Abstract
Ascorbate (vitamin C) is an essential antioxidant in fresh fruits and vegetables. To gain insight into the regulation of ascorbate metabolism in plants, we studied mutant tomato plants (Solanum lycopersicum) that produce ascorbate-enriched fruits. The causal mutation, identified by a mapping-by-sequencing strategy, corresponded to a knock-out recessive mutation in a class of photoreceptor named PAS/LOV protein (PLP), which acts as a negative regulator of ascorbate biosynthesis. This trait was confirmed by CRISPR/Cas9 gene editing and further found in all plant organs, including fruit that accumulated 2 to 3 times more ascorbate than in the WT. The functional characterization revealed that PLP interacted with the 2 isoforms of GDP-L-galactose phosphorylase (GGP), known as the controlling step of the L-galactose pathway of ascorbate synthesis. The interaction with GGP occurred in the cytoplasm and the nucleus, but was abolished when PLP was truncated. These results were confirmed by a synthetic approach using an animal cell system, which additionally demonstrated that blue light modulated the PLP-GGP interaction. Assays performed in vitro with heterologously expressed GGP and PLP showed that PLP is a noncompetitive inhibitor of GGP that is inactivated after blue light exposure. This discovery provides a greater understanding of the light-dependent regulation of ascorbate metabolism in plants.
The mechanism by which light stimulates the synthesis of ascorbate in plants involves a protein related to phototropins whose inhibitory action is repressed under blue light.
IN A NUTSHELL.
Background: Ascorbic acid, also known as vitamin C, is an essential antioxidant with key roles in several stress responses in living organisms. Since humans are not able to produce this molecule, we must obtain it through our diet, with plant products being the main source. Although the plant biosynthetic pathway of ascorbic acid is well known, the molecular mechanisms that regulate it are not yet understood. Among these regulatory factors, light occupies an important place, since it is vital for plants and it is well established that the more light there is, the more ascorbic acid plants produce and vice versa. However, the underlying mechanisms remain totally unknown.
Question: What are the factors that control ascorbic acid biosynthesis in plants and more particularly its accumulation in fruits?
Findings: Our work uncovers the mechanism by which plants use blue light to modulate ascorbate biosynthesis. From a tomato mutant, we identified a photoreceptor named PAS/LOV (PLP) and revealed that it functions as a negative regulator in vivo and in vitro by interacting with the GDP-galactose phosphorylase (GGP), which is known to be the most controlling enzyme of the pathway. We also demonstrated that blue light counteracts the inhibition of the GDP-galactose phosphorylase by the PAS/LOV photoreceptor. This is a major finding because it finally explains how plants protect themselves from too much light by synthesizing more of their main antioxidant.
Next steps: Future research should study the turnover of GGP in relation to PLP, particularly to better understand the adaptive role of PLP. Such research would open up new ways for developing plants that are more resistant to stress, fruits with prolonged “shelf-life”, and even plants with enhanced nutritional profiles.
Introduction
Ascorbate is an essential metabolite in living organisms. It has a leading role as an antioxidant by participating in eliminating reactive oxygen species (ROS) that are usually produced in response to biotic and abiotic stresses (Decros et al. 2019). Ascorbate also plays a crucial role in controlling the levels of ROS that are continuously produced under optimal conditions by cell metabolism, including photosynthesis. Due to its high antioxidant potential, ascorbate is one of the most important traits for the nutritional quality of fruits and vegetables. Indeed, evolution in humans and a few animal species has led to the loss of the L-gulono-γ-lactone oxidase activity, which catalyzes the last steps of the biosynthetic pathway (Burns 1957). Consequently, humans are unable to synthesize ascorbate, thus defined as vitamin C, and must have a daily intake through the consumption of fruits and vegetables. Paradoxically, the domestication of various fruit species has resulted in decreased ascorbate content, suggesting the occurrence of a trade-off between fruit yield and quality (Gest et al. 2013). Thus, understanding ascorbate metabolism is a critical issue in plant breeding, particularly for fleshy fruit species such as tomato (Solanum lycopersicum), which is considered one of the major sources of vitamin C in the human diet (Wheeler et al. 1998).
The map of plant ascorbate metabolism is well established since the discovery of the Smirnoff–Wheeler pathway, also called L-galactose pathway, although little is known about the regulatory mechanisms involved (Wheeler et al. 1998; Bulley and Laing 2016). The GGP protein, also known as VITAMIN C2 (VTC2) by analogy with Arabidopsis thaliana, corresponds to a GDP-L-galactose phosphorylase and is so far considered the controlling enzyme of the L-galactose pathway (Linster et al. 2008; Bulley et al. 2009; Li et al. 2013; Fenech et al. 2021). It catalyzes the first step, the conversion of GDP-L-galactose into L-galactose-1-phosphate, in ascorbate biosynthesis in plants. Arabidopsis knock-out vtc2 mutants display a drastic decrease in ascorbate, although a residual amount is still produced, due to the presence of another gene encoding a GDP-L-galactose phosphorylase, namely VITAMIN C5 (VTC5) (Dowdle et al. 2007). The VTC5 gene has a high sequence homology (∼66% identity) with its counterpart VTC2, but it was found to be 100 to 1,000 times less expressed. Other studies have hypothesized the existence of additional alternative pathways (Wheeler et al. 2015). Among them, only the galacturonate and myo-inositol pathways were considered relevant. However, these alternative routes have not been completely demonstrated, and some results tend to invalidate these assumptions. Indeed, the Arabidopsis vtc2/vtc5 double mutant is unable to grow without the addition of exogenous ascorbate (Dowdle et al. 2007). This suggests that there would be no other way than the L-galactose pathway to complement ascorbate deficiency. Among the enzymes involved in ascorbate synthesis, VTC2 is the only one to have a significant effect on ascorbate levels when overexpressed, although GDP-D-mannose 3′,5′-epimerase acts synergistically with it to increase ascorbate in leaves (Bulley et al. 2009; Fenech et al. 2021). Precise information on the regulation of VTC2 is lacking, with the exception of the activity of an upstream Open Reading Frame (uORF) in the 5′-UTR of the VTC2 gene, which was found to control the level of translation of the VTC2 protein (Laing et al. 2015). Interestingly, in the presence of high ascorbate concentration, the VTC2 protein was shown to be downregulated (Laing et al. 2015). This is to be linked to the fact that excess ascorbate can have deleterious effects, in particular male sterility (Deslous et al. 2021). At the cellular level, a fluorescent fusion protein approach emphasized that the VTC2 protein is localized in both cytoplasmic and nuclear compartments (Müller-Moulé 2008; Fenech et al. 2021). This unexpected nuclear localization for a metabolic enzyme suggests that GGP might also act as a dual-function protein: a regulatory factor as well as a catalytic enzyme.
Ascorbate levels are highly dependent on environmental conditions, such as salt stress, drought, and, in particular, intense light that induces ascorbate accumulation. The existence of regulators has been recently identified with the discovery of a few proteins acting at the transcriptional or posttranscriptional level that regulate specific genes and enzymes in the L-galactose pathway. These studies, mainly carried out in Arabidopsis leaves, identified Ascorbic acid Mannose pathway Regulator 1 (AMR1) (Zhang et al. 2009), Ethylene Response Factor 98 (ERF98) (Zhang et al. 2012), and COP9 Signalosome subunit 5B (CNS5B) (Wang et al. 2013) proteins as positive or negative regulators. However, for some of these effectors, the underlying mechanisms remain to be elucidated (Zhang and Huang 2010; Alimohammfadi et al. 2012; Zhang et al. 2012). Regarding the effect of light on plant development, previous works showed that light may directly or indirectly affect the expression of genes of the L-galactose pathway. For instance, Arabidopsis plants grown in the dark induced the degradation of GDP-mannose pyrophosphorylase (VTC1) by the CSN5B-Cop9 complex associated with the proteasome (Wang et al. 2013). Specifically, the AMR1/SCF complex has been shown to modulate the expression of all genes of the L-galactose pathway through an unknown mechanism (Zhang et al. 2009). Prolonged exposure to light increases the expression of the GGP gene (Dowdle et al. 2007), and some studies have shown that the expression of GGP1 (VTC2) and GGP2 (VTC5) genes is under circadian control (Tabata et al. 2002; Dowdle et al. 2007; Müller-Moulé 2008). All these studies demonstrate an apparent link between ascorbate metabolism and light signaling, but there is no evidence hitherto that light directly induces ascorbate biosynthesis or that overexpression or activation of the GGP enzyme results from oxidative stress related to light exposure.
It is well-established that there is a positive correlation between light intensity and ascorbate levels in photosynthetic tissues (Gautier et al. 2008; Bartoli et al. 2009). In tomato, light was found to impact ascorbate content more in the leaves than in fruit (Massot et al. 2012). Furthermore, Gautier et al. (2009) showed that fruit ascorbate content was not limited by leaf photosynthesis but was dependent on direct fruit irradiance. Surprisingly, the literature lacks information regarding the molecular mechanisms relating ascorbate and light sensing. The photoreceptor proteins that collect a light signal via the absorption of a photon to drive and govern biological activity are classified according to the wavelength and physiological processes involved. For instance, phytochromes (red and far-red) and UVR8 (UVB) play a crucial role in plant development, although UVR8 is also implicated in UV protection (Chen and Chory 2011; Jenkins 2014). Alternatively, plant response to blue light (390 to 500 nm) is extensive and mediated by 3 different classes of photoreceptors: cryptochromes, phototropins, and the members of the Zeitlupe family (Christie 2007; Chaves et al. 2011; Suetsugu and Wada 2013). Cryptochromes, which were the first blue light receptors to be characterized in plants (Ahmad and Cashmore 1993), are involved in the photoperiodic control of flowering, the photomorphogenic development of plants, and in setting up the circadian clock (Liu et al. 2011). Phototropins, named for their role in mediating higher plant phototropism, are the only blue light receptors identified to date that contain 2 Light Oxygen Voltage (LOV) domains; Christie et al. 1999; Briggs et al. 2001; Christie et al. 2002; Crosson et al. 2003; Christie 2007). These LOV domains belong to the Per-ARNT-SIM (PAS) superfamily that is involved in the sensing of a wide range of environmental signals. These blue light sensors are flavoproteins that require a flavin (FMN or FAD) cofactor to bind to their photo-activable domain, as well as to their effector domain. The LOV domains contain a very well-conserved cysteine involved in the formation of a covalent adduct with the FMN chromophore. This mechanism, induced by blue light and reversed in the darkness, has been named the photocycle (Christie 2007; Christie et al. 2015). There is another type of photoreceptor protein containing a unique LOV domain, named PAS/LOV Protein (PLP), for which the biological function remains to be established (Ogura et al. 2008).
In contrast to phototropins, the first domain (LOV1) of Arabidopsis and tomato PAS/LOV proteins was found to be nonfunctional as a result of the replacement of the conserved cysteine by a glycine, while the second domain (LOV2) was found to be functional (Kasahara et al. 2010). However, replacing the glycine located at this position with a cysteine restored the blue light-inducible function of the LOV1 domain (Kasahara et al. 2010). Consequently, the authors proposed to name this protein LOV/LOV (LLP) rather than PLP, although the term PLP remains prevalent in the literature (e.g. Li et al. 2018). Pending a consensual name that would consider that the LOV1 domain is no longer functional, we named this protein PLP in the present study. Interestingly, a yeast 2-hybrid screening performed with a cDNA bank from Arabidopsis leaves allowed the identification of AtGGP1 (VTC2) and AtGGP2 (VTC5) as being potential PLP interacting proteins (Ogura et al. 2008). Recently, Li and co-workers (2018) showed that in soybean (Glycine max), the expression of PLP can also be triggered when plants are cultured under darkness or red light. In their study, they also observed phenotypic alterations, especially hypocotyl growth, in Arabidopsis plp mutants over-expressing the soybean PLP. Nevertheless, no hypothesis was proposed to explain the putative function of the PLP protein in relation to such developmental processes (Li et al. 2018).
A population of EMS mutants obtained in the miniature Micro-Tom tomato cultivar was recently shown to exhibit a genetic and phenotypic variability far beyond the natural variation found in domesticated species (Just et al. 2013; Garcia et al. 2016). In a forward genetic strategy, this population proved valuable to screen for traits of interest (Garcia et al. 2016), including increased ascorbate (Deslous et al. 2021). The present study found a mutation causing a build-up in fruit ascorbate content and validated it within the gene encoding PLP. The interaction of PLP and GGP was confirmed in vivo and established in vitro, revealing that PLP inhibited GGP and that light promoted ascorbate synthesis by counteracting this inhibition.
