Abstract
SALT OVERLY SENSITIVE1 (SOS1) is a key component of plant salt tolerance. However, how SOS1 transcription is dynamically regulated in plant response to different salinity conditions remains elusive. Here, we report that C-type Cyclin1;1 (CycC1;1) negatively regulates salt tolerance by interfering with WRKY75-mediated transcriptional activation of SOS1 in Arabidopsis (Arabidopsis thaliana). Disruption of CycC1;1 promotes SOS1 expression and salt tolerance in Arabidopsis because CycC1;1 interferes with RNA polymerase II recruitment by occupying the SOS1 promoter. Enhanced salt tolerance of the cycc1;1 mutant was completely compromised by an SOS1 mutation. Moreover, CycC1;1 physically interacts with the transcription factor WRKY75, which can bind to the SOS1 promoter and activate SOS1 expression. In contrast to the cycc1;1 mutant, the wrky75 mutant has attenuated SOS1 expression and salt tolerance, whereas overexpression of SOS1 rescues the salt sensitivity of wrky75. Intriguingly, CycC1;1 inhibits WRKY75-mediated transcriptional activation of SOS1 via their interaction. Thus, increased SOS1 expression and salt tolerance in cycc1;1 were abolished by WRKY75 mutation. Our findings demonstrate that CycC1;1 forms a complex with WRKY75 to inactivate SOS1 transcription under low salinity conditions. By contrast, under high salinity conditions, SOS1 transcription and plant salt tolerance are activated at least partially by increased WRKY75 expression but decreased CycC1;1 expression.
CycC1;1 forms a transcriptional repression complex with the transcription factor WRKY75 to downregulate SOS1 expression, thereby negatively regulating salt stress tolerance in Arabidopsis.
Graphical Abstract
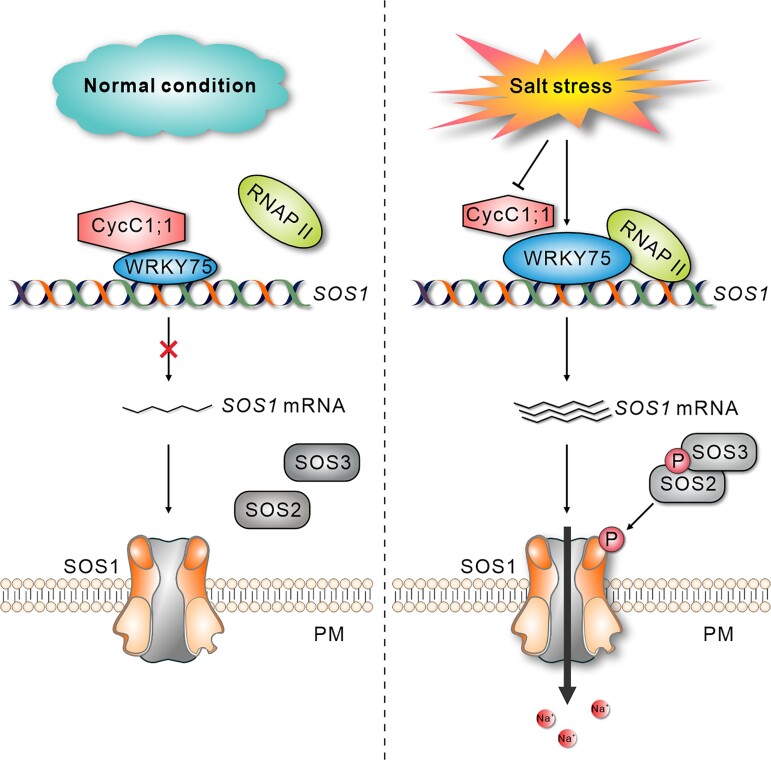
Graphical Abstract.
IN A NUTSHELL.
Background: Soil salinization is a major environmental hazard that severely affects plant growth and development. Plants have evolved sophisticated mechanisms that help them withstand elevated soil salinity, and Salt Overly Sensitive1 (SOS1) plays a crucial role in plant salt stress tolerance by facilitating the extrusion of excess Na+ from the cells. Although previous reports have demonstrated the important role of posttranslational regulation of SOS1 in plant salt stress tolerance, how SOS1 transcription is dynamically modulated in response to different salinity conditions remains unclear.
Question: What are the molecular mechanisms by which plants regulate SOS1 expression at the transcriptional level in response to salinity stress?
Findings: Disruption of the CycC1;1 subunit of the plant Mediator complex promotes salt-induced SOS1 expression and salt tolerance in Arabidopsis because CycC1;1 interferes with RNA polymerase II recruitment by occupying the SOS1 promoter. SOS1 mutation in the cycc1;1 mutant completely compromised its enhanced salt tolerance. Moreover, CycC1;1 can physically interact with the transcription factor WRKY75, which can directly bind to the SOS1 promoter and activate its expression. In contrast to the cycc1;1 mutant, the wrky75 mutant has attenuated SOS1 expression and salt tolerance, whereas overexpression of SOS1 can rescue the salt sensitivity of the mutant. Intriguingly, CycC1;1 inhibits WRKY75 transcriptional activation activity for SOS1 through their interaction; thus, increased SOS1 expression and salt tolerance in the cycc1;1 mutant were abolished by the WRKY75 mutation. In addition, CycC1;1 expression is repressed, and WRKY75 expression is stimulated in response to high salinity.
Next steps: In a future study, we will explore whether other components of the Mediator complex are coordinated with CycC1;1 to precisely control SOS1 transcription, and investigate how salinity affects CycC1;1 and WRKY75 expression in the plant's response to different salinity conditions.
Introduction
Soil salinization is one of the major abiotic stresses that severely affect plant growth and development, causing huge losses in crop production worldwide (Zhu 2001; Yang and Guo 2018a; Van Zelm et al. 2020). During evolution, plants have developed various adaptive strategies to cope with high salinity stress (Zhu 2002, 2003; Van Zelm et al. 2020). The Salt Overly Sensitive (SOS) pathway plays a crucial role in plant salt stress tolerance (Zhu 2001; Yang and Guo 2018a). Three core proteins have been identified in the SOS pathway in Arabidopsis (Arabidopsis thaliana): SOS1, SOS2, and SOS3 (Yang and Guo 2018b; Zhu 2001). SOS1 is a Na+/H+ antiporter located at the plasma membrane that acts downstream in the SOS pathway by facilitating the efflux of excess Na+ from cells and regulating long-distance transport of Na+ in plants (Shi et al. 2002; Zhu 2002). It is well-known that a salt stress–induced cytosolic calcium signal is perceived by SOS3, an EF-hand calcium-binding protein, which then interacts with and activates SOS2, a Ser/Thr protein kinase (Ishitani et al. 2000). The activated SOS2 phosphorylates SOS1, promoting its activity and thus increasing salt tolerance (Liu et al. 2000; Zhu 2002). SOS1 activity is tightly regulated in plants: stimulated under high salinity conditions while inactivated under low salinity conditions through multiple different regulatory mechanisms, enabling intracellular sodium homeostasis.
Besides the three core proteins in the SOS pathway, other factors are implicated in plant salt response and tolerance. CALCIUM BINDING PROTEIN8 (SCaBP8)/Calcineurin B-like10 (CBL10) is a SOS3-like protein that recruits SOS2 to the plasma membrane in a calcium-dependent manner, thereby activating SOS1 and promoting plant salt tolerance (Liu et al. 2000; Quan et al. 2007; Lin et al. 2009). Mitogen-activated protein kinase (MPK) signaling cascades are also involved in plant salt stress tolerance through MPK6-mediated phosphorylation of SOS1 (Yu et al. 2010). In addition, VPS23A, the component of the endosomal sorting complex required for transport (ESCRT) enhances SOS2–SOS3 interaction and their localization at the plasma membrane, thus increasing SOS1 activity and salt tolerance (Lou et al. 2020 ). Interestingly, salt stress increases the conversion of phosphatidylinositol into phosphatidylinositol 4-phosphate, enhancing plant salt tolerance by activating SOS1 activity at the plasma membrane (Yang et al. 2021). A recent study reported that a small peptide PAMPINDUCED SECRETED PEPTIDE 3 (PIP3) could bind to and activate a leucine-rich repeat receptor-like kinase RECEPTOR-LIKE KINASE 7 (RLK7), leading to activation of MPK3 and MPK6 as well as salt tolerance in Arabidopsis (Zhou et al. 2022). In contrast, some negative regulators of the SOS pathway have also been identified and documented. For example, the photoperiodic and circadian clock oscillator protein Gigantea (GI) interacts with and inhibits SOS2 to inactivate the SOS pathway under normal conditions, whereas high salinity stress decreases GI protein accumulation, resulting in the activation of the pathway (Kim et al. 2013). The 14-3-3 proteins λ and κ can also interact with and inhibit SOS2 activity in the absence of salt stress, while salt stress reduces the interaction between the 14-3-3 proteins and SOS2, leading to activation of the SOS pathway for higher salt tolerance in Arabidopsis (Zhou et al. 2014). In addition to posttranslational modulation, SOS1 activity is also significantly modulated at the transcriptional level. It has long been known that, upon salt stress treatment, the SOS1 transcripts are rapidly induced and accumulate, and their stability is promoted in Arabidopsis (Shi et al. 2000; Chung et al. 2008). Although a few transcription factors, including WRKY1 and MYB73, have been identified as positive or negative factors affecting SOS1 transcription in Arabidopsis (Kim et al. 2013; Wu et al. 2022), how SOS1 transcription is dynamically regulated to adjust plant responses to different salinity conditions remains to be further elucidated.
Mediator is a conserved protein complex in eukaryotes that significantly affects gene transcription by acting as a bridge between transcription factors and RNA polymerase II (RNAP II) (Asturias et al. 1999; Harper and Taatjes 2018; Agrawal et al. 2021). The Mediator complex is composed of 4 major components, including head, middle, tail, and kinase modules. The head, middle, and tail modules form the core part of the Mediator complex, providing interfaces for physical interaction between RNAP II and sequence-specific transcription factors (Jeronimo and Robert 2017). The kinase module is a separate part of the Mediator complex consisting of Cyclin-Dependent Kinase 8 (CDK8), C-type Cyclin (CycC), Mediator Complex Subunit12 (MED12), and MED13 (Wang and Chen 2004; Mathur et al. 2011; Maji et al. 2019) that significantly alters gene transcription by binding to the core part of Mediator (Poss et al. 2013). The Mediator complex functions in a variety of versatile roles in plant growth, development, and environmental responses (Agrawal et al. 2021; Chen et al. 2012; Chen et al. 2019; Guo et al. 2021; Maji et al. 2019), but whether and how it is involved in plant salt stress responses and tolerance remain unknown.
In this study, we identified C-type Cyclin1;1 (CycC1;1) as a negative regulator of salt stress tolerance in Arabidopsis by repressing SOS1 expression via disrupting the recruitment of RNAP II on SOS1 promoter. On the contrary, we found that WRKY75 positively regulated salt-induced SOS1 expression by binding to the SOS1 promoter. Furthermore, CycC1;1 interacted with WRKY75 and inhibited its transcriptional activation of SOS1 expression, and this inhibition was relieved both by high salinity-mediated repression of CycC1;1 expression and by induction of WRKY75 expression. Our results demonstrate an important role for the CycC1;1–WRKY75 complex in transcriptional regulation of SOS1, allowing plants to respond to different salinity conditions.
