Summary
To study the ancestry and phylogenetic relationships of native Korean dog breeds to other Asian dog populations, we analyzed nucleotide variations in whole-genome sequences of 205 canid individuals. Sapsaree, Northern Chinese indigenous dog, and Tibetan Mastiff were largely related to West Eurasian ancestry. Jindo, Donggyeongi, Shiba, Southern Chinese indigenous (SCHI), Vietnamese indigenous dogs (VIET), and Indonesian indigenous dogs were related to Southeast and East Asian ancestry. Among East Asian dog breeds, Sapsaree presented the highest haplotype sharing with German Shepherds, indicating ancient admixture of European ancestry to modern East Asian dog breeds. SCHI showed greater haplotype sharing with New Guinea singing dogs, VIET, and Jindo than with other Asian breeds. The predicted divergence time of East Asian populations from their common ancestor was approximately 2,000 to 11,000 years ago. Our results expand understanding of the genetic history of dogs in the Korean peninsula to the Asian continent and Oceanic region.
Subject areas: Animals, Evolutionary biology, Phylogenetics
Graphical abstract

Highlights
-
•
205 dog genomes were analyzed to address relationships of Asian dog populations
-
•
Divergence time of Korean dog breeds is approximately 2,000–11,000 years ago
-
•
Korean native breeds originate from both southeastern and West Eurasian ancestry
-
•
Sapsaree exhibits the high haplotype sharing with the West Eurasian ancestry
Animals; Evolutionary biology; Phylogenetics
Introduction
Dogs are large carnivores that have been domesticated and have migrated with humans as companion animals.1,2 Over 400 dog breeds with highly diverse phenotypic features exist owing to the development of numerous modern dog breeds (Canis lupus familiaris) by humans over the last 200 years.3 Recent advances in genome sequencing technology have greatly facilitated the genome sequencing of various canids, including wild canids, breed and indigenous dogs, and prehistoric dogs.1,4,5,6,7 Population genetic analyses using genome sequences have greatly expanded our understanding of the origin of domestic dogs and their divergence associated with human history, including parallel evolution between humans and dogs.1,4,6,7,8,9
Previous studies have suggested that domestication processes initiated from gray wolves 15,000–33,000 years ago (ya) in one or several wolf populations in East Asia, Southeast China, the Middle East, Europe, and the high Arctic, making dogs the earliest domesticated animal.4,5,10 However, a consensus on the history of modern dogs has not yet been achieved. Additional studies analyzing the genomes of indigenous dogs from various regions could be helpful for coalescing the different hypotheses regarding the history of modern dogs.
Evidence from archaeological sites dated between 5,500 and 2,000 BC suggests the practice of dog breeding in the Korean peninsula during the Neolithic period,11 highlighting the early history of native dogs in the Korean peninsula. In fact, the Korean Peninsula was contiguous to the Asian continent without the west shoreline of the Korean Peninsula during the ice age. Therefore, although archaeological evidence on the presence of dogs prior to the Neolithic period in Korea has not been discovered, the analysis of the genomes of native Korean dogs may expand the knowledge on the early history of modern dogs in the Korean peninsula.
Several native Korean dog breeds have been recognized, including Cheju, Donggyeongi, Jindo, Pungsan, and Sapsaree.12 Donggyeongi, Jindo, and Sapsaree are currently maintained as natural monuments in the country. A previous study using SNPs genotyping showed that short-haired native Korean dogs were phylogenetically clustered together with Japanese and Chinese dog breeds.12 Another study inferred that Sapsaree, a long-haired native Korean dog breed, could be related to long-haired breeds from Tibet.13 Although several recent studies have reported the genetic structure of diverse dog breeds and wild canids,4,14 the origin and ancestry of native Korean dogs related to the early ancestors of domestic dogs has not yet been addressed. In addition, an in-depth analysis of the ancestral and phylogenetic relationships of native Korean dog breeds using whole-genome sequencing (WGS) information has not been carried out to date.
In this study, we conducted WGS of 25 dogs, with two native Korean breed Sapsaree and Jindo, and three Chinese breeds, and analyzed their genetic relationships together with 180 genome sequences of 25 canids available from public databases to understand the ancestry of native Korean dog breeds. We investigated the origin of native Korean dog breeds in terms of their domestication and migration history in the Asian and Oceanic regions. Uncovering the genetic history of native Korean dog breeds through the analysis of WGS data could enrich our understanding of the history of dog breeds in Asia and Oceania.
Results
Preparing whole-genome sequencing data for 205 individuals of 23 dog breeds and 4 wild canid populations
We sequenced the genomes of five individuals each for five East Asian dog breeds, comprising two native Korean breeds, Sapsaree and Jindo, and three Chinese breeds, Tibetan Mastiff, Pekingese, and Pug, resulting in a mean coverage of 44.60×, ranging from ∼33× to ∼70× (BioProject: PRJNA782070). In addition, we retrieved the genome sequences of 180 canid individuals, including 83 belonging to 16 Asian breeds, 39 belonging to eight European breeds, 34 prehistoric dogs, five New Guinea singing dogs (Canis dingo hallstromi), five Dingoes (Canis lupus dingo), six gray wolves (Canis lupus), three Himalayan wolves (Canis lupus chanco), and four coyotes (Canis latrans) from public databases (Tables S1 and S2). A total of 1,983,163 variants were called, of which 1,772,390 (89.37%) were SNPs. From a dataset consisting of only 171 modern canid individuals, we obtained 35,319,513 variants, including 20,002,237 (56.63%) SNPs. The depth of variants or the number of mapped reads corresponding to observed variants ranged from 3.20× to 46.08× with the mean of 18.76× for the modern breed dataset (Table S1). Most variants (85.52%) were located in the noncoding region, and a small proportion (1.72%) of variants were in the genic region in the modern breed datasets.
Two different ancestral lineages for native Korean dog breeds
A UPGMA tree was constructed from clustering analysis using pairwise identity-by-state (IBS) distances among canids using 148 individuals from 23 dog and four wild canid populations including New Guinea singing dogs, wolves, and coyotes (Figure 1A). All dog populations except Northern Chinese indigenous dog, Southern Chinese indigenous dog, and Vietnamese indigenous dog formed monophyletic clusters (Figures 1A and S1). The clades of Jindo and Donggyeongi showed close genetic relatedness with Southern Asian dog breeds from Vietnam and Southern China. However, Sapsaree formed an outgroup to the other Korean dogs, indicating higher genetic similarity to Northern Chinese indigenous dog and Tibetan Mastiff. Surprisingly, Southern Chinese indigenous dog and Vietnamese indigenous dog were not clearly distinguishable as separate breeds (Figures 1A and S1). Tibetan Terrier and Pug formed a separate cluster from other Asian dog breeds such as Pekingese, Lhasa Apso, and Shih Tzu. Most prehistoric dogs were clustered with European breeds (Figure S1); thus, we included only seven distantly clustered prehistoric dogs in the tree to decrease redundancy. A maximum likelihood (ML) tree was also constructed (Figure S1), and results from UPGMA and ML trees were consistent except that Sapsaree and Shiba were clustered together with low confidence (bootstrap value = 24%) in the ML tree, in contrast to the distant clustering of the two breeds in the UPGMA tree.
Figure 1.
Genomic relationship and population structure of 205 individuals of 24 dog populations and wild canids
(A) IBS distance between 184 modern canid individuals was clustered using UPGMA. A maximum of five individuals were visualized per breed. The genetic distance is shown on the left.
(B) Principal component analysis of 23 dog breeds without wild canids.
(C) Genetic structure and admixture patterns. Maximum likelihood-based admixture analysis of 148 canids was conducted using the number of hypothetical cluster (K) ranging from 2 to 8, shown using different colors. The same individuals used for phylogenetic tree are presented. The admixture pattern corresponds to populations of the phylogenetic tree on the left.
