Abstract
Protein synthesis is a highly regulated essential process. As such, it is subjected to substantial regulation in response to stress. One hallmark of the Integrated Stress Response (ISR) is the immediate shutdown of most translation through phosphorylation of the alpha subunit of translation initiation factor eIF2 and activation of eIF4E binding proteins. While these posttranslational modifications largely inhibit cap-dependent translation, many mRNA resist this inhibition by alternative translation mechanisms involving cis-regulatory sequences and structures in 5′ transcript leaders, including upstream Open Reading Frames (uORFs), Internal Ribosome Entry Sites (IRESes), and Cap-Independent Translation Elements (CITEs). Studies of uORF and IRES activity are often performed on a gene-by-gene basis; however, high-throughput methods have recently emerged. Here, we describe a protocol for Polysome Library Sequencing (PoLib-Seq; Fig. 1), a multiplexed assay of reporter gene translation that can be used during the ISR. A designer library of reporter RNAs are transfected into tissue-culture cells, and their translation is assayed via sucrose gradient fractionation followed by high-throughput sequencing. As an example, we include PoLib-seq results simultaneously assaying translation of wildtype and uORF mutant human ATF4 reporter RNAs, recapitulating the known function of uORF1 in resisting translational inhibition during the ISR.
Keywords: mRNA translation, Polysome gradient fractionation, uORFs, Massively parallel reporter assay
1. Introduction
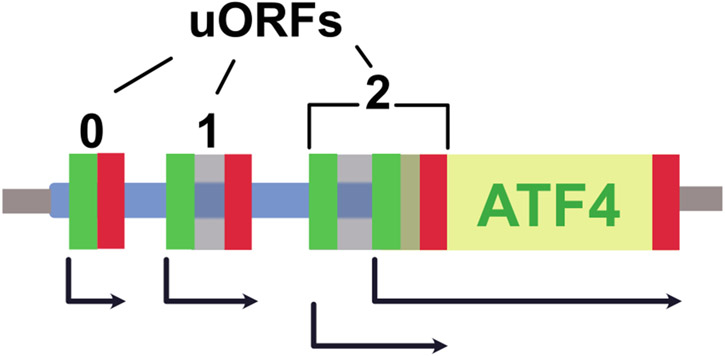
Inhibition of translation is a hallmark of the ISR. Multiple kinases, responding to a variety of stresses, phosphorylate the alpha subunit of eIF2, reducing the availability of ternary complex for translation initiation [1, 2]. While this globally represses mRNA translation, some stress-responsive transcripts continue to be translated through alternative initiation mechanisms controlled by cis-acting sequences and RNA structures located in 5′ transcript leaders [3]. For example, the transcript leader of human ATF4 encodes three uORFs (uORFs 0, 1, and 2; Fig. 1). Previous studies have shown that uORFs 1 and 2 in the homologous mouse ATF4 gene function to activate translation during the ISR [4]. After translation of the first uORF, which is only three amino acids in length, the small ribosomal subunit resumes scanning and quickly reacquires a new ternary complex. This allows subsequent reinitiation at uORF2, which overlaps the ATF4 CDS in an alternate reading frame and precludes translation of the ATF4 CDS. During the ISR, reduced levels of ternary complex delay translation reinitiation, allowing ribosomes to bypass uORF 2 and translate ATF4. Many other genes involved in the ISR are regulated via single uORFs, including CHOP, and GADD34 [1]. Thus, uORFs play a prominent role in the translational response to stress.
Fig. 1.
Human ATF4 transcript. The start (green boxes) and stop codons (red boxes) are indicated for the three ATF4 uORFs (transparent gray boxes) and the ATF4 CDS (yellow). The arrows indicate the location and frame of translation for the uORFs, and for ATF4, which is overlapped by the translation of uORF 2
Ribosome profiling studies have shown that uORFs are extremely common, being found at ~15% of yeast and −50% of human genes, respectively [5, 6]. Although the number of predicted uORFs has increased dramatically, relatively few have been experimentally tested for functions in regulating translation, especially during the ISR. Historically, uORF activity has been tested on a gene-by-gene basis primarily through the use of luciferase reporter assays. Usually, matched wild-type and uORF mutant transcript leaders are cloned upstream of luciferase and separately transfected to compare the amount of functional protein from each reporter, relative to reporter mRNA levels. Although effective, the relatively low throughput of these reporter assays is the chief reason why most uORFs have not been tested.
Over the past decade, several labs have developed Massively Parallel Reporter Assays (MPRAs) for high-throughput measurements of transcription [7, 8], splicing [9, 10], and translation [11-15]. Some translation MPRAs involve FACS-binning cells based on fluorescent reporter expression, while others separate reporter mRNAs based on their association with ribosomes through polysome sucrose-gradient fractionation. Separation by polysome fractionation has the advantage that it can be used to investigate acute translational responses to stress, as are seen in the ISR. However, this has not been reported previously. Here, we describe our protocol for Polysome Library Sequencing (PoLib-seq) in human tissue culture cells. We include PoLib-seq results using three ATF4 wild-type and uORF-mutant reporters as a proof-of-principle, recapitulating the established roles of uORFs 1 and 2 in the ATF4 translational response to stress (Fig. 2). By extension, large designer libraries cloned from pools of synthetic oligos [16] could be used to apply PoLib-seq to thousands of designer reporter RNAs simultaneously.
Fig. 2.
