Abstract
Ralstonia solanacearum is a devastating soil-borne bacterial pathogen capable of infecting many plant species, including tomato (Solanum lycopersicum). However, the perception of Ralstonia by the tomato immune system and the pathogen's counter-defense strategy remain largely unknown. Here, we show that PehC, a specific exo-polygalacturonase secreted by Ralstonia, acts as an elicitor that triggers typical immune responses in tomato and other Solanaceous plants. The elicitor activity of PehC depends on its N-terminal epitope, and not on its polygalacturonase activity. The recognition of PehC specifically occurs in tomato roots and relies on unknown receptor-like kinase(s). Moreover, PehC hydrolyzes plant pectin-derived oligogalacturonic acids (OGs), a type of damage-associated molecular pattern (DAMP), which leads to the release of galacturonic acid (GalA), thereby dampening DAMP-triggered immunity (DTI). Ralstonia depends on PehC for its growth and early infection and can utilize GalA as a carbon source in the xylem. Our findings demonstrate the specialized and dual functions of Ralstonia PehC, which enhance virulence by degrading DAMPs to evade DTI and produce nutrients, a strategy used by pathogens to attenuate plant immunity. Solanaceous plants have evolved to recognize PehC and induce immune responses, which highlights the significance of PehC. Overall, this study provides insight into the arms race between plants and pathogens.
Beyond cell wall degradation, the Ralstonia solanacerum exo-polygalacturonase PehC affects the arms race between host and pathogens.
Introduction
Ralstonia solanacearum is a soil-borne bacterial plant pathogen with a worldwide distribution and infects more than 250 plant species belonging to over 50 different families (Genin and Denny 2012; Mansfield et al. 2012). The pathogen infects Solanaceous plants, such as tomato (Solanum lycopersicum), tobacco (Nicotiana tabacum.), and potato (Solanum tuberosum), causing bacterial wilt disease, which leads to huge economic losses (Denny 2007). The main virulence determinants of Ralstonia include type II secretion system-related cell wall degrading enzymes, type III secretion system effectors, exopolysaccharides (EPS), and bacterial mobility (Peeters et al. 2013).
Ralstonia species thrive in the water-transporting xylem vessels of host plants. Although the xylem is a relatively nutrient-poor, high-flow environment, Ralstonia multiplies in the xylem sap despite this limited nutrition (Zuluaga et al. 2013). Notably, a quorum sensing system plays an important role in regulating the pathogen transition from a rapidly growing lifestyle, a characteristic of the early growth phase that consumes nutrients at a fast pace, to a slow-growing lifestyle, characteristic of the late growth phase, in which bacteria form a biofilm to adhere to plant surfaces and produce virulence factors to colonize effectively (Lowe-Power et al. 2018a). How Ralstonia flourishes inside the plant xylem and the defense mechanism of tomato against Ralstonia infection remain largely unknown.
Plants have evolved multiple layers of immunity to recognize and fend off pathogens. The plasma membrane-localized pattern recognition receptors (PRRs) sense the conserved microbe- and pathogen-associated molecular patterns (MAMPs and PAMPs, respectively) to initiate pattern-trigged immunity (PTI) (Jones and Dangl 2006). MAMPs derived from bacteria, oomycetes, and fungi can be proteins, carbohydrates, lipids, or lipopeptides. A highly conserved 22-amino acid (aa) fragment of bacterial flagellin (flg22) is sufficient to activate immunity in Arabidopsis thaliana and other plant species (Felix et al. 1999). An 18-aa peptide of the bacterial elongation factor-Tu, elf18, is recognized as a MAMP only in Brassicaceae species. According to their domain architecture, PRRs can be classified into 2 types: receptor-like kinases (RLKs) and receptor-like proteins (RLPs) (Wu and Zhou 2013). Both types of PRRs perceive MAMPs and transmit immune signals to activate a series of immune responses, such as reactive oxygen species (ROS) burst, calcium (Ca2+) influx, mitogen-activated protein kinase (MAPK) activation, and defense-related gene expression (Yu et al. 2017; Yuan et al. 2021). Many PRRs rely on BRASSINOSTEROID INSENSITIVE 1-ASSOCIATED RECEPTOR KINASE 1 (BAK1) family proteins as well as SUPPRESSOR OF BIR1-1 (SOBIR1) to trigger defense responses (Liebrand et al. 2014).
Besides recognizing the molecular components derived from pathogens, PRRs can also recognize damage-associated molecular patterns (DAMPs) of plant origin to activate DAMP-triggered immunity (DTI). DAMPs are divided into two categories: cell wall degradation fragments including oligogalacturonic acids (OGs) derived from pectin, and compounds released from the cytosol such as plant elicitor peptides and extracellular ATP (Gust et al. 2017). Immune responses triggered by plant DAMPs are considered to amplify and transmit PTI. Both pathogens and symbionts establish plant–microbe interactions using fascinating and intricate extracellular strategies to avoid recognition (Gong et al. 2020; Buscaill and van der Hoorn 2021). To subvert the host immune system, pathogens have evolved a variety of strategies such as masking themselves from host immune recognition, blocking immune signaling transduction, and reprograming immune responses (Buscaill and van der Hoorn 2021; Chen et al. 2021). Degradation of DAMPs would be a good strategy for pathogens, but whether pathogens utilize this strategy to counter plant defense remains unknown.
The immune recognition system employed by tomato to detect Ralstonia displays unique features. The two common bacterial MAMPs derived from Ralstonia, flg22 and elf18, cannot be recognized by tomato to induce PTI responses (Felix et al. 1999; Kunze et al. 2004). Flagellin from Ralstonia presents a polymorphic flg22 sequence that avoids perception in the host, although soybean (Glycine max), a nonhost, was found to recognize Ralstonia flg22 in a recent study (Wei et al. 2020). Conserved peptide CSP22 derived from a bacterial cold shock protein triggers immune responses only in Solanaceae species and is recognized by COLD SHOCK PROTEIN RECEPTOR in tomato (Hind et al. 2016; Wei et al. 2018). Ralstonia infection occurs under high-temperature conditions; thus, the function of cold shock protein in plant defense is unclear. Therefore, further research is needed to elucidate the mechanisms that allow tomato to perceive and defend against R. solanacearum infection.
Here, we report that PehC, an exo-polygalacturonase (exo-PG) from Ralstonia, can activate the early immune responses of tomato in plant roots. PehC can hydrolyze OGs to suppress OG-triggered DTI in tomato. Moreover, galacturonic acid (GalA) monomers released by PehC activity serve as a valuable carbon source for Ralstonia at the early growth stage. This study provides molecular insights into a distinct pathogen effector that exhibits dual functions to decrease plant immune activation and increase nutrient acquisition from the host, which is in turn recognized by plants as an immune elicitor.
Results
Ralstonia secreted proteins trigger an immune response in tomato
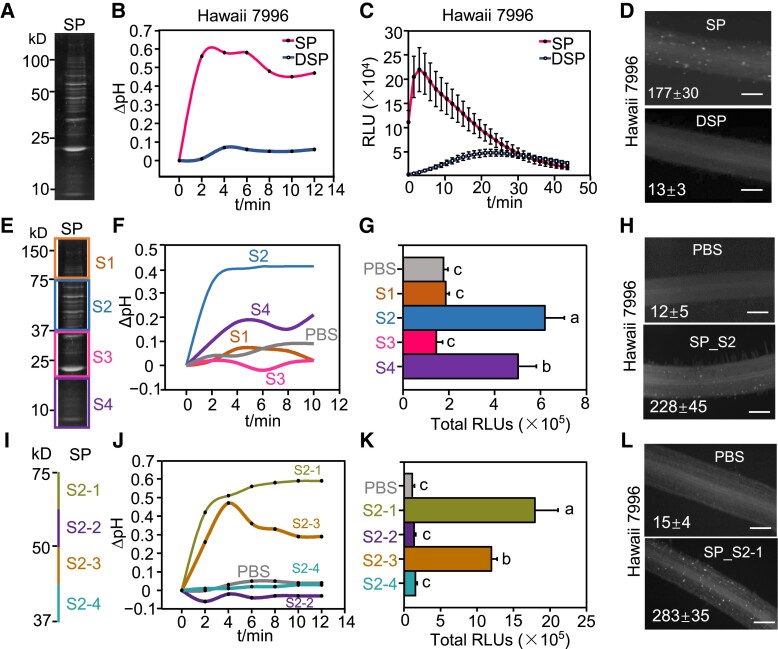
To identify proteinaceous elicitors from Ralstonia, we examined secreted proteins (SPs). To this end, a liquid culture of wild-type (WT) R. solanacearum strain GMI1000 was centrifuged and filtered through a membrane filter to remove bacterial cells. The suspension was precipitated by ammonium sulfate and dialyzed using a semipermeable membrane, and the total SPs were analyzed by SDS-PAGE (Fig. 1A). In the R. solanacearum resistant tomato cultivar Hawaii 7996, soaking of plant roots in the SP solution triggered an extracellular pH increase and ROS burst (Fig. 1, B and C). The immunogenic activity of SPs was abolished by pretreatment with Proteinase K (Fig. 1, B and C), suggesting that proteins or peptides are responsible for the immunogenic activity of the SPs. The R. solanacearum susceptible tomato cultivar Moneymaker also responded to SPs (Supplemental Fig. S1, A and B). In addition, the SPs activated the phosphorylation of MAPKs, expression of immune marker genes screened previously using the RNA-seq data of R. solanacearum culture filtrate-treated tomato roots, and deposition of callose in tomato roots of both Hawaii 7996 and Moneymaker cultivars (Fig. 1D; Supplemental Fig. S1, C to E). Together, these results indicate that SPs contain proteinaceous factor(s) that elicit typical tomato PTI responses.
Figure 1.
SPs from R. solanacearum trigger tomato immune responses. A) SPs extracted from WT strain GMI1000 were separated by SDS-PAGE. B) SPs trigger extracellular alkalization in Hawaii 7996 roots. SPs pretreated with proteinase K are represented as DSPs. The tomato roots were treated with 10 μg/mL SPs or DSPs of GMI1000 and the pH was recorded by a pH meter continuously. Root sections from three 2-wk-old seedlings were measured for each sample. C) SPs induce ROS burst in Hawaii 7996 roots. The relative light units (RLU) were detected post SPs or DSPs treatment. Error bars represent ± SE (n = 10). Replicates represent individual samples containing 4 root sections. D) Callose deposition triggered by SPs in Hawaii 7996 roots. The tomato roots were treated with 10 μg/mL SPs or DSPs for 24 h. E) SPs were separated by SDS-PAGE and divided into 4 segments based on MW: S1, S2, S3, and S4. F to H) S2 segment induces dramatic extracellular alkalization (F), ROS burst (G) and callose deposition (H) in Hawaii 7996 roots. PBS solution was used as a control. I) Schematic diagram of S2 segment that was divided into 4 segments as shown by different colors. J to L) S2-1 recognition induces dramatic extracellular alkalization (J), ROS burst (K) and callose deposition (L) in Hawaii 7996 roots. ROS production in (G) and (K) was presented as the total RLU during all time points upon treatment. Error bars represent ± SE (n = 7). The callose in each root section in (D), (H), and (L) was quantified using Image J and values represent the mean ± SE (n = 5, callose per mm2 in roots). Scale bars indicate 100 µm. Different letters in (G) and (K) represent significant differences by one-way ANOVA analysis (P < 0.05). Replicates represent individual samples containing 4 root sections. All the experiments were repeated at least 3 times with similar results.
To determine the responsible protein triggering tomato immunity, we separated the SPs by SDS-PAGE, cut the gel into 4 segments based on the molecular weight (MW) markers, and extracted the proteins (Fig. 1E). We found that SP_S2 and SP_S4 triggered a dramatic and mild increase in extracellular pH, respectively, upon soaking roots in the SP solution (Fig. 1F). Consistent with the pH change, we observed ROS burst triggered by SP_S2 and SP_S4, with SP_S2 inducing higher ROS production that SP_S4 (Fig. 1G). The expression of tomato immune marker genes ethylene response factors 2a (SlERF2a) and ethylene response factors 2b (SlERF2b) was upregulated by the SP_S2 treatment (Supplemental Fig. S1F). In addition, both SP_S2 and SP_S4 triggered callose deposition in tomato roots (Fig. 1H, Supplemental Fig. S1G).
