Abstract
Non-alcoholic liver disease (NAFLD) is a condition caused by excessive fat accumulation in the liver and developed via multiple pathways. miR-27b has been suggested to play crucial roles in the development of NAFLD, assuming via targeting genes involved in lipid catabolism and anabolism. However, other pathways regulated by miR-27b are largely unknown. Here we show that lipid accumulation was induced in miR-27b–transfected human and mouse hepatic cells and that knockdowns of three miR-27b–target genes, β-1,4-galactosyltransferase 3 (B4GALT3), matrix AAA peptidase interacting protein 1 (MAIP1) and PH domain and leucine rich repeat protein phosphatase 2 (PHLPP2), induced lipid accumulation. We also show that B4GALT3 and MAIP1 were direct targets of miR-27b and overexpression of MAIP1 ameliorated miR-27b−induced lipid accumulation. In addition, we show that hepatic Maip1 expression declined in mice fed a high-fat diet, suggesting the involvement of decreased Maip1 expression in the condition of fatty liver. Overall, we identified MAIP1/miR-27b axis as a mediator of hepatic lipid accumulation, a potential therapeutic target for NAFLD.
Subject terms: Mechanisms of disease, Fat metabolism, miRNAs
Gene expression studies identify a new miR-27b-mediated regulatory pathway that targets MAIP1 and leads to hepatic lipid accumulation in cultured human and mouse hepatic cells suggesting a potential role for MAIP1 as a therapeutic target for NAFLD
Introduction
Non-alcoholic fatty liver disease (NAFLD) is a highly prevalent hepatic manifestation of disordered metabolism, a cluster of conditions that includes obesity, insulin resistance, dyslipidemia and type 2 diabetes1–4, and 10–30% of NAFLD progresses to non-alcoholic steatohepatitis (NASH)1,5. The onset of NAFLD is attributed mainly to hepatic lipid accumulation, and NAFLD leads to NASH, cirrhosis and, eventually, hepatocellular carcinoma. Although the pathogenesis of NAFLD is not fully understood, the current “multiple hit” theory proposes that multiple insults synergistically and in parallel act to induce NAFLD6,7. The multiple hits include factors such as insulin resistance8,9, increased inflammatory cytokines10,11 and oxidative stress12–14, circulating adipokines15,16, intestinal microbiota17,18, and so on. In addition, crosstalk between them further accelerates progression of disease. However, a full understanding of NAFLD pathogenesis remains to be achieved.
MicroRNAs (miRNAs) are small (19–25 nucleotides long), single-stranded noncoding RNAs that regulate a variety of genes via binding to a perfectly or partially matched complementary sequence located within the 3’ untranslated region (UTR) of a target mRNA, resulting in translational suppression and/or mRNA degradation19–21. It is now widely recognized that miRNAs regulate a wide variety of biological pathways, including various cellular metabolisms. Liver tissue is rich in miRNAs that have important roles in liver functions22. Accmulative studies have demonstrated that several miRNAs regulate lipid metabolism-related genes that are presumably involved in the pathogenesis of NAFLD23–25. It was reported that some miRNAs were expressed in serum and liver tissues in good correlation with NAFLD progression26–29. The miR-27 family consists of miR-27a and miR-27b, which differ by one base difference at their 3’-terminal regions30,31. The upregulated expression of miR-27b was detected in NAFLD patients28,32. The miR-27b level in serum was identified as an accurate diagnostic marker for NAFLD. Also, overexpression of hepatitis C virus (HCV) proteins activates miR-27a/b expression; in turn, the overexpression of miR-27a/b in hepatic cell lines results in increased lipid droplets (LDs), suggesting that miR-27a/b expression contributes to the development of hepatic steatosis induced by HCV infection33. In animal models, it was demonstrated that miR-27b expression was upregulated in the livers of mice fed a high-fat diet34. Together, these previous studies strongly suggest that miR-27b−mediated regulation of lipid metabolism might be a causative factor for NAFLD development.
Hepatic lipid metabolism is tightly regulated by the crosstalk of multiple interrelated pathways. Excess lipid in hepatocytes induces the endoplasmic reticulum (ER) stress which activates altered lipid homeostasis 35–40. Oxidative stress is induced by impairment of lipid metabolism and in turn, it causes ER stresses and mitochondrial dysfunction which leads to disordered lipid metabolism41,42. In silico approach identified miR-27b as a regulatory hub in lipid metabolism due to the presence of predicted target sites in 3’-UTRs and open reading frames (ORFs) of many genes involved in lipid catabolism and anabolism43; some of those were experimentally implicated as the direct target of miR-27b43–47. Considering a complex manifestation of NAFLD and interrelated regulation of lipid metabolism by multiple pathways, however, it would also be possible that miR-27b contributes to NAFLD development via suppressing genes involved in such pathways, eventually leading to lipid accumulation. In this report, we aimed to uncover another pathway for miR-27b to induce lipid accumulation, the main characteristic of NAFLD, by identifying miR-27b target genes so far unidentified in our knowledge that mediate lipid accumulation.
Results
MiR-27b induces lipid accumulation in hepatic cells
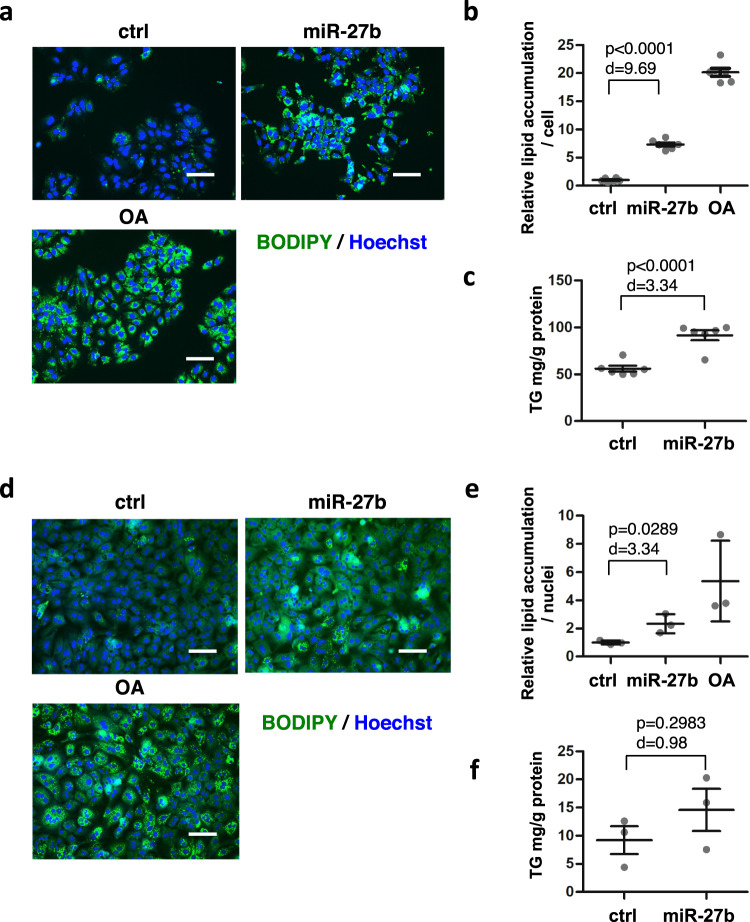
In a previous study, miR-27b and its ortholog miR-27a facilitated lipid accumulation in Huh-7 cells in the context of steatosis induced by HCV infection, where such infection upregulated miR-27a/b expression33. To confirm the effects of miR-27b on hepatic lipid accumulation, we transfected Huh-7 cells with miR-27b mimics and evaluated lipid accumulation by labeling lipids with fluorescent dyes, BODIPY 493/503. Lipid accumulation in miR-27b−transfected cells was approximately sevenfold that in control mimic-transfected cells (Fig. 1a and b). Consistent with this result, higher levels of triglyceride (TG) were detected in miR-27b-transfected cells compared to the control when lipids extracted from cells were analyzed (Fig. 1c). Similar phenomena were observed in HepG2 cells transfected with miR-27b mimics (Supplementary Fig. 1a and b). miR-27b mimics also stimulated lipid accumulation in mouse primary hepatocytes (Fig. 1d and e). An increased TG level was detected when miR-27b was transfected, although the difference was not significant (p > 0.05) (Fig. 1f). These results suggested that the lipid-related role of miR-27b was not restricted to hepatocellular carcinoma cell lines. Our observations, together with the previous ones33, revealed the potential of miR-27b to induce lipid accumulation in hepatic cells.
Fig. 1. miR-27b induces lipid accumulation in mouse and human hepatic cells.
a Huh-7 cells were transfected with control miRNA (ctrl) or miR-27b mimics at 10 nM followed by co-staining with BODIPY493/503 for the lipid and Hoechst 33342 for the nuclei 4 days after transfection. Untransfected cells were treated with oleic acid (OA) at 50 µM for the last 48 h. Scale bars, 100 μm. b The relative amount of lipid was estimated based on the images in (a). n = 6 independent experiments. Results are presented as means ± SE. p-values from Student’s t-test and Cohen’s d values are shown. c Triglycerides (TG) extracted from cells 4 days after transfection were quantitated. n = 6 independent experiments. Results are presented as means ± SE. p-values from Student’s t-test and Cohen’s d values are shown. d Mouse primary hepatocytes were transfected with the indicated mimics at 50 nM and stained 7 days after transfection as in (a). Untransfected cells were treated with OA at 50 µM for the last 48 h. Scale bars, 100 μm. e The relative lipid amount was estimated based on the images in (d). n = 3 biologically independent samples. Representative results from three independent experiments are presented as means ± SD. p-value from Student’s t-test and Cohen’s d value are shown. f TG extracted from cells 7 days after transfection were quantitated. n = 3 independent experiments. Results are presented as means ± SE. p-value from Student’s t-test and Cohen’s d value are shown.
