Abstract
OBJECTIVE:
To interrogate the circulating SARS-CoV-2 lineages and recombinant variants in persons living in migrant shelters and persons who inject drugs (PWID).
METHODS:
We combined data from two studies with marginalized populations (migrants in shelters and persons who inject drugs) in Tijuana, Mexico. SARS-CoV-2 variants were identified on nasal swabs specimens and compared to publicly available genomes sampled in Mexico and California.
RESULTS:
All but 2 of the 10 lineages identified were predominantly detected in North and Central America. Discrepancies between migrants and PWID can be explained by the temporal emergence and short time span of most of these lineages in the region.
DISCUSSION:
The results illustrate the temporo-spatial structure for SARS-CoV-2 lineage dispersal and the potential co-circulation of multiple lineages in high-risk populations with close social contacts. These conditions create the potential for recombination to take place in the California-Baja California border.
Keywords: SARS-CoV-2; lineages; recombination, genetic; drug users; emigrants and immigrants
RESUMEN
OBJECTIVO:
Investigar los linajes circulantes y variantes recombinantes de SARS-CoV-2 en migrantes en albergues y usuarios de drogas inyectables (UDI).
MÉTODOS:
Combinamos datos de dos estudios con poblaciones marginadas (migrantes en albergues y UDI) en Tijuana, México. Se identificaron variantes de SARS-CoV-2 en exudado nasal y se compararon con genomas disponibles públicamente de muestras de México y California.
RESULTADOS:
Ocho de 10 linajes identificados se detectaron principalmente en Norte y Centro América. Las discrepancias entre migrantes y UDI se pueden explicar por la emergencia temporal y la corta duración de la mayoría de estos linajes en la región.
DISCUSIÓN:
Los resultados ilustran la estructura temporo-espacial de la dispersión de linajes de SARS-CoV-2, y la circulación posible de linajes múltiples en poblaciones en riesgo con contactos sociales estrechos. Estas condiciones generan el potencial de recombinación en la frontera California-Baja California.
PALABRAS CLAVE: SARS-CoV-2, linajes, recombinación genética, usuarios de drogas, migrantes
INTRODUCTION
Mobility data can be used to predict the spread of SARS-CoV21-3 and other viruses.4 The successive emergence of SARS-CoV2 variants of concern (VOC) and their rapid spread around the world has highlighted the importance of genomic surveillance. For example, the B.1.1.529 variant aka ‘Omicron’ was first reported to the WHO from South Africa on 24 November 2021 and has been characterized by key mutations in the S protein, which play an integral role in viral transmission, immune evasion and virulence. Another mechanism for viral evolution is recombination. For recombination to occur, the parental lineages need to co-circulate in the same location to allow specific individuals to become co-infected. This scenario provides the circumstances during which chimeric genotypes can emerge. For SARS-CoV-2, the first recombinant lineage in the PANGO nomenclature system5 was designated as lineage XA after being identified in the UK. Since then, many other recombinants have been identified and led to limited or more global outbreaks. For example, XB variant6 (aka B.1.628), a recombinant lineage with parental lineage B.1.634 and B.1.631, was first identified in Mexico which also spread to the US, Guatemala, Honduras and El Salvador. The widespread circulation of lineage XB and other recombinants across multiple countries over an extended time span raised important questions regarding the role and potential effects of recombination on the evolution of SARS-CoV-2 during the ongoing COVID-19 pandemic.
The COVID-19 pandemic has disproportionately impacted socially marginalized populations.7 In this context, migrant populations living in temporary accommodations like camps and shelters may be at high risk for virus co-circulation and potential emergence of new VOC. Of particular concern are in-transit migrants, refugees, and asylum seekers, and those residing in camps and other overcrowded settings with limited access to resources for hygiene or physical distancing. Migrants in such settings typically have limited access to healthcare, employment, and social services, and are therefore vulnerable to the social and economic impacts of lockdown and other measures.8-10 We recently reported that the prevalence of SAR-CoV2 RNA positive samples among migrants to Tijuana living in shelters during 2021 was low (1.5%) but half (53.0%) were SARS-CoV2 seropositive.11 The high rate of social interactions within temporary accommodation and outside (e.g. in public transportation) and the mobility of these populations are important risk factors for co-circulation of different lineages and potential recombination.
