Abstract
The fields of genetics and genomics have greatly expanded across medicine through the development of new technologies that have revealed genetic contributions to a wide array of traits and diseases. Thus, the development of widely available educational resources for all healthcare providers is essential to ensure the timely and appropriate utilization of genetics and genomics patient care. In 2020, the National Human Genome Research Institute released a call for new proposals to develop accessible, sustainable online education for health providers. This paper describes the efforts of the six teams awarded to reach the goal of providing genetic and genomic training modules that are broadly available for busy clinicians.
Keywords: competency-based education, education, genomics, learning modules, providers
Increasing evidence demonstrates that utilizing genetic information in clinical decision-making can improve routine clinical care for a wide range of conditions [1–6]. Genetic testing and genomic sequencing have expanded across medical disciplines, with as many as ten new genetic tests introduced daily [7]. Despite the growing influence of genomic medicine in clinical practice, surveys continue to show that many clinicians, particularly primary care providers, feel inadequately prepared to order and interpret genetic tests or discuss genetic test results with patients [8–11]. Clinicians also report a lack of confidence in using genetic information to make clinical decisions [12]. These challenges are compounded by the rapidly expanding and changing knowledge base, the range of providers involved and the scope of care and the need for high-quality training that is adaptable to the skill level and time constraints of the busy clinician.
For more than 20 years, several collaborative projects have focused on enhancing education in genetics and genomics for nongeneticist practitioners, particularly in pediatrics, family and internal medicine, to insure widespread access and appropriate utilization of these tools [13–17]. In 2001, a group of geneticists and educators proposed a set of genetics core competencies for all health providers [18], which have been continually revised and updated [19]. Subsequently, other professional groups developed their own genetic competencies, including nurses [20–24], physician assistants [25], pharmacists [26,27], physicians [28] and genetic counselors [29]. These guidelines were assimilated into a National Human Genome Research Institute (NHGRI) repository [30] called the Genetics/Genomics Competency Center, or G2C2 [31].
The genetics and genomics community continues to engage with a broad range of partners to enhance provider knowledge and skills and develop continuing education opportunities [32–34]. Yet, the sheer complexity and volume of information appear to have outpaced efforts by individual educators and schools, and it remains uncertain if clinicians are aware of the diverse educational opportunities that are available. Furthermore, the delivery of continuing medical education has substantially shifted from traditional, in-person learning to remote learning on a much broader scale, with increasing evidence of its comparability to traditional modes of learning [35–37]. The COVID-19 pandemic has only accelerated the transition. Despite the conveniences of online learning, many hurdles remain to deliver education in genomics for clinicians, such as institutional support, the need for continual updating of content as the genomics landscape changes, competing priorities for busy clinicians and limited availability of engaging, just-in-time instruction.
The development of online modular genomic educational opportunities is one method to address some of these challenges. In 2020, NHGRI announced the availability of supplemental funding for its grantees “to support the development and implementation of modules aimed at providing healthcare professionals with genomic medicine training” [38]. The modules could be designed to stand alone or to build upon each other to form a certificate in genomic medicine or other recognition of healthcare professionals with specialized expertise and consultative capabilities. NHGRI funded six teams around the USA, recognized leaders in the fields of genomic medicine and education, to develop online scalable training modules for a wide range of healthcare professionals. The collection of teams was self-named the Competency-based Online Genomic Medicine Training (COGENT) group. Although each group’s educational projects were unique, there were several areas of potential synergy. Working together, the groups harmonized definitions, identified new methods for the delivery of educational materials and discussed ways to develop mechanisms to recognize healthcare professionals who complete the training, including through continuing education credits or certificates. In particular, six overlapping topics were identified across multiple teams (i.e., family health history; ethical, legal, and social implications; clinical testing and interpretation; basic biology; cardiovascular disease and pharmacogenomics [PGx]) and working groups were established to enable sharing of materials and harmonization where possible. The groups are discussing ways to support and coordinate these efforts over the long term to sustain high-quality genomic medicine modules that can provide healthcare professionals the competence and confidence they need to use genomics in patient care.
The following sections include a brief overview of each team’s objectives, the target audience, topics, assessment and types of recognition awarded upon completion (e.g., continuing education credits or certifications, digital badges; Figure 1).
Figure 1. . Main learning objective for each team.