Results
Detection of ascorbate-enriched mutants in an EMS Micro-Tom tomato population
A forward genetic approach was performed to discover regulators of ascorbate metabolism in plants. In that aim, the Ethyl Methanesulfonate (EMS) mutant collection generated in the Micro-Tom Tomato cultivar at INRAE Bordeaux (Just et al. 2013) was screened to find mutants producing ascorbate-enriched (also called AsA+) fruits (Deslous et al. 2021). A total of 500 M2 and M3 mutant families were cultivated in the greenhouse, in batches of 100 families representing a total of 6,000 plants screened. On each plant, at least 4 fruits at the red ripe stage were pooled and assayed for ascorbate content. The range of ascorbate content found in the mutants varied from 0.5 to 4 µmol.g−1 FW (Supplemental Figure S1A), whereas the WT mean values ranged from 1 to 1.3 µmol.g−1 FW depending on the period of the year. Although low, such variations in ascorbate content were expected in the WT, as it is well known that ascorbate content is regulated by environmental factors such as light irradiance (Gatzek et al. 2002; Gautier et al. 2008; Bartoli et al. 2009).
An ascorbate threshold value for selecting ascorbate-enriched mutants was set at 2 µmol.g−1 FW (approximately twice the value of the WT), resulting in the selection of 93 families (193 plants in total). Selected plants were cut to allow new growth and then used in a second screening for confirmation of the “AsA+” phenotype. Following this second screening, 5 families with 3 to 5 times higher fruit ascorbate content than the WT were selected. We present here the characterization of one of these mutants, named P21H6-3 (Supplemental Figure S1B). For this family, only 1 plant over 12 sown displayed the ascorbate-enriched phenotype. Interestingly, ascorbate content was 2.5 µmol.g−1 FW at the first screening performed at the end of autumn, whereas after the confirmation performed during the following spring season, the ascorbate content increased to up to 4 µmol.g−1 FW, thus suggesting an impact of the season on ascorbate pools. The P21H6-3 mutant was chosen as there was no apparent alteration in the phenotype of the vegetative and reproductive organs (Supplemental Figure S1B).
Identification of the causal mutation of the ascorbate-enriched P21H6 mutant
In order to identify the mutated locus responsible for the ascorbate-enriched phenotype, the genetic inheritance features of the mutation were first determined using classical Mendelian genetics. For this, the ascorbate-enriched phenotype was analyzed in the progeny after self-pollination (S1). For 12 S1 plants, all produced ascorbate-enriched fruits, whereas fruits from all plants of the backcross (BC1F1) displayed a WT-like ascorbate content phenotype (Supplemental Figure S2A). The resulting BC1F2 segregating population consisting of 440 individuals was then analyzed for the ascorbate-enriched phenotype (Supplemental Figure S2B). The analysis showed that 115 plants were defined as “AsA+” and 326 as “WT-like”, thus confirming a Mendelian 1:2:1 segregation, involving a single recessive mutation.
For the identification of the causal mutation, a mapping-by-sequencing strategy was used (Garcia et al. 2016). From the BC1F2 population, 2 bulked pools of 44 individual plants displaying either AsA+ or WT-like fruit phenotypes were generated (Supplemental Figure S2B). Pooled genomic DNA from each bulk was then sequenced to a tomato genome coverage depth of 39X, the trimmed sequences were mapped onto the tomato reference genome, and EMS mutation variants were filtered to exclude natural polymorphisms found in cv Micro-Tom (Kobayashi et al. 2014) compared with the cv Heinz 1706 reference genome (Supplemental Tables S1 and S2). Analysis of the allelic frequencies (AF) of variants in the 2 bulks led to the identification of chromosome 5 as the genome region carrying the causal mutation since it displayed high mutant AFs (AF > 0.95) in the AsA+ bulk and much lower frequencies (AF < 0.4) in the WT-like bulk (Fig. 1A). Analysis of the putative effects of the mutations on protein functionality highlighted 2 genes carrying mutations in exons. Among these, 1 affected an “unknown protein”, while the second affected a predicted PLP (Solyc05g007020) that was located at the top of the SNPs plot according to AF analysis. Given that a link between the closest homolog of PLP in Arabidopsis and the enzyme GGP had previously been found (Ogura et al. 2008), we considered PLP to be the most likely candidate. Unequivocally, to associate the plp mutation with the ascorbate-enriched trait, and to exclude any other mutated locus, recombinant plants selected from the BC1F2 progeny were used (Fig. 1B). To this end, we used the EMS-induced SNPs surrounding the PLP gene as genetic markers. It clearly indicated that a single G to A nucleotide transversion occurred in the fourth exon corresponding to the LOV domain of PLP (Fig. 1C), resulting in the knocking-out of the protein due to the appearance of a STOP codon instead of a glutamine codon (Supplemental Table S3).
Figure 1.
Identification of the mutation responsible for the ascorbate-enriched fruit phenotype. A) Identification of the chromosome associated with the ascorbate-enriched phenotype. Pattern of the mutation allelic frequencies obtained in the mutant and WT-like bulks are represented along tomato chromosomes. The plot represents allelic frequencies (y axis) against genome positions (x axis). A sliding window of 5 SNPs was used. The x axis displays the 12 tomato chromosomes; the arrow indicates the peak of allelic frequency (AF) of the chromosome 5 region carrying the putative causal mutations, since it displayed an AF > 0.95 in the ascorbate bulk and an AF < 0.4 in the WT-like bulk. B) Fine mapping of the causal mutation using the BC1F2 population. Recombinant analysis of 44 BC1F2 individuals displaying the ascorbate-enriched phenotype allowed us to locate the causal mutation at position 1,610,253 nucleotides. Marker positions are indicated by triangles. The number of recombinants is shown below the position of the markers. C) A single nucleotide transversion, G to A at position 1,610,253 in the Solyc05g007020 of the last codon of the fourth exon sequence, led to a change from glutamine to STOP codon. The sequence corresponding to the PAS and LOV domains is stained in the first exon and the third to the seventh exons, respectively.
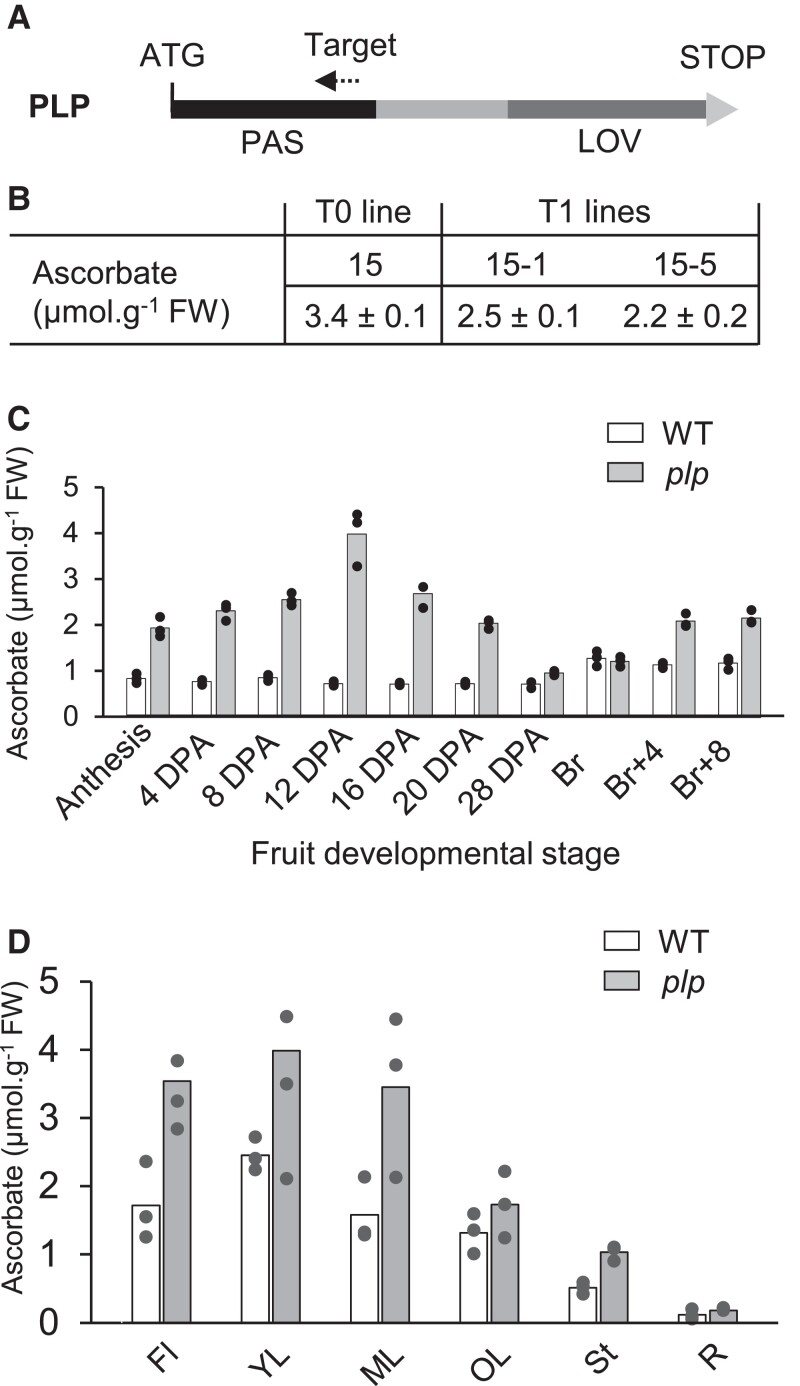
The functional validation of PLP as a negative regulator of ascorbate biosynthesis was next performed by generating plp mutants in the WT using clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9). Within the large family of photoreceptor proteins harboring a LOV domain, the latter has a very high level of sequence homology. Consequently, the PAS domain was targeted to produce knockout (KO) mutations in the 5′ terminal region of the PLP gene (Fig. 2A). Several T0 lines were generated, and mature fruits were analyzed for ascorbate content. Five T0 lines displaying at least a 2-fold increase in ascorbate were selected to produce the next T1 generation (Supplemental Table S4). The sequencing of the PLP gene for several T1 plants of lines 6, 15, 17, and 21 revealed various deletions that represented 1 to 8 nucleotides (Supplemental Fig. S3). All of these deletions resulted in a shift of the open reading frame and the appearance of a STOP codon in the downstream coding sequence of the PLP gene. For further studies, homozygous T2 plants of lines 15-1 and 15-5 were used indifferently as they harbored the same mutation (Fig. 2B; Supplemental Fig. S3). Moreover, a tomato genome investigation revealed the presence of a PLP-like protein (Solyc01g010480) displaying 65% peptide identity mainly distributed in the PAS and LOV domains characteristic of the phototropin proteins. Sequencing analysis showed that this PLP-like protein was not off-targeted by the chosen CRISPR/Cas9 strategy.
Figure 2.
Validation of PAS/LOV as the candidate gene involved in regulating ascorbate content in developing fruit and several tomato plant organs. A) Schematic representation of PLP showing its PAS and LOV domains. The dashed arrow in the PAS domain indicates the position of the target sequence for the CRISPR/Cas9 construct. B) Ascorbate in red ripe fruit (means ± SD, n = 4) of WT, T0 line 15, and progeny T1 lines 15-1 and 15-4. C) Ascorbate content in fruit of WT and plp mutant plants during development, from anthesis to ripeness. D) Ascorbate content in flowers, leaves at 3 stage of development, stem and roots of the 15-5 line, and control 1-mo-old plants. Data are the means (histograms) and 3 biological replicates representing 3 individual plants per organ and 3 organs per plant, except for anthesis (100 organs) and at 4 DPA (20 organs). PLP, PAS/LOV; DPA, days postanthesis; Br, breaker stage; FI, flowers; YL, young leaf; ML, mature leaf; OL, old leaf; St, stem; R, roots.
PAS/LOV is a repressor of ascorbate accumulation in tomato plants
As mentioned above, no noticeable morphological and physiological change was observed at the whole plant level in the mutant compared with the WT. To characterize the consequences of the knockout PLP gene on ascorbate metabolism, ascorbate assays were carried out during fruit development as well as in all tomato plant organs. As shown in Fig. 2C, growing fruits of the plp mutant had more ascorbate than the WT, with a peak at 12 DPA (days postanthesis). Interestingly, the difference in ascorbate content between mutant and WT declined to become negligible at the beginning of the maturation phase (breaker stage) but rose again during maturation. Finally, there was more ascorbate in all vegetative and reproductive organs of the mutant (Fig. 2D).