Similarly, the COVID-19 pandemic has disproportionally impacted other vulnerable and marginalized communities, such as people who use drugs (PWUD).12-14 In a study of people who inject drugs (PWID) in Tijuana and San Diego, California conducted from 2020-2021, SARS-CoV-2 seroprevalence was 37.%.15 Cross-border mobility and forced deportation has also been found to heighten vulnerability to a range of health and social harms in the Mexico-US border region, such as HIV and tuberculosis.16-20
Herein, we report upon SARS-CoV2 variants circulating in high risk and vulnerable populations of PWID and migrant populations living in temporary accommodations in the Mexico-US border region during the COVID-19 pandemic and interrogate the co-circulation of these variants in Mexico and neighboring California.
METHODS
Cohort description and sampling.
EPICO study.
A non-probability survey of adult migrants aged ≥18 years living in shelters was conducted from November–December 2020 and February–April 2021. Participants (N=598) underwent interviewer-administered surveys in Spanish, English or French, and provided anterior nasal swabs for SARS-CoV-2 RNA testing.
La Frontera Study.
Between October 2020 and September 2021, people who injected drugs within the last month who were aged ≥18 years were recruited through community outreach in Tijuana (N=200) and San Diego (N=400). The Tijuana sample and half of the San Diego sample were required to reside in either city but report not having ever crossed the US-Mexico border to inject drugs, whereas the remaining half of the San Diego sample was required to have injected drugs in Mexico within the last two years. Participants underwent interviewer-administered surveys in Spanish or English and provided anterior nasal swabs for SARS-CoV-2 RNA testing at enrolment and six months later.
Pre- and post-test counseling was provided following national guidelines in the U.S. and Mexico. For both studies, samples were batched and stored at −80 degrees Celsius and shipped weekly on dry ice for SARS-CoV-2 RNA detection.
SARS-CoV-2 detection and sequencing.
Qualitative tests for SARS-CoV-2 were performed on all collected nasal swabs specimen for SARS-CoV-2 RNA detection using the FluxErgy platform (Irvine, CA, USA). SARS-CoV-2 full genome sequencing was performed using the COVID-19 ARTIC v4 Illumina library construction and sequencing protocol (https://github.com/CDCgov/SARS-CoV-2_Sequencing ). Amplicons were generated with the NEBNext VarSkip VSS2b Primer kit ( https://github.com/nebiolabs/VarSkip). PCR conditions were 98°C for 30 s, followed by 35 cycles of 95°C for 15s and 63°C for 5 min. Libraries were generated with the NEBNext ARTIC SARS-CoV-2 FS Library prep kit (Illumina) with NEBNext Multiplex Oligos for Illumina. Samples were sequenced using a 2 x 75bp paired-end reads. Reads were processed with the CLC Genomics Workbench V22 (Qiagen). Briefly the workflow identifies individual SARS-CoV-2 sample variants by first trimming and mapping high quality reads (>20) to the reference genome and then calling variants to generate a full genome consensus for each sample.
Lineage Identification and analyses.
We used the Phylogenetic Assignment of Named Global Outbreak Lineages (Pangolin) (version 1.1.14) command-line tool and PANGO nomenclature system21 to determine the lineage of all SARS-CoV2 genomes. Publicly available genomes sampled in Mexico and California from the lineages identified in the LF and EPICO cohorts were also retrieved from the GISAID database.22 We employed the Wuhan lineage as the reference to characterize the isolates.
Ethical considerations.
These studies were reviewed and approved by the Ethics Committees of the University of California in San Diego (no. 201153), El Colegio de la Frontera Norte (no. 066-160720) and Xochicalco University. All participants gave written informed consent in their preferred language (Spanish, French or English).
RESULTS.
Cohort and sampling.
EPICO study.
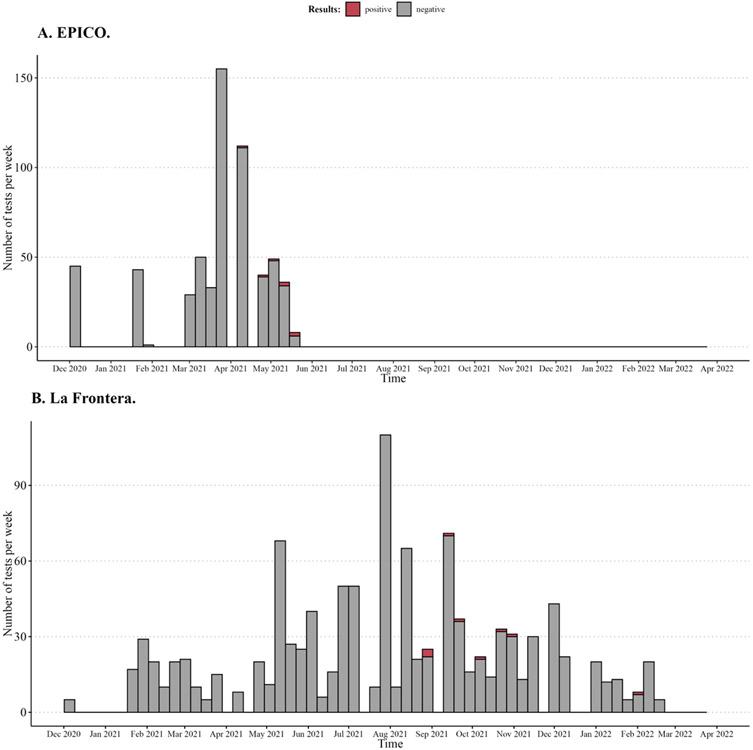
Samples were collected from November to December 2020, and from February to April 2021. Of 598 samples, seven (1.2%) were confirmed positive by RT-PCR and 6 were successfully sequenced (Figure 1A).
Figure 1. Overview of weekly sampling for the EPICO (A) and La Frontera (B) cohort.1.
1 Positive samples that were further sequenced are shown in red.
La Frontera study.
A total of 1159 samples (1.4 [Min-Max: 1 -3] samples per participant on average) were collected between December 2020 to January 2022. Nine of the 1159 (0.8%) tested positive and were sequenced (Figure 1B).
SARS-CoV-2 genomic analysis.
EPICO cohort.
During the spring of 2021, 3 distinct lineages were detected (from March 27th to April 12th (Figure 2). All 3 were most detected in the US and Mexico. XB recombinant lineage (n=4) 23 was first identified in July 2020 and March 2021 in California and Mexico respectively and last reported in September and August 2021. The 2 other B1.627 and B1.631 lineages were also most common in in the US and Mexico followed by Central American countries and were not detected after August 2021 (see Tables 1 and 2).
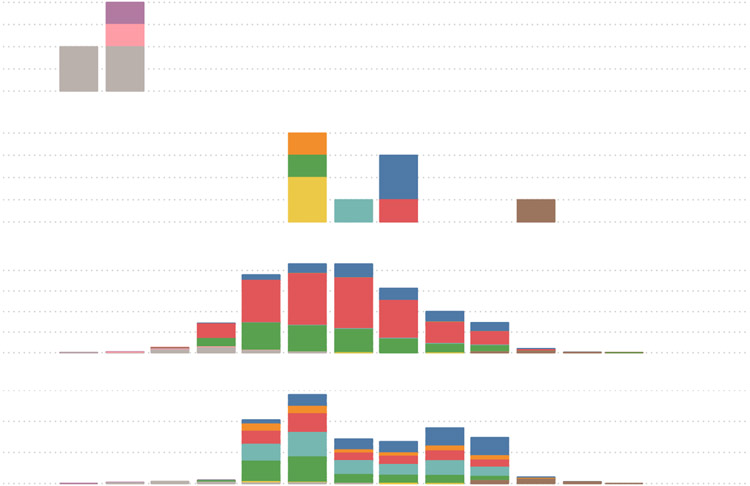
Figure 2. Monthly distribution of the 10 distinct lineages identified in California, Mexico and in the EPICO and La Frontera cohort.
Table 1.
Data from webpage cov-lineage.org 25 retrieved on May 22 2022, using the pangolin nomenclature5
| Lineage | N. sequences |
Most common countries |
Earliest date |
# designated |
# assigned |
Description |
|---|---|---|---|---|---|---|
| EPICO | ||||||
| B.1.627 | 1 | United States of America 72.0%, Mexico 11.0%, El_Salvador 7.0%, Honduras 5.0%, Guatemala 4.0% | 1/9/21 | 117 | 474 | USA, Mexico and Honduras lineage, from pango-designation issue #130 |
| B.1.634 | 1 | United States of America 64.0%, Mexico 34.0%, Honduras 1.0%, Guatemala 0.0%, Panama 0.0% | 3/2/21 | 71 | 289 | Mexico and USA lineage, from pango-designation issue #159 |
| XB | 4 | United States of America 77.0%, Mexico 16.0%, Guatemala 3.0%, Honduras 2.0%, El_Salvador 1.0% | 7/8/20 | 729 | 3186 | Recombinant lineage with parental lineages B.1.634 and B.1.631, Central and North America lineage, discussed in pango-designation issue #189. Formally B.1.628 |
| La Frontera | ||||||
| AY.100 | 2 | United States of America 87.0%, Mexico 4.0%, India 1.0%, Brazil 1.0%, Canada 1.0% | 7/16/20 | 16627 | 86622 | Alias of B.1.617.2.100, USA lineage |
| AY.119 | 1 | United States of America 95.0%, Germany 1.0%, Canada 1.0%, United Kingdom 0.0%, Dominican_Republic 0.0% | 10/29/20 | 2356 | 35639 | Alias of B.1.617.2.119, USA lineage |
| AY.20 | 1 | United States of America 58.0%, Mexico 32.0%, India 2.0%, Germany 1.0%, Colombia 1.0% | 1/8/21 | 2442 | 35116 | Alias of B.1.617.2.20, USA and Mexico lineage |
| AY.25.1 | 1 | United States of America 55.0%, Canada 40.0%, Chile 2.0%, Colombia 1.0%, Aruba 0.0% | 5/12/20 | 41309 | 124153 | Alias of B.1.617.2.25.1, Canada and USA lineage, from pango-designation issue #313 |
| AY.42 | 2 | Germany 19.0%, Spain 17.0%, France 15.0%, Switzerland 7.0%, Denmark 6.0% | 10/15/20 | 14252 | 26326 | Alias of B.1.617.2.42, European lineage, from pango-designation issue #239 |
| AY.26 | 1 | United States of America 83.0%, Mexico 12.0%, Brazil 1.0%, Israel 0.0%, Portugal 0.0% | 1/7/21 | 5011 | 43878 | Alias of B.1.617.2.26, USA and Mexico lineage, from pango-designation issue #188 |
| BA.1.17.2 | 1 | United Kingdom 71.0%, United States of America 5.0%, Canada 4.0%, Germany 3.0%, Denmark 3.0% | 10/7/21 | 30818 | 186393 | Alias of B.1.1.529.1.17.2, lineage from pango-designation issue #462 |
Table 2.
Lineage assignment of the 15 genomes from the EPICO (n=6) and La Frontera (n=9) and timespan of sampling distribution in the United States, California, USA and Mexico. Data from the GISAID initiative (Shu & McCauley, 2017).
| lineage | n | location | total | Cum. prevalence* |
First** | Last** |
|---|---|---|---|---|---|---|
| EPICO | ||||||
| B1.627 | 1 | Mexico | 51 | < 0.5% | 4-Mar-21 | 21-Jul-21 |
| California, US | 72 | < 0.5% | 3-Feb-21 | 21-Jul-21 | ||
| United States | 361 | < 0.5% | 16-Jan-21 | 15-Aug-21 | ||
| Worldwide | 493 | < 0.5% | 9-Jan-21 | 21-Aug-21 | ||
| B1.634 | 1 | Mexico | 96 | < 0.5% | 9-Apr-21 | 30-Jul-21 |
| California, US | 21 | < 0.5% | 12-Apr-21 | 11-Aug-21 | ||
| United States | 184 | < 0.5% | 2-Mar-21 | 16-Aug-21 | ||
| Worldwide | 284 | < 0.5% | 27-Dec-20 | 16-Aug-21 | ||
| X.B | 4 | Mexico | 513 | 1% | 19-Mar-21 | 24-Aug-21 |
| California, US | 876 | < 0.5% | 8-Jul-20 | 15-Sep-21 | ||
| United States | 2,627 | < 0.5% | 8-Jul-20 | 9-Jan-22 | ||
| Worldwide | 3,369 | < 0.5% | 8-Jul-20 | 15-Feb-22 | ||
| La Frontera | ||||||
| AY100 | 2 | Mexico | 3,107 | 5% | 22-Sep-20 | 16-Feb-22 |
| California, US | 12,480 | 2% | 31-Aug-20 | 29-Mar-22 | ||
| United States | 80,611 | 3% | 31-Aug-20 | 29-Mar-22 | ||
| Worldwide | 92,241 | 1% | 31-Aug-20 | 29-Mar-22 | ||
| AY119 | 1 | Mexico | 19 | < 0.5% | 2-Aug-21 | 17-Dec-21 |
| California, US | 5,849 | 1% | 12-Jun-21 | 7-Feb-22 | ||
| United States | 34,806 | 1% | 14-Jan-21 | 18-Apr-22 | ||
| Worldwide | 36,777 | < 0.5% | 29-Oct-20 | 18-Apr-22 | ||
| AY.20 | 1 | Mexico | 11,271 | 20% | 21-Jan-21 | 20-Mar-22 |
| California, US | 9,058 | 2% | 12-Apr-21 | 2-Feb-22 | ||
| United States | 20,870 | 1% | 14-Jan-21 | 31-Mar-22 | ||
| Worldwide | 35,985 | < 0.5% | 14-Jan-21 | 31-Mar-22 | ||
| AY.25.1 | 1 | California, US | 12,352 | 3% | 20-Apr-21 | 7-Feb-22 |
| Mexico | 163 | < 0.5% | 29-Jun-21 | 28-Feb-22 | ||
| United States | 75,346 | 2% | 26-Dec-20 | 24-Mar-22 | ||
| Worldwide | 134,23 | 1% | 11-Sep-20 | 28-Apr-22 | ||
| AY.42 | 2 | Mexico | 14 | < 0.5% | 11-Jul-21 | 20-Dec-21 |
| California, US | 111 | < 0.5% | 1-Jun-21 | 22-Dec-21 | ||
| United States | 525 | < 0.5% | 1-Jun-21 | 12-Jan-22 | ||
| Worldwide | 28,786 | < 0.5% | 15-Oct-20 | 11-Mar-22 | ||
| AY26 | 1 | Mexico | 14 | < 0.5% | 11-Jul-21 | 20-Dec-21 |
| California, US | 111 | < 0.5% | 1-Jun-21 | 22-Dec-21 | ||
| United States | 525 | < 0.5% | 1-Jun-21 | 12-Jan-22 | ||
| Worldwide | 28,786 | < 0.5% | 15-Oct-20 | 11-Mar-22 | ||
| BA.1.17.2 | 1 | Mexico | 102 | 1% | 16-Dec-21 | 31-Mar-22 |
| California, US | 1,230 | 1% | 1-Dec-21 | 29-Mar-22 | ||
| United States | 11,155 | < 0.5% | 31-Dec-20 | 27-Apr-22 | ||
| Worldwide | 196,031 | 2% | 23-Oct-20 | 6-May-22 | ||
apparent cumulative prevalence is the ratio of the sequences containing B.1.627 to all sequences collected since the identification of B.1.627 in that location.
Dates are based on the sample collection date;
genome key mutations from the outbreak.info26
La Frontera cohort.
Seven distinct lineages were identified from August 19th, 2021 (first detection), to January 2022 (Figure 2). All but AY.42 (considered a large European (especially Spain) Delta sub-lineage) and BA.1.17.2 (sublineage of wordwide spread BA.1.17 lineage with S:701V) were also most frequently reported in North and Central America and co-circulated in California and Mexico in the summer/fall of 2021.
To interrogate the co-circulation of these lineages in the Mexico and neighboring California region we retrieved all full-length genomes available on GISAID22 The spatial temporal distribution of these lineages in California and Mexico along with those from the EPICO and La Frontera cohort are presented in Figure 3 and under microreact project https://microreact.org/project/auby4ddjtcyrfy3q3dxdds. A detailed summary of the distribution of these 10 SARS-CoV2 lineages is provided in Tables 1 and 2 and Figure 3, along with timespan and cumulative prevalence in the United States, in Mexico and in California, USA and worldwide.
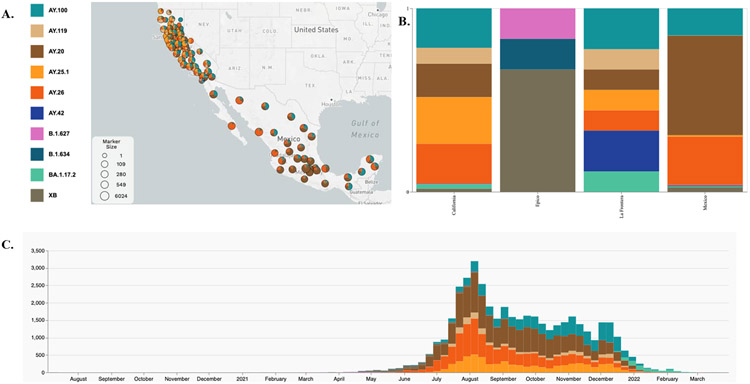
Figure 3. Spatial temporal distribution of the 10 LF/EPICO and La Frontera lineages in California and Mexico.
A. Microreact 24 display of the geolocation of all genomes with associated lineage inferred by the Pangolin assignment tool 21. B. Distribution of the lineages in California, in Mexico and in the EPICO and La Frontera cohorts. C. timeline of the 10 lineages. See also microreact project https://microreact.org/project/auby4ddjtcyrfy3q3dxdds
Data can be visualized under microreact project https://microreact.org/project/auby4ddjtcyrfy3q3dxdds
DISCUSSION
All but 2 of the 10 lineages reported in this study among PWID and migrants in shelters at the California-Mexico border were predominantly detected in North and Central America. The discrepancies between the 2 cohorts can be explained by the temporal emergence and short time span of most of these lineages in the region. Hence, the 3 lineages identified among migrants in shelters (EPICO study) were collected in spring 2021: The lineages B1.627, B1.634 emerged in March-April 2021 in both California and Mexico and were no longer detected worldwide after August 2021. Recombinant lineage X.B (originating from a recombination event between a B.1.631 major virus and a lineage B.1.634) was first reported in California during the summer 2020 but only in March 2021 in Mexico. While the widespread circulation of lineage XB across multiple countries lasted over a longer timespan worldwide, it was not reported after August/September 2021 in the region.
Seven distinct lineages were identified among PWID enrolled in the La Frontera cohort. These lineages were detected over a longer timespan than the previously mentioned lineages but all co-circulated in California and Mexico during 2021.
An important limitation of this observational study is that we had only a small number of isolates available for sequencing. Further larger scale studies would be required to better determine the prevalence trajectories of these co-circulating variants. However, these results illustrate the temporo-spatial structure for SARS-CoV-2 lineage dispersal and the potential co-circulation of multiple lineages in high-risk populations with close social contacts, conditions for a recombination event to take place.
Considering the successful worldwide dispersal of recombinant XB, our study underscores the need for strengthening the screening, diagnosis, and prevention measures to these vulnerable populations. It also highlights the potential benefit of genomic sequencing data for quick and informed public health decisions (e.g. appropriate vaccine response) and the importance of communication and partnerships at the regional level. Further studies are necessary to explore the role and potential effects of recombination on the evolution of SARS-CoV-2 during the COVID-19 pandemic.
ACKNOWLEDGMENTS
The authors want to thank the migrant shelters in Tijuana for facilitating this research, and the participants for generously contributing their time and experiences.
DATA ACKNOWLEDGEMENTS
We gratefully acknowledge the authors, originating and submitting laboratories of the genetic sequence and metadata made available through GISAID on which this research is based. A full acknowledgement table can be found with the following EPI_SET-IDs EPI_SET_20220606sv (https://doi.org/10.55876/gis8.220606sv); EPI_SET_20220606fw (https://doi.org/10.55876/gis8.220606fw) and EPI_SET_20220606wx (https://doi.org/10.55876/gis8.220606wx) in Data Acknowledgement Locator under GISAID resources (https://www.gisaid.org/).
FUNDING
This research was funded by the American Academy of Arts and Science, the University of California, San Francisco and the San Diego Center For AIDS Research (CFAR), which is funded by the National Institutes of Health (P30 AI036214). AC is supported by the James B. Pendleton Charitable Trust and by the San Diego Center for AIDS Research (SD CFAR), an NIH-funded program (P30 AI036214), which is supported by the following NIH Institutes and Centers: NIAID, NCI, NHLBI, NIA, NICHD, NIDA, NIDCR, NIDDK, NIGMS, NIMH, NIMHD, FIC, and OAR, the UCOP (R00RG2725) and by the NIH NIDA (R01DA055491). The study sponsors had no role in the study design; in the collection, analysis and interpretation of the data; in the writing of the report; or in the decision to submit the paper for publication.
Footnotes
COMPETING INTERESTS
None declared.
REFERENCES
- 1.Lemey P, Ruktanonchai N, Hong SL, Colizza V, Poletto C, Van den Broeck F, et al. Untangling introductions and persistence in COVID-19 resurgence in Europe. Nature. 2021;595(7869):713–7. 10.1038/s41586-021-03754-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kraemer MUG, Yang CH, Gutierrez B, Wu CH, Klein B, Pigott DM, et al. The effect of human mobility and control measures on the COVID-19 epidemic in China. Science. 2020;368(6490):493–7. 10.1126/science.abb4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodcroft EB, Zuber M, Nadeau S, Vaughan TG, Crawford KHD, Althaus CL, et al. Spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature. 2021;595(7869):707–12. 10.1038/s41586-021-03677-y. [DOI] [PubMed] [Google Scholar]
- 4.Lemey P, Rambaut A, Bedford T, Faria N, Bielejec F, Baele G, et al. Unifying viral genetics and human transportation data to predict the global transmission dynamics of human influenza H3N2. PloS pathog. 2014;10(2):e1003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rambaut A, Holmes EC, O’Toole Á, Hill V, McCrone JT, Ruis C, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nature microbiology. 2020;5(11):1403–7. 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutierrez B, Castelan Sanchez HG, Candido DDS, Jackson B, Fleishon S, Houzet R, et al. Emergence and widespread circulation of a recombinant SARS-CoV-2 lineage in North America. Cell Host Microbe. 2022;30(8):1112–23 e3. 10.1016/j.chom.2022.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Upshaw TL, Brown C, Smith R, Perri M, Ziegler C, Pinto AD. Social determinants of COVID-19 incidence and outcomes: A rapid review. PloS one. 2021;16(3):e0248336. 10.1371/journal.pone.0248336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayward SE, Deal A, Cheng C, Crawshaw A, Orcutt M, Vandrevala TF, et al. Clinical outcomes and risk factors for COVID-19 among migrant populations in high-income countries: A systematic review. J Migr Health. 2021;3:100041. 10.1016/j.jmh.2021.100041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lebano A, Hamed S, Bradby H, Gil-Salmeron A, Dura-Ferrandis E, Garces-Ferrer J, et al. Migrants' and refugees' health status and healthcare in Europe: a scoping literature review. BMC Public Health. 2020;20(1):1039. 10.1186/s12889-020-08749-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kondilis E, Papamichail D, McCann S, Orcutt M, Carruthers E, Veizis A, et al. The Impact of the COVID-19 Pandemic on Migrants, Refugees and Asylum Seekers in Greece: A Retrospective Analysis of National Surveillance Data (Feb-Nov 2020). SSRN; 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bojorquez-Chapela I, Strathdee SA, Garfein RS, Benson CA, Chaillon A, Ignacio C, et al. The impact of the COVID-19 pandemic among migrants in shelters in Tijuana, Baja California, Mexico. BMJ Global Health. 2022;7(3):e007202. 10.1136/bmjgh-2021-007202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubey MJ, Ghosh R, Chatterjee S, Biswas P, Chatterjee S, Dubey S. COVID-19 and addiction. Diabetes Metab Syndr. 2020;14(5):817–23. 10.1016/j.dsx.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aponte-Melendez Y, Mateu-Gelabert P, Fong C, Eckhardt B, Kapadia S, Marks K. The impact of COVID-19 on people who inject drugs in New York City: increased risk and decreased access to services. Harm reduction journal. 2021;18(1):118. 10.1186/s12954-021-00568-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang QQ, Kaelber DC, Xu R, Volkow ND. COVID-19 risk and outcomes in patients with substance use disorders: analyses from electronic health records in the United States. Mol Psychiatry. 2021;26(1):30–9. 10.1038/s41380-020-00880-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strathdee SA, Abramovitz D, Harvey-Vera A, Vera CF, Rangel G, Artamonova I, et al. Prevalence and correlates of SARS-CoV-2 seropositivity among people who inject drugs in the San Diego-Tijuana border region. PloS one. 2021;16(11):e0260286. 10.1371/journal.pone.0260286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deiss R, Garfein RS, Lozada R, Burgos JL, Brouwer KC, Moser KS, et al. Influences of cross-border mobility on tuberculosis diagnoses and treatment interruption among injection drug users in Tijuana, Mexico. Am J Public Health. 2009;99(8):1491–5. 10.2105/ajph.2008.142166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melo JS, Mittal ML, Horyniak D, Strathdee SA, Werb D. Injection Drug Use Trajectories among Migrant Populations: A Narrative Review. Substance use & misuse. 2018;53(9):1558–70. 10.1080/10826084.2017.1416404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldenberg S, Silverman J, Engstrom D, Bojorquez-Chapela I, Strathdee S. "Right Here is the Gateway": Mobility, Sex Work Entry and HIV Risk Along the Mexico-U.S. Border. Int Migr. 2014;52(4):26–40. 10.1111/imig.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strathdee SA, Lozada R, Ojeda VD, Pollini RA, Brouwer KC, Vera A, et al. Differential effects of migration and deportation on HIV infection among male and female injection drug users in Tijuana, Mexico. PLoS ONE. 2008;3(7):e2690. 10.1371/journal.pone.0002690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strathdee SA, Lozada R, Pollini RA, Brouwer KC, Mantsios A, Abramovitz DA, et al. Individual, social, and environmental influences associated with HIV infection among injection drug users in Tijuana, Mexico. J Acquir Immune Defic Syndr. 2008;47(3):369–76. 10.1097/QAI.0b013e318160d5ae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rambaut A, Holmes EC, O'Toole Á, Hill V, McCrone JT, Ruis C, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nature microbiology. 2020;5(11):1403–7. 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2017;22(13):30494. 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gutierrez B, Castelán Sánchez HG, da Silva Candido D, Jackson B, Fleishon S, Ruis C, et al. Emergence and widespread circulation of a recombinant SARS-CoV-2 lineage in North America. medRxiv. 2021:2021.11.19.21266601. 10.1101/2021.11.19.21266601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Argimón S, Abudahab K, Goater RJE, Fedosejev A, Bhai J, Glasner C, et al. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microbial Genomics. 2016;2(11). https://doi.org/ 10.1099/mgen.0.000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Toole Á, Hill V, Pybus OG, Watts A, Bogoch II, Khan K, et al. Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2 with grinch. Wellcome Open Res. 2021;6:121. 10.12688/wellcomeopenres.16661.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mullen GT Julia L., Latif Alaa Abdel, Alkuzweny Manar, Cano Marco, Haag Emily, Zhou Jerry, Zeller Mark, Hufbauer Emory, Matteson Nate, Andersen Kristian G., Wu Chunlei, Su Andrew I., Gangavarapu Karthik, Hughes Laura D., and the Center for Viral Systems Biology. outbreak.info 2022. [Available from: https://outbreak.info/. [Google Scholar]