Summary
Severe acute malnutrition (SAM) is a multifactorial disease affecting millions of children worldwide. It is associated with changes in intestinal physiology, microbiota, and mucosal immunity, emphasizing the need for multidisciplinary studies to unravel its full pathogenesis. We established an experimental model in which weanling mice fed a high-deficiency diet mimic key anthropometric and physiological features of SAM in children. This diet alters the intestinal microbiota (less segmented filamentous bacteria, spatial proximity to epithelium), metabolism (decreased butyrate), and immune cell populations (depletion of LysoDC in Peyer’s patches and intestinal Th17 cells). A nutritional intervention leads to a fast zoometric and intestinal physiology recovery but to an incomplete restoration of the intestinal microbiota, metabolism, and immune system. Altogether, we provide a preclinical model of SAM and have identified key markers to target with future interventions during the education of the immune system to improve SAM whole defects.
Subject areas: Immunology, Gastroenterology, Microbiome
Graphical abstract

Highlights
-
•
Mouse model of SAM mimics childhood SAM features
-
•
SAM alters the intestinal microbiota, its spatial organization, and metabolism
-
•
SAM rises fecal sIgA and Treg/Th17 ratio, but decreases LysoDC in Peyer’s patches
-
•
Dietary intervention rescues zoometry and physiology, but not microbiota and immunity
Immunology; Gastroenterology; Microbiome
Introduction
The first 1,000 days of life are critical for optimal growth, physiological, neurological and immune development, and establishment of symbiotic relationships with the microbiota.1 At weaning, the introduction of solid food and subsequent changes of gut microbiota are critical for inducing an immune reaction to microbiota that educates the immune system and protect from immunopathologies later in life.2,3 In low- and middle-income countries, weaning is a high-risk period for malnourished children because of the intake of foods low in protein and essential micronutrients. Undernutrition is often associated with altered immunity resulting in an elevated risk of infection and a reduced response to oral vaccines.4 Thus, severe acute malnutrition (SAM) is a life-threatening condition affecting 13.6 million children under five worldwide in 2020.5 SAM condition is defined by a weight-for-height Z score lower than −3 SD and a mid-upper arm circumference less than 115 mm.6 Despite advances in the management of SAM with therapeutic foods,5 considering only anthropometric and clinical criteria to assess nutritional recovery correlates poorly with optimal long-term recovery. This is reflected by the high post-discharge mortality associated with malnutrition relapse, recurrent infections, intestinal inflammation, and enteropathy.7,8,9,10
Children with SAM harbor an atypical immature microbiota composition that matches the one of younger healthy children.11,12 Furthermore, SAM is transmitted in germ-free mice on a malnutrition diet when receiving the microbiota or IgA-targeted bacterial taxa of children with SAM.13,14 These findings support the central role of microbiota composition in the pathogenesis of SAM. Moreover, increased susceptibility of children with SAM to diseases may be due to this abnormal microbiota unable to properly educate the mucosal immune system. The simultaneous assessment of physiological, microbial, and immune parameters at mucosal sites is limited in SAM children for obvious ethical reasons. Current preclinical models including inadequate diets, like dietary/calorie restricted or low protein ones, are pertinent to better understand the involvement of these parameters in SAM pathogenesis,15,16,17 but they do not recapitulate the full spectrum of SAM pathogenesis in children.
In this study, we developed a “SAM mouse” model starting at weaning and reproducing the main physiological features of SAM in children. Using this model, we investigated intestinal microbiota composition and location, metabolite production, and intestinal immunity. Nutritional intervention led to zoometric and physiological parameters recovery. However, microbiota, metabolic, and immune defects were prolonged, meaning that this murine model will not only help to dissect SAM pathogenesis but also to implement therapeutic foods with specific compounds restoring proper microbiota and immunity.
Results
Severe growth and intestinal barrier defects in malnourished weanling mice
We aimed to develop a preclinical mouse model of SAM that would mimic as closely as possible the physiopathology of SAM in children. According to the literature, the diets of children suffering from growth stunting are high in carbohydrates and low in protein and fat.13,18 However, severe forms of undernutrition, including kwashiorkor, are not only associated with protein deficiency19 but also with type II nutrients (e.g., essential amino acids, zinc, and potassium) and micronutrients deficiencies.18 Exclusive cereal-based diets, including corn, are deficient in type I and II nutrients, which are essential for metabolic processes and growth.20 In rats, protein deficiency associated with high concentration of sodium and/or potassium was associated with the development of edema, a clinical sign of kwashiorkor.21 Similar to the study by Lykke et al., who used a corn-based diet to reproduce severe growth stunting in a piglet model,18 and in order to mimic real-life situation in which young children are fed a low-variety and cereal-based diet deficient in type II nutrients, we designed a pure corn-based diet deficient in protein, fat, and most of essential minerals and vitamins to induce SAM in conventional mice (Table S1).
At weaning, male and female mice received either a regular control diet (Ctrl diet) or the isocaloric SAM diet during four weeks (Figure 1A). From the first week after weaning, males fed with the SAM diet gained significantly less weight than Ctrl males, reaching a 2.4 ± 1.1 times lower weight gain (Figure 1B) and a mean weight 1.8 ± 0.7 times lower (Figure S1A) four weeks after weaning. In line with previous studies15,22,23, only SAM male mice were affected by a severe growth delay comparable to SAM condition in children with a weight-for-length Z score (WL-Z) at −4.96 ± 1.43 (Figure 1C and Figure S1B). This growth delay occurred despite a calorie intake-to-weight gain ratio higher or equivalent in SAM mice compared to Ctrl mice (Figure S1C). The SAM males were also smaller (Figures 1C and S1D), had shorter tails (Figure 1C) and femurs (Figure S1E) than Ctrl males. Henceforth, only male mice were considered for the rest of the study.
Figure 1.
Severe growth and intestinal barrier defects in malnourished weanling mice and reversibility of defects upon nutritional intervention
(A) Diet composition and experimental procedure: at weaning (21 days old), mice are either on a regular (Ctrl) or on SAM diets for 4 weeks. Nutritional intervention (Int) was performed two weeks after SAM diet by switching to Ctrl diet.
(B) Weight gain (g) by age (days) of mice under different diets (6–9 independent experiments, Ctrl n = 31, SAM n = 24, and Int n = 28).
(C) Left: representative image of Ctrl, SAM, and Int mouse phenotype. Right: Weight-for-Length Z score (WL-Z) at 49 days old (5–9 independent experiments, Ctrl n = 27, SAM n = 20, and Int n = 23).
(D) Small intestine length (5–8 independent experiments, Ctrl n = 40, SAM n = 35, and Int n = 28).
(E) Length of ileal villi (left, 3 independent experiments, Ctrl n = 9, SAM n = 11, and Int n = 7) and crypt depth of the colon (right, 3 independent experiments, Ctrl n = 7, SAM n = 8, and Int n = 6).
(F) Representative micrographs of hematoxylin-eosin staining of terminal ileal villus and of nucleus staining of Peyer’s patches in the different diet conditions.
(G) Fecal neutrophil gelatinase-associated lipocalin (NGAL) concentration (5–8 independent experiments, Ctrl n = 35, SAM n = 29 and Int n = 28).
(H) FITC-dextran concentration in blood 4 h post-gavage (3 independent experiments, Ctrl n = 7, SAM n = 5, and Int n = 5).
(I) Representative staining of zonula occludens-1 (ZO-1) network in PPs and terminal ileum.
(J) Representative spectral confocal imaging projections of terminal ileum sections from 49-days-old Ctrl, SAM, and Int mice stained for MUC2 (magenta) and Vicia Villosa Agglutinin (VVA, cyan). All graphic values are represented as mean ± SD. Bars: 50 μm. Dunn’s multiple comparisons test, ∗p-value<0.05, ∗∗p-value<0.01, ∗∗∗p-value<0.001, ∗∗∗∗p-value<0.0001. See also Figure S1 and Table S1.
Children with SAM suffer from nutrient malabsorption associated with villus atrophy, increased intestinal permeability, and chronic intestinal inflammation in a subclinical condition termed environmental enteropathy.15 Our SAM mouse model also exhibited significant intestinal morphological changes characterized by a shorter small intestine (SI) (Figure 1D), an atrophy of ileal villi and of Peyer’s patches (PPs) (Figures 1E and 1F), and shorter colonic but not ileal crypts (Figures 1E, and S1F–S1G). SAM mice also showed a significant rise in the concentration of fecal neutrophil gelatinase-associated lipocalin (NGAL), a marker of intestinal inflammation (Figure 1G). This inflammation was accompanied by altered intestinal functions, highlighted by a significant increase in intestinal permeability (Figure 1H) and a decrease of the zonula occludens-1 (ZO-1) network at the follicle-associated epithelium of PPs and along the ileal villi (Figure 1I). Finally, the mucus layer in SAM mice was thicker and enriched with carbohydrate motifs recognized by the lectin VVA (α- or β-linked terminal N-acetylgalactosamine) (Figure 1J). Altogether, these data showed that our weanling model of SAM mimics most of the anthropometric and intestinal pathophysiological features of SAM in children suffering from environmental enteropathy.
Reversibility of growth and intestinal barrier defects of malnourished weanling mice
To test whether the effects of two weeks of severe undernutrition during this critical period of growth could be reversed, we carried out nutritional intervention by switching SAM to Ctrl diet (Int mice) (Figure 1A). This resulted in a gradual recovery of zoometric parameters within 2 weeks, with a final weight gain and growth slightly below that of the Ctrl mice (Δ = 1.45 g ± 1.83 g, p-value<0.05) (Figures 1B, and S1A), an increase in the size of the mice (Figures 1C, and S1D) and in the femur length (Figure S1E), and a WL-Z at −1.22 ± 1.02 (Figure 1C). Regarding the intestinal parameters, we observed a total recovery of the SI and ileal villus length (Figures 1D–1F) and of the size of PPs (Figure 1F). The colonic crypt depth (Figures 1E, and S1G) remained slightly below that of Ctrl mice although not significantly different. Fecal NGAL concentration (Figure 1G), intestinal permeability (Figure 1H), and ileal ZO-1 network (Figure 1I) were also restored. Finally, the overall aspect of the mucus layer appeared intermediate between Ctrl and SAM mice (Figure 1J). Therefore, return to a regular diet that met the nutritional requirements of the critical period of growth elicited zoometric and physiological recovery.
SAM alters the spatial organization and composition of the ileal microbiota
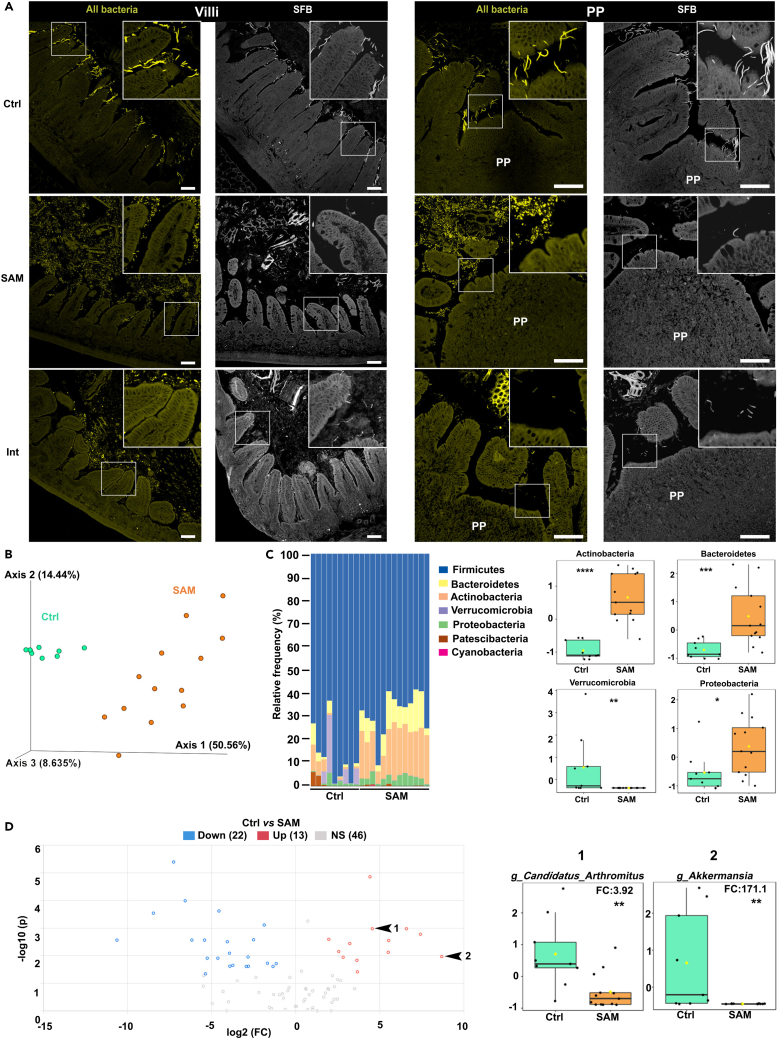
Since SAM is associated with an abnormal microbiota,11,12,14,24 we studied its spatial organization in the terminal part of the ileum by fluorescence in situ hybridization (FISH). In Ctrl mice, the microbiota was kept at bay from the epithelium of villi and PPs at the exception of segmented filamentous bacteria (SFB) known to anchor ileal villi and PPs25 (Figure 2A). Remarkably, in SAM mice, the microbiota was 3–4 times closer to the intestinal epithelium compared to the Ctrl group (Figures 2A, and S2A). An increased abundance of tissue-adherent bacteria was also detected in the space between villi and was not identified as SFB, as evidenced by FISH using the SFB-specific probe (Figure 2A). We also performed principal component analysis (PCA) of 16s rRNA sequencing data of the ileal microbiota, highlighting a clear separation in its composition between Ctrl and SAM mice (Figure 2B). Compared to Ctrl mice, Shannon diversity index increased in SAM mice (FigureS2B and Table S2A). SAM-associated microbiota was notably characterized by a significant increase in Actinobacteria, Bacteroidetes, and Proteobacteria and a significant decrease in Verrucomicrobia (Figure 2C and Table S2B). At the species level, 35 of the 81 amplicon sequence variants (ASVs) identified were significantly modified in SAM mice (Figure 2D and Table S2C). Among them, SFB (Candidatus Arthromitus) was decreased 4 times (Figure 2D and Table S2C), confirming our FISH data. Interestingly, we also detected an important depletion of Akkermansia in SAM mice (Figure 2D and Table S2C), in line with their altered mucus layer (Figure 1J). Nutritional intervention did not fully restore the spatial organization of the ileal microbiota, despite a gradual SFB re-colonization (Figures 2A and S2A). Together, these data showed that SAM mice exhibited an unusual organization and composition of the ileal microbiota characterized by depletion of key bacterial species involved in immune education and intestinal barrier integrity.26,27
Figure 2.
