Significance
We outline a structure-guided approach for generating conformationally stable, open MHC-I with enhanced ligand exchange kinetics spanning five HLA-A supertypes, all HLA-B supertypes, and oligomorphic HLA-Ib allotypes. We present direct evidence of positive allosteric cooperativity between peptide binding and β2m association with the heavy chain. We demonstrate that covalently linked β2m serves as a conformational chaperone to stabilize empty MHC-I molecules in a peptide-receptive state, by promoting an open conformation and preventing intrinsically unstable heterodimers from irreversible aggregation. Our study provides structural and biophysical insights into the conformational properties of MHC-I ternary complexes, improving the design of ultrastable, universal ligand exchange systems and the tool for characterizing TCRs against pathogen-, tumor-, or autoimmune-associated peptide epitopes in a pan-HLA allelic setting.
Keywords: human leukocyte antigen, peptide exchange, NMR, antigen processing and presentation, nonclassical MHC-I
Abstract
The polymorphic nature and intrinsic instability of class I major histocompatibility complex (MHC-I) and MHC-like molecules loaded with suboptimal peptides, metabolites, or glycolipids presents a fundamental challenge for identifying disease-relevant antigens and antigen-specific T cell receptors (TCRs), hindering the development of autologous therapeutics. Here, we leverage the positive allosteric coupling between the peptide and light chain (β2 microglobulin, β2m) subunits for binding to the MHC-I heavy chain (HC) through an engineered disulfide bond bridging conserved epitopes across the HC/β2m interface, to generate conformationally stable, peptide-receptive molecules named “open MHC-I.” Biophysical characterization shows that open MHC-I molecules are properly folded protein complexes of enhanced thermal stability compared to the wild type when loaded with low- to moderate-affinity peptides. Using solution NMR, we characterize the effects of the disulfide bond on the conformation and dynamics of the MHC-I structure, ranging from local changes in β2m-interacting sites of the peptide-binding groove to long-range effects on the α2-1 helix and α3 domain. The interchain disulfide bond stabilizes MHC-I molecules in an open conformation to promote peptide exchange across multiple human leukocyte antigen (HLA) allotypes, covering representatives from five HLA-A supertypes, six HLA-B supertypes, and oligomorphic HLA-Ib molecules. Our structure-guided design, combined with conditional β-peptide ligands, provides a universal platform to generate ready-to-load MHC-I systems of enhanced stability, enabling a range of approaches to screen antigenic epitope libraries and probe polyclonal TCR repertoires covering highly polymorphic HLA-I allotypes, as well as oligomorphic nonclassical molecules.
The proteins of the class I major histocompatibility complex (MHC-I) are essential components of adaptive immunity in all jawed vertebrates (1). They function by displaying a broad spectrum of self, aberrant, or foreign epitopic peptides, derived from the endogenous processing of cellular proteins on the cell surface, thereby enabling immune surveillance by cytotoxic T lymphocytes (CTL) and natural killer cells (2). Classical MHC-I molecules are comprised of a 8- to 15-amino acid peptide, an invariable light chain β2 microglobulin (β2m), and a highly polymorphic heavy chain (HC) that contains three extracellular domains (α1, α2, and α3) (3). The expansion of MHC-I genes in humans (human leukocyte antigens, or HLAs) has resulted in more than 35,000 alleles with polymorphic residues located on the peptide-binding groove, composed by the α1 and α2 helices and a β-sheet floor (4). The polymorphic nature of HLA allotypes leads to a diversity of displayed peptide repertoires, bound molecular chaperones and other components along the antigen-processing pathway, and T cell receptors (TCRs), which ultimately define immune responses and disease susceptibility. Classical HLA-A and HLA-B allotypes can be further divided into 12 supertypes according to their various peptide-binding specificity, determined by primary anchoring interactions with the peptide positions 2, 3, 5, and 9 (5, 6). Recombinant peptide-loaded MHC-I (pMHC-I) molecules are typically prepared by in vitro refolding with synthetic peptides, resulting in stable pMHC-I complexes (7). These soluble monomeric protein complexes can then be prepared as tetramers or multimers (8), which have become important tools for detecting, isolating, and stimulating CTLs, and screening, optimizing, and identifying immunodominant T cell epitopes for immunotherapy, diagnosis, and vaccine development (9).
The assembly and peptide loading of nascent pMHC-I molecules occur in the lumen of the endoplasmic reticulum and involve many molecular chaperones, including the peptide-loading complex (PLC)-restricted tapasin and the homologous PLC-independent transporter associated with antigen processing (TAP)-binding protein related (TAPBPR). The folding of the MHC-I HC and the formation of disulfide bonds in the α2 and α3 domains are assisted by calnexin and ERp57 (10). The HC then assembles with β2m to generate an empty heterodimer that is highly unstable for most MHC-I alleles, which is stabilized by association with tapasin, ERp57, calreticulin, and TAP in the PLC (11). Both chaperones tapasin and TAPBPR can facilitate the binding of high-affinity peptides to yield stable complexes and proper trafficking of MHC-I, which finally resides on the cell surface for hours to days (12). Loading of high-affinity peptides induces a “closed” conformation of the α2-1 helix via negative allosteric modulation between nonoverlapping peptide-binding sites and tapasin/TAPBPR-binding sites to release the chaperones (13). However, peptide loading and β2m binding to the HC are positively allosterically coupled, together stabilizing the ternary complex (14). The peptide-loading process is initiated by a rate-limiting step of β2m association with the HC, which yields HCs with enhanced peptide-binding affinity in the nanomolar range (15, 16). Consequently, maintaining a stable structure of pMHC-I requires not only the selection of a high-affinity peptide but also the proper function of β2m as a conformational chaperone (14, 15). Although under subphysiological temperatures, stable, peptide-deficient MHC-I HC/β2m heterodimers have been reported to express on the cell surface, these empty molecules can be rapidly internalized and have a short half-life (17, 18). This intrinsic instability of empty MHC-I can cause nonspecific exogenous peptide binding and irreversible denaturation in vitro, limiting its application as an off-the-shelf molecular probe for ligand screening and T cell detection.
A tremendous amount of work has been invested in the biophysical characterization and engineering of pMHC-I molecules, with specific efforts being made to understand the molecular mechanisms of peptide loading, and to develop tools for peptide exchange. Conditional ligands, bound to the MHC-I, can be cleaved by UV exposure or released by increasing the temperature-generating empty molecules, which can be loaded with a rescuing peptide (19–21). Dedicated MHC-I chaperones, including TAPBPR and its orthologs, have also been used to stabilize an array of different MHC-I allotypes in a peptide-receptive conformation with a preferred binding to HLA-A over HLA-B and HLA-C alleles, promoting the exchange of low- to intermediate-affinity peptides for high-affinity peptides in a process known as “peptide-editing” (22–26). In addition, molecular dynamics simulations combined with a tryptophan fluorescence assay have shown that empty MHC-I molecules are not molten globules like previously reported (27), but have varying degrees of structure in the α1 and α2 helices (28, 29). More recent studies have utilized these findings to stabilize peptide-deficient molecules by introducing a disulfide bond across the α1 and α2 helices, restricting the highly flexible F pocket of the peptide-binding groove to mimic the peptide-bound state for common alleles, such as HLA-A*02:01, HLA-A*24:02, and HLA-B*27:05 (30–34). Other studies have sought to stabilize the pMHC-I complex by characterizing the interaction of mutant or orthologous β2m variants. A functional study using human and murine β2m variants bound to HLA-A*02:01 and H-2Db demonstrated that human β2m has a greater affinity for H-2Db than that of murine β2m, resulting in enhanced complex stability due to a marked increase in polarity and the number of interchain hydrogen bonds (35). Another study identified a S55V mutant β2m characterized by its capability to stabilize pMHC-I molecules to a greater extent than the wild type (WT), enhancing peptide binding and CD8+ T cell recognition (36). These studies, altogether, have emphasized the importance and possibility of generating conformationally stable, receptive MHC-I heterodimers across different allotypes by manipulating the malleable HC/β2m interface.
