Summary
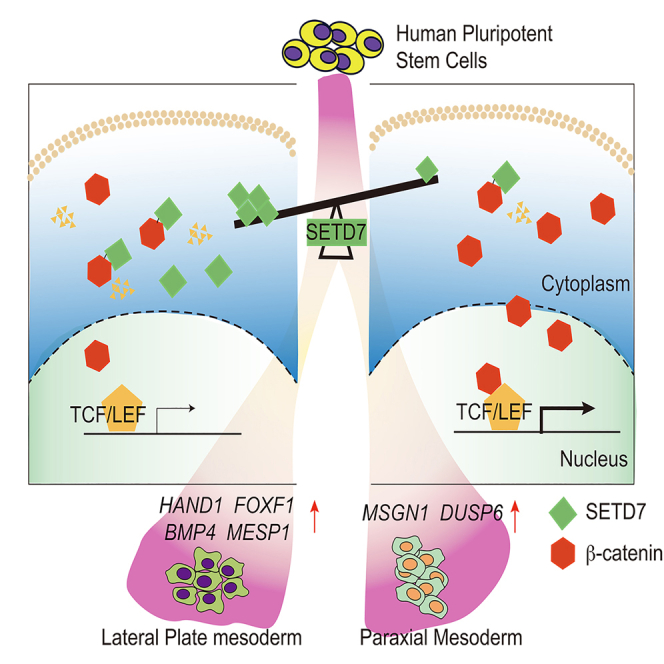
The role of SET domain containing 7 (SETD7) during human hematopoietic development remains elusive. Here, we found that deletion of SETD7 attenuated the generation of hematopoietic progenitor cells (HPCs) during the induction of hematopoietic differentiation from human embryonic stem cells (hESCs). Further analysis specified that SETD7 was required for lateral plate mesoderm (LPM) specification but dispensable for the generation of endothelial progenitor cells (EPCs) and HPCs. Mechanistically, rather than depending on its histone methyltransferase activity, SETD7 interacted with β-catenin at lysine residue 180 facilitated its degradation. Diminished SETD7 expression led to the accumulation of β-catenin and the consequent activation of the Wnt signaling pathway, which altered LPM patterning and facilitated the production of paraxial mesoderm (PM). Taken together, the findings indicate that SETD7 is related to LPM and PM patterning via posttranslational regulation of the Wnt/β-catenin signaling pathway, providing novel insights into mesoderm specification during hematopoietic differentiation from hESCs.
Subject areas: Biological sciences, Molecular biology, Developmental biology, Embryology
Graphical abstract
Highlights
-
•
SETD7 modulates later plate mesoderm (LPM) cell specification
-
•
The inhibitory effect of SETD7 on LPM is independent of the histone methylation
-
•
SETD7 promotes β-catenin degradation to facilitate LPM specification
Biological sciences; Molecular biology; Developmental biology; Embryology
Introduction
Human pluripotent stem cells (hPSCs) have the capacity to differentiate into cells of all three embryonic germ layers with indefinite self-renewal ability. In vitro generation of various blood cells from hPSCs provides a promising alternative strategy to overcome the critical shortage of blood supply and to meet the increasing demands of blood cells. Over the past decades, considerable advances have been made to develop various strategies to optimize conditions for hematopoietic differentiation of hPSCs.1,2,3 In general, the key steps related to hPSCs differentiation toward the hematopoietic lineage consist of mesodermal lateralization, endothelial progenitor cell (EPC) specification, and generation of hematopoietic progenitor cells (HPCs).4,5,6,7,8 Such an hPSCs differentiation process could be monitored by detecting the cells at each differentiation stage with specific cell surface marker(s), including APLNR+ (apelin receptor) for lateral plate mesodermal (LPM) cells, CD31+CD34+ for EPCs and CD43+ for HPCs.5,9,10,11,12 Despite great advances, the molecular mechanisms underlying the hematopoietic differentiation of hPSCs have yet to be fully elucidated.
Mesoderm is conventionally segregated into axial mesoderm, which organizes the axis of the body; paraxial mesoderm (PM), which forms somites and differentiates into muscle, bone and dermis; and LPM, which contributes to the formation of blood, heart and cardiovascular circulatory system, limb bud, and kidneys.13,14,15 Accumulating evidence shows that mesodermal patterning requires the tight orchestration of a variety of signaling pathways, including Wnt/β-catenin,16,17 TGFβ/BMP,18,19,20 FGF,21,22 etc. For example, the Wnt signaling pathway dictates the patterning of human PM vs. LPM by promoting PM and concomitantly repressing LPM.23 However, a high level of BMP signaling in the ventral domain is pivotal for LPM formation.24,25 Additionally, the FGF signaling pathway, by controlling Brachyury (T) and TBX6 expression, also plays an integral role in mesoderm patterning,26 and loss of FGF signaling in many vertebrate species results in a deficiency of PM induction.27,28,29,30,31 Although it is known that these signaling pathways interact with each other through temporal and spatial crosstalk and are highly coordinated to ensure proper mesoderm specification,23,32,33 it is still unclear how they are orchestrated during hematopoietic development.
Epigenetic regulation serves as another key player in inducing hematopoietic differentiation of hPSCs.34,35 Inhibiting EZH2 directs human embryonic stem cell (hESC) differentiation to the mesoderm and generates more mesenchymal stem/stromal cells.36 Loss of ezh1 promotes a developmental shift from arterial to hemogenic fate in the dorsal aorta in zebrafish.37 Reduced expression of EZH1 robustly favors lymphoid differentiation potential but has a limited impact on erythroid-myeloid potential during hematopoiesis induction from induced pluripotent stem cells (iPSCs).38 However, the interplay between epigenetic regulators and signaling pathways during hematopoietic differentiation has not been fully characterized.
SET domain containing 7 (SETD7) (also known as SET7/9, KMT7, SET7, and SET9), isolated from nuclear extracts of HeLa cells, was originally recognized as a lysine-specific methyltransferase for monomethylates histone 3 lysine 4 (H3K4me1) through a Su (var) 3–9, nhancer-of-zeste and trithorax (SET) domain 39.39,40,41 Recently, a growing body of studies has shown that SETD7 can also methylate various nonhistone substrates, such as P53,42 ERα,43 YAP,44 TAF-10,45 and STAT3,46 to modulate the DNA damage response, RNA polymerase II-dependent gene transcription, cell cycle, chromatin modulation, and cell differentiation. To date, except for the critical roles in cardiac differentiation,47 little is known about the role of SETD7 in hematopoietic differentiation derived from hESCs.
In this study, we revealed that depletion of SETD7 expression during hematopoietic differentiation derived from hESCs diminishes the generation of HPCs. Further analysis with temporally induced knockdown of SETD7 specified that SETD7 restricted its roles in the patterning of LPM. Mechanistically, we found that SETD7 modulates the Wnt signaling pathway by binding to β-catenin and thereby facilitating its degradation, which ultimately favors LPM formation at the expense of PM production.
Results
SETD7 expression peaks in APLNR+ LPM cells during the induction of hematopoietic differentiation from hESCs
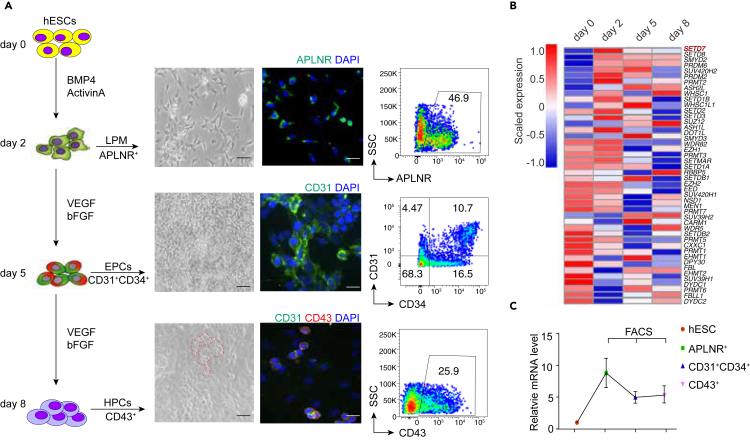
hESCs were subjected to hematopoietic differentiation in a previously described chemically defined system,4,5,48 which is capable of closely recapitulating primitive hematopoiesis. As shown in Figure 1A, during hematopoietic differentiation, hESCs exited from the pluripotent state and differentiated into the APLNR+ LPM at day 2. Then, specification of CD31+CD34+ EPCs occurred at day 5 and, ultimately, the emergence of CD43+ cobble-stone-like HPCs occurred at day 8 of differentiation. This hematopoietic differentiation process was also monitored with immunofluorescent staining.
Figure 1.
Dynamic expression of epigenetic modification genes during hematopoietic differentiation induced from hESCs
(A) Schematic illustration of the strategy for inducing hematopoietic differentiation from hESCs in vitro. APLNR+ LPM cells, CD31+CD34+ EPCs and CD43+ HPCs were monitored by morphology, immunofluorescence and flow cytometry during hematopoietic differentiation. Scale bars, 20 μm.
(B) Heatmap showing the dynamic expression of histone methyltransferase genes (42 genes) during hematopoietic differentiation (scaled by row). (n = 3).
(C) RT‒qPCR assay showing the relative expression of SETD7 in hESCs and FACS-sorted cells as indicated. (n = 3). Data are means ± SD.
Although epigenetic regulation plays a critical role during cell fate decisions,35 the functions of epigenetic modifications during hematopoiesis derived from hESCs remain incompletely understood. To identify the key epigenetic regulators affecting hematopoietic differentiation, we extracted the epigenetic modifiers directly involved in the DNA and histone epigenetic modifications and profiled their expression, utilizing our previously published time-course, high-throughput transcriptome data from cells differentiated at days 0, 2, 5, and 8 of hematopoietic differentiation.5,48 Based on their expression patterns, nearly all of these genes were dynamically expressed during differentiation. Some epigenetic modifiers, such as SETD7, SETD8, and SMYD2, were expressed at low levels at the hESC maintenance stage but increased at higher levels upon LPM generation (Figure 1B). In contrast, SETDB2, RPMT5, and CXXC1 were highly expressed during hESC self-renewal and were gradually downregulated during differentiation (Figure 1B). Among them, SETD7 was listed as the top dynamically expressed epigenetic gene by comparing its expression at day 2 to 0 as well as day 2 to day 5–8 (Figure 1B).
