Abstract
Background
Breast cancer 1 (BRCA1) and breast cancer 2 (BRCA2) tumor suppressor genes play crucial roles in DNA repair and regulation of transcription. Mutations in these genes are closely associated with the occurrence of cancers. However, the mutation status of BRCA gene in central south Chinese lung cancer patients remains unclear, and its expression levels in lung cancer also need to be further explored.
Methods
In this study, we use next‐generation sequencing (NGS) technology to analyze the BRCA genes mutations in 462 central south Chinese lung cancer patients. Public databases including cBioportal, Catalogue Of Somatic Mutations In Cancer (COSMIC), The Cancer Genome Atlas (TCGA), Human Protein Atlas (HPA) and Expression Profiling Interactive Analysis (GEPIA) are also applied to explore the expression level and mutation status of BRCA in lung cancer patients and their relationships with the prognosis.
Results
We found that the mutation rate of BRCA1/2 in central south Chinese lung cancer patients is 4.3% and 6.5% respectively, and missense mutations account for the majority in both BRCA1/2, which are similar to the international status of BRCA1/2 from public databases. In addition, 45 novel mutations of BRCA1/2 in lung cancer are reported in this study. Furthermore, we find that the BRCA2 mutations are negatively correlated with overall survival rate in lung cancer using cBioportal. Last, we demonstrate that both of the mRNA and protein levels of BRCA1/2 are upregulated in lung cancer, and the elevated mRNA expression levels are positively linked with poor prognosis.
Conclusion
In general, our study better complements knowledge of the BRCA1/2 mutation status in the Chinese lung cancer patients, and firstly reveals the association between BRCA1/2 expression levels and prognosis of lung cancer patients, which may provide great value for the early diagnosis and clinical treatment of lung cancer.
Keywords: BRCA, Chinese lung cancer patients, expression, mutation
We reported the mutation rates of Breast cancer 1 (BRCA1) and BRCA2 in 462 central south Chinese lung cancer patients to be 4.3% and 6.5%, respectively, with missense mutation as the most common mutation type, which is similar to the international status. In addition, 45 novel mutations of BRCA1/2 in lung cancer were detected, and BRCA2 mutation was negatively correlated with overall survival in lung cancer. Furthermore, the increased expression level of BRCA1/2 was positively linked with poor prognosis of lung cancer patients.
INTRODUCTION
Lung cancer is the second most commonly diagnosed cancer and the leading cause of cancer‐related deaths worldwide, with an estimated 2.2 million new cases and 1.8 million deaths in 2020. Of the global new cases of lung cancer in 2020, about 37% were in China, and among the deaths due to lung cancer, Chinese cases account for about 39.8%. 1 Due to lack of early detection procedures and rapid development of the disease, most patients are diagnosed with last‐stage disease with low survival rates, and the 5‐year survival rate remains under 20% despite improvements in surgery and chemotherapy. 2 Therefore, further exploration of the pathogenesis and drug resistance mechanisms of lung cancer to look for accurate methods for early diagnosis and precision treatment is urgently needed.
Gene mutations in cancer are becoming important biomarkers for cancer treatment selection and are well‐targeted for cancer drug resistance. 3 , 4 As the lung is the organ most exposed to exogenous DNA damaging factors, including tobacco smoking, lung cancer normally contains high somatic mutation loads. 5 There has been a lot of research on common mutations in lung cancer, such as EGFR, TP53, and KRAS, and these studies are promising for improving patient prognosis. 6 , 7 , 8 , 9 However, studies on the mutations of DNA repair genes such as homologous repair genes in lung cancer are still needed.
Breast cancer 1 (BRCA1) and breast cancer 2 (BRCA2) are important tumor‐suppressor genes that participate in the DNA repair processes through homologous recombination, rescuing stalled or damaged DNA replication forks, and regulation of cell cycle DNA damage checkpoints. 10 Their mutations lead to defects in homologous recombination that destabilize the genome and lead to the occurrence and development of many cancers, including breast, ovarian, pancreatic, prostate, and especially lung. 11 , 12 , 13 , 14 , 15 , 16 , 17 Thus, poly (ADP‐Ribose) polymerase 1 (PARP1) inhibitors such as olaparib, niraparib, rucaparib, and talazoparib, which block DNA damage repair, have excellent performance against BRCA‐mutated cancers, both germline and somatic. 18 , 19 , 20 , 21 , 22 , 23 Although the state of BRCA mutations is closely related to the effect of cancer treatment, few studies have explored the frequency and clinical significance of BRCA mutations in Chinese lung cancer patients, and the relationships between BRCA mutations and prognosis of lung cancer patients are also unknown. Moreover, BRCA shows high expression levels in many cancers such as breast cancer and ovarian cancer, 24 , 25 but its expression levels in lung cancer and its specific impact on the patient prognosis of lung cancer still remain unclear.
To address this limited knowledge, we performed this study using next‐generation sequencing (NGS) and clinical information to investigate the prevalence and clinical significance of the BRCA1/2 mutation in 462 lung cancer patients from central south China, and further used public databases to analyze the mutation state of BRCA1/2 and the expression levels of BRCA genes impact on the survival rate in lung cancer. Moreover, we utilized immunohistochemistry (IHC) from the Human Protein Atlas and TCGA database to explore the expression levels of BRCA genes in lung cancer and its effect on the prognosis of lung cancer patients.
PATIENTS AND METHODS
Patients and samples
Four hundred and sixty‐two patients who were diagnosed with lung cancer and received treatment in Xiangya Hospital of Central South University were nonselectively included in the BRCA1/2 mutation test. Clinical information including gender, age, and histology types was collected. Written informed consent was obtained from all patients.