Cincinnati Children’s Hospital Medical Center/University of Cincinnati
The goal of the University of Cincinnati and Cincinnati Children’s Hospital Medical Center (CCHMC) was to develop a genomics education program titled, “Reduce or Prolong the Diagnostic Odyssey: You Decide”. The CCHMC program sought to provide an opportunity for clinicians to increase their confidence in selecting genetic tests, interpreting results and identifying implications for patient care. Self-paced online learning modules were created that used unfolding pediatric cases.
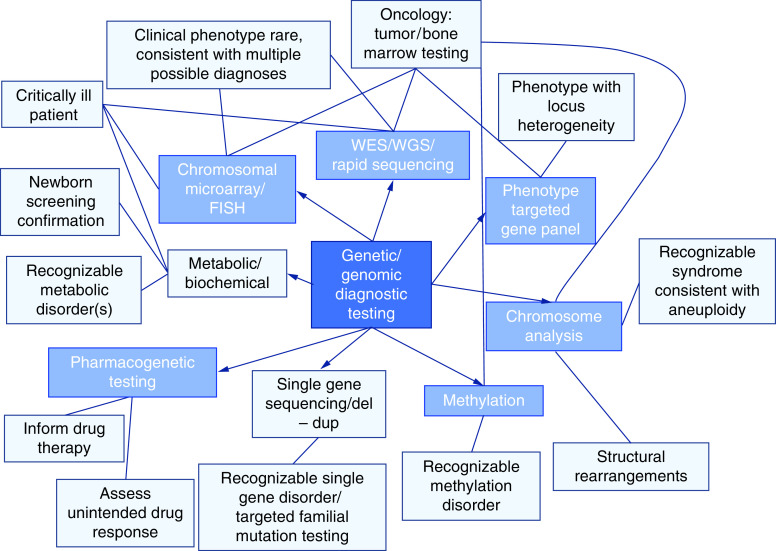
The CCHMC clinician target audience consists of physicians, advanced practice nurses and physician assistants. Learning objectives were primarily skills-based and map to existing genetics/genomic competencies to demonstrate relevance to practice [25,28,39]. Figure 2 shows an overview concept map for genetic/genomic diagnostic testing. Each type of test contains a link to currently available online information. In addition, basic genetic and genomic concepts, in the form of short 1 to 3 min videos, infographics and definitions appear as options as the learner clicks through each module. Many of these basic elements are reused per module, aligning with basic concepts. Each module ends with a short video of the clinician who submitted the case summarizing key points and lessons learned.
Figure 2. . Concept map for genetic/genomic diagnostic testing developed by the Cincinnati team.
Starting from white boxes around the outside, each case presents options for different types of testing, with the appropriate type of test indicated with a line. Short videos are presented describing the usefulness and limitations of the type of testing, as well as other basic concepts like inheritance, nomenclature and cascade testing.
del: Deletion; dup: Duplication; WES: Whole-exome sequencing; WGS: Whole-genome sequencing.
All learners will need to register and answer questions about demographics, use of genetics in the past month and 12 questions adapted from the Genetic Counseling Self-Efficacy Scale (GCSES) [40]. The GCSES asks the learner to rate how certain he/she is that each activity can be performed independently utilizing a continuous 0–100 Likert-like scale. Learners will be asked to complete the GCSES questions again after completing the full set of modules. Change in perceived ability to perform skills will be analyzed, as well as a comparison of the number of genetic tests ordered.
Before making modules widely available, the CCHMC team will randomize specialty divisions at CCHMC to receive invitations to select a division clinician to be part of a ‘genomic champions' group that includes completion of the online modules followed by monthly case discussion or receive advertising for the availability of the online modules only (a control group). All clinicians who complete the modules will receive continuing education credits. CCHMC hypothesizes that a larger proportion of clinicians from divisions with a champion will complete the full set of modules and demonstrate a greater gain in confidence and use of genetics than learners from divisions without a champion.
Columbia University Irving Medical Center
The overall goals of the Columbia University Irving Medical Center program were to develop educational materials on polygenic risk scores (PRS) for a wide range of nongeneticist providers including physicians, physician assistants, nurse practitioners and nurses. The online modules will also be incorporated into healthcare training programs. The educational modules build upon provider education materials developed in support of the NHGRI-funded Electronic Medical Records and Genomics (eMERGE) Network (phase IV) study on PRS, which will return PRS scores to over 25,000 participants and providers for select actionable diseases. Genomic medicine implementation has heretofore focused largely on rare genetic variants associated with high risk; however, most common diseases are largely polygenic and the feasibility of using PRS to identify high-risk patients was first demonstrated in 2018 for conditions including coronary artery disease and Type 2 diabetes [41]. In 2019, it was reported that individuals in the top 2–5% of PRS have risk equivalent to many of the rare variants currently tested for diseases such as breast cancer, colon cancer and coronary artery disease [42].
