Abstract
Background
Genomic surveillance of SARS-CoV-2 is crucial for monitoring the spread of COVID-19 and guiding public health decisions, but the capacity for SARS-CoV-2 testing and sequencing in Africa is low. We integrated SARS-CoV-2 surveillance into an existing influenza surveillance network with the aim of providing insights into SARS-CoV-2 transmission and genomics in Ghana.
Methods
In this molecular epidemiological analysis, which is part of a wider multifaceted prospective observational study, we collected national SARS-CoV-2 test data from 35 sites across 16 regions in Ghana from Sept 1, 2020, to Nov 30, 2021, via the Ghanaian integrated influenza and SARS-CoV-2 surveillance network. SARS-CoV-2-positive samples collected through this integrated national influenza surveillance network and from international travellers arriving in Accra were sequenced with Oxford Nanopore Technology sequencing and the ARTIC tiled amplicon method. The sequence lineages were typed with pangolin and the phylogenetic analysis was conducted with IQ-Tree2 and TreeTime.
Findings
During the study period, 5495 samples were submitted for diagnostic testing through the national influenza surveillance network (2121 [46·1%] of 4021 samples with complete demographic data were from female individuals and 2479 [53·9%] of 4021 samples were from male individuals). We also obtained 2289 samples from travellers who arrived in Accra and had a positive lateral flow test, of whom 1626 (71·0%, 95% CI 69·1–72·9) were confirmed to be SARS-CoV-2 positive. Co-circulation of influenza and SARS-CoV-2 in Ghana was detected, with increased cases of influenza in November, 2020, November, 2021, and January and June, 2021. In 4124 samples from individuals with influenza-like illness, SARS-CoV-2 was identified in 583 (14·1%, 95% CI 13·1–15·2) samples and influenza in 356 (8·6%, 7·8–9·5). Conversely, in 476 samples from individuals with of severe acute respiratory illness, SARS-CoV-2 was detected in 58 (12·2%, 9·5–15·5) samples and influenza in 95 (19·9%, 16·5–23·9). We detected four waves of SARS-CoV-2 infections in Ghana; each wave was driven by a different variant: B.1 and B.1.1 were the most prevalent lineages in wave 1, alpha (B.1.1.7) was responsible for wave 2, delta (B.1.617.2) and its sublineages (closely related to delta genomes from India) were responsible for wave 3, and omicron variants were responsible for wave 4. We detected omicron variants among 47 (32%) of 145 samples from travellers during the start of the omicron spread in Ghana (wave 4).
Interpretation
This study shows the value of repurposing existing influenza surveillance platforms to monitor SARS-CoV-2. Influenza continued to circulate in Ghana in 2020 and 2021, and remained a major cause of severe acute respiratory illness. We detected importations of SARS-CoV-2 variants into Ghana, including those that did or did not lead to onward community transmission. Investment in strengthening national influenza surveillance platforms in low-income and middle-income countries has potential for ongoing monitoring of SARS-CoV-2 and future pandemics.
Funding
The EDCTP2 programme supported by the EU.
Introduction
The first cases of SARS-CoV-2 infection in Ghana were reported in early March, 2020, among international travellers.1 Up to November, 2022, there were four waves of COVID-19 in Ghana, particularly in Accra and other major urban settings. As of April 21, 2023, Ghana had recorded 171 657 cases of SARS-CoV-2 infection with 1462 deaths, but serological surveillance suggests higher levels of transmission and substantial under-ascertainment of SARS-CoV-2 infection.2
To minimise further importation and transmission of SARS-CoV-2 in the country, the government introduced several measures, which initially included halting all flights into the country and closing land borders on March 22, 2020. Despite these interventions, transmission continued, leading the government to close schools and universities, announce a partial lockdown of Accra and Greater Kumasi (from March 30 to April 22, 2020), and enhance contact tracing. These restrictions were lifted by July, 2020, and air travel restarted on Sept 1, 2020. Between Sept 1, 2020, and March 28, 2022, air travellers were required to have a negative COVID-19 test 72 h before entering Ghana and to undergo an antigen test for SARS-CoV-2 at the Kotoka International Airport, regardless of their vaccination status. All SARS-CoV-2-positive travellers were quarantined for 1 week and samples were collected on day 3.3 Ghana's land borders were closed throughout the duration of the study.
Research in context.
Evidence before this study
Before the COVID-19 pandemic, many countries had established surveillance networks for monitoring influenza. At the start of the pandemic, the WHO Global Influenza Surveillance and Response System recommended the integration of the surveillance of SARS-CoV-2 and other respiratory pathogens into influenza surveillance. We searched PubMed, medRxiv, and Google Scholar using a combination of search terms (ie, “SARS-CoV-2”, “COVID-19”, “influenza surveillance”, “genomic surveillance”, “Ghana”, “Africa”, and “genomic sequencing”) to identify articles and preprints on the integration of influenza and SARS-CoV-2 surveillance, as well as those that described influenza activity during the pandemic (especially in low-income and middle-income countries). We also reviewed recommendations on SARS-CoV-2 and influenza surveillance and reports published by WHO. We reviewed all studies and latest recommendations published in English from database inception to March 1, 2023. Several reports, predominantly from Europe and Australia with one study in South Africa, suggest that in many settings, circulation of influenza has been substantially reduced during the COVID-19 pandemic. Despite the WHO recommendation of integrated influenza and SARS-CoV-2 surveillance, we did not identify any studies reporting on the roll-out of such surveillance in Africa.
Added value of this study
We were able to show that SARS-CoV-2 surveillance could be effectively included in an existing influenza surveillance network. Influenza continued to circulate in Ghana during the COVID-19 pandemic and was a major cause of both influenza-like illness and severe acute respiratory illness. By use of this national surveillance platform, we were able to distinguish between SARS-CoV-2 variants that were imported and resulted in onward community transmission and SARS-CoV-2 variants that did not establish ongoing transmission.
Implications of all the available evidence
Our study highlights the value of existing influenza surveillance networks with integrated genomic surveillance to track the spread and evolution of SARS-CoV-2. These networks can provide important and timely data to inform ministries of health on the evolving course of the SARS-CoV-2 pandemic. Investment in strengthening national influenza surveillance platforms in low-income and middle-income countries has potential for ongoing monitoring of SARS-CoV-2 and future pandemics due to respiratory viruses.
In keeping with other countries, Ghana had to rapidly establish platforms for diagnostic testing and genomic surveillance of SARS-CoV-2. Alongside establishing new testing facilities, a complementary strategy was the repurposing of existing surveillance networks. Ghana established a national influenza surveillance network in 2007, which was formally recognised by WHO in 2010.4 In collaboration with the Ghana Health Service (GHS), this network provides sentinel surveillance for influenza at 29 sites across ten regions in Ghana.
