Summary
Colorectal cancer (CRC) is a leading cause of morbidity and mortality worldwide. The majority of CRC deaths are caused by tumor metastasis, even following treatment. There is strong evidence for epigenetic changes, such as DNA methylation, accompanying CRC metastasis and poorer patient survival. Earlier detection and a better understanding of molecular drivers for CRC metastasis are of critical clinical importance. Here, we identify a signature of advanced CRC metastasis by performing whole genome-scale DNA methylation and full transcriptome analyses of paired primary cancers and liver metastases from CRC patients. We observed striking methylation differences between primary and metastatic pairs. A subset of loci showed coordinated methylation-expression changes, suggesting these are potentially epigenetic drivers that control the expression of critical genes in the metastatic cascade. The identification of CRC epigenomic markers of metastasis has the potential to enable better outcome prediction and lead to the discovery of new therapeutic targets.
Subject areas: Clinical genetics, Cancer, Transcriptomics, epigenomics, epigenetics, DNA methylation
Graphical abstract

Highlights
-
•
Epigenetic signature discriminates non-malignant, primary CRC and liver metastasis
-
•
Methylomes provide better separation of advanced CRC metastasis than transcriptomes
-
•
Altered CRC metastasis methylation associated with functional regulatory changes
-
•
Potential driver gene–methylation events of CRC metastasis identified
Clinical genetics; Cancer; Transcriptomics
Introduction
Colorectal cancer (CRC) is a leading cause of morbidity and mortality worldwide. Globally, more than 1.4 million individuals develop CRC and 0.6 million people die from CRC every year.1,2 Alarmingly, the global incidence of CRC has consistently risen at an annual rate of 3.2% over the last two decades and is predicted to continue to increase in the future.3 Furthermore, there is a particular trend of increasing rates of CRC in younger people (<50 years of age).4 CRC-related death occurs mainly because of metastasis of primary CRC (>90% cases), even after treatment,5 and patients with distant metastases have a 5-year survival rate of only ∼12%.6 The liver is the most common organ of distant metastasis in CRC – during the course of the disease ∼50% of CRC patients will develop liver metastasis and in ∼30% of patients with metastatic CRC, the liver is the only organ with metastases.7,8 Although significant efforts have been made to improve outcomes, CRC patients with distant metastasis who receive chemotherapy (such as fluoropyrimidines and oxaliplatin chemotherapy), eventually develop resistance.9 Earlier diagnosis or detection of metastatic potential would allow for earlier intervention and possibly improved prognosis. For example, adjuvant chemotherapy could be of significant benefit for early-stage patients with an increased risk of metastasis.10 However, improved risk stratification is necessary to identify early-stage patients who would benefit most from this approach.11 These observations clearly indicate that early detection and a better understanding of molecular drivers for CRC metastasis are of critical clinical importance.
Metastasis of CRC is a multi-step process and tumor cells require several different molecular or phenotypic changes to successfully complete the sequence of events.12,13 Remarkable progress has been made over the last few decades in understanding the genetic basis of primary cancer formation. However, even after extensive cancer genomic analyses, examples of causal genetic events for cancer metastasis are still limited.12,14 Very recently, non-mutational epigenetic reprogramming has been recognized as a key mechanistic determinant that enables tumor cells to achieve cancer hallmark capabilities.15 The key role of epigenetic mechanisms, such as DNA methylation, in driving metastatic progression is becoming established.16,17,18,19 Considering the dynamic nature of epigenetic changes, it is highly plausible that epigenetic events play a driver role in CRC metastasis. Indeed, recent studies have provided strong evidence for epigenetic changes accompanying CRC metastasis and poorer patient survival.20,21,22 Analysis of methylation and expression profiles of primary and metastatic CRCs at a single cell level revealed that methylation changes control expression of critical genes during metastasis.23 In addition, it is now well established that a subgroup of patients with CRC exhibits a particular methylation signature (referred to as CpG island methylation phenotype or CIMP), which possibly affects patient prognosis and response to specific therapies.24,25 Furthermore, pre-clinical data has demonstrated that DNA methylation-modifying drugs can reduce the proliferation of CRC-initiating cells.26 This evidence indicates that DNA methylation is a potential key regulator of CRC growth and also suggests the possibility of targeting methylation levels as a potential therapy.
One major issue in identifying epigenetic drivers of CRC metastasis is the extensive molecular heterogeneity exhibited in tumor cells. In this work, we performed whole-genome scale methylation and expression analysis in a well-defined set of paired patient samples (primary CRC and liver metastases). From our analyses, we aimed to identify functionally relevant epigenetic changes that occurred between primary cancers and liver metastases from the same patient, and to identify an epigenetic signature of advanced CRC metastasis. Furthermore, we utilized epigenetic data from normal colon to document progressive changes in potential epigenetic drivers of metastasis and we have validated our results and reproducibility with large additional cohorts of CRC. The development of a robust molecular signature that confers a high risk of metastasis could potentially be used to triage stage II and III patients who would benefit more significantly from adjuvant therapy. Furthermore, and possibly of greater clinical value, would be the development of new effective therapies that specifically target molecular drivers of metastasis.27
Results
Whole-genome scale DNA methylation profiles are conserved between primary CRC cancers and liver metastases but differ from normal colon
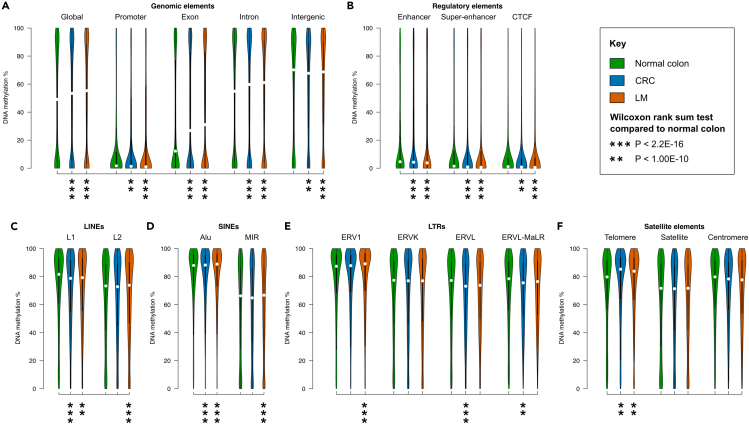
To investigate genome-scale DNA methylation changes of CRC metastasis, we performed reduced representation bisulfite sequencing (RRBS) on primary colonic cancers (CRC, n = 10, all adenocarcinomas) and matched liver metastases (n = 10, LM) from the same patients. These samples represented a diverse cohort, including a mixture of different primary tumor sites (left- and right-sided colon and rectum), and three patients received adjuvant chemotherapy before LM resection (Tables 1 and S1). In total, we obtained 1.4×109 sequence reads for the 20 cancer tissues, allowing for comprehensive genome-wide analysis (Table S2). For comparison, we analyzed our methylation maps alongside independent RRBS data of normal colon tissue (n = 10).28 Global methylome analysis revealed that genomic methylation levels were conserved between primary and matched metastatic tumors (median methylation = 53.37% and 53.32% for CRC and LM respectively), but increased compared to normal colon (median = 49.06%, Figure 1A and Table S3). Most genomic, regulatory and repeat elements had small methylation differences (<4%) in CRC and LM compared to normal colon, except for exons, introns and telomeres, which had increased median methylation of 16.75%, 5.51% and 4.79% respectively (Figures 1A–1F and Table S3). Compared to CRC, most elements had only small methylation differences (<2%) in LM, with the exception of exons, which were 4.16% more methylated (Figure 1A and Table S3).
Table 1.
Patient demographics and cancer characteristics
| Characteristics | Number of Patients |
|---|---|
| Age (years) | |
| 50–59 | 2 |
| 60–69 | 5 |
| 70–79 | 3 |
| Sex | |
| Male | 7 |
| Female | 3 |
| Primary cancer site | |
| Colon, left side | 4 |
| Colon, right side | 5 |
| Rectum | 1 |
| Treatment prior to liver resection | |
| Adjuvant chemotherapy | 3 |
| No treatment | 7 |
| Liver metastasis pathology | |
| Synchronous | 5 |
| Metachronous | 5 |
| Other metastases | |
| Lung metastases (metachronous) | 4 |
| Lymph node positive | 5 |
Figure 1.
Global differences in DNA methylation between normal colon, primary colorectal cancers and liver metastases
(A–F) Equal area violin plots of RRBS data show the distribution of methylation in different genomic elements of paired primary colorectal cancers (CRC, n = 10) and liver metastases (LM, n = 10) alongside normal colon (n = 10, Hanley et al. GSE95654). In all cases the Y axis represents DNA methylation level %. See also Table S3.
(A) Gene promoters (-5kb to +1kb from the TSS), exons, introns, and intergenic elements (>5kb upstream from the nearest TSS).
(B) Regulatory elements: enhancers, super-enhancers and CTCF binding sites.
(C) LINEs: L1 and L2.
(D) SINEs: Alu and MIR.
(E) LTRs: ERV1, ERVK, ERVL and ERV-MaLR.
(F) Satellite elements: telomere, satellite, centromere.