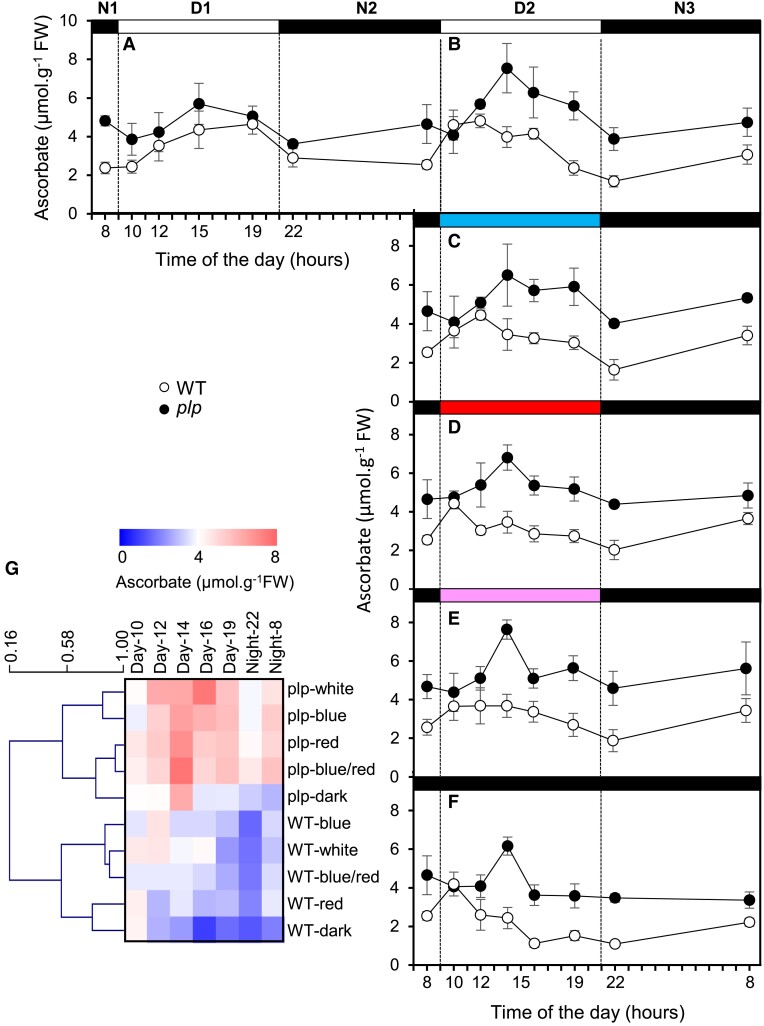
Next, we focused on leaves, which are easier to investigate than fruits. Since the mutated protein is a photosensor, the evolution of ascorbate across a day and night cycle was compared with those of the transcripts encoding PLP and GGP1. Photosynthetically active radiation (PAR) and temperature were recorded every 2 h, and plant material was harvested (Fig. 3A). As illustrated in Fig. 3B, while the level of ascorbate was always higher in the mutant, the daily variation in ascorbate content followed the same pattern in the T2 line 15-5 mutant and the WT. Thus, in both genotypes, ascorbate increased or plateaued during the first part of the day and then decreased toward the beginning of the night to rise again during the night. This was corroborated by the changes in GGP1 mRNA abundance, which decreased during the day and increased during the night, peaking 2 h before sunrise (Fig. 3D). Interestingly, PLP mRNA also peaked at 4 h in the night. However, PLP transcript levels were lower during the day than those encoding GGP1. Also, as expected, levels of PLP transcripts were significantly lower in the plp mutant compared with the WT (Fig. 3C).
Figure 3.
Changes in ascorbate and GDP-L-galactose phosphorylase and PAS/LOV mRNA during a day and night cycle in plp mutant and WT plants. The plp mutant (T2 line 15-5) and WT plants were cultured in the greenhouse for 1 mo. The night before the beginning of the experiment, all plants were moved outside and maintained under natural light conditions during 32 h. This experiment was carried out twice, on May 19 and July 10, 2018; they both lead remarkably to the same results. Here is presented the data obtained on July 10. A) Ambient temperature (diamonds) and light intensity (bars). B) Ascorbate content. C)PLP mRNA abundance. D)GGP1 mRNA abundance. Data are expressed as means ± SD of a total 3 mature leaves from 3 individual plants from the 15-5 plp line and WT control.
PAS/LOV interacts with GDP-L-galactose phosphorylase in the cytosol and the nucleus under the dependence of light signaling
In Arabidopsis, previous work has suggested a potential interaction between PLP and VTC2 and VTC5, which was governed by the light spectrum (Ogura et al. 2008). We first studied the subcellular localization of PLP by transient expression in Nicotiana benthamiana leaves. For this, a 35S-GFP-PLP construct was used either under its WT or truncated forms (Fig. 4A), as well as the 2 GGP isoforms, 35S-GFP-GGP1 and 35S-GFP-GGP2. Confocal microscopy analyses showed that the WT PLP, truncated PLP, GGP1, and GGP2, all localized in the nucleus and the cytoplasm (Fig. 4B). The dual localization of GGP1 is consistent with previous findings in Arabidopsis for VTC2 (Müller-Moulé 2008), which is a homolog to GGP1. However, according to previous literature, the subcellular localization of PLP, and more generally of blue light receptors, still needs to be established. To rule out the leakage of a cleaved GFP from PLP, we performed a liquid chromatography tandem mass spectrometry (LC–MS/MS) peptide analysis after sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) separation of a crude protein extracts from the stable transgenic tomato harboring the GFP-PLP construct. Peptides derived from the GFP-PLP fusion protein were detected in the gel band, corresponding to an apparent molecular mass of 73 kDa (Supplemental Fig. S4). This is consistent with the recovery of the nontruncated fused GFP-PLP protein after gel separation. No peptide derived from the GFP-PLP was detected in lower gel bands, in which cleaved products would have been expected to be recovered, confirming the integrity of the fused GFP-PLP after gel separation and, thus, during confocal microscopy analyses. Next, we tested the physical interactions of PLP with GGP1 by using the bimolecular fluorescence complementation (BiFC) technique in simple onion cell, with a combination of vectors for the GGP1, PLP, and truncated PLP (hereafter referred to as trPLP) sequences (Supplemental Table S5). The interaction between the 2 proteins was confirmed, but not with trPLP (Supplemental Fig. S5). Interestingly, the interaction occurred in both the cytoplasm and the nucleus. Since the mutation is characterized by a truncation of the blue light sensing LOV domain, a further verification of the protein–protein interactions and their light dependency was tested. For this, a heterologous system was used, thus allowing reconstructing and evaluating of the minimal protein interaction complex upon introduction of the individual components. Here, we implemented mammalian cells which are known to be suitable for expressing and assaying molecular mechanisms of regulation of plant proteins (Müller et al. 2014; Beyer et al. 2015). No additional plant components were used that might preclude a straightforward analysis of the interaction. In brief, a tetracycline-based split transcription factor approach was customized and used to test the light-regulated interaction of all GGP and PLP-combinations (GGP1, GGP2, PLP, and trPLP sequences, listed in Supplemental Table S6) (Müller et al. 2014). The PLP and trPLP were C-terminally fused to the tetracycline repressor (TetR) that binds to the tetracycline operator (tetO)-motif on the reporter plasmid. GGP1 and GGP2 were either C- or N-terminally coupled to the transactivation domain from the herpes simplex virus type 1 virion protein16 (VP16). Only the interaction of both proteins reconstitutes a functional transcription factor, capable of binding to the tetO-motif in close proximity to the PCMV minimal promoter and inducing gene expression of the reporter gene, via the VP16 transactivation domain. A strong interaction was found between WT PLP and both GGP1 and GGP2 in darkness, while the exposure to blue light (455 nm) minimized this interaction in all tested combinations (Fig. 4C). No interaction was found between the trPLP and GGP1 or GGP2, thus confirming the effect of the truncation (Fig. 4C). Interestingly, no additional factor was needed for the interaction of the WT PLP and GGP1 and GGP2. In summary, this experiment showed the light-controlled interaction between PLP and GGP proteins, while the mutant protein completely lost its ability to bind GGP1 or GGP2.
Figure 4.
Subcellular localization of PAS/LOV and GDP-L-galactose phosphorylase and their interaction. A) Diagram of the full-length PLP and the truncated PLP (trPLP). B) Localization of free GFP and 35S-GFP-fused proteins transiently expressed in N. benthamiana leaves 4 d after agroinfiltration, co-transformed with nuclear NLS-mcherry as nuclear marker. Scale bar = 50 µm. C) Analysis of PLP-GGP1/2 interactions and its light dependency in a heterologous mammalian split transcription factor system. Fifty thousand HEK-293 T cells were seeded in 24-well plates and transfected after 24 h with the plasmids pMZ1214, pMZ1215, pMZ1216, pMZ1217, pMZ1218, pMZ1219, pMZ1240, pMZ1241, pSAM, pRSET, and pKM006. Twenty-four hours posttransfection, the medium was exchanged by fresh medium and the cells were illuminated at 455 nm light (10 μmol m−2s−1) or kept in the dark for 24 h prior to SEAP quantification. Data are represented as means (histograms) and four biological replicates (dots). HEK-293 T, human embryonic kidney cells; SEAP, secreted alkaline phosphatase.
Repression of ascorbate accumulation via PAS/LOV is modulated by light
To investigate the physiological role of light signaling, we next analyzed the impact of blue, red, and white light, as well as darkness on ascorbate content in WT and plp mutants. One-month-old plants grown in the greenhouse were transferred to a growth chamber equipped with LEDs emitting white, blue, and/or red light. Plants were first transferred to a diel cycle of 12 h of white light at 250 to 260 µmol. m−2.s−1 and 12 h of darkness for 4 d. They were transferred under 4 light regimes at dawn, 100% blue light, 100% red light, or 50% blue/40% red light, respectively, and darkness or maintained under white light as the control. Light intensity was equally maintained for all conditions, except for darkness (Supplemental Fig. S6). In the WT under white light, leaf ascorbate content was increased during the first part of the photoperiod and decreased during the second part of the photoperiod (Fig. 5, A and B). In the mutant, a similar pattern was observed between minimum and maximum amplitude values. However, a higher basic level and a more sustained increase during the day were found. The same pattern was found for the mutant under the different light regimes, but not for the WT, in which ascorbate was no longer increased under darkness and red light (Fig. 5, C to F). Hierarchical clustering analysis showed a clear difference between WT and the mutant, which clustered separately (Fig. 5G). In the WT, 2 clusters were clearly distinguished, firstly darkness and red light and secondly white, blue/red, and blue light. Conversely, in the mutant, the red, blue/red, and darkness clustered together, probably because other factors were at play. These results suggest that blue light promotes ascorbate accumulation by counteracting the inhibitory effect of PLP on ascorbate synthesis. Given that GGP, one of the very few proteins described as interacting with PLP (Ogura et al. 2008), was found to interact in vivo with PLP (see above), we next investigated its effect on the activity of GGP in vitro.
Figure 5.
Effect of light on ascorbate evolution in WT and plp mutant leaves during a day–night cycle. A) In leaves of plants grown in a greenhouse then transferred to a growth chamber under a white light (with visible wavelengths from 420 nm to 760 nm) intensity of 260 to 270 µmol.m−2.s−1 for 24 h. B) Following cycle, still under white light. C) Following cycle under blue light (with a peak wavelength of 440 nm). D) Following cycle under red light (with peak wavelengths of 660 nm and 730 nm). E) Following cycle under blue (50%) and red (40%) light. F) Following cycle in the dark. G) Heat map representing a clustering analysis performed with mean values in MEV4.9. Columns correspond to time, and lines correspond to clustered content of ascorbate based on Pearson's correlation coefficient. All data shown in A to F) are expressed as means ± SD (n = 4).
Blue light prevents PAS/LOV inhibition of GDP-L-galactose phosphorylase activity in vitro
In order to study the in vitro interaction between PLP and GGP, the 2 tomato proteins were expressed heterologously (Fig. 6). In the case of GGP, a functional protein with characteristics reasonably close to those already published (Linster et al. 2008) was obtained with E. coli. Indeed, using GDP-alpha-glucose as a substrate, the Km and specific activity were respectively 17 µM and 37 U.mg−1 against 4 to 12 µM and 10 to 16 U.mg−1 in Arabidopsis (Linster et al. 2008). Notably, with Arabidopsis GGP, the Km and specific activity found for the commercially available GDP-glucose were close to those found for GDP-L-galactose (Linster et al. 2008), which is the precursor involved in ascorbate synthesis (Wheeler et al. 1998). In contrast, attempts to obtain a PLP capable of inhibiting GGP in vitro by expressing it in E. coli were unsuccessful, including accumulation of the protein in inclusion bodies that could only be resolubilized using chaotropic conditions. However, the latter did not make it possible to obtain a functional protein. Using a transient expression system in tobacco (Yamamoto et al. 2018), we obtained a PLP capable of inhibiting the activity of GGP (Fig. 6A). The inhibition of GGP by PLP set in quickly, probably within seconds (the time resolution of the spectrophotometer did not allow for more precise data) and was stable for hours (Supplemental Figure S7A). Strikingly, large amounts of PLP were necessary to inhibit GGP. Thus, for a GGP concentration of 3 nM, 90% inhibition was obtained with ∼300 nM of PLP, i.e. at a ratio of about 100. A kinetic study showed that the inhibition is noncompetitive (Fig. 6B) with PLP reducing the maximal activity but not the affinity for the substrate (here GDP-alpha-glucose). Blue light (445 ± 15 nm) had no effect when applied after mixing PLP and GGP (Supplemental Figure S7C). However, when applied to PLP before mixing with GGP, blue light counteracted its inhibitory effect at a level that depended on both the intensity and the duration of the illumination (Fig. 6C). In contrast, red light (2,000 µmol.s−1.m−2 at 653 ± 33 nm) had no effect on the interaction between PLP and GGP (Supplemental Figure S7C). Heating of PLP at 95 °C for 10 min suppressed the inhibitory effect of PLP (Supplemental Figure S7C). PLP appeared sensitive to blue light, with a response already significant at 25 µmol.s−1.m−2 and a plateau reached at around 250 µmol.s−1.m−2 for the rate of inactivation, confirming that the blue light intensities used in the in vivo experiment described above (Fig. 5) were effective. Finally, we tested the reversibility of the action of blue light by illuminating PLP with blue light at maximal intensity (2,000 µmol.m−2.s−1) during 2 h before transfer to darkness for 6 h, the time needed to fully recover the “dark” form (Ogura et al. 2008). While after 2 h, the inhibition was 86% and 16% for PLP incubated in the darkness and under blue light, respectively, it was 77% and 32% after 6 h of additional incubation in the dark (Supplemental Figure S7D). This confirms that PLP returns to its “dark” active form only very slowly following its exposure to blue light. Similar results were obtained with other preparations of both proteins (Supplemental Figure S7E and S7F).
