Abstract
Background
Growing evidence has revealed the critical regulatory role for circular RNAs (circRNAs) in cancer. This study aimed to explore the function of circ_0001387 in breast cancer (BC).
Methods
Circ_0001387, miR‐136‐5p, and spindle and kinetochore‐associated protein 2 (SKA2) levels were analyzed by quantitative real‐time polymerase chain reaction. Clone formation and 5‐ethynyl‐2′‐deoxyuridine assays were used to analyze cell proliferation. Cell apoptosis and cell migration and invasion abilities were analyzed using flow cytometry or transwell assay. Mechanism assay was used to confirm the association between miR‐136‐5p and circ_0001387 or SKA2. The effect of circ_0001387 on tumor growth in vivo was analyzed by the xenograft mice model.
Results
Circ_0001387 and SKA2 were expressed at high levels, whereas miR‐136‐5p was lowly expressed in BC tissues and cells. Meanwhile, the downregulation of circ_0001387 restrained BC cell progression in vitro and in vivo. Circ_0001387 competitively bound to miR‐136‐5p to regulate BC cell malignant behaviors. SKA2 was targeted by miR‐136‐5p, and SKA2 reinstated the suppressive effect of miR‐136‐5p upregulation in BC cells.
Conclusion
Our study indicated that circ_0001387 contributed to BC cell progression through miR‐136‐5p/SKA2 axis.
Keywords: breast cancer, circ_0001387, miR‐136‐5p, SKA2
Circ_0001387 is generated by the cyclization of exons 2–7 of the Wolf‐Hirschhorn syndrome candidate gene‐1 gene and its expression is upregulated in breast cancer cells. The increased circ_0001387 expression promotes SKA2 production by binding to miR‐136‐5p, further promoting breast cancer cell proliferation, migration, and invasion and inhibiting cell apoptosis.

INTRODUCTION
The latest cancer statistics reported in 2020 showed that there was more than 2 million new breast cancer (BC) patients worldwide, surpassing lung cancer. 1 The deaths of BC are as high as 680 000, making it the most reason for cancer death among women. 1 The combination of surgical resection and radiotherapy is the major treatment method for BC, which prolonged the survival of BC patients, 2 and the 5‐year survival rate for non‐metastatic BC is as high as 80%. 3 However, BC patients still have a high likelihood of metastasis even after surgical resection and other treatment measures, 4 and their 5‐year survival rates drops to 25%. 5 Therefore, an in‐depth study of BC pathogenesis and the search for promising molecular targets to enhance the effectiveness of BC treatment, as well as to alleviate the drug resistance phenomenon arising during chemotherapy, are essential to improve the survival of BC patients.
Derived from exon‐ or intron‐specific splicing of genes to form circular structures, circular RNAs (circRNAs) can be detected in all types of organisms. 6 Numberous references documented the role of circRNAs in various diseases. 7 CircRNAs are potential molecular markers for various diseases owing to their high stability. 8 In BC, a substantial quantity of dysregulated circRNAs were also identified. Cao et al. 9 disclosed that circRNF20 was notably elevated in BC and closely associated with the severity of breast cancer. Zeng et al. 10 also revealed that upregulation of circANKS1B in BC obviously contributed to BC metastasis, revealing the carcinogenic function of circANKS1B in BC. In the sequencing analysis of BC tissues, Xing et al. 11 found a noticeable upregulation of circ_0001387, but the function and regulatory mechanism of circ_0001387 in BC remains unclear.
MicroRNAs (miRNAs), a class of endogenous molecules, are commonly found in the genomes of higher organisms. 12 MiRNAs cannot encode proteins, but can act on the 3′‐untranslated regions (3'UTRs) of target genes to downregulate gene expression levels, therefore, regulating various life activities including tumorigenesis. 13 For example, miR‐429 was commonly dysregulated in different cancers and could modulate cancer cell proliferation, motility, and drug resistance phenomena. 14 Jiang et al. 15 also discovered that the high abundance of miR‐331 was associated with tumor metastasis BC. MiR‐144 was obviously downregulated in BC and could arrest BC progression in vitro by targeting centrosomal protein 55. 16 An early study identified the downregulation of miR‐136‐5p in BC, 17 but its detailed mechanism in BC still needs further investigation.
Spindle and kinetochore‐associated protein 2 (SKA2), closely associated with cell cycle processes, was found to be strongly related to the progression of tumors and psychiatric disorders. 18 Ren et al. 19 proved that SKA2 level was dramatically associated with tumor staging and metastasis of BC. Chen et al. 20 also revealed that SKA2 boosted esophageal cancer progression by facilitating cancer cell proliferation and metastasis.
The study was performed to comprehensively analyze the characteristics of circ_0001387/miR‐136‐5p/SKA2 in BC, to verify the effect of circ_0001387 on cell function using loss‐of‐function assays, and to analyze the downstream regulatory mechanism of circ_0001387.
MATERIALS AND METHODS
Tissue specimens
The Ethics Committee of the First Affiliated Hospital of Zhengzhou University approved all experimental projects in this study (institutional review board no., 2020‐KY‐449). Cancerous and normal paracancerous tissues were collected from 56 patients who were diagnosed with BC and not treated with chemotherapy or radiotherapy at the First Affiliated Hospital of Zhengzhou University. The collected samples were rapidly frozen in liquid nitrogen. All patients were informed about the use of tissue samples in this study and signed a written agreement.