(D) A boxplot for inbreeding coefficient of individuals for 11,443,767 SNPs. AFGH, Afghan Hound; BRDC, Bearded Collie; BOUV, Bouvier Des Flandres; BRIA, Briard; COYT, Coyote; DONG, Donggyeongi; GSD, German Shepherd; GREH, Greyhound; GRWF, gray wolf; HIWF, Himalayan wolf; IDNS, Indonesian indigenous dog; JIND, Jindo; LABR, Labrador Retriever; LHAS, Lhasa Apso; NCHI, Northern Chinese Indigenous; NGSD, New Guinea Singing Dog; OESD, Old English Sheepdog; PEKI, Pekingese; PUG, Pug; PWTD, Portuguese Water Dog; SAP, Sapsaree; SCHI, Southern Chinese indigenous; SHIB, Shiba; HUSK, Siberian Husky; SHIH, Shih Tzu; TIBM, Tibetan Mastiff; TIBT, Tibetan Terrier; and VIET, Vietnamese indigenous dog. See also Figures S1, S2, S3, S4, Tables S3, S4 and S5.
Genetic relationship between the Asian dog breeds was more clearly analyzed in principal component analysis (PCA) performed using the total breed dataset with and without outgroups (Figures 1B and S2). In the PCA plot with coyotes as an outgroup, the dog breeds were distributed widely in a cline between gray wolves and Portuguese water dog, where New Guinea singing dog, Dingo, Southern Chinese indigenous dog, Vietnamese indigenous dogs, and Indonesian indigenous dogs were closest to the wolves (Figure S2A). Without the coyote outgroup, Portuguese water dogs and New Guinea singing dogs were more distantly separated from other dog breeds than that in the result with outgroups (Figure S2B). The close genetic relationship of Sapsaree with Tibetan Mastiff and Siberian Husky was more clearly depicted in the PCA than in the phylogenetic tree (Figures 1A and 1B). In contrast, the other Korean breeds, Donggyeongi and Jindo, and the native Japanese breed Shiba consistently showed closer distances to Vietnamese indigenous dogs and Southern Chinese indigenous dog than to Sapsaree. Therefore, the Korean dog breeds could be descended from two ancestral lineages: a Southern Asian lineage and Northern Asian one. The Chinese toy dogs (Pekingese and Shih Tzu) and Tibetan breeds, including Lhasa Apso and Tibetan Terrier, were located close to European breeds, except for Pug, which is relatively unique, in the PCA plot (Figure 1B). The Tibetan Mastiff was distantly located from other Tibetan breeds, which is similar to relationships among native Korean dog breeds which show the existence of multiple lineages in the same region, indicating the existence of ancestral or genetic diversity within a close geographical boundary.
Population differentiation and admixture patterns of native Korean and East Asian dogs
The inter-breed genetic differentiation using Hudson’s pairwise FST using 11,443,767 autosomal SNPs was computed across 24 dog populations with coyote, gray wolf, Himalayan wolf, Dingo, and New Guinea singing dog (Figure S3). Among all the dog populations in this study, FST was the lowest between Southern Chinese indigenous dogs and Vietnamese indigenous dogs (FST = 0.016), suggesting the effect of common ancestry and/or admixture owing to their inhabitation of geographically close regions. Likewise, the FST values between New Guinea singing dogs and South Asian or East Asian dogs such as Indonesian indigenous dogs, Vietnamese indigenous dogs, Jindo, and Donggyeongi showed a difference of FST > 0.25 compared to the FST values between these and Northern Chinese indigenous dogs, which may reflect geographical influences of different ancestries (Figure S3 and Table S3).
Interestingly, the FST between Southern Chinese indigenous dogs and either Donggyeongi or Jindo was lower (0.051 and 0.090, respectively) than that between Southern Chinese indigenous dogs and Sapsaree (0.130). Sapsaree showed the lowest population differentiation with Northern Chinese indigenous dog (0.101) among all pairwise comparisons. The FST values between Southern Chinese and Vietnamese indigenous dogs, and Northern and Southern indigenous Chinese dogs were 0.015 and 0.060, respectively. Japanese Shiba showed a slightly higher range of FST with Jindo, Northern Chinese indigenous dog, and Southern Chinese indigenous dog (0.152–0.157). Vietnamese indigenous dog, Southern Chinese indigenous dog, Jindo, and Donggyeongi, which are clustered closely with South Asian ancestry or the Southern or Eastern Asian lineage, showed >0.05 lower FST with New Guinea singing dog than with coyotes (Table S4). In contrast, European dog breeds showed a slightly lower FST with the coyote than the New Guinea singing dog, the opposite case for the former group. This may reflect geographical influences, particularly on the Southeast and East Asian lineage. However, in the case of European breeds, the degree of difference was too small to be interpreted as a meaningful result. These results indicate that East Asian indigenous dogs are not genetically isolated. The New Guinea singing dog, as a representative dog population of the Oceanic lineage, showed the least population differentiation with both Southern Chinese indigenous dogs and Vietnamese indigenous dogs (0.351) among all breeds, and the value between the New Guinea singing dog and Jindo was also relatively low (0.381) compared to that between other dog breeds (0.406–0.651).
The clustered breeds in the PCA were further classified using ADMIXTURE analysis with 1,483,785 SNPs from the individuals used in the PCA and phylogenetic tree analyses (Figures 1A–1C). The optimal number of clusters was determined to be six from cross-validation errors (Figure S4). At K = 2 and 3, canids were divided into wild canids and dogs, and the contribution of wild canids is higher in Asian dogs than in European dogs. The population clustering in the phylogenetic tree (Figure 1A) was significantly consistent with the relative amounts of European and New Guinea singing dog admixtures in each population at K = 3. Pekingese and Shih Tzu were differentiated at K = 4 and 5, and varying degrees of admixture were observed in other Asian dog breeds, including the Tibetan Terrier, Lhasa Apso, Afghan Hound, and other Asian populations. At K = 5, New Guinea singing dogs are clearly differentiated from the rest of the dogs; however, a minor but noticeable influence of their lineage on Jindo, Southern Chinese indigenous dogs, and Vietnamese indigenous dogs was observed. Pugs were differentiated from other populations at K = 6, and more complex population-specific patterns were observed for each population at K = 7 and 8 in which the admixture pattern of Jindo among native Korean breeds was most similar to that of Southern Chinese and Vietnamese indigenous dogs. Admixture patterns of Sapsaree, Northern Chinese indigenous dog, Tibetan Terrier, and Siberian Husky were similar at K = 2–5. The analyzed admixture patterns were consistent with the results of phylogenetic and PCA analyses.
Higher genetic diversity of native or indigenous Asian dogs than that of breed dogs
The genetic diversity of dog populations was evaluated by the inbreeding coefficient (F) for dog populations constituting the modern breed dataset (Figure 1D and Table S5). The inbreeding coefficients of the 24 dog populations and five wild canids consisting of 3–22 individuals (average 5.9) for each breed were computed based on 11,443,767 SNPs. Interestingly, the two Korean breeds, Donggyeongi and Jindo, showed the lowest F (median F = 0.080 and 0.095, respectively) among all breeds (median F = 0.080–0.519), while New Guinea singing dogs showed the highest (median F = 0.804), which is most likely due to the very small founder population although two wild ones living in highland plateau of New Guinea were included in the analysis.15 Sapsarees also showed a relatively low F (median F = 0.194), similar to that of Southern Chinese indigenous dog (median F = 0.183) and lower than that of breed dogs from China and Europe (median F = 0.289–0.519) except Tibetan Mastiff (median F = 0.145). The high inbreeding coefficients of breed dogs including Chinese and European dog breeds (median F = 0.289–0.519) indicated that the genetic diversity of each dog population was strongly affected by the selection process during breed formation. Pugs, known to have a long breeding history, showed the highest F (median F = 0.416) among all the Asian dog breeds analyzed in this study although unrelated individuals from the sample list were selectively used. Linage disequilibrium (LD) decay analysis using the same number of individuals (n = 3) for the pairwise distance of the <300 kb region in the genome was also highly correlated with the inbreeding coefficients (Figure S5).