The PRS educational materials are adaptable to different target audiences. The modules incorporated the following competencies shared across different health professions: describe the major forms of genetic variability and contribution to disease risk; identify and describe monogenic and polygenic risk and understand how a PRS is developed including the limitations of its application. In addition, the modules addressed how to incorporate a PRS into clinical care including the recommended care and surveillance, effective communication of PRS risk to patients and patient-specific limitations and identification of and discussion about the patient-specific barriers to following recommendations.
All materials are accessible to the public and permitted to be used by other groups for educational purposes. In addition to self-learning through the online modules, the modules can also be incorporated into in-person presentations on continuing medical education courses and are included in a library of PRS educational materials hosted on the Columbia Precision Medicine website and the eMERGE websites. The team developed ACTion (ACT) Sheets, a one-page just-in-time reference resource for select high-risk PRS diseases (e.g., breast cancer, heart disease, diabetes) that can be adapted and integrated into clinical decision support tools in electronic health record systems.
Duke University School of Medicine
The Duke University School of Medicine team aimed to develop the Rapid Personalized Learning Platform for Genomic Medicine to promote knowledge about the science and enable the practice of genomic medicine. The platform includes a suite of short online modules that are designed to be free-standing but can also be combined to develop a comprehensive learning program for a range of health providers. In contrast to the other teams, the modules are intended to be introductory to genome sciences and medicine and include an overview of clinical applications across different specialties. Therefore, there is no specific target audience.
The modules are grouped into two levels and learners can choose to view them in any order (Level 1 topics will address general/foundation topics and Level 2 topics are divided by specialty and incorporate professional organization recommendations and case studies). As the field of genome medicine continues to rapidly develop, these short modules can be updated as needed with limited disruption and effort. Continuing education credits will be provided through the Duke University Health System Office of Clinical Education and Professional Development for physicians, pharmacists, physician assistants and nurses upon completion of the brief assessment at the conclusion of the module. Learners can also consent to receive a 60-day follow-up survey to gauge the impact of learning on attitudes and clinical practice behaviors (e.g., learner competence, performance and perceived barriers to implementing practice changes). A companion website will include resources to supplement continuing learning and clinical implementation. These online learning modules will serve as the foundation of a Certificate Program in Genomic Medicine.
Mayo Clinic
The Mayo Clinic Center for Individualized Medicine team’s goals were to create a sustainable, online, module-based certificate program with continuing education (CE) credits to meet genomic education needs specifically for the nursing community. Nursing professionals are a key component of the medical care team and play a significant role in both patient care and patient education. While nurses who have specific education and training in genetics and genomics exist, they are limited in number. To keep pace with the growth in genomic testing and the increasing use of genomics in clinical care, there is a critical need for updated, broad, accessible and practical genomics education for all nurses [43]. This education is needed to support direct clinical care and the role nurses have in supporting patients and patient education.
As there are no well-defined and agreed-upon competencies for genomics across nursing practices, the curriculum for the Mayo certificate program was developed based on the scope and standards of practice for genetics/genomics nursing published by the American Nurses Association with support from the International Society of Nurses in Genetics [44]. Among the key learning objectives of the certificate program are fundamentals of genomics, integrating genomics into practice, patient education and ethical issues, and will include primary care nursing along with specialty care. The effectiveness of the program will be assessed using a mixed-method analysis using pre/posteducation comprehension testing, online surveys, focus groups and quarterly content reviews by course directors and content experts. The results from these efforts will be used to make quarterly updates to topics, content and delivery mechanisms. The educational modules, while designed to be delivered as a single program specifically tailored to nurses, can be repurposed as individual modules for a range of learners.
University of Pittsburgh/Indiana University
The University of Pittsburgh School of Pharmacy and Indiana University School of Medicine partnered to develop online, modular, competency-based PGx education for multidisciplinary providers (pharmacists, physicians, nurse practitioners, physician assistants, nurses and genetic counselors). The overall goal was to accelerate the use of increasingly available PGx testing data in prescribing decisions. Currently, over 300 medications have genetic data in their FDA-approved labeling [45] and >50 drugs have level A prescribing recommendations in published Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines [46].
