Abstract
Exploring the fitness consequences of whole-genome multiplication (WGM) is essential for understanding the establishment of autopolyploids in diploid parental populations, but suitable model systems are rare. We examined the impact of WGM on reproductive traits in three major cytotypes (2x, 3x, 4x) of Pilosella rhodopea, a species with recurrent formation of neo-autopolyploids in mixed-ploidy populations. We found that diploids had normal female sporogenesis and gametogenesis, high fertility, and produced predominantly euploid seed progeny. By contrast, autopolyploids had highly disturbed developmental programs that resulted in significantly lower seed set and a high frequency of aneuploid progeny. All cytotypes, but particularly triploids, produced gametes of varying ploidy, including unreduced ones, that participated in frequent intercytotype mating. Noteworthy, the reduced investment in sexual reproduction in autopolyploids was compensated by increased production of axillary rosettes and the novel expression of two clonal traits: adventitious rosettes on roots (root-sprouting), and aposporous initial cells in ovules which, however, do not result in functional apomixis. The combination of increased vegetative clonal growth in autopolyploids and frequent intercytotype mating are key mechanisms involved in the formation and maintenance of the largest diploid-autopolyploid primary contact zone ever recorded in angiosperms.
Keywords: aneuploidy, apospory, clonality, mixed-ploidy, root-sprouting, unreduced gametes
1. Introduction
Polyploidization, the process of acquiring one or more additional haploid sets of chromosomes (whole-genome multiplication; hereafter 'WGM'), is a key evolutionary mechanism in angiosperms [1] (we prefer to use the term 'WGM' over the well-established 'whole-genome duplication' or WGD, as odd-ploids occur relatively frequently in nature; let our article be an example of that). WGM leads to an increase in cell size, and additional gene copies may affect gene expression [2]. This can result in changes in morphology and altered physiology, potentially providing a neo-autopolyploid organism with new phenotypic traits and adaptive functions [3–6]. Despite the widespread occurrence of polyploidy among vascular plants, we still know little about the direct phenotypic effects of WGM and its fitness consequences. This is largely because the majority of polyploids studied are allopolyploids, which are formed through hybridization between two or more species. In this case, hybridization rather than polyploidization per se leads to morphological and physiological divergence. Therefore, the direct consequences of WGM can only be studied in neo-autopolyploids, which arise within the same species [7,8]. However, because autopolyploids are morphologically and genetically like their diploid ancestors, they often go unnoticed [9].
Since autopolyploidization is always a sympatric process occurring in a primary contact zone through the fusion of reduced and unreduced gametes [10,11], the newly formed autopolyploid (also referred to as neo-autopolyploid hereafter) faces several developmental and density-dependent challenges that may reduce its fitness. Reduced fitness in neo-autopolyploids is often caused by multivalent chromosome pairing during meiosis, leading to aberrant chromosome segregations into gametes that cause aneuploidy and genetically unbalanced offspring [2,12]. In addition, minority neo-autopolyploids are subjected to hybridization with the more common parental cytotype [13,14]. Consequently, heteroploid crosses often result in seed abortion due to a shift in the ratio of maternal to paternal genomes in the endosperm (the triploid block [15]). Therefore, the emergence of any new trait that would improve the reproduction of the neo-autopolyploid or reduce the detrimental effects of interploidy crosses would enhance its successful establishment.
Alterations in reproductive strategies following WGM may counteract the disadvantages of a minority cytotype, e.g. by tolerating higher rates of autogamy [16] or by triggering/promoting asexual reproduction [17]. Asexuality can be achieved through the production of apomictically formed seeds, where meiosis and syngamy are bypassed [18] or through vegetative propagation [19–21]. Many apomicts are autopolyploids, particularly among grasses [22,23], and WGM is the primary cause of apomixis among these species, while hybridization seems to play a significant role in known allopolyploid apomicts [24,25].
It has been suggested that polyploids reproduce vegetatively more frequently than diploids [26–28]. Despite a positive association between ploidy level and intensity of vegetative propagation found in multispecies analyses [29,30], several studies using natural polyploids have provided conflicting evidence, either supporting [31,32] or refuting this hypothesis [33,34]. One reason for this discrepancy may be the fact that differences in clonal traits were assessed in established cytotypes that have undergone long-term post-polyploidization selection [35]. This may buffer or even erase the direct effects of WGM on clonal traits [4,36]. Synthetically produced neo-autopolyploids has been proposed as a solution to this problem [37,38]. While this approach seems straightforward [39], it also has disadvantages like strong directional selection towards fittest genotypes [40], chromosomal and physiological instability of synthetic neo-autopolyploids due to collateral effects of mitotic inhibitors [41,42], or production of neo-autotetraploid progeny, while natural WGM often produces autotriploids through the fusion of reduced and unreduced gametes [10]. Although natural autotriploids are highly sterile due to unbalanced meiosis, they are the first step of WGM, and as such they provide a ‘triploid bridge' in the formation of reproductively more stable autotetraploids [10,43–45].
Here, we use natural autopolyploids and explore the direct effects of WGM on reproductive traits in Pilosella rhodopea (Griseb.) Szela˛g (Asteraceae). In our previous studies, we found that: (i) the species consists of three major (2x, 3x, 4x) and two minor cytotypes (5x, 6x) [46,47]; (ii) the major cytotypes frequently occur in mixed-ploidy populations across the whole species range [47]; (iii) there is no ecological and spatial (above greater than 1 m) segregation of cytotypes [47]; (iv) the cytotypes form morphological and genetic clusters according to their population of origin [46–48]; (v) in contrast to many polyploid Pilosella species which often reproduce by autonomous apomixis [49], all P. rhodopea cytotypes produce seeds sexually after double fertilization and are self-incompatible [46]; and (vi) the three major cytotypes produce cytotypically variable seed progeny [46]. Altogether, the continuous production of ploidy-variable progeny and the co-occurrence of neo-autopolyploids with their diploid progenitors in the largest primary contact zone ever recorded in angiosperms [47] makes P. rhodopea an ideal model for studying the evolutionary and ecological consequences of WGM in natural populations.
In this study, we aim to examine the impact of WGM on reproductive traits which could determine the distributional success of P. rhodopea autopolyploids [46,47]. To do this, we analysed seed sets in three major cytotypes from mixed-ploidy populations, evaluated megasporogenesis and megagametogenesis in ovules, inferred the ploidy level of developed embryos and the pathways of their formation, and assessed the rate of vegetative growth and flowering in a greenhouse experiment. Using these data, we answer the following questions. (i) Does seed fertility differ between diploids and their autopolyploid derivates? (ii) How does WGM affect the development, viability and ploidy of female and male gametes and embryos in autopolyploids compared to their parental diploids? (iii) Does WGM alter the pattern of clonal reproduction in autopolyploids?
2. Material and methods
(a) . Study species
Pilosella rhodopea is a long-lived perennial herb (at least 5 years in cultivation) with a basal rosette forming also axillary rosettes growing from axils of rosette leaves or from axils of former rosette leaves on rhizome, i.e. bellow the actual rosette. The stem is 5–15 cm high and bears 1–3 inflorescences (capitula or flower heads) [46]. Plants produce one to several stems per rosette, and flowering takes place from July to September. This insect-pollinated species is strictly self-incompatible [46]. The achenes, i.e. dry single-seed fruits (hereafter 'seeds') with pappus, are adapted for long distance dispersal by wind. The species occurs on subalpine and alpine grasslands on acid bedrock in the Balkan Mountains [50].
(b) . Field sampling
The plant material was collected from natural populations during 2009–2011. To assess seed set, we sampled open and undamaged capitula containing ripe achenes from 396 plants occurring at several sites separated by tens of metres up to a few kilometres along four elevational, 4–8 km long transects situated in three mountain ranges in Bulgaria in 2009 and 2010 (electronic supplementary material, table S1) [47]. Of these 396 plants, 268 plants showed complete capitula, while the remaining ones had one or more incomplete capitula (in which some seeds were missing at the time of sampling). From each plant, we also sampled undamaged rosette leaves that were stored in silica gel for ploidy level determination. Seeds collected in 2010 were stored at 4°C until flow cytometry analyses and the greenhouse experiment. For embryological analysis, flower head buds were sampled from several 2x, 3x and 4x mother plants at GRA site in 2011. Details on provenance of material used for each analysis are given in electronic supplementary material, table S1.