In response to the introduction and circulation of SARS-CoV-2 in Ghana, we repurposed the influenza surveillance network to provide diagnostic testing for both influenza and SARS-CoV-2. As well as providing community surveillance for both influenza and SARS-CoV-2, this platform would allow surveillance for SARS-CoV-2 variants of interest and variants of concern that emerged since November, 2020, through genomic sequencing. We also aimed to combine targeted testing and sequencing of international traveller samples with our integrated influenza and SARS-CoV-2 surveillance platform. This strategy provided a way to assess introductions and community spread of SARS-CoV-2 variants throughout the COVID-19 pandemic. In this Article, we report our findings with the aim of providing insights into the epidemiology of influenza and COVID-19 and SARS-CoV-2 genomics in Ghana.
Methods
Study design and population
In this molecular epidemiological analysis, which is part of a wider multifaceted prospective observational study (appendix p 36), we analysed combined nasopharyngeal and oropharyngeal swabs collected from two groups of individuals in Ghana. The first group consisted of all samples collected through the integrated national influenza surveillance network between Sept 1, 2020, and Nov 30, 2021. During the period of the study, the surveillance network was expanded from 29 sites across ten regions in Ghana to a total of 35 sites, including six new regions (appendix p 5). Each week, the first five individuals who presented with an influenza-like illness or a severe acute respiratory illness at each site had samples collected by local health-care staff that were sent to the Noguchi Memorial Institute for Medical Research for combined influenza and SARS-CoV-2 testing. The second group consisted of international travellers arriving in Accra. All travellers who arrived in Accra were required to take a lateral flow test for SARS-CoV-2. In individuals who had a positive lateral flow test, a second swab was collected for PCR. Samples from individuals who had a positive PCR collected between Sept 1, 2020, and Nov 30, 2021, were included in this study (appendix p 6). All SARS-CoV-2-positive samples from the surveillance platform and all positive PCR tests of returning travellers between Sept 1, 2020, and Nov 30, 2021 were used for SARS-CoV-2 sequencing. There were no exclusion criteria. Historical data from the influenza surveillance network from 2017–19 were extracted for comparison. Data on sex, age, and region of samples were collected through the influenza surveillance network. Sex data were self-reported by individuals and the options male or female were provided.
This study was approved by the GHS (GHS-ERC 019/02/21), the Noguchi Memorial Institute for Medical Research (CPN 048/20–21 IORG 0000908), and the London School of Hygiene & Tropical Medicine ethics committee (22477). The sentinel sites for routine national influenza virus surveillance in Ghana were set up in collaboration with the GHS and the Ghana Armed Forces as part of the Integrated Disease Surveillance and Response system of the GHS. The study protocol is avaiable in the appendix (p 36).
Oral consent was sought from all participants before samples were collected. The requirement for specific consent for this study was waived as the study only used residual material that was already being collected for influenza surveillance.
Laboratory testing for SARS-CoV-2 and influenza
Data were anonymised before use and only laboratory identities were used during data analysis. Viral RNA was extracted from nasopharyngeal and oropharyngeal swabs with the QIAamp Viral RNA extraction kit (Qiagen, Hilden, Germany) following manufacturer instructions. Community samples from the influenza surveillance network underwent PCR testing for both influenza and SARS-CoV-2 via a multiplex primer set from the US Centers for Disease Control and Prevention targeting influenza A (eg, matrix gene), a non-structural gene segment of influenza B, and a nucleocapsid gene of SARS-CoV-2.5 Samples from travellers were tested only for SARS-CoV-2 with two commercial kits: Veri-Q nCoV-OM (MiCo BioMed, Gyeongju, South Korea), which targets ORF3a and nucleoprotein, and the Novel Coronavirus (2019-nCoV) Nucleic Acid Diagnostic Kit (Sansure Biotech, Changsha, China), which targets ORF1ab and nucleocapsid gene. Samples with a SARS-CoV-2 cycle threshold value below 30 were selected for whole-genome sequencing.
Library preparation and sequencing
The ARTIC amplicon sequencing protocol (ARTIC Network, Fareham, UK) and MinION (Oxford Nanopore Technologies, Oxford, UK) were used for SARS-CoV-2 sequencing. The manufacturer protocol for the QIAamp viral RNA extraction kit was modified to use 280 μL of starting material to isolate RNA, then elution in 30 μL of elution buffer. Complementary DNA was synthesised with Lunascript Reverse Transcriptase (New England Biolabs, Ipswich, MA, USA) and 400 bp amplicons were generated with the ARTIC 2019-nCoV version 3 primer sets.6 Products were quantified with the Qubit dsDNA High-Sensitivity Assay Kit (ThermoFisher Scientific, Waltham, MA, USA) and DNA concentrations were normalised to 15 ng/μL for further use. Amplicons were subjected to end repair with the Ultra II End Prep (New England Biolabs, Ipswich, MA, USA). DNA fragments were barcoded with the NBXX Barcode Kit (Oxford Nanopore Technologies), after which a clean-up was done with Agencourt AMPure XP (Beckman Coulter, Brea, CA, USA) beads. Adapters were then added via Adapter Mix II (Oxford Nanopore Technologies) with Quick T4 DNA Ligase (New England Biolabs). This solution was cleaned with Agencourt AMPure XP beads and loaded onto the MinION. Lineages were typed with pangolin version 3.1.17 and pangoLEARN version 2021–12–06. Sequences with more than 50% coverage were deposited into the Global Initiative on Sharing All Influenza Data (GISAID; appendix p 2).
Phylogenetic analysis of SARS-CoV-2 sequences
Sequences with more than 10 times the median read depth and more than 70% genome coverage were used for further analysis (appendix p 2). Two phylogenetic analyses were done. First, a time-scaled phylogeny including sequences from Ghana generated during our study was constructed to estimate the emergence and introduction time of SARS-CoV-2 lineages detected in the country. Second, maximum likelihood trees were built for each wave, combining sequences from Ghana and their closest-matching global sequences, to interrogate the phylogenetic relationships of SARS-CoV-2 lineages detected in Ghana to the global phylogeny.