The educational program is competency-based to promote the attainment of practical skills important for the integration of PGx into clinical care. These are strictly based on contemporary genomics competency standards for each discipline (Figure 3) and are comprised of stackable modules to allow learners to tailor their education to meet individual needs regarding the depth and duration of training. Topics covered include nomenclature, PGx testing, clinical decision-making, regulatory issues, reimbursement, communication and ethical, legal and social issues. The program will focus on clinically actionable PGx gene–drug pairs across a variety of diseases, such as oncology, cardiology, psychiatry, neurology, infection diseases and pain control.
Figure 3. . Competency mapping to inform development of a learning module on pharmacogenomic testing by the University of Pittsburgh/Indiana University team.
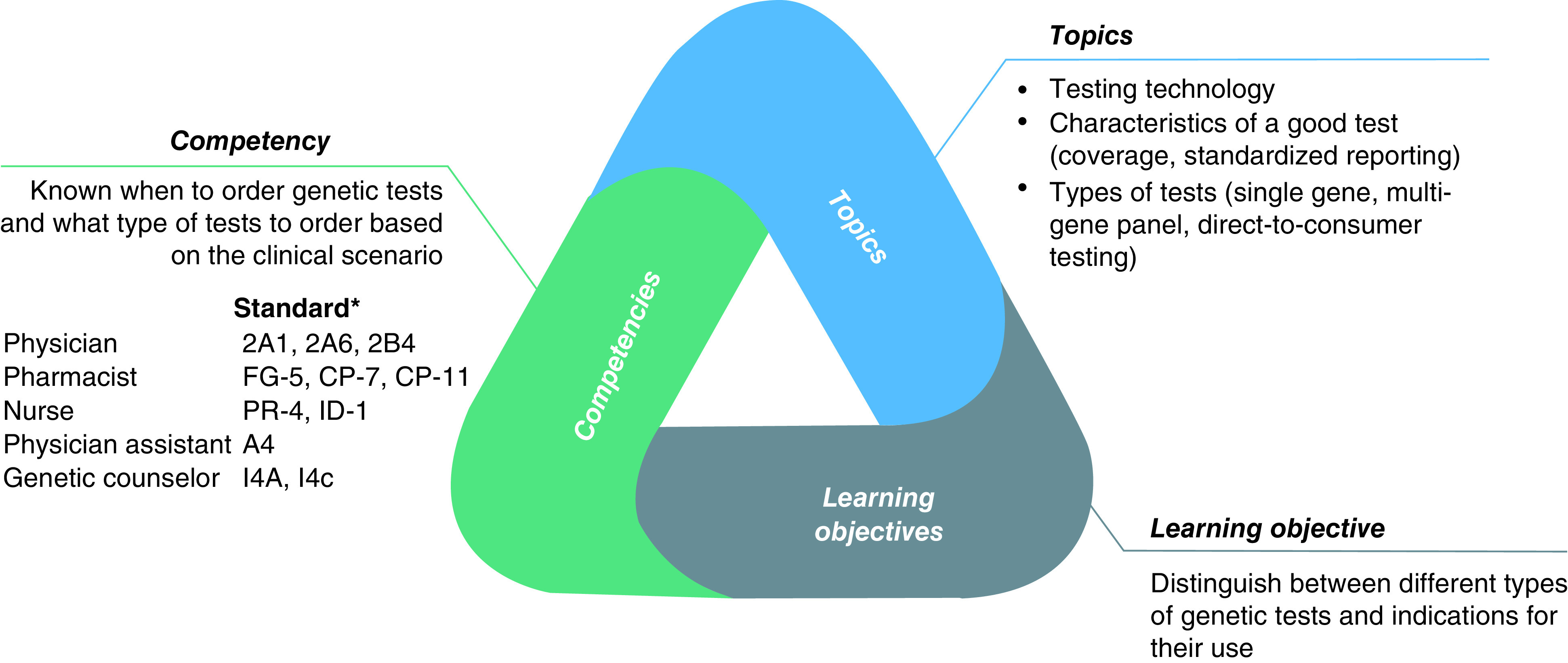
Genomics competency standards for each healthcare provider are aggregated and mapped to learning objectives to inform content development for a module topic.
*Competency standard numbers from the listing for each discipline on the G2C2 website.
The approach is highly innovative in that it leverages the Test2Learn platform [47] to deliver high-fidelity participatory education [48]. Trainees learn by undergoing testing personally (or using anonymous data if preferred) and using these data in interactive cases. Several groups have demonstrated participatory learning using real genomic data to be an engaging and effective approach [49–54] that produces superior learning outcomes. Source data can be from institutional laboratories, 23andMe, file upload or 1000 Genomes. Integrated online genomic resources, such as from CPIC, will further support the learning objectives. Microcredentialing (digital badges) and continuing education credits are awarded in a scalable model to achieve long-term sustainability. The project involves implementations at both institutions, pre/post and longitudinal assessments to evaluate the impact of training and sustainability end points.
