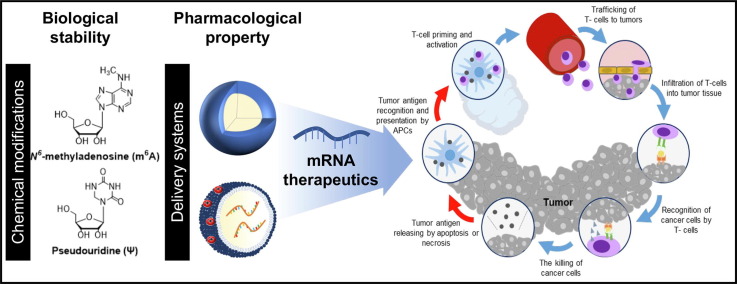
Graphical abstract
Keywords: mRNA, mRNA delivery, RNA drug, Chemical modification, Cancer immunotherapy
Abstract
RNA vaccines have demonstrated their ability to solve the issues posed by the COVID-19 pandemic. This success has led to the renaissance of research into mRNA and their nanoformulations as potential therapeutic modalities for various diseases. The potential of mRNA as a template for synthesizing proteins and protein fragments for cancer immunotherapy is now being explored. Despite the promise, the use of mRNA in cancer immunotherapy is limited by challenges, such as low stability against extracellular RNases, poor delivery efficiency to the target organs and cells, short circulatory half-life, variable expression levels and duration. This review highlights recent advances in chemical modification and advanced delivery systems that are helping to address these challenges and unlock the biological and pharmacological potential of mRNA therapeutics in cancer immunotherapy. The review concludes by discussing future perspectives for mRNA-based cancer immunotherapy, which holds great promise as a next-generation therapeutic modality.
1. Introduction
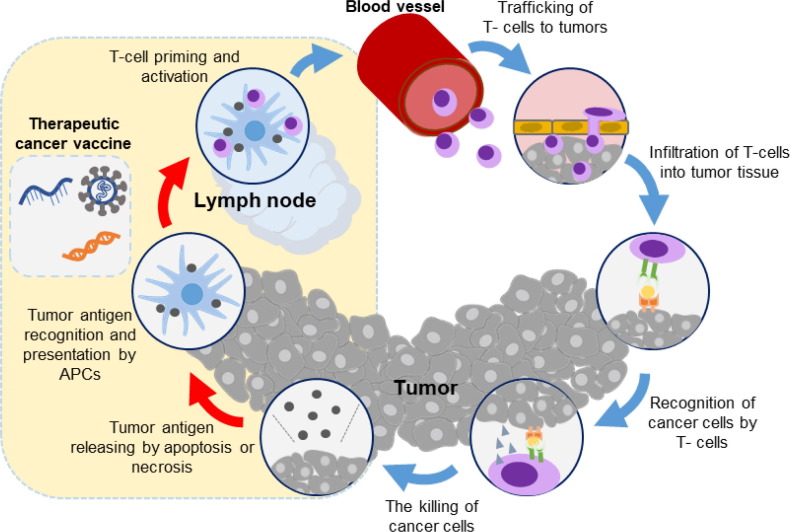
Cancer immunotherapy is a promising strategy for cancer treatment that aims to activate the host’s immune system against tumors, leading to immunological memory, reduced tumor burden, and increased the overall survival rates [1], [2], [3]. Cancer vaccines can deliver high-quality tumor antigens to antigen-presenting cells (APCs), process with the maturation of APCs, activate CD4+ helper T-cell and CD8+ cytotoxic T-cell responses, induce immune cell infiltration into the tumor microenvironment, and generate immunologic memory for chronic therapeutic effects [4], [5]. A schematic of typical cancer vaccine effects to elicit anti-cancer immune responses is illustrated in Fig. 1 . Cancer vaccines can selectively target tumor cells that expressing tumor-associated or tumor-specific antigens, stimulating the adaptive immune system to specifically eliminate them [6], [7], [8]. Among the various immunotherapeutic approaches, such as monoclonal antibodies, immune checkpoint inhibitors, cytokines, and chimeric antigen receptor (CAR) T-cell therapy, etc., cancer vaccines have the advantage of providing long-term chronic therapeutic effects accompanied with immunologic memory effect, making them an attractive therapeutic approach for cancer treatment [9]. Recent clinical trials of cancer vaccines have demonstrated their ability to activate anticancer immune responses and improve survival rates in cancer patients [10], [11]. However, challenges related to highly heterogenous tumor antigens, target-specific delivery, and low immune responses still need to be addressed for the clinical translation of cancer vaccines in cancer immunotherapy [12], [13].
Fig. 1.
The general action of therapeutic cancer vaccines and their effects on the cancer immunity cycle. Adapted with permission from reference [8].
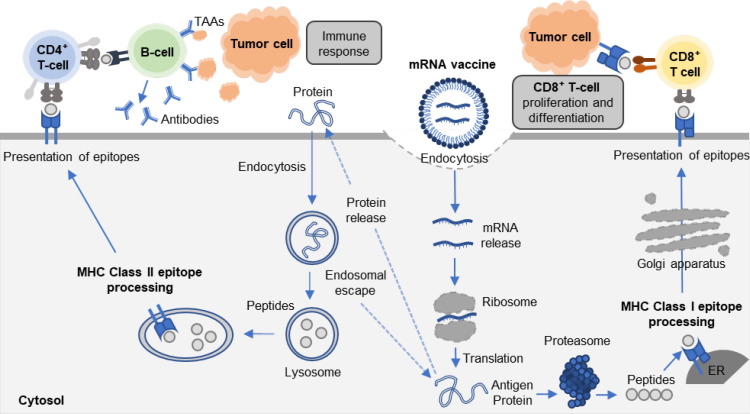
The recent COVID-19 pandemic has sparked an unprecedented interest in the potential of messenger RNA (mRNA) therapeutics, particularly in the context of the widespread application of mRNA vaccines such as BioNTech-Pfizer and Moderna [14]. mRNA, a single-stranded RNA molecule, plays a key role in gene expression by delivering genetic information from DNA in the nucleus to ribosomes in the cytoplasm, resulting in production of essential proteins for cellular function [15]. In recent years, mRNA has emerged as a promising therapeutic modality for treating various diseases, including cancer, infection diseases, and allergies, as well as for bioengineering purposes, such as cell reprogramming, genome editing, and protein replacement [16], [17]. mRNA-based therapeutic templates offer several benefits, including selective targeting of disease-related genes, rapid translation to clinical manufacturing [17]. Compared to conventional protein-based biologic agents, mRNA drug template offers better pharmacokinetic and pharmacodynamic properties and improved safety profiles of the target proteins [18], [19], [20]. Furthermore, exogenous mRNA inherently stimulates the innate immune system and triggers defense mechanisms in the living body [21], [22]. Combined with this inherent immunogenicity, mRNA encoding antigen-specific proteins, antibodies, and cytokines can induce adjuvant-like effects (Fig. 2 ) [23], [24]. Recently, adjuvant-like effects of mRNA have emerged as a fascinating approach for treating cancers and infectious diseases, where the immune system plays a critical role in disease progression [25]. RNA-based drugs offer a distinct advantage over DNA-based drugs as a cancer vaccine because they can rapidly translate into therapeutic proteins in the cytoplasm without the need for transportation into the nucleus [26], [27]. Although the development and applications of mRNA in cancer vaccines have some limitations, such as biological instability and poor pharmacokinetics of mRNA, and insufficient in vitro and in vivo delivery efficiency due to its negatively-charged and large-sized property [28], [29], researchers have developed various designs and delivery strategies to overcome these limitations and enhance the biological and pharmacological potential of mRNA over the past few decades. For example, several modifications of the mRNA backbone with untranslated regions or coding regions have been developed to enhance the stability and translation efficiency, thereby reducing susceptibility to enzymatic degradation [30], [31]. Furthermore, various non-viral mRNA delivery systems, including protamine, cationic lipids, polymers, exosomes, and gold nanoparticles, etc., have been designed for efficient in vitro and in vivo delivery, showing successful delivery and enhanced therapeutic efficacy [32], [33], [34], [35], [36], [37]. Although viral vectors can deliver mRNA with high efficacy, non-viral vectors are often preferred in RNA vaccines due to their safety, predictability, ease of manufacture, and greater flexibility in design. Non-viral vector avoids viral vector mediated immune responses and incorporation of additional foreign genomes. Therefore, this review will focus solely on non-viral vector mediated mRNA vaccine delivery. This review will present the recent progress in designs and delivery strategies of mRNA therapeutics for ameliorating the biological and pharmacological potential in cancer treatment. To achieve this goal, this review is structured as follows: the first section provides a summary of chemical modification of mRNA to improve stability and translation efficacy. The second section describes approaches for designing non-viral vector-based delivery systems to improve cytosolic delivery efficiency in vitro and in vivo conditions. The third section explores emerging approaches for using mRNA therapeutics in cancer immunotherapy. Finally, the fourth section discusses the future perspectives of mRNA therapeutics as novel therapeutic modalities in cancer immunotherapy.
Fig. 2.
The schematic illustration of adjuvant-like effects of mRNA vaccines, which encoded antigen-specific proteins. When mRNA is transfected into APCs, mRNA escapes from the endosome and is released into the cytosol. Subsequently, released mRNA is translated into encoded antigenic proteins. Antigenic protein is degraded in the proteasome and generates antigenic peptide epitope, which can be incorporated onto major histocompatibility (MHC) class I molecules in the endoplasmic reticulum (ER). Antigenic peptide epitope-loaded MHC class I complexes are presented on the cell surface, activating antigen-specific CD8+ T-cell responses by T-cell receptor (TCR) recognition and stimulation. On the other hand, secreted antigenic protein from the host cell can be taken up DCs. The exogenous antigenic protein is degraded in endosomes and lysosomes, generating antigenic peptide epitope and being loaded onto MHC class II. Antigenic peptide epitope-loaded MHC class II complexes are presented on the cell surface, eliciting CD4+ T-cell response. Exogenous antigenic protein also can be loaded onto MHC class I by cross-presentation mechanism after endosomal escape in the cytosol.
2. Chemical modification of mRNA
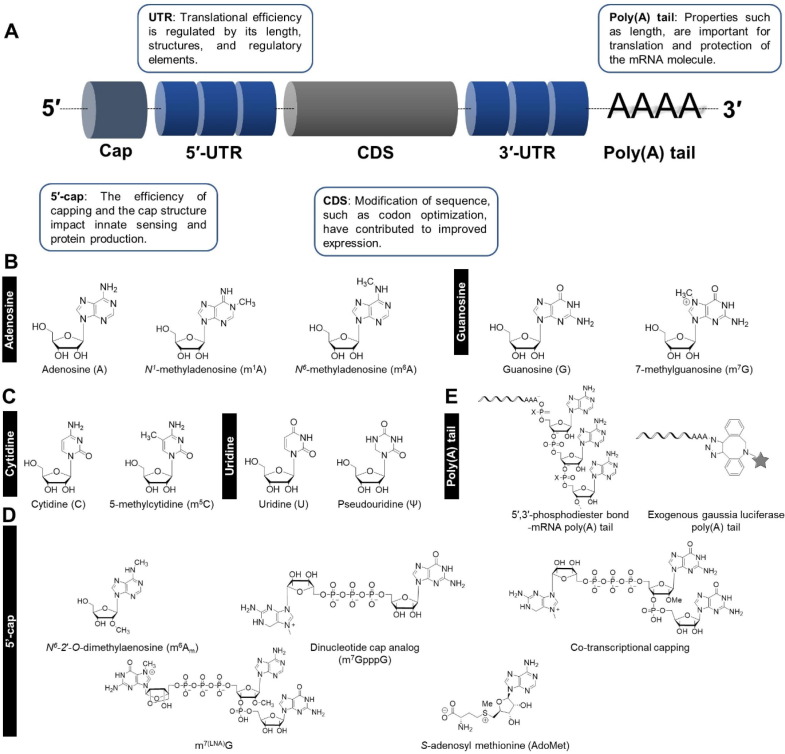
Various chemical modifications of mRNA are evolutionary products to control mRNA metabolic processes such as mRNA splicing, export, and stability [38]. These modifications can occur at various stages in the life cycle of mRNA, including during transcription, processing, transport, and translation [39]. Eukaryotic mRNA consists of a structure of 5′-cap, 5′-untranslated region (UTR), open reading frame (ORF), 3′-UTR, and poly(A) tail (Fig. 3 A). More than 100 chemical modifications of mRNA have been identified throughout this structure, representing a means of post-transcriptional regulations [40], [41] (Table 1 ). Enzymes known as 'writers,' 'readers,' and 'erasers' are associated with the chemical modifications of mRNA, which deposit, bind, and remove mRNA modifications, respectively, at mRNA base, cap, and tail. These modifications contribute to regulating body homeostasis, metabolism, embryonic development, and ultimately determine cell fate [42], [43].
Fig. 3.
Structure and modification type of mRNA. (A) Basic structure of mRNA. (B) Types of pyrimidine modification of mRNA. (C) Types of purine modification of mRNA. (D) Application of 5′-cap modification-containing mRNA. (E) Application of poly(A) tail modification-containing mRNA.
Table 1.
Types and characteristics of mRNA modifications.
| mRNA modification | Target | Application | Ref. | |
|---|---|---|---|---|
| Pyrimidine bases | m6A | Glioblastoma | Knockdown of METTL3 or METTL14 promotes glioblastoma stem cell (GSC) self-renewal and tumorigenesis. | [46] |
| m6A | Bladder cancer | METTL3 promotes cancer growth through pri-miR221/222 process in m6A-dependent manner. | [47] | |
| m6A | Hepatocellular carcinoma | Downregulation of METTL14 significantly promotes tumor metastasis. | [48] | |
| m6A | Cardiac hypertrophy | CHAPIR–METTL3–PARP10–NFATC4 signaling axis promotes cardiac hypertrophy. | [49] | |
| m1A | Breast and ovarian cancer cells | Up-regulation of ALKBH3 enhances the degree of CSF-1 expression and cancer cell invasion. | [56] | |
| m1A | Zebrafish embryos | ALKBH3 regulates the mRNA of Aurora A, a master suppressor of ciliogenesis. | [57] | |
| m7G | HUVEC | Improved METTL1 expression increases HUVECs migration, proliferation, and tube formation. | [65] | |
| m7G | Mammary epithelial cells | Cyclin D1 mRNA enhanced by RNMT regulates oncogene expression and cell transformation. | [67] | |
| Purine bases | m5C | Human colon carcinoma cells and HeLa cells | NSUN2 enhances p21 expression and cellular oxidative stress. | [71] |
| m5C | T lymphocytes | NSun2 regulates immune function by promoting the translation of IL-17A in T lymphocytes. | [73] | |
| Pseudouridine | Human embryonic kidney cells | mRNA with pseudouridine interacts less with PKR, resulting in reduced phosphorylation eIF-2α. | [82] | |
| 5′-cap | m7,3′-O-propargylGpppG | HeLa cells | Propargyl cap analog form a stable complex with the translation initiation factor than the standard 5′-cap, and the translation efficiency is 3.1-times higher. | [93] |
| Co-transcriptional capping | Human monocytic cells and human CD34+ cells | Co-transcriptional capping increases Cas9 mRNA activity and reduces immunogenicity. | [94] | |
| m7(LNA)GpppAmPG | Immortalized mouse immature dendritic cells | The combination of LNA and 5′-cap has 5-times more translation efficiency than standard 5′cap | [97] | |
| S-Adenosyl methionine (AdoMet) | Human embryonic kidney cells and HeLa cells |
AdoMet induces 3-times higher immune responses in human cells. | [98] | |
| Poly (A) tail | 5′,3′-phosphodiester bond poly (A) tail | HeLa cells | 5′,3′-phosphodiester bond poly (A) tail improves translation efficiency of Gaussia luciferase mRNA and suppresses mRNA degradation. | [99] |
| Azido-modified adenosine nucleotides in the poly(A) tail | HeLa cells | The covalent modification at the poly(A) tail leads to increase in translational efficiency without genetical editing. | [100] |
2.1. Base modifications
The nucleotide bases in mRNA are adenine (A), cytosine (C), guanine (G), and uracil (U) (Fig. 3B and 3C). Unlike DNA, the base adenine in mRNA forms a complementary base pair with uracil via hydrogen bonding, instead of thymine. The pairing of cytosine and guanine, on the other hand, involves the formation of three hydrogen bonds [43]. The sequence of these bases in mRNA carries the genetic code for the sequence of amino acids in a protein. Recent advances in RNA high-throughput sequencing and chemical analysis techniques have enabled deep research into the epitranscriptome and, therefore, there is a growing understanding of the roles of chemical modifications in regulating mRNA function and gene expression [44].
