Significance
Generation of mouse models for COVID-19 pathogenesis is of keen interest as they would allow for efficient investigation of disease mechanisms, as well as providing a vaccine and drug development platform. However, the current transgenic hACE2 (human angiotensin–converting enzyme II) mouse models have limitations regarding their ability to mimic the expression of ACE2 in human tissues, especially the alveolar AT2 (alveolar type II) cells. In this study, we generated an inducible transgenic mouse model that expresses hACE2 in any cell type of interest, which will be invaluable to model COVID-19-related pathologies and their underlying mechanisms.
Keywords: hACE2 transgenic model, COVID-19, SARS-CoV-2, pneumonia, lineage tracing
Abstract
The classical manifestation of COVID-19 is pulmonary infection. After host cell entry via human angiotensin–converting enzyme II (hACE2), the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus can infect pulmonary epithelial cells, especially the AT2 (alveolar type II) cells that are crucial for maintaining normal lung function. However, previous hACE2 transgenic models have failed to specifically and efficiently target the cell types that express hACE2 in humans, especially AT2 cells. In this study, we report an inducible, transgenic hACE2 mouse line and showcase three examples for specifically expressing hACE2 in three different lung epithelial cells, including AT2 cells, club cells, and ciliated cells. Moreover, all these mice models develop severe pneumonia after SARS-CoV-2 infection. This study demonstrates that the hACE2 model can be used to precisely study any cell type of interest with regard to COVID-19-related pathologies.
COVID-19 has been an ongoing global pandemic since early 2020 and has caused a severe threat to public health (1). The emergence of the Omicron variant of SARS-CoV-2, the virus that causes COVID-19, at the end of 2021 has contributed to a faster infection rate compared with other variants (2). The most common causes of deaths in patients with COVID-19 are diffuse alveolar damage and acute respiratory distress syndrome (1, 3). The development of mouse models for COVID-19 would not only enable more rapid and efficient efficacy testing for vaccines and drugs, but also assist in exploring the pathophysiological responses and mechanisms after SARS-CoV-2 infection (4, 5). Human angiotensin–converting enzyme II (hACE2), but not the mouse protein (mACE2), is a key cell entry receptor for SARS-CoV-2 (6). In humans, hACE2 is broadly expressed in multiple tissues, including the nasal airway, lung, esophagus, intestine, pancreas, kidney, liver, brain, arteries, and heart (7, 8). Notably, multiple types of human lung epithelial cells express hACE2, including alveolar type I (AT1), AT2, basal, secretory, and ciliated cells (6, 7). AT2 cells are alveolar epithelial progenitors and are responsible for homeostatic maintenance of the alveolar region (9, 10). Once the SARS-CoV-2 virus invades the lungs, the pulmonary epithelial cells become the top targets of attack, especially the AT2 cells. Virus infection induces capillary rupture, immune infiltration, and alveolar collapse, eventually leading to pneumonia and acute respiratory distress syndrome (2). Moreover, the virus can also invade other organs via the circulation, causing multiorgan failure and serious complications, such as hypertension, myocardial infarction, explosive myocarditis, arrhythmia, thrombosis, acute kidney injury, acute liver injury, and neurological diseases, which can also cause death if complications are sufficiently severe and if there is a lack of adequate intervention (1–3, 11–14). However, the pathogenesis and repair mechanisms after infection are largely unknown, and the applicability of current genetic tools for COVID-19 research has limitations, so genetic tools are necessary to develop cell- and tissue-specific research of this disease in an amenable preclinical animal model, such as the mouse.
Considering the direct role of hACE2 in assisting viral entry, transgenic mice ectopically expressing hACE2 are invaluable and have been used widely in research (15–25), such as transgenic knock-in mice that drive hACE2 in response to the Krt18, HFH4, CAG, or mACE2 promoters. However, previous mouse studies have shown that Krt18 is mainly expressed in the bronchiolar epithelium (26), HFH4 is expressed in bronchiolar ciliated cells of the lungs (19), and CAG is constitutively expressed in all cell types; thus, these promoters fail to offer the ability to perform cell-type specificity studies (23). Moreover, our work here reports that endogenous mACE2 is mainly expressed in bronchiolar club cells of the lung, pericytes of the heart and brain, and epithelium of some other organs. Therefore, the expression of hACE2 driven by the above promoters does not faithfully mimic its expression in human tissues, especially the alveolar AT2 cells. Therefore, previous studies utilizing conventional mouse models have not recapitulated many aspects of COVID-19 in humans (4), possibly owing to these limitations. Once the SARS-CoV-2 virus invades the human body, it infects a variety of organs and cell types, causing a complex infection process. Unraveling the distinct cellular roles and mechanisms of viral pathogenesis is critical for the development of cell-specific therapeutic strategies for COVID-19 treatment. Although many drugs and vaccines have been reported for COVID-19 treatment, we are concerned that some therapeutic strategies may need to be reevaluated due to the imperfection of the above transgenic hACE2 tools. Furthermore, delineating and exploring the response and pathological mechanism of specific cell types after infection will help to carry out cell-specific therapeutic research. It is therefore necessary to develop a transgenic hACE2 line that can be used to target any cell type of interest.
To address this unmet need, we generated in this study an inducible hACE2 transgenic mouse model that allows for the expression of hACE2 in any cell type of interest. We showcase here three examples for specifically inducing hACE2 expression; notably, in AT2, club, and ciliated cells. After SARS-CoV-2 infection, all the three of these epithelial-cell hACE2 mouse models successfully developed pneumonia, albeit with some differences in pathological features between them, though all of them recapitulated relevant pathological features found in patients. This inducible hACE2 transgenic model will be useful for precisely studying COVID-19-related pathologies in any cell type, while perhaps also acting as a vaccine and drug discovery platform.
Results
mAce2 Is Not a Suitable Locus for Driving hACE2 Expression.