BRCA1/2 testing
DNA isolation and targeted sequencing were performed at Burning Rock Biotech, a commercial clinical laboratory accredited by the College of American Pathologists (CAP) and certified by the Clinical Laboratory Improvement Amendments (CLIA). Tissue DNA was extracted from formalin‐fixed, paraffin‐embedded tumor tissues using a QIAamp DNA formalin‐fixed paraffin‐embedded tissue kit (Qiagen). Fragments between 200 and 400 bp from the sheared tissue DNA were purified (Agencourt AMPure XP Kit; Beckman Coulter), hybridized with capture probes baits, selected with magnetic beads, and amplified. Target capture was performed using a commercial panel consisting of 68/168/520 genes (OncoScreen Plus). The quality and the size of the fragments were assessed by high‐sensitivity DNA kit using Bioanalyzer 2100 (Agilent Technologies). Indexed samples were sequenced on Nextseq 500 (Illumina) with paired‐end reads and average sequencing depth of 1000× for tissue samples.
Mutation analysis from public databases
Catalogue Of Somatic Mutations In Cancer (COSMIC) and BioPortal were used to analyze the mutations of BRCA genes in lung cancer. All operations were performed according to the online instructions of cBioPortal and COSMIC.
Immunohistochemistry
The Human Protein Atlas was used to analyze differences in BRCA gene protein expression levels between lung cancer tissues and normal lung tissues.
Statistical analysis
Kaplan–Meier plots were used to analyze the prognostic values of BRCA genes in lung cancer, and statistical significance was assessed by log‐rank test. GraphPad Prism 8.3.0 software was used to perform statistical analyses. Differences in clinical information were determined using the chi‐square test and Student's t‐test was used to determine the significance of differences between two groups. p value <0.05 was considered to be statistically significant.
RESULTS
Patient characteristics
The patients were from Xiangya Hospital of Central South University. The total of 462 patients was composed of 286 (61.9%) males and 176 (38.1%) females with a median age of 60 years. The vast majority of patients (377, 81.6%) were diagnosed with adenocarcinoma, followed by squamous cell carcinoma (75, 16.2%), two patients (0.4%) had large cell carcinoma (LCC) or small‐cell lung cancer (SCLC), and the remaining patients (6, 1.3%) had unspecified types (Supporting Information Table S1). Among these 462 lung cancer patients, we identified 47 patients who had a median age of 61 years with BRCA mutations, 26 males and 21 females. Adenocarcinoma remained the highest proportion among these BRCAm carriers (Supporting Information Table S2).
Prevalence of BRCA1/2 mutations
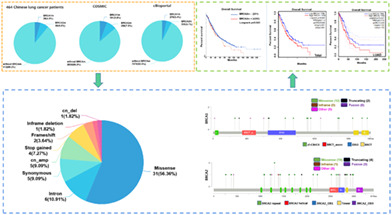
In the 462 Chinese patients with lung cancer, we detected 10.2% (47/462) of patients with BRCA mutations. The mutation rate of BRCA1 was 4.3% (20/462), which is lower than the 5.8% (191/3279) reported in COSMIC, but a little higher than the 3.4% (279/8187) reported in cBioportal. Similarly, the 6.5% (30/462) of patients with BRCA2 mutations was also lower than the 7.9% (258/3262) BRCA2 mutation rate from COSMIC, but higher than the 4.1% (338/8187) presented in cBioportal. These results indicate that lung cancer patients from central south China had similar BRCA mutation rates to the international level. In addition, there was 0.7% (3/462) of patients with both BRCA1 and BRCA2 mutations (Supporting Information Table S1). A predominance of BRCA2 mutations (63.8% vs. 42.6%) was found in central south Chinese patients (Supporting Information Table S2).
The distribution of mutations on BRCA1/2 is shown in Figure 1 and the detailed mutation status is presented with clinical information in Supporting Information Table S3. From the mutation distributions of the BRCA genes, we found no particular distribution hotspots, which was also observed in a previous study of pathogenic gBRCAm in Chinese non‐small‐cell lung cancer (NSCLC) (Figure 1a,b). 26 What surprised us was that, as far as we knew, we observed 45 novel mutations of BRCA not found previously in COSMIC, cBioportal, and related literature (Supporting Information Table S3). In addition, among the BRCA1 mutations, we observed five types of mutations, and the proportions of missense mutation (12/23), intro mutation (4/23), cn_amp (3/23), synonymous mutation (2/23), and frameshift mutation (2/23) were 52.17%, 17.39%, 13.04%, 8.7%, and 8.7%, respectively (Supporting Information Figure S1a). Seven types of mutations showed in the BRCA2 mutations, including missense mutation (19/32, 59.38%), stop gained mutation (4/32, 12.5%), synonymous mutation (3/32, 9.38%), cn_amp (2/32, 6.25%), intron mutation (2/32, 6.25%), cn_del (1/32, 3.12%), and inframe deletion (1/32, 3.12%) (Supporting Information Figure S1b). The missense mutation was the most frequent mutation in BRCA1, BRCA2 or BRCA1 and BRCA2 (Figure 1c). Moreover, four patients had multiple BRCA1/2 mutations (n ≥ 2), and one mutation in BRCA2, p.T1302A was detected in two different patients (Supporting Information Table S3).
FIGURE 1.