The local, time-scaled phylogeny was reconstructed with a maximum-likelihood-based method.7 To ensure the quality of the analysis, the sequences had to pass the initial quality control criteria. Sequences with more than 10 times the median read depth and more than 70% genome coverage were aligned to the SARS-CoV-2 reference strain Wuhan-Hu-1 (GenBank accession number MN908947.3) with Multiple Alignment using Fast Fourier Transform version 7.477 (European Molecular Biology Laboratory European Bioinformatics Institute, Cambridge, UK).8 Previously reported problematic positions,9 obvious sequencing error, and potential homoplasic positions were masked and removed from the alignment file by one author (SNH). The processed alignment was used to construct a maximum likelihood tree with IQ-Tree2 version 2.1.2.10 The IQ-Tree2 analysis was done with the generalised time-reversible (GTR)+G model (−m GTR+G) of nucleotide substitution,11, 12 the default ultrafast bootstrapping value of 1000 (−bb 1000), and the Shimodaira–Hasegawa-like approximate likelihood-ratio branch test (SH-aLRT; −alrt 1000).13 The ultrafast bootstrap feature in IQ-Tree2 provides a more robust approach to compute the support of phylogenetic groups in the maximum likelihood trees than the conventional non-parametric bootstrap approach.13 A maximum likelihood-based method, TreeTime, was used to infer a molecular clock phylogeny with a strict evolutionary rate of 0·001 substitutions per site per year (estimated by Duchene and colleagues)14 and a standard deviation (SD) of 0·00004. The tree was rerooted with least-squares criteria in TreeTime. From 692 total sequences, six were excluded from the analysis due to inconsistent temporal signals.
To evaluate the phylogenetic relationship of the SARS-CoV-2 cases detected in each wave in Ghana in a global context, we combined sequences from Ghana with closely related global sequences using Multiple Alignment using Fast Fourier Transform version 7.477. To identify global sequences closely related to the Ghana sequences, we aligned the GISAID dataset (accessed April 20, 2022) with uvaialign version 2.0.1 (GitHub). We then used uvaia version 2.0.1 (GitHub, San Francisco, CA, USA) to search the Ghanaian sequences against the aligned GISAID database. This process allowed us to identify the closest global sequences to the sequences from Ghana. The alignments were then used to create the initial maximum likelihood trees in IQ-Tree2, with the settings previously mentioned and with duplicate sequences removed. The resulting initial trees were visualised and cleaned with figtree version 1.4.4 (University of Edinburgh, Edinburgh, UK), in which manual curation was done to collapse nodes that only consisted of international sequences from the same country. Collapsing these international clusters would convey the same visual information when replaced by a single leaf. Tree pruning was with goalign version 0.3.5 and gotree version 0.4.3. To improve inference of the transmission direction, the cleaned maximum likelihood trees were then used to reconstruct the time-scaled phylogenies. The molecular clock phylogeny of these global phylogenies was inferred with TreeTime7 with the same settings mentioned previously.
Pangolin lineages were assigned to all genomes with pangolin version 2.3.2 and pangoLEARN version 2021–12–06. Visualisations of the phylogenetic tree were produced with R version 3.5.3, specifically ape version 5.6–2, ggtree version 3.0.4, ggplot2 version 3.3.6, ggtreeExtra version 1.2.3, dplyr version 1.0.9, phytools version 1.0–3, and tidytree version 0.3.9.
Statistical analysis
As this study used an ongoing national surveillance platform, no sample size calculation was done; we included all eligible samples during the study period. Only descriptive statistics are presented in this Article. 95% Wilson CIs were calculated. We report the proportion of influenza-like illness and severe acute respiratory illness, as recorded by regular data collection through the national influenza surveillance platform using standard WHO definitions, due to both influenza and SARS-CoV-2 stratified by time period and age. All descriptive analyses were done in R version 4.1. No imputation was used for missing data.
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
Between Sept 1, 2020, and Nov 30, 2021, 5495 samples were submitted for diagnostic testing through the national influenza surveillance network. 2199 (40·0%) of 5495 samples were received from the Greater Accra region and 673 (12·2%) of 5495 samples were received from the eastern region; 36 (0·7%) of 5495 samples were received from the Upper west region, the lowest number of samples. Complete data for age, sex, and region were available for 4602 (83·7%) of 5495 samples, of which 2121 (46·1%) were from female samples and 2479 (53·9%) were from male samples.
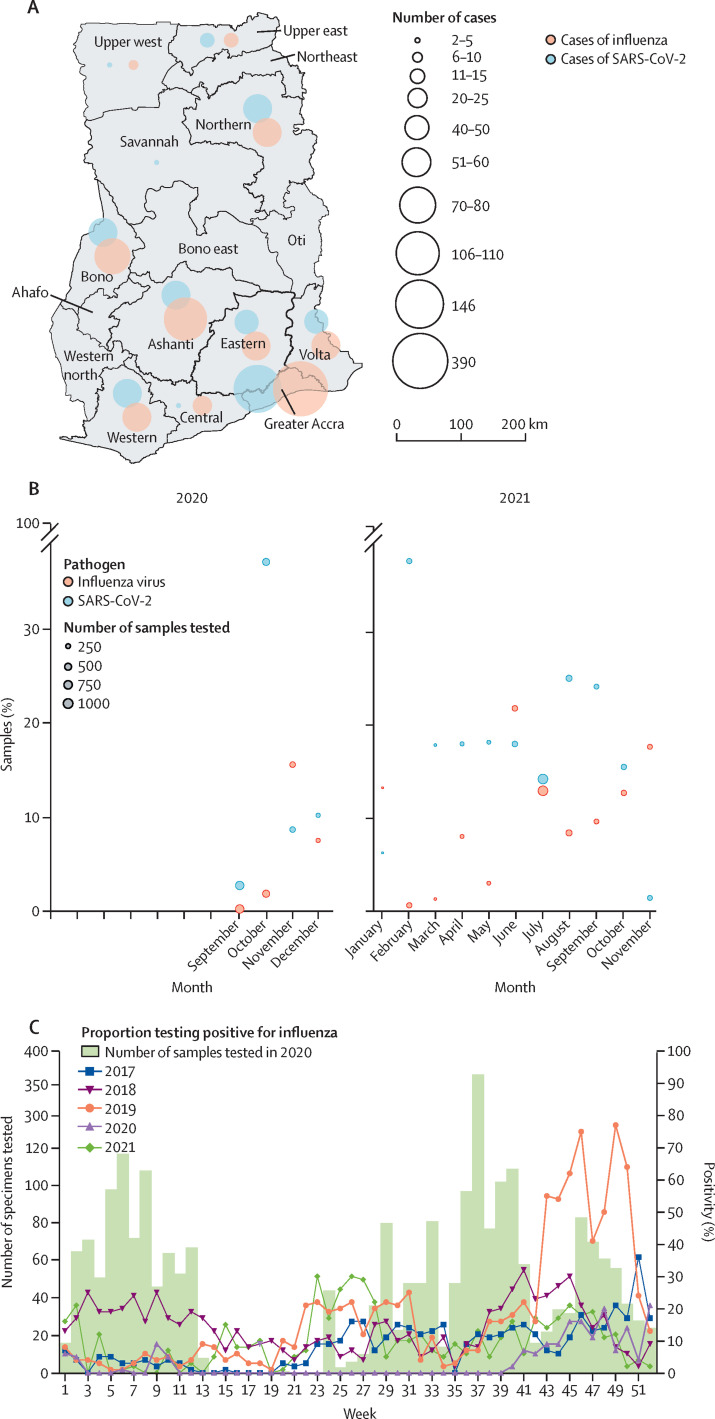