Results
CycC1;1 negatively modulates salt tolerance
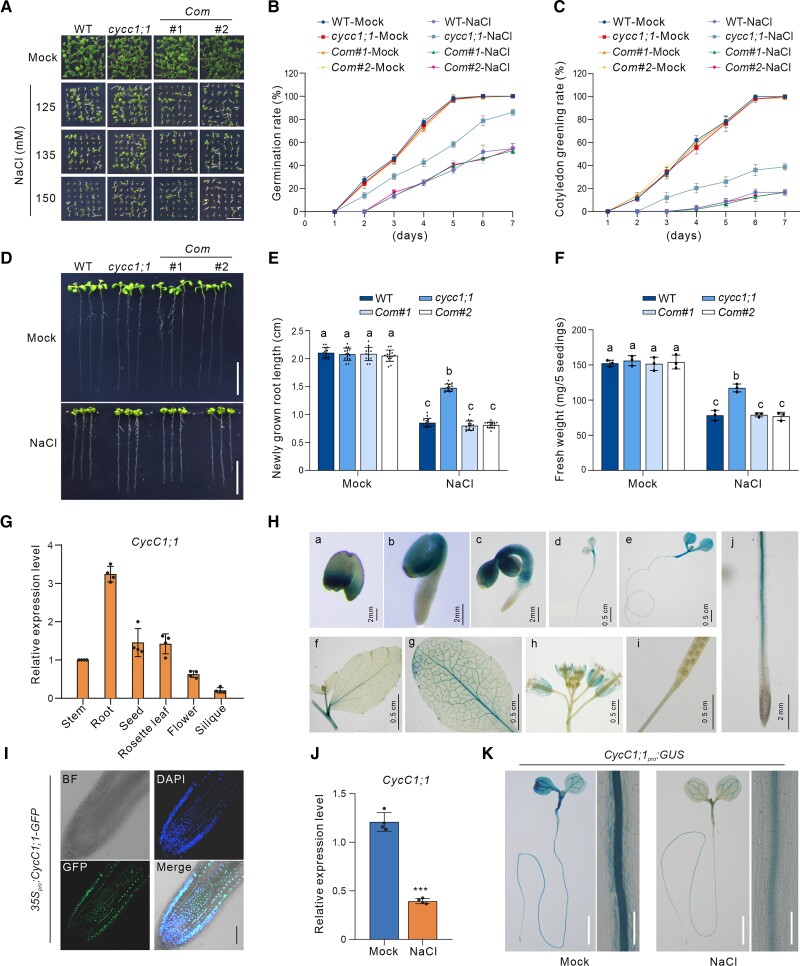
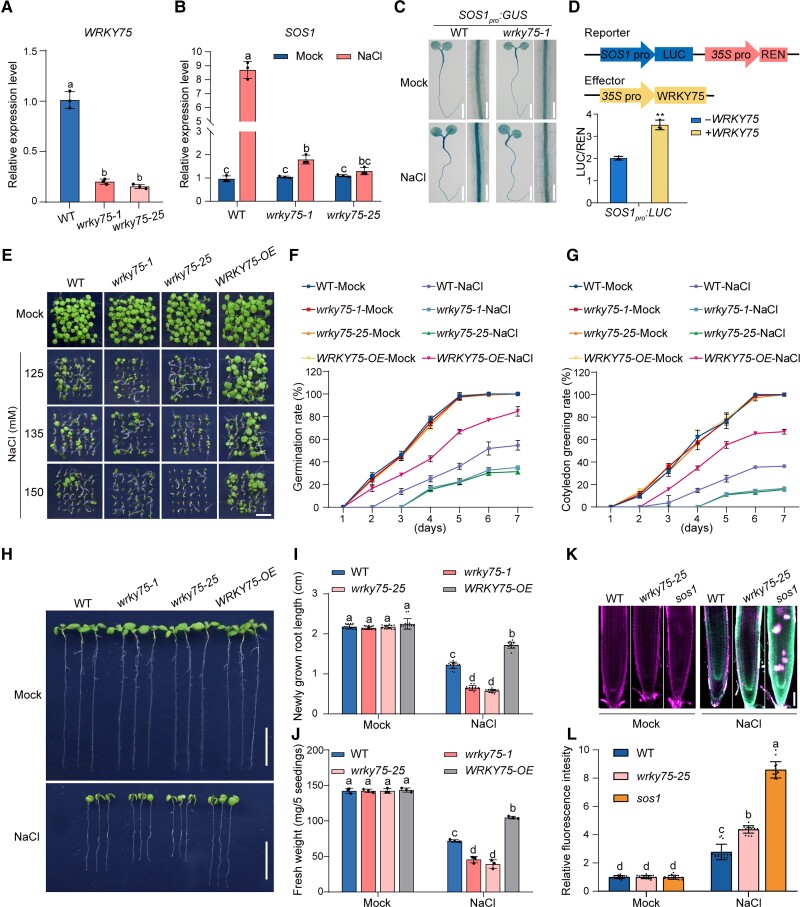
As a component of the Mediator complex, CycC is known to affect plant immunity by modulating the expression of defense-related genes (Zhu et al. 2014), but it has not been characterized in the context of abiotic stress responses in plants. We had previously identified cycc1;1 (SALK_053291), a T-DNA insertion mutant allele of CycC1;1, that showed decreased expression of CycC1;1 (Guo et al. 2022). We recently found that this mutant also showed lower sensitivity to high salinity compared with the wild-type (WT) as evidenced by increased seed germination and cotyledon greening rates after 5 d in high-salt (125 to 150 mM) conditions (Fig. 1, A to C), suggesting the involvement of CycC1;1 in the plant salt stress response. To test its salt tolerance, 5-d-old wild-type and the cycc1;1 mutant seedlings grown on 1/2× Murashige–Skoog (1/2× MS) medium were subjected to high salinity for 2 additional days. Our results showed that the cycc1;1 mutant had primary root length and fresh weight comparable to the wild-type under nonstress conditions, while the mutant displayed longer primary roots and a higher fresh weight than the wild-type under high salinity conditions (Fig. 1, D to F), revealing that the cycc1;1 mutant was more tolerant to the salt stress than the wild-type plant. To confirm whether the salt-tolerant phenotype of the cycc1;1 mutant was caused by the disruption of CycC1;1, transgenic complementary lines in which the expression of CycC1;1 was driven by its native promoter (Guo et al. 2022) were analyzed for salt sensitivity. As expected, the complementary lines had similar salt tolerance to the wild-type regarding seed germination, primary root elongation, and fresh weight (Fig. 1, A to F). Furthermore, we previously assayed the salt tolerance of the CycC1;1-overexpression transgenic lines, CaMV 35Spro:CycC1;1-GFP, and found that the expression of CycC1;1 was significantly higher than that in the wild-type plants (Guo et al. 2022). In contrast to the cycc1;1 mutant, when exposed to high salinity, the CycC1;1-overexpression plants had lower germination rates, shorter primary roots, and lower fresh weights than the wild-type plants (Supplemental Fig. S1). These results indicate that CycC1;1 is a negative regulator of plant salt stress tolerance.
Figure 1.
CycC1;1 negatively regulates plant salt tolerance. A to C) Phenotypes A) of the wild-type, cycc1;1, and complementation (Com) plants grown on 1/2× MS medium supplemented with 0 mM, 125 mM, 135 mM, or 150 mM NaCl for 5 d. Bar = 0.5 cm. Quantitative analysis of seed germination B) and cotyledon greening rates C) of plants grown on 1/2× MS medium supplemented with 0 mM or 125 mM NaCl for 7 d. Data are means ± Sd of 3 independent biological repeats. D to F) Root elongation and fresh weight analysis. Five-day-old wild-type, cycc1;1, and Com plants were transferred to 1/2× MS medium supplemented with 0 mM or 125 mM NaCl for additional growth. Photographs were taken 5 d after transfer D). Bar = 1 cm. The lengths of newly grown roots E) and the fresh weights F) of the seedlings were also analyzed. Data are means ± Sd (n = 15 for root length and n = 3 for fresh weight). Bars with different letters indicate significant differences at P < 0.05, determined using ANOVA with a Tukey's multiple comparison test (Supplemental Data Set 1). G) RT-qPCR results showing the expression of CycC1;1 in different plant tissues, including stem, root, seed, rosette leaf, flower, and silique. The experiment was repeated 3 times. Data are means ± Sd (n = 4). ACTIN2 was used as a reference gene. H) Glucuronidase (GUS) staining images of the CycC1;1pro:GUS transgenic reporter plants. a, seed; b, 1-d-old germinating seed; c, 3-d-old seedling; d, 5-d-old seedling; e, 7-d-old seedling; f, cauline leaf; g, rosette leaf; h, flower; i, silique; j, 5-d-old seedling root tip. a to c, j, bar = 2 mm; d to i, bar = 0.5 cm. I) Nuclear localization of CycC1;1-GFP in the 5-d-old 35Spro:CycC1;1-GFP transgenic plant root. DAPI was used to stain the nucleus. Bar = 100 μm. J) The expression of CycC1;1 in 5-d-old wild-type plant seedlings treated with or without 100 mM for 12 h. The experiment was repeated 3 times. Error bars indicate mean ± Sd (n = 4). Asterisks indicate significant differences determined by Student's t test (***P < 0.001). K) GUS staining images of the 5-d-old CycC1;1pro:GUS transgenic seedlings treated without or with 100 mM for 12 h. Bar = 0.5 cm.
To better understand the CycC1;1 expression pattern in Arabidopsis, we first measured the expression levels of CycC1;1 in different tissues and organs of the wild-type plants, including stem, root, seed, rosette leaf, flower, and silique, and found that CycC1;1 was highly expressed in root (Fig. 1G). To confirm this, we generated stable transgenic β-glucuronidase (GUS) reporter lines, CycC1;1pro:GUS, in which GUS expression was under the control of the CycC1;1 native promoter, enabling us to observe CycC1;1 expression levels by GUS staining. GUS activity was higher in the germinating seeds and seedlings than in the mature leaves, cauline leaves, flower, and siliques (Fig. 1H), suggesting that CycC1;1 is indeed highly expressed during the vegetative growth period, especially in the early stages. In addition to its expression pattern, we also examined the subcellular localization of CycC1;1 by observing the green fluorescence in the roots of 35Spro:CycC1;1-GFP transgenic plant seedlings (Fig. 1I). The location of CycC1;1-GFP fluorescence coincided well with the blue fluorescence resulting from staining with 4′,6-diamidino-2-phenylindole (DAPI), a fluorescent dye specific for nuclei. This indicated that CycC1;1 is a nuclear protein in Arabidopsis.
To determine how CycC1;1 responds to high salinity, we analyzed its expression levels in wild-type seedlings treated with or without high salinity. Our reverse transcription quantitative PCR (RT-qPCR) results showed that CycC1;1 expression was lower in the salt-treated plants than in the untreated control plants (Fig. 1J), implying that salt stress represses CycC1;1 expression. In agreement with this, when the CycC1;1pro:GUS seedlings were subjected to high salinity, both the GUS staining and activity were obviously reduced compared with the unstressed plants (Figs. 1K and S2), further supporting that salt stress suppresses CycC1;1 expression in Arabidopsis. Notably, CycC1;1 was highly expressed in the stele of the root elongation and mature zones based on the GUS staining results, while such staining was very much repressed by salt stress (Fig. 1K).
CycC1;1 negatively regulates salt-induced SOS1 expression
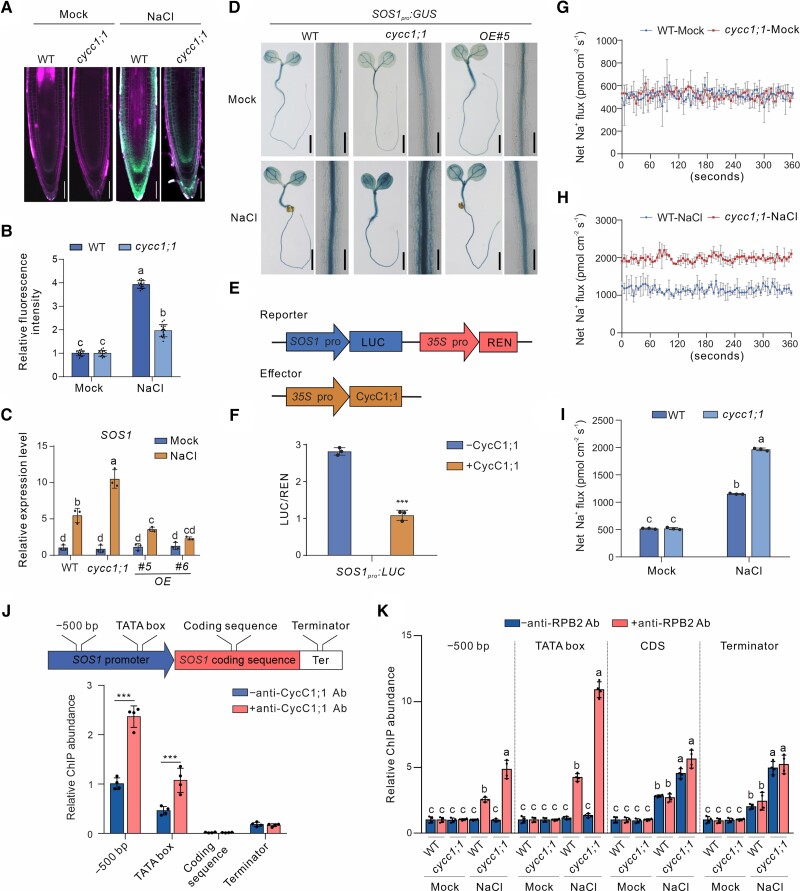
These results demonstrated that CycC1;1 negatively modulates salt tolerance, prompting us to analyze the Na+ content in roots of salt-treated cycc1;1 mutant plants. Using enhanced NaTrium Green-2 AM (ENG-2 AM), a sodium-specific fluorescent dye, our results showed that Na+ accumulation was higher in both the salt-treated wild-type and cycc1;1 mutant seedlings than in the untreated plants; nevertheless, the accumulation of Na+ was significantly lower in the mutant than in the wild-type when treated with high salinity (Fig. 2, A and B). Because the SOS pathway plays a predominant role in controlling plant salt stress tolerance by reducing Na+ overaccumulation, we then assayed the expression of SOS1, SOS2, and SOS3, three genes encoding core components of the SOS pathway in Arabidopsis, in the wild-type and cycc1;1 mutants. Our RT-qPCR results showed that the expression of these SOS genes was obviously induced by high salinity treatment in the wild-type, while such induction of the expression of SOS1 but not SOS2 or SOS3 was further enhanced in the cycc1;1 mutant (Figs. 2C and S3), suggesting that CycC1;1 greatly modulates the expression of SOS1, but not SOS2 or SOS3 expression. This finding was confirmed by results showing that salt-induced expression of SOS1, but not SOS2 or SOS3, was compromised in the CycC1;1-overexpression lines compared with the wild-type plants (Figs. 2C and S3). In addition, we generated stable SOS1pro:GUS transgenic reporter lines in the wild-type and cycc1;1 mutant backgrounds in which the expression of GUS was driven by the SOS1 promoter, enabling us to evaluate in fine detail the effect of CycC1;1 on salt-induced SOS1 expression. Consistent with the RT-qPCR results, GUS staining and activity were increased by the treatment of salt stress in SOS1pro:GUS plants, while such staining and activity were further enhanced in the cycc1;1 mutant but suppressed in the CycC1;1-overexpression line (Figs. 2D and S4). This further indicates that CycC1;1 negatively regulates salt-induced SOS1 expression in Arabidopsis. To further determine the role of CycC1;1 in the modulation of SOS1 expression, we performed dual-luciferase (LUC) reporter gene assays. With 35Spro:Renilla luciferase (REN) as an internal control, LUC driven by the SOS1 promoter as a reporter was coexpressed with 35Spro:CycC1;1-GFP as an effector in Nicotiana benthamiana leaves (Fig. 2E). Our results showed that the LUC activity was significantly repressed in the presence of CycC1;1 (Fig. 2F), revealing that CycC1;1 indeed plays a negative role in the regulation of SOS1 expression in Arabidopsis.
Figure 2.
