Abstract
NDRG1 is a member of the α/β hydrolase superfamily that resides in the cytoplasm and participates in the stress responses, hormone response, cell growth, and differentiation. Several studies have pointed to the importance of NDRG1 in the carcinogenesis. This gene has been found to be up-regulated in an array of cancer types such as bladder, esophageal squamous cell carcinoma, endometrial, lung and liver cancers, but being down-regulated in other types of cancers such as colorectal, gastric and ovarian cancers. The current study summarizes the evidence on the role of NDRG1 in the carcinogenic processes in different types of tissues.
Keywords: NDRG1, Cancer, Expression, Carcinogenesis, Biomarker
Introduction
N-myc downstream regulated 1 (NDRG1) is encoded by a gene located on chromosome 8q24.22, roughly 60 kb in size. This gene provides instructions for making a protein of 43 kDa (made up of 394 amino acids) that is extremely stable and is highly conserved among multicellular creatures. The mRNA for this gene is 3.0 kb in size [1] (Fig. 1). The NDRG1 gene is a part of the human NDRG family, which also includes the NDRG2, NDRG3, and NDRG4 genes. These genes are only 53–64% similar to one another [2]. As a member of the α/β hydrolase superfamily, this cytoplasmic protein takes part in the regulation of stress response, hormone response, cell growth, and differentiation. In addition, it has a crucial role in p53-mediated activation of caspase and apoptosis. Mutations in NDRG1 gene have been shown to cause Charcot-Marie-Tooth disease type 4D [3]. In addition to the cytoplasm, this protein can be found in the cell membrane and nucleus of the cells.
Fig. 1.
A graphical illustration of the component parts that make up the modular framework of NDRG1
More recently, studies have pointed to the importance of NDRG1 in the carcinogenesis. This gene has been found to be up-regulated in an array of cancer types, but being down-regulated in other types of cancers. The current study summarizes the evidence on the role of NDRG1 in the carcinogenic processes in different types of tissues. We have categorized this paper into distinct subtitles based on the obtained evidence from cell lines, xenograft models of cancer and expression assays in clinical samples.
Role of NDRG1 in cancer based on cell line studies
Cell line studies in triple-negative breast cancer (TNBC) have shown important effect of HJURP/YAP1/NDRG1 axis in these cells. Expression of HJURP has been found to be up-regulated in TNBC compared to other subtypes of breast cancer. This evolutionarily conserved chaperone can influence ubiquitination modification level of YAP1 protein, thus regulating its downstream transcriptional activities. YAP1 can induce transcription of NDRG1 through binding to its promoter. The HJURP/YAP1/NDRG1 axis can influence proliferation and chemosensitivity of TNBC cells [4].
Another study has re-analyzed the transcriptomic data from TNBC cybrids with a number of nuclear or mitochondrial donor cells. This study has shown up-regulation of 149 genes in the cybrids with mitochondria, among them being NDRG1. NDRG1 has been shown to be co-located with PVT1, and EXT1 on 8q24, a region that harbors amplification in breast cancer. NDRG1 has exhibited the most significant under-expression in the cybrids with benign mitochondria. NDRG1 silencing has led to reduction of proliferation of SUM-159 TNBC cells. Taken together, this study has demonstrated the role of mitochondria in facilitating over-expression of NDRG1 in TNBC [5].
Moreover, another study in breast cancer cells has shown that NDRG1 silencing leads to reduction of cell proliferation rate, abnormalities in lipid metabolism such as enhancement of incorporation of fatty acids into neutral lipids and lipid droplets. On the contrary, NDRG1 expression has diminished lipid droplet construction under nutrient supplied and starvation circumstances [6].
Besides, expression of NDRG1 can be affected by hormones. For instance, a cell line study has shown up-regulation of SGK1 and NDRG1 following treatment with progesterone. These observations have been accompanied by down-regulation of two miRNAs that target SGK1 3'UTR, namely miR-29a and miR-101–1. Authors have also demonstrated progesterone-mediated transcriptional and post-transcriptional control of SGK1 expression, resulting in over-expression of NDRG1 by a number of genes whose expressions are controlled by the transcription factor AP-1. NDRG1 can reduce activity of some kinases, thus reducing invasion and migratory potential of breast cancer cells [7].
In addition to hormones, expression of NDRG1 has been shown to be regulated by the long non-coding RNA (lncRNA) NDRG1 overlapping transcript 1 (NDRG1-OT1). Expression of this lncRNA is induced under hypoxic conditions. Notably, NDRG1-OT1 can inhibit expression of NDRG1 at both transcript and protein levels. NDRG1-OT1 enhances degradation of NDRG1 through ubiquitin-mediated proteolysis. Moreover, NDRG1-OT1 can decrease NDRG1 promoter activity. Most notably, different segments of NDRG1-OT1 have been shown to exert opposite impacts on NDRG1. While the first quarter segment of this lncRNA has no impact on the promoter of NDRG1, the second and third quarter segments represses and improves NDRG1 promoter activity, respectively. Finally, the fourth quarter segment of this lncRNA decreases activity of this promoter through reducing KHSRP under hypoxia [8].
5-Aza-2′-deoxycytidine is another agent that induces expression of NDRG1. While NDRG1 is expressed in the MDA-MB-231, this gene is expressed in T47D cells only following treatment with 5-Aza-2′-deoxycytidine. This observation shows the impact of DNA methylation on NDRG1 expression. NDRG1 gene promoter has a high level of methylation in T47D cells in spite of low level of methylation of this promoter in MDA-MB-231 cells [9].
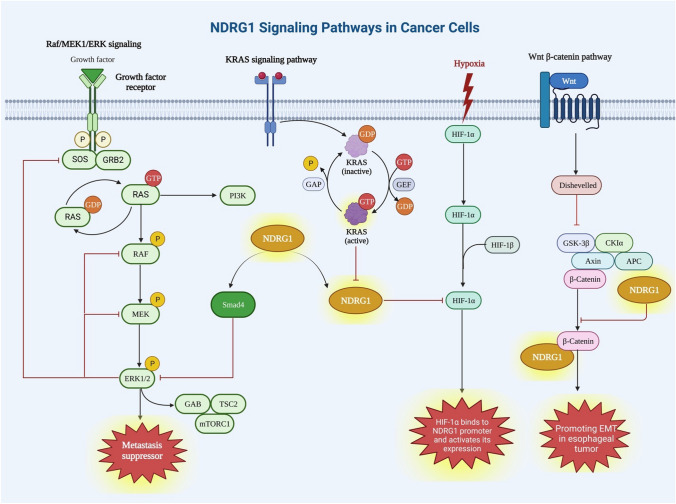
Furthermore, by modulating TLE2 and β-catenin expression, NDRG1 regulates Wnt pathway activation and EMT in esophageal cancer cells [10]. This suggests that NDRG1 in esophageal cancer cells plays a pro-oncogenic role through modulating tumor development (Fig. 2).