Genome-wide analysis identifies late-stage, liver metastasis-specific methylation changes compared to matched primary cancer and normal colon
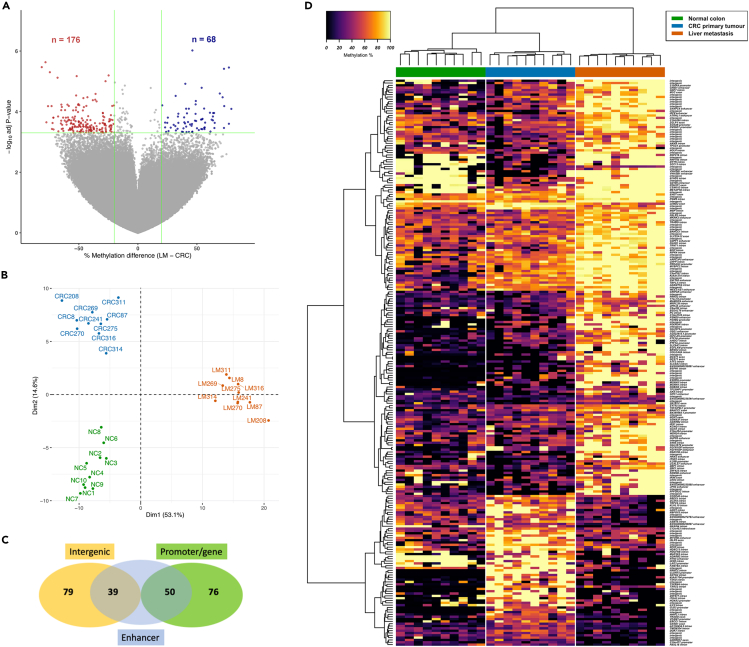
Locus-specific methylation analysis identified 244 differentially methylated CpGs (DMCs, P-value <5.0 × 10−4 with >20% methylation difference), of which 68 were hypomethylated and 176 were hypermethylated in LM compared to CRC (Figure 2A). Almost two-thirds of the DMCs showed large differences in methylation levels (>40%) and 17 CpGs had methylation differences >70% between CRC and LM (Figure S1A). We also observed that 20 of the DMCs were also differentially methylated between normal colon and CRC, and 44 were also differentially methylated between normal colon and LM. Approximately 50% of the DMCs within these two additional comparisons showed progressive changes in methylation from normal to CRC to LM. Therefore, the majority of identified DMCs were either metastasis-specific or were further altered in metastasis. Principal component analysis (PCA) of the top 1% CpG sites that varied most across all samples showed only segregation between tumor tissues (CRC, LM) and normal colon (Figure S1B). However, PCA of the DMCs showed distinct methylation patterns for each group. LM methylation was separated from CRC and normal colon in dimension 1, which accounted for 53.1% of the variance (Figure 2B). Unsupervised hierarchical clustering of the DMCs also showed that each group had distinct methylation patterns (Figure 2D and Table S4). Although imputation was used for missing methylation data values, the non-imputed methylation patterns of each group were also clearly segregated (Figure S1C). Of the 244 DMCs, 165 were in genes and/or regulatory regions (promoters/enhancers) associated with genes (Figure 2C and Table S4), indicating possible regulatory changes to these genes.
Figure 2.
Differential methylation patterns between normal colon, primary colorectal cancers and liver metastases
(A) Volcano plot showing DNA methylation differences between paired primary colorectal cancers (CRC, n = 10) and liver metastases (LM, n = 10). The x axis shows methylation differences of all analyzed CpG sites (LM - CRC) and the y axis shows the -log10 of P-values. Differentially methylated CpGs (DMCs, P-value <5.0 × 10−4 and methylation difference ≥20%) are shown in either red (increased methylation in LM compared to CRC) or blue (decreased methylation in LM).
(B) Principal component analysis (PCA) plot showing dimensionality reduction of the 244 DMCs alongside normal colon methylation data (n = 10, Hanley et al. GSE95654), with imputed missing methylation data values.
(C) Venn diagram of the number of DMCs in predicted enhancer elements, associated with genes (promoter/gene body) or in intergenic regions.
(D) Methylation heatmap of the 244 DMCs alongside normal colon (black = unmethylated, yellow = fully methylated). See also Figure S1 and Table S4.
Independent validation of methylation data and functional analysis of DMCs
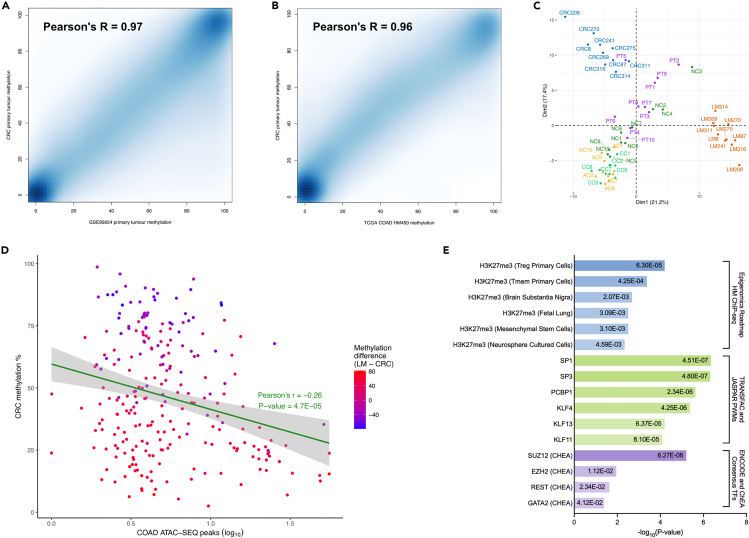
The methylation values of our CRC tissues were highly correlated with TCGA colorectal adenocarcinoma (TCGA-COAD) HumanMethylation450K (HM450K) data (Pearson’s r = 0.97, Figure 3A), and with primary colonic tumors from an independent RRBS dataset28 (Pearson’s r = 0.96, Figure 3A), indicating excellent reproducibility. Using this same RRBS data, PCA of the DMCs showed that the methylation patterns of the primary CRC tissues separated from non-cancerous tissues (normal colon, normal colonic crypt, aberrant crypt foci) and further separated from LM methylation (Figure 3C). To investigate our DMCs in a broader epigenomic context, we utilized several datasets: TCGA-COAD ATAC-seq chromatin accessibility, Epigenomics roadmap histone modification ChIP-Seq, TRANSFAC/JASPAR transcription factor binding profiles, and ENCODE/ChEA consensus target genes. The methylation values of the DMCs in our CRC tissues were negatively correlated with TCGA-COAD chromatin accessibility data (Pearson’s r = −0.26, Figure 3D and Table S5). These loci were highly enriched for H3K27me3, a repressive chromatin mark mediated by the polycomb repressive complex 2 (PRC2). They were also enriched for binding of transcription factors in the Sp/KLF family and of the PRC2 components, EZH2 and SUZ12 (Figure 3E and Table S6). Overall, these findings suggest that the differentially methylated CpG sites between primary colorectal tumors and liver metastases were associated with significant functional regulatory changes in these regions.
Figure 3.
Validation of genome-scale methylation and functional characterization of differentially methylated CpGs in primary colorectal cancers and liver metastases
(A and B) Smoothed scatterplot showing methylation correlation of primary colorectal adenocarcinomas (CRC, n = 10) compared to A) TCGA colorectal adenocarcinomas (TCGA-COAD, n = 410), and B) primary colorectal adenocarcinomas from an independent RRBS dataset (n = 10, Hanley et al. GSE95654).
(C) Principal component analysis (PCA) showing dimensionality reduction of the 244 differentially methylated CpGs (DMCs) alongside normal colon (NC, n = 10), normal colonic crypt (CC, n = 10), aberrant crypt foci (AC, n = 10) and primary colorectal adenocarcinomas (PT, n = 10; Hanley et al. GSE95654), with imputed missing methylation data values.
(D) Scatterplot showing correlation of CRC methylation and chromatin accessibility (TCGA-COAD ATAC-seq, n = 81) at the DMC loci.
(E) Enrichment analysis of histone modifications (Epigenomics Roadmap), transcription factor binding profiles (TRANSFAC/JASPAR) and consensus target genes (ENCODE/ChEA) associated with the DMCs, P-value <0.05. See also Tables S5 and S6.