2.1.1. Pyrimidine bases
2.1.1.1. Adenosine
N6-methyladenosine (m6A), the methylation at the sixth position of an adenine base, is the most prevalent internal modification of mRNA in eukaryotes and plays an important role in regulating gene expression. These modifications have been shown to be involved in a wide range of biological processes including DNA damage response, embryogenesis, pluripotency, heat shock responses, and reprogramming [45]. Dysregulation of the m6A variant is associated with various diseases including cancer and neurodegenerative disorders. Knockdown of METTL3, an m6A writer, has been shown to increase mRNA m6A enrichment and alter mRNA gene expression, leading to the dramatic induction of human glioblastoma [46]. In bladder cancer, METTL3 promotes cancer growth through pri-miR221/222 process in an m6A-dependent manner [47]. Downregulation of METTL14, another m6A writer, has been shown to promote tumor metastasis associated with the development of hepatocellular carcinoma [48]. m6A methylation has been also found to play a crucial role in cardiac health. For example, CHAPIR–PIWIL4 complexes inhibit m6A methylation of PARP10 mRNA transcripts by directly binding to METTL3 [49]. As a result, PARP10 mRNA promotes cardiac hypertrophy by blocking the antihypertrophic effect of GSK3β [49]. In addition, it has been reported that reduced fat mass and obesity-associated protein (FTO) expression in hypoxic cardiomyocytes and mammalian heart promotes m6A methylation and reduces cardiomyocyte contractility [50]. Thus, proper regulation of m6A in cardiomyocytes is important for the treatment of heart failure [51]. Although N1-methyladenosine modification (m1A), the methylation at the first position of an adenine base, is typically found in tRNA and rRNA, they are also present in mRNA at low levels [52], [53]. More specifically, it is known to account for less than 0.2 % of all adenosine in mammalian tissues and less than 0.1 % in cell lines [52], [53], [54]. m1A RNA modifications play a role in modulating mRNA stability, structure, and translation. For example, demethylation of m1A by the overexpressed ALKBH3, a m1A eraser, [55] enhances CSF-1 mRNA stability in breast and ovarian cancer cells, leading to increased CSF-1 expression and cancer cell invasion [56]. ALKBH3 also regulates the mRNA of Aurora A, a master suppressor of ciliogenesis [57]. By removing m1A modification of Aurora A mRNA, ALKBH3 inhibits mRNA degradation and promotes mRNA translation, which in turn suppresses ciliogenesis.
2.1.1.2. Guanosine
N7-methylguanosine modifications (m7G), the methylation at the seventh position of a guanosine base, are present in all species of rRNA and tRNA, and often found in eukaryotic miRNA, 5′-caps, and mRNAs [58], [59], [60], [61], [62]. This modification is crucial for regulating eukaryotic mRNA splicing, export and translation. This internal RNA processing under the influence of the m7G variant has been suggested to be associated with biological responses of mRNA [63], [64]. For example, overexpression of METTL1, accelerating mRNA m7G methylation, results in increased human umbilical vein endothelial cells (HUVECs) angiogenesis in an m7G post-transcription-dependent manner [65]. In particular, m7G modifications are known to be closely related to 5′-capping [59], [62]. The guanosine cap is methylated by RNA guanine-7 methyltransferase (RNMT). RNMT-Activating Mini (RAM) protein, a mammalian cap methyltransferase, contains an RNA-binding domain and an RNMT-activating domain [66]. In addition, RNMT can increase translation efficiency by forming an mRNA complex with the eIF4E co-factor [66]. V H Cowling et al. found that increasing RNMT expression leads to 5′-cap methylation of cyclin D1 mRNA, a dormant oncogene in epithelial tumors, and promotes the expression of tumor-associated protein [67]. This study revealed that 5′-cap methylation of cyclin D1 mRNA by RNMT regulates oncogene expression and cell transformation.
2.1.2. Purine bases
2.1.2.1. Cytosine
Research on N5-methylcytosine (m5C) modification of RNA has mainly focused on tRNA and rRNA, but new information on m5C modification of mRNA has recently emerged [68], [69]. According to cytosine methylomes analysis on mouse and HeLa cells, approximately 1500 mRNAs in mouse tissue and 2000 mRNAs in HeLa cells were modified with m5C [70]. This modification often acts as a regulator of protein translation. For example, NSUN2-mediated m5C modification promotes METTL3/METTL14-mediated m6A modification and vice versa, ultimately leading to increased expression of p21 protein and enhanced cellular oxidative stress [71]. In addition, NSUN2-mediated CKD1 methylation of mRNA enhances CDK1 translation and affects the cell division cycle [72]. NSUN2-mediated m5C modification also modulates T lymphocytes' immune function. m5C methylation of IL-17A mRNA by NSUN2 promotes the translation of IL-17A in T lymphocytes [73].
2.1.2.2. Uridine
Pseudouridine, an isomer of uridine, is the most abundant post-transcriptional RNA modification found in all organisms with an estimated pseudouridine/uridine ratio of 7–9 % [74], [75], [76]. It has been identified in various types of RNA, including tRNA, rRNA, snRNA, and mRNA. mRNA pseudouridylation has the potential to broadly affect gene expression processes related with immunity, metabolism, growth, and development [77], [78], [79], [80]. D. E. Eyler et al. found that specific mRNA pseudouridylation interferes with amino acid synthesis and EF-Tu GTPase activation, suggesting a possible role in regulating translation rate and mRNA decoding [81]. Moreover, RNA-dependent protein kinase (PKR), a protein that inhibits translation by interacting with mRNA, becomes inactive when uridine in mRNA is replaced with pseudouridine [82]. That is, pseudouridine may play a role in regulating mRNA translation by regulating PKR. Interestingly, higher pseudouridine ratios do not necessarily increase translational efficiency. In fact, adding three pseudouridine points to ErmCL mRNA in bacteria did not increase or even decreased the protein synthesis efficiency [83]. Therefore, detailed treatment direction setting and analysis at the biochemical level are essential when using pseudouridine for disease treatment purposes. Besides pseudouridine, H. Moradian et al. demonstrated the efficient uridine modification in vitro transcribed mRNA structure for the mRNA vaccine targeting macrophages. In their study, 5-methoxy-uridine exhibits 4-folds higher transgene expression and elicits moderate inflammatory responses compared to other modification, such as pseudouridine, N1-methyl-pseudouridine, 5-methylcytidine, as well as a combination of pseudouridine/5-methylcytidine.[84].
2.2. Cap and tail modification of mRNA
The 5′-cap and poly(A) tail of mRNA are evolutionary products of eukaryotes [85], [86]. The 5′-cap and poly(A) tail protects the transcriptome and enhances translation in ribosomes in eukaryotic cells [87], [88]. The 5′-cap of mRNA modified with m7G is added on the first nucleotide of the transcript to protect the mRNA from degradation by exonucleases [85], [88]. Also, eukaryotic 5′-cap is involved in splicing, translation, and initiation for efficient and correct mRNA processing [85], [89]. Structurally, the 5′-cap consists of a triphosphate group linking m7G to the mRNA, and compared to other parts of mRNA, the 5′-cap is subject to various chemical modifications (Fig. 3D). The 5′-cap is transported from the nucleus with the help of the cap binding complex (CBC), and mRNA translation is initiated with eukaryotic translation initiation factor 4E (eIF4E) [89], [90]. Therefore, for 5′-cap modification, not only synthesis efficiency and high translation efficiency of modified mRNA, but also binding ability with eIF4E should be considered [91]. In some case, the 5′-cap modification by replacing the base structure with a modified base or analogue can lead to new properties. J. Mauer et al. replaced the cap's m7G with N6,2′-O-dimethyladenosine (m6Am) to confirm the new function of the mRNA [92]. m6Am-modified caps are selectively demethylated by fat mass and obesity-associated proteins, resulting in reduced mRNA stability. In addition, the dinucleotide cap analog containing propargyl group exhibit excellent capping effect and in vitro transcription efficiency, forming a more stable complex with elF4E [93]. The 5′-cap modification is also used in the CRISPR-Cas9 system. Through a co-transcriptional capping method using an initiating capped trimer instead of an anti-reverse cap analog (ARCA), Cas9 mRNA reduced the innate immune response and reduced mRNA immunogenicity in human CD34+ cells, Cas9 mRNA reduced innate immune responses, resulting in less mRNA immunogenicity in human CD34+ cells [94]. Recently mRNA vaccines have gained a significant interest as a therapeutic agent, particularly with the onset of severe acute respiratory syndrome corona-virus (SARS-CoV-2), and numerous analogues for 5′-capping have been developed [95], [96]. The cap analog comprising of locked nucleic acid (LNA) and m7G has a 5-fold increase in translation efficiency compared to the naive form, making it a potential strategy for mRNA vaccines development [97]. Furthermore, the analogs such as naturally methylated S-adenosyl-l-methionine (AdoMet) elicited a three-fold higher immune response [98]. The AdoMet analog-cap showed efficacy consistent with the mRNA vaccine targeting the epitope of SARS-CoV-2.
The poly(A) tail plays a crucial role in mRNA stability and transportation from the nucleus to the cytoplasm (Fig. 3E). There are two ways to add a poly(A) tail. The first is a method of naturally adding 100 to 200 adenosines to the transcribed mRNA after the polyadenylation signal sequence, and the second is a method of adding artificially to the 3′-end of the mRNA using poly(A) polymerase [99]. It was found that a chemical modification of 5′,3′-phosphodiester bond in the mRNA poly(A) tail prevented mRNA degradation in eukaryotic cells [99]. The modification of poly(A) tail of exogenous Gaussia luciferase (GLuc) mRNA increases mRNA translation efficiency by nearly three times. In addition, azido-modified adenosine nucleotides in the poly(A) tail are a way to amplify the amount of the reporter protein without modifying the genetic information [100].
3. Non-viral vector-based mRNA delivery systems
Gene therapy using mRNA holds great potential to treat devastating diseases because it controls transiently the expression or repression of specific genes with high efficacy. Despite these attractions, there are several obstacles that must be overcome for the clinical application of mRNA to be commercially available. These hurdles include the inefficient translocation through the cell membrane's lipid layer, the risk of phagocytosis by intracellular endosome barrier macrophages, and the degradation by ribonucleases. To address these issues, it is essential to develop a biocompatible and efficient mRNA delivery system. Viral vectors, such as adenovirus and lentivirus, are capable to deliver transgenes into host cells with a high efficacy, but are gradually falling out of favor due to basic distrust of their origin, risk of genome integration, and immune response [101], [102]. Therefore, a number of non-viral vectors have been developed for mRNA delivery (Table 2 ).
Table 2.
Examples of application of non-viral vector-based mRNA delivery system.
| Types | Base | Structure | Application | Advantage | Disadvantage | Ref. |
|---|---|---|---|---|---|---|
| Lipid-based delivery system | Cationic lipids | |||||
| DOPE | Activation of DC maturation and inflammatory mechanisms through interferon-α (IFNα) release by functional RNA-LPX. |
Formation of colloidally stable nanoparticulate RNA-LPX with reproducible particle size and charge. |
Immediate formation of large aggregates in near-neutral RNA-LPX, rendering them unstable. |
[105] | ||
| DOTMA | ||||||
| RNA-LPX for HPV16 vaccine. | Suitability for RNA study attributed to the cationic net charge. | Depending on the mixing ratio of lipid and RNA, there is a range in which the carrier is unstable. | [106] | |||
| A potential DC-targeting delivery system for mRNA vaccine. | Safety in vitro. |
Limitation in the in vivo delivery profile and anti-tumor efficacy. |
[107] | |||
| DOTAP | ||||||
| mRNA-based vaccine for anti-tumor immunity. |
Dual function of protecting mRNA from degradation and enhancing DC uptake. |
High level of cytotoxicity from the DOTAP/Chol/DSPE-PEG-2000 formulation. |
[108] | |||
| ePC | ||||||
| Enhancing intracellular delivery mediated by shock waves. |
Capability of delivering mRNA to diverse cancer cell types in vitro. | Cell type-dependent transfection efficiency. |
[110] |
|||
| Anionic lipids | ||||||
| 18PA, 14PA, and 18BMP |
 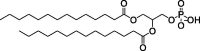 
|
Selective organ targeting for tissue-specific mRNA delivery and CRISPR-Cas gene editing. | Capacity for delivering therapeutic nucleic acids. | Inability to design nanoparticles for targeted tissue delivery. | [113] | |
| Ionizable lipids | ||||||
| DLin-MC3-DMA (MC3) |
Delivery of nucleic acid-based drugs. | Facilitation of endosomal escape for efficient nucleic acid delivery to the cytosol. | Depending on the mouse strain, toxicity of LNP to the fetus may occur. | [116] | ||
| Efficient transfection of retinal pigment epithelium (RPE). | Potential for rational design of optimal cell-specific gene delivery. |
Rapid elimination of dissociated components upon entry if nanoparticles exhibit instability. |
[117] | |||
| DSPC | Effective response to virus infection. | Favorable tolerability of the system compared to alternative non-viral delivery systems. | The effectiveness of vaccines against mutated viruses may be reduced. | [121], [122] | ||
| OF-02 | Promising delivery vehicle for therapeutic mRNA delivery to the liver. | Most potent mRNA delivery vehicle reported to date in the scientific literature. |
Absence of reports on the creation of a new series of ionizable lipids specifically designed for enhancing mRNA LNP delivery in vivo. |
[127] | ||
| A2-Iso5-2DC18 and A12-Iso5-2DC18 |
|
Optimal reduction of E7 mRNA expression, prevention of human papillomavirus, and stimulation of the STING pathway. |
Efficient delivery of mRNA. | LNP itself can trigger APC maturation. | [128] | |
| Lipid 5 |  |
Improved lipid delivery and rapid elimination in non-human primates. | Good balance of delivery efficiency and pharmacokinetics. |
The efficiency of LNP delivery to organs other than the liver is unknown. |
[131] | |
| Polymer-based delivery system | PEI | 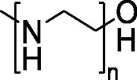 |
Efficient generation of “footprint-free” iPSCs using GO-PEI-RNA complexes for mRNA delivery. | Strong binding capacity to nucleic acids, effective uptake by cells, and excellent proton sponge effect for the endosomal release of DNA or RNA. | Significant decrease in cell viability and apparent increase in innate immune response gene expression. | [133] |
| PLGA | 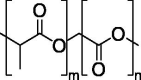 |
FDA-approved polyester type. | Carriers for DNA delivery. |
Absence of reports on intramuscular administration of PLGA-encapsulated plasmid DNA. |
[140] | |
| Poly-(β-amino ester) (PBAE) |
CAR or TCR mRNA delivery system for reprograming of circulating T cells | Less toxic than other nondegradable cationic polymers. |
A scale-up manufacturing process that can be applied clinically is essential. |
[142] | ||
| Chitosan | 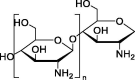 |
Promising therapeutic approach for cystic fibrosis. | Biodegradable characteristic. |
Depending on the route of administration, the efficiency of hCFTR expression varies. |
[149] | |
| Others | Gold nanoparticles | Rapid delivery of mRNA using VNB photoporation method. | Promising approach for safe and efficient intracellular mRNA delivery in cells. | Necessity to influence T cell homeostasis and therapeutic functionality. | [155] | |
| An indirect method of enhancing mRNA translation. | Enhancement of mRNA translation. |
Insufficiency of high cellular uptake and endosomal escape from endocytic vesicles. |
[157] | |||
| Silica nanoparticles | Analysis of mRNA delivery efficiency according to particle size and pore size of mesoporous SiNPs | SiNPs can be synthesized at room temperature. | The efficiency of mRNA delivery in vivo has not been confirmed. | [160] | ||
| By adding functional groups to SiNPs, new functions were added. |
As the tetrasulfide of SiNPs removed glutathione, the translation efficiency of the delivered mRNA increased. |
Cytotoxicity was observed at concentrations higher than 40 mg/ml of SiNPs. | [161] | |||
| Self-assembling mesoporous silica-cationic polymer-mRNA complex |
Tissue-specific delivery of mRNA to the pancreas and mesentery without toxicity. |
Target organs are limited. | [162] | |||
| Macrophage-targeted mRNA delivery | Silica shells protected mRNA from enzymatic degradation. |
The mechanism of macrophage-specific mRNA transfection has not been elucidated. |
[163] | |||
| Exosomes | Anti-inflammatory effects of IL-10 overexpression in atherosclerosis. | Potential to aid in disease diagnosis. | Unknown as physiological purpose of generating exosomes. | [172] | ||
| mRNA loading in exosomes via secreted after the endocytosis of LNP-mRNA system. |
Promising in vivo delivery carriers for siRNA-based therapies. |
Limitation of small size. | [173] | |||
| Potential therapeutic effects for stroke. | Low toxicity and immunogenicity. | Challenges in storage and transportation. | [174] | |||
|
Method for producing large-scale mRNA-encapsulating exosomes through cellular nanoporation. |
Favourable pharmacokinetic and immunological properties. |
Low yield observed when incorporating particularly large mRNA. |
[175] | |||
3.1. Lipid-based delivery systems
Since the first liposome was developed in 1965 [103], various lipid-based delivery systems have been developed for the transfection of exogenous mRNA. Amphiphilic lipids with a hydrophilic head and a hydrophobic tail can be engineered with specific residues to give them unique functions. Lipids are largely classified into cationic lipids, anionic lipids, and ionic lipids according to the nature of charge. Therefore, lipids used for mRNA delivery must be selected in consideration of cytotoxicity and delivery efficiency.
