ABSTRACT
Gemella species are core members of the human oral microbiome in healthy subjects and are regarded as commensals, although they can cause opportunistic infections.
Our objective was to evaluate the site-specialization of Gemella species among various habitats within the mouth by combining pangenomics and metagenomics. With pangenomics, we identified genome relationships and categorized genes as core and accessory to each species. With metagenomics, we identified the primary oral habitat of individual genomes. Our results establish that the genomes of three species, G. haemolysans, G. sanguinis and G. morbillorum, are abundant and prevalent in human mouths at different oral sites: G. haemolysans on buccal mucosa and keratinized gingiva; G. sanguinis on tongue dorsum, throat, and tonsils; and G. morbillorum in dental plaque.
The gene-level basis of site-specificity was investigated by identifying genes that were core to Gemella genomes at a specific oral site but absent from other Gemella genomes. The riboflavin biosynthesis pathway was present in G. haemolysans genomes associated with buccal mucosa but absent from the rest of the genomes. Overall, metapangenomics show that Gemella species have clear ecological preferences in the oral cavity of healthy humans and provides an approach to identifying gene-level drivers of site specificity.
KEYWORDS: oral, microbiome, Gemella, pangenome, metagenome, metapangenome
Introduction
Members of the genus Gemella are core species of the human oral microbiome [1] found in healthy subjects and are therefore regarded as commensals, although they are known to cause opportunistic infections. Compared to other members of the human microbiota, the genus has not attracted much attention: a search in PubMed for ‘Gemella’ yielded 634 publications (as of March 24, 2023), mostly about its presence in or isolation from clinical samples of patients with endocarditis, oral diseases, wound infections, vaginosis, or other conditions. The basic microbiology literature indicates that members of this genus are non-motile cocci, facultatively anaerobic, capnophilic, of low G+C content, catalase-negative, oxidase-negative, with a tendency to grow in pairs, tetrads, or short chains [2–4]. Gemella species are capable of fermenting glucose into lactate and acetate as major metabolic end products [5]. However, the roles that species of Gemella play in the human microbiome as a whole remain to be established.
The taxonomic position of the genus Gemella only became clear after several false starts. Gemella haemolysans was originally reported as Neisseria haemolysans [6] but was later proposed as the founding member of a new genus because its enzymatic properties deviated from those of most Neisseria species [7]. Because G. haemolysans easily decolorized, it was first thought to be Gram-negative, but it was later re-classified on the basis of cell wall structure and biochemistry as Gram-positive [4]. Before the advent of molecular taxonomy, Gemella species were frequently misidentified as streptococci because both groups are catalase-negative and Gram-positive [8]. Molecular taxonomy now places Gemella species together with staphylococci, lactobacilli, and streptococci in the class Bacilli of the phylum Firmicutes [1]. Currently, five species of human-associated Gemella are recognized in the Human Oral Microbiome Database (HOMD; www.homd.org), four named and one unnamed [9,10]; in addition, there are five more validly named species in the List of Prokaryotic Names with Standing in Nomenclature (LPSN) [11] (https://lpsn.dsmz.de/search?word=Gemella, accessed March 24, 2023). To date, all species of Gemella have been isolated from either humans or animals [2,3,12–15], primarily from the oral cavity, but also from blood, wound and vaginal samples.
In this study, we bring the information-harnessing power of pangenomics and metagenomics to bear on the role of Gemella species in the oral ecosystem. Our principal objective is to evaluate the site-specialization [16] of Gemella among the various habitats within the mouth. Many studies have established that oral sites are distinguishable from each other in terms of their microbial composition. However, analysis at genus or higher taxonomic levels has hidden the extent of the disparity between the communities. Analysis of human microbiome 16S rRNA gene sequence data at single-nucleotide resolution suggested that different species within common genera of the mouth, including Gemella, had different roles in oral ecology, as revealed by their dramatically different abundances in samples from different sites within the mouth, such as the teeth, the tongue, and the cheeks and gums [17]. Based on these findings and a review of the literature, we hypothesized [16] that most oral microbes are site-specialists: that they grow primarily in distinct microhabitats of the mouth. This is not a new idea, having been introduced almost 50 years ago [18] based on culture-dependent studies. We developed the idea further based on single-nucleotide analysis of the V1–V3 region of the 16S ribosomal RNA gene data [17], but such an important conclusion should not be based only on a small region of a single marker gene. With the advent of rapid, inexpensive DNA sequencing, it has become possible to revisit the site-specialist concept of microbial species at the genomic level.
Site-specialization can be evaluated using genome-level data by constructing pangenomes and analyzing metagenomes. The pangenome is the sum of all genes found in members of a given group. It reveals both the functional essence of the group – genes and functions shared by all its members (core genes) – and the diversity held within a genomic group – genes unique to one or a subset of its members (accessory genes) [19,20]. Complementary to pangenomics is metagenomics, the analysis of the totality of DNA sequences in a sample taken from a specific environment [21,22]. The Human Microbiome Project (HMP) [23] collected samples from nine different sites within the human mouth and generated short-read metagenomic sequence data from the samples. Mapping of these sequence data from different oral sites onto oral pangenomes can be used to determine natural groupings of strains that share the ability to thrive in a particular oral habitat. Combined with the pangenome, this mapping information enables a genomic and ecological framework for the analysis of microbiomes. This approach, termed metapangenomics, has been used to investigate microbial communities in the surface ocean and soil [24,25] and the human microbiome [25–28] and has recently been applied to examine habitat adaptation and cultivar diversity for Neisseria, Saccharibacteria, Rothia and Haemophilus parainfluenzae in the human mouth [29–31].
The strength of metapangenomics lies in its integration of genomic and ecological data. Through the pangenome, genomes are classified based on their gene content regardless of the habitat in which the organisms reside. Conversely, metagenomic data provides insights into genetic traits selected from different ecological niches. Here, we applied metapangenomics to investigate the site-specialist patterns of Gemella in the oral cavity.
Results
Gemella pangenome
To build a Gemella pangenome, we accessed the National Center for Biotechnology Information (NCBI) database and downloaded all available genomes for strains of any named or unnamed species within the genus (SI Table S1). The set included genomes of human oral-associated Gemella species as well as several genomes from non-human and/or non-oral-associated species and 10 genomes identified only to genus level. As a quality control measure, from the total of 35 genomes available at NCBI, we removed five genomes that were not in RefSeq, constructing the pangenome from the remaining 30 RefSeq genomes. These genomes were derived from nine named species (n = 22), one not validly published, and seven entries that were identified only to genus level. We then used the analysis and visualization tool for ‘omics data, anvi’o [32], to organize the genomes into similarity groups based on the presence of homologous genes. Briefly, for each genome, open reading frames were predicted, and the resulting hypothetical genes were translated into amino acid sequences and grouped into gene clusters (groups of putative homologous genes) based on the level of amino acid similarity among them. Then, to visualize the pangenome, the gene clusters were hierarchically grouped by representation (presence/absence) across genomes to produce a gene cluster dendrogram, and the genomes were hierarchically grouped based on the frequency of the homologous genes within gene clusters to produce a genome dendrogram. These two dendrograms organize the pangenome structure of the Gemella genus.
In the representation of the Gemella pangenome shown in Figure 1a, each genome is color-coded by its species designation at NCBI and is represented as a horizontal bar composed of vertical lines, each line representing a gene cluster (mean number of gene clusters/genome = 1605). Most gene clusters across the pangenome consist of genes present in a single copy of each genome. However, some genomes contain multiple copies of homologous genes; these additional genes were grouped into the same gene cluster. Thus, the mean number of genes per genome, 1686, is greater than the mean number of gene clusters. Gene cluster robustness was assessed by inspecting gaps and amino acid variations in each gene cluster using the geometric and functional homogeneity indices (average 0.96 ± 0.08 and 0.94 ± 0.08 – a value of 1 indicates no gaps or no amino acid variation, respectively) (see Methods). Metadata for whether the species is listed in HOMD, the sample is of human origin, and the sample was isolated from an oral site are indicated by the filled squares at the right of the pangenome. Sample descriptors that we considered to indicate an oral origin included mouth, teeth, submandibular abscess, saliva, and lung (bronchoalveolar fluid and sputum) (SI Table S1). Hierarchical clustering of genomes and gene clusters shows that the genomes for strains of human oral species G. sanguinis, G. haemolysans, and G. morbillorum are each sorted into separate groups distinguished by blocks of genes present in every genome of each species but not in the genomes of other species. Thus, the pangenome recapitulates and confirms the genomic distinctiveness of the three principal human oral Gemella species.
