SUMMARY
Normal vascular development includes the formation and specification of arteries, veins, and intervening capillaries. Vein of Galen malformations (VOGM) are among the most common and severe neonatal brain arterio-venous malformations, shunting arterial blood into the brain’s deep venous system through aberrant direct connections. Exome sequencing of 55 VOGM probands, including 52 parent-offspring trios, revealed enrichment of rare, damaging de novo mutations in chromatin modifier genes that play essential roles in brain and vascular development. Other VOGM probands harbored rare, inherited damaging mutations in Ephrin signaling genes, including a genome-wide significant mutation burden in EPHB4. Inherited mutations showed incomplete penetrance and variable expressivity, with mutation carriers often exhibiting cutaneous vascular abnormalities, suggesting a two-hit mechanism. Identified mutations collectively account for ~30% of studied VOGM cases. These findings provide insight into disease biology and may have clinical implications for risk assessment.
eTOC blurb
Vein of Galen malformations (VOGMs) are the most severe neonatal brain arteriovenous malformations. Duran et al. identify de novo chromatin modifier and rare inherited Ephrin signaling mutations in ~30% of VOGM cases. Data implicate disrupted arterio-venous specification in VOGM pathogenesis.
INTRODUCTIPON
Embryogenesis requires vascular development to meet hemodynamic and nutritive demands. Arterio-venous (A-V) specification in model organisms is genetically-determined and results in the differential expression of genes in arteries and veins prior to establishment of circulation (Fish and Wythe, 2015). For example, during development Ephrin-B2 and its receptor Eph-B4 are exclusively expressed in arteries or veins, respectively (Wang et al., 1998; Adams et al., 1999; Gerety et al., 1999). Murine deletion of Efnb2 or EphB4 impairs A-V specification and causes arterio-venous malformations (AVMs), high-flow vascular lesions characterized by direct connections of arteries to veins without intervening capillaries. A-V specification requires orchestrated activity of multiple signaling cascades (e.g., Eph-Ephrin, Hedgehog, VEGF, TGF-beta, and Notch) and transcriptional networks (e.g., HEY and HES, SOX factors, and COUP-TFII) (Fish and Wythe, 2015). Nonetheless, the genetic determinants of A-V specification in humans remain incompletely understood.
During normal brain development, primitive choroidal and subependymal arteries that perfuse deep brain structures are connected via an intervening capillary network to the embryonic precursor of the Vein of Galen (i.e., the median prosencephalic vein of Markowski [MPV]). The MPV returns deep cerebral venous blood to dural sinuses that drain into the internal jugular veins (Raybaud et al., 1989). Vein of Galen malformations (VOGMs), the most common and severe neontal brain AVMs (Long et al., 1974; Deloison et al., 2012), directly connect primitive choroidal or subependymal cerebral arteries to the MPV, exposing it to dangerously high blood flow and pressures that can result in high-output cardiac failure, hydrocephalus, and/or brain hemorrhage (Recinos et al., 2012). VOGMs are also often associated neurodevelopmental delay and congenital heart defects (CHDs) (McElhinney et al., 1998). While endovascular partial obliteration of anomalous A-V connections has improved VOGM outcomes (Mitchell et al., 2001) (Altschul et al., 2014), many VOGMs remain refractory to intervention, which are also are inaccessible to many patients.
Limited knowledge of the molecular pathophysiology of VOGMs has hindered development of early diagnostic and targeted therapeutic strategies. Considered isolated, sporadic congenital lesions, rare VOGMs have been associated with Mendelian disorders, including 8 cases of autosomal dominant (AD) capillary malformation-arteriovenous malformation (CM-AVM) syndrome type 1 (CM-AVM1) caused by RASA1 mutation (OMIM# 608354) (Revencu et al., 2013); 2 cases of AD CM-AVM syndrome type 2 (CM-AVM2) caused by EPHB4 mutation (Amyere et al., 2017), and single cases of AD Hereditary Hemorrhagic Telangiectasia (HHT) type 1 (HHT1) due to ENG mutation (OMIM# 187300) (Tsutsumi et al., 2011) and AD HHT type 2 (HHT2) caused by ACVRL1 mutation (OMIM# 600376; 1 case) (Chida et al., 2013).
Traditional genetic approaches have been limited in their ability to identify additional causative genes for VOGM because cases are rare and most often sporadic (Xu et al., 2010). This limitation motivates application of whole-exome sequencing (WES) to large numbers of affected subjects and their families, searching for genes mutated in probands more often than expected by chance. This unbiased approach has aided the study of other genetically heterogeneous neurodevelopmental disorders (Vissers et al., 2010; O’Roak et al., 2011; de Ligt et al., 2012; Iossifov et al., 2012; Neale et al., 2012; O’Roak et al., 2012; Rauch et al., 2012; Sanders et al., 2012; Epi et al., 2013; Iossifov et al., 2014; Krumm et al., 2015; Timberlake et al., 2016; Timberlake et al., 2017; Willsey et al., 2017), including those associated with brain malformations (Bilguvar et al., 2010; Barak et al., 2011; Mishra-Gorur et al., 2014; Nikolaev et al., 2018), CHDs (Zaidi et al., 2013; Homsy et al., 2015; Jin et al., 2017), and congenital hydrocephalus (Furey et al., 2018). We hypothesized that VOGMs might arise from damaging de novo mutation events or incomplete penetrance of rare transmitted variants.
RESULTS
VOGM cohort characteristics and whole exome sequencing
We recruited 55 probands with radiographically confirmed VOGMs (see STAR Methods) treated by endovascular therapy, including 52 parent-offspring trios with a single affected offspring and 3 singleton cases. 62% of probands were diagnosed prenatally or within one month after birth; only one was diagnosed after age 2. Common features at diagnosis included high output cardiac failure (62%), macrocephaly (64%), hydrocephalus (60%), and prominent face/scalp vasculature (49%). Among the CHDs found in 9% were partial anomalous pulmonary venous return, patent ductus arteriosus, and pulmonary valve stenosis. Supplementary Table 1 summarizes cohort demographics, clinical features, and radiographic classification, including angio-architectural subgroups (Lasjaunias et al., 2006). 37 of the 55 patients in our cohort presented with “choroidal” type lesions, defined as VOGMs with numerous feeder vessels and “pseudoniduses” that communicate with the MPV (Lasjaunias et al., 2006). The remaining 18 probands presented with “mural” VOGMs, characterized by fewer vessels of larger caliber that fistulize into the MPV (Lasjaunias et al., 2006). See Supplementary Figure 1 for representative VOGM images.
DNA was isolated and WES performed as described (Timberlake et al., 2016). WES from 1,789 control trios comprising parents and unaffected siblings of autism probands were analyzed (Fischbach and Lord, 2010; Krumm et al., 2015) by our in-house informatics pipeline. In both cases and controls, ≥ 94.6% of targeted bases had ≥ 8 independent reads and ≥ 89.8% had ≥ 15 independent reads. (See Supplementary Table 2 for exome metrics). Variant calling was by Genome Analysis Toolkit (GATK) HaplotypeCaller (McKenna et al., 2010; Van der Auwera et al., 2013), and allele frequency annotation by the Exome Aggregation Consortium (ExAC) and the Genome Aggregation Database (gnomAD) (Lek et al., 2016) (see STAR Methods). De novo mutation identification was by TrioDeNovo (Wei et al., 2015). MetaSVM was used to infer the impact of missense mutations (Dong et al., 2015). Missense variants were considered damaging if predicted deleterious by MetaSVM (D-mis). Inferred loss-of-function (LoF) mutations, including stop-gains, stop-losses, frameshift insertions/deletions, and canonical splice site mutations were also considered damaging. Mutations in genes of interest were validated by PCR amplification and Sanger sequencing (Supplementary Figure 2).
Enrichment in damaging de novo mutations in VOGM probands
The de novo mutation rate in probands was 1.56 × 10−8 per base pair, with 1.21 de novo coding region mutations per proband (Supplementary Table 3), consistent with expectation and prior results (Homsy et al., 2015; Ware et al., 2015; Timberlake et al., 2017). Total de novo mutation burden in controls was also as previously reported (Timberlake et al., 2017). The distribution of types of de novo coding mutations in probands was compared to that expected from the probability of mutation of each base in the coding region and flanking splice sites (Samocha et al., 2014). While synonymous mutations and inferred tolerated missense (T-mis) mutations in probands were not significantly enriched, de novo D-mis mutations in probands were marginally enriched (P = 0.01; enrichment = 2.05-fold; Supplementary Table 3). In contrast, control subjects displayed no enrichment in any class of coding region mutation (Supplementary Table 3). The observed excess of damaging de novo mutations in probands over that expected predicts these mutations contribute to ~13% of VOGM cases.
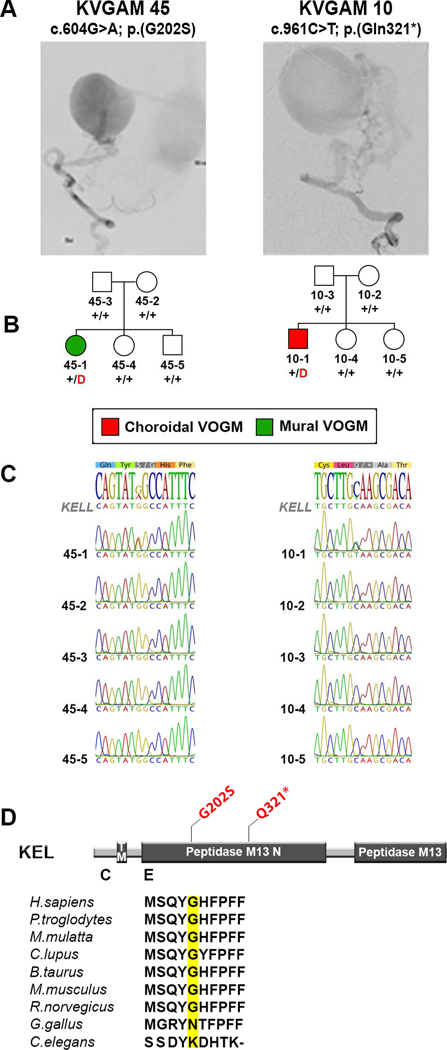
One gene, KEL, exhibited more than one protein-altering de novo mutation (Figure 1). Among the 52 trios analyzed, we observed a near-significant enrichment of protein-altering de novo mutations in KEL (one-tailed Poisson test P = 4.2 × 10−6; Benjamini-Hochberg false discovery rate [BH-FDR] = 0.08). One of these mutations caused premature termination (p.Gln321*) and the other was a missense mutation p.Gly202Ser (CADD = 22.6), both impacting the KELL polypeptide’s peptidase domain (Figure 1).
Figure 1. Protein-altering de novo KEL mutations in Vein of Galen malformation (VOGM).
(A) Representative digital subtraction angiography images demonstrating mural and choroidal VOGMs in KVGAM45–1 and KVGAM10–1, respectively.
(B) Pedigrees depicting kindred structures. Note that probands carrying de novo mutations in KEL are the only members of families KVGAM45 and KVGAM10 with VOGMs; none of the family members in these two families have any disease phenotypes or cutaneous manifestations. Red ‘D’ denotes protein-altering mutation, ‘+’ denotes wild type sequence.
(C) Mutations identified by exome sequencing were confirmed by direct PCR amplification with custom primers followed by Sanger sequencing.
(D) Linear representation of the KEL polypeptide, with functional domains as dark rectangles. Amino acid modifications are mapped (in red) on the protein structure. Conservation of the wild-type amino acid substituted by the missense mutations is depicted below. TM = Transmembrane domain.