3.1.1. Cationic lipids
Cationic lipids bind negatively charged mRNAs via electrostatic interactions. Liposomes that isolate mRNA from the external environment not only protect mRNA from loss by ribonucleases and phagocytosis, but also enhance endocytosis. Cationic lipids most commonly used in liposome formulations for mRNA delivery include various amine derivatives such as 1, 2-di-O-octadecenyl-3-trimethylammonium-propane (DOTMA), 2, 3-dioleyloxy-N-[2-sperminecarboxamido ethyl]-N, N-dimethyl-1-propanaminium trifluoro-acetate (DOSPA), 1, 2-dioleoyl-3-trimethylammonium-propane (DOTAP), Ethylphosphatidylcholine (ePC), etc [104]. For example, L.M. Kranz et al. found that mRNA and DOTMA/DOPE liposome complexes target splenic dendritic cells and trigger interferon-α (IFNα) release [105]. As a result, these complexes showed functionality for cancer vaccine by inducing immune activity against cancer and maturation of dendritic cells. Subsequently, this research group achieved sustained tumor suppression using DOTMA-antigen-encoding mRNA liposomes against cancers induced by human papillomavirus infection [106]. Liposomes with high ratios of DOTAP, DOPE, and mannose-cholesterol conjugates were assembled by self-assembly method, providing the capability of dendritic cell-specific delivery for mRNA vaccines by targeting the overexpressed mannose receptor (CD206) [107]. In addition, various mRNA transport systems using ePC variants-based liposome suggest the possibility of being an excellent gene therapy platform based on cationic lipids [108], [109], [110]. As introduced, optimizing the combinations and ratios of various cationic lipids is a fundamental and essential step in designing liposomes that are effective for mRNA delivery.
3.1.2. Anionic lipids
Ultimately, anionic lipid-mRNA complexes are extremely challenging to cross the cytoplasmic anionic lipid bilayer, and special mRNA transport pathways are required to overcome these obstacles. For example, the mRNA-loaded anionic phospholipid-based 1,2-dioleoyl-sn-glycero-3-phosphate (18PA) transporter preferentially targets the hepatic reticuloendothelial system (RES), significantly enhancing mRNA expression in cells within the RES [111], [112]. These studies demonstrate that it can be used as an alternative to cationic lipid carriers, using an independent delivery mechanism. In addition, 18PA, 1,2-dimyristoyl-sn-glycero-3-phosphate (14PA), and sn-(3-oleoyl-2-hydroxy)-glycerol-1-phospho-sn-3′-(1′,2′-dioleoyl)-glycero (18BMP) can selectively deliver mRNA to the spleen depending on the composition [113]. This target organ specific delivery platform can increase delivery efficiency and minimize the toxicity to non-target organs.
3.1.3. Ionizable lipids
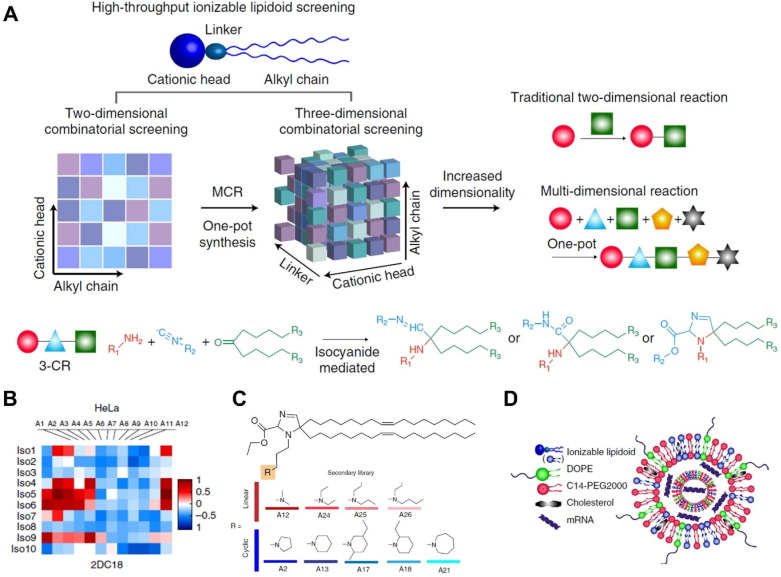
Ionizable lipids for mRNA delivery are neutral at physiological pH but become positive charge at low pH environment. This property enables the particle formation with negatively charged mRNAs and increase mRNA delivery efficiency by interacting with negatively charged endosome membranes, where pH is low, resulting in endosomal escape. In addition, neutral lipids present in the blood at neutral pH are relatively stable, preventing unexpected protein binding that may cause mRNA release and degradation [114]. Representatively, DLin-MC3-DMA (MC3) was the first ionizable lipid approved by the FDA as a siRNA delivery system and has great potential for mRNA delivery [115]. Studies by R. S. Riley et al. demonstrated that lipid nanoparticles (LNPs) composed of MC3 could deliver mRNA into mouse embryos in utero [116]. In addition, MC3-based ionizable lipids produced by various synthetic methods are widely applied as mRNA-mediated therapeutic agents [117], [118], [119], [120]. In response to concerns about potential infectious diseases, lipid-based mRNA vaccines are emerging as a rapidly producible and effective platform. DSPC-cholesterol-PEG lipid complex was developed as an ionizable lipid delivery system that can effectively respond to Zika virus infection and H10N8 and H7N9 influenza viruses [121], [122]. With the urgency of COVID-19, giant global biopharmaceutical companies such as Moderna, Pfizer-BioNTech, and AstraZeneca are taking the lead in developing mRNA delivery lipids [123], [124], [125], [126]. Anderson and colleagues have excelled successfully excessive expression of the human erythropoietin in the blood through the mRNA delivery using alkenyl amino alcohol lipid class [127]. Furthermore, this research group has developed the library of ionizable lipid combination using high-throughput synthesis method [128] (Fig. 4 A). Among numerous combinations of amine/isocyanide/ketone, they found two hydroimidazole-linked lipids optimally inducing target mRNA expression. These lipidoids showed anti-cancer effects by efficiently delivering mRNA to various types of cancer (Fig. 4B). The authors then compared the mRNA delivery efficiencies of heterocyclic amines and linear amines constituting LNP (Fig. 4C). Ultimately, combinations of heterocyclic lipidoid-mRNAs that represent the highest performing vaccines were obtained (Fig. 4D). The cytotoxicity of mRNA-containing LNPs is an important safety concern that determines the dose and interval of administration [129]. Recently, interest has increased in mRNA-containing LNPs with reduced cytotoxicity. Moderna has developed a wide range of ionizable lipids, such as Lipid 5, that are less immunostimulatory than MC3 in non-human primates [130], [131].
Fig. 4.
Optimal lipid formulations of mRNA vaccines via a high-throughput ionizable lipidoid screening system. (A) Schematic illustrating a three-dimensional combinatorial synthesis library for lipidoids. (B) Optimal lipidoids selection using mRNA transduction efficiency screening (C) Structures and candidates of heterocyclic amine combined lipidoids. (D) Schematic diagram of mRNA vaccine LNP-containing with heterocyclic lipid. Adapted with permission from reference [128].
3.2. Polymer-based delivery systems
Polymer-based mRNA delivery systems are widely used along with lipid delivery systems due to their customizable chemical modifications to the polymer end groups. In particular, cationic polymers, such as polyethyleneimine (PEI), electrically neutralize the negative charge of mRNA and aid cytosolic delivery through endocytosis and endosomal escape. However, PEI exhibits in vivo toxicity by interacting with various proteins, necessitating more biocompatible carrier design [132]. Graphene oxide (GO)-PEI coated mRNA complexes bind with negatively charge proteins, making a protein corona around the nanoparticles [133], [134]. This negatively charged protein-coated GO-PEI-mRNA complex showed less cytotoxicity due to reduced reactivity with surrounding positively charged proteins. They have been used to deliver mRNA to human induced pluripotent stem cells [133]. In addition, a polyplex of branched PEI with a succinylation (succPEI) containing disulfide building blocks showed the superior property in balancing endosomal escape and structural stability [135]. When the polyplex enters the cells, a disulfide block between the cationic backbone and the hydrophobic domain enhances the endosomal escape of mRNA in the presence of high concentrations of glutathione. Peptide-based poly (amino acid) has been used for a long time due to its high biocompatibility. For example, A. Dirisala et al. confirmed that poly(L-ornithine) based mRNA polyplexes showed higher stability than poly(lysine) [136]. In addition, protamine-complexed mRNA can be applied to prostate and lung cancer vaccine through the high antigen expression and immune stimulation by TLR 7/8 mechanism [137], [138], [139]. Poly(lactic-co-glycolic acid) (PLGA), an FDA-approved polyester, has strengths in biodegradability and biocompatibility due to hydrolysis of ester bonds, as well as high structural stability and economic feasibility [140]. PLGA-based mRNA complexes express nuclease-encoding mRNA and successfully complete genome editing at target sites [141]. Highly biodegradable poly (β-amino esters) (PBAEs) are also often used as mRNA delivery polyplexes. PBAEs are mRNA delivery platforms for T cell receptors (TCR) or chimeric antigen receptors (CAR) engineering to reprogram T cells [142]. Hyperbranched PBAEs (hPBAEs) mRNA polyplexes deliver mRNA drugs to the lungs via a non-invasive aerosol inhalation route. hPBAEs expressed homogeneous mRNA selectively in the lung and showed no toxicity with repeated administration [143]. Poly(amidoamine) (PAMAM) dendrimers have been utilized as non-viral vectors for mRNA due to high degree of functionality based on the primary amines and tertiary amines in the structure [144]. F. Joubert et al. explored cytosolic delivery efficiency of mRNA using end group modified PAMAM and poly(L-lysine) dendrimers [145]. They demonstrated that surface modifications could affect to form dendriplex complexes as small, stable, and well-encapsulating with mRNA. Additionally, the introduction of fusogenic groups to dendrimer could drive endosomal escape, resulting to achieve successful intracellular delivery and translation of mRNA in the cells.
Natural polysaccharides, such as chitosan, beta-glucan, and dextran, have excellent biodegradability, biocompatibility, and immune regulation, as well as various structures, charges, and sizes, making it easy to design carriers according to requirements [146], [147]. Since chitosan is a polysaccharide derived from chitin that can interact with nucleic acids, research results on the mRNA delivery potential of this natural material are abundant [148]. Chitosan-coated PLGA carrier via intratracheal (i.t.) or intravenous (i.v.) injection can be an effective treatment for mouse model of cystic fibrosis [149].
3.3. Inorganic nanoparticles
Gold nanoparticles (AuNPs) are attractive materials as versatile DNA/RNA carriers due to their facile surface modification and wide range of formulation options [150], [151]. In addition, AuNPs are inorganic nanoparticles that possess biological stability, a narrow particle size distribution, and a wide range of electromagnetic properties [152]. J. H. Yeom et al. hybridized the 5′-end of BCL2-associated X protein (BAX) mRNA to DNA oligonucleotides conjugated with 15 nm AuNP [153]. This AuNPs-RNA hybrid efficiently produced pro-apoptotic factors into tumor cells and suppressed xenograft tumor growth. When positively charged folic-acid (FA)-modified poly-amidoamine (PAMAM)-grafted AuNPs were nano-complexed with luciferase protein mRNA through electrostatic attraction, cells were transfected by the nano-complex and the expression of luciferase was promoted in vitro [154]. Unlike most existing RNA delivery methods through a direct attachment to AuNPs, a new gold nanoparticle-mediated vapor nanobubble (VNB) photoporation method has been developed [155]. This VNB photoporation method is a rapid delivery of mRNA by creating a nanobubble space through laser irradiation of AuNPs attached to the cell membrane surface. This method showed a transfection efficiency of up to 45 % in Jurkat T cells and significantly lower toxicity compared to electroporation. AuNPs have also been developed to increase protein production by increasing the translation efficiency for endogenous mRNAs in cells. For example, attachment of DNA oligomers with complementary sequences to target mRNAs on the surface of AuNPs increased the expression of insulin and green fluorescent protein in HeLa cells [156]. In addition, poly(thymine)-functionalized AuNPs (AuNP-p(T)DNA) were found to dramatically increase the expression of target mRNAs when polyadenylated target mRNA vectors were transfected into HeLa cells [157].
Amorphous silica nanoparticles (SiNPs) composed of silicon dioxide are well-established drug delivery carriers for small molecule drugs, therapeutic proteins, and gene materials, including mRNA, due to their convenient fabrication properties and excellent biocompatibility [158]. Non-porous (solid) SiNPs were initially developed in nanomedicine field with a simple fabrication process [159] and mesoporous SiNPs are widely used owing to their ability to accommodate large amounts of cargo, facile surface modification, and efficient biodegradation. Y. Wang et al. synthesized dendritic mesoporous SiNPs with homogenous and large pores and showed high mRNA transduction in vitro when the particle diameter was less than 50 nm and the pore size was larger than 20 nm [160]. This group also synthesized tetrasulfide incorporated large-pore dendritic SiNPs as mRNA delivery carriers [161]. This SiNPs can transport both mRNA and tetrasulfide moiety. This sulfide moiety act by depleting intracellular GSH, which in turn reduces the activity of GAPDH and activates mTORC1, consequently leading to enhance the translation efficiency of intracellularly delivered mRNA. S. Dong et al. showed the successful mRNA translation in mice after intraperitoneal injection of self-assembled mRNA/PEI/mesoporous SiNPs [162]. In contrast to the conventional approach of utilizing positively charged SiNPs to bind mRNA, R. Kamegawa et al. made mRNA-loaded SiNP by encapsulating mRNA/cationic polymer complex within a silica shell. This formulation demonstrated the potential for bioinspired macrophage-specific mRNA delivery [163]. Leveraging the ability of scavenger receptors on macrophages to recognize and bind negatively charged SiNPs, the researchers successfully achieved macrophage-specific delivery of SiNPs containing mRNA/polymer complexes. They compared the uptake efficiency of these particles in two cell lines: RAW 264.7 (murine macrophage cells) and CT26 (murine colorectal carcinoma cells). The silica shell surrounding the mRNA complex can protect mRNA payloads from enzymatic degradation and be degraded to silicate within the macrophage, thereby releasing the mRNA.