CycC1;1 negatively regulates salt-induced SOS1 expression in plants. A, B) Sodium accumulation in wild-type and cycc1;1 mutant seedling roots. Five-day-old wild-type and cycc1;1 seedlings were treated with 100 mM NaCl for 3 h and then stained in a 10 μM ENG-2 AM solution containing 0.05% Pluronic F-127 for 3 h. Fluorescence images A) were taken, and the ENG-2 AM fluorescence intensity B) was analyzed. Bar = 50 μm. Data are means ± Sd of 3 independent repeats (n = 15). Bars with different letters indicate significant differences at P < 0.05, revealed using ANOVA with a Tukey's multiple comparison test (Supplemental Data Set 1). C) The expression of SOS1 in the wild-type and cycc1;1 mutant seedlings subjected to salt stress. Seven-day-old wild-type and cycc1;1 mutant seedlings treated with or without 100 mM NaCl for 12 h. Data are means ± Sd (n = 3). Bars with different letters indicate significant differences at P < 0.05, revealed using ANOVA with a Tukey's multiple comparison test (Supplemental Data Set 1). D) GUS staining images of 5-d-old transgenic SOS1pro:GUS in the wild-type, cycc1;1, and OE backgrounds treated with 0 mM or 100 mM NaCl for 12 h. Bar = 0.5 cm. E, F) LUC reporter gene assay to examine the effect of CycC1;1 on SOS1 expression. The schematic diagram E) shows the reporters and effectors used in the assay. The relative LUC intensity F) represents the SOS1pro:LUC activity relative to the internal control (REN driven by 35Spro). The activity of SOS1pro:LUC without CycC1;1 expression was set to 1. Data are means ± Sd (n = 3). Asterisks indicate significant differences determined by Student's t test (***P < 0.001). G to I) NMT showing Na+ fluxes. Ten-day-old wild-type and cycc1;1 mutant seedlings cultured in 1/2× MS liquid medium were treated with 0 mM G) or 150 mM NaCl H) for 5 h, and then the continuous transient Na+ fluxes were recorded for about 6 min. Each point is the mean of 4 individual plants. Quantitative analysis of the means of net Na+ fluxes within a continuous period of 0 to 6 min I). Data are means ± Sd (n = 3). Bars with different letters indicate significant differences at P < 0.05, revealed using ANOVA with a Tukey's multiple comparison test (Supplemental Data Set S1). J) A diagram showing the positions of SOS1 gene primers used for ChIP-qPCR is shown at the top. The ChIP-qPCR results showing the association of CycC1;1 with SOS1 is shown at the bottom. Chromatin was extracted from 7-d-old wild-type seedlings and then precipitated with either an anti-CycC1;1 antibody (+Ab) or only IgG (−Ab). Data are means ± Sd (n = 3). Asterisks indicate significant differences determined by Student's t test (***P < 0.001). K) Occupancy of RNAP II at the SOS1 promoter in the wild-type and cycc1;1 mutants. Chromatin was extracted from 7-d-old wild-type and cycc1;1 mutant seedlings and precipitated with an anti-RPB2 antibody (+RPB2) or only IgG (−RPB2). Data are means ± Sd (n = 4). Bars with different letters indicate significant differences at P < 0.05, determined using ANOVA with a Tukey's multiple comparison test.
SOS1, a Na+/H+ antiporter localized at the plasma membrane, protects plants under high salinity conditions by facilitating Na+ efflux from the cells and regulating Na+ transport from roots to shoots (Shi et al. 2002; Munns and Tester 2008; Deinlein et al. 2014; Van Zelm et al. 2020). Because the mutant had higher SOS1 expression than the wild-type, we speculated that Na+ efflux and/or long distance of Na+ transport was affected in the cycc1;1 mutant. To test this hypothesis, we first measured the Na+ efflux at the root apex using noninvasive microtest technology (NMT). Our results showed that in the absence of a high salinity treatment, both the wild-type and mutant roots had similar low net Na+ efflux, but following salt stress, the net Na+ efflux in both plants markedly increased and was much higher in the mutant than in the wild-type (Fig. 2, G to I). This result reveals that Na+ excretion is higher in the cycc1;1 mutant than in the wild-type under salt stress. In addition, we measured the Na+ content in the xylem sap from the wild-type and cycc1;1 mutant plants. We found that both the wild-type and cycc1;1 mutants had a similar low level of Na+ in xylem sap under normal conditions, whereas the Na+ content was significantly higher in the salt-treated wild-type than in the treated mutant plants (Supplemental Fig. S5), implying a role of CycC1;1 in the regulation of Na+ accumulation in the xylem transpirational stream under high salinity conditions. These results further support the role of CycC1;1 in salt tolerance via changes of SOS1 expression in plants.
Previous reports documented that CycC1;1 affects transcription through the action of Mediator associating with the promoter of target genes (Agrawal et al. 2021; Guo et al. 2022). We thus asked whether CycC1;1 could be associated with the SOS1 promoter. To this end, the wild-type plant seedlings and an anti-CycC1;1 antibody (Guo et al. 2022) were used for a chromatin immunoprecipitation (ChIP) experiment. We used the immunoprecipitated DNA as templates for qPCR with the primers designed to bind at different positions of SOS1 genomic DNA, including −500 bp from the translation start site (ATG), the TATA box (the RNAP II binding site), the coding region, and the terminator (Fig. 2J). The DNA fragments of the −500 bp upstream and TATA box but not the coding region or terminator were significantly enriched in CycC1;1 antibody-treated samples vs. antibody-lacking controls (Fig. 2J), implying that CycC1;1 is associated with the SOS1 promoter in vivo in Arabidopsis.
The Mediator complex acts as a bridge connecting RNAPII and gene transcription factors, thus affecting the expression of associated genes (Agrawal et al. 2021). Whether CycC1;1 association with the SOS1 promoter affects RNAPII binding to the SOS1 genomic DNA was unknown. To address this, wild-type and cycc1;1 mutant seedlings were used for ChIP-qPCR analysis using a specific antibody raised against the RNAP II subunit B2 (anti-RPB2). Our results showed that the amounts of RNAP II-associated SOS1 genomic DNA fragments, including the −500 bp upstream, TATA box, coding region, and terminator, were similar in the untreated wild-type and cycc1;1 mutant seedlings, whereas after salt treatment, increased DNA fragments were detected in seedlings of both genotypes, but much more in the mutant than in the wild-type (Fig. 2K). These results reveal that salt-induced RNAP II association with SOS1 is promoted in the cycc1;1 mutant, in agreement with the increased SOS1 expression and salt tolerance of the mutant.
Disruption of SOS1 abolishes salt stress tolerance of the cycc1;1 mutant
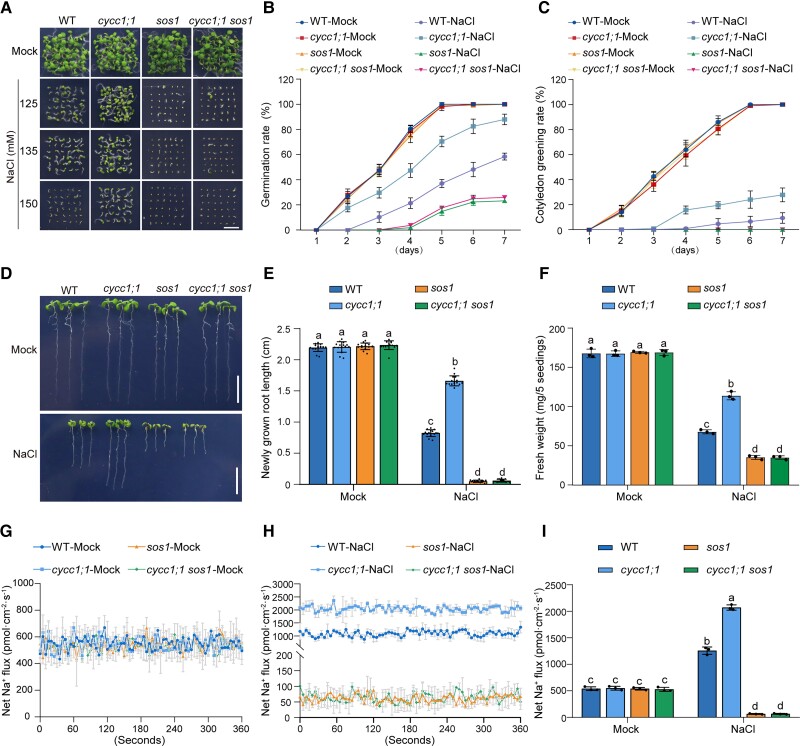
To analyze the genetic role of CycC1;1 in SOS1 expression for salt tolerance, we obtained a cycc1;1 sos1 double mutant by crossing cycc1;1 with the Col-0 background T-DNA insertion mutant of SOS1 (SALK_046400) and tested its salt stress tolerance. Similar to the above findings and a previous report (Shi et al. 2000), the cycc1;1 and sos1 mutants exhibited much lower and higher salt stress sensitivity than the wild-type, respectively, in terms of the germination and cotyledon greening rate (Fig. 3, A to C). By contrast, sensitivity of the cycc1;1 sos1 double mutant when subjected to high salinity was comparable to the sos1 mutant (Fig. 3, A to C), revealing that CycC1;1 acts upstream of SOS1 in plant salt sensitivity. Moreover, we also evaluated the salt tolerance of these plants by treating seedlings with high salinity and then assessing their primary root length and fresh weight. Our analysis results showed that the increased salt tolerance of the cycc1;1 mutant was completely compromised by the SOS1 mutation, as evidenced by similar primary short root lengths and low fresh weights in both the cycc1;1 sos1 double mutant and the sos1 mutant (Fig. 3, D to F). This further supports the idea that CycC1;1 acts upstream of SOS1 in plant salt stress tolerance.
Figure 3.
CycC1;1 affects plant salt tolerance through SOS1. A to C) Phenotypes A) of the wild-type, cycc1;1, sos1, and cycc1;1 sos1 plants grown on 1/2× MS medium supplemented with 0 mM, 125 mM, 135 mM, or 150 mM NaCl for 5 d. Quantitative analysis of seed germination B) and cotyledon greening rates C) of plants grown on 1/2× MS medium supplemented with 0 mM or 125 mM NaCl for 7 d. Data are means ± Sd of 3 independent experiments (n = 3). D to F) Root elongation and fresh weight analysis. Five-day-old wild-type, cycc1;1, sos1, and cycc1;1 sos1 plants were transferred to 1/2× MS medium supplemented with 0 mM or 125 mM NaCl for additional growth. The photographs were taken 5 d after transfer D). Bar = 1 cm. The lengths of newly grown roots E) and the fresh weights F) of the seedlings were also analyzed. Data are means ± Sd (n = 15 for root length and n = 3 for fresh weight). Bars with different letters indicate significant differences at P < 0.05, revealed using ANOVA with a Tukey's multiple comparison test (Supplemental Data Set 1). G to I) Net Na+ fluxes in root tips using NMT. Ten-day-old wild-type, cycc1;1, sos1, and cycc1;1 sos1 mutant seedlings cultured in 1/2× MS liquid medium were treated with 0 mM G) or 150 mM NaCl H) for 5 h, and then continuous transient Na+ fluxes were recorded for about 6 min. Each point is the mean of data from 4 individual plants. Quantitative analysis of the means of net Na+ fluxes within a continuous period of 0 to 6 min I). Data are means ± Sd (n = 3). Bars with different letters indicate significant differences at P < 0.05, revealed using ANOVA with a Tukey's multiple comparison test.
To verify the genetic relationship between CycC1;1 and SOS1, lower concentrations of NaCl were used to test the salt sensitivity of their single and double mutants. Consistent with the previous reports, the sos1 mutant was indeed very sensitive to salt stress, even though only 25 mM or 50 mM NaCl was used, while the cycc1;1 mutant was less sensitive to these salinity conditions than the wild-type in terms of seed germination, root length, and fresh weight (Supplemental Fig. S6). This clearly showed that the cycc1;1 sos1 double mutant displayed salt hypersensitive phenotypes similar to the sos1 single mutant under lower salt stress conditions (Supplemental Fig. S6). Consistent with this genetic evidence, the NMT analysis revealed that, upon salt stress treatment, the cycc1;1 and sos1 mutants had higher and lower net Na+ efflux than the wild-type, respectively, whereas the net Na+ efflux in salt-treated cycc1;1 sos1 double mutant was comparable to that in the treated sos1 mutant (Fig. 3, G to I). Moreover, the Na+ content in the xylem sap of the cycc1;1 mutant was similar to that in the cycc1;1 sos1 plant under either normal conditions or high salinity conditions (Supplemental Fig. S7). These data further show that the negative role of CycC1;1 in the regulation of salt tolerance is mainly dependent on SOS1 in Arabidopsis.
CycC1;1 interacts with WRKY75 for binding to the SOS1 promoter
Together, the above data demonstrated that CycC1;1 suppresses salt-induced SOS1 expression by interfering with RNAP II recruitment to the SOS1 promoter; however, the gene-specific transcription factors involved in CycC1;1-mediated repression of SOS1 expression remained unknown. To address the relationship between CycC1;1 and SOS1 transcription, we sought to identify transcription factors that not only can target the SOS1 promoter but also can interact with CycC1;1, thus providing a physical link between CycC1;1 and the SOS1 promoter. As a recent study reported that the transcription factor WRKY1 can bind to SOS1 promoter and activate its expression (Wu et al. 2022), we first examined whether CycC1;1 could interact with WRKY1 in a yeast 2-hybrid (Y2H) experiment. Regrettably, no interaction between CycC1;1 and WRKY1 was observed in yeast cells (Supplemental Fig. S8), suggesting that WRKY1 may not be involved in CycC1;1-mediated transcriptional regulation of SOS1.
