Abstract
Glioblastoma, IDH wild-type is the most common and aggressive form of glial tumors. The exact mechanisms of glioblastoma oncogenesis, including the identification of the glioma-initiating cell, are yet to be discovered. Recent studies have led to the hypothesis that glioblastoma arises from neural stem cells and glial precursor cells and that cell lineage constitutes a key determinant of the glioblastoma molecular subtype. These findings brought significant advancement to the comprehension of gliomagenesis. However, the cellular origin of glioblastoma with mesenchymal molecular features remains elusive. Mesenchymal stromal cells emerge as potential glioblastoma-initiating cells, especially with regard to the mesenchymal molecular subtype. These fibroblast-like cells, which derive from the neural crest and reside in the perivascular niche, may underlie gliomagenesis and exert pro-tumoral effects within the tumor microenvironment. This review synthesizes the potential roles of mesenchymal stromal cells in the context of glioblastoma and provides novel research avenues to better understand this lethal disease.
Keywords: Glioblastoma, Mesenchymal stromal cells, Perivascular fibroblasts, Neural crest, Microenvironment, Pericytes, Gliomagenesis
Introduction
Glioblastoma (GB), IDH wild-type (wt) is the most common and aggressive form of glial tumors, accounting for almost 50% of primary malignant central nervous system (CNS) tumors [1, 2]. It is classified as grade 4 in the World Health Organization (WHO) classification of tumors of the CNS. It belongs to the «adult-type diffuse glioma» family that also includes astrocytoma IDH-mutant (WHO grade 2, 3, or 4) and oligodendroglioma, IDH-mutant and 1p/19q-codeleted (WHO grade 2 or 3) [1].
As opposed to astrocytoma IDH-mutant WHO grade 4 (formerly, GB, IDH-mutant), GB, IDHwt arises de novo (without preexisting precursor lesion) and typically manifests rapidly after a short clinical history. Despite an aggressive multimodal therapeutic approach, GB IDHwt is associated with a dismal prognosis, showing a median survival of 8 months and an overall 5-year relative survival rate of 5.5% [2].
The exact mechanisms of glioblastoma oncogenesis are yet to be discovered. Over the last two decades, extensive and comprehensive molecular profiling of GB has brought new insights into gliomagenesis [3, 4]. The genomic and epigenomic landscape of GB have been thoroughly described, and biological subgroups have emerged, defining three molecular subtypes based on gene expression profiling signatures: proneural, classical, and mesenchymal [3, 5–7]. To date, it has not strongly impacted clinical practice, likely owing to marked intratumoral heterogeneity and differentiation plasticity of GB [8]. However, it has provided new research avenues to understand better the GB pathogenesis, including the identification of the glioma-initiating cell.
Recent studies have led to the hypothesis that GB may arise from neural stem cells (NSC) and glial precursor cells, such as oligodendrocyte and astrocytic precursor cells [9, 10]. In addition, it has been shown that the originating cell lineage is crucial to tumor molecular stratification, independently of the driver mutation that it initially harbors [11, 12]. While glial or neuronal progenitor cells have been suggested to initiate proneural and classical GB, the cellular origin of mesenchymal glioblastoma remains elusive. Studies have described a potential proneural to mesenchymal transition (PMT) that may illustrate the transcriptomic plasticity of GB upon treatment or recurrence. Recently, neural crest (NC)-derived cells have emerged as potential cells of origin in mesenchymal GB [13]. Herein, we discuss perivascular mesenchymal stromal cells (pMSC), also referred to as vascular fibroblasts (vFB), which originate from the NC, as potential candidates for the initiation of GB and their role in GB development [14].
Glioblastoma
General characteristics
Epidemiology
GB, IDHwt is the most common malignant CNS tumor in adults. It accounts for approximately 15% of all intracranial neoplasms and almost 50% of all malignant CNS tumors. It preferentially affects older adults, with a peak incidence in patients aged 55–85 years (median age of 64 years). In the United States of America, GB, IDHwt is more common in males compared to females (M: F ratio of 1.58: 1) [1, 2]. To date, the only validated risk factor is ionizing radiation to the head and neck [15, 16]. On the contrary, decreased risk has been observed among individuals with a history of allergies or atopic diseases [16]. Despite a multimodal therapeutic approach that includes surgery, radiotherapy, and chemotherapy, prognosis remains poor, with a 5-year survival rate of 5.5% [2, 17, 18].
Definition of GB
GB, IDHwt is a diffusely infiltrating high cellular glioma that characteristically shows microvascular proliferation and/or necrosis. As the former term « GB multiforme» suggests, GB morphology has remarkable inter-tumoral and intra-tumoral heterogeneity. Cellular pleomorphism includes small, undifferentiated, spindled, lipidized, granular, epithelioid, and/or giant cells. Secondary structures of Scherer illustrate the different routes that glioma cells can take to invade the brain: 1) the white matter tracts, 2) the vasculature (perivascular satellitosis), 3) the leptomeningeal space and 4) the brain parenchyma.
By definition, GB, IDHwt lacks mutations in IDH1 codon 132 and IDH2 codon 172. Molecularly, demonstration of TERT promoter mutations, EGFR gene amplification, and/or a gain of chromosome 7/ loss of chromosome 10 genotype is sufficient for the diagnosis of GB [1].
GB Molecular pathways
PI3K–AKT–mTOR and Ras/MAPK/ERK pathways
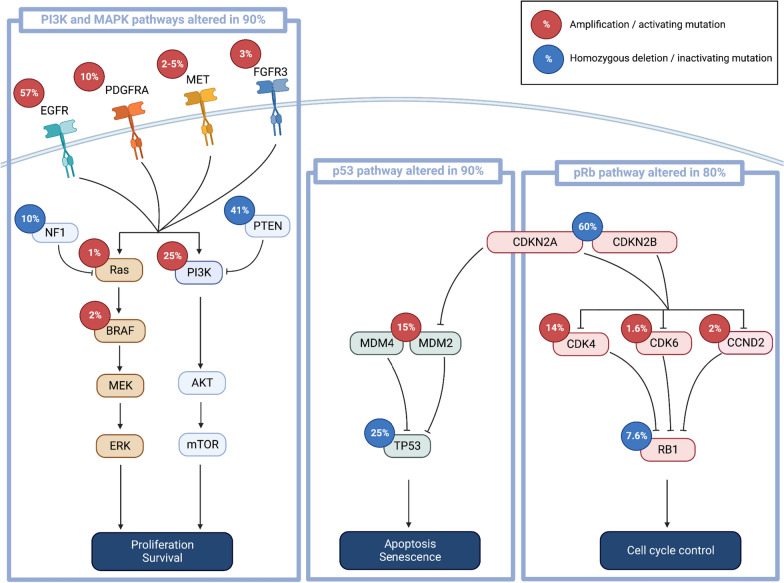
The PI3K and MAPK pathways, both activated by receptors tyrosine kinase (RTK), regulate many cellular processes, including cell proliferation. In approximately 90% of IDHwt GB, at least one activating alteration in the PI3K pathway is observed, including alterations of RTK genes, PI3K genes, and PTEN [3, 19]. Alterations of RTK genes are common in GB, involving EGFR (60%), PDGFRA (10–15%), MET (2–5%), or FGFR3 (~ 3%) [3, 5, 20]. PI3-kinase mutations are found in about 25% of GB [3]. In addition, the NF1 gene, which encodes neurofibromin that functions as a negative regulator of RAS signaling, is deleted or mutated in 10% of cases [3].
p14ARF–MDM2–MDM4–p53 pathway
The p53 pathway is altered in a large variety of cancer, including GB. Indeed, up to 90% of GB have an altered p53 signaling pathway, with mutation or deletion of TP53 in 20–25% of cases [3, 19]. In about 15% of GB, an amplification of MDM2 or MDM4 is observed, thus inhibiting p53 [3]. Homozygous deletion of CDKN2A locus, which encodes the p14ARF protein that inhibits MDM2, is detected in about 60% of GB, resulting in an inactivation of the p53 pathway and the pRB pathway (see below) [3].
CDK4/6–CDKN2A/B–RB1 cell-cycle pathway
The pRB pathway represents a critical cell cycle checkpoint, suppressing cell cycle entry (Fig. 1). CDK4 and CDK6 suppress the downstream inhibition of pRB, allowing the progression from G1 to S phase of the cell cycle. P16, encoded by CDKN2A, inhibits CDK4 and CDK6. Up to 80% of GB show at least one alteration of the pRB pathway, including CDKN2A deletions, amplifications of CDK4/CDK6, and inactivating alterations of RB1 [3, 19].
Fig. 1.
Signaling pathways involved in GB. Alteration rates are summarized for PI3K/MAPK, p53 and pRb regulatory pathways (created with Biorender.com)
GB molecular subtypes and the mesenchymal signature
Gene expression profiling has allowed the classification of GB into three distinct molecular subtypes: proneural, classical, and mesenchymal [3, 5–7]. Initially, this classification was based on the expression profile of 840 genes, but subsequent studies have shown that it can be simplified to rely on just 12 genes with good concordance (Table 1) [21]. However, despite their correlation with distinct genetic aberrations and clinical characteristics, these molecular subtypes have not gained clear significance in clinical practice.
Table 1.
Glioblastoma molecular subtypes
| GB subtypes | Marker genes |
|---|---|
| Proneural | P2RX7, STMN4, SOX10 and ERBB3 |
| Classical | ACSBG1 and KCNF1 |
| Mesenchymal | S100A, DAB2, TGFB1, THBS1, COL1A2, COL1A1 |
Glioblastomas can be classified into three molecular subtypes based on the expression profile of 12 genes
The proneural subtype is characterized by specific genetic alterations, including IDH1 mutation, TP53 mutation, PDGFRA amplification and/or mutation, and a glioma-CpG island methylator phenotype (G-CIMP) [5, 22]. Notably, both IDH1 mutations and G-CIMP are considered favorable prognostic factors. However, when excluding IDH-mutant tumors, the proneural subtype exhibits the worst prognosis among all subtypes [23].
The classical subtype is characterized by EGFR mutation/amplification and CDKN2A homozygous deletion.