To narrow down the responsible protein, proteins extracted from the gel slice corresponding to SP_S2 were further separated into 4 portions (Fig. 1I). An increase in extracellular pH was detected in tomato roots after treatment with both S2-1 and S2-3, although the increase observed with S2-1 was greater than that observed with S2-3 (Fig. 1J). Consistent with the pH change, a ROS burst was detected with S2-1 and S2-3, with S2-1 inducing higher ROS production that S2-3 (Fig. 1K). S2-1 treatment also induced callose deposition in tomato roots (Fig. 1L).
PehC is an immune elicitor for tomato
Since the S2-1 fraction showed the strongest induction of PTI in tomato, we performed liquid chromatography-tandem mass spectrometry (LC-MS/MS) to identify the immunogenic protein within this fraction. PehC was identified as the most abundant protein in the S2-1 fraction, based on protein coverage and peptide frequency data (Supplemental Table S1). Therefore, we hypothesized that PehC is the elicitor that triggers PTI in tomato.
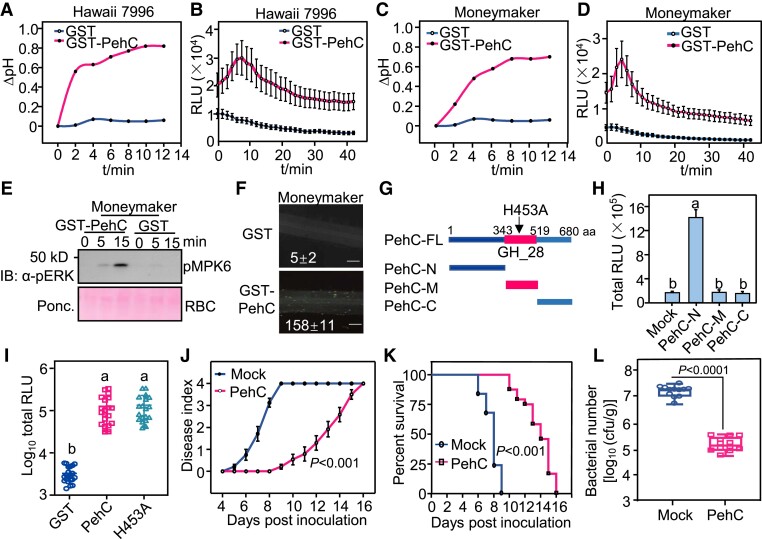
To test this hypothesis, we purified recombinant glutathione S-transferase (GST)-fused PehC protein from Escherichia coli and tested its immunogenic activity (Supplemental Fig. S2A). The GST-PehC protein, but not GST (negative control), induced an extracellular pH increase and ROS burst in Hawaii 7996 and Moneymaker plant roots (Fig. 2, A to D). GST-PehC also induced MAPK activation and callose deposition in Moneymaker roots (Fig. 2, E and F). Similar immune responses were detected in Hawaii 7996 roots (Supplemental Fig. S2, B and C). Together, these results suggest that PehC is an immune elicitor secreted by Ralstonia that induces typical PTI responses in tomato.
Figure 2.
PehC activates tomato immunity as an elicitor. A) PehC induces extracellular alkalization in Hawaii 7996 roots. The tomato roots were treated with 10 μg/mL GST-PehC or GST as a control. Root sections from three 2-wk-old seedlings were measured for each sample. B) PehC triggers a strong ROS burst in Hawaii 7996 roots. Error bars represent ± SE (n = 7). Replicates represent individual samples containing 4 root sections. C) PehC induces extracellular alkalization in Moneymaker roots. This assay was performed as in (A). D) PehC triggers strong ROS burst in Moneymaker roots. This assay was performed as in (B). E) PehC induces MAPK activation in Moneymaker roots. MAPK activation was detected by immunoblotting using an α-pERK1/2 antibody (Top). Protein loading was shown by Ponceau S staining for RuBisCO (RBC, bottom). F) PehC triggers callose deposition in Moneymaker roots. The callose in each root section was quantified using Image J and values represent the mean ± SE (n = 5, callose per mm2 in roots). Scale bars indicate 100 µm. G) Schematic diagram showing truncated variants of PehC and the H453 site which is the key residue for PehC enzymatic activity. PehC-N, N-terminus of PehC (1-343 aa). PehC-M, the middle part of PehC containing the enzymatic activation region (344-519 aa). PehC-C, C-terminus of PehC (520-680 aa). H) N-terminal PehC specifically induces ROS burst in Moneymaker roots. Error bars represent ± SE (n = 9). I) GST-PehC triggers ROS burst in Moneymaker roots independent of its enzymatic activity. Error bars represent ± SE (n = 8). ROS production in (H) and (I) was presented as the total RLU during all time points upon treatment. Replicates represent individual samples containing 4 root sections. J) PehC-pretreatment enhances Moneymaker resistance to R. solanacarum infection. The disease index was recorded based on a scale ranging from “0” (no symptoms) to “4” (complete wilting) at indicated time points. Data points represent the average disease index ± SE (n = 24 individual plants). The AUDPC was calculated and different letters represent significant differences by a LMMs analysis (P < 0.05). K) The survival ratio of Moneymaker upon R. solanacarum infection is increased by PheC pretreatment. The survival ratio was analyzed by comparing the number of survival plants to the total plants from the data in (J). The Kaplan Meier estimates survival analysis was performed and different letters represent significant differences by Log-rank (Mantel-Cox) test (P < 0.05). L) PehC-pretreatment inhibits the multiplication of R. solanacarum in Moneymaker stems. Tomato stems were collected and weighed at 3 dpi. The series of diluted samples were plated on triphenyl tetrazolium chloride (TTC) medium to count the CFU per gram of stem in fresh weight. Error bars represent ± SE (n = 12). Replicates represent individual samples containing one stem section from individual plants. Different letters in (H), (I), and (L) represent significant differences by one-way ANOVA analysis (P < 0.05). The above experiments were repeated 3 times with similar results.
PehC is annotated as an exo-PG and the conserved GH28 domain spans aa 343–419 (Fig. 2G) and can hydrolyze pectin and homogalacturonan to release GalA. In addition to PehC, the Ralstonia genome encodes 2 other PGs, PehA (endo-PG) and PehB (exo-PG) (Gonzalez and Allen 2003). Although PehA-C proteins contain similar functional domains and belong to the same superfamily, sequence similarities among these 3 proteins are quite low. Purified GST-PehA and GST-PehB fusion proteins did not trigger a ROS burst in tomato roots (Supplemental Fig. S2, D to F), suggesting that PehA and PehB do not exhibit immunogenic activity in tomato.
To identify the immunogenic region of PehC, its amino acid sequence was divided into 3 parts: N-terminal region (aa 1–343), middle region (aa 344–519; which carries the domain responsible for PehC enzymatic activity), and C-terminal region (aa 520–680) (Fig. 2G). Deletion mutagenesis of PehC revealed that the N-terminal region of PehC is required to induce the ROS burst in tomato roots (Fig. 2H), suggesting that the PTI-eliciting function of PehC is independent of its enzymatic activity. In support of this hypothesis, the PehCH453A variant protein carrying an amino acid substitution in the conserved GH28 domain, which is required for enzymatic activity, triggered similar ROS production as the WT PehC (Fig. 2I). These results indicate that the N-terminal region of PehC contains an immunogenic fragment.
PehC induces bacterial wilt resistance and global gene reprograming
Next, we tested the biological significance of PehC-triggered immunity. Moneymaker (susceptible) plants were pretreated with PehC or water (mock) 2 d before R. solanacearum inoculation. Compared with mock-pretreated plants, the PehC-pretreated plants showed a much lower disease index and significantly higher survival rate (Fig. 2, J and K, and Supplemental Fig. S2G). Meanwhile, the proliferation of the pathogen in roots and stems was assessed. Bacterial titer in both upper and lower parts of the stem was approximately 2 log10 values lower in PehC-pretreated plants than in mock-pretreated plants (Fig. 2L).
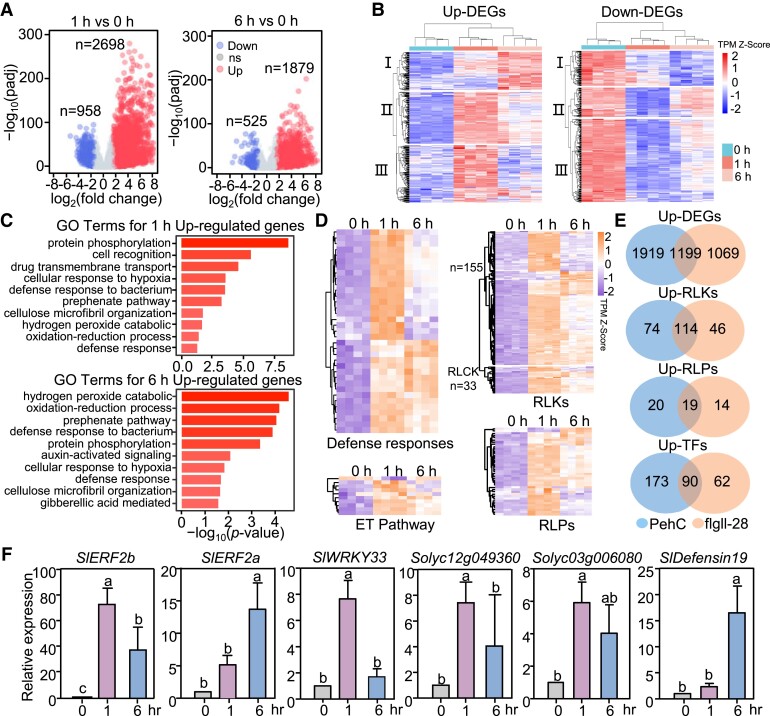
To further understand the PehC-induced transcriptional reprograming in tomato, we analyzed the transcriptomic changes in the tomato cultivar Moneymaker after elicitation with PehC. The roots of 2-wk-old tomato seedlings were treated with or without 10 μg/mL GST-PehC recombinant protein for 1 or 6 h, with 4 biological replications. Then, RNA was extracted from those samples and sequenced, resulting in, on average, 42 million 150-bp strand-specific paired-end reads per sample. Root samples in the same group showed good correlation and repeatability (Supplemental Fig. S3, A and B). A total of 3,656 differentially expressed genes (DEGs), including 2,698 upregulated and 958 down-regulated genes, were identified at 1 h post-PehC treatment (Fig. 3A, Supplemental Data Set 1). In comparison, fewer DEGs (2,414) were identified at 6 h post-PehC treatment (1,879 upregulated and 535 down-regulated genes) (Fig. 3A, Supplemental Data Set 2). Figure 3B shows a heatmap displaying the expression patterns of DEGs. Cluster analysis of DEGs showed that the genes in Clusters II and III were upregulated at 1 h post-PehC treatment but were less induced or downregulated at 6 h post-PehC treatment, indicating that the PehC-activated transcriptional reprograming of tomato was relatively transient, possibly owing to the rapid immune response of tomato at an early stage of infection.
Figure 3.
