ABSTRACT
Although changes in gut microbiome have been associated with the development of T2D and its complications, the role of the gut virome remains largely unknown. Here, we characterized the gut virome alterations in T2D and its complications diabetic nephropathy (DN) by metagenomic sequencing of fecal viral-like particles. Compared with controls, T2D subjects, especially those with DN, had significantly lower viral richness and diversity. 81 viral species were identified to be significantly altered in T2D subjects, including a decrease in some phages (e.g. Flavobacterium phage and Cellulophaga phaga). DN subjects were depleted of 12 viral species, including Bacteroides phage, Anoxybacillus virus and Brevibacillus phage, and enriched in 2 phages (Shigella phage and Xylella phage). Multiple viral functions, particularly those of phage lysing host bacteria, were markedly reduced in T2D and DN. Strong viral-bacterial interactions in healthy controls were disrupted in both T2D and DN. Moreover, the combined use of gut viral and bacterial markers achieved a powerful diagnostic performance for T2D and DN, with AUC of 99.03% and 98.19%, respectively. Our results suggest that T2D and its complication DN are characterized by a significant decrease in gut viral diversity, changes in specific virus species, loss of multiple viral functions, and disruption of viral-bacterial correlations. The combined gut viral and bacterial markers have diagnostic potential for T2D and DN.
KEYWORDS: Type 2 diabetes, gut virome, metagenomics, microbiota, biomarker
1. Introduction
Type 2 diabetes (T2D) is a common chronic metabolic disease. In recent years, the global prevalence of T2D is rising rapidly due to sedentary lifestyles and unhealthy diets.1 Chronic exposure to hyperglycemia in subjects with T2D will cause various microvascular complications, such as diabetic nephropathy (DN).2 DN has been reported to occur in 20% to 50% of diabetic subjects.3 It is the leading cause of end-stage renal disease worldwide, and is associated with high morbidity and mortality. Genetics, diet and lifestyle are considered major contributors to the development of T2D and DN. However, much other factors remain to be elucidated.
In recent years, growing evidence suggests that gut microbiota plays an important role in human health and disease.4 However, most studies have focused on gut bacteria, and the role of the enterovirus community (virome) has been largely unexplored. The gut virome is an important component of the gut microbiota. It is mainly composed of bacteriophages (phages), which are viruses that specifically attack bacteria.5,6 Several studies have suggested that gut virome signatures are associated with the development of some chronic diseases in humans, such as nonalcoholic fatty liver disease (NAFLD), colorectal cancer, alcoholic hepatitis, and inflammatory bowel disease.7–11 In 2018, a study first linked gut phages to T2D and observed a significant increase in gut phage numbers in T2D subjects.12 Another study showed that fecal virome transplantation from lean donors reduced weight gain and normalized glycemic tolerance in diet-induced obese mice.13 Furthermore, changes in the gut virome were reported in obese subjects with or without T2D, and obese subjects with T2D had a more perturbed gut viral dysbiosis than obese alone.14 These findings suggest the presence of disease-specific gut virome in obesity and T2D. However, the association between gut virome and T2D has not been fully elucidated. Moreover, the gut virome signature of DN has not been explored so far. In this study, we aimed to characterize changes in the gut virome and their correlation with gut bacteria in T2D subjects, and to explore whether alterations in viral composition were associated with DN complications. The information would increase our understanding of the important role of gut microbiota in the pathogenesis of T2D and its complications, and also provide new clues for the diagnosis and treatment of these metabolic diseases.
2. Methods
2.1. Study populations
This study recruited 90 adult T2D subjects with (n = 41) or without (n = 49) DN and 42 healthy controls. Ethical approval was obtained from the Medical Ethics Committee of the Affiliated Hospital of Chengdu University of Traditional Chinese Medicine (2020KL–060), and informed consent was obtained from all subjects. T2D were diagnosed by physicians according to the 1999 WHO criteria. Then, T2D subjects were divided into two groups based on urinary albumin/creatinine ratio (UACR) and/or estimated glomerular filtration rate (eGFR) levels: T2D without nephropathy (T2D-non-Ne, UACR <30 mg/g and/or eGFR >60 mL/min/1.73 m2) and T2D with nephropathy (T2D-Ne, UACR ≥30 mg/g and/or eGFR ≤60 mL/min/1.73 m2). Subjects were excluded if they had renal failure, kidney transplantation, gastrointestinal diseases, malignancy, infectious diseases, or other diseases known to affect gut microbes, or had received antibiotics, probiotics or immunosuppressive medications in the previous 3 months. Clinical data are presented in Table 1.
Table 1.
Clinical characteristics of the study cohort.
| Variables | Controls (n = 42) | T2D-non-Ne (n = 49) | T2D-Ne (n = 41) | P value |
|---|---|---|---|---|
| Age, years | 52.5 (47.0–56.3) | 51.0 (45.0–58.5) | 58.0 (49.5–62.5) | .048 |
| Female, n (%) | 18 (42.9) | 16 (32.7) | 10 (24.4) | .202 |
| Body mass index, kg/m2 | 24.8 (22.8–27.0) | 24.7 (21.5–28.2) | 25.0 (22.2–28.4) | .815 |
| Fasting glucose, mmol/L | 4.9 (4.5–5.4) | 7.1 (5.3–8.5) | 8.5 (5.9–12.0) | <0.001ab |
| HbA1c, % | NA | 9.1 (7.2–10.3) | 8.5 (7.4–9.8) | .571 |
| UACR, mg/g | NA | 13.0 (7.5–20.5) | 171.8 (106.4–297.0) | <0.001c |
| eGFR, mL/min/1.73 m2 | NA | 93.4 (79.4–110.3) | 53.4 (42.9–63.9) | <0.001c |
| TC, mmol/L | 4.7 (4.2–5.1) | 4.7 (3.7–5.9) | 4.6 (3.8–5.6) | .640 |
| TG, mmol/L | 1.4 (1.1–1.9) | 1.7 (1.1–2.7) | 2.0 (1.3–2.6) | 0.004ab |
| LDL-C, mmol/L | 3.0 (2.5–3.2) | 2.8 (2.3–3.2) | 2.5 (2.0–3.6) | .271 |
| HDL-C, mmol/L | 1.3 (1.0–1.5) | 1.1 (0.9–1.3) | 0.9 (0.8–1.2) | <0.001ab |
| AST, U/L | 23.0 (21.0–28.8) | 22.0 (17.0–33.0) | 22.0 (17.0–31.5) | .525 |
| ALT, U/L | 24.5 (17.8–38.5) | 28.0 (16.5–48.5) | 28.0 (20.0–37.5) | .536 |
Values are presented as median (interquartile range) for continuous variables or number (percentage) for categorical variables. NA, not available. Two groups were compared using the Student’s t-test for normally distributed variables or Mann – Whitney U test for non-normally distributed variables. Three groups were compared using one-way ANOVA with the Tukey post-hoc test for normally distributed variables or Kruskal-Wallis test with the Dunn post-hoc test for non-normally distributed variables. Categorical variables were compared by the χ2 test. aP < 0.05 for T2D-non-Ne vs controls, bP < 0.05 for T2D-Ne vs controls, and cP < 0.05 for T2D-Ne vs T2D-non-Ne. HbA1c, glycosylated hemoglobin; UACR, urinary albumin/creatinine ratio; eGFR, estimated glomerular filtration rate; TC, total cholesterol; TG, triglycerides; LDL-C, low-density lipoprotein cholesterol; HDL-C, high-density lipoprotein cholesterol; AST, aspartate aminotransferase; ALT, alanine aminotransferase.