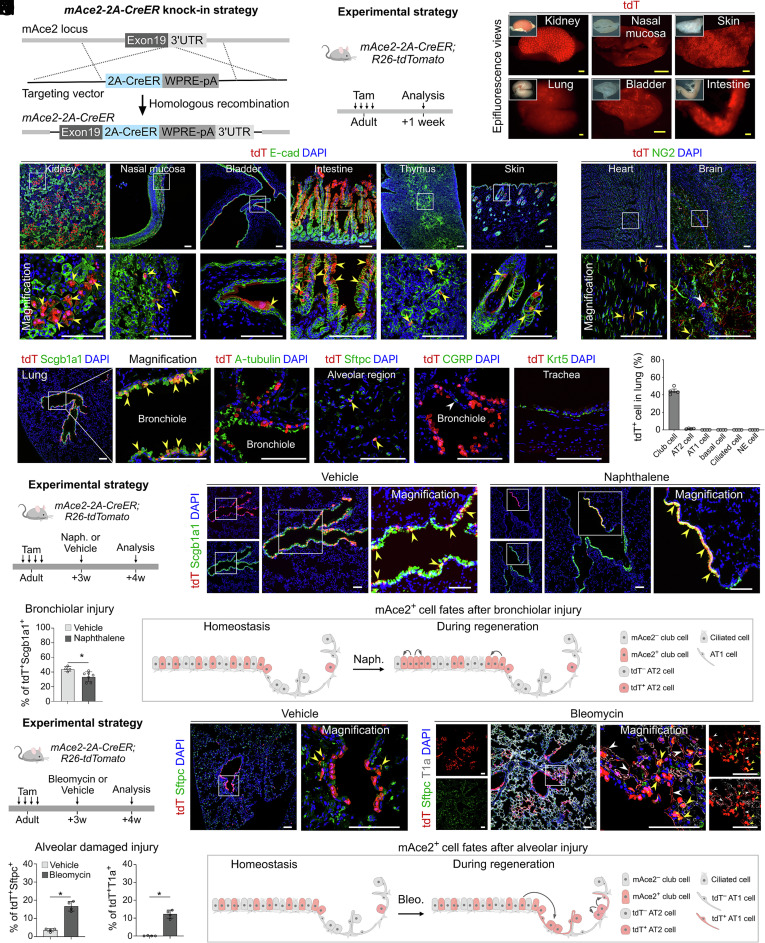
The mAce2 gene locus has been previously used to drive hACE2 expression for COVID-19 research (18, 20). However, the expression of mACE2 in vivo has not been fully characterized. Thus, we first generated an mAce2-2A-CreER mouse line to evaluate whether the mAce2 gene is suitable for driving hACE2 expression. This line was generated by inserting 2A-CreER after the endogenous mAce2 gene via homologous recombination (Fig. 1A). mAce2-2A-CreER mice were then crossed with Rosa26-loxP-stop-loxP-tdTomato (R26-tdTomato) mice to generate the mAce2-2A-CreER;R26-tdTomato mouse. We collected adult mouse tissues after tamoxifen (Tam) treatment for analysis (Fig. 1B). Whole-mount epi-fluorescence showed that tdTomato signals were mainly detected in the kidney, lung, nasal mucosa, bladder, skin, and intestine after Tam treatment (Fig. 1C and SI Appendix, Fig. S1 A and B). Immunostaining for tdTomato and E-cad on tissue sections showed that mAce2 was expressed in the E-cadlow epithelium of kidneys, E-cad+ epithelium of nasal mucosa, bladder, thymus, intestine, and hair follicle of skin (Fig. 1D and SI Appendix, Fig. S1C). Immunostaining for tdTomato and NG2 on tissue sections showed that mAce2 was expressed in a subpopulation of pericytes in the heart and brain (Fig. 1E). We could hardly detect tdTomato+ cells in mice without Tam treatment (SI Appendix, Fig. S1 D and E). Considering that the human lungs are the key targets for SARS-CoV-2 infection, we then examined the mAce2 expression in murine lungs. Immunostaining for tdTomato, Scgb1a1, A-tubulin, Sftpc, CGRP, or Krt5 on lung sections showed that mAce2 was expressed in 44.48 % ± 2.12 % of club cells and 1.36 % ± 0.21 % of AT2 cells, but not in AT1 cells, basal cells, ciliated cells, or neuroendocrine cells (Fig. 1 F and G). Moreover, tdTomato was not expressed in trachea epithelia (Fig. 1F). These data reveal that the expression profile of ACE2 is different in mouse and human lungs.
Fig. 1.
Generation and characterization of an mAce2-2A-CreER line. (A and B) Schematic diagrams illustrating the knock-in strategy of mAce2-2A-CreER by homologous recombination using CRISPR/Cas9 (A) and the experimental design (B). (C) Whole-mount fluorescent images of organs from mAce2-2A-CreER;R26-tdTomato mice after tamoxifen induction. (D and E) Immunostaining for tdTomato and E-cad (D) or NG2 (E) on tissue sections of mAce2-2A-CreER;R26-tdTomato mice after tamoxifen treatment. Yellow arrowheads, tdT+E-cad+ cells (D), tdT+NG2+ cells (E); white arrowheads, tdT+ neural cells. (F) Immunostaining for tdTomato and Scgb1a1, A-tubulin, Sftpc, CGRP, or Krt5 on lung sections. Yellow arrowheads, tdT+Scgb1a1+ club cells or tdT+Sftpc+ AT2 cells. White arrowheads, tdT+CGRP– cell. (G) Quantification of the percentage of club cells, AT2 cells, AT1 cells, basal cells, ciliated cells, and NE cells expressing tdTomato. Data are presented as mean ± SD; n = 4 mice per group. (H) Schematic showing the experimental strategy. (I) Immunostaining for tdTomato and Scgb1a1 on lung sections of mAce2-2A-CreER;R26-tdTomato mice after 4 wk of naphthalene (naph.) or vehicle (corn oil) treatment. Yellow arrowheads, tdT+Scgb1a1+ cells. (J) Quantification of the percentage of tdTomato+Scgb1a1+ club cells in bronchioles. Data are presented as mean ± SD; *P < 0.05. P value was calculated by unpaired two-tailed Student's t test. n = 4 mice in vehicle group and n = 7 mice in naphthalene group. (K) Cartoon image showing that mAce2+ progenitors contribute to bronchiolar epithelium after bronchiolar damage. (L) A schematic diagram illustrating the experimental strategy. (M) Immunostaining for tdTomato, Sftpc, and T1a on lung sections of mAce2-2A-CreER;R26-tdTomato mice after 4 wk of bleomycin or vehicle (PBS) treatment. Yellow arrowheads indicate tdT+Sftpc+ AT2 cells; White arrowheads indicate tdT+T1a+ AT1 cells. (N) Quantification of the percentage of tdTomato+Sftpc+ AT2 cells and tdTomato+T1a+ AT1 cells in the alveolar region. Data are presented as mean ± SD;*P < 0.05. P value was calculated by unpaired two-tailed Student's t test. n = 4 mice per group. (O) Cartoon image showing that mAce2+ progenitors contribute to alveolar epithelium after alveolar damage. Tam, tamoxifen. tdT, tdTomato. A-tubulin, Acetylated-tubulin. (Scale bars, yellow, 1 mm; white, 100 μm.)
Considering that mAce2 was mainly expressed in a subset of club cells, we further explored whether these mAce2+ club cells were progenitors. In naphthalene-induced bronchiolar injury model, we found that the mAce2+ club cells could replace part of the damaged bronchiolar epithelium during lung repair (Fig. 1 H–K). In bleomycin-induced alveolar injury model, we further revealed that these bronchiolar mAce2+ club cells could migrate to the alveolar region and contribute to a subset of AT2 and AT1 cells (Fig. 1 L–O). Collectively, these data demonstrate that mAce2 is not a suitable locus for driving the expression of hACE2 in most alveolar cells. We unexpectedly found that mAce2-expressing lung cells could serve as endogenous progenitors for the damaged epithelia after lung injuries.
Pneumonia Develops after SARS-CoV-2 Infection in the AT2-Specific hACE2 Model.
