Abstract
Formative asymmetric divisions produce cells with different fates and are critical for development. We show the maize (Zea mays) myosin XI protein, OPAQUE1 (O1), is necessary for asymmetric divisions during maize stomatal development. We analyzed stomatal precursor cells before and during asymmetric division to determine why o1 mutants have abnormal division planes. Cell polarization and nuclear positioning occur normally in the o1 mutant, and the future site of division is correctly specified. The defect in o1 becomes apparent during late cytokinesis, when the phragmoplast forms the nascent cell plate. Initial phragmoplast guidance in o1 is normal; however, as phragmoplast expansion continues o1 phragmoplasts become misguided. To understand how O1 contributes to phragmoplast guidance, we identified O1-interacting proteins. Maize kinesins related to the Arabidopsis thaliana division site markers PHRAGMOPLAST ORIENTING KINESINs (POKs), which are also required for correct phragmoplast guidance, physically interact with O1. We propose that different myosins are important at multiple steps of phragmoplast expansion, and the O1 actin motor and POK-like microtubule motors work together to ensure correct late-stage phragmoplast guidance.
The myosin XI protein OPAQUE1 is required for correct phragmoplast guidance during stomatal development and interacts with KINESIN12.
Introduction
Asymmetric divisions in plants and animals occur when a cell divides to give 2 daughters that differ from each other. Asymmetric divisions take place concomitantly with fate specification and are important in multicellular organisms to generate cell types of various fates, and to ensure the proper relative orientations of cells required for tissue patterning. Stomatal divisions from many plant species have served as a model for asymmetric cell division and have led to the discovery of a suite of proteins required for asymmetric division and fate regulation.
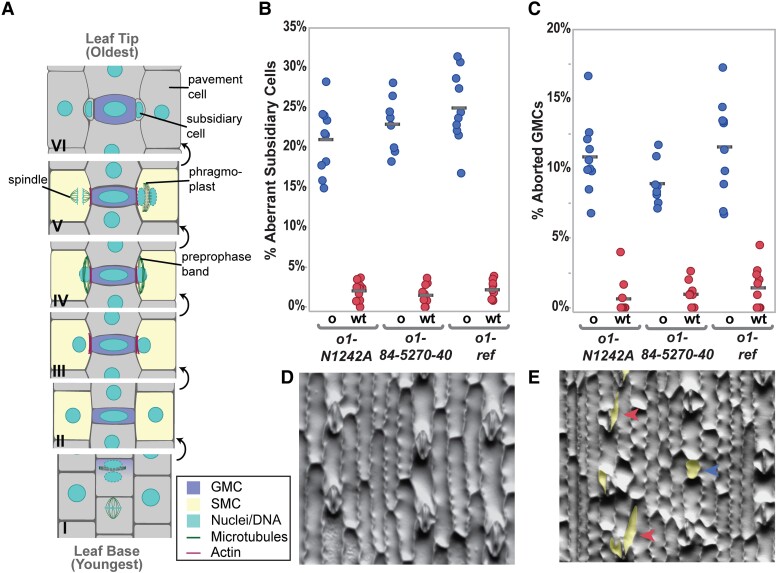
Stomata are always composed of 2 guard cells and may also consist of a variable number of subsidiary cells (Gray et al. 2020). Stomata from maize (Zea mays) and other grasses are made of 4 cells: 2 inner guard cells laterally flanked by a pair of outer subsidiary cells (Facette and Smith 2012; Hepworth et al. 2018; McKown and Bergmann 2020; Nunes et al. 2020). Three types of divisions are required to create a stomatal complex in grasses: (i) asymmetric division of a protodermal cell to form a guard mother cell (GMC) and an interstomatal cell (Fig. 1A, panel I); (ii) asymmetric divisions of 2 subsidiary mother cells (SMCs) flanking the GMC (Fig. 1A, panels III–V); and (iii) the symmetric but oriented division of the GMC (not shown in Fig. 1A). Subsidiary cells are believed to contribute to the rapid stomatal movements observed in grass species (Franks and Farquhar 2007; Gray et al. 2020; Nunes et al. 2020).
Figure 1.
OPAQUE1 is required for normal subsidiary cell formation in maize. A) Division sequence of stomatal development in maize and other grasses. B) Percentage of abnormal subsidiary cells and C) percentage of aborted GMCs in o1 mutants (o) and their wild-type siblings (wt). In (B and C) seeds were scored based on the appearance of their endosperm (o, seeds that are phenotypically opaque; wt, translucent seeds). Each data point represents the percentage of abnormal cells from 1 plant; between 100 and 200 cells were counted per plant. Gray horizontal bars indicate means. ANOVA comparing each mutant to wild-type sibling returned P < 0.0001 for all 3 alleles. D) Methacrylate impression of the leaf epidermis of a wild-type sibling showing normal stomatal complexes. E)o1-ref mutant showing abnormal subsidiary cells (red arrowheads) and aborted GMC (blue arrowhead). Abnormal cells in (E) are highlighted in yellow.
Stomatal development requires many coordinated cellular processes before and during the formative divisions. Mutants in grass species that fail to correctly form stomata have been identified in maize, rice (Oryza sativa), and purple false brome (Brachypodium distachyon). Grass mutants with abnormal stomatal development generally fall into 2 classes: mutants defective in genes important for cell fate specification (Liu et al. 2009; Raissig et al. 2016, 2017; Wang et al. 2019; Wu et al. 2019), or genes encoding proteins important for physically executing the asymmetric division. Mutants that fail to correctly execute stomatal divisions are classified based on which universal and temporally distinct phase of asymmetric division is defective: cell polarization, division plane establishment, division plane maintenance, or cytokinesis. The asymmetric division of the SMC, in particular, is useful as a model for dissecting the processes that occur during asymmetric division, as they have conspicuous polarization markers and the daughter cells are morphologically distinct.
Mutations in maize genes leading to defects in cell polarization include brick1 (brk1), brk2, and brk3 (Frank et al. 2003; Facette et al. 2015); pangloss2 (pan2) and pan1 (Cartwright et al. 2009; Zhang et al. 2012); and rho gtpase of plants2 (rop2), rop4, and rop9 (Humphries et al. 2011). These mutations lead to a nuclear polarization defect and aberrant division planes. In B. distachyon, BdPOLAR has the opposite localization of PAN1 (it is excluded from the GMC–SMC interface) and is required for correct SMC polarization (Zhang et al. 2022). Mutants in which SMCs polarize correctly, but fail in subsequent steps include the discordia (dcd) and tangled1 (tan1) mutants (Smith et al. 1996; Gallagher and Smith 1999; Martinez et al. 2017).
The microtubular preprophase band (PPB) is an early division site marker that disappears prior to metaphase. Identification of the microtubule-binding protein TANGLED1 (TAN1) answered a long-standing question of how the cortical division site was maintained throughout mitosis after the disappearance of the PPB (Smith et al. 1996; Walker et al. 2007). TAN1 and other division site markers are important for division plane maintenance and continuously mark the division site from prophase until telophase (Cleary and Smith 1998; Walker et al. 2007; Xu et al. 2008; Stöckle et al. 2016; Martinez et al. 2017). The TAN1-interacting partners PHRAGMOPLAST ORIENTING KINESIN1 (POK1) and POK2 are kinesin proteins that have been characterized in Arabidopsis (Arabidopsis thaliana) and mark the division site (Müller et al. 2006; Lipka et al. 2014; Herrmann et al. 2018). POK2 and TAN1 localize to the phragmoplast as well as the cortical division site (Herrmann et al. 2018; Buschmann and Müller 2019; Bellinger et al. 2021; Mills et al. 2022).
Cortical division site markers explain how the division site is maintained throughout mitosis. However, for division plane fidelity, the phragmoplast must be correctly guided to the correct division site. The phragmoplast is composed of membranes, microtubules, actin, and other accessory proteins, which starts as a disc in the center of the cell (Smertenko et al. 2018; Buschmann and Müller 2019). Vesicles targeted to the phragmoplast fuse to form the cell plate. The phragmoplast expands in circumference until it eventually meets the existing cell wall at the cortical division site marked by TAN1, POK, and other division site markers (Smertenko et al. 2018). Phragmoplast expansion (reviewed in Smertenko 2018; Smertenko et al. 2018; Buschmann and Müller 2019; Lee and Liu 2019)) requires microtubule turnover and many different mutants affect phragmoplast stability, morphology, or guidance.
Phragmoplast guidance occurs in stages. The initial rapid expansion of the disc-phragmoplast is likely guided by actin networks (Molchan et al. 2002; van Oostende-Triplet et al. 2017). Phragmoplast expansion slows when 1 edge meets the cell cortex and eventually, through expansion, the entire circumference of the phragmoplast reaches the cell cortex (van Oostende-Triplet et al. 2017). In this later stage, microtubules at the cell cortex are incorporated into the phragmoplast, where TAN1 (and probably other division site-localized proteins) are important for the incorporation of cortical telophase microtubules into the phragmoplast (Bellinger et al. 2021). After phragmoplast expansion is complete, the newly formed cell plate fuses with the existing cell wall (Smertenko et al. 2017; van Oostende-Triplet et al. 2017; Smertenko 2018; Lee and Liu 2019). Importantly, early phragmoplast guidance, late phragmoplast guidance, and phragmoplast integrity are distinct from one another. Despite recent progress using mutants and time-lapse imaging, the precise mechanism and protein–protein interactions that promote correct phragmoplast guidance remain unclear.
Using maize stomatal precursors as a model, we wanted to understand how plant cells correctly execute asymmetric divisions. Actin–myosin networks are prominent during plant cell division generally (Sadot and Blancaflor 2019), and specifically during the asymmetric division of the maize SMC (Facette et al. 2015). Actin accumulates in the PPB and the spindle of plant cells, and accumulates on either side of the division zone (Palevitz 1987; Sano et al. 2005; Yasuda et al. 2005; Van Damme et al. 2007; Panteris 2008; Kojo et al. 2013). F-actin, myosin VIII, and myosin XI localize to the spindle and phragmoplast (Wu and Bezanilla 2014; Abu-Abied et al. 2018; Sun et al. 2018). Actin–myosin networks play additional roles specific to asymmetric SMC divisions. Indeed, in grasses and monocots with similar stomatal development, nuclear migration is driven by actin networks (Cho and Wick 1991; Kennard and Cleary 1997; Apostolakos et al. 2018). Actin is polarized at the GMC–SMC interface, and the SCAR/WAVE complex—which promotes actin nucleation—is required for the polarization of PAN proteins (Facette et al. 2015). These observations indicate that the actin motors (i.e. myosins) are likely important for SMC divisions, and could potentially play roles during polarization, division plane establishment, and/or cytokinesis. Therefore, we investigated the role of the previously identified OPAQUE ENDOSPERM1 (O1) protein in asymmetric divisions.
The opaque class of mutants was originally identified based on their seed phenotype of opaque endosperm (Neuffer et al. 1968; Gibbon and Larkins 2005). O1 encodes a myosin XI protein required for normal endoplasmic reticulum (ER) and protein body morphology in developing seeds, although the gene is expressed throughout the plant (Wang et al. 2012). Notably, O1 is very similar to Arabidopsis MYOXI-I, which is required for nuclear movement and shape (Tamura et al. 2013; Zhou et al. 2015; Muroyama et al. 2020). In Arabidopsis stomatal precursors, pre-mitotic nuclear migration is driven by microtubule (rather than actin) networks, but postmitotic migration of the nucleus is driven by actin networks and MYOXI-I (Muroyama et al. 2020).
We hypothesized that O1 might play a role in the asymmetric division of maize SMCs, perhaps during pre-mitotic polarization of the nucleus. Indeed, we determined that asymmetric divisions of both SMCs and GMCs are abnormal in o1 mutants. However, division defects in o1 are not a result of cell polarization defects, but rather late-stage phragmoplast guidance defects. To gain insight into how O1 promotes correct phragmoplast guidance, we identified proteins that physically interact with O1, leading to the identification of the maize orthologs of POK1 and POK2, in addition to actin-binding proteins and other myosins. Given their physical interaction, and the similarity of pok mutant phenotypes in Arabidopsis to the phenotypes we observed in maize o1 mutants, we hypothesize these 2 cytoskeletal motors work together to promote phragmoplast guidance.