In this work, we outline an alternative, structure-guided approach to engineering conformationally stable, peptide-receptive MHC-I molecules, named “open MHC-I,” by introducing a disulfide bond bridging conserved sites across the HC/β2m interface. We exploit the allosteric mechanisms governing the assembly of MHC-I complexes by locking pMHC-I proteins in an open, peptide-receptive state via the mutations of G120C and H31C on flexible loop regions of the HC and β2m, respectively. We show that the interchain disulfide bond increases the thermostability of molecules loaded with low- and moderate-affinity peptide cargo. We use solution NMR and hydrogen–deuterium exchange-mass spectrometry (HDX-MS) to characterize the conformational difference between the peptide-loaded open and WT HLA-A*02:01 and between the open HLA-A*02:01 molecules in peptide-bound and -deficient states. We further demonstrate that engineered open MHC-I molecules have improved peptide exchange efficiency and overall stability across five HLA-A supertypes and all six HLA-B supertypes (37); oligomorphic HLA-Ib alleles; HLA-E, -F, and -G; and the MHC-like molecule MR1. Finally, we show that open MHC-I molecules are functionally competent in detecting antigen-specific cell populations, serving as a universal platform for identifying immunodominant peptide epitopes and probing T cell responses in research, preclinical, or diagnostic settings.
Results
Structure-Guided Disulfide Engineering Stabilizes Suboptimal MHC-I Ligands.
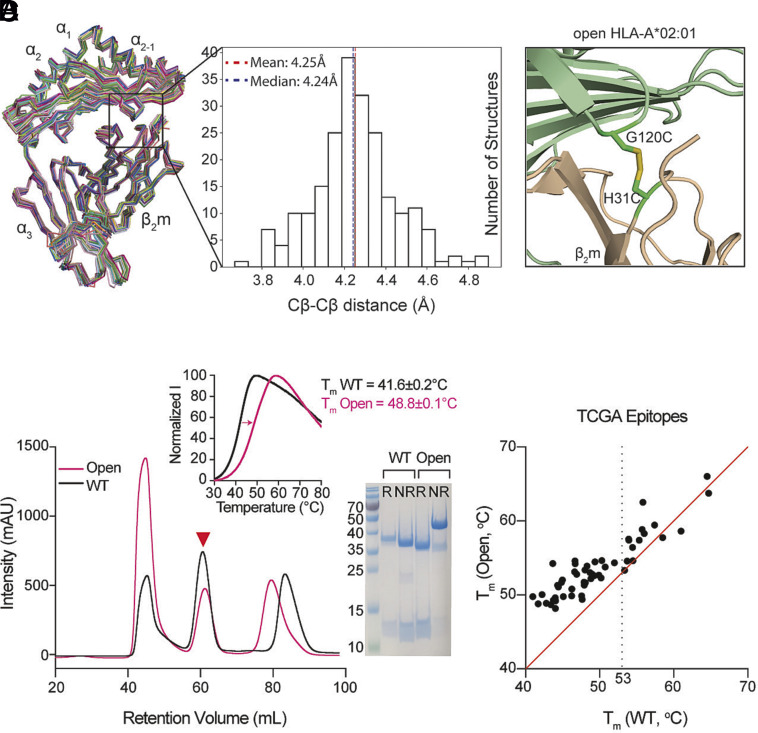
To engineer stable HLA molecules across different allotypes for rapid peptide exchange, we aimed to bridge the HC and the light chain β2m through a disulfide bond based on the positive cooperativity between peptide and β2m association with the HC (35, 38). We utilized a structure-guided approach by first aligning 215 high-resolution pMHC-I crystal structures that were curated in our previously developed database, HLA3DB (39) (Fig. 1A). We found an average distance of 4.25 Å (3.7 Å ≤ Cβ-Cβ ≤ 4.9 Å) between positions G120 of the HC and H31 of the β2m (Fig. 1A). The distances between the paired residues G120 and H31 on the HC and the β2m, respectively, fall within the molecular constraints (5.5 Å) for disulfide cross-linkage (40, 41). The structure of HLA-A*02:01/β2m (PDB:1DUZ) (42) shows that both regions are composed of flexible loops (Fig. 1B), which increase the probability of the two cysteine mutations forming a 90° dihedral angle necessary for disulfide bond formation (43). Additionally, a sequence alignment using 75 distinct HLA allotypes with a greater than 1% global population frequency revealed a conserved glycine at position 120, suggesting a potential generality of the design across distinct HLA allotypes, covering various HLA supertypes that can present diverse peptide repertoires (SI Appendix, Fig. S1). Selected residues G120 and H31 between the HC and β2m were further computationally validated using Disulfide by Design (44). Together, the structure and sequence alignments indicate the possibility of applying the interchain disulfide cross-linkage to a broad range of HLA allotypes, including oligomorphic HLA-Ib and monomorphic nonclassical MHC-I-related proteins, to stabilize their ligand-receptive conformations.
Fig. 1.
Structure-guided stabilization of suboptimal peptide-loaded HLA-A*02:01 by engineered disulfide between the HC and β2m. (A) Structure alignment and distribution of Cβ–Cβ distances between positions G120 of the HC and H31 of the β2m derived from 215 pMHC-I/β2m cocrystal structures with resolution values ≤3 Å. The structures of 52 distinct alleles are aligned by Cα atoms of α1, α2, and α3 domains as ribbons. (B) Structural model of HLA-A*02:01/β2m/TAX9 (PDB ID:1DUZ) with G120 and H31 mutated to cysteines. HLA-A*02:01 HC was colored in light green and β2m in wheat. (C) SEC traces of the WT (black) and the G120C/H31C open (pink) HLA-A*02:01/β2m/TAX8. The red triangle arrowhead indicates the complex peaks and is further confirmed by SDS/PAGE analysis in reduced (R) or nonreduced (NR) conditions. DSF shows thermal stability curves of the WT in black (Tm = 41.6 °C) and the open variant in pink (Tm = 48.8 °C). The average of three technical replicates (mean) is plotted. (D) Thermal stability correlation of the WT and open HLA-A*02:01/TAX8 loaded with each of 50 peptides from the Cancer Genome Atlas (TCGA) epitope library is shown in dots. The average of three technical replicates (mean) is plotted. The red line represents a conceptual 1:1 correlation (no difference in thermal stabilities).
We next sought to validate the design experimentally by expressing the G120C variant of one of the most common alleles HLA-A*02:01 in Escherichia coli, isolating denatured proteins from inclusion bodies, and refolding it in vitro with the H31C variant of the β2m in the presence of a low-affinity placeholder peptide, TAX8 (LFGYPVYV). Size-exclusion chromatography (SEC) and SDS/PAGE confirmed the formation of a G120C/H31C HLA-A*02:01/β2m complex (hereafter referred to as open HLA-A*02:01) and the interchain disulfide bond (Fig. 1C). We then performed differential scanning fluorimetry (DSF) and observed a substantial improvement in the thermal stability of the open HLA-A*02:01 compared to the WT with melting temperatures (Tm) of 48.8 °C and 41.6 °C, respectively (Fig. 1C). Furthermore, the WT and open HLA-A*02:01/β2m/TAX8 were further exchanged with 50 peptides from the Cancer Genome Atlas (TCGA) epitope library (SI Appendix, Table S1). While the resulting Tm values of WT and open HLA-A*02:01 loaded with high-affinity peptides (WT Tm ≥ 53 °C) were similar, a pronounced stabilizing effect was demonstrated on the open than WT HLA-A*02:01 when suboptimally loaded with low- to moderate-affinity peptides (WT Tm < 53 °C) (Fig. 1D). Therefore, evaluation of thermal stabilities for HLA-A*02:01-restricted epitopes spanning a broad range of affinities showed that disulfide linkage between the HC and β2m did not impede peptide binding and consistently supports the role of β2m in chaperoning and stabilizing the HC for peptide loading.
Disulfide-Engineered pMHC-I Shows Local and Long-Range Conformational Changes.