3.4. Exosomes
Exosomes are a type of approximately 30–150 nm extracellular vesicles secreted from parental cells [164]. Exosomes play a role in communication between cells and tissues by transporting macromolecules such as proteins, DNA, and RNA [165]. Due to the physiological characteristics of cell-derived exosomes, lipid bilayer exosomes have excellent biocompatibility and low immunogenicity compared to artificial carriers [166]. Exosomes are secreted from all types of cells and can be extracted from blood, urine, saliva, breast milk, etc., and the yield, stability, and biological properties are varied depending on the origin of exosomes [167]. However, generating clinically relevant amounts of exosomes requires several days of cell culture and loading large RNA molecules such as mRNA into exosomes is not easy [168], [169]. Electroporation is known to be an efficient method for loading short sequences of nucleic acids such as miRNAs and siRNAs, but is not entirely successful for mRNA [170]. Several mRNA loading methods have been developed to overcome the inefficiency of electroporation. One method is to load the complexes of negatively charged mRNA/cationic lipids into the exosomes by simple mixing [171]. Another method is to transfect the donor cells with plasmids encoding the target mRNA to increase the amount of mRNA loaded into exosomes. Since there is no modification in the external structure of exosomes, it has the advantage of maintaining the biological advantages of exosomes. For example, donor cells transfected with a plasmid encoding the internal ribosome entry site (IRES)-IL-10 mRNA secreted the exosomes passively loaded with IRES-IL-10 mRNA. These exosomes showed anti-inflammatory effects related to IL-10 overexpression in atherosclerosis [172]. In addition, M. Maugeri at al. used the endocytosis mechanism of LNP-mRNA to replace LNP with exosomes as an mRNA carrier [173]. The mRNA of LNP-mRNA escapes endosomes from donor cells and is eventually loaded into exosomes. Furthermore, inserting rabies virus glycoprotein (RVG) into exosomes of donor cells transformed with a plasmid encoding nerve growth factor (NGF) mRNA can efficiently deliver mRNA to ischemic cortex by targeting neurons [174]. Nanoporation method was developed in preparation for the limited yield of mRNA-loaded exosomes. This method resulted in the production of exosomes loaded with phosphatase and tensin homologue (PTEN) mRNA over 50 times more than electroporation [175]. Liposomes can easily encapsulate various drug molecules, but need the fine tuning of composition to avoid toxicity, and exosomes are biocompatible but have difficulties in RNA loading. To address the shortcomings of these two nanocarriers, a new carrier called liposome-exosome hybrid was developed, providing the possibility of improving mRNA loading efficiency and delivery rate in the future [176], [177], [178].
4. mRNA therapeutics for cancer immunotherapy
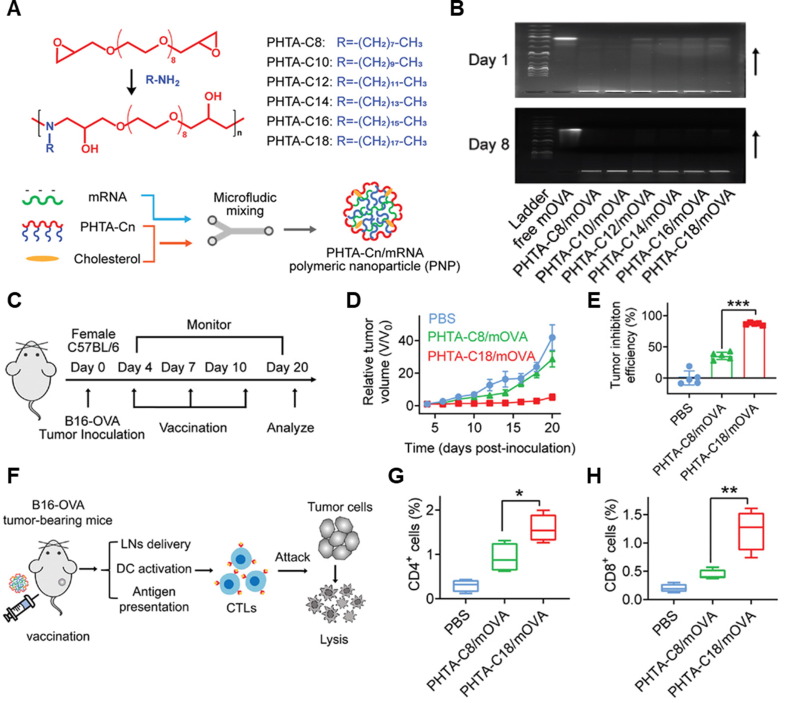
mRNA therapeutics that encoded genetic information of tumor-associated antigens (TAAs) can be utilized as a cancer vaccine. Relatively easy translation mechanism in the cytoplasm and transient expression of TAAs of mRNA cancer vaccines enables controlled antigen generation and repeated dosage, stimulating long-term immune responses effectively [179]. Early-stage mRNA cancer vaccine encodings melanoma-associated antigens, such as MAGE-A3, MAGE-C2, tyrosinase, and gp100, has shown tumor regression effects in melanoma patients by activating dendritic cells, CD4+ and CD8+ T-cells [180], [181]. Furthermore, mRNA therapeutics can be potent T-cell stimulators as well as immunosuppressive tumor microenvironment activators [182]. As described above, naked mRNA is biologically unstable and limited intracellular delivery efficiency due to negatively charged macromolecular structure. Thus, an efficient mRNA delivery strategy is important to achieve successful therapeutic effects for mRNA therapeutics [183]. M.A. Oberli et al. reported a lipid nanoparticle formulation of mRNA vaccines for eliciting CD8+ T-cell responses [36]. They prepared an LNP library with OVA-encoded mRNA for model antigen generation and optimized the vaccination effect using C57BL/6 mice by subcutaneous injection. The first optimization was conducted using different types of lipids (ionizable lipid, phospholipid, cholesterol, PEGylated lipid). Then, the molar compositions and combinations of the individual components, which were identified in the first optimization, were optimized. Under optimized composition of LNP, glycoprotein 100 (gp100) and a second tyrosinase-related protein (TRP-2)-encoded mRNA containing LNP showed tumor regression and increased survival rate of B16F10 melanoma model. These results showed that LNP formulations are promising non-viral vector for mRNA therapeutics-based eliciting T-cell response. LNPs are representative of a clinically advanced mRNA delivery vehicle but are among the limited options currently available. However, their utilization requires four intractable components, and often results in inflammatory side effects, limiting their therapeutic applications [184]. To address these issues, Huang et al. developed a low-inflammatory mRNA cancer vaccine using a series of alternating copolymers containing ortho-hydroxy tertiary amine (HTA) repeating unit called “PHTA” [185]. PHTA-Cn polymer, which was composed of a PEG backbone and hydrophobic alkyl side chain, was formulated with ovalbumin (OVA) encoding mRNA (mOVA) and cholesterols via microfluidic techniques (Fig. 5 A). PHTA-Cn/mOVA complexes showed nano-sized round shapes with effective encapsulation of mOVA. Furthermore, encapsulated mOVA was stable for 8 days, showing that the PHTA-Cn could inhibit mRNA degradation with the successful construction of PHTA-Cn/mOVA nanovaccines (Fig. 5B). After delivering PHTA Cn into mice through intradermal, intranasal, or intravenous injection, PHTA-Cn showed significantly low local- and systemic inflammatory reactions compared to typical LNP which was formulated with 1-octylnonyl 8-[(2hydroxyethyl) [6-oxo-6-(undecyloxy) hexyl] amino]-octanoate (SM-102), distearoyl phosphatidylcholine (DSPC), cholesterol, and 1,2-dimyristoyl-rac-glycero-3-methoxy poly (ethylene glycol)-2000 (DMG-PEG) at a molar ratio of 50:10:38.5:1.5. Among a series of PHTA-Cn, long alkyl side chains containing PHTA-C18 was selected as the most promising nanovaccine. The tumor growth inhibition effects of PHTA-C18/mOVA nanovaccine via subcutaneous injection were evaluated in the B16-OVA melanoma tumor-bearing mouse model, wherein PBS, PHTA-C8/mOVA were injected as controls (Fig. 5C). As a result, PHTA-C18/mOVA-treated mice showed a significant tumor growth inhibition effect compared to that of PBS- or PHTA-C8/mOVA-treated mice (Fig. 5D), showing 87 % (PHTA-C18/mOVA) and 36 % (PHTA-C8/mOVA) of tumor growth inhibition efficiency (Fig. 5E). To analyze the immune mechanisms relating the antitumor effect of nanovaccine, the tumor tissues were dissociated, and immune cells were analyzed (Fig. 5F). Consequently, PHTA-C18/mOVA-treated mice showed an increased population of maturated DCs, CD4+ T-cells, and CD8+ T-cells compared with the PBS and PHTA-C8/mOVA groups (Fig. 5G and 5H). Furthermore, high frequencies of central memory T-cells and effector memory T-cells in lymph nodes of PHTA-C18/mOVA-treated mice showed a successful generation of immune-memory effects, supporting that PHTA-C18/mOVA nanovaccine could elicit T-cell dependent antitumor immunity.
Fig. 5.
PHTA polymer-based mRNA cancer vaccine delivery system. (A) Synthetic scheme and preparation process of PHTA-Cn/mRNA nanovaccine. PHTA-Cn (n = 8, 10, 12, 14, 16, and 18) polymers were prepared via amino-epoxy polymerization. (B) Agarose gel electrophoresis images of PHTA-Cn/mRNA nanovaccine. (C) Therapeutic schedule of B16-OVA tumor-bearing mice using PHTA-C18/mRNA nanovaccine. (D) Tumor growth of PBS-, PHTA-C8/mRNA nanovaccine-, and PHTA-C18/mRNA nanovaccine-treated groups (n = 5 per group). (E) Tumor growth efficiency at the endpoint, compared with the PBS group (n = 5 per group). (F) Schematic illustration of therapeutic mechanism. Percentages of cells in B16-OVA tumor tissues on day 20: (G) CD4+ T-cells, and (H) CD8+ T-cells (n = 5 per group). Adapted with permission from reference [185].
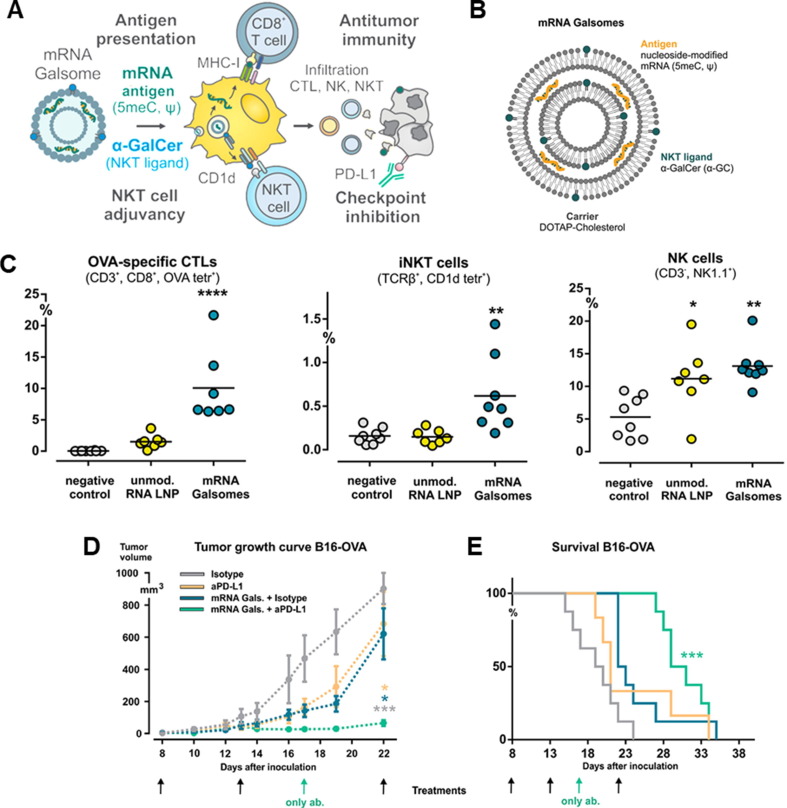
Recently, immune checkpoint pathway and immune-suppressive tumor microenvironment-related immune tolerance during tumor progression are major hurdles for cancer immunotherapy [186], [187]. To overcome this challenge, an alternative approach is to combine mRNA cancer vaccines with immune checkpoint inhibitors and/or adjuvants. Shi et al. developed adjuvant-pulsed mRNA vaccine nanoparticles which were composed of OVA-encoded mRNA and a palmitic acid-modified TLR7/8 agonist R848 (C16-R848), and lipid-polyethylene glycol (lipid-PEG) [188]. mRNA vaccine nanoparticles enhanced mRNA transfection efficiency (greater than95 %) and had adjuvant activity by C16-R848, resulting in mRNA-derived OVA antigen presentation on MHC class I of APCs. Furthermore, C16-R848 adjuvant-pulsed mRNA vaccine nanoparticles successfully activated OVA-specific CD8+ T-cells and significantly improved T-cell infiltration into tumor tissue, leading to anti-tumor immunity against OVA-expressing lymphoma and prostate cancer. Huang et al. reported MUC1 mRNA nanovaccine and CTLA-4 blockade combination immunotherapy of triple negative breast cancer (TNBC) [189]. MUC1-encoded mRNA was formulated with mannose-decorated Lipid/calcium/phosphate (LCP) nanoparticles to deliver TAA to DCs in lymph nodes. LCP-mRNA vaccine nanoparticle successfully delivered MUC1-mRNA to DCs through mannose receptor, expressing TAA in the DCs in lymph nodes and eliciting T-cell responses in 4 T1 tumor-bearing mice. In combination LCP-mRNA vaccine nanoparticle with anti-CTLA-4 (cytotoxic T-lymphocyte-associated protein-4) monoclonal antibody, anti-tumor immune responses were significantly improved in in 4 T1 tumor-bearing mice model. P. Huang et al. also developed an mRNA vaccine that could introduce an mRNA and immune checkpoint-specific siRNA into the APCs [190]. Mannose-functionalized LCP nanoparticles were formulated with TRP2-encoded mRNA and PD-L1-specific siRNA for eliciting robust antigen-specific T-cell responses and downregulating PD-L1 in the DCs, respectively. LCP mRNA vaccine nanoparticle successfully generated TRP2 as a TAA, inhibiting melanoma tumor growth in a B16F10 melanoma mouse model. Interestingly, co-delivery of PD-L1-specific siRNA with mRNA generated TAA and downregulated PD-L1 on DCs, enhancing T-cell responses with tumor growth inhibition effect. R. Verbeke et al. developed a nanovaccine formulated with nucleoside-modified mRNA and the glycolipid α-galactosylceramide (α-GC) for the antigen-specific T-cell responses by CD8+ T-cell, invariant natural killer T cell (iNKT cell), and natural killer cell (NK cell) activation (Fig. 6 A) [191]. The pseudouridine (Ψ) and 5-methylcytidine (5meC)-modified OVA-encoded mRNA was selected to minimize the immune-related toxicity of mRNA. Then, mRNA was formulated with 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP), cholesterol, and 1 mol % of α-GC, forming a 190 nm of nanovaccine, named mRNA Galsomes (Fig. 6B). α-GC in Galsomes is an immunopotentiator for antigen-presenting cells, enabling mRNA Galsomes to induce a pluripotent tumor-specific immune response. B16-OVA-bearing mice vaccinated twice using mRNA Galsomes showed 5 times higher CD8+ T-cells and significantly lower myeloid-derived suppressor cells and tumor-associated macrophages on day 14, compared to the untreated group. Likewise, OVA-specific CD8+ T-cell, iNKT cell, and NK cell in the tumor tissues were observed 7 times, 4 times, and 2.6 times higher than the untreated group, respectively (Fig. 6C). Based on the anti-tumor immune responses, mRNA Galsomes significantly inhibited tumor growth in the E. G7-OVA lymphoma mouse model, wherein 40 % of the tumor models showed complete tumor rejection. In addition, mRNA Galsomes showed effective therapeutic responsiveness and increased the overall survival rate in the B16-OVA melanoma mouse model in combination with an anti-PD-L1 antibody (Fig. 6D and 6E). These studies showed the potential of mRNA as a therapeutic cancer vaccine that can elicit antigen-specific immune responses and boost anti-tumor immune responses in combination with other immunotherapeutics.