939 SARS-CoV-2 infections (17·1%, 95% CI 16·1–18·1) were detected from 5495 samples. The overall monthly SARS-CoV-2 positivity rate from the influenza surveillance network was between 1·44% (0·5–3·9) and 37·7% (32·2–43·3), with the highest rate observed in February, 2021. We also detected evidence of ongoing influenza circulation in Ghana throughout the study period, with 500 (9·1%, 8·4–9·9) of 5495 samples positive for influenza (figure 1A, B ; appendix pp 3–4). The monthly influenza positivity rate varied between 0·3% (0·26–1·1) and 21·8% (17·6–26·7), with the highest rate observed in June, 2021. All regions in the country from which data were available had evidence of both influenza and SARS-CoV-2 during the study period (figure 1A). During November, 2020, the rate of influenza infection (15·6%, 12·0–20·0) was higher than the rate of SARS-CoV-2 infection (8·7%, 6·0–12·4); during November, 2021, the rate of influenza infection (17·7%, 13·5–22·8) was also higher than the rate of SARS-CoV-2 infection (1·4%, 0·5–3·9; figure 1B). Rates of influenza positivity that were higher than rates of SARS-CoV-2 were also observed in January and June, 2021 (figure 1B). We compared 2020 and 2021 influenza positivity rates with three pre-pandemic years, (2017–19; figure 1C). Influenza positivity rates were lower than previous years for most epidemiological weeks. 3·0% (2·4–3·8) of samples in 2020 and 7·3% (6·7–8·1) of samples in 2021 were positive for influenza compared with 22·4% (21·3–23·6) in 2019, 14·7% (13·7–15·7) in 2018, and 9·2% (8·4–10·1) in 2017 (figure 1C).
Figure 1.
SARS-CoV-2 and influenza detection in Ghana
(A) The distribution of influenza and SARS-CoV-2 cases in Ghana from Sept 1, 2020, to Nov 30, 2021, detected via the national influenza surveillance network. No data were submitted, and subsequently no cases were reported, during the time of the study for five regions (ie, northeast, Oti, Ahafo, Western north, and Bono east). (B) Positivity rates of influenza and SARS-CoV-2 from September, 2020, to November, 2021, detected via the national influenza surveillance network (appendix p 4). (C) Number of samples tested for influenza virus in 2020 by week and proportion testing positive for influenza by week and by year (data for 2017–19) were obtained from the National Influenza Centre Ghana, WHO, Accra, Ghana). Data for 2021 were collected via the national influenza surveillance network as part of this study. No samples were tested during weeks 14–23 as the platform had to be reactivated.
4602 (84·0%) of 5495 samples had complete data for age, sex, and region; of these 4602, there were 4124 (89·7%) cases of influenza-like illness and 476 (10·3%) cases of severe acute respiratory illness. SARS-CoV-2 was identified in 583 (14·1%, 95% CI 13·1–15·2) of 4124 cases of influenza-like illness and influenza was identified in 356 (8·6%, 7·8–9·5) of 4124 cases of influenza-like illness. Co-infections were identified in 14 (0·3%, 0·2–0·6) cases of influenza-like illness. SARS-CoV-2 was detected in 58 (12·2%, 9·5–15·5) of 476 cases of severe acute respiratory illness and influenza was detected in 95 (19·9%, 16·5–23·9) of 476 cases of severe acute respiratory illness, as defined by WHO. Both pathogens were identified in an additional two (0·4%, 0·1–1·7) cases of severe acute respiratory illness. Influenza was more common than SARS-CoV-2 as a cause of severe acute respiratory illness in individuals aged 40 years or younger and was a more common cause of influenza-like illness among children aged 0–10 years (table ).
Table.
Causes of influenza-like illness and severe acute respiratory illness by age
|
Influenza-like illness |
Severe acute respiratory illness |
|||||||
|---|---|---|---|---|---|---|---|---|
| Cases | SARS-CoV-2 | Influenza | Co-infection | Cases | SARS-CoV-2 | Influenza | Co-infection | |
| 0–10 years | 602 (13%) | 35/602 (6%, 4–8) | 120/602 (20%, 16–23) | 2/602 (0%, 0–1) | 155 (3%) | 6/155 (4%, 1–8) | 32/155 (21%, 14–28) | 0/155 (0%, 0–3) |
| 11–20 years | 937 (20%) | 71/937 (8%, 6–9) | 57/937 (6%, 4–7) | 0/937 (0%, 0–0) | 53 (1%) | 3/53 (6%, 1–16) | 13/53 (25%, 14–38) | 0/53 (0%, 0–8) |
| 21–30 years | 955 (21%) | 154/955 (16%, 13–18) | 65/955 (7%, 5–8) | 4/955 (0%, 0–1) | 88 (2%) | 12/88 (14%, 7–22) | 15/88 (17%, 10–26) | 1/88 (1%, 0–7) |
| 31–40 years | 841 (18%) | 173/841 (21%, 17–23) | 62/841 (7%, 5–9) | 4/841 (0%, 0–1) | 61 (1%) | 8/61 (13%, 6–24) | 17/61 (28%, 17–41) | 0/61 (0%, 0–7) |
| 41–50 years | 399 (9%) | 72/399 (18%, 14–22) | 32/399 (8%, 5–11) | 3/399 (1%, 0–2) | 44 (1%) | 10/44 (23%, 11–38) | 9/44 (20%, 10–35) | 0/44 (0%, 0–10) |
| 51–60 years | 200 (4%) | 46/200 (23%, 17–29) | 11/200 (6%, 2–9) | 1/200 (1%, 0–3) | 25 (1%) | 8/25 (32%, 15–53) | 3/25 (12%, 3–32) | 0/25 (0%, 0–17) |
| 61 years or older | 192 (4%) | 32/192 (17%, 11–22) | 9/192 (5%, 2–8) | 0/192 (0%, 0–2) | 50 (1%) | 11/50 (22%, 11–36) | 4/50 (8%, 2–20) | 1/50 (2%, 0–12) |
Data are n (%) or n/N (%, 95% CI). Based on 4602 individuals for whom sociodemographic data were available.
