Abstract
Background
Lung adenocarcinoma (LUAD) is a common type of lung cancer with a high risk of metastasis, but the exact molecular mechanisms of metastasis are not yet understood.
Methods
This study acquired single-cell transcriptomics profiling of 11 distal normal lung tissues, 11 primary LUAD tissues, and 4 metastatic LUAD tissues from the GSE131907 dataset. The lung multicellular ecosystems were characterized at a single-cell resolution, and the potential mechanisms underlying angiogenesis and metastasis of LUAD were explored.
Results
We constructed a global single-cell landscape of 93,610 cells from primary and metastatic LUAD and found that IGF2BP2 was specifically expressed both in a LUAD cell subpopulation (termed as LUAD_IGF2BP2), and an endothelial cell subpopulation (termed as En_IGF2BP2). The LUAD_IGF2BP2 subpopulation progressively formed and dominated the ecology of metastatic LUAD during metastatic evolution. IGF2BP2 was preferentially secreted by exosomes in the LUAD_IGF2BP2 subpopulation, which was absorbed by the En_IGF2BP2 subpopulation in the tumor microenvironment. Subsequently, IGF2BP2 improved the RNA stability of FLT4 through m6A modification, thereby activating the PI3K-Akt signaling pathway, and eventually promoting angiogenesis and metastasis. Analysis of clinical data showed that IGF2BP2 was linked with poor overall survival and relapse-free survival for LUAD patients.
Conclusions
Overall, these findings provide a novel insight into the multicellular ecosystems of primary and metastatic LUAD, and demonstrate that a specific LUAD_IGF2BP2 subpopulation is a key orchestrator promoting angiogenesis and metastasis, with implications for the gene regulatory mechanisms of LUAD metastatic evolution, representing themselves as potential antiangiogenic targets.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12943-023-01791-1.
Keywords: Lung adenocarcinoma, Single-cell RNA sequencing, Metastasis, N6-methyladenosine, Angiogenesis, IGF2BP2, Exosomes
Background
Lung cancer is the leading cause of cancer-related deaths worldwide and remains the most common cancer in China by 2022, according to the latest estimates of global cancer burden data published by the World Health Organization's International Agency for Research on Cancer (IARC) [1]. As the most common type of lung cancer, lung adenocarcinoma (LUAD) accounts for approximately 40% of all cases, and it mostly originates from precancerous lesions, such as atypical adenomatous hyperplasia, progressing to adenocarcinoma in situ and microinvasive adenocarcinoma, and eventually to invasive adenocarcinoma [2–4]. However, despite the great progress of multimodal treatment strategies including targeted therapy, immunotherapy, radiotherapy, and non-invasive surgical resection in recent decades, the five-year overall survival (OS) rate of LUAD is approximately 18% [5–7].
In long-term clinical practice, it has been found that many patients with LUAD have distant metastases at the time of initial diagnosis, and distant metastases in LUAD pose the greatest clinical challenge and may present with a different molecular staging and cellular profile than earlier cancers [8]. Currently, the antiangiogenic targeted therapy for LUAD is considered an effective strategy for the treatment of metastatic LUAD [9], and the monoclonal antibody bevacizumab has been shown to improve response rates and OS in combination with the standard first-line chemotherapy (such as paclitaxel, cisplatin, and carboplatin) for LUAD in phase II and III trials [10]. Other antiangiogenic approaches, such as vascular endothelial growth factor inducers and anticoagulants, are also being investigated [11–13]. Studies have shown that the formation of new blood vessels in the tumor microenvironment is a critical step in the growth of solid tumors and a prerequisite for tumor invasion and metastasis [14, 15]. One of the characteristics of the tumor angiogenesis process is that it stimulates pro-angiogenic factors in endothelial cells [16], which are activated and form various subpopulations. Therefore, a comprehensive and in-depth exploration of the cellular dynamics and molecular features associated with angiogenesis in the ecological niche of metastatic LUAD is required to effectively target key driver genes and develop personalized therapeutic strategies [3, 17, 18].
Insulin-like growth factor 2 (IGF2) mRNA-binding protein 2 (IGF2BP2) is an RNA-binding protein (RBP) with important post-transcriptional regulatory effects on mRNA localization, stability, and translational control [19, 20]. High expression of IGF2BP2 has been shown to be associated with poor prognosis in LUAD, and inhibition of IGF2BP2 expression mitigates LUAD growth, and angiogenesis [21, 22]. Here, this study focuses on exploring the IGF2BP2-mediated mechanism by which angiogenesis of LUAD promotes its metastasis based on single cell analysis. The results of the study may provide new targets for the treatment of LUAD.
Methods and materials
Data source
The raw expression matrix for single-cell RNA sequencing (scRNA-seq) of 11 cases of distal normal lung tissues, 11 cases of primary LUAD tissues, and 4 cases of metastatic LUAD tissues was acquired from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/), under the accession number GSE131907 [3]. Sequencing data from metastatic brain tissue samples, normal/metastatic lymph node tissue samples, and pleural effusion samples were excluded from this study.
Furthermore, this study collected 2 cases of LUAD tissue samples with high CD34 expression and 2 cases with low CD34 expression. This study was approved by the Ethics Committee of Yantai Yuhuangding Hospital of Qingdao University (2022–383) and written informed consent was obtained from all patients or their legal guardians who participated in the study.
High-throughput gene expression data from LUAD tissues and normal lung tissues were extracted from The Cancer Genome Atlas (TCGA) data portal (https://tcga-data.nci.nih.gov/tcga). The RNA-seq data from the Illumina HiSeq RNASeq platform (HTSeq-count) consisted of 502 LUAD samples and 49 adjacent non-cancerous samples. Clinical information of LUAD patients was collected from the TCGA database. From the GSE72094 dataset, expression profiling by array and survival information of 442 LUAD patients were acquired [23].
Tissue immunofluorescence
LUAD tissues with low or high CD34 expression were embedded and sectioned (6 μm thickness), then routinely dewaxed in xylene and alcohol, antigen repaired with citric acid and then washed three times with distilled water for 3 min each time. The sections were incubated with 1% bovine serum albumin (BSA) for 30 min at room temperature. The primary antibody against CD34, EPCAM, IGF2BP2, or FLT4 (Proteintech, Wuhan, China) was diluted with 1% BSA and incubated overnight at 4 ℃. The next day, the sections were washed three times with phosphate buffered saline (PBS) for 3 min each time. Then, the sections were diluted with 1% BSA as fluorescent secondary antibody (Proteintech) and incubated for 1 h at 37 ℃ in a temperature chamber away from light. The sections were washed 3 times with PBS under closed conditions for 3 min each time, and were blocked by adding glycerol diluted Hoechst 33342 (10 μg/mL) (Sigma-Aldrich, St. Louis, Missouri, USA) and photographed by a laser confocal microscope (Nikon, Tokyo, Japan). Fluorescently labeled primary antibodies diluted with 1% BSA were added between washing away the secondary antibodies and adding nucleation reagents, and then the sections were washed three times with PBS for 3 min each time. In the case of non-fluorescently labeled secondary antibodies with L-Lactyllysine, 488-labeled sheep anti-rabbit secondary antibodies were added after incubating the primary antibodies for another 30 min.
IG2BP2 mRNA in situ hybridization and immunofluorescence co-staining
RNA in situ hybridization was performed according to the kit instructions (Beyotime, Shanghai, China). Firstly, paraffin sections were routinely dewaxed, and proteinase K was briefly digested for 5 min in a 37 ℃ incubator, closed and denatured, then the probes (Sigma-Aldrich) were diluted and mixed with biotin according to the kit instructions, and then added dropwise to the sections for hybridization. Next, the sections were washed for 3 × 15 min, and finally the nuclei were stained with hoechst 33342. For co-staining with other proteins, after probe hybridization, the corresponding fluorescently labeled secondary antibodies were added, and since the corresponding protein would be reduced after proteinase K digestion, the antibody concentration was increased and incubated for 30 min at 37 ℃ in the incubator, followed by nucleation.
