Abstract
Background: Helicobacter pylori (H.pylori) infection is an important factor in the occurrence of human gastric diseases, but its pathogenic mechanism is not clear. N6-methyladenosine (m6A) is the most prevalent reversible methylation modification in mammalian RNA and it plays a crucial role in controlling many biological processes. However, there are no studies reported that whether H. pylori infection impacts the m6A methylation of stomach. In this study, we measured the overall level changes of m6A methylation of RNA under H. pylori infection through in vitro and in vivo experiment.
Methods: The total quantity of m6A was quantified in gastric tissues of clinical patients and C57 mice with H. pylori infection, as well as acute infection model [H. pylori and GES-1 cells were cocultured for 48 h at a multiplicity of infection (MOI) from of 10:1 to 50:1]. Furthermore, we performed m6A methylation sequencing and RNA-sequencing on the cell model and RNA-sequencing on animal model.
Results: Quantitative detection of RNA methylation showed that H. pylori infection group had higher m6A modification level. M6A methylation sequencing identified 2,107 significantly changed m6A methylation peaks, including 1,565 upregulated peaks and 542 downregulated peaks. A total of 2,487 mRNA was upregulated and 1,029 mRNA was downregulated. According to the comprehensive analysis of MeRIP-seq and RNA-seq, we identified 200 hypermethylation and upregulation, 129 hypermethylation but downregulation, 19 hypomethylation and downregulation and 106 hypomethylation but upregulation genes. The GO and KEGG pathway analysis of these differential methylation and regulatory genes revealed a wide range of biological functions. Moreover, combining with mice RNA-seq results, qRT- PCR showed that m6A regulators, METTL3, WTAP, FTO and ALKBH5, has significant difference; Two key genes, PTPN14 and ADAMTS1, had significant difference by qRT- PCR.
Conclusion: These findings provide a basis for further investigation of the role of m6A methylation modification in H. pylori-associated gastritis.
Keywords: gastritis, Helicobacter pylori, MeRIP-seq, M6A, N6-methyladenosine
Introduction
Helicobacter pylori (H. pylori), a Gram-negative microaerobic bacterium, is closely related to diseases such as gastritis, peptic ulcer and chronic gastritis (Cover and Blaser, 2009; Asano et al., 2016). It can initiate gastric carcinogenesis following the Correa cascade (Correa and Piazuelo, 2012). Once atrophy and intestinal metaplasia occur, there is still a lack of effective therapy to reverse the pathological changes, and some patients still progress to gastric cancer. Therefore, it is of great clinical significance to further explore the molecular mechanism of gastric diseases caused by H. pylori infection and find new intervention strategies and targets.
N6-Methyladenosine (m6A), involving methylation at the N6 position of RNA adenine, is the most prevalent RNA modification in eukaryotes (Meyer and Jaffrey, 2014; Huang et al., 2020a). In the 1970s, a study reported that there has m6A modification in mRNA and non-coding RNA of eucaryon (Desrosiers et al., 1974).
M6A is the most prevalent post-transcriptional modification of mRNAs and non-coding RNAs, which determines RNA fate, such as splicing, localization, stabilization, translation efficiency and nuclear export (Guo et al., 2021; Li et al., 2022a; Zhang et al., 2022). Recent years, more and more studies have reported that m6A plays different role during the growth and development of mammals, including embryonic development, circadian rhythm, neurogenesis, stress responses, sex determination and tumorigenesis (Pan et al., 2018; Chokkalla et al., 2020; Jiang et al., 2021; Xiao et al., 2022a). M6A modification mainly involves three enzymes, a family methyltransferase enzymes (writers), which including Methyltransferase Like 3 (METTL3), Methyltransferase Like 14 (METTL14), WT1 Associated Protein (WTAP) and et all, catalyze addition of m6A (Jiang et al., 2021) (Sun et al., 2022) (Sacco et al., 2022). The demethylase enzymes (erasers) that catalyze removal of m6A, such as alpha-ketoglutarate-dependent dioxygenase AlkB homolog 5 (ALKBH5) and fat mass and obesity-associated protein (FTO) (Roignant and Soller, 2017; Jiang et al., 2021). The m6A reader proteins can recognize the m6A-modified RNAs, which are divided into different protein families, such as IGF2 mRNA binding proteins (IGF2BP1/2/3) families, eukaryotic initiation factor (eIF) 3, the proteins contain the YT521-B homology (YTH) domain (YTHDF1/2/3 and YTHDC1/2) and et all (Jiang et al., 2021) (Shi et al., 2018) (Zhou et al., 2022). It is now clear that this reversible post-transcriptional modification is essential for gene regulation.
At present, research on stomach-related diseases m6A is mainly in gastric carcinoma and rarely in non-cancer disease. The role of m6A RNA modifications in diseases associated with H. pylori infection has not been investigated. In this study, we used high-throughput sequencing (MeRIP-seq) to identify the potential m6A modification of inflammation in gastric epithelial cells (GES-1) treated with H. pylori. Differential methylation genes (DMG), differential expression genes (DEG) and differential methylation and expression genes (DMEG) were analyzed by gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways to reveal the biological significance of genomes. In addition, combining with mice RNA-seq data, we used qRT- PCR tests to observe the expression of five common m6A regulatory and the three key gene, which were consistent in the sequencing results of cell model and animal model. These findings may provide new insights into the molecular mechanisms involved in H. pylori infection.
Materials and methods
Bacterial strains and cell lines
H. pylori was isolated from the gastric mucosa of gastric ulcer patient during gastroscopy as described (Xia et al., 2020). It was cultured in Columbia agar containing 10% sheep blood (Nanjing bianzhen Biological Technology Co., LTD., China) and antibiotics (5 mg/L cefsulodin, 5 mg/L amphotericin B, 5 mg/L trimethoprim, 10 mg/L vancomycin) (Oxoid, United Kingdom) at 37°C under microaerophilic conditions (5% O2, 10% CO2, and 85% N2) for 3–5 days. When the value of OD600 was 1, the bacterial concentration was approximately 2 × 108 CFU/mL.
GES-1 cells were obtained from Hybribio Biotech Ltd. (Guangdong, China). The GES-1 cells were cultured in RPMI-1640 medium (Gibco, United States), containing 10% fetal bovine serum (Biological Industries, Israel) and maintained at 37°C in humidified 5% CO2 incubator.
Clinical specimens
Four H. pylori-positive and four H. pylori-negative gastric tissues were collected from patients who underwent gastric biopsies at the Xiangya Third Hospital, Central South University (Changsha, China). The diagnoses were based on clinical and histological laboratory examination. All patients had signed informed consent for the study. The clinical information of patients was shown in Supplementary Table S1. This study was approved by the Ethics Committees of the Xiangya Third Hospital, Central South University.
H. pylori -infected animal model
Four to five weeks old male C57 BL/6 (18–22 g) were used. All the experimental animals were foster in the Department of Laboratory Animal Science of Central South University and were housed in an experimental animal room, which meets the specific pathogen-free (SPF)-class Meets the SPF standard, to ensure an environment with 12 h of light and 12 h of darkness. The eight mice were divided into two groups: control group (n = 4) and H. pylori infection group (n = 4). The mice were orally gavaged with 0.3 mL H. pylori suspension in phosphate buffered saline (PBS) (1 × 109) once daily for 9 days (repeat three times with 1 day off for three consecutive days) according to our previous study (Xia et al., 2020). The mice were only gavaged with sterile PBS in control group. The mice were sacrificed by cervical dislocation under CO2 narcosis at 2 weeks after last gavage. Rapid urease test (RUT) and Giemsa staining were used to verify whether mice were infected with H. pylori (Supplementary Figure S1).
Cell infection model
GES-1 cells were seeded in 6-well plates until the density reached 60%–80% (∼3 × 105) without H. pylori intervention and the cell culture medium containing no antibiotics. H. pylori was collected and re-suspended into antibiotic-free cell culture medium. The concentration of H. pylori suspension was adjusted to 1× 109/mL. Then, H. pylori suspension was added to GES-1 cells at a MOI of 10:1–50:1 and incubated for 48 h.
RNA extraction and qRT-PCR
Total RNA in tissues was extracted by the TRIzol reagent (Invitrogen, United States). Moreover, the extracted total RNA dissolved in RNase/DNase-free water. The ReverTra Ace qPCR RT Master Mix with gDNA Remover (Vazyme Biotech Co., Ltd., China) was used to reverse transcribe RNA in accordance with the manual. Primers for qRT-PCR were listed in Supplementary Table S2.
Quantification of the m6A modification
Total RNA was isolated as above. The quality of RNA was analyzed using a NanoDrop1000 (Thermo Fisher, United States). The EpiQuik m6A Methylation Quantification Kit (Epigentek, P-9005-96, United States) was used to measure the global m6A enrichment of mRNA. 200 ng RNA was coated in assay wells from each sample. The m6A levels are colorimetrical quantified at a wavelength of 450 nm absorbance.