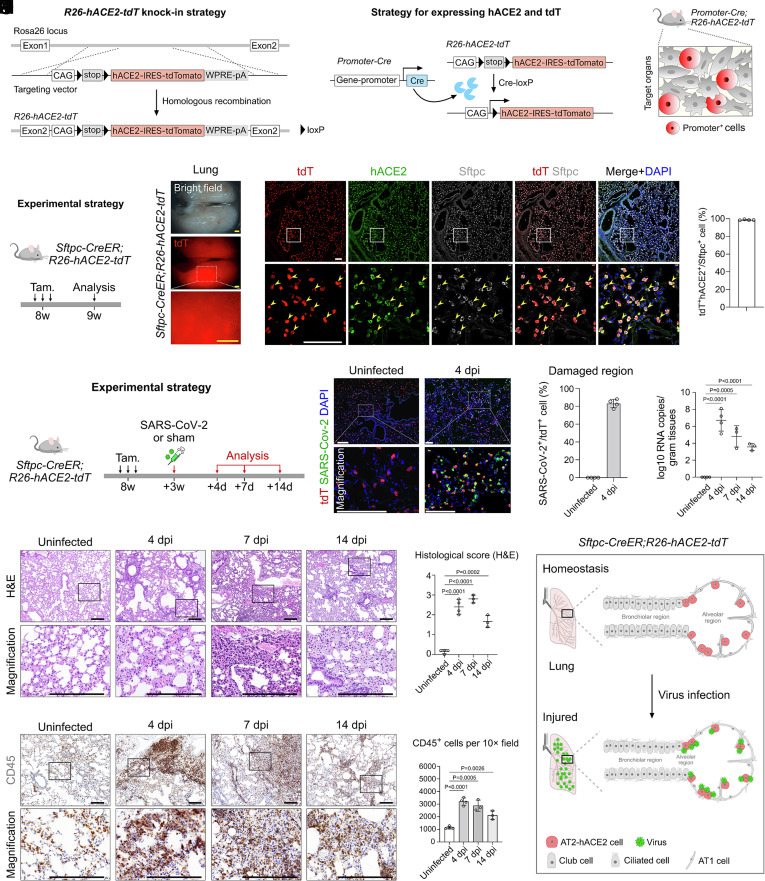
As Krt18, HFH4, and mAce2 were not appropriate loci to recapitulate the expression pattern of ACE2 in humans, we then generated an inducible hACE2 transgenic reporter line to enable induction of hACE2 expression specifically in any cell type of interest. The CAG-loxp-stop-loxp-hACE2-IRES-tdTomato sequence was knocked into the Rosa26 gene locus in mice by homologous recombination using CRISPR/Cas9 of the R26-hACE2-tdT mouse line (Fig. 2A). By design, only after Cre-loxP recombination, the Cre+ cells could specifically express hACE2 and could be simultaneously labeled by tdTomato for genetic tracking of these cells (Fig. 2 B and C). We next induced the expression of hACE2 in specific cell types of lung epithelium using R26-hACE2-tdT. As the AT2 cells were labeled inappropriately by previous hACE2 genetic tools, we first tested whether we can induce hACE2 expression in AT2 cells. We crossed the R26-hACE2-tdT line with the AT2 cell–specific Cre line Sftpc-CreER to generate Sftpc-CreER;R26-hACE2-tdT mice (hereafter called AT2-hACE2 mice, Fig. 2D). After Tam treatment, we collected tissues for analysis. By whole-mount epi-fluorescence, we found that tdT+ signals were readily detected in the lung (Fig. 2E). Immunostaining for tdT, hACE2, and Sftpc on lung sections showed that hACE2 and tdTomato were specifically and efficiently expressed in Sftpc+ AT2 cells (98.68 % ± 0.46 %) but not in other tissues (Fig. 2F and SI Appendix, Fig. S2 E and F). These data demonstrate that the R26-hACE2-tdT mouse could be used to specifically induce hACE2 expression specifically in AT2 cells.
Fig. 2.
Pathological changes after specific infection of AT2 cells by SARS-CoV-2. (A and B) Schematic diagrams illustrating the knock-in strategy of R26-hACE2-tdT mouse line by homologous recombination (A) and the strategy for inducing hACE2 and tdT expression (B). (C) Cartoon image showing that the R26-hACE2-tdT model could be combined with different genetic models for inducing expression of hACE2 in target cell type according to the research needs. (D) A schematic diagram illustrating the experimental design. (E) Whole-mount views of the lungs of Sftpc-CreER;R26-hACE2-tdT mice after tam treatment. (F) Immunostaining for tdTomato, hACE2, and Sftpc on lung sections of Sftpc-CreER;R26-hACE2-tdT mice. Yellow arrowheads indicate tdT+hACE2+Sftpc+ AT2 cells. Quantification of the percentage of AT2 cells expressing hACE2. Data are presented as mean ± SD; n = 4 mice per group. (G) A schematic diagram illustrating the virus infection strategy. (H) Immunostaining for tdT and SARS-CoV-2 S protein on lung sections at 4 days post infection (dpi) and uninfected mice. (I) Quantification of the percentage of tdT+ cells infected by SARS-CoV-2 in damaged regions. n = 4 mice per group. (J) RT-qPCR quantification of SARS-CoV-2 ORF gene expression in the lungs in uninfected, 4, 7, and 14 dpi. Data are presented as mean ± SD; n = 3 or 4 mice per group. Unpaired, two-tailed t test. (K) H&E staining of lung tissues in uninfected, 4, 7, and 14 dpi groups. (L) H&E score of the lung injuries in uninfected, 4, 7, and 14 dpi groups. P value was calculated by unpaired two-tailed Student's t test. Data are presented as mean ± SD; n = 3 or 4 mice per group. (M) IHC staining for CD45+ immune cells of lungs in uninfected, 4, 7, and 14 dpi groups. (N) Quantification of the cell number of CD45+ immune cells of infected lung tissues. P value was calculated by unpaired two-tailed Student's t test. Data are presented as mean ± SD; n = 3 or 4 mice per group. (O) Cartoon image showing that the Sftpc-CreER;R26-hACE2-tdT model could be specifically used for inducing the expression of hACE2 and tdT in AT2 cells. After host cell entry, SARS-CoV-2 virus could specifically infect AT2 cells via hACE2, resulting in epithelium-derived lung pneumonia. Tam, tamoxifen. tdT, tdTomato. A-tubulin, Acetylated-tubulin. (Scale bars, yellow, 1 mm; white, 100 μm; black, 200 μm.)
Next, we studied the susceptibility of AT2-hACE2 transgenic mice to SARS-CoV-2 infection in vivo. Notably, there was no tdTomato+ signal detected without Cre (SI Appendix, Fig. S2 A–D). Adult mice were intranasally infected with SARS-CoV-2 Delta variant at a dose of 2 × 104 Plaque Forming Unit (PFU) at 3 wk after Tam treatment and were then killed at 4 days, 7 days, and 14 days post infection (dpi) for analysis (Fig. 2G). The mice uninfected with virus were used as control. By immunostaining for tdTomato and SARS-CoV-2 spike (S) protein on lung sections of infected AT2-hACE2 mice, we found that SARS-CoV-2 efficiently and specifically bound to the tdTomato+ cells in the severe injury regions (Fig. 2 H and I). Mouse lung tissues were harvested in uninfected, 4, 7, and 14 dpi. Homogenized total RNA was extracted for RT-qPCR analysis and quantification to evaluate the level of SARS-CoV-2 infection. We showed that the viral RNA copies in the lung were significantly increased at 4 dpi and then gradually decreased at 7 dpi and 14 dpi, indicating that the virus was being cleared and lung injury was resolving (Fig. 2J).