SAM alters the spatial organization and composition of the ileal microbiota
(A) Representative spectral confocal imaging projections of ileal villus (left) and PPs (right) from 49-days-old Ctrl, SAM, and Int mice stained by FISH for all bacteria (Eub-338 probe, yellow) and SFB (SFB-1004 probe, gray). Bars, 50 μm.
(B) β-diversity distance analysis using unweight PCA based on a Bray Curtis similarity matrix of the Ctrl (green, n = 9) and SAM (orange, n = 13) groups, 3 independent experiments. PERMANOVA test, ∗∗∗p-value<0.001.
(C) Bar plot representation of individual composition of ileal content (left) and associated box and whisker plots (right) of significantly modified ASVs at the phylum level in Ctrl (n = 9) and SAM (n = 13) groups, 3 independent experiments.
(D) Volcano plot representation of significantly modified ASVs (35/81) at the species level in Ctrl (n = 9) compared to SAM group (n = 13) in the terminal ileum, 3 independent experiments. Among decreased species in SAM mice, arrows 1 and 2 pointing to (1) Candidatus Arthromitus (SFB) and (2) Akkermansia, are shown as box and whisker plots (right) that were generated after volcano plot analysis by combination of fold change (FC, threshold 2.0) and t-test (non-parametric, Wilcoxon rank-sum test) analyses using MetaboAnalyst 5.0 software.86 ∗p-value<0.05, ∗∗p-value<0.01, ∗∗∗p-value<0.001, ∗∗∗∗p-value<0.0001. See also Figure S2 and Table S2.
Nutritional intervention results in only partial recovery from SAM-induced cecal dysbiosis
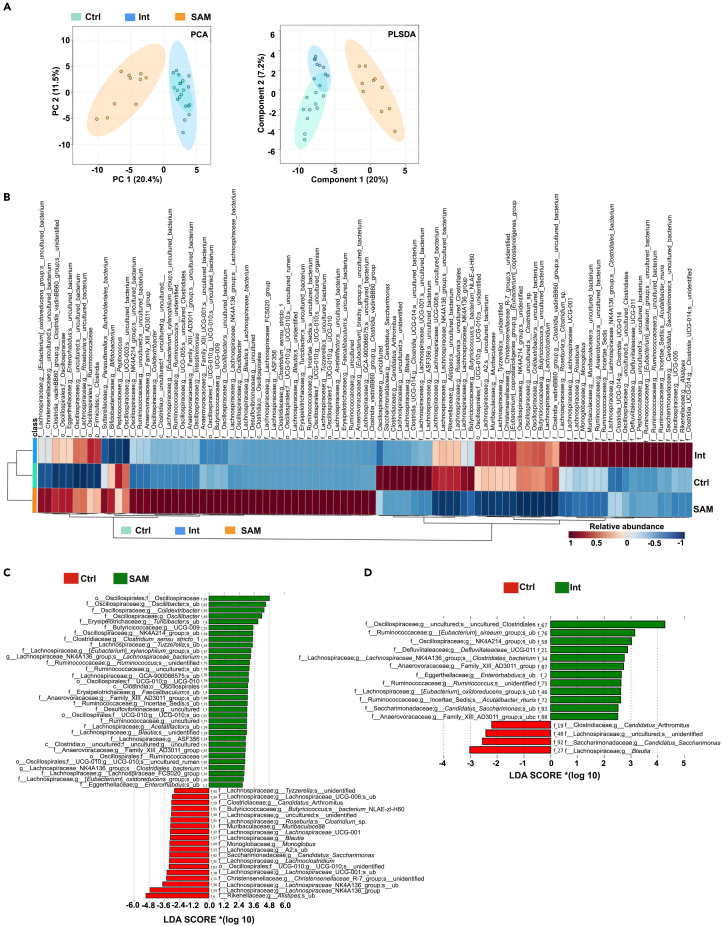
We observed a significant decrease in the weight and size of the ceca of SAM and Int mice compared to Ctrl mice (Figure S3A), likely indicative of microbial changes in the cecum.28,29 Indeed, unweighted PCA and weighted partial least squares discriminant analysis (PLSDA) separated cecal microbiota according to the diet (Figure 3A). Shannon 1 indexes were not significantly different between Ctrl and SAM mice (Figure S3B) but we observed a significant decrease in Bacteroidetes, whereas Firmicutes and Desulfobacterota, previously classified as proteobacterial classes Deltaproteobacteria,30 were significantly increased in SAM mice (Figure S3C and Table S3A). A clear separation between Ctrl and SAM mice was also observed at the species level (Figure 3B). Of the 52 ASVs significantly modified in SAM mice compared to Ctrl mice, 33 ASVs belonging to Oscillospiraceae (Oscillibacter, Colidextribacter, Oscillospiraceae_g_NK4A214 group), Erysipelotrichaceae (Turicibacter, Faecalibaculum), Lachnospiraceae (Tuzzerela, Lachnospiraceae NK4A136 group, Acetatifactor, Blautia), Clostridiaceae (Clostridium sensu stricto 1), Ruminococcaceae, Anaerovoracaceae (Family_XIII_AD3011_group), Desulfovibrionaceae, and Eggerthellaceae (Enterorhabdus) families had increased abundances in SAM mice (Figure 3C and Table S3B). Turicibacter, Clostridium sensu stricto 1 are known to be associated with weight loss and are increased during consumption of protein-deficient diets in animals and humans.14,31,32 SAM and stunting are also associated with enrichment in inflammogenic and opportunistic species such as Acetatifactor and Enterorhabdus,33,34,35 or members of Erysipelotrichaceae36 and Desulfovibrionaceae.13,37 Undernutrition is in general linked to depletion of several species from Lachnospiraceae and Ruminococceae families whose members are mostly short-chain fatty acids (SCFA) producers.38,39 However, some of them were both enriched and depleted in SAM mice compared to Ctrl mice (Figure 3C and Table S3B). Among the 19 ASVs diminished in SAM mice, Alistipes from Rikenellaceae family, Blautia, Lachnospiraceae NK4A136, Lachnoclostridium, and Roseburia from Lachnospiraceae family are also depleted in children with SAM39,40,41,42 and are butyrate producers associated with good health.38,43,44 The Christensenellaceae R-7 group was also decreased in SAM mice compared to Ctrl mice (Figure 3C and Table S3B). Consistent with our data, conditions associated with inflammation such as stunting,45 high fecal calprotectin,46 or inflammatory bowel diseases47 appear to deplete Christensenellaceae R-7 group, whose anti-inflammatory and SCFA-producer properties48 make it one of five taxa considered a signature of a healthy gut depleted in inflammatory bowel diseases.47 Finally, and in accordance with the ileal data (Figure 2), SAM mice were significantly depleted in Candidatus Arthromitus compared to the Ctrl mice (Figure 3C and Table S3B). Interestingly, Erysipelotrichaceae expansion and notably a Faecalibaculum species are able to displace SFB in the small intestine of malnourished mice.49
Figure 3.
Nutritional intervention results in only partial recovery from SAM-induced cecal dysbiosis
(A) Unweight PCA and Weight PLSDA of the 94 ASVs identified at the species level in the cecum content of the Ctrl, SAM, and Int mice.
(B) Heatmap of ASVs cecal abundance (rows) in the different diet conditions (columns).
(C and D) Histograms of linear discriminant analysis (LDA) score of cecal microbiota between groups. Bar length represents the effect size. Red color refers to the bacteria overexpressed in the Ctrl group, and green color refers to the bacteria overexpressed in the SAM group. N = 10/group, 3 independent experiments. Taxonomical features with p-value <0.05 and LDA effect size >2 were regarded as significant microbial signatures. ub: uncultured_bacterium, uo: uncultured_organism. See also Figure S3 and Tables S3.
Nutritional intervention resulted in a microbiota profile closer to Ctrl mice than to SAM mice (Figure 3A, S3C–S3D, and Table S3D), but with specific signatures such as increase in members of Oscillospiraceae, Ruminococcaceae, Defluviitaleaceae, Anaerovoracaceae, and Eggerthellaceae families, and decrease in Blautia and Candidatus Arthromitus compared to Ctrl mice (Figure 3D and Table S3C). Altogether, these results reveal the onset of a cecal dysbiosis in SAM mice that cannot be totally solved by this nutritional intervention.
Nutritional intervention results in only partial recovery from SAM-induced metabolic alterations
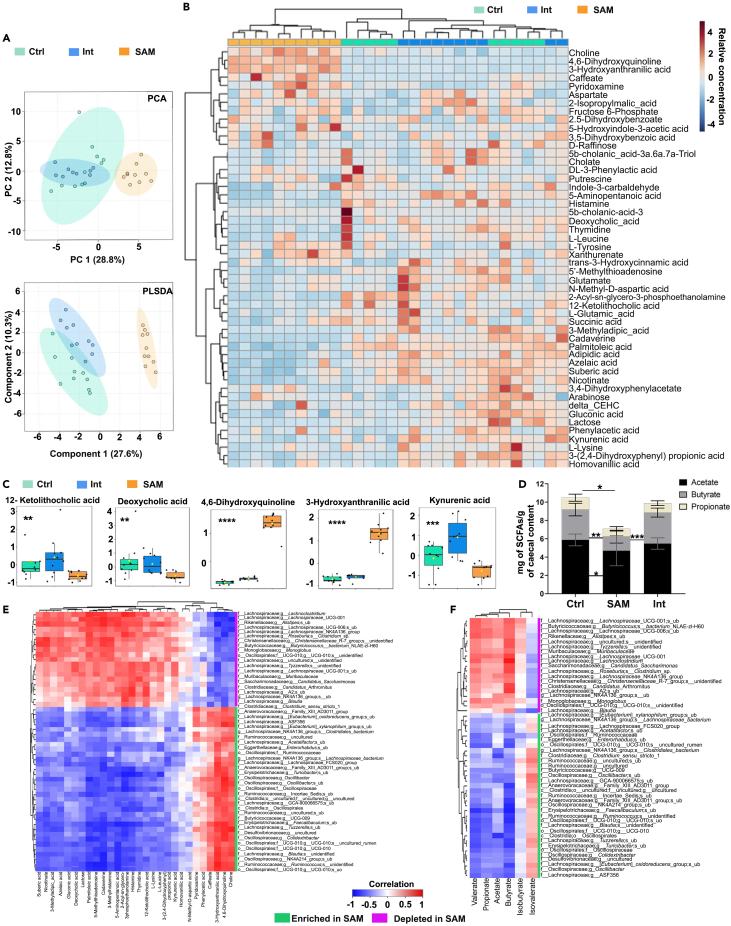
A wide variety of metabolites including vitamins and SCFAs are produced by the intestinal microbiota and drastic changes in microbiota induced by SAM may alter the profile of produced metabolites. Cecal contents of Ctrl, SAM, and Int groups were analyzed for changes in metabolite abundance using untargeted metabolomic analysis by UPLC-HRMS. The metabolomic profile of SAM mice was indeed distinct from the Ctrl mice as shown by PCA and PLSDA (Figure 4A). Among 50 identified metabolites, 5 were significantly increased and 23 decreased in SAM mice compared to Ctrl and Int mice (Figures 4B–4C, S4A and Table S4). As expected, from the higher protein content of the Ctrl diet, there was an overrepresentation of amino acid (AA) metabolism in Ctrl mice (Figure S4A). The significant reduction in L-Lysine, cadaverine (Lysine decarboxylation product), and 5-aminopentanoate (Lysine and cadaverine degradation product) converged to a shift toward the lysine degradation pathway in SAM mice, highlighting a crucial need for this essential AA (Figure S4A). The bile acid biosynthesis pathway (12-Ketolithocholic acid and deoxycholate) was also underrepresented in SAM mice (Figure 4C). The increase in 4,6-dihydroxyquinoline and 3-hydroxyanthranilate and decrease in kynurenic acid in SAM mice demonstrated a shift in the tryptophan metabolic pathway compared to the two other groups (Figure 4C). The increase in pyridoxamine (water soluble vitamin B6 form) in SAM mice pointed out a higher vitamin B6 concentration than in Ctrl mice (Figure S4A), consistent with the rise in the main phyla producing specific B-vitamins,50 Bacteroidetes, Actinobacteria, and Proteobacteria (Figure 2C). Since some butyrate producers were depleted in SAM mice (Figure 3C), we analyzed cecal SCFA concentrations by gas chromatography-mass spectrometry. There was a significant decrease in the production of SCFAs, especially in butyrate, in SAM mice (Figures 4D and S4B). Nutritional intervention resulted in a metabolomic profile closer to Ctrl mice than to SAM mice (Figures 4A, and S4A). SCFAs production and the ratio between acetate, butyrate, and propionate were completely restored (Figures 4D and S4B). Nevertheless, it induced specific signatures, including an increase in kynurenic acid and phenylacetic acid (Figures 4C, and S4A). Spearman’s rank correlation coefficients between bacterial relative abundance and metabolite concentrations showed strong positive correlation between the ASVs enriched in SAM mice and the increase of choline, 4,6-dihydroxyquinoline, 3-hydroxyanthranilic acid, caffeate, phenylacetic acid, and pyridoxamine (Figure 4E), and strong negative correlation between these ASVs and SCFAs at the exception of isovalerate (Figure 4F). Altogether, these data demonstrate that SAM mice had an unusual metabolic profile that cannot be fully restored by this nutritional intervention, except for SCFAs.