RNA-seq and m6A-RNA immunoprecipitation sequencing
Total RNA was isolated from GES-1 cells and gastric tissue of mouse by TRIzol reagent as above. The Poly (A) RNA was purified from 50 µg total RNA using Dynabeads Oligo (dT) (Thermo Fisher, Carlsbad CA, United States) and two rounds of purification were used. Next, a Magnesium RNA Fragmentation Module was used to fragment the captured mRNA at 86°C for 7 min. Cleaved RNA fragments were incubated with m6A-specific antibody (Synaptic Systems GmbH, Goettingen, Germany) for 2 h at 4°C in IP buffer which was consist of 750 mM NaCl, 50 mM Tris-HCl and 0.5% Igepal CA-630. After performing IP, the IP product was synthesized into cDNA using reverse tran-scriptase (Invitrogen SuperScript™ II Reverse Transcriptase, CA, United States). Escherichia coli DNA polymerase I (NEB, United States), RNase H (NEB, United States), and dUTP Solution (Thermo Fisher, United States) which assisted the synthesis of the double-stranded DNA and the ends of the double-stranded DNA were repaired to form blunt ends. The two strands were digested with the enzyme UDG (NEB, United States) after adding an A base to both blunt ends and using magnetic beads to screen and purify the fragments according to size. Through PCR experiment, a library with a fragment size of 300 ± 50 bp was established (Supplementary Table S2). Finally, an Illumina NovaSeq™ 6000 (LC- Bio Technology Co., Ltd., Hangzhou, China) was used to sequencing with PE150 (2 bp × 150 bp paired-end) sequencing mode.
Bioinformatics analysis
Fastp (https://github.com/OpenGene/fastp) was used for quality control on the original data and acquire clean data. HISAT2 package (http://daehwankim lab.github.io/hisat2) was used to compare the acquired clean data to the genome (human genome, version: hg19; and mus musculus genome, version: GRCm38). The R package exome-Peak (https://bioconductor.org/packages/exome Peak) was used to perform peak calling analysis and peak analysis of genetic difference. The IGV software (http://www.igv.org) visualized the results. HOMER (http://homer.ucsd.edu/homer/motif) and MEME2 (http://meme-suite.org) were used for motif analysis. StringTie (https://ccb.jhu.edu/softw are/stringtie) was used to determine the expression levels of all mRNAs in the input libraries. The different expression of mRNAs was selected according to thresholds of a p-value < 0.05 and a |log2 (fold change)| >1 with the R package edgeR (https://bioconduct or.org/packages/edgeR).
Statistical analysis
SPSS 22.0 and GraphPad Prism 7.0 were used for data processing. The t-test and χ2 test were used to analyze the differences among different samples. A p-value less than 0.05 was considered to indicate statistical significance (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Results
Establishment of H. pylori infection model in vivo and in vitro
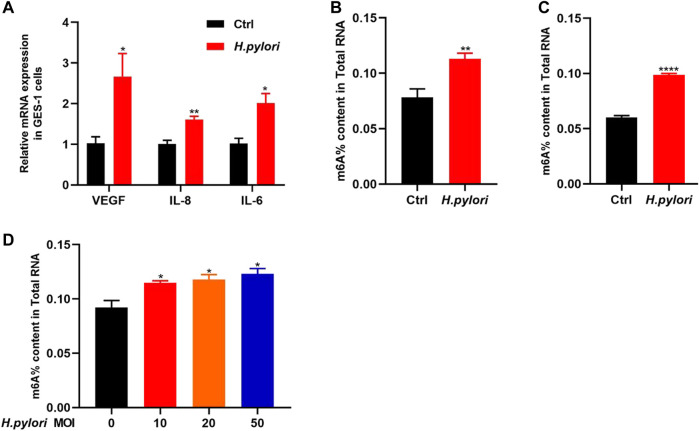
In this study, GES-1 cells were treated with H. pylori with a MOI of 10:1 for 48 h. We detected the mRNA expression of VEGF, IL-6 and IL-8 by qRT-PCR (Figure 1A). The results show that the expression levels of these proinflammatory factors were significantly increased in the GES-1 cells treated with H. pylori (p < 0.05). Next, we observed significant increase in the overall level of m6A methylation in H. pylori-infected patients and mice (Figures 1B, C), and mild significant increase in H. pylori-infected cells (Figure 1D).
FIGURE 1.
Establishment of H. pylori infection model and determination of total m6A (A) The RNA-level expression of inflammatory factors IL-8, IL-6, and VEGF after H. pylori infection of GES-1 cell. (B) The total m6A content in H. pylori negative and positive patients (n = 4). (C) The total m6A content in H. pylori negative and positive animal (n = 4). (D) The total m6A content in H. pylori-uninfected and H. pylori-infected cells in different MOI (n = 3). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Overview of methylation RNA immunoprecipitation sequencing
In the MeRIP-seq library, the two sets of samples obtained an average of 41,015,657 and 44,650,725 valid reads, while in the RNA-seq library the two groups obtained an average of 37,346,096 and 40,732,363 valid reads (Supplementary Table S3). Among the IP samples, the average matching rate of valid reads in the control group and H. pylori group was 97.2% and 97.1%, respectively. The mean matching rates for valid reads in the input samples were 97.5% and 97.7% (Supplementary Table S4). Clean reads that can be matched to the reference genome are defined as exons, introns and intergenic sequences according to the regional information of the reference genome. The mean rates of IP and exons in the input samples were 97.39% and 97.44% for the control group and 96.1% and 96.86% for the H. pylori-infected group, respectively (Supplementary Figure S2).
Profile of the m6A modification in GES-1 cells treated with H. pylori
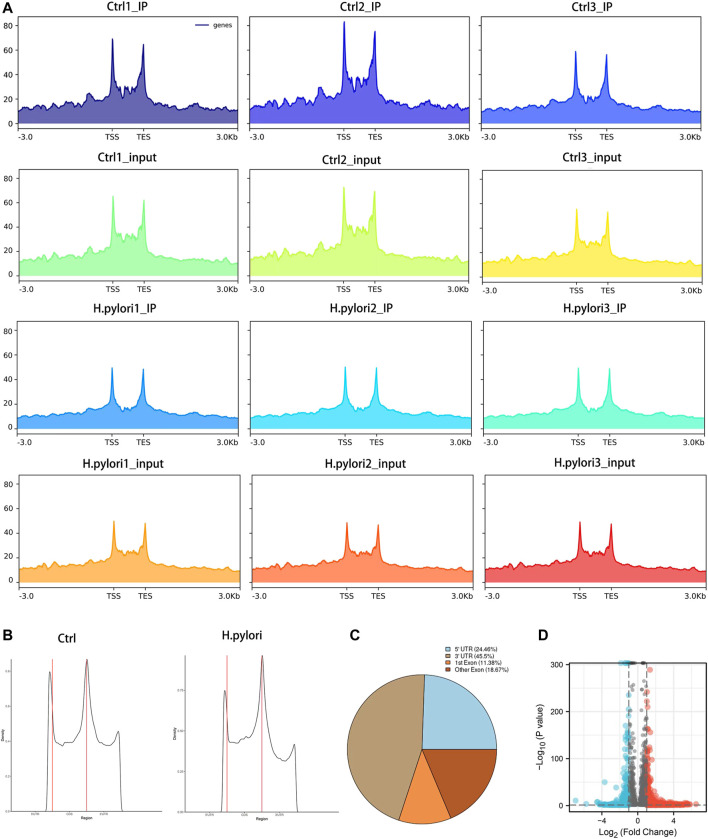
To obtain a map of m6A modifications in gastric epithelial cells infected with H. pylori, we used meRIP-seq to performed a transcriptome analysis of m6A modification. Combining all the peak reads, we found that the enrichment of reads was located near the transcription start site (TSS) and the transcription end site (TES) (Figure 2A). To further understand the distribution of the differential peak on the functional elements of the gene, we divided it into three regions: the 5′ untranslated region (5′ UTR), the first exon, the other exons and the 3′ UTR (Figure 2B). Meanwhile, we analyzed the distribution pattern of differential m6A methylation peaks. A total of 24.46% of the m6A methylation peaks were contained in the 5′UTR, 45.5% were enriched in the 3′ UTR, while 11.38% were enriched in the exons (Figure 2C). Under the screening conditions of |log2 (fold change)| > 1 and p-value < 0.05, a total of 9,097 peaks were identified in both groups. The results showed 2,107 significantly different peaks compared to the control group, of which 1,565 peaks were upregulated and 542 peaks were downregulated (Figure 2D). The top 20 distinct m6A methylation peaks are shown in Supplementary Table S5.
FIGURE 2.
Significant dysregulation of the m6A peak during H. pylori infection of gastric epithelial cells. (A) Enrichment of peaks near the gene transcription start site. (B,C) Distribution of differentially methylated m6A peaks in control and H. pylori groups (D) Volcano plot of genes with differential m6A peaks (|log2(FC)|>1 and p-value < 0.05).
