The sequencing libraries were strongly correlated among three biological replicates (Pearson correlation coefficient > 0.995 for RNA-Seq and > 0.985 for Ribo-Seq) (SI Appendix, Fig. S3C). The RPFs have a narrow size distribution with a peak of approximately 33 nucleotides, a typical length for MNase-digested RPFs (19, 20), whereas the RNA fragments have a broad length distribution ranging from 30 to 50 nucleotides (SI Appendix, Fig. S3D). The RPFs were predominantly (> 90%) enriched in the coding sequence (CDS), while the RNA fragments were distributed in CDS, 3′-UTR (UTR3), and 5′-UTR (UTR5) in order of enrichment (SI Appendix, Fig. S3E). Three-nucleotide periodicity was not observed for the RPFs (SI Appendix, Fig. S3F) because of the use of MNase (18). Metagene analysis showed that the RPFs were enriched for CDS, especially near the start codon (SI Appendix, Fig. S3G), showing that our libraries are well-suited for further analysis.
We analyzed translation efficiency (TE) using the Z-score transformation method (21). The differential gene analysis based on Z-score displayed six genes that passed the arbitrary threshold (|Z-score| > 2.576) (Fig. 1C and Dataset S1)—Two genes displayed increased TE: Serine and Arginine Rich Splicing Factor 3 (Srsf3), Tyrosine 3-Monooxygenase/Tryptophan 5-Monooxygenase Activation Protein Zeta (Ywhaz), four genes exhibited decreased TE: ATP Synthase Membrane Subunit G (Atp5l), Eif4g1, Y-Box Binding Protein 1 (Ybx1), and ATP synthase subunit e, mitochondrial (Atp5k). Strikingly, the average TE of 135 genes involved in OXPHOS (green dots) was significantly decreased in the eIF4G1-1D brains, while the average TE of 67 genes involved in glycolysis (cyan dots) was not (Fig. 1D). Moreover, the decreased TE of OXPHOS genes was due to the mitochondria-encoded OXPHOS genes (orange dots), but not to the nuclear-encoded OXPHOS genes (purple dots), indicating that translation of mRNAs encoding mitochondrial OXPHOS proteins is modulated by cytosolic eIF4G1 (Discussion). To bolster this result, we analyzed the polysomal distribution of nuclear- vs. mitochondria-encoded genes for OXPHOS complex I, III, IV, and V (OXPHOS complex II, succinate dehydrogenase, is composed of four nuclear-encoded proteins). The polysomal distribution of nuclear-encoded OXPHOS mRNAs of all four OXPHOS complexes was almost identical between the genotypes (Fig. 1 E, Right). In contrast, the polysomal distribution of mitochondria-encoded OXPHOS mRNAs in the eIF4G1-1D brains showed a reproducible shift from fraction #9 to #8 (Fig. 1 E, Left).
To directly examine whether mitochondrial translation is affected by eIF4G1, we performed fluorescent noncanonical amino acid tagging (FUNCAT) (22) using Neuro2A cells, a mouse neuroblastoma cell line, stably expressing control shRNA or eIF4G1 shRNA (hereafter referred to as shCon and sh4G1, respectively) (Fig. 1 F, Lower Right). To visualize mitochondria-specific translation, cytosolic translation was selectively inhibited by eukaryotic translation inhibitor cycloheximide (CHX) (23, 24). Mitochondrial translation was not altered by CHX since mitochondrial ribosomal proteins (mitoRPs) have a long half-life (approximately 7 d in cultured neurons) (25), while it was completely suppressed by mitochondrial translation inhibitor chloramphenicol (CHL) (Fig. 1 F, Upper). The intensity of the mitochondrial FUNCAT signal was 40% decreased in sh4G1 compared to shCon cells (Fig. 1 F, Lower Left). These data demonstrate that eIF4G1 affects mitochondrial translation.
Mitochondrial Respiration, Not Glycolysis, Is Altered in eIF4G1-Silenced Cells.