We next performed histological assays to test for the development of pneumonia. Hematoxylin & eosin (H&E) staining showed that parenchymal wall expansion, alveolar wall thickness, alveolar congestion, inflammatory cell infiltration, and even the extravasation of blood cells in chambers were detected at 4, 7, or 14 dpi compared to those of the control group (Fig. 2K). We then evaluated the histopathology at different time points after infection. We found that the histological score was significantly elevated at 4 dpi compared to the uninfected group, with alveolar wall thickening, fibrotic deposition, and extravasation of blood cells (Fig. 2 K and L). The histological score was further increased at 7 dpi, and at this time point, we even detected the extravasation of blood cells in the bronchioles. However, after long-term repair at 14 dpi, the histological score was lower compared with those of 4 dpi and 7 dpi, but was still higher than that in control group (Fig. 2 K and L). Immunohistochemical staining (IHC) for CD45 on lung sections also demonstrated immune infiltration after infection (Fig. 2M). Quantification of the CD45+ immune cells per 10× field showed that the infiltrated immune cells dramatically increased at 4 dpi and then gradually decreased at 7 dpi and 14 dpi (Fig. 2N). The pathological phenotype was significantly improved at 14 dpi, indicating that the recovery process was initiated in the lung, which was consistent with the results of RT-qPCR (Fig. 2J). Moreover, by histopathology analysis, we found that no significant toxicity and immune effects were induced by Tam treatment (SI Appendix, Fig. S3). Collectively, these data indicate that our transgenic mice could be used for efficient and specific induction of hACE2 expression and tdTomato staining of specific cell types, which enables induction of interstitial pneumonia after SARS-COV-2 infection and recapitulates relevant pathological features found in patients (Fig. 2O).
Pneumonia Develops after SARS-CoV-2 Infection in the Club-Specific hACE2 Model.
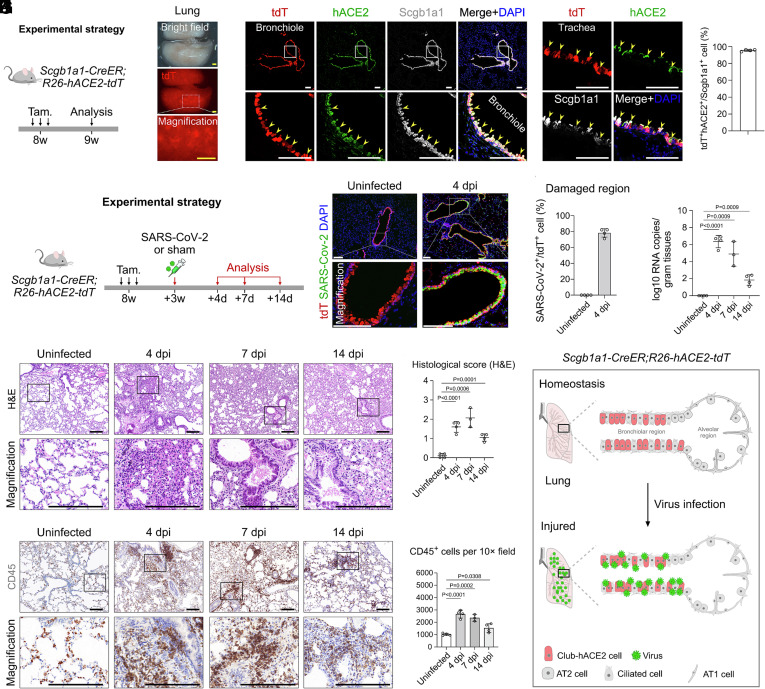
In addition to alveolar epithelium, SARS-CoV-2 also targets the tracheobronchial epithelium in human lungs, mainly including club cells, ciliated cells, and basal cells (6, 7, 27, 28). We next wanted to explore whether all respiratory club cells can be labeled by using a club cell genetic tool (Scgb1a1-CreER) (29). We then generated Scgb1a1-CreER;R26-hACE2-tdT mice (hereafter called club-hACE2 mice) for analysis (Fig. 3A). Whole-mount epi-fluorescence and immunostaining for tdT, hACE2, and Scgb1a1 on lung sections showed that hACE2 and tdTomato were specifically and efficiently expressed in 97.47 % ± 1.05 % Scgb1a1+ club cells of both trachea and bronchiole regions, which mimicked the expression pattern of the humans (Fig. 3 B and C). There were no tdTomato+ signal detected in other tissues (SI Appendix, Fig. S2 G and H).
Fig. 3.
Pathological changes after specific infection of bronchiolar club cells by SARS-CoV-2. (A) A schematic diagram illustrating the experimental design. (B) Whole-mount views of the lungs of Scgb1a1-CreER;R26-hACE2-tdT mice after tam treatment. (C) Immunostaining for tdTomato, hACE2, and Scgb1a1 on lung sections of Scgb1a1-CreER;R26-hACE2-tdT mice. Yellow arrowheads indicate tdT+hACE2+Scgb1a1+ club cells. Quantification of the percentage of club cells expressing hACE2. Data are presented as mean ± SD; n = 4 mice per group. (D) A schematic diagram illustrating the virus infection strategy. (E) Immunostaining for tdT and SARS-CoV-2 S protein on lung sections at 4 dpi and uninfected mice. (F) Quantification of the percentage of tdT+ cells infected by SARS-CoV-2 in damaged regions. n = 4 mice per group. (G) RT-qPCR quantification of SARS-CoV-2 ORF gene expression in the lungs in uninfected, 4, 7, and 14 dpi groups. Data are presented as mean ± SD; n = 3 or 4 mice per group. P value was calculated by unpaired two-tailed Student's t test. (H) H&E staining of the lung tissues in uninfected, 4, 7, and 14 dpi groups. (I) H&E score of the lung injuries in uninfected, 4, 7, and 14 dpi groups. P value was calculated by unpaired two-tailed Student's t test. Data are presented as mean ± SD; n = 3 or 4 mice per group. (J) IHC staining for CD45+ immune cells of lungs in uninfected, 4, 7, and 14 dpi groups. (K) Quantification of the cell number of CD45+ immune cells of infected lung tissues. P value was calculated by unpaired two-tailed Student's t test. Data are presented as mean ± SD; n = 3 or 4 mice per group. (L) Cartoon image showing that specific expression of hACE2 in club cells results in lung pneumonia after SARS-CoV-2 infection. Tam, tamoxifen. tdT, tdTomato. A-tubulin, Acetylated-tubulin. (Scale bars, yellow, 1 mm; white, 100 μm; black, 200 μm.)
Next, we investigated the susceptibility of club-hACE2 transgenic mice to SARS-CoV-2 infection in vivo. We collected tissues in 4, 7, and 14 dpi after SARS-CoV-2 infection for analysis, and the uninfected mice were used as the control group (Fig. 3D). Immunostaining for tdTomato and SARS-CoV-2 spike (S) protein on lung sections of infected club-hACE2 mice showed that SARS-CoV-2 efficiently and specifically bound to the tdTomato+ club cells in the severely damaged airway epithelial regions (Fig. 3 E and F). RT-qPCR analysis showed that the viral RNA copies in the lungs were significantly elevated at 4 dpi, then gradually decreased from 7 dpi to 14 dpi, indicating that the lung injury resolved and viremia was reduced (Fig. 3G). We then performed histological assays to test for the development of pneumonia. H&E staining and histological scores showed that increased alveolar wall thickness and congestion and inflammatory cell infiltration were observed at 4 dpi, and these pathological reactions were more severe at peribronchiolar regions (Fig. 3 H and I). At 7 dpi, the extravasation of blood cells was detected at the bronchiolar lumen (Fig. 3H). Moreover, although the extravasation of blood cells was still observed at the bronchiolar lumen, the histological score was decreased at 14 dpi with reduced alveolar wall thickness, congestion, and parabronchial fibrosis (Fig. 3 H and I). IHC staining for CD45 on lung sections also showed that there was immune cell infiltration into the lungs after infection that was aggregated at peribronchiolar regions, which was consistent with the pathological features observed by H&E staining (Fig. 3J). Further quantification of CD45+ immune cells showed that the infiltrated immune cells were dramatically elevated at 4 dpi and then gradually decreased from 7 dpi to 14 dpi, indicating a recovery period after infection (Fig. 3K), which was consistent with the RT-qPCR results (Fig. 3G). Collectively, these data indicate that club-hACE2 mice could be used for efficient and specific labeling of club cells of both trachea and bronchioles by hACE2 and tdTomato, which enables induction of pneumonia after SARS-COV-2 infection (Fig. 3L).