Differential m6A modification is involved in important biological pathways
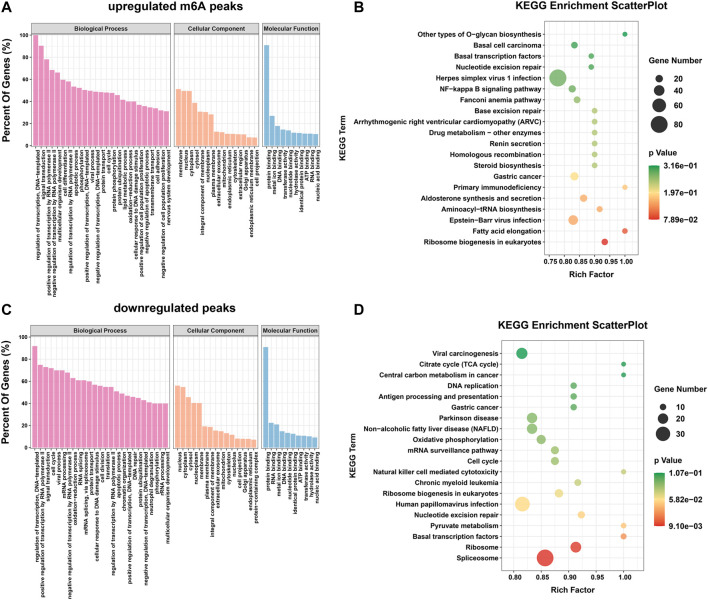
To explore the important functions of m6A modification in H. pylori-induced gastric epithelial cells, GO and KEGG enrichment analyses were performed for the above m6A differential peak (DMG) genes. The GO results were classified into three categories: cellular component (CC) and biological process (BP) and molecular function (MF) categories. It can be observed that both hypermethylated and hypomethylated genes are associated with “regulation of transcription, DNA template,” “signal transduction,” “apoptotic process,” " regulation of transcription by RNA polymerase II,” “cell cycle” and “RNA splicing” (ontology: biological processes); “nucleus,” " membrane,” “cytoplasm” and “cytoplasm” (ontology: cellular components); and “protein binding,” “RNA binding,” “metal ion binding” (ontology: molecular function) (Figures 3A, C). In addition, the results of the KEGG signaling pathway analysis showed that the genes upregulated by the m6A peak were mainly enriched in “fatty acid elongation,” “primary immunodeficiency,” “Epstein-Barr virus infection,” “drug metabolism-other enzymes,” “NF-κB signaling pathway,” and “basic transcription factors” (Figure 3B); The genes downregulated by the m6A peak were mainly concentrated in “natural killer cell mediated cytotoxicity,” “basal transcription factors,” “cell cycle,” “mRNA surveillance pathway,” and “pyruvate metabolism” (Figure 3D).
FIGURE 3.
Differential m6A modifications are involved in important biological pathways. (A,B) GO enrichment and KEGG pathway analysis of the hypermethylated peaks. (C,D) GO enrichment and KEGG pathway analysis of the hypomethylated peaks.
Analysis of RNA-seq differential expression genes
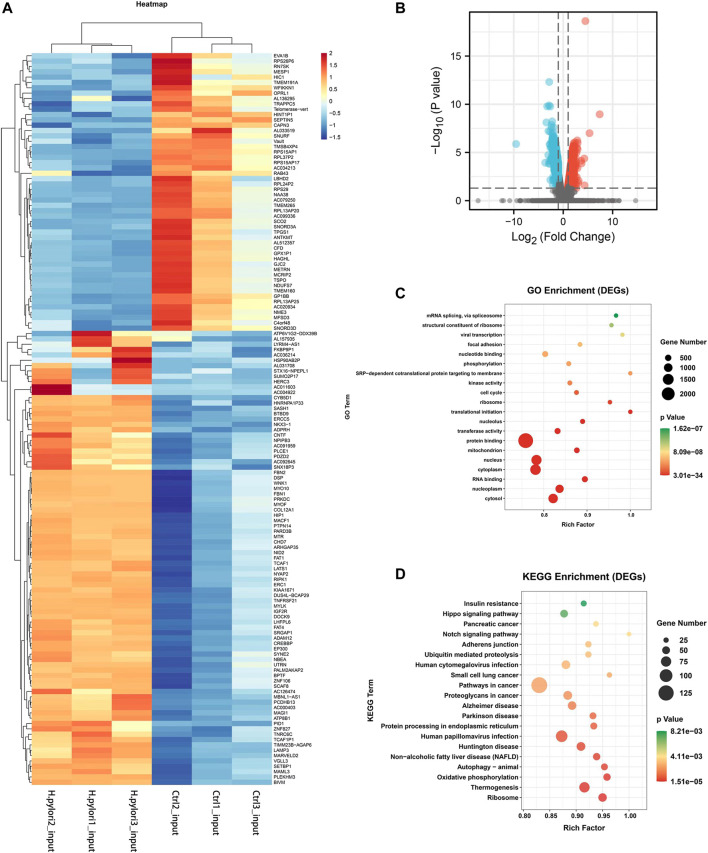
To explore the potential relationship between m6A modifications and gene expression, differential expression gene analysis was performed using input sequencing data. By hierarchical clustering of RNA-seq data, we detected significantly different expression between the control and H. pylori groups (Figure 4A). We then screened the RNA-seq database for a total of 3,516 differential genes (|log2(FC)|>1 and p-value < 0.05) compared to control samples. Among them, 2,487 upregulated genes and 1,029 downregulated genes were identified (Figure 4B). These differential expression genes were then used for GO enrichment and KEGG pathway analysis. GO enrichment results showed these genes were significantly related to “translation initiation,” “SRP-dependent cotranslation protein targeting membranes,” “viral transcription” and “mRNA splicing, via spliceosomes” (Figure 4C). KEGG analysis showed that these genes were mainly enriched in “Notch signaling pathway,” “adherens junctions,” “Hippo signaling pathway,” “protein processing in endoplasmic reticulum,” “ubiquitin mediated proteolysis” and “oxidative phosphorylation” (Figure 4D).
FIGURE 4.
Differentially expressed gene analysis by RNA-Seq (A) Heat map showing differentially expressed mRNAs in three H. pylori input samples and three control input samples. (B) Volcano plot showing the differentially expressed mRNAs between H. pylori and control groups with statistical significance (fold change ≥ 2.0 and p < 0.05). (C) GO enrichment analysis of differential genes. (D) KEGG pathway analysis of differential genes.
Combined analysis between m6A-seq and RNA-seq
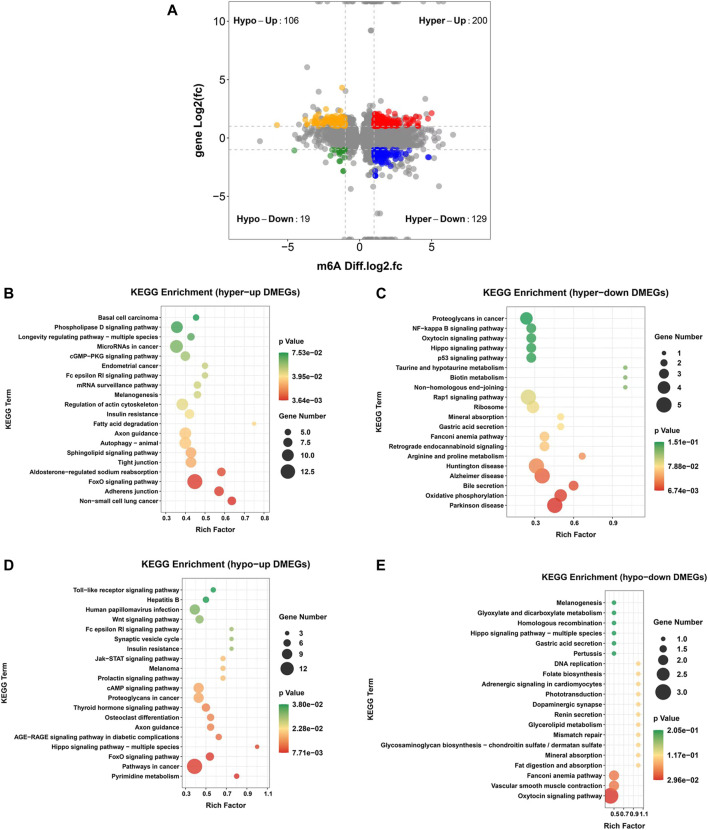
To further explore the functional significance of m6A modifications in H. pylori-infected gastric epithelial cells, we investigated whether m6A methylation underlies the observed differences in expression. For this purpose, DMGs and DEGs were detected using m6A-seq data and RNA-seq data. Thereafter, a combination of m6A-seq and RNA-seq analysis classified all genes into four major groups: including 200 hypermethylated and upregulated (hypo-up), 129 hypermethylated but downregulated (hypo-down), 19 hypermethylated and downregulated (hypo-down) and 106 hypomethylated but upregulated genes or transcripts (hypo-up) (Figure 5A). Four groups of DMEG were further investigated by KEGG analysis. The results showed that hyper-up genes were mainly enriched in “Adherens junctions,” “FoxO signaling pathway” and “Fatty acid degradation” pathways (Figure 5B); in contrast, hyper-down genes were mainly enriched in “Bile secretion,” “Gastric acid secretion,” “Oxidative phosphorylation” and “NF-κappa B signaling pathway” (Figure 5C). In addition, hypo-up genes were mainly enriched in “Toll-like receptor signaling pathway,” “Wnt signaling pathway,” “Jak-STAT signaling pathway,” “cAMP signaling pathway,” “pathway in cancer” and “Hippo signaling pathway-multi-species” (Figure 5D), while hypo-down genes were mainly enriched in “Vascular smooth muscle contraction” and “DNA replication” (Figure 5E). Moreover, we list the top 20 transcripts of differential m6A modification and mRNA expression between control group and H. pylori group based on diff.log2.fc (Table 1).