Transcriptomic mapping revealed large expression changes and distinct pathways in liver metastasis
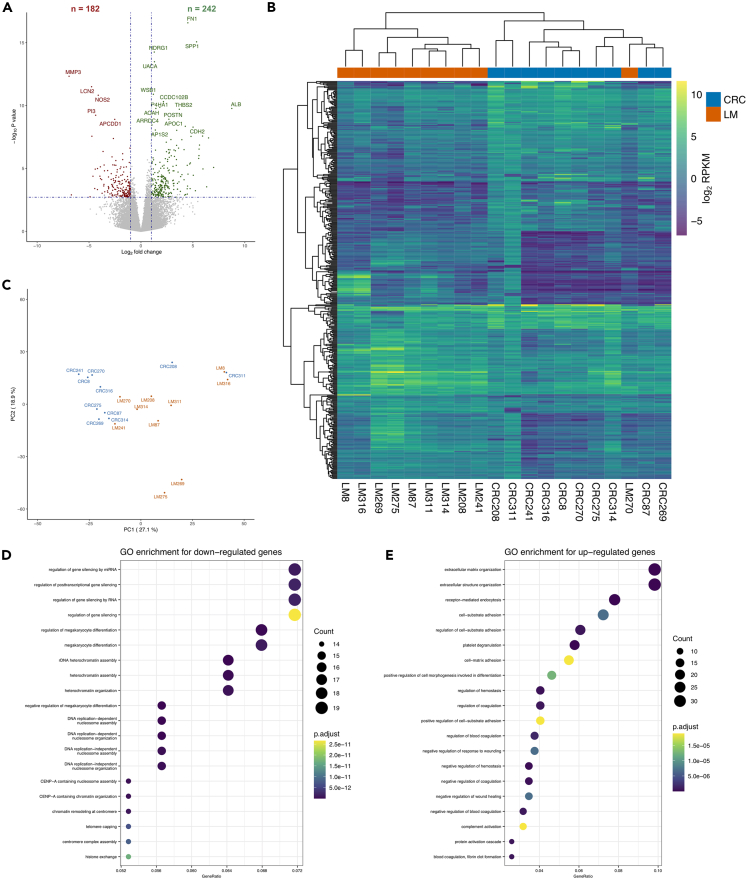
To analyze transcriptomic changes of CRC metastasis, we performed RNA-Seq on the same CRC and LM tissues (Table S7). We identified 424 genes that were significantly differentially expressed (DEG) between CRC and LM (FDR-corrected P-value <0.05 and fold-change of mean RPKM ≥2). Of these DEGs, 182 genes were down-regulated, whereas the remaining 242 genes were up-regulated in LM (Figure 4A and Table S8). With the exception of one sample (LM270), unsupervised hierarchical clustering of the DEGs separated the expression patterns of the two groups (Figure 4B). PCA of the 500 most variable genes showed only partial segregation of the CRC and LM groups across principal components 1 and 2, which in total accounted for 46% of the variance (Figure 4C).
Figure 4.
Differential gene expression patterns between paired primary colorectal cancers and liver metastases
(A) Volcano plot showing gene expression differences between paired primary colorectal cancers (CRC, n = 10) and liver metastases (LM, n = 10). The x axis shows log2 fold change of gene expression values (RPKM) and the y axis show the -log10 of P-values. Differentially expressed genes (DEGs, FDR-adjusted P-value <0.05 and expression difference ≥2-fold) are shown in green (increased expression in LM compared to CRC) or red (decreased expression in LM).
(B) Heatmap of the 424 DEGs (blue-to-yellow = low-to-high gene expression).
(C) Principal component analysis (PCA) plot showing dimensionality reduction of the 500 most variable genes.
(D) Gene ontology (GO) enrichment of genes with decreased expression in LM compared to CRC.
(E) GO enrichment of genes with increased expression in LM. See also Tables S7, S8, S9, and S10.
Functional gene enrichment analysis revealed that the 182 genes down-regulated in LM compared to CRC were mainly involved in gene silencing mechanisms and chromatin organization. These genes were also enriched for regulation of cell differentiation (Figure 4D and Table S9). In contrast, the 242 up-regulated genes were significantly implicated in extracellular matrix organization and cell adhesion. In addition, these genes were enriched for regulation of hemostasis and response to wound healing (Figure 4E and Table S10).
Immune cell composition and tumor microenvironment in paired primary colorectal cancers and liver metastases
Using expression-based deconvolution, we investigated the fractions of 22 infiltrated immune cell types in the paired CRC and LM tissues. In CRC, CD4 memory resting T cells, M0 macrophages, resting mast cells and resting NK cells were the most infiltrating fractions compared with other immune cells. This pattern is generally what has been previously observed in primary CRCs.29 In LM, the largest fractions of infiltrating immune cells were M0 macrophages, CD4 memory resting T cells, M2 macrophages and monocytes. The immune cell composition between the paired CRC and LM tumor tissues was mostly similar, although LM had significantly larger fractions of monocytes and M2 macrophages, but fewer resting mast cells than CRC (P-value <0.05, Figures 5A and 5B, and Table S11).
Figure 5.
Expression-based immune composition of paired primary colorectal cancers and liver metastases
(A) Boxplots of the immune cell composition of paired primary colorectal cancers (CRC, n = 10) and liver metastases (LM, n = 10), estimated by expression-based CIBERSORT deconvolution.
(B) Differences in immune cell composition of LM compared to CRC, based on CIBERSORT scores of paired samples. P-value <0.05 shown in red.
(C) Boxplots showing immune scores of CRC and LM compared to TCGA colorectal adenocarcinomas (TCGA-COAD, n = 263), based on ESTIMATE analysis. See also Table S11.
To further investigate immune cell composition and tumor microenvironment (TME) in advanced CRC and assess its relationship with the epigenetic signature, we performed a series of additional analyses. We used the ESTIMATE algorithm to obtain immune, stromal and purity scores of each analyzed sample. To benchmark these data from our cohort (CRC and LM), we have analyzed and directly compared them with colorectal adenocarcinoma patients (TCGA-COAD RNA-Seq version 2 dataset) as the standard. We found that the overall immune scores of CRC and LM were not significantly different (P-value = 0.39, Figure 5C and Table S11), which further validates the findings from CIBERSORT analysis that the global immune cell composition of CRC and LM were mostly similar. The immune scores in TCGA CRC patients were higher than for CRC (P-value = 0.04), but not significantly different compared to LM (P-value = 0.63; Figure 5C).
For the stromal scores, we also found that the difference between CRC and LM was not significant (P-value = 0.075, Table S11). The TCGA-COAD stromal scores were similar to LM (P-value = 0.39), but significantly higher than the CRC samples (P-value = 0.0097). The tumor purity analysis (ESTIMATE score) also showed that the purity of CRC and LM samples were not significantly different (P-value = 0.11, Table S11) and the purity of our cohort was similar to TCGA – LM scores were not significantly different (P-value = 0.84) whereas CRC samples had higher tumor purity than TCGA (P-value = 0.011). We also found highly significant negative correlations of tumor purity with both immune scores (Pearson r = −0.988, P-value = 6.42 × 10−16) and stromal scores (Pearson r = −0.917, P-value = 1.36 × 10−8). These data are consistent with previous TME analysis using multiple CRC cohorts, which demonstrated significant negative correlations of tumor purity with immune and stromal scores.30,31 Finally, we analyzed our identified signature in the context of an immune and stromal gene signature related to CRC metastasis. A well-validated 292 gene expression signature associated with immune and stromal components of CRC tumors was previously described using data from TCGA.32 This immune/stromal gene signature did not overlap with any of the 165 genes associated with the 244 CpG sites in our methylation signature, but did overlap with 44 of the 424 differentially expressed genes between CRC and LM.
Coordinated methylome-transcriptomic changes of colon-to-liver metastasis
To identify potential driver-gene methylation events of CRC metastasis to the liver, we performed integrative analysis of the 165 DMCs that were in genes and/or regulatory regions (promoters/enhancers) and the corresponding expression levels of these genes. As a result, we obtained a list of 21 DMCs related to 20 protein-coding genes that showed strong correlations between methylation and expression changes (P-value <0.05, either Spearman or Pearson correlation coefficient <0.4; Table S12). These included 16 hypermethylation and 5 hypomethylation events. Of the 21 DMCs, 17 were in predicted enhancer elements and 18 had negative correlations between methylation and expression. In most cases, the differences in methylation and expression showed a pairwise pattern between the matched CRC and LM tissues, of which the most significant are shown in Figure 6. The proteins encoded by these genes have a variety of different functions, including: regulation of nucleic acid processing and/or binding (KDM4B, SWI5, ZBP1, ZBTB7C, ZC3H3); intracellular signaling and transport (KLHL10, MAP2K3, NKAIN4, PDGFRB, PRKCZ); ECM/membrane and cell adhesion (EFHD2, EHD1, KCNQ1, SEC13, VCAM1); and lipid/carbohydrate metabolism (B3GALT6, GAL3ST2, TNFAIP8L1). Some of these have been described previously in CRC metastasis to distant organs, such as PDGFRB, PRKCZ, VCAM1 and ZBP1 (aka IMP1).33,34,35,36 Several more have been identified as markers for CRC diagnosis or prognosis, including GAL3ST2, KCNQ1, KDM4B, TNFAIP8L1 (aka TIPE1) and ZBTB7C.37,38,39,40,41 We have also identified several novel putative epigenetic drivers that have not been previously reported in CRC (C8orf82, EFHD2, SEC13, SWI5 and ZC3H3). Furthermore, we validated the reproducibility of the correlated methylation and expression relationship of six genes from TCGA-COAD data. All of them exhibited significantly negative correlated methylation and expression (Figure S2 and Table S12).
Figure 6.
Concordant DNA methylation and expression differences in paired primary colorectal cancers and liver metastases
(A–I) Scatterplots and boxplots of the most differentially methylated CpGs (DMCs, P-value <5.0 × 10−4 and methylation difference ≥20%) with correlated changes in gene expression (Spearman, P-value <0.05). A) B3GALT6 enhancer, chr1:1176934-1176935.