Figure 4.
Nutritional intervention results in only partial recovery from SAM-induced metabolic alterations
(A) Unweight PCA and Weight PLSDA of the 50 metabolites identified in the cecum content of the Ctrl, SAM, and Int groups.
(B) Heatmap of individual metabolite concentration (rows) organized by distance measured in the three groups of diet (columns).
(C) Box and whisker plots of selected significantly increased or decreased metabolites in the SAM group. N = 10/group, 3 independent experiments. Non-parametric Kruskal Wallis Test, ∗p-value<0.5, ∗∗p-value<0.01, ∗∗∗p-value<0.001, ∗∗∗∗p-value<0.0001.
(D–F) SCFAs concentration (mg/g of cecal content) in Ctrl (n = 10), SAM (n = 8) and Int (n = 10) groups. Dunn’s multiple comparisons test, ∗p-value<0.5, ∗∗p-value<0.01, ∗∗∗p-value<0.001. All values are represented as mean ± SD, 3 to 4 independent experiments. Heatmaps of Spearman’s correlations between cecal ASVs and (E) untargeted metabolites or (F) SCFAs (mg/g cecum). Rows are hierarchically clustered based on pairwise distances calculated from Pearson’s correlation values. Green and magenta bars to the right of each heatmap indicate significant genus enrichment and depletion in SAM mice, respectively (p < 0.05; Student’s t test). See also Figure S4 and Tables S4.
Alteration of the IgA response in Peyer’s patches and villi of SAM mice
Continuously exposed to food-derived antigens, microbiota, and pathogens, PPs are primary immune response initiation sites of the SI, notably inducing the generation of IgA-secreting plasma cells, which play a key role in controlling pathogen invasion and ensuring microbiota regulation.51,52,53 Yet, the impact of malnutrition on PPs is understudied.54 Due to PP atrophy, the numbers of total B cells, IgA-expressing B cells (IgA+ B cells), germinal center B cells (GL-7+ B cells), and naive B cells (IgD+ B cells) were significantly reduced in SAM mice compared to Ctrl mice (Figure 5A and S5A). However, there was also a significant decrease in frequency and proportion among B cells of IgA+ B cells in SAM mice (Figures 5A, 5B, and S5A). IgA-secreting cells generated in PPs home into villus lamina propria of the SI. In PPs, the large majority of IgA+ cells are B220+CD19+ cells (Figure S5B) whereas the IgA+ cells lose the B220 marker in the SI (Figure S5C). We found in the SI of SAM mice a significant decrease in the percentage of both total B and IgA+ cells, although their absolute numbers were not significantly modified (Figure 5C). Accordingly, the ratio between IgA+ and IgM+ cells showed a decrease in IgA+ cells (Figures 5D, and S5D). By contrast, the fecal concentration of total sIgA was significantly higher in SAM mice than in Ctrl mice (Figure 5E). Nutritional intervention tended to restore the number of total B cells and IgA+ cells in the PPs and in the villi, but without reaching the percentages observed in Ctrl mice (Figures 5A–5D, and S5A–S5D). However, the fecal concentration of sIgA of Int mice dropped to the level of Ctrl mice (Figure 5E). These data reveal that SAM diet impacts the intestinal immune system by limiting the expansion of IgA+ cells in PPs and villi although fecal sIgA increase.
Figure 5.
Alteration of the IgA response in Peyer’s patches and villi of SAM mice
(A) Absolute numbers of total and IgA+ B cells, and frequencies of total, IgA+, IgD+, and IgM+ B cells in PPs of Ctrl, SAM, and Int mice.
(B) Ratio (%) between IgA+, IgM+, and IgD+ among total B cells in PPs.
(C) Absolute numbers and associated frequencies of total B and IgA+ cells in villi of Ctrl, SAM, and Int groups.
(D) Ratio (%) between IgA+ and IgM+ in villi of Ctrl, SAM, and Int groups.
(E) Fecal sIgA concentration (ng/mL/100 mg of feces). All values are represented as mean ± SD of 5 independent experiments. For PPs analyses, Ctrl n = 4–5, SAM n = 4–5, and Int n = 5. For villus analyses, Ctrl n = 10, SAM n = 12, and Int n = 9. Dunn’s multiple comparisons test, ∗p-value<0.05, ∗∗p-value<0.01. NS: non-significant. See also Figure S5.
Imbalance of the Th17/Treg ratio in the intestinal mucosa of SAM mice
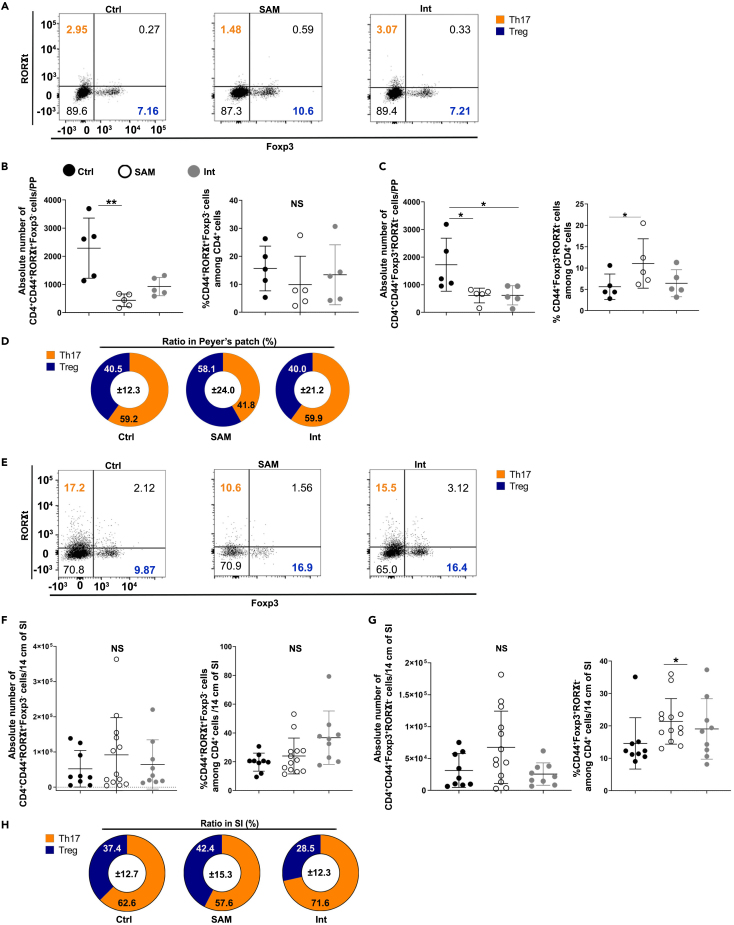
As for B cells, the absolute numbers of T cells and their subsets significantly diminished in PPs of SAM mice compared to those of Ctrl mice, except for the RORγt+Foxp3+ cells (Figures 6A–6C and S6A). However, a significant increase in the frequency of regulatory T cells (Treg, CD4+CD44+ Foxp3+RORγt− cells) and a trend to decrease for Th17 cells (CD4+CD44+RORγt+Foxp3- cells) were observed in SAM mice (Figures 6A–6C, and S6A–S6B). This generated an imbalance in the Th17/Treg ratio in SAM mice compared to Ctrl mice (Figure 6D). Nutritional intervention did not fully restore the numbers of T cells and their subsets (Figures 6A–6C, and S6A), although the Th17/Treg ratio returned to normal (Figure 6D). In villi, absolute numbers and frequencies of CD3+, CD4+, CD8+, activated CD4+CD44+, RORγt+Foxp3+, CD8+CD44+ T cells (Figure S6C), and Th17 cells (Figures 6E and 6F) and the balance between CD4+/CD8+ cells were not altered by the diet (Figure S6D). Nonetheless, the frequency of Treg rose in the villi of SAM mice compared to Ctrl mice (Figures 6E and 6G). Consequently, the SAM diet induced an imbalance in the Th17/Treg ratio in the villi but to a lesser extent than in PPs (Figure 6H). Nutritional intervention increased the Th17/Treg ratio in villi as compared to Ctrl mice (Figures 6E–6H). These data show that SAM diet favors Treg at the expanse of Th17 cells in both PPs and villi with a capacity for nutritional intervention to restore Th17/Treg ratio in PPs and to favor Th17 cell population in villi.
Figure 6.
Imbalance of the Th17/Treg ratio in the intestinal mucosa of SAM mice
(A–H) Representative dot plots (A), absolute numbers and frequencies of (B) Th17 (CD4+CD44+RORγt+Foxp3- cells), and (C) Treg (CD4+CD44+Foxp3+RORγt− cells), and (D) ratio (%) between Th17 and Treg cells in PPs of Ctrl, SAM, and Int mice. Representative dot plots (E), absolute numbers and frequencies of (F) Th17 (CD4+CD44+RORγt+Foxp3- cells) cells and (G) Treg (CD4+CD44+Foxp3+RORγt− cells), and (H) ratio (%) between Th17 and Treg cells in villi of Ctrl, SAM, and Int mice. All values are represented as mean ± SD, with 5 independent experiments. For PPs analyses, Ctrl n = 5, SAM n = 5, and Int n = 5. For villi analyses, Ctrl n = 9, SAM n = 12, and Int n = 9. Eight thousand of cells of the parent population, CD4+CD44+ cells, were included to generate each representative dot plots for the three groups. Dunn’s multiple comparisons test, ∗p-value<0.05, ∗∗p-value<0.01. NS: non-significant. See also Figure S6.
Alteration of Peyer’s patch phagocyte distribution in SAM mice
We have previously characterized the PP mononuclear phagocyte system (Figure S7A) pinpointing notably the pivotal roles of the lysozyme-expressing dendritic cells termed LysoDC, in bacterial and particulate antigen sampling, innate defense against pathogens as well as Th cell priming.55,56,57,58 Noteworthily, SAM diet induced a 3.2-fold diminution in absolute number and a reduction by half in the frequency of LysoDC among MCHIIhiCD11c+ cells when compared to Ctrl mice (Figure 7A); in contrast, other PP phagocytes, i.e. macrophages (LysoMac) and conventional dendritic cells (cDC1 and cDC2), were not significantly affected, either in absolute numbers or frequencies (Figure 7A). This resulted in an imbalance of the CD11c+ phagocyte distribution within PPs mainly favoring cDC2 at the expense of LysoDC (Figures 7B, and S7B). By microscopy, we also noticed in SAM mice that although LysoDC were still present at the top of the domes formed by the follicles, they were less frequent on their side (Figure 7C). Nutritional intervention allowed a recovery of LysoDC number, frequency, and location on the side of domes (Figure 7A and S7B). However, an imbalance of phagocyte distribution was still observed after two weeks of nutritional intervention (Figures 7B–7C and S7B). In conclusion, SAM diet significantly alters the composition and distribution of phagocytes in PPs, especially limiting the number of LysoDC. This defect is mitigated by nutritional intervention.
Figure 7.
Alteration of Peyer’s patch phagocyte distribution in SAM mice
(A) Absolute numbers and frequencies of monocyte-derived cells (LysoDC and LysoMac), and conventional DC (cDC1 and cDC2) in PPs of Ctrl, SAM, and Int mice. All values are represented as mean ± SD; Ctrl n = 10, SAM n = 7, and Int n = 4, 4 to 10 independent experiments. Dunn’s multiple comparisons test, ∗∗p-value<0.01, ∗∗∗p-value<0.001. NS, non-significant.
(B) Relative proportion (%) of LysoDC, LysoMac, cDC1, and cDC2 among MHCIIhiCD11chi cells of PP.