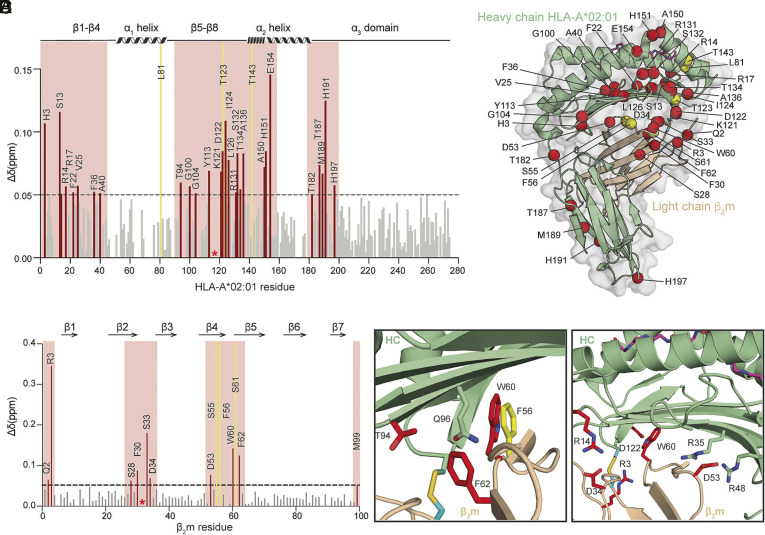
Conformational plasticity and dynamics have been previously shown to be important for several aspects of MHC-I function, including peptide loading, chaperone recognition, and TCR activation (2, 45–47). To elucidate differences in the conformational landscapes of peptide-loaded WT and open HLA-A*02:01, we used established solution NMR methods (48). First, we refolded WT and open HLA-A*02:01/β2m complexes with a high-affinity MART-1 peptide (ELAGIGILTV), which were isotopically labeled with 15N, 13C, and 2H at either the MHC-I heavy or the β2m light chain, followed by reintroduction of exchangeable protons during complex refolding. After independently assigning each protein subunit using a suite of TROSY-based triple-resonance experiments (49) (SI Appendix, Figs. S2 and S3), we measured differences in backbone amide chemical shifts between the WT and open HLA-A*02:01/β2m. We then calculated chemical shift perturbations (CSPs), capturing two-dimensional chemical shift changes of the amide 1H and 15N. Residues showing CSPs above 0.05 ppm (five times the sensitivity relative to 1H) were mapped on the complex structure to highlight sites undergoing conformational changes based on the difference in the local magnetic environment (Fig. 2).
Fig. 2.
Disulfide-engineered pHLA-A*02:01 shows induced conformational adaptations in solution. (A and B) Calculated CSPs between the WT and open HLA-A*02:01/β2m/MART1 are plotted as bar graphs across (A) HC and (B) β2m amide backbone. A significance threshold of 0.05 ppm is determined that is fivefold higher than the 1H sensitivity of the NMR instrument. Residues with significant CSPs are highlighted in red, and exchange-broadened residues in the open HLA-A*02:01 relative to the WT are colored in yellow. Cysteine mutations (G120C and H31C) are indicated by a red asterisk. (C) Residues with CSPs above the significance threshold and exchange broadened in the open HLA-A*02:01/β2m/MART1 are plotted as red and yellow spheres for the amide, respectively, on a representative HLA-A*02:01/β2m/MART1 crystal structure (PDB ID: 3MRQ). (D and E) Enlarged images of (D) hydrophobic residues near the disulfide bond and (E) residues within the hydrogen network. Side chains are displayed and highlighted in red for significant CSPs and yellow for exchange broadening.
In total, we identified 38 residues significantly affected by the interchain disulfide bond formation, revealing substantial global differences between the conformational ensembles sampled by open and WT peptide-loaded HLA-A*02:01 in solution (Fig. 2 A and B). As expected, most of the impacted residues were found near the HC and β2m interface in the region surrounding the disulfide linkage (G120C and H31C) (Fig. 2C). Particularly, the β-sheet floor of the peptide binding groove, including the C, D, E, and F pockets, showed high CSP values (Fig. 2 A and C). Arginine at position 3, located on the flexible loop region close to the engineered disulfide bond, was the most affected residue on the β2m subunit (Fig. 2 B and C). These effects indicated local structural rearrangements, induced at the vicinity of the disulfide linkage. Remarkably, our NMR data also showed CSPs at residue W60 in β2m, while F56 displayed exchange broadening in the open but not in the WT, indicating altered microsecond-to-millisecond timescale dynamics (Fig. 2D). As shown in previous studies, the species-conserved F56 and W60 in β2m play a central role in stabilizing the interface with the α1α2 domain, acting as a conformational switch that controls peptide binding and release (35, 50–52). Additionally, the HLA allele–conserved residues F8, T10, Q96, and M98 form the central part of a hydrophobic pocket together with F56, W60, and F62 from β2m (35). Therefore, the conformational changes observed for these residues upon covalently associating the HC and β2m may contribute to the overall stabilization of the peptide-loaded MHC-I, given the known role of β2m in promoting an allosteric enhancement of peptide binding (53, 54).
Moreover, residues H31 and W60 in β2m are also known to participate in a hydrogen bond network together with residues Q96, G120, and D122 in the α1α2 domains (35, 50–52). Our NMR data showed that disulfide bond formation rearranged this network, including R3, D34, D53, and W60 on β2m and corresponding R14, R35, R48, and D122 on the HC (Fig. 2E). In addition, we observed long-range CSPs on the α2-1 helix and the far end of the β-sandwich fold on the α3 domain (Fig. 2 A and C). This long-range effect supports our hypothesis that the interchain disulfide can trigger substantial global changes in protein dynamics and conformation, since the α2-1 helix has been previously shown to transition between open and closed states of the MHC-I groove for peptide loading (2, 55). Similarly, our CSP data showed a pronounced long-range effect suggesting a repacking of residues T187, M189, H191, and H197 within the α3 domain, via a lever arm effect transduced through residue T182 located on the loop joining the α2 and α3 domains. Thus, our NMR data demonstrate extensive local and long-range structural changes introduced by the bridging disulfide between the HC and β2m. Further, these results collectively show that the engineered disulfide bond induces an allosteric conformational change of the peptide-binding groove that can potentially enhance peptide loading.
Interchain Disulfide Bond Formation Induces a Peptide-Receptive MHC-I Conformation.
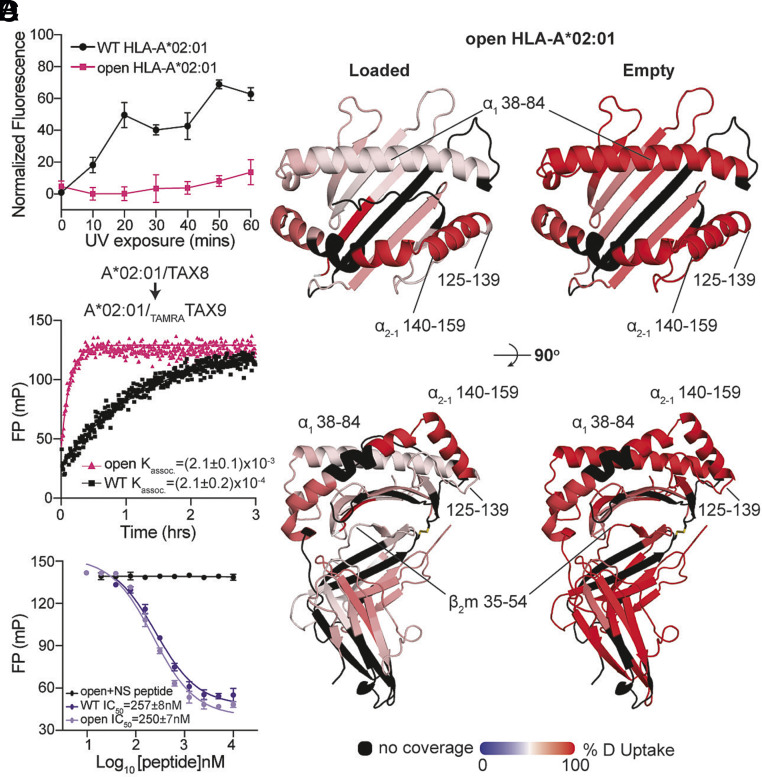
To further test our hypothesis that the covalent association between the β2m and α1α2 interface can improve the overall stability of empty MHC-I molecules, we used DSF to compare the stability of WT and open HLA-A*02:01 refolded with a photocleavable peptide upon varying periods of UV exposure. In the absence of a rescuing peptide, UV-irradiated WT HLA-A*02:01 molecules showed increased binding to the hydrophobic fluorescent dye SYPRO orange as quantified by normalized fluorescence at 25 °C, indicating an increasing amount of protein denaturation (Fig. 3A and SI Appendix, Fig. S4). However, empty open HLA-A*02:01 molecules showed no substantial increases in normalized fluorescence, which was approximately fivefold lower than the WT upon 1-h exposure (Fig. 3A and SI Appendix, Fig. S4). We demonstrated that peptide-deficient, open MHC-I also showed enhanced stability, consistent with our previous DSF data that open molecules loaded with low- to moderate-affinity peptides exhibited higher thermal stabilities compared to the WT (Fig. 1D).