Cell culture and transfection
Human normal mammary epithelial cell line MCF‐10A and BC cell lines (MCF‐7, MDA‐MB‐231, and MDA‐MB‐468) were obtained from the China center for type culture collection. Cells were placed in Dulbecco's modified Eagle's medium (DMEM) medium (Invitrogen) containing 10% fetal bovine serum (FBS) (Invitrogen) in an incubator at 37°C with 5% CO2. Short hairpin RNA (shRNA) targeting circ_0001387 (sh‐circ_0001387) and negative control sh‐NC, miR‐136‐5p mimic (miR‐136‐5p), miR‐136‐5p inhibitor (in‐miR‐136‐5p), and the corresponding controls (mimic NC and in‐miR‐NC), SKC2 overexpression vector (SKC2) and empty plasmid cloning DNA (pcDNA) were purchased from Sangon. The plasmids were transfected into cells by using Lipofectamine 2000 (Invitrogen), referring to the standard instructions.
RNA extraction and quantitative real‐time polymerase chain reaction
RNA extraction was implemented as per the TRIzol reagent (Solarbio, Beijing, China) instructions. Complementary DNA (cDNA) was obtained by reverse transcription with AMV Reverse Transcriptase and M‐MLV Reverse Transcriptase (Solarbio). These cDNA were then quantified using SYBR Green (Solarbio). All primers were purchased from Sangon and the sequences were presented in Table 1. The data were analyzed using the 2−ΔΔCt method with Homo sapiens actin β (β‐actin) and RNA U6 (U6) as a internal reference.
TABLE 1.
Primers sequences used for PCR.
| Name | Primers for PCR (5′–3′) | |
|---|---|---|
| hsa_circ_0001387 | Forward | AGATGCTGAAGCTGAGGACAC |
| Reverse | GGGGACTCTGCTTGATGCTAA | |
| WHSC1 | Forward | CATGGCGAGAATCAAGCACG |
| Reverse | CACACGAGTGTCCCCATTCA | |
| miR‐136‐5p | Forward | GCCGAGACTCCATTTGTTTTGAT |
| Reverse | CTCAACTGGTGTCGTGGAG | |
| SKA2 | Forward | AGAGCCGCATTTGTGCTACT |
| Reverse | TAGTCAGTGGTGACAGCTCCA | |
| β‐actin | Forward | CTTCGCGGGCGACGAT |
| Reverse | CCACATAGGAATCCTTCTGACC | |
| U6 | Forward | CTCGCTTCGGCAGCACA |
| Reverse | AACGCTTCACGAATTTGCGT |
Abbreviations: PCR, polymerase chain reaction; SKA2, spindle and kinetochore‐associated protein 2; WHSC1, Wolf‐Hirschhorn syndrome candidate gene‐1.
RNase R and actinomycin D treatment
Enzyme‐free centrifuge tubes were added with 2 μg of RNA and 3 U/μg RNase R reagent (ZhongBei LinGe) and incubated at 37°C for 1 h. BC cells were then incubated with actinomycin D (Sigma) for the defined time, then RNA was extracted from the cells. Circ_0001387 and linear Wolf‐Hirschhorn syndrome candidate gene‐1 (WHSC1) were subsequently analyzed using quantitative real‐time polymerase chain reaction (qRT‐PCR).
Clone formation assay
Approximately 1000 transfected cells were plated in 9.6‐cm petri dishes supplemented with complete medium and cultured for 2 weeks until obvious clones appeared. After cells were stained with crystal violet (Pinkeyan), clone formation ability was observed under a microscope (magnification, ×10).
5‐Ethynyl‐2′‐deoxyuridine assay
The analysis of cell proliferation was performed according to the guidebook of Cell Proliferation Assay Kit (Beyotime). A total of 10 μL of 5‐ethynyl‐2′‐deoxyuridine (EdU) solution was added in each well for 2 h. Subsequently, cells were fixed with paraformaldehyde, mixed with 100 μL of click reaction solution, followed by nuclear staining with 100 μL of 2‐(4‐amidinophenyl)‐6‐indolecarbamidine dihydrochloride solution. Subsequently, cells were photographed with a fluorescent inverted microscope.
Flow cytometry
Apoptosis was determined using Annexin V‐fluorescein isothiocyanate (Annexin V‐FITC) Apoptosis Detection reagents (Solarbio). After centrifugation, the cells were mixed with binding buffer and then stained with 5 μL of Annexin V‐FITC and propidium iodide (PI). Apoptosis was measured using the flow cytometer.
Western blot
Radioimmunoprecipitation assay lysis buffer (Beyotime) was used to extract proteins. Bicinchoninic Acid Assay Protein Assay Kit (Beyotime) was then implemented with reference to the instructions of guidebook to quantify protein. Next, the proteins were separated by SurePAGE gels (Beyotime) and transferred onto polyvinylidene fluoride membranes (Beyotime). The blocking solution (Beyotime) was added until the membrane was completely covered. After 2 h, the membrane was co‐incubated with primary antibodies for Bcl‐2‐like protein 4 (Bax) (ab32503, 1:1000, Abcam), B‐cell lymphoma 2 (Bcl‐2) (ab32124, 1:1000, Abcam), β‐actin (ab8226, 1:1000, Abcam), and SKA2 (ab75345, 1:1000, Abcam) overnight at 4°C, and then mixed with secondary antibody goat anti‐rabbit immunoglobulin G (IgG) (ab205718, 1:20000, Abcam) at 37°C for 2 h. Enhanced chemiluminescence (Beyotime) was then used for quantitative analysis of protein levels.