(c) . Ploidy level determination and flow cytometric seed screening analyses
Ploidy level of plants was estimated from silica-gel dried leaf tissue using a Partec ML Cyflow cytometer (Partec GmbH., Münster, Germany) equipped with a mercury lamp. Samples were prepared in a two-step procedure using a staining buffer containing 4′,6-diamidino-2-phenylindole [51,52] and Bellis perennis (2C = 3.38 pg; [53]) as an internal standard. The DNA ploidy level of seedlings from the greenhouse experiment was determined using a Partec CyFlow SL instrument (Partec GmbH, Münster, Germany) equipped with an argon-ion laser. In this case samples were prepared using a general-purpose buffer [54] with propidium iodide (PI) as a fluorescent stain and tetraploid Centaurea stoebe s.l. (2C = 3.0 pg; P. Mráz 2010, unpublished data) as an internal standard.
We also used flow cytometric analyses of individual mature seeds (hereafter FCSS) to infer the ploidy of embryos (euploid embryos have a genome size that is an exact multiple of the haploid genome value, while aneuploid embryos have a genome size either greater or smaller than an exact multiple of the haploid genome value) and reconstruct the functional reproductive pathways of seed formation in the three major cytotypes. A total of 1500 seeds, 500 for each cytotype, were analysed (electronic supplementary material, table S1). FCSS analyses were performed with a Partec CyFlow SL instrument using the protocol and internal standard used for ploidy level assessment in seedlings (see above). Histograms with at least 1500 particles (nuclei) and CV of up to 6% were accepted. We considered embryos to be euploid when the relative DNA content was within the range expected for euploid genome (see electronic supplementary material, text S1), while when it was either lower or exceeding threshold values, the embryo was classified as aneuploid. Aneuploidy of selected offspring grown from seeds was verified by karyological approach following [55] (electronic supplementary material, figure S4).
Reproductive pathways were inferred by comparing the endosperm to embryo ploidy ratio, and only for those euploid seeds with sufficient number of endosperm nuclei. In the case of apomictic reproduction (i.e. simultaneous omission of meiosis and gamete syngamy), the above ratio is always equal to 2 because the embryo sacs (hereafter 'ESs') originating from unreduced aposporic initial cells (hereafter 'AIs') in Pilosella develop the embryo parthenogenetically from a 2n egg cell and the endosperm autonomously from a 4n central cell [56]. By contrast, in sexual reproduction between homoploid partners the endosperm-to-embryo ploidy ratio is always equal to 1.5 because the embryo and endosperm develop after double fertilization of a reduced ES by two reduced pollen spermatic cells [57]. If the endosperm-to-embryo ploidy ratio is between 1.5 and 2 this still indicates a sexual pathway but with participation of an unreduced or partially reduced gamete [46]. In addition to the differentiation between sexual and parthenogenetic reproduction, FCSS allows to infer the ploidy of the gametes that participate in the formation of the embryo when the ploidy of the maternal plant is known [57].
(d) . Embryology
Flower buds collected in situ were fixed in a mixture of 96% ethanol and glacial acetic acid (3 : 1 v/v) for 24–48 h and stored in 70% ethanol at 4°C until analyses (for further details on sample preparation see electronic supplementary material, text S1) Ovaries and anthers were dissected and mounted on glass slides, and both female and male sporogenesis and gametogenesis were analysed with a Zeiss Axiophot microscope with Nomarski DIC optics (Carl Zeiss GmbH). Different developmental stages were evaluated in 19 individuals and about 600 ovules (table 1). Images were taken using a digital image analysis system (AxioVision 4.8.1; Carl Zeiss GmbH).
Table 1.
Results of embryological analyses in diploid, autotriploid and autotetraploid cytotypes of Pilosella rhodopea. Abbreviations used: Nor, normal; Abn, abnormal; ES, embryo sac.
| mother ploidy | plants (N) | ovules (N) | male meiosis | female meiosis | functional megaspores (%) | aposporic initials (%) | mature ES (%) | Aposporic activity (N) |
|---|---|---|---|---|---|---|---|---|
| 2x | 6 | 225 | Nor | Nor | 92.86 | 0.00 | 84.75 | 0.0 |
| 3x | 8 | 188 | Abn | Abn | 44.44 | 16.67 | 52.38 | 23.81 |
| 4x | 5 | 167 | Abn | Abna | 92.6 | 3.7 | 43.75 | 0.0 |
aDespite the high proportion of functional megaspores, meiosis in tetraploid individuals was irregular compared to diploids (see details in the main text).
(e) . Potential and realized seed set in plants from natural populations
The potential seed set per capitulum and plant was determined as a total number of ovules (i.e. the sum of full and empty seeds collected from a plant) divided by number of sampled capitula per plant. The realized seed set per plant was determined as a proportion of full (well-developed, electronic supplementary material, figure S1) seeds from all seeds sampled per plant.
(f) . Reproductive traits in plants from the greenhouse experiment and field survey
We used seedlings grown from seeds collected in 2010 in two mixed-ploidy transects (electronic supplementary material, table S1). Germination was carried out during November and December 2011 in Petri dishes placed in a growth chamber (Sanyo, Japan) with 12/12 hour light/dark photoperiod and 20°C/10°C temperature regime. Germinating seedlings were removed regularly and transplanted into 2 × 2 × 4 cm bedding cells with a mixture of nutrient-poor TKS1 soil (Floragard, Oldenburg, Germany) and fine-grained granite sand (in proportions 3 : 1). After transplantation, seedlings were cultivated in the greenhouse for subsequent ploidy level determination. Euploid seedlings with known ploidy level belonging to each of the three cytotypes were subsequently re-potted into 270 ml pots of 9 cm diameter filled with mixture of granite sand, sterilized compost and peat bog substrate in proportions 1 : 1 : 1. The pots were placed on two tables (representing two blocks), and their positions within each table were changed five times during the experiment. The experiment was performed in a greenhouse at the University of Fribourg under controlled light (16 h day/8 h night; EYE Clean ArcTM 400 W lamps) and temperature (22°C/15°C, day/night) from 9 February 2012 to 6 June 2012. In total, we used 63 plants, i.e. 3 cytotypes × 21 replicates (electronic supplementary material, table S2). During the experiment, we recorded the formation and number of flowering stems and capitula (electronic supplementary material, table S2). At the end of the experiment, we counted the number of axillary rosettes, and adventitious rosettes originated from the adventitious root buds (root-sprouting; electronic supplementary material, table S2).
In order to verify results related to root-sprouting from the greenhouse experiment (see Results), in August 2021 we visited the mixed-ploidy GRA population. Based on previous knowledge on microspatial distribution of cytotypes [47], we randomly selected 10 plants from permanent 5 × 5 m plots and sampled leaf material for ploidy level verification using flow cytometry. Subsequently, we carefully removed a soil layer around each selected plant to assess the presence/absence of root buds or adventitious rosettes formed from these buds (electronic supplementary material, figure S2).
(g) . Data analyses
All statistical analyses and plotting were performed within the R environment, version 3.6.2 [58]. The putative effect of ploidy on potential seed sets in plants with complete capitula was assessed using a linear mixed-effect model (LMM) with the lme function implemented in the nlme package, version 3.1–142 [59]. The putative effect of ploidy on realized seed sets was assessed using a generalized linear mixed-effect model (GLMM) using the glmer function with binomial distribution implemented in the lme4 package, version 1.1–23 [60]. The models included potential and realized seed sets as a response variable, and ploidy as an explanatory variable. Sampling site, nested in one of the four transects, was treated as a random factor. We used GLMMs with binomial distribution to test the effect of ploidy on flowering probability and root-sprouting as explanatory variables, and GLMM with quasi-Poisson distribution to test the effect of ploidy on number of root sprouts.
The anova function with likelihood ratio tests (LRT), comparing fully fitted LMMs and GLMMs to the models from which the tested term (i.e. ploidy) was removed, were used to test the statistical significance of ploidy. Between-cytotype differences in tested variables were subsequently assessed by post-hoc Tukey test using the glht function from the multcomp package [61] suitable for mixed models, and Wilcoxon and Fischer tests. We used χ2-test to assess the effect of ploidy on production of unreduced euploid gametes. The proportion of explained variance of a fixed factor (ploidy) for LMM was computed using the partR2 function [62].