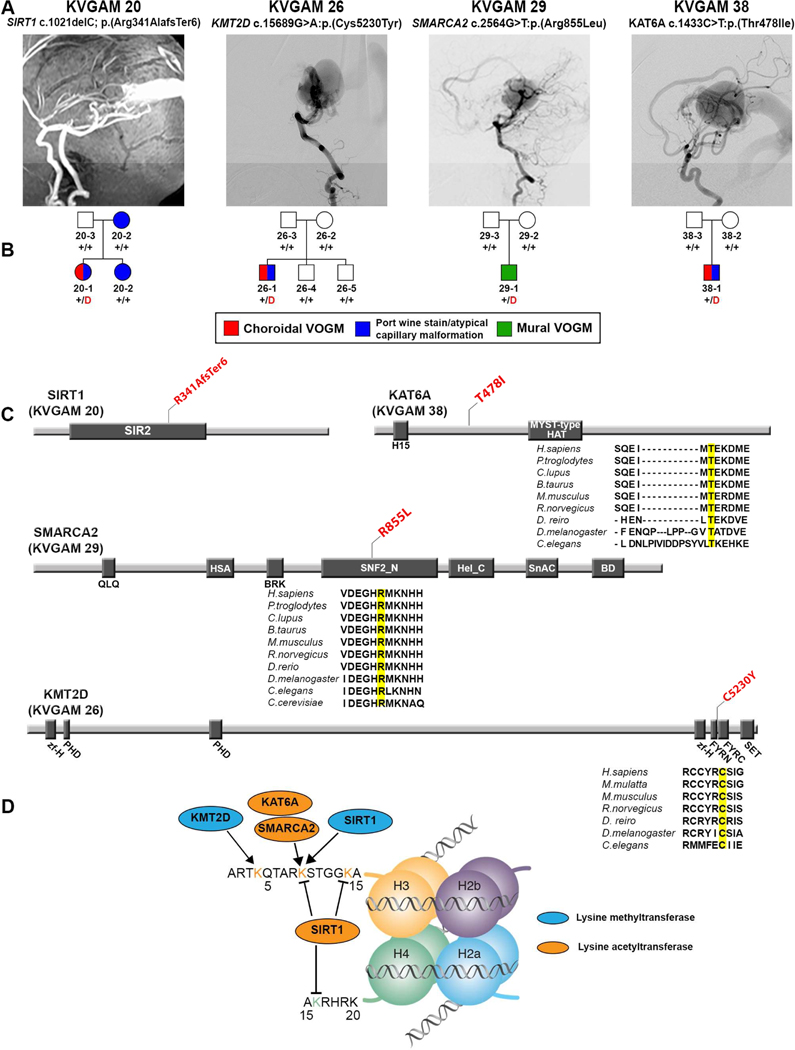
We analyzed the burden of de novo mutations in genes highly intolerant to heterozygous LoF mutation (LoF-intolerant genes; pLI ≥ 0.9) (Lek et al., 2016), consistent with loss of one gene copy markedly impairing reproductive fitness. de novo D-mis mutations were marginally enriched (6 mutations; P = 0.02; enrichment = 2.82-fold; Table 1), of which four (3 D-mis and the only LoF mutation) occurred in genes encoding chromatin modifiers (KMT2D, SMARCA2, SIRT1, and KAT6A) (Figure 2). The 547 genes in the chromatin modifier gene ontology term GO:0016569 include 272 LoF-intolerant chromatin modifier genes; mutations in this set are enriched (P = 8.9 × 10−4; enrichment = 9.63-fold; Table 1), but de novo mutations in the LoFintolerant chromatin modifier gene set were not enriched in controls (Table 1). A case-control analysis using the two-tailed binomial exact test (Sanders et al., 2012; Willsey et al., 2017) further supported this result (Supplementary Table 4). In an orthogonal analysis of all genes using the permutation-based test (1 million iterations), the probability of finding 4 or more damaging de novo mutations in LoF-intolerant chromatin modifiers among a total of only 19 damaging de novo mutations in all genes was also low (empirical P = 3.5 × 10−3; expected number = 0.66; see STAR Methods). Damaging de novo mutations in chromatin modifiers were thus identified in ~7% of all VOGM probands (Supplementary Table 5). Missense mutations occur in key functional domains of the encoded proteins at positions conserved through worm (KAT6A and KMT2D) and yeast (SMARCA2) (Figure 2, panel C; Supplementary Table 5). Clinical characteristics of probands harboring these mutations are shown in Figure 2 and Supplementary Table 5.
Table 1.
Enrichment of damaging de novo mutations in chromatin modifiers intolerant to LoF mutation in VOGM
| Cases, N = 52 | Controls, N=1,789 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Observed | Expected | Enrichment | p | Observed | Expected | Enrichment | p | ||||||
| N | Rate | N | Rate | N | Rate | N | Rate | ||||||
| LoF-intolerant genes (n = 3,230) | LoF-intolerant genes (n = 3,230) | ||||||||||||
| Total | 14 | 0.27 | 15.6 | 0.30 | 0.90 | 0.69 | Total | 504 | 0.28 | 527 | 0.29 | 0.96 | 0.85 |
| Syn | 2 | 0.04 | 4.3 | 0.08 | 0.46 | 0.93 | Syn | 127 | 0.07 | 145.6 | 0.08 | 0.87 | 0.95 |
| Mis | 11 | 0.21 | 9.8 | 0.19 | 1.12 | 0.39 | Mis | 339 | 0.19 | 331.7 | 0.19 | 1.02 | 0.33 |
| D-Mis | 6 | 0.12 | 2.1 | 0.04 | 2.82 | 0.02 | D-Mis | 64 | 0.04 | 72.7 | 0.04 | 0.88 | 0.86 |
| LoF | 1 | 0.02 | 1.5 | 0.03 | 0.69 | 0.77 | LoF | 38 | 0.02 | 49.7 | 0.03 | 0.76 | 0.96 |
| Damaging | 7 | 0.13 | 3.6 | 0.07 | 1.96 | 0.07 | Damaging | 102 | 0.06 | 122.4 | 0.07 | 0.83 | 0.97 |
| All chromatin genes (n = 547) | All chromatin genes (n = 547) | ||||||||||||
| Total | 4 | 0.08 | 2.3 | 0.04 | 1.73 | 0.20 | Total | 65 | 0.04 | 78.3 | 0.04 | 0.83 | 0.94 |
| Syn | 0 | 0.00 | NA | NA | NA | NA | Syn | 12 | 0.01 | 21.3 | 0.01 | 0.56 | 0.99 |
| Mis | 3 | 0.06 | 1.5 | 0.03 | 2.06 | 0.18 | Mis | 47 | 0.03 | 49.4 | 0.03 | 0.95 | 0.65 |
| D-Mis | 3 | 0.06 | 0.3 | 0.01 | 8.80 | 5.12×10−03 | D-Mis | 12 | 0.01 | 11.6 | 0.01 | 1.03 | 0.50 |
| LoF | 1 | 0.02 | 0.2 | 3.85×10−03 | 4.53 | 0.20 | LoF | 6 | 3.35×10−03 | 7.6 | 4.25×10−03 | 0.80 | 0.76 |
| Damaging | 4 | 0.08 | 0.6 | 0.01 | 7.12 | 2.66×10−03 | Damaging | 18 | 0.01 | 19.2 | 0.01 | 0.94 | 0.64 |
| Intolerant chromatin genes (n = 272) | Intolerant chromatin genes (n = 272) | ||||||||||||
| Total | 4 | 0.08 | 1.6 | 0.03 | 2.56 | 0.07 | Total | 47 | 0.03 | 52.7 | 0.03 | 0.89 | 0.80 |
| Syn | 0 | 0.00 | NA | NA | NA | NA | Syn | 9 | 0.01 | 14.2 | 0.01 | 0.63 | 0.95 |
| Mis | 3 | 0.06 | 1 | 0.02 | 3.04 | 0.08 | Mis | 35 | 0.02 | 33.3 | 0.02 | 1.05 | 0.41 |
| D-Mis | 3 | 0.06 | 0.3 | 0.01 | 11.40 | 2.48×10−03 | D-Mis | 7 | 3.91×10−03 | 9 | 0.01 | 0.78 | 0.79 |
| LoF | 1 | 0.02 | 0.2 | 3.85×10−03 | 6.55 | 0.14 | LoF | 3 | 1.68×10−03 | 5.2 | 2.91×10−03 | 0.58 | 0.89 |
| Damaging | 4 | 0.08 | 0.4 | 0.01 | 9.63 | 8.91×10−04 | Damaging | 10 | 0.01 | 14.2 | 0.01 | 0.71 | 0.90 |
LoF: loss-of-function; N: the number of de novo mutations; Rate: the number of de novo mutations divided by the number of individuals in the cohort; Enrichment: ratio of observed to expected numbers of mutations; Intolerant genes: Genes with a pLI ≥ 0.9; D-Mis: Damaging missense mutations as predicted by MetaSVM; Damaging: D-Mis + LoF. Chromatin genes used for analysis were extracted from the Biomart database using GO:0016569 as the input.
Figure 2. Damaging de novo mutations in chromatin modifiers in VOGM.
(A) Magnetic resonance angiographies and a digital subtraction angiography reconstruction demonstrating VOGM in probands from four pedigrees.
(B) Pedigree structures of VOGM kindreds. For each kindred, the gene and mutation, the angiographic image, and pedigree structure are shown. Subjects with atypical capillary malformations are denoted by blue symbols. Red ‘D’ denotes damaging mutation, ‘+’ denotes wild type sequence.
(C) Linear representation of functional domains of SIRT1, KMT2D, SMARCA2, and KAT6A, with location of VOGM mutations. Functional domains are represented by dark rectangles. Amino acid changes (red) are located on the protein structure. For missense mutations, phylogenetic conservation of the wild-type amino acid is shown, with the mutated amino acid in yellow. SIR2 = Sirtuin catalytic domain, SIR2 Domain; PHD = Zinc Finger PHD type; MOZ_SAS = Histone acetyltransferase domain, MYST-type; zf-H = PHD-like zinc binding domain; FYRN = F/Y-rich domain - F/Y-rich N-terminus motif; FYRC = F/Y-rich domain - F/Y-rich C-terminus motif; SET = Su(var)3–9, Enhancer-of-zeste and Trithorax; QLQ = Glutamine-Leucine-Glutamine domain; HAS = Helicase-SANT associated domain; BRK = BRK domain; SNF2_N = SNF2-related, N-terminal domain; Hel_C = Helicase C-terminal domain; SnAC = Snf2-ATP coupling, chromatin remodeling complex; BD = Bromodomain.
(D) Schematic of histone mark modifications by SIRT1, KMT2D, SMARCA2, and KAT6A.
Enrichment of rare damaging transmitted mutations in EPHB4
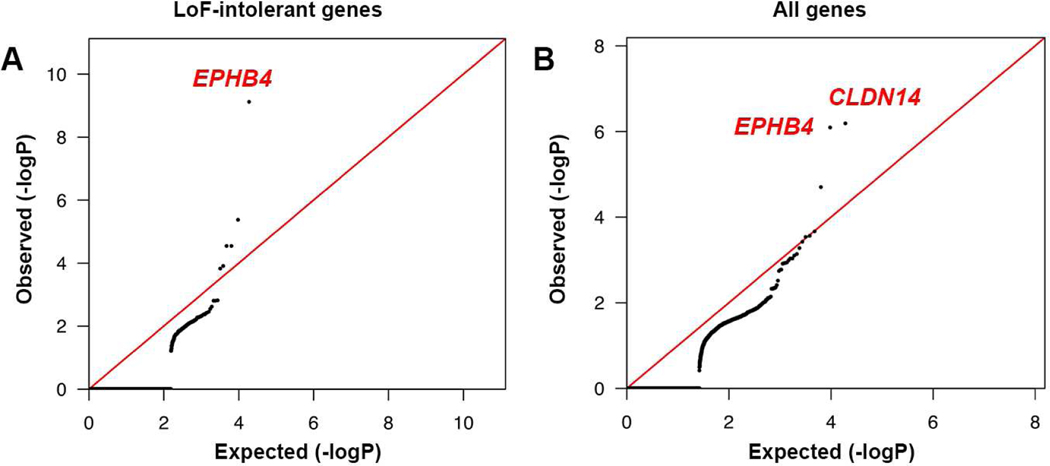
We next assessed the total burden in all probands of rare (minor allele frequency [MAF] ≤ 2 × 10−5) de novo and transmitted D-mis and LoF mutations in LoF-intolerant genes (see STAR Methods). The probability of the observed number of rare variants in each gene occurring by chance was calculated by comparing observed to the expected burden, adjusting for gene lengths (Besse et al., 2017). Analysis of damaging variants in LoF-intolerant genes revealed genome-wide significant enrichment (Bonferroni multiple testing threshold = 2.63 × 10−6) of mutations only in EPHB4 (pLI = 0.99), with one LoF and three independent D-mis mutations (one-tailed binomial P = 7.47 × 10−10; BH-FDR = 2.40 × 10−6; enrichment = 341.13-fold; Figure 3, panel A; Table 2).