(B) C8orf82 intron, chr8:145753492-145753493.
(C) GAL3ST2 promoter, chr2:242715169-242715170.
(D) SEC13 intron, chr3:10346500-10346501.
(E) TNFAIP8L1 promoter, chr19:4638477-4638478.
(F) VCAM1 promoter, chr1:101184331-101184332.
(G) ZBP1 intron, chr20:56194194-56194195.
(H) ZBTB7C exon, chr18:45663647-45663648. I) ZC3H3 intron, chr8:144553096-144553097. See also Figure 6 and Table S12.
Discussion
Metastasis is a complex process which involves several biological and cellular steps that a cancer cell uses to successfully spread12,13; it is plausible that epigenetic changes play a driving role in this process.17,42 Although many studies have provided DNA methylation data for primary CRCs, epigenomic data for tumor metastases remains limited and particularly for paired tumor samples with well-defined clinical features. To our knowledge, here we provide the first sequencing-based genome-wide RRBS methylation and whole transcriptomic map of paired primary CRCs and matched liver metastases (LM) to identify a signature of advanced CRC metastasis. Furthermore, by utilizing multiple patient cohorts we provide extensive independent validation of our results.
We observe that in terms of the global methylation landscape, primary cancers and liver metastases show global hypermethylation when compared to normal colon. The methylome of primary cancers and liver metastases were very similar to each other, suggesting conservation of methylome architecture between primary and matched distant metastases. Global hypomethylation in metastasis compared to primary tumor has been reported for several cancer types,43,44 however, we did not observe that in our study. The global DNA methylation level is highly dependent on the technical platform used. We have used RRBS, which profiles dynamic and functional regions of the genome, but the true reflection of global methylation status can be most accurately obtained by WGBS (Whole Genome Bisulfite Sequencing), as it allows profiling of large repeat regions which are often hypomethylated in cancer. We have validated our global RRBS methylome by comparison of all common CpGs in independent RRBS data from CRC as well as with Illumina 450K methylation array data from the cancer genome atlas (containing >400 colorectal tumors). In both cases and platforms, we found excellent validation, providing strong evidence for the robustness of our methylation profiles across platforms and cohorts.
Although global methylation levels showed conservation, we have identified strong site-specific methylation changes (244 DMCs) associated with LM in colon cancer and the majority (>70%) of these were hypermethylated in LM. The comparison of normal colon, primary CRC and LM methylomes enabled us to determine the nature of methylation change and whether these DMCs were LM-specific. We revealed that the majority of DMCs showed similar and hypomethylated states in normal colon and primary CRC but became heavily hypermethylated in LM. A relatively small number of DMCs showed dynamic change as these became hypermethylated in primary tumor compared to normal colon and then lost methylation to become hypomethylated in LM. Integration of the methylation profiles of our identified DMCs with chromatin accessibility data (using ATAC-Seq platform) from CRC patients revealed a highly significant correlation of methylation levels and chromatin activity. These data suggest that the methylation levels of the DMCs have a possible functional genomic role, which warrants further investigation. When we analyzed multiple histone modification marks and transcription factor binding profiles, we found that the DMCs were heavily enriched for the repressive chromatin mark H3K27me3 mediated by the PRC2 complex. This was highly consistent with the fact that the DMC signature also showed high enrichment for transcription factors of the core PRC2 complex such as EZH2 and SUZ12. The expression of EZH2 and H3K27me3 were shown to be up-regulated in CRC and associated with advanced tumor stage. H3K27me3 and EZH2 levels were also shown to be positively correlated with the metastasis-free survival of CRC patients.45,46,47 We have demonstrated the cooperation of DNA methylation with EZH2 and H3K27me3 levels in other cancers.48,49 Our consistent results provide further strong evidence of specific and coordinated multilayer epigenetic deregulation in CRC metastasis. This also provides a rationale to explore combinatorial epigenetic therapy (such as DNA methylation inhibitor plus EZH2 inhibitors) for the treatment of CRC in the future.
Metastasis is a complex process that involves multiple molecular steps and is broadly explained by two potential models. In the initiation or founder model, a small population of cells harboring specific genetic or epigenetic profile becomes metastatic over time, whereas in the second and progressive model, the acquisition of specific changes over time provides a selective advantage to the tumor cells to successfully metastasize.50,51,52 Our results demonstrate similar global methylomes but highly discriminatory, LM-specific methylation changes in functionally important regions with a role in genomic regulation. These data provide support toward the progressive model where a specific methylation signature is gained and required during the metastasis cascade. Future studies, particularly involving other metastatic sites and appropriate models, could provide important further insights into explaining the possible model of metastasis, which remains a big question in the field of cancer biology. For example, DNA methylation analysis of circulating tumor cells would reveal dynamic epigenetic changes that are required for the cell to survive in circulation but change again to survive the new metastatic niche.53
Our identified signature consisting of 244 DMCs was able to completely segregate LMs from primary CRC and normal colon in PCA analysis. To test the generalizability of our identified signature, we have performed rigorous validation with additional cohorts and analyses. For further validation of our signature, we have included an additional independent cohort (generated by the same RRBS platform),28 consisting of normal colonic crypt, aberrant crypt foci and additional primary colonic cancers, the 244 CpG signature was able to clearly segregate LMs from all the other groups. Our cohort of tumors consists of matched pairs of primary cancers from both the left and right side of the colon and three patients in the cohort received adjuvant chemotherapy (FOLFOX) before liver resection, whereas the others did not. Therefore, a major finding from this study is our identified methylation signature is able to separate liver metastasis from normal colon, crypt and primary colon adenocarcinoma irrespective of the side of colon cancer and also suggests that the methylation signature may not be affected by treatment pressure. These results provide evidence that our methylation signature represents a universal molecular marker of the CRC metastatic process to the liver irrespective of the specific biology of CRC subtypes. This opens new opportunities to further test these signatures in a clinical setting to predict metastasis. These findings also provide a new avenue to conduct future studies in other cancer types (such as in lung and peritoneal metastasis) to reveal similar true methylation markers that could be identified in cancers originating from other anatomical sites. If this advanced metastasis signature is conserved across different subtypes of CRC, then it is plausible it is the same for other cancer types, which is worth investigating further. Future whole genome-scale studies with a larger number of well-curated and paired metastatic samples are likely to further elucidate the utility of epigenetic signatures in the metastasis process.
Our transcriptomic studies revealed large gene expression changes in LM compared to the matched primary tumors and these changes were associated with metastasis-related and gene regulatory pathways. Further confirming our evidence of multilayer epigenetic deregulation in CRC metastasis, there was an enrichment of differentially expressed genes related to epigenetic regulation of gene expression and chromatin remodeling – particularly H3 and H4 histones. There was a substantial decrease in the expression of different subunit components of the electron transport chain (e.g., complex I subunit gene NDUFS3, complex III subunit gene CYC1 and complex IV subunit gene COX8A). This is indicative of metabolic reprogramming, a cancer hallmark known to enhance cancer cell dissemination through EMT activation.54 Also consistent with alterations to support survival of metastatic tumor cells, there were expression changes in genes involved in extracellular matrix organization (e.g., ADAMTS10, TIMP1),55 regulation of wound healing (FGB, SERPINE1),56 and regulation of cell-substrate adhesion (e.g., MACF1, NRP1).57 However, our PCA analysis of expression profiles was unable to clearly segregate LMs from primary CRC, unlike the DNA methylation signature. Therefore, we provide evidence that DNA methylation is likely to provide a sustained and distinct signature for metastasis in contrast to the transcriptome, which is likely to represent acute and dynamic expression changes in cancer metastasis. A similar observation has been demonstrated in the context of a longitudinal personal multi-omic study, where DNA methylation was temporally stable and changes associated with chronic conditions but transcriptomic changes were related to acute events.58 By combining the DMCs and their associated gene expression levels from the transcriptome data, we were able to identify 21 loci that each showed a strong methylation-expression relationship in primary and LM tumors. The genes PDGFRB, PRKCZ, VCAM1, and ZBP1 (aka IMP1) have been described previously in CRC metastasis to distant organs.33,34,35,36 It appears that PDGFRB,33 PRKCZ,34 and VCAM159 all promote liver metastasis in CRC via an EMT-related mechanism. In addition, the genes GAL3ST2, KCNQ1, KDM4B, TNFAIP8L1 (aka TIPE1), and ZBTB7C have been identified as markers for CRC diagnosis or prognosis.37,38,39,40,41 As a histone demethylase, KDM4B is an important epigenetic regulator that is more highly expressed in advanced stages of CRC, is associated with poor prognosis, and is regulated by a variety of different cellular stimuli.60 Notably, we have identified several novel candidates that have not been previously reported in CRC (C8orf82, EFHD2, SEC13, SWI5 and ZC3H3). Our work provides a strong basis for future functional characterization of these candidate epigenetic drivers. Although the candidates described here have gene-specific methylation changes (ie. in genes and/or regulatory regions), we and others have shown previously that methylation changes at intergenic regions and repeat elements can also strongly influence gene expression.61 Functional studies with in vitro and in vivo models utilizing cutting-edge epigenomic editing tools17,62,63 could provide direct evidence for the role of methylation drivers in the metastatic cascade and identify new targets for treating advanced CRC.