Pneumonia Develops after SARS-CoV-2 Infection in the Ciliated-Specific hACE2 Model.
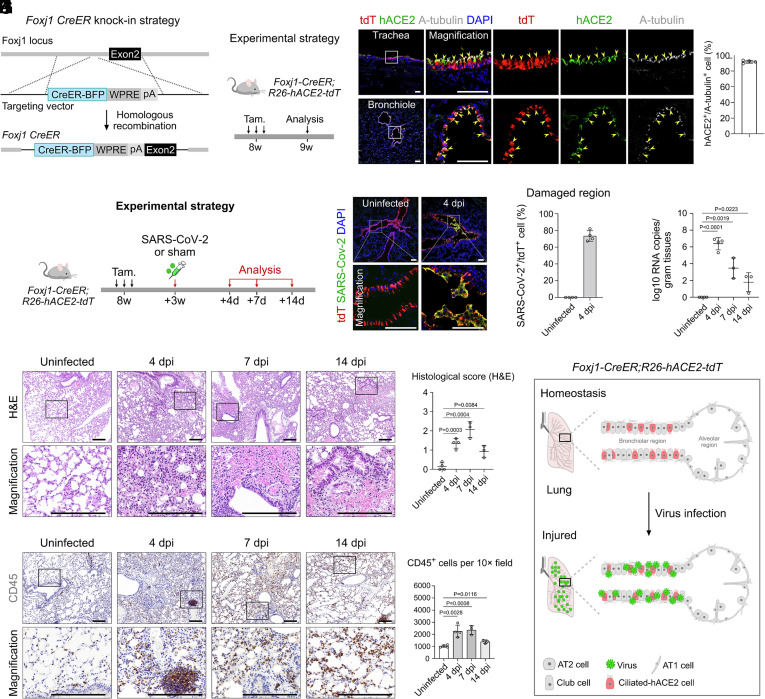
Ciliated cells of human lungs also express hACE2 (6, 7). To develop a ciliated-specific hACE2 model, we first generated a Foxj1-CreER line by inserting the CreER-BFP component after the endogenous Foxj1 gene via homologous recombination (Fig. 4A). We then generated Foxj1-CreER;R26-hACE2-tdT mice (hereafter called ciliated-hACE2 mice) to study the pathological features after SARS-CoV-2 infection (Fig. 4B). By immunostaining for tdT, hACE2, and acetylated-tubulin (A-tubulin) on lung sections, we found that hACE2 and tdTomato were specifically and efficiently expressed in 97.13% ± 1.12% acetylated-tubulin+ ciliated cells of both trachea and bronchiole regions (Fig. 3C). No tdTomato+ signal was detected in other tissues, except for the brain (SI Appendix, Fig. S2 I and J). Next, we tested the susceptibility of ciliated-hACE2 transgenic mice to SARS-CoV-2 infection in vivo. We collected and analyzed the tissues in 4, 7, and 14 dpi after SARS-CoV-2 infection (Fig. 4D). Immunostaining for tdTomato and SARS-CoV-2 spike (S) protein on lung sections showed that SARS-CoV-2 bound to the tdTomato+ ciliated cells in the severely damaged airway epithelial regions, suggesting that the hACE2+ ciliated cells were targeted by SARS-CoV-2 (Fig. 4 E and F). RT-qPCR analysis showed that the viral RNA copies in the lungs were increased at 4 dpi and then gradually decreased from 7 dpi to 14 dpi (Fig. 4G).
Fig. 4.
Pathological changes after specific infection of bronchiolar ciliated cells by SARS-CoV-2. (A and B) Schematic diagrams illustrating the knock-in strategy of Foxj1-CreER-BFP by homologous recombination (A) and the experimental design (B). (C) Immunostaining for tdTomato, hACE2, and A-tubulin on lung sections of Foxj1-CreER;R26-hACE2-tdT mice. Yellow arrowheads indicate tdT+hACE2+A-tubulin+ ciliated cells. Quantification of the percentage of ciliated cells expressing hACE2. Data are presented as mean ± SD; n = 4 mice per group. (D) A schematic diagram illustrating the virus infection strategy. (E) Immunostaining for tdT and SARS-CoV-2 S protein on lung sections at uninfected and 4 dpi mice. (F) Quantification of the percentage of tdT+ cells infected by SARS-CoV-2 in damaged regions. n = 4 mice per group. (G) RT-qPCR quantification of SARS-CoV-2 ORF gene expression in the lungs in uninfected, 4, 7, and 14 dpi groups. Data are presented as mean ± SD; n = 3 or 4 mice per group. (H) H&E staining of the lung tissues in uninfected, 4, 7, and 14 dpi groups. (I) H&E score of the lung injuries in uninfected, 4, 7, and 14 dpi groups. P value was calculated by unpaired two-tailed Student's t test. Data are presented as mean ± SD; n = 3 or 4 mice per group. (J) IHC staining for CD45+ immune cells of lungs in uninfected, 4, 7, and 14 dpi groups. (K) Quantification of the cell number of CD45+ immune cells of uninfected and infected lung tissues. P value was calculated by unpaired two-tailed Student's t test. Data are presented as mean ± SD; n = 3 or 4 mice per group. (L) Cartoon image showing that specific expression of hACE2 in ciliated cells results in lung pneumonia after SARS-CoV-2 infection. Tam, tamoxifen. tdT, tdTomato. A-tubulin, Acetylated-tubulin. (Scale bars, yellow, 1 mm; white, 100 μm; black, 200 μm.)
To test for the development of pneumonia in this strain, we performed histological analysis. H&E staining and histological scores showed that increased alveolar wall thickness, alveolar congestion, and fibrosis were observed at 4 dpi (Fig. 4 H and I). These pathological features were more severe in peribronchiolar regions, especially the extravasation of blood cells detected at 7 dpi (Fig. 4 H and I). While the pathological symptoms were significantly reduced at 14 dpi, the blood cells were still leaky in the bronchiolar lumen (Fig. 4H). Furthermore, quantification of the IHC staining data showed that the immune cells significantly infiltrated the lungs at 4 dpi and then gradually decreased from 7 dpi to 14 dpi, indicating a recovery period after infection, which was consistent with the pathological features observed by H&E staining and by RT-qPCR analysis (Fig. 4 J and K). The above data demonstrate that the ciliated-hACE2 mice could mimic the hACE2 expression of human ciliated cells and could be used to induce pneumonia after SARS-COV-2 infection (Fig. 4L).
Comparison of Ubiquitous and Tissue-Specific Expression of hACE2 Mouse Models.