FIGURE 5.
Combined analysis between m6A-Seq and RNA-Seq (A) Four-quadrant diagram of hyper-up, hyper-down, hypo-up, and hypo-down DMEG. (B) KEGG pathway analysis of hyper-up DMEG (C) KEGG pathway analysis of hyper-down DMEG (D) KEGG pathway analysis of hypo-up DMEG (E) KEGG pathway analysis of hypo-down DMEG.
TABLE 1.
Top 20 transcripts of differential m6A modification and mRNA expression between control group and H. pylori group.
| Gene name | Change | Seqnames | m6A modification change | mRNA expression change | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Peak start | Peak end | Width | Peak region | logFC | p-value | logFC | p-value | |||
| PTPN14 | Hyper-up | chr1 | 214,532,254 | 214,532,432 | 179 | Exon | 5 | 4.57E-02 | 2.13 | 1.75E-06 |
| FRY | Hyper-up | chr13 | 32,298,645 | 32,298,945 | 301 | Exon | 4.75 | 1.74E-04 | 1.65 | 9.74E-04 |
| TRANK1 | Hyper-up | chr3 | 36,857,290 | 36,857,679 | 390 | Exon | 4.14 | 9.55E-04 | 1.79 | 4.63E-03 |
| HSPA12A | Hyper-up | chr10 | 116,827,654 | 116,827,984 | 331 | 3′ UTR | 4.07 | 1.70E-02 | 1.02 | 4.62E-02 |
| ADAM10 | Hyper-up | chr15 | 58,748,838 | 58,749,018 | 181 | 3′ UTR | 4.06 | 5.75E-03 | 1.29 | 1.96E-02 |
| BOLA2 | Hyper-down | chr16 | 29,454,886 | 29,455,005 | 120 | 5′ UTR | 4.81 | 3.72E-03 | −1.65 | 5.48E-06 |
| AL136038 | Hyper-down | chr14 | 63,642,540 | 63,642,600 | 61 | Exon | 4.75 | 4.07E-02 | −1.64 | 3.21E-04 |
| QPCTL | Hyper-down | chr19 | 45,703,571 | 45,703,690 | 120 | 3′ UTR | 3.44 | 1.74E-03 | −1.07 | 2.95E-04 |
| PIDD1 | Hyper-down | chr11 | 804,682 | 805,011 | 330 | 5′ UTR | 3.20 | 3.24E-03 | −1.38 | 9.14E-05 |
| RPS9 | Hyper-down | chr19 | 54,224,672 | 54,224,881 | 210 | Exon | 2.91 | 1.17E-02 | −1.28 | 7.32E-05 |
| NSF | Hypo-up | chr17 | 46,640,090 | 46,643,106 | 3,017 | 3′ UTR | −5.75 | 2.34E-04 | 1.10 | 2.92E-02 |
| ADAMTS1 | Hypo-up | chr21 | 26,843,766 | 26,844,363 | 598 | 5′ UTR | −3.78 | 2.00E-03 | 1.57 | 2.25E-03 |
| MAPRE2 | Hypo-up | chr18 | 34,977,018 | 34,978,391 | 1,374 | 5′ UTR | −3.65 | 8.91E-03 | 1.15 | 1.58E-02 |
| CEP78 | Hypo-up | chr9 | 78,276,633 | 78,276,783 | 151 | 3′ UTR | −3.63 | 4.37E-03 | 1.17 | 1.91E-02 |
| LAMA3 | Hypo-up | chr18 | 23,899,378 | 23,899,528 | 151 | Exon | −3.31 | 1.17E-02 | 1.42 | 4.32E-03 |
| SLX1B | Hypo- down | chr16 | 29,457,636 | 29,458,189 | 554 | 3′ UTR | −4.54 | 1.95E-08 | −1.07 | 2.50E-04 |
| RPS3AP47 | Hypo- down | chr15 | 43,115,761 | 43,115,907 | 147 | Exon | −2.03 | 2.88E-03 | −1.52 | 5.21E-05 |
| RPL29P11 | Hypo- down | chr3 | 37,016,898 | 37,017,014 | 117 | Exon | −1.91 | 8.71E-06 | −1.01 | 5.54E-04 |
| WTIP | Hypo- down | chr19 | 34,504,119 | 34,504,179 | 61 | 3′ UTR | −1.73 | 4.47E-02 | −1.04 | 1.35E-03 |
| PPP1R12C | Hypo- down | chr19 | 55,092,432 | 55,093,073 | 642 | Exon | −1.47 | 5.01E-13 | −1.06 | 2.37E-04 |
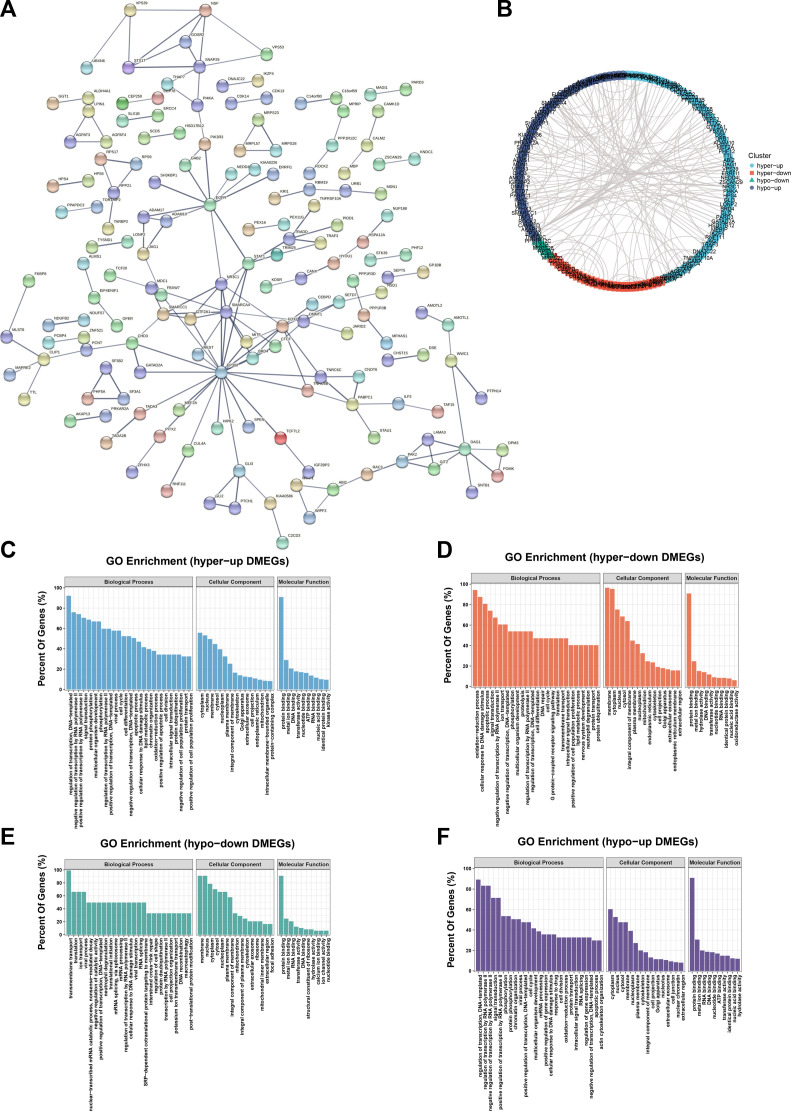
PPI network and hub genes were identified in DMEG
The PPI network of DMEG is carried out by the STRING database (Figure 6A) and Cytoscape. As above, the network was divided into four clusters, which respectively are hyper-up, hyper-down, hypo-down and hypo-up DMEGs (Figure 6B). GO enrichment analysis was performed for each DMEG cluster to elucidate its biological functions (Figures 6C–F). PPI network interaction data are listed in Supplementary Table S6.
FIGURE 6.
PPI networks and hub genes were found in DMEG. (A) PPI network of DMEG constructed from STRING database (B) Cytoscape was performed divided into four clusters. Blue represents hyper-up genes, orange represents hyper-down genes, green represents hypo-down genes, and purple represents hypo-up genes. (C) GO enrichment analysis of the hyper-up cluster in this DMEG. (D) GO enrichment analysis of hyper-down clusters in this DMEG. (E) GO enrichment analysis of the hypo-down cluster in this DMEG. (F) GO enrichment analysis of the hypo-up cluster in this DMEG.