2.2. Viral-like particles (VLPs) enrichment and sequencing
Fecal samples (300–400 mg) were suspended in 2 mL of saline-magnesium buffer, vortexed for 10 minutes, and then centrifuged at 5,000× g for 20 minutes. Clarified suspension was passed through a 0.45 μm filter to remove residual host and bacteria cells. Additionally, to degrade the remaining bacterial and host cell membranes, the filtrate was treated with 1 mg/ml lysozyme for 30 min at 37°C, followed by 0.2× volume of chloroform for 10 min at room temperature. Non-viral nucleic acid was digested by treatment with 200 U Benzonase (Millipore) and 0.1 mg/ml RNase A (Sangon Biotech), followed by heat inactivation at 65°C for 10 min. Viral DNA was extracted by Qiagen MinElute Virus Spin Kit following the manufacturer’s instructions. Shotgun libraries were constructed using VAHTS® Universal Plus DNA Library Prep Kit for Illumina (Vazyme Biotech Co., Ltd). Sequencing was performed on an Illumina NovaSeq 6000 platform with 2 × 150 bp pair-end reads. The detailed procedures for sequencing data processing, viral contigs identification and taxonomy annotation are presented in the Supplementary Methods.
2.3. Virome function analysis
Functional annotation of virus sequences was performed using the HUMANN3 pipeline based on Gene ontology (GO) and Pfam protein family databases. Predictive functions were collapsed by gene family identity, and abundance values were expressed as RPK (read per kilobase). The RPK threshold value > 10 was used to define the presence of a function in a sample. In addition, LEfSe analysis was performed (http://huttenhower.sph.harvard.edu/galaxy/) to determine differences in virus function between groups, with the threshold of FDR-adjusted p < .05 and LDA score > 2.
2.4. Bacterial DNA extraction and 16s rRNA sequencing
Bacterial DNA was extracted from stool samples using the CTAB/SDS method. The V4 region of the bacterial 16S rRNA gene was amplified by PCR using primers 515F and 806 R. Sequencing libraries were generated using TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, USA). Sequencing was performed on an Illumina NovaSeq 6000 platform. Quality control and data analysis of raw reads were performed with the DADA2 pipeline implemented in QIIME2.15,16 Briefly, the DADA2 plugin was used to generate amplicon sequencing variants (ASVs) of individual sample and construct an ASVs abundance table. Taxonomical assignment of the preprocessed ASVs was performed using the RDP classifier.17 To obtain the 16S relative microbiome profiling matrix, ASVs abundance information was obtained by normalizing the sequence number corresponding to the sample with the least sequences.
2.5. Statistical analysis
The abundance tables of both virome and bacteriome were imported into R environment v4.0.5 for statistical analysis. Alpha diversity was calculated using the phyloseq package in R. Two-tailed Wilcoxon’s rank sum test was used to determine statistically significant differences for several alpha diversity indices between 2 groups or 3 groups. For beta diversity, principal coordinate analysis (PCoA) was evaluated based on Bray-Curtis distance using the vegan package in R. Statistical significance was determined by permutational multivariate analysis of variance (PERMANOVA) with permutations done 999 times. Multivariate association with linear models (MaAsLin2) was used to identify differential microbial taxa between groups while controlling for confounders. Only the taxa with relative abundance higher than 0.01% and prevalence higher than 10% were selected for MaAsLin2 analysis. To characterize the relationship between gut virome and bacteriome, Spearman’s correlations were calculated and heatmaps were constructed using OriginPro 9.8.0.200 software. Moreover, eXtreme Gradient Boosting (XGBoost) was used to construct a classification model to explore the potential of gut virome in disease diagnosis and prediction. It was performed in R using the XGBoost package. In each case, 80% of the samples were used for model training and the remaining 20% were used to test the performance of the model. Parameters were optimized using 5-fold cross validation. The receiver operating characteristic (ROC) curves of two classification and three classification models were plotted using the plotROC and multiROC packages, respectively.
3. Results
3.1. Clinical characteristics of the study population
A total of 132 participants were enrolled. Most of the study subjects were male. There were significant differences in age between the three groups, and T2D-Ne subjects had an older median age (58.0 years). Compared with healthy controls, subjects in the T2D-non-Ne and T2D-Ne groups showed significantly higher levels of fasting glucose and TG, while lower levels of HDL-C. Furthermore, T2D-Ne subjects had significantly higher UACR levels and lower eGFR levels compared with T2D-non-Ne subjects. The median UACR and eGFR values in T2D-Ne subjects were 171.8 mg/g and 53.4 mL/min/1.73 m2, respectively, indicating kidney damage. Other clinical parameters were comparable between groups.
3.2. Diversity and compositional alterations of gut virome in T2D
On average, 75674,916 clean reads were obtained from the VLP metagenomic sequencing (Table S1, Supporting Information). Alpha diversity refers to intra-community diversity, which can reflect the richness and diversity of microbial community.18 Beta diversity focuses on inter-community diversity and can be used to evaluate the differences of microbial communities between different groups.18,19 We first compared viral alpha diversity in T2D and healthy subjects. T2D subjects had a significantly lower viral diversity (Shannon and Simpson indices) and richness (Chao1 index) compared with healthy controls at the contig level (Figure 1). Furthermore, using principal coordinates analysis (PCoA) based on the Bray-Curtis distance, we assessed the differences in viral communities (i.e., beta diversity) between T2D and controls. The results showed that T2D and healthy subjects were significantly divided into 2 distinct clusters (R2 = 0.026, P = 0.001; Figure 1D). These data suggest that the gut virome profiles of T2D subjects are significantly different from those of healthy controls.
Figure 1.
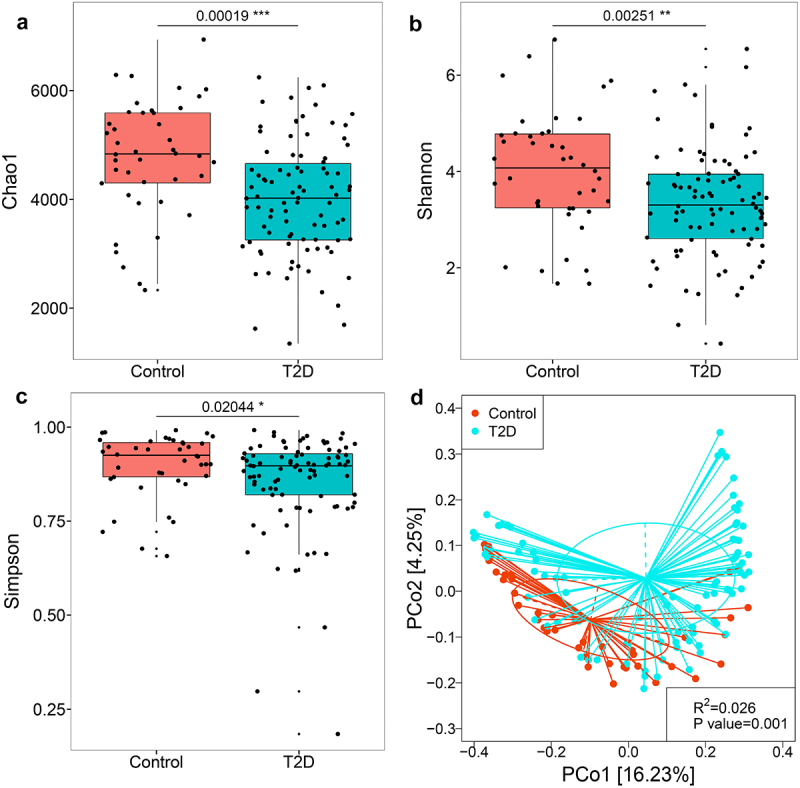
Alterations of the gut virome in T2D subjects compared with healthy controls. Differences between groups in viral richness based on the (a) Chao1 index and viral diversity based on the (b) Shannon and (c) Simpson indices at the contig level. For the box plots, the boxes extend from the first to the third quartile (25th to 75th percentiles), with the center line indicating the median. (d) PCoA analysis based on Bray-Curtis distance showing differences in gut viral community between T2D and healthy controls.