PehC triggers global transcriptional reprograming in tomato roots. A) Volcano plot of whole-genome transcripts in tomato treated with PehC 1 h versus 0 h (left panel) and 6 h versus 0 h (right panel). The x-axis indicates log2 (fold change) and the y-axis indicates –log10 (padj). Padj is the adjusted P-value. Red circles indicate up-regulated genes, blue circles indicate down-regulated genes, and gray circles indicate genes without significant expression change (ns). The up-regulated genes and down-regulated genes were calculated in accordance with log2 (fold change) ≥ 1.5 and padj < 0.05. B) Cluster of DEGs in tomato treated with PehC for 0, 1, and 6 h. Longitudinal clustering was performed according to the transcripts per kilobase of the exon model per million mapped reads (TPM) of all DEGs. C) Selected top GO terms enriched in up-regulated genes in tomato treated with PehC for 1 and 6 h. The GO enrichment analysis was performed on the agriGO website. D) Heatmaps showing the expression changes of up-regulated genes that related to defense response (top left panel), ET pathway (bottom left panel), RLKs (top right panel), and RLPs (bottom right panel) upon PehC treatment. Longitudinal clustering was performed according to the original TPM. “n” indicates the number of genes listed in the corresponding group. E) Venn diagrams of up-DEGs, up-RLKs, up-RLPs, and up-TFs with the comparison between PehC- and flgll-28-induced genes. F) The expression of immune-related genes was upregulated upon PehC treatment. Two-week-old Moneymaker seedlings were treated with GST-PehC for 1 h or 6 h. Data were normalized to the expression of SlACTIN2 in RT-qPCR analysis. Error bars represent ± SD (n = 3 technical repeats from one independent experiment). Different letters represent significant differences by one-way ANOVA analysis (P < 0.05). These experiments were repeated at least 3 times with similar results.
To better understand the changes in specific biological pathways after the PehC treatment, we performed a Gene Ontology (GO) enrichment analysis of the DEGs. Genes upregulated at 1 and 6 h post-PehC treatment were associated with defense response and oxidation–reduction reactions, which are related to the classic PTI immune response (Fig. 3C). For instance, the typical ethylene signaling pathway-related marker genes, including SlERF2a and SlERF2b, were upregulated (Fig. 3D), and GO terms such as protein phosphorylation, cell recognition, and drug transmembrane transport were enriched, which is consistent with signal transduction under stress (Fig. 3C). In addition, the up-regulated genes were associated with the secondary metabolites related to immunity, such as lipid, phenylpropanoid, and fatty acid metabolic processes (Fig. 3C, Supplemental Fig. S3E, and Supplemental Data Set 3). By contrast, genes downregulated by the PehC treatment were associated with photosynthesis, metabolites, and energy-related to vegetative growth (Supplemental Fig. S3C). Collectively, GO enrichment analysis suggested that tomato defense responses were activated upon the PehC treatment (Fig. 3D).
Previous reports indicate that flgII-28 is the major MAMP responsible for immunity-associated transcriptional changes in tomato (Cai et al. 2011; Pombo et al. 2017). Therefore, we compared the genes differentially expressed by the PehC treatment (PehC-DEGs) with those differentially expressed by the flgII-28 treatment (flgII-28-DEGs) to determine the change in immune-responsive gene expression due to the PehC treatment. For convenience, we combined the 1 and 6 h post-PehC treatment datasets to form the PehC-DEG dataset. A total of 188 RLKs, 39 RLPs, and 263 transcription factors (TFs) were identified in the PehC-upDEG dataset; these included some cysteine-rich RLKs and a number of diseases resistance-related WRKY, MYB, and ERF family TFs (Fig. 3E, Supplemental Fig. S3D, and Supplemental Data Set 4). The percentage of RLKs, RLPs, and TFs in the PehC-DEG and flgII-28-DEG datasets was found to be similar (Supplemental Data Set 5). A total of 1,199 up-regulated genes were common to both the flgII-28-upDEG and PehC-upDEG datasets, including 114 RLKs, 19 RLPs, and 90 TFs (Fig. 3E). Among the known immunity-related PRRs, such as tomato systemin receptor 1 (SlSYR1, Solyc03g082470) and systemin receptor 2 (SlSYR2, Solyc03g082450), the receptors for plant peptide hormone systemin, were upregulated upon the PehC treatment (Wang et al. 2018). These data indicate that PehC is an immune elicitor that triggers defense responses and bacterial wilt resistance in tomato plants.
PehC triggers immune responses in multiple hosts in a SERK coreceptor-dependent manner
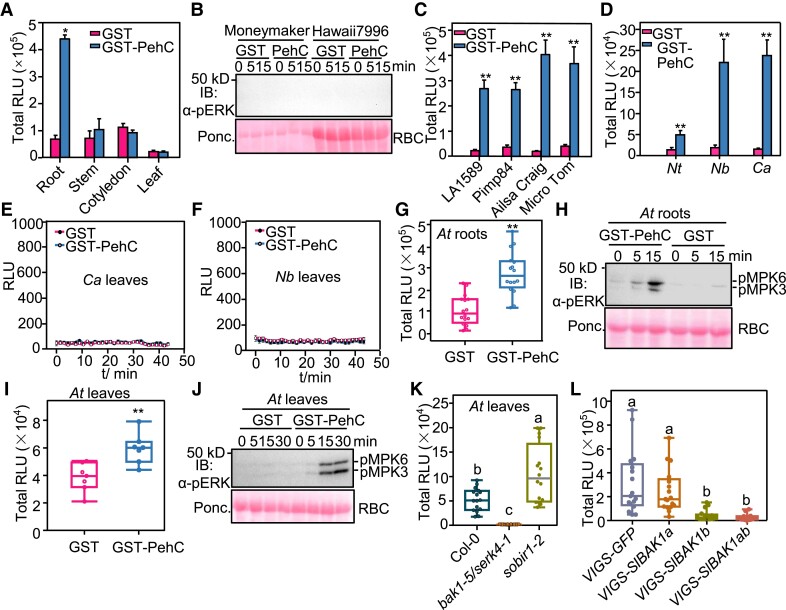
Next, we investigated the PehC recognition ability of different tissues and plant species. First, since Ralstonia is a soil-borne pathogen and enters the plant through its roots, we tested whether PehC-triggered PTI responses are root-specific. Interestingly, among the roots, leaves, cotyledons, and stems of tomato plants, only the roots responded to the PehC treatment and produced ROS (Fig. 4A). PehC failed to activate MAPK in tomato leaves (Fig. 4B). Second, we tested whether PehC triggers immunity in wild tomato (Solanum pimpinellifolium) and other tomato cultivars. Results showed that the PehC treatment triggered a ROS burst in the roots of wild tomato accessions LA1589 and Pimp84 as well as in the roots of the tomato cultivars Ailsa Craig and Micro Tom (Fig. 4C). Furthermore, PehC-induced ROS burst in the roots, but not in the leaves, of other Solanaceous species, including N. benthamiana, N. tabacum, and pepper (Capsicum annuum) (Fig. 4, D, E, and F). These findings demonstrate that PehC triggers immunity in the roots of Solanaceae plants.
Figure 4.
PehC triggers immune responses dependent on the SERK coreceptors. A) PehC triggers ROS burst specifically in tomato roots. Different tomato tissues from 2-wk-old tomato seedings were treated with 10 μg/mL GST-PehC. Error bars represent ± SE (n = 8). Replicates represent individual samples containing 4 root sections, 2 stem sections, or 1 leaf disc. B) PehC cannot trigger MAPK activation in tomato leaves. Leaf discs from 4-wk-old Moneymaker or Hawaii 7996 were treated with 10 μg/mL GST-PehC or GST as a control. Total protein extracts were detected by immunoblotting using an α-pERK1/2 antibody. C) ROS burst can be induced by PehC in roots of different tomato accessions. Roots of 2-wk-old LA1589, Pimp84, Micro Tom, and Alisa Craig seedings were treated with 10 μg/mL GST-PehC. Error bars represent ± SE (n = 12). D) PehC induces ROS burst in roots of different Solanaceous crops Nicotiana tabacum (Nt), Nicotiana benthamiana (Nb), and Capsicum annuum (Ca). Error bars represent ± SE (n = 10). E and F) PehC cannot activate ROS burst in C. annuum and N. benthamiana leaves. Error bars represent ± SE (n = 9, one leaf disc for each sample). G and H) PehC activates ROS burst (G) and MAPK activation (H) in Arabidopsis thaliana (At) roots. Data for ROS level are presented as box plots displaying all the points from min to max, split by the median (n = 16). I and J) PehC induces ROS burst (I) and MAPK activation (J) in Col-0 leaves. Leaf discs of 4-wk-old Col-0 were treated with 10 μg/mL GST-PehC or GST. Data for ROS level are presented as box plots displaying all the points from min to max, split by the median (n = 7, one leaf disc for each sample). K) Compromised PehC-induced ROS bust in the leaves of bak1-5/serk4-1 mutant. Measurements were plotted as box plots displaying all the points from min to max, split by the median (n = 16, one leaf disc for each sample). L) Compromised PehC-induced ROS burst in SlBAK1-silenced tomatoes. Hawaii 7996 seedlings were infiltrated with Agrobacteria containing pTRV-GFP, pTRV-SlBAK1a, pTRV-SlBAK1b, or pTRV-SlBAK1ab, respectively. Root sections of indicated tomato plants were collected 2 wks after VIGS and were treated with 10 μg/mL GST-PehC. Measurements were plotted as box plots displaying all the points from min to max, split by the median (n = 17). Replicates in (C), (D), (G), and (L) represent individual sample containing 4 root sections. Asterisk in (A), (C to D), (G), and (I) indicates a significant difference compared to control (Student's one-tailed t-test, *P < 0.05, **P < 0.01). Different letters in (K) and (L) represent significant differences by one-way ANOVA analysis (P < 0.05). The above experiments were repeated 3 times with similar results.
R. solanacearum also infects the model plant Arabidopsis, which belongs to the Brassicaceae family. We found that PehC triggered ROS burst and activated MAPKs in Arabidopsis roots (Fig. 4, G and H). Surprisingly, strong ROS production and MAPK phosphorylation were detected in Arabidopsis leaves upon PehC treatment (Fig. 4, I and J), which was in contrast to the results obtained in the tested Solanaceae species. Since Arabidopsis is not a natural host of Ralstonia, it is possible that the potential PehC receptor is universally expressed in both the roots and leaves of Arabidopsis but only in the roots of Solanaceous plants.
Given that BAK1 and SOBIR1 are two key coreceptors of plant RLKs and RLPs responsible for MAMP recognition (Liebrand et al. 2014), we tested the PehC-mediated immunity of bak1 and sobir1 Arabidopsis mutants. Notably, the sobir1 mutant exhibited WT-like PehC-induced ROS burst in both leaves and roots (Fig. 4K, Supplemental Fig. S4A), implying that PehC may be not recognized by RLPs. By contrast, the PehC-triggered ROS burst was almost abolished in the bak1-5 serk4-1 double mutant (Fig. 4K, Supplemental Fig. S4A). MAPK activation triggered by PehC in both leaves and roots was also largely reduced in the bak1-5 serk4-1 double mutant compared with the WT Arabidopsis (Col-0) plants (Supplemental Fig. S4, B and C), indicating that 1 or more RLK acts as the PehC receptor in Arabidopsis. Tomato possesses 2 BAK1 orthologs: SlBAK1a and SlBAK1b. We transiently silenced SlBAK1a and SlBAK1b genes in tomato through Agrobacterium tumefaciens-mediated transformation of tobacco rattle virus vectors and found that PehC failed to trigger a ROS burst in SlBAK1b- and SlBAK1a/b-silenced plants (Fig. 4L). Collectively, these results indicate that PehC recognition in plants is mediated by an RLK-type receptor and is dependent on SERK coreceptors.