To further clarify the expression of SETD7 at each differentiation stage during hematopoietic development induced by hESCs, we FACS-sorted APLNR+ LPM cells, CD31+CD34+ EPCs, and CD43+ HPCs at days 2, 5, and 8 and performed RT-qPCR. Consistently, SETD7 expression was low in hESCs, peaked at the mesoderm lateralization stage and began to be downregulated in EPCs and HPCs (Figure 1C). In accordance with it, we also found the expression of SETD7 in mesoderm during in vivo embryonic development of humans and mice based on previous studies (Figures S1A and S1B).49,50 Collectively, this result suggests that SETD7 might be important for mesoderm lateralization and hematopoietic lineage commitment.
Deletion of SETD7 impairs hematopoietic differentiation
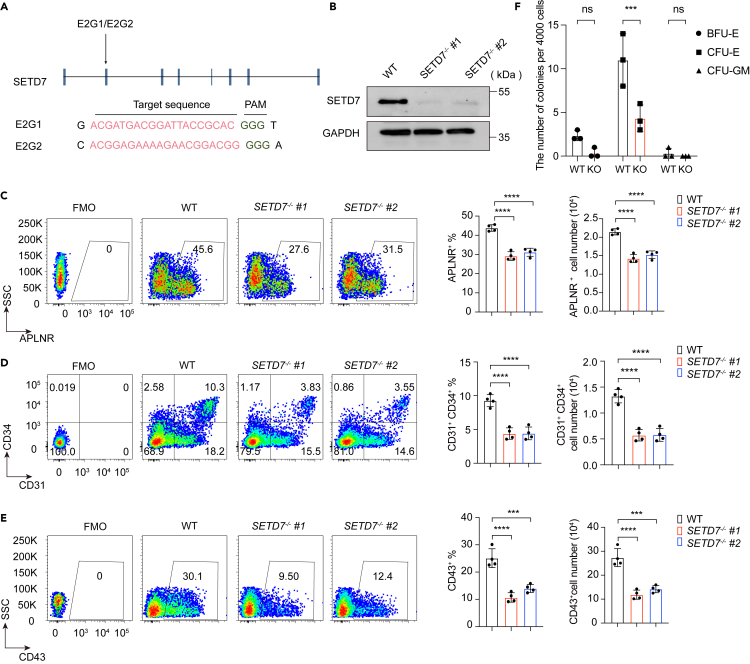
To reveal the potential function of SETD7 in hematopoietic lineage commitment during hESC differentiation, we deleted SETD7 expression using the lentiviral-mediated CRISPR/Cas9 genome editing system, with guide RNA sequences targeted in exon 2 (Figure 2A). Based on the single-cell colonies, we established two SETD7−/− cell lines (SETD7−/− #1 and #2) that harbored SETD7 mRNA frameshifts with consequently impaired expression (Figures 2B and S1C). Both of the genetically engineered SETD7−/− cell lines maintained typical hESC morphology (Figure S1D). Transcriptomic analysis indicated that SETD7 deletion did not alter the total expression of pluripotency genes, including core pluripotency genes such as POU5F1, NANOG, and SOX2 (Figures S1E–S1H). Thus, SETD7 deletion does not evidently influence pluripotency.
Figure 2.
Deletion of SETD7 does not influence the pluripotency of hESCs
(A) Schematic illustration of the strategy to target SETD7 exon 2 by CRISPR/Cas9 technology. E2G1 and E2G2 refer to two sgRNAs with targeting sequences (red) and protospacer adjacent motifs (PAMs) (green).
(B) Western blot showing the expression of SETD7 in WT, SETD7−/− #1 and #2 cell lines. GAPDH served as the loading control.
(C–E) Representative flow cytometry plots (left) and bar graphs showing the proportion (middle) and the cell number (right) of APLNR+ LPM cells at day 2 (C), CD31+CD34+ EPCs at day 5 (D) and CD43+ HPCs (E) at day 8 in WT, SETD7−/− #1 and SETD7−/− #2 cells during hematopoietic differentiation. (n = 4, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, ordinary one-way ANOVA). Data are means ± SD. (F) The number of colonies (BFU-E, CFU-E and CFU-GM) generated from day 12 differentiated HPCs of WT or SETD7−/− cells. (n = 3, ∗∗∗p < 0.001, ns, no significance, ordinary one-way ANOVA). Data are means ± SD. See also Figure S1 and Table S2.
We then induced wild-type (WT) and SETD7−/− cells to undergo hematopoietic differentiation. On day 2 of differentiation, we observed that SETD7 deletion significantly reduced the average production of APLNR+ LPM cells from 45.60% to 27.60%. With differentiation processing, we further detected decreased generation of CD31+CD34+ EPCs and CD43+ HPCs, from 10.30% to 3.55% and 30.10%–9.50%, respectively (Figures 2C–2E). In line with the reduced frequencies, we found that the numbers of APLNR+ LPM cells, CD31+CD34+ EPCs, and CD43+ HPCs were significantly decreased upon SETD7 deficiency (Figures 2C–2E). We further excluded the possibility of the reduced cell number arising from cell apoptosis and cell-cycle arrest (Figures S1I–S1L).
To reveal the underlying biological processes contributing to the hematopoietic differentiation defects upon SETD7 depletion, we performed RNA sequencing (RNA-seq) from the differentiated cells harvested on day 8. Indeed, we detected that the expression of hematopoiesis functional gene categories was downregulated in SETD7−/− cells (Figure S1M). To support this hypothesis, we carried out a colony-formation assay on HPCs at day 12 of differentiation and revealed that SETD7 depletion significantly attenuated the clonogenic capacities of CFU-E (Figure 2F). These results suggest that SETD7 is involved in inducing hematopoietic differentiation from hESCs.
SETD7 is indispensable for generating LPM cells but not EPCs and HPCs
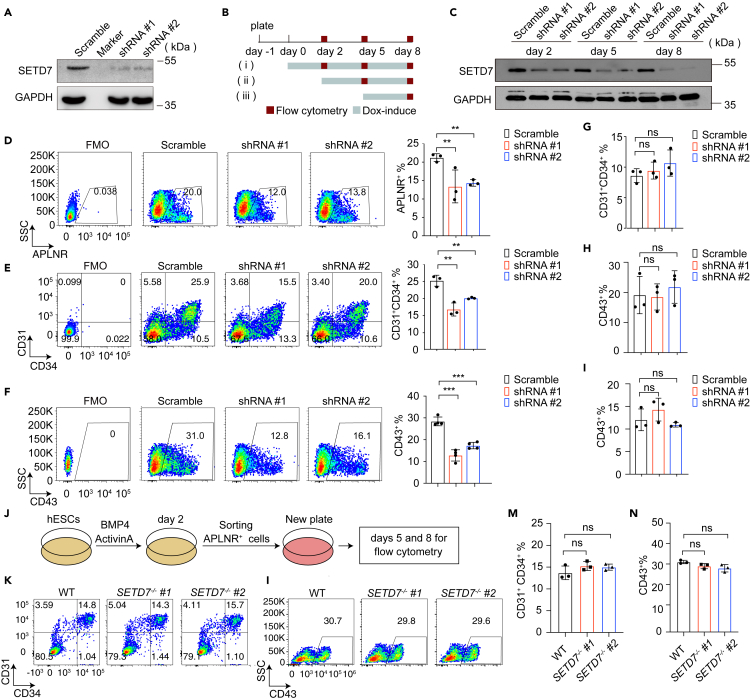
SETD7 deletion attenuated EPC specification and HPC generation. However, it remained unclear whether it was directly involved in such processes or was the consequence of defects inherited from LPM cells. To clarify, we introduced a doxycycline (Dox)-dependent inducible shRNA system to hESCs and generated stable clones carrying shRNAs targeting SETD7. After two days of Dox treatment, we detected significantly reduced SETD7 protein levels by Western blotting in the stable cell lines (Figure 3A).
Figure 3.
SETD7 deletion impairs LPM generation
(A) Western blot showing SETD7 protein levels after Dox induction for 2 days in hESCs harboring scramble control, Dox-inducible SETD7 KD #1 and #2 expression vectors. GAPDH served as the loading control.
(B) Experimental schematic showing the period of Dox exposure (gray lines) and the time points for flow cytometry analysis (red square). Dox was supplemented either throughout the culture period from day 0–8 (i), after mesoderm specification from day 2–8 (ii) or after EPC generation from day 5–8 (iii).
(C) Western blot showing SETD7 protein levels after Dox induction throughout the differentiation course. GAPDH served as the loading control.
(D–F) Representative flow cytometry plots (left) and bar graphs (right) showing the percentages of APLNR+ LPM cells (D), CD31+CD34+ EPCs (E) and CD43+ HPCs (F) treated with Dox from days 0–8. (n = 3, ∗∗p < 0.01, ∗∗∗p < 0.001, unpaired two-tailed Student’s t test). Data are means ± SD.
(G–H) Bar graphs showing the percentage of CD31+CD34+ EPCs (G) and CD43+ HPCs (H) with Dox treatment from days 2–8. (n = 3, ns, no significance, unpaired two-tailed Student’s t test). Data are means ± SD.
(I) Bar graph showing the percentage of CD43+ HPCs with Dox treatment from days 5–8. (n = 3, ns, no significance, unpaired two-tailed Student’s t test). Data are means ± SD.
(J) Experimental design. APLNR+ cells derived from WT and SETD7−/− cells on day 2 of differentiation were FACS-sorted and further differentiated toward the hematopoietic lineage.