We next compared our hACE2 models with a previously reported CAG promoter–driven hACE2 model (24, 25). The main component of this tool was the CAG-hACE2-IRES-Luciferase sequence (hereafter called CAG-hACE2 mice) (SI Appendix, Fig. S4A). We then performed a side-by-side comparison of viral titers and lung pathology of the CAG-hACE2 mice with our AT2-hACE2, club-hACE2, and ciliated-hACE2 models at 4 dpi. Immunostaining for SARS-CoV-2 spike (S) protein on lung sections showed that the SARS-CoV-2 virus infected the cells of the bronchiolar and alveolar regions broadly in CAG-hACE2 mice, while virus more specifically infected distinct epithelial cells of our three hACE2 mouse models (Figs. 2H, 3E, and 4E and SI Appendix, Fig. S4C). H&E staining and IHC staining for CD45 on lung sections showed that increased alveolar wall thickness, alveolar congestion, fibrosis, and inflammatory cell infiltration were detected in the CAG-hACE2 mice (SI Appendix, Fig. S4C). Statistical analysis showed that the viral RNA copies, histological score, and inflammatory cell infiltration were highest in CAG-hACE2 mice compared with our three hACE2 mice, especially the alveolar congestion (SI Appendix, Fig. S4 B, D, and E). Therefore, combined with cell-specific Cre drivers, our hACE2 tool could be used to induce hACE2 expression in any cell type of interest and can be applied to study the inflammatory responses on specific cell types.
Discussion
The classical manifestation of COVID-19 is pulmonary infection. After entry into the host cell via hACE2, the SARS-CoV-2 virus can replicate in pulmonary epithelial cells, especially the AT2 cells that are crucial for maintaining normal lung function. hACE2 mouse genetic tools based on the Krt18 and HFH4 promoters could only express the transgene in limited epithelial cell types, while CAG is active in all cell types, both of which do not fully mimic ACE2 expression in humans. In the first part of this study, we first generated an mAce2-2A-CreER line and revealed that mAce2 was not a suitable locus for driving the expression of hACE2. We found that the mAce2 was mainly expressed in bronchiolar club cells of the lung, pericytes of the heart and brain, and a subset of epithelium in other organs, but not expressed in the trachea, stomach, esophagus, eye, liver, aortic, and muscle, which has been reported to express ACE2 in humans (6–8, 12, 28, 30–32). We further found that the mAce2+ cells were progenitors and participated in the regeneration of the damaged epithelium after bronchiolar injury and alveolar injury. The proportion of mAce2-CreER-labeled club cells was reduced after naphthalene injury, which was more likely due to the compensation of other progenitor-derived club cells, such as the mAce2– basal cells and mAce2– neuroendocrine cells (33–36).
We then generated an inducible hACE2 transgenic mouse model that enables hACE2 expression in any cell type of interest. To characterize our hACE2 model, we generated three genetic lines for targeting AT2, club, and ciliated cells and revealed that hACE2 and tdTomato were specifically and efficiently expressed in these three type cells. For functional verification of our model, we performed SARS-CoV-2 infection experiments and then evaluated pneumonia at multiple time points by RT-qPCR and histological analysis. We detected that the SARS-CoV-2 spike protein (S) was specifically localized to the targeted cells of the AT2-hACE2, club-hACE2, and ciliated-hACE2 mice, respectively. Moreover, we showed that the pathological features of viral RNA copies, histological scores, and inflammatory cell infiltration were significantly increased in the early stage of infection, and then gradually decreased from 7 dpi to 14 dpi, indicating that viral load was being eliminated and the pneumonia was resolving, which is consistent with the progression of COVID-19 in most patients (3, 5, 14).
Although the pathological development trends were consistent, we still found differences between these three cell type–specific hACE2 mouse models. The histological score and infiltrated immune cells were highest in AT2-hACE2 mice, indicating that the alveolar region was more susceptible to SARS-CoV-2 infection and more severely damaged, which was consistent with diffuse alveolar damage in patients (3, 14, 37). However, in club-hACE2 and ciliated-hACE2 mice, the pathological features were more noticeable in peribronchiolar regions, which is consistent with a few reported clinical cases but was not highlighted in the previous hACE2 mouse model (38). This feature also suggests that the phenotype of pneumonia could be affected by injured cell types or injured regions. The proportion of hACE2 expression in the human lung epithelium ranged from 1% to 7% in scRNA-seq data and was detected more by RNA in situ hybridization (~40% in AT2 cells)(6, 7, 28), while hACE2 expression in our mouse model was significantly higher (>97%). This was very likely due to the hACE2 expression in our mice being controlled by the ubiquitously strong CAG promoter after induction. Moreover, we compared our model with a published CAG promoter–driven hACE2 model (CAG-hACE2) and showed that the viral titers were highest and the pneumonia was most severe in CAG-hACE2 mice after infection, compared with our three epithelially expressed hACE2 mouse models. Of note, a structurally similar hACE2 mouse as ours have been reported (39), but the previous study lacks the functional assessment by SARS-CoV-2 infection, and also has shortcomings caused by the transgenic generation strategy with low induction efficiency and uncertain copy numbers (39).
SARS-CoV-2 can infect a variety of tissue cells and its infection can result in multiple organ failures in patients because of the broad expression of the ACE2 receptor (14). However, many cell types express ACE2 in humans, but some of these cell types in mouse do not express endogenous Ace2 (7, 8, 11–13, 40). Lacking the ability to study these cells with existing hACE2 tools makes us unable to accurately elucidate the complex infection mechanism caused by SARS-CoV-2 at the organ or cellular levels. Combined with cell-specific Cre drivers, our mouse line could be used for inducing hACE2 expression in any cell type of interest. As the expression of hACE2 and tdTomato was mediated by Cre-loxP recombination, the Cre+ cells and their progenies could also be permanently tracked by reporters, such as tdTomato. Coupled with a genetic lineage tracing strategy, our hACE2 transgenic model may allow researchers to model COVID-19-related pathologies in any cell type for in-depth studying of the underlying disease mechanism and for therapeutic screening in vivo.
Materials and Methods
Mice.
All mouse experiments were strictly performed within the guidelines of the Institutional Animal Care and Use Committee (IACUC) of the Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences and the Key Laboratory of Medical Molecular Virology (Ministry of Education/National Health Commission/ Chinese Academy of Medical Science), Shanghai Institute of Infectious Disease and Biosecurity, School of Basic Medical Sciences, Shanghai Frontiers Science Center of Pathogenic Microbes and Infection, Fudan University. The animal protocol number is SIBCB-S374-1702-001-C1. Sftpc-CreER, Scgb1a1-CreER, R26-tdTomato, and CAG-hACE2 mouse lines were described previously (25, 29, 41, 42). mAce2-2A-CreER mouse line was generated by inserting 2A-CreER component after endogenous mAce2 gene via homologous recombination. The R26-hACE2-tdT mouse line was generated by knocking CAG-loxp-stop-loxp-hACE2-IRES-tdTomato into intron 1 of Rosa26 gene locus through homologous recombination. mAce2-2A-CreER, R26-hACE2-tdT, and Foxj1-CreER mouse lines were generated by Shanghai Model Organisms Center, Inc. All mice were kept at C57BL6/ICR mixed backgrounds. All mice for SARS-CoV-2 infection were maintained in animal BSL3 facility at Fudan University. Tam (Sigma, T5648) treatment was carried out by oral gavage (0.2 mg/g).
SARS-CoV-2 Infection.