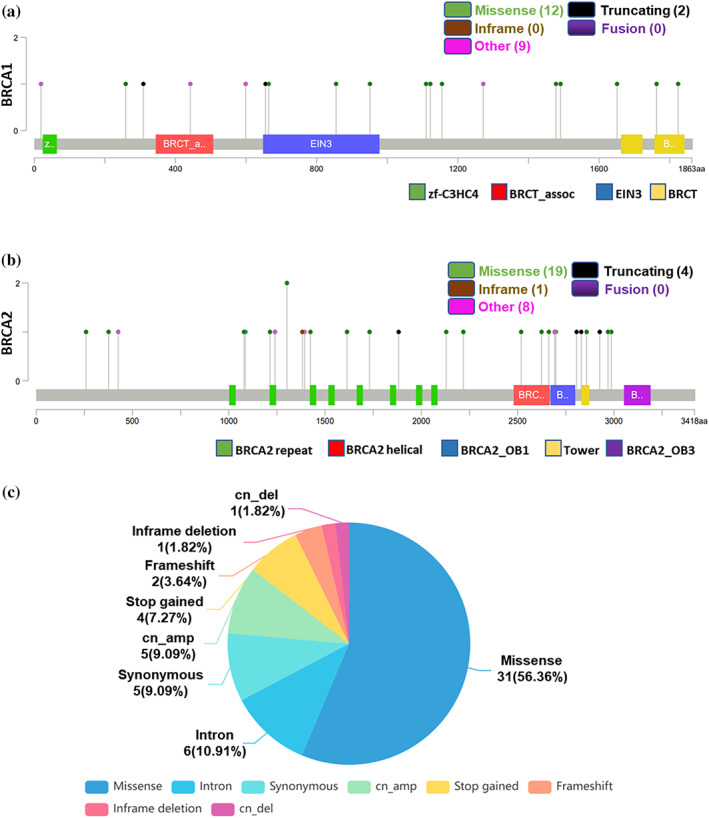
Analysis of BRCA1/2 mutations in Chinese lung cancer patients. (a, b) The distributions of mutations on BRCA1 (a) and BRCA2 (b). (c) The proportion of different types of mutations in BRCA1/2
Clinicopathological characteristics
The relationship between BRCAm and clinical characteristics is summarized in Table 1. When all histologies were analyzed together, females showed a little higher frequency of BRCA mutation compared with males (11.9% vs. 9.1%). Within different age groups, the highest mutation rate was found in patients aged 40–49 years, reaching 18.2%, and patients with an onset of disease before 50 were more likely to have BRCA mutation than those 50 years old or older patients (15.7% vs. 9.0%), consistent with earlier reports. 26 , 27 The lung adenocarcinoma (LUAD) subgroup showed lower mutations than any other histology subgroup (9.5% vs. 12.9%). However, all these analyses had no statistically significant difference, and this problem possibly be solved by increasing the sample sizes.
TABLE 1.
Relationship between BRCA mutations and clinical characteristics
| Characteristics | N | BRCA mutations | ||
|---|---|---|---|---|
| Wild | Mutant | p | ||
| Gender | ||||
| Male | 286 | 260 (90.9%) | 26 (9.1%) | 0.327 |
| Female | 176 | 155 (88.1%) | 21 (11.9%) | |
| Age (years) | ||||
| ≤40 | 17 | 16 (94.1%) | 1 (5.9%) | 0.087 |
| 40–49 | 66 | 54 (81.8%) | 12 (18.2%) | |
| 50–59 | 138 | 130 (94.2%) | 8 (5.8%) | |
| 60–69 | 175 | 157 (89.7%) | 18 (10.3%) | |
| ≥70 | 66 | 58 (87.9%) | 8 (12.1%) | |
| Histology | ||||
| Adenocarcinoma | 377 | 341 (90.5%) | 36 (9.5%) | 0.245 |
| Squamous cell carcinoma | 75 | 66 (88%) | 9 (12%) | |
| Large cell carcinoma | 2 | 1 (50%) | 1 (50%) | |
| Small cell lung cancer | 2 | 2 (100%) | 0 (0%) | |
| Not specified | 6 | 5 (83.3%) | 1 (16.7%) | |
BRCA1/2 mutations in lung cancer from public databases
The COSMIC database was used to analyze the BRCA gene mutation, and the specific mutation type ratio is shown in Figure 2. It can be seen that the missense mutation of BRCA1/2 is the most common mutation type in lung cancer, with proportions of 70.16% in BRCA1 and 71.71% in BRCA2, which is similar to what we found in our study. In addition, the BRCA1 mutation rate was 5.8% (191/3279) and 7.9% (258/3262) of lung cancer samples had BRCA2 mutations (Figure 2a,b). We analyzed the alteration frequencies of BRCA genes in lung cancer using cBioportal. A total of 29 studies on lung cancer with 8187 samples were included in this database. The overall mutation rate of BRCA1 was 3.4% (279/8187), and 4.1% (338/8187) of these samples showed BRCA2 mutations. Among 279 mutations in BRCA1, 228 of which were missense mutations, 30 were truncating and inframe mutations and nine were other types of mutations. There were 338 mutations in BRCA2, of which missense mutations (256/338) accounted for the majority and 54 were truncating mutations (Figure 3a,b). Furthermore, we investigated the effects of BRCA mutations on the prognosis of lung cancer patients using cBioportal, and the results showed that BRCA2 mutation was closely associated with poor prognosis (Figure 3c,d).
FIGURE 2.