Functional screening of target genes of miR-27b mediating lipid accumulation
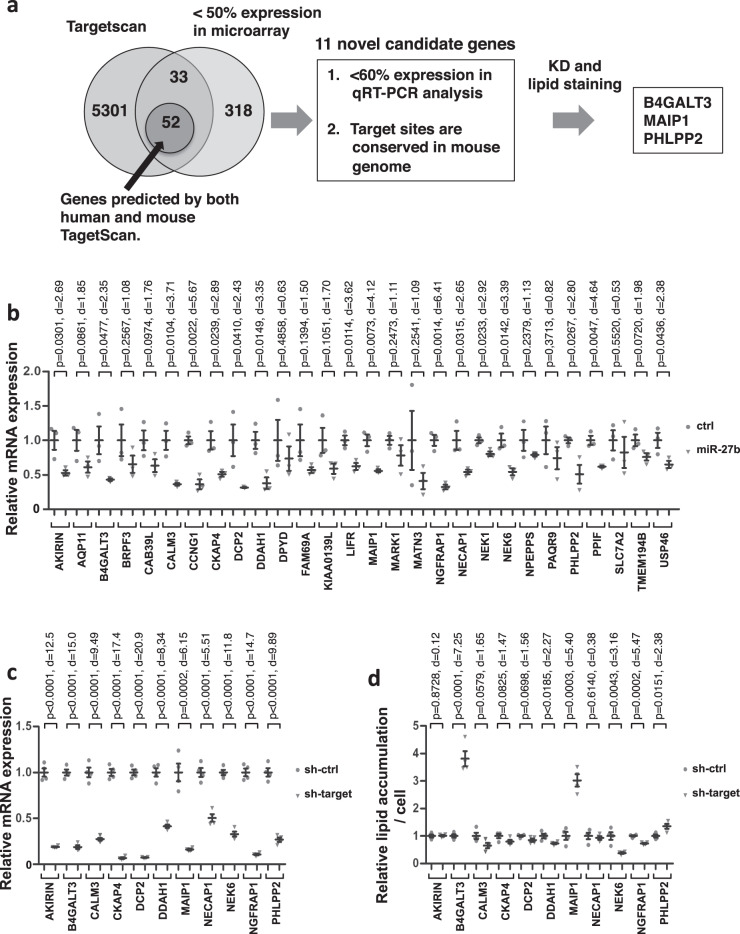
Next, we aimed to identify genes that mediate lipid accumulation in Huh-7 cells as targets of miR-27b (Fig. 2a). Microarray analyses were performed to detect changes in gene expression in miR-27b-transfected Huh-7 cells. This revealed 749 and 403 genes with twofold up- and down-regulated expression in miR-27b-transfected cells, respectively (Supplementary Data 1 and 2). Functional annotation clustering analyses using DAVID, a functional annotation tool (https://david.ncifcrf.gov/home.jsp), revealed that the up-regulated genes were enriched in multiple pathways with significance (Supplementary Fig. 2a). Enrichment in the hexose metabolic pathway suggests a compensatory activation of the glycolysis pathway for the presumably compromised lipid metabolism (Supplementary Fig. 2a). In fact, several known lipid metabolism-related genes, including fatty acyl CoA reductase 2 (FAR2)48, glycerol-3-phosphate acyltransferase 2 (GPAT2)49, apolipoprotein L5 (APOL5)50, ELOVL fatty acid elongase 4 (ELOVL4)51 and fatty acid desaturase 2 (FADS2)52, were up-regulated when individual genes were assessed (Supplementary Table 1). Down-regulated genes were also enriched in multiple pathways, although none of these pathways was related to energy metabolism (Supplementary Fig. 2b). Several lipid-related genes, including acyl-CoA oxidase-like (ACOXL)53, acyl-CoA dehydrogenase, long chain (ACADL)54 appeared, however, when the genes were assessed (Supplementary Table 1). ACADL was particularly interesting because it encodes long-chain acyl-CoA dehydrogenase (LCAD), an enzyme that catalyzes the initial step of fatty acid ß-oxidation in mitochondria54, and the ACADL knockout mice developed more severe fasting-induced hepatic steatosis55. Peroxisome proliferator-activated receptor gamma (PPARG) was also interesting because it is a regulator of adipocyte differentiation56 and is involved in hepatic steatosis56,57. Among the down-regulated genes, 85 genes were predicted as human targets of miR-27b according to Targetscan 7.0 (http://www.targetscan.org), a target prediction tool (Supplementary Table 2). Given the conservation of miR-27b across species and the similar effects of miR-27b on mouse hepatocytes to human hepatic cell lines regarding lipid accumulation, it would be highly possible that the human target genes of miR-27b were conserved as target genes in the mouse genome. Fifty-two genes were also predicted as miR-27b targets in the mouse genome (Supplementary Table 2). Unfortunately, the miR-27b target site in ACADL was not conserved in the mouse genome. PPARG, a known direct target of miR-27b43,45,46 was included in the list. However, hepatocyte-specific knockout experiments as well as overexpression experiments revealed that the hepatic PPARG expression positively regulated hepatic lipid accumulation58–60, suggesting that suppression of PPARG by miR-27b would reduce lipid accumulation. Therefore we decided to continue the screening, with PPARG excluded from it, for genes that mediate lipid accumulation when suppressed. We examined the remaining 51 genes for target site conservation and expression by qRT-PCR. The miR-27b–target sites in 32 out of the 51 genes were conserved in the mouse genome (Supplementary Table 3). Four out of the 32 genes could not be analyzed for expression presumably due to their low expression, while twelve genes were suppressed by miR-27b to below 60% of control mimics (Fig. 2b and Supplementary Table 3). Noticeably, cyclin G1 (CCNG1), a previously reported target gene of miR-27b as a negative regulator of p53 in various types of cancer cells61, was included, indicating that the screening was reliable. The remaining 11 genes were further examined.
Fig. 2. Screening of miR-27b target genes by in silico analyses and knockdown experiment.
a The schema for identifying the miR-27b target genes. In silico target prediction and microarray analysis identified 85 genes as target gene candidates. The mouse orthologs of 52 of the 85 genes were also predicted as miR-27b target genes. Then 32 of the 52 genes were further validated for target site conservation in human and mouse and for suppression by miR-27b mimics using qRT-PCR. Eleven genes were finally identified as candidate genes that miR-27b suppressed to below 60 % and that had conserved target sites between human and mouse. Knockdown experiments were carried out and lipid accumulation was evaluated, raising the three genes as the candidates. b Huh-7 cells were transfected with control miRNA (ctrl) or miR-27b mimics at 10 nM and mRNA expression of the candidate genes with conserved target sites was analyzed. n = 3 independent experiments. Results are presented as means ± SE. p-values from Student’s t-test and Cohen’s d values are shown. Gray circles, ctrl; gray triangles, miR-27b. c Huh-7 cells were transduced with lentiviral (LV) vectors expressing an shRNA for each candidate gene at 30 MOI. mRNA was prepared 3 days after transduction and mRNA levels of the target genes were analyzed by qRT-PCR. Relative mRNA expression levels are shown compared to those when the cells were transduced with the control shRNA-expressing LV vector, pLKO.1-puro-shRNA-control. n = 4 independent experiments. Results are presented as means ± SE. p-values from Student’s t-test and Cohen’s d values are shown. Gray circles, sh-ctrl; gray triangles, sh-target. d Lipids were stained with Bodipy493/503 4 days after transduction with LV vectors, and lipid amounts were evaluated based on the images. The relative lipid amount compared to the control LV vector was shown. n = 4 independent experiments. Results are presented as means ± SE. p-values from Student’s t-test and Cohen’s d values are shown. Gray circles, sh-ctrl; gray triangles, sh-target. sh-ctrl, LV vector expressing the control shRNA; sh-target, LV vectors expressing shRNAs for target genes.
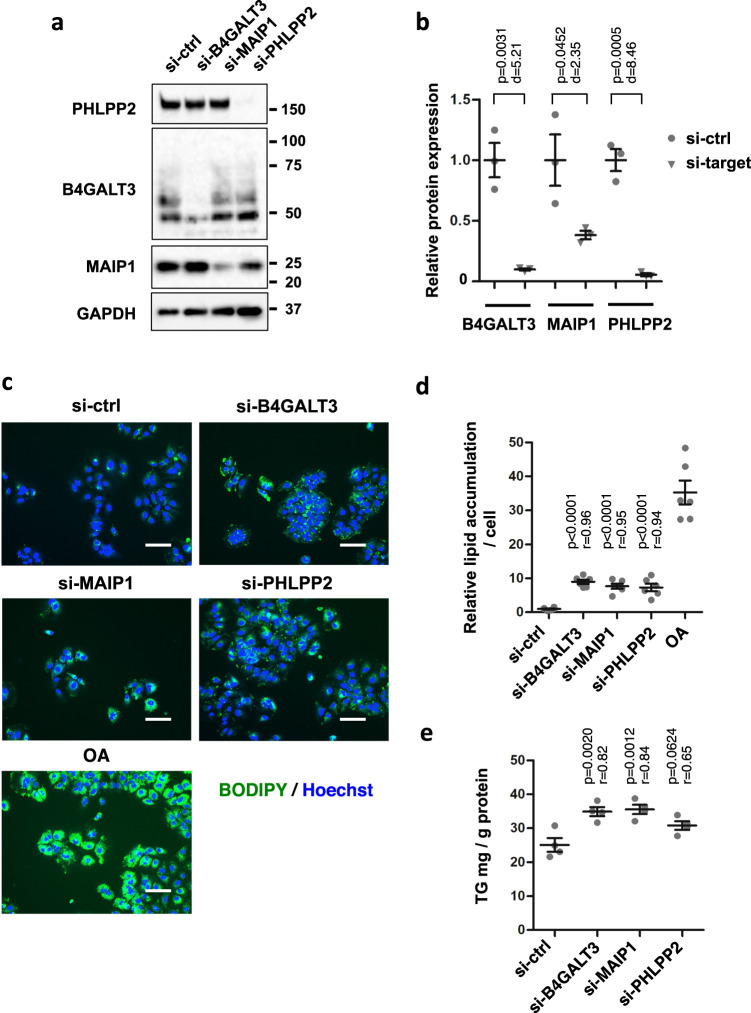
To explore genes related to lipid metabolism, the expression of the 11 genes was individually suppressed by shRNAs for each of the candidate genes, and the lipid accumulation was evaluated in Huh-7 cells. The knockdown efficiency was confirmed (Fig. 2c). Then, lipid accumulation was assessed by staining cells with BODIPY493/503, demonstrating that the knockdown of three genes, β-1,4-galactosyltransferase 3 (B4GALT3), matrix AAA peptidase interacting protein 1 (MAIP1, previously known as C2orf47) and PH domain and leucine rich repeat protein phosphatase 2 (PHLPP2), enhanced lipid accumulation (Fig. 2d).
Knockdown of B4GALT3 and MAIP1 induces lipid accumulation
To confirm that the decreased expression of these three genes was responsible for lipid accumulation, we performed knockdown experiments using siRNAs (Fig. 3). Huh-7 cells were transfected with siRNAs for the three genes and the knockdown efficiencies were assessed, confirming that they were efficiently suppressed (Fig. 3a and b). Under these conditions, lipid accumulation was evaluated, demonstrating that lipid accumulation was stimulated by knockdown of B4GALT3 and of MAIP1. The knockdown of PHLPP2 also caused an increase in lipid accumulation (Fig. 3c and d). Similarly, higher levels of triglyceride (TG) were detected in cells transfected with siRNAs for B4GALT and MAIP1 compared to the control. Transfection of cells with si-PHLPP2 also resulted in higher levels of TG (Fig. 3e).