Fig. 3.
Engineered disulfide stabilizes MHC-I at an open, peptide-receptive conformation. (A) Normalized fluorescence measured at 25 °C for the WT or open HLA-A*02:01/KILGFVFJV upon UV exposure. The duration of UV irradiation is shown on the x axis. Results of three technical replicates (mean ± σ) are plotted. (B) Deuterium uptake resolved to individual peptide fragments upon 600-second deuterium labeling for peptide-loaded (Left) and empty (Right) states of open HLA-A*02:01 is mapped onto the WT HLA-A*02:01 crystal structure (PDB ID: 1DUZ) for visualization. Red- and blue-colored regions indicate segments containing peptides with 100% ΔHDX (red—more deuteration) or 0% ΔHDX (blue—less deuteration), respectively; black indicates regions where peptides were not obtained for peptide-loaded and empty protein states. (C) Association profiles of the fluorophore-conjugated peptide TAMRATAX9 to the WT or open HLA-A*02:01/TAX8, as indicated. Results of three replicates (mean) are plotted. (D) Competitive binding of TAMRATAX9 to the WT or open HLA-A*02:01/TAX8 as a function of increasing peptide concentration, measured by fluorescence polarization. An irrelevant peptide, p29 (YPNVNIHNF), was used as a negative control.
We next performed hydrogen–deuterium exchange-mass spectrometry (HDX-MS) (56) to identify differences in solvent accessibility and conformational plasticity between the peptide-bound and empty states of the open HLA-A*02:01. Tandem mass spectrometry (MS/MS) analysis of peptide-loaded or UV-exposed empty open HLA-A*02:01 protein samples revealed exchange saturation within 600 s when plotting the percent deuterium uptake as a function of exchange reaction time for different peptide fragments (SI Appendix, Fig. S5 and Dataset S1). Overall, we observed a higher deuterium uptake profile for empty, open HLA-A*02:01, relative to peptide-loaded molecules (Fig. 3B). Particularly, we observed increased levels of deuterium uptake for regions corresponding to the α1 helix and β-sheet floor of the peptide-binding groove (Fig. 3B). This result is consistent with previously published HDX-MS studies of WT HLA-B*08:01 and chicken MHC-I BF2*15:01 and BF2*19:01 molecules, in which peptide release causes a significant increase of deuterium uptake throughout the peptide-binding groove, most notably in the α2-1 helix (26, 57). The α2-1 helix plays a key role in peptide loading by acting as a conformational switch to facilitate the transition from a closed to an open, peptide-receptive state of the peptide-binding groove as seen in crystal structures and solution NMR studies (13, 46, 47, 58). Notably, our HDX data further highlight that the α2-1 helix in peptide-loaded, open HLA-A*02:01 has an equally high deuterium uptake levels as in empty molecules (Fig. 3B and SI Appendix, Fig. S5). These results suggest that the engineered interchain disulfide enhances the plasticity of the α2-1 helix in the peptide-bound form. Placed within the context of our NMR data (Fig. 2), we conclude that the disulfide linkage induces allosteric changes in the dynamics of peptide-loaded MHC-I, to enhance transitions of the α2-1 helix between “open” and “closed” states of the peptide-binding groove.
To examine whether the increased conformational plasticity of the MHC-I groove revealed by our NMR and HDX data bore any functional consequences in promoting peptide exchange and the overall stability of the final pMHC-I, we compared the binding traces of a fluorophore-labeled TAX9 (TAMRATAX9) peptide to HLA-A*02:01 refolded with the placeholder peptide TAX8. We observed a 10-fold higher apparent association rate, Kassoc., of TAMRATAX9 for open relative to WT TAX8/HLA-A*02:01 (Fig. 3C). In a separate FP assay, we used unlabeled, high-affinity TAX9 to outcompete TAMRATAX9 for binding to either open or WT TAX8/HLA-A*02:01 molecules (Fig. 3D). While having different apparent association rates, TAX9 showed identical IC50 values (approx. 250 nanomolar range) toward both WT and open HLA-A*02:01 molecules (Fig. 3 C and D). This finding is consistent with our previous results that open and WT high-affinity peptide–loaded HLA-A*02:01 showed identical thermal stabilities (Fig. 1D). Taken together with our NMR and HDX data, these results suggest that tethering the MHC-I heavy and light chains enhances peptide exchange kinetics by facilitating conformational transitions between open and closed states of peptide-loaded molecules, without impacting the final stability of the exchanged pMHC-I molecules.
Open MHC-I Molecules Promote Ligand Exchange on a Broad Repertoire of HLA Allotypes.
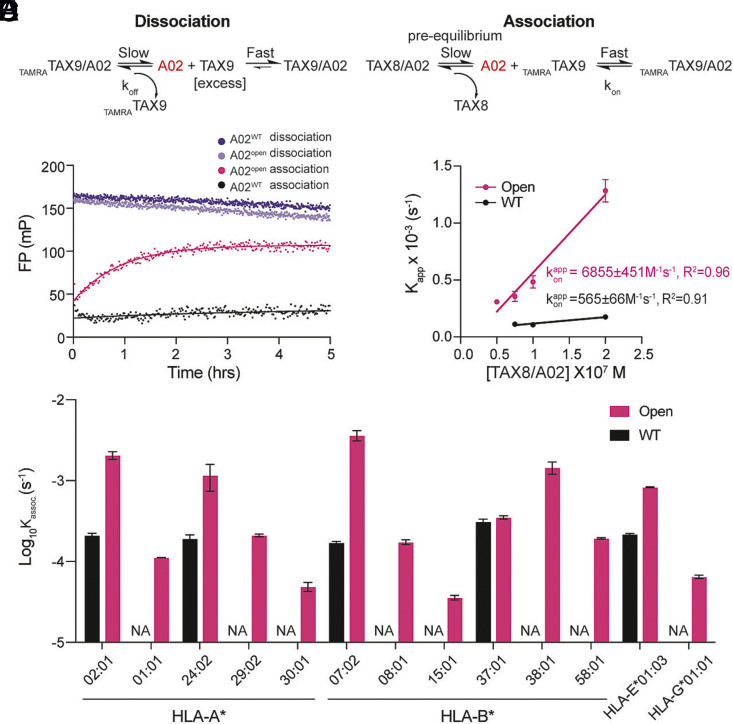
We next sought to investigate how stabilizing the open MHC-I conformation and accelerating the transition between the open and closed states can enhance peptide exchange kinetics. To do this, we developed independent fluorescence polarization (FP) assays under conditions that allowed us to monitor the dissociation or association of TAMRATAX9 to WT versus open HLA-A*02:01 (Fig. 4 A and B). When HLA-A*02:01 was refolded with the high-affinity probe TAMRATAX9 and mixed with a large molar excess of competing TAX9 peptide, open molecules demonstrated a minor decrease in FP relative to the WT, indicating accelerated unloading of TAMRATAX9 to generate empty HLA-A*02:01 that was rapidly occupied by excess unlabeled TAX9 (Fig. 4C). We also observed an accelerated association of TAMRATAX9 to open HLA-A*02:01 refolded with the moderate-affinity placeholder peptide TAX8 relative to WT molecules (Fig. 4C). An increased level of stable, peptide-receptive molecules was also observed for the open construct, indicated by a higher plateau and Kassoc of the FP curve at the same protein concentration (Fig. 4C and SI Appendix, Fig. S6 and Table S2). To quantitatively compare the effect of preexisting receptive molecules, we measured the apparent association rate constant, kappon, defined as the slope of the linear correlation between Kassoc. as a function of TAX8/HLA-A*02:01 protein concentration. Assuming a first-order reaction scheme, this rate is proportional to the concentration of empty, receptive HLA-A*02:01 molecules in the system (Fig. 4B). Open HLA-A*02:01, compared to the WT, exhibited a greater than 10-fold enhancement of kappon, indicating an increased amount of preexisting empty molecules (Fig. 4D). Taken together, open HLA-A*02:01 demonstrated faster kinetics of peptide exchange, which can be attributed to both the increased dissociation rate of placeholder peptide and the overall stabilizing effect on empty molecules originating from the covalent association between the heavy chain and β2m.
Fig. 4.