Accounting for approximately 34% of GB, the mesenchymal subtype displays an expression profile characterized by mesenchymal markers, such as CHI3L1 and MET [5–7]. Mesenchymal subtype tumors are predominantly IDHwt and G-CIMP- and commonly harbor NF1 mutation [5, 22, 24]. In addition, they tend to correlate with poor response to radiation therapy and relatively poor outcome [7, 24]. The mesenchymal subtype is also characterized by high levels of angiogenic markers, such as CD31/PECAM-1, VEGF, flt1/VEGFR1, and kdr/VEGFR2 [6]. Furthermore, it exhibits high expressions of immune-related genes, particularly proinflammatory genes and immunosuppressive genes [3, 5, 6, 25]. Several of these genes are involved in the recruitment of monocytes/macrophages (CSF-1, CCL2, CCL-22, TREM1, and TREM2) and in the macrophage-polarization towards an immunosuppressive M2-phenotype (CD163, CD204) [25]. Notably, the mesenchymal subtype shows enrichment of macrophages and microglial cells, constituting the largest stroma cell population in GB [26–28].
It is important to note that while an initial neural subtype was described in this classification, it was later considered to be the result of contamination with normal cells [5, 21, 23].
The origin of GB
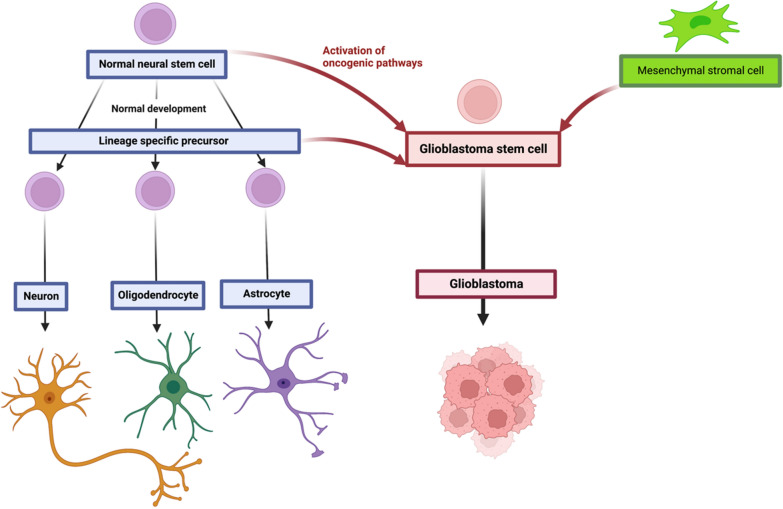
The exact cell of origin of GB has yet to be definitively identified. Several CNS cell types within the CNS, including neural precursor cells (NPC), oligodendrocyte precursor cells (OPC), and astrocytic precursor cells (APC), have been proposed as potential candidates for initiating GB (Fig. 2) [9, 29, 30]. Moreover, emerging evidence indicates that the cell lineage plays a crucial role in determining the molecular subtype of GB. Indeed, the introduction of identical driver mutations in different precursor cells leads to the development of distinct molecular subtypes [11, 12]. Studies have demonstrated that neural stem cells (NSC) in the subventricular zone carry the driver mutations responsible for GB, suggesting them as a potential cell of origin [10, 31, 32]. Further supporting this notion, single-cell RNA-sequencing (scRNAseq) studies have identified profiles resembling NPC, OPC, and APC, providing evidence for a neuronal/glial origin of GB [8, 33]. However, the cellular origin of GB with mesenchymal features, despite the well-described mesenchymal transcriptomic profile, remains elusive. The hypothesis of pMSC as GB-initiating cells will be discussed further (Fig. 2).
Fig. 2.
The origin of glioblastoma. During normal embryonic development and in the adult brain, normal neural stem cells generate glial and neuronal cells. Glioblastoma stem cells may arise from neural stem cells and/or glial precursor cells through the activation of oncogenic pathways. They may also originate from neural crest (NC)-derived, pMSC. During development, the NC arises from the neural tube and its component cells migrate and invade virtually all tissues, giving rise to numerous differentiated cells, such as pMSC, melanocytes, chondrocytes, peripheral neuronal and glial cells, thyroid C cells, and adrenergic cells (created with Biorender.com)
To support the hierarchical development model, CD133+ glioma stem cells (GSC) have been identified within GB tumors. These GSC exhibit remarkable proliferation capacity, self-renewal abilities, and differentiation potential [34, 35]. They are characterized by the expression of CD133, OCT4, CD44, nestin, and SOX2, although a specific marker exclusive to GSC has not yet been identified [34, 36, 37]. GSC are described as slow-dividing or quiescent cells that reside in protective microenvironments called GSC niches, contributing to intratumoral heterogeneity and therapy resistance [34, 38, 39]. GSC preferentially reside in the perivascular niche, interacting with endothelial cells in intricate bidirectional crosstalk, and in the perinecrotic niche (Fig. 3) [40].
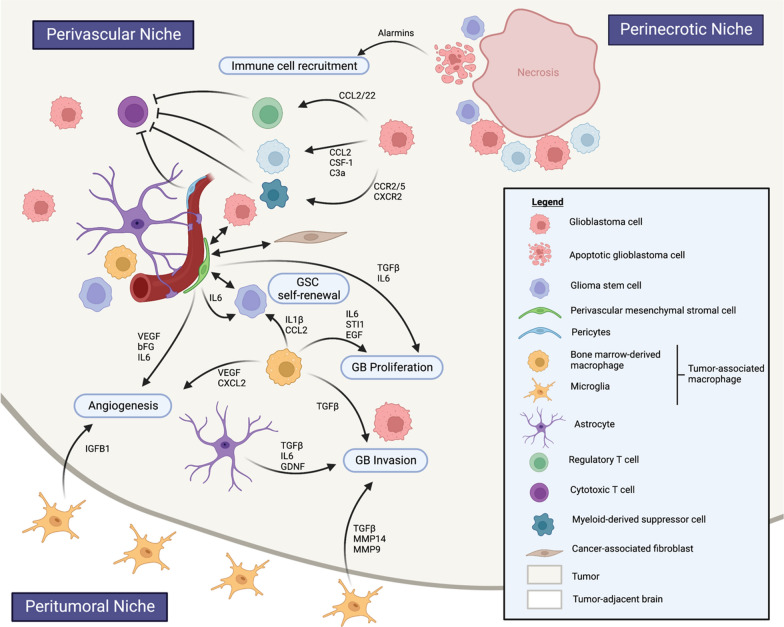
Fig. 3.
Glioblastoma tumor micro-environment. GB TME is compartmentalized in perivascular, perinecrotic and peritumoral niches. Tumor-associated macrophages (Bone marrow-derived macrophages and microglia) and mesenchymal stromal cells play key roles in supporting GB proliferation, invasion and angiogenesis (created with Biorender.com)
GB immune microenvironment
GB is a highly complex tissue composed of tumor cells and their surrounding microenvironment which supports tumor growth through a permissive neighborhood. The tumor microenvironment (TME) consists of cells (including immune cells, vascular cells, glial and neuronal cells, and stem cells), soluble factors, signaling molecules, and an extracellular matrix. It is a dynamic milieu considered to play an active role in tumorigenesis through reciprocal communication with cancer cells [41]. GB TME is compartmentalized in tumor niches which are critical regions where interactions between cancer cells and host cell populations are promoted (Fig. 3).
Tumor vasculature
One of the main features of GB is microvascular proliferation. The tumor vasculature plays a crucial role in supporting tumor growth through various mechanisms, including:
Angiogenesis This is the primary process involved in GB vascularization, triggered by the release of pro-angiogenic factors, such as vascular endothelial growth factor (VEGF), by tumor cells.
Vessel co-option Tumor cells possess the ability to migrate along existing blood vessels, enabling invasion of the brain through vascular routes [42].
Vasculogenesis This process involves the recruitment of endothelial cell progenitors derived from the bone marrow, contributing to the assembly of neo-vessels [43].
Transdifferentiating process GSC demonstrate the capacity to differentiate into tumor-derived endothelial cells, a phenomenon known as the transdifferentiating process [44, 45].
Vascular mimicry This refers to the presence of vascular structures within the tumor that result from GSC differentiating into vascular smooth muscle cells (vSMC) or pericytes (PC) [46, 47].
Macrophages
Immune cells may represent up to 50% of the GB tumor bulk [48]. Among these immune cells, tumor-associated macrophages (TAM) are the predominant population and are characterized by their origin, localization, and functions, encompassing both microglia and bone marrow-derived macrophages (BMDM) [49–51]. Notably, BMDMs represent approximately 85% of TAM and are primarily found in perivascular regions within the tumor, while microglia are localized in peri-tumoral areas [51, 52].
TAM can exhibit different activation states depending on environmental cues, polarizing into either type I response (M1 TAM) or type II response (M2 TAM) through classical or alternative activation, respectively. M1 TAM promote inflammation by producing pro-inflammatory cytokines, such as IL- 12, IL-1β, TNF-α, IL-6, and IL-23, while M2 TAM suppress inflammation by producing ARG1, IL-10 and IL-4 [53]. Initially, once recruited within the GB TME, TAM were considered to polarize toward an M2-like phenotype that promotes invasion, angiogenesis and immunosuppression [54–56]. However, recent studies have revealed that TAM encompass a dynamic entity that includes antitumoral M1-like, pro-tumoral M2-like, and non-polarized M0 phenotypes [27, 57].
Regarding GB molecular subtypes, mesenchymal GB exhibit higher AIF1 expression (encoding for IBA1), a marker associated with TAM [28]. These findings is consistent with previous studies showing increased infiltration of TAM in NF1-altered GB [7, 58]. In addition, it has been suggested that PMT was associated with increased TAM infiltration [24].
Functionally, TAM play a pivotal role in gliomagenesis through complex cross-talk with tumor and TME cells, contributing to tumor progression, immunosuppression, and cerebral edema [59]. TAM release factors such as TGFβ, IL-1β, IL-6, stress-inducible protein 1 (STI1), and epidermal growth factor (EGF) that stimulate tumor growth and invasion (Fig. 3) [60–63]. In addition, their immunosuppressive role includes the recruitment of CD4+ /FOXP3+ T regulatory (Treg) cells and myeloid-derived suppressor cells (Fig. 3) [64, 65]. Furthermore, due to their perivascular localization, TAM have been investigated for their involvement in cerebral edema. Studies have shown that dexamethasone, commonly used for the management of cerebral edema, inhibits TAM production of IL-1β, and genetic ablation of IL-1α/β or IL-1β in a murine GB model or the administration of a potential IL-1β inhibitor (Sulfasazaline) reduces cerebral edema [66, 67]. The potential role of TAM in vasogenic cerebral edema underscores the need for further investigations into the complex interaction of TAM with the components of the blood–brain barrier (BBB).