Introgression of West Eurasian and Southeast and East Asian ancestral lineages to native Korean dogs
To study the demographic histories of the domestic dogs inhabiting the Asian continent and Korean peninsula, we conducted admixture analysis allowing 0–10 migratory tracks for 139 individuals of 19 canids using the TreeMix program (Figure 2A). In the maximum likelihood tree with admixture edges, the ancestry of the modern dog lineage in East Asia and Siberia was largely traceable to two different lineages according to their relatedness to New Guinea singing dogs (Figures 2A and S6). Among the TreeMix graphs with different numbers of migration tracks, the tree with eight tracks most closely fit our results in PCA and admixture analyses, which showed the same topology with a tree without a migration edge (Figure S6). The result indicates the large influence of genetic introgression of the Oceanic lineage to Southern and Northern Chinese indigenous dog, Shiba, and native Korean breeds. Based on the estimated residuals from TreeMix analysis, the goodness of fit of the residuals in the graph is that the magnitude can be more closely related to the Vietnam indigenous dog and the New Guinea singing dog than the visualized tree topology (Figure S7).
Figure 2.
Admixture graph of East Eurasian dogs
(A) The maximum-likelihood graph was inferred with eight migration edges indicated by colored arrows. Three clades were indicated based on the similarity of genetic structure and geological origin. Each clade is marked with a circled number. (B) Depiction of outgroup f3-statistics for each breed (“X”) to Northern Chinese indigenous dog (NCHI) and Southern Chinese indigenous dog (SCHI) with gray wolf (GRWF) as common ancestor. The distance from the diagonal line indicates skewness in ancestry to either NCHI at the upper diagonal region or SCHI breeds at the lower diagonal region. See also Figures S6, S7 and S8.
In the admixture tree, the ancestral lineages differentiated into sublineages to form four clades. The clade I consisted of Southern Chinese indigenous dog, Vietnamese indigenous dog, and New Guinea singing dog. The clade II consisted of the native Korean dog breeds and Shiba, a Japanese breed. The clade III consisted of Tibetan and Chinese breeds, except for Southern Chinese indigenous dog. Siberian Husky forms the clade IV. Three different major introgression routes were predicted, from Himalayan wolf to New Guinea singing dogs, from the Oceanic lineage represented by New Guinea singing dogs to all three clades, and from the ancestor of Chinese toy breeds to Sapsaree. The five migration edges from the New Guinea singing dog lineage to each clade indicate that dogs in the clades I and II were more strongly influenced by the lineage than other clades when the migration weight was considered. This was also supported by outgroup f3-statistics showing shared genetic drift between New Guinea singing and Southern Chinese indigenous dogs in the clade II (Figure S8). The morphological resemblance among Shiba, Jindo, Donggyeongi, Vietnamese indigenous dogs, and New Guinea singing dogs could be attributed to both the presence of ancestral relationships among the populations and the introgression of gene flux from the Oceanic lineage (Figure S9).
Outgroup f3-statistics can be used to evaluate the shared genetic drift of two populations relative to an outgroup population in the format of f3 (outgroup; population 1, population 2). In outgroup f3 analyses of f3 (gray wolf; New Guinea singing dogs, other breeds) among seven European and 14 Asian dog breeds, Jindo, Donggyeongi, Shiba, Southern Chinese indigenous dog, and Vietnamese indigenous dog were differentiated from other dog breeds, and Sapsaree was between the five differentiated breeds and the rest (Figure S8). This indicates that the Southern Chinese indigenous dog, New Guinea singing dog, Vietnamese indigenous dog, Shiba, Donggyeongi, and Jindo have a more similar ancestry than other tested breeds. The outgroup f3-statistics also indicated genetic similarity between Sapsaree and Tibetan Mastiff. These results, together with outgroup f3-statistics with Southern and Northern Chinese indigenous dogs, indicate a difference in the ancestral lineage between the Donggyeongi and Jindo group and Sapsaree among native Korean dogs (Figure 2B). Therefore, Donggyeongi and Jindo are more Oceanic lineage-related than Sapsaree, which is more West Eurasian lineage-related, consistent with the ancestry of dogs around the world indicated by Bergstrom et al.1 We putatively named the ancestry of native populations with New Guinea singing dog-related genetic structures inhabiting Southern China, Vietnam, Korea, and Japan as the Southeast and East Asian (SEA) lineage, and the ancestry of native populations in the same region with a large genetic composition of European breeds as West Eurasian (WE) ancestry.
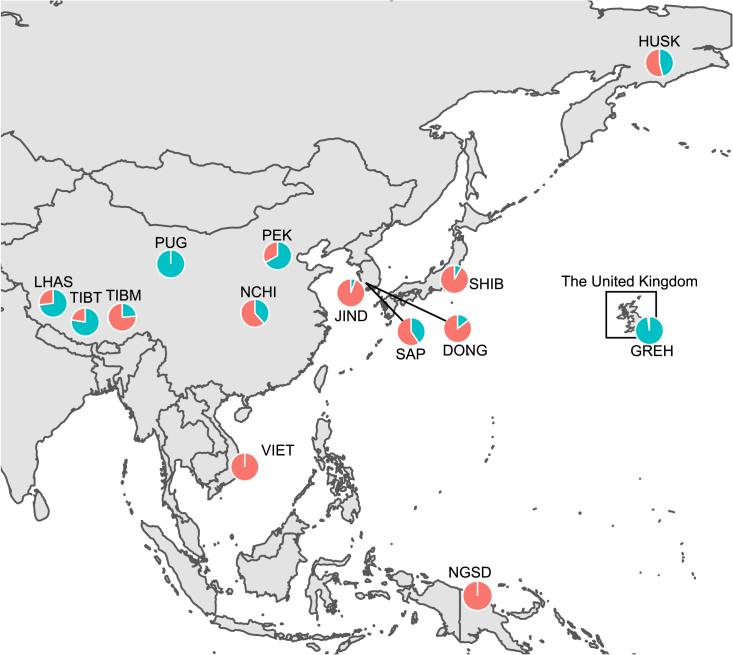
When the contributing proportion of SEA and Tibetan Terrier lineages to the breeds belonging to the clade II in the admixture analysis (Table S6) was calculated using qpAdm, SEA ancestry was higher in Jindo, Donggyeongi, and Shiba (81.8%–93.2%) than Tibetan Terrier ancestry. In contrast, Sapsaree was admixed with 47.3% and 52.7% of SEA and Tibetan Terrier ancestry, respectively. When the introgression of SEA and WE ancestries each to Asian dog breeds was analyzed with Southern Chinese indigenous dog and German Shepherd as representative populations of each lineage, New Guinea singing dog and Vietnamese indigenous dog were entirely made up of SEA ancestry (Figure 3 and Table S7). Likewise, Jindo, Shiba, and Donggyeongi had 94.5%, 92.0%, and 85.5% of the SEA lineage proportion, respectively. The proportions of the WE lineages to Northern Chinese dog and Tibetan Mastiff were 38.4% and 23.9%, respectively. Sapsaree was contributed by 59.6% of SEA and 40.4% WE ancestral lineages (Figure 3 and Table S7). Siberian Husky showed a similar admixture proportion to that of Sapsaree. Tibetan Terrier, Lhasa Apso, Pekingese, and Pug are consisted of a large fraction of the WE lineage (67%–100%).
Figure 3.
Relative proportions of the Southeast and East Asian and Eurasian ancestry of Asian dogs
Ancestral proportions of the Southeast and East Asian (SEA) and West Eurasian (WE) ancestries were estimated by the best-fitting qpAdm model adopting Southern Chinese indigenous dog and German Shepherd as the source populations and coyote, gray wolf, Himalayan wolf, and 5,000-year-old dog genomes found at Frälsegården, Sweden as the outgroup populations. See also Tables S6 and S7.