Open MHC-I improves peptide exchange efficiency on a broad repertoire of HLA allotypes. (A and B) Schematic summary of fitted kinetics obtained from FP analyses of peptide exchange, showing (A) the dissociation of 40 nM TAMRATAX9/A02 in the presence of 1 μM unlabeled TAX9 and (B) 40 nM TAMRATAX9 in different concentrations of TAX8/A02 (50, 75, 100, and 200 nM). (C) The dissociation profiles of 40 nM WT or open TAMRATAX9/A02 in the presence of 1 μM unlabeled TAX9, and association profiles of 40 nM WT or open TAMRATAX9 in 50 nM TAX8/A02, as indicated. (D) Linear correlations between the apparent rate constants Kassoc. and the concentrations of TAX8/A02. The extrapolation of the slope between Kassoc. and the concentrations of TAX8/A02 determine the apparent association rate kon. (E) Log-scale comparison of Kassoc. for the WT (black) or the open (pink) HLA-A*02:01, A*01:01, A*24:02, A*29:02, A*30:01, B*07:02, B*08:01, B*15:01, B*37:01, B*38:01, B*58:01, E*01:03, and G*01:01. The apparent rate constant Kassoc. was determined by fitting the raw trace to a monoexponential association model. NA indicates no fitted Kassoc.. Results of three technical replicates (mean ± σ) are plotted.
Placeholder peptide–bound open MHC-Is generate stable empty molecules, allowing a rapid association with high-affinity peptide ligands in a ready-to-load manner. We, therefore, hypothesized that the interchain disulfide engineering could be applied to different HLA allotypes, resulting in a universal, open MHC-I platform for antigen-screening experiments. To quantitatively compare peptide exchange rates Kassoc across different alleles, we performed a series of FP experiments using optimized placeholder peptides, pHLA concentrations, and the previously described FP protocol (26), where the exchange of high-affinity fluorophore-labeled peptides was monitored through an increase in FP (SI Appendix, Fig. S7). Representatives covering five HLA-A supertypes and all HLA-B supertypes (37) (A01, A02, A0103, A0124, A24, and B07, B08, B27, B44, B58, B62) were selected based on their global allelic frequency (SI Appendix, Tables S3 and S4). Additionally, we extended the study to cover the oligomorphic class Ib molecules, namely HLA-E*01:03 and HLA-G*01:01 (SI Appendix, Table S3).
Our FP results showed that open MHC-I molecules demonstrate improved peptide exchange efficiency compared to the WT across different HLA allotypes. Like open HLA-A*02:01 molecules, open HLA-B*07:02 exhibited a more than 20-fold increase in Kassoc. (Fig. 4E and SI Appendix, Fig. S7E). Both open HLA-A*24:02 and HLA-E*01:03 displayed enhanced peptide exchange kinetics by approximately 6- and 4-fold, respectively (Fig. 4E and SI Appendix, Fig. S7 B and K). The remaining allotypes, HLA-A*01:01, A*29:02, A*30:01, B*08:01, B*15:01, B*38:01, B*58:01, and G*01:01, showed a fitted Kassoc. only in their open forms rather than in their WT counterparts (Fig. 4E and SI Appendix, Fig. S7). Overall, we consistently observed a stabilizing effect on low- to moderate-affinity peptide-loaded molecules across alleles (WT Tm < 53 °C) (SI Appendix, Table S2). When loaded with a high-affinity peptide (WT Tm ≥ 53 °C), Tm values generally stayed the same between the open and the WT, except for HLA-G*01:01 (SI Appendix, Table S2). Suboptimally loaded HLA-B*37:01 in open or WT format exhibited similar thermal stabilities and peptide exchange kinetics, revealing that receptive, empty molecules were preexisting in both proteins for peptide binding. Although open MHC-I demonstrated fast exchange kinetics, we showed that two additional high-affinity type 1 diabetes (T1D) peptides, HLVEALYLV and ALIDVFHQY, have the same IC50 toward both the WT and open variants encompassing HLA-A*02:01 and HLA-A*29:02 (SI Appendix, Fig. S8). Our observation of identical IC50s (Fig. 3D and SI Appendix, Fig. S8) consistently indicates that the corresponding binding free energy (ΔGB) remains very similar between open and WT MHC-I. Altogether, our FP results demonstrate that a wide range of open HLA allotypes exhibits accelerated peptide exchange efficiency and enhanced thermal stabilities when loaded with placeholder peptides. These results provide additional evidence to support our hypothesis that the interchain disulfide bond offers an adaptable structural feature (35, 50), which stabilizes a receptive MHC-I state for the spontaneous loading of peptide ligands across polymorphic HLA allotypes.
Application of Open MHC-I as Molecular Probes for T Cell Detection and Ligand Screening.
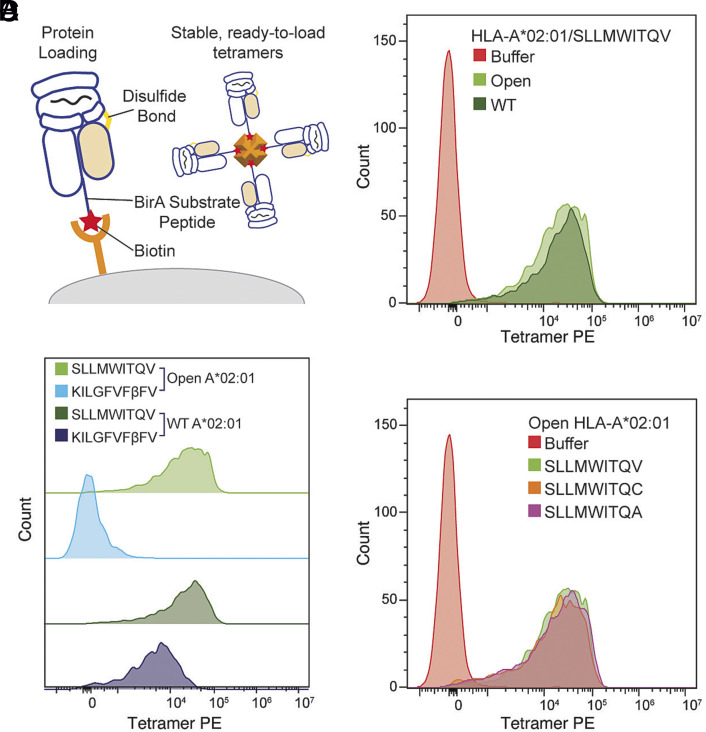
Finally, we evaluated the use of open MHC-I molecules as ready-to-load reagents in tetramer-based T cell detection strategies. We conducted 2-hour peptide exchange reactions at room temperature for both WT and open HLA-A*02:01 loaded with a placeholder peptide (KILGIVFβFV) (26) for an established tumor-associated antigen, NY-ESO-1 (SLLMWITQV). BSP-tagged HLA-A*02:01 was purified, biotinylated, and tetramerized using streptavidin labeled with predefined fluorochromes (Fig. 5A and SI Appendix, Fig. S9). We then stained primary CD8+ T cells transduced with the TCR 1G4 that recognized the NY-ESO-1 peptide displayed by HLA-A*02:01 (59). Compared to WT HLA-A*02:01/NY-ESO-1, tetramers generated with open molecules exhibited a similar staining level (Fig. 5B). We used nonexchanged HLA-A*02:01/KILGIVFβFV molecules as negative controls. Analysis by flow cytometry showed minimal levels of background staining using the open tetramers (Fig. 5C), showing that the 1G4 was not able to recognize HLA-A*02:01/KILGIVFβFV. However, the WT tetramers demonstrated a higher level of background staining, exhibiting only one order of magnitude lower intensity relative to the WT HLA-A*02:01/NY-ESO-1 tetramer staining levels (Fig. 5C), likely due to the loss of β2m and the formation of empty MHC-I heterodimers, which can interact with the CD8 coreceptor (60). Using SDS-PAGE analysis, we confirmed that WT tetramer contained significant amounts of β2m subunit (SI Appendix, Fig. S10). While the partial loss of β2m for some molecules can contribute to an increased level of protein aggregates, this effect would be undetected since large protein aggregates would not enter the gel. Despite having rapid peptide exchange kinetics, disulfide-engineered open MHC-I had similar IC50 values for high-affinity epitopes as the WT and enhanced heterodimer stabilization in a conformation that is peptide receptive (Fig. 3D and SI Appendix, Fig. S8). In addition, open HLA-A*02:01 loaded with moderate-affinity peptides, SLLMWITQC and SLLMWITQA (NYESO C and A), via exchange reaction shows consistent T cell staining as SLLMWITQV (NYESO V) (Fig. 5D). Thus, the engineered disulfide does not interfere with the peptide binding and interactions with T cell receptors. Having a reduced level of background staining allows the use of higher concentrations of tetramers to study interactions with low-affinity TCRs, as seen, for example, in the case of autoimmune peptide epitopes (61). Using open MHC-I as a ready-to-load system can help elucidate the intrinsic peptide selector function across different alleles to optimize peptide-binding motifs, but also has important ramifications for developing combinatorial barcoded libraries of pHLA antigens toward TCR repertoire characterization (24).