The effect on translation of the 5′ leader sequence of ATF4 during stress. HEK 293T cells were simultaneously transfected with wild-type (WT) and start codon mutants of the second (M2) and third (M3) uORFs (red Xs, uORFs 1 and 2) upstream of MGFP (green box). The cells were either subjected to sodium arsenite stress (orange 40 μM; 30 min) or left untreated (blue). The number of reads for each variant were counted in each fraction and normalized by the number of total reads in each fraction. Data shown are the averages of two biological replicates for three transcript variants assayed. As expected, the wild-type ATF4 transcript leader shifts into the polysome in response to stress (top, compare blue to gold). This translational induction does not occur in transcript leaders mutated for uORF1 (middle). Mutation of the inhibitory uORF2 (bottom) increases translation under both unstressed and stressed conditions (compare to WT, top)
2. Materials
Pipettes with filter tips (P1000, P200, P20, P2).
Monster Green® Fluorescent Protein phMGFP vector (Promega).
Custom primers (Table 1).
1.5-ml and 2.0-ml microcentrifuge tubes.
0.2-ml tubes for PCR.
Q5 Polymerase (New England Biolabs).
10 mM dNTP mix.
Nuclease-free water.
Thermocycler, any model.
Agarose.
TAE buffer (Tris–acetate EDTA buffer for gel electrophoresis).
SYBR safe DNA gel stain (or equivalent).
DpnI enzyme and CutSmart® buffer (New England Biolabs).
DNA Clean and Concentrator kit (Zymo Research).
NanoDrop™ 2000 Spectrophotometer or similar.
HiScribe T7 Quick High Yield RNA Synthesis kit (New England Biolabs).
3 M sodium acetate pH 5.2.
Acid-Phenol:Chloroform pH 4.5 (with IAA, 125:24:1).
Chloroform:Isoamyl alcohol 24:1.
Ethanol, absolute 200 proof (EtOH, molecular biology grade).
80% Ethanol: 80% absolute ethanol, 20% nuclease-free water.
Refrigerated centrifuge with rotor for 1.5- to 2.0-ml microcentrifuge tubes.
Thermomixer with 1.5-ml tube capacity (or similar).
Vaccinia Capping System (New England Biolabs).
2-propanol (isopropanol).
HEK 293T cell line.
Fetal bovine serum (FBS).
Dulbecco’s Modified Eagle Media (DMEM).
Disposable pipettes (25 ml, 10 ml, 5 ml).
10-cm tissue culture–treated culture dish.
Incubator suitable for tissue culture, supplied with 5% CO2.
50-ml conical tube.
Steriflip-GP sterile 50-ml centrifuge tube top filter unit (MilliporeSigma™).
Polysome Lysis Buffer (PLB): 20 mM Tris–HCl pH 7.5, 250 mM NaCl, 15 mM MgCl2, 0.5% Triton™ X-100. Prepare 50 ml in nuclease-free water. Filter sterilize using Steriflip-GP sterile 50-ml centrifuge tube top filter unit according to the manufacturer’s instructions. Store at 4 °C.
Opti-MEM Reduced Serum Media.
Lipofectamine® MessengerMAX™ transfection reagent (Life Technologies).
50 mM sodium arsenite: Mix 0.065 g sodium arsenite in 10 ml of nuclease-free water. Store at room temperature.
50 mg/ml cycloheximide: Mix 50 mg cycloheximide in 1 ml ethanol. Store at −20 °C.
1 M dl-dithiothreitol (DTT).
SUPERase-In™ RNase inhibitor, 20 U/μl (Invitrogen™).
TURBO™ DNase, 2 U/ml (Invitrogen™).
Phosphate-buffered saline (PBS).
Cell scraper.
10-ml syringe with Luer Tip.
20 gauge needle.
TRIzol™ Reagent.
RNA Clean and Concentrator kit (Zymo Research).
RNA reagents ScreenTape (Agilent Technologies).
TapeStation Instrument (Agilent Technologies, model 4400 or similar).
Sucrose.
10% Polysome Sucrose-Gradient Solution (10% PLB): 5 g of sucrose, 1 ml of 1 M Tris-HCl pH 7.5, 3.125 ml of 4 M NaCl, 750 μl of 1 M MgCl2, 250 μl of Triton™ X-100, and nuclease-free water to 50 ml. Agitate until the sucrose is fully dissolved into solution. Filter sterilize using Steriflip-GP sterile 50-ml centrifuge tube top filter unit according to the manufacturer’s instructions. Store at 4 °C.
50% Polysome Sucrose-Gradient Solution (50% PLB): 25 g of sucrose, 1 ml of 1 M Tris–HCl pH 7.5, 3.125 ml of 4 M NaCl, 750 μl of 1 M MgCl2, 250 μl of Triton™ X-100, and nuclease-free water to 50 ml. Agitate until the sucrose is fully dissolved into solution. Filter sterilize using Steriflip-GP sterile 50-ml centrifuge tube top filter unit according to the manufacturer’s instructions. Store at 4 °C.
Polyclear™ open top ultracentrifuge tubes 9/16″ × 3 ¾″ (Seton Scientific 7031).
Silicone grease.
Stainless steel 304 syringe needle.
Lint-free wipes.
Ultracentrifuge with Beckman Coulter SW 40 Ti swinging bucket rotor (Beckman Coulter).
Gradient station™ with gradient forming attachments (Biocomp Instruments).
Gilson Fraction collection system with Triax flow cell and fractionating tube mount (Biocomp Instruments).
Sodium dodecyl sulfate (SDS).
70% ethanol (70% EtOH): 70% ethanol, 30% ddH2O.
SuperScript™ IV Reverse Transcriptase (Thermo Fisher).
Magnetic separation rack for 0.2-ml tubes.
AMPure® XP magnetic PCR purification beads (Beckman Coulter).
100 μM forward N primer mix: 10 μl of each of the following: 100 μM ATF4_PCR1_N0-F, 100 μM ATF4_PCR1_N1-F, 100 μM ATF4_PCR1_N2-F, 100 μM ATF4_PCR1_N3-F, 100 μM ATF4_PCR1_N4-F, 100 μM ATF4_PCR1_N5-F, 100 μM ATF4_PCR1_N6-F, and 100 μM ATF4_PCR1_N7-F (Table 1).