(K–N) Representative flow cytometry plots (K, L) and the percentage (M, N) of CD31+CD34+ EPCs on day 5 and CD43+ HPCs on day 8. (n = 3, ns, no significance, unpaired two-tailed Student’s t test). Data are means ± SD. See also Figure S2 and Table S3.
We next induced these stable clones, harboring SETD7 shRNAs and scramble shRNAs, to differentiate toward the hematopoietic lineage. During the differentiation process, these cells were treated with Dox in a stepwise manner (Figure 3B). When supplementing Dox throughout the differentiation course (Figures 3B–3I), we detected diminished SETD7 protein levels in shRNA-expressing cells at days 2, 5, and 8 of differentiation (Figure 3C). Consequently, we revealed that the generation of APLNR+ LPM cells, CD31+CD34+ EPCs, and CD43+ HPCs (Figures 3D–3F) was dampened, phenocopying the SETD7−/− cells. Next, we began to treat the cells with Dox on day 2 of differentiation and onward (Figure 3C-(ii)), when cells had already completed mesoderm lateralization and started to differentiate into EPCs. Perturbation of SETD7 expression did not affect EPC specification or HPC production (Figures 3G–3H, S2B and S2C). Additionally, when treating the cells with Dox from days 5–8 of differentiation (Figure 3C-(iii)), we also failed to detect any alteration in the production of CD43+ HPCs in SETD7-deficient cells (Figures 3I, S2A and S2D).
Next, to discriminate whether the observed hematopoietic deficiency was LPM cell autonomous, we FACS-sorted APLNR+ LPM cells from WT and SETD7−/− cells, seeded an equal number of cells and induced them to further differentiate into EPCs and HPCs. There were no marked differences in the generation of EPCs and HPCs on days 5 and 8, respectively. This result indicated that it was likely that SETD7 depletion impaired the emergence rather than the quality of APLNR+ LPM cells, highlighting the possibility that SETD7 participates in the lineage choice of mesoderm specification rather than directly impairing the potential of APLNR+ LPM cells (Figures 3J–3N).
Collectively, these results indicate that SETD7 predominantly modulates LPM cell specification and exerts minor effects on subsequent EPC and HPC generation, in accordance with the elevated expression of SETD7 in LPM cells and the downregulation of SETD7 expression afterward during differentiation.
Ectopic expression of SETD7 partially restores the differentiation defects in SETD7−/− cells
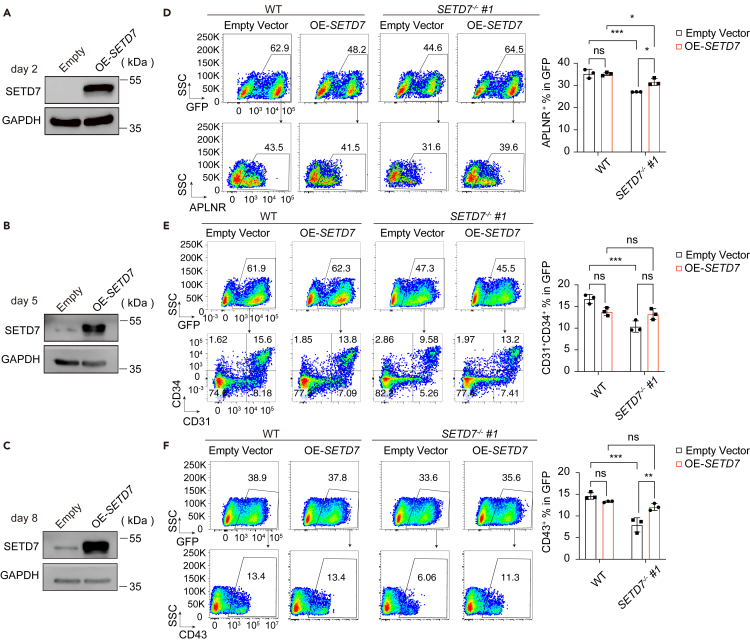
Since SETD7 reduction attenuated mesoderm lateralization and consequently impaired hematopoietic generation, we then wondered whether ectopic expression of SETD7 could rescue such differentiation defects in SETD7−/− cells. To test this possibility, we established Dox-induced SETD7 overexpression cell lines harboring a GFP expression cassette in WT and SETD7-knockout cells. We confirmed that SETD7 was overexpressed by Western blotting in WT cells at days 2, 5, and 8 of differentiation (Figures 4A–4C). Next, we induced the WT and SETD7−/− cells harboring either a control empty vector or a SETD7 overexpression vector, respectively, to undergo hematopoietic differentiation with Dox administered throughout. SETD7 overexpression rescued, at least in part, the yield of APLNR+ LPM cells and the subsequent generation of CD31+CD34+ EPCs and CD43+ HPCs (Figures 4D–4G). Interestingly, the overexpression of SETD7 in WT cells throughout differentiation did not affect the generation of LPM cells, EPCs and HPCs (Figures 4D–4F). Taken together, our results show that overexpression of SETD7 in SETD7−/− cells can partially rescue the defects in LPM generation, whereas excessive SETD7 expression seems redundant during normal hematopoietic differentiation.
Figure 4.
Overexpression of SETD7 can partially rescue hematopoietic differentiation deficiency in SETD7−/− cells
(A–C) Western blot showing the SETD7 protein levels in cells harboring an empty or a Dox-inducible SETD7 overexpression vector at days 2, 5 and 8 of hematopoietic differentiation. GAPDH served as a loading control.
(D–F) Representative flow cytometry plots (left) and bar graphs (right) showing the percentage of APLNR+ LPM cells at day 2 of differentiation (D), CD31+CD34+ EPCs at day 5 of differentiation (E) and CD43+ HPCs at day 8 of differentiation (F) in GFP+ WT and SETD7−/− #1 cells without or with SETD7 overexpression. (n = 3, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns, no significance, ordinary one-way ANOVA). Data are means ± SD. See also Table S1.
The inhibitory effect of SETD7 on APLNR+ LPM cells is independent of histone methylation
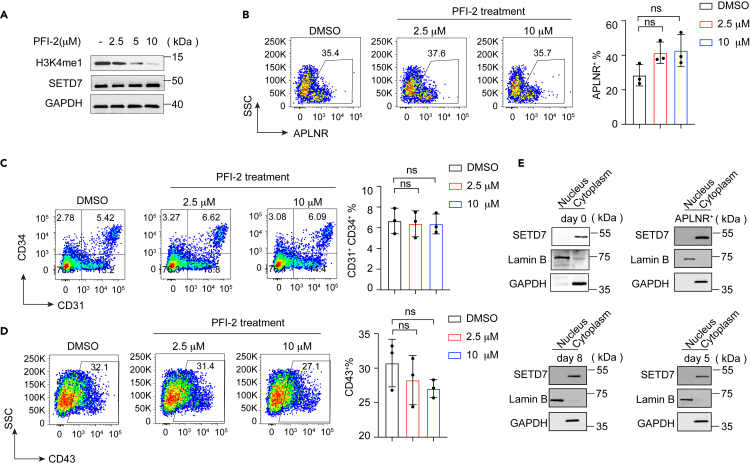
To determine whether the inhibitory effects of SETD7 on APLNR+ LPM cells arose from altered H3K4me1 modifications, we treated WT cells with (R)-PFI-2. (R)-PFI-2 is a selective and potent small-molecule inhibitor targeting the catalytic methyltransferase activity of SETD7, and it efficiently occupies the substrate lysine-binding groove of SETD7 to prevent its methylating histone.51 We found that supplementation with (R)-PFI-2 indeed reduced the global levels of H3K4me1 in a dose-dependent manner in H1 cells (Figure 5A).
Figure 5.
(R)-PFI-2 exhibited negligible effects on hematopoietic differentiation induced from hESCs
(A) Western blot showing H3K4me1 and SETD7 protein levels after treatment with (R)-PFI-2 in hESCs. GAPDH was used as a loading control.
(B–D) Representative flow cytometry plots (left) and bar graphs (right) showing the percentage of APLNR+ LPM cells (B), CD31+CD34+ EPCs (C) and CD43+ HPCs (D) in cells at days 2, 5 and 8 of differentiation with or without (R)-PFI-2 treatment, respectively. (n = 3, ns, no significance, unpaired two-tailed Student’s t test). Data are means ± SD.
(E) Western blot showing the nuclear and cytoplasmic expression of SETD7 in hESCs, FACS-sorted APLNR+ cells on day 2, and cells on day 5 and day 8 of hematopoietic differentiation. GAPDH served as the cytoplasmic loading control, and Lamin B served as the nuclear loading control.
Then (R)-PFI-2 was supplemented throughout the differentiation course at concentrations of 2.5 and 10 μM. Unexpectedly, (R)-PFI-2 exhibited minor effects on hematopoietic differentiation, including the generation of LPM cells (Figures 5B–5D). Further analysis elucidated that such negligible effects of (R)-PFI-2, in part, were attributable to the nearly complete absence of SETD7 expression in the nucleus during hematopoietic differentiation (Figure 5E). Therefore, it appears likely that SETD7 modulates hematopoietic differentiation, especially the generation of LPM cells, is independent of alterations in H3K4me1 in these cells.