Results
Opaque1 is required for stomatal divisions
To determine if the myosin OPAQUE1 was involved in stomatal divisions, we examined the morphology of subsidiary cells in fully expanded juvenile leaves (leaf 4). We phenotyped segregating F2 plants for opaque seeds and calculated the corresponding frequency of abnormal subsidiary cells in wild-type (both o1/+ and +/+) and o1 sibling plants. We examined 3 different o1 mutant alleles. Between 20% to 30% of subsidiary cells were abnormal in o1, while siblings with translucent seed (i.e. wild type) had less than 5% abnormal subsidiary cells (Fig. 1B). This phenotypic penetrance was consistent with that seen in other mutants such as brk, pan, and dcd, which also show ∼25% abnormal subsidiary cells (Gallagher and Smith 1999; Cartwright et al. 2009; Zhang et al. 2012; Facette et al. 2015). All 3 alleles also displayed an increased frequency of aborted GMCs (Fig. 1C). We classified GMCs as aborted when a single cell with the same morphology as a GMC was present instead of a normal 4-celled stomatal complex (blue arrow, Fig. 1E).
Aberrant subsidiary cells and aborted GMCs in o1 suggest a role for this myosin in both types of stomatal formative asymmetric divisions. To confirm the defect, we compared o1 and wild-type siblings at the early stages of leaf development, when stomatal divisions are taking place. We stained cell walls and nuclei with propidium iodide (PI), and plasmodesmata and new cell plates with aniline blue. In o1 mutant plants, recently formed GMCs showed abnormal division planes (Fig. 2, A and B). We propose that aborted GMCs are present in expanded leaves when the GMC progenitor divides abnormally, resulting in failed GMC fate specification. We also observed abnormal division planes in recently formed SMCs (Fig. 2, C and D). Importantly, we noticed no multinucleate cells or cell wall stubs, as we only observed abnormal division planes. This result suggests that this specific myosin is not required for cell plate integrity, but rather it is the division plane that is affected. These data indicate a role for O1 during the asymmetric division of stomatal precursors.
Figure 2.
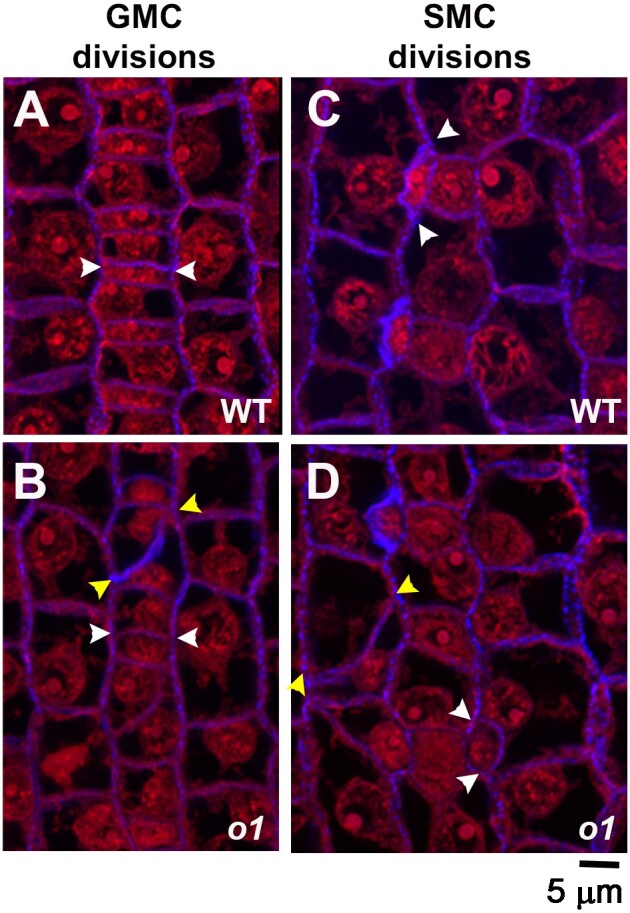
Stomatal lineage cells have abnormal division planes in o1. The region of developing leaf 4 undergoing stomatal divisions was dissected, fixed, and stained with PI (red) and aniline blue (blue). Segregating wild-type (WT; A, B) and homozygous sibling o1-N1242A mutants (o1;B, D) are shown. White arrowheads mark correct divisions; yellow arrowheads mark incorrect divisions. A, B) Recently formed GMCs. C, D) Recently formed subsidiary cells, formed from SMCs. Z-projection of 3 confocal images. Scale bar, 5 µm.
At what point does O1 play a role in determining the division plane: cell polarization, division plane establishment, division plane maintenance, or cytokinesis? We examined o1 SMCs to determine if failed polarization caused the division plane defect. Polarization in SMCs occurs in a series of ordered steps with BRK1, PAN2, PAN1, and ROP proteins each becoming polarized sequentially, and each protein is required for the next to become polarized (Cartwright et al. 2009; Humphries et al. 2011; Zhang et al. 2012; Facette et al. 2015). Actin patch formation and nuclear migration are the last steps of cellular polarization (Facette et al. 2015). We assayed PAN1-YFP polarization, actin patch formation, and nuclear migration in o1 and wild-type siblings to determine if cell polarization was normal. PAN1-YFP becomes polarized in SMCs prior to nuclear migration and remains polarized throughout division until after the subsidiary cell is formed (Cartwright et al. 2009). We analyzed recently divided SMCs for PAN1-YFP polarization and classified the daughter as having correct or incorrect division planes. PAN1-YFP polarized normally in all recently divided SMCs in o1 (209/209), regardless of whether the SMC divided normally (136 cells) or abnormally (73 cells) (Fig. 3, Supplemental Table S1). When a GMC progenitor cell divided abnormally and failed to form a morphologically normal GMC, adjacent cells did not polarize PAN1-YFP—presumably because GMC fate was not correctly specified (Fig. 3D).
Figure 3.
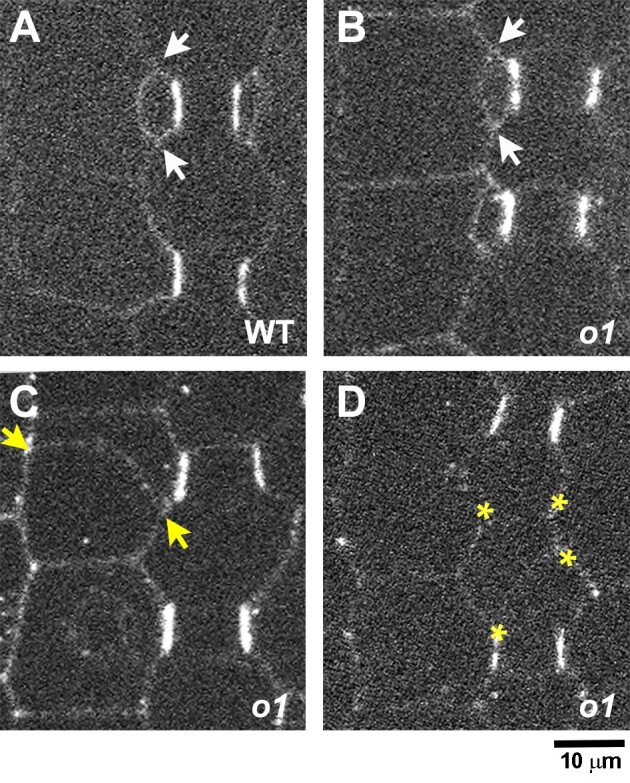
PAN1-YFP polarizes correctly in o1. Recently formed subsidiary cells from o1-ref plants and wild-type siblings expressing PAN1-YFP were assayed for PAN1-YFP polarization. Arrows in (A–C) indicate correctly (white) or incorrectly (yellow) oriented cell walls generated from an SMC division. A) Recently divided SMC with polarized PAN1-YFP at the GMC-subsidiary cell interface. B) Correctly formed subsidiary cell from o1. PAN1-YFP is correctly polarized. C) Incorrectly oriented cell wall generated from an aberrant SMC division. PAN1-YFP is correctly polarized. D) Incorrectly oriented cell wall generated from an aberrant division of the GMC progenitor cell. Yellow asterisks in (D) indicate 4 corners of a cell formed from an aberrant GMC-generating division. Scale bar, 10 µm.
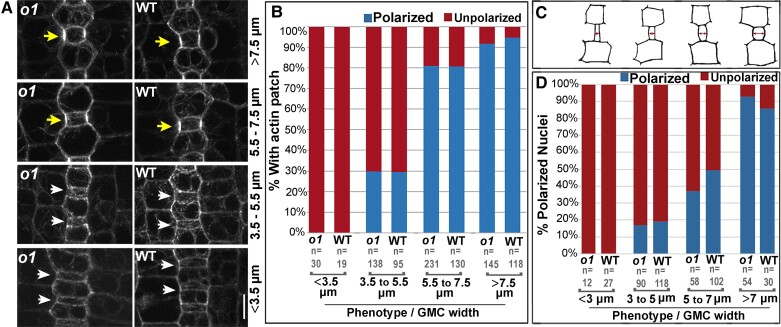
We also assayed whether the polar accumulation of actin and nuclear migration to the division site occurred normally in o1 (Fig. 4, Supplemental Fig. S1). We hypothesized that nuclear migration, in particular, may be affected in o1 mutants, as different myosin XI isoforms have been previously shown to be important for nuclear positioning (Tamura et al. 2013; Ali et al. 2020; Muroyama et al. 2020). We assayed actin and nuclear polarization at different developmental stages. As stomatal development proceeds, GMC width increases. Therefore, GMC width can be used as a proxy for the developmental state (Fig. 4, A and C). We quantified the number of SMCs with polarized actin patches (Fig. 4, A and B) and polarized nuclei (Fig. 4D, Supplemental Fig. S1) in o1 and wild-type siblings and observed normal polarization in o1. Actin patch formation and nuclear polarization are the last known steps of polarization and depend on the polarization of earlier factors, implying earlier factors that were not examined (BRK1, PAN2, and ROP) also polarized normally. Together, the data indicate that polarization is normal in o1, and the defect that leads to abnormal asymmetric divisions in o1 SMCs occurs post-polarization.
Figure 4.
Actin patch formation and nuclear migration is normal in o1. A) The stomatal division zone was examined in leaf 4 of o1-N1242A mutant plants and wild-type siblings expressing the ABD2-YFP marker. GMCs in early developmental stages, found at the leaf base (lower panels), are narrow and width increases towards the leaf tip as development proceeds (upper panels). Early SMCs flanking narrow GMCs do not form an actin patch (white arrows). SMCs at later developmental stages, flanking wider GMCs, have an actin patch (yellow arrows). B) Percentage of SMCs with a polarized actin patch at progressive developmental stages in o1-N1242A mutant plants and their corresponding wild-type siblings. C) Cartoons depicting representative cell outlines at increasing GMC widths. Red arrows indicate where GMC width was measured. D) Percentage polarized nuclei in SMCs at progressive developmental stages in o1-N1242A mutant plants and their corresponding wild-type siblings. Fisher's exact tests comparing o1 mutants to their respective wild-type siblings indicate no differences between mutants and wild type at each developmental stage (P > 0.05 in all cases).