3. Results
(a) . Whole-genome multiplication distorts megaspore and embryo sac formation and stimulates the expression of aposporic initial cells
Diploids showed normal development of male and female gametophytes. After male meiosis, four meiocytes (microspores) were individualized and developed into pollen grains. After female meiosis, only one meiocyte (megaspore) remained functional (figure 1a) and developed into an ES containing the female gametes (an egg cell and a central cell) showing regular polarity (table 1 and figure 1b). The proportion of functional megaspores and mature ESs was high in diploids (table 1). Antipodal cells in mature ES had an ephemeral life (they showed early signs of abortion or were not seen). No signs of aposporic initial cells were observed.
Figure 1.
Megasporogenesis and megagametogenesis in (a,b) diploid, (c,d) autotriploid and (e–g) autotetraploid Pilosella rhodopea cytotypes. An interpretative drawing is presented for each stage. (a) Functional and aborted megaspores at the end of female meiosis. (b) Mature embryo sac carrying two synergid cells (sc, at the bottom), an egg cell (ec), a central cell (cc) with a secondary nucleus (and two nucleoli) and two visible antipodal cells (ac; at the top). (c) Chromosomes aligned in the metaphase plate during meiosis I (black arrow) and a vacuolized, metabolically active initial of apospory cell (white arrow). (d) Aborted embryo sac during megagametogenesis. (e) Functional and aborted megaspores at the end of female meiosis (two nucleoli are visible in the diploid nucleus). (f) Mature embryo sac carrying two synergid cells (sc; at the bottom), an egg cell (ec), a central cell (cc) with one of the two polar nucleus and two visible antipodal cells (ac; at the top). (g) Aborting embryo sac during megagametogenesis (the two nuclei of the central cell are still visible). For each cytotype the scale bar = 10 µm.
Triploids exhibited significant meiotic irregularities in male and female tissues, and AIs were observed in ovules by the end of meiosis (figure 1c). However, a high proportion of ovules had no functional megaspores (table 1), and only a low proportion of ESs developed further and reached maturity (table 1 and figure 1d). Two to three antipodal cells were regularly observed in these mature ESs. Triploids showed a relatively high proportion of aposporous development compared to 4x, but all megagametogenesis was altered showing asynchronous nuclear divisions, irregular polarity and variable ES sizes (figure 1c,d).
Tetraploids showed slightly more regular meiosis than triploids in both male and female tissues, but not as regular as in diploids. A high proportion of apparently functional megaspores was observed in ovules (table 1 and figure 1e). However, megagametogenesis was unstable in ovules of these individuals. In addition to ESs showing signs of abortion or aborting (figure 1g), AIs appeared at a low proportion during this stage (table 1). These AI cells did not develop further or, if they developed into ESs, it was not possible to distinguish them from the regular reduced mature ESs (figure 1f). In one ovule, we observed a pro-embryo in a mature ES. As in triploids, functional ESs of tetraploids had antipodals cells.
(b) . Whole-genome multiplication strongly reduces seed set in autopolyploids
In the field survey, three cytotypes differed in their potential seed sets (LMM: LRT = 73.3, p < 0.001, ploidy explained 17% of total variance) with diploids producing more ovules per capitulum than both triploids and tetraploids (mean ± s.d.: 2x − 57 ± 16, 3x − 41 ± 14, 4x − 49 ± 15; figure 2a). Importantly, diploids produced a considerably higher percentage of well-developed seeds per capitulum compared to both autopolyploids (GLMM, χ2 = 1288.1, p < 0.001, and post-hoc Tukey test based on this model; mean ± s.d.; 2x: 48.8 ± 26.7%, 3x: 7.6 ± 9.5%, 4x: 26.7 ± 26.3%) (figure 2b). Furthermore, almost 42% of triploids were completely seed sterile, while only 8.3% and 18.1% of diploids and tetraploids, respectively, produced capitula without well-developed seed (χ2 = 36.6, d.f. = 2, p < 0.001).
Figure 2.
(a) Potential seed set (i.e. total number of ovules per capitulum) and (b) realized seed set (i.e. percentage of well-developed seeds per capitulum) in diploid (2x), autotriploid (3x) and autotetraploid (4x) plants of Pilosella rhodopea assessed in primary contact zones. Statistically significant differences (p = 0.05) between cytotypes indicated by different lower case letters were assessed using Tukey post-hoc tests applied on LMM and GLM model, respectively. Number of analysed plants per cytotype: 2x − 122, 3x − 110, 4x − 37.
(c) . Whole-genome multiplication enhances the production of aneuploid and unreduced gametes, and high cytotypic variation in seed progenies
Out of 1500 seeds analysed using FCSS, the ploidy level of the embryo was successfully inferred in 1420 seeds (table 2). Euploid embryos were detected in 762 seeds (54%). The majority of euploid embryos were diploid (63%), while triploid and tetraploid embryos were less common (20% and 11%, respectively), and high-ploidy cytotypes (5x, 6x, 7x, 8x) were rare (table 2). All three major cytotypes also produced seeds with aneuploid embryos, with frequencies strongly differing among cytotypes. While 2x plants produced 11% of aneuploid seeds, this proportion exceeded 60% in both 3x and 4x plants (χ2 = 373.3, p < 0.001; table 2; electronic supplementary material, figures S3 and S4).
Table 2.
Cytotype structure, ploidy of euploid gametes, and reproduction pathways of seed progenies produced by diploid, autotriploid and autotetraploid plants of Pilosella rhodopea co-occurring in mixed-ploidy populations.
| ploidy mother plants | ploidy embryo (euploid) | euploid embryo [N] | pathway | endo [N] | ploidy embryo (aneuploid) | aneuploid embryo [N] |
|---|---|---|---|---|---|---|
| 2x | 2x | 420 | x+x | 380 | <2x | 18 |
| 3x | 14 | x + 2x | 10 | 2x − 3x | 24 | |
| 2x + x | 1 | 3x − 4x | 13 | |||
| total 2x | 434 [89%] | 55 [11%] | ||||
| 3x | 2x | 59 | x + x | 45 | <2x | 8 |
| 3x | 28 | x + 2x | 6 | 2x − 3x | 51 | |
| 2x + x | 6 | |||||
| 4x | 43 | x + 3x | 4 | 3x − 4x | 54 | |
| 3x + x | 5 | |||||
| 2x + 2x | 5 | |||||
| 5x | 12 | 3x + 2x | 3 | 4x − 5x | 124 | |
| 6x | 23 | 3x + 3x | 5 | 5x − 6x | 79 | |
| >6x | 8 | |||||
| total 3x | 165 [34%] | 324 [66%] | ||||
| 4x | 3x | 107 | 2x + x | 33 | 2x − 3x | 51 |
| 4x | 41 | 2x + 2x | 17 | 3x − 4x | 44 | |
| 5x | 8 | 2x + 3x | 4 | 4x − 5x | 173 | |
| 6x | 3 | 4x + 2x | 3 | 5x − 6x | 8 | |
| 7x | 3 | – | 6x − 7x | 1 | ||
| 8x | 1 | 4x + 4x | 1 | 7x − 8x | 2 | |
| total 4x | 163 [38%] | 280 [62%] | ||||
| total | 762 | 528 | 658 |
Euploid embryo [N], number of euploid embryos (seeds) in particular ploidy group within each cytotype; Pathway, egg ploidy + pollen ploidy; unreduced egg cells, i.e. female gametes, are in italics; Endo [N], number of seeds showing also peaks corresponding to endospermatic tissue necessary for accurate evaluation of reproduction pathway; Aneuploid embryo [N], number of aneuploid embryos (seeds) in particular ploidy group within each cytotype.
Among 762 seeds with euploid embryos, a total of 234 did not produce a visible (measurable) endosperm peak. Therefore, the reproductive pathway could be reliably identified in 528 seeds (table 2). In all cases, the endosperm to embryo ploidy ratio varied between 1.5 and 1.8, indicating that all evaluated seeds were formed through the sexual pathway.