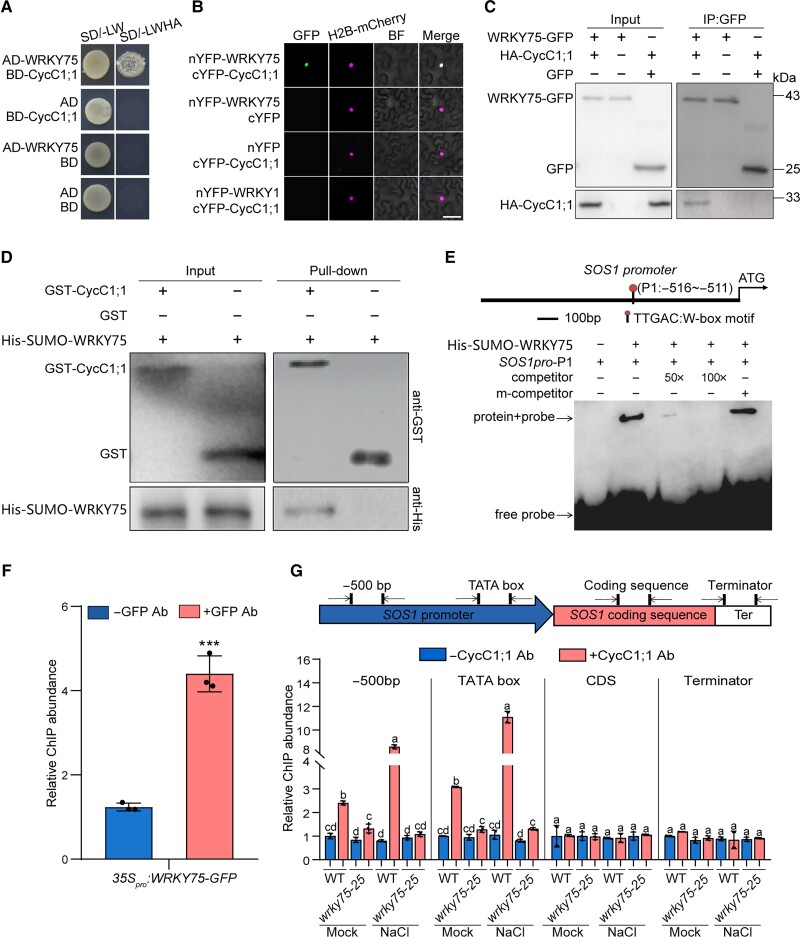
Therefore, we screened CycC1;1-interacting proteins by Y2H from a yeast library, and unexpectedly, another WRKY family member WRKY75 was identified as a possible CycC1;1-interacting partner (Fig. 4A). To confirm their interaction, we performed a bimolecular fluorescence complementation (BiFC) assay. When we coexpressed WRKY75 fused to the N-terminal half of yellow fluorescent protein (YFP) and CycC1;1 fused to the C-terminal half of YFP in N. benthamiana leaves, with H2B-RFP as a nuclear marker (Rosa et al. 2014), we observed reconstituted YFP fluorescence in the nucleus but not in the negative controls (Fig. 4B), indicating that CycC1;1 interacts with WRKY75 in planta. The interaction between CycC1;1 and WRKY75 was verified by a co-immunoprecipitation (Co-IP) experiment in which HA-tagged CycC1;1 and GFP-tagged WRKY75 were coexpressed in N. benthamiana leaves and immunoprecipitated using an anti-GFP antibody. Our results showed that HA-CycC1;1 could be specifically detected by an anti-HA antibody in the protein precipitants immunoprecipitated by the anti-GFP antibody (Fig. 4C), further supporting the previous result that CycC1;1 interacts with WRKY75 in vivo. To test their interaction in vitro, we purified glutathione S-transferase (GST)-tagged CycC1;1 and 6×His-tagged WRKY75 proteins that were expressed in Escherichia coli and performed a GST pull-down assay using Glutathione Sepharose beads. Our results showed that His-WRKY75 was specifically pulled down by GST-CycC1;1, but not by a construct containing the GST tag alone (Fig. 4D), clearly indicating that CycC1;1 has physical interaction with WRKY75 in vitro. These results reveal that CycC1;1 interacts with WRKY75 both in vivo and in vitro.
Figure 4.
CycC1;1 associates with SOS1 promoter by interacting with WRKY75. A) Interaction between WRKY75 and CycC1;1 examined by Y2H assay. Protein interactions were examined based on the growth of yeast cells on selective media. SD indicates synthetic dropout medium. −L/W indicates Leu and Trp dropout plates. −L/W/H/A indicates Trp, Leu, His, and Ade dropout plates. B) BiFC showing CycC1;1 interaction with WRKY75. nYFP-WRKY75 and cYFP-CycC1;1 were transiently coexpressed in N. benthamiana leaves, and YFP fluorescence was observed under confocal microscopy. H2B-mCherry (Rosa et al. 2014) was used as a nuclear marker. Bar = 20 μm. C) Interaction between WRKY75 and CycC1;1 assayed by Co-IP. GFP or GFP-tagged WRKY75 and HA-tagged CycC1;1 were transiently coexpressed in N. benthamiana leaves, and proteins were immunoprecipitated using anti-GFP antibody-conjugated agarose beads. The resulted precipitates were detected using anti-GFP and anti-HA antibodies, respectively. D) GST pull-down for the analysis of in vitro interaction between WRKY75 and CycC1;1. 6×His-SUMO-WRKY75 were mixed with GST-CycC1;1 or GST and immobilized on Glutathione Sepharose beads. After washing, the eluted proteins were subjected to immunoblot analysis with anti-GST or anti-His antibodies, respectively. E) The EMSA experiment showing that WRKY75 can bind to the SOS1 promoter in vitro. A diagram showing the SOS1 promoter has a typical W-box motif (−516∼−511 bp) recognized by WRKY75 transcription factor is shown at the top. Purified 6×His-SUMO-WRKY75 was incubated with biotin-labeled SOS1 promoter probes, and unlabeled probes (mutated or not mutated) were used as competitors. F) ChIP-qPCR showing that WRKY75 can bind to the SOS1 promoter in vivo. Chromatin was extracted from 7-d-old 35Spro:WRKY75-GFP transgenic seedlings and then precipitated with either an anti-GFP antibody (+Ab) or only IgG (−Ab). Data are means ± Sd (n = 3). Asterisks indicate significant differences determined by Student's t test (***P < 0.001). G) A diagram showing the positions of SOS1 gene primers used for ChIP-qPCR is shown at the top. The ChIP-qPCR results showing the changes of CycC1;1 association with SOS1 in the wild-type and wrky75-25 mutant are shown at the bottom. Chromatin was extracted from 7-d-old wild-type and wrky75-25 mutant seedlings and precipitated with anti-CycC1;1 antibody (+Ab) or only IgG (−Ab). Data are means ± Sd (n = 4). Bars with different letters indicate significant differences at P < 0.05, determined using ANOVA with a Tukey's multiple comparison test.
To investigate whether WRKY75 can target the SOS1 promoter, we analyzed the cis-elements of the SOS1 promoter and identified a typical W-box motif in the SOS1 promoter (Fig. 4E), raising the possibility that WRKY75 binds to the SOS1 promoter. As expected, results from an electrophoresis mobility shift assay (EMSA) revealed that 6×His-SUMO-tagged WRKY75 purified form E. coli bound to a biotin-labeled P1 probe containing the W-box of the SOS1 promoter (Fig. 4E). This binding was blocked by an unlabeled P1 probe but not by an unlabeled P1 probe with mutations in the W-box motif (Fig. 4E), indicating the specific binding of WRKY75 to SOS1 promoter. In addition, a 35Spro:WRKY75-GFP transgenic plant (Guo et al. 2017) and an anti-GFP antibody were used for ChIP-qPCR, and our results showed that the amounts of DNA fragments enriched by the anti-GFP antibody were much higher than those in the antibody-lacking control (Fig. 4F), further supporting that WRKY75 can bind to the SOS1 promoter in Arabidopsis.
To test whether WRKY75 links CycC1;1 and the SOS1 promoter in plants, we performed ChIP-qPCR by isolating CycC1;1-associated SOS1 DNA fragments from the wild-type and wrky75-25 mutant using an anti-CycC1;1 antibody. Our results showed that SOS1 promoter DNA fragments including −500 bp upstream and the TATA box were enriched in samples from the untreated wild-type plant, and high salinity treatment significantly increased such enrichment. However, this enrichment was greatly repressed in the wrky75-25 mutant (Fig. 4G), suggesting that CycC1;1 associates with SOS1 promoter in a WRKY75-dependent manner. Together, these results reveal CycC1;1 association with SOS1 promoter through its physical interaction with WRKY75.
WRKY75 confers plant salt tolerance by increasing SOS1 expression
Our results so far have revealed that WRKY75 can bind to the SOS1 promoter, leading us to further study its role in SOS1 transcription. Two T-DNA insertion mutant alleles of WRKY75, wrky75-1 and wrky75-25 (Fig. 5A; Chen et al. 2021; Guo et al. 2017), were identified and used to measure the expression of SOS1. In contrast to the cycc1;1 mutant, salt-induced SOS1 expression was greatly inhibited in both wrky75-1 and wrky75-25 mutants compared with that in the wild-type (Fig. 5B), revealing the involvement of WRKY75 in salt-induced SOS1 expression. To confirm this, we obtained SOS1pro:GUS wrky75-1 plants by crossing the SOS1pro:GUS line with the wrky75-1 mutant. We found that GUS staining and activity in the SOS1pro:GUS seedlings were highly induced by salt stress, whereas this induction was dampened in the SOS1pro:GUS wrky75-1 plant (Figs. 5C and S9), indicating that WRKY75 plays an important role in salt-induced SOS1 expression. In addition, using a dual LUC reporter gene assay in which both SOS1pro:LUC as a reporter and 35Spro:REN as an internal control were coexpressed with GFP or 35Spro:WRKY75-GFP as an effector in N. benthamiana leaves (Fig. 5D), we found that WRKY75-GFP, but not GFP alone, could activate LUC activity (Fig. 5D). These results demonstrate that WRKY75 can bind to the SOS1 promoter and activate its expression in response to high salinity. Consistent with the above findings, the wrky75 mutants and WRKY75-overexpression transgenic plants (Guo et al. 2017) displayed lower and higher tolerance to salt stress than the wild-type, respectively, in terms of germination and cotyledon greening rates, primary root length, and fresh weight (Fig. 5, E to J), indicating that WRKY75 is a positive regulator of salt stress tolerance in Arabidopsis. Consistently, Na+ accumulation in the wrk75-25 mutant root and xylem sap was significantly higher than in the wild-type plants when subjected to high salinity (Figs. 5, K and L, and S10).
Figure 5.
WRKY75 is required for salt-induced SOS1 expression. A) Expression of WRKY75 in 5-d-old wild-type, wrky75-1, and wrky75-25 mutant plants. Data are means ± Sd (n = 3). Bars with different letters indicate significant differences at P < 0.05, determined using ANOVA with a Tukey's multiple comparison test. B) Expression of SOS1 in 5-d-old wild-type, wrky75-1, and wrky75-25 mutant plants treated with 0 mM (Mock) or 100 mM NaCl for 12 h. Data are means ± Sd (n = 3). Bars with different letters indicate significant differences at P < 0.05, revealed using ANOVA with a Tukey's multiple comparison test. C) GUS staining images of 5-d-old SOS1pro:GUS and SOS1pro:GUS wrky75-1 seedlings treated with 0 mM or 100 mM NaCl for 12 h. Bar = 0.5 cm. D) LUC reporter gene assay showing WRKY75-mediated activation of SOS1 expression. The schematic diagrams at the top show the reporters and effectors used in the assay. The relative LUC intensity represents the SOS1pro:LUC activity relative to the internal control (REN driven by the 35S promoter). The activity of SOS1pro:LUC without WRKY75 expression was set to 1. Data are means ± Sd (n = 3). Asterisks indicate significant differences determined by Student's t test (**P < 0.01). E to G) Phenotypes E) of the wild-type, wrky75-1, wrky75-25, and 35Spro:WRKY75-GFP (WRKY75-OE) plants grown on 1/2× MS medium supplemented with 0 mM, 125 mM, 135 mM, or 150 mM NaCl for 5 d. Quantitative analysis of seed germination F) and cotyledon greening G) of plants grown on 1/2× MS medium supplemented with 0 mM or 125 mM NaCl for 7 d. Data are means ± Sd of 3 independent repeats. H to J) Root elongation and fresh weight analysis. Five-day-old wild-type, wrky75-1, wrky75-25, and WRKY75-OE plants were transferred to 1/2× MS medium supplemented with 0 mM or 125 mM NaCl for additional growth. The photographs in H) were taken 5 d after transfer. Bar = 1 cm. The length of newly grown roots I) and the fresh weight J) of the seedlings were also analyzed. Data are means ± Sd of 3 independent repeats (n = 15 for root length and n = 3 for fresh weight). Bars with different letters indicate significant differences at P < 0.05, determined using ANOVA with a Tukey's multiple comparison test. K, L) Sodium accumulation in wild-type, wrky75-25, and sos1 mutant seedling roots. Five-day-old plant seedlings were treated with 100 mM NaCl for 12 h and then stained by 10 μM ENG-2 AM solution containing 0.05% Pluronic F-127 for 3 h. Fluorescence images K) were taken, and relative ENG-2 AM fluorescence intensity L) was analyzed. Data are means ± Sd of 3 independent repeats (n = 15). Bars with different letters indicate significant differences at P < 0.05, revealed using ANOVA with a Tukey's multiple comparison test.