Figure 1.
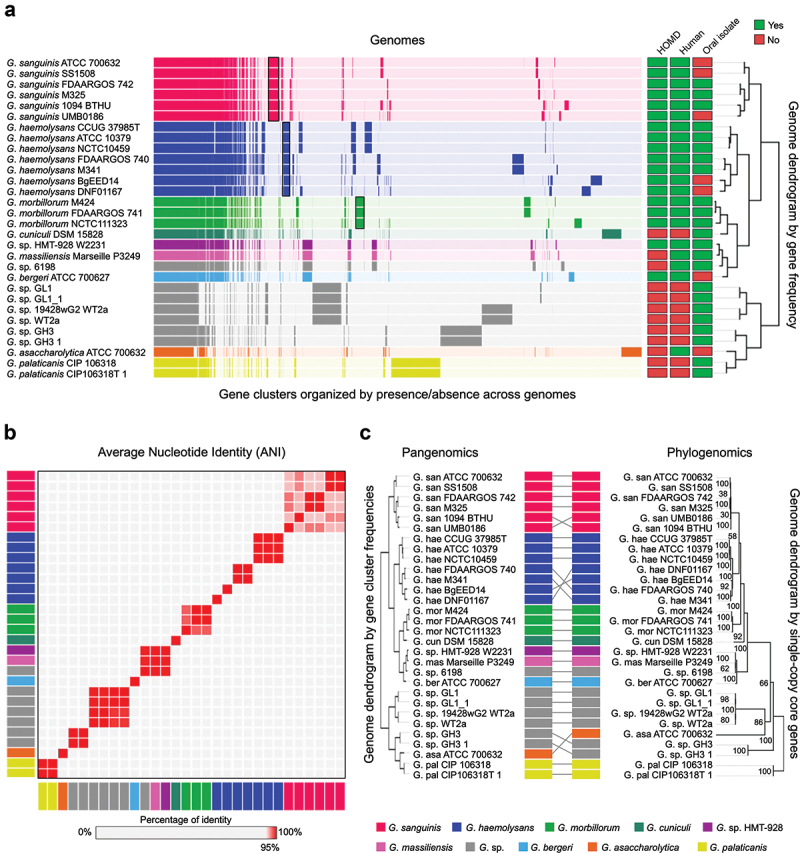
Gemella genomes cluster into species-level groups. A) Pangenome constructed from all (n = 30) available NCBI RefSeq Gemella genomes. Gene clusters are colored by species and arranged based on their presence or absence across the genomes. Genomes are hierarchically clustered based on gene cluster frequency, i.e. the number of representatives of each gene cluster (n = 6,922) present in each genome. Metadata columns indicate whether the species is included in HOMD, if the genome is of human origin, and if it was isolated from an oral site. Black boxes show gene clusters unique to G. sanguins, G. haemolysans and G. morbillorum. b) Average nucleotide identity (ANI) comparison of genomes in pangenome. ANI represents the genome-level similarity at the nucleotide level between any two genomes. Rectangle color indicates species as in A. Order of genomes is same as in the pangenome dendrogram. Color scale denotes genome similarity; 100% is red; below 95% is light grey. c) Pangenomic and phylogenomic tree comparison. Rectangle color indicates species as in A. The phylogenomic tree was constructed using maximum-likelihood with 17 concatenated single-copy core genes.
In addition to showing the similarity of gene content within identified species, the pangenome shows similarities and differences across both named species and unnamed strains. The genome of G. cuniculi, which was isolated from a rabbit, is similar to G. morbillorum, isolated from a human, illustrating divergent evolution of related bacterial species that co-evolved with different mammalian hosts. The genomes of human-associated species G. sp. HMT−928, ‘G. massiliensis’ (not validly published), G. sp. strain 6198, and G. bergeri form a distinct group. The genome of a human vaginal species, G. asaccharolytica, is very different from the oral genomes. The genomes of G. palaticanis, isolated from a dog, and other genomes identified only to genus level but obtained from mice were very different from human oral genomes. Three genomes of G. sanguinis and two genomes of G. haemolysans were isolated from non-oral sites, including blood, urinary tract, vagina and duodenum. The isolation of oral Gemella genomes from non-oral sites indicates the potential for systemic spread of oral bacteria.
The construction of the pangenome is based on the similarity of amino acid sequences of predicted open reading frames. As a check on the grouping of genomes, we also evaluated groupings at the DNA level by average nucleotide identity (ANI), which measures the genome-scale similarity between genomes and has been used to identify genomes from the same species [33,34]. The results (Figure 1b, SI Table S2) confirmed the groups of the principal Gemella oral species, as well as indicating that the three genomes variously named Gemella sp. HMT−928, ‘G. massiliensis’, and Gemella sp. strain 6198 could be taken together as a distinct group (>98% ANI) and are highly similar (94.8–94.9% ANI) to G. bergeri; these findings replicate and confirm the results of a prior analysis of Gemella genomes by ANI [35]. The genomes of the human vaginal G. asaccharolytica and the animal genomes were distinctive as expected. Overall, the ANI did not show any discrepancy from the pangenome dendrogram.
As a third estimate of the grouping of genomes within the pangenome, we considered their evolutionary relationships as determined by estimating the phylogeny of core, single-copy genes within the Gemella genus. Based on the sequence similarity of gene clusters in all genomes (see Methods), we identified 17 informative, single-copy core genes (SI Table S3) that were used to build a phylogenomic tree using IQ-TREE [36]. The phylogenomic tree of all Gemella genomes so constructed showed no difference in genome grouping at the species level in comparison to the genome dendrogram of the pangenome (Figure 1c), although the organization of genomes within certain clades was slightly different. Thus, all three measures of genome evaluation – gene content, ANI and phylogenomics – agreed on the basic grouping of oral Gemella genomes.
Close inspection of the pangenome indicated that some genomes were nearly identical in terms of both gene clusters and ANI. In several cases, the genomes are of the same strain deposited in different culture collections, and the replicated sequences provide increased confidence in the maintenance of the strain over time and the replicability of sequencing and assembly. The type strain of G. haemolysans, for example, is deposited in CCUG as ‘37985T’ and in ATCC as ‘10379’, and the assemblies of these strains showed 99.9% ANI with one another. In other cases, the high identity of sequence makes clear that the strains, although apparently different, have the same origin. The G. haemolysans strain NCTC 10459 has an ANI of 99.9% with the two type strains but is listed as not being a type strain at NCTC. However, the older literature [37] indicates the identity of the strain with type strain ATCC 10379. Other examples of 99.9% identical strains include G. haemolysans genomes FDAARGOS 740 and M341; two pairs of G. sanguinis genomes (ATCC 700632 and SS1507 and FDAARGOS 742 and M325); and two G. morbillorum genomes (FDAARGOS 741 and M424). In addition, four pairs of genomes showed 100% identity by ANI: three pairs of mouse genomes (GL1 and GL1 1; WT2a and 1942wG2 WT2a; GH3 and GH3 1) and one pair of canine genomes (CIP 106318 and CIP106318T 1). These genomes represent duplicate depositions in NCBI of the same microbial genome (e.g. GL1 and GL1 1, WT2a 1942wG2 WT2a; GH3 and GH3 1, CIP 106318 and CIP106318T 1). The fact that genomes deposited with different names had the same origin was discovered only with considerable sleuthing, as the depositions are listed with unique Assembly, BioSample, BioProject, WGS, and Strain designations. Their degree of identity was established only after pangenomic and ANI analysis. Pangenomes of each of the human oral Gemella species individually as well as taken together as a group are presented in SI Fig. S1.
Distribution of Gemella genomes across human oral sites
Isolates of Gemella species have been obtained mainly from the mouth of humans but also from other mucosal sites and blood. The site of isolation is indicative but may be misleading, as cultivation can result from the presence of as little as a single cell even if that cell represents a transient or low-abundance population at that site. To understand the distribution of Gemella species across the oral cavity of healthy subjects, we used metagenomic sequence data from the HMP for individual sites and mapped them onto the Gemella genomes as a way of testing the true oral habitat of the Gemella species.