Concerning the formation of euploid cytotypes in seed progeny (table 2; electronic supplementary material, figure S5), diploids produced mostly diploid progenies through the fusion of a haploid egg and a haploid pollen, as well as a few triploid progenies through the fusion of a haploid egg cell with a diploid male gamete, and a diploid unreduced egg cell and a haploid male gamete. Triploids produced five euploid cytotypes ranging from 2x to 6x, with the participation of haploid, diploid but also frequent unreduced triploid egg cells. Tetraploids produced six euploid cytotypes ranging from 3x to 8x, with the participation of diploid and tetraploid egg cells. The proportion of unreduced euploid female gametes in the evaluated seed progenies significantly varied among ploidies: 0.24% in 2x (one unreduced egg cell out of 391 euploid ones), 16.46% in 3x (13 out of 79), and 6.9% in 4x (4 out of 58) (χ2 = 54.79, p < 0.001).
As for the ploidy of pollen involved in formation of euploid seeds in natural mixed-cytotype populations, we recorded haploid, diploid, triploid and tetraploid pollen grains (table 2).
(d) . Whole-genome multiplication stimulates vegetative clonal growth quantitatively and qualitatively
Initially, we intended to assess the production of axillary rosettes arising from axillary meristems (figure 3a,d). However, after approximately three months of cultivation, we observed that certain plants formed another type of small rosettes growing up from the soil, distant from the main rosette to which they were not visibly connected (figure 3b). After a detail inspection of the root systems, we found that these rosettes originated from adventitious buds on roots (root-sprouting; figure 3c,e) that were formed in autopolyploids, while they were completely missing in diploids (GLMM, χ2 = 42.2, p < 0.001; figure 4a). Probability of root-sprouting and the quantity of adventitious buds and derived adventitious rosettes per individual plant were higher in autotetraploids than in autotriploids (86% in 4x and 62% in 3x; figure 4a; 31 ± 24 in 4x and 25 ± 23 in 3x, respectively) but these trends were statistically not significant (Fischer test, p = 0.159; Wilcoxon test, p = 0.395, respectively).
Figure 3.
Vegetative clonal organs in diploid (2x) and autotriploid (3x) plants of Pilosella rhodopea. (a) Axillary rosettes formed from rosette leaf axils. (b) Adventitious root rosettes around main rosette. (c) Root system of diploid (2x) and autotriploid (3x) plants. (d) Axillary rosettes at the base of main rosette. (e) Adventitious root bud of cultivated autotriploid plant.
Figure 4.
Vegetative clonal traits and probability of flowering in diploid (2x), autotriploid (3x) and autotetraploid (4x) plants of Pilosella rhodopea assessed in the greenhouse experiment. (a) Probability of adventitious root buds formation (root-sprouting). (b) Variation in number of axillary rosettes. (c) Probability of flowering. Number of analysed plants (N) was 21 per each of the cytotypes. Bars in (a) and (c) represent 95% confidence intervals of means derived from GLM models. Statistically significant differences (p = 0.05) between cytotypes indicated by different lower case letters were assessed using Tukey post-hoc tests applied on GLM models.
Interestingly, we recorded the formation of adventitious rosettes in one 3x offspring (D10-3C) derived from the 3x mother plant (S10-D3), and in one 4x offspring (D7-5C) derived from another 3x mother (R8-A1), while six 2x offspring produced by the same mothers (with three progenies per mother, respectively) produced no adventitious root buds or rosettes. Two neo-autotriploids derived from two different 2x mother plants were also assessed for root-sprouting. While one 3x offspring (D2_2D) from 2x mother (R6-A1) produced numerous adventitious rosettes, the other 3x offspring (D3_9C) from 2x mother (S10_C3) did not produce these structures. A field survey at the GRA mixed-ploidy population revealed that all ten evaluated 2x plants lacked adventitious root buds or rosettes, while seven out of ten inspected 3x plants (70%), and all ten tetraploids (100%) produced at least one adventitious root rosette or bud (electronic supplementary material, figure S2).
Regarding axillary rosettes, we recorded a positive but non-significant trend between the number of axillary rosettes per plant and the ploidy level (GLMM, χ2 = 5.71, p = 0.057; mean ± s.d.; 2x: 1.33 ± 1.96, 3x: 1.52 ± 1.72, 4x: 2.24 ± 1.37; figure 4b), with tetraploids producing significantly more axillary rosettes than diploids (Tukey post-hoc based on the GLMM model: z = 2.391, p = 0.05; figure 4b).
The probability of flowering within six months after germination significantly differed among cytotypes (GLMM, χ2 = 18.4, d.f. = 2, p < 0.001), with almost one half of diploids showing an induction of flowering (9 plants; 43%), while only one triploid (5%) and no tetraploid showing flowering shoots during that period (figure 4c).
4. Discussion
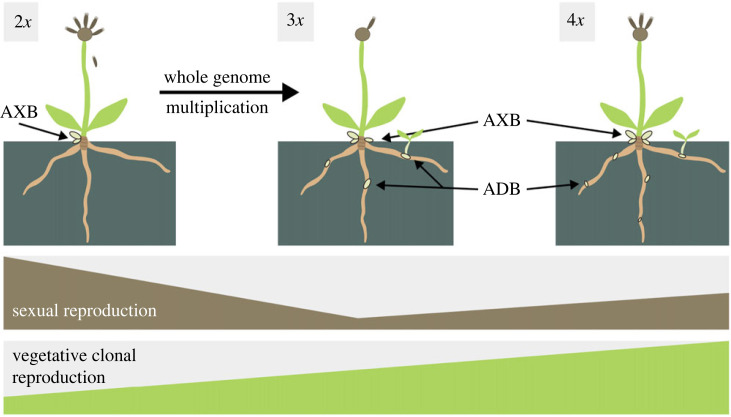
We studied the effect of WGM on reproductive traits in Pilosella rhodopea—a species in which coexisting diploids and autopolyploids share the same genetic background and they are exposed to the same environment. The results of our study show that, compared to diploids, WGM disturbs female sporogenesis and gametogenesis with considerably lowered seed set and a high proportion of aneuploid seeds. We further show that free intercytotype mating that includes reduced, unreduced and aneuploid gametes results in cytotypically variable progenies. Reduced fertility in autopolyploids is, however, compensated by enhanced vegetative propagation through axillary rosettes and the novel expression of two asexual traits never observed in diploids, i.e. aposporous initial cells in ovules and adventitious rosettes on roots (figure 5). While the former trait does not result in functional apomixis, the latter one is translated into substantially increased clonal growth of autopolyploids.
Figure 5.
Graphical summary of qualitative and quantitative effects of WGM on sexual and vegetative reproduction in natural Pilosella rhodopea autopolyploids in the largest primary contact zone among polyploid lineages ever recorded in angiosperms. This summary integrates the results on realized seed set assessed in natural populations (figure 2b) and vegetative propagation assessed in the greenhouse experiment and one natural population (figure 4a,b; electronic supplementary material, figure S2). AXB—axillary buds which are formed on rhizomes (shown) and in the axils of rosette leaves (not shown), and from which axillary rosettes are formed. ADB—adventitious buds on roots from which adventitious rosettes (root-sprouting) are formed. Note that the expression of aposporous initial cells in ovules of autopolyploids (figure 1)—a novel trait, when compared to ancestral diploids, but non-functional—is not depicted in this figure.
(a) . Whole-genome multiplication and sexual reproduction
Natural autopolyploids of P. rhodopea showed substantially lower seed set when compared to coexisting diploids. Our results suggest that there are at least two possible factors contributing to this pattern. First, the predominant multivalent pairing of homologous chromosomes during meiosis (instead of bivalent pairing in diploids or allopolyploids), which leads to abnormal chromosome segregation and the production of aneuploid gametes and zygotes with reduced viability [2,3,11,62]. Using FCSS, we found that P. rhodopea autopolyploids produced aneuploid seed progeny approximately six times more often than diploids. This pattern suggests that unbalanced meiosis is likely responsible for the observed developmental abnormalities and abortion of ESs in autopolyploids. Although we observed relatively high frequency of aneuploid seeds also in diploids (ca 11%), this pattern can be easily explained by the fertilization of normal euhaploid ES in diploids by aneuploid pollen from co-occurring autopolyploids, since diploids showed normal female meiosis with a high proportion of mature ESs. Second, in addition to multivalent paring, the odd chromosome number of autotriploids causes also unbalanced meiosis, further exacerbating the detrimental effect on seed production in this cytotype. Such abnormal meioses are likely the main cause for the 40–50% reduction in ES formation observed in autopolyploids compared to diploids. In addition, crosses involving gametes of different ploidy levels may negatively affect seed fertility through the triploid block [15]. This process may explain the absence or rarity of interploid hybrids in various diploid-polyploid contact zones [63–65], or in controlled heteroploid crosses [63–68]. In contrast to the above examples, the triploid block seems to be relatively weak in Pilosella rhodopea since we recorded frequent formation of functional seeds from heteroploid crosses. Similarly, the absence of a strong triploid-block effect was found in artificial crosses between synthetic 4x Arabidopsis lyrata and 2x A. arenosa, but not in the reciprocal crosses suggesting species-specific sensitivity to endosperm-based hybridization barrier [69].