Fig. 2.
An illustration of the individual components of the architecture of NDRG1 signaling pathways in cancer cells
Additionally, K-Ras is critical for controlling in vitro NDRG1 protein level in pancreatic ductal adenocarcinoma (PDAC) cancer cells through ERK signaling [11]. Notably, NDRG1 expression reduces the amount and activity of HIF1 by inhibiting K-Ras downstream Akt/mTOR signaling. In particular, NDRG1 plays an especial part in controlling PDAC cancer metabolism. NDRG1 mediates downregulation of many glycolysis essential enzymes, including GLUT1, HK2, LDHA, and PDK1 [11]. Furthermore, it appears that NDRG1 inhibits glycolysis enzymes via decreasing the activity of HIF1, which may be the cause of this phenomenon.
Moreover, NDRG1 communicates with its target cells and other cells via signaling pathways and molecular motors. For instance, NDRG1 is associated with cancer metastasis via its function in Raf/MEK1/ERK signaling. Ras is bound to GTP upon activation of GFRs [12]. The downstream targets are activated when the active Ras:GTP phosphorylates Raf/MEK1/ERK1/2. Smad4 is upregulated by NDRG1, which in turn blocks the pathway (Fig. 2).
Prostate cancer cell lines have been other sources for identification of the function of NDRG1. A chemical genetic screening method for identification of substrates for the oncogenic serine/threonine kinase PIM1 has led to identification of NDRG1 as an important substrate for this kinase in prostate cancer cells. PIM1 can phosphorylate pS330 of NDRG1, leading to reduction of stability NDRG1, its nuclear localization, and interaction with androgen receptor (AR), which consequently enhances cell migration and invasion [13]. Another study in prostate cancer has shown that N-cadherin induces expression of c-Jun and inhibits expression of NDRG1 to increase invasive properties and migratory potential of prostate cancer cells via affecting epithelial to mesenchymal transition (EMT). Moreover, c-Jun, AR, and DNMT1 establish a complex in the TPA response elements region of the NDRG1 promoter, which inhibits transcription of NDRG1 via induction of hypermethylation of DNA [14]. Two other studies have revealed a tumor suppressor role for NDRG1 in prostate cancer in association with the role of this protein in modulation of AR activity. First, NDRG1 has been shown to directly regulate AR signaling in this type of cancer [15]. Second, the impact of MLL5α on activation of AR/NDRG1 signaling has been found to result in the suppression of prostate cancer progression [16]. Table 1 summarizes the results of studies on the role of NDRG1 in different cancer cell lines.
Table 1.
Role of NDRG1 in cancer cell lines
| Tumor type | Interactions/regulatory mechanisms | Cell line | Function | References |
|---|---|---|---|---|
| Breast cancer (TNBC) | HJURP/YAP1 axis |
T47D, ZR-75–1, BT549, HCC1937, BCAP-37, MCF-7, HS578T, SK-BR-3 and MDA-MB-231 |
↑ NDRG1: ↑ tumor cell proliferation YAP1 can induce transcription of NDRG1 through binding with its promoter |
[4] |
| Breast cancer (TNBC) | mitochondria | MDA-MB-231 and SUM-159 | Δ NDRG1: ↓ tumor cell proliferation | [5] |
| Breast cancer | Neutral lipid metabolism | SKBR3, MCF7, HCC1569, BT474, MDA-MB-231, MDA-MB-468 and HEK293T | Δ NDRG1: ↓ tumor cell proliferation | [6] |
| Breast cancer (PR ±) | AP-1/NDRG1 axis, kinase gene SGK1 | T47D, BT474, MDA-MB-231, ZR-75–1,MCF7 and 184A1 | ↑ NDRG1: ↑tumor cell proliferation | [7] |
| Breast cancer | lncRNA NDRG1-OT1 | MCF-7 and HEK293T | NDRG1-OT1: ↓NDRG1 promoter activities | [8] |
| Breast cancer | NDRG1-OT1_v4 | MCF-7, SKBR3, MDAMB-231, ZR75-30, MDAMB-361 and MDAMB-453 and MCF-10A |
NDRG1-OT1_v4 inhibits expression of NDRG1 at mRNA and protein levels NDRG1-OT1_v4: destabilizes NDRG1 |
[17] |
| Breast cancer | aryl hydrocarbon receptor (AHR) | MCF-7 |
↑↑AHR: ↑cell proliferation and migration via up-regulation of NDRG1 ↑ NDRG1: ↑tumor cell proliferation |
[18] |
| Breast cancer | miR-769-3p | MCF-7 |
miR-769-3p inhibits the expression of NDRG1 ↑↑ miR-769-3p: ↓ cell proliferation and ↑ apoptosis |
[19] |
| Breast cancer | methylation | MDA-MB-231 and T47D | ↓NDRG1: ↑ tumor cell invasion | [9] |
| Breast cancer | β-casein | SK-BR-3, MDA-MB-231, T47D and MCF-7 | Correlation between endogenous expression levels of NDRG1 and differentiation status | [2] |
| Prostate cancer | YWHAZ | – | ↑NDRG1: ↑ tumor cell proliferation | [20] |
| Prostate cancer | PIM1 | LNCaP-95, LNCaP-abl, LAPC-4, C4–2, 293 T, VCaP and 22Rv1 | NDRG1 phosphorylation: ↑ cell migration and invasion | [13] |
| Prostate cancer | N-cadherin/c-Jun/NDRG1 axis | LNCaP and PC3 | ↓NDRG1: ↑ tumor cell invasion and migration | [14] |
| Prostate cancer | androgen receptor signaling | LNCaP, C4-2B, and 22Rv1 |
↓NDRG1: ↑ tumor cell growth and metastasis NDRG1 suppresses AR activation |
[15] |
| Prostate cancer | MLL5α, AR/NDRG1 signaling | LNCaP, C4-2, 22RV1, PC3 and 293 T |
↓NDRG1: ↑ tumor cell invasion and migration MLL5α suppresses cancer progression by inducing AR/NDRG1 signaling |
[16] |
| Prostate Cancer | E6AP-NDRG1 axis | DU145, PC3, LNCaP and BPH | ↑↑E6AP: ↓NDRG1 | [21] |
| methylation of CpG islands of NDRG1 promoter | LNCap, PC-3, DU145, 22Rv1 and RWPE-1 | ↓NDRG1: ↑ tumor cell proliferation and invasion | [22] | |
| Prostate cancer | Proteolytic cleavage | DU145, PC3, LNCaP, PC3MM/Tet-Flag-Drg-1 and PrECs |