The Wnt signaling pathway participates in SETD7-mediated LPM cell generation
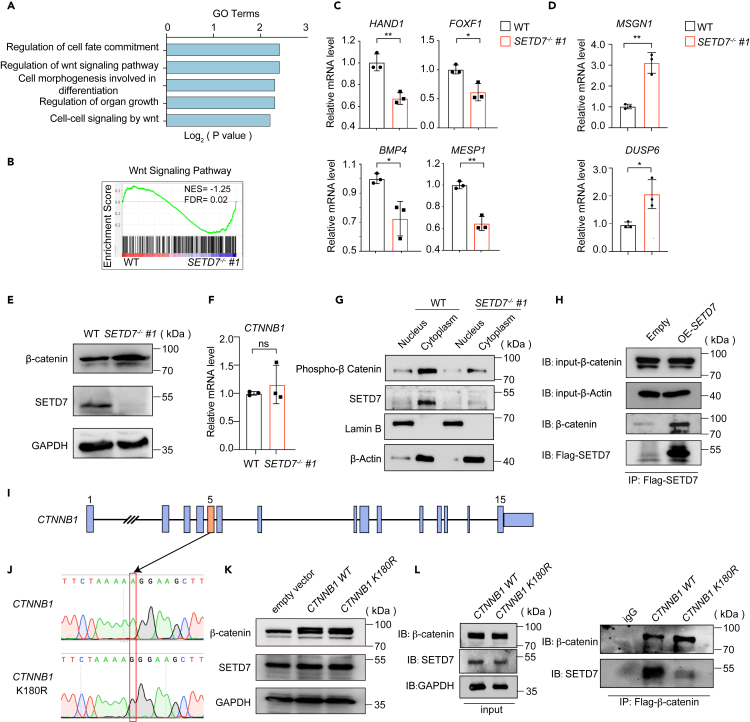
To determine the underlying molecular mechanisms mediated by SETD7, we performed transcriptomic analysis of SETD7−/− cells on day 2 of differentiation. Gene Ontology (GO) analysis of differentially expressed genes (DEGs) showed that the Wnt signaling pathway was significantly enriched in SETD7−/− cells compared with WT cells (Figure 6A), in line with gene set enrichment analysis (GSEA) (Figure 6B). Furthermore, recent studies have revealed that the delicate balance of the Wnt signal ensures the proper formation of PM and LPM.23,52 Enhancing the Wnt signal tilts differentiation toward the PM, while reducing its activity facilitates the formation of LPM.23 Consistent with this observation, we also detected reduced expression of LPM marker genes, such as HAND1,53 FOXF1,54 BMP4,55 and MESP156 (Figure 6C), but elevated expression of genes correlated with the PM, such as MSGN1 and DUSP6 in SETD7−/− cells23,57 (Figure 6D).
Figure 6.
Linking β-catenin with SETD7-mediated LPM cell generation
(A) GO enrichment of the upregulated genes in SETD7−/− #1 cells at day 2 of hematopoietic differentiation.
(B) GSEA plot depicting the enrichment of the Wnt signaling pathway in WT and SETD7−/− #1 cells at day 2 of hematopoietic differentiation.
(C–D) RT-qPCR assay showing the relative mRNA expression of designed genes in WT and SETD7−/− #1 cells at day 2 of differentiation. (n = 3, ∗p < 0.05, ∗∗p < 0.01, unpaired two-tailed Student’s t test). Data are means ± SD.
(E) Western blot showing the protein levels of β-catenin and SETD7 in WT and SETD7−/− #1 hESCs. GAPDH was used as a loading control.
(F) RT-qPCR assay representing the relative mRNA expression of CTNNB1 in WT and SETD7−/− #1 cells at day 2 of differentiation. (n = 3, ns, no significance, unpaired two-tailed Student’s t test). Data are means ± SD.
(G) Western blot showing the expression of phosphorated β-catenin and SETD7 in nuclear and cytoplasmic of WT and SETD7−/− #1 cells at day 2 of differentiation. Lamin B was used as the nuclear loading control, and β-actin was used as the cytoplasmic loading control.
(H) Coimmunoprecipitation of flag-tagged SETD7 and endogenous β-catenin in control (empty vector) and SETD7-overexpressing hESCs. Ten percent of the input was used as the total protein control.
(I) Schematic illustration of the strategy used to construct the CTNNB1 K180R mutant expression vector.
(J) Sanger sequencing confirmed the K180R mutation.
(K) Western blot showing the expression of β-catenin and SETD7 in hESCs infected by the empty vector, CTNNB1 WT and CTNNB1 K180R-mutant vectors. GAPDH was used as the loading control.
(L) Coimmunoprecipitation of β-catenin with anti-FLAG antibody and endogenous SETD7 in control (empty vector), CTNNB1 WT and CTNNB1 K180R mutant hESCs. Ten percent of the input was used as the total protein control. See also Figure S3 and Table S4.
Emerging evidence indicates that LPM differentiates from mid-primitive streak progenitors, while PM differentiates from anterior primitive streak progenitors.23 To further trace the potential influence of SETD7 on the specification of primitive steak, we tested genes critical for mid and anterior primitive streak progenitors on day one of differentiation. We found that marker genes of the anterior primitive streak, such as TBX6, GSC, and EMOES, were upregulated, whereas signature genes of the mid/anterior primitive streak, such as FOXF1, MESP1, MESP2, and EVX1, were downregulated upon SETD7 depletion (Figure S3A). These results suggested that SETD7 is involved in primitive streak development and the resultant bifurcation of the LPM and PM patterning.
To investigate how SETD7 modulates the Wnt signaling pathway, we examined its key mediator β-catenin and found that β-catenin protein was more abundant in SETD7−/− cells than in WT control cells at day 2 of differentiation (Figure 6E), whereas no noticeable difference was identified at the transcript level (Figure 6F). This result led us to speculate that SETD7 might exert posttranslational regulation of β-catenin. To test this hypothesis, we examined the level of phosphorylated β-catenin. Phosphorylated β-catenin can be recognized by phosphokinase glycogen synthase kinase (GSK)-3β for its degradation.58 After separating the proteins from the nucleus and cytoplasm, we found that the level of phosphorylated β-catenin was much lower in the cytoplasm of SETD7−/− cells than in the cytoplasm of its WT control counterparts, indicating that the absence of SETD7 results in the accumulation of β-catenin (Figure 6G). Prior studies have demonstrated that SETD7 can methylate nonhistone proteins,43,45,59 including β-catenin, where SETD7 methylates β-catenin to facilitate (GSK)-3β-mediated degradation in 293T and HeLa cells.60 We conducted an immunoprecipitation assay and confirmed the endogenous and exogenous interaction between SETD7 and β-catenin in SETD7-overexpressing H1 cells as well as cells on day 2 of hematopoietic differentiation when APLNR+ LPM cells composed nearly 50% of the cell population (Figures 6H, S3B and S3C).
SETD7 protein monomethylates β-catenin at lysine residue 180 in both 293T and HeLa cells.60 Inspired by this evidence, we introduced the CTNNB1 (encoding β-catenin) K180R mutant expression vector (Figures 6I and 6J). Compared with that of the WT β-catenin protein, the interaction of K180R mutant β-catenin with SETD7 was greatly abolished (Figures 6K and 6L), indicating that the lysine residue 180 of β-catenin is essential for SETD7-mediated β-catenin methylation.
Taken together, our results demonstrate that SETD7 protein interacts with β-catenin at lysine residue 180 to promote its degradation, probably by methylation, and consequently decreases the activity of the Wnt signaling pathway, dictating the formation of PM and LPM.
Discussion
In this study, we revealed the role of SETD7 in LPM specification during hematopoiesis inducing from hESCs. Intriguingly, instead of massive alteration of H3K4me1 modifications, we found that SETD7 finely balances the level of Wnt signaling to modulate PM and LPM patterning. Deletion of SETD7 leads to the accumulation of β-catenin, the key mediator of the Wnt pathway, and contributes to the defects in LPM formation and hematopoiesis.
During mesoderm patterning, PM is primarily responsible for the formation of somites, muscle, bone, and dermis, while LPM contributes to forming blood, the heart, and the cardiovascular circulatory system.13,14,15 Prior studies have demonstrated that the Wnt signaling pathway is a crucial mediator for the specification between PM and LPM.17,23,61 Genetic depletion of Wnt3a results in the absence of PM in mouse embryos.28,62 Wnt-1 and Wnt7a promote myogenesis from PM by activating Myf5 and MyoD, respectively.63,64 During in vitro induction of hPSCs differentiation, Wnt activation by GSK3 inhibition induces paraxial signatures while suppressing lateral signatures. In contrast, Wnt inhibition elicits LPM while blocking paraxial fate.23 However, how the Wnt pathway is modulated in mesoderm specification is still not fully understood. Here, we revealed that Wnt signaling, at least in part, is regulated by SETD7 via the posttranslational modification of β-catenin, which aids in mediating PM and LPM patterning upon induction of hESC hematopoietic differentiation.
The pleiotropic roles of SETD7 have been explored during development and differentiation. In agreement with a prior study,65 we also found that SETD7 is expressed at a low level in hESCs and that its abrogation minimally affects the pluripotency of hESCs, which might be relevant to the suppression of the SETD7 promoter region by the pluripotent proteins POU5F1, NANOG, and SOX2.47,65 In accordance with hESCs, SETD7 is also not expressed in quiescent adult muscle stem cells.44 Nonetheless, SETD7 could participate in the differentiation process. Upon mesoderm and endoderm specification, SETD7 facilitates mesoderm formation by interacting with components of the SWI/SNF chromatin-remodeling complex (e.g., BRG1) and recruiting them to the promoters of mesoderm-specific genes (e.g., T, EVX1, EOMES).47 During further mesoderm patterning, we illustrated that SETD7 is important for PM and LPM specification. The deletion of SETD7 attenuated LPM formation, which can further differentiate into blood and heart. Interestingly, during the induction of hematopoietic differentiation from hESCs, except for the mesodermal stage, we discovered that SETD7 is dispensable for EPC specification and HPC production. In contrast, SETD7 is necessary for cardiac lineage differentiation derived from hESCs.47 SETD7 can sequentially bind to well-known cardiac transcription factors, such as NKX2.5, TBX5, and GATA4, and cardiac maturation/morphogenesis genes, such as TNNT1, TNNT2, ACTC1, and MYH6, in cardiac progenitors and terminally differentiated cardiomyocytes.47 These results suggest that in the lineage differentiation of lateral plate mesoderm, SETD7 plays distinct roles in hematopoietic and cardiac lineage differentiation. Intriguingly, overexpression of SETD7 impacts neither hematopoietic nor cardiac differentiation, suggesting that SETD7 is important but insufficient to drive either blood or cardiac differentiation from hESCs.