(C) Representative spectral confocal imaging projections (10–14 optical sections) of PPs from Ctrl (first row), SAM (second row), and Int (third row) mice stained for MerTK (green), lysozyme (yellow), CD11c (red), CD4 (blue), SIRPα (magenta), and EpCAM (gray). In SAM mice, LysoDC (MerTK+Lysozyme+CD11c+SIRPα+CD4− cells, arrowheads) but not LysoMac (MerTK+Lysozyme+CD11c+SIRPα+CD4+ cells, asterisks) were depleted from the side but not from the top of the subepithelial dome (SED). In the follicle (Fol), LysoDC number seems to be reduced, too, although caution should be taken in this regard due to the atrophy of PPs. Numbers of LysoDC and LysoMac in each region delimited by a dotted line (top and side of SED and follicle, fol.) are indicated on the right of each image. DAV, dome-associated villus. All images are shown at the same magnification. Bars, 20 μm. See also Figure S7.
Discussion
Despite nutritional therapeutic treatments, the mortality among SAM children remains high, emphasizing the need to unravel the complexity of SAM pathology and improve the current treatments. We have developed a physiological weanling mouse model of SAM that mimics key SAM features in children, including villus atrophy rarely observed in other animal models of undernutrition23,59,60 in the absence of infection15,22,54 or damage to the intestinal barrier.16 Therefore, type II nutrient deficiency at the time of weaning may be critical in SAM onset, in accordance with piglet experiments showing that a similar diet induces severe stunting accompanied by villus atrophy.18
Our model displays impaired ileal ZO-1 network, enhanced intestinal permeability, and NGAL production, which have been positively correlated with intestinal inflammation in undernourished children.61,62 In malnutrition situations, this inflammation can be triggered by an increase of tissue-adherent bacteria,15,63 which we have indeed observed in our model. Mucus structure is also dependent on microbiota composition64 and we indeed observed a thicker layer of ileal mucus in SAM mice. A sticky mucus64 as well as protein- and iron-limiting conditions65 promote the expansion of Bacteroidetes in the ileum. Accordingly, we observed a significant rise in Bacteroidetes but also in Proteobacteria in the ileum of SAM mice, mimicking their increase in children with SAM, whose growth synergy is associated with inflammatory-driving properties causing enteropathy and contributing to undernutrition.15 At the genus level, we observed a significant decrease of the mucin degrader Akkermansia. Although poorly characterized,66 the role in intestinal homeostasis of Akkermansia-like spp. in the ileum may be similar to that in colon, i.e. competition with other bacteria, strengthening of the gut epithelial barrier, and immune signaling.67 Their decrease may therefore contribute to the intestinal barrier integrity loss observed in our model.
We further reported a cecal dysbiosis in SAM mice associated with an unusual metabolomic profile. SAM mice harbor specific microbiota signatures characterized by enrichment in taxa associated with leanness, protein-deficient and low-fat content diets, inflammation, and opportunism (Turicibacter, Acetatifactor, Enterorhabdus, Faecalibaculum, and members of Desulfovibrionaceae family),13,14,15,22,32,33,34,35,36,37,54 whereas anti-inflammatory and SCFA-producing bacteria (Blautia, Lachnospiraceae NK4A136, Christensenellaceae R-7 group, Lachnoclostridium, and Roseburia) and butyrate production are decreased in SAM mice. These data mimic those of undernourished children, where butyrate producers are discriminatory taxa between stunted and non-stunted children,45,46 and a low fecal butyrate concentration is predictive of non-recovery and death.68 In our model, the combined effect of food iron deficiency (8.8-fold reduction) and iron sequestration by NGAL might explain the decrease of Roseburia-related butyrate producers sensitive to iron availability69 and the promotion of bacteria with minimal iron requirements such as Proteobacteria, lactic acid bacteria, and Turicibacter.69,70 Consistently with iron depletion/repletion studies,69,70,71 the return to a regular diet leads to a full recovery of butyrate-producing bacteria and SCFAs production. The SAM diet also produced other striking changes in host metabolism such as alterations in essential AA (lysine and tryptophan) and bile acid pathways, which are major metabolic changes observed during protein starvation and contribute to stunting.1,15,72,73 As observed in undernourished children1,11,74 and in animal models11,75 following nutritional rehabilitation, AA and bile acid metabolism were improved in Int group during the re-nutrition phase. However, despite catch-up growth, the cecal metabolome and associated microbiota of the Int mice were partially restored, suggesting that these alterations could be either time-dependent or long-lasting, even after adequate feeding74,75 due to epigenetic alterations that may occur during weaning to adapt to nutritional deficiencies.73,76
An unbalanced exposure to the intestinal microbiota11,12,77 as well as an inadequate education of the immune system4,78 are set in undernourished children. Those with low DC numbers, impaired DC maturation, and T cell priming have a bad prognosis.79 In our model, the composition of phagocytes is altered in PPs with a decrease of LysoDC number, which is with LysoMac, the main phagocytes in the subepithelial region of PPs, playing pivotal roles in innate antiviral and antibacterial defenses.55,56,57 Among PP phagocytes, LysoDC and LysoMac express the highest levels of Clec4e encoding Mincle,55 a C-type lectin involved in the detection of commensals in PPs and in the differentiation of Th17 cells.80 In addition to SFB depletion, the well-known inducers of Th17 cells,81,82 the low numbers of LysoDC could therefore contribute to the Th17/Treg ratio imbalance of our model. Here, the mechanism by which Treg cells are favored remains to be fully explored, but we hypothesize that the immunosuppressive response through Treg cells may be beneficial to the host in the context of the inflammation present in SAM.54 The decrease of IgA+ cells in PPs and villi might be also caused by phagocyte distribution alteration83 in addition to Tfh-reduced number84 and decrease of SFB.85 By contrast, increased secretion of sIgA is consistent with increased tissue-adherent bacteria and enrichment in Proteobacteria and Bacteroidetes, two IgA-bound phyla enriched in SAM children and promoting SAM pathology in recipient mice.13,14
Strikingly, our model exhibits a delayed recovery of the immunological, metabolic, and microbiota profiles after nutritional intervention despite positive zoometric changes, mirroring malnourished food-complemented children who harbor a sustained alteration of their fecal microbiota despite anthropometric improvement. In this regard, our model allowed the identification of critical biomarkers to target for nutritional intervention, including SFB, Akkermansia, and SCFA-producing bacteria as well as LysoDC and Th17 cells. In conclusion, our model not only led to the identification of the multiple defects induced by SAM but also provides a preclinical model, which allows an integrated view of the contribution of gut physiology, immunity, and microbiota in malnutrition.
Limitations of the study
We report a new preclinical model of diet-induced severe chronic undernutrition in conventional mice. In children, SAM is often associated with infections that worsen malnutrition. Our model recapitulates without any bias introduced by infection many aspects of SAM with environmental enteropathy in children. However, the physiology and gut microbiota of the mouse undoubtedly differ from those of humans, some of which may be important. As such, the impact of a burst of enteropathogens (Enterobacteriaceae) or infectious agents will be one of the next steps to investigate. Although colonization of a human microbiota at the species and strain levels is not complete in rodents, humanized mice with the microbiota of children with SAM have proven to be a powerful tool in identifying biomarkers and/or microbial targets in humans. In our study, SAM mice are not humanized; yet, they mimic many of the microbial characteristics of SAM in children and may pave the way to the identification of crucial microbial targets. In our system, like in many institutes in the world, normo-nourished mice harbor SFB, a highly immunogenic and diet-sensitive commensal, unlike the SAM mice. Therefore, the straightforward implementation of the SAM model in various institutes should take into consideration the initial composition of the in-house microbiota. Our study was focused on the early stages of nutritional intervention (2 weeks). Nutritional intervention resulted in a fast recovery of zoometry and intestinal physiology, but in a partial restoration of the intestinal microbiota, metabolism, and immunity. To determine whether the effects of SAM on microbiota and intestinal immunity at weaning are reversible, longer duration and/or different types of intervention may be required. Nevertheless, our study presents a new animal model that phenocopies most features of a complex human disease, which we believe will be useful to the research community for investigating the underlying mechanisms and testing sustainable alternative therapies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-MUC2 | Santa Cruz Biotechnology | Cat#sc-15334; AB_2146667 |
| anti-ZO-1 | Invitrogen | Cat # 61–7300; RRID:AB_2533938 |
| XCR1-BV421 (clone ZET) | Biolegend | Cat #148216; RRID:AB_2565230 |
| CD172α-FITC (clone P84) | BD Biosciences | Cat #560316; RRID:AB_1645240 |
| CD4-PeCy5.5 (clone RM4-5) | eBioscience | Cat #35-0042-82; RRID:AB_11218300 |
| CD11c-PeCy7 (clone N418) | Biolegend | Cat #117318; RRID:AB_493568 |
| BST2-APC (clone 927) | Biolegend | Cat #127016; RRID:AB_1967127 |
| CMHII-A700 (clone M5/114.15.2) | eBioscience | Cat #56-5321-82; RRID:AB_494009 |
| CD11b-APC/Cy7 (clone M1/70) | Biolegend | Cat #101226; RRID:AB_830642 |
| CD45-BUV395 (clone 30-F11) | BD Biosciences | Cat #564279; RRID:AB_2651134 |
| PD-1-BV605 (clone J43) | BD Biosciences | Cat #563059; RRID:AB_2737980 |
| CD62L-BV786 (clone MEL-14) | BD Biosciences | Cat #564109; RRID:AB_2738598 |
| CD8α-AF488 (clone 53-6.7) | Biolegend | Cat #100723; RRID:AB_389304 |
| CXCR5-PE (clone 2G8) | BD Biosciences | Cat #551959; RRID:AB_394300 |
| RORγt-Pe-eFluor610 (clone B2D) | eBioscience | Cat #61-6981-80; RRID:AB_2574649 |
| Foxp3-APC (clone FJK-16S) | eBioscience | Cat #17-5773-82; RRID:AB_469457 |
| CD44-A700 (clone IM7) | eBioscience | Cat #56-0441-82; RRID:AB_494011 |
| CD43-BUV737 (clone S7) | BD Biosciences | Cat #564398; RRID:AB_2738790 |
| CD3-eFluor450 (clone 17A2) | eBioscience | Cat #48-0032-80; RRID:AB_1272229 |
| Siglec-F-BV421 (clone E50-2440) | BD Biosciences | Cat #565934; RRID:AB_2739398 |
| CD19-BV605 (clone 6D5) | Biolegend | Cat #115539; RRID:AB_11203538 |
| B220-BV786 (clone RA3-6B2) | BD Biosciences | Cat #563894; RRID:AB_2738472 |
| IgM-biotin (clone II/41) | eBioscience | Cat #13-5790-82; RRID:AB_466675 |
| IgA-PE (clone mA-6E1) | eBioscience | Cat #12-4204-81; RRID:AB_465916 |
| IgD-Pe/Cy7 (clone 11-26c.2a) | Biolegend | Cat #405719; RRID:AB_2561875 |
| GL-7-eF660 (GL-7) | eBioscience | Cat #50-5902-82; RRID:AB_2574252 |
| Streptavidin PE-CF594 | BD Biosciences | Cat #562284; RRID:AB_11154598 |
| Rat anti-MerTK (clone DS5MMER) | eBioscience | Cat #14-5751-82; RRID:AB_2688282 |
| armenian hamster anti-CD11c (clone N418) | Biolegend | Cat #117301; RRID:AB_313771 |
| rabbit anti-lysozyme (polyclonal) | Abcam | Cat# ab2408; RRID:AB_303050 |
| SIPR-α-AF594 (clone P84) | Biolegend | Cat #144020; RRID:AB_2629588 |
| CD4-eFluor450 (clone RM4-5) | eBioscience | Cat #48-0042-82; RRID:AB_1272194 |
| CD4-eF570 (clone RM4-5) | Invitrogen | Cat #41-0042-82; RRID:AB_2573595 |
| EpCAM-eF450 (clone G8.8) | eBioscience | Cat #48-5791-82; RRID:AB_10717090 |
| Donkey anti-rat IgG Cy3 | Jackson Immuno-research | Cat #712-165-153; RRID:AB_2340667 |
| Donkey anti-rat IgG AF488 | Invitrogen | Cat #A21208; RRID:AB_2535794 |
| Goat anti-Armenian hamster IgG AF594 | Jackson Immuno-research | Cat #127-585-160; RRID:AB_2338999 |
| Goat anti-Armenian hamster IgG Cy3 | Jackson Immuno-research | Cat #127-165-160; RRID:AB_2338989 |
| Goat anti-rabbit IgG AF514 | Invitrogen | Cat #A-31558; RRID:AB_2536173 |
| CD11c microbeads | Miltenyi Biotec | Cat #130-125-835 |
| Chemicals, peptides, and recombinant proteins | ||
| 4-kDA Fluorescein isothiocyanate–dextran | Sigma-Aldrich | Cat #60842-46-8 |
| Critical commercial assays | ||
| IgA mouse uncoated ELISA kit | Invitrogen | Cat #88-50450-88 |
| Mouse NGAL ELISA kit, ELISAGenie | Ozyme | Cat #MOFI00997 |
| E.Z.N.A Tissue DNA Kit | Omega bio-tek | Cat #D3396-00S |
| Deposited data | ||
| Row fastq files 16s rRNA sequencing | This study | Zenodo https://doi.org/10.5281/zenodo.7107337 |
| Workflow microbiome profiling of the terminal ileum and caecal contents | This study | Zenodo https://doi.org/10.5281/zenodo.7079338 |
| Experimental models: Organisms/strains | ||
| C57BL6/J mouse 21 days old | This study | Parents from Janvier |
| Oligonucleotides | ||
| pan-bacteria probe Eub338-Alexa 555 5-GCTGCCTCCCGTAGGAGT-3′ |
Invitrogen | https://doi.org/10.1128/aem.56.6.1919-1925.1990 |
| Segmented Filamentous Bacteria (SFB) 1008-AF594 probe 5-GCGAGCTTCCCTCATTACAAGG-3′ | Invitrogen | https://doi.org/10.1099/00207713-45-4-780 |
| Software and algorithms | ||
| PRISM 9 | GraphPad | N/A |
| Photoshop CC 2019 | Adobe | N/A |
| MetaboAnalyst 5.0 | https://doi.org/10.1093/nar/gkq329 | https://www.metaboanalyst.ca/home.xhtml |
| QIIME 2 | https://doi.org/10.1038/s41587-019-0209-9 | https://qiime2.org/ |
| ZEN Lite 2.3 SP1 | Carl Zeiss Microscopy | https://www.zeiss.com/microscopy/us/RRID:SCR_013672 |
| Other | ||
| Safe® A03, regular diet | SAFE | Cat #U8200G10R |
| MAIS +1800 mg/kg Sodium, SAFE R8960A01R 00128, SAM diet | SAFE | This study |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Julie Tomas (tomas@ciml.univ-mrs.fr).