Fig. 6.
Nanovaccine co-loaded with mRNA and α-galactosylceramide (α-GC) for eliciting antitumor immunity via activation of invariant natural killer T-cells (iNKT) and NK T-cells. (A) Therapeutic mechanism illustration of mRNA Galsomes. (B) Schematic illustration of mRNA Galsomes. Immune cell infiltration in B16-OVA tumor tissues on day 14, wherein B16-OVA tumor-bearing mice were two vaccinations with OVA mRNA Galsomes.: (C) Percentage of OVA-specific CD8+ T-cells, iNKT cells, and NK cells. (D) Tumor growth and (E) Survival rate of B16-OVA tumor-bearing mice in respective treatment groups. Black arrows indicate treatment with mRNA Galsomes and antibody via intravenous and intraperitoneal injection, respectively. Adapted with permission from reference [191].
Bispecific antibodies (bsAbs), binding between tumor antigen and T-cell surface molecule, offer a promising therapeutic strategy by addressing the limited response observed in current T-cell-based cancer immunotherapy, which is hampered by poor T-cell infiltration into tumor tissue [192]. However, the manufacturing of most bsAbs encounters challenges, including issues related to poor long-term stability, aggregates formation, and various impurities. Moreover, the short half-life of bsAbs in patients (less than 2 h) necessitates the use of continuous delivery system, such as an infusion pump, for effective treatment [193]. C.R. Stadler et al. proposed a method to generate bsAb (RiboMABs) directly in the patient’s body by transfecting chemically modified in vitro transcribed (IVT) mRNA. This mRNA modification aimed to prevent immune activation and enhance molecular stability. The mRNA was modified with 1-methylpseudouridine (m1Ψ) and poly(A) tail-containing mRNAs [194]. Singly dose intravenous injection of mRNA-loaded polymer/lipid-based particles (TransIT-mRNA Transfection kit) resulted the rapid production of bsAbs targeting CD3 and three TAAs (claudin 6, claudin 18.2, and EpCAM). Furthermore, this approach led to tumor elimination without any observed toxicities such as proinflammatory cytokine release, non-specific T-cell activation, or liver toxicity.
Chimeric antigen receptor T (CAR T) cell therapy, a highly effective form of personalized adoptive cell transfer (ATC) cancer immunotherapy, has emerged as a successful method in the clinical setting [195]. CARs are engineered to target specific TAAs in the receptor domain, while simultaneously engaging the T-cell receptor [196]. Recently, there has been a growing focus on mRNA electroporation-based expression of CARs on T-cells due to its cost- and time-efficiency, along with its ability to achieve successful antitumor activity compared to viral vector-based gene transduction. However, it is important to note that the mRNA electroporation-based CAR T system has potential limitations, including transient CAR expression and inefficient in vivo proliferation [197]. To achieve a potent antitumor effect, J.B. Foster et al. used modified IVT mRNA CAR T with pseudouridine and 1-methylpseudouridine (m1Ψ) [198]. These modifications were implemented to prevent immune stimulation and increase the stability of transfected mRNA. By utilizing RNase III-based purified double-strand RNA, the duration of mRNA transduction was prolonged. T cells treated with 1-methylpseudouridine (m1Ψ) and purified mRNA exhibited a significant increase in CD19 CAR expression on the cell surface, resulting in enhanced cytotoxicity against the leukemia cells up to 5 days. Furthermore, the leukemic burden in the leukemia mouse model was effectively suppressed by purified mRNA-electroporated T cells. However, it is important to note that despite the significant improvement in therapeutic outcomes with this approach, the risk of non-specific cleavage of single-strand RNA by RNase III remains a concern.
5. Conclusions and perspectives
mRNA is a promising therapeutic drug entity for treating and preventing various diseases, including cancer, due to its controllable and transient protein expression properties. Compared to DNA-based therapeutics, mRNA offers a relatively higher safety profile as it does not require localization to the nucleus and thereby avoid the risk of genomic mutation. However, RNA’s biological instability, immunogenicity, and low delivery efficiency due to negatively charged structure limits in cancer treatment. Recent advances in chemical modification strategies and non-viral delivery systems have addressed these challenges. The chemical modification of mRNA, including base, 5′-cap, and poly(A) tail modification, increase the biological stability and functionality of mRNA. Additionally, recent studies discovered naturally occurring mRNA modifications in the coding sequence region, offering opportunities for site-specific modification of mRNA using synthetic technology. However, considerable efforts to fully understand the biology of mRNA modifications are necessary to optimize the functionality and metabolism of mRNA-based therapeutics. Non-viral such as protamine, cationic lipids, polymers, exosomes, and gold nanoparticles, have improved the stability, pharmacokinetic properties, and delivery efficiency of mRNA, enabling further clinical therapeutic approaches. Nevertheless, challenges such as encapsulation efficiency, biocompatibility, transfection efficiency, target tissue or cell delivery efficiency, and toxicity remain to be optimized in mRNA/non-viral vector formulation. For example, mRNA/LNP formulation is representative strategy in current mRNA delivery but pharmacological properties including tissue-specific delivery, cellular uptake, endosomal escape with releasing of mRNA are remained complex issues and unoptimized stage. On the other hand, considerable efforts are needed to fully comprehend the diverse genomic information-based heterogenous tumor antigens, which are critical for anti-tumor immune responses and the development of effective mRNA-based cancer vaccines. Furthermore, combination therapy with mRNA-based cancer vaccines and other treatment regimes, including immune checkpoint therapy, chemotherapy, and radiotherapy, can improve therapeutic response and efficacy. For synergistic therapeutic efficacy, optimal dosage, treatment schedule, and administration route need to be optimized with the consideration of the different antigen presentation efficiency in different recipient cells during the treatment stage. Finally, toxicity and safety are major concerns for clinical translation of new mRNA-based cancer vaccines. It is important that synthetic mRNA and impurities avoid detection by the innate immune system. This is because the stimulation of the innate immune system can cause recognize mRNA therapeutics as foreign nucleic acid and lead to unwanted immune responses, limiting therapeutic efficacy with repeated treatment.
The mRNA has vast biological potential as a therapeutic tool for various applications, including cancer vaccines. Recent studies have demonstrated remarkable advancements in mRNA therapeutics for diverse therapeutic approaches such as cancer immunotherapy, vaccines, protein replacement, and gene editing. Despite certain biological- and pharmacological obstacles that need to be overcome for clinical applications, recent achievements such as COVID-19 mRNA vaccines and other mRNA therapeutics in the clinical stages highlight the potential of mRNA as a groundbreaking treatment for various diseases. We expect that continuous efforts towards innovation and optimization of mRNA technologies will lead to novel and effective outcomes in life science and medical research.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the Intramural Research Program of the Korea Institute of Science and Technology (KIST).
Data availability
No data was used for the research described in the article.
References
- 1.Khodaei T., Inamdar S., Suresh A.P., Acharya A.P. Drug delivery for metabolism targeted cancer immunotherapy. Adv. Drug Deliv. Rev. 2022;184 doi: 10.1016/j.addr.2022.114242. [DOI] [PubMed] [Google Scholar]
- 2.Kyu Shim M., Yang S., Sun I.-C., Kim K. Tumor-activated carrier-free prodrug nanoparticles for targeted cancer Immunotherapy: preclinical evidence for safe and effective drug delivery. Adv. Drug Deliv. Rev. 2022;183 doi: 10.1016/j.addr.2022.114177. [DOI] [PubMed] [Google Scholar]
- 3.Manspeaker M.P., Thomas S.N. Lymphatic immunomodulation using engineered drug delivery systems for cancer immunotherapy. Adv. Drug Deliv. Rev. 2020;160:19–35. doi: 10.1016/j.addr.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saxena M., van der Burg S.H., Melief C.J.M., Bhardwaj N. Therapeutic cancer vaccines. Nat. Rev. Cancer. 2021;21:360–378. doi: 10.1038/s41568-021-00346-0. [DOI] [PubMed] [Google Scholar]
- 5.Shemesh C.S., Hsu J.C., Hosseini I., Shen B.-Q., Rotte A., Twomey P., Girish S., Wu B. Personalized cancer vaccines: clinical landscape, challenges, and opportunities. Mol. Ther. 2021;29:555–570. doi: 10.1016/j.ymthe.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aikins M.E., Xu C., Moon J.J. Engineered nanoparticles for cancer vaccination and immunotherapy. Acc. Chem. Res. 2020;53:2094–2105. doi: 10.1021/acs.accounts.0c00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel R.B., Ye M., Carlson P.M., Jaquish A., Zangl L., Ma B., Wang Y., Arthur I., Xie R., Brown R.J. Development of an in situ cancer vaccine via combinational radiation and bacterial-membrane-coated nanoparticles. Adv. Mater. 2019;31:1902626. doi: 10.1002/adma.201902626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniel S., Chen I. Mellman, oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 9.Donninger H., Li C., Eaton J.W., Yaddanapudi K. Cancer vaccines: promising therapeutics or an unattainable dream. Vaccines. 2021;9:668. doi: 10.3390/vaccines9060668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morse M.A., Crosby E.J., Force J., Osada T., Hobeika A.C., Hartman Z.C., Berglund P., Smith J., Lyerly H.K. Clinical trials of self-replicating RNA-based cancer vaccines. Cancer Gene Ther. 2023;30:803–811. doi: 10.1038/s41417-023-00587-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lorentzen C.L., Haanen J.B., Met Ö., Svane I.M. Clinical advances and ongoing trials on mRNA vaccines for cancer treatment. Lancet Oncol. 2022;23:e450–e458. doi: 10.1016/S1470-2045(22)00372-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowen W.S., Svrivastava A.K., Batra L., Barsoumian H., Shirwan H. Current challenges for cancer vaccine adjuvant development. Expert Rev. Vaccines. 2018;17:207–215. doi: 10.1080/14760584.2018.1434000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang X., Kong N., Zhang X., Cao Y., Langer R., Tao W. The landscape of mRNA nanomedicine. Nat. Med. 2022;28:2273–2287. doi: 10.1038/s41591-022-02061-1. [DOI] [PubMed] [Google Scholar]
- 14.Barbier A.J., Jiang A.Y., Zhang P., Wooster R., Anderson D.G. The clinical progress of mRNA vaccines and immunotherapies. Nat. Biotechnol. 2022;40:840–854. doi: 10.1038/s41587-022-01294-2. [DOI] [PubMed] [Google Scholar]
- 15.Mascola J.R., Fauci A.S. Novel vaccine technologies for the 21st century. Nat. Rev. Immunol. 2020;20:87–88. doi: 10.1038/s41577-019-0243-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Billingsley M.M., Singh N., Ravikumar P., Zhang R., June C.H., Mitchell M.J. Ionizable lipid nanoparticle-mediated mRNA delivery for human CAR T cell engineering. Nano Lett. 2020;20:1578–1589. doi: 10.1021/acs.nanolett.9b04246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics — developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 18.Rohner E., Yang R., Foo K.S., Goedel A., Chien K.R. Unlocking the promise of mRNA therapeutics. Nat. Biotechnol. 2022;40:1586–1600. doi: 10.1038/s41587-022-01491-z. [DOI] [PubMed] [Google Scholar]
- 19.S. Ramaswamy, N. Tonnu, K. Tachikawa, P. Limphong, J.B. Vega, P.P. Karmali, P. Chivukula, I.M. Verma, Systemic delivery of factor IX messenger RNA for protein replacement therapy, Proc. Natl. Acad. Sci. 114 (2017) E1941–E1950. [DOI] [PMC free article] [PubMed]
- 20.Khalil A.S., Yu X., Umhoefer J.M., Chamberlain C.S., Wildenauer L.A., Diarra G.M., Hacker T.A., Murphy W.L. Single-dose mRNA therapy via biomaterial-mediated sequestration of overexpressed proteins. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aba2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linares-Fernández S., Lacroix C., Exposito J.-Y., Verrier B. Tailoring mRNA vaccine to balance innate/adaptive immune response. Trends Mol. Med. 2020;26:311–323. doi: 10.1016/j.molmed.2019.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Kranz L.M., Diken M., Haas H., Kreiter S., Loquai C., Reuter K.C., Meng M., Fritz D., Vascotto F., Hefesha H. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 23.De Beuckelaer A., Grooten J., De Koker S. Type I interferons modulate CD8+ T cell immunity to mRNA vaccines. Trends Mol. Med. 2017;23:216–226. doi: 10.1016/j.molmed.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 24.Kobiyama K., Ishii K.J. Making innate sense of mRNA vaccine adjuvanticity. Nat. Immunol. 2022;23:474–476. doi: 10.1038/s41590-022-01168-4. [DOI] [PubMed] [Google Scholar]
- 25.Miao L., Zhang Y., Huang L. mRNA vaccine for cancer immunotherapy. Mol. Cancer. 2021;20:41. doi: 10.1186/s12943-021-01335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jahanafrooz Z., Baradaran B., Mosafer J., Hashemzaei M., Rezaei T., Mokhtarzadeh A., Hamblin M.R. Comparison of DNA and mRNA vaccines against cancer. Drug Discov. Today. 2020;25:552–560. doi: 10.1016/j.drudis.2019.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cafri G., Gartner J.J., Zaks T., Hopson K., Levin N., Paria B.C., Parkhurst M.R., Yossef R., Lowery F.J., Jafferji M.S. mRNA vaccine–induced neoantigen-specific T cell immunity in patients with gastrointestinal cancer. J. Clin. Invest. 2020;130:5976–5988. doi: 10.1172/JCI134915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Son S., Nam J., Zenkov I., Ochyl L.J., Xu Y., Scheetz L., Shi J., Farokhzad O.C., Moon J.J. Sugar-nanocapsules imprinted with microbial molecular patterns for mRNA vaccination. Nano Lett. 2020;20:1499–1509. doi: 10.1021/acs.nanolett.9b03483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hajj K.A., Whitehead K.A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017;2:1–17. [Google Scholar]
- 30.Chen Q., Zhang Y., Yin H. Recent advances in chemical modifications of guide RNA, mRNA and donor template for CRISPR-mediated genome editing. Adv. Drug Deliv. Rev. 2021;168:246–258. doi: 10.1016/j.addr.2020.10.014. [DOI] [PubMed] [Google Scholar]
- 31.Kwon H., Kim M., Seo Y., Moon Y.S., Lee H.J., Lee K., Lee H. Emergence of synthetic mRNA: In vitro synthesis of mRNA and its applications in regenerative medicine. Biomaterials. 2018;156:172–193. doi: 10.1016/j.biomaterials.2017.11.034. [DOI] [PubMed] [Google Scholar]
- 32.Weide B., Pascolo S., Scheel B., Derhovanessian E., Pflugfelder A., Eigentler T.K., Pawelec G., Hoerr I., Rammensee H.-G., Garbe C. Direct injection of protamine-protected mRNA: results of a phase 1/2 vaccination trial in metastatic melanoma patients. J. Immunother. 2009;32:498–507. doi: 10.1097/CJI.0b013e3181a00068. [DOI] [PubMed] [Google Scholar]