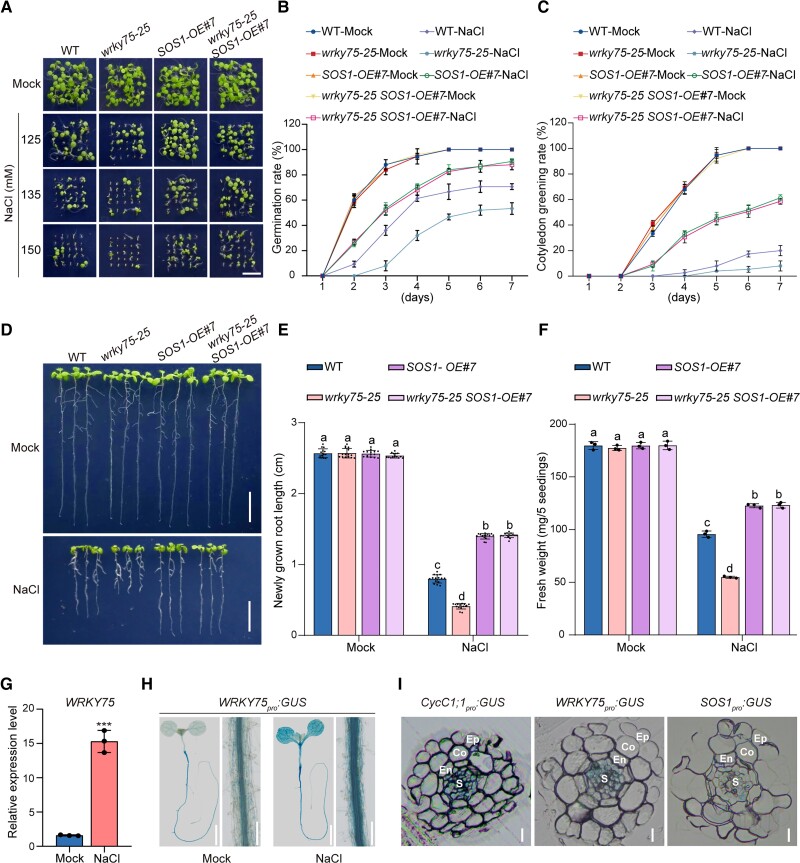
To provide genetic evidence supporting that WRKY75 confers salt tolerance by acting upstream of SOS1, we first obtained the SOS1-overexpression (SOS1-OE) transgenic line and then crossed it with the wrky75-25 mutant, producing wrky75-25 SOS1-OE plants. Our results showed that the SOS1-OE plants indeed had stronger tolerance to the salt stress than the wild-type, while the wrky75-25 SOS1-OE plant exhibited similar salt tolerance to the SOS1-OE line (Fig. 6, A to F), implying that the salt-sensitive phenotype of the wrky75-25 mutant could be rescued by the overexpression of SOS1.
Figure 6.
WRKY75 acts upstream of SOS1 in the regulation of plant salt stress tolerance. A to C) Phenotypes A) of the wild-type, wrky75-25, 35Spro:SOS1-GFP#7 (SOS1-OE#7), and wrky75-25 SOS1-OE#7 plants grown on 1/2× MS medium supplemented with 0 mM, 125 mM, 135 mM, or 150 mM NaCl for 5 d. Quantitative analysis of seed germination B) and cotyledon greening rates C) of plants grown on 1/2× MS medium supplemented with 0 mM or 125 mM NaCl for 7 d. Data are means ± Sd (n = 3). D to F) Root elongation and fresh weight analysis. Five-day-old wild-type, wrky75-25, 35Spro:SOS1-GFP#7 (SOS1-OE#7), and wrky75-25 SOS1-OE#7 plants were transferred to 1/2× MS medium supplemented with 0 mM or 125 mM NaCl for additional growth. The photographs were taken 5 d after transfer D). Bar = 1 cm. The lengths of newly grown roots E) and the fresh weights F) of the seedlings were also analyzed. Data are means ± Sd (n = 15 for root length and n = 3 for fresh weight). Bars with different letters indicate significant differences at P < 0.05, determined using ANOVA with a Tukey's multiple comparison test. G) The expression of WRKY75 in 5-d-old wild-type plants treated with 0 mM or 100 mM NaCl for 12 h. Data are means ± Sd (n = 3). Asterisks indicate significant differences determined by Student's t test (***P < 0.001). H) GUS staining images of 5-d-old WRKY75pro:GUS transgenic plant seedlings and longitudinal sections of roots of plants treated with 0 mM or 100 mM NaCl for 12 h. Bar = 0.5 cm. I) GUS staining images showing transverse sections of 7-d-old CycC1;1pro:GUS, WRKY75pro:GUS, and SOS1pro:GUS seedling roots. Ep, epidermis; Co, cortex; En, endodermis; S, stele. Bar = 10 μm.
To assess how WRKY75 responds to salt stress, we first assayed its transcription levels in the wild-type seedlings treated with or without high salinity and showed that WRKY75 expression was significantly higher in salt-treated plants than in the untreated plant (Fig. 6G). We also obtained a previously reported WRKY75pro:GUS line (Guo et al. 2017) and subjected its seedlings to high salinity treatment. We found that the GUS was highly active in seedling roots, while salt treatment further increased such activity in both root and shoot (Figs. 6H and S11). These results clearly indicate that salt stress induces WRKY75 expression in Arabidopsis. We also noticed that WRKY75 was highly expressed in the stele of root elongation and mature zones, based on the GUS staining experiment (Fig. 6H), and that this pattern likely overlapped with the staining pattern of CycC1;1pro:GUS and SOS1pro:GUS seedling roots (Figs. 1K and 2D). To exactly study their expression pattern in roots, transverse sections of the GUS-stained roots from the three GUS reporter lines were analyzed under a microscope, revealing that CycC1;1, WRKY75, and SOS1 were highly expressed in the root stele parenchyma cells (Fig. 6I).
CycC1;1 interferes with WRKY75 transcriptional activation activity for SOS1
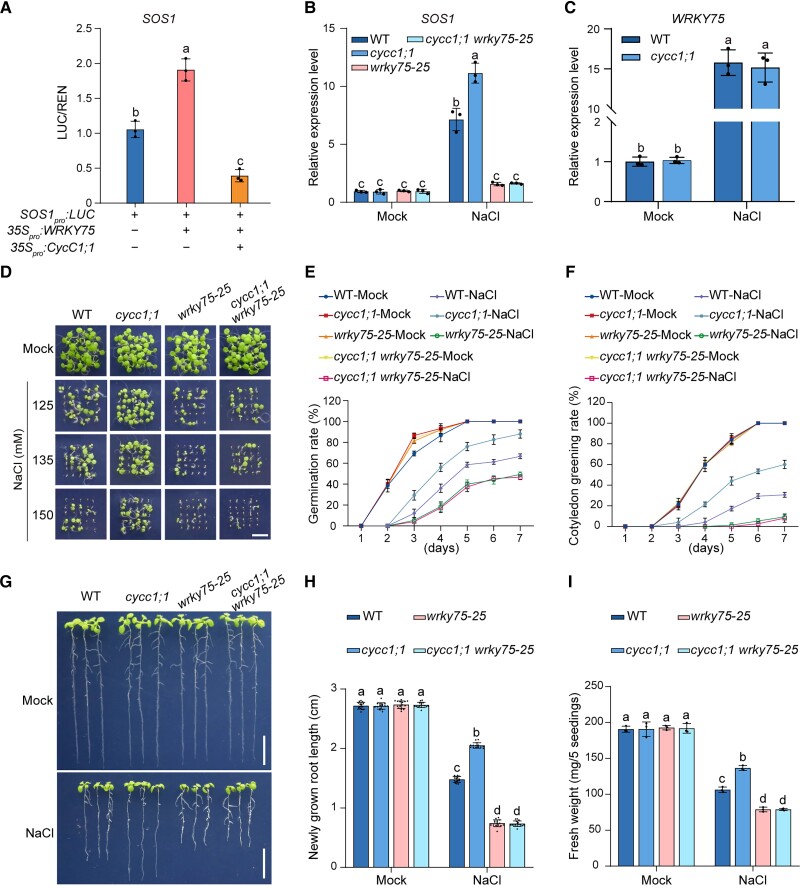
We further asked whether CycC1;1 involves WRKY75-mediated transcriptional regulation of SOS1 through their interaction. We performed dual LUC reporter gene assays, in which SOS1pro:LUC as a reporter and 35Spro:WRKY75-GFP as an effector were coexpressed with or without CycC1;1 in N. benthamiana leaves. Our results showed that LUC activity was activated when WRKY75-GFP was expressed, whereas WRKY75-promoted LUC activity was significantly suppressed in the presence of CycC1;1 (Fig. 7A). This revealed that CycC1;1 interferes with WRKY75-mediated transcriptional activation of SOS1. Therefore, mutation of WRKY75 should dampen both higher SOS1 expression and salt tolerance of the cycc1;1 mutant. To test this, we crossed the cycc1;1 mutant with the wrky75-25 mutant, producing a cycc1;1 wrky75-25 double mutant. The cycc1;1 wrky75-25 double mutant had lower SOS1 expression in the presence of high salinity, a level similar to that in the wrky75-25 mutant (Fig. 7B). In addition, we analyzed the expression levels of WRKY75 in the wild-type and cycc1;1 mutant seedlings and found that WRKY75 expression in the wild-type was similar to that in the cycc1;1 mutant, either in the presence or absence of high salinity (Fig. 7C), revealing that CycC1;1 does not affect WRKY75 expression in response to salt stress. Furthermore, the cycc1;1 wrky75-25 double mutant displayed a salt-sensitive phenotype similar to that of the wrky75-25 mutant regarding seed germination, cotyledon greening, primary root length, and fresh weight (Fig. 7, D to I), results consistent with measurements of the Na+ content in the xylem sap of the wild-type and mutant plants treated with high salinity (Supplemental Fig. S12). These results indicate that CycC1;1 interferes WRKY75-mediated transcriptional activation of SOS1 in plant response to high salinity.
Figure 7.
CycC1;1 interferes WRKY75-mediated transcriptional activation for SOS1. A) LUC reporter gene assay showing the effect of CycC1;1 on WRKY75-mediated activation of SOS1 expression. The relative LUC intensity represents the SOS1pro:LUC activity relative to the internal control (REN driven by the 35S promoter). The activity of SOS1pro:LUC without WRKY75 expression was set to 1. Data are means ± Sd (n = 3). B) The expression of SOS1 in 5-d-old wild-type, cycc1;1, wrky75-25, and cycc1;1 wrky75-25 mutant seedlings treated with 0 mM or 100 mM NaCl for 6 h. Data are means ± Sd (n = 3). Bars with different letters indicate significant differences at P < 0.05, determined using ANOVA with a Tukey's multiple comparison test. C) The expression of WRKY75 in 5-d-old wild-type and cycc1;1 mutant seedlings treated without or with 100 mM NaCl for 12 h. Data are means ± Sd (n = 3). ns, no significant differences. D to F) Phenotypes D) of the wild-type, cycc1;1, wrky75-25, and cycc1;1 wrky75-25 mutant plants grown on 1/2× MS medium supplemented with 0 mM, 125 mM, 135 mM, or 150 mM NaCl for 5 d. Quantitative analysis of seed germination E) and cotyledon greening rates F) of plants grown on 1/2× MS medium supplemented with 0 mM or 125 mM NaCl for 7 d. Data are means ± Sd (n = 3). G to I) Root elongation and fresh weight analysis. Five-day-old wild-type, cycc1;1, wrky75-25, and cycc1;1 wrky75-25 mutant plants were transferred to 1/2× MS medium supplemented with 0 mM or 125 mM NaCl for additional growth. The photographs were taken 5 d after transfer G). Bar = 1 cm. The lengths of newly grown roots H) and the fresh weights I) of the seedlings were also analyzed. Data are means ± Sd (n = 15 for root length and n = 3 for fresh weight). Bars with different letters indicate significant differences at P < 0.05, determined using ANOVA with a Tukey's multiple comparison test.
Discussion
Due to their sessile lifestyle, plants cannot avoid adverse environments such as high salinity by changing location; thus, they must precisely perceive and respond to environmental stresses to survive. SOS1 plays a critical role in salt tolerance in Arabidopsis by pumping excess Na+ out of the cell (Zhou et al. 2018; Chai et al. 2020). Numerous reports have documented that SOS1 expression is induced by salt stress (Shi et al. 2000, 2002; Wu et al. 2022), but the underlying mechanism remains unclear. Here, we report that CycC1;1 negatively regulates plant salt tolerance by interfering with WRKY75-mediated transcriptional activation of SOS1. Biochemical and genetic analyses revealed that CycC1;1 negatively regulates SOS1 expression and salt tolerance by interacting with WRKY75 to interfere with the association of RNAP II with the SOS1 promoter. Thus, our study reveals the critical role of CycC1;1 as a component of the Mediator complex in linking gene-specific transcription factor WRKY75 and RNAP II during the dynamic regulation of SOS1 transcription in plants. We propose a model illustrating this in Fig. 8.
Figure 8.
A proposed model showing the mechanism of CycC1;1–WRKY75 complex-mediated transcriptional regulation of SOS1 in response to different salinity conditions in Arabidopsis. Under the normal condition, CycC1;1 interacts with WRKY75 to form a transcriptional repression complex that inactivates SOS1 expression by interfering RNAP II occupancy on the promoter of SOS1 in WT seedlings. When plants are subjected to high salinity stress, the expression of CycC1;1 is suppressed while WRKY75 expression is stimulated, leading to increased recruitment of RNAP II to the SOS1 promoter, thereby activating SOS1 expression and enhancing salt tolerance in WT seedlings. When CycC1;1 is disrupted in the cycc1;1 mutant, WRKY75 transcriptional activation of SOS1 is further enhanced under high salinity conditions, thus leading to higher salt stress tolerance in the mutant than in the WT. In contrast to the cycc1;1 mutant, the wrky75 mutant has impaired salt-induced SOS1 transcription and thus attenuated salt stress tolerance. WT, wild-type; PM, plasma membrane; cycc1;1, CycC1;1 loss-of-function mutant; wrky75, WRKY75 loss-of-function mutant.
It has been widely accepted that activation of SOS1 at the posttranslational level is important for the plant salt stress response, and several components key to this response have been identified, especially the SOS2–SOS3 kinase complex (Qiu et al. 2002; Quintero et al. 2002, 2011). In fact, a potential role of SOS1 transcription in plant salt tolerance was proposed when this gene was initially cloned and analyzed, as salt stress could highly induce SOS1 expression in both plant roots and shoots (Shi et al. 2000, 2002). This finding was later confirmed by numerous reports, and further work supports the key role of SOS1 transcription in plant salt tolerance based on the fact that overexpression of Arabidopsis SOS1 or its homologous genes from other plant species, such as Crossostephium chinense, Artemisia japonica, and Chrysanthemum, can significantly enhance salt tolerance in plants (Shi et al. 2002; Yang et al. 2009; Gao et al. 2016). These previous reports, as well as this study, show that dynamic regulation of SOS1 activity is likely achieved by precise transcriptional as well as posttranslational regulation of SOS1 during the response of plants to different salinity conditions.