Figure 6.
In vitro inhibition of GDP-L-galactose phosphorylase by PAS/LOV and effect of blue light on PAS/LOV. A) Relation between the PLP/GGP ratio and the inhibition of GGP. B) Hanes–Woolf plot GDP-L-galactose phosphorylase inhibition by PLP. GDP-glucose at concentrations of 12, 30, 60, 120, and 300 µM was used as substrate. C) Effect of blue light exposure duration on GGP inhibition by PLP. The light was applied before mixing the 2 proteins. All data shown are expressed as means (dashed lines) and technical duplicates (dots).
Discussion
Given the importance of ascorbate for human health and plant performance, the study of ascorbate metabolism in plants holds great interest from both agronomic and economic perspectives. Since the discovery of the main ascorbate biosynthetic pathway (Wheeler et al. 1998) and the characterization of the enzymes involved (Conklin et al. 1999, 2006; Linster et al. 2007), many studies have tried to decipher the regulatory mechanisms involved. Although it has long been known that light modulates ascorbate metabolism in plants, the underlying mechanisms were far from understood. In the present study, we identified a major player in the regulation of ascorbate metabolism by identifying the causal mutation leading to a 2- to 4-fold increase in the ascorbate content in an EMS Micro-Tom mutant. The recessive mutation corresponded to a stop codon in the Solyc05g007020 gene that encodes a photoreceptor PAS/LOV protein. Our work allowed (i) to undoubtedly define PLP as a negative regulator of ascorbate biosynthesis, (ii) to confirm the interaction of PLP with the GGP protein, (iii) to provide in vitro evidence that this interaction results in the inhibition of the activity of GGP, and (iv) to demonstrate that blue light counteracts this inhibition.
Why blue light as a regulator of ascorbate synthesis?
The function of PLP, which was first reported 20 yr ago (Crosson et al. 2003), has remained unknown despite the demonstration of its interaction with GGP (Ogura et al. 2008) and conformational change induced by blue light (Kasahara et al. 2010). The presence of a LOV domain links it to phototropins, which have 2 LOV domains (LOV1 and LOV2) associated with a kinase domain and initiate various developmental responses to blue light (phototropism, opening of stomata, chloroplast movements, leaf expansion. and movements) via autophosphorylation or even transphosphorylation (Christie 2007). Members of the Zeitlupe family, which, like PLP, have only 1 LOV domain, are involved in the photo-control of flowering and regulating the circadian clock (Ito et al. 2012). More generally, blue light stimulates shoot compactness by repressing the growth of the hypocotyl and internodes, promoting leaf thickness, flowering, production of secondary compounds such as carotenoids and flavonoids, and photosynthesis (Huche-Thelier et al. 2016). Plants would therefore use blue light to perceive the amount of light energy available in order to optimize their developmental program, but also to protect themselves from an excess of energy, in response to the different levels of photosensitivity of the photoreceptors (Gyula et al. 2003; Christie 2007). This idea is reinforced by the fact that flavins that generate ROS when excited by blue light are precisely the cofactors of these photoreceptors (Losi and Gartner 2012). The promoting effect of blue light on ascorbate synthesis would enable it to cope with the increase in ROS production when light intensity augments. In addition, higher ROS content tends to increase the proportion of the oxidized form of ascorbate, which is less stable (Bulley and Laing 2016; Truffault et al. 2017).
Blue light stimulation of the phototropins via the activation of 2 LOV domains results in the transduction of the signal through serine–threonine kinase activity leading to their autophosphorylation as well as phosphorylation of substrate targets (Christie et al. 2015). In contrast, the opposite is observed with PLP, which targets an enzyme of the ascorbate pathway but no longer acts or acts very weakly once exposed to blue light. PLP would therefore represent a peculiar evolution among photoreceptors, although favored by a modular nature that has allowed the appearance of numerous combinations of domains and, therefore, of neo-functionalization during evolution (Moglich et al. 2010). Strikingly, the photocycle rate is highly variable in photoreceptors containing the LOV domain. Thus, while photoexcitation only lasts a few tens of seconds in phototropins (Kasahara et al. 2002), it is maintained much longer in members of the ZTL/ADO family with more than 60 h for FKF1 (Zikihara et al. 2006). This is also seen in PLP, for which several hours were necessary for a return to the dark form (Kasahara et al. 2010), a result confirmed in the present work via the in vitro measurement of its inhibitory action. The irradiation of a LOV domain causes the formation, within a few microseconds, of a covalent bond between FMN and a nearby cysteine residue (Christie et al. 2015). While after 10 s of exposure to blue light, a significant change in the absorption spectrum of tomato PLP expressed in E. coli reflecting the change of state of FMN was found (Kasahara et al. 2010), several tens of minutes were necessary to reach minimum effect of PLP on GGP. However, PLP responded to low light intensity, with the inactivation rate increasing almost linearly until it plateaued at an intensity corresponding to the fraction of blue light in direct sunlight (∼200 µmol.m−2.s−1). These results are not necessarily contradictory if we consider that higher intensity increases the probability that PLP inactivation would occur, with inactivation being different from the change of state of FMN itself. Moreover, PLP expressed in E. coli was not able to inhibit GGP but was expressed in N. benthamiana. It is therefore possible that a difference in folding would affect its photosensitivity.
How blue light promotes ascorbate synthesis in a diel cycle in leaves
The results obtained here introduce an additional layer of complexity to the general regulation of ascorbate metabolism but specifically to GGP. The latter, identified as being the most controlling enzyme of the ascorbate synthesis pathway (Fenech et al. 2021), is known to be regulated by a range of factors, including light and stress, at the transcriptional (Bulley and Laing 2016) and translational (Laing et al. 2015) levels. Additionally, it appears to be regulated posttranslationally. This also extends the list of processes associating ascorbate synthesis and light, such as those involving the AMR1 protein (Zhang et al. 2009), and the COP9 signalosome complex (Mach 2013). The apparent irreversible inhibition of GGP by PLP found in vitro (Supplemental Figure S7A and S7B) suggests that the dark form of PLP formed a stable complex with GGP that could scarcely be dissociated by blue light (Fig. 7). The latter would therefore only act on newly formed copies of PLP, preventing them from interacting with GGP. The timing and amplitude of the expression of these 2 proteins would be essential to condition their interaction. It is shown in Figs. 3 and 5 that ascorbate exhibited relatively large fluctuations during day–night cycles and was amplified by the loss of PLP. In both cases, ascorbate increased during the day, dropped at the end of the day or the beginning of the night, increased again during the night, and then dropped again at the beginning of the day. These fluctuations can be attributed to the amount of substrate available for ascorbate synthesis, the rate of ascorbate turnover, and the amount of active GGP (Bulley and Laing 2016).
Figure 7.
Schematic model describing the activation of ascorbate synthesis by blue light. Newly synthesized PAS/LOV protein binds GDP-L-galactose phosphorylase unless deactivated by blue light. Its deactivated form is stable for several hours, while its active form irreversibly inhibits its target, possibly leading to its degradation. PLP, PAS/LOV; GGP, GDP-L-galactose phosphorylase.
The expression of genes encoding GGP1, which is by far the most strongly expressed of the 2 isoforms of GGP in tomato, and PLP increased at night to both peaks toward the end of the night. Interestingly, GGP1 expression was lower in the mutant (Fig. 5), a result that seems to confirm that ascorbate exerts a negative feedback on GGP expression. This contradicts the absence of effect of ascorbate itself as reported in Arabidopsis (Laing et al. 2015; Bulley et al. 2021), suggesting that another factor could be at play. The fact that ascorbate increased indicates that there were insufficient copies of PLP to neutralize all of the GGP formed overnight. Thereafter, GGP expression dropped to 0 or very low for most of the day, while PLP expression was retained and even increased during the day. Consequently, the number of copies of PLP would then become sufficient to block GGP, as evidenced by the decrease in ascorbate observed in the WT, but not in the mutant during the photoperiod under red light. In contrast, this is not true under white or blue light under which PLP would be deactivated (Fig. 5). Taken together, these elements suggest that the stoichiometry between GGP and PLP plays an essential role, the interaction between PLP and blue light making it possible to adjust the production of ascorbate to light intensity and ultimately to the production of light-dependent ROS. Therefore, it will be interesting to study the turnover of both proteins in diel cycles and under various light regimes, particularly in fruit, where PLP also modulates ascorbate synthesis (Fig. 2).
Regulation of ascorbate synthesis by PAS/LOV as an unusual and costly mechanism
Here, we investigated whether transcriptomic and proteomic data would be obtainable for these 2 genes. Available transcriptomic data indicate that the expression levels of GGP and PLP are elevated (Supplemental Figure- S8A and S8B). Thus, in the Arabidopsis leaves, during a day–night cycle (Bläsing et al. 2005), VTC2 was among the 2% to 3% and PLP among the 10% to 30% of the most highly expressed genes based on their average expression. Interestingly, the expression of PLP was strongly stimulated during the night in a starchless mutant, likely related to carbon starvation occurring at night in the mutant (Bläsing et al. 2005). Indeed, PLP is one of the most strongly repressed genes by sucrose and glucose (Bläsing et al. 2005). Yet, neither PLP nor VTC2 (GGP1) was found among the almost 5,000 proteins recently detected in a day–night cycle in the leaves of the same Arabidopsis accession (Uhrig et al. 2021). Similar results were found during tomato fruit development (Belouah et al. 2020), where the expression levels of GGP1 and PLP were also high, 2% and 22%, respectively, of the most highly expressed genes based on the average calculated during fruit development (Supplemental Figure S8C and S8D).
The encoded proteins were not among the more than 2,800 proteins detected. Additionally, these proteins were not present in a larger dataset of almost 8,000 proteins detected in tomato fruit (Szymanski et al. 2017). These observations are supported by the fact that the activity of GGP measured in the leaves of Arabidopsis is very low (Yoshimura et al. 2014). Among the explanations for such a discrepancy, one could invoke “inefficient” translation, in particular for GGP whose translation is jointly repressed by uORF and ascorbate (Laing et al. 2015), and/or high instability of the protein. It therefore seems relevant to ask whether the interaction between the 2 proteins leads to their degradation. Indeed, we found in vitro that the interaction was stable for at least several hours and could not be reversed even by the application of intensive blue light. The ratio of 100 found between PLP and GGP to block the activity of the latter may be overestimated, given that the proportion of functional protein among purified PLP was unknown. In order to learn more about the stoichiometry between these 2 proteins, it will be interesting to carry out crystallography studies.
Apart from blocking copies of PLP and GGP, the interaction in vivo between GGP and PLP could lead to the degradation of GGP or even that of PLP. The dual localization between the cytosol and the nucleus observed by confocal microscopy could be linked to the turnover of these proteins. Indeed, LOV Kelch protein 2 (LKP2), a member of the Zeitlupe family located in the nucleus, has been shown to form a complex that functions as an ubiquitin E3 ligase and interacts with the circadian clock (Yasuhara et al. 2004), possibly by addressing 1 or more components of the latter to the 26S proteasome. It would therefore be interesting to search for a similar mechanism that would involve PLP and GGP. It is also worth mentioning that 2 members of the TALE homeodomain protein transcription factor family, which is involved in numerous developmental processes (Hackbusch et al. 2005), have been identified as interacting with PLP by a double-hybrid screening of an Arabidopsis cDNA library (Ogura et al. 2008).
Finally, the mechanism regulating ascorbate synthesis shown here appears to be very costly with respect to GGP expression. However, the very high expression of GGP can also be seen as providing a great flexibility in response to stress. For example, an increase in sugars, often observed in response to various stresses, would repress the expression of PLP, thus making it possible to quickly increase the activity of the most controlling enzyme of the ascorbate synthesis pathway. These valuable data on the regulation of ascorbate metabolism will also make it possible to envisage new strategies for improving the nutritional quality of cultivated plants and their ability to withstand environmental stresses. It will be important to study in detail the turnover of GGP in relation to PLP to better understand the adaptive role of PLP. Indeed, while ascorbate is often seen as beneficial, extremely high concentrations can also have deleterious effects (Deslous et al. 2021).