Cell clustering and differentially expressed genes (DEGs)
Firstly, data preprocessing and quality control of scRNA-seq were conducted. Cells without any gene expression, or with > 10% mitochondrial gene expression and < 10% ribosomal gene expression were removed. The Seurat package [24] was then used for cell clustering of scRNA-seq data and to visualize clusters using the uniform manifold approximation and projection (UMAP) package [25]. Cell clusters characterized by similar marker genes were combined into one cell type and subsequently marker genes for known cell types were used to define the partitioned cell clusters. In addition, DEGs in each cell type of control, primary LUAD, and metastatic LUAD tissues were identified by the "FindAllMarkers" function of the Seurat package. Differences associated with adjusted P < 0.05 were considered significant.
Gene regulatory network analysis
The Single Cell Regulatory Network Inference and Clustering (SCENIC) algorithm was developed to evaluate regulatory network analysis associated with transcription factors (TFs) and to discover regulators (i.e., TFs and their target genes) in individual cells. To reconstruct gene regulatory networks for control lung, primary, and metastatic LUAD from scRNA-seq data, we performed SCENIC analysis, which utilizes co-expression modules between TFs and candidate target genes, as well as a database of DNA binding patterns of TFs to infer important gene regulation of TFs [26, 27]. Regulon modules based on regulon crosstalk (correlation between regulons and regulators) were determined by the connection specificity index (CSI), which ranks the importance of regulons and mitigates the effect of non-specific interactions, and were visualized based on the ComplexHeatmap package [28].
Pseudotime trajectory analysis
The Monocle 3 package (https://cole-trapnell-lab.github.io/monocle3) was applied to sort cells along the pseudotime trajectory [29]. After clustering the cells using the above mentioned method, the dimensionality was reduced and the results were visualized using the UMAP method. Subsequently, cells were sorted according to their progression through the developmental program. In this study, single-cell trajectory analysis of cell subtypes was performed as needed.
Functional enrichment analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was used to determine the biological significance of each cell type. GO and KEGG pathway analyses were performed using the clusterProfiler package based on marker genes for each cell subpopulation [30]. In addition, Gene Set Enrichment Analysis (GSEA) [31] was performed on the DEGs using “c2.cp.kegg.v7.0.symbols.gmt” from the MSigDB database [32] as a background set. GSEA was performed using the clusterProfiler package and P < 0.05 was considered significant.
Cell culture and transfection
Human LUAD cell lines A549 and NCI-H1299 (American Type Culture Collection, Manassas, Virginia, USA) were maintained in Dulbecco’s modified Eagle medium (GIBCO, Gaithersburg, MD, USA) with 10% fetal bovine serum (FBS; GIBCO) and 5% penicillin streptomycin double antibody (GIBCO). Human umbilical vein endothelial cells (HUVECs) (ATCC) were cultivated in EGM™-2 endothelial cell growth medium (Lonza, Basel, Switzerland). All the cells were grown in a 5% CO2 incubator at 37 °C.
Small interfering RNA (siRNA) of IGF2BP2 (si-IGF2BP2; sequence: 5’-GGGACCAAGAUAACAAUCUTT-3’ (sense); 5’-AGAUUGUUAUCUUGGUCCCTT-3’ (antisense)) and negative control (si-NC) (GenePharma, Shanghai, China) were synthesized and transfected into LUAD cells through Lipofectamine RNAiMAX transfection reagent (ThermoFisher Scientific, Waltham, Massachusetts, USA).
Reverse transcription quantitative PCR (RT-qPCR)
Total RNA was extracted via TRZOL reagent (Invitrogen, Carlsbad, California, USA), and complementary DNA was synthesized by a reverse transcription kit (Invitrogen). RT-qPCR was implemented using SYBR Green Master Mix (Solarbio, Beijing, China) and ABI QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems, Carlsbad, USA). Primer sequences were as follows: IGF2BP2, 5’-AGTGGAATTGCATGGGAAAATCA-3’ (forward), 5’-CAACGGCGGTTTCTGTGTC-3’ (revere); GAPDH, 5’-GGAGCGAGATCCCTCCAAAAT-3’ (forward), 5’-GGCTGTTGTCATACTTCTCATGG-3’ (revere). Relative expression was calculated with 2−ΔΔCt method.
Wound healing assay
LUAD cells were seeded onto 6-well culture plates. Following monolayer formation, a 10 μL pipette tip was used to make a straight scratch. The cells were gently washed by PBS and subsequently replenished by serum-free medium. Images were photographed at 0 h, 24 h, and 48 h under a microscope (Olympus, Tokyo, Japan).
Transwell assay
Cell invasion assay was conducted utilizing Transwell chambers (Corning Costar, Tewksbury, MA, USA). The upper chamber membrane was coated with Matrigel (BD Bioscience, San Diego, CA, USA). 3 × 104 LUAD cells were plated to the upper chamber of each insert with 300 μL serum-free medium. In addition, the bottom chamber was added with 500 μL medium containing 10% FBS. After 48-h incubation, invasive cells in the bottom chamber were fixed by 10% methanol and stained by 0.1% crystal violet. Invasive cells were photographed under a microscope (Olympus).
Tube formation assay
Culture medium from LUAD cells was harvested as conditioned medium and utilized for HUVEC culture. HUVECs were plated to Matrigel-coated 96-well plates (1 × 104 cells/well) and cultivated with serum-free medium for 6 h. Next, the HUVECs were maintained with conditioned medium for 6 h. Branch points and capillary length were calculated by use of ImageJ software.
Analysis of intercellular communication
To further explore the interaction relationships between different cell subpopulations, a cellular communication analysis was performed on endothelial cell subpopulations and LUAD cell subpopulations using the iTALK package [33]. This package matched and paired these genes through the ligand-receptor database by identifying genes that were differentially expressed in cell clusters to find important intercellular communication events. Receptor-ligand interactions were determined using the protein–protein interactions from the STRING database [34].
Molecular docking
Possible protein-mRNA binding sequences were predicted using the catRAPID omics v2.0 [35]. Molecular docking was performed using the Hex 8.0.0 [36] to test the feasibility of ligand-receptor interactions. Docking models were visualized using Pymol [37]. Docking energies less than 0 indicate that the two have binding potential, and the lower the energy, the higher the binding potential.
m.6A methylated RNA immunoprecipitation-qPCR (MeRIP-qPCR)
Total RNA was extracted from LUAD cells, followed by treatment with DNase (Sigma-Aldrich) to remove genomic DNA. Next, RNA was purified and fragmentated, and the fragments were incubated with m6A primary antibody for immunoprecipitation by use of a Magna MeRIP™ m6A kit (Millipore, Boston, Massachusetts, USA). Enriched m6A modified mRNA was measured via RT-qPCR.
Western blot
Cells were lysed in RIPA buffer (Beyotime), and protein was fractionated via sodium dodecyl sulfate–polyacrylamide gel electrophoresis, followed by transference onto polyvinylidene fluoride membrane (Millipore). The membrane was blocked in 5% nonfat milk, and subsequently probed with primary antibody against IGF2BP2 (1/2000; ab124930; Abcam, Massachusetts, USA), FLT4 (1/1000; ab243232; Abcam), PI3K (1/1000; ab302958; Abcam), AKT (1/10000; ab179463; Abcam) or β-actin (1/1000; ab8227; Abcam) overnight at 4 °C. Next, the membrane was incubated in horseradish peroxidase-labeled secondary antibody for 2 h at room temperature. Protein band was developed through enhanced chemiluminescence (Millipore), and grey value was quantified with ImageJ software.