Figure 3. Exome-wide significant enrichment of rare damaging transmitted mutations in EPHB4 and CLDN14.
(A) Quantile-quantile plots of observed vs. expected binomial test p-values for rare damaging (D-mis+LoF) variants with MAF ≤ 2 × 10−5 in the Genome Aggregation database (gnomAD) in LoF-intolerant genes (pLI ≥ 0.9).
(B) Quantile-quantile plots of observed vs. expected binomial test p-values for rare damaging (D-mis+LoF) variants with MAF ≤ 2 × 10−5 in gnomAD for all genes. MAF = Minor allele frequency; D-mis = Missense mutations predicted to be deleterious per MetaSVM; LoF = Canonical loss-of-function mutations (stop-gains, stop-losses, frameshift insertions or deletions, and canonical splice site mutations).
Table 2.
Transmitted mutations in EPHB4 and CLDN14 in VOGM
| Family | Type of VOGM | Ethnicity | Gene | Mutation | Domain affected | ExAC MAF* | gnomAD MAF* | pLI | MetaSVM | CADD |
|---|---|---|---|---|---|---|---|---|---|---|
| VGAM115 | Choroidal | European | EPHB4 | p.(Glu432fs1) | N/A | < 9.06E-06 | < 4.76E-06 | 0.99 | N/A | N/A |
| KVGAM25 | Choroidal | European | EPHB4 | p.(Ala509Gly) | Fibronectin III | 3.30E-05 | 1.13E-05 | 0.99 | D | 25 |
| KVGAM33 | Choroidal | Mexican | EPHB4 | p.(Lys650Asn) | Tyrosine kinase | < 8.24E-06 | < 4.06E-06 | 0.99 | D | 29.9 |
| KVGAM18 | Choroidal | European | EPHB4 | p.(Phe867Leu) | Tyrosine kinase | < 9.03E-06 | < 5.17E-06 | 0.99 | D | 31 |
| VGAM100 | Choroidal | Mexican | CLDN14 | p.(Ala113Pro) | Second intracellular segment | < 8.37E-06 | 8.28E-06 | 0 | D | 24.2 |
| KVGAM20 | Choroidal | European | CLDN14 | p.(Val143Met) | Second extracellular segment | 9.43E-06 | 1.24E-05 | 0 | D | 31 |
| KVGAM51 | Mural | European | CLDN14 | p.(Val143Met) | Second extracellular segment | 9.43E-06 | 1.24E-05 | 0 | D | 31 |
For variants not observed in public databases, their minor allele frequency is calculated as less than 1 out of total number of alleles sampled at the closest locus with allele number available
Independent case-control gene burden analyses for damaging variants in all probands versus a cohort of 3,578 autism parental and ExAC controls showed a significant mutation burden in EPHB4 vs. autism parental controls (one-tailed Fisher’s P = 1.68 × 10−6, odds ratio = 89.97, 95% CI [19.29, Inf]) and vs. ExAC controls (one-tailed Fisher’s P = 1.98 × 10−6, odds ratio = 49.76, 95% CI [16.39, Inf]); Supplementary Table 6). The Bonferroni-corrected threshold for the 3,230 LoF-intolerant genes, 1.55 × 10−5, was also surpassed by the CAD gene (one-tailed binomial P = 4.17 × 10−6; BH-FDR = 0.01; enrichment = 101.08-fold).
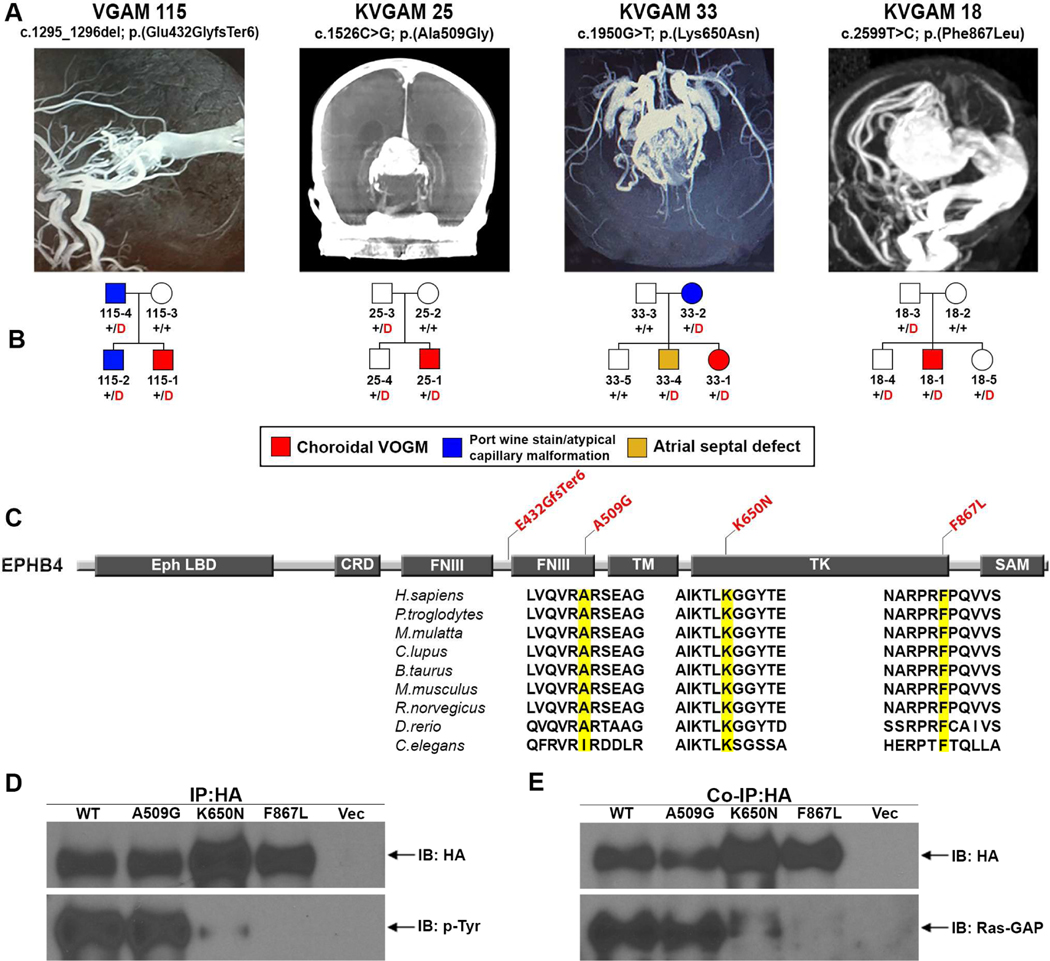
All EPHB4 mutations were transmitted; three of these have allele frequencies of zero in ExAC and gnomAD, while one has a MAF of 1.13 × 10−5 in gnomAD. All D-mis mutations in EPHB4 alter highly conserved amino acid residues (Figure 4, panel C; Table 2). Mutations p.Lys650Asn and p.Phe867Leu lie in the tyrosine kinase domain of the vein-specific (Gerety et al., 1999) Eph-B4 receptor. p.Ala509Gly lies in one of two EphB4 extracellular fibronectin III domains believed to bind extracellular matrix (Figure 4, panel C).
Figure 4. Damaging EPHB4 mutations in choroidal VOGM.
(A) Vascular imaging of probands from coronal reconstruction of computed tomography and magnetic resonance angiography demonstrating VOGMs.
(B) Pedigree structures of the kindreds, showing gene, mutation, and angiographic image. A carrier with atrial septal defect in family KVGAM 33 is in yellow. Red ‘D’ denotes damaging mutation, ‘+’ denotes wild type sequence.
(C) Eph-B4 functional domains (dark rectangles) with location of VOGM mutations (red), and phylogenetic conservation of wild-type amino acid (yellow) at each mutated position. LB = ligand binding domain; CRD = Cysteine-rich domain; FNIII = Fibronectin III domain; TM = Transmembrane domain; TK = Tyrosine kinase domain; SAM = Sterile alpha motif.
(D) Representative immunoblots showing effects of Ala509Gly, Lys650Asn, and Phe867Leu mutations on resting state Eph-B4 tyrosine phosphorylation in HEK 293T cells, analyzed by immunoprecipitation (IP) with HA-Tag antibody followed by immunoblot (IB) with anti–p-tyrosine (p-Tyr) and HA-Tag antibodies. Blot demonstrates reduced p-Tyr content in Lys650Asn, with none detectable in Phe867Leu.Representative immunoblot demonstrating binding of Eph-B4 constructs by Ras-GAP.
Ras-GAP protein was co-immunoprecipitated Ras-GAP protein was co-immunoprecipitated with Eph-B4 mutants. Ras-GAP binding to Lys650Asn and to Phe867 was markedly reduced and abrogated, respectively.
7 additional family members without diagnosed VOGM in these kindreds carried the same mutations. However, three EPHB4 mutation carriers exhibited uncommon cutaneous vascular lesions. Thus in kindred VGAM-115, the mutation-carrying father had an abdominal port wine stain, and the proband’s mutation carrier sibling had atypical left cheek and posterior thigh capillary malformations (Supplementary figure 3, panels A & B). Similarly, the mutation-carrying mother in kindred KVGAM-33 had an atypical capillary malformation on her left arm, while the proband’s mutation carrier sibling had an atrial septal defect (Figure 4, panel B). Vascular and cardiac abnormalities were absent among non-mutation carriers in these families. All VOGMs associated with EPHB4 mutations were of the choroidal subtype (Figure 4, panels A & B). Together, these findings provide evidence of incomplete penetrance and variable expressivity of EPHB4 mutations. Interestingly, 2 of 54 CM-AVM2 patients with EPHB4 mutations also had VOGM (Amyere et al., 2017).
Wild type and VOGM-mutant Eph-B4 (p.Ala509Gly; p.Lys650Asn; p.Phe867Leu) were expressed in mammalian cells (see STAR Methods), and levels of Eph-B4 tyrosine (Tyr)-phosphorylation, an index of tyrosine kinase activity (Lisabeth et al., 2013; Ferguson et al., 2015), were compared. Eph-B4 kinase domain mutants p.Lys650Asn and p.Phe867Leu showed reduced or absent phosphorylation, respectively (Figure 4, panel D). In contrast, the extracellular fibronectin III domain mutant p.Ala509Gly showed unchanged phosphorylation.
Eph-B4 Tyr phosphorylation creates docking sites for signaling molecules via a phosphotyrosine-SH2 domain interaction, facilitating Eph-B4 downstream signaling (Pawson and Scott, 1997; Wang et al., 2002). RASA1-encoded Ras GTPase-activating protein 1 (Ras-GAP) binds to these docking sites and regulates downstream signaling pathways (Kawasaki et al., 2014; Roth Flach et al., 2016). We immunoprecipitated wild type or VOGM-mutant Eph-B4 from mammalian cells co-expressing Ras-GAP. Ras-GAP co-immunoprecipation with EPHB4 mutant p.Lys650Asn was reduced, and Ras-GAP failed to bind Eph-B4 mutant p.Phe867Leu (Figure 4, panel E). In contrast, wild type Eph-B4 and Eph-B4 p.Ala509Gly bound Ras-GAP with similar affinity. These results are consistent with the cytoplasmic mutations impairing receptor kinase activity. The extracellular mutation alters the fibronectin domains, likely to affect signaling by altering binding to extracellular matrix ligands (a hypothesis not yet tested).