We also examined the association of age, gender, obesity, and several commonly used drugs with gut viral alpha diversity. There was no significant correlation between age and the 3 alpha diversity indices (Figure S1, Supporting Information). Similarly, no significant differences were observed for the 3 indices between obese and lean subjects. Furthermore, there was no significant difference in viral diversity (Shannon and Simpson indices) between men and women in T2D subjects, although their Chao1 richness was different. Metformin, sulfonylureas, non-steroidal anti-inflammatory drugs (NSAIDs), and statins are commonly used drugs in patients with T2D. We found no significant difference in viral diversity between subjects taking these drugs and those not taking them (Figure S2, Supporting Information).
The compositional differences in the gut virome of T2D and controls were further investigated by using MaAsLin2. At the order level, Caudovirales which comprise phages were the predominant viruses, and 6 viral taxa were found to be significantly decreased in T2D subjects compared with healthy controls (Figure S3, Supporting Information). At the family level, Microviridae were the dominant viruses, and 12 viral families showed significant differences between groups. Likewise, at the species level, 81 viral species were identified to be significantly altered in T2D subjects (Table S2, Supporting Information). Notably, 78% of them are phages. For example, Flavobacterium phage and Staphylococcus phage were significantly decreased in T2D subjects compared with healthy controls (Figure 2). Collectively, the above results indicate that T2D subjects have a significant gut viral dysbiosis, including changes in both diversity and taxonomy.
Figure 2.
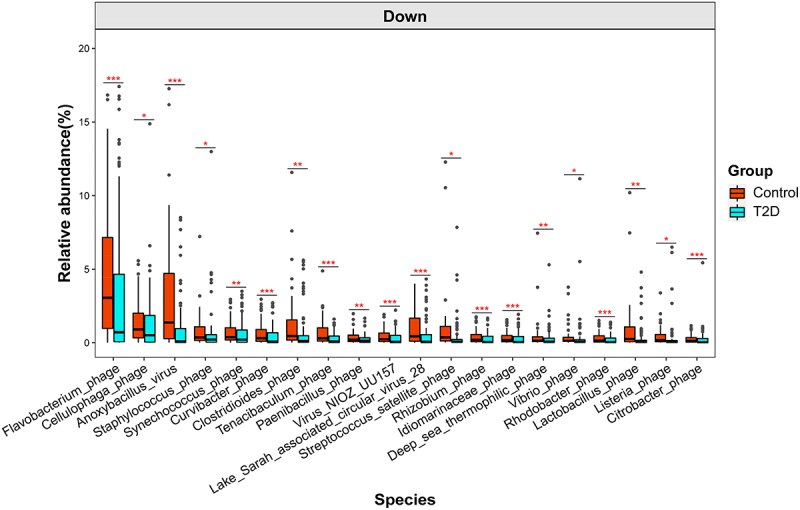
Differential viral species between T2D and healthy controls as determined by MaAsLin2 analysis and adjustment for confounders, including age, gender and DN (only the top 20 most abundant species are shown). q < 0.05 was considered to be statistically significant. *q <0.05, **q <0.01 and ***q <0.001. For the box plots, the boxes extend from the first to the third quartile (25th to 75th percentiles), with the center line indicating the median.
3.3. Diversity and compositional alterations of gut virome in DN
To determine whether gut viral dysbiosis occurs in DN complications, we further divided T2D subjects into T2D-non-Ne and T2D-Ne groups based on UACR and/or eGFR levels. Analysis of alpha diversity revealed a marked decrease in viral Chao1 richness in both subjects with and without DN compared with healthy controls (Figure 3). However, no significant differences were observed in viral diversity as measured by the Shannon and Simpson indices between T2D-non-Ne and controls. Interestingly, T2D-Ne had a significantly lower viral diversity when compared with controls or even T2D-non-Ne (Shannon index). Moreover, PCoA analysis showed that T2D-Ne samples were significantly separated from T2D-non-Ne and controls (Figure 3D). Notably, most of the T2D-Ne samples were further away from health controls than T2D-non-Ne samples. These results indicate that alterations in gut viral diversity are more pronounced in T2D-Ne subjects compared with T2D-non-Ne.
Figure 3.
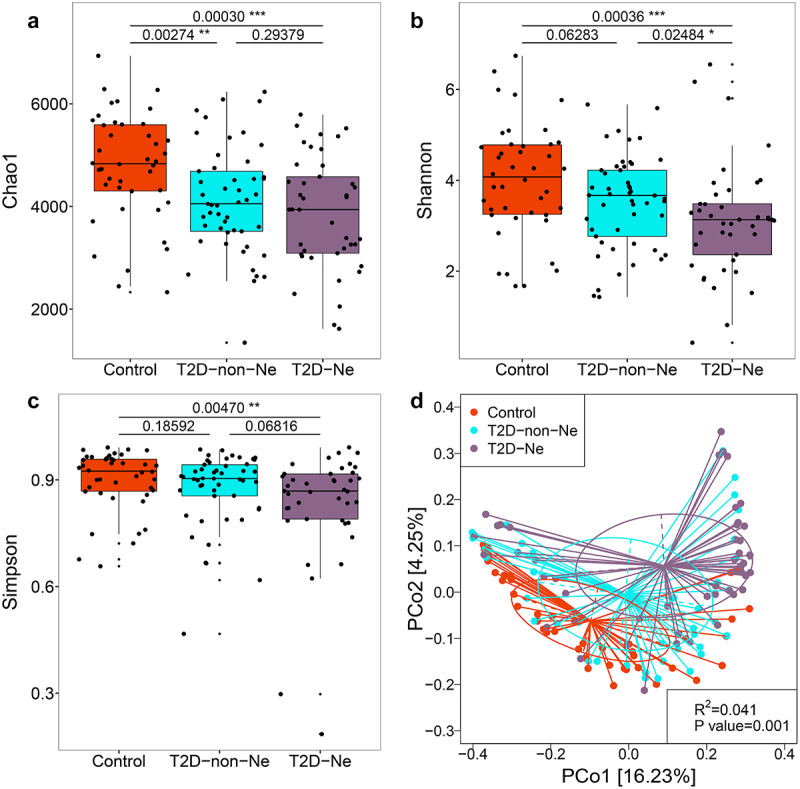
Alterations of the gut virome in T2D-Ne and T2D-non-Ne compared with healthy controls. Differences between the 3 groups in viral richness based on the (a) Chao1 index and viral diversity based on the (b) Shannon and (c) Simpson indices at the contig level. For the box plots, the boxes extend from the first to the third quartile (25th to 75th percentiles), with the center line indicating the median. (d) PCoA analysis based on Bray-Curtis distance between T2D-Ne, T2D-non-Ne and healthy controls.
We also investigated the effect of several commonly used drugs, including metformin, NSAIDs, statins and angiotensin receptor blockers, on viral alpha diversity in T2D-Ne. No significant differences were observed in the 3 diversity indices between subjects taking and not taking these drugs (Figure S4, Supporting Information), indicating that these medications had little effect on the gut virome.
MaAsLin2 analysis was used to identify gut viral signatures associated with DN complications. 2 viral orders, 9 families and 8 genera were significantly decreased, while 9 genera were increased in T2D-Ne compared with healthy controls (Figure S5, Supporting Information). At the species level, 14 viral species were identified to be associated with T2D-Ne, and 85% of them belong to phages (Figure 4; Table S3, Supporting Information). Among them, 12 species (e.g., Bacteroides phage and Anoxybacillus virus) were significantly decreased, while 2 species (Shigella phage and Xylella phage) were increased in T2D-Ne compared with controls. These data suggest that DN subjects have significant gut viral disturbances.
Figure 4.