These findings point out the crucial role of the perivascular niche in gliomagenesis, by promoting angiogenesis, modulating the immune response, supporting tumor cell invasion, and providing a stem cell niche. Within this niche, pMSC are also present, and their role in GB development and progression will be discussed below.
Perivascular mesenchymal stromal in the CNS
Definition
First identified in the bone marrow (BM) and termed colony-forming unit fibroblasts (CFU-F), MSC are characterized in vitro by a spindle-shaped, fibroblast-like, plastic-adherent appearance [68, 69]. They are multipotent progenitor cells that have the ability to differentiate into adipocytes, chondrocytes, and osteoblasts [70–72]. Their multipotency has raised much interest in tissue engineering research for using culture-expanded MSC to replace injured or damaged mesenchymal tissue [73]. MSC express CD105 (Endoglin), CD73, and CD90 (Thy1) and lack the expression of CD45, CD34, CD14 or CD11b, CD79a or CD19, and HLA-DR [70, 71, 74]. Other markers, such as CD140b (PDGFRB), CD271 (Low-affinity NGF receptor), CD146 (Muc18), and CD248 have been suggested to identify MSC [75–78] but these markers are equally expressed by PC. As the concept of MSC was initially defined as a multipotent cell residing within the BM, it has evolved over the years to a wide concept that includes multipotent perivascular cells of any organ, including the CNS, as discussed below.
Origin of pMSC and PC in the CNS vasculature
MSC in adult tissues have two main embryonic origins, deriving either from the mesoderm or the NC [79–83]. During embryogenesis, MSC migrate along vessels and then reside in the perivascular niche of all adult tissues, adopting similar features to that of PC [80, 84].
The term ‘PC’ is often used in the literature to refer to microvascular periendothelial cells [85]. The accepted definition of a PC is a cell that is embedded within the vascular basement membrane, as observed by electron microscopy. However, because ultrastructural analyses are impractical, most published papers may not differentiate PC from other periendothelial cells, including vSMC and pMSC [14, 86–91]. It is now clear that different cell types exist in the periendothelial compartment but accurately identifying their phenotypes remains a challenge. PC cannot be definitively identified and distinguished from vSMC or pMSC using a single molecular marker. Commonly applied markers or genes (Table 2), such as NG2/Cspg4, CD13/Anpep, and desmin, are not specific and their expression is not stable, particularly in disease conditions. Other markers, such as CD248 (endosialin) and CD90 (Thy-1), are highly expressed by PC but recent investigations revealed that they are also expressed by pMSC, especially in the context of GBM [86]. In addition, it has been demonstrated that PC, originally defined by their vascular mural localization, have the same osteogenic, adipogenic, and myogenic potential as MSC and also express surface markers of MSC, such as CD44, CD73, CD90, and CD105 in vitro [80, 92].
Table 2.
Identification and markers (genes) of endothelial and perivascular cells in the CNS
| Cell types | Markers | Comments | Reference |
|---|---|---|---|
| Endothelial cells | CD31 | PECAM-1, cell adhesion | [86] |
| CD93 | CD248 family member | [86, 95] | |
| CLDN5 | Claudin 5 | [86] | |
| CDH5 | Cadherin 5, also expressed by CNS fibroblasts | [86] | |
| Pericytes (PC) | PDGFRβ | Receptor for platelet derived growth factor | [85] |
| NG2 | Encoded by Chondroitin sulfate proteoglycan 4 CSPG4. Also expressed by oligodendrocyte progenitor cells | [85] | |
| Desmin | [85] | ||
| CD13 | Aminopeptidase N | ||
| CD248 | Endosialin (TEM-1), highly expressed in glioma (GBM) (PC > pMSC) | [85, 96, 97] | |
| Vascular smooth muscle cells (vSMC) | PDGFRβ | Levels in PC > vSMC | [98] |
| NG2 | [98] | ||
| Desmin | Muscle class III intermediate filament | [98] | |
| CD13 | |||
| RGS5 | Regulator of G protein signaling 5 GTPase activating protein | [85, 99] | |
| CD146 | Melanoma cell adhesion molecule (MCAM) | [85, 99] | |
| αSMA | Alpha smooth muscle cell actin encoded by ACTA2. Level of expression in vSMC > > PC | [85] | |
| TAGLN | Trangelin, smooth muscle protein 22 alpha (SM22) | [85] | |
| pMSC fibroblast-like | PDGFRβ | ||
| PDGFRα | [86] | ||
| CD13 | |||
| COL1A1 | pMSC also express high levels of COL1A2, COL3A1, COL4A1. Expressed by PC (less than 2%) | ||
| LAMA-1 | Laminin subunit alpha 1, also expressed by epithelial cells | [86] | |
| LUM | Lumican | [86] | |
| DCN | Decorin | [86] | |
| MPZL2 | Myelin protein zero-like 2, adhesion | [86] | |
| SRPX2 | Sushi Repeat Containing Protein X-Linked 2 | [86] | |
| FN | Fibronectin, also expressed by PC / scar tissue | [100] | |
| FBLN1 | Fibulin-1, Type I Hu pMSC/fibroblast | [14] | |
| CEMIP | Cell migration-inducing protein, Type II Hu FB | [14] | |
| KCNMA1 | Potassium channel, Type III Hu pMSC/Fib | [14] | |
| Macrophages (M) | CD11b / CD18 | Complement receptor type 3 involved in phagocytosis of host cell debris | [87, 101] |
| Border-associated M (BAM) | CD163 | Scavenger receptor, M2 antiinflammatory | [102] |
| CD206 | Mannose receptor | [88] | |
| LYVE1 | Hyaluronan receptor | [88] | |
| Astrocytes (AS) | GFAP | Glial fibrillary acidic protein | [87] |
Several markers are shared between the different subsets of periendothelial cells while others (in bold) are restricted (but not exclusive) to specific subsets. TEM1: Tumor endothelial marker 1
Recent studies utilizing cell lineage tracing and single-cell RNA sequencing experiments have provided insights into the role of pMSC cells in the CNS. Garcia et al. have identified 11 cell subtypes within the human CNS vasculature, including three distinct subtypes of pMSC (referred to as vFB in this study), with specific markers (Table 2) [14]. Type I pMSC in humans appear to be primarily involved in extracellular matrix (ECM) organization and fibrosis, while type III cells express various growth factors, including VEGFA. Interestingly, the gradient of gene expression from type I to type II pMSC was continuous with a subpopulation of pericytes, suggesting a potential lineage from type I to type II to PC [14]. Two of the pMSC subtypes align with the subtypes previously identified in mice by Vanlandewijck et al. (referred to as vFB in this study, type I and II) [86]. Similar findings were observed in a zebrafish study, which demonstrated the stem cell potential of pMSCs to transdifferentiate into PC [93].
These findings substantiate the affiliation of PC and pMSC, which are also referred to vFB, within a continuum of differentiation [72, 94].
Physiological functions of PC and pMSC in the CNS.
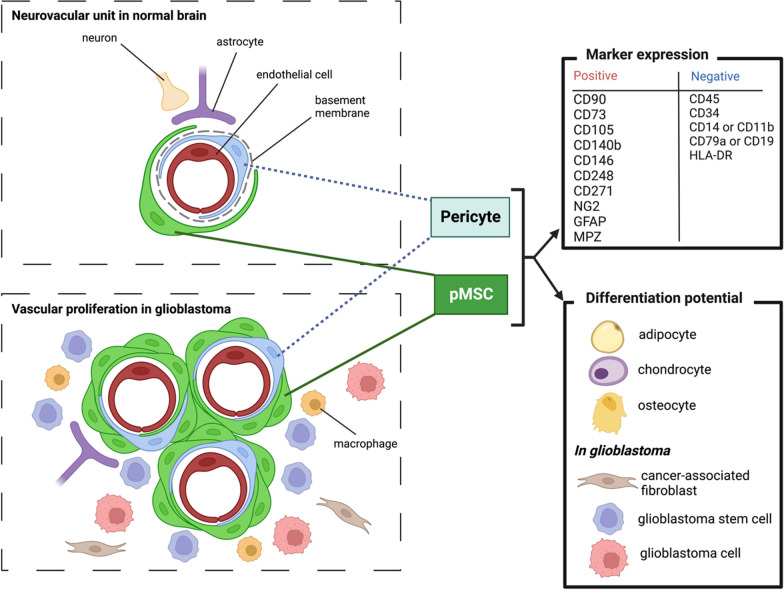
The close association between PC and endothelial cells contributes to the formation of the BBB, the maintenance of vascular stability, and the regulation of vascular tone [103–105]. Other functions have been described, including a role in angiogenesis and immune regulation properties, making pMSC key players in brain homeostasis and disease. Together with endothelial cells, astrocytes and neurons, they form the neurovascular unit that supplies nutrients and oxygen through the BBB and provides an optimal environment for NSC (as well as GSC) homing and proliferation (Fig. 4) [106, 107].
Fig. 4.
Perivascular mesenchymal stromal cells (pMSC) in normal brain and in glioblastoma. In normal brain, pMSC form the neurovascular unit, together with endothelial cells, astrocytes, and neurons. The neurovascular unit supplies nutrients and oxygen through the blood brain barrier. In glioblastoma (GB), resident pMSC and glioma stem cell-differentiated pMSC participate in vascular proliferation. Leaving the vessel, pMSC may give rise to GB stem cells, GB cells, and cancer-associated fibroblasts. (MPZ: Myelin P zero) (created with Biorender.com)
Injury repair
Many studies have demonstrated the ability of MSC to differentiate toward a neuronal/glial phenotype in vitro and therefore, have suggested a potential role of MSC in brain repair [108–112]. However, while transplantation of MSC in brain and spinal cord injury models tends to improve the functional outcome, the transformation of MSC into neurons/glial cells in vivo is rare and partly results from the fusion of MSC with brain cells [113–116]. Consequently, it has been suggested that the role of MSC in brain injury mostly relies on their immune regulation properties rather than their neuronal differentiation ability. In fact, this paradigm shift in which MSCs exert healing effects not through their differentiation abilities but rather through their immune modulation functions, has been observed in many therapeutic contexts [117–120].