Preferred habitats for Gemella in the mouth were evaluated by concatenating all 30 Gemella genomes and then competitively mapped the metagenomic short reads onto the concatenated sequence sample by sample (see Methods). The goal was to assess the similarity of the available genomes to the natural population in the mouth by using the reference genomes to capture closely related sequences from the metagenomic data. In total, we mapped approximately 50 billion quality-filtered metagenomic short reads from 1,215 samples collected from all nine oral sites (SI Table S4) described in the HMP. Genomes that were not isolated from humans served as outgroup controls. Mapping results by strain (genomes sharing ≥98% ANI) were combined and transformed into a heat map representing the relative proportion of reads recruited by each Gemella genome at each site (SI Figs S2 and S3). However, as raw read recruitment data can give misleading estimates of taxon abundance, we carried out additional analysis steps to accurately reflect the proportions of distinct strains present in each habitat.
A confounding factor that influences genome detection via read mapping is the presence of short regions of high identity across different genomes. This problem is particularly acute when the pangenome contains genomes with high similarity to one another. To adjust for the presence of such closely related genomes in the pangenome, we combined the abundance values for genomes of the same strain and labeled the results according to the highest quality strain genome as judged by having the fewest contigs. Animal genomes (except for the type strain genome of G. palaticanis) were removed from the display because they were not detected in any sample. Cross-mapping of reads from unrelated genomes to the target Gemella genomes can distort apparent abundance values, suggesting a false-positive result – that a genome is present when, in fact, only a few highly conserved genes may have attracted reads. Therefore, a breadth of coverage criterion was employed for determining whether a genome was classified as detected in a sample. Following the recently developed procedures [24,38] (https://instrain.readthedocs.io/en/latest/important_concepts.html), we used the criterion that at least 50% of the nucleotides in a genome had to be covered by at least 1× for the genome to be considered detected in a metagenomic sample (SI Table S5). We then calculated the proportion of reads recruited by genomes that passed the detection criterion.
Results for the three sites sampled most frequently (Figure 2) showed that each of the three sites was inhabited primarily by a different Gemella species. Tongue dorsum was dominated by G. sanguinis, buccal mucosa by G. haemolysans, and supragingival plaque by G. morbillorum. Statistical tests (SI Table S6) indicated that this tropism was significant for all three taxa. The number of samples available for other oral sites was smaller, thus allowing only limited conclusions. Nevertheless, the mapping results from them indicated that G. sanguinis could be found in the throat and palatine tonsils; G. haemolysans on keratinized gingiva; and G. morbillorum on subgingival plaque (SI Fig. S3, SI Table S7). Thus, Gemella species seem to be specialized to sets of ecologically related sites: G. haemolysans for buccal mucosa and keratinized gingiva; G. sanguinis for tongue dorsum, throat and tonsils; and G. morbillorum for plaque, both supra- and subgingival. The gender of the donor did not correlate with any of the mapping results. Overall, the mapping results show with genome-scale information that Gemella species have clear ecological preferences in the oral cavity of healthy humans.
Figure 2.
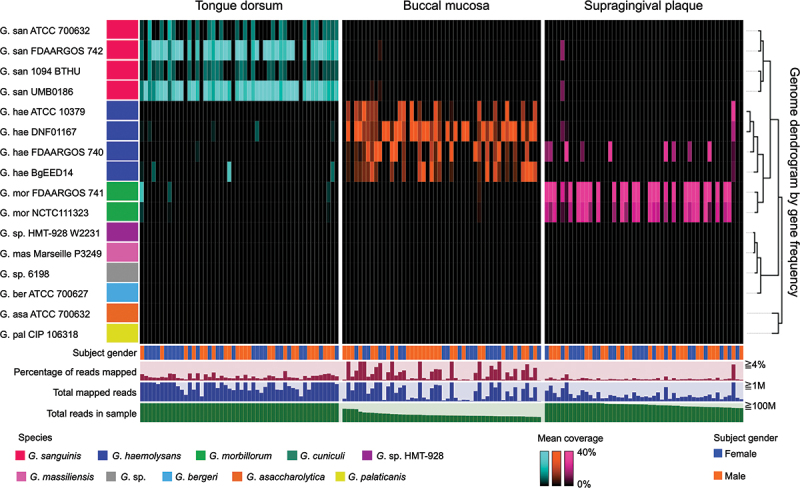
Distribution of Gemella strains across human oral sites. Heatmap shows the relative proportion of detected Gemella genomes across samples from three oral sites (tongue dorsum, buccal mucosa, and supragingival plaque). Replicate genomes and animal genomes not detected in any sample were removed from the pangenome. Color of genomes is the same as in the pangenome. The dendrogram was obtained from a pangenome which included the 16 strains shown in this figure. Samples are grouped by oral site and ordered by decreasing number of total reads. For each site, 50 samples containing the greatest number of reads are presented. Additional data are shown for gender, total mapped reads, total sample reads, and percentage of mapped reads, which may be taken as a measure of genus abundance.
Gene-level analysis across human oral sites
Metapangenomics identified the genomes of three species, G. haemolysans, G. sanguinis, and G. morbillorum, as particularly abundant and prevalent in human mouths at different oral sites. Having fully sequenced genomes presents an opportunity to identify specific genes that are associated with organisms in each habitat and that may encode key functions permitting the organism to thrive in that habitat. To identify these candidate gene-level drivers of site specialization, we constructed a revised Gemella pangenome that included only the dereplicated human genomes plus one canine genome (n = 16) and then carried out metagenomic mapping against this smaller pangenome in order to determine the abundance and prevalence in the oral environment of each gene in these genomes across all samples by site. Gene detection was determined by a breadth metric at the nucleotide level analogous to the metric used for genome detection but more stringent. Our criterion for a gene to be detected in a sample was that mapping from that sample resulted in at least 1× coverage of at least 90% of the nucleotides of the gene.
Gene-level detection data were displayed for each of the Gemella genomes as a radial heat map by site across the 50 metagenomic samples containing the greatest number of total reads. The gene-level maps for the three genomes showing the greatest prevalence across their respective preferred sites, tongue dorsum (TD), buccal mucosa (BM) and supragingival plaque (SUPP) are shown in Figure 3, and the displays for all 16 genomes are shown in SI Fig. S4. Genes within the displays were ordered according to category in the pangenome – genus core, species core, accessory, and singleton. For each genome, almost all of its genes were detected in only one of the three major oral sites – genomes of G. sanguinis in TD, G. haemolysans in BM and G. morbillorum in SUPP. This pattern of detection supports the concept of site-specificity of Gemella species at the gene level. Some genes from each of the strain genomes were not detected in any of the oral sites. This indicates a disparity between the cultivar genomes and oral environmental strains of the same species. Clearly, each cultivar genome contains some genes that are absent or rare in the oral environment. Of note, the genomes for the type strains of the three Gemella species were not the most prevalent nor the most representative of the genes in their respective oral environments (SI Table S7). For each genome, the pattern of detection of genes (presence and absence) was similar across samples, indicating that Gemella genomes in different individuals at a specific oral site contained a highly similar set of genes. Exceptions to this pattern were a few genes that were detected in the majority of samples at two or all three sites, suggesting that these genes were highly conserved in the oral environment across sites. Not surprisingly, annotation associated some of these genes with highly conserved functions such as DNA replication, 16S rRNA gene, transcriptional regulators, and mobile elements.
Figure 3.
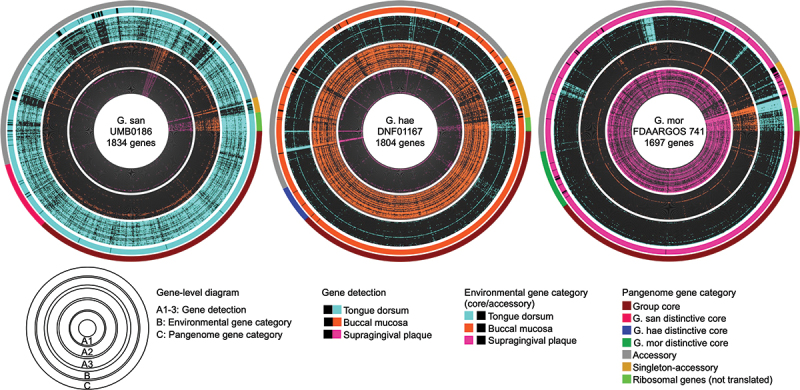
Gene-level detection in Gemella genomes across three oral sites. Circular heatmap for three genomes (G. sanguinis UMB0186, G. haemolysans DNF01167, and G. morbillorum FDAARGOS 741) shows the presence/absence of genes across samples from three oral sites (tongue dorsum, buccal mucosa, and supragingival plaque). The criterion for a gene to be present in a sample is that 90% of the nucleotides be covered by at least 1 × . Genes are ordered according to the gene-cluster category given by the pangenome of detected genomes. Within each oral site, samples are ordered by decreasing number of total reads in the same order as Figure 2. The outermost layer links each individual gene to the amino acid gene cluster category given in the pangenome (Group core, G. san distinctive core, G. hae distinctive core, G. mor distinctive core, accessory, and singletons). Ribosomal genes (non-translated genes) are indicated in bright green.