Recurrent rare damaging transmitted mutations in CLDN14
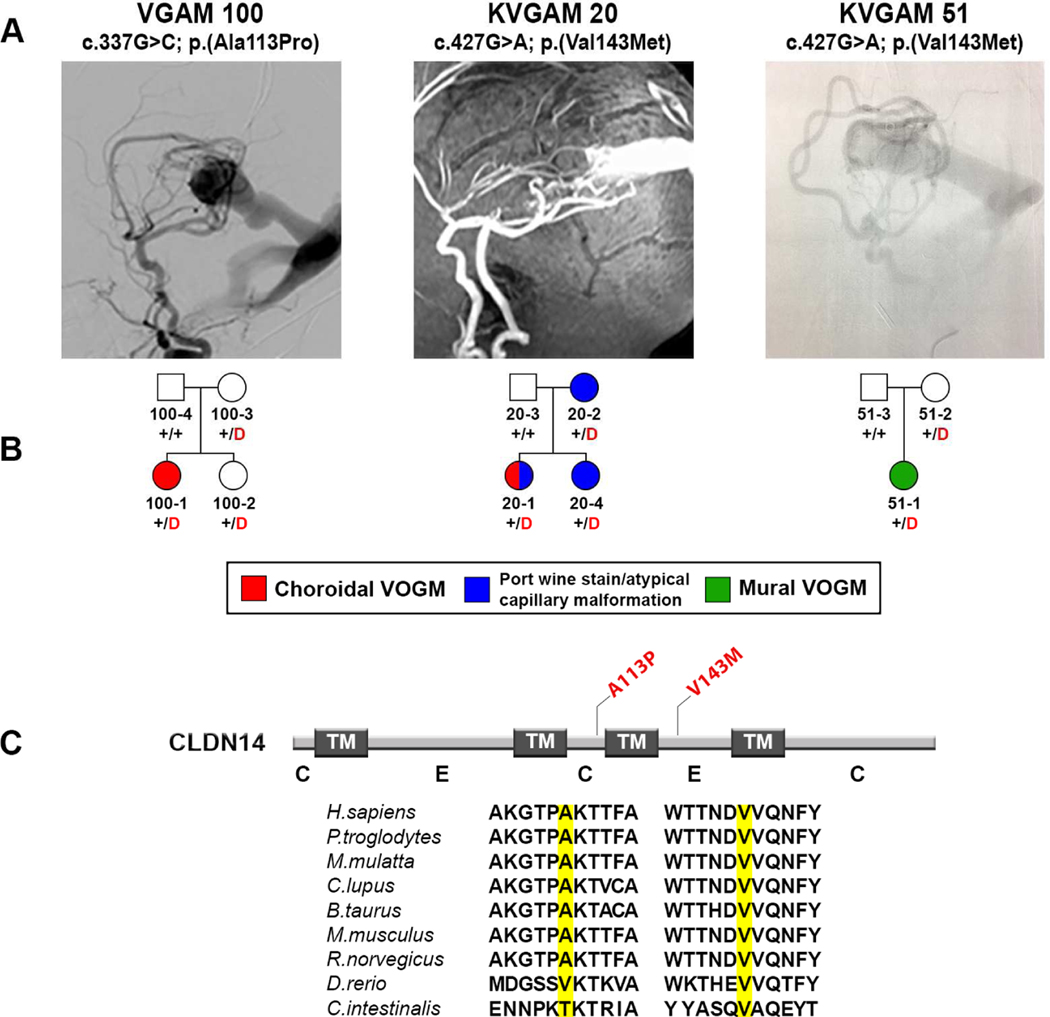
Expansion of the analysis to include the burden of rare (MAF ≤ 2 × 10−5) D-mis and LoF mutations in all genes identified one additional gene, CLDN14, with genome-wide significant damaging mutation burden (one-tailed binomial P = 6.44 × 10−7; BH-FDR = 0.01; enrichment = 190-fold; Figure 3, panel B; Table 2). Comparing CLDN14 mutation burden by case-control analysis against both autism parental controls (odds ratio = Inf, 95% CI [38.42, Inf]; one-tailed Fisher’s P = 3.38 × 10−6; Supplementary Table 6a) and ExAC controls, odds ratio = 67.82, 95% CI [17.66, Inf]; one-tailed Fisher’s P = 1.65 × 10−5; Supplementary Table 6b) supported the significance of this finding. The three rare damaging CLDN14 mutations were all heterozygous transmitted missense mutations (5.5% of probands; Figure 5). Two are identical (p.Val143Met) and present in unrelated VOGM probands, with MAF of 0 in Non-Finnish Europeans in ExAC. Beagle v3.3.2 kinship analysis (Browning and Browning, 2011; Stuart et al., 2015) and trio analysis using 139 phased genotypes flanking the mutation revealed that p.Val143Met variant lies on a segment shared identically by-descent from a common ancestor by the two probands, with a minimum shared segment of 0.34 Mb. Nonetheless, these probands shared no other rare variants, indicating that they do not share a recent common ancestor. The other rare damaging variant, p.Ala113Pro, is absent in ExAC, with MAF 8.28 × 10−6 in gnomAD.
Figure 5. Damaging CLDN14 mutations in VOGM.
(A) Representative images from digital subtraction angiographies and magnetic resonance angiography demonstrating VOGMs of the probands.
(B) Pedigree structures indicating genotypes and phenotypes as described in Figure 2.
(C) Linear representation of Claudin-14 functional domains (dark rectangles) with location of VOGM mutations (in red). Conservation of the wild-type amino acid is shown. TM = Transmembrane domain; C = cytoplasmic loop; E = extracellular loop.
CLDN14 encodes Claudin-14, a tight junction protein expressed in epithelia and endothelial cells of brain and kidney (Kniesel and Wolburg, 2000; Wattenhofer et al., 2005). Claudin extracellular loops make homo- or heterotypic interactions with adjacent cells to form the tight junction barrier (Van Itallie and Anderson, 2013). p.Val143Met alters a highly conserved residue in the second extracellular loop of Claudin-14, and is predicted to be deleterious by MetaSVM, CADD, Polyphen2, and SIFT. p.Ala113Pro lies in the second intracellular loop of Claudin-14(Figure 5, panel C; Supplementary Figure 4).
Interestingly, in one of the families (KVGAM20) harboring the recurrent Claudin-14 mutation p.Val143Met, cutaneous vascular lesions segregated with the mutation among family members. The mother who transmitted the mutation had a port wine stain on her left thigh, while her children (proband and carrier sibling) had similar atypical capillary malformations in the nuchal area. This VOGM proband also had a midline atypical capillary malformation on her lower back (Supplementary Figure 3, panels G–J).
Enrichment of mutations in genes in the Ephrin signaling pathway
To search for pathways enriched for rare damaging mutations in VOGM, LoF-intolerant genes harboring damaging de novo and/or rare (MAF ≤ 2 × 10−5) damaging transmitted mutations, along with KEL and CLDN14, were input into Ingenuity Pathway Analysis (IPA; May 2018 version; total 128 input genes) (Kramer et al., 2014). Axonal guidance signaling, essential for vascular patterning and regulated by Ephrin-Eph receptor signaling (Adams and Eichmann, 2010), was the most significantly enriched canonical pathway (P = 6.61 × 10−8; BH-FDR = 1.33 × 10−5), including Ephrin receptor signaling pathway genes (also significant). Nine additional IPA pathways were significant after B-H correction (Supplementary Table 7). Because IPA does not adjust for gene length, we tested each of these pathways in the case-control analysis (see STAR Methods) using ethnicity-matched autism parents and ExAC controls. Axonal guidance and Ephrin receptor signaling pathways showed significant enrichment in cases (Supplementary Tables 8).
We further analyzed mutation burden in genes in these two pathways by binomial test, comparing observed to expected values as corrected for gene size (see STAR Methods). Significant enrichment in both axonal guidance and Ephrin receptor signaling pathways was observed in cases, whereas analyses of synonymous variants in cases as well as rare damaging variants in autism parents and ExAC controls showed no significant enrichment (Supplementary Tables 9). Lastly, we found that most of the signal in the axonal guidance pathway was attributable to the Ephrin receptor signaling pathway because, after removal from analysis of the latter genes, the axonal guidance pathway was no longer significantly enriched (Supplementary Table 10). Detailed variant information of damaging mutations in genes that contributed to the significant result is described in Supplementary Table 11.
Of note, EFNB2 (pLI = 0.94), encoding the Eph-B4 ligand Ephrin-B2, harbored the rare D-mis mutation p.Arg277His in a neonate with a choroidal VOGM (Supplementary Figure 5). EFNB2 is represented in the IPA axonal guidance and Ephrin receptor signaling gene sets, but this specific EFNB2 variant was not included as input in our pathway analysis because its MAF slightly exceeded our threshold of 2 × 10−5 (MAF of 1.88 × 10−5 and 2.89 × 10−5 in ExAC and gnomAD, respectively). The proband’s EFNB2 mutation-carrying mother, while not having VOGM, exhibited multiple atypical nuchal capillary malformations. Of note, Ephrin-b2 knockout mice phenocopy eph-b4 knockout mice, exhibiting CNS and systemic AVMs, defects in cerebral angiogenesis, and embryonic lethality (Wang et al., 2010b).
Rare damaging mutations in genes implicated in Mendelian AVM syndromes
VOGM is a rare feature in patients with CM-AVM types 1 and 2 due to mutation in RASA1 (Revencu et al., 2013; Duran et al., 2018) and EPHB4 (Amyere et al., 2017), respectively. VOGM has also previously been associated with single cases of Hereditary Hemorrhagic Telangiectasia (HHT) type 1 due to ENG mutation (Tsutsumi et al., 2011) and HHT type 2 due to ACVRL1 mutation (OMIM# 600376) (Chida et al., 2013).
Our VOGM cohort included no ENG mutations. We found one damaging mutation in RASA1 (p.Arg709*, Supplementary Table 11), encoding the Eph-B4 binding partner and effector Ras GTPase-activating protein 1 (Ras-GAP) (Kawasaki et al., 2014), in a patient with a mural VOGM. This patient and mutation has been previously reported (Revencu et al., 2013) and was independently ascertained in the present study.
A single patient with a choroidal VOGM had a damaging p.Arg484Gln mutation in ACVRL1 (Supplementary Table 12), encoding ALK1, a receptor kinase in the TGF-β signaling pathway highly expressed in developing human vasculature (Zhang et al., 2017). This mutation altering a conserved residue in the ALK1 kinase domain was reported in HHT2 with isolated pulmonary hypertension (ClinVar accession RCV000198604.1; rs863223408) (Harrison et al., 2005). However, neither the ACVRL1-mutant VOGM proband nor family members carrying the mutation exhibited HHT-associated findings (e.g., epistaxis or telangiectasia) or other vascular abnormalities. ACVRL1 (ALK1) is a known regulator of Ephrin-B2-Eph-B4 signaling (Zhang, 2009; Kim et al., 2012; Roman and Hinck, 2017), and acvrl1 deficient mice exhibit markedly enlarged cerebral vessels with arteriovenous shunting and altered Eph-B4 expression (Walker et al., 2011).
Unrelated probands with mural VOGMs carried two rare damaging mutations (p.Gly39Ser and p.Asn373Ser) in the ACVRL1 paralog, ACVR1 (pLI = 0.96); Supplementary Table 12; Supplementary Figure 6, panels A & B), a gene not previously implicated in VOGM. The ACVR1 p.Asn373Ser VOGM proband also had an atrial septal defect and partial anomalous pulmonary venous return (Supplementary Figure 6, panel B), and two mutation carriers in the family had cutaneous capillary malformations (Supplementary Figure 6, panel B). Neither the proband nor other family members had common HHT features (Abdalla et al., 2003). ACVR1 encodes the receptor serine-threonine receptor kinase ALK2 coordinating with TGF-beta type 2 receptors and co-receptors such as Endoglin (ENG) (Chen et al., 1998; Barbara et al., 1999; Wolfe and Myers, 2010). p.Gly39Ser alters a conserved residue in the extracellular ligand binding domain (Supplementary Figure 6, panel C). p.Asn373Ser alters a conserved residue predicted to be structurally critical to the cytoplasmic serine-threonine kinase (Supplementary Figure 6, panels D & E).