Data analysis and statistics
In this study, all the analyses were performed based on the Bioinforcloud platform (http://www.bioinforcloud.org.cn) and GraphPad Prism software (version 9.0.1; San Diego, CA, USA). Significant difference between two groups was evaluated with Student’s t-test, with one- or two-way ANOVA followed by the Bonferroni post hoc test for comparing between ≥ 3 groups. Correlation analysis was carried out using Pearson or Spearman correlation test. The "Survival" package was performed for survival analysis. Time-independent receiver operating characteristic (ROC) curves were conducted via the "timeROC" package. A nomogram model was built by use of the "rms" package, and was evaluated through calibration curves. P < 0.05 was set as statistically significant.
Results
A global single-cell landscape of primary and metastatic LUAD
In this study, CD34 expression was used to characterize the pathological status of tumor angiogenesis [38]. Immunofluorescence results showed that CD34 expression was higher in metastatic LUAD than in primary LUAD (Fig. 1A). The analytical flow of this study is illustrated in Fig. 1B. We obtained scRNA-seq data of 11 distal normal lung tissues, 11 primary LUAD tissues, and 4 metastatic LUAD tissues from the GSE131907 dataset to further explore the ecological atlas both in primary and metastatic LUAD. After data pre-processing and quality control (Supplementary Fig. 1A-C), a total of 93,610 high-quality single-cells were captured and clustered into 19 cell types, including LUAD cells (LUAD), endothelial cells (En), macrophages (Mac), and others (Fig. 1C). The markers positively expressed in cell clusters were consistent with the gene signatures published by recent scRNA-seq and laboratory research, amongst others [3], and were consistent with the phenotypic characteristics of the corresponding cells (Fig. 1D). Multiple or low copy number events (i.e., malignant copy number variation events) occurred in LUAD tumor cells (Fig. 1E). Further analysis revealed that metastatic LUAD represented a multicellular ecosystem distinct from those of control lung and primary LUAD (Fig. 1F). Notably, a dramatic increase in LUAD cell abundance was observed in metastatic LUAD. In summary, we initially constructed a global single-cell landscape of primary and metastatic LUAD by single-cell analysis and explored the altered cellular ecology in different sample types.
Fig. 1.
Construction of a global single-cell landscape of primary and metastatic LUAD. A Multiplex immunofluorescence map demonstrating CD34 expression in primary and metastatic LUAD tissues. Bar, 50 μm. B Flow chart of this study. The global single-cell landscape of primary and metastatic LUAD was constructed based on single-cell technology. C The single-cell atlas mapping cell types. D Bubble map showing cell marker genes for cell annotations. E Heat map showing copy number variation results to further validate the accuracy of cell annotation in the presence of multi-copy or low-copy number events in LUAD cells. F The difference in cell abundance between control, primary and metastatic LUAD
Clonal evolutionary trajectory of metastatic LUAD tumor cells
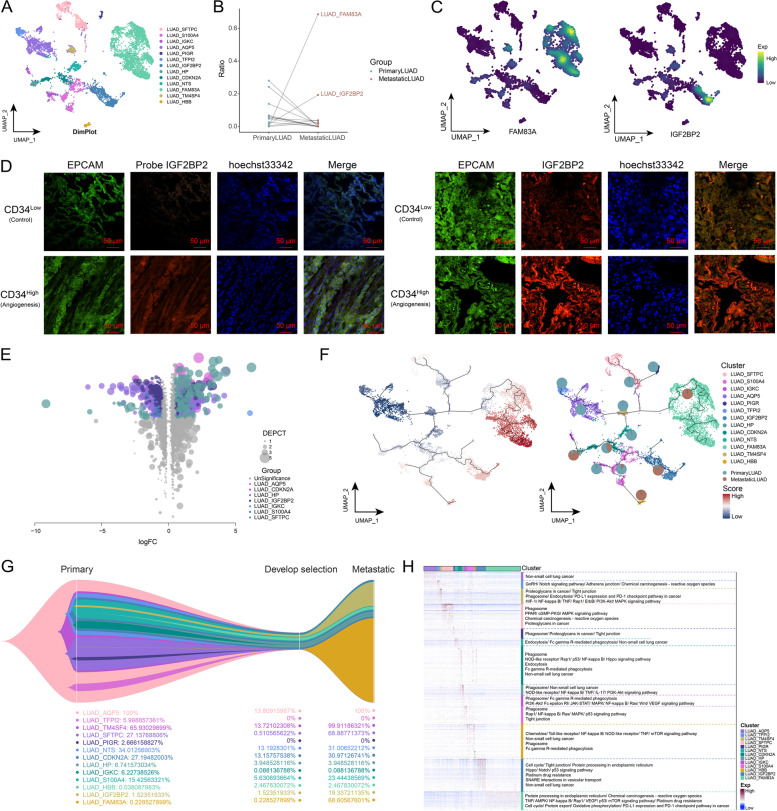
The process of clonal evolution of tumor cells is often critical in cancer development [39]. Therefore, to explore the clonal evolutionary trajectory of metastatic LUAD cells, LUAD cell types were first analyzed. Notably, LUAD cells were highly abundant in metastatic LUAD compared to primary tumors, so we performed subpopulation analyses of LUAD cells and constructed a single-cell atlas (Fig. 2A). In exploring the cellular ecology of LUAD cell subpopulations in primary and metastatic LUAD, the abundance of the LUAD_FAM83A and the LUAD_IGF2BP2 subpopulations was found to be significantly higher in metastatic LUAD than in primary tumors (Fig. 2B). The expression of FAM83A and IGF2BP2 was subsequently demonstrated in single-cell profiles (Fig. 2C), and fluorescence results proved that IGF2BP2 expression was significantly higher in LUAD tumors with high CD34 expression than those with low CD34 expression (Fig. 2D), revealing the close relationships between IGF2BP2 and angiogenesis. To further explore the potential molecular mechanisms of metastasis, DEGs in LUAD cell subpopulations between primary and metastatic LUAD were identified (Fig. 2E). According to pseudotime trajectory analysis on LUAD cells (Fig. 2F), an evolutionary trend from primary to metastatic LUAD was identified, which demonstrated the potential structural changes of LUAD cell subpopulations from primary LUAD to natural selection and metastatic disease (Fig. 2G). The signaling pathways and biological functions involved in the proposed time-related DEGs were explored by the functional enrichment analysis, and the LUAD_IGF2BP2 subpopulation was significantly enriched by vesicle synthesis, and secretion of signals (Fig. 2H). In summary, this study inferred the origin and clonal evolution trajectory of metastatic LUAD cell subpopulations and identified the genes involved in the evolutionary process and their biological functions.
Fig. 2.
Clonal evolutionary trajectory of metastatic LUAD cells. A The single cell atlas showing tumor cell subpopulations. B Dotted line graph showing the ratio of distinct tumor cell subpopulations in control, primary and metastatic LUAD. C The single-cell atlas mapping the expression of FAM83A and IGF2BP2 in tumor cells. D Fluorescence assay-fluorescent probe assay showing IGF2BP2 in LUAD tissues with high or low CD34 expression. Bar, 50 μm. E Volcano map showing differentially expressed genes in malignant cells and their subpopulations between metastatic and primary LUAD. F The single-cell atlas mapping the pseudotime values and evolutionary trajectory. G Evolutionary tree showing the evolutionary trajectory of tumor cells. H Heat map-pathway showing signaling pathways involved in temporally relevant genes
Endothelial cell landscape of metastatic LUAD
Cancer progression is characterized by the activation of endothelial cells which in turn promotes tumor angiogenesis. Therefore, endothelial cell subpopulations were identified in the single-cell atlas (Fig. 3A), and the unique cellular ecology of each endothelial cell subpopulation was observed both in primary and metastatic LUAD. Notably, the significant increase in En_S100A9, En_KRT19, and En_IGF2BP2 subpopulations was observed during the progression of primary LUAD to metastasis (Fig. 3B). The simultaneous expression of IGF2BP2 both in the LUAD_IGF2BP2 and the En_IGF2BP2 subpopulations (Fig. 3C, D), indicated the interactions between the two subpopulations. Subsequently, marker genes shared by the LUAD_IGF2BP2 and En_IGF2BP2 subpopulations were further identified, including IGF2BP2, IGF2BP3, SOX4, ID1, WASF2, ADGRG6, and FKBP1A (Fig. 3E). Enrichment analysis (Fig. 3F, G) revealed that these specific cell subsets exhibited the significant activation of critical biological functions such as endocytosis. Further GSEA analysis revealed that angiogenesis, exosome, epithelial mesenchymal transition (EMT), and N6-methyladenosine (m6A) were notably enriched in the specific cell subpopulations (Fig. 3H). Based on these findings, we tentatively constructed the endothelial cell landscape of metastatic LUAD.