In the greenhouse experiment, we also recorded significantly reduced rates of flowering in autopolyploids when compared to diploids. Because all cytotypes in natural populations produce flowering heads and show overlapping flowering, our results suggest a delayed flowering in young autopolyploid plants. Cultivation of seedlings for a longer period would be needed to test this hypothesis. In any case, the observed delayed induction of flowering is likely a direct effect of WGM, which may be caused by slower cell division due to a larger cell size [26], or by an antagonistic interaction of phytohormones involved in regulation of sexual and vegetative reproduction [70]. This interaction could temporarily inhibit the allocation of resources into flowering stems in P. rhodopea autopolyploids.
(b) . Whole-genome multiplication and asexual seed reproduction (apomixis)
Among natural apomicts (approx. 1500 species with documented apomixis [71]), the occurrence of both diploid sexual and polyploid apomictic cytotypes is frequently observed. In such agamic complexes, to which Pilosella genus belong [49], diploid sexuals may (or may not) show potential for apomixis through the rare formation of AIs or unreduced ESs, while polyploids exhibit high proportions of AIs and unreduced ESs. However, in all cases (except for some Boechera species [72]), apomixis is only functional in polyploids where the unreduced egg cell develops parthenogenetically and produces a seed with a clonal embryo [71]. Our embryological observations agree with this pattern of diploid sexuals, but revealed a new feature in P. rhodopea autopolyploids. While enlarged AIs originating from somatic tissues were not observed in diploids (as expected), those frequently observed in ovules of 3x and 4x cytotypes were unexpectedly non-functional, as revealed from our flow cytometric analysis. Unlike other apomictic systems where the formation of AIs and unreduced ESs are functional and produce apomictic seeds in polyploids [but see 73], in P. rhodopea autopolyploids the apomictic pathway is not functionally established. Consequently, relatively few developed seeds found in autopolyploids were of sexual origin. These results support the view that apomixis in natural populations can be transiently activated during polyploidization and established (or not) in the new polyploids depending upon a combination of genetically controlled reproductive components that are individually non-functional [74]. In P. rhodopea, WGM induced the formation of AIs as an evolutionary novelty in autopolyploids, but the other components of apomixis, particularly the parthenogenetic competence needed for fertilization-free development of the egg cell into an embryo, are missing (in Pilosella apomicts, endosperm development is independent from fertilization [75–77]). This pattern corroborates the independence of apomeiotic and parthenogenetic components of apomixis in the genus [78], agrees with the theorized dynamics of the establishment of apomixis in natural populations [74,79], and represents (together with P. brzovecensis [73]) a rare case of species within an apomictic genus that fails either to functionally establish polyploid apomixis or to restore sexuality.
(c) . Whole-genome multiplication and vegetative reproduction
Our greenhouse experiment revealed that autopolyploids invest significantly more to vegetative reproduction than diploids. On average, autotriploids produced ca 14% and autotetraploids ca 68% more axillary rosettes per plant than diploids. Autopolyploids also produced adventitious rosettes from root buds, a trait that has never been observed in diploids. While many Pilosella species can propagate vegetatively through axillary rosettes, or underground or aboveground stolons [80], root-sprouting seems to be rare in the genus [81]. The fact that all natural P. rhodopea diploids, including those produced by autotriploids, lack adventitious root buds unlike autopolyploids strongly suggests that this trait is a direct consequence of WGM. Theoretically, it could also be a trait introgression from other Pilosella species, but this scenario is highly unlikely because root-sprouting has only been observed in polyploid P. piloselloides [81], and P. rhodopea does not co-occurs with this species. Furthermore, our published [47,48] and more recent studies (B. Šingliarová, J. Lihová & P. Mráz 2016, unpublished results) using a wide range of multilocus molecular markers rule out any sign of interspecific hybridization, with parental diploids being genetically identical with autopolyploids.
In addition to this qualitative difference, we also observed quantitative differences in the extent of vegetative production of both axillary and adventitious rosettes, which were positively associated with increasing ploidy level. These multiple lines of evidence suggest that the quantitative and qualitative shifts in vegetative growth observed in P. rhodopea autopolyploids are triggered by WGM. While there are several examples of quantitative shifts in clonality among diploids and autopolyploids (e.g. [36,38]; reviewed in [82]), to our knowledge, there is no other example of a similar qualitative novelty in autopolyploids, such as the emergence of adventitious root sprouts. We hypothesize that the altered patterns in vegetative growth are likely caused by the deregulation of genes involved in metabolic pathways of phytohormones responsible for plant organogenesis and development. Our data show that the novel expression of root-sprouting can happen in one generation only, but the extent of its manifestation likely depends on the level of gene-deregulation. Root-sprouting is principally regulated by cytokinins and their interaction with auxin [83–86]. Therefore, the deregulation of such metabolic pathway as a direct effect of WGM is probably playing a central role in enhancing vegetative propagation in P. rhodopea autopolyploids. In agreement with this view, our pilot analyses assessing concentration of phytohormones and the expression of genes in roots revealed striking differences among diploids and autopolyploids (P. Mráz, Y. Betrand, I. Dobrev, A. Soukup & B. Šingliarová 2021, unpublished results).
(d) . Origin, establishment and coexistence of autopolyploids in a primary contact zone
Neo-autotriploids can be formed on diploids through the fusion of one reduced and one unreduced gamete. Although the rate of unreduced female gamete formation in diploids was low, the cumulative importance of neo-autopolyploidization could be substantially higher when accounting also for unreduced male gametes, and relatively large population sizes of P. rhodopea.
Once formed, autotriploids will benefit from reproductive assurance through WGM-driven root-sprouting and enhanced production of axillary rosettes. The vegetative propagation of autotriploids, in turn, will have a cumulatively positive effect on sexual reproduction through enhanced production of flowering stems and seeds. Over time, axillary rosettes, but especially rosettes from adventitious root buds, could detach from maternal plant and form independent individual ramets. Because there is a variation in root-sprouting in autopolyploids, we suppose that the phenotype with adventitious formation of rosettes on roots, thus assuring higher survival, will be favoured. This could be a reason of higher incidence of root-sprouting in the field when compared to the greenhouse experiment (3x: 70% in the field versus 62% in the greenhouse; 4x: 100% versus 86%, respectively). As autotriploids produce gametes of different ploidies, these can participate in within- and among-cytotype gene flow resulting in a variety of new cytotypes, mostly autotetraploids as observed in our study. Since autotetraploids are also ‘armed' with an enhanced capacity for vegetative propagation, and as their sexual reproduction via seeds is more stable, they further contribute to the span of the cytotype diversity observed in the field. We assume that autopolyploidization through unreduced gametes and subsequent reproductive stabilization through vegetative propagation occurs in every diploid population of P. rhodopea, and that this interplay between sexual and vegetative reproduction in autopolyploids allows cytotype coexistence and explains the present-day cytogeographical pattern [46–48].