Fig. 5.
Open HLA-A*02:01 molecules enable effective T cell detection by reducing nonspecific background staining compared to the WT. (A) A schematic summary of the disulfide-linked HLA-I molecules with the desired BSA tag enables biotinylation and sets the stage for tetramerization. (B) Staining of 1G4-transduced primary CD8+ T cells with PE-tetramers of open and WT HLA-A*02:01/NY-ESO-1(V), light and dark green, respectively. (C) Staining of 1G4-transduced primary CD8+ T cells with PE-tetramers of open and WT HLA-A*02:01/NY-ESO-1(V), light and dark green, respectively, compared to open and WT HLA-A*02:01 loaded with a nonspecific peptide, light and dark blue, respectively. (D) Staining of 1G4-transduced primary CD8+ T cells with PE-tetramers of open HLA-A*02:01 loaded with different NY-ESO-1 peptides SLLMWITQV (light green), SLLMWITQVC (orange), and SLLMWITQA (purple).
Finally, we extended the design of open HLA-I to nonclassical MHC-I, MR1, which can present small-molecule metabolites via both noncovalent and covalent loading by the A pocket (SI Appendix, Fig. S11A). We demonstrated that the open MR1 C262S could be refolded in vitro with covalently linked molecules Ac-6-FP and the noncovalent molecule DCF with an improvement in protein yield (SI Appendix, Fig. S11 B and C). Consistently, we observed a substantial upward shift of the Tm by more than 10 °C for the open MR1/Ac-6-FP than the WT (SI Appendix, Fig. S11D). HLA-F*01:01 molecules known to be stable in their empty form and accommodating long peptides with a length range from 7 to >30 amino acids averaging at 12 residues (62) can also adapt the same structural design to generate open heterodimers (SI Appendix, Fig. S11E). These results further support the universality of the open platform, covering not only classical HLA-Ia allotypes but also the oligomorphic HLA-Ib and nonclassical MHC-I.
Discussion
The inherent instability of peptide-deficient MHC-I heterodimers is a major pitfall of current peptide exchange technologies, limiting screening applications for important therapeutic antigens. Our combined biochemical and biophysical characterization outlines a universal design strategy for generating ready-to-load MHC-I conformers across various disease-relevant HLA allotypes. Compared to the UV- or heat-induced peptide exchange methods (19, 21), open MHC-I molecules combined with β-peptide “goldilocks” ligands introduce mild exchange conditions suitable for large-scale screening applications. Previous work has highlighted the potential of chaperone-mediated exchange in various settings (24, 61, 63). While tapasin has shown preferential binding to HLA-B alleles, TAPBPR preferably interacts with HLA-A alleles but mainly covers the A02 and A24 supertypes (47, 64). More recent work has expanded the TAPBPR-mediated peptide exchange on a broad repertoire of allotypes using TAPBPR orthologs and engineered variants (26). However, compared to the open MHC-I platform, the approach requires optimized placeholder peptides and recombinant chaperone proteins to stabilize empty, receptive molecules. Ready-to-load MHC-I molecules have been derived by introducing an engineered disulfide bond between WT residues Tyr 84 and Ala 139 (Y84C and A139C), linking the α1 and α2 helices at the F pocket to stabilize MHC-I molecules in an empty, peptide-loaded conformation (32). However, this approach has been applied to a restricted set of HLA allotypes, while restriction of MHC-I groove plasticity has been demonstrated to abrogate chaperone binding (30–34), and modifying the F-pocket region could impact interactions with T cell receptors. Instead, open MHC-I molecules exploit the positive cooperativity between peptide association and β2m binding to the HC (16, 38) to stabilize the peptide-binding groove in an open conformation without directly altering the properties of the MHC-I peptide–binding groove or the TCR interface. Our approach leverages the known allosteric switch connecting W60 from β2m with the floor of the MHC-I groove and α2-1 helix, to generate molecules with favorable exchange properties. We show the generality of the design by aligning both sequences and structures and demonstrating peptide exchange applications for representatives from five HLA-A supertypes and all HLA-B supertypes, as well as the oligomorphic HLA-Ib and nonclassical MHC-I.
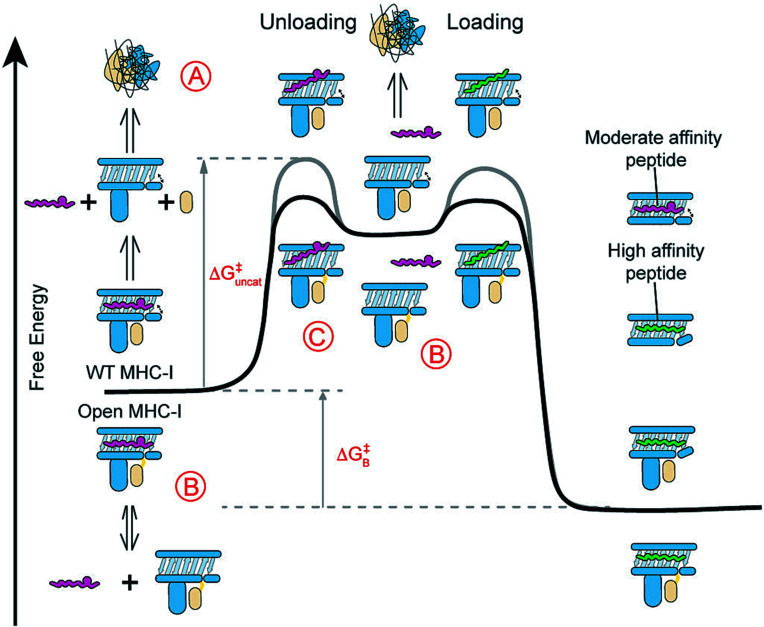
Both WT and open MHC-I molecules loaded with moderate-affinity placeholder peptides undergo a slow, spontaneous peptide-unloading process to generate empty molecules (Fig. 6A). While empty WT MHC-I heterodimers are intrinsically unstable and susceptible to β2m loss and irreversible heavy chain aggregation through the exposure of hydrophobic surfaces, empty open MHC-I molecules maintain a soluble reservoir of receptive molecules (Fig. 6B), as shown by our DSF and FP data (Figs. 3 and 4 C–E). Previous studies have demonstrated that the flexibility of the α2-1 helix allows the peptide-binding groove to dynamically shift between an open and closed state for peptide loading (6, 55). Binding to high-affinity peptides stabilizes the closed conformation and promotes the dissociation of molecular chaperones, which are known to recognize an open conformational epitope at the α2-1 helix (13, 47, 65, 66). Using solution NMR, we have demonstrated that our open MHC-I molecules undergo a similar allosteric conformational change of the α2-1 helix (Fig. 2 A and C), enabling the rapid capture of incoming peptides without perturbing the global stability of the resulting pMHC-I product. Consistently, our HDX-MS data reveal a high solvent exposure and structural plasticity for the α2-1 helix in both peptide-loaded and empty states of open MHC-I molecules (Fig. 3B). These results suggest that the interchain disulfide bond enhances the transition between the open and closed states modulated via the α2-1 helix, toward generating receptive molecules. As a result, the energy barrier required to transition from the closed to open state is reduced, lowering the activation free energy for peptide unloading (Fig. 6C). Therefore, the covalently linked β2m subunit functions as a conformational chaperone to stabilize empty HC/β2m heterodimers and allosterically induces the open conformation of the peptide binding groove. These two mechanisms contribute to rapid peptide loading and unloading in favor of high affinity over placeholder peptides.
Fig. 6.