Accumulating reports demonstrated that SETD7, as a lysine methyltransferase, mediates nonhistone gene methylation, including that of NFκB-p65,66,67 INS1/2,68 ER,43 and YY169 in various tissues and cells. In this study, we also elucidated that (R)-PFI-2, a selective and potent small-molecule inhibitor of the catalytic methyltransferase activity of SETD7,51 did not affect hematopoietic differentiation, suggesting that the mechanism by which SETD7 drives hematopoietic differentiation might be independent of its particular histone enzymatic activity. Consistently, (R)-PFI-2 also failed to influence cardiac mesodermal progenitor cells.47 Thus, it is likely that the H3K4me1 enzymatic activity of SETD7 is dispensable in either mesoderm formation47 or LPM patterning.
Instead, SETD7 mediates the posttranslational regulation of β-catenin. We found that SETD7 is able to bind with β-catenin at lysine residue 180 and that abolishing SETD7 expression leads to the accumulation of β-catenin protein but not its mRNA transcripts. Cytoplasmic β-catenin is usually degraded by GSK3β in the absence of the Wnt ligand.58 Given the accumulation of cytoplasmic β-catenin in mesodermal cells with abrogated SETD7 expression, we propose that SETD7 dictates LPM patterning mainly by controlling the protein level of β-catenin. Conversely, a recent study linked SETD7-Hippo/YAP-Wnt/β-catenin signaling to epithelial proliferation.70 SETD7-dependent methylation of YAP facilitates the nuclear translocation of β-catenin.70 Analogously, during skeletal muscle differentiation from muscle stem cells, SETD7 also stimulates β-catenin nuclear localization to activate the muscle differentiation transcriptional program.44 Therefore, it seems likely that the SETD7-mediated Wnt/β-catenin signaling pathway is context and cellular-dependent.
In summary, we have identified the pivotal role of SETD7 in LPM generation during hESC hematopoietic differentiation, which is mediated by β-catenin in a posttranslational modification manner. This study provides insights into the molecular regulation of hematopoiesis, especially mesoderm specification, from hPSCs.
Limitations of the study
Our findings indicate a SETD7-related LPM and PM patterning via PTM of β-catenin. Diminished SETD7 expression leads to the accumulation of β-catenin and consequently activates the Wnt signaling pathway. However, several limitations were encountered in this study. Firstly, our monolayer differentiation system of hESCs is biased for yolk sac-like/primitive hematopoiesis, whether SETD7 has a similar role during definitive hematopoiesis requires further studies. Secondly, we revealed SETD7 could interact with β-catenin at lysine residue 180 and whether there are other potential interacting sites awaits further investigations.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Human APJ Antibody | R&D | Cat# MAB8561; RRID: AB_2058383 |
| Anti-CD31 | Abcam | Cat# ab32457; RRID: AB_726369 |
| Anti-CD34 | Abcam | Cat# ab81289; RRID: AB_1640331 |
| Anti-CD43 | Abcam | Cat# ab101533; RRID: AB_10710687 |
| Alexa Fluor 488 | Invitrogen | Cat# A11034; RRID: AB_2576217 |
| Alexa Fluor 488 | Life Technologies | Cat# A21202; RRID: AB_141607 |
| Alexa Fluor 594 | Life Technologies | Cat# A-21207; RRID: AB_141637 |
| Anti-SETD7 | Abcam | Cat# ab124708; RRID: AB_10975432 |
| Anti-β-Catenin | CST | Cat# 9562s; RRID: AB_331149 |
| Anti-GAPDH | Proteintech | Cat# 60004-1-1g; RRID: AB_2107436 |
| Anti-Histone H3 (mono methyl K4) | Abcam | Cat# ab8895; RRID: AB_306847 |
| Anti-POU5F1 | CST | Cat# 2750S; RRID: AB_823583 |
| Anti-NANOG | CST | Cat# 3580S; RRID: AB_2150399 |
| Anti-SOX2 | CST | Cat# 3579S; RRID: AB_2195767 |
| Phospho-β-Catenin | CST | Cat# 4176T; RRID: AB_1903923 |
| HRP-conjugated AffiniPure Goat Anti-mouse IgG | Proteintech | Cat# SA00001-1; RRID: AB_2722565 |
| Anti-rabbit IgG HRP-linked Antibody | CST | Cat# 7074P2; RRID: AB_2099233 |
| Lamin B1 | Proteintech | Cat# 66095-1; RRID: AB_11232208 |
| Human APJ-APC-conjugated Antibody | R&D | Cat# FAB8561A |
| Anti-FLAG | Sigma | Cat# F1804; RRID: AB_262044 |
| PE Mouse Anti-Human CD31 | BD Biosciences | Cat# 555446; RRID: AB_395839 |
| APC Mouse Anti-Human CD34 | BD Biosciences | Cat# 55582; RRID: AB_2228972 |
| Mouse IgG3 APC-conjugated Antibody | R&D | Cat# 1C007A |
| PE Mouse IgG1, κ Isotype Control | BD Biosciences | Cat# 559320; RRID: AB_397218 |
| propidium iodide (PI) | Solarbio | Cat# C0080 |
| DAPI | Solarbio | Cat# C0060 |
| APC Mouse Anti-Human CD43 | BD Biosciences | Cat# 560198; RRID: AB_1645460 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE228287 |
| Critical commercial assays | ||
| EasyScript® One-Step gDNA Removal and cDNA Synthesis SuperMix | transgene | Cat# N10613 |
| Hieff UNICON® Universal Blue qPCR SYBR Green Master Mix | YESEN | Cat# 11184ES03 |
| Nuclear and Cytoplasmic Protein Extraction Kit | Beyotime | Cat# P0028 |
| EasyScript® One-Step gDNA Removal and cDNA Synthesis SuperMix | transgene | Cat# N10613 |
| apoptosis detection kit | BD Biosciences | Cat# 556547 |
| protein G superparamagnetic beads | Invitrogen | Cat# 10003D |
| Lipofectamine Stem regent | Invitrogen | Cat# STEM00001 |
| Experimental models: Cell lines | ||
| hESC: H1 | WiCell | RRID: CVCL_9771 |
| SETD7 KO H1 | This paper | N/A |
| SETD7 KD H1 | This paper | N/A |
| SETD7 OE H1 | This paper | N/A |
| b-catenin OE H1 | This paper | N/A |
| b-catenin K180R OE H1 | This paper | N/A |
| Scramble control H1 | This paper | N/A |
| Empty Vector control H1 | This paper | N/A |
| Oligonucleotides | ||
| RT-qPCR primers | This paper | Table S4 |
| SETD7 gRNA1 | This paper | Table S2 |
| SETD7 gRNA2 | This paper | Table S2 |
| SETD7 inducible OE primers | This paper | Table S1 |
| SETD7 inducible KD primers | This paper | Table S3 |
| inducible KD Scramble primers | This paper | Table S3 |
| CTNNB1 inducible OE primers | This paper | Table S1 |
| CTNNB1 inducible K180R primers | This paper | Table S1 |
| Software and algorithms | ||
| FlowJo | FlowJo | https://www.flowjo.com |
| GSEA v4.0 | Subramanian et al.71 | http://software.broadinstitute.org/gsea/index.jsp |
| HISAT2-StringTie | Pertea et al.72 | http://ccb.jhu.edu/software.shtml |
| R v3.6 | R-Project | https://www.r-project.org/ |
| GraphPad Prism | GraphPad | https://graphpad.com |
| Chemicals, peptides, and recombinant proteins | ||
| Activin A | PeproTech | Cat# 120-14-1000 |
| BMP4 | PeproTech | Cat# 120-05ET-1000 |
| bFGF | PeproTech | Cat# 100-18B-100 |
| VEGF | PeproTech | Cat# 100-20-100 |
| TPO | PeproTech | Cat# 30018 |
| EPO | Prospec | Cat# 10064 |
| SCF | Prospec | Cat# CYT-255 |
| IL11 | PeproTech | Cat# 200-11 |
| IL6 | STEMCELL Technologies | Cat# 78050.1 |
| IL3 | Sigma | Cat# I1646 |
| GM-CSF | PeproTech | Cat# HZ-1002 |
| GCSF | PeproTech | Cat# HZ-1207 |
| Flt-3L | PeproTech | Cat# 300-19 |
| Y27632 | Calbiochem | Cat# 688000 |
| hydrocortisone | Sigma | Cat# H0888 |
| L-ascorbic acid | Sigma | Cat# A8960-5G |
| heparin | Sigma | Cat# H3149-500KU-9 |
| mTeSR+supplement | Stem Cell | Cat# 85852 |
| DMEM/F-12(1;1) basic | Gibco | Cat# C11330500BT |
| Fetal bovine serum | Gibco | Cat# 16000-044 |
| Penicillin-Streptomycin | Gibco | Cat# 15140-122 |
| Accutase | Gibco | Cat# AB11105-01 |
| Costom TESR+supplement | Stem Cell | Cat# 05892 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Lihong Shi (shilihongxys@ihcams.ac.cn).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Induction of hematopoietic differentiation from hESCs
Human embryonic stem cells (H1) were purchased from ATCC.4 Cells were maintained on Matrigel-coated plates using mTESR1 medium (STEMCELL Technologies) consisting of 1% penicillin-streptomycin. To induce hematopoietic differentiation, H1 cells were plated on growth factor-reduced (GFR) Matrigel-coated plates and cultured overnight in mTESR1 with 10 μM Y27632. The next day, the culture medium was replaced by mTESR1 without select factors (STEMCELL Technologies), which was supplemented with 50 ng/ml Activin A (PeproTech) and 40 ng/ml BMP4 (PeproTech) from day 0 to day 2 and 50 ng/ml bFGF (PeproTech) and 40 ng/ml VEGF (PeproTech) from day 2 to day 8 of culture.