The detection of each gene across all samples by site was compared with the pangenome in order to determine whether genes that are core in the pangenome are also common in the oral environment. Following the definitions established in anvi’o [32], a gene was considered ‘environmentally core’ at a site if its median coverage across samples from that site was equal to or greater than 25% of the median coverage of its genome across samples from that site (see Methods). Below the 25% threshold, genes were considered ’environmentally accessory’ at that site. The binary categorization of genes as environmentally core or accessory at a site is displayed in layer B of Figure 3, SI Fig. S2, while layer C displays the status of the gene in the pangenome – genus core, species core, accessory, or singleton. As expected, almost all pangenome genus core genes were environmentally core across oral sites. In contrast, species core genes were environmentally core at a specific oral site – although some were environmentally accessory, perhaps reflecting a small sample size from the relatively limited set of genomes included in the pangenome; this observation leads to the hypothesis that adding additional genomes would shrink the species core genome and that many of these environmentally accessory genes would prove to be absent from the species core calculated from a larger set of genomes. Pangenome accessory and singleton genes were mixed in status – some environmentally core and some environmentally accessory, reflecting the genetic diversity of microbial genomes that are present in the population.
Functional analysis
Metapangenomics presents an opportunity to analyze the gene-level basis of site specificity. In particular, it enables identification of genes that are core to a group of genomes at a specific oral site but absent from the genomes of closely related groups. Such genes may hold information associated with adaptation to that site. A complicating factor in this analysis is that the same or a similar function may be encoded by genes in different gene clusters. Therefore, we made use of a method developed to identify statistically significant enrichment or depletion of functions in one set of genomes compared to another [30].
We carried out functional enrichment analysis on all gene clusters (n = 5,036 gene clusters containing a total of 27,204 genes) from all genomes (n = 16) using three different annotation datasets (see Methods). Each gene cluster was associated with a function by annotating each gene in the gene cluster and assigning the consensus function to the gene cluster as a whole. Based on their distribution and prevalence across oral sites (Figure 2, SI Table S6, SI Table S7), the genomes of the three human oral species were defined, for purposes of this analysis, as associated with three distinct sites: G. sanguinis with tongue dorsum, G. haemolysans with buccal mucosa, and G. morbillorum with supragingival plaque. To determine which functions were not merely associated with but required for residence in the individual sites, tongue dorsum, buccal mucosa, or supragingival plaque, we applied the stringent criterion that a function must be present in all genomes abundant in that site and absent from the rest of the genomes (SI Table S6).
Following this procedure, we identified 31 functions that were unique to a specific oral site: TD (n = 8), BM (n = 15) and SUPP (n = 8). Unique functions in TD, associated with G. sanguinis, were linked to acid resistance, antibiotic synthesis, and virulence; unique functions in SUPP, associated with G. morbillorum, were linked primarily to DNA conjugation and transport; and unique functions in BM, associated with G. haemolysans, corresponded to riboflavin biosynthesis, antibiotic resistance, and adhesion/immune evasion. Within the G. haemolysans/BM group, we also found a gene encoding immunoglobulin A (IgA) metalloendopeptidase which cleaves the heavy and light chain of the IgA. This activity may be important in enabling G. haemolysans to avoid the immune response.
Among these functions unique to Gemella species at a specific oral site, the riboflavin (vitamin B2) biosynthesis pathway [39,40] stands out as a complete vitamin synthesis pathway present in all BM associated genomes (G. haemolysans) and absent in the rest of the genomes (G. morbillorum and G. sanguinis) (Figure 4). This pathway is comprised of seven enzymes that catalyze 10 steps leading to the production of riboflavin, as shown in Figure 4: ribBA, ribD, ycsE, ribH, ribE, ribF, and yigB. Additionally, a riboflavin transporter homologous to the one in Bacillus subtilis was found. RibBA, RibD, RibH and RibE are responsible for the key steps in riboflavin synthesis and are found only in G. haemolysans within a single biosynthetic operon (q < 0.05 in each case). YcsE and YigB, which belong to the haloacid dehalogenase-like phosphatase family, catalyze the removal of phosphate groups and are found in multiple copies across all genomes. RibF, also found in all genomes, is a bifunctional enzyme that phosphorylates and adenylates riboflavin to generate flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD). YigB is also involved in dephosphorylation of FMN to produce riboflavin. Finally, the riboflavin transporter FmnP was also identified in all Gemella genomes.
Figure 4.
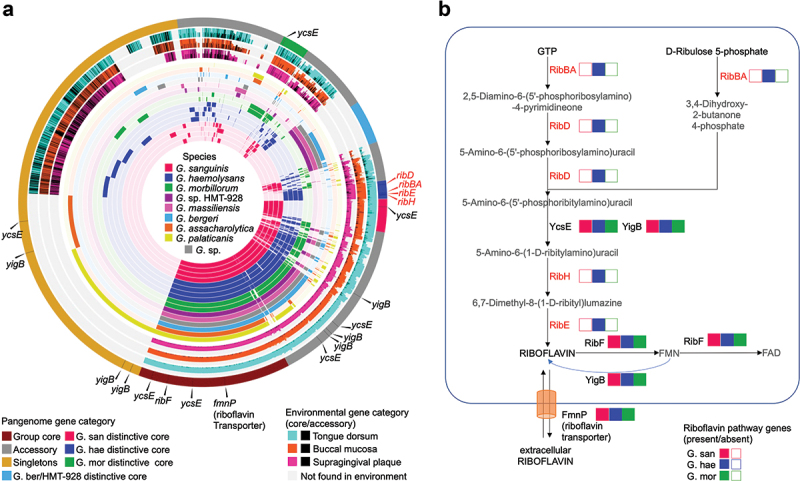
Human oral Gemella metapangenome and riboflavin biosynthetic pathway. (A) Metapangenome of oral Gemella constructed by metagenomic read recruitment from three oral sites (tongue dorsum, buccal mucosa and supragingival plaque) onto the pangenome gene-clusters from genomes of G. sanguinis, G. haemolysans, G. morbillorum, G. sp. HMT − 928, ‘G. massiliensis’, G. sp. 6198, G. bergeri, G. asaccharolytica, G. palaticanis. Starting from the innermost layer, the first 16 layers represent the pangenome depicting 5,036 gene clusters containing one or more genes from one or more genomes. Gene clusters are arranged by hierarchical clustering based on their presence or absence across all genomes. Genomes are color coded by species and hierarchically clustered based on gene cluster frequency, i.e. the number of representatives of each gene cluster present in each genome. The next three layers link gene clusters to the environment, represented by the ratio of environmentally core versus environmentally accessory genes, for supragingival plaque, buccal mucosa, and tongue dorsum, respectively. Environmentally core genes are color coded by the oral site they represent while environmentally accessory genes are shown in black when a genome passes the detection criterion in at least one sample; grey areas indicate genes not found in the environment when a genome fails the detection criterion in all samples. The outermost layer indicates the category of a gene-cluster within the pangenome (Group core, G. san distinctive core, G. hae distinctive core, G. mor distinctive core, G. sp. ber/HMT −928 distinctive core, accessory, and singletons). Riboflavin-pathway associated genes are indicated with a black line in the pangenome-category layer and pointed out by a black triangle. B) Riboflavin biosynthetic pathway genes in Gemella oral species. The image shows the enzymes, metabolites and transporters involved in the riboflavin pathway. Presence of genes associated with each enzyme is indicated with a colored box; empty boxes mean no gene found. The colors designate each oral Gemella species. The enzymes in the pathway are: ribBA, 3,4-dihydroxy 2-butanone 4-phosphate synthase/GTP cyclohydrolase II [EC:4.1.99.12 3.5.4.25]; ribD, diaminohydroxyphosphoribosylaminopyrimidine deaminase/5-amino −6-(5-phosphoribosylamino)uracil reductase [EC:3.5.4.26 1.1.1.193]; ycsE, 5-amino −6-(5-phospho-D-ribitylamino)uracil phosphatase [EC:3.1.3.104]; yigB, FMN and 5-amino −6-(5-phospho-D-ribitylamino)uracil phosphatase; ribH, 6,7-dimethyl −8-ribityllumazine synthase [EC:2.5.1.78]; ribE, riboflavin synthase [EC:2.5.1.9]; ribF, riboflavin kinase/FMN adenylyltransferase [EC:2.7.1.26 2.7.7.2]; fmnP, riboflavin transporter (fmnP – Bacillus subtilis).