Our study showed that CycC1;1 forms a transcriptional repression complex with WRKY75 via their physical interaction and that this complex hinders the transcription of SOS1 by interfering with RNAP II recruitment to the SOS1 promoter when plants are grown under normal conditions. The relief of this transcriptional repression of SOS1 is at least partially caused by decreased CycC1;1 expression, but increased WRKY75 expression, under high salinity conditions, thereby leading to increased SOS1 transcription and salt tolerance in plants. Thus, we infer that the protein complex formed by CycC1;1 and WRKY75 and its transcriptional regulation of SOS1 is dynamically affected by the different expression levels of CycC1;1 and WRKY75 in plant response to different salinity conditions.
Nevertheless, many more biochemical experiments must be performed in the future to further elucidate whether and how the CycC1;1–WRKY75 complex is associated with other components of the Mediator complex. It will also be important to determine their distinct status in plant responses to different salinity conditions. In addition to the impacts of differential CycC1;1 and WRKY75 expression on the CycC1;1–WRKY75 complex in plants when challenged by salinity, we cannot rule out another possibility that salt stress may abolish formation of the CycC1;1–WRKY75 complex to upregulate SOS1 transcription by dampening their interaction. Although it is difficult to directly investigate the changes of CycC1;1–WRKY75 interaction in planta, it is certainly worth exploring the role of salinity on the interaction between CycC1;1 and WRKY75 and further investigating these effects on the transcriptional activation of SOS1 and plant salt stress tolerance.
It is also worth noting that CycC1;1 association with the SOS1 promoter was nearly completely abolished when WRKY75 expression was largely disrupted in the wrky75 mutant, implying that CycC1;1 associates with SOS1 in a WRKY75-dependent manner in planta. This is consistent with the current model that the Mediator complex functions in gene transcription as a bridge connecting transcription factors with RNAP II. Whether CycC1;1 can repress SOS1 expression by inhibiting WRKY75 binding to the SOS1 promoter in addition to interfering WRKY75 transcriptional activation activity remains to be further elucidated and is worthy of further experimental exploration.
SOS1 is specifically expressed in the xylem parenchyma cells and the epidermis of plant roots (Shi et al. 2002; Chung et al. 2008). In our study, we generated a GUS reporter line where the GUS activity is under the control of the SOS1 native promoter. This enabled us to observe the changes of SOS1 expression patterns in roots of plants of in the wild-type, cycc1;1, and wrky75-25 mutant backgrounds. Our results showed that SOS1 is indeed specifically expressed in epidermal and xylem parenchyma cells (Fig. 6I), consistent with the previous report (Shi et al. 2002). Our biochemical and genetic analyses revealed that WRKY75 functions in salt-induced SOS1 expression by directly binding to the W-box in the SOS1 promoter. Indeed, the GUS staining and activity of SOS1pro:GUS were lower in the wrky75-25 mutant roots than in the wild-type roots (Figs. 6I and S11), especially in the stele of the seedling roots. This further supports the positive role of WRKY75 in the regulation of SOS1 expression in xylem parenchyma cells. However, observations using the WRKY75pro:GUS reporter line showed obvious GUS activity only in the stele, but not in the epidermis in roots (Guo et al. 2017), demonstrating that WRKY75 is specifically expressed in the stele. Since a previous report confirmed that WRKY75 RNA or protein can move into the epidermal cells from the cells where it is transcribed (Rishmawi et al. 2014), we infer that salt stress increases WRKY75 transcription in stele and then WRKY75 moves into the epidermis to promote SOS1 expression, thus conferring plant enhanced salt tolerance. By contrast, under normal conditions, WRKY75-regulated SOS1 expression is inhibited by CycC1;1 through its interaction, as CycC1;1 is widely distributed in the whole root tissues, including epidermal and xylem parenchyma cells (Fig. 6I). Investigation of these complex and dynamic expression patterns of CycC1;1–WRKY75 signaling should contribute to our understanding of the precise regulation of SOS1 expression in a proper plant response to low or high salt conditions.
In addition to the expression in roots, our GUS staining experiments using the SOS1pro:GUS reporter line revealed that SOS1 is also expressed in shoots (Fig. 2D), consistent with previous reports and the prediction that SOS1 functions to expel Na+ from the xylem parenchyma cells into the apoplastic space of mesophyll cells in leaves (Shi et al. 2002; Zhu et al. 2016). Interestingly, salt-induced GUS staining in shoots was also increased in the cycc1;1 mutant but decreased in the wrky75-25 mutant (Figs. 2D and 5C), implying a similar regulatory role of CycC1;1 and WRKY75 in SOS1 expression in both shoots and roots. How shoot-expressed SOS1 functions in plant salt tolerance remains unclear and is worthy of experimental exploration, which will provide more insights into the role of SOS1 in long-distance transport of Na+ in plants.
Emerging evidence has revealed that many subunits of the Mediator complex participate in plant growth, development, and stress responses (Zhu et al. 2014; Guo et al. 2021). It is reported that the Mediator complex tail module can interact with different transcription factors to affect gene expression. For example, MED25 interacts with MYC2 and ABSCISIC ACID INSENSITIVE5 (ABI5), 2 master transcription factors in JA and ABA signaling pathways, respectively, thus differentially regulating plant responses to JA and ABA (Chen et al. 2012). In addition to the tail module, the kinase module can also interact with transcription factors to modulate the expression of associated genes. CDK8, a subunit of the kinase module, regulates plant immunity and drought stress tolerance by interacting with the transcription factor WAX INDUCER1 and ERF/AP2 transcription factor RAP2.6, respectively (Zhu et al. 2014; Zhu et al. 2020). Unlike the well-studied CDK8 subunit in the kinase module, our knowledge of CycC regulation of plant stress responses is poor as is its role in resistance to necrotrophic pathogens in Arabidopsis (Zhu et al. 2020). This study showed that CycC1;1 plays a critical role in plant salt tolerance by interacting with and inhibiting WRKY75, a key transcription factor positively regulating SOS1, shedding light on how CycC1;1 is involved in the plant salt stress response and tolerance. However, we have noticed that although CDK8 promotes drought tolerance by increasing the expression of RAP2.6-targeted genes such as RD29A and COR15A, as documented in a previous study (Zhu et al. 2020), CycC1;1 suppresses ABA signaling and the expression of ABA-responsive genes as shown in our recent report (Guo et al. 2022). Also, the cdk8 and cycc1 mutants exhibit similar susceptibility to the necrotrophic pathogen Alternaria brassicicola but show different resistance against Botrytis cinerea, another necrotrophic pathogen (Zhu et al. 2014). However, flowering and a prolonged reproductive phase are similarly delayed in both mutants of CDK8 and CycC in pea (Pisum sativum L.) (Hasan et al. 2020). Based on these findings that different subunits of Mediator complex exert similar or different effects on plant growth and stress responses, we hypothesize that CycC elicits its role in these processes in both CDK8-dependent and CDK8-independent manners. However, it is still important to perform additional experiments to investigate the role of CDK8 in the plant response to salt stress and possible relationships with CycC1;1.
In summary, our study reveals how the CycC1;1–WRKY75 signaling module is a key component of the precise and dynamic regulation of SOS1 expression and elucidates the underlying mechanism by which CycC1;1 interferes WRKY75-mediated transcriptional activation of SOS1 under normal conditions. It also reveals how CycC1;1 expression is suppressed but WRKY75 expression is upregulated to promote SOS1 expression in Arabidopsis, leading to enhanced salt tolerance under salt stress conditions (Fig. 8).
Materials and methods
Plant materials and growth conditions
Arabidopsis (A. thaliana) ecotype Columbia-0 was used as the WT. The cycc1;1 (SALK_053291) and sos1 (SALK_046400) mutants were obtained from the Arabidopsis Biological Resource Center. The complementation (Com) lines of the cycc1;1 mutant and CycC1;1-overexpressing transgenic plants (OE) were generated in our previous report (Guo et al. 2022). The wrky75-1, wrky75-25 (Guo et al. 2017; Zhang et al. 2018), and WRKY75pro:GUS (Guo et al. 2017) seeds were previously reported and verified by genomic DNA PCR. The cycc1;1 sos1 and cycc1;1 wrky75-25 double mutants were obtained by genetic crossing. The transgenic plants CycC1;1pro:GUS, 35Spro:SOS1 (SOS1-OE), and SOS1pro:GUS were generated as described below in methods. Subsequently, the SOS1pro:GUS cycc1;1, SOS1pro:GUS CycC1;1-OE, SOS1pro:GUS wrky75-1, and wrky75-25 SOS1-OE plants were obtained by genetic crossing.
Seeds were surface sterilized using 10% (w/v) sodium hypochlorite (NaClO) for 5 min, washed 5 times with sterile water, and sown on a 1/2× MS medium (pH 5.8, containing 1% [w/v] sucrose and 0.8% [w/v] agar). Plates were kept at 4 °C for 3 d, and then seeds were germinated and grown under a 16-/8-h light/dark photoperiod at 100 μmol m−2 s−1 at 22 °C.
Construction of transgenic plants
To generate the CycC1;1pro:GUS transgenic reporter line, the genomic sequence containing 1,030 bp upstream of the CycC1;1 translation start codon (ATG) was amplified and cloned into pCambia1381 at the EcoRI site according to our previous report (Wang et al. 2022). The resulting plasmid was introduced into the wild-type plant via Agrobacterium tumefaciens-mediated floral transformation using the floral dip method (Clough and Bent 1998). Com T4 transgenic plants were used for GUS staining analysis.
To generate SOS1-overexpression lines, the full-length coding sequence (CDS) of SOS1 was cloned into pCambia1300 at the SalI site under the control of the cauliflower mosaic virus 35S promoter, and the resulting plasmid was used to transform the wild-type Arabidopsis plants by A. tumefaciens-mediated transformation as described above. At least 8 T4 homozygous 35Spro:SOS1-GFP transgenic lines were selected, and one of them (SOS1-OE #7) with higher SOS1 transcription and salt tolerance than the wild-type was used for our study. The wrky75-25 SOS1-OE line was obtained by genetic crossing wrky75-25 with the selected SOS1-OE #7 transgenic plant.
To generate the SOS1pro:GUS transgenic reporter line, the genomic sequence containing 1,386 bp upstream of the SOS1 translation start codon (ATG) was amplified and cloned into pCambia1381 at the BamHI site. The resulting plasmid was introduced into the wild-type plant via A. tumefaciens-mediated floral transformation. T4 transgenic plants were used for GUS staining analysis. The SOS1pro:GUS cycc1;1, SOS1pro:GUS CycC1;1-OE, and SOS1pro:GUS wrky75-25 plants were obtained by genetic crossing the SOS1pro:GUS line with the cycc1;1, CycC1;1-OE#5, and wrky75-25 plants, respectively. The primers used for genotyping, plasmid construction, and RT-qPCR are listed in Supplemental Table S1.
Analysis of seed germination and cotyledon greening rates
Mature Arabidopsis seeds were harvested and dried at room temperature for 3 wk and then used for the germination assays. Sterilized seeds were plated on 1/2× MS medium without or with the indicated concentrations of NaCl. Following stratification at 4 °C for 3 d in the dark, the plates were transferred to a growth chamber with a 16-h light/8-h dark cycle at 100 μmol m−2 s−1 at 23 °C. The germination and cotyledon greening rates were analyzed from the first day to the seventh day after the plates were transferred to the light.
Analysis of root elongation and fresh weight
Five-day-old wild-type, mutant, or overexpression transgenic seedlings grown on 1/2× MS medium were transferred to 1/2× MS containing 0 mM or 125 mM NaCl for another 5 d, and then lengths of the newly grown roots were measured using ImageJ (https://imagej.nih.gov/ij/). At least 15 seedling roots were analyzed per group for each experiment. For the fresh weight analysis, a group of 5 seedlings was used as 1 biological sample, and at least 3 biological replicates were used for the fresh weight analysis.
RT-qPCR analysis
Tissues from treated or untreated wild-type, mutant, or overexpression transgenic seedlings were collected for total RNA isolation, first-strand cDNA synthesis, and RT-qPCR as we described previously (Wang et al. 2022). The constitutively expressed ACTIN2 gene was used as an internal control. The primers used for RT-qPCR are listed in Supplemental Table S1.
Measurement of net Na+ flux using NMT
The wild-type and mutant seedlings were grown on 1/2× MS medium for 10 d, incubated in liquid medium or 150 mM NaCl for 5 h, and then subjected to measurement of Na+ fluxes using NMT (NMT150; YoungerUSA, Amherst, MA, USA; Xuyue [Beijing] Sci. & Tech., Beijing, China) according to the previous reported method (Lou et al. 2020). The continuous transient Na+ fluxes were recorded for about 6 min. Each point is the mean of 4 individual plants.
Determination of sodium content by ENG-2 AM staining
The sodium-specific fluorescent dye, enhanced NaTrium Green-2 AM (ENG-2 AM, MX4514, Shanghai Maokang Biotechnology, Shanghai, China), was used to determine sodium accumulation in roots. Five-day-old plant seedlings grown on 1/2× MS medium were treated with or without 100 mM NaCl for 12 h, and then the seedlings were stained with a 10 μM ENG-2 AM solution containing 0.05% Pluronic F-127 (MS4301, Shanghai Maokang Biotechnology, Shanghai, China) for 3 h. The Pluronic F-127 is a nonionic surfactant that can promote the dissolution capacity of the fluorescent dye ENG-2 AM. The seedlings were stained with 20 μg/mL propidium iodide (PI) solution for 3 min, after which the green fluorescence of ENG-2 AM (488 nm excitation, 505 to 530 nm emission, and 5% laser intensity) and the red fluorescence of PI (514 nm excitation, 575 to 615 nm emission, and 2% laser intensity) were detected using a confocal laser scanning microscope equipped with Plan-Apochromat 20×/0.8 objective (ZEISS LSM 980, Zeiss, https://www.zeiss.com). The fluorescence intensities were analyzed using ImageJ. At least 15 seedling roots were analyzed per group for each experiment.