DISCUSSION
The rarity and the sporadic nature of VOGM have hindered its genetic understanding. This study, the largest trio-based genomic analysis of VOGM to date, has provided the following novel insights. First, probands exhibit an excess of damaging de novo mutations (~13% of cases); among these, mutations in chromatin modifier genes with essential roles in brain and heart development are enriched, and inferred to impact ~8% of cases. Second, there is a prevalence of rare inherited damaging mutations in the Ephrin signaling genes, including a genome-wide significant burden in EPHB4, (another ~16% of probands). Third, inherited mutations show incomplete penetrance and variable expressivity, with mutation carriers often exhibiting cutaneous vascular lesions, suggesting a two-hit mechanism. Thus, while rare mutations of large effect contribute to a significant fraction of VOGM cases, mutations in many additional genes likely contribute to disease pathogenesis. Our results support this hypothesis, suggesting potential pathogenic roles for de novo KEL mutations and rare, inherited CLDN14 mutations. However, the small number of observations and lack of replication studies require validation and extension by larger follow-up studies. Of note, analysis of rare homozygous and compound heterozygous genotypes (MAF ≤ 0.001) revealed no genes with more than one such genotype. See Supplementary Table 13 for proband information and identified likely pathogenic mutations.
Genes encoding covalent histone modifiers and chromatin remodelers have been implicated in autism (De Rubeis et al., 2014), CHD (Jin et al., 2017), congenital hydrocephalus (Furey et al., 2018), and other congenital disorders (Feinberg, 2018). In our cohort, 4/55 probands had de novo mutations in chromatin modifiers. Enrichment of these mutations in our cohort and the conservation of the mutated residues in critical domains of protein function are consistent with each of these mutations contributing to VOGM pathogenesis. All four genes (KMT2D, SMARCA2, SIRT1, KAT6A) are highly expressed in the developing human and murine brain (Machida et al., 2001; Ogawa et al., 2011; Pollen et al., 2015; Tham et al., 2015) and essential for neuronal and/or vascular development (Potente et al., 2007; Griffin et al., 2008; Van Laarhoven et al., 2015). SMARCA2, KAT6A, KMT2D are mutated in Mendelian diseases that feature intellectual disability and/or epilepsy (Morin et al., 2003; Dentici et al., 2015). Mendelian phenotypes associated with KAT6A and KMT2D mutations include vascular defects and CHD (Arboleda et al., 2015; Van Laarhoven et al., 2015). Multiple mutated chromatin modifiers are shared among patients with CHD and autism (Zaidi et al., 2013; Homsy et al., 2015; Jin et al., 2017). ~87% of CHD patients with LoF de novo mutations in chromatin modifier genes exhibit neurodevelopmental phenotypes (Jin et al., 2017). These observations suggest that neurodevelopmental phenotypes in VOGM patients currently attributed to secondary CNS damage may instead reflect primary impairment from genetic mutation.
The role of Eph-B4 in A-V specification is well established in model systems (Zhang and Hughes, 2006; Mosch et al., 2010). Heterozygous missense variants in EPHB4 have also been reported in two families with non-immune hydrops fetalis and/or atrial septal defect (HFASD; OMIM # 617300) (Martin-Almedina et al., 2016) and 54 families with CM-AVM2, featuring isolated cutaneous capillary malformations (63%) and associated AVMs (35%) (Amyere et al., 2017). We found damaging mutations in EPHB4 in 7% of VOGM probands in this cohort. Two of 52 prior CM-AVM2 patients with EPHB4 mutations were reported to have VOGMs (Amyere et al., 2017). During preparation of this manuscript, (Vivanti et al., 2018) reported 3 transmitted damaging mutations in EPBH4 among whole exome sequences from 19 VOGM case-parent trios, and two additional mutations from targeted sequencing of 32 other singleton VOGM probands. Eph-b4 antisense morpholino knockdown in zebrafish embryos disrupts angioarchitecture of the dorsal longitudinal vein, homolog of the human Vein of Galen precursor (Aurboonyawat et al., 2007). We conclude that heterozygous EPHB4 germline mutations contribute to a spectrum of vascular pathology, and EPHB4 is a bona fide VOGM risk gene.
Our analysis also demonstrated enrichment of rare heterozygous damaging mutations in Ephrin signaling genes (Supplementary Table 11). These genes are expressed in the embryonic human brain and vasculature (Guo et al., 2012), regulate neurovascular development, and can be mapped into a single experimentally-supported STRING interactome (Supplementary Figure 7, see STAR Methods). Cutaneous vascular lesions are a common hallmark of developmental vascular disorders such as RASA1- and EPHB4-mutated CM-AVM (Revencu et al., 2013; Amyere et al., 2017); ENG1- and ACVRL1-mutated HHT (Chida et al., 2013); and RASA1-mutated Parkes Weber syndrome (Brouillard and Vikkula, 2007). We found similar cutaneous vascular lesions in VOGM kindreds harboring mutations in EFNB2, EPHB4, EPHA4, ACVR1, and CLDN14. These findings further implicate these genes in a Eph-B4-RASA1 signaling network (Supplementary Figure 8).
Transmitted VOGM-associated mutations show incomplete penetrance. Moreover, many of the identified VOGM-associated genes harboring damaging de novo and inherited mutations have been implicated in other Mendelian diseases, sometimes producing different phenotypes. These observations highlight the pleiotropy with variable expressivity resulting from these mutations.
These features have been described in other diseases. For example, haploinsufficiency for the identical chromatin modifier genes results in CHD (Zaidi et al., 2013) or autism (Iossifov et al., 2014), or both. Variable expressivity of VOGM and associated features could arise from environmental modifiers (Garcia et al., 2015) in concert with the rare mutations identified and/or specific genetic modifiers (Timberlake et al., 2016). Co-mutation of genes in KVGAM20, VGAM100, and KVGAM45 could be important in this regard (see Supplementary Table 13). Sequencing additional exomes from VOGM kindreds will help clarify this issue.
The mechanisms by which syndromes characterized by abnormal A-V specification present with multifocal distributions of lesions remains poorly understood. Since 68% of VOGM families with full clinical data had capillary malformations or other uncommon cutaneous vascular lesions, and that identified mutations in probands were found in all family members with these cutaneous lesions, provide evidence linking VOGM and the cutaneous lesions to the same mutations (Supplementary Table 13). This is consistent with a two-hit mechanism, in which phenotypic expression relies upon an inherited mutation and a second, post-zygotic mutation in the other wild-type allele (Brouillard et al., 2002; Pagenstecher et al., 2009). This mechanism has been shown for other hereditary multifocal vascular malformations such as RASA1-mutated CM-AVM1 (Revencu et al., 2013), glomuvenous malformations (OMIM# 138000), cutaneomucosal venous malformation (OMIM# 600195), and cerebral cavernous malformations (OMIM# 116860) (Pagenstecher et al., 2009). In this context, phenotypic expression depends on the cell types in which somatic mutations occur, and could explain the low penetrance of VOGM arising from transmitted mutations. Further work, including exome sequencing of lesional VOGM tissue, will test this hypothesis.
Although the identified de novo KEL mutations and inherited CLDN14 mutations will require further validation by WES of additional VOGM patients and functional studies, several observations suggest the importance of the current findings. KEL, encoding the Kell blood group transmembrane glycoprotein, was the only gene in our study that harbored more than one protein-altering de novo mutation (see Figure 1). Both the premature termination and p.Gly202Ser mutations in Kell alter its peptidase domain, shown to generate vasoactive endothelin peptides via cleavage of the endothelin-3 pro-protein (Lee et al., 1999). Endothelins provide vascular-derived axonal guidance cues (Makita et al., 2008) involved in Ephrin-dependent vascular patterning (Adams and Eichmann, 2010).
CLDN14 was the only other gene besides EPHB4 with genome-wide significant enrichment of transmitted damaging mutations. Recessive LoF genotypes in CLDN14 cause sensorineural deafness type 29 (OMIM # 614035); in contrast, VOGM-associated CLDN14 mutations are heterozygous and D-mis, and include a recurrent missense mutation, suggesting gain-of-function or neomorphic effects, and phenotypic heterogeneity. Claudin-14 is a tight junction protein in brain epithelia and endothelial cells (Kniesel and Wolburg, 2000; Wattenhofer et al., 2005). The regulation of tight junction formation by Claudins can impact endothelial cell permeability, integrity, and proliferation (Morita et al., 1999; Gonzalez-Mariscal et al., 2007). The recurrent VOGM-associated Claudin-14 mutation in lies in the large 2nd extracellular loop that likely plays a role in tight junction formation. Endothelial cells heterozygous, but not homozygous, for CLDN14 exhibit disruption of ZO-1-positive cell-cell junctions, abnormal distribution of basement membrane laminin, increased VEGF-stimulated angiogenesis, and significantly enhanced cell proliferation – suggesting a gene dosage effect (Baker et al., 2013). Functional interactions have been reported between Claudins and Ephrin-B2-EphB4 bi-directional signaling (Tanaka et al., 2005). In support of a possible Ephrin-Claudin-14 interaction is the fact that one of the families harboring the recurrent Claudin-14 mutation p.Val143Met had CM-AVM-like cutaneous vascular lesions that segregated with the mutation in family members (see Figure 5, panel B). The role of Claudin-14 in potential EphB4-dependent A-V specification will be a topic of future investigation.
These findings suggest mutation carrier offspring may be at increased risk of VOGMs, as well as capillary malformations and potentially other AVMs. However, not all mutation carriers develop capillary malformations, making absence of capillary malformations an unreliable clinical marker for transmission risk in affected families. These observations suggest the importance of family history and mutation-based screening for risk assessment. The narrow developmental window of gestational weeks 6–11 during which PVM fistulas form (Raybaud et al., 1989) poses a challenge to improved early therapeutic strategies for VOGM. Thus, attempted diagnosis with intention to treat must occur before the safe gestational age threshold for amniocentesis (Shulman et al., 1994). These difficulties highlight the need for continued genetic research on VOGM, with focus on mechanistic implications of recently discovered VOGM-associated mutations.
Eph-B4 kinase domain mutations remove inhibition of downstream RAS/MAPK/ERK1/2 and PI3K/AKT/mTORC1 signaling cascades (Kim et al., 2002; Salaita and Groves, 2010; Xiao et al., 2012). We showed select VOGM-associated Eph-B4 mutations result in decreased binding of Eph-B4 to RASA1(see Figure 4, panel E). PI3K/AKT/mTORC1 up-regulation has been noted in capillary malformations of RASA1-mutant CM-AVM1 patients (Kawasaki et al., 2014). Therapy targeting Eph-B4-Ras-GAP-mTOR signaling may represent a viable therapeutic approach for VOGM and, perhaps, CM-AVM spectrum lesions.
STAR∗METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Kristopher T. Kahle (kristopher.kahle@yale.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Pateint Subjects
All procedures in this study comply with Yale University’s Human Investigation Committee (HIC) and are Human Research Protection Program. Written informed consent was obtained from all adult participants. Parent or legal guardian authorization was obtained in writing for sample collection of all minors in this study. Inclusion criteria included male or female patients with clearly defined mural or choroidal VOGMs, radiographically-confirmed by both a neurosurgeon and neuroradiologist from an angiogram or magnetic resonance angiogram, along with their family members. Fifty-five probands diagnosed with VOGM were included in this study, of which 52 were parent-offspring trios. Among these 55 VOGM probands, 65.5% were female, 76.4% were self-reported Europeans, and 56.4% had a family history of cutaneous vascular abnormalities. (Supplementary Table 1).
Controls consist of 1,789 unaffected sublings of autism cases and unaffected parents from the Simons Foundation Autism Research Initiative Simplex Collection (SSC)(Fischbach and Lord, 2010; O’Roak et al., 2011; Sanders et al., 2012; Iossifov et al., 2014; Krumm et al., 2015). Only the unaffected siblings and parents, as designated by SSC, were included in the analysis and served as controls for this study. Permission to access to the genomic data in the SCC on the National Institute of Mental Health Data Repository was obtained. Written informed consent for all participants was provided by the Simons Foundation Autism Research Initiative.
Whole Exome Sequencing and Variant Calling
Exon capture was performed on genomic DNA samples derived from saliva or blood using Roche SeqCap EZ MedExome Target Enrichment kit or IDT xGen target capture kit followed by 99 base paired-end sequencing on the Illumina HiSeq 2500 platform. Sequence reads were aligned to the human reference genome GRCh37/hg19 using BWA-MEM (Li, 2014) and further processed to call variants following the GATK Best Practices workflow (McKenna et al., 2010). Variants were annotated with ANNOVAR (Wang et al., 2010a) and MetaSVM (Dong et al., 2015) was used to predict the deleteriousness of non-synonymous variants (herein referred to as D-mis). All variants covered by independent aligned sequencing reads with a depth of 8x or greater were visualized in silico to eliminate false positives.