Materials and methods
Plant material and culture conditions
S. lycopersicum L. cv. Micro-Tom was used for all experiments performed in this study. Plant culture conditions in the greenhouse were natural sunlight with addition of LED white light source when needed, 25 °C/20 °C for day/night and about 65% humidity as described in Rothan et al. (2016). To study the effect of light on ascorbate, experiments were carried out in spring or summer with 1-mo-old plants grown in the greenhouse. Thereafter, the plants remained either in the greenhouse in the case of the analysis of the ascorbate change in the time course of fruit development or the whole plant organs. For the “light assay”, the harvesting time of the plant materials was designed according to the light period (https://jekophoto.eu/tools/twilight-calculator-blue-hour-golden-hour/). This experiment was carried out twice in optimal weather conditions, on May 19 and July 10, 2018. One-month-old plants cultured in the greenhouse were moved out the night before the beginning of the experiment which began at 10 Am, with 3 leaves per plant from 3 plants harvested every 2 h. Concurrently, temperature, hygrometry, and photosynthetically active radiation (PAR, in µmol of photons m−2s−1) were measured with a Quantum photometer with a light sensor LI-190R (LI-COR Corporate, Lincoln Nebraska, USA), while the light spectrum was recorded with a JAZ Spectrometer (Ocean Optics Inc, Largo, Florida, USA).
To investigate the effect of the light spectrum, 1-mo-old plants were cultured in growth chambers (HiPoint Bionef Montreuil, France) as shown in Supplemental Fig. S6. To avoid stress, plants were moved from the greenhouse to the growth chambers at night. The plants were then acclimatized for 3 d under a 12 h photoperiod of white light with a photosynthetic photon flux density of around 260 to 265 µmol photons m−2 s−1 at 10 cm of the LED source and corresponding to the height of the top leaves. The day/night temperature and relative hygrometry were 25/20 °C and 65% to 60%, respectively. The first hour of the photoperiod (9 Am to 10 Am) was programmed as a ramp resulting in a linear increase of the light intensity to the required set-up. This was reversed at night (8 Pm to 9 Pm) returning to darkness, to mimic sunrise and sunset. On the fourth day, under the same culture conditions, and at the initial harvesting time (8 Am), 3 leaves per plant on 4 plants was were collected just before the beginning of the photoperiod and then every 2 to 3 h thereafter. On the fifth day, the plants were subjected to 4 light conditions, white (as control), blue, red, blue and red, and darkness, and leaves were harvested up to the end of the following night. For each light condition, the photon flux density was maintained at 260 to 270 µmol photons m−2 s−1 (Supplemental Fig. S6).
For the transient transformation experiments by agroinfiltration, WT N. benthamiana L. plants were cultured in soil for 1 mo in the greenhouse prior to the experiment. Watering was carried out 3 times per week: once with a Liquoplant fertilizing solution (1.85 g L−1, Plantin SARL Courthezon, France) and twice using tap water at pH 6. For the bimolecular fluorescence complementation (BiFC) analyses by confocal microscopy, freshly harvested onion (Allium cepa) was purchased from the local market.
To determine ascorbate content, leaves or fruits at several stages of development were collected, cut into small pieces, and immediately placed into liquid nitrogen. Samples were stored at −80 °C until extraction.
Mapping-by-sequencing
Mapping-by-sequencing was performed as described in Garcia et al. (2016). A mapping BC1F2 population of 440 plants was created by crossing the P21H6 cv Micro-Tom ascorbate-enriched mutant line with a WT cv Micro-Tom parental line. Two bulks were then constituted by pooling 44 plants displaying either a mutant fruit phenotype (AsA+ bulk) or 44 plants with a WT phenotype (WT-like bulk). To this end, 25 leaf discs (5 mm diameter each) were collected from each BC1F2 plant (∼60 mg fresh weight) and pooled into the AsA+ bulk and in the WT-like bulk. The same amount of plant material was also collected from the WT parental line. Genomic DNA was extracted from each bulk and the parental line using a cetyl-trimethyl-ammonium bromide method as described by Garcia et al. (2016). DNA was suspended in 200 mL of distilled water and quantified by fluorometric measurement with a Quant-it dsDNA assay kit (Invitrogen). Illumina paired-end shotgun-indexed libraries were prepared using the TruSeq DNA PCRF-Free LT Sample Preparation Kit according to the manufacturer's instructions (Illumina). The libraries were validated using an Agilent High Sensitivity DNA chip (Agilent Technologies) and sequenced using an Illumina HiSeq 2000 at the INRA EPGV facility, operating in a 100-bp paired-end run mode. Raw fastq files were mapped to the tomato reference genome sequence S. lycopersicum build release SL3.0 (ftp://ftp. solgenomics.net) using BWA version 0.7.12 (Li et al. 2009; http://bio-bwa.sourceforge.net/). Variant calling (SNPs and INDELs) was performed using SAMtools version 1.2 (Li et al. 2009; http://htslib.org). As the tomato reference genome (cv Heinz 1706) used to map the reads is distinct from that of cv Micro-Tom, the variants identified would include both cv Heinz 1706/cv Micro-Tom natural polymorphisms in addition to EMS mutations. In this context, additional sequencing to a minimum depth of 203 of the cv Micro-Tom line was performed to take into account and further remove the cv Heinz 1706/cv Micro-Tom natural polymorphism. The output file included various quality parameters relevant to sequencing and mapping that were subsequently used to filter the variants. The cv Micro-Tom line output file (.vcf) included all variants (SNPs plus INDELs) corresponding to natural polymorphisms between cv Micro-Tom and cv Heinz 1706. The two.vcf output files obtained from the AsA+ and WT-like bulks included variants (SNPs plus INDELs) corresponding to natural polymorphisms between cv Micro-Tom and cv Heinz 1706 and also to EMS mutations. The.vcf files were annotated using SnpEff version 4.1 (http://snpeff.sourceforge.net/SnpEff.html; Cingolani et al. 2012) using ITAG2.40 gene models (ftp://ftp. solgenomics.net). SNP allelic frequencies between AsA+ and WT-like bulks and the cv Micro-Tom parental line were compared using a custom Python script version 2.6.5 (https://www.python.org). Once the putative causal mutation was detected using the mapping-by-sequencing procedure, the EMS-induced SNPs flanking the putative mutation were used as markers for genotyping the BC1F2 individuals using a KASP assay (Smith and Maughan 2015). Specific primer design was performed using batchprimer3 software (Smith and Maughan 2015; http://probes.pw.usda. gov/batchprimer), and genotyping was done using KASP procedures (LGC Genomics).
CRISPR/Cas9 gene editing of PLP and stable tomato transformation
CRISPR/Cas9 gene editing was performed as described in Fauser et al. (2014). A construction comprising a single sgRNA alongside the Cas9 endonuclease gene was designed to induce target deletions in the PAS/LOV-coding sequence. The sgRNA target sequence was designed using CRISPR-P 2.0 web software (http://crispr.hzau.edu.cn/CRISPR2/) (Lei et al. 2014). Since targeting-RNAs are inserted into pDECAS9 vector, the final plasmid was used to transform Micro-Tom tomato cotyledons which were infected through Agrobacterium as described in Fernandez et al. (2009). The T0 plants resulting from the regeneration of the cotyledons were genotyped, and their fruits were phenotyped for ascorbate content. T1 seeds from selected ascorbate-enriched T0 plants were sown for further characterization. The CRISPR/Cas9 positive lines were further genotyped for indel mutations using primers flanking the target sequence. To obtain tomato plants overexpressing the GFP-PLP fusion protein, the Pro35S:eGFP-PLP construct in pK7WGF2 vector was used for the transformation as described above. All primers used for cloning are shown in Supplemental Table S6.
Ascorbate assay
Samples were ground in liquid nitrogen to a fine powder using a TissueLyser II (Qiagen). Ascorbate content was assayed using a protocol that takes advantage of the specificity of ascorbate oxidase, adapted from Bergmeyer (1987), using 40 and 100 mg FW for leaves and fruits, respectively, extracted in 400 µL 0.1 M HCl at 4 °C. For the determination of total ascorbate, 20 µL of extract was first incubated 10 min at 25 °C in 0.15 M HEPES/KOH pH 7.5 and 0.75 mM DTT. N-Ethyl maleimide was added to reach 0.035% w/v. After 10 min, 1 unit.mL−1 of ascorbate oxidase was added. After 20 min, phenazine ethosulfate and thiazolyl blue mix were added for final concentrations of respectively 0.3 and 0.6 mM. The thiazolyl blue mix was prepared as follows: 10 mM thiazolyl blue, 0.2 M Na2HPO4, 0.2 M citric acid, 2 mM EDTA, and 0.3% v/v Triton X100 at pH 3.5. All steps were performed in a polystyrene microplate at room temperature. Measurements were performed at 570 nm in MP96 readers (SAFAS, Monaco). For reduced ascorbate, the same protocol was used, except steps involving DTT and N-ethylmaleimide were omitted.
RT–qPCR analysis
Total RNA was extracted from leaves using Trizol reagent (Invitrogen) and purified with a RNeasy Plant Mini Kit (Qiagen). Relative transcript levels were determined as described previously (Deslous et al. 2021) using gene-specific primers and eIF4A and β-tubulin as an internal control. The primer sequences are shown in Supplemental Table S6.
Protein subcellular localization
All constructs used were generated using Gateway® technology (Invitrogen). The cDNA without STOP codon (NS) of GGP1, GGP2, PLP, and trPLP were synthesized by GeneArt® Gene Synthesis (Invitrogen) and directly provided into the entry vector pDONR201™ (Supplemental Table S5). The mutation of the PLP construct was the same as the one identified in the EMS mutant. In order to obtain fusion proteins with fluorescent tag either in C-terminal or N-terminal, specific primers (listed in Supplemental Table S6) were used to add a STOP codon and the flanking AttB sequences by PCR reaction. Classical BP recombination reactions allowed to insert the new sequences into a pDONR201™, and then LR reaction permitted to transfer our sequence of interest into the different destination vectors. The Agrobacterium tumefaciens electro-competent strain GV3101 was transformed with the above fluorescent fusion constructs. Transformed agrobacteria were selected on LB medium supplemented with suitable antibiotics and conserved at −80 °C. Prior to agroinfiltration, inoculated LB cultures were incubated overnight at 28 °C until 0.6 to 0.8 OD600nm. For subsequent infiltration, the culture was centrifuged and the pellet suspended in water to reach 0.2 OD600nm in the case of sub-cellular localization. Then, 50 to 100 µL of this bacterial solution was infiltrated in the leaf epidermis of 3-wk-old N. benthamiana plants and wounded by a 1 mL syringe needle to improve infiltration. The plants were maintained in normal culture conditions (12 h photoperiod, white LED at 260 to 270 µmol photons m−2 s−1, 25 °C day/20 °C night) for 48 h, and the observation was carried out on the abaxial leaf epidermis.
Protein interactions in plant cells
To assess the interaction of PLP and GGP1 in plant cells, a BiFC (Bimolecular Fluorescence Complementation) (Walter et al. 2004) experiment was performed in onion epidermal cells by biolistic transformation. Each cDNA of the genes of interest was inserted using the Gateway® technique in different vectors in order to test all possible orientations of the protein fusions (Supplemental Table S5). Then, 2.5 μg of plasmid DNA of each construct was mixed with 25 μL of a suspension (250 μg/μL) of gold micro-particles (diameter = 0.6 μm) in 50% ethanol (v/v); then, 25 μL of 2.5 M CaCl2 and 10 μL of 0.1 M spermidine are added. The micro-particles were left to sediment for 10 min before being washed with 70% and 100% (v/v) ethanol. Eight microliters of the micro-particle suspension (30 μL) was used for the transformation of epidermal onion cells using the PDE-1000He particle gun (Bio-Rad). Before transformation, the onion epidermis was peeled from the innermost scales of the onion bulb and placed with the adaxial side in contact with the MS medium. Transformation of the onion epidermal cells was performed at a pressure of 710 mm Hg at a helium pressure of 1100 psi and at a distance of 6 cm.
Imaging
Live imaging was performed in the plant imaging division of the BIC platform (Bordeaux Imaging Centre), using a Zeiss LSM 880 confocal laser scanning microscopy system equipped with 40× objectives. The excitation wavelengths used for the eGFP (or YFP) and mCherry were 514 and 543 nm, respectively. The emission windows defined for their observation were respectively between 525 and 600 nm for the eGFP (or YFP) and between 580 and 650 nm for the mCherry.
Light-mediated protein–protein interactions in mammalian cells
We employed an orthogonal mammalian, namely human embryonic kidney cells (HEK-293 T), to study the light-mediated protein–protein interactions. HEK-293 T cells were transfected with a combination of plasmids (Supplemental Table S6) and cultured as described by Müller et al. (2013). For the experimental set-up, 50,000 cells were seeded into 24-well plates. Twenty-four hours after seeding, cells were transfected using a polyethylenimine-based (PEI, linear, MW: 25 kDa, Polyscience) method, as described elsewhere (Müller et al. 2013). If co-transfected, plasmids were applied in equal-weight-based volume. Four hours posttransfection, the cell-culture medium was exchanged by pre-warmed fresh medium under green safelight conditions. Twenty hours later, cells were illuminated, using LED panels emitting blue light of a wavelength of 455 nm for 24 h (reporter gene activity), while control cells were kept in the dark. The quantitative determination of the activity of the secreted alkaline phosphatase (SEAP) in the cell culture medium was performed by using a previously described colorimetric assay (Müller et al. 2014; Beyer et al. 2015).