Fig. 3. Knockdowns of B4GALT3, MAIP1, and PHLPP2 induce lipid accumulations in Huh-7 cells.
a Huh-7 cells were transfected with control siRNA (si-ctrl) or siRNA for each gene at 10 nM. Protein expression was examined 3 days after transfection by Western blotting. Molecular weight markers are shown on the right. GAPDH is for the internal control. b Protein expression levels were evaluated based on the images in (a). n = 3 independent experiments. Results are presented as means ± SE. p-values from one-way ANOVA followed by Bonferroni’s post-hoc comparisons tests for si-ctrl and Pearson’s r values are shown. si-ctrl, negative control siRNA; si-target, the corresponding siRNA for the target gene analyzed. Gray circles, si-ctrl; gray triangles, si-target. c Huh-7 cells were transfected with siRNAs at 10 nM. Cells were co-stained with BODIPY493/503 for the lipid and Hoechst 33342 for the nuclei 4 days after transfection. Untransfected cells were treated with oleic acid (OA) at 50 µM for the last 48 h. Scale bars, 100 μm. d The amount of lipids was evaluated based on the images in (c). The relative lipid accumulation of each sample was compared to that of the control siRNA. n = 6 independent experiments. Results are presented as means ± SE. e Triglycerides (TG) extracted from cells 4 days after transfection were quantitated. n = 4 independent experiments. Results are presented as means ± SE. p-values from one-way ANOVA followed by Bonferroni’s post-hoc comparisons tests for si-ctrl and Pearson’s r values are shown in (d) and (e).
MiR-27b directly regulates expression of B4GALT3 and MAIP1, but not PHLPP2
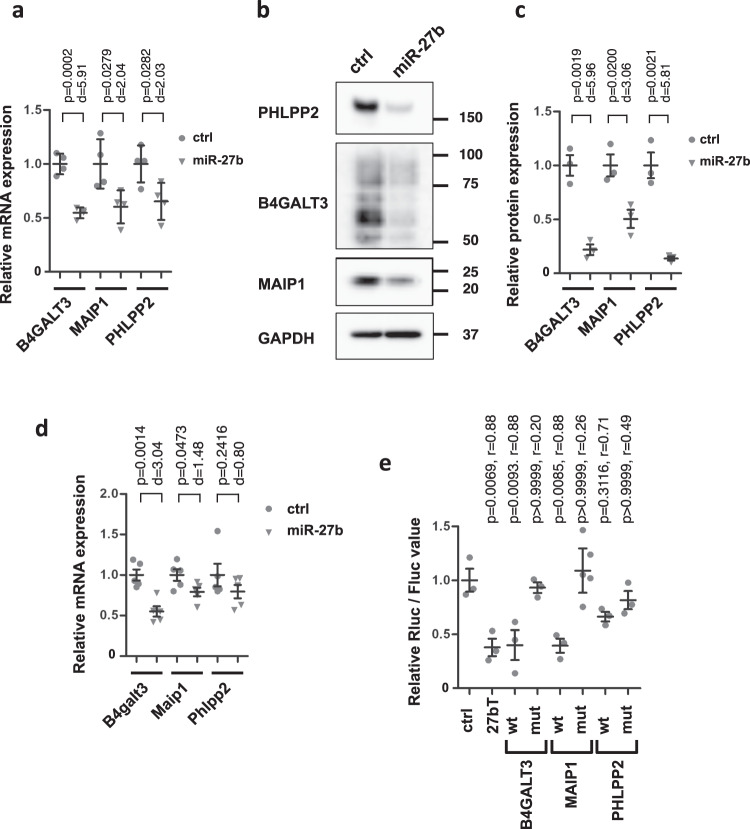
Next we examined whether B4GALT3, MAIP1, and PHLPP2 were actually regulated by miR-27b. Huh-7 cells were transfected with miR-27b mimics, and mRNA and protein expressions were analyzed. The mRNA expression levels of all three genes were suppressed by ectopic expression of miR-27b, which was well consistent with the results obtained in Fig. 2b (Fig. 4a). Similar suppression was observed in HepG2 cells transfected with miR-27b mimics (Supplementary Fig. 1c). Suppression of the protein expression was also confirmed (Fig. 4b and c). Further, expression of Maip1 and B4galt3 was suppressed in mouse primary hepatocytes when transfected with miR-27b mimics (Fig. 4d). These results clearly indicated that miR-27b suppressed the expression of MAIP1 and B4GALT3, and presumably PHLPP2, both in human and mouse hepatic cells.
Fig. 4. miR-27b suppresses the expression of B4GALT3 and MAIP1 as direct targets.
a Huh-7 cells were transfected with control miRNA (ctrl) and miR-27b mimics at 10 nM, and mRNA levels were examined 2 days after transfection by qRT-PCRs. n = 4 biologically independent samples. Representative results from three independent experiments are shown. Results are presented as means ± SD. p-values from Student’s t-test and Cohen’s d values are shown. Gray circles, ctrl; gray triangles, miR-27b mimics. b Protein expression was examined 3 days after transfection by Western blotting. Molecular weight markers are shown on the right. GAPDH is for the internal control. c Quantitation of protein expression. n = 3 independent experiments. Results are presented as means ± SE. p-values from Student’s t-test and Cohen’s d values are shown. Gray circles, ctrl; gray triangles, miR-27b mimics. d Primary mouse hepatocytes were transfected with control miRNA (ctrl) and miR-27b mimics at 50 nM, and mRNA levels were examined 2 days after transfection by qRT-PCRs. n = 5 independent experiments. Results are presented as means ± SE. p-values from Student’s t-test and Cohen’s d values are shown. Gray circles, ctrl; gray triangles, miR-27b mimics. e Huh-7 cells were co-transfected with reporter plasmids containing wild-type (wt) or mutated (mut) 3’UTRs with control miRNA and miR-27b mimics (10 nM). The empty reporter plasmid and the reporter plasmid containing the complementary sequence to miR-27b were used as a negative (ctrl) and a positive (27bT) control, respectively. Luciferase assay was performed 2 days after transfection. The renilla luciferase (Rluc) activity was normalized by firefly luciferase (Fluc) activity. The relative Rluc/Fluc values of miR-27b transfectants to the control miRNA mimic transfectants are shown. n = 3 independent experiments. Results are presented as means ± SE. The relative Rluc/Fluc of each reporter plasmid was compared to that of the control reporter plasmid. p-values from one-way ANOVA followed by Bonferroni’s post-hoc comparisons tests for ctrl and Pearson’s r values are shown.
Since these three genes have the target sequences, it is highly possible that they were directly suppressed by miR-27b. Therefore, we examined the direct interaction between miR-27b and the 3’UTRs of the three targets by a reporter assay (Fig. 4e). Two miR-27b target sequences were predicted in the 3’UTR of B4GALT3 and of MAIP1, while three were predicted in the 3’UTR of PHLPP2. The 3’UTRs were cloned into the reporter plasmids downstream of the luciferase genes, and mutations were introduced in the target sites. Huh-7 cells were co-transfected with miR-27b mimics and the wild-type or mutant reporter plasmids. miR-27b suppressed the luciferase gene expressions from wild-type reporter plasmids of B4GALT3 and MAIP1. Conversely, the suppressions were canceled when mutations were introduced in all the target sites. Meanwhile, the suppressive effect of miR-27b on the PHLPP2 reporter plasmid was relatively mild and the effects of the mutations were subtle. These results suggested that miR-27b suppressed B4GALT3 and MAIP1 directly, and PHLPP2 rather in an indirect manner.
Taken together, these results clearly demonstrated that miR-27b reduced the expression levels of B4GALT3 and MAIP1 directly via binding to the target sequences in their 3’UTRs.
Overexpression of MAIP1 ameliorated miR-27b-induced lipid accumulation
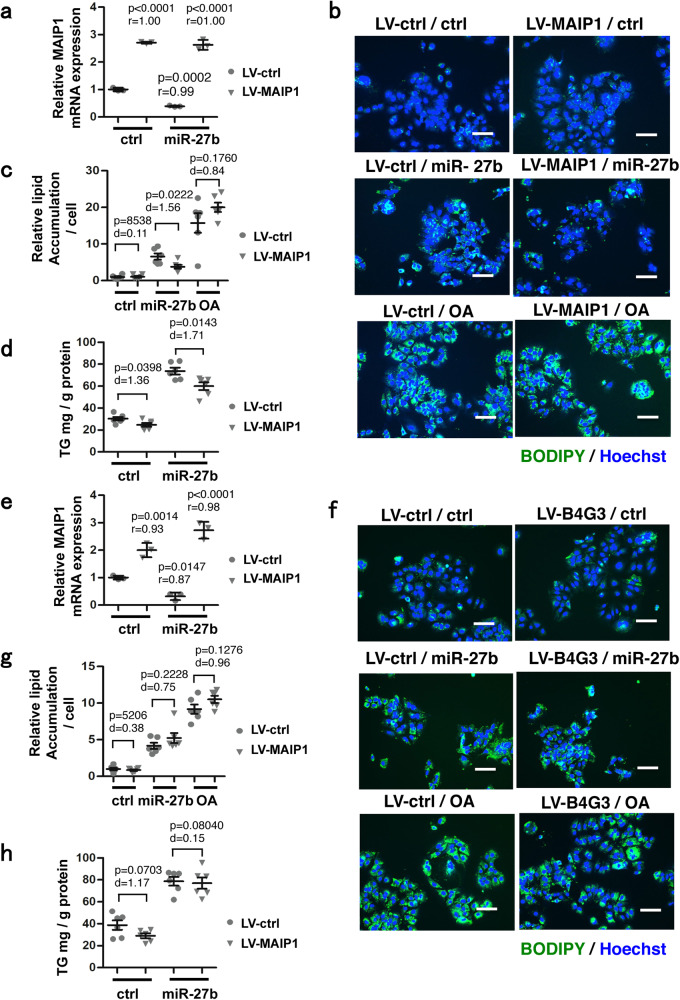
As demonstrated above, a decrease in the B4GALT3 and MAIP1 expression levels caused lipid accumulation and these genes were direct targets of miR-27b. Therefore, we examined whether miR-27b induces lipid accumulation via the suppression of B4GALT3 and MAIP1 expression (Fig. 5). Huh-7 cells were transduced with LV vectors expressing B4GALT3 or MAIP1 and then transfected with miR-27b mimics. The expression levels of B4GALT3 and MAIP1, which miR-27b suppressed, were restored to approximately two to threefold the endogenous expression levels by LV vector-mediated overexpression (Fig. 5a and d). Under these conditions, the overexpression of MAIP1 partially repressed the miR-27b-induced lipid accumulation (Fig. 5b and c). The overexpression of MAIP1 slightly repressed the TG level compared to the LV-ctrl (Fig. 5d). On the other hand, overexpression of B4GALT3 did not cancel the lipid accumulation induced by miR-27b (Fig. 5e–h).