De novo mutations were called using TrioDeNovo (Venugopal, 2014). Candidate de novo mutations were further filtered based on the following criteria: (1) exonic or splice-site variants; (2) read depth (DP) of 10 in the proband and both parents; (3) genotype quality (GQ) score ≥ 20; (4) minium proband alternative read depth of 5; (5) proband alternative allele ratio ≥ 28% if having < 10 alternative reads or ≥ 20% if having ≥ 10 alternative reads; (6) alternative allele ratio in both parents ≤ 3.5%; (8) in-cohort allele frequency ≤ 4 × 10−4 for controls and MAF ≤ 4 × 10−4 in gnomAD for cases due to limited cohort size.
For rare transmitted dominant variants, only LoF mutations (stop-gains, stop-losses, canonical splice-sites, and frameshift indels) and D-mis mutations (missense mutations predicted deleterious by MetaSVM) were considered potentially damaging for subsequent one-tailed binomial analysis and filtered using the following criteria to reduce false positives: (1) GATK variant quality score recalibration (VQSR) of PASS, (2) MAF ≤ 2 × 10−5 in gnomAD (calculated based on combined dataset of WES and WGS data from gnomAD database, Lek et al. 2016), (3) DP ≥ 8 independent reads, and (4) GQ score ≥ 20. Transmitted recessive variants were filtered for rare (MAF ≤ 10−3 in gnomAD) homozygous and compound heterozygous variants using the same criteria described above.
Candidate mutations were confirmed by PCR amplification followed by Sanger sequencing (primer sequences available on request).
METHOD DETAILS
Kinship Analysis
Pairwise proband relatedness and pedigree information of trios were confirmed using KING (Manichaikul et al., 2010) by estimating kinship coefficient and calculating identity-by-descent (IBD). The shared pairwise IBD segments in 45 European probands were detected using Beagle v3.3.2 (Browning and Browning, 2011).
De Novo Mutation Expectation Model
We used a sequence context probability model to derive the per-gene probability of observing a de novo mutation by chance as previously described (Samocha et al., 2014). In brief, for each base in the exonic region, the probability of observing each of the three possible single nucleotide changes was determined. The coding consequence of each possible mutation was determined, and then these probabilities of mutations were summed for each variant functional class (synonymous, missense, nonsense, canonical splice site, frameshift, and stop-lost) to create a per-gene probability of mutations. The probability of a frameshift mutation was determined by multiplying the probability of a nonsense mutation by 1.25 as described previously (Samocha et al., 2014). In-frame insertions and deletions are not currently accounted for by this framework and were not included in the analysis. The per-gene probability for each functional class was adjusted to control for sequencing coverage. Due to the difference in exome capture kits, DNA sequencing platforms, and variable sequencing coverage between case and control cohorts, the expected number of de novo mutations was estimated by adjusting for sequencing depth in 52 case trios and 1,789 autism control trios separately.
De Novo Enrichment Analysis and Variant Stratification
Rather than using the variant calls in controls published in the SSC study (Krumm et al., 2015), we downloaded the bam files from the SSC, reanalyzed the data, and filtered the control vcf file using the same filtering criteria as what was used in our case cohort. A one-tail Poisson test was used to compare observed number of de novo mutations across each variant class to expected number under the null hypothesis. R package ‘denovolyzeR’ (Ware et al., 2015) was used to perform the analyses. The Benjamini-Hochberg method was used to correct for multiple testing while taking into account all genes (N = 18,989) and calculate adjusted p-values. A gene was considered significant if Benjamini-Hochberg adjusted p-value is ≤ 0.05. All genes represented in this dataset were annotated with artery-specific and brain-specific expression values in a form of reads per kilobase transcript per million reads (RPKM) from the GTEx database (https://gtexportal.org/home/). Genes harboring de novo mutations were also annotated with human brain specific expression data obtained during the first four weeks of development (Gerrard et al., 2016) in the form of quantile rank based on transcript per kilobase million (TPM), indicating the relative rank of expression level within human genome. The final dataset was analyzed for recurrently affected genes, and all variants in genes affected by a single de novo mutation were stratified. LoF variants were ranked based on pLI (from highest to lowest).
QUANTIFICATION AND STATISTICAL ANALYSIS
Estimation of the Number of Damaging De Novo Mutations in LoF-intolerant Chromatin Modifiers
One million permutations were performed to derive the empirical distribution of the number of damaging de novo mutation in the LoF-intolerant chromatin modifier genes. For each permutation, the number of observed damaging de novo mutations in all genes (n = 19) was randomly distributed across the exome, weighted according to the de novo probabilities of damaging mutations. The empirical p-value is calculated as the proportion of times that the number of damaging de novo mutations in the LoF-intolerant chromatin modifier genes is greater than or equal to the observed number (n = 4). The average number of damaging de novo mutations in the LoF-intolerant chromatin modifier genes is also calculated.
Binomial Analysis
Independent binomial tests were used to compare the expected and observed counts of rare variants in each gene. The expected number of rare damaging variants is determined by taking the fractional length of a gene (in base pairs) relative to the entire exome and multiplying this by the total number of rare damaging variants. This represents the expected occurrence of sporadic mutations in each gene without considering the influences of selection pressure or precedents of ethnic background. Inputs for this test were those with inferred pathogenicity, including missense mutations called deleterious per MetaSVM and inferred LoF mutations (stop-gains, stop-losses, frameshift insertions/deletions, or canonical splice site mutations). Binomial analysis for mutational enrichment did not include non-frameshift insertions or deletions. The Benjamini-Hochberg method was performed taking into account all genes (N = 18,989 for CLDN14) or all LoF-intolerant genes (N = 3,230 for EPHB4 and CAD) as described above and the significance cutoff was 0.05. We reported genes that reached a more stringent Bonferroni multiple testing cutoff of 2.63 × 10−6 (= 0.05/18,989) or 1.55 × 10−5 for binomial testing of all genes or LoF-intolerant genes, respectively.
Pathway Analysis
Inputs for this analysis were LoF-intolerant genes (pLI ≥ 0.9) harboring damaging de novo mutations and/or rare (MAF ≤ 2 × 10−5) damaging transmitted mutations, as well as genes with significant burden of de novo (KEL) or transmitted mutation (EPHB4 and CLDN14) (n = 128 genes) into Ingenuity Pathway Analysis (IPA, Apr 2018). Core analysis using Ingenuity Knowledge Base (Gene Only) as the reference set was performed. P-value was calculated using a one-tailed Fisher’s exact test reflecting the likelihood that the overlap between the input and a given gene set is due to random chance. In individual based case-control analysis, ethnicity-matched case and control samples were filtered using the same criteria. Individuals carrying variants of interest in case and control groups were tallied separately, and the p-value was obtained from a one-tailed Fisher’s exact test. In binomial pathway analysis, the observed number of rare damaging variants in LoF-intolerant genes that belong to statistically significant canonical pathways of interest were compared to the expected number of mutations in each set using a one-tailed binomial test. Gene sets of canonical pathways were obtained from IPA. The expected number of mutations in a given gene set is calculated as the formula below:
Where N denotes total number of rare damaging de novo and transmitted mutations in intolerant genes as well as genes with significant burden of de novo and transmitted mutations.
Interactome Construction
We input all genes contributing to the significantly enriched pathway to String (version 10.5) (Szklarczyk et al., 2015). For organism, Homo sapiens was selected. For each displayed interaction, active interaction sources were restricted to experiments, and the maximum number of interactors was limited to 50.
In Silico Modeling of Mutational Effects on Protein Structure
The sequence for all available modeled human proteins was downloaded from Uniprot (Apweiler et al., 2004). The stereochemical parameters of VOGM-associated mutations were analyzed using PROCHECK (Laskowski et al., 1993) and PROSA (Wiederstein and Sippl, 2007), and the final models were chosen based on the lowest energy function score (Dope) within the modeling program. The mutations were constructed and the free energy of change calculated (ΔΔG) in silico using the ICM mutagenesis (Abagyan et al., 1994).
Cell Culture
HEK 293T cells were passaged at 80–90% confluence on high glucose DMEM (Dulbecco’s modified Eagle’s medium, Gibco Life Technologies, Waltham MA, USA) supplemented with 10% fetal bovine serum (FBS, Gibco Life Technologies Waltham MA, USA), L-glutamine, and penicillin/streptomycin.
Mutagenesis of Eph-B4 and Plasmid Transfection
A wild-type mouse EPHB4 cDNA was sub-cloned into the pShuttle-IRES-hrGFP-2 plasmid vector with HA-Tag sequence (Protack et al., 2017). The QuikChange II Site-Directed Mutagenesis Kit (Aligent Technologies, Santa Clara CA, USA) was used to generate isolated single amino-acid changes within the EPHB4 ORF (A509G, K650N, F867L). All mutant constructs were sequenced to confirm successful mutagenesis. The wild-type EPHB4, the mutant constructs, and empty vector were individually transiently transfected into HEK 293T cells using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA) according to its standard protocol. DNA complexes were removed after 5 h and replaced with fresh complete medium. After 48 hours, the medium was aspirated and the cells starved for 18 h in serum-free conditions.
Co-immunoprecipitation and Western Blotting
Cell lysates were prepared using NP40 lysis buffer (50 mM Tris, pH7.5; 1% Nonidet P-40; 150 mM NaCl; 10% Glycerol) containing protease and phosphatase inhibitor cocktail (Roche, Basel, Switzerland). Protein concentrations were measured with DC Protein Assay Reagents (Bio-Rad, Hercules CA, USA). For immunoprecipitation, equal amounts of cell lysates were incubated with Sepharose beads linked to anti-HA-Tag antibody (Cell Signaling Technology, Danvers, MA, USA) overnight at 4 °C. Immunoprecipitated protein complexes were separated on SDS-PAGE gel and analyzed by Western blotting using following antibodies: anti-phosphotyrosine P-Tyr-100, anti-HA-Tag, (Cell Signaling Technology, Danvers MA, USA), anti-Ras GAP (Santa Cruz Biotechnology, Dallas TX, USA). Band intensities were quantified using ImageJ software (Schneider et al., 2012). Statistical analyses were performed using Prism 7 software (GraphPad Software, La Jolla CA, USA).
DATA AND SOFTWARE AVAILABILITY
WES data for all VOGM parent-offspring trios reported in this study have been deposited in the NCBI database of Genotypes and Phenotypes under the accession number phs000744.v4.p2
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited Data | ||
| Whole exome sequencing data of 52 VOGM trios and 3 singletons | This paper | Accession no. phs000744.v4.p2 http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000744 |
| Whole exome sequencing data of 1,789 control trios from the Simon Simplex Collection | Iossifov et al., 2014 | NDAR: DOI:10.15154/1149697 https://ndar.nih.gov/study.html?id=352 |
| Software and Algorithms | ||
| Genome Analysis Tool Kit (GATK) | DePristo et al., 2011; McKenna et al., 2010; Van der Auwera et al., 2013 |
https://software.broadinstitute.org/gatk/best-practices/
|
| BWA-mem | Li and Durbin, 2009 |
http://bio-bwa.sourceforge.net/
|
| Annovar | Wang et al., 2010 |
http://annovar.openbioinformatics.org/en/latest/
|
| PLINK/SEQ | Fromer et al., 2014 |
https://atgu.mgh.harvard.edu/plinkseq/
|
| Antibodies | ||
| Anti-HA-Tag antibody | Cell Signaling Technology, Danvers, MA, USA | AB_10691311 |
| Other | ||
| 1000 Genomes GRCh37 h19 genome build | 1000 Genomes Project |
http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/reference/human_g1k_v37.fasta.gz
|
| RefSeq hg19 gene annotation | UCSC Genome Browser |
http://genome.ucsc.edu/cgi-bin/hgTables?command=start
|
| Intervals file for IDT xGen v1.0 | Integrated DNA Technologies |
https://www.idtdna.com/pages/products/next-generation-sequencing/hybridization-capture/lockdown-panels/xgen-exome-research-panel
|
| ExAC Browser (Beta) | Exome Aggregation Consortium | http://exac.broadinstitute.org/ |
| gnomAD Browser | genome Aggregation Database | http://gnomad-old.broadinstitute.org/ |
Highlights.