Methods details
Colony-formation assays
Day-8 differentiated cells were induced to generate CD45+ HPCs with StemPro-34 basal medium (Gibco) supplemented with 50 ng/ml TPO (PeproTech), 6 U/ml EPO (Prospec), 20 ng/ml SCF (Prospec), 20 ng/ml IL11 (PeproTech), 15 ng/ml IL6 (STEMCELL Technologies), 15 ng/ml IL3 (Sigma), 20 ng/ml GM-CSF (PeproTech), 20 ng/ml GCSF (PeproTech), and 20 ng/ml Flt-3L (PeproTech) for 4 days. Then, the cells were seeded in MethoCult™ semisolid medium for colony-formation assays (STEMCELL Technologies). The CFU-E colonies were counted on day 7, while the BFU-E colonies and other CFUs were counted on day 14 after seeding. Colonies were identified according to the manufacturer’s instructions.
Establishment of SETD7-/- H1 cell lines
CRISPOR (http://crispor.tefor.net/)71 was used to design small guide RNAs targeting SETD7. Then, the oligonucleotides were cloned into the CRISPR-Cas9-Lenti-V2 vector. After infection, we treated the cells with 1 μg/ml puromycin for 2 weeks, and small colonies emerging from single cells were picked and expanded for Sanger sequencing. The primers used are listed in Table S1.
Establishment of the SETD7-overexpressing cell line
SETD7 cDNA was amplified from the cDNA of H1 cells and then cloned into a Tet-On inducible piggyBac transposon expression vector. The plasmids were cotransfected with transposase to H1 with Lipofectamine Stem regent (Invitrogen). After 10 days of puromycin selection, nearly all live cells exhibited puromycin resistance. Cells were induced by 1 μg/ml Dox for 2 days, and the expression was confirmed by Western blotting. The primers used are listed in Table S1.
Generation of stable Dox-inducible shRNA-mediated SETD7-knockdown cell lines
SETD7 shRNAs and a scrambled shRNA (Scr) were cloned into a Dox-inducible pLKO-Tet-On-shRNA lentiviral vector, and then the plasmids were transfected into 293T cells for viral packaging. Next, 1×105 H1 cells were seeded as single cells on Matrigel-coated plates for 48 hours before lentiviral infection with 8 ng/ml polybrene. After 1 μg/ml puromycin selection for approximately 10 days, the cells were induced by 1 μg/ml Dox for 2 days and collected for Western blot assay. The primers used are listed in Table S1.
Flow cytometry analysis and cell sorting
Cells were pretreated with Accutase (Invitrogen) for 3 minutes at 37°C and then washed with cold PBS. After 300×g centrifugation, 2×105 cells were resuspended in 100 ml of PBS and incubated with primary antibodies for 30 minutes at 4°C. Then, the cells were washed by using PBS with 2% FBS and 0.2 mM EDTA and used for flow cytometry analysis or sorting. The antibodies were as follows: Human APJ-APC-conjugated Antibody (R&D), PE Mouse Anti-Human CD31 (BD Biosciences), APC Mouse Anti-Human CD34 (BD Biosciences), APC Mouse Anti-Human CD43 (BD Biosciences), Mouse IgG3 APC-conjugated Antibody (R&D), PE Mouse IgG1, κ Isotype Control (BD Biosciences).
Cell cycle and apoptosis analysis
For the cell cycle assay, cells were fixed with 100% cold ethanol and incubated at 4°C overnight. The next day, the cells were washed with cold PBS supplemented with 1% BSA (Sigma) and stained using 200 μl propidium iodide (PI) staining buffer (1% BSA + 100 μg/ml RNase A (Tiangen Biotechnology) + 50 μg/ml PI (Solarbio) + 0.1% Triton X-100) at 37°C in the dark for 45 minutes. The cells were then subjected to flow cytometric analysis.
A total of 1×105 cells were prepared for apoptosis analysis using an apoptosis detection kit (BD Biosciences) according to the manufacturer’s instructions. The cells were then subjected to flow cytometric analysis.
RNA purification and RT-qPCR
Total RNA was extracted with TRIzol® Reagent (Invitrogen). We conducted reverse transcription with EasyScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (transgene) according to the manufacturer’s instructions. qPCR assays were performed using Hieff UNICON® Universal Blue qPCR SYBR Green Master Mix (YESEN) with a QuantStudio 5 real-time PCR system. All primers used are described in Table S1.
Nuclear and cytoplasmic protein extraction
A Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime) was used for nuclear and cytoplasmic protein extraction. Briefly, 2×106 cells were lysed in cytoplasmic protein extraction buffer A for 8 minutes. Then, cytoplasmic protein extraction buffer B was added for approximately 1 minute until greater than 90% of cells had ruptured membranes. After 12,000×g centrifugation, the upper liquid was harvested for cytoplasmic protein. Then, the nuclear pellets were washed 3 times to remove cytoplasmic residues. We used RIPA lysis buffer (Solarbio) to extract nuclear protein.
Immunofluorescence staining
Cells were fixed with 4% paraformaldehyde for 20 minutes at room temperature, washed and permeabilized with 0.2% Triton-X-100 for 15 minutes at room temperature. Then, they were blocked with 1% bovine serum albumin (BSA, Sigma) for 1 hour and incubated with primary antibodies overnight at 4°C. The next day, the cells were incubated with secondary antibody for 1 hour and 1 μg/ml DAPI for 5 minutes at room time and observed under a microscope. The antibodies were as follows: Human APJ Antibody (R&D), Anti-CD31 (Abcam), Anti-CD34 (Abcam), Anti-CD43 (Abcam), Alexa Fluor 488 (Invitrogen), Alexa Fluor 594 (Life Technologies), and Alexa Fluor 488 (Life Technologies).
Western blot analysis
A total of 1×106 cells were first lysed in 100 ml of RIPA lysis buffer (Solarbio) containing 1×protease and phosphatase inhibitor cocktail (Beyotime) on ice for 30 minutes. Second, SDS‒PAGE loading buffer was added to the lysate before heating at 100°C for 10 minutes (CWBIO). The lysate was loaded onto 10% SDS‒PAGE gels for running. After transferring, the membrane was blocked in 5% nonfat milk in PBS with 1% Tween-20 and incubated with primary antibodies overnight at 4°C. HRP-conjugated anti-rabbit or HRP-conjugated anti-mouse antibodies were used as the secondary antibody. The antibodies were as follows: Anti-SETD7 (Abcam), Anti-β-Catenin (CST), Anti-GAPDH (Proteintech), Anti-Histone H3 (mono methyl K4) (Abcam), Anti-POU5F1 (CST), Anti-NANOG (CST), Anti-SOX2 (CST), Phospho-β-Catenin (CST), Beta Actin (Proteintech), HRP-conjugated AffiniPure Goat Anti-mouse IgG (Proteintech), and an Anti-rabbit IgG HRP-linked Antibody (CST).
Coimmunoprecipitation
The cells were lysed in 0.5% NP40 buffer with 50 mM pH 7.5 Tris-HCl, 150 mM NaCl, 1×Protease and the phosphatase inhibitor cocktail for 30 minutes. After centrifugation, the supernatant was incubated with the antibody at 4°C overnight. The next day, 30 μl of protein G superparamagnetic beads (Invitrogen) were added and further incubated for 2 hours at 4°C before washing four times. Finally, the precipitated components were analyzed by Western blotting. The antibodies were as follows: Anti-SETD7 (Abcam), Anti-FLAG (Sigma), Lamin B1 (Proteintech), and β-Catenin (CST).
RNA sequencing and data processing
WT and SETD7-/- cells were collected at day 0, day 2 and day 8 of hematopoietic differentiation induction. Total RNA was extracted according to the manufacturer’s instructions (Invitrogen). The library was constructed and sequenced by Novogene Co., Ltd. (Beijing, China) using the Illumina HiSeq platform. After quality control, the clean data were used as input files to map the human reference genome (GRCh38/hg38) using the HISAT2-StringTie analysis pipeline with default parameters.
GSEA
GSEA software (version 4.0) was downloaded from the Broad Institute (https://www.gsea-msigdb.org/gsea/downloads.jsp) and used for analysis. Gene sets were obtained through the Molecular Signatures Database (http://www.gsea-msigdb.org/gsea/msigdb/search.jsp), and we used the normalized expression dataset as the input file.
Quantification and statistical analysis
All data are expressed as the mean ± SD. For multiple comparisons, we performed one-way ANOVA followed by Dunnett’s test. For others, we performed a two-tailed Student’s t-test. P < 0.05 was considered to indicate statistical significance, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001, ns, no significance. All experiments were conducted with at least 3 biological replicates.
Acknowledgments
This work was supported by the grants from National Key Research and Development Program of China (2022YFA1103503 to L.S.), the National Natural Science Foundation of China (82225003 to L.S., 81870089 to L.S., 82100152 to J.T., 81890990 to J.T.), the CAMS Innovation Fund for Medical Sciences (2021-I2M-1-040 to L.S., 2022-I2M-JB-015 to L.S., 2021-I2M-1-073 to J.T.), the Haihe Laboratory of Cell Ecosystem Innovation Fund (22HHXBSS00017 to L.S.), the Nonprofit Central Research Institute Fund of Chinese Academy of Medical Sciences (2022-RC180-04 to L.S.).
Author contributions
L.S. designed and supervised the study; D.W. performed the all experiments with the help of Y.L., X.H., H.W., C.X., J.G., J.L., and J.T; C.X performed the bioinformatics analysis; L.S. wrote the manuscript with the help from D.W. and X.J.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: May 19, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106917.