University of Texas Health Science Center at Houston (UTHealth)
The goals of UTHealth’s Cardiovascular Genomics Certificate (UTHCGC) program were: to fulfill an unmet need for adult cardiovascular genetics education, to increase awareness of heritable cardiovascular diseases by all health professionals with a focus on primary care providers who are gatekeepers and partners in the diagnosis and treatment of these patients and to improve the recognition and referral of patients with genetically triggered cardiovascular diseases. In the last 20 years, genomics research has identified thousands of genetic variants that influence cardiovascular disease risk. Adult-onset cardiovascular diseases are influenced by highly penetrant variants that cause early onset disease and common variants that modify the timing or severity of disease. The development of clinical genetic tests for cardiovascular diseases has enabled the use of genetic information to improve outcomes and save lives by identifying patients at risk before they develop life-threatening cardiovascular complications and by altering treatment strategies based on the genetic variant.
The target audience for the UTHCGC program is primary care providers, nurses, nurse practitioners and cardiovascular clinicians (i.e., cardiologists, cardiovascular surgeons and cardiovascular anesthetists) who do not have specialized training in genetics. The team identified three core competencies as essential end points for training in genomic medicine: to obtain and interpret genomic information, including family history and pedigree data; to order genetic tests or to provide referrals for genomic medicine consultations and to understand and apply appropriate treatment decisions based on genomic data. The courses are designed to appeal to busy clinicians by emphasizing a concise, case-based format focusing on genetic issues that providers are likely to encounter in clinical practice and allowing learners to customize their experience by providing a range of learning formats including short presentations on basic genetic concepts and terminology (Tier 1), workshop-style activities to apply these concepts (Tier 2) and longer clinical cases where learners can demonstrate that they are able to contextualize these concepts in the practice of genomic medicine (Tier 3). Learners can revisit and master a set of core competencies as they progress through the tiers of modules.
The case presentations in Tier 2 and Tier 3 modules feature relatively common hereditary diseases such as cardiomyopathies, hyperlipidemias and arrhythmias, as well as aortic aneurysms and pharmacogenetic questions that can impact clinical management. By the end of the course, learners will be able to recognize genetically triggered diseases based on clinical features and family history, distinguish actionable results on laboratory and direct-to-consumer genetic test reports and identify and monitor individuals who are at risk for disease based on genetic information. The modules are designed to stand alone, with individualized quizzes and pre- and post-assessments, but may also be combined flexibly according to specialties and interests. The UTHCGC will be accredited for continuing medical and nursing education credit. In addition, learners who complete at least 3.5 h will be eligible to receive a UTHealth Cardiovascular Genomics Certificate.
The UTHCGC team will continually revise and restructure the modules to provide more interactive and engaging learning opportunities based on learner feedback (collected through anonymous Qualtrics questionnaires). Target benchmarks will include >90% of modules being rated as helpful and >90% of participants rating the program as moderately or highly impactful. Baseline and annual follow-up surveys will also be sent to those who complete the entire certificate program to assess changes in clinical practice. These surveys will provide data on the long-term effectiveness of the certificate program to improve the recognition and referral of patients with genetically triggered cardiovascular diseases.
Conclusion
Funding from NHGRI has brought together six innovative educational programs to create a highly interactive and crossdisciplinary approach to genomic education for healthcare providers. The result is a set of modular and flexible web-accessible learning resources for health professionals, clinicians, nurses, pharmacists, physician assistants, genetic counselors and even students of the profession. Each institution will roll out the modules and make them accessible to learners outside of their institutions. The work by the teams represents a concerted effort to develop depth in the online modules across a wide range of topics linked to published competencies that will be widely accessible to health providers at all levels and specialties.
Future perspective
The COGENT platform will address a critical need and gap at the intersection of genetics, genomics and clinical care and a series of tools will be generated that would not have otherwise been developed had the six groups not partnered. This collaborative effort was effective in providing a mechanism for the exchange of content as well as innovative ideas and best practices for education. Future collaborations will aim to increase access through a single interface to multiple genomic medicine training modules. Each institution will continue to provide updates and expand to new topics while being seamless to learners, particularly regarding evaluation tools (e.g., validated measurements of e-learning) and the impact of learning modules on provider behavior (pre/postlearning).