Sequencing was attempted in 851 samples, taken from 306 individuals presenting to the national influenza surveillance network with influenza-like illness and 80 with severe acute respiratory illness, and from 465 travellers arriving in Accra who had tested positive on a lateral flow assay. 692 (81%) of the 851 samples had sufficient genome coverage (>70%) for lineage typing and 83 unique lineages were detected. On the basis of routine reporting data, Ghana had four waves of COVID-19 between April, 2020, and November, 2021. Each wave was driven by different variants of the SARS-CoV-2 virus. For example, the first wave was April to December, 2020, and was characterised by a combination of early SARS-CoV-2 lineages (70 [23%] of 306 samples were of lineage B.1 and 121 [40%] of 306 were of lineage B.1.1; figure 2 ). The alpha variant (B.1.1.7) was the most dominant lineage in the second wave, January to April, 2021, after the government reduced or removed existing COVID-19 protocols by December, 2020. The delta variant (B.1.617.2) and delta sublineages were responsible for the third wave in Ghana between May and November, 2021. The fourth wave was predominantly caused by the omicron variant (B.1.1.529).
Figure 2.
SARS-CoV-2 lineage distribution across four COVID-19 waves in Ghana
Number of cases caused by each lineage during each week of 2020 (starting at week 10) and each week of 2021. Data are from a subset of 692 samples sequenced in the current study, and 1123 samples collected in Ghana by Morang’a and colleagues.15 Wave 1 (April to December, 2020) was driven by early variants (eg, B.1 and B.1.1 were most dominant), whereas wave 2 (January to April, 2021), wave 3 (May to November, 2021), and wave 4 (November to December, 2021) were driven by variants of concern and variants of interest; alpha (B.1.1.7), beta (1.351), delta (B.1.617.2), and omicron (B.1.1.529) were designated as variants of concern, and eta (B.1.525), iota (B.1.526), and kappa (B.1.617.1) as variants of interest. All lineages that were not variants of concern or variants of interest are included in the Other category.
Each SARS-CoV-2 lineage that we detected was categorised into one of four categories (ie, A, B, C, or D), depending on the characteristics of the patient in whom the lineage was first identified and whether there was a community spread after the first identification (appendix p 4). Included in the type A category were the lineages first detected in travellers and subsequently detected in influenza-like illness samples. Included in the type B category were the lineages first detected in travellers but not subsequently detected in influenza-like illness samples. Included in the type C category were the lineages first detected in influenza-like illness samples with subsequent community spread, and included in the type D category were the lineages first detected in influenza-like illness samples without subsequent community spread. 57 (69%) of all 83 lineages were first identified among travellers. The earliest alpha variant was detected in influenza-like illness surveillance during the first wave, rather than in a returning traveller, which could indicate cross-border introductions of this variant.
We detected SARS-CoV-2 in 1626 (71·0%, 69·1–72·9) of 2289 samples from travellers who arrived in Ghana from Sept 1, 2020, to Nov 30, 2021, and had tested positive on a lateral flow assay. We were able to successfully sequence and analyse 392 samples with a cycle threshold value below 30. 241 (61·5%) of these 392 samples were from travellers arriving from other African countries and 102 (26·0%) were from travellers coming from Asia. 69 (18%) of the 392 samples were from travellers from Nigeria and had enough sequence coverage to identify their variants, which made up the largest group of importations of SARS-CoV-2 into Ghana from a single country. Of 241 travellers who arrived from Africa, 27 (11%) arrived to Ghana from neighbouring countries (19 [8%] from Côte d’Ivoire and eight [3%] from Burkina Faso). During the study period, 33 (32%) of 104 travellers arriving from Nigeria were positive for the SARS-CoV-2 omicron variant of concern and 21 (20%) were positive for the SARS-CoV-2 delta variant of concern. 22 (60%) of 37 travellers arriving from South Africa were positive for omicron variants. 39 (43%) of 91 travellers arriving from Asia were positive for the kappa (B.1.617.1) and eta (B.1.525) variants of interest and the delta variant (figure 3 ).
Figure 3.
Lineages of SARS-CoV-2 imported into Ghana by arriving travellers, by continent
Data are from 484 samples sequenced from travellers arriving in Accra from September, 2020, to November, 2021. Each circle represents the number of cases. Travellers from Africa were the highest proportion of travellers into Ghana (appendix p 8–10). Alpha (B.1.1.7), beta (B.1.351 and B.1.351.2), delta (B.1.617.2 and AY.x), gamma (P.1 and P.1.1), and omicron (BA.1 and B.1.1.529) were all designated as variants of concern.
A time-scaled phylogenetic tree was inferred to estimate the emergence and introduction timeline of variants of concern and variants of interest in Ghana (appendix p 7). Five lineages (ie, B.617.2, AY.109, AY.59, AY.88, and B.1.1.318) were first identified from the travellers entering Ghana and subsequently detected in influenza-like illness surveillance samples. Phylogenetic clusters of both AY.88 and B.1.1.318 sequences from community samples have ancestral sequences from returning travellers, providing phylogenetic support for the hypothesis that returning travellers introduced lineages to Ghana that subsequently spread in the community (appendix p 7). Delta was first detected in a traveller arriving from India in early May, 2021. B.1.1.318 was first detected in a traveller arriving from the neighbouring country Côte d’Ivoire on April 10, 2021. B.1.1.318 was most prevalent in travellers arriving from west African countries (eg, Benin, Gabon, São Tomé and Príncipe, and Ghana; appendix pp 8–11). The estimated time of the most recent common ancestor (tMRCA) of Ghanaian B.1.1.318 was Feb 25, 2021 (95% CI Feb 9 to March 16), 45 days before it was first detected in Ghana, indicating a possible cryptic transmission. AY.88, a delta sublineage that is predominantly seen in Ghana, has a tMRCA of April 1, 2021 (95% CI March 12 to April 1), which is close to when we detected the first AY.88 sample, suggesting that we might have captured one of the earliest AY.88 samples in Ghana.