OPAQUE1 localizes to the phragmoplast
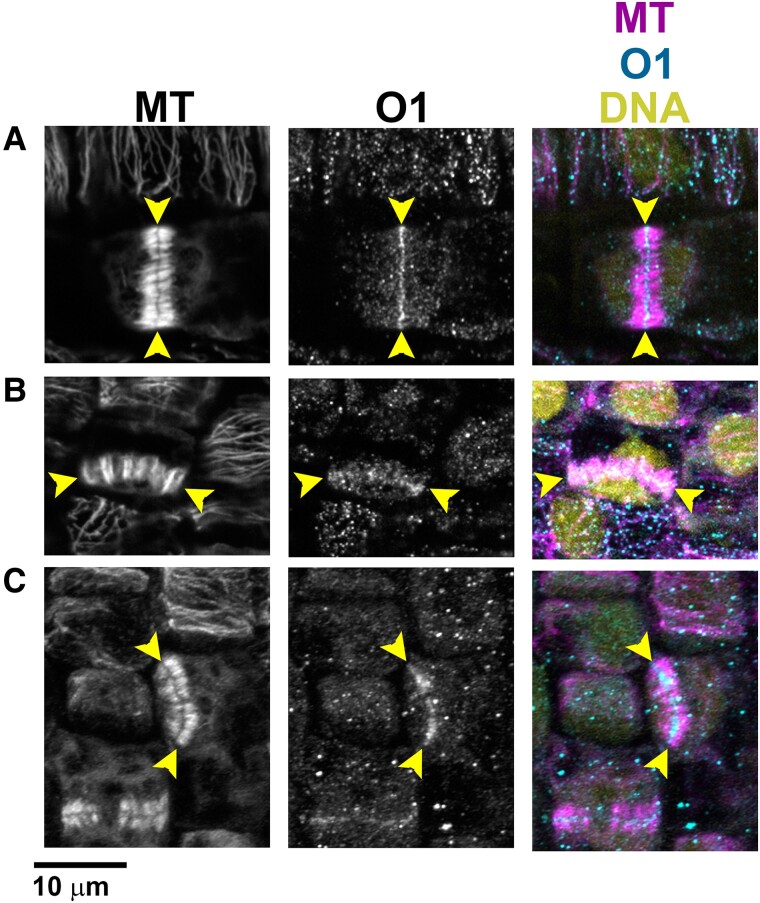
If O1 is not required for cell polarization, it must be required at a later stage of asymmetric cell division—specifically during division plane establishment, division plane maintenance, and/or cytokinesis. We used immunofluorescence to determine O1 localization during cell division, to ascertain at what stage of the cell cycle it might be important. We generated an O1-specific peptide antibody and performed co-immunostaining for O1 and microtubules (Fig. 5, A to C; Supplemental Fig. S2). We observed specific staining, present only in wild-type but not mutant siblings, at the phragmoplast midline. O1 localized to phragmoplasts in all dividing cells we examined, including symmetrically dividing pavement cells (Fig. 5A), asymmetrically dividing GMC progenitor cells (Fig. 5B), and asymmetrically dividing SMCs (Fig. 5C). Previously, myosin XI isoforms were shown to localize to the phragmoplast, spindle, and cell cortex (Abu-Abied et al. 2018; Sun et al. 2018); phragmoplast localization was previously shown for a myosin VIII protein (Wu and Bezanilla 2014). We did not observe reproducible or specific staining in the PPB, spindle, or at the cortical division zone in dividing cells (Supplemental Fig. S2). However, the background signal was high and, therefore, any faint staining (such as in the spindle midzone or cell cortex) would be difficult to detect. These data suggest that O1 plays a role during cell division, especially in the phragmoplast during cytokinesis.
Figure 5.
O1 localizes to phragmoplasts. Immunofluorescence detection of microtubules and O1 in wild-type cells (A to C). All samples are from the division zone of developing leaf 4. O1 is detected in phragmoplasts of symmetrically dividing cells (A), asymmetrically dividing stomatal lineage cells that will form GMCs (B), and asymmetrically dividing SMCs (C). Yellow arrowheads indicate phragmoplast ends. Scale bar, 10 µm.
Phragmoplasts are misguided in o1 mutants
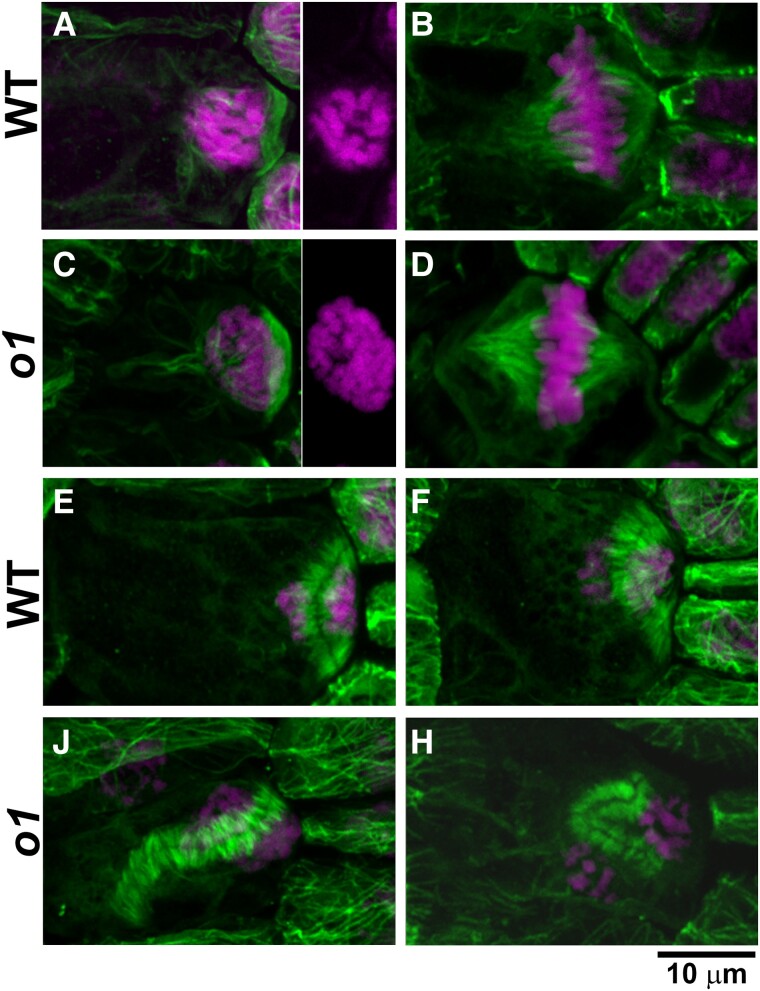
Since O1 localizes to the phragmoplast and the mutant has a post-polarization defect, we wanted to know if any division structures—especially the phragmoplast—were abnormal in o1 mutants. We examined microtubule division structures using immunofluorescence on SMCs from developing leaf 4 (Fig. 6, D to K). We observed no abnormal PPBs in immunostained o1 cells (0/55) (Fig. 2, D and F). Spindles persist only briefly and, therefore, were rare, but were always normal in o1 (0/8) (Fig. 2, E and G). We observed abnormal late-stage phragmoplasts in some o1 SMCs (18/52 = 35%) (Fig. 2, H to K). Abnormal phragmoplasts were misguided and did not localize at the expected site of division. However, the phragmoplast midline and microtubule alignment within the phragmoplast appeared normal and we saw no evidence for destabilized or fragmented phragmoplasts. This result is consistent with the absence of cell wall stubs in o1 and suggests normal phragmoplast assembly. The misguided phragmoplasts showed either small deviations or large deviations in the division plane. Notably, all misguided phragmoplasts appeared to be correctly oriented at 1 edge, i.e. 1 edge of the phragmoplast was always anchored at the expected SMC division site. Because O1 is an actin motor, we also examined actin in o1 mutants. The maize ACTIN-BINDING DOMAIN 2 (ABD2)-YFP marker used to assess actin patch formation does not localize to phragmoplasts (Sutimantanapi et al. 2014); therefore, we used fluorescently labeled phalloidin staining on fixed cells. Phragmoplast structure and orientation in phalloidin-stained cells was similar to microtubule-stained cells—while some were normal, a subset of phragmoplasts were misguided (Supplemental Fig. S3).
Figure 6.
O1 is required for normal phragmoplast guidance. Immunofluorescence detection of microtubules (green) and DAP-stained nuclei (magenta) in wild-type (A, B, E, F) and o1 cells (C, D, J, K). PPBs in SMCs (A, C) appear similar in wild-type siblings and o1-N1242A. Side panels in (A and C) show DAPI channel only to show condensed chromosomes. Spindles in SMCs (B, D) were similar in wild-type and o1 SMCs. In o1, SMC phragmoplasts appeared as in wild type (E, F) or misguided (J, H). Z-projections of 40 to 60 images. Scale bar, 10 µm applies to all images.
The discordia class of mutants displays post-polarization defects during SMC divisions (Gallagher and Smith 1999). Both dcd1 and dcd2 mutants are similar to o1 in that they have aberrant GMC divisions, and normal nuclear polarization in SMCs (Gallagher and Smith 1999, 2000). The Dcd2 gene has not yet been identified, but was mapped to chromosome 4, and O1 lies within the mapping interval (Supplemental Fig. S4). Examination of dcd2 seeds revealed that their endosperm is opaque. Complementation crosses between dcd2 and 2 o1 mutant alleles indicated that dcd2, which was identified based on subsidiary cell defects (Gallagher and Smith 1999), is allelic to o1 (Supplemental Fig. S4).
Live cell imaging of opaque1 mutants indicates that phragmoplasts are misguided
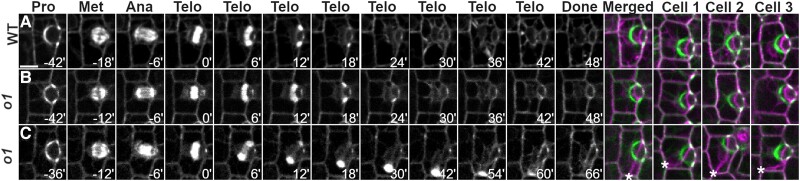
To confirm that the abnormally divided cells seen in o1 were a result of abnormal phragmoplast guidance, and to determine when phragmoplasts become misguided, we performed time-lapse imaging of dividing SMCs. CFP-TUBULIN (CFP-TUB) or YFP-TUB were crossed into o1 mutants. We directly compared the location of the PPB, phragmoplast, and newly formed cell wall in dividing SMCs. These live markers also allowed us to observe PPBs and spindles to confirm our immunostaining results. We imaged cells in developing leaf 5 or 6 from 2 o1 alleles and their corresponding wild-type siblings from prophase until the end of cytokinesis (Fig. 7 and Supplemental Fig. S5). In wild type, all dividing SMCs formed new cell walls that aligned with the former location of the PPB (n = 76) (Fig. 7A and Supplemental Movie S1). Division proceeded normally (including normal PPBs and spindles) in all o1 mutant cells until telophase. In o1-N1242A mutants, ∼35% (n = 28/81) of divisions displayed misguided phragmoplasts (Fig. 7B). In all cases, the initial site of contact between the phragmoplast edge was always aligned with the site predicted by the PPB. This finding was consistent with our immunofluorescence data (Fig. 6), where 1 edge of the phragmoplast was always correctly oriented. However, as the phragmoplast continued to expand, the phragmoplast would “fall off track” in some cells and become misguided. The most severe division plane defects occurred when the phragmoplast became misguided shortly after initial contact with the existing cell wall (Supplemental Movie S2); small defects occurred when the phragmoplast became misguided near the completion of expansion (Supplemental Movie S3). We obtained similar results with the o1-5270-84 allele (Supplemental Fig. S5). These data indicate that (i) initial phragmoplast guidance, prior to first contact with the cortex, is separable from late-stage guidance and (ii) in o1, only late-stage phragmoplast guidance is defective.
Figure 7.
Time-lapse imaging confirms a phragmoplast guidance defect in o1. CFP-TUB was used to observe progression of cell division SMCs from leaf 5 or 6 in o1-N1242A and corresponding wild-type siblings. A) Wild-type cell division. B) Correctly oriented o1 cell division. C) Misoriented o1 cell division. Pro, prophase; Met, metaphase; Ana, anaphase; Telo, telophase; Done, completed division; Merged, overlay of prophase (green) and completed division. Cells 1 to 3 show 3 additional representative cells. Time (min) is listed at the bottom of each image. Misoriented cell walls are indicated by asterisks. Z-projections of 6 images. All cells displayed at the same magnification; scale bar in (A), 10 µm.
The myosin O1 interacts with the maize orthologs of the kinesins POK1 and POK2
To gain insight into how O1 might be influencing phragmoplast guidance, we identified proteins that interact with O1 using co-immunoprecipitation followed by mass spectrometry (co-IP/MS). We extracted membrane and membrane-associated proteins from the stomatal division zone of o1 plants and their wild-type siblings. We performed 3 biological replicates using the same antibody used in immunostaining. We also performed a second set of 3 independent biological replicates using a second anti-O1 antibody generated from an independent rabbit. Both antibodies identified 2 protein bands present in wild type but not in their o1 mutant siblings (Supplemental Fig. S6). Our MS analyses isolated 2 isoforms of O1 that have predicted sizes corresponding to these 2 bands. We considered proteins among the immunoprecipitates showing a more than 2-fold increase in abundance in the wild-type samples relative to those from their o1 siblings, using both antibodies, as likely O1-interactors (Supplemental Data Set 1).