Groups of hACE2 mice were treated with tam at adult stage, then infected intranasally with 2 × 104 PFU of SARS-CoV-2 Delta variant (B.1.617.2) after 3 wk. R26-hACE2-tdT mice were control group and intranasally treated with sterile (phosphate-buffered saline) PBS. Tissues were collected 4, 7, and 14 d after infection for measurement of virus RNA loading and inflammation by using RT-qPCR and immunohistology. The virus was propagated in Vero-E6 cells and stored at −80 °C. All experiments involving SARS-CoV-2 were performed in the BSL3 facility at Fudan University.
Bronchiolar Injury.
The bronchiolar injury was induced by naphthalene (Sigma 84679) as previously described (43). Naphthalene was dissolved in sterile corn oil at a concentration of 25 mg/mL. The mAce2-2A-CreER;R26-tdTomato mice were treated with tam at 7 wk, and then treated with 250 mg/kg naphthalene or vehicle (corn oil) at 10 wk for bronchiolar injury. Then, the lung tissues of naphthalene-treated mice were collected after 4 wk for analysis.
Alveolar Injury.
The alveolar injury was induced by bleomycin (Sigma B8416) as previously described (43). Bleomycin was dissolved in sterile PBS at a concentration of 10 U/mL. The 10 U/mL bleomycin was stored at –80 °C and diluted to 1 U/mL with PBS before use. The mAce2-2A-CreER;R26-tdTomato mice were treated with tam at 7 wk, and then treated with 2 U/kg bleomycin or vehicle (PBS) at 10 wk for alveolar injury. Then, the lung tissues of bleomycin-treated mice were collected after 4 wk for analysis.
Tissue Whole-Mount Fluorescence Microscopy.
After washing off the blood in PBS, the tissues were fixed in 4% PFA (paraformaldehyde) at 4 °C for 1 h, then washed several times in PBS. Whole-mount bright-field and fluorescence images of the tissues were acquired by Zeiss stereoscope (Axio Zoom.V16) and analyzed by ImageJ software.
Tissue Collection and Immunofluorescent Staining.
The immunofluorescent staining protocol was performed as described previously by Liu et al. (43). Briefly, mice were killed and tissues were collected in 12-well plates, then fixed in 4% PFA for 1 h at 4 °C. After washing in PBS, the tissues were then dehydrated in 30% sucrose (dissolved in 1× PBS) overnight at 4 °C. Then, the tissues were embedded in OCT (Sakura) and stored at −80 °C. Ten micrometers of frozen sections was collected on slides. For immunofluorescent staining, the slides were first air-dried at room temperature (RT) and then washed in PBS to remove OCT. Next, the slides were blocked in 5% PBSST block buffer (0.1% Triton X-100 and 5% donkey serum in 1× PBS) for 30 min at RT, then incubated with primary antibodies at 4 °C overnight. Primary antibodies used were E-cadherin (R&D system, AF748, 1:500), NG2 (Millipore, AB5320, 1:500), human ACE2 (R&D system, AF933, 1:300), Scgb1a1/Uteroglobin (Abcam, ab213203, 1:300), tdTomato (Rockland, 600-401-379), acetylated-tubulin (Sigma, T7451, 1:500), T1a (DSHB, 8.1.1, 1:100), and Sftpc (Millipore, AB3786, 1:200). The next day, the slides were washed several times in PBS to remove primary antibodies and then incubated with secondary antibodies for 30 min at RT. Next, after washing several times in PBS, the slides were mounted on a mounting medium. The secondary antibodies were Alexa donkey anti-rabbit 555 (Invitrogen, A31572, 1:1,000), Alexa donkey anti-goat 488 (Invitrogen, A11055, 1:1,000), Alexa donkey anti-goat 647 (Invitrogen, A21447, 1:1,000), Alexa donkey anti-mouse 647 (Invitrogen, A31571; 1:1,000, 1:1,000), and Alexa donkey anti-rat 647 (Invitrogen, A21247, 1:1,000). Immunostaining images were acquired by Olympus FV1200 confocal system and Nikon A1 confocal system. The ImageJ (NIH) software was used to analyze images.
Histology and Immunohistochemistry Staining.
The tissues of SARS-CoV-2-infected mice were first fixed in 4% PFA for 5 d at 4 °C. Then, the tissues washed in PBS for several times and dehydrated with a series of increasing concentrations of ethanol. After ethanol dehydration, the samples were placed in a transparent agent (xylene). Next, the tissues were embedded in paraffin. Sections (4 μm) were cut from embedded blocks and collected on negatively charged slides, which were sequentially baked at 62 °C for 2 h and deparaffinized in xylene and rehydrated using ethanol with concentration gradient.
For H&E, the staining was performed using standard methods. Briefly, the sections were washed in PBS and then stained in hematoxylin solution for 5 min to stain the nuclear. The sections were cleared with 1% hydrochloric acid in 70% ethanol and 1% ammonia for 1 min each followed by washing in water. Then, the sections were rinsed in 95% ethanol for 10 s followed by staining in eosin for 10 s to stain the cytoplasm. Dehydrate procedure included fixing in 95% ethanol for 10 s, in 100% ethanol for 2 min twice, and in xylene for 5 min twice. Finally, the sections were covered by neutral resins for visualization.
For IHC, the sections were incubated in 1× ethylenediamine tetraacetic Acid (EDTA) repair solution and heated at 95 to 98 °C for 20 min, then incubated with 3% hydrogen peroxide at RT for 10 min to inactive the endogenous peroxidase. Next, the sections were blocked with 5% normal donkey serum in PBST (0.2% Triton X-100 in PBS). After that, the sections were stained with primary antibodies at RT for 1 h. Primary antibodies used were tdTomato (Rockland, 600-401-379, 1:1,000) and SARS-CoV Spike (Sinobiological, 40150-T52, 1:1,000). Then, the sections were washed with PBS for three times and incubated with peroxidase-conjugated horseradish peroxidase (HRP) secondary antibody for 30 min at RT. After washing with PBS for three times, the signals were detected by using 3,3′ Diaminobenzidine Tetrahydrochloride (DAB) for about 5 min. Then, the sections were counterstained with hematoxylin to label the nuclei for about 15 min. For fluorescence, the signals were detected by using Tyramide Signal Amplification (TSA) reagents (Novo-light TSA kit, M-D110031). Then, the sections were mounted on a mounting medium after washing for confocal analysis.
Histopathological Analysis.
The histopathological score was used for assessing the degree of lung damage by H&E staining and inflammatory cell infiltration. The lung damage includes parenchymal wall expansion, alveolar wall thickness, alveolar congestion, hemorrhage, and immune cell infiltration. The degrees are as follows: score 0, no damage; score 1, the area of damage was less than 25%; score 2, the area of damage was 25 to 50%; score 3, the area of damage was 50 to 75%; and score 4, the area of damage was over 75%. Five random fields of each section were scored under the microscope at a magnification of 10× field for histopathological assessment. The average score of five fields was the individual H&E score of the lung injury.
RNA Extraction and Quantitative Reverse Transcription PCR.
Viral RNA of individual tissues from SARS-CoV-2-infected mice was extracted with TRIzol reagent (Invitrogen, 10296010) according to the protocols. Quantitative Reverse Transcription PCR (RT-qPCR) targeting the orf gene of SARS-CoV-2 was performed to quantify the viral RNA with the following primers and probes:
SARS-CoV-2-ORF1ab-F: CCCTGTGGGTTTTACACTTAA;
SARS-CoV-2-ORF1ab-R: ACGATTGTGCATCAGCTGA;
SARS-CoV-2-ORF1ab-probe:5′-FAM-CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1-3′.