The transcriptome profile also allowed us to deconvolute immune cell and tumor microenvironment (TME) composition of primary and matched LMs. The results broadly show that the immune cell make-up is similar in paired CRC and LMs, except for tumor-associated macrophages (TAMs). Orthogonal analysis using ESTIMATE also confirmed the overall similarity of immune scores between CRC and LM. Although TAMs represent a highly heterogeneous population, they are thought to pave the way for tissue invasion and intravasation and to provide favorable conditions for metastasis.64 We found both classically activated M1 (immune-stimulating and tumor-suppressive) and alternatively activated M2 macrophages (immune-suppressive and tumor-promoting) were increased in LM compared to primary CRC. However, only the increase in M2 macrophages was statistically significant. The proportion of M2/M1 type TAMs has been shown to be positively correlated with liver metastasis in patients with CRC.65 Our analysis suggests that LM contains elevated levels of mixed subtypes of TAMs compared to matched primary CRC, supporting a line of work that proposes polarizing immune-suppressive and metastasis-promoting macrophages (M2) into immune-stimulating and tumor-suppressive (M1) macrophages as a possible modulator of TME and therapeutic strategy in CRC.66 Future experimental studies focusing on specific immune cells and their phenotype in the CRC microenvironment are likely to provide an experimental basis for this hypothesis. Our ESTIMATE analysis demonstrated high tumor purity of CRC and LM, which we benchmarked against TCGA data and also showed no significant difference between CRC and LM in terms of purity and stromal score. Furthermore, our identified methylation signature did not show any overlap with the previously established immune/stromal CRC gene signature. These multiple lines of investigation provide evidence that our identified methylation signature and the differences we observed in methylation status are very likely to be characteristics of tumor cells within CRC and LM rather than the influence of infiltrating immune and stromal cells.
In conclusion, we have identified a signature of advanced CRC metastasis by performing unbiased whole genome-scale DNA methylation and full transcriptome analyses of paired primary and liver metastases from CRC patients. Although our genome-wide analyses were performed in well-defined, discrete clinical samples, we show high technical validation as well as excellent validation of our methylation signature in independent cohorts. We also show that the methylation signature is independent of immune and stromal components. Our analysis uncovered universal markers of CRC metastasis to the liver that are independent of clinical subtype and future large-scale studies will aid the assessment of clinical utility of these markers. The metastasis signature was associated with functionally relevant changes to genomic regulation. Furthermore, these markers could represent epigenetic changes that facilitate a more generalized mechanism of metastasis. A more comprehensive validation of this signature in other cancer types and metastatic sites could provide a target for inhibiting metastatic spread.
Limitations of the study
A limitation of our study is that we have derived the epigenetic signature based on the analysis of only 20 tumors. Paired primary and metastatic tumor samples (especially with good clinical information) are challenging to obtain and therefore limited data is available for paired epigenetic analysis. To test the generalizability of our identified signature, we have performed rigorous validation with additional cohorts and analyses.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Tissue samples of primary colorectal adenocarcinomas and liver metastases from the same patients | Cancer Society Tissue Bank (University of Otago, Christchurch, New Zealand) | https://www.otago.ac.nz/mackenzie-cancer/tissue-bank/ |
| Critical commercial assays | ||
| TruSeq DNA Nano Low Throughput Library Prep Kit | Illumina | Cat#: TG-202-1001 |
| EZ DNA Methylation Kit | Zymo Research | Cat#: D5001 |
| TruSeq RNA Library Preparation Kit v2 | Illumina | Cat#: RS-122-2001 |
| Deposited data | ||
| Raw and analyzed RRBS and RNA-seq data of primary colorectal adenocarcinomas and liver metastases from the same patients | This paper | GEO: GSE213402 |
| Human reference genome NCBI build 37, GRCh37 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| Raw RRBS data of primary colorectal adenocarcinomas, adjacent normal colon, normal colonic crypt, and aberrant crypt foci | Hanley et al.1 | GEO: GSE95654 |
| GeneHancer | Fishilevich et al.2 | https://genome.ucsc.edu/ |
| H3K27Ac ChIP-seq data of HCT116 cells | Orouji et al.3 | GEO: GSE136888 |
| CTCF ChIP-seq data of HCT116 cells | Wang et al.4 | GEO: GSE30263 |
| RepeatMasker | Smit et al.5 | https://genome.ucsc.edu/ |
| HumanMethylation450K methylation data from colon adenocarcinoma and rectum adenocarcinoma cohorts | TCGA | https://portal.gdc.cancer.gov |
| ATAC-Seq data from colon adenocarcinoma cohort | TCGA | https://portal.gdc.cancer.gov/ |
| Expression-based ESTIMATE scores from colon adenocarcinoma cohort (RNA-Seq version 2) | TCGA | https://bioinformatics.mdanderson.org/estimate/ |
| Software and algorithms | ||
| Bismark | Krueger et al.6 | https://www.bioinformatics.babraham.ac.uk/projects/bismark/ |
| DMAP | Stockwell et al.7 | https://www.otago.ac.nz/chatterjee-lab/tools/ |
| R packages: hclust, prcomp, methyLImp, TCGAbiolinks, deeptools, featureCounts, DESeq2, ClusterProfiler | RStudio | https://posit.co |
| BEDTools | Quinlan et al.8 | https://bedtools.readthedocs.io/en/latest/ |
| Wanderer | Díez-Villanueva et al.9 | http://maplab.cat/wanderer |
| Enrichr | Xie et al.10 | https://maayanlab.cloud/Enrichr/ |
| HISAT2 | Kim et al.11 | http://daehwankimlab.github.io/hisat2/ |
| TIMER2.0 | Li et al.12 | http://timer.cistrome.org |
| ESTIMATE | Yoshihara et al.13 | https://bioinformatics.mdanderson.org/public-software/estimate/ |
Resource availability
Lead contact
Further information and requests should be directed to and will be fulfilled by the lead contact, Aniruddha Chatterjee: aniruddha.chatterjee@otago.ac.nz.
Materials availability
This study did not generate any new unique materials or reagents.
Experimental model and study participant details
Sample collection and analysis
Tissue samples of primary colorectal adenocarcinomas (n=10) and liver metastases from the same patients (n=10) were accessed from the Cancer Society Tissue Bank (University of Otago, Christchurch). Ethical approval was granted for the study from the University of Otago, Human Ethics Committee (Approval number: H16/037). Patients provided written, informed consent prior to biobanking of specimens. Patient data, including staging, recurrence, metastases, treatment and histology were retrospectively collected from patient medical records (Table S1). Routine clinical testing of MMR, BRAF and KRAS was not undertaken at the time these patients underwent resection and therefore this data is not available. Exclusion criteria included patients with hereditary CRC, and patients who had received pre-operative chemotherapy or radiation therapy, prior to resection of their primary cancers. Patients were included if resection specimens were available from their liver metastases. Tumour core samples were dissected from surgical specimens and immediately frozen in liquid nitrogen and initially stored at −80°C.
Method details
RRBS library preparation and sequencing
We used reduced representation bisulfite sequencing (RRBS) to map genome-scale DNA methylation at single nucleotide resolution as we have described previously.67,68 Briefly, genomic DNA was extracted from <20 mg of tissue using the Qiagen QIAamp DNA Mini Kit. The DNA was digested with NEB MspI enzyme followed by end-repair and ligation of Illumina TruSeq sequencing adaptors. Fragments were then size-selected and bisulfite-converted using the Zymo EZ DNA Methylation Kit prior to 16–18 rounds of PCR amplification. The quality and size distribution of the libraries was determined using an Agilent 2100 Bioanalyzer and libraries were sequenced on an Illumina HiSeq2500 machine (100 bp reads, single-ended). Base-calling of the reads was performed with Illumina Real Time Analyzer (RTA) software.
RNA isolation and sequencing
RNA extraction and sequencing were carried out as detailed previously.69 Briefly, RNA was extracted from <20 mg of tissue using the RNeasy Plus Mini Kit, including DNAse treatment, following tissue disruption using a Retsch Mixer Mill. RNA-seq libraries were prepared using Illumina TruSeq V2 reagents, including ribosomal RNA depletion using RiboZero Gold. Sequencing was performed using the Illumina HiSeq 2500 V4 platform to produce 125 bp paired-end reads.
Quantification and statistical analysis
DNA methylation data analysis
The quality check and processing of the sequenced RRBS reads was performed using in-house developed bioinformatics tools as previously described.70,71 The sequencing coverage and quality statistics for each sample are summarised in Table S2. Processed sequence reads were aligned to the reference human genome (GRCh37) using Bismark72 with stringent mapping criteria by allowing only one mismatch (default = 2) in the seed. The median non-CpG DNA methylation of all samples was 0.41%, indicating effective bisulfite conversion and low levels of true non-CpG methylation. CpG sites overlapping with SNPs (dbSNP build 151) were filtered out. For CpGs with at least 5× coverage in at least eight paired samples, a two-sample t-test with Welch correction identified 244 differentially methylated CpGs (DMCs) with at least 20% difference in methylation and P-value < 5.0×10-04. Gene information for all CpGs, including location of gene promoters, introns and exons, was derived using the identgeneloc program of the DMAP tool.71 R packages were used for unsupervised hierarchical clustering of CpG site methylation, using ward linkage (hclust method = "ward.D2") and Euclidean distance (dist method = "euclidean"), and Principal Component Analysis (PCA) using default parameters (prcomp). The methyLImp package was used to impute missing methylation data values (maximum 2 per group for each CpG).