Materials availability
This study did not generate new unique reagents.
Experimental model details
Animals
Animal experiments were approved by the French Ethics Committee and the French Research Ministry (APAFIS #31437). Mice were maintained in our animal house facility in ventilated cages under animal biosafety level 2 conditions at an ambient temperature of 22°C with a 12h light/dark cycle. At weaning (21 days), C57BL6/J male mice separated from normo-nourished mothers (Janvier Labs, Saint-Berthevin, France) were fed during one month either with regular diet (A03; Safe, France), or with a SAM diet based on a porcine model of infant malnutrition18 (MAIS +1800 mg/kg Sodium, SAFE R8960A01R 00128; Safe, France). The detailed composition is shown in Table S1. Nutritional intervention was performed on weanling mice fed with SAM diet for two weeks by switching to the regular diet for another two weeks. Weight and food consumption of mice were followed once a week. Mice were euthanized at 49 days old and length of the mice, small intestine (SI) and femur length were measured. Weight/Length was calculated for each mouse (W/L). The length without the tail was used to calculate the Weight-for-Length Z score (WL-Z). The Z score is calculated by deducting the mean of the referent population (μreferent) from the given variable (X) and then dividing the result by the standard deviation of the referent population (SDreferent) leading to the following formulation (Z = X-μreferent)/SDreferent. In the case of the WL-Z, the variable X = W/L, and the control group (Ctrl, n = 27) chosen as the referent group to calculate the μreferent (W/Lreferent) and the SDreferent leading to the formulation: WL-Z= (W/L- W/Lreferent)/SDreferent. Caecum (N = 10/group of diet), terminal ileal content (N = 8 for Ctrl, N = 13 for SAM) and feces were weighted and immediately store at −80°C for metagenomic, metabolomic and ELISA studies. Peyer’s patches and small intestine were collected for flow cytometry and histology studies. Due to the limited number of male mice generated from our own breeding, the smallest cohorts were generally used to assess physiological parameters, while the largest cohorts were dedicated to immune studies. A minimum of 3 mice per group is indeed required to analyze Peyer’s patches phagocyte populations. Furthermore, we first compared the two groups Ctrl and SAM, and only later included the third group, Int. Therefore, the number of mice and the number of replicates differ between the groups. For instance, 5–10 independent experiments means that five independent experiments were performed including all groups, while the other five included only the Ctrl and SAM groups. Therefore, the legend indicates 5 to 10 independent experiments.
Method details
Peyer’s patch and villus cell extraction for flow cytometry analyses
Peyer’s patches (PPs) were digested for 40 min at room temperature with collagenase/DNase as previously described.55 CD11c+ cells were enriched using anti-CD11c microbeads and an autoMACS magnetic cell separator according to manufacturer’s instructions (Miltenyi Biotec, France).
After removal of adherent adipose tissue and resection of PPs, 14 cm of the SI was cut into representative pieces of each part of the SI (duodenum-jejunum and ileum), opened longitudinally, washed in cold PBS 1X and placed in complete medium (RPMI 5% FCS, 1% Penicillin/Streptamycin, 1% L-Glutamine, 10 mM HEPES and 50 μM β-mercaptoethanol) containing 1 mM dithiothreitol. SI was cut into 2 mm pieces, washed in cold HBSS (Thermo Fischer) containing 2% FCS and filtered with a 100 μm cell strainer. Epithelial cells were removed by incubating tissues in HBSS+2 mM EDTA buffer at 37°C for 20 min at 180 rpm. This step was repeated twice before digestion at 37°C in complete medium with collagenase VIII (1 mg/mL, Sigma) and DNAse I (200 μg/mL, Sigma) for 15 min at 180 rpm. Digestion was stopped by adding 20 mL of cold PEF 2% buffer (PBS1X + 5 mM EDTA +2% FCS). Debris were removed by filtration. The cells were washed with cold PEF 2% before staining.
Flow cytometry analyses
After autoMACS separation, CD11c+ (positive fraction) and CD11c− (negative fraction) cells were pre-incubated on ice for 10 min with the 2.4G2 antibody to block Fc receptors. Cell mortality was evaluated using LIVE/DEAD Fixable Blue Dead Cell Stain Kit (Life Technologies). Intracellular staining was performed using eBioscience Intracellular Fixation & Permeabilization Buffer set according to manufacturer’s instructions (Invitrogen). Multiparametric flow cytometry was performed using a FACS LSRII UV. Data were analyzed with Flow Jo software. Three panels were designed to study the phagocyte population in PPs, T and B cells in PPs and SI.
Antibodies used for FACS analyses
Phagocyte panel: XCR1-BV421 (clone ZET), CD172α-FITC (clone P84), CD4-PeCy5.5 (clone RM4-5), CD11c-PeCy7 (clone N418), BST2-APC (clone 927), CMHII-A700 (clone M5/114.15.2) and CD11b-APC/Cy7 (clone M1/70). T cell panel: CD45-BUV395 (clone 30-F11), PD-1-BV605 (clone J43), CD62L-BV786 (clone MEL-14), CD8α-AF488 (clone 53-6.7), CXCR5-PE (clone 2G8), CD4-PeCy5.5 (clone RM4-5), RORγt-Pe-eFluor610 (clone B2D), Foxp3-APC (clone FJK-16S) and CD44-A700 (clone IM7). B cell panel: CD45-BUV395 (clone 30-F11), CD43-BUV737 (clone S7), CD3-eFluor450 (clone 17A2), Siglec-F-BV421 (clone E50-2440), XCR1-BV421 (clone ZET), CD19-BV605 (clone 6D5), B220-BV786 (clone RA3-6B2), IgM-biotin (clone II/41), IgA-PE (clone mA-6E1), IgD-Pe/Cy7 (clone 11-26c.2a), GL-7-eF660 (GL-7), Streptavidin PE-CF594.
Immunofluorescence staining and confocal microscopy
The three-last ileal PPs were removed and fixed with Antigenfix (Diapath) for 1h. After washed in 1X PBS, PPs were incubated overnight in 34% sucrose at 4°C. After inclusion in OCT (Invitrogen), PPs were cut and stained as previously described.56 After immunofluorescence staining, the slides were observed with a Zeiss LSM 780 confocal microscope using the spectral imaging mode.87 Quantification and images processing were done using Adobe Photoshop CC 2019. The following antibodies were used for phagocyte population: Rat anti-MerTK (clone DS5MMER), armenian hamster anti-CD11c (clone N418, Biolegend) and rabbit anti-lysozyme (polyclonal, Dako), SIPR-α-AF594 (clone P84, Biolegend), CD4-eFluor450 (clone RM4-5, eBioscience) or CD4-eF570 (clone RM4-5, Invitrogen), EpCAM-eF450 (clone G8.8, eBioscience). Secondary antibodies were used: donkey anti rat-Cy3 (Jackson Immuno-research) or -AF488 (Invitrogen), goat anti hamster-AF594 or Cy3 (Jackson Immuno-research), goat anti-rabbit AF514 (Invitrogen). SYTOX blue for nuclei acid stain (ThermoFisher Scientific).
Fluorescence in situ hybridization (FISH)
Unflushed distal PPs and SI were collected in Carnoy (absolute ethanol/chloroform/acetic acid 60:30:10) and incubated overnight at 4°C. Samples were dehydrated, and embedded in paraffin according to the standard protocol for hematoxylin and eosin stain (HE) or immunofluorescence (IF). All the staining was done on dewaxed 8-μm sections. In IF experiments, antigen retrieval was performed in citric acid buffer 2 mM pH 6 for 45 min at 96 °C and fluorescence-labeled secondary antibodies were used and nuclei stained with DAPI. IF assay was performed with the anti-MUC2 antibody (1/500; sc-15334, Santa Cruz Biotechnology) and anti-ZO-1 (1/100, Invitrogen). Procedure used for FISH experiments are described in Pedron et al., 2012.88 The pan-bacteria probe Eub338-Alexa 555 5′-GCTGCCTCCCGTAGGAGT-3′′,89 and the specific Segmented Filamentous Bacteria (SFB) 1008-AF594 probe 5′-GCGAGCTTCCCTCATTACAAGG-3′′90 were used in embedded paraffin sections of PPs and ileum. DNA was stained with Sytoxblue revealing both eukaryotic and bacterial cells.
Spectral confocal imaging projections of ileal villus and Peyer’s patches from 49-day old Ctrl, SAM and Int mice stained by FISH for all bacteria (Eub-338 probe) and specific probe to identify SFB (SFB-1004 probe) were used to quantify the distance between the microbiota and the intestinal epithelium. Only Eub-338 specific signal that was not identified as positive for SFB-1004 specific signal was used to measure the proximity of the luminal microbiota to the epithelium (top of villi or follicle-associated epithelium, FAE) in each diet group, using ZEN Lite 2.3 SP1 software line tool (Carl Zeiss Microscopy).
Intestinal permeability assessment
The gut permeability was tested using 4-kDA FITC-dextran (Sigma-Aldrich). Mice were fasted 4 h before gavage. Each mouse was gavaged with 100 μL of FITC-dextran solution at 80 mg/mL diluted in sterile 1X PBS. Four hours after gavage, mice were anesthetized with isoflurane and blood was collected retro-orbitallly in EDTA tubes. The blood was centrifuged at 5000 rpm during 10 min at 4°C. FITC-dextran fluorescence was read in the microplate reader Infinite M1000 PRO using the i-Control software (Tecan) with excitation of 485 nm and emission of 530 nm (20 nm emission bandwidth). Diluted or not diluted samples were added in 96-well black plate in duplicate when possible. Plasma from ungavaged mice as well as 1X PBS wells were added for the calculation of the concentration. A standard curve was used to convert values to concentration.
Quantification of fecal IgA by ELISA
Stools were weighted and diluted in an inhibitor cocktail at 100 mg/mL containing PMSF 0.35 mg/mL (Sigma-Aldrich), EDTA-Na2 0.75 mg/mL (Sigma-Aldrich), pepstatine 1 μg/mL (Sigma-Aldrich) and aprotinine 2,000 times diluted (Sigma-Aldrich). After mechanical disaggregation, stools were incubated for 20 min in the inhibitor cocktail at 4°C. Stools were then vortexed and centrifuged at 14, 000 rpm for 10 min. Supernatants were collected and conserved at −20°C for IgA quantification. Total IgA detection was determined according to the manufacturer’s protocol (IgA mouse uncoated ELISA kit, Invitrogen). The NUNC MaxiSorp high protein-binding capacity 96 well ELISA plates were used. Colorimetric reaction was measured at OD = 450 nm by microplate reader Infinite M1000 PRO using the i-Control software (Tecan). Corresponding immunoglobulin isotype was used as standard after serial dilutions.
Quantification of fecal NGAL (neutrophil gelatinase associated lipocalin)
Stools were weighted and mechanically disaggregated in 300 μL of sample dilution buffer provided by the manufacturer (Mouse NGAL ELISA kit, ELISAGenie, Ozyme). The supernatants were removed after centrifugation (5,000 rpm, 10 min) and diluted for ELISA quantification following manufacturer’s instructions.