Executive summary.
The rapid pace of developments in genetics and genomics and advances in clinical practice across disciplines has created a challenging scenario for educators.
Over the past several decades, multiple initiatives have resulted in the creation of disparate genetic educational resources and learning opportunities.
Online modules are convenient and widely accessible, allowing for educational opportunities in genetics and genomics for multiple types of providers that are geographically diverse.
Collaborations between institutions represent an effective and efficient way to innovate in genomic education and should be supported in the future.
Acknowledgments
The authors would like to acknowledge the support of additional COGENT team members: Cincinnati Children’s Hospital Medical Center/University of Cincinnati team: CA Prows, LDM Peña and H Aungst; Mayo Clinic team: SA Johnson, NR Nicholas, C Berg, C Scott and LA Schimmenti; University of Pittsburgh/Indiana University: P Dexter, E Tillman, L Berenbrok, C Fulton, M Massart, V Pratt, V Murthy and S Prucka; UTHealth Adult Cardiovascular Genomics Certificate Program team: S Abu Dayeh, V Huff, A Cecchi and D Milewicz, with contributions from D An, L Kurzlechner, A Landstrom, Y Liang, M Moulik, M Wanat, E Wang and TC Wong.
Footnotes
Financial & competing interests disclosure
SB Haga’s contribution was funded by the National Human Genome Research Institute (5T32HG008955-04S1 and 3U01HG010225-04S1). WK Chung’s contribution was funded by the National Human Genome Research Institute (U01HG008680). LA Cubano has no conflicts of interest to disclose. TB Curry’s contribution was funded by the National Human Genome Research Institute (3 UL1 TR002377-04S4). PE Empey’s contribution was funded by U01HG010245. PE Empey has received support from the RK Mellow Foundation and the National Association of Chain Drug Stores related to the contributions described in this manuscript. The University of Pittsburgh licenses other work related to the educational platform (Test2Learn). GS Ginsburg’s contribution was funded by the National Human Genome Research Institute (5T32HG008955-04S1). K Mangold’s contribution was funded by the National Human Genome Research Institute (3 UL1 TR002377-04S4). CY Miyake’s contribution was funded by the National Human Genome Research Institute (3UL1TR003167-02S4). SK Prakash’s contribution was funded by the National Human Genome Research Institute (3UL1TR003167-02S4). LB Ramsey’s contribution was funded by the National Human Genome Research Institute (3UL1TR001425-05A1S1). R Rowley has no conflicts of interest to disclose. CRR Vitek’s contribution was funded by the National Human Genome Research Institute (3 UL1 TR002377-04S4). TC Skaar’s contribution was funded by the National Human Genome Research Institute (U01HG010245). J Wynn’s contribution was funded by the National Human Genome Research Institute (U01HG008680). TA Manolio has no conflicts of interest to disclose. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Manolio TA, Chisholm RL, Ozenberger B et al. Implementing genomic medicine in the clinic: the future is here. Genet. Med. 15(4), 258–267 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Relling MV, Evans WE. Pharmacogenomics in the clinic. Nature 526(7573), 343–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Empey PE, Pratt VM, Hoffman JM, Caudle KE, Klein TE. Expanding evidence leads to new pharmacogenomics payer coverage. Genet. Med. 23(5), 830–832 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milewicz DM, Guo D, Hostetler E, Marin I, Pinard AC, Cecchi AC. Update on the genetic risk for thoracic aortic aneurysms and acute aortic dissections: implications for clinical care. J. Cardiovasc. Surg. 62(3), 203–210 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Regalado ES, Guo DC, Prakash S et al. Aortic disease presentation and outcome associated with ACTA2 mutations. Circ. Cardiovasc. Genet. 8(3), 457–464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson D, Dissanayake VH, Korf BR, Towery M, Haspel RL. An international genomics health workforce education priorities assessment. Per. Med. 19(4), 299–306 (2022). [DOI] [PubMed] [Google Scholar]