High-confidence interactors included many actin-associated proteins such as other myosins (including both myosin VIII and myosin XI family members), Actin Related Protein 2/3 complex (ARP2/3) proteins, and villin (Supplemental Data Set 1). We did not identify other actin-associated proteins such as fimbrin, NETWORKED, formins, or cofilin/actin-depolymerizing factors, suggesting that the observed interactions are likely specific. Notably, we identified the known myosin interactor MadA1 (myosin adaptors of family A) (Kurth et al. 2017).
While we identified many actin-associated proteins, the only microtubule-associated proteins identified were 2 paralogous kinesins, KIN12C and KIN12D. We detected a third closely related kinesin, KIN12E, in the immunoprecipitates from 1 antibody, but was just below the threshold cutoff for the second antibody, making it another but lower-confidence potential interactor (Table 1). These 3 proteins are related to Arabidopsis POK1, POK2, and KIN12E (Müller et al. 2006; Lipka et al. 2014; Herrmann et al. 2018; Herrmann et al. 2021). Notably, these 3 maize proteins were all previously identified as direct interactors of maize TAN1 (Müller et al. 2006). TAN1 (in both Arabidopsis and maize), AtPOK1, and AtPOK2 all positively mark the cortical division site during cell division. These 3 proteins all localize to the phragmoplast and have phragmoplast guidance defects (Müller et al. 2006; Walker et al. 2007; Martinez et al. 2017; Herrmann et al. 2018; Mills et al. 2022). It is possible that the interaction between O1 and maize POK orthologs is important for phragmoplast guidance.
Table 1.
Co-IP/MS results for O1 as bait protein and 3 related kinesin-like proteins
| Accession | Name | Antibody 1 | Antibody 2 | ||
|---|---|---|---|---|---|
| WT peptides | o1 peptides | WT peptides | o1 peptides | ||
| Zm00001d052110_P042 | OPAQUE1 | 546/435/546 | 0/0/0 | 474/493/458 | 0/0/0 |
| Zm00001d052110_P007 | OPAQUE1 | 258/229/268 | 0/0/0 | 230/232/213 | 0/0/0 |
| Zm00001d022276_P012 | KINESIN-LIKE PROTEIN 12C | 29/3/18 | 0/0/0 | 36/56/8 | 1/0/0 |
| Zm00001d041353_P002 | KINESIN-LIKE PROTEIN 12D | 12/6/9 | 0/0/0 | 16/24/5 | 0/0/0 |
| Zm00001d034030_P017 | KINESIN-LIKE PROTEIN 12E | 3/3/3 | 0/0/0 | 7/9/6 | 4/2/0 |
Co-IPs were performed using two independently generated antibodies against O1. For each antibody, 3 biological replicates were conducted using wild-type plants and o1-N1242A mutants as a negative control. The number of peptides identified for each replicate are separated by a slash. At least 2 isoforms of O1 are present in developing leaves; no peptide corresponding to O1 was identified in homozygous o1 mutants. The POK-like kinesins KIN12C and KIN12D were found only in wild-type samples and are considered plausible interactors. Fewer peptides were found for the related KIN12E and did not meet the threshold for a probable high-confidence interactor.
Although we replicated the interaction of O1 and POK-like KIN12 proteins with 2 different antibodies, we also determined if these proteins can interact using a yeast 2-hybrid (Y2H) assay (Supplemental Fig. S7). Myosin and kinesin proteins are large; in our hands, the full-length cDNA of O1 was lethal to E. coli. The KIN12C and KIN12D coding sequences are also large with 5,766 and 8,163 bp, respectively. Therefore, we used the C-terminal tails of O1, KIN12C, and KIN12D, which excludes the N-terminal motor domains. However, the C-terminal portion of KIN12C autoactivated in our Y2H assay, and the C-terminal portion of KIN12D did not show an interaction with O1. We used the full-length version of KIN12E, which is much smaller than KIN12C and KIN12D. O1 and KIN12E showed a positive interaction in our Y2H conditions (Supplemental Fig. S7).
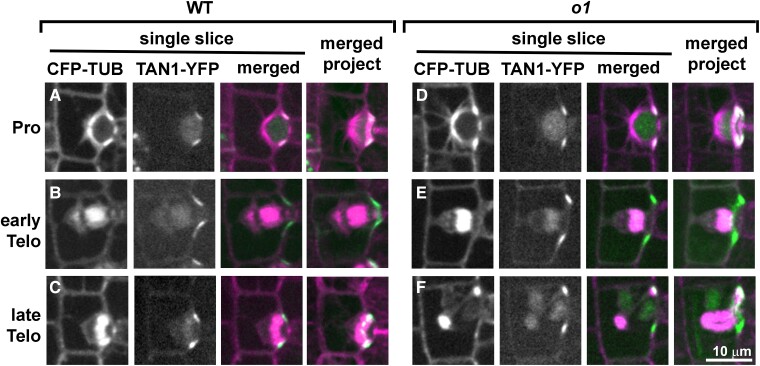
Since POK proteins are division site markers, we wanted to know if the division site was being correctly maintained in o1. A failure to maintain the division site might explain the late-stage phragmoplast guidance defect. We used a previously characterized ZmTAN1-YFP marker line (Martinez et al. 2017) co-expressed with CFP-TUBULIN to determine if division plane maintenance was normal in o1 mutants in different phases of mitosis. Importantly, in Arabidopsis, TAN1 localization to the division site is dependent on POK (Lipka et al. 2014). Therefore, if TAN1 localization is normal, we speculate that POK localization and division site specification in general is also normal. We separated telophase cells into early telophase, before the phragmoplast initially meets the cortex; and late telophase, when the defect in o1 occurs. In wild-type SMCs, TAN1-YFP always correctly marked the predicted division plane throughout mitosis from prophase (Fig. 8A) through early telophase (Fig. 8B) and late telophase (Fig. 8C; Supplemental Table S2; n = 154 total cells). In o1 mutants, TAN1-YFP always marked the correct division site from prophase (Fig. 8D) through early telophase (Fig. 8E) and late telophase (Fig. 8F), even in 44 late telophase cells where the phragmoplast became misguided (Supplemental Table S2; n = 231 total cells). In rare cases, we observed TAN1-YFP at an additional site during early telophase (2/231 cells; Supplemental Fig. S8). Since this additional TAN1-YFP localization occurred only rarely, and we always saw correct TAN1-YFP localization in late telophase cells (when we see the phragmoplast guidance defect), ectopic TAN1 localization cannot be the primary cause of the o1 phragmoplast guidance defect. Since the asymmetric division of GMC progenitor cells is also abnormal, we also examined TAN1 and phragmoplast localization during these divisions. Similar to SMC divisions, the division plane was correctly marked in divisions of GMC progenitor cells (Supplemental Fig. S9). These data indicate that O1 is not required for correct division site maintenance and specification. Rather, O1 is required for the phragmoplast to be guided to the specified division site during cytokinesis.
Figure 8.
TAN1-YFP correctly marks the division plane during normal and abnormal o1 SMC divisions. Dividing SMCs from leaf 5 or 6 in wild-type siblings (A to C) or o1-N1242A(D to F) cells co-expressing CFP-TUB (magenta) and TAN1-YFP (green). Single planes are shown in the first 3 panels and a full projection is shown in the last panel. In wild-type cells, TAN1-YFP correctly marked the predicted division plane throughout mitosis including prophase (A; n = 33/33), metaphase (n = 30/30), anaphase (n = 12/12), early telophase (B; n = 21/21), and late telophase (C; n = 58/58). In o1-N1242A mutant cells, TAN1-YFP always correctly marked the division plane in prophase (D; n = 85/85), metaphase (n = 20/20), anaphase (n = 13/13), and early telophase (E; n = 32/32). In 2/32 cases, TAN1-YFP was also seen at an additional site during early telophase (see Supplemental Fig. S10). During late telophase, TAN1-YFP was at the cortical division site in o1 SMCs with correctly oriented phragmoplasts (n = 38/38) and incorrectly oriented phragmoplasts (F; n = 44/44). Scale bar, 10 µm applies to all images.
Discussion
We characterized the myosin XI protein OPAQUE1/DCD2 as essential for late-stage phragmoplast guidance during maize stomatal asymmetric divisions. Myosin XI family proteins are implicated in organelle trafficking and motility, cytoplasmic streaming, tip growth, auxin response, gravitropism, growth and division plane orientation (Sparkes et al. 2008; Avisar et al. 2009; Ueda et al. 2010; Vidali et al. 2010; Madison et al. 2015; Talts et al. 2016; Abu-Abied et al. 2018; Nebenführ and Dixit 2018; Olatunji and Kelley 2020). O1 was originally isolated as a mutant with opaque seed endosperm (Wang et al. 2012), which has abnormal protein bodies that are potentially caused by defects in ER motility. Recently, it was discovered that o1 also has defects in plant height and tassel branching (Zebosi 2022). Together with the data presented here, this suggests O1 influences multiple cellular processes. Although myosin XIs participate in nuclear positioning in other cell types (Tamura et al. 2013; Muroyama et al. 2020), O1 was not required for pre-mitotic nuclear migration in maize SMCs—either because O1 does not play a role, or its role is masked by genetic redundancy. The asymmetric division plane defect we observed in o1 was attributable to a late-stage phragmoplast guidance defect, consistent with localization of O1 to the phragmoplast midline. This phragmoplast guidance defect is phenotypically similar to Arabidopsis pok1 pok2 double mutants (Lipka et al. 2014).
In addition to O1, many other proteins localize to the phragmoplast. Cell plate or phragmoplast localization has been observed for both myosin VIII and XI proteins—some of these myosins also localize to the PPB, cortical division site, spindles, nuclei, and other organelles (Miller et al. 1995; Reisen and Hanson 2007; Sattarzadeh et al. 2008; Wu and Bezanilla 2014; Zhou et al. 2015; Abu-Abied et al. 2018; Duan and Tominaga 2018). It was previously observed that Arabidopsis MYOSIN XI-K localizes to the cortical division site, spindle, and phragmoplast; in the same article, a knockout in 3 myosin XI genes led to aberrant division planes in roots (Abu-Abied et al. 2018). However, no clear mechanism was advanced to explain how XI-K might lead to aberrant division planes. Indeed, given the complex localization pattern of this protein family, there are several potential roles for myosins during division. We showed here that in the case of O1, the division plane defect was specifically due to late-stage phragmoplast guidance. Although the role for O1 appears to be specific, it is plausible that myosins have other roles in cell division and that multiple myosins might fulfil the same role. Like XI-K, a myosin VIII from moss (Physcomitrium patens) localized to multiple mitotic structures, and the corresponding mutants also had a phragmoplast guidance defect, including slower vesicle delivery (Wu and Bezanilla 2014). Treatment with a general myosin inhibitor alters division planes (Molchan et al. 2002), cellular polarity (Holweg et al. 2003), and even resulted in incomplete divisions (Molchan et al. 2002). We did not observe polarity defects or failed divisions; together, these data suggest multiple roles for myosins during division and cytokinesis. Interestingly, we observed physical interactions between O1 and multiple other myosins, including both class VIII and XI myosins. Careful phenotypic analyses of different myosin mutants will help unravel the roles that each actin–myosin network plays during cell division.
Our data confirm that phragmoplast guidance is dynamic and that different proteins likely play different roles at different stages. Dividing cells treated with caffeine are unaffected in early-stage phragmoplast guidance, but the phragmoplasts disintegrate at later stages (Valster and Hepler 1997), often resulting in multinucleate cells. This observation led to the conclusion that phragmoplast guidance occurs in (at least) 2 steps (Valster and Hepler 1997) and is coupled with the observation that actin filaments connect the phragmoplast leading edge and the cell cortex (Valster and Hepler 1997; Wu and Bezanilla 2014). Indeed, studies of phragmoplast guidance in tobacco (Nicotiana tabacum) BY-2 cells indicate that rates of phragmoplast expansion vary, and slow considerably at the last stage of expansion when the leading edge first strikes the cortex (van Oostende-Triplet et al. 2017). At this final stage, the phragmoplast is more sensitive to actin depolymerization via latrunculin B (van Oostende-Triplet et al. 2017). It is also during this late stage that cortical telophase microtubules are incorporated into the phragmoplast (Mills et al. 2022). Since the phragmoplast always correctly meets the cortical division site at the initial site of contact in o1 mutants, it is this last stage of expansion where O1 plays its role.