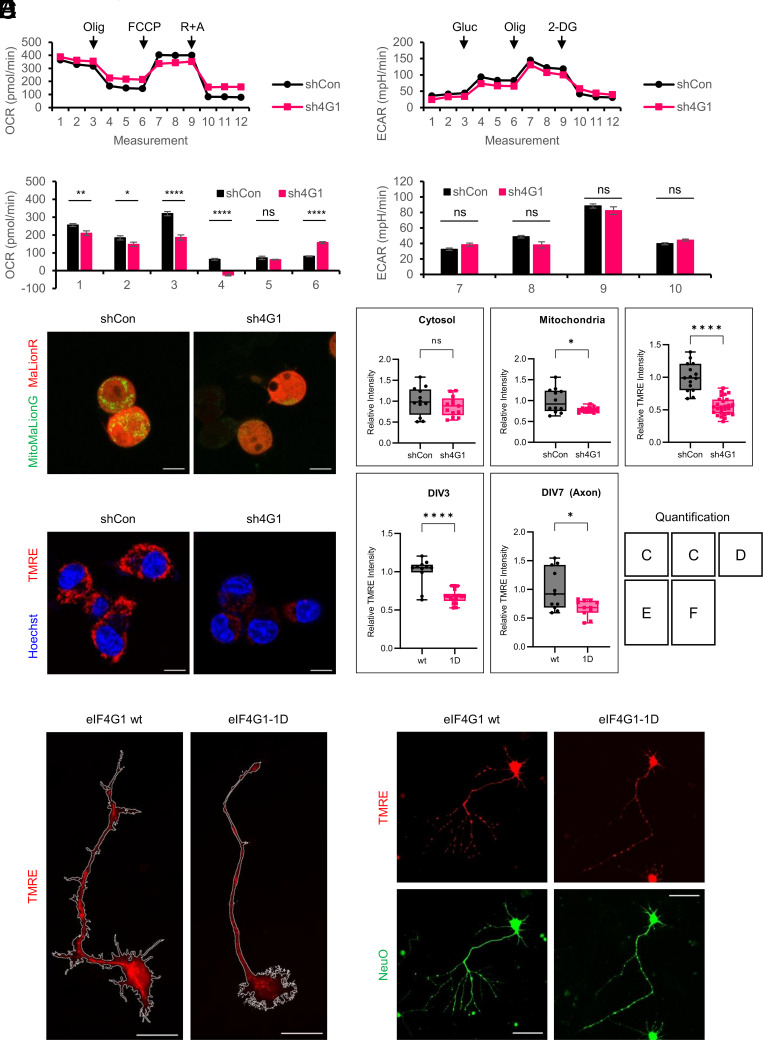
Mitochondria possess 13 protein-coding genes that encode crucial components of the OXPHOS complexes. It is therefore predicted that the reduction of mitochondrial translation should result in diminished mitochondrial respiration. Oxygen consumption rate (OCR) measured in shCon and sh4G1 cells showed that basal and ATP-linked mitochondrial respiration in eIF4G1-deficient cells were reduced by 18% and 20%, respectively (Fig. 2A). Importantly, reduced maximal respiration and impaired spare capacity in sh4G1 cells indicate that eIF4G1-deficient cells cannot properly respond to energy demand and/or have deficits in the energy-producing machinery. In contrast, extracellular acidification rate (ECAR) mediated by glycolysis, glycolytic capacity, glycolytic reserve, and non-glycolytic acidification was not altered in sh4G1 cells (Fig. 2B), which is consistent with the ribosome profiling data (Fig. 1D). As expected, mitochondrial ATP levels monitored by fluorescence protein-based ATP sensors in the cytosol or mitochondria were 20% decreased in sh4G1 compared to shCon cells (Fig. 2C).
Fig. 2.
Mitochondrial OXPHOS is decreased in eIF4G1-silenced cells and eIF4G1-1D primary hippocampal pyramidal neurons. (A) Seahorse mito stress test. (A, Upper) Oxygen consumption rate (OCR) was measured for shCon and sh4G1 cells after treatment with the indicated compounds. Olig, oligomycin; FCCP, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone; R, rotenone; A, antimycin A. (A, Lower) Each point represents the mean ± SEM. n = 15 for shCon (black) and 15 for sh4G1 (red); 1, basal respiration; 2, ATP-linked respiration; 3, maximal respiration capacity; 4, reserve capacity; 5, proton leak; 6, non-mitochondrial respiration; ns, not significant; *P < 0.05; **P < 0.01; ****P < 0.0001 (two-way ANOVA followed by Sidak’s post hoc test). (B) Seahorse glycolytic stress test. (B, Upper) Extracellular acidification rate (ECAR) was measured for shCon and sh4G1 cells after treatment with the indicated drugs. Gluc, glucose; Olig, oligomycin; 2-DG, 2-deoxy-d-glucose. (B, Lower) Each point represents the mean ± SEM. n = 15 for shCon (black) and 15 for sh4G1 (red); 7, non-glycolytic acidification; 8, glycolysis; 9, glycolytic capacity; 10, glycolytic reserve; ns, not significant (two-way ANOVA followed by Sidak’s post hoc test). (C) ATP levels in eIF4G1 stable knockdown cells. Cells were co-transfected with MaLionR and MitoMaLionG to represent cytosolic (red) and mitochondrial (green) ATP levels, respectively. (Scale bar, 10 µm.) (Quantification) Relative intensity of cytosolic (C, Left) and mitochondrial (C, Right) fluorescence. wt (n = 12; cytosol, 1.000 ± 0.098; mitochondria, 1.000 ± 0.084) vs. 1D (n = 13; cytosol, 0.876 ± 0.065; mitochondria, 0.785 ± 0.018); ns, not significant; *P < 0.05 (two-tailed Student’s t-test). (D) Mitochondrial membrane potential (ΔΨm) in eIF4G1 stable knockdown cells. shCon and sh4G1 cells were treated with tetramethylrhodamine ethyl ester (TMRE, red) and Hoechst (blue). (Scale bar, 10 µm.) (Quantification) Relative TMRE intensity. Each point indicates the intensity of the TMRE signal from a single image. wt (n = 15; 1.000 ± 0.056) vs. 1D (n = 27; 0.551 ± 0.026); ****P < 0.0001 (two-tailed Student’s t-test). (E) Mitochondrial membrane potential (ΔΨm) in DIV3 primary hippocampal pyramidal neurons. eIF4G1 wt (E, Left) or eIF4G1-1D (E, Right) neurons were treated with TMRE (red) at DIV3. The boundary of the neuron is indicated in a white line. (Scale bar, 20 µm.) (Quantification) wt (n = 10; 1.000 ± 0.054) vs. 1D (n = 18; 0.675 ± 0.021); ****P < 0.0001 (two-tailed Student’s t-test). (F) ΔΨm in DIV7 primary hippocampal pyramidal neurons. eIF4G1 wt (F, Left) or eIF4G1-1D (F, Right) neurons were co-treated with TMRE (red) and NeuO (green) at DIV7. (Scale bar, 50 µm.) (Quantification) wt (n = 11; 1.000 ± 0.109) vs. 1D (n = 11; 0.676 ± 0.043); *P < 0.05 (two-tailed Student’s t-test).