- 7.Phillips KA, Deverka PA, Hooker GW, Douglas MP. Genetic test availability and spending: Where are we now? Where are we going? Health Aff. (Millwood) 37(5), 710–716 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arora NS, Davis JK, Kirby C et al. Communication challenges for nongeneticist physicians relaying clinical genomic results. Per. Med. 14(5), 423–431 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubanovich CK, Cheung C, Mandel J, Bloss CS. Physician preparedness for big genomic data: a review of genomic medicine education initiatives in the United States. Hum. Mol. Genet. 27(R2), R250–R258 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauser D, Obeng AO, Fei K, Ramos MA, Horowitz CR. Views of primary care providers on testing patients for genetic risks for common chronic diseases. Health Aff. (Millwood) 37(5), 793–800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamilton JG, Abdiwahab E, Edwards HM, Fang ML, Jdayani A, Breslau ES. Primary care providers’ cancer genetic testing-related knowledge, attitudes, and communication behaviors: a systematic review and research agenda. J. Gen. Intern. Med. 32(3), 315–324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen KD, Vassy JL, Jamal L et al. Are physicians prepared for whole genome sequencing? a qualitative analysis. Clin. Genet. 89(2), 228–234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mcinerney J. Education in a genomic world. J. Med. Philos. 27(3), 369–390 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Collins FS. Preparing health professionals for the genetic revolution. JAMA 278(15), 1285–1286 (1997). [PubMed] [Google Scholar]
- 15.Guttmacher AE, Jenkins J, Uhlmann WR. Genomic medicine: who will practice it? A call to open arms. Am. J. Med. Genet. 106(3), 216–222 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Rohrer Vitek CR, Abul-Husn NS, Connolly JJ et al. Healthcare provider education to support integration of pharmacogenomics in practice: the eMERGE Network experience. Pharmacogenomics 18(10), 1013–1025 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manolio TA, Murray MF. The growing role of professional societies in educating clinicians in genomics. Genet. Med. 16(8), 571–572 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Jenkins J, Blitzer M, Boehm K et al. Recommendations of core competencies in genetics essential for all health professionals. Genet. Med. 3(2), 155–159 (2001). [DOI] [PubMed] [Google Scholar]
- 19.National Coalition for Health Professional Education in Genetics. Core Competencies in Genetics Essential for All Health-Care Professionals (3rd Edition).(2007). www.jax.org/education-and-learning/clinical-and-continuing-education/clinical-topics/core-competencies-for-health-care-professionals [Google Scholar]
- 20.Greco K, Tinley S, Seibert D. Development of the essential genetic and genomic competencies for nurses with graduate degrees. Annu. Rev. Nurs. Res. 29, 173–190 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Lewis JA, Calzone KM, Jenkins J. Essential nursing competencies and curricula guidelines for genetics and genomics. MCN Am. J. Matern. Child Nurs. 31(3), 146–153, Quiz, 154–145, (2006). [DOI] [PubMed] [Google Scholar]
- 22.Jenkins JF, Calzone K. Consensus Panel on Genetic/Genomic Nursing Competencies (2009). Essentials of Genetic and Genomic Nursing: Competencies, Curricula Guidelines, and Outcome Indicators (2nd Edition). American Nurses Association, Silver Spring, MD, USA: (2008). www.genome.gov/Pages/Careers/HealthProfessionalEducation/geneticscompetency.pdf [Google Scholar]
- 23.Howington L, Riddlesperger K, Cheek DJ. Essential nursing competencies for genetics and genomics: implications for critical care. Crit. Care Nurse 31(5), e1–7 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Jenkins J. Essential genetic and genomic nursing competencies for the oncology nurse. Semin. Oncol. Nurs. 27(1), 64–71 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldgar C, Michaud E, Park N, Jenkins J. Physician assistant genomic competencies. J. Physician Assist. Educ. 27(3), 110–116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roederer MW, Kuo GM, Kisor DF et al. Pharmacogenomics competencies in pharmacy practice: a blueprint for change. J. Am. Pharm. Assoc. 57(1), 120–125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gammal RS, Lee YM, Petry NJ et al. Pharmacists leading the way to precision medicine: updates to the core pharmacist competencies in genomics. Am. J. Pharm. Educ. 86(4), 8634 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korf BR, Berry AB, Limson M et al. Framework for development of physician competencies in genomic medicine: report of the Competencies Working Group of the Inter-Society Coordinating Committee for Physician Education in Genomics. Genet. Med. 16(11), 804–809 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Doyle DL, Awwad RI, Austin JC et al. 2013 Review and Update of the Genetic Counseling Practice Based Competencies by a task force of the Accreditation Council for Genetic Counseling. J. Genet. Couns. 25(5), 868–879 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Calzone KA, Jerome-D'Emilia B, Jenkins J et al. Establishment of the genetic/genomic competency center for education. J. Nurs. Scholarsh. 43(4), 351–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.US National Human Genome Research Institute. Genetics/Genomics Competency Center (2021). (Accessed 14 September 2021). https://genomicseducation.net/competency