The identification of O1-interacting partners provides clues as to O1 functions. Since co-IP data will report both direct and indirect interactions, some observed interactors may be indirect. O1 interacted with many proteins, including actin-binding proteins, confirmed myosin binding partners, and other myosins. Our data are consistent with prior observations of interactions between myosin and MadA1 (Kurth et al. 2017) myosin and (super)villin (Smith et al. 2013) and myosin and calmodulin/calmodulin-like (Shen et al. 2016). We also observed interactions between O1 and 2 closely related kinesin 12 proteins that are similar to Arabidopsis POK1. Even though many kinesins localize to the phragmoplast midline (Smertenko et al. 2018), we only identified KIN12D and KIN12E as O1-interactors. Moreover, the only other 2 microtubule-associated binding proteins we observed were IQ-domain proteins, recently shown in Arabidopsis to interact with POKs (Kumari et al. 2021). Mutations in pok1 and pok2 lead to misguided phragmoplasts similar to those observed in o1 (Lipka et al. 2014; Herrmann et al. 2018). Mutations in other kinesins, such as Arabidopsis PAKRP1 (PHRAGMOPLAST-ASSOCIATED KINESIN-RELATED PROTEIN 1, also named KIN12a) and PAKRP2 (orphan kinesin) and P. patens KINID1a (KINESIN FOR INTERDIGITATED MICROTUBULES 1a) and KINID1b (orphan kinesins) lead to severe phragmoplast structural defects not seen in pok or o1, despite similar localization of all these proteins to the phragmoplast (Pan et al. 2004; Lee et al. 2007; Hiwatashi et al. 2008). Indeed, mutations in several genes encoding phragmoplast-localized proteins result in defects that alter phragmoplast structure, often leading to cell wall stubs and multinucleate cells (Müller et al. 2004; Bannigan et al. 2007; Ho et al. 2011; Zhang et al. 2018, 3; Schmidt and Smertenko 2019). The similar phenotypes of pok and o1 coupled with their physical interaction suggest that they work together to ensure correct phragmoplast guidance.
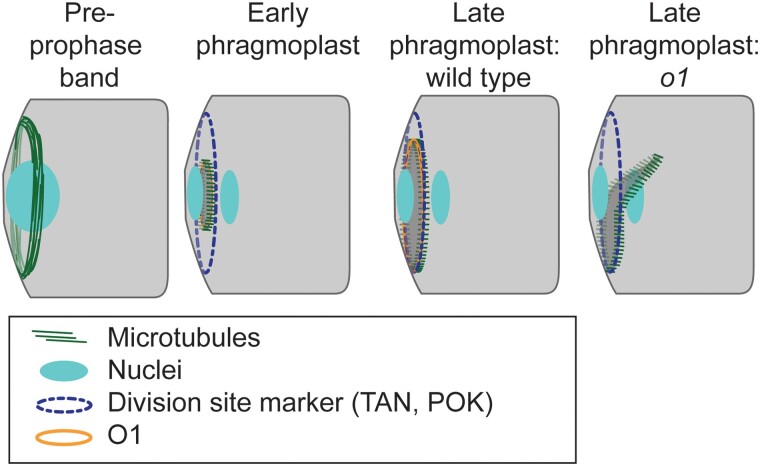
How might O1 and the POK-like kinesins work together to ensure proper phragmoplast guidance and disassembly? During late and slow phragmoplast expansion (van Oostende-Triplet et al. 2017), the phragmoplast falls “off-track” and becomes misguided in o1 (Fig. 9). Since POK proteins mark the cortical division site, and O1 localizes to the phragmoplast midline, a plausible model is that physical interactions between POKs at the division site and O1 at the phragmoplast midline help the phragmoplast “zip-up” around the cell cortex at the time of cell plate fusion. O1 and POK proteins may mediate interactions between the actin and microtubule cytoskeletons, to promote fusion of the phragmoplast at the cell wall. An alternative (non-mutually exclusive) model is that POK-like kinesins interact with O1 within the phragmoplast to coordinate microtubule and actin functions that promote slow phragmoplast expansion or phragmoplast disassembly once it reaches the cell cortex.
Figure 9.
Participation of O1 in late-stage phragmoplast guidance. The cortical division site is initially marked by the PPB and later by division site markers such as TAN1 and POK proteins (which are also present in the phragmoplast). In wild-type cells, the phragmoplast is guided to the division site by actin filaments and myosin VIII (Wu and Bezanilla 2014). After meeting the cortex, the phragmoplast continues to expand and interactions are stabilized by microtubules (Bellinger et al. 2021). In wild type, the phragmoplast fuses with the existing cell wall along the established division site, which is mediated by POK, TAN, and O1. In o1 mutants, after initial contact the phragmoplast becomes misguided, resulting in abnormal division planes.
Materials and methods
Plant material and growth conditions
The 3 maize (Z. mays) o1 mutant alleles (o1-ref, o1-N1242A, and o1-84-5270-40) used for this study were obtained from the Maize Genetics Cooperation stock center. Mutant alleles were backcrossed to the maize inbred line B73 1 to 4 times and then selfed. In all experiments, segregating o1 mutants were analyzed and compared with their corresponding wild-type siblings (grown side-by-side) as controls. The progeny from segregating families were classified based on their seed phenotype.
Mutant o1 plants were crossed with various fluorescent protein-tagged maize lines generated by the Maize Cell Genomics Project (described at http://maize.jcvi.org/cellgenomics/index.php). PAN1-YFP (Humphries et al. 2011), the actin marker line YFP-ABD2-YFP (Mohanty et al. 2009), TAN1-YFP (Martinez et al. 2017), or the tubulin marker lines CFP-β-tubulin and YFP-α-tubulin (Mohanty et al. 2009) were crossed to o1 homozygous plants, and the F1 progeny were backcrossed to o1 homozygous plants to obtain progeny expressing the fluorescent marker and segregating homozygous mutant (o1/o1) and phenotypically wild-type heterozygous (o1/+) individuals for experiments.
Plants used for phenotypic analysis and imaging were grown for 10 to 14 d in a greenhouse maintained between 22°F and 32°F under natural light illumination in greenhouses at the University of New Mexico, the University of Massachusetts at Amherst, or the University of California, Riverside. Plants were grown in Pro-mix Professional soil supplemented with Peters Excel 15-5-15 Cal Mag (weekly) and chelated iron (Southern Ag) as needed.
Stomatal defects in expanded leaves
To quantify subsidiary cell and GMC defects, o1 homozygous plants and their wild-type siblings from self-pollinated o1-ref/+, o1-N1243/+, and o1-N1478A/+ were classified via their seed phenotypes. Impressions of fully expanded leaf 4 of o1 homozygous and corresponding wild-type siblings were prepared using cyanoacrylate glue (Allsman et al. 2019) and imaged on a Nikon stereo microscope.
Confocal microscopy
Instruments are described here; details on specific experimental protocols are given below.
For O1 immunostaining and nuclear polarization:
Images were collected using a Zeiss LSM710 with a 63× (1.4 NA) oil immersion objective. Aniline blue was excited at 405 nm with a violet blue laser, and PI was excited using the 568-nm laser line and emission filter 620/60.
For PAN-YFP and ABD2-YFP localization:
ABD2-YFP-ABD2 and PAN1-YFP images were acquired with a custom spinning-disk confocal microscope (3i) equipped with a Yokagawa W1 spinning disk with 50-µm pinholes, iXon Life 888 EM-CCD camera (Andor) using 150 EM-CCD intensification, ASI piezo stage, and solid-state lasers. YFP Images were acquired using a 60× (1.2NA) silicone immersion objective, YFP fluorescence was excited by a 514-nm 100 mW solid-state laser at 6% with a dichroic excitation filter (Chroma), and a 542/27 emission filter (Semrock).
For microtubule immunostaining and phalloidin staining:
Immunolocalization and actin localization experiment images were collected with a Nikon A1R with a 60× (1.40 NA) oil immersion objective. Alexa Fluor 488 and Alexa Fluor 568 were excited at the appropriate wavelengths of 488 and 568 nm, respectively, emission filters were 525/50 nm for Alexa Fluor 488 and 595/50 for Alexa Fluor 568.
For live imaging of CFP-TUBULIN, YFP-TUBULIN, and TAN1-YFP:
Time-lapse imaging was performed using a custom-built spinning-disk confocal microscope (Solamere Technology) with a Yokogawa W1 spinning disk (Yokogawa), EM-CCD camera (Hamamatsu 910 °c), and an Eclipse Ti-U (Nikon) inverted microscope. A 60× water immersion lens (1.2 NA) was used with perfluorocarbon immersion liquid (RIAAA-678, Cargille). The stage was controlled by Micromanager software (www.micromanager.org) with ASI Piezo (300 µm range) and 3 axis DC servo motor controller. Solid-state lasers (Obis from 40 to 100 mW) and standard emission filters (Chroma Technology) were used. For CFP-TUBULIN, a 445-nm laser with an emission filter 480/40 was used. For YFP-TUBULIN and TAN1-YFP, a 514-nm laser with emission filter 540/30 was used.
All image analyses and figure preparations including cell measurements and processing were performed using ImageJ/FIJI (Schindelin et al. 2012) Adobe Photoshop CS6 or GIMP using only linear adjustments.
Polarization measurements
To analyze PAN1 polarization, the basal 0.5 to 2.5 cm of leaf 4 of o1-ref homozygous and heterozygous wild-type sibling plants expressing PAN1-YFP were examined. Z-stacks of the stomatal division zone were collected using a spinning-disk confocal microscope (3i) described above using a 60× silicone-oil immersion lens, YFP settings, and a 500-ms exposure. Recently formed subsidiary cells adjacent to a GMC that had not yet divided were assayed for polarity. PAN1-YFP polarization was scored by eye by comparing fluorescence intensity at the GMC–SMC interface and the adjacent SMC cell membrane. Cells were scored as having divided “normally” if both ends of the newly formed cell wall met the angled wall of the subsidiary cell.
To analyze F-actin polarization, the basal 0.5 to 2.5 cm of leaf 4 of o1-N1242A/o1-N1242A and wild-type o1/+ expressing YFP-ABD2-YFP (Mohanty et al. 2009) were examined using a spinning-disk confocal microscope (3i) described below using YFP settings and a 100-ms exposure. Actin polarization was scored by eye by comparing fluorescence intensity at the GMC–SMC interface and the adjacent SMC cell membrane. GMC widths were measured using FIJI (Schindelin et al. 2012). Cell counts were then binned by GMC width and the % of cells with an actin was calculated.
To analyze nuclear polarization, double staining using aniline blue and PI of fixed tissue was performed. The stomatal division zone (basal 0.5 to 2.5 cm of unexpanded leaves) from leaf 4 of o1 plants and wild-type siblings was isolated and fixed with FAA (3.7% formaldehyde, 5% acetic acid, 50% ethanol, w/v/v) for 1 h. Tissues were stained with 0.1% (w/v) aniline blue in phosphate-buffered saline (PBS) at pH 11 for 30 min. After rinsing with PBS, the tissues were stained with PI (10 µg mL−1 in water) and mounted onto slides.
Anti-O1 antibody generation
Custom rabbit antibodies were obtained from Pacific Immunology (Ramona, CA). Two peptides (Cys-NSEPKHIYESPTPTK and NSEPKHIYESPTPTK-Cys) were co-injected into 2 separate rabbits. The resulting sera were affinity-purified against both peptides, according to Cartwright et al. (2009). The 2 antibodies were named O1-11759 (Antibody 1) and O1-11760 (Antibody 2).
Immunolocalization and phalloidin staining
Dual labeling of O1 and microtubules was performed as previously described, with minor modifications (Cartwright et al. 2009; Nan et al. 2019). The basal 0.5 to 2.5 cm of leaf 4 from o1-N1242A and wild-type siblings or o1-N1242A and wild-type siblings was used. Immunolocalization and phalloidin staining were performed separately. For immunolocalization, the dilutions used for rabbit anti-O1 (antibody O1-11759) and mouse anti-tubulin (Sigma Aldrich) antibodies were 1:1,000. Alexa Fluor 488-conjugated anti-mouse and Alexa Fluor 568-conjugated anti-rabbit (Invitrogen) were used at a dilution of 1:500. Samples were mounted in ProLong Gold Antifade with DAPI (ThermoFisher). For phalloidin staining, the basal 0.5 to 2.5 cm of leaf 4 from o1 and wild-type siblings was fixed and stained with Alexa fluor 488-phalloidin (ThermoFisher) as described previously (Cartwright et al. 2009; Nan et al. 2019). Nuclei and cell walls were stained using 10 µg mL−1 PI (ThermoFisher).