Proteomic analysis
Protein samples from tomato leaves were prepared by grinding 200 mg of leaves in 1 mL of 2× Laemmli buffer for 5 min in liquid nitrogen. Proteins were further solubilized by heating at 80 °C for 20 min. The insoluble material was removed by centrifugation (20 min at 13,000 × g), and the proteins of the supernatant were separated by SDS-PAGE (Laemmli 1970) using 4% stacking gel and 10% running gel. After colloidal blue staining, 3 bands were excised from the SDS-PAGE 10% gel and subsequently cut in 1 mm × 1 mm gel pieces. Gel pieces were destained in 25 mM ammonium bicarbonate 50% acetonitrile, rinsed twice in ultrapure water, and shrunk in acetonitrile for 10 min. After acetonitrile removal, gel pieces were dried at room temperature, covered with the trypsin solution (10 ng/µL in 40 mM NH4HCO3 and 10% acetonitrile), rehydrated at 4 °C for 10 min, and finally incubated overnight at 37 °C. Spots were then incubated for 15 min in 40 mM NH4HCO3 and 10% acetonitrile at room temperature with rotary shaking. The supernatant was collected, and an H2O/acetonitrile/HCOOH (47.5:47.5:5) extraction solution was added onto gel slices for 15 min. The extraction step was repeated twice. Supernatants were pooled and concentrated in a vacuum centrifuge to a final volume of 30 µL of 0.01% HCOOH. Digests were finally stored at −20 °C.
The peptide mixture was analyzed on an Ultimate 3000 nanoLC system (Dionex, Amsterdam, The Netherlands) coupled to an Electrospray Q-Exactive quadrupole Orbitrap benchtop mass spectrometer (Thermo Fisher Scientific, San Jose, CA). Ten microliters of peptide digests was loaded onto a 300-µm-inner diameter × 5-mm C18 PepMapTM trap column (LC Packings) at a flow rate of 30 µL/min. The peptides were eluted from the trap column onto an analytical 75-mm id × 25-cm C18 Pep-Map column (LC Packings) with a 4% to 40% linear gradient of solvent B in 108 min (solvent A was 0.1% formic acid in 5% acetonitrile, and solvent B was 0.1% formic acid in 80% acetonitrile). The separation flow rate was set at 300 nL/min. The mass spectrometer operated in positive ion mode at a 1.8-kV needle voltage. Data were acquired using Xcalibur 2.2 software in a data-dependent mode. MS scans (m/z 350 to 1600) were recorded at a resolution of R = 70,000 (@ m/z 200) and an AGC target of 3 × 106 ions collected within 100 ms. Dynamic exclusion was set to 30 s and top 12 ions were selected from fragmentation in HCD mode. MS/MS scans with a target value of 1 × 105 ions were collected with a maximum fill time of 100 ms and a resolution of R = 17,500. Additionally, only +2 and +3 charged ions were selected for fragmentation. Others settings were as follows: no sheath nor auxiliary gas flow, heated capillary temperature of 250 °C, normalized HCD collision energy of 25%, and an isolation width of 2 m/z.
Heterologous expression and purification of PLP and GGP1
The codon-optimized PLP was chemically synthesized by Proteogenix (Schiltigheim, France) and then cloned into the pET32a vector (Novagen). The PLP was expressed as a fusion protein with thioredoxin and His6 at the N-terminus in E. coli BL21(DE3)pLysS (Novagen) host cell. A bacterial culture was performed as described in Kasahara et al. (2010). The BL21(DE3)pLysS transformant harboring the PLP construct was grown at 25 °C in M9 medium supplemented with ampicillin (100 µg mL−1) and chloramphenicol (25 µg mL−1). When the culture reached 0.35 OD600nm, the PLP expression was induced in the presence of 1 mM isopropyl β-D-thiogalactopyranoside for 18 h at 25 °C. The cells were harvested by centrifugation; the pellet was suspended in 30 mL of lysis buffer [Tris-HCl 50 mM pH8, NaCl 0.5 M, glycerol 2% (v/v), 5 mM β-mercaptoethanol, 0.2% Sarkosyl, imidazole 50 mM, protease inhibitor cocktail EDTA free (Roche), and lysozyme 100 µg mL−1] and kept at room temperature for 20 min. Then, the suspension was frozen in nitrogen liquid and thawed at 25 °C twice before sonication for 15 min on ice. The lysate was centrifuged at 30,000 × g for 30 min at 4 °C, and the supernatant was filtered on 0.45 µm units before loading onto a nickel-Sepharose Fast Flow column (1 mL of bed volume) using an ÄKTATM Start Fast Protein Liquid Chromatography system (GE Healthcare). The PLP protein was eluted at 160 mM imidazole. The fractions containing the PLP peak were pooled and desalted on PD10 column; equilibrated with HEPES/KOH 50 mM pH7.5, glycerol 2% and Sarkosyl 0.2% (w/v), and 5 mM β-mercaptoethanol; and then concentrated using Vivaspin® 6 concentrators (Sartorius Stedim Lab Ltd, UK) and stored at −80 °C before use.
The full-length coding sequence of GGP1 was amplified by PCR and cloned into the pET28a vector (Novagen). The GGP1 enzyme was expressed in E. coli BL21(DE3)pLysS strain as a fusion protein with His6 at the N-terminus. The BL21(DE3)pLysS transformant harboring the GGP1 construct was grown at 37 °C in LB (500 mL) medium supplemented with kanamycin (50 µg mL−1) and chloramphenicol (25 µg mL−1). When the culture reached 0.5 OD600nm, the GGP1 expression was induced in the presence of 1 mM isopropyl β-D-thiogalactopyranoside for 6 h at 37 °C. The protein extraction and purification were performed as described above without adding β−mercaptoethanol and Sarkosyl in the lysis and desalting elution buffers.
PLP expression in tobacco and purification
The expression of PLP in N. benthamiana plants was carried out as described by Yamamoto et al. (2018). Briefly, the full-length coding sequence of PLP with His6 at the N-terminus was amplified by PCR and inserted into SalI-digested pBYR2HS vector using the In-Fusion Snap Assembly Master Mix (Takara Bio). The leaves of 4-wk-old N. benthamiana plants were infiltrated with the A. tumefaciens GV3101, harboring pBYR2HS-PLP with OD600nm adjusted approximately at 0.5. Once infiltrated, the plants were sprayed with a 200 mM ascorbate solution containing 0.1% Triton X-100 as described by Nosaki et al. (2021). Finally, the plants were grown under 16 h photoperiod of red light with a photosynthetic photon flux density of around 260 to 265 µmol photons m−2 s−1 in a growth chamber with a day/night temperature and relative hygrometry of 25/20 °C and 65% to 60%, respectively. After 4 d, whole infiltrated leaves were harvested and stored at −80 °C until protein extraction.
The infiltrated N. benthamiana leaves were ground using mortar and pestle in liquid nitrogen. All the following steps were carried out at 4 °C. Five grams of leaf powder was homogenized in 40 mL of extraction buffer [50 mM Na-phosphate pH 7.5, 0.5 Mm NaCl, 2% glycerol (v/v), 0.2% Tween 20 (v/v), 50 mM imidazole, and 5 mM β-mercaptoethanol] using a Polytron PT2100 for 20 s. The homogenate was centrifuged at 30,000 × g for 30 min, and the supernatant was clarified using 0.45 µm filters before loading onto a nickel-Sepharose Fast Flow column (5 mL of bed volume) using an ÄKTATM Start Fast Protein Liquid Chromatography system (GE Healthcare). The column was washed with 40 mL of extraction buffer, followed by 15 mL of extraction buffer with 90 mM imidazole. Proteins were then eluted with a linear imidazole gradient (0.09 to 0.5 M). The 3 mL fractions were analyzed by 10% SDS-PAGE gel electrophoresis, and those containing the protein of interest were desalted on PD10 column equilibrated with 50 mM HEPES/KOH pH 7.5, 2% glycerol (v/v), and 0.2% Tween 20 (v/v), then concentrated using Vivaspin® 6 concentrators (Sartorius Stedim Lab Ltd, UK), and stored at −80 °C before use.
Assay of GDP-L-galactose phosphorylase activity
For the determination of GGP activity under substrate-saturating conditions, a continuous assay was used in which GGP extract was incubated at 25 °C in the presence of 50 mM HEPES/KOH pH 7.5, 10 mM MgCl2, 2 mM EDTA, 1 mM GDP-glucose, 20 mM phosphate, 2 mM phosphoenolpyruvate, 0.5 mM NADH, 1 unit.mL−1 pyruvate kinase, and 1 unit.mL−1 lactate dehydrogenase. Changes in absorbance were measured at 340 nm in a filter-based MP96 microplate reader (SAFAS, Monaco) until rates were stabilized.
The effect of light was tested by incubating purified PLP in 50 mM HEPES/KOH pH 7.5, 0.2% (v/v) Tween 20, 2% (v/v) glycerol, and 1 µM FMN, under a LedHUB light source (Omicron Laserage, Laserprodukte GmbH, Rodgau-Dudenhofen, Germany) emitting at 445 ± 15 nm or 653 ± 33 nm, at intensities ranging from 0 to 2,000 µmol.m−2.s−1.
Database search and results processing
Data were searched by SEQUEST through Proteome Discoverer 2.2 (Thermo Fisher Scientific Inc.) against the S. lycopersicum protein database downloaded from the SGN website (version ITAG3.2; 35768 entries) in which the sequences of the 3 constructs were added. Spectra from peptides higher than 5,000 Da or lower than 350 Da were rejected. The search parameters were as follows: mass accuracy of the monoisotopic peptide precursor and peptide fragments was set to 10 ppm and 0.02 Da, respectively. Only b- and y-ions were considered for mass calculation. Oxidation of methionines (+16 Da) was considered as variable modification and carbamidomethylation of cysteines (+57 Da) as fixed modification. Two missed trypsin cleavages were allowed. Peptide validation was performed using Percolator algorithm (Käll et al. 2007), and only “high confidence” peptides were retained corresponding to a 1% False Positive Rate at peptide level.
Accession numbers
Sequence data from key genes in this article can be found in the Sol Genomics Network (https://solgenomics.net/) or The Arabidopsis Information Resource (https://www.arabidopsis.org/) under the following accession numbers: Solyc06g073320 (SlGGP1), Solyc02g091510 (SlGGP2), Solyc05g07020 (SlPLP), Solyc01g010480 (SlPLP-like), AT4G26850 (AtGGP1/VTC2), AT5G55120 (AtGGP2/VTC5), and AT2G02710 (AtPLP).
Supplementary Material
Acknowledgments
The authors are grateful to C. Cheniclet and L. Brocard from the Bordeaux Imaging Center for their assistance in the cytological analysis, S. Claverol from the Plateforme Protéome of the Centre Génomique Fonctionnelle Bordeaux for the proteomic analysis, and all the Master’s students, C. Cerruti, S. Barré, M. Alonso, H. El Ouarrat, R. Gomez, D. Taillis, J. Hunziker, J. Paillassa, and M. Zion, who helped in the screening and the phenotypic characterization of the mutants. We also are grateful to J. Christie for helpful discussion about how to name the protein and M. Kasahara for sharing detailed bacterial culture protocols for the production of the PLP protein. A special thanks to I. Atienza who took care of the plants during all these years.
Contributor Information
Céline Bournonville, UMR 1332 Biologie du Fruit et Pathologie, Univ. Bordeaux, INRAE, 33883 Villenave d'Ornon, France.
Kentaro Mori, UMR 1332 Biologie du Fruit et Pathologie, Univ. Bordeaux, INRAE, 33883 Villenave d'Ornon, France.
Paul Deslous, UMR 1332 Biologie du Fruit et Pathologie, Univ. Bordeaux, INRAE, 33883 Villenave d'Ornon, France.
Guillaume Decros, UMR 1332 Biologie du Fruit et Pathologie, Univ. Bordeaux, INRAE, 33883 Villenave d'Ornon, France.
Tim Blomeier, Institute of Synthetic Biology—CEPLAS—Faculty of Mathematics and Natural Sciences, Heinrich-Heine-Universität Düsseldorf, Dusseldorf 40225, Germany.
Jean-Philippe Mauxion, UMR 1332 Biologie du Fruit et Pathologie, Univ. Bordeaux, INRAE, 33883 Villenave d'Ornon, France.
Joana Jorly, UMR 1332 Biologie du Fruit et Pathologie, Univ. Bordeaux, INRAE, 33883 Villenave d'Ornon, France.
Stéphanie Gadin, UMR 1332 Biologie du Fruit et Pathologie, Univ. Bordeaux, INRAE, 33883 Villenave d'Ornon, France.
Cédric Cassan, UMR 1332 Biologie du Fruit et Pathologie, Univ. Bordeaux, INRAE, 33883 Villenave d'Ornon, France.
Mickael Maucourt, UMR 1332 Biologie du Fruit et Pathologie, Univ. Bordeaux, INRAE, 33883 Villenave d'Ornon, France.