100 μM reverse N primer mix: 10 μl of each of the following: 100 μM ATF4_PCR1_N0_R, 100 μM ATF4_PCR1_N2_R, 100 μM ATF4_PCR1_N4_R, and 100 μM ATF4_PCR1_N6_R (Table 1).
NEBNext® Multiplex Oligos for Illumina (unique dual index primer pairs, New England Biolabs).
D1000 ScreenTape and sample buffer (Agilent).
Qubit™ dsDNA HS Assay Kit (Invitrogen).
0.5-ml optically clear individual PCR tube.
Table 1.
Custom primers
| Primer name | Sequence (5′–3′) |
|---|---|
| phMGFP-T7-F | TAATACGACTCACTATAGG |
| phMGFP-GFP3p-R | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT TTTTTTTTTTTTTTTTTTTTTTTTTTTTGCTCGAAGCATTAACCCTC |
| MGFP-52-72-R | ACGAATTTGTGGCCGTTCAC |
| ATF4_PCR1_N0_F | TTCAGACGTGTGCTCTTCCGATCTTTTCTACTTTGCCCGCCCAC |
| ATF4_PCR1_N1_F | TTCAGACGTGTGCTCTTCCGATCTNTTTCTACTTTGCCCGCCCAC |
| ATF4_PCR1_N2_F | TTCAGACGTGTGCTCTTCCGATCTNNTTTCTACTTTGCCCGCCCAC |
| ATF4_PCR1_N3_F | TTCAGACGTGTGCTCTTCCGATCTNNNTTTCTACTTTGCCCGCCCAC |
| ATF4_PCR1_N4_F | TTCAGACGTGTGCTCTTCCGATCTNNNNTTTCTACTTTGCCCGCCCAC |
| ATF4_PCR1_N5_F | TTCAGACGTGTGCTCTTCCGATCTNNNNNTTTCTACTTTGCCCGCCCAC |
| ATF4_PCR1_N6_F | TTCAGACGTGTGCTCTTCCGATCTNNNNNNTTTCTACTTTGCCCGCCCAC |
| ATF4_PCR1_N7_F | TTCAGACGTGTGCTCTTCCGATCTNNNNNNNTTTCTACTTTGCCCGCCCAC |
| ATF4_PCR1_N0_R | GCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC TAGCAACGCTGCTGCTGAATG |
| ATF4_PCR1_N2_R | GCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC TNNAGCAACGCTGCTGCTGAATG |
| ATF4_PCR1_N4_R | GCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC TNNNNAGCAACGCTGCTGCTGAATG |
| ATF4_PCR1_N6_R | GCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC TNNNNNNAGCAACGCTGCTGCTGAATG |
3. Methods
3.1. In Vitro RNA Transcription Template Preparation
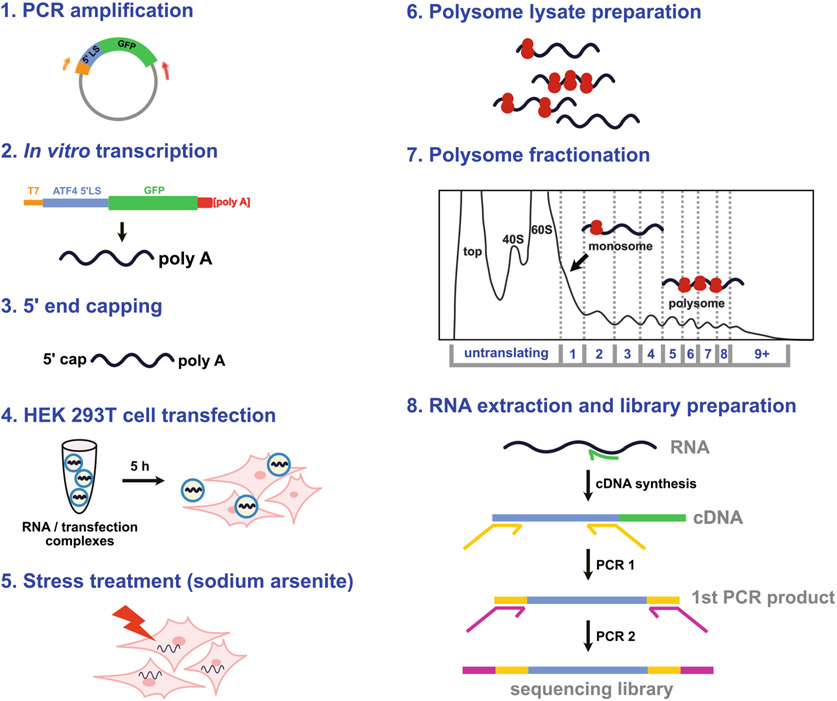
In this section, a template for in vitro transcription is produced by PCR amplification of the phMGFP vector (Fig. 3). The forward primer, phMGFP-T7-F (Table 1), anneals to the T7 polymerase site upstream of the 5′ LS, and the reverse primer, phMGFP-GFP3p-R (Table 1), anneals downstream of the GFP (green fluorescent protein) sequence. The reverse primer also incorporates a 70-nucleotide-long poly A tail sequence into the PCR product. For this step, the vector template can contain either a single 5′ leader sequence or a library of up to thousands of leader sequences.
Fig. 3.