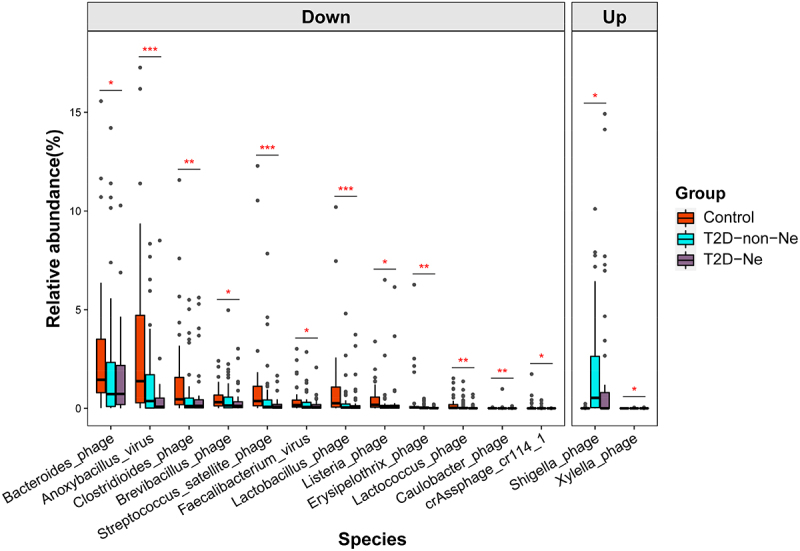
Differential viral species between T2D-Ne and healthy controls as determined by MaAsLin2 analysis and adjustment for confounders, including age and gender. q < 0.05 was considered to be statistically significant. *q <0.05, **q <0.01 and ***q <0.001. For the box plots, the boxes extend from the first to the third quartile (25th to 75th percentiles), with the center line indicating the median.
We also compared the compositional differences in the gut virome between T2D-non-Ne and healthy subjects, and 75 virus species with obvious differences were identified (Table S4, Supporting Information). Among them, 68 species did not belong to the differential viruses identified in “T2D-Ne vs Control” (Figure 5), indicating that these 68 viruses are associated with T2D only but not with DN. Furthermore, 6 species, including Erysipelothrix phage, Lactococcus phage, Faecalibacterium virus, Brevibacillus phage, Bacteroides phage, and crAssphage cr114_1, were identified as differential markers only in “T2D-Ne vs Control” but not in “T2D-non-Ne vs Control” and “T2D vs Control” (Figure 5), suggesting that DN has unique gut virome profiles and that these 6 viruses may play a potential role in DN progression. Altogether, our study revealed dysregulation of gut virome in DN subjects, including significantly decreased diversity and altered viral composition.
Figure 5.

Venn diagram of differential viral species for T2D subjects vs healthy controls, T2D-Ne subjects vs healthy controls, and T2D-non-Ne subjects vs healthy controls.
3.4. Functional alterations of gut virome in T2D and DN
To evaluate functional changes in the gut virome, HUMANN3 analysis was performed on the viral sequences against Gene ontology (GO) and Pfam protein family databases. We first compared the predictive gut virome function of T2D with that of healthy controls. Overall, diverse viral functions were lost in T2D subjects (Figure S6, Supporting Information). Moreover, differential virus functions were further screened by LEfSe analysis. We found that a large number of viral functions were down-regulated in T2D subjects compared with healthy controls (134 vs 36 for GO database, 339 vs 71 for Pfam database, Tables S5 and S6, Supporting Information). Notably, some functions related to viral replication and integration, phage lysis of bacteria host, virus-host biology, and bacterial susceptibility were decreased in T2D subjects, including ABC transporter, integrase core domain, DNA replication, transcription DNA-templated, translation, ribosome, beta-lactamase superfamily domain, glutamine metabolic process, and glutamine amidotransferase domain (Figure 6).20,21
Figure 6.
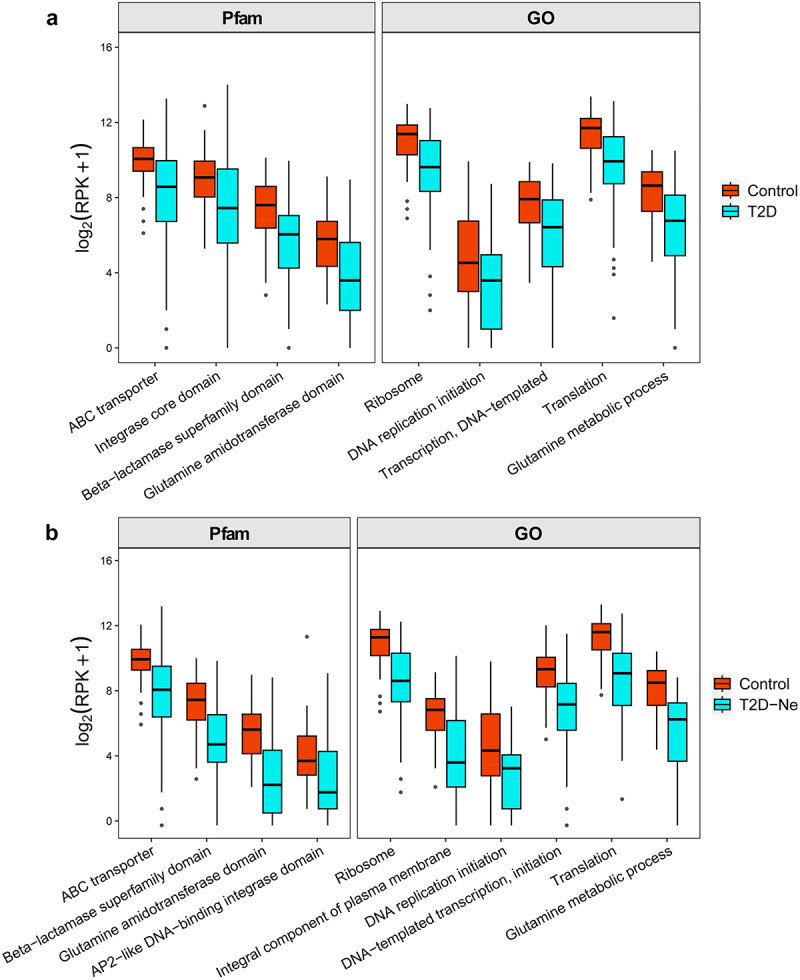
Differential viral functions between (a) T2D subjects and healthy controls and (b) T2D-Ne subjects and healthy controls against GO and Pfam protein family databases. The differential functions were determined by LEfSe analysis. For the box plots, the boxes extend from the first to the third quartile (25th to 75th percentiles), with the center line indicating the median.
We further compared the predictive viral function of T2D-Ne with that of healthy subjects. As shown in the heatmap (Figure S7, Supporting Information), T2D-Ne subjects showed loss of multiple viral functions in terms of GO and Pfam protein functions. Specifically, compared with healthy subjects, 158 and 428 viral functions were determined to be downregulated in T2D-Ne for GO and Pfam databases, respectively (Tables S7 and S8, Supporting Information). Similar to T2D, these down-regulated functions include ABC transporter, integral component of plasma membrane, DNA-templated transcription initiation, AP2-like DNA-binding integrase domain, DNA replication initiation, beta-lactamase superfamily domain, translation, ribosome, glutamine amidotransferase domain, and glutamine metabolic process (Figure 6). Taken together, multiple viral functions, especially those of phage lysing host bacteria, were markedly reduced in both T2D and DN, which may affect bacterial microbiome ecology.