Fig. 3.
Endothelial cell landscape in metastatic LUAD. A The single-cell atlas showing endothelial cell subpopulations. B Dotted line graph showing changes in abundance of endothelial cell subpopulations in control, primary and metastatic LUAD. C The single-cell atlas mapping the expression of IGF2BP2 in endothelial cells. D Immunofluorescence assay-fluorescent probe assay showing IGF2BP2 expression in endothelial cell subpopulations with high and low CD34 expression. Bar, 50 μm. E Shared markers of specific cell subpopulations (LUAD_IGF2BP2 and En_IGF2BP2). F Cluster-bubble plots demonstrating biological functions shared by specific cell subpopulations. G Cluster-bubble plots demonstrating KEGG pathways shared by specific cell subpopulations. H GSEA plots demonstrating the significant activation of biological signals related to secretion, secretion and synthesis of vesicles and phagocytosis of contents diffusion in specific cell subpopulations En_IGF2BP2 (left) and LUAD_IGF2BP2 (right)
Targeting IGF2BP2 mitigates migration, invasion, and angiogenesis in LUAD cells
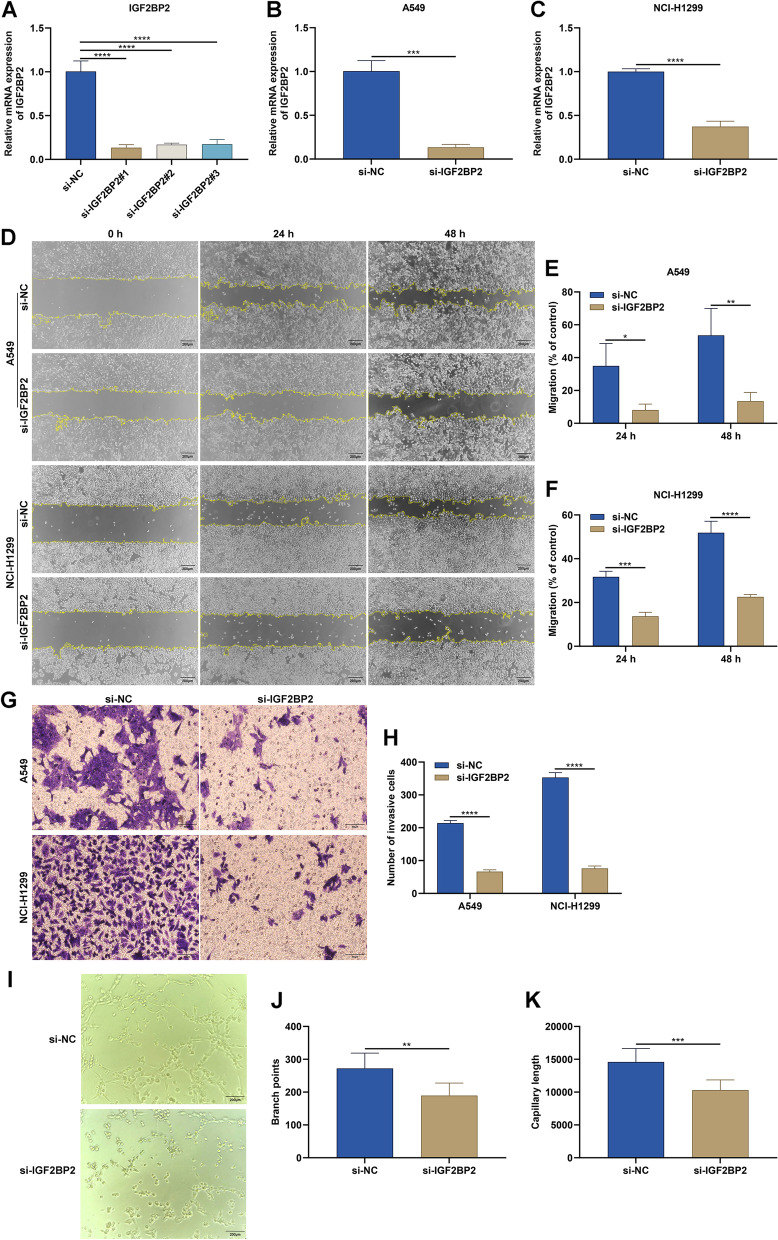
Next, we investigated the influence of IGF2BP2 on biological behaviors of LUAD cells. For transient transfection, three specific siRNAs targeting IGF2BP2 were designed, and among them, si-IGF2BP2#1 had the optimal interference effect in A549 and NCI-H1299 cells (Fig. 4A-C). According to wound healing assay, migration of A549 and NCI-H1299 cells was significantly decreased by si-IGF2BP2 (Fig. 4D-F). Moreover, our transwell assay results demonstrated that si-IGF2BP2 significantly suppressed invasion of A549 and NCI-H1299 cells (Fig. 4G, H). After HUVECs cultured with conditioned medium from si-IGF2BP2-transfeced A549 cells, both branch points and capillary length were significantly attenuated (Fig. 4I-K), indicating that IGF2BP2 knockdown in LUAD cells inhibited angiogenesis.
Fig. 4.