Polib-seq protocol overview. (Step 1) The cloned library is amplified to generate a template for in vitro transcription. A T7 polymerase site and a poly A sequence are incorporated during this step. (Step 2) Uncapped in vitro transcribed RNA is produced using the PCR product as the template. (Step 3) A 5′ cap is enzymatically added to the RNA. (Step 4) HEK 293T cells are transfected for 5 h with the RNA. (Step 5) The cells are subjected to sodium arsenite for 30 min to induce the integrated stress response. (Step 6) Cell polysome lysates are prepared and fractionated over a sucrose gradient into 10 fractions (step 7). (Step 8) RNA is extracted from the fractions and sequencing libraries are prepared by targeted RT-PCR
Dilute the plasmid vector containing the 5′ leader sequence (s) of interest to 1 ng/μl in nuclease-free water.
- Set up the following reaction in a 0.2-ml PCR tube on ice.
Reagent Volume (μl) phMGFP-5′LS plasmid (1 ng/μl) 2 Nuclease-free water 64 5× Q5 reaction buffer 20 10 mM dNTPs 4 10 μM phMGFP-T7-F 4 10 μM phMGFP-GFP3p-R 4 Q5 polymerase 3 Mix the reaction by setting a pipettor to 50 μl and pipetting up and down several times.
- Put the reaction in a thermocycler and run the program:
- 98 °C for 30 s.
- 35 cycles of 98 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s (or 40 s per kb of product).
Run 10 μl of the PCR product on a 1% agarose gel in 1× TAE to check for successful amplification.
Purify the PCR product using the Zymo DNA clean & concentrator kit (or similar) according to the manufacturer’s instructions. Elute the PCR product in 45 μl of nuclease-free water.
- Set up the following reaction on ice.
Reagent Volume (μl) PCR product (step 6) 44 10× CutSmart buffer 5 DpnI (20 U/μl 1 Mix the reaction by setting pipettor to 25 μl and pipetting up and down several times.
Incubate the reaction for 1 h at 37 °C in a thermocycler.
Purify the PCR product using the Zymo DNA clean & concentrator kit according to the manufacturer’s instructions. Elute the PCR product in 10 μl of nuclease-free water.
Measure the concentration of the PCR product using a Nano-Drop™ spectrophotometer or something similar.
3.2. In Vitro RNA Transcription
In a 0.2-ml PCR tube, dilute the PCR product from Subheading 3.1, step 12 to 1 μg in 8 μl of nuclease-free water.
Add 10 μl of NTP Buffer mix followed by 2 μl of T7 RNA polymerase mix from the HiScribe™ T7 Quick High Yield RNA Synthesis Kit.
Mix by setting a pipettor to 10 μl and pipetting up and down several times. The total volume of the reaction should be 20 μl.
Incubate the reaction for 2 h at 37 °C in a thermocycler.
Add 30 μl of water to the reaction, followed by 2 μl DNase I (supplied in the kit) to the reaction. Incubate for 15 min at 37 °C in a thermocycler.
Transfer the reaction to a 1.5-ml tube. Add 150 μl of nuclease-free water, and 20 μl of 3 M sodium acetate, pH 5.2. Mix well using a pipettor, or briefly vortex.
Add 250 μl of acid-phenol:chloroform pH 4.5 (with IAA, 125: 24:1). Vortex on high for 15 s.
Centrifuge at 14,000 × g for 2 min at room temperature. Remove the supernatant to a new 1.5-ml tube.
Add 300 μl of chloroform:isoamyl alcohol 24:1 to the supernatant. Vortex on high for 15 s. Repeat step 8.
Add 600 μl of ethanol to the supernatant. Briefly vortex to mix and precipitate at −20 °C overnight, or for at least several hours.
Centrifuge the RNA at 20,000 × g for 30 min at 4 °C.
Carefully remove the supernatant as to not disturb the pellet.
Wash the pellet by adding 1 ml of 80% ethanol. Briefly vortex and centrifuge the RNA at 20,000 × g for 10 min at 4 °C.
Carefully remove the supernatant as to not disturb the pellet. Leave the lid open and let the remaining ethanol evaporate (roughly 2–5 min). A clear or slightly white pellet should be visible at the bottom of the tube.
Resuspend the pellet in 50 μl of nuclease-free water.
Measure the concentration of the RNA using a nanodrop. This transcription reaction produces up to about 180 μg of RNA.
Save the RNA at −20 °C overnight or continue on to Subheading 3.3.
3.3. 5′ Capping Reaction
Combine 100 μg of RNA (step 17) with nuclease-free water in a 1.5-ml tube to a final volume of 150 μl.
Denature the RNA by heating the RNA for 5 min at 65 °C. After the incubation is complete, immediately place the tube on ice for 5 min.
Proceed with the Vaccina Capping System kit. While the RNA is incubating on ice, prepare a 2 mM solution of SAM by adding 2 μl of 32 mM SAM to 30 μl nuclease-free water. Save the 2 mM SAM on ice.
- Set up the following reaction on ice in a 1.5-ml tube in the order specified.
Reagent Volume (μl) 100 μg denatured RNA (from step 2) 150 10× capping buffer 20 10 mM GTP 10 2 mM SAM (from step 3) 10 Vaccinia capping enzyme 10 Mix by setting a pipettor to 50 μl and pipetting up and down several times.
Aliquot the reaction into five 0.2-ml PCR tubes with 50 μl in each tube. Incubate the tubes at 37 °C in a thermocycler for 30 min.
Combine the reactions into one 1.5-ml tube. Add 700 μl of nuclease-free water, and 100 μl of 3 M sodium acetate, pH 5.2 to each tube. Briefly vortex.
Divide the reaction into two 1.5-ml tubes by adding 500 μl of the reaction to each tube.
To each tube, add 500 μl of acid-phenol:chloroform pH 4.5 (with IAA, 125:24:1). Vortex on high for 15 s.
Centrifuge at 14,000 × g for 2 min at room temperature. Remove the supernatants to new 1.5-ml tubes.