Transwell assay
The transfected cells were placed into the upper chamber of the transwell chambers (Corning) added with 200 μL of DMEM without FBS. Twenty‐four hours later, the cells in the lower chamber were stained with crystal violet. Results were analyzed under a microscope at the magnification 100×. For invasion analysis, the upper chamber of transwell was precoated with Matrigel matrix (Corning).
Dual‐luciferase reporter assay
The complementary sites of miR‐136‐5p with circ_0001387 or SKA2 were predicted by Starbase (http://starbase.sysu.edu.cn/). The wild‐type (WT) circ_0001387 and SKA2 3'UTR sequences containing the miR‐136‐5p complementary sites were cloned, whereas mutant (MUT) circ_0001387 and SKA2 3'UTR sequences were also obtained by the overlap extension PCR. These fragments were recombined into psi‐check2 vector (Youbio) to produce luciferase reporter plasmids, named as circ_0001387 WT, circ_0001387 MUT, SKA2 3'UTR WT, and SKA2 3'UTR MUT, respectively. Next, these plasmids were co‐transfected into MCF‐7 and MDA‐MB‐231 cells with miR‐136‐5p mimic or miR‐NC and continued to culture for 24 h. Finally, the luciferase activity of each group was measured with the assistance of Dual‐Lucy Assay Kit (Solarbio).
RNA immunoprecipitation assay
RNA Immunoprecipitation (RIP) Kit (Sigma) was implemented for RIP assay. BC cells were harvested and mixed with 500 μL of RIP lysis buffer. Cell lysates were then incubated with magnetic beads coupled with argonaute2 (Ago2) and IgG antibodies at 4°C for 12 h. Proteinase K was used to digest proteins in the complexes induced by the antibodies. Last, qRT‐PCR was used to analyze the miR‐136‐5p and circ_0001387 content.
Immunohistochemistry assay
Tumor sections were inactivated with hydrogen peroxide (Solarbio), and then mixed with 5% serum (Solarbio). Next, the sections were incubated with SKA2 (ab75345, 1:1000, Abcam) and nuclear proliferation marker (ki‐67) (ab15580, 1:2000, Abcam) primary antibodies. Secondary antibody (ab6721, 1:500, Abcam) was then added. After 1 h, the sections were stained according to the instructions of the DBA kit (Solarbio) and then viewed at 100× magnification under a microscope.
Xenograft mice model
One‐month‐old nude mice (n = 10) were purchased from Vital River Laboratory Animal Technology and randomized into two groups. The second mammary fat pad of mice in both groups were injected with MCF‐7 cells stably transfected with sh‐circ_0001387 or sh‐NC, respectively. Tumor volume was measured once a week following the formula: width 2 × length/2. After 35 days, all mice were euthanized and subsequently the tumor tissue was weighed and the abundance of relevant genes was analyzed. Animal experiments for this study were undertaken with the approval of the First Affiliated Hospital of Zhengzhou University Animal Committee.
Statistical analysis
The results of three independent experiments were used for statistical analysis. The data were analyzed by using SPSS 23.0 and expressed as mean ± standard deviation. For the analysis of differences between the two groups, Student's t‐test was applied. Analysis of variance was used to analyze the differences between more than two groups. The value of p < 0.05 was deemed as statistically significant.
RESULTS
Circ_0001387 was highly expressed in BC
Circ_0001387 was located on chromosome 4 and derived from exon 2 and exon 7 of WHSC1 (Figure 1(a)). Next, we observed a significant upregulation of circ_0001387 in BC tissues (n = 56) compared with adjacent tissue samples (n = 56) (Figure 1(b)). As shown in Figure 1(c), the high abundance of circ_0001387 was also detected in BC cells, and the most evident upregulation of circ_0001387 was observed in MCF‐7 cells and MDA‐MB‐231 cells. Therefore, MCF‐7 and MDA‐MB‐231 were selected for the follow‐up study (Figure 1(c)). In addition, as shown in Figure S1, BC patients with high circ_0001387 expression had a poorer prognosis than the BC patients with low circ_0001387 expression. Circ_0001387 was apparently resistant to RNase R digestion when compared with linear WHSC1 (Figure 1(d),(e)). Simultaneously, we detected that circ_0001387 level had no change in actinomycin D‐exposed BC cells, whereas linear WHSC1 was notably downregulated (Figure 1(f),(g)). All findings indicated circ_0001387 was a highly abundant and stable circRNA in BC.
FIGURE 1.

The expression profile of circ_0001387 in breast cancer (BC) was determined. (a) The structure diagram of circ_0001387. (b), (c) The abundance of circ_0001387 in BC tissues and cells was detected with the implementation of quantitative real‐time polymerase chain reaction (qRT‐PCR.) (d), (e) After RNase R treatment, circ_0001387 and WHSC1 contents were determined by qRT‐PCR. (f), (g) The levels of circ_0001387 and linear WHSC1 in MCF‐7 and MDA‐MB‐231 cells treated with actinomycin D were assayed using qRT‐PCR. **p < 0.01, ***p < 0.001. circRNAs, circular RNAs; WHSC1, Wolf‐Hirschhorn syndrome candidate gene‐1.