Targeting IGF2BP2 mitigates migration, invasion, and angiogenesis in LUAD cells. A Selection of the optimal siRNAs targeting IGF2BP2 through RT-qPCR. B, C Verification of IGF2BP2 mRNA expression in A549 and NCI-H1299 cells after transfection with si-NC or si-IGF2BP2. D-F Wound healing scratches at 0 h, 24 h, and 48 h for A549 and NCI-H1299 cells transfected with si-NC or si-IGF2BP2. Bar, 200 μm. G, H Transwell assay for detecting invasive A549 and NCI-H1299 cells after transfection with si-NC or si-IGF2BP2. Bar, 50 μm. I-K Images of HUVECs cultured with conditioned medium from A549 cells transfected with si-NC or si-IGF2BP2, and quantification of branch points and capillary length. Bar, 200 μm. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001
LUAD cell-derived exosomes mediate IGF2BP2 transfer to microenvironmental endothelial cells to activate the PI3K-Akt signaling for angiogenesis
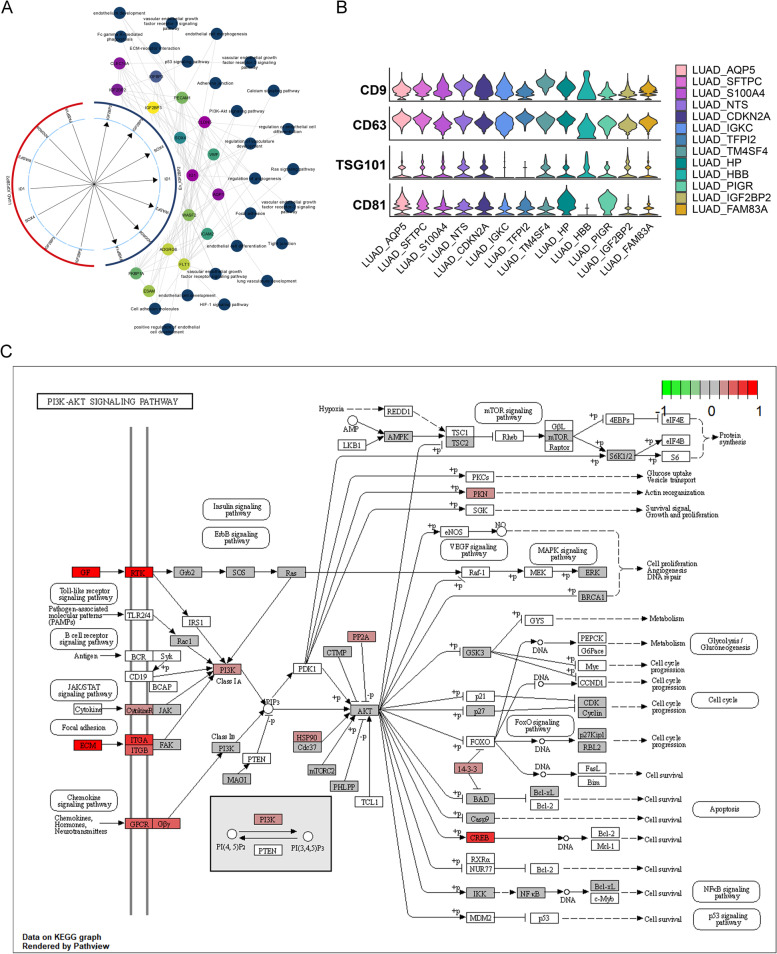
Our above study indicated that metastatic LUAD might achieve angiogenesis through exosomes. Thus, to investigate the mechanism of LUAD cell-derived exosomes transfer to endothelial cells in the tumor microenvironment, we constructed a global regulatory landscape of angiogenesis-specific ecotypes and obtained an integrated regulatory network from the LUAD_IGF2BP2 to the En_IGF2BP2 subpopulations (Fig. 5A). Subsequently, exosome marker genes (CD9, CD63, TSG01 and CD81) were all found to be highly expressed in all LUAD subpopulations, proving the formation of exosomes in LUAD cells (Fig. 5B). Combining the expression of IGF2BP2 both in the LUAD_IGF2BP2 and the En_IGF2BP2 subpopulations, cellular internalization of LUAD cells-derived exosomal IGF2BP2 into endothelial cells was inferred. Furthermore, the PI3K-Akt signaling pathway that confers LUAD angiogenesis and metastasis was found to be activated in the En_IGF2BP2 subpopulation (Fig. 5C). These results indicated that LUAD cells might transmit exosomal IGF2BP2 to endothelial cells to activate the PI3K-Akt signaling and ultimately promote angiogenesis.
Fig. 5.
Overexpression of IGF2BP2 in endothelial cells activates the PI3K-Akt signaling to promote angiogenesis. A Circle diagram showing the relationships between LUAD_IGF2BP2 ligand genes and En_IGF2BP2 receptor genes. The gene colors characterize their logFC and the pathway colors are aligned with the circles. The first loop genes are shared markers for the LUAD_IGF2BP2 and the En_IGF2BP2 subpopulations, the second loop is a non-shared marker, and the third and fourth loops are signaling pathways. B Violin diagram showing the expression pattern of exosomal markers in the LUAD subpopulations. C Pathway map showing the PI3K-Akt signaling pathway activated in the En_IGF2BP2 subpopulation
IGF2BP2 mediates the m6A modification of FLT4 and activates the PI3K-Akt signaling pathway
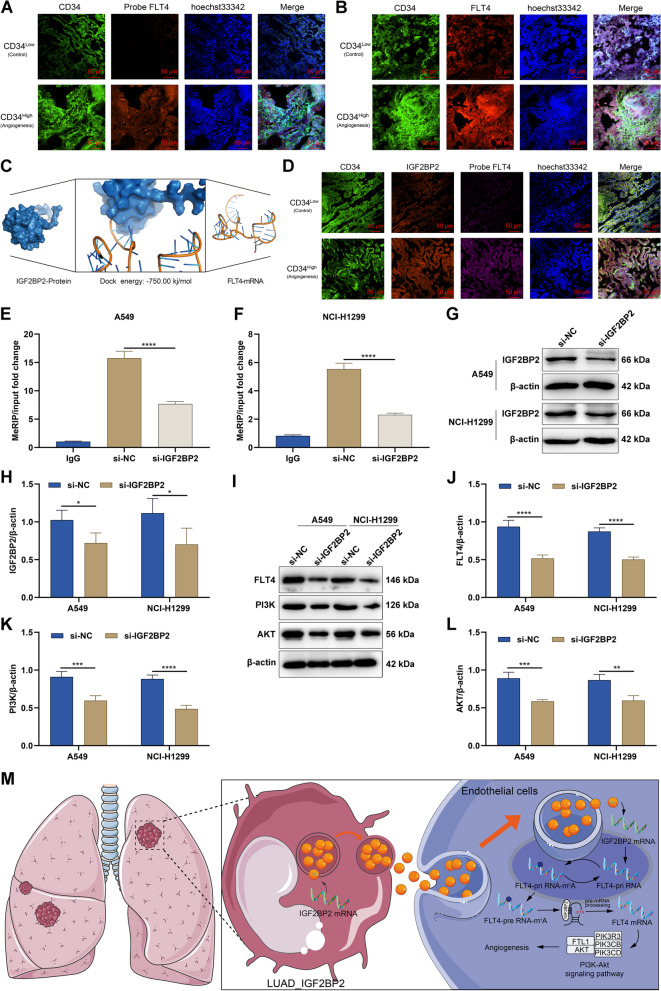
How IGF2BP2 activates the PI3K-Akt signaling and thus promotes angiogenesis was further probed. FLT4 was identified as a possible gene targeted by IGF2BP2 to activate the PI3K-Akt signaling. Immunofluorescence experiments showed that RNA levels (Fig. 6A) and protein levels (Fig. 6B) of FLT4 were significantly higher in focal endothelial cells of LUAD samples with high CD34 expression than in those with low CD34 expression. The binding potential of IGF2BP2 protein to the mRNA of FLT4 was predicted by molecular docking, and docking energy of -750 kj/mol indicated a great potential for targeted binding between them (Fig. 6C). Multiplex immunofluorescence also demonstrated the co-expression of FLT4 RNA with IGF2BP2 protein (Fig. 6D). Knockdown of IGF2BP2 significantly decreased m6A levels of FLT4 both in A549 and NCI-H1299 cells (Fig. 6E, F). In addition, FLT4 protein levels were significantly down-regulated in IGF2BP2-knockout A549 and NCI-H1299 cells (Fig. 6G-J). This demonstrated that IGF2BP2 could up-regulate FLT4 expression through mediating the m6A modification of FLT4. Moreover, PI3K and AKT protein levels were significantly decreased in A549 and NCI-H1299 cells with IGF2BP2 knockdown (Fig. 6K, L), indicating that IGF2BP2 was involved in activating the PI3K-Akt signaling pathway. These results suggested that during the clonal evolution of metastatic LUAD, the IGF2BP2-overexpressing LUAD cell subpopulation diffuses IGF2BP2 into the tumor microenvironment and is taken up by endothelial cells by cellular internalization, which subsequently targets to enhance the RNA stability of FLT4 through m6A modification, thereby activating the PI3K-Akt signaling pathway, and promoting angiogenesis (Fig. 6M).
Fig. 6.