Validation of differential expression genes
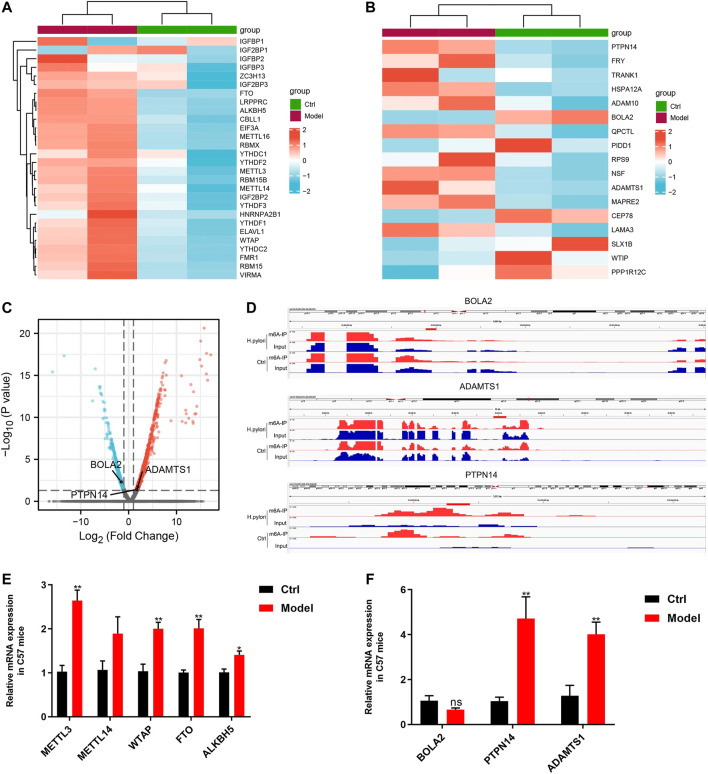
In the RNA-seq data of mice, we analyzed the mRNA levels of 28 m6A regulators, except IGF2BP1, 27 of 28 m6A regulators showed an increasing tendency (Figure 7A). Furthermore, qRT-PCR was used to detect the expression levels of five common regulators, including METTL3, METTL14, WTAP, FTO and ALKBH5; The change trends in those genes revealed by qRT- PCR were consistent with the with the RNA-seq results, those gene all did arrive significant difference except METTL14 (Figure 7E).
FIGURE 7.
Validation of differential expression genes (A) Heatmap of m6A regulators in sequencing of mice samples. (B) RNA-seq data of mice to validate the top 20 genes from cell sequencing by Heatmap. (C) Volcanic map of differentially expressed genes sequencing of mice samples. (D) IGV visualization show the three key m6A-modified genes. (E) qRT- PCR results for five common m6A regulators. (F) qRT- PCR results of the three key m6A-modified genes.
We used sequencing of mice samples results to validate the expression of top 20 genes of cell sequencing (Table 1), which shows that only 3 genes were consistent, including PTPN14, BOLA2 and ADAMTS1 (Figures 7B, C). We performed IGV visualization for the three genes and all found significantly different m6A levels (Figure 7D). Moreover, except BOLA2, PTPN14 and ADAMTS1 had significant difference by qRT- PCR (Figure 7F).
Discussion
H. pylori infection can damage the stomach mucosa, leading to the development of various stomach diseases which involve many pathophysiological changes. Abnormalities in m6A modifying enzymes can cause a series of diseases (Jiang et al., 2021) (Zhang et al., 2019). However, the mechanism of m6A modification in H. pylori-induced gastric epithelial infection remains unclear. In this study, the relationship between m6A modification profile and H. pylori-induced gastric epithelial infection was analyzed for the first time.
In the beginning, we found that H. pylori infection increase the level of m6A modification in vitro and in vivo. After that, we obtained an overview of m6A modification in H. pylori infection associated gastritis through MeRIP-seq. The total peak numbers of m6A revealed significant differences in m6A modification between the control and H. pylori groups. Therefore, we assume that m6A modification may be related to H. pylori -induced gastritis.
As is known to all, m6A modification of mRNA often affects the occurrence and development of the disease. In this study, we identified 2,107 significantly different peaks compared to the control, of which 1,565 peaks were upregulated and 542 peaks were downregulated. From this, we found that H. pylori can alter the methylation peak of GES-1. Therefore, we hypothesized that m6A modification may be associated with H. pylori-induced gastric epithelial infection. M6A peaks were mostly enriched near the 3′ UTR region, and these sites were m6A specific and consistent with previous studies (Huang et al., 2020a). The 3′ UTR regulates mRNA stability, localization, expression and translation of mRNA. Multiple RNA-binding proteins bind in this region to perform regulatory functions and regulate the interaction between proteins (Mayr, 2019). In addition, in H. pylori-infected gastric epithelial cells, differential methylation peaks were significantly enriched in “transcriptional regulation,” “RNA splicing,” “signal transduction,” “apoptotic processes” and “cell cycle.” Previous studies indicated that H. pylori involve the regulation of apoptosis, proliferation and the cell cycle (Hirata et al., 2001; Nozawa et al., 2002; Ding et al., 2008). This suggests a conserved and fundamental role of m6A in the regulation of development and cell fate specification. The hypermethylation peaks were mainly concentrated in “fatty acid elongation,” “EBV infection,” “drug metabolism,” “NF-κB signaling pathway” “EBV infection,” “drug metabolism,” “NF-κB signaling pathway” and “basic transcription factor” pathways. NF-κB is a key regulator of the immune response against H. pylori infection and is known to modulate genes involved in the control of inflammation, cell proliferation and apoptosis (Lamb and Chen, 2010; Chaturvedi et al., 2011; Shu et al., 2022). The hypomethylation peaks were mainly enriched in “natural killer cell-mediated cytotoxicity,” “basic transcription factors,” “cell cycle,” “mRNA surveillance pathway,” “basic transcription factors.” The mRNA surveillance pathway” and “pyruvate metabolism” pathways. This evidence indicates that m6A modification is probably associated with H. pylori -associated gastritis.
In order to clarify the mechanism of m6A affecting the process of H. pylori infection in gastric epithelial cells, we combined m6A methylation group with transcription group to find the key signaling pathways affected by m6A modification. Previous studies have shown that during the time course of H. pylori infection, H. pylori infection destroys the integrity of the gastric mucosa. H. pylori induces classical and alternative NF-κB signaling pathways through its effector ADP-L-glycero-β-D-manno-heptose (ADP-heptose), leading to deleterious gastric pathophysiology (Maubach et al., 2022). It has also been shown that H. pylori can induce a signaling cascade by activating the Toll-like receptor pathway, which ultimately leads to the transcription of pro-and anti-inflammatory cytokines and type I interferons (Peek et al., 2010). The Hippo signaling pathway appears to be a protective pathway in the host-pathogen conflict that generates an inflammatory environment, cellular injury, and epithelial renewal and differentiation, limiting the loss of gastric epithelial properties prior to adenocarcinoma development, which may be beneficial for H. pylori colonization and chronic infection (Molina-Castro et al., 2020). As in previous studies, some classical pathways regarding H. pylori causing gastric disease were significantly enriched in the present study. These include NF-κB signaling pathway (Keates et al., 1997; Sasaran et al., 2021), p53 signaling pathway (Cai et al., 2021; Imai et al., 2021), Hippo signaling pathway, Toll-like receptor signaling pathway (Lam et al., 2022), and Wnt signaling pathway (Abdi et al., 2021). This evidence also suggests that m6A modification may be associated with H. pylori-induced gastric disease.
In the past few years, numerous studies have illustrated the biological effects of m6A modification on RNA. On the one hand, the m6A methylation process is reversible, and this mark on RNA can be written or erased under various stimuli and biological factors (Feng et al., 2022; Liu et al., 2022; Wang et al., 2022). On the other hand, m6A can affect RNA processing and metabolism through a variety of mechanisms, including selective polyadenylation, selective splicing, RNA stability, RNA export, RNA degradation, and translation (Wang et al., 2014; Zhao et al., 2014; Coots et al., 2017; Hong et al., 2022). Thus, m6A up- or downregulates gene expression in a complex and context-dependent manner. For this reason, we observed four groups of DMEGs in the present study, which are hyper-up, hyper-down, hypo-up, and hypo-down. Our functional enrichment analysis showed that these four groups of DMEGs are associated with essential and different biological processes. Many previous studies reported that m6A modification is involved in different biological processes, such as transcriptional regulation, signal transduction, and the DNA damage response (Jia et al., 2011; Zheng et al., 2013; Hong et al., 2022). Our results are corresponded to previously these published studies.