Fig. 5. Restoring MAIP1-expression ameliorates the lipid accumulation induced by miR-27b.
a Huh-7 cells were transduced with LV-vectors expressing MAIP1 (20 MOI) followed by transfection with control miRNA (ctrl) and miR-27b mimics (10 nM) 24 h later. The mRNA levels of MAIP1 were analyzed 48 h after transfection by qRT-PCRs, and the relative expression of MAIP1 was evaluated. n = 3 biologically independent samples. Representative results from three independent experiments are presented as means ± SD. p-values from one-way ANOVA followed by Bonferroni’s post-hoc comparisons tests for LV-ctrl-transduced and ctrl-transfected samples and Pearson’s r values are shown. Gray circles, LV-ctrl; gray triangles, LV-MAIP1. b Huh-7 cells transduced with LV-vectors expressing MAIP1 (20 MOI) were transfected with control miRNA (ctrl) or miR-27b mimics (10 nM) and co-stained with BODIPY493/503 for the lipid and Hoechst 33342 for the nuclei 4 days after transfection. Untransfected cells were treated with oleic acid (OA) at 50 µM for the last 48 h. Scale bars, 100 μm. c The relative amount of lipid was evaluated based on the images in (b). n = 6 independent experiments. Results are presented as means ± SE. p-values from Student’s t-test and Cohen’s d values are shown. Gray circles, LV-ctrl; gray triangles, LV-MAIP1. d Triglycerides (TG) extracted from cells 4 days after transfection were quantitated. n = 6 independent experiments. Results are presented as means ± SE. p-values from Student’s t-test and Cohen’s d values are shown. Gray circles, LV-ctrl; gray triangles, LV-MAIP1. e Huh-7 cells were transduced with LV-vectors expressing B4GALT3 (50 MOI) followed by transfection with control miRNA (ctrl) and miR-27b mimics (10 nM) 24 h later. The mRNA levels of B4GALT3 were evaluated. n = 3 biologically independent samples. Representative results from three independent experiments are presented as means ± SD. p-values from one-way ANOVA followed by Bonferroni’s post-hoc comparisons tests for LV-ctrl–transduced and ctrl–transfected samples and Pearson’s r values are shown. Gray circles, LV-ctrl; gray triangles, LV-B4GALT3. f Huh-7 cells transduced with LV-vectors expressing B4GALT3 (50 MOI) were transfected with control miRNA (ctrl) or miR-27b mimics (10 nM) and co-stained with BODIPY493/503 and Hoechst 33342 4 days after transfection as in (b). Untransfected cells were treated with OA at 50 µM for the last 48 h. Scale bars, 100 μm. g The relative amount of lipid was evaluated based on the images in (f). n = 6 independent experiments. Results are presented as means ± SE. p-values from Student’s t-test and Cohen’s d values are shown. Gray circles, LV-ctrl; gray triangles, LV-B4GALT3. h TG extracted from cells 4 days after transfection were quantitated as in (d). n = 6 independent experiments. Results are presented as means ± SE. p-values from Student’s t-test and Cohen’s d values are shown. Gray circles, LV-ctrl; gray triangles, LV-B4GALT3. LV-ctrl, control LV vector; LV-MAIP1, MAIP1-expressing LV vector; LV-B4G3, B4GALT3-expressing LV vector.
Taken together, these results indicated that MAIP1 was a direct target of miR-27b and mediated, at least in part, miR-27b−induced lipid accumulation.
Decreased expression of MAIP1 did not inactivate AMP-activated protein kinase (AMPK)
Next, we attempted to elucidate the pathway by which MAIP1 affected lipid accumulation. Previous studies reported that essential MCU regulator (EMRE), a component of the calcium uniporter complex that includes the mitochondrial calcium uniporter (MCU), was assisted by MAIP1 to localize to the mitochondrial membrane62 and that hepatocyte-specific defects of MCU induced steatosis in mice, which was implicated in inactivation of Ampk, a master regulator of energy expenditure, in a cytoplasmic Ca2+-dependent manner63. Therefore, it was possible that MAIP1 depletion induced inactivation of AMPK, which could have been the cause of the lipid accumulation. To test this hypothesis, MAIP1 was knocked down in Huh-7 cells and the phosphorylated state of AMPK was examined. However, neither the phosphorylated protein level nor the phosphorylated and total AMPK protein ratio was suppressed by the depletion of MAIP1 protein, suggesting that AMPK was not the mediator of lipid accumulation by MAIP1 depletion (Supplementary Fig. 3).
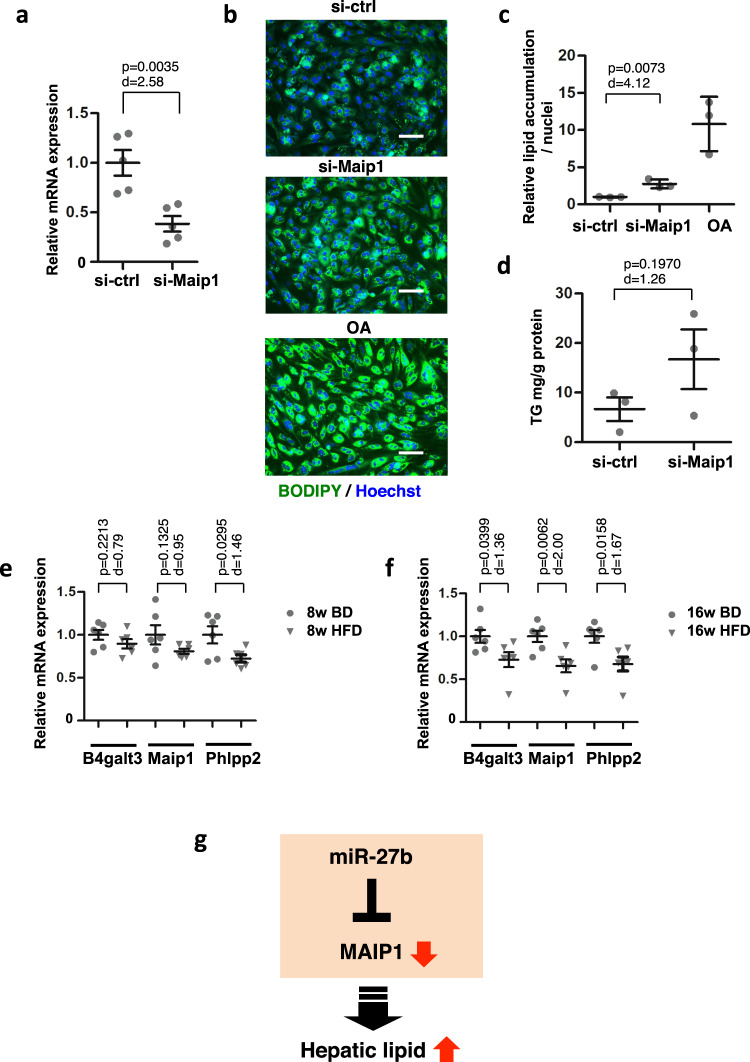
Decreased expression of Maip1 induced lipid accumulation in primary hepatocytes and was observed in the livers of HFD-fed mice
Next, we evaluated the association between mouse Maip1 expression and hepatic lipid accumulation. To examine the effect of decreased Maip1 expression on liver lipid accumulation, primary mouse hepatocytes were treated with siRNA for Maip1. Moderately down-regulated expression of Maip1 was confirmed (Fig. 6a). Under these conditions, lipid accumulation was stimulated in hepatocytes treated with si-Maip1, suggesting that down-regulated expression of Maip1 induces lipid accumulation in primary hepatic cells as observed in Huh-7 cells (Fig. 6b and c). The TG level revealed a upward trend when si-MAIP1 was transfected (Fig. 6d). The high-fat diet-fed mouse is a widely used animal model for fatty liver diseases. In order to examine the Maip1 expression in fatty livers, C57B/6 mice were fed either a basic or high-fat diet for 8 and 16 weeks followed by an analysis of the Maip1 expression. A slight decline in Maip1 expression was observed in the livers of HFD-fed mice at 8 weeks of feeding (Fig. 6e), and this decline became more prominent at 16 weeks of HFD feeding (Fig. 6f), suggesting that Maip1 expression decreased in association with the fatty liver progression. Of note, similar reductions in B4galt3 and Phlpp2 expression were observed, indicating that these expressions were also associated with in vivo fatty liver conditions.
Fig. 6. Decreased expression of Maip1 is associated with lipid accumulation in primary mouse hepatocytes and livers in high-fat diet-fed mice.
a Primary mouse hepatocytes were transfected with control siRNA (si-ctrl) or si-Maip1 at 50 nM. The mRNA levels were analyzed 48 h after transfection by qRT-PCRs, and the relative expression of Maip1 was evaluated. n = 5 independent experiments. Results are presented as means ± SE. The p-value from Student’s t-test and Cohen’s d value are shown. b Primary mouse hepatocytes were transfected with the indicated mimics at 50 nM followed by co-staining with BODIPY493/503 for the lipid and Hoechst 33342 for the nuclei 7 days after transfection. Untransfected cells were treated with oleic acid (OA) at 50 µM for the last 48 h. Scale bars, 100 μm. c The relative amount of lipid was evaluated based on the images in (b). n = 3 biologically independent samples. Representative results from three independent experiments are presented as means ± SD. The p-value from Student’s t-test and Cohen’s d value are shown. d Triglycerides (TG) extracted from cells 7 days after transfection were quantitated. n = 3 independent experiments. Results are presented as means ± SE. The p-value from Student’s t-test and Cohen’s d value are shown. e and f Livers from mice fed a basic diet (BD) or a high-fat diet (HFD) for 8 weeks (e) or 16 weeks (f) were analyzed by qRT-PCR. n = 6 mice. Data are presented as means ± SE. p-values from Student’s t-test and Cohen’s d values are shown. Gray circles, BD; gray triangles, HFD. g Schematic presentation of the miR-27b/MAIP1 axis-mediated hepatic lipid accumulation.
Gene expression analysis of MAIP1, B4GALT3 and PHLPP2 in liver biopsies form NASH patients
We also performed a separate analysis using a public database of RNA-seq data from the livers of NASH patients (GEO, GE225740)64,65, in which we searched for the expressions of MAIP1, B4GALT3, and PHLPP2 (Supplementary Fig. 4). The RNA samples were categorized into three groups (0, no steatohepatitis; 1, borderline steatohepatitis; 2, definite steatohepatitis) according to Powell et al.65. The expression of MAIP1 tended to decline with increasing level of steatohepatitis, suggesting that MAIP1 was involved in the manifestation.