One-Step PrimeScript RT-PCR Kit (Takara, Japan, RR064) was used for performing RT-qPCR.
Statistical Analysis.
For quantification the percentage of tdT+SARS-CoV-2+ cells among tdT+ cells, five 20 × fields were analyzed per mouse. All data were obtained from at least three independent experiments as indicated in figure legends and presented as mean values ± SD. Two-tailed unpaired Student’s t test was used for statistical comparison between two groups. P < 0.05 was accepted as statistically significant.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This study was supported by the National Key Research & Development Program of China (2019YFA0802000, 2019YFA0110403), NSF of China (82088101, 32050087, 32100648, 32200661), Shanghai Municipal Science and Technology Major Project under Grant number (ZD2021CY001 to L.L.), Research Funds of Hangzhou Institute for Advanced Study (2022ZZ01015), Shanghai Municipal Science and Technology Major Project, and by the New Cornerstone Science Foundation through the New Cornerstone Investigator Program and the XPLORER PRIZE. We thank the help of the Core Facility of Microbiology and Parasitology of Shanghai Medical College (SHMC) and the Biosafety Level 3 Laboratory of Fudan University. We thank Shanghai Model Organisms Center, Inc., for mouse generation.
Author contributions
K.L., M.T., W.X., R. Sun, L.L., and B.Z. designed research; K.L., M.T., W.X., J.P., Y.L., F.J., and L.L. performed research; W.X., R. Sun, R. Shen, L.L., and B.Z. contributed new reagents/analytic tools; K.L., M.T., W.X., X.M., H.J., M.H., K.O.L., L.L., and B.Z. analyzed data; and K.L. and B.Z. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Lu Lu, Email: lul@fudan.edu.cn.
Bin Zhou, Email: zhoubin@sibs.ac.cn.
Data, Materials, and Software Availability
This paper does not report original code, and all study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Robinson P. C., et al. , COVID-19 therapeutics: Challenges and directions for the future. Proc. Natl. Acad. Sci. U.S.A. 119, e2119893119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeGrace M. M., et al. , Defining the risk of SARS-CoV-2 variants on immune protection. Nature 605, 640–652 (2022), 10.1038/s41586-022-04690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan F., et al. , Time course of lung changes at chest CT during recovery from Coronavirus Disease 2019 (COVID-19). Radiology 295, 715–721 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munoz-Fontela C., et al. , Animal models for COVID-19. Nature 586, 509–515 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murakami N., et al. , Therapeutic advances in COVID-19. Nat. Rev. Nephrol. 19, 38–52 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ziegler C. G. K., et al. , SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 181, 1016–1035.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muus C., et al. , Single-cell meta-analysis of SARS-CoV-2 entry genes across tissues and demographics. Nat. Med. 27, 546–559 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L., Li X., Chen M., Feng Y., Xiong C., The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovasc Res. 116, 1097–1100 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barkauskas C. E., et al. , Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123, 3025–3036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nabhan A. N., Brownfield D. G., Harbury P. B., Krasnow M. A., Desai T. J., Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 359, 1118–1123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villapol S., Gastrointestinal symptoms associated with COVID-19: Impact on the gut microbiome. Transl. Res. 226, 57–69 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicin L., et al. , Cell type-specific expression of the putative SARS-CoV-2 receptor ACE2 in human hearts. Eur. Heart J. 41, 1804–1806 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee I. C., Huo T. I., Huang Y. H., Gastrointestinal and liver manifestations in patients with COVID-19. J. Chin Med. Assoc. 83, 521–523 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russell C. D., Lone N. I., Baillie J. K., Comorbidities, multimorbidity and COVID-19. Nat. Med. 29, 334–343 (2023). [DOI] [PubMed] [Google Scholar]
- 15.Sun J., et al. , Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell 182, 734–743.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sefik E., et al. , A humanized mouse model of chronic COVID-19. Nat. Biotechnol. 40, 906–920 (2021), 10.1038/s41587-021-01155-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bao L., et al. , The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 583, 830–833 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Sun S. H., et al. , A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe 28, 124–133.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang R. D., et al. , Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell 182, 50–58.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu F.-L., et al. , Rapid generation of ACE2 humanized inbred mouse model for COVID-19 with tetraploid complementation. Natl. Sci. Rev. 8, nwaa285 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCray P. B. Jr., et al. , Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 81, 813–821 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng J., et al. , COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature 589, 603–607 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asaka M. N., et al. , Highly susceptible SARS-CoV-2 model in CAG promoter-driven hACE2-transgenic mice. JCI Insight 6, e152529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue S., et al. , A novel cyclic gamma-AApeptide-based long-acting pan-coronavirus fusion inhibitor with potential oral bioavailability by targeting two sites in spike protein. Cell Discov. 8, 88 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia S., et al. , Structural and functional basis for pan-CoV fusion inhibitors against SARS-CoV-2 and its variants with preclinical evaluation. Signal Transduct. Target Ther. 6, 288 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chow Y. H., et al. , Development of an epithelium-specific expression cassette with human DNA regulatory elements for transgene expression in lung airways. Proc. Natl. Acad. Sci. U.S.A. 94, 14695–14700 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sims A. C., et al. , Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: Role of ciliated cells in viral spread in the conducting airways of the lungs. J. Virol. 79, 15511–15524 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zou X., et al. , Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 14, 185–192 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rawlins E. L., et al. , The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell 4, 525–534 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamming I., et al. , Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 203, 631–637 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hikmet F., et al. , The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 16, e9610 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M. Y., Li L., Zhang Y., Wang X. S., Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis. Poverty 9, 45 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rock J. R., et al. , Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. U.S.A. 106, 12771–12775 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y., et al. , Spatial-temporal lineage restrictions of embryonic p63(+) progenitors establish distinct stem cell pools in adult airways. Dev. Cell 44, 752–761.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song H., et al. , Functional characterization of pulmonary neuroendocrine cells in lung development, injury, and tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 109, 17531–17536 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ouadah Y., et al. , Rare pulmonary neuroendocrine cells are stem cells regulated by Rb, p53, and notch. Cell 179, 403–416.e23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerard L., et al. , Increased angiotensin-converting enzyme 2 and loss of alveolar type II cells in COVID-19-Related acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 204, 1024–1034 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cellina M., Orsi M. A., Oliva G., Peribronchial consolidation with surrounding ground-glass opacity in COVID-19 pneumonia: 3D reconstruction of a chest computed tomography. Am. J. Trop Med. Hyg. 103, 7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bruter A. V., et al. , Novel transgenic mice with Cre-dependent co-expression of GFP and human ACE2: A safe tool for study of COVID-19 pathogenesis. Transgenic Res. 30, 289–301 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burrell L. M., et al. , Myocardial infarction increases ACE2 expression in rat and humans. Eur. Heart J. 26, 369–375 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Rock J. R., et al. , Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc. Natl. Acad. Sci. U.S.A. 108, E1475–E1483 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madisen L., et al. , Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–958 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Q., et al. , Lung regeneration by multipotent stem cells residing at the bronchioalveolar-duct junction. Nat. Genet. 51, 728–738 (2019), 10.1038/s41588-019-0346-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
This paper does not report original code, and all study data are included in the article and/or SI Appendix.