In the RNA-seq data of mice, we found numerous m6A regulators were found to have changes. Except IGF2BP1, other m6A regulators showed an increasing tendency. To further verify the results of sequencing, we observed the expression of five common regulators by qRT- PCR which showed similar results to sequencing. Those genes (METTL3, WTAP, FTO and ALKBH5) all did arrive significant difference except METTL14. METTL3, as one of the core components of the m6A methyltransferase complex, has been found to be closely related to multiple signaling pathways, such as the JAK/STAT (Yao et al., 2019), MAPK/NF-κB (Li et al., 2020a), PI3K/AKT (Bi et al., 2021), and Wnt/β-catenin pathways (Cui et al., 2020). WTAP has been reported to be associated with a number of signaling pathways, such as TGFβ (Li et al., 2020b), hippo (Hu et al., 2020), NF-κB (Li et al., 2021), and Hedgehog pathways (Wei et al., 2022). FTO is associated with various signaling pathways, for example, PKA/CREB (Hu et al., 2022), TNF-α (Li et al., 2022b), ERK (Xiao et al., 2022b), WNT (Kim et al., 2022) and JAK2/STAT3 pathways (Shen et al., 2021). ALKBH5 is involved in many signaling pathways, including WNT (Lin et al., 2022), PTEN/AKT (He et al., 2021), NF-κB (Qu et al., 2022), AKT (Wang et al., 2020). Interestingly, many studies have shown that H. pylori infection is closely related to these signaling pathways. H. pylori can active the expression of STAT1 and PD-L1 which may prevent immune surveillance in the gastric mucosa, allowing premalignant lesions to progress to gastric cancer (Li et al., 2022c). H. pylori can induce injuries to the stomach through MAPK/NF-κB pathway (Shu et al., 2022). H. pylori can induce the occurrence of gastric carcinogenesis at the early stage by activating the PI3K/Akt signaling pathway (Xu et al., 2018). H. pylori infection activated WNT/β-catenin signaling pathway by upregulating to induce gastritis (Zuo et al., 2022). Judging from these, m6A regulators may also involve in regulating different signaling pathways in H. pylori-associated gastritis.
Moreover, the results of sequencing of mice samples were used to validate the expression of top 20 genes of cell sequencing, which found three genes, PTPN14, BOLA2 and ADAMTS1, that are consistent. Furthermore, qRT- PCR showed PTPN14 and ADAMTS1 had significant difference. Although there was no significant difference in BOLA2 expression, there was a downward trend. H. pylori was able to significantly upregulate PTPN14 and ADAMTS1 mRNA expression levels. At present, there are no studies relationship between these three genes and H. pylori-infected diseases. Many studies showed that PTPN14 has different function, such as suppressing the occurrence and development of tumor (Hatterschide et al., 2022), blunting the formation of atherosclerosis (Yang et al., 2021a) and promoting inflammation and fibrosis (Fu et al., 2020; Lin et al., 2021). Previous studies confirmed that ADAMTS1 is involved in inhibiting the proliferation, polarization and migration of tumor (Li et al., 2015; de Assis Lima et al., 2021), affecting the quality of oocytes and embryonic development potential (Yang et al., 2021b) and promoting collagen production (Toba et al., 2016). In addition, a recent study revealed that YTHDF2 inhibited ADAMTS1 expression and promoted sperm adhesion through m6A/mRNA pathway (Huang et al., 2020b). Above all, these results suggest that m6A is likely to exhibit an as-yet- unknown function in the process by H. pylori -induced gastritis.
In summary, we can infer that m6A methylation was shown to play a role in H. pylori-induced gastritis. Though the mechanism of m6A-regulated gastritis is not clearly understood, we provide the first m6A transcriptome profile of gastritis and an initial map revealing the function of m6A modification in gastritis using advanced technologies, thereby contributing critical insights for further research on the role of m6A in gastritis. These findings provide a basis for further investigation of the role of m6A methylation modification in H. pylori infection of the gastric mucosa. However, the finding into clinical scenario may be limited by the lack of verification of the expression and distribution of m6A related regulatory molecules in clinical samples of H. pylori-associated gastritis, which should be further studied in future investigations.
Funding Statement
This study was supported by Changsha Science and Technology Project (kq2202118), Natural Science Foundation of Hunan Province (2022JJ30906) and Hunan Provincial Innovation Foundation For Postgraduate (QL20220061).
Data availability statement
The data presented in the study are deposited in the GEO repository, accession number GSE230869 and GSE231337.
Ethics statement
The studies involving human participants were reviewed and approved by the Ethics Committee of the Third Xiangya Hospital of Central South University. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Ethics Committee of the Third Xiangya Hospital of Central South University.
Author contributions
All of the authors contributed to the conception of the article. The main experimental conception and design: CX and XL; performed the experiments: HL and JiL; analyzed the data and contributed reagents: SC, JC, and JuL; collected samples: YT, WZ, and YS; writing the manuscript: HL and JiL. All authors read and approved the final manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1136096/full#supplementary-material
References
- Abdi E., Latifi-Navid S., Abedi Sarvestani F., Esmailnejad M. H. (2021). Emerging therapeutic targets for gastric cancer from a host-Helicobacter pylori interaction perspective. Expert Opin. Ther. Targets 25 (8), 685–699. [published Online First: 2021/08/20]. 10.1080/14728222.2021.1971195 [DOI] [PubMed] [Google Scholar]
- Asano N., Imatani A., Watanabe T., Fushiya J., Kondo Y., Jin X., et al. (2016). Cdx2 expression and intestinal metaplasia induced by H. pylori infection of gastric cells is regulated by NOD1-mediated innate immune responses. Cancer Res. 76 (5), 1135–1145. [published Online First: 2016/01/14]. 10.1158/0008-5472.CAN-15-2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X., Lv X., Liu D., Guo H., Yao G., Wang L., et al. (2021). METTL3-mediated maturation of miR-126-5p promotes ovarian cancer progression via PTEN-mediated PI3K/Akt/mTOR pathway. Cancer Gene Ther. 28 (3-4), 335–349. [published Online First: 2020/09/18]. 10.1038/s41417-020-00222-3 [DOI] [PubMed] [Google Scholar]
- Cai Q., Shi P., Yuan Y., Peng J., Ou X., Zhou W., et al. (2021). Inflammation-associated senescence promotes Helicobacter pylori-induced atrophic gastritis. Cell. Mol. Gastroenterol. Hepatol. 11 (3), 857–880. [published Online First: 2020/11/09]. 10.1016/j.jcmgh.2020.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi M. M., Sung B., Yadav V. R., Kannappan R., Aggarwal B. B. (2011). NF-κB addiction and its role in cancer: 'one size does not fit all'. Oncogene 30 (14), 1615–1630. [published Online First: 2010/12/21]. 10.1038/onc.2010.566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokkalla A. K., Mehta S. L., Vemuganti R. (2020). Epitranscriptomic regulation by m(6)A RNA methylation in brain development and diseases. J. Cereb. Blood Flow. Metab. 40 (12), 2331–2349. [published Online First: 2020/09/25]. 10.1177/0271678X20960033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coots R. A., Liu X. M., Mao Y., Dong L., Zhou J., Wan J., et al. (2017). m6A Facilitates eIF4F-Independent mRNA Translation. Mol. Cell. 68 (3), 504–514. [published Online First: 2017/11/07]. 10.1016/j.molcel.2017.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa P., Piazuelo M. B. (2012). The gastric precancerous cascade. J. Dig. Dis. 13 (1), 2–9. [published Online First: 2011/12/23]. 10.1111/j.1751-2980.2011.00550.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover T. L., Blaser M. J. (2009). Helicobacter pylori in health and disease. Gastroenterology 136 (6), 1863–1873. [published Online First: 2009/05/22]. 10.1053/j.gastro.2009.01.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., Wang Z., Li J., Zhu J., Ren Z., Zhang D., et al. (2020). Cross talk between RNA N6-methyladenosine methyltransferase-like 3 and miR-186 regulates hepatoblastoma progression through Wnt/β-catenin signalling pathway. Cell. Prolif. 53 (3), e12768. [published Online First: 2020/01/23]. 10.1111/cpr.12768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Assis Lima M., da Silva S. V., Serrano-Garrido O., Hülsemann M., Santos-Neres L., Rodríguez-Manzaneque J. C., et al. (2021). Metalloprotease ADAMTS-1 decreases cell migration and invasion modulating the spatiotemporal dynamics of Cdc42 activity. Cell. Signal 77, 109827. [published Online First: 2020/11/09]. 10.1016/j.cellsig.2020.109827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R., Friderici K., Rottman F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 71 (10), 3971–3975. [published Online First: 1974/10/01]. 10.1073/pnas.71.10.3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S. Z., Smith M. F., Jr., Goldberg J. B. (2008). Helicobacter pylori and mitogen-activated protein kinases regulate the cell cycle, proliferation and apoptosis in gastric epithelial cells. J. Gastroenterol. Hepatol. 23 (7), e67–e78. [published Online First: 2008/08/16]. 10.1111/j.1440-1746.2007.04912.x [DOI] [PubMed] [Google Scholar]