3.5. Alterations of gut bacteriome in T2D and DN
Bacterial 16S rRNA sequencing was performed to evaluate alterations in the gut bacteriome in T2D and DN (Table S9, Supporting Information). No significant differences were observed in gut bacterial diversity between T2D and controls (Figure S8, Supporting Information). However, in contrast to the gut virome, bacterial richness was significantly increased in T2D subjects compared with healthy controls, which may indicate some bacterial expansion in T2D. PCoA analysis showed significant separation of T2D and controls (R2 = 0.018, P = 0.001). MaAsLin2 analysis found that some bacteria were significantly changed in T2D subjects (Figure S8 and Table S10, Supporting Information). Notably, several pathogens (e.g., Eggerthella lenta, Clostridioides difficile, Clostridium innocuum, and Streptococcus mutans) were significantly enriched in T2D subjects compared with healthy controls, which was consistent with findings from other studies.22
We further compared the differences in gut bacteriome between T2D-Ne, T2D-non-Ne, and controls. Similarly, no significant differences were observed in bacterial diversity among the three groups (Figure S9, Supporting Information). PCoA analysis revealed significant changes in the gut bacterial profiles of T2D-Ne (R2 = 0.034, P = 0.001). Moreover, MaAsLin2 analysis identified some bacterial species associated with T2D-Ne (Figure S9 and Table S11, Supporting Information), including an increase in several opportunistic pathogens (e.g., Bacteroides caccae, Eggerthella lenta and Clostridium innocuum). Collectively, these results suggest that gut bacterial disorders are present in both T2D and DN subjects.
3.6. Transkingdom correlations between gut virome and bacteriome are disrupted in T2D and DN
Gut viral-bacterial transkingdom correlations play important roles in health and disease.23,24 We first investigated the correlation between the alpha diversity of the gut virome and bacteriome. In controls, there were no significant correlations between the three indicators of virome and bacteriome (Figure S10, Supporting Information). However, significant positive correlations of viral richness and bacterial diversity were observed in T2D and T2D-Ne. Moreover, we also evaluated the correlation between viral species and bacterial species. The number of significant positive and negative correlations between groups changed significantly. Compared with healthy controls, a decreased number of negative correlations (36 vs 52, P < 0.01) but an increased number of positive correlations (32 vs 18, P < 0.01) were observed in T2D subjects (Figure 7). Similarly, T2D-non-Ne showed a reduction in the number of negative correlations compared with controls (28 vs 52, P < 0.01). Likewise, T2D-Ne subjects exhibited a significantly reduced number of negative correlations (19 vs 52, P < 0.001) and a significantly increased number of positive correlations (39 vs 18, P < 0.001) compared with healthy controls.
Figure 7.
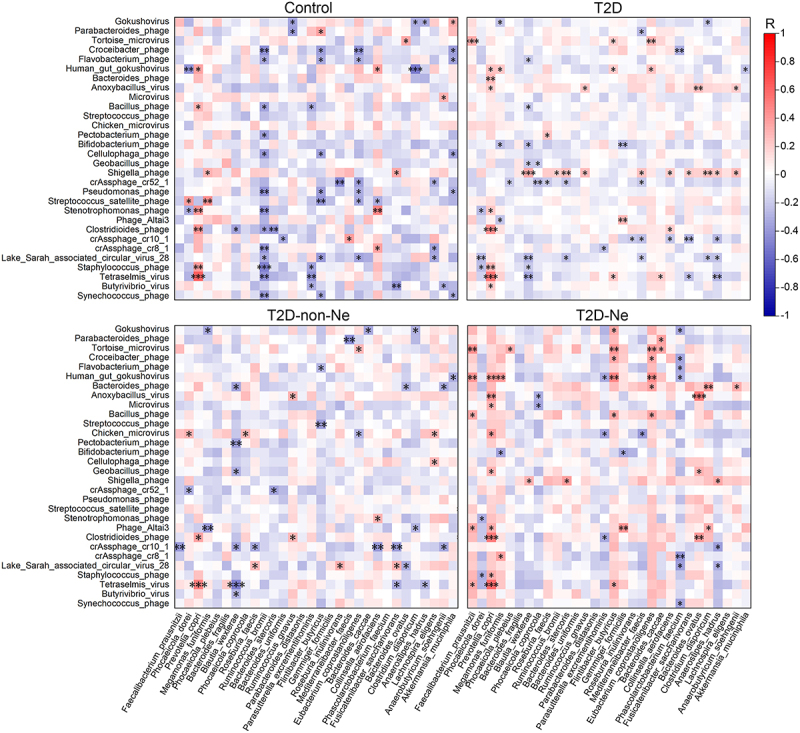
Alterations of transkingdom correlations between the gut virome and bacteriome in T2D, T2D-non-Ne and T2D-Ne compared with healthy controls. Heatmap shows color-coded Spearman’s correlations of the most abundant 30 virus species with the most abundant 30 bacteria species. Red color indicates positive correlation and blue color indicates negative correlation. Significant correlations are displayed with an asterisk. *P <.05, **P <.01 and ***P <.001.
These changes in virus-bacteria correlations were mainly attributed to the loss of some associations and the emergence of some new ones (Figure 7). For example, Streptococcus satellite phage showed significant correlation with five bacteria (Phocaeicola dorei, Megamonas funiformis, Flintibacter butyricus, Eubacterium coprostanoligenes, and Collinsella aerofaciens) in healthy controls. However, these associations disappeared in T2D, T2D-non-Ne, and T2D-Ne subjects. Similarly, significant associations between Pseudomonas phage and four bacteria (Ruminococcus bromii, F. butyricus, E coprostanoligenes, and Akkermansia muciniphila) in healthy controls were all lost in T2D, T2D-non-Ne, and T2D-Ne subjects. Furthermore, new associations between Shigella phage and three bacteria emerged in T2D and T2D-Ne subjects compared with healthy controls. Taken together, these findings suggest that strong viral-bacterial interactions in healthy subjects are disrupted in T2D and DN.
3.7. Combining gut virome and bacteriome improves diagnostic performance for T2D and DN
To evaluate the potential of gut virome as diagnostic markers for T2D and DN, we constructed a machine learning classifier model based on XGBoost algorithm to distinguish disease subjects from healthy controls. Our analysis identified a viral taxa signature consisting of 47 species as the optimal marker set between T2D and healthy controls, with an area under the curve (AUC) of 84.61% (Figure 8; Table S12, Supporting Information). Among the contributing features, Erwinia phage had the greatest impact as indicated by the gain value, followed by Anoxybacillus virus and Streptococcus satellite phage. Notably, 19 of the top 20 contributors belong to phages. Moreover, the model based on bacterial features also performed well, with an AUC of 98.07% (Figure 8; Table S13, Supporting Information). Interestingly, the combination of virome and bacteriome could classify T2D from healthy controls with better accuracy (AUC = 99.03%) than either virome or bacteriome alone (Figure 8; Table S14, Supporting Information).
Figure 8.
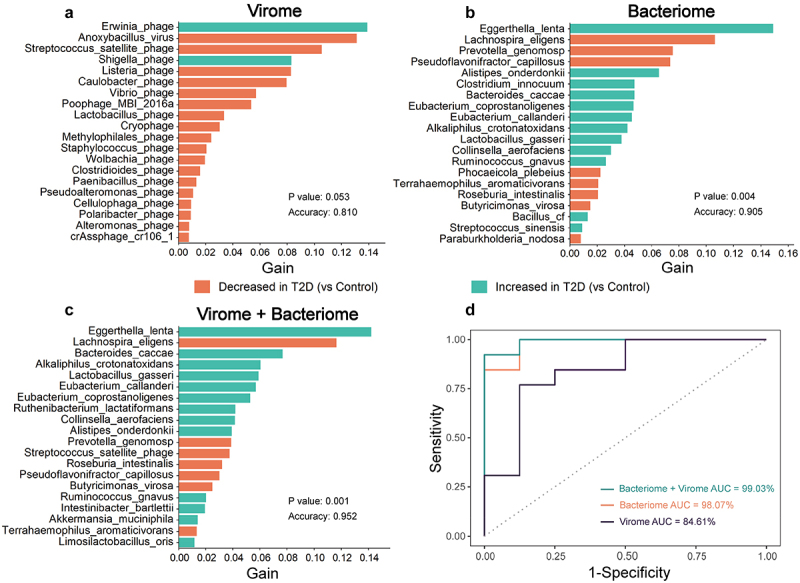
The combination of gut virome and bacteriome improves diagnostic performance in discriminating T2D subjects from healthy controls based on the XGBoost model. The top 20 important features for (a) virome, (b) bacteriome, and (c) combined virome and bacteriome. Gain refers to the relative contribution of the feature to the model. The color of the bars corresponds to differential abundance between groups. (d) the ROC curve analysis for each of the three models, including their AUC values.