Mitochondrial membrane potential (ΔΨm), generated by a proton gradient across the inner mitochondrial membrane, is a critical driving force for synthesizing ATP in the OXPHOS process and is widely used as a marker for mitochondrial activity (26). ΔΨm, measured by tetramethylrhodamine ethyl ester (TMRE), a cationic fluorescent dye sequestered by active mitochondria, was 45% decreased in sh4G1 compared to shCon cells (Fig. 2D). Furthermore, ΔΨm was 30% decreased in eIF4G1-1D neurons at 3 d in vitro (DIV3) (Fig. 2E) and in the axon of eIF4G1-1D neurons at DIV7 (Fig. 2F). Taken together, these results demonstrate that the reduction in eIF4G1-mediated OXPHOS translation results in suppression of mitochondrial respiration and ATP production.
Reduced Mitochondrial Respiration Alters the Morphology of eIF4G1-1D Neurons.
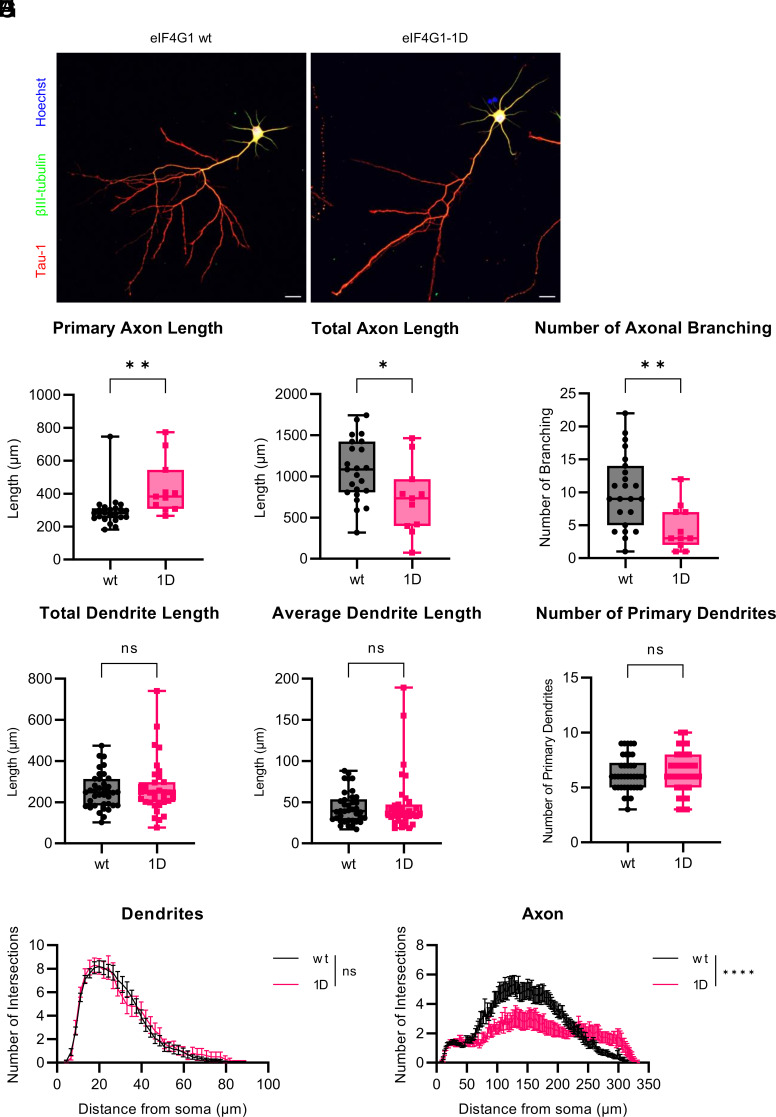
Since ATP is important for axonal branch formation (27, 28), we investigated the axonal morphology of in vitro cultured primary neurons (SI Appendix, Fig. S4A). eIF4G1 protein levels in eIF4G1-1D primary hippocampal pyramidal neurons were 80% decreased compared to eIF4G1 wt neurons (SI Appendix, Fig. S4B). Axonal morphology was measured in DIV3 neurons (SI Appendix, Fig. S4C), focusing on primary and total axon length, the number of axonal branching, and the distance from the soma to the 1st branch. No apparent difference between genotypes (SI Appendix, Fig. S4 D–G). Next, axonal arborization and dendritic outgrowth were measured in DIV7 neurons. Strikingly, the axonal morphology of eIF4G1-1D neurons was dramatically disrupted (Fig. 3A). The primary axon of eIF4G1-1D neurons was 50% longer than that of eIF4G1 wt neurons (Fig. 3B), whereas the total axon length of eIF4G1-1D neurons was reduced by 50% compared to eIF4G1 wt neurons (Fig. 3C). This was because the number of axonal branching was halved in eIF4G1-1D compared to eIF4G1 wt neurons (Fig. 3D). In contrast, total dendritic length (Fig. 3E), average dendritic length (Fig. 3F), and the number of primary dendrites (Fig. 3G) were not significantly altered in eIF4G1-1D neurons. Sholl analysis revealed consistent morphological alteration, demonstrating that axonal complexity was dramatically decreased in eIF4G1-1D neurons (Fig. 3 H, Right), whereas dendritic outgrowth was almost identical between the genotypes (Fig. 3 H, Left). These data indicate that eIF4G1 shapes the axonal morphology of hippocampal neurons by affecting mitochondrial respiration.