IGF2BP2 activates the PI3K-Akt signaling pathway through mediating the m6A modification of FLT4. A Fluorescent probe assay for detecting RNA levels of FLT4 in focal endothelial cells of LUAD patients with high or low CD34 expression. Bar, 50 μm. B Multiplex immunofluorescence assay for measuring protein levels of FLT4 in focal endothelial cells of LUAD patients with high or low CD34 expression. Bar, 50 μm. C Molecular docking for predicting the docking potential of IGF2BP2 protein with mRNA of FLT4. D Multiplex immunofluorescence assay for verifying protein levels of IGF2BP2 and RNA levels of FLT4 in focal endothelial cells of LUAD patients with high or low CD34 expression. Bar, 50 μm. E, F The m.6A levels of FLT4 in A549 and NCI-H1299 cells after transfection with si-NC or si-IGF2BP2. G, H IGF2BP2 protein levels in A549 and NCI-H1299 cells with si-NC or si-IGF2BP2 transfection. I-L FLT4, PI3K, and AKT expression levels in A549 and NCI-H1299 cells with si-NC or si-IGF2BP2 transfection. M The diagram for IGF2BP2-mediated FLT4-PI3K-Akt signaling pathway in regulating angiogenesis during LUAD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001
Construction of an IGF2BP2-based prognostic scoring system for LUAD
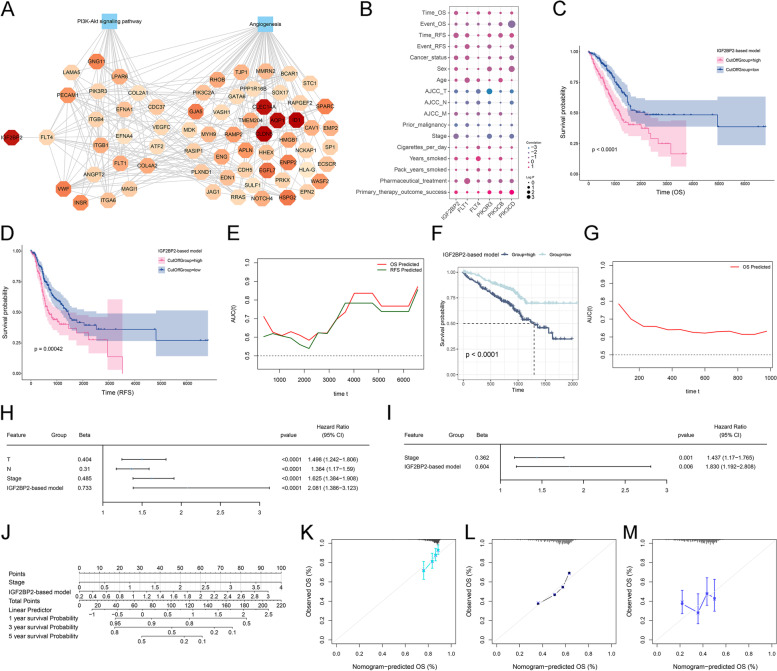
We further constructed a global regulatory network of IGF2BP2-FLT4-PI3K-Akt signaling-angiogenesis (Fig. 7A). The potential of these network genes in predicting LUAD prognosis was then evaluated. The association of the key network genes with clinical indicators was explored in the TCGA-LUAD dataset and they were found to be significantly associated with clinical parameters, especially OS and relapse-free survival (RFS) (Fig. 7B). A multivariate cox regression model termed as IGF2BP2-based prognostic scoring system was established based on the key network genes (including IGF2BP2, FLT1, FLT4, PIK3R3, PIK3CB, and PIK3CD), and patients were stratified into low- or high-score groups according to the median value. Survival analyses showed that OS and RFS outcomes were significantly better in the low-score than in the high-score group (Fig. 7C, D). Time-independent ROC curves demonstrated the excellent performance of the model in predicting OS and RFS (Fig. 7E). The IGF2BP2-based prognostic scoring system was proven in an external validation dataset (GSE72094) (Fig. 7F, G). Combining uni- and multivariate-cox regression analyses, it was found that the IGF2BP2-based model and stage were independent risk factors of LUAD prognosis (Fig. 7H, I). We established a nomogram comprising the two independent risk factors (Fig. 7J). Calibration curves demonstrated that the nomogram could accurately predict one-, three- and five-year OS outcomes (Fig. 7K-M). These results indicate the IGF2BP2-FLT4-PI3K-Akt signaling-angiogenesis network plays a crucial role in LUAD prognosis, which provides a theoretical basis for accurate risk stratification of LUAD patients.
Fig. 7.
Development of the IGF2BP2-based prognostic scoring system for LUAD. A The global regulatory network of IGF2BP2-FLT4-PI3K-Akt signaling-angiogenesis. B Bubble plots demonstrating the correlation between the IGF2BP2-FLT4-PI3K-Akt signaling-angiogenesis key genes and clinical indicators in the TCGA-LUAD dataset. C, D Survival curves of OS and RFS outcomes for the low and high-score patients stratified by the IGF2BP2-based model in the TCGA-LUAD dataset. E Time-independent ROC curves for evaluating the performance of the IGF2BP2-based model in predicting OS and RFS outcomes in the TCGA-LUAD dataset. F, G External validation of OS analysis and time-independent ROC curves in the GSE72094 dataset. H, I Uni- and multivariate cox regression results on the IGF2BP2-based model and clinical parameters with patient survival in the TCGA-LUAD dataset. J Construction of the nomogram composed of the IGF2BP2-based model and stage. K-M Calibration curves for the nomogram-predicted and actual one-, three- and five-year OS outcomes
Screening potential small molecule inhibitors of IGF2BP2
Based on the GDSC, and CTRP databases, we predicted potential small molecule inhibitors of IGF2BP2 according to the threshold of correlation coefficient < -0.1 and adjusted p < 0.05. Consequently, three small molecule inhibitors shared by the GDSC, and CTRP databases were screened, including trametinib, selumetinib, and dasatinib (Supplementary Fig. 2).
Discussion
In this study, a global single-cell landscape of primary and metastatic LUAD was constructed and two specific LUAD malignant cell subpopulations (LUAD_IGF2BP2, LUAD_FAM83A) with positive expression of IGF2BP2 and FAM83A, respectively, were captured, and both were highly abundant in metastatic LUAD. In addition, we proposed the mechanism of LUAD cell-derived exosomes mediating IGF2BP2 transfer to microenvironmental endothelial cells to activate the PI3K-Akt signaling for angiogenesis. The genes involved in the mechanisms had prognostic potential and may be potential prognostic markers for LUAD, which will provide a theoretical basis for a more precise assessment of LUAD prognosis.
The family member A gene with sequence similarity 83 (FAM83A) was initially identified as a novel tumor-specific gene, and previous studies demonstrated that it is highly expressed in LUAD and positively associated with poor prognosis, and that it plays an important role in the regulation of LUAD progression [40, 41]. Furthermore, many studies have shown that tumor heterogeneity is responsible for the progression of advanced tumors, and that tumor cells are constantly mutated and differentiated into different subclonal populations during natural selection [14]. The IGF2BP2 and FAM83A expression-positive malignant cell subpopulations identified in this study provide cytological insights into the development of LUAD, where the LUAD_IGF2BP2 subpopulation predominates in metastatic LUAD and is involved in vesicle synthesis, signaling molecule secretion, and exosome-related biological functions, from which we suggested that tumor microenvironment reprogramming during the malignant evolution of metastatic LUAD involves intercellular communication with exosomes as the main means.