Determination of Na+ content in xylem sap
Determination of Na+ accumulation in xylem sap was performed according to a previously reported method (Shi et al. 2002). Briefly, plants were grown in soil for 3 wk in a growth chamber under a 16-/8-h light/dark photoperiod at 100 μmol m−2 s−1 at 22 °C and then irrigated with water or 100 mM NaCl solution for 1 d. The plants were then kept in a chamber with 100% humidity, and their rosette leaves and inflorescence stems were cut at the base. The water droplet on the cut surface of the inflorescence stem was collected, and the Na+ content of this xylem sap was analyzed using inductively coupled plasma-optical emission spectrometer (ICP-OES, Agilent 5110).
Y2H assays
pGBKT7-CycC1;1 was constructed in our previous report (Guo et al. 2022). The full-length CDSs of WRKY1 and WRKY75 were cloned into pGADT7 (Clontech) at the BamHI site, respectively. Yeast transformation and growth were carried out using the Matchmaker system (Clontech) according to the manufacturer’s protocols. Yeast transformants were selected on double dropout medium lacking Leu and Trp (−LW), and protein interactions were analyzed on quadruple dropout medium lacking Leu, Trp, His, and Ade (−LWHA). Primer sequences are listed in Supplemental Table S1.
BiFC assay
The CDSs of CycC1;1, WRKY75, and WRKY1 were cloned into the pSPYCE or pSPYNE vector (Walter et al. 2004) containing the C-terminal of YFP (cYFP) and or the N-terminal of YFP (nYFP), respectively. The nuclear marker H2B-mCherry (Rosa et al. 2014), nYFP-WRKY75 (N-terminal tag), and cYFP-CycC1;1 (C-terminal tag) were coexpressed in N. benthamiana leaves for 3 d, after which YFP (excitation 488 nm and emission 520 to 560 nm) and mCherry (excitation 561 nm and emission 600 to 630 nm) fluorescence signals were detected using a laser scanning confocal microscope (Zeiss LSM980, Zeiss, Germany). Primer sequences are listed in Supplemental Table S1.
Co-IP assays
To perform Co-IP assays, the CDS of WRKY75 was cloned into pCambia1300 at the SalI site, including sequences encoding a GFP tag fused to its C-terminus and under the control of the 35S promoter. The Co-IP experiment was performed according to the previously reported methods with some modifications (Nie et al. 2022; Liu et al. 2022a). Briefly, GFP-tagged WRKY75 and HA-tagged CycC1;1 were transformed into Agrobacterium GV3101 and infiltrated into 3-wk-old N. benthamiana leaves. After 3 d, total proteins were extracted from the infected leaves by homogenization in IP buffer (150 mM NaCl, 25 mM Tris-HCl, pH 7.5, 0.2% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktail [Roche]) and then immunoprecipitated by an anti-GFP antibody (ABclonal, AE012, diluted 1:1,000). The resulted precipitates were resuspended and detected using an anti-GFP (ABclonal, AE012) and anti-HA (Sungene Biotech, KM8004) antibodies, respectively.
EMSA assay
The CDS of WRKY75 was cloned into pPSUMO (pET28a-6×His-SUMO, Wang et al. 2022) at the BamHI site, where sequences encoding a 6×His-SUMO tag allowed fusion to its N-terminus. The resulted pPSUMO-His-WRKY75 vector was transformed and induced in the E. coli BL21 (DE3) strain, and then the 6×His-SUMO-tagged WRKY75 protein was purified using NTA-Ni beads (Ni-NTA Purose 6 Fast Flow, A41002-06, Qianchun Bio, Jiaxing, China) according to our previous report (Wang et al. 2022). The EMSA assay was performed using a Light Shift Chemiluminescent EMSA kit (Thermo Fisher Scientific), according to the manufacturer's instructions. Briefly, biotin-labeled probes were incubated with or without purified 6×His-WRKY75 in binding buffer (2.5% glycerol, 50 mM KCl, 5 mM MgCl2, and 10 mM EDTA) for 30 min at room temperature. For the EMSA competition experiments, unlabeled unmutated and mutated probes used as competitors were added to the binding reactions, respectively. The probe sequences are listed in Supplemental Table S1.
GST pull-down assay
The CDSs of full-length WRKY75 and CycC1;1 were cloned into the pPSUMO (6×His-SUMO) and pGEX4T-1 vectors, respectively. The GST pull-down assay was performed according to our previous reports (Guo et al. 2022; Liu et al. 2022a). Briefly, GST-CycC1;1 and 6×His-SUMO-WRKY75 were expressed and purified from E. coli. 6×His-SUMO-WRKY75 was mixed with GST-CycC1;1 or GST alone on ice for 1 h and then incubated with GST-Sefinose Resin 4FF (Settled Resin) (Sangon Biotech, C600031) at 4 °C for 3 h. After washing, the eluted protein with Elution Buffer (10 mM GSH in 50 mM Tris-HCl, pH 8.0) was detected with anti-His (EnoGene, #E12-004-3) and anti-GST (ABclonal, AE006) antibodies, respectively. Primer sequences are listed in Supplemental Table S1.
LUC reporter gene assay
The LUC reporter gene assay was performed according to previous reports (Liu et al. 2022b). The SOS1 promoter was cloned into pGreen0800 (Zhang et al. 2020) at the BamHI site, and LUC expression was controlled by SOS1 promoter activity. The resulted reporter and the internal control (35Spro:REN in pGreen0800 vector) were transiently coexpressed with or without the effector pEGAD-CycC1;1 (Guo et al. 2022) in N. benthamiana leaves for 3 d, and then the activities of LUC and REN were detected using a Multimode Reader Platform (Tecan Spark, Tecan Group Ltd., Zurich, Switzerland) according to our previous reports (Guo et al. 2022; Liu et al. 2022b). Primer sequences are listed in Supplemental Table S1.
ChIP-qPCR analysis
To test the association of CycC1;1 with SOS1 genomic DNA, 7-d-old wild-type and wrky75-25 mutant seedlings were used for ChIP assays according to a previously reported method (Guo et al. 2022). The chromatin was immunoprecipitated from the wild-type plants with or without the anti-CycC1;1 antibody (Guo et al. 2022), and then both the input and the immunoprecipitated DNA fragments were quantified by qPCR with specific primers. ACTIN7 was used as a reference gene. The primers used for ChIP-qPCR are listed in Supplemental Table S1.
To assay the binding of WRKY75 to the SOS1 promoter, chromatin was extracted from 7-d-old 35Spro:WRKY75-GFP (Guo et al. 2017) seedlings and precipitated using GFP antibody-conjugated agarose beads (ABclonal, AE074). DNA fragments in both input and immunoprecipitated samples were quantified by qPCR. ACTIN7 was used as a reference gene. The primers used for ChIP-qPCR are listed in Supplemental Table S1.
To examine the effect of CycC1;1 on RNAP II recruitment to the SOS1 promoter, chromatin was extracted from 7-d-old wild-type and cycc1;1 mutant seedlings and precipitated with an anti-RPB2 antibody (ABclonal, A5928, diluted 1:1,000). DNA fragments in both input and immunoprecipitated samples were quantified by qPCR. ACTIN7 was used as a reference gene. The primers used for ChIP-qPCR are listed in Supplemental Table S1.
GUS staining and GUS activity
Untreated and salt-treated CycC1;1pro:GUS, SOS1pro:GUS, and WRKY75pro:GUS transgenic plant seedlings or tissues were stained in GUS staining solution (100 mM sodium phosphate buffer, pH 7.5, 10 mM EDTA, 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide, 1 mM 5-bromochloro-3-indolyl-b-D-glucuronide, and 0.1% Triton X-100). The samples were rinsed with 70% ethanol several times to remove the chlorophyll, and images were then taken under a microscope (Leica DMI8, Leica Microsystems, Germany). The GUS activities of the treated and untreated promoter-GUS transgenic seedlings were assayed according to a method described previously (Li et al. 2015).
Statistical analysis
Data are means ± Sd of 3 biological replicates, and the asterisks indicate statistically significant differences (*P < 0.05, **P < 0.01, and ***P < 0.001, Student's t test). Bars with different letters indicate significant differences at P < 0.05 by ANOVA with Tukey's multiple comparison test. Detailed statistical analysis results can be found in Supplemental Data Set 1.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CycC1;1 (AT5G48640), SOS1 (AT2G01980), SOS2 (AT5G35410), SOS3 (AT5G24270), ACTIN2 (AT3G18780), ACTIN7 (AT5G09810), WRKY1 (AT2G04880), and WRKY75 (AT5G13080).
Supplementary Material
Acknowledgments
We thank the anonymous reviewers for their thoughtful reviews and constructive suggestions which have improved this paper. We thank Prof. Hongwei Guo (Southern University of Science and Technology, China) and Prof. Diqiu Yu (Yunnan University, China) for kindly providing the WRKY75pro:GUS, 35Spro:WRKY75-GFP, wrky75-1, and wrky75-25 seeds. We thank Man-cang Zhang (Electrophysiology platform, Henan University) for performing the NMT experiment.
Contributor Information
Kai-Kai Lu, State Key Laboratory of Crop Stress Adaptation and Improvement, Collaborative Innovation Center of Crop Stress Biology, College of Life Sciences, Henan University, Kaifeng 475004, China.
Ru-Feng Song, State Key Laboratory of Crop Stress Adaptation and Improvement, Collaborative Innovation Center of Crop Stress Biology, College of Life Sciences, Henan University, Kaifeng 475004, China.
Jia-Xing Guo, State Key Laboratory of Crop Stress Adaptation and Improvement, Collaborative Innovation Center of Crop Stress Biology, College of Life Sciences, Henan University, Kaifeng 475004, China.
Yu Zhang, State Key Laboratory of Crop Stress Adaptation and Improvement, Collaborative Innovation Center of Crop Stress Biology, College of Life Sciences, Henan University, Kaifeng 475004, China.
Jia-Xin Zuo, State Key Laboratory of Crop Stress Adaptation and Improvement, Collaborative Innovation Center of Crop Stress Biology, College of Life Sciences, Henan University, Kaifeng 475004, China.
Hui-Hui Chen, State Key Laboratory of Crop Stress Adaptation and Improvement, Collaborative Innovation Center of Crop Stress Biology, College of Life Sciences, Henan University, Kaifeng 475004, China.
Cai-Yi Liao, State Key Laboratory of Crop Stress Adaptation and Improvement, Collaborative Innovation Center of Crop Stress Biology, College of Life Sciences, Henan University, Kaifeng 475004, China.
Xiao-Yu Hu, State Key Laboratory of Crop Stress Adaptation and Improvement, Collaborative Innovation Center of Crop Stress Biology, College of Life Sciences, Henan University, Kaifeng 475004, China.
Feng Ren, Hubei Key Laboratory of Genetic Regulation and Integrative Biology, School of Life Sciences, Central China Normal University, Wuhan 430079, China.
Ying-Tang Lu, State Key Laboratory of Hybrid Rice, College of Life Sciences, Wuhan University, Wuhan 430072, China.
Wen-Cheng Liu, State Key Laboratory of Crop Stress Adaptation and Improvement, Collaborative Innovation Center of Crop Stress Biology, College of Life Sciences, Henan University, Kaifeng 475004, China.
Author contributions
W.-C.L. and Y.-T.L. designed and directed the project. K.-K.L. performed the experiments with assistance of R.-F.S., J.-X.G., Y.Z., H.-H.C., J.-X.Z., C.-Y.L., and X.-Y.H. K.-K.L., W.-C.L., and F.R. analyzed the data. W.-C.L. wrote the manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. CycC1;1-overexpressing plants are sensitive to salt stress.
Supplemental Figure S2. Relative GUS activity of the CycC1;1pro:GUS transgenic plants subjected to salt stress.
Supplemental Figure S3. SOS2 and SOS3 expression in the cycc1;1 mutant.
Supplemental Figure S4. Relative GUS activity of the SOS1pro:GUS transgenic plants subjected to salt stress.
Supplemental Figure S5. Na+ accumulation in the xylem sap of the wild-type and cycc1;1 mutant plants.
Supplemental Figure S6. Sensitivity of single and double mutants of cycc1;1 and sos1 to mild salt stress.
Supplemental Figure S7. Na+ accumulation in the xylem sap of single and double mutants of cycc1;1 and sos1.
Supplemental Figure S8. CycC1;1 does not interact with WRKY1.
Supplemental Figure S9. Relative GUS activity of the SOS1pro:GUS and SOS1pro:GUS wrky75-1 transgenic plants subjected to salt stress.
Supplemental Figure S10. Na+ accumulation in the xylem sap of the wrky75 mutant.
Supplemental Figure S11. Relative GUS activity of WRKY75pro:GUS transgenic plants subjected to salt stress.
Supplemental Figure S12. Na+ accumulation in the xylem sap of single and double mutants of cycc1;1 and wrky75.
Supplemental Table S1. Primers used in this study.
Supplemental Data Set 1. Statistical analysis results.
Funding
This work was supported by the National Natural Science Foundation of China (#32000150 and #31971814) and the Natural Science Foundation of Henan Province (#222300420401).