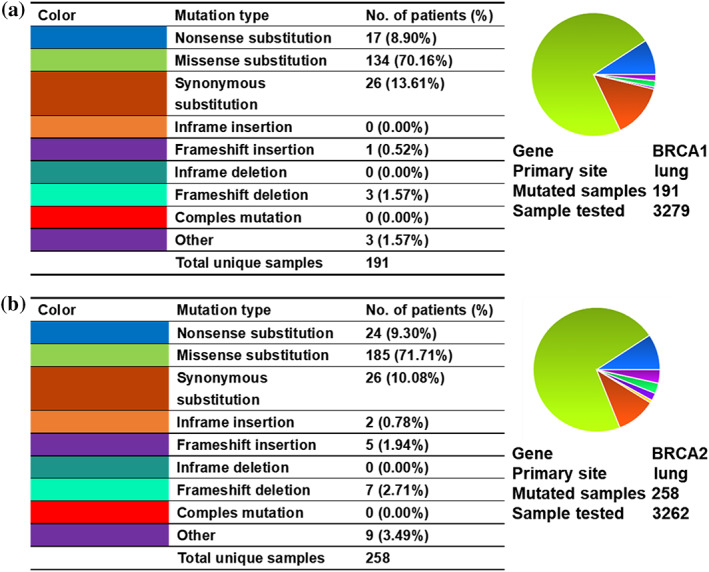
Analysis of BRCA1/2 mutations using COSMIC. (a, b) The proportion of different types of mutations in BRCA1 (a) or BRCA2 (b) in lung cancer samples from COSMIC
FIGURE 3.
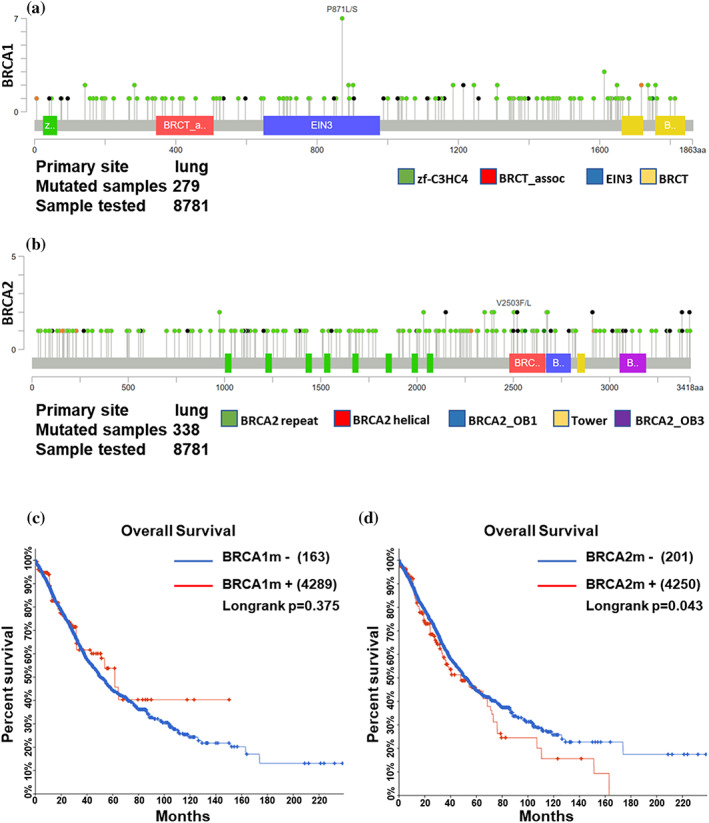
Analysis of BRCA1/2 mutations according to cBioportal. (a, b) The distributions of mutations on BRCA1 (a) and BRCA2 (b) in lung cancer samples from cBioportal. (c, d) Kaplan–Meier curve showing the overall survival rate of cBioportal lung cancer samples classified by mutation of BRCA1 (c) or BRCA2 (d)
Expression level and prognostic significance analysis of BRCA1/2 in lung cancer
The mRNA expression levels of BRCA1/2 in 31 kinds of cancers were analyzed by GEPIA, and the results showed that BRCA expression levels were higher in most cancers (Supporting Information Figure S2a,b), including LUAD and lung squamous cell carcinoma (LUSC) (Figure 4a,b), than in corresponding normal tissues. Moreover, the expression of BRCA was positively correlated with the pathological stage of LUAD samples (Figure 4c), but not LUSC samples (Supporting Information Figure S3a,b). We used data from GSE6044 (Supporting Information Figure S4a) and GSE40275 (Supporting Information Figure S4b) to demonstrate that the expression levels of BRCA1/2 were also elevated in SCLC. In addition, IHC experiments on clinical samples obtained from lung cancer patients further confirmed that the protein expression level of BRCA1/2 was elevated in lung cancer (Figure 5). We performed survival analysis to investigate the impact of BRCA1/2 on the survival rate of lung cancer patients. The results showed that the high expression levels of BRCA1/2 were negatively correlated with overall survival rate of LUAD patients (Figure 6a), but not in LUSC (Figure 6b), and BRCA1 was linked with poor prognosis of all lung cancer patients (Figure 6c).
FIGURE 4.