Greater haplotype sharing of Sapsaree with other Western and Asian breeds than that of Jindo and Donggyeongi
The genome-wide shared haplotype length of Sapsaree, Jindo, Donggyeongi, and Southern Chinese indigenous dog with 26 canids, including 14 Asian and eight European populations, and wolves was estimated using 15,202,681 phased SNPs on autosomes (Figure 4). The within-population shared haplotype length was the largest for New Guinea singing dogs, as expected from their extreme F (Figure 4A). The average mean value of shared haplotype length between a breed and the others, excluding wild canids, was 169.00, 142.06, and 126.55 Mbp for Jindo, Donggyeongi, and Southern Chinese indigenous dog, respectively, highlighting the higher haplotype sharing of Sapsaree (mean of 189.48 Mbp) to other breeds (Figure 4B). In contrast, Southern Chinese indigenous dog showed the lowest haplotype sharing with other breeds and higher haplotype sharing with New Guinea singing dogs, Vietnamese indigenous dogs, and Jindo than others (Figure 4C). Among the analyzed Asian dog breeds, Sapsaree showed the highest haplotype sharing with German Shepherds (Figure S10) and the lowest with Afghan Hound and New Guinea singing dogs (Figure 4D), indicating ancient admixture of European ancestry to Sapsaree.1 For Jindo, the sharing was the greatest with Donggyeongi and Sapsaree, but lowest with Afghan Hound (Figure 4E). These results are consistent with those of our phylogenetic, PCA, and admixture analyses. The shared haplotype length estimated for each breed from WGS data in this study was slightly larger than that determined in previous studies that used genotyping data using SNP chips.16
Figure 4.
Amount of haplotype sharing of native Korean breeds across other dog populations
Boxplots show distribution of haplotype length shared between two dogs of (A) the same breed and (B) different breeds across autosomes. The shared haplotype length with other breeds for (C) Southern Chinese indigenous dog, (D) Sapsaree, (E) Jindo, and (F) Donggyeongi is shown on the y axis. The breed names are shown on the x axis. See also Figure S10.
Estimated divergence time of Asian dog populations including native Korean breeds
To estimate the divergence time of five East Asian dog breeds, including three native Korean breeds, Sapsaree, Jindo, and Donggyeongi, we performed an SNAPP analysis with 500,000 Markov chain Monte Carlo iterations using 303,488 biallelic SNPs, with a 1 kb minimum length of inter-SNP distance in neutral regions (size of 276,974,635 bp). Because the exact divergence time between dogs and wolves is somewhat controversial, we estimated the divergence time of native Korean dogs with a root constraint of 15 and 35 thousand years ago (kya), respectively. We did not consider multiple domestication events and migrations of Asian dogs to other parts of the world in our analysis.
When the node age was assumed to be 15 kya, the divergence time of German Shepherd representing European ancestry from the Asian dog lineage was estimated to be ∼6.78 kya (95% highest posterior density (HPD): 6.68–6.89 kya) (Figure 5A). Subsequently, Tibetan Mastiff was diverged before other Asian indigenous dog breeds ∼5.98 kya (95% HPD: 5.89–6.09 kya). Among the Korean dog breeds, Sapsaree was diverged from the Tibetan Mastiff lineage 4.66 kya (95% HPD: 4.56–4.74 kya). Jindo and Donggyeongi showed the most recent divergence with 2.60 kya (95% HPD: 2.45–2.67 kya) after the divergence of ∼3.00 kya for Southern Chinese indigenous dog (95% HPD: 2.93–3.08 kya). Assuming a node age of 35 kya, the tree topology was consistent, but the divergence time of all nodes increased by a factor of 2.33 (Figure 5B).
Figure 5.
Divergence time of Asian dogs
We used SNAPP to predict divergence time with 500,000 MCMC iterations. The root age for the analysis was constrained to 15,000 (A) and 35,000 years ago (B). The approximate divergence time together with 95% highest posterior density is denoted on each node. (C) Estimation of effective population size of Sapsaree and Jindo using PSMC.
In addition, we performed pairwise sequential Markovian coalescent (PSMC) analysis using a mutation rate of 4.0 × 10−9 (Figure 5C). Differences in effective population size of gray wolves showed that their divergence from dogs could be at least older than approximately 30 kya. Similarly, Jindo and Sapsaree showed an increase in their effective population sizes from ∼2 kya. The results of PSMC are inconclusive in supporting which of the two root ages used in SNAPP was more likely than the other. Therefore, considering the results of both PSMC and SNAPP analyses, the divergence time between Sapsaree and other Korean breeds was determined to be between 2 and 11 kya.
Discussion
Several studies have reported the results of population genetic analyses of the ancestry of dogs globally.1,2,4,6,14,16,17 However, the population relationships between native dog breeds in East Asia and the Korean peninsula have not been thoroughly investigated. To improve our understanding of the ancestry and phylogenetic relationships of native Korean dog breeds with other Asian dog populations, we conducted population genetic analyses using the genome data of 205 canid individuals, including 25 newly sequenced Northeastern Asian dogs in this study and 186 publicly available dog genomes. Our results showed that native Korean dog breeds fall into two different ancestries and have been differentially admixed by the SEA lineage represented by New Guinea singing dogs. The relatively close clustering in PCA and TreeMix analyses and the similarity in morphological appearance among several Korean, Japanese, Southern Chinese, Vietnamese indigenous, and New Guinea singing dogs indicate either a common genetic ancestry or hybridization among them by migration events, possibly during the early history of Asian and Oceanic dogs (Figures 1 and 3). The phylogenetic relationships of commonly assessed dog breeds in this study and those in other studies were consistent.
Prior research on the ancestry of native Korean dogs using microsatellite and high-density SNP array genotyping has suggested a close genetic relationship between native Korean dog breeds and Siberian Husky or Chinese breeds.12,18 However, the results were somewhat limited by the number of loci or breeds, particularly for East Asian dogs. Our results on the genetic relationships of native Korean dogs to other native Asian breeds consistently indicate that native Korean dogs can be differentiated into two subgroups: WE lineage-related, such as Sapsaree, and SEA lineage-related, including Donggyeongi and Jindo. Furthermore, we speculate that other native Korean dog breeds, such as Cheju and Pungsan, which were not included in this study, could also be related to the SEA lineage considering their morphological similarity to Donggyeongi and Jindo. Interestingly, the results of genome-wide haplotype sharing analysis showed the greatest haplotype sharing between Sapsaree and German Shepherds (Figure 4). A previous study predicted the admixture of German Shepherds with a large number of dog breeds,1 which supports the introgression of the WE lineage to Sapsaree. In addition, PCA and outgroup f3-statistics showed a close relationship between SEA-lineage type Korean breeds, Vietnamese indigenous dogs, and Shiba, a native Japanese breed, and New Guinea singing dogs, suggesting the possible introgression of the Oceanic lineage to these canids, which was supported by the TreeMix analysis (Figures 2A and S8).
A recent study by Bergstrom et al. (2020) showed the comprehensive ancestry of dogs globally using large-scale WGS data, including prehistoric dogs.1 In their study, they showed that several modern Chinese dog populations displayed evidence of being the product of an admixture between populations related to the New Guinea singing dogs and West Eurasian dogs. They also reported a high proportion of New Guinea singing dog ancestry in Jindo, Shiba, and Vietnamese indigenous dogs and no or minimal New Guinea singing dog-related ancestry from populations in Siberia. Similar with these results, we found a substantial amount of admixture of Southern Chinese indigenous dogs in both Donggyeongi and Jindo (Table S7). However, we observed a much lower admixture of New Guinea singing dogs in Sapsaree, which is more closely clustered with Tibetan Mastiff and Siberian Husky than the other two Korean breeds (Figure 3). This is consistent with the results of a previous study that predicted a separation between Baikal and New Guinea singing dog-related ancestries in Asia at > 7 kya.1 The ancestral difference between Sapsaree and other native Korean dogs could be explained by this hypothesis.