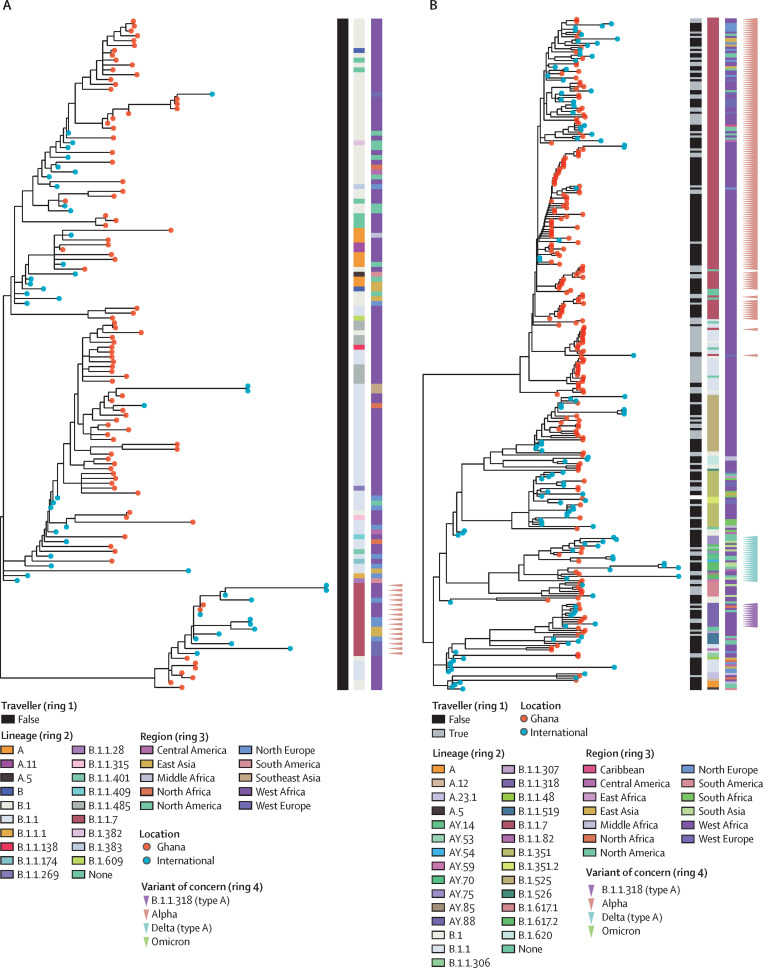
To contextualise the sequences from Ghana within the global SARS-CoV-2 phylogeny, maximum likelihood phylogenetic trees were produced for each separate wave, with all sequences from the current study and the closest related non-Ghanaian sequences. Alpha was first detected in wave 1 and most prevalent in wave 2 (figure 4A, B ; appendix pp 12–13). Two alpha samples (nCoV-20–133709 and nCoV-20–134569) detected in wave 1 were both most closely related to samples from Côte d’Ivoire, suggesting a possible cross-border transmission. The wave 2 phylogeny revealed several different clusters of alpha variants circulating in Ghana. One of the alpha clusters, consisting of only non-traveller samples, is closely related to the cases in Côte d’Ivoire. Other alpha sequences detected in Ghana are mainly related to 17 European and 11 North American genomes. At least four independent B.1.1.318 introductions can be inferred from the wave 2 phylogeny, and the most closely related international genomes are from other African countries including Côte d’Ivoire, Mauritania, and Tunisia (figure 4B; appendix p 13).
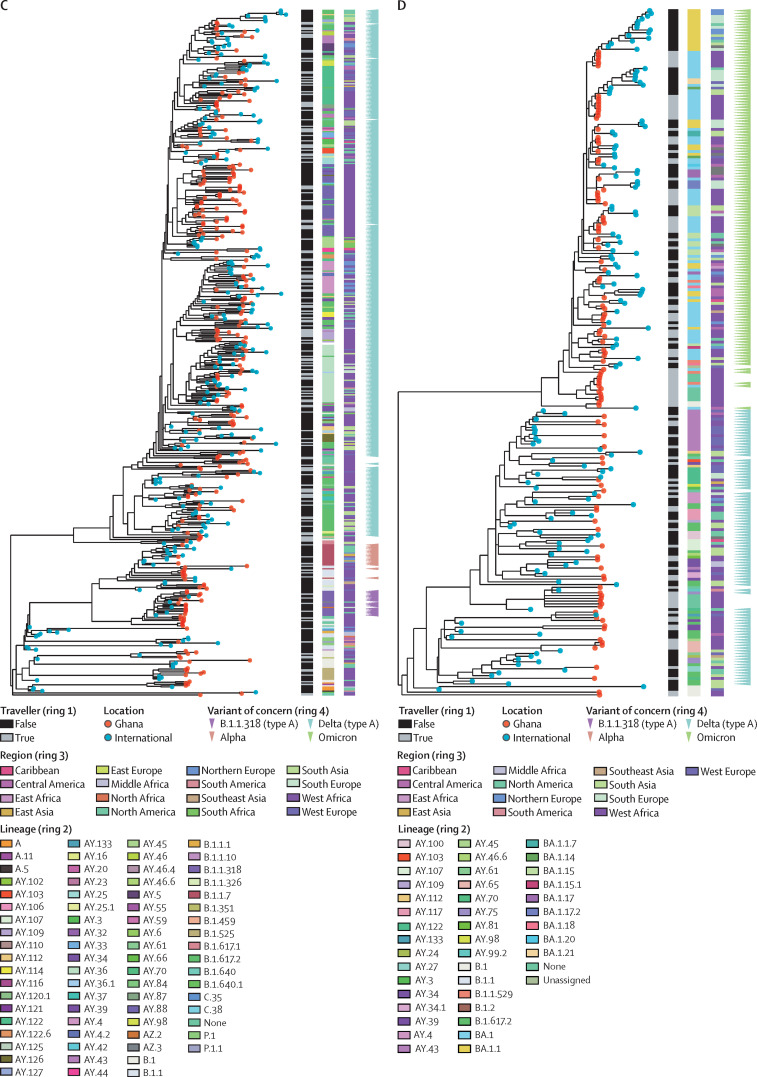
Figure 4.
Phylogeny of SARS-CoV-2 sequences in Ghana and closely related genomes from international samples
(A) Maximum likelihood phylogenetic tree of wave 1. (B) Maximum likelihood phylogenetic tree of wave 2. (C) Maximum likelihood phylogenetic tree of wave 3. (D) Maximum likelihood phylogenetic tree of wave 4. The lineages and origin of samples are indicated in the rings. Type A lineages are lineages that were first detected in travellers and possibly precede community waves. 48 global sequences were included for the wave 1 analysis, 121 in the wave 2 analysis, 248 in the wave 3 analysis, and 159 in the wave 4 analysis.