- 33.Mai Y., Guo J., Zhao Y., Ma S., Hou Y., Yang J. Intranasal delivery of cationic liposome-protamine complex mRNA vaccine elicits effective anti-tumor immunity. Cell. Immunol. 2020;354 doi: 10.1016/j.cellimm.2020.104143. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X., Men K., Zhang Y., Zhang R., Yang L., Duan X. Local and systemic delivery of mRNA encoding survivin-T34A by lipoplex for efficient colon cancer gene therapy. Int. J. Nanomed. 2019;14:2733–2751. doi: 10.2147/IJN.S198747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lei S., Zhang X., Men K., Gao Y., Yang X., Wu S., Duan X., Wei Y., Tong R. Efficient colorectal cancer gene therapy with IL-15 mRNA nanoformulation. Mol. Pharm. 2020;17:3378–3391. doi: 10.1021/acs.molpharmaceut.0c00451. [DOI] [PubMed] [Google Scholar]
- 36.Oberli M.A., Reichmuth A.M., Dorkin J.R., Mitchell M.J., Fenton O.S., Jaklenec A., Anderson D.G., Langer R., Blankschtein D. Lipid nanoparticle assisted mRNA delivery for potent cancer immunotherapy. Nano Lett. 2017;17:1326–1335. doi: 10.1021/acs.nanolett.6b03329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang W., Mixich L., Boonstra E., Cabral H. Polymer-based mRNA delivery strategies for advanced therapies. Adv. Healthc. Mater. 2023;12:2202688. doi: 10.1002/adhm.202202688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilbert W.V., Bell T.A., Schaening C. Messenger RNA modifications: form, distribution, and function. Science. 2016;352:1408–1412. doi: 10.1126/science.aad8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaccara S., Ries R.J., Jaffrey S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019;20:608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 40.Boo S.H., Kim Y.K. The emerging role of RNA modifications in the regulation of mRNA stability. Exp. Mol. Med. 2020;52:400–408. doi: 10.1038/s12276-020-0407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fisher A.J., Beal P.A. Structural basis for eukaryotic mRNA modification. Curr. Opin. Struct. Biol. 2018;53:59–68. doi: 10.1016/j.sbi.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 42.Jaffrey S.R. An expanding universe of mRNA modifications. Nat. Struct. Mol. Biol. 2014;21:945–946. doi: 10.1038/nsmb.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harcourt E.M., Kietrys A.M., Kool E.T. Chemical and structural effects of base modifications in messenger RNA. Nature. 2017;541:339–346. doi: 10.1038/nature21351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rengaraj P., Obrdlik A., Vukic D., Varadarajan N.M., Keegan L.P., Vanacova S., O'Connell M.A. Interplays of different types of epitranscriptomic mRNA modifications. RNA Biol. 2021;18:19–30. doi: 10.1080/15476286.2021.1969113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin Z., Tong M.H. m(6)A mRNA modification regulates mammalian spermatogenesis. Biochim. Biophys. Acta. 1862;2019:403–411. doi: 10.1016/j.bbagrm.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 46.Cui Q., Shi H., Ye P., Li L., Qu Q., Sun G., Sun G., Lu Z., Huang Y., Yang C.G., Riggs A.D., He C., Shi Y. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han J., Wang J.Z., Yang X., Yu H., Zhou R., Lu H.C., Yuan W.B., Lu J.C., Zhou Z.J., Lu Q., Wei J.F., Yang H. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer. 2019;18:110. doi: 10.1186/s12943-019-1036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma J.Z., Yang F., Zhou C.C., Liu F., Yuan J.H., Wang F., Wang T.T., Xu Q.G., Zhou W.P., Sun S.H. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65:529–543. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 49.Gao X.Q., Zhang Y.H., Liu F., Ponnusamy M., Zhao X.M., Zhou L.Y., Zhai M., Liu C.Y., Li X.M., Wang M., Shan C., Shan P.P., Wang Y., Dong Y.H., Qian L.L., Yu T., Ju J., Wang T., Wang K., Chen X.Z., Wang Y.H., Zhang J., Li P.F., Wang K. The piRNA CHAPIR regulates cardiac hypertrophy by controlling METTL3-dependent N(6)-methyladenosine methylation of Parp10 mRNA. Nat. Cell Biol. 2020;22:1319–1331. doi: 10.1038/s41556-020-0576-y. [DOI] [PubMed] [Google Scholar]
- 50.Mathiyalagan P., Adamiak M., Mayourian J., Sassi Y., Liang Y., Agarwal N., Jha D., Zhang S., Kohlbrenner E., Chepurko E., Chen J., Trivieri M.G., Singh R., Bouchareb R., Fish K., Ishikawa K., Lebeche D., Hajjar R.J., Sahoo S. FTO-dependent N(6)-methyladenosine regulates cardiac function during remodeling and repair. Circulation. 2019;139:518–532. doi: 10.1161/CIRCULATIONAHA.118.033794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao B.S., Roundtree I.A., He C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X., Xiong X., Wang K., Wang L., Shu X., Ma S., Yi C. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat. Chem. Biol. 2016;12:311–316. doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- 53.Dominissini D., Nachtergaele S., Moshitch-Moshkovitz S., Peer E., Kol N., Ben-Haim M.S., Dai Q., Di Segni A., Salmon-Divon M., Clark W.C., Zheng G., Pan T., Solomon O., Eyal E., Hershkovitz V., Han D., Dore L.C., Amariglio N., Rechavi G., He C. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441–446. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Legrand C., Tuorto F., Hartmann M., Liebers R., Jacob D., Helm M., Lyko F. Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs. Genome Res. 2017;27:1589–1596. doi: 10.1101/gr.210666.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aas P.A., Otterlei M., Falnes P.O., Vagbo C.B., Skorpen F., Akbari M., Sundheim O., Bjoras M., Slupphaug G., Seeberg E., Krokan H.E. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 56.Woo H.H., Chambers S.K. Human ALKBH3-induced m(1)A demethylation increases the CSF-1 mRNA stability in breast and ovarian cancer cells. Biochim. Biophys. Acta. 1862;2019:35–46. doi: 10.1016/j.bbagrm.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 57.Kuang W., Jin H., Yang F., Chen X., Liu J., Li T., Chang Y., Liu M., Xu Z., Huo C., Yan X., Yang Y., Liu W., Shu Q., Xie S., Zhou T. ALKBH3-dependent m(1)A demethylation of Aurora A mRNA inhibits ciliogenesis. Cell Discov. 2022;8:25. doi: 10.1038/s41421-022-00385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kouzarides T., Pandolfini L., Barbieri I., Bannister A.J., Andrews B. Further evidence supporting N7-methylation of guanosine (m(7)G) in human microRNAs. Mol. Cell. 2020;79:201–202. doi: 10.1016/j.molcel.2020.05.023. [DOI] [PubMed] [Google Scholar]
- 59.Furuichi Y. Discovery of m(7)G-cap in eukaryotic mRNAs. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2015;91:394–409. doi: 10.2183/pjab.91.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alexandrov A., Martzen M.R., Phizicky E.M. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA. 2002;8:1253–1266. doi: 10.1017/s1355838202024019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zueva V.S., Mankin A.S., Bogdanov A.A., Baratova L.A. Specific fragmentation of tRNA and rRNA at a 7-methylguanine residue in the presence of methylated carrier RNA. Eur. J. Biochem. 1985;146:679–687. doi: 10.1111/j.1432-1033.1985.tb08704.x. [DOI] [PubMed] [Google Scholar]
- 62.Muthukrishnan S., Both G.W., Furuichi Y., Shatkin A.J. 5'-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature. 1975;255:33–37. doi: 10.1038/255033a0. [DOI] [PubMed] [Google Scholar]
- 63.Luo Y., Yao Y., Wu P., Zi X., Sun N., He J. The potential role of N(7)-methylguanosine (m7G) in cancer. J. Hematol. Oncol. 2022;15:63. doi: 10.1186/s13045-022-01285-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang L.S., Liu C., Ma H., Dai Q., Sun H.L., Luo G., Zhang Z., Zhang L., Hu L., Dong X., He C. Transcriptome-wide mapping of internal N(7)-methylguanosine methylome in mammalian mRNA. Mol. Cell. 2019;74:1304–1316 e1308. doi: 10.1016/j.molcel.2019.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao Y., Kong L., Pei Z., Li F., Li C., Sun X., Shi B., Ge J. m7G methyltransferase METTL1 promotes post-ischemic angiogenesis via promoting VEGFA mRNA translation. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.642080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gonatopoulos-Pournatzis T., Dunn S., Bounds R., Cowling V.H. RAM/Fam103a1 is required for mRNA cap methylation. Mol. Cell. 2011;44:585–596. doi: 10.1016/j.molcel.2011.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cowling V.H. Enhanced mRNA cap methylation increases cyclin D1 expression and promotes cell transformation. Oncogene. 2010;29:930–936. doi: 10.1038/onc.2009.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chan C.T., Pang Y.L., Deng W., Babu I.R., Dyavaiah M., Begley T.J., Dedon P.C. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 2012;3:937. doi: 10.1038/ncomms1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen Y., Sierzputowska-Gracz H., Guenther R., Everett K., Agris P.F. 5-Methylcytidine is required for cooperative binding of Mg2+ and a conformational transition at the anticodon stem-loop of yeast phenylalanine tRNA. Biochemistry. 1993;32:10249–10253. doi: 10.1021/bi00089a047. [DOI] [PubMed] [Google Scholar]
- 70.Yang X., Yang Y., Sun B.F., Chen Y.S., Xu J.W., Lai W.Y., Li A., Wang X., Bhattarai D.P., Xiao W., Sun H.Y., Zhu Q., Ma H.L., Adhikari S., Sun M., Hao Y.J., Zhang B., Huang C.M., Huang N., Jiang G.B., Zhao Y.L., Wang H.L., Sun Y.P., Yang Y.G. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27:606–625. doi: 10.1038/cr.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Q., Li X., Tang H., Jiang B., Dou Y., Gorospe M., Wang W. NSUN2-mediated m5C methylation and METTL3/METTL14-mediated m6A methylation cooperatively enhance p21 translation. J. Cell. Biochem. 2017;118:2587–2598. doi: 10.1002/jcb.25957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xing J., Yi J., Cai X., Tang H., Liu Z., Zhang X., Martindale J.L., Yang X., Jiang B., Gorospe M., Wang W. NSun2 promotes cell growth via elevating cyclin-dependent kinase 1 translation. Mol. Cell Biol. 2015;35:4043–4052. doi: 10.1128/MCB.00742-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang N., Tang H., Wang X., Wang W., Feng J. Homocysteine upregulates interleukin-17A expression via NSun2-mediated RNA methylation in T lymphocytes. Biochem. Biophys. Res. Commun. 2017;493:94–99. doi: 10.1016/j.bbrc.2017.09.069. [DOI] [PubMed] [Google Scholar]
- 74.Charette M., Gray M.W. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 75.Schwartz S., Bernstein D.A., Mumbach M.R., Jovanovic M., Herbst R.H., Leon-Ricardo B.X., Engreitz J.M., Guttman M., Satija R., Lander E.S., Fink G., Regev A. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Carlile T.M., Rojas-Duran M.F., Zinshteyn B., Shin H., Bartoli K.M., Gilbert W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karijolich J., Yu Y.T. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature. 2011;474:395–398. doi: 10.1038/nature10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu G., Huang C., Yu Y.T. Pseudouridine in mRNA: incorporation, detection, and recoding. Methods Enzymol. 2015;560:187–217. doi: 10.1016/bs.mie.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kariko K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen C., Zhao X., Kierzek R., Yu Y.T. A flexible RNA backbone within the polypyrimidine tract is required for U2AF65 binding and pre-mRNA splicing in vivo. Mol. Cell Biol. 2010;30:4108–4119. doi: 10.1128/MCB.00531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Eyler D.E., Franco M.K., Batool Z., Wu M.Z., Dubuke M.L., Dobosz-Bartoszek M., Jones J.D., Polikanov Y.S., Roy B., Koutmou K.S. Pseudouridinylation of mRNA coding sequences alters translation. PNAS. 2019;116:23068–23074. doi: 10.1073/pnas.1821754116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., Kariko K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hoernes T.P., Clementi N., Faserl K., Glasner H., Breuker K., Lindner H., Huttenhofer A., Erlacher M.D. Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res. 2016;44:852–862. doi: 10.1093/nar/gkv1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moradian H., Roch T., Anthofer L., Lendlein A., Gossen M. Chemical modification of uridine modulates mRNA-mediated proinflammatory and antiviral response in primary human macrophages. Mol. Ther. Nucleic Acids. 2022;27:854–869. doi: 10.1016/j.omtn.2022.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ramanathan A., Robb G.B., Chan S.H. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016;44:7511–7526. doi: 10.1093/nar/gkw551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gallie D.R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 87.Munroe D., Jacobson A. mRNA poly(A) tail, a 3' enhancer of translational initiation. Mol. Cell Biol. 1990;10:3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moteki S., Price D. Functional coupling of capping and transcription of mRNA. Mol. Cell. 2002;10:599–609. doi: 10.1016/s1097-2765(02)00660-3. [DOI] [PubMed] [Google Scholar]
- 89.Cowling V.H. Regulation of mRNA cap methylation. Biochem. J. 2009;425:295–302. doi: 10.1042/BJ20091352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kyrieleis O.J., Chang J., de la Pena M., Shuman S., Cusack S. Crystal structure of vaccinia virus mRNA capping enzyme provides insights into the mechanism and evolution of the capping apparatus. Structure. 2014;22:452–465. doi: 10.1016/j.str.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Muttach F., Muthmann N., Rentmeister A. Synthetic mRNA capping. Beilstein J. Org. Chem. 2017;13:2819–2832. doi: 10.3762/bjoc.13.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mauer J., Luo X., Blanjoie A., Jiao X., Grozhik A.V., Patil D.P., Linder B., Pickering B.F., Vasseur J.J., Chen Q., Gross S.S., Elemento O., Debart F., Kiledjian M., Jaffrey S.R. Reversible methylation of m(6)A(m) in the 5' cap controls mRNA stability. Nature. 2017;541:371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shanmugasundaram M., Charles I., Kore A.R. Design, synthesis and biological evaluation of dinucleotide mRNA cap analog containing propargyl moiety. Bioorg. Med. Chem. 2016;24:1204–1208. doi: 10.1016/j.bmc.2016.01.048. [DOI] [PubMed] [Google Scholar]
- 94.Vaidyanathan S., Azizian K.T., Haque A., Henderson J.M., Hendel A., Shore S., Antony J.S., Hogrefe R.I., Kormann M.S.D., Porteus M.H., McCaffrey A.P. Uridine depletion and chemical modification increase Cas9 mRNA activity and reduce immunogenicity without HPLC purification. Mol. Ther. Nucleic Acids. 2018;12:530–542. doi: 10.1016/j.omtn.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kwon S., Kwon M., Im S., Lee K., Lee H. mRNA vaccines: the most recent clinical applications of synthetic mRNA. Arch. Pharm. Res. 2022;45:245–262. doi: 10.1007/s12272-022-01381-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shanmugasundaram M., Senthilvelan A., Kore A.R. Recent advances in modified cap analogs: synthesis, biochemical properties, and mRNA based vaccines. Chem. Rec. 2022;22 doi: 10.1002/tcr.202200005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Senthilvelan A., Vonderfecht T., Shanmugasundaram M., Pal I., Potter J., Kore A.R. Trinucleotide cap analogue bearing a locked nucleic acid moiety: synthesis, mRNA modification, and translation for therapeutic applications. Org. Lett. 2021;23:4133–4136. doi: 10.1021/acs.orglett.1c01037. [DOI] [PubMed] [Google Scholar]