In addition, Sapsaree might have been affected by the spread of steppe-related ancestry occurring at <5 kya, and a substantial amount of steppe-related ancestry admixture has also been detected in Tibetan Mastiff and Siberian Husky.1 Our results indicate that Sapsaree displayed a similar admixture pattern to the Tibetan Mastiff and Siberian Husky (Figure 1C).
A recent study on the origin and composition of Korean ethnicity that analyzed ancient and present-day human genome sequences inferred that the foundation of Koreans may have been established through a rapid admixture with ancient Southern Chinese populations associated with Iron Age Cambodians around 5,000–4,000 years before the present,19 which seems to be consistent to the genetic relationships of indigenous dogs in the region predicted in this study. The introgression of the New Guinea singing dog-related Oceanic lineage might also be associated with the migration of humans.
A previous study speculated that the ancestor of Sapsaree could be related to the Tibetan Terrier and Lhasa Apso.13 However, our study indicated that Sapsaree is more closely related to Tibetan Mastiff and Siberian Husky than to Tibetan Terrier and Lhasa Apso, which is also supported by the higher haplotype sharing of dog leukocyte antigen class II genes between Sapsaree and Tibetan Mastiff than that between other breeds.20 Recently, we reported that the 167 bp repeat sequence insertion in the RSPO2 3′ UTR, which is identical to the previously reported mutation for furnishing, is responsible for the long hair phenotype of Sapsaree, and the hair length-related RSPO2 mutation is identical across all modern dog breeds with long hair.20 This is consistent with our result that predicted the admixture between the Tibetan Terrier and Sapsaree in the Treemix analysis (Figure 2A). The low population differentiation (FST) among the native Korean breeds suggests gene flow among native Korean dog breeds or they were not isolated. However, admixture analysis showed that the genetic structure of Sapsaree was more complex than that of other native Korean breeds (Figure 1C).
Archaeological evidence supports the existence of dogs in Oceania at approximately 3.5 kya.21 The influence of oceanic migration on South Asian dogs has been reported in other studies.6 Migration from the New Guinea singing dog lineage to Donggyeongi and Jindo and other breeds in Japan and Vietnam has also been observed. New Guinea singing dogs are genetically similar to Dingo (C. lupus dingo) but represent a distinct population within the distinct evolutionary lineage of Oceanic canids. A recent study reported that village dogs in New Guinea and Vietnam share 13% and 11% of their genome haplotypes, respectively, with New Guinea singing dogs.6 The finding that the Korean peninsula harbors native breeds with origins from both West Eurasian and Southern Asian lineages is interesting and suggests multiple introgressions of different ancestries to the Korean peninsula. However, it is also plausible that, alternatively, gradual differentiation and isolated by distance (with differential gene flow from the neighboring regions) would have produced the observed admixture patterns between European-like and New Guinea singing dog-like ancestors.
Several studies have reported a higher level of nuclear and mitochondrial genome diversity in East Asian dogs than in dogs of other geographical origins, serving as evidence to support the East Asian origin of domestic dogs.4,14 Wang et al. (2016) suggested the Southeast Asian origin of domestic dogs approximately 33,000 years ago, based on the significantly higher genetic diversity of dogs.4 Interestingly, the level of genetic diversity estimated by the inbreeding coefficient and linkage disequilibrium was higher in native Korean breeds than that in other East Asian breeds (Figures 1D and S5). The increased genetic diversity of native dogs on the Korean peninsula may be due to the introduction of multiple lineages to the Korean peninsula, such as the West Eurasian and Oceanic lineages. However, this may also be because of the early history of native Korean breeds, considering that the current west coast of the Korean peninsula was contiguous with the Asian continent when the ancestral lineages of modern dogs diverged. Further studies using the genome sequences of indigenous dogs from other Asian regions should greatly contribute to understanding the phylogenetic relationships and gene flow among Asian dogs.
Consistent with the higher genetic diversity of native Asian breeds, their genomes showed a lower LD (r2) than those of breed dogs, indicating that there was a time in the history of the native Asian breeds when the population size was large. All three native Korean dog breeds were restored from a limited number of individuals.18,22,23 In a previous study, the high genetic diversity of Sapsaree was attributed to the large effective population size (Ne) before population reduction and restoration.13,24 In our analysis, the estimated Ne was the greatest in Southern Chinese indigenous dogs, followed by that in other native Asian breeds, and the lowest for New Guinea singing dogs (Figure S11).
The estimated divergence time can vary depending on the applied methods and parameters. The date of divergence between wolves and dogs has been estimated to be between 35,000 and 15,000 ya in previous studies,4,10,25 and thus we conducted SNAPP analysis using both time points, respectively. The structure of the divergence-time tree was consistent with that of the phylogenetic tree based on the IBS distance in Figure 1A. To the best of our knowledge, this is the first attempt to estimate the divergence time of the native Korean dog lineage together with other Asian dogs.
In summary, we presented the genetic characteristics and relationships of East Asian dogs, including Tibetan, Chinese, Korean, Japanese, Vietnamese, and New Guinea singing dog populations, via population genetic analyses of WGS data. Our results showed that Asian dogs can be largely divided into two groups according to the amount of genetic contribution from WE and SEA lineages. Similarly, Sapsaree was largely related to WE ancestry and Donggyeongi and Jindo were related to SEA ancestry, which allowed the origin and history of native Korean dog lineages to be delineated. Therefore, our results extend the genetic history and ancestral relationships of dogs in the Asian continent to include the Korean peninsula and highlight the presence of genetic relatedness between South and East Asian and Oceanic dog lineages. The results of population genetic analyses of the genomes of native or indigenous dogs of East Asia and the newly reported genome sequence of 25 Asian dogs of five different breeds provide new insights into the origins and migration history of dogs in Asia.
Limitation of the study
In this study, we analyzed genetic relationships of Asian dogs and their ancestry based on a large-scale whole-genome sequencing data. However, the lack of ancient dog DNA, difficulty in estimating haplotype, and limited breed range and population size of East Asian native dogs may be limitations of this study. Additional genomes of indigenous dogs near or within the Korean Peninsula, geographically including North Korea, Japan, Mongolia, and Siberia will be needed to further clarify East Asian canine ancestry.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Whole genome sequences of 25 Asian dogs | This paper | BioProject:PRJNA782070 |
| Dog reference genome, CanFam3.1 | Dog Genome Sequencing Consortium | RefSeq:GCF_000002285.3 |
| Software and algorithms | ||
| fasterq-dump v2.10.8 | N/A | https://github.com/ncbi/sra-tools |
| BWA MEM v0.7.17-r1188 | Li and Durbin,26 | https://github.com/lh3/bwa |
| samtools v1.10 | Li et al.27 | https://github.com/samtools/samtools |
| Picard v2.21.3 | N/A | https://broadinstitute.github.io/picard/ |
| Genome analysis toolkit (GATK) package v4.1.9.0 | Van der Auwera et al.28 | https://gatk.broadinstitute.org/hc/en-us |
| PMDtools v.0.60 | Skoglund et al.29 | https://github.com/pontussk/PMDtools |
| Vcftools v0.1.17 | Danecek et al.30 | https://vcftools.github.io/index.html |
| SNPRelate (R package) v1.20.1 | Zheng et al.31 | https://www.bioconductor.org/packages/release/bioc/html/SNPRelate.html |
| R v3.6.3 | N/A | https://www.r-project.org/ |
| RAxML-NG v.1.1.0 | Kozlov et al.32 | https://github.com/amkozlov/raxml-ng |
| Vcf2phylip v2.8 | Ortiz,33 | https://github.com/edgardomortiz/vcf2phylip |
| PLINK v1.90b6.12 | Purcell et al.34 | https://www.cog-genomics.org/plink/ |
| PLINK v2.00a3LM | Purcell et al.34 | https://www.cog-genomics.org/plink/2.0/ |
| PopLDdecay v3.41 | Zhang et al.35 | https://github.com/BGI-shenzhen/PopLDdecay |
| ADMIXTURE v1.3.0 | Alexander et al.36 | https://dalexander.github.io/admixture/ |
| pong v1.4.9 | Behr et al.37 | https://github.com/ramachandran-lab/pong |
| Treemix v1.13 | Pickrell and Pritchard,38 | https://bitbucket.org/nygcresearch/treemix/wiki/Home |
| Admixtools v7.0.2 | Patterson et al.39 | https://github.com/DReichLab/AdmixTools |
| Beagle v5.2 | Browning et al.40 | https://faculty.washington.edu/browning/beagle/b5_2.html |
| SNP and AFLP Package for Phylogenetic analysis (SNAPP) v1.5.2 | Bryant et al.41 | https://www.beast2.org/snapp/ |
| Snapp_prep.rb | Stange et al.42 | https://github.com/mmatschiner/snapp_prep |
| TreeAnnotator v2.6.6 | Drummond and Rambaut,43 | https://beast.community/treeannotator |
| FigTree v1.4.4 | Rambaut,43 | http://tree.bio.ed.ac.uk/software/figtree/ |
| bcftools v1.16 | Li,44 | https://github.com/samtools/bcftools |
| PSMC v0.6.5-r67 | Li and Durbin,45 | https://github.com/lh3/psmc |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Chankyu Park (chankyu@konkuk.ac.kr).