Fig. 3.
Axonal branching is disrupted in eIF4G1-1D hippocampal pyramidal neurons. (A) Representative images of DIV7 eIF4G1 wt (A, Left) or eIF4G1-1D (A, Right) hippocampal pyramidal neuron. Axons were stained using Tau-1 (red), and dendrites were stained using βIII-tubulin (green). Nuclei were stained by Hoechst (blue). (Scale bar, 20 µm.) (B) Primary axon length. wt (n = 23; 296.382 ± 22.331 μm) vs. 1D (n = 11; 435.421 ± 48.890 μm); **P < 0.01 (two-tailed Student’s t-test). (C) Total axon length. wt (1076.248 ± 77.833 μm) vs. 1D (724.806 ± 128.100 μm); *P < 0.05 (two-tailed Student’s t-test). (D) Number of axonal branching. wt (10.174 ± 1.159) vs. 1D (4.636 ± 1.038); **P < 0.01 (two-tailed Student’s t-test). (E) Total dendrite length. wt (n = 38; 255.365 ± 13.802 μm) vs. 1D (n = 35; 269.662 ± 21.789 μm); ns, not significant; P = 0.579 (two-tailed Student’s t-test). (F) Average dendrite length. wt (43.596 ± 3.102 μm) vs. 1D (47.597 ± 5.995 μm); ns, not significant; P = 0.550 (two-tailed Student’s t-test). (G) Number of primary dendrites. wt (6.263 ± 0.258) vs. 1D (6.371 ± 0.313); ns, not significant; P = 0.791 (two-tailed Student’s t-test). (H) Sholl analysis of dendrites (H, Left) or axon (H, Right) of eIF4G1 wt (black) or eIF4G1-1D (red) DIV7 neurons. The graph is shown as the mean ± SEM of neurons for each genotype. n = 23 (dendrites and axon) for eIF4G1 wt (black) and 10 (dendrites) or 11 (axon) for eIF4G1-1D (red). ns, not significant; ****P < 0.0001 (two-way ANOVA followed by Sidak’s post hoc test).
eIF4G1-1D Mice Are Impaired in Hippocampus-Dependent Learning and Memory.
We next examined the behavioral features of eIF4G1-1D mice (29). The self-grooming test measures spontaneous repetitive behavior, which is often increased in autism spectrum disorder (30). eIF4G1-1D mice showed normal self-grooming levels (SI Appendix, Fig. S5A). The open-field test showed that locomotor activity and total time spent in the center of eIF4G1-1D mice were not altered (SI Appendix, Fig. S5B). The elevated plus maze and the light-dark transition test revealed no difference in anxiety levels between the genotypes (SI Appendix, Fig. S5 C and D). The rotarod test showed normal motor learning in eIF4G1-1D mice (SI Appendix, Fig. S5E). The body weight was similar between the genotypes (SI Appendix, Fig. S5F).
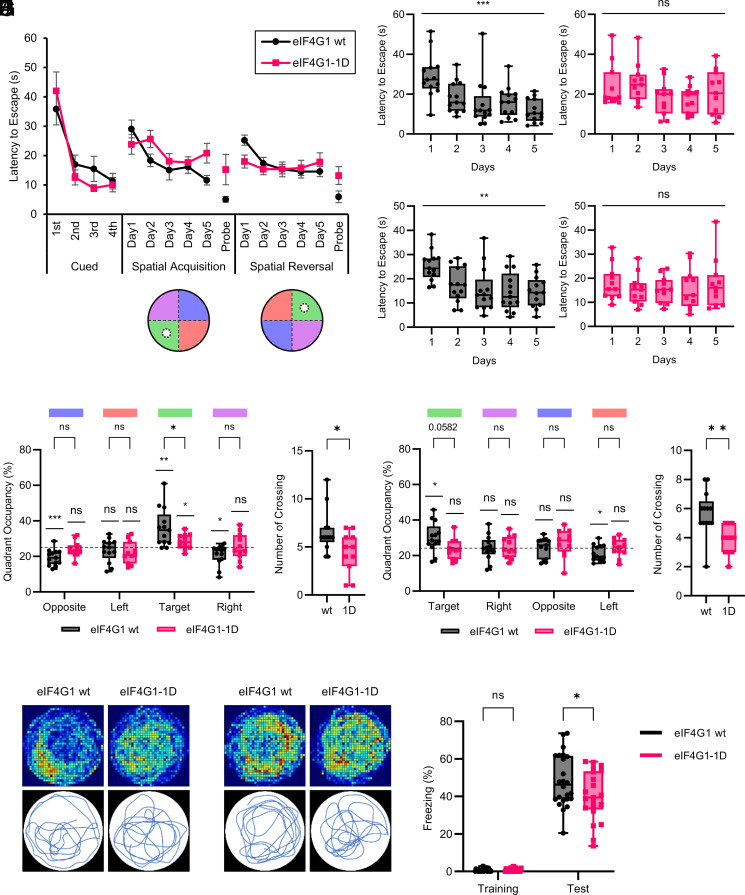
The Morris water maze test was performed to investigate hippocampus-dependent spatial learning and memory (31). The latency to escape during a cued learning phase using a visible platform was similar between the genotypes (Fig. 4A), indicating no impairment in motivation, vision, or motor ability in eIF4G1-1D mice. Using a hidden platform, eIF4G1 wt mice exhibited spatial and reversal learning as their latency to escape was decreased during the 5-d training in a spatial memory acquisition phase (Fig. 4 B, Left; from 29.06 ± 3.03 to 11.62 ± 1.60 s) and a reversal memory acquisition phase (Fig. 4 C, Left; from 25.23 ± 1.72 to 14.58 ± 1.77 s). Strikingly eIF4G1-1D mice displayed impairment in finding the hidden platform in both phases (Fig. 4 B, Right; from 25.61 ± 2.94 to 20.82 ± 3.34 s; and Fig. 4 C, Right; from 15.36 ± 1.90 to 17.80 ± 3.12 s). Likewise, eIF4G1-1D mice were defective in the retrieval of spatial memory as their time spent in the target quadrant (Fig. 4 D and E; 36.09 ± 2.99% vs. 28.40 ± 1.28%) and the number of platform crossings (Fig. 4F; 6.54 ± 0.62 vs. 4.45 ± 0.64) were significantly reduced compared to eIF4G1 wt mice during a probe test for spatial memory acquisition. The impairment of reversal memory retrieval in eIF4G1-1D mice was less severe since the difference in time spent in the target quadrant between the genotypes was not statistically significant (Fig. 4 G and H; 30.14 ± 2.35% vs. 23.57 ± 1.73%; P = 0.0582). Nevertheless, during a probe test for reversal memory acquisition, eIF4G1-1D mice did not show a clear preference for the target quadrant (Fig. 4G), and the number of platform crossings was significantly decreased compared to eIF4G1 wt mice (Fig. 4I; 5.69 ± 0.43 vs. 3.82 ± 0.30).