Supplemental information
Data and code availability
-
•
The datasets generated during this study are available at the Gene Expression Omnibus (GEO) under the accession number GSE228287. The following public datasets were downloaded from GEO and reanalyzed: GSE92245.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Ditadi A., Sturgeon C.M., Keller G. A view of human haematopoietic development from the Petri dish. Nat. Rev. Mol. Cell Biol. 2017;18:56–67. doi: 10.1038/nrm.2016.127. [DOI] [PubMed] [Google Scholar]
- 2.Choi K.D., Vodyanik M., Slukvin I.I. StemBook. Harvard Stem Cell Institute; 2008. Hematopoietic differentiation. Copyright: © 2012 Kyung-Dal Choi, Maxim Vodyanik, and Igor I Slukvin. [PubMed] [Google Scholar]
- 3.Slukvin I.I. Hematopoietic specification from human pluripotent stem cells: current advances and challenges toward de novo generation of hematopoietic stem cells. Blood. 2013;122:4035–4046. doi: 10.1182/blood-2013-07-474825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y., Wang D., Wang H., Huang X., Wen Y., Wang B., Xu C., Gao J., Liu J., Tong J., et al. A splicing factor switch controls hematopoietic lineage specification of pluripotent stem cells. EMBO Rep. 2021;22:e50535. doi: 10.15252/embr.202050535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H., Liu C., Liu X., Wang M., Wu D., Gao J., Su P., Nakahata T., Zhou W., Xu Y., et al. MEIS1 regulates hemogenic endothelial generation, megakaryopoiesis, and thrombopoiesis in human pluripotent stem cells by targeting TAL1 and FLI1. Stem Cell Rep. 2018;10:447–460. doi: 10.1016/j.stemcr.2017.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Y., Liu B., Xu J., Wang J., Wu J., Shi C., Xu Y., Dong J., Wang C., Lai W., et al. Derivation of pluripotent stem cells with in vivo embryonic and extraembryonic potency. Cell. 2017;169:243–257.e25. doi: 10.1016/j.cell.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang H., Mesquitta W.T., Jung H.S., Moskvin O.V., Thomson J.A., Slukvin I.I. GATA2 is dispensable for specification of hemogenic endothelium but promotes endothelial-to-hematopoietic transition. Stem Cell Rep. 2018;11:197–211. doi: 10.1016/j.stemcr.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu S.J., Feng Q., Caballero S., Chen Y., Moore M.A.S., Grant M.B., Lanza R. Generation of functional hemangioblasts from human embryonic stem cells. Nat. Methods. 2007;4:501–509. doi: 10.1038/nmeth1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H., Wang M., Wen Y., Xu C., Chen X., Wu D., Su P., Zhou W., Cheng T., Shi L., Zhou J. Biphasic regulation of mesenchymal genes controls fate switches during hematopoietic differentiation of human pluripotent stem cells. Adv. Sci. 2020;7:2001019. doi: 10.1002/advs.202001019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ditadi A., Sturgeon C.M., Tober J., Awong G., Kennedy M., Yzaguirre A.D., Azzola L., Ng E.S., Stanley E.G., French D.L., et al. Human definitive haemogenic endothelium and arterial vascular endothelium represent distinct lineages. Nat. Cell Biol. 2015;17:580–591. doi: 10.1038/ncb3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramos-Mejía V., Navarro-Montero O., Ayllón V., Bueno C., Romero T., Real P.J., Menendez P. HOXA9 promotes hematopoietic commitment of human embryonic stem cells. Blood. 2014;124:3065–3075. doi: 10.1182/blood-2014-03-558825. [DOI] [PubMed] [Google Scholar]
- 12.Choi K.D., Vodyanik M.A., Togarrati P.P., Suknuntha K., Kumar A., Samarjeet F., Probasco M.D., Tian S., Stewart R., Thomson J.A., Slukvin I.I. Identification of the hemogenic endothelial progenitor and its direct precursor in human pluripotent stem cell differentiation cultures. Cell Rep. 2012;2:553–567. doi: 10.1016/j.celrep.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawson K.A., Meneses J.J., Pedersen R.A. Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development. 1991;113:891–911. doi: 10.1242/dev.113.3.891. [DOI] [PubMed] [Google Scholar]
- 14.Tam P.P., Beddington R.S. The formation of mesodermal tissues in the mouse embryo during gastrulation and early organogenesis. Development. 1987;99:109–126. doi: 10.1242/dev.99.1.109. [DOI] [PubMed] [Google Scholar]
- 15.Rosenquist G.C. Location and movements of cardiogenic cells in the chick embryo: the heart-forming portion of the primitive streak. Dev. Biol. 1970;22:461–475. doi: 10.1016/0012-1606(70)90163-6. [DOI] [PubMed] [Google Scholar]
- 16.Kishimoto K., Furukawa K.T., Luz-Madrigal A., Yamaoka A., Matsuoka C., Habu M., Alev C., Zorn A.M., Morimoto M. Bidirectional Wnt signaling between endoderm and mesoderm confers tracheal identity in mouse and human cells. Nat. Commun. 2020;11:4159. doi: 10.1038/s41467-020-17969-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marvin M.J., Di Rocco G., Gardiner A., Bush S.M., Lassar A.B. Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev. 2001;15:316–327. doi: 10.1101/gad.855501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki A., Kaneko E., Maeda J., Ueno N. Mesoderm induction by BMP-4 and -7 heterodimers. Biochem. Biophys. Res. Commun. 1997;232:153–156. doi: 10.1006/bbrc.1997.6219. [DOI] [PubMed] [Google Scholar]
- 19.Zhang P., Li J., Tan Z., Wang C., Liu T., Chen L., Yong J., Jiang W., Sun X., Du L., et al. Short-term BMP-4 treatment initiates mesoderm induction in human embryonic stem cells. Blood. 2008;111:1933–1941. doi: 10.1182/blood-2007-02-074120. [DOI] [PubMed] [Google Scholar]
- 20.Nomura M., Li E. Smad2 role in mesoderm formation, left-right patterning and craniofacial development. Nature. 1998;393:786–790. doi: 10.1038/31693. [DOI] [PubMed] [Google Scholar]
- 21.Amaya E., Stein P.A., Musci T.J., Kirschner M.W. FGF signalling in the early specification of mesoderm in Xenopus. Development. 1993;118:477–487. doi: 10.1242/dev.118.2.477. [DOI] [PubMed] [Google Scholar]
- 22.Schulte-Merker S., Smith J.C. Mesoderm formation in response to Brachyury requires FGF signalling. Curr. Biol. 1995;5:62–67. doi: 10.1016/s0960-9822(95)00017-0. [DOI] [PubMed] [Google Scholar]
- 23.Loh K.M., Chen A., Koh P.W., Deng T.Z., Sinha R., Tsai J.M., Barkal A.A., Shen K.Y., Jain R., Morganti R.M., et al. Mapping the pairwise choices leading from pluripotency to human bone, heart, and other mesoderm cell types. Cell. 2016;166:451–467. doi: 10.1016/j.cell.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferretti E., Hadjantonakis A.K. Mesoderm specification and diversification: from single cells to emergent tissues. Curr. Opin. Cell Biol. 2019;61:110–116. doi: 10.1016/j.ceb.2019.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishimatsu S., Thomsen G.H. Ventral mesoderm induction and patterning by bone morphogenetic protein heterodimers in Xenopus embryos. Mech. Dev. 1998;74:75–88. doi: 10.1016/s0925-4773(98)00070-7. [DOI] [PubMed] [Google Scholar]
- 26.Ciruna B., Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev. Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- 27.Griffin K., Patient R., Holder N. Analysis of FGF function in normal and no tail zebrafish embryos reveals separate mechanisms for formation of the trunk and the tail. Development. 1995;121:2983–2994. doi: 10.1242/dev.121.9.2983. [DOI] [PubMed] [Google Scholar]
- 28.Chalamalasetty R.B., Garriock R.J., Dunty W.C., Jr., Kennedy M.W., Jailwala P., Si H., Yamaguchi T.P. Mesogenin 1 is a master regulator of paraxial presomitic mesoderm differentiation. Development. 2014;141:4285–4297. doi: 10.1242/dev.110908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng C., Wynshaw-Boris A., Zhou F., Kuo A., Leder P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 30.Mitrani E., Ziv T., Thomsen G., Shimoni Y., Melton D.A., Bril A. Activin can induce the formation of axial structures and is expressed in the hypoblast of the chick. Cell. 1990;63:495–501. doi: 10.1016/0092-8674(90)90446-l. [DOI] [PubMed] [Google Scholar]
- 31.Amaya E., Musci T.J., Kirschner M.W. Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell. 1991;66:257–270. doi: 10.1016/0092-8674(91)90616-7. [DOI] [PubMed] [Google Scholar]
- 32.Fürthauer M., Van Celst J., Thisse C., Thisse B. Fgf signalling controls the dorsoventral patterning of the zebrafish embryo. Development. 2004;131:2853–2864. doi: 10.1242/dev.01156. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka M., Jokubaitis V., Wood C., Wang Y., Brouard N., Pera M., Hearn M., Simmons P., Nakayama N. BMP inhibition stimulates WNT-dependent generation of chondrogenic mesoderm from embryonic stem cells. Stem Cell Res. 2009;3:126–141. doi: 10.1016/j.scr.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 34.Gökbuget D., Blelloch R. Epigenetic control of transcriptional regulation in pluripotency and early differentiation. Development. 2019;146 doi: 10.1242/dev.164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boland M.J., Nazor K.L., Loring J.F. Epigenetic regulation of pluripotency and differentiation. Circ. Res. 2014;115:311–324. doi: 10.1161/circresaha.115.301517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu Y., Deng P., Yu B., Szymanski J.M., Aghaloo T., Hong C., Wang C.Y. Inhibition of EZH2 promotes human embryonic stem cell differentiation into mesoderm by reducing H3K27me3. Stem Cell Rep. 2017;9:752–761. doi: 10.1016/j.stemcr.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soto R.A., Najia M.A.T., Hachimi M., Frame J.M., Yette G.A., Lummertz da Rocha E., Stankunas K., Daley G.Q., North T.E. Sequential regulation of hemogenic fate and hematopoietic stem and progenitor cell formation from arterial endothelium by Ezh1/2. Stem Cell Rep. 2021;16:1718–1734. doi: 10.1016/j.stemcr.2021.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vo L.T., Kinney M.A., Liu X., Zhang Y., Barragan J., Sousa P.M., Jha D.K., Han A., Cesana M., Shao Z., et al. Regulation of embryonic haematopoietic multipotency by EZH1. Nature. 2018;553:506–510. doi: 10.1038/nature25435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao B., Jing C., Wilson J.R., Walker P.A., Vasisht N., Kelly G., Howell S., Taylor I.A., Blackburn G.M., Gamblin S.J. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421:652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- 40.Wilson J.R., Jing C., Walker P.A., Martin S.R., Howell S.A., Blackburn G.M., Gamblin S.J., Xiao B. Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell. 2002;111:105–115. doi: 10.1016/s0092-8674(02)00964-9. [DOI] [PubMed] [Google Scholar]