Metagenomics
DNA extraction and library preparation
Terminal ileal content
Bacterial DNA was extracted by a manual treatment with acid washed glass beads (G4649-500g Sigma) and 0.5 mm glass beads (Scientific Industries, Inc) using a FastPrep-24 5G Grinder (mpBio) at maximum speed (6.5 m/s) for 90s. To optimize DNA extraction from all the bacteria present in the stools, two protocols were applied: the former using E.Z.N.A Tissue DNA Kit (Omega bio-tek, Norcross, USA) and the latter based on glycan degradation using EZ1 Advanced XL device (Qiagen, Courtaboeuf, France).91 For each extraction protocol, genomic DNA was amplified for the 16S “V3-V4” regions by PCR for 45 cycles, using the Kapa HiFi Hotstart ReadyMix 2x (Kapa Biosystems Inc,Wilmington, MA U.S.A), and the surrounding conserved region V3_V4 primers with overhang adapters (FwOvAd_341FTCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG; RevOvAd_785RGTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC). After purification on AMPure XP beads (Beckman Coulter Inc, Fullerton, CA, USA), the concentration was measured using High sensitivity Qubit technology (Beckman Coulter Inc, Fullerton, CA, USA). Library of each extraction (440–470 pb) method were pooled (1:1) at 3.5 ng/μL and Illumina sequencing adapters and dual-index barcodes were added to amplicons. After purification on AMPure XP beads (Beckman Coulter Inc, Fullerton,CA,USA), this library was pooled with 92 others multiplexed samples. The global concentration was quantified by a Qubit assay with the high sensitivity kit (Life technologies, Carlsbad, CA, USA). Purified amplicons were pooled in equimolar concentration to obtain a 7 pm library containing 15% of PhiX control. The samples were then sequenced for 16S rRNA sequencing on MiSeq technology (Illumina, Inc, San Diego CA 92121, USA) with the paired-end 300 bases pair protocol according to the 16S Metagenomic Sequencing Library Preparation (Illumina) at the Institute Hospitalo-Universitaire Méditerranée Infection, Marseille, France. Automated cluster generation and paired-end sequencing with dual index reads were performed in single 39-h run in a 2x250bp.
Caecum content
The extraction was performed using the protocol developed and standardized by Genoscreen, as well as the recommendations of the IHMS. An optimized and standardized DNA extraction protocol dedicated to bacterial DNA extraction from stools samples has been used (GenoScreen, Lille, France). Genomic DNA extraction from stools samples was done with the QIAamp Fast DNA stool mini kit (Qiagen, Germany) with optimized protocol for lysis step. After DNA extraction, the concentration was quantified with the SybrGreen assay Kit (Life Technologies, USA).
16S rRNA gene sequencing and microbiome profiling
Terminal ileal content
Twenty-two samples were sequenced including 9 from the Ctrl group and 13 from the SAM group with PCR mix control. A total of 147,711,748 read pairs were generated (mean 120,045 read pairs per sample with median of 120,401). The reads were first demultiplexed according to their dual barcode by samples. Quality of the raw reads were assessed using FastQC v0.11.9 toolkit92 and low-quality reads were trimmed using Trimmomatic v0.39.93 V3_V4 primer with overhang adapters (FwOvAd_341F and RevOvAd_785R) were removed using cutadapt v3.2. Amplicon Sequence Variants (ASVs) were produce using DADA294 in QIIME 2.95 We obtained 6475 ASVs with a length ≥ 400 nt to avoid non v3-v4 ribosomal sequence. A Naive Bayes classifiers trained on the Silva (release 138) 99% OTUs full-length sequences were applied to obtain the pre-trained taxonomy classifiers used for the taxonomic assignation of the features with the classify-sklearn method.96 The table was rarefied to 7500 sequences and filtered to exclude samples with less than 7500 sequences. A rooted tree for phylogenetic diversity analysis was generated with SEPP.97 The resulting phylogenetic tree was further processed to calculate core diversity metrics, including β-diversity based on weighted and unweighted UniFrac distances98 and α-diversity measures calculated in QIIME 2. To test for significant differences in taxonomic abundances we used the Shannon index (in this case, Faith’s Phylogenetic Diversity) and the non-parametric Kruskal–Wallis test. Differences were considered significant when the false-discovery rate correction is at p < 0.05. The intergroup high similarity and intra group low similarity of microbiota are proven by β-diversity.
Caecum content
Thirty samples were sequenced including 10 for each Ctrl, SAM and Int groups. Microbial diversity was determined for each sample by targeting part of ribosomal genes. A 16S rRNA gene fragment comprising V3 and V4 hypervariable regions was amplified using an optimized and standardized 16S-amplicon-library preparation protocol (Metabiote, GenoScreen, Lille, France). Briefly, 16S rRNA gene PCR was carried out using 5 ng of genomic DNA according to Metabiote protocole (or maximal of DNA volume) instructions using 192 bar-coded primers (Metabiote MiSeq Primers, GenoScreen, Lille, France) at final concentrations of 0.2 μM and an annealing temperature of 50°C for 30 cycles. PCR products were cleaned up with Agencourt AMPure XP-PCR Purification system (Beckman Coulter, Brea, USA), quantified according to the manufacturer’s protocol, and multiplexed at equal concentration. Sequencing was performed using a 250-bp paired-end sequencing protocol on the Illumina MiSeq platform (Illumina, San Diego, USA) at GenoScreen, Lille, France. Raw paired-end reads were subjected to the following process 1) quality filtering with the PRINSEQ-lite PERL script99 by truncation of bases from the 3′end not with quality <30 based on the Phred algorithm, 2) paired-end read assembly using FLASH100 with a minimum length overlap of 30 bases and 97% overlap identity and 3) the search and removal of both forward and reverse primer sequences using CutAdapt, with no mismatches allowed in primers sequences. Assembled sequences for which perfect forward and reverse primers are not found are eliminated. Amplicon Sequence Variants (ASVs) were produce using DADA294 in QIIME 2.95 We obtained 1549 ASVs with a length ≥ 400 nt to avoid non v3-v4 ribosomal sequence. The following analyses was realized as described for terminal ileum content.
Untargeted metabolomic approach
Short chain fatty acids (SCFAs) analyses
Acetate, propionate, butyrate, isobutyrate, valerate, and isovalerate were quantified in 35–115 mg of caecal content by gas chromatography-mass spectrometry (GC-MS) following the procedure described in Apper et al., 2020.101
Caecal preparation
Forty-five mg of each cecal samples were weight in Eppendorff and content was homogenized in cooled methanol (3 μL/mg feces) at −20 °C. Samples were vortexed for 1 min and incubated at −20 °C for 30 min. Samples were then centrifuged for 15 min (11,000 × g, 4 °C). The supernatant recovered from each sample was filtered through 10 kDa filter tubes by centrifuging for 45 min (11,000 × g, 4 °C). The extracts obtained were then dried using a stream of nitrogen. The dry extracts were reconstituted with 125 μL acetonitrile/water (50:50; v/v) and filtered through 0.45 μm filters by centrifugation for 35 min (11,000 × g, 4 °C). Fifty μL of sample extract were transferred into vial for Liquid Chromatography Mass Spectrometry (LCMS) metabolomic analyses. The samples were separated using ultrahigh-performance liquid chromatography (UPLC) ultimate 3000 (Thermo Scientific), coupled to a high-resolution mass spectrometer (HRMS), Q-Exactive Plus quadrupole-orbitrap hybrid equipped with electrospray ionization source (H-ESI II). A reverse-phase C18 Hypersil Gold (100 mm × 2.1 mm × 1.9 μm) (Thermo Scientific, France) column was used for compound separation in the samples. The injection volume was 5 μL. Here, 0.1% formic acid solutions in water and acetonitrile were used as solvents A and B, respectively, as mobile phases. The elution gradient for sample analysis was as follows: Solvent B was maintained at 5% during the first minute. A linear gradient then raised solvent B to 50%, maintained it for 2 min, before raising it again to 97% of B for the next 6 min. Then, 97% of solvent B was held in isocratic conditions for 2 min, after which initial conditions were restored, and the column re-equilibrated for 4 min. The separated molecules were analyzed in both positive and negative ionization modes in the same run. The repeatability of the analysis was checked by analyzing interspaced (1 out of every 5 samples) quality control samples (QC).
Data processing and molecule identification
All the raw data generated by the UPLC- HRMS were converted to mzXML by ProteoWizard (Version 2.0), and then processed by MZmine 2.26. The identification of the metabolites was performed by using an in-house database referencing more than 800 metabolites with their chromatographic retention time and their exact mass and MSMS spectra obtained in positive and negative ionization modes, including their adducts and neutral losses. Statistical analyses were performed using MetaboAnalyst 5.0 software.86
Quantification and statistical analysis
Statistical analysis was conducted using Prism 9 (GrahPad, San Diego, CA) (Figures 1, Figure 5, Figure 6, Figure 7) or using MetaboAnalyst 5.0 software for metabolomic and metagenomic studies (Figure 2, Figure 3, Figure 4). The data are expressed as the mean ± the standard deviation (SD). For the analysis in Figures 1, 4D, 5, 6, and 7 we performed a non-parametric one-way ANOVA, followed by Dunn test to assess the significance among pairs of conditions. For the analyses in Figures 2C, 3C and 3D and 4C a non-parametric Kruskal Wallis Test was performed.
For the analysis in Figures 3B and 4B, 4A T-test/Anova was performed. For the analysis in Figure 2D, the non-parametric Wilcoxon rank-sum test was used to generate the volcano plot. The details for each experiment including the number of animals and the number of independent experiments are specified in the figure legends. In the Figures 3C and 3D, Linear discriminant analysis (LDA) effect size (LEfSe) was applied to discern significant differences in the relative abundance of microbial taxa among all the groups, which included the non-parametric Kruskal–Wallis test and LDA2.102 The groups were compared in pairs. Taxonomical features with p-value <0.05 and LDA effect size >2 were regarded as significant microbial signatures. In the Figures 4E and 4F, Spearman’s rank correlation analyses were performed in the R environment using stats package and cor function. Heatmap where generated with gplots-3.0.1.2 package and heatmap.2 function using dist and hclust arguments to compute the distance and the hierarchical clustering based on pairwise distances calculated from Spearman correlation values.
Acknowledgments
We thank Sylvie Mémet for helpful discussions and improvement in the manuscript. We acknowledge the PICSL imaging facility of the CIML (ImagImm), member of the national infrastructure France-BioImaging supported by the French National Research Agency (ANR-10-INBS-04), and the cytometry core facility of the CIML for expert assistance. This work was supported by institutional funding from Centre National de la Recherche Scientifique and Institut National de la Santé et de la Recherche Médicale, by the Agence Nationale de la Recherche ANR-20-CE15-0016, and by Fondation pour la Recherche Médicale (FRM), grant number DEQ20170336745. The project leading to this publication has received funding from France 2030, the French Government program managed by the Agence Nationale de la Recherche (ANR-16-CONV-0001) and from Excellence Initiative of Aix-Marseille University – A∗MIDEX. C.A.P. was supported by the University of Costa Rica. F.H.-V. was supported by the Infectiopôle Sud at Marseille and the University of Honduras.
Author contributions
Conceptualization, J.T., H.L., and M.M.; Methodology, J.T., H.L., and F.H.-V.; Investigation, F.H.-V., M.M., M.L., C.W., C.A.P., L.C., C.L., H.L., and J.T. Metagenomics, M.M., T.V., V.B., and J.T.; Metabolomics, C.D., L.S., F.H.-V., and J.T.; Writing-Original Draft, F.H.-V. and J.T.; Writing-Review & Editing, F.H.-V., H.L., J.T., and J.P.G.; Supervision, H.L., J.P.G., and J.T.; Project Administration and Funding Acquisition, H.L. and J.P.G.
Declaration of interests
The authors declare no competing interests.
Published: May 19, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106910.
Supplemental information
Data and code availability
- Data
- Raw fastq files 16s rRNA sequencing data have been deposited at Zenodo and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
- Workflow data on the parameters used for the microbiome profiling of the terminal ileum and caecal contents have been deposited at Zenodo and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
- Code
- This paper does not report original code.
- Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Mayneris-Perxachs J., Swann J.R. Metabolic phenotyping of malnutrition during the first 1000 days of life. Eur. J. Nutr. 2019;58:909–930. doi: 10.1007/s00394-018-1679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al Nabhani Z., Dulauroy S., Marques R., Cousu C., Al Bounny S., Déjardin F., Sparwasser T., Bérard M., Cerf-Bensussan N., Eberl G. A weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity. 2019;50:1276–1288.e5. doi: 10.1016/j.immuni.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 3.Wagner C., Torow N., Hornef M.W., Lelouard H. Spatial and temporal key steps in early-life intestinal immune system development and education. FEBS J. 2022;289:4731–4757. doi: 10.1111/febs.16047. [DOI] [PubMed] [Google Scholar]
- 4.Bourke C.D., Jones K.D.J., Prendergast A.J. Current understanding of innate immune cell dysfunction in childhood undernutrition. Front. Immunol. 2019;10:1728. doi: 10.3389/fimmu.2019.01728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.UNICEF . UNICEF; 2021. RUTF-technical-expert-meeting-report-02-03092019. [Google Scholar]
- 6.WHO/UNICEF . 2009. Child Growth Standard and the Identification of Severe Acute Malnutrition in Infants and Children. [Google Scholar]
- 7.Bwakura-Dangarembizi M., Amadi B., Bourke C.D., Robertson R.C., Mwapenya B., Chandwe K., Kapoma C., Chifunda K., Majo F., Ngosa D., et al. Health outcomes, pathogenesis and epidemiology of severe acute malnutrition (HOPE-SAM): rationale and methods of a longitudinal observational study. BMJ Open. 2019;9:e023077. doi: 10.1136/bmjopen-2018-023077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerac M., Bunn J., Chagaluka G., Bahwere P., Tomkins A., Collins S., Seal A. Follow-up of post-discharge growth and mortality after treatment for severe acute malnutrition (FuSAM study): a prospective cohort study. PLoS One. 2014;9:e96030. doi: 10.1371/journal.pone.0096030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prendergast A., Kelly P. Enteropathies in the developing world: neglected effects on global health. Am. J. Trop. Med. Hyg. 2012;86:756–763. doi: 10.4269/ajtmh.2012.11-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiens M.O., Pawluk S., Kissoon N., Kumbakumba E., Ansermino J.M., Singer J., Ndamira A., Larson C. Pediatric post-discharge mortality in resource poor countries: a systematic review. PLoS One. 2013;8:e66698. doi: 10.1371/journal.pone.0066698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gehrig J.L., Venkatesh S., Chang H.W., Hibberd M.C., Kung V.L., Cheng J., Chen R.Y., Subramanian S., Cowardin C.A., Meier M.F., et al. Effects of microbiota-directed foods in gnotobiotic animals and undernourished children. Science. 2019;365:eaau4732. doi: 10.1126/science.aau4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanian S., Huq S., Yatsunenko T., Haque R., Mahfuz M., Alam M.A., Benezra A., DeStefano J., Meier M.F., Muegge B.D., et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith M.I., Yatsunenko T., Manary M.J., Trehan I., Mkakosya R., Cheng J., Kau A.L., Rich S.S., Concannon P., Mychaleckyj J.C., et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kau A.L., Planer J.D., Liu J., Rao S., Yatsunenko T., Trehan I., Manary M.J., Liu T.-C., Stappenbeck T.S., Maleta K.M., et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci. Transl. Med. 2015;7:276ra24. doi: 10.1126/scitranslmed.aaa4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown E.M., Wlodarska M., Willing B.P., Vonaesch P., Han J., Reynolds L.A., Arrieta M.-C., Uhrig M., Scholz R., Partida O., et al. Diet and specific microbial exposure trigger features of environmental enteropathy in a novel murine model. Nat. Commun. 2015;6:7806. doi: 10.1038/ncomms8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salameh E., Jarbeau M., Morel F.B., Zeilani M., Aziz M., Déchelotte P., Marion-Letellier R. Modeling undernutrition with enteropathy in mice. Sci. Rep. 2020;10:15581. doi: 10.1038/s41598-020-72705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salameh E., Morel F.B., Zeilani M., Déchelotte P., Marion-Letellier R. Animal models of undernutrition and enteropathy as tools for assessment of nutritional intervention. Nutrients. 2019;11:2233. doi: 10.3390/nu11092233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lykke M., Hother A.L., Hansen C.F., Friis H., Mølgaard C., Michaelsen K.F., Briend A., Larsen T., Sangild P.T., Thymann T. Malnutrition induces gut atrophy and increases hepatic fat infiltration, studies in a pig model of chilhood malnutrition. Am. J. Transl. Res. 2013;15;5:543–554. [PMC free article] [PubMed] [Google Scholar]
- 19.Golden M. 1985. The Consequences of Protein Deficiency in Man and its Relationship to the Clinical Features of Kwashiorkor. [DOI] [Google Scholar]
- 20.Golden M.H.N. Specific deficiencies versus growth failure: type I and type II nutrients. J. Nutr. Environ. Med. 2009;6:301–308. doi: 10.3109/13590849609007256. [DOI] [PubMed] [Google Scholar]
- 21.McGuire E.A., Young V.R. Nutritional edema in a rat model of protein deficiency significance of the dietary potassium and sodium content. J. Nutr. 1986;116:1209–1224. doi: 10.1093/jn/116.7.1209. [DOI] [PubMed] [Google Scholar]
- 22.Costa L.B., JohnBull E.A., Reeves J.T., Sevilleja J.E., Freire R.S., Hoffman P.S., Lima A.A.M., Oriá R.B., Roche J.K., Guerrant R.L., Warren C.A. Cryptosporidium-malnutrition interactions: mucosal disruption, cytokines, and TLR signaling in a weaned murine model. J. Parasitol. 2011;97:1113–1120. doi: 10.1645/GE-2848.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soni K.G., Dike P.N., Suh J.H., Halder T., Edwards P.T., Foong J.P.P., Conner M.E., Preidis G.A. Early-life malnutrition causes gastrointestinal dysmotility that is sexually dimorphic. Neuro Gastroenterol. Motil. 2020;32:e13936. doi: 10.1111/nmo.13936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raman A.S., Gehrig J.L., Venkatesh S., Chang H.W., Hibberd M.C., Subramanian S., Kang G., Bessong P.O., Lima A.A.M., Kosek M.N., et al. A sparse covarying unit that describes healthy and impaired human gut microbiota development. Science. 2019;365:eaau4735. doi: 10.1126/science.aau4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis C.P., Savage D.C. Habitat, succession, attachment, and morphology of segmented, filamentous microbes indigenous to the murine gastrointestinal tract. Infect. Immun. 1974;10:948–956. doi: 10.1128/iai.10.4.948-956.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Derrien M., Belzer C., de Vos W.M. Akkermansia muciniphila and its role in regulating host functions. Microb. Pathog. 2017;106:171–181. doi: 10.1016/j.micpath.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 27.Schnupf P., Gaboriau-Routhiau V., Sansonetti P.J., Cerf-Bensussan N. Segmented filamentous bacteria, Th17 inducers and helpers in a hostile world. Curr. Opin. Microbiol. 2017;35:100–109. doi: 10.1016/j.mib.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 28.Hill D.A., Hoffmann C., Abt M.C., Du Y., Kobuley D., Kirn T.J., Bushman F.D., Artis D. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010;3:148–158. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson G.R., Trexler P.C. Gastrointestinal structure and function in germ-free or gnotobiotic animals. Gut. 1971;12:230–235. doi: 10.1136/gut.12.3.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waite D.W., Chuvochina M., Pelikan C., Parks D.H., Yilmaz P., Wagner M., Loy A., Naganuma T., Nakai R., Whitman W.B., et al. Proposal to reclassify the proteobacterial classes Deltaproteobacteria and Oligoflexia, and the phylum Thermodesulfobacteria into four phyla reflecting major functional capabilities. Int. J. Syst. Evol. Microbiol. 2020;70:5972–6016. doi: 10.1099/ijsem.0.004213. [DOI] [PubMed] [Google Scholar]
- 31.Fan P., Liu P., Song P., Chen X., Ma X. Moderate dietary protein restriction alters the composition of gut microbiota and improves ileal barrier function in adult pig model. Sci. Rep. 2017;7:43412. doi: 10.1038/srep43412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navarro G., Sharma A., Dugas L.R., Forrester T., Gilbert J.A., Layden B.T. Gut microbial features can predict host phenotype response to protein deficiency. Phys. Rep. 2018;6:e13932. doi: 10.14814/phy2.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagkouvardos I., Joseph D., Kapfhammer M., Giritli S., Horn M., Haller D., Clavel T. IMNGS: a comprehensive open resource of processed 16S rRNA microbial profiles for ecology and diversity studies. Sci. Rep. 2016;6:33721. doi: 10.1038/srep33721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yusufu I., Ding K., Smith K., Wankhade U.D., Sahay B., Patterson G.T., Pacholczyk R., Adusumilli S., Hamrick M.W., Hill W.D., et al. A tryptophan-deficient diet induces gut microbiota dysbiosis and increases systemic inflammation in aged mice. Int. J. Mol. Sci. 2021;22:5005. doi: 10.3390/ijms22095005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X.L., Chen L., Yang J., Zhao S.S., Jin S., Ao N., Yang J., Liu H.X., Du J. Vitamin D alleviates non-alcoholic fatty liver disease via restoring gut microbiota and metabolism. Front. Microbiol. 2023;14:1117644. doi: 10.3389/fmicb.2023.1117644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mancabelli L., Milani C., Lugli G.A., Turroni F., Cocconi D., van Sinderen D., Ventura M. Identification of universal gut microbial biomarkers of common human intestinal diseases by meta-analysis. FEMS Microbiol. Ecol. 2017;93 doi: 10.1093/femsec/fix153. [DOI] [PubMed] [Google Scholar]
- 37.Dinh D.M., Ramadass B., Kattula D., Sarkar R., Braunstein P., Tai A., Wanke C.A., Hassoun S., Kane A.V., Naumova E.N., et al. Longitudinal analysis of the intestinal microbiota in persistently stunted young children in south India. PLoS One. 2016;11:e0155405. doi: 10.1371/journal.pone.0155405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Million M., Tomas J., Wagner C., Lelouard H., Raoult D., Gorvel J.-P. New insights in gut microbiota and mucosal immunity of the small intestine. Human Microbiome Journal. 2018;7–8:23–32. doi: 10.1016/j.humic.2018.01.004. [DOI] [Google Scholar]
- 39.Pekmez C.T., Dragsted L.O., Brahe L.K. Gut microbiota alterations and dietary modulation in childhood malnutrition - the role of short chain fatty acids. Clin. Nutr. 2019;38:615–630. doi: 10.1016/j.clnu.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 40.Castro-Mejía J.L., O'Ferrall S., Krych Ł., O'Mahony E., Namusoke H., Lanyero B., Kot W., Nabukeera-Barungi N., Michaelsen K.F., Mølgaard C., et al. Restitution of gut microbiota in Ugandan children administered with probiotics (Lactobacillus rhamnosus GG and Bifidobacterium animalis subsp. lactis BB-12) during treatment for severe acute malnutrition. Gut Microb. 2020;11:855–867. doi: 10.1080/19490976.2020.1712982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huus K.E., Rodriguez-Pozo A., Kapel N., Nestoret A., Habib A., Dede M., Manges A., Collard J.M., Sansonetti P.J., Vonaesch P., et al. Immunoglobulin recognition of fecal bacteria in stunted and non-stunted children: findings from the Afribiota study. Microbiome. 2020;8:113. doi: 10.1186/s40168-020-00890-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tidjani Alou M., Million M., Traore S.I., Mouelhi D., Khelaifia S., Bachar D., Caputo A., Delerce J., Brah S., Alhousseini D., et al. Gut bacteria missing in severe acute malnutrition, can we identify potential probiotics by culturomics? Front. Microbiol. 2017;8:899. doi: 10.3389/fmicb.2017.00899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamanai-Shacoori Z., Smida I., Bousarghin L., Loreal O., Meuric V., Fong S.B., Bonnaure-Mallet M., Jolivet-Gougeon A. Roseburia spp. a marker of health. Future Microbiol. 2017;12:157–170. doi: 10.2217/fmb-2016-0130. [DOI] [PubMed] [Google Scholar]
- 44.Vacca M., Celano G., Calabrese F.M., Portincasa P., Gobbetti M., De Angelis M. The controversial role of human gut Lachnospiraceae. Microorganisms. 2020;8:573. doi: 10.3390/microorganisms8040573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vonaesch P., Morien E., Andrianonimiadana L., Sanke H., Mbecko J.R., Huus K.E., Naharimanananirina T., Gondje B.P., Nigatoloum S.N., Vondo S.S., et al. Stunted childhood growth is associated with decompartmentalization of the gastrointestinal tract and overgrowth of oropharyngeal taxa. Proc. Natl. Acad. Sci. USA. 2018;115:E8489–E8498. doi: 10.1073/pnas.1806573115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vonaesch P., Araújo J.R., Gody J.C., Mbecko J.R., Sanke H., Andrianonimiadana L., Naharimanananirina T., Ningatoloum S.N., Vondo S.S., Gondje P.B., et al. Stunted children display ectopic small intestinal colonization by oral bacteria, which cause lipid malabsorption in experimental models. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2209589119. e2209589119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waters J.L., Ley R.E. The human gut bacteria Christensenellaceae are widespread, heritable, and associated with health. BMC Biol. 2019;17:83. doi: 10.1186/s12915-019-0699-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kropp C., Le Corf K., Relizani K., Tambosco K., Martinez C., Chain F., Rawadi G., Langella P., Claus S.P., Martin R. The Keystone commensal bacterium Christensenella minuta DSM 22607 displays anti-inflammatory properties both in vitro and in vivo. Sci. Rep. 2021;11:11494. doi: 10.1038/s41598-021-90885-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawano Y., Edwards M., Huang Y., Bilate A.M., Araujo L.P., Tanoue T., Atarashi K., Ladinsky M.S., Reiner S.L., Wang H.H., et al. Microbiota imbalance induced by dietary sugar disrupts immune-mediated protection from metabolic syndrome. Cell. 2022;185:3501–3519.e20. doi: 10.1016/j.cell.2022.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshii K., Hosomi K., Sawane K., Kunisawa J. Metabolism of dietary and microbial vitamin B family in the regulation of host immunity. Front. Nutr. 2019;6:48. doi: 10.3389/fnut.2019.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Macpherson A.J., Yilmaz B., Limenitakis J.P., Ganal-Vonarburg S.C. IgA function in relation to the intestinal microbiota. Annu. Rev. Immunol. 2018;36:359–381. doi: 10.1146/annurev-immunol-042617-053238. [DOI] [PubMed] [Google Scholar]