PehC plays a positive role in R. solanacearum infection
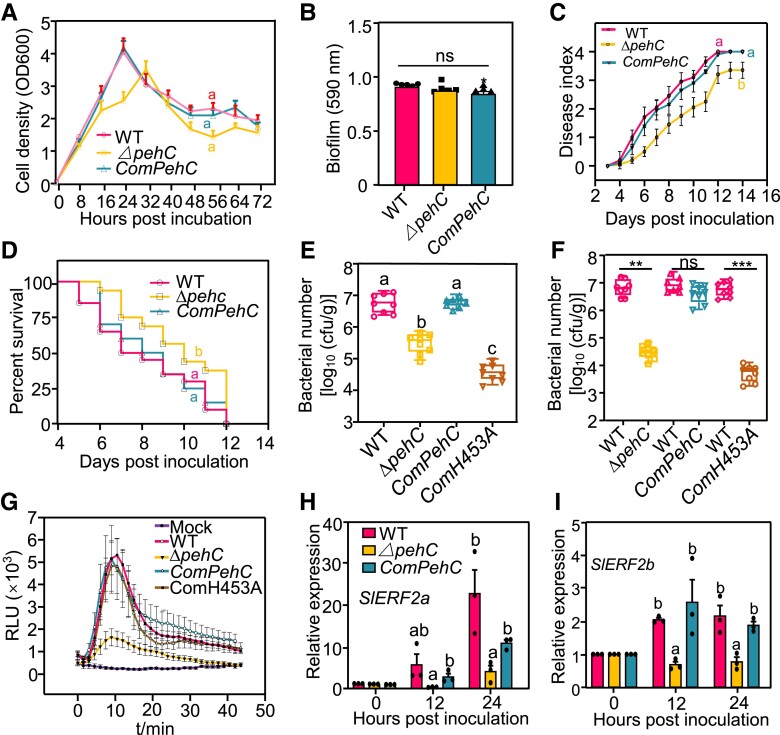
Next, to investigate the biological function of PehC in Ralstonia infection, we generated a pehC deletion mutant (ΔpehC) by homologous recombination in the WT strain GMI1000 (Supplemental Fig. S5A). A PehC complementation strain (ComPehC) was generated by expressing the native promoter-driven PehC labeled with an HA-tag, and confirmed by immunoblotting (Supplemental Fig. S5B). We also generated the complementation strain ComH453A expressing PehCH543A, which encodes a catalytically inactive variant of PehC, to verify the importance of PehC enzymatic function (Supplemental Fig. S5C). The growth dynamics of ΔpehC, ComPehC, ComH453A, and WT strains showed no significant difference in rich medium (Fig. 5A), and the colony size, color, and morphology of these strains were comparable on triphenyl tetrazolium chloride (TTC) plates (Supplemental Fig. S5D). Furthermore, the pathogenicity-related biofilm formation and EPS secretion ability as well as the motility of ΔpehC mutant, ComPehC and ComH453A complementation strains, and WT strain were also similar (Fig. 5B, Supplemental Fig. S5, E and F).
Figure 5.
PehC is required for R. solanacearum virulence and multiplication in plant xylem. A) Bacterial growth curve of GMI1000, ΔpehC mutant, and complementation strain ComPehC in rich medium Casamino Acids-Peptone-Glucose (CPG). The cell density was determined by measuring the OD at 600 nm at indicated time points. Data points represent average values and error bars represent ± SD (n = 3). B) Biofilm formation of GMI1000, ΔpehC, and CompehC in CPG medium. The biofilm formation was stained by crystal violet and quantitated at 590 nm. Error bars represent ± SE (n = 7). C) Compromised virulence of ΔpehC mutant compared to the WT strain. Soil-drenching inoculation assays were performed with GMI1000, ΔpehC, and ComPehC on 4-wk-old Moneymaker plants. The disease index was recorded based on a scale ranging from “0” (no symptoms) to “4” (complete wilting) at indicated time points. Data points represent the average disease index ± SE (n = 20 individual plants). The AUDPC was calculated and different letters represent significant differences by LMMs analysis (P < 0.05). D) Increased survival rate of Moneymaker plants post the ΔpehC mutant inoculation compared with GMI1000. The survival ratio was analyzed by comparing the number of survival plants to total plants from the data in (C). The Kaplan Meier estimates survival analysis was performed and different letters represent significant differences by Log-rank (Mantel-Cox) test (P < 0.05). E) Bacterial multiplication of GMI1000, ΔpehC mutant, and complementation strains in Moneymaker xylem. Soil-drenching inoculation assays were performed with different strains on 4-wk-old Moneymaker plants. Error bars represent ± SD (n = 8, stem from one infected plant as a replicate). F) Compromised growth competition of ΔpehC mutants in Moneymaker stems. Injection inoculation was performed with a 1:1 mixture of 1 × 108 CFU/mL cell suspension of GMI1000 and ΔpehC mutants, or other indicated pairs. The bacterial titer was calculated 24 h post inoculation. Error bars represent ± SD (n = 12, stem from one infected plant as a replicate). G) SPs of ΔpehC strain induce weaker ROS burst in Moneymaker roots than that of GMI1000. The SPs of GMI1000, ΔpehC mutant, and complementation strains were separated by a 30 kD ultrafiltration centrifugal tube. ROS was measured upon the treatment by the >30 kD fraction. Error bars represent ± SE (n = 8). Replicates represent individual samples containing 4 root sections. H and I) The ΔpehC mutant induces weaker expression of immune-related genes than GMI1000 in Moneymaker roots. The tomato roots were soaked in the suspension of GMI1000 or ΔpehC mutant strain for 30 min and samples were collected at 12 and 24 hpi. The expression of SlERF2a and SlERF2b was normalized to the expression of SlACTIN2. Values represent ± SD (n = 3). Different letters in (A), (E), and (H to I) represent significant differences by one-way ANOVA analysis (P < 0.05). Asterisk in (B) and (F) indicates a significant difference compared to the control (Student's one-tailed t-test, **P < 0.01, ***P < 0.005, ns, not significant). The above experiments were repeated 3 times with similar results.
To test the contribution of PehC to virulence, we inoculated tomato Moneymaker plants with the WT (GMI1000), ΔpehC mutant, and ComPehC and ComH453A complementation strains by the soil soaking method. The ΔpehC mutant strain displayed lower virulence and a relatively lower disease index than the WT strain, while the ComPehC strain showed restored virulence (Fig. 5C). Consistently, plants infected with the ΔpehC mutant strain showed a higher survival rate than those inoculated with the WT strain (Fig. 5D). However, the ComH453A complementation strain could not restore the virulence of the ΔpehC mutant to the WT level (Supplemental Fig. S6, A and B). Then, we quantified the bacterial population in infected tomato stems at 3 d postinoculation (dpi). The bacterial titer of the ΔpehC mutant was 1,000-fold lower than that of the WT strain in tomato stems, and the population of the ComPehC strain was restored to the level of the WT strain (Fig. 5E). Notably, the population of the ComPehC and ComH453A strains was significantly lower than that of the ΔpehC mutant, indicating that the catalytically inactive variant PehCH543A acts only as an immune elicitor. Furthermore, we competed for the ΔpehC mutant against the WT strain in planta by injecting a mixture of the WT and ΔpehC mutant cell suspensions into Moneymaker stems. The WT strain showed more competitive growth than the ΔpehC mutant strain, and the ComPehC strain, but not the ComH453A strain, could recover the growth defect of ΔpehC in tomato xylem (Fig. 5F). Our data suggest that PehC is required for R. solanacearum virulence and growth in tomato xylem.
Since PehC functions as an important immune elicitor for the Ralstonia–tomato interaction, we hypothesized that the total SPs of the ΔpehC mutant could evade recognition by plants. To test this hypothesis, SPs were isolated from the WT, ΔpehC, ComPehC, and ComH453A strains. Figure 1, F and G show that a fraction of SPs (MW < 25 kD) also showed immunogenic activity; therefore, to avoid the influence of this fraction, the SPs were separated using the 30-kD cutoff Ultra centrifugal filters. Indeed, an SP fraction (MW > 30 kD) extracted from the WT, ComPehC, and ComH453A strains, but not from the of ΔpehC mutant strain, triggered a strong ROS burst in Moneymaker roots (Fig. 5G). Additionally, the SP fraction of both WT and ΔpehC mutant strains that passed through the Ultra Centrifugal Filters triggered a ROS burst, with no significant difference (Supplemental Fig. S6C).
To further verify whether the ΔpehC mutant strain triggers lower immunity in planta, we monitored immune-responsive gene expression and callose deposition in tomato roots post Ralstonia infection. The ΔpehC mutant induced SlERF2a and SlERF2b expression and callose deposition to much lower levels than the WT, ComPehC, and ComH453A strains in Moneymaker roots (Fig. 5H, Supplemental Fig. S6, D and E). The weaker immune responses may also partially be due to the low cell density of the ΔpehC mutant in tomato roots. These results suggest that PehC contributes to Ralstonia virulence, but the protein itself is recognized as an immune elicitor by tomato.
Immune evasion by PehC during R. solanacearum infection
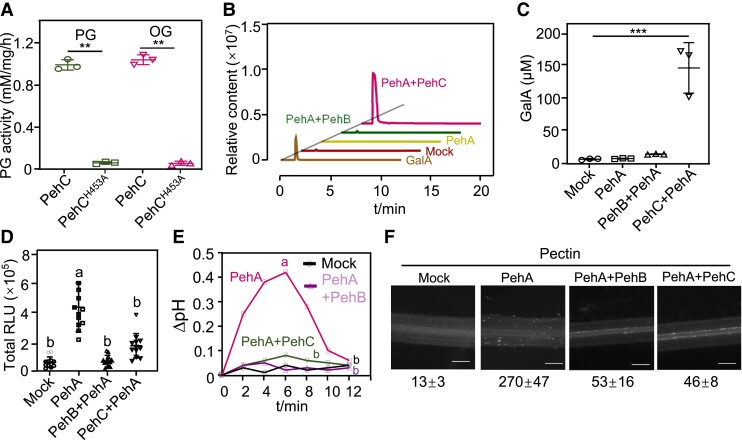
The ΔpehC mutant evaded plant recognition and immunity, which could increase virulence. However, the mutant showed reduced virulence and fitness in tomato, indicating that PehC may perform other biological roles during the plant–Ralstonia interaction. To test this possibility, the enzymatic activity of PehC and the amount of reducing sugars were measured by the 3,5-dinitrosalicylic acid (DNS) assay. The WT PehC protein, but not the PehCH453A variant, could hydrolyze pectin and OGs (Fig. 6A). Among the three PGs in R. solanacearum, PehA (endo-PG) hydrolyzes the cell wall pectin homogalacturonan backbone, leading to the production of OGs. We performed in vitro pectin degradation assays using the different R. solanacearum PGs. Through LC-MS analysis, we showed that PehC (exo-PG) could further hydrolyze PehA-produced OGs, leading to the release of the monosaccharide GalA (Fig. 6B). The LC-MS quantitative data also indicated that the enzymatic activity of PehC, but not that of PehA and PehB, could lead to higher accumulation of GalA (Fig. 6C). These data confirmed that PehC is an exo-PG that hydrolyzes long-chain pectin into the monosaccharide GalA.
Figure 6.
The immune evasion by PehC during R. solanacearum infection. A) PehC hydrolyzes pectin and OGs dependent on its enzyme activity. The concentration of GalA was measured using the DNS method. Values represent data from 3 independent reaction and error bars represent ± SD. B) PehC mediates the production of monosaccharide GalA from pectin. LC-MS/MS analysis of the GalA concentration in the indicated hydrolysis reactions. C) Quantitative analysis of GalA hydrolyzed from pectin by PehC in (B). Error bars represent ± SD (n = 3). D) PehC abolishes the ROS burst triggered by PehA/pectin reaction products OGs. Moneymaker roots were treated with indicated pectin hydrolysis reactions or pectin as a control. Error bars represent ± SE (n = 9). Replicates represent individual samples containing 4 root sections. E) PehC inhibits the extracellular alkalization in tomato roots induced by PehA/pectin reaction products OGs. Roots from three 2-wk-old Moneymaker seedlings were monitored as one biological repeat. F) PehC inhibits the PehA/pectin reaction products-induced callose deposition. The callose in the root section was quantified using Image J, and values represent the mean ± SE (n = 5, callose per mm2 in roots). Asterisk in (A) and (C) indicates a significant difference compared to the control (Student's one-tailed t-test, **P < 0.01, ***P < 0.005). Different letters in (D) and (E) represent significant differences by one-way ANOVA analysis (P < 0.05). The above experiments were repeated 3 times with similar results.