Materials availability
No new material was generated from this study.
Experimental models and study participants
For in vitro DNA extraction, blood samples were obtained from a total of 25 adult dogs, consisting of five Sapsarees, five Jindos, five Tibetan Mastiffs, five Pugs, and five Pekingeses. Each animal was housed in hygienic cages under veterinary supervision. The results were not influenced by sex, as the analyses throughout the study excluded sex chromosomes.
Method details
Animals and DNA preparation
Blood samples from five Sapsarees and five Jindos were collected from the Sapsaree Research Foundation in Gyeongsan, Korea and Jindo Dog Theme Park in Jindo, Korea, respectively, according to the protocols of the Institutional Animal Care and Use Committee (IACUC) of Konkuk University. Blood samples from 15 Chinese dogs, including five Tibetan Mastiffs, five Pekingeses, and five Pugs, were collected by a veterinarian using the protocol approved by the IACUC of Jilin University. Genomic DNA of Sapsarees and Jindos was isolated from 300 μL of whole blood using the ExgeneTM Blood SV mini (GeneAll Biotechnology, Seoul, Korea) or DNeasy Blood & Tissue kit (Qiagen, MD, USA), according to the manufacturer’s protocol.
Whole genome sequencing and variant analysis
Two micrograms of genomic DNA was sheared using the Covaris S2 instrument (Covaris, MA, USA) and a paired-end DNA sequencing library was prepared using a DNA PCR-Free Library prep kit (Illumina, CA, USA) according to the manufacturer’s protocol. WGS was performed using a NovaSeq 6000 instrument (Illumina). In addition, the whole-genome sequence of 186 canids was downloaded from the NCBI SRA database using fasterq-dump v2.10.8 (Table S1). The reads were mapped to the dog reference genome CanFam3.1 (RefSeq accession no. GCF_000002285.3) using BWA MEM v0.7.17-r1188.26 The mapped reads were converted to the binary format using samtools v1.10.27 PCR duplicates were marked using Picard MarkDuplicates v2.21.3,46 and the base qualities were recalibrated using BaseRecalibrator and ApplyBQSR in Genome analysis toolkit (GATK) package v4.1.9.0.28 Per-sample genomic variants were called using GATK HaplotypeCaller,47 the variants of all individuals were consolidated using GATK GenomicsDBImport, and a joint variant was created using GATK GenotypeGVCFs. Variants with high strand bias (Fisher Strand (FS) > 30.0), low quality (QualityOfDepth (QD) < 2), and high complexity, wherein three SNPs were placed within a 35 bp window (-window 35 -cluster 3)), were removed using GATK VariantFiltration. Insertions and deletions (INDELs) were removed from the variants using GATK SelectVariants. To call prehistoric dog variants, we removed reads with postmortem damage (PMD score <3) using PMDtools v.0.60.29 The subsequent processes using Picard and GATK is identical to calling variants of other samples. A total of 2,984,901 and 20,002,237 SNPs from 34 prehistoric dogs and 171 modern canid individuals were called, respectively. Shared SNPs were consolidated using bcftools isec (-n = 2 -w 1, 2) and merge.48
Phylogenetic analysis
Biallelic SNPs were selected using Vcftools v0.1.1730 and subjected to SNPRelate package v1.20.1 in R v3.6.3.31 SNPs were additionally filtered when the missing genotype rate was >0.05 and the minor allele frequency (MAF) was <0.01. An identity-by-state (IBS)-based pairwise distance matrix was generated and used to construct a phylogenetic tree with 205 individuals using the unweighted pair group method with arithmetic mean (UPGMA) clustering method.31 Principal component analysis (PCA) was conducted using the snpgdsPCA function, which calculates the genetic covariance matrix and computes correlation coefficients between samples.31 To construct the maximum likelihood tree, Vcf2phylip v2.833 was used to generate multiple sequence alignments with 116,017 SNPs (genotype ratio >0.8) from 205 canine individuals, including 34 prehistoric dogs. Ten randomized maximum parsimony trees were generated to search for starting trees. A maximum likelihood tree was constructed with GTR+I+G model49 using RAxML-NG v.1.1.032 and 50 bootstrap replicates.
Genetic diversity analysis
Inbreeding coefficients (F) were calculated using 11,443,767 SNPs from 171 modern canid individuals using PLINK v1.90b6.12 with parameter ‘--het small-sample’.34 Subsequently, breed-level F was calculated by averaging individual F values in the same canid group. LD decay analysis was conducted for the same SNPs (miscalling <0.05 and MAF >0.01) using PopLDdecay v3.41.35 Hudson’s pairwise FST was calculated using PLINK v2.00a3LM with parameter ‘--fst’.34,50,51
Admixture and graph analyses
A total of 148 canids consisting of 130 individuals from 23 dog breeds (n = 3–10), five New Guinea singing dogs, six gray wolves, three Himalayan wolves, and four coyotes were subjected to admixture analysis using ADMIXTURE v1.3.0, with hypothetical cluster (K) ranging from 2 to 8.36 Linkage disequilibrium-based pruning was performed to reduce computational burden with the ‘--indep-pairwise 50 10 0.1’ parameter using PLINK v1.90b6.12, which selects the pair of SNPs with the greatest pairwise r2 within a 50 kb window over an r2 threshold of 0.1 with a step size of 10 variants. Bootstrapping was performed five times and the cross-validation error was calculated for each hypothetical cluster. The admixture bar plot was visualized using pong v1.4.9.37 Migration events were analyzed for 139 individuals from 19 canid groups consisting of 126 individuals from 14 Asian dog breeds, with Siberian Husky, New Guinea singing dog, gray wolf, Himalayan wolf, and coyote as outgroups (-root coyote) using Treemix v1.13.38 Maximum likelihood graphs with 0–10 migration edges were generated with the frequency of SNP blocks consisting of 10 SNPs using 15,664,509 biallelic autosomal SNPs (-k 10) with bootstrapping (-bootstrap). Sample size correction was not performed. Graphs were generated for each number of edges ten times with a random seed number (-seed).