- Feng H., Yuan X., Wu S., Yuan Y., Cui L., Lin D., et al. (2022). Effects of writers, erasers and readers within miRNA-related m6A modification in cancers. Cell. Prolif. 56, e13340. [published Online First: 2022/09/27]. 10.1111/cpr.13340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu B., Yin S., Lin X., Shi L., Wang Y., Zhang S., et al. (2020). PTPN14 aggravates inflammation through promoting proteasomal degradation of SOCS7 in acute liver failure. Cell. Death Dis. 11 (9), 803. [published Online First: 2020/09/27]. 10.1038/s41419-020-03014-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Zheng J., Zhang H., Tong J. (2021). RNA m6A methylation regulators in ovarian cancer. Cancer Cell. Int. 21 (1), 609. [published Online First: 2021/11/20]. 10.1186/s12935-021-02318-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatterschide J., Castagnino P., Kim H. W., Sperry S. M., Montone K. T., Basu D., et al. (2022). YAP1 activation by human papillomavirus E7 promotes basal cell identity in squamous epithelia. Elife 11, e75466. [published Online First: 2022/02/17]. 10.7554/eLife.75466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Yue H., Cheng Y., Ding Z., Xu Z., Lv C., et al. (2021). ALKBH5-mediated m(6)A demethylation of KCNK15-AS1 inhibits pancreatic cancer progression via regulating KCNK15 and PTEN/AKT signaling. Cell. Death Dis. 12 (12), 1121. [published Online First: 2021/12/03]. 10.1038/s41419-021-04401-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata Y., Maeda S., Mitsuno Y., Akanuma M., Yamaji Y., Ogura K., et al. (2001). Helicobacter pylori activates the cyclin D1 gene through mitogen-activated protein kinase pathway in gastric cancer cells. Infect. Immun. 69 (6), 3965–3971. [published Online First: 2001/05/12]. 10.1128/IAI.69.6.3965-3971.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J., Xu K., Lee J. H. (2022). Biological roles of the RNA m(6)A modification and its implications in cancer. Exp. Mol. Med. 54 (11), 1822–1832. [published Online First: 2022/11/30]. 10.1038/s12276-022-00897-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C., Yu M., Li C., Wang Y., Li X., Ulrich B., et al. (2020). miR-550-1 functions as a tumor suppressor in acute myeloid leukemia via the hippo signaling pathway. Int. J. Biol. Sci. 16 (15), 2853–2867. [published Online First: 2020/10/17]. 10.7150/ijbs.44365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Chen J., Wang Y., Sun J., Huang P., Feng J., et al. (2022). Fat mass and obesity-associated protein alleviates Aβ1-40 induced retinal pigment epithelial cells degeneration via PKA/CREB signaling pathway. Cell. Biol. Int. 47, 584–597. [published Online First: 2022/11/16]. 10.1002/cbin.11959 [DOI] [PubMed] [Google Scholar]
- Huang H., Weng H., Chen J. (2020). The biogenesis and precise control of RNA m(6)A methylation. Trends Genet. 36 (1), 44–52. [published Online First: 2019/12/08]. 10.1016/j.tig.2019.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T., Liu Z., Zheng Y., Feng T., Gao Q., Zeng W. (2020). YTHDF2 promotes spermagonial adhesion through modulating MMPs decay via m(6)A/mRNA pathway. Cell. Death Dis. 11 (1), 37. [published Online First: 2020/01/22]. 10.1038/s41419-020-2235-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Ooki T., Murata-Kamiya N., Komura D., Tahmina K., Wu W., et al. (2021). Helicobacter pylori CagA elicits BRCAness to induce genome instability that may underlie bacterial gastric carcinogenesis. Cell. Host Microbe 29 (6), 941–958.e10. [published Online First: 2021/05/15]. 10.1016/j.chom.2021.04.006 [DOI] [PubMed] [Google Scholar]
- Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7 (12), 885–887. [published Online First: 2011/10/18]. 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Liu B., Nie Z., Duan L., Xiong Q., Jin Z., et al. (2021). The role of m6A modification in the biological functions and diseases. Signal Transduct. Target Ther. 6 (1), 74. [published Online First: 2021/02/22]. 10.1038/s41392-020-00450-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keates S., Hitti Y. S., Upton M., Kelly C. P. (1997). Helicobacter pylori infection activates NF-kappa B in gastric epithelial cells. Gastroenterology 113 (4), 1099–1109. [published Online First: 1997/10/10]. 10.1053/gast.1997.v113.pm9322504 [DOI] [PubMed] [Google Scholar]
- Kim H., Jang S., Lee Y. S. (2022). The m6A(m)-independent role of FTO in regulating WNT signaling pathways. Life Sci. Alliance 5 (5). [published Online First: 2022/02/17]. 10.26508/lsa.202101250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S. Y., Mommersteeg M. C., Yu B., Broer L., Spaander M. C. W., Frost F., et al. (2022). Toll-like receptor 1 locus Re-examined in a genome-wide association study update on anti-Helicobacter pylori IgG titers. Gastroenterology 162 (6), 1705–1715. [published Online First: 2022/01/16]. 10.1053/j.gastro.2022.01.011 [DOI] [PubMed] [Google Scholar]
- Lamb A., Chen L. F. (2010). The many roads traveled by Helicobacter pylori to NFκB activation. Gut Microbes 1 (2), 109–113. [published Online First: 2011/02/18]. 10.4161/gmic.1.2.11587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Liu L., Zang W., Wang Y., Du Y., Chen X., et al. (2015). miR-365 overexpression promotes cell proliferation and invasion by targeting ADAMTS-1 in breast cancer. Int. J. Oncol. 47 (1), 296–302. [published Online First: 2015/05/23]. 10.3892/ijo.2015.3015 [DOI] [PubMed] [Google Scholar]
- Li D., Cai L., Meng R., Feng Z., Xu Q. (2020). METTL3 modulates osteoclast differentiation and function by controlling RNA stability and nuclear export. Int. J. Mol. Sci. 21 (5), 1660. [published Online First: 2020/03/04]. 10.3390/ijms21051660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Chen Y. X., Yang B., Liao J. Y., Peng J. W., Zhu S. (2020). The crosstalk between RNA m6A epitranscriptome and TGFβ signaling pathway contributes to the arrest of cell cycle. Gene 738, 144483. [published Online First: 2020/02/20]. 10.1016/j.gene.2020.144483 [DOI] [PubMed] [Google Scholar]
- Li Q., Wang C., Dong W., Su Y., Ma Z. (2021). WTAP facilitates progression of endometrial cancer via CAV-1/NF-κB axis. Cell. Biol. Int. 45 (6), 1269–1277. [published Online First: 2021/02/10]. 10.1002/cbin.11570 [DOI] [PubMed] [Google Scholar]
- Li P., Wang Y., Sun Y., Jiang S., Li J. (2022). N (6)-methyladenosine RNA methylation: From regulatory mechanisms to potential clinical applications. Front. Cell. Dev. Biol. 10, 1055808. [published Online First: 2022/11/22]. 10.3389/fcell.2022.1055808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Du M., Sun Q., Cao Z., He H. (2022). m6 A demethylase Fto regulates the TNF-alpha-induced inflammatory response in cementoblasts. Oral Dis. [published Online First: 2022/10/14]. 10.1111/odi.14396 [DOI] [PubMed] [Google Scholar]
- Li X., Pan K., Vieth M., Gerhard M., Li W., Mejías-Luque R. (2022). JAK-STAT1 signaling pathway is an early response to Helicobacter pylori infection and contributes to immune escape and gastric carcinogenesis. Int. J. Mol. Sci. 23 (8), 4147. [published Online First: 2022/04/24]. 10.3390/ijms23084147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Shao Z., Zhao M., Li J., Xu X. (2021). PTPN14 deficiency alleviates podocyte injury through suppressing inflammation and fibrosis by targeting TRIP6 in diabetic nephropathy. Biochem. Biophys. Res. Commun. 550, 62–69. [published Online First: 2021/03/09]. 10.1016/j.bbrc.2020.12.030 [DOI] [PubMed] [Google Scholar]
- Lin C., Wang Y., Dong Y., Lai S., Wang L., Weng S., et al. (2022). N6-methyladenosine-mediated SH3BP5-AS1 upregulation promotes GEM chemoresistance in pancreatic cancer by activating the Wnt signaling pathway. Biol. Direct 17 (1), 33. [published Online First: 2022/11/19]. 10.1186/s13062-022-00347-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Zou H., Dang Q., Xu H., Liu L., Zhang Y., et al. (2022). Biological and pharmacological roles of m(6)A modifications in cancer drug resistance. Mol. Cancer 21 (1), 220. [published Online First: 2022/12/15]. 10.1186/s12943-022-01680-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maubach G., Vieth M., Boccellato F., Naumann M. (2022). Helicobacter pylori-induced NF-κB: Trailblazer for gastric pathophysiology. Trends Mol. Med. 28 (3), 210–222. [published Online First: 2022/01/12]. 10.1016/j.molmed.2021.12.005 [DOI] [PubMed] [Google Scholar]