Open MHC-I molecules modulate the free energy landscape of peptide exchange. (A) WT MHC-I molecules loaded with moderate-affinity placeholder peptides can spontaneously generate empty molecules via peptide unloading, leading to β2m loss and irreversible protein aggregation. The incoming high-affinity peptides rescue these empty molecules, resulting in stable ternary complexes. The binding free energy (ΔGB) of the moderate-affinity peptide exchanged for the high-affinity peptide is unchanged between the open and WT molecules. (B and C) Open MHC-I enhances peptide exchange by (B) stabilizing empty molecules to prevent their aggregation and (C) lowering the activation free energy (ΔGuncat) for peptide unloading via a stabilized open conformation. The gray and black lines indicate the free energy curves for the WT and open MHC-I, respectively.
The open MHC-I platform allows for minimal protein modifications, leading to enhanced exchange reactions across HLA allotypes. Thus, these molecules could be a versatile tool for screening antigenic epitopes, enabling the detection of low-frequency receptors. Further, it is necessary to confirm that open MHC-I molecules are not interfering with peptide repertoire selection or interactions with cognate TCRs. A more detailed study of thermodynamic parameters relevant to peptide binding using isothermal titration calorimetry will provide additional insights into how the presence of the interchain disulfide modulates the peptide-free energy landscape. Additionally, a follow-up study using the open MHC-I focusing on detecting antigen-specific T cells across different HLA allotypes is required to demonstrate the broad usage of this platform as an off-the-shelf reagent. In summary, we outline an alternative structure-guided design of open MHC-I molecules that are conformationally stable and ligand receptive across five HLA-A supertypes and all HLA-B supertypes; oligomorphic HLA-Ib alleles; HLA-E, -G, and -F; and nonclassical MHC-like molecules, MR1. Our data provide a framework for exploring the allosteric networks that exist in the structures of native MHC-I molecules to further guide the design of ultrastabilized, universal ligand exchange technologies, which can be used to address highly polymorphic HLA allotypes.
Ethical Approval and Informed Consent.
The Human Immunology Core at the University of Pennsylvania provided all primary cells used in this study. The studies involving human participants were reviewed and approved by the University of Pennsylvania review board. A written informed consent to participate in this study was provided by the participants.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Acknowledgments
This work was supported through grants by National Institute of Allergy and Infectious Diseases (NIAID, 5R01AI143997), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK, 5U01DK112217), and National Institute of General Medical Sciences (NIGMS, 5R35GM125034) to N.G.S. and NIGMS (R35GM142505) to G.M.B. Additional support was provided through a Fox Chase Cancer Center pilot project grant, and NIH grants P30CA006927 and T32GM132039. We are thankful to Drs. Andy J. Minn and Devin Dersh (University of Pennsylvania) for providing the primary CD8+ cell lines for cell staining and flow cytometry. We acknowledge Nicolas Marotta for performing MR1 protein refolding. We thank Andrew C. McShan and Tyler J. Florio’s assistance using the Disulfide by Design web server, Ananya Majumdar for assistance with data collection on the 800 MHz spectrometer at Johns Hopkins University, Leland Mayne for advising on the HDX-MS experiments, and the Human Immunology Core at the University of Pennsylvania for providing all primary cells used in this study.
Author contributions
Y.S., M.C.Y., J.F.-B., G.M.B., and N.G.S. designed research; Y.S., M.C.Y., C.H.W., J.N.D., H.V.T., and J.F.-B. performed research; Y.S., M.C.Y., J.N.D., H.V.T., and T.J.W. contributed new reagents/analytic tools; Y.S., M.C.Y., C.H.W., J.N.D., H.V.T., S.G., and J.F.-B. analyzed data; and Y.S., M.C.Y., C.H.W., and N.G.S. wrote the paper.
Competing interests
Y.S., M.C.Y., and N.G.S. are listed as co-inventors in a provisional patent application related to this work.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
NMR assignments for the wild-type and open A*02/MART1 complexes (for both the heavy and β2m light chains) have been deposited into the Biological Magnetic Resonance Data Bank (http://www.bmrb.wisc.edu) under accession numbers 51101 and 51781 (67, 68), respectively. All study data are included in the article and/or supporting information.
Supporting Information
References
- 1.Parham P., Ohta T., Population biology of antigen presentation by MHC class I molecules. Science 272, 67–74 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Wieczorek M., et al. , Major histocompatibility complex (MHC) class I and MHC class II proteins: Conformational plasticity in antigen presentation. Front. Immunol. 8, 292 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjorkman P. J., et al. , Structure of the human class I histocompatibility antigen, HLA-A2. Nature 329, 506–512 (1987). [DOI] [PubMed] [Google Scholar]
- 4.Barker D. J., et al. , The IPD-IMGT/HLA database. Nucleic Acids Res. 51, D1053–D1060 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harndahl M., et al. , Peptide-MHC class I stability is a better predictor than peptide affinity of CTL immunogenicity: Antigen processing. Eur. J. Immunol. 42, 1405–1416 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Bailey A., et al. , Selector function of MHC I molecules is determined by protein plasticity. Sci. Rep. 5, 14928 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garboczi D. N., Hung D. T., Wiley D. C., HLA-A2-peptide complexes: Refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc. Natl. Acad. Sci. U.S.A. 89, 3429–3433 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altman J. D., et al. , Phenotypic analysis of antigen-specific T lymphocytes. Science 274, 94–96 (1996). [PubMed] [Google Scholar]
- 9.Hadrup S. R., et al. , “High-Throughput T-Cell Epitope Discovery Through MHC Peptide Exchange” in Epitope Mapping Protocols: Second Edition, Methods in Molecular BiologyTM , Schutkowski M., Reineke U., Eds. (Humana Press, 2009), pp. 383–405. [DOI] [PubMed] [Google Scholar]
- 10.Cresswell P., Ackerman A. L., Giodini A., Peaper D. R., Wearsch P. A., Mechanisms of MHC class I-restricted antigen processing and cross-presentation. Immunol. Rev. 207, 145–157 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Wearsch P. A., Cresswell P., The quality control of MHC class I peptide loading. Curr. Opin. Cell Biol. 20, 624–631 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blum J. S., Wearsch P. A., Cresswell P., Pathways of antigen processing. Annu. Rev. Immunol. 31, 443–473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McShan A. C., et al. , Peptide exchange on MHC-I by TAPBPR is driven by a negative allostery release cycle. Nat. Chem. Biol. 14, 811–820 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L., Dong M., Wang X.-G., The implication and significance of beta 2 microglobulin: A conservative multifunctional regulator. Chinese Med. J. 129, 448–455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binz A.-K., Rodriguez R. C., Biddison W. E., Baker B. M., Thermodynamic and kinetic analysis of a peptide−class I MHC interaction highlights the noncovalent nature and conformational dynamics of the class I heterotrimer. Biochemistry 42, 4954–4961 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Gakamsky D. M., Bjorkman P. J., Pecht I., Peptide interaction with a class I major histocompatibility complex-encoded molecule: Allosteric control of the ternary complex stability. Biochemistry 35, 14841–14848 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Ljunggren H. G., et al. , Empty MHC class I molecules come out in the cold. Nature 346, 476–480 (1990). [DOI] [PubMed] [Google Scholar]
- 18.Schumacher T. N. M., et al. , Direct binding of peptide to empty MHC class I molecules on intact cells and in vitro. Cell 62, 563–567 (1990). [DOI] [PubMed] [Google Scholar]
- 19.Rodenko B., et al. , Generation of peptide-MHC class I complexes through UV-mediated ligand exchange. Nat. Protoc. 1, 1120–1132 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Toebes M., et al. , Design and use of conditional MHC class I ligands. Nat. Med. 12, 246–251 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Luimstra J. J., et al. , A flexible MHC class I multimer loading system for large-scale detection of antigen-specific T cells. J. Exp. Med. 215, 1493–1504 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hermann C., Strittmatter L. M., Deane J. E., Boyle L. H., TAPBPR and tapasin binding to MHC class I is mutually exclusive. J. Immunol. 191, 5743–5750 (2013), 10.4049/jimmunol.1300929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morozov G. I., et al. , Interaction of TAPBPR, a tapasin homolog, with MHC-I molecules promotes peptide editing. Proc. Natl. Acad. Sci. U.S.A. 113, E1006–E1015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Overall S. A., et al. , High throughput pMHC-I tetramer library production using chaperone-mediated peptide exchange. Nat. Commun. 11, 1909 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lan H., et al. , Exchange catalysis by tapasin exploits conserved and allele-specific features of MHC-I molecules. Nat. Commun. 12, 1–13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Y., et al. , Xeno interactions between MHC-I proteins and molecular chaperones enable ligand exchange on a broad repertoire of HLA allotypes. Sci. Adv. 9, eade7151 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouvier M., Wiley D. C., Structural characterization of a soluble and partially folded class I major histocompatibility heavy chain/beta 2m heterodimer. Nat. Struct. Biol. 5, 377–384 (1998). [DOI] [PubMed] [Google Scholar]