Taking these results together, we identified a miR-27b-mediated regulatory pathway that targets MAIP1 and that eventually leads to hepatic lipid accumulation, as illustrated in Fig. 6g.
Disccussion
NAFLD is characterized by excessive lipid accumulation in the liver. A growing body of evidence suggests that miR-27b is involved in hepatic steatosis by altering lipid metabolisms. In this study, we screened for miR-27b target genes, whose knockdowns induced lipid accumulation, and identified B4GLAT3, MAIP1, and PHLPP2. PHLPP2 has been implicated in repressing the AKT serine/threonine kinase (AKT)/mechanistic target of rapamycin complex (mTORC) 1 pathway, which regulates de novo lipogenesis, as discussed below66–68. However, B4GALT3 and MAIP1 have not been directly implicated as regulators of lipid accumulation. Therefore, the latter two genes seem to be yet identified effectors whose altered expression induces lipid accumulation.
B4GALT3 is a resident of the Golgi apparatus in cells, which is where glycosylation processes occur and where the glycosylated proteins are then sorted. B4GALT3, one of seven β-1,4-galactosyltransferases, catalyzes the synthesis of the first N-acetyllactosamine unit of poly-N-acetyllactosamine chains and further glycosylates N-linked glycosylated proteins transported from ER69,70. Integrin β1 is so far the only reported substrate of B4GALT3, and impaired expression of B4GALT3 in tumors causes modifications in the behavior of cancer cells69,71,72. Proteins critical for lipid metabolism, such as lipases and proteins involved in LD formation, are the substrates for N-glycosylation69,73,74. Besides compromising the glycosylation of lipid metabolism-related proteins, disrupted glycosylation interrupts protein folding in the ER, inducing the accumulation of misfolded proteins in the ER, a condition of ER stress75,76. ER stress stimulates LD synthesis through the unfolded protein response (UPR) by activating ER stress sensors, endoplasmic reticulum to nucleus signaling 1 (IRE1a), eukaryotic translation initiation factor 2α kinase 3 (PERK), and activating transcription factor 6 (ATF6)35–40. In a yeast study, James et al. demonstrated that N-glycosylation deficiency induced ER stress accompanied by increased levels of LDs77. Further studies are required to elucidate the regulatory mechanisms of lipid accumulation by B4GALT3.
MAIP1 has been reported to be a mitochondrial matrix protein and implicated in protecting EMRE, a component of the calcium uniporter complex, MCU, from proteolysis by assisting membrane insertion62. Mitochondrial calcium uptake is involved in the regulation of a variety of cellular bioenergetic events78–80. Tomar et al. reported that a liver-specific MCU knockout in mice caused an increase in hepatic lipid accumulation, which was implicated as a result of an elevated cytoplasmic calcium level63. An increased cytoplasmic calcium level activates Ca-dependent protein phosphatase 4 (PP4) which, in turn, dephosphorylates and thus inactivates AMP-activated protein kinase (AMPK), resulting in decreased energy expenditures63. In addition, the impairment of MCU functions, which might be caused by MAIP1 depletion, may directly delay energy consumption, that is to say, suspended fatty acid oxidation, since oxidative phosphorylation depends on mitochondrial Ca2+81,82. Although neither decreased Ca2+ influx into mitochondria nor any changes in the cytoplasmic Ca2+ level were detected in the MAIP1-depleted non-hepatic cell line in the previous report62, such changes involving Ca2+ might still occur in hepatic cells. However, inactivation of AMPK was not observed in MAIP1-depleted Huh-7 cells (Supplementary Fig. 2), suggesting that massive changes in Ca2+ influx into mitochondria, such as would be sufficient to change the cytoplasmic Ca2+ levels, also did not occur by knockdown of MAIP1 in hepatic cells. Alternatively, the localization of enzymes involved in β-oxidation to the mitochondrial membrane might require the assistance of MAIP1, similarly to the case with EMRE; thus, depletion of MAIP1 might disturb fatty acid degradation, in turn inducing lipid accumulation62. It is also possible that MAIP1 would be required to maintain normal mitochondrial functions, probably via assisting a wide variety of mitochondrial proteins to be transported into mitochondria or integrated into the mitochondrial membrane; thus MAIP1-depletion would induce overall mitochondrial dysfunctions with increased oxidative stresses which might cause the lipid accumulation42. From the pathological point of view, morphological abnormality in mitochondria was observed in the livers of NASH patients83 and mitochondrial dysfunction was suggested to play a central role in the development and progression of NAFLD84–87. These possibilities, in particular the contribution of MAIP1-depletion to the development of NAFLD, need to be elucidated in future studies.
PHLPP2 is a regulator of AKT and protein kinase C (PKC) together with PHLPP1 and has been reported to play suppressive roles in cancers88. PHLPP2 also has been implicated in terminating insulin signaling by dephosphorylating AKT and thus inactivating mTORC167. PHLPP2 knockout in hepatoma cells actually demonstrated prolonged effects on insulin signals68. Since insulin generally stimulates de novo lipogenesis through the AKT/mTORC1 pathway in the liver89,90, a decrease in PHLPP2 expression might induce accelerated lipogenesis, leading to increased lipid accumulation. In fact, Kim et al. have reported that liver-specific PHLPP2-knockout mice exhibited hepatic steatosis68. They also provided evidence that the degradation of PHLPP2 by the ubiquitin pathway was involved in the downregulated expression of PHLPP2 in NAFLD68. Although the extent of lipid accumulation caused by PHLPP2 knockdown was not large in the short periods in our knockdown experiments (Fig. 3), our observation that miR-27b overexpression clearly suppressed PHLPP2 expression (Fig. 4) presents miR-27b as an unidentified player, in our knowledge, in the AKT/mTORC1-dependent network of lipid metabolisms.
In conclusion, our study provides additional insights into the molecular mechanism underlying hepatic steatosis caused by miR-27b and identifies B4GALT3 and MAIP1 as potential therapeutic targets for NAFLD.
Methods
Cell culture
Huh-7 and HepG2 cells (human hepatocarcinoma cell lines, obtained from the JCRB Cell Bank, Tokyo, Japan) and HEK293T cells (a transformed human embryonic kidney cell line) were cultured with Dulbecco’s modified Eagle’s medium (DMEM, 4.5 g/ml glucose; WAKO, Osaka, Japan) supplemented with 10% fetal bovine serum (FBS), streptomycin (100 μg/ml), and penicillin (100 U/ml). Cell cultures were maintained at 37 °C under 5% CO2 and fresh culture medium was supplemented every 48 h.
Plasmids
Luciferase reporter plasmids were generated as follows. The genomic fragments containing the 3’UTRs of B4GALT3 (nt 1800-2330), MAIP1 (nt 1065-1235) and PHPPL2 (nt 5074-5906) were amplified from genomic DNA of Huh-7 cells using the primers listed in Supplementary Table 4 and integrated between XhoI and NotI of psiCHECK2 (Promega, Madison, WI). Nucleotide substitutions (ACTGTGAA to ATTATAAA) were introduced into each seed sequence by the inverse PCR method91, generating the mutant reporter plasmids. psiCHECK2-27bT was generated by cloning the double-stranded oligonucleotides complementary to the sequence of miR-27b into the XhoI and NotI sites. shRNA-expressing LV vectors, TRC1.0 Lentiviral Plasmid Vector pLKO.1-puro-shRNA (MISSION shRNA, Sigma Aldrich, St. Louis, MO), was obtained from the Center for Medical Research and Education, Osaka University. A negative control shRNA-expressing LV vector, TRC1.0 Lentiviral Plasmid Vector pLKO.1-puro-shRNA-control, was purchased from Sigma Aldrich. To generate B4GALT3- and MAIP1-expressing LV vectors, coding regions were amplified from the cDNA of Huh-7 cells using the primers listed in Supplementary Table 4 and cloned between NotI and XbaI of CSII-EF-MCS-IRES2-Venus92. Their eukaryotic translation elongation factor (EF)-1α promoter regions were replaced with the thymidine kinase (TK) promoter by cloning DNA fragments of the TK promoter region amplified from psiCHECK2 between AgeI and XhoI, generating CSII-TK-B4GALT3 and CSII-TK-MAIP1, respectively. CSII-TK-MCS was similarly generated using CSII-EF-MCS93 as a control vector. CSII-EF-IRES2-Venus, CSII-EF-MCS, pCMV-VSVG-RSV-Rev and pCAG-HIVgp were provided by RIKEN BRC.
miRNA and siRNA transfection experiments
MirVana miRNA mimic negative control #1 (a random sequence mimic molecule), mirVana miRNA mimic hsa-miR-27b-3p (MC10750), silencer select siRNA negative control #1 (a siRNA with no sequence similarity to mouse, rat, or human genes), and silencer select pre-designed siRNA-human B4GALT3, -human MAIP1 and -human PHLPP2, and -mouse Maip1 were purchased from Thermo Fisher Scientific (Grand Island, NY). MiRNA and siRNA transfection was performed using Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. Four hours after transfection, the culture medium was replaced with DMEM (1.0 g/ml glucose; Sigma Aldrich) supplemented with 10% FBS, streptomycin (100 μg/ml), and penicillin (100 U/ml).
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Huh-7 or HepG2 cells were seeded in 48-well plates at a density of 1 × 104 cells/well and transfected with miRNA mimics or siRNA at 10 nM the next day. For the overexpression experiments, cells were transduced with LV vectors right after seeding and transfected with miRNA mimics at 10 nM 24 h post-transduction. Total RNA was isolated using ISOGEN (Nippon Gene, Tokyo, Japan). Complementary DNA (cDNA) was synthesized using a Superscript VILO cDNA synthesis kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Quantitative PCR (qPCR) was performed using a StepOnePlus real-time PCR system (Thermo Fisher Scientific) using THUNDERBIRD SYBR qPCR Mix (Toyobo, Osaka, Japan). The primers used in this study are listed in Supplementary Table 4. Values were normalized by GAPDH expression.
Western blotting
Protein samples were prepared 72 h after transfection using RIPA lysis buffer (Thermo Fisher Scientific) containing a proteinase inhibitor cocktail (Nacalai Tesque, Kyoto, Japan) and a phosphatase inhibitor cocktail (Nacalai Tesque). Whole-cell lysates (15–20 µg) were subjected to Western blotting. The following antibodies were used: rabbit anti-G3PDH polyclonal antibody (1:2000, Trevigen, Gaithersburg, MD); rabbit anti-MAIP1 polyclonal antibody (1:1000) (Atlas Antibodies, Stockholm, Sweden); rabbit anti-PHLPP2 polyclonal antibody (1:1000) (Abcam, Cambridge, UK); rabbit anti-B4GALT3 polyclonal antibody (1:500) (Atlas Antibodies); rabbit anti-AMPK polyclonal antibody (1:1000) (Cell Signaling Technology, Danvers, MA); rabbit anti-phosphorylated AMPK polyclonal antibody (1:1000) (Cell Signaling Technology). Signals were developed using Chemi-Lumi One Super (Nacalai Tesque) and visualized using ImageQuant LAS3000 (Fujifilm, Tokyo, Japan). Signal intensities were quantified using MultiGauge ver.3 software (Fujifilm). Samples prepared in three independent experiments were analyzed on the same membrane.