A recent study evaluated the acquisition of proteins by horizontal transfer in the genus Gemella and concluded that genes encoding proteins core to a single species, but not to the whole genus, tended to have an unusual GC content [35]. We therefore evaluated the GC content of the RibBA, RibD, RibH, and RibE genes, which form a single operon. We found no significant difference in GC% of these genes compared to all genes in G. haemolysans genomes (32.9% ± 1.9% and 31.5% ± 5.5%, respectively). A BLAST search showed no significant similarity to any other bacterial taxon, although it did reveal high similarity (E-value = 2.5E−158 with 99% coverage on average) of two genes to a partial virus assembly from human metagenomes. While this finding is suggestive, it leaves the source of the operon as an open question.
Discussion
The power of metapangenomics lies in its combination of two orthologous classes of information – genomic and ecological [24,25,27]. The pangenome provides a grouping of genomes based on their gene content, irrespective of where the organisms containing those genes happen to live. In contrast, mapping of metagenomic short reads to the pangenome identifies which genes are prevalent in particular oral habitats irrespective of the genomes in which they are contained. Our results show that the grouping of Gemella genomes by their gene content corresponds well to established taxonomy, to phylogeny, and also to habitat specificity. Pangenome construction identified genomes that comprised distinct genomic groups within the Gemella genus and metagenomic mapping identified which of these genomes were present in the oral cavity and at which site.
The results obtained here are consistent with previous indications from 16S ribosomal RNA gene datasets that Gemella species were site-specialists [16,17], but whereas the 16S data provided only a small piece of a marker gene indicating the presence of a species, the metapangenomic approach provides a higher resolution view directly into the gene complement and functional capacity of the taxa. Using this approach, we have obtained results that support the site-specialist hypothesis [16] in that each taxon shows a preference for a subset of oral habitats. The three oral sites for which the most metagenome data were available, tongue dorsum, buccal mucosa, and supragingival plaque, represent three distinct habitats: keratinized mucosa, non-keratinized mucosa, and non-mucosal surfaces, respectively. Each of these habitats supports predominantly a single Gemella species – TD supports G. sanguinis, BM supports G. haemolysans, and SUPP supports G. morbillorum, consistent with the site-specialist hypothesis. However, each of these species can also be found at certain other oral sites. G. sanguinis genomes can be found in palatine tonsils and throat, G. haemolysans genomes in keratinized gingiva, and G. morbillorum genomes in subgingival plaque. The strong association of G. morbillorum with both supra- and subgingival plaque suggests that it is a specialist for a non-mucosal habitat. In contrast, G. haemolysans associates with both a non-keratinized mucosal surface (BM) and a keratinized mucosal surface (KG) suggesting that a mucosal surface is important but that keratinization itself is not the defining characteristic of the habitat conducive to G. haemolysans.
Although each of the major habitats, TD, BM and SUPP, supports a single species of Gemella that is dominant at that site, strains of these species can also be detected in a small fraction of subjects at sites where the species are not dominant. For example, G. haemolysans, dominant in BM, can be detected, albeit infrequently, in SUPP. There are several possible explanations for the detection of strains in sites where they are generally not dominant. One possibility is that they could represent cross-contamination during the sampling process. Another possibility is that different subpopulations of a strain could have the capacity to occupy a non-preferred site. Finally, a strain’s tropism need not be absolute; a strain that is dominant in one site could nevertheless exist as a minor component of other sites as well. Further research will be required to distinguish between these alternatives.
Microbial taxonomy is a challenging field that is increasingly being informed by whole-genome sequences. For the major Gemella species abundant in the healthy human microbiome – G. haemolysans, G. morbillorum, and G. sanguinis – existing species names are consonant with overall similarity at the nucleotide level (ANI), evolutionary relatedness estimated by phylogenomics, and gene content as shown in our pangenome. Thus, our analysis confirms that these species are well defined and well validated. By contrast, the genomes of G. sp. HMT−928, ‘G. massiliensis’, and G. sp. 6198 not only are highly similar to one another in gene content but also are nearly identical by ANI, with 99.5% to 99.7% pairwise identity. Therefore, these three genomes, although isolated from donors from three different geographic regions, represent the same species. The genome representing G. bergeri is close to this grouping in both gene content and ANI, with approximately 94.9% pairwise identity to each of the other three genomes and clustered tightly with them in the phylogenomic tree, and thus may be regarded as a sister taxon. Our findings are completely consistent with a recent ANI analysis of Gemella isolates in connection with virulence factors for opportunistic infections [35].
The failure of some Gemella genomes from the human microbiome (G. bergeri, G. sp. HMT−928, ‘G. massiliensis’, G. sp. 6198, and G. asaccharolytica) to recruit mapped reads from any of the oral sites may have several explanations. G. asaccharolytica recruited reads from supragingival plaque but did not satisfy our detection criterion; it may be present in dental plaque but at levels below the detection threshold. The HMP data were derived from healthy subjects; species that recruit few reads from these samples could be characteristic of diseased sites. However, it should be noted that the Gemella species that are characteristic of oral sites may also be found at other body sites in association with disease. For example, all three species of human oral Gemella have been isolated from endocarditis patients [41–43]. The HMP data were derived from nine specific oral sites; species that recruit few reads could be characteristic of an oral site not sampled. Finally, the primary habitat of these species might be elsewhere in the human body. For example, for G. asaccharolytica, our mapping data (not presented) suggests that its primary habitat is vaginal. Although further investigation is required to understand the basis for species adaptation to habitats in the oral environment, it is clear that individual species and strains are specialists for a small set of sites and that sub-specialization of a subset of strains to non-canonical habitats also occurs.
Analysis of the genomic differences between G. haemolysans, G. sanguinis and G. morbillorum provided gene-level insights into the basis for their site-specialization. Functional annotation identified a limited number of genes and functions that correlated with the site-specificity. One pathway, namely that of riboflavin synthesis, emerged from this analysis as a distinctive feature of G. haemolysans that was lacking in G. sanguinis and G. morbillorum. Riboflavin is an essential vitamin – required for life. In bacteria, riboflavin is the main precursor for the cofactors flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD) involved in redox metabolism. Organisms must either synthesize riboflavin themselves or obtain it from external sources [44]. The presence of the complete riboflavin biosynthetic pathway in G. haemolysans indicates that G. haemolysans is capable of making riboflavin for itself, which suggests that neither the buccal mucosa host tissue nor the other microbes living on buccal mucosa reliably supply riboflavin to G. haemolysans. In contrast, both G. sanguinis and G. morbillorum lack a functioning riboflavin biosynthetic pathway. However, G. sanguinis and G. morbillorum do contain a riboflavin transporter gene, consistent with the idea that these organisms secure their riboflavin primarily from the environment. Finally, the presence of all three species in YigB, which act on FMN when FMN levels are high to produce riboflavin, provides a pathway for riboflavin salvage for G. sanguinis and G. morbillorum. This pathway also provides G. haemolysans with redundancy in the production of riboflavin and FMN.
The riboflavin genes that encode the key steps in riboflavin synthesis (RibBA, RibD, RibH, and RibE) in G. haemolysans genomes are encoded in the same operon. This organization is similar to the ones found in Bacillus subtilis and some species of oral Streptococcus [45] and thus could have been acquired horizontally from one of these genera. However, neither an examination of GC content nor a BLAST search provided evidence for the source of any such horizontal transfer. The strong inference that can be drawn from our results is that G. haemolysans is self-reliant for making riboflavin, whereas G. sanguinis and G. morbillorum, lacking a synthetic pathway, must acquire riboflavin from external sources or excess FMN.