↓NDRG1: ↑ tumor cell growth Cleavage of NDRG1 results in loss of tumor suppressor activity of this protein |
[23] |
| Prostate cancer | AKT, TGF-β and ERK pathways | PrECs, DU145 and PC-3 |
Dp44mT: ↑ NDRG1 and PTEN expression Δ NDRG1: ↑phosphorylation of AKT, ERK1/2 and SMAD2L and ↓ PTEN level |
[24] |
| Prostate cancer | miR-182 | LNCap, PC-3, DU145, 22Rv1 and RWPE-1 |
miR-182: ↓NDRG1 ↓NDRG1: ↑ tumor cell proliferation |
[25] |
| Gastric cancer | DNMT family (DNA methylation) | SGC7901, MKN45,GES1 |
DNA methylation: ↓NDRG1 expression ↓NDRG1: ↑ tumor cell proliferation |
[26] |
| Gastric cancer | DNA methylation | - |
DNA methylation: ↓NDRG1 expression ↓NDRG1: ↑ tumor cell invasion and migration |
[27] |
| Gastric cancer | miR-223-3p/NDRG1 axis, Hsa_circ_0003159 | GES-1, NUGC-3, AGS, HS-746 T and N87 |
Hsa_circ_0003159 suppresses proliferation, migration and invasion but promotes apoptosis by decreasing miR-223-3p and increasing NDRG1 ↓NDRG1: ↑ tumor cell growth |
[28] |
| Gastric carcinoma | Several cellular signaling pathways | – | ↓NDRG1: ↑ tumor aggressiveness | [29] |
| Gastric cancer | MMP-9 | SGC7901 |
Δ NDRG1: ↑ cell proliferation and invasion |
[30] |
| Colorectal cancer | CLDN2/ZO1/ZONAB-NDRG1 axis | Lovo, SW480, SK-CO15, HCT116, LIM1215, SW620, Caco2, HT29, SW480, NCM460 and HEK293T | CLDN2 depletion significantly promotes NDRG1 transcription, leading to termination of tumor growth and metastasis | [31] |
| Colorectal cancer | CDC42 | HCT116, RKO |
Δ NDRG1: ↑ dissemination of CRC cells ↓NDRG1: ↑formation of filopodia and invasiveness of cancer |
[32] |
| Colorectal cancer | EGFR trafficking | RKO and HCT116 |
NDRG1 enhances the sensitivity colorectal cancer to CTX NDRG1 can increase CTX activity in metastatic cancer |
[33] |
| Colorectal cancer | MORC2 | HT‐29, SW‐480, SW‐620 and HEK‐293 | MORC2 downregulates NDRG1 mRNA, protein levels, and decreases its promoter activity | [34] |
| Colorectal cancer | caveolin-1 ubiquitylation | HT29, SW480 and SW1116 |
↓NDRG1: ↑ EMT, migration and invasion NDRG1 decreases expression of caveolin-1 protein via enhancing its ubiquitylation and consequent degradation |
[35] |
| Colorectal cancer | nuclear β-catenin and CD44 | HT29 and HCT116 |
The anti-metastatic activity of NDRG1 in CRC is mediated via decreasing expression of nuclear β-catenin Δ NDRG1: ↑tumorigenic ability and stem cell-like properties |
[36] |
| Rectal cancer | Genes inducing resistance to ionizing radiation | SNU-61, SNU-283, SNU-503, SNU-977, SNU-977R80Gy, SNU-1411, and SNU-1411R80Gy | NDRG1 levels were found to be increased in radio-resistant cell lines | [37] |
| Osteosarcoma | PI3K/AKT pathway, miR-96-5p | MG63,U2OS, HOS, 143B and hFOB 1.19 | ↑ LncRNA NDRG1: ↑ tumor cell proliferation and migration | [38] |
| Osteosarcoma | mitochondrial function and CSCs differentiation |
hFOB, U2OS, and MG63 |
↓NDRG1: ↓ cell viability, invasion ability and ↑ cell apoptosis increased Δ NDRG1: promotes the CSCs differentiation and decreases cancer progression |
[39] |
| Osteosarcoma | HER4 |
Saos2, MCF-7, CCHO and MG63.2 |
↑↑NDRG1: ↑ cell growth and ↓ apoptosis Δ HER4: ↓ cell growth and tumorigenesis, and ↑ cell senescence and apoptosis and ↑chemosensitivity |
[40] |
| Ovarian cancer | CD105 | OVCAR3 (OC3), PTX-resistant OC3/TAX300 |
Δ CD105: ↑ NDRG1 expression ↓NDRG1: ↑ tumor cell proliferation |
[41] |
| Cervical cancer | ANXA2-NDRG1-STAT1 | CaSki and C-4I | ↓ANXA2 and NDRG1, and ↑ STAT1 predict sensitivity of patients to concomitant CRT | [42] |
| Cervical and ovarian cancer | LPXN, DDR2, COL6A1, IL6, IL8, FYN, PTP4A3, PAPPA, ETV5, CYGB and CLCA2 | CaSki and HO-8910PM | Δ NDRG1: ↑ tumor cell adhesion, migration and invasion activities without affecting cell proliferation | [43] |
| Endometrial carcinoma | PTEN | – | ↑↑NDRG1 and ↓PTEN: ↑tumor cell proliferation | [44] |
| Lung cancer | ATF3 | Human bronchial epithelial (HBE) cells, lung carcinoma cell lines |
↑↑NDRG1: ↓ cisplatin-induced cytotoxicity in lung cancer A549 cells ↑↑NDRG1: ↓ATF3 |
[45] |
| Lung cancer | HIF-1α | A549 |
↑↑NDRG1: ↑ tumor cell proliferation and ↓apoptosis HIF-1α binds to NDRG1 promoter and activates its expression |
[46] |
| Lung adenocarcinoma | Digoxin, HIF-1α | A549 |
↑↑NDRG1: ↑tumor cell proliferation Hypoxia-induced up-regulation of VEGF, NDRG1, and HIF-1α is suppressed by digoxin |
[47] |
| Non-small-cell lung cancer | NDRG1/Cap43/Drg-1 axis | A549, PC9, 11_18, LK87, LC-1, QG56, LC-Sq-1, and RERF-LC-AI | Δ NDRG1/Cap43: ↓ tumor growth and angiogenesis | [48] |
| Bladder cancer | epithelial-mesenchymal transition (EMT) | 5637, T24 and UMUC3 |
↑↑NDRG1: ↑cell proliferation, migration, invasion and ↓apoptotic cell numbers Δ NDRG1: ↓ cell proliferation, migration, invasion and ↑apoptotic cell numbers |
[49] |
| Hepatocellular carcinoma | LINC00844 | HCCLM9, SMMC-7721, SK-Hep1, and HepG2 |
↑NDRG1: ↑ tumor cell proliferation ↑↑ LINC00844: proliferation, migration, and invasion LINC00844 can suppress expression of NDRG1 |
[50] |
| Hepatocellular Carcinoma | miR-188-3p and miR-133b | HepG2 | ↑miR-188-3p and miR-133b: ↓ NRDG1 expression and ↓ cell growth and cell migration | [51] |
| Hepatocellular carcinoma | LINC01419 |
SMMC7721 and SK-Hep1 |
↑ LINC01419: ↑ NDRG1 promoter activity and cell proliferation, migration and invasion | [52] |