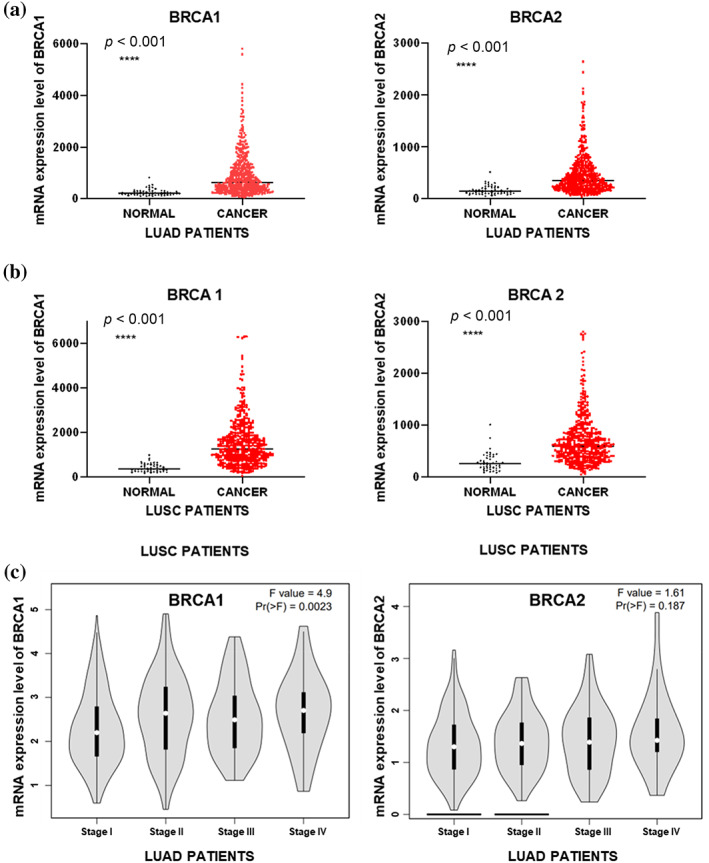
Evaluation of the mRNA expression levels of BRCA1/2 in lung cancer. (a, b) TCGA analysis of the mRNA expression level of BRCA1/2 in LUAD samples (n = 535) and corresponding normal lung tissues (n = 59) (a) or LUSC samples (502) and corresponding normal lung tissues (n = 49) (b). (c) GEPIA analysis of the mRNA expression level of BRCA1/2 in LUAD based on pathological stage
FIGURE 5.
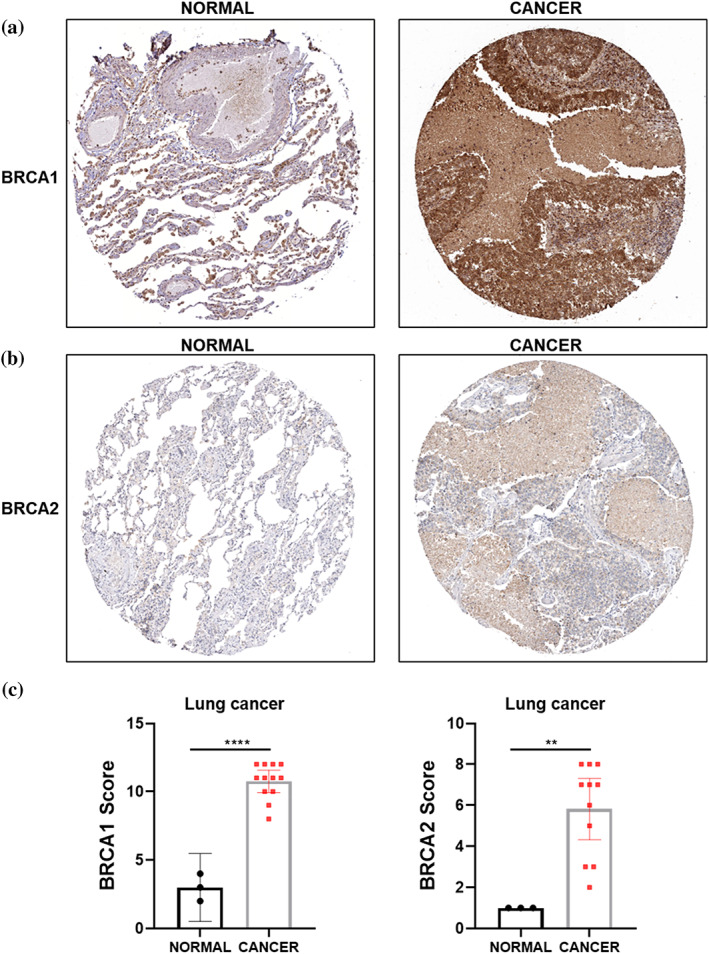
Immunohistochemical expression of BRCA1/2 protein in lung cancer. (a, b) IHC test of BRCA1 (a) or BRCA2 (b) protein expression in lung cancer and normal lung tissues from the Human Protein Atlas. (c) IHC scores indicating BRCA1 (normal n = 3, cancer n = 12) or BRCA2 (normal n = 3, cancer n = 11) expression levels in lung cancer tissues versus normal tissues. **, p < 0.01; ****, p < 0.001
FIGURE 6.