Outgroup f3-statistics and qpAdm analysis
The f3-statistics were calculated for 184 individuals belonging to 26 canids based on 13,793,410 biallelic SNPs (missing genotyping rate <0.05 and MAF >0.01) using the qp3Pop software included in the Admixtools v7.0.2 package.39 The number of jackknife blocks was 327. The admixture proportion was computed with 1,372,698 autosomal SNPs from 212 canids using the qpAdm v1520 program of Admixtools v7.0.2.39,52 Coyote was used as a base outgroup population together with gray wolf, Himalayan wolf, and a prehistoric dog genome, C88, found in Frälsegården, Sweden 5,000 ya.1
Haplotype sharing
The haplotypes of 184 modern canids were phased with 15,725,060 SNPs (MAF >0.01 and missing genotyping rate <0.05) using Beagle v5.240 with the parameters of a 10 Mb sliding window and 0.5 Mb overlapped length between windows. We defined 100 consecutive SNPs as a unit of haplotype blocks. The pairwise shared haplotype length between two individuals was computed with the average value of the length of identical haplotype blocks between individuals:
where corresponds to the shared haplotype length of a haplotype combination among four possible cases between two diploid genomes with phased haplotypes.
Estimation of divergence time
Neutral regions on the genome were selected after excluding gapped regions and genic regions, including 30 kb neighboring regions of coding sequences (CDSs), repeat elements, and conserved non-coding elements (CNEs). The repeat element annotation of the dog reference genome CanFam3.1 was obtained using Table Browser implemented in the UCSC Genome Browser. The information of CNEs across 21 placental mammalian species was obtained in coordinates of the mouse genome assembly GRCm38 (RefSeq accession no. GCF_000001635.2) from the phastCons60wayPlacental table using the UCSC Table Browser and converted to those of the dog reference genome using UCSC LiftOver by setting the minimum ratio of bases that must be remapped to 0.5.53 Additionally, the minimum distance between SNPs was set to 1 kb to reduce the number of SNPs, resulting in 302,436 biallelic sites out of 1,536,434 SNPs called from variant analysis using a single individual with the highest sequence depth for each breed. Divergence time was estimated using the SNP and AFLP Package for Phylogenetic analysis (SNAPP) v1.5.2, included in BEAST v2.6.6, while constraining the root divergence times to 35,000 ± 5,000 and 15,000 ± 5,000 years ago in log-normal distribution by running 500,000 iterations.41 The input XML file was generated using ruby script snapp_prep.rb.42 In addition, 20% of burn-in iterations were removed using TreeAnnotator v2.6.6.43 Inferred trees with divergence time were visualized using FigTree v1.4.4.44
PSMC analysis
Consensus sequences with base quality >30 and mapping quality >30 were called using bcftools v1.16 mpileup and call48 from the recalibrated BAM file. Minimum and maximum depths were limited to 10 and 100 respectively using vcftutils.pl in bcftools. Consensus sequences were converted to PSMC FASTA format using fq2psmcfa included in PSMC v0.6.5-r67.45 PSMC was run with parameters “-N30 -t15 -p “4 + 25∗2 + 4+6”.
Acknowledgments
This paper was supported by Konkuk University in 2022. We acknowledge Rogar Sargent for allowing usage of the copyrighted photograph of Vietnamese indigenous dogs.
Author contributions
Conceptualization, B.A. and C.P.; Methodology, B.A.; Software, B.A.; Resources, M.K., H.J., and J.-S.K.; Investigation, B.A. and C.P.; Data Curation, B.A.; Writing – Original draft, B.A. and C.P.; Writing – Review & Editing, B.A., C.P., and J.H.; Visualization, B.A. and M.K.; Project Administration, C.P. and H.J.; Supervision, C.P., J.H., and H.J.; Funding acquisition, C.P.
Declaration of interests
We have no conflict of interest to declare.
Published: May 28, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106982.
Supplemental information
Data and code availability
-
•
The whole genome sequence of the 25 East Asian dogs have been deposited at Short Read Archive (https://www.ncbi.nlm.nih.gov/sra) and are publicly available under bioproject accession no. PRJNA782070.
-
•
This paper does not report original code.
References
- 1.Bergström A., Frantz L., Schmidt R., Ersmark E., Lebrasseur O., Girdland-Flink L., Lin A.T., Storå J., Sjögren K.G., Anthony D., et al. Origins and genetic legacy of prehistoric dogs. Science. 2020;370:557–564. doi: 10.1126/science.aba9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perri A.R., Feuerborn T.R., Frantz L.A.F., Larson G., Malhi R.S., Meltzer D.J., Witt K.E. Dog domestication and the dual dispersal of people and dogs into the Americas. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2010083118. e2010083118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plassais J., Kim J., Davis B.W., Karyadi D.M., Hogan A.N., Harris A.C., Decker B., Parker H.G., Ostrander E.A. Whole genome sequencing of canids reveals genomic regions under selection and variants influencing morphology. Nat. Commun. 2019;10:1489. doi: 10.1038/s41467-019-09373-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang G.-D., Zhai W., Yang H.-C., Wang L., Zhong L., Liu Y.-H., Fan R.-X., Yin T.-T., Zhu C.-L., Poyarkov A.D., et al. Out of southern East Asia: the natural history of domestic dogs across the world. Cell Res. 2016;26:21–33. doi: 10.1038/cr.2015.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skoglund P., Ersmark E., Palkopoulou E., Dalén L. Ancient wolf genome reveals an early divergence of domestic dog ancestors and admixture into high-latitude breeds. Curr. Biol. 2015;25:1515–1519. doi: 10.1016/j.cub.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 6.Surbakti S., Parker H.G., McIntyre J.K., Maury H.K., Cairns K.M., Selvig M., Pangau-Adam M., Safonpo A., Numberi L., Runtuboi D.Y.P., et al. New Guinea highland wild dogs are the original New Guinea singing dogs. Proc. Natl. Acad. Sci. USA. 2020;117:24369–24376. doi: 10.1073/pnas.2007242117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frantz L.A.F., Mullin V.E., Pionnier-Capitan M., Lebrasseur O., Ollivier M., Perri A., Linderholm A., Mattiangeli V., Teasdale M.D., Dimopoulos E.A., et al. Genomic and archaeological evidence suggest a dual origin of domestic dogs. Science. 2016;352:1228–1231. doi: 10.1126/science.aaf3161. [DOI] [PubMed] [Google Scholar]
- 8.Feuerborn T.R., Carmagnini A., Losey R.J., Nomokonova T., Askeyev A., Askeyev I., Askeyev O., Antipina E.E., Appelt M., Bachura O.P., et al. Modern Siberian dog ancestry was shaped by several thousand years of Eurasian-wide trade and human dispersal. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2100338118. e2100338118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang G.-d., Zhai W., Yang H.-c., Fan R.-x., Cao X., Zhong L., Wang L., Liu F., Wu H., Cheng L.-g., et al. The genomics of selection in dogs and the parallel evolution between dogs and humans. Nat. Commun. 2013;4:1860. doi: 10.1038/ncomms2814. [DOI] [PubMed] [Google Scholar]
- 10.Freedman A.H., Gronau I., Schweizer R.M., Ortega-Del Vecchyo D., Han E., Silva P.M., Galaverni M., Fan Z., Marx P., Lorente-Galdos B., et al. Genome sequencing highlights the dynamic early history of dogs. PLoS Genet. 2014;10:e1004016. doi: 10.1371/journal.pgen.1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J.J. Interpreting roles of domestic dogs in the neolithic to the three kingdoms periods in Korea. J. Korean Ancient Historical Soc. 2013;81:5–34. [Google Scholar]