Comparison with external datasets
We utilised previously described RRBS data from primary colorectal adenocarcinomas (n=10), adjacent normal colon (n=10), normal colonic crypt (n=10) and aberrant crypt foci (n=10; GSE95654).28 There were 4,733,522 CpGs in common with our data. We used BEDTools73 to overlap the methylation data with several genomic elements and regulatory features: enhancers (GeneHancer74), super-enhancers (H3K27Ac ChIP-seq data of HCT116 cells, GSE136888),75 CTCF binding sites (CTCF ChIP-seq data of HCT116 cells, GSE30263)76 and specific repeat classes (RepeatMasker).77 Level 2 normalised TCGA Infinium HumanMethylation450K (HM450K) methylation data from colon adenocarcinoma (TCGA-COAD) and rectum adenocarcinoma (TCGA-READ) projects (n = 410) were obtained using the R package TCGAbiolinks. There were 136,838 CpGs in common with our data. The Wanderer tool78 was used to obtain level 3 TCGA-COAD methylation array data of the HM450K probes closest to each of the DMCs and corresponding RNA-Seq gene expression data (n=249). ATAC-Seq bigwig files from the TCGA-COAD cohort (n=81) were downloaded from the National Cancer Institute Genomic Data Commons (https://portal.gdc.cancer.gov/). The multiBamSummary function from the deeptools package was used to profile the chromatin accessibility at the loci of the 244 DMCs. The online platform Enrichr79 was used to infer the enrichment of the DMCs within several datasets (modified Fisher’s exact test, P-value <0.05): Epigenomics Roadmap histone modifications, TRANSFAC/JASPAR transcription factor binding profiles and ENCODE/ChEA consensus target genes.
Analysis of transcriptomic data
The quality check and processing of the RNA-seq data was performed as previously described.80 The sequencing coverage and quality statistics for each sample are summarised in Table S7. Adaptor and low quality (<Q20) sequences were removed using fastq-mcf tools. Cleaned reads were then aligned to the human genome (GRCh37) using HISAT2 version 2.0.5.81 Read counts were retrieved using featureCounts based on Ensembl gene annotation. Read counts were normalised and differential expression analysis was conducted using DESeq2. The design formula integrated tumour type, controlling for “patient-specific effect”. A Wald test identified 424 genes significantly differentially expressed genes (DEGs) with Benjamini-Hochberg (BH) adjusted P-value <0.05 and fold-change of mean RPKM ≥2.
Pathway, tumour microenvironment and tumour purity analysis
Pathway analysis was performed using an over-representation approach. DEGs were tested against Gene Ontology terms using ClusterProfiler and all annotated genes as background (BH-adjusted P-value <0.05). The online platform TIMER2.082 was used to perform expression-based deconvolution, estimating the proportion of 22 different immune cell types in each tissue sample, using the CIBERSORT-ABS values from the estimation matrix for comparisons (Wilcoxon signed-rank test, P-value <0.05). The tidyestimate R package (URL: https://cran.r-project.org/web/packages/tidyestimate/vignettes/using-tidyestimate.html; based on ESTIMATE algorithm83) was used to calculate tumour purity, immune and stromal scores from expression data. To compare our results with TCGA data, tumour purity, immune and stromal scores were downloaded from MD Anderson Cancer Centre curated TCGA database (URL: https://bioinformatics.mdanderson.org/estimate/disease.html), using RNA-Seq version 2 to match our platform of data. As recommended, we used ESTIMATE as a measure of tumour purity – higher purity is indicated by a lower ESTIMATE score. For consistent comparisons, a Wilcoxon signed-rank test (P-value <0.05) was used between different groups to calculate the significance of immune, stromal and purity components.
Acknowledgments
The authors are grateful to the patients and their families for contributing the samples used in this study, The Cancer Society Tissue Bank (CSTB), and the Otago Genomics Facility at the University of Otago for providing the sequencing service and assistance. This research was funded by the Royal Society of New Zealand Te Apārangi (for a Rutherford Discovery Fellowship to A.C. and Marsden Fund to E.J.R. and A.C.), and grants from Lottery Health Research New Zealand, the University of Otago (A.C.), Maurice and Phyllis Paykel Trust, and Gut Cancer Foundation NZ (to R.V.P.). We are also grateful for additional support from the T.D. Scott Chair in Urology and Department of Surgical Sciences (to A.C. and E.J.R.), Hugh Green Foundation, Colorectal Surgical Society of Australia and New Zealand (to R.V.P.), Maurice Wilkins Centre for Molecular Biodiscovery, Cancer Society of New Zealand, Health Research Council of New Zealand, New Zealand Institute for Cancer Research Trust (to M.R.E. – salary support for A.C., E.J.R., S.A., A.L.), and John Gavin Postdoctoral Fellowship from the Cancer Research Trust New Zealand (to A.A.).
Author contributions
Conceptualization: E.R., M.E., S.P., J.M., R.P., and A.C.; Formal analysis: E.R., G.G., P.A., P.S., and S.A.; Investigation: E.R., S.A., S.B., A.L., A.A., and S.S.; Resources: M.E., F.F., R.P, and A.C.; Data curation (access and verification of the data): E.R., G.G., P.A., P.S., and S.S.; Writing–original draft: E.R. and A.C.; Writing–review and editing: All authors. Visualization: E.R., G.G., and P.A.; Supervision: M.E., R.P., and A.C.; Project Administration: E.R., R.P., and A.C.; Funding acquisition: R.P. and A.C. All authors read and approved the final version of the manuscript.
Declaration of interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: May 28, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106986.
Contributor Information
Euan J. Rodger, Email: euan.rodger@otago.ac.nz.
Rachel V. Purcell, Email: rachel.purcell@otago.ac.nz.
Aniruddha Chatterjee, Email: aniruddha.chatterjee@otago.ac.nz.
Supplemental information
Data and code availability
-
•
RRBS and RNA-seq data have been deposited at GEO and are publicly available as of the date of publication; the series accession number is listed in the key resources table. De-identified patient data is available in this paper’s supplemental information. This paper also analyses existing, publicly available data. The accession numbers for these datasets are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
References
- 1.Verma M., Kumar V. Epigenetic biomarkers in colorectal cancer. Mol. Diagn. Ther. 2017;21:153–165. doi: 10.1007/s40291-016-0244-x. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA A Cancer J. Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 4.Stoffel E.M., Murphy C.C. Epidemiology and mechanisms of the increasing incidence of colon and rectal cancers in young adults. Gastroenterology. 2020;158:341–353. doi: 10.1053/j.gastro.2019.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel R., Naishadham D., Jemal A. Cancer statistics, 2013. CA A Cancer J. Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 6.Bupathi M., Wu C. Biomarkers for immune therapy in colorectal cancer: mismatch-repair deficiency and others. J. Gastrointest. Oncol. 2016;7:713–720. doi: 10.21037/jgo.2016.07.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tauriello D.V.F., Calon A., Lonardo E., Batlle E. Determinants of metastatic competency in colorectal cancer. Mol. Oncol. 2017;11:97–119. doi: 10.1002/1878-0261.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vatandoust S., Price T.J., Karapetis C.S. Colorectal cancer: metastases to a single organ. World J. Gastroenterol. 2015;21:11767–11776. doi: 10.3748/wjg.v21.i41.11767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou H., Liu Z., Wang Y., Wen X., Amador E.H., Yuan L., Ran X., Xiong L., Ran Y., Chen W., Wen Y. Colorectal liver metastasis: molecular mechanism and interventional therapy. Signal Transduct. Targeted Ther. 2022;7:70. doi: 10.1038/s41392-022-00922-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casadaban L., Rauscher G., Aklilu M., Villenes D., Freels S., Maker A.V. Adjuvant chemotherapy is associated with improved survival in patients with stage II colon cancer. Cancer. 2016;122:3277–3287. doi: 10.1002/cncr.30181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan G.H.J., Chee C.E. Making sense of adjuvant chemotherapy in colorectal cancer. J. Gastrointest. Oncol. 2019;10:1183–1192. doi: 10.21037/jgo.2019.06.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lambert A.W., Pattabiraman D.R., Weinberg R.A. Emerging biological principles of metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen M., Kang Y. Stresses in the metastatic cascade: molecular mechanisms and therapeutic opportunities. Genes Dev. 2020;34:1577–1598. doi: 10.1101/gad.343251.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makohon-Moore A.P., Zhang M., Reiter J.G., Bozic I., Allen B., Kundu D., Chatterjee K., Wong F., Jiao Y., Kohutek Z.A., et al. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat. Genet. 2017;49:358–366. doi: 10.1038/ng.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. [DOI] [PubMed] [Google Scholar]