Add 500 μl of chloroform:isoamyl alcohol 24:1 to the supernatants. Vortex on high for 15 s.
Centrifuge at 14,000 × g for 2 min at room temperature. Remove each supernatant to one 2.0-ml tube. The volume of the supernatants is typically about 800–900 μl at this step.
Add one volume of isopropanol. Briefly vortex to mix and precipitate at −20 °C overnight, or for at least several hours.
Centrifuge the RNA at 20,000 × g for 30 min at 4 °C.
Carefully remove the supernatant as to not disturb the pellet.
Wash the pellet by adding 1 ml of 80% ethanol. Briefly vortex and centrifuge the RNA at 20,000 × g for 10 min at 4 °C.
Carefully remove the supernatant as to not disturb the pellet. Leave the lid open and let the remaining ethanol evaporate (roughly 2–5 min).
Resuspend the pellet in 50 μl of nuclease-free water.
Measure the concentration of the RNA using a nanodrop. 10 μg of RNA is required for each transfection. It may be necessary to repeat Subheadings 3.2 and 3.3 to obtain enough capped RNA for multiple transfections.
Save the RNA at −20 °C overnight or continue on to Subheading 3.4.
3.4. HEK 293 T Cell In Vitro RNA Transfection
For each transfection, seed two million cells in a 10-cm tissue culture dish in 10 ml DMEM + 10% FBS. Incubate at 37 °C in an incubator supplemented with 5% CO2 for 24 h.
Prepare 50 ml of Polysome Lysis Buffer (PLB, see Subheading 2).
For each transfection, add 30 μl of Lipofectamine® MessengerMAX™ transfection reagent to 500 μl of Opti-MEM Reduced Serum Media in a 1.5-ml tube. Incubate at room temperature for 10 min.
In a separate 1.5-ml tube, add 10 μg of RNA (Subheading 3.3, step 20) with 500 μl of Opti-MEM Reduced Serum Media.
Combine the Lipofectamine complexes (step 3) with the RNA (step 4) into one tube. Briefly mix with a pipettor set to 500 μl. Incubate the transfection mixture at room temperature for 5 min.
Remove the cells from the CO2 incubator and add the transfection mixture dropwise to the HEK 293T cells while gently swirling the dish. Place the cells back in the incubator. Incubate for 5.5 h for non-stress conditions, or 5 h for assays under stressed conditions.
For the stress condition, after 5 h of incubation, remove the tissue culture dish from the incubator. Add 8 μl of 50 mM sodium arsenite to the dish for a final sodium arsenite concentration of 40 μM. Briefly swirl the media in the dish. Place the tissue culture dish back into the incubator and incubate for 30 min.
While the cells are incubating, prepare enough PLB for each transfection (500 μl per transfection) on ice by adding 1 μl of 1 M DTT, 12 μl of TURBO™ DNase, 2 μl of 50 mg/ml cycloheximide, and 1 μl of SUPERase-In™ RNase inhibitor to 1 ml of PLB. Save the PLB on ice.
Prepare one 10-ml syringe with a 20 G needle attached per transfection.
Prepare 10 ml of ice-cold PBS per transfection. This can be done by aliquoting 10 ml of PBS into a 15-ml conical and burying the conical in ice in an ice bucket.
- Harvest the cells for each transfection as follows.
- Remove the tissue culture dish containing the cells from the incubator into a biosafety cabinet.
- Remove 0.5 ml of the media from the dish into a 1.5-ml tube.
- Add 20 μl of 50 mg/ml cycloheximide to the 0.5 ml of media and briefly vortex the tube.
- Add the media containing the cycloheximide to the cells in the tissue culture dish. Briefly swirl the media in the dish. Place the cells back into the incubator and incubate for 2 min.
- Remove the cells to a biosafety cabinet. Using a 10-ml pipette, quickly pipette all of the media off of the cells.
- Gently wash the cells by adding 10 ml of ice-cold PBS to the cells by pipetting down the side of the dish, taking care to not disturb the cells.
- Gently swirl the dish and place the dish onto ice.
- Quickly remove all of the PBS using a 10-ml pipette.
- Add 500 μl of the prepared PLB (step 8) dropwise to the cells.
- Tilt the dish to about a 20° angle while keeping the dish on ice.
- Using a cell scraper, scrape all of the cells down to the bottom of the dish that is closest to the ice.
- Titrate the cells in the dish by passing the cells through a 20 G needle about ten times.
- Remove the lysed cells to a 1.5-ml tube on ice. Incubate the lysate on ice for 10 min.
Once all of the lysates are collected, centrifuge the lysates for 10 min at 20,000 × g at 4 °C.
The cell debris should be collected at the bottom of the tube. Remove the lysate without disturbing the cell debris to a new tube. The total volume of the lysate at this point is usually about 0.6 ml.
For the total RNA sample, remove 50 μl of lysate to a new 1.5-ml tube. Proceed to Subheading 3.6.
Freeze and store the lysates at −80 °C. The lysates can be stored overnight or for up to several months.
3.5. Polysome Fractionation
Turn on and cool the ultracentrifuge to 4 °C. Also cool the SW40 rotor to 4 °C (keep in a cold room or refrigerator overnight).
- Prepare 6.5 ml each of 10% and 50% polysome sucrose-gradient solution (10% PSG and 50% PSG, see Subheading 2) per gradient by adding the reagents to the solutions as follows. Save the solutions on ice.
Reagent Final concentration DTT 1 mM Cycloheximide 0.1 mg/ml SUPERase-In™ RNase inhibitor 20 U/ml Place each ultracentrifuge tube into the marker block and mark the tube along the upper marking edge of the block. Lightly grease the inside of the tube with silicone grease using the provided bolt and piston tip. Label the ultracentrifuge tube with the sample name.
