Soil salinity is a major threat to agricultural productivity. Plants have evolved sophisticated mechanisms to cope with salinity. The Salt Overly Sensitive (SOS) pathway plays an important role in salt tolerance by facilitating the extrusion of excess Na+ from cells. The core components of the SOS pathway in Arabidopsis include SOS1, SOS2, and SOS3. A salt-mediated increase in cytosolic calcium is perceived by the SOS3 EF-hand calcium-binding protein, leading to SOS2 activation. SOS2 is a Ser/Thr protein kinase that phosphorylates and activates SOS1, a Na+/H+ antiporter that facilitates Na+ efflux from the cells. How SOS1 is transcriptionally regulated in response to salt remained elusive. In this issue, Kai-Kai Lu and colleagues (Lu et al. 2023) found that C-type Cyclin1; 1 (CycC1; 1) forms a transcriptional repression complex with WRKY75 to downregulate SOS1, thus acting as a negative regulator of salt stress response in Arabidopsis (see Figure).
Figure.
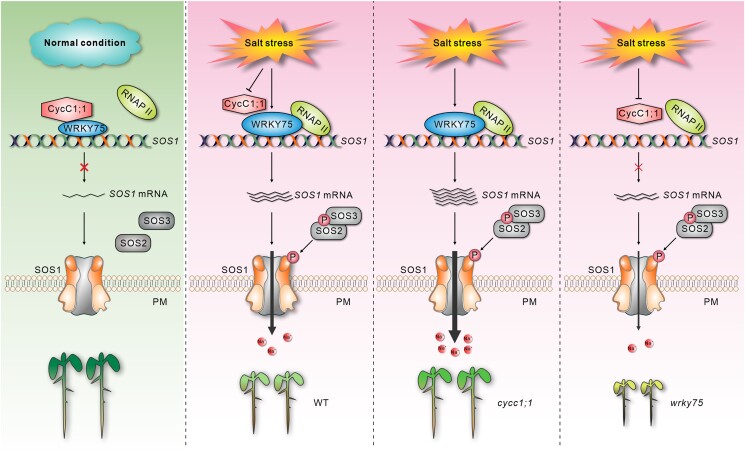
The proposed model describing CycC1; 1-WRKY75 mediated transcriptional regulation of SOS1. Under normal conditions, CycC1:1 interacts with WRKY75 to form a transcriptional repression complex that inhibits SOS1 gene expression. However, salt stress inhibits CycC1; 1 expression and enhances WRKY75 expression, leading to increased recruitment of RNAP II to the SOS1 promoter, activating its expression and enhancing salt tolerance. Adapted from Lu et al. (2023), Figure 8.
CycC1; 1 is a plant mediator protein complex subunit that influences gene transcription by acting as a bridge between transcription factors and RNA polymerase II (RNAP II) (Agrawal et al. 2021). CycC1; 1 is known to play a role in plant immunity by regulating defense-related genes (Zhu et al. 2014). Lu et al. (2023) found that a cycc1; 1 T-DNA insertion mutant was less sensitive to high salt than wild-type plants as measured by seed germination, cotyledon greening, and seedling growth parameters, suggesting that CycC1; 1 plays a role in the salt stress response. Using gene expression and GUS reporter assays, the authors showed that CycC1; 1 was expressed mainly in the root stele, similar to SOS1, during early developmental stages and that salt treatment decreased its expression, implying that CycC1; 1 acts as a repressor of the salt stress response.