Many studies have shown that OGs produced through the hydrolysis of pectin by endo-PGs act as DAMPs in various plant systems (Ferrari et al. 2013). R. solanacearum PehC was able to degrade OGs into the GalA monomer; we hypothesized that this may abolish the immunogenic activation of OGs in the host plant. To test this hypothesis, we monitored the immunogenic activity of different pectin degradation products. Indeed, the products of pectin hydrolysis catalyzed by the endo-PG PehA triggered ROS burst and extracellular pH increase in Moneymaker roots (Fig. 6, D and E). Interestingly, extracellular alkalization and the ROS burst in tomato roots were blocked when an exo-PG (PehB or PehC) was added to the pectin degradation reaction, together with PehA. Consistent with these changes, PehB or PehC also inhibited PehA/pectin hydrolysis product-induced callose deposition in tomato roots (Fig. 6F, Supplemental Fig. S7A). Taken together, these results suggest that PehC degrades the DAMP OGs, which are hydrolyzed by PehA to prevent the plant immune response during infection.
PehC-produced GalA acts as a carbon source for Ralstonia
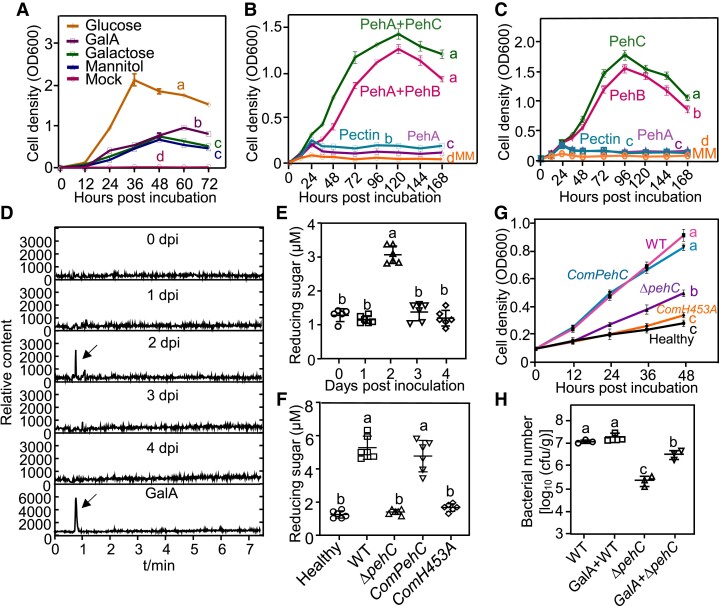
Given that Ralstonia colonizes the xylem, the pectin and related components of the cell wall are degraded by bacterial PGs and likely serve as potential carbon sources for Ralstonia. The phylogenetic analysis of bacterial PG proteins indicated that the endo-PG PehA homologs are widespread in many bacteria species; however, exo-PGs occur in only a limited number of bacterial species (Supplemental Fig. S8). Thus, we tested the bacterial growth rate in minimal medium (MM) supplemented with different carbon sources. Glucose promoted vigorous Ralstonia growth. Galactose and mannitol, which were previously demonstrated to serve as carbon sources in the tomato xylem, supported Ralstonia growth at a relatively lower rate. However, GalA, as the pectin monomer, was able to support Ralstonia multiplication as the sole carbon source (Fig. 7A). Next, we performed a growth assay with various pectin hydrolysis products as the sole carbon source in the MM. Pectin and OGs produced by PehA could not be utilized by Ralstonia (Fig. 7, B and C). However, the WT strain GMI1000 showed vigorous growth using products digested together by PehA and PehC or together by PehA and PehB as well as using products digested independently by PehC or PehB, suggesting that Ralstonia could utilize GalA monomers or dimers for growth and multiplication.
Figure 7.
PehC produces GalA as a carbon source for Ralstonia infection. A) Bacterial growth dynamic of R. solanacearum in MM supplemented with different carbon resources. The cell density was measured at OD600 at indicated time point. Error bars represent ± SD (n = 3). B and C) The pectin hydrolysis products catalyzed by PehB and PehC promote the growth of GMI1000, respectively. Error bars represent ± SD (n = 3). D) GalA production in Moneymaker xylem sap post R. solanacarum inoculation was analyzed by LC-MS/MS. Base peak chromatograms indicate signal intensity at each retention time (min). A representative chromatogram of purified GalA was shown at the bottom. E) Total concentration of reducing sugars in the Moneymaker xylem sap post R. solanacarum inoculation. Petiole inoculation assays were performed with GMI1000 and the concentration of reducing sugars in xylem sap was measured using the DNS method at indicated time points. Error bars represent ± SE (n = 6, xylem sap from one infected plant as a sample). F) The increase of reducing sugars in the tomato xylem sap is dependent on PehC. Four-week-old Moneymaker was petiole-inoculated with indicated bacterial strains and the reducing sugar was measured using DNS assay. Error bars represent ± SD (n = 6, xylem sap from one infected plant as a sample). G) Xylem sap from Ralstonia-infected Moneymaker facilitates bacterial multiplication in MM. The tomato xylem sap was collected from 6 healthy plants or plants infected by GMI1000, ΔpehC strain, CompehC, or ComH453A at 2 dpi. Error bars represent mean ± SD (n = 3). H) GalA promotes ΔpehC growth in Moneymaker xylem. Petiole inoculation assays were performed with GMI1000 and ΔpehC supplemented with or without 5 μM GalA. Bacterial multiplication in the xylem of each infected plant was monitored at 2 dpi. Error bars represent ± SE (n = 6). The AUC was calculated for statistical analysis in (A to C) and (G) and different letters represent significant differences by Log-rank test (P < 0.05). Different letters in (E to F) and (H) represent significant differences by one-way ANOVA analysis (P < 0.05). The above experiments were repeated at least 3 times with similar results.
Further, we tested whether GalA could be produced in the tomato xylem during R. solanacearum infection. The petioles of Moneymaker plants were inoculated with GMI1000 to ensure uniform infection of the xylem, and the xylem sap was collected at the indicated time points postinoculation. LC-MS analysis revealed a transient increase in GalA concentration in the xylem sap at 2 dpi, which dropped to a normal concentration subsequently (Fig. 7D). We also measured the total concentration of reducing sugars (including GalA) in the xylem sap as another readout of PehC activity (Fig. 7E). The peak of concentrations both GalA and total reducing sugars in the xylem sap were observed at 2 dpi. Based on this observation, at 2 dpi, the disease index of tomato plants showing relatively uniform wilt symptoms was rated as 1. Then, we used the soil drench inoculation method to inoculate plants and collected the xylem sap samples from plants showing different disease indexes. Consistently, reducing sugars showed the highest concentration in the xylem sap of plants with a disease index of 1, which is equivalent to the early stage of wilt symptom development (Supplemental Fig. S7B). These results suggested that Ralstonia infection rapidly increases the concentration of reducing sugars in the xylem.
To study the biological relevance of PehC-mediated GalA production in the xylem sap, the ΔpehC mutant and ComPehC and ComH453A strains were analyzed. Interestingly, unlike the WT strain, the ΔpehC mutant strain could not increase the concentration of reducing sugars. The ability to increase the concentration of reducing sugars in the xylem was restored by ComPehC but not by ComH453A (Fig. 7F). To test the relationship between reducing sugars in the xylem and Ralstonia growth, we used the xylem sap of Moneymaker plants infected with different strains to supplement the MM. The growth curve indicated that the WT GMI1000 strain grew much faster with the xylem sap from WT-infected plants than with that from ΔpehC mutant-infected plants (Fig. 7G). Consistently, compared with the WT strain, propagation of the ΔpehC mutant strain in tomato stem was impaired (Fig. 5E), and GalA supplementation could promote ΔpehC growth in the tomato xylem (Fig. 7H). These data indicate that GalA produced by PehC from pectin or OGs serves as an important carbon source for Ralstonia in the xylem at the early stage of infection.
Discussion
The activation of plant innate immunity relies on the perception of pathogen-derived molecules. Detection of MAMPs by PRRs results in PTI, which constitutes the first layer of defense against most microbial pathogens (Tang et al. 2017). Here, we revealed that Ralstonia employs the exo-PG PehC to degrade the plant cell wall-derived DAMP OGs, which are used to avoid plant immune activation and produce GalA as an important carbon source, thereby increasing virulence. The significance of PehC for bacterial virulence is likely reflected by plants acquiring the ability to recognize PehC as an elicitor upon Ralstonia infection (Fig. 8). Collectively, our findings illuminate the multiple roles of PehC at the interface of plant–pathogen interaction. PehC degrades pectin and OGs to not only decrease plant immunity but also increase the carbon nutrient GalA, thereby contributing to Ralstonia virulence. Consistent with the finding that PehA and PehB are required for rapid colonization by and the full virulence of Ralstonia (Huang and Allen 2000), our findings showed the virulence of the ΔpehC mutant was lower than that of the WT GMI1000 strain. However, a previous study showed that the virulence of ΔpehC is statistically indistinguishable from that of the parent strain K60 (Gonzalez and Allen 2003). These contradictory conclusions could be due to the differences in the parent strains (GMI1000 and K60) or the different inoculation methods.
Figure 8.
A working model for the multiple roles of PehC during R. solanacearum infection. During the infection, R. solanacearum secretes PehA to hydrolyze pectin, an important component of cell wall of host plants, into OGs. OGs functioning as a DAMP could be recognized by plant WAK1-related receptor to trigger DTI. In order to avoid the recognition, R. solanacearum evolved PehB and PehC to degrade OGs which generate GalA2 dimer and GalA monomer. These monosaccharide sugars can be utilized by Ralstonia as the carbon sources to support its growth and colonization at early infection stage. To counter the pathogenicity of R. solanacearum, tomatoes have further evolved the unknown pattern recognition receptor(s) to recognize PehC, leading to the activation of PTI.
Ralstonia causes severe bacterial wilt on tomato and many other plant species, but the MAMPs from Ralstonia perceived by tomato are not well characterized. Early studies found that the PG-deficient pehA/B/C triple mutant exhibits stronger pathogenicity than the pehA/B double mutant (Gonzalez and Allen 2003). These results indicate that PehC may trigger plant immunity, because in the pehA/B double mutant background, no DAMPs or nutrient precursors are produced and the virulence component of PehC is minimal, totally overcome by the immunity responses it triggers. In the current study, we demonstrated that PehC secreted by R. solanacearum is indeed an immune elicitor that could be perceived by Solanaceae plants, specifically in root tissues. PehC triggers typical immune responses, and pretreatment with PehC enhances resistance against Ralstonia infection in tomato plants. Furthermore, complementation with the catalytically inactive form of PehC (PehCH453A) further compromised the virulence of ΔpehC. The root-specific recognition of the pathogen indicates that the receptor(s) for PehC is expressed only in the roots of Solanaceae plants. Surprisingly, both the roots and leaves of Arabidopsis plants responded to the PehC treatment. This difference between plant species is probably related to different expression patterns of the potential receptor(s). Moreover, although Ralstonia infects Arabidopsis, it does so weakly. This may explain why Arabidopsis is not a natural host for Ralstonia. During the screening, we found that more than 1 fraction of the SPs conferred immunogenic function to tomato; the other MAMP components will be explored in future studies.
Secreted endo-PGs recognized by plants as MAMPs have been reported in fungal pathogens. However, in our assay, Ralstonia PehA (endo-PG) did not trigger an immune response in tomato, suggesting that PehA is not a MAMP in tomato. The endo-PG of Botrytis cinerea, BcPG1, activates defense responses in grapevine (Vitis vinifera), and the immunogenic activity of BcPG1 does not require its enzymatic activity (Poinssot et al. 2003). This is reminiscent of Ralstonia PehC, whose immunogenic activity was independent of the enzymatic activity. Arabidopsis receptor-like protein 42 (RLP42) was identified as a receptor to recognize fungal PGs (Zhang et al. 2014). More recently, a conserved 9-aa fragment, pg9, was identified as the minimal region of Botrytis PGs perceived by RLP42 (Zhang et al. 2021). However, PehC-mediated immunity is likely activated through the RLK-type receptor(s) in tomato and Arabidopsis. Furthermore, additional immunogenic epitopes within BcPGs sensed by Brassicaceae species are localized in the GH28 domain of BcPG6 and are not conserved in Ralstonia PehC (Zhang et al. 2021). Thus, the receptor of PehC awaits identification.