- Mayr C. (2019). What are 3' UTRs doing? Cold Spring Harb. Perspect. Biol. 11 (10), a034728. [published Online First: 2018/09/06]. 10.1101/cshperspect.a034728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. D., Jaffrey S. R. (2014). The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell. Biol. 15 (5), 313–326. [published Online First: 2014/04/10]. 10.1038/nrm3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Castro S. E., Tiffon C., Giraud J., Boeuf H., Sifre E., Giese A., et al. (2020). The hippo kinase LATS2 controls Helicobacter pylori-induced epithelial-mesenchymal transition and intestinal metaplasia in gastric mucosa. Cell. Mol. Gastroenterol. Hepatol. 9 (2), 257–276. [published Online First: 2019/11/02]. 10.1016/j.jcmgh.2019.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa Y., Nishihara K., Peek R. M., Nakano M., Uji T., Ajioka H., et al. (2002). Identification of a signaling cascade for interleukin-8 production by Helicobacter pylori in human gastric epithelial cells. Biochem. Pharmacol. 64 (1), 21–30. [published Online First: 2002/07/11]. 10.1016/s0006-2952(02)01030-4 [DOI] [PubMed] [Google Scholar]
- Pan Y., Ma P., Liu Y., Li W., Shu Y. (2018). Multiple functions of m(6)A RNA methylation in cancer. J. Hematol. Oncol. 11 (1), 48. [published Online First: 2018/03/29]. 10.1186/s13045-018-0590-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peek R. M., Jr., Fiske C., Wilson K. T. (2010). Role of innate immunity in Helicobacter pylori-induced gastric malignancy. Physiol. Rev. 90 (3), 831–858. [published Online First: 2010/07/29]. 10.1152/physrev.00039.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J., Hou Y., Chen Q., Chen J., Li Y., Zhang E., et al. (2022). RNA demethylase ALKBH5 promotes tumorigenesis in multiple myeloma via TRAF1-mediated activation of NF-κB and MAPK signaling pathways. Oncogene 41 (3), 400–413. [published Online First: 2021/11/12]. 10.1038/s41388-021-02095-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant J. Y., Soller M. (2017). m(6 A in mRNA: An ancient mechanism for fine-tuning gene expression. Trends Genet. 33 (6), 380–390. [published Online First: 2017/05/14]. 10.1016/j.tig.2017.04.003 [DOI] [PubMed] [Google Scholar]
- Sacco M. T., Bland K. M., Horner S. M. (2022). WTAP targets the METTL3 m(6)a-methyltransferase complex to cytoplasmic hepatitis C virus RNA to regulate infection. J. Virol. 96 (22), e0099722. [published Online First: 2022/11/01]. 10.1128/jvi.00997-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaran M. O., Melit L. E., Dobru E. D. (2021). MicroRNA modulation of host immune response and inflammation triggered by Helicobacter pylori . Int. J. Mol. Sci. 22 (3), 1406. [published Online First: 2021/02/13]. 10.3390/ijms22031406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Liu P., Sun Q., Li Y., Acharya R., Li X., et al. (2021). FTO inhibits UPR(mt)-induced apoptosis by activating JAK2/STAT3 pathway and reducing m6A level in adipocytes. Apoptosis 26 (7-8), 474–487. [published Online First: 2021/07/03]. 10.1007/s10495-021-01683-z [DOI] [PubMed] [Google Scholar]
- Shi H., Zhang X., Weng Y. L., Lu Z., Liu Y., Lu Z., et al. (2018). m(6 A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 563 (7730), 249–253. [published Online First: 2018/11/08]. 10.1038/s41586-018-0666-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu C., Tian J., Si X., Xie X. (2022). Blueberry anthocyanin extracts protect against Helicobacter pylori-induced peptic epithelium injuries both in vitro and in vivo: The key role of MAPK/NF-κB pathway. Eur. J. Nutr. 61 (5), 2749–2759. [published Online First: 2022/03/16]. 10.1007/s00394-022-02830-1 [DOI] [PubMed] [Google Scholar]
- Sun Z., Chen W., Wang Z., Wang S., Zan J., Zheng L., et al. (2022). Matr3 reshapes m6A modification complex to alleviate macrophage inflammation during atherosclerosis. Clin. Immunol. 245, 109176. [published Online First: 2022/11/12]. 10.1016/j.clim.2022.109176 [DOI] [PubMed] [Google Scholar]
- Toba H., de Castro Bras L. E., Baicu C. F., Zile M. R., Lindsey M. L., Bradshaw A. D. (2016). Increased ADAMTS1 mediates SPARC-dependent collagen deposition in the aging myocardium. Am. J. Physiol. Endocrinol. Metab. 310 (11), E1027–E1035. [published Online First: 2016/05/05]. 10.1152/ajpendo.00040.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Lu Z., Gomez A., Hon G. C., Yue Y., Han D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505 (7481), 117–120. [published Online First: 2013/11/29]. 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. F., Kuang M. J., Han S. J., Wang A. B., Qiu J., Wang F., et al. (2020). BMP2 modified by the m(6)A demethylation enzyme ALKBH5 in the ossification of the ligamentum flavum through the AKT signaling pathway. Calcif. Tissue Int. 106 (5), 486–493. [published Online First: 2020/01/04]. 10.1007/s00223-019-00654-6 [DOI] [PubMed] [Google Scholar]
- Wang S., Lv W., Li T., Zhang S., Wang H., Li X., et al. (2022). Dynamic regulation and functions of mRNA m6A modification. Cancer Cell. Int. 22 (1), 48. [published Online First: 2022/01/31]. 10.1186/s12935-022-02452-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei A., Zhao F., Hao A., Liu B., Liu Z. (2022). N-acetyl-seryl-aspartyl-lysyl-proline (AcSDKP) mitigates the liver fibrosis via WTAP/m(6)A/Ptch1 axis through Hedgehog pathway. Gene 813, 146125. [published Online First: 2021/12/19]. 10.1016/j.gene.2021.146125 [DOI] [PubMed] [Google Scholar]
- Xia X., Zhang L., Chi J., Liu X., Li H., Hu T., et al. (2020). Helicobacter pylori infection impairs endothelial function through an exosome-mediated mechanism. J. Am. Heart Assoc. 9 (6), e014120. [published Online First: 2020/03/17]. 10.1161/JAHA.119.014120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Chen J., Yang S., Sun H., Xie L., Li J., et al. (2022). Maternal mRNA deadenylation and allocation via Rbm14 condensates facilitate vertebrate blastula development. EMBO J. 42, e111364. [published Online First: 2022/12/09]. 10.15252/embj.2022111364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q., Lei L., Ren J., Peng M., Jing Y., Jiang X., et al. (2022). Mutant NPM1-regulated FTO-mediated m(6)A demethylation promotes leukemic cell survival via PDGFRB/ERK signaling Axis. Front. Oncol. 12, 817584. [published Online First: 2022/02/26]. 10.3389/fonc.2022.817584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Huang Y., Yang Z., Hu Y., Shu X., Xie C., et al. (2018). Helicobacter pylori promotes gastric epithelial cell survival through the PLK1/PI3K/Akt pathway. Onco Targets Ther. 11, 5703–5713. [published Online First: 2018/09/27]. 10.2147/OTT.S164749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Ma Q., Li Z., Wang H., Zhang C., Liu Y., et al. (2021). Harmine alleviates atherogenesis by inhibiting disturbed flow-mediated endothelial activation via protein tyrosine phosphatase PTPN14 and YAP. Br. J. Pharmacol. 178 (7), 1524–1540. [published Online First: 2021/01/22]. 10.1111/bph.15378 [DOI] [PubMed] [Google Scholar]
- Yang G., Yao G., Xu Z., Fan H., Liu X., He J., et al. (2021). Expression level of ADAMTS1 in granulosa cells of PCOS patients is related to granulosa cell function, oocyte quality, and embryo development. Front. Cell. Dev. Biol. 9, 647522. [published Online First: 2021/04/30]. 10.3389/fcell.2021.647522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Bi Z., Wu R., Zhao Y., Liu Y., Liu Q., et al. (2019). METTL3 inhibits BMSC adipogenic differentiation by targeting the JAK1/STAT5/C/EBPβ pathway via an m6A-YTHDF2-dependent manner. FASEB J. 33 (6), 7529–7544. [published Online First: 2019/03/14]. 10.1096/fj.201802644R [DOI] [PubMed] [Google Scholar]
- Zhang S. Y., Zhang S. W., Fan X. N., Meng J., Chen Y., Gao S. J., et al. (2019). Global analysis of N6-methyladenosine functions and its disease association using deep learning and network-based methods. PLoS Comput. Biol. 15 (1), e1006663. [published Online First: 2019/01/03]. 10.1371/journal.pcbi.1006663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Yang C., Ruan X., Liu X., Wang D., Liu L., et al. (2022). CPEB2 m6A methylation regulates blood-tumor barrier permeability by regulating splicing factor SRSF5 stability. Commun. Biol. 5 (1), 908. [published Online First: 2022/09/07]. 10.1038/s42003-022-03878-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Yang Y., Sun B. F., Shi Y., Xiao W., Yang X., et al. (2014). FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell. Res. 24 (12), 1403–1419. [published Online First: 2014/11/22]. 10.1038/cr.2014.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G., Dahl J. A., Niu Y., Fedorcsak P., Huang C. M., Li C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 49 (1), 18–29. [published Online First: 2012/11/28]. 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., Ni W., Qin C., Zhou Y., Li Y., Huo J., et al. (2022). A functional loop between YTH domain family protein YTHDF3 mediated m(6)A modification and phosphofructokinase PFKL in glycolysis of hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 41 (1), 334. [published Online First: 2022/12/07]. 10.1186/s13046-022-02538-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo W., Yang H., Li N., Ouyang Y., Xu X., Hong J. (2022). Helicobacter pylori infection activates Wnt/β-catenin pathway to promote the occurrence of gastritis by upregulating ASCL1 and AQP5. Cell. Death Discov. 8 (1), 257. [published Online First: 2022/05/11]. 10.1038/s41420-022-01026-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in the study are deposited in the GEO repository, accession number GSE230869 and GSE231337.