- 52.Da Silva C., Wagner C., Bonnardel J., Gorvel J.P., Lelouard H. The peyer's patch mononuclear phagocyte system at steady state and during infection. Front. Immunol. 2017;8:1254. doi: 10.3389/fimmu.2017.01254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung C., Hugot J.P., Barreau F. Peyer's patches: the immune sensors of the intestine. Int. J. Inflamm. 2010;2010:823710. doi: 10.4061/2010/823710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhattacharjee A., Burr A.H.P., Overacre-Delgoffe A.E., Tometich J.T., Yang D., Huckestein B.R., Linehan J.L., Spencer S.P., Hall J.A., Harrison O.J., et al. Environmental enteric dysfunction induces regulatory T cells that inhibit local CD4+ T cell responses and impair oral vaccine efficacy. Immunity. 2021;54:1745–1757.e7. doi: 10.1016/j.immuni.2021.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonnardel J., Da Silva C., Henri S., Tamoutounour S., Chasson L., Montañana-Sanchis F., Gorvel J.P., Lelouard H. Innate and adaptive immune functions of peyer's patch monocyte-derived cells. Cell Rep. 2015;11:770–784. doi: 10.1016/j.celrep.2015.03.067. [DOI] [PubMed] [Google Scholar]
- 56.Lelouard H., Fallet M., de Bovis B., Méresse S., Gorvel J.P. Peyer's patch dendritic cells sample antigens by extending dendrites through M cell-specific transcellular pores. Gastroenterology. 2012;142:592–601.e3. doi: 10.1053/j.gastro.2011.11.039. [DOI] [PubMed] [Google Scholar]
- 57.Lelouard H., Henri S., De Bovis B., Mugnier B., Chollat-Namy A., Malissen B., Méresse S., Gorvel J.P. Pathogenic bacteria and dead cells are internalized by a unique subset of Peyer's patch dendritic cells that express lysozyme. Gastroenterology. 2010;138:173–184.e1. doi: 10.1053/j.gastro.2009.09.051. [DOI] [PubMed] [Google Scholar]
- 58.Wagner C., Bonnardel J., Da Silva C., Spinelli L., Portilla C.A., Tomas J., Lagier M., Chasson L., Masse M., Dalod M., et al. Differentiation paths of peyer's patch LysoDCs are linked to sampling site positioning, migration, and T cell priming. Cell Rep. 2020;31:107479. doi: 10.1016/j.celrep.2020.03.043. [DOI] [PubMed] [Google Scholar]
- 59.Maghraby M.K., Li B., Chi L., Ling C., Benmoussa A., Provost P., Postmus A.C., Abdi A., Pierro A., Bourdon C., Bandsma R.H.J. Extracellular vesicles isolated from milk can improve gut barrier dysfunction induced by malnutrition. Sci. Rep. 2021;11:7635. doi: 10.1038/s41598-021-86920-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwarzer M., Gautam U.K., Makki K., Lambert A., Brabec T., Joly A., Šrůtková D., Poinsot P., Novotná T., Geoffroy S., et al. Microbe-mediated intestinal NOD2 stimulation improves linear growth of undernourished infant mice. Science. 2023;379:826–833. doi: 10.1126/science.ade9767. [DOI] [PubMed] [Google Scholar]
- 61.Guerrant R.L., Leite A.M., Pinkerton R., Medeiros P.H., Cavalcante P.A., DeBoer M., Kosek M., Duggan C., Gewirtz A., Kagan J.C., et al. Biomarkers of environmental enteropathy, inflammation, stunting, and impaired growth in children in northeast Brazil. PLoS One. 2016;11:e0158772. doi: 10.1371/journal.pone.0158772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Naylor C., Lu M., Haque R., Mondal D., Buonomo E., Nayak U., Mychaleckyj J.C., Kirkpatrick B., Colgate R., Carmolli M., et al. Environmental enteropathy, oral vaccine failure and growth faltering in infants in Bangladesh. EBioMedicine. 2015;2:1759–1766. doi: 10.1016/j.ebiom.2015.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tomas J., Mulet C., Saffarian A., Cavin J.B., Ducroc R., Regnault B., Kun Tan C., Duszka K., Burcelin R., Wahli W., et al. High-fat diet modifies the PPAR-gamma pathway leading to disruption of microbial and physiological ecosystem in murine small intestine. Proc. Natl. Acad. Sci. USA. 2016;113:E5934–E5943. doi: 10.1073/pnas.1612559113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johansson M.E.V., Jakobsson H.E., Holmén-Larsson J., Schütte A., Ermund A., Rodríguez-Piñeiro A.M., Arike L., Wising C., Svensson F., Bäckhed F., Hansson G.C. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe. 2015;18:582–592. doi: 10.1016/j.chom.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huus K.E., Hoang T.T., Creus-Cuadros A., Cirstea M., Vogt S.L., Knuff-Janzen K., Sansonetti P.J., Vonaesch P., Finlay B.B. Cross-feeding between intestinal pathobionts promotes their overgrowth during undernutrition. Nat. Commun. 2021;12:6860. doi: 10.1038/s41467-021-27191-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Geerlings S.Y., Kostopoulos I., de Vos W.M., Belzer C. Akkermansia muciniphila in the human gastrointestinal tract: when, where, and how? Microorganisms. 2018;6:75. doi: 10.3390/microorganisms6030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cani P.D., Depommier C., Derrien M., Everard A., de Vos W.M. Akkermansia muciniphila: paradigm for next-generation beneficial microorganisms. Nat. Rev. Gastroenterol. Hepatol. 2022;19:625–637. doi: 10.1038/s41575-022-00631-9. [DOI] [PubMed] [Google Scholar]
- 68.Attia S., Versloot C.J., Voskuijl W., van Vliet S.J., Di Giovanni V., Zhang L., Richardson S., Bourdon C., Netea M.G., Berkley J.A., et al. Mortality in children with complicated severe acute malnutrition is related to intestinal and systemic inflammation: an observational cohort study. Am. J. Clin. Nutr. 2016;104:1441–1449. doi: 10.3945/ajcn.116.130518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dostal A., Chassard C., Hilty F.M., Zimmermann M.B., Jaeggi T., Rossi S., Lacroix C. Iron depletion and repletion with ferrous sulfate or electrolytic iron modifies the composition and metabolic activity of the gut microbiota in rats. J. Nutr. 2012;142:271–277. doi: 10.3945/jn.111.148643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dostal A., Lacroix C., Bircher L., Pham V.T., Follador R., Zimmermann M.B., Chassard C. Iron modulates butyrate production by a child gut microbiota in vitro. mBio. 2015;6:e01415–e01453. doi: 10.1128/mBio.01453-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pu Y., Li S., Xiong H., Zhang X., Wang Y., Du H. Iron promotes intestinal development in neonatal piglets. Nutrients. 2018;10:726. doi: 10.3390/nu10060726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu G., Ling C., Chi L., Thind M.K., Furse S., Koulman A., Swann J.R., Lee D., Calon M.M., Bourdon C., et al. The role of the tryptophan-NAD + pathway in a mouse model of severe malnutrition induced liver dysfunction. Nat. Commun. 2022;13:7576. doi: 10.1038/s41467-022-35317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Preidis G.A., Keaton M.A., Campeau P.M., Bessard B.C., Conner M.E., Hotez P.J. The undernourished neonatal mouse metabolome reveals evidence of liver and biliary dysfunction, inflammation, and oxidative stress. J. Nutr. 2014;144:273–281. doi: 10.3945/jn.113.183731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Di Giovanni V., Bourdon C., Wang D.X., Seshadri S., Senga E., Versloot C.J., Voskuijl W., Semba R.D., Trehan I., Moaddel R., et al. Metabolomic changes in serum of children with different clinical diagnoses of malnutrition. J. Nutr. 2016;146:2436–2444. doi: 10.3945/jn.116.239145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Preidis G.A., Ajami N.J., Wong M.C., Bessard B.C., Conner M.E., Petrosino J.F. Microbial-derived metabolites reflect an altered intestinal microbiota during catch-up growth in undernourished neonatal mice. J. Nutr. 2016;146:940–948. doi: 10.3945/jn.115.229179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Preidis G.A., Ajami N.J., Wong M.C., Bessard B.C., Conner M.E., Petrosino J.F. Composition and function of the undernourished neonatal mouse intestinal microbiome. J. Nutr. Biochem. 2015;26:1050–1057. doi: 10.1016/j.jnutbio.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 77.Blanton L.V., Charbonneau M.R., Salih T., Barratt M.J., Venkatesh S., Ilkaveya O., Subramanian S., Manary M.J., Trehan I., Jorgensen J.M., et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science. 2016;351 doi: 10.1126/science.aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bourke C.D., Berkley J.A., Prendergast A.J. Immune dysfunction as a cause and consequence of malnutrition. Trends Immunol. 2016;37:386–398. doi: 10.1016/j.it.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hughes S.M., Amadi B., Mwiya M., Nkamba H., Tomkins A., Goldblatt D. Dendritic cell anergy results from endotoxemia in severe malnutrition. J. Immunol. 2009;183:2818–2826. doi: 10.4049/jimmunol.0803518. [DOI] [PubMed] [Google Scholar]
- 80.Martinez-Lopez M., Iborra S., Conde-Garrosa R., Mastrangelo A., Danne C., Mann E.R., Reid D.M., Gaboriau-Routhiau V., Chaparro M., Lorenzo M.P., et al. Microbiota sensing by mincle-syk Axis in dendritic cells regulates interleukin-17 and -22 production and promotes intestinal barrier integrity. Immunity. 2019;50:446–461.e449. doi: 10.1016/j.immuni.2018.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gaboriau-Routhiau V., Rakotobe S., Lécuyer E., Mulder I., Lan A., Bridonneau C., Rochet V., Pisi A., De Paepe M., Brandi G., et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 82.Ivanov I.I., Atarashi K., Manel N., Brodie E.L., Shima T., Karaoz U., Wei D., Goldfarb K.C., Santee C.A., Lynch S.V., et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reboldi A., Arnon T.I., Rodda L.B., Atakilit A., Sheppard D., Cyster J.G. IgA production requires B cell interaction with subepithelial dendritic cells in Peyer's patches. Science. 2016;352:aaf4822. doi: 10.1126/science.aaf4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kato L.M., Kawamoto S., Maruya M., Fagarasan S. Gut TFH and IgA: key players for regulation of bacterial communities and immune homeostasis. Immunol. Cell Biol. 2014;92:49–56. doi: 10.1038/icb.2013.54. [DOI] [PubMed] [Google Scholar]
- 85.Lécuyer E., Rakotobe S., Lengliné-Garnier H., Lebreton C., Picard M., Juste C., Fritzen R., Eberl G., McCoy K.D., Macpherson A.J., et al. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity. 2014;40:608–620. doi: 10.1016/j.immuni.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 86.Xia J., Wishart D.S. MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010;38:W71–W77. doi: 10.1093/nar/gkq329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lelouard H., Mailfert S., Fallet M. 2018. A Ten-Color Spectral Imaging Strategy to Reveal Localization of Gut Immune Cell Subsets.https://www.zeiss.fr/microscopie/service-assistance/notes-application/a-ten-color-spectralimaging-strategy.html [Google Scholar]
- 88.Pédron T., Mulet C., Dauga C., Frangeul L., Chervaux C., Grompone G., Sansonetti P.J. A crypt-specific core microbiota resides in the mouse colon. mBio. 2012;3 doi: 10.1128/mBio.00116-12. 001166-e212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Amann R.I., Binder B.J., Olson R.J., Chisholm S.W., Devereux R., Stahl D.A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Snel J., Heinen P.P., Blok H.J., Carman R.J., Duncan A.J., Allen P.C., Collins M.D. Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats, and chickens and proposal of “Candidatus arthromitus”. Int. J. Syst. Bacteriol. 1995;45:780–782. doi: 10.1099/00207713-45-4-780. [DOI] [PubMed] [Google Scholar]
- 91.Angelakis E., Bachar D., Henrissat B., Armougom F., Audoly G., Lagier J.C., Robert C., Raoult D. Glycans affect DNA extraction and induce substantial differences in gut metagenomic studies. Sci. Rep. 2016;6:26276. doi: 10.1038/srep26276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Andrews S. 2010. FastQC: A Quality Control Tool for High Throughput Sequence Data.http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 93.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 97.Mirarab S., Nguyen N., Warnow T. SEPP saté-enabled phylogenetic placement. Pac. Symp. Biocomput. 2012:247–258. doi: 10.1142/9789814366496_0024. [DOI] [PubMed] [Google Scholar]
- 98.Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schmieder R., Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Magoč T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Apper E., Privet L., Taminiau B., Le Bourgot C., Svilar L., Martin J.C., Diez M. Relationships between gut microbiota, metabolome, body weight, and glucose homeostasis of obese dogs fed with diets differing in prebiotic and protein content. Microorganisms. 2020;8:513. doi: 10.3390/microorganisms8040513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
- Data
- Raw fastq files 16s rRNA sequencing data have been deposited at Zenodo and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
- Workflow data on the parameters used for the microbiome profiling of the terminal ileum and caecal contents have been deposited at Zenodo and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
- Code
- This paper does not report original code.
- Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.