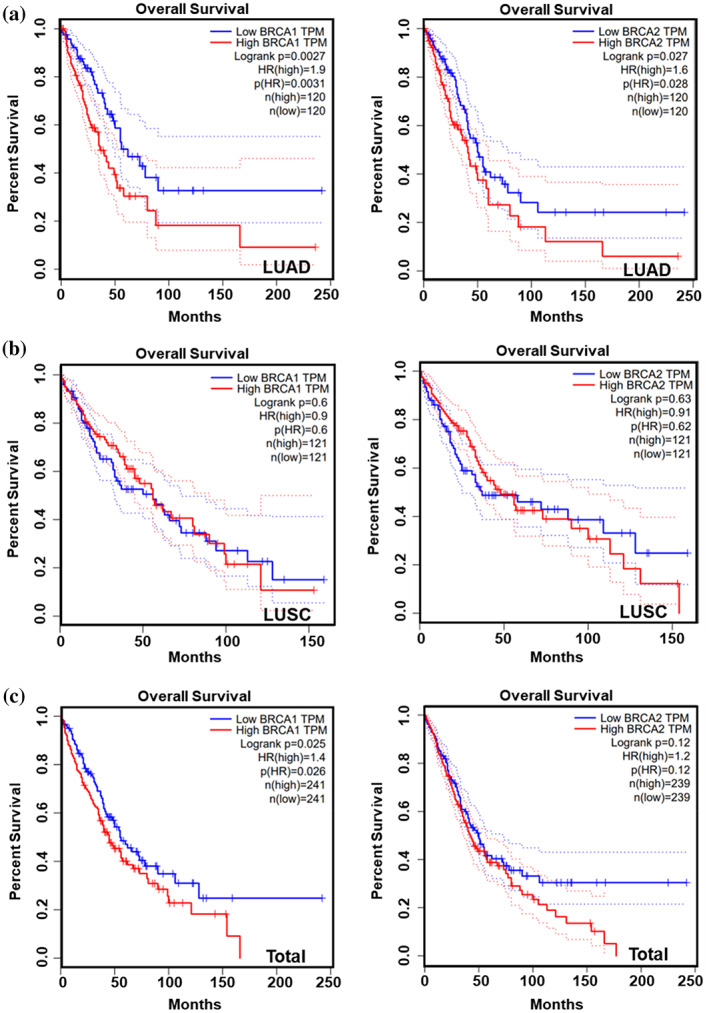
Survival analysis of BRCA1/2 in lung cancer. (a–c) Kaplan–Meier curve showing the overall survival rate of LUAD (a), LUSC (b) or lung cancer samples (c) classified by expression of BRCA1 or BRCA2 by GEPIA.
DISCUSSION
To the best of our knowledge, our study is the first and largest NGS‐based BRCA1/2 mutation prevalence study with detailed clinical information in lung cancer patients from central south China. In this study, we found that 10.2% of cases had BRCA1/2 mutations, with a predominance of BRCA2. The mutation rate was similar to a previous study of BRCA1/2 mutation (12.3%) in Chinese NSCLC patients, and the predominance of BRCA2 also occurred in that study. 26 In addition, the BRCA1/2 mutation frequency in central south Chinese lung cancer patients was similar to the international level. The public databses also showed that among all kinds of mutations, the missense mutation accounted for the largest proportion in these lung cancer patients. Moreover, to our knowledge, we found 45 novel mutations which had not been found in literature and public databases and may be unique to the central south Chinese population. We reveal that the mutation of BRCA2 is associated with poor prognosis of lung cancer patients using cBioportal, indicating that BRCA2 may play an important role in the tumorigenesis and development of lung cancer.
Previous studies had shown that BRCA mutations, which are strongly linked to the development of many cancers, such as breast, ovarian and pancreatic cancers, can lead to younger onset and be treated as therapeutic targets. 28 , 29 , 30 However, the role of BRCA mutations in lung cancer patients from central south China was unclear. This study showed that patients with BRCA mutations tended to be younger, and up to 15.7% of lung cancer patients aged 40–49 had BRCA1/2 mutations, consistent with earlier studies in the literature. 26 , 27 Patients with adenocarcinoma exhibited the lowest rate of BRCA mutations. Although these conclusions are not statistically significant, they have certain reference value for the diagnosis and clinical treatment of lung cancer.
As crucial genes in DNA repair and transcriptional regulation, BRCA genes are significantly upregulated in various cancer types, such as breast cancer, ovarian cancer, and glioma, and are closely associated with poor prognosis. 31 , 32 However, the expression level of BRCA in lung cancer and its impacts on the prognosis of lung cancer patients needs to be further explored. In the present study, we demonstrated that BRCA1/2 are highly expressed in lung cancer. Moreover, the high mRNA expression levels of BRCA were negatively correlated with overall survival rate in lung cancer, as showed by survival analysis, indicating that BRCA1/2 may serve as potentially important targets for cancer therapy.
There are some deficiencies in this study that need to be improved. First, there are many other identified cancer susceptibility genes whose mutations played a key role in the progression of lung cancer, but we were limited to studying the role of BRCA mutations in lung cancer. Second, due to the lack of NGS results of corresponding normal tissue samples, we were unable to identify somatic and germline mutations of BRCA1/2. Finally, more clinical information, such as smoking status, treatment, and family history, needs to be collected to improve this study.
In conclusion, our study is probably the largest survey of the mutation status of BRCA genes in central south Chinese lung cancer patients. We reveal that the state of BRCA gene mutation in these patients is similar to the international level, and the increased expression levels of BRCA genes in lung cancer are associated with poor prognosis. This study complements knowledge of the BRCA1/2 mutation rate in Chinese lung cancer patients, deepening our understanding of BRCA mutations, and may provide a research basis for the treatment of lung cancer by targeting BRCA mutations.