Fig. 4.
eIF4G1-1D mice are impaired in hippocampus-dependent learning and memory. (A) Latency to reach the platform in the Morris water maze test. Mice were subjected to four trainings per day during the training phase. Each point is shown as the mean ± SEM. The position of the platform (white circle) and quadrants (target, green; opposite, blue; left, orange; right, purple) used in each phase are shown. n = 13 for eIF4G1 wt (black) and 11 for eIF4G1-1D (red). (B) Latency to reach the hidden platform for each genotype during a spatial memory acquisition phase. wt (black, left; day 1, 29.06 ± 3.03 s; day 2, 18.37 ± 2.14 s; day 3, 15.06 ± 3.35 s; day 4, 16.12 ± 2.14 s; day 5, 11.62 ± 1.60 s) vs. 1D (red, right; day 1, 23.80 ± 3.34 s; day 2, 25.61 ± 2.94 s; day 3, 18.07 ± 2.64 s; day 4, 17.61 ± 1.94 s; day 5, 20.82 ± 3.34 s); ns, not significant; ***P < 0.001 (one-way ANOVA followed by a test for linear trend). (C) Latency to reach the hidden platform for each genotype during a reversal memory acquisition phase. wt (black, left; day 1, 25.23 ± 1.72 s; day 2, 17.37 ± 2.01 s; day 3, 15.31 ± 2.54 s; day 4, 14.56 ± 2.22 s; day 5, 14.58 ± 1.77 s) vs. 1D (red, right; day 1, 17.98 ± 2.19 s; day 2, 15.36 ± 1.90 s; day 3, 15.39 ± 1.63 s; day 4, 15.82 ± 2.57 s; day 5, 17.80 ± 3.12 s); ns, not significant; **P < 0.01 (one-way ANOVA followed by a test for linear trend). (D) Time spent in each of the four quadrants of each genotype during a spatial memory acquisition phase. wt (black; opposite, 19.41 ± 1.23%; left, 23.44 ± 1.80%; target, 36.09 ± 2.99%; right, 20.95 ± 1.59%) vs. 1D (red; opposite, 24.09 ± 1.36%; left, 22.00 ± 1.90%; target, 28.40 ± 1.28%; right, 25.39 ± 2.24%); ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001 [two-way ANOVA followed by Sidak’s post hoc test, one sample t-test against 25]. (E) Travel path of each genotype during a spatial memory acquisition phase. (E, Upper) Heatmap of the cumulative travel path of all mice of each genotype. (E, Lower) Representative travel path of a single mouse of each genotype. (F) Number of platform crossings in a probe test for spatial memory acquisition. wt (black; 6.54 ± 0.62) vs. 1D (red; 4.45 ± 0.64); *P < 0.05 (two-tailed Student’s t-test). (G) Time spent in each of the four quadrants of each genotype during a reversal memory acquisition phase. wt (black; target, 30.14 ± 2.35%; right, 24.16 ± 1.96%; opposite, 24.37 ± 1.57%; left, 21.21 ± 1.28%) vs. 1D (red; target, 23.57 ± 1.73%; right, 24.94 ± 1.87%; opposite, 27.05 ± 2.31%; left, 24.32 ± 1.55%); ns, not significant; *P < 0.05 [two-way ANOVA followed by Sidak’s post hoc test, one sample t-test against 25]. (H) Travel path of each genotype during a reversal memory acquisition phase. (H, Upper) Heatmap of the cumulative travel path of all mice of each genotype. (H, Lower) Representative travel path of a single mouse of each genotype. (I) Number of platform crossings in a probe test for reversal memory acquisition. wt (black; 5.69 ± 0.43) vs. 1D (red; 3.82 ± 0.30); **P < 0.01 (two-tailed Student’s t-test). (J) Percentage of freezing of mice during a training and test phase in the contextual fear conditioning test. Each point represents data from an individual mouse, shown as the mean ± SEM n = 24 for eIF4G1 wt (black) and 21 for eIF4G1-1D (red); wt (training, 0.64 ± 0.17%; test, 48.30 ± 2.82%) vs. 1D (training, 0.66 ± 0.18%; test, 40.20 ± 2.91%); ns, not significant; *P < 0.05 (two-way ANOVA followed by Sidak’s post hoc test).