Prepare two 10-ml syringes with stainless steel syringe needles. To fill one syringe with approximately 7 ml of 10% PSG, pull the plunger up and down a few times while the syringe is submerged in the 10% PSG. This removes any air bubbles. Gently fill an ultracentrifuge tube with ~6.5 ml of 10% PSG, just so the liquid reaches the marked line.
Fill the other syringe with approximately 7 ml of 50% PSG (again, pull the plunger up and down in the PSG to remove the bubbles) and dry the outside of the tip using a lint-free wipe. Gently place the tip of the metal syringe tip at the bottom of the ultracentrifuge and slowly press the syringe plunger down. Observe the heavier, 50% PSG solution, displace the 10% PSG solution as it moves the 10% solution up the tube. Keep pressing the plunger down until the top of the 50% solution is at the marker line. Carefully remove the syringe as to not disrupt the layers. Repeat steps 4 and 5 for all of the gradients.
Place the rubber lids onto the ultracentrifuge tubes so that the small hole in the side of the lid is the last to be placed into the tube.
Turn on the gradient station and level the platform and tube holder where the gradients will be placed.
Carefully transfer the ultracentrifuge tubes to the gradient station tube holder. Run the 10–50% gradient program on the gradient station (see Note 1).
While the gradient station is running, thaw the lysates on ice if they were previously frozen.
Once the gradient station has finished the program, remove the ultracentrifuge tubes from the tube holder and gently place them on ice. Be sure that the tubes are sitting in the ice so that they are upright and not at an angle.
Remove the rubber lids. Using a pipette, remove the equivalent volume of lysate plus an additional 100 μl. For example, if the volume of the lysate is 600 μl, remove 700 μl of the sucrose gradient from the top of the tube.
Place the gradients into the SW40 buckets. Be sure to record which ultracentrifuge tube was placed into each bucket.
Using a pipettor, gently layer the lysate on top of the gradients by pipetting the sample down the side of the tube. This avoids dropping the sample directly on top of the gradient which could disrupt the layers.
Weigh each bucket with the gradients in them (along with the caps to the buckets) and balance the buckets that will be across from each other in the rotor using the PLB stock solution.
Cap the buckets and place them into the rotor. If there are empty buckets, be sure to load those as well. If there are an odd number of samples, prepare a mock gradient and place it in the bucket opposite of the odd tube.
Load the rotor into the centrifuge. Set the acceleration and de-acceleration to 1 and 7, respectively. Centrifuge the gradients at 35,600 rpm (160,000 × g) for 2.5 h at 4 °C with the vacuum on.
While the centrifuge is running, clean the metal syringe tips and rubber caps by soaking in 10% bleach for 30 min, followed by rinsing with distilled water.
Label thirty 1.5-ml microcentrifuge tubes per sample and set aside.
See Note 2. Zero the Triax flow cell using ddH2O. Leave the fraction collection system on to warm up for at least 30 min before use.
Remove the rotor from the ultracentrifuge and save the buckets with the gradients on a stable surface as to not tilt the buckets—in an ice bucket, a cold room, or a fridge.
Place thirty 1.5-ml labeled tubes with the lids open, in the tube holder for the fractionator in the correct orientation (see fractionator’s user manual).
Put a silicone-greased piston tip onto the piston and load one sample under the piston.
Set up the fractionator to fractionate the sample into 30 fractions using the default settings. Each fraction will be about 400 ml, with the exception of the last fraction which will be less.
After the fractionator has finished with fractionating one gradient, rinse the system following the prompts, with ddH2O. Repeat steps 21–23 for each gradient.
After the last fraction, follow the manufacturer’s suggested protocol for cleaning and storing the instrument. We run mock samples filled with warm 0.5% SDS followed by ddH2O. Then, the instrument lines are rinsed with 70% EtOH and air is pumped through to dry.
At this point, the fractions can be saved at −20 °C, or they can be combined as desired (next steps).
Examine the polysome traces to determine which fractions are representative of the peaks of interest. Combine the fractions as needed so that the total volume of the combined fractions is 400 μl. For example, if fraction numbers 9 and 10 are where the monosome peak is on the polysome trace, combine 200 μl of fraction 9 and 200 μl of fraction 10 into one 1.5-ml ultracentrifuge tube.
Either save the combined fractions at −20 °C or proceed to Subheading 3.6.
3.6. RNA Extraction
Perform steps 1, 2, and 4 in a fume hood. Dispose of TRIzol™ reagent and chloroform as chemical hazardous wastes.
Add 800 μl of TRIzol™ reagent to each of the 400 μl of combined fractions (or 50 μl of total unfractionated lysate, Subheading 3.4, step 14) and vortex briefly. Incubate at room temperature for 5 min.
Add 300 μl of chloroform:isoamyl alcohol 24:1 to the TRIzol™ Reagent/lysate mix and briefly vortex. Incubate at room temperature for 2 min.
Centrifuge at 12,000 × g for 15 min at 4 °C.
Carefully remove the supernatant (roughly 900 μl) to a new 2.0-ml tube.
Add one volume of ethanol and briefly vortex.
Purify the RNA using a Zymo RNA clean and concentrator-5 kit as per the manufacturer’s instructions, and as listed in the subsequent steps. All of the centrifuge spins are at 12,000 × g at room temperature.
Transfer up to 700 μl of the sample to the spin column and centrifuge for 30 s. Discard flow through. Repeat this step as necessary until all of the sample from step 5 has been added to the column.
Add 400 μl of RNA Prep Buffer and centrifuge for 30 s. Discard the flow through.
Add 700 μl of RNA Wash Buffer and centrifuge for 30 s. Discard the flow through.