- 32.Campion M, Goldgar C, Hopkin RJ, Prows CA, Dasgupta S. Genomic education for the next generation of health-care providers. Genet. Med. 21(11), 2422–2430 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Talwar D, Tseng TS, Foster M, Xu L, Chen LS. Genetics/genomics education for nongenetic health professionals: a systematic literature review. Genet. Med. 19(7), 725–732 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Weitzel KW, Aquilante CL, Johnson S, Kisor DF, Empey PE. Educational strategies to enable expansion of pharmacogenomics-based care. Am. J. Health Syst. Pharm. 73(23), 1986–1998 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinclair P, Kable A, Levett-Jones T. The effectiveness of internet-based e-learning on clinician behavior and patient outcomes: a systematic review protocol. JBI Database System Rev. Implement. Rep. 13(1), 52–64 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Tuma F, Nituica C, Mansuri O, Kamel MK, Mckenna J, Blebea J. The academic experience in distance (virtual) rounding and education of emergency surgery during COVID-19 pandemic. Surg. Open Sci. 5, 6–9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng L, Seow KC, Macdonald L et al. eLearning in physical therapy: lessons learned from transitioning a professional education program to full eLearning during the COVID-19 pandemic. Phys. Ther. 101(4), pzab082 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.US NIH. Notice of Special Interest (NOSI): Administrative Supplements for Development of Training Modules in Genomic Medicine for Health Care Professionals (2021). (Accessed 14 September 2021). https://grants.nih.gov/grants/guide/notice-files/NOT-HG-20-020.html
- 39.Genetics/genomics competencies for RNs and nurses with graduate degrees. Nurs. Manage. 50(1), 1–3 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Caldwell S, Wusik K, He H, Yager G, Atzinger C. Development and validation of the Genetic Counseling Self-Efficacy Scale (GCSES). J. Genet. Couns. 27(5), 1248–1257 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Khera AV, Chaffin M, Aragam KG et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 50(9), 1219–1224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Läll K, Lepamets M, Palover M et al. Polygenic prediction of breast cancer: comparison of genetic predictors and implications for risk stratification. BMC Cancer 19(1), 557 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitley KV, Tueller JA, Weber KS. Genomics education in the era of personal genomics: academic, professional, and public considerations. Int. J. Mol. Sci. 21(3), 768 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.International Society of Nurses in Genetics. Genetics/Genomics Nursing: Scope and Standards of Practice/International Society of Nurses in Genetics. American Nurses Association, Silver Springs, MD, USA: (2016). [Google Scholar]
- 45.USFDA. Table of Pharmacogenomic Biomarkers in Drug Labeling (2021) (6 June Accessed 2021). www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling
- 46.Relling MV, Klein TE, Gammal RS, Whirl-Carrillo M, Hoffman JM, Caudle KE. The Clinical Pharmacogenetics Implementation Consortium: 10 years later. Clin. Pharmacol. Ther. 107(1), 171–175 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.University of Pittsburgh School of Pharmacy. Test2Learn. (2022). (Accessed 24 June 2022). www.test2learn.org
- 48.Adams SM, Anderson KB, Coons JC et al. Advancing pharmacogenomics education in the core PharmD curriculum through student personal genomic testing. Am. J. Pharm. Educ. 80(1), 3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burghardt KJ, Ward KM, Howlett BH, Burghardt PR. Personal genotyping and student outcomes in genetic and pharmacogenetic teaching: a systematic review and meta-analysis. Pharmacogenomics 22(7), 423–433 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Linderman MD, Sanderson SC, Bashir A et al. Impacts of incorporating personal genome sequencing into graduate genomics education: a longitudinal study over three course years. BMC Med. Genomics 11(1), 5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frick A, Benton C, Suzuki O et al. Implementing clinical pharmacogenomics in the classroom: student pharmacist impressions of an educational intervention including personal genotyping. Pharmacy (Basel) 6(4), 115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frick A, Benton CS, Scolaro KL et al. Transitioning pharmacogenomics into the clinical setting: training future pharmacists. Front. Pharmacol. 7, 241 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salari K, Karczewski KJ, Hudgins L, Ormond KE. Evidence that personal genome testing enhances student learning in a course on genomics and personalized medicine. PLOS ONE 8(7), e68853 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weitzel KW, McDonough CW, Elsey AR, Burkley B, Cavallari LH, Johnson JA. Effects of using personal genotype data on student learning and attitudes in a pharmacogenomics course. Am. J. Pharm. Educ. 80(7), 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]