AUTHOR CONTRIBUTIONS
Yongguang Tao and Desheng Xiao designed and supervised this research. Bokang Yan, Bin Xie, and Jiaxing Guo performed the experiments and data analysis. Meiyuan Huang, Jingyue Sun, and Jielin Chen provided helpful discussions. Bokang Yan wrote the paper with the help of Yongguang Tao and Desheng Xiao. The manuscript was approved by all authors.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
Supporting information
TABLE S1. Demographics of 462 patients with lung cancer
TABLE S2. Demographics of 47 patients with BRCA1/2 mutations
TABLE S3. Mutational profile of the patients with mutations in BRCA1/2
FIGURE S1. Proportion of different types of mutations in BRCA1 (a) or BRCA2 (b)
FIGURE S2. Analysis of mRNA expression profiles of BRCA1 (a) or BRCA2 (b)
FIGURE S3. The expression of BRCA1 (a) or BRCA2 (b) in LUSC based on pathological stage
FIGURE S4. The mRNA expression levels of BRCA1 or BRCA2 in SCLC. (a) The mRNA expression levels of BRCA1 or BRCA2 in GSE6044. (b) The mRNA expression levels of BRCA1 or BRCA2 in GSE40275
ACKNOWLEDGMENTS
This manuscript is not considered for publication elsewhere. We would like to thank all members for their contributions to this work and apologize to those not mentioned due to space limitations. This work was supported by the National Natural Science Foundation of China (82073136 [Desheng Xiao], 82072594 [Yongguang Tao], 82073097 [Jingyue Sun]) and the Hunan Natural Science Foundation (S2021JJMSXM2846 [Desheng Xiao]).
Yan B, Xie B, Huang M, Guo J, Sun J, Chen J, et al. Mutations and expressions of breast cancer 1/2 in lung cancer. Thorac Cancer. 2023;14(18):1753–1763. 10.1111/1759-7714.14920
Contributor Information
Yongguang Tao, Email: taoyong@csu.edu.cn.
Desheng Xiao, Email: xdsh96@csu.edu.cn.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are partly available from The Cancer Genome Atlas: http://cancergenome.nih.gov/, and partly available from the corresponding author upon reasonable request.
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2. Zeng H, Chen W, Zheng R, Zhang S, Ji JS, Zou X, et al. Changing cancer survival in China during 2003‐15: a pooled analysis of 17 population‐based cancer registries. Lancet Glob Health. 2018;6:e555–67. 10.1016/s2214-109x(18)30127-x [DOI] [PubMed] [Google Scholar]
- 3. Li YQ, Zheng Z, Liu QX, Lu X, Zhou D, Zhang J, et al. Moesin as a prognostic indicator of lung adenocarcinoma improves prognosis by enhancing immune lymphocyte infiltration. World J Surg Oncol. 2021;19:109. 10.1186/s12957-021-02229-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Feng L, Wang Z, Jing L, Zhou Z, Shi S, Deng R, et al. Recombinant human endostatin combined with chemotherapy for advanced squamous cell lung cancer: a meta‐analysis. World J Surg Oncol. 2021;19:64. 10.1186/s12957-021-02161-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. La Fleur L, Falk‐Sörqvist E, Smeds P, Berglund A, Sundström M, Mattsson JS, et al. Mutation patterns in a population‐based non‐small cell lung cancer cohort and prognostic impact of concomitant mutations in KRAS and TP53 or STK11. Lung Cancer. 2019;130:50–8. 10.1016/j.lungcan.2019.01.003 [DOI] [PubMed] [Google Scholar]
- 6. Reck M, Carbone DP, Garassino M, Barlesi F. Targeting KRAS in non‐small‐cell lung cancer: recent progress and new approaches. Ann Oncol. 2021;32:1101–10. 10.1016/j.annonc.2021.06.001 [DOI] [PubMed] [Google Scholar]
- 7. Lu S, Yu Y, Li Z, Yu R, Wu X, Bao H, et al. EGFR and ERBB2 germline mutations in Chinese lung cancer patients and their roles in genetic susceptibility to cancer. J Thorac Oncol. 2019;14:732–6. 10.1016/j.jtho.2018.12.006 [DOI] [PubMed] [Google Scholar]
- 8. Costa DB. TP53 mutations are predictive and prognostic when co‐occurring with ALK rearrangements in lung cancer. Ann Oncol. 2018;29:2028–30. 10.1093/annonc/mdy339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Skoulidis F, Heymach JV. Co‐occurring genomic alterations in non‐small‐cell lung cancer biology and therapy. Nat Rev Cancer. 2019;19:495–509. 10.1038/s41568-019-0179-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao W, Steinfeld JB, Liang F, Chen X, Maranon DG, Jian Ma C, et al. BRCA1‐BARD1 promotes RAD51‐mediated homologous DNA pairing. Nature. 2017;550:360–5. 10.1038/nature24060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rosenberg SM, Ruddy KJ, Tamimi RM, Gelber S, Schapira L, Come S, et al. BRCA1 and BRCA2 mutation testing in young women with breast cancer. JAMA Oncol. 2016;2:730–6. 10.1001/jamaoncol.2015.5941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alkassis S, Yazdanpanah O, Philip PA. BRCA mutations in pancreatic cancer and progress in their targeting. Expert Opin Ther Targets. 2021;25:547–57. 10.1080/14728222.2021.1957462 [DOI] [PubMed] [Google Scholar]
- 13. Talwar V, Rauthan A. BRCA mutations: implications of genetic testing in ovarian cancer. Indian J Cancer. 2022;59:S56–s67. 10.4103/ijc.IJC_1394_20 [DOI] [PubMed] [Google Scholar]
- 14. Oh M, Alkhushaym N, Fallatah S, Althagafi A, Aljadeed R, Alsowaida Y, et al. The association of BRCA1 and BRCA2 mutations with prostate cancer risk, frequency, and mortality: a meta‐analysis. Prostate. 2019;79:880–95. 10.1002/pros.23795 [DOI] [PubMed] [Google Scholar]