CFP-TUBULIN, YFP-TUBULIN, and TAN1-YFP imaging
Time-lapse imaging was performed by taking a Z-stack every 6 min and assessing the morphology of the mitotic structure. The start of metaphase was counted from the first time the spindle was observed until the anaphase spindle was observed. This time point became the first time point for anaphase. Telophase timing was measured from the first time point when a phragmoplast was observed until the phragmoplast was completely disassembled. Leaf 5 or 6 from 12- to 14-d-old maize seedlings were used. Samples were prepared as described before (19).
Dcd2-O mapping
dcd2-O was mapped to chromosome 4 using a near-isogenic line analysis after 4 backcrosses to B73. dcd2 mutants in the B73 background were crossed to Mo17 and W64 inbred lines creating mapping populations for positional cloning. Markers on chromosome 4 were evaluated in over 1,300 dcd2 mutants from the 2 mapping populations. Markers used for fine-mapping on chromosome 4 included single sequence repeat (SSR) markers from Sigma's Maize SSR Polymorphic Primer Set (umc2038, umc1620, bnlg1189, umc1871, and bnlg2162) and discovered single nucleotide polymorphism (SNP) markers (N19-SNP: amplify with 5′cggagagaaaggtttggttg and 5′ctcatcgttccgtttggttt and cut with MboII; F07-SNP: amplify with 5′tggaataaacccagctttgc and 5′gccaaccagatgctcttctc and cut with StuI: and AC185621: amplify with 5′aagtcaacctgttgcgttcc and 5′cgccttctgattcaccatct and cut with PvuII).
Co-IP/MS
Co-IP/MS experiments were performed as previously described with some modifications (Facette et al. 2015). Families segregating o1-N1242A and wild-type siblings were used, with 3 biological replicates per genotype. The experiment was entirely conducted twice, once with the O1-11759 antibody and once with the o1-117560 antibody. The cell division zone (0.5 to 2.5 cm from the leaf base) was isolated from unexpanded leaves 4 to 6 of 10- to 14-d-old seedlings; 4 to 10 seedlings were pooled per replicate to obtain 1.5 g of tissue. The leaf tissues were ground in liquid nitrogen. Extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EGTA, 5 mM EDTA, 0.3% [v/v] ß-mercaptoethanol, 1% [v/v] Plant Protease Inhibitor Cocktail) was added as 1 mL per 0.25 g of tissue and the mixture was homogenized for 3 × 15 s, with 30-s breaks in between. Extracts were centrifuged at 25,000 × g at 4 °C in a microcentrifuge 2 times, then the supernatant was transferred and centrifuged at 110,000 × g at 4 °C for 45 min in an ultracentrifuge. After spinning, the supernatant was removed and the pellet was resuspended in 500 µL (per 0.25 g of starting tissue) of solubilization buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% [v/v] NP-40, 10% [v/v] glycerol, 0.05% [w/v] sodium deoxycholate). Samples were sonicated 2 × 15 s on ice and left rotating at 4 °C for 1 to 2 h. The extracts were centrifuged again at 110,000 × g at 4 °C for 45 min, and the supernatant was transferred to a new tube. Dynabeads coupled with anti-O1 antibody 11759 or 117560 were prepared according to the Dynabeads kit (Thermo Fisher) and added to the supernatant. The sample was incubated by rotating at room temperature for 30 min. The Dynabeads-Co-IP complex was washed according to the instructions of the Dynabeads kit.
The Dynabeads-Co-IP complexes were digested overnight at 37 °C with 400 ng of trypsin (Promega) per sample in 50 mM NH4CO3 buffer. After digestion, peptides were reduced with 1 mM dithiothreitol at room temperature for 30 min and then alkylated with 5 mM iodoacetamide at room temperature in the dark for 30 min. Formic acid was added to a final concentration of 0.1% (v/v) and peptides were extracted from the beads and desalted using the C18-Stage-Tip method and then vacuum-dried. The dried peptides were reconstituted in 20 µL of 5% (v/v) formic acid and 5% (v/v) acetonitrile and 3 μL of each sample was injected into an LC column with a run time of 60 min of the gradient method for each run for MS analysis. Samples were run in technical triplicates on a Q-Exactive mass spectrometer with instrument and chromatography settings as described previously (Markmiller et al. 2018). The RAW files were analyzed using Andromeda/MaxQuant (version 1.6.0.16) (Cox and Mann 2008) with default settings except that match between the runs and label-free quantification settings were enabled. Data were searched against a concatenated target-decoy database comprised of forward and reverse FASTA peptide sequences from the B73 RefGen_v4, AGPv4 (Jiao et al. 2017). Data generated from samples from Experiment 1 (using antibody O1-11759) and Experiment 2 (using O1-11760) were searched against the database independently. A similar number of peptides was mapped for each sample from the different experiments. Experiment 1 resulted in 112,927 mapped peptides, with 21,900, 18,310, and 22,319 total in the 3 wild-type samples and 20,624, 14,777, and 14,977 in the 3 o1 samples. Experiment 2 resulted in 158,803 mapped peptides, with 29,038, 29,219, and 28,476 total in the 3 wild-type samples and 24,262, 24,423, and 23,385 in the 3 o1 samples.
Y2H analysis
Total RNA was isolated from the stomatal division zone and cDNA was prepared using a Protoscript II First Strand DNA synthesis kit (NEB). A cDNA fragment encoding the C-terminal region of O1 was PCR-amplified with primers O1CTAIL_NO_INTRO_GWF and O1_NOSTOP_GWR. The full-length KIN12E cDNA was cloned using primers KIN12E_GWF and KIN12E_GWR. Fragments of KIN12C and KIN12D were cloned using primers KIN12C_Cterm_GWF and KIN12C_GW_R or KIN12D_Cterm_GW_F and KIN12D_GW_R, respectively. Primer sequences are listed in Supplemental Table S3. The cDNA fragments were subcloned into pDONR221 (ThermoFisher) using BP Clonase II, and then cloned into either pAS-GW or pACT-GW (Nakayama et al. 2002) using LR Clonase II (ThermoFisher). Sanger sequence-verified plasmids were co-transformed into yeast strain AH109 according to Gietz and Woods 92002). Co-transformants were selected on a synthetic defined (SD) medium without leucine and tryptophan (SD–LT) (Clontech). Between 12 and 80 colonies from each independent transformation were randomly picked and replated on SD medium without leucine, tryptophan, histidine, and adenine (SD–LTHA). Plates were incubated at 28 °C. Growth was assayed after 3 d.
Accession numbers
The accession number for O1 is Zm00001d052110 (B73 RefGen_v4, AGPv4) or Zm00001eb193160 (B73 RefGen_v5). The accession number for Kin12C is Zm00001d022276 (B73 RefGen_v4, AGPv4) or Zm00001eb328750 (B73 RefGen_v5). The accession number for Kin12D is Zm00001d041353 (B73 RefGen_v4, AGPv4) or Zm00001eb135060 (B73 RefGen_v5). The accession number for Kin12E is Zm00001d034030 (B73 RefGen_v4, AGPv4) or Zm00001eb057370 (B73 RefGen_v5). The accession number for ZmTan1 is Zm00001d038060 (B73 RefGen_v4, AGPv4) or Zm00001eb286860 or (B73 RefGen_v5).
Supplementary Material
Acknowledgments
We would like to thank Miguel Vasquez, Lindy Allsman, Laurie Smith, and Anne Sylvester for performing maize crosses.
Contributor Information
Qiong Nan, Department of Biology, University of Massachusetts, Amherst, MA 01003, USA.
Hong Liang, Department of Botany and Plant Sciences, University of California, Riverside, CA 92521, USA.
Janette Mendoza, Department of Botany, University of New Mexico, Albuquerque, NM 87131, USA.
Le Liu, Department of Biology, University of Massachusetts, Amherst, MA 01003, USA.
Amit Fulzele, Division of Biological Sciences, University of California, Riverside, CA 92093, USA.
Amanda Wright, Department of Biological Sciences, University of North Texas, Denton, TX 76203, USA.
Eric J Bennett, Division of Biological Sciences, University of California, Riverside, CA 92093, USA.
Carolyn G Rasmussen, Department of Botany and Plant Sciences, University of California, Riverside, CA 92521, USA.
Michelle R Facette, Department of Biology, University of Massachusetts, Amherst, MA 01003, USA.
Author contributions
Q.N., H.L., J.M., A.J.W., E.J.B., C.G.R., and M.R.F. designed the research. Q.N., H.L., J.M., L.L., A.F., and M.R.F. performed the research; Q.N., H.L., J.M., L.L., A.F., E.J.B., C.G.R., and M.R.F. analyzed the data; A.W. contributed the dcd2 allele; M.R.F., C.G.R., H.L., and Q.N. wrote the paper with input from other authors; M.R.F., C.G.R., and E.J.B. provided funding.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nuclear migration is normal in o1.
Supplemental Figure S2. O1 localizes to phragmoplasts.
Supplemental Figure S3. Actin in phragmoplasts of wild-type and o1 mutant cells.
Supplemental Figure S4. Dcd2 is allelic to O1.
Supplemental Figure S5. Time-lapse imaging of wild-type and o1-84-5270-40 mutant subsidiary mother cell division.
Supplemental Figure S6. Validation of anti-O1 antibodies and co-IP/immunoblot results.
Supplemental Figure S7. The C-terminal region of O1 interacts with KIN12E.
Supplemental Figure S8. Unexpected TAN1-YFP localization in o1 telophase SMCs.
Supplemental Figure S9. TAN1-YFP localization during division of GMC progenitor cells.
Supplemental Table S1. PAN1-YFP is polarized in o1 subsidiary mother cells that have divided normally and abnormally.
Supplemental Table S2. Total number of SMCs examined for TAN1-YFP localization at different steps of the cell cycle.
Supplemental Table S3. Primers used in this study for yeast 2-hybrid analysis.
Supplemental Data Set 1. Peptides identified in co-IP/MS experiments in the o1 mutant and wild-type siblings using 2 different antibodies.
Supplemental Data Set 2. Variables, degrees of freedom, and P-values for statistical tests.
Supplemental Movie S1. Normal asymmetric division of a wild-type SMC.
Supplemental Movie S2. Division of an o1-N1242A maize leaf subsidiary mother cell with a misguided phragmoplast.
Supplemental Movie S3. Division of an o1-N1242A maize leaf subsidiary mother cell with a misguided phragmoplast.
Supplemental Movie S4. Localization of TAN1-YFP in o1-N1242A SMC with phragmoplast
Funding
Research reported in this publication was supported in part by National Science Foundation, Division of Integrative Organismal Systems 1754665 to M.R.F., National Science Foundation, Division of Molecular and Cellular Biosciences 1716972 and 1942734 to C.G.R and National Institute of Health, National Institute of General Medical Sciences P30 GM110907. We thank Steve Eyles at the UMass Institute of Applied Life Sciences Mass Spectrometry Facility, RRID:SCR_019063. We thank Jim Chambers at the UMass Institute of Applied Life Sciences Light Microscopy Facility, RRID:SCR_021148. Microscope image data using the Zeiss LSM710 was gathered in the UNM CETI Cell Biology Core with support from National Institute of Health, National Institute of General Medical Sciences P30 GM110907.