We explored subpopulations of endothelial cells and found that subpopulations of endothelial cells positive for S100A9, KRT19, and IGF2BP2 expression (En_S100A9, En_KRT19, and En_IGF2BP2 subpopulations) were highly enriched in metastatic LUAD. Surprisingly, the LUAD_IGF2BP2 subpopulation expressed the same subpopulation marker IGF2BP2 as the En_IGF2BP2 subpopulation, and co-expressed some m6A modifiers (IGF2BP3, SOX4, ID1, WASF2, ADGRG6 and FKBP1A); moreover, enrichment analysis revealed that phagocytosis was significantly activated in the En_IGF2BP2 subpopulation. It was inferred that LUAD cell-derived exosomes mediated the delivery of IGF2BP2 to microenvironmental endothelial cells. Further information combined with downstream regulatory levels confirmed that IGF2BP2 acts as an m6A methylation reader, enhancing the RNA stability of FLT4, which in turn promotes the PI3K-Akt signaling-endothelial activation cascade. FLT4 encodes the tyrosine kinase receptor for vascular endothelial growth factors C and D, and its expression is significantly upregulated in microvessels of tumors and wounds, however, its role has not been investigated in LUAD [42]. IGF2BP2 is a unique m6 A reader that targets a large number of mRNA transcripts and promotes the stability and storage of targeted mRNAs in oncogenic effects [20]. It is well known that m6A modification induces oncogenic protein expression, cancer cell proliferation, survival, tumorigenesis and progression [43], and thus the overexpression of m6A modification reader (IGF2BP2) is highly likely to promote the progression of LUAD [39]. Although m6A modification is one of the common post-transcriptional epigenetic modalities with important roles in tumor development [44], it remains largely uncharted territory [45]. The present study put forward the novel theory that the m6A modification regulatory factor (IGF2BP2) is transmitted from malignant cell subclones to microenvironmental endothelial cells via exosomes and is subsequently taken up and forms a distinct endothelial subpopulation, which in turn promotes LUAD angiogenesis and metastasis.
In addition, based on the global regulatory network of IGF2BP2-FLT4 axis regulating the PI3K-Akt signaling pathway to promote angiogenesis, this study constructed an IGF2BP2-based clinical model and found that it was significantly associated with LUAD prognosis. Based on the above findings, it was confirmed that the IGF2BP2-based prognostic scoring model not only helps to assess the prognostic outcomes of patients, but also contributes to an in-depth understanding of the cancer metastasis mechanism, and also provides a profound theoretical basis and scientific foundation for the development of anti-LUAD strategies by targeting IGF2BP2-based m6A modification.
Conclusion
In summary, this study constructs a global single-cell landscape of primary and metastatic LUAD through single-cell transcriptomics analysis and reveals an IGF2BP2-based mechanism in LUAD angiogenesis and metastasis, which provides an opportunity to elucidate the relevant cellular dynamics and molecular features of LUAD metastasis.
Supplementary Information
Additional file 1: Supplementary Fig. 1. Data preprocessing and quality control of scRNA-seq. A, B. Violin plots showing the number of genes detected in each cell (nFeature_RNA), the total number of mRNA molecules detected in the cells (nCount_RNA), and the percentage of mitochondrial gene expression in the total gene expression (percent.mt) (A) before and (B) after quality control. C. Scatter plots showing the distribution of nCount_RNA and percent.mt as well as nCount_RNA and nFeature_RNA to filter low-quality cells.
Additional file 2: Supplementary Fig. 2. Prediction of potential small molecule inhibitors of IGF2BP2 based on the GDSC, and CTRP databases.
Acknowledgements
Not applicable.
Abbreviations
- LUAD
Lung adenocarcinoma
- OS
Overall survival
- IGF2BP2
Insulin-like growth factor 2 (IGF2) mRNA-binding protein 2
- scRNA-seq
Single-cell RNA sequencing
- TCGA
The Cancer Genome Atlas
- DEGs
Differentially expressed genes
- UMAP
Uniform manifold approximation and projection
- SCENIC
Single Cell Regulatory Network Inference and Clustering
- TFs
Transcription factors
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GSEA
Gene Set Enrichment Analysis
- siRNA
Small interfering RNA
- RT-qPCR
Reverse transcription quantitative PCR
- MeRIP-qPCR
M6A methylated RNA immunoprecipitation-qPCR
- ROC
Receiver operating characteristic
- RFS
Relapse-free survival
Authors’ contributions
Xicheng Song, Xiaofeng Yu, Ping Sun and Ke Mo are responsible for the design of this study; Han Fang, Qi Sun, Jin Zhou and Ke Mo are responsible for the writing of this manuscript; Han Fang, Qi Sun, Jin Zhou, Huijuan Zhang, Qiong Song, Hua Zhang, Guohua Yu, Ying Guo, Chengyu Huang and Yakui Mou are responsible for the data analysis; Chuanliang Jia, Yingjian Song, Aina Liu, Kaiyu Song, Congxian Lu, Ruxian Tian, Yixuan Chen, Ting Li, Han Fang and Qi Sun are responsible for the experimental testing; Kejian Wang, Yilan Yu and Yufeng Lv are responsible for sample collection; Xicheng Song support the funding for this study. All authors reviewed and approved the manuscript.
Funding
This work was funded by Taishan Scholars Project (ts20190991), The Natural Science Fund of Shandong Province grant (ZR2021MH323), The Science and Technology Innovation Development Plan of Yantai (2022YD037), China Scholarship Council (201909370036), The National Natural Science Fund of China grant (82103646), the Key Research and Development Program of Shandong Province (Major Science and Technology Innovation Project) (2022CXPT023).
Availability of data and materials
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Yantai Yuhuangding Hospital of Qingdao University (2022–383).
Consent for publication
All patients provided written informed consent.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Han Fang, Qi Sun, Jin Zhou and Huijuan Zhang contributed equally to this work.
Contributor Information
Ke Mo, Email: kemo@ydlife.org.
Ping Sun, Email: sunping20039@hotmail.com.
Xiaofeng Yu, Email: 15615085999@163.com.
Xicheng Song, Email: drxchsong@163.com.