Some delta sublineages (eg, AY.88) were detected in late wave 2; however, B.1.617·2 was not detected until wave 3, suggesting that we probably missed B.1.617.2 during earlier waves in our sampling and sequencing strategy. The delta lineage sequences from Ghana were most closely related to 21 genomes from India, eight from the USA, seven from France, and six from Côte d’Ivoire (figure 4C; appendix p 14). The delta samples from India were estimated to be the ancestral genomes of some of the delta cases in Ghana, suggesting a possible importation of the delta lineage from India in wave 3. AY.88 was first detected in a traveller in wave 2 and shared a common ancestor with a sequence from India. Other delta sublineages were mainly seen in a single or few travel-associated cases and not in infections detected via the national influenza surveillance network. Delta and its sublineage sequences from Ghana detected during wave 4 have 15 ancestral international genomes from India, 14 from France, 12 from the USA, and nine from Côte d’Ivoire, suggesting suggested multiple importations and exportations to and from Ghana (figure 4D; appendix p 15).
In late 2021, the new SARS-CoV-2 omicron variant of concern was initially detected in South Africa and subsequently displaced delta globally. Currently, omicron comprises four lineages including B.1.1.529, BA.1, BA.2, and BA.3. As part of this study, 145 SARS-CoV-2-positive samples from travellers entering Ghana from November to December, 2021, were sequenced successfully, with omicron detected in 47 (32%). 15 (32%) of these 47 travellers arrived from South Africa, 29 (62%) from Nigeria, one (2%) from Eswatini, one (2%) from Portugal, and one (2%) from an unknown region. Other major variants of concern detected from travellers from November to December, 2021, included 15 (10%) delta cases from the 145 samples and 58 (40%) delta sublineages from the 145 samples. Of community samples from the same period that were successfully sequenced, 17 (81%) of 21 variants were identified as delta or its sublineages. The omicron variant subsequently spread in Ghana. Omicron sequences in Ghana were closely related to 19 genomes from India, 15 from Slovenia, and eight from the USA, with possible exportation from Ghana (appendix p 15).
Discussion
COVID-19 has had a devastating public health impact globally. Low-income and middle-income countries, such as Ghana, continue to experience the severe effects associated with the pandemic. Thus, ensuring adequate monitoring for SARS-CoV-2, including genomic surveillance, is crucial. As of April 21, 2023, Ghana had recorded 171 657 cases of SARS-CoV-2 infection and 1462 deaths. To our knowledge, our study is the first to show the value of an existing influenza surveillance network to track SARS-CoV-2 epidemiology in a low-income or middle-income country. By testing for both SARS-CoV-2 and influenza, we showed that influenza remained an important cause of severe acute respiratory illnesses, especially in children aged 0–10 years. Between Sept 1, 2020, and Nov 30, 2021, 500 (9%) of 5495 samples of submitted samples to the National Influenza Centre tested positive for influenza viruses, showing that sustained influenza transmission continued in Ghana. Each of the four SARS-CoV-2 waves that occurred during our study period have been driven by distinct lineages, which were initially detected among travellers entering the country, with subsequent community spread detected among the population being tested via the national influenza surveillance platform. The cases in travellers captured by our surveillance platform might provide useful insight in understanding the importation of some of these lineages.
Early in the COVID-19 pandemic, the Global Influenza Surveillance and Response System, which represents a network of laboratories, recommended the integration of COVID-19 surveillance into influenza surveillance networks, but we are not aware of any other published data that show the implementation of this approach. We successfully integrated SARS-CoV-2 testing into the existing Ghanaian influenza-like illness surveillance, allowing prompt reporting to the national surveillance platforms for contact tracing and follow-up; this network generated almost a third of all SARS-CoV-2 genomes from Ghana. A notable finding in our study was ongoing transmission of influenza, with co-circulation with SARS-CoV-2. Several countries16, 17, 18 have reported reduced or no cases of influenza during the COVID-19 pandemic. Ghana has, however, consistently detected influenza throughout the study period. Overall influenza activity in 2020 was lower than previous years (2017–19), although it increased again in 2021. We detected approximately 950 cases of COVID-19 (symptomatic individuals with SARS-CoV-2) and approximately 500 cases of influenza (symptomatic individuals with influenza virus). Influenza was more common than SARS-CoV-2 as a cause of severe acute respiratory illness, as defined by WHO, in our study, especially in adults aged 40 years or younger and children aged 0–10 years. A key finding of our study is the value of existing influenza-like illness surveillance networks as a sustainable platform for broader disease surveillance activities, especially in settings where more extensive community SARS-CoV-2 testing programmes are not feasible.
Our work builds on previous genomic surveillance studies from Ghana. First, a previous study15 used a convenience selection of available SARS-CoV-2 samples, whereas we had access to a representative sampling platform. Second, we were able to combine this sampling platform with cases from travellers. Third, our data collection period expands genomic surveillance in Ghana across the entire study period including introduction, alpha, delta, and omicron waves. We used our combined community and traveller surveillance platform to successfully track the introduction of SARS-CoV-2 into Ghana and monitor the circulation of these imported strains in the general population. The four waves identified from our study are compatible with previous research,15 with wave 1 dominated by B.1.1, wave 2 dominated by alpha, and wave 3 dominated by delta.
Our data highlight the key role of importation of SARS-CoV-2 variants. More than two-thirds of all SARS-CoV-2 lineages detected in Ghana were first identified among international travellers. Of these lineages, five variants (ie, B.1.1.318, delta, A.88, AY.59, and AY.109) subsequently spread in the local population. In Ghana, local transmission of the alpha variant led to wave 2, with mean daily rates of 800 cases per day. The beta B.1.351 variant was initially identified in our traveller population but did not spread into the local population, which is consistent with other data from west Africa and Europe. Delta B.1.617·2 and delta sublineages were first detected in our traveller population and spread throughout Ghana, resulting in the third wave of SARS-CoV-2 infections in the country. In Novermber 2021, omicron variants were initially detected in our traveller population within 2 weeks of the declaration of omicron as a variant of concern by WHO. By combining this surveillance with phylogenetic analyses, we were also able to identify variants that were probably circulating within the community before their first detection. The B.1.1.318 lineage was first detected in a traveller returning from Côte d’Ivoire; this sublineage spread widely throughout Ghana and other west African countries. The B.1.1.318 lineage (which contains the Glu484Lys mutation that affects antibody recognition) was also reported to have caused an outbreak in Mauritius and other west African countries, including Nigeria, Gabon, and Cameroon.16 Studies from Rwanda and Uganda have also highlighted the importance of neighbouring countries in the introduction of SARS-CoV-2 lineages into countries.19, 20 Circulation of B.1.1.318 between wave 2 and wave 3 in Ghana was also reported by Morang’a and colleagues.15 We did not detect any B.1.1.359 or B.1.623 cases in our study, which might be due to a bias in the sampling location as both of these lineages were only detected outside Greater Accra in the cohort from Morang’a and colleagues.