Microarray analysis
Huh-7 cells were transfected with control miRNA or miR-27b mimics at 10 nM, and total RNA samples were extracted 48 h post-transfection. Hybridization, signal acquisition, and data analysis were performed by Milteny Biotechnology (Bergisch Gladbach, Germany). Briefly, 100 ng of the total RNA sample was used to produce Cy3-labeled cRNA using the Agilent Low Input Quick Amp Labeling Kit (Agilent Technologies, Santa Clara, CA). Hybridization was performed using Agilent Whole Human Genome Oligo Microarrays 8x60K V2. Fluorescence signals were detected using Agilent’s Microarray Scanner System (Agilent Technologies). All the microarray data in this work have been deposited at GEO under accession number GSE174075.
Lipid droplet staining
Huh-7 or HepG2 cells were seeded in 48-well plates at a density of 1 × 104 cells/well or 0.2 × 104 cells/well, respectively, and transfected with miRNA mimics or siRNA at 10 nM the next day. For overexpression experiments, cells were transduced with LV vectors right after seeding and transfected with miRNA mimics at 10 nM 24 h post-transduction. As a control for each experiment, cells were cultured in the presence of 50 µM bovine serum albumin (BSA)-conjugated oleic acid (OA) for the last 48 h. The BSA-conjugated OA stock solution was prepared by vigorous agitation of 10 mM OA (Sigma Aldrich) in 20% fatty acid-free BSA (Wako)/PBS. Cells were fixed with 4% paraformaldehyde for 15 min at room temperature 96 h post-transfection. Lipid droplets were stained with 1 µg/mL of BODIPY 493/503 (Thermo Fisher Scientific) and cell nuclei were counterstained with 5 μg/mL Hoechst 33342 (Thermo Fisher Scientific) for 30 min at room temperature. Four images per well were taken by fluorescence microscopy (BZ-9000, Keyence, Osaka, Japan). Signal intensity was measured using a BZ-II analyzer (Keyence).
Quantitation of cellular triglycerides (TGs)
Huh-7 cells (1.5 × 105 cells/well) and primary mouse hepatocytes (1 × 105 cells/well) were seeded in 6- and 24-well plates and prepared for lipid extraction at days 4 and 7 of transfection, respectively. Cells were homogenized in the lysis buffer (1% Triton X-100/150 mM NaCl/1 mM EDTA/50 mM Tris–HCl, pH 7.5) and lipids were extracted according to the Folch method94. Briefly, 4 volume of chloroform (Fujifilm)/methanol (Nacalai Tesque) (1:2) was added to the homogenates and mixed well. After adding 1.25 volumes each of chloroform and distilled water and mixing, the lower phase was separated by centrifugation. The collected lower phase was dried-up and the residual lipids were resolved in 2-propanol (Nacalai tesque). The TG levels were measured using the enzymatic assay kit Cholestest TG (Sekisui Medical, Tokyo, Japan). TG values were normalized by the protein concentration of the homogenates.
Primary mouse hepatocytes
Hepatocytes were isolated from perfused livers of 4–9 week-old C57B/6N mice as previously described95. Briefly, livers in anesthetized mice were serially perfused with the warmed pre-perfusion buffer (Hank’s balanced salt solution (HBSS, Thermo Fisher Scientific)/0.5 mM EDTA (Dojindo, Kumamoto, Japan)/25 mM HEPES (Nacalai tesque)) and the perfusion buffer (0.5 µg/ml Collagenase D (Sigma Aldrich)/HBSS with Ca2+, Mg2+ (Thermo Fischer Scientific)/25 mM HEPES) via the inferior vena cava. Hepatocytes were purified by repeated centrifugation at 50×g for 5 min and viable cells were further purified by centrifugation in a Percol solution (45% Percol plus (GE Healthcare Bio-Sciences AB, Uppsala, Sweden)/5% 10xHBSS/50% DMEM) at 100×g for 20 min. Purified hepatocytes were plated in a collagen-coated 96-well and a 24-well plate (Iwaki Glass, Tokyo, Japan) at a density of 2.5 × 104 and 1 × 105 cells/well, respectively. After three-hour culture in William’s medium E (Thermo Fisher Scientific) supplemented with 10% FBS, streptomycin (100 μg/ml), penicillin (100 U/ml) and insulin (5 μg/ml, WAKO), the culture medium was replaced with Cellartice Power Primary HEP Medium (Takara Bio, Otsu, Japan). Cells were transfected with miRNA mimics or siRNAs at 50 nM using RNAiMAX on the next day of seeding. RNA preparation was performed on day 2 and staining with BODIPY493/503 and TG preparation was performed on day 7 of transfection.
Mice fed a high fat-diet (HFD)
Six-week-old male C57B/6J mice were fed a standard chow (Oriental Yeast Co., Osaka, Japan) or an HFD (HFD32; 5.1 kcal/g, 23% kcal from carbohydrate, 57% kcal from fat, 20% kcal from protein; CLEA Japan Inc., Tokyo, Japan) for either 8 or 16 weeks.
Luciferase assay
Huh-7 cells (1 × 104/well) were seeded in 96-well plates on the day before transfection. The cells were co-transfected with 25 ng of reporter plasmids carrying wild-type or mutant 3ʹ UTRs and miRNA mimics (10 nM) using Lipofectamin3000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. The luciferase activity was measured 48 h post-transfection using the dual-luciferase reporter assay (Promega) according to the manufacturer’s instructions. Luminescence was measured using a luminometer (Orion II Microplate Luminometer, Berthold Detection Systems, Pforzheim, Germany). All assays were performed in triplicate, and three independent experiments were evaluated.
Lentiviral vectors
LV vectors were prepared according to the RIKEN BRC protocol (http://cfm.brc.riken.jp/). Briefly, LV vector plasmids, together with pCMV-VSVG-RSV-Rev and pCAG-HIVgp, were transfected into HEK293T cells using polyethylenimine transfection reagents (Polysciences, Warrington, PA), and viral particles were purified from the culture supernatants using ultracentrifugation (50,000×g, 2 h). Vector titers were determined by transducing Huh-7 cells and subsequent genomic qPCR using CSII-EF-MCS-IRES2-Venus as a standard.
Animal experiments
All procedures involving animals and their care were approved by the Osaka University Institutional Animal Care and Use Committee in accordance with Fundamental Guidelines for Proper Conduct of Animal Experiment and Related Activities in Academic Research Institutions under the jurisdiction of the Ministry of Education, Culture, Sports, Science and Technology of Japan. We have complied with all the ethical regulations for animal experiments related to the approval.
Statistics and reproducibility
Statistical analyses were performed using GraphPad Prism 5.04 or 9.5.1. For quantitation of mRNA expression in Figs. 4a, 5a, 5e and Supplementary Fig. 1c, and lipid accumulation in Fig. 1e and Supplementary Fig. 1b, data from biologically independent samples were plotted as means ± SD. For the mouse and biopsy analysis, data from individuals were plotted as means ± SE. For the rest analyses, data from independent experiments were plotted as means ± SE. The exact sample size for each graph was presented in the corresponding figure legend. Comparisons between two groups and multiple groups were performed using unpaired two-tailed Student’s t-test and one-way ANOVA followed by Bonferroni’s post-hoc comparisons tests, respectively. P < 0.05 was considered to indicate significance. For effect size estimation, Cohen’s d and Pearson’s r values were calculated for the Student’s t-tests and the multiple-comparison tests, respectively.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This research was financially supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (grant numbers 18K06656, 21K18247) and by Bristol-Myers Squibb Foundation.
Author contributions
E.S. designed experiments, collected and analyzed data and wrote the manuscript. T.I., R.S., M.T.-G. and T.F. collected and analyzed data. F.S. supervised the study. H.M. supervised the study and edited the manuscript.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Joao Valente. A peer review file is available.
Data availability
The Data that supports the findings of this study are included in this article and its supplementary files. The source data behind the graph in the paper is available in Supplementary Data 3. Uncropped scans are available in Supplementary Fig. 5. Data from microarray analysis have been deposited in GEO under accession number GSE174075. Included data IDs are GSM5285801, GSM5285802, GSM5285803 GSM5285804 and GSM5285805. The accesion code for GSE174075 is obkxakwovbcxvcv. The following plasmids are available from Addgene: CSII-TK-MCS (#203597); CSII-TK-B4GALT3 (#203598); CSII-TK-MAIP1 (#203599); psicheck2-27bT (#203600); psicheck2-B4GALT3 (#203601); psicheck2-B4GALT3mut (#203602); psicheck2-MAIP1 (#203603); psicheck2-MAIP1mut (#203604); psicheck2-PHLPP2 (#203605); psicheck2-PHLPP2mut (#203606).
Code availability
The Agilent Feature Extraction Software (Agilent Technologies) was used to read out and process the microarray image files. The data files were further analyzed using Rosetta Resolver gene expression data analysis system (Rosetta Biosoftware, Seattle, USA). DAVID 2023 was used for the pathway analysis in Supplementary Fig. 2 and is available at https://david.ncifcrf.gov.
Ethics declarations
Animal experiments.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Eiko Sakai, Tsutomu Imaizumi.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-023-05049-w.