Add 400 μl of RNA Wash Buffer and centrifuge for 30 s. Discard the flow through.
Centrifuge the column for an additional 2 min to dry the membrane.
Transfer the column to a clean 1.5-ml tube with the cap removed. Add 45 μl of nuclease-free water to the column membrane.
Elute the RNA by centrifuging the column at room for 1 min.
Transfer the eluate to a fresh 1.5-ml tube.
For total RNA (unfractionated) samples, check the quality of the lysate preparation by assaying RNA integrity. Run 1 μl of the total RNA on an RNA ScreenTape using an Agilent TapeStation, following the manufacturer’s instructions. The RNA Integrity Value (RIN) should be at least 9.0 for high-quality RNA. The typical range of RIN numbers for this assay ranges between 9.5 and 10.0.
Save the RNA at −20 °C or proceed to Subheading 3.7.
3.7. DNase Treatment
- Set up a DNase treatment reaction in a 0.2-ml PCR tube as follows.
Reagent Volume (μl) RNA (Subheading 3.6, step 14) 43 10× TURBO™ DNase buffer 5 SUPERase-In™ RNase inhibitor 1 TURBO™ DNase 1 Pipette up and down several times with a P200 pipettor set to 45 μl to mix.
Incubate the reaction at 37 °C for 30 min.
Purify the RNA over an RNA clean and concentrator—5 column following the manufacturer’s instructions. Elute the RNA in 15 μl of nuclease-free water.
Quantify the RNA using a nanodrop.
Either save the RNA at −20 °C or proceed to Subheading 3.8.
3.8. Reverse Transcription
- In a 0.2-ml tube, set up the reaction on ice as follows.
Reagent Volume RNA (Subheading 3.7, step 8) 250 ng to 2.5 μg (max 11 μl) 10 mM dNTPs 1 μl 2 μM MGFP-52-72-R primer 1 μl Nuclease-free water To 13 μl Incubate the reaction for 5 min at 65 °C in a thermocycler. Place the reaction on ice immediately after the incubation is complete.
- To the reaction, add the following.
Reagent Volume (μl) 5× Superscript™ IV buffer 4 100 mM DTT 1 SUPERase-In™ RNase inhibitor 1 Superscript™ IV reverse transcriptase 1 In a thermocycler, incubate the reaction for 30 min at 50 °C. Heat inactivate at 80 °C for 10 min.
Add 30 μl of nuclease-free water to the cDNA.
Prepare the bottle of AMPure XP beads by vortexing until the beads are fully resuspended.
Add 50 μl of AMPure XP of Agencourt beads to the cDNA. Mix well by setting a pipettor to 50 μl and pipetting up and down.
Incubate the cDNA and beads at room temperature for 5 min.
Place the tube on a magnetic separation rack. Incubate at room temperature for 2 min. All of the beads should move to one side and the supernatant should be clear.
Pipette off the supernatant and discard.
While the tubes are still on the magnetic rack, wash the beads by adding 180 μl of 80% EtOH to the side of the tube opposite of the beads. Incubate at room temperature for 30 s.
Pipette off the 80% EtOH and discard.
While the tube is still on the rack with the lids open, pipette off the ethanol that has been collected at the bottom of the tube and let the remaining ethanol evaporate. The bead pellet should appear to be dry (not shiny) and more matte in appearance.
Resuspend the beads by removing the tube from the magnet and adding 20 μl of nuclease-free water to the pellet. Pipette up and down several times so that the beads are fully resuspended. Incubate at room temperature for 2 min.
Place the tube back on the magnet and wait for the supernatant to become clear (approximately 30 s to 1 min).
Remove the supernatant which now contains the cDNA to a new 1.5-ml tube.
Either save the cDNA at −20 °C or continue with Subheading 3.9.
3.9. Sequencing Library Preparation
Prepare a 10 μM F/R primer mix by adding 10 μl of 100 μM forward N primer mix and 10 μl of 100 μM reverse N primer mix (see Subheading 2), to 80 μl of nuclease-free water. The primer mix can be stored at −20 °C.
- In a 0.2-ml tube, set up the reaction on ice as follows.
Reagent Volume Purified cDNA (Subheading 3.8, step17) 3 Nuclease-free water 32 5× Q5 buffer 10 10 mM dNTPs 2 10 μM F/R primer mix (step 1) 2 Q5 polymerase 1 - Put the reaction in a thermocycler and run the program:
- 98 °C for 30 s.
- 6 cycles of 98 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s.
Prepare the bottle of AMPure XP beads by vortexing until the beads are fully resuspended.
Add 50 μl of AMPure XP of Agencourt beads to the amplification reaction. Mix well by setting a pipettor to 50 μl and pipetting up and down.
Incubate amplification reaction with the beads at room temperature for 5 min.
Place the tube on a magnetic separation rack. Incubate at room temperature for 2 min. All of the beads should move to one side, and the supernatant should be clear.
Pipette off the supernatant and discard.
While the tubes are still on the magnetic rack, wash the beads by adding 180 μl of 80% EtOH to the side of the tube opposite of the beads. Incubate at room temperature for 30 s.
Pipette off the 80% EtOH and discard.
While the tube is still on the rack with the lids open, pipette off the ethanol that has been collected at the bottom of the tube and let the remaining ethanol evaporate. The bead pellet should appear to be dry (not shiny) and more matte in appearance.
Resuspend the beads by removing the tube from the magnet and adding 50 μl of nuclease-free water to the pellet. Pipette up and down several times so that the beads are fully resuspended. Incubate at room temperature for 2 min.
Place the tube back on the magnet and wait for the supernatant to become clear (approximately 30 s to 1 min).
Remove the supernatant which now contains the library to a new 1.5-ml tube.
Either save the library at −20 °C or continue with the subsequent steps.