Recent findings suggest that pMSC and PC may have a unique ability to monitor the microenvironment of injured tissues. Indeed, it has been demonstrated that they secrete a large number of chemokines, cytokines, and other soluble factors [120–123]. Their role in immune regulation has initially been highlighted by the observations of prolonged skin graft survival, improvement in severe graft-versus-host disease, and therapeutic effects in an experimental autoimmune encephalomyelitis mouse model [124–126]. Indeed, MSC can modulate effector T-cell activation and proliferation, directly through soluble factors or indirectly by controlling the activity of regulatory T-cells (Treg). MSC are also able to control the proliferation and the activation of monocytes/macrophages, natural killer T-cells, dendritic cells, B-cells, and neutrophils, by the secretion of soluble factors such as IFN gamma, nitric oxide (NO), indoleamine 2,3-dioxygenase (IDO), prostaglandin E2 (PGE2), TGFβ, and IL10 [127, 128]. Consequently, MSC have a major role in the coordination of healing responses and the prevention of autoimmunity [119, 127–129].
Scar formation is a ubiquitous healing mechanism that is preserved throughout the CNS. Initially, the CNS scar has been referred to as the glial scar as a whole. The glial scar predominantly consists of reactive astrocytes and proteoglycans (heparan sulphate proteoglycan, dermatan sulphate proteoglycan, keratan sulphate proteoglycan and chondroitin sulphate proteoglycan) that stabilize injured CNS tissue by modulating the inflammatory response, yet prevent tissue regeneration [130–132]. The glial scar circumscribes the lesion core where the inflammatory response leads to a fibrotic scar, composed of immune cells, fibroblasts, fibronectin, collagen and laminin [133]. It is commonly accepted that fibroblasts are absent in the CNS parenchyma and it has been suggested that they are restricted to the vascular and meningeal niches [134]. Several studies have underscored the role of pMSC and PC in generating the fibrotic scar in the CNS [100, 135–137]. In response to spinal cord injury, PC proliferate locally and give rise to myofibroblasts, generating the fibrotic scar [100]. A rapid pMSC/PC loss after cerebral ischemia in human stroke has been observed, with subsequent proliferation of resident PDGFRβ + CD13 + stromal cells that transform to αSMA + CD105 + myofibroblasts [135]. These findings suggest the critical role of the endothelial cell-pMSC/PC interaction to maintain pMSC and PC in a quiescent state to prevent fibrosis.
pMSC in GB
In GB, pMSC can be recruited either from local brain sources, in the perivascular niches, or from the BM by MSC homing to the GB TME [138, 139]. pMSC may also result from GSC differentiation. As discussed above, GSC predominantly reside in perivascular niches and interact with endothelial cells in a bidirectional manner [40]. First reports have suggested that GSC may transdifferentiate into endothelial cells but it has been shown that endothelial cells do not harbor molecular alterations of GB [44, 140–142]. In addition, the ability of GSC to undergo mesenchymal differentiation has raised the hypothesis of GSC transdifferentiating into pMSC rather than endothelial cells [143, 144]. Furthermore, it has been demonstrated that GSC generate PC, which may carry the same genetic alterations of GB, such as EGFR amplification, chr 10 loss and PTEN loss [145].
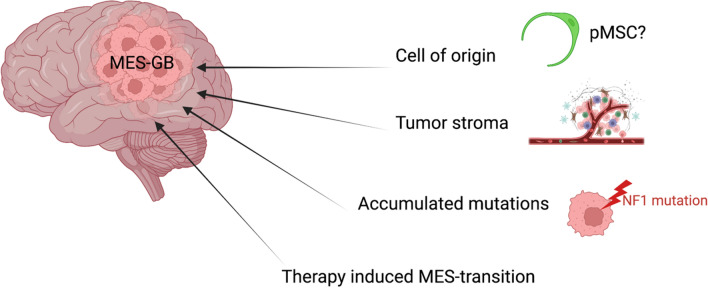
As discussed above, the origin of mesenchymal GB remains elusive and until recently, an alternative non-neural progenitor cell has not been explored. Indeed, deep scRNAseq of GB progenitor cells uncovered two principal cell-lineage profiles, NC perivascular and radial glia (and its progenies) [13]. Consistently, introducing driver mutations in perivascular cells was sufficient to initiate brain tumors in vivo. In addition, it has been shown that GB of a perivascular lineage represent 44% of the mesenchymal GB subtype and showed significant poorer survival than those of radial glia-lineage [13]. These results suggest that the mesenchymal signature results, at least partially, from pMSC transformation. Indeed, the mesenchymal subtype can be induced by other factors such as the influence TME, the accumulation of mutations in tumor cells (particularly NF1 mutation) and the therapy-induced mesenchymal transition (Fig. 5) [146].
Fig. 5.
Origin of mesenchymal glioblastoma subtype (created with Biorender.com)
Several studies have demonstrated the involvement of PC and pMSC in GB tumor vasculature development through a vascular mimicry mechanism [46, 47, 139, 147, 148]. It has also been shown that pMSC overexpress several proteins involved in the promotion of tumor angiogenesis, including CSPG4/NG2, CRYAB, CNN1, CALD1, and VASP, and secrete high levels of angiogenic factors such as SDF-1/CXCL12 and HGF [149].
The role of pMSC in immune regulation during GB progression was demonstrated by the high levels of anti-inflammatory cytokines (IL-10 and TGFβ) detected in vitro and in vivo in pMSC (referred as to PC in this study) that interact with GB cells [150]. In contrast, after activation by GB cells, pMSC did not produce proinflammatory cytokines, such as IL-1, IL-23, and IL-12 [150]. These observations suggest an immunosuppressive response of pMSC to interaction with GB cells.pMSC also have a tumor growth-enhancing and tumor invasiveness-increasing role [138, 151]. It has been demonstrated that pMSC secrete TGFβ1, stimulating GB cell proliferation and viability through paracrine effect [152]. pMSC are also capable of enhancing GB cell proliferation under direct cell–cell contact, independently of TGFβ1 levels, in vitro and in vivo [152]. Similarly, it has been shown that pMSC secrete IL-6, increasing proliferation and self-renewal of GSC in vitro and enhancing GSC tumorigenicity in vivo (Fig. 3) [153].
Studies have isolated two subpopulations of pMSC (CD90high pMSC and CD90low pMSC) and have described specific roles in GB progression [154, 155]. It has been observed that CD90low pMSC are more abundant than CD90high pMSC and that CD90low pMSC contribute to angiogenesis and CD90high pMSC promote GB cell growth both in vivo and in vitro [155, 156]. Indeed, CD90low pMSC were shown to produce higher levels of angiogenic factors, such as VEGF, bFGF and IL-6, and CD90high pMSC to produce higher levels of growth factors, such as SDF-1α, CCL5 and MMP9 [155].
Perivascular and intratumoral cells that co-express PDGFRβ and fibroblast activation protein α (FAP), a common marker used to identify cancer-associated fibroblasts (CAF), were identified in GB [157]. Proteomic quantitative analysis has also demonstrated that pMSC expressed high levels of CAF markers, such as CD146, S100A4/FSP1, nestin, and NG2 [149]. These findings suggest that pMSC, mirroring their transition to myofibroblasts in the context of fibrotic scar, may give rise to CAF that support tumor progression with the GB TME, as described in other solid cancers [158, 159].
Concluding remarks
pMSC exert pro-tumoral effects, promoting angiogenesis, tumor proliferation and invasiveness, and immunosuppression, in agreement with the observation that increased percentages of pMSC within high-grade gliomas are associated with worse clinical outcome [160]. Recent studies suggest that, in addition to their activities to support GB growth, pMSC may be the cell of origin of GB, particularly the mesenchymal GB subtype [13]. This alternative paradigm provides exciting new research avenues to characterize pMSC in the context of GB and understand better the gliomagenesis.
Acknowledgements
Figures were created with Biorender.com
Abbreviations
- ACSBG1
Acyl-CoA synthetase bubblegum family member 1
- AIF1
Allograft inflammatory factor 1
- APC
Astrocytic precursor cells
- BBB
Blood–brain barrier
- BMDM
Bone marrrow-derived macrophage
- CAF
Cancer-associated fibroblast
- CALD1
Caldesmon
- CCL
C-C motif chemokine ligand
- CDK4
Cyclin-dependent kinase 4
- CDK6
Cyclin-dependent kinase 6
- CDKN2A
Cyclin-dependent kinase inhibitor 2A
- CFU-F
Fibroblastic colony forming units
- CHI3L1
Chitinase 3 like 1
- CNN1
Calponin 1
- CNS
Central nervous system
- COL1A1
Collagen type I alpha 1
- COL1A2
Collagen type I alpha 2
- CRYAB
Crystallin alpha B
- CSF-1
Colony stimulating factor 1
- DAB2
Disabled-2
- EGF
Epidermal growth factor
- EGFR
Epidermal growth factor receptor
- ERBB3
Erb-b2 receptor tyrosine kinase 3
- FAP
Fibroblast activation protein-α
- FGFR3
Fibroblast growth factor receptor 3
- FOXP3
Forkhead box P3
- GB
Glioblastoma
- G-CIMP
Glioma-CpG island methylator phenotype
- GSC
Glioma stem cell
- HGF
Hepatocyte growth factor
- IBA1
Ionized calcium binding adaptor molecule 1
- IDH
Isocitrate dehydrogenase
- IDO
Indoleamine 2,3-dioxygenase
- IFN
Interferon
- IL
Interleukin
- KCNF1
Potassium voltage-gated channel modifier subfamily F member 1
- MAPK
Mitogen-activated protein kinase
- MDM2
Mouse double minute 2
- MDM4
Mouse double minute 4
- MET
Mesenchymal-epithelial transition factor
- MMP9
Matrix metallopeptidase 9
- NF1
Neurofibromatosis type 1
- NG2
Neural/glial antigen 2
- NO
Nitric oxide
- NPC
Neural precursor cells
- NSC
Neural stem cells
- OCT4
Octamer-binding transcription factor 4
- OPC
Oligodendrocyte precursor cells
- P2RX7
Purinergic receptor P2X 7
- PC
Pericytes
- PDGFR
Platelet-derived growth factor receptor
- PECAM1
Platelet endothelial cell adhesion molecule 1
- PGE2
Prostaglandin E2
- PI3K
Phosphoinositide 3-kinase
- pMSC
Perivascular mesenchymal stromal cell
- PMT
Proneural to mesenchymal transition
- pRB
Retinoblastoma protein
- S100A
S100 calcium-binding protein A
- scRNAseq
Single-cell RNA-sequencing
- SDF-1
Stromal cell-derived factor 1
- SMA
Smooth muscle actin
- SOX10
SRY-box transcription factor 10
- SOX2
SRY-box transcription factor 2
- STMN4
Stathmin 4
- TAM
Tumor-associated macrophage
- TERT
Telomerase reverse transcriptase
- TGFβ
Transforming growth factor beta
- THBS1
Thrombospondin 1
- TME
Tumor microenvironment
- TREM1
Triggering receptor expressed on myeloid cells 1
- TREM2
Triggering receptor expressed on myeloid cells 2
- VASP
Vasodilator-stimulated phosphoprotein
- VEGF
Vascular endothelial growth factor
- VEGFR
Vascular endothelial growth factor receptor
- vFB
Vascular fibroblast (or perivascular fibroblast)
- WHO
World Health Organization
Author contributions
FA, MK, YB, YS, MD, and PG contributed to writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Regional Council of La Réunion, French State and Europe (CPER‐FEDER, GURDTI 2017‐1198‐0002583 VIROPAM) and (CPER‐FEDER 20192211‐0022768, EPIGEN).