5. Conclusion
Our study provides empirical evidence for a strong, WGM-induced trade-off between sexual and vegetative reproduction in Pilosella rhodopea. The direct effect of WGM on phenotype and fitness of natural autopolyploids is still poorly explored owing to the scarcity of suitable model systems. Given the recurrent formation of autopolyploids holding the same genetic background and environmental context, P. rhodopea is an ideal species to study the impact of WGM on plant phenotype, fitness and cytotype coexistence in the wild. Although WGM strongly reduces fertility in autopolyploids, this negative effect is compensated by enhanced vegetative growth through increased production of axillary rosettes and production of adventitious rosettes from roots—a trait never observed in parental diploids. Although quantitative differences in vegetative reproduction among cytotypes have already been reported [32,33,36,38], our study is the first one showing the novel expression of a vegetative clonal trait in autopolyploids. This qualitative and quantitative shift to enhanced clonality literally changes ‘the game' by giving autopolyploids the chance for their establishment and maintenance with putatively strong impact on their spatial clonal structure and ramets's age. Altogether, our study shows how WGM may immediately and substantially alter the phenotype of a plant and provides a perspective for further in-depth exploration of underlying (epi)genetic and physiological mechanisms of this fascinating phenotypic change.
Acknowledgements
We thank Róbert Lakoštík for valuable help in the field, Laura Castellani for help with FCSS analyses, Barbora Mrázová and Samuel Mráz for constructing a graphical summary, two anonymous referees and Prof. Gary Carvalho, associate editor, for very constructive and valuable suggestions.
Data accessibility
The data are provided in electronic supplementary material [87].
Authors' contributions
B.Š.: conceptualization, data curation, formal analysis, funding acquisition, investigation, visualization, writing—review and editing; D.H.: data curation, formal analysis, investigation, visualization, writing—review and editing; H.M.-S.: conceptualization, writing—review and editing; P.M.: conceptualization, data curation, formal analysis, investigation, supervision, visualization, writing—original draft.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
Study was supported by the Slovak Academy of Sciences (grant no. Mob-Open-20-05). Greenhouse experiment was carried out in the frame of a post-doctoral fellowship of BS at University of Fribourg (Exchange Programme Sciex-NMSch, project ASIPOL—Adaptive Significance of Polyploidy).
References
- 1.Ren R, Wang H, Guo C, Zhang N, Zeng L, Chen Y, Ma H, Qi J. 2018. Widespread whole genome duplications contribute to genome complexity and species diversity in angiosperms. Molec. Plant 11, 414-428. ( 10.1016/j.molp.2018.01.002) [DOI] [PubMed] [Google Scholar]
- 2.Comai L. 2005. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836-846. ( 10.1038/nrg1711) [DOI] [PubMed] [Google Scholar]
- 3.Parisod C, Holderegger R, Brochmann C. 2010. Evolutionary consequences of autopolyploidy. New Phytol. 186, 5-17. ( 10.1111/j.1469-8137.2009.03142.x) [DOI] [PubMed] [Google Scholar]
- 4.Ramsey J. 2011. Polyploidy and ecological adaptation in wild yarrow. Proc. Natl Acad. Sci. USA 108, 7096-7101. ( 10.1073/pnas.1016631108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker MS, Husband BC, Pires JC. 2016. Spreading winge and flying high: the evolutionary importance of polyploidy after a century of study. Amer. J. Bot. 103, 1139-1145. ( 10.3732/ajb.1600272) [DOI] [PubMed] [Google Scholar]
- 6.Van de Peer Y, Mizrachi E, Marchal K. 2017. The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411-424. ( 10.1038/nrg.2017.26) [DOI] [PubMed] [Google Scholar]
- 7.Ramsey J, Ramsey TS. 2014. Ecological studies of polyploidy in the 100 years following its discovery. Proc. R. Soc. B 369, 20130352. ( 10.1098/rstb.2013.0352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spoelhof JP, Soltis PS, Soltis DE. 2017. Pure polyploidy: closing the gaps in autopolyploid research. J. Syst. Evol. 55, 340-352. ( 10.1111/jse.12253) [DOI] [Google Scholar]
- 9.Soltis DE, Soltis PS, Schemske DW, Hancock JF, Thompson JN, Husband BC, Judd WS. 2007. Autopolyploidy in angiosperms: have we grossly underestimated the number of species? Taxon 56, 13-30. ( 10.2307/25065732) [DOI] [Google Scholar]
- 10.Ramsey J, Schemske DW. 1998. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Evol. Syst. 29, 467-501. ( 10.1146/annurev.ecolsys.29.1.467) [DOI] [Google Scholar]
- 11.Ramsey J, Schemske DW. 2002. Neopolyploidy in flowering plants. Annu. Rev. Ecol. Evol. Syst. 33, 589-639. ( 10.1146/annurev.ecolsys.33.010802.150437) [DOI] [Google Scholar]
- 12.Cifuentes M, Grandont L, Moore G, Chèvre AM, Jenczewski E. 2010. Genetic regulation of meiosis in polyploid species: new insights into an old question. New Phytol. 186, 29-36. ( 10.1111/j.1469-8137.2009.03084.x) [DOI] [PubMed] [Google Scholar]
- 13.Levin D. 1975. Minority cytotype exclusion in local plant populations. Taxon 24, 35-43. ( 10.2307/1218997) [DOI] [Google Scholar]
- 14.Husband BC. 2000. Constraints on polyploid evolution: a test of the minority cytotype exclusion principle. Proc. R. Soc. Lond. B 267, 217-223. ( 10.1098/rspb.2000.0990) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Köhler C, Mittelsten Scheid O, Erilova A. 2010. The impact of the triploid block on the origin and evolution of polyploid plants. Trends Genet. 26, 142-148. ( 10.1016/j.tig.2009.12.006) [DOI] [PubMed] [Google Scholar]
- 16.Eliášová A, Trávníček P, Mandák B, Münzbergová Z. 2014. Autotetraploids of Vicia cracca show a higher allelic richness in natural populations and a higher seed set after artificial selfing than diploids. Ann. Bot. 113, 159-170. ( 10.1093/aob/mct252) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Drunen WE, Friedman J. 2022. Autopolyploid establishment depends on life history strategy and the mating outcomes of clonal architecture. Evolution 76, 1953-1976. ( 10.1111/evo.14582) [DOI] [PubMed] [Google Scholar]
- 18.Koltunow M. 1993. Apomixis: embryo sacs and embryos formed without meiosis or fertilization in ovules. Plant Cell 5, 1425-1437. ( 10.1105/tpc.5.10.1425) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klimeš L, Klimešová J, Hendriks R, van Groenendael J. 1997. Clonal plant architectures: a comparative analysis of form and function. In The ecology and evolution of clonal plants (eds de Kroon H., van Groenendael J), pp. 1-29. Leiden, The Netherlands: Backhuys Publishers. [Google Scholar]
- 20.Vallejo-Marín M, Dorken ME, Barrett SCH. 2010. The ecological and evolutionary consequences of clonality for plant mating. Annu. Rev. Ecol. Evol. Syst. 41, 193-213. ( 10.1146/annurev.ecolsys.110308.120258) [DOI] [Google Scholar]
- 21.Chrtek J, Herben T, Rosenbaumová R, Münzbergová Z, Dočkalová Z, Zahradníček J, Krejčíková J, Trávníček P. 2017. Cytotype coexistence in the field cannot be explained by inter-cytotype hybridization alone: linking experiments and computer simulations in the sexual species Pilosella echioides (Asteraceae). BMC Evol. Biol. 17, 87. ( 10.1186/s12862-017-0934-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortiz JP, et al. 2013. Harnessing apomictic reproduction in grasses: what we have learned from Paspalum. Ann. Bot. 112, 767-787. ( 10.1093/aob/mct152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karunarathne P, Hojsgaard D. 2021. Single independent autopolyploidization events from distinct diploid gene pools and residual sexuality support range expansion of locally adapted tetraploid genotypes in a South American grass. Front. Genet. 12, 736088. ( 10.3389/fgene.2021.736088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beck BB, Windham MD, Pryer KM. 2011. Do asexual polyploid lineages lead short evolutionary lives? A case study from the fern genus Astrolepis. Evolution 65, 3217-3229. ( 10.1111/j.1558-5646.2011.01362.x) [DOI] [PubMed] [Google Scholar]