References
- Agrawal R, Jiří F, Thakur JK. Kinase module of Mediator complex: important signalling processor for development and survival of plants. J Exp Bot. 2021:72(2):224–240. 10.1093/jxb/eraa439 [DOI] [PubMed] [Google Scholar]
- Asturias FJ, Jiang YW, Myers LC, Gustafsson CM, Kornberg RD. Conserved structures of mediator and RNA polymerase II holoenzyme. Science. 1999:283(5404):985–987. 10.1126/science.283.5404.985 [DOI] [PubMed] [Google Scholar]
- Barajas-Lopez JD, Moreno JR, Gamez-Arjona FM, Pardo JM, Punkkinen M, Zhu JK, Quintero FJ, Fujii H. Upstream kinases of plant SnRKs are involved in salt stress tolerance. Plant J. 2018:93(1):107–118. 10.1111/tpj.13761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai H, Guo J, Zhong Y, Hsu CC, Zou C, Wang P, Zhu JK, Shi H. The plasma-membrane polyamine transporter PUT3 is regulated by the Na+/H+ antiporter SOS1 and protein kinase SOS2. New Phytol. 2020:226(3):785–797. 10.1111/nph.16407 [DOI] [PubMed] [Google Scholar]
- Chen J, Mohan R, Zhang Y, Li M, Fu ZQ. NPR1 promotes its own and target gene expression in plant defense by recruiting CDK8. Plant Physiol. 2019:181(1):289–304. 10.1104/pp.19.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhang L, Xiang S, Chen Y, Zhang H, Yu D. The transcription factor WRKY75 positively regulates jasmonate-mediated plant defense to necrotrophic fungal pathogens. J Exp Bot. 2021:72(4):1473–1489. 10.1093/jxb/eraa529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Jiang H, Li L, Zhai Q, Qi L, Zhou W, Liu X, Li H, Zheng W, Sun J, et al. The Arabidopsis Mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell. 2012:24(7):2898–2916. 10.1105/tpc.112.098277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JS, Zhu JK, Bressan RA, Hasegawa PM, Shi H. Reactive oxygen species mediate Na+-induced SOS1 mRNA stability in Arabidopsis. Plant Journal. 2008:53(3):554–565. 10.1111/j.1365-313X.2007.03364.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal. 1998:16(6):735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Deinlein U, Stephan AB, Horie T, Luo W, Xu G, Schroeder JI. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014:19(6):371–379. 10.1016/j.tplants.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Sun J, Cao P, Ren L, Liu C, Chen S, Chen F, Jiang J. Variation in tissue Na+ content and the activity of SOS1 genes among two species and two related genera of Chrysanthemum. BMC Plant Biol. 2016:16(1):98. 10.1186/s12870-016-0781-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JX, Song RF, Lu KK, Zhang Y, Chen HH, Zuo JX, Li TT, Li XF, Liu WC. Cycc1; 1 negatively modulates ABA signaling by interacting with and inhibiting ABI5 during seed germination. Plant Physiol. 2022:190(4):2812–2827. 10.1093/plphys/kiac456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Chong L, Wu F, Hsu CC, Li C, Zhu JK, Zhu Y. Mediator tail module subunits MED16 and MED25 differentially regulate abscisic acid signaling in Arabidopsis. J Integr Plant Biol. 2021:63(4):802–815. 10.1111/jipb.13062 [DOI] [PubMed] [Google Scholar]
- Guo P, Li Z, Huang P, Li B, Fang S, Chu J, Guo H. A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell. 2017:29(11):2854–2870. 10.1105/tpc.17.00438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper TM, Taatjes DJ. The complex structure and function of Mediator. Journal of Biological Chemistry. 2018:293(36):13778–13785. 10.1074/jbc.R117.794438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan ASMM, Vander Schoor JK, Hecht V, Weller JL. The cyclin-dependent kinase module of the Mediator complex promotes flowering and reproductive development in pea. Plant Physiol. 2020:182(3):1375–1386. 10.1104/pp.19.01173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Liu J, Halfter U, Kim CS, Shi W, Zhu JK. SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell. 2000:12(9):1667–1678. 10.1105/tpc.12.9.1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, Robert F. The Mediator complex: at the nexus of RNA polymerase II transcription. Trends in Cellular Biology. 2017:27(10):765–783. 10.1016/j.tcb.2017.07.001 [DOI] [PubMed] [Google Scholar]
- Kim WY, Ali Z, Park HJ, Park SJ, Cha JY, Perez-Hormaeche J, Quintero FJ, Shin G, Kim MR, Qiang Z. Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat Commun. 2013:4(1):273–275. 10.1038/ncomms2846 [DOI] [PubMed] [Google Scholar]
- Li J, Xu HH, Liu WC, Zhang XW, Lu YT. Ethylene inhibits root elongation during alkaline stress through AUXIN1 and associated changes in auxin accumulation. Plant Physiol. 2015:168(4):1777–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Yang Y, Quan R, Mendoza I, Wu Y, Du W, Zhao S, Schumaker KS, Pardo JM, Guo Y. Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell. 2009:21(5):1607–1619. 10.1105/tpc.109.066217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA. 2000:97(7):3730–3734. 10.1073/pnas.97.7.3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WC, Song RF, Qiu YM, Zheng SQ, Li TT, Wu Y, Song CP, Lu YT, Yuan HM. Sulfenylation of ENOLASE2 facilitates H2O2-conferred freezing tolerance in Arabidopsis. Dev Cell. 2022b:57(15):1883–1898. 10.1016/j.devcel.2022.06.012 [DOI] [PubMed] [Google Scholar]
- Liu WC, Song RF, Zheng SQ, Li T-T, Zhang BL, Gao X, Lu YT. Coordination of plant growth and abiotic stress responses by tryptophan synthase β subunit1 through modulating tryptophan and ABA homeostasis in Arabidopsis. Mol Plant. 2022a:15(6):1–18. 10.1016/j.molp.2022.04.009 [DOI] [PubMed] [Google Scholar]
- Lou L, Yu F, Tian M, Liu G, Wu Y, Wu Y, Xia R, Pardo JM, Guo Y, Xie Q. ESCRT-I component VPS23A sustains salt tolerance by strengthening the SOS module in Arabidopsis. Mol Plant. 2020:13(8):1134–1148. 10.1016/j.molp.2020.05.010 [DOI] [PubMed] [Google Scholar]
- Maji S, Dahiya P, Waseem M, Dwivedi N, Bhat DS, Dar TH, Thakur JK. Interaction map of Arabidopsis Mediator complex expounding its topology. Nucleic Acids Res. 2019:47(8):3904–3920. 10.1093/nar/gkz122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur S, Vyas S, Kapoor S, Tyagi AK. The Mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress. Plant Physiol. 2011:157(4):1609–1627. 10.1104/pp.111.188300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R, Tester M. (2008). Mechanisms of salinity tolerance. Annu Rev Plant Biol. 59(1):651–681. 10.1146/annurev.arplant.59.032607.092911 [DOI] [PubMed] [Google Scholar]
- Nie K, Zhao H, Wang X, Niu Y, Zhou H, Zheng Y. The MIEL1-ABI5/MYB30 regulatory module fine tunes abscisic acid signaling during seed germination. J Integr Plant Biol. 2022:64(4):930–941. 10.1111/jipb.13234 [DOI] [PubMed] [Google Scholar]
- Poss ZC, Ebmeier CC, Taatjes DJ. The Mediator complex and transcription regulation. Crit Rev Biochem Mol Biol. 2013:48(6):575–608. 10.3109/10409238.2013.840259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q-S, Guo Y, Dietrich MA, Schumaker KS, Zhu J-K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA. 2002:99(12):8436–8441. 10.1073/pnas.122224699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan R, Lin H, Mendoza I, Zhang Y, Cao W, Yang Y, Shang M, Chen S, Pardo JM, Guo Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell. 2007:19(4):1415–1431. 10.1105/tpc.106.042291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Martinez-Atienza J, Villalta I, Jiang X, Kim W-Y, Ali Z, et al. Activation of the plasma membrane Na/H antiporter salt-overly-sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc Natl Acad Sci USA. 2011:108(6):2611–2616. 10.1073/pnas.1018921108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Ohta M, Shi H, Zhu J-K, Pardo JM. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA. 2002:99(13):9061–9066. 10.1073/pnas.132092099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishmawi L, Pesch M, Juengst C, Schauss AC, Schrader A, Hulskamp M. Non-cell-autonomous regulation of root hair patterning genes by WRKY75 in Arabidopsis. Plant Physiol. 2014:165(1):186–195. 10.1104/pp.113.233775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa S, Ntoukakis V, Ohmido N, Pendle A, Abranches R, Shaw P. Cell differentiation and development in Arabidopsis are associated with changes in histone dynamics at the single-cell level. Plant Cell. 2014:26(12):4821–4833. 10.1105/tpc.114.133793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA. 2000:97(12):6896–6901. 10.1073/pnas.120170197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu JK. The putative plasma membrane Na(+)/H(+) antiporter SOS1 controls long-distance Na(+) transport in plants. Plant Cell. 2002:14(2):465–477. 10.1105/tpc.010371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zelm E, Zhang Y, Testerink C. Salt tolerance mechanisms of plants. Annu Rev Plant Biol. 2020:71(1):403–433. 10.1146/annurev-arplant-050718-100005 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant Journal. 2004:40(3):428–438. 10.1111/j.1365-313X.2004.02219.x [DOI] [PubMed] [Google Scholar]
- Wang LF, Lu KK, Li TT, Zhang Y, Guo JX, Song RF, Liu WC. Maize PHYTOMELATONIN RECEPTOR1 functions in plant osmotic and drought stress tolerance. J Exp Bot. 2022:73(17):5961–5973. 10.1093/jxb/erab553 [DOI] [PubMed] [Google Scholar]
- Wang W, Chen X. HUA ENHANCER3 reveals a role for a cyclin-dependent protein kinase in the specification of floral organ identity in Arabidopsis. Development. 2004:131(13):3147–3156. 10.1242/dev.01187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Xu J, Meng X, Fang X, Xia M, Zhang J, Cao S, Fan T. Linker histone variant HIS1-3 and WRKY1 oppositely regulate salt stress tolerance in Arabidopsis. Plant Physiol. 2022:189(3):1833–1847. 10.1093/plphys/kiac174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Chen ZZ, Zhou XF, Yin HB, Li X, Xin XF,Hong XH, Zhu JK, Gong Z. Overexpression of SOS (Salt Overly Sensitive) genes increases salt tolerance in transgenic Arabidopsis. Molecular. Plant. 2009:2(1):22–31. 10.1093/mp/ssn058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Guo Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytologist. 2018a:217(2):523–539. 10.1111/nph.14920 [DOI] [PubMed] [Google Scholar]
- Yang Y, Guo Y. Unraveling salt stress signaling in plants. J Integr Plant Biol. 2018b:60(9):796–804. 10.1111/jipb.12689 [DOI] [PubMed] [Google Scholar]
- Yang Y, Han X, Ma L, Wu Y, Liu X, Fu H, Liu G, Lei X, Guo Y. Dynamic changes of phosphatidylinositol and phosphatidylinositol 4-phosphate levels modulate H+-ATPase and Na+/H+ antiporter activities to maintain ion homeostasis in Arabidopsis under salt stress. Mol Plant. 2021:14(12):2000–2014. 10.1016/j.molp.2021.07.020 [DOI] [PubMed] [Google Scholar]
- Yu L, Nie J, Cao C, Jin Y, Yan M, Wang F, Liu J, Xiao Y, Liang Y, Zhang W. Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytologist. 2010:188(3):762–773. 10.1111/j.1469-8137.2010.03422.x [DOI] [PubMed] [Google Scholar]
- Zhang L, Chen L, Yu D. Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiol. 2018:176(1):790–803. 10.1104/pp.17.00657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ji TT, Li TT, Tian YY, Wang LF, Liu WC. Jasmonic acid promotes leaf senescence through MYC2-mediated repression of Catalase2 expression in Arabidopsis. Plant Sci. 2020:299:110604. 10.1016/j.plantsci.2020.110604 [DOI] [PubMed] [Google Scholar]
- Zhou H, Lin H, Chen S, Becker K, Yang Y, Zhao J, Kudla J, Schumaker KS, Guo Y. Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell. 2014:26(3):1166–1182. 10.1105/tpc.113.117069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Wang C, Tan T, Cai J, He J, Lin H. Patellin1 negatively modulates salt tolerance by regulating PM Na+/H+ antiport activity and cellular redox homeostasis in Arabidopsis. Plant Cell Physiol. 2018:59(10):2165. 10.1093/pcp/pcy172 [DOI] [PubMed] [Google Scholar]
- Zhou H, Xiao F, Zheng Y, Liu G, Zhuang Y, Wang Z, Zhang Y, He J, Fu C, Lin H. PAMP-INDUCED SECRETED PEPTIDE 3 modulates salt tolerance through RECEPTOR-LIKE KINASE 7 in plants. Plant Cell. 2022:34(2):927–944. 10.1093/plcell/koab292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Cell signaling under salt, water and cold stresses. Curr Opin Plant Biol. 2001:4(5):401–406. 10.1016/S1369-5266(00)00192-8 [DOI] [PubMed] [Google Scholar]
- Zhu JK. Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol. 2003:6(5):441–445. 10.1016/S1369-5266(03)00085-2 [DOI] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002:53(1):247–273. 10.1146/annurev.arplant.53.091401.143329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Huang P, Guo P, Chong L, Yu G, Sun X, Hu T, Li Y, Hsu CC, Tang K, et al. CDK8 is associated with RAP2.6 and SnRK2.6 and positively modulates abscisic acid signaling and drought response in Arabidopsis. New Phytol. 2020:228(5):1573–1590. [DOI] [PubMed] [Google Scholar]
- Zhu M, Shabala L, Cuin TA, Huang X, Zhou M, Munns R, Shabala S. Nax loci affect SOS1-like Na+/H+ exchanger expression and activity in wheat. J Exp Bot. 2016:67(3):835–844. 10.1093/jxb/erv493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Schluttenhoffer CM, Wang P, Fu F, Thimmapuram J, Zhu JK, Lee SY, Yun DJ, Mengiste T. CYCLIN-DEPENDENT KINASE8 differentially regulates plant immunity to fungal pathogens through kinase-dependent and -independent functions in Arabidopsis. Plant Cell. 2014:26(10):4149–4170. 10.1105/tpc.114.128611 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.