- 98.van Dulmen M., Muthmann N., Rentmeister A. Chemo-enzymatic modification of the 5' cap maintains translation and increases immunogenic properties of mRNA. Angew. Chem. Int. Ed. Engl. 2021;60:13280–13286. doi: 10.1002/anie.202100352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Strzelecka D., Smietanski M., Sikorski P.J., Warminski M., Kowalska J., Jemielity J. Phosphodiester modifications in mRNA poly(A) tail prevent deadenylation without compromising protein expression. RNA. 2020;26:1815–1837. doi: 10.1261/rna.077099.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Anhauser L., Huwel S., Zobel T., Rentmeister A. Multiple covalent fluorescence labeling of eukaryotic mRNA at the poly(A) tail enhances translation and can be performed in living cells. Nucleic Acids Res. 2019;47:e42. doi: 10.1093/nar/gkz084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wadhwa A., Aljabbari A., Lokras A., Foged C., Thakur A. Opportunities and challenges in the delivery of mRNA-based vaccines. Pharmaceutics. 2020;12:102. doi: 10.3390/pharmaceutics12020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shirley J.L., de Jong Y.P., Terhorst C., Herzog R.W. Immune responses to viral gene therapy vectors. Mol. Ther. 2020;28:709–722. doi: 10.1016/j.ymthe.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bangham A.D., Standish M.M., Watkins J.C. Diffusion of univalent ions across the lamellae of swollen phospholipids. J. Mol. Biol. 1965;13:238–252. doi: 10.1016/s0022-2836(65)80093-6. [DOI] [PubMed] [Google Scholar]
- 104.Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021;6:1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kranz L.M., Diken M., Haas H., Kreiter S., Loquai C., Reuter K.C., Meng M., Fritz D., Vascotto F., Hefesha H., Grunwitz C., Vormehr M., Husemann Y., Selmi A., Kuhn A.N., Buck J., Derhovanessian E., Rae R., Attig S., Diekmann J., Jabulowsky R.A., Heesch S., Hassel J., Langguth P., Grabbe S., Huber C., Tureci O., Sahin U. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 106.Grunwitz C., Salomon N., Vascotto F., Selmi A., Bukur T., Diken M., Kreiter S., Tureci O., Sahin U. HPV16 RNA-LPX vaccine mediates complete regression of aggressively growing HPV-positive mouse tumors and establishes protective T cell memory. Oncoimmunology. 2019;8:e1629259. doi: 10.1080/2162402X.2019.1629259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang F., Xiao W., Elbahnasawy M.A., Bao X., Zheng Q., Gong L., Zhou Y., Yang S., Fang A., Farag M.M.S., Wu J., Song X. Optimization of the linker length of mannose-cholesterol conjugates for enhanced mRNA delivery to dendritic cells by liposomes. Front. Pharmacol. 2018;9:980. doi: 10.3389/fphar.2018.00980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Persano S., Guevara M.L., Li Z., Mai J., Ferrari M., Pompa P.P., Shen H. Lipopolyplex potentiates anti-tumor immunity of mRNA-based vaccination. Biomaterials. 2017;125:81–89. doi: 10.1016/j.biomaterials.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kapoor R., Peyear T.A., Koeppe R.E., 2nd, Andersen O.S. Antidepressants are modifiers of lipid bilayer properties. J. Gen. Physiol. 2019;151:342–356. doi: 10.1085/jgp.201812263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang J., Shrivastava S., Cleveland R.O., Rabbitts T.H. Lipid-mRNA nanoparticle designed to enhance intracellular delivery mediated by shock waves. ACS Appl. Mater. Interfaces. 2019;11:10481–10491. doi: 10.1021/acsami.8b21398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pattipeiluhu R., Arias-Alpizar G., Basha G., Chan K.Y.T., Bussmann J., Sharp T.H., Moradi M.A., Sommerdijk N., Harris E.N., Cullis P.R., Kros A., Witzigmann D., Campbell F. Anionic lipid nanoparticles preferentially deliver mRNA to the hepatic reticuloendothelial system. Adv. Mater. 2022;34:e2201095. doi: 10.1002/adma.202201095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Campbell F., Bos F.L., Sieber S., Arias-Alpizar G., Koch B.E., Huwyler J., Kros A., Bussmann J. Directing nanoparticle biodistribution through evasion and exploitation of Stab2-dependent nanoparticle uptake. ACS Nano. 2018;12:2138–2150. doi: 10.1021/acsnano.7b06995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cheng Q., Wei T., Farbiak L., Johnson L.T., Dilliard S.A., Siegwart D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol. Ther. 2019;27:710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Akinc A., Maier M.A., Manoharan M., Fitzgerald K., Jayaraman M., Barros S., Ansell S., Du X., Hope M.J., Madden T.D., Mui B.L., Semple S.C., Tam Y.K., Ciufolini M., Witzigmann D., Kulkarni J.A., van der Meel R., Cullis P.R. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019;14:1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 116.Riley R.S., Kashyap M.V., Billingsley M.M., White B., Alameh M.G., Bose S.K., Zoltick P.W., Li H., Zhang R., Cheng A.Y., Weissman D., Peranteau W.H., Mitchell M.J. Ionizable lipid nanoparticles for in utero mRNA delivery. Sci. Adv. 2021;7 doi: 10.1126/sciadv.aba1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ryals R.C., Patel S., Acosta C., McKinney M., Pennesi M.E., Sahay G. The effects of PEGylation on LNP based mRNA delivery to the eye. PLoS ONE. 2020;15 doi: 10.1371/journal.pone.0241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Robinson E., MacDonald K.D., Slaughter K., McKinney M., Patel S., Sun C., Sahay G. Lipid nanoparticle-delivered chemically modified mRNA restores chloride secretion in cystic fibrosis. Mol. Ther. 2018;26:2034–2046. doi: 10.1016/j.ymthe.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ramaswamy S., Tonnu N., Tachikawa K., Limphong P., Vega J.B., Karmali P.P., Chivukula P., Verma I.M. Systemic delivery of factor IX messenger RNA for protein replacement therapy. PNAS. 2017;114:E1941–E1950. doi: 10.1073/pnas.1619653114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Patel S., Ryals R.C., Weller K.K., Pennesi M.E., Sahay G. Lipid nanoparticles for delivery of messenger RNA to the back of the eye. J. Control. Release. 2019;303:91–100. doi: 10.1016/j.jconrel.2019.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bahl K., Senn J.J., Yuzhakov O., Bulychev A., Brito L.A., Hassett K.J., Laska M.E., Smith M., Almarsson O., Thompson J., Ribeiro A.M., Watson M., Zaks T., Ciaramella G. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol. Ther. 2017;25:1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C., Ciaramella G., Diamond M.S. Modified mRNA vaccines protect against zika virus infection. Cell. 2017;168:1114–1125 e1110. doi: 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Falsey A.R., Sobieszczyk M.E., Hirsch I., Sproule S., Robb M.L., Corey L., Neuzil K.M., Hahn W., Hunt J., Mulligan M.J., McEvoy C., DeJesus E., Hassman M., Little S.J., Pahud B.A., Durbin A., Pickrell P., Daar E.S., Bush L., Solis J., Carr Q.O., Oyedele T., Buchbinder S., Cowden J., Vargas S.L., Guerreros Benavides A., Call R., Keefer M.C., Kirkpatrick B.D., Pullman J., Tong T., Brewinski Isaacs M., Benkeser D., Janes H.E., Nason M.C., Green J.A., Kelly E.J., Maaske J., Mueller N., Shoemaker K., Takas T., Marshall R.P., Pangalos M.N., Villafana T., Gonzalez-Lopez A. Phase 3 safety and efficacy of AZD1222 (ChAdOx1 nCoV-19) Covid-19 vaccine. N. Engl. J. Med. 2021;385:2348–2360. doi: 10.1056/NEJMoa2105290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.A.B. Vogel, I. Kanevsky, Y. Che, K.A. Swanson, A. Muik, M. Vormehr, L.M. Kranz, K.C. Walzer, S. Hein, A. Guler, J. Loschko, M.S. Maddur, A. Ota-Setlik, K. Tompkins, J. Cole, B.G. Lui, T. Ziegenhals, A. Plaschke, D. Eisel, S.C. Dany, S. Fesser, S. Erbar, F. Bates, D. Schneider, B. Jesionek, B. Sanger, A.K. Wallisch, Y. Feuchter, H. Junginger, S.A. Krumm, A.P. Heinen, P. Adams-Quack, J. Schlereth, S. Schille, C. Kroner, R. de la Caridad Guimil Garcia, T. Hiller, L. Fischer, R.S. Sellers, S. Choudhary, O. Gonzalez, F. Vascotto, M.R. Gutman, J.A. Fontenot, S. Hall-Ursone, K. Brasky, M.C. Griffor, S. Han, A.A.H. Su, J.A. Lees, N.L. Nedoma, E.H. Mashalidis, P.V. Sahasrabudhe, C.Y. Tan, D. Pavliakova, G. Singh, C. Fontes-Garfias, M. Pride, I.L. Scully, T. Ciolino, J. Obregon, M. Gazi, R. Carrion, Jr., K.J. Alfson, W.V. Kalina, D. Kaushal, P.Y. Shi, T. Klamp, C. Rosenbaum, A.N. Kuhn, O. Tureci, P.R. Dormitzer, K.U. Jansen, U. Sahin, BNT162b vaccines protect rhesus macaques from SARS-CoV-2, Nature 592 (2021) 283–289. [DOI] [PubMed]
- 125.Wu K., Werner A.P., Koch M., Choi A., Narayanan E., Stewart-Jones G.B.E., Colpitts T., Bennett H., Boyoglu-Barnum S., Shi W., Moliva J.I., Sullivan N.J., Graham B.S., Carfi A., Corbett K.S., Seder R.A., Edwards D.K. Serum neutralizing activity elicited by mRNA-1273 vaccine. N. Engl. J. Med. 2021;384:1468–1470. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Muik A., Wallisch A.K., Sanger B., Swanson K.A., Muhl J., Chen W., Cai H., Maurus D., Sarkar R., Tureci O., Dormitzer P.R., Sahin U. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021;371:1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fenton O.S., Kauffman K.J., McClellan R.L., Appel E.A., Dorkin J.R., Tibbitt M.W., Heartlein M.W., DeRosa F., Langer R., Anderson D.G. Bioinspired alkenyl amino alcohol ionizable lipid materials for highly potent in vivo mRNA delivery. Adv. Mater. 2016;28:2939–2943. doi: 10.1002/adma.201505822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Miao L., Li L., Huang Y., Delcassian D., Chahal J., Han J., Shi Y., Sadtler K., Gao W., Lin J., Doloff J.C., Langer R., Anderson D.G. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat. Biotechnol. 2019;37:1174–1185. doi: 10.1038/s41587-019-0247-3. [DOI] [PubMed] [Google Scholar]
- 129.Lv H., Zhang S., Wang B., Cui S., Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 130.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T., Mihai C., Lynn A., McFadyen I., Moore M.J., Senn J.J., Stanton M.G., Almarsson O., Ciaramella G., Brito L.A. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sabnis S., Kumarasinghe E.S., Salerno T., Mihai C., Ketova T., Senn J.J., Lynn A., Bulychev A., McFadyen I., Chan J., Almarsson O., Stanton M.G., Benenato K.E. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang X., Niu D., Hu C., Li P. Polyethyleneimine-based nanocarriers for gene delivery. Curr. Pharm. Des. 2015;21:6140–6156. doi: 10.2174/1381612821666151027152907. [DOI] [PubMed] [Google Scholar]
- 133.Choi H.Y., Lee T.J., Yang G.M., Oh J., Won J., Han J., Jeong G.J., Kim J., Kim J.H., Kim B.S., Cho S.G. Efficient mRNA delivery with graphene oxide-polyethylenimine for generation of footprint-free human induced pluripotent stem cells. J. Control. Release. 2016;235:222–235. doi: 10.1016/j.jconrel.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 134.Chong Y., Ge C., Yang Z., Garate J.A., Gu Z., Weber J.K., Liu J., Zhou R. Reduced cytotoxicity of graphene nanosheets mediated by blood-protein coating. ACS Nano. 2015;9:5713–5724. doi: 10.1021/nn5066606. [DOI] [PubMed] [Google Scholar]
- 135.Krhac Levacic A., Berger S., Muller J., Wegner A., Lachelt U., Dohmen C., Rudolph C., Wagner E. Dynamic mRNA polyplexes benefit from bioreducible cleavage sites for in vitro and in vivo transfer. J. Control. Release. 2021;339:27–40. doi: 10.1016/j.jconrel.2021.09.016. [DOI] [PubMed] [Google Scholar]
- 136.Dirisala A., Uchida S., Li J., Van Guyse J.F.R., Hayashi K., Vummaleti S.V.C., Kaur S., Mochida Y., Fukushima S., Kataoka K. Effective mRNA protection by poly(l-ornithine) synergizes with endosomal escape functionality of a charge-conversion polymer toward maximizing mRNA introduction efficiency. Macromol. Rapid Commun. 2022;43 doi: 10.1002/marc.202100754. [DOI] [PubMed] [Google Scholar]
- 137.Kubler H., Scheel B., Gnad-Vogt U., Miller K., Schultze-Seemann W., Vom Dorp F., Parmiani G., Hampel C., Wedel S., Trojan L., Jocham D., Maurer T., Rippin G., Fotin-Mleczek M., von der Mulbe F., Probst J., Hoerr I., Kallen K.J., Lander T., Stenzl A. Self-adjuvanted mRNA vaccination in advanced prostate cancer patients: a first-in-man phase I/IIa study. J. Immunother. Cancer. 2015;3:26. doi: 10.1186/s40425-015-0068-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rausch S., Schwentner C., Stenzl A., Bedke J. mRNA vaccine CV9103 and CV9104 for the treatment of prostate cancer. Hum. Vaccin. Immunother. 2014;10:3146–3152. doi: 10.4161/hv.29553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sebastian M., Schroder A., Scheel B., Hong H.S., Muth A., von Boehmer L., Zippelius A., Mayer F., Reck M., Atanackovic D., Thomas M., Schneller F., Stohlmacher J., Bernhard H., Groschel A., Lander T., Probst J., Strack T., Wiegand V., Gnad-Vogt U., Kallen K.J., Hoerr I., von der Muelbe F., Fotin-Mleczek M., Knuth A., Koch S.D. A phase I/IIa study of the mRNA-based cancer immunotherapy CV9201 in patients with stage IIIB/IV non-small cell lung cancer. Cancer Immunol. Immunother. 2019;68:799–812. doi: 10.1007/s00262-019-02315-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Holvold L.B., Fredriksen B.N., Bogwald J., Dalmo R.A. Transgene and immune gene expression following intramuscular injection of Atlantic salmon (Salmo salar L.) with DNA-releasing PLGA nano- and microparticles. Fish Shellfish Immunol. 2013;35:890–899. doi: 10.1016/j.fsi.2013.06.030. [DOI] [PubMed] [Google Scholar]
- 141.Mahiny A.J., Dewerth A., Mays L.E., Alkhaled M., Mothes B., Malaeksefat E., Loretz B., Rottenberger J., Brosch D.M., Reautschnig P., Surapolchai P., Zeyer F., Schams A., Carevic M., Bakele M., Griese M., Schwab M., Nurnberg B., Beer-Hammer S., Handgretinger R., Hartl D., Lehr C.M., Kormann M.S. In vivo genome editing using nuclease-encoding mRNA corrects SP-B deficiency. Nat. Biotechnol. 2015;33:584–586. doi: 10.1038/nbt.3241. [DOI] [PubMed] [Google Scholar]
- 142.Parayath N.N., Stephan S.B., Koehne A.L., Nelson P.S., Stephan M.T. In vitro-transcribed antigen receptor mRNA nanocarriers for transient expression in circulating T cells in vivo. Nat. Commun. 2020;11:6080. doi: 10.1038/s41467-020-19486-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Patel A.K., Kaczmarek J.C., Bose S., Kauffman K.J., Mir F., Heartlein M.W., DeRosa F., Langer R., Anderson D.G. Inhaled nanoformulated mRNA polyplexes for protein production in lung epithelium. Adv. Mater. 2019;31 doi: 10.1002/adma.201805116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mignani S., Rodrigues J., Tomas H., Roy R., Shi X., Majoral J.-P. Bench-to-bedside translation of dendrimers: reality or utopia? A concise analysis. Adv. Drug Del. Rev. 2018;136–137:73–81. doi: 10.1016/j.addr.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 145.Joubert F., Munson M.J., Sabirsh A., England R.M., Hemmerling M., Alexander C., Ashford M.B. Precise and systematic end group chemistry modifications on PAMAM and poly(l-lysine) dendrimers to improve cytosolic delivery of mRNA. J. Control. Release. 2023;356:580–594. doi: 10.1016/j.jconrel.2023.03.011. [DOI] [PubMed] [Google Scholar]