Although our study has substantially contributed to knowledge about imported and circulating SARS-CoV-2 variants in Ghana, our community population was restricted to influenza-like illness sentinel sites; this community-based surveillance platform is still dependent on individuals seeking care. A wider selection of cases (eg, contact tracing samples or data) could have showed the extent of community spread of imported variants. Despite the design of our study, we cannot exclude the possibility that our sample of cases of severe acute respiratory illness is not completely representative as we were missing data on either the age or sex of approximately 15% of individuals tested in the study. These missing data might have resulted in incorrect estimates of the proportion of cases of severe acute respiratory illness caused by SARS-CoV-2 and influenza, as data were excluded from the analysis if they had missing data on age or sex. We had little data about the study population, including only age, sex, region, and time of testing. Additional data on vaccination status would enhance ongoing surveillance efforts. We do not have data on the relative proportion of inbound flights to Ghana from different countries, which makes directly attributing importations to travellers from specific locations difficult. Furthermore, we only tested for SARS-CoV-2 and influenza virus and did not identify a cause in a large proportion of cases. Expanding the influenza-like illness network further to allow testing for other pathogens would be of substantial value.
Our study has highlighted the value of existing influenza surveillance networks with integrated genomic surveillance to track the spread and evolution of SARS-CoV-2. These platforms can provide important and timely data to inform ministries of health, policy makers, and researchers on the evolving course of the SARS-CoV-2 pandemic. Investment in strengthening national influenza surveillance platforms in low-income and middle-income countries has potential for ongoing monitoring of SARS-CoV-2 and future pandemics.
Data sharing
All genomic data have been deposited in the GISAID and are available for re-use without restrictions. Accession numbers and the study protocol are provided in the appendix. Raw data from the influenza surveillance platform are restricted to the Ghana Health Service and, therefore, are not available for re-use.
Declaration of interests
We declare no competing interests.
Acknowledgments
Acknowledgments
This study is part of the EDCTP2 programme supported by the EU (RIA2020EF-2983-CSIGN). We thank the Ghana Health Service, all influenza sentinel sites, and Kotoka International Airport (Accra, Ghana). We acknowledge the COVID-19 Presidential Task Force, who allowed us to access the traveller samples.
Editorial note: The Lancet Group takes a neutral position with respect to territorial claims in institutional affiliations.
Acknowledgments
Contributors
MM, TIdS, and WKA conceptualised and set up the study. IAA and SNH wrote the manuscript. LB, BHF, and LK did the sequencing. SNH, LdO-M, TIdS, and MDP analysed the sequences. MM, IAA, SNH, TIdS, and WKA accessed and verified the data. All authors reviewed and approved the final version of the manuscript. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Supplementary Material
References
- 1.Asante IA, Adusei-Poku M, Bonney HK, et al. Molecular diagnosis for the novel coronavirus SARS-CoV-2: lessons learnt from the Ghana experience. Ghana Med J. 2020;54(suppl):77–85. doi: 10.4314/gmj.v54i4s.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quashie PK, Mutungi JK, Dzabeng F, et al. Trends of SARS-CoV-2 antibody prevalence in selected regions across Ghana. medRxiv. 2021 doi: 10.1101/2021.04.25.21256067. published online April 29. (preprint). [DOI] [Google Scholar]
- 3.A3M Global Monitoring COVID-19 pandemic—Ghana. 2023. https://global-monitoring.com/gm/page/events/epidemic-0002023.G61q45qnqj0N.html?lang=en
- 4.Bonney JHK, Kronmann KC, Lindan CP, et al. Virological surveillance of influenza-like illness among children in Ghana, 2008–2010. J Infect Dis. 2012;206(suppl 1):S108–S113. doi: 10.1093/infdis/jis577. [DOI] [PubMed] [Google Scholar]
- 5.Shu B, Kirby MK, Davis WG, et al. Multiplex real-time reverse transcription PCR for influenza A virus, influenza B virus, and severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2021;27:1821–1830. doi: 10.3201/eid2707.210462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quick J. nCoV-2019 sequencing protocol v3 (LoCost) 2020. https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye
- 7.Sagulenko P, Puller V, Neher RA. TreeTime: maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4 doi: 10.1093/ve/vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.GitHub ProblematicSites_SARS-CoV2. 2022. https://github.com/W-L/ProblematicSites_SARS-CoV2
- 10.Minh BQ, Schmidt HA, Chernomor O, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tavaré S. Some probabilistic and statistical problems in the analysis of DNA sequences. 1986. https://www.semanticscholar.org/paper/Some-probabilistic-and-statistical-problems-in-the-Tavar%C3%A9/55e3359cd05b1903ffd8f633eb5aa6156791b364
- 12.Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- 13.Rambaut A, Holmes EC, O'Toole Á, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duchene S, Featherstone L, Haritopoulou-Sinanidou M, Rambaut A, Lemey P, Baele G. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evol. 2020;6 doi: 10.1093/ve/veaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morang'a CM, Ngoi JM, Gyamfi J, et al. Genetic diversity of SARS-CoV-2 infections in Ghana from 2020–2021. Nat Commun. 2022;13 doi: 10.1038/s41467-022-30219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olsen SJ, Azziz-Baumgartner E, Budd AP, et al. Decreased influenza activity during the COVID-19 pandemic—United States, Australia, Chile, and South Africa, 2020. Am J Transplant. 2020;20:3681–3685. doi: 10.1111/ajt.16381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soo RJJ, Chiew CJ, Ma S, Pung R, Lee V. Decreased influenza incidence under COVID-19 control measures, Singapore. Emerg Infect Dis. 2020;26:1933–1935. doi: 10.3201/eid2608.201229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakamoto H, Ishikane M, Ueda P. Seasonal influenza activity during the SARS-CoV-2 outbreak in Japan. JAMA. 2020;323:1969–1971. doi: 10.1001/jama.2020.6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butera Y, Mukantwari E, Artesi M, et al. Genomic sequencing of SARS-CoV-2 in Rwanda reveals the importance of incoming travelers on lineage diversity. Nat Commun. 2021;12 doi: 10.1038/s41467-021-25985-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bugembe DL, Phan MVT, Ssewanyana I, et al. Emergence and spread of a SARS-CoV-2 lineage A variant (A.23.1) with altered spike protein in Uganda. Nat Microbiol. 2021;6:1094–1101. doi: 10.1038/s41564-021-00933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genomic data have been deposited in the GISAID and are available for re-use without restrictions. Accession numbers and the study protocol are provided in the appendix. Raw data from the influenza surveillance platform are restricted to the Ghana Health Service and, therefore, are not available for re-use.