Cell wall degrading enzymes act as critical virulence factors for diverse pathogens. Pathogens break down plant cell wall to facilitate infection and to move freely inside plant tissues. On the other hand, fragments degraded from the cell wall including high MW compounds, such as cutins, cellobioses, and OGs, act as DAMPs, giving a danger warning (Gust et al. 2017). The plant cell membrane receptor wall-associated receptor kinase 1 (WAK1) perceives OGs and induces the host defense response to inhibit pathogen growth and propagation (Brutus et al. 2010). In turn, plant pathogens have evolved a variety of strategies to evade plant recognition, such as masking or modifying MAMPs from host immune recognition (Buscaill and van der Hoorn 2021; Mart Nez-Cruz et al. 2021). In addition, the degradation of DAMPs by pathogens may serve as another efficient strategy for inhibiting plant immune activation. Ralstonia PehB and PehC are both exo-PGs, which release GalA monomers or dimers from the polygalacturonic acid backbone. During infection, endo-PG PehA cleaves the pectin polymer internally to produce oligomer OGs, and then PehB and PehC further degrade OGs to avoid recognition by WAK1. Our study shows that pathogens interfere with plant DAMP recognition, adding another dimension to pathogen-mediated plant immune suppression.
We also found that PehC functions as a producer of carbon sources. Xylem sap has long been described as a nutrient-poor environment consisting of water and minerals since the mature vessel cells are dead (Zuluaga et al. 2013). Pathogens employ different mechanisms to actively elevate nutrient concentration in the apoplast or xylem sap. Ralstonia effector RipI interacts with plant glutamate decarboxylases to promote the biosynthesis of gamma-aminobutyric acid and support bacterial growth (Xian et al. 2020). Moreover, type III effectors of Xanthomonas spp. induce the expression of host sugar transporters, likely causing an efflux of sugar to feed bacteria in the xylem or the apoplast (Cox et al. 2017). Ralstonia replicates fast and reaches a high cell density in the xylem, implying the presence of additional virulence activities to establish a nutrient-sufficient environment. Upon infection, compounds in the xylem sap change considerably, especially for providing nutrition (Zuluaga et al. 2013).
Eight metabolites have been shown to promote Ralstonia growth as the sole carbon source (Lowe-Power et al. 2018b). Pectin is a complex heteropolysaccharide present in the primary cell wall of plants, and its breakdown products serve as potential nutrients for pathogens. We showed that products depolymerized by the endo-PG PehA are not utilized by GMI1000. However, further breakdown products, such as GalA monomer (produced by PehC) and GalA dimer (produced by PehB), are utilized by Ralstonia as the sole carbon source, which is consistent with previous reports (Gonzalez and Allen 2003). PehC hydrolyzes pectin to release GalA at an early stage of Ralstonia infection when the bacterial titer is low. With the multiplication of Ralstonia, the rate of GalA utilization exceeds the rate of GalA production. However, the amount of pectin in the xylem is limited. Therefore, GalA concentration peaked at the early stage of infection and then decreased. Notably, we found that PehC activity is critical for early nutrient acquisition, potentially driving pathogen growth by increasing nutrient availability. However, the exuT mutant, which cannot uptake PG degradation products, showed similar virulence in tomato compared with the WT strain (Gonzalez and Allen 2003). The importance of GalA for virulence in planta and the uptake pathway need further dissection. Collectively, our findings illuminate the distinct property of a pathogen virulence factor, PehC, which exerts dual functions (i.e. decrease plant immunity and increase nutrient availability) and is monitored by plants.
Materials and methods
Plant materials and growth condition
Tomato (Solanum lycopersicum) cultivars Hawaii 7996, Moneymaker, LA1589, Pimp84, Micro Tom, and Ailsa Craig were kindly provided by Dr. Taotao Wang. Arabidopsis thaliana plants were in the Columbia-0 (Col-0) background, sobir1-12 and bak1-5/serk4-1 mutants were reported in previous studies (Meng et al. 2015). Plants were grown in soil at 22 °C, 45% relative humidity, and 75 µE/m2/s1 (T5 LED tube lights, 4000 K) with 12 h-light/12 h-dark cycles. Seedlings were grown on half-strength (1/2) MS plates containing 1% (w/v) sucrose and 0.8% (w/v) agar for about 2 weeks at 22 °C in the same growth chamber.
Generation of PehC knockout and complementation strains
To generate the PehC knockout mutants, about 500 bp upstream and downstream sequences flanking PehC were amplified from the GMI1000 genomic DNA. The fragments were inserted into the flanking regions of a Kanamycin resistance gene in pC007 vector to generate pC007-Up-Kan-Down. The resulting construct was verified by PCR analysis and transformed into GMI1000 competent cells by electroporation. The putative mutants were selected with expected antibiotic resistances and verified by PCR. The primers used are listed in Supplemental Table S2.
To complement PehC in the Δpehc mutant, as previous described (Gonzalez and Allen 2003) the native promoter (300 bp upstream of the start codon) of PehC was amplified by PCR using specific primers listed in Supplemental Table S2 and was cloned into pBBR1-HA vector through In-Fusion assembly. Then the PehC coding region was introduced to the above plasmid to create pBBR1-proPehC-PehC-HA or pBBR1-proPehC-H453A-HA plasmid (Supplemental Table S3). The resulting constructs were introduced into Δpehc mutant using electroporation. The complementation strains were selected using gentamycin (25 µg/mL) and kanamycin (25 µg/mL), confirmed by immunoblotting with α-HA antibody (Roche, Cat. 12013819001,1:2000 dilution).
Characterization of bacterial-related phenotypes
For analysis of strains growth, GMI1000, Δpehc, and complementation strains were cultured overnight in a Casamino Acids-Peptone-Glucose (CPG) medium. The bacterial cells were collected and resuspended with a fresh CPG medium. The initial density was unified to OD600 = 0.05, and the OD600 was measured every 8 h as an index to evaluate the growth of the strains.
To measure the biofilm formation of R. solanacearum, We conducted crystal violet staining based on previously reported studies (Yao and Allen 2007). Briefly, bacterial cells were inoculated into 96-well polystyrene plates of CPG medium at a concentration of OD600 = 0.1. After incubation at 28 °C for 48 h, the cells were stained with 0.1% (w/v) crystal violet. The stained cells were dissolved in 95% ethanol. The optical density (OD) of the solution at 590 nm was measured to indicate the biofilm formation ability.
Mobility was measured on a semi-solid MM plate (FeSO4 0.5 mg/L; (NH4)2SO4 0.5 g/L; MgSO4 0.05 g/L, and KH2PO4 3.4 g/L; pH = 7.0) (Song et al. 2022). The bacteria were inoculated in the center of the MM plate containing 0.3% (w/v) agar. The colony diameter was measured with a vernier caliper after the plate was cultured at 28 °C for 2 d.
In order to quantify the production of EPS, the bacteria were precultured overnight and inoculated into a CPG liquid medium. The original OD600 was maintained at 0.1, and the bacteria were cultured in a shaker at 28 °Cfor 72 h. The supernatant was collected and stained with acetylacetone-Na2CO3 and P-diaminobenzaldehyde (1.5%, w/v). The OD of the solution at 525 nm was measured to represent the EPS production capacity.
Purification of SPs from R. solanacearum
Bacteria were cultured in a CPG liquid medium at 28 °C for 48 h. The suspension was collected by centrifugation and then filtered with a 0.22 μm filter to eliminate bacterial pellets. Total proteins in the culture were precipitated by (NH4)2SO4. The precipitate was collected by centrifugation and then resuspended in 10 mM Tris–HCl (pH 7.5). SPs were separated by SDS-PAGE and stained with Coomassie-blue. The gel was divided into different segments following frozen with liquid nitrogen and then ground into powder, followed by the extraction with 8 M urea. Finally, the proteins were tested for activation of PTI responses in tomato roots.
Expression vectors construction and protein purification
The genomic DNA of GMI1000 was used as a template to amplify the coding sequences of PehA, PehB, and PehC. Fragments were cloned into the pGST prokaryotic expression vector with a C-terminal GST tag. pGST-PehA, pGST-PehB, and pGST-PehC (Supplemental Table S3) plasmids were transformed into Escherichia coli strain BL21 and induced for 12 h at 28 °C with 0.25 mM IPTG. Purification of recombinant GST protein from the culture supernatant was performed by affinity chromatography using glutathione agarose resin. Diafiltration and ultrafiltration were used for glutathione removal.
PehC exo-PG activity analysis
Enzymatic activities of PehC were measured following the 3,5-DNS method as previously described (Evans et al. 2002). The commercial pectin (Coolaber, China) was used as substrates and the PG activity detection kit (Solarbio, BC2665, China) was used to measure PehC enzymatic activity. The standard reaction mixture containing 25 μg GST-PehC protein and 1 mg pectin in phosphate buffered saline (PBS) buffer (pH 5.2) was incubated for 2 h at 40 °C, then an equal amount of DNS reagent was added. The mixture was boiled for 5 min and the color intensity was determined at 540 nm. The GalA concentration was calculated with the standard curve.
Detection of plant immune responses
For extracellular alkalinization, root samples were incubated with ddH2O for overnight recovery. After the recovery, water was replaced with indicated elicitor solution (10 μg/mL), and the extracellular pH was continuously measured using a Micro pH Probe. Root sections from three 2-wk-old tomato seedlings were analyzed as 1 replicate. For ROS burst, the tomato seedling roots were cut into 5 mm-sections. Each sample containing 4 root sections was floated on 200 μL ddH2O in a 96-well plate for overnight recovery. Before detection, the water was replaced with a solution containing 5 μM luminol L012 (Wako Chemicals, Japan) and 10 μg/mL peroxidase from horseradish (Sigma) with or without indicated elicitors. Luminescence was measured continuously over a 60 min period by a multimode reader platform (Tecan, SPARK 10 M). For MAPK assays, 2 tomato seedlings as 1 sample were soaked with indicated elicitor solution, and root tissues were collected at different time points. The total proteins were extracted and subjected to immunoblotting analysis. Phosphorylated MPK proteins were detected by an antiphospho-p44/42 antibody (Cell Signaling Technology, Cat. 9101S, 1:2000 dilution).
For callose deposition, tomato roots treated with elicitors were fixed in ethanol: acetic acid (3:1) solution, followed by rehydration in 70% ethanol and 50% ethanol. After being washed with water, the roots were incubated in a staining solution containing 150 mM K2HPO4 (pH 9.5) and 0.01% (w/v) aniline blue. The samples were then observed using a Leica microscope under UV light (Olympus BX51). Immune gene induction: Tomato roots were treated with elicitors and then collected at indicated time points. Total RNA was extracted using Trizol reagent (Invitrogen) according to the manufacturer's protocol. One microgram of RNA was used for reverse transcription using the HiScript III 1st strand cDNA synthesis kit. RT-qPCR analysis was carried out with the SYBR Green qPCR Master Mix on a CFX96 real-time machine (Bio-Rad). SlACTIN2 was used as the reference gene for normalization and the primers were listed in Supplemental Table S2.
RNA-seq and data analysis
Two-week-old tomato Moneymaker seedlings were treated with or without 10 μg/mL GST-PehC for 0 h, 1 h, or 6 h before harvested. Six seedlings were treated as 1 biological replicate and total of 4 biological replicates were performed. RNA extraction, library construction, and sequencing were performed by Shanghai Personalbio Technology. After sequencing, the reads with low sequencing quality or sequencing connectors in the raw data were filtered. Total RNA-seq reads were mapped to tomato reference genome (https://solgenomics.net/) using HISAT2 software (Kim et al. 2015). Transcriptome quantification based on gene level was conducted with Feature Recounts after comparison, and DEGs were searched using DESeq2 (Love et al. 2014). The criteria for screening DEGs were log2fold change ≥ 1.5 and false discovery rate <0.05. GO enrichment was performed using agri GO v2.0. The subsequent comparison and plotting were finished in R.