A contextual fear conditioning test was performed to assess context-dependent fear memory, which is hippocampus-dependent. eIF4G1-1D mice displayed reduced freezing time compared to eIF4G1 wt mice during a test phase, 24 h after training (48.30 ± 2.82% vs. 40.20 ± 2.91%), demonstrating that contextual fear memory was impaired in eIF4G1-1D mice (Fig. 4J). Taken together, the results demonstrate that eIF4G1-1D mice are impaired in hippocampus-dependent learning and memory.
Discussion
We documented the behavioral deficits of eIF4G1 haploinsufficient mice as a consequence of eIF4G1 reduction. The mice exhibited hippocampal memory impairment (Fig. 4). Notably, depletion of eIF4G1 phenocopies the inhibition of mTOR, a critical regulator of anabolic and catabolic processes (10), indicating that eIF4G1-mediated mRNA translational control plays a crucial role in cellular processes. Neuro/gliogenesis in the brain mostly occur during the late embryonic days (32), as eIF4G1 protein levels in the brain are high throughout embryo development and up to 2 wk after birth (SI Appendix, Fig. S1C), pointing to a critical function of eIF4G1 during early brain development, including neuronal morphogenesis.
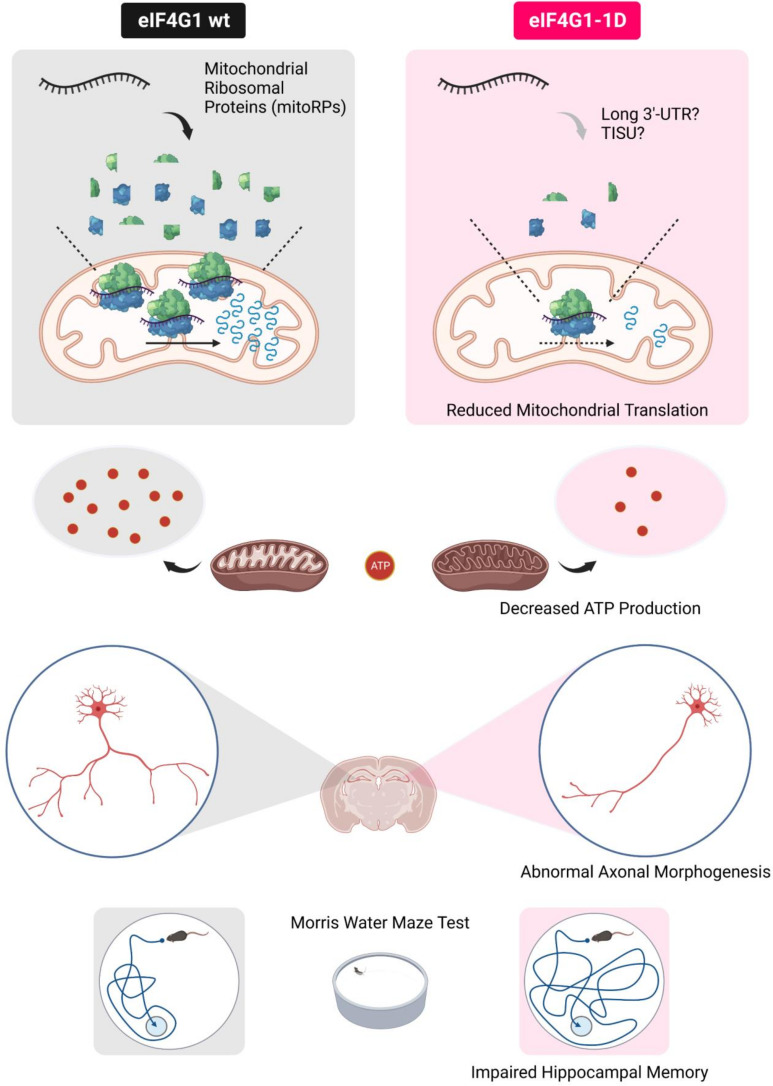
eIF4G1 is not present in mitochondria. Mitochondrial mRNAs do not possess a 5′ cap and most do not contain a 5′-UTR (33, 34). Thus, the direct control of mitochondrial protein synthesis by eIF4G1 is precluded. How does eIF4G1 affect mitochondrial translation? A previous study using eIF4G1-silenced cells showed that mRNAs decreased in eIF4G1-depleted polysomes contain longer 3′-UTR than those that are increased (10). Ribosome recycling and reinitiation in mRNAs containing long 3′-UTR requires eukaryotic initiation factors for maintaining closed loop formation (35), and eIF4G1 plays a crucial role in this process by interacting with eIF4E and PABP (3). Notably, the 3′-UTR of mitoRP mRNAs is approximately twice longer than that of cytosolic ribosomal protein (cytoRP) mRNAs (768 ± 135 nt vs. 349 ± 75.6 nt), whereas there is no significant difference in the length of 5′-UTR (87.5 ± 9.87 nt vs. 88.9 ± 6.42 nt) and CDS (652 ± 31.9 nt vs. 507 ± 24.5 nt). In addition, mRNAs containing translation initiator of short 5′-UTR (TISU) element in their 5′-UTR require eIF4G1 to initiate translation under energy deprivation conditions (36). TISU is enriched in mRNAs encoding proteins involved in mitochondrial energy metabolism and in mitoRPs (36). TISU is twofold enriched in mitoRP mRNAs (26/77) relative to cytoRP mRNAs (13/82) in mice (36). These observations are consistent with the model that mitochondrial translation is decreased in eIF4G1-deficient cells because of mitoRPs reduction (Fig. 5).