References
- Abu-Abied M, Belausov E, Hagay S, Peremyslov V, Dolja V, Sadot E. Myosin XI-K is involved in root organogenesis, polar auxin transport, and cell division. J Exp Bot. 2018:69(12):2869–2881. 10.1093/jxb/ery112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali MF, Fatema U, Peng X, Hacker SW, Maruyama D, Sun M-X, Kawashima T. ARP2/3-independent WAVE/SCAR pathway and class XI myosin control sperm nuclear migration in flowering plants. Proc Natl Acad Sci U S A. 2020:117(51):32757–32763. 10.1073/pnas.2015550117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allsman LA, Dieffenbacher RN, Rasmussen CG. Glue impressions of maize leaves and their use in classifying mutants. Bio-101. 2019:e3209. 10.21769/BioProtoc.3209 [DOI] [Google Scholar]
- Apostolakos P, Livanos P, Giannoutsou E, Panteris E, Galatis B. The intracellular and intercellular cross-talk during subsidiary cell formation in Zea mays: existing and novel components orchestrating cell polarization and asymmetric division. Ann Bot. 2018:122(5):679–696. 10.1093/aob/mcx193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisar D, Abu-Abied M, Belausov E, Sadot E, Hawes C, Sparkes IA. A comparative study of the involvement of 17 Arabidopsis myosin family members on the motility of Golgi and other organelles. Plant Physiol. 2009:150(2):700–709. 10.1104/pp.109.136853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannigan A, Scheible WR, Lukowitz W, Fagerstrom C, Wadsworth P, Somerville C, Baskin TI. A conserved role for kinesin-5 in plant mitosis. J Cell Sci. 2007:120(16):2819–2827. 10.1242/jcs.009506 [DOI] [PubMed] [Google Scholar]
- Bellinger MA, Uyehara AN, Allsman L, Martinez P, McCarthy MC, Rasmussen CG. Cortical microtubules contribute to division plane positioning during telophase in maize. Plant Cell. 2023:35(5):1496–1512. 10.1093/plcell/koad033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann H, Müller S. Update on plant cytokinesis: rule and divide. Curr Opin Plant Biol. 2019:52:97–105. 10.1016/j.pbi.2019.07.003 [DOI] [PubMed] [Google Scholar]
- Cartwright HN, Humphries JA, Smith LG. PAN1: a receptor-like protein that promotes polarization of an asymmetric cell division in maize. Science. 2009:323(5914):649–651. 10.1126/science.1161686 [DOI] [PubMed] [Google Scholar]
- Cho S-O, Wick SM. Actin in the developing stomatal complex of winter rye: a comparison of actin antibodies and Rh-phalloidin labeling of control and CB-treated tissues. Cell Motil Cytoskeleton. 1991:19(1):25–36. 10.1002/cm.970190105 [DOI] [Google Scholar]
- Cleary AL, Smith LG. The Tangled1 gene is required for spatial control of cytoskeletal arrays associated with cell division during maize leaf development. Plant Cell. 1998:10(11):1875–1888. 10.1105/tpc.10.11.1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. Maxquant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008:26(12):1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Duan Z, Tominaga M. Actin–myosin XI: an intracellular control network in plants. Biochem Biophys Res Commun. 2018:506(2):403–408. 10.1016/j.bbrc.2017.12.169 [DOI] [PubMed] [Google Scholar]
- Facette MR, Park Y, Sutimantanapi D, Luo A, Cartwright HN, Yang B, Bennett EJ, Sylvester AW, Smith LG. The SCAR/WAVE complex polarises PAN receptors and promotes division asymmetry in maize. Nat Plants. 2015:1(2):14024. 10.1038/nplants.2014.24 [DOI] [PubMed] [Google Scholar]
- Facette MR, Smith LG. Division polarity in developing stomata. Curr Opin Plant Biol. 2012:15(6):585–592. 10.1016/j.pbi.2012.09.013 [DOI] [PubMed] [Google Scholar]
- Frank MJ, Cartwright HN, Smith LG. Three Brick genes have distinct functions in a common pathway promoting polarized cell division and cell morphogenesis in the maize leaf epidermis. Development. 2003:130(4):753–762. 10.1242/dev.00290 [DOI] [PubMed] [Google Scholar]
- Franks PJ, Farquhar GD. The mechanical diversity of stomata and its significance in gas-exchange control. Plant Physiol. 2007:143(1):78–87. 10.1104/pp.106.089367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher K, Smith LG. Discordia mutations specifically misorient asymmetric cell divisions during development of the maize leaf epidermis. Development. 1999:126(20):4623–4633. 10.1242/dev.126.20.4623 [DOI] [PubMed] [Google Scholar]
- Gallagher K, Smith LG. Roles for polarity and nuclear determinants in specifying daughter cell fates after an asymmetric cell division in the maize leaf. Curr Biol. 2000:10(19):1229–1232. 10.1016/S0960-9822(00)00730-2 [DOI] [PubMed] [Google Scholar]
- Gibbon BC, Larkins BA. Molecular genetic approaches to developing quality protein maize. Trends Genet. 2005:21(4):227–233. 10.1016/j.tig.2005.02.009 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. In: Christine G, Gerald RF, editors. Methods in enzymology. Cambridge (MA): Academic Press; 2002. p. 87–96. [DOI] [PubMed] [Google Scholar]
- Gray A, Liu L, Facette M. Flanking support: how subsidiary cells contribute to stomatal form and function. Front Plant Sci. 2020:11:881. 10.3389/fpls.2020.00881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth C, Caine RS, Harrison EL, Sloan J, Gray JE. Stomatal development: focusing on the grasses. Curr Opin Plant Biol. 2018:41:1–7. 10.1016/j.pbi.2017.07.009 [DOI] [PubMed] [Google Scholar]
- Herrmann A, Livanos P, Lipka E, Gadeyne A, Hauser M-T, Van Damme D, Müller S. Dual localized kinesin-12 POK2 plays multiple roles during cell division and interacts with MAP65-3. EMBO Rep. 2018:19(9):e46085. 10.15252/embr.201846085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann A, Livanos P, Zimmermann S, Berendzen K, Rohr L, Lipka E, Müller S. KINESIN-12E regulates metaphase spindle flux and helps control spindle size in Arabidopsis. Plant Cell. 2021:33(1):27–43. 10.1093/plcell/koaa003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiwatashi Y, Obara M, Sato Y, Fujita T, Murata T, Hasebe M. Kinesins are indispensable for interdigitation of phragmoplast microtubules in the moss Physcomitrella patens. Plant Cell. 2008:20(11):3094–3106. 10.1105/tpc.108.061705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C-MK, Hotta T, Guo F, Roberson RW, Lee Y-RJ, Liu B. Interaction of antiparallel microtubules in the phragmoplast is mediated by the microtubule-associated protein MAP65-3 in Arabidopsis. Plant Cell. 2011:23(8):2909–2923. 10.1105/tpc.110.078204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holweg C, Honsel A, Nick P. A myosin inhibitor impairs auxin-induced cell division. Protoplasma. 2003:222(3–4):193–204. 10.1007/s00709-003-0025-3 [DOI] [PubMed] [Google Scholar]
- Humphries JA, Vejlupkova Z, Luo A, Meeley RB, Sylvester AW, Fowler JE, Smith LG. ROP GTPases act with the receptor-like protein PAN1 to polarize asymmetric cell division in maize. Plant Cell. 2011:23(6):2273–2284. 10.1105/tpc.111.085597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Peluso P, Shi J, Liang T, Stitzer MC, Wang B, Campbell MS, Stein JC, Wei X, Chin, C.-S., et al. Improved maize reference genome with single-molecule technologies. Nature. 2017:546(7659):524–527. 10.1038/nature22971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennard JL, Cleary AL. Pre-mitotic nuclear migration in subsidiary mother cells of Tradescantia occurs in G1 of the cell cycle and requires F-actin. Cell Motil Cytoskeleton. 1997:36(1):55–67. [DOI] [PubMed] [Google Scholar]
- Kojo KH, Higaki T, Kutsuna N, Yoshida Y, Yasuhara H, Hasezawa S. Roles of cortical actin microfilament patterning in division plane orientation in plants. Plant Cell Physiol. 2013:54(9):1491–1503. 10.1093/pcp/pct093 [DOI] [PubMed] [Google Scholar]
- Kumari P, Dahiya P, Livanos P, Zergiebel L, Kölling M, Poeschl Y, Stamm G, Hermann A, Abel S, Müller S, et al. IQ67 DOMAIN proteins facilitate preprophase band formation and division-plane orientation. Nat Plants. 2021:7(6):739–747. 10.1038/s41477-021-00923-z [DOI] [PubMed] [Google Scholar]
- Kurth EG, Peremyslov VV, Turner HL, Makarova KS, Iranzo J, Mekhedov SL, Koonin EV, Dolja VV. Myosin-driven transport network in plants. Proc Natl Acad Sci U S A. 2017:114(8):E1385–E1394. 10.1073/pnas.1620577114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-RJ, Li Y, Liu B. Two Arabidopsis phragmoplast-associated kinesins play a critical role in cytokinesis during male gametogenesis. Plant Cell. 2007:19(8):2595–2605. 10.1105/tpc.107.050716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-RJ, Liu B. Microtubule nucleation for the assembly of acentrosomal microtubule arrays in plant cells. New Phytol. 2019:222(4):1705–1718. 10.1111/nph.15705 [DOI] [PubMed] [Google Scholar]
- Lipka E, Gadeyne A, Stöckle D, Zimmermann S, De Jaeger G, Ehrhardt DW, Kirik V, Van Damme D, Müller S. The phragmoplast-orienting kinesin-12 class proteins translate the positional information of the preprophase band to establish the cortical division zone in Arabidopsis thaliana. Plant Cell. 2014:26(6):2617–2632. 10.1105/tpc.114.124933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Ohashi-Ito K, Bergmann DC. Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development. 2009:136(13):2265–2276. 10.1242/dev.032938 [DOI] [PubMed] [Google Scholar]
- Madison SL, Buchanan ML, Glass JD, McClain TF, Park E, Nebenführ A. Class XI myosins move specific organelles in pollen tubes and are required for normal fertility and pollen tube growth in Arabidopsis. Plant Physiol. 2015:169(3):1946–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, Luo E-C, Krach F, Yang D, et al. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell. 2018:172(3):590–604.e13. 10.1016/j.cell.2017.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P, Luo A, Sylvester A, Rasmussen CG. Proper division plane orientation and mitotic progression together allow normal growth of maize. Proc Natl Acad Sci U A. 2017:114(10):2759–2764. 10.1073/pnas.1619252114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKown KH, Bergmann DC. Stomatal development in the grasses: lessons from models and crops (and crop models). New Phytol. 2020:227(6):1636–1648. 10.1111/nph.16450 [DOI] [PubMed] [Google Scholar]
- Miller DD, Scordilis SP, Hepler PK. Identification and localization of three classes of myosins in pollen tubes of Lilium longiflorum and Nicotiana alata. J. Cell Sci. 1995:108(7):2549–2563. 10.1242/jcs.108.7.2549 [DOI] [PubMed] [Google Scholar]
- Mills AM, Morris VH, Rasmussen CG. The localization of PHRAGMOPLAST ORIENTING KINESIN1 at the division site depends on the microtubule-binding proteins TANGLED1 and AUXIN-INDUCED IN ROOT CULTURES9 in Arabidopsis. Plant Cell. 2022:34(11):4583–4599. 10.1093/plcell/koac266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty A, Luo A, DeBlasio S, Ling X, Yang Y, Tuthill DE, Williams KE, Hill D, Zadrozny T, Chan A, et al. Advancing cell biology and functional genomics in maize using fluorescent protein-tagged lines. Plant Physiol. 2009:149(2):601–605. 10.1104/pp.108.130146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molchan TM, Valster AH, Hepler PK. Actomyosin promotes cell plate alignment and late lateral expansion in Tradescantia stamen hair cells. Planta. 2002:214(5):683–693. 10.1007/s004250100672 [DOI] [PubMed] [Google Scholar]
- Müller S, Han S, Smith LG. Two kinesins are involved in the spatial control of cytokinesis in Arabidopsis thaliana. Curr Biol. 2006:16(9):888–894. 10.1016/j.cub.2006.03.034 [DOI] [PubMed] [Google Scholar]
- Müller S, Smertenko A, Wagner V, Heinrich M, Hussey PJ, Hauser M-T. The plant microtubule-associated protein AtMAP65-3/PLE is essential for cytokinetic phragmoplast function. Curr Biol. 2004:14(5):412–417. 10.1016/j.cub.2004.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroyama A, Gong Y, Bergmann DC. Opposing, polarity-driven nuclear migrations underpin asymmetric divisions to pattern Arabidopsis stomata. Curr Biol. 2020:30(22):4549–4552. 10.1016/j.cub.2020.09.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M, Kikuno R, Ohara O. Protein-protein interactions between large proteins: two-hybrid screening using a functionally classified library composed of long cDNAs. Genome Res. 2002:12:1773–1784. 10.1101/gr.406902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan Q, Mendoza J, Facette M. Double labeling of microtubules and actin filaments in maize leaf division zone. Bio-101. 2019:e3262. 10.21769/BioProtoc.3262 [DOI] [Google Scholar]
- Nebenführ A, Dixit R. Kinesins and myosins: molecular motors that coordinate cellular functions in plants. Annu Rev Plant Biol. 2018:69(1):329–361. 10.1146/annurev-arplant-042817-040024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuffer MG, Jones L, Zuber MS. The mutants of maize. Madison (WI): Crop Science Society of America);1968.