| Esophageal squamous cell carcinoma | GC-GR pathway, GR /Sgk1/NDRG1 axis | – | ↑activity of the GC-GR pathway with↑ induction of Sgk1 and NDRG1: tumor progression and development of chemoresistance | [53] |
| Nasopharyngeal cancer | Smad2 signaling | 5–8F and 5–8F-LN |
ΔNDRG1: ↑ tumor cell proliferation, migration, and invasion and induced EMT ↓NDRG1: ↑ tumor cell proliferation and metastasis |
[54] |
| Pancreatic cancer | Wnt/tenascin C | AsPC-1, PANC-1, and MIAPaCa-2 |
NDRG1 inhibits Wnt/TnC (antioncogenic activity) ↓NDRG1: ↑ tumor cell proliferation |
[55] |
| Pancreatic Cancer | Histone Deacetylase inhibitor | Capan-1 and PANC-1 | ↓NDRG1: ↑ tumor cell proliferation and invasion | [56] |
| Clear cell renal cell carcinoma | HIF-1/2α | 786‐O, Caki‐1, RCC4/EV and RCC4/VHL | ↓NDRG1: ↑ tumor cell proliferation, metastasis and invasion | [57] |
| Kaposi’s sarcoma-associated herpesvirus | PCNA | MM, KMM, SLK, iSLK.RGB, iSLK.LANAstop, and HEK293T, BCBL1, JSC1, BC3, DG75, Raji, Loukes, Ramous, HUVECs |
↑NDRG1: ↑ tumor cell proliferation ΔNDRG1: ↓ viral genome copy number in tumor cells |
[58] |
Animal studies
The bulk of evidence from animal studies has indicated a tumor suppressor role for NDRG1 in colorectal, gastric, nasopharyngeal and renal cell cancers (Table 2). However, in osteosarcoma models, up-regulation of NDRG1 has been associated with higher rate of tumor growth [40]. Similarly, in non-small cell carcinoma, NDRG1 silencing has attenuated tumor growth and reduced angiogenesis [48].
Table 2.
Animal studies on the role of NDRG1
| Tumor type | Results | References |
|---|---|---|
| Colorectal cancer | ΔNDRG1: ↑tumor growth and liver metastasis | [31] |
| Colorectal cancer | ΔNDRG1: ↑invasion and metastasis | [32] |
| Colorectal cancer | ΔNDRG1: ↑tumor growth and tumor weight | [33] |
| Colorectal cancer | ΔNDRG1: ↑tumor growth and lung metastasis | [34] |
| Colorectal cancer | ΔNDRG1: ↑invasion and metastasis | [35] |
| Colorectal cancer | ΔNDRG1: ↑tumor growth | [36] |
| Breast cancer (TNBC) |
ΔYAP1(interaction with NDRG1): ↓tumor growth ↑↑ NDRG1: ↑ cell growth |
[4] |
| Breast cancer |
NDRG1 was co-expressed with and β-casein or MFP Up-regulation of NDRG1 resulted in the expansion of the differentiated area |
[2] |
| Gastric cancer | ΔNDRG1: ↑tumor weight and tumor volume | [28] |
| Nasopharyngeal cancer | ΔNDRG1: ↑cell proliferation | [54] |
| Non-small-cell lung cancer | ΔNDRG1: ↓tumor growth, tumor volume and angiogenesis | [48] |
| Osteosarcoma | Δ LncRNA NDRG1: ↓Tumor mass and volume and lung metastasis | [38] |
| Osteosarcoma |
ΔHER4 (interaction counterpart of NDRG1): ↓ tumor growth ↑↑NDRG1: ↑ tumor growth |
[40] |
| Prostate cancer |
↓ NDRG1: ↑ tumor growth Δ N-cadherin (interaction counterpart of NDRG1): ↓invasion and metastasis |
[14] |
| Prostate cancer | ↑↑ MLL5α (interaction counterpart of NDRG1): ↓tumor growth | [16] |
| Prostate cancer | Δ E6AP (interaction counterpart of NDRG1): ↓tumor growth | [21] |
| Clear cell renal cell carcinoma | Δ NDRG1: ↑tumor growth, ↑tumor volume and lung metastasis | [57] |
Studies in clinical samples
NDRG1 has been suggested as a prognostic marker in patients with inflammatory breast cancer. Based on the results of univariate analyses, expression level of NDRG1, tumor grade, clinical stage and estrogen receptor (ER) status have been associated with overall and disease-free survival times. Patient with over-expression of NDRG1 have exhibited poor survival times. Particularly, those having over-expression of NDRG1 and ER-negative status have been shown to have worse outcome [59].
A meta-analysis of NDRG1 levels in numerous publicly available databases has shown correlation between NDRG1 up-regulation and expression of glycolytic and hypoxia-associated genes. Moreover, over-expression of NDRG1 has been associated with enhancement of metastasis and increase in patients' mortality [6]. Moreover, another study has revealed methylation of the NDRG1 promoter in about one third of primary breast cancer specimens. Moreover, methylation of promoter of this gene has been correlated with the TNM stage, metastasis, lymph node involvement, moderate and poor histological grade in these patients [9].
Proteomic analysis in human prostate cancer and benign prostatic hyperplasia samples have revealed association between over-expression of YWHAZ and NDRG1 and poor prognosis based on Gleason scores. In fact, YWHAZ and NDRG1 expression levels could define two groups of prostate cancer patients with high and intermediate risks of mortality. Based on the multivariable analyses, expression levels of these genes predict prognosis in an independent manner from Gleason scores [20].
Expression of NDRG1 has been found to be down-regulated in gastric cancer samples. Notably, its expression has been negatively correlated with the expression levels of DNMT1, DNMT3A and DNMT3B. Consistent with this finding, DNA methylation status of NDRG1 has been positively correlated to DNMT family. NDRG1 levels have been inversely correlated with invasion depth. However, levels of NDRG1 and DNMT1 have not been associated with prognosis of gastric cancer [26]. The latter finding is in contrast with another study in gastric cancer which has reported association between down-regulation of NDRG1 and poor clinical outcome [27].