In addition, IgA metalloendopeptidase, considered a virulence factor, was found uniquely in G. haemolysans. This gene has been proposed to arise from a horizontal gene transfer event from a Streptococcus genome suggesting other genes might also have been acquired through a similar process [46]. However, its presence in all genomes suggests that this event is not recent. In a healthy human oral mucosa, it could favor adhesion to host cells.
The differences in biosynthetic requirements, immune evasion, and other physiological functions for Gemella species can be interpreted as an adaptation to their host habitat. Bacteria living on tongue dorsum or supragingival plaque grow in dense, complex biofilms. Bacteria in these communities are adjacent to many other bacteria and can obtain factors and metabolites from their neighbors. Presumably, G. sanguinis and G. morbillorum obtain riboflavin in this manner from the other microbes inhabiting these sites. In contrast, bacteria living on buccal epithelial cells often form single-layer biofilms and are more exposed. Thus, if G. haemolysans requires riboflavin, it may not be able to reliably get it from other members of the buccal mucosal community and so must synthesize it itself. Additionally, G. haemolysans because of its exposure on buccal mucosa may be more vulnerable to immune surveillance. By encoding an IgA metalloendopeptidase, it may mitigate the immune response. The potential relationship between the pathogenicity of Gemella and its oral tropisms is unclear. However, genes such as IgA metalloendopeptidase that play a role in their tropism and maintenance in the oral cavity could be harmful in the development of endocarditis.
In summary, using cultivar sequences and metagenomic data we have analyzed the genomes, gene content, and distribution of commensal species of the genus Gemella. We have shown that gene content, nucleotide-level similarity, and ecology all indicate a consistent assignment of genomes to species for the three abundant and prevalent species of this genus. Each of these species showed site-specialization to a different part of the mouth, while the detection of some genomes in lower abundance at alternate sites indicates the likely complexity of microhabitat characteristics underlying the overall site specificity. Analysis of the complement of functional genes revealed a biosynthetic pathway present in only one of the three major species and illustrates how metapangenomics can lead to the identification of genes associated with adaptation to an oral site. The extension of metapangenomic analysis to other oral taxa promises to provide further insights into the ecology of the oral microbiome.
Materials and Methods
Genomes
All Gemella genomes were obtained from the National Center for Biotechnology Information (NCBI). We used the prokaryotes genome browser from NCBI to retrieve the genus report (https://www.ncbi.nlm.nih.gov/genome/browse/#!/prokaryotes/Gemella) as of March 1, 2021. Then, using the browser links (Strain, BioSample and BioProject), we searched for information (isolation host, isolation site, RefSeq status, isolation/metagenome-assembled genome, type strain, Human Microbiome Project reference, association to disease and submitter) for all 35 deposited genomes. Additionally, we used the GenBank Assembly ID to cross-check the genomes with HOMD genomes and added the Human Microbiome Taxon ID when available for a genome. This information can be found in SI Table S1. We used an internal script to download the assembly files of NCBI RefSeq genomes (n = 30) and simplify the files and header names to an alphanumeric code.
Individual contigs-database
We used anvi’o [32] v7 as our analysis and visualization platform. The first step in working with anvi’o is building a contigs database (contigsDB). Each genome was processed individually. We ran the anvi’o script anvi-script-reformat-fasta to remove contigs of <300 nt and renamed the contigs’ deflines (definition line in headers) to ensure a common name (prefix) plus an increment number. Because sequences sometimes contained non-canonical nucleotide letters, we substituted N for any letter other than A, T, C, or G. A record file containing the changes in the deflines was produced for traceability. Then, we used anvi-gen-contigs-database to generate the contigsDB where contig information was stored (i.e. open reading frames, k-mer frequencies, GC content, genome length, and functional annotation). Open reading frames (ORF) were obtained using prodigal (v2.6.3) [47].
Annotation of ORFs in the contigs-database
ORFs from the contigsDB were annotated using programs built into anvi’o. We used the scripts anvi-run-hmms to find bacterial single copy genes (Bacteria71 SCG set) [48,49] and ribosomal RNA genes (Ribosomal RNAs set) [50] through hidden Markov Model (HMM) profiles; anvi-run-ncbi-cogs using blastp (v2.10.1+) to annotate with the cluster of orthologous genes (COGs) database (version COG20) [51]; and anvi-run-pfams and anvi-run-kegg-kofams with hmmscan from HMMER (v3.3.1) to functionally annotate with Pfams (v34.0) [52] and KOfams/KEGG Modules (97.0) [53–55], respectively.
Pangenome and ANI
We used anvi-gen-genomes-storage and anvi-pan-genome to generate the pangenome. Briefly, the first script generates a file indicating the location of the contigsDB and ensures the contigsDBs were built with the same parameters (i.e. the functional annotation was done using the same databases). The second script performed a local alignment of all amino acid sequences (blastp) [56], followed by the removal of weak matches between two sequences with the minbit parameter (minbit = 0.5). Then, sequences were clustered using the Markov Clustering Algorithm (mcl = 10) [57]. Amino acid sequences within each cluster were aligned with MUSCLE (v3.8.1551) [58]. Finally, hierarchical clustering was performed across gene clusters and genomes (distance = euclidean and linkage = ward). We estimated the average nucleotide identify (ANI) between genomes using anvi-compute-genome-similarity with the program pyANI (v0.2.10) with ANIb which used the blastn method [59].
Phylogenomic tree
To construct the phylogenomic tree, we selected 17 informative single-copy core genes from the pangenome. Briefly, we extracted genes that were present in each genome of the pangenome as a single copy (13,350 genes grouped into 445 gene clusters). Next, we performed multiple alignment (MUSCLE) on the amino acid sequences within each gene cluster. Then, we kept those gene clusters in which the aligned sequences had differing residues and no gaps using the functional and genometrical homogeneity indices, respectively. Finally, the resulting genes (n = 510; from 30 genomes) were concatenated, and the tree was built using IQ-TREE (v2.2’.0.3) [36] with the ModelFinder algorithm [60] and visualized with FigTree (v1.4.4) [61].
Concatenated contigs-database
We followed the anvi’o metapangenomic pipeline where reference genome fasta files (in this case, 30 genomes) are combined into a single concatenated file. The deflines were renamed, and a record file was made to link each contig to its original genome (deconstruction Table). Then, a unique contigsDB was generated using the same parameters as done for individual contigsDBs. The genes were annotated as described above.
Oral metagenomes
We obtained the manifest and metadata file from the HMP portal site (https://portal.hmpdacc.org/search/s?facetTab=cases) by selecting the oral sites (buccal mucosa, gingiva, dorsum of tongue, hard palate, palatine tonsil, throat and portion of saliva), Healthy Human Study (HHS), fastq files (FASTQ), and whole-genome sequencing (wgs_raw_seq_set). To download the files, we used the program portal client [62] with the manifest file and 20 retries. We used an in-house script to i) count the number of reads in each sample in parallel and in batch and ii) verify the oral site from where the sample was taken by comparing the metadata file with the deposited data in NCBI (BioSample) using E-utils [63]. Samples identified as gingiva in the metadata file consisted of three sites supragingival plaque (SUPP), subgingival plaque (SUBP) and keratinized gingiva (KG) when searched in NCBI. Date of download: February 10, 2020. We downloaded 1,275 samples that contained ~90 billion total reads. Quality filtering was performed on all samples using Minoche criteria [64,65] (iu-filter-quality-minoche) and kept any QC-sample with at least 2 million reads (1 million paired reads). A total of 1,215 samples containing ~50 billion quality-filtered reads met our criteria.
Competitive mapping
Individual metagenomic samples were mapped to the concatenated genome file using Bowtie2 (v2.4.2) [66] allowing each read to map to the single best-matching sequence across all genomes, for better discrimination between closely related genomes. The minimum alignment score was −0.6 + −0.6 * read length (average read length = 98.8 ± 7.2). The alignment options for seed substring were -D 20 -R 3 -N 0 -L 20 -i S,1,0.50 (–very-sensitive); where D is the number of tries for extending the seed, R is the number of tries for repetitive seeds, N is the number of mismatches in seed alignment, L is the length of the seed substrings, and i is the interval between seed substrings. Mapped reads were extracted, binarized and sorted with SAMtools (v1.11) [67]. On average 68% ± 9.8% of reads mapped with ≥95% percent identity to the target region.
Profiling
Using anvi-profile, individual mapping results (i.e. mean coverage, detection, and abundance) were linked to the contigsDB generating single-profile databases. Then, we combined single-profiles by oral site with anvi-merge. This produces a merged-profile database (mergedDB) for each of the nine oral sites.