Daniel Just, UMR 1332 Biologie du Fruit et Pathologie, Univ. Bordeaux, INRAE, 33883 Villenave d'Ornon, France.
Cécile Brès, UMR 1332 Biologie du Fruit et Pathologie, Univ. Bordeaux, INRAE, 33883 Villenave d'Ornon, France.
Christophe Rothan, UMR 1332 Biologie du Fruit et Pathologie, Univ. Bordeaux, INRAE, 33883 Villenave d'Ornon, France.
Carine Ferrand, UMR 1332 Biologie du Fruit et Pathologie, Univ. Bordeaux, INRAE, 33883 Villenave d'Ornon, France.
Lucie Fernandez-Lochu, UMR 1332 Biologie du Fruit et Pathologie, Univ. Bordeaux, INRAE, 33883 Villenave d'Ornon, France.
Laure Bataille, UMR 1332 Biologie du Fruit et Pathologie, Univ. Bordeaux, INRAE, 33883 Villenave d'Ornon, France.
Kenji Miura, Tsukuba Innovation Plant Research Center, University of Tsukuba, 1-1-1 Tennodai, 305-8577 Ibaraki, Tsukuba, Japan.
Laure Beven, UMR 1332 Biologie du Fruit et Pathologie, Univ. Bordeaux, INRAE, 33883 Villenave d'Ornon, France.
Matias D Zurbriggen, Institute of Synthetic Biology—CEPLAS—Faculty of Mathematics and Natural Sciences, Heinrich-Heine-Universität Düsseldorf, Dusseldorf 40225, Germany.
Pierre Pétriacq, UMR 1332 Biologie du Fruit et Pathologie, Univ. Bordeaux, INRAE, 33883 Villenave d'Ornon, France.
Yves Gibon, UMR 1332 Biologie du Fruit et Pathologie, Univ. Bordeaux, INRAE, 33883 Villenave d'Ornon, France.
Pierre Baldet, UMR 1332 Biologie du Fruit et Pathologie, Univ. Bordeaux, INRAE, 33883 Villenave d'Ornon, France.
Author contributions
C.Bo. screened the EMS population and contributed to the identification of the causal mutation. P.D. validated the candidate gene and performed microscopy experiments. M.D.Z. and P.B. designed experiments to assess light-mediated molecular regulation. T.B. performed the analysis in animal cells. K.Mo., J.-P.M., and J.J. performed the generation of CRISPR lines and the stable transgenic line. S.G. performed the transcriptional analysis. C.Br. and L.F. performed the genetic analysis. L.Be. performed the LC–MS proteomic analysis. K.Mo., K. Mi., L.Ba., and P.B. performed the protein expression and purification experiments. C.C., M.M., and Y.G. performed the enzymatic analysis. G.D., C.F., and D.J. participated in the set-up of the plant culture and growth chambers. C.Bo., K.Mo., P.D., Y.G., and P.B. wrote the manuscript with input from the other authors. P.P. contributed to the scientific input and English editing of the manuscript. All authors read and approved the manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Screening of the EMS Micro-Tom population (supports Fig. 1).
Supplemental Figure S2. Identification of the plp mutation responsible for the ascorbate-enriched fruit phenotype by mapping-by-sequencing (supports Fig. 1).
Supplemental Figure S3. CRISPR/Cas9 strategy for the PLP gene (supports Fig. 2).
Supplemental Figure S4. Subcellular localization of PLP in tomato and proteomic analysis of the leaf extract (supports Fig. 4).
Supplemental Figure S5. In vivo protein–protein interaction of PLP and GGP1 (supports Fig. 5).
Supplemental Figure S6. Photos of tomato plants cultured in growth chamber under different light regimes and the corresponding light spectra measured at INRAE Bordeaux (supports Fig. 3).
Supplemental Figure S7. Reversibility of the effect of blue light on GDP-L-galactose phosphorylase (GGP) inhibition by PAS/LOV (PLP) (supports Fig. 6).
Supplemental Figure S8. Continuous time assay of transcripts encoding GDP-L-galactose phosphorylase (GGP) and the PAS/LOV protein (supports Fig. 7).
Supplemental Table S1. Illumina sequencing of BC1F2 bulk individuals displaying an ascorbate-enriched mutant fruit or a WT-like fruit (supports Fig. 1).
Supplemental Table S2. Number of SNPs in the mutant and the WT-like bulks for the P21H6-3 mutant (supports Fig. 1).
Supplemental Table S3. Protein sequences of the Tomato PAS/LOV in WT and P21H6 mutant (supports Fig. 1).
Supplemental Table S4. Ascorbate content in rep ripe fruits of T0 and T1 CRISPR plp plants (supports Fig. 2).
Supplemental Table S5. Result of the different combinations tested for the BiFC experiments between GGP1 and PLP or trPLP.
Supplemental Table S6. Set of oligos and plasmids used in this study.
Funding
This work was supported by Région Aquitaine [Con. 20111201002] (C.B. and P.D.), Syngenta Seeds SAS [CA Tom AsA INRA MD 0502-TG_SYN-MD1505] (C.B.), the Plant Biology and Breeding Division of INRAE (to P.D.), and the Département Sciences de l’Environnement at the University of Bordeaux (Projet Emergent FRUITO), PHENOME (ANR-11-INBS-0012), INRAE BAP RARE and LIA FreQUenCE INRAE-Tsukuba University (2020-2024), and Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy CEPLAS— EXC-2048/1—Project no. 390686111, and NEXTplant (GRK 2466) to M.D.Z.
Data availability
The data supporting the findings of this study are available from the corresponding author (pierre.baldet@inrae.fr) upon request.
References
- Ahmad M, Cashmore AR. HY4 gene of A-Thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 1993:366(6451):162–166. 10.1038/366162a0 [DOI] [PubMed] [Google Scholar]
- Alimohammfadi M, de Silva K, Ballu C, Ali N, Khodakovskaya MV. Reduction of inositol (1,4,5)–trisphosphate affects the overall phosphoinositol pathway and leads to modifications in light signalling and secondary metabolism in tomato plants. J Exp Bot. 2012:63(2):825–835. 10.1093/jxb/err306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli CG, Tambussi EA, Diego F, Foyer CH. Control of ascorbic acid synthesis and accumulation and glutathione by the incident light red/far red ratio in Phaseolus vulgaris leaves. FEBS Lett. 2009:583(1):118–122. 10.1016/j.febslet.2008.11.034 [DOI] [PubMed] [Google Scholar]
- Belouah I, Bénard C, Denton A, Blein-Nicolas M, Balliau T, Teyssier E, Gallusci P, Bouchez O, Usadel B, Zivy M, et al. Transcriptomic and proteomic data in developing tomato fruit. Data Brief 2020:28:105015. 10.1016/j.dib.2019.105015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmeyer HU. Methods of enzymatic analysis. Weinheim: VCH. 1987 [Google Scholar]
- Beyer HM, Juillot S, Herbst K, Samodelov SL, Müller K, Schamel WW, Römer W, Schäfer E, Nagy F, Strähle U, et al. Red light-regulated reversible nuclear localization of proteins in mammalian cells and zebrafish. ACS Synth Biol. 2015:4(9):951–958. 10.1021/acssynbio.5b00004 [DOI] [PubMed] [Google Scholar]
- Bläsing OE, Gibon Y, Günther M, Höhne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible W-R, Stitt M. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 2005:17(12):3257–3281. 10.1105/tpc.105.035261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs WR., Beck CF, Cashmore AR, Christie JM, Hughes J, Jarillo JA, Kagawa T, Kanegae H, Liscum E, Nagatani A, et al. The phototropin family of photoreceptors. Plant Cell 2001:13(5):993–997. 10.1105/tpc.13.5.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulley SM, Cooney JM, Laing W. Elevating ascorbate in Arabidopsis stimulates the production of abscisic acid, phaseic acid, and to a lesser extent auxin (IAA) and jasmonates, resulting in increased expression of DHAR1 and multiple transcription factors associated with abiotic stress tolerance. Int J Mol Sci. 2021:22(13):6743. 10.3390/ijms22136743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulley S, Laing W. The regulation of ascorbate biosynthesis. Curr Opin Plant Biol. 2016:33:15–22. 10.1016/j.pbi.2016.04.010 [DOI] [PubMed] [Google Scholar]
- Bulley SM, Rassam M, Hoser D, Otto W, Schünemann N, Wright M, MacRae E, Gleave A, Laing W. Gene expression studies in kiwifruit and gene over-expression in Arabidopsis indicates that GDP-L-galactose guanyltransferase is a major control point of vitamin C biosynthesis. J Exp Bot. 2009:60(3):765–778. 10.1093/jxb/ern327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JJ. Missing step in man, monkey and Guinea pig required for the biosynthesis of L-ascorbic acid. Nature. 1957:180(4585):553. 10.1038/180553a0 [DOI] [PubMed] [Google Scholar]
- Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GTJ, Batschauer A, Ahmad M. The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol. 2011:62(1):335–366. 10.1146/annurev-arplant-042110-103759 [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J. Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 2011:21(11):664–671. 10.1016/tcb.2011.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM. Phototropin blue-light receptors. Annu Rev Plant Biol. 2007:58:21–45. 10.1146/annurev.arplant.58.032806.103951 [DOI] [PubMed] [Google Scholar]
- Christie JM, Blackwood L, Petersen J, Sullivan S. Plant flavoprotein photoreceptors. Plant Cell Physiol. 2015:56(3):401–413. 10.1093/pcp/pcu196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Salomon M, Nozue K, Wada M, Briggs WR. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc Natl Acad Sci USA 1999:96(15):8779–8783. 10.1073/pnas.96.15.8779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Swartz TE, Bogomolni RA, Briggs WR. Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. Plant J. 2002:32(2):205–219. 10.1046/j.1365-313X.2002.01415.x [DOI] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly. 2012:6(2):80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Gatzek S, Wheeler GL, Dowdle J, Raymond MJ, Rolinski S, Isupov M, Littlechild JA, Smirnoff N. Arabidopsis thaliana VTC4 encodes L-galactose-1-P phosphatase, a plant ascorbic acid biosynthetic enzyme. J Biol Chem. 2006:281(23):15662–15670. 10.1074/jbc.M601409200 [DOI] [PubMed] [Google Scholar]
- Conklin PL, Norris SR, Wheeler GL, Williams EH, Smirnoff N, Last RL. Genetic evidence for the role of GDP-D-mannose in plant ascorbic acid (vitamin C) biosynthesis. Proc Natl Acad Sci SA 1999:96(7):4198–4203. 10.1073/pnas.96.7.4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosson S, Rajagopal S, Moffat K. The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry 2003:42(1):2–10. 10.1021/bi026978l [DOI] [PubMed] [Google Scholar]
- Decros G, Baldet P, Beauvoit B, Stevens R, Flandin A, Colombie S, Gibon Y, Pétriacq P. Get the balance right: ROS homeostasis and redox signalling in fruit. Front Plant Sci. 2019:10:1–16. 10.3389/fpls.2019.01091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslous P, Bournonville C, Decros G, Okabe Y, Mauxion J-P, Jorly J, Gadin S, Brès C, Mori K, Ferrand C, et al. Overproduction of ascorbic acid impairs pollen fertility in tomato. J Exp Bot. 2021:72(8):3091–3107. 10.1093/jxb/erab040 [DOI] [PubMed] [Google Scholar]
- Dowdle J, Ishikawa T, Gatzek S, Rolinski S, Smirnoff N. Two genes in Arabidopsis thaliana encoding GDP-L-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. Plant J. 2007:52(4):673–689. 10.1111/j.1365-313X.2007.03266.x [DOI] [PubMed] [Google Scholar]
- Fauser F, Schiml S, Puchta H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 2014:79(2):348–359. 10.1111/tpj.12554 [DOI] [PubMed] [Google Scholar]
- Fenech M, Amorim-Silva V, Esteban del Valle A, Arnaud D, Ruiz-Lopez N, Castillo AG, Smirnoff N, Botella MA. The role of GDP-L-galactose phosphorylase in the control of ascorbate biosynthesis. Plant Physiol. 2021:185(4):1574–1594. 10.1093/plphys/kiab010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AI, Viron N, Alhagdow M, Karimi M, Jones M, Amsellem Z, Sicard A, Czerednik A, Angenent G, Grierson D, et al. Flexible tools for gene expression and silencing in tomato. Plant Physiol. 2009:151(4):1729–1740. 10.1104/pp.109.147546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Bres C, Just D, Fernandez L, Tai FWJ, Mauxion JP, Le Paslier MC, Bérard A, Brunel D, Aoki K, et al. Rapid identification of causal mutations in tomato EMS populations via mapping-by-sequencing. Nat Protoc. 2016:11(12):2401–2418. 10.1038/nprot.2016.143 [DOI] [PubMed] [Google Scholar]
- Gatzek S, Wheeler GL, Smirnoff N. Antisense suppression of L-galactose dehydrogenase in Arabidopsis thaliana provides evidence for its role in ascorbate synthesis and reveals light modulated L-galactose synthesis. Plant J. 2002:30(5):541–553. 10.1046/j.1365-313X.2002.01315.x [DOI] [PubMed] [Google Scholar]