Studies in other types of cancers have also revealed dysregulation of protein or mRNA levels of NDRG1 and associations between this abnormal expression pattern and malignant behavior of cancer as reflected in survival time of patients (Table 3).
Table 3.
Expression studies in clinical samples on the role of NDRG1
| Tumor type | Samples | Expression (tumor vs. normal) | Kaplan–Meier analysis (impact of NDRG1 dysregulation) | Univariate/multivariate cox regression | Association of dysregulation of NDRG1 with clinical data | References |
|---|---|---|---|---|---|---|
| Breast cancer (TNBC) | Up | Shorter OS | – | – | [4] | |
| Breast cancer (TNBC) | TCGA database (963 patients) | Up | Shorter OS | – | – | [5] |
| Breast cancer (IBC) | 64 patients | Up | Shorter OS and DFS | NDRG1 was an independent prognostic factor for OS and DFS | Negative HER2 status | [59] |
| Breast cancer | TCGA database | Up | Shorter OS | – | – | [6] |
| Breast cancer | 389 patients | Down | – | – | NDRG1 promoter methylation was correlated with TNM at stage III/IV, metastasis, lymph invasion, moderate and poor histological grade | [9] |
| Breast cancer | 45 patients | Down | – | – | Differentiation status | [2] |
| Bladder cancer | 100 patients | Up | Shorter OS | NDRG1 expression was an independent prognostic factor | Lymph node metastasis, TNM stage | [49] |
| Cervical cancer | 40 patients | – | – | [42] | ||
| Colorectal cancer | 104 CRC tumors and 85 adjacent normal mucosa | Down | Shorter OS | – | – | [31] |
| Colorectal cancer | 86 PTN | Down | – | – | Advanced T stage | [32] |
| Colorectal cancer | 65 patients | Down | Poor OS | – | – | [33] |
| Colorectal cancer | 64 PTN | Down | – | – | – | [35] |
| Colorectal cancer | 116 patient | Down | – | – | Positive lymph node metastasis | [36] |
| Colorectal cancer | 119 CRC tissues and 36 Non-tumor colon tissues | Down | Shorter OS | – | Lymph node metastasis and poor pTNM stage | [34] |
| Endometrial carcinoma | 103 patient | Up | – | – | – | [44] |
| Esophageal squamous cell carcinoma | 98 patient | Up | Shorter OS and DFS | Univariate analysis showed association between patients' survival and expression of NDRG1 | pT, pStage, and lymphovascular invasion | [53] |
| Gastric cancer | 34 PTN | Down | Shorter OS | – | NDRG1 expression was negatively associated with tumor size, depth of invasion, lymph node metastasis, lymphatic invasion and differentiation | [27] |
| Gastric cancer |
TCGA database 407 patients |
Down | – | – | Invasion depth | [26] |
| Gastric cancer | 101 patients | Down | – | – | Degree of tumor cell differentiation, invasion depth, lymph node metastasis and TNM stage | [30] |
| Gastric cancer | 228 patients | Down | Shorter OS | Univariate Cox: VM, HER2, tumor size, TNM stage, lymphatic metastasis, distant metastasis and metastasis and recurrence Multivariate Cox: Only HER2, metastasis and recurrence were independent risk factors in gastric cancer | Tumor histological differentiation, TNM, Lauren type, lymph node metastasis, distant metastasis, recurrence and metastasis, and HER2 expression | [29] |
| Gastric cancer | 55 PTN | Down | – | – | – | [28] |
| Hepatocellular carcinoma | 40 PTN | Up | – | – | Histological grade | [50] |
| Hepatocellular carcinoma | 43 PTN | Up | – | – | – | [51] |
| Nasopharyngeal cancer | 83 patients | Down | Shorter OS | Lymphatic metastasis | – | [54] |
| Non-small-cell lung cancer | 182 patients | Up | Shorter OS | – | Age and cytoplasmic NDRG1/Cap43 expression | [48] |
| Osteosarcoma | 18 patients | Up | Shorter OS | – | Enneking stage and distant metastasis | [38] |
| Ovarian cancer | 53 patients | Down | – | – | – | [41] |
| Prostate cancer | TCGA database | Up | Shorter OS | YWHAZ, NDRG1, GS, and age are independent predictors of mortality | GS, age group, and TMPRSS2-ERG fusion | [20] |
| Prostate cancer | 60 patients | Down | Shorter OS | – | – | [14] |
| Prostate cancer | 31 patients | Down | – | – | – | [15] |
| Prostate cancer | 45 patients | Down | Shorter OS | – | – | [16] |
| Clear cell renal cell carcinoma | 645 tumor samples and 260 adjacent normal tissues | Down | – | – | – | [57] |
OS overall survival, DFS disease free survival, PTN parried tumor and non-tumoral samples
Discussion
The role of NDRG1 has been vastly assessed in the contexts of breast, prostate, gastric and colorectal cancers. NDRG1 is an example of proteins with different roles in the carcinogenesis. These various and opposite effects can be at least partly explained by cell- or tissue-specific roles. However, experiments in a certain type of cancer have sometimes reported opposite effects for NDRG1. This is particularly true in breast and prostate cancers, two types of cancer that have been the focus of several independent studies. This discrepancy can be explained by differences in the stage, grade or other pathological features of cancer cells. Since tumor tissues are heterogeneous in terms of gene expression patterns, single cell gene expression profiling is needed to elaborate expression of NDRG1 in relation with pathological state.
The lncRNA encoded by the same region (NDRG1-OT1) has been shown to effectively influence expression of NDRG1. Aryl hydrocarbon receptor and a number of miRNAs mediate other mechanisms for regulation of expression NDRG1. Finally, methylation status of NDRG1 promoter has an established role in the regulation of its expression based on the results of studies in different tissues/ cell types.
Most importantly, the lncRNA encoded from NDRG1 locus has been shown to serve as a spong for miR-96-5p [38, 60, 61]. Since competing endogenous RNA function is an important mechanism for the regulation of expression of genes by lncRNAs, further studies should find other miRNAs that are sponged by this lncRNA to unravel additional parts from the regulatory network of NDRG1.
Expression profiles of NDRG1 in clinical samples have revealed association between dysregulation of NDRG1 and poor clinical outcomes, demonstrating the prognostic impact of this protein in the context of cancer. However, the role of this protein in the early diagnosis of cancer, particularly based on expression assays in biofluids should be discovered in future.
NDRG1 has been shown to interact with a number of transcription factors and signaling pathways such as YAP1, AP-1, aryl hydrocarbon receptor, β-casein, androgen receptor, N-cadherin/c-Jun, MLL5α, E6AP, PTEN, ATF3, HIF-1α and PI3K/AKT, Smad2, Wnt/tenascin C, TGF-β and ERK pathways. Moreover, miR-769-3p, miR-182, miR-223-3p, miR-96-5p, miR-188-3p, miR-133b, hsa_circ_0003159, LINC00844 and LINC01419 are among non-coding RNAs that interact with NDRG1. Therefore, the oncogenic versus tumor suppressor roles of NDRG1 in different tissues should be interpreted considering the extensive number of NDRG1 counterparts in each situation.