Virus-induced gene silencing (VIGS) assay on tomato
Plasmids of binary vectors containing pTRV-RNA1 and pTRV-RNA2 derivatives pTRV-SlBAK1a, pTRV-SlBAK1b, and pTRV-GFP (Supplemental Table S3) were transformed into Agrobacterium tumefaciens GV3101. Bacterial cultures were grown in an Luria-Bertani medium containing 50 mg/mL kanamycin and 25 mg/mL gentamycin. The bacterial suspensions were adjusted to OD600 = 1.5 with a solution containing 10 mM MgCl2, 10 mM MES, and 200 mM acetosyringone. The Agrobacterium suspensions containing pTRV-RNA1 and pTRV-RNA2 derivatives were mixed at 1:1 as indicated, respectively. Then bacterial suspensions were injected into tomato cotyledons by leaf injection. Root sections were collected 2 wks postinoculation and treated with 10 μg/mL GST-PehC to detect the ROS burst in genes silenced tomatoes.
Bacterial inoculation and population analysis
For R. solanacearum soil-drenching inoculation, about 20∼25 4-wk-old tomato plant were used for each bacterial strain in 1 independent experiment. Each plant root was wounded by a razor blade and was soaked with 10 mL bacterial suspension at the OD600 = 0.1. Plants were maintained in a growth chamber with a 12 h photoperiod at 28 °C. The visual disease symptoms of each plant were scored based on a 0 to 4 disease index scale (Yao and Allen 2007), where 0 represented no symptoms, 1 corresponded to 1% to 25% leaf area wilting, 2 for 26% to 49%, 3 for 50% to 74%, and 4 for complete wilting). To assess the virulence difference among strains, the area under the disease progression curve (AUDPC), which is defined by the curve and the x-axis, was analyzed using linear mixed effect models (LMMs) with Tukey’s honestly significant difference for multiple comparisons by R according to previous described (Schandry 2017). Survival analysis was performed using Log-rank (Mantel-Cox) test of the Kaplan Meier estimate with Tukey's honestly significant difference for multiple comparisons and Bonferroni adjusted. Different letters denote a significant difference (P < 0.05, Supplemental Data Set 6).
For the PehC-triggered resistance to bacterial wilt, 4-wk-old Moneymaker plants were pretreated by pipetting 10 mL GST-PehC protein solution (10 μg/mL) to the soil around each plant and incubated for 2 d. The soil-drenching inoculation was performed with 10 mL GMI1000 suspension at the OD600 = 0.1 for each plant. The disease progress of each inoculated plant was rated as above.
For bacteria population quantification in tomato stems. The midstems which cotyledon of infected tomato plant were collected and weighted at 3 d postinoculation (dpi). The stems were surface sterilized by 70% ethanol for 30 s and sterilized in water for 1 min. Stem sample were soaked in ddH2O and were ground with a microelectronic drill. The bacterial suspension was subjected to a serial of 1:10 dilution by ddH2O, and the series of dilution samples were plated on TTC medium to count CFU per gram of stem fresh weight.
For the bacterial competition test, the stem injection assay was performed with 8∼12 tomato plants for each strain in 1 independent experiment. Briefly, the WT, ΔpehC mutants, ComPehc complementation strain bacterial suspension was adjusted to 106 cfu/mL. An equal amount of indicated paired bacterial suspension was mixed and 5 μL was injected into the tomato stem. The stems around the site of inoculation were collected from each infected plants and bacterial population size was monitored as above described.
Xylem sap collection from tomato stems
Xylem sap was harvested by cutting the tomato stem at 1 cm above the rough leaf with a razor blade and allowing the sap to pool on the stump. The first droplet of xylem sap would flow out for about 2 to 3 min after the cut, which was discarded to prevent contamination of the exudate with the content of damaged cells or phloem vessels. The incision was rinsed with sterilized water, the subsequent xylem sap was collected into a sterilized tube. The pooled xylem sap from 1 plant was centrifuged at 12,000 × g for 10 min to remove bacterial pellet and plant tissue residue.
Pectin hydrolysis assay with PehA, PehB, and PehC
Commercial pectin from plant cell wall extract was dissolved in Tris–HCl buffer (50 mM, pH 7.5) to a final concentration of 10 mg/mL. The pectin solution was treated with GST-PehA (5 μg/mL) and kept at 37 °C for 8 h. After treatment, 50 mg/mL Proteinase K was added and incubated at 58 °C for 2 h to digest GST-PehA and then treated at 72 °C for 10 min for inactivation. The resulting solution was further treated with 5 μg/mL GST-PehB or GST-PehC for 8 h at 37 °C. After GST-PehB or GST-PehC treatment, a similar Proteinase K digestion was performed as described above.
Detection of GalA by LC-MS/MS
The GalA concentration in tomato xylem sap was analyzed using an LC-ESI-MS/MS system (UPLC, Shim-pack UFLC SHIMADZU CBM A system; MS, API 4000 Q TRAP) as previously described (Chen et al. 2013). The analytical conditions were as follows: column, Waters ACQUITY UPLC HSS T3 C18 (1.8 µm, 2.1 mm × 100 mm); flow rate, 0.40 mL/min; solvent system, water (0.04% acetic acid): acetonitrile (0.04% acetic acid); gradient program, 95:5 V/V at 0 min, 5:95 V/V at 10.0 min, 5:95 V/V at 11.0 min, 95:5 V/V at 11.1 min, and 95:5 V/V at 15.0 min. injection volume, 2 μL; column temperature, 40 °C. The ESI source operation parameters were as follows: source temperature 500 °C; ion source gas I, gas II, curtain gas at 55, 60, and 25.0 psi, respectively; ion spray voltage −5500 V (negative); the collision gas (CAD) was high. An MRM transition of 193 to 71 was monitored for GalA detection. The Analyst software 1.6.3 was used to quantify the content of GalA in each sample.
Bacterial growth assay with different carbon sources
R. solanacearum was cultured in a CPG medium overnight and collected by centrifugation. The bacterial pellet was resuspended in sterile water and adjusted to a final OD600 of 0.01 before inoculation in liquid MM (0.125 mg FeSO4·7H2O, 0.5 g (NH4)2SO4, 0.05 g MgSO4·7H2O, and 3.4 g KH2PO4 in 1 L, the pH was adjusted to 7 with KOH) without carbon source. For the effects of oligosaccharides on R. solanacearum growth, the oligosaccharide materials were added into MM as the single carbon source, and the final concentration was adjusted to 5 mg/mL. For the effect of pectin hydrolyzed product, 50 μL hydrolysis product was added into 2 mL MM as the single carbon source, respectively. For the effect of tomato xylem sap, 100 μL different sources of xylem sap were into 2 mL MM, respectively. The OD600 of each sample was monitored per 12 h by the spectrophotometer for drawing the growth curve. The area under the curve (AUC) for Ralstonia growth rate curve was calculated using GraphPad Prism. Statistical analyses between 2 sets of data were performed by Student's one-tailed t-test and 3 or more groups of data were analyzed by one-way ANOVA with Tukey's significant difference for multiple comparisons. Different letters represent significant differences (P < 0.05, Supplemental Data Set 6).
Phylogenetic analysis
The amino acid sequences of the PG proteins were obtained from the Integrated Microbial Genomes and Microbiomes (IMG/M) website (https://img.jgi.doe.gov/) and NCBI website (https://www.ncbi.nlm.nih.gov/). Sequence alignment was performed with ClustalW and results have been provided as supplemental files in FASTA format. The phylogenetic tree was generated with MEGA6.0 software using the neighbor-joining method (Tamura et al. 2013). Three cellulase proteins (Pme, cbhA, and Egl) were selected as outgroups. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. The tree branch lengths represent the number of variants per 100 amino acids. The alignment and machine-readable tree files are provided as Supplemental Files S1 and S2.
Accession numbers
Sequence data in this article can be found in the R. solanacearum Database, the Integrated Microbial Genomes and Microbiomes (IMG/M) website, and Sol Genomics Network website under the following accession numbers: PehA (RSp0880), PehB (RSc1756), PehC (RSp0833), Egl (RSp0162), Pme (RSp0138), cbhA (RSp0583), SlACTIN2 (Solyc11g005330), SlERF2a (Solyc12g042210), SlERF2b (Solyc04g051360), SlWRKY33 (Solyc09g014990), SlDefensin19 (Solyc07g009090), SlPR1b (Solyc00g174340), SlPDF1 (Solyc04g009590), SlBAK1a (Solyc10g047140), and SlBAK1b (Solyc01g104970). The RNA-seq was deposited in the Gene Expression Omnibus (accession number GSE204888) at the National Center for Biotechnology Information.
Supplementary Material
Acknowledgments
We thank Dr. Taotao Wang for sharing the tomato seeds, Dr. Huilan Chen for helping with the bacterial gene knockout. We thank the Center for Protein Research, Huazhong Agricultural University for technical support. We are grateful to Dr. Jianmin Zhou and Dr. Kenichi Tsuda for the critical reading and constructive suggestions on the manuscript.
Contributor Information
Jingjing Ke, State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; The Provincial Key Lab of Plant Pathology of Hubei Province, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; Hubei Hongshan Laboratory, Wuhan, Hubei 430070, China.
Wanting Zhu, State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; The Provincial Key Lab of Plant Pathology of Hubei Province, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; Hubei Hongshan Laboratory, Wuhan, Hubei 430070, China.
Ying Yuan, State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; The Provincial Key Lab of Plant Pathology of Hubei Province, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; Hubei Hongshan Laboratory, Wuhan, Hubei 430070, China.
Xinya Du, State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; The Provincial Key Lab of Plant Pathology of Hubei Province, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; Hubei Hongshan Laboratory, Wuhan, Hubei 430070, China.
Ai Xu, State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; The Provincial Key Lab of Plant Pathology of Hubei Province, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; Hubei Hongshan Laboratory, Wuhan, Hubei 430070, China.
Dan Zhang, State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; The Provincial Key Lab of Plant Pathology of Hubei Province, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; Hubei Hongshan Laboratory, Wuhan, Hubei 430070, China.
Sen Cao, State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; The Provincial Key Lab of Plant Pathology of Hubei Province, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; Hubei Hongshan Laboratory, Wuhan, Hubei 430070, China.
Wei Chen, Hubei Hongshan Laboratory, Wuhan, Hubei 430070, China; National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Yang Lin, The Provincial Key Lab of Plant Pathology of Hubei Province, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Jiatao Xie, State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; The Provincial Key Lab of Plant Pathology of Hubei Province, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; Hubei Hongshan Laboratory, Wuhan, Hubei 430070, China.
Jiasen Cheng, State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; The Provincial Key Lab of Plant Pathology of Hubei Province, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Yanping Fu, The Provincial Key Lab of Plant Pathology of Hubei Province, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Daohong Jiang, State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; The Provincial Key Lab of Plant Pathology of Hubei Province, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; Hubei Hongshan Laboratory, Wuhan, Hubei 430070, China.
Xiao Yu, State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; The Provincial Key Lab of Plant Pathology of Hubei Province, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; Hubei Hongshan Laboratory, Wuhan, Hubei 430070, China.
Bo Li, State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; The Provincial Key Lab of Plant Pathology of Hubei Province, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China; Hubei Hongshan Laboratory, Wuhan, Hubei 430070, China.
Author contributions
J.K. purified the protein and analyzed immune responses; W.Z. performed experiments and organized the figures; Y.Y. analyzed the RNA-seq data; A.X. conducted VIGS assay and protein purification; D.Z. and X.D. generated the bacterial mutants and complementation strains; S.C. performed gene expression assay; W.C. conducted the LC/MS analysis; J.X., Y.F., J.C., D.J., and X.Y. analyzed data, provided critical feedback. B.L. conceived the project, designed experiments, wrote the manuscript with input from all co-authors.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. SPs from R. solanacearum induce immune responses in susceptible tomato susceptible cultivars.
Supplemental Figure S2. PehC, but not PehA and PehB induces tomato immune responses.
Supplemental Figure S3. PehC triggers global gene transcription reprograming in tomato roots.
Supplemental Figure S4. SERK coreceptors are required for PehC-induced immune responses.