- 15. Wang C, Lin Y, Zhu H, Zhou Y, Mao F, Huang X, et al. Breast‐conserving therapy for breast cancer with BRCA mutations: a meta‐analysis. Breast Cancer. 2022;29:314–23. 10.1007/s12282-021-01312-2 [DOI] [PubMed] [Google Scholar]
- 16. Lee YC, Lee YC, Li CY, Lee YL, Chen BL. BRCA1 and BRCA2 gene mutations and lung cancer sisk: a meta‐analysis. Medicina. 2020;56:212. 10.3390/medicina56050212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ji W, Weng X, Xu D, Cai S, Lou H, Ding L. Non‐small cell lung cancer cells with deficiencies in homologous recombination genes are sensitive to PARP inhibitors. Biochem Biophys Res Commun. 2020;522:121–6. 10.1016/j.bbrc.2019.11.050 [DOI] [PubMed] [Google Scholar]
- 18. Mirza MR, Pignata S, Ledermann JA. Latest clinical evidence and further development of PARP inhibitors in ovarian cancer. Ann Oncol. 2018;29:1366–76. 10.1093/annonc/mdy174 [DOI] [PubMed] [Google Scholar]
- 19. Litton JK, Rugo HS, Ettl J, Hurvitz SA, Gonçalves A, Lee KH, et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N Engl J Med. 2018;379:753–63. 10.1056/NEJMoa1802905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu H, Wei M, Xu J, Hua J, Liang C, Meng Q, et al. PARP inhibitors in pancreatic cancer: molecular mechanisms and clinical applications. Mol Cancer. 2020;19:49. 10.1186/s12943-020-01167-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Senkus E, Delaloge S, Domchek SM, Conte P, Im SA, Xu B, et al. Olaparib efficacy in patients with germline BRCA‐mutated, HER2‐negative metastatic breast cancer: subgroup analyses from the phase III OlympiAD trial. Int J Cancer. 2023;1–12. 10.1002/ijc.34525 [DOI] [PubMed] [Google Scholar]
- 22. González‐Martín A, Pothuri B, Vergote I, DePont Christensen R, Graybill W, Mirza MR, et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2019;381:2391–402. 10.1056/NEJMoa1910962 [DOI] [PubMed] [Google Scholar]
- 23. Hobbs EA, Litton JK, Yap TA. Development of the PARP inhibitor talazoparib for the treatment of advanced BRCA1 and BRCA2 mutated breast cancer. Expert Opin Pharmacother. 2021;22:1825–37. 10.1080/14656566.2021.1952181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Satyananda V, Oshi M, Endo I, Takabe K. High BRCA2 gene expression is associated with aggressive and highly proliferative breast cancer. Ann Surg Oncol. 2021;28:7356–65. 10.1245/s10434-021-10063-5 [DOI] [PubMed] [Google Scholar]
- 25. Wang Z, Zhang J, Zhang Y, Deng Q, Liang H. Expression and mutations of BRCA in breast cancer and ovarian cancer: evidence from bioinformatics analyses. Int J Mol Med. 2018;42:3542–50. 10.3892/ijmm.2018.3870 [DOI] [PubMed] [Google Scholar]
- 26. Hu X, Yang D, Li Y, Li L, Wang Y, Chen P, et al. Prevalence and clinical significance of pathogenic germline BRCA1/2 mutations in Chinese non‐small cell lung cancer patients. Cancer Biol Med. 2019;16:556–64. 10.20892/j.issn.2095-3941.2018.0506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peng W, Li B, Li J, Chang L, Bai J, Yi Y, et al. Clinical and genomic features of Chinese lung cancer patients with germline mutations. Nat Commun. 2022;13:1268. 10.1038/s41467-022-28840-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Boussios S, Rassy E, Moschetta M, Ghose A, Adeleke S, Sanchez E, et al. BRCA mutations in ovarian and prostate cancer: bench to bedside. Cancer. 2022;14:3888. 10.3390/cancers14163888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rosen MN, Goodwin RA, Vickers MM. BRCA mutated pancreatic cancer: a change is coming. World J Gastroenterol. 2021;27:1943–58. 10.3748/wjg.v27.i17.1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Copson E, Maishman TC, Tapper WJ, Cutress RI, Greville‐Heygate S, Altman DG, et al. Germline BRCA mutation and outcome in young‐onset breast cancer (POSH): a prospective cohort study. Lancet Oncol. 2018;19:169–80. 10.1016/s1470-2045(17)30891-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Paul A, Paul S. The breast cancer susceptibility genes (BRCA) in breast and ovarian cancers. Front Biosci. 2014;19:605–18. 10.2741/4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meimand SE, Pour‐Rashidi A, Shahrbabak MM, Mohammadi E, Meimand FE, Rezaei N. The prognostication potential of BRCA genes expression in gliomas: a genetic survival analysis study. World Neurosurg. 2022;157:e123–8. 10.1016/j.wneu.2021.09.107 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TABLE S1. Demographics of 462 patients with lung cancer
TABLE S2. Demographics of 47 patients with BRCA1/2 mutations
TABLE S3. Mutational profile of the patients with mutations in BRCA1/2
FIGURE S1. Proportion of different types of mutations in BRCA1 (a) or BRCA2 (b)
FIGURE S2. Analysis of mRNA expression profiles of BRCA1 (a) or BRCA2 (b)
FIGURE S3. The expression of BRCA1 (a) or BRCA2 (b) in LUSC based on pathological stage
FIGURE S4. The mRNA expression levels of BRCA1 or BRCA2 in SCLC. (a) The mRNA expression levels of BRCA1 or BRCA2 in GSE6044. (b) The mRNA expression levels of BRCA1 or BRCA2 in GSE40275
Data Availability Statement
The data that support the findings of this study are partly available from The Cancer Genome Atlas: http://cancergenome.nih.gov/, and partly available from the corresponding author upon reasonable request.