- 16.Chen J.F., Yan Q. The roles of epigenetics in cancer progression and metastasis. Biochem. J. 2021;478:3373–3393. doi: 10.1042/BCJ20210084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerjee R., Smith J., Eccles M.R., Weeks R.J., Chatterjee A. Epigenetic basis and targeting of cancer metastasis. Trends Cancer. 2022;8:226–241. doi: 10.1016/j.trecan.2021.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Pomerantz M.M., Qiu X., Zhu Y., Takeda D.Y., Pan W., Baca S.C., Gusev A., Korthauer K.D., Severson T.M., Ha G., et al. Prostate cancer reactivates developmental epigenomic programs during metastatic progression. Nat. Genet. 2020;52:790–799. doi: 10.1038/s41588-020-0664-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Z., Zou J., Li S., Topper M.J., Tao Y., Zhang H., Jiao X., Xie W., Kong X., Vaz M., et al. Epigenetic therapy inhibits metastases by disrupting premetastatic niches. Nature. 2020;579:284–290. doi: 10.1038/s41586-020-2054-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beggs A.D., Jones A., El-Bahrawy M., Abulafi M., Hodgson S.V., Tomlinson I.P.M. Whole-genome methylation analysis of benign and malignant colorectal tumours. J. Pathol. 2013;229:697–704. doi: 10.1002/path.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez-Cardús A., Moran S., Musulen E., Moutinho C., Manzano J.L., Martinez-Balibrea E., Tierno M., Élez E., Landolfi S., Lorden P., et al. Epigenetic homogeneity within colorectal tumors predicts shorter relapse-free and overall survival times for patients with locoregional cancer. Gastroenterology. 2016;151:961–972. doi: 10.1053/j.gastro.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Vidal E., Sayols S., Moran S., Guillaumet-Adkins A., Schroeder M.P., Royo R., Orozco M., Gut M., Gut I., Lopez-Bigas N., et al. A DNA methylation map of human cancer at single base-pair resolution. Oncogene. 2017;36:5648–5657. doi: 10.1038/onc.2017.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bian S., Hou Y., Zhou X., Li X., Yong J., Wang Y., Wang W., Yan J., Hu B., Guo H., et al. Single-cell multiomics sequencing and analyses of human colorectal cancer. Science. 2018;362:1060–1063. doi: 10.1126/science.aao3791. [DOI] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia M., Gao X., Zhang Y., Hoffmeister M., Brenner H. Different definitions of CpG island methylator phenotype and outcomes of colorectal cancer: a systematic review. Clin. Epigenet. 2016;8:25. doi: 10.1186/s13148-016-0191-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roulois D., Loo Yau H., Singhania R., Wang Y., Danesh A., Shen S.Y., Han H., Liang G., Jones P.A., Pugh T.J., et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162:961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson R.L., Balasas T., Callaghan J., Coombes R.C., Evans J., Hall J.A., Kinrade S., Jones D., Jones P.S., Jones R., et al. A framework for the development of effective anti-metastatic agents. Nat. Rev. Clin. Oncol. 2019;16:185–204. doi: 10.1038/s41571-018-0134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanley M.P., Hahn M.A., Li A.X., Wu X., Lin J., Wang J., Choi A.H., Ouyang Z., Fong Y., Pfeifer G.P., et al. Genome-wide DNA methylation profiling reveals cancer-associated changes within early colonic neoplasia. Oncogene. 2017;36:5035–5044. doi: 10.1038/onc.2017.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge P., Wang W., Li L., Zhang G., Gao Z., Tang Z., Dang X., Wu Y. Profiles of immune cell infiltration and immune-related genes in the tumor microenvironment of colorectal cancer. Biomed. Pharmacother. 2019;118:109228. doi: 10.1016/j.biopha.2019.109228. [DOI] [PubMed] [Google Scholar]
- 30.Wu Y., Jia H., Zhou H., Liu X., Sun J., Zhou X., Zhao H. Immune and stromal related genes in colon cancer: analysis of tumour microenvironment based on the cancer genome atlas (TCGA) and gene expression omnibus (GEO) databases. Scand. J. Immunol. 2022;95:e13119. doi: 10.1111/sji.13119. [DOI] [PubMed] [Google Scholar]
- 31.Chen W., Huang J., Xiong J., Fu P., Chen C., Liu Y., Li Z., Jie Z., Cao Y. Identification of a tumor microenvironment-related gene signature indicative of disease prognosis and treatment response in colon cancer. Oxid. Med. Cell. Longev. 2021;2021:6290261. doi: 10.1155/2021/6290261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y., Cheng L., Li C., Zhang C., Wang L., Zhang J. Identification of tumor microenvironment-related prognostic genes in colorectal cancer based on bioinformatic methods. Sci. Rep. 2021;11:15040. doi: 10.1038/s41598-021-94541-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steller E.J.A., Raats D.A., Koster J., Rutten B., Govaert K.M., Emmink B.L., Snoeren N., van Hooff S.R., Holstege F.C.P., Maas C., et al. PDGFRB promotes liver metastasis formation of mesenchymal-like colorectal tumor cells. Neoplasia. 2013;15:204–217. doi: 10.1593/neo.121726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shelton P.M., Duran A., Nakanishi Y., Reina-Campos M., Kasashima H., Llado V., Ma L., Campos A., García-Olmo D., García-Arranz M., et al. The secretion of miR-200s by a PKCzeta/ADAR2 signaling Axis promotes liver metastasis in colorectal cancer. Cell Rep. 2018;23:1178–1191. doi: 10.1016/j.celrep.2018.03.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexiou D., Karayiannakis A.J., Syrigos K.N., Zbar A., Kremmyda A., Bramis I., Tsigris C. Serum levels of E-selectin, ICAM-1 and VCAM-1 in colorectal cancer patients: correlations with clinicopathological features, patient survival and tumour surgery. Eur. J. Cancer. 2001;37:2392–2397. doi: 10.1016/s0959-8049(01)00318-5. [DOI] [PubMed] [Google Scholar]
- 36.Hamilton K.E., Noubissi F.K., Katti P.S., Hahn C.M., Davey S.R., Lundsmith E.T., Klein-Szanto A.J., Rhim A.D., Spiegelman V.S., Rustgi A.K. IMP1 promotes tumor growth, dissemination and a tumor-initiating cell phenotype in colorectal cancer cell xenografts. Carcinogenesis. 2013;34:2647–2654. doi: 10.1093/carcin/bgt217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snow S.M., Matkowskyj K.A., Maresh M., Clipson L., Vo T.N., Johnson K.A., Deming D.A., Newton M.A., Grady W.M., Pickhardt P.J., Halberg R.B. Validation of genetic classifiers derived from mouse and human tumors to identify molecular subtypes of colorectal cancer. Hum. Pathol. 2022;119:1–14. doi: 10.1016/j.humpath.2021.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.den Uil S.H., Coupé V.M.H., Linnekamp J.F., van den Broek E., Goos J.A.C.M., Delis-van Diemen P.M., Belt E.J.T., van Grieken N.C.T., Scott P.M., Vermeulen L., et al. Loss of KCNQ1 expression in stage II and stage III colon cancer is a strong prognostic factor for disease recurrence. Br. J. Cancer. 2016;115:1565–1574. doi: 10.1038/bjc.2016.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen B., Zhu Y., Chen J., Feng Y., Xu Y. Activation of TC10-like transcription by lysine demethylase KDM4B in colorectal cancer cells. Front. Cell Dev. Biol. 2021;9:617549. doi: 10.3389/fcell.2021.617549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye T., Yang B., Wang C., Su C., Luo J., Yang X., Yu H., Yuan Z., Meng Z., Xia J. TIPE1 impairs stemness maintenance in colorectal cancer through directly targeting beta-catenin. Carcinogenesis. 2020;41:25–35. doi: 10.1093/carcin/bgz079. [DOI] [PubMed] [Google Scholar]
- 41.Chen X., Jiang Z., Pu Y., Jiang X., Xiang L., Jiang Z. Zinc finger and BTB domain-containing 7C (ZBTB7C) expression as an independent prognostic factor for colorectal cancer and its relevant molecular mechanisms. Am. J. Transl. Res. 2020;12:4141–4159. [PMC free article] [PubMed] [Google Scholar]