References
- 1.Williams CD, et al. Prevalence of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: a prospective study. Gastroenterology. 2011;140:124–131. doi: 10.1053/j.gastro.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 2.Portillo-Sanchez P, et al. High prevalence of nonalcoholic fatty liver disease in patients with type 2 diabetes mellitus and normal plasma aminotransferase levels. J. Clin. Endocrinol. Metab. 2015;100:2231–2238. doi: 10.1210/jc.2015-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeFilippis AP, et al. Nonalcoholic fatty liver disease and serum lipoproteins: the Multi-Ethnic Study of Atherosclerosis. Atherosclerosis. 2013;227:429–436. doi: 10.1016/j.atherosclerosis.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eslam M, et al. MAFLD: a consensus-driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology. 2020;158:1999–2014. doi: 10.1053/j.gastro.2019.11.312. [DOI] [PubMed] [Google Scholar]
- 5.Wanless IR, Lentz JS. Fatty liver hepatitis (steatohepatitis) and obesity: an autopsy study with analysis of risk factors. Hepatology. 1990;12:1106–1110. doi: 10.1002/hep.1840120505. [DOI] [PubMed] [Google Scholar]
- 6.Buzzetti E, Pinzani M, Tsochatzis EA. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD) Metabolism. 2016;65:1038–1048. doi: 10.1016/j.metabol.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 7.Tilg H, Moschen AR. Evolution of inflammation in nonalcoholic fatty liver disease: The multiple parallel hits hypothesis. Hepatology. 2010;52:1836–1846. doi: 10.1002/hep.24001. [DOI] [PubMed] [Google Scholar]
- 8.Sanyal AJ, et al. Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology. 2001;120:1183–1192. doi: 10.1053/gast.2001.23256. [DOI] [PubMed] [Google Scholar]
- 9.Utzschneider KM, Kahn SE. The role of insulin resistance in nonalcoholic fatty liver disease. J. Clin. Endocrinol. Metab. 2006;91:4753–4761. doi: 10.1210/jc.2006-0587. [DOI] [PubMed] [Google Scholar]
- 10.Crespo J, Fern P, Hern M, Mayorga M, Pons-Romero F. Gene expression of tumor necrosis factor [alpha] and TNF-receptors, p55 and p75, in nonalcoholic steatohepatitis patients. Hepatology. 2001;34:1158–1163. doi: 10.1053/jhep.2001.29628. [DOI] [PubMed] [Google Scholar]
- 11.Kern PA, Ranganathan S, Li C, Wood L, Ranganathan G. Adipose tissue tumor necrosis factor and interleukin-6 expression in human obesity and insulin resistance. Am. J. Physiol. Metab. 2001;280:E745–E751. doi: 10.1152/ajpendo.2001.280.5.E745. [DOI] [PubMed] [Google Scholar]
- 12.Gornicka A, et al. Transcriptional profile of genes involved in oxidative stress and antioxidant defense in a dietary murine model of steatohepatitis. Antioxid. Redox Signal. 2011;15:437–445. doi: 10.1089/ars.2010.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Videla LA, Rodrigo R, Araya J, Poniachik J. Oxidative stress and depletion of hepatic long-chain polyunsaturated fatty acids may contribute to nonalcoholic fatty liver disease. Free Radic. Biol. Med. 2004;37:1499–1507. doi: 10.1016/j.freeradbiomed.2004.06.033. [DOI] [PubMed] [Google Scholar]
- 14.Takaki A, Kawai D, Yamamoto K. Multiple hits, including oxidative stress, as pathogenesis and treatment target in non-alcoholic steatohepatitis (NASH) Int. J. Mol. Sci. 2013;14:20704–20728. doi: 10.3390/ijms141020704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feldman A, et al. Clinical and metabolic characterization of lean Caucasian subjects with non-alcoholic fatty liver. Off. J. Am. Coll. Gastroenterol. 2017;112:102–110. doi: 10.1038/ajg.2016.318. [DOI] [PubMed] [Google Scholar]
- 16.Xu A, et al. The fat-derived hormone adiponectin alleviates alcoholic and nonalcoholic fatty liver diseases in mice. J. Clin. Investig. 2003;112:91–100. doi: 10.1172/JCI200317797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boursier J, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63:764–775. doi: 10.1002/hep.28356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Roy T, et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2013;62:1787–1794. doi: 10.1136/gutjnl-2012-303816. [DOI] [PubMed] [Google Scholar]
- 19.Wightman B, et al. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 20.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 22.Laudadio I, et al. A feedback loop between the liver-enriched transcription factor network and miR-122 controls hepatocyte differentiation. Gastroenterology. 2012;142:119–129. doi: 10.1053/j.gastro.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Gjorgjieva M, et al. Genetic ablation of miR-22 fosters diet-induced obesity and NAFLD development. J. Pers. Med. 2020;10:170. doi: 10.3390/jpm10040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J, et al. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. J. Biol. Chem. 2010;285:12604–12611. doi: 10.1074/jbc.M109.094524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chai C, et al. Metabolic circuit involving free fatty acids, microRNA 122, and triglyceride synthesis in liver and muscle tissues. Gastroenterology. 2017;153:1404–1415. doi: 10.1053/j.gastro.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 26.He Z, Hu C, Jia W. miRNAs in non-alcoholic fatty liver disease. Front. Med. 2016;10:389–396. doi: 10.1007/s11684-016-0468-5. [DOI] [PubMed] [Google Scholar]
- 27.Pirola CJ, et al. Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut. 2015;64:800–812. doi: 10.1136/gutjnl-2014-306996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.López-Riera M, et al. Non-invasive prediction of NAFLD severity: a comprehensive, independent validation of previously postulated serum microRNA biomarkers. Sci. Rep. 2018;8:10606. doi: 10.1038/s41598-018-28854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ando Y, et al. Association of circulating miR-20a, miR-27a, and miR-126 with non-alcoholic fatty liver disease in general population. Sci. Rep. 2019;9:18856. doi: 10.1038/s41598-019-55076-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagos-Quintana M, et al. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/S0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 31.Michael MZ, O’Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer. Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 32.Tan Y, Ge G, Pan T, Wen D, Gan J. A pilot study of serum microRNAs panel as potential biomarkers for diagnosis of nonalcoholic fatty liver disease. PLoS ONE. 2014;9:e105192–e105192. doi: 10.1371/journal.pone.0105192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singaravelu R, et al. Hepatitis C virus induced up-regulation of microRNA-27: a novel mechanism for hepatic steatosis. Hepatology. 2014;59:98–108. doi: 10.1002/hep.26634. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi K, et al. High-fat diet increases vulnerability to atrial arrhythmia by conduction disturbance via miR-27b. J. Mol. Cell. Cardiol. 2016;90:38–46. doi: 10.1016/j.yjmcc.2015.11.034. [DOI] [PubMed] [Google Scholar]
- 35.Lee A-H, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang K, et al. The unfolded protein response transducer IRE1α prevents ER stress-induced hepatic steatosis. EMBO J. 2011;30:1357–1375. doi: 10.1038/emboj.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, et al. The role of CCAAT enhancer-binding protein homologous protein in human immunodeficiency virus protease-inhibitor-induced hepatic lipotoxicity in mice. Hepatology. 2013;57:1005–1016. doi: 10.1002/hep.26107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao G, et al. ATF4 protein deficiency protects against high fructose-induced hypertriglyceridemia in mice. J. Biol. Chem. 2013;288:25350–25361. doi: 10.1074/jbc.M113.470526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oyadomari S, Harding HP, Zhang Y, Oyadomari M, Ron D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008;7:520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rutkowski DT, et al. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev. Cell. 2008;15:829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fang D, et al. Endoplasmic reticulum stress leads to lipid accumulation through upregulation of SREBP-1c in normal hepatic and hepatoma cells. Mol. Cell. Biochem. 2013;381:127–137. doi: 10.1007/s11010-013-1694-7. [DOI] [PubMed] [Google Scholar]
- 42.Lee J, Homma T, Kurahashi T, Kang ES, Fujii J. Oxidative stress triggers lipid droplet accumulation in primary cultured hepatocytes by activating fatty acid synthesis. Biochem. Biophys. Res. Commun. 2015;464:229–235. doi: 10.1016/j.bbrc.2015.06.121. [DOI] [PubMed] [Google Scholar]
- 43.Vickers KC, et al. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. 2013;57:533–542. doi: 10.1002/hep.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kida K, et al. PPARα is regulated by miR-21 and miR-27b in human liver. Pharm. Res. 2011;28:2467–2476. doi: 10.1007/s11095-011-0473-y. [DOI] [PubMed] [Google Scholar]
- 45.Jennewein C, von Knethen A, Schmid T, Brüne B. MicroRNA-27b contributes to lipopolysaccharide-mediated peroxisome proliferator-activated receptor gamma (PPARgamma) mRNA destabilization. J. Biol. Chem. 2010;285:11846–11853. doi: 10.1074/jbc.M109.066399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J-J, Drakaki A, Iliopoulos D, Struhl K. MiR-27b targets PPARγ to inhibit growth, tumor progression and the inflammatory response in neuroblastoma cells. Oncogene. 2012;31:3818–3825. doi: 10.1038/onc.2011.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goedeke L, et al. miR-27b inhibits LDLR and ABCA1 expression but does not influence plasma and hepatic lipid levels in mice. Atherosclerosis. 2015;243:499–509. doi: 10.1016/j.atherosclerosis.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng JB, Russell DW. Mammalian wax biosynthesis: I. Identification of two fatty acyl-coenzyme A reductases with different substrate specificities and tissue distributions. J. Biol. Chem. 2004;279:37789–37797. doi: 10.1074/jbc.M406225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang S, et al. Cloning and functional characterization of a novel mitochondrial N-ethylmaleimide-sensitive glycerol-3-phosphate acyltransferase (GPAT2) Arch. Biochem. Biophys. 2007;465:347–358. doi: 10.1016/j.abb.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Page NM, Butlin DJ, Lomthaisong K, Lowry PJ. The human apolipoprotein L gene cluster: identification, classification, and sites of distribution. Genomics. 2001;74:71–78. doi: 10.1006/geno.2001.6534. [DOI] [PubMed] [Google Scholar]
- 51.Agbaga M-P, et al. Role of Stargardt-3 macular dystrophy protein (ELOVL4) in the biosynthesis of very long chain fatty acids. Proc. Natl. Acad. Sci. USA. 2008;105:12843–12848. doi: 10.1073/pnas.0802607105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cho HP, Nakamura MT, Clarke SD. Cloning, expression, and nutritional regulation of the mammalian Δ-6 desaturase. J. Biol. Chem. 1999;274:471–477. doi: 10.1074/jbc.274.1.471. [DOI] [PubMed] [Google Scholar]
- 53.Reubsaet FAG, Veerkamp JH, Bukkens SGF, Trijbels JMF, Monnens LAH. Acyl-CoA oxidase activity and peroxisomal fatty acid oxidation in rat tissues. Biochim. Biophys. Acta (BBA)-Lipids Lipid Metab. 1988;958:434–442. doi: 10.1016/0005-2760(88)90229-9. [DOI] [PubMed] [Google Scholar]