- Gautier H, Diakou-Verdin V, Bénard C, Reich M, Buret M, Bourgaud F, Poëssel JL, Caris-Veyrat C, Génard M. How does tomato quality (sugar, acid, and nutritional quality) vary with ripening stage, temperature, and irradiance? J Agric Food Chem. 2008:56(4):1241–1250. 10.1021/jf072196t [DOI] [PubMed] [Google Scholar]
- Gautier H, Massot C, Stevens R, Sérino S, Génard M. Regulation of tomato fruit ascorbate content is more highly dependent on fruit irradiance than leaf irradiance. Ann Bot. 2009:103(3):495–504. 10.1093/aob/mcn233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gest N, Gautier H, Stevens R. Ascorbate as seen through plant evolution: the rise of a successful molecule? J Exp Bot. 2013:64(1):33–53. 10.1093/jxb/ers297 [DOI] [PubMed] [Google Scholar]
- Gyula P, Schäfer E, Nagy F. Light perception and signaling in higher plants. Curr Opin Plant Biol. 2003:6(5):446–452. 10.1016/S1369-5266(03)00082-7 [DOI] [PubMed] [Google Scholar]
- Hackbusch J, Richter K, Muller J, Salamini F, Uhrig JF. A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc Natl Acad Sci USA 2005:102(13):4908–4912. 10.1073/pnas.0501181102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huche-Thelier L, Crespel L, Le Gourrierec J, Morel P, Sakr S, Leduc N. Light signaling and plant responses to blue and UV radiations-perspectives for applications in horticulture. Environ Exp Bot. 2016:121:22–38. 10.1016/j.envexpbot.2015.06.009 [DOI] [Google Scholar]
- Ito S, Song YH, Imaizumi T. LOV domain-containing F-box proteins: light-dependent protein degradation modules in Arabidopsis. Mol Plant. 2012:5(3):573–582. 10.1093/mp/sss013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI. Structure and function of the UV-B photoreceptor UVR8. Curr Opin Struct Biol. 2014:29:52–57. 10.1016/j.sbi.2014.09.004 [DOI] [PubMed] [Google Scholar]
- Just D, Garcia V, Fernandez L, et al. Micro-Tom mutants for functional analysis of target genes and discovery of new alleles in tomato. Plant Biotechnol. 2013:30(3):225–231. 10.5511/plantbiotechnology.13.0622a [DOI] [Google Scholar]
- Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets, Nat Methods. 2007:4(1):923–925. 10.1038/nmeth1113 [DOI] [PubMed] [Google Scholar]
- Kasahara M, Swartz TE, Olney MA, Onodera A, Mochizuki N, Fukuzawa H, Asamizu E, Tabata S, Kanegae H, Takano M, et al. Photochemical properties of the flavin mononucleotide-binding domains of the phototropins from Arabidopsis, rice, and Chlamydomonas reinhardtii. Plant Physiol. 2002:129(2):762–773. 10.1104/pp.002410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M, Torii M, Fujita A, Tainaka K. FMN Binding and photochemical properties of plant putative photoreceptors containing two LOV domains, LOV/LOV proteins. J Biol Chem. 2010:285(45):34765–34772. 10.1074/jbc.M110.145367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Nagasaki H, Garcia V, et al. Genome-wide analysis of intraspecific DNA polymorphism in ‘Micro-Tom’, a model cultivar of tomato (Solanum lycopersicum). Plant Cell Physiol. 2014:55(2):445–454. 10.1093/pcp/pct181 [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970:227(5259):680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Laing WA, Martinez-Sanchez M, Wright MA, Bulley SM, Brewster D, Dare AP, Rassam M, Wang D, Storey R, Macknight RC, et al. An upstream open reading frame is essential for feedback regulation of ascorbate biosynthesis in Arabidopsis. Plant Cell 2015:27(3):772–786. 10.1105/tpc.114.133777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Lu L, Liu H-Y, Li S, Xing F, Chen L-L. CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant. 2014:7(9):1494–1496. 10.1093/mp/ssu044 [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup . The sequence alignment/map format and SAMtools. Bioinformatics 2009:25(16):2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liang D, Li M, Ma F. Light and abiotic stresses regulate the expression of GDP-L-galactose phosphorylase and levels of ascorbic acid in two kiwifruit genotypes via light-responsive and stress-inducible cis-elements in their promoters. Planta 2013:238(3):535–547. 10.1007/s00425-013-1915-z [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang Y, Li M, Luo Q, Mallano AI, Jing Y, Zhang Y, Zhao L, Li W. GmPLP1, a PAS/LOV protein, functions as a possible new type of blue light photoreceptor in soybean. Gene 2018:645:170–178. 10.1016/j.gene.2017.12.016 [DOI] [PubMed] [Google Scholar]
- Linster CL, Adler LN, Webb K, Christensen KC, Brenner C, Clarke SG. A second GDP-L-galactose phosphorylase in Arabidopsis en route to vitamin C. Covalent intermediate and substrate requirements for the conserved reaction. J Biol Chem. 2008:283(27):18483–18492. 10.1074/jbc.M802594200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linster CL, Gomez TA, Christensen KC, Adler LN, Young BD, Brenner C, Clarke SG. Arabidopsis VTC2 encodes a GDP-L-galactose phosphorylase, the last unknown enzyme in the smirnoff-wheeler pathway to ascorbic acid in plants. J Biol Chem. 2007:282(26):18879–18885. 10.1074/jbc.M702094200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X, Lin C. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 2011:25(10):1029–1034. 10.1101/gad.2025011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losi A, Gartner W. The evolution of flavin-binding photoreceptors: an ancient chromophore serving trendy blue-light sensors. Annu Rev Plant Biol. 2012:63(1):49–72. 10.1146/annurev-arplant-042811-105538 [DOI] [PubMed] [Google Scholar]
- Mach J. COP9 signalosome-regulated proteolysis: turning off ascorbic acid synthesis when the lights go out. Plant Cell. 2013:25(2):359–359. 10.1105/tpc.113.250212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massot C, Stevens R, Génard M, Longuenesse J-J, Gautier H. Light affects ascorbate content and ascorbate-related gene expression in tomato leaves more than in fruits. Planta 2012:235(1):153–163. 10.1007/s00425-011-1493-x [DOI] [PubMed] [Google Scholar]
- Moglich A, Yang XJ, Ayers RA, Moffat K. Structure and function of plant photoreceptors. Annu Rev Plant Biol. 2010:61(1):21–47. 10.1146/annurev-arplant-042809-112259 [DOI] [PubMed] [Google Scholar]
- Müller-Moulé P. An expression analysis of the ascorbate biosynthesis enzyme VTC2. Plant Mol Biol. 2008:68(1-2):31–41. 10.1007/s11103-008-9350-4 [DOI] [PubMed] [Google Scholar]
- Müller K, Engesser R, Schulz S, Steinberg T, Tomakidi P, Weber CC, Ulm R, Timmer J, Zurbriggen MD, Weber W. Multi-chromatic control of mammalian gene expression and signaling. Nucleic Acids Res. 2013:41(12):e124. 10.1093/nar/gkt340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K, Siegel D, Rodriguez Jahnke F, Gerrer K, Wend S, Decker EL, Reski R, Weber W, Zurbriggen MD. A red light-controlled synthetic gene expression switch for plant systems. Mol Biosyst. 2014:10(7):1679–1688. 10.1039/C3MB70579J [DOI] [PubMed] [Google Scholar]
- Nosaki S, Kaneko MK, Tsuruta F, Yoshida H, Kato Y, Miura K. Prevention of necrosis caused by transient expression in Nicotiana benthamiana by application of ascorbic acid. Plant Physiol. 2021:186(2):832–835. 10.1093/plphys/kiab102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Komatsu A, Zikihara K, Nanjo T, Tokutomi S, Wada M, Kiyosue T. Blue light diminishes interaction of PAS/LOV proteins, putative blue light receptors in Arabidopsis thaliana, with their interacting partners. J Plant Res. 2008:121(1):97–105. 10.1007/s10265-007-0118-8 [DOI] [PubMed] [Google Scholar]
- Rothan C, Just D, Fernandez L, Atienza I, Ballias P, Lemaire-Chamley M. Culture of the tomato micro-tom cultivar in greenhouse. In: Botella JR, Botella MA editors. Plant signal transduction: methods and protocols. New York: (NY: ): Springer New York; 2016. p. 57–64. [DOI] [PubMed] [Google Scholar]
- Smith SM, Maughan PJ. SNP Genotyping using KASPar assays. Methods Mol Biol. 2015:1245:243–256. 10.1007/978-1-4939-1966-6_18 [DOI] [PubMed] [Google Scholar]
- Suetsugu N, Wada M. Evolution of three LOV blue light receptor families in green plants and photosynthetic stramenopiles: phototropin, ZTL/FKF1/LKP2 and aureochrome. Plant Cell Physiol. 2013:54(1):8–23. 10.1093/pcp/pcs165 [DOI] [PubMed] [Google Scholar]
- Szymanski J, Levin Y, Savidor A, Breitel D, Chappell-Maor L, Heinig U, Töpfer N, Aharoni A. Label-free deep shotgun proteomics reveals protein dynamics during tomato fruit tissues development. Plant J. 2017:90(2):396–417. 10.1111/tpj.13490 [DOI] [PubMed] [Google Scholar]
- Tabata K, Takaoka T, Esaka M. Gene expression of ascorbic acid-related enzymes in tobacco. Phytochemistry 2002:61(6):631–635. 10.1016/S0031-9422(02)00367-9 [DOI] [PubMed] [Google Scholar]
- Truffault V, Fry SC, Stevens RG, Gautier H. Ascorbate degradation in tomato leads to accumulation of oxalate, threonate and oxalyl threonate. Plant J. 2017:89(5):996–1008. 10.1111/tpj.13439 [DOI] [PubMed] [Google Scholar]
- Uhrig RG, Echevarría-Zomeño S, Schlapfer P, Grossmann J, Roschitzki B, Koerber N, Fiorani F, Gruissem W. Diurnal dynamics of the Arabidopsis rosette proteome and phosphoproteome. Plant Cell Environ. 2021:44(3):821–841. 10.1111/pce.13969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004:40(3):428–438. 10.1111/j.1365-313X.2004.02219.x [DOI] [PubMed] [Google Scholar]
- Wang J, Yu Y, Zhang Z, Quan R, Zhang H, Ma L, Deng XW, Huang R. Arabidopsis CSN5B interacts with VTC1 and modulates ascorbic acid synthesis. Plant Cell 2013:25(2):625–636. 10.1105/tpc.112.106880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler G, Ishikawa T, Pornsaksit V, Smirnoff N. Evolution of alternative biosynthetic pathways for vitamin C following plastid acquisition in photosynthetic eukaryotes. Elife 2015:4:e06369. 10.7554/eLife.06369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler GL, Jones M, Smirnoff N. Vitamin C in higher plants. Nature 1998:393(6683):365–369. 10.1038/30728 [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Hoshikawa K, Ezura K, Okazawa R, Fujita S, Takaoka M, Mason HS, Ezura H, Miura K. Improvement of the transient expression system for production of recombinant proteins in plants. Sci Rep. 2018:8(1):4755. 10.1038/s41598-018-23024-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara M, Mitsui S, Hirano H, Takanabe R, Tokioka Y, Ihara N, Komatsu A, Seki M, Shinozaki K, Kiyosue T. Identification of ASK and clock-associated proteins as molecular partners of LKP2 (LOV kelch protein 2) in Arabidopsis. J Exp Bot. 2004:55(405):2015–2027. 10.1093/jxb/erh226 [DOI] [PubMed] [Google Scholar]
- Yoshimura K, Nakane T, Kume S, Shiomi Y, Maruta T, Ishikawa T, Shigeoka S. Transient expression analysis revealed the importance of VTC2 expression level in light/dark regulation of ascorbate biosynthesis in Arabidopsis. Biosci Biotechnol Biochem. 2014:78(1):60–66. 10.1080/09168451.2014.877831 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Huang R. Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol Biol. 2010:73(3):241–249. 10.1007/s11103-010-9609-4 [DOI] [PubMed] [Google Scholar]
- Zhang W, Lorence A, Gruszewski HA, Chevone BI, Nessler CL. AMR1, An arabidopsis gene that coordinately and negatively regulates the mannose/L-galactose ascorbic acid biosynthetic pathway. Plant Physiol. 2009:150(2):942–950. 10.1104/pp.109.138453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang J, Zhang R, Huang R. The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J. 2012:71(2):273–287. 10.1111/j.1365-313X.2012.04996.x [DOI] [PubMed] [Google Scholar]
- Zikihara K, Iwata T, Matsuoka D, Kandori H, Todo T, Tokutomi S. Photoreaction cycle of the light, oxygen, and voltage domain in FKF1 determined by low-temperature absorption spectroscopy. Biochemistry 2006:45(36):10828–10837. 10.1021/bi0607857 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available from the corresponding author (pierre.baldet@inrae.fr) upon request.