Circ_0001387 knockdown evidently retarded BC cell malignancy
The good knockdown efficiency of sh‐circ_0001387 was demonstrated in MCF‐7 and MDA‐MB‐231 cells (Figure 2(a)). Circ_0001387 silence reduced the number of clones (Figure 2(b)) and EdU‐positive cells (Figure 2(c)), displaying that cell proliferation was apparently repressed by circ_0001387 silencing. Meanwhile, cell apoptosis in sh‐circ_0001387 group was increased (Figure 2(d)). Downregulation of circ_0001387 obviously increased Bax and decreased Bcl‐2 expression (Figure 2(e), (f)). Transwell assay revealed that cell migration and invasion abilities were remarkably restrained by sh‐circ_0001387 transfection (Figure 2(g), (h)). These data exhibited circ_0001387 knockdown dramatically constrained the progression of BC.
FIGURE 2.

Effects of circ_0001387 silencing on the malignant phenotypes of breast cancer. (a)–(h) The sh‐circ_0001387 or sh‐NC were transfected into MCF‐7 and MDA‐MB‐231 cells. (a) The knockdown efficiency of sh‐circ_0001387. (b) Clone formation assay was used to assess cell proliferation. (c) 5‐Ethynyl‐2′‐deoxyuridine assay was performed to detect cell proliferation. (d) Flow cytometry was used to analyze apoptosis. (e), (f) Bax and Bcl‐2 levels were measured under the execution of western blot. (g), (h) The transwell assay was implemented to analyze migration and invasion. **p < 0.01, ***p < 0.001. shRNA, short hairpin RNA; sh‐NC, short hairpin RNA negative control; Bax, Bcl‐2‐like protein 4; Bcl‐2, B‐cell lymphoma 2.
MiR‐136‐5p targeted circ_0001387
Figure 3(a) showed the binding sites of circ_0001387 within miR‐136‐5p and the mutant sites of circ_0001387. Meanwhile, miR‐136‐5p levels were greatly upregulated after transfection of miR‐136‐5p mimic (Figure 3(b)). Dual‐luciferase activity assay displayed that the luciferase activity was dramatically reduced after co‐transfection with miR‐136‐5p mimic and circ_0001387 WT, whereas we did not observe this phenomenon in the miR‐136‐5p mimic and circ_0001387 MUT co‐transfection group (Figure 3(c), (d)). The study showed both circ_0001387 and miR‐136‐5p abundance were higher in the Anti‐Ago2 group than in the Anti‐IgG group (Figure 3(e), (f)). Moreover, miR‐136‐5p expression was dramatically lower in BC tissues and cells in comparison with normal tissues and MCF‐10A cells (Figure 3(g), (h)). Meanwhile, miR‐136‐5p and circ_0001387 levels exhibited a negative correlation in BC tissues (Figure 3(i)). Overall, circ_0001387 was identified as a sponge for miR‐136‐5p.
FIGURE 3.
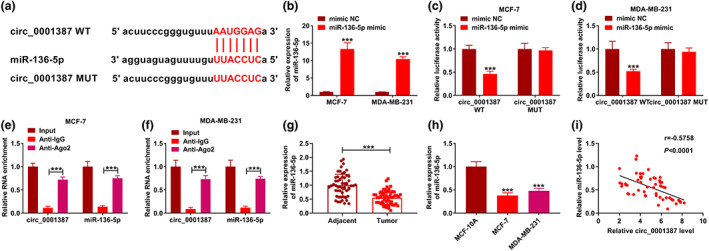
The association between circ_0001387 and miR‐136‐5p was established. (a) The binding sites between circ_0001387 and miR‐136‐5p were exhibited. (b) The upregulation efficiency of miR‐136‐5p mimic was analyzed under the use of quantitative real‐time polymerase chain reaction. (c)–(f) The interaction of circ_0001387 and miR‐136‐5p was analyzed. (g), (h) The abundance of miR‐136‐5p in breast cancer (BC) tissues and cells. (i) The correlation between miR‐136‐5p and circ_0001387 levels in BC tissues. ***p < 0.001. circRNAs, circular RNAs; miRNAs, microRNAs.
MiR‐136‐5p silencing impaired the effect of circ_0001387 knockdown on BC cell function
In in‐miR‐136‐5p‐transfected cells, miR‐136‐5p level was clearly reduced (Figure 4(a)). Co‐transfection of in‐miR‐136‐5p attenuated the effect on miR‐136‐5p caused by circ_0001387 silencing (Figure 4(b)). MiR‐136‐5p knockdown rescued the suppression impact of circ_0001387 downregulation on cell proliferation (Figure 4(c),(d)). Moreover, the effects of circ_0001387 silencing on apoptosis and apoptosis‐related protein (Bax and Bcl‐2) levels were attenuated with co‐transfection of in‐miR‐136‐5p (Figure 4(e)–(g)). Finally, circ_0001387 knockdown‐mediated the inhibition effects on migration and invasion were abolished via decreasing expression of miR‐136‐5p (Figure 4(h),(i)). Hence, circ_0001387 sponged miR‐136‐5p to modulate BC progression in vitro.