- 54.Le W, Abbas AS, Sprecher H, Vockley J, Schulz H. Long-chain acyl-CoA dehydrogenase is a key enzyme in the mitochondrial β-oxidation of unsaturated fatty acids. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids. 2000;1485:121–128. doi: 10.1016/s1388-1981(00)00034-2. [DOI] [PubMed] [Google Scholar]
- 55.Kurtz DM, et al. Targeted disruption of mouse long-chain acyl-CoA dehydrogenase gene reveals crucial roles for fatty acid oxidation. Proc. Natl. Acad. Sci. USA. 1998;95:15592–15597. doi: 10.1073/pnas.95.26.15592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y-L, et al. Aberrant hepatic expression of PPARγ2 stimulates hepatic lipogenesis in a mouse model of obesity, insulin resistance, dyslipidemia, and hepatic steatosis. J. Biol. Chem. 2006;281:37603–37615. doi: 10.1074/jbc.M604709200. [DOI] [PubMed] [Google Scholar]
- 57.Yamazaki T, Shiraishi S, Kishimoto K, Miura S, Ezaki O. An increase in liver PPARγ2 is an initial event to induce fatty liver in response to a diet high in butter: PPARγ2 knockdown improves fatty liver induced by high-saturated fat. J. Nutr. Biochem. 2011;22:543–553. doi: 10.1016/j.jnutbio.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 58.Matsusue K, et al. Liver-specific disruption of PPARgamma in leptin-deficient mice improves fatty liver but aggravates diabetic phenotypes. J. Clin. Investig. 2003;111:737–747. doi: 10.1172/JCI200317223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolf Greenstein A, et al. Hepatocyte-specific, PPARγ-regulated mechanisms to promote steatosis in adult mice. J. Endocrinol. 2017;232:107–121. doi: 10.1530/JOE-16-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu S, et al. J. Adipocyte-specific gene expression and adipogenic steatosis in the mouse liver due to peroxisome proliferator-activated receptor gamma1 (PPARgamma1) overexpression. J. Biol. Chem. 2003;278:498–505. doi: 10.1074/jbc.M210062200. [DOI] [PubMed] [Google Scholar]
- 61.Shang Y, et al. The miR27b-CCNG1-P53-miR-508-5p axis regulates multidrug resistance of gastric cancer. Oncotarget. 2016;7:538–549. doi: 10.18632/oncotarget.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.König T, et al. The m-AAA protease associated with neurodegeneration limits MCU activity in mitochondria. Mol. Cell. 2016;64:148–162. doi: 10.1016/j.molcel.2016.08.020. [DOI] [PubMed] [Google Scholar]
- 63.Tomar D, et al. Blockade of MCU-mediated Ca(2+) uptake perturbs lipid metabolism via PP4-dependent AMPK dephosphorylation. Cell Rep. 2019;26:3709–3725.e7. doi: 10.1016/j.celrep.2019.02.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Powell, N. R. et al. Clinically important alterations in pharmacogene expression in histologically severe nonalcoholic fatty liver disease. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE225740 (2023). [DOI] [PMC free article] [PubMed]
- 65.Powell NR, et al. Clinically important alterations in pharmacogene expression in histologically severe nonalcoholic fatty liver disease. Nat. Commun. 2023;14:1474. doi: 10.1038/s41467-023-37209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brognard J, Sierecki E, Gao T, Newton AC. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol. Cell. 2007;25:917–931. doi: 10.1016/j.molcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 67.Kim K, et al. MTORC1-independent Raptor prevents hepatic steatosis by stabilizing PHLPP2. Nat. Commun. 2016;7:10255. doi: 10.1038/ncomms10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim K, et al. Degradation of PHLPP2 by KCTD17, via a glucagon-dependent pathway, promotes hepatic steatosis. Gastroenterology. 2017;153:1568–1580.e10. doi: 10.1053/j.gastro.2017.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo S, Sato T, Shirane K, Furukawa K. Galactosylation of N-linked oligosaccharides by human β-1,4-galactosyltransferases I, II, III, IV, V, and VI expressed in Sf-9 cells. Glycobiology. 2001;11:813–820. doi: 10.1093/glycob/11.10.813. [DOI] [PubMed] [Google Scholar]
- 70.Bydlinski N, et al. The contributions of individual galactosyltransferases to protein specific N-glycan processing in Chinese Hamster Ovary cells. J. Biotechnol. 2018;282:101–110. doi: 10.1016/j.jbiotec.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 71.Chen C-H, et al. β-1,4-Galactosyltransferase III suppresses β1 integrin-mediated invasive phenotypes and negatively correlates with metastasis in colorectal cancer. Carcinogenesis. 2014;35:1258–1266. doi: 10.1093/carcin/bgu007. [DOI] [PubMed] [Google Scholar]
- 72.Liao W-C, et al. β-1,4-galactosyltransferase III suppresses extravillous trophoblast invasion through modifying β1-integrin glycosylation. Placenta. 2015;36:357–364. doi: 10.1016/j.placenta.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 73.Wölle J, Jansen H, Smith LC, Chan L. Functional role of N-linked glycosylation in human hepatic lipase: asparagine-56 is important for both enzyme activity and secretion. J. Lipid Res. 1993;34:2169–2176. doi: 10.1016/S0022-2275(20)35357-8. [DOI] [PubMed] [Google Scholar]
- 74.Fan H, Chen S, Sun Y, Xu S, Wu L. Seipin mutation at glycosylation sites activates autophagy in transfected cells via abnormal large lipid droplets generation. Acta Pharmacol. Sin. 2015;36:497–506. doi: 10.1038/aps.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Riederer MA, Hinnen A. Removal of N-glycosylation sites of the yeast acid phosphatase severely affects protein folding. J. Bacteriol. 1991;173:3539–3546. doi: 10.1128/jb.173.11.3539-3546.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marquardt T, Helenius A. Misfolding and aggregation of newly synthesized proteins in the endoplasmic reticulum. J. Cell Biol. 1992;117:505–513. doi: 10.1083/jcb.117.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.William James A, Ravi C, Srinivasan M, Nachiappan V. Crosstalk between protein N-glycosylation and lipid metabolism in Saccharomyces cerevisiae. Sci. Rep. 2019;9:14485. doi: 10.1038/s41598-019-51054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jouaville LS, Pinton P, Bastianutto C, Rutter GA, Rizzuto R. Regulation of mitochondrial ATP synthesis by calcium: evidence for a long-term metabolic priming. Proc. Natl. Acad. Sci. USA. 1999;96:13807–13812. doi: 10.1073/pnas.96.24.13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brini M, et al. A calcium signaling defect in the pathogenesis of a mitochondrial DNA inherited oxidative phosphorylation deficiency. Nat. Med. 1999;5:951–954. doi: 10.1038/11396. [DOI] [PubMed] [Google Scholar]
- 80.Mallilankaraman K, et al. MCUR1 is an essential component of mitochondrial Ca2+ uptake that regulates cellular metabolism. Nat. Cell Biol. 2012;14:1336–1343. doi: 10.1038/ncb2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Territo PR, Mootha VK, French SA, Balaban RS. Ca2+ activation of heart mitochondrial oxidative phosphorylation: role of the F0/F1-ATPase. Am. J. Physiol. Physiol. 2000;278:C423–C435. doi: 10.1152/ajpcell.2000.278.2.C423. [DOI] [PubMed] [Google Scholar]
- 82.Murphy AN, Kelleher JK, Fiskum G. Submicromolar Ca2+ regulates phosphorylating respiration by normal rat liver and AS-30D hepatoma mitochondria by different mechanisms. J. Biol. Chem. 1990;265:10527–10534. doi: 10.1016/S0021-9258(18)86979-4. [DOI] [PubMed] [Google Scholar]
- 83.Caldwell SH, et al. Mitochondrial abnormalities in non-alcoholic steatohepatitis. J. Hepatol. 1999;31:430–434. doi: 10.1016/S0168-8278(99)80033-6. [DOI] [PubMed] [Google Scholar]
- 84.Rector RS, et al. Mitochondrial dysfunction precedes insulin resistance and hepatic steatosis and contributes to the natural history of non-alcoholic fatty liver disease in an obese rodent model. J. Hepatol. 2010;52:727–736. doi: 10.1016/j.jhep.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koliaki C, et al. Adaptation of hepatic mitochondrial function in humans with non-alcoholic fatty liver is lost in steatohepatitis. Cell Metab. 2015;21:739–746. doi: 10.1016/j.cmet.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 86.Moore MP, et al. Compromised hepatic mitochondrial fatty acid oxidation and reduced markers of mitochondrial turnover in human NAFLD. Hepatology. 2022;76:1452–1465. doi: 10.1002/hep.32324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pérez-Carreras M, et al. Defective hepatic mitochondrial respiratory chain in patients with nonalcoholic steatohepatitis. Hepatology. 2003;38:999–1007. doi: 10.1002/hep.1840380426. [DOI] [PubMed] [Google Scholar]
- 88.Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol. Cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 89.Yecies JL, et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 2011;14:21–32. doi: 10.1016/j.cmet.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li S, Brown MS, Goldstein JL. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc. Natl. Acad. Sci. USA. 2010;107:3441–3446. doi: 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ochman H, Gerber AS, Hartl DL. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–3. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hayashi S, Yano M, Igarashi M, Okano HJ, Okano H. Alternative role of HuD splicing variants in neuronal differentiation. J. Neurosci. Res. 2015;93:399–409. doi: 10.1002/jnr.23496. [DOI] [PubMed] [Google Scholar]
- 93.Yamaguchi T, Morikawa A, Miyoshi H. Comparison of gene-trapping efficiency between retroviral and lentiviral vectors in mouse embryonic stem cells. Biochem. Biophys. Res. Commun. 2012;425:297–303. doi: 10.1016/j.bbrc.2012.07.085. [DOI] [PubMed] [Google Scholar]
- 94.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957;226:497–509. doi: 10.1016/S0021-9258(18)64849-5. [DOI] [PubMed] [Google Scholar]
- 95.Charni-Natan M, Goldstein I. Protocol for primary mouse hepatocyte isolation. STAR Protoc. 2020;1:100086. doi: 10.1016/j.xpro.2020.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The Data that supports the findings of this study are included in this article and its supplementary files. The source data behind the graph in the paper is available in Supplementary Data 3. Uncropped scans are available in Supplementary Fig. 5. Data from microarray analysis have been deposited in GEO under accession number GSE174075. Included data IDs are GSM5285801, GSM5285802, GSM5285803 GSM5285804 and GSM5285805. The accesion code for GSE174075 is obkxakwovbcxvcv. The following plasmids are available from Addgene: CSII-TK-MCS (#203597); CSII-TK-B4GALT3 (#203598); CSII-TK-MAIP1 (#203599); psicheck2-27bT (#203600); psicheck2-B4GALT3 (#203601); psicheck2-B4GALT3mut (#203602); psicheck2-MAIP1 (#203603); psicheck2-MAIP1mut (#203604); psicheck2-PHLPP2 (#203605); psicheck2-PHLPP2mut (#203606).
The Agilent Feature Extraction Software (Agilent Technologies) was used to read out and process the microarray image files. The data files were further analyzed using Rosetta Resolver gene expression data analysis system (Rosetta Biosoftware, Seattle, USA). DAVID 2023 was used for the pathway analysis in Supplementary Fig. 2 and is available at https://david.ncifcrf.gov.