Availability of data and materials
Not applicable
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
F. Ah-Pine, Email: franck.ah-pine@chu-reunion.fr
M. Khettab, Email: mohamed.khettab@chu-reunion.fr
Y. Bedoui, Email: yosra.bedoui@chu-reunion.fr
Y. Slama, Email: youssef.slama@clinifutur.net
M. Daniel, Email: matthieu.daniel@chu-reunion.fr
B. Doray, Email: berenice.doray@chu-reunion.fr
P. Gasque, Email: philippe.gasque@chu-reunion.fr, Email: philippe.gasque@gmail.com
References
- 1.WHO Classification of Tumours Editorial Board (2021) World Health Organization Classification of Tumours of the Central Nervous System [Internet]. 5th ed. International Agency for Research on Cancer, Lyon [cited 2022 Aug 17]. Available from: https://publications.iarc.fr/Book-And-Report-Series/Who-Classification-Of-Tumours/Central-Nervous-System-Tumours-2021
- 2.Ostrom QT, Cioffi G, Waite K, Kruchko C, Barnholtz-Sloan JS (2021) CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2014-2018. Neuro-Oncol 23: iii1–iii105 [DOI] [PMC free article] [PubMed]
- 3.Brennan CW, Verhaak RGW, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of Glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sturm D, Bender S, Jones DTW, Lichter P, Grill J, Becher O, et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat Rev Cancer. 2014;14:92–107. doi: 10.1038/nrc3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 7.Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2017;32:42–56.e6. doi: 10.1016/j.ccell.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178:835–849.e21. doi: 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu C, Sage JC, Miller MR, Verhaak RGW, Hippenmeyer S, Vogel H, et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146:209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alcantara Llaguno S, Chen J, Kwon C-H, Jackson EL, Li Y, Burns DK, et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15:45–56. doi: 10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alcantara Llaguno S, Sun D, Pedraza AM, Vera E, Wang Z, Burns DK, et al. Cell-of-origin susceptibility to glioblastoma formation declines with neural lineage restriction. Nat Neurosci. 2019;22:545–555. doi: 10.1038/s41593-018-0333-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Sun D, Chen Y-J, Xie X, Shi Y, Tabar V, et al. Cell lineage-based stratification for glioblastoma. Cancer Cell. 2020;38:366–379.e8. doi: 10.1016/j.ccell.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y, Jiang Y, Behnan J, Ribeiro MM, Kalantzi C, Zhang M-D, et al. Neural network learning defines glioblastoma features to be of neural crest perivascular or radial glia lineages. Sci Adv. 2022;8:eabm6340. doi: 10.1126/sciadv.abm6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia FJ, Sun N, Lee H, Godlewski B, Mathys H, Galani K, et al. Single-cell dissection of the human brain vasculature. Nature. 2022;603:893. doi: 10.1038/s41586-022-04521-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wrensch M, Minn Y, Chew T, Bondy M, Berger MS. Epidemiology of primary brain tumors: current concepts and review of the literature. Neuro-Oncology. 2002;4:278–299. doi: 10.1093/neuonc/4.4.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro- Oncology. 2014;16:896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 18.Tan AC, Ashley DM, López GY, Malinzak M, Friedman HS, Khasraw M. Management of glioblastoma: state of the art and future directions. CA Cancer J Clin. 2020;70:299–312. doi: 10.3322/caac.21613. [DOI] [PubMed] [Google Scholar]
- 19.McLendon R, Friedman A, Bigner D, Van Meir EG, Brat DJ, M. Mastrogianakis G,, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–1235. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Behnan J, Stangeland B, Hosainey SA, Joel M, Olsen TK, Micci F, et al. Differential propagation of stroma and cancer stem cells dictates tumorigenesis and multipotency. Oncogene. 2017;36:570–584. doi: 10.1038/onc.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG Island methylator phenotype that defines a distinct subgroup of Glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sturm D, Witt H, Hovestadt V, Khuong-Quang D-A, Jones DTW, Konermann C, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 24.Bhat KPL, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24:331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doucette T, Rao G, Rao A, Shen L, Aldape K, Wei J, et al. Immune heterogeneity of glioblastoma subtypes: extrapolation from the cancer genome atlas. Cancer Immunol Res. 2013;1:112–122. doi: 10.1158/2326-6066.CIR-13-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engler JR, Robinson AE, Smirnov I, Hodgson JG, Berger MS, Gupta N, et al. Increased microglia/macrophage gene expression in a subset of adult and pediatric astrocytomas. PLoS One. 2012;7:e43339. doi: 10.1371/journal.pone.0043339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabrusiewicz K, Rodriguez B, Wei J, Hashimoto Y, Healy LM, Maiti SN, et al. (2016) Glioblastoma-infiltrated innate immune cells resemble M0 macrophage phenotype. JCI Insight [Internet]. [cited 2022 Aug 19]. Available from: https://insight-jci-org.proxy.insermbiblio.inist.fr/articles/view/85841 [DOI] [PMC free article] [PubMed]
- 28.Kaffes I, Szulzewsky F, Chen Z, Herting CJ, Gabanic B, Velázquez Vega JE, et al. Human mesenchymal glioblastomas are characterized by an increased immune cell presence compared to proneural and classical tumors. OncoImmunology. 2019;8:e1655360. doi: 10.1080/2162402X.2019.1655360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holland EC, Hively WP, DePinho RA, Varmus HE. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes Dev. 1998;12:3675–3685. doi: 10.1101/gad.12.23.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O, et al. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080–1084. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N Engl J Med. 2005;353:811–822. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- 32.Lee JH, Lee JE, Kahng JY, Kim SH, Park JS, Yoon SJ, et al. Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature. 2018;560:243–247. doi: 10.1038/s41586-018-0389-3. [DOI] [PubMed] [Google Scholar]
- 33.Bhaduri A, Di Lullo E, Jung D, Müller S, Crouch EE, Espinosa CS, et al. Outer radial glia-like cancer stem cells contribute to heterogeneity of glioblastoma. Cell Stem Cell. 2020;26:48–63.e6. doi: 10.1016/j.stem.2019.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 35.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 36.Alameda F, Velarde JM, Carrato C, Vidal N, Arumí M, Naranjo D, et al. Prognostic value of stem cell markers in glioblastoma. Biomarkers. 2019;24:677–683. doi: 10.1080/1354750X.2019.1652345. [DOI] [PubMed] [Google Scholar]
- 37.Arai H, Ikota H, Sugawara K, Nobusawa S, Hirato J, Nakazato Y. Nestin expression in brain tumors: its utility for pathological diagnosis and correlation with the prognosis of high-grade gliomas. Brain Tumor Pathol. 2012;29:160–167. doi: 10.1007/s10014-012-0081-5. [DOI] [PubMed] [Google Scholar]
- 38.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 39.Chen J, Li Y, Yu T-S, McKay RM, Burns DK, Kernie SG, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, et al. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11:69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 41.Broekman ML, Maas SLN, Abels ER, Mempel TR, Krichevsky AM, Breakefield XO. Multidimensional communication in the microenvirons of glioblastoma. Nat Rev Neurol. 2018;14:482–495. doi: 10.1038/s41582-018-0025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seano G, Jain RK. Vessel co-option in glioblastoma: emerging insights and opportunities. Angiogenesis. 2020;23:9–16. doi: 10.1007/s10456-019-09691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lyden D, Hattori K, Dias S, Costa C, Blaikie P, Butros L, et al. Impaired recruitment of bone-marrow-derived endothelial and hematopoietic precursor cells blocks tumor angiogenesis and growth. Nat Med. 2001;7:1194–1201. doi: 10.1038/nm1101-1194. [DOI] [PubMed] [Google Scholar]