We further used the XGBoost model to identify gut microbial markers capable of discriminating between T2D-non-Ne, T2D-Ne and controls. 14 viral markers were selected as the optimal marker set with an AUC of 78.46% (Figure 9; Table S15, Supporting Information). Shigella phage was the largest contributor, followed by Listeria phage and Streptococcus satellite phage. Furthermore, the model based on bacteriome (AUC = 91.49%) had a greater predictive power than the virome model (Figure 9; Table S16, Supporting Information). However, the combination of virome and bacteriome achieved the largest AUC of 98.19% in discriminating the three groups (Figure 9; Table S17, Supporting Information). Taken together, our results suggest that gut virome can be used to discriminate T2D and DN from controls, and the combination of viral and bacterial markers enhances diagnostic performance for T2D and DN.
Figure 9.
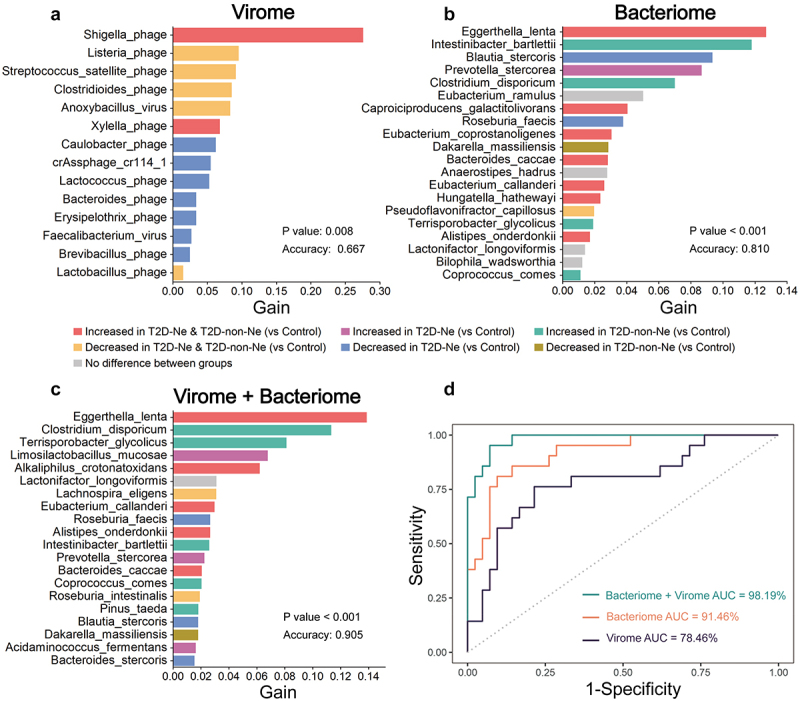
The combination of gut virome and bacteriome improves diagnostic performance in discriminating between T2D-Ne, T2D-non-Ne, and healthy controls based on the XGBoost model. The important features for (a) virome (only 14), (b) bacteriome (top 20), and (c) combined virome and bacteriome (top 20). Gain refers to the relative contribution of the feature to the model. The color of the bars corresponds to differential abundance between groups. (d) the ROC curve analysis for each of the three models, including their AUC values.
4. Discussion
The association between gut bacteria and T2D has been well-acknowledged, while the role of gut virome is poorly understood. Here, we revealed significant alterations in the gut virome in T2D by metagenomic sequencing of VLP-enriched DNA, characterized by decreased alpha diversity, altered viral composition, and loss of multiple viral functions. Moreover, we found for the first time that DN, a major complication of T2D, also showed significant gut viral dysbiosis. Notably, the decrease in viral alpha diversity was more pronounced in T2D-Ne than in T2D-non-Ne. These findings raise the interesting question of whether the gut virome is involved in the development of T2D and its complications.
T2D is a common metabolic disease. In recent years, several studies have been conducted to investigate gut virome changes in other metabolic diseases, including obesity, NAFLD and metabolic syndrome.7,14,25 Similar to our results, significant reductions in alpha diversity and alterations in viral composition and taxa abundance were also found in these three diseases. These common features suggest that gut virome may play an important role in the development of metabolic diseases. In the present study, we identified 81 significantly altered viral species in T2D subjects. However, to date, little is known about the specific role of these viral markers in the development of T2D. It is speculated that several mechanisms may be involved. On the one hand, some viruses might directly affect disease by stimulating host immune responses.23,24 Phages can be transported into the systemic circulation through intestinal epithelial cells and then affect the human immune system by regulating the release of cytokines and the activities of T and B cells.23,26 A recent study has shown that Escherichia and Bacteroides phages can activate IFN-γ mediated immune responses via the TLR9-dependent pathway and exacerbate colitis.27 In our study, four viral species, such as Shigella phage, were identified to be significantly enriched in T2D subjects compared with healthy controls. Whether they affect the immune system to mediate the development of T2D deserves further investigation. On the other hand, some viruses, particularly phages, might also indirectly mediate disease by affecting gut bacteria.23 As natural predators of bacteria, phages can regulate the metabolism, evolution and viability of bacteria in the gut, thus affecting their composition and abundance.23,26 Notably, the gut bacteriome have been revealed to play a key role in the pathogenesis of T2D.4,28 In our study, the significant reduction in viral diversity and specific viral species, as well as the loss of multiple viral functions, especially those of phage lysing host bacteria, lead to a quantitative increase of certain pathogenic bacteria, which may mediate the development of T2D. For example, Clostridioides phage and Streptococcus satellite phage were significantly reduced, while their respective host bacteria, Clostridioides difficile and Streptococcus mutans, were significantly increased in T2D subjects compared with healthy controls. C. difficile is a common pathogen, and its exotoxin can activate macrophages to promote inflammation and epithelial damage.29 S. mutans, a major pathogen of dental caries, has been linked to inflammation in cardiovascular disease. It can induce the production of inflammatory cytokines in human aortic endothelial cells.30 Moreover, a specific S. mutans strain was found to aggravate nonalcoholic steatohepatitis in a mouse model fed a high-fat diet.31 These data indicate that gut phages might influence T2D by regulating bacterial microbes. However, phage-bacteria interactions in the gut are complex. In addition to the empirical “predator-prey” dynamics, there are “kill-the-winner” and “arms-race” patterns among them.20 This may be the reasons for our findings that virus-bacteria interactions presented in healthy subjects were significantly disrupted in T2D, including the loss of some associations and the appearance of some new associations. Taken together, the gut virome might play a role in the development of T2D by stimulating immune responses and/or influencing gut bacteria. Given the complexity of the human situation, additional animal experiments are required to determine the contribution of gut viruses, especially phages, to T2D development.
DN is one of the most important complications in diabetic patients. Several studies have revealed changes in gut bacteria in DN subjects.32,33 However, it remains unclear whether gut viral dysbiosis occurs in DN. In this study, we found significant changes in the gut virome of DN subjects and identified some viral species associated with DN. Notably, our study found that Bacteroides phage was significantly reduced, while its host bacterium, Bacteroides caccae, was significantly enriched in DN subjects compared with healthy controls. B. caccae is an opportunistic pathogen with inflammatory properties that can destroy the colonic mucus barrier by secreting mucus-degrading proteases.34 It is also involved in the production of several uremic toxins, including phenol and p-cresol.35 These metabolites are nephrotoxic and can cause kidney damage and fibrosis.36 These data suggest that gut phages might play a role in the development of DN by regulating gut bacteria and their metabolites. In addition, Shigella phage, Xylella phage, Erysipelothrix phage, Lactococcus phage, Faecalibacterium virus, Brevibacillus phage, and crAssphage cr114_1 were also characteristic viruses associated with DN. Further functional studies are needed to assess the role of these viral markers in the development of DN.