Fig. 5.
eIF4G1-mediated mitochondrial translation is required for learning and memory. The role of eIF4G1-mediated mRNA translation in cognitive function. Translation of mRNAs encoding mitochondrial ribosomes is selectively reduced in the brain of eIF4G1 haploinsufficient (eIF4G1-1D) mice. This leads to a decrease in mitochondrial translation and ATP production, which consequently result in diminished complexity of axonal arborization of hippocampal pyramidal neurons. The disruption of hippocampal neuron morphology interdicted hippocampus-dependent learning and memory tasks, such as the Morris water maze test.
This study presents molecular, metabolic, and cellular consequences of eIF4G1 deficiency. However, most of our experiments were carried out using in vitro cultured primary neurons and a neuronal cell line which provides better transfection efficiency and consistency to counter environmental stresses. In vivo studies, including electrophysiology, are required to fully understand hippocampus-dependent memory impairment. Recent techniques, such as in vivo imaging of ATP levels in the brain (37) and visualization of in vivo hippocampal neurons using retrovirus (38), will inform on the mechanisms of behavioral deficits in eIF4G1-1D mice. It is necessary to investigate whether an infusion of BDNF and netrin, which promote axonal arborization (39), into the eIF4G1-1D hippocampus during the early postnatal days could rescue neural connectivity and behavioral deficits.
Materials and Methods
Mice.
Genotyping of eIF4G1-1D mice was carried out as previously described (9). Male mice aged 3 to 4 mo were used for experiments. Mice were cared for in compliance with the guidelines of the Canadian Council on Animal Care and approved by the McGill University Animal Care Committee.
Generation of eIF4G1 Stable Knockdown Cells.
eIF4G1 stable knockdown cells were generated in Neuro2A cells by lentiviral transduction (40). Briefly, lentivirus expressing control shRNA (SHC002) or Eif4g1 shRNA (TRCN0000096812) was produced in HEK293T cells with lentiviral packaging plasmids (pLP-1, pLP-2, and pLP/VSVG) (Invitrogen) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After 48 h, the medium containing lentivirus was collected, filtered using a 0.45 µm syringe filter (Fisher Scientific), and stored at −80°C. Frozen lentivirus was thawed on ice and added to Neuro2A cells with 5 µg/mL of polybrene. Positive colonies were selected using 5 µg/mL of puromycin and maintained in the normal culture medium containing 2 µg/mL of puromycin.
Further information is available in SI Appendix, Materials and Methods.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Acknowledgments
We thank Karim Nader and Claudio Cuello for providing access to behavioral testing rooms, Arkady Khoutorsky, Wayne Sossin, and Heidi McBride for critical reading of the manuscript, Annie Sylvestre and Annik Lafrance for animal care, and Eva Migon and Meena Vipparti for technical assistance. We acknowledge technical assistance from the Rosalind and Morris Goodman Cancer Institute Histology Innovation Platform, McGill Platform for Cellular Perturbation, and Metabolomics Innovation Resource. This work was supported by the Canadian Institute of Health Research Foundation grants FDN-148423 (to N.S.) and Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education to S.-H.K (2019R1A6A3A03034060) and J.-H.C. (2020R1A6A3A03040141). S.-H.K. was supported by Canderel Postdoctoral Fellowship. A.Y. was supported by Grant-in-Aid for Scientific Research (C) (18K06975) from the Japan Ministry of Education, Culture, Sports, Science and Technology. Fig. 5 and SI Appendix, Fig. S3A were created with BioRender.com.
Author contributions
S.-H.K. and N.S. designed research; S.-H.K., J.-H.C., L.M.-G., and M.A. performed research; S.-H.K. and A.Y. contributed new reagents/analytic tools; S.-H.K., J.-H.C., L.M.-G., and M.A. analyzed data; and S.-H.K. and N.S. wrote the paper.
Competing interests
N.S. and R.J.S. were coauthors on a 2019 review article. All other authors declare no competing interests.