Based on the importance of NDRG1 in the pathogenesis of cancer and induction or modulation of chemo-/radio-resistance in different types of cancers, future studies are required to unravel the exact role of this protein in the tissue-specific carcinogenic processes and develop specific therapies for each type of cancer.
Taken together, the data presented above casts doubt on the previously supposed "anti-metastatic" role for NDRG1 and suggests a tissue- or cell- or stage-specific role for this protein in the carcinogenesis.
Acknowledgements
The authors would like to thank the clinical Research Development Unit (CRDU) of Loghman Hakim Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran for their support, cooperation and assistance throughout the period of study.
Author contributions
SGF: wrote the draft and revised it. MT: designed and supervised the study. SAT, BMH and GS: collected the data and designed the figures and tables. All the authors read the submitted version and approved it.
Funding
Open Access funding enabled and organized by Projekt DEAL. Not applicable.
Data availability
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Declarations
Conflict of interest
The authors declare they have no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mohammad Taheri, Email: Mohammad.taheri@uni-jena.de.
Guive Sharifi, Email: gibnow@yahoo.com.
References
- 1.Ellen TP, Ke Q, Zhang P, Costa M. NDRG1, a growth and cancer related gene: regulation of gene expression and function in normal and disease states. Carcinogenesis. 2008;29(1):2–8. doi: 10.1093/carcin/bgm200. [DOI] [PubMed] [Google Scholar]
- 2.Fotovati A, Abu-Ali S, Kage M, Shirouzu K, Yamana H, Kuwano M. N-myc downstream-regulated gene 1 (NDRG1) a differentiation marker of human breast cancer. Pathol Oncol Res. 2011;17(3):525–533. doi: 10.1007/s12253-010-9342-y. [DOI] [PubMed] [Google Scholar]
- 3.Echaniz-Laguna A, Degos B, Bonnet C, Latour P, Hamadouche T, Lévy N, et al. NDRG1-linked Charcot-Marie-Tooth disease (CMT4D) with central nervous system involvement. Neuromuscul Disord : NMD. 2007;17(2):163–168. doi: 10.1016/j.nmd.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Mao M, Jia Y, Chen Y, Yang J, Xu L, Zhang X, et al. HJURP regulates cell proliferation and chemo-resistance via YAP1/NDRG1 transcriptional axis in triple-negative breast cancer. Cell Death Dis. 2022;13(4):396. doi: 10.1038/s41419-022-04833-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mishra A, Bonello M, Byron A. Evaluation of gene expression data from cybrids and tumours highlights elevated NDRG1-driven proliferation in triple-negative breast cancer. Breast Cancer: Basic Clin Res. 2020;14:1178223420934447. doi: 10.1177/1178223420934447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sevinsky CJ, Khan F, Kokabee L, Darehshouri A, Maddipati KR, Conklin DS. NDRG1 regulates neutral lipid metabolism in breast cancer cells. Breast Cancer Res. 2018;20(1):55. doi: 10.1186/s13058-018-0980-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Godbole M, Togar T, Patel K, Dharavath B, Yadav N, Janjuha S, et al. Up-regulation of the kinase gene SGK1 by progesterone activates the AP-1-NDRG1 axis in both PR-positive and -negative breast cancer cells. J Biol Chem. 2018;293(50):19263–19276. doi: 10.1074/jbc.RA118.002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeh CC, Luo JL, Nhut Phan N, Cheng YC, Chow LP, Tsai MH, et al. Different effects of long noncoding RNA NDRG1-OT1 fragments on NDRG1 transcription in breast cancer cells under hypoxia. RNA Biol. 2018;15(12):1487–1498. doi: 10.1080/15476286.2018.1553480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han LL, Hou L, Zhou MJ, Ma ZL, Lin DL, Wu L, et al. Aberrant NDRG1 methylation associated with its decreased expression and clinicopathological significance in breast cancer. J Biomed Sci. 2013;20(1):52. doi: 10.1186/1423-0127-20-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ai R, Sun Y, Guo Z, Wei W, Zhou L, Liu F, et al. NDRG1 overexpression promotes the progression of esophageal squamous cell carcinoma through modulating Wnt signaling pathway. Cancer Biol Ther. 2016;17(9):943–954. doi: 10.1080/15384047.2016.1210734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu W, Zhang B, Hu Q, Qin Y, Xu W, Shi S, et al. A new facet of NDRG1 in pancreatic ductal adenocarcinoma: Suppression of glycolytic metabolism. Int J Oncol. 2017;50(5):1792–1800. doi: 10.3892/ijo.2017.3938. [DOI] [PubMed] [Google Scholar]
- 12.Sun J, Zhang D, Bae D-H, Sahni S, Jansson P, Zheng Y, et al. Metastasis suppressor, NDRG1, mediates its activity through signaling pathways and molecular motors. Carcinogenesis. 2013;34(9):1943–1954. doi: 10.1093/carcin/bgt163. [DOI] [PubMed] [Google Scholar]
- 13.Ledet RJ, Ruff SE, Wang Y, Nayak S, Schneider JA, Ueberheide B, et al. Identification of PIM1 substrates reveals a role for NDRG1 phosphorylation in prostate cancer cellular migration and invasion. Commun Biol. 2021;4(1):36. doi: 10.1038/s42003-020-01528-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quan Y, Zhang X, Butler W, Du Z, Wang M, Liu Y, et al. The role of N-cadherin/c-Jun/NDRG1 axis in the progression of prostate cancer. Int J Biol Sci. 2021;17(13):3288–3304. doi: 10.7150/ijbs.63300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim SC, Geleta B, Maleki S, Richardson DR, Kovačević Ž. The metastasis suppressor NDRG1 directly regulates androgen receptor signaling in prostate cancer. J Biol Chem. 2021;297(6):101414. doi: 10.1016/j.jbc.2021.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quan Y, Cui Y, Wahafu W, Liu Y, Ping H, Zhang X. MLL5α activates AR/NDRG1 signaling to suppress prostate cancer progression. Am J Cancer Res. 2020;10(5):1608–1629. [PMC free article] [PubMed] [Google Scholar]