Supplemental Figure S5. Verification and phenotype characterization of ΔpehC mutant strains.
Supplemental Figure S6. Role of PehC-triggered immunity in the interaction with plants.
Supplemental Figure S7. Function of PehC in immune evasion and nutrition supplementation.
Supplemental Figure S8. Phylogenetic relationship of PGs in important plant pathogenic bacteria.
Supplemental Table S1. Proteins identified in the S2-1 fraction by LC-MS/MS.
Supplemental Table S2. Primers used in this study.
Supplemental Table S3. Summary of the vectors used in this study.
Supplemental File 1. Multiple protein sequence alignment from PGs in plant pathogenic bacteria.
Supplemental File 2. Newick file format of the phylogenetic tree show in Supplemental Fig. S8.
Supplemental Data Set 1. DEGs at 1 h post-PehC treatment.
Supplemental Data Set 2. DEGs at 6 h post-PehC treatment.
Supplemental Data Set 3. List of the PehC up-regulated genes involved in immune-related secondary metabolites process.
Supplemental Data Set 4. List of the PehC up-regulated genes encoding RLKs and transcriptional factors.
Supplemental Data Set 5. The percentage of immune-related genes in flgII-28 and PehC regulated gene groups.
Supplemental Data Set 6. Results of statistical test for the data in this study.
Funding
This research was supported by the National Key R&D program of China (Grant no. 2022YFA1304400), National Natural Science Foundation of China (Grant no. 31970125), Pests and Diseases Green Prevention and Control Major Special Project (Grant no. 110202101045, LS-05), and Fundamental Research Funds for the Central Universities, Huazhong Agricultural University Scientific & Technological Self-innovation Foundation (Grant no. 2017RC001).
References
- Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci U S A. 2010:107(20):9452–9457. 10.1073/pnas.1000675107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaill P, van der Hoorn RAL. Defeated by the nines: nine extracellular strategies to avoid microbe-associated molecular patterns recognition in plants. Plant Cell. 2021:33(7):2116–2130. 10.1093/plcell/koab109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai RM, Lewis J, Yan SC, Liu HJ, Clarke CR, Campanile F, Almeida NF, Studholme DJ, Lindeberg M, Schneider D, et al. The plant pathogen Pseudomonas syringae pv. tomato is genetically monomorphic and under strong selection to evade tomato immunity. PLoS Pathog. 2011:7(8):e1002130. 10.1371/journal.ppat.1002130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Gong L, Guo ZL, Wang WS, Zhang HY, Liu XQ, Yu SB, Xiong LZ, Luo J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol Plant. 2013:6(6):1769–1780. 10.1093/mp/sst080 [DOI] [PubMed] [Google Scholar]
- Chen H, Raffaele S, Dong S. Silent control: microbial plant pathogens evade host immunity without coding sequence changes. FEMS Microbiol Rev. 2021:45(4):fuab002. 10.1093/femsre/fuab002 [DOI] [PubMed] [Google Scholar]
- Cox KL, Meng FH, Wilkins KE, Li FJ, Wang P, Booher NJ, Carpenter SCD, Chen LQ, Zheng H, Gao XQ, et al. TAL effector driven induction of a SWEET gene confers susceptibility to bacterial blight of cotton. Nat Commun. 2017:8(1):15588. 10.1038/ncomms15588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny T. Plant pathogenic Ralstonia species. In: Gnanamanickam SS, editor. Plant-associated bacteria. Dordrecht: Springer Netherlands; 2007. p. 573–644. [Google Scholar]
- Evans JD, Akin DE, Foulk JA. Flax-retting by polygalacturonase-containing enzyme mixtures and effects on fiber properties. J Biotechnol. 2002:97(3):223–231. 10.1016/S0168-1656(02)00066-4 [DOI] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999:18(3):265–276. 10.1046/j.1365-313X.1999.00265.x [DOI] [PubMed] [Google Scholar]
- Ferrari S, Savatin DV, Sicilia F, Gramegna G, Cervone F, De Lorenzo G. Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Front Plant Sci. 2013:4:49. 10.3389/fpls.2013.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin S, Denny TP. Pathogenomics of the Ralstonia solanacearum species complex. Annu Rev Phytopathol. 2012:50(1):67–89. 10.1146/annurev-phyto-081211-173000 [DOI] [PubMed] [Google Scholar]
- Gong BQ, Wang FZ, Li JF. Hide-and-seek: chitin-triggered plant immunity and fungal counterstrategies. Trends Plant Sci. 2020:25(8):805–816. 10.1016/j.tplants.2020.03.006 [DOI] [PubMed] [Google Scholar]
- Gonzalez ET, Allen C. Characterization of a Ralstonia solanacearum operon required for polygalacturonate degradation and uptake of galacturonic acid. Mol Plant Microbe Interact. 2003:16(6):536–544. 10.1094/MPMI.2003.16.6.536 [DOI] [PubMed] [Google Scholar]
- Gust AA, Pruitt R, Nurnberger T. Sensing danger: key to activating plant immunity. Trends Plant Sci. 2017:22(9):779–791. 10.1016/j.tplants.2017.07.005 [DOI] [PubMed] [Google Scholar]
- Hind SR, Strickler SR, Boyle PC, Dunham DM, Bao Z, O’Doherty IM, Baccile JA, Hoki JS, Viox EG, Clarke CR, et al. Tomato receptor FLAGELLIN-SENSING 3 binds flgII-28 and activates the plant immune system. Nat Plants. 2016:2(9):16128. 10.1038/nplants.2016.128 [DOI] [PubMed] [Google Scholar]
- Huang Q, Allen C. Polygalacturonases are required for rapid colonization and full virulence of Ralstonia solanacearum on tomato plants. Physiol Mol Plant Pathol. 2000:57(2):77–83. 10.1006/pmpp.2000.0283 [DOI] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006:444(7117):323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015:12(4):357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell. 2004:16(12):3496–3507. 10.1105/tpc.104.026765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand TWH, van den Burg HA, Joosten MHAJ. Two for all: receptor-associated kinases SOBIR1 and BAK1. Trends Plant Sci. 2014:19(2):123–132. 10.1016/j.tplants.2013.10.003 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014:15(12):550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe-Power TM, Hendrich CG, von Roepenack-Lahaye E, Li B, Wu D, Mitra R, Dalsing BL, Ricca P, Naidoo J, Cook D, et al. Metabolomics of tomato xylem sap during bacterial wilt reveals Ralstonia solanacearum produces abundant putrescine, a metabolite that accelerates wilt disease. Environ Microbiol. 2018b:20(4):1330–1349. 10.1111/1462-2920.14020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe-Power TM, Khokhani D, Allen C. How Ralstonia solanacearum exploits and thrives in the flowing plant xylem environment. Trends Microbiol. 2018a:26(11):929–942. 10.1016/j.tim.2018.06.002 [DOI] [PubMed] [Google Scholar]
- Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer SV, Machado MA, et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol. 2012:13(6):614–629. 10.1111/j.1364-3703.2012.00804.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mart Nez-Cruz JS, Romero D, Hierrezuelo JS, Thon M, de Vicente A, Rez-Garc A AP. Effectors with chitinase activity (EWCAs), a family of conserved, secreted fungal chitinases that suppress chitin-triggered immunity. Plant Cell. 2021:33(4):1319–1340. 10.1093/plcell/koab011 [DOI] [PubMed] [Google Scholar]
- Meng X, Chen X, Mang H, Liu C, Yu X, Gao X, Torii KU, He P, Shan L. Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr Biol. 2015:25(18):2361–2372. 10.1016/j.cub.2015.07.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters N, Guidot A, Vailleau F, Valls M. Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Mol Plant Pathol. 2013:14(7):651–662. 10.1111/mpp.12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinssot B, Vandelle E, Bentejac M, Adrian M, Levis C, Brygoo Y, Garin J, Sicilia F, Coutos-Thevenot P, Pugin A. The endopolygalacturonase 1 from Botrytis cinerea activates grapevine defense reactions unrelated to its enzymatic activity. Mol Plant Microbe Interact. 2003:16(6):553–564. 10.1094/MPMI.2003.16.6.553 [DOI] [PubMed] [Google Scholar]
- Pombo MA, Zheng Y, Fei ZJ, Martin GB, Rosli HG. Use of RNA-seq data to identify and validate RT-qPCR reference genes for studying the tomato-Pseudomonas pathosystem. Sci Rep. 2017:7(1):44905. 10.1038/srep44905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schandry N. A practical guide to visualization and statistical analysis of R. solanacearum infection data using R. Front Plant Sci. 2017:8:623. 10.3389/fpls.2017.00623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SH, Sun XY, Guo Q, Cui BB, Zhu Y, Li X, Zhou JN, Zhang LH, Deng YY. An anthranilic acid-responsive transcriptional regulator controls the physiology and pathogenicity of Ralstonia solanacearum. PLoS Pathog. 2022:18(5):e1010562. 10.1371/journal.ppat.1010562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013:30(12):2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Wang G, Zhou JM. Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell. 2017:29(4):618–637. 10.1105/tpc.16.00891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Einig E, Almeida-Trapp M, Albert M, Fliegmann J, Mithofer A, Kalbacher H, Felix G. The systemin receptor SYR1 enhances resistance of tomato against herbivorous insects. Nat Plants. 2018:4(3):152–156. 10.1038/s41477-018-0106-0 [DOI] [PubMed] [Google Scholar]
- Wei Y, Balaceanu A, Rufian JS, Segonzac C, Zhao A, Morcillo RJL, Macho AP. An immune receptor complex evolved in soybean to perceive a polymorphic bacterial flagellin. Nat Commun. 2020:11(1):3763. 10.1038/s41467-020-17573-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Caceres-Moreno C, Jimenez-Gongora T, Wang K, Sang Y, Lozano-Duran R, Macho AP. The Ralstonia solanacearum csp22 peptide, but not flagellin-derived peptides, is perceived by plants from the Solanaceae family. Plant Biotechnol J. 2018:16(7):1349–1362. 10.1111/pbi.12874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhou JM. Receptor-like kinases in plant innate immunity. J Integr Plant Biol. 2013:55(12):1271–1286. 10.1111/jipb.12123 [DOI] [PubMed] [Google Scholar]
- Xian L, Yu G, Wei Y, Rufian JS, Li Y, Zhuang H, Xue H, Morcillo RJL, Macho AP. A bacterial effector protein hijacks plant metabolism to support pathogen nutrition. Cell Host Microbe. 2020:28(4):548–557.e7. 10.1016/j.chom.2020.07.003 [DOI] [PubMed] [Google Scholar]
- Yao J, Allen C. The plant pathogen Ralstonia solanacearum needs aerotaxis for normal biofilm formation and interactions with its tomato host. J Bacteriol. 2007:189(17):6415–6424. 10.1128/JB.00398-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Feng B, He P, Shan L. From chaos to harmony: responses and signaling upon microbial pattern recognition. Annu Rev Phytopathol. 2017:55(1):109–137. 10.1146/annurev-phyto-080516-035649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Jiang Z, Bi G, Nomura K, Liu M, Wang Y, Cai B, Zhou JM, He SY, Xin XF. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature. 2021:592(7852):105–109. 10.1038/s41586-021-03316-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Hua C, Pruitt RN, Qin S, Wang L, Albert I, Albert M, van Kan JAL, Nurnberger T. Distinct immune sensor systems for fungal endopolygalacturonases in closely related Brassicaceae. Nat Plants. 2021:7(9):1254–1263. 10.1038/s41477-021-00982-2 [DOI] [PubMed] [Google Scholar]
- Zhang LS, Kars I, Essenstam B, Liebrand TWH, Wagemakers L, Elberse J, Tagkalaki P, Tjoitang D, van den Ackerveken G, van Kan JAL. Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the Arabidopsis receptor-like protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol. 2014:164(1):352–364. 10.1104/pp.113.230698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuluaga AP, Puigvert M, Valls M. Novel plant inputs influencing Ralstonia solanacearum during infection. Front Microbiol. 2013:4:349. 10.3389/fmicb.2013.00349 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.