Using a sodium-specific fluorescent dye to measure Na+ content in roots, the authors showed that Na+ accumulation increased upon salt treatment in cycc1; 1 mutant and wild-type plants, but the mutant accumulated significantly less Na+ than the wild-type plants. Whereas gene expression of all 3 core components of the SOS pathway (SOS1, 2, 3) was induced upon salt treatment in wild-type plants, SOS1 expression alone was induced to a much greater extent in the cycc1; 1 mutant, indicating that CycC1; 1 mainly influences SOS1 expression and the accumulation of SOS1 transcripts. Chromatin immunoprecipitation assays showed that CycC1; 1 binds to the SOS1 promoter. In addition, the amount of RNAP II associated with SOS1 genomic DNA after salt treatment was greater in cycc1; 1 than in wild type, suggesting that CycC1; 1 inhibits salt-induced SOS1 expression by interfering with the RNAP II association to the SOS1 promoter. The salt sensitivity phenotype of a cycc1; 1 sos1 double mutant further suggested that CycC1; 1 acts upstream of SOS1 in plant salt stress response.
Next, the authors used yeast 2-hybrid, bimolecular fluorescence complementation, and co-immunoprecipitation assays to identify WRKY75 as a CycC1; 1 interactor and electrophoretic mobility shift assay to show that WRKY75 binds to the W-box motif in the SOS1 promoter. In contrast to cycc1; 1 mutant plants, wrky75 mutants were salt sensitive and showed decreased expression of SOS1, indicating that WRKY75 also plays a role in salt-induced SOS1 expression. The authors also found that WRKY75 has a similar expression pattern as CycC1; 1 and SOS1, that is, mainly in the root stele. Using a dual LUC reporter assay, the authors found that CycC1; 1 interferes with WRKY75-mediated transcriptional activation of SOS1.
Together, Lu et al. (2023) show that CycC1; 1 and WRKY75 form a signaling module that facilitates the dynamic regulation of SOS1 expression. Under normal conditions, CycC1; 1 blocks WRKY75-mediated transcriptional activation of SOS1, whereas CycC1; 1 expression is suppressed and WRKY75 expression is upregulated under high salt, promoting SOS1 expression and leading to enhanced salt tolerance (see Figure). Further work is needed to identify the receptor and signaling components leading to the salt-induced downregulation of CycC1; 1. Also of interest, 2 more newly published studies shed light on the SOS2 component of the SOS signaling pathway and its connection to shade avoidance as well as salt tolerance through interaction with phytochrome A and B light receptors (Han et al. 2023; Ma et al. 2023). Thus the SOS pathway is emerging as a signaling hub in plant response to a variety of environmental conditions and stresses.
References
- Agrawal R, Jiří F, Thakur JK. The kinase module of the mediator complex: an important signalling processor for the development and survival of plants. J Exp Bot. 2021:72(2):224–240. 10.1093/jxb/eraa439 [DOI] [PubMed] [Google Scholar]
- Han R, Ma L, Lv Y, Qi L, Peng J, Li H, Zhou Y, Song P, Duan J, Li J, et al. The salt tolerance regulator SALT OVERLY SENSITIVE2 promotes shade avoidance by stabilizing the growth-promoting factors PIF4 and PIF5. Plant Cell. 2023. 10.1093/plcell/koad119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KK, Song RF, Guo JX, Zhang Y, Zuo JX, Chen HH, Liao CY, Hu XY, Ren F, Lu YT, et al. Cycc1; 1-WRKY75 complex-mediated transcriptional regulation of SOS1 controls salt stress tolerance in Arabidopsis. Plant Cell. 2023:35(7):2570–2591. 10.1093/plcell/koad105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Han R, Yang Y, Liu X, Li H, Zhao X, Li J, Fu H, Huo Y, Sun L, et al. Phytochromes promote plant salt tolerance by enhancing SOS2-mediated phosphorylation and degradation of PIF1 and PIF3 in the light. Plant Cell. 2023. 10.1093/plcell/koad117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Schluttenhoffer CM, Wang P, Fu F, Thimmapuram J, Zhu J-K, Lee SY, Yun D-J, Mengiste T. CYCLIN-DEPENDENT KINASE8 differentially regulates plant immunity to fungal pathogens through kinase-dependent and-independent functions in Arabidopsis. Plant Cell. 2014:26(10):4149–4170. 10.1105/tpc.114.128611 [DOI] [PMC free article] [PubMed] [Google Scholar]