- 17.Lin HC, Yeh CC, Chao LY, Tsai MH, Chen HH, Chuang EY, et al. The hypoxia-responsive lncRNA NDRG-OT1 promotes NDRG1 degradation via ubiquitin-mediated proteolysis in breast cancer cells. Oncotarget. 2018;9(12):10470–10482. doi: 10.18632/oncotarget.23732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li EY, Huang WY, Chang YC, Tsai MH, Chuang EY, Kuok QY, et al. Aryl hydrocarbon receptor activates NDRG1 transcription under hypoxia in breast cancer cells. Sci Rep. 2016;6:20808. doi: 10.1038/srep20808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo EC, Chang YC, Sher YP, Huang WY, Chuang LL, Chiu YC, et al. MicroRNA-769-3p down-regulates NDRG1 and enhances apoptosis in MCF-7 cells during reoxygenation. Sci Rep. 2014;4:5908. doi: 10.1038/srep05908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lage-Vickers S, Bizzotto J. The expression of YWHAZ and NDRG1 predicts aggressive outcome in human prostate cancer. Commun Biol. 2021;4(1):103. doi: 10.1038/s42003-020-01645-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gamell C, Bandilovska I, Gulati T, Kogan A, Lim SC, Kovacevic Z, et al. E6AP promotes a metastatic phenotype in prostate cancer. iScience. 2019;22:1–15. doi: 10.1016/j.isci.2019.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Pan P, Qiao P, Liu R. Downregulation of N-myc downstream regulated gene 1 caused by the methylation of CpG islands of NDRG1 promoter promotes proliferation and invasion of prostate cancer cells. Int J Oncol. 2015;47(3):1001–1008. doi: 10.3892/ijo.2015.3086. [DOI] [PubMed] [Google Scholar]
- 23.Ghalayini MK, Dong Q, Richardson DR, Assinder SJ. 2013. Proteolytic cleavage and truncation of NDRG1 in human prostate cancer cells, but not normal prostate epithelial cells. Biosci Rep. [DOI] [PMC free article] [PubMed]
- 24.Dixon KM, Lui GY, Kovacevic Z, Zhang D, Yao M, Chen Z, et al. Dp44mT targets the AKT, TGF-β and ERK pathways via the metastasis suppressor NDRG1 in normal prostate epithelial cells and prostate cancer cells. Br J Cancer. 2013;108(2):409–419. doi: 10.1038/bjc.2012.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu R, Li J, Teng Z, Zhang Z, Xu Y. Overexpressed microRNA-182 promotes proliferation and invasion in prostate cancer PC-3 cells by down-regulating N-myc downstream regulated gene 1 (NDRG1) PLoS ONE. 2013;8(7):e68982. doi: 10.1371/journal.pone.0068982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang X, Ma J, Xue X, Wang G, Yan T, Su L, et al. DNMT family induces down-regulation of NDRG1 via DNA methylation and clinicopathological significance in gastric cancer. PeerJ. 2021;9:e12146. doi: 10.7717/peerj.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao XJ, Zheng HC. NDRG1 was downregulated and worked as favorable biomarker in the development of gastric cancer. Transl Cancer Res. 2020;9(1):210–221. doi: 10.21037/tcr.2019.12.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Lv W, Lin Z, Wang X, Bu J, Su Y. Hsa_circ_0003159 inhibits gastric cancer progression by regulating miR-223-3p/NDRG1 axis. Cancer Cell Int. 2020;20:57. doi: 10.1186/s12935-020-1119-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong X, Hong Y, Sun H, Chen C, Zhao X, Sun B. NDRG1 suppresses vasculogenic mimicry and tumor aggressiveness in gastric carcinoma. Oncol Lett. 2019;18(3):3003–3016. doi: 10.3892/ol.2019.10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang X, Xu X, Xue X, Ma J, Li Z, Deng P, et al. NDRG1 controls gastric cancer migration and invasion through regulating MMP-9. Pathol Oncol Res. 2016;22(4):789–796. doi: 10.1007/s12253-016-0071-8. [DOI] [PubMed] [Google Scholar]
- 31.Wei M, Zhang Y, Yang X, Ma P, Li Y, Wu Y, et al. Claudin-2 promotes colorectal cancer growth and metastasis by suppressing NDRG1 transcription. Clin Transl Med. 2021;11(12):e667. doi: 10.1002/ctm2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aikemu B, Shao Y, Yang G, Ma J, Zhang S, Yang X, et al. NDRG1 regulates Filopodia-induced colorectal cancer invasiveness via modulating CDC42 activity. Int J Biol Sci. 2021;17(7):1716–1730. doi: 10.7150/ijbs.56694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang G, Huang L. NDRG1 enhances the sensitivity of cetuximab by modulating EGFR trafficking in colorectal cancer. Oncogene. 2021;40(41):5993–6006. doi: 10.1038/s41388-021-01962-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Shao Y, He Y, Ning K, Cui X, Liu F, et al. MORC2 promotes development of an aggressive colorectal cancer phenotype through inhibition of NDRG1. Cancer Sci. 2019;110(1):135–146. doi: 10.1111/cas.13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mi L, Zhu F, Yang X, Lu J, Zheng Y, Zhao Q, et al. The metastatic suppressor NDRG1 inhibits EMT, migration and invasion through interaction and promotion of caveolin-1 ubiquitylation in human colorectal cancer cells. Oncogene. 2017;36(30):4323–4335. doi: 10.1038/onc.2017.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wangpu X, Yang X, Zhao J, Lu J, Guan S, Lu J, et al. The metastasis suppressor, NDRG1, inhibits “stemness” of colorectal cancer via down-regulation of nuclear β-catenin and CD44. Oncotarget. 2015;6(32):33893–33911. doi: 10.18632/oncotarget.5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim SC, Shin YK, Kim YA, Jang SG, Ku JL. Identification of genes inducing resistance to ionizing radiation in human rectal cancer cell lines: re-sensitization of radio-resistant rectal cancer cells through down regulating NDRG1. BMC Cancer. 2018;18(1):594. doi: 10.1186/s12885-018-4514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z, Wei Y, Zhu H, Yu L, Zhu J, Han Q, et al. LncRNA NDRG1 aggravates osteosarcoma progression and regulates the PI3K/AKT pathway by sponging miR-96-5p. BMC Cancer. 2022;22(1):728. doi: 10.1186/s12885-022-09833-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Zhao T, Meng Y, Wang Y, Wang W. NDRG1 regulates osteosarcoma cells via mediating the mitochondrial function and CSCs differentiation. J Orthop Surg Res. 2021;16(1):364. doi: 10.1186/s13018-021-02503-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, Sun W, Sun M, Fu Z, Zhou C, Wang C, et al. (2018) HER4 promotes cell survival and chemoresistance in osteosarcoma via interaction with NDRG1. Biochim et Biophys Acta Mol Basis Dis. 2018;1864(5 Pt A):1839–1849. doi: 10.1016/j.bbadis.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, Sang X, Zhang R, Chi J, Bai W. CD105 expression is associated with invasive capacity in ovarian cancer and promotes invasiveness by inhibiting NDRG1 and regulating the epithelial-mesenchymal transition. Am J Transl Res. 2021;13(11):12461–12479. [PMC free article] [PubMed] [Google Scholar]