- 44.Soda Y, Marumoto T, Friedmann-Morvinski D, Soda M, Liu F, Michiue H, et al. Transdifferentiation of glioblastoma cells into vascular endothelial cells. Proc Natl Acad Sci U S A. 2011;108:4274–4280. doi: 10.1073/pnas.1016030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao C, Gomez GA, Zhao Y, Yang Y, Cao D, Lu J, et al. ETV2 mediates endothelial transdifferentiation of glioblastoma. Signal Transduct Target Ther. 2018;3:4. doi: 10.1038/s41392-018-0007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El Hallani S, Boisselier B, Peglion F, Rousseau A, Colin C, Idbaih A, et al. A new alternative mechanism in glioblastoma vascularization: tubular vasculogenic mimicry. Brain J Neurol. 2010;133:973–982. doi: 10.1093/brain/awq044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scully S, Francescone R, Faibish M, Bentley B, Taylor SL, Oh D, et al. Transdifferentiation of glioblastoma stem-like cells into mural cells drives vasculogenic mimicry in glioblastomas. J Neurosci. 2012;32:12950–12960. doi: 10.1523/JNEUROSCI.2017-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang P, Miska J, Lee-Chang C, Rashidi A, Panek WK, An S, et al. Therapeutic targeting of tumor-associated myeloid cells synergizes with radiation therapy for glioblastoma. Proc Natl Acad Sci. 2019;116:23714–23723. doi: 10.1073/pnas.1906346116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klemm F, Maas RR, Bowman RL, Kornete M, Soukup K, Nassiri S, et al. Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell. 2020;181:1643–1660.e17. doi: 10.1016/j.cell.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bowman RL, Klemm F, Akkari L, Pyonteck SM, Sevenich L, Quail DF, et al. Macrophage ontogeny underlies differences in tumor-specific education in brain malignancies. Cell Rep. 2016;17:2445–2459. doi: 10.1016/j.celrep.2016.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Z, Feng X, Herting CJ, Garcia VA, Nie K, Pong WW, et al. Cellular and molecular identity of tumor-associated macrophages in glioblastoma. Cancer Res. 2017;77:2266–2278. doi: 10.1158/0008-5472.CAN-16-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Müller S, Kohanbash G, Liu SJ, Alvarado B, Carrera D, Bhaduri A, et al. Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol. 2017;18:1–14. doi: 10.1186/s13059-017-1362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 54.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–555. doi: 10.1016/S1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 55.Gutmann DH, Kettenmann H. Microglia/brain macrophages as central drivers of brain tumor pathobiology. Neuron. 2019;104:442–449. doi: 10.1016/j.neuron.2019.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ah-Pine F, Malaterre-Septembre A, Bedoui Y, Khettab M, Neal JW, Freppel S, et al. Complement activation and up-regulated expression of anaphylatoxin C3a/C3aR in glioblastoma: deciphering the links with TGF-β and VEGF. Cancers. 2023;15:2647. doi: 10.3390/cancers15092647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Landry AP, Balas M, Alli S, Spears J, Zador Z. Distinct regional ontogeny and activation of tumor associated macrophages in human glioblastoma. Sci Rep. 2020;10:19542. doi: 10.1038/s41598-020-76657-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sørensen MD, Dahlrot RH, Boldt HB, Hansen S, Kristensen BW. Tumour-associated microglia/macrophages predict poor prognosis in high-grade gliomas and correlate with an aggressive tumour subtype. Neuropathol Appl Neurobiol. 2018;44:185–206. doi: 10.1111/nan.12428. [DOI] [PubMed] [Google Scholar]
- 59.Buonfiglioli A, Hambardzumyan D. Macrophages and microglia: the cerberus of glioblastoma. Acta Neuropathol Commun. 2021;9:1–21. doi: 10.1186/s40478-021-01156-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coniglio SJ, Eugenin E, Dobrenis K, Stanley ER, West BL, Symons MH, et al. Microglial stimulation of glioblastoma invasion involves epidermal growth factor receptor (EGFR) and colony stimulating factor 1 receptor (CSF-1R) signaling. Mol Med. 2012;18:519–527. doi: 10.2119/molmed.2011.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carvalho da Fonseca AC, Wang H, Fan H, Chen X, Zhang I, Zhang L, et al. Increased expression of stress inducible protein 1 in glioma-associated microglia/macrophages. J Neuroimmunol. 2014;274:71–77. doi: 10.1016/j.jneuroim.2014.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ye X, Xu S, Xin Y, Yu S, Ping Y, Chen L, et al. Tumor-associated microglia/macrophages enhance the invasion of glioma stem-like cells via TGF-β1 signaling pathway. J Immunol. 2012;189:444–453. doi: 10.4049/jimmunol.1103248. [DOI] [PubMed] [Google Scholar]
- 63.Feng X, Szulzewsky F, Yerevanian A, Chen Z, Heinzmann D, Rasmussen RD, et al. Loss of CX3CR1 increases accumulation of inflammatory monocytes and promotes gliomagenesis. Oncotarget. 2015;6:15077–15094. doi: 10.18632/oncotarget.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gielen PR, Schulte BM, Kers-Rebel ED, Verrijp K, Petersen-Baltussen HMJM, ter Laan M, et al. Increase in both CD14-positive and CD15-positive myeloid-derived suppressor cell subpopulations in the blood of patients with glioma but predominance of CD15-positive myeloid-derived suppressor cells in glioma tissue. J Neuropathol Exp Neurol. 2015;74:390–400. doi: 10.1097/NEN.0000000000000183. [DOI] [PubMed] [Google Scholar]
- 65.Chang AL, Miska J, Wainwright DA, Dey M, Rivetta CV, Yu D, et al. CCL2 produced by the glioma microenvironment is essential for the recruitment of regulatory T cells and myeloid-derived suppressor cells. Cancer Res. 2016;76:5671–5682. doi: 10.1158/0008-5472.CAN-16-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Herting CJ, Chen Z, Maximov V, Duffy A, Szulzewsky F, Shayakhmetov DM, et al. Tumour-associated macrophage-derived interleukin-1 mediates glioblastoma-associated cerebral oedema. Brain. 2019;142:3834–3851. doi: 10.1093/brain/awz331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sehm T, Fan Z, Ghoochani A, Rauh M, Engelhorn T, Minakaki G, et al. Sulfasalazine impacts on ferroptotic cell death and alleviates the tumor microenvironment and glioma-induced brain edema. Oncotarget. 2016;7:36021–36033. doi: 10.18632/oncotarget.8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Caplan A. Mesenchymal stem-cells. J Orthop Res. 1991;9:641–650. doi: 10.1002/jor.1100090504. [DOI] [PubMed] [Google Scholar]
- 69.Friedenstein AJ, Chailakhyan RK, Latsinik NV, Panasyuk AF, Keiliss-Borok IV. Stromal cells responsible for transferring the microenvironment of the hemopoietic tissues. Cloning in vitro and retransplantation in vivo. Transplantation. 1974;17:331–40. doi: 10.1097/00007890-197404000-00001. [DOI] [PubMed] [Google Scholar]
- 70.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 71.Dominici M, Blanc KL, Mueller I, Slaper-Cortenbach I, Marini FC, Krause DS, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The international society for cellular therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 72.da Silva ML, Chagastelles PC, Nardi NB. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J Cell Sci. 2006;119:2204–2213. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- 73.DiMarino AM, Caplan AI, Bonfield TL. Mesenchymal stem cells in tissue repair. Front Immunol. 2013;4:201–201. doi: 10.3389/fimmu.2013.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deans RJ, Moseley AB. Mesenchymal stem cells: biology and potential clinical uses. Exp Hematol. 2000;28:875–884. doi: 10.1016/S0301-472X(00)00482-3. [DOI] [PubMed] [Google Scholar]
- 75.Bedoui Y, Lebeau G, Guillot X, Dargai F, Guiraud P, Neal JW, et al. Emerging roles of perivascular mesenchymal stem cells in synovial joint inflammation. J Neuroimmune Pharmacol. 2020 doi: 10.1007/s11481-020-09958-z. [DOI] [PubMed] [Google Scholar]
- 76.El Agha E, Kramann R, Schneider RK, Li X, Seeger W, Humphreys BD, et al. Mesenchymal stem cells in fibrotic disease. Cell Stem Cell. 2017;21:166–177. doi: 10.1016/j.stem.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 77.Duffield JS, Lupher M, Thannickal VJ, Wynn TA (2013) host responses in tissue repair and fibrosis. In: Abbas AK, Galli SJ, Howley PM (eds.) Annu Rev Pathol Mech Dis Vol 8 [Internet]. Palo Alto: Annual Reviews; [cited 2022 Jul 16]. p. 241–76. Available from: http://www.webofscience.com/api/gateway?GWVersion=2&SrcAuth=DOISource&SrcApp=WOS&KeyAID=10.1146%2Fannurev-pathol-020712-163930&DestApp=DOI&SrcAppSID=EUW1ED0DFEpvTyL7e8DlqaU8zUdSS&SrcJTitle=ANNUAL+REVIEW+OF+PATHOLOGY%3A+MECHANISMS+OF+DISEASE%2C+VOL+8&DestDOIRegistrantName=Annual+Reviews [DOI] [PMC free article] [PubMed]
- 78.Payet M, Ah-Pine F, Guillot X, Gasque P. Inflammatory mesenchymal stem cells express abundant membrane-bound and soluble forms of C-type lectin-like CD248. Int J Mol Sci. 2023;24:9546. doi: 10.3390/ijms24119546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.da Meirelles LS, Caplan AI, Nardi NB. In search of the in vivo identity of mesenchymal stem cells. Stem Cells. 2008;26:2287–2299. doi: 10.1634/stemcells.2007-1122. [DOI] [PubMed] [Google Scholar]
- 80.Crisan M, Yap S, Casteilla L, Chen C-W, Corselli M, Park TS, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 81.Takashima Y, Era T, Nakao K, Kondo S, Kasuga M, Smith AG, et al. Neuroepithelial cells supply an initial transient wave of MSC differentiation. Cell. 2007;129:1377–1388. doi: 10.1016/j.cell.2007.04.028. [DOI] [PubMed] [Google Scholar]
- 82.Le Douarin NM, Creuzet S, Couly G, Dupin E. Neural crest cell plasticity and its limits. Development. 2004;131:4637–4650. doi: 10.1242/dev.01350. [DOI] [PubMed] [Google Scholar]
- 83.Majesky MW. Developmental basis of vascular smooth muscle diversity. Arterioscler Thromb Vasc Biol. 2007;27:1248–1258. doi: 10.1161/ATVBAHA.107.141069. [DOI] [PubMed] [Google Scholar]
- 84.Hungerford JE, Little CD. Developmental biology of the vascular smooth muscle cell: building a multilayered vessel wall. J Vasc Res. 1999;36:2–27. doi: 10.1159/000025622. [DOI] [PubMed] [Google Scholar]
- 85.Armulik A, Genové G, Betsholtz C. Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev Cell. 2011;21:193–215. doi: 10.1016/j.devcel.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 86.Vanlandewijck M, He L, Mae MAA, Andrae J, Ando K, Del Gaudio F, et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 2018;554:475. doi: 10.1038/nature25739. [DOI] [PubMed] [Google Scholar]