- 41.Wang H., Cao R., Xia L., Erdjument-Bromage H., Borchers C., Tempst P., Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 42.Kurash J.K., Lei H., Shen Q., Marston W.L., Granda B.W., Fan H., Wall D., Li E., Gaudet F. Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol. Cell. 2008;29:392–400. doi: 10.1016/j.molcel.2007.12.025. [DOI] [PubMed] [Google Scholar]
- 43.Subramanian K., Jia D., Kapoor-Vazirani P., Powell D.R., Collins R.E., Sharma D., Peng J., Cheng X., Vertino P.M. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol. Cell. 2008;30:336–347. doi: 10.1016/j.molcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Judson R.N., Quarta M., Oudhoff M.J., Soliman H., Yi L., Chang C.K., Loi G., Vander Werff R., Cait A., Hamer M., et al. Inhibition of methyltransferase Setd7 allows the in vitro expansion of myogenic stem cells with improved therapeutic potential. Cell Stem Cell. 2018;22:177–190. doi: 10.1016/j.stem.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kouskouti A., Scheer E., Staub A., Tora L., Talianidis I. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol. Cell. 2004;14:175–182. doi: 10.1016/s1097-2765(04)00182-0. [DOI] [PubMed] [Google Scholar]
- 46.Yang J., Huang J., Dasgupta M., Sears N., Miyagi M., Wang B., Chance M.R., Chen X., Du Y., Wang Y., et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc. Natl. Acad. Sci. USA. 2010;107:21499–21504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee J., Shao N.Y., Paik D.T., Wu H., Guo H., Termglinchan V., Churko J.M., Kim Y., Kitani T., Zhao M.T., et al. SETD7 drives cardiac lineage commitment through stage-specific transcriptional activation. Cell Stem Cell. 2018;22:428–444.e5. doi: 10.1016/j.stem.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang M., Wang H., Wen Y., Chen X., Liu X., Gao J., Su P., Xu Y., Zhou W., Shi L., Zhou J. MEIS2 regulates endothelial to hematopoietic transition of human embryonic stem cells by targeting TAL1. Stem Cell Res. Ther. 2018;9:340. doi: 10.1186/s13287-018-1074-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pijuan-Sala B., Griffiths J.A., Guibentif C., Hiscock T.W., Jawaid W., Calero-Nieto F.J., Mulas C., Ibarra-Soria X., Tyser R.C.V., Ho D.L.L., et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature. 2019;566:490–495. doi: 10.1038/s41586-019-0933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tyser R.C.V., Mahammadov E., Nakanoh S., Vallier L., Scialdone A., Srinivas S. Single-cell transcriptomic characterization of a gastrulating human embryo. Nature. 2021;600:285–289. doi: 10.1038/s41586-021-04158-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barsyte-Lovejoy D., Li F., Oudhoff M.J., Tatlock J.H., Dong A., Zeng H., Wu H., Freeman S.A., Schapira M., Senisterra G.A., et al. (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells. Proc. Natl. Acad. Sci. USA. 2014;111:12853–12858. doi: 10.1073/pnas.1407358111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kyba M. Mesoderm, cooked up fast and served to order. Cell Stem Cell. 2016;19:146–148. doi: 10.1016/j.stem.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernandez-Teran M., Piedra M.E., Rodriguez-Rey J.C., Talamillo A., Ros M.A. Expression and regulation of eHAND during limb development. Dev. Dynam. 2003;226:690–701. doi: 10.1002/dvdy.10271. [DOI] [PubMed] [Google Scholar]
- 54.Mahlapuu M., Ormestad M., Enerbäck S., Carlsson P. The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development. 2001;128:155–166. doi: 10.1242/dev.128.2.155. [DOI] [PubMed] [Google Scholar]
- 55.Pourquié O., Fan C.M., Coltey M., Hirsinger E., Watanabe Y., Bréant C., Francis-West P., Brickell P., Tessier-Lavigne M., Le Douarin N.M. Lateral and axial signals involved in avian somite patterning: a role for BMP4. Cell. 1996;84:461–471. doi: 10.1016/s0092-8674(00)81291-x. [DOI] [PubMed] [Google Scholar]
- 56.Chan S.S.K., Shi X., Toyama A., Arpke R.W., Dandapat A., Iacovino M., Kang J., Le G., Hagen H.R., Garry D.J., Kyba M. Mesp1 patterns mesoderm into cardiac, hematopoietic, or skeletal myogenic progenitors in a context-dependent manner. Cell Stem Cell. 2013;12:587–601. doi: 10.1016/j.stem.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nowotschin S., Ferrer-Vaquer A., Concepcion D., Papaioannou V.E., Hadjantonakis A.K. Interaction of Wnt3a, Msgn1 and Tbx6 in neural versus paraxial mesoderm lineage commitment and paraxial mesoderm differentiation in the mouse embryo. Dev. Biol. 2012;367:1–14. doi: 10.1016/j.ydbio.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu C., Li Y., Semenov M., Han C., Baeg G.H., Tan Y., Zhang Z., Lin X., He X. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 59.Chuikov S., Kurash J.K., Wilson J.R., Xiao B., Justin N., Ivanov G.S., McKinney K., Tempst P., Prives C., Gamblin S.J., et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 60.Shen C., Wang D., Liu X., Gu B., Du Y., Wei F.Z., Cao L.L., Song B., Lu X., Yang Q., et al. SET7/9 regulates cancer cell proliferation by influencing β-catenin stability. Faseb. J. 2015;29:4313–4323. doi: 10.1096/fj.15-273540. [DOI] [PubMed] [Google Scholar]
- 61.Sweetman D., Wagstaff L., Cooper O., Weijer C., Münsterberg A. The migration of paraxial and lateral plate mesoderm cells emerging from the late primitive streak is controlled by different Wnt signals. BMC Dev. Biol. 2008;8:63. doi: 10.1186/1471-213x-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoshikawa Y., Fujimori T., McMahon A.P., Takada S. Evidence that absence of Wnt-3a signaling promotes neuralization instead of paraxial mesoderm development in the mouse. Dev. Biol. 1997;183:234–242. doi: 10.1006/dbio.1997.8502. [DOI] [PubMed] [Google Scholar]
- 63.Stern H.M., Brown A.M., Hauschka S.D. Myogenesis in paraxial mesoderm: preferential induction by dorsal neural tube and by cells expressing Wnt-1. Development. 1995;121:3675–3686. doi: 10.1242/dev.121.11.3675. [DOI] [PubMed] [Google Scholar]
- 64.Tajbakhsh S., Borello U., Vivarelli E., Kelly R., Papkoff J., Duprez D., Buckingham M., Cossu G. Differential activation of Myf5 and MyoD by different Wnts in explants of mouse paraxial mesoderm and the later activation of myogenesis in the absence of Myf5. Development. 1998;125:4155–4162. doi: 10.1242/dev.125.21.4155. [DOI] [PubMed] [Google Scholar]
- 65.Castaño J., Morera C., Sesé B., Boue S., Bonet-Costa C., Martí M., Roque A., Jordan A., Barrero M.J. SETD7 regulates the differentiation of human embryonic stem cells. PLoS One. 2016;11:e0149502. doi: 10.1371/journal.pone.0149502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Y., Reddy M.A., Miao F., Shanmugam N., Yee J.K., Hawkins D., Ren B., Natarajan R. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J. Biol. Chem. 2008;283:26771–26781. doi: 10.1074/jbc.M802800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brasacchio D., Okabe J., Tikellis C., Balcerczyk A., George P., Baker E.K., Calkin A.C., Brownlee M., Cooper M.E., El-Osta A. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes. 2009;58:1229–1236. doi: 10.2337/db08-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deering T.G., Ogihara T., Trace A.P., Maier B., Mirmira R.G. Methyltransferase Set7/9 maintains transcription and euchromatin structure at islet-enriched genes. Diabetes. 2009;58:185–193. doi: 10.2337/db08-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang W.J., Wu X.N., Shi T.T., Xu H.T., Yi J., Shen H.F., Huang M.F., Shu X.Y., Wang F.F., Peng B.L., et al. Regulation of transcription factor yin yang 1 by SET7/9-mediated lysine methylation. Sci. Rep. 2016;6:21718. doi: 10.1038/srep21718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oudhoff M.J., Braam M.J.S., Freeman S.A., Wong D., Rattray D.G., Wang J., Antignano F., Snyder K., Refaeli I., Hughes M.R., et al. SETD7 controls intestinal regeneration and tumorigenesis by regulating wnt/β-catenin and hippo/YAP signaling. Dev. Cell. 2016;37:47–57. doi: 10.1016/j.devcel.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 71.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The datasets generated during this study are available at the Gene Expression Omnibus (GEO) under the accession number GSE228287. The following public datasets were downloaded from GEO and reanalyzed: GSE92245.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.