- Nunes TDG, Zhang D, Raissig MT. Form, development and function of grass stomata. Plant J. 2020:101(4):780–799. 10.1111/tpj.14552 [DOI] [PubMed] [Google Scholar]
- Olatunji D, Kelley DR. A role for Arabidopsis myosins in sugar-induced hypocotyl elongation. MicroPubl Biol. 2020. 10.17912/micropub.biology.000276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palevitz BA. Actin in the preprophase band of Allium cepa. J Cell Biol. 1987:104(6):1515–1519. 10.1083/jcb.104.6.1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan R, Lee Y-RJ, Liu B. Localization of two homologous Arabidopsis kinesin-related proteins in the phragmoplast. Planta. 2004:220(1):156–164. 10.1007/s00425-004-1324-4 [DOI] [PubMed] [Google Scholar]
- Panteris E. Cortical actin filaments at the division site of mitotic plant cells: a reconsideration of the actin-depleted zone. New Phytol. 2008:179(2):334–341. 10.1111/j.1469-8137.2008.02474.x [DOI] [PubMed] [Google Scholar]
- Raissig MT, Abrash E, Bettadapur A, Vogel JP, Bergmann DC. Grasses use an alternatively wired bHLH transcription factor network to establish stomatal identity. Proc Natl Acad Sci U S A. 2016:113(29):8326–8331. 10.1073/pnas.1606728113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig MT, Matos JL, Anleu Gil MX, Kornfeld A, Bettadapur A, Abrash E, Allison HR, Badgley G, Vogel JP, Berry JA, et al. Mobile MUTE specifies subsidiary cells to build physiologically improved grass stomata. Science. 2017:355(6330):1215–1218. 10.1126/science.aal3254 [DOI] [PubMed] [Google Scholar]
- Reisen D, Hanson MR. Association of six YFP-myosin XI-tail fusions with mobile plant cell organelles. BMC Plant Biol. 2007:7(1):6. 10.1186/1471-2229-7-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadot E, Blancaflor EB. The actomyosin system in plant cell division: lessons learned from microscopy and pharmacology. In: Sahi VP, Baluška F, editors. The cytoskeleton: diverse roles in a plant's life (plant cell monographs). Cham: Springer International Publishing; 2019. p. 85–100. [Google Scholar]
- Sano T, Higaki T, Oda Y, Hayashi T, Hasezawa S. Appearance of actin microfilament “twin peaks” in mitosis and their function in cell plate formation, as visualized in tobacco BY-2 cells expressing GFP-fimbrin. Plant J. 2005:44(4):595–605. 10.1111/j.1365-313X.2005.02558.x [DOI] [PubMed] [Google Scholar]
- Sattarzadeh A, Franzen R, Schmelzer E. The Arabidopsis class VIII myosin ATM2 is involved in endocytosis. Cell Motil Cytoskeleton. 2008:65(6):457–468. 10.1002/cm.20271 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012:9(7):676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Smertenko A. Identification and characterization of the land-plant-specific microtubule nucleation factor MACET4. J Cell Sci. 2019:132:jcs232819. 10.1242/jcs.232819 [DOI] [PubMed] [Google Scholar]
- Shen M, Zhang N, Zheng S, Zhang W-B, Zhang H-M, Lu Z, Su QP, Sun Y, Ye K, Li X. Calmodulin in complex with the first IQ motif of myosin-5a functions as an intact calcium sensor. Proc Natl Acad Sci U S A. 2016:113(40):E5812–E5820. 10.1073/pnas.1607702113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko A. Phragmoplast expansion: the four-stroke engine that powers plant cytokinesis. Curr Opin Plant Biol. 2018:46:130–137. 10.1016/j.pbi.2018.07.011 [DOI] [PubMed] [Google Scholar]
- Smertenko A, Assaad F, Baluška F, Bezanilla M, Buschmann H, Drakakaki G, Hauser M-T, Janson M, Mineyuki Y, Moore I, et al. Plant cytokinesis: terminology for structures and processes. Trends Cell Biol. 2017:27(12):885–894. 10.1016/j.tcb.2017.08.008 [DOI] [PubMed] [Google Scholar]
- Smertenko A, Hewitt SL, Jacques CN, Kacprzyk R, Liu Y, Marcec MJ, Moyo L, Ogden A, Oung HM, Schmidt S, et al. Phragmoplast microtubule dynamics - a game of zones. J Cell Sci. 2018:131:jcs203331. 10.1242/jcs.203331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LG, Hake S, Sylvester AW. The tangled-1 mutation alters cell division orientations throughout maize leaf development without altering leaf shape. Development. 1996:122(2):481–489. 10.1242/dev.122.2.481 [DOI] [PubMed] [Google Scholar]
- Smith TC, Fridy PC, Li Y, Basil S, Arjun S, Friesen RM, Leszyk J, Chait BT, Rout MP, Luna EJ. Supervillin binding to myosin II and synergism with anillin are required for cytokinesis. Mol Biol Cell. 2013:24(23):3603–3619. 10.1091/mbc.e12-10-0714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Teanby NA, Hawes C. Truncated myosin XI tail fusions inhibit peroxisome, Golgi, and mitochondrial movement in tobacco leaf epidermal cells: a genetic tool for the next generation. J Exp Bot. 2008:59(9):2499–2512. 10.1093/jxb/ern114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöckle D, Herrmann A, Lipka E, Lauster T, Gavidia R, Zimmermann S, Müller S. Putative RopGAPs impact division plane selection and interact with kinesin-12 POK1. Nat Plants. 2016:2(9):1–6. 10.1038/nplants.2016.120 [DOI] [PubMed] [Google Scholar]
- Sun H, Furt F, Vidali L. Myosin XI localizes at the mitotic spindle and along the cell plate during plant cell division in Physcomitrella patens. Biochem Biophys Res Commun. 2018:506(2):409–421. 10.1016/j.bbrc.2018.01.082 [DOI] [PubMed] [Google Scholar]
- Sutimantanapi D, Pater D, Smith LG. Divergent roles for maize PAN1 and PAN2 receptor-like proteins in cytokinesis and cell morphogenesis. Plant Physiol. 2014:164(4):1905–1917. 10.1104/pp.113.232660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talts K, Ilau B, Ojangu E-L, Tanner K, Peremyslov VV, Dolja VV, Truve E, Paves H. Arabidopsis myosins XI1, XI2, and XIK are crucial for gravity-induced bending of inflorescence stems. Front Plant Sci. 2016:7:1932. 10.3389/fpls.2016.01932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Iwabuchi K, Fukao Y, Kondo M, Okamoto K, Ueda H, Nishimura M, Hara-Nishimura I. Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis. Curr Biol. 2013:23(18):1776–1781. 10.1016/j.cub.2013.07.035 [DOI] [PubMed] [Google Scholar]
- Ueda H, Yokota E, Kutsuna N, Shimada T, Tamura K, Shimmen T, Hasezawa S, Dolja VV, Hara-Nishimura I. Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proc Natl Acad Sci U S A. 2010:107(15):6894–6899. 10.1073/pnas.0911482107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valster AH, Hepler PK. Caffeine inhibition of cytokinesis: effect on the phragmoplast cytoskeleton in living Tradescantia stamen hair cells. Protoplasma. 1997:196(3–4):155–166. 10.1007/BF01279564 [DOI] [Google Scholar]
- Van Damme D, Vanstraelen M, Geelen D. Cortical division zone establishment in plant cells. Trends Plant Sci. 2007:12(10):458–464. 10.1016/j.tplants.2007.08.011 [DOI] [PubMed] [Google Scholar]
- van Oostende-Triplet C, Guillet D, Triplet T, Pandzic E, Wiseman PW, Geitmann A. Vesicle dynamics during plant cell cytokinesis reveals distinct developmental phases. Plant Physiol. 2017:174(3):1544–1558. 10.1104/pp.17.00343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L, Burkart GM, Augustine RC, Kerdavid E, Tüzel E, Bezanilla M. Myosin XI is essential for tip growth in Physcomitrella patens. Plant Cell. 2010:22(6):1868–1882. 10.1105/tpc.109.073288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker KL, Muller S, Moss D, Ehrhardt DW, Smith LG. Arabidopsis TANGLED identifies the division plane throughout mitosis and cytokinesis. Curr Biol. 2007:17(21):1827–1836. 10.1016/j.cub.2007.09.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Wang F, Wang G, Wang F, Zhang X, Zhong M, Zhang J, Lin D, Tang Y, Xu Z, et al. Opaque1 encodes a myosin XI motor protein that is required for endoplasmic reticulum motility and protein body formation in maize endosperm. Plant Cell. 2012:24(8):3447–3462. 10.1105/tpc.112.101360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Guo S, Qiao X, Guo J, Li Z, Zhou Y, Bai S, Gao Z, Wang D, Wang P, et al. BZU2/ZmMUTE controls symmetrical division of guard mother cell and specifies neighbor cell fate in maize. PLoS Genet. 2019:15(8):e1008377. 10.1371/journal.pgen.1008377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S-Z, Bezanilla M. Myosin VIII associates with microtubule ends and together with actin plays a role in guiding plant cell division. eLife. 2014:3:e03498. 10.7554/eLife.03498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Chen L, Yu Q, Zhou W, Gou X, Li J, Hou S. Multiple transcriptional factors control stomata development in rice. New Phytol. 2019:223(1):220–232. 10.1111/nph.15766 [DOI] [PubMed] [Google Scholar]
- Xu XM, Zhao Q, Rodrigo-Peiris T, Brkljacic J, He CS, Muller S, Meier I. RanGAP1 is a continuous marker of the Arabidopsis cell division plane. Proc Natl Acad Sci U S A. 2008:105(47):18637–18642. 10.1073/pnas.0806157105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda H, Kanda K, Koiwa H, Suenaga K, Kidou S, Ejiri S. Localization of actin filaments on mitotic apparatus in tobacco BY-2 cells. Planta. 2005:222(1):118–129. 10.1007/s00425-005-1522-8 [DOI] [PubMed] [Google Scholar]
- Zebosi B. Functional characterization of ramosa suppressor locus-12.2995 that shows the opaque1 gene regulates plant architecture in maize [masters thesis]. Order No. 29320061. [Ames (IA)]: Iowa State University;2022. ProQuest. Web. 4 April 2023.
- Zhang D, Abrash EB, Nunes TDG, Prados IH, Gil MXA, Jesenofsky B, Lindner H, Bergmann DC, Raissig MT. Opposite polarity programs regulate asymmetric subsidiary cell divisions in grasses. eLife. 2022:11:e79913. 10.7554/eLife.79913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Deng X, Sun B, Lee Van S, Kang Z, Lin H, Lee Y-RJ, Liu B. Role of the BUB3 protein in phragmoplast microtubule reorganization during cytokinesis. Nat Plants. 2018:4(7):485–494. 10.1038/s41477-018-0192-z [DOI] [PubMed] [Google Scholar]
- Zhang X, Facette M, Humphries JA, Shen Z, Park Y, Sutimantanapi D, Sylvester AW, Briggs SP, Smith LG. Identification of PAN2 by quantitative proteomics as a leucine-rich repeat-receptor-like kinase acting upstream of PAN1 to polarize cell division in maize. Plant Cell. 2012:24(11):4577–4589. 10.1105/tpc.112.104125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Groves NR, Meier I. Plant nuclear shape is independently determined by the SUN-WIP-WIT2-myosin XI-i complex and CRWN1. Nucleus. 2015:6(2):144–153. 10.1080/19491034.2014.1003512 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.