FIGURE 4.
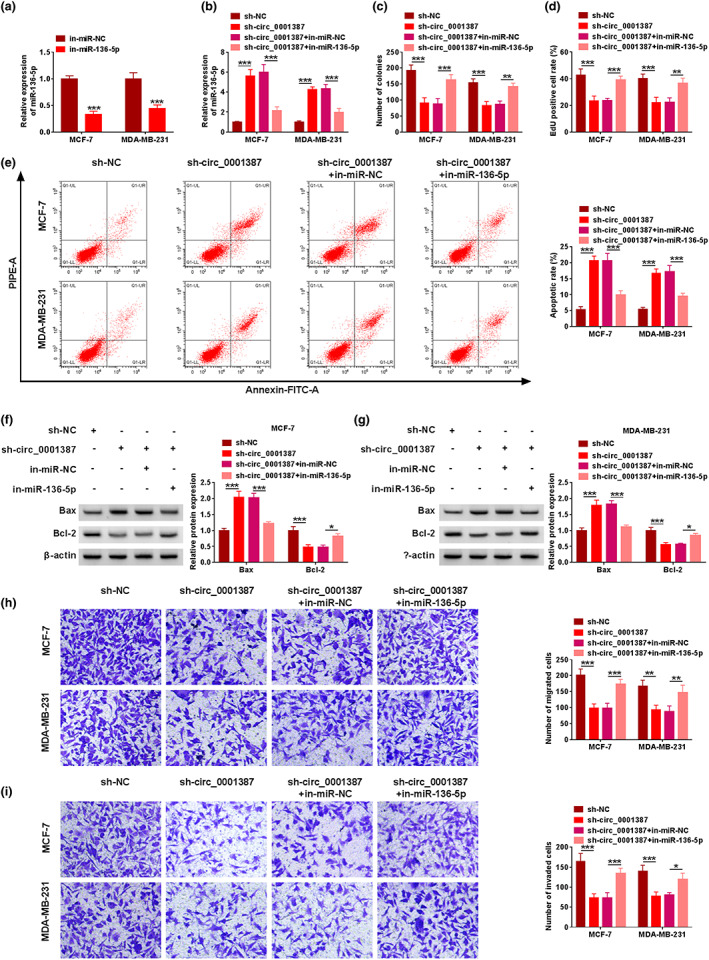
Circ_0001387 regulated breast cancer cell malignancy by sponging miR‐136‐5p. (a) The silencing efficiency of in‐miR‐136‐5p. (b)–(i) MCF‐7 and MDA‐MB‐231 cells were transfected with sh‐NC, sh‐circ_0001387, sh‐circ_0001387 + in‐miR‐NC, or sh‐circ_0001387 + in‐miR‐136‐5p, respectively. (b) MiR‐136‐5p level was analyzed using quantitative real‐time polymerase chain reaction. (c), (d) Cell proliferation was detected under the application of clone formation and 5‐ethynyl‐2′‐deoxyuridine assays. (e) Apoptosis was detected using flow cytometry. (f), (g) The western blot analysis for Bax and Bcl‐2. (h), (i) Migration and invasion analysis by transwell assay. *p < 0.05, **p < 0.01, ***p < 0.001. circRNAs, circular RNAs; miRNAs, microRNAs; sh‐NC, short hairpin RNA negative control; Bax, Bcl‐2‐like protein 4; Bcl‐2, B‐cell lymphoma 2.
SKA2 was targeted by miR‐136‐5p
The complementary binding sequence between SKA2 and miR‐136‐5p was presented in Figure 5(a). The phenomenon that miR‐136‐5p only downregulated the luciferase activity of SKA2 3'UTR WT group was shown in Figure 5(b),(c). The high abundance of SKA2 in BC tissue and cell samples was presented in Figure 5(d)–(g). Additionally, western blot result also indicated that circ_0001387 knockdown‐mediated decrease of SKA2 level was resumed by the introduction of in‐miR‐136‐5p (Figure 5(h),(i)). Finally, SKA2 in BC tissues exhibited a negative correlation with miR‐136‐5p level and a positive correlation with circ_0001387 level (Figure 5(j),(k)). Collectively, SKA2 was targeted by miR‐136‐5p.
FIGURE 5.

The relationship between SKA2 and miR‐136‐5p was verified. (a) The complementary binding sequence of SKA2 and miR‐136‐5p. (b), (c) The relationship between SKA2 and miR‐136‐5p was revealed. (d)–(f) The SKA2 content in breast cancer (BC) tissues. (g) SKA2 abundance in BC cells was assessed by western blot. (h), (i) Western blot analysis for SKA2 level. (j), (k) Pearson correlation analysis was used to assess the correlation between SKA2 and miR‐136‐5p or circ_0001387 levels. *p < 0.05, **p < 0.01, ***p < 0.001. SKA2, spindle and kinetochore‐associated protein 2; miRNAs, microRNAs; circRNAs, circular RNAs.