Footnotes
Reviewers: J.D.R., University of Massachusetts Medical School; and R.J.S., New York University Grossman School of Medicine.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Costa-Mattioli M., Sossin W. S., Klann E., Sonenberg N., Translational control of long-lasting synaptic plasticity and memory. Neuron 61, 10–26 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sonenberg N., Hinnebusch A. G., Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136, 731–745 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gingras A.-C., Raught B., Sonenberg N., eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68, 913–963 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Gkogkas C. G., et al. , Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature 493, 371–377 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santini E., et al. , Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature 493, 411–415 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao R., et al. , Light-regulated translational control of circadian behavior by eIF4E phosphorylation. Nat. Neurosci. 18, 855–862 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aguilar-Valles A., et al. , Translational control of depression-like behavior via phosphorylation of eukaryotic translation initiation factor 4E. Nat. Commun. 9, 2459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park E.-H., Zhang F., Warringer J., Sunnerhagen P., Hinnebusch A. G., Depletion of eIF4G from yeast cells narrows the range of translational efficiencies genome-wide. BMC Genomics 12, 68 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sénéchal P., et al. , Assessing eukaryotic initiation factor 4F subunit essentiality by CRISPR-induced gene ablation in the mouse. Cell. Mol. Life Sci. 78, 6709–6719 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramírez-Valle F., Braunstein S., Zavadil J., Formenti S. C., Schneider R. J., eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J. Cell Biol. 181, 293–307 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong Z., et al. , CUL3 deficiency causes social deficits and anxiety-like behaviors by impairing excitation-inhibition balance through the promotion of cap-dependent translation. Neuron 105, 475–490.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonatopoulos-Pournatzis T., et al. , Autism-misregulated eIF4G microexons control synaptic translation and higher order cognitive functions. Mol. Cell 77, 1176–1192.e16 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Magistretti P. J., Allaman I., A cellular perspective on brain energy metabolism and functional imaging. Neuron 86, 883–901 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Bonvento G., Bolaños J. P., Astrocyte-neuron metabolic cooperation shapes brain activity. Cell Metab. 33, 1546–1564 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Gallo G., The bioenergetics of neuronal morphogenesis and regeneration: Frontiers beyond the mitochondrion. Dev. Neurobiol. 80, 263–276 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirazi P., Mel B. W., Impact of active dendrites and structural plasticity on the memory capacity of neural tissue. Neuron 29, 779–796 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Spruston N., Pyramidal neurons: Dendritic structure and synaptic integration. Nat. Rev. Neurosci. 9, 206–221 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Gerashchenko M. V., Gladyshev V. N., Ribonuclease selection for ribosome profiling. Nucleic Acids Res. 45, e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darnell A. M., Subramaniam A. R., O’Shea E. K., Translational control through differential ribosome pausing during amino acid limitation in mammalian cells. Mol. Cell 71, 229–243.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reid D. W., Shenolikar S., Nicchitta C. V., Simple and inexpensive ribosome profiling analysis of mRNA translation. Methods 91, 69–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andreev D. E., et al. , Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. ELife 4, e03971 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dieterich D. C., et al. , In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat. Neurosci. 13, 897–905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yousefi R., et al. , Monitoring mitochondrial translation in living cells. EMBO Rep. 22, e51635 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zorkau M., Albus C. A., Berlinguer-Palmini R., Chrzanowska-Lightowlers Z. M. A., Lightowlers R. N., High-resolution imaging reveals compartmentalization of mitochondrial protein synthesis in cultured human cells. Proc. Natl. Acad. Sci. U.S.A. 118, e2008778118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dörrbaum A. R., Kochen L., Langer J. D., Schuman E. M., Local and global influences on protein turnover in neurons and glia. ELife 7, e34202 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zorova L. D., et al. , Mitochondrial membrane potential. Anal. Biochem. 552, 50–59 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spillane M., Ketschek A., Merianda T. T., Twiss J. L., Gallo G., Mitochondria coordinate sites of axon branching through localized intra-axonal protein synthesis. Cell Rep. 5, 1564–1575 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith G. M., Gallo G., The role of mitochondria in axon development and regeneration. Dev. Neurobiol. 78, 221–237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wahlsten D., “Chapter 3 – tests of mouse behavior” in Mouse Behavioral Testing: How to Use Mice in Behavioral Neuroscience, Wahlsten D., Ed. (Academic Press, 2011), pp. 39–51. [Google Scholar]
- 30.Kim H., Lim C. S., Kaang B. K., Neuronal mechanisms and circuits underlying repetitive behaviors in mouse models of autism spectrum disorder. Behav. Brain Funct. 12, 3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vorhees C. V., Williams M. T., Morris water maze: Procedures for assessing spatial and related forms of learning and memory. Nat. Protoc. 1, 848–858 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chini M., Hanganu-Opatz I. L., Prefrontal cortex development in health and disease: Lessons from rodents and humans. Trends Neurosci. 44, 227–240 (2021). [DOI] [PubMed] [Google Scholar]
- 33.Smits P., Smeitink J., van den Heuvel L., Mitochondrial translation and beyond: Processes implicated in combined oxidative phosphorylation deficiencies. J. Biomed. Biotechnol. 2010, 737385 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Temperley R. J., Wydro M., Lightowlers R. N., Chrzanowska-Lightowlers Z. M., Human mitochondrial mRNAs-like members of all families, similar but different. Biochim. Biophys. Acta 1797, 1081–1085 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miettinen T. P., Björklund M., Modified ribosome profiling reveals high abundance of ribosome protected mRNA fragments derived from 3′ untranslated regions. Nucleic Acids Res. 43, 1019–1034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinvani H., et al. , Translational tolerance of mitochondrial genes to metabolic energy stress involves TISU and eIF1-eIF4GI cooperation in start codon selection. Cell Metab. 21, 479–492 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Kitajima N., et al. , Real-time in vivo imaging of extracellular ATP in the brain with a hybrid-type fluorescent sensor. ELife 9, e57544 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cole J. D., et al. , Adult-born hippocampal neurons undergo extended development and are morphologically distinct from neonatally-born neurons. J. Neurosci. 40, 5740–5756 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalil K., Dent E. W., Branch management: Mechanisms of axon branching in the developing vertebrate CNS. Nat. Rev. Neurosci. 15, 7–18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim S.-H., et al. , Mitochondrial threonyl-tRNA synthetase TARS2 is required for threonine-sensitive mTORC1 activation. Mol. Cell 81, 398–407.e4 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Data Availability Statement
All study data are included in the article and/or SI Appendix.