- 28.Saini S. K., et al. , Not all empty MHC class I molecules are molten globules: Tryptophan fluorescence reveals a two-step mechanism of thermal denaturation. Mol. Immunol. 54, 386–396 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Kurimoto E., et al. , Structural and functional mosaic nature of MHC class I molecules in their peptide-free form. Mol. Immunol. 55, 393–399 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Hein Z., et al. , Peptide-independent stabilization of MHC class I molecules breaches cellular quality control*. J. Cell Sci. 127, 2885–2897 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Hein Z., Borchert B., Abualrous E. T., Springer S., Distinct mechanisms survey the structural integrity of HLA-B*27:05 intracellularly and at the surface. PLoS One 13, e0200811 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saini S. K., et al. , Empty peptide-receptive MHC class I molecules for efficient detection of antigen-specific T cells. Sci. Immunol. 4, eaau9039 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Moritz A., et al. , High-throughput peptide-MHC complex generation and kinetic screenings of TCRs with peptide-receptive HLA-A*02: 01 molecules. Sci. Immunol. 4, eaav0860 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Anjanappa R., et al. , Structures of peptide-free and partially loaded MHC class I molecules reveal mechanisms of peptide selection. Nat. Commun. 11, 1314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Achour A., et al. , Structural basis of the differential stability and receptor specificity of H-2Db in complex with murine versus human β2-microglobulin. J. Mol. Biol. 356, 382–396 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Malonis R. J., Lai J. R., Vergnolle O., Peptide-based vaccines: Current progress and future challenges. Chem. Rev. 120, 3210–3229 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sidney J., Peters B., Frahm N., Brander C., Sette A., HLA class I supertypes: A revised and updated classification. BMC Immunol. 9, 1 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gakamsky D. M., et al. , An allosteric mechanism controls antigen presentation by the H-2Kb complex. Biochemistry 38, 12165–12173 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Gupta S., Nerli S., Kandy S. K., Mersky G. L., Sgourakis N. G., HLA3DB: Comprehensive annotation of peptide/HLA complexes enables blind structure prediction of T cell epitopes. bioRxiv [Preprint] (2023). 10.1101/2023.03.20.533510 (Accessed 23 March 2023). [DOI] [PMC free article] [PubMed]
- 40.Dombkowski A. A., Sultana K. Z., Craig D. B., Protein disulfide engineering. FEBS Lett. 588, 206–212 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Liu Y. C., et al. , A molecular basis for the interplay between T cells, viral mutants, and human leukocyte antigen micropolymorphism. J. Biol. Chem. 289, 16688–16698 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan A. R., Baker B. M., Ghosh P., Biddison W. E., Wiley D. C., The structure and stability of an HLA-A*0201/Octameric tax peptide complex with an empty conserved peptide-N-terminal binding site1. J. Immunol. 164, 6398–6405 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Van Wart H. E., Lewis A., Scheraga H. A., Saeva F. D., Disulfide bond dihedral angles from Raman spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 70, 2619–2623 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Craig D. B., Dombkowski A. A., Disulfide by Design 2.0: A web-based tool for disulfide engineering in proteins. BMC Bioinformatics 14, 346 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Natarajan K., et al. , The role of molecular flexibility in antigen presentation and T cell receptor-mediated signaling. Front. Immunol. 9, 1657 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas C., Tampé R., Proofreading of peptide—MHC complexes through dynamic multivalent interactions. Front. Immunol. 8, 65 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McShan A. C., et al. , Molecular determinants of chaperone interactions on MHC-I for folding and antigen repertoire selection. Proc. Natl. Acad. Sci. U.S.A. 116, 25602–25613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sgourakis N. G., et al. , A novel MHC-I surface targeted for binding by the MCMV m06 immunoevasin revealed by solution NMR. J. Biol. Chem. 290, 28857–28868 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salzmann M., Pervushin K., Wider G., Senn H., Wüthrich K., TROSY in triple-resonance experiments: New perspectives for sequential NMR assignment of large proteins. Proc. Natl. Acad. Sci. U.S.A. 95, 13585–13590 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Z., et al. , The mechanism of β2m molecule-induced changes in the peptide presentation profile in a bony fish. iScience 23, 101119 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okamura K., et al. , Discovery of an ancient MHC category with both class I and class II features. Proc. Natl. Acad. Sci. U.S.A. 118, e2108104118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beerbaum M., et al. , NMR spectroscopy reveals unexpected structural variation at the protein–protein interface in MHC class I molecules. J. Biomol. NMR 57, 167–178 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Esposito G., et al. , The controlling roles of Trp60 and Trp95 in β2-microglobulin function, folding and amyloid aggregation properties. J. Mol. Biol. 378, 887–897 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Ricagno S., et al. , DE loop mutations affect β2-microglobulin stability and amyloid aggregation. Biochem. Biophys. Res. Commun. 377, 146–150 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Mage M. G., et al. , The peptide-receptive transition state of MHC-I molecules: Insight from structure and molecular dynamics. J. Immunol. 189, 1391–1399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Masson G. R., et al. , Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments. Nat. Methods 16, 595–602 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Hateren A., et al. , Direct evidence for conformational dynamics in major histocompatibility complex class I molecules. J. Biol. Chem. 292, 20255–20269 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang J., et al. , Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation. Science 358, 1064–1068 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen J.-L., et al. , Structural and kinetic basis for heightened immunogenicity of T cell vaccines. J. Exp. Med. 201, 1243–1255 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Geng J., Altman J. D., Krishnakumar S., Raghavan M., Empty conformers of HLA-B preferentially bind CD8 and regulate CD8+ T cell function. ELife 7, e36341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ilca F. T., Neerincx A., Wills M. R., de la Roche M., Boyle L. H., Utilizing TAPBPR to promote exogenous peptide loading onto cell surface MHC I molecules. Proc. Natl. Acad. Sci. U.S.A. 115, E9353–E9361 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dulberger C. L., et al. , Human leukocyte antigen F (HLA-F) presents peptides and regulates immunity through interactions with NK-cell receptors. Immunity 46, 1018–1029.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hermann C., TAPBPR alters MHC class I peptide presentation by functioning as a peptide exchange catalyst. ELife 4, e09617 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ilca F. T., Drexhage L. Z., Brewin G., Peacock S., Boyle L. H., Distinct polymorphisms in HLA class I molecules govern their susceptibility to peptide editing by TAPBPR. Cell Rep. 29, 1621–1632.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McShan A. C., et al. , TAPBPR promotes antigen loading on MHC-I molecules using a peptide trap. Nat. Commun. 12, 3174 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Hateren A., Elliott T., The role of MHC I protein dynamics in tapasin and TAPBPR-assisted immunopeptidome editing. Curr. Opin. Immunol. 70, 138–143 (2021). [DOI] [PubMed] [Google Scholar]
- 67.Truong H. V., Sgourakis N. G., Backbone assignments of the heavy and light chains from the HLA-A*02:01/b2m/ELAGIGILTV(MART-1) human MHC-I protein complex. BMRB. https://bmrb.io/data_library/summary/index.php?bmrbId=51101. Deposited 24 September 2021.
- 68.Woodward C. H., Sgourakis N. G., Backbone assignments of the heavy and light chains from the disulfide mutant HLA-A*02:01-G121C/b2m-H32C/ELAGIGILTV(MART-1) human MHC-I protein complex. BMRB. https://bmrb.io/data_library/summary/index.php?bmrbId=51781. Deposited 10 January 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Data Availability Statement
NMR assignments for the wild-type and open A*02/MART1 complexes (for both the heavy and β2m light chains) have been deposited into the Biological Magnetic Resonance Data Bank (http://www.bmrb.wisc.edu) under accession numbers 51101 and 51781 (67, 68), respectively. All study data are included in the article and/or supporting information.