References
- 1.Xia C, Dong X, Li H, Cao M, Sun D, He S, Yang F, Yan X, Zhang S, Li N, Chen W. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J (Engl) 2022;135:584–590. doi: 10.1097/CM9.0000000000002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gridelli C, Rossi A, Carbone DP, Guarize J, Karachaliou N, Mok T, Petrella F, Spaggiari L, Rosell R. Non-small-cell lung cancer. Nat Rev Dis Primers. 2015;1:15009. doi: 10.1038/nrdp.2015.9. [DOI] [PubMed] [Google Scholar]
- 3.Kim N, Kim HK, Lee K, Hong Y, Cho JH, Choi JW, Lee JI, Suh YL, Ku BM, Eum HH, et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun. 2020;11:2285. doi: 10.1038/s41467-020-16164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inamura K. Clinicopathological Characteristics and Mutations Driving Development of Early Lung Adenocarcinoma: Tumor Initiation and Progression. Int J Mol Sci. 2018;19(4):1259. doi: 10.3390/ijms19041259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 6.Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ, Jr, Wu YL, Paz-Ares L. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 7.Gettinger S, Horn L, Jackman D, Spigel D, Antonia S, Hellmann M, Powderly J, Heist R, Sequist LV, Smith DC, et al. Five-Year Follow-Up of Nivolumab in Previously Treated Advanced Non-Small-Cell Lung Cancer: Results From the CA209-003 Study. J Clin Oncol. 2018;36:1675–1684. doi: 10.1200/JCO.2017.77.0412. [DOI] [PubMed] [Google Scholar]
- 8.Riihimaki M, Hemminki A, Fallah M, Thomsen H, Sundquist K, Sundquist J, Hemminki K. Metastatic sites and survival in lung cancer. Lung Cancer. 2014;86:78–84. doi: 10.1016/j.lungcan.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Nie J, Dai L, Hu W, Zhang J, Han J, Ma X, Tian G, Han S, Wu D, et al. Comparison of endostatin combined with PT-DC versus bevacizumab combined with PT-DC in the first-line treatment of advanced lung adenocarcinoma: a retrospective propensity score-matched cohort study. Ann Palliat Med. 2021;10:7847–7856. doi: 10.21037/apm-21-1401. [DOI] [PubMed] [Google Scholar]
- 10.Soria JC, Mauguen A, Reck M, Sandler AB, Saijo N, Johnson DH, Burcoveanu D, Fukuoka M, Besse B, Pignon JP. Systematic review and meta-analysis of randomised, phase II/III trials adding bevacizumab to platinum-based chemotherapy as first-line treatment in patients with advanced non-small-cell lung cancer. Ann Oncol. 2013;24:20–30. doi: 10.1093/annonc/mds590. [DOI] [PubMed] [Google Scholar]
- 11.Wheatley-Price P, Shepherd FA. Targeting angiogenesis in the treatment of lung cancer. J Thorac Oncol. 2008;3:1173–1184. doi: 10.1097/JTO.0b013e318187220f. [DOI] [PubMed] [Google Scholar]
- 12.Noroxe DS, Wallerek S, Sorensen JB. Platinum-based doublet chemotherapy plus bevacizumab without bevacizumab maintenance in advanced non-small cell lung cancer (NSCLC) Anticancer Res. 2013;33:3275–3278. [PubMed] [Google Scholar]
- 13.Hu W, Fang J, Nie J, Dai L, Zhang J, Chen X, Ma X, Tian G, Wu D, Han S, et al. Efficacy and safety of extended use of platinum-based doublet chemotherapy plus endostatin in patients with advanced nonsmall cell lung cancer. Medicine (Baltimore) 2016;95:e4183. doi: 10.1097/MD.0000000000004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makrilia N, Lappa T, Xyla V, Nikolaidis I, Syrigos K. The role of angiogenesis in solid tumours: an overview. Eur J Intern Med. 2009;20:663–671. doi: 10.1016/j.ejim.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Park JH, Yoon J, Park B. Pomolic acid suppresses HIF1alpha/VEGF-mediated angiogenesis by targeting p38-MAPK and mTOR signaling cascades. Phytomedicine. 2016;23:1716–1726. doi: 10.1016/j.phymed.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 16.Ushio-Fukai M, Nakamura Y. Reactive oxygen species and angiogenesis: NADPH oxidase as target for cancer therapy. Cancer Lett. 2008;266:37–52. doi: 10.1016/j.canlet.2008.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lavin Y, Kobayashi S, Leader A, Amir ED, Elefant N, Bigenwald C, Remark R, Sweeney R, Becker CD, Levine JH, et al. Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell. 2017;169(750–765):e717. doi: 10.1016/j.cell.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Li Z, Zhou K, Wang C, Jiang L, Zhang L, Yang Y, Luo W, Qiao W, Wang G, et al. Deciphering cell lineage specification of human lung adenocarcinoma with single-cell RNA sequencing. Nat Commun. 2021;12:6500. doi: 10.1038/s41467-021-26770-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu X, Peng WX, Zhou H, Jiang J, Zhou X, Huang D, Mo YY, Yang L. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 2020;27:1782–1794. doi: 10.1038/s41418-019-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia M, Shi Y, Xie Y, Li W, Deng J, Fu D, Bai J, Ma Y, Zuberi Z, Li J, Li Z. WT1-AS/IGF2BP2 Axis Is a Potential Diagnostic and Prognostic Biomarker for Lung Adenocarcinoma According to ceRNA Network Comprehensive Analysis Combined with Experiments. Cells. 2021;11(1):25. doi: 10.3390/cells11010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma YS, Shi BW, Guo JH, Liu JB, Yang XL, Xin R, Shi Y, Zhang DD, Lu GX, Jia CY, et al. microRNA-320b suppresses HNF4G and IGF2BP2 expression to inhibit angiogenesis and tumor growth of lung cancer. Carcinogenesis. 2021;42:762–771. doi: 10.1093/carcin/bgab023. [DOI] [PubMed] [Google Scholar]
- 22.Yu J, Hou M, Pei T. FAM83A Is a Prognosis Signature and Potential Oncogene of Lung Adenocarcinoma. DNA Cell Biol. 2020;39:890–899. doi: 10.1089/dna.2019.4970. [DOI] [PubMed] [Google Scholar]
- 23.Schabath MB, Welsh EA, Fulp WJ, Chen L, Teer JK, Thompson ZJ, Engel BE, Xie M, Berglund AE, Creelan BC, et al. Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene. 2016;35:3209–3216. doi: 10.1038/onc.2015.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177(1888–1902):e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, Newell EW. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2018;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 26.Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC, Geurts P, Aerts J, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van de Sande B, Flerin C, Davie K, De Waegeneer M, Hulselmans G, Aibar S, Seurinck R, Saelens W, Cannoodt R, Rouchon Q, et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat Protoc. 2020;15:2247–2276. doi: 10.1038/s41596-020-0336-2. [DOI] [PubMed] [Google Scholar]
- 28.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 29.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Wang R, Zhang S, Song S, Jiang C, Han G, Wang M, Ajani J, Futreal A, Wang L: iTALK: an R Package to Characterize and Illustrate Intercellular Communication. bioRxiv 2019:507871.
- 34.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Armaos A, Colantoni A, Proietti G, Rupert J, Tartaglia GG. catRAPID omics v2.0: going deeper and wider in the prediction of protein-RNA interactions. Nucleic Acids Res. 2021;49:W72–W79. doi: 10.1093/nar/gkab393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macindoe G, Mavridis L, Venkatraman V, Devignes MD, Ritchie DW. HexServer: an FFT-based protein docking server powered by graphics processors. Nucleic Acids Res. 2010;38:W445–449. doi: 10.1093/nar/gkq311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mooers BHM. Shortcuts for faster image creation in PyMOL. Protein Sci. 2020;29:268–276. doi: 10.1002/pro.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN, Chen ZH, Zeng ZL, Wang F, Zheng J, et al. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18:112. doi: 10.1186/s12943-019-1038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang G, Li X, Yao Y, Jiang Z, Zhou H, Xie K, Luo J, Shen Y. FAM83A and FAM83A-AS1 both play oncogenic roles in lung adenocarcinoma. Oncol Lett. 2021;21:297. doi: 10.3892/ol.2021.12558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R, Rosenthal R, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 42.Lan Q, Liu PY, Haase J, Bell JL, Huttelmaier S, Liu T. The Critical Role of RNA m(6)A Methylation in Cancer. Cancer Res. 2019;79:1285–1292. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 43.Tian Y, Xiao H, Yang Y, Zhang P, Yuan J, Zhang W, Chen L, Fan Y, Zhang J, Cheng H, et al. Crosstalk between 5-methylcytosine and N(6)-methyladenosine machinery defines disease progression, therapeutic response and pharmacogenomic landscape in hepatocellular carcinoma. Mol Cancer. 2023;22:5. doi: 10.1186/s12943-022-01706-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niu X, Xu J, Liu J, Chen L, Qiao X, Zhong M. Landscape of N(6)-Methyladenosine Modification Patterns in Human Ameloblastoma. Front Oncol. 2020;10:556497. doi: 10.3389/fonc.2020.556497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu ZX, Li LM, Sun HL, Liu SM. Link Between m6A Modification and Cancers. Front Bioeng Biotechnol. 2018;6:89. doi: 10.3389/fbioe.2018.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Fig. 1. Data preprocessing and quality control of scRNA-seq. A, B. Violin plots showing the number of genes detected in each cell (nFeature_RNA), the total number of mRNA molecules detected in the cells (nCount_RNA), and the percentage of mitochondrial gene expression in the total gene expression (percent.mt) (A) before and (B) after quality control. C. Scatter plots showing the distribution of nCount_RNA and percent.mt as well as nCount_RNA and nFeature_RNA to filter low-quality cells.
Additional file 2: Supplementary Fig. 2. Prediction of potential small molecule inhibitors of IGF2BP2 based on the GDSC, and CTRP databases.
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.