- 146.Serrano-Sevilla I., Artiga A., Mitchell S.G., De Matteis L., de la Fuente J.M. Natural polysaccharides for siRNA delivery: nanocarriers based on chitosan, hyaluronic acid, and their derivatives. Molecules. 2019;24:2570. doi: 10.3390/molecules24142570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Benchamas G., Huang S., Huang G. The influence of traditional and new processing technologies on the structure and function of food polysaccharide. Food Funct. 2020;11:5718–5725. doi: 10.1039/d0fo00854k. [DOI] [PubMed] [Google Scholar]
- 148.Chuan D., Jin T., Fan R., Zhou L., Guo G. Chitosan for gene delivery: methods for improvement and applications. Adv. Colloid Interface Sci. 2019;268:25–38. doi: 10.1016/j.cis.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 149.Haque A., Dewerth A., Antony J.S., Riethmuller J., Schweizer G.R., Weinmann P., Latifi N., Yasar H., Pedemonte N., Sondo E., Weidensee B., Ralhan A., Laval J., Schlegel P., Seitz C., Loretz B., Lehr C.M., Handgretinger R., Kormann M.S.D. Chemically modified hCFTR mRNAs recuperate lung function in a mouse model of cystic fibrosis. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-34960-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lopes T.S., Alves G.G., Pereira M.R., Granjeiro J.M., Leite P.E.C. Advances and potential application of gold nanoparticles in nanomedicine. J. Cell. Biochem. 2019;120:16370–16378. doi: 10.1002/jcb.29044. [DOI] [PubMed] [Google Scholar]
- 151.Chen Y., Xianyu Y., Jiang X. Surface modification of gold nanoparticles with small molecules for biochemical analysis. Acc. Chem. Res. 2017;50:310–319. doi: 10.1021/acs.accounts.6b00506. [DOI] [PubMed] [Google Scholar]
- 152.Ding Y., Jiang Z., Saha K., Kim C.S., Kim S.T., Landis R.F., Rotello V.M. Gold nanoparticles for nucleic acid delivery. Mol. Ther. 2014;22:1075–1083. doi: 10.1038/mt.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yeom J.H., Ryou S.M., Won M., Park M., Bae J., Lee K. Inhibition of Xenograft tumor growth by gold nanoparticle-DNA oligonucleotide conjugates-assisted delivery of BAX mRNA. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0075369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mbatha L.S., Maiyo F., Daniels A., Singh M. Dendrimer-coated gold nanoparticles for efficient folate-targeted mRNA delivery in vitro. Pharmaceutics. 2021;13:900. doi: 10.3390/pharmaceutics13060900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Raes L., Stremersch S., Fraire J.C., Brans T., Goetgeluk G., De Munter S., Van Hoecke L., Verbeke R., Van Hoeck J., Xiong R., Saelens X., Vandekerckhove B., De Smedt S., Raemdonck K., Braeckmans K. Intracellular delivery of mRNA in adherent and suspension cells by vapor nanobubble photoporation. Nanomicro Lett. 2020;12:185. doi: 10.1007/s40820-020-00523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chan K.P., Gao Y., Goh J.X., Susanti D., Yeo E.L., Chao S.H., Kah J.C. Exploiting the protein corona from cell lysate on DNA functionalized gold nanoparticles for enhanced mRNA translation. ACS Appl. Mater. Interfaces. 2017;9:10408–10417. doi: 10.1021/acsami.6b15269. [DOI] [PubMed] [Google Scholar]
- 157.Chan K.P., Chao S.H., Kah J.C.Y. Universal mRNA translation enhancement with gold nanoparticles conjugated to oligonucleotides with a poly(T) sequence. ACS Appl. Mater. Interfaces. 2018;10:5203–5212. doi: 10.1021/acsami.7b16390. [DOI] [PubMed] [Google Scholar]
- 158.Yang Y., Zhang M., Song H., Yu C. Silica-based nanoparticles for biomedical applications: from nanocarriers to biomodulators. Acc. Chem. Res. 2020;53:1545–1556. doi: 10.1021/acs.accounts.0c00280. [DOI] [PubMed] [Google Scholar]
- 159.Huang Y., Li P., Zhao R., Zhao L., Liu J., Peng S., Fu X., Wang X., Luo R., Wang R., Zhang Z. Silica nanoparticles: biomedical applications and toxicity. Biomed. Pharmacother. 2022;151 doi: 10.1016/j.biopha.2022.113053. [DOI] [PubMed] [Google Scholar]
- 160.Wang Y., Song H., Yu M., Xu C., Liu Y., Tang J., Yang Y., Yu C. Room temperature synthesis of dendritic mesoporous silica nanoparticles with small sizes and enhanced mRNA delivery performance. J. Mater. Chem. B. 2018;6:4089–4095. doi: 10.1039/c8tb00544c. [DOI] [PubMed] [Google Scholar]
- 161.Wang Y., Tang J., Yang Y., Song H., Fu J., Gu Z., Yu C. Functional nanoparticles with a reducible tetrasulfide motif to upregulate mRNA translation and enhance transfection in hard-to-transfect cells. Angew. Chem. Int. Ed. Engl. 2020;59:2695–2699. doi: 10.1002/anie.201914264. [DOI] [PubMed] [Google Scholar]
- 162.Dong S., Feng Z., Ma R., Zhang T., Jiang J., Li Y., Zhang Y., Li S., Liu X., Liu X., Meng H. Engineered design of a mesoporous silica nanoparticle-based nanocarrier for efficient mRNA delivery in vivo. Nano Lett. 2023;23:2137–2147. doi: 10.1021/acs.nanolett.2c04486. [DOI] [PubMed] [Google Scholar]
- 163.Kamegawa R., Naito M., Uchida S., Kim H.J., Kim B.S., Miyata K. Bioinspired silicification of mRNA-loaded polyion complexes for macrophage-targeted mRNA delivery. ACS Appl. Biol. Mater. 2021;4:7790–7799. doi: 10.1021/acsabm.1c00704. [DOI] [PubMed] [Google Scholar]
- 164.Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367 doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Moon B., Chang S. Exosome as a delivery vehicle for cancer therapy. Cells. 2022;11:316. doi: 10.3390/cells11030316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Duan L., Xu L., Xu X., Qin Z., Zhou X., Xiao Y., Liang Y., Xia J. Exosome-mediated delivery of gene vectors for gene therapy. Nanoscale. 2021;13:1387–1397. doi: 10.1039/d0nr07622h. [DOI] [PubMed] [Google Scholar]
- 167.Amiri A., Bagherifar R., Ansari Dezfouli E., Kiaie S.H., Jafari R., Ramezani R. Exosomes as bio-inspired nanocarriers for RNA delivery: preparation and applications. J. Transl. Med. 2022;20:125. doi: 10.1186/s12967-022-03325-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Usman W.M., Pham T.C., Kwok Y.Y., Vu L.T., Ma V., Peng B., Chan Y.S., Wei L., Chin S.M., Azad A., He A.B., Leung A.Y.H., Yang M., Shyh-Chang N., Cho W.C., Shi J., Le M.T.N. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat. Commun. 2018;9:2359. doi: 10.1038/s41467-018-04791-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yeo R.W., Lai R.C., Zhang B., Tan S.S., Yin Y., Teh B.J., Lim S.K. Mesenchymal stem cell: an efficient mass producer of exosomes for drug delivery. Adv. Drug Deliv. Rev. 2013;65:336–341. doi: 10.1016/j.addr.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 170.Wang J.H., Forterre A.V., Zhao J., Frimannsson D.O., Delcayre A., Antes T.J., Efron B., Jeffrey S.S., Pegram M.D., Matin A.C. Anti-HER2 scFv-directed extracellular vesicle-mediated mRNA-based gene delivery inhibits growth of HER2-positive human breast tumor xenografts by prodrug activation. Mol. Cancer Ther. 2018;17:1133–1142. doi: 10.1158/1535-7163.MCT-17-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tsai S.J., Atai N.A., Cacciottolo M., Nice J., Salehi A., Guo C., Sedgwick A., Kanagavelu S., Gould S.J. Exosome-mediated mRNA delivery in vivo is safe and can be used to induce SARS-CoV-2 immunity. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.101266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bu T., Li Z., Hou Y., Sun W., Zhang R., Zhao L., Wei M., Yang G., Yuan L. Exosome-mediated delivery of inflammation-responsive Il-10 mRNA for controlled atherosclerosis treatment. Theranostics. 2021;11:9988–10000. doi: 10.7150/thno.64229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Maugeri M., Nawaz M., Papadimitriou A., Angerfors A., Camponeschi A., Na M., Holtta M., Skantze P., Johansson S., Sundqvist M., Lindquist J., Kjellman T., Martensson I.L., Jin T., Sunnerhagen P., Ostman S., Lindfors L., Valadi H. Linkage between endosomal escape of LNP-mRNA and loading into EVs for transport to other cells. Nat. Commun. 2019;10:4333. doi: 10.1038/s41467-019-12275-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yang J., Wu S., Hou L., Zhu D., Yin S., Yang G., Wang Y. Therapeutic effects of simultaneous delivery of nerve growth factor mRNA and protein via exosomes on cerebral ischemia. Mol. Ther. Nucleic Acids. 2020;21:512–522. doi: 10.1016/j.omtn.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yang Z., Shi J., Xie J., Wang Y., Sun J., Liu T., Zhao Y., Zhao X., Wang X., Ma Y., Malkoc V., Chiang C., Deng W., Chen Y., Fu Y., Kwak K.J., Fan Y., Kang C., Yin C., Rhee J., Bertani P., Otero J., Lu W., Yun K., Lee A.S., Jiang W., Teng L., Kim B.Y.S., Lee L.J. Large-scale generation of functional mRNA-encapsulating exosomes via cellular nanoporation. Nat. Biomed. Eng. 2020;4:69–83. doi: 10.1038/s41551-019-0485-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lin Y., Wu J., Gu W., Huang Y., Tong Z., Huang L., Tan J. Exosome-liposome hybrid nanoparticles deliver CRISPR/Cas9 system in MSCs. Adv. Sci. (Weinh.) 2018;5 doi: 10.1002/advs.201700611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Evers M.J.W., van de Wakker S.I., de Groot E.M., de Jong O.G., Gitz-Francois J.J.J., Seinen C.S., Sluijter J.P.G., Schiffelers R.M., Vader P. Functional siRNA delivery by extracellular vesicle-liposome hybrid nanoparticles. Adv. Healthc. Mater. 2022;11 doi: 10.1002/adhm.202101202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sato Y.T., Umezaki K., Sawada S., Mukai S.A., Sasaki Y., Harada N., Shiku H., Akiyoshi K. Engineering hybrid exosomes by membrane fusion with liposomes. Sci. Rep. 2016;6 doi: 10.1038/srep21933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ulmer J.B., Mason P.W., Geall A., Mandl C.W. RNA-based vaccines. Vaccine. 2012;30:4414–4418. doi: 10.1016/j.vaccine.2012.04.060. [DOI] [PubMed] [Google Scholar]
- 180.Van Lint S., Goyvaerts C., Maenhout S., Goethals L., Disy A., Benteyn D., Pen J., Bonehill A., Heirman C., Breckpot K. Preclinical evaluation of TriMix and antigen mRNA-based antitumor therapy mRNA-based antitumor vaccination. Cancer Res. 2012;72:1661–1671. doi: 10.1158/0008-5472.CAN-11-2957. [DOI] [PubMed] [Google Scholar]
- 181.Jansen Y., Kruse V., Corthals J., Schats K., van Dam P.-J., Seremet T., Heirman C., Brochez L., Kockx M., Thielemans K. A randomized controlled phase II clinical trial on mRNA electroporated autologous monocyte-derived dendritic cells (TriMixDC-MEL) as adjuvant treatment for stage III/IV melanoma patients who are disease-free following the resection of macrometastases. Cancer Immunol. Immunother. 2020;69:2589–2598. doi: 10.1007/s00262-020-02618-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Huff A.L., Jaffee E.M., Zaidi N. Messenger RNA vaccines for cancer immunotherapy: progress promotes promise. J. Clin. Invest. 2022;132 doi: 10.1172/JCI156211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Qiu M., Li Y., Bloomer H., Xu Q. Developing biodegradable lipid nanoparticles for intracellular mRNA delivery and genome editing. Acc. Chem. Res. 2021;54:4001–4011. doi: 10.1021/acs.accounts.1c00500. [DOI] [PubMed] [Google Scholar]
- 184.Parhiz H., Brenner J.S., Patel P.N., Papp T.E., Shahnawaz H., Li Q., Shi R., Zamora M.E., Yadegari A., Marcos-Contreras O.A. Added to pre-existing inflammation, mRNA-lipid nanoparticles induce inflammation exacerbation (IE) J. Control. Release. 2022;344:50–61. doi: 10.1016/j.jconrel.2021.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Huang P., Jiang L., Pan H., Ding L., Zhou B., Zhao M., Zou J., Li B., Qi M., Deng H., Zhou Y., Chen X. An integrated polymeric mRNA vaccine without inflammation side effects for cellular immunity mediated cancer therapy. Adv. Mater. 2023;35 doi: 10.1002/adma.202207471. [DOI] [PubMed] [Google Scholar]
- 186.Xiong W., Gao Y., Wei W., Zhang J. Extracellular and nuclear PD-L1 in modulating cancer immunotherapy. Trends Cancer. 2021;7:837–846. doi: 10.1016/j.trecan.2021.03.003. [DOI] [PubMed] [Google Scholar]
- 187.Pardoll D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Islam M.A., Rice J., Reesor E., Zope H., Tao W., Lim M., Ding J., Chen Y., Aduluso D., Zetter B.R., Farokhzad O.C., Shi J. Adjuvant-pulsed mRNA vaccine nanoparticle for immunoprophylactic and therapeutic tumor suppression in mice. Biomaterials. 2021;266 doi: 10.1016/j.biomaterials.2020.120431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Liu L., Wang Y., Miao L., Liu Q., Musetti S., Li J., Huang L. Combination immunotherapy of MUC1 mRNA nano-vaccine and CTLA-4 blockade effectively inhibits growth of triple negative breast cancer. Mol. Ther. 2018;26:45–55. doi: 10.1016/j.ymthe.2017.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang Y., Zhang L., Xu Z., Miao L., Huang L. mRNA vaccine with antigen-specific checkpoint blockade induces an enhanced immune response against established melanoma. Mol. Ther. 2018;26:420–434. doi: 10.1016/j.ymthe.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Verbeke R., Lentacker I., Breckpot K., Janssens J., Van Calenbergh S., De Smedt S.C., Dewitte H. Broadening the message: a nanovaccine co-loaded with messenger RNA and alpha-GalCer induces antitumor immunity through conventional and natural killer T cells. ACS Nano. 2019;13:1655–1669. doi: 10.1021/acsnano.8b07660. [DOI] [PubMed] [Google Scholar]
- 192.Zhou S., Liu M., Ren F., Meng X., Yu J. The landscape of bispecific T cell engager in cancer treatment. Biomarker Res. 2021;9:38. doi: 10.1186/s40364-021-00294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Garber K. Bispecific antibodies rise again: Amgen's blinatumomab is setting the stage for a bispecific-antibody revival, enabled by new formats that may solve the field's long-standing problems. Nat. Rev. Drug Discov. 2014;13:799–802. doi: 10.1038/nrd4478. [DOI] [PubMed] [Google Scholar]
- 194.Stadler C.R., Bähr-Mahmud H., Celik L., Hebich B., Roth A.S., Roth R.P., Karikó K., Türeci Ö., Sahin U. Elimination of large tumors in mice by mRNA-encoded bispecific antibodies. Nat. Med. 2017;23:815–817. doi: 10.1038/nm.4356. [DOI] [PubMed] [Google Scholar]
- 195.Vitale C., Strati P. CAR T-cell therapy for B-cell non-Hodgkin lymphoma and chronic lymphocytic leukemia: clinical trials and real-world experiences. Front. Oncol. 2020;10:849. doi: 10.3389/fonc.2020.00849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.June C.H., Sadelain M. Chimeric antigen receptor therapy. N. Engl. J. Med. 2018;379:64–73. doi: 10.1056/NEJMra1706169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Schutsky K., Song D.-G., Lynn R., Smith J.B., Poussin M., Figini M., Zhao Y., Powell D.J. Rigorous optimization and validation of potent RNA CAR T cell therapy for the treatment of common epithelial cancers expressing folate receptor. Oncotarget. 2015;6:28911. doi: 10.18632/oncotarget.5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Foster J.B., Choudhari N., Perazzelli J., Storm J., Hofmann T.J., Jain P., Storm P.B., Pardi N., Weissman D., Waanders A.J., Grupp S.A., Karikó K., Resnick A.C., Barrett D.M. Purification of mRNA encoding chimeric antigen receptor is critical for generation of a robust T-cell response. Hum. Gene Ther. 2019;30:168–178. doi: 10.1089/hum.2018.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.