Several factors have been reported to influence the gut virome, such as age, gender, and diet. Gregory et al. found that gut viral diversity is age-dependent in healthy Westerners.37 Similarly, there is a significant positive correlation between age and gut viral diversity among Japanese adults.38 Moreover, gender has been shown to be significantly associated with 68 viral operational taxonomic units and 24 viral clusters in the Japanese adult cohort.38 However, Yang et al. found that age and gender did not show significant associations with gut viral diversity in obese subjects,14 which is consistent with our data. Heterogeneity between these studies may be related to differences in disease phenotype, population and/or geographic location. Future multicenter, large-scale studies are needed to further evaluate the effects of age and gender on the gut virome of T2D and DN. In addition, diet is also one of the environmental factors affecting the human gut virome.39 A recent study found that the frequency of consuming meat and vegetables and alcohol intake had significant effects on the gut virome.40 However, dietary information was not obtained in our cross-sectional study, which prevented us to from assessing the effect of diet on the gut virome differences between T2D/DN and healthy subjects. Further prospective studies with dietary controls or detailed dietary information are needed to evaluate the effects of diet on the gut virome of T2D and DN.
Bacteria and viruses coexist in the human gut, and their interactions have been shown to be associated with health and disease.14,20 Our results demonstrate the presence of strong virus-bacteria interactions in healthy subjects. However, these transkingdom interactions were markedly disrupted in T2D and DN, characterized by a decrease in negative correlations and an increase in positive correlations. The significant changes in transkingdom correlations between gut virome and bacteriome may suggest a perturbance of the whole ecosystem in T2D and DN, and highlight the potential role of the virome in shaping healthy gut microecology.
In this study, we used a XGBoost classification model to explore the potential of gut virome as diagnostic markers for T2D and DN. XGBoost is an ensemble learning method, which can prevent the model from overfitting by using some regularization methods.41 It has been proven to outperform other machine learning algorithms in classifying human microbiome data.41,42 Previous studies have revealed the potential of gut bacterial markers for the diagnosis of T2D.22,43 Here, we demonstrated for the first time that specific gut viruses could be used as biomarkers to distinguish T2D subjects from healthy controls. Notably, the combined use of viral and bacterial markers yielded the best diagnostic performance, with an AUC of 99.03%. In general, the diagnostic value of single omics data is limited. Our results highlight the importance of integrating multi-omics data (gut virome and bacteriome), which can improve the classification accuracy. Diabetes complications should be paid more attention than diabetes itself because of their high disability and mortality.44 In our study, gut virome also showed a good ability to distinguish between T2D-Ne, T2D-non-Ne, and controls. A panel combining viral and bacterial markers achieved a powerful diagnostic performance with an AUC of 98.19%. Collectively, our results suggest that the combination of gut virome and bacteriome improves the diagnostic performance of T2D and DN.
Despite the important findings of our study, there are some limitations that should be acknowledged. First, we conducted a cross-sectional study that could not reveal the causal relationship between gut virome and the two diseases. In the future, some key methods, such as Mendelian randomization analysis, are required to further elucidate their causality. Additional preclinical studies are urgently needed to determine the contribution of specific viruses, particularly phages, to the progression of T2D and DN. Second, in addition to age and sex, other confounding factors such as diet and lifestyle are known to affect the gut virome.14,39 The impact of these confounders on the gut virome was not evaluated in this study. In addition, although the combined viral and bacterial markers achieved strong diagnostic performance in our cohort, additional multicenter independent cohorts are still needed to validate their clinical diagnostic potential for T2D and DN.
In conclusion, our study provides evidence that T2D and DN are characterized by disturbances in gut viral diversity and taxonomic composition compared with healthy controls. The significant decrease in diversity, changes in specific virus species, loss of multiple viral functions, and disruption of viral-bacterial correlations suggest that gut virome may play an important role in the development of T2D and DN. The combination of specific viral and bacterial markers has good diagnostic potential for T2D and DN, which may improve the performance of currently only bacterial-based biomarkers. Although more clinical validation and mechanistic studies are needed, this study would increase our understanding of the role of gut virome in the pathogenesis of diabetes and its complications, and provide new avenues for the development of novel therapeutic strategies for T2D and DN.
Supplementary Material
Funding Statement
The work was supported by the Key Research and Development Program of Sichuan Province of China [2022YFS0434], the National Key Research and Development Program of China [2017YFC1703900 and 2019YFC1712302], and the Sichuan Provincial Scientific and Technological Innovation Seedling Project [2022036].
Data availability statement
The data that support the findings of this study are openly available in the National Center for Biotechnology Information through BioProject number PRJNA891875 https://www.ncbi.nlm.nih.gov/.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplemental material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2023.2226925.
References
- 1.Zheng Y, Ley SH, Hu FB.. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat Rev Endocrinol. 2018;14(2):88–17. doi: 10.1038/nrendo.2017.151. [DOI] [PubMed] [Google Scholar]
- 2.Harding JL, Pavkov ME, Magliano DJ, Shaw JE, Gregg EW. Global trends in diabetes complications: a review of current evidence. Diabetologia. 2019;62(1):3–16. doi: 10.1007/s00125-018-4711-2. [DOI] [PubMed] [Google Scholar]
- 3.Selby NM, Taal MW. An updated overview of diabetic nephropathy: diagnosis, prognosis, treatment goals and latest guidelines. Diabetes Obes Metab. 2020;22(S1):3–15. doi: 10.1111/dom.14007. [DOI] [PubMed] [Google Scholar]
- 4.Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 2021;19(1):55–71. doi: 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- 5.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466(7304):334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rasmussen TS, Koefoed AK, Jakobsen RR, Deng L, Castro-Mejía JL, Brunse A, Neve H, Vogensen FK, Nielsen DS. Bacteriophage-mediated manipulation of the gut microbiome – promises and presents limitations. FEMS Microbiol Rev. 2020;44(4):507–521. doi: 10.1093/femsre/fuaa020. [DOI] [PubMed] [Google Scholar]