- 25.Chrtek J, et al. 2020. Evolutionary history and genetic diversity of apomictic allopolyploids in Hieracium s.str. morphological versus genomic features. Amer. J. Bot. 107, 66-90. ( 10.1002/ajb2.1413) [DOI] [PubMed] [Google Scholar]
- 26.Müntzing A. 1936. The evolutionary significance of autopolyploidy. Hereditas 21, 363-378. ( 10.1111/j.1601-5223.1936.tb03204.x) [DOI] [Google Scholar]
- 27.Gustafsson A. 1948. Polyploidy, life-form and vegetative reproduction. Hereditas 34, 1-22. ( 10.1111/j.1601-5223.1948.tb02824.x) [DOI] [Google Scholar]
- 28.Obeso JR. 2002. The costs of reproduction in plants. New Phytol. 155, 321-348. ( 10.1046/j.1469-8137.2002.00477.x) [DOI] [PubMed] [Google Scholar]
- 29.Herben T, Suda J, Klimešová J. 2017. Polyploid species rely on vegetative reproduction more than diploids: a re-examination of the old hypothesis. Ann. Bot. 120, 341-349. ( 10.1093/aob/mcx009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Drunen WE, Husband BC. 2019. Evolutionary associations between polyploidy, clonal reproduction, and perenniality in the angiosperms. New Phytol. 224, 1266-1277. ( 10.1111/nph.15999) [DOI] [PubMed] [Google Scholar]
- 31.Eckert CG, Lui K, Bronson K, Corradini P, Bruneau A. 2003. Population genetic consequences of extreme variation in sexual and clonal reproduction in an aquatic plant. Molec. Ecol. 12, 331-344. ( 10.1046/j.1365-294X.2003.01737.x) [DOI] [PubMed] [Google Scholar]
- 32.Fialová M, Jandová M, Ohryzek J, Duchoslav M. 2014. Biology of the polyploid geophyte Allium oleraceum (Amaryllidaceae): variation in size, sexual and asexual reproduction and germination within and between tetra-, penta- and hexaploid cytotypes. Flora 209, 312-324. ( 10.1016/j.flora.2014.04.001) [DOI] [Google Scholar]
- 33.Baldwin SJ, Husband BC. 2013. The association between polyploidy and clonal reproduction in diploid and tetraploid Chamerion angustifolium. Molec. Ecol. 22, 1806-1819. ( 10.1111/mec.12217) [DOI] [PubMed] [Google Scholar]
- 34.Martínková J, Klimešová J, Doležal J, Kolář F. 2015. Root sprouting in Knautia arvensis (Dipsacaceae): effects of polyploidy, soil origin and nutrient availability. Plant Ecol. 216, 901-911. ( 10.1007/s11258-015-0477-5) [DOI] [Google Scholar]
- 35.Weiss-Schneeweiss H, Emadzade K, Jang TS, Schneeweiss GM. 2013. Evolutionary consequences, constraints and potential of polyploidy in plants. Cytogenet. Genome Res. 140, 137-150. ( 10.1159/000351727) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Drunen WE, Husband BC. 2018. Immediate vs. evolutionary consequences of polyploidy on clonal reproduction in an autopolyploid plant. Ann. Bot. 122, 195-205. ( 10.1093/aob/mcy071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Husband BC, Ozimec B, Martin SL, Pollock L. 2008. Mating consequences of polyploid evolution in flowering plants: current trends and insights from synthetic polyploids. Int. J. Plant Sci. 169, 195-206. ( 10.1086/523367) [DOI] [Google Scholar]
- 38.Van Drunen WE, Husband BC. 2018. Whole genome duplication decreases clonal stolon production and genet size in the wild strawberry Fragaria vesca. Amer. J. Bot. 105, 1712-1724. ( 10.1002/ajb2.1159) [DOI] [PubMed] [Google Scholar]
- 39.Blakeslee A, Avery A. 1937. Methods of inducing doubling of chromosomes in plants by treatment with colchicine. J. Hered. 28, 393-411. ( 10.1093/oxfordjournals.jhered.a104294) [DOI] [Google Scholar]
- 40.Münzbergová Z. 2017. Colchicine application significantly affects plant performance in the second generation of synthetic polyploids and its effects vary between populations. Ann. Bot. 120, 329-339. ( 10.1093/aob/mcx070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaskani MJ, Kwon SW, Kim DH. 2005. Comparative study on vegetative, reproductive and qualitative traits of seven diploid and tetraploid watermelon lines. Euphytica 145, 259-268. ( 10.1007/S10681-005-1644-X) [DOI] [Google Scholar]
- 42.Cohen H, Fait A, Tel-Zur N. 2013. Morphological, cytological and metabolic consequences of autopolyploidization in Hylocereus (Cactaceae) species. BMC Plant Biol. 13, 173. ( 10.1186/1471-2229-13-173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bretagnolle F, Thompson JD. 1995. Gametes with the somatic chromosome number: mechanisms of their formation and role in the evolution of autopolyploid plants. New Phytol. 129, 1-22. ( 10.1111/j.1469-8137.1995.tb03005.x) [DOI] [PubMed] [Google Scholar]
- 44.Yamauchi A, Hosokawa A, Nagata H, Shimoda M. 2004. Triploid bridge and role of parthenogenesis in the evolution of autopolyploidy. Amer. Nat. 164, 101-112. ( 10.1086/421356) [DOI] [PubMed] [Google Scholar]
- 45.Mason AS, Pires JC. 2015. Unreduced gametes: meiotic mishap or evolutionary mechanism? Trends Genet. 31, 5-10. ( 10.1016/j.tig.2014.09.011) [DOI] [PubMed] [Google Scholar]
- 46.Šingliarová B, Hodálová I, Mráz P. 2011. Biosystematic study of the diploid-polyploid Pilosella alpicola complex with variation in breeding system: patterns and processes. Taxon 60, 450-470. ( 10.1002/TAX.602014) [DOI] [Google Scholar]
- 47.Šingliarová B, Zozomová-Lihová J, Mráz P. 2019. Polytopic origin and scale-dependent spatial segregation of cytotypes in primary diploid–autopolyploid contact zones of Pilosella rhodopea (Asteraceae). Biol. J. Linn. Soc. 127, 173. ( 10.1093/biolinnean/blz035) [DOI] [Google Scholar]
- 48.Šingliarová B, Mráz P, Chrtek J, Plačková I. 2011. Allozyme variation in diploid, polyploid and ploidy-mixed populations of the Pilosella alpicola group (Asteraceae): relation to morphology, origin of polyploids and breeding system. Folia Geobot. 46, 387-410. ( 10.1007/s12224-011-9102-0) [DOI] [Google Scholar]
- 49.Krahulcová A, Krahulec F, Chapman HM. 2000. Variation in Hieracium subgen. Pilosella (Asteraceae): What do we know about its sources? Folia Geobot. 35, 319-338. ( 10.1007/BF02803122) [DOI] [Google Scholar]
- 50.Šingliarová B, Šuvada R, Mráz P. 2013. Allopatric distribution, ecology and conservation status of the Pilosella alpicola species group (Asteraceae). Nord. J. Bot. 31, 122-128. ( 10.1111/j.1756-1051.2012.01603.x) [DOI] [Google Scholar]
- 51.Otto F. 1990. DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. In Methods in cell biology (eds Crissman H, Darzynkiewicz Z), pp. 105-110. New York, NY: Academic Press. [DOI] [PubMed] [Google Scholar]
- 52.Doležel J, Göhde W. 1995. Sex determination in dioecious plants Melandrium album and M. rubrum using high resolution flow cytometry. Cytometry 19, 103-106. ( 10.1002/cyto.990190203) [DOI] [PubMed] [Google Scholar]
- 53.Schönswetter P, Suda J, Popp M, Weiss-Schneeweiss H, Brochmann C. 2007. Circumpolar phylogeography of Juncus biglumis (Juncaceae) inferred from AFLP fingerprints, cpDNA sequences, nuclear DNA content and chromosome numbers. Molec. Phylogen. Evol. 42, 92-103. ( 10.1016/j.ympev.2006.06.016) [DOI] [PubMed] [Google Scholar]