MiR‐136‐5p targeted SKA2 to arrest BC cell progression
Transfection of SKA2 upregulated intracellular SKA2 level and also resumed the reduction of SKA2 level by miR‐136‐5p introduction (Figure 6(a),(b)). Functionally, miR‐136‐5p introduction‐caused cell proliferation inhibition was recuperated after SKA2 upregulation (Figure 6(c),(d)). SKA2 overexpression also diminished the effects of miR‐136‐5p upregulation on apoptosis and intracellular Bax and Bcl‐2 protein levels (Figure 6(e)–(g)). Finally, the inhibitory impacts of miR‐136‐5p overexpression on cell migration and invasion were demonstrated by transwell assay; however, these effects were overturned by SKA2 overexpression (Figure 6(h),(i)). Conclusively, miR‐136‐5p curbed BC progression by downregulating SKA2 level.
FIGURE 6.

Effects of miR‐136‐5p and SKA2 on the malignant phenotypes of breast cancer cells. (a) The efficiency of SKA2 overexpression. (b)–(i) The mimic NC, miR‐136‐5p mimic, miR‐136‐5p+pcDNA, or miR‐136‐5p+SKA2 were transfected into MCF‐7 and MDA‐MB‐231 cells. (b) SKA2 protein level was detected after the implementation of western blot. (c), (d) The assessment of cell proliferation was achieved using clone formation and 5‐ethynyl‐2′‐deoxyuridine assays. (e) Determination of apoptosis was conducted by using flow cytometry. (f), (g) Bax and Bcl‐2 protein level analysis by western blot assay. (h), (i) The transwell assay was used to measure migration and invasion. *p < 0.05, **p < 0.01, ***p < 0.001. miRNAs, microRNAs; SKA2, spindle and kinetochore‐associated protein 2; NC, negative control; Bax, Bcl‐2‐like protein 4; Bcl‐2, B‐cell lymphoma 2.
Knockdown of circ_0001387 restrained tumor growth in vivo
As presented in Figure 7(a)–(c), compared with the sh‐NC transfected group, tumor growth was considerably slower in nude mice injected with MCF‐7 cells stably transfected with sh‐circ_0001387, as evidenced by smaller tumor size and lighter weight (Figure 7(a)–(c)). Moreover, the abundance of circ_0001387 and SKA2 in tumor tissues of sh‐circ_0001387 group was lower, whereas miR‐136‐5p level was higher than that in sh‐NC group (Figure 7(d), (e)). Finally, immunohistochemistry (IHC) results exhibited that SKA2 and ki‐67 levels were decreased by circ_0001387 downregulation in xenograft tumor tissues (Figure 7(e)). These data illustrated the inhibitory impact of circ_0001387 knockdown on tumor growth.
FIGURE 7.

Effect of circ_0001387 knockdown on breast cancer progression in vivo. (a) The representative pictures of xenograft tumor tissues were shown. (b), (c) Tumor volume and weight of the xenograft tumor tissues were measured. (d) The contents of circ_0001387 and miR‐136‐5p in xenograft tumor tissues. (e) SKA2 level in xenograft tumor tissues. (f) Immunohistochemistry assay analysis for SKA2 and ki‐67 levels. **p < 0.01, ***p < 0.001. circRNAs, circular RNAs; miRNAs, microRNAs; SKA2, spindle and kinetochore‐associated protein 2.
DISCUSSION
BC poses a serious threat to people's quality of life. In addition to improving early diagnosis of BC as much as possible through regular screening, the development of targeted molecule‐related drugs for BC therapy is an important means to mitigate the increased mortality. 21 Available evidence has confirmed the potential of circRNA as a promising biomarker. 22 For example, circ_0006156 was associated with greater differentiation of cancer cell and poorer prognosis in gastric cancer (GC) patients, revealing that circ_0006156 could be used as a desirable molecular marker for GC prognosis. 23 Yang et al. 24 also evidenced that the high abundance of circ‐LDLRAD3 in pancreatic cancer and cell metastasis was closely correlated with the circRNA, which also displayed the potential of circ‐LDLRAD3 as a good diagnostic marker.
The high abundance of circ_0001387 in BC was detected in our study, indicating the potential of circ_0001387 as a molecular marker for BC diagnosis. In addition, the functional effect of circ_0001387 in cancer has also been unveiled in previous studies. In ovarian cancer (OC), circ_0001387 was expressed at high level, whereas OC progression was apparently accelerated by circ_0001387 upregulation and retarded by circ_0001387 silencing. 25 , 26 Additionally, upregulation of circ_0001387 was also observed in endometrial cancer, and circ_0001387 overexpression facilitated the cancer progression. 27 However, our study verified the impacts of circ_0001387 on BC cell proliferation, apoptosis, and motility and displayed that circ_0001387 knockdown distinctly blocked BC development in vitro, and in vivo assays similarly confirmed the remarkable blocking effect of circ_0001387 knockdown on BC tumor growth. Although some circRNAs were involved in regulating BC progression, such as circ‐DNMT1, which was remarkably upregulated in BC and accelerated BC progression. 28 Our findings also further provided circRNAs‐related data for the elucidation of BC pathogenesis.