Dilute the NEBNext® Multiplex Oligos for Illumina to 2 μM by adding 5 μl of undiluted primer to 20 μl of nuclease-free water. This can be done in 0.2-ml PCR strip tubes or 1.5-ml tubes. Take care to record which well in the plate in the undiluted primers originated from.
- In a 0.2-ml tube, set up the reaction on ice as follows. Be sure to add a different primer pair to each sample, and record which sample received which primer pair.
Reagent Volume (μl) Purified library PCR 1 (from step 15) 2 Nuclease-free water 11.5 5× Q5 buffer 5 10 mM dNTPs 1 2 μM NEB primer (step 16) 5 Q5 polymerase 0.5 - Put the reaction in a thermocycler and run the program:
- 98 °C for 30 s.
- 15 cycles of 98 °C for 10 s, 64 °C for 10 s, and 72 °C for 30 s.
Check for amplification and library quality using a DNA ScreenTape (D1000 or similar). Load 1 μl of the PCR product onto the TapeStation (Fig. 4). If the PCR products are the correct size and the no RT control is negative for a band, proceed to with the next steps.
Add 25 μl of AMPure XP of Agencourt beads to the amplification reaction. Mix well by setting a pipettor to 50 μl and pipetting up and down.
Continue with the bead cleanup procedure (steps 6–11 above).
Resuspend the beads by removing the tube from the magnet and adding 15 μl of nuclease-free water to the pellet. Pipette up and down several times so that the beads are fully resuspended. Incubate at room temperature for 2 min.
Place the tube back on the magnet and wait for the supernatant to become clear (approximately 30 s to 1 min).
Remove the supernatant which now contains the library to a new 1.5-ml tube.
Either save the library at −20 °C or continue with the subsequent steps.
Quantify the concentration using the Qubit™ dsDNA HS Assay Kit according to the manufacturer’s instructions. Typically, 1–2 μl of library is sufficient for accurate quantification. At this point, the libraries can be pooled and run on an Illumina sequencer.
Fig. 4.
TapeStation gel image of sequencing library final PCR products (Lanes A1, B1, C1). A standard DNA ladder is shown in lane A0, and molecular size standards are at 1500 bp (purple) and 25 bp (green). The libraries range from 355 to 368 bp due to the incorporation of up to 13 random bases during the first PCR step. The PCR product will sometimes contain a small amount of non-specific product (~700 bp) which does not affect the quality of the sequencing library. Non-specific products and excess primers less than 100 bp are removed during the subsequent bead clean up step
4. Notes
- We use the gradient (10–50%) program as described in Aboulhouda et al. [17] (described below). Depending on the gradient platform being used, other preset programs for 10–50% sucrose gradients can be used.
- 05/85/35.
- 01/77/0.
- 04/86/35.
- 03/86.5/35.
- 20/81/14.
-
07/86/20.Sequence = abcbdbabcbdbef.
The protocol for fractionation may be different depending on the fractionator being used. The steps in this procedure are for the Gilson fraction collection system. Refer to the user’s manual for the Gilson fraction collection system for more detailed instructions or to your particular fraction collector for alternative instructions.
References
- 1.Ryoo HD, Vasudevan D (2017) Two distinct nodes of translational inhibition in the integrated stress response. BMB Rep 50:539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wek RC (2018) Role of eIF2α kinases in translational control and adaptation to cellular stress. Cold Spring Harb Perspect Biol 10:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pakos-Zebrucka K, Koryga I, Mnich K et al. (2016) The integrated stress response. EMBO Rep 17:1374–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vattem KM, Wek RC (2004) Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A 101:11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, Wang Y, Lu J (2019) Function and evolution of upstream ORFs in eukaryotes. Trends Biochem Sci 44:782–794 [DOI] [PubMed] [Google Scholar]
- 6.McGillivray P, Ault R, Pawashe M et al. (2018) A comprehensive catalog of predicted functional upstream open reading frames in humans. Nucleic Acids Res 46:3326–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melnikov A, Murugan A, Zhang X et al. (2012) Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol 30:271–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharon E, Kalma Y, Sharp A et al. (2012) Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat Biotechnol 30:521–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soemedi R, Cygan KJ, Rhine CL et al. (2017) Pathogenic variants that alter protein code often disrupt splicing. Nat Genet 49:848–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adamson SI, Zhan L, Graveley BR (2018) Vex-seq: high-throughput identification of the impact of genetic variation on pre-mRNA splicing efficiency. Genome Biol 19:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sample PJ, Wang B, Reid DW et al. (2019) Human 5′ UTR design and variant effect prediction from a massively parallel translation assay. Nat Biotechnol 37:803–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dvir S, Velten L, Sharon E et al. (2013) Deciphering the rules by which 5′-UTR sequences affect protein expression in yeast. Proc Natl Acad Sci U S A 110:E2792–E2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin Y, May GE, Kready H et al. (2019) Impacts of uORF codon identity and position on translation regulation. Nucleic Acids Res 2:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia L, Mao Y, Ji Q et al. (2020) Decoding mRNA translatability and stability from the 5′ UTR. Nat Struct Mol Biol 27:814–821 [DOI] [PubMed] [Google Scholar]
- 15.Noderer WL, Flockhart RJ, Bhaduri A et al. (2014) Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol Syst Biol 10:748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.May GE, McManus CJ (2022) High-throughput quantitation of yeast uORF regulatory impacts using FACS-uORF. Methods Mol Biol 2404:331–351. 10.1007/978-1-0716-1851-6_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aboulhouda S, Di Santo R, Therizols G, Weinber D (2017) Accurate, Streamlined Analysis of mRNA Translation by Sucrose Gradient Fractionation. Bio Protoc 7(19):e2573. 10.21769/BioProtoc.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]