- 42.Chatterjee A., Rodger E.J., Eccles M.R. Epigenetic drivers of tumourigenesis and cancer metastasis. Semin. Cancer Biol. 2018;51:149–159. doi: 10.1016/j.semcancer.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 43.Vidal E., Sayols S., Moran S., Guillaumet-Adkins A., Schroeder M.P., Royo R., Orozco M., Gut M., Gut I., Lopez-Bigas N., et al. A DNA methylation map of human cancer at single base-pair resolution. Oncogene. 2017;36:5648–5657. doi: 10.1038/onc.2017.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chatterjee A., Stockwell P.A., Ahn A., Rodger E.J., Leichter A.L., Eccles M.R. Genome-wide methylation sequencing of paired primary and metastatic cell lines identifies common DNA methylation changes and a role for EBF3 as a candidate epigenetic driver of melanoma metastasis. Oncotarget. 2017;8:6085–6101. doi: 10.18632/oncotarget.14042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Q., Chen X., Jiang Y., Liu S., Liu H., Sun X., Zhang H., Liu Z., Tao Y., Li C., et al. Elevating H3K27me3 level sensitizes colorectal cancer to oxaliplatin. J. Mol. Cell Biol. 2020;12:125–137. doi: 10.1093/jmcb/mjz032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang C.G., Ye Y.J., Yuan J., Liu F.F., Zhang H., Wang S. EZH2 and STAT6 expression profiles are correlated with colorectal cancer stage and prognosis. World J. Gastroenterol. 2010;16:2421–2427. doi: 10.3748/wjg.v16.i19.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vilorio-Marqués L., Martín V., Diez-Tascón C., González-Sevilla M.F., Fernández-Villa T., Honrado E., Davila-Batista V., Molina A.J. The role of EZH2 in overall survival of colorectal cancer: a meta-analysis. Sci. Rep. 2017;7:13806. doi: 10.1038/s41598-017-13670-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tiffen J., Gallagher S.J., Filipp F., Gunatilake D., Emran A.A., Cullinane C., Dutton-Register K., Aoude L., Hayward N., Chatterjee A., et al. EZH2 cooperates with DNA methylation to downregulate key tumour suppressors and interferon gene signatures in melanoma. J. Invest. Dermatol. 2020;140:2442–2454.e5. doi: 10.1016/j.jid.2020.02.042. [DOI] [PubMed] [Google Scholar]
- 49.Emran A.A., Chatterjee A., Rodger E.J., Tiffen J.C., Gallagher S.J., Eccles M.R., Hersey P. Targeting DNA methylation and EZH2 activity to overcome melanoma resistance to immunotherapy. Trends Immunol. 2019;40:328–344. doi: 10.1016/j.it.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 50.Nowell P.C. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 51.Ramaswamy S., Ross K.N., Lander E.S., Golub T.R. A molecular signature of metastasis in primary solid tumors. Nat. Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 52.Fares J., Fares M.Y., Khachfe H.H., Salhab H.A., Fares Y. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct. Targeted Ther. 2020;5:28. doi: 10.1038/s41392-020-0134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vasantharajan S.S., Eccles M.R., Rodger E.J., Pattison S., McCall J.L., Gray E.S., Calapre L., Chatterjee A. The Epigenetic landscape of Circulating tumour cells. Biochim. Biophys. Acta Rev. Canc. 2021;1875:188514. doi: 10.1016/j.bbcan.2021.188514. [DOI] [PubMed] [Google Scholar]
- 54.Sun L., Zhang H., Gao P. Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell. 2022;13:877–919. doi: 10.1007/s13238-021-00846-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winkler J., Abisoye-Ogunniyan A., Metcalf K.J., Werb Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020;11:5120. doi: 10.1038/s41467-020-18794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deyell M., Garris C.S., Laughney A.M. Cancer metastasis as a non-healing wound. Br. J. Cancer. 2021;124:1491–1502. doi: 10.1038/s41416-021-01309-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Janiszewska M., Primi M.C., Izard T. Cell adhesion in cancer: beyond the migration of single cells. J. Biol. Chem. 2020;295:2495–2505. doi: 10.1074/jbc.REV119.007759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen R., Xia L., Tu K., Duan M., Kukurba K., Li-Pook-Than J., Xie D., Snyder M. Longitudinal personal DNA methylome dynamics in a human with a chronic condition. Nat. Med. 2018;24:1930–1939. doi: 10.1038/s41591-018-0237-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang D., Bi J., Liang Q., Wang S., Zhang L., Han F., Li S., Qiu B., Fan X., Chen W., et al. VCAM1 promotes tumor cell invasion and metastasis by inducing EMT and transendothelial migration in colorectal cancer. Front. Oncol. 2020;10:1066. doi: 10.3389/fonc.2020.01066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson C., Krieg A.J. KDM4B: a nail for every hammer? Genes. 2019;10:134. doi: 10.3390/genes10020134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chatterjee A., Rodger E.J., Ahn A., Stockwell P.A., Parry M., Motwani J., Gallagher S.J., Shklovskaya E., Tiffen J., Eccles M.R., Hersey P. Marked global DNA hypomethylation is associated with constitutive PD-L1 expression in melanoma. iScience. 2018;4:312–325. doi: 10.1016/j.isci.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith J., Banerjee R., Weeks R.J., Chatterjee A. Editing of DNA methylation patterns using CRISPR-based tools. Methods Mol. Biol. 2022;2458:63–74. doi: 10.1007/978-1-0716-2140-0_4. [DOI] [PubMed] [Google Scholar]
- 63.Urbano A., Smith J., Weeks R.J., Chatterjee A. Gene-specific targeting of DNA methylation in the mammalian genome. Cancers. 2019;11:1515. doi: 10.3390/cancers11101515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cortese N., Soldani C., Franceschini B., Barbagallo M., Marchesi F., Torzilli G., Donadon M. Macrophages in colorectal cancer liver metastases. Cancers. 2019;11:633. doi: 10.3390/cancers11050633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cui Y.L., Li H.K., Zhou H.Y., Zhang T., Li Q. Correlations of tumor-associated macrophage subtypes with liver metastases of colorectal cancer. Asian Pac. J. Cancer Prev. APJCP. 2013;14:1003–1007. doi: 10.7314/apjcp.2013.14.2.1003. [DOI] [PubMed] [Google Scholar]
- 66.Wang H., Tian T., Zhang J. Tumor-associated macrophages (TAMs) in colorectal cancer (CRC): from mechanism to therapy and prognosis. Int. J. Mol. Sci. 2021;22:8470. doi: 10.3390/ijms22168470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Al Momani S., Rodger E.J., Stockwell P.A., Eccles M.R., Chatterjee A. Generating sequencing-based DNA methylation maps from low DNA input samples. Methods Mol. Biol. 2022;2458:3–21. doi: 10.1007/978-1-0716-2140-0_1. [DOI] [PubMed] [Google Scholar]
- 68.Chatterjee A., Rodger E.J., Stockwell P.A., Weeks R.J., Morison I.M. Technical considerations for reduced representation bisulfite sequencing with multiplexed libraries. J. Biomed. Biotechnol. 2012;2012:741542. doi: 10.1155/2012/741542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Purcell R.V., Visnovska M., Biggs P.J., Schmeier S., Frizelle F.A. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci. Rep. 2017;7:11590. doi: 10.1038/s41598-017-11237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chatterjee A., Stockwell P.A., Rodger E.J., Morison I.M. Comparison of alignment software for genome-wide bisulphite sequence data. Nucleic Acids Res. 2012;40:e79. doi: 10.1093/nar/gks150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stockwell P.A., Chatterjee A., Rodger E.J., Morison I.M. DMAP: differential methylation analysis package for RRBS and WGBS data. Bioinformatics. 2014;30:1814–1822. doi: 10.1093/bioinformatics/btu126. [DOI] [PubMed] [Google Scholar]
- 72.Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fishilevich S., Nudel R., Rappaport N., Hadar R., Plaschkes I., Iny Stein T., Rosen N., Kohn A., Twik M., Safran M., et al. GeneHancer: Genome-wide Integration of Enhancers and Target Genes in GeneCards. Database. 2017 doi: 10.1093/database/bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Orouji E., Raman A.T., Singh A.K., Sorokin A., Arslan E., Ghosh A.K., Schulz J., Terranova C., Jiang S., Tang M., et al. Chromatin state dynamics confers specific therapeutic strategies in enhancer subtypes of colorectal cancer. Gut. 2022;71:938–949. doi: 10.1136/gutjnl-2020-322835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang H., Maurano M.T., Qu H., Varley K.E., Gertz J., Pauli F., Lee K., Canfield T., Weaver M., Sandstrom R., et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–1688. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smit A.F.A., Hubley R., Green P. 2013. RepeatMasker Open-4.0. [Google Scholar]
- 78.Díez-Villanueva A., Mallona I., Peinado M.A. Wanderer, an interactive viewer to explore DNA methylation and gene expression data in human cancer. Epigenet. Chromatin. 2015;8:22. doi: 10.1186/s13072-015-0014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xie Z., Bailey A., Kuleshov M.V., Clarke D.J.B., Evangelista J.E., Jenkins S.L., Lachmann A., Wojciechowicz M.L., Kropiwnicki E., Jagodnik K.M., et al. Gene set knowledge discovery with Enrichr. Curr. Protoc. 2021;1:e90. doi: 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gimenez G., Stockwell P.A., Rodger E.J., Chatterjee A. Strategy for RNA-seq experimental design and data analysis. Methods Mol. Biol. 2023;2588:249–278. doi: 10.1007/978-1-0716-2780-8_16. [DOI] [PubMed] [Google Scholar]
- 81.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li T., Fu J., Zeng Z., Cohen D., Li J., Chen Q., Li B., Liu X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., Treviño V., Shen H., Laird P.W., Levine D.A., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
RRBS and RNA-seq data have been deposited at GEO and are publicly available as of the date of publication; the series accession number is listed in the key resources table. De-identified patient data is available in this paper’s supplemental information. This paper also analyses existing, publicly available data. The accession numbers for these datasets are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.