- 12.Choi B.H., Wijayananda H.I., Lee S.H., Lee D.H., Kim J.S., Oh S.I., Park E.W., Lee C.K., Lee S.H. Genome-wide analysis of the diversity and ancestry of Korean dogs. PLoS One. 2017;12:e0188676. doi: 10.1371/journal.pone.0188676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gajaweera C., Kang J.M., Lee D.H., Lee S.H., Kim Y.K., Wijayananda H.I., Kim J.J., Ha J.H., Choi B.H., Lee S.H. Genetic diversity and population structure of the Sapsaree, a native Korean dog breed. BMC Genet. 2019;20:66. doi: 10.1186/s12863-019-0757-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shannon L.M., Boyko R.H., Castelhano M., Corey E., Hayward J.J., McLean C., White M.E., Abi Said M., Anita B.A., Bondjengo N.I., et al. Genetic structure in village dogs reveals a Central Asian domestication origin. Proc. Natl. Acad. Sci. USA. 2015;112:13639–13644. doi: 10.1073/pnas.1516215112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang S.-j., Wang G.-D., Ma P., Zhang L.-l., Yin T.-T., Liu Y.-h., Otecko N.O., Wang M., Ma Y.-p., Wang L., et al. Genomic regions under selection in the feralization of the dingoes. Nat. Commun. 2020;11:671. doi: 10.1038/s41467-020-14515-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker H.G., Dreger D.L., Rimbault M., Davis B.W., Mullen A.B., Carpintero-Ramirez G., Ostrander E.A. Genomic analyses reveal the influence of geographic origin, migration, and hybridization on modern dog breed development. Cell Rep. 2017;19:697–708. doi: 10.1016/j.celrep.2017.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergström A., Stanton D.W.G., Taron U.H., Frantz L., Sinding M.-H.S., Ersmark E., Pfrengle S., Cassatt-Johnstone M., Lebrasseur O., Girdland-Flink L., et al. Grey wolf genomic history reveals a dual ancestry of dogs. Nature. 2022;607:313–320. doi: 10.1038/s41586-022-04824-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim K.S., Tanabe Y., Park C.K., Ha J.H. Genetic variability in East Asian dogs using microsatellite loci analysis. J. Hered. 2001;92:398–403. doi: 10.1093/jhered/92.5.398. [DOI] [PubMed] [Google Scholar]
- 19.Kim J., Jeon S., Choi J.-P., Blazyte A., Jeon Y., Kim J.-I., Ohashi J., Tokunaga K., Sugano S., Fucharoen S., et al. The origin and composition of Korean ethnicity analyzed by ancient and present-day genome sequences. Genome Biol. Evol. 2020;12:553–565. doi: 10.1093/gbe/evaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang M., Ahn B., Youk S., Lee Y.-M., Kim J.-J., Ha J.-H., Park C. Tracing the origin of the RSPO2 long-hair allele and epistatic interaction between FGF5 and RSPO2 in sapsaree dog. Genes. 2022;13:102. doi: 10.3390/genes13010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milham P., Thompson P. Relative antiquity of human occupation and extinct fauna at madura cave, Southeastern Western Australia. Mankind. 2010;10:175–180. doi: 10.1111/j.1835-9310.1976.tb01149.x. [DOI] [Google Scholar]
- 22.Lee C.G., Lee J.I., Lee C.Y., Sun S.S. A Review of the Jindo, Korean native dog - Review. Asian-Australas. J. Anim. Sci. 2000;13:381–389. doi: 10.5713/ajas.2000.381. [DOI] [Google Scholar]
- 23.Jang G., Hong S., Kang J., Park J., Oh H., Park C., Ha J., Kim D., Kim M., Lee B. Conservation of the sapsaree (Canis familiaris), a Korean natural monument, using somatic cell nuclear transfer. J. Vet. Med. Sci. 2009;71:1217–1220. doi: 10.1292/jvms.71.1217. [DOI] [PubMed] [Google Scholar]
- 24.Alam M., Han K.I., Lee D.H., Ha J.H., Kim J.J. Estimation of effective population size in the sapsaree: a Korean native dog (Canis familiaris) Asian-Australas. J. Anim. Sci. 2012;25:1063–1072. doi: 10.5713/ajas.2012.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galeta P., Lázničková-Galetová M., Sablin M., Germonpré M. Morphological evidence for early dog domestication in the European Pleistocene: new evidence from a randomization approach to group differences. Anat. Rec. 2021;304:42–62. doi: 10.1002/ar.24500. [DOI] [PubMed] [Google Scholar]
- 26.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., et al. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;43:11.10.11–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skoglund P., Northoff B.H., Shunkov M.V., Derevianko A.P., Pääbo S., Krause J., Jakobsson M. Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. Proc. Natl. Acad. Sci. USA. 2014;111:2229–2234. doi: 10.1073/pnas.1318934111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng X., Levine D., Shen J., Gogarten S.M., Laurie C., Weir B.S. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28:3326–3328. doi: 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozlov A.M., Darriba D., Flouri T., Morel B., Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ortiz E.M. 2019. vcf2phylip v2.0: Convert a VCF Matrix into Several Matrix Formats for Phylogenetic Analysis (Zenodo) [Google Scholar]
- 34.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C., Dong S.-S., Xu J.-Y., He W.-M., Yang T.-L. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics. 2019;35:1786–1788. doi: 10.1093/bioinformatics/bty875. [DOI] [PubMed] [Google Scholar]
- 36.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Behr A.A., Liu K.Z., Liu-Fang G., Nakka P., Ramachandran S. pong: fast analysis and visualization of latent clusters in population genetic data. Bioinformatics. 2016;32:2817–2823. doi: 10.1093/bioinformatics/btw327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pickrell J.K., Pritchard J.K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., Zhan Y., Genschoreck T., Webster T., Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Browning B.L., Tian X., Zhou Y., Browning S.R. Fast two-stage phasing of large-scale sequence data. Am. J. Hum. Genet. 2021;108:1880–1890. doi: 10.1016/j.ajhg.2021.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bryant D., Bouckaert R., Felsenstein J., Rosenberg N.A., RoyChoudhury A. Inferring species trees directly from biallelic genetic markers: bypassing gene trees in a full coalescent analysis. Mol. Biol. Evol. 2012;29:1917–1932. doi: 10.1093/molbev/mss086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stange M., Sánchez-Villagra M.R., Salzburger W., Matschiner M. Bayesian divergence-time estimation with genome-wide single-nucleotide polymorphism data of sea catfishes (ariidae) supports miocene closure of the Panamanian isthmus. Syst. Biol. 2018;67:681–699. doi: 10.1093/sysbio/syy006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drummond A.J., Rambaut A. BEAST: bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rambaut A. 2009. FigTree.https://github.com/rambaut/figtree [Google Scholar]
- 45.Li H., Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Institute, B. 2019. Picard Toolkit.https://broadinstitute.github.io/picard [Google Scholar]
- 47.Poplin R., Ruano-Rubio V., DePristo M.A., Fennell T.J., Carneiro M.O., Van der Auwera G.A., Kling D.E., Gauthier L.D., Levy-Moonshine A., Roazen D., et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv. 2017 doi: 10.1101/201178. Preprint at. [DOI] [Google Scholar]
- 48.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abadi S., Azouri D., Pupko T., Mayrose I. Model selection may not be a mandatory step for phylogeny reconstruction. Nat. Commun. 2019;10:934. doi: 10.1038/s41467-019-08822-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hudson R.R., Slatkin M., Maddison W.P. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhatia G., Patterson N., Sankararaman S., Price A.L. Estimating and interpreting FST: the impact of rare variants. Genome Res. 2013;23:1514–1521. doi: 10.1101/gr.154831.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haak W., Lazaridis I., Patterson N., Rohland N., Mallick S., Llamas B., Brandt G., Nordenfelt S., Harney E., Stewardson K., et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., David The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The whole genome sequence of the 25 East Asian dogs have been deposited at Short Read Archive (https://www.ncbi.nlm.nih.gov/sra) and are publicly available under bioproject accession no. PRJNA782070.
-
•
This paper does not report original code.