- 87.Dorrier CE, Aran D, Haenelt EA, Sheehy RN, Hoi KK, Pintaric L, et al. CNS fibroblasts form a fibrotic scar in response to immune cell infiltration. Nat Neurosci. 2021;24:234–244. doi: 10.1038/s41593-020-00770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dorrier CE, Jones HE, Pintarić L, Siegenthaler JA, Daneman R. Emerging roles for CNS fibroblasts in health, injury and disease. Nat Rev Neurosci. 2022;23:23–34. doi: 10.1038/s41583-021-00525-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu J, He L, Muhl L, Mocci G, Gustavsson S, Buyandelger B, et al. A human cell type similar to murine central nervous system perivascular fibroblasts. Exp Cell Res. 2021;402:112576. doi: 10.1016/j.yexcr.2021.112576. [DOI] [PubMed] [Google Scholar]
- 90.Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, et al. Molecular diversity and specializations among the cells of the adult mouse brain. Cell. 2018;174:1015. doi: 10.1016/j.cell.2018.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, et al. Molecular architecture of the mouse nervous system. Cell. 2018;174:999–1014.e22. doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Covas DT, Panepucci RA, Fontes AM, Silva WA, Orellana MD, Freitas MCC, et al. Multipotent mesenchymal stromal cells obtained from diverse human tissues share functional properties and gene-expression profile with CD146+ perivascular cells and fibroblasts. Exp Hematol. 2008;36:642–654. doi: 10.1016/j.exphem.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 93.Rajan AM, Ma RC, Kocha KM, Zhang DJ, Huang P. Dual function of perivascular fibroblasts in vascular stabilization in zebrafish. PLoS Genet. 2020;16:e1008800. doi: 10.1371/journal.pgen.1008800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Caplan AI. All MSCs are pericytes? Cell Stem Cell. 2008;3:229–230. doi: 10.1016/j.stem.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 95.Griffiths MR, Botto M, Morgan BP, Neal JW, Gasque P. CD93 regulates central nervous system inflammation in two mouse models of autoimmune encephalomyelitis. Immunology. 2018;155:346–355. doi: 10.1111/imm.12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brady J, Neal J, Sadakar N, Gasque P. Human endosialin (tumor endothelial marker 1) is abundantly expressed in highly malignant and invasive brain tumors. J Neuropathol Exp Neurol. 2004;63:1274–1283. doi: 10.1093/jnen/63.12.1274. [DOI] [PubMed] [Google Scholar]
- 97.Teicher BA. CD248: a therapeutic target in cancer and fibrotic diseases. Oncotarget. 2019;10:993–1009. doi: 10.18632/oncotarget.26590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bergers G, Song S. The role of pericytes in blood-vessel formation and maintenance. Neuro-Oncology. 2005;7:452–464. doi: 10.1215/S1152851705000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Berger M, Bergers G, Arnold B, Hämmerling GJ, Ganss R. Regulator of G-protein signaling-5 induction in pericytes coincides with active vessel remodeling during neovascularization. Blood. 2005;105:1094–1101. doi: 10.1182/blood-2004-06-2315. [DOI] [PubMed] [Google Scholar]
- 100.Goritz C, Dias DO, Tomilin N, Barbacid M, Shupliakov O, Frisen J. A pericyte origin of spinal cord scar tissue. Science. 2011;333:238–242. doi: 10.1126/science.1203165. [DOI] [PubMed] [Google Scholar]
- 101.Lapenna A, De Palma M, Lewis CE. Perivascular macrophages in health and disease. Nat Rev Immunol. 2018;18:689–702. doi: 10.1038/s41577-018-0056-9. [DOI] [PubMed] [Google Scholar]
- 102.Van Hove H, Martens L, Scheyltjens I, De Vlaminck K, Pombo Antunes AR, De Prijck S, et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat Neurosci. 2019;22:1021–1035. doi: 10.1038/s41593-019-0393-4. [DOI] [PubMed] [Google Scholar]
- 103.Díaz-Flores L, Gutiérrez R, Madrid JF, Varela H, Valladares F, Acosta E, et al. Pericytes. Morphofunction, interactions and pathology in a quiescent and activated mesenchymal cell niche. Histol Histopathol. 2009;24:909–69. doi: 10.14670/HH-24.909. [DOI] [PubMed] [Google Scholar]
- 104.Hall CN, Reynell C, Gesslein B, Hamilton NB, Mishra A, Sutherland BA, et al. Capillary pericytes regulate cerebral blood flow in health and disease. Nature. 2014;508:55–60. doi: 10.1038/nature13165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Peppiatt CM, Howarth C, Mobbs P, Attwell D. Bidirectional control of CNS capillary diameter by pericytes. Nature. 2006;443:700–704. doi: 10.1038/nature05193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Goldman SA, Chen Z. Perivascular instruction of cell genesis and fate in the adult brain. Nat Neurosci. 2011;14:1382–1389. doi: 10.1038/nn.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tavazoie M, Van der Veken L, Silva-Vargas V, Louissaint M, Colonna L, Zaidi B, et al. A specialized vascular niche for adult neural stem cells. Cell Stem Cell. 2008;3:279–288. doi: 10.1016/j.stem.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hermann A, Gastl R, Liebau S, Popa MO, Fiedler J, Boehm BO, et al. Efficient generation of neural stem cell-like cells from adult human bone marrow stromal cells. J Cell Sci. 2004;117:4411–4422. doi: 10.1242/jcs.01307. [DOI] [PubMed] [Google Scholar]
- 109.Zhang H-T, Liu Z-L, Yao X-Q, Yang Z-J, Xu R-X. Neural differentiation ability of mesenchymal stromal cells from bone marrow and adipose tissue: a comparative study. Cytotherapy. 2012;14:1203–1214. doi: 10.3109/14653249.2012.711470. [DOI] [PubMed] [Google Scholar]
- 110.Zhao L-R, Duan W-M, Reyes M, Keene CD, Verfaillie CM, Low WC. Human bone marrow stem cells exhibit neural phenotypes and ameliorate neurological deficits after grafting into the ischemic brain of rats. Exp Neurol. 2002;174:11–20. doi: 10.1006/exnr.2001.7853. [DOI] [PubMed] [Google Scholar]
- 111.Safford KM, Safford SD, Gimble JM, Shetty AK, Rice HE. Characterization of neuronal/glial differentiation of murine adipose-derived adult stromal cells. Exp Neurol. 2004;187:319–328. doi: 10.1016/j.expneurol.2004.01.027. [DOI] [PubMed] [Google Scholar]
- 112.Paul G, Özen I, Christophersen NS, Reinbothe T, Bengzon J, Visse E, et al. The adult human brain harbors multipotent perivascular mesenchymal stem cells. PLoS One. 2012;7:e35577. doi: 10.1371/journal.pone.0035577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hofstetter CP, Schwarz EJ, Hess D, Widenfalk J, El Manira A, Prockop DJ, et al. Marrow stromal cells form guiding strands in the injured spinal cord and promote recovery. Proc Natl Acad Sci U S A. 2002;99:2199–2204. doi: 10.1073/pnas.042678299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chopp M, Li Y. Treatment of neural injury with marrow stromal cells. Lancet Neurol. 2002;1:92–100. doi: 10.1016/S1474-4422(02)00040-6. [DOI] [PubMed] [Google Scholar]
- 115.Borlongan CV, Lind JG, Dillon-Carter O, Yu G, Hadman M, Cheng C, et al. Bone marrow grafts restore cerebral blood flow and blood brain barrier in stroke rats. Brain Res. 2004;1010:108–116. doi: 10.1016/j.brainres.2004.02.072. [DOI] [PubMed] [Google Scholar]
- 116.Weimann JM, Charlton CA, Brazelton TR, Hackman RC, Blau HM. Contribution of transplanted bone marrow cells to Purkinje neurons in human adult brains. Proc Natl Acad Sci U S A. 2003;100:2088–2093. doi: 10.1073/pnas.0337659100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lebeau G, Ah-Pine F, Daniel M, Bedoui Y, Vagner D, Frumence E, et al. Perivascular mesenchymal stem/stromal cells, an immune privileged niche for viruses? Int J Mol Sci. 2022;23:8038. doi: 10.3390/ijms23148038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Prockop DJ. Repair of tissues by adult stem/progenitor cells (MSCs): controversies, myths, and changing paradigms. Mol Ther. 2009;17:939–946. doi: 10.1038/mt.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Caplan AI, Correa D. The MSC: an injury drugstore. Cell Stem Cell. 2011;9:11–15. doi: 10.1016/j.stem.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Prockop DJ. “Stemness” does not explain the repair of many tissues by mesenchymal stem/multipotent stromal cells (MSCs) Clin Pharmacol Ther. 2007;82:241–243. doi: 10.1038/sj.clpt.6100313. [DOI] [PubMed] [Google Scholar]
- 121.Kovac A, Erickson MA, Banks WA. Brain microvascular pericytes are immunoactive in culture: cytokine, chemokine, nitric oxide, and LRP-1 expression in response to lipopolysaccharide ». J Neuroinflamm. 2011;8:139. doi: 10.1186/1742-2094-8-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.ElAli A, Thériault P, Rivest S. The role of pericytes in neurovascular unit remodeling in brain disorders. Int J Mol Sci. 2014;15:6453–6474. doi: 10.3390/ijms15046453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jansson D, Rustenhoven J, Feng S, Hurley D, Oldfield RL, Bergin PS, et al. A role for human brain pericytes in neuroinflammation. J Neuroinflamm. 2014;11:104. doi: 10.1186/1742-2094-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Le Blanc K, Frassoni F, Ball L, Locatelli F, Roelofs H, Lewis I, et al. Mesenchymal stem cells for treatment of steroid-resistant, severe, acute graft-versus-host disease: a phase II study. Lancet Lond Engl. 2008;371:1579–1586. doi: 10.1016/S0140-6736(08)60690-X. [DOI] [PubMed] [Google Scholar]