Exome sequencing identifies genetic drivers of Vein of Galen malformations (VOGMs)
Mutations in chromatin modifier and Ephrin genes account for ~30% of VOGM cases.
Probands often exhibit vasculo-cutaneous lesions, suggesting a two-hit mechanism.
These data implicate impaired arterio-venous specification in VOGM pathogenesis.
ACKNOWLEDGEMENTS
We thank the patients and families who participated in this research. We acknowledge support from the Yale-NIH Center for Mendelian Genomics (5U54HG006504) and an NIH NRCDP award to K.T.K. S.C.J. was supported by the James Hudson Brown-Alexander Brown Coxe Postdoctoral Fellowship and the American Heart Association Postdoctoral Fellowship. J.G. was supported by the Howard Hughes Institute Medical Research Fellowship. Lastly, we acknowledge Collin Whitmore and his family for their inspiration, courage, and generous support.
Footnotes
DECLARATION OF INTERESTS
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abagyan R, Totrov M, and Kuznetsov D.(1994). Icm - a New Method for Protein Modeling and Design - Applications to Docking and Structure Prediction from the Distorted Native Conformation. J Comput Chem 15, 488–506. [Google Scholar]
- Abdalla SA, Geisthoff UW, Bonneau D, Plauchu H, McDonald J, Kennedy S, Faughnan ME, and Letarte M.(2003). Visceral manifestations in hereditary haemorrhagic telangiectasia type 2. J Med Genet 40, 494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams RH, and Eichmann A.(2010). Axon guidance molecules in vascular patterning. Cold Spring Harb Perspect Biol 2, a001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul D, Paramasivam S, Ortega-Gutierrez S, Fifi JT, and Berenstein A.(2014). Safety and efficacy using a detachable tip microcatheter in the embolization of pediatric arteriovenous malformations. Childs Nerv Syst 30, 1099–1107. [DOI] [PubMed] [Google Scholar]
- Amyere M, Revencu N, Helaers R, Pairet E, Baselga E, Cordisco M, Chung W, Dubois J, Lacour JP, Martorell L, et al. (2017). Germline Loss-of-Function Mutations in EPHB4 Cause a Second Form of Capillary Malformation-Arteriovenous Malformation (CM-AVM2) Deregulating RAS-MAPK Signaling. Circulation 136, 1037–1048. [DOI] [PubMed] [Google Scholar]
- Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. (2004). UniProt: the Universal Protein knowledgebase. Nucleic Acids Res 32, D115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurboonyawat T, Suthipongchai S, Pereira V, Ozanne A, and Lasjaunias P.(2007). Patterns of cranial venous system from the comparative anatomy in vertebrates. Part I, introduction and the dorsal venous system. Interv Neuroradiol 13, 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M, Reynolds LE, Robinson SD, Lees DM, Parsons M, Elia G, and Hodivala-Dilke K.(2013). Stromal Claudin14-heterozygosity, but not deletion, increases tumour blood leakage without affecting tumour growth. PloS one 8, e62516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak T, Kwan KY, Louvi A, Demirbilek V, Saygi S, Tuysuz B, Choi M, Boyaci H, Doerschner K, Zhu Y, et al. (2011). Recessive LAMC3 mutations cause malformations of occipital cortical development. Nature genetics 43, 590–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbara NP, Wrana JL, and Letarte M.(1999). Endoglin is an accessory protein that interacts with the signaling receptor complex of multiple members of the transforming growth factor-beta superfamily. The Journal of biological chemistry 274, 584–594. [DOI] [PubMed] [Google Scholar]
- Besse W, Dong K, Choi J, Punia S, Fedeles SV, Choi M, Gallagher AR, Huang EB, Gulati A, Knight J, et al. (2017). Isolated polycystic liver disease genes define effectors of polycystin-1 function. J Clin Invest 127, 1772–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilguvar K, Ozturk AK, Louvi A, Kwan KY, Choi M, Tatli B, Yalnizoglu D, Tuysuz B, Caglayan AO, Gokben S, et al. (2010). Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature 467, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouillard P, Boon LM, Mulliken JB, Enjolras O, Ghassibe M, Warman ML, Tan OT, Olsen BR, and Vikkula M.(2002). Mutations in a novel factor, glomulin, are responsible for glomuvenous malformations (“glomangiomas”). American journal of human genetics 70, 866–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouillard P, and Vikkula M.(2007). Genetic causes of vascular malformations. Human molecular genetics 16 Spec No. 2, R140–149. [DOI] [PubMed] [Google Scholar]
- Browning BL, and Browning SR (2011). A fast, powerful method for detecting identity by descent. American journal of human genetics 88, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YG, Hata A, Lo RS, Wotton D, Shi Y, Pavletich N, and Massague J.(1998). Determinants of specificity in TGF-beta signal transduction. Genes Dev 12, 2144–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chida A, Shintani M, Wakamatsu H, Tsutsumi Y, Iizuka Y, Kawaguchi N, Furutani Y, Inai K, Nonoyama S, and Nakanishi T.(2013). ACVRL1 gene variant in a patient with vein of Galen aneurysmal malformation. J Pediatr Genet 2, 181–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, Vulto-van Silfhout AT, Koolen DA, de Vries P, Gilissen C, et al. (2012). Diagnostic exome sequencing in persons with severe intellectual disability. The New England journal of medicine 367, 1921–1929. [DOI] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, et al. (2014). Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloison B, Chalouhi GE, Sonigo P, Zerah M, Millischer AE, Dumez Y, Brunelle F, Ville Y, and Salomon LJ (2012). Hidden mortality of prenatally diagnosed vein of Galen aneurysmal malformation: retrospective study and review of the literature. Ultrasound Obstet Gynecol 40, 652–658. [DOI] [PubMed] [Google Scholar]
- Dong C, Wei P, Jian X, Gibbs R, Boerwinkle E, Wang K, and Liu X.(2015). Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Human molecular genetics 24, 2125–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran D, Karschnia P, Gaillard JR, Karimy JK, Youngblood MW, DiLuna ML, Matouk CC, Aagaard-Kienitz B, Smith ER, Orbach DB, et al. (2018). Human genetics and molecular mechanisms of vein of Galen malformation. J Neurosurg Pediatr, 1–8. [DOI] [PubMed] [Google Scholar]
- Epi KC, Epilepsy Phenome/Genome, P., Allen AS, Berkovic SF, Cossette P, Delanty N, Dlugos D, Eichler EE, Epstein MP, Glauser T, et al. (2013). De novo mutations in epileptic encephalopathies. Nature 501, 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP (2018). The Key Role of Epigenetics in Human Disease Prevention and Mitigation. The New England journal of medicine 378, 1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson BD, Tan YH, Kanteti RS, Liu R, Gayed MJ, Vokes EE, Ferguson MK, Iafrate AJ, Gill PS, and Salgia R.(2015). Novel EPHB4 Receptor Tyrosine Kinase Mutations and Kinomic Pathway Analysis in Lung Cancer. Sci Rep 5, 10641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach GD, and Lord C.(2010). The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron 68, 192–195. [DOI] [PubMed] [Google Scholar]
- Fish JE, and Wythe JD (2015). The molecular regulation of arteriovenous specification and maintenance. Developmental dynamics : an official publication of the American Association of Anatomists 244, 391–409. [DOI] [PubMed] [Google Scholar]
- Gerety SS, Wang HU, Chen ZF, and Anderson DJ (1999). Symmetrical mutant phenotypes of the receptor EphB4 and its specific transmembrane ligand ephrin-B2 in cardiovascular development. Mol Cell 4, 403–414. [DOI] [PubMed] [Google Scholar]
- Gerrard DT, Berry AA, Jennings RE, Piper Hanley K, Bobola N, and Hanley NA (2016). An integrative transcriptomic atlas of organogenesis in human embryos. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Mariscal L, Lechuga S, and Garay E.(2007). Role of tight junctions in cell proliferation and cancer. Prog Histochem Cytochem 42, 1–57. [DOI] [PubMed] [Google Scholar]
- Griffin CT, Brennan J, and Magnuson T.(2008). The chromatin-remodeling enzyme BRG1 plays an essential role in primitive erythropoiesis and vascular development. Development 135, 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Zhou Y, Xing C, Lok J, Som AT, Ning M, Ji X, and Lo EH (2012). The vasculome of the mouse brain. PloS one 7, e52665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison RE, Berger R, Haworth SG, Tulloh R, Mache CJ, Morrell NW, Aldred MA, and Trembath RC (2005). Transforming growth factor-beta receptor mutations and pulmonary arterial hypertension in childhood. Circulation 111, 435–441. [DOI] [PubMed] [Google Scholar]
- Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, DePalma SR, McKean D, Wakimoto H, Gorham J, et al. (2015). De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 350, 1262–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, et al. (2014). The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, et al. (2012). De novo gene disruptions in children on the autistic spectrum. Neuron 74, 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, Zeng X, Qi H, Chang W, Sierant MC, et al. (2017). Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nature genetics 49, 1593–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki J, Aegerter S, Fevurly RD, Mammoto A, Mammoto T, Sahin M, Mably JD, Fishman SJ, and Chan J.(2014). RASA1 functions in EPHB4 signaling pathway to suppress endothelial mTORC1 activity. J Clin Invest 124, 2774–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Ryu YS, Kwak HJ, Ahn SY, Oh JL, Yancopoulos GD, Gale NW, and Koh GY (2002). EphB ligand, ephrinB2, suppresses the VEGF- and angiopoietin 1-induced Ras/mitogen-activated protein kinase pathway in venous endothelial cells. FASEB J 16, 1126–1128. [DOI] [PubMed] [Google Scholar]
- Kim JH, Peacock MR, George SC, and Hughes CC (2012). BMP9 induces EphrinB2 expression in endothelial cells through an Alk1-BMPRII/ActRII-ID1/ID3-dependent pathway: implications for hereditary hemorrhagic telangiectasia type II. Angiogenesis 15, 497–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniesel U, and Wolburg H.(2000). Tight junctions of the blood-brain barrier. Cell Mol Neurobiol 20, 57–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Green J, Pollard J Jr., and Tugendreich S.(2014). Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, Raja A, Coe BP, Stessman HA, He ZX, et al. (2015). Excess of rare, inherited truncating mutations in autism. Nature genetics 47, 582–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasjaunias PL, Chng SM, Sachet M, Alvarez H, Rodesch G, and Garcia-Monaco R.(2006). The management of vein of Galen aneurysmal malformations. Neurosurgery 59, S184–194; discussion S183–113. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Macarthur MW, Moss DS, and Thornton JM (1993). Procheck - a Program to Check the Stereochemical Quality of Protein Structures. J Appl Crystallogr 26, 283–291. [Google Scholar]
- Lee S, Lin M, Mele A, Cao Y, Farmar J, Russo D, and Redman C.(1999). Proteolytic processing of big endothelin-3 by the kell blood group protein. Blood 94, 1440–1450. [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.(2014). Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 30, 2843–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisabeth EM, Falivelli G, and Pasquale EB (2013). Eph receptor signaling and ephrins. Cold Spring Harb Perspect Biol 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long DM, Seljeskog EL, Chou SN, and French LA (1974). Giant arteriovenous malformations of infancy and childhood. J Neurosurg 40, 304–312. [DOI] [PubMed] [Google Scholar]
- Machida Y, Murai K, Miyake K, and Iijima S.(2001). Expression of chromatin remodeling factors during neural differentiation. J Biochem 129, 43–49. [DOI] [PubMed] [Google Scholar]