Genome deconstruction of mergedDB and summary
Using the deconstruction table (see Concatenated contigs-database), we recovered the mapping properties of individual genomes from the mergedDB (anvi-import-collection). Multiple summary tables, including gene coverage, were produced per oral site (anvi-summarize).
Detection and prevalence of genomes
Breadth of genome coverage was defined as the proportion of nucleotides covered at least 1 ×. A genome was considered to be detected if the breadth of coverage was at least 0.5. The prevalence of genomes or strains was defined as the proportion of samples in which the genome or strain was detected. Each genome/strain was assigned to oral sites based on their prevalence.
Strain representation of genes in oral sites
Here, we combined coverage and detection at the gene level. On the one hand, gene-detection matrices from oral sites were merged by genome and used to generate a gene-detection database (geneDB). On the other hand, we used anvi-script-gen-distribution-of-genes-in-a-bin to analyze the distribution of genes in each environment (oral site) and classify them as environmental core or accessory genes, ECG and EAG, respectively [24]. If the total depth of coverage of a gene in all metagenomes is more than 0.25 multiplied by the median depth of coverage of all genes in all metagenomes, the gene is classified as ECG, otherwise, it is EAG. However, if the genome being analyzed is not detected (breadth of coverage <0.5) in any metagenome, the gene could not be analyzed and was classified as UNKNOWN.
Functional enrichment
We made use of anvi’o script anvi-compute-functional-enrichment to identify functional annotations that are differentially enriched or depleted in one set of genomes compared to another [30]. First, each genome was assigned to a group (oral site). Then, the script associated each gene cluster with the most frequently annotated function and generated a frequency table of functions across genomes. Finally, the enrichment test was done using a generalized linear model with a logit linkage function to obtain the enrichment score and a p-value. This analysis was performed for COG20, Pfams, and KEGG annotations independently, and the results were combined based on the gene cluster id.
Gene representation in the pangenome
We used anvi-meta-pan-genome to represent the ratio of environmentally core versus environmentally accessory genes for each genome across the pangenome for each habitat (oral site). Only genes present in the detected genomes (–min-detection = 0.5) were classified as environmentally common or accessory, A gene was considered common if the sum of its mean depth of coverage across samples from a site was equal to or greater than a quarter (–fraction-of-median-coverage = 0.25) of the median of the sums of coverages for all genes across samples from that site. The height of the ratio varies depending on the number of genomes that pass or fail the detection criterion in at least one sample for a given habitat [24].
Two-tail t-test
We performed a two-tail t-test (Welch’s t-test) with an ɑ = 0.05 to test (i) if Gemella species are non-randomly distributed across oral sites and (ii) to test whether the species show site preferences of at least 10× strength. We used the 50 samples with the largest number of reads from each site, assessed the relative abundance of each Gemella species, and calculated the mean and standard deviation of relative abundance in each site for all samples in which at least one species of Gemella was detected. Finally, we evaluate the site preference for each species by comparing its abundance in the preferred site to the non-preferred sites for our two hypotheses.
Supplementary Material
Acknowledgments
This work was supported by the NIH under Grants DE 030136 and DE 022586 and under Forsyth collaborative pilot project grant FSI_CP04. We thank Anthony McLean for critical insights in the analysis and Tabita Ramirez-Puebla for reading the manuscript. We thank A. Murat Eren and the anvi’o team for their assistance, consultations, and support in the use of anvi’o.
Funding Statement
The work was supported by the National Institute of Dental and Craniofacial Research [Grant DE 022586]; National Institute of Dental and Craniofacial Research [Grant DE 030136]; Forsyth collaborative pilot project [Grant FSI_CP04]
Disclosure statement
No potential conflict of interest was reported by the authors.
Author contributions
J.T.M., J.M.W., F.E.D., and G.G.B. designed the study; J.T.M. acquired the data and performed the analysis; G.G.B., J.T.M., and J.M.W. wrote the manuscript. All authors reviewed the manuscript and approved the final version.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/20002297.2023.2225261
References
- [1].Dewhirst FE, Chen T, Izard J, et al. The human oral microbiome. J Bacteriol. 2010;192(19):5002–15. doi: 10.1128/JB.00542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Collins MD, Hutson RA, Falsen E, et al. Description of Gemella sanguinis sp. nov., isolated from human clinical specimens. J Clin Microbiol. 1998;36(10):3090–3093. doi: 10.1128/JCM.36.10.3090-3093.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Collins MD, Hutson RA, Falsen E, et al. Gemella bergeriae sp. nov., isolated from human clinical specimens. J Clin Microbiol. 1998;36(5):1290–1293. doi: 10.1128/JCM.36.5.1290-1293.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Stackebrandt E, Wittek B, Seewaldt E, et al. Physiological, biochemical and phylogenetic studies on Gemella haemolysans. FEMS Microbiol Lett. 1982;13(4):361–365. doi: 10.1111/j.1574-6968.1982.tb08288.x [DOI] [Google Scholar]
- [5].Kilpper-Balz R, Schleifer KH.. Transfer of streptococcus morbillorum to the genus gemella as Gemella morbillorum comb. nov. Int J Syst Bacteriol. 1988;38(4):442–443. doi: 10.1099/00207713-38-4-442 [DOI] [Google Scholar]
- [6].Thjotta T, Boe J. Neisseria haemolysans. A haemolytic species of Neisseria trevisan. Acta Path Microbiol Scand. 1938;37:527–531. [Google Scholar]
- [7].Berger U. A proposed new genus of gram-negative cocci: gemella. Int Bull Bacteriol Nomencl Taxon. 1961;11(1):17–19. doi: 10.1099/0096266X-11-1-17 [DOI] [Google Scholar]
- [8].Ruoff KL. Miscellaneous catalase-negative, Gram-positive cocci: emerging opportunists. J Clin Microbiol. 2002;40(4):1129–1133. doi: 10.1128/JCM.40.4.1129-1133.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chen T, Yu WH, Izard J, et al. The human oral microbiome database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford). 2010;2010:1–10. doi: 10.1093/database/baq013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Escapa IF, Chen T, Huang Y, et al. New insights into human nostril microbiome from the expanded human oral microbiome database (eHOMD): a resource for the microbiome of the human aerodigestive tract. Msystems. 2018;3(6):1–20.9. doi: 10.1128/mSystems.00187-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Parte AC, Sardà Carbasse J, Meier-Kolthoff JP, et al. List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol. 2020;70(11):5607–5612. doi: 10.1099/ijsem.0.004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hoyles L, Foster G, Falsen E, et al. Characterization of a Gemella-like organism isolated from an abscess of a rabbit: description of Gemella cuniculi sp. nov. Int J Syst Evol Microbiol. 2000;50:2037–2041. doi: 10.1099/00207713-50-6-2037 [DOI] [PubMed] [Google Scholar]
- [13].Ulger-Toprak N, Summanen PH, Liu C, et al. Gemella asaccharolytica sp. nov., isolated from human clinical specimens. Int J Syst Evol Microbiol. 2010;60(5):1023–1026. doi: 10.1099/ijs.0.001966-0 [DOI] [PubMed] [Google Scholar]
- [14].Hung WC, Chen HJ, Tsai JC, et al. Gemella parahaemolysans sp. nov. and Gemella taiwanensis sp. nov., isolated from human clinical specimens. Int J Syst Evol Microbiol. 2014;64(Pt_6):2060–2065. doi: 10.1099/ijs.0.052795-0 [DOI] [PubMed] [Google Scholar]
- [15].Fonkou MD, Bilen M, Cadoret F, et al. Gemella massiliensis’ sp. nov., new bacterial species isolated from the human respiratory microbiome. New Microbe And New Infect. 2017;22:37–43. doi: 10.1016/j.nmni.2017.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mark Welch JL, Dewhirst FE, Borisy GG. Biogeography of the oral microbiome: the site-specialist hypothesis. Annu Rev Microbiol. 2019;73(1):335–358. doi: 10.1146/annurev-micro-090817-062503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Eren AM, Borisy GG, Huse SM, et al. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci, USA. 2014;111(28):E2875–84. doi: 10.1073/pnas.1409644111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Socransky SS, Manganiello SD. The oral microbiota of man from birth to senility. J Periodontol. 1971;42(8):485–496. doi: 10.1902/jop.1971.42.8.485 [DOI] [PubMed] [Google Scholar]