- 7.Lang S, Demir M, Martin A, Jiang L, Zhang XL, Duan Y, Gao B, Wisplinghoff H, Kasper P, Roderburg C, et al. Intestinal virome signature associated with severity of nonalcoholic fatty liver disease. Gastroenterology. 2020;159(5):1839–1852. doi: 10.1053/j.gastro.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakatsu G, Zhou HK, Wu KK, Wong SH, Coker OO, Dai ZW, Li XC, Szeto CH, Sugimura N, Lam TY, et al. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology. 2018;155(2):529. doi: 10.1053/j.gastro.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 9.Jiang L, Lang SJ, Duan Y, Zhang XL, Gao B, Chopyk J, Schwanemann LK, Ventura-Cots M, Bataller R, Bosques-Padilla F, et al. Intestinal virome in patients with alcoholic hepatitis. Hepatology. 2020;72(6):2182–2196. doi: 10.1002/hep.31459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, Kambal A, Monaco CL, Zhao GY, Fleshner P, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160(3):447–460. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clooney AG, Sutton TDS, Shkoporov AN, Holohan RK, Daly KM, O’Regan O, Ryan FJ, Draper LA, Plevy SE, Ross RP, et al. Whole-virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host & Microbe. 2019;26(6):764. doi: 10.1016/j.chom.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 12.Ma YF, You XY, Mai GQ, Tokuyasu T, Liu CL. A human gut phage catalog correlates the gut phageome with type 2 diabetes. Microbiome. 2018;6(1):6. doi: 10.1186/s40168-018-0410-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rasmussen TS, Mentzel CMJ, Kot W, Castro-Mejía JL, Zuffa S, Swann JR, Hansen LH, Vogensen FK, Hansen AK, Nielsen DS. Faecal virome transplantation decreases symptoms of type 2 diabetes and obesity in a murine model. Gut. 2020;69(12):2122–2130. doi: 10.1136/gutjnl-2019-320005. [DOI] [PubMed] [Google Scholar]
- 14.Yang KL, Niu JK, Zuo T, Sun Y, Xu ZL, Tang W, Liu Q, Zhang JW, Enders KW, Wong S, et al. Alterations in the gut virome in obesity and type 2 diabetes mellitus. Gastroenterology. 2021;161(4):1257. doi: 10.1053/j.gastro.2021.06.056. [DOI] [PubMed] [Google Scholar]
- 15.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581. doi: 10.1038/Nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thukral AK. A review on measurement of Alpha diversity in biology. Intern J Contemp Microbiol. 2007;54(1):1. doi: 10.5958/2395-146X.2017.00001.1. [DOI] [Google Scholar]
- 19.Pelissier R, Couteron P, Dray S. Analyzing or explaining beta diversity? Comment Ecology. 2008;89(11):3232–3237. doi: 10.1890/08-1247.1. [DOI] [PubMed] [Google Scholar]
- 20.Zuo T, Lu XJ, Zhang Y, Cheung CP, Lam S, Zhang F, Tang W, Ching JYL, Zhao R, Chan PKS, et al. Gut mucosal virome alterations in ulcerative colitis. Gut. 2019;68(7):1169. doi: 10.1136/gutjnl-2018-318131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong C, Liu G, Kalady MF, Jin T, Ma Y. Dysbiosis of the stool DNA and RNA virome in Crohn’s disease. J Med Virol. 2023;95(2):e28573. doi: 10.1002/jmv.28573. [DOI] [PubMed] [Google Scholar]
- 22.Qin JJ, Li YR, Cai ZM, Li SH, Zhang JF, Zhang F, Liang SS, Zhang WW, Guan YL, Shen DQ, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 23.Cao ZR, Sugimura N, Burgermeister E, Ebert MP, Zuo T, Lan P. The gut virome: a new microbiome component in health and disease. Ebiomedicine. 2022;81:81. doi: 10.1016/j.ebiom.2022.104113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang G, Bushman FD. The human virome: assembly, composition and host interactions. Nat Rev Microbiol. 2021;19(8):514–527. doi: 10.1038/s41579-021-00536-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Jonge PA, Wortelboer K, Scheithauer TPM, van den Born BJH, Zwinderman AH, Nobrega FL, Dutilh BE, Nieuwdorp M, Herrema H. Gut virome profiling identifies a widespread bacteriophage family associated with metabolic syndrome. Nat Commun. 2022;13(1):13. doi: 10.1038/s41467-022-31390-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Popescu M, Van Belleghem JD, Khosravi A, Bollyky PL. Bacteriophages and the immune system. Annu Rev Virol. 2021;8(1):415–435. doi: 10.1146/annurev-virology-091919-074551. [DOI] [PubMed] [Google Scholar]
- 27.Gogokhia L, Buhrke K, Bell R, Hoffman B, Brown G, Hanke-Gogokhia C, Ajami N, Wonge MC, Ghazaryan A, JF V, et al. Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host & Microbe. 2019;25(2):285–299.e8. doi: 10.1016/j.chom.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du L, Li Q, Yi H, Kuang T, Tang Y, Fan G. Gut microbiota-derived metabolites as key actors in type 2 diabetes mellitus. Biomed Pharmacother. 2022;149:112839. doi: 10.1016/j.biopha.2022.112839. [DOI] [PubMed] [Google Scholar]
- 29.Chan H, Li Q, Wang XS, Liu WY, Hu W, Zeng JD, Xie C, Kwong TNY, IHT H, Liu XD, et al. Vitamin D3 and carbamazepine protect against Clostridioides difficile infection in mice by restoring macrophage lysosome acidification. Autophagy. 2022;18(9):2050–2067. doi: 10.1080/15548627.2021.2016004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagata E, Oho T. Invasive Streptococcus mutans induces inflammatory cytokine production in human aortic endothelial cells via regulation of intracellular toll-like receptor 2 and nucleotide-binding oligomerization domain 2. Mol Oral Microbiol. 2016;32(2):131. doi: 10.1111/omi.12159. [DOI] [PubMed] [Google Scholar]
- 31.Naka S, Nomura R, Takashima Y, Okawa R, Ooshima T, Nakano K. A specific Streptococcus mutans strain aggravates non-alcoholic fatty liver disease. Oral Dis. 2014;20(7):700–706. doi: 10.1111/odi.12191. [DOI] [PubMed] [Google Scholar]
- 32.Du X, Liu J, Xue Y, Kong XY, Lv CX, Li ZQ, Huang YH, Wang BH. Alteration of gut microbial profile in patients with diabetic nephropathy. Endocrine. 2021;73(1):71–84. doi: 10.1007/s12020-021-02721-1. [DOI] [PubMed] [Google Scholar]
- 33.Tao SB, Li LZ, Ma L, Fu P. Understanding the gut-kidney axis among biopsy-proven diabetic nephropathy, type 2 diabetes mellitus and healthy controls: an analysis of the gut microbiota composition. Nephrol Dial Transpl. 2019;34(Supplement_1):216. doi: 10.1007/s00592-019-01316-7. [DOI] [PubMed] [Google Scholar]
- 34.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 2016;167(5):1339. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saito Y, Sato T, Nomoto K, Tsuji H. Identification of phenol- and p-cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. FEMS Microbiol Ecol. 2020;96. doi: 10.1093/femsec/fiy125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pletinck A, Vanholder R, Glorieux G. Uremic toxins. Hoboken, N.J: John Wiley & Sons; 2012. [Google Scholar]
- 37.Gregory AC, Zablocki O, Zayed AA, Howell A, Sullivan MB, Sullivan, M B.. The gut virome database reveals age-dependent patterns of virome diversity in the human gut. Cell Host & Microbe. 2020;28(5):724–740.e8. doi: 10.1016/j.chom.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishijima S, Nagata N, Kiguchi Y, Kojima Y, Miyoshi-Akiyama T, Kimura M, Ohsugi M, Ueki K, Oka S, Mizokami M, et al. Extensive gut virome variation and its associations with host and environmental factors in a population-level cohort. Nat Commun. 2022;13(1):5252. doi: 10.1038/s41467-022-32832-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, Lewis JD, Bushman FD. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21(10):1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuo T, Sun Y, Wan YT, Yeoh YK, Zhang F, Cheung CP, Chen N, Luo J, Wang W, Sung JJY, et al. Human-gut-DNA virome variations across geography, ethnicity, and urbanization. Cell Host & Microbe. 2020;28(5):741–751.e4. doi: 10.1016/j.chom.2020.08.005. [DOI] [PubMed] [Google Scholar]
- 41.Volkova A, Ruggles KV. Predictive metagenomic analysis of autoimmune disease identifies robust autoimmunity and disease specific microbial signatures. Front Microbiol. 2021;12:12. doi: 10.3389/fmicb.2021.621310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang XW, Liu YY. Comparative study of classifiers for human microbiome data. Med Microecol. 2020;4:4. doi: 10.1016/j.medmic.2020.100013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yassour M, Lim MY, Yun HS, Tickle TL, Sung J, Song YM, Lee K, Franzosa EA, Morgan XC, Gevers D, et al. Sub-clinical detection of gut microbial biomarkers of obesity and type 2 diabetes. Genome Med. 2016;8(1). doi: 10.1186/s13073-016-0271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young BA, Lin E, Von Korff M, Simon G, Ciechanowski P, Ludman EJ, Everson-Stewart S, Kinder L, Oliver M, Boyko EJ, et al. Diabetes complications severity index and risk of mortality, hospitalization, and healthcare utilization. Am J Manag Care. 2008;14:15–23. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available in the National Center for Biotechnology Information through BioProject number PRJNA891875 https://www.ncbi.nlm.nih.gov/.