Numerous reports shed light on the mechanism by which circRNAs competitively bound miRNAs, and in turn modulated downstream mRNA expression to regulate disease progression. 29 Our study suggested that miR‐136‐5p was a downstream molecule of circ_0001387. The miRNA has been reported to function in diseases, including inflammatory response. MiR‐136‐5p possessed the ability to regulate inflammatory factor secretion. 30 In cardiovascular disease, Zhong et al. 31 also revealed that lnc‐SNHG14 upregulated rho associated coiled‐coil containing protein kinase 1 to suppress miR‐136‐5p content in oxygen–glucose deprivation and reoxygenation‐stimulated PC‐12 cells, which in turn exacerbated cerebral ischemia‐induced neural injury and stroke. Chen et al. 32 found a substantial downregulation of miR‐136‐5p in osteoarthritis (OA), whereas it had a positive effect on chondrocyte development, indicating that this miRNA was a high promising target in OA therapy. Furthermore, miR‐136‐5p also had a vital function in cancer research. In lung cancer, miR‐136‐5p was lower expressed and was closely related to lung carcinogenesis. 33 MiR‐136‐5p was competitively bound by circ_0110389 and repressed GC cell proliferation and metastasis. 34 In this article, the remarkable downregulation of miR‐136‐5p and the suppressive effects of miR‐136‐5p on BC cell proliferation, migration, and invasion and the pro‐apoptotic role in BC were revealed, which were similar to the previous findings. 33 , 34 Overall, the positive role of miR‐136‐5p in BC was revealed, which may be a hopeful target for future BC treatment.
SKA2 was suggested to be a target of miR‐136‐5p, and high abundance of SKA2 was observed in BC tissues. Dysregulation of SKA2 has also been reported in past studies. Wang et al. 35 revealed the apparent upregulation of SKA2 in hepatocellular carcinoma tissues, confirming that SKA2 had promotion effects on tumor cell metastasis. Zhuang et al. 36 also uncovered that SKA2 combined with cyclic‐AMP response binding protein and contributed to the proliferation of renal cell carcinoma cells and tumor growth in renal cell carcinoma. Upregulation of SKA2 was also found in GC, and SKA2 introduction attenuated miR‐520a‐3p‐induced GC cell progression inhibition in vitro. 37 Our study also evidenced that SKA2 upregulation abrogated the suppressive impact of miR‐136‐5p upregulation, demonstrating a positive role of SKA2 in BC progression. To conclude, this study disclosed that circ_0001387 upregulated SKA2 through competitive binding of miR‐136‐5p, thereby exerting a facilitative effect on BC cell malignancy. However, the clinical use of circ_0001387 in BC need to be addressed in the future.
CONCLUSION
Conclusively, circ_0001387 boosted BC progression by acting on miR‐136‐5p/SKA2 axis, offering a promising target for BC‐targeted therapy.
AUTHOR CONTRIBUTIONS
Youyi Xiong and Yuanting Gu designed the experiments.Youyi Xiong, Lin Li and Nan Wang performed the cell experiments and drafted the manuscript. Youyi Xion and Fang Wang are responsible for the mice experiments. Zhuo Chen, Luhong Han and Shan Jiang analyzed the data and plotted the figures. All authors approved the final version.
FUNDING INFORMATION
This work was supported by Henan Province Medical Science and Technology Research Program Joint Construction Project (LHGJ20200333).
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflicts of interest.
Supporting information
FIGURE S1. Kaplan–Meier method was performed to analyze the prognosis of BC patients with high or low circ_0001387 expression.
Xiong Y, Li L, Wang N, Wang F, Chen Z, Han L, et al. Circ_0001387 regulates SKA2 to accelerate breast cancer progression through miR‐136‐5p. Thorac Cancer. 2023;14(18):1707–1718. 10.1111/1759-7714.14916
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 2. Pearce L. Breast cancer. Nurs Stand. 2016;30:15. [DOI] [PubMed] [Google Scholar]
- 3. Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Nikšić M, et al. Global surveillance of trends in cancer survival 2000‐14 (CONCORD‐3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population‐based registries in 71 countries. Lancet. 2018;391:1023–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cancer Genome Atlas Network . Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR‐7 in cancer. Cancer Res. 2013;73:5609–12. [DOI] [PubMed] [Google Scholar]
- 8. Shafabakhsh R, Mirhosseini N, Chaichian S, Moazzami B, Mahdizadeh Z, Asemi Z. Could circRNA be a new biomarker for pre‐eclampsia? Mol Reprod Dev. 2019;86:1773–80. [DOI] [PubMed] [Google Scholar]
- 9. Cao L, Wang M, Dong Y, Xu B, Chen J, Ding Y, et al. Circular RNA circRNF20 promotes breast cancer tumorigenesis and Warburg effect through miR‐487a/HIF‐1alpha/HK2. Cell Death Dis. 2020;11:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zeng K, He B, Yang BB, Xu T, Chen X, Xu M, et al. The pro‐metastasis effect of circANKS1B in breast cancer. Mol Cancer. 2018;17:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xing L, Yang R, Wang X, Zheng X, Yang X, Zhang L, et al. The circRNA circIFI30 promotes progression of triple‐negative breast cancer and correlates with prognosis. Aging. 2020;12:10983–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–22. [DOI] [PubMed] [Google Scholar]