- [19].Tettelin H, Masignani V, Cieslewicz MJ, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci, USA. 2005;102(39):13950–13955.doi: 10.1073/pnas.0506758102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Vernikos G, Medini D, Riley DR, et al. Ten Years of pan-genome analyses. Curr Opin Microbiol. 2015;23:148–154. doi: 10.1016/j.mib.2014.11.016 [DOI] [PubMed] [Google Scholar]
- [21].Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68(4):669–685. doi: 10.1128/MMBR.68.4.669-685.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Quince C, Walker AW, Simpson JT, et al. Shotgun metagenomics, from sampling to analysis. Nature Biotechnol. 2017;35(9):833–844. doi: 10.1038/nbt.3935 [DOI] [PubMed] [Google Scholar]
- [23].Human Microbiome Project Consortium . Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402): 207–214. doi: 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Delmont TO, Eren AM. Linking pangenomes and metagenomes: the prochlorococcus metapangenome. PeerJ. 2018;6:e4320. doi: 10.7717/peerj.4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Nayfach S, Rodriguez-Mueller B, Garud N, et al. An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res. 2016;26(11):1612–1625. doi: 10.1101/gr.201863.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kraal L, Abubucker S, Kota K, et al. The prevalence of species and strains in the human microbiome: a resource for experimental efforts. PLoS ONE. 2014;9(5):e97279. doi: 10.1371/journal.pone.0097279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lloyd-Price J, Mahurkar A, Rahnavard G, et al. Strains, functions and dynamics in the expanded human microbiome project. Nature. 2017;550(7674):61. doi: 10.1038/nature23889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Truong DT, Tett A, Pasolli E, et al. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res. 2017;27(4):626–638. doi: 10.1101/gr.216242.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Donati C, Zolfo M, Albanese D, et al. Uncovering oral Neisseria tropism and persistence using metagenomic sequencing. Nat Microbiol. 2016;1(7):16070. doi: 10.1038/nmicrobiol.2016.70 [DOI] [PubMed] [Google Scholar]
- [30].Shaiber A, Willis AD, Delmont TO, et al. Functional and genetic markers of niche partitioning among enigmatic members of the human oral microbiome. 2020. Genome Biol. 2020;21(1):292. doi: 10.1186/s13059-020-02195-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Utter DR, Borisy GG, Eren AM, et al. Metapangenomics of the oral microbiome provides insights into habitat adaptation and cultivar diversity. Genome Bio. 2020;21(1):293. doi: 10.1186/s13059-020-02200-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Eren AM, Öc E, Quince C, et al. Anvi’o: an advanced analysis and visualization platform for ‘Omics Data. Peer J. 2015;3:e1319. doi: 10.7717/peerj.1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci, USA. 2005;102(7):2567–2572. doi: 10.1073/pnas.0409727102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kim M, Oh HS, Park SC, et al. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64(Pt_2):346–351. doi: 10.1099/ijs.0.059774-0 [DOI] [PubMed] [Google Scholar]
- [35].García López E, Martín-Galiano AJ. The versatility of opportunistic infections caused by gemella isolates is supported by the carriage of virulence factors from multiple origins. Front Microbiol. 2020;11(March):1–16. doi: 10.3389/fmicb.2020.00524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Nguyen L-T, Schmidt HA, von Haeseler A, et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Reyn A, Birch-Andersen A, Berger U. Fine structure and taxonomic position of Neisseria haemolysans (Thjotta and Boe 1938) or Gemella haemolysans (Berger 1960). Acta Pathol Microbiol Scand B Microbiol Immunol. 1970;78(3):375–389. doi: 10.1111/j.1699-0463.1970.tb04317.x [DOI] [PubMed] [Google Scholar]
- [38].Olm MR, Crits-Christoph A, Bouma-Gregson K, et al. In Strain profiles population microdiversity from metagenomic data and sensitively detects shared microbial strains. Nature Biotechnol. 2021;39(6):727–736. doi: 10.1038/s41587-020-00797-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bacher A, Eberhardt S, Fischer M, et al. Biosynthesis of vitamin b2 (riboflavin). Annu Rev Nutr. 2000;20:153–167. doi: 10.1146/annurev.nutr.20.1.153 [DOI] [PubMed] [Google Scholar]
- [40].Rodionov DA, Arzamasov AA, Koroshkin MS, et al. Micronutrient requirements and sharing capabilities of the human gut microbiome. Front Microbiol. 2019;10. doi: 10.3389/fmicb.2019.01316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sideris AC, Zimmermann E, Ogami T, et al. A rare case of isolated mitral valve endocarditis by Gemella sanguinis: case report and review of the literature. Int J Surg Case Rep. 2020. [Epub 2020 Mar 7];69:51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Agrawal T, Irani M, Fuentes Rojas S, et al. A rare case of infective endocarditis caused by Gemella haemolysans. Cureus. 2019. Nov 26;11(11):e6234. doi: 10.7759/cureus.6234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Shahani L. Gemella morbillorum prosthetic aortic valve endocarditis.BMJ Case Rep. 2014. [Nov 18; 2014];2014(nov18 1):bcr2014207304. doi: 10.1136/bcr-2014-207304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].García-Angulo VA. Overlapping riboflavin supply pathways in bacteria. Crit Rev Microbiol. 2017;43(2):196–209. doi: 10.1080/1040841X.2016.1192578 [DOI] [PubMed] [Google Scholar]
- [45].Vitreschak AG, Rodionov DA, Mironov AA, et al. Evidence for an ABC-type riboflavin transporter system in pathogenic spirochetes. Nucleic Acid Research. 2002;30(14):3141–3151. doi: 10.1093/nar/gkf433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Takenouchi-Ohkubu N, Mortensen LM, Drasbek KR, et al. Horizontal transfer of the immunoglobulin A1 protease gene (iga) from Streptococcus to Gemella haemolysans. Microbiology. 2006;152(7):2171–2180. doi: 10.1099/mic.0.28801-0 [DOI] [PubMed] [Google Scholar]
- [47].Hyatt D, Chen GL, LoCascio PF, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 2010;11:119. doi: 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lee MD, Ponty Y. GToTree: a user-friendly workflow for phylogenomics. Bioinformatics. 2019. Oct 15;35(20):4162–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hug LA, Baker BJ, Anantharaman K, et al. A new view of the tree of life. Nat Microbiol. 2016. Apr 11;1(5):16048. [DOI] [PubMed] [Google Scholar]
- [50].Seemann T. https://github.com/tseemann/barrnap/blob/master/README.md#barrnap
- [51].Galperin MY, Wolf YI, Makarova KS, et al. COG database update: focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res. 2021. [2021 Jan 8];49(D1):D274–D281. doi: 10.1093/nar/gkaa1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Mistry J, Chuguransky S, Williams L, et al. Pfam: the protein families database in 2021. Nucleic Acids Res. 2020;49(D1). doi: 10.1093/nar/gkaa913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kanehisa M, Furumichi M, Sato Y, et al. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–D551. doi: 10.1093/nar/gkaa970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Camacho C, Coulouris G, Avagyan V, et al. BLAST+: architecture and applications. BMC Bioinf. 2009;10:421. doi: 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002. 1 April 2002;30(7):1575–1584. 10.1093/nar/30.7.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 2004;5:113. doi: 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Pritchard L, Glover RH, Humphris S, et al. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods. 2016;8:12–24. doi: 10.1039/C5AY02550H [DOI] [Google Scholar]
- [60].Kalyaanamoorthy S, Minh BQ, Wong TKF, et al. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods. 2017;14(6):587–589. doi: 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Rambaut A. https://github.com/rambaut/figtree
- [62].IGS . https://github.com/IGS/portal_client
- [63].Entrez Programming Utilities Help [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010. https://www.ncbi.nlm.nih.gov/books/NBK25501/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Minoche AE, Dohm JC, Himmelbauer H. Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and genome analyzer systems. Genome Biol. 2011;12(11):R112. doi: 10.1186/gb-2011-12-11-r112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Eren AM, Vineis JH, Morrison HG, et al. A Filtering Method to Generate High Quality Short Reads Using Illumina Paired-End Technology. PLoS ONE. 2013;8(6):e66643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. 2012. Nature Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Danecek BP, Liddle J, Marshall J, et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021. February;10(2):giab008. doi: 10.1093/gigascience/giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