- 54.Loureiro J, Rodriguez E, Doležel J, Santos C. 2007. Two new nuclear isolation buffers for plant DNA flow cytometry: a test with 37 species. Ann. Bot. 100, 875-888. ( 10.1093/aob/mcm152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mráz P, Šingliarová B, Urfus T, Krahulec F. 2008. Cytogeography of Pilosella officinarum (Compositae): altitudinal and longitudinal differences in ploidy level distribution in the Czech Republic and Slovakia and the general pattern in Europe. Ann. Bot. 101, 59-71. ( 10.1093/aob/mcm282) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hand ML, Vít P, Krahulcová A, Johnson SD, Oelkers K, Siddons H, Chrtek J, Fehrer J, Koltunow AM. 2015. Evolution of apomixis loci in Pilosella and Hieracium (Asteraceae) inferred from the conservation of apomixis-linked markers in natural and experimental populations. Heredity 114, 17-26. ( 10.1038/hdy.2014.61) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matzk F, Meister A, Schubert I. 2000. An efficient screen for reproductive pathways using mature seeds of monocots and dicots. Plant J. 21, 97-108. ( 10.1046/j.1365-313x.2000.00647.x) [DOI] [PubMed] [Google Scholar]
- 58.R Core Team. 2023. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. (http://www.r-project.org). [Google Scholar]
- 59.Pinheiro J, Bates D, DebRoy S, Sarkar D, Core Team R. 2017. Nlme: linear and nonlinear mixed effects models. R package version 3.1-131. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 60.Bates D, Maechler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1-48. ( 10.18637/jss.v067.i01) [DOI] [Google Scholar]
- 61.Hothorn T, Bretz F, Westfall P. 2008. Simultaneous inference in general parametric models. Biom. J. 50, 346-363. ( 10.1002/bimj.200810425) [DOI] [PubMed] [Google Scholar]
- 62.Stoffel MA, Nakagawa S, Schielzeth H. 2020. partR2: Partitioning R2 in generalized linear mixed models. bioRxiv. ( 10.1101/2020.07.26.221168) [DOI]
- 63.Kolář F, Štech M, Trávníček P, Rauchová J, Urfus T, Vít P, Kubešová M, Suda J. 2009. Towards resolving the Knautia arvensis agg. (Dipsacaceae) puzzle: primary and secondary contact zones and ploidy segregation at landscape and microgeographic scales. Ann. Bot. 103, 963-974. ( 10.1093/aob/mcp016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Castro S, Münzbergová Z, Raabová J, Loureiro J. 2011. Breeding barriers at a diploid–hexaploid contact zone in Aster amellus. Evol. Ecol. 25, 795-814. ( 10.1007/s10682-010-9439-5) [DOI] [Google Scholar]
- 65.Mráz P, Španiel S, Keller A, Bowmann G, Farkas A, Šingliarová B, Rohr RP, Broennimann O, Müller-Schärer H. 2012. Anthropogenic disturbance as a driver of microspatial and microhabitat segregation of cytotypes of Centaurea stoebe and cytotypes interactions in secondary contact zones. Ann. Bot. 110, 615-627. ( 10.1093/aob/mcs120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hardy OJ, De Loose M, Vekemans X, Meerts P. 2001. Allozyme segregation and inter-cytotype reproductive barriers in the polyploid complex Centaurea jacea. Heredity 87, 136-145. ( 10.1046/j.1365-2540.2001.00862.x) [DOI] [PubMed] [Google Scholar]
- 67.Koutecký P, Baďurová T, Štech M, Košnar J, Karásek J. 2011. Hybridization between diploid Centaurea pseudophrygia and tetraploid C. jacea (Asteraceae): the role of mixed pollination, unreduced gametes, and mentor effects. Biol. J. Linn. Soc. 104, 93-106. ( 10.1111/j.1095-8312.2011.01707.x) [DOI] [Google Scholar]
- 68.Morgan EJ, Čertner M, Lučanová M, Deniz U, Kubíková K, Venon A, Kovářík O, Lafon Placette C, Kolář F. 2021. Disentangling the components of triploid block and its fitness consequences in natural diploid–tetraploid contact zones of Arabidopsis arenosa. New Phytol. 232, 1449-1462. ( 10.1111/nph.17357) [DOI] [PubMed] [Google Scholar]
- 69.Lafon-Placette C, et al. 2017. Endosperm-based hybridization barriers explain the pattern of gene flow between Arabidopsis lyrata and Arabidopsis arenosa in Central Europe. Proc. Natl Acad. Sci. USA 114, E1027-E1035. ( 10.1073/pnas.1615123114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Santner A, Calderon-Villalobos LI, Estelle M. 2009. Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 5, 301-307. ( 10.1038/nchembio.165) [DOI] [PubMed] [Google Scholar]
- 71.Hojsgaard D, Pullaiah T. 2022. Apomixis in angiosperms: mechanisms, occurrences and biotechnology. Boca Raton, FL: CRC Press. [Google Scholar]
- 72.Böcher T. 1951. Cytological and embryological studies in the amphi-apomictic Arabis holboellii complex. Biol. Skr. 6, 1-58. [Google Scholar]
- 73.Janas AB, Szeląg Z, Musiał K. 2021. In search of female sterility causes in the tetraploid and pentaploid cytotype of Pilosella brzovecensis (Asteraceae). J. Plant Res. 134, 803-810. ( 10.1007/s10265-021-01290-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hojsgaard D. 2018. Transient activation of apomixis in sexual neotriploids may retain genomically altered states and enhance polyploid establishment. Front. Plant Sci. 9, 230. ( 10.3389/fpls.2018.00230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rosenberg O. 1906. Über die Embryobildung in der Gattung Hieracium. Ber. Deutsch. Bot. Ges. 24, 157-161. ( 10.1111/j.1438-8677.1906.tb06494.x) [DOI] [Google Scholar]
- 76.Rosenberg O. 1907. Cytological studies on the apogamy in Hieracium. Bot. Tiddskr. 28, 143-170. [Google Scholar]
- 77.Koltunow AMG, Johnson SD, Okada T. 2011. Apomixis in hawkweed: Mendel's experimental nemesis. J. Exp. Bot. 62, 1699-1170. ( 10.1093/jxb/err011) [DOI] [PubMed] [Google Scholar]
- 78.Catanach AS, Erasmuson SK, Podivinsky E, Jordan BR, Bicknell R. 2006. Deletion mapping of genetic regions associated with apomixis in Hieracium. Proc. Natl Acad. Sci. USA 103, 18 650-18 655. ( 10.1073/pnas.0605588103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hojsgaard D, Hörandl E. 2019. The rise of apomixis in natural plant populations. Front. Plant Sci. 10, 358. ( 10.3389/fpls.2019.00358) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zahn KH. 1930. Hieracium L. In Synopsis der mitteleuropäischen Flora XXII/1 (eds Graebner P, Graebner P). Liepzig, Germany: Borntraeger. [Google Scholar]
- 81.Peterson RL, Thomas AG. 1971. Buds on the roots of Hieracium florentinum (hawkweed). Botany 49, 53-54. [Google Scholar]
- 82.Bomblies K. 2020. When everything changes at once: finding a new normal after genome duplication. Proc. R. Soc. B. 287, 2020215420202154. ( 10.1098/rspb.2020.2154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Atta R, Laurens L, Boucheron-Dubuisson E, Guivarc'h A, Carnero E, Giraudat-Pautot V, Rech P, Chriqui D. 2009. Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J. 57, 626-644. ( 10.1111/j.1365-313X.2008.03715.x) [DOI] [PubMed] [Google Scholar]
- 84.Rosspopoff O, et al. 2017. Direct conversion of root primordium into shoot meristem relies on timing of stem cell niche development. Development 144, 1187-1200. ( 10.1242/dev.142570) [DOI] [PubMed] [Google Scholar]
- 85.Martínková J, et al. 2023. What determines root-sprouting ability: injury or phytohormones? Am. J. Bot. 110, e16102. ( 10.1002/ajb2.16102) [DOI] [PubMed] [Google Scholar]
- 86.Martínková J, et al. 2023. Why is root-sprouting not more common among plants? Phytohormonal clues and ecological correlates. Environ. Exp. Bot. 205, 105147. ( 10.1016/j.envexpbot.2022.105147) [DOI] [Google Scholar]
- 87.Šingliarová B, Hojsgaard D, Müller-Schärer H, Mráz P. 2023. The novel expression of clonality following whole-genome multiplication compensates for reduced fertility in natural autopolyploids. Figshare. ( 10.6084/m9.figshare.c.6700004) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data are provided in electronic supplementary material [87].