- Makita T, Sucov HM, Gariepy CE, Yanagisawa M, and Ginty DD (2008). Endothelins are vascular-derived axonal guidance cues for developing sympathetic neurons. Nature 452, 759–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, and Chen WM (2010). Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Almedina S, Martinez-Corral I, Holdhus R, Vicente A, Fotiou E, Lin S, Petersen K, Simpson MA, Hoischen A, Gilissen C, et al. (2016). EPHB4 kinase-inactivating mutations cause autosomal dominant lymphatic-related hydrops fetalis. J Clin Invest 126, 3080–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElhinney DB, Halbach VV, Silverman NH, Dowd CF, and Hanley FL (1998). Congenital cardiac anomalies with vein of Galen malformations in infants. Arch Dis Child 78, 548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, and DePristo MA (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra-Gorur K, Caglayan AO, Schaffer AE, Chabu C, Henegariu O, Vonhoff F, Akgumus GT, Nishimura S, Han W, Tu S, et al. (2014). Mutations in KATNB1 cause complex cerebral malformations by disrupting asymmetrically dividing neural progenitors. Neuron 84, 1226–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PJ, Rosenfeld JV, Dargaville P, Loughnan P, Ditchfield MR, Frawley G, and Tress BM (2001). Endovascular management of vein of Galen aneurysmal malformations presenting in the neonatal period. AJNR Am J Neuroradiol 22, 1403–1409. [PMC free article] [PubMed] [Google Scholar]
- Morita K, Sasaki H, Furuse M, and Tsukita S.(1999). Endothelial claudin: claudin-5/TMVCF constitutes tight junction strands in endothelial cells. J Cell Biol 147, 185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosch B, Reissenweber B, Neuber C, and Pietzsch J.(2010). Eph receptors and ephrin ligands: important players in angiogenesis and tumor angiogenesis. J Oncol 2010, 135285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, et al. (2012). Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485, 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev SI, Vetiska S, Bonilla X, Boudreau E, Jauhiainen S, Rezai Jahromi B, Khyzha N, DiStefano PV, Suutarinen S, Kiehl TR, et al. (2018). Somatic Activating KRAS Mutations in Arteriovenous Malformations of the Brain. The New England journal of medicine 378, 250–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, Mackenzie AP, Ng SB, Baker C, et al. (2011). Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nature genetics 43, 585–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. (2012). Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Wakai C, Saito T, Murayama A, Mimura Y, Youfu S, Nakamachi T, Kuwagata M, Satoh K, and Shioda S.(2011). Distribution of the longevity gene product, SIRT1, in developing mouse organs. Congenit Anom (Kyoto) 51, 70–79. [DOI] [PubMed] [Google Scholar]
- Pagenstecher A, Stahl S, Sure U, and Felbor U.(2009). A two-hit mechanism causes cerebral cavernous malformations: complete inactivation of CCM1, CCM2 or CCM3 in affected endothelial cells. Human molecular genetics 18, 911–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T, and Scott JD (1997). Signaling through scaffold, anchoring, and adaptor proteins. Science 278, 2075–2080. [DOI] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, Shuga J, Liu SJ, Oldham MC, Diaz A, et al. (2015). Molecular identity of human outer radial glia during cortical development. Cell 163, 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potente M, Ghaeni L, Baldessari D, Mostoslavsky R, Rossig L, Dequiedt F, Haendeler J, Mione M, Dejana E, Alt FW, et al. (2007). SIRT1 controls endothelial angiogenic functions during vascular growth. Genes Dev 21, 2644–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protack CD, Foster TR, Hashimoto T, Yamamoto K, Lee MY, Kraehling JR, Bai H, Hu H, Isaji T, Santana JM, et al. (2017). Eph-B4 regulates adaptive venous remodeling to improve arteriovenous fistula patency. Sci Rep 7, 15386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, Albrecht B, Bartholdi D, Beygo J, Di Donato N, et al. (2012). Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 380, 1674–1682. [DOI] [PubMed] [Google Scholar]
- Raybaud CA, Strother CM, and Hald JK (1989). Aneurysms of the vein of Galen: embryonic considerations and anatomical features relating to the pathogenesis of the malformation. Neuroradiology 31, 109–128. [DOI] [PubMed] [Google Scholar]
- Recinos PF, Rahmathulla G, Pearl M, Recinos VR, Jallo GI, Gailloud P, and Ahn ES (2012). Vein of Galen malformations: epidemiology, clinical presentations, management. Neurosurg Clin N Am 23, 165–177. [DOI] [PubMed] [Google Scholar]
- Revencu N, Boon LM, Mendola A, Cordisco MR, Dubois J, Clapuyt P, Hammer F, Amor DJ, Irvine AD, Baselga E, et al. (2013). RASA1 mutations and associated phenotypes in 68 families with capillary malformation-arteriovenous malformation. Hum Mutat 34, 1632–1641. [DOI] [PubMed] [Google Scholar]
- Roman BL, and Hinck AP (2017). ALK1 signaling in development and disease: new paradigms. Cell Mol Life Sci 74, 4539–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth Flach RJ, Guo CA, Danai LV, Yawe JC, Gujja S, Edwards YJ, and Czech MP (2016). Endothelial Mitogen-Activated Protein Kinase Kinase Kinase Kinase 4 Is Critical for Lymphatic Vascular Development and Function. Mol Cell Biol 36, 1740–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salaita K, and Groves JT (2010). Roles of the cytoskeleton in regulating EphA2 signals. Commun Integr Biol 3, 454–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, Kosmicki JA, Rehnstrom K, Mallick S, Kirby A, et al. (2014). A framework for the interpretation of de novo mutation in human disease. Nature genetics 46, 944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, et al. (2012). De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nature methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman LP, Elias S, Phillips OP, Grevengood C, Dungan JS, and Simpson JL (1994). Amniocentesis performed at 14 weeks’ gestation or earlier: comparison with first-trimester transabdominal chorionic villus sampling. Obstet Gynecol 83, 543–548. [DOI] [PubMed] [Google Scholar]
- Stuart BD, Choi J, Zaidi S, Xing C, Holohan B, Chen R, Choi M, Dharwadkar P, Torres F, Girod CE, et al. (2015). Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nature genetics 47, 512–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43, D447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Kamata R, and Sakai R.(2005). EphA2 phosphorylates the cytoplasmic tail of Claudin-4 and mediates paracellular permeability. The Journal of biological chemistry 280, 42375–42382. [DOI] [PubMed] [Google Scholar]
- Tham E, Lindstrand A, Santani A, Malmgren H, Nesbitt A, Dubbs HA, Zackai EH, Parker MJ, Millan F, Rosenbaum K, et al. (2015). Dominant mutations in KAT6A cause intellectual disability with recognizable syndromic features. American journal of human genetics 96, 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timberlake AT, Choi J, Zaidi S, Lu Q, Nelson-Williams C, Brooks ED, Bilguvar K, Tikhonova I, Mane S, Yang JF, et al. (2016). Two locus inheritance of non-syndromic midline craniosynostosis via rare SMAD6 and common BMP2 alleles. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timberlake AT, Furey CG, Choi J, Nelson-Williams C, Yale Center for Genome, A., Loring E, Galm A, Kahle KT, Steinbacher DM, Larysz D, et al. (2017). De novo mutations in inhibitors of Wnt, BMP, and Ras/ERK signaling pathways in non-syndromic midline craniosynostosis. Proceedings of the National Academy of Sciences of the United States of America 114, E7341-E7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi Y, Kosaki R, Itoh Y, Tsukamoto K, Matsuoka R, Shintani M, Nosaka S, Masaki H, and Iizuka Y.(2011). Vein of Galen aneurysmal malformation associated with an endoglin gene mutation. Pediatrics 128, e1307–1310. [DOI] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. (2013). From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43, 11 10 11–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Itallie CM, and Anderson JM (2013). Claudin interactions in and out of the tight junction. Tissue Barriers 1, e25247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laarhoven PM, Neitzel LR, Quintana AM, Geiger EA, Zackai EH, Clouthier DE, Artinger KB, Ming JE, and Shaikh TH (2015). Kabuki syndrome genes KMT2D and KDM6A: functional analyses demonstrate critical roles in craniofacial, heart and brain development. Human molecular genetics 24, 4443–4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal A.(2014). Disseminated intravascular coagulation. Indian J Anaesth 58, 603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, van Lier B, Arts P, Wieskamp N, del Rosario M, et al. (2010). A de novo paradigm for mental retardation. Nature genetics 42, 1109–1112. [DOI] [PubMed] [Google Scholar]
- Vivanti A, Ozanne A, Grondin C, Saliou G, Quevarec L, Maurey H, Aubourg P, Benachi A, Gut M, Gut I, et al. (2018). Loss of function mutations in EPHB4 are responsible for vein of Galen aneurysmal malformation. Brain : a journal of neurology 141, 979–988. [DOI] [PubMed] [Google Scholar]
- Walker EJ, Su H, Shen F, Choi EJ, Oh SP, Chen G, Lawton MT, Kim H, Chen Y, Chen W, and Young WL (2011). Arteriovenous malformation in the adult mouse brain resembling the human disease. Annals of neurology 69, 954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, and Hakonarson H.(2010a). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38, e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Nakayama M, Pitulescu ME, Schmidt TS, Bochenek ML, Sakakibara A, Adams S, Davy A, Deutsch U, Luthi U, et al. (2010b). Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature 465, 483–486. [DOI] [PubMed] [Google Scholar]
- Wang Z, Miura N, Bonelli A, Mole P, Carlesso N, Olson DP, and Scadden DT (2002). Receptor tyrosine kinase, EphB4 (HTK), accelerates differentiation of select human hematopoietic cells. Blood 99, 2740–2747. [DOI] [PubMed] [Google Scholar]
- Ware JS, Samocha KE, Homsy J, and Daly MJ (2015). Interpreting de novo Variation in Human Disease Using denovolyzeR. Current protocols in human genetics / editorial board, Jonathan L Haines [et al] 87, 7 25 21–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattenhofer M, Reymond A, Falciola V, Charollais A, Caille D, Borel C, Lyle R, Estivill X, Petersen MB, Meda P, and Antonarakis SE (2005). Different mechanisms preclude mutant CLDN14 proteins from forming tight junctions in vitro. Hum Mutat 25, 543–549. [DOI] [PubMed] [Google Scholar]
- Wei Q, Zhan X, Zhong X, Liu Y, Han Y, Chen W, and Li B.(2015). A Bayesian framework for de novo mutation calling in parents-offspring trios. Bioinformatics 31, 1375–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederstein M, and Sippl MJ (2007). ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35, W407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsey AJ, Fernandez TV, Yu D, King RA, Dietrich A, Xing J, Sanders SJ, Mandell JD, Huang AY, Richer P, et al. (2017). De Novo Coding Variants Are Strongly Associated with Tourette Disorder. Neuron 94, 486–499 e489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe JM, and Myers L.(2010). Fur in the midst of the waters: visual search for material type is inefficient. J Vis 10, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Carrasco R, Kinneer K, Sabol D, Jallal B, Coats S, and Tice DA (2012). EphB4 promotes or suppresses Ras/MEK/ERK pathway in a context-dependent manner: Implications for EphB4 as a cancer target. Cancer Biol Ther 13, 630–637. [DOI] [PubMed] [Google Scholar]
- Xu DS, Usman AA, Hurley MC, Eddleman CS, and Bendok BR (2010). Adult presentation of a familial-associated vein of galen aneurysmal malformation: case report. Neurosurgery 67, E1845–1851; discussion 1851. [DOI] [PubMed] [Google Scholar]
- Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, et al. (2013). De novo mutations in histone-modifying genes in congenital heart disease. Nature 498, 220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, and Hughes S.(2006). Role of the ephrin and Eph receptor tyrosine kinase families in angiogenesis and development of the cardiovascular system. The Journal of pathology 208, 453–461. [DOI] [PubMed] [Google Scholar]
- Zhang J, Schwartz MP, Hou Z, Bai Y, Ardalani H, Swanson S, Steill J, Ruotti V, Elwell A, Nguyen BK, et al. (2017). A Genome-wide Analysis of Human Pluripotent Stem Cell-Derived Endothelial Cells in 2D or 3 D Culture. Stem Cell Reports 8, 907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE (2009). Non-Smad pathways in TGF-beta signaling. Cell Res 19, 128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
WES data for all VOGM parent-offspring trios reported in this study have been deposited in the NCBI database of Genotypes and Phenotypes under the accession number phs000744.v4.p2