- 125.Bartholomew A, Sturgeon C, Siatskas M, Ferrer K, McIntosh K, Patil S, et al. Mesenchymal stem cells suppress lymphocyte proliferation in vitro and prolong skin graft survival in vivo. Exp Hematol. 2002;30:42–48. doi: 10.1016/S0301-472X(01)00769-X. [DOI] [PubMed] [Google Scholar]
- 126.Zappia E, Casazza S, Pedemonte E, Benvenuto F, Bonanni I, Gerdoni E, et al. Mesenchymal stem cells ameliorate experimental autoimmune encephalomyelitis inducing T-cell anergy. Blood. 2005;106:1755–1761. doi: 10.1182/blood-2005-04-1496. [DOI] [PubMed] [Google Scholar]
- 127.Singer NG, Caplan AI. Mesenchymal stem cells: mechanisms of inflammation. Annu Rev Pathol Mech Dis. 2011;6:457–478. doi: 10.1146/annurev-pathol-011110-130230. [DOI] [PubMed] [Google Scholar]
- 128.Nauta AJ, Fibbe WE. Immunomodulatory properties of mesenchymal stromal cells. Blood. 2007;110:3499–3506. doi: 10.1182/blood-2007-02-069716. [DOI] [PubMed] [Google Scholar]
- 129.Daniel M, Bedoui Y, Vagner D, Raffray L, Ah-Pine F, Doray B, et al. Pathophysiology of sepsis and genesis of septic shock: the critical role of mesenchymal stem cells (MSCs) Int J Mol Sci. 2022;23:9274. doi: 10.3390/ijms23169274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Silver J, Miller JH. Regeneration beyond the glial scar. Nat Rev Neurosci. 2004;5:146–156. doi: 10.1038/nrn1326. [DOI] [PubMed] [Google Scholar]
- 131.Johnson-Green PC, Dow KE, Riopelle RJ. Characterization of glycosaminoglycans produced by primary astrocytes in vitro. Glia. 1991;4:314–321. doi: 10.1002/glia.440040309. [DOI] [PubMed] [Google Scholar]
- 132.McKeon RJ, Jurynec MJ, Buck CR. The chondroitin sulfate proteoglycans neurocan and phosphacan are expressed by reactive astrocytes in the chronic CNS glial scar. J Neurosci Off J Soc Neurosci. 1999;19:10778–10788. doi: 10.1523/JNEUROSCI.19-24-10778.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schreiber J, Schachner M, Schumacher U, Lorke DE. Extracellular matrix alterations, accelerated leukocyte infiltration and enhanced axonal sprouting after spinal cord hemisection in tenascin-C-deficient mice. Acta Histochem. 2013;115:865–878. doi: 10.1016/j.acthis.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 134.Wyss-Coray T, Feng L, Masliah E, Ruppe MD, Lee HS, Toggas SM, et al. Increased central nervous system production of extracellular matrix components and development of hydrocephalus in transgenic mice overexpressing transforming growth factor-beta 1. Am J Pathol. 1995;147:53–67. [PMC free article] [PubMed] [Google Scholar]
- 135.Fernandez-Klett F, Potas JR, Hilpert D, Blazej K, Radke J, Huck J, et al. Early loss of pericytes and perivascular stromal cell-induced scar formation after stroke. J Cereb Blood Flow Metab. 2013;33:428–439. doi: 10.1038/jcbfm.2012.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Soderblom C, Luo X, Blumenthal E, Bray E, Lyapichev K, Ramos J, et al. Perivascular fibroblasts form the fibrotic scar after contusive spinal cord injury. J Neurosci. 2013;33:13882. doi: 10.1523/JNEUROSCI.2524-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fernandez-Klett F, Priller J. The fibrotic scar in neurological disorders. Brain Pathol. 2014;24:404–413. doi: 10.1111/bpa.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Behnan J, Isakson P, Joel M, Cilio C, Langmoen IA, Vik-Mo EO, et al. Recruited brain tumor-derived mesenchymal stem cells contribute to brain tumor progression. Stem Cells. 2014;32:1110–1123. doi: 10.1002/stem.1614. [DOI] [PubMed] [Google Scholar]
- 139.Birnbaum T, Roider J, Schankin CJ, Padovan CS, Schichor C, Goldbrunner R, et al. Malignant gliomas actively recruit bone marrow stromal cells by secreting angiogenic cytokines. J Neurooncol. 2007;83:241–247. doi: 10.1007/s11060-007-9332-4. [DOI] [PubMed] [Google Scholar]
- 140.Rodriguez FJ, Orr BA, Ligon KL, Eberhart CG. Neoplastic cells are a rare component in human glioblastoma microvasculature. Oncotarget. 2012;3:98–106. doi: 10.18632/oncotarget.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kulla A, Burkhardt K, Meyer-Puttlitz B, Teesalu T, Asser T, Wiestler OD, et al. Analysis of the TP53 gene in laser-microdissected glioblastoma vasculature. Acta Neuropathol (Berl) 2003;105:328–332. doi: 10.1007/s00401-003-0681-6. [DOI] [PubMed] [Google Scholar]
- 142.Ricci-Vitiani L, Pallini R, Biffoni M, Todaro M, Invernici G, Cenci T, et al. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468:824–830. doi: 10.1038/nature09557. [DOI] [PubMed] [Google Scholar]
- 143.deCarvalho AC, Nelson K, Lemke N, Lehman NL, Arbab AS, Kalkanis S, et al. Gliosarcoma stem cells undergo glial and mesenchymal differentiation in vivo. Stem Cells. 2010;28:181–190. doi: 10.1002/stem.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ricci-Vitiani L, Pallini R, Larocca LM, Lombardi DG, Signore M, Pierconti F, et al. Mesenchymal differentiation of glioblastoma stem cells. Cell Death Differ. 2008;15:1491–1498. doi: 10.1038/cdd.2008.72. [DOI] [PubMed] [Google Scholar]
- 145.Cheng L, Huang Z, Zhou W, Wu Q, Donnola S, Liu JK, et al. Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell. 2013;153:139–152. doi: 10.1016/j.cell.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Behnan J, Finocchiaro G, Hanna G. The landscape of the mesenchymal signature in brain tumours. Brain. 2019;142:847–866. doi: 10.1093/brain/awz044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Birnbaum T, Hildebrandt J, Nuebling G, Sostak P, Straube A. Glioblastoma-dependent differentiation and angiogenic potential of human mesenchymal stem cells in vitro. J Neurooncol. 2011;105:57–65. doi: 10.1007/s11060-011-0561-1. [DOI] [PubMed] [Google Scholar]
- 148.Kong BH, Shin H-D, Kim S-H, Mok H-S, Shim J-K, Lee J-H, et al. Increased in vivo angiogenic effect of glioma stromal mesenchymal stem-like cells on glioma cancer stem cells from patients with glioblastoma. Int J Oncol. 2013;42:1754–1762. doi: 10.3892/ijo.2013.1856. [DOI] [PubMed] [Google Scholar]
- 149.Clavreul A, Guette C, Faguer R, Tétaud C, Boissard A, Lemaire L, et al. Glioblastoma-associated stromal cells (GASCs) from histologically normal surgical margins have a myofibroblast phenotype and angiogenic properties. J Pathol. 2014;233:74–88. doi: 10.1002/path.4332. [DOI] [PubMed] [Google Scholar]
- 150.Valdor R, García-Bernal D, Bueno C, Ródenas M, Moraleda JM, Macian F, et al. Glioblastoma progression is assisted by induction of immunosuppressive function of pericytes through interaction with tumor cells. Oncotarget. 2017;8:68614–68626. doi: 10.18632/oncotarget.19804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Breznik B, Motaln H, Vittori M, Rotter A, Lah TT. Mesenchymal stem cells differentially affect the invasion of distinct glioblastoma cell lines. Oncotarget. 2017;8:25482–25499. doi: 10.18632/oncotarget.16041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rodini CO, da Silva PBG, Assoni AF, Carvalho VM, Okamoto OK. Mesenchymal stem cells enhance tumorigenic properties of human glioblastoma through independent cell-cell communication mechanisms. Oncotarget. 2018;9:24766–24777. doi: 10.18632/oncotarget.25346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hossain A, Gumin J, Gao F, Figueroa J, Shinojima N, Takezaki T, et al. Mesenchymal stem cells isolated from human gliomas increase proliferation and maintain stemness of glioma stem cells through the IL-6/gp130/STAT3 pathway. Stem Cells. 2015;33:2400–2415. doi: 10.1002/stem.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Svensson A, Ramos-Moreno T, Eberstål S, Scheding S, Bengzon J. Identification of two distinct mesenchymal stromal cell populations in human malignant glioma. J Neuro-Oncol. 2017;131:245–254. doi: 10.1007/s11060-016-2302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang Q, Yi D-Y, Xue B-Z, Wen W-W, Lu Y-P, Abdelmaksou A, et al. CD90 determined two subpopulations of glioma-associated mesenchymal stem cells with different roles in tumour progression. Cell Death Dis. 2018;9:1101. doi: 10.1038/s41419-018-1140-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yi D, Xiang W, Zhang Q, Cen Y, Su Q, Zhang F, et al. Human glioblastoma-derived mesenchymal stem cell to pericytes transition and angiogenic capacity in glioblastoma microenvironment. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol. 2018;46:279–290. doi: 10.1159/000488429. [DOI] [PubMed] [Google Scholar]
- 157.Li M, Li G, Kiyokawa J, Tirmizi Z, Richardson LG, Ning J, et al. Characterization and oncolytic virus targeting of FAP-expressing tumor-associated pericytes in glioblastoma. Acta Neuropathol Commun. 2020;8:1–13. doi: 10.1186/s40478-020-01096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ligorio M, Sil S, Malagon-Lopez J, Nieman LT, Misale S, Di Pilato M, et al. Stromal microenvironment shapes the intratumoral architecture of pancreatic cancer. Cell. 2019;178:160–175.e27. doi: 10.1016/j.cell.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Turley SJ, Cremasco V, Astarita JL. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat Rev Immunol. 2015;15:669–682. doi: 10.1038/nri3902. [DOI] [PubMed] [Google Scholar]
- 160.Shahar T, Rozovski U, Hess KR, Hossain A, Gumin J, Gao F, et al. Percentage of mesenchymal stem cells in high-grade glioma tumor samples correlates with patient survival. Neuro-Oncol. 2017;19:660–668. doi: 10.1093/neuonc/now239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable