ABSTRACT
Freshwater network ecosystems consist of interconnected lotic and lentic environments within the same catchment area. Using Plitvice Lakes as an example, we studied the changes in environmental conditions and microbial communities (bacteria and fungi) that occur with downstream flow. Water samples from tributaries, interlake streams, connections of the cascading lakes, and the Korana River, the main outflow of the system, were characterized using amplicon sequencing of bacterial 16S rRNA and fungal ITS2 genes. Our results show that different environmental conditions and bacterial and fungal communities prevail among the three stream types within the freshwater network ecosystem during multiple sampling seasons. Microbial community differences were also confirmed along the longitudinal gradient between the most distant sampling sites. The higher impact of “mass effect” was evident during spring and winter, while “species sorting” and “environmental selection” was more pronounced during summer. Prokaryotic community assembly was majorly influenced by deterministic processes, while fungal community assembly was highly dominated by stochastic processes, more precisely by the undominated fraction, which is not dominated by any process. Despite the differences between stream types, the microbial community of Plitvice Lakes is shown to be very stable by the core microbiome that makes up the majority of stream communities. Our results suggest microbial community succession along the river-lake continuum of microbial communities in small freshwater network ecosystems with developed tufa barriers.
IMPORTANCE Plitvice Lakes represent a rare freshwater ecosystem consisting of a complex network of lakes and waterfalls connecting them, as well as rivers and streams supplying water to the lake basin. The unique geomorphological, hydrological, biogeochemical, and biological phenomenon of Plitvice Lakes lies in the biodynamic process of forming tufa barriers. In addition to microbial communities, abiotic water factors also have a major influence on the formation of tufa. Therefore, it is important to understand how changes in environmental conditions and microbial community assembly affect the functioning of the ecosystem of a freshwater network with developed tufa barriers.
KEYWORDS: microbial communities, spatiotemporal, freshwater ecosystem, mass effects, species sorting, community assembly, tufa barriers
INTRODUCTION
Unique freshwater ecosystems consisting of lotic systems, such as streams and rivers, and lentic environments, such as lakes, connected within a catchment area, represent a spatial and temporal continuum from the source system to the mouth. Significant hydrologic and biogeochemical changes occur with community succession during runoff within lotic systems. Rivers and streams are primary receivers of nutrients and organic matter due to input from groundwater (1), soil, and surface runoff (2) but also from anthropogenic point sources (3). All this input with hydrology can significantly influence the microbial community in the ecosystem (4).
The streams’ water flow contributes to the structuring of microbial communities by massive advection of microbes from other systems through so-called “mass effects” (5, 6). Reaching lakes, microbial communities are massively influenced by the local hydrological and geochemical conditions, and through “environmental selection” and “species sorting,” the allochthonous communities can be displaced by more competitive species (7). Therefore, hydrology, the system position in a network and local environmental conditions, alongside community assembly processes (6) are the main influencers shaping freshwater microbial communities in network ecosystems.
Understanding the mechanisms underlying microbial community assembly, it is necessary to capture community composition across time and space. Temporal history shapes local communities, and unilateral water flow links temporal and spatial history. The control of the microbial community distribution patters is still an open box (8), but from the macroecological studies, we can identify four fundamental categories: selection, dispersal, diversification, and drift (9); the categories are shaped by the deterministic or stochastich microbial assembly processes (10). These processes are not mutually exclusive, and their relative importance alter the microbial diversity and its biological function (11).
Environmental filtering and interactions among species are regarded as deterministic selection processes (9, 12), whereas stochastic selection processes include random colonization, demographic coincidences, and ecological drift (13). The microbial community is formed as a net result of the upstream assembly processes due to the hydrological conditions in each of the subsystems within the aquatic network (14). In these networks, shifts in community compositions are not so much marked by the presence or absence of species but by changes in relative abundances (15). The community’s structure and thus the connection of the aquatic network are additionally shaped by seasonal hydrological fluctuations (16, 17). Thus, spatiotemporal studies are particularly important for a more detailed understanding of the functioning of freshwater network ecosystems. The main reason for the functioning of such network ecosystems is probably the existence of the core microbiome (15, 18, 19).
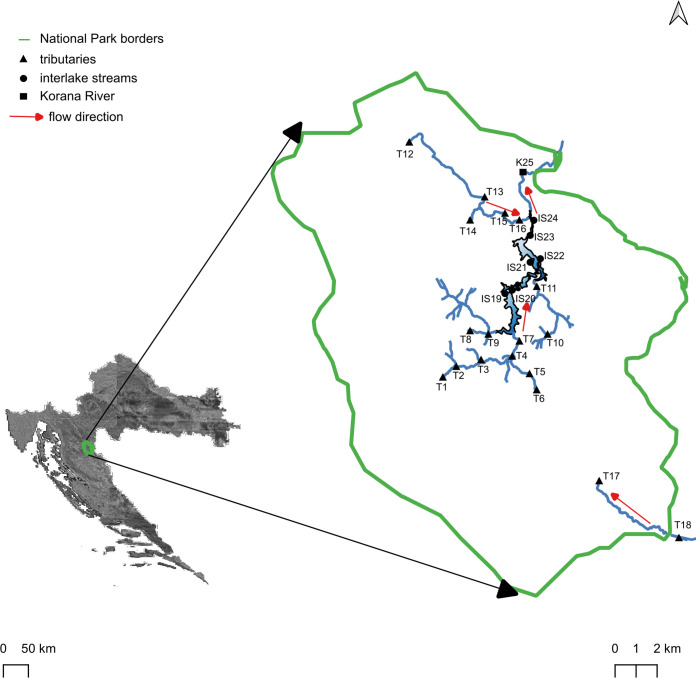
Plitvice Lakes represents a complex and biodiverse freshwater network ecosystem of lakes and waterfalls, as well as rivers and streams, providing the lake basin with water. Between the lakes are tufa barriers that form natural lake outlet habitats (20). In this complex environment (Fig. 1), we investigated the role of the spatial-temporal processes in the microbial community’s assembley and their response to different environmental conditions alongside the catchment area. Bacterial and fungal communities were examined throughout three different seasons during a period of 2 years. Our primary hypothesis was that there is a difference between microbial communities and assembley processes in tributaries, interlake streams, and the main outflow, the Korana River.
FIG 1.
Locations of sampling points at streams in Plitvice Lakes catchment area. The map was generated using the software QGIS 3.24.
Considering the positions of streams in the network ecosystem, we expected the occurrence of the so-called mass effect downstream of the entire system. Similarly, the various microbial communities from forested tributaries were expected to undergo significant changes as they entered the lakes. These changes were expected to be due to species sorting, longer water residence time (WTR) in lakes, and different environmental conditions in the lakes. Finally, in order to better understand the functioning of the entire freshwater network ecosystem, the core microbiome was determined, which included taxa present in all stream types throughout all seasons.
RESULTS
Environmental variables.
All the physical and chemical parameters obtained at 25 sampling locations within the network ecosystem through different seasons are either shown in Fig. S1 or listed in Table S1. Water temperatures measured within tributaries were more stable and varied from 2°C (winter 2021) to 12°C (summer 2020) compared to interlake streams, which varied from 2°C (winter 2021) to 22°C (spring 2019; Fig. S1). The lowest measured temperature in the Korana River was 3°C (winter 2021), and the highest was 17°C (summer 2019). The lowest measured O2 concentration in tributaries was 4.8 mg/liter (summer 2019), and the highest was 12.9 mg/liter (winter 2021; Fig. S1). Similarly, in interlake streams and the Korana River, the lowest measured O2 concentrations were 6.9 and 4.7 mg/liter (summer 2019), and the highest were 12.6 (winter 2020) and 13.3 mg/liter (winter 2021). Measured dissolved organic (DOC) concentrations within all areas from spring 2019 to 2020 varied between 0.8 and 3.3 mg/liter (Fig. S1). The measured DOC concentration in summer 2020 in the Korana River was 6.9 mg/liter. The DOC concentrations peaked in winter 2021 in all streams, where they were up to 17.3 mg/liter in tributaries, 18.9 mg/liter in interlake streams, and 15.5 mg/liter in the Korana River, respectively. Generally, the lowest Ca2+ concentrations were measured in spring 2019 (Fig. S1). Highest Ca2+ concentrations in tributaries were measured in winter 2020 (89.4 mg/liter) and in interlake streams in winter 2021 (69.4 mg/liter). concentrations were very low and varied between 0.6 mg/liter (in tributaries in winter 2020) and 6.8 mg/liter (in tributaries in winter 2021). One higher concentration was measured within interlake streams in spring 2019 (14.9 mg/liter). The rainfall amounts differed in the same periods of the different years (Table S1). Spring samples were taken during the same period in May. However, it was rainy in May 2019, with a rainfall of 421.2 mm, while in May 2020, the rainfall was 161.5 mm. The summer samples in 2019 were taken in early September during dry season, with a rainfall of 52.3 mm, while the summer samples in 2020 were taken in the mid-September 2020, when rainfall already reached 217.8 mm. Winter samples were taken in February, which was a dry season in 2020 with a rainfall amount of 44.8 mm, while in 2021, it snowed during the same period, and the rainfall amount was 90.4 mm.
Environmental parameters measured at the sampling locations through six seasons. Download FIG S1, TIF file, 1.0 MB (1,022.5KB, tif) .
Copyright © 2023 Čačković et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Environmental parameters measured at 25 locations through 6 seasons. Download Table S1, DOCX file, 0.03 MB (31.4KB, docx) .
Copyright © 2023 Čačković et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
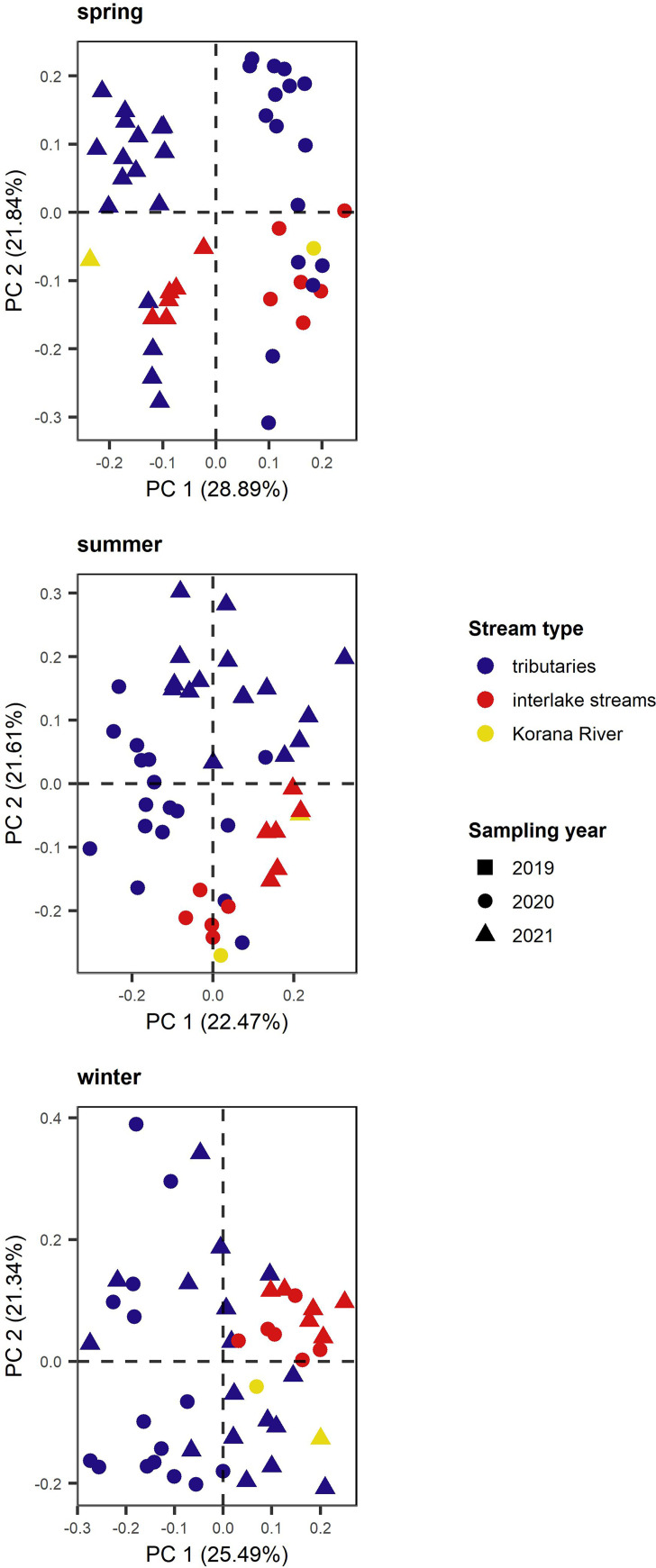
Based on Pearson correlation coefficient temperature showed a negative correlation with DOC (R2 = −0.39, P < 0.05), O2 (R2 = −0.38, P < 0.05), and Ca2+ (R2 = −0.45, P < 0.05), while DOC and O2 (R2 = 0.40, P < 0.05) showed a positive correlation. Based on the principal component analysis (PCA) results, examined separately by season, the samples from spring and summer are divided on account of sampling year, while the samples from winter showed no clustering based on sampling year (Fig. 2). In the spring, samples from interlake streams were not separate from samples from tributaries, and in the summer and winter, little separation of samples from interlake streams and tributaries was evident. In general, interlake stream samples clustered together more than tributaries, which were scattered. The Korana River samples did not separate from the groups from the same years in the spring and were clustered with the interlake streams samples in the summer and the tributary samples in the winter.
FIG 2.
Principal component analysis (PCA) ordination on the environmental variables during the investigated period divided by sampling seasons. Different colors represent stream types.
Microbial composition of freshwater network ecosystem.
α-Diversity of prokaryotic communities reached higher values in spring and winter in samples from tributaries than in samples from interlake stream or the Korana River, while values in summer were relatively the same for all stream types (Fig. S2A). Tukey’s honestly significant difference (HSD) test showed significant differences between tributaries and interlake streams and the Korana River in spring (P < 0.05), whereas samples from interlake samples and the Korana River were not significantly different from each other in the same season (Table S2). For the winter samples, Tukey’s HSD test showed significant differences between all stream types, while no significant differences were detected for the summer samples (Table S2).
α-Diversity indices of prokaryotic (A) and fungal (B) communities determined by Pieolu Evenness, Chao1, and Shannon index. The samples are divided based on sampling season and then by stream type and color marked as follows: blue, tributaries; red, interlake streams; and yellow, Korana River. The sample abbreviations refer to different sampling locations. Download FIG S2, TIF file, 0.9 MB (892.1KB, tif) .
Copyright © 2023 Čačković et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Tukey’s honestly significant difference (HSD) test results. Download Table S2, DOCX file, 0.01 MB (15.5KB, docx) .
Copyright © 2023 Čačković et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
For the fungal communities, higher values for α-diversity were observed in the tributary samples in winter, while the values in spring and summer were similar to those observed in the interlake streams and Korana River samples (Fig. S2B). Tukey’s HSD test showed significant differences only among tributaries and interlake streams in winter (P < 0.05; Table S2).
The taxonomic composition revealed, altogether, 54 prokaryotic (Fig. S3) and 12 different fungal phyla (Fig. S4). The most abundant bacterial phyla in all stream types were Actinobacteriota, Bacteroidota, Cyanobacteria, Patescibacteria, Proteobacteria, and Verrucomicrobiota. Archaea were overall rare. Among fungal phyla, Ascomycota, Basidiomycota, and Chytridiomycota were relatively most abundant.
Relative abundance of the total bacterial community structure (phylum level), divided by sampling seasons. The sample abbreviations refer to sampling locations. The “Other” group contains phyla with relative abundance of less than 1%. Download FIG S3, TIF file, 1.0 MB (1MB, tif) .
Copyright © 2023 Čačković et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundance of the total fungal community structure (phylum level), divided by sampling seasons. The sample abbreviations refer to sampling locations. Download FIG S4, TIF file, 0.6 MB (587.3KB, tif) .
Copyright © 2023 Čačković et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial amplicon sequence variants (ASVs) affiliated with Actinobacteriota, Bacteroidota, Cyanobacteria, and Verrucomicrobiota systematically increased in relative abundance from tributaries toward interlake streams (Fig. S3). Within tributaries, alongside the above-mentioned most abundant phyla, Bdellovibrionota appeared in higher relative abundances through all seasons. Higher relative abundances of Cyanobacteria in spring samples, Firmicutes and Planctomycetota in summer 2020, and Campylobacterota in winter 2021 were observed. In interlake streams, the relative abundances of Bacteroidota were lower in summer, while those of Cyanobacteria were higher. The Korana River samples showed similar bacterial community composition to interlake streams and followed the changes throughout the seasons (Fig. S3).
The fungal phylum Chytridiomycota dominated in the winter throughout almost all stream types, while Ascomycota- and Basidiomycota-related ASVs appeared in higher abundance in summer samples in all stream types (Fig. S4). In spring seasons, Chytridiomycota dominated in tributaries, while in spring 2020, Ascomycota and Basidiomycota dominated in interlake streams and the Korana River.
Spatial-temporal shifts of microbial communities across the freshwater network ecosystem.
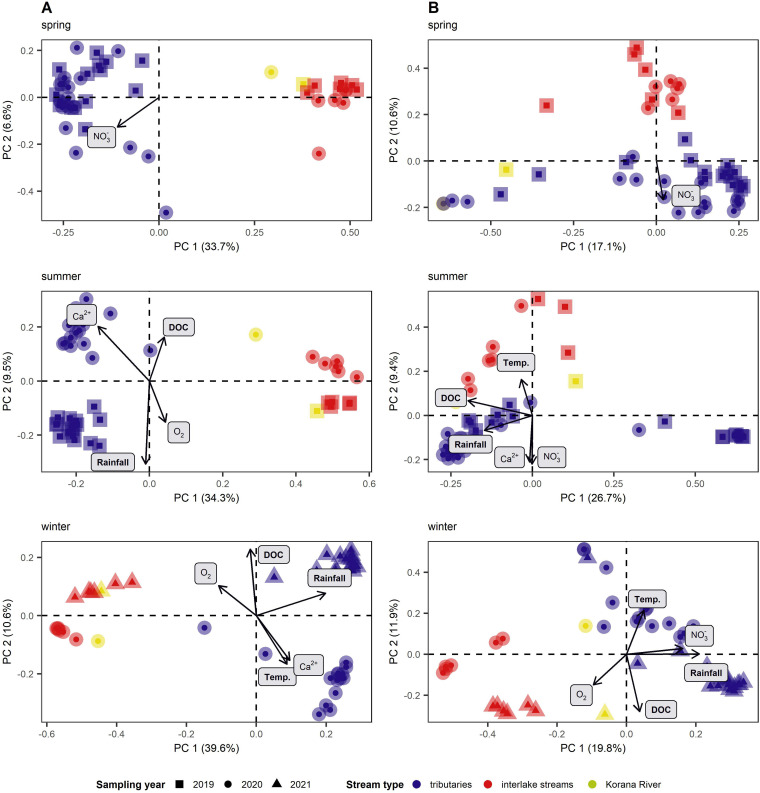
Multivariate analysis revealed differences among microbial communities based primarily on the stream types, when all samples were examined across different sampling seasons. Clustering was also observed within the prokaryotic community in summer and winter between samples of the same stream type based on sampling year, while there was no difference between samples from different years in spring (Fig. 3A; Table S3). Prokaryotic communities of the Korana River clustered with the interlake stream communities. Within the fungal community, clustering between samples of the same stream type based on sampling year was most pronounced in winter, while in spring and summer, the influence of sampling year was significant but minor (Fig. 3B; Table S3). Fungal communities of the Korana River clustered with the communities of the tributaries in spring and winter, while in summer they clustered with the communities of interlake streams.
FIG 3.
Principal coordinate analysis (PCoA) of bacterial (A) and fungal (B) communities’ β-diversity based on Bray-Curtis dissimilarity divided by sampling season. The influence of different stream type and sampling season on the sample clustering was tested by permutational multivariate analysis of variance (PERMANOVA). Different colors represent stream types, and different shapes represent sampling years.
Permutational multivariate analysis of variance (PERMANOVA) test results. Download Table S3, DOCX file, 0.01 MB (13.3KB, docx) .
Copyright © 2023 Čačković et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Main drivers of microbial community composition across the freshwater network ecosystem.
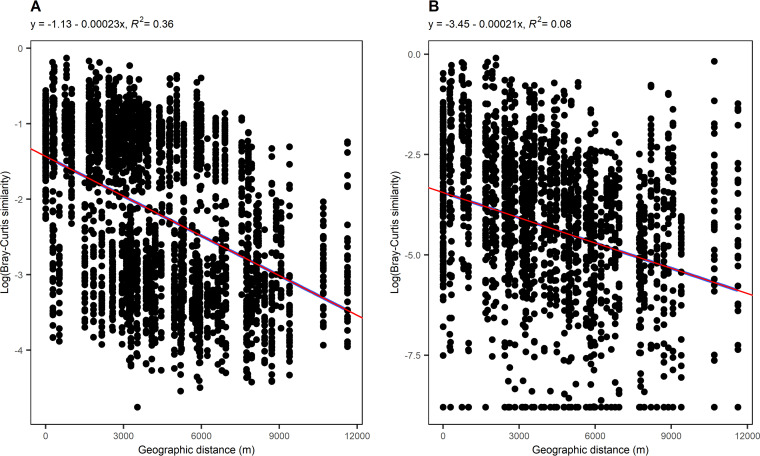
To explore biogeographic differences of microbial communities along the longitudinal gradient, the community similarity was plotted as a function of geographic distance between sampling points (Fig. 4). The geographic distance between farthest sampling points was 12 km. Generally, microbial community similarity between samples decreased with increasing geographic distance (prokaryotic community slope, −0.00023; fungal community slope, −0.00021). Linear regression of the DDR showed a higher decrease in community similarity within the prokaryotic communities with increasing geographic distance (Fig. 4A), while the same observation was noticed for the fungal communities with slightly smaller values (Fig. 4B).
FIG 4.
Pairwise Bray-Curtis community similarity between samples with respect to geographic distance (m). (A) Bacterial community. (B) Fungal community. For the evolution with distance, pairwise community similarity was evaluated exclusively between samples of the longest tributaries: Bijela Rijeka with interlake streams and Korana River. Blue lines illustrate linear models computed for the subset of samples considered, and red lines represent the overall linear regression when including all the samples.
In order to discern the main drivers of the microbial distribution of the downstream flow within the freshwater ecosystem network depicted by the PCoA plots, the measured environmental variables were fitted on the separated ordinations (Fig. 3). Both prokaryotic and fungal community compositions were influenced by temperature, DOC, O2, Ca2+, , or rainfall. influenced both communities of tributaries samples in spring. Tributary prokaryotic communities in summer were influenced by rainfall in 2019 and Ca2+ in 2020. In winter, temperature and Ca2+ influenced tributary prokaryotic communities in 2020, while rainfall amounts and DOC influenced tributary communities in 2021. Interlake stream prokaryotic communities were influenced by O2 in summer 2019, DOC in summer 2020, and O2 in winter 2021. Tributary fungal communities were influenced by and Ca2+ and rainfall in summer, by temperature and in winter 2020, and by rainfall and DOC in winter 2021. Fungal communities in interlake streams were influenced by temperature in summer 2020 and by O2 concentrations in winter 2021. The results were confirmed with permutational multivariate analysis of variance (PERMANOVA) (Table S3).
Relative influence and quantitative analysis of assembly processes between different seasons.
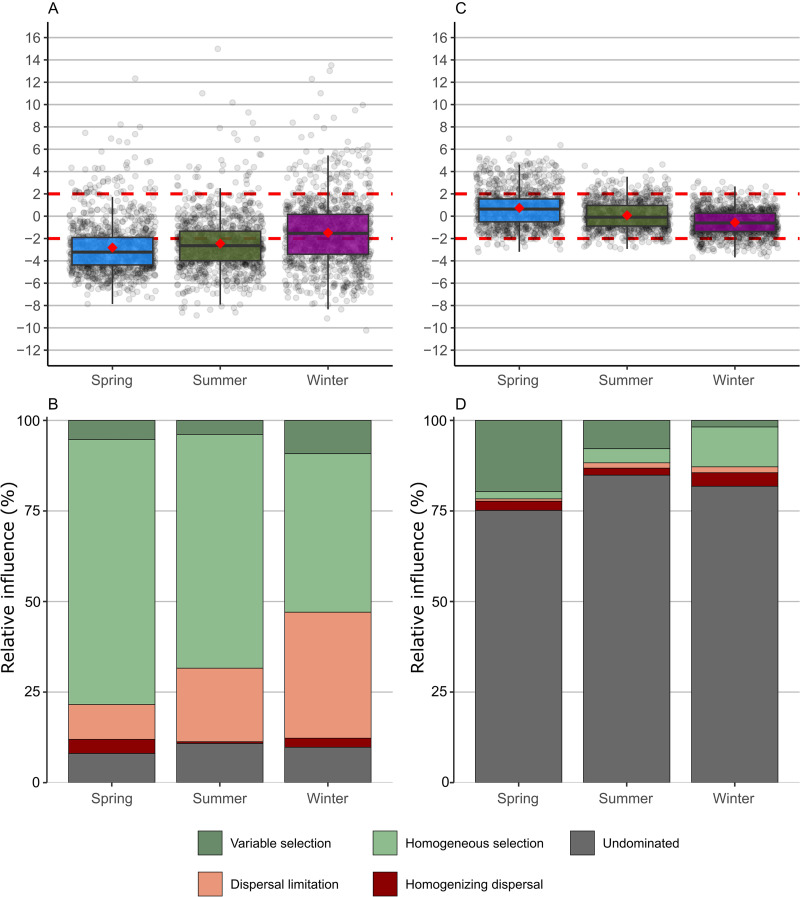
Partitioning the phylogenetic distance between prokaryotic ASVs using the null model, we found βNTI to be below −2 in spring and summer and below −2 in winter in some cases and above −2 in others, suggesting that stochastic assembly effected prokaryotic communities in winter (Fig. 5A). Homogeneous selection contributed to a large extent to the assembly of prokaryotic communities in all seasons (Fig. 5B). The highest contributions of homogeneous selection were found for prokaryotic communities in spring. Dispersal limitations also influenced prokaryotic community assembly in all seasons, with the greatest influence on prokaryotic communities in winter. Variable selection influenced prokaryotic communities more strongly in winter than in spring and summer. When we partitioned the phylogenetic distance between fungal ASVs using the null model, we also found that most βNTI ranged from −2 to +2 from site to site, suggesting that deterministic assembly had little influence on fungal community structuring (Fig. 5C). The processes that influenced fungal communities were mainly undominated. Homogeneous selection contributed to the assembly of fungal communities in spring, whereas variable selection influenced fungal communities in winter (Fig. 5D).
FIG 5.
Community assembly processes for prokaryotic and fungal communities. Boxplots illustrating variation in β-nearest taxonomic index (βNTI) for prokaryotic (A) and fungal (C) communities from different seasons. The percentage of turnover in prokaryotic (B) and fungal (D) community assembly through different seasons governed primarily by various deterministic, including homogeneous and variable selection, and stochastic processes, including dispersal limitations and homogenizing dispersal, as well as the fraction that was not dominated by any single process (“Undominated”).
Core microbiome of the freshwater network ecosystem.
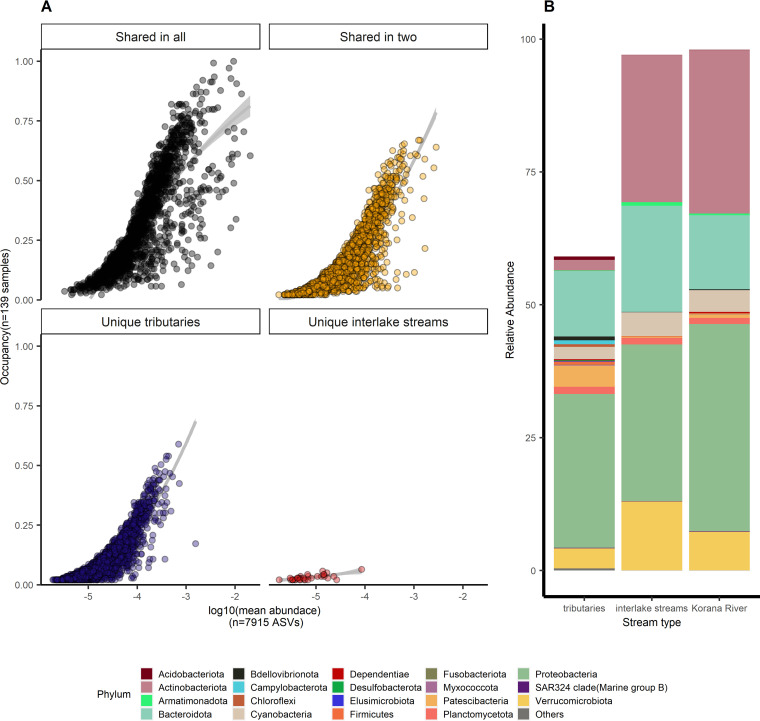
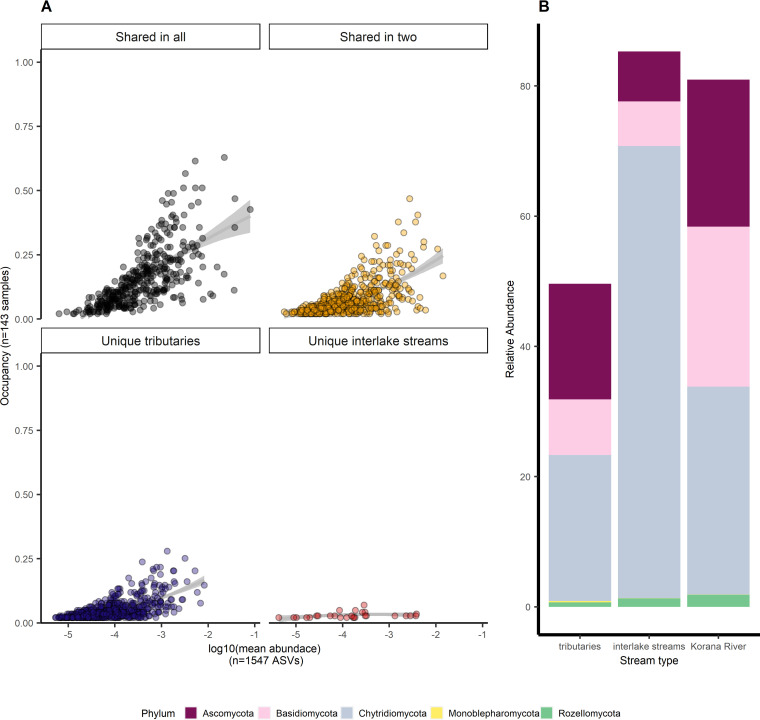
An analysis of the overlap between prokaryotic (Fig. S5A) and fungal (Fig. S5B) communities in the different sampling seasons showed that the majority of the taxa were unique to the tributaries. A considerable number of ASVs were shared by all three stream types or two stream types in all seasons. A minor number of ASVs was unique to interlake streams, while the Korana River had a maximum of 11 unique ASVs. The abundance-occupancy distributions of both bacterial (Fig. 6A) and fungal (Fig. 7A) communities showed the highest occupancy among samples of ASVs shared in all samples. Although the majority of ASVs were unique for tributaries, they were relatively rare. ASVs shared in all three stream types were also relatively most abundant in all samples and defined as core microbiome of this specific freshwater network ecosystem. The core microbiome covered 90% of the interlake stream and the Korana River and 70% of the tributary prokaryotic community (Fig. 6B). The most abundant ASVs were affiliated with the phyla Proteobacteria, Bacteroidota, and Actinobacteria. By exploring deeper taxonomic levels within the core microbiome, 195 different families have been discovered. In tributaries, bacteria from the Comamonadaceae family were most dominant, and bacteria from the Flavobacteriaceae and Sphingomonadaceae family were present in greater relative abundance than others. Downstream, i.e., in interlake streams and the Korana River, bacteria from the family Sporichthyaceae were dominant. Bacteria from the families Clade III, Comamonadaceae, Microbacteriaceae, and Rubritaleaceae were present in greater relative abundance (Fig. S6). In the fungal community, the core microbiome comprised 80% to –85% of interlake and Korana River communities and about 50% of tributary communities (Fig. 7B). The core microbiome of the fungal community was comprised of phyla Ascomycota, Basidiomycota, Chytridiomycota, Monoblepharomycota, and Rozellomycota. Among the ASVs unique for tributaries, Proteobacteria- and Bacteroidota-related ASVs in prokaryotic communities and Chytridiomycota-related ASVs in fungal communities prevailed. To better identify the uniqueness of tributaries, the 30 most abundant and rare families were investigated. The families Chitinophagaceae, Leptolyngbyaceae, Saprospiraceae, and Spirosomaceae were the most abundant, while Erysipelatoclostridiaceae, Pelotomaculaceae, and Vibrionaceae stood out among the rare taxa (Fig. S7). Although less numerous, ASVs shared in two stream types in bacterial communities were more abundant than unique ones and made 25% of bacterial and fungal communities of tributaries, while in interlake streams and the Korana River, ASVs made up less than 5% of bacterial and less than 20% of fungal communities.
FIG 6.
(A) Abundance-occupancy distributions were used to identify core members of the freshwater network ecosystem for bacteria. Taxa exclusive to stream types are indicated in blue (tributaries) or red (interlake streams), taxa shared between two stream types are indicated in yellow, and taxa shared across all systems are indicated in black. (B) Relative abundance of core microbiome taxa, grouped by stream type and color-coded by phylum. ASV, amplicon sequence variant.
FIG 7.
(A) Abundance-occupancy distributions were used to identify core members of the freshwater network ecosystem for fungi. Taxa exclusive to stream types are indicated in blue (tributaries) or red (interlake streams), taxa shared between two stream types are indicated in yellow, and taxa shared across all systems are indicated in black. (B) Relative abundance of core microbiome taxa, grouped by stream type and color-coded by phylum.
Venn diagram representing the number of shared and unique bacterial (A) and fungal (B) ASVs between three different stream types: tributaries (green), interlake streams (red), and the Korana River (blue). Download FIG S5, TIF file, 0.8 MB (810.4KB, tif) .
Copyright © 2023 Čačković et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundance of 30 most abundant core microbiome bacterial taxa, grouped by stream type and color-coded by family. Download FIG S6, TIF file, 0.5 MB (505.7KB, tif) .
Copyright © 2023 Čačković et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundance of 30 most abundant and 30 most rare bacterial taxa unique for tributaries. The panels are color-coded by family. Download FIG S7, TIF file, 0.7 MB (679.3KB, tif) .
Copyright © 2023 Čačković et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
In this study, we were primarily interested in determining changes in bacterial and fungal communities along the interconnected lotic systems during different seasons. Environmental parameters of the entire system were also determined during six sampling seasons.
In general, environmental parameters in samples from interlake streams showed overall uniformity, whereas the tributary samples were more scattered (Fig. 2). Accordingly, we expected differences in microbial communities among stream types, as previously shown in similarly studied aquatic network ecosystems (6, 21). Both bacterial and fungal communities (Fig. 3) showed differences in community composition between tributaries and downstream sites (interlake streams and the Korana River). Further confirmation of the different microbial communities in the tributaries and downstream can be provided by examining the biogeographic patterns of the microorganisms (Fig. 4). Indeed, when we examined the longest water flow in the Plitvice Lakes freshwater network ecosystem, it became clear that both bacterial and fungal communities differed with increasing geographic distance, that is, both microbial communities showed strong distance decay (21). The diversity of microbial communities was already evident at the level of phylum taxonomy, such that the strong dominance of phyla Proteobacteria and Bacteroidota in the tributaries was replaced by the sudden increase in the relative abundance of Actinobacteriota downstream in the network (22). Similarly, the dominance of Chytridiomycota was visible in the lower reaches, proving that rivers are dispersal pathways for terrestrial fungi (23–25), where fungi enter tributaries and flow downstream.
Based on environmental parameters, spring samples differed between sampling years but not between stream types (Fig. 2), while PCoA analysis indicated that there was a difference between microbial communities by stream type rather than sampling year (Fig. 3). Deterministically determined prokaryotic communities are mostly formed by the mass effect in spring (4). Namely, homogeneous selection influences community assembly when environmental conditions are relatively predictable (26). Under a low pressure of environmental changes (Table S3), bacterial communities that enter shallow tributaries from surrounding land, with water flow due to the strong mass effect, lose their abundance and diversity (6, 27) (Fig. S2). The only abiotic factor that affected bacterial community composition was the elevated concentrations in tributaries (Fig. 3), which reach higher levels in karst aquifer systems during the wet season (28). For this very reason, we can conclude that species sorting and environmental selection had little effect on bacterial community formation. Fungal community assembly was largely stochastically influenced due to the undominated fraction (Fig. 5C and D). When we consider that most fungal ASVs were unclassified, this information is not surprising. Nonetheless, the small fraction of detected processes was dominated by variable selection, which causes high compositional turnover with large variations in environmental factors (26). Fungal communities were therefore more affected by environmental changes in spring (Table S3) but still mostly formatted by mass effect.
As might be expected, larger changes in environmental conditions occurred in the summer between the relatively sheltered, forested tributaries and the lower reaches of the system, which are subject to greater seasonal fluctuations (Fig. 2). The tributaries were obviously still strongly influenced by the surrounding soil, and the mass effect also shaped the entire community composition. However, depending on the environmental conditions during the summer, the influences of species sorting and environmental selection were stronger, especially in the lower reaches (4). Bacterial community assembly composition was still influenced by deterministic processes. Environmental factors had changed in terms of seasonality, but there have been no great leaps, so that the community composition was still influenced by homogeneous selection (26). Fungal community assembly was, again, largely stochastically influenced due to the undominated fraction (Fig. 5D).
In winter, samples from interlake streams were uniform according to environmental factors, whereas samples from tributaries were scattered (Fig. 2). A major impact on tributaries was a large jump in the concentration of DOC, which was elevated in 2021 due to snow (29). In winter, bacterial community assembly was dominated by deterministic processes, i.e., homogeneous selection. Nevertheless, the influence of stochastic processes was stronger here compared to other seasons (Fig. 5B). In winter, the movement of microorganisms to a new location is more limited; therefore, they showed a stronger dispersal limitation (8). The lesser influence of environmental factors (Table S3) and the differences in α- and β-diversity confirmed the greater impact of mass effect and more stochastic influence on bacterial community formation. The fungal communities were again not conditioned by any process (Fig. 5D), and homogeneous selection stood out from the detected processes. Changes in environmental parameters were not surprising, and therefore, communities were formed by homogeneous selection, but the parameters still had an effect on community changes (Table S3).
Overall, the Plitvice Lakes’ freshwater network ecosystem was characterized by relatively stable microbial communities with small seasonal changes due to communities present in all stream types, described as the core microbiome. Although the majority of bacterial ASVs were unique to the tributaries, they were rare and present in a smaller relative abundance (Fig. 6A), and the most significant part of the prokaryotic communities in tributaries was the core microbiome, also present in more than 90% of other stream types (Fig. 6B). Proteobacteria, already recognized earlier as transitional elements in a network ecosystem from streams to lakes (30), represented the most abundant phylum in the core microbiome. The phylum Proteobacteria was dominated by the chemotrophic Comamonadaceae, which are commonly found in groundwater and karst waters (reference 31 and references therein), primarily because of their role in utilizing various carbon sources (32). Other commonly occurring phyla were Bacteroidota and Verrucomicrobiota, which, together with Proteobacteria, represent the most profilic phyla in the bacterial communities of freshwater ecosystems (33). The most abundant families within the aforementioned phyla, Flavobacteriaceae (Bacteroidota) and Rubritaleaceae (Verrucomicrobiota), were bacteria normally associated with phytoplankton blooms and the degradation of polymeric matter (34). Flavobacteriace were more abundant in upstream samples because they are capable of degrading all organic matter present in the environment and are therefore important for biodegradation processes in subsurface karst environments (35). Actinobacteriota has already been associated with karst ecosystems (36), so it is not surprising that they are ubiquitous in Plitvice Lake’s network. Although they were present in low abundance in the tributaries, they experienced a sudden growth downstream, confirming their competitiveness (37). The most dominant family within the phylum Actinobacteriota, Sporichthyaceae, possesses phototrophic properties and is able to survive in oligotrophic environments (38). As photoheterotrophic bacteria, they use biodegradable DOC as a carbon source and produce their own DOC in combination with solar energy, which is why they dominated in downstream parts (38). Some unexpected phyla were present in the core microbiome, such as Patescibacteria, which are originally found in groundwater and aquifers, nutrient-poor habitats, where they make up as much as 20% of the microbial community (39, 40). Their high abundance in tributaries was not surprising as it is known that microbial communities are discharged with karst water to the surface (41). Still, their presence in lakes indicates their competitiveness and ecological roles in oxygen-rich environments (42). As for the core microbiome of the fungal community, a smaller percentage was present in all stream types (Fig. 7B) with a predominance of phylum Chytridiomycota, commonly found in aquatic habitats (43, 44) with various functionality from litter decomposition (45) to algae parasitism, when they potentially force cell death and cell wall disruption for organic matter release to decompose it directly (46). Representatives of the phyla Ascomycota and Basidiomycota were present in higher abundances in tributary and Korana River samples, probably due to their involvement in lichen symbioses (47). Also, the higher abundance of Ascomycota is associated with their important role in ecosystem succession (48). Dominance among taxa unique for tributaries in bacterial community was led by widespread forest soil bacteria from families Chitinophagaceae and Saprospiraceae (49) and Leptolyngbyaceae, which among other functions have the function of nitrogen fixation (50). Among rare taxa, bacteria from families associated with aromatic decomposition can be found, for instance Pelotomaculaceae (51). Taxa unique for tributaries and more diverse taxa shared in two stream types, constituted larger part of the fungal community (Fig. 7A). Various phyla, like Mortierellomycota and Neocallimastigomycota, showed a higher influence of soil and sediment on tributaries (44), same as Olpidiomycota, species that are typically internal parasites of algae, fungi, rotifers, and plant roots (52).
Conclusions.
In summary, our study revealed the environmental and microbial differences between different stream types and sampling seasons within the freshwater network ecosystem. We have confirmed the stronger impact of soil on shallow tributaries, and we have shown that interlake streams show the condition of the lakes. Microbial communities in freshwater network ecosystems differed among stream types. In spring and winter, the mass effect had a greater influence on microbial community formation, while in summer, it was species sorting and environmental selection. Prokaryotic community assembly was influenced by deterministic processes throughout the system and in all seasons, while a lesser influence of stochastic processes was evident in winter. Fungal community assembly was dominated by stochastic processes. Despite minor changes in the communities through the seasons, we have shown that the microbial community of the Plitvice Lakes is very stable by determining the core microbiome. Still, due to the increasing anthropogenic influence and inevitable climate changes, it requires more profound research and monitoring.
MATERIALS AND METHODS
Study area.
The Plitvice Lakes National Park occupies 29.630,8 ha within the area of the Dinaric karst in west continental Croatia (Fig. 1). Less than 1% of this area is surface water. The aquatic ecosystem of the Plitvice Lakes consists of 16 cascading lakes, formed by the continuous biodynamic growth process of the tufa barriers that cut through this former river valley and enabled the formation of cascades.
Water samples were collected from all streams in the surface catchment area of Plitvice Lakes at 25 different locations (Fig. 1). The area of interest included three different stream types: tributaries (samples T1 to T18), interlake streams (samples IS19 to IS24), and the Korana River (sample K25). Tributaries included a total of 18 sampling locations, including several down the two leading water suppliers, the Bijela River, along its tributaries (Vukmirovića and Ljeskovac streams), and the Crna River. Tributaries also included locations along Sušanj and Rječica streams, which flow into the two largest lakes in the system, Prošćansko and Kozjak Lakes, as well as the more remote Korenička stream in Drakulić Rijeka and Rijeka Korenička. At last, three locations along the Plitvice stream and its affluent Sartuk stream were sampled. Together, they flow into the Korana River via the 76-m-high waterfall.
Interlake streams are the streams interconnecting the 16 lakes into a unique system. A total of six sampling locations were sampled at interlake streams: Labudovačke barriers, the exit barriers of Gradinsko Lake, a canal on Kozjački bridges, and the entrance barrier in Novakovića Brod Lake. Finally, a sample from the Korana River was taken as the central outflow of the entire Plitvice Lakes water system. Notably, the most distant tributaries are only 20 km separated from each other, and the geographic distance between the tributary of the longest flow, Bijela River, and the outflow of lakes, Korana River, is only 12 km.
At each sampling point, a total of 2 liters of water was collected in sterile polycarbonate (PC) bottles. Sampling was conducted during spring and summer in 2019 and 2020 and during winter in 2020 and 2021. The samplings in spring and winter were carried out in the same period both years; however, samplings in summer were done at the end of August 2019 and September 2020, respectively. The water samples were filtered onto a 0.22-μm-pore size PC filters (Whatman Nuclepore Track-Etch membrane; diameter, 47 mm) with a peristaltic pump. The filters were immediately stored at −20°C until DNA extraction, while the filtrates were preserved at 4°C for physicochemical analysis.
Physicochemical analysis.
A Multisensor probe (EXO3, YSI, USA) was used to measure dissolved oxygen (O2), temperature, and pH in situ. Concentrations of the cations (Ca2+, Mg2+, and Na+) and anions (Cl−, , ) in filtered water samples were measured on a Dionex ICS-6000 DC (Thermo Fisher Scientific, Waltham, MA, USA). DOC and dissolved inorganic carbon (DIC) were analyzed using the HACH QBD1200 analyzer in filtered water samples. For DOC analysis, DIC was first removed (i.e., converted to CO2 and outgassed) by the addition of H3PO4. All analyses were performed in the Hydrochemical Laboratory of the Croatian Geological Survey.
DNA extraction, amplification, and sequencing.
Total genomic DNA from filters was extracted with the DNeasy PowerWater kit (Qiagen, Inc., Valencia, CA, USA) following the manufacturer’s protocol. The hypervariable V4 region of the prokaryotic 16S rRNA gene was amplified by PCR using primer pair 515F Parada (5′-GTG YCA GCM GCC GCG GTA A-3′) (53) and 806R Apprill (5′-GGA CTA CNV GGG TWT CTA AT-3′) (54). The transcribed intergenic spacer 2 (ITS2) region of the fungal rRNA gene was amplified by PCR using primer pair ITS3-Mix1-Mix2 (TCCTCCGCTTAyTgATAtGc), a modified ITS3 Mix2 forward primer from Tedersoo48 named ITS3-mkmix2 CAWCGATGAAGAACGCAG, and a reverse primer ITS4 (equimolar mix of cwmix1 TCCTCCGCTTAyTgATAtGc and cwmix2 TCCTCCGCTTAtTrATAtGc) (44). As described in detail in reference 55, all samples were amplified, barcoded, purified, and prepared for sequencing on an Illumina MiSeq System (v3 chemistry, 2 × 300 bp) at the Joint Microbiome Facility of the Medical University of Vienna and the University of Vienna.
Individual amplicon pools were extracted from the raw sequencing data using the FASTQ workflow in BaseSpace (Illumina) with default parameters, allowing one mismatch for the 6-nucleotide (nt) library indexes. The input data were filtered for PhiX contamination with BBDuk (BBTools) (56). Further demultiplexing of each amplicon pool library into single amplicon libraries was performed with the python package demultiplex (Laros JFJ, github.com/jfjlaros/demultiplex), allowing one mismatch for barcodes and two mismatches for linkers and primer sequences, respectively.
ASVs were inferred using the DADA2 R package version 1.14.1 (57) with R version 3.6.1 (58) applying the recommended workflow (59) in pooled mode using all amplicon libraries per sequencing run. 16S rRNA region V4/V3-4 amplicon FASTQ reads were trimmed at 150/220 nt with allowed expected error 2. rRNA region ITS2 amplicon FASTQ reads were trimmed at 230/230 nt with allowed expected errors 4 and 6. Taxonomy was assigned to ASVs based on SILVA database SSU Ref NR 99 release 138.1 (https://www.ncbi.nlm.nih.gov/pubmed/23193283) and UNITE all eukaryotes general FASTA version 8.2 (60) using SINA version 1.6.1 (https://www.ncbi.nlm.nih.gov/pubmed/22556368).
The sequencing of V4 16S rRNA resulted in 1,187,393 reads, and the ITS sequencing resulted in a total of 359,819 reads. After filtering, the remaining 1,047,022 reads of the V4 16S rRNA gene clustered and affiliated with 7,915 prokaryotic ASVs and 316,582 reads of ITS2 fungal rRNA gene clustered and affiliated with 1,547 fungal ASVs were further analyzed. Due to a low number of reads, 7 samples were excluded from of the prokaryotic data set (sample T4_spring 2019, T2_summer 2019, T5_winter 2020, T18_ spring 2020, samples T14_summer 2020, and T16_summer2020, and T14_winter 2021), and further analysis was conducted on a total of 139 samples. A total of three samples (sample T4_spring 2019, IS19_summer 2019, and T14_winter 2021) were removed from the fungal data set because of a low number of reads. The analysis was conducted on 143 samples. In the prokaryotic data set, the lowest number of reads was determined within tributaries in spring 2020 (1,165 reads), and the highest was in interlake streams in summer 2019 (17,803 reads). The lowest number of reads within fungal data set was determined in tributaries in spring 2020 (105 reads), and the highest number was in interlake streams in winter 2021 (9,732 reads).
Statistical analysis.
Statistical analyses were performed in the R environment (version 4.1.1) (58) using the packages phyloseq (61), vegan (62), dplyr (63), and ggplot2 (64). The β-diversity of environmental parameters was calculated by performing a PCA on a distance matrix of Z-score-normalized data using vegan.
Prior to statistical analysis, ASVs classified as eukaryotes, mitochondria, or chloroplasts in the 16S rRNA gene amplicon data set were removed. In addition, unassigned ASVs at the phylum level, singletons, and doubletons were removed from both data sets.
For estimation of α-diversity, rarefaction was computed on the data set by subsampling libraries to the smallest library size. α-Diversity was estimated as richness according to Chao1 (65), evenness was estimated according to reference 66, and diversity was estimated according to the Shannon index (67). The analysis of variance (ANOVA) test and Tukey’s HSD post hoc test were used to test for differences in microbial communities’ α-diversity among stream types. Taxonomic abundance was examined by removing unassigned taxa and by forming an “others” group of all the taxa with relative abundance less than 1%. Visualization of the distance decay relationship (DDR) relied on community similarity calculated using a Bray-Curtis index after normalization of the data set through cumulative sum scaling with the metagenomeSeq package (68). Geographic distance was measured using a “Vincenty” (ellipsoid) great circle distance to take into account Earth curvature, relying on packages enmSdm (69) and geosphere (v1.5.10) (70). The DDR was calculated between the samples belonging to the tributary with the longest flow, the Bijela River, all interlake stream samples, and the Korana River, the main outflow. Abundance-occupancy analysis was used to detect core taxa of entire freshwater network ecosystem (71). Each taxon’s mean relative abundance was calculated across the data set, log-transformed, and plotted against the proportion of discrete samples in which it occurred (with occupancy of 1 to be found in all samples). Taxa found in all sampling points were considered core taxa. Shared and unique ASVs of prokaryotic and fungal communities were depicted in a Venn diagram using the package ggVennDiagram (72). To capture the difference between prokaryotic and fungal β-diversities on a spatiotemporal scale between different stream types, sampling season, and sampling year, a PERMANOVA test was carried out on distance matrices based on Bray-Curtis dissimilarity and visualized via principal coordinate analysis (PCoA). The function envfit of the package vegan was applied to the results of PCoA to visualize the correlations with environmental factors.
A biodiversity ecological null model was used to evaluate processes driving microbial community composition (11, 73). Based on the rarified abundance ASV tables and amplicon phylogenetic trees, we calculated the β-nearest taxon index (βNTI) of prokaryotic and fungal communities. In order to test whether there was a significant difference between molecular and phylogenetic turnover between the observed microbial assemblages, the β-mean nearest taxon index (βMNTD) was calculated. Further, the βNTI was calculated as the difference between the observed βMNTD and the null distribution. Deterministic processes (variable or homogeneous selection) dominated when βNTI is greater than 2 or less than −2. Values within the range of 2 > NTI > −2 indicate the dominance of stochastic processes (homogenizing dispersal or dispersal limitation) or random drift. On the basis of the abundance of microbial communities, we calculated the Raup-Crick (RC) β-diversity to distinguish stochastic processes. Assemblies were structured by dispersal limitation if RC > +0.95, homogenizing dispersal if RC < −0.95, or random processes acting alone (ecological drift) if RC falls between −0.95 and +0.95.
Data availability.
Raw sequence reads were deposited in the EBI-EMBL ENA database, project PRJEB57627.
ACKNOWLEDGMENTS
This research was partially supported under project STIM-REI contract KK.01.1.1.01.0003; a project funded by the European Union through the European Regional Development Fund and the Operational Program Competitiveness and Cohesion 2014-2020 (contract KK.01.1.1.01); by DNKVODA project contract KK.01.2.1.02.0335; and by the Croatian Science Foundation through grants HRZZ IP-2020-02-9021 and DOK-2018-09-1550.
We acknowledge Jasmin Schwarz, Gudrun Kohl, and Bela Hausmann from the Joint Microbiome Facility for their support with sample and data processing. We also acknowledge Maja Mitrović and Ema Kostešić for their support with sampling and DNA extraction.
Contributor Information
Sandi Orlić, Email: sorlic@irb.hr.
Michael J. Imperiale, University of Michigan
REFERENCES
- 1.Sorensen JPR, Maurice L, Edwards FK, Lapworth DJ, Read DS, Allen D, Butcher AS, Newbold LK, Townsend BR, Williams PJ. 2013. Using boreholes as windows into groundwater ecosystems. PLoS One 8:e70264. doi: 10.1371/journal.pone.0070264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crump BC, Adams HE, Hobbie JE, Kling GW. 2007. Biogeography of bacterioplankton in lakes and streams of an arctic tundra catchment. Ecology 88:1365–1378. doi: 10.1890/06-0387. [DOI] [PubMed] [Google Scholar]
- 3.Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. 2011. A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev 75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stadler M, del Giorgio PA. 2021. Terrestrial connectivity, upstream aquatic history and seasonality shape bacterial community assembly within a large boreal aquatic network. ISME J 16:937–947. doi: 10.1038/s41396-021-01146-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindström ES, Kamst-Van Agterveld MP, Zwart G. 2005. Distribution of typical freshwater bacterial groups is associated with pH, temperature, and lake water retention time. Appl Environ Microbiol 71:8201–8206. doi: 10.1128/AEM.71.12.8201-8206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niño-García JP, Ruiz-González C, A del Giorgio P. 2016. Interactions between hydrology and water chemistry shape bacterioplankton biogeography across boreal freshwater networks. ISME J 10:1755–1766. doi: 10.1038/ismej.2015.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staley C, Gould TJ, Wang P, Phillips J, Cotner JB, Sadowsky MJ. 2016. Sediments and soils act as reservoirs for taxonomic and functional bacterial diversity in the Upper Mississippi River. Mol Ecol 71:814–824. doi: 10.1007/s00248-016-0729-5. [DOI] [PubMed] [Google Scholar]
- 8.Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JBH. 2012. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol 10:497–506. doi: 10.1038/nrmicro2795. [DOI] [PubMed] [Google Scholar]
- 9.Vellend M. 2016. The theory of ecological communities. Princeton University Press, Princeton, NJ. [Google Scholar]
- 10.Zhou J, Ning D. 2017. Stochastic community assembly: does it matter in microbial ecology? Microbiol Mol Biol Rev 81:e00002-17. doi: 10.1128/MMBR.00002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stegen JC, Lin X, Fredrickson JK, Chen X, Kennedy DW, Murray CJ, Rockhold ML, Konopka A. 2013. Quantifying community assembly processes and identifying features that impose them. ISME J 7:2069–2079. doi: 10.1038/ismej.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leibold MA, Chase JM. 2018. Metacommunity ecology. Princeton University Press, Princeton, NJ. [Google Scholar]
- 13.Chase JM, Myers JA. 2011. Disentangling the importance of ecological niches from stochastic processes across scales. Philos Trans R Soc Lond B Biol Sci 366:2351–2363. doi: 10.1098/rstb.2011.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson CE, Sadro S, Melack JM. 2009. Contrasting the influences of stream inputs and landscape position on bacterioplankton community structure and dissolved organic matter composition in high-elevation lake chains. Limnol Oceanogr 54:1292–1305. doi: 10.4319/lo.2009.54.4.1292. [DOI] [Google Scholar]
- 15.Staley C, Unno T, Gould TJ, Jarvis B, Phillips J, Cotner JB, Sadowsky MJ. 2013. Application of Illumina next-generation sequencing to characterize the bacterial community of the Upper Mississippi River. J Appl Microbiol 115:1147–1158. doi: 10.1111/jam.12323. [DOI] [PubMed] [Google Scholar]
- 16.Caillon F, Besemer K, Peduzzi P, Schelker J. 2021. Soil microbial inoculation duringfloodevents shapes headwater stream microbial communities and diversity. Microb Ecol 82:591–601. doi: 10.1007/s00248-021-01700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Melo ML, Bertilsson S, Amaral JHF, Barbosa PM, Forsberg BR, Sarmento H. 2019. Flood pulse regulation of bacterioplankton community composition in an Amazonian floodplain lake. Freshwater Biol 64:108–120. doi: 10.1111/fwb.13198. [DOI] [Google Scholar]
- 18.Read DS, Gweon HS, Bowes MJ, Newbold LK, Field D, Bailey MJ, Griffiths RI. 2015. Catchment-scale biogeography of riverine bacterioplankton. ISME J 9:516–526. doi: 10.1038/ismej.2014.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savio D, Sinclair L, Ijaz UZ, Parajka J, Reischer GH, Stadler P, Blaschke AP, Blöschl G, Mach RL, Kirschner AKT, Farnleitner AH, Eiler A. 2015. River bacterioplankton diversity. Environ Microbiol 17:4994–5007. doi: 10.1111/1462-2920.12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Šemnički P, Prevušić A, Ivković M, Čmrlec K, Mihaljević Z. 2012. Tufa barriers from a caddisfly’s point of view: streams or lake outlets? Int Rev Hydrobiol 97:465–484. doi: 10.1002/iroh.201101500. [DOI] [Google Scholar]
- 21.Tang X, Xie G, Shao K, Hu Y, Cai J, Bai C, Gong Y, Gao G. 2020. Contrast diversity patterns and processes of microbial community assembly in a river-lake continuum across a catchment scale in northwestern China. Environ Microbiome 15:10. doi: 10.1186/s40793-020-00356-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, Wang P, Wang C, Wang X, Miao L, Liu S, Yuan Q, Sun S. 2020. Fungal community demonstrates stronger dispersallimitation and less network connectivity than bacterialcommunity in sediments along a large river. Environ Microbiol 22:832–849. doi: 10.1111/1462-2920.14795. [DOI] [PubMed] [Google Scholar]
- 23.Deiner K, Fronhofer EA, Mächler E, Walser JC, Altermatt F. 2016. Environmental DNA reveals that rivers are conveyer belts of biodiversity information. Nat Commun 7:12544. doi: 10.1038/ncomms12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LeBrun ES, Taylor DL, King RS, Back JA, Kang S. 2018. Rivers may constitute an overlooked avenue of dispersal for terrestrial fungi. Fungal Ecol 32:72–79. doi: 10.1016/j.funeco.2017.12.003. [DOI] [Google Scholar]
- 25.Matsuoka S, Sugiyama Y, Sato H, Katano I, Harada K, Doi H. 2019. Spatial structure of fungal DNA assemblages revealed with eDNA metabarcoding in a forest river network in western Japan. Metabarcoding Metagenom 3:e36335. doi: 10.3897/mbmg.3.36335. [DOI] [Google Scholar]
- 26.Zhang H, Yan Y, Lin T, Xie W, Hu J, Hou F, Han Q, Zhu X, Zhang D. 2022. Disentangling the mechanisms shaping the prokaryotic communities in a eutrophic bay. Microbiol Spectr 10:e01481-22. doi: 10.1128/spectrum.01481-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruiz-González C, Niño-García J, Kembel S, A del Giorgio P. 2017. Identifying the core seed bank of a complex boreal bacterial metacommunity. ISME J 11:2012–2021. doi: 10.1038/ismej.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yue F-J, Waldron S, Li S-L, Wang Z-J, Zeng J, Xu S, Zhang Z-C, Oliver D. 2019. Land use interacts with changes in catchment hydrology to generate chronic nitrate pollution in karst waters and strong seasonality in excess nitrate export. Science of The Total Environment 696:134062. doi: 10.1016/j.scitotenv.2019.134062. [DOI] [Google Scholar]
- 29.Ylla I, Peter H, Romaní AM, Tranvik LJ. 2013. Different diversity–functioning relationship in lake and stream bacterial communities. FEMS Microbiol Ecol 85:95–103. doi: 10.1111/1574-6941.12101. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Qu X, Elser JJ, Peng W, Zhang M, Ren Z, Zhang H, Zhang Y, Yang H. 2019. Impact of nutrient and stoichiometry gradients on microbial assemblages in Erhai Lake and its input streams. Water 11:1711. doi: 10.3390/w11081711. [DOI] [Google Scholar]
- 31.Kostanjšek R, Pašić L, Daims H, Sket B. 2013. Structure and community composition of sprout-like bacterial aggregates in a Dinaric karst subterranean stream. Microb Ecol 66:5–18. doi: 10.1007/s00248-012-0172-1. [DOI] [PubMed] [Google Scholar]
- 32.Madigan MT, Jung DO, Woese CR, Achenbach LA. 2000. Rhodoferax antarcticus sp. nov., a moderately psychrophilic purple nonsulfur bacterium isolated from an Antarctic microbial mat. Arch Microbiol 173:269–277. doi: 10.1007/s002030000140. [DOI] [PubMed] [Google Scholar]
- 33.Bandh SA, Shafi S, Shameem N. 2019. Freshwater microbiology. Perspectives of Bacterial Dynamics in Lake Ecosystems. P.G. Department of Environmental Science, Sri Pratap College Campus, Cluster University, Srinagar, India. [Google Scholar]
- 34.Espín Y, Menchén A, Moreno JL, Sanz D, Álvarez-Ortí M, Fernández JA, Gómez-Alday JJ. 2021. Water and sediment bacterial communities in a small Mediterranean, oxygen-stratified, saline lake (Lake Alboraj, SE Spain). Applied Sciences 11:6309. doi: 10.3390/app11146309. [DOI] [Google Scholar]
- 35.Brad T, Bizic M, Ionescu D, Chiriac CM, Kenesz M, Roba C, Ionescu A, Fekete A, Mirea IC, Moldovan OT. 2022. Potential for natural attenuation of domestic and agricultural pollution in karst groundwater environments. Water 14:1597. doi: 10.3390/w14101597. [DOI] [Google Scholar]
- 36.Xiao D, He X, Zhang W, Hu P, Sun M, Wang K. 2022. Comparison of bacterial and fungal diversity and network connectivity in karst and non-karst forests in southwest China. Sci Total Environ 822:153179. doi: 10.1016/j.scitotenv.2022.153179. [DOI] [PubMed] [Google Scholar]
- 37.Chen Q, Chen Y, Yang J, Maberly SC, Zhang J, Ni J, Wang G, Tonina D, Xiao L, Ma H. 2021. Bacterial communities in cascade reservoirs along a large river. Limnol Oceanogr 66:4363–4374. doi: 10.1002/lno.11967. [DOI] [Google Scholar]
- 38.He Q, Xiao Q, Fan J, Zhao H, Cao M, Zhang C, Jiang Y. 2022. The impact of heterotrophic bacteria on recalcitrant dissolved organic carbon formation in a typical karstic river. Sci Total Environ 815:152576. doi: 10.1016/j.scitotenv.2021.152576. [DOI] [PubMed] [Google Scholar]
- 39.Kantor RS, Wrighton KC, Handley KM, Sharon I, Hug LA, Castelle CJ, Thomas BC, Banfield JF. 2013. Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. mBio 4:e00708-13. doi: 10.1128/mBio.00708-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luef B, Frischkorn KR, Wrighton KC, Holman HYN, Birarda G, Thomas BC, Singh A, Williams KH, Siegerist CE, Tringe SG, Downing KH, Comolli LR, Banfield JF. 2015. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat Commun 6:6372. doi: 10.1038/ncomms7372. [DOI] [PubMed] [Google Scholar]
- 41.Smart C, Worthington SRH. 2004. Springs, p. 1495–1505. In Gunn J. (ed). Encyclopedia of caves and karst science. Taylor and Francis, New York, NY. [Google Scholar]
- 42.David GM, López-García P, Moreira D, Alric B, Deschamps P, Bertolino P, Restoux G, Rochelle-Newall E, Thébault E, Simon M, Jardillier L. 2021. Small freshwater ecosystems with dissimilar microbial communities exhibit temporal patterns. Mol Ecol 30:2162–2177. doi: 10.1111/mec.15864. [DOI] [PubMed] [Google Scholar]
- 43.Gleason FH, Marano AV, Digby AL, Al-Shugairan N, Lilje O, Steciow MM, Barrera MD, Inaba S, Nakagiri A. 2011. Patterns of utilization of different carbon sources by Chytridiomycota. Hydrobiologia 659:55–64. doi: 10.1007/s10750-010-0461-y. [DOI] [Google Scholar]
- 44.Wurzbacher C, Warthmann N, Bourne EC, Attermeyer K, Allgaier M, Powell JR, Detering H, Mbedi S, Grossart HP, Monaghan MT. 2016. High habitat-specificity in fungal communities in oligo-mesotrophic, temperate Lake Stechlin (North-East Germany). MycoKeys 16:17–44. doi: 10.3897/mycokeys.16.9646. [DOI] [Google Scholar]
- 45.Seena S, Bärlocher F, Sobral O, Gessner MO, Dudgeon D, McKie BG, Chauvet E, Boyero L, Ferreira V, Frainer A, Bruder A, Matthaei CD, Fenoglio S, Sridhar KR, Albariño RJ, Douglas MM, Encalada AC, Garcia E, Ghate SD, Giling DP, Gonçalves V, Iwata T, Landeira-Dabarca A, McMaster D, Medeiros AO, Naggea J, Pozo J, Raposeiro PM, Swan CM, Tenkiano NSD, Yule CM, Graça MAS. 2019. Biodiversity of leaf litter fungi in streams along a latitudinal gradient. Sci Total Environ 661:306–315. doi: 10.1016/j.scitotenv.2019.01.122. [DOI] [PubMed] [Google Scholar]
- 46.Senga Y, Yabe S, Nakamura T, Kagami M. 2018. Influence of parasitic chytrids on the quantity and quality of algal dissolved organic matter (AOM). Water Res 145:346–353. doi: 10.1016/j.watres.2018.08.037. [DOI] [PubMed] [Google Scholar]
- 47.Muggia L, Grube M. 2018. Fungal diversity in lichens: from extremotolerance to interactions with algae. Life 8:15. doi: 10.3390/life8020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang N, Li Y, Wubet T, Bruelheide H, Liang Y, Purahong W, Buscot F, Ma K. 2018. Tree species richness and fungi in freshly fallen leaf litter: unique patterns of fungal species composition and their implications for enzymatic decomposition. Soil Biol Biochem 127:120–126. doi: 10.1016/j.soilbio.2018.09.023. [DOI] [Google Scholar]
- 49.Madhaiyan M, Poonguzhali S, Senthilkumar M, Pragatheswari D, Lee J-S, Lee K-C. 2015. Arachidicoccus rhizosphaerae gen. nov., sp. nov., a plant-growth-promoting bacterium in the family Chitinophagaceae isolated from rhizosphere soil. Int J Syst Evol Microbiol 65:578–586. doi: 10.1099/ijs.0.069377-0. [DOI] [PubMed] [Google Scholar]
- 50.Li Y, Naman CB, Alexander KL, Guan H, Gerwick WH. 2020. The chemistry, biochemistry and pharmacology of marine natural products from Leptolyngbya, a chemically endowed genus of Cyanobacteria. Mar Drugs 18:508. doi: 10.3390/md18100508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuroda K, Narihiro T, Shinshima F, Yoshida M, Yamaguchi H, Kurashita H, Nakahara N, Nobu MK, Noguchi TQP, Yamauchi M, Yamada M. 2022. High-rate cotreatment of purified terephthalate and dimethyl terephthalate manufacturing wastewater by a mesophilic upflow anaerobic sludge blanket reactor and the microbial ecology relevant to aromatic compound degradation. Water Res 219:118581. doi: 10.1016/j.watres.2022.118581. [DOI] [PubMed] [Google Scholar]
- 52.Mehrotra RS, Aneja KR. 1990. An Introduction to Mycology, New Delhi, India: Wiley Eastern Ltd. [Google Scholar]
- 53.Parada AE, Needham DM, Fuhrman JA. 2016. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 54.Apprill A, McNally S, Parsons R, Weber L. 2015. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol 75:129–137. doi: 10.3354/ame01753. [DOI] [Google Scholar]
- 55.Pjevac P, Hausmann B, Schwarz J, Kohl G, Herbold CW, Loy A, Berry D. 2021. An economical and flexible dual barcoding, two-step PCR approach for highly multiplexed amplicon sequencing. Front Microbiol 12:669776. doi: 10.3389/fmicb.2021.669776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bushnell B. 2014. BBMap: A fast, accurate, splice-aware aligner. sourceforge.net/projects/bbmap.
- 57.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.R Core Team. 2021. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- 59.Callahan BJ, Sankaran K, Fukuyama JA, McMurdie PJ, Holmes SP. 2016. Bioconductor workflow for microbiome data analysis: from raw reads to community analyses. F1000Res 5:1492. doi: 10.12688/f1000research.8986.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abarenkov K, Zirk A, Piirmann T, Pöhönen R, Ivanov F, Nilsson RH, Kõljalg U. 2020. UNITE general FASTA release for eukaryotes 2, version 04.02.2020. UNITE Commun doi: 10.15156/BIO/786371. [DOI] [Google Scholar]
- 61.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P. 2017. vegan: community ecology package. R package version 2.5-7. https://CRAN.R-project.org/package=vegan.
- 63.Wickham H, François R, Henry L, Müller K. 2020. Dplyr: A grammar of data manipulation. https://CRAN.R-project.org/package=dplyr.
- 64.Wickham H. 2016. Ggplot2: elegant graphics for data analysis. Springer International Publishing, New York, NY. [Google Scholar]
- 65.Chao A. 1984. Non-parametric estimation of the number of classes in a population. Scand J Stat 11:265–270. https://www.jstor.org/stable/4615964. [Google Scholar]
- 66.Pielou EC. 1966. The measurement of diversity in different types of biological collections. J Theor Biol 13:131–144. doi: 10.1016/0022-5193(66)90013-0. [DOI] [Google Scholar]
- 67.Shannon CE. 1948. A mathematical theory of commun1ication. Bell System Tech J 27:379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x. [DOI] [Google Scholar]
- 68.Paulson JN, Stine OC, Bravo HC, Pop M. 2013. Differential abundance analysis for microbial marker-gene surveys. Nat Methods 10:1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morelli TL, Smith AB, Mancini AN, Balko EA, Borgenson C, Dolch R, Farris Z, Federman S, Golden CD, Holmes SM, Irwin M, Jacobs RL, Johnson S, King T, Lehman SM, Louis EE, Jr, Murphy A, Randriahaingo HNT, Randrianarimanana HLL, Ratsimbazafy J, Razafindratsima OH, Baden AL. 2020. The fate of Madagascar’s rainforest habitat. Nat Clim Chang 10:89–96. doi: 10.1038/s41558-019-0647-x. [DOI] [Google Scholar]
- 70.Hijmans RJ. 2019. geosphere: spherical trigonometry. R package version 1.5.10. https://CRAN.R-project.org/package=geosphere.
- 71.Shade A, Stopnisek N. 2019. Abundance-occupancy distributions to prioritize plant core microbiome membership. Curr Opin Microbiol 49:50–58. doi: 10.1016/j.mib.2019.09.008. [DOI] [PubMed] [Google Scholar]
- 72.Gao CH, Yu G, Cai P. 2021. ggVennDiagram: an intuitive, easy-to-use, and highly customizable R package to generate Venn diagram. Front Genet 12:706907. doi: 10.3389/fgene.2021.706907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stegen JC, Lin X, Fredrickson JK, Konopka AE. 2015. Estimating and mapping ecological processes influencing microbial community assembly. Front Microbiol 6:370. doi: 10.3389/fmicb.2015.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Environmental parameters measured at the sampling locations through six seasons. Download FIG S1, TIF file, 1.0 MB (1,022.5KB, tif) .
Copyright © 2023 Čačković et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Environmental parameters measured at 25 locations through 6 seasons. Download Table S1, DOCX file, 0.03 MB (31.4KB, docx) .
Copyright © 2023 Čačković et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
α-Diversity indices of prokaryotic (A) and fungal (B) communities determined by Pieolu Evenness, Chao1, and Shannon index. The samples are divided based on sampling season and then by stream type and color marked as follows: blue, tributaries; red, interlake streams; and yellow, Korana River. The sample abbreviations refer to different sampling locations. Download FIG S2, TIF file, 0.9 MB (892.1KB, tif) .
Copyright © 2023 Čačković et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Tukey’s honestly significant difference (HSD) test results. Download Table S2, DOCX file, 0.01 MB (15.5KB, docx) .
Copyright © 2023 Čačković et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundance of the total bacterial community structure (phylum level), divided by sampling seasons. The sample abbreviations refer to sampling locations. The “Other” group contains phyla with relative abundance of less than 1%. Download FIG S3, TIF file, 1.0 MB (1MB, tif) .
Copyright © 2023 Čačković et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundance of the total fungal community structure (phylum level), divided by sampling seasons. The sample abbreviations refer to sampling locations. Download FIG S4, TIF file, 0.6 MB (587.3KB, tif) .
Copyright © 2023 Čačković et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Permutational multivariate analysis of variance (PERMANOVA) test results. Download Table S3, DOCX file, 0.01 MB (13.3KB, docx) .
Copyright © 2023 Čačković et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Venn diagram representing the number of shared and unique bacterial (A) and fungal (B) ASVs between three different stream types: tributaries (green), interlake streams (red), and the Korana River (blue). Download FIG S5, TIF file, 0.8 MB (810.4KB, tif) .
Copyright © 2023 Čačković et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundance of 30 most abundant core microbiome bacterial taxa, grouped by stream type and color-coded by family. Download FIG S6, TIF file, 0.5 MB (505.7KB, tif) .
Copyright © 2023 Čačković et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundance of 30 most abundant and 30 most rare bacterial taxa unique for tributaries. The panels are color-coded by family. Download FIG S7, TIF file, 0.7 MB (679.3KB, tif) .
Copyright © 2023 Čačković et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Raw sequence reads were deposited in the EBI-EMBL ENA database, project PRJEB57627.