- 42.Buttarelli M, Babini G, Raspaglio G, Filippetti F, Battaglia A, Ciucci A, et al. A combined ANXA2-NDRG1-STAT1 gene signature predicts response to chemoradiotherapy in cervical cancer. J Exp Clin Cancer Res: CR. 2019;38(1):279. doi: 10.1186/s13046-019-1268-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao G, Chen J, Deng Y, Gao F, Zhu J, Feng Z, et al. Identification of NDRG1-regulated genes associated with invasive potential in cervical and ovarian cancer cells. Biochem Biophys Res Commun. 2011;408(1):154–159. doi: 10.1016/j.bbrc.2011.03.140. [DOI] [PubMed] [Google Scholar]
- 44.Chen J, Li S, Yang Z, Lu G, Hu H. Correlation between NDRG1 and PTEN expression in endometrial carcinoma. Cancer Sci. 2008;99(4):706–710. doi: 10.1111/j.1349-7006.2008.00749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du A, Jiang Y, Fan C. NDRG1 downregulates ATF3 and inhibits cisplatin-induced cytotoxicity in lung cancer A549 cells. Int J Med Sci. 2018;15(13):1502–1507. doi: 10.7150/ijms.28055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Q, Li LH, Gao GD, Wang G, Qu L, Li JG, et al. HIF-1α up-regulates NDRG1 expression through binding to NDRG1 promoter, leading to proliferation of lung cancer A549 cells. Mol Biol Rep. 2013;40(5):3723–3729. doi: 10.1007/s11033-012-2448-4. [DOI] [PubMed] [Google Scholar]
- 47.Wei D, Peng JJ, Gao H, Li H, Li D, Tan Y, et al. Digoxin downregulates NDRG1 and VEGF through the inhibition of HIF-1α under hypoxic conditions in human lung adenocarcinoma A549 cells. Int J Mol Sci. 2013;14(4):7273–7285. doi: 10.3390/ijms14047273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azuma K, Kawahara A, Hattori S, Taira T, Tsurutani J, Watari K, et al. NDRG1/Cap43/Drg-1 may predict tumor angiogenesis and poor outcome in patients with lung cancer. J Thorac Oncol. 2012;7(5):779–789. doi: 10.1097/JTO.0b013e31824c92b4. [DOI] [PubMed] [Google Scholar]
- 49.Li A, Zhu X, Wang C, Yang S, Qiao Y, Qiao R, et al. Upregulation of NDRG1 predicts poor outcome and facilitates disease progression by influencing the EMT process in bladder cancer. Sci Rep. 2019;9(1):5166. doi: 10.1038/s41598-019-41660-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou W, Huang K, Zhang Q, Ye S, Zhong Z, Zeng C, et al. LINC00844 promotes proliferation and migration of hepatocellular carcinoma by regulating NDRG1 expression. PeerJ. 2020;8:e8394. doi: 10.7717/peerj.8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo Z, Fan Y, Liu X, Liu S, Kong X, Ding Z, et al. MiR-188–3p and miR-133b suppress cell proliferation in human hepatocellular carcinoma via post-transcriptional suppression of NDRG1. Technol Cancer Res Treat. 2021;20:15330338211033074. doi: 10.1177/15330338211033074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dang H, Chen L, Tang P, Cai X, Zhang W, Zhang R, et al. LINC01419 promotes cell proliferation and metastasis in hepatocellular carcinoma by enhancing NDRG1 promoter activity. Cell Oncol. 2020;43(5):931–947. doi: 10.1007/s13402-020-00540-6. [DOI] [PubMed] [Google Scholar]
- 53.Ueki S, Fujishima F, Kumagai T, Ishida H, Okamoto H, Takaya K, et al. GR, Sgk1, and NDRG1 in esophageal squamous cell carcinoma: their correlation with therapeutic outcome of neoadjuvant chemotherapy. BMC Cancer. 2020;20(1):161. doi: 10.1186/s12885-020-6652-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu ZY, Xie WB, Yang F, Xiao LW, Wang XY, Chen SY, et al. NDRG1 attenuates epithelial-mesenchymal transition of nasopharyngeal cancer cells via blocking Smad2 signaling. Biochem Biophys Acta. 2015;1852(9):1876–1886. doi: 10.1016/j.bbadis.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geleta B, Tout FS, Lim SC, Sahni S, Jansson PJ, Apte MV, et al. Targeting Wnt/tenascin C-mediated cross talk between pancreatic cancer cells and stellate cells via activation of the metastasis suppressor NDRG1. J Biol Chem. 2022;298(3):101608. doi: 10.1016/j.jbc.2022.101608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tiffon C. Histone deacetylase inhibition restores expression of hypoxia-inducible protein NDRG1 in pancreatic cancer. Pancreas. 2018;47(2):200–207. doi: 10.1097/MPA.0000000000000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang ZY, Zhang SL, Chen HL, Mao YQ, Li ZM, Kong CY, et al. The up-regulation of NDRG1 by HIF counteracts the cancer-promoting effect of HIF in VHL-deficient clear cell renal cell carcinoma. Cell Prolif. 2020;53(7):e12853. doi: 10.1111/cpr.12853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang F, Liang D, Lin X, Zou Z, Sun R, Wang X, et al. NDRG1 facilitates the replication and persistence of Kaposi’s sarcoma-associated herpesvirus by interacting with the DNA polymerase clamp PCNA. PLoS Pathog. 2019;15(2):e1007628. doi: 10.1371/journal.ppat.1007628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Villodre ES, Gong Y, Hu X, Huo L, Yoon EC. NDRG1 expression is an independent prognostic factor in inflammatory breast cancer. Cancers. 2020 doi: 10.3390/cancers12123711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghafouri-Fard S, Shoorei H, Taheri M. Non-coding RNAs are involved in the response to oxidative stress. Biomed Pharmacother. 2020 doi: 10.1016/j.biopha.2020.110228. [DOI] [PubMed] [Google Scholar]
- 61.Ghafouri-Fard S, Abak A, Shoorei H, Mohaqiq M, Majidpoor J, Sayad A, et al. Regulatory role of microRNAs on PTEN signaling. Biomed Pharmacother. 2021 doi: 10.1016/j.biopha.2020.110986. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Ghalayini MK, Dong Q, Richardson DR, Assinder SJ. 2013. Proteolytic cleavage and truncation of NDRG1 in human prostate cancer cells, but not normal prostate epithelial cells. Biosci Rep. [DOI] [PMC free article] [PubMed]
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.