- 14. Guo CM, Liu SQ, Sun MZ. miR‐429 as biomarker for diagnosis, treatment and prognosis of cancers and its potential action mechanisms: a systematic literature review. Neoplasma. 2020;67:215–28. [DOI] [PubMed] [Google Scholar]
- 15. Jiang F, Zhang L, Liu Y, Zhou Y, Wang H. Overexpression of miR‐331 indicates poor prognosis and promotes progression of breast cancer. Oncol Res Treat. 2020;43:441–8. [DOI] [PubMed] [Google Scholar]
- 16. Yin Y, Cai J, Meng F, Sui C, Jiang Y. MiR‐144 suppresses proliferation, invasion, and migration of breast cancer cells through inhibiting CEP55. Cancer Biol Ther. 2018;19:306–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Han C, Fu Y, Zeng N, Yin J, Li Q. LncRNA FAM83H‐AS1 promotes triple‐negative breast cancer progression by regulating the miR‐136‐5p/metadherin axis. Aging. 2020;12:3594–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xie M, Bu Y. SKA2/FAM33A: a novel gene implicated in cell cycle, tumorigenesis, and psychiatric disorders. Genes Dis. 2019;6:25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ren Z, Yang T, Zhang P, Liu K, Liu W, Wang P. SKA2 mediates invasion and metastasis in human breast cancer via EMT. Mol Med Rep. 2019;19:515–23. [DOI] [PubMed] [Google Scholar]
- 20. Chen J, Yang HM, Zhou HC, Peng R‐R, Niu Z‐X, Kang C‐Y. PRR11 and SKA2 promote the proliferation, migration and invasion of esophageal carcinoma cells. Oncol Lett. 2020;20:639–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang X, Ji C, Hu J, Deng X, Zheng W, Yu Y, et al. Hsa_circ_0005273 facilitates breast cancer tumorigenesis by regulating YAP1‐hippo signaling pathway. J Exp Clin Cancer Res. 2021;40:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–91. [DOI] [PubMed] [Google Scholar]
- 23. He YX, Ju H, Li N, Jiang YF, Zhao WJ, Song TT, et al. Association between hsa_circ_0006156 expression and incidence of gastric cancer. Eur Rev Med Pharmacol Sci. 2020;24:3030–6. [DOI] [PubMed] [Google Scholar]
- 24. Yang F, Liu DY, Guo JT, Ge N, Zhu P, Liu X, et al. Circular RNA circ‐LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol. 2017;23:8345–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zong ZH, Du Y‐P, Guan X, Chen S, Zhao Y. CircWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR‐145 and miR‐1182. J Exp Clin Cancer Res. 2019;38:437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang X, Mei J, Wang H, Gu D, Ding J, Liu C. The emerging roles of circular RNAs in ovarian cancer. Cancer Cell Int. 2020;20:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu Y, Chen S, Zong ZH, Guan X, Zhao Y. CircRNA WHSC1 targets the miR‐646/NPM1 pathway to promote the development of endometrial cancer. J Cell Mol Med. 2020;24:6898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Du WW, Yang W, Li X, Awan FM, Yang Z, Fang L, et al. A circular RNA circ‐DNMT1 enhances breast cancer progression by activating autophagy. Oncogene. 2018;37:5829–42. [DOI] [PubMed] [Google Scholar]
- 29. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52:710–8. [DOI] [PubMed] [Google Scholar]
- 30. Deng G, Gao Y, Cen Z, He J, Cao B, Zeng G, et al. miR‐136‐5p regulates the inflammatory response by targeting the IKKbeta/NF‐kappaB/A20 pathway after spinal cord injury. Cell Physiol Biochem. 2018;50:512–24. [DOI] [PubMed] [Google Scholar]
- 31. Zhong Y, Yu C, Qin W. LncRNA SNHG14 promotes inflammatory response induced by cerebral ischemia/reperfusion injury through regulating miR‐136‐5p /ROCK1. Cancer Gene Ther. 2019;26:234–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen X, Shi Y, Xue P, Ma X, Li J, Zhang J. Mesenchymal stem cell‐derived exosomal microRNA‐136‐5p inhibits chondrocyte degeneration in traumatic osteoarthritis by targeting ELF3. Arthritis Res Ther. 2020;22:256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xie ZC, Li TT, Gan BL, Gao X, Gao L, Chen G, et al. Investigation of miR‐136‐5p key target genes and pathways in lung squamous cell cancer based on TCGA database and bioinformatics analysis. Pathol Res Pract. 2018;214:644–54. [DOI] [PubMed] [Google Scholar]
- 34. Liang M, Yao W, Shi B, Zhu X, Cai R, Yu Z, et al. Circular RNA hsa_circ_0110389 promotes gastric cancer progression through upregulating SORT1 via sponging miR‐127‐5p and miR‐136‐5p. Cell Death Dis. 2021;12:639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang D, Suo YJ, Gong L, Lv SQ. SKA2 promotes proliferation and invasion of hepatocellular carcinoma cells via activating the beta‐catenin signaling pathway. Neoplasma. 2020;67:743–50. [DOI] [PubMed] [Google Scholar]
- 36. Zhuang H, Meng X, Li Y, Wang X, Huang S, Liu K, et al. Cyclic AMP responsive element‐binding protein promotes renal cell carcinoma proliferation probably via the expression of spindle and kinetochore‐associated protein 2. Oncotarget. 2016;7:16325–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Su H, Ren F, Jiang H, Chen Y, Fan X. Upregulation of microRNA‐520a‐3p inhibits the proliferation, migration and invasion via spindle and kinetochore associated 2 in gastric cancer. Oncol Lett. 2019;18:3323–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1. Kaplan–Meier method was performed to analyze the prognosis of BC patients with high or low circ_0001387 expression.


